
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 04 August 2021
Sec. Virology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.686025
This article is part of the Research TopicPlant Viruses, Volume II: Molecular Plant Virus Epidemiology and its ManagementView all 17 articles
Orthotospoviruses are responsible for serious crop losses worldwide. Orthotospoviral diseases have spread rapidly in China over the past 10 years and are now found in 19 provinces. Currently, 17 Orthotospovirus species have been reported in China, including eight newly identified species from this genus. The number of new highly pathogenic Orthotospovirus strains or species has increased, likely because of the virus species diversity, the wide range of available hosts, adaptation of the viruses to different climates, and multiple transmission routes. This review describes the distribution of Orthotospovirus species, host plants, typical symptoms of infection under natural conditions, the systemic infection of host plants, spatial clustering characteristics of virus particles in host cells, and the orthotospoviral infection cycle in the field. The evolutionary relationships of orthotospoviruses isolated from China and epidemiology are also discussed. In order to effectively manage orthotospoviral disease, future research needs to focus on deciphering the underlying mechanisms of systemic infection, studying complex/mixed infections involving the same or different Orthotospovirus species or other viruses, elucidating orthotospovirus adaptative mechanisms to multiple climate types, breeding virus-resistant plants, identifying new strains and species, developing early monitoring and early warning systems for plant infection, and studying infection transmission routes.
Orthotospoviruses have a worldwide distribution and cause serious economic losses in a variety of crops (Komoda et al., 2017). Before 2017, orthotospoviruses were considered to be part of the genus Tospovirus in the family Bunyaviridae and were divided into seven or nine serogroups based on serology (Chen et al., 2012b, 2016). However, in 2020, the International Committee on Taxonomy of Viruses (ICTV) announced that the genus Orthotospovirus belongs to the family Tospoviridae, order Bunyavirales, class Ellioviricetes, realm Riboviria, subphylum Polyploviricotina, phylum Negarnaviricota, and kingdom Orthornavirae (Hong et al., 2020).
As with other members of the order Bunyavirales, orthotospovirus particles are spherical, and multi-virions form aggregations in host-derived vesicles. Mature virions range in diameter from 80 to 120 nm. The surface of the virions is composed mainly of two glycoproteins (Gn and Gc), which are responsible for the virus acquisition and transmission by the thrips vectors. The core of the virion contains three viral RNA fragments, named L RNA, M RNA, and S RNAs according to their lengths, encapsulated by the nucleocapsid protein (N). The orthotospovirus are single-negative-stranded, ambisense RNA viruses. The L RNA (∼8.9 kb) encodes the RNA-dependent RNA polymerase (RdRp) in the complementary (vcRNA) strand; M RNA (∼4.8 kb) encodes the movement protein (NSm) in the viral (vRNA) strand and the Gn and Gc proteins in the vcRNA strand; and RNA S (∼2.9 kb) encodes the N protein in the vcRNA and silencing suppressor (NSs) in the vRNA strand (Oliver and Whitfield, 2016).
To date, many articles have illustrated details of the global occurrence, epidemiology, and molecular interactions between orthotospoviruses and their thrips vectors (Pappu et al., 2009; Oliver and Whitfield, 2016). In this article, we will summarize the latest research progress on species diversity, occurrence, distribution, epidemiology, and management of orthotospoviruses in China.
Symptoms of orthotospoviral disease in host plants are very similar, with only minor differences between species of virus. The major symptoms are ringspots (including chlorotic, yellow, necrotic, and zonate spots), bud necrosis, silver mottle, and vein banding. Zonate spots are characteristic of orthotospovirus infection. Although symptoms vary between disease stages, chlorotic, yellow, and necrotic ringspots can occur at all stages (early, middle, and late). Herbaceous plants with severe disease die in the late stage. The symptoms occur in the leaves and fruit, with a few cases of stem necrosis (Figure 1).

Figure 1. Typical symptoms of host plants infected with orthotospoviruses under natural field conditions. (A) Zonate spot in leaf of chili pepper (Capsicum annuum L.) infected with tomato zonate spot orthotospovirus (TZSV). (B) Necrotic spot in leaf of macadamia nut (Macadamia ternifolia F. Muell.) infected with watermelon silver mottle virus (WSMoV) serogroup member. (C) Yellow spot and ringspot in tomato (Solanum lycopersicum L.) infected with tomato spotted wilt orthotospovirus (TSWV). (D) Necrotic zonate spot in tomato infected with tomato necrotic spot-associated orthotospovirus (TNSaV). (E) Chlorotic spot in sweet pepper (C. annuum L.) infected with TZSV. (F) Necrotic ringspot in potato (Solanum tuberosum L.) infected with TZSV.
The host range of orthotospovirus has expanded from crops to other plants, including weeds such as Bidens bipinnata (Zhang et al., 2020) and even to woody plants such as kiwifruit, mulberry, and macadamia nut (Fang et al., 2013; Meng et al., 2015; Wang et al., 2016). Orthotospoviral diseases in China mainly involve diseases of vegetables, fruits, tobacco, groundnuts, and ornamental plants, including crops of the families Solanaceae, Cucurbitaceae, Asteraceae, Brassicaceae, Fabaceae, Orchidaceae, and Amaryllidaceae (Alliaceae), weeds (such as B. bipinnata L.), and woody plants (such as kiwifruit, macadamia nut, and mulberry) (Table 1).
There are currently 30 Orthotospovirus species known worldwide, comprising 11 definitive species (written in italics) and 19 tentative species (written in upright letters) (Figure 2A; International Committee on Taxonomy of Viruses Executive Committee [ICTVEC], 2020; Zheng et al., 2020). Orthotospoviruses can be divided into five phylogenetic clades based on the amino acid sequence of nucleocapsid (N) protein (Figure 2A), namely the tomato spotted wilt orthotospovirus (TSWV) clade, watermelon silver mottle orthotospovirus (WSMoV) clade, soybean vein necrosis-associated orthotospovirus (SVNV) clade, iris yellow spot orthotospovirus (IYSV) clade, and groundnut yellow spot orthotospovirus (GYSV) clade (Peng et al., 2011; Oliver and Whitfield, 2016). Based on the geography of the recent epidemics, viruses from the WSMoV and IYSV clades are mainly found in Asia and Europe (Figure 3A). Viruses from the WSMoV clade are most commonly found in East Asia and mainly infect crops in the families Solanaceae, Cucurbitaceae, and Asteraceae, with several new species from this clade reported in recent years (Dong et al., 2008; Yin et al., 2014b; Zheng et al., 2017, 2020). Viruses from the IYSV clade are mainly found in Central Asia and Europe. Some of the species in this clade, including IYSV and tomato yellow ring orthotospovirus (TYRV), were originally isolated from plants in the Middle East but have become endemic in Europe in recent years (Bag et al., 2015; Zarzynska-Nowak et al., 2016). Viruses in the GYSV clade, which is also Asian, have been reported from Taiwan province, China, and India (Satyanarayana et al., 1996; Chao et al., 2001). The SVNV and TSWV clades belong to the group found in the Americas. Soybean vein necrosis-associated orthotospovirus (SVNV) and bean necrotic mosaic orthotospovirus (BeNMV), two species in the SVNV clade, have been reported to infect bean plants in the United States and Brazil (Zhou et al., 2011; de Oliveira et al., 2012). Several members of the TSWV clade were originally found in the Americas (Torres et al., 2012; Webster et al., 2015). However, TSWV, which belongs to the TSWV clade, is now widely distributed throughout the world, and indeed is considered to be the most harmful of the Orthotospovirus species, causing great damage and large crop losses globally (Pappu, 2008; Pappu et al., 2009).
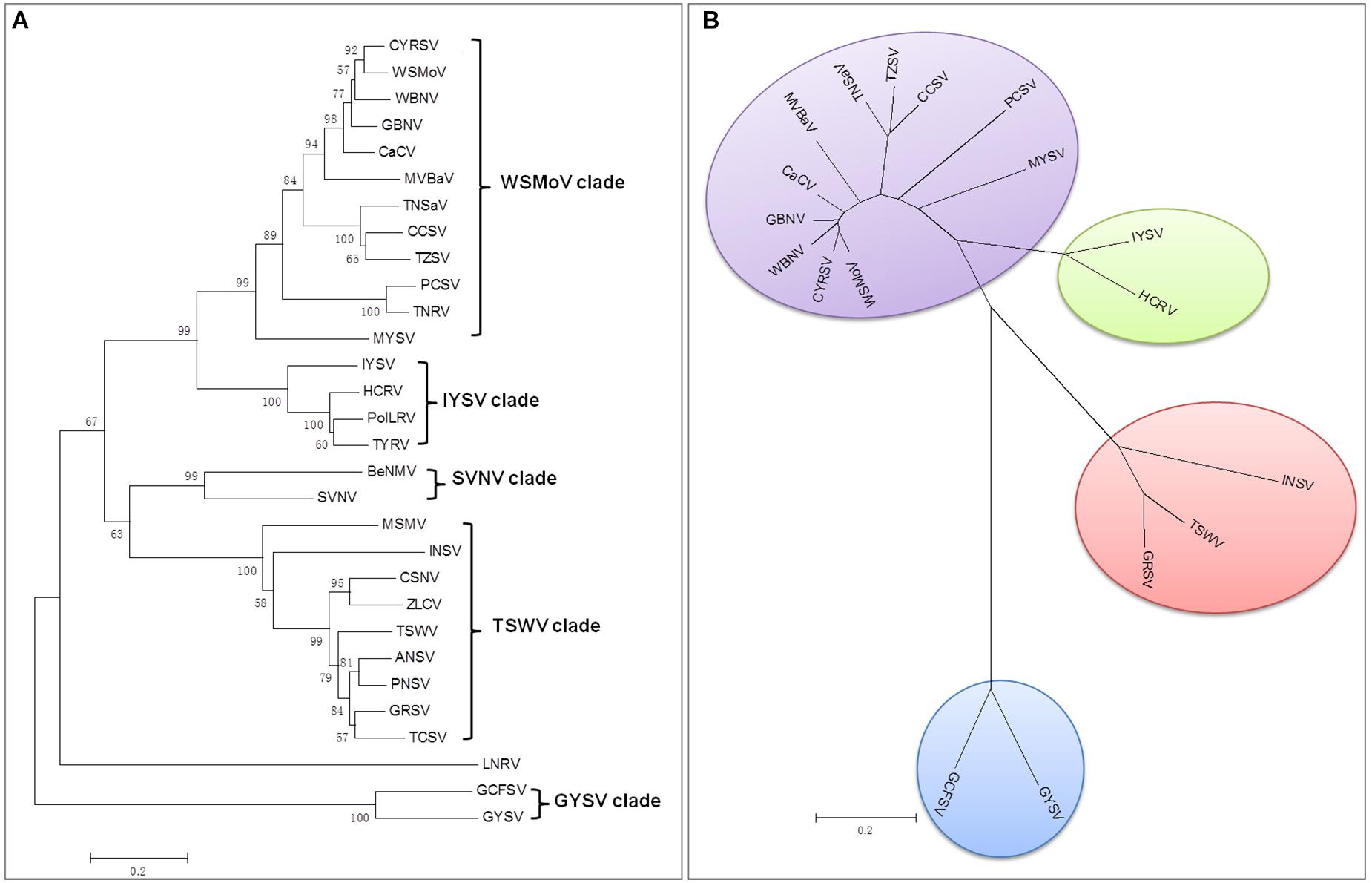
Figure 2. Phylogenetic tree of orthotospoviruses based on the amino acid sequence of nucleocapsid (N) protein. (A) Phylogenetic tree of 30 orthotospoviruses reported worldwide. (B) Phylogenetic tree of the 17 orthotospoviruses reported from China. Bootstrap values on the branches represent support for the branches based on 1,000 bootstrap replicates. Definitive species are written in italics and tentative species are written in upright letters. Abbreviations (and NCBI no.): ANSV, Alstroemeria necrotic spot orthotospovirus (GQ478668); BeNMV, Bean necrotic mosaic orthotospovirus (NC_018071); CaCV, Capsicum chlorosis orthotospovirus (NC_008301); CCSV, Calla lily chlorotic spot orthotospovirus (AY867502); CSNV, Chrysanthemum stem necrosis orthotospovirus (NC_027719); CYRSV, Chili yellow ringspot orthotospovirus (MH779495); GBNV, Groundnut bud necrosis orthotospovirus (NC_003619); GRSV, Groundnut ringspot orthotospovirus; GYSV, Groundnut yellow spot orthotospovirus (AF013994); HCRV, Hippeastrum chlorotic ringspot orthotospovirus (KC290943); INSV, Impatiens necrotic spot orthotospovirus (NC_003624); IYSV, Iris yellow spot orthotospovirus (AF001387); LNRV, Lisianthus necrotic ringspot orthotospovirus (AB852525); MSMV, Melon severe mosaic orthotospovirus (EU275149); MVBaV, Mulberry vein banding associated orthotospovirus (KM819701); MYSV, Melon yellow spot orthotospovirus (AB038343); GCFSV, Groundnut chlorotic fan-spot orthotospovirus (AF080526); PCSV, Pepper chlorotic spot orthotospovirus (KF383956); PolRSV, Polygonum ringspot orthotospovirus (KF383956); PNSV, Pepper necrotic spot orthotospovirus (HE584762); SVNV, Soybean vein necrosis-associated orthotospovirus (HQ728387); TCSV, Tomato chlorotic spot orthotospovirus (S54325); TSWV, Tomato spotted wilt orthotospovirus (NC_002051); TNRV, Tomato necrotic ringspot orthotospovirus (FJ489600); TNSaV, Tomato necrotic spot-associated orthotospovirus (KM355773); TYRV, Tomato yellow ring orthotospovirus (AY686718); TZSV, Tomato zonate spot orthotospovirus (NC_010489); WBNV, Watermelon bud necrosis orthotospovirus (EU249351); WSMoV, Watermelon silver mottle orthotospovirus (NC_003843); and ZLCV, Zucchini lethal chlorosis orthotospovirus (AF067069).
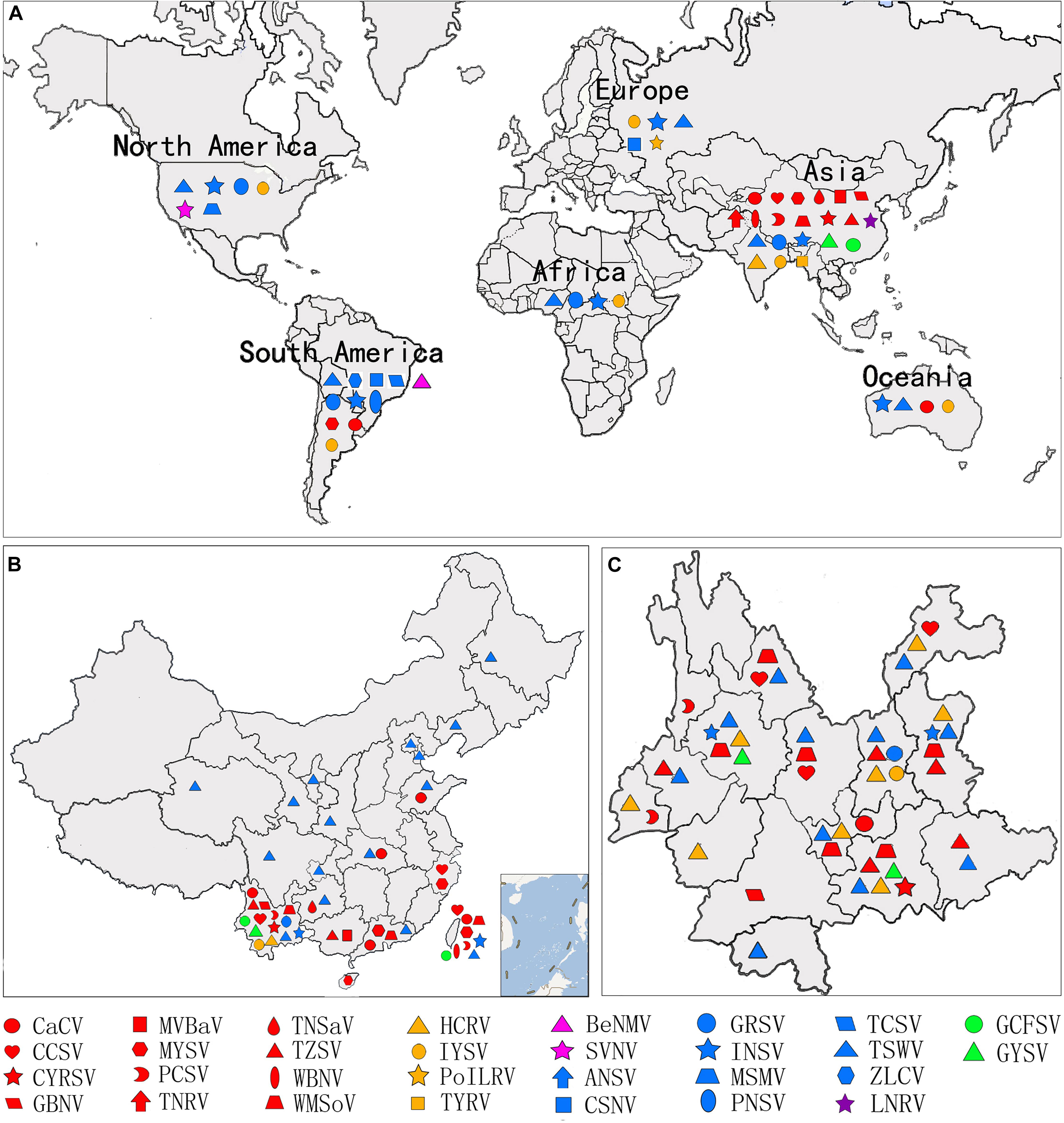
Figure 3. Geographic distribution of orthotospoviruses. (A) Worldwide geographic distribution of orthotospoviruses. (B) Geographic distribution of orthotospoviruses in China. (C) Geographic distribution of orthotospoviruses in Yunnan province, China.
The country from which the largest number of Orthotospovirus species has been reported is China. A total of 17 Orthotospovirus species are known in China to date (Table 1), comprising 8 definitive and 10 tentative species. These species belong to four phylogenetic clades (TSWV, WSMoV, IYSV, and GYSV) (Figure 2B). Of the Orthotospovirus clades found in China, the WSMoV clade is the most diverse. Several new virus species in this clade have been reported for the first time in China (Dong et al., 2008; Yin et al., 2014b; Zheng et al., 2017, 2020), and it has been speculated that the WSMoV clade originated in China. TSWV, which belongs to the Americas group (Pappu et al., 2009), has become the virus posing the largest threat to agricultural production in China. Compared with native WSMoV clade viruses, TSWV has wider geographical adaptability (Figure 3B), and evidence of adaptive evolution of TSWV in China can be found in the phylogenetic analysis of TSWV N gene diversity (Mao et al., 2019; Liu et al., 2021).
Before 2000, only two Orthotospovirus species had been reported from China. The earliest record of TSWV symptoms in peanuts was published in 1986 by Xu in Guangdong and Guangxi provinces (Xu et al., 1986). Subsequently, TSWV-like virus particles were observed using transmission electron microscopy in tomato and tobacco from Sichuan and Yunnan provinces (Su et al., 1987; Yao, 1992; Zhang et al., 1998). WSMoV was first reported in Taiwan and identified as a new Orthotospovirus species through sequencing analysis (Chu and Yeh, 1998).
After 2000, the new Orthotospovirus species tomato zonate spot orthotospovirus (TZSV) was reported in tomatoes from Yunnan province, based on whole-genome sequencing and viral particle clustering characteristics in host cells (Dong et al., 2008). TZSV has a wide host range, causing harm in tomato, pepper, tobacco, and other crops, and it is now the dominant Orthotospovirus species found in these crops in Yunnan province (Huang et al., 2015; Zheng et al., 2015c).
Different Orthotospovirus species have been reported from different provinces, but TSWV is currently spreading rapidly from the south to the north of China. A total of 17 Orthotospovirus species have been reported from 19 Chinese provinces (Table 1 and Figure 3B). Yunnan has extremely high Orthotospovirus diversity, with 13 species identified, while most other Chinese provinces harbor only one or two Orthotospovirus species. Of all the Orthotospoviruses, TSWV has the most extensive distribution, occurring in 18 provinces to date (Table 1 and Figure 3B).
Yunnan has a high incidence of orthotospoviral disease, with diverse species of viruses and host plants, and a wide distribution. We identified 13 Orthotospovirus species from areas with very different climates (including tropical, subtropical, temperate, and cold temperate) (Figure 3C) and more than 20 species of natural host plants in Yunnan. The natural host range included most of the plants in China known to become infected with orthotospoviruses (Table 1). The orthotospoviruses isolated from Yunnan belonged to four phylogenetic clades, with the majority (seven species) belonging to the WSMoV clade. Three new Orthotospovirus species were first reported in Yunnan: TZSV (Dong et al., 2008), hippeastrum chlorotic ringspot orthotospovirus (HCRV; Dong et al., 2013), and chili yellow ringspot virus (CYRSV; Zheng et al., 2020). TSWV and TZSV were the dominant species, based on disease epidemics and severity (Zhang et al., 2016). The orthotospoviruses in Yunnan province have caused serious harm to crops including tomato, chili pepper, potato, and tobacco (Solanaceae); zucchini, watermelon, and squash (Cucurbitaceae); Phalaenopsis and Dendrobium (Orchidaceae); lettuce and chrysanthemum (Asteraceae); soybean and groundnut (Fabaceae), and more than 10 weed species.
Orthotospoviruses are becoming more and more harmful due to their multiple transmission routes and wide host ranges. The main transmission routes under natural conditions are via the vector thrips, which are insects in the order Thysanoptera. More than 14 species of thrips are known to act as vectors for orthotospoviruses.
The thrips that are known to transmit orthotospovirus include Ceratothrip oidesclaratris, Dictyothrips betae, Frankliniella occidentalis, Frankliniella schultzei, Frankliniella gemina, Frankliniella intonosa, Frankliniella cephalica, Frankliniella bispinosa, Frankliniella fusca, Frankliniella zucchini, Scirtothrips dorsalis, Neohydatothrips variabilis, Thrips palmi, and Thrips tabaci (Pappu et al., 2009; Whitfield et al., 2015). A single Orthotospovirus species can be transmitted by one or many species of thrips (e.g., TSWV is transmitted by nine species of thrips). A single species of thrips can transmit one or many Orthotospovirus species (e.g., F. occidentalis can transmit five Orthotospovirus species) (Whitfield et al., 2015).
Besides transmission by thrips, other routes of transmission facilitate spread of orthotospoviruses. Orthotospoviruses such as SVNV spread through seeds (Groves et al., 2016). In addition, spreading seedings infected with orthotospoviruses is also an important source for the virus to spread in different places (Zhang et al., 2020). Weeds, as an important primary infection source, provide potential conditions for the secondary infection and outbreak of orthotospoviruses in the field.
Orthotospoviruses have spread rapidly through China in recent years. Before 2015, orthotospoviral diseases mainly occurred in Southwest and Southeast China. However, over the last 5 years, orthotospoviral diseases have also occurred in Central, Northwest, and Northeast China. Although TSWV is the main epidemical Orthotospovirus species, we have recently found WSMoV, TZSV, and HCRV in Tibet, Hainan, and other Chinese provinces. Orthotospoviruses tend to quickly replace other viral pathogens (such as tobacco mosaic virus and cucumber mosaic virus) or form complex/mixed infections with other viruses (such as potato virus Y and whitefly transmitted geminivirus) in solanaceous crops, based on our research results over the past 20 years in Yunnan province.
The main reasons for the rapid expansion of Orthotospovirus species, especially TSWV, in China are as follows: (1) Over the last 20 years, the rapid popularization and development of greenhouses in China has provided perfect conditions for the propagation of the vector insects, thrips. Particularly in the north of China, greenhouses provide suitable temperatures for the overwintering of the virus-transmitting thrips, and indeed the orthotospoviral diseases in most northern provinces occur mainly in greenhouses (Jie et al., 2017; Liu et al., 2021). In southern China, as well as the greenhouse-thrips-vegetables infection pathway, a secondary infection cycle can be formed by weeds and other intermediate hosts, and weeds also provide thrips with overwintering habitats (Zhang et al., 2020). (2) Changes in planting structure have also led to the rapid expansion of TSWV. With the popularization of greenhouses, monocultures of a single crop variety with susceptibility to orthotospoviruses, such as pepper, tomato, and other Solanaceous crops, have been planted in large quantities (Miao, 2018). This is particularly obvious in Yunnan province, where the planting structure, which usually includes flowers, tomatoes, peppers, tobacco, and potatoes, provides very favorable conditions for the reproduction of the thrips population and susceptibility to orthotospoviruses (Zhao et al., 2021).
Orthotospoviruses in host cells have distinct histopathological characteristics that mainly relate to the virus species rather than to the host plant species, and these characteristics have important diagnostic value. The histopathological characteristics differ between the different stages of infection. In general, at 3 days post inoculation (dpi), there are a large number of vesicles in the host cells. At 7 dpi, virus particles form in the vesicles. At this stage (in the early stages of infection), double-enveloped virions (DEV) are also observed in host cells. At 9 dpi, virus particle aggregates appear in the host cells. By 12 dpi (in the late stages of infection), the cells are filled with virus particles (Zhang, 2015).
The virus particle clustering characteristics differ among Orthotospovirus species. TSWV virus particles always cluster in the endoplasmic reticulum (ER) or in special vesicles (based on transmission electron microscopy of ultrathin sections or negative staining). TZSV virus particles usually cluster in a moniliform structure in the ER membrane (vertical section) or as double-enveloped virions (cross-section) (Zhang et al., 2016). WSMoV virus particles always cluster in vesicles connected to other empty vesicles or in host cell vacuoles (unpublished data). Impatiens necrotic spot orthotospovirus (INSV) virus particles usually cluster in the lumen of the ER (Zhang Z. K. et al., 2010; Figure 4). Although these virus particle clustering characteristics are not necessarily unique to these viruses in the host cells, histopathological ultrastructure characteristics can help to identify Orthotospovirus species.
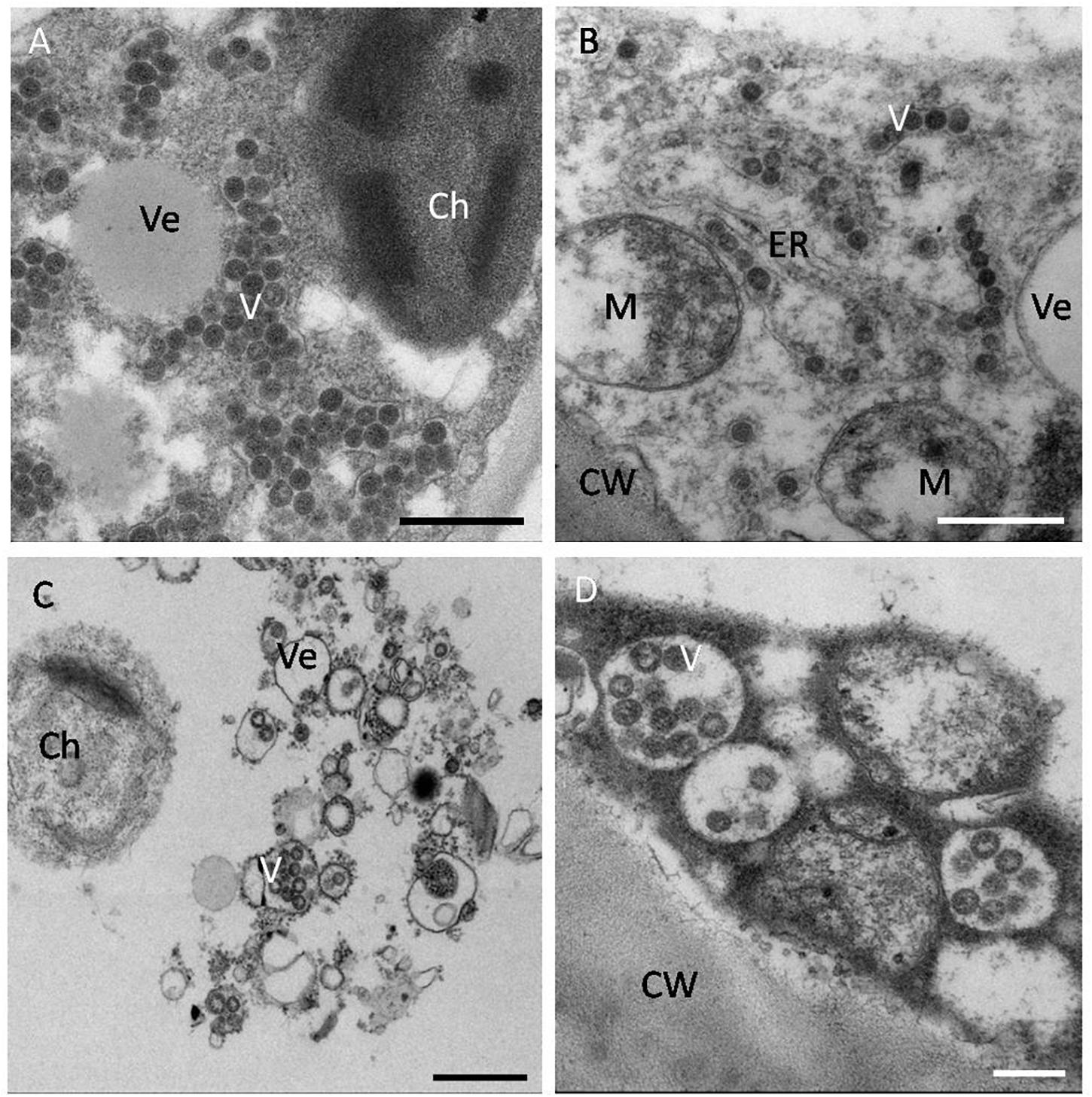
Figure 4. Histopathological characteristics of orthotospoviruses in host cells. (A) TSWV virus particles clustered in the cytoplasm of a tomato leaf cell. Bar = 500 nm. (B) TZSV virus particles clustered in the endoplasmic reticulum (ER) membrane of a tomato fruit cell. Bar = 500 nm. (C) WSMoV virus particles clustered in vesicles in a tomato fruit cell. Bar = 500 nm. (D) INSV virus particles clustered in the lumen of the ER in a tomato fruit cell. Bar = 20 nm. Ve, vesicle; V, virus particle; Ch, chloroplast; M, mitochondrion; ER, endoplasmic reticulum; CW, cell wall.
To establish a systemic infection in a host plant, the TSWV ribonucleoprotein (RNP) complex migrates along the ER membrane. The N protein wraps with TSWV RNAs to form the RNP complex, and these RNPs are subsequently driven along the ER membrane and actin by the action of myosin, after which the viral RNPs enter into the Golgi body, where they form mature virus particles (Feng et al., 2013; Ribeiro et al., 2013). To infect a new cell, the RNP complexes move to the plasmodesmata, traversing the plasmodesmata through the action of the viral movement protein NSm to reach a neighboring cell. This process involves the interaction of the N-terminal of the NSm protein with nucleocapsid protein N and assists the movement of the RNPs toward the plasmodesmata using the actin microfilament/ER transport system (Leastro et al., 2015; Tripathi et al., 2015; Feng et al., 2016). The viral movement protein NSm plays a decisive role in the intercellular and long-distance movement of viral RNPs. However, research also shows that nucleocapsid protein N plays a key role in long-distance movement, and it has been confirmed that both TSWV N and NSm are necessary for the long-distance movement of movement-deficient TMV (Lewandowski and Adkins, 2005; Zhang et al., 2011). Deletion mutations have demonstrated that NSm mediates virus intercellular movement and long-distance movement using different domains, suggesting that the mechanisms by which NSm mediates virus intercellular movement and long-distance movement are different (Li et al., 2009). Interestingly, TSWV and TZSV virus particles with vesicles have been also found in plant vascular tissue, suggesting that viral particles can also load/unload from vascular tissues and establish systematic infection through long-distance movement (Zhang, 2015; Wen et al., 2020).
Biological control is an effective method to control thrips-borne orthotospovirus disease, which includes diversified prevention and control measures. These include a reasonable rotation or continuous replanting mode (Bokil et al., 2019); release of predatory mites, mirids, and other natural enemies to the greenhouse environment (Bouagga et al., 2020); and adding beneficial microorganisms to the soil environment to help plants enhance disease resistance (Beris et al., 2018; Bonanomi et al., 2020). In disease management, integrated pest management (IPM) has been proved to be more effective than chemical control (Rodríguez et al., 2019).
Growing highly virus-resistant plant varieties can be an effective way to prevent and control viral diseases. TSWV-resistant tomato and pepper varieties have been bred by Bayer, Syngenta, and other companies. These varieties possess the Sw-5 and Tsw resistance genes, respectively. After selection trials, virus-resistant plants were grown in areas of high TSWV occurrence in China. However, the resistance of these plants was poor, due to single-gene resistance combined with the existence of complex/mixed infections. To date, there are no varieties with high levels of comprehensive resistance.
Transgene technology is a rapid method for plants to acquire resistance to orthotospoviruses. So far, transgenic tobacco with broad-spectrum resistance against four other serologically unrelated Orthotospovirus TSWV, GYSV, INSV, and GCFCV has been developed by transforming the conserved motifs of the RdRp gene of WSMoV through hagrobacterium-mediated transformation (Kung et al., 2012; Peng et al., 2014).
Natural chemicals have attracted increasing attention and have become the most promising strategy in the defense against pathogenic infections. These naturally occurring chemicals are popular because they are environmentally friendly and leave little residue, they are highly identifiable, and they have low toxicity for the plant hosts. Because of the serious damage to agricultural crops caused by TSWV infection, natural products able to limit TSWV have attracted a great deal of interest. One potential natural derivative is atin-3-acetonyl-3-hydroxyoxindole (AHO), isolated from Strobilanthescusia, which is able to up-regulate PR-10 genes in the salicylic acid (SA) pathway, as well as up-regulate the levels of miRNAs (miR156, miR172f, miR172g, miR408a) that contribute to inhibiting TSWV infection (Chen et al., 2014, 2017). Actigard, imidacloprid, and Bacillus amyloliquefaciens strain MBI600 have also been found to induce the SA signaling pathway and to prevent TSWV infection (Csinos et al., 2001; McPherson et al., 2005; Beris et al., 2018). The terpenoid compound 3α-angeloyloxy-9β-hydroxy-ent-kaur-16-en-19-oic acid (AHK) has been isolated from Wedeliatrilobata and found to defend against TSWV activity and infection, with an inhibition rate of 62.4% in curative effects assays and 76.5% in protective effects assays, mainly through activation of the jasmonic acid (JA) signaling pathway and inhibition of NSs, NSm, and RdRp gene expression (Zhao et al., 2019). Tagitinin A is a sesquiterpene isolated from Tithonia diversifolia and was found to have even higher curative and protective effects against TSWV, with an inhibition rate of over 75%. Furthermore, the expression of the genes NSs and NSm was inhibited in inoculated and systemic leaves in the protective assay, with an inhibition rate of more than 85% in systemic leaves (Zhao et al., 2017, 2020). It is therefore possible that these natural products can be used as chemical elicitors to trigger systemic acquired resistance (SAR), stimulating natural plant immunity. There is a wide variety of potential applications for such chemicals in agriculture.
Viral infection at the seedling stage (a highly susceptible stage) is the main reason for disease outbreaks in the middle and late plant growth periods. To obtain virus-free seeds and effectively reduce the occurrence and loss caused by orthotospovirus diseases, seedlings should be grown in greenhouses with insect-proof netting or maximal barrier precautions, yellow or blue sticky plates should be used to attract and trap the vector insects (thrips), and the virus carriage rate of the seedlings should be regularly monitored. Comprehensive measures to reduce the vector insects (thrips) should also be applied.
Orthotospoviruses are expected to further expand worldwide because of their wide host range, multiple transmission routes, and ability to adapt to diverse climates. Additionally, increases in agricultural trade have accelerated global transmission. In China, new species or strains with high pathogenicity (because of mutation, reassortment, and recombination within and/or among Orthotospovirus species) have increased the frequency of disease. Orthotospoviruses are therefore a threat to crop production because of their high pathogenicity and the complex/mixed infections involving different Orthotospovirus species or other viruses.
It is necessary to enhance the monitoring of orthotospovirus infections and set up early warning systems in high-incidence areas; to allow the rapid diagnosis of symptoms caused by Orthotospovirus species in host plants; to assess virus carriage rates in seeds, seedlings, thrips, and weeds; and to allow dynamic monitoring of thrips levels. A deeper understanding of the mechanisms of systemic infection, of the pathogenesis of complex/mixed infections involving the same or different Orthotospovirus species or other viruses, and of Orthotospovirus adaptation mechanisms to multiple climate types is required, as is the screening and breeding of virus-resistant plant varieties. Furthermore, to ensure environmentally friendly prevention and control of orthotospoviral diseases, highly effective biological antiviral agents should be developed, and several techniques should be popularized, including the use of virus-free seeds and standardized seedling propagation technology (such as building greenhouses with insect-proof nets, sterilizing tools before raising seedlings, controlling the number of insects by using yellow or blue sticky plates, and releasing predatory mites, mirids, or the other natural enemies of thrips, and monitoring of virus incidence on seedlings before transplantation).
ZZ conceived and designed the review and wrote most of the manuscript. KZ wrote the text on orthotospoviruses evolutionary relationships, conducted the statistical analyses shown in Figure 2 and Table 1, and revised the manuscript. LZ wrote the text on the screening of natural anti-orthotospoviral products. XS conducted the statistical analyses shown in Figure 3. XZ wrote the text on orthotospoviruses transmission by thrips. TW conducted the transmission electron microscopy procedures. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (U1802235), the Yunling Scholar Programme of the Yunnan Provincial Government (2016-2020), and Science and Technology Program of Yunnan Province (2018FB028).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the reviewer XT (Nanjing Agricultural University, China) and Sijun Zheng (Yunnan Academy of Agricultural Science, China/Alliance of Bioversity International and CIAT) for critically reading and revising the manuscript.
Adam, G., Yeh, S. D., Reddy, D. V., and Green, S. K. (1993). Serological comparison of tospovirus isolates from Taiwan and India with impatiens necrotic spot virus and different Tomato spotted wilt virus isolates. Arch. Virol. 130, 237–250. doi: 10.1007/BF01309657
Bag, S., Schwartz, H. F., Cramer, C. S., Havey, M. J., and Pappu, H. R. (2015). Iris yellow spot virus (Tospovirus: bunyaviridae): from obscurity to research priority. Mol. Plant Pathol. 16, 224–237. doi: 10.1111/mpp.12177
Beris, D., Theologidis, L., Skandalis, N., and Vassilakos, N. (2018). Bacillus amyloliquefaciens strain MBI600 induces salicylic acid dependent resistance in tomato plants against Tomato spotted wilt virus and Potato virus Y. Sci. Rep. 8:10320. doi: 10.1038/s41598-018-28677-3
Bokil, V. A., Allen, L. J. S., Jeger, M. J., and Lenhart, S. (2019). Optimal control of a vectored plant disease model for a crop with continuous replanting. J. Biol. Dyn. 13, 325–353. doi: 10.1080/17513758.2019.1622808
Bonanomi, G., Alioto, D., Minutolo, M., Marra, R., Cesarano, G., and Vinale, F. (2020). Organic amendments modulate soil microbiota and reduce virus disease incidence in the TSWV-tomato pathosystem. Pathogens 9:379. doi: 10.3390/pathogens9050379
Bouagga, S., Urbaneja, A., Depalo, L., Rubio, L., and Pérez-Hedo, M. (2020). Zoophytophagous predator-induced defences restrict accumulation of the tomato spotted wilt virus. Pest Manag. Sci. 76, 561–567. doi: 10.1002/ps.5547
Cai, J. H., Qin, B. X., Wei, X. P., Huang, J., Zhou, W. L., Lin, B. S., et al. (2011). Molecular Identification and Characterization of Tomato zonate spot virus in Tobacco in Guangxi, China. Plant Dis. 95:1483. doi: 10.1094/PDIS-06-11-0486
Chao, C. H., Chen, T. C., Kang, Y. C., Li, J. T., Huang, L. H., and Yeh, S. D. (2010). Characterization of Melon yellow spot virus infecting cucumber (Cucumissativus L.) in Taiwan. Plant Pathol. Bull. 19, 41–52.
Chao, J. C. J., Chu, K. F., Peng, Y. Y., Lin, S. C., Yeh, E. S., and Chen, C. (2001). Serological and Molecular Characterization of Peanut chlorotic fan-spot virus, a New Species of the Genus Tospovirus. Phytopathology 91, 856–863. doi: 10.1094/PHYTO.2001.91.9.856
Chen, C. C., Chen, T. C., Lin, Y. H., Yeh, S. D., and Hsu, H. T. (2005). A chlorotic spot disease on calla lilies (Zantedeschia spp.) is caused by a Tospovirus serologically but distantly related to Watermelon silver mottle virus. Plant Dis. 89, 440–445. doi: 10.1094/PD-89-0440
Chen, K., Xu, Z., Yan, L., and Wang, G. (2007). Characterization of a new strain of capsicumchlorosis virus from peanut (Arachishypogaea l.) in China. J. Phytopathol. 155, 178–181. doi: 10.1111/j.1439-0434.2007.01217.x
Chen, K. R., Xu, Z. Y., Yan, L. Y., and Wang, G. P. (2006). Complete genome sequence of mulberry vein banding associated virus, a new Tospovirus infecting mulberry. PLoS One 10:e0136196. doi: 10.1371/journal.pone.0136196
Chen, T. C., Chang, C. A., Kang, Y. C., Yeh, S. D., Huang, C. H., and Chen, C. C. (2012a). Identification of capsicum chlorosis virus causing chlorotic spots and stripes on calla lily. Agric. Res. Taiwan 61, 64–74. doi: 10.6156/JTAR/2012.06101.06
Chen, T. C., Li, J. T., Lin, Y. P., Yeh, Y. C., Kang, Y. C., Huang, L. H., et al. (2012b). Genomic characterization of calla lily chlorotic spot virus and design of broad: pectrum primers for detection of tospoviruses. Plant Pathol. 61, 183–194.
Chen, T. C., Lu, Y. Y., Cheng, Y. H., Li, J. T., Yeh, Y. C., Kang, Y. C., et al. (2010). Serological relationship between Melon yellow spot virus and Watermelon silver mottle virus and differential detection of the two viruses in cucurbits. Arch. Virol. 155, 1085–1095. doi: 10.1007/s00705-010-0688-y
Chen, Y. D, Dong, J. H, Bennetzen, J. L., Zhong, M., Yang, J., Zhang, J., et al. (2017). Integrating transcriptome and microRNA analysis identifies genes and microRNAs for AHO-induced systemic acquired resistance in N. tabacum. Sci. Rep. 7:12504. doi: 10.1038/s41598-017-12249-y
Chen, Y. D, Zhang, J., Wu, K., Liu, C. M., Xiao, J. H., Li, X. Y., et al. (2014). Field control effect of plant-derived natural product AHO on tospovirus disease. J. Southern Agric. 45, 2167–2171. doi: 10.3969/j:issn.2095-1191.2014.12.2167
Chen, Y. H., Dong, J., Chien, W. C., Zheng, K. Y., Wu, K., Yeh, S. D., et al. (2016). Monoclonal antibodies for differentiating infections of three serological-related tospoviruses prevalent in Southwestern China. Virol. J. 13:72. doi: 10.1186/s12985-016-0525-3
Cheng, Y. H., Zheng, Y. X., Tai, C. H., Yen, J. H., Chen, Y. K., and Jan, F. J. (2013). Identification, characterisation and detection of a new tospovirus on sweet pepper. Ann. Appl. Biol. 164, 107–115. doi: 10.1111/aab.12084
Chu, F. H., Chao, C. H., Peng, Y. C., Lin, S. S., Chen, C. C., and Yeh, S. D. (2001). Serological and molecular characterization of Peanut chlorotic fan-spot virus, a new species of the genus Tospovirus. Phytopathology 91, 856–863.
Chu, F. H., and Yeh, S. D. (1998). Comparison of ambisense M RNA of watermelon silver mottle virus with other tospoviruses. Phytopathology 88, 351–358.
Csinos, A. S., Pappu, H. R., McPherson, R. M., and Stephenson, M. G. (2001). Management of Tomato spotted wilt virusin flue-cured tobacco with acibenzolar-S-methyl and imidacloprid. Plant Dis. 85, 292–296. doi: 10.1094/PDIS.2001.85.3.292
de Oliveira, A. S., Melo, F. L., Inoue-Nagata, A. K., Nagata, T., Kitajima, E. W., and Resende, R. O. (2012). Characterization of Bean necrotic mosaic virus: a member of a novel evolutionary lineage within the genus Tospovirus. PLoS One 7:e38634. doi: 10.1371/journal.pone.0038634
Ding, M., Zhang, L. Z., Fang, Q., Li, T., Su, T., X, X., et al. (2004). Identification, purification and antiserum preparation of tospovirus isolate affecting potato. Southwest Chin. J. Agric. Sci. S1, 160–162. doi: 10.16213/j.cnki.scjas.2004.s1.038
Dong, J. H., Cheng, X. F., Yin, Y. Y., Fang, Q., Ding, M., Li, T. T., et al. (2008). Characterization of Tomato zonate spot virus, a new tospovirus in China. Arch. Virol. 153, 855–864. doi: 10.1007/s00705-008-0054-5
Dong, J. H., Yin, Y. Y., Fang, Q., McBeath, J. H., and Zhang, Z. K. (2013). A new tospovirus causing chlorotic ringspot on Hippeastrum sp. in China. Virus Genes 46, 567–570. doi: 10.1007/s11262-012-0873-z
Du, X., Wu, K., Liu, X., Zhang, L. Z., Su, X. X., Zhang, H. R., et al. (2020). The occurrence trends of dominant species of potato viruses and thrips in Yunnan Province. Sci. Agric. Sin. 53, 551–562. doi: 10.3864/j.issn.0578-1752.2020.03.008
Fang, Q., Ding, M., Dong, J. H., Yin, Y. Y., Zhang, L., Su, X. X., et al. (2013). Preliminary report of tospovirus infecting macadamia seedlings in Yunnan, China. Acta Hortic. Sin. 40, 350–354. doi: 10.16420/j.issn.0513-353x.2013.02.004
Feng, Z., Chen, X., Bao, Y., Dong, J., Zhang, Z., and Tao, X. R. (2013). Nucleocapsid of Tomato spotted wilt tospovirus forms mobile particles that traffic on an actin/endoplasmic reticulum network driven by myosin XI-K. New Phytol. 200, 1212–1224. doi: 10.1111/nph.12447
Feng, Z., Xue, F., Xu, M., Chen, X. J., Zhao, W. Y., Garcia-Murria, M. J., et al. (2016). The ER-membrane transport system is critical for intercellular trafficking of the NSm movement protein and Tomato spotted wilt tospovirus. PLoS Pathog. 12:e1005443. doi: 10.1371/journal.ppat.1005443
Gao, W., Wang, Y., Zhang, C. X., Zhang, A. S., and Zhu, X. P. (2016). Investigation and Pathogen Preliminary Identification of Pepper Virus Disease in Tianjin. Shandong Agric. Sci 48, 91–94.
Gao, X., Wang, S., Li, Z., Yang, B., and Liu, Y. T. (2020). Investigation and identification of orthotospovirus on tobacco in Yunnan province, china. Acta Tabacaria Sin. 26, 84–90. doi: 10.16472/j.chinatobacco.2019.164
Groves, C., German, T., Dasgupta, R., Mueller, D., and Smith, D. L. (2016). Seed transmission of soybean vein necrosis virus: the first tospovirus implicated in seed transmission. PLoS One. 11:e0147342. doi: 10.1371/journal.pone.0147342
Han, S., Zhang, Q. H., Wu, J., Li, H. H., and Xi, Y. D. (2020). Identification and detection of Tomato spotted wilt virus and pepper mild mottle virus infecting pepper in Sichuan using RNA sequencing. Acta Phytopathol. Sin. 50, 147–154. doi: 10.13926/j.cnki.apps.000417
Hong, J., Xie, L., Zhang, Z. K., and Zhou, X. P. (2020). Plant viruses in the new 15-rank taxonomic system of ICTV. Acta Phytopathol. Sin. 51, 143–162. doi: 10.13926/j.cnki.apps.000554
Huang, C., Yong, L., Yu, H., and Li, B. (2015). Occurrence of Tomato zonate spot virus on potato in China. Plant Dis. 99:733. doi: 10.1094/PDIS-08-14-0851-PDN
Huang, C. H., Zheng, Y. X., Cheng, Y. H., Lee, W. S., and Jan, F. J. (2010). First report of capsicum chlorosis virus infecting tomato in Taiwan. Plant Dis. 94:1263. doi: 10.1094/PDIS-04-10-0275
International Committee on Taxonomy of Viruses Executive Committee [ICTVEC]. (2020). The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 5, 668–674. doi: 10.1038/s41564-020-0709-x
Jie, L., Chi, S., Yang, Q., Ding, T., and Dong, C. (2017). Identification of Tomato spotted wilt virus disease in Shandong, China. Plant Prot. 43, 228–232. doi: 10.3969/j.issn.0529-1542.2017.01.043
Jin, F. M., Song, J., Xue, J., Chen, D. L., Wang, S., Zhang, Y., et al. (2020). Molecular detection and partial genome sequence analysis of Tomato spotted wilt virus infected chrysanthemum in Beijing. Acta Agric. Boreali Sin. 35, 175–184.
Komoda, K., Narita, M., Yamashita, K., Tanaka, I., and Yao, M. (2017). Asymmetric trimeric ring structure of the nucleocapsid protein of tospovirus. J. Viro. 91, e01002–17. doi: 10.1128/JVI.01002-17
Kung, Y. J., Lin, S. S., and Huang, Y. L. (2012). Multiple artificial micrornas targeting conserved motifs of the replicase gene confer robust transgenic resistance to negative-sense single-stranded rna plant virus. Mol. Plant Pathol. 13, 303–317. doi: 10.1111/j.1364-3703.2011.00747.x
Leastro, M. O., Pallás, V., Resende, R. O., and Sánchez-Navarro, J. A. (2015). The movement proteins (NSm) of distinct tospoviruses peripherally associate with cellular membranes and interact with homologous and heterologous NSm and nucleocapsid proteins. Virology 478, 39–49. doi: 10.1016/j.virol.2015.01.031
Lewandowski, D. J., and Adkins, S. (2005). The tubule-forming Nsm protein from Tomato spotted wilt virus complements cell-to-cell and long-distance movement of tobacco mosaic virus hybrids. Virology 342, 26–37. doi: 10.1016/j.virol.2005.06.050
Li, J. T., Yeh, Y. C., Yeh, S. D., Raja, J. A., Rajagopalan, P. A., Liu, L. Y., et al. (2011). Complete genomic sequence of watermelon bud necrosis virus. Arch. Virol. 156, 359–362. doi: 10.1007/s00705-010-0881-z
Li, Q. F., Zhi, L., Li, M., Xu, Y., Huang, Y. N., and Liu, Y. T. (2014). Tomato zonate spot virus as a Pathogen Found in Iris tectorum. J. Yunnan Agric. Univ. 29, 167–172. doi: 10.3969/j.issn.1004-390X(n).2014.02.004
Li, W., Lewandowski, D. J., Hilf, M. E., and Adkins, S. (2009). Identification of domains of the Tomato spotted wilt virus Nsm protein involved in tubule formation, movement and symptomatology. Virology 390, 110–121. doi: 10.1016/j.virol.2009.04.027
Liu, J., Chen, D., Liang, Y., Liu, D., and Huang, C. (2021). Identification of tomato spotted wilt virus on Beijing pepper and tomato. Plant Quar. 35, 44–48. doi: 10.19662/j.cnki.issn1005-2755.2021.00.009
Liu, Z., Li, Y., Wu, Y., Rao, X. (2010). Identification of tospovirus on pepper in Guangzhou. Acta Phytophylacica Sin. 37, 383–384. doi: 10.13802/j.cnki.zwbhxb.2010.04.016
Liu, Y., Li, F., Li, Y. Y., Zhang, B. S., Gao, X. W., Xie, Y., et al. (2019). Identification, distribution and occurrence of viruses in the main vegetables of China. Entia Agric. Sin. 52, 239–261. doi: 10.3864/j.issn.0578-1752.2019.02.005
Liu, Y. T., Zhi, X. P., and Zheng, Y. X. (2012). Calla lily chlorotic spot virus from spider lily (Hymenocallislitteralis) and tobacco (Nicotianatabacum) in the south-west of China. J. Phytopathol. 160, 201–205. doi: 10.1111/j.1439-0434.2011.01873.x
Mao, L., Zhao, K., Deng, M., Yang, Z., Tang, D., and Zhu, H. (2019). Variation analysis of nucleocapsid protein and movement protein of Tomato spotted wilt virus around Kunming area. Acta Botanica Boreali Occidentalia Sin. 39, 1929–1934. doi: 10.7606/j.issn.1000-4025.2019.11.1929
McPherson, R. M., Stephenson, M. G., Lahue, S. S., and Mullis, S. W. (2005). Impact of early-season thrips management on reducing the risks of spotted wilt virus and suppressing aphid population in flue-cured tobacco. J. Econ. Entomol. 98, 129–134. doi: 10.1603/0022-0493-98.1.129
Meng, J., Liu, P., Zhu, L., Zou, C., Li, J., and Chen, B. (2015). Complete genome sequence of mulberry vein banding associated virus, a new tospovirus infecting mulberry. PLoS One 10:e0136196.
Miao, J. (2018). Problems and Development Countermeasures of Facility Vegetable Industry in China From Macroscopic and Microscopic Perspectives. Northern Hortic. 2018, 185–190. doi: 10.11937/bfyy.20172601
Oliver, J. E., and Whitfield, A. E. (2016). The genus Tospovirus: emerging Bunyaviruses that threaten food security. Annu. Rev. Virol. 3, 101–124. doi: 10.1146/annurev-virology-100114-055036
Pappu, H. R. (2008). “Tomato spotted wilt virus (Bunyaviridae),” in Encyclopedia of Virology, eds B. W. J. Mahy and M. H. V. Van Regenmortel (Oxford, UK: Elsevier Ltd), 133–138.
Pappu, H. R., Jones, R. A., and Jain, R. K. (2009). Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 141, 219–236. doi: 10.1016/j.virusres.2009.01.009
Peng, J. C., Chen, T. C., Raja, J., Yang, C. F., Wan-Chu, C., and Lin, C. H. (2014). Broad-spectrum transgenic resistance against distinct tospovirus species at the genus level. PLoS One 9:e96073. doi: 10.1371/journal.pone.0096073
Peng, J. C., Yeh, S. D., Huang, L. H., Li, J. T., and Chen, T. C. (2011). Emerging threat of thrips-borne melon yellow spot virus on melon and watermelon in Taiwan. Eur. J. Plant Pathol. 130, 205–214. doi: 10.1007/s10658-011-9746-x
Rao, X., Wu, Z., and Li, Y. (2013). Complete genome sequence of a Watermelon silver mottle virus isolate from China. Virus Genes 46, 576–580. doi: 10.1007/s11262-013-0885-3
Ren, X., Liu, Y., Tan, Y., Sun, C., Huang, Y., and Liu, Z. (2014). Distribution characteristics of main tobacco virus diseases in Guizhou. Guizhou Agric. Sci. 42, 117–120. doi: 10.6041/j.issn.1000-1298.2014.07.017
Ribeiro, D., Jung, M., Moling, S., Borst, J. W., Goldbach, R., and Kormelink, R. (2013). The cytosolic nucleoprotein of the plant-infecting Bunyavirus tomato spotted wilt recruits endoplasmic reticulum-resident proteins to endoplasmic reticulum export sites. Plant Cell 25, 3602–3614. doi: 10.2307/23598370
Rodríguez, E., Téllez, M. M., and Janssen, D. (2019). Whitefly Control Strategies against Tomato Leaf Curl New Delhi Virus in Greenhouse Zucchini. Int. J. Environ. Res. Public Health 16:2673. doi: 10.3390/ijerph16152673
Satyanarayana, T., Reddy, K. L., Ratna, A. S., Deom, C. M., and Reddy, D. V. R. (1996). Peanut yellow spot virus: a distinct tospovirus species based on serology and nucleic acid hybridisation. Ann. App. Biol. 129, 237–245. doi: 10.1111/j.1744-7348.1996.tb05748.x
Song, X., Liu, Y., Chen, J., Shi, X., Zhang, D., and Branch, L. (2019). Systemic infection of Chinese garlic by Tomato spotted wilt tospovirus. Plant Prot. 45, 149–151. doi: 10.16688/j.zwbh.2018266
Su, D. K., Yuan, X. Z., Xie, Y. H., Wang, S. R., and Ding, H. (1987). Tomato spotted wilt virus in tomato in Chengdu and Dukou. Acta Phytopathol. Sin. 14, 255–256. doi: 10.13926/j.cnki.apps.1987.04.023
Sun, M., Jing, C., Chu, C., Wu, G., and Qing, L. (2017). Serological detection and molecular identification of tomato spotted wilt virus in pepper in Chongqing. Acta Hortic. Sin. 44, 487–494. doi: 10.16420/j.issn.0513-353x.2016-0733
Sun, S. E., Wang, J. Q., Chen, S., Zhang, S. B., Zhang, D. Y., and Liu, Y. (2018). First report of Capsicumchloros is orthotospovirus Infecting Zucchini (Cucurbitapepo) in China. Plant Dis. [Epub Online ahead of print]. doi: 10.1094/PDIS-12-17-1876-PDN
Torres, R., Larenas, J., Fribourg, C., and Romero, J. (2012). Pepper necrotic spot virus, a new tospovirus infecting solanaceous crops in Peru. Arch. Virol. 157, 609–615. doi: 10.1007/s00705-011-1217-3
Tripathi, D., Raikhy, G., and Pappu, H. R. (2015). Movement and nucleocapsid proteinscoded by two tospovirus species interact through multiple binding regions in mixed infections. Virology. 478, 137–147. doi: 10.1016/j.virol.2015.01.009
Wang, K., Zhan, B., and Zhou, X. (2019). Detection and partial biological characterization of Tomato spotted wilt tospovirus in Heilongjiang Province. Plant Prot. 45, 37–43. doi: 10.16688/j.zwbh.2018117
Wang, Y., Yang, Z., Wang, G., Wang, L., and Ni, H. (2016). First report of the tospovirus Tomato necrotic spot associated virus infecting kiwifruit (Actinidia sp.) in China. Plant Dis 100, 2539–2539. doi: 10.1094/PDIS-05-16-0629-PDN
Webster, C. G., Frantz, G., Reitz, S. R., Funderburk, J. E., and Mellinger, H. C. (2015). Emergence of Groundnut ringspot virus and Tomato chlorotic spot virus in vegetables in Florida and the southeastern United States. Phytopathology 105, 388–398. doi: 10.1094/PHYTO-06-14-0172-R
Wen, Z. D., Ding, Y., Shang, W. N., Wang, T. T., Wu, S. Z., Zhang, J., et al. (2020). Distribution characteristics of TSWV virions in vascular tissues of the systemic host tobacco K326. J. Southwest For. Univ. 40, 124–131. doi: 10.11929/j.swfu.202001020
Whitfield, A. E., Falk, B. W., and Rotenberg, D. (2015). Insect vector-mediated transmission of plant viruses. Virology 479, 278–289. doi: 10.1016/j.virol.2015.03.026
Wu, B., and Liu, Y. (2017). First report of Hippeastrum chlorotic ringspot virus infecting Zephyranthescandida in China. Plant Dis. 101:1960. doi: 10.1094/PDIS-12-16-1837-PDN
Wu, S. H., Xu, L. Q., Xian, W. R., Ren, J. H., Gao, D. N., Ji, Y., et al. (2020). Detection and Identification of Tomato spotted wilt tospovirus on pepper from Qinghai Province. Acta Hortic. Sin. 47, 1391–1400. doi: 10.16420/j.issn.0513-353x.2019-0920
Wu, X., Wu, X., Li, W., and Cheng, X. (2018). Molecular characterization of a divergent strain of calla lily Chlorotic spot virus infecting lettuce (Lactuca sativa var. augustana) in China. Arch. Virol. 163, 1375–1378. doi: 10.1007/s00705-018-3743-8
Xu, Y., Lou, S. G., Li, X. L., Zheng, Y. X., and Wang, W. C. (2013). The complete S RNA and M RNA nucleotide sequences of a Hippeastrum chlorotic ringspot virus (HCRV) isolate from Hymenocallislittoralis (Jacq.) Salisb. in China. Arch. Virol. 158, 2597–2601. doi: 10.1007/s00705-013-1756-x
Xu, Z. Y., Reddy, D. V. R., Rajeshwari, R., Zhang, Z. Y., and Huang, L. X. (1986). A new peanut virus disease caused by tomato spotted wilt virus in southern China. Chin. J. Virol. 03, 271–274.
Yao, G. (1992). Tomato spotted wilt virus (TSWV) found in Sichuan tobacco. Chin. J. Tob. 6, 39–40. doi: 10.13496/j.issn.1007-5119.1992.04.001
Yin, Y., Li, T., Xun, L., Qi, F., and Zhang, Z. K. (2015). First report of capsicum chlorosis virus infecting tomato in Yunnan, Southwest of China. Plant Dis. 100:230. doi: 10.1094/PDIS-01-15-0088-PDN
Yin, Y. Y., Fang, Q., Lu, X., Li, T. T., and Zhang, Z. K. (2014a). Detection of Watermelon silver mottle virus infecting watermelon in Yunnan, southwest of China. J. Plant. Pathol. 96:S4.123. doi: 10.1002/chin.199620258
Yin, Y. Y., Zheng, K. Y., Dong, J. H., Fang, Q., Wu, S., Wang, L. S., et al. (2014b). Identification of a new tospovirus causing necrotic ringspot on tomato in China. Virol. J. 11:213. doi: 10.1186/s12985-014-0213-0
Zarzynska-Nowak, A., Rymelska, N., Borodynko, N., and Hasiow-Jaroszewska, B. (2016). The occurrence of Tomato yellow ring virus on tomato in Poland. Plant Dis. 100:234. doi: 10.1094/PDIS-05-15-0521-PDN
Zhang, Q., Ding, Y. M., and Li, M. (2010). First report of Impatiens necrotic spot virus infecting Phalaenopsis and Dendrobium orchids in Yunnan Province. Chin. Plant Dis. 94:915. doi: 10.1094/PDIS-94-7-0915A
Zhang, W. H., and Feng, K. (2020). The infection of TSWV was first found in Shandong tobacco growing areas. Chin Tob. Sci. 41, 87–91. doi: 10.13496/j.issn.1007-5119.2020.05.011
Zhang, Y., Hou, M., and Li, C. (2017). Detection of main viral species of vegetable virus diseases in Hubei Province. J. Huazhong Agric. Univ. 36, 1–38. doi: 10.13300/j.cnki.hnlkxb.2017.06.005
Zhang, Y., Zhang, C., and Li, W. (2011). The nucleocapsid protein of an enveloped plant virus, tomato spotted wilt virus, facilitates long-distance movement of Tobacco mosaic virus hybrids. Virus Res. 163, 246–253. doi: 10.1016/j.virusres.2011.10.006
Zhang, Z. K. (2015). The Mechanism of Assembly and Movement of Tospoviruses Virion in Host Plant. Beijing: Graduate School of Chinese Academy of Agricultural Sciences.
Zhang, Z. K., Fang, Q., Wu, Z. Q., He, Y. K., Li, Y. H., and Huang, X. Q. (1998). The occurrence and distribution of main Solanacae crop viruses in Yunnan. J. Yunnan Univ. 20, 128–131.
Zhang, Z. K., He, Y. F., Fang, Q., Cheng, X. F., Dong, J. H., Yin, Y. Y., et al. (2010). Cytopathological characterization of three distinct tospovirus species in infected host plants. Southwest Chin. J. Agric. Sci. 23, 1522–1524. doi: 10.3724/SP.J.1142.2010.40491
Zhang, Z. K., Ma, X. Y., Wu, K., Zheng, K. Y., Pei, W. H., Chen, Y. D., et al. (2020). Infection cycle and epidemic characteristics of spotted wilt diseases on genera Lactuca vegetables in Luliang county, Yunnan province, southwest China. J. Agric. Sci. 33, 2827–2832.
Zhang, Z. K., Zheng, K. Y., Dong, J. H., Hong, J., and Wang, X. F. (2016). Clustering and cellular distribution characteristics of virus particles of Tomato spotted wilt virus and Tomato zonate spot virus in different plant hosts. Virol. J. 13:11.
Zhao, J., Liu, Y., Li, Y., Dong, L., Xie, J., Wang, L., et al. (2021). Sales status and development countermeasures of vegetable products in yunnan province in 2019. Yangtze River Vegetables 4, 72–76. doi: 10.3865/j.issn.1001-3547.2021.04.023
Zhao, L. H., Dong, J. H., Su, X. X., Li, S. L., Zhang, J., Xu, T., et al. (2017). Inhibition effects of sesquiterpenoids from Tithoniadiversifolia on Tomato spotted wilt virus. Tob. Sci. Technol. 50, 21–25. doi: 10.16135/j.issn1002-0861.2016.0553
Zhao, L. H., Hu, Z. H., Li, S. L., Zhang, L. Z., Yu, P., Zhang, J., et al. (2020). Tagitinin A from Tithoniadiversifolia provides resistance to Tomato spotted wilt orthotospovirus by inducing systemic resistance. Pestic. Biochem. Phys. 169:104654. doi: 10.1016/j.pestbp.2020.104654
Zhao, L. H., Hu, Z. H., Li, S. L., Zhou, X. P., Li, J. P., Su, X. X., et al. (2019). Diterpenoid compounds from Wedeliatrilobata induce resistance to Tomato spotted wilt virus via the JA signal pathway in tobacco plants. Sci. Rep. 9:2763. doi: 10.1038/s41598-019-39247-6
Zheng, K., Chen, T. C., Yeh, S. D., Rahman, M. S., Su, X., Wu, K., et al. (2017). Characterization of a new isolate of pepper chlorotic spot virus from Yunnan province, China. Arch. Virol. 162, 2809–2814. doi: 10.1007/s00705-017-3402-5
Zheng, K. Y., Chen, T. C., Wu, K., Kang, Y. C., Yeh, S. D., Zhang, Z. K., et al. (2020). Characterization of a new orthotospovirus from chilli pepper in Yunnan Province. Chin. Plant Dis. 104, 1175–1182. doi: 10.1094/PDIS-09-19-1925-RE
Zheng, K. Y., Dong, J. H., Fang, Q., Li, T. T., Yin, Y. Y., and Zhang, Z. K. (2015a). Detection of Tomato zonate spot virus by immunogold labeling electron microscopy. J. Chin. Electron. Microsc. Soc. 34, 67–70.
Zheng, K. Y., Liu, H. Y., Yin, Y. Y., Chen, T. C., Yeh, S. D., Zhang, Z. K., et al. (2016). Full-length M and L RNA sequences of tospovirus isolate 2009-GZT, which causes necrotic ringspot on tomato in China. Arch. Virol. 161, 1411–1414. doi: 10.1007/s00705-016-2788-9
Zheng, K. Y., Wu, K., Dong, J. H., Fang, Q., and Zhang, Z. K. (2015b). Infection and damage of Tomato spotted wilt virus on lettuce vegetables in Yunnan. Plant Prot. 41, 174–178. doi: 10.3969/j.issn.0529-1542.2015.05.033
Zheng, X., Zhang, J., Chen, Y. D., Wu, K., Dong, J. H., and Zhang, Z. K. (2015c). Occurrence dynamics and control status of Tospoviruses in vegetables of Yunnan in 2014. Shandong Agric. Sci. 47, 83–87. doi: 10.14083/j.issn.1001-4942.2015.10.021
Zheng, Y. X., Chen, C. C., Yang, C. J., Yeh, S. D., and Jan, F. J. (2008). Identification and characterization of a tospovirus causing chloroticringspots on Phalaenopsis orchids. Eur. J. Plant Pathol. 120, 199–209. doi: 10.1007/s10658-007-9208-7
Zhou, J., Kantartzi, S. K., Wen, R. H., Newman, M., Hajimorad, M. R., Rupe, J. C., et al. (2011). Molecular characterization of a new tospovirus infecting soybean. Virus Genes 43, 289–295. doi: 10.1007/s11262-011-0621-9
Keywords: Orthotospovirus, species diversity, systemic infection, multiple transmission routes, monitoring and early warning
Citation: Zhang Z, Zheng K, Zhao L, Su X, Zheng X and Wang T (2021) Occurrence, Distribution, Evolutionary Relationships, Epidemiology, and Management of Orthotospoviruses in China. Front. Microbiol. 12:686025. doi: 10.3389/fmicb.2021.686025
Received: 26 March 2021; Accepted: 25 June 2021;
Published: 04 August 2021.
Edited by:
Xiaofei Cheng, Northeast Agricultural University, ChinaReviewed by:
Xiaorong Tao, Nanjing Agricultural University, ChinaCopyright © 2021 Zhang, Zheng, Zhao, Su, Zheng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongkai Zhang, emhvbmdrYWk5OUBzaW5hLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.