
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 July 2021
Sec. Microbial Symbioses
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.694466
This article is part of the Research Topic Insect Microbiome: From Diversity To Applications View all 35 articles
 Yong Zhang1,2†
Yong Zhang1,2† Song Liu1†
Song Liu1† Rui Jiang1†
Rui Jiang1† Chen Zhang1
Chen Zhang1 Tian Gao1
Tian Gao1 Yun Wang3
Yun Wang3 Cui Liu1
Cui Liu1 Yanhua Long2*
Yanhua Long2* Yinglao Zhang2*
Yinglao Zhang2* Yunqiu Yang1*
Yunqiu Yang1*Members of the Wolbachia genus manipulate insect–host reproduction and are the most abundant bacterial endosymbionts of insects. The tea Geometrid moth Ectropis grisescens (Warren) (Lepidoptera: Geometridae) is the most devastating insect pest of tea plants [Camellia sinensis (L.) O. Kuntze] in China. However, limited data on the diversity, typing, or phenotypes of Wolbachia in E. grisescens are available. Here, we used a culture-independent method to compare the gut bacteria of E. grisescens and other tea Geometridae moths. The results showed that the composition of core gut bacteria in larvae of the three Geometridae moth species was similar, except for the presence of Wolbachia. Moreover, Wolbachia was also present in adult female E. grisescens samples. A Wolbachia strain was isolated from E. grisescens and designated as wGri. Comparative analyses showed that this strain shared multilocus sequence types and Wolbachia surface protein hypervariable region profiles with cytoplasmic incompatibility (CI)-inducing strains in supergroup B; however, the wGri-associated phenotypes were undetermined. A reciprocal cross analysis showed that Wolbachia-uninfected females mated with infected males resulted in 100% embryo mortality (0% eggs hatched per female). Eggs produced by mating between uninfected males and infected females hatched normally. These findings indicated that wGri induces strong unidirectional CI in E. grisescens. Additionally, compared with uninfected females, Wolbachia-infected females produced approximately 30–40% more eggs. Together, these results show that this Wolbachia strain induces reproductive CI in E. grisescens and enhances the fecundity of its female host. We also demonstrated that wGri potential influences reproductive communication between E. grisescens and Ectropis obliqua through CI.
Wolbachia are Gram-negative bacteria that were first identified in the oophoron of Culex pipiens. Their prevalence in arthropods and nematodes worldwide and stunning arsenal of parasitic and mutualistic adaptations (Zug and Hammerstein, 2012). It has been recently estimated that ∼50% of arthropod and several filarial nematode species harbor Wolbachia (Weinert et al., 2015; Lefoulon et al., 2016). Moreover, Wolbachia is estimated to infect 40–60% of insect species (Mateos et al., 2020). In fact, Wolbachia is the most common and widespread facultative symbiont of insects (Zug and Hammerstein, 2015). As parasites, Wolbachia manipulate insect–host reproduction in various ways, via cytoplasmic incompatibility (CI), male-killing, feminization, and parthenogenesis, in order to facilitate their maternal transmission (Yen and Barr, 1971; Rousset et al., 1992). Of these, CI is the most prevalent host phenotype caused by the presence of Wolbachia (Stouthamer et al., 1999). In contrast to Wolbachia’s well-established reproductive role, recent years have extensive evidence that Wolbachia can benefit their arthropod host as mutualists (Teixeira et al., 2008; Brownlie et al., 2009; Nikoh et al., 2014; Zug and Hammerstein, 2015). In some insects, Wolbachia strains function as mutualists to provide benefits to the host, including supplementing essential nutrients, protection of the host against pathogens, increasing successful egg development, and enhancing female insect host fecundity (Dedeine et al., 2001; Martijn and Ellers, 2008; Hosokawa et al., 2010; Iturbe-Ormaetxe et al., 2011; Guo et al., 2018). For example, Wolbachia provides riboflavin (vitamin B2) to all its insect host, and Wolbachia’s riboflavin provisioning certainly contributes to growth, survival, and reproduction in its insect host (Moriyama et al., 2015). For instance, in the two planthopper species Nilaparvata lugens and Laodelphax striatellus, intracellular Wolbachia strains wLug and wStriCN (both in supergroup B) provide biotin and riboflavin to enhance reproduction of their female hosts (Ju et al., 2020). Accurate identification of Wolbachia strains is a fundamental component of all Wolbachia-related research. However, Wolbachia cannot be cultured outside host cells, and identification of Wolbachia strain is solely based on molecular typing.
The genus Wolbachia is currently divided into at least 17 possible phylogenetic supergroups, named A–F, H–Q, and S (O’Neill et al., 1992; Werren et al., 1995; Bandi et al., 1998; Ros et al., 2009; Lefoulon et al., 2020). Supergroups A and B are primarily found in arthropods and often function as reproductive parasites that manipulate host reproduction to increase their own spread through the matriline (Kaur et al., 2021). An earlier phylogenetic analysis based on Wolbachia surface protein (wsp) gene sequences further divided the A and B supergroups into 12 groups (Mel, AlbA, Pip, Uni, Riv, Pap, Aus, Ori, Dei, Haw, Con, and Mors) (Zhou et al., 1998). Later, seven more groups were added (Kue, Dro, Sib, Kay, Div, For, and Vul) (van Meer et al., 1999). However, the wsp gene in different strains of Wolbachia has undergone extensive recombination (Werren and Bartos, 2001). Thus, this single-locus identification system is unreliable as a research tool. A multilocus sequence typing (MLST) system was developed to type Wolbachia strains of diverse origins (Baldo et al., 2006). This system is based on five conserved genes (gatB, coxA, hcpA, ftsZ, and fbpA) and four WSP hypervariable regions (HVRs): HVR1 (52–84 aa), HVR2 (85–134 aa), HVR3 (135–185 aa), and HVR4 (186–222 aa). Data generated using the five gene-based MLST system have been deposited in a web-accessible database1 that facilitates the storage, management, and analysis of sequence data and isolate information.
Tea plant [Camellia sinensis (L.) O. Kuntze], a perennial evergreen woody plant, is one of the most important economic crops in almost 30 countries such as China, Japan, India, Kenya, and Indonesia. The young leaves and buds are processed into a variety of tea, including green, white, yellow, oolong, black, and dark tea, which is a popularly daily beverage (Jiang et al., 2019). Tea has numerous health and medical benefits for humans due to its many characteristic secondary plant metabolites, including catechins, caffeine, polyphenols, and theanine (Too et al., 2015; Ji et al., 2016). Tea production plays a role in the development of agricultural economy in China. Unfortunately, leaf herbivory on tea plants by tea Geometridae moths can cause severe yield loss and quality damage for tea.
Tea Geometridae moths are the most destructive chewing insects for tea plant by feeding on the young tea leaves in China (Zhang, 2001). Among them, Ectropis grisescens (Warren) (Lepidoptera: Geometridae, Ennominae) is the most serious Geometridae pest of tea in China because of its wide distribution and destructive nature (Zhang et al., 2019; Pan et al., 2021). This pest infests thousands of hectares of tea plants per year, severely reducing tea plant growth and negatively impacting tea production (Jiang et al., 2014). Compared with Ectropis obliqua and Scopula subpunctaria, the other main Geometridae pests of tea, E. grisescens, has a wider distribution and stronger fecundity (Chen L. L. et al., 2020). Little is known about why E. grisescens is the most harmful pest in tea gardens, and there is no explanation from the perspective of insect symbionts. Recently, extensive evidence shows that insect symbionts do affect essential physiological functions in their lepidoptera host, including degrading complex dietary polymers, supplementing essential nutrients, overcoming plant anti-herbivore defenses, or strengthening of immune responses for protection against pathogens (Paniagua et al., 2018). A study reported that mundticin KS (bacteriocin) produced by gut bacteria Enterococcus mundtii inhibits pathogen colonization in its host Spodoptera littoralis (Lepidoptera: Noctuidae) (Shao et al., 2017). Besides, Lei et al. (2020) found that Wolbachia reduce the susceptibility of striped stem borer, Chilo suppressalis (Lepidoptera: Crambidae), to two insecticides (fipronil and avermectin).
In a previous study, we detected Wolbachia in the gut of E. grisescens larvae (Zhang et al., 2019). However, the Wolbachia strain in E. grisescens was not typed, and its resulting phenotypes were not elucidated. Here, we use a culture-independent method to investigate and compare the diversity and richness of gut bacterial communities among three species of Geometridae moths and found that the presence of Wolbachia was the most obvious difference. MLST- and wsp-based phylogenetic analyses revealed that the Wolbachia from different populations of E. grisescens had the same Wolbachia, which is from supergroup B. Reciprocal cross analyses revealed that Wolbachia enhances the fecundity of E. grisescens. This may be one of the reasons why its host E. grisescens is the most damaging leaf-eating tea pest in China.
The larvae were collected from tea gardens in the five main tea-producing provinces of China (Table 1) and were maintained at the State Key Laboratory of Tea Plant Biology and Utilization, Anhui Agricultural University, Hefei, China (31.86°N, 117.27°E). The collected larvae were reared on tea leaves in transparent boxes in a controlled climate room (22°C ± 1°C; relative humidity 75% ± 10%; 16-h light:8-h dark photoperiod). The tea leaves used in the experiments were cut from branches of tea plants using scissors and inserted into floral foam for storage.
For Geometrid larvae, 10 fifth instar larvae were pooled to provide a biological replicate and five biological replicates were established per group. For E. grisescens adults, five female adults were pooled to provide a biological replicate and five biological replicates were established per group. Note that the DNA used for Wolbachia-typing was extracted from single female moth.
Larvae were surface-sterilized by dipping in 70% ethanol for 15 s and then rinsing twice with sterile water for 15 s each time. Dissecting scissors were used to cut laterally behind the head capsule, and the gut was removed from the cuticle with larval forceps. The whole gut including the gut contents was collected and placed in a 2.0-ml micro-centrifuge tube for DNA extraction.
Wings were removed from female moths, and other tissues were surface-sterilized by dipping in 70% ethanol for 15 s, followed by rinsing twice with sterile water for 15 s each time. The tissues were then ground for 2 min and placed in a 2.0-ml micro-centrifuge tube for DNA extraction.
Total genomic DNA was extracted from samples using QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). The quality of the extracted DNA was assessed by electrophoresis on a 1.2% (w/v) agarose gel. The genomic DNA extracted from bacteria in the larvae gut was subjected to amplification and sequencing.
The Genomic DNA extracted from bacteria in adult female moths was subjected to metagenomic and Wolbachia-typing analyses.
Gut bacteria from three Geometridae larvae were analyzed by sequencing the V3–V4 region of the 16S ribosomal RNA gene (16S rRNA), using the Illumina NovaSeq platform.
Genomic DNA samples were subjected to PCRs for amplification of the V3–V4 regions of the 16S rRNA with the universal primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). All PCR reactions consisted of 15 μl of Phusion ®High-Fidelity PCR Master Mix (New England Biolabs, Beverly, MA, United States), 2 μM forward and reverse primers, and 10 ng of template DNA. The thermal cycling conditions were as follows: initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, elongation at 72°C for 30 s, and final extension at 72°C for 5 min. Then, the mixture of PCR products was purified with a Gel Extraction Kit (Qiagen).
After PCR amplification, the samples were sequenced on the Illumina NovaSeq platform (Illumina, San Diego, CA, United States) and 250-bp paired-end reads were generated. All sequences are available as SRA files at the National Center for Biotechnology Information Sequence Read Archive database (NCBI-SRA) under bioProject PRJNA720281 (SRA accession numbers: SAMN18644841-843).
The sequences were analyzed using the QIIME software package and used to compare the relative abundance of bacterial taxa (Caporaso et al., 2010b). Operational taxonomic units (OUTs) were assigned at 97% similarity cutoff using UCLUST version 1.2.22 (Edgar, 2010). The representative sequence (the sequence with the highest relative abundance) of each OTU was selected to build the overall OTU table. The taxonomic classification of each microbial OTU was assigned by Ribosomal Database Project classifier PyNast using SILVA and UNITE as bacterial 16S rRNA databases (Caporaso et al., 2010a). Abundance values of OTUs were normalized using a standard value for the sample with the least sequences. Subsequent analyses of alpha diversity were performed using this normalized output.
The alpha- and beta-diversity indexes were calculated using QIIME V1.7.0. The alpha-diversity indexes were the Pielou evenness index (J) and the ACE richness index (Shannon and Weaver, 1949; Pielou, 1966; Chao et al., 1993). Rarefaction curves and Shannon index were used to verify the quality and depth of sampling. Principal coordinate analysis (PCoA) with weighted and unweighted UniFrac distance metrics was performed to detect differences in microbial community structures among the three Geometridae larvae (Lozupone and Knight, 2005). Analysis of similarity (ANOSIM) was carried out based on Bray–Curtis dissimilarity to assess the difference of microbial community structures (Clarke, 1993). Microbes that showed significant differences in relative abundance among different groups were identified using Metastats software2 with false discovery rate (FDR) (Paulson et al., 2011).
The DNA sample was fragmented by sonication to produce 350-bp fragments; then, these DNA fragments were end-polished, A-tailed, and ligated with the full-length adaptor for Illumina sequencing with further PCR amplification. Finally, PCR products were purified using the AMPure XP system. The libraries were analyzed to determine size distribution with an Agilent2100 Bioanalyzer and quantified using real-time PCR.
Clustering of the index-coded samples was performed using the cBot Cluster Generation System according to the manufacturer’s instructions. After cluster generation, the libraries were sequenced on the Illumina NovaSeq 6000 platform and paired-end reads were generated.
The raw data obtained from the Illumina NovaSeq sequencing platform using Readfq (V8)3 were processed to obtain clean data for subsequent analyses. The processing steps removed reads containing low-quality bases (default quality threshold value ≤ 38, default length 40 bp), reads containing a certain proportion of N (unspecified) bases (default length 10 bp), and reads containing adapter sequences (default length 15 bp). The raw data have been submitted to the NCBI-SRA under bioProject PRJNA720281 (SRA accession number: SAMN18644841).
Considering the possibility that samples may be polluted with host DNA, the clean data were used in BLAST searches against the host database using Bowtie2.2.4 software (Bowtie2.2.4)4 to filter reads of host origin. The parameters were as follows : –end-to-end, –sensitive, -I 200, -X 400.
Due to serious host pollution, we use a read-based (mapping) approach for metagenomic analysis. Species annotations were based on clean data and the RPKM method was used to calculate abundance. The MetaPhalAn2 method was used to analyze bacterial diversity (Segata et al., 2012).
The WSP locus was amplified using the wsp primers (wsp_F1: 5′-GTCCAATARSTGATGARGAAAC-3′ and wsp_R1: 5′-CYGCACCAAYAGYRCTRTAAA-3′) to confirm single infections. The PCR reactions were performed in 25-μl final volumes containing 9.7 μl of ddH2O, 12.5 μl of Premix Taq (LA Taq Version 2.0, TaKaRa, Dalian, China), 0.8 μl of DNA template, and 1 μl of each primer (10 μM). The thermal cycling conditions were as follows: 95°C for 3 min; 35 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 90 s; 72°C for 10 min. The products were stored at 4°C.
The MLST loci were amplified in accordance with previously published protocols (see text footnote 1). The PCR reactions were performed in 25-μl final volumes containing 9.7 μl of ddH2O, 12.5 μl of Premix Taq (LA Taq Version 2.0, TaKaRa), 0.8 μl of DNA template, and 1 μl of each primer (10 μM). The thermal cycling conditions were as follows: 94°C for 4 min; 35 cycles of 94°C for 30 s, appropriate Tm (Table 2) for 45 s and 72°C for 90 s; 72°C for 10 min. The products were stored at 4°C.
Each sample was mixed with the same volume of 1× loading buffer containing SYBR green and then separated by electrophoresis on 1.2% (v/v) agarose gels for amplicon detection. All the PCR products were sequenced by Sangon Biotech (Shanghai, China). The sequences of all products were used in BLAST or MLST analyses at the GenBank and PubMLST databases, respectively.
The amplified wsp gene sequences were subjected to BLAST analysis at the GenBank database. A list of Wolbachia isolates, including those reported by Zhou et al. (1998) and van Meer et al. (1999) and those with wsp gene sequences in GenBank, was created (Supplementary Table 1). These isolates were used as references to classify the type of Wolbachia strain isolated from E. grisescens.
A phylogenetic tree was constructed using the Wolbachia wsp genes and the maximum likelihood (ML) approach implemented in MEGA X. The substitution model was Tamura 3-parameter and gamma distributed (G) with 1000 bootstrap replications.
The sequences of gatB, coxA, hcpA, ftsZ, fbpA, and wsp were submitted to the PubMLST database for sequence typing to generate MLST allelic profiles and WSP HVR profiles. A list of Wolbachia isolates was generated by searching PubMLST and downloading isolate sequences from GenBank (Supplementary Table 2). Isolates with a complete set of MLST and HVR profiles, as well as phenotypic data, were selected. These isolates had been used by Baldo et al. (2006) to establish the MLST typing system for Wolbachia. The ML trees were constructed using the concatenated MLST and wsp sequences, independently, in MEGA X.
The alignments of concatenated MLST alleles and wsp alleles were generated using ClustalW with 1000 bootstrap iterations. For phylogenetic reconstructions, the nucleotide substitution model was determined by the BIC value calculated by MEGAX software. The best-fit substitution models were as follows: GTR + G for MLST and T92 + G for wsp. The sequences of the wsp gene and the five genes used for MLST of the Wolbachia strain from E. grisescens have been deposited in the GenBank database (Accession number: MW630082-MW630111).
To analyze the effects of Wolbachia on its host E. grisescens, the fecundity (egg production) of infected (W+) and uninfected (W–) insects was recorded. All the insects used in the experiments were confirmed to be infected with Wolbachia by amplifying the wsp gene. E. grisescens was collected from Lu’an, Anhui Province, and successfully subcultured for more than 40 generations.
The F1 generation of W+ E. grisescens, as larvae, were reared on tea leaves until they reached the third instar stage. Then, 140 healthy third-instar larvae were divided into two groups of 70 larvae, which were placed individually in 90-mm plastic Petri dishes. Each dish contained food for the insects and had a tight lid. The control group was fed an agar-based insect diet, and the antibiotic group was fed the insect diet supplemented with 300 μg/ml each of rifampicin, gentamicin, penicillin, and streptomycin (Zhang et al., 2019). The insect diet comprised 60 ml of sterile water, 20 g of tea powder, and 2 g of wheat agar. Fresh tea leaves were collected, freeze-dried, and ground to make the tea powder.
To ensure that the effects on reproduction were due to the absence of Wolbachia and not due to antibiotics, the same antibiotics were used to treat E. obliqua and S. subpunctaria.
Ectropis obliqua and E. grisescens are two closely related moth species. Reciprocal cross analyses were conducted to determine whether Wolbachia affects reproduction between these two moths. E. grisescens was collected from Luan, Anhui Province, and successfully subcultured for more than 40 generations. E. obliqua was collected from Xuancheng, Anhui Province, and successfully subcultured for more than three generations.
We selected newly emerged moths of E. obliqua and E. grisescens for reciprocal crosses. The egg hatching rate was counted.
GraphPad Prism 7.0, R, or Origin 9.0 was used for statistical analyses and to construct figures.
The Illumina NovaSeq sequencing of the bacterial 16S rRNA amplicons from larvae of E. grisescens (EG), E. obliqua (EO), and S. subpunctaria (SS) yielded 951,717 raw reads in total. After quality filtering and read merging, a total of 876,703 high-quality sequences remained for bacteria. Rarefaction curves (Figure 1A) and Shannon index (Figure 1B) clearly demonstrated that the sampling efforts were adequate to fully represent the richness of the gut communities analyzed.
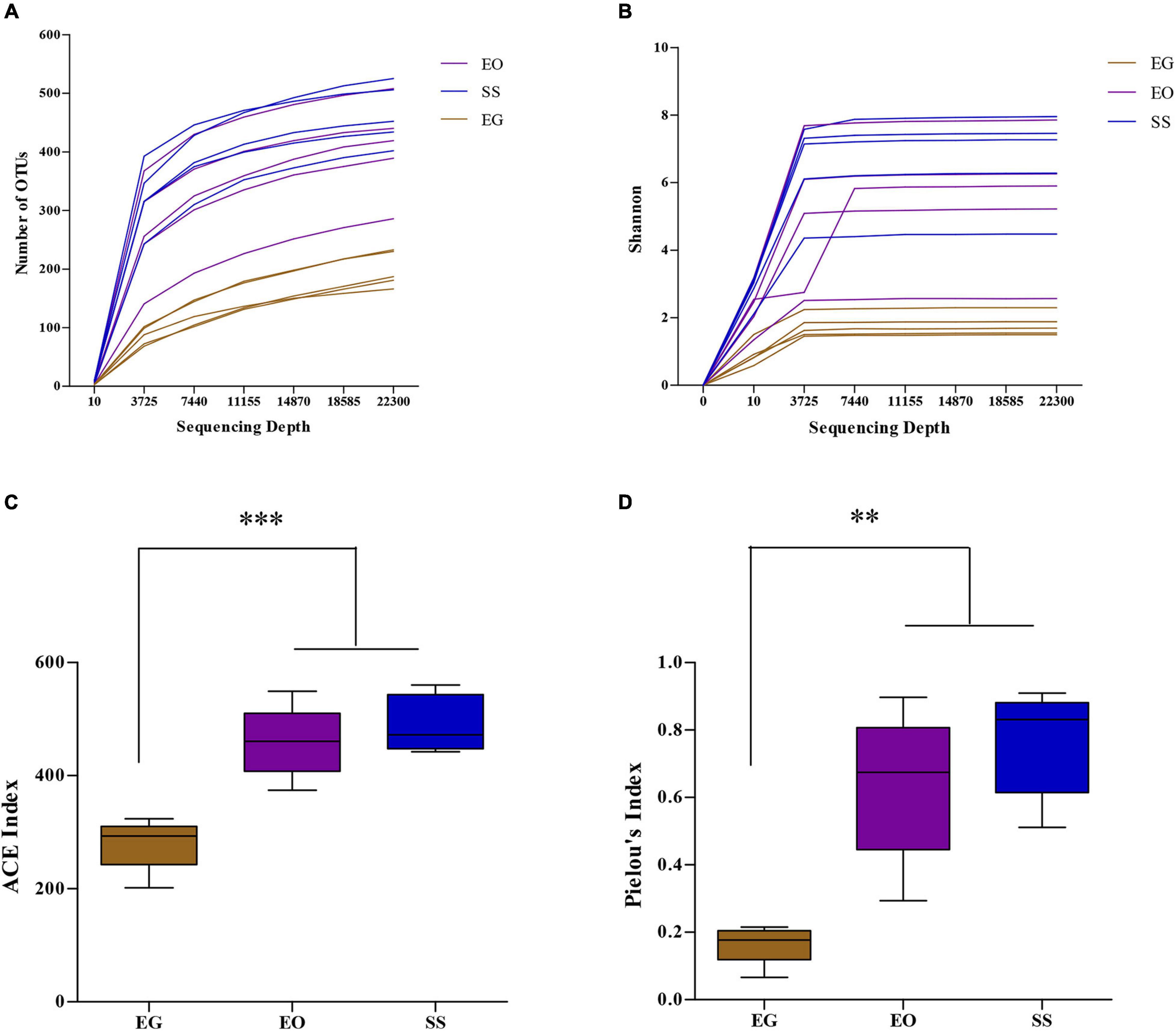
Figure 1. Rarefaction curves, based on number of operational taxonomic units (OTUs) observed (A), and Shannon index (B), and the alpha-diversity, based on ACE index (C), and Pielou’s index (D), of the microbial community in the larval gut of E. grisescens, E. obliqua, and S. subpunctaria (EG, EO, and SS, respectively). Significant differences were detected by unpaired two-tailed t-test; **p < 0.001; ***p < 0.0001.
The OTU richness (ACE index) of the gut bacteria of EG (280.04 ± 41.54) differed from those of EO (458.75 ± 55.42; t = 6.91; df = 7; p < 0.0001) and SS (490.30 ± 45.15; t = 6.91; df = 7; p < 0.0001) (Figure 1C). The OTU evenness (Pielou index; J) of the gut bacteria of EG (J = 0.16 ± 0.05) differed from those of EO (J = 0.64 ± 0.19; t = 4.62; df = 8; p < 0.001) and SS (J = 0.76 ± 0.14; t = 7.98; df = 8; p < 0.0001) (Figure 1D). Thus, the OTU richness and evenness of the gut bacteria were lowest in E. grisescens among the three species analyzed.
The PCoA analysis using the distances based on weighted and unweighted UniFrac showed that the gut bacteria communities of E. grisescens larvae were the most distinctive (Figures 2A,B). A non-parametric test (ANOSIM), which is based on Bray–Curtis dissimilarity, indicated that the gut bacterial beta-diversity of EG was significantly different from those of EO (R = 0.664; p < 0.05) and SS (R = 0.732; p < 0.001) (Figures 2C,D). These results showed that the gut bacterial beta-diversity of E. grisescens larvae was significantly distinctive compared with those of E. obliqua and S. subpunctaria larvae.
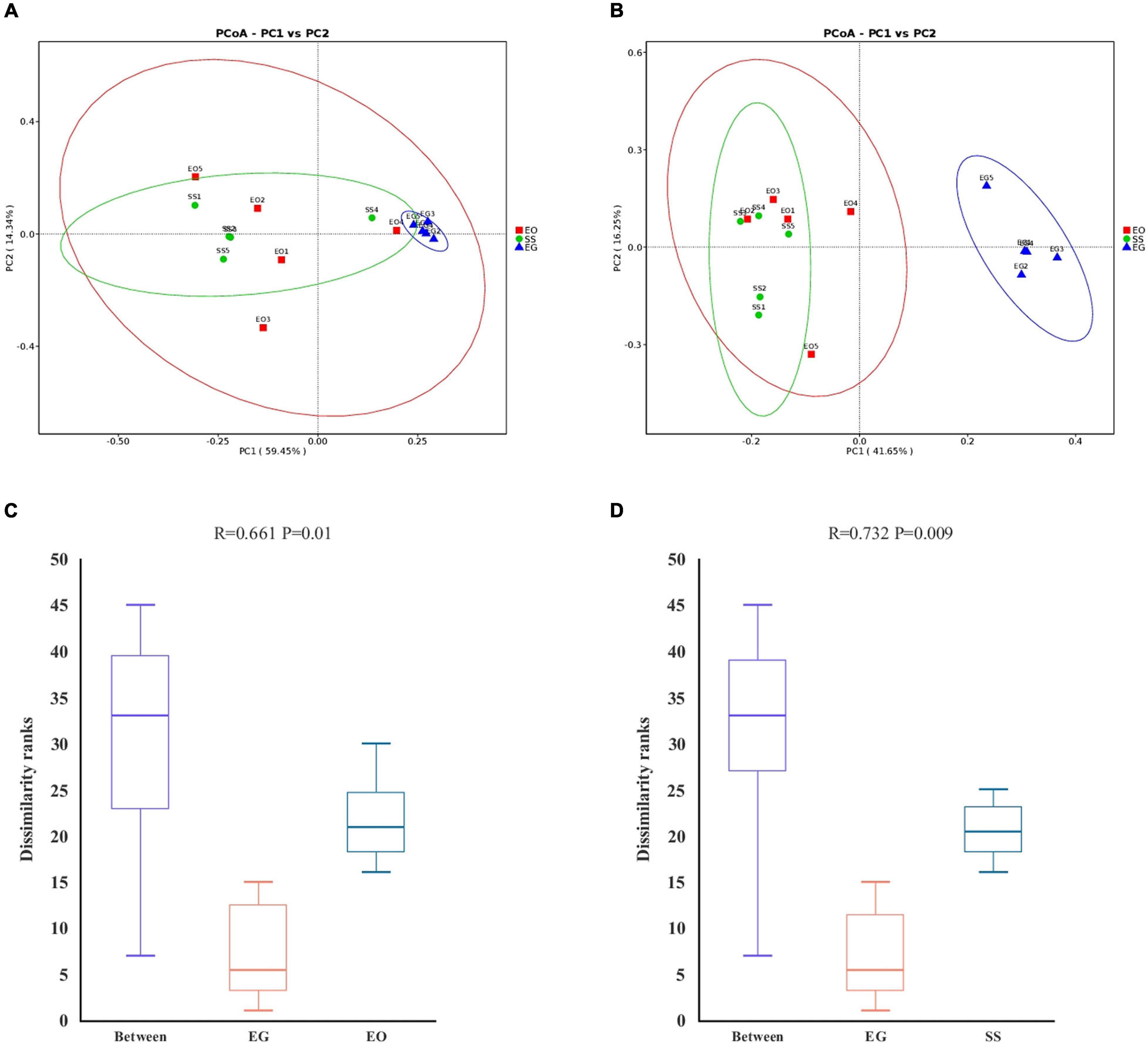
Figure 2. Principal component and non-parametric test analysis of bacterial diversity. Principal coordinate analysis (PCoA) of bacteria using distance based on weighted (A) and unweighted (B) UniFrac values. Non-parametric test (ANOSIM) of bacterial diversity in EG compared with EO (C) and SS (D). EG, EO, and SS refer to E. grisescens, E. obliqua, and S. subpunctaria, respectively.
As shown in the Venn diagram in Figure 3A, 379 OTUs were shared by all groups, and accounted for 95.47, 59.78, and 58.58% of the total OTUs in EG, EO, and SS, respectively. The petal diagram in Figure 3B shows that a total of 44 OTUs were shared among all samples. The relative abundance of these 44 OTUs in EG, EO, and SS was 83.41% ± 6.95%, 60.01% ± 11.02%, and 65.46% ± 13.21%, respectively (Supplementary Figure 1). The alpha- and beta-diversity indexes were lowest for bacterial community in the gut of E. grisescens larvae, but overall, the composition of core gut bacteria in the three geometrids was similar.
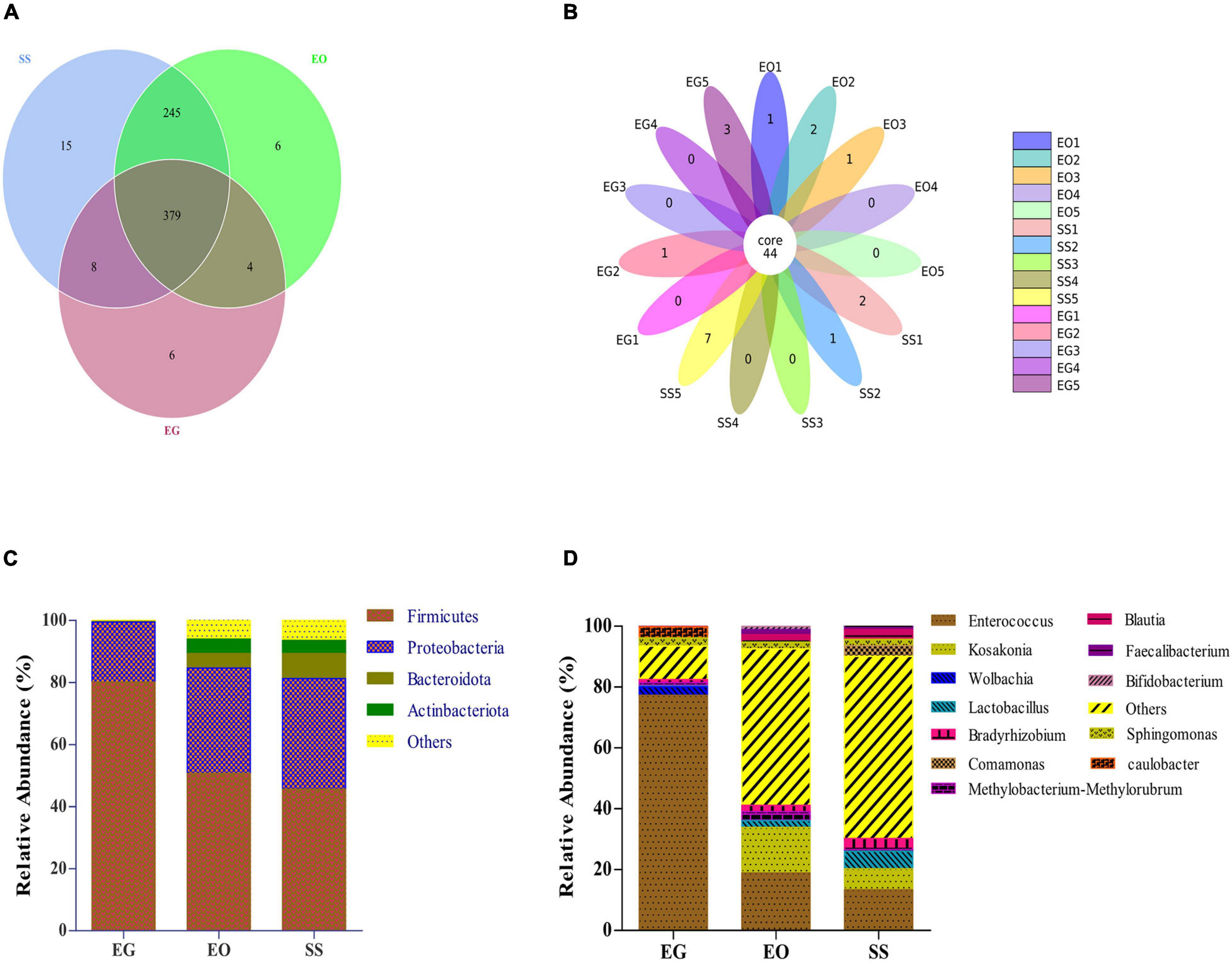
Figure 3. Venn diagram (A) and petal diagram (B) of operational taxonomic units (OTUs) in EG, EO, and SS. Relative abundances are shown for bacteria with relative abundance of >1% in larval gut of at least one species at the phylum level (C) and three species at the genus level (D). EG, EO, and SS refer to E. grisescens, E. obliqua, and S. subpunctaria, respectively.
The dominant bacterial phyla in each group were Firmicutes (45.64% ± 12.25% to 80.13% ± 8.63%) and Proteobacteria (19.26% ± 4.22% to 33.92% ± 4.22%) (Figure 3C). The dominant bacterial genera in EG were Enterococcus (77.29% ± 16.53%), Caulobacter (3.58% ± 1.6%), and Wolbachia (2.8% ± 1.52%). The dominant bacterial genera in EO and SS were Enterococcus (13.40% ± 5.51% to 18.8% ± 6.20%), Kosakonia (6.87% ± 2.33% to 14.99% ± 4.16%), and Lactobacillus (2.27% ± 2.33% to 5.74% ± 4.16%) (Figure 3D). These results showed that Enterococcus was the dominant gut bacterial genus in tea Geometridae larvae.
The heat map of the top 35 abundant gut bacteria illustrates differences in microbial community structures at the genus level (Figure 4). The relative abundance of Caulobacter and Wolbachia in E. grisescens was significantly different from that in E. obliqua and S. subpunctaria as determined by MetaStat analysis. Among these three bacterial species, Wolbachia was only detected in E. grisescens larvae, with a relative abundance of up to 7.6% (Supplementary Figure 2).
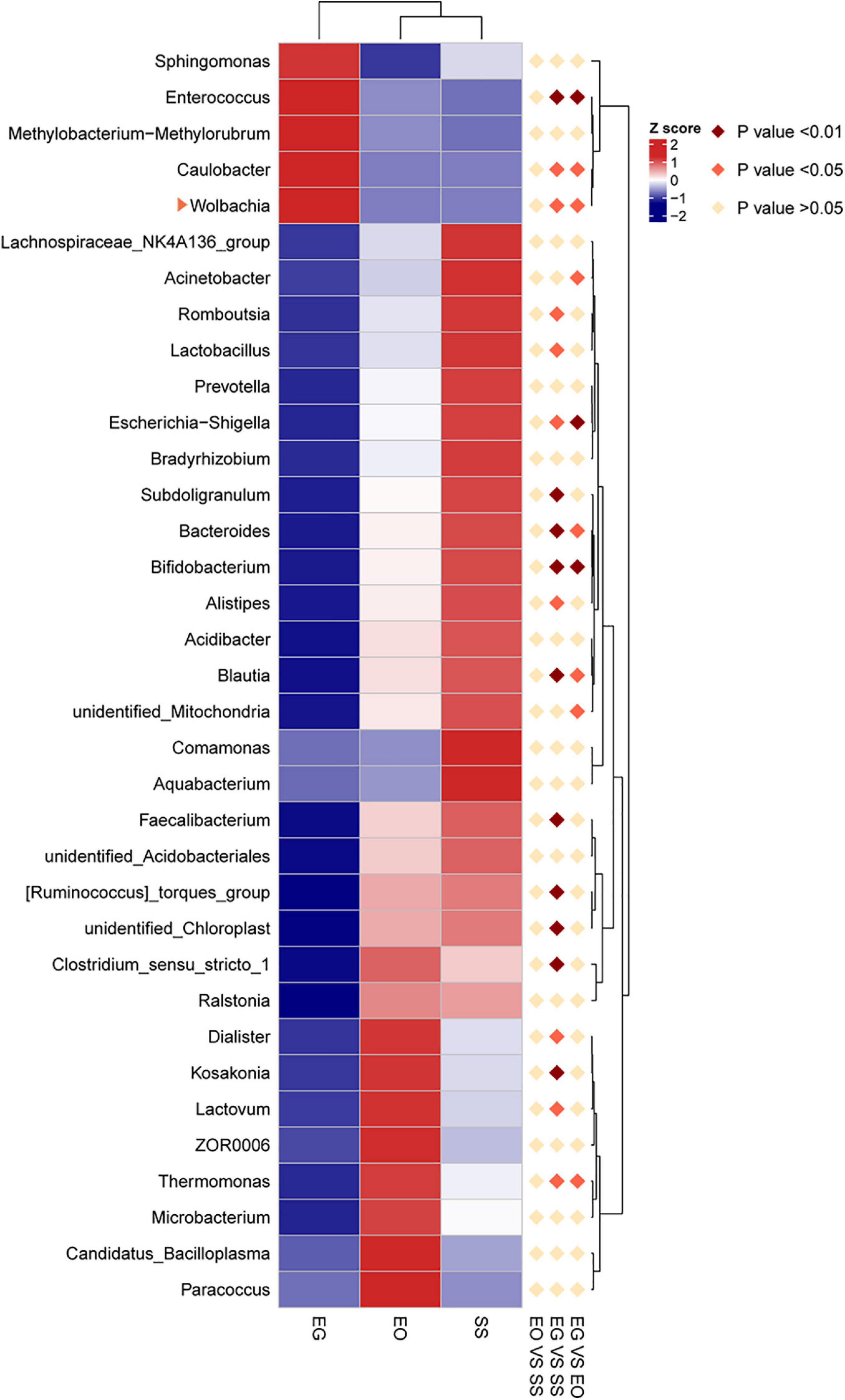
Figure 4. Hierarchical clustering analysis of three groups based on the relative abundance of the top 30 microbiota genera identified in EG, EO, and SS. EG, EO, and SS refer to E. grisescens, E. obliqua, and S. subpunctaria, respectively.
The Illumina NovaSeq sequencing of bacteria from female adults of E. grisescens yielded about 6.33–6.73 G raw data per sample. After quality filtering and read merging, a total of 6.32–6.73 G clean data remained per sample. After removing the host’s genomic data, only 0.32–0.39 G data per sample remained for analysis. We also used a read-based (mapping) approach for metagenomic analysis.
The dominant bacterial species in EG female moths were Wolbachia endosymbiont (64.98%) and E. mundtii (29.35%) (Figure 5A). Wolbachia spp. were detected in all five EG samples and were the most abundant bacteria in EG1, EG2, and EG5 (Figure 5B).
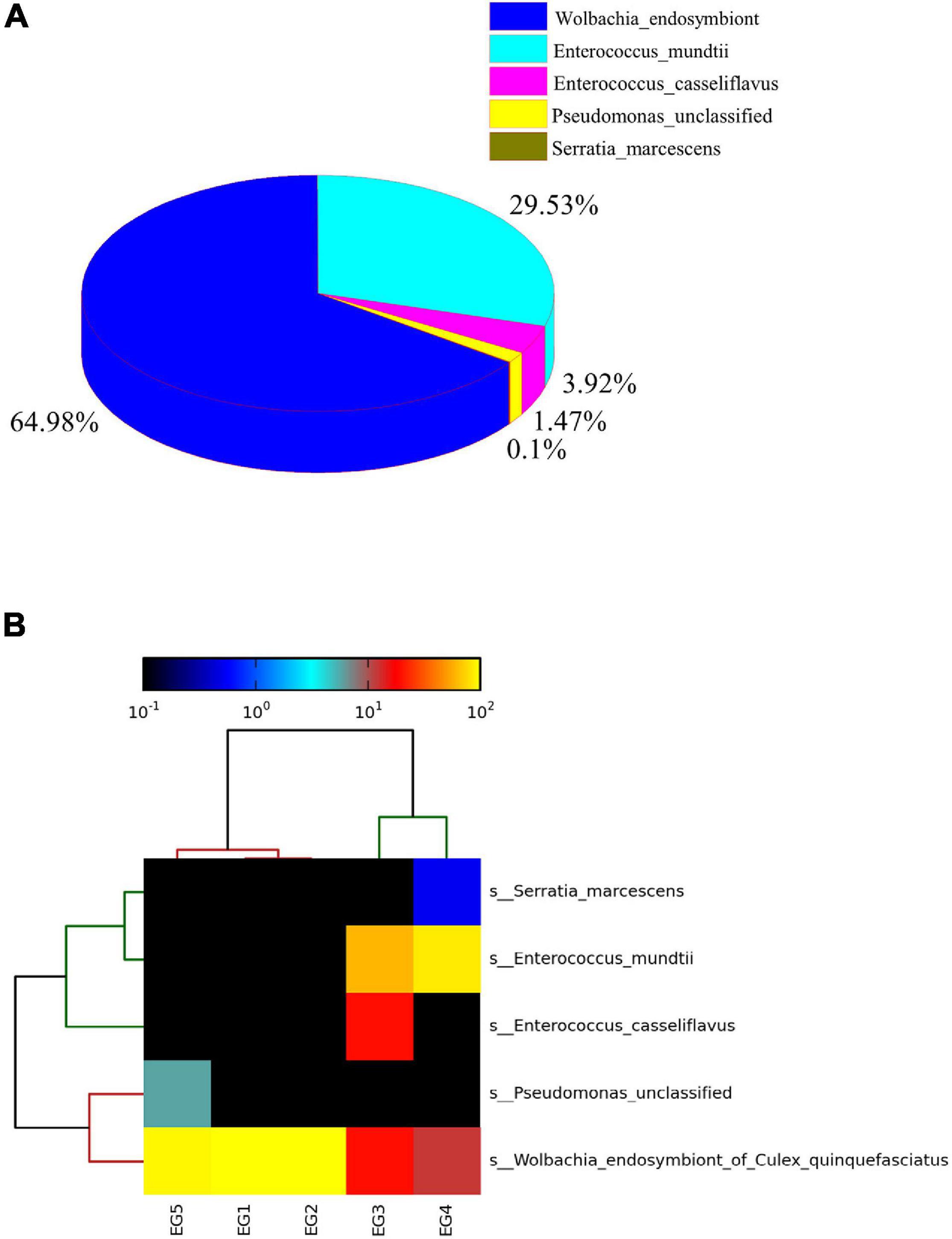
Figure 5. Relative abundances of microbiota species in E. grisescens female moths (A). Hierarchical clustering analysis of five samples identified in E. grisescens female moths (B).
Next, molecular typing analyses were conducted to identify the Wolbachia strain in E. grisescens. Five Wolbachia strains were isolated from E. grisescens collected from Anhui, Hubei, Zhejiang, Fujian, and Yunnan (the five representative tea-producing provinces in China) and were designated as wGriAH, wGriHB, wGriZJ, wGriFJ, and wGriYN, respectively.
Figure 6 shows the tree based on the general data set using the ML implementation after bootstrapping 1000 times. The topology shows the division of Wolbachia into four supergroups, A, B, D, and F. wGriAH, wGriHB, wGriZJ, wGriFJ, and wGriYN were in supergroup B, subgroup Pip. All the reference members of this group are CI-inducing strains (Figure 6).
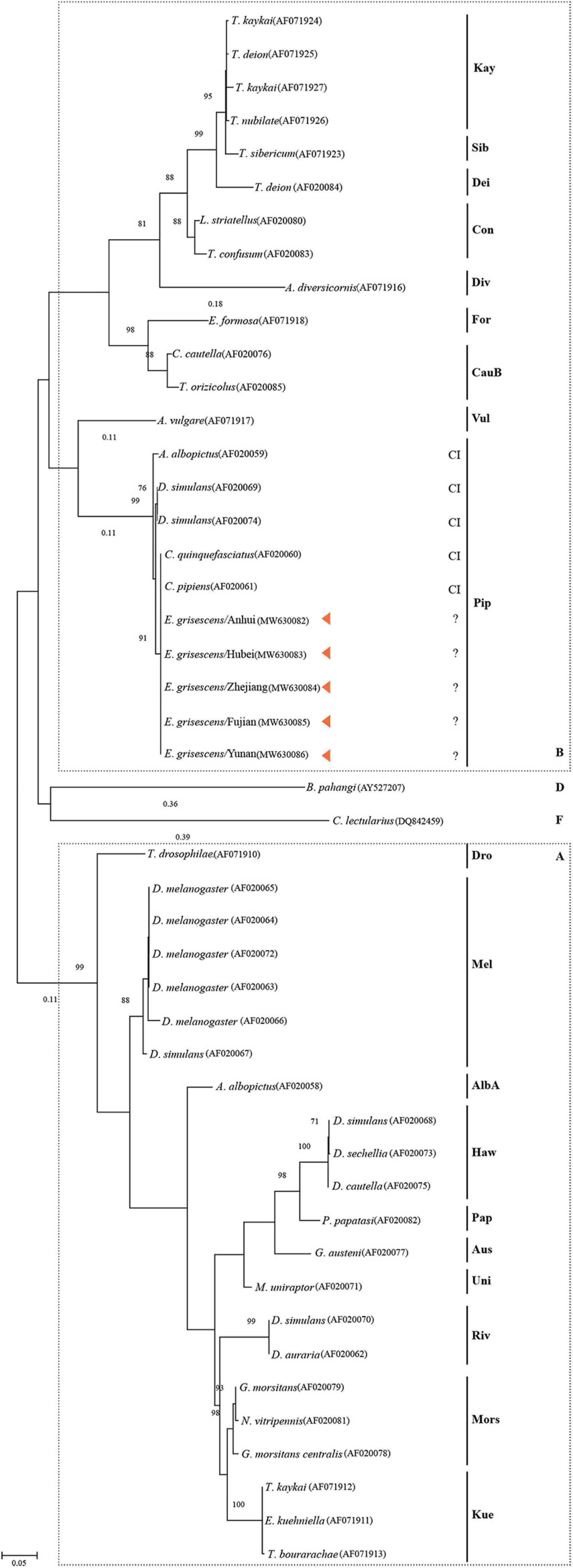
Figure 6. Phylogenetic tree of Wolbachia based on the wsp sequences. The tree has been constructed by the Maximum likelihood method. The name of the host arthropod species is listed in Supplementary Table 1. Bar represents amino acid substitutions per site. Bootstrap values are shown at each node. CI, cytoplasmic incompatibility; A, supergroup A; B, supergroup B; D, supergroup D; F, supergroup F.
In the MLST typing analyses, wGriAH, wGriHB, wGriZJ, wGriFJ, and wGriYN had the same allelic profile and HVR profiles. The allelic profile for gatB, coxA, hcpA, ftsZ, and fbpA was 39, 14, 40, 36, and 4 (ST-41), respectively, and the HVR profiles for HVR1, 2, 3, and 4 were 10, 8, 10, and 8 (wsp-10), respectively (Table 3).
The MLST-based ML tree separated the isolates into three major clusters: A, B, and D + F (Figure 7A). Within supergroup B, wGriAH, wGriHB, wGriZJ, wGriFJ, and wGriYN, as one subclade, formed a cluster with Ekue_B, Cpip_B, and Cqui_B, all of which are CI-inducing strains. Differently, the HVR-based ML tree separated the isolates into five major clusters: A, B-I, B-II, and B-III, and a cluster containing D and F (Figure 7B). wGriAH, wGriHB, wGriZJ, wGriFJ, and wGriYN formed a subclade with Cpip_B and Cqui_B and placed in B-I. All the available reference Wolbachia strains shared the same HVR and had CI-inducing phenotypes (Figure 7B).
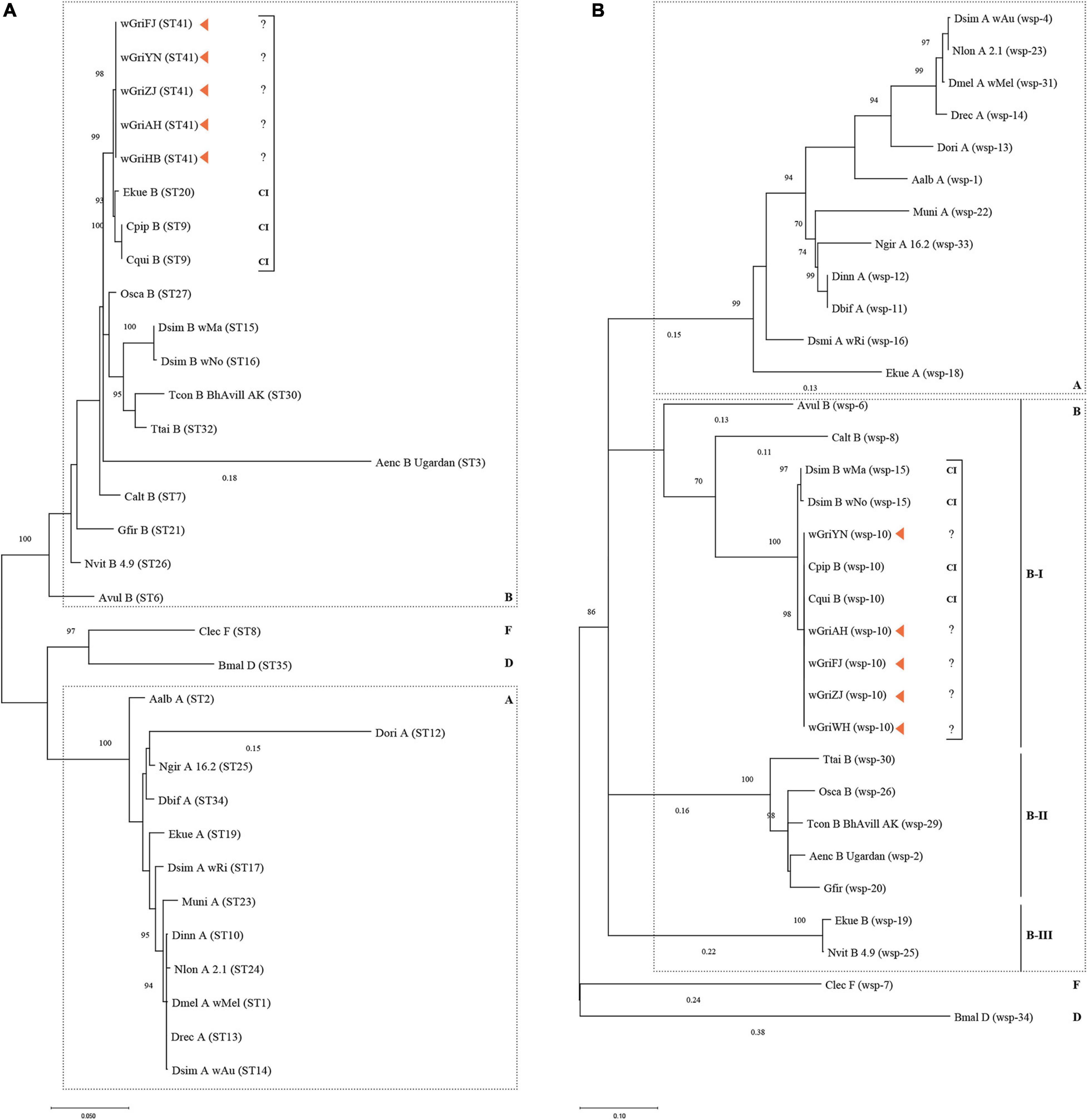
Figure 7. Phylogenetic analysis based on Wolbachia MLST and wsp sequences. Maximum likelihood trees constructed using the MLST allelic (A) and WSP HVR (B) profiles. For strain names, see Supplementary Table 2. Bar represents amino acid substitutions per site. Bootstrap values are shown at each node. CI, cytoplasmic incompatibility; A, supergroup A; B, supergroup B; D, supergroup D; F, supergroup F.
Based on this, Wolbachia isolates wGriAH, wGriHB, wGriZJ, wGriFJ, and wGriYN were identified as the same strain, designated as wGri. Thus, wGri was classified as a potentially CI-inducing Wolbachia strain in supergroup B.
To explore the effects of Wolbachia strain wGri on E. grisescens reproduction, we treated E. grisescens with antibiotics to eliminate wGri and carried out reciprocal cross analyses. To eliminate the effects of antibiotic interference on the cross results, E. obliqua and S. subpunctaria were treated with the same antibiotics.
The survival rates of E. grisescens infected with Wolbachia (EG-CK) or treated with antibiotics (EG-T) showed no significant difference (unpaired and two-tailed t-tests; p > 0.5) (Figure 8A). Egg production (fecundity) was significantly greater in infected females (♂ × ♀; EG-CK × EG-CK: 320.56 ± 35.13 eggs per female and EG-T × EG-CK: 322.56 ± 77.48 eggs per female) than in uninfected females (♂ × ♀; EG-T × EG-CK: 225.67 ± 54.99 eggs per female and EG-T × EG-T: 189.12 ± 26.64 eggs per female) in both tests (Figure 8D). This indicated that the presence of wGri increased the number of eggs produced.
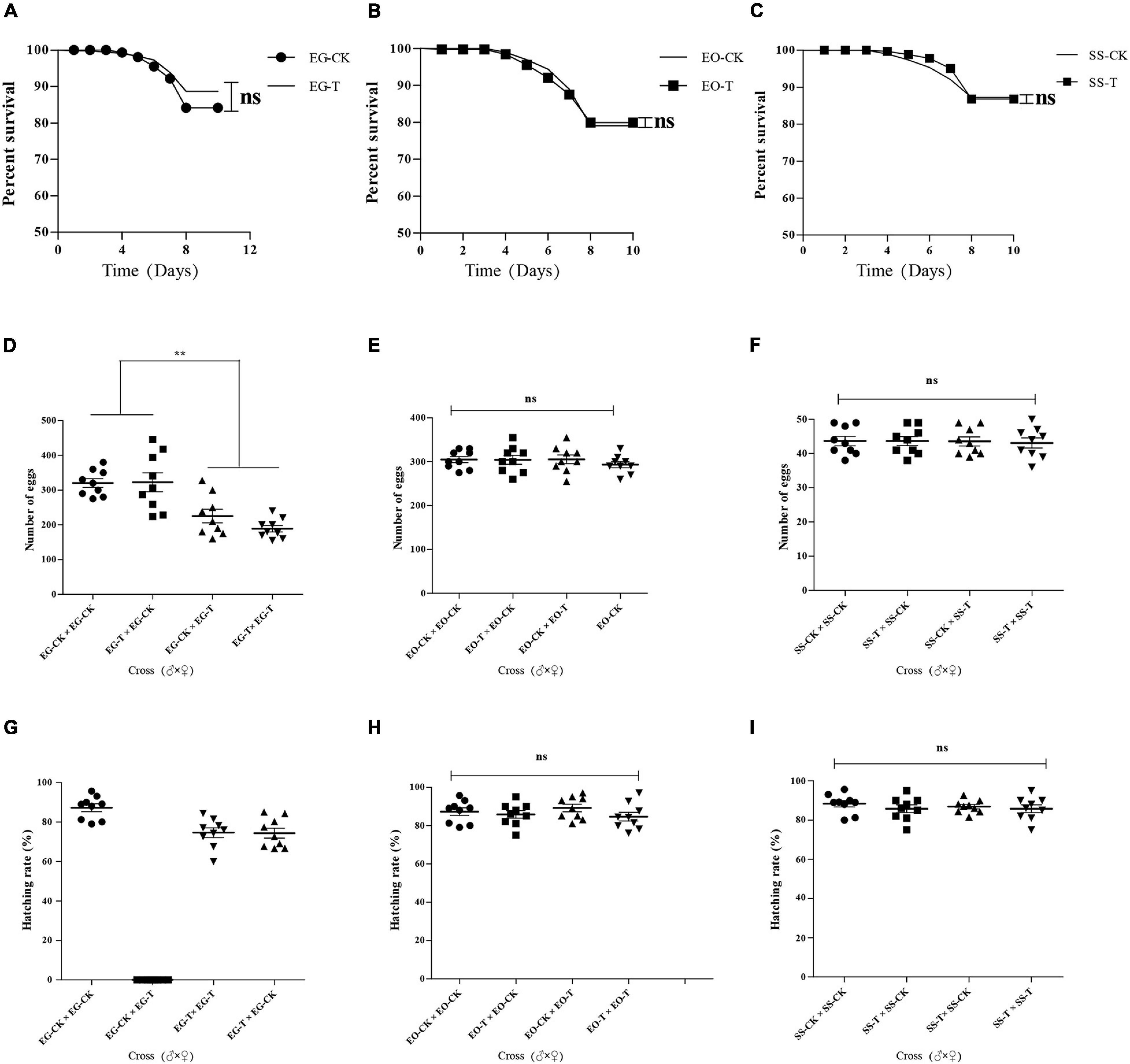
Figure 8. Effect of antibiotic on the survival rates of EG (A), EO (B), and SS (C); fecundity of EG (D), EO (E), and SS (F); and fertility of EG (G), EO (H), and SS (I). EG, EO, and SS refer to E. grisescens, E. obliqua, and S. subpunctaria, respectively. Significant differences were detected by unpaired two-tailed t-test; ns: no significant difference; **p < 0.001.
The fertility (total number of eggs hatched) rates of E. grisescens infected with Wolbachia (EG-CK) or treated with antibiotics (EG-T) were not significantly different (unpaired and two-tailed t-tests; p > 0.5). However, the uninfected females mated with infected males showed 0% eggs hatched per female (100% embryo mortality). Infected females mated with uninfected males had high fertility rates (Figure 8G). These results indicated that Wolbachia strain wGri induces strong unidirectional CI in E. grisescens.
The survival (Figures 8B,C), fertility (Figures 8E,F), and fecundity rates (Figures 8H,I) of E. obliqua and S. subpunctaria showed no significant difference between the control and the antibiotic treatments, confirming that antibiotics did not affect the results of the crossing analyses.
These data indicated that wGri resulted in a high CI level in E. grisescens and enhanced the fecundity of the female hosts.
Crossing experiments were conducted using E. grisescens and E. obliqua. The hatching rate of the eggs produced by E. grisescens females mated with E. obliqua males was 75%, while the eggs produced by E. obliqua females mated with E. grisescens males did not hatch, or very few hatched (Figure 9).
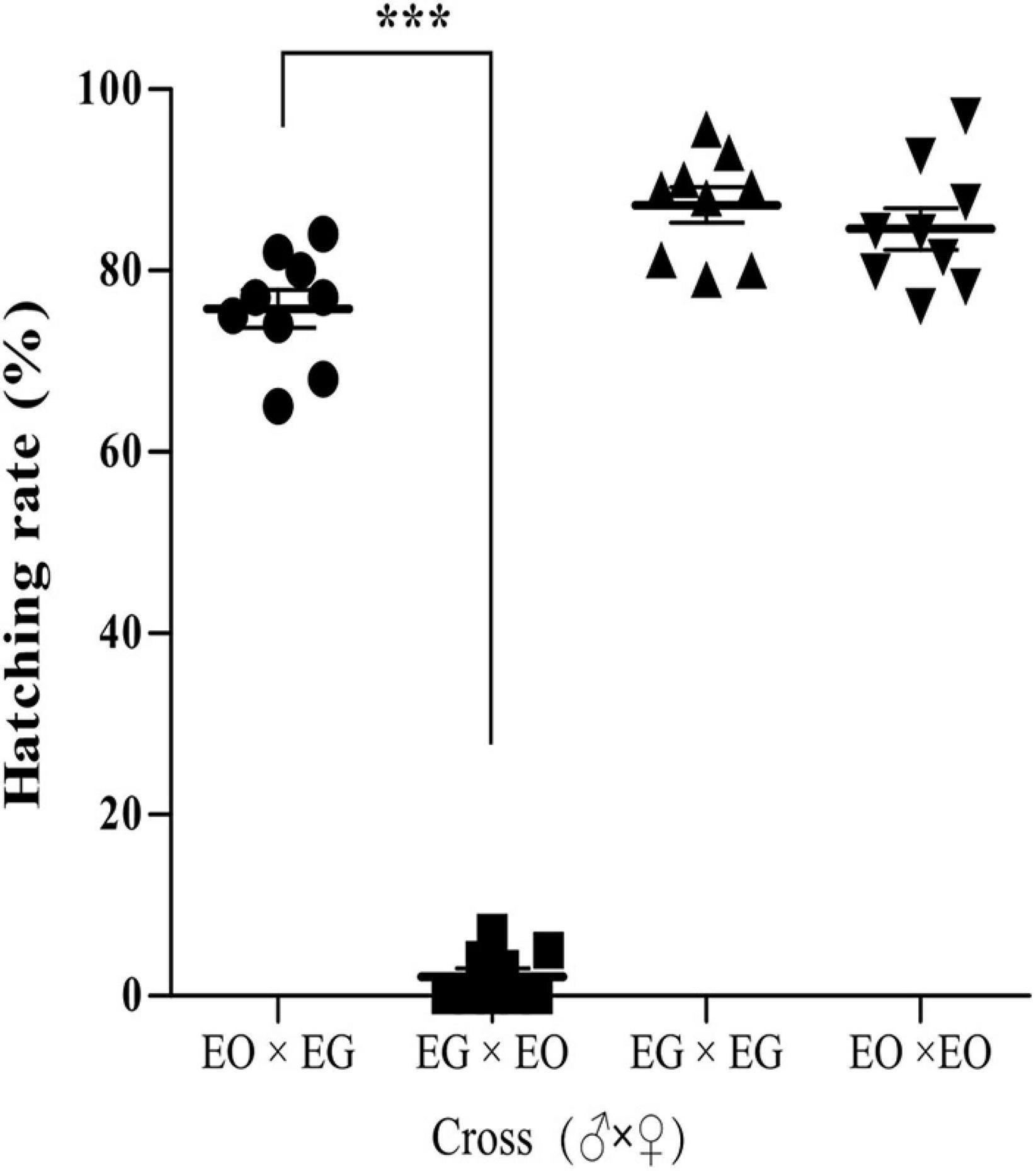
Figure 9. Crossing experiments between E. grisescens (EG) and E. obliqua (EO). Significant differences were detected by unpaired two-tailed t-test; ***p < 0.0001.
These findings indicated that Wolbachia strain wGri influences reproductive communication between E. grisescens and E. obliqua through CI.
The microbiomes of insects play important roles in mediating their health and fitness, and hence, microbiomes are attracting the attention of entomologists (Oliver et al., 2010; Engel and Moran, 2013; Blow and Douglas, 2019). In this study, a culture-independent approach was used to compare the composition and diversity of gut bacteria among three tea Geometridae moths, E. grisescens, E. obliqua, and S. subpunctaria. The results revealed that Enterococcus was the dominant bacterial genus in the gut of the three Geometridae larvae. Many studies have reported that Enterococcus is active in the gut throughout the larval life cycle of most Lepidoptera and probably plays a key role in insect defense against potentially harmful microorganisms (Dillon and Dillon, 2004; Chen B. S. et al., 2020). The results of the metagenomic analyses demonstrated that E. mundtii was the main Enterococcus species in the gut of E. grisescens.
Although the alpha- and beta-diversity indexes of gut bacteria were lowest in E. grisescens larvae, the composition of core gut bacteria (except for Wolbachia) was similar among the three Geometridae species. Wolbachia are the most widespread endosymbiotic bacteria among arthropods. In these hosts, they are notorious for their reproductive parasitism, which ensures their spread, even though it may lower host fitness (Vavre et al., 2002). For instance, CI provides the infected females with a reproductive advantage in the population, allowing Wolbachia to rapidly expand in the host population (Werren et al., 2008). In this study, we detected Wolbachia in all the tested E. grisescens samples collected from five different representative tea plantations. Molecular typing analyses demonstrated that these Wolbachia were the same strain, designed as wGri. Our analyses showed that wGri has a strong capability to induce CI. Besides, in a previous study, Wolbachia were detected in all the tested E. grisescens samples that had been collected from eight different geographical tea-producing areas in China (Wang et al., 2020). Consequently, we speculate that the 100% wGri infection rate of E. grisescens in nature has resulted in the spread of wGri in E. grisescens via induced unidirectional CI.
Wolbachia also have the potential to engage in mutualistic relationships with their hosts (Zug and Hammerstein, 2015). In insects, there are a few cases whereby Wolbachia act as an obligate mutualist. For example, in the bed bug Cimex lectularius, elimination of Wolbachia (supergroup F) through antibiotic treatment renders abnormal development of eggs, which can be restored by dietary supplementation of synthesizes biotin and riboflavin (Hosokawa et al., 2010; Nikoh et al., 2014). Wolbachia also act as facultative mutualists whereby hosts benefit from infection; they do not depend on Wolbachia for survival or fecundity (Kaur et al., 2021). The results of the mating experiment showed that wGri could enhance the fecundity of its host, indicative of a mutually beneficial symbiosis between wGri and E. grisescens. Meanwhile, elimination of wGri through antibiotic treatment did not affect its host survival rate. Based on this, we hypothesize that Wolbachia strain wGri as the facultative symbiont of its host E. grisescens has contributed to its rapid spread in tea gardens by enhancing its fecundity.
The tea Geometrid moths E. grisescens and E. obliqua are closely related species that feed on tea leaves. In China, E. grisescens and E. obliqua are treated as the same species in tea garden management because their morphology is so similar that it is difficult to tell them apart (Wang et al., 2019). According to the records, E. obliqua has always been one of the main tea pests in China. Previous data showed that it was distributed in the main tea-producing areas, while E. grisescens was mainly distributed in southern tea-producing areas (Luo et al., 2017). However, recent surveys have found that the distribution of E. obliqua, which was once distributed in all tea-producing areas, has greatly reduced. In contrast, E. grisescens, once a relatively minor pest species, is now distributed in almost all tea-producing provinces in China (Jiang et al., 2014; Li et al., 2019). These sibling species can mate. Our results showed that mating between E. grisescens and E. obliqua conformed to the phenomenon of unidirectional CI. In other words, the eggs produced by female E. grisescens mated with male E. obliqua are able to hatch. The characteristics of Wolbachia maternal inheritance have promoted rapid evolution of its host. We suggest that the gradual decrease in the distribution of E. obliqua and the gradual increase in the distribution of E. grisescens is related to the presence of Wolbachia strain wGri.
The results of this study show that Wolbachia strain wGri results in a high level of CI in E. grisescens and enhances the fecundity of its female hosts. We propose that the presence of wGri in its host E. grisescens has contributed to host rapid spread among tea gardens by improving the host’s fertility and interfering with the reproduction of its competitors (E. obliqua).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MW630082-MW630111 and https://www.ncbi.nlm.nih.gov/, PRJNA720281.
YY, YZ, and YlZ designed the research. YL supervised the study. YZ, SL, RJ, CZ, and TG performed the experiments. YZ analyzed and wrote the manuscript. YZ, RJ, and CL revised the manuscript. All authors approved the final version for submission.
This study was supported by the National Natural Science Foundation of China (Grants 31870635 and 32072421), Anhui Provincial Important Science & Technology Specific Projects (201903a06020019), National Key Research and Development Programs (2019yfd1001601), the Anhui provincial key R&D projects (202004e11020006), and the Open Fund of State Key Laboratory of Tea Plant Biology and Utilization (SKLTOF20200104).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.694466/full#supplementary-material
Baldo, L., Dunning Hotopp, J. C., Jolley, K. A., Bordenstein, S. R., Biber, S. A., and Choudhury, R. R. (2006). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 72, 7098–7110. doi: 10.1128/AEM.00731-06
Bandi, C., Anderson, T. J., Genchi, C., and Blaxter, M. L. (1998). Phylogeny of Wolbachia in filarial nematodes. Proc. Biol. Sci. 265, 2407–2413. doi: 10.1098/rspb.1998.0591
Blow, F., and Douglas, A. E. (2019). The hemolymph microbiome of insects. J. Insect Physiol. 115, 33–39. doi: 10.1016/j.jinsphys.2019.04.002
Brownlie, J. C., Cass, B. N., Riegler, M., Witsenburg, J. J., Iturbe-Ormaetxe, I., McGraw, E. A., et al. (2009). Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog. 5:e1000368. doi: 10.1371/journal.ppat.1000368
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chao, A., Ma, M. C., and Yang, M. C. K. (1993). Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika 80, 193–201. doi: 10.2307/2336768
Chen, B. S., Zhang, N., Xie, S., Zhang, X. C., and Shao, Y. Q. (2020). Gut bacteria of the silkworm Bombyx mori facilitate host resistance against the toxic effects of organophosphate insecticides. Environ. Int. 143:105886. doi: 10.1016/j.envint.2020.105886
Chen, L. L., Zhou, H., and Zhao, J. (2020). Potential climate-suitable distribution of Ectropis grisescens in China based on the CLIMEX and ArcGIS prediction. Chin. J. Tea Sci. 40, 817–829. doi: 10.13305/j.cnki.jts.2020.06.009
Clarke, K. R. (1993), Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Dedeine, F., Vavre, F., Fleury, F., Loppin, B., Hochberg, M. E., and Boulé-treau, M. (2001). Removing symbiotic Wolbachia bacteria specififically inhibits oogenesis in a parasitic wasp. Proc. Natl. Acad. Sci. U.S.A. 98, 6247–6252. doi: 10.1073/pnas.101304298
Dillon, R. J., and Dillon, V. M. (2004). The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49, 71–92.
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Guo, Y., Hoffmann, A. A., Xu, X. Q., Zhang, X., and Hong, X. Y. (2018). Wolbachia -induced apoptosis associated with increased fecundity in Laodelphax striatellus (Hemiptera: Delphacidae). Insect Mol. Biol. 27(Suppl. 1), 769–807. doi: 10.1111/imb.12518
Hosokawa, T., Koga, R., Kikuchi, Y., Meng, X. Y., and Fukatsu, T. (2010). Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. U.S.A. 107, 769–774. doi: 10.1073/pnas.0911476107
Iturbe-Ormaetxe, I., Walker, T., and O’ Neill, S. L. (2011). Wolbachia and the biological control of mosquito-borne disease. EMBO Rep. 12, 508–518. doi: 10.1038/embor.2011.84
Ji, H. G., Lee, Y. R., Lee, M. S., Hwang, K. H., Kim, E. H., Park, J. S., et al. (2016). Metabolic phenotyping of various tea (Camellia sinensis L.) cultivars and understanding oftheir intrinsic metabolism. Food Chem. 233, 321–330. doi: 10.1016/j.foodchem.2017.04.079
Jiang, H., Yu, F., Qin, L., Zhang, N., Cao, Q., Schwab, W., et al. (2019). Dynamic change in amino acids, catechins, alkaloids, and gallic acid in six types of tea processed from the same batch of fresh tea (Camellia sinensis l.) leaves. J. Food Compos. Anal. 77, 28–38. doi: 10.1016/j.jfca.2019.01.005
Jiang, N., Liu, S. X., Xue, D. Y., Tang, M. J., Xiao, Q., and Han, H. X. (2014). External morphology and molecular identification of two tea Geometrid moth from southern China. Chin. J. Appl. Entomol. 51, 987–1002. doi: 10.7679/j.issn.2095-1353.2014.118
Ju, J. F., Bing, X. L., Zhao, D. S., Guo, Y., Xi, Z., Hoffmann, A. A., et al. (2020). Wolbachia supplement biotin and riboflavin to enhance reproduction in planthoppers. ISME J. 14, 676–687. doi: 10.1038/s41396-019-0559-9
Kaur, R., Shropshire, J. D., Cross, K., Leigh, B., Mansueto, A., Stewart, V., et al. (2021). Living in the endosymbiotic world of Wolbachia: a centennial review. Cell Host Microbe 29, 879–893. doi: 10.1016/j.chom.2021.03.006
Lefoulon, E., Bain, O., Makepeace, B. L., Haese, C., Uni, S., Martin, C., et al. (2016). Breakdown of coevolution between symbiotic bacteria Wolbachia and their filarial hosts. PeerJ 4:e1840. doi: 10.7717/peerj.1840
Lefoulon, E., Clark, T., Borvetõ, F., Perriat-Sanguinet, M., Moulia, C., Slatko, B., et al. (2020). Pseudoscorpion Wolbachia symbionts: diversity and evidence for a new supergroup S. BMC Microbiol. 20:188. doi: 10.21203/rs.2.22118/v2
Lei, S., Zhang, F., Yun, Y., Zhou, W., and Peng, Y. (2020). Wolbachia bacteria affect rice striped stem borer (Chilo suppressalis) susceptibility to two insecticides. Bull. Insectol. 73, 39–44.
Li, Z. Q., Cai, X. M., Luo, Z. X., Bian, L., Xin, Z. J., Liu, Y., et al. (2019). Geographical distribution of Ectropis grisescens (Lepidoptera: Geometridae) and Ectropis obliqua in China and description of an efficient identification method. J. Econ. Entomol. 1, 277–283. doi: 10.1093/jee/toy358
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/aem.71.12.8228-8235.2005
Luo, Z. X., Li, Z. Q., Cai, X., Lei, B., and Chen, Z. (2017). Evidence of premating isolation between two sibling moths: Ectropis grisescens and Ectropis obliqua (Lepidoptera: Geometridae). J. Econ. Entomol. 110, 2364–2370. doi: 10.1093/jee/tox216
Martijn, T., and Ellers, J. (2008). Wolbachia endosymbiont is essential for egg hatching in a parthenogenetic arthropod. Evol. Ecol. 23:931. doi: 10.1007/s10682-008-9282-0
Mateos, M., Montoya, H. M., Lanzavecchia, S. B., Conte, C., and Tsiamis, G. (2020). Wolbachia pipientis associated with tephritid fruit fly pests: from basic research to applications. Front. Microbiol. 11:1080. doi: 10.3389/fmicb.2020.01080
Moriyama, M., Nikoh, N., Hosokawa, T., and Fukatsu, T. (2015). Riboflavin provisioning underlies Wolbachia’s fitness contribution to its insect host. mBio 6:e01732-15. doi: 10.1128/mBio.01732-15
Nikoh, N., Hosokawa, T., Moriyama, M., and Fukatsu, T. (2014). Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. U.S.A. 111, 10257–10262. doi: 10.1073/pnas.1409284111
O’Neill, S. L., Giordano, R., Colbert, A. M., Karr, T. L., and Robertson, H. M. (1992). 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. U.S.A. 89, 2699–2702. doi: 10.1073/pnas.89.7.2699
Oliver, K. M., Degnan, P. H., Burke, G. R., and Moran, N. A. (2010). Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55, 247–266. doi: 10.1146/annurev-ento-112408-085305
Pan, Y., Fang, G., Wang, Z., Cao, Y., and Zhan, S. (2021). Chromosome—evel genome reference and genome editing of the tea geometrid. Mol. Ecol. Resour. doi: 10.1111/1755-0998.13385 [Epub ahead of print].
Paniagua, V., Frago, E., Martin, K., Monika, H., and Fatouros, N. E. (2018). Bacterial symbionts in lepidoptera: their diversity, transmission, and impact on the host. Front. Microbiol. 9:556. doi: 10.3389/fmicb.2018.00556
Paulson, J. N., Pop, M., and Bravo, H. C. (2011). Metastats: an improved statistical method for analysis of metagenomic data. Genome Biol. 12:17.
Pielou, E. C. (1966). The measurement of diversity in diferent types of biological collections. J. Theor. Biol. 13, 131–144. doi: 10.1016/0022-5193(66)90013-0
Ros, V. I. D., Fleming, V. M., Feil, E. J., and Breeuwer, J. A. J. (2009). How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae). Appl. Environ. Microbiol. 75, 1036–1043. doi: 10.1128/AEM.01109-08
Rousset, F., Bouchon, D., Pintureau, B., Juchault, P., and Solignac, M. (1992). Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proc. R. Soc. B Biol. Sci. 250, 91–98. doi: 10.1098/rspb.0135
Segata, N., Waldron, L., Ballarini, A., Narasimhan, V., Jousson, O., and Huttenhower, C. (2012). Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 9, 811–814. doi: 10.1038/nmeth.2066
Shannon, C., and Weaver, W. (1949). The mathematics of communication. Sci. Am. 181, 11–15. doi: 10.2307/24967225
Shao, Y., Chen, B., Sun, C., Ishida, K., Hertweck, C., and Boland, W. (2017). Symbiont-derived antimicrobials contribute to the control of the lepidopteran gut microbiota. Cell Chem. Biol. 24, 66–75. doi: 10.1016/j.chembiol.2016.11.015
Stouthamer, R., Breeuwer, J., and Hurst, G. (1999). Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53, 71–102. doi: 10.1146/annurev.micro.53.1.7
Teixeira, L., Ferreira, A., and Ashburner, M. (2008). The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6:e2. doi: 10.1371/journal.pbio.1000002
Too, J. C., Kinyanjui, T., Wanyoko, J. K., and Wachira, F. N. (2015). Effect of sunlight exposure and different withering durations on theanine levels in tea (Camellia sinensis). Food Nutr. Sci. 6, 1014–1021. doi: 10.4236/fns.2015.611105
van Meer, M. M. M., Witteveldt, J., and Stouthamer, R. (1999). Phylogeny of the arthropod endosymbiont Wolbachia based on the wsp gene. Insect Mol. Biol. 8, 399–408. doi: 10.1046/j.1365-2583.1999.83129.x
Vavre, F., Fleury, F., Varaldi, J., Fouillet, P., and Boulétreau, M. (2002). Infection polymorphism and cytoplasmic incompatibility in Hymenoptera Wolbachia associations. Heredity 88, 361–365. doi: 10.1038/sj.hdy.6800063
Wang, Z. B., Bai, J. H., Liu, Y. J., Li, H., Zhan, S., and Xiao, Q. (2019). Transcriptomic analysis reveals insect hormone biosynthesis pathway involved in desynchronized development phenomenon in hybridized sibling species of tea Geometrids (Ectropis grisescens and Ectropis obliqua). Insects 10:381. doi: 10.3390/insects10110381
Wang, Z. B., Li, H., Zhou, X. G., Tang, M., Sun, L., Zhan, S., et al. (2020). Comparative characterization of microbiota between the sibling species of tea geometrid moth Ectropis obliqua Prout and E. grisescens Warren. Bull. Entomol. Res. 110, 684–693. doi: 10.1017/S0007485320000164
Weinert, L. A., Araujo-Jnr, E. V., Ahmed, M. Z., and Welch, J. (2015). The incidence of bacterial endosymbionts in terrestrial arthropods. Proc. Biol. Sci. 282:20150249. doi: 10.1098/rspb.2015.0249
Werren, J. H., and Bartos, J. D. (2001). Recombination in Wolbachia. Curr. Biol. 11, 431–435. doi: 10.1016/S0960-9822(01)00101-4
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Werren, J. H., Zhang, W., and Guo, L. R. (1995). Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. R. Soc. Lond. B 261, 55–63. doi: 10.1098/rspb.1995.0117
Yen, J. H., and Barr, A. R. (1971). New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiens L. Nature 232, 657–658. doi: 10.1038/232657a0
Zhang, H. H. (2001). Fauna of tea geometrid in China. Chin. J. Tea Sci. 2, 157–160. doi: 10.13305/j.cnki.jts.2001.02.019
Zhang, Y., Zhao, T. Y., Deng, J. D., Long, Y. H., Zhang, L. W., and Yang, Y. Q. (2019). Positive effects of the tea catechin (-)-epigallocatechin-3-gallate on gut bacteria and fitness of Ectropis obliqua Prout (Lepidoptera: Geometridae). Sci. Rep. 9:5021. doi: 10.1038/s41598-019-41637-9
Zhou, W., Rousset, F., and O’Neill, S. (1998). Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. 265, 509–515. doi: 10.1098/rspb.1998.0324
Zug, R., and Hammerstein, P. (2012). Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7:e38544. doi: 10.1371/journal.pone.0038544
Keywords: cytoplasmic incompatibility, culture-independent, mutualism, reciprocal cross, multilocus sequence types
Citation: Zhang Y, Liu S, Jiang R, Zhang C, Gao T, Wang Y, Liu C, Long Y, Zhang Y and Yang Y (2021) Wolbachia Strain wGri From the Tea Geometrid Moth Ectropis grisescens Contributes to Its Host’s Fecundity. Front. Microbiol. 12:694466. doi: 10.3389/fmicb.2021.694466
Received: 13 April 2021; Accepted: 07 June 2021;
Published: 19 July 2021.
Edited by:
George Tsiamis, University of Patras, GreeceReviewed by:
Aggeliki Saridaki, Technical University of Crete, GreeceCopyright © 2021 Zhang, Liu, Jiang, Zhang, Gao, Wang, Liu, Long, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunqiu Yang, eXlxX2x5aEBhaGF1LmVkdS5jbg==; YinglaoZhang, emhhbmd5bEBhaGF1LmVkdS5jbg==; Yanhua Long, bG9uZ3lhbmh1YUBhaGF1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.