
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 17 May 2021
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.687260
This article is part of the Research TopicRoles of Regulatory RNAs in Bacterial PathogensView all 13 articles
The outer membrane protein OmpA is a virulence factor in many mammalian pathogens. In previous global RNA structure probing studies, we found evidence for a temperature-modulated RNA structure in the 5'-untranslated region (5'-UTR) of the Yersinia pseudotuberculosis ompA transcript suggesting that opening of the structure at host-body temperature might relieve translational repression. Here, we support this hypothesis by quantitative reverse transcription PCR, translational reporter gene fusions, enzymatic RNA structure probing, and toeprinting assays. While ompA transcript levels decreased at 37°C compared to 25°C, translation of the transcript increased with increasing temperature. Biochemical experiments show that this is due to melting of the RNA structure, which permits ribosome binding to the 5'-UTR. A point mutation that locks the RNA structure in a closed conformation prevents translation by impairing ribosome access. Our findings add another common virulence factor to the growing list of pathogen-associated genes that are under RNA thermometer control.
OmpA is a highly abundant and conserved outer membrane protein in Gram-negative bacteria (Smith et al., 2007). It has a barrel-like structure that confers porin activity. Apart from its function in the influx and efflux of various compounds, OmpA is a multifaceted protein with various other functions, hence its designation as a molecular Swiss army knife. For instance, it plays an important role in envelope stability. In Acinetobacter baumannii, it was shown that two highly conserved residues anchor OmpA to the peptidoglycan layer (Park et al., 2012). In addition, OmpA forms complexes with the outer membrane protein Pal, which also associates with the peptidoglycan layer via a conserved α-helical interaction motif (Clavel et al., 1998).
OmpA and OmpA-like proteins have been recognized as virulence factors and are considered as potential vaccine candidates (Confer and Ayalew, 2013). The mode of action by which OmpA contributes to virulence ranges from adherence to epithelial cells and invasion (Smith et al., 2007; Gaddy et al., 2009) to biofilm formation (Weiser and Gotschlich, 1991; Azghani et al., 2002). In Escherichia coli, OmpA contributes to the resistance against serum killing by binding to the C4b-binding protein, which inhibits excessive activation of the complement system (Weiser and Gotschlich, 1991; Prasadarao et al., 2002). A. baumannii OmpA confers serum resistance through the acquisition of factor H to the cell surface (Kim et al., 2009). In addition, secreted A. baumannii OmpA is able to facilitate apoptosis by inducing mitochondrial damage and the release of proapoptotic molecules, leading to epithelial cell death (Choi et al., 2005). In a mouse model, A. baumannii OmpA is important to induce death as demonstrated by a killing defect of an ompA deletion strain (Sánchez-Encinales et al., 2017). In E. coli K1, OmpA is required to cause neonatal meningitis by enabling invasion of brain microvascular endothelial cells and crossing the blood-brain barrier (Prasadarao et al., 1996). OmpA in Yersinia pestis and Yersinia pseudotuberculosis is important for intracellular survival within macrophages as shown by reduced survival of strains lacking ompA (Bartra et al., 2012).
The amount of OmpA and other porins in the outer membrane is tightly regulated in response to numerous external conditions. Like many other genes coding for outer membrane proteins, E. coli ompA is subject to posttranscriptional regulation by small RNAs (sRNAs; Guillier et al., 2006). The paradigm of this type of control is the regulation of the ompF mRNA by the MicF sRNA (Mizuno et al., 1984). The 5'-untranslated region (5'-UTR) of ompA base-pairs with the sRNA MicA, which destabilizes the ompA transcript and requires the help of the RNA chaperone Hfq (Rasmussen et al., 2005; Udekwu et al., 2005). A second Hfq-dependent sRNA involved in ompA regulation in E. coli is RseX, which also regulates the expression of another outer membrane protein gene called ompC (Douchin et al., 2006).
A fundamentally different mode of riboregulation was recently reported for the Shigella dysenteriae ompA gene, which is regulated by a fourU-type RNA thermometer (RNAT; Murphy et al., 2020). Shigella dysenteriae is a serious foodborne pathogen causing the diarrheal disease shigellosis, and the OmpA protein is involved in intracellular spreading of the pathogen (Ambrosi et al., 2012). The last 20 residues of the 133-nucleotides long 5'-UTR of the ompA transcript fold into a simple hairpin structure, in which four uridines pair with GGAG, a part of the Shine-Dalgarno (SD) sequence. This structure is sufficiently strong to inhibit ribosome access at environmental temperatures but labile enough to denature at a host-body temperature in order to facilitate efficient translation resulting in an elevated level of the virulence factor (Murphy et al., 2020).
Regulating the production of virulence factors like OmpA in response to temperature is a sensible strategy since a shift to 37°C is among the most consistent changes upon arrival in the warm-blooded host. Therefore, many pathogens have established various mechanisms to measure the ambient temperature at the DNA, RNA, or protein level (Steinmann and Dersch, 2013; Mandin and Johansson, 2020). A widely used strategy in numerous pathogenic bacteria is the translational control of virulence-associated genes by RNATs (Loh et al., 2018). The sequence and structure of these regulatory elements are surprisingly diverse. Even orthologous genes in related organisms are controlled by unrelated RNATs, and this is also the case for the ompA gene. While the Shigella ompA transcript is regulated by a fourU thermometer (Murphy et al., 2020), the RNAT upstream of ompA in Y. pseudotuberculosis presented here is structurally disparate but functionally equivalent. Like Shigella, Y. pseudotuberculosis is a notorious foodborne pathogen causing gut-associated diseases. A considerable number of Yersinia virulence genes are temperature regulated, often by riboregulatory processes (Knittel et al., 2018). We identified a very promising RNAT upstream of ompA by a global RNA structuromics approach, in which Y. pseudotuberculosis RNA structures were probed at three different temperatures (Righetti et al., 2016). Here, we set out to study the structure-function relationship of this temperature-labile element and show that it is a functional RNAT allowing induction of OmpA levels at increasing temperature.
Bacterial strains used in this study are listed in Table 1. Cells were grown in lysogeny broth (LB; 1% NaCl, 1% tryptone, and 0.5% yeast extract) at indicated temperatures. Cultures were supplemented with 150 μg/ml ampicillin when necessary. For the induction of the PBAD promoter, the medium was supplemented with L-arabinose to a final concentration of 0.01% (E. coli) or 0.1% (Y. pseudotuberculosis), respectively.
All utilized oligonucleotides and plasmids are summarized in Tables 2 and 3, respectively. Point mutations were generated by site-directed mutagenesis according to the instruction manual of the QuikChange® mutagenesis kit (Agilent Technologies).
The ompA RNAT-bgaB fusion plasmid (pBO4435) was constructed by first amplifying the YPK_2630 (ompA) 5'-UTR including 30 bp of the ompA coding region (108 bp) with primer pairs ompA_UTR_fw/ompA_UTR_rev, digested with NheI and EcoRI and ligated into pBAD2-bgaB-His. The repressed ompA RNAT-bgaB (U38UC) fusion was constructed by site-directed mutagenesis with primer pair ompA_rep_fw/ompA_rep_rev using pBO4435 as a template.
The runoff plasmid for in vitro transcription of the ompA RNAT (pBO4920) was constructed by blunt-end ligation of a PCR-amplified DNA fragment (primer pair ompA_ro_fw/ompA_ro_rev), comprising the T7 RNA polymerase promoter, and the ompA RNAT including 30 bp of the ompA coding region, into the EcoRV restriction site of pUC18. Insertion of the repressive mutation (U38UC) into pBO4920 was achieved by site-directed mutagenesis (primer pair ompA_rep_fw/ompA_rep_rev).
Escherichia coli DH5α or Y. pseudotuberculosis YPIII cells carrying the ompA RNAT-bgaB fusion plasmids were grown overnight in LB supplemented with ampicillin at 25°C. Before being inoculated with an overnight culture to an optical density at 600 nm (OD600) of 0.1, LB media supplemented with ampicillin were pre-warmed to 25°C. After growth to an OD600 of 0.5, transcription was induced with 0.01% (E. coli) or 0.1% for (Y. pseudotuberculosis) L-arabinose, respectively. The culture was split and shifted to pre-warmed 100 ml flasks. After incubation for 30 min, 400 μl samples was subsequently taken for β-galactosidase assay, 2 ml samples for Western blotting, and 4 ml samples for RNA isolation. The β-galactosidase assay was carried out as described previously (Gaubig et al., 2011; Righetti et al., 2016).
Cell pellets were resuspended in 1 × SDS sample buffer (2% SDS, 0.1% bromophenol blue, 1% 2-mercaptoethanol, 25% glycerol, 50 mM Tris/HCl, and pH 6.8) according to their optical density (100 μl per OD600 of 1). After boiling for 10 min at 95°C, samples were centrifugated (10 min, 13,000 rpm) and the supernatant was separated by SDS gel electrophoresis in 5% stacking and 12% separating gels. Size-separated proteins were transferred by tank blotting onto a nitrocellulose membrane (Hybond-C Extra, GE Healthcare). An anti-His-HRP conjugate antibody (Bio-Rad) was used in a 1:4,000 dilution. Luminescence signals were detected by incubating membranes with Immobilon Forte Western HRP substrate (Millipore) and the FluorChem SP (Alpha Innotec).
Total RNA was extracted using the peqGOLD TriFast reagent according to the manufacturer’s protocol. RNA samples were treated with Turbo™ DNase (TURBO DNA-free™ Kit, Invitrogen) to remove DNA contamination. cDNA synthesis was performed using the iScript™ cDNA synthesis Kit (Bio-Rad) according to the manufacturer’s protocol with 1 μg RNA per reaction. 2 μl of 1:10 diluted cDNA was mixed with 250 nM of each primer, 5 μl of 2× iTaq Universal SYBR Green Supermix, and 2.5 μl sterile water (Carl Roth). Amplification and detection were performed in a CFX Connect™ Real-Time System (Bio-Rad). Standard curves were used to calculate primer efficiency and determine the linear range of amplification. Relative transcript amounts were calculated using the primer efficiency corrected method (Pfaffl, 2001). The non-thermoregulated reference genes gyrB and nuoB were used for normalization.
RNA for RNA structure probing and primer extension inhibition experiments were synthesized in vitro by runoff transcription with T7 RNA polymerase (Thermo Scientific) from EcoRV-linearized plasmids (listed in Table 3) as previously described (Righetti et al., 2016).
RNA structure probing of the 5'-UTR and 30 nt of ompA was performed as described previously (Twittenhoff et al., 2020ab). Briefly, in vitro transcribed and 5'-[32P]-labeled RNA (3,000 cpm) was mixed with buffer and tRNAs, preincubated for 5 min at the respective temperature, and treated with T1 (0.0017U; Invitrogen) or T2 (0.056U; MoBiTec) RNases for 5 min. For digestion with RNase T1 and RNase T2, a 5 × TN buffer (100 mM Tris acetate, pH 7, and 500 mM NaCl) was used. An alkaline hydrolysis ladder was prepared as described in Brantl and Wagner (1994), meaning 60,000 cpm of labeled RNA was mixed with tRNAs and ladder buffer (1M Na2CO3 and 1M NaHCO3; pH 9) incubated for 2 min at 90°C. The T1 ladder was generated by using 30,000 cpm-labeled RNA. The samples were heated with 2 μl sequencing buffer (provided with RNase T1) at 90°C. Afterward, the RNA was incubated with the T1 RNase for 5 min at 37°C. All reactions were stopped by the addition of formamide loading dye and denaturation at 95°C.
Toeprinting analysis was performed with 30S ribosomal subunits, in vitro transcribed RNA, and tRNAfMet (Sigma-Aldrich) according to a protocol as described before (Hartz et al., 1988). The 5'-[32P]-labeled oligonucleotide ompA_ro_rv, complementary to the 3' end of the ompA mRNA, was used as a primer for cDNA synthesis. The radiolabeled primer (0.16 pmol) was annealed to the ompA mRNA (0.08 pmol) and incubated with 30S ribosomal subunits (24 pmol) or Watanabe buffer [60 mM HEPES/KOH, 10.5 mM Mg(CH3COO)2, 690 mM NH4COO, 12 mM 2-mercaptoethanol, 10 mM spermidine, and 0.25 mM spermine] in the presence of tRNAfMet (8 pmol) at 25, 37, or 42°C for 10 min. After the addition of 2 μl MMLV-Mix [VD+Mg2+ buffer, BSA, dNTPs, and 800 U MMLV reverse transcriptase (Invitrogen)], cDNA synthesis was performed for 10 min at 37°C. The reaction was stopped by the addition of formamide loading dye, and the samples were separated on an 8% denaturing polyacrylamide gel. The Thermo Sequenase Cycle Sequencing Kit (Applied Biosystems) was used for sequencing reactions with plasmid pBO4920 as a template and radiolabeled primer ompA_ro_rv.
A previously conducted global in vitro RNA structure probing analysis (Righetti et al., 2016) provided strong evidence for the existence of temperature-sensitive RNA structures in the 5'-UTRs of numerous mRNAs. One of the top candidates (rank #3 of chromosomally encoded mRNAs) discovered by this parallel analysis of RNA structures approach was the 5'-UTR of ompA. The RNA structure observed at 25°C was destabilized at higher temperatures (Figure 1A). This was especially prominent in the region around the SD sequence and in the early coding region. Here, the PARS profile dropped from positive to negative values indicative of a transition from a double-stranded (ds) to single-stranded (ss) conformation. This behavior is typical of zipper-like RNATs that facilitate ribosome binding at elevated temperatures (Kortmann and Narberhaus, 2012). The PARS analysis revealed an almost completely folded ompA 5'-UTR that is engaged in several stem-loop structures (Figure 1B). Strikingly, the SD sequence (53-GGAG-56) is imperfectly paired and contains a bulged adenine. It is very likely that this mismatched nucleotide in concert with four unpaired nucleotides (58-GUAA-61) between the SD sequence and the AUG start codon is key to the temperature sensitivity of this 5'-UTR.
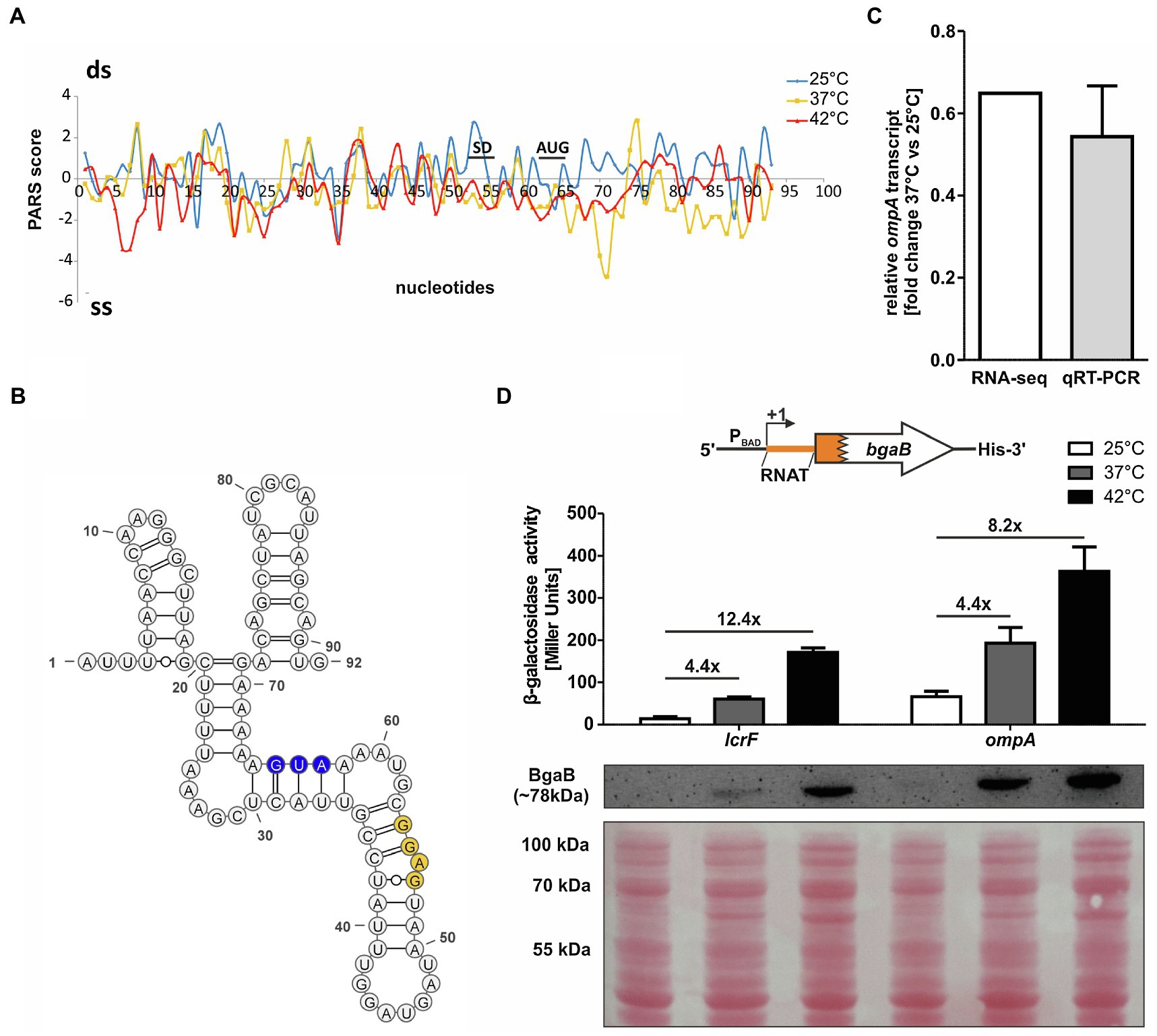
Figure 1. The ompA 5'-UTR contains a thermoresponsive RNA structure. (A) PARS profiles of the ompA RNAT (−62 nt to +30 nt from AUG) at 25, 37, and 42°C (Righetti et al., 2016). The potential SD sequence and the AUG are shown. A positive PARS score is indicative of a double-stranded (ds) conformation, whereas a negative PARS score suggests a single-stranded (ss) conformation. A drop from a positive score at 25°C to a negative score at 37°C suggests melting of a secondary structure. (B) PARS-derived secondary structure of the ompA RNAT at 25°C. The potential SD sequence is highlighted in yellow and its corresponding AUG in blue. –, AU pair; =, GC pair; and ○, GU pair. (C) Comparison of ompA transcript amount between 37 and 25°C by RNA sequencing (Nuss et al., 2015) and qRT-PCR (this study) of exponentially grown YPIII cells at 25 and 37°C, respectively. For qRT-PCR, the obtained data were normalized to gyrB and nuoB. qRT-PCR was prepared in biological triplicates and technical triplicates. (D) Translational control was measured by bgaB fusions. A schematic representation of the reporter gene fusion is displayed. The ompA RNAT was translationally fused to bgaB under control of the PBAD promotor. As a control, the lcrF RNAT fusion was used. The fusion plasmids were introduced into E. coli cells and grown to an OD600 of 0.5 at 25°C. Subsequently, transcription was induced by the addition of 0.01% L-arabinose and the cultures were split and transferred into pre-warmed flask at 25, 37, and 42°C, respectively. After 30 min of incubation, samples were taken for β-galactosidase assays and Western blot analysis. Experiments were carried out in biological triplicates. Mean and corresponding standard deviations are shown. Western blot membranes were stained with Ponceau S as a loading control. One representative Western blot is shown. Analysis was carried out in biological triplicates.
To examine the regulation of ompA in detail, we first checked, whether transcription of the gene is temperature-controlled by performing qRT-PCR on total RNA samples from Y. pseudotuberculosis YPIII after growth at 25 and 37°C (Figure 1C). Consistent with already existing RNA-seq results (Nuss et al., 2015), we observed a fold change around 0.6 (37 vs. 25°C) showing a reduction in ompA mRNA at a higher temperature.
The RNA structurome data, in contrast, suggested an upregulation of OmpA protein at 37°C due to the thermoresponsive nature of the ompA 5'-UTR. To test this hypothesis, we translationally fused the ompA 5'-UTR to the bgaB gene coding for a heat-stable His-tagged β-galactosidase downstream of the arabinose-inducible PBAD promotor and measured the β-galactosidase activity and protein amounts at 25°C and after a shift to 37 or 42°C. The well-studied lcrF RNAT of Y. pseudotuberculosis served as a positive control (Figure 1D). A clear increase in β-galactosidase activity and protein amount after the temperature shift supported the existence of a functional RNAT upstream of ompA able to confer translational repression at 25°C and induction at 37 or 42°C.
One way of verifying the contribution of a temperature-sensitive RNA structure in translational control is the construction of a stabilized version. Based on the PARS-derived secondary structure, we introduced a cytosine between U38 and A39 (called U38UC or “rep” for repressed variant) in the anti-SD sequence leading to a perfectly paired SD sequence without any mismatches (Figure 2A). The increased thermodynamic stability should prevent the RNA structure from melting at higher temperatures and thereby abolish RNAT functionality. Indeed, reporter gene assays revealed the absence of β-galactosidase protein and activity at 25 and 37°C in E. coli as well as in Y. pseudotuberculosis (Figure 2B). Apparently, the introduction of just one additional base pair was sufficient to abolish the temperature regulation. To solidify our claim that the 5'-UTR regulates translation (and not transcription or mRNA degradation), we analyzed the bgaB mRNA levels in all three tested Y. pseudotuberculosis strains by qRT-PCR and found that the transcript levels were the same at 25 and 37°C (Figure 2C).
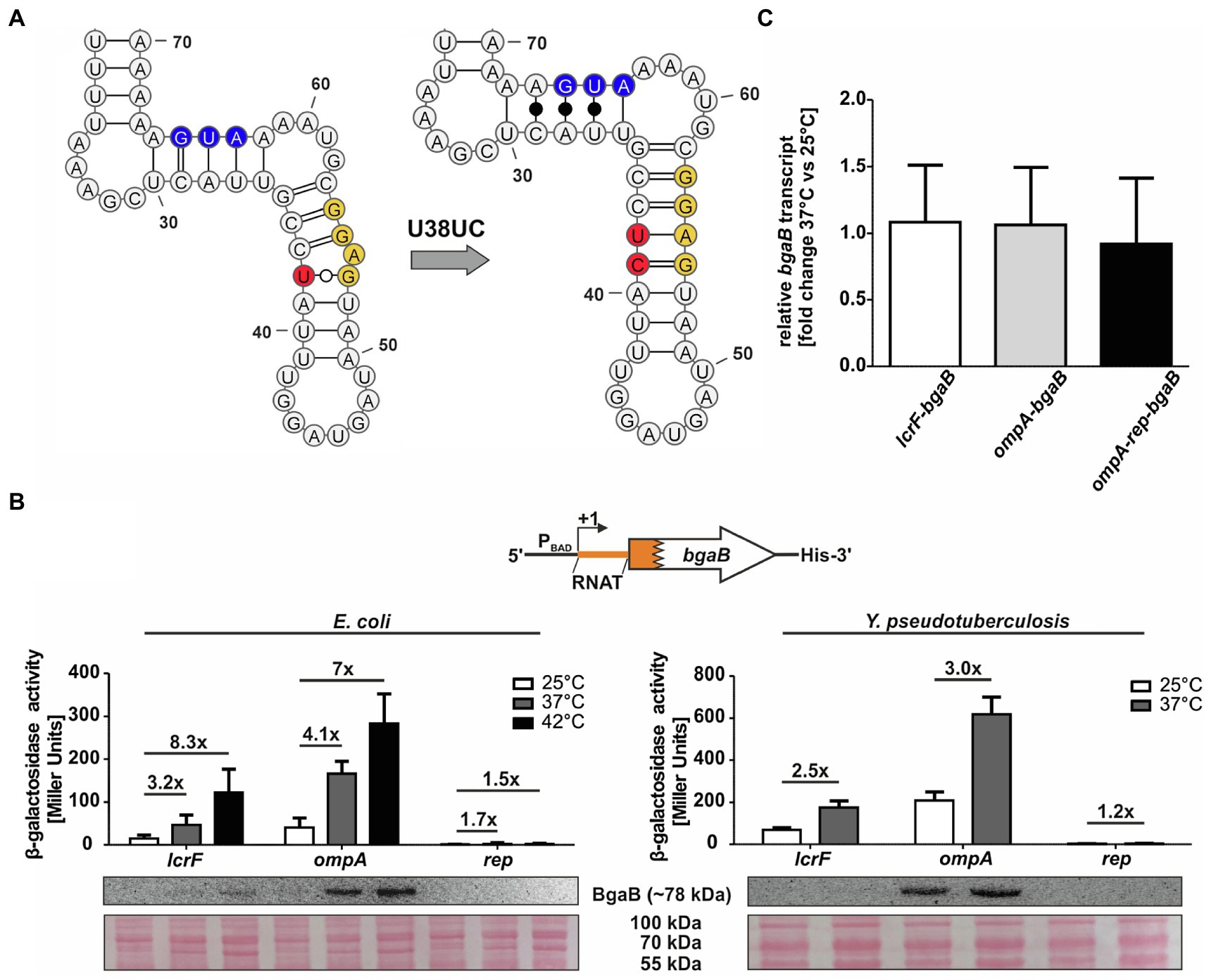
Figure 2. Mutation of the anti-SD sequence locks the RNAT in a closed conformation. (A) PARS-derived secondary structure of the ompA RNAT stem and the predicted stabilized structure at 25°C. The potential SD sequence is highlighted in yellow, the mutation site of the anti-SD sequence in red and the AUG in blue. (B) Translational control was measured by bgaB fusions. A schematic representation of the reporter gene fusion is displayed. The ompA RNAT was translationally fused to bgaB under control of the PBAD promotor. Additionally, a point mutation (U38UC) was introduced to stabilize the RNAT (rep). As a control, the lcrF RNAT fusion was used. The fusion plasmids were introduced into E. coli and Y. pseudotuberculosis YPIII cells and grown to an OD600 of 0.5 at 25°C. Subsequently, transcription was induced by the addition of 0.01% for E. coli or 0.1% for Y. pseudotuberculosis L-arabinose and the cultures were split and transferred into pre-warmed flask at 25 and 37°C, respectively. After 30 min of incubation, samples were taken for β-galactosidase assays, Western blot analysis, and qRT-PCR. Experiments were carried out in biological triplicates. Mean and corresponding standard deviations are shown. Western blot membranes were stained with Ponceau S as a loading control. One representative Western blot is shown. Analysis was carried out in biological triplicates. (C) Comparison of bgaB transcript amount between 37 and 25°C by qRT-PCR of exponentially grown YPIII cells harboring the ompA RNAT-bgaB fusion plasmids at 25 and 37°C, respectively. For qRT-PCR, the obtained data were normalized to gyrB and nuoB. qRT-PCR was prepared in biological triplicates and technical triplicates.
Next, we examined the structure and temperature-induced conformational changes in the RNAT by enzymatic RNA structure probing using in vitro transcribed and 5' end-labeled ompA RNA at 25, 37, and 42°C treated with RNases T1 (preferentially cleaves at unpaired guanines) and T2 (preferentially cleaves at unpaired adenines but also other ss residues; Figure 3A). The cleavage pattern supported the overall structure of the ompA 5'-UTR by prominent cleavage at 25°C in apical loops (e.g., C80 and G81). In accordance with the dynamic PARS-derived secondary structure (Figures 1A, 3B), residues around the SD sequence were sensitized against RNase cleavage at higher temperatures due to melting of the secondary structure. For example, nucleotides G53 and G55 were almost untouched by T1 at 25°C but efficiently cleaved at 37 and 42°C. Residual cleavage of the bulged A54 by T2 at 25°C massively increased at 37 and 42°C making it the most prominent cleavage product at high temperatures. Fully consistent with the reporter gene assays (Figure 2), the SD sequence of the rep variant was almost completely inaccessible to RNase cleavage demonstrating that melting of the RNAT is prevented by the point mutation, which locks the structure in a closed conformation.
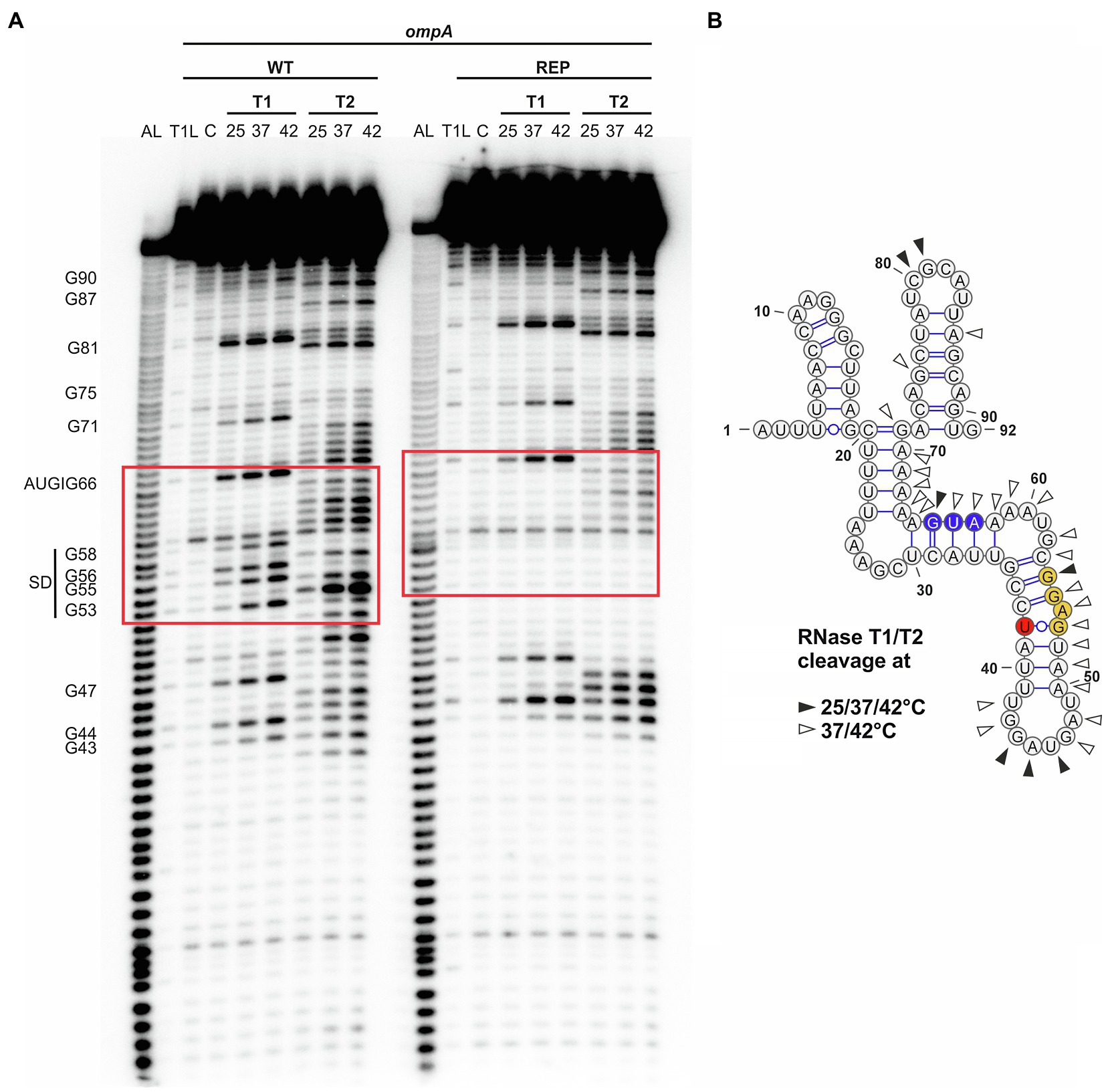
Figure 3. Accessibility of the ribosome-binding site of ompA is increased at higher temperatures. (A) Enzymatic structure probing of the ompA RNAT (WT) and its stabilized version (REP) at 25, 37, and 42°C. In vitro transcribed RNA was 5'-[32P] labeled and treated with RNases T1 and T2, respectively. AL, alkaline ladder; T1L, RNase T1 cleavage ladder in sequencing buffer at 37°C; C, RNA treated with water instead of RNases – cleavage control. Ribosome-binding site is highlighted by a red box. Experiment was carried out at least three times. (B) PARS-derived secondary structure of the ompA RNAT at 25°C. The potential SD sequence is highlighted in yellow, the mutation site of the anti-SD sequence in red, and the AUG in blue. Distinctive nucleotide cleavage is indicated by black and white arrows.
The observed melting of the ompA 5'-UTR at higher temperatures (Figures 1A, 3) together with the reporter gene studies (Figures 1D, 2) strongly suggested increased translation initiation at higher temperatures. We employed primer extension inhibition assays (toeprinting) to demonstrate ribosome binding. In vitro transcribed ompA RNA was reversely transcribed from a radiolabeled oligonucleotide in the presence or absence of 30S ribosomal subunits at 25 and 37°C. Ribosomes bound to the mRNA are stabilized in a ternary complex with tRNAfMet and act as a roadblock for reverse transcription. This leads to premature termination of reverse transcription and thus shorter fragments (toeprints). As expected, in the presence of ribosomes, the toeprint signals in the appropriate distance from the AUG start codon were weak at 25°C but strong at 37°C (Figure 4). No toeprint signal, i.e., no ribosome binding, was found when the stabilized rep version was assayed. Instead, termination products independent of the presence of ribosomes appeared within the codon sequence, a phenomenon frequently observed downstream (considering the direction of the reverse transcriptase) of stable structures in stabilized RNATs (Böhme et al., 2012; Twittenhoff et al., 2020ab).
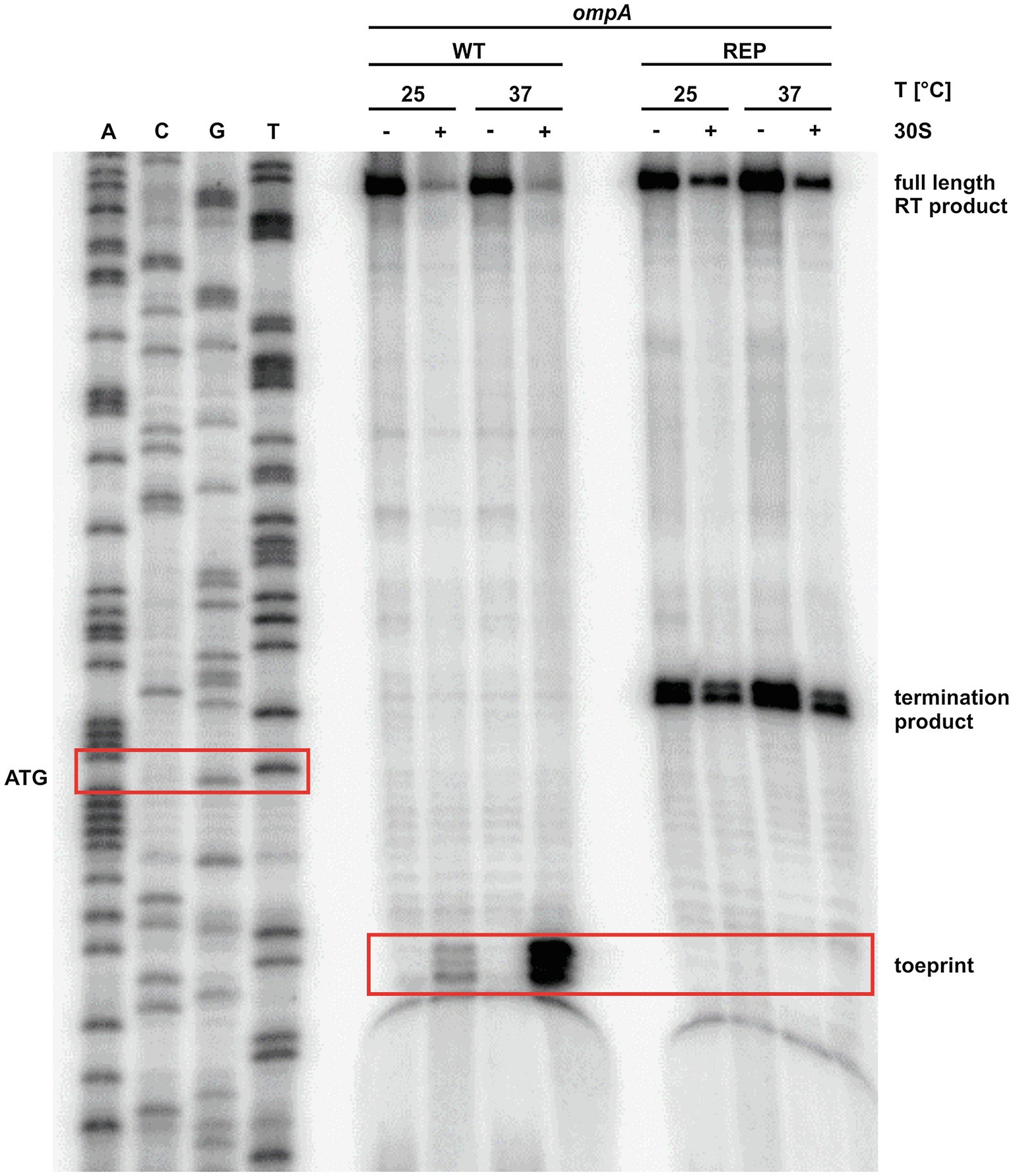
Figure 4. The ribosome binds to the ompA transcript at 37°C. Primer extension inhibition of the ompA RNAT (WT) and its stabilized version (REP) at 25 and 37°C with (+) and without (−) the addition of 30S ribosomal subunits. The signals represent the full-length reverse transcription (RT) product, the premature termination product, and the toeprint signal (caused by ribosome binding). ACGT lanes indicate sequencing reactions. Position of ATG and the toeprint signal is highlighted by red boxes. Experiment was carried out at least three times.
Sensing of and rapidly responding to the sudden external changes associated with host infection is crucial for enteric pathogens like Y. pseudotuberculosis. One parameter that predictably and reversibly changes during the transitions from the environment to a warm-blooded host and back into the environment is temperature. A fast and efficient way to reversibly modulate gene expression in response to temperature shifts is zipper-like RNATs because the mRNA is already available for translation (Grosso-Becera et al., 2015; Wei and Murphy, 2016; Loh et al., 2018). This is certainly the reason why RNATs are extremely widespread posttranscriptional control elements in numerous bacterial pathogens (Hoe and Goguen, 1993; Johansson et al., 2002; Böhme et al., 2012; Kouse et al., 2013; Loh et al., 2013; Weber et al., 2014; Wei et al., 2017; Twittenhoff et al., 2020a,b; Brewer et al., 2021).
Here, we describe the structural and functional features of an RNAT in the Y. pseudotuberculosis ompA transcript that was one of the prime candidates in our previous PARS study (Righetti et al., 2016). We were particularly interested in this candidate because a recent report demonstrated RNAT-controlled translation of ompA in S. dysenteriae (Murphy et al., 2020). Despite regulating the same gene, it is important to note that the Yersinia and Shigella RNATs are very different with respect to length, sequence, and structure. First, the S. dysenteriae ompA 5'-UTR is 133 nucleotides long, and the last 35 residues are sufficient to confer translational control. In contrast, the RNAT from Y. pseudotuberculosis is 92 nucleotides long and more densely folded. Second, the S. dysenteriae RNAT belongs to the fourU family showing the characteristic complementarity of the SD sequence to four consecutive uracil residues (Waldminghaus et al., 2007). The Y. pseudotuberculosis RNAT is not a member of this family. The inherent temperature lability is due to a bulged adenine residue in the center of the SD sequence, which pairs imperfectly with an anti-SD sequence comprised of CCUA. Introducing one additional residue that pairs with the exposed adenine completely eliminated the expression and heat induction even when sufficient ompA mRNA was present. This demonstrates the relevance of precisely balanced structural features in a functional RNAT to respond within the virulence-relevant temperature regime. Overall, the ompA thermometer is another interesting example of convergent evolution of RNATs as has previously been proposed for the regulation of CnfY-type toxins (Twittenhoff et al., 2020a b).
Many enterobacterial outer membrane protein genes are under tight posttranscriptional regulation (Guillier et al., 2006; Vogel and Papenfort, 2006). A highly conserved sRNA also believed to control ompA in Yersinia species is MicA (Udekwu et al., 2005). It binds across the SD sequence and prevents ribosome binding, which facilitates RNaseE-mediated cleavage. The temperature-induced structural transition of the ompA 5'-UTR from a closed to an open conformation potentially adds another layer of posttranscriptional control. Under circumstances when MicA is abundant, the sRNA might interfere with translation initiation. The exact interplay between the ompA thermosensor, MicA (and maybe other sRNAs), the RNA chaperone Hfq, and RNases is not yet understood. The situation, however, is somewhat reminiscent of the Listeria prfA thermometer that – in its open form – is able to interact with the two sRNAs, SreA and SreB, which decreases the level of PrfA and thereby links virulence gene expression to nutrient availability (Loh et al., 2009).
Having OmpA under temperature regulation supports its role as a virulence factor. There is growing evidence that the outer membrane protein is involved in many pathogenesis-related processes in diverse Gram-negative bacteria (Krishnan and Prasadarao, 2012). It is involved in adherence to epithelial cells (Smith et al., 2007; Gaddy et al., 2009), mouse mortality (Sánchez-Encinales et al., 2017), and biofilm formation (Weiser and Gotschlich, 1991; Azghani et al., 2002). OmpA of pathogenic Yersinia species is a highly immunogenic protein and may be an excellent vaccine candidate, due to its cross-immunogenicity and intense immune response among Yersinia species (Chen et al., 2015). Furthermore, Y. pestis and Y. pseudotuberculosis ompA mutants are defective in surviving in macrophages, and the Y. pestis mutant is outcompeted by the wild-type strain in a mouse-infection model (Bartra et al., 2012).
Given the importance of OmpA for efficient pathogenesis, it may be a promising target for treatment of infections caused by Gram-negative bacteria. A recently identified OmpA inhibitor, cyclic hexapeptide AOA-2, inhibits adherence to eukaryotic cells and biofilm formation (Vila-Farrés et al., 2017). The compound attenuates virulence by reducing the dissemination between organs and decreases mouse mortality after A. baumannii, E. coli, and Pseudomonas aeruginosa infections. In combination with other antimicrobial agents, OmpA inhibitors might be helpful for combating multidrug-resistant bacteria and thereby reducing mortality caused by infections with Gram-negative pathogens.
A recent study on OmpA in Acinetobacter sp. linked ompA expression to oxidative stress (Shahryari et al., 2021). The authors suggested a protective effect by the poor permeability of the slow OmpA porin compared to other outer membrane proteins, such as OmpC, which is known to produce larger pores, at least in Salmonella (van der Heijden et al., 2016). A connection between OmpA and oxidative stress is interesting since pathogens are typically exposed to a multitude of reactive oxygen species (ROS) upon infection of a host. An intriguing finding of our RNA structurome analyses is the presence of potential RNATs upstream of various oxidative stress response genes (Righetti et al., 2016; Twittenhoff et al., 2020ab). This is unlikely to be a coincidence and suggests parallel ways of host-body temperature-induced protective measures against ROS attack. Following up on RNAT-mediated control of genes combating ROS in pathogenic bacteria might be a worthwhile subject of future studies.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
DS: conceptualization, data curation, formal analysis, investigation, validation, supervision, methodology, and writing original draft. CT: conceptualization, data curation, formal analysis, investigation, validation, supervision, and methodology. FB and MH: investigation, formal analysis, and methodology. FN: conceptualization, data curation, formal analysis, supervision, funding acquisition, project administration, writing original draft, review, and editing. All authors contributed to the article and approved the submitted version.
This study was financially supported by the German Research Foundation (DFG, NA240/10–2). We acknowledge support by the Open Access publication funds of the Ruhr-Universität Bochum.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the RNA group for continuous discussions and reading earlier versions of the manuscript.
Ambrosi, C., Pompili, M., Scribano, D., Zagaglia, C., Ripa, S., and Nicoletti, M. (2012). Outer membrane protein A (OmpA): a new player in Shigella flexneri protrusion formation and inter-cellular spreading. PLoS One 7:e49625. doi: 10.1371/journal.pone.0049625
Azghani, A. O., Idell, S., Bains, M., and Hancock, R. E. (2002). Pseudomonas aeruginosa outer membrane protein F is an adhesin in bacterial binding to lung epithelial cells in culture. Microb. Pathog. 33, 109–114. doi: 10.1006/mpat.2002.0514
Bartra, S. S., Gong, X., Lorica, C. D., Jain, C., Nair, M. K. M., Schifferli, D., et al. (2012). The outer membrane protein A (OmpA) of Yersinia pestis promotes intracellular survival and virulence in mice. Microb. Pathog. 52, 41–46. doi: 10.1016/j.micpath.2011.09.009
Böhme, K., Steinmann, R., Kortmann, J., Seekircher, S., Heroven, A. K., Berger, E., et al. (2012). Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 8:e1002518. doi: 10.1371/journal.ppat.1002518
Bölin, I., Norlander, L., and Wolf-Watz, H. (1982). Temperature-inducible outer membrane protein of Yersinia pseudotuberculosis and Yersinia enterocolitica is associated with the virulence plasmid. Infect. Immun. 37, 506–512. doi: 10.1128/IAI.37.2.506-512.1982
Brantl, S., and Wagner, E. G. (1994). Antisense RNA-mediated transcriptional attenuation occurs faster than stable antisense/target RNA pairing: an in vitro study of plasmid pIP501. EMBO J. 13, 3599–3607. doi: 10.1002/j.1460-2075.1994.tb06667.x
Brewer, S. M., Twittenhoff, C., Kortmann, J., Brubaker, S. W., Honeycutt, J., Massis, L. M., et al. (2021). A Salmonella Typhi RNA thermosensor regulates virulence factors and innate immune evasion in response to host temperature. PLoS Pathog. 17:e1009345. doi: 10.1371/journal.ppat.1009345
Chen, Y., Duan, R., Li, X., Li, K., Liang, J., Liu, C., et al. (2015). Homology analysis and cross-immunogenicity of OmpA from pathogenic Yersinia enterocolitica, Yersinia pseudotuberculosis and Yersinia pestis. Mol. Immunol. 68, 290–299. doi: 10.1016/j.molimm.2015.09.016
Choi, C. H., Lee, E. Y., Lee, Y. C., Park, T. I., Kim, H. J., Hyun, S. H., et al. (2005). Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell. Microbiol. 7, 1127–1138. doi: 10.1111/j.1462-5822.2005.00538.x
Clavel, T., Germon, P., Vianney, A., Portalier, R., and Lazzaroni, J. C. (1998). TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol. Microbiol. 29, 359–367. doi: 10.1046/j.1365-2958.1998.00945.x
Confer, A. W., and Ayalew, S. (2013). The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet. Microbiol. 163, 207–222. doi: 10.1016/j.vetmic.2012.08.019
Douchin, V., Bohn, C., and Bouloc, P. (2006). Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J. Biol. Chem. 281, 12253–12259. doi: 10.1074/jbc.M600819200
Gaddy, J. A., Tomaras, A. P., and Actis, L. A. (2009). The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77, 3150–3160. doi: 10.1128/IAI.00096-09
Gaubig, L. C., Waldminghaus, T., and Narberhaus, F. (2011). Multiple layers of control govern expression of the Escherichia coli ibpAB heat-shock operon. Microbiology 157, 66–76. doi: 10.1099/mic.0.043802-0
Grosso-Becera, M. V., Servin-González, L., and Soberón-Chávez, G. (2015). RNA structures are involved in the thermoregulation of bacterial virulence-associated traits. Trends Microbiol. 23, 509–518. doi: 10.1016/j.tim.2015.04.004
Guillier, M., Gottesman, S., and Storz, G. (2006). Modulating the outer membrane with small RNAs. Genes Dev. 20, 2338–2348. doi: 10.1101/gad.1457506
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/S0022-2836(83)80284-8
Hartz, D., McPheeters, D. S., Traut, R., and Gold, L. (1988). Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 164, 419–425. doi: 10.1016/s0076-6879(88)64058-4
Hoe, N. P., and Goguen, J. D. (1993). Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J. Bacteriol. 175, 7901–7909. doi: 10.1128/JB.175.24.7901-7909.1993
Johansson, J., Mandin, P., Renzoni, A., Chiaruttini, C., Springer, M., and Cossart, P. (2002). An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110, 551–561. doi: 10.1016/S0092-8674(02)00905-4
Kim, S. W., Choi, C. H., Moon, D. C., Jin, J. S., Lee, J. H., Shin, J. H., et al. (2009). Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins. FEMS Microbiol. Lett. 301, 224–231. doi: 10.1111/j.1574-6968.2009.01820.x
Knittel, V., Vollmer, I., Volk, M., and Dersch, P. (2018). Discovering RNA-based regulatory systems for Yersinia virulence. Front. Cell. Infect. Microbiol. 8:378. doi: 10.3389/fcimb.2018.00378
Kortmann, J., and Narberhaus, F. (2012). Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 10, 255–265. doi: 10.1038/nrmicro2730
Kouse, A. B., Righetti, F., Kortmann, J., Narberhaus, F., and Murphy, E. R. (2013). RNA-mediated thermoregulation of iron-acquisition genes in Shigella dysenteriae and pathogenic Escherichia coli. PLoS One 8:e63781. doi: 10.1371/journal.pone.0063781
Krishnan, S., and Prasadarao, N. V. (2012). Outer membrane protein A and OprF: versatile roles in gram-negative bacterial infections. FEBS J. 279, 919–931. doi: 10.1111/j.1742-4658.2012.08482.x
Loh, E., Dussurget, O., Gripenland, J., Vaitkevicius, K., Tiensuu, T., Mandin, P., et al. (2009). A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139, 770–779. doi: 10.1016/j.cell.2009.08.046
Loh, E., Kugelberg, E., Tracy, A., Zhang, Q., Gollan, B., Ewles, H., et al. (2013). Temperature triggers immune evasion by Neisseria meningitidis. Nature 502, 237–240. doi: 10.1038/nature12616
Loh, E., Righetti, F., Eichner, H., Twittenhoff, C., and Narberhaus, F. (2018). RNA thermometers in bacterial pathogens. Microbiol. Spectr. 6. doi: 10.1128/microbiolspec.RWR-0012-2017
Mandin, P., and Johansson, J. (2020). Feeling the heat at the millennium: thermosensors playing with fire. Mol. Microbiol. 113, 588–592. doi: 10.1111/mmi.14468
Mizuno, T., Chou, M. Y., and Inouye, M. (1984). A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc. Natl. Acad. Sci. U. S. A. 81, 1966–1970. doi: 10.1073/pnas.81.7.1966
Murphy, E. R., Roßmanith, J., Sieg, J., Fris, M. E., Hussein, H., Kouse, A. B., et al. (2020). Regulation of OmpA translation and Shigella dysenteriae virulence by an RNA thermometer. Infect. Immun. 88:e00871–19. doi: 10.1128/IAI.00871-19
Nuss, A. M., Heroven, A. K., Waldmann, B., Reinkensmeier, J., Jarek, M., Beckstette, M., et al. (2015). Transcriptomic profiling of Yersinia pseudotuberculosis reveals reprogramming of the Crp regulon by temperature and uncovers Crp as a master regulator of small RNAs. PLoS Genet. 11:e1005087. doi: 10.1371/journal.pgen.1005087
Park, J. S., Lee, W. C., Yeo, K. J., Ryu, K. S., Kumarasiri, M., Hesek, D., et al. (2012). Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J. 26, 219–228. doi: 10.1096/fj.11-188425
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Prasadarao, N. V., Blom, A. M., Villoutreix, B. O., and Linsangan, L. C. (2002). A novel interaction of outer membrane protein A with C4b binding protein mediates serum resistance of Escherichia coli K1. J. Immunol. 169, 6352–6360. doi: 10.4049/jimmunol.169.11.6352
Prasadarao, N. V., Wass, C. A., Weiser, J. N., Stins, M. F., Huang, S. H., and Kim, K. S. (1996). Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 64, 146–153. doi: 10.1128/IAI.64.1.146-153.1996
Rasmussen, A. A., Eriksen, M., Gilany, K., Udesen, C., Franch, T., Petersen, C., et al. (2005). Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol. 58, 1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x
Righetti, F., Nuss, A. M., Twittenhoff, C., Beele, S., Urban, K., Will, S., et al. (2016). Temperature-responsive in vitro RNA structurome of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 113, 7237–7242. doi: 10.1073/pnas.1523004113
Sánchez-Encinales, V., Álvarez-Marín, R., Pachón-Ibáñez, M. E., Fernández-Cuenca, F., Pascual, A., et al. (2017). Overproduction of outer membrane protein A by Acinetobacter baumannii as a risk factor for nosocomial pneumonia, bacteremia, and mortality rate increase. J. Infect. Dis. 215, 966–974. doi: 10.1093/infdis/jix010
Shahryari, S., Talaee, M., Haghbeen, K., Adrian, L., Vali, H., Shahbani Zahiri, H., et al. (2021). New provisional function of OmpA from Acinetobacter sp. strain SA01 based on environmental challenges. mSystems 6:e01175–20. doi: 10.1128/mSystems.01175-20
Smith, S. G., Mahon, V., Lambert, M. A., and Fagan, R. P. (2007). A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 273, 1–11. doi: 10.1111/j.1574-6968.2007.00778.x
Steinmann, R., and Dersch, P. (2013). Thermosensing to adjust bacterial virulence in a fluctuating environment. Future Microbiol. 8, 85–105. doi: 10.2217/fmb.12.129
Twittenhoff, C., Brandenburg, V. B., Righetti, F., Nuss, A. M., Mosig, A., Dersch, P., et al. (2020a). Lead-seq: transcriptome-wide structure probing in vivo using lead(II) ions. Nucleic Acids Res. 48:e71. doi: 10.1093/nar/gkaa404
Twittenhoff, C., Heroven, A. K., Mühlen, S., Dersch, P., and Narberhaus, F. (2020b). An RNA thermometer dictates production of a secreted bacterial toxin. PLoS Pathog. 16:e1008184. doi: 10.1371/journal.ppat.1008184
Udekwu, K. I., Darfeuille, F., Vogel, J., Reimegård, J., Holmqvist, E., and Wagner, E. G. (2005). Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 19, 2355–2366. doi: 10.1101/gad.354405
van der Heijden, J., Reynolds, L. A., Deng, W., Mills, A., Scholz, R., Imami, K., et al. (2016). Salmonella rapidly regulates membrane permeability to survive oxidative stress. MBio 7:e01238. doi: 10.1128/mBio.01238-16
Vila-Farrés, X., Parra-Millán, R., Sánchez-Encinales, V., Varese, M., Ayerbe-Algaba, R., Bayó, N., et al. (2017). Combating virulence of gram-negative bacilli by OmpA inhibition. Sci. Rep. 7:14683. doi: 10.1038/s41598-017-14972-y
Vogel, J., and Papenfort, K. (2006). Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 9, 605–611. doi: 10.1016/j.mib.2006.10.006
Waldminghaus, T., Heidrich, N., Brantl, S., and Narberhaus, F. (2007). FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 65, 413–424. doi: 10.1111/j.1365-2958.2007.05794.x
Weber, G. G., Kortmann, J., Narberhaus, F., and Klose, K. E. (2014). RNA thermometer controls temperature-dependent virulence factor expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 111, 14241–14246. doi: 10.1073/pnas.1411570111
Wei, Y., Kouse, A. B., and Murphy, E. R. (2017). Transcriptional and posttranscriptional regulation of Shigella shuT in response to host-associated iron availability and temperature. Microbiology 6:e00442. doi: 10.1002/mbo3.442
Wei, Y., and Murphy, E. R. (2016). “Temperature-dependent regulation of bacterial gene expression by RNA thermometers” in Nucleic Acids – From Basic Aspects to Laboratory Tools. eds. M. Larramendy and S. Soloneski (London:IntechOpen).
Weiser, J. N., and Gotschlich, E. C. (1991). Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect. Immun. 59, 2252–2258. doi: 10.1128/IAI.59.7.2252-2258.1991
Keywords: gene expression, thermosensor, RNA structure, pathogen, virulence, outer membrane protein, porin
Citation: Scheller D, Twittenhoff C, Becker F, Holler M and Narberhaus F (2021) OmpA, a Common Virulence Factor, Is Under RNA Thermometer Control in Yersinia pseudotuberculosis. Front. Microbiol. 12:687260. doi: 10.3389/fmicb.2021.687260
Received: 29 March 2021; Accepted: 21 April 2021;
Published: 17 May 2021.
Edited by:
Olga Soutourina, Institut de Biologie Intégrative de la Cellule (I2BC), FranceReviewed by:
Pierre Mandin, Laboratoire de chimie bactérienne (LCB), FranceCopyright © 2021 Scheller, Twittenhoff, Becker, Holler and Narberhaus. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Franz Narberhaus, ZnJhbnoubmFyYmVyaGF1c0BydWIuZGU=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.