- 1College of Grassland and Resources and Environment, Inner Mongolia Agricultural University, Hohhot, China
- 2Grassland Research Institute, Chinese Academy of Agricultural Sciences, Hohhot, China
- 3College of Agriculture, Inner Mongolia University for Nationalities, Tongliao, China
This study investigated the fermentation quality of alfalfa grown in different salt stress regions in China. Following the production of silage from the natural fermentation of alfalfa, the interplay between the chemical composition, fermentation characteristics, and microbiome was examined to understand the influence of these factors on the fermentation quality of silage. The alfalfa was cultivated under salt stress with the following: (a) soil content of <1%0 (CK); (b) 1–2%0 (LS); (c) 2–3%0 (MS); (d) 3–4%0 (HS). The pH of the silage was high (4.9–5.3), and lactic acid content was high (26.3–51.0 g/kg DM). As the salt stress increases, the NA+ of the silages was higher (2.2–5.4 g/kg DM). The bacterial alpha diversities of the alfalfa silages were distinct. There was a predominance of desirable genera including Lactococcus and Lactobacillus in silage produced from alfalfa under salt stress, and this led to better fermentation quality. The chemical composition and fermentation characteristics of the silage were closely correlated with the composition of the bacterial community. Furthermore, NA+ was found to significantly influence the microbiome of the silage. The results confirmed that salt stress has a great impact on the quality and bacterial community of fresh alfalfa and silage. The salt stress and plant ions were thus most responsible for their different fermentation modes in alfalfa silage. The results of the study indicate that exogenous epiphytic microbiota of alfalfa under salt stress could be used as a potential bioresource to improve the fermentation quality.
Introduction
Alfalfa (Medicago sativa L.) is one of the most important forage, with high protein and highly digestible fiber contents and salinization tolerance. Not only is alfalfa showing high salt tolerance to saline-alkali soil, but appropriate salt stress would improve the quality of alfalfa as well, including amino acids, proteins, and other important nutrients (Liu et al., 2019). Soil salinization has become an important factor in constraining global food and forage crops (Lu et al., 2021). According to statistics from UNESCO and FAO, soil salinization has a presence in more than 100 countries around the world, with an area of 932.2 Mha (Rengasamy, 2006). Soil salinization is an issue affecting the development of global agriculture and animal husbandry (Xiao et al., 2019).
In the salinization land, alfalfa can not only supply adequate forage resources but also provide a large amount of high-quality protein feed for animal husbandry (Guo et al., 2013). In particular, competition for ionic factors such as K+, Na+, and Cl− would lead to plant nutrient imbalance (Mahmoud et al., 2017). It has been documented that excessive concentrations of Na+ in plant tissues prevent nutrient and osmotic balance, leading to specific ionic toxicity (Hariadi et al., 2010). Moreover, from the perspective of plant chemical composition metabolism, with the increase of salt stress, the content of proline in crops also gradually increased (Sanada et al., 1995). In the study by Xue et al. (2018) tomato's soluble sugar and soluble protein eventually got an increasing trend in response to salt stress. Soil salinization forage is a potential and huge source of forage. The demand for alfalfa is large, but production is affected by seasonal harvest. In order to combat this and meet the year-round demand, alfalfa needs proper pretreatment and conservation.
Ensiling has been recognized as a good way of keeping forage nutrition, and as this method preserves protein, it improves forage palatability for livestock as well (McDonald et al., 1991). The quality of silage mainly depends on LAB (lactic acid bacteria). LAB could inhibit not only the growth of spoilage bacteria but also WSC (catabolize water-soluble carbohydrates) to lactic acid and decrease pH (Li et al., 2018). Lower pH would inhibit spoilage microorganisms, including primary members of Enterobacteriaceae, clostridial spores, yeast, and molds (Bolsen et al., 1996). From a silage preservation perspective, the loss of dry matter (DM) is also critical. Therefore, making use of microbial resources to carry out fermentation and consume the least energy to preserve nutrients is very important.
However, the biotechnological potential of LAB involved in silage fermentation remains poorly explored. To seek the better fermentation effect as we provided, we need to excavate the microbial resources in different habitats to obtain LAB. Indigenous strains could be isolated from the habitat in which they will be inoculated, which can confer competitive advantages because the efficacy of silage inoculant bacteria is based on their ability to compete effectively with the epiphytic microbiota in forages (Duffner and O'Connell, 1995; Fabiszewska et al., 2019). Besides, the microbial diversity in silages has mainly been studied using culture-independent techniques [polymerase chain reaction (PCR)-based techniques], revealing a wide diversity of unidentified microorganisms in the fermentation (McAllister et al., 2018). To the best of our knowledge, few researchers have investigated the potential of alfalfa nutrition and microbiota under salt stress. Thus, the aim of this study was to investigate the potential of alfalfa microbiota under salt stress and the effects of ions on silage fermentation characteristics and microbial community, supplying the theoretical basis for controlling silage fermentation.
Materials and Methods
Site Description
The experiment was conducted at the Baotou Experimental Station for Forage Processing and High Efficient Utilization, Inner Mongolia Agricultural University. The experimental station was located in Baotou City in Inner Mongolia, China, which was a typical area with high salinity in the Hetao Plain. The land spans between 110°37″ −110°27″ E and 40°05″ −40°17″ N. The climate is a north temperate continental climate with arid and windy conditions. The dominant wind direction throughout the year is northwest wind. The annual average temperature is 6.8°C, and the frost-free period is about 165 days. The annual average rainfall is 330 mm, and the annual average evaporation is 2,094 mm.
Description of Raw Materials
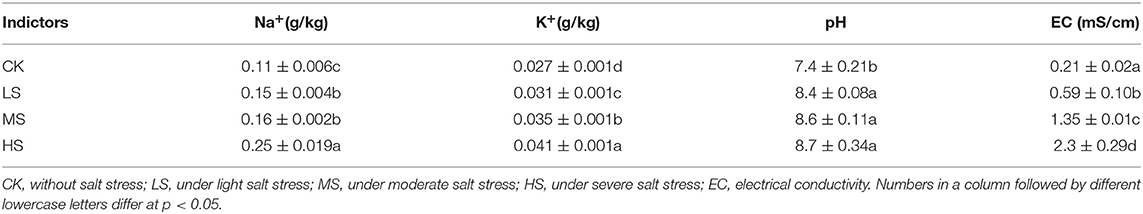
The field experiments were conducted in 2019, and the variety of alfalfa tested was the ZhongMu No.3, provided by the Beijing Institute of Animal Science and Veterinary Medicine of the Chinese Academy of Agricultural Sciences. The characteristics of alfalfa were strong salt resistance, good palatability, high nutrition, and rich in value. Alfalfa was sown in May 2018, the sowing method was a drill, and the row-to-row distance was 10 cm. Four positions were selected to represent various salt stress levels, i.e., non-salt stress (CK), light salt stress (LS), moderate stress (MS), and severe stress (HS). The salt stress contents in CK, LS, MS, and HS sites were <1, 1–2, 2–3, and 3–4%0, respectively. Each treatment was repeated three times. The physical properties of the soil are shown in Table 1.
Alfalfa was harvested in the initial flowering stage, then wilted for 5 h to obtain a targeted DM content, and immediately chopped into 2- to 3-cm lengths by a fodder chopper. Each material was treated separately to prevent crossing contaminations. Two kilograms of the prepared alfalfa were packed in polyethylene plastic bags and sealed with a vacuum sealer in each group. All bags were assigned without additives. Triplicates for each treatment were opened after 30 and 60 days of ensiling, respectively.
Chemical Composition and Organic Acid
The DM of the fresh alfalfa and silage was determined by oven drying at 65°C for 72 h. The neutral detergent fiber (NDF) and acid detergent fiber (ADF) were measured according to Van Soest's procedures (Van et al., 1991). Colorimetry after reaction with anthrone reagent was used to determine the starch and water-soluble carbohydrate (WSC) content (Lei et al., 2015). Non-structural carbohydrates (NSC) are the sum of WSC and starch. The crude protein (CP = total N × 6.25) was determined using a Kjeldahl apparatus (Gerhart Vapodest 50 s, Germany) according to Patrica (1997). The concentrations of Na+ and K+ ions of alfalfa were measured relative to standard solutions using a model 425 flame photometer (Sherwood Scientific Ltd., UK).
To determine the fermentation traits of forage, a sample (10 g) of silage was mixed with 90 g of deionized water kept at a 4°C fridge for 24 h. The liquid extract was filtered through four layers of cheesecloth and filtered paper. The prepared filtrates were determined for measuring pH, ammonia nitrogen (ammonia-N), and organic acids. The pH was measured immediately with a glass electrode pH meter (LEICI pH S-3C, Shanghai, China). The content of ammonia-N was followed by the phenol-hypochlorite procedure of Broderick and Kang (1980). The concentration of organic acids was determined by high-performance liquid chromatography (HPLC, Waters e2695, Massachusetts USA; column: Waters Symmetry C18; oven temperature, 50°C; mobile phase, 3 mmol L−1 perchlorate solution; flow rate, 1.0 ml min−1; flame photometric detector, 210 nm; sample size, 5.0 μl) of Ping et al. (2017). Buffering capacity (BC) was determined by the hydrochloric acid sodium hydroxide method (Lin et al., 1992).
Microorganisms Enumeration
For enumeration of the microorganisms, 10 g of fresh sample or silage was shaken with 90 ml of sterile distilled water at 120 rpm for 2 h. Then, 1 ml of solution was used for 10-fold serial dilution for microorganism enumeration and then the remaining solution was filtered and stored in the −80°C refrigerator for DNA extraction. The colonies of LAB (lactic acid bacteria) were counted on MRS agar medium after incubation in an anaerobic incubator (Heal Force Instrument Manufacturing Co., Ltd., Shanghai, China) at 37°C for 48 h. Aerobic bacteria were cultured and counted on nutrient agar medium (Nissui seiyaku Ltd., Tokyo, Japan). Yeasts were counted on potato dextrose agar (Nissui seiyaku Ltd., Tokyo, Japan). Enterobacteriaceae was counted on the Violet Red Bile Glucose Agar medium after 48 h of incubation at 37°C under aerobic conditions. The microbial data were obtained as colony-forming units (CFU) and were transformed to a logarithmic scale on a fresh matter (FM) basis.
High-Throughput Sequencing of Microbial Population
Microbial DNA of alfalfa samples was extracted using the E.Z.N.A.® soil DNA Kit (Omega Bio-Tek, Norcross, GA, U.S.) according to the manufacturer's protocols. The final DNA concentration and purification were determined by NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA), and DNA quality was checked by 1% agarose gel electrophoresis. The V3–V4 hypervariable regions of the bacteria 16S rRNA gene were amplified with primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) by thermocycler PCR system (GeneAmp 9700, ABI, USA). The PCR reactions were conducted using the following program: 3 min of denaturation at 95°C, 27 cycles of 30 s at 95°C, 30 s for annealing at 55°C, and 45 s for elongation at 72°C, and a final extension at 72°C for 10 min. PCR reactions were performed in triplicate 20-μl mixture containing 4 μl of 5 × FastPfu Buffer, 2 μl of 2.5 mM dNTPs, 0.8 μl of each primer (5 μM), 0.4 μl of FastPfu Polymerase, and 10 ng of template DNA. The resulting PCR products were extracted from a 2% agarose gel and further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluor™-ST (Promega, USA) according to the manufacturer's protocol.
Raw fastq files were demultiplexed, quality-filtered by Trimmomatic, and merged by FLASH with the following criteria: (a) the reads were truncated at any site receiving an average quality score <20 over a 50-bp sliding window; (b) primers were exactly matched allowing two nucleotide mismatching, and reads containing ambiguous bases were removed; (c) sequences whose overlap was longer than 10 bp were merged according to their overlap.
Operational taxonomic units (OTUs) were clustered with 97% similarity cutoff using UPARSE, and chimeric sequences were identified and removed using UCHIME. The taxonomy of each 16S rRNA gene sequence was analyzed by the RDP Classifier algorithm against the Silva (SSU123) 16S rRNA database using a confidence threshold of 70%.
Statistical Analysis
The statistical data were analyzed by the procedure of SAS (version 9.3, SAS Institute Inc., Cary, NC, USA). Duncan's multiple range tests were used to evaluate differences among treatments. High-throughput sequencing data were performed using an online platform of Majorbio I-Sanger Cloud Platform (www.i-sanger.com).
Results
The Chemical Composition of Fresh Alfalfa
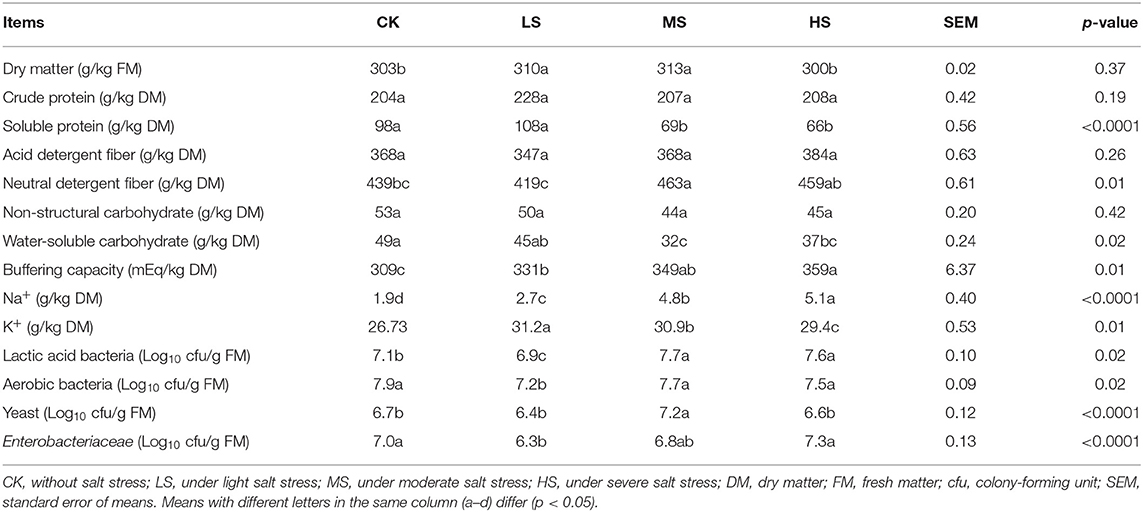
The chemical composition and microbial populations of fresh materials and stressed by salt are described in Table 2. The alfalfa DM content ranged from 300 g/kg to 313 g/kg; the MS and HS samples had the highest and lowest DM contents, respectively (p < 0.05). There was no significant difference between the CP, ADF, and NSC (p > 0.05). The SP content (108 g/kg) of LS was significantly higher than the others (p < 0.05). There was no significant difference among the treatments (p > 0.05), but the WSC content (49 g/kg) in CK was significantly higher than in other groups (p < 0.05). Also, salt stress significantly affected the Na+ and K+ content of alfalfa (p < 0.05). The K+ content in the HS (5.1 g/kg) was significantly highest (p < 0.05). Furthermore, It proved that the microbial population on plants could be affected by salt stress. The content of LAB (lactic acid bacteria) in MS (7.7 Log10 cfu/g) and HS (7.6 Log10 cfu/g) was significantly higher than that in CK (7.1 Log10 cfu/g) and LS (6.91 Log10 cfu/g).
Fermentation Characteristics and Chemical Composition of Alfalfa Silage
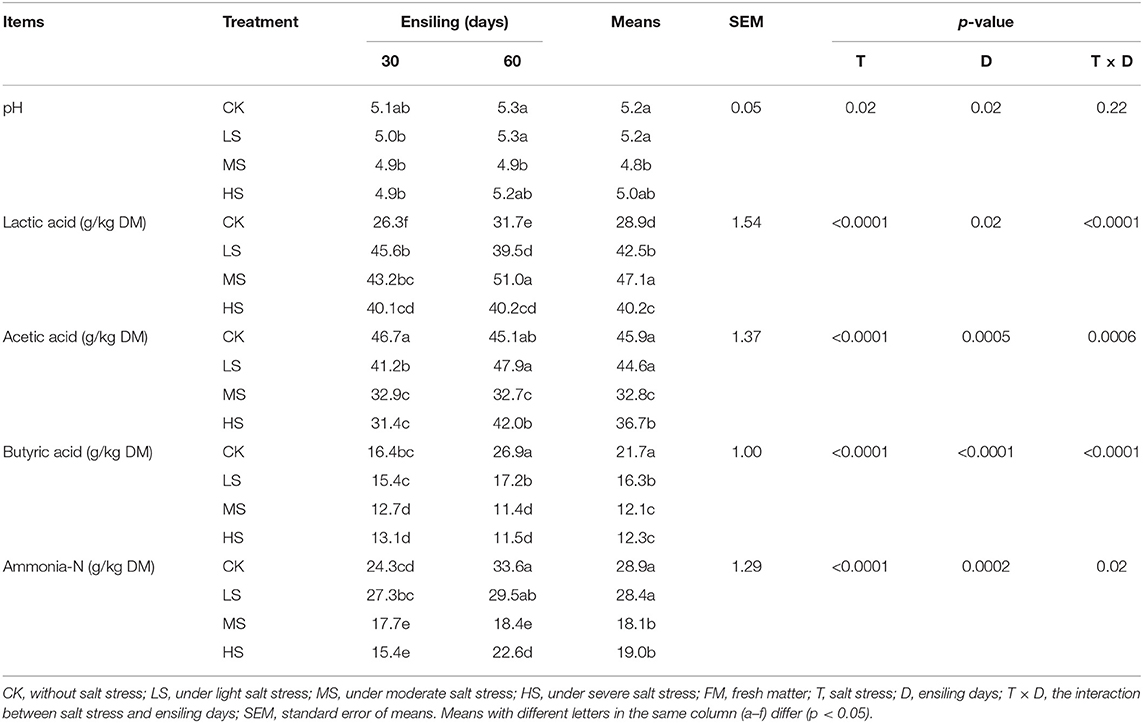
The fermentation characteristics of alfalfa silage on different days are shown in Table 3. Factor analysis illustrated that salt stress (T), the ensiling day (D), and the interaction of salt stress and the ensiling day had significantly (p < 0.05) affected LA (Lactic acid), AA (Acetic acid), BA (Butyric acid), and ammonia-N (Ammonia nitrogen) contents. Salt stress significantly affects the content of LA, AA, BA, and ammonia-N (p < 0.0001). After 60 days of ensiling, the pH of the silages ranged from 4.9 to 5.3, and the MS silage achieved significantly lower pH values and higher LA concentration as compared to other silage (p < 0.05). With the rapid decrease in pH, the WSC (17 g/kg) content of MS at 60 days was reduced by 15 g/kg compared to 0 days (32 g/kg). The AA of the silages ranged from 31.4 to 45.1; as the salt stress increases, the accumulation of AA presents a trend of increase and then decrease throughout ensiling of four groups, resulting in the highest ratio of highest (p < 0.05) AA concentration after 60 days of ensiling in the MS. The BA of the silages ranged from 11.4 to 26.9; the content of BA in CK was significantly highest at 60 days (21.7 g/kg) of fermentation (p < 0.05), and the content of LA (31.7 g/kg) was also significantly lowest (p < 0.05). After 60 days of silage, the ammonia-N content in the MS and HS group was notably (p < 0.05) higher than LS and CK. In this research, the Na+ content in MS and HS groups was also significantly (p < 0.05) higher than LS and CK.
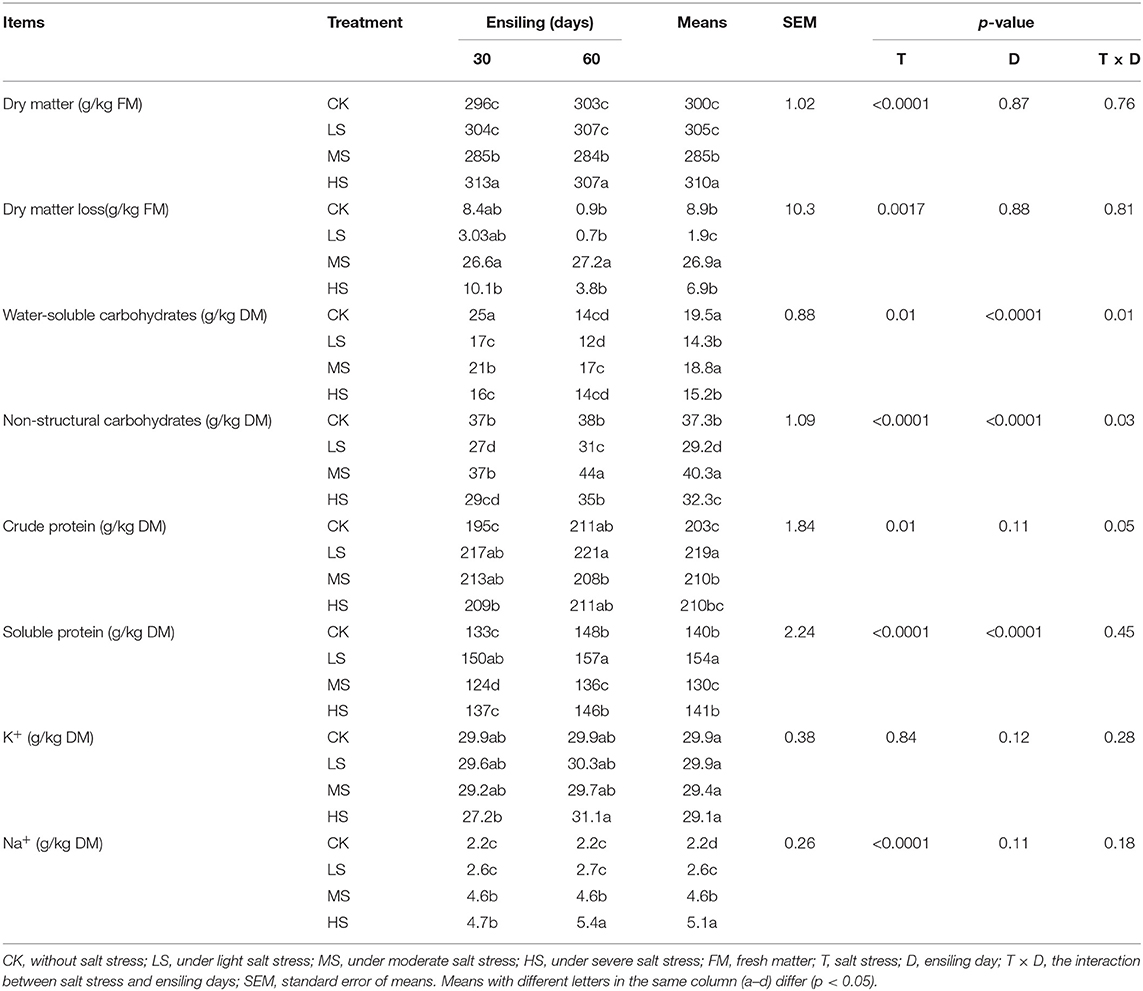
The chemical composition of alfalfa silage on different days are shown in Table 4. Factor analysis illustrated that salt stress (T), the ensiling day (D), and the interaction of salt stress and ensiling day had a significant (p < 0.05) effect on WSC and NSC contents. Salt stress significantly (p < 0.05) affects the content of DM, NSC, SP, and NA+. After 60 days of ensiling, the DM losses ranged from 0.7 g/kg to 27.2 g/kg. The WSC content in the four groups was consumed more (43.8%−73.3%) than the NSC content, while the NSC content was consumed between 22.2% and 38.0%. It was the large consumption of WSC that leads to the rapid reproduction of LAB. In contrast, starch was less consumed during ensiling. Factor analysis illustrated that salt stress (T) and ensiling day (D) had a significant (p < 0.0001) effect on SP content. Compared with the alfalfa raw materials of each group, the SP content increased by 45.4–121.2%. After 60 days of ensiling, the SP content in LS (157 g/kg) silage was significantly higher than in other groups. At the same time, the content of ammonia-N was also the lowest. This may inhibit the further decomposition of CP by protease and then eventually leads to the accumulation of SP content. After 60 days of ensiling, NA+ content (5.1 g/kg) was significantly higher than CK, LS, and MS (p < 0.0001). The NA+ content changes a little after ensiling.
Microbial Community of Alfalfa Silage
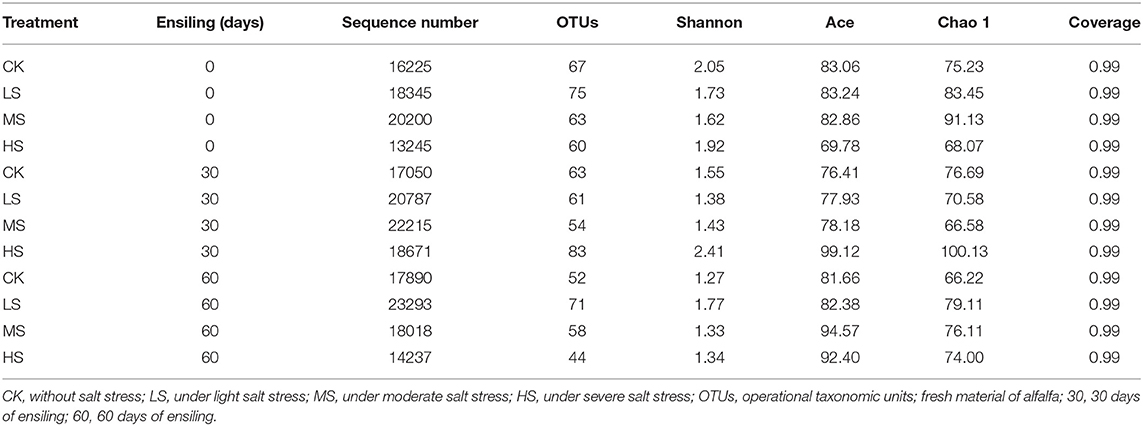
A high-throughput sequencing method was performed in variable regions 3 and 4 of 16s rDNA to calculate and evaluate bacterial diversity after ensiling (Table 5). Based on the coverage values, >99% of coverage values had adequately obtained most of the bacterial communities. The OTUs, Shannon, Ace, and Chao 1 values ranged from 44 to 83, 1.27 to 2.41, 69.78 to 99.12, and 66.58 to 100.13, respectively. In conclusion, salt stress greatly affected the microbiome of the resultant silage.
Bacterial community dynamics at the phylum level during the ensiling of alfalfa are shown in Figure 1. The dominant phyla in CK microbiota were Proteobacteria (92.98%), Actinobacteria (5.9%), and Firmicutes (0.64%), whereas the LS microbiota was dominated by Proteobacteria (86.6%), Actinobacteria (4.47%), and Firmicutes (1.72%). The MS microbiota was dominated by Proteobacteria (96.5%), Actinobacteria (6.4%), and Bacteroidetes (8.27%). The HS microbiota was dominated by Proteobacteria (85.1%), Actinobacteria (5.3%), and Bacteroidetes (6.42%). The dominant phyla in fresh alfalfa microbiota were Proteobacteria. The proportions of Firmicutes increased while the relative abundance of Proteobacteria decreased with the extension of time.
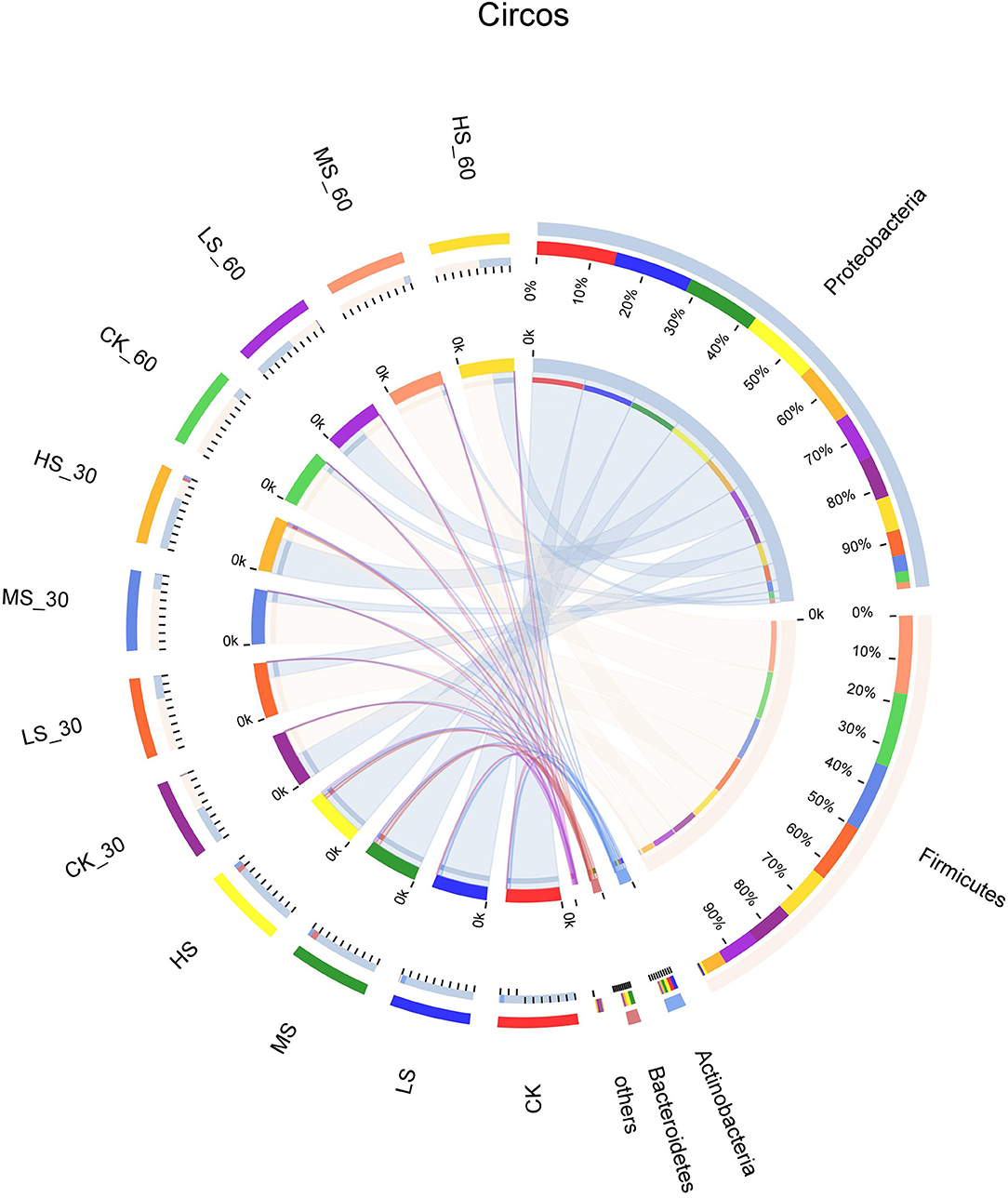
Figure 1. Differences in relative abundance of microbial community on phylum level shown by using a Circos plot. The size of the bars from each phylum shows the relative abundance of that phylum in the sample. CK, fresh material of alfalfa without salt stress; LS, alfalfa under light salt stress; MS, alfalfa under moderate salt stress; HS, alfalfa under severe salt stress; 30, 30 days of ensiling; 60, 60 days of ensiling.
To further comprehend the succession of bacterial communities of alfalfa silage under salt stress, bacterial communities at the genus level were evaluated in Figures 2, 3. It shows that the alfalfa under salt stress used to produce the silage had a dramatic impact on the resultant microbial community. Pantoea were the dominant genera in CK (46.32%), LS (56.42%), MS (51.68%), and HS (45.98%); what is more, after 30 days and 60 days of ensiling, the relative abundance of Pantoea significantly decreased. After the fermentation process, Enterococcus, Lactococcus, and Lactobacillus were the dominant genera. Lower pH inhibited the growth and breeding of Pantoea, Pseudomonas, and Methylobacterium. Interestingly, with the increase of salt stress, the relative abundance of Pantoea decreased gradually, while that of Pseudomonas increased gradually. In the absence of oxygen, Enterococcus became the dominant genera in the CK_30 (38.99%) and LS_30 (59.75%). Enterococcus in CK_60 (14.6%) and LS_60 (26.17%) also decreased. Sphingomonas (20.15%) was found in HS_30. After 60 days of ensiling, Lactococcus became the dominant genera in the CK_60 and HS_60, Lactobacillus and Lactococcus became the dominant genera in the MS_60, and Enterococcus was still the dominant genera in the LS_60. With the decrease of pH, the acidic environment restricted the existence and relative abundance of Enterococcus, while the relative abundance of Lactococcus and Lactobacillus increased in the four groups.
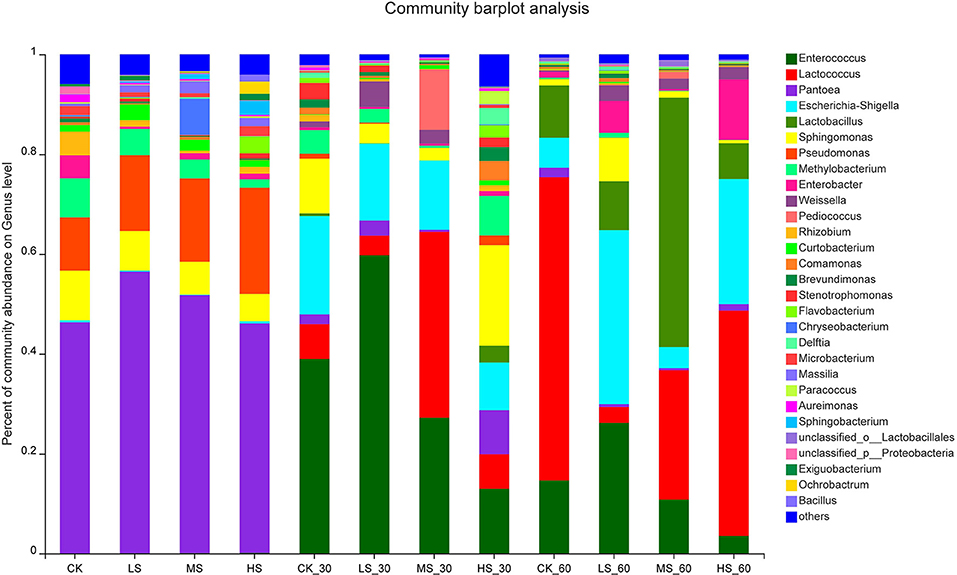
Figure 2. Bacterial community composition on genus level in alfalfa silage. CK, fresh material of alfalfa without salt stress; LS, alfalfa under light salt stress; MS, alfalfa under moderate salt stress; HS, alfalfa under severe salt stress; 30, 30 days of ensiling; 60, 60 days of ensiling.
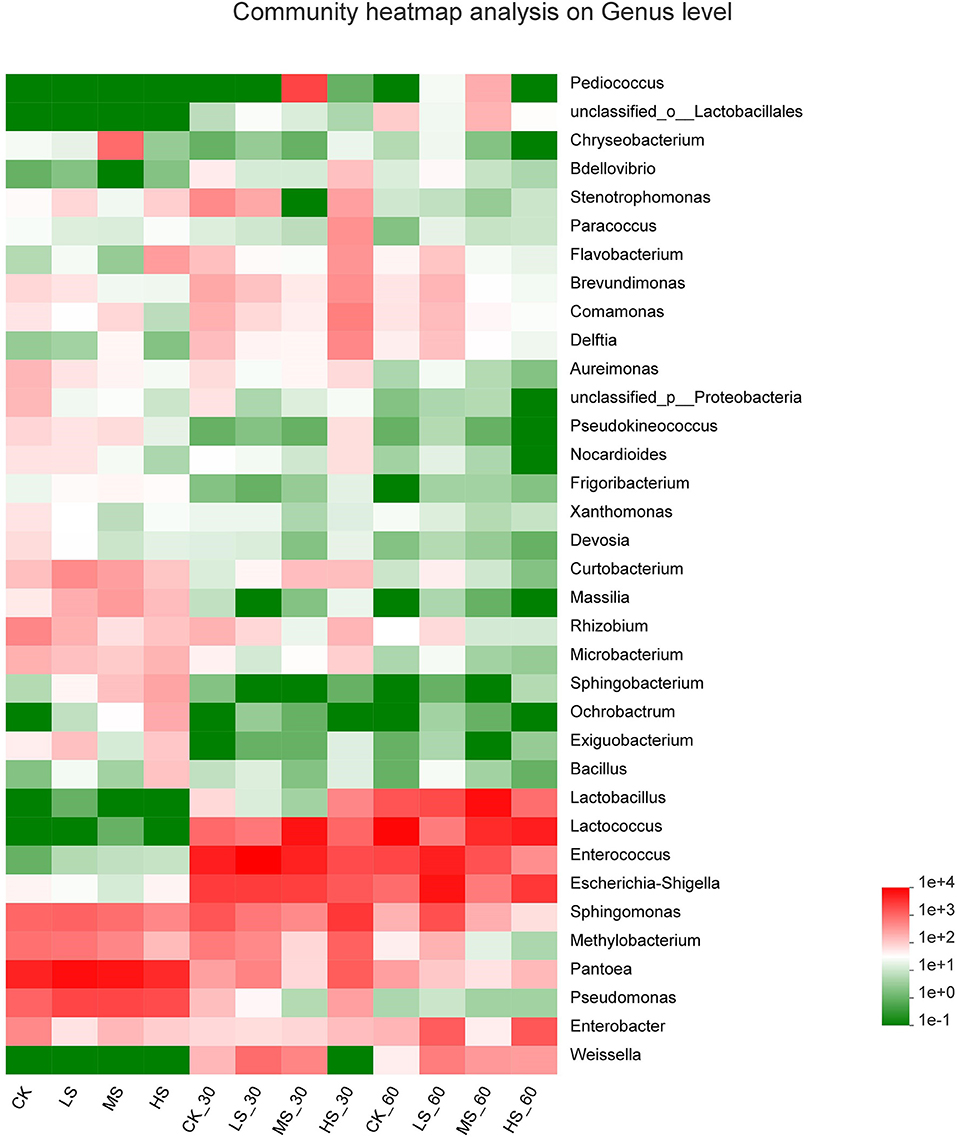
Figure 3. Heatmap of prominent bacterial genera (35 most abundant genera) for alfalfa silage. CK, fresh material of alfalfa without salt stress; LS, alfalfa under light salt stress; MS, alfalfa under moderate salt stress; HS, alfalfa under severe salt stress; 30, 30 days of ensiling; 60, 60 days of ensiling.
To evaluate the bacterial communities of different silage groups under salt stress, the bacterial communities at the genus level were evaluated in Figure 4. With the gradual increase in salt stress, Pseudomonas, Sphingomonas, and Methylobacterium showed strong response patterns. In contrast, Sphingomonas, Methylobacterium, and Aureimonas decreased with increasing salt stress and showed salt-tolerant properties. After 30 days of ensiling, the abundance of Weissella and Escherichia would decrease with the increase of salt stress. Besides, the content of ammonia-N in MS_30 and HS_30 would be the lowest significantly (p < 0.05), and the same rules and patterns were also reflected in 60 days.
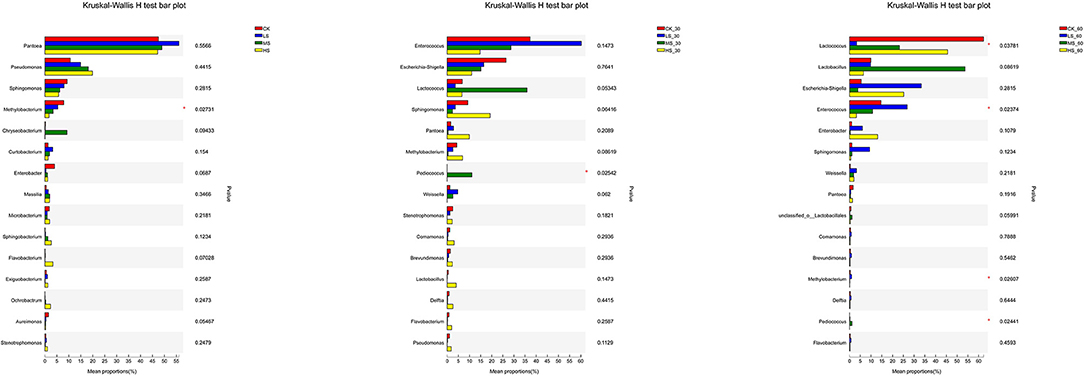
Figure 4. Comparison of microbial variations using the Kruskal–Wallis H test for alfalfa silage. *p < 0.05. CK, fresh material of alfalfa without salt stress; LS, alfalfa under light salt stress; MS, alfalfa under moderate salt stress; HS, alfalfa under severe salt stress; 30, 30 days of ensiling; 60, 60 days of ensiling.
The Relationships Among the Samples, Microbial Community, and Characteristics Products in Alfalfa Silage
The variations in the microbial communities of the silage were further verified by β-diversity analysis using principal coordinates analysis (PCoA) (Figure 5). The perspicuous separation among the microbiota indicated that different epiphytic microbial communities exist in alfalfa under different salt stress. As seen in Figure 5A, salt stress had a great influence on the microbial community of fresh plants. The results further validate the differences between the microbial communities in Figures 2, 3. By analyzing the composition of the microbial community structure, we would reveal the changes in microbial community structure under different salt stress. There were obvious differences in microbial community composition among different habitat types in Figure 5A. It is not easy to understand and master the distribution characteristics of microorganisms. Intriguingly, the microbial community structure was different at different fermentation stages, and small changes were found in LS_30 and LS_60, which might be due to the fact that Lactococcus and Lactobacillus were used as the dominant bacteria in the 60-day stage of the fermentation process.
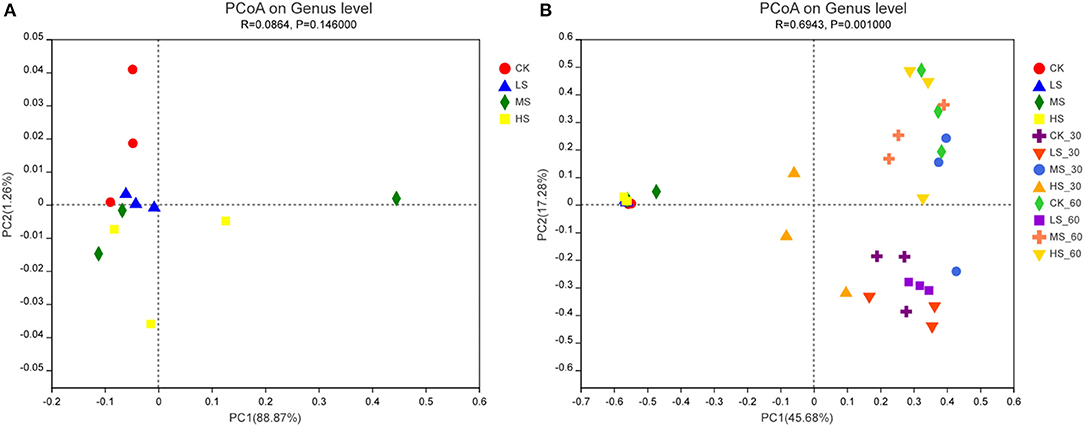
Figure 5. (A,B) Principal co-ordinates analysis (PCoA) of bacterial communities in alfalfa silage. CK, fresh material of alfalfa without salt stress; LS, alfalfa under light salt stress; MS, alfalfa under moderate salt stress; HS, alfalfa under severe salt stress; 30, 30 days of ensiling; 60, 60 days of ensiling.
As seen in Figure 6, the obvious relationships among high-throughput sequencing data on the genus level and major fermentation products are visualized in a relevance analysis plot. Silage pH was positively correlated with the abundance of Lactococcus, Escherichia–Shigella, and Pantoea in the silage microbiome, while it was negatively correlated with the abundance of Weissella. The silage DM was negatively correlated with the abundance of Enterococcus and Methylobacterium (p < 0.01). The silage CP and SP were negatively correlated with the abundance of Lactobacillales, Lactococcus, and Lactobacillus. The silage WSC was negatively correlated with the abundance of Escherichia–Shigella (p < 0.01). The silage Na+ was positively correlated with the abundance of Bifidobacterium (p < 0.01), while it was negatively correlated with the abundance of Bdellovibrio, Brevundimonas, Comamonas, Curtobacterium, Enterococcus, Flavobacterium, Methylobacterium, and Sphingomonas. The silage LA was positively correlated with the abundance of Lactobacillus (p < 0.05). The silage BA was positively correlated with the abundance of Enterococcus, Bdellovibrio, Brevundimonas, Delftia, Flavobacterium, Leuconostoc, Methylobacterium, and Rhizobium (p < 0.05).
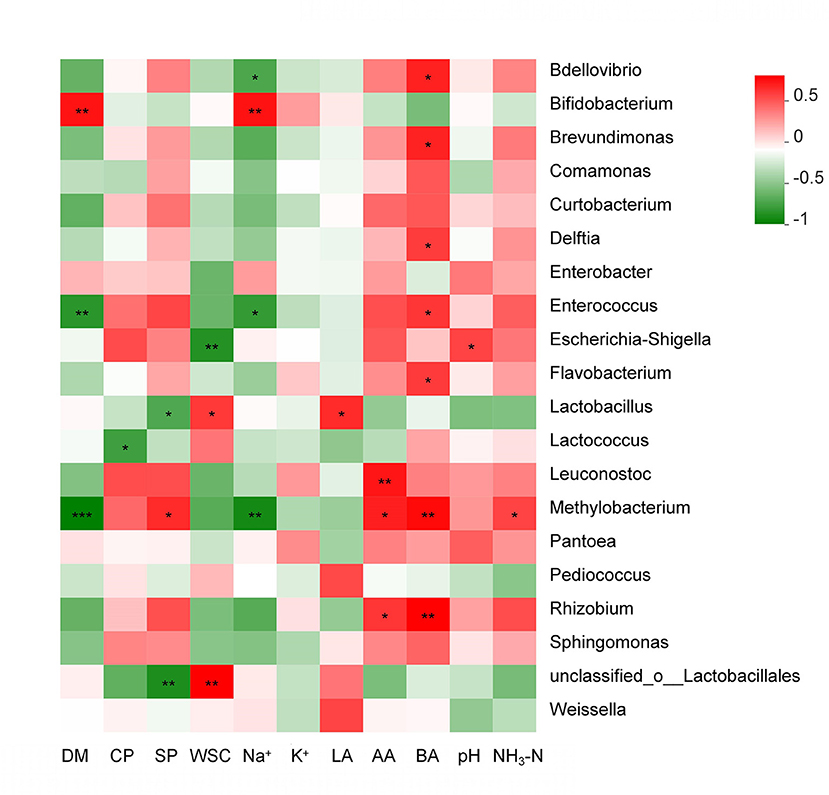
Figure 6. Relevance analysis of high-throughput sequencing data related to the genus level (20 most abundant); DM, dry matter; CP, crude protein; SP, soluble protein; LA, lactic acid; AA, acetic acid; BA, butyric acid; NH3-N, ammonia nitrogen; WSC, water-soluble carbohydrates. The legend on the right is the color interval of different R values. * 0.01 < p ≤ 0.05, ** 0.001 < p ≤ 0.01, *** p ≤ 0.001.
Discussion
The Chemical Composition of Fresh Alfalfa
Salt stress affects the normal growth and development of plants and affects the chemical composition of plants. Differences between SP in different groups can be considered as a detoxification effect of plants and there was a also significant difference in SP between LS and MS groups. SP is an important factor in relieving osmotic pressure in response to salt stress (Kanwal et al., 2019). The SP content in a low NaCl group increased significantly (Lacramioara et al., 2016); this is congruent with the findings of Agastian et al. (2000), who reported that the SP content in a high salinity growth environment was decreased (Agastian et al., 2000). The main storage form of energy in plants is non-structural carbohydrates (NSC), which include soluble carbohydrates (sucrose and fructose, etc.) and starch, which are the important sources of energy in the process of plant growth and metabolism. The WSC content (49 g/kg) in CK was significantly higher than in other groups, because salt stress inhibits the metabolism and accumulation of carbohydrates in plants (Morais et al., 2019). Salt stress is a likely factor to cause chemical composition changes. In the study, the colonies of LAB alfalfa provided enough bacteria to initiate the silage fermentation reported by Oliveira et al. (2017).
Fermentation Characteristics and Chemical Composition of Alfalfa Silage
Silage pH is a very important evaluation indicator to consider when evaluating the fermentation efficacy; well-fermented silage should have a pH of 4.2 or lower (McDonald et al., 1991). In the study, the results presented that all silages had a pH of higher than 4.2 and showed a poor fermentation quality. The variety of pH might be caused by different alfalfa under salt stress, or differences in the silage microbiome. The differing chemical compositions of the alfalfa samples may have contributed to the substantial variation in silage fermentation characteristics; Xue et al. (2021) reported this phenomenon previously, albeit in Pennisetum sinese silage. The lower pH and high LA concentration of MS and HS silage were attributed to the LAB, which quickly metabolized WSC into LA, thus reducing silage pH and stability in a short time (Nazar et al., 2020). AA is derived from the decomposition and fermentation of sugars by heterofermentative LAB, Enterobacteriaceae, and Clostridium (Nazar et al., 2020).
In this study, there were Weissella, Enterobacter, and Sphingobium bacteria in the fermentation for 30 and 60 days. Weissella and Enterobacter could not only consume a large amount of WSC content but also have a low utilization rate of WSC (Blajman and Vinderola, 2020). It was also possible that with the increase of salt stress, more heterogeneous LAB was attached to alfalfa plants. BA and other acids reflect the efficiency of fermentation or secondary fermentation (Wang et al., 2020). Besides, the amount of BA also depends on the amount of LA; this may be caused by the secondary fermentation of heteromorphic LAB and yeast (Lei et al., 2015). The presence of ammonia-N in silage is an important indicator of the degree of proteolysis. Li et al. (2018) found that the process was mainly driven by plant protease activity and proteolytic Clostridia. It might be the reason that Na+ had impacted the activity of protease enzymes throughout the ensiling. When Na+ content is below 60 mg/L, Na+ could promote the activity of protease enzymes, while above 60 mg/L, it could inhibit the activity of protease enzymes instead. It might be that the NA+ content in MS and HS groups in this study inhibited the activity of protease enzymes.
From a perspective of the ammonia-N content of alfalfa silage, The ammonia-N is considered as a representation of proteolytic activity (Li et al., 2018). The inhibiting effect on ammonia-N accumulation of MS and HS suggested an enhancement in protein preservation during ensiling. A typical reason for ammonia-N accumulation was the proteolytic enzymes (Kung and Shaver, 2001). Even most plant proteolytic enzymes in alfalfa silage showed greater activities at pH 5.0–6.0 (Yang et al., 2019), but excess ions can inhibit proteolytic enzymes (Jaci et al., 2014), which might be an explanation for the reduction in ammonia-N formation in the MS and HS silage. After alfalfa fermentation, the content of SP increased from 66–108 to 124–150, which may be related to the hydrolysis of protein during the fermentation process. The protein of macromolecules was decomposed into small molecular proteins with water-soluble characteristics (Liu et al., 2021), which might be an explanation for the increase of SP.
Microbial Community of Alfalfa Silage
In the present study, Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria were dominant in all alfalfa silage. These findings are consistent with previous reports examining alfalfa silage and even corn silage (Lei et al., 2015; Blajman and Vinderola, 2020). Firmicutes and Proteobacteria were the most abundant phyla in the fermentation silage (Jie et al., 2020). The transformation of the bacterial community from Proteobacteria to Firmicutes was reported by Nazar et al. (2020), who revealed that the anaerobic and acidic environment quite sustained the growth of Firmicutes. Besides, with the increase of salt stress, the relative abundance of Proteobacteria decreased regularly. Luttenton and Rada, 2010 had stated that the same plants have different epiphytes flora as if they grow under different environmental conditions; salt stress could be the reason for the changes of microbial communities in this study. The response of plant microorganisms to different salt stresses was very interesting and beyond expectation. With the gradual increase in salt stress, Pseudomonas, Sphingomonas, and Methylobacterium showed strong response patterns. Pseudomonas increased with salt stress increased, which may be since these two genera prefer to survive and reproduce in salinity environments (Nidhi et al., 2013).
Compared to raw materials, the anaerobic environment causes the changes of microbial habitat, and the silage process inhibited the aerobic microorganisms, which contributed to the differences in microbial communities of the fresh alfalfa and silages (Nazar et al., 2020); the diversity and richness of bacteria decreased in CK, LS, MS, and HS silages, whether in 30 or 60 days. Similarly, Jie et al. (2020) reported that silage bacterial diversity decreased due to the increase in the abundance of the predominant genus (Lactococcus and Enterococcus). When the dominant bacteria were abundant, the microbial community of the silage would be decreased (Nazar et al., 2020). With the increase of salt stress, the bacteria diversity and richness of raw alfalfa were decreased and then increased. This phenomenon could be caused by competition between bacteria due to antibacterial activity. Heinz et al. (2019) revealed a hypothesis that halophilic bacteria like Planococcus halocryophilus manifested different phenotypic responses under high salt stress (>8%Nacl). Planococcus halocryophilus can develop perchlorate-specific stress adaptations that were not (or only to a lower extent) used to counteract high Na+ concentrations.
After the fermentation process, Enterococcus, Lactococcus, and Lactobacillus were the dominant genera. Lower pH inhibited the growth and breeding of Pantoea, Pseudomonas, and Methylobacterium. Interestingly, with the increase of salt stress, the relative abundance of Pantoea decreased gradually, while that of Pseudomonas increased gradually. It was reported that Pantoea and Pseudomonas, as PGPR (plant-growing-promoting Rhizobacteria), could produce IAA and promote biomass to help plants survive better under salt stress (Sun et al., 2020), but Pantoea's adaptation to the halophytic environment was limited, so its abundance would gradually decrease, while Pseudomonas was more adaptive to the halophytic environment than Pantoea (Ching and Deivanai, 2015). In the absence of oxygen, Enterococcus became the dominant genera in the CK_30 (38.99%) and LS_30 (59.75%). Enterococcus was often used to enhance the fermentation characteristics reported by Wang et al. (2020). Enterococcus could rapidly produce LA and establish an acidic anaerobic environment to promote LAB growth in the early fermentation stage, but Enterococcus is not an acid-resistant genera (Cai et al., 1998). Similarly, the same conclusion was also verified in this study; Enterococcus in CK_60 and LS_60 also decreased significantly. A large abundance of Sphingomonas has existed in HS_30. Sphingobium could cause further hydrolysis of CP and SP (Zhou et al., 2019). Sphingobium might cause an increase in ammonia-N.
Lactococcus generally existed in naturally fermented silages. This was not consistent with the conclusion of Wang et al. (2020), possibly because no additives were used in this study and only rely on the microorganisms attached to alfalfa under salt stress. It is undeniable that Lactobacillus and Lactococcus are important in pH decline and LA accumulation, and in the whole process of silage, they are generally present. Wang et al. (2020) stated that the composition of microorganisms in silage was quite different at the initial stage of the fermentation. Lactobacillus could grow vigorously at the end of the fermentation stage due to their stronger acid resistance than cocci LAB. Lactobacillus could become the dominant bacteria at the end of the fermentation stage because of its stronger acid resistance than Lactococcus. Generally, LA in silage is mainly produced by Lactobacillus, while with the decrease of pH, Lactococcus and Enterococcus could not survive with lower pH due to weaker acid resistance.
This study found that there was a significant correlation between the chemical composition of silage and silage microorganisms. The chemical composition and microbial community of alfalfa under different salt stress were different, so the characteristics of the silage microbial community were also different. This may be explained by the fact that these microorganisms are chemoorganotrophic bacteria that produce energy through the oxidation of organic matter such as starch and organic acids. Pseudomonas, Bacteroides, and Stenotrophomonas all consume protein, while Megamonas, Raoultella, Citrobacter, and Enterococcus ferment carbohydrates (Tian et al., 2019; Xue et al., 2021). The results of this study further compound the significant relationship between the chemical composition of the raw material and the resultant silage.
Intriguingly, Na+ and K+ would inhibit the growth and reproduction of most microorganisms; although ions are not directly responsible for the growth of microorganisms, bacteria can be able to look at potassium ion channel–electrical signaling to achieve cell–cell communication (Jing et al., 2020). This could explain the fact that the number and diversity of microbes were low under salt stress (Table 5). Ion stress inhibited not only the activity and abundance of microorganisms but also the breeding of harmful microorganisms. The composition and distribution of microorganisms could be clearly defined, and it would be of great significance to the improvement and development of ecology and biogeography as well as the protection and utilization of microbial resources.
Conclusion
The study presented herein examined the microbiome and quality of silages from the natural fermentation of alfalfa under salt stress. Salt stress has a great impact on the quality and bacterial community of fresh alfalfa and silage. There was a predominance of desirable genera including Lactococcus and Lactobacillus in silage produced from alfalfa under salt stress, and this led to better fermentation quality. There was a strong correlation between the chemical composition and bacterial community of the silage. The salt stress and plant ions were thus most responsible for their different fermentation modes in alfalfa silage. The results of the study indicate that exogenous epiphytic microbiota of alfalfa under salt stress could be used as a potential bioresource to improve the fermentation quality.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: NCBI PRJNA753242.
Author Contributions
QL: writing original draft. DS and ZJW: writing review and editing. GG and ZW: data curation. YJ: methodology. MH: software. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Inner Mongolia Autonomous Region Science and Technology Project (201802069), the National Natural Science Foundation of China (42077054), and the National Technical System of Forage Industry for Dry Grass Storage (CARS-34).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely thank the Key Laboratory of Grassland Resources of the Ministry of Education and the Key Laboratory of Forage Cultivation, Processing, and High Efficient Utilization of the Ministry of Agriculture for their technical support for this research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.688695/full#supplementary-material
References
Agastian, P., Kingsley, S., and Vivekanandan, M. (2000). Effect of salinity on photosynthesis and biochemical characteristics in mulberry genotypes. Photosynthetica 38, 287–290. doi: 10.1023/A:1007266932623
Blajman, J. E., and Vinderola, G. R. B. (2020). The role of homofermentative and heterofermentative lactic acid bacteria for alfalfa silage: a meta-analysis. J. Agric. Sci. 158, 107–118. doi: 10.1017/S0021859620000386
Bolsen, K. K., Ashbell, G., and Weinberg, Z. G. (1996). Silage fermentation and silage additives—review. Asian-australas. J. Anim. Sci. 9, 483–493. doi: 10.5713/ajas.1996.483
Broderick, G. A., and Kang, J. H. (1980). Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 63, 64–75. doi: 10.3168/jds.S0022-0302(80)82888-8
Cai, Y., Benno, Y., Ogawa, M., Ohmomo, S., Kumai, S., and Nakase, T. (1998). Influence of Lactobacillus spp. from an inoculant and of Weissella and Leuconostoc spp. from forage crops on silage fermentation. Appl. Environ. Microbiol. 64, 82–87 doi: 10.1128/AEM.64.8.2982-2987.1998
Ching, C. J., and Deivanai, S. (2015). Ameliorative effect of plant growth promoting bacterial endophyte Pantoea agglomerans on salt stress at early stage of growth in rice. Malays. J. Microbiol. 11, 341–351. doi: 10.21161/mjm.68014
Duffner, F., and O'Connell, M. (1995). Comparative evaluation of plasmid profiling and ribotyping in the analysis of Lactobacillus plantarum strain heterogeneity in silage. J. Appl. Bacteriol. 78, 20–27. doi: 10.1111/j.1365-2672.1995.tb01668.x
Fabiszewska, A. U., Zielińska, K.J., and Wróbel, B. (2019). Trends in designing microbial silage quality by biotechnological methods using lactic acid bacteria inoculants: a minireview. World J. Microbiol. Biotecnol. 35:76. doi: 10.1007/s11274-019-2649-2
Guo, A. J., Liu, C. Q., Wang, J. X., Zhang, Y. J., An, T. W., and Liu, G. X. (2013). The effects of land utilization type on soil property in coastal saline alkali wetland. J. Chongqing Norm. Univ. 30, 95–110. doi: 10.11721/cqnuj20130121
Hariadi, Y., Marandon, K., Tian, Y., Jacobsen, S. E., and Shabala, S. (2010). Ionic and osmotic relations in quinoa plants grown at various salinity levels. J. Exp. Bot. 62, 185–193. doi: 10.1093/jxb/erq257
Heinz, J., Waajen, A. C., Airo, A., Alibrandi, A., Schirmack, J., and Schulze-Makuch, D. (2019). Bacterial growth in chloride and perchlorate brines: halotolerances and salt stress responses of planococcus halocryophilus[J]. Astrobiology. 19, 77–87. doi: 10.1089/ast.2019.2069
Jaci, V. N., Diego, M., Márcio, B., Jaqueline, S., Antnio, C. N., Ana, L., et al. (2014). Effect of salt stress on the activity of bromelain in pineapple plants grown in vitro. BMC Proceed. 8:199. doi: 10.1186/1753-6561-8-S4-P199
Jie, B., Dong, M. X., Musen, W., Zi, Q. L., and Xu, S. G. (2020). Effects of antibacterial peptide-producing Bacillus subtilis and Lactobacillus buchneri on fermentation, aerobic stability, and microbial community of alfalfa silage. Bioresour. Technol. 315:123881. doi: 10.1016/j.biortech.2020.123881
Jing, X., Yang, Y., Ai, Z., Chen, S., and Zhou, S. (2020). Potassium channel blocker inhibits the formation and electroactivity of Geobacter biofilm. Sci. Total Environ. 705, 61–66. doi: 10.1016/j.scitotenv.2019.135796
Kanwal, M., Razzaq, A., and Maqbool, A. (2019). Characterization of phytase transgenic wheat under salt stress. Biol. Bull. 46, 371–380. doi: 10.1134/S106235901904006X
Kung, L. J., and Shaver, R. (2001). Interpretation and use of silage fermentation analysis reports. Focus Forage 3, 1–5. Available online at: https://fyi.extension.wisc.edu/forage/files/2016/10/Fermentation2.pdf
Lacramioara, O., Oscar, V., Monica, B., and Marius, N. (2016). Enzymatic activity and soluble protein content in seedlings of calendula officinalis l. under salt stress. J. Plant Develop. 23, 71–79. Available online at: https://plant-journal.uaic.ro/docs/2016/7.pdf
Lei, C., Gang, G., Chengqun, Y., Jie, Z., Masataka, S., and Tao, S. (2015). The effects of replacement of whole-plant corn with oat and common vetch on the fermentation quality, chemical composition and aerobic stability of total mixed ration silage in Tibet. Anim. Sci. J. 86, 69–76. doi: 10.1111/asj.12245
Li, X., Tian, J., Zhang, Q., Jiang, Y., Wu, Z., and Yu, Z. (2018). Effects of mixing red clover with alfalfa at different ratios on dynamics of proteolysis and protease activities during ensiling. J. Dairy Sci. 101, 54–64. doi: 10.3168/jds.2018-14763
Lin, C., Bolsen, K. K., Brent, B. E., Hart, R. A., Dickerson, J. T., Feyerherm, A. M., et al. (1992). Epiphytic microflora on alfalfa and whole-plant corn. J. Dairy Sci. 75, 84–93. doi: 10.3168/jds.S0022-0302(92)78010-2
Liu, J., Tang, L., Gao, H., Zhang, M., and Guo, C. H. (2019). Enhancement of alfalfa yield and quality by plant growth-promoting rhizobacteria under saline-alkali conditions. J. Sci. Food Agric. 99, 281–289. doi: 10.1002/jsfa.9185
Liu, J. H., Chu, B., Yang, X., and Jin, Y. L. (2021). Relationship between the index of protein modification (Kolbach index) and degradation of macromolecules in wheat malt. J. Food Sci. 86, 2300–2311. doi: 10.1111/1750-3841.15701
Lu, Q., Ge, G., Sa, D., Wang, Z., Hou, M., and Jia, Y. S. (2021). Effects of salt stress levels on nutritional quality and microorganisms of alfalfa-influenced soil. Peer J. 9:e11729. doi: 10.7717/peerj.11729
Luttenton, M. R., and Rada, R. G. (2010). Effects of disturbance on epiphytic community architecture. J. Phycol. 22, 320–326. doi: 10.1111/j.1529-8817.1986.tb00030.x
Mahmoud, S., Talaat, B., Rania, S., and Ali, M. (2017). Effect of plant growth stimulants on alfalfa response to salt stress. Agric. Sci. 8, 267–291. doi: 10.4236/as.2017.84020
McAllister, T. A., Duniere, L., Drouin, P., Xu, S., Wang, Y., Munns, K., et al. (2018). Silage review: using molecular approaches to define the microbial ecology of silage. J. Dairy Sci. 101, 60–74. doi: 10.3168/jds.2017-13704
McDonald, P., Henderson, A. R., and Heron, S. J. E. (1991). The biochemistry of silage. 2nd ed. Marlow: Chalcombe Publications.
Morais, M. B., Barbosa-Neto, A. G., Willadino, L., Ulisses, C., and Junior, T. C. (2019).al. Salt Stress induces increase in starch accumulation in duckweed (Lemna aequinoctialis, L.): biochemical and physiological aspects. J. Plant Grow. Regul. 38, 683–700. doi: 10.1007/s00344-018-9882-z
Nazar, M., Wang, S., Zhao, J., Dong, Z., Li, J., Kaka, N. A., et al. (2020). The feasibility and effects of exogenous epiphytic microbiota on the fermentation quality and microbial community dynamics of whole crop corn. Bioresour. Technol. 306:123106. doi: 10.1016/j.biortech.2020.123106
Nidhi, B., Savita, B., Deepti, B., Anju, Y., and Alok, K. (2013). The greater effectiveness of glomus mosseae and glomus intraradices in improving productivity, oil content and tolerance of salt-stressed menthol mint (Mentha arvensis). J. Sci. Food Agric. 93, 54–61. doi: 10.1002/jsfa.6021
Oliveira, A. S., Weinberg, Z. G., Ogunade, I. M., Cervantes, A. A. P., Arriola, K. G., et al. (2017). Meta-analysis of effects of inoculation with homofermentative and facultative heterofermentative lactic acid bacteria on silage fermentation, aerobic stability, and the performance of dairy cows. J. Dairy Sci. 100, 4587–4603. doi: 10.3168/jds.2016-11815
Ping, L., Shuren, J., Qian, W., Meng, Z. Q., Chen, H., and Yi, X. S. (2017). Adding sweet potato vines improve the quality of rice straw silage. Anim. Sci. J. 88, 625–632. doi: 10.1111/asj.12690
Rengasamy, P. (2006). World salinization with emphasis on Australia. J. Exp. Bot. 57, 1017–1023. doi: 10.1093/jxb/erj108
Sanada, Y., Kuribayashi, K., Andoh, T., Hayashi, F., Tamai, N., Wada, K., et al. (1995). Novel light-dark change of proline levels in halophyte (Mesembryanthemum crystallinum L.) and glycophytes (Hordeum vulgare L. and triticum aestivum L.) leaves and roots under salt stress. Plant Cell Physiol. 36, 965–970. doi: 10.1093/oxfordjournals.pcp.a078867
Sun, L., Lei, P., Wang, Q., Ma, J., Zhan, Y., Jiang, K., et al. (2020).The endophyte pantoea alhagi NX-11 alleviates salt stress damage to rice seedlings by secreting exopoly saccharides. Front. Microbiol. 10:3112. doi: 10.3389/fmicb.2019.03112
Tian, D., Xu, X., Peng, Q., Wen, Z., Zhang, Y., Wei, C., et al. (2019). In vitro fermentation of arabinoxylan from oat (Avena sativa L.) by Pekin duck intestinal microbiota. Biotech 9:54doi: 10.1007/s13205-019-1571-5
Van, S. P. V., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 83–97. doi: 10.3168/jds.S0022-0302(91)78551-2
Wang, S., Zhao, J., Dong, Z., Li, J., Kaka, N. A., and Shao, T. (2020). Sequencing and microbiota transplantation to determine the role of microbiota on the fermentation type of oat silage. Bioresour. Technol. 309:123371. doi: 10.1016/j.biortech.2020.123371
Xiao, Y., Zhao, G., Li, T., Zhou, X., and Li, J. (2019). Soil salinization of cultivated land in Shandong Province, China dynamics during the past 40 years. Land Degrad. Develop. 30, 426–436. doi: 10.1002/ldr.3236
Xue, F. L., Hong, T. X., Mu, K. Y., and Chen, S. W. (2018). Morphological and physiological response and salt-tolerance differences of three coastal plants under salt stress[J]. For. Res. 31, 95–103. doi: 10.13275/j.cnki.lykxyj.2018.03.013
Xue, J. Z., Mao, L., Dao, G. Y., Jun, T., Han, L. Z., and Ye, Y. C. (2021). Natural fermentation quality and bacterial community of 12 Pennisetum sinese varieties in Southern China. Front. Microbiol. 2021:627820. doi: 10.3389/fmicb.2021.627820
Yang, L. L., Yuan, X. J., Li, J. F., Dong, Z. H., and Shao, T. (2019). Dynamics of microbial community and fermentation quality during ensiling of sterile and nonsterile alfalfa with or without Lactobacillus plantarum inoculant. Bioresour. Technol. 275, 280–287. doi: 10.1016/j.biortech.2018.12.067
Keywords: alfalfa, salt stress, silage, fermentation quality, microbial community
Citation: Lu Q, Wang Z, Sa D, Hou M, Ge G, Wang Z and Jia Y (2021) The Potential Effects on Microbiota and Silage Fermentation of Alfalfa Under Salt Stress. Front. Microbiol. 12:688695. doi: 10.3389/fmicb.2021.688695
Received: 01 April 2021; Accepted: 06 September 2021;
Published: 11 October 2021.
Edited by:
Sanket J. Joshi, Sultan Qaboos University, OmanReviewed by:
Ricardo Andrade Reis, São Paulo State University, BrazilYimin Cai, Japan International Research Center for Agricultural Sciences (JIRCAS), Japan
Copyright © 2021 Lu, Wang, Sa, Hou, Ge, Wang and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yushan Jia, anlzX25tQHNpbmEuY29t
†These authors have contributed equally to this work
 Qiang Lu
Qiang Lu Zhen Wang
Zhen Wang Duowen Sa1
Duowen Sa1 Yushan Jia
Yushan Jia