
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 June 2021
Sec. Infectious Agents and Disease
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.685343
 Chuan Chiang-Ni1,2,3,4*
Chuan Chiang-Ni1,2,3,4* Yen-Shan Liu2
Yen-Shan Liu2 Chieh-Yu Lin2
Chieh-Yu Lin2 Chih-Yun Hsu1
Chih-Yun Hsu1 Yong-An Shi2
Yong-An Shi2 Yi-Ywan M. Chen1,2,3
Yi-Ywan M. Chen1,2,3 Chih-Ho Lai1,2,3
Chih-Ho Lai1,2,3 Cheng-Hsun Chiu2,3,5
Cheng-Hsun Chiu2,3,5The acquisition of the phage-encoded superantigen ssa by scarlet fever-associated group A Streptococcus (Streptococcus pyogenes, GAS) is found in North Asia. Nonetheless, the impact of acquiring ssa by GAS in invasive infections is unclear. This study initially analyzed the prevalence of ssa+ GAS among isolates from sterile tissues and blood. Among 220 isolates in northern Taiwan, the prevalence of ssa+ isolates increased from 1.5% in 2008–2010 to 40% in 2017–2019. Spontaneous mutations in covR/covS, which result in the functional loss of capacity to phosphorylate CovR, are frequently recovered from GAS invasive infection cases. Consistent with this, Phostag western blot results indicated that among the invasive infection isolates studied, 10% of the ssa+ isolates lacked detectable phosphorylated CovR. Transcription of ssa is upregulated in the covS mutant. Furthermore, in emm1 and emm12 covS mutants, ssa deletion significantly reduced their capacity to grow in human whole blood. Finally, this study showed that the ssa gene could be transferred from emm12-type isolates to the emm1-type wild-type strain and covS mutants through phage infection and lysogenic conversion. As the prevalence of ssa+ isolates increased significantly, the role of streptococcal superantigen in GAS pathogenesis, particularly in invasive covR/covS mutants, should be further analyzed.
Streptococcus pyogenes [group A Streptococcus (GAS)] is a gram-positive bacterium that causes diseases like pharyngitis, pyoderma, scarlet fever, necrotizing fasciitis, and toxic shock syndrome. In 2011, a scarlet fever outbreak was reported in Hong Kong (Hsieh and Huang, 2011; Tse et al., 2012). The scarlet fever isolates typically harbor phage-associated superantigen genes ssa and speC, and the DNase gene spd1 (Tse et al., 2012; Ben Zakour et al., 2015). Luk et al. (2012) found insufficient evidence to support the association between increased scarlet fever incidence or severity and a particular emm type, virulence gene profile, or the presence of specific foreign genetic elements. Nonetheless, Davies et al. (2015) showed that ssa is absent in the clade not associated with scarlet fever, but variably present (95% isolates) in scarlet fever-associated clades, suggesting that ssa acquisition could be potentially related to the expansion of scarlet fever-associated emm12-type clones within the Hong Kong population.
An recent increased incidence of scarlet fever has also been reported in South Korea, Singapore, England, and Germany (Turner et al., 2016; Park et al., 2017; Brockmann et al., 2018; Kim and Cheong, 2018; Lamagni et al., 2018; Walker and Brouwer, 2018; You et al., 2018; Yung and Thoon, 2018). Park et al. (2017) showed that emm4, emm28, emm1, and emm3 contributed to scarlet fever in South Korea. Notably, antibiotic resistance was uncommon in these scarlet fever-associated isolates (Park et al., 2017). Interestingly in England, the most prevalent isolates recovered during a national increase in scarlet fever incidence in 2016 were an emergent clone of the emm1 M1T1 genetic lineage, designated M1T1UK, which was characterized by increased production of the phage-encoded SpeA superantigen (Lynskey et al., 2019). This contrasts with the results in Hong Kong and Beijing where during an increased incidence of scarlet fever cases in 2011. The most prevalent isolates recovered were polyclonal emm12 strains and the vast majority of which were characterized by the presence of the phage-encoded streptococcal superantigen (SSA; Davies et al., 2015). These findings show that emm type, antibiotic-resistant phenotype, and superantigen gene in scarlet fever-associated isolates vary from country to country; therefore, the specific factors leading to scarlet fever resurgence are not completely understood.
Streptococcal superantigen is a 260-residue protein with 60% sequence identify to that of staphylococcal enterotoxin B (SEB; Reda et al., 1994). Brouwer et al. (2020) showed that glutathione from streptolysin O (SLO)-lysed host cells not only enhances SSA production but also activates its superantigen activity. ssa has been detected in the toxic shock syndrome related M3 isolates (Mollick et al., 1993), suggesting it to be a potential virulence factor of GAS. As the prevalence of ssa+ GAS isolates was >70% in certain geographic areas (Li et al., 2020a), the effects of ssa acquisition or ssa+ isolate dissemination on GAS diseases need to be further clarified.
Previous studies have reported that isolates with spontaneous mutations in the covR/covS operon are highly related to severe diseases such as necrotizing fasciitis and toxic shock syndrome (Ikebe et al., 2010; Friaes et al., 2015). CovR/CovS is a two-component regulatory system in GAS, and intracellular CovR is phosphorylated by the sensor kinase CovS (Trevino et al., 2009; Tran-Winkler et al., 2011). Mutations in the covR/covS operon increase the production of virulence factors such as hyaluronic acid capsule, M protein, SLO, and streptokinase (Tran-Winkler et al., 2011; Chiang-Ni et al., 2017; Horstmann et al., 2018). Furthermore, CovR/CovS also regulates the expression of phage-related genes such as DNase sda1 (Walker et al., 2007). Increased Sda1 expression by a covS mutant promoted bacterial escape from immune clearance by degrading the DNA structure of the neutrophil extracellular traps (Walker et al., 2007).
In this study, we found that four ssa+ isolates lacked phosphorylated CovR, and ssa transcription was upregulated in these isolates. In line with the repression in ssa and slo transcription by the CovR/CovS system, ssa deletion in the emm1 and emm12 covS mutants attenuated bacterial growth activity in human blood, suggesting that ssa acquisition by covS mutants or spontaneous mutations in the covR/covS operon of ssa+ isolates could enhance bacterial survival during infection. As the prevalence of ssa+ isolates dramatically increased in northern Taiwan, the impact of the dissemination of ssa+ isolates on our society should be continuously monitored and studied.
Group A Streptococcus isolates from sterile tissues [ascites (2), blood (199), deep tissue (14), pleural fluid (3), and synovial fluid (2)] collected during 2008–2019 (220 isolates) at the LinKou Chang Gung Memorial Hospital (Taiwan) were included in this study. The emm1 wild-type A20 strain, its covR mutant, covS mutant AP3, and the CovS kinase-inactivated (CovSH280A) mutant were described previously (Chiang-Ni et al., 2016, 2017). GAS strains were cultured on trypticase soy agar with 5% sheep blood or in tryptic soy broth (Becton, Dickinson and Company; Sparks, MD, United States) supplemented with 0.5% yeast extract (TSBY). Escherichia coli DH5α was purchased from Yeastern Biotech Co., LTD. (Taipei, Taiwan) and was cultured in Luria-Bertani (LB; Becton, Dickinson and Company; Sparks, MD, United States) broth at 37°C with vigorous aeration. When appropriate, chloramphenicol (25 μg/ml and 3 μg/ml for E. coli and GAS, respectively) and spectinomycin (100 μg/ml) were used for selection. This study was approved by the Institutional Review Board (201900274B0 and 202000479B0) of Chang Gung Memorial Hospital, Taiwan.
The susceptibility of GAS isolates to erythromycin and clindamycin was determined using a disk diffusion assay according to the Clinical and Laboratory Standards Institute Guideline (Clinical and Laboratory Standards Institute, 2018). Inducible clindamycin resistance in GAS isolates was determined using the D test with erythromycin and clindamycin disks. A flattened inhibition zone around the clindamycin disk proximal to the erythromycin disk was considered a positive result.
Genomic DNA was extracted using a previously described method (Chiang-Ni et al., 2019a), and emm typing, PCR amplification, and DNA sequencing were performed according to the protocol from the Centers for Disease Control and Prevention.1 The ssa and erythromycin resistance genes (mefA, ermB, and ermTR) were screened for by PCR amplification using previously described primers (Table 1; Meisal et al., 2010; Villasenor-Sierra et al., 2012).
Group A Streptococcus chromosomal DNA was digested using HindIII, and the DNA fragments were resolved on 0.9% agarose gel. The PCR product of the chloramphenicol cassette (716 bp; Table 1) was labeled with alkaline phosphatase as the probe, and DNA hybridization was performed according to the manufacturer’s instructions (AlkPhos Direct Labeling and Detection System; GE Healthcare UK Limited; Amersham, United Kingdom). The signal was detected using a Gel Doc XR+ system (Bio-Rad; Hercules, CA, United States).
ssa and flanking upstream and downstream regions (1805 bp) were amplified using primers ssa-BamHI-F and ssa-BamHI-R (Table 1), and the PCR product was ligated into a T-A cloning vector (Yeastern Biotech). ssa was removed by PCR with reverse primers ssa-SacII-F and ssa-SacII-R (Table 1) and replaced by the chloramphenicol cassette from Vector78 (Tsou et al., 2010) at the SacII site. The ssa knockout DNA fragment was sub-cloned into the temperature-sensitive vector pCN143 (Chiang-Ni et al., 2016) at the BamHI site (designed as pCN211). The plasmids pCN211 and pCN160 (Chiang-Ni et al., 2019a) were transformed into emm12 SPY128 and emm1 SPY131, respectively, by electroporation for allelic exchange, and the transformants were selected using 3 μg/ml chloramphenicol as described previously (Chiang-Ni et al., 2019b). ssa and covS deletion in the selected transformants was confirmed by Sanger sequencing. To construct covS and ssa double mutants, pCN160 was transformed into ssa mutants. covS deletion in these ssa mutants was selected according to the encapsulated phenotype and confirmed by Sanger sequencing.
RNA extraction and reverse transcription were performed as previously described (Chiang-Ni et al., 2009). The bacterial strains were cultured for 6 h (exponential phase) and 8 h (stationary phase), and total RNA was extracted for analysis. Quantitative PCR was performed in a 20 μl mixture containing 1 μl cDNA, 0.8 μl primer (10 μM), and 10 μl Sensifast Lo-ROX premixture (Bioline, Ltd.; London, United Kingdom) according to the manufacturer’s instructions. Three biological replicates were performed, and ssa expression level was normalized to that of gyrA and analyzed using the threshold cycle (∆∆CT) method (Roche LightCycler® 96 System; Roche Molecular Systems, Inc.; Pleasanton, CA, United States). All values from the control and experimental groups were divided by the mean value of the control samples before statistical analysis (Valcu and Valcu, 2011). Primers used to detect ssa (Table 1) and gyrA were designed using Primer3 (v.0.4.0) according to the HKU360 sequence (NCBI accession no. CP003901.1).
Phostag western blotting was performed as previously described (Chiang-Ni et al., 2019a). Briefly, 10 μg bacterial total protein was mixed with 6× loading dye (without boiling) and separated using 10% SDS-PAGE containing 10 μM Phostag (Wako Pure Chemical Industries, Ltd.; Richmond, VA, United States) and 0.5 M MnCl2 for 120–140 min at 100 V at 4°C. For detecting SLO, 30 μl bacterial culture supernatants were mixed with 6× protein loading dye and separated by 12% SDS-PAGE. The separated proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore; Billerica, CA, United States), which was blocked with 5% skim milk PBST buffer (PBS containing 0.2% Tween 20) at 37°C for 1 h. CovR was detected by the anti-CovR serum (Chiang-Ni et al., 2016), and SLO was detected by the anti-SLO antibody (GeneTex; Irvine, CA, United States). The phosphorylated and nonphosphorylated CovR and SLO were visualized using a previously described method (Chiang-Ni et al., 2020).
The growth of GAS strains in human whole blood was studied according to a previous study with modifications (Brouwer et al., 2020). Freshly drawn heparinized venous blood from healthy adults was aliquoted (360 μl) into wells of a 24-well plate. GAS strains were grown to the exponential phase (OD600 = 0.6) in TSBY, washed, resuspended in 1× PBS buffer at ~5 × 105 colony forming unit (CFU)/ml, and added to whole blood in a final volume of 400 μl (~5 × 104 CFU/ml). After incubating for 1.5 h at 37°C, the growth of GAS strains was analyzed by plating serial dilutions on TSBY plates. Experiments were performed with blood from three different donors.
Mitomycin C (0.2 μg/ml; Sigma-Aldrich; St. Louis, MO, United States) was added to the culture of donor strain grown to the early exponential phase and incubated at 37°C for another 4 h. Culture supernatants were passed through a 0.45 μm filter (Millipore Ireland Ltd., Co.; Cork, Ireland) to remove bacteria and large fragments, and phage particles were collected from the filtrate by ultra-centrifugation at 112,000 × g for 2 h at 10°C. The recipients were grown to the logarithmic phase in the presence of 5 mM CaCl2 and then co-incubated with the phage particles at 37°C for 3 h. The bacterial suspension was plated on agar plates supplemented with chloramphenicol (3 μg/ml) and resistant convertants were collected for Southern blotting analysis.
Statistical analysis was performed using Prism software, version 5 (GraphPad; San Diego, CA, United States). Significant differences between multiple groups were determined using the ANOVA. Post-test for ANOVA was analyzed using Tukey’s honestly significant difference test. Statistical significance was set at p < 0.05.
A total of 34 different emm types were identified among 220 isolates, among which emm1 (37/220; 16.8%), emm12 (32/220; 14.5%), emm113 (20/220; 9.1%), emm102 (16/220; 7.3%), emm11 (14/220; 6.4%), and emm90 (14/220; 6.4%) accounted for 60.5% total isolates. Phage-encoded ssa was detected by PCR. One isolate was ssa+ in 2008–2010 (1/65, Figure 1A); however, ssa+ isolate prevalence increased to 6.5% (3/46) in 2011–2013, 24.5% (12/49) in 2014–2016, and 40% (24/60) in 2017–2019 (Figure 1B). The most prevalent emm types of ssa+ isolates were emm12 (18/40; 45%) and emm1 (12/40; 30%). Although emm1 and emm12 isolate prevalence did not dramatically increase during 2008–2019 (Figure 1B), that of ssa+ emm1 isolates increased to 58.3% (7/12) in 2017–2019 compared to 6.3% (1/16) in 2008–2010 and that of ssa+ emm12 isolates increased to 100% (9/9) in 2017–2019 compared to 0% (0/5) in 2008–2010 (Figure 1C).
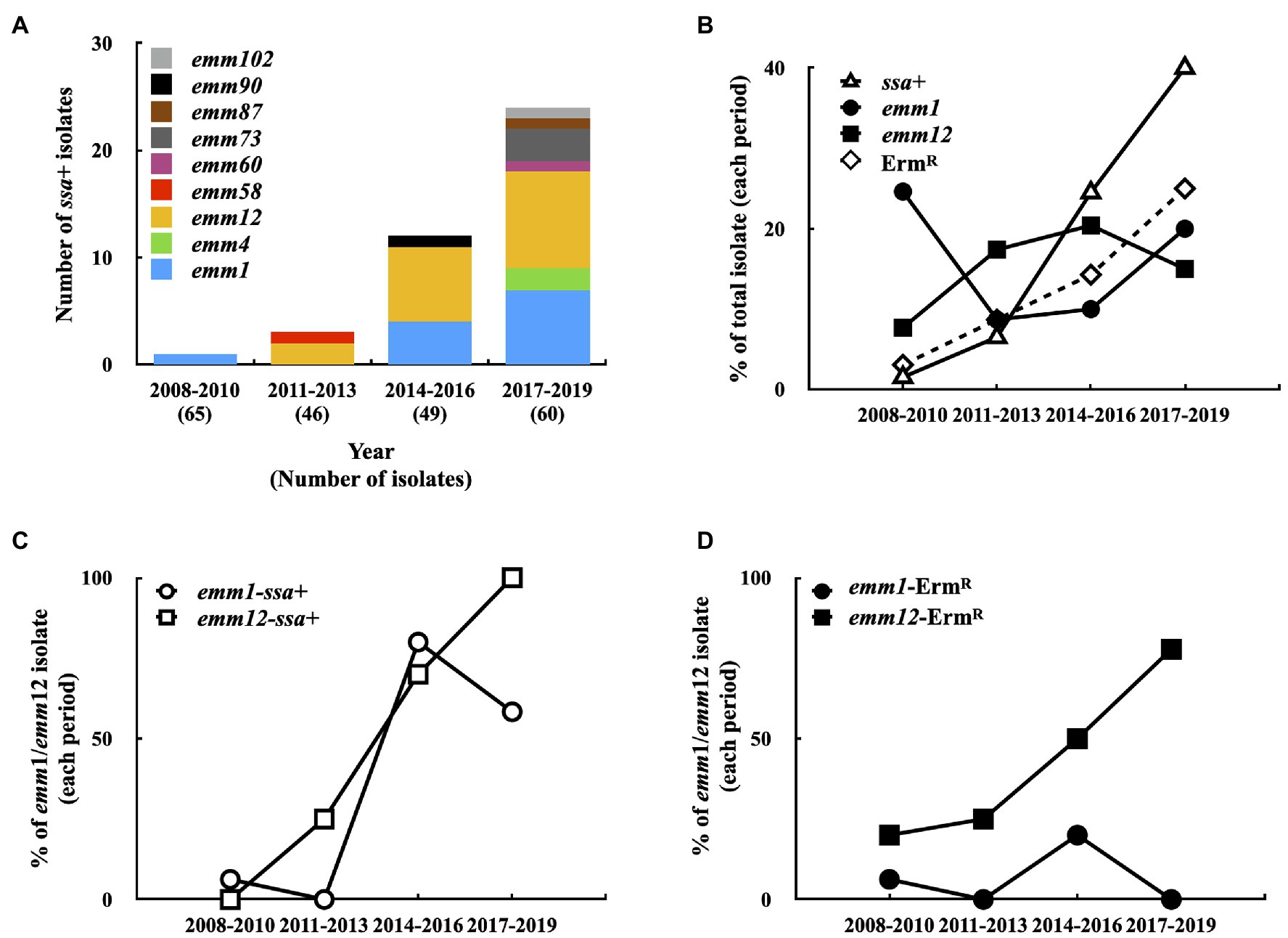
Figure 1. Prevalence of emm type, ssa+, and erythromycin-resistant (ErmR) isolates in 2008–2019. (A) The emm type of ssa+ isolates in 2008–2019. (B) Prevalence of emm1-type, emm12-type, ssa+, and ErmR isolates in 2008–2019. (C) Prevalence of emm1 and emm12 ssa+ isolates in 2008–2019. (D) Prevalence of emm1 and emm12 ErmR isolates in 2008–2019.
Among the 220 isolates, 28 (12.7%) were erythromycin-resistant. The prevalence of erythromycin-resistant isolates gradually increased from 3.1% (2/65) in 2008–2010 to 8.7% (4/46) in 2011–2013, 14.3% (7/49) in 2014–2016, and 25% (15/60) in 2017–2019 (Figure 1B). PCR analysis showed that mefA was detected in one erythromycin-resistant isolate (emm12 type), and the remaining 27 erythromycin-resistant isolates were either ermB+ (22 isolates) or ermTR+ (5 isolates), indicating that these isolates were also clindamycin-resistant (Supplementary Table S1). In addition, 53.6% (15/28) of erythromycin-resistant isolates were type emm12; the prevalence of erythromycin-resistant emm12 isolates increased from 20% (1/5) in 2008–2010 to 77.8% (7/9) in 2017–2019 (Figure 1D). Moreover, 77.8% (14/18) ssa+ emm12 isolates were erythromycin-resistant and 92.9% (13/14) ssa− emm12 isolates were erythromycin-susceptible. Although the prevalence of the ssa+ emm1 isolates was increased (Figure 1C), only two isolates (total 37 isolates) were erythromycin-resistant. These results indicate that the prevalence of ssa+ isolates, particularly emm1 and emm12 isolates, increased. Most of the erythromycin-resistant emm12 isolates were ssa+ (14/15); however, 94.6% (35/37) ssa+ emm1 isolates were erythromycin-susceptible.
Brouwer et al. (2020) showed that SSA is a thiol-activated superantigen, and its release and activity are promoted by the pore-forming toxin SLO. Moreover, the isolates from patients with invasive infection obtained spontaneous mutations in the covR/covS operon more frequently than those from patients with pharyngeal/tonsil infection (Ikebe et al., 2010; Friaes et al., 2015). Spontaneous inactivating mutations in covR/covS cause loss of CovR phosphorylation resulting in derepression of multiple secreted virulence factors including SLO (Sumby et al., 2006; Chiang-Ni et al., 2019b). In this study, after analyzing CovR phosphorylation level in 220 isolates using Phostag western blot assay, phosphorylated CovR was absent in 29 isolates (data not shown; Supplementary Table S1). Among the 40 ssa+ isolates, we identified four isolates that did not express the phosphorylated CovR protein (Figure 2A). Also, these four isolates expressed higher levels of SLO than those with the phosphorylated CovR protein (Figure 2B).
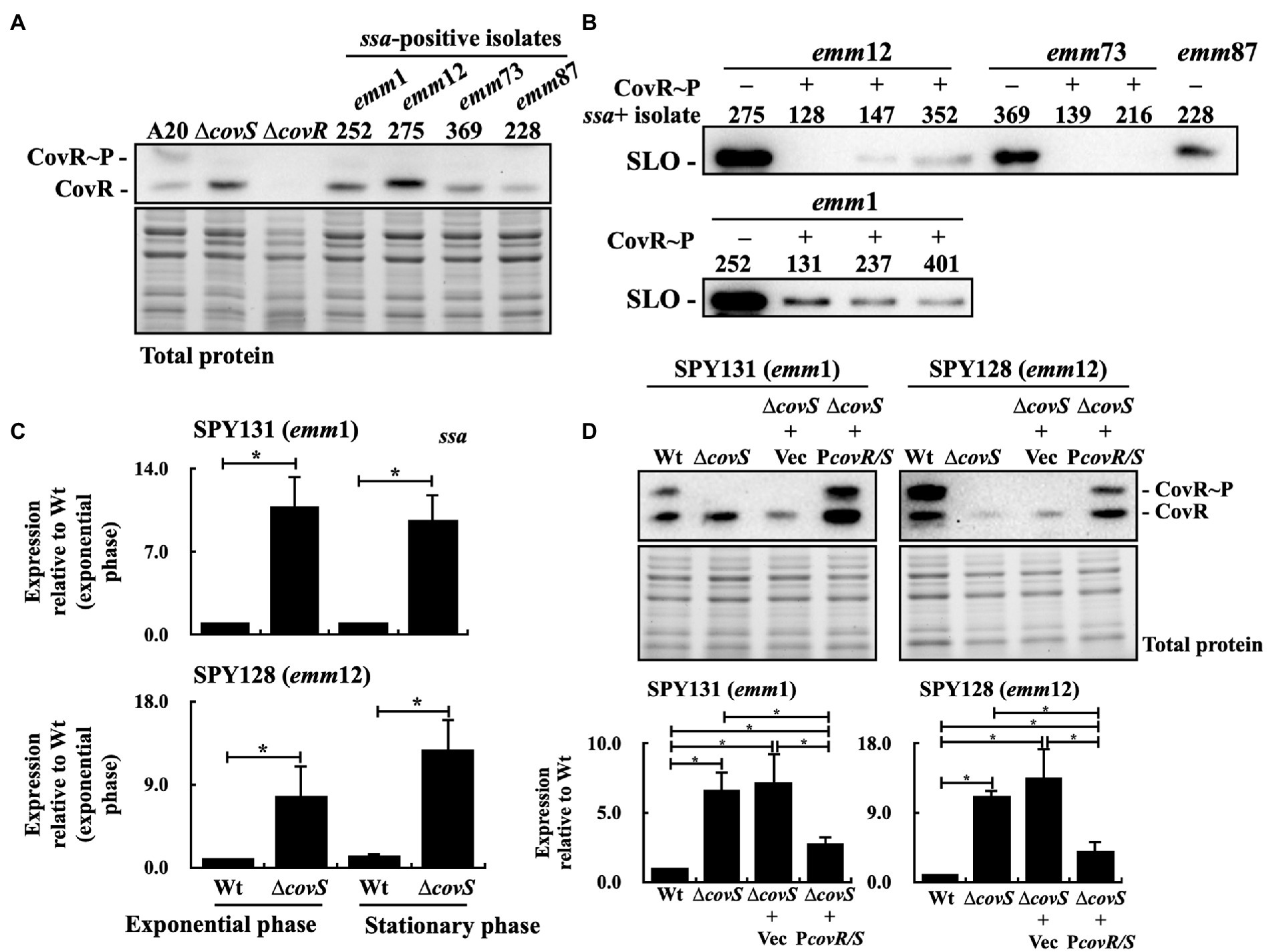
Figure 2. Phosphorylated CovR and streptolysin O (SLO) expression and ssa transcription in selected clinical isolates, covS isogenic mutants, vector-control strains, and covR/covS trans-complementary strains. (A) Detection of the phosphorylated CovR in the selected clinical isolates using Phostag western blot assay. A20 and its covS (∆covS) and covR (∆covR) mutants were utilized as experimental controls. (B) SLO secretion in selected ssa-positive isolates. SLO was detected in the culture supernatants using western blot analysis. (C) ssa transcription in the SPY131 (emm1) and SPY128 (emm12; Wt) and their covS mutants (∆covS). (D) Phosphorylated CovR expression and ssa transcription in vector-control (∆covS+Vec) and the covR/covS trans-complementary (∆covS+PcovR/S) strains. RNA was extracted for reverse transcription-PCR (RT-qPCR) analysis. *p < 0.05. CovR~P, phosphorylated CovR; CovR, nonphosphorylated CovR. Total protein is served as the internal loading control.
To investigate the effect of CovR phosphorylation inactivation on the ssa expression, isogenic covS mutants of emm1-type (SPY131) and emm12-type (SPY128) isolates were constructed, and ssa transcription was analyzed by quantitative PCR. The results showed that ssa expression in the covS mutants was 7–12-fold higher than that in the parental strains (Figure 2C). To further verify that ssa expression is repressed by phosphorylated CovR, the covR/covS trans-complementary strains of the covS mutants were constructed, and the phosphorylated CovR and ssa transcription in the covR/covS trans-complementary strains were analyzed. Phostag western blot analysis showed that phosphorylated CovR was detected in the trans-complementary strains, but not in the vector-control strains and covS mutants (Figure 2D). In addition, ssa transcription was repressed in the covR/covS trans-complementary strains compared to that in the covS mutants (Figure 2D). These results indicate that ssa transcription is repressed by phosphorylated CovR.
Brouwer et al. (2020) showed that the wild-type strain and ssa mutant had similar growth in human whole blood. ssa transcription and SLO production were upregulated in the covS mutants compared to the wild-type strains (Figures 2B,C); therefore, in this study, the role of SSA in covS mutant survival in human whole blood was further investigated. The ssa and covS double mutants of emm1-type SPY131 and emm12-type SPY128 were constructed. The growth of the wild-type strains, covS, and ssa mutants in the culture broth were similar (data not shown). covS mutants were more resistant to phagocytic killing (Sumby et al., 2006); in agreement with this, we observed that the covS mutants of SPY131 and SPY128 had better growth in human blood than the wild-type strains and ssa mutants (Figure 3). Furthermore, the results showed that ssa deletion in the covS mutants significantly attenuated bacterial growth in blood compared to the covS mutants (Figure 3), suggesting that SSA contributes to the survival of covS mutants in human whole blood.
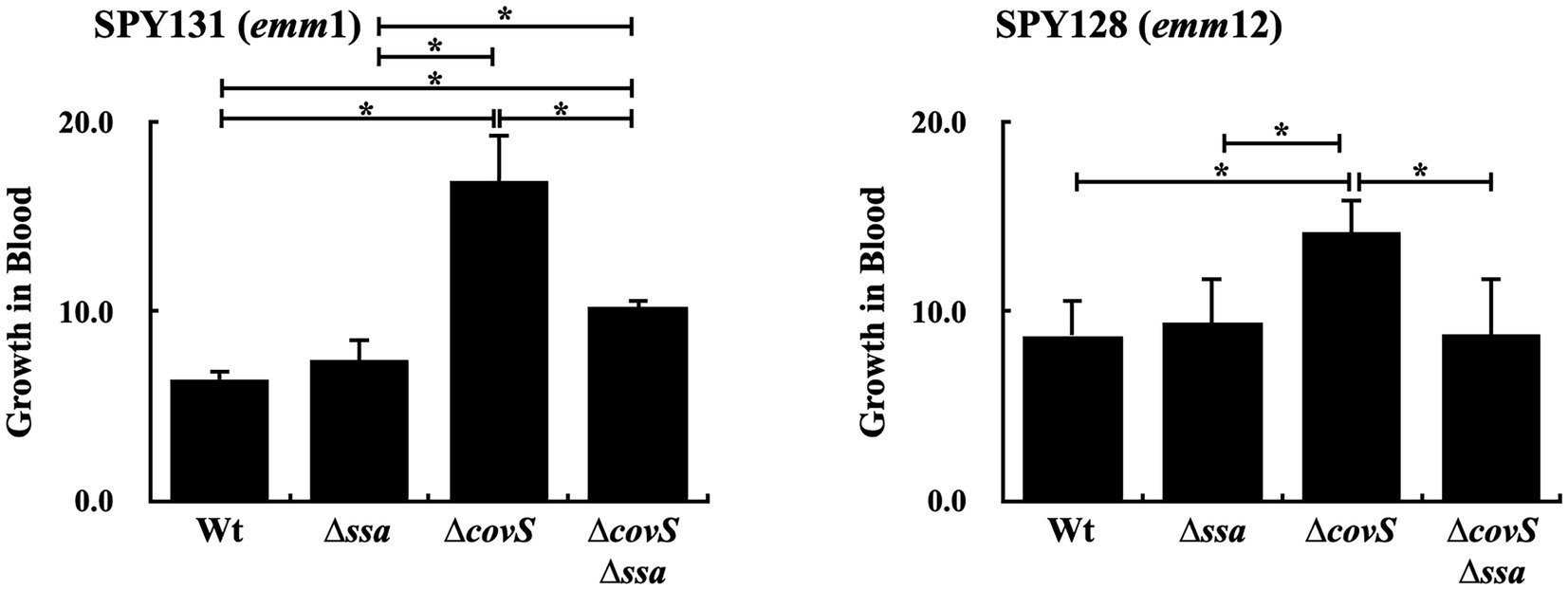
Figure 3. Growth activity of SPY131 (emm1), SPY128 (emm12), their ssa mutants (∆ssa), covS mutants (∆covS), and covS and ssa double mutants (∆covS∆ssa) in human whole blood. Group A Streptococcus (GAS) strains were incubated with whole blood from donors for 1.5 h. The number of surviving bacteria in human blood was determined by plating and enumerating the colony forming units (CFUs), which determined growth relative to the initial inoculum. *p < 0.05.
The ssa+ isolates that did not express the phosphorylated CovR protein were identified (Figure 2A). These isolates could acquire inactivating spontaneous mutations in covR/covS during infection. Nonetheless, the ssa gene is carried by phage and the possibility of covR/covS mutants directly acquired ssa by phage infection could not be excluded. Therefore, whether covS mutants could acquire ssa through phage infection and lysogenic conversion was further investigated. To select the lysogenic convertants after phage infection, ssa was replaced with the chloramphenicol (cm) cassette in the emm12-type SPY128 (SCN279), which was utilized as the ssa-encoding phage donor. After mitomycin C induction, the cm cassette, but not chromosomal speB, was detected in the filtered supernatant from SCN279 (Figure 4A), indicating that the target phage was released from SCN279. Next, the recipients, including the emm1-type A20 strain, its covS mutant AP3, and the CovS kinase-inactivated CovSH280A mutant (Chiang-Ni et al., 2016, 2019b), were incubated with the collected phage particles. We found that lysogenic convertants can be obtained. The cm cassette in the lysogenic convertants was integrated to the same insertion site of the chromosome and located on the same phage in donor strain SCN279 (Figure 4B). These results indicate that not only emm1 wild-type strain but also its covS mutants could acquire ssa by lysogenic conversion.
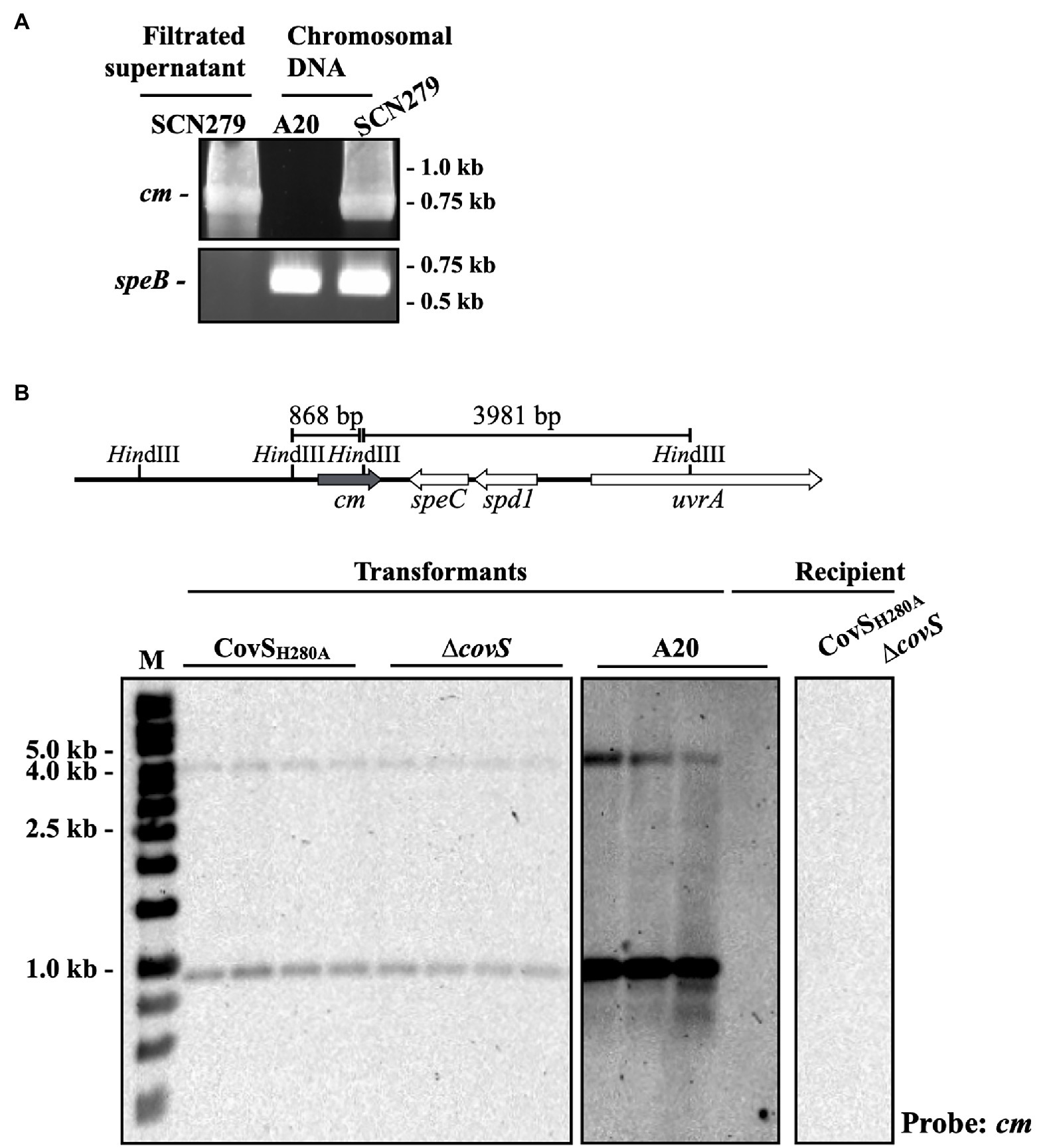
Figure 4. PCR and Southern blot analyses of the ssa mutant and lysogenic convertants. (A) Detection of the chloramphenicol cassette (cm) and the chromosomal speB gene in the chromosomal DNA and filtrated supernatant after mitomycin C treatment using PCR. SCN279, the phage donor. A20 (emm1-type), the recipient. (B) Southern blot hybridization of lysogenic convertants. The upper panel shows the HindIII site and the predicted sizes for cm probe hybridization according to ΦHKU.vir (HKU360, NCBI accession no. CP009612) and A20 (NCBI accession no. CP003901.1) sequence. The recipient strains (A20, CovSH280A, and ∆covS) showed no signal for cm probe hybridization and used as the experimental negative control. M, DNA marker.
This study showed that the prevalence of ssa+ GAS isolates increased from 1.5% (1/65) in 2008–2010 to 40% (24/60) in 2017–2019 in northern Taiwan. A total of 29 isolates could not produce phosphorylated CovR protein, and four of these isolates were ssa+. We found that ssa transcription is repressed by phosphorylated CovR. Moreover, ssa gene deletion attenuated the growth activity of the covS mutants in human blood, suggesting that the acquisition of ssa by covR/covS mutants or spontaneous mutations in the covR/covS operon in the ssa+ isolates could be related to the increase in bacterial survival during infection.
In this study, 10% ssa+ isolates did not produce phosphorylated CovR under the conditions tested. Spontaneous inactivating mutations in the covR/covS operon increases SLO expression in GAS (Sumby et al., 2006; Chiang-Ni et al., 2019a). In addition, some clinical isolates with spontaneous mutations or a truncated allele of rocA (regulator of cov; upstream regulator of CovR/CovS) also increased the SLO expression (Feng et al., 2017; Jain et al., 2017; Horstmann et al., 2018; Chiang-Ni et al., 2020). SSA is a thio-activated superantigen, and the GAS secreted SLO, which triggers glutathione release from host cells to activate SSA in vivo (Brouwer et al., 2020). Brouwer et al. (2020) showed that ssa deletion did not attenuate bacterial survival in human blood; however, our results showed that the covS and ssa double mutants had significantly decreased growth in human blood compared to the covS mutants (Figure 3). As expected (Sumby et al., 2006), the encapsulated covS mutant was resistant to phagocytosis (Figure 3), thus not cleared despite high expression (promoted by SLO) of the superantigen SSA, which might activate T cells and phagocytic cells. Okamoto et al. (2001) showed that macrophages in SEB-pretreated mice were less phagocytic than those in non-pretreated mice. SSA sequence is 60% identity to SEB. Based on these observations, although covS mutant and covS and ssa double mutant were both encapsulated, the covS and ssa double mutant might encounter higher pressure from macrophages than that of covS mutant. Nonetheless, these mutants were co-cultured with human whole blood for 1.5 h; whether SSA could act to macrophages like SEB during this short incubation period is not clear. Therefore, the role of SSA in GAS survival in human blood needs to be further elucidated. Whether the ssa transcription is directly regulated by phosphorylated CovR is not clear; however, our results suggest that SSA could potentially contribute to the pathogenesis of invasive covR/covS mutants.
Lynskey et al. (2019) showed that the toxigenic M1T1 clone (M1T1UK) is related to the increased incidence of invasive GAS disease in the United Kingdom. Thereafter, the M1T1UK strain was identified in Canada, the Netherlands, and the United States (Demczuk et al., 2019; Li et al., 2020b; Rumke et al., 2020), demonstrating the possibility of clonal expansion of the M1T1UK clone. In Taiwan, Yan et al. (2000) showed that 64.3% erythromycin-resistant isolates were clindamycin-susceptible and harbored mefA before 1998. From 2001 to 2010, the erythromycin resistance rate decreased from 53.1 to 0% in southern Taiwan (Chuang et al., 2015). In this study, we found that the erythromycin resistance rate in GAS increased from 3.1% in 2008–2010 to 25% in 2017–2019. Notably, only one isolate harbored mefA (1/28). The changes in phenotype and genotype of erythromycin-resistant GAS suggested that the expansion of erythromycin-resistant (ermB and ermTR) and ssa+ emm12 clones could occur in Taiwan. In the phage infection assay, ssa was replaced with a chloramphenicol cassette as the selection marker, and an identical genetic element was found in the emm12-type donor and emm1-type recipient strains using Southern blot (Figure 4B and data not shown), suggesting that ssa could be transferred by phage. Furthermore, we also found that the chloramphenicol cassette could be transferred between emm12-type and emm73-type isolates (data not shown). These results suggest that clonal expansion and phage infection could be involved in the increased prevalence of ssa+ isolates in Taiwan. The emerged clones in the United States (M1T1 clone) and in Asia (Hong Kong and Taiwan, emm12 clone) are different, but both possess superantigen genes such as speA and ssa. Nonetheless, the exact role of superantigens in the survival fitness of GAS during infection needs to be addressed.
In summary, this study showed an increased prevalence of ssa+ and erythromycin-resistant GAS isolates in northern Taiwan, and demonstrated that ssa acquisition by invasive covS mutants or spontaneous mutations in the covR/covS operon of ssa-positive isolates could enhance bacterial growth in human blood. The role of SSA in GAS pathogenesis, particularly in invasive covR/covS mutants, should be further investigated.
The original contributions presented in the study are included in the article/Supplementary Material , further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by the Institutional Review Board (201900274B0 and 202000479B0) of the Chang Gung Memorial Hospital, Taiwan. The patients/participants provided their written informed consent to participate in this study.
CC-N, Y-SL, and C-YL contributed to the conceptualization, methodology, formal analysis, and writing the original draft. Y-SL, C-YL, C-YH, and Y-AS contributed to the methodology and investigation. Y-YC and C-HL contributed to the conceptualization and methodology. CC-N contributed to the writing – review and editing. CC-N and C-HC contributed to the funding acquisition. All authors contributed to the manuscript revision, read, and approved the submitted version.
This work was supported by parts of grants from the Chang Gung Memorial Hospital, Linkou, Taiwan (CMRPD1J0031-3 and CMRPD1K0331) and Ministry of Science and Technology, Taiwan (MOST 109-2320-B-182-036).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to the Bacterial Bank of Chang Gung Memorial Hospital, Linkou (Taiwan) for providing clinical GAS isolates.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.685343/full#supplementary-material
Ben Zakour, N. L., Davies, M. R., You, Y., Chen, J. H., Forde, B. M., Stanton-Cook, M., et al. (2015). Transfer of scarlet fever-associated elements into the group A Streptococcus M1T1 clone. Sci. Rep. 5:15877. doi: 10.1038/srep15877
Brockmann, S. O., Eichner, L., and Eichner, M. (2018). Constantly high incidence of scarlet fever in Germany. Lancet Infect. Dis. 18, 499–500. doi: 10.1016/S1473-3099(18)30210-X
Brouwer, S., Barnett, T. C., Ly, D., Kasper, K. J., De Oliveira, D. M. P., Rivera-Hernandez, T., et al. (2020). Prophage exotoxins enhance colonization fitness in epidemic scarlet fever-causing Streptococcus pyogenes. Nat. Commun. 11:5018. doi: 10.1038/s41467-020-18700-5
Chiang-Ni, C., Chiou, H. J., Tseng, H. C., Hsu, C. Y., and Chiu, C. H. (2020). RocA regulates phosphatase activity of virulence sensor CovS of group A Streptococcus in growth phase‐ and pH-dependent manners. mSphere 5, e00361–e00320. doi: 10.1128/mSphere.00361-20
Chiang-Ni, C., Chu, T. P., Wu, J. J., and Chiu, C. H. (2016). Repression of Rgg but not upregulation of LacD.1 in emm1-type covS mutant mediates the SpeB repression in group A Streptococcus. Front. Microbiol. 7:1935. doi: 10.3389/fmicb.2016.01935
Chiang-Ni, C., Kao, C. Y., Hsu, C. Y., and Chiu, C. H. (2019a). Phosphorylation at the D53 but not T65 residue of CovR determines the repression of rgg and speB transcription in emm1‐ and emm49-type group A streptococci. J. Bacteriol. 201, e00681–e00618. doi: 10.1128/JB.00681-18
Chiang-Ni, C., Shi, Y. A., Lai, C. H., and Chiu, C. H. (2018). Cytotoxicity and survival fitness of invasive covS mutant of group A Streptococcus in phagocytic cells. Front. Microbiol. 9:2592. doi: 10.3389/fmicb.2018.02592
Chiang-Ni, C., Tseng, H. C., Hung, C. H., and Chiu, C. H. (2017). Acidic stress enhances CovR/S-dependent gene repression through activation of the covR/S promoter in emm1-type group A Streptococcus. Int. J. Med. Microbiol. 307, 329–339. doi: 10.1016/j.ijmm.2017.06.002
Chiang-Ni, C., Tseng, H. C., Shi, Y. A., and Chiu, C. H. (2019b). Effect of phosphatase activity of the control of virulence sensor (CovS) on clindamycin-mediated streptolysin O production in group A Streptococcus. Infect. Immun. 87, e00583–e00519. doi: 10.1128/IAI.00583-19
Chiang-Ni, C., Zheng, P. X., Ho, Y. R., Wu, H. M., Chuang, W. J., Lin, Y. S., et al. (2009). emm1/sequence type 28 strains of group A streptococci that express covR at early stationary phase are associated with increased growth and earlier SpeB secretion. J. Clin. Microbiol. 47, 3161–3169. doi: 10.1128/JCM.00202-09
Chuang, P. K., Wang, S. M., Lin, H. C., Cho, Y. H., Ma, Y. J., Ho, T. S., et al. (2015). The trend of macrolide resistance and emm types of group A streptococci from children at a medical center in southern Taiwan. J. Microbiol. Immunol. Infect. 48, 160–167. doi: 10.1016/j.jmii.2013.08.015
Clinical and Laboratory Standards Institute (2018). “Performance standards for antimicrobial susceptibility testing,” in CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute.
Davies, M. R., Holden, M. T., Coupland, P., Chen, J. H., Venturini, C., Barnett, T. C., et al. (2015). Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance. Nat. Genet. 47, 84–87. doi: 10.1038/ng.3147
Demczuk, W., Martin, I., Domingo, F. R., MacDonald, D., and Mulvey, M. R. (2019). Identification of Streptococcus pyogenes M1UK clone in Canada. Lancet Infect. Dis. 19, 1284–1285. doi: 10.1016/S1473-3099(19)30622-X
Feng, W., Minor, D., Liu, M., Li, J., Ishaq, S. L., Yeoman, C., et al. (2017). Null mutations of group A Streptococcus orphan kinase RocA: selection in mouse infection and comparison with CovS mutations in alteration of in vitro and in vivo protease SpeB expression and virulence. Infect. Immun. 85, e00790–e00716. doi: 10.1128/IAI.00790-16
Friaes, A., Pato, C., Melo-Cristino, J., and Ramirez, M. (2015). Consequences of the variability of the CovRS and RopB regulators among Streptococcus pyogenes causing human infections. Sci. Rep. 5:12057. doi: 10.1038/srep12057
Horstmann, N., Tran, C. N., Brumlow, C., DebRoy, S., Yao, H., Nogueras Gonzalez, G., et al. (2018). Phosphatase activity of the control of virulence sensor kinase CovS is critical for the pathogenesis of group A Streptococcus. PLoS Pathog. 14:e1007354. doi: 10.1371/journal.ppat.1007354
Hsieh, Y. C., and Huang, Y. C. (2011). Scarlet fever outbreak in Hong Kong, 2011. J. Microbiol. Immunol. Infect. 44, 409–411. doi: 10.1016/j.jmii.2011.07.003
Ikebe, T., Ato, M., Matsumura, T., Hasegawa, H., Sata, T., Kobayashi, K., et al. (2010). Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS Pathog. 6:e1000832. doi: 10.1371/journal.ppat.1000832
Jain, I., Miller, E. W., Danger, J. L., Pflughoeft, K. J., and Sumby, P. (2017). RocA is an accessory protein to the virulence-regulating CovRS two-component system in group A Streptococcus. Infect. Immun. 85, e00274–e00317. doi: 10.1128/IAI.00274-17
Kim, J. H., and Cheong, H. K. (2018). Increasing number of scarlet fever cases, South Korea, 2011-2016. Emerg. Infect. Dis. 24, 172–173. doi: 10.3201/eid2401.171027
Lamagni, T., Guy, R., Chand, M., Henderson, K. L., Chalker, V., Lewis, J., et al. (2018). Resurgence of scarlet fever in England, 2014-16: a population-based surveillance study. Lancet Infect. Dis. 18, 180–187. doi: 10.1016/S1473-3099(17)30693-X
Li, Y., Nanduri, S. A., Van Beneden, C. A., and Beall, B. W. (2020b). M1UK lineage in invasive group A Streptococcus isolates from the USA. Lancet Infect. Dis. 20, 538–539. doi: 10.1016/S1473-3099(20)30279-6
Li, H., Zhou, L., Zhao, Y., Ma, L., Xu, J., Liu, Y., et al. (2020a). Epidemiological analysis of group A Streptococcus infections in a hospital in Beijing, China. Eur. J. Clin. Microbiol. Infect. Dis. 39, 2361–2371. doi: 10.1007/s10096-020-03987-5
Luk, E. Y., Lo, J. Y., Li, A. Z., Lau, M. C., Cheung, T. K., Wong, A. Y., et al. (2012). Scarlet fever epidemic, Hong Kong, 2011. Emerg. Infect. Dis. 18, 1658–1661. doi: 10.3201/eid1810.111900
Lynskey, N. N., Jauneikaite, E., Li, H. K., Zhi, X., Turner, C. E., Mosavie, M., et al. (2019). Emergence of dominant toxigenic M1T1 Streptococcus pyogenes clone during increased scarlet fever activity in England: a population-based molecular epidemiological study. Lancet Infect. Dis. 19, 1209–1218. doi: 10.1016/S1473-3099(19)30446-3
Meisal, R., Andreasson, I. K., Hoiby, E. A., Aaberge, I. S., Michaelsen, T. E., and Caugant, D. A. (2010). Streptococcus pyogenes isolates causing severe infections in Norway in 2006 to 2007: emm types, multilocus sequence types, and superantigen profiles. J. Clin. Microbiol. 48, 842–851. doi: 10.1128/JCM.01312-09
Mollick, J. A., Miller, G. G., Musser, J. M., Cook, R. G., Grossman, D., and Rich, R. R. (1993). A novel superantigen isolated from pathogenic strains of Streptococcus pyogenes with aminoterminal homology to staphylococcal enterotoxins B and C. J. Clin. Invest. 92, 710–719. doi: 10.1172/JCI116641
Okamoto, S., Kawabata, S., Nakagawa, I., and Hamada, S. (2001). Administration of superantigens protects mice from lethal Listeria monocytogenes infection by enhancing cytotoxic T cells. Infect. Immun. 69, 6633–6642. doi: 10.1128/IAI.69.11.6633-6642.2001
Park, D. W., Kim, S. H., Park, J. W., Kim, M. J., Cho, S. J., Park, H. J., et al. (2017). Incidence and characteristics of scarlet fever, South Korea, 2008-2015. Emerg. Infect. Dis. 23, 658–661. doi: 10.3201/eid2304.160773
Reda, K. B., Kapur, V., Mollick, J. A., Lamphear, J. G., Musser, J. M., and Rich, R. R. (1994). Molecular characterization and phylogenetic distribution of the streptococcal superantigen gene (ssa) from Streptococcus pyogenes. Infect. Immun. 62, 1867–1874. doi: 10.1128/IAI.62.5.1867-1874.1994
Rumke, L. W., de Gier, B., Vestjens, S. M. T., van der Ende, A., van Sorge, N. M., Vlaminckx, B. J. M., et al. (2020). Dominance of M1UK clade among Dutch M1 Streptococcus pyogenes. Lancet Infect. Dis. 20, 539–540. doi: 10.1016/S1473-3099(20)30278-4
Sumby, P., Whitney, A. R., Graviss, E. A., DeLeo, F. R., and Musser, J. M. (2006). Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2:e5. doi: 10.1371/journal.ppat.0020005
Tran-Winkler, H. J., Love, J. F., Gryllos, I., and Wessels, M. R. (2011). Signal transduction through CsrRS confers an invasive phenotype in group A Streptococcus. PLoS Pathog. 7:e1002361. doi: 10.1371/journal.ppat.1002361
Trevino, J., Perez, N., Ramirez-Pena, E., Liu, Z., Shelburne, S. A., Musser, J. M., et al. (2009). CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect. Immun. 77, 3141–3149. doi: 10.1128/IAI.01560-08
Tse, H., Bao, J. Y., Davies, M. R., Maamary, P., Tsoi, H. W., Tong, A. H., et al. (2012). Molecular characterization of the 2011 Hong Kong scarlet fever outbreak. J. Infect. Dis. 206, 341–351. doi: 10.1093/infdis/jis362
Tsou, C. C., Chiang-Ni, C., Lin, Y. S., Chuang, W. J., Lin, M. T., Liu, C. C., et al. (2010). Oxidative stress and metal ions regulate a ferritin-like gene, dpr, in Streptococcus pyogenes. Int. J. Med. Microbiol. 300, 259–264. doi: 10.1016/j.ijmm.2009.09.002
Turner, C. E., Pyzio, M., Song, B., Lamagni, T., Meltzer, M., Chow, J. Y., et al. (2016). Scarlet fever upsurge in England and molecular-genetic analysis in North-West London, 2014. Emerg. Infect. Dis. 22, 1075–1078. doi: 10.3201/eid2206.151726
Valcu, M., and Valcu, C. M. (2011). Data transformation practices in biomedical sciences. Nat. Methods 8, 104–105. doi: 10.1038/nmeth0211-104
Villasenor-Sierra, A., Katahira, E., Jaramillo-Valdivia, A. N., Barajas-Garcia Mde, L., Bryant, A., Morfin-Otero, R., et al. (2012). Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains isolated from invasive and non-invasive infections from Mexico and the USA during 1999-2010. Int. J. Infect. Dis. 16, e178–e181. doi: 10.1016/j.ijid.2011.11.005
Walker, M. J., and Brouwer, S. (2018). Scarlet fever makes a comeback. Lancet Infect. Dis. 18, 128–129. doi: 10.1016/S1473-3099(17)30694-1
Walker, M. J., Hollands, A., Sanderson-Smith, M. L., Cole, J. N., Kirk, J. K., Henningham, A., et al. (2007). DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat. Med. 13, 981–985. doi: 10.1038/nm1612
Yan, J. J., Wu, H. M., Huang, A. H., Fu, H. M., Lee, C. T., and Wu, J. J. (2000). Prevalence of polyclonal mefA-containing isolates among erythromycin-resistant group A streptococci in southern Taiwan. J. Clin. Microbiol. 38, 2475–2479. doi: 10.1128/JCM.38.7.2475-2479.2000
You, Y., Davies, M. R., Protani, M., McIntyre, L., Walker, M. J., and Zhang, J. (2018). Scarlet fever epidemic in China caused by Streptococcus pyogenes serotype M12: epidemiologic and molecular analysis. EBioMedicine 28, 128–135. doi: 10.1016/j.ebiom.2018.01.010
Keywords: group A Streptococcus, streptococcal superantigen, SSA, CovR/CovS, superantigen, invasive GAS infection
Citation: Chiang-Ni C, Liu Y-S, Lin C-Y, Hsu C-Y, Shi Y-A, Chen Y-YM, Lai C-H and Chiu C-H (2021) Incidence and Effects of Acquisition of the Phage-Encoded ssa Superantigen Gene in Invasive Group A Streptococcus . Front. Microbiol. 12:685343. doi: 10.3389/fmicb.2021.685343
Received: 25 March 2021; Accepted: 12 May 2021;
Published: 04 June 2021.
Edited by:
Mattias Collin, Lund University, SwedenReviewed by:
Aftab Jasir, Independent Researcher, Stockholm, SwedenCopyright © 2021 Chiang-Ni, Liu, Lin, Hsu, Shi, Chen, Lai and Chiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuan Chiang-Ni, ZW50Y2h1YW5AZ2FwLmNndS5lZHUudHc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.