- 1Department of Viroscience, Erasmus University Medical Center, Rotterdam, Netherlands
- 2Department of Immunology and Allergy, Lausanne University Hospital, Lausanne, Switzerland
The presence of a stable HIV-1 reservoir persisting over time despite effective antiretroviral suppression therapy precludes a cure for HIV-1. Characterizing and quantifying this residual reservoir is considered an essential prerequisite to develop and validate curative strategies. However, a sensitive, reproducible, cost-effective, and easily executable test is still needed. The quantitative viral outgrowth assay is considered the gold standard approach to quantify the reservoir in HIV-1-infected patients on suppressive ART, but it has several limitations. An alternative method to quantify the viral reservoir following the reactivation of latent HIV-1 provirus detects multiply-spliced tat/rev RNA (msRNA) molecules by real-time PCR [tat/rev induced limiting dilution assay (TILDA)]. This article provides a perspective overview of the clinical relevance, various applications, recent advancements of TILDA, and how the assay has contributed to our understanding of the HIV-1 reservoir.
Introduction
Over the past decade, multiple interventions to reduce or eliminate the latent HIV-1 reservoir have been proposed. A prominent strategy is to pharmacologically reactivate latently infected cells through so called latency reversal agents (LRAs) in conjunction with immune-modulating interventions to enhance extrinsic immune-based clearance or induce intrinsic apoptosis of reactivated cells (Kim et al., 2018; Abner and Jordan, 2019; Collins et al., 2020). Other eradication strategies include genome editing of target molecules such as the HIV co-receptor CCR5 (Almeida and Matos, 2019) or on the HIV-1 genome itself using CRISPR/Cas9 (Zhu et al., 2015) (reviewed in Wang et al., 2018) or Zinc finger nucleases (Qu et al., 2013). “Block and lock” strategies using pharmacological compounds (Ahlenstiel et al., 2015; Mousseau et al., 2015) or via therapeutic vaccination using broadly neutralizing monoclonal anti-HIV-1 antibodies (bNAbs) (Mendoza et al., 2018) to permanently silence the provirus have also been proposed. The advent of these strategies has prompted the development of several methods to quantify the HIV-1 reservoir and validate the effectiveness of potential cure interventions. However, existing cure strategies require further optimization and new assays with higher sensitivity, accuracy, precision, and reproducibility (Falcinelli et al., 2019) are urgently needed. A panel of latent HIV-1 reservoir assays have been recommended for use in HIV cure intervention trials (reviewed in Abdel-Mohsen et al., 2020). The quantitative viral outgrowth assay (QVOA), currently considered the reference assay for quantifying the replication-competent HIV-1 reservoir (Eriksson et al., 2013), quantifies the number of resting CD4+ T cells releasing infectious virus after in vitro stimulation and subsequent co-culture with feeder cells (Siliciano and Siliciano, 2005; Laird et al., 2016). However, the widespread use of QVOA in clinical trial settings is precluded by several factors related to the complexity of the assay {large blood draws, high variability [recently evaluated in the RAVEN studies (Rosenbloom et al., 2019; Stone et al., 2020)], labor intensity, and execution time (14–21 days)}. Moreover, QVOA underestimates the size of the reservoir and has a low dynamic range (Ho et al., 2013). Several efforts to improve QVOA (sensitivity and dynamic range, assay turn-around time, and overall costs) have been implemented (Passaes et al., 2017; Massanella et al., 2018; Stuelke et al., 2020; Zhang et al., 2020). A general concern, however, is that QVOA protocol modifications further complicate assay standardization (Stone et al., 2020). Alternatively, real-time or digital PCR-based methods, which are better suited for large clinical trials, have widely been used to detect HIV-1 DNA in infected cells from peripheral blood and lymphoid tissue cells (Vandergeeten et al., 2014; Alidjinou et al., 2015; Avettand-Fenoel et al., 2016; Anderson and Maldarelli, 2018). Although these methods are relatively easy to perform, fast, and cost-effective, they overestimate the reservoir size because most integrated HIV-1 genomes are not replication-competent (Ho et al., 2013). Advanced HIV-1 DNA quantification methods such as the intact proviral DNA assay (IPDA) (Bruner et al., 2019) and Q4PCR (Gaebler et al., 2019), distinguish intact proviruses from defective ones thereby providing a better resolution for studying the dynamics of defective and intact HIV-1 proviral DNA (Falcinelli et al., 2020; Peluso et al., 2020; Simonetti et al., 2020). However, proviral intactness does not guarantee virion production (Ho et al., 2013; Siliciano and Siliciano, 2021), and the fraction of intact proviruses that can be induced to produce virions cannot be determined by these assays, which is a critical limitation given that many intact proviruses exhibit low inducibility (Ho et al., 2013; Siliciano and Siliciano, 2021). In long-term ART-suppressed people living with HIV (PLWH), intact HIV-1 proviruses are preferentially integrated in an opposite orientation to host genes, in relative proximity or increased distance from active transcriptional start sites and accessible chromatin regions (Einkauf et al., 2019). The transcription-competent (inducible) HIV-1 reservoir can be quantified by several methods (Bullen et al., 2014; Cillo et al., 2014; Yucha et al., 2017; Massanella et al., 2018) including an approach developed by Procopio et al. (2015), which detects multiply-spliced tat/rev RNA (msRNA) molecules by real-time PCR [tat/rev induced limiting dilution assay (TILDA)], and reduces the likelihood of quantifying defective genomes (Procopio et al., 2015; Imamichi et al., 2016; Pollack et al., 2017). The percentage of intact proviruses capable of producing tat/rev msRNA transcripts is uncertain but a recent study showed no association between inducible HIV measures (TILDA, p24 SIMOA) and IPDA-intact proviruses but with IPDA-total measures (Papasavvas et al., 2021). TILDA correlates poorly with QVOA (Procopio et al., 2015; Stone et al., 2020), which may be explained by several factors including: technical (inherent assay variation and poor sensitivity); differences in provirus inducibility (Ho et al., 2013; Siliciano and Siliciano, 2021); the biological targets measured as readouts [tat/rev msRNA (TILDA) and HIV-1 p24 gag RNA or Gag protein (QVOA)] (Massanella et al., 2018); tat/rev msRNA is measured after 12–18 h of activation, whereas QVOA measurements are taken after 7–14 days of culture and virus production. TILDA probes total CD4+ T cells, whereas QVOA uses sorted resting CD4+ T cells. Furthermore, due to post-transcription blocks in RNA processing (Yukl et al., 2018), not all cells producing tat/rev msRNA transcripts will yield infectious virus (Hong et al., 2018), and therefore quantifying these cells overestimates the replication-competent HIV-1 reservoir size. Despite these discrepancies and limitations, TILDA is a well-established approach to quantify the inducible HIV-1 reservoir as a proxy for replication competence, and a technically feasible alternative to QVOA for routine execution in large scale HIV cure intervention trials [requires less blood, shorter assay turn-around time (2 days), and medium throughput]. In this article we provide a perspective overview of the clinical relevance, various applications, and recent advancements of TILDA, and how the assay has contributed to our understanding of the HIV-1 reservoir.
Principle of the tat/rev Induced Limiting Dilution Assay
The fundamental steps in performing TILDA as published by Procopio et al. (2015) include: (i) isolation of CD4+ T cells from 10 mL blood collected from HIV-1-infected individuals; (ii) in vitro stimulation for 12 h with 100 ng/mL phorbol 12-myristate 13-acetate (PMA) and 1 μg/mL ionomycin to induce production of tat/rev msRNA; (iii) distribution of cells in a limiting dilution scheme (24 replicates of 1.8 × 104 to 1.0 × 103 cells per well) in a 96-well plate containing a one-step reverse transcription PCR (RT-PCR) master mix, which bypasses RNA extraction, allowing simultaneous RT and pre-amplification of tat/rev msRNA; (iv) quantification of tat/rev msRNA using a diluted fraction of the pre-amplification product; (v) estimation of the frequency of tat/rev msRNA+ cells using maximum likelihood method (Hu and Smyth, 2009). TILDA has a limit of detection of ∼1 cell expressing tat/rev msRNA per million CD4+ T cells, a coefficient of variation of ∼20% (Procopio et al., 2015; Lungu et al., 2020), and correlates with various assays to measure latency, reactivation and reservoir (total and integrated HIV DNA, p24 SIMOA, and HIV FISH-Flow) (Procopio et al., 2015; Pardons et al., 2019; Papasavvas et al., 2021). In-depth comparisons of these assays have been reviewed elsewhere (Eriksson et al., 2013; Siliciano and Siliciano, 2018; Falcinelli et al., 2019; Abdel-Mohsen et al., 2020).
Clinical Relevance of Inducible HIV-1 Reservoir Quantification by TILDA
In ART-suppressed PLWH, the median frequency of latently infected CD4+ T cells estimated by TILDA is ∼24 per million CD4+ T cells (Procopio et al., 2015; Pardons et al., 2019; Lungu et al., 2020), 48 times larger than the frequency measured with QVOA. Nevertheless, the median frequency of intact, non-induced proviruses is reported to be 60-fold higher than the frequency of induced proviruses detected in QVOA (Ho et al., 2013). The chromosomal integration site is a critical factor determining provirus inducibility and there is recent evidence that long-term ART-suppressed PLWH harbor intact proviruses with features of deeper viral latency (Battivelli et al., 2018; Einkauf et al., 2019). TILDA readouts therefore serve as intermediate, surrogate measures for the frequency of latently infected cells harboring replication-competent HIV-1. Procopio et al. (2015) demonstrated that the size of the inducible HIV-1 reservoir was smaller in subjects who initiated ART during acute HIV infection (AHI) when compared to those who began therapy at a later stage of the disease (median frequencies of 11 and 29 cells per million CD4+ T cells in acute and chronic infection, respectively). This trend has also been shown in other studies using QVOA and HIV-1 DNA (total and integrated) (Buzon et al., 2014; Ananworanich and Mellors, 2015). Notably, the frequency of cells with inducible msRNA in most samples obtained from individuals who initiated ART during Fiebig stages I to III is even lower (Leyre et al., 2020), nearing or below the TILDA limit of detection (Henrich et al., 2017; Colby et al., 2018; Leyre et al., 2020). Comparing TILDA estimates before and after stimulation of CD4+ T cells obtained from ART-naïve individuals, most infected cells (81.4%) were latently infected and produced msRNA only after stimulation. In contrast, merely 18.6% of the infected cells spontaneously produced msRNA (Procopio et al., 2015). Therefore, similarly to ART-suppressed PLWH, ART-naïve individuals harbor a large pool of latently infected CD4+ T cells. Moreover, the frequency of latently infected CD4+ T cells in ART-naïve individuals positively correlated with the duration of HIV-1 infection, which suggests that the size of the latent and inducible reservoir increases over time in ART-naïve individuals as a result of continuous seeding of the HIV-1 reservoir (Procopio et al., 2015). A recent study reported a correlation between the inducible reservoir measured by TILDA and the CD4/CD8 ratio (Bertoldi et al., 2020), an important clinical parameter reflecting the immune system’s functionality and linked to reservoir size (Chun et al., 2002; Lu et al., 2015).
Application of TILDA in HIV-1 Cure Interventions
Evaluating Latent HIV-1 Reservoir Reactivation and Elimination Using TILDA
Transcriptional activation of HIV-1 provirus from latently infected cells using LRAs and subsequent immune-mediated clearance is viewed as one of the most promising approaches toward HIV-1 eradication. During the last years, multiple LRAs have been tested in vitro and ex vivo, and in combination with other interventions in clinical trials (reviewed in Stoszko et al., 2019; Zerbato et al., 2019). In two clinical trials evaluating depsipeptide Romidepsin’s latency reversal potential in vivo, no substantial changes in HIV-1 reservoir size could be observed by total HIV-1-DNA quantification, QVOA, or TILDA (Sogaard et al., 2015; McMahon et al., 2020), despite detectible HIV-1 viral reactivation measured using standard plasma HIV-1 RNA assays. In another in vivo clinical study, the histone deacetylase inhibitor Vorinostat induced a sustained increase of CA unspliced HIV-1 RNA from CD4+ T cells from the blood. Still, there was no significant change in plasma HIV-1 RNA or HIV-1 reservoir size measured by TILDA and HIV-1 DNA (total and integrated) (Elliott et al., 2014). These and other studies highlight that while it is possible to disrupt HIV-1 latency in patients on suppressive ART, the LRA interventions so far have not significantly impacted the latent HIV-1 reservoir size or time to viral rebound (reviewed in Zerbato et al., 2019).
All considered, TILDA could be valuable in LRA studies; the initial step of viral reactivation is the early expression of tat and rev RNAs, the assay’s primary targets. Therefore, TILDA would be an ideal screening tool to validate promising compounds. For instance, TILDA contributed to defining a new class of novel pure phorbol ester-class compounds, isolated from an African medicinal plant (Mukungulu) (Tietjen et al., 2018), with the capacity to promote HIV-1 latency reversal ex-vivo in J-Lat cells containing an HIV-1-GFP provirus (Tietjen et al., 2019). TILDA results could also be used to differentiate T cell populations with large and small inducible HIV-1 reservoirs. In multiple studies, TILDA revealed that CD4+ T effector memory cells harbor the largest inducible HIV-1 reservoir in peripheral blood when compared to CD4+ T central memory cells and CD4+ T transcriptional memory cells (Fromentin et al., 2016; Kulpa et al., 2019). This finding suggests the possibility to induce the differentiation toward effector memory cells as a strategy to increase the HIV-1 reservoir susceptibility to LRAs (Hiener et al., 2017; Wonderlich et al., 2019; Kwon et al., 2020; Rabezanahary et al., 2020), and TILDA may be useful for measuring the effects of such novel approaches.
Furthermore, innovative strategies to enhance killing mechanisms in different T cell populations are gaining more attention (reviewed in Kim et al., 2018). Several recent studies have demonstrated the use of pharmacological compounds that selectively induce cell apoptosis in HIV-1-infected cells as a strategy to eliminate these cells (Campbell et al., 2018; Matsuda et al., 2019; Zhang et al., 2019). In a study by Rao et al. (2021), CD4+ T cells obtained from ART-suppressed HIV-1-infected individuals were cultured for 5 days in the absence or presence of DDX3 inhibitors. The addition of DDX3 inhibitors resulted in a reduction (∼50%) of the inducible HIV-1 reservoir, determined by quantifying CA HIV-1 RNA, the frequency of cells expressing tat/rev msRNA (TILDA), and the frequency of cells positive for gag RNA using FISH-flow (Rao et al., 2021).
Characterizing Latently Infected Cells Using TILDA
The elimination or reduction of HIV-1-infected cells is a priority toward an HIV cure. Unfortunately, there is not a specific biomarker for the identification of infected cells. It is well known that CD4+ memory T cells (Chomont et al., 2009) and CD4+ T cells with stem cell-like properties (Buzon et al., 2014) in peripheral blood, and follicular helper T (Tfh) cells (PD-1high/CXCR5high) and memory CD4+ T cells (Banga et al., 2016) in lymph nodes are enriched in cells harboring replication-competent HIV-1. These cell populations share multiple co-inhibitory molecules (immune checkpoints, ICs), which down-modulate the immune response and prevent hyper-immune activation. ICs are typically upregulated after T-cell activation, and their overexpression is associated with T-cell exhaustion in cancer and chronic viral infections, including HIV-1 (Day et al., 2006; Petrovas et al., 2006; Trautmann et al., 2006). A recent study identified PD-1, TIGIT, and LAG-3 as ICs, which are positively associated with the frequency of CD4+ T cells harboring integrated HIV-1 DNA (Fromentin et al., 2016). Notably, the same populations were threefold more positive for tat/rev msRNA when compared to the PD-1neg, TIGITneg, and LAG-3neg CD4+ T cell populations, suggesting that resting or exhausted cell status favors HIV-1 persistence. Integrated HIV-1 DNA, TILDA, and plasma HIV-1 RNA measurements were also used to characterize the HIV-1 reservoir in another study that aimed to identify molecular signatures that may support the role of Tfh cells as a significant compartment for HIV-1 persistence (Aid et al., 2018). Notably, BCL6 gene expression, which is the master regulator of Tfh cell differentiation and modulator of a series of other transcription factors and their downstream targets, positively correlated with integrated HIV-1 DNA as well as TILDA and plasma viral load (Aid et al., 2018). Multiple studies demonstrate that α4β7-expressing cells represent early targets for HIV-1 and that pre-infection frequencies of α4β7 on circulating CD4+ T cells may predict the risk of HIV-1 acquisition and disease progression (Cicala et al., 2009; Kader et al., 2009; Martinelli et al., 2013; Sivro et al., 2018). In a phase I clinical trial to study the impact of anti-α4β7 monoclonal infusion on HIV-1 reservoir size TILDA was used to determine change in inducible reservoir size after treatment. A reduction in msRNA post-treatment was observed in two out of four donors, but the low sample size impaired the test’s statistical significance (Uzzan et al., 2018).
TILDA Advancements
Broadening the Detection of tat/rev msRNA Beyond HIV-1 Clade B
High sequence diversity in the tat/rev region demands the modification of primers and probes to increase TILDA’s specificity for different HIV-1 clades. In its original design, TILDA optimally quantifies the HIV-1 reservoir in individuals infected with clade B (Procopio et al., 2015), the dominant clade in North America and Europe. This primer and probe restriction is a limitation considering that 46.6% of HIV-1 infections worldwide are clade C, the most dominant HIV-1 clade in Africa (Hemelaar et al., 2019). Given this, C-TILDA was recently developed with primers and probes modified specifically for clade C viruses (Bertoldi et al., 2020), which extends the possibility of screening other HIV-1-infected populations and implementing the assay in eradication strategies in regions where clade C is more abundant. In two other studies, primers and probes were designed to efficiently amplify tat/rev msRNA for clade A/E, A1, and A/G viruses as well (Colby et al., 2018; Dhummakupt et al., 2020). We aligned the published TILDA primer and probe sequences to a consensus sequence generated from 2695 global HIV-1 sequences downloaded from the Los Alamos HIV-1 sequence database (Figure 1). The highest degree of variability is evident in the Tat 2.0 forward primer in which 9/34 positions have <80% (39.2–63%) identity to consensus. Further studies should validate a generalized approach using combinations of primer and probe sets for these and other circulating HIV-1 subtypes, broadening the detection and increasing the assay’s applicability.

Figure 1. Sequence of TILDA primers and probes specific for different HIV-1 subtypes. (A–D) TILDA primer and probe sequences were retrieved from the literature [HIV-1 subtype: A (Dhummakupt et al., 2020), B (Procopio et al., 2015), C (Bertoldi et al., 2020; Dhummakupt et al., 2020), A/E (Colby et al., 2018), A/D, and A/G (Dhummakupt et al., 2020)] and aligned to a consensus sequence generated from an alignment of 2695 HIV-1 sequences downloaded from the Los Alamos HIV-1 sequence database. Nucleotide substitutions are colored blue, green, yellow, and red. Nucleotide positions with degenerate bases are colored gray. The least conserved nucleotide positions (<80% identity) are marked in red.
Indeed, non-human primate (NHP) models are critical to test novel therapeutic strategies and better understand the HIV-1 reservoir (Whitney et al., 2014; Nixon et al., 2017). Considering the limited number of cells available, performing QVOA is impractical in NHP studies, therefore Frank et al. (2019) developed a simian version of TILDA (termed SIV/SHIV-1 TILDA) to detect msRNA of SIVmac251/mac239 and SHIV-1AD80E viruses, which are widely employed in NHP studies. The SIV/SHIV-1 TILDA could detect msRNA also in the setting of very low/undetectable viremia. Interestingly, in animals that started ART early after infection, the results from reservoir estimation by TILDA correlated with the viral rebound post ART interruption (Frank et al., 2019).
Improving TILDA Sensitivity
A major advantage of TILDA is the direct amplification of tat/rev msRNA from whole cell lysate. However, the cell lysate complexity itself can reduce the signal to noise ratio impacting overall tat/rev msRNA amplification efficiency. To improve msRNA detection, a new TILDA protocol that includes an RNA extraction step was developed, the results of which strongly correlated with those of the original protocol (Pezzi et al., 2017). In a recent study, activating cells with phytohemagglutinin for an extended period (18 h instead of 12 h) enhanced the expression of tat/rev msRNA in CD4+ T cells isolated from infants living with HIV-1 (Dhummakupt et al., 2020). Several studies have shown that the frequency of cells that could be induced to express HIV-1 msRNA is small in most individuals treated during AHI (Fiebig stages 1-III) (Colby et al., 2018; Leyre et al., 2020). Furthermore, some long-term suppressed patients or elite HIV-1 controllers have significantly lower latent viral reservoirs (Siliciano et al., 2003; Autran et al., 2011; Bertoldi et al., 2020; Galvez et al., 2020). In these cases, reservoir estimation using TILDA is technically challenging. A higher cell input is required to increase the assay’s sensitivity, which can be achieved by increasing the number of replicates through processing multiple assay plates (Procopio et al., 2015). However, processing multiple TILDA plates per sample reduces assay throughput and increases the overall cost. To circumvent these limitations, maximizing the number of cells that can be probed per reaction is essential. In the recently developed C-TILDA (Bertoldi et al., 2020), the authors increased cell input (up to 5.4 × 104 cells per reaction instead of 1.8 × 104) to maximize detection of tat/rev msRNA. In another study, which assessed assay robustness and amenability for routine use in clinical studies of the latent reservoir, TILDA was performed using alternative RT-qPCR reagents according to a modified (TILDA v2.0) protocol (Lungu et al., 2020). Particularly, tat/rev msRNA pre-amplification reactions were carried out in larger volumes (10 μl instead of 1 μl) to minimize pipetting imprecisions. Furthermore, the pre-amplification products were directly added to the real-time PCR mix to detect msRNA, which improves assay throughput by reducing the number of pipetting steps and minimizes the risk of cross-contamination associated with diluting PCR products (Lungu et al., 2020). In our preliminary assessment of TILDA v2.0 performance, using a higher input number of ACH-2 or HIV-1-infected CD4+ T cells per reaction (7.2 × 104 cells to 1.4 × 105 cells), resulting in six to eightfold more cells per assay, improved target detection and precision (narrower 95% confidence intervals) when quantifying small reservoirs (Figure 2). Further studies are necessary to assess TILDA or TILDA v2.0 reproducibility with these higher cell inputs, which require an increased total blood volume of 30–50 mL, and the capacity thereof to quantify inducible reservoirs below the current detection limits. Indeed, such validation studies would reveal more insight into TILDA’s suitability for evaluating HIV-1 reservoir-reduction clinical interventions.
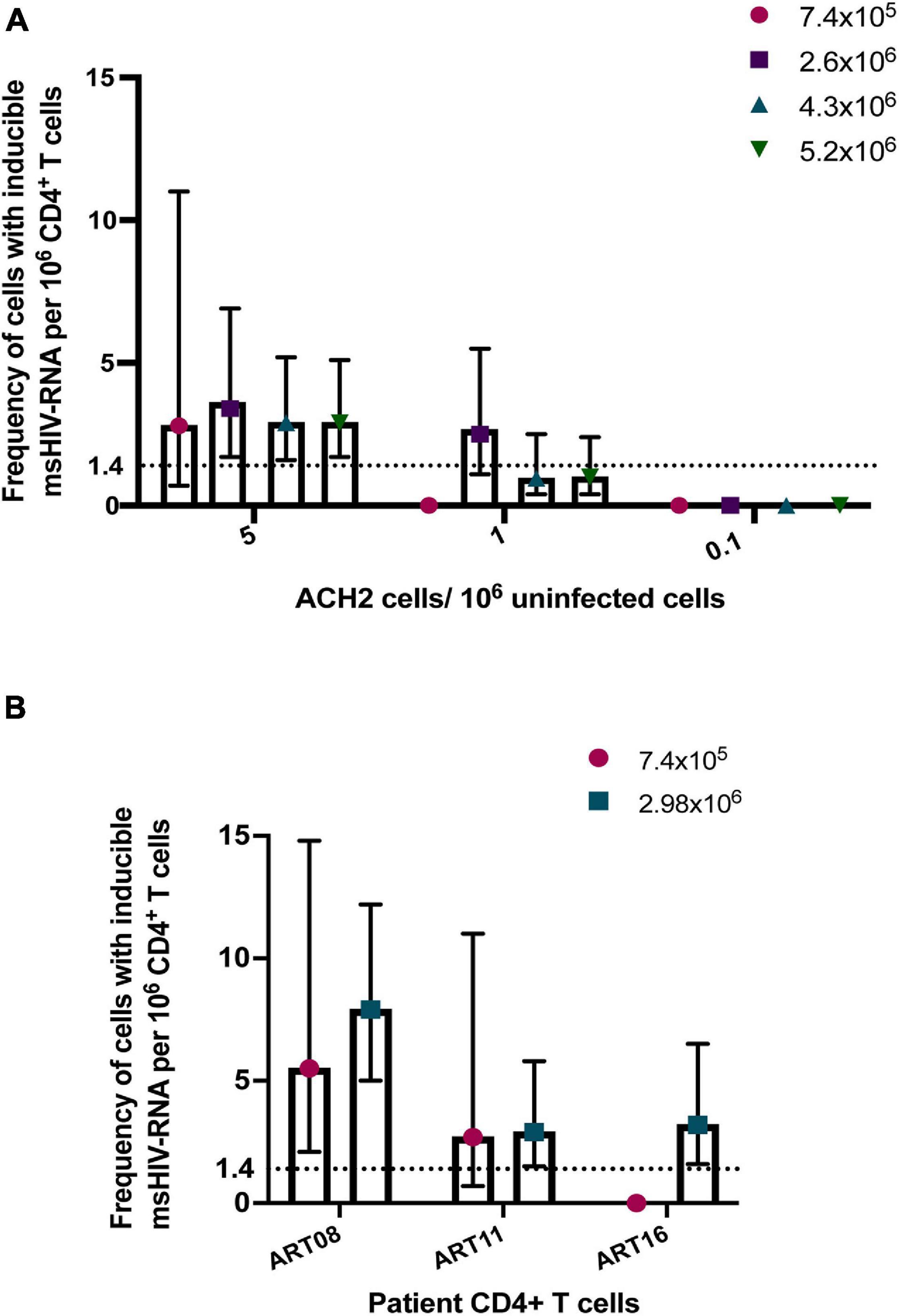
Figure 2. Detection of tat/rev msRNA in low reservoir samples. (A) Target detection in three low concentrations of ACH-2 cells mixed with one million uninfected CD4+ T cells. The total number of cells tested per plate ranged from 7.4 × 105 cells to 5.2 × 106 cells (B) CD4+ T cells obtained from three ART-suppressed individuals (plasma HIV RNA <20 c/mL) were stimulated with PMA/ionomycin for 12–14 h. The frequency of cells expressing tat/rev msRNA was determined using TILDA v2.0 (Lungu et al., 2020) with standard (7.4 × 105 cells) or higher cell inputs (colored symbols). The mean estimates of inducible cells/106 CD4+ T cells are plotted in both graphs. The error bars represent the 95% confidence interval upper and lower limits. The horizontal line represents the assay limit of detection (LOD) based on the standard cell input (7.4 × 105 cells).
Conclusion
As we advance toward large, scalable clinical interventions to significantly deplete or permanently restrict inducible HIV-1 reservoirs in well-suppressed PLWH, TILDA stands out as a suitable option for implementation in the aforementioned HIV cure clinical trials. Within 2 days, inducible HIV-1 reservoir measurements can be generated using less than 1 × 106 viable target cells per condition with the possibility to increase cell input [six to eightfold (TILDA v2.0)] without significantly impacting overall assay costs. The assay is also very robust, as demonstrated in multiple studies to test intra- and inter-laboratory reproducibility, which is important for the cross-validation of results in multi-center, multi-laboratory clinical trials.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the Erasmus MC Medical Ethics Committee MEC-2016-148 (July 22, 2016). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
CL and FP conceptualized the perspective article, researched the literature, and wrote the first draft of the manuscript. CL performed the experiments and analyzed the data. All authors read and approved the final manuscript.
Funding
This work was supported by EHVA, funded by the European Union’s Horizon 2020 Research and Innovation Program (grant number 681032) and the Swiss Government (grant number 150337).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
In loving memory of Charles A.B. Boucher, who strongly supported HIV cure-related research endeavors and the implementation of innovative tools to evaluate HIV persistence in clinical settings. We thank Tokameh Mahmoudi and Shringar Rao for helpful discussions.
References
Abdel-Mohsen, M., Richman, D., Siliciano, R. F., Nussenzweig, M. C., Howell, B. J., Martinez-Picado, J., et al. (2020). Recommendations for measuring HIV reservoir size in cure-directed clinical trials. Nat. Med. 26, 1339–1350. doi: 10.1038/s41591-020-1022-1
Abner, E., and Jordan, A. H. I. V. (2019). “shock and kill” therapy: In need of revision. Antiviral. Res. 166, 19–34. doi: 10.1016/j.antiviral.2019.03.008
Ahlenstiel, C., Mendez, C., Lim, S. T., Marks, K., Turville, S., Cooper, D. A., et al. (2015). Novel RNA Duplex Locks HIV-1 in a latent state via Chromatin-mediated transcriptional silencing. Mol. Ther. Nucleic Acids. 4:e261. doi: 10.1038/mtna.2015.31
Aid, M., Dupuy, F. P., Moysi, E., Moir, S., Haddad, E. K., Estes, J. D., et al. (2018). Follicular CD4 T helper cells as a major HIV reservoir compartment: A molecular perspective. Front. Immunol. 9:895. doi: 10.3389/fimmu.2018.00895
Alidjinou, E. K., Bocket, L., and Hober, D. (2015). Quantification of viral DNA during HIV-1 infection: A review of relevant clinical uses and laboratory methods. Pathol. Biol. (Paris). 63, 53–59. doi: 10.1016/j.patbio.2014.07.007
Almeida, M. J., and Matos, A. (2019). Designer nucleases: gene-editing therapies using CCR5 as an emerging target in HIV. Curr. HIV Res. 17, 306–323. doi: 10.2174/1570162x17666191025112918
Ananworanich, J., and Mellors, J. W. (2015). How much HIV is Alive? The challenge of measuring replication competent HIV for HIV Cure research. EBioMedicine 2, 788–789. doi: 10.1016/j.ebiom.2015.07.036
Anderson, E. M., and Maldarelli, F. (2018). Quantification of HIV DNA using droplet digital PCR techniques. Curr. Protoc. Microbiol. 51:e62. doi: 10.1002/cpmc.62
Autran, B., Descours, B., Avettand-Fenoel, V., and Rouzioux, C. (2011). Elite controllers as a model of functional cure. Curr. Opin. HIV AIDS. 6, 181–187. doi: 10.1097/coh.0b013e328345a328
Avettand-Fenoel, V., Hocqueloux, L., Ghosn, J., Cheret, A., Frange, P., Melard, A., et al. (2016). Total HIV-1 DNA, a marker of viral reservoir dynamics with clinical implications. Clin. Microbiol. Rev. 29, 859–880. doi: 10.1128/cmr.00015-16
Banga, R., Procopio, F. A., Noto, A., Pollakis, G., Cavassini, M., Ohmiti, K., et al. (2016). PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat. Med. 22, 754–761. doi: 10.1038/nm.4113
Battivelli, E., Dahabieh, M. S., Abdel-Mohsen, M., Svensson, J. P., Tojal Da Silva, I., Cohn, L. B., et al. (2018). Distinct chromatin functional states correlate with HIV latency reactivation in infected primary CD4(+) T cells. Elife 7:e34655.
Bertoldi, A., D’Urbano, V., Bon, I., Verbon, A., Rokx, C., Boucher, C., et al. (2020). Development of C-TILDA: A modified TILDA method for reservoir quantification in long term treated patients infected with subtype C HIV-1. J. Virol. Methods. 276:113778. doi: 10.1016/j.jviromet.2019.113778
Bruner, K. M., Wang, Z., Simonetti, F. R., Bender, A. M., Kwon, K. J., Sengupta, S., et al. (2019). A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature. 566, 120–125. doi: 10.1038/s41586-019-0898-8
Bullen, C. K., Laird, G. M., Durand, C. M., Siliciano, J. D., and Siliciano, R. F. (2014). New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 20, 425–429. doi: 10.1038/nm.3489
Buzon, M. J., Sun, H., Li, C., Shaw, A., Seiss, K., Ouyang, Z., et al. (2014). HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat. Med. 20, 139–142.
Campbell, G. R., Bruckman, R. S., Chu, Y. L., Trout, R. N., and Spector, S. A. (2018). SMAC Mimetics induce autophagy-dependent apoptosis of HIV-1-infected resting memory CD4+ T Cells. Cell Host Microbe. 24, 689–702. doi: 10.1016/j.chom.2018.09.007
Chomont, N., El-Far, M., Ancuta, P., Trautmann, L., Procopio, F. A., Yassine-Diab, B., et al. (2009). HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 15, 893–900. doi: 10.1038/nm.1972
Chun, T. W., Justement, J. S., Pandya, P., Hallahan, C. W., McLaughlin, M., Liu, S., et al. (2002). Relationship between the size of the human immunodeficiency virus type 1 (HIV-1) reservoir in peripheral blood CD4+ T cells and CD4+:CD8+ T cell ratios in aviremic HIV-1-infected individuals receiving long-term highly active antiretroviral therapy. J. Infect. Dis. 185, 1672–1676. doi: 10.1086/340521
Cicala, C., Martinelli, E., McNally, J. P., Goode, D. J., Gopaul, R., Hiatt, J., et al. (2009). The integrin alpha4beta7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proc. Natl. Acad. Sci. U S A. 106, 20877–20882. doi: 10.1073/pnas.0911796106
Cillo, A. R., Sobolewski, M. D., Bosch, R. J., Fyne, E., Piatak, M. Jr., Coffin, J. M., et al. (2014). Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. U S A. 111, 7078–7083. doi: 10.1073/pnas.1402873111
Colby, D. J., Trautmann, L., Pinyakorn, S., Leyre, L., Pagliuzza, A., Kroon, E., et al. (2018). Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat. Med. 24, 923–926. doi: 10.1038/s41591-018-0026-6
Collins, D. R., Gaiha, G. D., and Walker, B. D. (2020). CD8(+) T cells in HIV control, cure and prevention. Nat. Rev. Immunol. 20, 471–482. doi: 10.1038/s41577-020-0274-9
Day, C. L., Kaufmann, D. E., Kiepiela, P., Brown, J. A., Moodley, E. S., Reddy, S., et al. (2006). PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443, 350–354.
Dhummakupt, A., Rubens, J. H., Anderson, T., Powell, L., Nonyane, B. A., Siems, L. V., et al. (2020). Differences in inducibility of the latent HIV reservoir in perinatal and adult infection. JCI Insight. 5:e134105.
Einkauf, K. B., Lee, G. Q., Gao, C., Sharaf, R., Sun, X., Hua, S., et al. (2019). Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J. Clin. Invest. 129, 988–998. doi: 10.1172/jci124291
Elliott, J. H., Wightman, F., Solomon, A., Ghneim, K., Ahlers, J., Cameron, M. J., et al. (2014). Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog. 10:e1004473. doi: 10.1371/journal.ppat.1004473
Eriksson, S., Graf, E. H., Dahl, V., Strain, M. C., Yukl, S. A., Lysenko, E. S., et al. (2013). Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 9:e1003174. doi: 10.1371/journal.ppat.1003174
Falcinelli, S. D., Ceriani, C., Margolis, D. M., and Archin, N. M. (2019). New frontiers in measuring and characterizing the HIV reservoir. Front. Microbiol. 10:2878. doi: 10.3389/fmicb.2019.02878
Falcinelli, S. D., Kilpatrick, K. W., Read, J., Murtagh, R., Allard, B., Ghofrani, S., et al. (2020). Longitudinal dynamics of intact HIV proviral DNA and outgrowth virus frequencies in a cohort of ART-treated individuals. J. Infect. Dis. 2020:jiaa718.
Frank, I., Acharya, A., Routhu, N. K., Aravantinou, M., Harper, J. L., Maldonado, S., et al. (2019). A Tat/Rev induced limiting dilution assay to measure viral reservoirs in non-human primate models of HIV infection. Sci. Rep. 9:12078.
Fromentin, R., Bakeman, W., Lawani, M. B., Khoury, G., Hartogensis, W., DaFonseca, S., et al. (2016). CD4+ T cells expressing PD-1, TIGIT and LAG-3 Contribute to HIV Persistence during ART. PLoS Pathog. 12:e1005761. doi: 10.1371/journal.ppat.1005761
Gaebler, C., Lorenzi, J. C. C., Oliveira, T. Y., Nogueira, L., Ramos, V., Lu, C. L., et al. (2019). Combination of quadruplex qPCR and next-generation sequencing for qualitative and quantitative analysis of the HIV-1 latent reservoir. J. Exp. Med. 216, 2253–2264. doi: 10.1084/jem.20190896
Galvez, C., Urrea, V., Dalmau, J., Jimenez, M., Clotet, B., Monceaux, V., et al. (2020). Extremely low viral reservoir in treated chronically HIV-1-infected individuals. EBioMedicine 57:102830. doi: 10.1016/j.ebiom.2020.102830
Hemelaar, J., Elangovan, R., Yun, J., Dickson-Tetteh, L., Fleminger, I., Kirtley, S., et al. (2019). Global and regional molecular epidemiology of HIV-1, 1990-2015: a systematic review, global survey, and trend analysis. Lancet Infect. Dis. 19, 143–155.
Henrich, T. J., Hatano, H., Bacon, O., Hogan, L. E., Rutishauser, R., Hill, A., et al. (2017). HIV-1 persistence following extremely early initiation of antiretroviral therapy (ART) during acute HIV-1 infection: An observational study. PLoS Med. 14:e1002417. doi: 10.1371/journal.pmed.1002417
Hiener, B., Horsburgh, B. A., Eden, J. S., Barton, K., Schlub, T. E., Lee, E., et al. (2017). Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Rep. 21, 813–822. doi: 10.1016/j.celrep.2017.09.081
Ho, Y. C., Shan, L., Hosmane, N. N., Wang, J., Laskey, S. B., Rosenbloom, D. I., et al. (2013). Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551. doi: 10.1016/j.cell.2013.09.020
Hong, F., Jacobs, J. L., Aga, E., Cillo, A. R., Fyne, E., Koontz, D. L., et al. (2018). Associations between HIV-1 DNA copy number, proviral transcriptional activity, and plasma viremia in individuals off or on suppressive antiretroviral therapy. Virology. 521, 51–57. doi: 10.1016/j.virol.2018.05.018
Hu, Y., and Smyth, G. K. (2009). ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 347, 70–78. doi: 10.1016/j.jim.2009.06.008
Imamichi, H., Dewar, R. L., Adelsberger, J. W., Rehm, C. A., O’Doherty, U., Paxinos, E. E., et al. (2016). Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc. Natl. Acad. Sci. U S A. 113, 8783–8788. doi: 10.1073/pnas.1609057113
Kader, M., Wang, X., Piatak, M., Lifson, J., Roederer, M., Veazey, R., et al. (2009). Alpha4(+)beta7(hi)CD4(+) memory T cells harbor most Th-17 cells and are preferentially infected during acute SIV infection. Mucosal. Immunol. 2, 439–449. doi: 10.1038/mi.2009.90
Kim, Y., Anderson, J. L., and Lewin, S. R. (2018). Getting the “Kill” into “Shock and Kill”: strategies to eliminate latent HIV. Cell Host Microbe. 23, 14–26. doi: 10.1016/j.chom.2017.12.004
Kulpa, D. A., Talla, A., Brehm, J. H., Ribeiro, S. P., Yuan, S., Bebin-Blackwell, A. G., et al. (2019). Differentiation into an effector memory phenotype potentiates HIV-1 latency reversal in CD4(+) T Cells. J. Virol. 93:e00969–19.
Kwon, K. J., Timmons, A. E., Sengupta, S., Simonetti, F. R., Zhang, H., Hoh, R., et al. (2020). Different human resting memory CD4(+) T cell subsets show similar low inducibility of latent HIV-1 proviruses. Sci. Transl. Med. 12:eaax6795. doi: 10.1126/scitranslmed.aax6795
Laird, G. M., Rosenbloom, D. I., Lai, J., Siliciano, R. F., and Siliciano, J. D. (2016). Measuring the frequency of latent HIV-1 in resting CD4(+) T cells using a limiting dilution coculture assay. Methods Mol. Biol. 1354, 239–253. doi: 10.1007/978-1-4939-3046-3_16
Leyre, L., Kroon, E., Vandergeeten, C., Sacdalan, C., Colby, D. J., Buranapraditkun, S., et al. (2020). Abundant HIV-infected cells in blood and tissues are rapidly cleared upon ART initiation during acute HIV infection. Sci. Transl. Med. 12:eaav3491. doi: 10.1126/scitranslmed.aav3491
Lu, W., Mehraj, V., Vyboh, K., Cao, W., Li, T., and Routy, J. P. (2015). CD4:CD8 ratio as a frontier marker for clinical outcome, immune dysfunction and viral reservoir size in virologically suppressed HIV-positive patients. J. Int. AIDS Soc. 18:20052. doi: 10.7448/ias.18.1.20052
Lungu, C., Procopio, F. A., Overmars, R. J., Beerkens, R. J. J., Voermans, J. J. C., Rao, S., et al. (2020). Inter-laboratory reproducibility of inducible HIV-1 reservoir quantification by TILDA. Viruses 12:973. doi: 10.3390/v12090973
Martinelli, E., Veglia, F., Goode, D., Guerra-Perez, N., Aravantinou, M., Arthos, J., et al. (2013). The frequency of alpha(4)beta(7)(high) memory CD4(+) T cells correlates with susceptibility to rectal simian immunodeficiency virus infection. J. Acquir. Immune Defic. Syndr. 64, 325–331. doi: 10.1097/qai.0b013e31829f6e1a
Massanella, M., Yek, C., Lada, S. M., Nakazawa, M., Shefa, N., Huang, K., et al. (2018). Improved assays to measure and characterize the inducible HIV reservoir. EBioMedicine. 36, 113–121. doi: 10.1016/j.ebiom.2018.09.036
Matsuda, K., Kobayakawa, T., Tsuchiya, K., Hattori, S. I., Nomura, W., Gatanaga, H., et al. (2019). Benzolactam-related compounds promote apoptosis of HIV-infected human cells via protein kinase C-induced HIV latency reversal. J. Biol. Chem. 294, 116–129. doi: 10.1074/jbc.ra118.005798
McMahon, D. K., Zheng, L., Cyktor, J. C., Aga, E., Macatangay, B. J., Godfrey, C., et al. (2020). A phase I/II randomized, placebo-controlled trial of romidepsin in persons with HIV-1 on suppressive antiretroviral therapy to assess safety and activation of HIV-1 expression (A5315). J. Infect. Dis. 2020:jiaa777. doi: 10.1093/infdis/jiaa777
Mendoza, P., Gruell, H., Nogueira, L., Pai, J. A., Butler, A. L., Millard, K., et al. (2018). Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 561, 479–484.
Mousseau, G., Kessing, C. F., Fromentin, R., Trautmann, L., Chomont, N., and Valente, S. T. (2015). The tat inhibitor Didehydro-Cortistatin A prevents HIV-1 reactivation from latency. mBio. 6:e00465.
Nixon, C. C., Mavigner, M., Silvestri, G., and Garcia, J. V. (2017). In vivo models of human immunodeficiency virus persistence and cure strategies. J. Infect. Dis. 215, S142–S151.
Papasavvas, E., Azzoni, L., Ross, B. N., Fair, M., Yuan, Z., Gyampoh, K., et al. (2021). Intact Human Immunodeficiency Virus (HIV) reservoir estimated by the intact proviral DNA assay correlates with levels of total and integrated DNA in the blood during suppressive antiretroviral therapy. Clin. Infect. Dis. 72, 495–498. doi: 10.1093/cid/ciaa809
Pardons, M., Baxter, A. E., Massanella, M., Pagliuzza, A., Fromentin, R., Dufour, C., et al. (2019). Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection. PLoS Pathog. 15:e1007619. doi: 10.1371/journal.ppat.100761
Passaes, C. P. B., Bruel, T., Decalf, J., David, A., Angin, M., Monceaux, V., et al. (2017). Ultrasensitive HIV-1 p24 assay detects single infected cells and differences in reservoir induction by latency reversal agents. J. Virol. 91:e002296–16.
Peluso, M. J., Bacchetti, P., Ritter, K. D., Beg, S., Lai, J., Martin, J. N., et al. (2020). Differential decay of intact and defective proviral DNA in HIV-1-infected individuals on suppressive antiretroviral therapy. JCI Insight. 5:e132997.
Petrovas, C., Casazza, J. P., Brenchley, J. M., Price, D. A., Gostick, E., Adams, W. C., et al. (2006). PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J. Exp. Med. 203, 2281–2292. doi: 10.1084/jem.20061496
Pezzi, H. M., Berry, S. M., Beebe, D. J., and Striker, R. (2017). RNA-mediated TILDA for improved cell capacity and enhanced detection of multiply-spliced HIV RNA. Integr. Biol. (Camb) 9, 876–884. doi: 10.1039/c7ib00112f
Pollack, R. A., Jones, R. B., Pertea, M., Bruner, K. M., Martin, A. R., Thomas, A. S., et al. (2017). Defective HIV-1 proviruses are expressed and can be recognized by Cytotoxic T lymphocytes, which shape the proviral landscape. Cell Host Microbe. 21, 494–506. doi: 10.1016/j.chom.2017.03.008
Procopio, F. A., Fromentin, R., Kulpa, D. A., Brehm, J. H., Bebin, A. G., Strain, M. C., et al. (2015). A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine 2, 874–883. doi: 10.1016/j.ebiom.2015.06.019
Qu, X., Wang, P., Ding, D., Li, L., Wang, H., Ma, L., et al. (2013). Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res. 41, 7771–7782. doi: 10.1093/nar/gkt571
Rabezanahary, H., Moukambi, F., Palesch, D., Clain, J., Racine, G., Andreani, G., et al. (2020). Despite early antiretroviral therapy effector memory and follicular helper CD4 T cells are major reservoirs in visceral lymphoid tissues of SIV-infected macaques. Mucosal. Immunol. 13, 149–160. doi: 10.1038/s41385-019-0221-x
Rao, S., Lungu, C., Crespo, R., Steijaert, T. H., Gorska, A., Palstra, R. J., et al. (2021). Selective cell death in HIV-1-infected cells by DDX3 inhibitors leads to depletion of the inducible reservoir. Nat. Commun. 12:2475.
Rosenbloom, D. I. S., Bacchetti, P., Stone, M., Deng, X., Bosch, R. J., Richman, D. D., et al. (2019). Assessing intra-lab precision and inter-lab repeatability of outgrowth assays of HIV-1 latent reservoir size. PLoS Comput. Biol. 15:e1006849. doi: 10.1371/journal.pcbi.1006849
Siliciano, J. D., Kajdas, J., Finzi, D., Quinn, T. C., Chadwick, K., Margolick, J. B., et al. (2003). Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9, 727–728. doi: 10.1038/nm880
Siliciano, J. D., and Siliciano, R. F. (2005). Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol. Biol. 304, 3–15.
Siliciano, J. D., and Siliciano, R. F. (2018). Assays to measure latency, reservoirs, and reactivation. Curr. Top Microbiol. Immunol. 417, 23–41. doi: 10.1007/82_2017_75
Siliciano, J. D., and Siliciano, R. F. (2021). Low inducibility of latent human immunodeficiency virus Type 1 proviruses as a major barrier to cure. J. Infect. Dis. 223, 13–21.
Simonetti, F. R., White, J. A., Tumiotto, C., Ritter, K. D., Cai, M., Gandhi, R. T., et al. (2020). Intact proviral DNA assay analysis of large cohorts of people with HIV provides a benchmark for the frequency and composition of persistent proviral DNA. Proc. Natl. Acad. Sci. U S A. 117, 18692–18700. doi: 10.1073/pnas.2006816117
Sivro, A., Schuetz, A., Sheward, D., Joag, V., Yegorov, S., Liebenberg, L. J., et al. (2018). Integrin alpha4beta7 expression on peripheral blood CD4(+) T cells predicts HIV acquisition and disease progression outcomes. Sci. Transl. Med. 10:eaam6354. doi: 10.1126/scitranslmed.aam6354
Sogaard, O. S., Graversen, M. E., Leth, S., Olesen, R., Brinkmann, C. R., Nissen, S. K., et al. (2015). The depsipeptide romidepsin reverses HIV-1 latency in vivo. PLoS Pathog. 11:e1005142. doi: 10.1371/journal.ppat.1005142
Stone, M., Rosenbloom, D., Bacchetti, P., Deng, X., Dimapasoc, M., Keating, S., et al. (2020). Assessing suitability of next-generation viral outgrowth assays as proxies for classic QVOA to measure HIV-1 latent reservoir size. J. Infect. Dis. 2020:jiaa089.
Stoszko, M., Ne, E., Abner, E., and Mahmoudi, T. (2019). A broad drug arsenal to attack a strenuous latent HIV reservoir. Curr. Opin. Virol. 38, 37–53. doi: 10.1016/j.coviro.2019.06.001
Stuelke, E. L., James, K. S., Kirchherr, J. L., Allard, B., Baker, C., Kuruc, J. D., et al. (2020). Measuring the inducible, replication-competent HIV reservoir using an ultra-sensitive p24 readout, the digital ELISA viral outgrowth assay. Front. Immunol. 11:1971. doi: 10.3389/fimmu.2020.0197
Tietjen, I., Ngwenya, B. N., Fotso, G., Williams, D. E., Simonambango, S., Ngadjui, B. T., et al. (2018). The Croton megalobotrys Mull Arg. traditional medicine in HIV/AIDS management: documentation of patient use, in vitro activation of latent HIV-1 provirus, and isolation of active phorbol esters. J. Ethnopharmacol. 211, 267–277. doi: 10.1016/j.jep.2017.09.038
Tietjen, I. R. K., Williams, D., Rivera-Ortiz, J., Cai, Y., Pagliuzza, A., Chomont, N., et al. (2019). Phorbol esters isolated from Croton megalobotrys reverse HIV latency ex vivo. J. Virus Eradic. 5:44. doi: 10.1016/s2055-6640(20)30191-6
Trautmann, L., Janbazian, L., Chomont, N., Said, E. A., Gimmig, S., Bessette, B., et al. (2006). Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat. Med. 12, 1198–1202. doi: 10.1038/nm1482
Uzzan, M., Tokuyama, M., Rosenstein, A. K., Tomescu, C., SahBandar, I. N., Ko, H. M., et al. (2018). Anti-alpha4beta7 therapy targets lymphoid aggregates in the gastrointestinal tract of HIV-1-infected individuals. Sci. Transl. Med. 10:eaau4711. doi: 10.1126/scitranslmed.aau4711
Vandergeeten, C., Fromentin, R., Merlini, E., Lawani, M. B., DaFonseca, S., Bakeman, W., et al. (2014). Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J. Virol. 88, 12385–12396. doi: 10.1128/jvi.00609-14
Wang, G., Zhao, N., Berkhout, B., and Das, A. T. (2018). CRISPR-Cas based antiviral strategies against HIV-1. Virus Res. 244, 321–332. doi: 10.1016/j.virusres.2017.07.020
Whitney, J. B., Hill, A. L., Sanisetty, S., Penaloza-MacMaster, P., Liu, J., Shetty, M., et al. (2014). Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 512, 74–77. doi: 10.1038/nature13594
Wonderlich, E. R., Subramanian, K., Cox, B., Wiegand, A., Lackman-Smith, C., Bale, M. J., et al. (2019). Effector memory differentiation increases detection of replication-competent HIV-l in resting CD4+ T cells from virally suppressed individuals. PLoS Pathog. 15:e1008074. doi: 10.1371/journal.ppat.1008074
Yucha, R. W., Hobbs, K. S., Hanhauser, E., Hogan, L. E., Nieves, W., Ozen, M. O., et al. (2017). High-throughput Characterization of HIV-1 reservoir reactivation using a single-cell-in-droplet PCR assay. EBioMedicine 20, 217–229. doi: 10.1016/j.ebiom.2017.05.006
Yukl, S. A., Kaiser, P., Kim, P., Telwatte, S., Joshi, S. K., Vu, M., et al. (2018). HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med. 10:eaa9927.
Zerbato, J. M., Purves, H. V., Lewin, S. R., and Rasmussen, T. A. (2019). Between a shock and a hard place: challenges and developments in HIV latency reversal. Curr. Opin. Virol. 38, 1–9. doi: 10.1016/j.coviro.2019.03.004
Zhang, G., Luk, B. T., Wei, X., Campbell, G. R., Fang, R. H., Zhang, L., et al. (2019). Selective cell death of latently HIV-infected CD4(+) T cells mediated by autosis inducing nanopeptides. Cell Death Dis. 10:419.
Zhang, Y., Planas, D., Raymond Marchand, L., Massanella, M., Chen, H., Wacleche, V. S., et al. (2020). Improving HIV outgrowth by optimizing cell-culture conditions and supplementing with all-trans retinoic acid. Front. Microbiol. 11:902. doi: 10.3389/fmicb.2020.00902
Keywords: HIV-1 latency, HIV-1 cure, reservoir quantification, inducible reservoir, TILDA
Citation: Lungu C, Banga R, Gruters RA and Procopio FA (2021) Inducible HIV-1 Reservoir Quantification: Clinical Relevance, Applications and Advancements of TILDA. Front. Microbiol. 12:686690. doi: 10.3389/fmicb.2021.686690
Received: 27 March 2021; Accepted: 21 May 2021;
Published: 15 June 2021.
Edited by:
Miguel Angel Martinez, IrsiCaixa, SpainReviewed by:
Adam M. Spivak, The University of Utah, United StatesNicolas Sluis-Cremer, University of Pittsburgh, United States
Copyright © 2021 Lungu, Banga, Gruters and Procopio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesco A. Procopio, ZnJhbmNlc2NvLnByb2NvcGlvQGNodXYuY2g=
 Cynthia Lungu
Cynthia Lungu Riddhima Banga
Riddhima Banga Rob A. Gruters
Rob A. Gruters Francesco A. Procopio
Francesco A. Procopio