- 1Department of Translational Research and New Technologies in Medicine and Surgery, University of Pisa, Pisa, Italy
- 2Research Center Nutraceuticals and Food for Health-Nutrafood, University of Pisa, Pisa, Italy
On the worldwide market, a great number of probiotic formulations are available to consumers as drugs, dietary supplements, and functional foods. For exerting their beneficial effects on host health, these preparations should contain a sufficient amount of the indicated living microbes and be pathogen-free to be safe. Therefore, the contained microbial species and their amount until product expiry are required to be accurately reported on the labels. While commercial formulations licensed as drugs are subjected to rigorous quality controls, less stringent regulations are generally applied to preparations categorized as dietary supplements and functional foods. Many reports indicated that the content of several probiotic formulations does not always correspond to the label claims in terms of microbial identification, number of living organisms, and purity, highlighting the requirement for more stringent quality controls by manufacturers. The main focus of this review is to provide an in-depth overview of the microbiological quality of probiotic formulations commercialized worldwide. Many incongruences in the compositional quality of some probiotic formulations available on the worldwide market were highlighted. Even if manufacturers carry at least some of the responsibility for these inconsistencies, studies that analyze probiotic products should be conducted following recommended and up-to-date methodologies.
Introduction
Although the term probiotic made its first appearance in 1965, the first universally accepted definition was introduced by the Joint Food and Agriculture Organization of the United Nations (FAO) and the World Health Organization (WHO) in 2001 (FAO/WHO, 2001). For grammatical reasons, this definition was revisited in 2014 and definitively turned into “live microorganisms that, when administered in adequate amounts, confer health benefits on the host” (Hill et al., 2014).
A great number of studies highlight the ability of orally administered probiotics to improve the gut-barrier function, modulate the gut microbiota, enhance host immune response, and exert antimicrobial activities (Bermudez-Brito et al., 2012; Bron et al., 2017; Hu et al., 2017; Maldonado Galdeano et al., 2019). In addition, probiotics can be able to synthetize vitamins, food-degrading enzymes, and molecules contributing to cellular metabolism, thus ameliorating host health (LeBlanc et al., 2017; Oak and Jha, 2019).
Probiotics support physiological bodily functions and may reduce risk or shorten the duration or severity of many diseases. In particular, probiotic drugs are frequently used to prevent or help in the treatment of many gastrointestinal diseases due to infections, microbiome dysbiosis, and gut barrier perturbation. These include traveler, antibiotic-associated, and acute-infectious diarrhea, ulcerative colitis, necrotizing enterocolitis, inflammatory bowel diseases, irritable bowel syndrome, and Clostridium difficile and Helicobacter pylori infections (Wilkins and Sequoia, 2017). In the last decades, the use of probiotics has been extended to a variety of other disorders, including lactose intolerance, respiratory and urinary infections, asthma, atopic dermatitis, osteoporosis, allergy, metabolic syndromes, as well as liver, neurological, cardiovascular, and autoimmune diseases (Stavropoulou and Bezirtzoglou, 2020).
Microbes used as probiotics must be safe for consumption (FAO/WHO, 2002). To this regard, probiotic microbes intended for human use may have a “generally recognized as safe” (GRAS) notification for specific intended use to the US Food and Drug Administration (FDA, 2019) or the “qualified presumption of safety” (QPS) status (European Food Safety Authority: EFSA, 2007, 2018; Mattia and Merker, 2008). In addition, the antibiotic resistance profile of probiotics should be determined to highlight the presence of acquired resistance genes that can potentially be transmitted to pathogens (FAO/WHO, 2002; Cohen, 2018).
Several lactic acid bacteria (LAB) belonging to the Lacticaseibacillus, Lactiplantibacillus, Lactobacillus, Lentilactobacillus, Levilactobacillus, Ligilactobacillus, Limosilactobacillus, Bifidobacterium, and Streptococcus genera are traditionally used as probiotics, constituting the vast majority of commercial products available in the worldwide market (Williams, 2010; Hanchi et al., 2018; Zheng et al., 2020). Spore-forming bacteria of the genus Bacillus have progressively taken on, since they can be administered as spores that are extremely resistant to the harsh gastrointestinal conditions (Cutting, 2011; Elshaghabee et al., 2017; Jeżewska-Fra̧ckowiak et al., 2018; Lee et al., 2019). Among yeast, the species Saccharomyces cerevisiae var. boulardii exhibited a variety of beneficial properties, being adopted as probiotic microbe for several decades (Sen and Mansell, 2020). Several other microorganisms isolated from the human gut (e.g., Akkermansia muciniphila, Faecalibacterium prausnitzii) have been proposed as next generation probiotic candidates to face up specific diseases (Chang et al., 2019; Zhai et al., 2019).
Categorization of Probiotics and Notes on Regulation
In different countries, both regulatory aspects that govern probiotics and probiotic categories profoundly differ (for detailed regulatory aspects see Arora and Baldi, 2015; Kumar et al., 2015; de Simone, 2019; Domínguez Díaz et al., 2020; Koirala and Anal, 2021). Probiotics are generally classified as drugs (i.e., medicinal products and pharmaceuticals), dietary supplements (also referred as food supplements), and functional foods depending on the intended use (Halsted, 2003; Sreeja and Prajapati, 2013; de Simone, 2019; Koirala and Anal, 2021). Probiotic drugs are used for the prevention, treatment, and mitigation of human diseases and are subjected to the stringent regulations applied to other drugs, thus requiring approval before marketing and continuous pre- and post-marketing safety and quality controls (Venugopalan et al., 2010; Sreeja and Prajapati, 2013; Kolaček et al., 2017).
Dietary supplements can be administered as tablets, capsules, liquid suspensions, or powders and are intended to complement diet by ensuring the intake of specific dietary components (Taylor, 2004; Domínguez Díaz et al., 2020). In Europe, probiotic food supplements fall under the Food Products Directive and Regulation and each health claim need to be accurately scrutinized by the EFSA before being authorized (de Simone, 2019; Fusco et al., 2021). Good Manufacturing Practice that manufacturers of dietary supplements should follow have been developed (Food Supplements Europe, 2014). In the US, dietary supplements are regulated by the FDA under the Dietary Supplement Health and Education Act (DSHEA) and the Federal Trade Commission (FTC) and need to comply the Good Manufacturing Practice guidelines (Brown, 2017; de Simone, 2019; Fusco et al., 2021).
Although a statutory definition of functional foods does not exist in several countries, these products are intended to provide health benefits being consumed as part of the usual diet (Gul et al., 2016; Domínguez Díaz et al., 2020). This category meets the general laws applicable to foods in many countries (Kumar et al., 2015; Domínguez Díaz et al., 2020; Koirala and Anal, 2021).
Probiotic Labeling
Probiotic formulations possess specific label requirements that take into account the current laws and regulatory specifications of each country. FAO/WHO reported a list of information that manufacturers are recommended to state on the product label. In particular, labels should clearly indicate the minimum number of each strain until the expiry, the identification of each microorganism contained in the formulation based on the current nomenclature, and the claimed beneficial effects. According to the FAO/WHO guidelines, labels should also contain the suggested serving size to obtain health benefits, the appropriate storage conditions, and corporate contact details (FAO/WHO, 2002).
In 2017, the International Probiotics Association, in partnership with the Council for Responsible Nutrition, added new details to the FAO/WHO indications on the basis of the US regulation and proposed new guidelines for probiotic labeling that worldwide manufacturers were recommended to comply (Council for Responsible Nutrition and International Probiotics Association, 2017). These guidelines were aimed at improving transparency, consistency, and consumer understanding, thus promoting comparisons between different probiotic formulations and improved consumer awareness. Product labels should indicate the quantitative amount of alive microorganisms expressed as colony forming units (CFUs) and the expiration date. For formulations containing multiple species and/or strains, information on the total amount of microorganisms was recommended and, when technically feasible, the amount of each species as well.
Compositional Quality
Although publications on probiotics rapidly expanded in the last years, most of the studies have been focused on the beneficial properties exhibited by probiotic microbes. A relatively low number of studies analyzed the real-time composition of probiotic formulations that consumers find on the market.
Some reports denouncing poor quality of commercial probiotic products already populated the scientific literature before general guidelines were developed (Gilliland and Speck, 1977; Hamilton-Miller et al., 1996; Yaeshima et al., 1996; Canganella et al., 1997; Hamilton-Miller and Gibson, 1999; Hamilton-Miller et al., 1999; Schillinger, 1999; Hoa et al., 2000). With the introduction of guidelines for probiotics in 2002 (FAO/WHO, 2002), some investigations were conducted to evaluate the compliance between label claims and effective microbial composition, amount of viable cells, and absence of contaminant microorganisms of probiotic formulations available on the market.
In this review, we took into consideration only studies that were published after the introduction of the FAO/WHO guidelines. The probiotic formulations analyzed in different studies were divided into the three category drugs, dietary supplements, and functional foods (Tables 1–3). For simplicity, probiotic formulations indicated as foods with specific medical purposes (FSMP) were included in the category dietary supplements. We decided to incorporate in the tables only products for which name, category, country of origin, and possibly, manufacturer were known. For formulations produced by unmentioned manufacturers, product name and country of origin were used to conduct a careful research for retrieving the missing data. In addition, we did not include commercial products for which the concordance with label claims of amount of living cells, contained species, and purity were not clearly stated by the Authors. For completeness, studies not clearly stating product names, manufacturers, categorization, or origin (further referred to as unidentified probiotics) will be separately discussed in the main text.
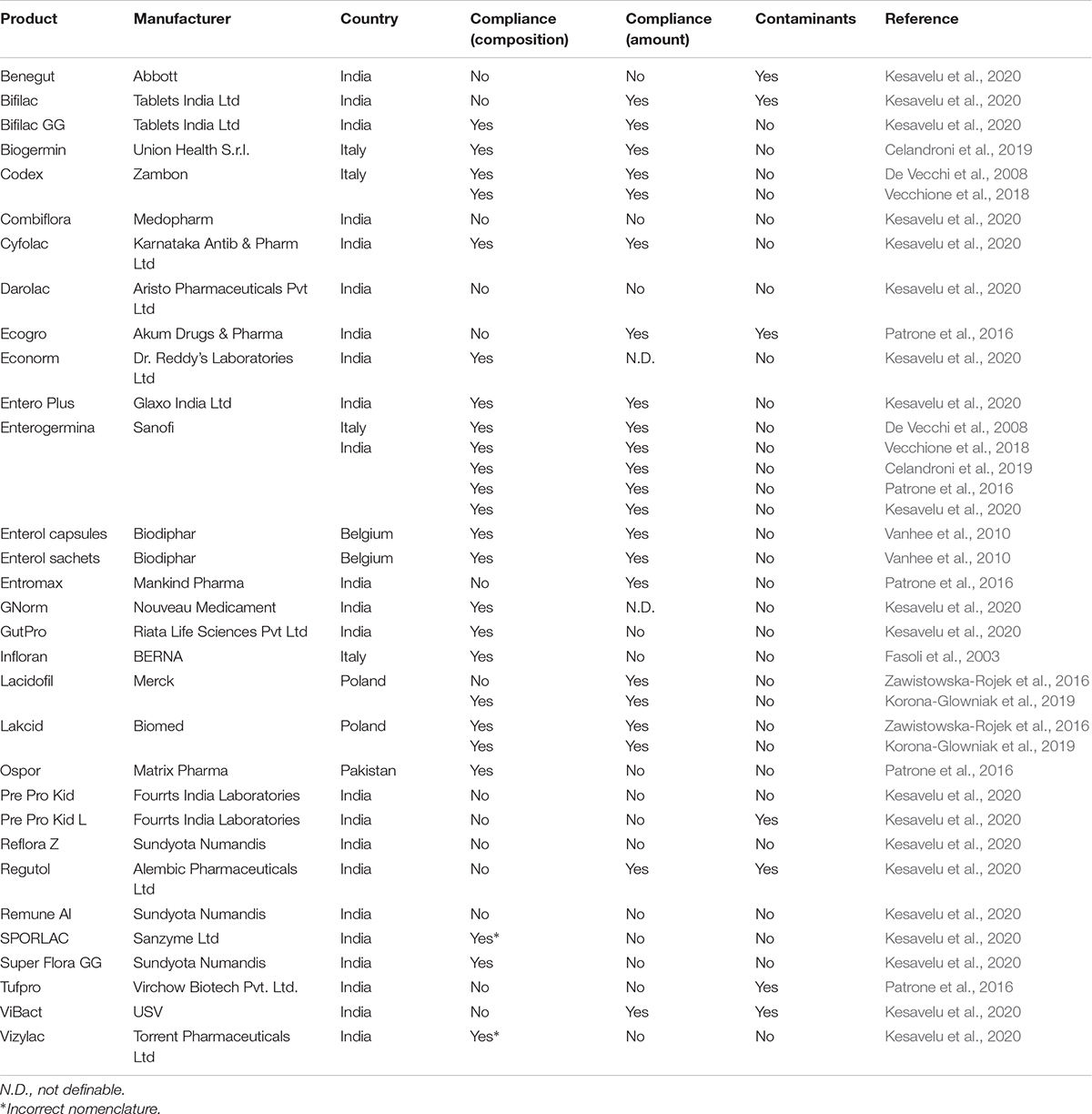
Table 1. Compliance with the label claims of probiotic drugs marketed worldwide: focus on the microbial composition, amount of living cells, and presence of contaminant microorganisms.
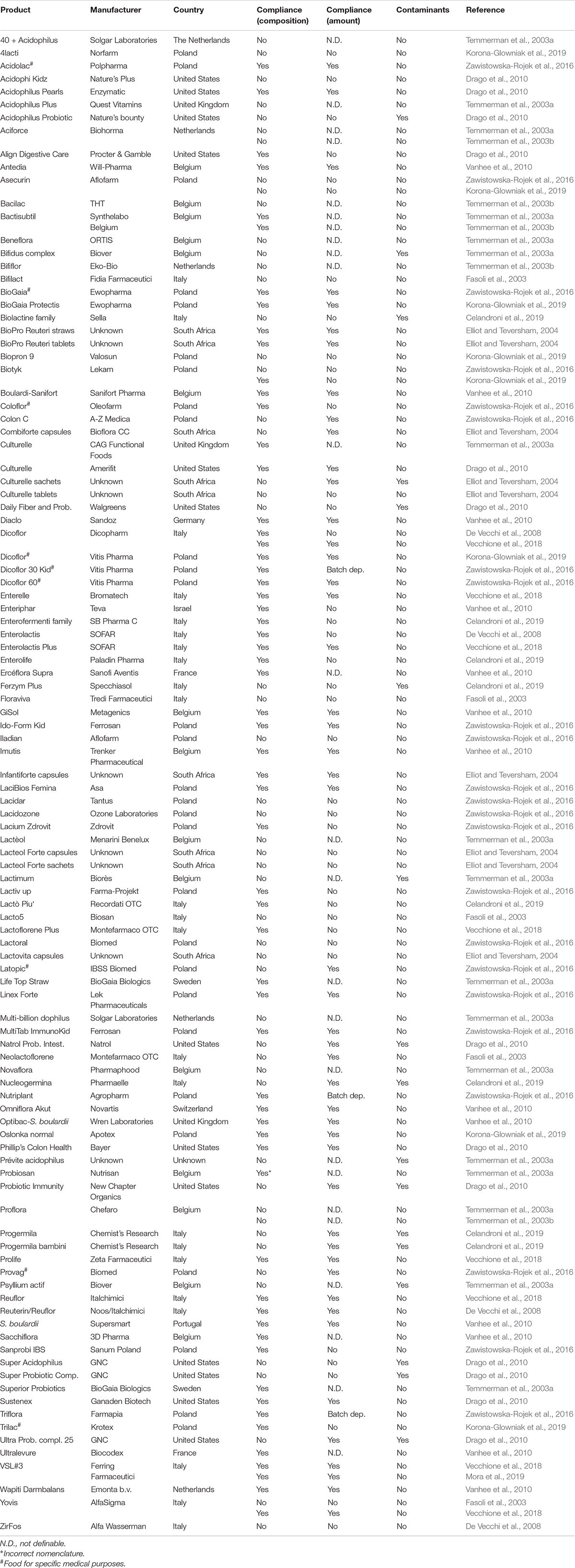
Table 2. Compliance with the label claims of probiotic dietary supplements marketed worldwide: focus on the microbial composition, amount of living cells, and presence of contaminant microorganisms.
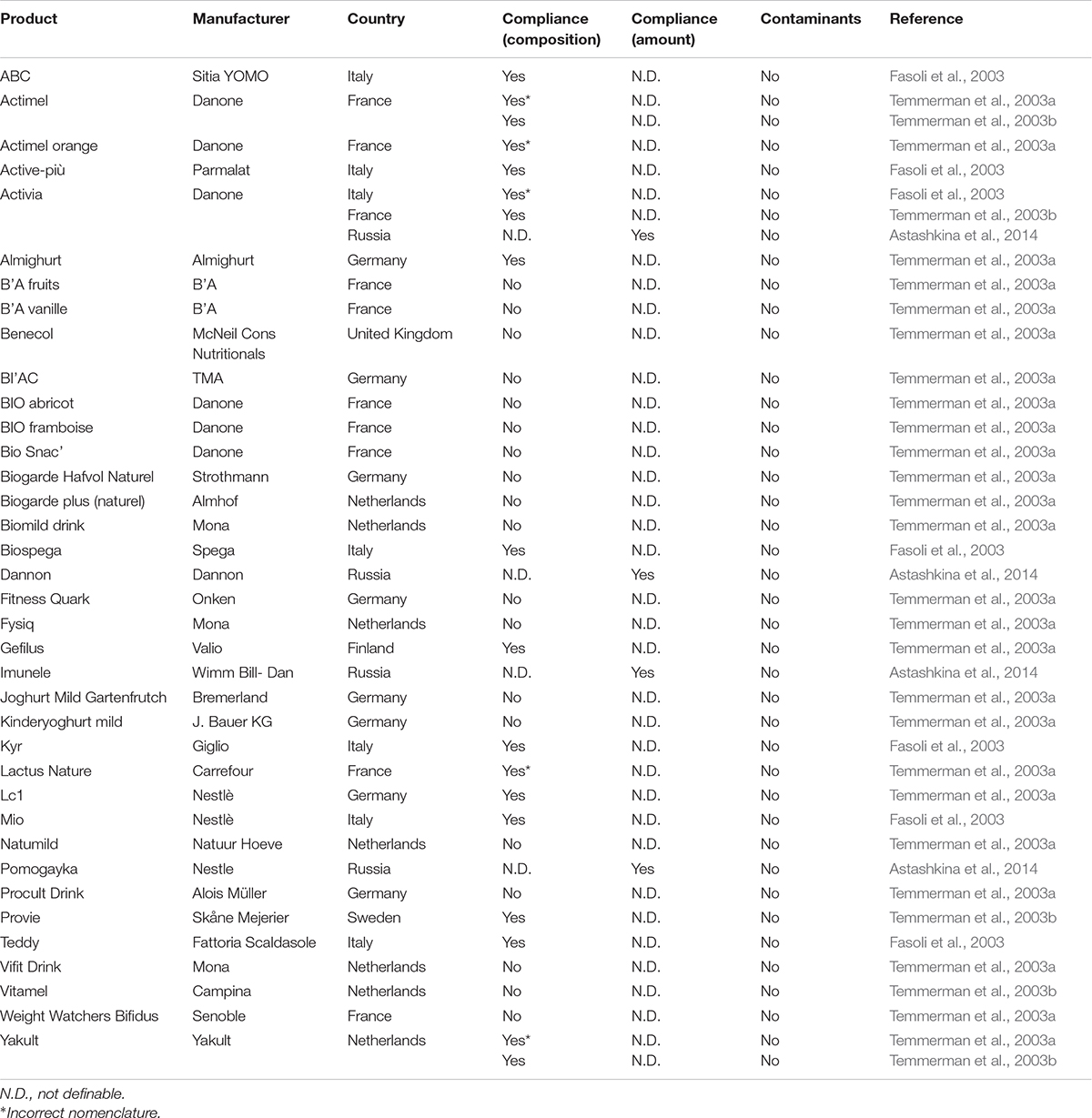
Table 3. Compliance with the label claims of probiotic functional foods marketed worldwide: focus on the microbial composition, amount of living cells, and presence of contaminant microorganisms.
Microbial Composition
The identity of microbes (i.e., at genus, species, and strain level) included in commercial probiotic formulations should be stated by the manufacturer on the product label and follow the current accepted nomenclature (FAO/WHO, 2002; Council for Responsible Nutrition and International Probiotics Association, 2017). To this regard, Weese analyzed the labels of 21 dietary supplements intended for human use to verify label’s accuracy (Weese, 2003). The author showed that nine products reported adequate label information in terms of microbial composition with only two indicating microbes at the strain level. On the other products, microorganisms were indicated with a name not concordant with the actual nomenclature or misspelled (Weese, 2003). Similarly, label analysis of the top 10 probiotic formulations sold on the Indian market revealed that seven did not specify the contained species, but only gave an unsuitable explanation (i.e., Lactic Acid Bacillus; Ghattargi et al., 2018).
In this study, we also evidenced that the labels of some formulations, particularly functional foods, did not comply general guidelines for probiotic labeling, since not indicating the contained microbes or only reporting a general and inaccurate description (e.g., “LAB,” “probiotics,” “lactic ferment,” “lactic acid bacillus,” “live probiotic AB cultures,” “active bifidus,” “special live cultures,” “viable LAB,” “bifidus essensis,” “life yeast”) (Fasoli et al., 2003; Temmerman et al., 2003a,b; Elliot and Teversham, 2004; Masco et al., 2005; Theunissen et al., 2005; Lin et al., 2006; Perea Vélez et al., 2007; Angelakis et al., 2011; Chen et al., 2014; Kesavelu et al., 2020). In addition, we found that some preparations only indicated the contained microbes at the genus level, thus resulting in disagreement with the FAO/WHO guidelines (Fasoli et al., 2003; Temmerman et al., 2003a,b; Masco et al., 2005; Theunissen et al., 2005; Lin et al., 2006; Perea Vélez et al., 2007; Chen et al., 2014; Yonkova Marinova et al., 2019).
Regarding probiotic drugs, the microbial composition of 31 products available in Europe (Italy, Belgium, and Poland), India, and Pakistan is reported in Table 1 (column 4). Only four products among these 31 (Codex, Enterogermina, Lacidofil, Lakcid) were analyzed by independent studies, with three of them showing concordant results. In fact, Lactobacillus acidophilus declared to be contained in Lacidofil was detected only in one study by species-specific PCR (Korona-Glowniak et al., 2019) but not in the other, which used biochemical methods and matrix assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) (Zawistowska-Rojek et al., 2016). Overall, our analysis highlighted that 17 formulations (56.67%) contained the species stated on the labels. Bacillus coagulans in two products (SPORLAC, Vizylac) was labeled with an obsolete nomenclature (i.e., Lactobacillus sporogenes). Thirteen formulations (43.33%) did not comply the label claims. In particular, six formulations (Darolac, Ecogro, Entromax, Pre Pro Kid L, Regutol, Tufpro) contained more species than those declared, while three (Combiflora, Reflora Z, Remune Al) lacked one or more species. In addition, Benegut, Bifilac, Pre Pro Kid, and Vibact possessed different species than those labeled (Table 1, column 4).
Regarding dietary supplements, the microbial composition of a total of 106 products marketed around the world is reported in Table 2 (column 4). Byotik and Yovis were analyzed in two studies by using different methods, but the recovered species were found to be concordant with the label claims only in one study each. The discordant results obtained for Yovis could depend on the different years in which the studies were performed (Fasoli et al., 2003; Vecchione et al., 2018). In fact, it can not be excluded the product underwent changes in 15 years and the manufacturer’s controls had increased over time. Aciforce, Asecurin, Bactisubtil, Dicoflor (Dicopharm), Proflora, and VSL#3 were also analyzed by independent studies with concordant results. Globally, among 104 products, 56 (53.85%) contained the same species declared on the labels, with the exception of Lactobacillus crispatus in Probiosan that was indicated with an obsolete nomenclature. The other 48 products (46.15%) were not compliant for the microbial composition, since lacking one or more species or containing more or different species than those declared on the labels (Table 2, column 4).
As regards products categorized as functional foods, a total of 37 formulations is reported in Table 3 (column 4). For three products (Dannon, Imunele, Pomogayka), the correspondence between contained species and label information could not be established, since the identification of microbes isolated from these products was only performed at the genus level (Astashkina et al., 2014). Actimel, Activia, and Yakult were analyzed by independent studies, all showing concordant results with label information. Globally, the microbial composition was concordant with the label claims for 15 functional foods (44.12%), with 5 of them reporting an obsolete nomenclature (Actimel, Actimel Orange, Activia, Lactus Nature, Yakult). In the other 19 products (55.88%), no concordance between microbial composition and labeled information was found.
As regards studies on unidentified probiotics, Weese reported concordant results with label claims in only 2 of 5 products intended for human use (Weese, 2002). Analysis of the microbial composition of 58 products obtained from 13 Countries indicated that many formulations were mislabeled (Masco et al., 2005). Similarly, a study conducted on 10 products sold in Europe indicated that three were not concordant with the label claims and four were concordant at the species level but contained different strains than those labeled (Coeuret et al., 2004). Aureli et al. (2010) verified the compliance to the label claims of 41 dietary supplements sold in Italy showing that 20 were constituted by the species reported on the labels. Toscano et al. (2013) investigated the microbial composition of 24 dietary supplements available on the Italian market by using PCR, pyrosequencing of the 16S rRNA gene, and biochemical methods. They showed that 21 products were compliant with the label claims. Nevertheless, seven of these supplements did not contain all the labeled microbes in the amount declared by manufacturers (Toscano et al., 2013). Di Pierro et al. (2019) evaluated the compositional quality of nine formulations declaring to contain alive L. crispatus alone or in combination with other species. The presence of L. crispatus in the formulations was investigated by using selective media and by species-specific PCR (Di Pierro et al., 2019). The authors found that all products contained L. crispatus by culture-independent methods, but only six formulations carried living cells (Di Pierro et al., 2019). The identification of species declared to be contained in five Italian oral products revealed concordant microbial composition to the label claims for four formulations (Blandino et al., 2016). Identification of species contained in 13 French functional foods by MALDI-TOF MS, PCR and sequencing of tuf and 16S rRNA genes revealed different species than those stated for two products (Angelakis et al., 2011). The analysis of different lots of a probiotic formulation intended for preterm infant in The Netherlands evidenced the absence of one or more species claimed on the label depending on the tested lot (Vermeulen et al., 2020). The microbial composition of five Polish medicinal products was analyzed by biochemical methods (Szajewska et al., 2004). This study showed that only three products completely complied the labeled information in terms of composition, while the other two contained other species than those declared (Szajewska et al., 2004). Investigation of 26 Bulgarian dietary supplements by two-step multiple PCR indicated that none contained all the species claimed on the labels (Yonkova Marinova et al., 2019).
By using PCR-based denaturing gradient gel electrophoresis (DGGE) and species-specific PCR to decipher the microbial composition of 20 products commercialized in South Africa, Theunissen et al. (2005) found that 54.6% of functional foods and 33.3% of dietary supplements contained the microorganisms stated on the labels.
In a study aimed at evaluating the composition of seven functional foods available on the Columbian market, the authors showed that the content of many dairy products did not comply the label claims (Perea Vélez et al., 2007). Compliance for microbial composition was found for 1 of 5 products available in the US (Drisko et al., 2005). The analysis of the microbial compositions of 14 dietary supplements commercialized in the US by terminal restriction fragment length polymorphism (T-RFLP) and species-specific PCR revealed that 12 products contained additional species than those declared (Marcobal et al., 2008). By studying the bacterial diversity of 16 Bifidobacterium-containing products available in the US market, Lewis et al. (2016) showed that only 1 product completely fulfill the label claims. The other formulations lacked one or more Bifidobacterium species and/or contained additional species. In addition, a pill-to-pill and lot-to-lot variation was highlighted by the authors for most formulations (Lewis et al., 2016). In a complex study aimed at evaluating the microbiological quality of 52 dietary supplements sold in North Carolina, the authors showed that 30 products effectively contained the labeled species (Morovic et al., 2016). By culture-independent metagenomic sequencing of 10 dietary supplements marked in the US, Patro et al. (2016) showed that five products were mislabeled (Patro et al., 2016).
The analysis of 28 products available on the Chinese market revealed that some preparations respected the labeled species (Chen et al., 2014). Lastly, 4 of 17 products marketed in China were not compliant with the labeled information for microbial composition (Ullah et al., 2019).
Amount of Viable Cells
According to the guidelines for probiotics, manufacturers are required to declare the amount of microbes contained in commercial formulation by the expiration date. In the analysis of labels of 21 products for human use conducted by Weese, 16 were shown to report the contained amount of cells (Weese, 2003). In this analysis of the published data, the amount of cells was not labeled for two drugs (Econorm, GNorm; Table 1, column 5) and three dietary supplements (Ercèflora Supra, Sacchiflora, Ultralevure; Table 2, column 5). Two dietary supplements (Acidophilus Probiotic Gold, Natrol Probiotic Intestinal) declared the total amount of microbes at the production time and not at the expiration date (Table 2, column 5).
As regards studies on unidentified probiotics, the microbial amount was not declared for one Italian dietary supplement, 4 European functional foods, 5 Indian products, 4 products sold in Taiwan, and 15 dairy products available on the Chinese market (Coeuret et al., 2004; Lin et al., 2006; Chen et al., 2014; Ghattargi et al., 2018; Di Pierro et al., 2019).
The majority of multispecies or multistrain products were found specifies on the labels only the total amount of microorganisms. In fact, species/strain amount declaration was only found for six drugs (Bifilac, Pre Pro Kid, Pre Pro Kid L, Reflora Z, Regutol, Vibact; Table 1) and five dietary supplements (Biolactine family, Infloran, Ferzym Plus, Floraviva, Yovis; Table 2). Three drugs and two dietary supplements sold in China, one functional food available in South Africa, as well as two drugs and nine supplements marketed in Italy indicated the specific amount of the contained species or strain (Theunissen et al., 2005; Aureli et al., 2010; Toscano et al., 2013; Chen et al., 2014; Di Pierro et al., 2019; Ullah et al., 2019).
Microbial counts in probiotic formulations were conventionally performed by the plate count method. This culture-based analysis may underestimate the real number of alive microbes if methodologies which are not up to current standards were used. Recommended methods for the analysis of probiotics in foods and supplements have been published (Champagne et al., 2011; Schoeni, 2015). In the analysis of probiotic formulations the use of an appropriate medium, period, and temperature for rehydration, homogenization procedure, plating medium, and incubation time, temperature, and redox level should be adopted (Champagne et al., 2011; Schoeni, 2015). Regarding the evaluation of the amount of viable cells compared with the number declared on the label, we found that 15 out of 29 probiotic drugs (51.72%) were compliant (Table 1, column 5). Among them, all the products investigated in different studies (Codex, Enterogermina, Lacidofil, Lakcid) provided similar results. The other 14 probiotic drugs (48.28%) contained less microbes than those stated on the labels.
As regards to dietary supplements (Table 2, column 5), microbial viability was not definable for 22 products, since the amount of cells was not declared. In addition, contrasting outputs were obtained for Dicoflor and Yovis in different studies (Fasoli et al., 2003; Vecchione et al., 2018) and some variations due to different product batches were observed for Dicoflor 30 Kid, Nutriplant, and Triflora (Table 2, column 5). Among the remaining 79 supplements, 44 (55.70%) were compliant for the number of cells. The total CFU amount obtained for 35 products (44.30%) was lower than the number declared on the labels. In particular, the analysis of Asecurin and Byotik in two independent studies led to the same incongruent results (Zawistowska-Rojek et al., 2016; Korona-Glowniak et al., 2019).
As shown in Table 3 (column 5), the compliance with the labeled amount of microbes could not be established for 33 out of 37 functional foods, since the amount of cells was not declared (Fasoli et al., 2003; Temmerman et al., 2003a,b). The remaining four products analyzed for microbial viability (Activia, Dannon, Imunele, Pomogayka) contained the amount of cells declared by manufacturers on labels.
As regards studies on unidentified probiotics, Masco et al. (2005) showed that many of the investigated formulations did not contain viable cells. An incongruent microbial amount was found in 5 among 10 investigated European products (Coeuret et al., 2004). Thirty among 46 analyzed Italian products contained the amount of living microbes declared on the labels (Aureli et al., 2010; Blandino et al., 2016). Among 24 Italian products tested by Toscano et al. (2013), 10 were found to contain an incorrect amount of cells. On a total of nine Italian analyzed formulations, only two contained the amount of viable cells declared by manufacturers (Di Pierro et al., 2019). All the Polish probiotic formulations included in the study by Szajewska et al. (2004) complied with the label claims for content of viable cells. The analysis of the content of living microbes in 26 supplements sold in Bulgaria revealed that for 10 there was concordance with the label claims (Yonkova Marinova et al., 2019).
By analyzing the amount of viable cells of five dietary supplements available in South Africa, three of them resulted to contain a lower microbial load than that labeled (Brink et al., 2005). Determination of viable Bifidobacterium conducted in 58 yogurts from 7 manufacturers sold in North Carolina revealed that 14 did not contain viable cells (Ibrahim and Carr, 2006). Morovic et al. (2016) found that 17 of 52 supplements had a CFU count drastically below the labeled amount. On a total of 10 formulations analyzed, one was found to contain a lower amount of viable cells than that declared by the manufacturer (Patro et al., 2016).
In one study aimed at investigating the presence of LAB in eight probiotic products available in Taiwan, the authors found that two products did not contain the labeled amount of LAB since no viable cells were recovered from these formulations (Lin et al., 2006). For 28 Chinese products, concordance with the labeled number of total living microbes was evidenced for 13 products (Chen et al., 2014). Nevertheless, the authors showed that other 12 products in which the contained amount of cells was not labeled met the minimum quantitative requirement of living cells established in China (Chen et al., 2014). In addition, Ullah and coauthors showed that 5 among 17 tested Chinese dietary supplements contained a number of viable cells that was not concordant with the amount declared on the labels. In particular, four products were found to contain a lower microbial load and no viable cells were detected in one product (Ullah et al., 2019). Among four analyzed dietary supplements available in Bangladesh, none contained the labeled amount of cells (Begum et al., 2015).
Microbiological Purity
Commercial probiotic formulations should be free of pathogenic organisms (further reported as contaminants) that can constitute a health risk for humans. While all the analyzed functional foods were shown to lack contaminants (Table 3, column 6), 7 drugs (22.58%; Table 1, column 6) and 17 dietary supplements (16.04%; Table 2, column 6) contained potential pathogens.
As regards to drugs, the presence of Bacillus thuringiensis was revealed in Benegut and Regutol, while Enterococcus hirae was detected in Bifilac, Pre Pro Kid L, and Vibact (Table 1). Interestingly, while DGGE detected Bacillus cereus, Alcaligenes faecalis, Xanthomonas spp., and Lysinibacillus spp. in Tufpro, culture-dependent analysis identified only B. cereus in the product, thus indicating the presence of viable cells of this organism (Table 1). Culture-independent DGGE analysis of Ecogro (Table 1) revealed several contaminants, e.g., B. cereus, Bacillus licheniformis, Acinetobacter spp., Xanthomonas/Pseudomonas spp. Staphylococcus/Lysinibacillus spp. Regarding dietary supplements, high loads of B. cereus (about 1.0 × 1010 CFU/dose) and Lysinibacillus fusiformis (about 1.3 × 1011 CFU/dose) were detected in Progermila bambini (Table 2). In addition, B. cereus was found to contaminate 1 unidentified dietary supplement (4.5 × 106 CFU/g) available on the Italian market (Aureli et al., 2010). The products Biolactine family and Ferzym Plus were found to contain more than 1.0 × 103 CFU/dose of B. licheniformis and Bacillus badius, respectively (Table 2). Super Probiotic Complex was contaminated by Bacillus lentus (9.0 × 109 CFU/dose) and Bacillus spp. (1.0 × 102 CFU/dose). Contamination by Bacillus spp. was also found in Daily Fiber and Probiotics (3.5 × 102 CFU/dose) and Natrol Probiotic Intestinal (1.3 × 108 CFU/dose). Drago et al. (2010) showed the presence of 1.0 × 102 CFU/dose of Staphylococcus spp. in Acidophilus Probiotic Gold and a mold in Ultra Probiotic Complex. L. fusiformis was detected in high amount (about 7.0 × 1011 CFU/dose) in Progermila and Acinetobacter baumannii (about 1.2 × 1011 CFU/dose) was isolated from Nucleogermina (Table 2).
Enterococcus faecium was the most common contaminant found in dietary supplements. The microorganism was found in Bifidus complex, Culturelle sachets, Lactimum, Prévite acidophilus, and Psyllium actif (Table 2). E. faecium was also detected in high amount in Probiotic Immunity (4.2 × 109 CFU/dose), Super Acidophilus (4.1 × 105 CFU/dose), and Super Probiotic Complex (9.9 × 104 CFU/dose) (Table 2). Additionally, the microbe was found in several unidentified products. In particular, E. faecium was revealed in one US and two Italian dietary supplements, as well as in most of the Chinese products analyzed by Chen et al. (2014; Toscano et al., 2013; Patro et al., 2016). The presence of E. faecium and Enterococcus faecalis was denounced in some batches of a Dutch product for infants (Vermeulen et al., 2020).
Other potential pathogens such as B. cereus, B. licheniformis, Enterobacter cloacae, Enterobacter spp., Staphylococcus epidermidis, Klebsiella spp. and Serratia spp. were revealed by DGGE analysis in some Chinese probiotics (Chen et al., 2014). Nevertheless, none of these microbes were isolated from the products by using culture-dependent methods, thus suggesting that they could be present in a killed form (Chen et al., 2014). Microorganisms belonging to the genus Weissella were isolated from some products marketed in Bulgaria (Yonkova Marinova et al., 2019). Lastly, contamination of a dietary supplement for preterm newborns by Rhizopus oryzae was evidenced by culture-dependent methods (Vallabhaneni et al., 2015).
Discussion
According to their definition, probiotics must be alive and administered in adequate amount to benefit host health (FAO/WHO, 2001, 2002; Hill et al., 2014). The administration of probiotic preparations containing lower amounts of living microbes may lead to reduced or even absent beneficial effect (Ouwehand, 2017). The daily amount of living probiotics should derive from in vitro and in vivo experiments and be opportunely determined for each strain and product. Therefore, a universal dose to administer is not established a priori for all products. However, some countries (e.g., Italy) require that the daily dose of probiotic formulations contains at least 109 CFU (Ministero della Salute, 2018). Manufacturers are recommended to clearly state the total content of microbes on the product label and this amount should be guaranteed until the expiration date at the declared handling and storage conditions (FAO/WHO, 2002; Guarner et al., 2012; Council for Responsible Nutrition and International Probiotics Association, 2017; World Gastroenterology Organisation [WGO], 2017).
The analysis of commercial probiotic products included in this review highlighted a critical scenario. Many probiotics, particularly functional foods, do not indicate the contained amount of microbes on the label and discrepancies between label information and real content emerged. Even if manufacturers carry at least some of the responsibility for these incongruences, it should be mentioned that the included studies have adopted different methodologies that were sometimes not rigorous and not adherent to recommended methods for the analysis of probiotic products. In fact, papers published before 2015 could have suffered of the lack of guidelines for good practices (Schoeni, 2015) and the adopted methods led to erroneous (underestimated) or incomplete results. It should be mentioned that many studies analyzed products off the shelf, and did not store them until the expiration (“best before”) date prior to analysis. It could be that some products judged to be “compliant” were not at the expiration date, particularly if the manufacturers allow storage at room temperature. Thus many conclusions in Table 2 are only “presumptively compliant.”
Among the formulations that declare the total CFU number, more than 40% included in the category drugs and dietary supplements carried a lower amount of viable cells than stated. In the analyzed studies, determination of the amount of living microbes was performed by the plate count method and the number of cells for dose or gram of product was generally provided (Jackson et al., 2019). Although some culture-independent technologies (e.g., flow cytometry, direct imaging and enumeration, nucleic acid amplification techniques) for quantifying the total microbial content (i.e., alive and dead cells) of probiotic formulations are available, the plate count method currently represents the gold-standard applied in probiotic industry for quality controls (International Standards Organisation [ISO], 2006; International Standards Organisation [ISO], 2010; Davis, 2014; Jackson et al., 2019). However, some limits on the use of the plate count method for the analysis of probiotic formulations have been evidenced. In addition to the fact that not all microbes are culturable by traditional methods and that different microorganisms can require extremely variegate cultural conditions, this technique is unable to detect viable but non-culturable (VBNC) cells that can be generated by the stressful manufacturing procedures (Davis, 2014; Jackson et al., 2019). To obtain the correct enumeration of microbes contained in probiotic products, both plate-count and culture-independent techniques should be applied, as performed in a limited number studies included in this review (Vanhee et al., 2010; Morovic et al., 2016; Patro et al., 2016; Ullah et al., 2019). In culture-dependent analyses, enumeration of different species in multi-species formulations can be critical. To overcome this limit, selective media are commonly used to discriminate species with different metabolic features (Fasoli et al., 2003; Elliot and Teversham, 2004; Aureli et al., 2010; Drago et al., 2010; Toscano et al., 2013; Chen et al., 2014; Di Pierro et al., 2019; Korona-Glowniak et al., 2019; Yonkova Marinova et al., 2019; Kesavelu et al., 2020). Nevertheless, this aim remains challenging, particularly for Lactobacillus and Bifidobacterium species. In fact, some selective media can impair growth of the target microbe or allow growth of other microorganisms, thus resulting not completely selective (Roy, 2001; Davis, 2014; Hayek et al., 2019; Margolles and Ruiz, 2021).
The viability of probiotic microbes in the finished product is influenced by manufacturing, packaging, and handling. To this regard, overage amounts of microbes are commonly included by manufacturers in probiotic supplements to guarantee the presence of the labeled dose until the expiration date (Fenster et al., 2019). In addition, probiotic products are often produced in a dried or microencapsulated form to ensure stability of microbes over time (Fenster et al., 2019; Grumet et al., 2020). Adequate packing is also crucial to prevent humidification, which can affect cell viability (Fenster et al., 2019).
An additional factor that affects the stability of microbes is the duration of the product shelf life. In fact, while for functional foods the expiration date is generally measured in days or months, for dietary supplements and drugs it is commonly considerably longer (i.e., up to 24 months) (Fenster et al., 2019). Different studies demonstrated a progressive and time-related decrease in the number of viable cells in probiotics investigated at different times before the expiry (De Vecchi et al., 2008; Toscano et al., 2013; Blandino et al., 2016). Stability controls should be performed by manufacturers to guarantee the presence of living cells, since some probiotic organisms can enter in a viable but non-culturable status or lose viability (Forssten et al., 2011; Jackson et al., 2019).
Beneficial health effects of probiotics are generally species- and often strain-specific (Hill et al., 2014; Vitetta et al., 2017). Therefore, the correct identification of microorganisms contained in commercial probiotic formulations is of crucial importance (FAO/WHO, 2002; Council for Responsible Nutrition and International Probiotics Association, 2017).
In this work, we highlighted that more than 40% of the formulations included in the category drugs, dietary supplements, and functional foods do not comply the label claims for microbial composition. Analyzing all the investigated papers over time and looking at the percentages of compliant formulations, no substantial improvement in the quality of probiotic products was found (Supplementary Table 1). Nevertheless, due to the fact that many of the analyzed papers were published many years ago, we cannot exclude the current situation of marketed probiotics is improved.
The evidenced lack of concordance between label claims and microbial composition could be due to manufacturer-dependent biases and flaws in quality controls. The FAO/WHO guidelines recommended to identify microbes included in probiotic formulations by using molecular techniques, such as DNA-DNA hybridization or 16S rRNA gene sequencing (FAO/WHO, 2002). In a comprehensive study analyzing 213 microbial cultures intended for production of probiotic formulations, more than 28% were incorrectly identified for the application of unsuitable identification methods (Huys et al., 2006).
It is remarkable to underline that the use of different methods for the identification of microbes can lead to divergent results, particularly for multispecies products. Contrasting results in the composition of the same probiotic brands were evidenced in studies using different methods (Fasoli et al., 2003; Zawistowska-Rojek et al., 2016; Vecchione et al., 2018; Korona-Glowniak et al., 2019). In addition, we cannot exclude that some of the analyzed papers could have produced erroneous conclusions, since not applying rigorous and up-to-date methodologies in the analysis of probiotic formulations. In most of the included studies, the microbial composition of probiotic products was analyzed by a combination of culture-dependent methods (e.g., biochemical tests, MALDI-TOF MS, species-specific PCR, sodium dodecyl sulfate polyacrylamide gel electrophoresis, sequencing of target genes, microsatellite-based typing) applied to microbes isolated from the formulations (Temmerman et al., 2003a; Coeuret et al., 2004; Szajewska et al., 2004; Perea Vélez et al., 2007; De Vecchi et al., 2008; Aureli et al., 2010; Drago et al., 2010; Vanhee et al., 2010; Blandino et al., 2016; Zawistowska-Rojek et al., 2016; Vecchione et al., 2018; Celandroni et al., 2019; Korona-Glowniak et al., 2019; Vermeulen et al., 2020; Yonkova Marinova et al., 2019). However, culture-dependent methods are strictly bound to the ability of the investigators to grow in vitro all microbes contained in probiotic products and to phenotypically discriminate colonies belonging to different species. This aim is particularly challenging for multispecies formulations and can result in the inability to detect some species, as humbly reported in some studies (Temmerman et al., 2003a; Drago et al., 2010; Vecchione et al., 2018). In addition, culture-dependent methods fail in the detection of VBNC cells that do not grow on selective media and some biochemical tests (e.g., API system) have been shown to misidentify closely related species and identify Bifidobacterium only at the genus level (Blandino et al., 2016; Zawistowska-Rojek et al., 2016). Similarly, sequencing of the 16S rRNA gene can result unable to distinguish closely related species that share an almost identical sequence of this gene (Fusco et al., 2021). On the other hand, culture-independent techniques (DGGE, T-RFLP, species-specific PCR, high-throughput sequencing, shotgun metagenomic sequencing) directly applied to the whole probiotic products were primarly adopted for microbial identification in some papers (Elliot and Teversham, 2004; Drisko et al., 2005; Theunissen et al., 2005; Marcobal et al., 2008; Chen et al., 2014; Morovic et al., 2016; Mora et al., 2019). These methods lead to detect all the species effectively contained in the products (including VBNC) but are unable to establish if the identified species are viable. Although some probiotic cells have been shown to maintain their beneficial properties also in a dead form (Taverniti and Guglielmetti, 2011; de Almada et al., 2016), probiotics are required to be alive to benefit host health (FAO/WHO, 2001). Considering the limits of culture-dependent and culture-independent techniques, we believe that a combination of both methods should be reasonably applied to definitely decipher the microbial composition of commercial probiotic products, as performed in some of the included studies (Fasoli et al., 2003; Temmerman et al., 2003b; Masco et al., 2005; Angelakis et al., 2011; Toscano et al., 2013; Lewis et al., 2016; Patro et al., 2016; Patrone et al., 2016; Di Pierro et al., 2019; Ullah et al., 2019; Kesavelu et al., 2020). Recently, whole genome sequence analysis has been proposed as a promising method for the identification of microbes contained in probiotic formulations (Fusco et al., 2021).
Purity of commercial probiotic formulations represents a major issue in product testing and quality controls. The introduction of potential pathogenic microorganisms in probiotics can accidentally occur during the entire manufacturing and handling steps and constitute a dangerous health threat, particularly for susceptible individuals. A case of fatal gastrointestinal mucormycosis in a preterm infant due to the assumption of probiotic dietary supplement contaminated by the mold Rhizopus oryzae has been reported (Vallabhaneni et al., 2015). Although some Enterococcus strains are conventionally used as probiotics, FAO/WHO recommended to keep careful attention in the inclusion of Enterococcus microbes in commercial formulations. In fact, some strains can exhibit transmissible vancomycin resistance, acquire other resistance genes, and show a certain degree of pathogenic potential (FAO/WHO, 2001, 2002; Ben Braïek and Smaoui, 2019).
Enterococcus hirae, E. faecium, and E. faecalis were found to contaminate some probiotic products, particularly dietary supplements. However, Temmerman et al. (2003a) declared than none of E. faecium strains isolated in their study as contaminant was resistant to vancomycin. Due to their marked antibiotic resistance and ability to cause urinary tract and wound infections, endocarditis, and bacteremia, E. faecium and E. faecalis are well known as important nosocomial pathogens (Madsen et al., 2017; Gao et al., 2018; Ben Braïek and Smaoui, 2019; Zhou et al., 2020). Differently, E. hirae is generally considered a zoonotic pathogen and is rarely isolated from clinical samples derived from humans. Nevertheless, some studies highlighted this microorganism as responsible with urinary tract infections, endocarditis, and bacteremia (Bourafa et al., 2015; Dicpinigaitis et al., 2015; Ebeling and Romito, 2019; Pinkes et al., 2019; Winther et al., 2020).
Other products resulted to be contaminated with high Bacillus loads. Although some Bacillus strains are traditionally used as probiotics, others are sufficiently equipped of virulence determinants and cause infections in humans (Cutting, 2011; Celandroni et al., 2016; Elshaghabee et al., 2017; Jeżewska-Fra̧ckowiak et al., 2018; Lee et al., 2019). In fact, B. cereus is well known as causative agent of two-types of food poisoning diseases. While the emetic syndrome is due to the ingestion of food contaminated by the pre-formed cereulide, the diarrhoic syndrome follows the consumption of food containing high doses of B. cereus spores. Once reached the gut, spores germinate and vegetative cells produce some enterotoxins that are responsible of the symptomatology (Ehling-Schulz et al., 2019). In addition, many other opportunistic infections such as endophthalmitis, endocarditis, bacteremia, as well as wound, respiratory, urinary, and central nervous system infections have been associated with this organism (Bottone, 2010; Celandroni et al., 2016; Ehling-Schulz et al., 2019). The ability of B. thuringiensis to cause food-poisoning diseases, periodontitis, wound infections, and bacteremia in humans has occasionally been reported (Celandroni et al., 2014). B. licheniformis can be responsible of serious diseases such as peritonitis, food poisoning, and bacteremia, particularly in immunocompromised patients (Park et al., 2006; Celandroni et al., 2016). B. badius and B. lentus are generally not considered human pathogens, but some strains of B. lentus have been shown to possess many B. cereus virulence factors (Beattie and Williams, 1999).
Lysinibacillus fusiformis was also detected in some products. This species can behave as opportunistic pathogen in humans, particularly immunocompromised patients, being responsible of a variety of infections (Wenzler et al., 2015). Isolation of the opportunistic nosocomial pathogen A. baumannii, potentially causing severe infections, was also reported (Antunes et al., 2014; Harding et al., 2018; Celandroni et al., 2019).
In conclusion, in our review of the published data we found a great number of inconsistencies in the compositional quality of many probiotic formulations available on the worldwide market. In general, no improvement in the quality of products over time was evidenced (Supplementary Table 1). Several discrepancies were also observed for probiotic drugs, although this category is subjected to different quality controls compared to dietary supplements and functional foods (de Simone, 2019). As regards quality of probiotic formulations, the ESPGHAN Working Group for Probiotics and Prebiotics already published a society paper stressing the need to improve quality controls for commercial formulations (Kolaček et al., 2017). More recently, another international expert panel emphasized the requirement of more transparency by manufacturers on the quality of probiotics (Jackson et al., 2019). We also believe manufacturers should be imposed to perform more accurate quality controls and adopt innovative methods for producing preparations that are microbiologically pure and qualitatively and quantitatively compliant with the label claims. Scientists who examine compliance of products should wait until the expiration date to carry out the analysis and at the storage temperature indicated on the label; if there is no mention of the need to refrigerate the product during storage, it should be tested after storage at room temperature. Furthermore, it should be stressed that the use of adequate methods for enumerating and identifying microbes contained in commercial formulations by the investigators is crucial to obtain an accurate and objective overview of the product quality. In addition, it should be underlined the urgent need for more specific and shared regulatory guidelines that govern the global market of these widely used products.
Author Contributions
DM and EG reviewed the literature and wrote the manuscript. MC, FC, and AL contributed to the analysis and discussion. All the authors revised the manuscript and approved its submission.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.693973/full#supplementary-material
References
Angelakis, E., Million, M., Henry, M., and Raoult, D. (2011). Rapid and accurate bacterial identification in probiotics and yoghurts by MALDI-TOF mass spectrometry. J. Food Sci. 76, M568–M572. doi: 10.1111/j.1750-3841.2011.02369.x
Antunes, L. C., Visca, P., and Towner, K. J. (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 71, 292–301. doi: 10.1111/2049-632X.12125
Arora, M., and Baldi, A. (2015). Regulatory categories of probiotics across the globe: a review representing existing and recommended categorization. Indian J. Med. Microbiol. 33, 2–10. doi: 10.4103/0255-0857.150868
Astashkina, A. P., Khudyakova, L. I., and Kolbysheva, Y. V. (2014). Microbiological quality control of probiotic products. Proc. Chem. 10, 74–79. doi: 10.1016/j.proche.2014.10.014
Aureli, P., Fiore, A., Scalfaro, C., Casale, M., and Franciosa, G. (2010). National survey outcomes on commercial probiotic food supplements in Italy. Int. J. Food Microbiol. 137, 265–273. doi: 10.1016/j.ijfoodmicro.2009.12.016
Beattie, S. H., and Williams, A. G. (1999). Detection of toxigenic strains of Bacillus cereus and other Bacillus spp. with an improved cytotoxicity assay. Lett. Appl. Microbiol. 28, 221–225. doi: 10.1046/j.1365-2672.1999.00498.x
Begum, A. A., Jakaria, D. M., Anisuzzaman, S. M., Islam, M., and Mahmud, S. A. (2015). Market assessment and product evaluation of probiotic containing dietary supplements available in bangladesh market. J. Pharm. 2015:763796. doi: 10.1155/2015/763796
Ben Braïek, O., and Smaoui, S. (2019). Enterococci: between emerging pathogens and potential probiotics. Biomed. Res. Int. 2019:5938210. doi: 10.1155/2019/5938210
Bermudez-Brito, M., Plaza-Díaz, J., Muñoz-Quezada, S., Gómez-Llorente, C., and Gil, A. (2012). Probiotic mechanisms of action. Ann. Nutr. Metab. 61, 160–174. doi: 10.1159/000342079
Blandino, G., Fazio, D., Petronio, G. P., Inturri, R., Tempera, G., and Furneri, P. M. (2016). Labeling quality and molecular characterization studies of products containing Lactobacillus spp. strains. Int. J. Immunopathol. Pharmacol. 29, 121–128. doi: 10.1177/0394632015600534
Bottone, E. J. (2010). Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 23, 382–398. doi: 10.1128/CMR.00073-09
Bourafa, N., Loucif, L., Boutefnouchet, N., and Rolain, J. M. (2015). Enterococcus hirae, an unusual pathogen in humans causing urinary tract infection in a patient with benign prostatic hyperplasia: first case report in Algeria. New Microbes New Infect. 8, 7–9. doi: 10.1016/j.nmni.2015.08.003
Brink, M., Senekal, M., and Dicks, L. M. (2005). Market and product assessment of probiotic/prebiotic-containing functional foods and supplements manufactured in South Africa. S. Afr. Med J. 95, 114–119.
Bron, P. A., Kleerebezem, M., Brummer, R. J., Cani, P. D., Mercenier, A., MacDonald, T. T., et al. (2017). Can probiotics modulate human disease by impacting intestinal barrier function? Br. J. Nutr. 117, 93–107. doi: 10.1017/S0007114516004037
Brown, A. C. (2017). An overview of herb and dietary supplement efficacy, safety and government regulations in the United States with suggested improvements. Part 1 of 5 series. Food Chem. Toxicol. 107, 449–471. doi: 10.1016/j.fct.2016.11.001
Canganella, F., Paganini, S., Ovidi, M., Vettraino, A. M., Bevilacqua, L., Massa, S., et al. (1997). A microbiology investigation on probiotic pharmaceutical products used for human health. Microbiol. Res. 152, 171–179. doi: 10.1016/s0944-5013(97)80009-2
Celandroni, F., Salvetti, S., Gueye, S. A., Mazzantini, D., Lupetti, A., Senesi, S., et al. (2016). Identification and pathogenic potential of clinical Bacillus and Paenibacillus isolates. PLoS One 11:e0152831. doi: 10.1371/journal.pone.0152831
Celandroni, F., Salvetti, S., Senesi, S., and Ghelardi, E. (2014). Bacillus thuringiensis membrane-damaging toxins acting on mammalian cells. FEMS Microbiol. Lett. 361, 95–103. doi: 10.1111/1574-6968.12615
Celandroni, F., Vecchione, A., Cara, A., Mazzantini, D., Lupetti, A., and Ghelardi, E. (2019). Identification of Bacillus species: implication on the quality of probiotic formulations. PLoS One 14:e0217021. doi: 10.1371/journal.pone.0217021
Champagne, C. P., Ross, R. P., Saarela, M., Hansen, K. F., and Charalampopoulos, D. (2011). Recommendations for the viability assessment of probiotics as concentrated cultures and in food matrices. Int. J. Food Microbiol. 149, 185–193. doi: 10.1016/j.ijfoodmicro.2011.07.005
Chang, C. J., Lin, T. L., Tsai, Y. L., Wu, T. R., Lai, W. F., Lu, C. C., et al. (2019). Next generation probiotics in disease amelioration. J. Food Drug Anal. 27, 615–622. doi: 10.1016/j.jfda.2018.12.011
Chen, T., Wu, Q., Li, S., Xiong, S., Jiang, S., Tan, Q., et al. (2014). Microbiological quality and characteristics of probiotic products in China. J. Sci. Food Agric. 94, 131–138. doi: 10.1002/jsfa.6221
Coeuret, V., Gueguen, M., and Vernoux, J. P. (2004). Numbers and strains of lactobacilli in some probiotic products. Int. J. Food Microbiol. 97, 147–156. doi: 10.1016/j.ijfoodmicro.2004.04.015
Cohen, P. A. (2018). Probiotic Safety-No Guarantees. JAMA Intern. Med. 178, 1577–1578. doi: 10.1001/jamainternmed.2018.5403
Council for Responsible Nutrition and International Probiotics Association (2017). Best Practices Guidelines for Probiotics. Available online at: https://www.crnusa.org/sites/default/files/pdfs/CRN-IPA-Best-Practices-Guidelines-for-Probiotics.pdf (accessed April 7, 2021).
Cutting, S. M. (2011). Bacillus probiotics. Food Microbiol. 28, 214–220. doi: 10.1016/j.fm.2010.03.007
Davis, C. (2014). Enumeration of probiotic strains: review of culture-dependent and alternative techniques to quantify viable bacteria. J. Microbiol. Methods 103, 9–17. doi: 10.1016/j.mimet.2014.04.012
de Almada, C. N., Almada, C. N., Martinez, R. C. R., and Sant’Ana, A. S. (2016). Paraprobiotics: evidences on their ability to modify biological responses, inactivation methods and perspectives on their application in foods. Trends Food Sci. Technol. 58, 96–114. doi: 10.1016/j.tifs.2016.09.011
de Simone, C. (2019). The unregulated probiotic market. Clin. Gastroenterol. Hepatol. 17, 809–817. doi: 10.1016/j.cgh.2018.01.018
De Vecchi, E., Nicola, L., Zanini, S., and Drago, L. (2008). In vitro screening of probiotic characteristics of some Italian products. J. Chemother. 20, 341–347. doi: 10.1179/joc.2008.20.3.341
Di Pierro, F., Polzonetti, V., Patrone, V., and Morelli, L. (2019). Microbiological assessment of the quality of some commercial products marketed as Lactobacillus crispatus-containing probiotic dietary supplements. Microorganisms 7:524. doi: 10.3390/microorganisms7110524
Dicpinigaitis, P. V., De Aguirre, M., and Divito, J. (2015). Enterococcus hirae bacteremia associated with acute pancreatitis and septic shock. Case Rep. Infect. Dis. 2015:123852. doi: 10.1155/2015/123852
Domínguez Díaz, L., Fernández-Ruiz, V., and Cámara, M. (2020). The frontier between nutrition and pharma: the international regulatory framework of functional foods, food supplements and nutraceuticals. Crit. Rev. Food Sci. Nutr. 60, 1738–1746. doi: 10.1080/10408398.2019.1592107
Drago, L., Rodighiero, V., Celeste, T., Rovetto, L., and De Vecchi, E. (2010). Microbiological evaluation of commercial probiotic products available in the USA in 2009. J. Chemother. 22, 373–377. doi: 10.1179/joc.2010.22.6.373
Drisko, J., Bischoff, B., Giles, C., Adelson, M., Rao, R. V., and McCallum, R. (2005). Evaluation of five probiotic products for label claims by DNA extraction and polymerase chain reaction analysis. Dig. Dis. Sci. 50, 1113–1117. doi: 10.1007/s10620-005-2714-6
Ebeling, C. G., and Romito, B. T. (2019). Aortic valve endocarditis from Enterococcus hirae infection. Proceedings 32, 249–250. doi: 10.1080/08998280.2018.1551698
EFSA (2007). Introduction of a Qualified Presumption of Safety (QPS) Approach for Assessment of Selected Microorganisms Referred to EFSA. Parma: EFSA.
EFSA (2018). Update of the List of QPS-Recommended Biological Agents Intentionally Added to Food or Feeds as Notified to EFSA 8: Suitability of Taxonomic Units Notified to EFSA Until March 2018. Parma: EFSA.
Ehling-Schulz, M., Lereclus, D., and Koehler, T. M. (2019). The Bacillus cereus group: Bacillus species with pathogenic potential. Microbiol. Spectr. 7:18. doi: 10.1128/microbiolspec.GPP3-0032-2018
Elliot, E., and Teversham, K. (2004). An evaluation of nine probiotics available in South Africa, August 2003. S. Afr. Med. J. 94, 121–124.
Elshaghabee, F. M. F., Rokana, N., Gulhane, R. D., Sharma, C., and Panwar, H. (2017). Bacillus as potential probiotics: status, concerns, and future perspectives. Front. Microbiol. 8:1490. doi: 10.3389/fmicb.2017.01490
FAO/WHO (2001). Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation. Rome: FAO.
Fasoli, S., Marzotto, M., Rizzotti, L., Rossi, F., Dellaglio, F., and Torriani, S. (2003). Bacterial composition of commercial probiotic products as evaluated by PCR-DGGE analysis. Int. J. Food Microbiol. 82, 59–70. doi: 10.1016/s0168-1605(02)00259-3
FDA (2019). Generally Recognized as Safe (GRAS). Available online at: https://www.fda.gov/food/food-ingredients-packaging/generally-recognized-safe-gras (accessed April 7, 2021).
Fenster, K., Freeburg, B., Hollard, C., Wong, C., Rønhave Laursen, R., and Ouwehand, A. C. (2019). The production and delivery of probiotics: a review of a practical approach. Microorganisms 7:83. doi: 10.3390/microorganisms7030083
Food Supplements Europe (2014). Good Manufacturing Practice for Manufacturers of Food Supplements. Available online at: https://foodsupplementseurope.org/wp-content/themes/fse-theme/documents/publications-and-guidelines/good-manufacturing-practice-for-manufacturers-of-food-supplements.pdf (accessed May 17, 2021).
Forssten, S. D., Sindelar, C. W., and Ouwehand, A. C. (2011). Probiotics from an industrial perspective. Anaerobe 17, 410–413. doi: 10.1016/j.anaerobe.2011.04.014
Fusco, V., Fanelli, F., and Chieffi, D. (2021). Authenticity of probiotic foods and dietary supplements: a pivotal issue to address. Crit. Rev. Food Sci. Nutr. 5, 1–18. doi: 10.1080/10408398.2021.1907300
Gao, W., Howden, B. P., and Stinear, T. P. (2018). Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr. Opin. Microbiol. 41, 76–82. doi: 10.1016/j.mib.2017.11.030
Ghattargi, V. C., Kalam, S. H., Pandit, S. S., Pawar, S. P., and Shouche, Y. S. (2018). Survey of probiotic preparations and labeling practices in Indian market. Indian J. Med. Microbiol. 36, 116–118. doi: 10.4103/ijmm.IJMM_18_63
Gilliland, S. E., and Speck, M. L. (1977). Enumeration and identity of Lactobacilli in dietary products. J. Food Prot. 40, 760–762. doi: 10.4315/0362-028X-40.11.760
Grumet, L., Tromp, Y., and Stiegelbauer, V. (2020). The development of high-quality multispecies probiotic formulations: from bench to market. Nutrients 12:2453. doi: 10.3390/nu12082453
Guarner, F., Khan, A. G., Garisch, J., Eliakim, R., Gangl, A., Thomson, A., et al. (2012). World Gastroenterology Organization. World Gastroenterology Organisation Global Guidelines: probiotics and prebiotics October 2011. J. Clin. Gastroenterol. 46, 468–481. doi: 10.1097/MCG.0b013e3182549092
Gul, K., Singh, A. K., and Jabeen, R. (2016). Nutraceuticals and functional foods: the foods for the future world. Crit. Rev. Food Sci. Nutr. 56, 2617–2627. doi: 10.1080/10408398.2014.903384
Halsted, C. H. (2003). Dietary supplements and functional foods: 2 sides of a coin? Am. J. Clin. Nutr. 77, 1001S–1007S. doi: 10.1093/ajcn/77.4.1001S
Hamilton-Miller, J. M., and Gibson, G. R. (1999). Efficacy studies of probiotics: a call for guidelines. Br. J. Nutr. 82, 73–75. doi: 10.1017/s0007114599001142
Hamilton-Miller, J. M., Shah, S., and Smith, C. T. (1996). “Probiotic” remedies are not what they seem. BMJ 312, 55–56. doi: 10.1136/bmj.312.7022.55c
Hamilton-Miller, J. M., Shah, S., and Winkler, J. T. (1999). Public health issues arising from microbiological and labelling quality of foods and supplements containing probiotic microorganisms. Public Health Nutr. 2, 223–229. doi: 10.1017/s1368980099000282
Hanchi, H., Mottawea, W., Sebei, K., and Hammami, R. (2018). The genus Enterococcus: between probiotic potential and safety concerns-an update. Front. Microbiol. 9:1791. doi: 10.3389/fmicb.2018.01791
Harding, C. M., Hennon, S. W., and Feldman, M. F. (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 16, 91–102. doi: 10.1038/nrmicro.2017.148
Hayek, S. A., Gyawali, R., Aljaloud, S. O., Krastanov, A., and Ibrahim, S. A. (2019). Cultivation media for lactic acid bacteria used in dairy products. J. Dairy Res. 86, 490–502. doi: 10.1017/S002202991900075X
Hill, C., Guarner, F., Reid, G., Gibson, G. R., and Merenstein, D. J. (2014). Expert consensus document. The international scientific association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Hoa, N. T., Baccigalupi, L., Huxham, A., Smertenko, A., Van, P. H., Ammendola, S., et al. (2000). Characterization of Bacillus species used for oral bacteriotherapy and bacterioprophylaxis of gastrointestinal disorders. Appl. Environ. Microbiol. 66, 5241–5247. doi: 10.1128/aem.66.12.5241-5247.2000
Hu, S., Wang, L., and Jiang, Z. (2017). Dietary additive probiotics modulation of the intestinal microbiota. Protein Pept. Lett. 24, 382–387. doi: 10.2174/0929866524666170223143615
Huys, G., Vancanneyt, M., D’Haene, K., Vankerckhoven, V., Goossens, H., and Swings, J. (2006). Accuracy of species identity of commercial bacterial cultures intended for probiotic or nutritional use. Res. Microbiol. 157, 803–810. doi: 10.1016/j.resmic.2006.06.006
Ibrahim, S. A., and Carr, J. P. (2006). Viability of bifidobacteria in commercial yogurt products in North Carolina. Int. J. Dairy Tech. 59, 272–277. doi: 10.1111/j.1471-0307.2006.00282.x
International Standards Organisation [ISO] (2006). Milk Products – Enumeration of Presumptive Lactobacillus acidophilus on a Selective Medium – Colony- Count Technique at 37 Degrees C. ISO 20128:2006 (IDF 192:2006). Geneva: International Standards Organisation.
International Standards Organisation [ISO] (2010). Milk Products – Enumeration of Presumptive Bifidobacteria – Colony Count Technique at 37 Degrees C. ISO 29981:2010 (IDF 220:2010). Geneva: International Standards Organisation.
Jackson, S. A., Schoeni, J. L., Vegge, C., Pane, M., Stahl, B., Bradley, M., et al. (2019). Improving end-user trust in the quality of commercial probiotic products. Front. Microbiol. 10:739. doi: 10.3389/fmicb.2019.00739
Jeżewska-Fra̧ckowiak, J., Seroczyńska, K., Banaszczyk, J., Jedrzejczak, G., Żylicz-Stachula, A., and Skowron, P. M. (2018). The promises and risks of probiotic Bacillus species. Acta Biochim. Pol. 65, 509–519. doi: 10.18388/abp.2018_2652
Kesavelu, D. Sr., Rohit, A., Karunasagar, I., and Karunasagar, I. (2020). Composition and laboratory correlation of commercial probiotics in India. Cureus 12:e11334. doi: 10.7759/cureus.11334
Koirala, S., and Anal, A. K. (2021). Probiotics-based foods and beverages as future foods and their overall safety and regulatory claims. Future Foods 3:100013. doi: 10.1016/j.fufo.2021.100013
Kolaček, S., Hojsak, I., Berni Canani, R., Guarino, A., Indrio, F., Orel, R., et al. (2017). ESPGHAN Working Group for probiotics and prebiotics. commercial probiotic products: a call for improved quality control. a position paper by the ESPGHAN Working Group for probiotics and prebiotics. J. Pediatr. Gastroenterol. Nutr. 65, 117–124. doi: 10.1097/MPG.0000000000001603
Korona-Glowniak, I., Siwiec, R., Luszczewska-Sierokowska, I., Maciejewski, R., Wrabel, R., and Malm, A. (2019). Microbiological evaluation of 10 commercial probiotic products available in Poland. Curr. Issue Pharm. Med. Sci. 32, 121–124. doi: 10.2478/cipms-2019-0022
Kumar, H., Salminen, S., Verhagen, H., Rowland, I., Heimbach, J., Bañares, S., et al. (2015). Novel probiotics and prebiotics: road to the market. Curr. Opin. Biotechnol. 32, 99–103. doi: 10.1016/j.copbio.2014.11.021
LeBlanc, J. G., Chain, F., Martín, R., Bermúdez-Humarán, L. G., Courau, S., and Langella, P. (2017). Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Fact. 16:79. doi: 10.1186/s12934-017-0691-z
Lee, N. K., Kim, W. S., and Paik, H. D. (2019). Bacillus strains as human probiotics: characterization, safety, microbiome, and probiotic carrier. Food Sci. Biotechnol. 28, 1297–1305. doi: 10.1007/s10068-019-00691-9
Lewis, Z. T., Shani, G., Masarweh, C. F., Popovic, M., Frese, S. A., Sela, D. A., et al. (2016). Validating bifidobacterial species and subspecies identity in commercial probiotic products. Pediatrics 79, 445–452. doi: 10.1038/pr.2015.244
Lin, W. H., Hwang, C. F., Chen, L. W., and Tsen, H. Y. (2006). Viable counts, characteristic evaluation for commercial lactic acid bacteria products. Food Microbiol. 23, 74–81. doi: 10.1016/j.fm.2005.01.013
Madsen, K. T., Skov, M. N., Gill, S., and Kemp, M. (2017). Virulence factors associated with Enterococcus faecalis infective endocarditis: a mini review. Open Microbiol. J. 11, 1–11. doi: 10.2174/1874285801711010001
Maldonado Galdeano, C., Cazorla, S. I., Lemme Dumit, J. M., Vélez, E., and Perdigón, G. (2019). Beneficial effects of probiotic consumption on the immune system. Ann. Nutr. Metab. 74, 115–124. doi: 10.1159/000496426
Marcobal, A., Underwood, M. A., and Mills, D. A. (2008). Rapid determination of the bacterial composition of commercial probiotic products by terminal restriction fragment length polymorphism analysis. J. Pediatr. Gastroenterol. Nutr. 46, 608–611. doi: 10.1097/MPG.0b013e3181660694
Margolles, A., and Ruiz, L. (2021). Methods for isolation and recovery of bifidobacteria. Methods Mol. Biol. 2278, 1–12. doi: 10.1007/978-1-0716-1274-3_1
Masco, L., Huys, G., De Brandt, E., Temmerman, R., and Swings, J. (2005). Culture-dependent and culture-independent qualitative analysis of probiotic products claimed to contain bifidobacteria. Int. J. Food Microbiol. 102, 221–230. doi: 10.1016/j.ijfoodmicro.2004.11.018
Mattia, A., and Merker, R. (2008). Regulation of probiotic substances as ingredients in foods: premarket approval or “generally recognized as safe” notification. Clin. Infect. Dis. 46(Suppl. 2), S115–S118. doi: 10.1086/523329
Ministero della Salute (2018). Direzione generale per l’igiene e la sicurezza degli alimenti e la nutrizione - ufficio 4. Guidelines on Probiotics and Prebiotics. Available online at: http://www.salute.gov.it/imgs/C_17_pubblicazioni_1016_allegato.pdf (accessed March 18, 2019).
Mora, D., Filardi, R., Arioli, S., Boeren, S., Aalvink, S., and de Vos, W. M. (2019). Development of omics-based protocols for the microbiological characterization of multi-strain formulations marketed as probiotics: the case of VSL#3. Microb. Biotechnol. 12, 1371–1386. doi: 10.1111/1751-7915.13476
Morovic, W., Hibberd, A. A., Zabel, B., Barrangou, R., and Stahl, B. (2016). Genotyping by PCR and high-throughput sequencing of commercial probiotic products reveals composition biases. Front. Microbiol. 7:1747. doi: 10.3389/fmicb.2016.01747
Oak, S. J., and Jha, R. (2019). The effects of probiotics in lactose intolerance: a systematic review. Crit. Rev. Food Sci. Nutr. 59, 1675–1683. doi: 10.1080/10408398.2018.1425977
Ouwehand, A. C. (2017). A review of dose-responses of probiotics in human studies. Benef. Microbes 8, 143–151. doi: 10.3920/BM2016.0140
Park, D. J., Yun, J. C., Baek, J. E., Jung, E. Y., Lee, D. W., Kim, M. A., et al. (2006). Relapsing Bacillus licheniformis peritonitis in a continuous ambulatory peritoneal dialysis patient. Nephrology 11, 21–22. doi: 10.1111/j.1440-1797.2006.00539.x
Patro, J. N., Ramachandran, P., Barnaba, T., Mammel, M. K., Lewis, J. L., and Elkins, C. A. (2016). Culture-independent metagenomic surveillance of commercially available probiotics with high-throughput next-generation sequencing. mSphere 1:e00057-16. doi: 10.1128/mSphere.00057-16
Patrone, V., Molinari, P., and Morelli, L. (2016). Microbiological and molecular characterization of commercially available probiotics containing Bacillus clausii from India and Pakistan. Int. J. Food Microbiol. 237, 92–97. doi: 10.1016/j.ijfoodmicro.2016.08.012
Perea Vélez, M., Hermans, K., Verhoeven, T. L., Lebeer, S. E., Vanderleyden, J., and De Keersmaecker, S. C. (2007). Identification and characterization of starter lactic acid bacteria and probiotics from Columbian dairy products. J. Appl. Microbiol. 103, 666–674. doi: 10.1111/j.1365-2672.2007.03294.x
Pinkes, M. E., White, C., and Wong, C. S. (2019). Native-valve Enterococcus hirae endocarditis: a case report and review of the literature. BMC Infect. Dis. 19:891. doi: 10.1186/s12879-019-4532-z
Roy, D. (2001). Media for the isolation and enumeration of bifidobacteria in dairy products. Int. J. Food Microbiol. 69, 167–182. doi: 10.1016/s0168-1605(01)00496-2
Schillinger, U. (1999). Isolation and identification of lactobacilli from novel-type probiotic and mild yoghurts and their stability during refrigerated storage. Int. J. Food Microbiol. 47, 79–87. doi: 10.1016/s0168-1605(99)00014-8
Schoeni, J. L. (2015). “Probiotics,” in Compendium of Methods for the Microbiological Examination of Foods, 5th Edn, eds Y. Salfinger and M. L. Tortorello (Washington, DC: American Public Health Association), 232–286.
Sen, S., and Mansell, T. J. (2020). Yeasts as probiotics: mechanisms, outcomes, and future potential. Fungal Genet. Biol. 137:103333. doi: 10.1016/j.fgb.2020.103333
Sreeja, V., and Prajapati, J. B. (2013). Probiotic formulations: application and status as pharmaceuticals-a review. Probiotics Antimicrob. Proteins. 5, 81–91. doi: 10.1007/s12602-013-9126-2
Stavropoulou, E., and Bezirtzoglou, E. (2020). Probiotics in medicine: a long debate. Front. Immunol. 11:2192. doi: 10.3389/fimmu.2020.02192
Szajewska, H., Fordymacka, A., Bardowski, J., Górecki, R. K., Mrukowicz, J. Z., and Banaszkiewicz, A. (2004). Microbiological and genetic analysis of probiotic products licensed for medicinal purposes. Med. Sci. Monit. 10, BR346–BR350.
Taverniti, V., and Guglielmetti, S. (2011). The immunomodulatory properties of probiotic microorganisms beyond their viability (ghost probiotics: proposal of paraprobiotic concept). Genes Nutr. 6, 261–274. doi: 10.1007/s12263-011-0218-x
Taylor, C. L. (2004). Regulatory frameworks for functional foods and dietary supplements. Nutr. Rev. 62, 55–59. doi: 10.1111/j.1753-4887.2004.tb00024.x
Temmerman, R., Pot, B., Huys, G., and Swings, J. (2003a). Identification and antibiotic susceptibility of bacterial isolates from probiotic products. Int. J. Food Microbiol. 81, 1–10. doi: 10.1016/s0168-1605(02)00162-9
Temmerman, R., Scheirlinck, I., Huys, G., and Swings, J. (2003b). Culture-independent analysis of probiotic products by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69, 220–226. doi: 10.1128/aem.69.1.220-226.2003
Theunissen, J., Britz, T. J., Torriani, S., and Witthuhn, R. C. (2005). Identification of probiotic microorganisms in South African products using PCR-based DGGE analysis. Int. J. Food Microbiol. 98, 11–21. doi: 10.1016/j.ijfoodmicro.2004.05.004
Toscano, M., de Vecchi, E., Rodighiero, V., and Drago, L. (2013). Microbiological and genetic identification of some probiotics proposed for medical use in 2011. J. Chemother. 25, 156–161. doi: 10.1179/1973947812Y.0000000068
Ullah, M., Raza, A., Ye, L., and Yu, Z. (2019). Viability and composition validation of commercial probiotic products by selective culturing combined with next-generation sequencing. Microorganisms 7:188. doi: 10.3390/microorganisms7070188
Vallabhaneni, S., Walker, T. A., Lockhart, S. R., Ng, D., Chiller, T., Melchreit, R., et al. (2015). Notes from the field: fatal gastrointestinal mucormycosis in a premature infant associated with a contaminated dietary supplement–Connecticut, 2014. MMWR Morb. Mortal. Wkly. Rep. 64, 155–156.
Vanhee, L. M., Goemé, F., Nelis, H. J., and Coenye, T. (2010). Quality control of fifteen probiotic products containing Saccharomyces boulardii. J. Appl. Microbiol. 109, 1745–1752. doi: 10.1111/j.1365-2672.2010.04805.x
Vecchione, A., Celandroni, F., Mazzantini, D., Senesi, S., Lupetti, A., and Ghelardi, E. (2018). Compositional quality and potential gastrointestinal behavior of probiotic products commercialized in Italy. Front. Med. 5:59. doi: 10.3389/fmed.2018.00059
Venugopalan, V., Shriner, K. A., and Wong-Beringer, A. (2010). Regulatory oversight and safety of probiotic use. Emerg. Infect. Dis. 16, 1661–1665. doi: 10.3201/eid1611.100574
Vermeulen, M. J., Luijendijk, A., van Toledo, L., van Kaam, A. H., and Reiss, I. K. M. (2020). Quality of probiotic products for preterm infants: contamination and missing strains. Acta Paediatr. 109, 276–279. doi: 10.1111/apa.14976
Vitetta, L., Coulson, S., Thomsen, M., Nguyen, T., and Hall, S. (2017). Probiotics, D-Lactic acidosis, oxidative stress and strain specificity. Gut Microbes 8, 311–322. doi: 10.1080/19490976.2017.1279379
Weese, J. S. (2002). Microbiologic evaluation of commercial probiotics. J. Am. Vet. Med. Assoc. 220, 794–797. doi: 10.2460/javma.2002.220.794
Weese, J. S. (2003). Evaluation of deficiencies in labeling of commercial probiotics. Can. Vet. J. 44, 982–983.
Wenzler, E., Kamboj, K., and Balada-Llasat, J. M. (2015). Severe sepsis secondary to persistent Lysinibacillus sphaericus, Lysinibacillus fusiformis and Paenibacillus amylolyticus Bacteremia. Int. J. Infect. Dis. 35, 93–95. doi: 10.1016/j.ijid.2015.04.016
Wilkins, T., and Sequoia, J. (2017). Probiotics for gastrointestinal conditions: a summary of the evidence. Am. Fam. Phys. 96, 170–178.
Winther, M., Dalager-Pedersen, M., Tarpgaard, I. H., and Nielsen, H. L. (2020). First Danish case of infective endocarditis caused by Enterococcus hirae. BMJ Case Rep. 13:e237950. doi: 10.1136/bcr-2020-237950
World Gastroenterology Organisation [WGO] (2017). Probiotics and Prebiotics. Available online at: http://medi-guide.meditool.cn/ymtpdf/24B36233-2CB9-27AE-C86B-A55F6003C9AE.pdf (Accessed April 7, 2021).
Yaeshima, T., Takahashi, S., Ishibashi, N., and Shimamura, S. (1996). Identification of bifidobacteria from dairy products and evaluation of a microplate hybridization method. Int. J. Food Microbiol. 30, 303–313. doi: 10.1016/0168-1605(96)00956-7
Yonkova Marinova, V., Kirilova Rasheva, I., Krasimirova Kizheva, Y., Dimitrova Dermenzhieva, Y., and Koitcheva Hristova, P. (2019). Microbiological quality of probiotic dietary supplements. Biotechnol. Biotechnol. Equip. 1, 834–841. doi: 10.1080/13102818.2019.1621208
Zawistowska-Rojek, A., Zareba, T., Mrówka, A., and Tyski, S. (2016). Assessment of the microbiological status of probiotic products. Pol. J. Microbiol. 65, 97–104. doi: 10.5604/17331331.1197281
Zhai, Q., Feng, S., Arjan, N., and Chen, W. (2019). A next generation probiotic, Akkermansia muciniphila. Crit. Rev. Food Sci. Nutr. 59, 3227–3236. doi: 10.1080/10408398.2018.1517725
Zheng, J., Wittouck, S., Salvetti, E., Franz, C. M. A. P., Harris, H. M. B., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Keywords: probiotics, compositional quality, viable cells, microbial species, purity, label claims
Citation: Mazzantini D, Calvigioni M, Celandroni F, Lupetti A and Ghelardi E (2021) Spotlight on the Compositional Quality of Probiotic Formulations Marketed Worldwide. Front. Microbiol. 12:693973. doi: 10.3389/fmicb.2021.693973
Received: 12 April 2021; Accepted: 29 June 2021;
Published: 20 July 2021.
Edited by:
Francesca Fanelli, Institute of Sciences of Food Production, Italian National Research Council, ItalyReviewed by:
Arthur C. Ouwehand, Danisco, FinlandClaude P. Champagne, Saint-Hyacinthe Research and Development Center, Agriculture and Agri-Food Canada, Canada
Copyright © 2021 Mazzantini, Calvigioni, Celandroni, Lupetti and Ghelardi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emilia Ghelardi, ZW1pbGlhLmdoZWxhcmRpQG1lZC51bmlwaS5pdA==
 Diletta Mazzantini
Diletta Mazzantini Marco Calvigioni
Marco Calvigioni Francesco Celandroni1
Francesco Celandroni1 Antonella Lupetti
Antonella Lupetti Emilia Ghelardi
Emilia Ghelardi