- 1Ecosystems and Environment Research Program, University of Helsinki, Helsinki, Finland
- 2SUGAR, X-star, Japan Agency of Marine-Earth Science and Technology (JAMSTEC), Yokosuka, Japan
- 3UMR 8187 - LOG - Laboratoire d’Océanologie et de Géosciences, Université de Lille - CNRS, Université du Littoral Côte d’Opale, Station Marine de Wimereux, Lille, France
- 4Evolution, Cell Biology, and Symbiosis Unit, Okinawa Institute of Science and Technology, Okinawa, Japan
- 5Research Institute for Global Change (RIGC), Japan Agency of Marine-Earth Science and Technology (JAMSTEC), Yokosuka, Japan
Foraminifera are unicellular eukaryotes that are an integral part of benthic fauna in many marine ecosystems, including the deep sea, with direct impacts on benthic biogeochemical cycles. In these systems, different foraminiferal species are known to have a distinct vertical distribution, i.e., microhabitat preference, which is tightly linked to the physico-chemical zonation of the sediment. Hence, foraminifera are well-adapted to thrive in various conditions, even under anoxia. However, despite the ecological and biogeochemical significance of foraminifera, their ecology remains poorly understood. This is especially true in terms of the composition and diversity of their microbiome, although foraminifera are known to harbor diverse endobionts, which may have a significant meaning to each species’ survival strategy. In this study, we used 16S rRNA gene metabarcoding to investigate the microbiomes of five different deep-sea benthic foraminiferal species representing differing microhabitat preferences. The microbiomes of these species were compared intra- and inter-specifically, as well as with the surrounding sediment bacterial community. Our analysis indicated that each species was characterized with a distinct, statistically different microbiome that also differed from the surrounding sediment community in terms of diversity and dominant bacterial groups. We were also able to distinguish specific bacterial groups that seemed to be strongly associated with particular foraminiferal species, such as the family Marinilabiliaceae for Chilostomella ovoidea and the family Hyphomicrobiaceae for Bulimina subornata and Bulimina striata. The presence of bacterial groups that are tightly associated to a certain foraminiferal species implies that there may exist unique, potentially symbiotic relationships between foraminifera and bacteria that have been previously overlooked. Furthermore, the foraminifera contained chloroplast reads originating from different sources, likely reflecting trophic preferences and ecological characteristics of the different species. This study demonstrates the potential of 16S rRNA gene metabarcoding in resolving the microbiome composition and diversity of eukaryotic unicellular organisms, providing unique in situ insights into enigmatic deep-sea ecosystems.
Introduction
Benthic foraminifera are single-celled eukaryotes, widespread in global oceans and particularly abundant in deep-sea sediments, accounting for up to 50% or more of the total eukaryotic biomass in places (Snider et al., 1984; Gooday et al., 1992). They are important consumers of phytodetritus (Gooday, 1988; Ohga and Kitazato, 1997; Moodley et al., 2002), experimentally shown to consume carbon faster than metazoans (Nomaki et al., 2005). Part of their ecological success in the benthic environment may be attributed to their ability to thrive in the low-oxygen environments (e.g., Moodley and Hess, 1992; Bernhard, 1993; Jorissen et al., 1995; Moodley et al., 1997; Langlet et al., 2013). Adaptations to hypoxia may include, for example, the wide-spread ability of foraminifera to accumulate and respire nitrate (Risgaard-Petersen et al., 2006; Piña-Ochoa et al., 2009), as well as their capability to reduce metabolism, suspend reproduction, and enter a state of dormancy (LeKieffre et al., 2017; Koho et al., 2018; Richirt et al., 2020).
A yet unresolved link in deep-sea foraminiferal ecology and environmental adaptation is the composition and role of their associated microbiomes. Benthic foraminifera are known to develop endobiotic relationships and close associations with prokaryotes (e.g., Bernhard, 2003; Bernhard et al., 2006, 2018), which may include denitrifiers (Bernhard et al., 2011), sulphur cycle-related bacteria (Tsuchiya et al., 2015; Salonen et al., 2019), Cyanobacteria (Prazeres et al., 2017), or even methanotrophs (Bernhard and Panieri, 2018). The endobiotic relationships between foraminifera and bacteria have been hypothesized to provide the host with metabolic flexibility, alternative carbon assimilation pathways, and enhanced nutrient/organic compound cycling, enabling them to not only survive but also flourish in the harsh benthic environment (Bernhard et al., 2011; Bird et al., 2017; Prazeres et al., 2017; Tsuchiya et al., 2018; Salonen et al., 2019). Despite the ecological importance of microbiome in foraminiferal adaptation, very little is known of its prevalence, significance, and evolution. Some studies indicate that the prokaryotic composition of the microbiome may be flexible and display a considerable amount of local variation (Prazeres et al., 2017) whereas evidence of species-specific microbiomes have also been observed in shallow intertidal areas with no indication of spatial variability (Salonen et al., 2019).
Foraminiferal microbiome may also be directly linked to their microhabitat preferences (Fontanier et al., 2005). In sediments, foraminifera display species-specific preferences in their vertical distribution, which may depend on the geochemical zonation of the sediment and the availability of oxygen, other electron acceptors (e.g., nitrate), and organic matter (e.g., Corliss, 1985; Jorissen et al., 1995; Fontanier et al., 2002; Koho and Piña-Ochoa, 2012). Microhabitat preferences are typically reflected in foraminifera’s trophic behavior and survival strategies (e.g., Kitazato, 1988; Tsuchiya et al., 2018). For example, some deep-sea epifaunal and shallow-infaunal species, living above or within the surface sediments, respectively, have been suggested to entirely depend on an opportunistic lifestyle specialized in rapid consumption of phytodetritus in response to phytoplankton blooms occurring in the ocean surface (Gooday, 1988). They also typically migrate faster in sediments than the deep-dwellers (Kitazato, 1988; Ernst et al., 2002). In contrast, deep infaunal species, which can live below oxygen penetration depth, react slower to the fresh organic matter inputs (Nomaki et al., 2005, 2006). This may be due to their suspected longer life cycle, during which they are likely to rely on deposit-feeding and consumption of degraded organic matter instead of fresh phytodetritus (Goldstein and Corliss, 1994; Ohga and Kitazato, 1997; Schmiedl et al., 2004). However, the exact feeding preferences of foraminifera, especially on species level, are poorly resolved due to the difficulty of examining them in situ. New molecular approaches, such as metabarcoding, can provide new insights on foraminiferal ecology and their feeding preferences (Chronopoulou et al., 2019).
In marine science, the research on microbiomes has rapidly increased during the last decade, but host-associated microbiome research is still mainly focused on multicellular and larger animals, such as sponges and corals, instead of single-celled organisms (Trevathan-Tackett et al., 2019). In the case of foraminifera, previous studies on their intracellular bacterial communities have been mainly based on transmission electron microscopy (TEM) observations (e.g., Bernhard et al., 2018; Koho et al., 2018). However, the current molecular ecology tools, such as DNA metabarcoding, offer new, efficient ways to investigate the diversity and composition of microbiomes of the unicellular foraminifera (e.g., Bird et al., 2017; Prazeres et al., 2017; Salonen et al., 2019).
In this study, we applied the definition of microbiome by Trevathan-Tackett et al. (2019), stating that microbiome is a consortium of intracellular bacteria derived from genetic material, to investigate the microbiome of deep-sea foraminifera with variable microhabitat distributions. We utilized a DNA metabarcoding approach based on 16S rRNA gene to investigate the microbiome characteristics of deep-sea foraminifera in terms of intracellular bacterial and chloroplast composition. Comparison was also made with the surrounding bacterial sediment community to detect and identify bacteria that are enriched in relative abundance (RA) inside the foraminifera. The bacterial composition of the foraminiferal microbiome is likely reflecting the ecological characteristics of the given foraminiferal species, and therefore, the overall aim of the study was to gain insights into individual ecological strategies of the studied foraminiferal species and identify bacteria-foraminifera interactions that may be endobiotic in nature. In addition, the foraminiferal chloroplast composition was used to identify potential differences in trophic strategies.
Materials and Methods
Sediment Sampling
The sampling was carried out onboard the research vessel R/V Seiyo Maru in early June 2017 at Sagami Bay, Japan (35°02ꞌ52ꞌꞌN 139°19ꞌ46ꞌꞌE). Samples were taken with a mini-multiple corer equipped with four cores (inner diameter 8.2 cm), allowing the collection of undisturbed sediments. Altogether, three casts were deployed and eight cores from 734 to 735 m water depth were collected.
Pore water sampling (see below for details) from two cores (SB4 and SB5) was carried out immediately after sampling, on board the research vessel (Table 1). Afterwards all cores were kept cool and transported into the JAMSTEC laboratory within 8 h of sampling for core slicing and further processing. Cores SB4 and SB5 were sliced for Rose Bengal staining of foraminifera (see Supplementary Additional Figure 1). Five cores (SB1–SB3, X2, and X3) were used for the collection of living benthic foraminifera of which three (SB1–SB3) were also sampled for sediment bacterial community (see more details below). A core (SB6) was also used for dissolved oxygen profiling. In addition, a ninth core (SB19) was collected from the same site at Sagami Bay in September 2019 during the R/V Kaimei KM19-07 cruise and was used for additional oxygen profiling and pore water nutrients (nitrate, nitrite, and ammonium) analysis (Table 1).
Pore Water Sampling and Analyses
Pore water was extracted on board the research vessel using Rhizons™ (Rhizosphere Research Products B.V., The Netherlands). Prior to the deployment, cores (SB4, SB5; Table 1) with predrilled holes were covered using water resistant tape. Immediately after retrieving the cores, the tape was removed, and Rhizons™ with connected 10 ml plastic syringes were used to extract pore water with 1 cm intervals starting from 0.5 cm sediment depth down to 10 cm. After, the samples were acidified with suprapur 1 M HNO3 (10 μl per 1 ml of sample) and stored for elemental analyses (namely for analyses of iron and manganese) with inductively coupled plasma-optical emission spectrometry (ICP-OES) at the Geolab of Utrecht University, the Netherlands.
Core SB6 (Table 1) was used for oxygen profiling. Five oxygen profiles were retrieved using 100 μm tip oxygen microsensor (Unisense, Denmark) with a 100 μm vertical resolution. The oxygen microsensor was two-point calibrated in an anoxic sodium-ascorbate solution and a saturated solution of sea water. Overlying water temperature was 11–14°C.
During the KM19-07 cruise in 2019, onboard analyses of a core (SB19; Table 1) for both dissolved pore water oxygen and nutrients (nitrite, nitrate, and ammonium) were carried out as described in Langlet et al. (2020). In brief, oxygen micro-profiling was carried out in a cold room at 4°C immediately after sediment sampling. The overlying water for nutrient analyses was sampled prior to the oxygen profiling. After the oxygen profiling, sediments were sliced at every 1 or 2 cm thickness and centrifuged at 2,600 × g for 5 min. The supernatant was sampled for porewater, filtered together with the overlaying water sample over a 0.45 μm syringe filter, and subsequently measured for dissolved nitrogen species with a continuous-flow analyzer (BL-Tech QUAATRO 2-HR system).
Sediment Processing and Extraction of Living Foraminifera
At JAMSTEC laboratory, three of the cores (cores SB1, SB2, and SB3; Table 1) were sliced down to 5 cm depth, first 2 cm with 0.5 cm intervals, and then with 1 cm intervals (Supplementary Additional Figure 2). From each sediment slice, a subsample of approximately 1–2 g (wet weight) of sediment was taken with a sterile plastic spatula and placed into a sterile vial for the analysis of the surrounding sediment bacterial community. The bacterial samples were immediately frozen in liquid nitrogen and stored in −20°C until DNA extraction. Two additional cores (X2 and X3) were used to retrieve living foraminifera from which only a top slice of 0–1 cm was examined (Table 1; Supplementary Additional Figure 2).
The sediment slices for foraminifera were subsequently sieved using filtered natural seawater through 150 μm mesh, and living foraminifera were picked in a chilled-petri dish under binocular microscope. Only specimens leaving a displacement track on a thin layer of sediment and confirmed active were selected, as described in Koho et al. (2018). After morphological identification under the stereomicroscope, each specimen was thoroughly washed three times with sterile artificial nitrate-free sea water (ASW, prepared with 35 g of RedSea salt per litter of MilliQ ultrapure water, and autoclaved before use).
The foraminifera selected for microbiome analysis were placed into a sterile vial filled with RNAlater solution, which dissolves the calcite shell of foraminifera while preserving the DNA. The samples were stored in RNAlater solution in +8°C until DNA extraction (see Bird et al., 2017). Before extraction, the naked foraminiferal cells were again washed three times with sterile ASW to obviate any remaining shell material or residual RNAlater solution from hampering the downstream analysis (Bird et al., 2017; Salonen et al., 2019). By dissolving the shell and performing repeated washing prior and after that, we ensured that the extracted bacterial community was indeed intracellular and not shell associated.
The 16S rRNA gene of altogether 50 specimens was successfully (see Supplementary Additional Table 1) sequenced for this study: four Bulimina striata, 18 Bulimina subornata, 16 Chilostomella ovoidea, 11 Globobulimina pacifica, and two Nonionella labradorica. The five species studied here have contrasting microhabitat preferences, as B. subornata and B. striata are shallow-infaunal species (Kitazato, 1989; Supplementary Additional Figure 1), N. labradorica is a shallow- and intermediate-infaunal species (Kitazato, 1989; Fontanier et al., 2014; Supplementary Additional Figure 1), and C. ovoidea and G. pacifica are deep-infaunal species (Corliss, 1985; Jorissen et al., 1995; Ohga and Kitazato, 1997; Supplementary Additional Figure 1).
Denitrification and Nitrate Pool Measurements
After the foraminifera were identified and cleaned, 5–11 individuals of the same species were pooled together to test denitrification activity and determine intracellular nitrate quantity. Nitrate respiration rates were then determined based on Fick’s first law of diffusion from steady-state N2O profiles measured with a N2O micro electrode (Andersen et al., 2001) after acetylene inhibition of N2O reduction (Smith et al., 1978; Risgaard-Petersen et al., 2006; Høgslund et al., 2008) in anoxic and nitrate free ASW at 11°C.
Specimens selected for intracellular nitrate determination were placed in 1 ml centrifugation microtubes and frozen at −80°C to break the foraminiferal cell and conserve the samples until further nutrient content analysis by following Nomaki et al. (2015) and Langlet et al. (2020). Nitrate and nitrite extraction from the foraminiferal cells was achieved by freezing at −80°C, and the frozen cells were then dissolved in 200 μl of MilliQ water and homogenized in the microtube using a plastic pestle. NO3− + NO2− was determined by the vanadium (III) chloride reduction method adapted from Doane and Horwáth (2003) where 160 μl of the aliquot was mixed with 20 μl of nitrate reductant (8 g of VCl3 per liter of 0.6 mol/l HCl) and 20 μl of color reagent (prepared from 2 g sulfanilamine and 100 mg n-1-naphthyl ethylenediamine dihydrochloride per liter of 0.6 mol/l HCl) and heated at 60°C for 2 h. Calibration was performed using 0.1–50 μmol/l NO3− + NO2− standards prepared with KNO3 and MilliQ water. The NO3− + NO2− concentration was determined from the absorbance at a wavelength of 540nm with a UV–VIS spectrophotometer (UV-1800, Shimadzu corp.) and normalized to the number of individuals to calculate individual NO3− + NO2− content (in pmol per individual).
DNA Extraction, Amplification, and Illumina MiSeq Sequencing
The DNA from within each foraminiferal specimen was extracted within 2 months of collection using the DOC method (Holzmann and Pawlowski, 1996). Sediment DNA from the three replicates was extracted using the DNeasy PowerSoil® DNA Isolation Kit (Qiagen, Germany). The DNA from foraminifera and sediment was amplified using universal bacterial primers 27F (5ꞌAGAGTTTGATCMTGGCTCAG) and 519R (5ꞌGTATTACCGCGGCTGCTG) targeting the variable regions V1–V3 of the 16S rRNA gene, in a similar way as described in Salonen et al. (2019). Duplicate PCR reactions were performed and pooled in equal volumes, in order to ensure adequate amplicon volume for Illumina MiSeq library preparations and minimize PCR amplification bias. All amplicons were checked with agarose gel electrophoresis. Sequencing was carried out on the Illumina MiSeq platform in the laboratory of DNA sequencing and Genomics in the Institute of Biotechnology, at the University of Helsinki. Prior to sequencing, the amplicons were purified and multiplexed as described in Salava et al. (2017). Barcodes for later sample de-multiplexing were selected using BARCOSEL (Somervuo et al., 2018).
As single-cell DNA extractions are susceptible to contamination, careful measures were taken to monitor and exclude any bacterial signal non-related to foraminifera. Blank samples were taken for all the steps and reagents involved in the foraminiferal processing, including RNAlater (two blanks), ASW used for cleaning of the foraminifera (three blanks) and the DOC extraction buffer (one blank sample). Negative controls were routinely checked in the PCR step. Regardless of whether the blank samples produced a visible band on the agarose gel or not, they were sequenced and analyzed alongside the rest of the samples. Additionally, one negative control that produced a faint band on the agarose gel was also sequenced and analyzed. The blank and negative control samples were of low sequencing depth and resulted in a distinct community where the most abundant bacterial groups included Propionibacterium, Sphingomonas, Corynebacterium, and Pseudomonas, which are common contaminants in DNA extraction kits and laboratory reagents (Salter et al., 2014). In addition, we observed a Halomonas contamination particularly associated with the ASW blank samples. The operational taxonomic units (OTUs) associated with the blank samples and negative control (in total 1.4% of all reads) were bioinformatically removed from the final dataset. In addition, mitochondrial sequence reads and unclassified OTUs were removed from the dataset.
Illumina MiSeq sequencing produced 4,789,674 sequence reads in the sediment dataset (23 sediment samples) and 9,660,770 in the foraminiferal dataset (50 specimens in total). Trimming, quality-checking, and removal on chimeric sequences reduced the datasets to 2,558,292 and 5,534,414 reads for sediment and foraminifera, respectively. These reads were clustered into 449,494 and 141,217 OTUs. To avoid overestimating diversity, the OTU number was further reduced by plotting the total number of reads against estimated cutoffs (see Chronopoulou et al., 2019; Salonen et al., 2019). A cutoff of 20 OTUs per sample reduced the number of reads to 1,883,492 (95.9% of original reads) in the sediment dataset, consisting of 9,056 unique OTUs in total. In the foraminiferal data, a cutoff of five OTUs was used, which reduced the amount of reads to 2,834,713 (95.8% of original reads) and the number of OTUs to 8,489 in total. The number of OTUs and reads per individual samples are reported in the Supplementary Additional Table 2.
Full-Length 16S rRNA Gene Sequencing
In addition, DNA extracts from four B. subornata, one B. striata, one G. pacifica, two C. ovoidea, and one sediment sample (0–0.5 cm sediment depth, core SB1) were amplified with primers 27F (5ꞌ-AGAGTTTGATCMTGGCTCAG-3ꞌ) and 1492R (5ꞌ-GGTTACCTTGTTACGACTT-3ꞌ) to obtain full-length 16S rRNA gene sequences (Lane, 1991). A fragment of approximately 1,465 bp was sequenced with PacBio RS II sequencing instrument (Pacific Biosciences) in the Institute of Biotechnology, University of Helsinki, Finland. These long 16S rRNA gene sequences were analyzed in a similar way to shorter MiSeq reads, except adjusting the maximum length to 1,465 bp. The full-length sequencing of the 16S rRNA gene produced in total 20,480 sequences that formed 3,298 OTUs.
Bioinformatical and Statistical Analysis
Raw sequences were de-multiplexed based on barcodes. Then, MiSeq overhangs, primers, and barcodes were removed, and sequences were quality-checked, trimmed (maximum length adjusted to 550 bp), and aligned using Mothur, following the standard operating procedure (version 1.41.1; Schloss et al., 2009). Chimeric sequences were removed in Mothur using the UCHIME algorithm (Edgar et al., 2011). Taxonomy was assigned against the SILVA reference database (release 132). OTU tables created in Mothur were analyzed in R (version 3.6.3; R Core Team 2020) using the package phyloseq (version 1.30.0; McMurdie and Holmes, 2013). Non-metric multidimensional scaling (nMDS) was used to visualize the separation of foraminiferal species and the five most common intracellular bacterial groups in the ordination space. Permutational multivariate ANOVA (PERMANOVA) test using the adonis2 function in Vegan (Oksanen et al., 2015) was used to analyze the significant variables (e.g., species and sediment depth) influencing the sediment bacterial community and the foraminiferal microbiome. For the nMDS, Hellinger transformed (Legendre and Gallagher, 2001) data were used. Alpha diversity estimates are dependent on singletons and doubletons, and therefore, untrimmed and non-normalized data (as recommended by McMurdie and Holmes, 2014) were used to calculate Shannon diversity (Hꞌ) and Pielou’s evenness (Jꞌ) indices in both sediment and foraminiferal datasets using the Vegan package (version 2.5-6; Oksanen et al., 2015).
Phylogenetic Analysis
Closest relatives for the phylogenetic trees were obtained using a BLAST search against the NCBI GenBank database (Altschul et al., 1990). Representative sequences of the most common chloroplast OTUs, including Cyanobacteria, in foraminifera (24 OTUs in total) and sediment (six OTUs in total) were aligned with their closest matches from NCBI GenBank using the MUSCLE algorithm (v. 3.8.31; Edgar, 2004). Aligned sequences were manually checked, and Maximum likelihood phylogenetic trees were constructed with MEGA7 (version 7.0.26; Kumar et al., 2016).
Results
Porewater Geochemistry
In the 2017 measurement, the overlying water oxygen (O2) concentration ranged from 100 to 130 μmol/l and O2 penetration depth in sediment varied from 0.52 to 0.85 cm (Figure 1). The results were consistent with measurements of 2019 when the overlying water O2 concentration was 100 μmol/l and the penetration depth 0.6 cm (Figure 1). As the measured O2 conditions in the 2 years were similar (Figure 1), an assumption was made that the pore water ammonium (NH4+), nitrite (NO2−), and nitrate (NO3−) concentrations in year 2017 resembled the 2019 situation.
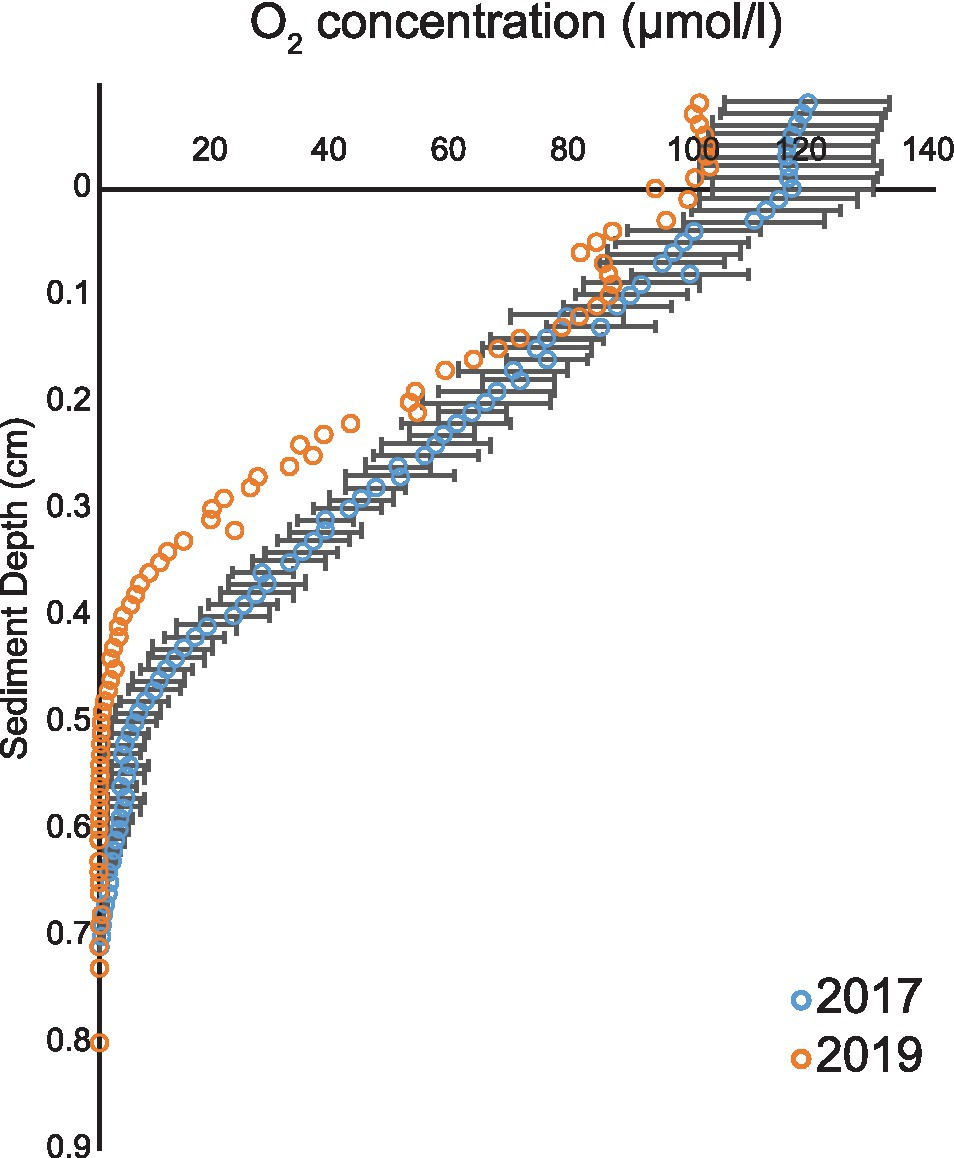
Figure 1. Oxygen profiles from years 2017 and 2019. Average (blue open circles) and SEM (black line) of five O2 profiles during 2017 and 2019 cruises (orange open circles).
Below oxygen and nitrate penetration depth manganese (Mn2+) and iron (Fe3+) reduction was detected. The concentration of Mn2+ reached up to 11.5 μmol/l at approximately 1.5 cm sediment depth and concentration of Fe3+ up to 80 μmol/l at approximately 3.5 cm sediment depth (Figure 2). Based on measurements from 2019 (Table 1), a NO3− reduction zone was present in the sediment down to approximately 1.5 cm depth (Figure 2), which is consistent with the Mn reduction taking place at similar sediment depths. The concentration of NH4+ increased steadily below the O2 penetration depth, reaching 47 μmol/l at 10 cm sediment depth (Figure 2).
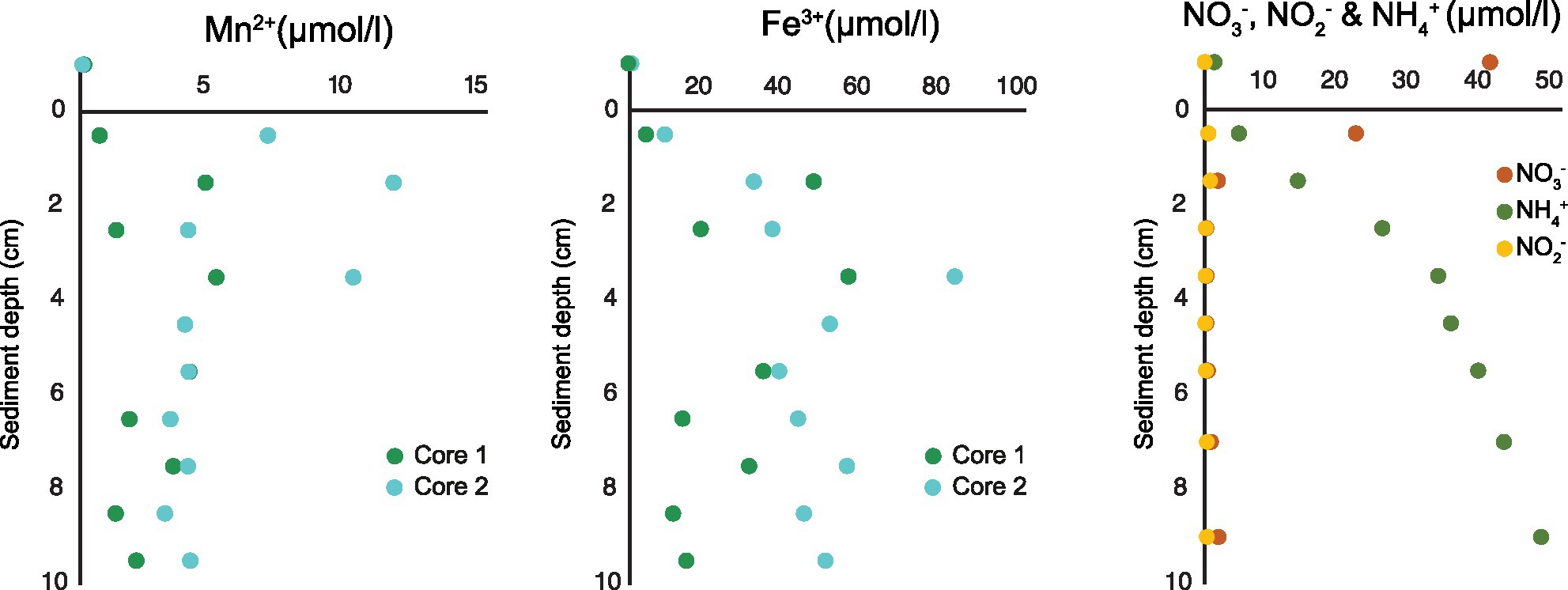
Figure 2. Porewater profiles of manganese and iron (2017) and nitrate, nitrite, and ammonium (2019) in 0–10 cm sediment depth.
Intracellular Nitrate Content and Nitrate Respiration
Intracellular nitrate + nitrite pool was measured for three species, including B. subornata, C. ovoidea, and G. pacifica. The largest intracellular nitrate pool was found in G. pacifica (678.8 ± 143.5 pmol per individual; Table 2) followed closely C. ovoidea, having intracellular nitrate concentration of 624.8 pmol/individual (±143.5; three replicates), and B. subornata 305.7 ± 259.2 pmol/individual (eight replicates; Table 2). The denitrification rate for B. subornata was 39.9 pmol per individual per day (average of three replicates ±16.2; Table 2).
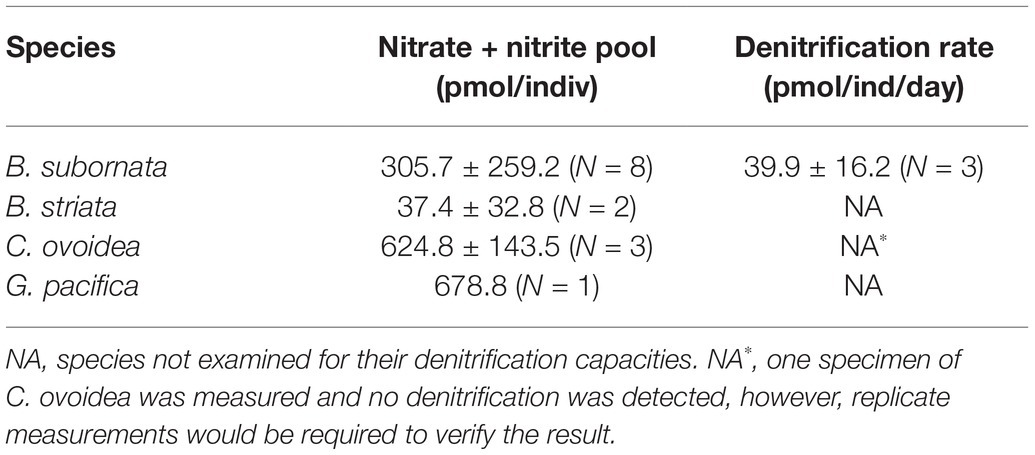
Table 2. Average (±SD) intracellular nitrate content per foraminiferal cell and denitrification rates for all tested species.
Sediment Bacterial Community
Sediment bacterial community was dominated by Deltaproteobacteria [up to 39.5% relative abundance (RA)], followed by Gammaproteobacteria (up to 30% RA) and Bacteroidia (up to 20.7% RA; Figure 3A). The class Deltaproteobacteria was dominated by the order Desulfobacterales (up to 48% RA), Myxococcales (up to 24.8% RA), and NB1-j group (up to 24.6% RA). The sediment bacterial community changed with depth (Figure 3A); for example, the class Deltaproteobacteria (from average RA 19.5% ±0.8 to 38.5% ±1.3) and Anaerolinae (from average RA 1% ±0.1 to 4.3% ±0.3) increased in relative abundance with increasing sediment depths whereas classes Bacteroidia (16.6% ±3.5 to 8.9% ±0.8) and Gammaproteobacteria (27.8% ±2.3 to 11.8% ±1.6) decreased. The full-length 16S rRNA sequencing of the surface sediment (0–0.5 cm) suggested that bacterial community was dominated by Gammaproteobacteria (30.6% RA), followed by Deltaproteobacteria (RA 20.4%), Chloroplasts (10.5%), and Alphaproteobacteria (9.1%; Figure 3B). PERMANOVA test using the adonis2 function in vegan including two variables (depth and core) confirmed that depth was a significant factor influencing the sediment community variance (p = 0.001, F = 9.29). The three different cores had similar dominant groups and similar depth-related community changes but displayed some heterogeneity reflected within their varying communities (PERMANOVA p = 0.044, F = 2.02). The cores had similar Shannon-Wiener (Hꞌ) indices, Hꞌ ranging from 7.4 ± 0.1 (SB1) to 7.5 ± 0.1 (SB2 and SB3). Core SB1 also had the lowest Pielou’s evenness (Jꞌ) index 1.6 ± 0.1, whereas SB3 (1.7 ± 0.4) and SB2 (1.8 ± 0.4) were higher.
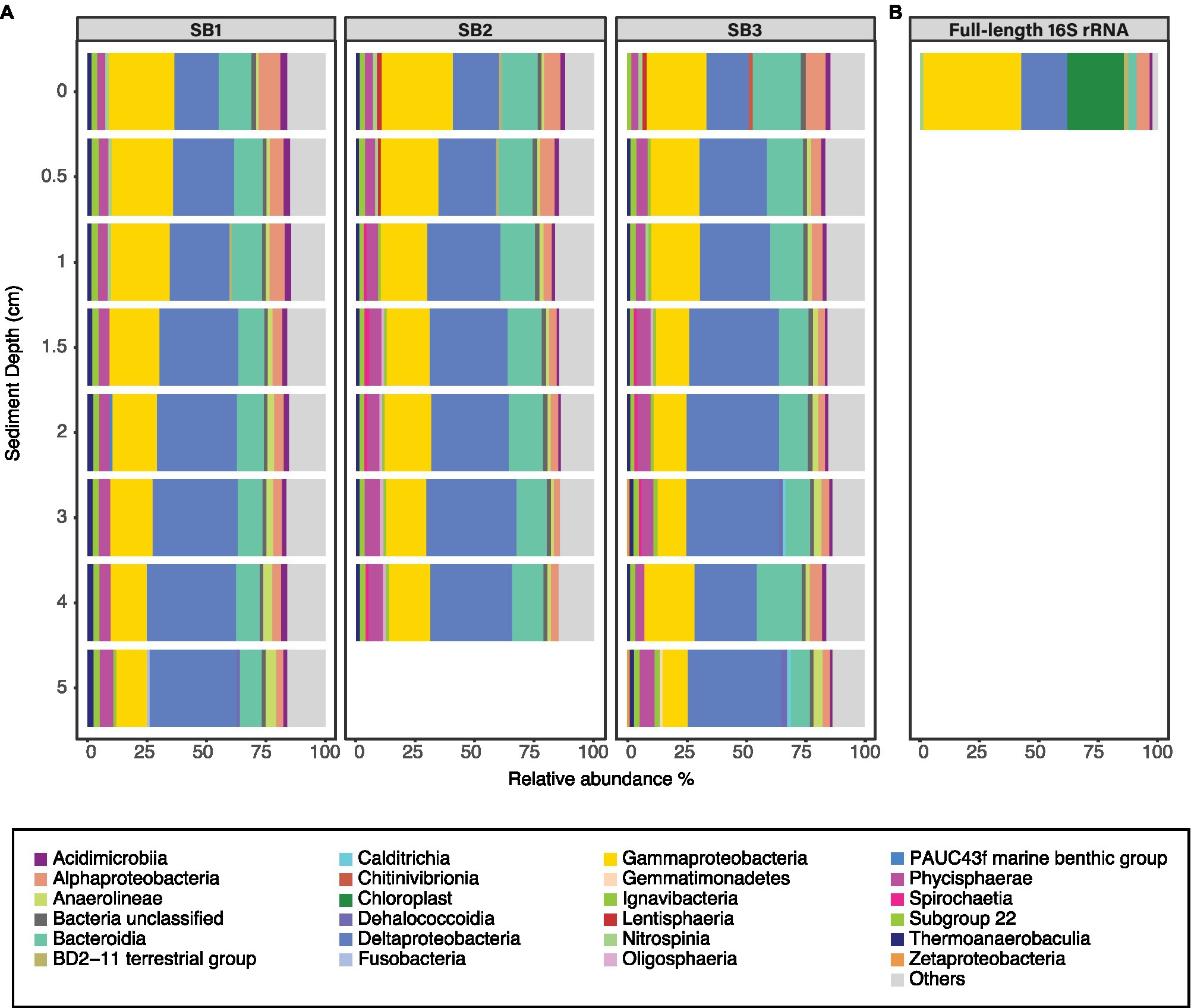
Figure 3. Relative abundance of bacterial classes (RA > 2%) in sediment samples based on Illumina MiSeq (A) and full-length (B) sequencing of the 16S rRNA gene. The three horizontal facets (A) indicate the community in cores SB1, SB2, and SB3. Those bacterial classes that are present <2% RA are grouped in the group “others.”
Bacterial Microbiome of Foraminifera: Diversity and Composition
Compared to sediment, the alpha diversity (i.e., Hꞌ and Jꞌ) of the foraminiferal microbiome index was lower (Figure 4). Median Hꞌ value for sediment samples was 7.5 (Figure 4), with highest values recorded at sediment depth 1–1.5 (Hꞌ 7.6) and lowest at sediment depth 4–5 cm (Hꞌ 7.2). Median Jꞌ for sediment was 1.6 (Figure 4). Among foraminiferal species, highest Hꞌ and Jꞌ index was recorded for G. pacifica with median 4 and 0.8, respectively (Figure 4). Median Hꞌ and Jꞌ index values for B. striata were 2.1 and 0.4 and for B. subornata 2.4 and 0.6 (Figure 4), whereas C. ovoidea had the lowest Hꞌ and Jꞌ value of the studied species with medians of 1.5 and 0.3, respectively (Figure 4). PERMANOVA test including three variables (species, sediment depth, and core) indicated that the species was the strongest factor influencing the foraminiferal microbiome (p = 0.001, F = 5.74), whereas sediment depth (p = 0.033, F = 1.93) and core (p = 0.789, F = 0.86) seemed not to have a significant effect on the microbiome composition. As visualized in the nMDS plot, species C. ovoidea, B. subornata, B. striata, and N. labradorica were separated into distinguishable group in the ordination space due to intraspecific homogeneity whereas G. pacifica specimens depicted more intraspecific variation (Figure 5) as indicated also by the higher Hꞌ and Jꞌ median (Figure 4). Of the five most abundant bacterial families, Cyanobiaceae and Hyphomicrobiaceae correlated with B. striata and B. subornata whereas the family Marinilabiliaceae correlated with C. ovoidea in the ordination space (Figure 5).
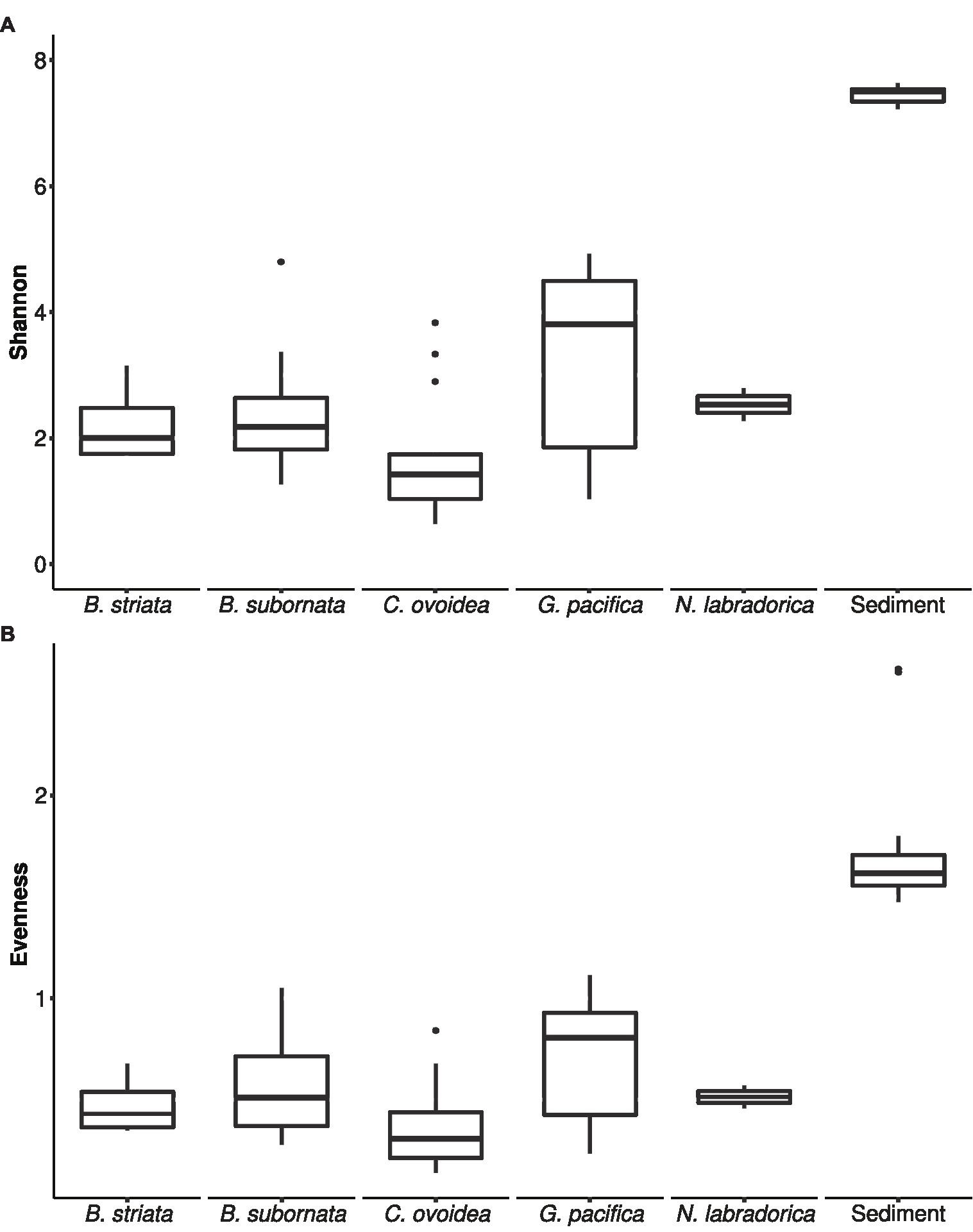
Figure 4. Box and whiskers plot of microbiome alpha-diversity in foraminiferal species and sediment based on Shannon H' index (A) and Pielou’s evenness index J' (B). The lower and upper hinges of the box correspond to the first and third quartiles, the line in between the median, and the whiskers extend to 1.5 times the interquartile range. Values outside this range are represented by black circles.
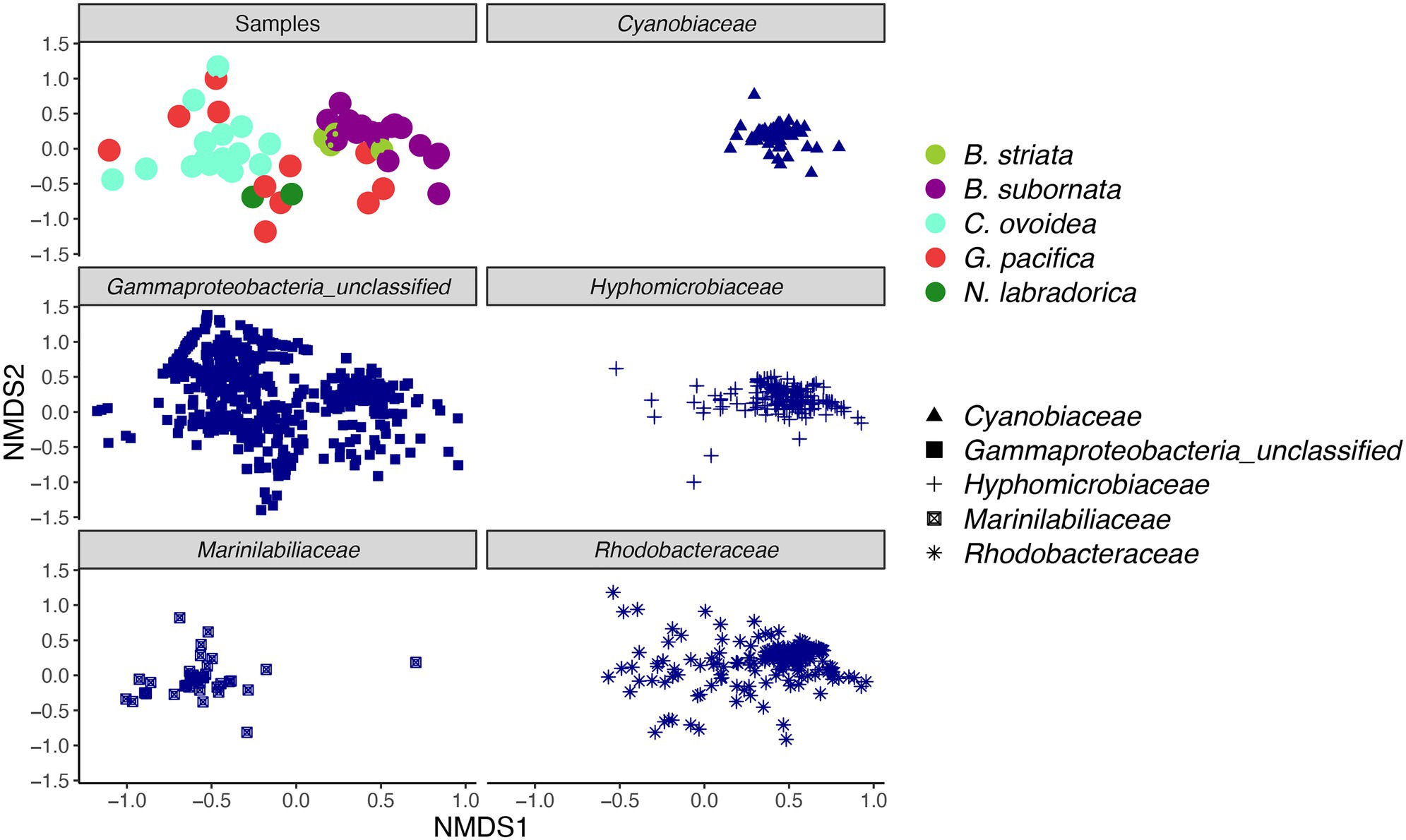
Figure 5. Non-metric multidimensional scaling (nMDS) plot of intracellular bacterial microbiome of foraminiferal species (top left) and five most abundant bacterial families. Foraminiferal species marked in different colors and bacterial families with different shape. nMDS was based on a Bray-Curtis distance (stress 0.198).
In total, four B. striata and 18 B. subornata specimens were analyzed. Typically, living B. subornata displayed a dark brown, sometimes almost purple color (Supplementary Additional Figure 3). The most abundant bacterial OTUs in both of these species belonged to the family Hyphomicrobiaceae [phylum Alphaproteobacteria; average across samples 24.9% RA, ±18 (B. striata), and 34.8% ±24.5 (B. subornata); Figure 6A]. This OTU was present in the majority of the specimens in high relative abundance (up to 71.6% RA), whereas in the surrounding sediments, it had RA < 0.1%, indicating a clear enrichment in the microbiome of these species (Supplementary Additional Table 3). The full-length 16S rRNA gene sequences of four B. subornata and one B. striata samples confirmed that the most abundant OTU (up to 70% RA) was the same for these two species; a member of the family Hyphomicrobiaceae (Figure 6B). In these species, this OTU had the average RA of 45.8% ±17.6 (Supplementary Additional Table 3). Additional BLAST search of the full-length 16S rRNA gene sequence of this OTU indicated that its closest relative was an unclassified member of the Hyphomicrobiaceae family with 94.75% ID. Phylogenetic analysis of the Hyphomicrobiaceae OTU and its closest relatives along with some other members of representative sequences of Hyphomicrobiaceae family revealed, that the Sagami Bay Hyphomicrobiaceae OTU groups close to the genera Hyphomicrobium (Figure 7).
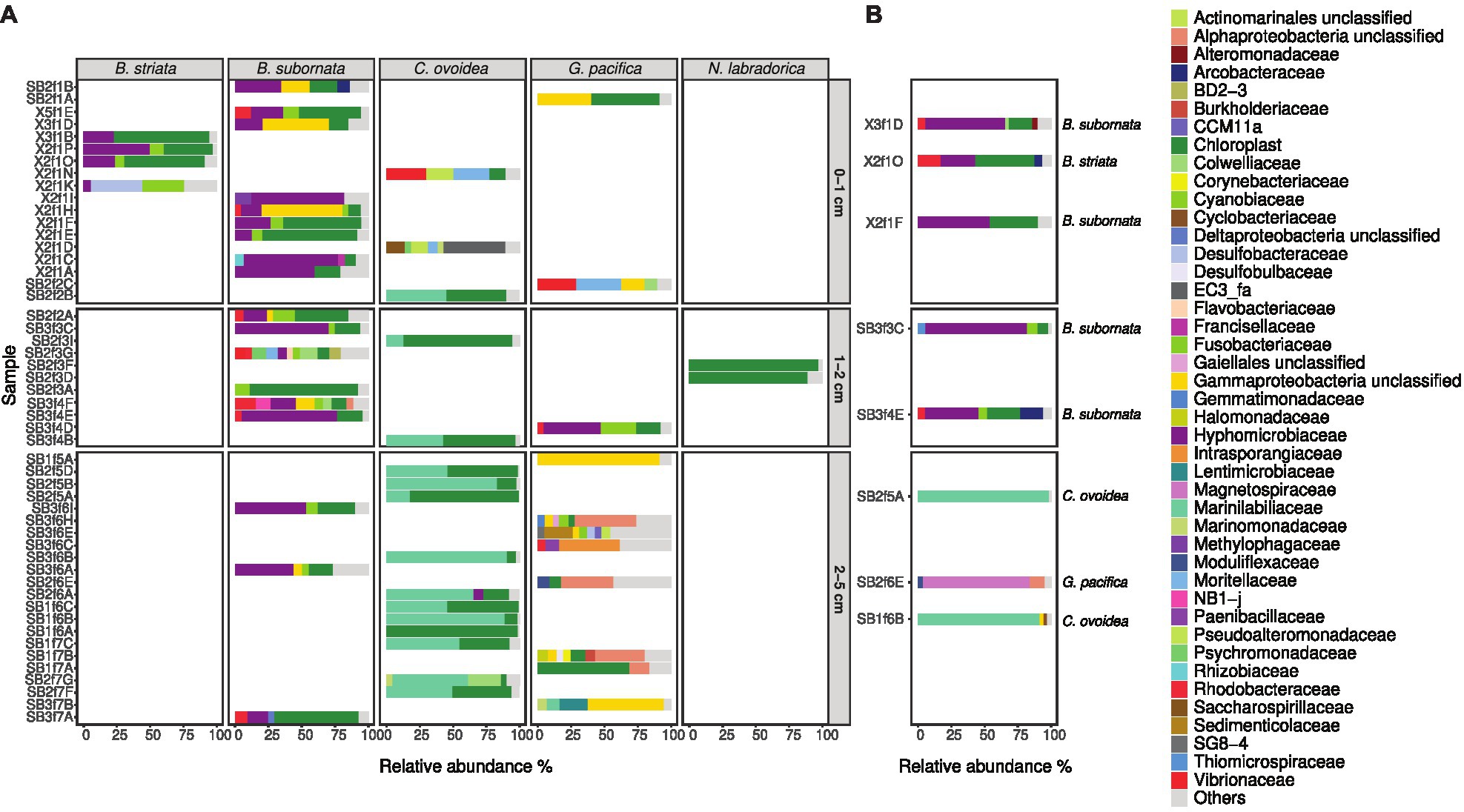
Figure 6. Relative abundance of bacterial families in foraminiferal samples at different sampling depths based on Illumina MiSeq sequencing (A) and full-length sequencing (B) of the 16S rRNA gene. Bacterial families with RA over 4% (A) and 2% (B) are shown; taxa present with lower RA are grouped in “Others.”
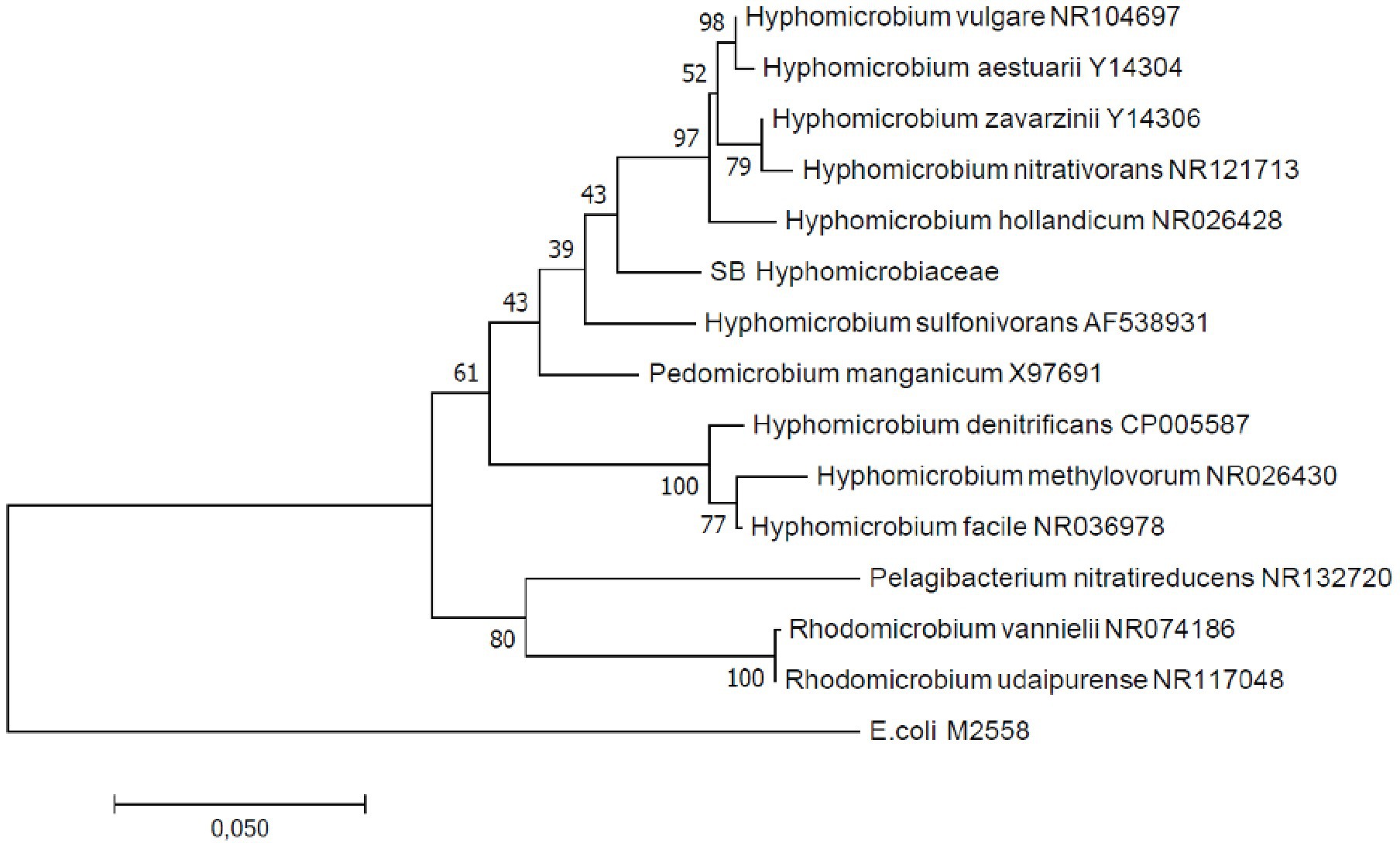
Figure 7. Maximum likelihood phylogenetic tree based on full-length 16S rRNA gene sequences indicating the position of the Hyphomicrobium operational taxonomic unit (OTU; SB Hyphomicrobiaceae). GenBank accession numbers are given for the closest relatives after the taxonomic name.
The total number of G. pacifica specimens analyzed was 11. The most abundant OTUs in G. pacifica were an unclassified Gammaproteobacteria OTU (average RA 18.2% ±30.7) followed by an unclassified Alphaproteobacteria OTU (average RA 8.9% ±12.9; Supplementary Additional Table 3; Figure 6A). In the microbiome of a specimen (SB2f6E) of G. pacifica, which was subjected to the full-length sequencing of the 16S rRNA gene, 92.8% of the reads belonged to class Alphaproteobacteria, of which 85.2% of reads belonged to the family Magnetospiraceae (Figure 6B). The most common OTU in the family Magnetospiraceae accounted for 45.6% of the reads (Supplementary Additional Table 3).
In total, 16 C. ovoidea specimens were analyzed. The most abundant OTU (average across samples 43.4% ±30.4) belonged to group Bacteroidetes, family Marinilabiliaceae (Figure 6A; Supplementary Additional Table 3). Bacteroidetes were also present in the sediment (average RA 13.3% ±2.9; Figure 3A) but dominated by the bacterial families Flavobacteriaceae, Cyclobacteriaceae, and the group BD2-2 (comprising 23,3, 22,6, and 20.1% of reads in all Bacteroidetes OTUs, respectively) whereas only 0.4% of all Bacteroidetes reads in the sediment were assigned to the family Marinilabiliaceae and <0.01% to the most common Marinilabiliaceae OTU in C. ovoidea (Supplementary Additional Table 3). Full-length sequencing of the 16S rRNA gene of two C. ovoidea specimens confirmed the presence of this bacterial groups, as in average, 83.6% of reads belonged to one OTU of the family Marinilabiliaceae, order Bacteroidales (Figure 6B; Supplementary Additional Table 3). Additional BLAST search of the full-length 16S rRNA gene sequence indicated that the closest match (93.14% ID) was Saccharicrinis fermentas in the family Marinilabiliaceae.
Intracellular Chloroplasts in Foraminifera
Compared to surrounding sediment, where chloroplasts and Synechococcales had a low relative abundance of <1 and <0.1%, respectively (Figure 3A), they were clearly enriched in foraminifera. In B. striata, chloroplast OTUs accounted for 42.4% RA (±31.7) in average and OTUs belonging to the order Synechococcales had the average relative abundance of 12.3% (±13.3; Figure 6A). The most common chloroplast OTUs in B. striata were OTU7 [average RA of all chloroplast reads 40.8% (±36.5)] followed by OTU5 [average RA 22.4% (±24.6; Table 3)]. The closest BLAST match of OTU7 was Picochlorum (94.55% ID) and of OTU5 was Pycnococcus (98.99% ID), both of which are green algae. In the phylogenetic tree, these OTUs along with OTU95 grouped in the coccoid green algae branch (Figure 8). In B. subornata chloroplasts (average 30.2% ±24.2) and Synechococcales (average 5.8% ±4.2) were also abundant (Figure 6A). The most common chloroplast OTU (40.4% ±36.5 of all chloroplast reads) in B. subornata, was OTU5 followed by OTU7 (average RA of chloroplast reads 31.9% ±19; Table 3). According to the full-length 16S rRNA sequence data, chloroplasts had an average, RA of 26.4% ±14.2 in both Bulimina species (Figure 6B).
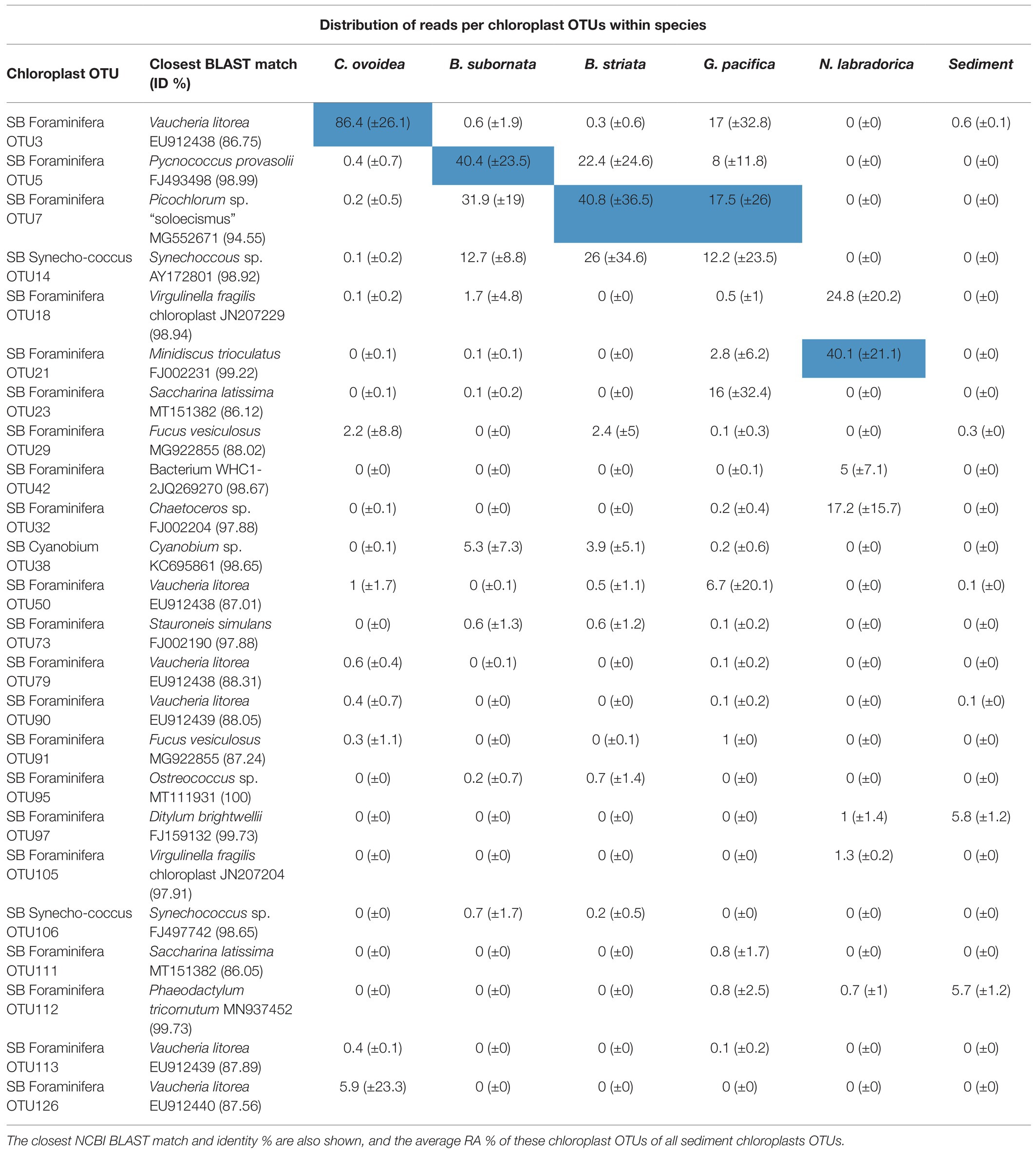
Table 3. The most abundant chloroplast OTUs in foraminifera, including Cyanobacteria, based on the average distribution of chloroplast sequence reads (%) per OTU within each species [the most abundant OTU per species is highlighted in blue and SD (±) is presented in parenthesis].
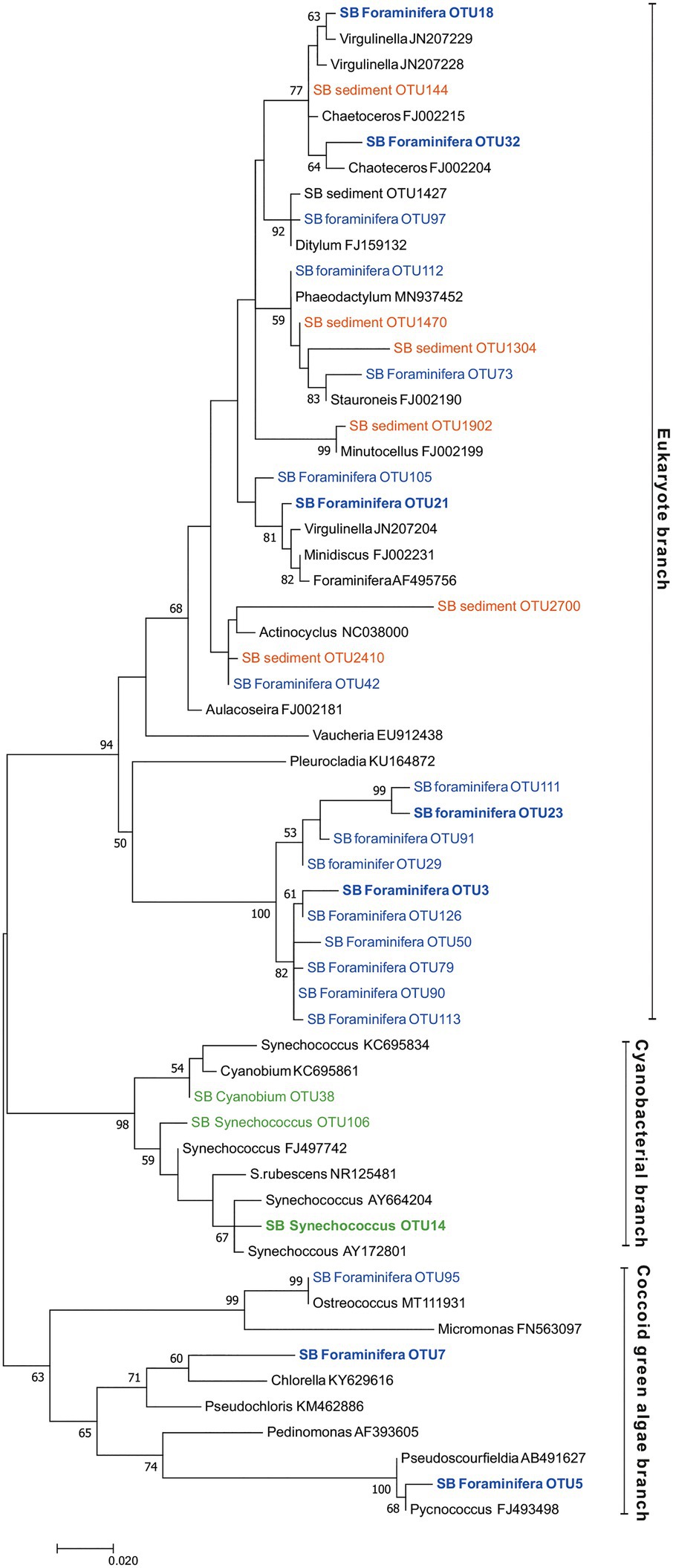
Figure 8. Maximum likelihood tree based on Illumina MiSeq-derived 16S rRNA gene sequences indicating the position of chloroplast OTUs of foraminifera specimens (SB foraminifera OTU, blue color) and sediment samples (SB sediment OTU, orange color) as well as cyanobacterial OTUs (green color). Bootstrap values greater than 50% based on 1000 resamplings are shown at branch nodes. GenBank accession numbers are given for the closest relatives (in black color) after the taxonomic name. The most abundant OTUs are highlighted in bold.
On average, 92.9% ±4.1 of all intracellular 16S rRNA gene sequences of N. labradorica were taxonomically assigned to chloroplasts (Figure 6A). The three most abundant chloroplast OTUs in N. labradorica microbiome were OTU21 (average RA of chloroplast reads 40.1% ±21.1), OTU18 (24.8% ±20.2), and OTU32 (17.2% ±15.7; Table 3). These OTUs were not dominant in any other foraminiferal species (RA < 3%), and in the phylogenetic tree, they were found in the eukaryotic branch implying an algal instead of cyanobacterial origin for these OTUs (Figure 8).
Chloroplasts were found to be abundant in G. pacifica were they accounted for 13.3% ±21.6 in average of the intracellular OTUs (Figure 6A). Most common chloroplast in G. pacifica was OTU7 (average RA 17.5% ±26) followed by OTU3, OTU23, OTU14, and OTU5 (Table 3).
In C. ovoidea, chloroplasts accounted on average for 38.7% ±30.8 of the intracellular 16S rRNA gene derived OTUs of (Figure 6). Of these OTUs, on average, 86.4% ±26.1 belonged to a single OTU, OTU3 (Table 3). Additional BLAST search indicated that the closest match of OTU3 (86.75% ID, excluding uncultured/environmental samples) was Vaucheria litorea chloroplast, which is a species of yellow-green algae (ID 86.75; Table 3). In sediments and other foraminiferal species, except for some G. pacifica specimens, this OTU was present in very low numbers (Table 3). In the phylogenetic tree (Figure 8), OTU3 grouped together with other chloroplasts found in lower abundance within the foraminiferal samples, in particular C. ovoidea. Conversely, the full-length 16S rRNA gene data contained only a low relative abundance (<1%) of chloroplasts in C. ovoidea specimens (Figure 6B).
Discussion
Differences of Bacterial Community Structure in Sediments and in Foraminiferal Microbiome
According to our results, the microbiome of the deep-sea benthic foraminifera differed from the surrounding sediment bacterial community. For example, all sediment cores had relatively similar alpha diversity values, whereas foraminiferal species displayed higher degree of intra- and inter-specific variation, as for example, the median Shannon diversity index for G. pacifica was 4 but for C. ovoidea only 1.5. Previously, shallow-water foraminifera from intertidal areas have also been observed to have a microbiome, which is distinct from the surrounding sediment communities (Salonen et al., 2019). In contrast, some larger benthic foraminifera have been discovered to have a site-specific microbiome (Prazeres et al., 2017). In this study, no spatial analyses were carried out and any depth-related changes are likely to reflect the associated changes in pore water redox chemistry and associated changes in sediment bacterial community. Instead, our results clearly demonstrated that in deep-sea foraminifera, the driving factor of the variability of the foraminiferal microbiome was solely the species (p = 0.001). Compared to sediments, the intracellular microbiome of foraminifera was less diverse, and typically dominated by few key bacterial groups, with the exception of Nonionella labradorica, where bacterial OTUs had the RA of <10% as their microbiome was quasi-solely dominated by chloroplasts. In general, bacterial groups abundant in sediments, such as Deltaproteobacteria, comprised only a small part of the foraminiferal microbiomes. Bacteria could represent a food source for foraminifera, however, bacterivory is not likely to be the preferred trophic strategy of these species or benthic foraminifera in general; instead, it is likely to occur randomly during deposit-feeding (van Oevelen et al., 2006; Nomaki et al., 2008). If the foraminiferal microbiome would primarily result from bacterivory or deposit-feeding, it would be expected to closely resemble the sediment bacterial community, which was not the case here. Instead, specific bacterial taxa, typically rare in sediments, were found to constitute a significant proportion of the foraminiferal microbiomes, suggesting that there are species-characteristic, selective mechanisms, and potential endobiotic interactions influencing the bacterial composition of the foraminiferal microbiome. The successfully amplification of long DNA fragments (>1,400 bp) of the key intracellular bacterial groups from single foraminiferal cells, further corroborates that bacterial DNA was relatively intact and not degraded due to digestion.
Species-Characteristic Foraminiferal Bacterial Microbiome
Endobiotic relationships with bacteria could provide the foraminiferal host, an ecological advantage in the hypoxic sediments, by for example providing the host with alternative respiration and/or carbon and nitrogen assimilation pathways (Bernhard et al., 2011; Tsuchiya et al., 2015). In B. striata and B. subornata, the full-length 16S rRNA gene sequence reads confirmed the presence of associated bacterial family Hyphomicrobiaceae, also suggested by the shorter sequence data. The family Hyphomicrobiaceae usually displays a variety of chemo-organotrophic metabolic pathways ranging from sulfide oxidation and metal cycling to denitrification (Stein et al., 2001; Osaka et al., 2006; Gliesche et al., 2015). Some species belonging to this bacterial family have been reported to have a substantial role in the marine nitrogen cycle (Anderson et al., 2011). A key adaptation of foraminifera to hypoxic sediments is the ability to collect nitrate and denitrify (Risgaard-Petersen et al., 2006), a respiration pathway which has also been confirmed for B. subornata (Table 2). Some foraminiferal species are able to perform denitrification themselves (Risgaard-Petersen et al., 2006; Woehle et al., 2018), however, it is possible that some species rely on denitrifying endobionts (Bernhard et al., 2011). For example, if B. subornata does not perform denitrification itself, the endobiotic bacterial family Hyphomicrobiaceae could be involved in this process as members of this bacterial family have denitrifying capabilities (Osaka et al., 2006). Alternatively, the family Hyphomicrobiaceae could be involved in metal, such as manganese, oxidation (Stein et al., 2001), or play another yet unknown biochemical role in this foraminiferal species.
Chilostomella ovoidea is a deep-infaunal species, typically found in anoxic sediments in 3–5 cm sediment depth (Corliss, 1985; Ohga and Kitazato, 1997; Nomaki et al., 2005). Relatively high intracellular nitrate content has been measured in C. ovoidea (Table 2; Piña-Ochoa et al., 2009), however, further experiments would be needed to investigate the potential nitrate reduction through denitrification in this species (Table 2). Here, C. ovoidea specimens displayed a very distinct low-diversity microbiome, where a bacterial taxonomic group belonging to family Marinilabiliaceae, class Bacteroidetes, was the most abundant. In sediments, the bacterial family Marinilabiliaceae was rare compared to other Bacteroidetes families, and it was not abundant in the microbiome of any other foraminiferal species. Moreover, as the sediment sample used for bacterial DNA extractions was intact, it may have contained also a low number of foraminifera. Therefore, it cannot be excluded that the sediment signal of Marinilabiliaceae could be also derived from foraminifera. The group Bacteroidetes is commonly found in marine sediments including in the deep-sea, where they typically grow anaerobically, degrading particulate matter such as proteins (Fernández-Gómez et al., 2013). In this class, bacteria belonging to the family Marinilabiliaceae are Gram-negative, motile rods that display saccharolytic growth, and some genera have been isolated previously from marine mud containing decaying algae (Ludwig et al., 2015). The species Saccharicrinis fermentas, to which the Marinilabiliaceae group was closest related according to BLAST search (ID 93.14%), is a facultatively anaerobic chemo-organotroph that does not have denitrifying capacities (Yang et al., 2014).
Globobulimina pacifica is also a deep-infaunal species known to thrive in low-oxygen conditions (e.g., Corliss, 1985; Jorissen et al., 1995; Fontanier et al., 2002). This species can collect nitrate and denitrify (Table 2; Langlet et al., 2020). In previous studies, genus Globobulimina has been shown to perform complete denitrification itself (Woehle et al., 2018) and contain intracellular bacteria only in low abundance, suggesting a lack of bacterial endobionts (Risgaard-Petersen et al., 2006). Here, bacterial signal was retrieved from G. pacifica, but in contrast to other foraminiferal species, the microbiome composition seemed to be more heterogenous, which was also reflected in its Shannon diversity, ranging from 1.1 to 5.1 between specimens. Yet, G. pacifica microbiome composition was also significantly different from the surrounding sediment community. Four specimens of G. pacifica collected from the deeper (2–5 cm) sediment layers contained abundant Alphaproteobacterial OTUs, which in the specimen (SB26E) was confirmed by the analyses of the full-length 16S rRNA gene to belong to bacteria family Magnetospiracea. This family includes metal reducing magnetotactic and microaerophilic/anaerobic bacteria that have been suggested to have a role in Fe3+ mobilisation and bioavailability (Schüler and Frankel, 1999; Molari et al., 2020). Previously, magnetotactic bacteria have been observed to form symbiotic relationships with other protists, Euglenozoa, potentially helping the host to navigate in anoxic sediments toward optimal chemical niches (Monteil et al., 2019). The genus Globobulimina has been observed to distribute in sediments tightly linked with Fe2+ oxidation zone, which could be an indication of a specialized lifestyle and a potential trophic or symbiotic relationship with chemolithoautotrophic prokaryotes (Fontanier et al., 2005). However, due to variability in the microbiome composition and limited full-length sequence data, further research is required to verify the amount and potential meaning of these bacterial groups in G. pacifica. Potentially, the variability observed in the microbiome could also reflect the trophic strategy of this species. In previous experiments from Sagami Bay, Globobulimina affinis showed a selective uptake of algae, suggesting a preference for phytodetritus as a primary food source when available (Nomaki et al., 2006), although the sterol compositions of G. affinis and surrounding sediments suggest that in general it possesses a deposit-feeding strategy (Nomaki et al., 2009). Potentially, G. pacifica has a more opportunistic lifestyle, which might include various ecological behaviors, leading to the variability in the microbiome.
To resolve the ecological significance and potential function of intracellular bacteria, such as of Marinilabiliaceae for C. ovoidea and Hyphomicrobiacea for B. subornata, more data are required on the activity and intracellular location of these bacterial groups. In addition, the method of symbiont transmission and acquisition for benthic foraminifera remains to be resolved. Previously, it has been suggested that some foraminiferal species may retrieve their putative endobionts from the surrounding environment (Bird et al., 2017; Salonen et al., 2019). In the case of C. ovoidea and both Bulimina species, the putatively endobiotic bacterial groups were also present in the surrounding sediment, but in very low abundance (<0.1% RA). Alternatively, foraminifera could acquire the endobionts via asexual reproduction and co-occurring vertical transmission of endobiotic bacteria (Tsuchiya et al., 2009, 2015; Takagi et al., 2020).
Although depth was not a significant parameter explaining the foraminiferal microbiome composition, in this study, we observed that two C. ovoidea specimens (X2f1N and X2f1D) retrieved from the surface sediment (0–1 cm depth) had a distinct microbiome compared to the specimen SB2Bf2B (retrieved from 0.5–1 cm sediment depth) and all specimens from deeper sediment depths. Two surface specimens also had higher Shannon diversity indices (median 3.3 and 3.9) compared to deeper specimens (median 1.4 below 1 cm depth), and interestingly, the surface specimens lacked the anaerobic bacterial group Marinilabiliaceae, which was dominant in the other specimens. However, due to the low number of specimens recovered from certain depth intervals, we are not able to drive secure conclusions. Future studies may resolve the possibility that vertical distribution and availability of oxygen could be linked to the composition of the microbiome of C. ovoidea – perhaps either by providing the suitable environmental conditions for its sustenance or the putative endobionts themselves. In addition, further studies are needed to resolve the stability of the foraminiferal microbiome in temporal and spatial scale.
Intracellular Chloroplasts
In the surrounding sediment, chloroplasts accounted for less than 1% RA of all OTUs at all sampling depths whereas majority of foraminiferal specimens of all studied species had a significantly higher intracellular chloroplast OTUs RA (ranging from 92.9 ± 6% RA in N. labradorica to 15.3 ± 23% in G. pacifica). In sediment, the chloroplasts originated mainly from diatoms, with the most common OTU (SB Sediment OUT 144; Figure 8) that accounted 59.4 ± 12.8% of the chloroplast reads assigned to Chaoteceros calcitrans (BLAST ID 99.73). According to the phylogenetic analysis, the chloroplast OTUs in foraminifera originated from several sources, of which some were bacterial and some eukaryote-related (Figure 8). In sediments, the most common chloroplasts originated from eukaryote, namely diatom sources. Some chloroplast OTUs were tightly linked to a specific foraminiferal species (Table 3), implying species-characteristic preferences in terms of chloroplast sources.
A large abundance of chloroplasts was observed in the intracellular microbiomes of the two Nonionella labradorica specimens where they accounted for >90% of total OTU abundance. Although chloroplasts strongly dominated the microbiome of N. labradorica, relatively high Hꞌ and Jꞌ indices (2.4, 2.9, and 0.5, 0.6, respectively) can be explained by the variability in the chloroplast OTUs (Table 3) and the rare (bacteria had the average RA 7% ±5.7) yet diverse intracellular bacterial community (median Hꞌ 5.5 excluding chloroplasts). Based on the phylogenetic analysis, the most common chloroplasts of N. labradorica originated from eukaryote sources (mainly diatoms), which were common also in the surrounding sediment (Figure 8). The intracellular chloroplast OTUs of foraminifera may be linked to herbivorous feeding behavior as feeding experiments have shown that many benthic foraminiferal species ingest algae (e.g., Moodley et al., 2000; Nomaki et al., 2005, 2006). If the chloroplasts retained by the N. labradorica are used for nutrition, their high enrichment in the cytoplasm could indicate that part of the survival strategy of this species is to store food, in this case chloroplasts, intracellularly. Alternatively, foraminifera living in the photic zone have a well-documented ability to collect and retain chloroplasts, which they then utilize in a mixotrophic life strategy called kleptoplasty (Lee et al., 1989; Bernhard and Bowser, 1999; Cesbron et al., 2017). Curiously, some deep-sea foraminifera within the genus of Nonionella have been found to retain active chloroplasts in the non-photic zone, even at depths up to 600 m (Bernhard and Bowser, 1999; Jauffrais et al., 2018). Although the exact reason why these specimens retain chloroplasts in the non-photic zone remains unclear, it has been suggested that they may be used for ammonium or sulfate assimilation pathways similarly to non-photosymbiontic algae (Grzymski et al., 2002; Jauffrais et al., 2018).
Previously, B. subornata and B. striata have shown δ13C values typical for phytophagous species, implying a preference for algal diet (Nomaki et al., 2008). Here, the phylogenetic analysis of the most common chloroplast OTUs inside B. subornata and B. striata revealed that they were closely associated to Pycnococcus provasolii (98.99% ID) and Picochlorum sp. (94.55% ID; Table 3). Both of these species are small, coccoid green algae commonly found in marine pelagic environments (Campbell et al., 1994; Henley et al., 2004), some of them able to accumulate high amounts of lipids (Unkefer et al., 2017). As the most common chloroplasts in the sediment samples belonged to diatoms, the chloroplast composition of B. subornata and B. striata suggests that these species could potentially favor these green algae as a nutrition source over diatoms and selectively ingest them. Therefore, the large intracellular abundance of these chloroplasts (>30% RA, on average) in these species is most likely reflecting their phytodetrivorous trophic preference, which may also be linked to their shallow infaunal distribution in sediment.
Unlike, in other studied species, the chloroplast sequences detected in C. ovoidea was highly specific and more than 85% of them belonged to only one OTU (OTU 3). The closest BLAST hit (86.8% ID) of this OTU was a filamentous green-yellow algae Vaucheria litorea, the chloroplasts of which are used by the sea slug Elysia chlorotica as intracellular photosymbionts (Mujer et al., 1996). However, this closest match was of low identity (only 86.8%) corroborated by the phylogenetic analysis which clustered this chloroplast in a phylogenetically separate branch rather than close to V. litorea. This is a well-supported branch (100% support based on 1,000 bootstrap replicates) unique to the foraminiferal specimens of this study. This foraminiferal branch was part of the eukaryote branch of the phylogenetic tree (Figure 8), neighboring with chloroplasts from algal origins (Vaucheria and Pleurocladia), suggesting a eukaryotic origin of this OTU rather than a cyanobacterial one. Chloroplast OTUs in this branch (>90% of reads comprising this branch) were associated with C. ovoidea (except OTUs 23 and 111, >98% associated with G. pacifica), implying that similar microhabitat preference of C. ovoidea might explain their association to these chloroplast OTUs.
Unlike those foraminiferal species that prefer surface sediment as their microhabitat, C. ovoidea does not usually display strong seasonality or response to phytodetritus input, and thus it has been thought to be a slow-growing species with a preference for more refractory organic matters (Ohga and Kitazato, 1997; Nomaki et al., 2005). In feeding experiments, C. ovoidea has shown almost no or random ingestion of algae, DOM, and bacteria, suggesting either non-selective deposit-feeding or that their food preferences and life strategy are very specific and still unknown, and thus not well-captured in experimental studies (Nomaki et al., 2005, 2006). Potentially, the unique chloroplast OTUs in C. ovoidea could be resulting from a very specific type of selective herbivory that was uncaptured in feeding studies where a different type of algae was used. However, the OTUs of the chloroplast branch associated with C. ovoidea were extremely rare in sediments (0.001% RA), challenging the plausibility of the uptake of these chloroplasts directly from the surroundings. Alternatively, this OTU could belong to an algal symbiont, which the foraminifera potentially transfer asexually, as suggested for some foraminiferal species (Tsuchiya et al., 2015). Recently, the ultrastructure of C. ovoidea was found to contain abundant chitinous structures that are not commonly observed in other foraminifera, highlighting that this particular species has unique ecological characteristics (Nomaki et al., 2020). The function and origin of these structures are still unknown. They may be synthetized either by the foraminifera itself or alternatively linked to specific type of feeding or symbiosis (Nomaki et al., 2020). If the origin of the chitinous structures is algal, the distinct chloroplast signal in C. ovoidea could be linked to the same phenomenon. However, in contrary to chloroplast reads in both Bulimina species, the chloroplast OTUs of two C. ovoidea (SB1 6B and SB2 5A) did not amplify with full length 16S rRNA gene sequencing, although the specimens seemed to contain intracellular chloroplasts according to the shorter amplicon data. This could, for example, be due to potential degradation of the DNA, which would preclude the extraction and amplification of long DNA fragments. However, as the universal bacterial 16S rRNA gene amplicons provide only limited information on intracellular chloroplasts, further studies using for example 18S rRNA gene metabarcoding (Jauffrais et al., 2018; Chronopoulou et al., 2019) or genomic approaches would be advisable in elucidating the source and meaning of this chloroplast OTU for C. ovoidea.
Conclusion
16S rRNA gene metabarcoding provides a powerful tool for investigating and elucidating the microbiomes of even unicellular eukaryotes, such as the foraminifera. Here, deep-sea foraminifera, representing various microhabitats, were found to harbor microbiomes that clearly differed from one species to another in terms of the relative abundance of the dominant intracellular bacteria and chloroplasts, as well as diversity and degree of intra-specific variation. Based on these observations, we were able to identify two bacterial groups that were strongly associated to specific foraminiferal species; the Marinilabiliaceae bacterial group for C. ovoidea and the Hyphomicrobiaceae for B. subornata and B. striata. The high relative abundance and consistent occurrence of these bacteria in these species could insinuate a stable and putatively endobiotic relationship, therefore calling for further research focusing on these bacterial groups. In addition, the origins of chloroplasts differed between species, most likely reflecting the divergent ecological and trophic strategies of the studied species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repositories and accession numbers can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA679187/.
Author Contributions
KK conceived the study. IS, P-MC, KK, and HN carried out the 2017 sampling campaign and sample processing in the field. DL carried out the oxygen profiling and the foraminiferal nitrate pool and denitrification measurements. IS, P-MC, HN, DL, and KK identified and picked the living foraminifera. HN carried out the 2019 sampling campaign including oxygen profiling and porewater nutrient analysis. IS carried out the molecular laboratory work including DNA extractions and amplifications, completed bioinformatical, statistical, and phylogenetic analysis of 16S rRNA gene sequence data, and drafted the manuscript. P-MC assisted with the phylogenetic and bioinformatical analysis. All authors contributed to the interpretation of the results and the final version of the manuscript.
Funding
Funding for this research has been provided by the Academy of Finland (grant numbers: 278827, 283453, and 312495). In addition, IS was supported by the International Research Fellow Program of Japan Society for the Promotion of Science (Postdoctoral Fellowships for Research in Japan P20770). DL was supported by the International Research Fellow Program of Japan Society for the Promotion of Science (Postdoctoral Fellowships for Research in Japan P16708). The open access publication fees have been covered by the Helsinki University Library.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge the captains, the crew members, chief scientists (Michiyo Kawai and Shinsuke Kawagucci), and the onboard scientists of the research vessel R/V Seiyo-Maru and R/V Kaimei to their help during sampling campaigns SE-17-06 and KM19-07 in 2017 and 2019, respectively. We would like to thank Fuminori Hashihama, Toshiro Yamanaka, and Jota Kanda for their help on the sampling campaign of Seiyo-Maru. We want to acknowledge JAMSTEC Laboratories in Yokosuka, Japan as well as Molecular Ecology and Systematics (MES) Laboratory at the Viikki Campus in University of Helsinki, Finland for providing laboratory space and equipment for molecular analysis. The DNA Sequencing and Genomics Laboratory in the Institute of Biotechnology at the University of Helsinki was acknowledged for the help with Illumina MiSeq and Single Molecule, Real-Time (SMRT) sequencing. We would also like to thank Soila Silvonen from the University of Helsinki for assistance with the Rose-Bengal counting. We wish to acknowledge Geolab in Utrecht University and Helen de Waard for the analysis of ICP-OES samples. We also wish to thank the reviewers of this manuscript for their constructive feedback.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.694406/full#supplementary-material
References
Altschul, S., Gish, W., Miller, W., Myers, E., and Lipman, D. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/s0022-2836(05)80360-2
Andersen, K., Kjær, T., and Revsbech, N. (2001). An oxygen insensitive microsensor for nitrous oxide. Sens. Actuators B Chem. 81, 42–48. doi: 10.1016/S0925-4005(01)00924-8
Anderson, C., Condron, L., Clough, T., Fiers, M., Stewart, A., Hill, R., et al. (2011). Biochar induced soil microbial community change: implications for biogeochemical cycling of carbon, nitrogen and phosphorus. Pedobiologia 54, 309–320. doi: 10.1016/j.pedobi.2011.07.005
Bernhard, J. (1993). Experimental and field evidence of Antarctic foraminiferal tolerance to anoxia and hydrogen sulfide. Mar. Micropaleontol. 20, 203–213. doi: 10.1016/0377-8398(93)90033-t
Bernhard, J. (2003). Potential symbionts in bathyal foraminifera. Science 299: 861. doi: 10.1126/science.1077314
Bernhard, J., and Bowser, S. (1999). Benthic foraminifera of dysoxic sediments: chloroplast sequestration and functional morphology. Earth Sci. Rev. 46, 149–165. doi: 10.1016/s0012-8252(99)00017-3
Bernhard, J., Edgcomb, V., Casciotti, K., McIlvin, M., and Beaudoin, D. (2011). Denitrification likely catalyzed by endobionts in an allogromiid foraminifer. ISME J. 6, 951–960. doi: 10.1038/ismej.2011.171
Bernhard, J., Habura, A., and Bowser, S. (2006). An endobiont-bearing allogromiid from the Santa Barbara Basin: implications for the early diversification of foraminifera. J. Geophys. Res. 111:G03002. doi: 10.1029/2005jg000158
Bernhard, J., and Panieri, G. (2018). Keystone Arctic paleoceanographic proxy association with putative methanotrophic bacteria. Sci. Rep. 8:10610. doi: 10.1038/s41598-018-28871-3
Bernhard, J., Tsuchiya, M., and Nomaki, H. (2018). Ultrastructural observations on prokaryotic associates of benthic foraminifera: food, mutualistic symbionts, or parasites? Mar. Micropaleontol. 138, 33–45. doi: 10.1016/j.marmicro.2017.09.001
Bird, C., Darling, K., Russell, A., Davis, C., Fehrenbacher, J., Free, A., et al. (2017). Cyanobacterial endobionts within a major marine planktonic calcifier (Globigerina bulloides, foraminifera) revealed by 16S rRNA metabarcoding. Biogeosciences 14, 901–920. doi: 10.5194/bg-14-901-2017
Campbell, L., Shapiro, L., and Haugen, E. (1994). Immunochemical characterization for eukaryotic ultraplankton from the Atlantic and Pacific oceans. J. Plankton Res. 16, 35–51. doi: 10.1093/plankt/16.1.35
Cesbron, F., Geslin, E., Kieffre, C., Jauffrais, T., Nardelli, M., Langlet, D., et al. (2017). Sequestered chloroplasts in the benthic foraminifer Haynesina Germanica: cellular organization, oxygen fluxes and potential ecological implications. J. Foraminifer. Res. 47, 268–278. doi: 10.2113/gsjfr.47.3.268
Chronopoulou, P., Salonen, I., Bird, C., Reichart, G., and Koho, K. (2019). Metabarcoding insights into the trophic behavior and identity of intertidal benthic foraminifera. Front. Microbiol. 10:1169. doi: 10.3389/fmicb.2019.01169
Corliss, B. (1985). Microhabitats of benthic foraminifera within deep-sea sediments. Nature 314, 435–438. doi: 10.1038/314435a0
Doane, T., and Horwáth, W. (2003). Spectrophotometric determination of nitrate with a single reagent. Anal. Lett. 36, 2713–2722. doi: 10.1081/al-120024647
Edgar, R. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5:113. doi: 10.1186/1471-2105-5-113
Edgar, R., Haas, B., Clemente, J., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Ernst, S., Duijnstee, I., and van der Zwaan, B. (2002). The dynamics of the benthic foraminiferal microhabitat: recovery after experimental disturbance. Mar. Micropaleontol. 46, 343–361. doi: 10.1016/s0377-8398(02)00080-4
Fernández-Gómez, B., Richter, M., Schüler, M., Pinhassi, J., Acinas, S., González, J., et al. (2013). Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. 7, 1026–1037. doi: 10.1038/ismej.2012.169
Fontanier, C., Duros, P., Toyofuku, T., Oguri, K., Koho, K., Buscail, R., et al. (2014). Living (stained) deep-sea foraminifera off Hachinohe (NE Japan, Western Pacific): environmental interplay in oxygen-depleted ecosystems. J. Foraminiferal Res. 44, 281–299. doi: 10.2113/gsjfr.44.3.281
Fontanier, C., Jorissen, F., Chaillou, G., Anschutz, P., Grémare, A., and Griveaud, C. (2005). Live foraminiferal faunas from a 2800m deep lower canyon station from the Bay of Biscay: faunal response to focusing of refractory organic matter. Deep-Sea Res. I Oceanogr. Res. Pap. 52, 1189–1227. doi: 10.1016/j.dsr.2005.01.006
Fontanier, C., Jorissen, F., Licari, L., Alexandre, A., Anschutz, P., and Carbonel, P. (2002). Live benthic foraminiferal faunas from the Bay of Biscay: faunal density, composition, and microhabitats. Deep-Sea Res. I Oceanogr. Res. Pap. 49, 751–785. doi: 10.1016/s0967-0637(01)00078-4
Gliesche, C., Fesefeldt, A., and Hirsch, P. (2015). “Hyphomicrobium,” in Bergey's Manual of Systematics of Archaea and Bacteria. ed. W. B. Whitman (John Wiley & Sons, Inc.), 1–34.
Goldstein, S., and Corliss, B. H. (1994). Deposit feeding in selected deep-sea and shallow-water benthic foraminifera. Deep-Sea Res. I Oceanogr. Res. Pap. 41, 229–241. doi: 10.1016/0967-0637(94)90001-9
Gooday, A. (1988). A response by benthic foraminifera to the deposition of phytodetritus in the deep sea. Nature 332, 70–73. doi: 10.1038/332070a0
Gooday, A., Levin, L., Linke, P., and Heeger, T. (1992). “The role of benthic foraminifera in deep-sea food webs and carbon cycling,” in Deep-Sea Food Chains and the Globsal Carbon Cycle. eds. G. T. Rowe and V. Pariente (Springer), 63–91.
Grzymski, J., Schofield, O., Falkowski, P., and Bernhard, J. (2002). The function of plastids in the deep-sea benthic foraminifer, Nonionella Stella. Limnol. Oceanogr. 47, 1569–1580. doi: 10.4319/lo.2002.47.6.1569
Henley, W., Hironaka, J., Guillou, L., Buchheim, M., Buchheim, J., Fawley, M., et al. (2004). Phylogenetic analysis of the ‘Nannochloris-like’ algae and diagnoses of Picochlorum oklahomensis gen. Et sp. nov. (Trebouxiophyceae, Chlorophyta). Phycologia 43, 641–652. doi: 10.2216/i0031-8884-43-6-641.1
Høgslund, S., Revsbech, N. P., Cedhagen, T., Nielsen, L. P., and Gallardo, V. A. (2008). Denitrification, nitrate turnover, and aerobic respiration by benthic foraminiferans in the oxygen minimum zone off Chile. J. Exp. Mar. Biol. Ecol. 359, 85–91. doi: 10.1016/j.jembe.2008.02.015
Holzmann, M., and Pawlowski, J. (1996). Preservation of foraminifera for DNA extraction and PCR amplification. J. Foraminiferal Res. 26, 264–267. doi: 10.2113/gsjfr.26.3.264
Jauffrais, T., LeKieffre, C., Schweizer, M., Geslin, E., Metzger, E., Bernhard, J., et al. (2018). Kleptoplastidic benthic foraminifera from aphotic habitats: insights into assimilation of inorganic C, N and S studied with sub-cellular resolution. Environ. Microbiol. 21, 125–141. doi: 10.1111/1462-2920.14433
Jorissen, F., de Stigter, H., and Widmark, J. (1995). A conceptual model explaining benthic foraminiferal microhabitats. Mar. Micropaleontol. 26, 3–15. doi: 10.1016/0377-8398(95)00047-x
Kitazato, H. (1988). Locomotion of some benthic foraminifera in and on sediments. J. Foraminiferal Res. 18, 344–349. doi: 10.2113/gsjfr.18.4.344
Kitazato, H. (1989). Vertical distribution of benthic foraminifera within sediments (preliminary report). Benthos Res. 1989, 41–51. doi: 10.5179/benthos1981.1989.41
Koho, K., LeKieffre, C., Nomaki, H., Salonen, I., Geslin, E., Mabilleau, G., et al. (2018). Changes in ultrastructural features of the foraminifera Ammonia spp. in response to anoxic conditions: field and laboratory observations. Mar. Micropaleontol. 138, 72–82. doi: 10.1016/j.marmicro.2017.10.011
Koho, K. A., and Piña-Ochoa, E. (2012). “Benthic foraminifera: inhabitants of low-oxygen environments,” in Anoxia Evidence for Eukaryote Survival and Paleontological Strategies Series (Anoxia, Cellular Origin, Life in Extreme Habitats and Astrobiology). eds. A. Altenbach, J. M. Bernhard, and J. Seckbach (Netherlands: Springer), 249–285.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic acid Techniques in Bacterial Systematics. eds. E. Stackebrandt and M. Goodfellow (New York, NY: John Wiley and Sons), 115–175.
Langlet, D., Bouchet, V., Riso, R., Matsui, Y., Suga, H., Fujiwara, Y., et al. (2020). Foraminiferal ecology and role in nitrogen benthic cycle in the hypoxic southeastern bering sea. Front. Mar. Sci. 7:582818. doi: 10.3389/fmars.2020.582818
Langlet, D., Geslin, E., Baal, C., Metzger, E., Lejzerowicz, F., Riedel, B., et al. (2013). Foraminiferal survival after long-term in situ experimentally induced anoxia. Biogeosciences 10, 7463–7480. doi: 10.5194/bg-10-7463-2013
Lee, J., McEnery, M., Kuile, B., Erez, J., Röttger, R., Rockwell, R. Jr., et al. (1989). Identification and distribution of endosymbiotic diatoms in larger foraminifera. Micropaleontology 35, 353–366. doi: 10.2307/1485677
Legendre, P., and Gallagher, E. D. (2001). Ecologically meaningful transformations for ordination of species data. Oecologia 129, 271–280. doi: 10.1007/s004420100716
LeKieffre, C., Spangenberg, J., Mabilleau, G., Escrig, S., Meibom, A., and Geslin, E. (2017). Surviving anoxia in marine sediments: the metabolic response of ubiquitous benthic foraminifera (Ammonia tepida). PLoS One 12:e0177604. doi: 10.1371/journal.pone.0177604
Ludwig, W., Euzéby, J., and Whitman, W. (2015). “Marinilabiliaceae fam. Nov.,” in Bergey’s Manual of Systematics of Archaea and Bacteria. ed. W. B. Whitman (John Wiley & Sons, Inc.), 1–2.
McMurdie, P., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
McMurdie, P., and Holmes, S. (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 10:e1003531. doi: 10.1371/journal.pcbi.1003531
Molari, M., Janssen, F., Vonnahme, T., Wenzhöfer, F., and Boetius, A. (2020). The contribution of microbial communities in polymetallic nodules to the diversity of the deep-sea microbiome of the Peru Basin (4130–4198m depth). Biogeosciences 17, 3203–3222. doi: 10.5194/bg-17-3203-2020
Monteil, C., Vallenet, D., Menguy, N., Benzerara, K., Barbe, V., Fouteau, S., et al. (2019). Ectosymbiotic bacteria at the origin of magnetoreception in a marine protist. Nat. Microbiol. 4, 1088–1095. doi: 10.1038/s41564-019-0432-7
Moodley, L., Boschker, H., Middelburg, J., Pel, R., Herman, P., de Deckere, E., et al. (2000). Ecological significance of benthic foraminifera: 13C Labelling experiments. Mar. Ecol. Prog. Ser. 202, 289–295. doi: 10.3354/meps202289
Moodley, L., and Hess, C. (1992). Tolerance of infaunal benthic foraminifera for low and high oxygen concentrations. Biol. Bull. 183, 94–98. doi: 10.2307/1542410
Moodley, L., Middelburg, J., Boschker, H., Duineveld, G., Pel, R., Herman, P., et al. (2002). Bacteria and foraminifera: key players in a short-term deep-sea benthic response to phytodetritus. Mar. Ecol. Prog. Ser. 236, 23–29. doi: 10.3354/meps236023
Moodley, L., van der Zwaan, G., Herman, P., Kempers, L., and van Breugel, P. (1997). Differential response of benthic meiofauna to anoxia with special reference to foraminifera (Protista: Sarcodina). Mar. Ecol. Prog. Ser. 158, 151–163. doi: 10.3354/meps158151
Mujer, C., Andrews, D., Manhart, J., Pierce, S., and Rumpho, M. (1996). Chloroplast genes are expressed during intracellular symbiotic association of Vaucheria litorea plastids with the sea slug Elysia chlorotica. Proc. Natl. Acad. Sci. 93, 12333–12338. doi: 10.1073/pnas.93.22.12333
Nomaki, H., Chen, C., Oda, K., Tsuchiya, M., Tame, A., Uematsu, K., et al. (2020). Abundant chitinous structures in Chilostomella (foraminifera, Rhizaria) and their potential functions. J. Eukaryot. Microbiol. 68:e12828. doi: 10.1111/jeu.12828
Nomaki, H., Chikaraishi, Y., Tsuchiya, M., Toyofuku, T., Suga, H., Sasaki, Y., et al. (2015). Variation in the nitrogen isotopic composition of amino acids in benthic foraminifera: implications for their adaptation to oxygen-depleted environments. Limnol. Oceanogr. 60, 1906–1916. doi: 10.1002/lno.10140
Nomaki, H., Heinz, P., Nakatsuka, T., Shimanaga, M., and Kitazato, H. (2005). Species-specific ingestion of organic carbon by deep-sea benthic foraminifera and meiobenthos: in situ tracer experiments. Limnol. Oceanogr. 50, 134–146. doi: 10.4319/lo.2005.50.1.0134
Nomaki, H., Heinz, P., Nakatsuka, T., Shimanaga, M., Ohkouchi, N., Ogawa, N., et al. (2006). Different ingestion patterns of 13C-labeled bacteria and algae by deep-sea benthic foraminifera. Mar. Ecol. Prog. Ser. 310, 95–108. doi: 10.3354/meps310095
Nomaki, H., Ogawa, N., Ohkouchi, N., Suga, H., Toyofuku, T., Shimanaga, M., et al. (2008). Benthic foraminifera as trophic links between phytodetritus and benthic metazoans: carbon and nitrogen isotopic evidence. Mar. Ecol. Prog. Ser. 357, 153–164. doi: 10.3354/meps07309
Nomaki, H., Ohkouchi, N., Heinz, P., Suga, H., Chikaraishi, Y., Ogawa, N., et al. (2009). Degradation of algal lipids by deep-sea benthic foraminifera: an in situ tracer experiment. Deep-Sea Res. I Oceanogr. Res. Pap. 56, 1488–1503. doi: 10.1016/j.dsr.2009.04.013
Ohga, T., and Kitazato, H. (1997). Seasonal changes in bathyal foraminiferal populations in response to the flux of organic matter (Sagami Bay, Japan). Terra Nova 9, 33–37. doi: 10.1046/j.1365-3121.1997.d01-6.x
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., Minchin, P. R., et al. (2015). Vegan: Community Ecology Package. R Package Version 2.3–1, 264.
Osaka, T., Yoshie, S., Tsuneda, S., Hirata, A., Iwami, N., and Inamori, Y. (2006). Identification of acetate- or methanol-assimilating bacteria under nitrate-reducing conditions by stable-isotope probing. Microb. Ecol. 52, 253–266. doi: 10.1007/s00248-006-9071-7
Piña-Ochoa, E., Hogslund, S., Geslin, E., Cedhagen, T., Revsbech, N., Nielsen, L., et al. (2009). Widespread occurrence of nitrate storage and denitrification among foraminifera and gromiida. Proc. Natl. Acad. Sci. 107, 1148–1153. doi: 10.1073/pnas.0908440107
Prazeres, M., Ainsworth, T., Roberts, T., Pandolfi, J., and Leggat, W. (2017). Symbiosis and microbiome flexibility in calcifying benthic foraminifera of the great barrier reef. Microbiome 5:38. doi: 10.1186/s40168-017-0257-7
R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: https://www.R-project.org/
Richirt, J., Riedel, B., Mouret, A., Schweizer, M., Langlet, D., Seitaj, D., et al. (2020). Foraminiferal community response to seasonal anoxia in Lake Grevelingen (the Netherlands). Biogeosciences 17, 1415–1435. doi: 10.5194/bg-17-1415-2020
Risgaard-Petersen, N., Langezaal, A., Ingvardsen, S., Schmid, M., Jetten, M., Op den Camp, H., et al. (2006). Evidence for complete denitrification in a benthic foraminifer. Nature 443, 93–96. doi: 10.1038/nature05070
Salava, A., Aho, V., Lybeck, E., Pereira, P., Paulin, L., Nupponen, I., et al. (2017). Loss of cutaneous microbial diversity during first 3 weeks of life in very low birthweight infants. Exp. Dermatol. 26, 861–867. doi: 10.1111/exd.13312
Salonen, I., Chronopoulou, P., Bird, C., Reichart, G., and Koho, K. (2019). Enrichment of intracellular Sulphur cycle—associated bacteria in intertidal benthic foraminifera revealed by 16S and aprA gene analysis. Sci. Rep. 9:11692. doi: 10.1038/s41598-019-48166-5
Salter, S., Cox, M., Turek, E., Calus, S., Cookson, W., Moffatt, M., et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12:87. doi: 10.1186/s12915-014-0087-z
Schloss, P., Westcott, S., Ryabin, T., Hall, J., Hartmann, M., Hollister, E., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/aem.01541-09
Schmiedl, G., Pfeilsticker, M., Hemleben, C., and Mackensen, A. (2004). Environmental and biological effects on the stable isotope composition of recent deep-sea benthic foraminifera from the western Mediterranean Sea. Mar. Micropaleontol. 51, 129–152. doi: 10.1016/j.marmicro.2003.10.001
Schüler, D., and Frankel, R. (1999). Bacterial magnetosomes: microbiology, biomineralization and biotechnological applications. Appl. Microbiol. Biotechnol. 52, 464–473. doi: 10.1007/s002530051547
Smith, M. S., Firestone, M. K., and Tiedje, J. M. (1978). The acetylene inhibition method for short-term measurement of soil denitrification and its evaluation using nitrogen-131. Soil Sci. Soc. Am. J. 42:611. doi: 10.2136/sssaj1978.03615995004200040015x
Snider, L., Burnett, B., and Hessler, R. (1984). The composition and distribution of meiofauna and nanobiota in a central North Pacific deep-sea area. Deep Sea Res. Part I Oceanogr. Res. Pap. 31, 1225–1249. doi: 10.1016/0198-0149(84)90059-1
Somervuo, P., Koskinen, P., Mei, P., Holm, L., Auvinen, P., and Paulin, L. (2018). BARCOSEL: a tool for selecting an optimal barcode set for high-throughput sequencing. BMC Bioinform. 19:257. doi: 10.1186/s12859-018-2262-7
Stein, L., La Duc, M., Grundl, T., and Nealson, K. (2001). Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediments. Environ. Microbiol. 3, 10–18. doi: 10.1046/j.1462-2920.2001.00154.x
Takagi, H., Kurasawa, A., and Kimoto, K. (2020). Observation of asexual reproduction with symbiont transmission in planktonic foraminifera. J. Plankton Res. 42, 403–410. doi: 10.1093/plankt/fbaa033
Trevathan-Tackett, S., Sherman, C., Huggett, M., Campbell, A., Laverock, B., Hurtado-McCormick, V., et al. (2019). A horizon scan of priorities for coastal marine microbiome research. Nat. Ecol. Evol. 3, 1509–1520. doi: 10.1038/s41559-019-0999-7
Tsuchiya, M., Chikaraishi, Y., Nomaki, H., Sasaki, Y., Tame, A., Uematsu, K., et al. (2018). Compound-specific isotope analysis of benthic foraminifer amino acids suggests microhabitat variability in rocky-shore environments. Ecol. Evol. 8, 8380–8395. doi: 10.1002/ece3.4358
Tsuchiya, M., Grimm, G., Heinz, P., Stögerer, K., Ertan, K., Collen, J., et al. (2009). Ribosomal DNA shows extremely low genetic divergence in a world-wide distributed, but disjunct and highly adapted marine protozoan (Virgulinella fragilis, Foraminiferida). Mar. Micropaleontol. 70, 8–19. doi: 10.1016/j.marmicro.2008.10.001
Tsuchiya, M., Toyofuku, T., Uematsu, K., Brüchert, V., Collen, J., Yamamoto, H., et al. (2015). Cytologic and genetic characteristics of endobiotic bacteria and kleptoplasts of Virgulinella fragilis (foraminifera). J. Eukaryot. Microbiol. 62, 454–469. doi: 10.1111/jeu.12200
Unkefer, C., Sayre, R., Magnuson, J., Anderson, D., Baxter, I., Blaby, I., et al. (2017). Review of the algal biology program within the National Alliance for advanced biofuels and bioproducts. Algal Res. 22, 187–215. doi: 10.1016/j.algal.2016.06.002
van Oevelen, D., Moodley, L., Soetaert, K., and Middelburg, J. (2006). The trophic significance of bacterial carbon in a marine intertidal sediment: results of an in situ stable isotope labeling study. Limnol. Oceanogr. 51, 2349–2359. doi: 10.4319/lo.2006.51.5.2349
Woehle, C., Roy, A., Glock, N., Wein, T., Weissenbach, J., Rosenstiel, P., et al. (2018). A novel eukaryotic denitrification pathway in foraminifera. Curr. Biol. 28, 2536.e5–2543.e5. doi: 10.1016/j.cub.2018.06.027
Yang, S., Seo, H., Woo, J., Oh, H., Jang, H., Lee, J., et al. (2014). Carboxylicivirga gen. Nov. in the family Marinilabiliaceae with two novel species, Carboxylicivirga mesophila sp. nov. and Carboxylicivirga taeanensis sp. nov., and reclassification of Cytophaga fermentans as Saccharicrinis fermentans gen. Nov., comb. nov. Int. J. Syst. Evol. Microbiol. 64, 1351–1358. doi: 10.1099/ijs.0.053462-0
Keywords: foraminifera, unicellular eukaryotes, sediment, deep sea, endobionts, metabarcoding
Citation: Salonen IS, Chronopoulou P-M, Nomaki H, Langlet D, Tsuchiya M and Koho KA (2021) 16S rRNA Gene Metabarcoding Indicates Species-Characteristic Microbiomes in Deep-Sea Benthic Foraminifera. Front. Microbiol. 12:694406. doi: 10.3389/fmicb.2021.694406
Edited by:
Francisco J. A. Nascimento, Stockholm University, SwedenReviewed by:
Amir Szitenberg, Dead Sea and Arava Science Center – Dead Sea Branch, IsraelFatma Gomaa, Woods Hole Oceanographic Institution, United States
Copyright © 2021 Salonen, Chronopoulou, Nomaki, Langlet, Tsuchiya and Koho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Iines S. Salonen, aWluZXMuc2Fsb25lbkBoZWxzaW5raS5maQ==
 Iines S. Salonen
Iines S. Salonen Panagiota-Myrsini Chronopoulou1
Panagiota-Myrsini Chronopoulou1 Hidetaka Nomaki
Hidetaka Nomaki Dewi Langlet
Dewi Langlet Masashi Tsuchiya
Masashi Tsuchiya Karoliina A. Koho
Karoliina A. Koho