- 1College of Agronomy and Biotechnology, Southwest University, Chongqing, China
- 2Engineering Research Center of South Upland Agriculture, Ministry of Education, Chongqing, China
Sclerotinia sclerotiorum is a devastating necrotrophic fungal pathogen and has a substantial economic impact on crop production worldwide. Magnaporthe appressoria-specific (MAS) proteins have been suggested to be involved in the appressorium formation in Magnaporthe oryzae. Sscnd1, an MAS homolog gene, is highly induced at the early infection stage of S. sclerotiorum. Knock-down the expression of Sscnd1 gene severely reduced the virulence of S. sclerotiorum on intact rapeseed leaves, and their virulence was partially restored on wounded leaves. The Sscnd1 gene-silenced strains exhibited a defect in compound appressorium formation and cell integrity. The instantaneous silencing of Sscnd1 by tobacco rattle virus (TRV)-mediated host-induced gene silencing (HIGS) resulted in a significant reduction in disease development in tobacco. Three transgenic HIGS Arabidopsis lines displayed high levels of resistance to S. sclerotiorum and decreased Sscnd1 expression. Production of specific Sscnd1 siRNA in transgenic HIGS Arabidopsis lines was confirmed by stem-loop qRT-PCR. This study revealed that the compound appressorium-related gene Sscnd1 is required for cell integrity and full virulence in S. sclerotiorum and that Sclerotinia stem rot can be controlled by expressing the silencing constructs of Sscnd1 in host plants.
Introduction
Sclerotinia sclerotiorum (Lib.) de Bary is a well-known necrotrophic phytopathogenic fungus with a broad host range, including many economically important crops, such as oilseed rape (Brassica napus), sunflowers, soybeans, peanuts and lentils (Boland and Hall, 1994; Bolton et al., 2006). Sclerotinia stem rot caused by S. sclerotiorum often causes significant losses in crop production.
As a necrotrophic parasite, S. sclerotiorum has evolved a sophisticated infection process to effectively infect hosts (Kabbage et al., 2015). To adhere to the host surface, the tips of its hyphae become swollen and extensive branch prior to penetration and then develop a multicellular, melanin-rich hyphal penetration structure, called compound appressorium (Jamaux et al., 2007; Huang et al., 2008; Uloth et al., 2016). This formation of compound appressorium is essential for the process in which fungi penetrate the host cuticle and form infectious hyphae to spread horizontally beneath the host cuticle (Liang and Rollins, 2018).
Multiple S. sclerotiorum genes are required for the formation and development of compound appressorium. The disruption of GATA-type transcription factors (SsAMS2 and SsNSD1) impairs compound appressorium formation and virulence (Li et al., 2017; Liu et al., 2018). The secretory proteins SsCaf1 and SsRhs1 are highly induced in the initial infection stage, and their gene-silenced strains exhibit defects compound appressorium formation (Xiao et al., 2014; Yu et al., 2017). Additionally, oxalic acid (OA) accumulation, the cAMP-PKA signaling pathway and other genes, such as the genes γ-glutamyl transpeptidase (Ssggt1) and survival factor 1 (Sssvf1), have also been shown to be associated with the development of compound appressorium (Jurick and Rollins, 2007; Li et al., 2012; Liang et al., 2015a,b; Yu et al., 2019). Similar to S. sclerotiorum, Magnaporthe oryzae also produces appressoria to penetrate host plant cells and initiate infection (Dean, 1997; Hamer and Talbot, 1998; Zhang et al., 2016a). GAS1 and GAS2, encoding the Magnaporthe appressoria-specific (MAS) proteins, function in the appressorium formation and fungal virulence in M. oryzae (Xu and Hamer, 1996; Xue et al., 2002). BcGAS2, a homolog gene of the M. oryzae GAS2, is required for appressorial function but is not essential for the growth and infection of Botrytis cinerea (Schamber et al., 2010). Although two MAS homologs are overexpressed in the infection cushion of S. sclerotiorum (Sexton et al., 2006), there is no experimental evidence of their functions.
RNA interference (RNAi) is a universal gene regulation mechanism in eukaryotes that involves exogenous double-stranded RNA (dsRNA) (Baulcombe, 2005). RNA-silencing technology has been exploited extensively to knock down fungal genes to improve resistance in plants (Duan et al., 2012). Interestingly, some insect pests and nematodes can be successfully controlled by merely feeding dsRNAs (RNAi constructs) of their genes (Huang et al., 2006; Baum et al., 2007; Huvenne and Smagghe, 2010). Recently, this strategy, named host-induced gene silencing (HIGS), has been applied to reduce pathogen aggressiveness, such as Puccinia (Panwar et al., 2013; Yin et al., 2015; Zhu et al., 2017; Qi et al., 2018) and Fusarium (Ghag et al., 2014; Cheng et al., 2015; Chen et al., 2016) in wheat, Rhizoctonia solani in tall fescue (Zhou et al., 2016), Phytophthora infestans in potato (Jahan et al., 2015; Sanju et al., 2015), Bremia lactucae in lettuce (Govindarajulu et al., 2015) and Verticillium in Arabidopsis, tomato and cotton (Zhang et al., 2016b; Song and Thomma, 2018; Xu et al., 2018).
Thus, the blockage of compound appressorium differentiation by interfering with the S. sclerotiorum MAS genes may be an efficient strategy for decreasing the disease phenotype. In this study, Sscnd1 encoding a MAS homolog, was characterized in S. sclerotiorum. The function of Sscnd1 in the compound appressorium formation and pathogenicity of S. sclerotiorum was determined. We further explored the potential of improving plant resistance via target silencing of Sscnd1 by gene silencing. Our data suggest that Sscnd1 is required for compound appressorium formation, cell integrity and full virulence in S. sclerotiorum.
Materials and Methods
Fungal Strains, Plants, and Culture Conditions
The S. sclerotiorum isolate 1980 (Godoy et al., 1990) was used as the wild-type strain and cultured on potato dextrose agar (PDA) (Difco Laboratories, Detroit). Sscnd1 gene-silenced strains were cultured on PDA supplemented with hygromycin B at 100 μg/mL (Calbiochem, San Diego, CA). The wild-type Arabidopsis thaliana Col-0 (Columbia zero background ecotype) and its transgenic lines were grown in a controlled environment chamber with 16-h/23°C days and 8-h/16°C nights at 100 μmol/m2/s light intensity.
Bioinformatic Analysis of Sscnd1
The sequences of Sscnd1 were obtained from the genomic sequence database of S. sclerotiorum genome (http://fungidb.org/common/downloads/Current_Release/Ssclerotiorum_/). BlastP analysis was performed on the website of NCBI (http://www.ncbi.nlm.nih.gov/). The signal peptide sequence and transmembrane domain were predicted using SignalP 5.0 Server (http://www.cbs.dtu.dk/services/SignalP/), TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and TMpred (http://www.ch.embnet.org/software/TMPRED_form.html). Multiple sequence alignment was implemented with DNAMAN6.0 (Lynnon BioSoft, Quebec, Canada) and CLUSTALX2.0 (Chenna et al., 2003). The phylogenetic tree was constructed with MEGA 6.0 software (Tamura et al., 2013) using the maximum likelihood method, and the bootstrap test was replicated 1,000 times.
RNA Extraction, cDNA Synthesis and qRT-PCR
To evaluate the Sscnd1 expression levels during hyphal development, the wild-type strain was cultured on cellophane over PDA, and mycelia were harvested at 1 and 2 days post-inoculation (dpi) (hyphae), 3 and 4 dpi (initial sclerotia), 5, 6, and 7 dpi (developing sclerotia), and 8 dpi (mature sclerotia). To examine the Sscnd1 expression levels during infection stages, the wild-type strain was cultured in potato dextrose broth (PDB) for 2 days and the mycelia were harvested and ground into fragments. The hyphal fragments were suspended in ddH2O and then sprayed on the leaves of rapeseed, as well as on the cellophane placed on PDA plates as controls. The inoculated leaves and hyphae growing on PDA plates were harvested at 0, 3, 6, 9, 12, 24, and 48 h post inoculation (hpi). Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA), and first-strand cDNA was synthesized for quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR). qRT-PCR was performed using Bio-Rad CFX96 Real-Time System (America) and Quantitect SYBR Green PCR master mix (Bio-Rad, USA) according to the manufacturer's instructions. The β-tubulin gene Sstub1 (SS1G_04652) was used as the internal reference for normalization. The transcript level of the gene of interest was calculated from the threshold cycle using the 2−ΔΔCT method (Livak and Schmittgen, 2001) with three replicates, and the data were analyzed using CFX Manager™ v3.0. The primers were listed in Supplementary Table 1.
Binary Constructs
The Sscnd1 gene-silencing vector was constructed based on the plasmid pCIT (Yu et al., 2017). The sense and antisense fragments of Sscnd1 with a length of 418 bp were cloned into the corresponding clone sites of pCIT, and the hygromycin resistance gene cassette from pSKH (Hamid et al., 2013) was subsequently inserted into it. The resulting Sscnd1-RNAi construct pSicnd1 was transformed into the S. sclerotiorum wild-type strain 1980 according to the method of Rollins (2003). Meanwhile, the plasmid pCIT containing a hygromycin resistance gene cassette was used as the empty vector pRNAi. The strain containing the empty vector pRNAi was used as the control in whole experiments.
The sense and antisense fragments of Sscnd1 were cloned and flanked with the malate synthase gene intron 3 from A. thaliana (i3). The cassette was cloned into the plasmid pBinGlyRed3, which contained a red fluorescent protein (DS Red). The resulting HIGS construct HIGS-Sscnd1 was introduced into the Agrobacterium tumefaciens strain GV3101 by electroporation (Wise et al., 2006) and then transformed into A. thaliana Col-0 using the floral dip method (Clough and Bent, 1998).
Pathogenicity Assays
The pathogenicity of the S. sclerotiorum wild-type, empty vector, and Sscnd1 gene-silenced strains was evaluated in the unwounded and wounded (wounded with a dissecting needle) leaves of B. napus (Zhongshuang 11). The 0.6-cm mycelium-colonized agar plugs obtained from actively growing colony edges were used to inoculate onto the leaves. The inoculated leaves were kept in 90% relative humidity at 20°C. The lesions were measured at 48 hpi. Each strain was evaluated with three leaves in one replicate and the experiments were performed five times.
The 4-week-old HIGS-Sscnd1 transgenic A. thaliana lines were inoculated with 0.2-cm mycelium-colonized agar plugs of the S. sclerotiorum wild-type strain 1980 from actively growing colony edges. Lesion area was measured at 24 hpi for in vitro inoculation and 4 dpi for in vivo inoculation. The experiments were performed at least five times with five leaves or plants for every line in one replicate.
To evaluate the resistance of HIGS-Sscnd1 transgenic A. thaliana lines to B. cinerea, the 0.2-cm mycelium-colonized agar plugs of B. cinerea strain B05.10 from actively growing colony edges were used to inoculate the detached leaves of HIGS-Sscnd1 transgenic A. thaliana lines. Lesion area was measured at 24 hpi. The experiments were performed at least five times with five leaves for every strain in one replicate.
The lesion area (S, cm2) was calculated with the formula S = π*a*b/4, where a and b represent the long and short diameter of an approximately elliptical lesion.
Detection of siRNA in HIGS-Sscnd1 Transgenic A. thaliana Lines
To detect Sscnd1 siRNA production in HIGS-Sscnd1 transgenic A. thaliana lines, stem-loop qRT-PCR was performed against the Sscnd1 gene of S. sclerotiorum according to Mahto et al. (2020) with some modifications. For this purpose, a putative siRNA sequence (UAACUUGAGGAAGAGUUUCAC) was identified within the 418 bp Sscnd1 sequence employed for the construction of RNAi vector via siDirect version 2.0 (http://sidirect2.rnai.jp/), which is functionally appropriate for knocking down the Sscnd1 gene expression. Low molecular weight RNA was isolated and then utilized to synthesize cDNA using stem-loop primer (ST-Sscnd1). Subsequently, stem-loop qRT-PCR was performed using siRNA specific primers. The primers were listed in Supplementary Table 1. qRT-PCR was performed using Bio-Rad CFX96 Real-Time System (America) and Quantitect SYBR Green PCR master mix (Bio-Rad, USA) according to the manufacturer's instructions. The A. thaliana U6 gene AtU6-26 was used as the internal reference for normalization. The transcript level of the gene of interest was calculated from the threshold cycle using the 2−ΔΔCT method (Livak and Schmittgen, 2001) with three replicates, and the data were analyzed using CFX Manager™ v3.0.
High Osmotic Stress Assay
To calculate the inhibition rate of hyphal growth when cultured with high osmotic stress and membrane damage stress, the 0.6-cm mycelium-colonized agar plugs of Sscnd1 gene-silenced strains, wild-type and empty vector strain obtained from actively growing colony edges were inoculated on the center of PDA plates supplemented with 2% sorbose, 5% sorbose, 1 M sorbitol, 1.2 M sucrose and 0.02% sodium dodecylsulphate (SDS), respectively. Colony radius was measured every 12 h before the colony reached margins of the plates. The colony phenotype photographs were taken at 3 dpi. Each experiment was repeated three times with five plates for every treatment in one replicate.
Compound Appressorium Assay
Compound appressorium formation of Sscnd1 gene-silenced strains was observed according to Yu et al. (2019). The 0.6-cm mycelial plugs were inoculated onto parafilm-overlaid PDA plates and rapeseed leaves. The plugs were removed at 8 hpi. the parafilm surface was stained with 5% trypan blue. The compound appressorium on parafilm was observed using a microscope. The inoculated rapeseed leaves were stained with 5% trypan for 12 h and then cleared with ethanol/acetic acid (3:1 v/v) solution for 12 h. The compound appressoria on rapeseed leaves were observed using an electron microscope (JEOL JEM-6390LV). The experiment was repeated for three independent times.
TRV-Based Sscnd1 Gene Silencing in N. benthamiana
To determine the role of Sscnd1 during infection, a TRV-based gene-silencing system was applied in N. benthamiana. A 218-bp fragment of Sscnd1 named VIGS-Sscnd1-1 and a 296-bp fragment of Sscnd1 named VIGS-Sscnd1-2 were amplified with primers Sscnd1-VIGS-1F/R and Sscnd1-VIGS-2F/R, respectively. The amplicons were inserted into TRV2 vector (Liu and Page, 2008) to produce the VIGS constructs TRV2:: Sscnd1-1 and TRV2:: Sscnd1-2. The recombinant virus TRV2:: GFP was applied as a control. The infiltration of N. benthamiana plants was performed according to Liu and Page (2008). The upper leaves from the infiltrated N. benthamiana leaf were inoculated with S. sclerotiorum wild-type strain 1980 seven days after infiltration. The experiment was repeated for five independent times and every construct was infiltrated with at least five plants in one replicate. Lesion phenotypes were recorded at 48 hpi.
RNA Sequencing and Data Analysis
The mycelia of wild-type stain 1980 and Sscnd1 gene-silenced strains (Sicnd1-9 and Sicnd1-20) on the PDA plate were collected at 48 hpi. The sequencing library of three samples with two replicates was generated using the Illumina RNA Library Prep Kit (NEB, USA) following the manufacturer's recommendation, and sequenced on an Illumina Hiseq 2000 platform that yields 100-bp paired-end reads. The raw reads were filtered to obtain high-quality clean reads by removing adaptor sequences, duplicated sequences, reads containing more than 5% “N” (i.e., ambiguous bases in reads), and reads in which more than 50% of the bases showed a Q-value (i.e., Bonferroni-adjusted P-value) ≤ 5. Clean reads were aligned to the reference genome of S. sclerotiorum (http://fungidb.org/common/downloads/Current_Release/Ssclerotiorum_/) by using the TopHat2 (http://ccb.jhu.edu/software/tophat/index.shtml) with default parameters except that the Q value was set to 100. Gene expression was quantified using Salmon (https://combine-lab.github.io/salmon/). The raw counts were normalized by TPM (Transcripts Per Million reads) and the differential expression analysis was performed using the DESeq2 (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html). The threshold determining the significance of differentially expressed genes (DEGs) among multiple tests was set at a p-adjust < 0.05 and |log2 ratio| ≥ 1 (Mao et al., 2018). GO enrichment analyses were analyzed on the free online platform of Majorbio Cloud Platform (www.majorbio.com).
Results
Identification and Expression Patterns of the Genes Encoding MAS Proteins in S. sclerotiorum
A total of five MAS homologs were identified in the genome of S. sclerotiorum (Figure 1). Among them, SS1G_11468 had the highest expression level during the infection of both Brassica oleracea leaves and stems, as revealed by RNA-seq in the previous study (Mei et al., 2016; Ding et al., 2019) (Supplementary Figures 1A,B). Additionally, SS1G_11468 was the most highly expressed gene during infecting Brassica oleracea leaves (Supplementary Figure 1C). Furthermore, the sequence of SS1G_11468 showed best matches to a specific expressed sequence tag (EST) DV643832 from the infection cushion library of S. sclerotiorum. SS1G_11468 contains a 747-bp ORF with two exons and encodes a protein with a length of 248 amino acids. The N-terminus of SS1G_11468 was predicted to contain a typical signal peptide with SignalP 5.0 Server and an extracellular and non-membrane location with TMHMM 2.0 or TMpred. The predicted cleavage site was between amino acid positions 18 and 19. Sequence comparison and phylogenetic tree analysis showed that SS1G_11468 exhibited high sequence similarity with B. cinerea CND1 (BC1G_08931) (86.29% identity in amino acid sequence, E-value: 7e-122) and M. oryzae MAS3 (MGG_11595) (58.20% identity in amino acid sequence, E-value: 6e-074) (Figures 1A,B). Therefore, SS1G_11468 was named Sscnd1.
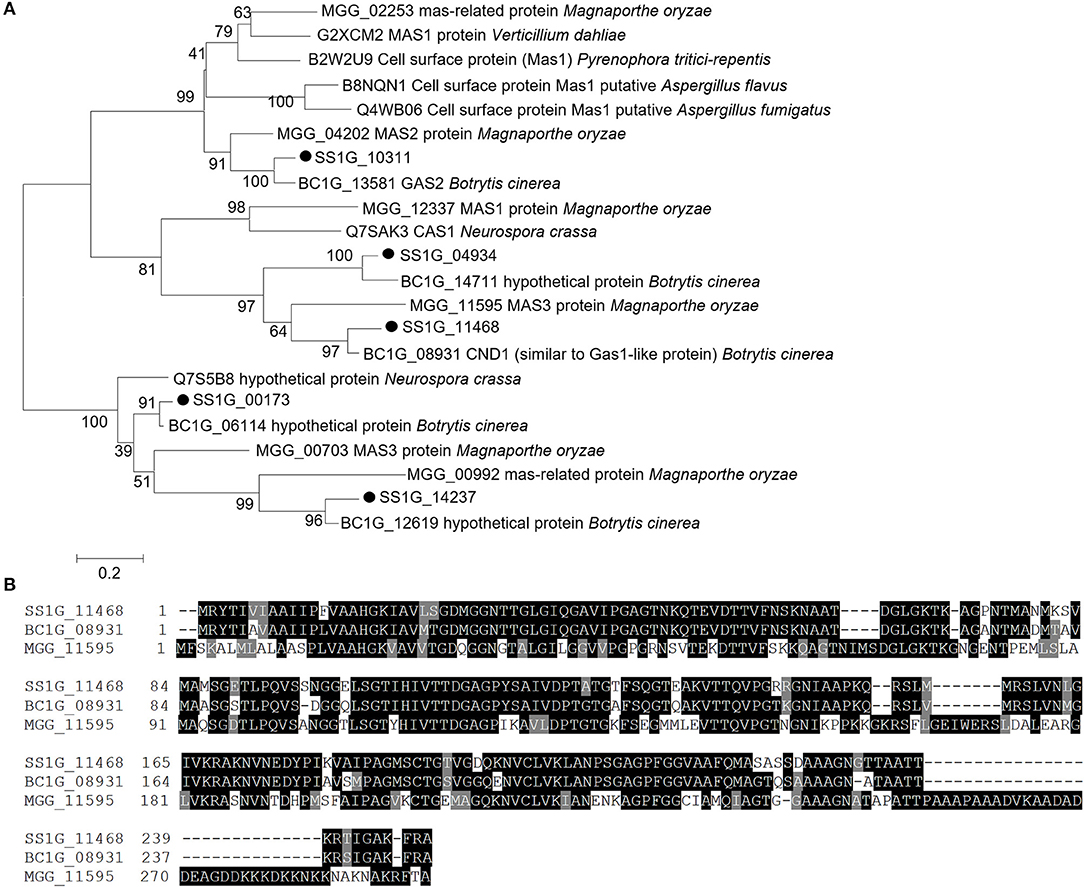
Figure 1. Sequence and phylogenetic analysis of MAS-related genes of Sclerotinia sclerotiorum. (A) Phylogenetic analysis of S. sclerotiorum SS1G_11468 and other homologous MAS-related proteins from Botrytis cinerea, Magnaporthe oryzae, Pyrenophora tritici-repentis, Aspergillus flavus, Verticillium dahliae, Aspergillus fumigatus, and Neurospora crassa. (B) Multiple alignment of the protein sequences of SS1G_11468, CND1 in Botrytis cinerea (BC1G_08931) and MAS3 in Magnaporthe oryzae (MGG_11595). Phylogenetic analysis was performed using MEGA 6.0 software using the maximum likelihood method.
The expression patterns of Sscnd1 during the different developmental stages and infection processes were determined via qRT-PCR. The results showed that Sscnd1 was highly expressed during the hyphal growth stage (Figure 2A). When inoculated on B. napus leaves, Sscnd1 expression dramatically increased from 3 hpi to 12 hpi by almost 39-fold (Figure 2B). However, the other four homologous MAS-related genes showed significantly lower expression than Sscnd1 during infection processes (Supplementary Figure 2). These results suggest that Sscnd1 is strongly induced during the infection of S. sclerotiorum, especially at the initial infection stage.
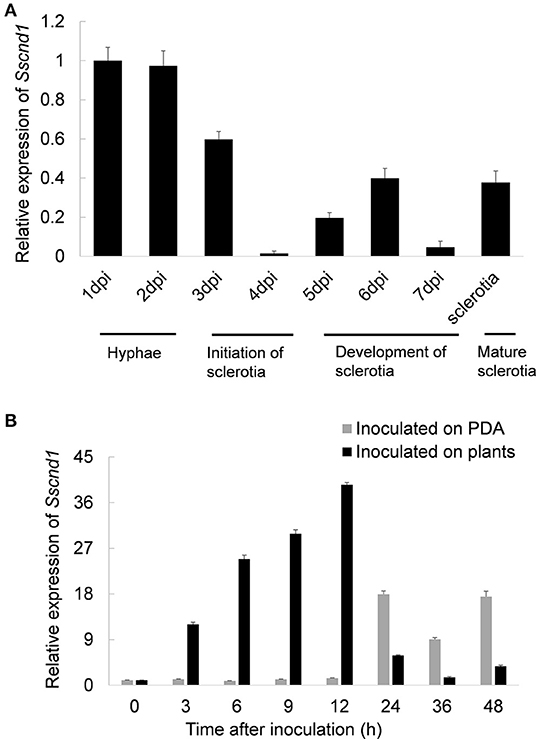
Figure 2. Expression analysis of Sscnd1 during various stages. (A) Quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) analysis of the Sscnd1 gene expression during different developmental stages of sclerotia. (B) Expression analysis of Sscnd1 in S. sclerotiorum after contact with rapeseed leaves and growing on potato dextrose agar (PDA) plates. The quantity of Sstub1 was used to normalize the expression levels of Sscnd1 in different samples. The relative abundance of Sscnd1 cDNA in the hyphae on PDA at 1 dpi (A) and in the hyphae inoculated on plants at 0 h (B) was set as one, respectively. Error bars indicate the standard deviation of three independent assays.
Sscnd1 Gene-Silenced Strains Showed Impaired Mycelial Growth
To determine the possible roles of Sscnd1 in the development of the S. sclerotiorum mycelium, Sscnd1-knockdown strains were obtained via RNAi. The RNAi vector pSicnd1 was transformed into the S. sclerotiorum wild-type strain 1980 (Figure 3A). Two strains, Sicnd1-9 and Sicnd1-20, with reduced Sscnd1 expression levels were chosen (Figure 3B). Multiple sequence alignment showed that the amplified fragment of Sscnd1 exhibited low similarity with the other four S. sclerotiorum MAS-related genes in nucleic acid sequence (SS1G_10311: 1.20% identity, SS1G_14237: 1.40% identity, SS1G_00173: 2.20% identity, SS1G_04934: 1.80% identity) (Supplementary Figure 3A). We found that the expression of the other four MAS-related genes was unimpaired in Sicnd1-9 and Sicnd1-20 (Supplementary Figure 3B).
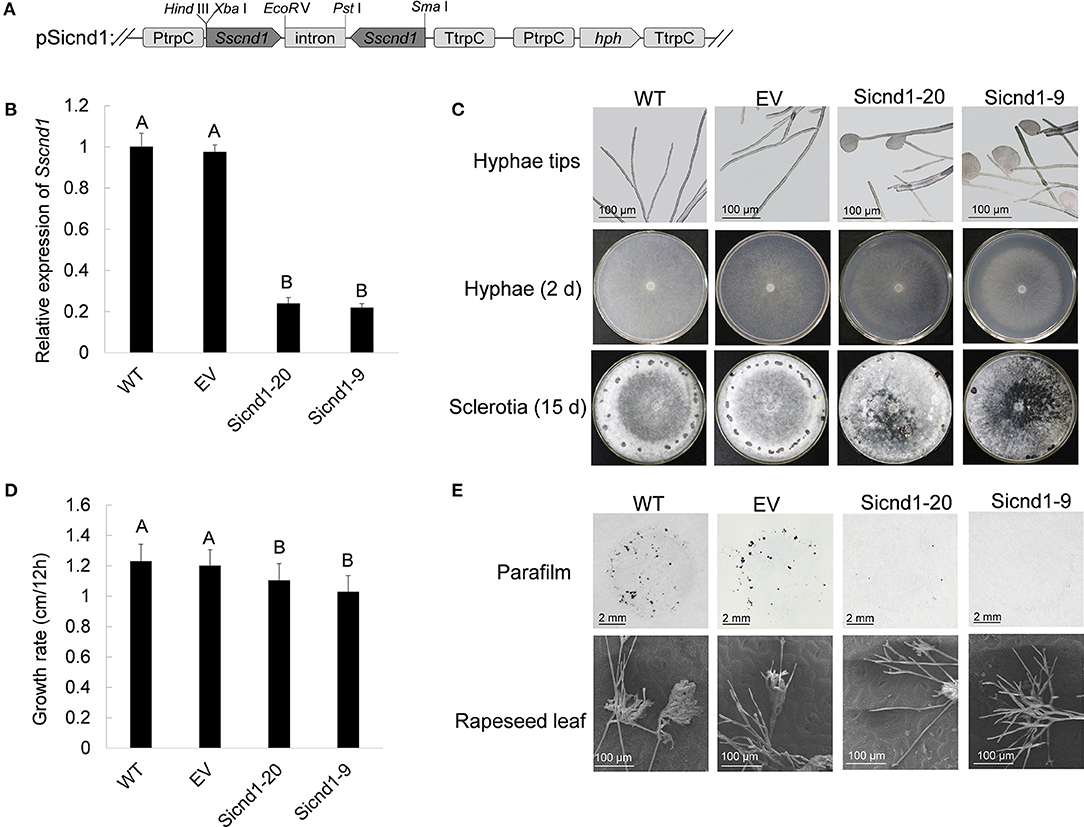
Figure 3. Phenotype of Sscnd1 gene-silenced strains. (A) Construction of the Sscnd1 RNAi vector pSicnd1. (B) Relative expression level of Sscnd1 in different isolates containing pSicnd1, as well as in the wild-type strain (WT) and empty vector strain (EV), as determined by qRT-PCR. The quantity of Sstub1 was used to normalize the expression levels of Sscnd1 in different samples. The relative abundance of Sscnd1 in WT was set as one. Error bars indicate the standard deviation of three independent assays. Differences were assessed using Tukey's HSD test. Different letters indicate statistical significance at the 0.05 level (P < 0.05). (C) Phenotypes of the WT, EV, Sicnd1-9, and Sicnd1-20. One representative biological replicate is shown. (D) Radial growth of Sscnd1 gene-silenced strains on PDA. Error bars indicate the standard deviation of three independent assays. Differences were assessed using Tukey's HSD test. Different letters indicate statistical significance at the 0.05 level (P < 0.05). (E) Compound appressorium formation of WT, EV, Sicnd1-9, and Sicnd1-20 on parafilm-overlaid PDA (8 hpi [hours post inoculation]) as revealed by the microscope and on rapeseed leaves (8 hpi) as revealed by the scanning electron microscope. One representative biological replicate out of three is shown.
Morphological analysis showed that both Sicnd1-9 and Sicnd1-20 exhibited frequent cytoplasmic bleeding at hyphal tips by microscopy (Figure 3C). The proportion of hyphal tips with cytoplasmic bleeding for Sicnd1-9 and Sicnd1-20 was 34.02 and 30.37%, respectively, which was higher than the wild-type strain (2.38%) and control strain (1.59%). In addition to the aberrant morphology of hyphal tips, Sicnd1-9 and Sicnd1-20 exhibited significantly reduced mycelial growth and sclerotia formation on PDA plates (Figure 3C). The growth rate was 1.23 cm/12 hpi for the wild-type strain and 1.20 cm/12 hpi for the empty vector strain, but 1.03 cm/12 hpi and 1.10 cm/12 hpi for Sicnd1-9 and Sicnd1-20, respectively (Figure 3D). The results indicate that Sscnd1 is associated with mycelial growth in S. sclerotiorum.
Sscnd1 Is Required for Compound Appressorium Formation and Full Virulence in S. sclerotiorum
To explore whether Sscnd1 was involved in compound appressorium development, the wild-type, empty vector strain and Sscnd1 gene-silenced strains were inoculated on parafilm-overlaid PDA and on the leaves of B. napus. We found that both Sicnd1-9 and Sicnd1-20 produced fewer compound appressoria on parafilm or rapeseed leaves than the wild-type strain and empty vector strain (Figure 3E). Additionally, the number of compound appressorium was positively related to the expression of Sscnd1 (r = 0.920, P < 0.05) (Supplementary Figure 4), indicating that Sscnd1 is associated with compound appressorium formation in S. sclerotiorum.
Furthermore, to examine whether Sscnd1 is involved in the pathogenicity of S. sclerotiorum, the detached B. napus leaves were inoculated with agar plugs derived from the wild-type, empty vector strain and Sscnd1 gene-silenced strains, and we found that the lesion areas of B. napus leaves infected with Sicnd1-9 and Sicnd1-20 were reduced to 0.52- and 0.58-fold of the wild-type strain and to 0.53- and 0.59-fold of the empty vector strain, respectively (Figures 4A,B). These results indicate that the infection capacity of the Sscnd1 gene-silenced strains is highly impaired. We further monitored the rescue of penetration events by inoculation on the wounded leaves of B. napus. We found that Sscnd1 gene-silenced strains induced larger lesions on the wounded leaves than on the intact leaves, but the lesion area was still significantly smaller than wild-type and empty vector strain (Figures 4A,B). These results suggest that Sscnd1 may contribute toward full virulence at the penetration phase of S. sclerotiorum.
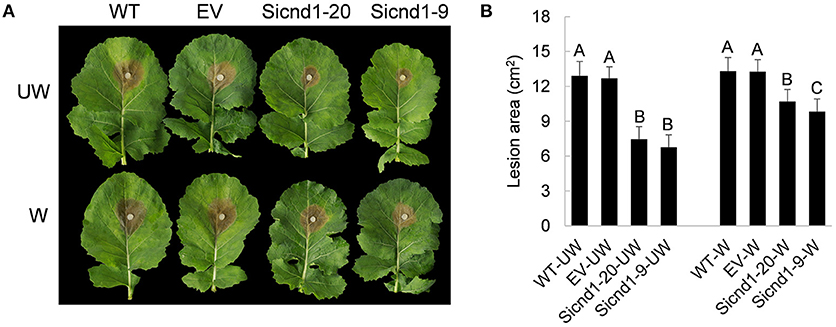
Figure 4. Pathogenicity assays of the Sscnd1 gene-silenced strains. (A) Disease phenotype of the wild-type strain (WT), empty vector strain (EV), Sicnd1-9 and Sicnd1-20 on the unwounded (UW) and wounded (W, wounded with a dissecting needle) leaves of rapeseed. Photographs were taken at 48 hpi. One representative biological replicate is shown. (B) Statistical analysis of the lesion area in panels (A). Error bars indicate the standard deviation for five replicates. Differences in the UW group and W group were assessed using Tukey's HSD test, respectively. Different letters indicate statistical significance at the 0.05 level (P < 0.05).
Sscnd1 Is in Association With Cell Integrity of S. sclerotiorum
To further investigate the role of Sscnd1 in the growth and virulence of S. sclerotiorum, we performed whole genome expression profiling analysis of the hyphae in wild-type and Sscnd1 gene-silenced strains through RNA sequencing (RNA-Seq). The raw data were stored in NCBI BioProject database with the accession ID PRJNA744751. Gene expression of Sscnd1 in the silenced strains was reduced to 0.3 fold of wild-type strain, the other four MAS-related genes showed no significant changes (Sicnd1 [Sicnd1-9 and Sicnd1-20]_vs_ WT) (Supplementary Figure 5). Additionally, there were 662 differentially expressed genes (DEGs) (Sicnd1 [Sicnd1-9 and Sicnd1-20]_vs_WT), consisting of 269 up-regulated DEGs and 393 down-regulated DEGs (Figure 5A and Supplementary Table 2). To validate the data obtained by RNA-seq, we performed qRT-PCR analysis by choosing 15 S. sclerotiorum genes of interest, and found a high consistence of gene expression between qRT-PCR and RNA-seq (r = 0.952, P < 0.01) (Supplementary Figure 6). Gene ontology (GO) enrichment analysis showed that these down-regulated DEGs were categorized into 14 GO terms (q < 0.01). Of these, the most significantly enriched GO terms were associated with the intrinsic component of membrane and integral component of membrane (Figure 5B). These results indicate that Sscnd1 is associated with the cell membrane integrity.
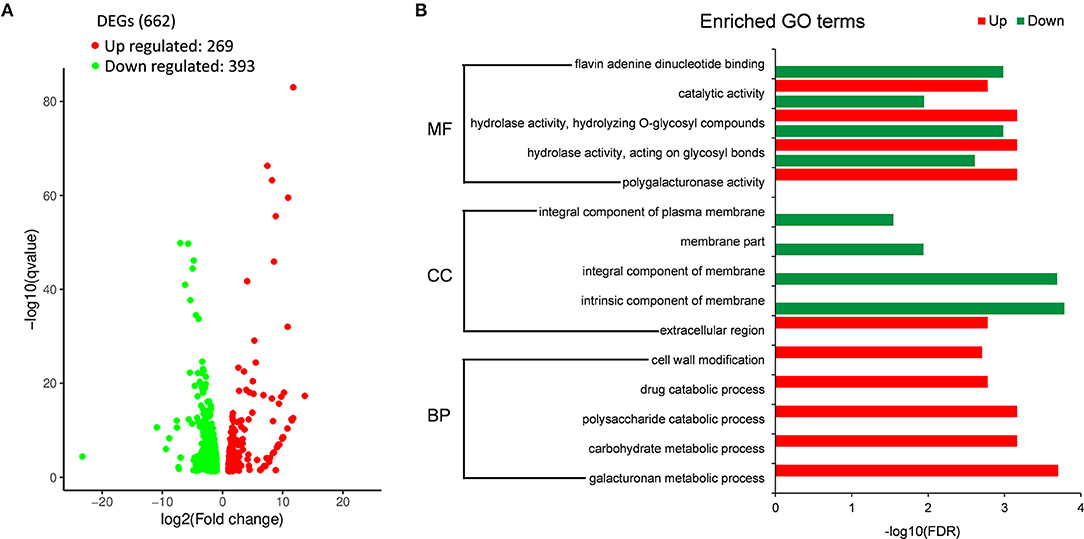
Figure 5. RNA-Seq analysis of wild-type and Sscnd1 gene-silenced strains. RNA sequencing was performed between wild-type strain (WT) and Sscnd1 gene-silenced strains. Experiments contain two biological replicates. (A) The number of differentially expressed genes (DEGs) (Sscnd1 gene-silenced strains vs. WT). (B) Significantly enriched Gene Ontology (GO) terms for up- and down-regulated DEGs.
To explore whether the suppression of Sscnd1 affects the cell membrane integrity in S. sclerotiorum. The tolerance to high osmotic stresses was assessed. Growth on PDA plates supplemented with 2% sorbose, 5% sorbose, 1 M sorbitol or 1.2 M sucrose among the wild-type, empty vector strain and Sscnd1 gene-silenced strains was investigated. The results showed that the inhibition of hyphal growth was significantly greater in Sscnd1 gene-silenced strains than in the wild-type strain and empty vector strain (Figures 6A,B). Furthermore, in the presence of 0.02% SDS, which damages cell membrane of organisms (Temme et al., 2012), the growth rate of the Sscnd1 gene-silenced strains was almost completely suppressed, while the wild-type strain and empty vector strain could slowly grow, suggesting that the Sscnd1-silenced strains were more sensitive to SDS. These results indicate that Sscnd1 is involved in the response to high osmotic stresses and cell integrity.
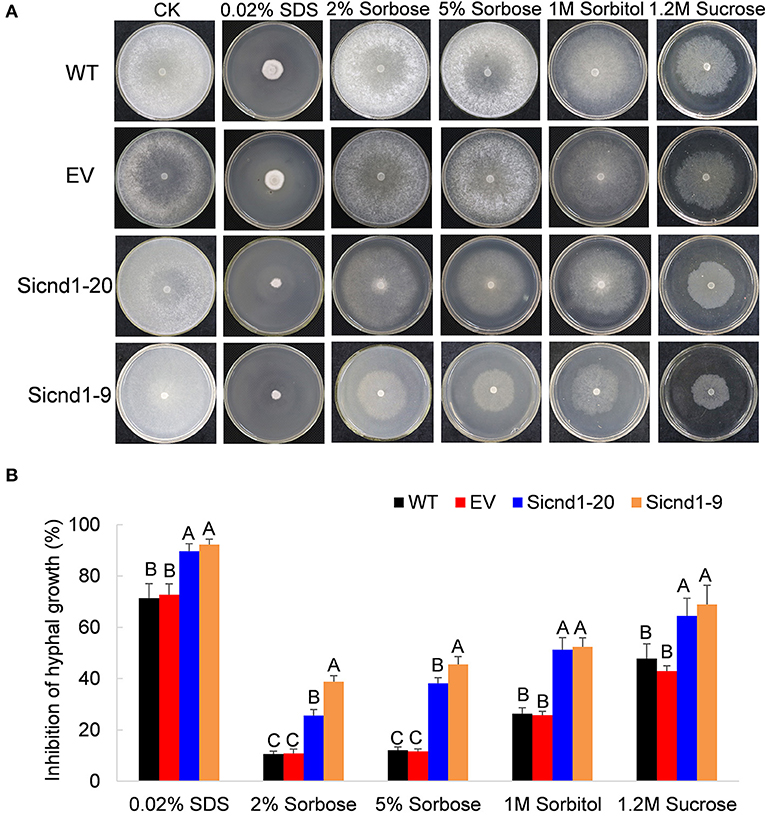
Figure 6. The growth of Sscnd1 gene-silenced strains in the presence of various stressors. Phenotypes (A) and growth inhibition (B) of the wild-type strain (WT), empty vector strain (EV), Sicnd1-9 and Sicnd1-20 grown on PDA supplemented with 0.02% SDS, 2% sorbose, 5% sorbose, 1 M sorbitol, and 1.2 M sucrose, respectively. Photographs were taken at 3 dpi (days post inoculation). One representative biological replicate is shown. Error bars indicate the standard deviation for three replicates. Differences in every stressor group were assessed using Tukey's HSD test. Different letters indicate statistical significance at the 0.05 level (P < 0.05).
HIGS of Sscnd1 in the Host Enhances S. sclerotiorum Resistance
Sequence alignment and phylogenetic tree analysis revealed no genes homologous to Sscnd1 in plants, indicating that Sscnd1 could be a target gene for the application of HIGS to control Sclerotinia stem rot disease. A tobacco rattle virus (TRV)-mediated transient silencing of Sscnd1 was performed in Nicotiana benthamiana. Seven days after TRV treatment, plants were challenged with the S. sclerotiorum wild-type strain 1980. The relative expression of Sscnd1 in TRV:: Sscnd1-1- and TRV:: Sscnd1-2-infected leaves was reduced by 63 and 66%, as determined by qRT-PCR, compared with that in the control leaves (TRV:: GFP), and the lesion area on TRV:: Sscnd1-1- and TRV:: Sscnd1-2-infected leaves was reduced by 51 and 56% at 48 hpi, respectively, compared with that on the control plants (TRV:: GFP) (Supplementary Figure 7).
To assess whether the resistance against S. sclerotiorum can be improved by expressing an RNAi construct targeting Sscnd1 in stable transgenic plants, we transferred a HIGS vector containing the RNAi cassette of Sscnd1 into wild-type A. thaliana Col-0 (Figure 7A). All the fifty transgenic lines in T1 generation exhibited smaller lesion areas than the wild-type Col-0 and plants carrying empty vector (EV plants, positive control) (Supplementary Table 3). Of which three transgenic HIGS-Sscnd1 lines (HIGS-Sicnd1-25, HIGS-Sicnd1-39, and HIGS-Sicnd1-42) with the smallest lesion area were continuously self-crossed to develop homozygous lines (Figure 7 and Supplementary Table 3). No significant difference was observed in growth between transgenic and control A. thaliana lines (Supplementary Figure 8). The wild-type Col-0, EV plants, and these homozygous lines of three transgenic HIGS-Sscnd1 in T3 generations were challenged with the S. sclerotiorum wild-type strain 1980 in vitro and in vivo. At 24 hpi, the lesion areas on the leaves of HIGS-Sicnd1-25, HIGS-Sicnd1-39, and HIGS-Sicnd1-42 were reduced by 35, 45, and 33% in vitro and 61, 71, and 53% in vivo compared with those on the leaves of the EV lines, respectively (Figures 7B,C). The in vivo lesion areas were significantly correlated with the in vitro lesion areas (r = 0.985, P < 0.05). The expression of Sscnd1 at 9, 12, and 24 hpi was clearly suppressed in these transgenic HIGS-Sscnd1 lines compared with the Col-0 and EV plants (Figure 7D). To verify the production of specific siRNA (Sscnd1-siRNA) in transgenic HIGS-Sscnd1 A. thaliana lines, stem-loop qRT-PCR was carried out with cDNA of leaf tissues (Supplementary Table 1). The results showed that Sscnd1-siRNA was highly expressed in HIGS-Sscnd1 A. thaliana lines, but no expression of Sscnd1-siRNA was detected in Col-0 and EV plants (Supplementary Figure 9). To rule out the effect that expression of HIGS-Sscnd1 construct activates plant defense responses in plants, several defense-related marker genes (AtPR1, AtPR2, AtPR5, and AtPDF1.2) were performed the expression analysis in the HIGS-Sscnd1 lines. There was no significant difference in these genes in HIGS-Sscnd1 lines compared with Col-0 and EV (Supplementary Figure 10A). Additionally, the expression of four Sscnd1 homologous genes showed no significant changes in the inoculated HIGS-Sscnd1 lines (Supplementary Figure 10B).
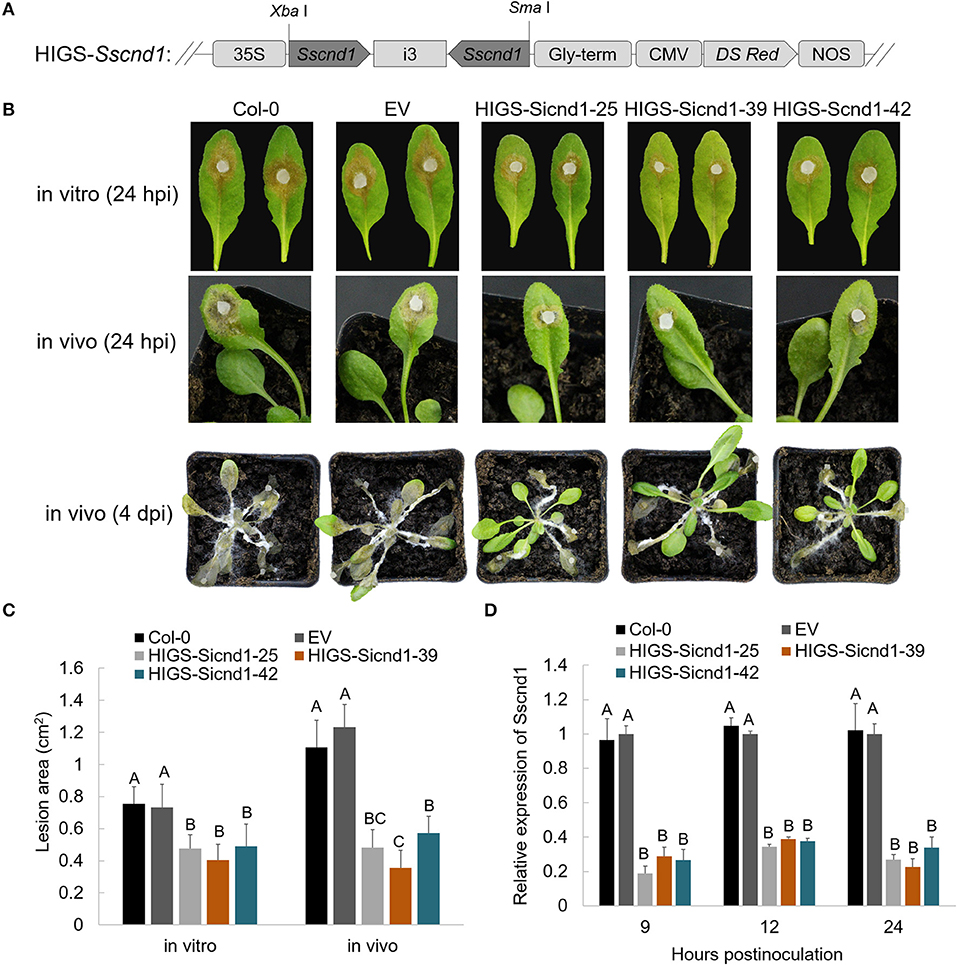
Figure 7. Expression of the HIGS-Sscnd1 construct in Arabidopsis thaliana enhances resistance to S. sclerotiorum. (A) Diagram representing the construct of the HIGS-Sscnd1 vector. (B) Disease phenotypes of the leaves of Col-0 (wild-type A. thaliana), EV (A. thaliana lines containing the empty vector) and HIGS-Sscnd1 transgenic A. thaliana lines (HIGS-Sicnd1-25, HIGS-Sicnd1-39, and HIGS-Sicnd1-42) in vitro (at 24 hpi) and in vivo (at 24 hpi and at 4 dpi) after inoculation with S. sclerotiorum wild-type strain 1980. One representative biological replicate is shown. (C) Quantification of the lesion area in (A) at 24 hpi in vitro and in vivo. Error bars indicate the standard deviation for five replicates. Differences in the in vitro group and in vivo group were assessed using Tukey's HSD test, respectively. Different letters indicate statistical significance at the 0.05 level (P < 0.05). (D) Relative transcript levels of Sscnd1 in the leaves at 9, 12, and 24 hpi with the S. sclerotiorum wild-type strain 1980. The quantity of Sstub1 was used to normalize the expression levels of Sscnd1 in different samples. Error bars indicate the standard deviation of three independent assays. At every time point, the relative abundance of Sscnd1 in EV was set as one. Differences in every time point group were assessed using Tukey's HSD test, respectively. Different letters indicate statistical significance at the 0.05 level (P < 0.05).
Considering that the sequence of Sscnd1 exhibits 87.01% identity in nucleic acid sequence with B. cinerea Bccnd1 (BC1G_08931), the HIGS-Sscnd1, Col-0 and EV lines were challenged with B. cinerea B05.10. We found that the lesion areas on HIGS-Sicnd1-25, HIGS-Sicnd1-39, and HIGS-Sicnd1-42 were reduced at 24 hpi compared with those on the Col-0 and EV lines (Supplementary Figures 11A,B). The relative expression of BC1G_08931 in HIGS-Sscnd1 lines decreased at 24 hpi compared with that in the Col-0 and EV lines (Supplementary Figure 11C).
Discussion
As multicellular infectious structures, compound appressoria are formed unless penetration occurs directly via stomata and are essential for S. sclerotiorum to successfully penetrate hosts (Uloth et al., 2016; Liang and Rollins, 2018). Sscnd1 coding a protein with 58.20% identity to M. oryzae MAS3. In this study, the expression of Sscnd1 was upregulated in the early infection stage. The Sscnd1 gene-silenced strains showed a drastic reduction in virulence and compound appressorium formation. The virulence of Sscnd1 gene-silenced strains was partially restored on wounded leaves, but still significantly lower than control strains. These findings indicate that Sscnd1 is associated with compound appressorium formation and fungal full virulence at penetration phase in S. sclerotiorum. In B. cinerea and M. oryzae, when the disruption of the genes coding MAS proteins (BC1G_13581 in B. cinerea, MGG_12337 and MGG_04202 in M. oryzae), though the mutants present impaired virulence, there are no significant changes in the hyphae growth (Xue et al., 2002; Schamber et al., 2010). In contrast with B. cinerea and M. oryzae, Sscnd1 gene-silenced strains showed a reduction in the hyphae growth. The protein sequence alignment showed that Sscnd1 coding a protein with 27.84% identity to BC1G_13581, 37.55% identity to MGG_12337, and 29.9% identity to MGG_04202. The sequence specificity may give new functions of Sscnd1. Homologous genes originate from the same ancestral gene, but they may lose their original functions or evolve new functions in the process of evolution (Li et al., 2005). Therefore, the role of Sscnd1 revealed both common and unique properties compared with those of other plant pathogenic fungi.
S. sclerotiorum secretes OA, enzymes, and effector proteins to induce the necrosis of host cells and absorbs nutrients from dying host cells (Amselem et al., 2011; Williams et al., 2011; Bashi et al., 2012; Kabbage et al., 2013; Guyon et al., 2014; Seifbarghi et al., 2017; Yang et al., 2018). However, the cytoplasmic exudate of the dying cells causes osmotic stress, which inhibits fungal survival, germling differentiation and penetration (Kamamura et al., 2002; Skamnioti and Gurr, 2007). The hyphal tips of Sscnd1 gene-silenced strains exhibited frequent cytoplasmic bleeding. Down-regulated expression of genes in modulating component of membrane and high sensitivity to osmotic stress, suggesting that the Sscnd1 gene-silenced strains have a defect in cell integrity. The association of cell integrity and osmotic stress response with the appressorium formation has been shown in many important pathogenic fungi, such as M. oryzae (Jeon et al., 2008; Deng et al., 2019), Colletotrichum fructicola (Liang et al., 2019), Colletotrichum graminicola (Albarouki and Deising, 2013) and B. cinerea (Liu et al., 2019). These findings indicate that the Sscnd1 gene-silenced strains exhibit a defect in the compound appressorium formation possibly due to a defect in cell integrity, which caused a high sensitivity to environmental stressors. A mutant with a disruption in the secretory protein SsCaf1 failed to form compound appressoria and was severely inhibited by osmotic stress conditions (Xiao et al., 2014). Similar to SsCaf1, Sscnd1 was predicted to contain a signal peptide and an extracellular and non-membrane location. Additional studies are needed to explore whether the molecular role of Sscnd1 in compound appressorium formation is the same as that of SsCaf1.
HIGS is a RNAi technology where small RNAs produced in plants can specifically silence the pathogen genes. It has been suggested to be an efficient tool for the potential control of various fungi in crops (Chen et al., 2016; Zhu et al., 2017; Qi et al., 2018, 2019; Xu et al., 2018). Excavation and functional analysis of the virulence factors in S. sclerotiorum, such as OA (Cessna et al., 2000; Liang et al., 2015a,b), cell wall-degrading enzymes (CWDEs) (Yajima et al., 2009; Yu et al., 2016), secretory proteins (Guyon et al., 2014; Yang et al., 2018) and ROS (Kim et al., 2011; Xu and Chen, 2013), supply key HIGS targets for enhancing Sclerotinia resistance. Andrade et al. (2015) first proved that the HIGS-mediated chitin synthase gene (CHS) in tobacco enhanced resistance to S. sclerotiorum in the T1 generation. McCaghey et al. (2021) provided evidence supporting that S. sclerotiorum can uptake environmental RNAs and RNAi of oxaloacetate acetylhydrolase (Ssoah1) using HIGS reduced the pathogen aggressiveness. In this study, we selected Sscnd1 as the target gene to apply HIGS in A. thaliana. We transiently expressed the RNAi construct of Sscnd1 in N. benthamiana and stably expressed it in A. thaliana. The transgenic lines showed significantly enhanced resistance to S. sclerotiorum. The limitation of RNAi technologies is the potential off-target effect (Lundgren and Duan, 2013). Although there were four other homologous genes in S. sclerotiorum, Sscnd1 was sequence specific among its homologs in S. sclerotiorum at the nucleotide level. Meanwhile, the sequence of Sscnd1 revealed relatively low similarity with A. thaliana genome. This sequence-specific prevents silencing of other homologous non-target genes by the application of RNAi (Nakayashiki and Nguyen, 2008). The random insertion rather than site-specific insertion of RNAi construct may interfere with the expression of related genes (Jia et al., 2017). However, different Sscnd1 RNAi strains and HIGS-Sscnd1 lines showed similar phenotypes for the pathogen virulence and compound appressorium formation, which makes the role of Sscnd1 persuasive.
Compound appressoria are hyphal tip-differentiated multicellular infection structures formed by many plant-pathogenic fungi on the host surface and is critical for penetrating into the host cells (Boenisch and Schäfer, 2011). Hu et al. (2020) proved that genes involved in urediniospore germination or appressorium formation can be used to manage Asian soybean rust (ASR) through HIGS in soybean. Mahto et al. (2020) found that silencing the CgCOM1 in chili and tomato suppressed the appressoria formation and mycelial growth of Colletotrichum gloeosporioides, resulting in reduced infection of plant tissues. The infection stages of C. gloeosporioides switches from biotrophic (conidia germination, formation of appressoria, penetration peg and primary hyphae) and necrotrophic (formation of secondary hyphae) phases (O'Connell et al., 2012). Similar to C. gloeosporioides, there may be a transition from a biotrophic to necrotrophic lifestyle in S. sclerotiorum (Kabbage et al., 2015; Liang and Rollins, 2018). Sscnd1 encoding a appressorium- related protein and function in the fungal full virulence in S. sclerotiorum. Silencing Sscnd1 in A. thaliana limited the compound appressorium formation during infection, and enhanced the host resistance. Meanwhile, the expression of several defense-related marker genes (AtPR1, AtPR2, AtPR5, and AtPDF1.2) showed no significant difference in HIGS-Sscnd1 lines from that in control lines, indicating that the reduced pathogenicity in HIGS-Sscnd1 lines is indeed caused by silencing of Sscnd1. These results prove the role of Sscnd1 in conferring resistance to S. sclerotiorum to host plants.
Many studies evident that target ds/siRNAs were presented in HIGS lines (Dou et al., 2020; Hu et al., 2020; Mahto et al., 2020; Singh et al., 2020; McCaghey et al., 2021), indicating that the uptake of ds/siRNA is likely a common occurrence in the fungal kingdom. It was shown that host Arabidopsis cells secrete exosome-like extracellular vesicles to deliver sRNAs into B. cinerea, and these sRNA-containing vesicles accumulate at the infection sites and are taken up by fungal cells, resulting in the silencing of fungal pathogenicity genes (Cai et al., 2018). Koch et al. (2020) also found that HIGS involves the transfer of dsRNA-derived siRNA via extracellular vesicles in Arabidopsis. Several studies suggest that host-derived siRNA is thought to translocate into pathogens via haustoria or similar structures, and silencing the highly expressed haustoria genes have been proved be more effective in HIGS application (Nowara et al., 2010; Yin et al., 2011; Panwar et al., 2013). However, the mechanism of host-derived RNA translocation across the plant cells to S. sclerotiorum cells is yet to be determined.
In conclusion, we found that Sscnd1 is required for compound appressorium formation, cell integrity and fungal full virulence in S. sclerotiorum and is a potential target for improving Sclerotinia resistance in crops via HIGS.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
WQ planned and designed the research. YD, YC, BY, HL, and MD performed research. XM conducted RNA-seq analyses. WQ, HW, and YD analyzed and validated the data. YD and WQ wrote the original draft of the manuscript. All authors discussed the data, edited, and approved the manuscript.
Funding
This study was financially supported by the National Nature Science Foundation of China (31801395 and 31971978), the Project of Chongqing Science and Technology Commission (cstc2017shms-xdny80050, cstc2019jcyj-zdxmX0012, and cstc2019jcyj-msxm0486) and the Fundamental Research Funds for the Central Universities (XDJK2018AA004, XDJK2018B022, and SWU120075).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are indebted to Dr. Yang Yu for providing the pCIT and pSKH vector.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.693334/full#supplementary-material
Supplementary Figure 1. Expression of MAS-related genes in S. sclerotiorum during infection. Expression of five MAS-related genes in S. sclerotiorum during the inoculation of Brassica oleracea leaves (A) and stems (B), as revealed by RNA-seq in our previous study (Mei et al., 2016; Ding et al., 2019). (C) Top 10 expressed S. sclerotiorum genes during the inoculation of B. oleracea leaves. RL, resistant B. oleracea leaf; SL, susceptible B. oleracea leaf; RS, resistant B. oleracea stem; SL, susceptible B. oleracea stem; 0 h, pre-inoculation; 6 h, 6 h post-inoculation; 12 h, 12 h post-inoculation; 24 h, 24 h post-inoculation.
Supplementary Figure 2. Expression analysis of Sscnd1 and its homologous genes during the inoculation of rapeseed leaves. The quantity of Sstub1 was used to normalize the expression levels of every gene in different samples. The relative expression of SS1G_11468 at 0 h was set as one. Mean values from three independent assays were used to construct the heatmap.
Supplementary Figure 3. Expression analysis of the Sscnd1 homologous genes in Sscnd1 gene-silenced strains. (A) Multiple alignment of the amplified 418 bp of Sscnd1 with the other four homologous genes in S. sclerotiorum. (B) Relative transcript levels of the four homologous genes in the Sscnd1 gene-silenced strains. The quantity of Sstub1 was used to normalize the expression levels of Sscnd1 in different samples. Error bars indicate the standard deviation of three independent assays. Differences in every gene group were assessed using Tukey's HSD test, respectively. Different letters indicate statistical significance at the 0.05 level (P < 0.05).
Supplementary Figure 4. The correlation between the Sscnd1 expression and the compound appressorium numbers. Sicnd1-9, Sicnd1-10, Sicnd1-15, Sicnd1-16, Sicnd1-17, and Sicnd1-20 were six Sscnd1 gene-silenced strains.
Supplementary Figure 5. Expression changes of Sscnd1 and its homologous genes in Sicnd1-9 and Sicnd1-20, as revealed by RNA-seq. FC: Fold change (Sicnd1-9 or Sicnd1-20 vs. wild-type strain).
Supplementary Figure 6. Expression changes as estimated by RNA-Seq (black bars) and qRT-PCR (gray bars) for 15 S. sclerotiorum genes of interest.
Supplementary Figure 7. Functional assessment of Sscnd1 in S. sclerotiorum pathogenicity determined by TRV-mediated HIGS. (A) Disease phenotypes of Mock (mock-inoculated with FES buffer)-, TRV::GFP-, TRV::Sscnd1-1- and TRV::Sscnd1-2-inoculated N. benthamiana leaves at 48 hpi with S. sclerotiorum strain 1980. (B) Quantification of the lesion area in (A). Error bars indicate the standard deviation for five replicates. Differences were assessed using Tukey's HSD test. Different letters indicate statistical significance at the 0.05 level (P < 0.05). (C) Relative transcript levels of Sscnd1 in the Mock-, TRV::GFP-, TRV::Sscnd1-1- and TRV::Sscnd1-2-inoculated N. benthamiana leaves 48 hpi with S. sclerotiorum strain 1980. The quantity of Sstub1 was used to normalize the expression levels of Sscnd1 in different samples. Error bars indicate the standard deviation of three independent assays. Differences were assessed using Tukey's HSD test. Different letters indicate statistical significance at the 0.05 level (P < 0.05).
Supplementary Figure 8. Morphological characterizations of transgenic and control A. thaliana lines.
Supplementary Figure 9. The expression of specific siRNA of Sscnd1 gene (Sscnd1-siRNA) in HIGS-Sscnd1 transgenic A. thaliana lines as determined by stem-loop qRT-PCR analysis. The cDNA harboring the loop sequence was used for stem-loop qRT-PCR analysis in Col-0 (wild-type A. thaliana), EV (A. thaliana lines containing the empty vector) and HIGS-Sscnd1 A. thaliana lines. The quantity of AtU6-26 was used to normalize the expression levels of Sscnd1 siRNA in different samples. Error bars indicate the standard deviation of three independent assays.
Supplementary Figure 10. Gene expression analysis. (A) Relative transcript levels of AtPR1, AtPR2, AtPR5, and AtPDF1.2 genes in the Col-0 (wild-type A. thaliana), EV (A. thaliana lines containing the empty vector) and HIGS-Sscnd1 transgenic A. thaliana lines. The quantity of AtActin8 was used to normalize the expression levels of every gene in different samples. Error bars indicate the standard deviation of three independent assays. The relative expression of every gene in EV was set as one. (B) Relative transcript levels of Sscnd1 homologous genes in the S. sclerotiorum-infected HIGS-Sscnd1 lines at 24 hpi. The quantity of Sstub1 was used to normalize the expression levels of every gene in different samples. Error bars indicate the standard deviation of three independent assays. The relative expression of every gene in EV was set as one. Differences in every gene group were assessed using Tukey's HSD test, respectively. Different letters indicate statistical significance at the 0.05 level (P < 0.05).
Supplementary Figure 11. Expression of the HIGS-Sscnd1 construct in A. thaliana Col-0 enhances resistance against Botrytis cinerea. (A) Disease phenotypes of Col-0 (wild-type A. thaliana), EV (empty vector line) and HIGS transgenic lines (HIGS-Sicnd1-25, HIGS-Sicnd1-39, and HIGS-Sicnd1-42) after inoculation with B. cinerea strain B05.10. Photographs were taken at 24 hpi. (B) Quantification of the lesion area in (A). Error bars indicate the standard deviation for five replicates. Differences were assessed using Tukey's HSD test. Different letters indicate statistical significance at the 0.05 level (P < 0.05). (C) Relative transcript levels of BC1G_08931 in the leaves of Col-0, EV, and HIGS transgenic lines 24 hpi with B. cinerea strain B05.10. Total BC1G_08931 cDNA abundance in the samples was normalized using BcActin gene as a control. Error bars indicate the standard deviation of three independent assays. Differences were assessed using Tukey's HSD test. Different letters indicate statistical significance at the 0.05 level (P < 0.05).
Supplementary Table 1. Information on the primers used in this study.
Supplementary Table 2. Information of the differentially expressed genes (DEGs) (Sicnd1 [Sicnd1-9 and Sicnd1-20]_vs_ WT).
Supplementary Table 3. Segregation analyses and lesion areas of Col-0 (wild-type A. thaliana), EV (empty vector line) and HIGS transgenic lines (HIGS-Sicnd1-25, HIGS-Sicnd1-39, and HIGS-Sicnd1-42) after inoculation with S. sclerotiorum.
References
Albarouki, E., and Deising, H. B. (2013). Infection structure-specific reductive iron assimilation is required for cell wall integrity and full virulence of the maize pathogen Colletotrichum graminicola. Mol. Plant-Microbe Interact. 26, 695–708. doi: 10.1094/MPMI-01-13-0003-R
Amselem, J., Cuomo, C. A., van Kan, J. A. L., Viaud, M., Benito, E. P., Couloux, A., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230. doi: 10.1371/journal.pgen.1002230
Andrade, C., Tinoco, M., Rieth, A., Maia, F., and Aragão, F. (2015). Host-induced gene silencing in the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Pathol. 65, 626–632. doi: 10.1111/ppa.12447
Bashi, Z. D., Rimmer, S. R., Khachatourians, G. G., and Hegedus, D. D. (2012). Factors governing the regulation of Sclerotinia sclerotiorum cutinase A and polygalacturonase 1 during different stages of infection. Can. J. Microbiol. 58, 605–616. doi: 10.1139/w2012-031
Baulcombe, D. (2005). RNA silencing. Trends Biochem. Sci. 30, 290–293. doi: 10.1016/j.tibs.2005.04.012
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi: 10.1038/nbt1359
Boenisch, M. J., and Schäfer, W. (2011). Fusarium graminearum forms mycotoxin producing infection structures on wheat. BMC Plant Biol. 11:110. doi: 10.1186/1471-2229-11-110
Boland, G. J., and Hall, R. (1994). Index of host plants of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 16, 93–108. doi: 10.1080/07060669409500766
Bolton, M. D., Thomma, B. P., and Nelson, B. D. (2006). Sclerotinia sclerotiorum (lib.) de bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. doi: 10.1111/j.1364-3703.2005.00316.x
Cai, Q., Qiao, L., Wang, M., He, B., Lin, F. M., Palmquist, J., et al. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129. doi: 10.1126/science.aar4142
Cessna, S. G., Sears, V. E., Dickman, M. B., and Low, P. S. (2000). Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 12, 2192–2200. doi: 10.2307/3871114
Chen, W., Kastner, C., Nowara, D., Oliveira-Garcia, E., Rutten, T., Zhao, Y., et al. (2016). Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J. Exp. Bot. 67, 4979–4991. doi: 10.1093/jxb/erw263
Cheng, W., Song, X. S., Li, H. P., Cao, L. H., Sun, K., Qiu, X. L., et al. (2015). Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 13, 1335–1345. doi: 10.1111/pbi.12352
Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. G., et al. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500. doi: 10.1093/nar/gkg500
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dean, R. A. (1997). Signal pathways and appressorium morphogenesis. Annu. Rev. Phytopathol. 35, 211–234. doi: 10.1146/annurev.phyto.35.1.211
Deng, S., Sun, W., Dong, L., Cui, G., and Deng, Y. Z. (2019). MoGT2 is essential for morphogenesis and pathogenicity of Magnaporthe oryzae. mSphere 4, e00309–e00319. doi: 10.1128/mSphere.00309-19
Ding, Y., Mei, J., Chai, Y., Yu, Y., Shao, C., Wu, Q., et al. (2019). Simultaneous transcriptome analysis of host and pathogen highlights the interaction between Brassica oleracea and Sclerotinia sclerotiorum. Phytopathology 109, 542–550. doi: 10.1094/PHYTO-06-18-0204-R
Dou, T., Shao, X., Hu, C., Liu, S., Sheng, O., Bi, F., et al. (2020). Host-induced gene silencing of Foc TR 4 ERG 6/11 genes exhibits superior resistance to Fusarium wilt of banana. Plant Biotechnol. J. 18, 11–13. doi: 10.1111/pbi.13204
Duan, C. G., Wang, C. H., and Guo, H. S. (2012). Application of RNA silencing to plant disease resistance. Silence 3:5. doi: 10.1186/1758-907X-3-5
Ghag, S. B., Shekhawat, U. K., and Ganapathi, T. R. (2014). Host-induced posttranscriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 12, 541–553. doi: 10.1111/pbi.12158
Godoy, G., Steadman, J. R., Dickman, M. B., and Dam, R. (1990). Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol. Mol. Plant Pathol. 37, 179–191. doi: 10.1016/0885-5765(90)90010-U
Govindarajulu, M., Epstein, L., Wroblewski, T., and Michelmore, R. W. (2015). Host-induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnol. J. 13, 875–883. doi: 10.1111/pbi.12307
Guyon, K., Balagu,é, C., Roby, D., and Raffaele, S. (2014). Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics 15:336. doi: 10.1186/1471-2164-15-336
Hamer, J. E., and Talbot, N. J. (1998). Infection-related development in the rice blast fungus Magnaporthe grisea. Curr. Opin. Microbiol. 1, 693–697. doi: 10.1016/s1369-5274(98)80117-3
Hamid, M. I., Zeng, F., Cheng, J., Jiang, D., and Fu, Y. (2013). Disruption of heat shock factor 1 reduces the formation of conidia and thermotolerance in the mycoparasitic fungus Coniothyrium minitans. Fungal Genet. Biol. 53, 42–49. doi: 10.1016/j.fgb.2012.12.002
Hu, D., Chen, Z. Y., Zhang, C., and Ganiger, M. (2020). Reduction of Phakopsora pachyrhizi infection on soybean through host-and spray-induced gene silencing. Mol. Plant Pathol. 21, 794–807. doi: 10.1111/mpp.12931
Huang, G., Allen, R., Davis, E. L., Baum, T. J., and Hussey, R. S. (2006). Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. U.S.A. 103, 14302–14306. doi: 10.1073/pnas.0604698103
Huang, L., Buchenauer, H., Han, Q., Zhang, X., and Kang, Z. (2008). Ultrastructural and cytochemical studies on the infection process of Sclerotinia sclerotiorum in oilseed rape. J. Plant Dis. Protect. 115, 9–16. doi: 10.1007/bf03356233
Huvenne, H., and Smagghe, G. (2010). Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J. Insect Physiol. 56, 227–235. doi: 10.1016/j.jinsphys.2009.10.004
Jahan, S. N., Åsman, A. K., Corcoran, P., Fogelqvist, J., Vetukuri, R. R., and Dixelius, C. (2015). Plant-mediated gene silencing restricts growth of the potato late blight pathogen Phytophthora infestans. J. Exp. Bot. 66, 2785–2794. doi: 10.1093/jxb/erv094
Jamaux, I., Gelie, B., and Lamarque, C. (2007). Early stages of infection of rapeseed petals and leaves by Sclerotinia sclerotiorum revealed by scanning electron microscopy. Plant Pathol. 44, 22–30. doi: 10.1111/j.1365-3059.1995.tb02712.x
Jeon, J., Goh, J., Yoo, S., Chi, M. H., Choi, J., Rho, H. S., et al. (2008). A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Mol. Plant-Microbe Interact. 21, 525–534. doi: 10.1094/MPMI-21-5-0525
Jia, R., Zhao, H., Huang, J., Kong, H., Zhang, Y., Guo, J., et al. (2017). Use of RNAi technology to develop a PRSV-resistant transgenic papaya. Sci. Rep. 7:12636. doi: 10.1038/s41598-017-13049-0
Jurick, W. M., and Rollins, J. A. (2007). Deletion of the adenylate cyclase (sac1) gene affects multiple developmental pathways and pathogenicity in Sclerotinia sclerotiorum. Fungal Genet. Biol. 44, 521–530. doi: 10.1016/j.pmpp.2004.07.004
Kabbage, M., Yarden, O., and Dickman, M. B. (2015). Pathogenic attributes of Sclerotinia sclerotiorum: Switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 233, 53–60. doi: 10.1016/j.plantsci.2014.12.018
Kabbage, M, Williams, B., and Dickman, M. B. (2013). Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 9:el003287. doi: 10.1371/journal.ppat.1003287
Kamamura, T., Yamaguchi, S., Saitoh, K., Teraoka, T., and Yamaguchi, I. (2002). A novel gene, CBP1, encoding a putative extracellular chitin-binding protein, may play an important role in the hydrophobic surface sensing of Magnaporthe grisea during appressorium differentiation. Mol. Plant-Microbe Interact. 15, 437–444. doi: 10.1094/MPMI.2002.15.5.437
Kim, H. J., Chen, C., Kabbage, M., and Dickman, M. B. (2011). Identification and characterization of Sclerotinia sclerotiorum NADPH oxidases. Appl. Environ. Microb. 77, 7721–7729. doi: 10.1128/AEM.05472-11
Koch, A., Schlemmer, T., Hoefle, L., Werner, B., Preusser, C., Hardt, M., et al. (2020). Host-induced gene silencing involves transfer of dsRNA-derived siRNA via extracellular vesicles. bioRxiv [Preprint]. doi: 10.1101/2020.02.12.945154
Li, J., Mu, W., Veluchamy, S., Liu, Y., Zhang, Y., Pan, H., et al. (2017). The GATA-type IVb zinc-finger transcription factor SsNsd1 regulates asexual-sexual development and appressoria formation in Sclerotinia sclerotiorum. Mol. Plant Pathol. 19, 1679–1689. doi: 10.1111/mpp.12651
Li, M. Y., Liang, X. F., and Rollins, J. A. (2012). Sclerotinia sclerotiorum gamma-glutamyl transpeptidase (Ss-ggt1) is required for regulating glutathione accumulation and development of sclerotia and compound appressoria. Mol. Plant-Microbe Interact. 25, 412–420. doi: 10.1094/Mpmi-06-11-0159
Li, W. H., Yang, J., and Gu, X. (2005). Expression divergence between duplicate genes. Trends Genet. 21, 602–607. doi: 10.1016/j.tig.2005.08.006
Liang, X., Liberti, D., Li, M., Kim, Y. T., Hutchens, A., Wilson, R., et al. (2015a). Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 16, 559–571. doi: 10.1111/mpp.12211
Liang, X., Moomaw, E. W., and Rollins, J. A. (2015b). Fungal oxalate decarboxylase activity contributes to Sclerotinia sclerotiorum early infection by affecting both compound appressoria development and function. Mol. Plant Pathol. 16, 825–836. doi: 10.1111/mpp.12239
Liang, X., and Rollins, J. A. (2018). Mechanisms of broad host range necrotrophic pathogenesis in Sclerotinia sclerotiorum. Phytopathology 108, 1128–1140. doi: 10.1094/PHYTO-06-18-0197-RVW
Liang, X., Wei, T., Cao, M., Zhang, X., Liu, W., Kong, Y., et al. (2019). The MAP kinase CfPMK1 is a key regulator of pathogenesis, development, and stress tolerance of Colletotrichum fructicola. Front. Microbiol. 10:1070. doi: 10.3389/fmicb.2019.01070
Liu, E., and Page, J. E. (2008). Optimized cDNA libraries for virus-induced gene silencing (VIGS) using tobacco rattle virus. Plant Methods 4:5. doi: 10.1186/1746-4811-4-5
Liu, L., Wang, Q., Zhang, X., Liu, J., Zhang, Y., and Pan, H. (2018). Ssams2, a gene encoding GATA transcription factor, is required for appressoria formation and chromosome segregation in Sclerotinia sclerotiorum. Front. Microbiol. 9:3031. doi: 10.3389/fmicb.2018.03031
Liu, Y., Liu, J. K., Li, G. H., Zhang, M. Z., Zhang, Y. Y., Wang, Y. Y., et al. (2019). A novel Botrytis cinerea-specific gene BcHBF1 enhances virulence of the grey mould fungus via promoting host penetration and invasive hyphal development. Mol. Plant Pathol. 20, 731–747. doi: 10.1111/mpp.12788
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lundgren, J. G., and Duan, J. J. (2013). RNAi-based insecticidal crops: Potential effects on nontarget species. Bioscience 63, 657–665. doi: 10.1525/bio.2013.63.8.8
Mahto, B. K., Singh, A., Pareek, M., Rajam, M. V., Dhar-Ray, S., and Reddy, P. M. (2020). Host-induced silencing of the Colletotrichum gloeosporioides conidial morphology 1 gene (CgCOM1) confers resistance against Anthracnose disease in chili and tomato. Plant Mol. Biol. 104, 381–395. doi: 10.1007/s11103-020-01046-3
Mao, Z., Shi, Y., Cao, Q., Chen, Y., Sun, Y., Liu, Z., et al. (2018). Transcriptional regulation on the gene expression signature in combined allergic rhinitis and asthma syndrome. Epigenomics 10, 119–131. doi: 10.2217/epi-2017-0072
McCaghey, M., Shao, D., Kurcezewski, J., Lindstrom, A., Ranjan, A., Whitham, S. A., et al. (2021). Host-induced gene silencing of a Sclerotinia sclerotiorum oxaloacetate acetylhydrolase using bean pod mottle virus as a vehicle reduces disease on Soybean. Front. Plant Sci. 12:677631. doi: 10.3389/fpls.2021.677631
Mei, J., Ding, Y., Li, Y., Tong, C., Du, H., Yu, Y., et al. (2016). Transcriptomic comparison between Brassica oleracea and rice (Oryza sativa) reveals diverse modulations on cell death in response to Sclerotinia sclerotiorum. Sci. Rep. 10:34900. doi: 10.1038/srep33706
Nakayashiki, H., and Nguyen, Q. B. (2008). RNA interference: Roles in fungal biology. Curr. Opin. Microbiol. 11, 494–502. doi: 10.1016/j.mib.2008.10.001
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
O'Connell, R. J., Thon, M. R., Hacquard, S., Amyotte, S. G., Kleemann, J., Torres, M. F., et al. (2012). Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 44, 1060–1065. doi: 10.1038/ng.2372
Panwar, V., McCallum, B., and Bakkeren, G. (2013). Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol. Biol. 81, 595–608. doi: 10.1007/s11103-013-0022-7
Qi, T., Guo, J., Peng, H., Liu, P., Kang, Z., and Guo, J. (2019). Host-Induced Gene Silencing: A powerful strategy to control diseases of wheat and barley. Int. J. Mol. Sci. 20:206. doi: 10.3390/ijms20010206
Qi, T., Zhu, X., Tan, C., Liu, P., Guo, J., Kang, Z., et al. (2018). Host-induced gene silencing of an important pathogenicity factor PsCPK1 in Puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust. Plant Biotechnol. J. 16, 797–807. doi: 10.1111/pbi.12829
Rollins, J. A. (2003). The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant-Microbe Interact. 16, 785–795. doi: 10.1094/Mpmi.2003.16.9.785
Sanju, S., Siddappa, S., Thakur, A., Shukla, P. K., Srivastava, N., Pattanayak, D., et al. (2015). Host-mediated gene silencing of a single effector gene from the potato pathogen Phytophthora infestans imparts partial resistance to late blight disease. Funct. Integr. Genomics 15, 697–706. doi: 10.1007/s10142-015-0446-z
Schamber, A., Leroch, M., Diwo, J., Mendgen, K., and Hahn, M. (2010). The role of mitogen-activated protein (MAP) kinase signalling components and the Ste12 transcription factor in germination and pathogenicity of Botrytis cinerea. Mol. Plant Pathol. 11, 105–119. doi: 10.1111/j.1364-3703.2009.00579.x
Seifbarghi, S., Borhan, M. H., Wei, Y., Coutu, C., Robinson, S. J., and Hegedus, D. D. (2017). Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus. BMC Genomics 18:266. doi: 10.1186/s12864-017-3642-5
Sexton, A. C., Cozijnsen, A. J., Keniry, A., Jewell, E., Love, C. G., Batley, J., et al. (2006). Comparison of transcription of multiple genes at three developmental stages of the plant pathogen Sclerotinia sclerotiorum. FEMS Microbiol. Lett. 258, 150–160. doi: 10.1111/j.1574-6968.2006.00212.x
Singh, N., Mukherjee, S. K., and Rajam, M. V. (2020). Silencing of the ornithine decarboxylase gene of Fusarium oxysporum f. sp. lycopersici by host-induced RNAi confers resistance to Fusarium wilt in tomato. Plant Mol. Bio. Rep. 38, 419–429. doi: 10.1007/s11105-020-01205-2
Skamnioti, P., and Gurr, S. J. (2007). Magnaporthe grisea cutinase2 mediates appressorium differentiation and host penetration and is required for full virulence. Plant Cell 19, 2674–2689. doi: 10.1105/tpc.107.051219
Song, Y., and Thomma, B. P. (2018). Host-induced gene silencing compromises Verticillium wilt in tomato and Arabidopsis. Mol. Plant Pathol. 19, 77–89. doi: 10.1111/mpp.12500
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Temme, N., Oeser, B., Massaroli, M., Heller, J., Simon, A., Collado, I. G., et al. (2012). BcAtf1, a global regulator, controls various differentiation processes and phytotoxin production in Botrytis cinerea. Mol. Plant Pathol. 13, 704–718. doi: 10.1111/j.1364-3703.2011.00778.x
Uloth, M. B., Clode, P. L., You, M. P., and Barbetti, M. J. (2016). Attack modes and defence reactions in pathosystems involving Sclerotinia sclerotiorum, Brassica carinata, B. juncea and B. napus. Ann. Bot. 117, 79–95. doi: 10.1093/aob/mcv150
Williams, B., Kabbage, M., Kim, H. J., Britt, R., and Dickman, M. B. (2011). Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7:e1002107. doi: 10.1371/journal.ppat.1002107
Wise, A. A., Liu, Z., and Binns, A. N. (2006). Three methods for the introduction of foreign DNA into Agrobacterium. Methods Mol. Biol. 343, 43–53. doi: 10.1385/1-59745-130-4:43
Xiao, X., Xie, J., Cheng, J., Li, G., Yi, X., Jiang, D., et al. (2014). Novel secretory protein Ss-caf1 of the plant-pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Mol. Plant-Microbe Interact. 27, 40–55. doi: 10.1094/MPMI-05-13-0145-R
Xu, J., Wang, X., Li, Y., Zeng, J., Wang, G., Deng, C., et al. (2018). Host-induced gene silencing of a regulator of G protein signalling gene (VdRGS1) confers resistance to Verticillium wilt in cotton. Plant Biotechnol. J. 16, 1629–1643. doi: 10.1111/pbi.12900
Xu, J. R., and Hamer, J. E. (1996). MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10, 2696–2706. doi: 10.1101/gad.10.21.2696
Xu, L., and Chen, W. (2013). Random T-DNA mutagenesis identifies a Cu/Zn superoxide dismutase gene as a virulence factor of Sclerotinia sclerotiorum. Mol. Plant-Microbe Interact. 26, 431–441. doi: 10.1094/MPMI-07-12-0177-R
Xue, C., Park, G., Choi, W., Zheng, L., Dean, R. A., and Xu, J. R. (2002). Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell 14, 2107–2119. doi: 10.1105/tpc.003426
Yajima, W., Liang, Y., and Kav, N. N. (2009). Gene disruption of an arabinofuranosidase/β-xylosidase precursor decreases Sclerotinia sclerotiorum virulence on canola tissue. Mol. Plant-Microbe Interact. 22, 783–789. doi: 10.1094/MPMI-22-7-0783
Yang, G., Tang, L., Gong, Y., Xie, J., Fu, Y., Jiang, D., et al. (2018). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 217, 739–755. doi: 10.1111/nph.14842
Yin, C., Jurgenson, J. E., and Hulbert, S. H. (2011). Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant-Microbe. Interact. 24, 554–561. doi: 10.1094/mpmi-10-10-0229
Yin, C. T., Downey, S. I., Klages-Mundt, N. L., Ramachandran, S., Chen, X., Szabo, L. J., et al. (2015). Identification of promising host-induced silencing targets among genes preferentially transcribed in haustoria of Puccinia. BMC Genomics 16:579. doi: 10.1186/s12864-015-1791-y
Yu, Y., Du, J., Wang, Y., Zhang, M., Huang, Z., Cai, J., et al. (2019). Survival factor 1 contributes to the oxidative stress response and is required for full virulence of Sclerotinia sclerotiorum. Mol. Plant Pathol. 20, 895–906. doi: 10.1111/mpp.12801
Yu, Y., Xiao, J., Du, J., Yang, Y., Bi, C., and Qing, L. (2016). Disruption of the gene encoding endo-β-1, 4-xylanase affects the growth and virulence of Sclerotinia sclerotiorum. Front. Microbiol. 7:1787. doi: 10.3389/fmicb.2016.01787
Yu, Y., Xiao, J., Zhu, W., Yang, Y., Mei, J., Bi, C., et al. (2017). Ss-Rhs1, a secretory Rhs repeat-containing protein, is required for the virulence of Sclerotinia sclerotiorum. Mol. Plant Pathol. 18, 1052–1061. doi: 10.1111/mpp.12459
Zhang, H., Zheng, X., and Zhang, Z. (2016a). The Magnaporthe grisea species complex and plant pathogenesis. Mol. Plant Pathol. 17, 796–804. doi: 10.1111/mpp.12342
Zhang, T., Jin, Y., Zhao, J. H., Gao, F., Zhou, B. J., Fang, Y. Y., et al. (2016b). Host-induced gene silencing of the target gene in fungal cells confers effective resistance to the cotton wilt disease pathogen Verticillium dahliae. Mol. Plant 9, 939–942. doi: 10.1016/j.molp.2016.02.008
Zhou, B., Bailey, A., Niblett, C., and Qu, R. (2016). Control of brown patch (Rhizoctonia solani) in tall fescue (Festuca arundinacea Schreb.) by host induced gene silencing. Plant Cell Rep. 35, 791–802. doi: 10.1007/s00299-015-1921-7
Keywords: compound appressorium, pathogenicity, host-induced gene silencing, Sclerotinia sclerotiorum, Sscnd1
Citation: Ding Y, Chen Y, Yan B, Liao H, Dong M, Meng X, Wan H and Qian W (2021) Host-Induced Gene Silencing of a Multifunction Gene Sscnd1 Enhances Plant Resistance Against Sclerotinia sclerotiorum. Front. Microbiol. 12:693334. doi: 10.3389/fmicb.2021.693334
Received: 12 April 2021; Accepted: 08 September 2021;
Published: 08 October 2021.
Edited by:
Daohong Jiang, Huazhong Agricultural University, ChinaReviewed by:
Chenggang Wang, University of Florida, United StatesYuemin Pan, Anhui Agricultural University, China
Copyright © 2021 Ding, Chen, Yan, Liao, Dong, Meng, Wan and Qian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Qian, cWlhbndlaTY2NiYjeDAwMDQwO2hvdG1haWwuY29t
 Yijuan Ding
Yijuan Ding Yangui Chen1,2
Yangui Chen1,2 Huafang Wan
Huafang Wan Wei Qian
Wei Qian