- 1College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 2State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Institute of Plant Virology, Ningbo University, Ningbo, China
- 3Key Laboratory of Biotechnology in Plant Protection of MOA of China and Zhejiang Province, Institute of Virology and Biotechnology, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
The light-harvesting chlorophyll a/b complex protein 3 (LHCB3) of photosystem II plays important roles distributing the excitation energy and modulating the rate of state transition and stomatal response to abscisic acid. However, the functions of LHCB3 in plant immunity have not been well investigated. Here, we show that the expression of LHCB3 in Nicotiana benthamiana (NbLHCB3) was down-regulated by turnip mosaic virus (TuMV) infection. When NbLHCB3 was silenced by tobacco rattle virus-induced gene silencing, systemic infection of TuMV was inhibited. H2O2 was over-accumulated in NbLHCB3-silenced plants. Chemical treatment to inhibit or eliminate reactive oxygen species (ROS) impaired the resistance of the NbLHCB3-silenced plants to TuMV infection. Co-silencing of NbLHCB3 with genes involved in ROS production compromised the resistance of plants to TuMV but co-silencing of NbLHCB3 with genes in the ROS scavenging pathway increased resistance to the virus. Transgenic plants overexpressing NbLHCB3 were more susceptible to TuMV. These results indicate that downregulation of NbLHCB3 is involved in defense against TuMV by inducing ROS production.
Introduction
Chloroplasts are not only the organelles where photosynthesis takes place, but also the biosynthetic site of defense-related molecules like reactive oxygen species (ROS) and phytohormones like jasmonic acid (JA) (Fonseca et al., 2009) and salicylic acid (SA) (Strawn et al., 2007). ROS are able to kill pathogens directly by oxidation. The accumulation of ROS is known as the pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) response (Park et al., 2012). Increasing evidence shows that ROS produced by plant plasma membrane respiratory burst oxidase homologs (RBOHs) act as secondary messages which are involved in different signal transduction pathways triggering a physiological or programmed pathway for cell death (Baxter et al., 2014; Schieber and Chandel, 2014; Adachi et al., 2015; Waszczak et al., 2018). Recently Wu et al. (2020) found that a leucine-rich-repeat receptor kinase named hydrogen-peroxide-induced Ca2+ increases (HPCA) is a sensor to perceive extracellular H2O2 and induce activation of Ca2+ channels to trigger an influx of Ca2+ ions in Arabidopsis.
To efficiently harvest solar energy, light-harvesting antenna protein complexes bind chlorophylls and carotenoids and then create a number of light-harvesting chlorophyll a/b binding proteins named LHCBs or LHCAs (Jansson, 1999; Klimmek et al., 2005, 2006). LHCB proteins are associated with the light-harvesting complexes of photosystem II (PS II), while LHCA proteins are associated with those of photosystem I (PS I). PS I is central to the light-driven conversion of water to molecular oxygen, and consists of a PS II core dimer (C2) bound by four trimeric LHCBs (LHCB1–3) that are surrounded by the monomeric LHCB4 (CP29), LHCB5 (CP26), and LHCB6 (CP24) (Caffarri et al., 2009). By modulating the rate of state transitions, LHCB3 is involved in excitation energy transfer and charge separation and is also required for stomatal response to abscisic acid (Damkjaer et al., 2009; Xu et al., 2012; Adamiec et al., 2015). In Arabidopsis, LHCB3 affects the macrostructure of photosystem II and the rate of state transitions (Damkjaer et al., 2009; Kouřil et al., 2013; Pietrzykowska et al., 2014; Longoni et al., 2015), and in tomato and oil palm, the LHCB3 gene confers continuous light tolerance and enhances yield (Velez-Ramirez et al., 2014; Neoh et al., 2019). LHCB3 is involved in cyclic electron flow and then affects state transition by interacting with PGR5a in cucumber (Wu et al., 2021b). However, it is not known whether LHCB3 functions in plant immunity.
We now report that the expression of Nicotiana benthamiana LHCB3 (NbLHCB3) is downregulated in plants infected with turnip mosaic virus (TuMV). When NbLHCB3 was silenced, systemic infection of TuMV was inhibited. Further analysis showed that the accumulated ROS in the NbLHCB3-silenced plants form an essential part of the plant defense against TuMV infection. The results indicate a mechanism in which the downregulation of NbLHCB3 is involved in defense against TuMV by inducing ROS.
Materials and Methods
Plant Materials and Growth Conditions
Nicotiana benthamiana plants were grown in pots with premixed soil in a climate chamber at 26°C, with 60% humidity and under a 16-/8-h photoperiod (light daily from 08:00 to 24:00) with an average light intensity of 130 μmol m–2 s–1.
Viral Inoculation and Detection
Agroinfiltration was done as described before (Li et al., 2019) using Agrobacterium tumefaciens transformed with viral infectious clones infiltrated into N. benthamiana leaves at the stage when three or five true leaves were fully expanded. For mechanical inoculation, N. benthamiana leaves previously infiltrated with agrobacteria carrying an infectious clone and displaying viral symptoms were homogenized in 0.1 M phosphate buffer (pH 7.2) using a sterile pestle and mortar and then filtered. Standardized volumes of infectious sap (or phosphate buffer for controls) were then applied to the upper surface of fully expanded leaves which had been sprayed with carborundum. The distribution of virus in infected tissues or plants was first determined by the intensity of visible green fluorescence under a handheld UV lamp. The transcriptional or translational levels of viral capsid protein were then determined by qRT-PCR, northern blot and western blot using the procedures described below.
Agrobacterium-Mediated Transient Expression in N. benthamiana Leaves
The cDNA fragment of NbLHCB3 was cloned into expression vectors with different tags using ligation-independent cloning (LIC) exactly as described previously (Wang et al., 2019). Recombinant clones were transformed into Agrobacterium GV3101 by electroporation, grown at 28°C overnight, collected by centrifugation, resuspended in infiltration buffer [10 mM MgCl2, 10 mM 2−(N−morpholino) ethanesulfonic acid (MES), and 200 μM acetosyringone, pH 5.6] and kept at room temperature for 4 h. Fully expanded true leaves of N. benthamiana were infiltrated with A. tumefaciens resuspension solution (OD600 = 1.0) and were harvested at 36∼72 hours post-infiltration (hpi) for further research. For co-expression, equal concentrations and equal volumes of individual Agrobacterium cultures were mixed.
Virus-Induced Gene Silencing
The tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) system was used to silence genes in N. benthamiana. This system includes TRV vectors, pYL196 (TRV1) and pYL156 (TRV2), both of which were kindly provided by Prof. Yule Liu at Tsinghua University (Beijing, China) (Liu et al., 2002). Specific primers were designed and used to amplify a ∼300 bp cDNA fragment of the target gene from the N. benthamiana cDNA library. For virus-induced double gene silencing, two fragments were amplified and then linked by homologous recombination. The fragment was then cloned into the vector pYL156 by the LIC method (Wang et al., 2019) to yield the VIGS vector. The negative control vector was prepared using a fragment of the gene encoding glucuronidase. Equal volumes of Agrobacteria carrying pYL196 and pYL156 constructs at OD600 = 1 were mixed and co-infiltrated into N. benthamiana plants. At least 12 plants were used for each silencing treatment and the same number of plants were used as controls. The infiltrated plants were returned to the climate chamber and used for further analysis at about 15 days post-infiltration (dpi).
Total RNA Extraction and RNA Analysis
Total RNA was extracted from leaf tissues of N. benthamiana using Trizol reagent (Thermo Fisher, Waltham, MA, United States) as instructed. DNA contamination was removed and cDNA synthesized using ReverTra Ace® qPCR RT Master Mix with gDNA Remover (TOYOBO, Japan). Real-time PCR was performed in a 10 μL volume containing 2 μL of 25-fold diluted cDNA, 5 μM each primer, and 1 × SYBR Green PCR Mix (Vazyme, China) in a LightCycler®480 II Real-Time PCR System (Roche, Switzerland) and the results were analyzed by the ΔΔCT method (Livak and Schmittgen, 2001). The N. benthamiana Ubiquitin C gene (UBC, AB026056.1) (Rotenberg et al., 2006; Shi et al., 2016) was used as an internal reference for analysis. Four individual plants per treatment were sampled and each treatment was repeated three times independently.
Northern blot of TuMV RNAs was performed as described before (Wang et al., 2019). The genomic RNA of TuMV was detected by a 260-bp fragment of TuMV CP gene labeled with digoxin (DIG) according to the DIG Oligonucleotide 3′-End Labeling Kit protocol (Roche, Switzerland). Ten micrograms of total RNA per sample was loaded on 1.5% formaldehyde-agarose gels and was separated by electrophoresis at 50 V for about 2 h in 1 × MOPS running buffer, and blotted to Hybond-NX nylon membranes (Amersham Biosciences, United Kingdom) for more than 16 h by capillary transfer. Subsequent procedures including pre-hybridization and hybridization were done following the protocol of the DIG High Primer DNA Labeling and Detection Starter Kit II (Roche).
Western Blot
Equal weight samples of N. benthamiana leaves were ground to powder in liquid nitrogen, then resuspended in lysis buffer [100 mM Tris–HCl (pH 8.8), 6% sodium dodecyl sulfate (SDS), 2% β-mercaptoethanol] and centrifuged at 4°C for 12,000 rpm. Total proteins in the supernatants were separated in 12% SDS-PAGE (TGX Stain-Free FastCast Acrylamide Kit, Bio-Rad) and transferred onto 0.45 μm nitrocellulose membrane (Amersham Biosciences, United Kingdom) using semi-dry electroblotting, then incubated with relevant primary antibodies including anti-NbLHCB3 (YouLong Biotech, China), anti-TuMV CP (NEOGEN, United Kingdom), anti-GFP (Transgen Biotech, China) and anti-c-Myc (Gene Tex, Irvine, CA, United States). Secondary antibodies were AP-linked anti-rabbit or anti-mouse (Sigma-Aldrich, St Louis, MO, United States). The antigen-antibody complex was visualized by adding nitrotetrazolium blue chloride/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) (Sigma-Aldrich) at room temperature for a few minutes and results were captured with a Canon digital camera.
ROS Detection and Treatment With Chemical Inhibitors
To detect H2O2 or O2– by histochemical staining, six leaves of each plant were collected at 22:00, immersed and vacuum-infiltrated in freshly prepared solutions of either 3,3′-diaminobenzidine (DAB) [1 mg/ml DAB in Tris–HCl buffer (pH 3.8)] or NBT [0.2% (W/V) NBT in 50 mM PBS (pH 7.5)]. After incubation overnight in darkness at room temperature they were destained in 95% (W/V) ethanol by heating in a boiling water-bath to remove the chlorophyll. Images were captured with a Canon digital camera.
Diphenylene iodonium (DPI) (Mei et al., 2017; Zhang et al., 2020) is a chemical inhibitor of RBOHs. A concentrated stock solution of DPI (10 mM) was prepared by adding 10 mg DPI to 3.179 ml anhydrous dimethylsulphoxide (DMSO). This stock solution was then diluted with Agrobacterium infiltration buffer to provide a final working concentration of 50 μM for infiltration into N. benthamiana leaves.
Dimethylthiourea (DMTU) (Jiang and Zhang, 2002; Li et al., 2016) is a chemical inhibitor of H2O2 and responsible for scavenging H2O2. A concentrated stock solution of DMTU (150 mM) was prepared by adding 0.1563 g DMTU to 10 ml ddH2O. This stock solution was then diluted with Agrobacterium infiltration buffer to provide a final working concentration of 5 mM for infiltration into N. benthamiana leaves.
Results
The Expression of NbLHCB3 Was Downregulated in TuMV-Infected N. benthamiana
In our previous work, we described the transcriptome of N. benthamiana plants infected with TuMV (Wu et al., 2021a). One gene with downregulated expression had the highest identity to AtLHCB3. For further analysis, we therefore first identified this gene from the genomic sequence of N. benthamiana. Using AtLHCB3 (At5G54270) as query, NbLHCB3 (Sequence ID Niben101Scf07510g00016.1) was identified in the Solanaceae Genomics Network.1 It was predicted to encode a protein with 265 amino acids. NbLHCB3 had 88.8% protein identity and 79.9% nucleotide identity to AtLHCB3 (Supplementary Figure 1). To confirm its expression response to TuMV infection, N. benthamiana leaves were mechanically inoculated with GFP-labeled TuMV so that viral infection could easily be monitored by detecting GFP fluorescence (Figure 1A). At 4 dpi, the inoculated leaves (IL) were collected and the expression of NbLHCB3 was determined (Figure 1). There was a significant decrease in accumulation of both NbLHCB3 transcripts and NbLHCB3 protein (Figures 1B,C) compared to mock-inoculated controls, suggesting that NbLHCB3 was downregulated in the TuMV-infected plants.
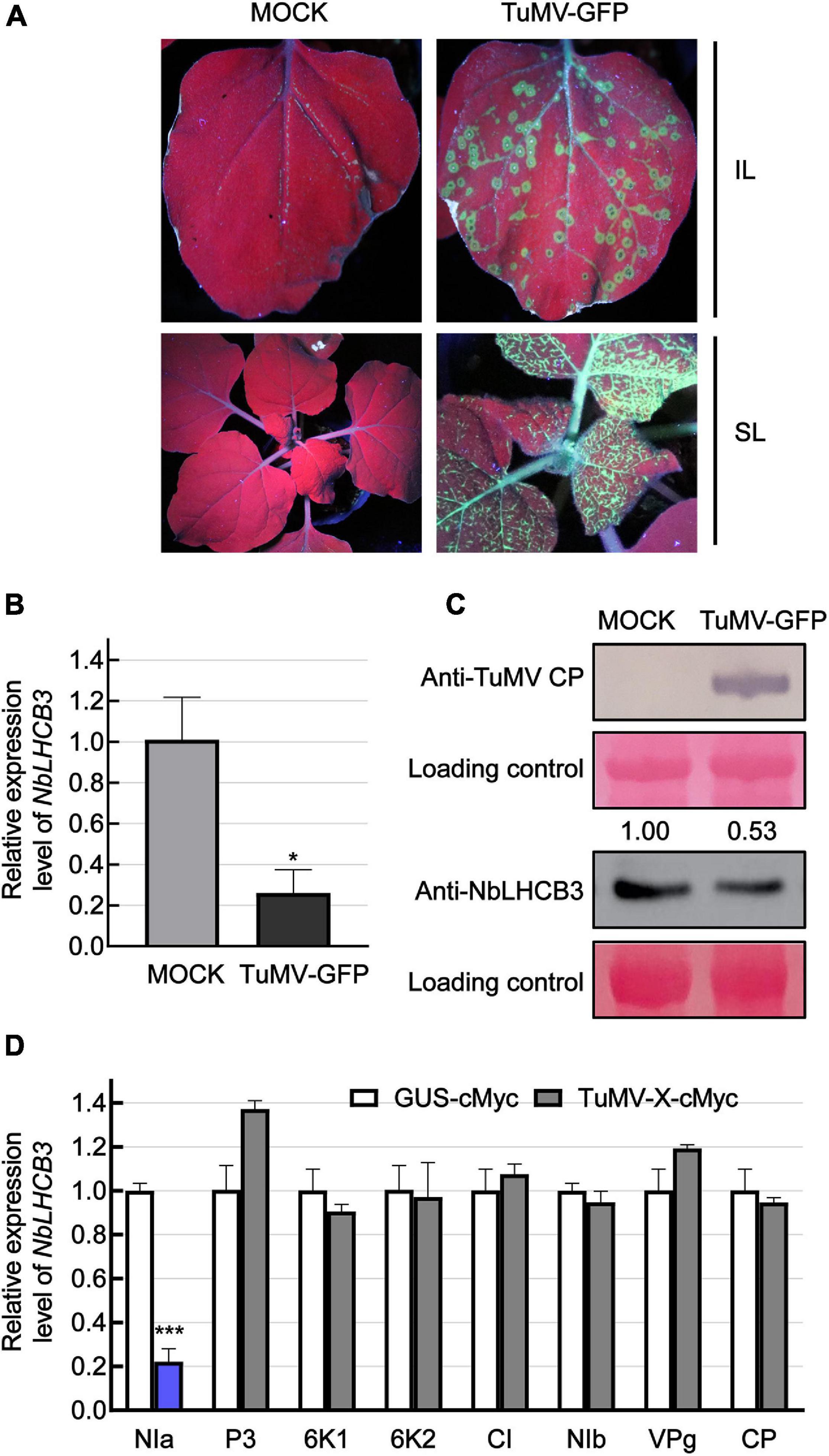
Figure 1. Expression of NbLHCB3 was downregulated in N. benthamiana infected with TuMV-GFP. (A) Fluorescence under UV showing TuMV-GFP infection on inoculated leaves (IL) and systemically infected leaves (SL). Leaves of 2-week-old plants were mechanically inoculated with an extract of TuMV-GFP-infected leaves. Phosphate buffer was used as mock control. Pictures were captured at 4 dpi under UV. (B) qRT-PCR showing the transcript levels of NbLHCB3 in the inoculated leaves of three mock and three TuMV-GFP-infected N. benthamiana plants at 4 dpi. Experiments were repeated three times. (C) Western blot analysis showing TuMV CP and NbLHCB3 accumulation on the systemically infected leaves of TuMV-GFP-infected plants at 4 dpi. The relative intensity of the blot signal quantified by IMAGE J software is shown on the lanes. (D) Results of qRT-PCR showing NbLHCB3 expression in zones from four leaves of four plants expressing different cMyc-fused TuMV proteins. cMyc-fused beta-glucuronidase protein (GUS-cMyc) was used as control. Bars represent the standard errors of the means from three biological repeats. A two-sample unequal variance directional Student’s t-test was used to test the significance of the differences (*P < 0.05; ***P < 0.001).
To determine which viral protein is responsible for downregulated expression of NbLHCB3, we transiently expressed cMyc-fused TuMV proteins individually by agroinfiltration and detected the expression level of NbLHCB3 in leaves at 2 dpi. In infiltrated leaf areas or regions expressing NIa-cMyc, expression of NbLHCB3 was downregulated significantly, but not in those areas or regions expressing the other TuMV proteins, indicating that TuMV NIa is responsible for downregulation of NbLHCB3 in the infected plants (Figure 1D).
Silencing of NbLHCB3 Reduced the Infection of TuMV in N. benthamiana
To investigate the function of NbLHCB3 in plants infected by TuMV, we used the TRV-induced gene silencing system (VIGS), creating a VIGS vector (TRV:LHCB3) to silence NbLHCB3 in at least 12 plants and then mechanically inoculated these plants with TuMV-GFP. At 12 dpi of TRV:LHCB3, the expression of NbLHCB3 was decreased to 30% of the normal (TRV: 00 control) level (Figures 2A,B). We also determined the expression of other members of the NbLHCB family in the TRV:LHCB3-infected plants but found no obvious difference in their expression compared to that in the control TRV:00-infected plants, indicating that NbLHCB3 had been specifically silenced in the TRV:LHCB3-infected plants (Supplementary Figure 2). The silenced plants showed mild yellowing with reduced chlorophyll content (Figures 2A,C). The plants inoculated with TRV:LHCB3 and TRV:00 were then mechanically inoculated with TuMV-GFP. At 4 dpi of TuMV-GFP, there were fewer fluorescent infection foci on the IL of the NbLHCB3-silenced plants than on non-silenced plants (Figure 2A). TuMV RNAs and CP protein in both IL and systemically infected leaves accumulated less in the NbLHCB3-silenced plants than in non-silenced plants (Figure 2D), showing that silencing of NbLHCB3 inhibited TuMV-GFP infection.
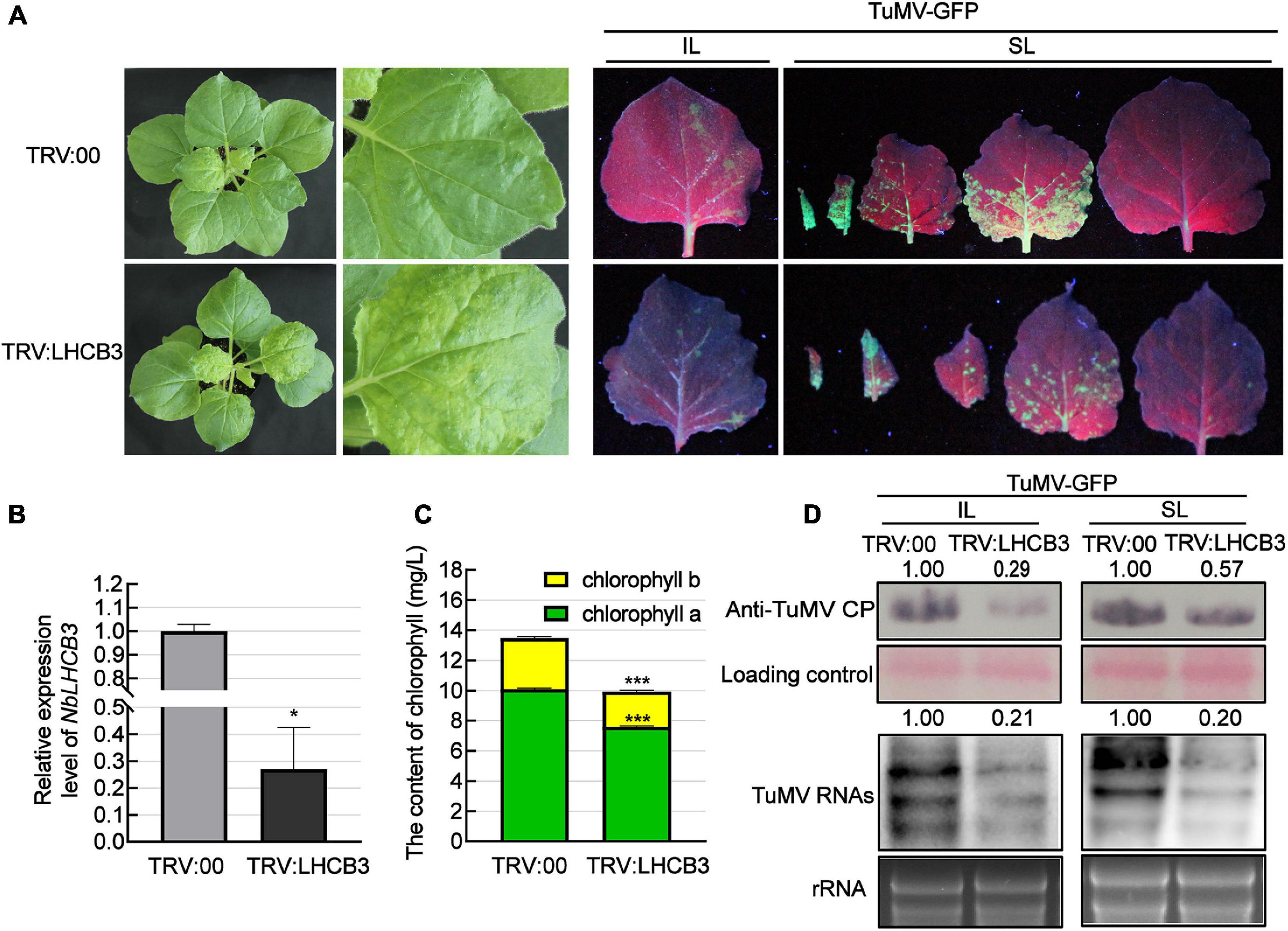
Figure 2. Silencing of NbLHCB3 inhibited the infection of TuMV in N. benthamiana. (A) The phenotype of plants inoculated with TRV:LHCB3 or TRV:00 (Left). Examples of LHCB3-silenced and control plants mechanically inoculated with TuMV-GFP at 12 dpi and photographed at 4 dpi under UV light (Right). Systemically infected leaves (SL) at the corresponding location on both plants were collected for monitoring under UV. (B) Relative expression level of NbLHCB3 in the NbLHCB3-silenced plants. (C) The chlorophyll contents [measured as described before (Yang et al., 2020)] were reduced in the NbLHCB3-silenced plants. (D) Detection of TuMV RNAs and CP protein accumulation in plants inoculated with TRV:LHCB3 or TRV:00 at 4 dpi. Polyclonal antibody to TuMV CP and a CP probe tagged with DIG were used for analysis. Bars represent the standard errors of the means from three biological repeats. The relative intensity of the blot signal quantified by IMAGE J software is shown on the lanes. A two-sample unequal variance directional Student’s t-test was used to test the significance of the differences (*P < 0.05; ***P < 0.001).
ROS Accumulated in the NbLHCB3-Silenced Plants
Light-harvesting chlorophyll a/b binding proteins play a key role in the electron transport chain in chloroplasts, which are the major sites of ROS production (Galvez-Valdivieso and Mullineaux, 2010). In previous reports, ROS levels increased in all the Arabidopsis thaliana lhcb T-DNA insertion mutants (Xu et al., 2012). We next used DAB and NBT staining to determine the H2O2 and O2– levels in the NbLHCB3-silenced plants before and after inoculation with TuMV. H2O2 and O2– accumulated significantly in the NbLHCB3-silenced plants regardless of TuMV infection (Figures 3A–D). We also determined the expression of genes involved in ROS production or scavenging by qRT-PCR in the NbLHCB3-silenced plants and found that genes encoding AO (aldehyde oxidase) (Yergaliyev et al., 2016; Wu et al., 2017) and respiratory burst oxidase homolog C (RBOHC) (Pei et al., 2003) were all expressed more in the NbLHCB3-silenced plants (Figure 3E). By contrast, expression of genes encoding CAT (catalase) (Lee et al., 2017), SOD(CuZn) [superoxide dismutase (CuZn)], SOD(Fe), SOD(Mn) (Miller, 2012), and Glutathione reductase (GR) (Mittler et al., 2004), the major components of ROS scavenging systems, were all significantly downregulated (Figure 3E). These results demonstrate that ROS accumulated in the NbLHCB3-silenced plants.
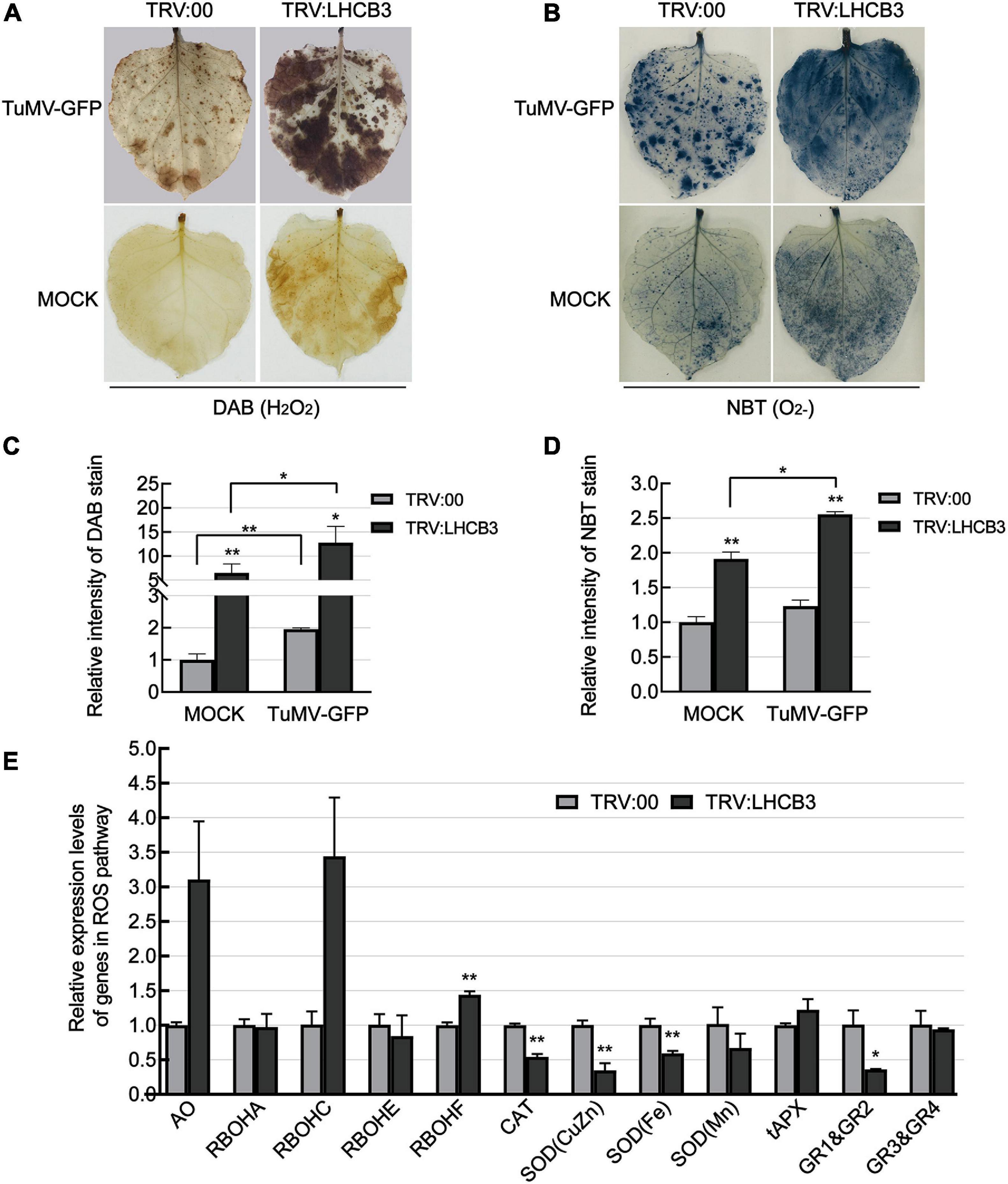
Figure 3. Reactive oxygen species accumulated in the NbLHCB3-silenced plants. (A) Detection of H2O2 by DAB staining in plants inoculated with TRV:LHCB3 or TRV:00 with or without TuMV infection. (B) Detection of O2− by NBT staining in the plants inoculated with TRV:LHCB3 or TRV:00 with our without TuMV infection. (C) The relative DAB stain intensity of the NbLHCB3-silenced and control plants. (D) The relative NBT stain intensity of the NbLHCB3-silenced and control plants. (E) Identification of transcriptional levels of genes in ROS production and scavenging pathways. The relative DAB and NBT stain intensity was quantified by IMAGE J software. Bars represent the standard errors of the means from three biological repeats. A two-sample unequal variance directional Student’s t-test was used to test the significance of the differences (*P < 0.05; **P < 0.01).
Suppressing ROS Accumulation Facilitated TuMV Infection in NbLHCB3-Silenced Plants
To determine whether the accumulation of ROS plays a role against TuMV infection in the NbLHCB3-silenced plants, we treated the silenced plants with 50 μM DPI and 5 mM dimethylthiourea (DMTU) to suppress the production of ROS and eliminate existing ROS, and then examined its effect on TuMV infection (Figure 4). The amount of ROS in the NbLHCB3-silenced leaves was indeed decreased after being treated with DPI and DMTU (Figure 4A). When TuMV-GFP was inoculated, more fluorescent infection foci appeared and there was stronger GFP fluorescence on the IL of the NbLHCB3-silenced plants treated with DPI and DMTU, compared to those of the non-treated NbLHCB3-silenced plants (Figures 4B–D). Furthermore, TuMV RNAs and CP protein accumulated at higher levels in the NbLHCB3-silenced plants treated with DPI and DMTU than in the non-treated NbLHCB3-silenced plants (Figure 4E), indicating that the accumulated ROS in the NbLHCB3-silenced plants plays a role against TuMV infection.
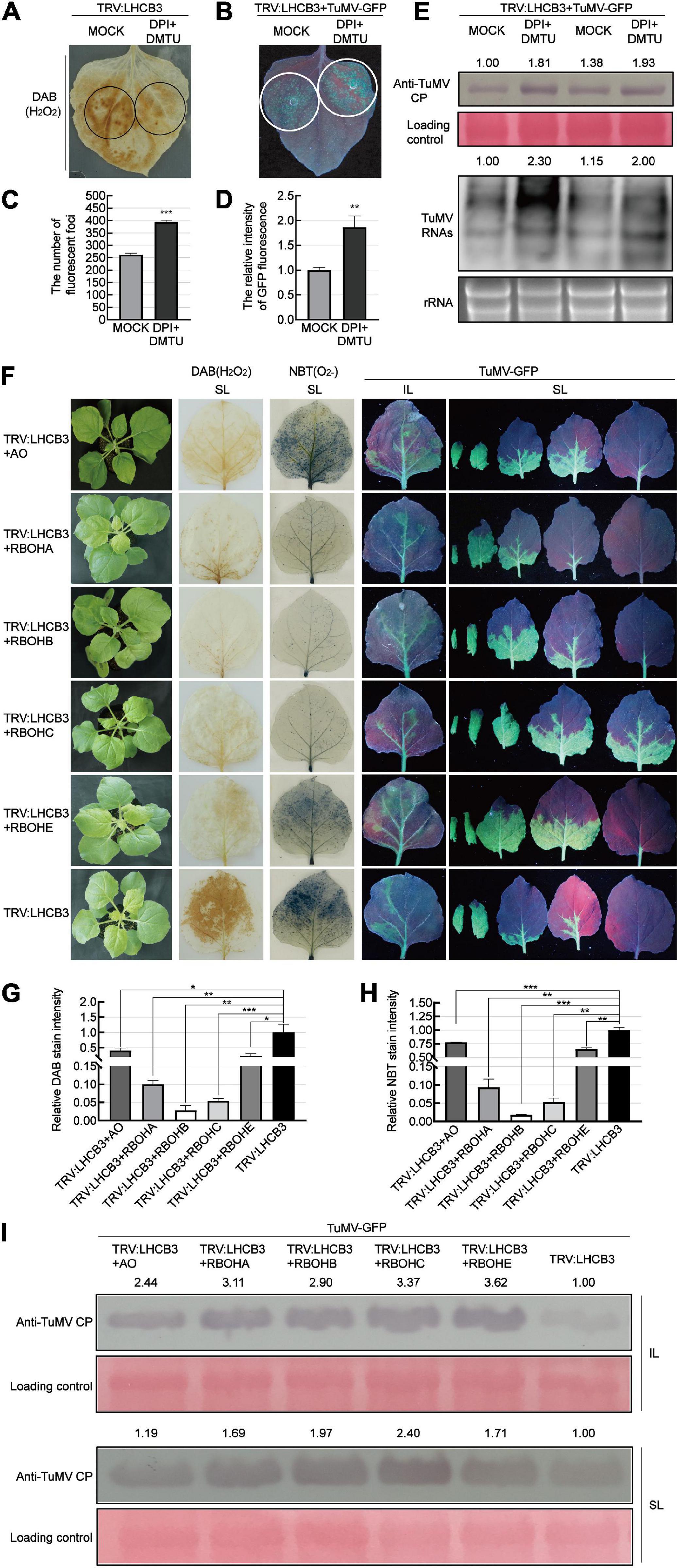
Figure 4. Suppressing ROS accumulation facilitated TuMV infection in the NbLHCB3-silenced plants. (A) DAB staining of ROS accumulation in the TRV:LHCB3 plants after being treated with DPI and DMTU. Treated zones are circled in black. (B) Treatment of DPI and DMTU facilitated TuMV-GFP infection in the NbLHCB3-silenced plants. Treated zones are circled in white. GFP fluorescence is shown under UV light at 4 dpi. (C) The number of fluorescent foci in the zones of NbLHCB3-silenced leaves treated or not with chemical inhibitors of ROS. (D) The relative intensity of GFP fluorescence in the zones of NbLHCB3-silenced leaves treated or not with chemical inhibitors of ROS. (E) Accumulation of TuMV CP protein and RNAs in the TRV:LHCB3 plants treated with DPI or DMTU. (F) TuMV-GFP infection in plants where NbLHCB3 was co-silencing with different genes in the ROS production pathway. The phenotype of plants under bright light is shown at the left. DAB staining and NBT staining show ROS accumulation in the silenced plants. TuMV-GFP infection in the inoculated leaves (IL) and systemically infected leaves (SL) was monitored under UV light at 5 dpi. SL at the corresponding location in plants were collected for analysis. (G) The relative DAB stain intensity calculated by IMAGE J software. (H) The relative NBT stain intensity calculated by IMAGE J software. (I) Western blotting showing the accumulation of TuMV-GFP CP in the plants at 5 dpi. The relative intensity of the blot signal was quantified by IMAGE J software. Bars represent the standard errors of the means from three biological repeats. A two-sample unequal variance directional Student’s t-test was used to test the significance of the differences (*P < 0.05; **P < 0.01; ***P < 0.001).
To further confirm this, we co-silenced NbLHCB3 and each of the individual genes involved in ROS production: AO, RBOHA, RBOHB, RBOHC, and RBOHE. The downregulated expression of these genes in the co-silenced plants was confirmed by qRT-PCR (Supplementary Figures 5A–E). It was expected that silencing of these genes would impair the accumulation of ROS in the NbLHCB3-silenced plants. Indeed, DAB and NBT staining showed a significantly lower accumulation of ROS in these co-silenced plants than in those where only NbLHCB3 was silenced (Figures 4F–H). TuMV-GFP was then mechanically inoculated onto these plants. At 5 dpi of TuMV-GFP infection, GFP fluorescence was stronger on the systemic leaves (SL) of TRV:LHCB3 + AO, TRV:LHCB3 + RBOHA, TRV:LHCB3 + RBOHB, TRV:LHCB3 + RBOHC, and TRV:LHCB3 + RBOHE compared with the TRV:LHCB3 plants (Figure 4F). Consistently, TuMV RNAs and CP protein accumulated at higher levels in both the IL and SL of these co-silenced plants (Figure 4I and Supplementary Figure 3). These results further demonstrate that the accumulated ROS in the NbLHCB3-silenced plants plays a role against TuMV infection.
Co-silencing of Genes in the ROS Scavenging Pathway With NbLHCB3 Increased the Resistance of NbLHCB3-Silenced Plants to TuMV Infection
To further confirm the relationship between ROS and defense against TuMV in the NbLHCB3-silenced plants, we co-silenced genes in the ROS degradative pathway, CAT (CAT functions in H2O2 scavenging), SOD(CuZn), SOD(Fe), SOD(Mn) (SODs play pivotal roles in metabolizing O2– and producing H2O2), tAPX (encodes ascorbate peroxidase which has a similar function to CAT and exists in thylakoids), GR1&GR2 and GR3&GR4, with NbLHCB3, and examined TuMV infection on the silenced plants. The expression of these genes was shown to be significantly downregulated in the co-silenced plants by qRT-PCR (Supplementary Figures 5F–L). We expected that ROS would accumulate more in these co-silenced plants than in those where only NbLHCB3 was silenced, and this was indeed confirmed by DAB and NBT staining (Figures 5A–C). TuMV-GFP was then inoculated onto these plants. At 8 dpi of inoculation, there were fewer fluorescent spots on the IL of TRV:LHCB3 + CAT, TRV:LHCB3 + SOD(CuZn), TRV:LHCB3 + SOD(Fe), TRV:LHCB3 + SOD(Mn), TRV:LHCB3 + tAPX, TRV:LHCB3 + GR1&GR2, and TRV:LHCB3 + GR3&GR4 plants than on the control TRV:LHCB3 plants. Consistently, fluorescent areas on the systemically infected leaves of these plants were small, while the fluorescence intensity was not affected significantly. TuMV RNAs and CP also accumulated at lower levels in these co-silenced plants (Figure 5D and Supplementary Figure 4). These results give further support to the conclusion that the accumulated ROS in the NbLHCB3-silenced plants plays roles against TuMV infection.
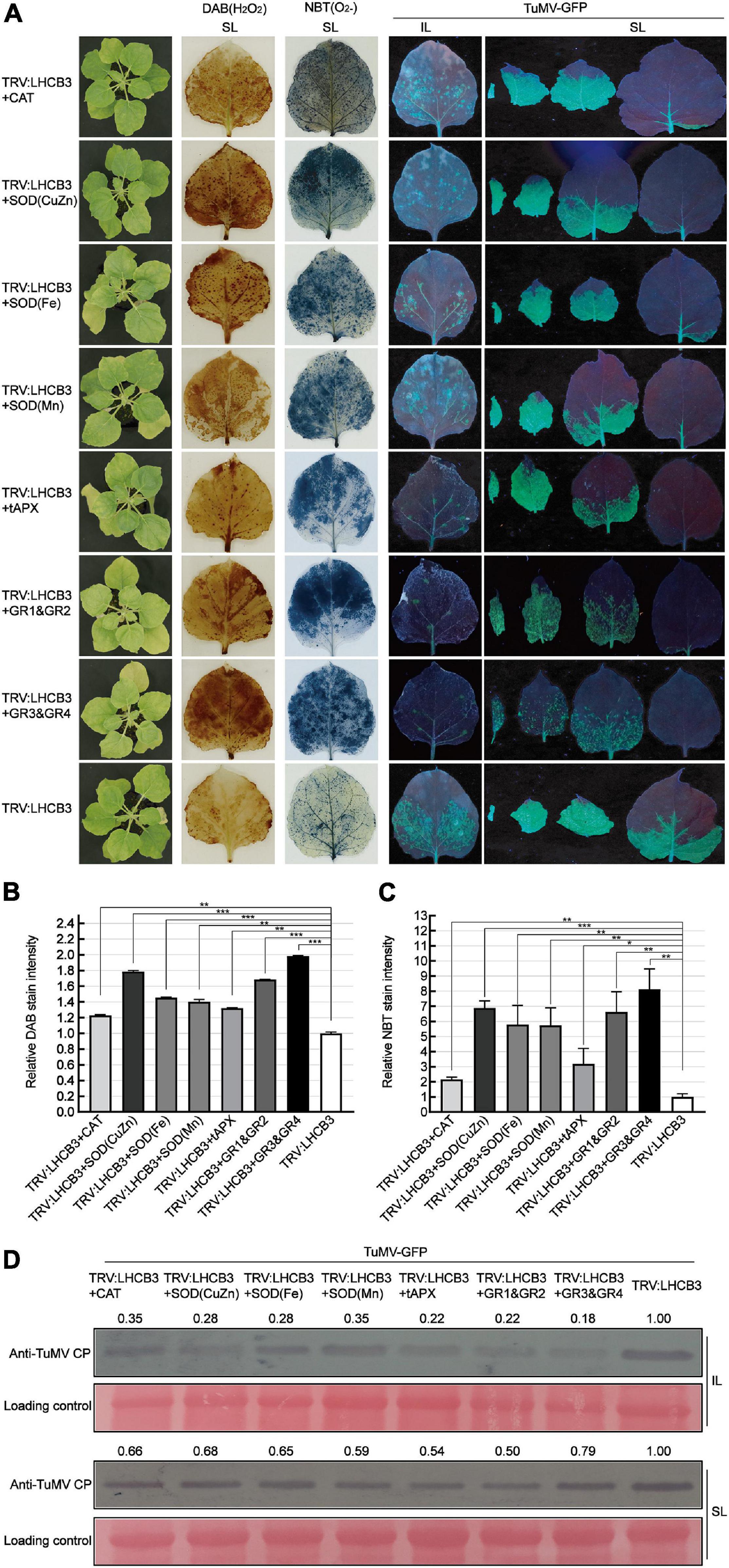
Figure 5. Co-silencing of NbLHCB3 and genes in the ROS scavenging pathway further suppressed TuMV-GFP infection. (A) The phenotype of plants under bright light is shown at the left. DAB and NBT staining show ROS accumulation in the silenced plants. TuMV-GFP infection in the inoculated leaves (IL) and systemically infected leaves (SL) was monitored under UV light at 8 dpi. SL at the corresponding location in plants were collected for analysis. (B) The relative DAB stain intensity calculated by IMAGE J software. (C) The relative NBT stain intensity calculated by IMAGE J software. (D) Western blotting showed the accumulation of TuMV-GFP CP in the plants at 8 dpi. The relative intensity of the blot signal quantified by IMAGE J software is shown on the lanes. Bars represent the standard errors of the means from three biological repeats. A two-sample unequal variance directional Student’s t-test was used to test the significance of the differences (*P < 0.05; **P < 0.01; ***P < 0.001).
Overexpression of NbLHCB3 in N. benthamiana Increased the Accumulation of TuMV RNAs and CP
To further explore the biological function of NbLHCB3 in TuMV-GFP infection, we obtained transgenic N. benthamiana overexpressing NbLHCB3, and detected the accumulation of TuMV in these plants. These transgenic plants contained the full-length ORF of NbLHCB3 driven by the 35S promoter of cauliflower mosaic virus (CaMV) and were obtained by agrobacterium-mediated transformation. Three independent transgenic lines (OE:NbLHCB3 −4, −8 and −9) were identified with about three times increased expression of NbLHCB3 compared to wild type N. benthamiana (Figure 6B) with no obvious developmental phenotype compared to WT (Figure 6A). In these plants, ROS levels were decreased (Figures 6A,C,D). After inoculation with TuMV-GFP, there were more fluorescent spots on the IL at 5 dpi than on the controls (Figure 6A) and fluorescence was also more extensive on their SL (Figure 6A). Blotting analysis showed a significantly increased accumulation of TuMV RNAs and CP on the three transgenic lines, further demonstrating that decreased ROS together with increased expression levels of NbLHCB3 resulted in greater susceptibility to TuMV (Figures 6E,F).
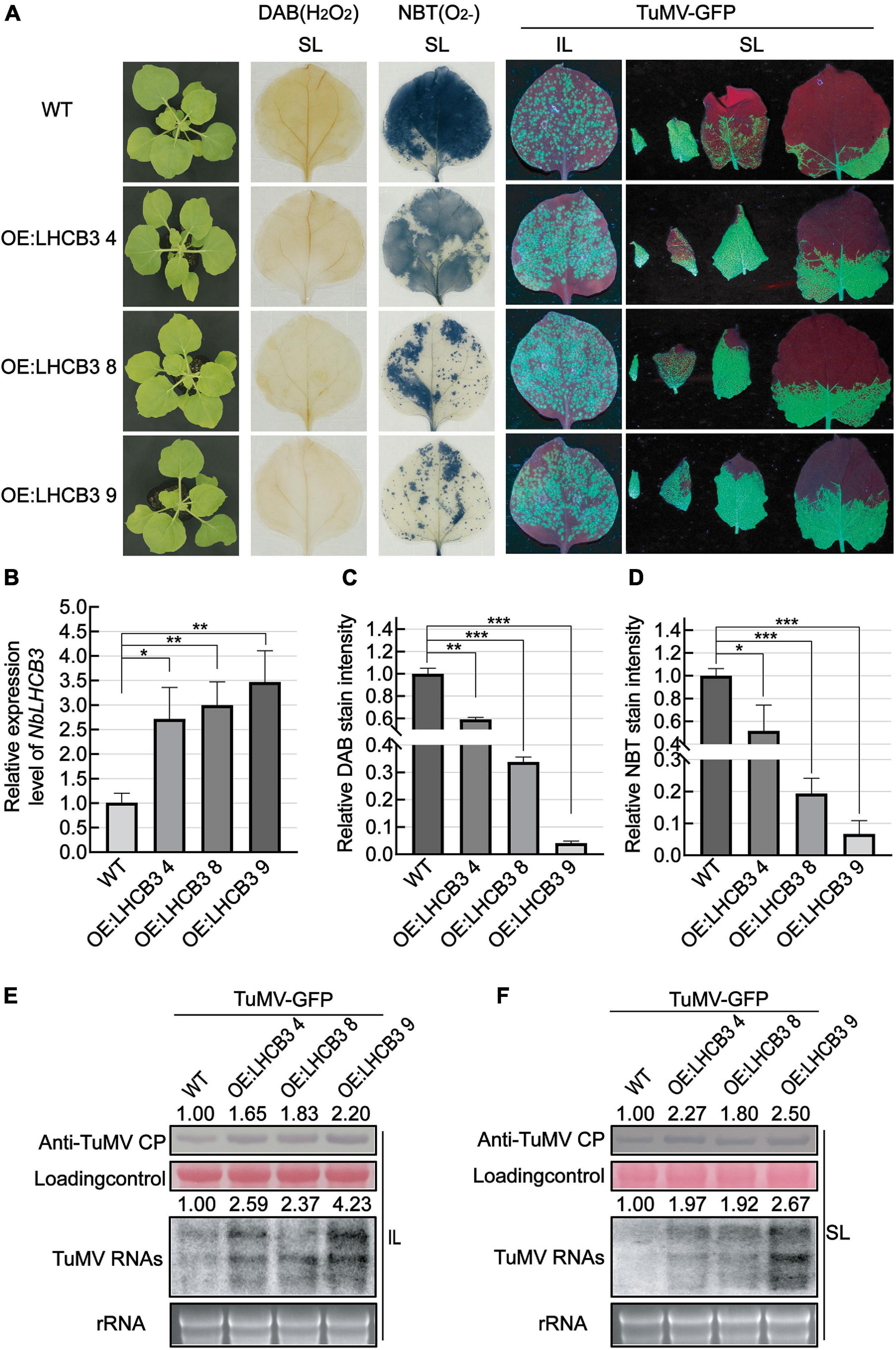
Figure 6. Overexpression of NbLHCB3 in N. benthamiana increased the accumulation of TuMV RNAs and CP. (A) The phenotype of plants under bright light is shown at the left. DAB and NBT staining show less ROS accumulation in the transgenic plants. Fluorescence arising from TuMV-GFP infection on three transgenic lines and wild type (WT) plants: IL and SL were photographed at 5 dpi under UV light. (B) qRT-PCR assay of expression of NbLHCB3 in these three transgenic lines compared to WT. (C) The relative DAB stain intensity in these three transgenic lines compared with WT calculated by IMAGE J software. (D) The relative NBT stain intensity in these three transgenic lines compared with WT calculated by IMAGE J software. (E,F) Accumulation of TuMV RNAs and CP in IL (E) and SL (F). The relative intensity of the blot signal quantified by IMAGE J software is shown on the lanes. Bars represent the standard errors of the means from three biological repeats. A two-sample unequal variance directional Student’s t-test was used to test the significance of the differences (*P < 0.05; **P < 0.01; ***P < 0.001).
Discussion
Viral infections usually cause plant chlorosis, stunting, necrosis, or other symptoms (Shi et al., 2016). Chlorophyll pigmentation can be reduced following virus infection (Grisham et al., 2010), chloroplast structures and functions may be changed (Otulak et al., 2015; Rong et al., 2018), and the expression of nuclear-encoded chloroplast photosynthesis-related genes repressed (Mochizuki et al., 2014; Das et al., 2019; Zhao et al., 2019). All these suggest that viruses and chloroplasts are inextricably linked. Many viral proteins interact directly with chloroplast proteins thus linking chloroplasts to viruses (Ma et al., 2008; Sun et al., 2013; Balasubramaniam et al., 2014; Kong et al., 2014; Gnanasekaran et al., 2019). TuMV also associates with chloroplasts and its successful infection depends on the fusion of virus-induced vesicles with chloroplasts (Wei et al., 2013). In this study we found that chloroplast antenna protein NbLHCB3 was downregulated by TuMV infection and played roles in the interaction between TuMV and plants, providing further evidence of the association of chloroplasts with TuMV infection (Figures 1B,C).
Many studies have shown that viruses from different genera significantly downregulate chloroplast-related genes (ChRGs) including light harvesting genes (Bhattacharyya et al., 2015; Shi et al., 2016; Das et al., 2019; Zhao et al., 2019; Leonetti et al., 2021). The mechanism by which viruses regulate expression of these ChRGs is not well understood. One reported mechanism is RNA silencing, in which small RNAs derived from the viral genome or satellite RNAs target ChRGs for silencing (Shimura et al., 2011; Smith et al., 2011). We here found that the NIa protein of TuMV was responsible for NbLHCB3 reduction, indicating a potential role for NIa in regulating chloroplast function (Figure 1D).
Light-harvesting chlorophyll a/b complex protein 3 interacts with PsbS and affects the macrostructure of photosystem II (Damkjaer et al., 2009; Kouřil et al., 2013; Pietrzykowska et al., 2014; Gerotto et al., 2015; Longoni et al., 2015). PsbS (NPQ4) plays a major role in activating the photoprotection mechanism known as “non-photochemical quenching (NPQ)” which dissipates chlorophyll excited states exceeding the capacity for photosynthetic electron transport (Fan et al., 2015; Gerotto et al., 2015; Correa-Galvis et al., 2016; Tibiletti et al., 2016; Sacharz et al., 2017). The latest research finds that PsbS can also activate photoprotection by some mechanism independent of NPQ (Redekop et al., 2020). OsPsbS-deficient rice generated more superoxide and hydrogen peroxide (Zulfugarov et al., 2014). Consistent with this, silencing of NbLHCB3 here caused ROS accumulation and it seems possible that this was a consequence of the impaired NPQ in the NbLHCB3-silenced plants. Meanwhile, the NbLHCB3-silenced plants also had upregulated expression of genes in ROS production and downregulated expression of genes in the ROS scavenging pathway (Figure 3E). Although the mechanism by which NbLHCB3 regulates the expression of these genes is not yet clear, there is clearly some indirect mechanism to cause ROS accumulation in the NbLHCB3-silenced plants.
Reactive oxygen species produced by RBOHs, and especially RBOHD, appear to be involved in plant immune responses (Sagi and Fluhr, 2001; Yoshioka et al., 2003; Torres et al., 2005; Asai et al., 2008; Lee et al., 2020; Ngou et al., 2021; Yuan et al., 2021). RBOHD is highly responsive to MAMP or pathogen treatments (Morales et al., 2016) and is negatively regulated through phosphorylation and degradation by ubiquitination (Lee et al., 2020). NbRBOHC is homologous to AtRBOHD. Here, the expression of NbRBOHC was upregulated in the NbLHCB3-silenced plants (Figure 3E). Co-silencing of NbRBOHC significantly impaired the inhibition of TuMV in the NbLHCB3-silenced plants, compared to other genes in the ROS production pathway, which indicates that NbRBOHC plays an essential role in the accumulation of ROS in the NbLHCB3-silenced plants (Figure 4).
It has been reported that light deficiency and photosystem impairment increased the susceptibility of N. benthamiana to TuMV infection, suggesting that both light and optimal chloroplast function influence virus infection by limiting the cellular resources needed by TuMV to establish replication complexes (Manfre et al., 2011). However, the effect of the photosystem on viral infection is likely to be complicated since many essential molecules, including ROS and various sugars that all play roles in plant defense, are produced from the photosystem (Bhattacharyya and Chakraborty, 2018; Littlejohn et al., 2021). Deficiencies of different components in the photosystem would have different effects on these molecules, which might result in plants showing different responses to pathogens. In our previous report, silencing of genes in the Calvin cycle limited rice stripe virus (RSV) infection (Bi et al., 2019). We supposed that the energy produced in the Calvin cycle was necessary for normal RSV infection. Here, silencing of NbLHCB3 caused the accumulation of ROS, which was demonstrated to regulate TuMV infection negatively (Figures 3, 4). Both results suggest the different roles of photosystem products in response to viral infection and reflect the complicated functions of chloroplasts in the interaction between viruses and plants.
Reactive oxygen species themselves have also been reported to be harnessed by viral infection and transmission. Red clover necrotic mosaic virus and brome mosaic virus both harness host NbRBOHB-dependent O2– for robust genome replication (Hyodo et al., 2017a,b). For TuMV, ROS is reported to be beneficial to aphid-borne spread (Berthelot et al., 2019; Guo et al., 2019). Taken together with the results of the present study, this suggests an interesting hypothesis that ROS accumulates in TuMV-infected N. benthamiana by down-regulation of NbLHCB3 to inhibit TuMV infection, while the accumulated ROS enhances its transmission by aphids. We will examine this hypothesis next.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
SQ, JC, and FY designed the experiments. SQ, XC, YZ, WC, XA, and SR performed the experiments and interpreted the data. SQ and FY drafted the manuscript. FY and JC revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the Chinese Agriculture Research System (CARS-24-C-04) and the K. C. Wong Education Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mike Adams for manuscript correction.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.690988/full#supplementary-material
Supplementary Figure 1 | Sequence analysis and phylogenetic tree of LHCB homologs from different plant species. (A) Protein sequence alignment of NbLHCB3 and AtLHCB3. (B) Nucleotide sequence alignment of NbLHCB3 and AtLHCB3 cDNA fragments. (C) Phylogenetic tree of LHCB homologs from different plant species. NbLHCB3 is marked with red lines.
Supplementary Figure 2 | Phylogenetic tree of NbLHCBs and relative expression levels of NbLHCBs in the TRV:LHCB3-inoculated plants. (A) Comparison of cDNA sequences and construction of phylogenetic tree of NbLHCBs. NbLHCB3 is marked with red lines. (B) Relative expression levels of NbLHCB family members in the TRV:LHCB3-inoculated plants. Bars represent the standard errors of the means from three biological repeats. A two-sample unequal variance directional Student’s t-test was used to test the significance of the differences (∗P < 0.05).
Supplementary Figure 3 | Northern blotting showing the accumulation of TuMV-GFP RNAs at 5 dpi in the plants where NbLHCB3 was co-silenced with individual genes in the ROS production pathway, AO, RBOHA, RBOHB, RBOHC, and RBOHE. The relative intensity of the blot signal quantified by IMAGE J software is shown on the lanes.
Supplementary Figure 4 | Northern blotting showing the accumulation of TuMV-GFP RNAs at 8 dpi in the plants where NbLHCB3 was co-silenced with individual genes in the ROS scavenging pathway, CAT, SOD(CuZn), SOD(Fe), SOD(Mn), tAPX, GR1&GR2, and GR3&GR4. The relative intensity of the blot signal quantified by IMAGE J software is shown on the lanes.
Supplementary Figure 5 | The expression of LHCB3 and genes in the ROS production and scavenging pathways were all downregulated in these co-silenced plants. (A–E) The expression of LHCB3 and genes in the ROS production pathway (AO, RBOHA, RBOHB, RBOHC, and RBOHE) were downregulated in these co-silenced plants. (F–L) The expression of LHCB3 and genes in the ROS scavenging pathway [CAT, SOD(CuZn), SOD(Fe), SOD(Mn), tAPX, GR1&GR2, and GR3&GR4] were downregulated in these co-silenced plants.
Supplementary Table 1 | Primers used in the experiments.
Supplementary Table 2 | Accession numbers of Nicotiana benthamiana genes.
Footnotes
References
Adachi, H., Nakano, T., Miyagawa, N., Ishihama, N., and Yoshioka, M. (2015). WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27, 2645–2663. doi: 10.1105/tpc.15.00213
Adamiec, M., Gibasiewicz, K., Luciński, R., Giera, W., and Chełminiak, P. (2015). Excitation energy transfer and charge separation are affected in Arabidopsis thaliana mutants lacking light-harvesting chlorophyll a/b binding protein Lhcb3. J. Photochem. Photobiol. B 153, 423–428. doi: 10.1016/j.jphotobiol.2015.11.002
Asai, S., Ohta, K., and Yoshioka, H. (2008). MAPK signaling regulates nitric oxide and NADPH Oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20, 1390–1406. doi: 10.1105/tpc.107.055855
Balasubramaniam, M., Kim, B., Hutchens-Williams, H. M., and Loesch-Fries, L. S. (2014). The photosystem II Oxygen-Evolving complex protein PsbP interacts with the coat protein of Alfalfa mosaic virus and inhibits virus replication. Mol. Plant Microbe Interact 27, 1107–1118. doi: 10.1094/MPMI-02-14-0035-R
Baxter, A., Mittler, R., and Suzuki, N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. doi: 10.1093/jxb/ert375
Berthelot, E., Ducousso, M., Macia, J. L., Bogaert, F., and Baecker, V. (2019). Turnip mosaic virus is a second example of a virus using transmission activation for Plant-to-Plant propagation by aphids. J. Virol. 93, e01822–18. doi: 10.1128/JVI.01822-18
Bhattacharyya, D., and Chakraborty, S. (2018). Chloroplast: the Trojan horse in plant-virus interaction. Mol. Plant Pathol. 19, 504–518. doi: 10.1111/mpp.12533
Bhattacharyya, D., Gnanasekaran, P., Kumar, R. K., Kushwaha, N. K., and Sharma, V. K. (2015). A geminivirus betasatellite damages the structural and functional integrity of chloroplasts leading to symptom formation and inhibition of photosynthesis. J. Exp. Bot. 66, 5881–5895. doi: 10.1093/jxb/erv299
Bi, J., Yang, Y., Chen, B., Zhao, J., and Chen, Z. (2019). Retardation of the calvin cycle contributes to the reduced CO2 assimilation ability of Rice stripe virus-infected N. benthamiana and suppresses viral infection. Front. Microbiol. 10:568. doi: 10.3389/fmicb.2019.00568
Caffarri, S., Kouril, R., Kereïche, S., Boekema, E. J., and Croce, R. (2009). Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28, 3052–3063. doi: 10.1038/emboj.2009.232
Correa-Galvis, V., Poschmann, G., Melzer, M., Stühler, K., and Jahns, P. (2016). PsbS interactions involved in the activation of energy dissipation in Arabidopsis. Nat. Plants 2:15225. doi: 10.1038/nplants.2015.225
Damkjaer, J. T., Kereïche, S., Johnson, M. P., Kovacs, L., and Kiss, A. Z. (2009). The photosystem II Light-Harvesting protein lhcb3 affects the macrostructure of photosystem II and the rate of state transitions in Arabidopsis. Plant Cell 21, 3245–3256. doi: 10.1105/tpc.108.064006
Das, P. P., Lin, Q., and Wong, S. M. (2019). Comparative proteomics of Tobacco mosaic virus-infected Nicotiana tabacum plants identified major host proteins involved in photosystems and plant defence. J. Proteomics 194, 191–199. doi: 10.1016/j.jprot.2018.11.018
Fan, M., Li, M., Liu, Z., Cao, P., and Pan, X. (2015). Crystal structures of the PsbS protein essential for photoprotection in plants. Nat. Struct. Mol. Biol. 22, 729–735. doi: 10.1038/nsmb.3068
Fonseca, S., Chini, A., Hamberg, M., Adie, B., and Porzel, A. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. doi: 10.1038/nchembio.161
Galvez-Valdivieso, G., and Mullineaux, P. M. (2010). The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol. Plant 138, 430–439. doi: 10.1111/j.1399-3054.2009.01331.x
Gerotto, C., Franchin, C., Arrigoni, G., and Morosinotto, T. (2015). In vivo identification of photosystem II light harvesting complexes interacting with PHOTOSYSTEM II SUBUNIT s. Plant Physiol. 168, 1747–1761. doi: 10.1104/pp.15.00361
Gnanasekaran, P., Ponnusamy, K., and Chakraborty, S. (2019). A geminivirus betasatellite encoded βC1 protein interacts with PsbP and subverts PsbP-mediated antiviral defence in plants. Mol. Plant Pathol. 20, 943–960. doi: 10.1111/mpp.12804
Grisham, M. P., Johnson, R. M., and Zimba, P. V. (2010). Detecting Sugarcane yellow leaf virus infection in asymptomatic leaves with hyperspectral remote sensing and associated leaf pigment changes. J. Virol. Methods 167, 140–145. doi: 10.1016/j.jviromet.2010.03.024
Guo, H., Gu, L., Liu, F., Chen, F., and Ge, F. (2019). Aphid-borne viral spread is enhanced by virus-induced accumulation of plant reactive oxygen species. Plant Physiol. 179, 143–155. doi: 10.1104/pp.18.00437
Hyodo, K., Hashimoto, K., Kuchitsu, K., Suzuki, N., and Okuno, T. (2017a). Harnessing host ROS-generating machinery for the robust genome replication of a plant RNA virus. Proc. Natl. Acad. Sci. U. S. A. 114, E1282–E1290. doi: 10.1073/pnas.1610212114
Hyodo, K., Suzuki, N., Mise, K., and Okuno, T. (2017b). Roles of superoxide anion and hydrogen peroxide during replication of two unrelated plant RNA viruses in Nicotiana benthamiana. Plant Signal. Behav. 12:e1338223. doi: 10.1080/15592324.2017.1338223
Jansson, S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4, 236–240. doi: 10.1016/s1360-1385(99)01419-3
Jiang, M., and Zhang, J. (2002). Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 53, 2401–2410. doi: 10.1093/jxb/erf090
Klimmek, F., Ganeteg, U., Ihalainen, J. A., van Roon, H., and Jensen, P. E. (2005). Structure of the higher plant light harvesting complex I: in vivo characterization and structural interdependence of the Lhca proteins. Biochemistry 44, 3065–3073. doi: 10.1021/bi047873g
Klimmek, F., Sjödin, A., Noutsos, C., Leister, D., and Jansson, S. (2006). Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol. 140, 793–804. doi: 10.1104/pp.105.073304
Kong, L., Wu, J., Lu, L., Xu, Y., and Zhou, X. (2014). Interaction between Rice stripe virus Disease-Specific Protein and Host PsbP Enhances Virus Symptoms. Mol. Plant 7, 691–708. doi: 10.1093/mp/sst158
Kouřil, R., Wientjes, E., Bultema, J. B., Croce, R., and Boekema, E. J. (2013). High-light vs. Low-light: effect of light acclimation on photosystem II composition and organization in Arabidopsis thaliana. Biochim. Biophys. Acta 1827, 411–419. doi: 10.1016/j.bbabio.2012.12.003
Lee, D., Lal, N. K., Lin, Z. D., Ma, S., and Liu, J. (2020). Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat. Commun. 11:1838. doi: 10.1038/s41467-020-15601-5
Lee, D. J., Choi, H. J., Moon, M. E., Chi, Y. T., and Ji, K. Y. (2017). Superoxide serves as a putative signal molecule for plant cell division: overexpression of CaRLK1 promotes the plant cell cycle via accumulation of O2- and decrease in H2O2. Physiol. Plant. 159, 228–243. doi: 10.1111/ppl.12487
Leonetti, P., Ghasemzadeh, A., Consiglio, A., Gursinsky, T., and Behrens, S. E. (2021). Endogenous activated small interfering RNAs in virus-infected Brassicaceae crops show a common host gene-silencing pattern affecting photosynthesis and stress response. New Phytol. 229, 1650–1664. doi: 10.1111/nph.16932
Li, Q., Wang, Z., Zhao, Y., Zhang, X., and Zhang, S. (2016). Putrescine protects hulless barley from damage due to UV-B stress via H2S- and H2O2-mediated signaling pathways. Plant Cell Rep. 35, 1155–1168. doi: 10.1007/s00299-016-1952-8
Li, S., Zhao, J., Zhai, Y., Yuan, Q., and Zhang, H. (2019). The hypersensitive induced reaction 3 (HIR3) gene contributes to plant basal resistance via an EDS1 and salicylic acid-dependent pathway. Plant J. 98, 783–797. doi: 10.1111/tpj.14271
Littlejohn, G. R., Breen, S., Smirnoff, N., and Grant, M. (2021). Chloroplast immunity illuminated. New Phytol. 229, 3088–3107. doi: 10.1111/nph.17076
Liu, Y., Schiff, M., and Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313x.2002.01394.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using Real-Time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Longoni, P., Douchi, D., Cariti, F., Fucile, G., and Goldschmidt-Clermont, M. (2015). Phosphorylation of the Light-Harvesting complex II isoform lhcb2 is central to state transitions. Plant Physiol. 169, 2874–2883. doi: 10.1104/pp.15.01498
Ma, Y., Zhou, T., Hong, Y., Fan, Z., and Li, H. (2008). Decreased level of ferredoxin I in Tobacco mosaic virus-infected tobacco is associated with development of the mosaic symptom. Physiol. Mol. Plant 72, 39–45. doi: 10.1016/j.pmpp.2008.05.004
Manfre, A., Glenn, M., Nuñez, A., Moreau, R. A., and Dardick, C. (2011). Light quantity and photosystem function mediate host susceptibility to Turnip mosaic virus via a salicylic acid-independent mechanism. Mol. Plant Microbe Interact 24, 315–327. doi: 10.1094/MPMI-08-10-0191
Mei, Y., Chen, H., Shen, W., Shen, W., and Huang, L. (2017). Hydrogen peroxide is involved in hydrogen sulfide-induced lateral root formation in tomato seedlings. BMC Plant Biol. 17:162. doi: 10.1186/s12870-017-1110-7
Miller, A. (2012). Superoxide dismutases: ancient enzymes and new insights. FEBS Lett. 586, 585–595. doi: 10.1016/j.febslet.2011.10.048
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Mochizuki, T., Ogata, Y., Hirata, Y., and Ohki, S. T. (2014). Quantitative transcriptional changes associated with chlorosis severity in mosaic leaves of tobacco plants infected with Cucumber mosaic virus. Mol. Plant Pathol. 15, 242–254. doi: 10.1111/mpp.12081
Morales, J., Kadota, Y., Zipfel, C., Molina, A., and Torres, M. A. (2016). The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J. Exp. Bot. 67, 1663–1676. doi: 10.1093/jxb/erv558
Neoh, B. K., Wong, Y. C., Teh, H. F., Ng, T., and Tiong, S. H. (2019). Diurnal biomarkers reveal key photosynthetic genes associated with increased oil palm yield. PLoS One 14:e213591. doi: 10.1371/journal.pone.0213591
Ngou, B. P. M., Ahn, H., Ding, P., and Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
Otulak, K., Chouda, M., Bujarski, J., and Garbaczewska, G. (2015). The evidence of Tobacco rattle virus impact on host plant organelles ultrastructure. Micron 70, 7–20. doi: 10.1016/j.micron.2014.11.007
Park, C. H., Chen, S., Shirsekar, G., Zhou, B., and Khang, C. H. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24, 4748–4762. doi: 10.1105/tpc.112.105429
Pei, Z., Bloom, R. E., Schroeder, J. I., Torres, M. A., and Jones, J. D. G. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22, 2623–2633. doi: 10.1093/emboj/cdg277
Pietrzykowska, M., Suorsa, M., Semchonok, D. A., Tikkanen, M., and Boekema, E. J. (2014). The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell 26, 3646–3660. doi: 10.1105/tpc.114.127373
Redekop, P., Rothhausen, N., Rothhausen, N., Melzer, M., and Mosebach, L. (2020). PsbS contributes to photoprotection in Chlamydomonas reinhardtii independently of energy dissipation. Biochim. Biophys. Acta Bioenerg. 1861:148183. doi: 10.1016/j.bbabio.2020.148183
Rong, W., Wang, X., Wang, X., Massart, S., and Zhang, Z. (2018). Molecular and ultrastructural mechanisms underlying yellow dwarf symptom formation in wheat after infection of Barley yellow dwarf virus. Int. J. Mol. Sci. 19:1187. doi: 10.3390/ijms19041187
Rotenberg, D., Thompson, T. S., German, T. L., and Willis, D. K. (2006). Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J. Virol. Methods 138, 49–59. doi: 10.1016/j.jviromet.2006.07.017
Sacharz, J., Giovagnetti, V., Ungerer, P., Mastroianni, G., and Ruban, A. V. (2017). The xanthophyll cycle affects reversible interactions between PsbS and light-harvesting complex II to control non-photochemical quenching. Nat. Plants 3:16225. doi: 10.1038/nplants.2016.225
Sagi, M., and Fluhr, R. (2001). Superoxide production by plant homologues of the gp91phox NADPH oxidase. Modulation of activity by calcium and by tobacco mosaic virus infection1. Plant Physiol. 126, 1281–1290. doi: 10.1104/pp.126.3.1281
Schieber, M., and Chandel, N. S. (2014). ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–R462. doi: 10.1016/j.cub.2014.03.034
Shi, B., Lin, L., Wang, S., Guo, Q., and Zhou, H. (2016). Identification and regulation of host genes related to Rice stripe virus symptom production. New Phytol. 209, 1106–1119. doi: 10.1111/nph.13699
Shimura, H., Pantaleo, V., Ishihara, T., Myojo, N., and Inaba, J. (2011). A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 7:e1002021. doi: 10.1371/journal.ppat.1002021
Smith, N. A., Eamens, A. L., and Wang, M. B. (2011). Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 7:e1002022. doi: 10.1371/journal.ppat.1002022
Strawn, M. A., Marr, S. K., Inoue, K., Inada, N., and Zubieta, C. (2007). Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J. Biol. Chem. 282, 5919–5933. doi: 10.1074/jbc.M605193200
Sun, X., Li, Y., Shi, M., Zhang, N., and Wu, G. (2013). In vitro binding and bimolecular fluorescence complementation assays suggest an interaction between tomato mosaic virus coat protein and tobacco chloroplast ferredoxin I. Arch. Virol. 158, 2611–2615. doi: 10.1007/s00705-013-1778-4
Tibiletti, T., Auroy, P., Peltier, G., and Caffarri, S. (2016). Chlamydomonas reinhardtii PsbS Protein is functional and accumulates rapidly and transiently under high light. Plant Physiol. 171, 2717–2730. doi: 10.1104/pp.16.00572
Torres, M. A., Jones, J. D., and Dangl, J. L. (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37, 1130–1134. doi: 10.1038/ng1639
Velez-Ramirez, A. I., van Ieperen, W., Vreugdenhil, D., van Poppel, P. M. J. A., and Heuvelink, E. (2014). A single locus confers tolerance to continuous light and allows substantial yield increase in tomato. Nat. Commun. 5:4549. doi: 10.1038/ncomms5549
Wang, S., Han, K., Peng, J., Zhao, J., and Jiang, L. (2019). NbALD1 mediates resistance to Turnip mosaic virus by regulating the accumulation of salicylic acid and the ethylene pathway in Nicotiana benthamiana. Mol. Plant Pathol. 20, 990–1004. doi: 10.1111/mpp.12808
Waszczak, C., Carmody, M., and Kangasjärvi, J. (2018). Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 69, 209–236. doi: 10.1146/annurev-arplant-042817-040322
Wei, T., Zhang, C., Hou, X., Sanfaçon, H., and Wang, A. (2013). The SNARE protein Syp71 is essential for Turnip mosaic virus infection by mediating fusion of virus-induced vesicles with chloroplasts. PLoS Pathog. 9:e1003378. doi: 10.1371/journal.ppat.1003378
Wu, F., Chi, Y., Jiang, Z., Xu, Y., and Xie, L. (2020). Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 578, 577–581. doi: 10.1038/s41586-020-2032-3
Wu, J., Yang, R., Yang, Z., Yao, S., and Zhao, S. (2017). ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants 3:16203. doi: 10.1038/nplants.2016.203
Wu, X., Lai, Y., Rao, S., Lv, L., and Ji, M. (2021a). Genome-wide identification reveals that Nicotiana benthamiana hypersensitive response (HR)-like lesion inducing protein 4 (NbHRLI4) mediates cell death and salicylic acid-dependent defense responses to Turnip mosaic virus. Front. Plant Sci. 12:627315. doi: 10.3389/fpls.2021.627315
Wu, X., Wu, J., Wang, Y., He, M., and He, M. (2021b). The key cyclic electron flow protein PGR5 associates with cytochrome b6f, and its function is partially influenced by the LHCII state transition. Hortic. Res. 8:55. doi: 10.1038/s41438-021-00460-y
Xu, Y., Liu, R., Yan, L., Liu, Z., and Jiang, S. (2012). Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 63, 1095–1106. doi: 10.1093/jxb/err315
Yang, X., Lu, Y., Wang, F., Chen, Y., and Tian, Y. (2020). Involvement of the chloroplast gene ferredoxin 1 in multiple responses of Nicotiana benthamiana to Potato virus X infection. J. Exp. Bot. 71, 2142–2156. doi: 10.1093/jxb/erz565
Yergaliyev, T. M., Nurbekova, Z., Mukiyanova, G., Akbassova, A., and Sutula, M. (2016). The involvement of ROS producing aldehyde oxidase in plant response to Tombusvirus infection. Plant Physiol. Biochem. 109, 36–44. doi: 10.1016/j.plaphy.2016.09.001
Yoshioka, H., Numata, N., Nakajima, K., Katou, S., and Kawakita, K. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15, 706–718. doi: 10.1105/tpc.008680
Yuan, M., Jiang, Z., Bi, G., Nomura, K., and Liu, M. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-6
Zhang, X. N., Liao, Y. W., Wang, X. R., Zhang, L., and Ahammed, G. J. (2020). Epigallocatechin-3-gallate enhances tomato resistance to Tobacco mosaic virus by modulating RBOH1-dependent H2O2 signaling. Plant Physiol. Biochem. 150, 263–269. doi: 10.1016/j.plaphy.2020.03.008
Zhao, J., Xu, J., Chen, B., Cui, W., and Zhou, Z. (2019). Characterization of proteins involved in chloroplast targeting disturbed by Rice stripe virus by novel protoplast–chloroplast proteomics. Int. J. Mol. Sci. 20:253. doi: 10.3390/ijms20020253
Keywords: turnip mosaic virus, TuMV, LHCB3, reactive oxygen species, ROS
Citation: Qiu S, Chen X, Zhai Y, Cui W, Ai X, Rao S, Chen J and Yan F (2021) Downregulation of Light-Harvesting Complex II Induces ROS-Mediated Defense Against Turnip Mosaic Virus Infection in Nicotiana benthamiana. Front. Microbiol. 12:690988. doi: 10.3389/fmicb.2021.690988
Received: 06 April 2021; Accepted: 14 June 2021;
Published: 05 July 2021.
Edited by:
Fangfang Li, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Xian Zhang, Hangzhou Normal University, ChinaJianzhong Liu, Zhejiang Normal University, China
Yongliang Zhang, China Agricultural University, China
Copyright © 2021 Qiu, Chen, Zhai, Cui, Ai, Rao, Chen and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Yan, eWFuZmVpQG5idS5lZHUuY24=; Jianping Chen, amlhbnBpbmdjaGVuQG5idS5lZHUuY24=
 Shiyou Qiu
Shiyou Qiu Xuwei Chen2
Xuwei Chen2 Weijun Cui
Weijun Cui Jianping Chen
Jianping Chen Fei Yan
Fei Yan