
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 June 2021
Sec. Infectious Agents and Disease
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.684473
This article is part of the Research Topic Gram-Negative Pathogenesis View all 9 articles
 Valentina Lucchini1,2
Valentina Lucchini1,2 Adeline Sivignon3
Adeline Sivignon3 Michel Pieren1
Michel Pieren1 Marc Gitzinger1
Marc Gitzinger1 Sergio Lociuro1
Sergio Lociuro1 Nicolas Barnich3
Nicolas Barnich3 Christian Kemmer1
Christian Kemmer1 Vincent Trebosc1*
Vincent Trebosc1*Gut microbiota dysbiosis toward adherent-invasive Escherichia coli (AIEC) plays an important role in Crohn’s disease (CD). The OmpR transcriptional regulator is required for the AIEC LF82 prototype strain to adhere and invade intestinal epithelial cells. In this study, we explored the role of OmpR in AIEC pathogenesis using a panel of eight Escherichia coli strains isolated from CD patients and identified as AIEC. The deletion of ompR together with the implementation of two cell-based assays revealed that the role of OmpR in adhesion in vitro was not conserved in AIEC clinical strains. Nevertheless, we showed that OmpR was required for robust gut colonization of transgenic mice expressing human CEACAM receptors, suggesting that OmpR is involved in alternative virulence mechanisms in AIEC strains. We found that deletion of ompR compromised the ability of AIEC strains to cope with the stress induced by bile salts, which may be key for AIEC pathogenesis. More specifically, we demonstrated that OmpR was involved in a tolerance mechanism toward sodium deoxycholate (DOC), one of bile salts main component. We showed that the misregulation of OmpF or the loss of outer membrane integrity are not the drivers of OmpR-mediated DOC tolerance, suggesting that OmpR regulates a specific mechanism enhancing AIEC survival in the presence of DOC. In conclusion, the newly discovered role of OmpR in AIEC bile tolerance suggests that OmpR inhibition would interfere with different aspects of AIEC virulence arsenal and could be an alternative strategy for CD-treatment.
Crohn’s disease (CD) is a type of inflammatory bowel disease (IBD), which is a complex chronic disorder that primarily disturbs the digestive system (Crohn’s disease, 2020). CD affects more than 2.5 million individuals in Western countries and it has an increasing incidence in the newly industrialized world (Ng et al., 2017). Interactions between the altered intestinal microbiota, the abnormal mucosal immune system of the host and environmental factors contribute to the onset and progression of IBD diseases (Elhenawy et al., 2018; Chervy et al., 2020).
Adherent-invasive E. coli (AIEC) is a functionally distinct group of resident mucosa-associated bacteria that is enriched in CD patients (Darfeuille-Michaud, 2002). In the absence of common identifying genetic determinants, AIEC strains have been characterized by their ability to adhere and invade intestinal epithelial cells (Camprubí-Font et al., 2019; Camprubí-Font and Martinez-Medina, 2020). AIEC adhesion and invasion is mainly mediated by the type 1 pili and their adhesin FimH that bind to the glycoproteins, such as carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) that are enriched at the surface of ileal epithelial cells of CD patients (Boudeau et al., 2001; Barnich et al., 2007). In vitro studies have demonstrated that AIEC strains are able to extensively survive and replicate within monocytes-derived macrophages (Glasser et al., 2001). AIEC strains have been described as moderate to strong in vitro biofilm producers (Martinez-Medina et al., 2009). Altogether, these attributes potentially contribute to AIEC mediated exacerbation of CD through their enrichment in ileal mucosa leading to chronic intestinal inflammation.
Two-component signal transduction systems (TCSs) have evolved to coordinate bacterial communication with the host as an important aspect of both symbiosis and pathogenesis (Calva and Oropeza, 2006). OmpR, the transcriptional regulator of the OmpR/EnvZ TCS, was shown to be essential for FimH-mediated in vitro adhesion and invasion of intestinal epithelial cells in the prototype LF82 AIEC strain (Rolhion et al., 2007). These results suggest that OmpR inhibition may be an interesting approach to prevent AIEC-mediated exacerbation of CD.
The goal of the present study was to uncover the contribution of OmpR in AIEC pathogenesis using diverse clinical strains isolated from CD patients to evaluate the clinical relevance of OmpR as a drug target. We found that, despite a non-conserved OmpR-mediated in vitro adhesion phenotype, OmpR was required for robust AIEC colonization of CEABAC10 mouse gut. In addition, we demonstrated that OmpR plays a role in AIEC bile tolerance, which may be central for AIEC pathogenesis.
Eight adherent-invasive Escherichia coli (AIEC) clinical isolates and the reference K12 MG1655 Escherichia coli strain were used in this study (Table 1). Multiple locus sequence type was determined according to the Pasteur scheme (Clermont et al., 2015). The microdilution method was used to determine the minimum inhibitory concentration (MICs) in cation-adjusted Mueller-Hinton broth (CA-MHB) according to the CLSI guidelines (CLSI, 2020). Bacteria were grown using Luria-Bertani (LB) broth or agar at 37°C unless otherwise stated.
Scarless deletions of ompR were performed using a two-step recombination method previously described (Trebosc et al., 2016). Briefly, DNA fragments corresponding to 700-bp up- and downstream genomic regions of ompR were amplified by PCR using oCK647/oCK648 and oCK649/oCK650, respectively (Supplementary Table 1). The obtained fragments were cloned using NEBuilder HiFi DNA assembly (New England Biolabs) in the multiple cloning site of the E. coli knockout plasmid pCK452. The pCK452 plasmid is a derivative of the pVT77 plasmid where the ColE1 origin of replication has been replaced by the E. coli non-replicative R6K origin of replication and the IPTG regulated thymidine kinase expression cassette has been replaced by an anhydrotetracycline (aTC) inducible SceI expression cassette, corresponding to the counter selection system (Trebosc et al., 2016). The cloned knockout plasmid was transformed in E. coli conjugative strain MFDpir to proceed with the construction of markerless deletion in AIEC. Briefly, after conjugation, genomic plasmid integration was selected on LB agar plates containing 100 μg/ml sodium tellurite. Clones were screened for up- or downstream integration by PCR using primer oCK576, which anneals on the genome upstream of the ompR flanking region, and oCK354, which anneals on the plasmid. Clones containing up- and downstream plasmid integrations were transferred on LB agar plates containing 2 μg/ml aTC to select for plasmid removal from the genome. Clones were screened for gene deletion and plasmid removal by PCR using primers oCK576/oCK578 (Supplementary Table 1). The genomic gene deletions were finally confirmed by DNA sequencing (Microsynth AG, Balgach, Switzerland). The ompR gene was PCR amplified using oVT559/oVT331 and cloned using NEBuilder HiFi DNA assembly in the multiple cloning site of the pSEVA324 expression plasmid to construct the OmpR complementation plasmid.
Commercial baker’s yeast was diluted in phosphate-buffered saline (PBS) to 0.5 mg/ml. AIEC cultures incubated at 37°C in LB for 24 h without shaking were collected and diluted in PBS to OD600 = 5. Equal volumes (50 μL) of a fixed concentration of yeast suspension and decreasing concentrations of E. coli suspension were mixed in a 96-well plate (Claret et al., 2007). After 2 h, the aggregation was monitored visually and the titer was recorded as the lowest dilution of bacteria giving a positive aggregation reaction (Supplementary Figure 1).
The human intestinal epithelial T84 cell line was obtained from the American Type Culture Collection (ATCC® CCL248). T84 cells were cultured in Dulbecco’s Modified Eagle Medium: Nutrient mixture F-12 supplemented with 2 mM L-Glutamine, 0.1 M HEPES buffer, 10% fetal bovine serum and antibiotics-antimycotics to a final concentration of 100 U/ml penicillin G, 100 μg/ml streptomycin, and 0.25 μg/ml of amphotericin B. T84 monolayers were seeded in 24-well tissue culture plates with 4 × 105 cells/well and incubated for 48 h. Cell monolayers were washed 2 times with PBS and infected with AIEC bacteria for 3 h at 37°C, in an atmosphere containing 5% of CO2 at a multiplicity of infection (MOI) estimated at 10 bacteria per cell. For that, bacterial suspensions were prepared at a concentration of 1.6 × 108 bacteria/ml in culture medium and 25 μL were added onto the cells to reach ≈4 × 106 bacteria/well (=MOI 10). Cells were washed 4 times with PBS and the T84 cells were lysed with X-100 Triton 1% for 5 min at room temperature, which is a lysis treatment that does not affect bacterial cells (Supplementary Figure 3). Finally, the lysates were appropriately diluted and spread on LB agar plates and incubated at 37°C, overnight.
Bacteria were placed for 5 min on carbon-formvar copper grids and negatively stained during 30 s with Uranyless (Delta Microscopies, France). Samples were observed using a Hitachi transmission electron microscope (H-7650, Japan) at 80 kV acceleration voltage. Micrographs were made using a Hamamatsu camera placed in a side position.
Eight-week-old CEABAC10 male mice [heterozygous, (Chan and Stanners, 2004)] were used to perform in vivo experiment for testing wild-type (WT) and ompR deleted (ΔompR) AIEC strains. Mice were housed in specific-pathogen-free conditions (21–22°C, 12:12-h light-dark cycle), with access to food and water ad libitum, in the animal facility of the University Clermont Auvergne (EOPS animal care facility, Clermont-Ferrand, France). This study was carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the University of Clermont Auvergne (Clermont-Ferrand, France). The animal protocol was approved by the Committee for Research and Ethical Issues of the Department of Auvergne (CEMEA Auvergne; Permit Number: CEMEAA, 2018103015295515). CEABAC10 mice were treated with streptomycin in drinking water (1.5 g/L) and a low concentration of 0.5% of DSS (dextran sulfate sodium salt colitis grade, MP Biomedicals) for 3 days to disrupt normal resident bacterial intestinal microbiota and to induce a low-grade inflammation. DSS/Streptomycin was replaced by DSS 0.5% 24 h before AIEC infection. The day of infection (day 0), overnight bacterial cultures of AIEC strains were harvested by centrifugation at 2500 × g for 10 min. The bacterial pellet was resuspended in PBS at 1.5 × 1010 bacteria/ml. Mice were challenged with 3 × 109 bacteria (0.2 ml), immediately after an oral administration of 0.1 ml of sodium bicarbonate at 0.2 M. At day 7 post-infection, fresh fecal pellets (≈100 mg) were collected and resuspended in PBS. After appropriate serial dilutions, AIEC bacteria were enumerated by plating on LB agar medium containing 16 μg/ml vancomycin.
AIEC WT, ΔompR, and complemented mutants were grown for 24 h at 37°C in LB without shaking. The cultures were normalized to an OD600 of 1 and 10-fold serial dilutions were plated on selective MacConkey and Drigalski agar plates (Condalab) and on non-selective LB agar plates. Colony forming units (CFU) were counted after 24 h incubation at 37°C.
Overnight LB cultures of AIEC WT and ΔompR mutants were diluted to OD600 = 0.05 in 10 ml of fresh LB broth in presence of bile salts at the specified concentrations. The cultures were incubated at 37°C with shaking at 200 rpm and samples were taken at 1, 2, 4, 6, 8, and 24 h to determine the OD600 and the CFU/ml. Samples were plated on LB agar and CFU were counted after 24 h incubation at 37°C.
The expression of ompR, ompC, ompF, and mdtE was quantified using qRT-PCR. The strains were grown in LB broth at 37°C to mid-log (OD600 of 0.4), late-log (OD600 of 1.2), or late stationary (OD600 of 4.5) growth phase and total RNA was extracted using a PureLink RNA minikit (Ambion) according to the manufacturer’s recommendations. Residual DNA contaminations were removed using a Turbo DNA-free kit (Ambion). Quantitative reverse transcription-PCR (qRT-PCR) was performed using a GoTaq 1-Step RT-qPCR System kit (Promega, Madison, WI, United States) on a StepOne Real-Time PCR LightCycler (Applied Biosystems, Foster City, CA, United States). The expression of the target genes was normalized to that of the rpoD housekeeping gene using the comparative ΔΔCT (where CT is threshold cycle) method. The primers used in qRT-PCR can be found in Supplementary Table 1.
A panel of 8 diverse AIEC clinical strains isolated from 8 different CD patients was used to evaluate the clinical relevance of OmpR as a drug target (Table 1). All the strains belonged to distinct sequence types and encoded different FimH variants, reflecting the genotypic variability of the AIEC group (Desvaux et al., 2020). Except for the strain S179, all AIEC strains belonged to the S70/N78 FimH clade, conferring higher ability to adhere to intestinal epithelial cells (Dreux et al., 2013). The non-pathogenic K12 MG1655 E. coli strain was included as a control strain. The ompR gene was deleted in all the AIEC strains using a markerless recombination method (Trebosc et al., 2016) leading to clean gene deletion. The deletion of ompR did not significantly affect the in vitro activity of standard of care antibiotics, with a maximum of one out of the 8 AIEC strains tested showing a 4-fold MIC shift (Supplementary Table 2). To evaluate the role of OmpR in AIEC adhesion properties we first used a yeast aggregation assay that allows to study FimH-mediated binding to the highly mannosylated yeast cell wall structure. The deletion of ompR abolished the yeast aggregation ability of 3 out of the 8 AIEC strains, namely S136, S135, and S162 (Table 2). Alternatively, we assessed the ability of the AIEC strains and their isogenic ΔompR mutants to adhere to T84 intestinal epithelial cells expressing the specific CEACAM6 receptor (Dreux et al., 2013). Deletion of ompR significantly impaired the adhesion abilities of the S136 and S162 strains (Figure 1 and Supplementary Figure 4). Together, the results from two orthogonal bacterial adhesion methods indicate that the previously described role of OmpR in AIEC adhesion properties is not conserved in all AIEC strains.
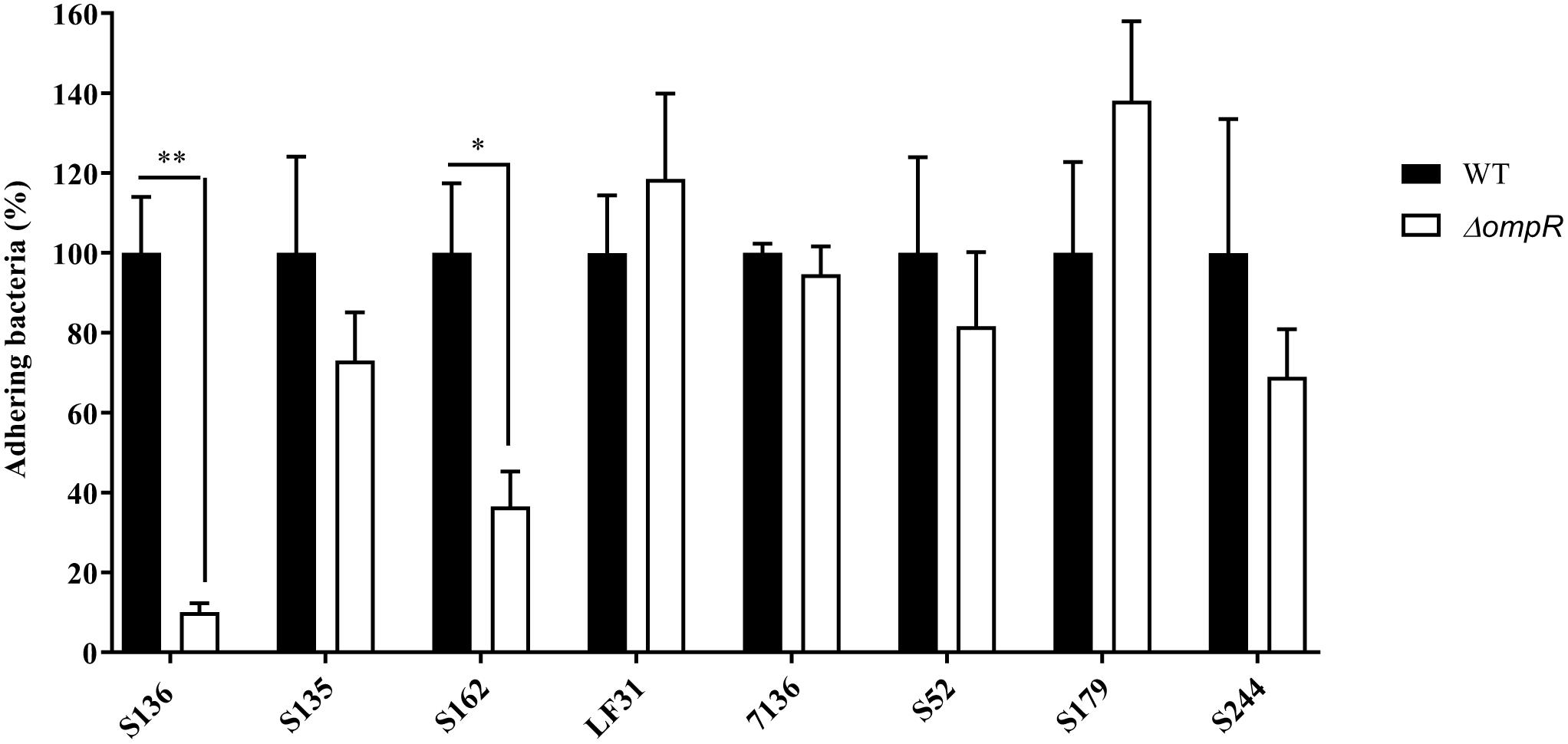
Figure 1. Adhesion levels of AIEC WT and ΔompR mutants to intestinal epithelial cells T84. Adhesion assay was performed with T84 intestinal epithelial cells infected with WT (black bars) and ΔompR mutants (white bars) at a MOI of 10 bacteria/cell for 3 h. Results are expressed in %, considering 100% as adhesion level for the WT strain (means ± SEM, 5 independent experiments). ∗p < 0.05, ∗∗p < 0.01. Unpaired t test.
Mutation in OmpR may affect the expression of regulated genes and explain the difference in adhesion properties between AIEC strains (Vidal et al., 1998). However, none of the AIEC strains studied encoded a mutation in OmpR. In addition, we sequenced the promoter of the fimB and fimE genes encoding recombinases that control type I pili expression through the inversion of the fimS element encoding the promoter of fimA (Schwan, 2011). No mutations were found in the P1 and P2 promoters of fimB while only the AIEC strain S162 encoded a point mutation in the P1 promoter of fimE (Supplementary Figure 2). Together, neither OmpR mutation nor fimB/E promoter mutation can explain the divergent role of OmpR in AIEC adhesion phenotype.
CEABAC10 transgenic mice were used to determine the contribution of OmpR in the ability of AIEC bacteria to colonize the gastrointestinal tract. This mouse model expresses the human CEACAM6 glycoprotein that is enriched in CD ileal epithelial cells and favors AIEC gut colonization through type 1 pili mediated recognition of mannose residues harbored by CEACAM6 (Carvalho et al., 2009). We selected two AIEC strains (S136 and S162) strongly impaired in their in vitro adhesion abilities in absence of ompR and one AIEC strain (S244) not significantly affected in its adhesion phenotype to evaluate the role of OmpR in AIEC virulence. At day 7 post-infection, the bacterial loads in the feces of CEABAC10 mice were significantly lower in the three ΔompR mutants compared to their WT ancestors (Figure 2). These results indicate that OmpR is required for AIEC robust colonization of mouse gut in CD conditions. Interestingly, the AIEC strain S244, whose in vitro adhesion properties were not affected by ompR deletion, showed the most pronounced impairment in gut colonization upon loss of OmpR. The lack of correlation with epithelial cell adhesion phenotype suggests that OmpR regulates alternative virulence mechanisms that are required for AIEC pathogenesis.
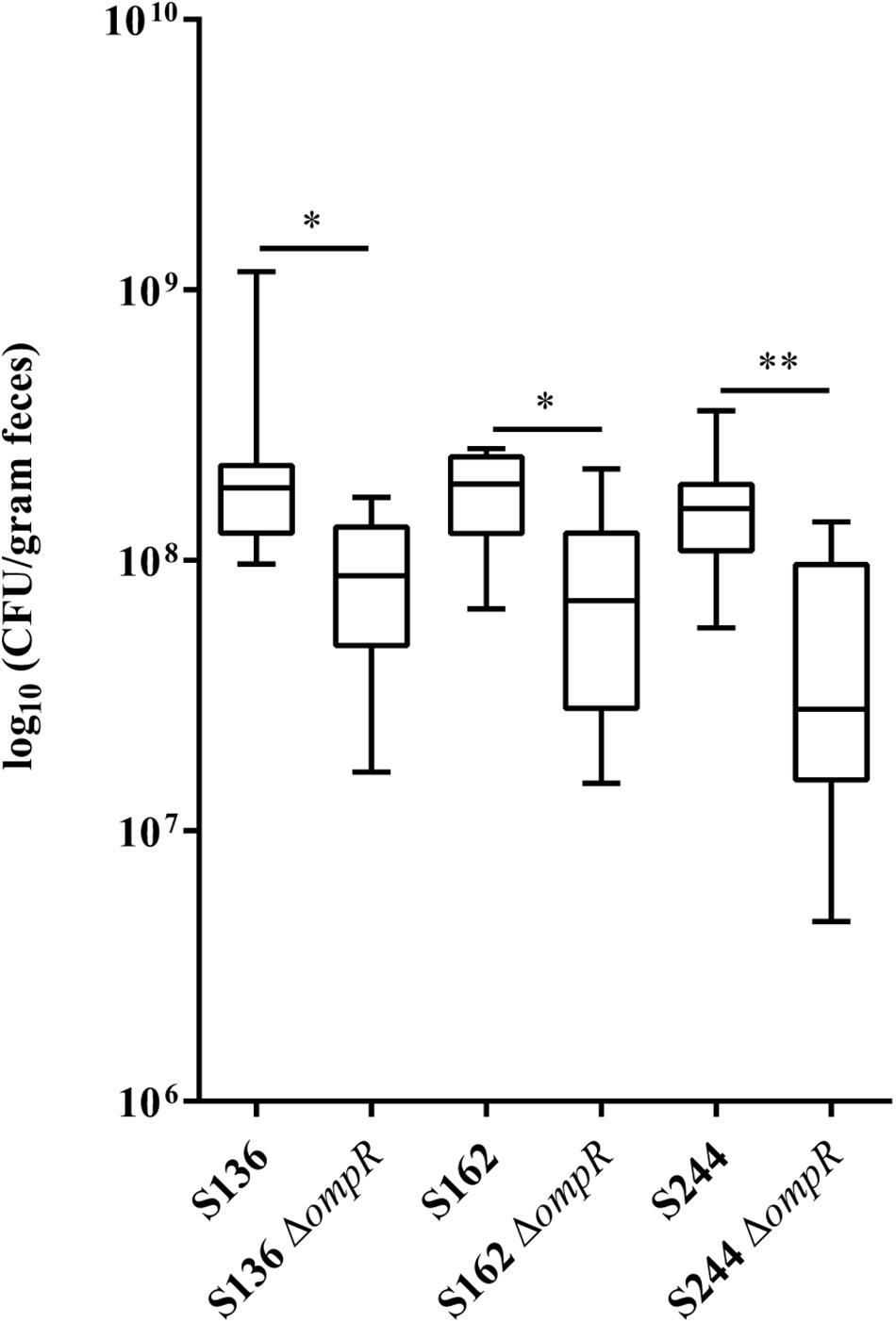
Figure 2. Ability of AIEC WT and ΔompR mutants to colonize the intestine of CEABAC10 mice. Bacterial load in the feces of CEABAC10 mice (8 mice per group) infected with WT and ΔompR AIEC strains at 7-day post-infection. The quantification of AIEC bacteria in the feces of CEABAC10 mice are expressed in CFU/gram feces. Representation in box and whiskers (Min to Max); Mann Whitney test realized between WT and ompR mutant groups; ∗p < 0.05 and ∗∗p < 0.01.
Bile salts and their detergent properties represent one of the major bacterial selecting agents in the gut, suggesting that adaptation to bile induced stress plays an important role in AIEC pathogenesis (Urdaneta and Casadesús, 2017; Delmas et al., 2019). Interestingly, the ΔompR mutants showed impaired growth in MacConkey and Drigalski media, which both contain bile salts as a selective agent, whereas growth on selective medium was partially restored in a ΔompR complemented strain (Figure 3). The two most abundant bile salts are sodium cholate and sodium deoxycholate (DOC), which are found at equivalent concentrations in bile while DOC is prevalent in the feces due to the biotransformation by the gut microbiota (Ridlon et al., 2006; Pavlidis et al., 2015). Physiological concentrations of bile salts and DOC specifically inhibited the growth of the ΔompR AIEC mutants, while the growth of the ΔompR AIEC mutants was still observed in the presence of the same concentration of sodium cholate (Figure 4). To note, increased concentrations of cholate may lead to similar growth inhibition than the one observed with DOC, however, this would be less relevant in the context of AIEC physiological niche where DOC is prevalent. In addition, a DOC derivative (chenodeoxycholate) exerted the same growth inhibition as DOC on the ΔompR AIEC mutants. These data indicate that OmpR is involved in the regulation of DOC stress response in AIEC strains, which may be a driver of the attenuated virulence observed in the AIEC ΔompR mutants.
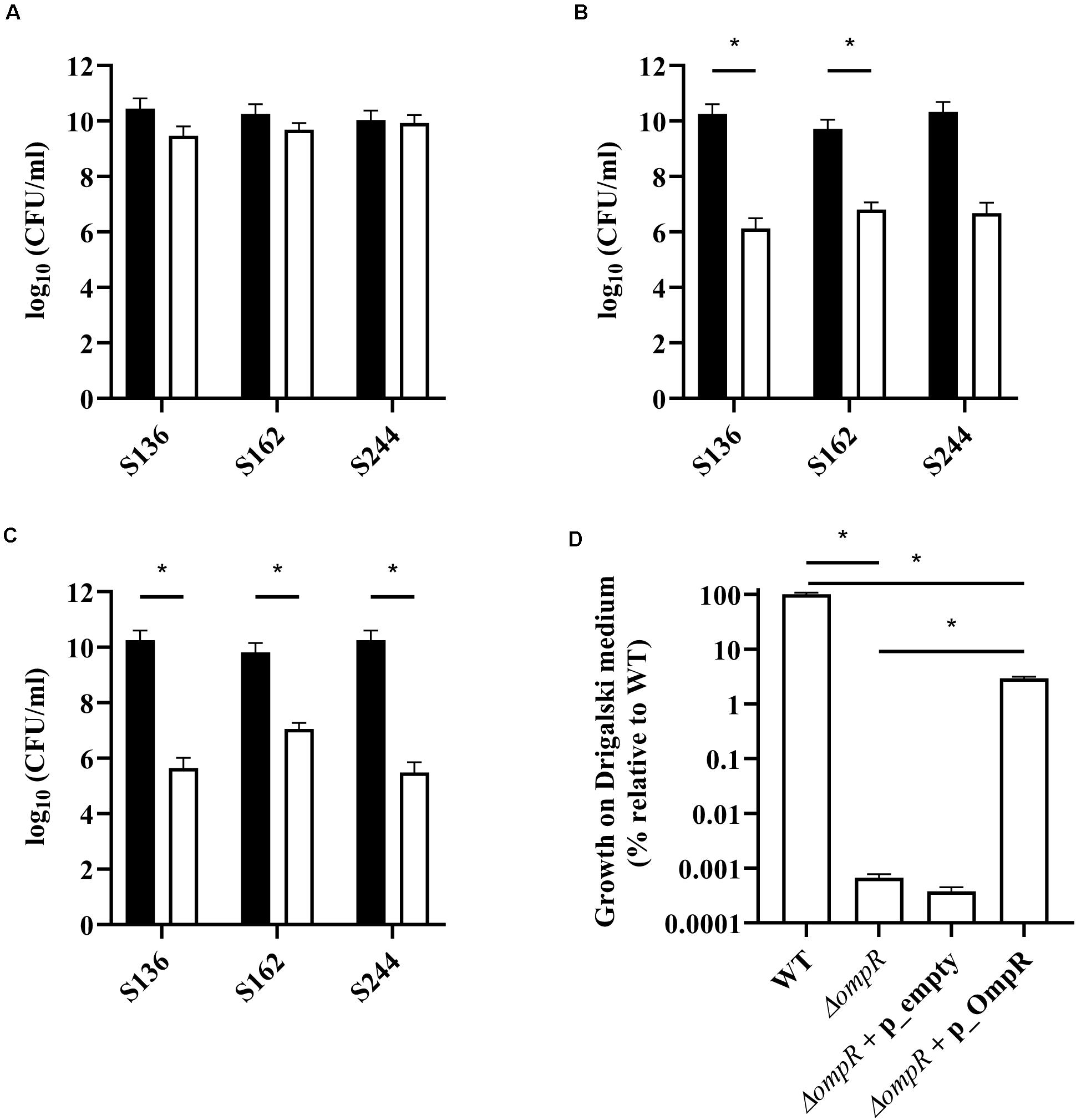
Figure 3. Quantification of AIEC WT and ΔompR mutants in different agar media. WT (black bars) and ΔompR (white bars) AIEC strains were inoculated on (A) LB, (B) MacConkey, and (C) Drigalski agar media and the CFU/ml were determined after overnight growth at 37°C (means ± SEM, 2 independent experiments). (D) The WT AIEC S244 strain and its respective ΔompR mutant complemented with an empty plasmid (ΔompR + p_empty) or a plasmid expressing OmpR (ΔompR + p_OmpR) were inoculated on Drigalski agar and the CFU/ml were determined after overnight growth at 37°C (means ± SEM, 3 technical replicates). *p < 0.05, unpaired t test.
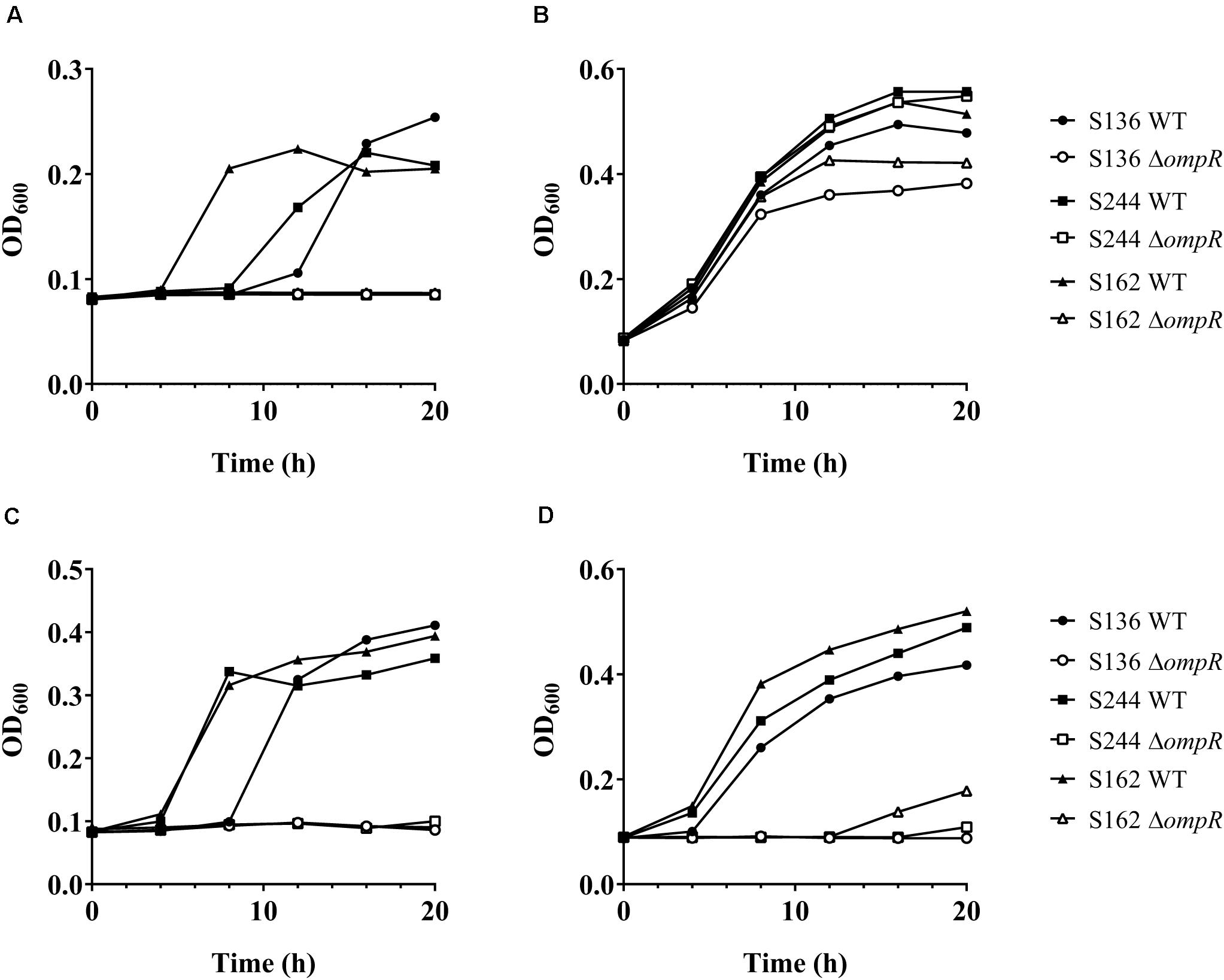
Figure 4. Growth of AIEC WT and ΔompR mutants in presence of bile salts. The WT (filled symbols) AIEC strains S136 (circle), S244 (square), and S162 (triangle) and their ΔompR mutants (empty symbols) were grown in LB supplemented with (A) 0.5% bile salts, (B) 0.1% sodium cholate, (C) 0.1% DOC, and (D) 0.1% chenodeoxycholate. Data representative of at least two independent experiments.
Time kill curve experiments were performed to confirm the inability of the ΔompR AIEC mutants to respond to the stress induced by DOC. Unexpectedly, both the AIEC WT and the ΔompR mutant showed initial CFU reduction in the presence of DOC followed by a regrowth after 6 h (Figures 5A,B). These data indicate that DOC is bactericidal against AIEC strains independently from the presence of a functional OmpR. Nevertheless, the killing rate against the ΔompR mutant was more pronounced than against the WT, allowing only the WT to regrow in the presence of 0.5 and 1% DOC. These data indicate that OmpR is involved in the regulation of AIEC DOC tolerance mechanisms, since bacterial tolerance is characterized by a reduced killing rate (Balaban et al., 2019). In addition, we investigated if the regrowth was due to decay in the concentration of DOC or to the acquisition of resistance. We evaluated the ability of the WT and ΔompR mutant recovered after regrowth (at 24 h) to grow again in the presence of the same DOC concentration. DOC-mediated killing was not observed anymore on DOC pre-exposed strains for both the WT and the ΔompR mutant (Figures 5C,D). A passage on non-selective media of the pre-exposed strains led to the same results (data not shown). These results indicate that the regrowth observed after DOC-mediated killing is due to the development of a stable resistance mechanism and that OmpR is not involved in the acquisition of this mechanism.
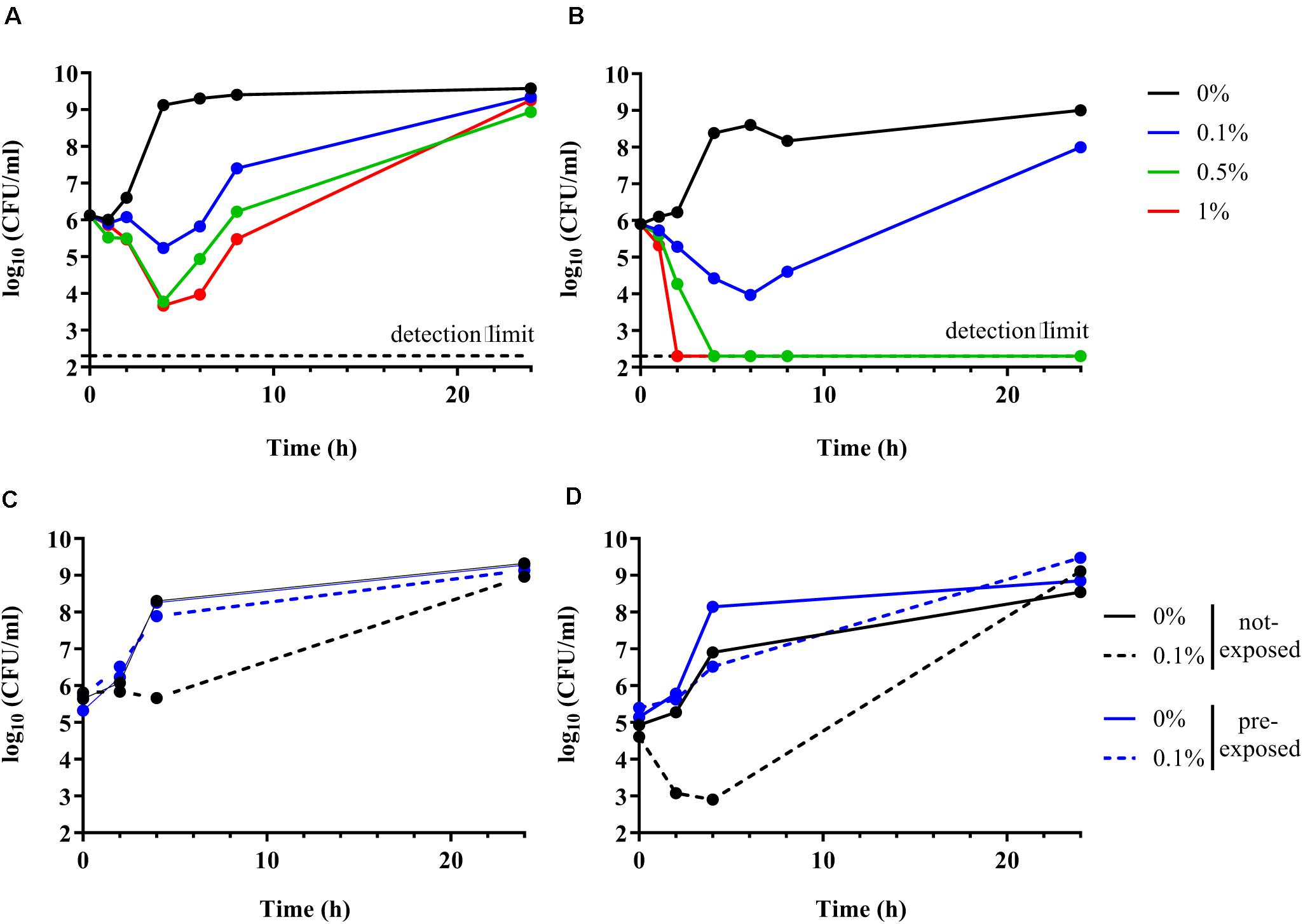
Figure 5. Time kill curve of AIEC WT and ΔompR mutant in presence of DOC. (A,B) The WT AIEC strain S136 (A) and its ΔompR mutant (B) were grown in LB without DOC (black line) or with 0.1% (blue line), 0.5% (green line), and 1% DOC (red line). (C,D) The S136 WT (C) and ΔompR mutant (D) recovered after 24 h of an LB culture without DOC (not-exposed, black line) and with 0.1% DOC (pre-exposed, blue line) (see Figures 5A,B) were grown again in LB without DOC (filled line) and with 0.1% DOC (dotted line). Data representative of at least two independent experiments.
Antibiotics that do not cross the Gram-negative outer membrane were used to exclude that the inability to respond to DOC stress was caused by a non-specific loss of outer membrane integrity in the ΔompR mutant strains. A 4-fold reduction in vancomycin MIC was observed in two of the eight ΔompR mutant strains, while the MIC of rifampicin remained unchanged in all the strains (Supplementary Table 3). Moreover, the sensitivity to polymyxin B, a known membrane permeabilizing drug, did not increase in the ΔompR mutants, indicating that the observed increased DOC sensitivity cannot be generalized to all membrane permeabilizing agents (Supplementary Table 2). Together, these data suggest that OmpR regulates a specific DOC tolerance mechanism in AIEC strains, as opposed to a non-specific loss of cell envelope integrity. This was confirmed using transmission electron microscopy excluding membrane defects on the ΔompR AIEC mutants (Figure 6A).
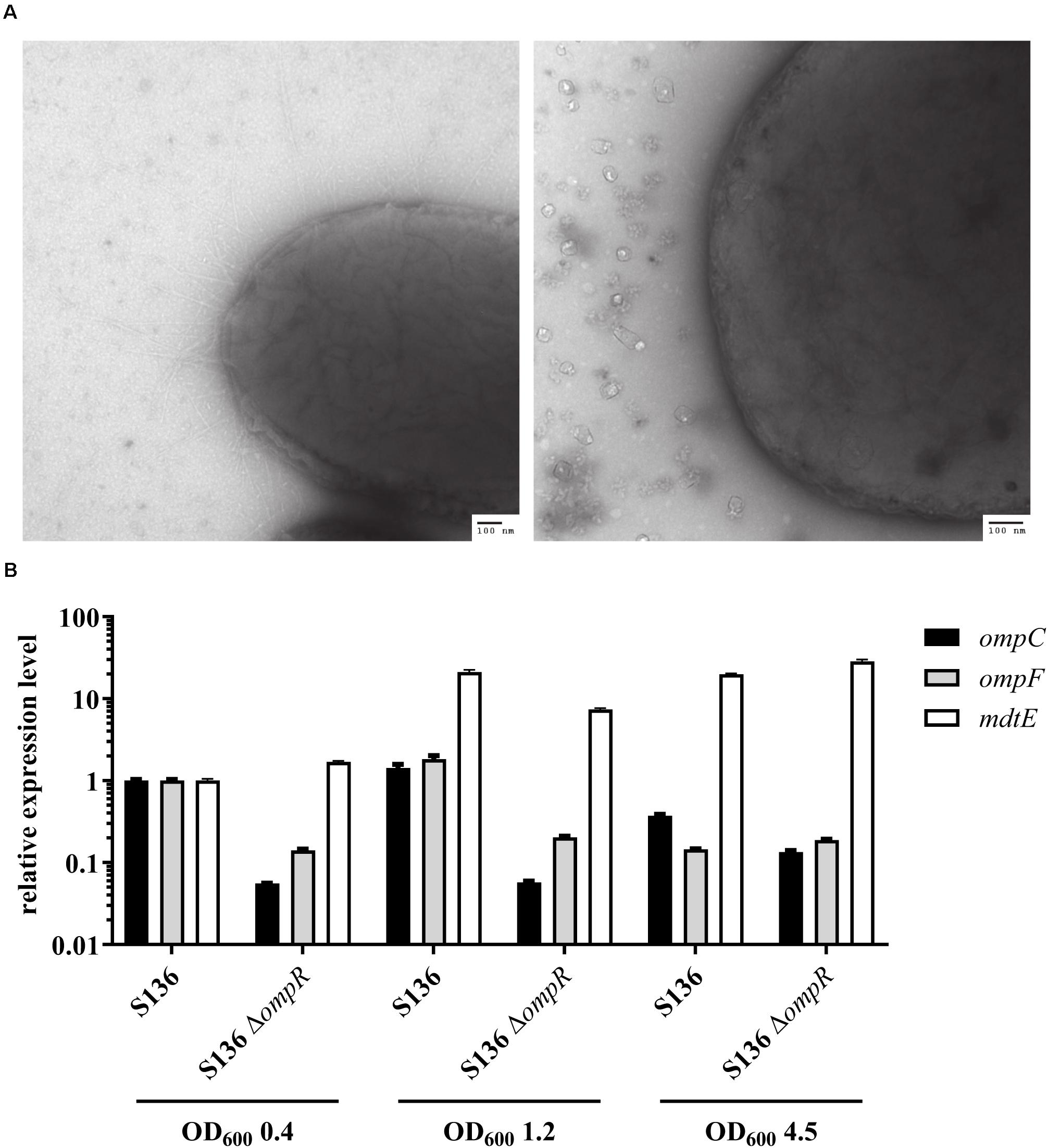
Figure 6. Investigation of potential OmpR-mediated DOC tolerance mechanisms. (A) Micrographs of AIEC S136 (left) and its related ΔompR mutant (right) obtained by transmission electronic microscopy. (B) Expression levels of ompC (black bar), ompF (gray bar), and mdtE (white bar) were quantified by qRT-PCR in WT and ΔompR S136 strains at mid-log (OD600 0.4), late-log (OD600 1.2), and late-stationary (OD600 4.5) growth phases. The expression levels were normalized to the S136 WT strain at mid-log phase (means ± SEM, 2 technical replicates).
Previous studies showed that downregulation of the outer membrane porin OmpF may be involved in adaptation to bile salt mediated stress (Thanassi et al., 1997). Since OmpR regulates the expression of outer membrane porins, we investigated whether OmpC and OmpF misregulation may be the driver of the observed OmpR-mediated DOC tolerance mechanism. Considering the complexity of OmpR-mediated regulation of porin expression (De la Cruz and Calva, 2010), we assessed the expression of ompC and ompF at three different growth phases, namely mid-log, late-log and late stationary phase. Both ompC and ompF genes were downregulated in the S136 ΔompR mutant at all growth phases, indicating that the increased DOC sensitivity in the ΔompR strains is not due to OmpC or OmpF upregulation (Figure 6B). In addition, the growth of the ΔompCΔompF double mutant was not affected by DOC compared to the growth of the ΔompR mutant (Supplementary Figure 5).
Besides porin downregulation, the expression of efflux pumps has been associated with increased resistance to DOC (Nishino and Yamaguchi, 2001). Interestingly, the MdtEF (previously known as YhiUV) efflux pump was shown to be moderately downregulated in a transcriptomic study performed on the ompR deleted E. coli K-12 MG1655 mutant (Seo et al., 2017), suggesting that MdtEF might play a role in OmpR-mediated DOC tolerance mechanism. We showed that mdtE expression is not regulated by OmpR in the S136 AIEC strain when tested at mid-log and late-stationary growth phases (Figure 6B). We observed a moderate downregulation (3-fold) of mdtE expression in the ΔompR mutant compared to the WT strain when tested at late-log growth phase, however, mdtE remained overexpressed compared to mid-log growth phase. To note, DOC supplementation did not affect the level of ompC, ompF or mdtE expression at mid-log growth phase (Supplementary Figure 6).
Signal transduction systems, such as TCSs, are essential for bacteria to communicate and adapt within the harsh gut environment, ultimately leading to the modulation of their fitness and pathogenicity (Breland et al., 2017). The specific enrichment of AIEC strains in the intestinal ecosystem of CD patients is poorly understood (Kittana et al., 2019). The results from a previous study showed that OmpR is involved in in vitro adhesion and invasion capacities of the LF82 AIEC reference strain, suggesting that OmpR may contribute to AIEC-mediated exacerbation of CD (Rolhion et al., 2007).
In this study, we demonstrated the key role of OmpR in AIEC pathogenesis in CEABAC10 mouse infection model. However, we also showed that the role of OmpR in adhesion in vitro is not necessarily conserved in all AIEC strains, suggesting that other virulence mechanisms may be regulated by OmpR. We uncovered that OmpR is involved in AIEC tolerance toward the secondary bile salt DOC. DOC is present in large quantity in the gastrointestinal tract as it is produced by the microbiota from the conversion of the primary bile salt cholic acid (Ridlon et al., 2006; Pavlidis et al., 2015). This supports an important role for OmpR-mediated bile tolerance in AIEC survival and subsequent colonization of the gastrointestinal tract, ultimately leading to dysbiosis and enhanced chronic inflammation.
Bacterial cell-envelope structure maintenance and remodeling are required to resist to the action of bile salts, which is consistent with the role of the OmpR/EnvZ TCS in bile response (Merritt and Donaldson, 2009; Hernández et al., 2015; Sistrunk et al., 2016). Bile salts induce membrane damages that are likely to trigger OmpR activation through the envelope stress response pathway (Hews et al., 2019). Alternatively, it has been suggested that bile salts induce DNA supercoiling and it has been established that the OmpR/EnvZ TCS responds to DNA supercoiling, suggesting that OmpR/EnvZ may indirectly sense bile salts (Begley et al., 2005; Cameron and Dorman, 2012). It has been recently shown that OmpR from Vibrio cholerae is activated by bile salts (Kunkle et al., 2020). In contrast, using ompC and ompF expression levels as a surrogate for OmpR activation, we did not observe DOC-mediated activation of OmpR in AIEC. Another TCS from the OmpR family, namely PhoPQ, is required for Salmonella spp. to resist the action of bile (Velkinburgh and Gunn, 1999; Prouty and Gunn, 2000). Interestingly, as for OmpR in AIEC, PhoPQ conferred enhanced resistance preferentially toward DOC over other bile salts, suggesting that the two distinct TCSs regulate common DOC-induced stress response mechanisms in the two species. However, the exact mechanism by which PhoPQ enhances bile resistance in Salmonella remains to be elucidated (Villarreal et al., 2014).
We showed that loss of DOC tolerance was not due to loss of membrane integrity, suggesting that OmpR regulates a specific mechanism allowing AIEC to cope with DOC-induced stress. One of the DOC resistance mechanisms in E. coli consists in micF-mediated downregulation of OmpF porin synthesis to exclude charged molecules such as DOC from entering bacterial cells (Bernstein et al., 1999). Since OmpR is a known regulator of micF and ompF expression, we hypothesized that ompF downregulation might be the driver of OmpR-mediated DOC tolerance (Thanassi et al., 1997). However, we demonstrated that OmpR-mediated DOC tolerance in AIEC was not related to ompF porin downregulation. Another DOC resistance mechanism consists in the overexpression of specific efflux pumps (Nishino and Yamaguchi, 2001), among which the MdtEF was recently suggested to be regulated by OmpR in E. coli K-12 (Seo et al., 2017). However, we showed that the MdtEF efflux is only transiently regulated by OmpR resulting in limited downregulation of the efflux pump in the studied AIEC strain.
Alternatively, exposure to bile salts triggers a profound metabolic remodeling that favors intracellular accumulation of acetyl-CoA, ultimately conferring AIEC strains with a competitive advantage over commensal bacteria, including commensal E. coli, to colonize the gastrointestinal tract (Delmas et al., 2019). Interestingly, OmpR has been described as a key regulator of E. coli metabolism, with for instance the regulation of the GltA citrate synthase that controls the entry of acetyl-CoA into the tricarboxylic acid cycle (Chakraborty and Kenney, 2018). Therefore, an altered regulation of GltA expression in the ompR deleted AIEC mutants may be involved in the observed loss of bile salt tolerance and gut colonization defect of these strains. Further studies are needed to shed light on the OmpR-mediated bile salt tolerance mechanism.
Our work provides additional insights into the role of OmpR in AIEC pathogenesis. The newly discovered role of OmpR in AIEC DOC tolerance suggests that OmpR inhibition would interfere with different aspects of the AIEC virulence arsenal. Therefore, OmpR inhibition could be an alternative strategy for CD-treatment enabling a limited impact on commensal gut microbiota compared to standard antimicrobial treatments.
The original contributions presented in the study are included in the manuscript/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by The Committee for Research and Ethical Issues of the Department of Auvergne (CEMEA Auvergne; Permit Number: CEMEAA, 2018103015295515).
VL: investigation, methodology, validation, formal analysis, visualization, and writing – original draft. AS: investigation, methodology, validation, formal analysis, visualization, and writing – review and editing. MP, MG, and SL: conceptualization, funding acquisition, and writing – review and editing. NB: conceptualization, project administration, funding acquisition, supervision, and writing – review and editing. CK: conceptualization, project administration, funding acquisition, supervision, visualization, and writing – review and editing. VT: conceptualization, project administration, supervision, formal analysis, visualization, writing – original draft, and writing review and editing. All authors contributed to the article and approved the submitted version.
This project has received funding from the European Union’s Horizon 2020 Research and Innovation Program for VL under the Marie Skłodowska-Curie grant agreement number 721484 and was supported by the Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation, Inserm (UMR1071), INRAE (USC 2018).
MP, MG, SL, CK, and VT own equity in BioVersys.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Christelle Blavignac, Centre Imagerie Cellulaire Santé - UCA PARTNER, (Clermont-Ferrand, France) for her technical support and expertise in TEM analysis. Furthermore, we thank Birgit Schellhorn and Julian Schill, BioVersys AG, for technical assistance. We also thank Dirk Bumann, Biozentrum, University of Basel, for his general supervision and his valuable insights.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.684473/full#supplementary-material
Balaban, N. Q., Helaine, S., Lewis, K., Ackermann, M., Aldridge, B., Andersson, D. I., et al. (2019). Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448. doi: 10.1038/s41579-019-0196-3
Barnich, N., Carvalho, F. A., Glasser, A.-L., Darcha, C., Jantscheff, P., Allez, M., et al. (2007). CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Invest. 117, 1566–1574. doi: 10.1172/JCI30504
Begley, M., Gahan, C. G. M., and Hill, C. (2005). The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651. doi: 10.1016/j.femsre.2004.09.003
Bernstein, C., Bernstein, H., Payne, C. M., Beard, S. E., and Schneider, J. (1999). Bile salt activation of stress response promoters in Escherichia coli. Curr. Microbiol. 39, 68–72. doi: 10.1007/s002849900420
Boudeau, J., Barnich, N., and Darfeuille-Michaud, A. (2001). Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn’s disease is involved in bacterial invasion of intestinal epithelial cells. Mol. Microbiol. 39, 1272–1284. doi: 10.1111/j.1365-2958.2001.02315.x
Breland, E. J., Eberly, A. R., and Hadjifrangiskou, M. (2017). An overview of two-component signal transduction systems implicated in extra-intestinal pathogenic E. coli infections. Front. Cell. Infect. Microbiol. 7:162. doi: 10.3389/fcimb.2017.00162
Calva, E., and Oropeza, R. (2006). Two-component signal transduction systems, environmental signals, and virulence. Microb. Ecol. 51, 166–176. doi: 10.1007/s00248-005-0087-1
Cameron, A. D. S., and Dorman, C. J. (2012). A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet. 8:2615. doi: 10.1371/journal.pgen.1002615
Camprubí-Font, C., and Martinez-Medina, M. (2020). Why the discovery of adherent-invasive Escherichia coli molecular markers is so challenging? World J. Biol. Chem. 11, 1–13. doi: 10.4331/wjbc.v11.i1.1
Camprubí-Font, C., Ewers, C., Lopez-Siles, M., and Martinez-Medina, M. (2019). Genetic and phenotypic features to screen for putative adherent-invasive Escherichia coli. Front. Microbiol. 10:108. doi: 10.3389/fmicb.2019.00108
Carvalho, F. A., Barnich, N., Sivignon, A., Darcha, C., Chan, C. H. F., Stanners, C. P., et al. (2009). Crohn’s disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J. Exp. Med. 206, 2179–2189. doi: 10.1084/jem.20090741
Chakraborty, S., and Kenney, L. J. (2018). A new role of OmpR in acid and osmotic stress in Salmonella and E. coli. Front. Microbiol. 9:2656. doi: 10.3389/fmicb.2018.02656
Chan, C. H. F., and Stanners, C. P. (2004). Novel mouse model for carcinoembryonic antigen-based therapy. Mol. Ther. J. Am. Soc. Gene Ther. 9, 775–785. doi: 10.1016/j.ymthe.2004.03.009
Chervy, M., Barnich, N., and Denizot, J. (2020). Adherent-Invasive E. coli: update on the lifestyle of a troublemaker in Crohn’s disease. Int. J. Mol. Sci. 21, 734. doi: 10.3390/ijms21103734
Claret, L., Miquel, S., Vieille, N., Ryjenkov, D. A., Gomelsky, M., and Darfeuille-Michaud, A. (2007). The flagellar sigma factor FliA regulates adhesion and invasion of Crohn disease-associated Escherichia coli via a Cyclic dimeric GMP-dependent pathway. J. Biol. Chem. 282, 33275–33283. doi: 10.1074/jbc.M702800200
Clermont, O., Gordon, D., and Denamur, E. (2015). Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology 161, 980–988. doi: 10.1099/mic.0.000063
CLSI (2020). M100Ed30 | Performance Standards for Antimicrobial Susceptibility Testing, 30th Edn. Clinical and Laboratory Standards Institute. Available online at: https://clsi.org/standards/products/microbiology/documents/m100/ (accessed October 14, 2020).
Crohn’s disease (2020). Crohn’s disease. Nat. Rev. Dis. Primer 6, 1–2. doi: 10.1038/s41572-020-0166-0
Darfeuille-Michaud, A. (2002). Adherent-invasive Escherichia coli: a putative new E. coli pathotype associated with Crohn’s disease. Int. J. Med. Microbiol. 292, 185–193. doi: 10.1078/1438-4221-00201
Darfeuille-Michaud, A., Boudeau, J., Bulois, P., Neut, C., Glasser, A.-L., Barnich, N., et al. (2004). High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127, 412–421. doi: 10.1053/j.gastro.2004.04.061
De la Cruz, M. A., and Calva, E. (2010). The complexities of porin genetic regulation. J. Mol. Microbiol. Biotechnol. 18, 24–36. doi: 10.1159/000274309
Delmas, J., Gibold, L., Faïs, T., Batista, S., Leremboure, M., Sinel, C., et al. (2019). Metabolic adaptation of adherent-invasive Escherichia coli to exposure to bile salts. Sci. Rep. 9:2175. doi: 10.1038/s41598-019-38628-1
Desvaux, M., Dalmasso, G., Beyrouthy, R., Barnich, N., Delmas, J., and Bonnet, R. (2020). Pathogenicity factors of genomic islands in intestinal and Extraintestinal Escherichia coli. Front. Microbiol. 11:2065. doi: 10.3389/fmicb.2020.02065
Dreux, N., Denizot, J., Martinez-Medina, M., Mellmann, A., Billig, M., Kisiela, D., et al. (2013). Point mutations in FimH adhesin of Crohn’s disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog. 9:141. doi: 10.1371/journal.ppat.1003141
Elhenawy, W., Oberc, A., and Coombes, B. K. (2018). A polymicrobial view of disease potential in Crohn’s-associated adherent-invasive E. coli. Gut Microbes 9, 166–174. doi: 10.1080/19490976.2017.1378291
Glasser, A.-L., Boudeau, J., Barnich, N., Perruchot, M.-H., Colombel, J.-F., and Darfeuille-Michaud, A. (2001). Adherent invasive Escherichia coli strains from patients with Crohn’s disease survive and replicate within macrophages without inducing host cell death. Infect. Immun. 69, 5529–5537. doi: 10.1128/IAI.69.9.5529-5537.2001
Hernández, S. B., Cava, F., Pucciarelli, M. G., Portillo, F. G., Pedro, M. A., and de Casadesús, J. (2015). Bile-induced peptidoglycan remodelling in Salmonella enterica. Environ. Microbiol. 17, 1081–1089. doi: 10.1111/1462-2920.12491
Hews, C. L., Cho, T., Rowley, G., and Raivio, T. L. (2019). Maintaining integrity under stress: envelope stress response regulation of pathogenesis in gram-negative bacteria. Front. Cell. Infect. Microbiol. 9:313. doi: 10.3389/fcimb.2019.00313
Kittana, H., Gomes-Neto, J. C., Heck, K., Sughroue, J., Xian, Y., Mantz, S., et al. (2019). Establishing the phenotypic basis of adherent-invasive Escherichia coli (AIEC) pathogenicity in intestinal inflammation. Microbiology [Preprint]. doi: 10.1101/772012
Kunkle, D. E., Bina, T. F., Bina, X. R., and Bina, J. E. (2020). Vibrio cholerae OmpR represses the ToxR regulon in response to membrane intercalating agents that are prevalent in the human gastrointestinal tract. Infect. Immun. 88:e00912-19. doi: 10.1128/IAI.00912-19
Martinez-Medina, M., Naves, P., Blanco, J., Aldeguer, X., Blanco, J. E., Blanco, M., et al. (2009). Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC). BMC Microbiol. 9:202. doi: 10.1186/1471-2180-9-202
Merritt, M. E., and Donaldson, J. R. (2009). Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J. Med. Microbiol. 58, 1533–1541. doi: 10.1099/jmm.0.014092-0
Ng, S. C., Shi, H. Y., Hamidi, N., Underwood, F. E., Tang, W., Benchimol, E. I., et al. (2017). Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 390, 2769–2778. doi: 10.1016/S0140-6736(17)32448-0
Nishino, K., and Yamaguchi, A. (2001). Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183, 5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001
Pavlidis, P., Powell, N., Vincent, R. P., Ehrlich, D., Bjarnason, I., and Hayee, B. (2015). Systematic review: bile acids and intestinal inflammation-luminal aggressors or regulators of mucosal defence? Aliment. Pharmacol. Ther. 42, 802–817. doi: 10.1111/apt.13333
Prouty, A. M., and Gunn, J. S. (2000). Salmonella enterica Serovar Typhimurium invasion is repressed in the presence of bile. Infect. Immun. 68, 6763–6769. doi: 10.1128/IAI.68.12.6763-6769.2000
Ridlon, J. M., Kang, D.-J., and Hylemon, P. B. (2006). Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–259. doi: 10.1194/jlr.R500013-JLR200
Rolhion, N., Carvalho, F. A., and Darfeuille-Michaud, A. (2007). OmpC and the σE regulatory pathway are involved in adhesion and invasion of the Crohn’s disease-associated Escherichia coli strain LF82. Mol. Microbiol. 63, 1684–1700. doi: 10.1111/j.1365-2958.2007.05638.x
Schwan, W. R. (2011). Regulation of fim genes in uropathogenic Escherichia coli. World J. Clin. Infect. Dis. 1, 17–25. doi: 10.5495/wjcid.v1.i1.17
Seo, S. W., Gao, Y., Kim, D., Szubin, R., Yang, J., Cho, B.-K., et al. (2017). Revealing genome-scale transcriptional regulatory landscape of OmpR highlights its expanded regulatory roles under osmotic stress in Escherichia coli K-12 MG1655. Sci. Rep. 7:2181. doi: 10.1038/s41598-017-02110-7
Sistrunk, J. R., Nickerson, K. P., Chanin, R. B., Rasko, D. A., and Faherty, C. S. (2016). Survival of the fittest: how bacterial pathogens utilize bile to enhance infection. Clin. Microbiol. Rev. 29, 819–836. doi: 10.1128/CMR.00031-16
Thanassi, D. G., Cheng, L. W., and Nikaido, H. (1997). Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179, 2512–2518. doi: 10.1128/JB.179.8.2512-2518.1997
Trebosc, V., Gartenmann, S., Royet, K., Manfredi, P., Tötzl, M., Schellhorn, B., et al. (2016). A novel genome-editing platform for drug-resistant Acinetobacter baumannii reveals an AdeR-unrelated tigecycline resistance mechanism. Antimicrob. Agents Chemother. 60, 7263–7271. doi: 10.1128/AAC.01275-16
Urdaneta, V., and Casadesús, J. (2017). Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. 4:163. doi: 10.3389/fmed.2017.00163
Velkinburgh, J. C. V., and Gunn, J. S. (1999). PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67:9.
Vidal, O., Longin, R., Prigent-Combaret, C., Dorel, C., Hooreman, M., and Lejeune, P. (1998). Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180, 2442–2449.
Villarreal, J. M., Becerra-Lobato, N., Rebollar-Flores, J. E., Medina-Aparicio, L., Carbajal-Gómez, E., Zavala-García, M. L., et al. (2014). The Salmonella enterica serovar Typhi ltrR-ompR-ompC-ompF genes are involved in resistance to the bile salt sodium deoxycholate and in bacterial transformation. Mol. Microbiol. 92, 1005–1024. doi: 10.1111/mmi.12610
Keywords: OmpR, AIEC, virulence, CEABAC10 mice, sodium deoxycholate, tolerance
Citation: Lucchini V, Sivignon A, Pieren M, Gitzinger M, Lociuro S, Barnich N, Kemmer C and Trebosc V (2021) The Role of OmpR in Bile Tolerance and Pathogenesis of Adherent-Invasive Escherichia coli. Front. Microbiol. 12:684473. doi: 10.3389/fmicb.2021.684473
Received: 23 March 2021; Accepted: 31 May 2021;
Published: 28 June 2021.
Edited by:
Hidetada Hirakawa, Gunma University, JapanReviewed by:
Paolo Landini, University of Milan, ItalyCopyright © 2021 Lucchini, Sivignon, Pieren, Gitzinger, Lociuro, Barnich, Kemmer and Trebosc. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vincent Trebosc, dmluY2VudC50cmVib3NjQGJpb3ZlcnN5cy5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.