- 1Department of Clinical Sciences, Institute of Tropical Medicine Antwerp, Antwerp, Belgium
- 2Laboratory of Medical Microbiology, Vaccine and Infectious Disease Institute, University of Antwerp, Antwerp, Belgium
- 3Department of Medicine, University of Cape Town, Cape Town, South Africa
Horizontal gene transfer (HGT) in the penA and multidrug efflux pump genes has been shown to play a key role in the genesis of antimicrobial resistance in Neisseria gonorrhoeae. In this study, we evaluated if there was evidence of HGT in the genes coding for the ribosomal proteins in the Neisseria genus. We did this in a collection of 11,659 isolates of Neisseria, including N. gonorrhoeae and commensal Neisseria species (N. cinerea, N. elongata, N. flavescens, N. mucosa, N. polysaccharea, and N. subflava). Comparative genomic analyses identified HGT events in three genes: rplB, rplD, and rplY coding for ribosomal proteins L2, L4 and L25, respectively. Recombination events were predicted in N. gonorrhoeae and N. cinerea, N. subflava, and N. lactamica were identified as likely progenitors. In total, 2,337, 2,355, and 1,127 isolates possessed L2, L4, and L25 HGT events. Strong associations were found between HGT in L2/L4 and the C2597T 23S rRNA mutation that confers reduced susceptibility to macrolides. Whilst previous studies have found evidence of HGT of entire genes coding for ribosomal proteins in other bacterial species, this is the first study to find evidence of HGT-mediated chimerization of ribosomal proteins.
Introduction
Neisseria gonorrhoeae is a sexually transmitted pathogen that causes the disease gonorrhea (Tapsall et al., 2009; Quillin and Seifert, 2018). Treatment guidelines typically recommend dual antimicrobial therapy, including ceftriaxone and azithromycin (Unemo et al., 2020). The rapid emergence of antimicrobial resistance to azithromycin in numerous populations threatens this treatment approach and has recently led to the dropping of azithromycin from treatment guidelines in countries such as the United States (Martin et al., 2016; Wan et al., 2018; St Cyr et al., 2020). Azithromycin [9-deoxo-9a-aza-9a-methyl-9a-homoerythromycin], a 15-membered macrolide, binds to the bacterial large (50S) ribosomal subunit, which consists of 23S rRNA, 5S rRNA, and ribosomal proteins. It interferes with protein synthesis via various mechanisms, including inhibiting the transpeptidation/translocation step, blocking the 50S peptide exit tunnel, and causing ribosomes to release incomplete peptides (Douthwaite and Champney, 2001; Champney and Miller, 2002; Unemo and Shafer, 2014).
The molecular mechanisms that cause azithromycin resistance in N. gonorrhoeae include an alteration of the drug target either by methylation of a single adenine in 23S rRNA (rrl) or chromosomal mutations (Roberts et al., 1999; Chisholm et al., 2010). Three such mutations have been found in domain V of the 23S rRNA: C2597T, A2045G, and A2046G (Chisholm et al., 2010; Pham et al., 2020). In addition, overexpressed efflux pumps [multidrug efflux pump (MtrCDE), MacAB, and mef-encoded pumps] may reduce azithromycin susceptibility (Unemo and Shafer, 2014). The MtrCDE efflux pump plays an essential role in azithromycin resistance. The mtr locus contains three genes (mtrC – mtrD – mtrE) within an operon that forms a MtrCDE and contains genes for a transcriptional repressor (mtrR) upstream of mtrCDE (Golparian et al., 2014). Mutations upstream of mtrC, including those within the MtrR binding region and mutations in the promoter region and mosaic-like sequences within the mtrD gene, contribute to reduced susceptibility to azithromycin (Hagman and Shafer, 1995; Zarantonelli et al., 1999; Shafer, 2018).
The above well-characterized resistance-associated mutations (RAMs) are, however, unable to explain all the variation in gonococcal susceptibility to macrolides (Ma et al., 2020). A point mutation in the 50S ribosomal protein, L4 (G70D), has been implicated in macrolide resistance in N. gonorrhoeae and other bacteria (Gregory and Dahlberg, 1999; Tait-Kamradt et al., 2000; Ma et al., 2020). Recently, a genome-wide association study conditioned on the known resistance mechanisms, with experimental validation, confirmed that the G70D mutation resulted in reduced susceptibility to azithromycin. In addition, the study identified other L4 mutations at amino acid positions 68 (G68D, G68C), 69 (T69I), and 70 (G70S, G70A, G70R, and G70duplication) associated with reduced azithromycin susceptibility (Ma et al., 2020). These mutations are all situated at the end of the L4 loop close to the macrolide binding site (Gregory and Dahlberg, 1999; Tait-Kamradt et al., 2000; Ma et al., 2020). In a recent in vitro study investigating the molecular pathways to high-level azithromycin resistance in N. gonorrhoeae, we found mutations in 50S ribosomal proteins L4, L22, and L34 that play a prominent role in the genesis of azithromycin resistance (Laumen et al., 2020). Similar mutations in L4 and L22 have been shown to cause macrolide resistance in several other bacterial species (Gregory and Dahlberg, 1999; O’Connor et al., 2004; Halling and Jensen, 2006; Diner and Hayes, 2009).
Horizontal gene transfer (HGT) occurs via conjugation, transformation and transduction. In Neisseria, natural transformation is the primary mode of HGT which occurs with remarkable frequency and efficiency and is dependent on type IV pilus complex (Biswas et al., 1977; Koomey, 1998; Chen and Dubnau, 2004). DNA uptake sequence (DUS), 5’-GCCGTCTGAA-3’ is required for efficient transformation and is both strain and strand specific in N. gonorrhoeae (Goodman and Scocca, 1988; Smith et al., 1999; Ambur et al., 2007; Duffin and Seifert, 2010).
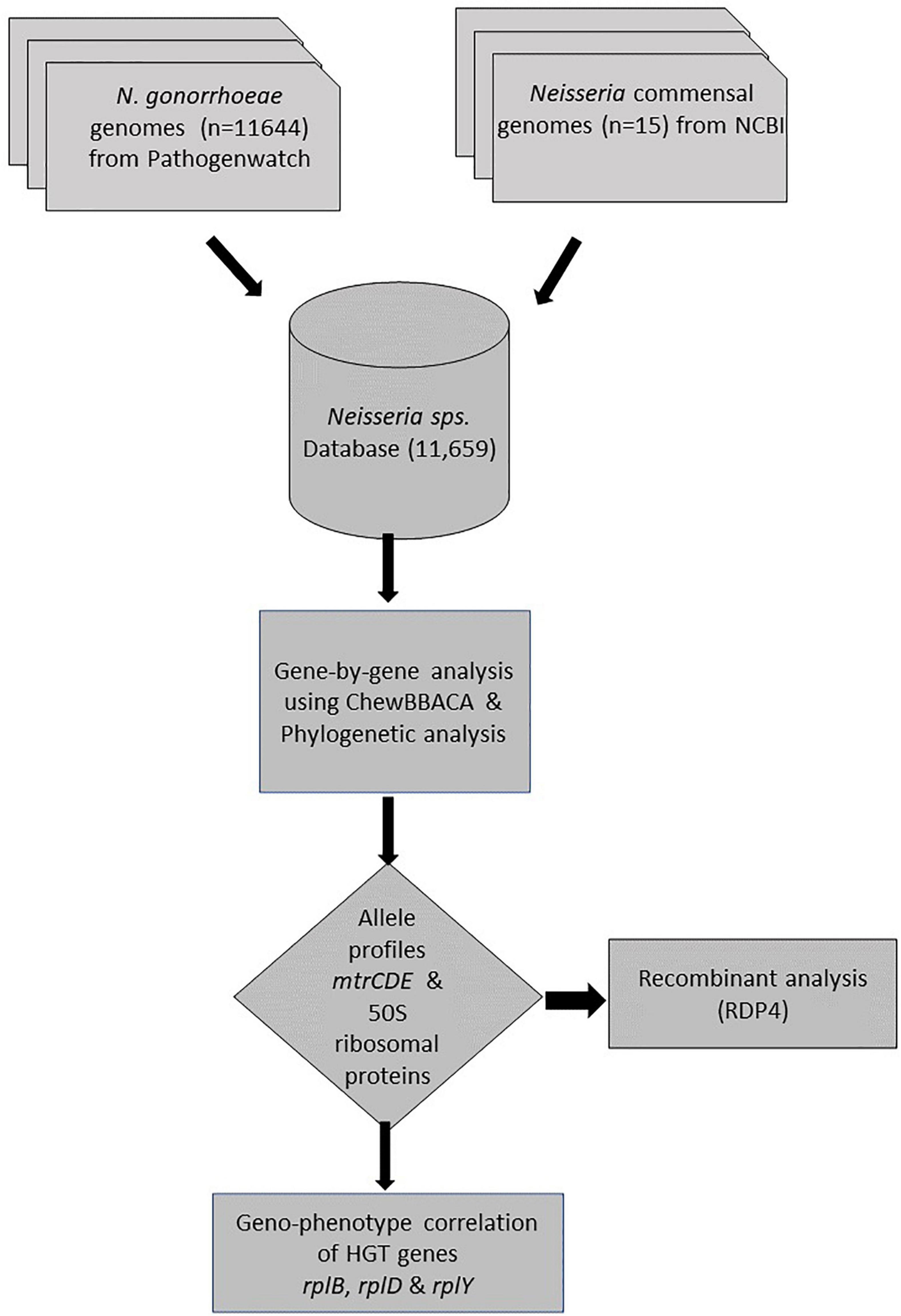
Both chromosomal mutations and HGT have been shown to play an important role in the genesis of aminocyclitol (spectinomycin), macrolide (azithromycin), and cephalosporins (ceftriaxone and cefixime) resistance (Ilina et al., 2013; Grad et al., 2016). HGT has been particularly important in the genesis of resistance at the rpsE, mtrCDE, and penA loci (Bowler et al., 1994; Ilina et al., 2013; Wadsworth et al., 2018; Yahara et al., 2018). We hypothesized that HGT in the 50S ribosomal genes might play a similar role in generating macrolide resistance in N. gonorrhoeae. Several studies suggest that ribosomal proteins that are both essential to function (e.g., L12 and L2) and not essential to function are amenable to HGT in other bacterial species (Weglöhner et al., 1997; Ühlein et al., 1998; Garcia-Vallvé et al., 2002; Miyazaki and Tomariguchi, 2019). As such, we assessed if there was any evidence of HGT in the gonococcal 50S ribosomal genes in N. gonorrhoeae. We considered all N. gonorrhoeae isolates (11,659 genomes available from Pathogenwatch at the time of data collection during April 2020). We set-out by analyzing these species that may have undergone HGT in the 50S ribosomal genes with the Neisseria commensal species (N. cinerea, N. elongata, N. flavescens, N. mucosa, N. polysaccharea, and N. subflava).
Materials and Methods
Data Extraction
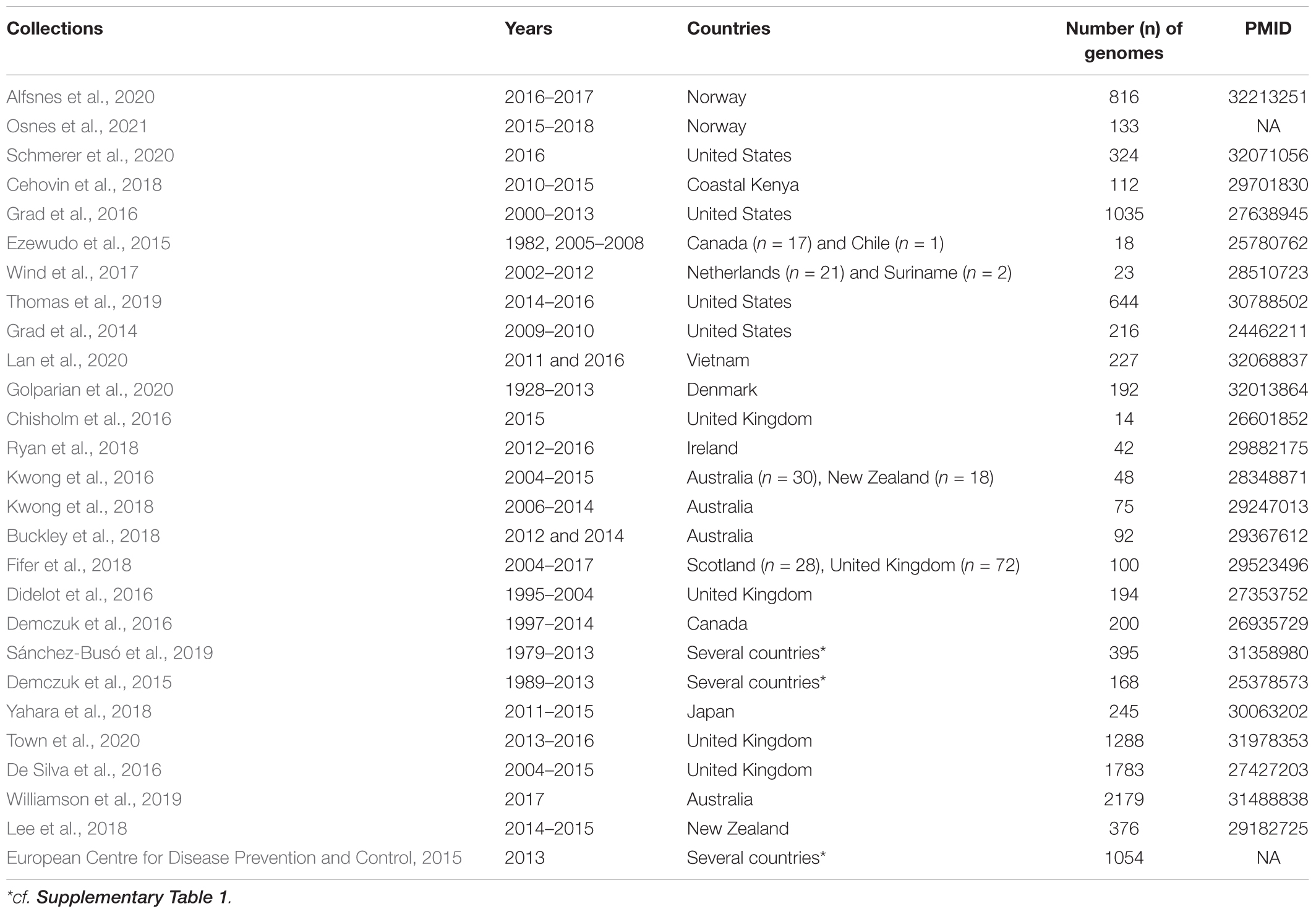
As of April 2020, all 27 collections of N. gonorrhoeae whole genome sequences (WGS) that were available from the global platform for genomic surveillance of microbial pathogens, Pathogenwatch1, were downloaded. These comprised a total of 11,993 genomes. Plausibility checks were carried out to remove duplicates (n = 349), and in total, 11,659 WGS [N. gonorrhoeae (n = 11,644) from pathogenwatch and commensal Neisseria spp. (n = 15) from NCBI (GenBank)] were used in the study (Tables 1, 2 and Supplementary Table 1). The following outcomes were extracted from the metadata: azithromycin MIC, azithromycin SIR (susceptible/intermediate/resistant), collection label, country, genogroup, MLST, NG-STAR type, SNPs (23S and mtrR), and year. When the azithromycin MIC was only reported as S (susceptible) or R (resistant), we assigned an azithromycin value of 0.25 mg/L for S, and 2 mg/L for R isolates. Also, we defined isolates as azithromycin susceptible, intermediate, and resistant if their MICs were <0.5 mg/L, ≥0.5 and <1 mg/L, and ≥1 mg/L, respectively. The isolates were divided into three distinct eras: “pre-antibiotic” (pre-1950s), “golden” (1950–1970s), and “post-modern” (1980-twenty-first century) following the topology of Golparian et al., 2020.
Gene by Gene Analysis
A gene-by-gene approach on whole-genome sequences (WGS; n = 11659) followed by recombination analysis was carried out (Figure 1). A study-specific Neisseria scheme was created from 35 complete Neisseria genomes [N. gonorrhoeae reference genomes (n = 20) and commensal Neisseria sps. (n = 15), Table 2] using the Blast Score Ratio Based Allele Calling Algorithm (chewBBACA; Silva et al., 2018). The complete genome of N. gonorrhoeae FA1090 was used to create a training file using Prodigal, which was used in subsequent steps (Hyatt et al., 2010). Firstly, coding sequences (CDS) for each genome were defined and compared in a pairwise manner to generate a single FASTA file containing the selected CDS. Secondly, AlleleCall implemented in chewBBACCA was carried out and followed by filtering out paralogous alleles, thus creating the wgMLST profiles used to define the cgMLST profiles. The SchemaEvaluator option allows for multiple sequence alignments of the alleles of each locus using MAFFT (Katoh et al., 2002) and the construction of a neighbor-joining tree using ClustalW2 (Larkin et al., 2007). The UniProtFinder2 was used to retrieve the functional information of the CDS. 50S ribosomal and mtrCDE genes were selected based on the UniProt identifier, and the corresponding multiple sequence alignments of the alleles and neighbor-joining trees were extracted from the schema evaluator. The trees and the corresponding metadata were visualized using microreact (Argimón et al., 2016).
Allelic Profiling
Alleles were identified for the ribosomal genes. The FASTA files of the ribosomal genes that were generated from cgMLST were used as input sequences in MEGAX (Kumar et al., 2018). The CDS were translated, and all the variable amino acid sites that were singleton or parsimony-informative were exported. If a non-synonymous substitution (variant) site was present only in a commensal, it was excluded from variant calling and further analysis. The presence or absence of a variant for each site was denoted as “0” and “1,” respectively. The geometric mean (GM) AZM MIC was calculated for each allele.
Recombination Analysis
To identify the donors of mutation-harboring ribosomal genes (n = 23) in N. gonorrhoeae isolates, complete genomes of commensal Neisseria spp., including N. cinerea, N. elongata, N. flavescens, N. lactamica, N. mucosa, N. polysaccharea, and N. subflava were analyzed. The nucleotide alignment of each ribosomal gene derived from cgMLST were screened for the presence of recombination events using the Recombination Detection Program (RDP4) program (Martin et al., 2015). Recombinant events supported by at least two of the seven algorithms implemented in RDP software: RDP, GENECONV, Bootscan, Maxchi, Chimera, SiSscan, and 3Seq were used with default settings except for window size that was increased to 60 nt in RDP, to 120 in MaxChi and Chimera, and to 500 in BootScan and SiScan (Lemey et al., 2009; Gomez-Valero et al., 2011). Neighbor-joining phylogenetic trees were constructed using 1,000 bootstrap replicates (Salminen et al., 1995). We defined the minor parent as the one contributing the smaller fraction of the recombinant, and the major parent as the one contributing the larger fraction of the recombinant (Martin et al., 2015). Additionally, sequences in the order of 5’ to 3’: 100 bp upstream of rplD (621 bp), rplW (321 bp), rplB (834 bp) and 100 bp downstream of rplB (length −1,971 nt) from Neisseria commensals (n = 15) and subset of N. gonorrhoeae sequences, ECDC isolates (n = 1,054) were aligned and followed by recombination analysis using RDP4 program.
Statistical Analysis
Statistical analyses were performed using JMP Pro V14.0.0 (SAS Institute, NC, United States) and XLSTAT (Statistical and data analysis solution, New York, United States). MIC variables were log2-transformed for use as continuous outcome variables (Ma et al., 2020). Differences between variant and wildtype were analyzed by the Mann–Whitney test. Only mutations present in >10 isolates were used in these analyses. To evaluate the correlation between multiple pairs of variables (SNPs) and to assess the correlations between phenotypic and genotypic patterns of resistance to azithromycin, statistical correlation tests including Kendall’s tau-b and Spearman’s rank correlation were used (Elhadidy et al., 2020). Differences in categorical variables were evaluated by Chi-square/Fischer exact tests. A p-value of <0.001 was considered statistically significant.
Results
Neisseria gonorrhoeae Isolates Used in This Study
A total of 11,993 N. gonorrhoeae genome sequences were available at Pathogenwatch. Of these, 349 were duplicates represented in more than one study, and thus 11,644 unique N. gonorrhoeae genomes were included in the study. The isolates were from 68 countries (Supplementary Figure 1A) and the period of sampling spanned over 90 years (Supplementary Figure 1B). A total of 456 different sequence types (STs) characterized the available population. The most frequent ST was ST1901 (n = 1,549). Twenty-nine AZM high-level resistant (AZM-HLR, MIC ≥ 256 mg/L) isolates were detected and belonged to the following ST types: 1,579 (n = 2), 1,580 (n = 8), 1,901 (n = 7), 7,822 (n = 3), 7,823 (n = 1), 9,363 (n = 4), 10,314 (n = 1), 10,899 (n = 1), 10,931 (n = 1), and 12,039 (n = 1). Further details pertaining to distribution of MLST and AZM MICs are provided in Supplementary Table 2.
Prevalence of Known RAMs
The MICs of azithromycin and the molecular determinants of known RAMs of the N. gonorrhoeae isolates analyzed in this study are summarized in Figure 2. Of the 11,644 N. gonorrhoeae isolates, AZM MIC value was not available for 2,520 (21.64%) isolates; 6,124 (52.59%) isolates were susceptible, 3,000 (25.76%) isolates were non-susceptible to azithromycin, including 1,593 (13.68%) resistant, and 1,407 (12.08%) intermediate isolates. The MIC’s of azithromycin ranged from 0.008 to 512 mg/L (Figure 2).
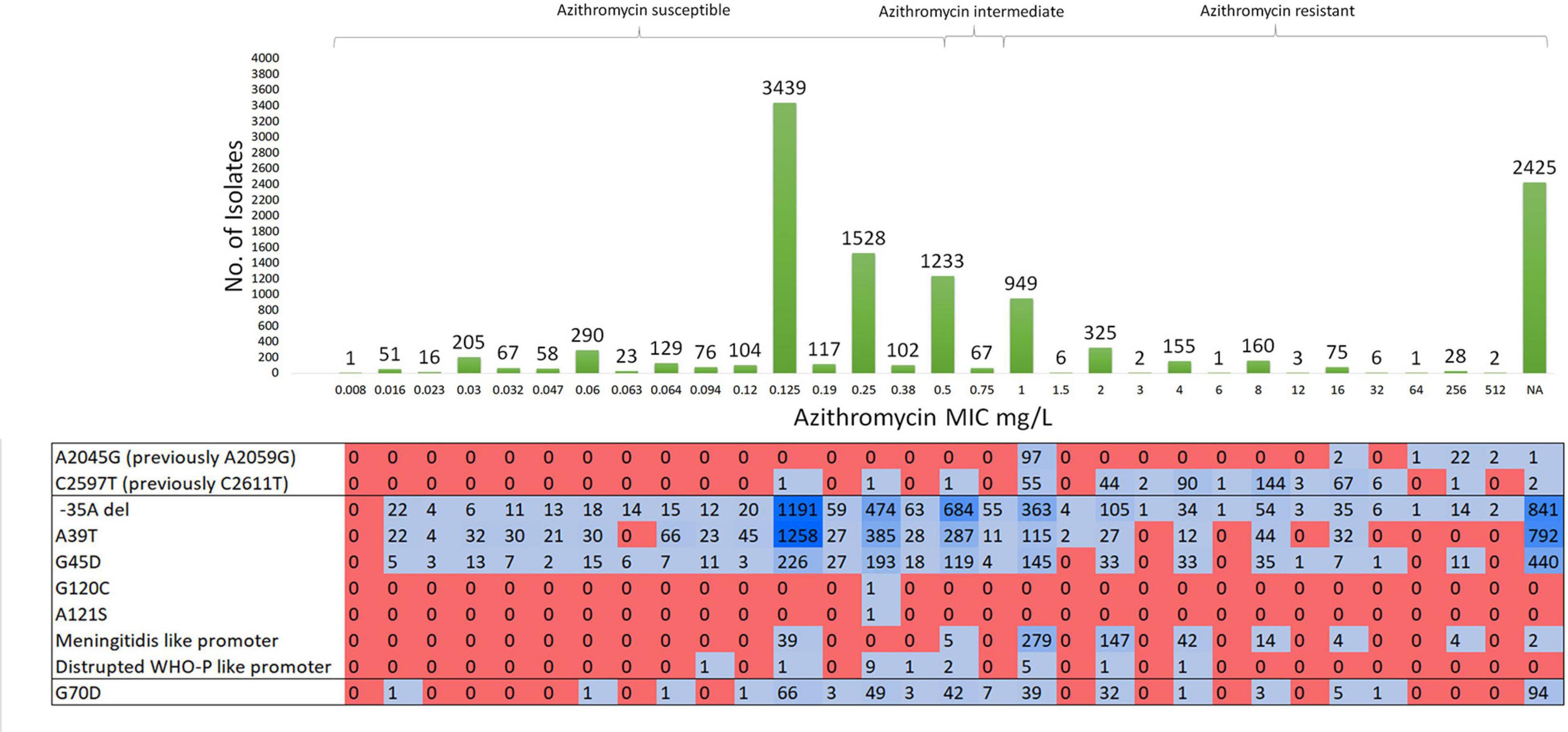
Figure 2. Molecular characterization of susceptible, intermediate-, and resistant-azithromycin N. gonorrhoeae isolates and their known resistance associated mutations. Total number of isolates – 11644.
Mutations in domain V of 23S rRNA were detected in 540 (4.63%) isolates. Azithromycin-resistant N. gonorrhoeae isolates contained A2045G (n = 124) and C2597T (n = 416) substitutions with MICs of azithromycin ranging from 1 to 512 mg/L. Of note, two isolates with C2597T mutation were susceptible to azithromycin with an MIC of 0.125 mg/L (ERR3325414) and 0.25 mg/L (SRR8560311).
Mutations in the mtrR promoter (-35A deletion) were present in 4,125 (35.42%) isolates. Of these, AZM MIC was not available for 835 (20.24%) isolates, and 1,934 (46.8%), 744 (18.03%), and 612 (14.83%) of these isolates were susceptible, intermediate, and resistant to AZM, respectively. Non-synonymous SNPs (A39T, G45D, G120C, and A121S) in MtrR were present in 4,660 (40.02%) isolates (2,509 azithromycin susceptible, 406 azithromycin intermediate, and 513 azithromycin resistant). The G120C and A121S amino acid substitutions of MtrR were each observed in only one susceptible isolate. N. meningitidis-like or WHO-P-like mtrR promoter mutations were present in 557 isolates with a high proportion of resistant isolates (n = 490) containing the N. meningitidis-like mtrR promoter mutation. Non-synonymous mutations in rplD at positions G68C/D/V and G70A/D/R/S were found in 2,760 N. gonorrhoeae isolates which were all significantly associated with higher AZM MICs (Supplementary Table 3 and Supplementary Figure 3).
Gene-by-Gene Analysis Reveals the Diversity of 50S Ribosomal Variants
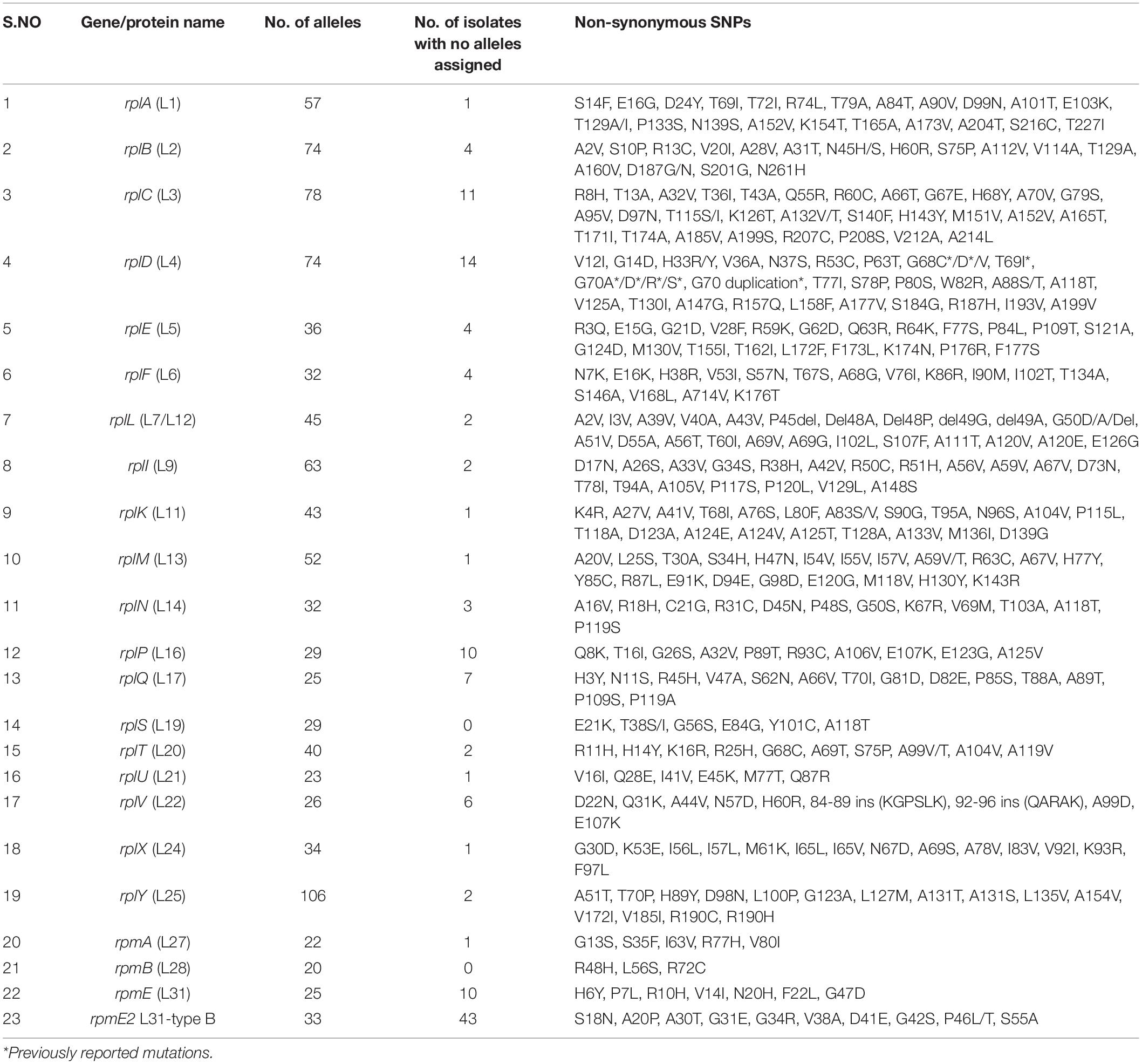
The 50S subunit of the N. gonorrhoeae genome (FA1090) consists of 36 ribosomal proteins, designated L1 to L36 (Figure 3C). Alleles were identified for the following 23 ribosomal genes: rplA, rplB, rplC, rplD, rplE, rplF, rplL, rplI, rplK, rplM, rplN, rplP, rplQ, rplS, rplT, rplV, rplU, rplX, rplY, rpmA, rpmB, rpmE, and rpmE2 which encodes L1, L2, L3, L4, L5, L6, L7/L12, L9, L11, L13, L14, L16, L17, L19, L20, L21, L22, L24, L25, L27, L28, L31, and L31-type B ribosomal proteins, respectively. A total of 336 non-synonymous mutations were determined for the 23 ribosomal genes (Table 3). A number of the newly described mutations had raised GM AZM MICs compared to the wild type: T129I mutation in L1 (rplA, n = 158, Supplementary Figure 3A), R157Q mutation in L4 (rplD, n = 2,335, Figure 4E), and A66V in L17 (rplQ, n = 47, Supplementary Figure 3B).
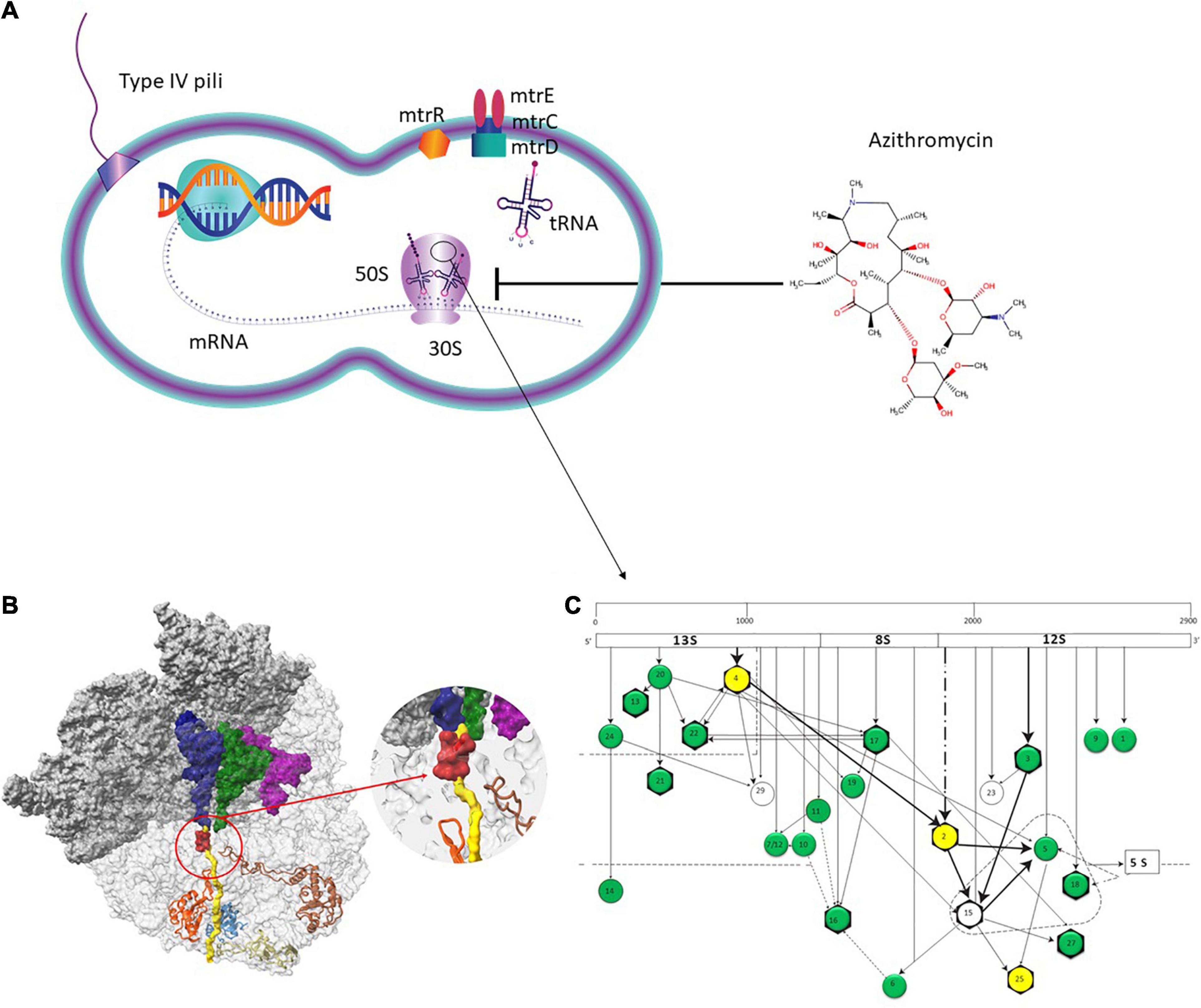
Figure 3. Azithromycin action pathway. (A) Azithromycin targets the protein synthesis of bacteria by binding to the 50S subunit of the bacterial ribosome near the polypeptide exit tunnel. Efflux pumps, such as MtrCDE protect N. gonorrhoeae from antimicrobials. (B) The ribosome nascent chain tunnel environment in Staphylococcus aureus. Left: The 70S S. aureus (SA_WT) ribosome where the large subunit is shown in light gray and the small subunit is shown in dark gray (PDBID 5TCU). The A-site, P-site, and E-site docked tRNA molecules (from PDBID 5JTE) are shown in blue, green, and magenta, respectively. The surface of a nascent chain within the peptide exit tunnel is shown in yellow. Bound macrolide (erythromycin) is shown in red, uL4, uL22, uL23, and uL24 are shown in brown, orange, teal, and khaki, respectively. Right: zoom into the macrolide binding site at the upper tunnel. Figure reproduced with permission from Halfon et al. (2019). This image is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International license (CC-BY-NC), a copy of which is available at https://creativecommons.org/licenses/by/4.0/. (C) Assembly map of 50S subunit of E. coli ribosomes. The main fragments of 23S rRNA (13S, 8S, and 12S) are indicated (top bar) as well as the ribosomal proteins (circles). The arrows denote the direction of the dependence of ribosomal proteins. Proteins in the dotted triangle are important for mediating the binding of 5S rRNA to 23S rRNA. Proteins colored in green and yellow were analyzed in the present study. Proteins with evidence of horizontal gene transfer (HGT) are colored in yellow. L31 and L31-Type B proteins are not depicted. Figure adapted with permission from Rohl and Nierhaus (1982).
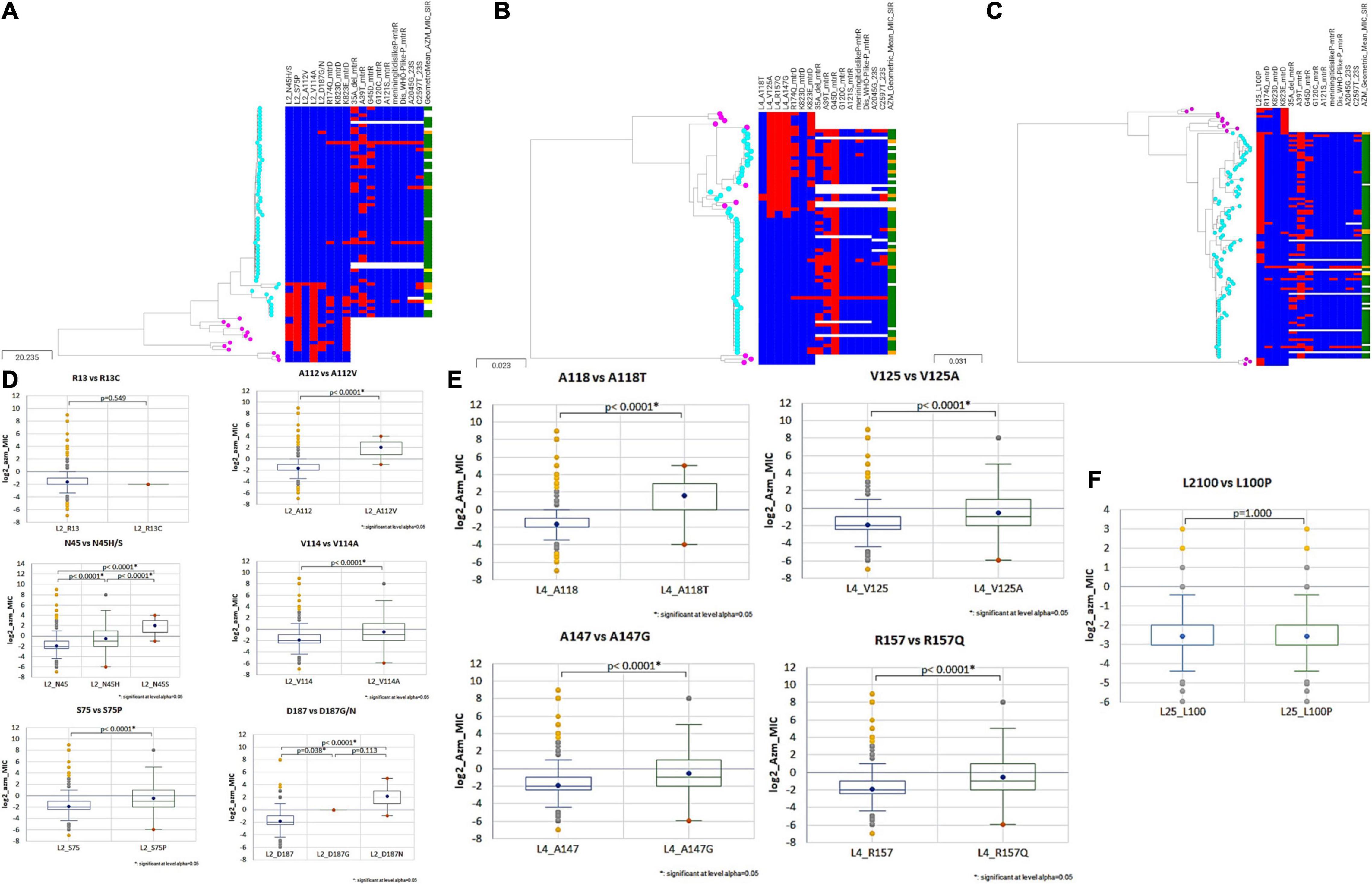
Figure 4. Phylogenetic trees of (A) rplB (B) rplD (C) rplY based on Neisseria sps. (N. gonorrhoeae, N. lactamica, N. subflava, N. cinerea, and N. mucosa) cgMLST allelic profiles determined by the chewBACCA software. The tree was created with Microreact. Cyan node denotes N. gonorrhoeae, and pink nodes denote Neisseria commensals. Blue and red color denote the absence and presence of non-synonymous SNPs, respectively. The geometric mean azithromycin MIC SIR profiles are denoted as green, yellow, and orange, respectively. (D–F) Statistical significance between non-synonymous SNPs and wildtype log2 AZM MIC distributions as assessed by Mann–Whitney U Test. The line inside the box marks the median. The upper and the lower hinges corresponds to the 25th and 75th percentiles. Statistical significance between variants and wildtype MIC distributions are depicted, p < 0.0001 in all cases, except rplA (R13) wildtype and variant (R13S), and rplY (L100) wildtype and variant (L100P).
The Occurrence of HGT in 50S Ribosomal Genes in N. gonorrhoeae and Association With Azithromycin MICs
Between 20 and 106 different alleles were found for each of the 23–50S ribosomal proteins median number of alleles 34 [IQR 26–57]. Probable HGT was predicted in three ribosomal genes rplB, rplD, and rplY. RplB, and rplD genes were assigned to 74 alleles, and the rplY gene was assigned to 106 alleles. Unique recombination events were supported by at least 2 out of 7 detection methods. For rplB we found that N. gonorrhoeae SRR8071393 (allele-71; n = 1) was recombinant, and the sequences with the same recombinant event were present in 2,280 N. gonorrhoeae isolates and had corresponding minor parent N. cinerea NZ_LS483369 (allele-59; n = 1; region – 12 to 486 bases) and major parents N. gonorrhoeae ERR439316, ERR439342 and ERR775150 (allele-41; n = 3; region – 1 to 11 and 487 to 834 nt), which were supported by 6 recombination methods (Table 4 and Supplementary Figure 2A). The gene, rplD had N. gonorrhoeae ECDC_ES052 (allele-12; n = 1) as the recombinant with the same recombination event present in N. gonorrhoeae (n = 2,320), N. mucosa (n = 2), N. lactamica (n = 3), and N. polysacchareae (n = 1). The minor and major parents were N. subflava NZ_CP031251 (allele-53; n = 1; region – 350 to 571 nt) and N. gonorrhoeae ECDC_GC_095 (allele-14; n = 1; region – 1 to 349 and 572–624 nt), respectively. This event was only supported by 2 methods (Table 4 and Supplementary Figure 2B). For rplY the recombinant event was supported by 4 methods, and the recombinant was N. gonorrhoeae ERR388419 (allele-59; n = 1) with the same recombinant event occurring in 10442, 5, and 2 N. gonorrhoeae, N. lactamica, and N. mucosa isolates, respectively. The corresponding minor parent was N. lactamica NZ_CP019894 (allele 82, n = 1; region – 222 to 370 nt), and the major parent was N. gonorrhoeae SRR5827245 (allele-102; n = 1; region – 1 – 221 and 371 – 573 nt; Table 4, Supplementary Figure 2C). The start and end breakpoints are provided in Table 4.
Additionally, recombination analysis were carried out for the closely linked rplD and rplB genes. We found two unique recombination events. The first event was supported by 6 out of 7 detection methods. The major and minor parent were N. gonorrhoeae ERR1528156 (region – 1 to 277 and 959 to 1,971 nt) and N. cinerea NZ_LS483369 (region – 278 to 958 nt), respectively, with N. gonorrhoeae ERR1560943 as the recombinant. The second event was supported by all the 7 methods. N. gonorrhoeae ERR155164 was the recombinant, and the sequences with the same recombinant event were present in 180 N. gonorrhoeae ECDC isolates and had corresponding minor parent N. gonorrhoeae ERR1528156 (region – 1 to 4 and 1,423 to 1,971 nt) and major parent N. lactamica NZ_LS483369 (region – 5 to 1,422 nt) and minor parent N. gonorrhoeae ERR1528156 (Figure 6). The non-synonymous SNPs in HGT regions of rplB, rplD, and rplY were further investigated to assess the population-wide prevalence and association with azithromycin MIC. Mutations were identified in 16 amino acid positions in L2, 5 in L4 and 1 in L25. The following mutations at amino acids positions R13C, N45H/S, S75P, A112V, V114A, A160V, and D187G/N for L2; A118T, V125A, T130I, A147G, and R157Q for L4; and L100P for L25 were analyzed. All the above mutations except L2 (R13C) and L25 (L100P) were associated with significantly higher azithromycin MICs (Figures 4D–F) across varied genetic backgrounds (Figures 4A–C). Some of these mutations were associated with established macrolide RAMs, meaning the association between the ribosomal protein mutations and elevated MICs may not be causal. Statistically significant associations were found between the presence of C2597T in 23S rRNA and mutations in L2 (N45H/S, S75P, A112V, and V114A) and L4 (A118T, V125A, A147G, and R157Q; all p < 0.001; Supplementary Tables 4, 5). Likewise, significant correlations were found between mutations in MtrR (-35A del, A39T, G45D) and both L2 (N45H/S, S75P, and V114A) and L4 (A118T, V125A, A147G, and R157Q, p < 0.001; Supplementary Tables 4, 5). Alignments of L2, L4, and L25 alleles are provided as Supplementary Tables 6–8.
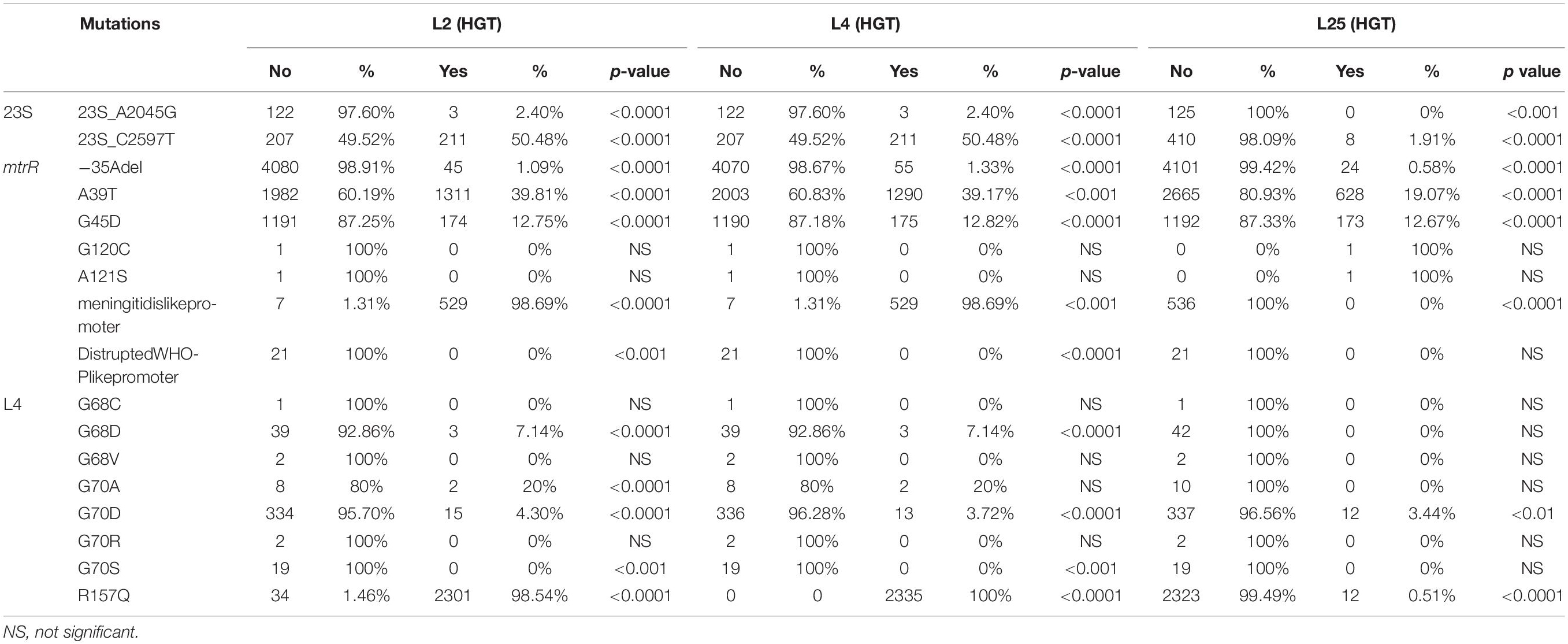
Table 5. Overview of the number of isolates with L2, L4, and L25 HGT events and known AMR mutations.
Distribution of Horizontally Acquired Ribosomal Genes
A total of 66 (0.56%), 59 (0.5%), and 11,432 (98.17%) isolates belonged to pre-antibiotic (pre-1950s), golden (1950–1970s), and post-modern era (1980s to 21st century), respectively. Eighty-seven (0.74%) isolates did not have any information on the isolation year. Of these, AZM MICs were available for 49 isolates of the pre-antibiotic and golden era and 8988 isolates from the post-modern era (Supplementary Table 9). The proportion of isolates with HGT in rplB increased from 7.5% in the pre-antibiotic era to 8.4% in golden era and 20.2% in post-modern era. A similar trend was seen for rplD with 1.5%/1.6% in the pre-antibiotic/golden eras to 20.3% in the post-modern era. The opposite trend was observed for isolates with HGT in rplY with 51%/50.8% in the pre-antibiotic/golden eras and 9.08% in postmodern era.
The first observation of gene transfer occurred in 1930 in 1 isolate involving the rplD gene, followed by rplB gene transfer in 1931 in one isolate and rplY gene transfer in 1932 in two isolates (Figure 5 and Supplementary Table 9). In total, 2337, 2355, and 1127 isolates had rplB, rplD, and rplY HGT events, respectively. Out of 68 countries from where the isolates were available, evidence of HGT in rplB, rplD, and rplY were observed in 46 countries.
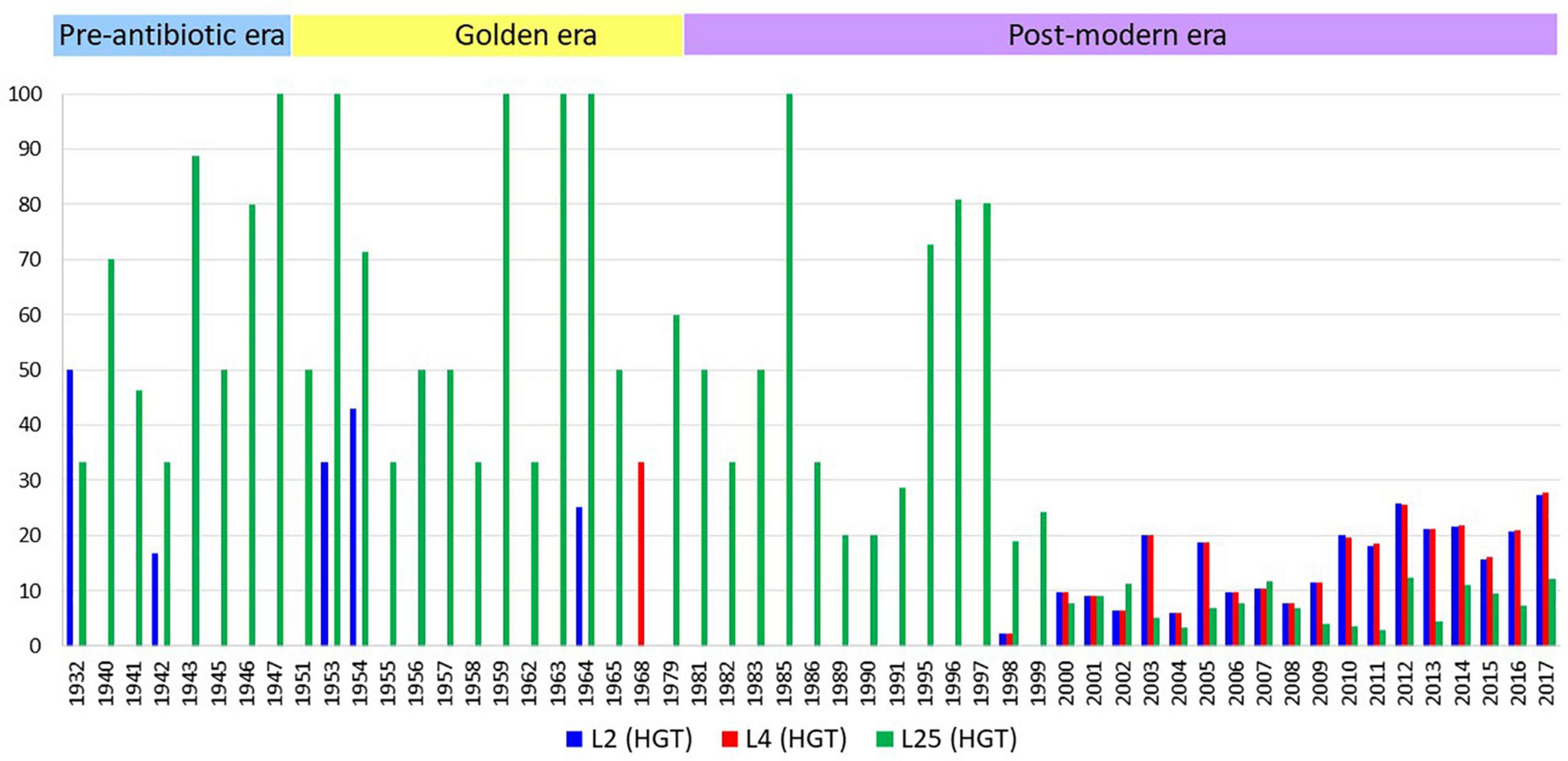
Figure 5. The number of isolates with HGT events per year. X-axis represents years, y-axis represents the number of isolates in percentage adjusted according to the total number of isolates by year. The number of isolates n = 1 and the years with no HGT events are not depicted.
There was a strong positive correlation between alleles with HGT in L2 and L4 (rho = 0.97; p < 0.0001) and a negative correlation between HGT in L25 and L2 (rho = -0.15 p < 0.0001), and L25 and L4 (rho = -0.15 p < 0.0001). The positive association between HGT in L2 and L4 was most noticeable in ST11428 where 100% of the isolates had HGT in both these loci (n = 384) and ST9363 where both L2/L4 HGT were present in 1,163 out of 1,169 isolates (99.5%; Supplementary Table 10). These two sequence types contained 66.4%/65.7% L2/L4 HGT events and had a negligible amount of L25 HGT event (ST9363 – 0.35% and ST11428 – 0%).
All isolates from ST7359 and ST1918 had L25 HGT alleles and together these sequence types comprised 531 out of 1127 L25 HGT alleles (47.1%). These sequence type had no isolates with HGT in L2 or L4. Among the ST9363 isolates (n = 1,169), 3 and 87 isolates had the 23S (A2045G) and 23S (C2597T) mutations, respectively. ST11428 (n = 384) and ST7359 (n = 417) isolates had 23S (C2597T) mutation in 3 and 1 isolates, respectively. Out of 418 isolates, 211 (50.4%) isolates from these two sequence types with the C2597T mutation had L2 or L4 HGT alleles.
Interestingly, almost all of the isolates with HGT in L2 (2301/2335; 98.5%) and L4 (2335/2335; 100%) had the L4 (R157Q) mutation, whereas only 0.5% of isolates with L25 HGT had this L4 mutation (Table 5). The first observation of an isolate with a combined L2 and L4 HGT event was in the year 2001 (ERR1082180, ST1580, AZM MIC – 1 mg/L). This isolate also had 23S (C2597T) and mtrR (G45D) mutations but no L25 HGT event.
Discussion
We found evidence of HGT in three gonococcal 50S ribosomal genes rplB, rplD, and rplY. Our analyses suggest that the horizontally transferred DNA was acquired from three commensal Neisseria species: N. cinerea, N. subflava, and N. lactamica. In Neisseria, the genetic organization of 50S ribosomal protein in the order from 5’ to 3’ is rplD (621 bp), rplW (321 bp) and rplB (834 bp). In order to discern the linkage between rplD and rplB about 1970 nt including rplD, rplW, rplB and 100 bp downstream and upstream of the genes were analyzed. Recombination with N. cinerea generated the rplD recombinant, followed by another recombination event with N. lactamica generating the rplB recombinant. The lengths of the recombined sequence were approximately 678 bases and 544 bases, respectively, which is within the recent estimate (2.5 kb) of the mean of the geometrically distributed DNA tract lengths transferred between donors and recipients in N. gonorrhoeae (Arnold et al., 2018; Yahara et al., 2021). It should be noted that the AT-DUS was present in N. cinerea NZ_LS483369 at 815 bp upstream of rplD (Figure 6).
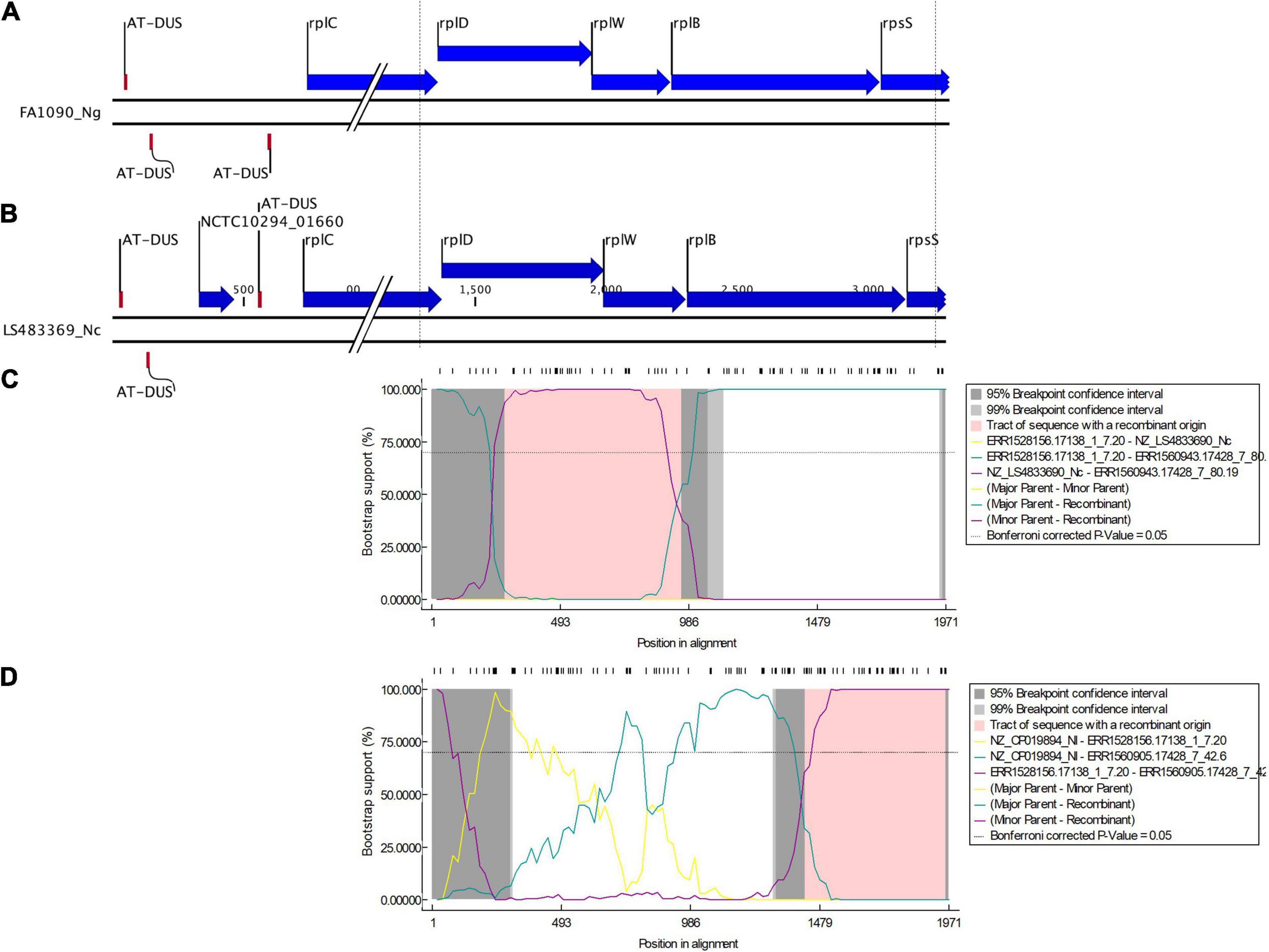
Figure 6. Genetic organization of 50S ribosomal protein in the order from 5’ to 3’: rplD (621 bp), rplW (321 bp), and rplB (834 bp) of (A) N. gonorrhoeae (FA1090) and (B) N. cinerea (LS483369). Dotted lines denote the region of sequence used in the multiple alignment. DNA uptake sequence (DUS), here AT-DUS (56’-ATGCCGTCTGAA-3’) is depicted. Bootscan analysis (by RDP4) of the sequences of ECDC (n = 1054) and Neisseria commensals (n = 15). The X axis shows the nucleotide position number and Y axis shows the bootstrap support. The dashed line denotes a bootstrap cutoff of 70% (C) a recombination event starting at position 278 and ending at position 958 was predicted with N. gonorrhoeae ERR1528156 being the major parent isolate and N. cinerea NZ_LS483369 being the minor parent isolate (D) a recombination event starting at position 1423 and ending at position 1971 was predicted with N. lactamica NZ_LS483369 being the major parent isolate and N. gonorrhoeae ERR1528156 being the minor parent isolate.
The transferred DNA contained mutations associated with reduced susceptibility to azithromycin: 7, 5 and 1 mutations in rplB, rplD, and rplY, respectively. These mutations were, however, also associated with other mutations that are well established determinants of macrolide resistance. Confounding could thus explain the association we observed between these novel mutations in rplB, rplD, rplY, and azithromycin MIC. It is also possible that these associations could be explained by the ribosomal mutations compensating for the fitness costs of the RAMs such as those in the 23S rRNA (Melnyk et al., 2015).
The strong association between HGT in L2/L4 and the 23S C2597T mutation could be explained by such a fitness restoring effect. Of note, studies of clinical isolates of Mycoplasma genitalium have found associations between the presence of mutations in L4 and L22 and macrolide resistance conferring mutations in 23S rRNA (Cao et al., 2010; Jensen et al., 2014). Likewise, mutations in the S5 and S12 ribosomal protein can confer resistance to spectinomycin and streptomycin, respectively, via structural changes in its rRNA target (Allen and Noller, 1989; Ilina et al., 2013). Mutations in L2 in Bacillus subtilis have also been found to confer resistance to bactobolin via altering its interaction with the 23S rRNA (Chandler et al., 2012). Whilst these hypotheses will require experimental validation, recent studies have illustrated the complexity of the effects of ribosomal mutations on bacterial phenotypes. Multiple studies have, for example, found various mutations in L4 and L22 in a range of bacteria, including N. gonorrhoeae, leading to macrolide resistance (Gregory and Dahlberg, 1999; Tait-Kamradt et al., 2000; Ma et al., 2020). Many of these studies have assumed that these mutations reduce macrolide susceptibility by narrowing the peptide exit tunnel so that macrolides are not able to bind to the 50S ribosomal subunit (Figures 3A,B) (Moore and Sauer, 2008). However, further experimental evidence in Escherichia coli showed that the mechanism was not simple blockage of the exit-tunnel but rather due to reduced intracellular erythromycin concentration via increased expression of AcrAB-TolC efflux pumps. This effect was, in turn, thought to be mediated by increased translation of the relevant mRNAs by reducing programmed ribosome stalling. Similarly, in Mycobacterium smegmatis, antibiotic-induced mutations in four ribosomal proteins resulted in large alterations in the transcriptome and proteome, which facilitated the acquisition of mutations in other genes that in turn conferred higher-level resistance (Gomez et al., 2017). These findings suggest that mutations in the ribosomal proteins and their interactions with mutations in other genes (such as the 23S rRNA) may be more important and complex than previously appreciated.
Previous studies in other organisms have found evidence of HGT in the ribosomal proteins, but in all cases, this involved uptake of the entire ribosomal gene (Makarova et al., 2001; Garcia-Vallvé et al., 2002). HGT-mediated chimerization has been previously reported for 16S rRNAs but not for ribosomal proteins (Miyazaki and Tomariguchi, 2019). To the best of our knowledge, this is the first study showing chimerization of ribosomal proteins. This finding provides further support for Miyazaki and Tomariguchi’s “cradle model” of ribosomal evolution which posits that the functionally essential interactions between rRNA and ribosomal proteins (the cradle) were locked-in early on in ribosomal mutation and therefore amenable to HGT events (Miyazaki and Tomariguchi, 2019). Gonococci have a fundamentally non-clonal population structure due to high rates of intraspecies HGT (O’Rourke and Stevens, 1993). In addition, HGT from a number of commensal species has led to mosaicism of a number of gonococcal and meningococcal genes such as penA and mtrCDE (Bowler et al., 1994; Wadsworth et al., 2018; Yahara et al., 2018). HGT in rplB, rplD, and rplY genes was first detected between 1930 and 1932, which means the original events cannot plausibly be linked with antimicrobial pressure. In fact, the prevalence of L25 HGT alleles declined over time. The increase in the prevalence of isolates with L2/L4 HGT over time could, however, be explained by antimicrobial pressure.
An important limitation of our study is that it only included 15 commensal Neisseria isolates and no N. meningitidis isolates. The inclusion of a greater number of commensals would be expected to increase the probability of detecting HGT events. Moreover, HGT events that resulted in no or few nucleotide changes would not have been detected by our methodology (Nielsen et al., 2013; Overballe-Petersen et al., 2013). Our methodology also did not allow us to confirm what phenotypic effects the variations in ribosomal proteins had. Nevertheless, we were able to characterize the extensive allelic variations present in Neisserial ribosomal proteins. For the first time, we also established that HGT can contribute to this variation via chimerization of ribosomal proteins between different Neisseria species.
Data Availability Statement
The data we used is publicly available from pathogenwatch and NCBI. The datasets analyzed for this study can be found in the microreact link: https://microreact.org/project/ucwXprtUMAAxXzKyVJwd72, https://microreact.org/project/dxnKsuFhRSAf7BRL6U1cZ3, https://microreact.org/project/hTZbP4fVvNSu3QWLbtaCXf.
Author Contributions
SSM-B and CK conceptualized the study, interpreted the data, performed the statistical analysis, and wrote the first draft. SSM-B was responsible for data collection, bioinformatic analysis, and Figure 3A. All the authors read and approved the final draft.
Funding
The study was funded by SOFI 2021 grant – “PReventing the Emergence of untreatable STIs via radical Prevention” (PRESTIP).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.683901/full#supplementary-material
Supplementary Figure 1 | Global distribution of N. gonorrhoeae genomes used in the study. (A) Distribution by country. (B) Distribution by year.
Supplementary Figure 2 | Neighbor-joining trees of (A) rplB (Major parent: 1–11 nt and 487–834 nt; Minor parent: 12–486 nt; B) rplD (Major parent: 1–349 nt and 572–624 nt; Minor parent: 350–571 nt) and (C) rplY (Major parent: 1–221 nt and 371–573 nt; Minor parent: 222–370 nt) nucleotide regions inferred from the major (left) and minor regions (right), showing evidence for the recombination events. All branch lengths are drawn to a scale of nucleotide substitutions per site. The sequences marked with red, salmon red, blue and green colors indicate the recombinant, the sequences with same recombinant event, the minor parent and major parent, respectively. Ng, denotes N. gonorrhoeae; Nc, N. cinerea; Ne, N. elongata; Nf, N. flavescens; Nl, N. lactamica; Nmu, N. mucosa; Ns, N. subflava; and Np, N. polysaccharea.
Supplementary Figure 3 | Statistical significance between (A) L1 (T129I), (B) L17 (A66V), and (C) L4 (G70 D/A/R/S) and their respective wildtype log2 AZM MIC distributions as assessed by Mann–Whitney U Test. The line inside the box marks the median. The upper and the lower hinges corresponds to the 25th and 75th percentiles. Statistical significance between variants and wildtype MIC distributions are depicted, p < 0.0001 in all cases.
Supplementary Table 1 | Collections used in the study.
Supplementary Table 2 | MLST and azithromycin MIC distribution.
Supplementary Table 3 | Molecular characterization of susceptible, intermediate-, and resistant-azithromycin N. gonorrhoeae isolates with WT 23S rRNA (n = 9527).
Supplementary Table 4 | Multivariate correlation probability of horizontally acquired 50 ribosomal gene mutations and the known mutations.
Supplementary Table 5 | Kendall’s tau-b and Spearman’s rank correlations.
Supplementary Table 6 | Alignment of L2 alleles.
Supplementary Table 7 | Alignment of L4 alleles.
Supplementary Table 8 | Alignment of L25 alleles.
Supplementary Table 9 | Overview of the number of isolates, the period of isolation, and distribution of azithromycin MIC with L2, L4, and L25 HGT events.
Supplementary Table 10 | Overview of the number of isolates and ST-types with L2, L4, and L25 HGT events.
Footnotes
References
Alfsnes, K., Eldholm, V., Olsen, A. O., Brynildsrud, O. B., Bohlin, J., Steinbakk, M., et al. (2020). Genomic epidemiology and population structure of Neisseria gonorrhoeae in norway, 2016–2017. Microb. Genom. 6:e000359. doi: 10.1099/mgen.0.000359
Allen, P. N., and Noller, H. F. (1989). Mutations in ribosomal proteins S4 and S12 influence the higher order structure of 16 S ribosomal RNA. J. Mol. Biol. 208, 457–468. doi: 10.1016/0022-2836(89)90509-3
Ambur, O. H., Frye, S. A., and Tønjum, T. (2007). New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J. Bacteriol. 189, 2077–2085. doi: 10.1128/JB.01408-06
Argimón, S., Abudahab, K., Goater, R. J. E., Fedosejev, A., Bhai, J., Glasner, C., et al. (2016). Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom. 2:e000093. doi: 10.1099/mgen.0.000093
Arnold, B. J., Gutmann, M. U., Grad, Y. H., Sheppard, S. K., Corander, J., Lipsitch, M., et al. (2018). Weak epistasis may drive adaptation in recombining bacteria. Genetics 208, 1247–1260. doi: 10.1534/genetics.117.300662
Biswas, G. D., Sox, T., Blackman, E., and Sparling, P. F. (1977). Factors affecting genetic transformation of Neisseria gonorrhoeae. J. Bacteriol. 129, 983–992. doi: 10.1128/jb.129.2.983-992.1977
Bowler, L. D., Zhang, Q. Y., Riou, J. Y., and Spratt, B. G. (1994). Interspecies recombination between the penA genes of Neisseria meningitidis and commensal Neisseria species during the emergence of penicillin resistance in N. meningitidis: natural events and laboratory simulation. J. Bacteriol. 176, 333–337. doi: 10.1128/jb.176.2.333-337.1994
Buckley, C., Forde, B. M., Trembizki, E., Lahra, M. M., Beatson, S. A., and Whiley, D. M. (2018). Use of whole genome sequencing to investigate an increase in Neisseria gonorrhoeae infection among women in urban areas of Australia. Sci. Rep. 8:1503. doi: 10.1038/s41598-018-20015-x
Cao, B., Zhao, C. J., Yin, Y. D., Zhao, F., Song, S. F., Bai, L., et al. (2010). High prevalence of macrolide resistance in mycoplasma pneumoniae isolates from adult and adolescent patients with respiratory tract infection in China. Clin. Infect. Dis. 51, 189–194. doi: 10.1086/653535
Cehovin, A., Harrison, O. B., Lewis, S. B., Ward, P. N., Ngetsa, C., Graham, S. M., et al. (2018). Identification of novel Neisseria gonorrhoeae lineages harboring resistance plasmids in coastal Kenya. J. Infect. Dis. 218, 801–808. doi: 10.1093/infdis/jiy240
Champney, W. S., and Miller, M. (2002). Inhibition of 50S ribosomal subunit assembly in Haemophilus influenzae cells by azithromycin and erythromycin. Curr. Microbiol. 44, 418–424. doi: 10.1007/s00284-001-0016-6
Chandler, J. R., Truong, T. T., Silva, P. M., Seyedsayamdost, M. R., Carr, G., Radey, M., et al. (2012). Bactobolin resistance is conferred by mutations in the L2 ribosomal protein. MBio 3:e00499-12. doi: 10.1128/mBio.00499-12
Chen, I., and Dubnau, D. (2004). DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2, 241–249. doi: 10.1038/nrmicro844
Chisholm, S. A., Dave, J., and Ison, C. A. (2010). High-level azithromycin resistance occurs in Neisseria gonorrhoeae as a result of a single point mutation in the 23S rRNA genes. Antimicrob. Agents Chemother. 54, 3812–3816. doi: 10.1128/AAC.00309-10
Chisholm, S. A., Wilson, J., Alexander, S., Tripodo, F., Al-Shahib, A., Schaefer, U., et al. (2016). An outbreak of high-level azithromycin resistant Neisseria gonorrhoeae in England. Sex. Transm. Infect. 92, 365–367. doi: 10.1136/sextrans-2015-052312
De Silva, D., Peters, J., Cole, K., Cole, M. J., Cresswell, F., Dean, G., et al. (2016). Whole-genome sequencing to determine transmission of Neisseria gonorrhoeae: an observational study. Lancet Infect. Dis. 16, 1295–1303. doi: 10.1016/S1473-3099(16)30157-8
Demczuk, W., Lynch, T., Martin, I., Van Domselaar, G., Graham, M., Bharat, A., et al. (2015). Whole-genome phylogenomic heterogeneity of Neisseria gonorrhoeae isolates with decreased cephalosporin susceptibility collected in Canada between 1989 and 2013. J. Clin. Microbiol. 53, 191–200. doi: 10.1128/JCM.02589-14
Demczuk, W., Martin, I., Peterson, S., Bharat, A., Van Domselaar, G., Graham, M., et al. (2016). Genomic epidemiology and molecular resistance mechanisms of azithromycin-resistant Neisseria gonorrhoeae in Canada from 1997 to 2014. J. Clin. Microbiol. 54, 1304–1313. doi: 10.1128/JCM.03195-15
Didelot, X., Dordel, J., Whittles, L. K., Collins, C., Bilek, N., Bishop, C. J., et al. (2016). Genomic analysis and comparison of two gonorrhea outbreaks. MBio 7:e00525-16. doi: 10.1128/mBio.00525-16
Diner, E. J., and Hayes, C. S. (2009). Recombineering reveals a diverse collection of ribosomal proteins L4 and L22 that confer resistance to macrolide antibiotics. J. Mol. Biol. 386, 300–315. doi: 10.1016/j.jmb.2008.12.064
Douthwaite, S., and Champney, W. S. (2001). Structures of ketolides and macrolides determine their mode of interaction with the ribosomal target site. J. Antimicrob. Chemother. 48, 1–8. doi: 10.1093/jac/48.suppl_2.1
Duffin, P. M., and Seifert, H. S. (2010). DNA uptake sequence-mediated enhancement of transformation in Neisseria gonorrhoeae is strain dependent. J. Bacteriol. 192, 4436–4444. doi: 10.1128/JB.00442-10
Elhadidy, M., Ali, M. M., El-Shibiny, A., Miller, W. G., Elkhatib, W. F., Botteldoorn, N., et al. (2020). Antimicrobial resistance patterns and molecular resistance markers of Campylobacter jejuni isolates from human diarrheal cases. PLoS One. 15:e0227833. doi: 10.1371/journal.pone.0227833
European Centre for Disease Prevention and Control (2015). Gonococcal antimicrobial susceptibility surveillance in Europe, 2013. Stockholm: ECDC.
Ezewudo, M. N., Joseph, S. J., Castillo-Ramirez, S., Dean, D., Del Rio, C., Didelot, X., et al. (2015). Population structure of Neisseria gonorrhoeae based on whole genome data and its relationship with antibiotic resistance. PeerJ 3:e806. doi: 10.7717/peerj.806
Fifer, H., Cole, M., Hughes, G., Padfield, S., Smolarchuk, C., Woodford, N., et al. (2018). Sustained transmission of high-level azithromycin-resistant Neisseria gonorrhoeae in England: an observational study. Lancet Infect. Dis. 18, 573–581. doi: 10.1016/S1473-3099(18)30122-1
Garcia-Vallvé, S., Simó, F. X., Montero, M. A., Arola, L., and Romeu, A. (2002). Simultaneous horizontal gene transfer of a gene coding for ribosomal protein L27 and operational genes in Arthrobacter sp. J. Mol. Evol. 55, 632–637. doi: 10.1007/s00239-002-2358-5
Golparian, D., Harris, S. R., Sánchez-Busó, L., Hoffmann, S., Shafer, W. M., Bentley, S. D., et al. (2020). Genomic evolution of Neisseria gonorrhoeae since the preantibiotic era (1928-2013): antimicrobial use/misuse selects for resistance and drives evolution. BMC Genomics 21:116. doi: 10.1186/s12864-020-6511-6
Golparian, D., Shafer, W. M., Ohnishi, M., and Unemo, M. (2014). Importance of multidrug efflux pumps in the antimicrobial resistance property of clinical multidrug-resistant isolates of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 58, 3556–3559. doi: 10.1128/AAC.00038-14
Gomez, J. E., Kaufmann-Malaga, B. B., Wivagg, C. N., Kim, P. B., Silvis, M. R., Renedo, N., et al. (2017). Ribosomal mutations promote the evolution of antibiotic resistance in a multidrug environment. eLife 6:e20420. doi: 10.7554/eLife.20420
Gomez-Valero, L., Rusniok, C., Jarraud, S., Vacherie, B., Rouy, Z., Barbe, V., et al. (2011). Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics 12:536. doi: 10.1186/1471-2164-12-536
Goodman, S. D., and Scocca, J. J. (1988). Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U.S.A. 85, 6982–6986. doi: 10.1073/pnas.85.18.6982
Grad, Y. H., Harris, S. R., Kirkcaldy, R. D., Green, A. G., Marks, D. S., Bentley, S. D., et al. (2016). Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000-2013. J. Infect. Dis. 214, 1579–1587. doi: 10.1093/infdis/jiw420
Grad, Y. H., Kirkcaldy, R. D., Trees, D., Dordel, J., Harris, S. R., Goldstein, E., et al. (2014). Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect. Dis. 14, 220–226. doi: 10.1016/S1473-3099(13)70693-5
Gregory, S. T., and Dahlberg, A. E. (1999). Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J. Mol. Biol. 289, 827–834. doi: 10.1006/jmbi.1999.2839
Hagman, K. E., and Shafer, W. M. (1995). Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J. Bacteriol. 177, 4162–4165. doi: 10.1128/jb.177.14.4162-4165.1995
Halfon, Y., Matzov, D., Eyal, Z., Bashan, A., Zimmerman, E., Kjeldgaard, J., et al. (2019). Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant. Sci. Rep. 9:11460. doi: 10.1038/s41598-019-48019-1
Halling, S. M., and Jensen, A. E. (2006). Intrinsic and selected resistance to antibiotics binding the ribosome: analyses of Brucella 23S rrn, L4, L22, EF-Tu1, EF-Tu2, efflux and phylogenetic implications. BMC Microbiol. 6:84. doi: 10.1186/1471-2180-6-84
Hyatt, D., Chen, G. L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11:119. doi: 10.1186/1471-2105-11-119
Ilina, E. N., Malakhova, M. V., Bodoev, I. N., Oparina, N. Y., Filimonova, A. V., and Govorun, V. M. (2013). Mutation in ribosomal protein S5 leads to spectinomycin resistance in Neisseria gonorrhoeae. Front. Microbiol. 4:186. doi: 10.3389/fmicb.2013.00186
Jensen, J. S., Fernandes, P., and Unemo, M. (2014). In vitro activity of the new fluoroketolide solithromycin (CEM-101) against macrolide-resistant and -susceptible Mycoplasma genitalium strains. Antimicrob. Agents Chemother. 58, 3151–3156. doi: 10.1128/AAC.02411-14
Katoh, K., Misawa, K., Kuma, K. I., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Koomey, M. (1998). Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal genetransfer. APMIS Suppl. 84, 6–61. doi: 10.1111/j.1600-0463.1998.tb05649.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kwong, J. C., Chow, E. P. F., Stevens, K., Stinear, T. P., Seemann, T., Fairley, C. K., et al. (2018). Whole-genome sequencing reveals transmission of gonococcal antibiotic resistance among men who have sex with men: an observational study. Sex. Transm. Infect. 94, 151–157. doi: 10.1136/sextrans-2017-053287
Kwong, J. C., Gonçalves da Silva, A., Dyet, K., Williamson, D. A., Stinear, T. P., Howden, B. P., et al. (2016). NGMASTER:in silico multi-antigen sequence typing for Neisseria gonorrhoeae. Microb. Genomics 2:e000076. doi: 10.1099/mgen.0.000076
Lan, P. T., Golparian, D., Ringlander, J., Van Hung, L., Van Thuong, N., and Unemo, M. (2020). Genomic analysis and antimicrobial resistance of Neisseria gonorrhoeae isolates from Vietnam in 2011 and 2015-16. J. Antimicrob. Chemother. 75, 1432–1438. doi: 10.1093/jac/dkaa040
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Laumen, J. G. E., Manoharan-Basil, S. S., Verhoeven, E., Abdellati, S., De Baetselier, I., Crucitti, T., et al. (2020). Molecular pathways to high-level azithromycin resistance in Neisseria gonorrhoeae. BioRxiv [Preprint] doi: 10.1101/2020.12.02.409193 2020.12.02.409193,
Lee, R. S., Seemann, T., Heffernan, H., Kwong, J. C., Gonçalves da Silva, A., Carter, G. P., et al. (2018). Genomic epidemiology and antimicrobial resistance of Neisseria gonorrhoeae in New Zealand. J. Antimicrob. Chemother. 73, 353–364. doi: 10.1093/jac/dkx405
Lemey, P., Salemi, M., and Vandamme, A. Eds (2009). The Phylogenetic Handbook: A Practical Approach to Phylogenetic Analysis and Hypothesis Testing. Cambridge: Cambridge University Press. doi: 10.5860/choice.47-1986
Ma, K. C., Mortimer, T. D., Duckett, M. A., Hicks, A. L., Wheeler, N. E., Sánchez-Busó, L., et al. (2020). Increased power from conditional bacterial genome-wide association identifies macrolide resistance mutations in Neisseria gonorrhoeae. Nat. Commun. 11:5374. doi: 10.1038/s41467-020-19250-6
Makarova, K. S., Ponomarev, V. A., and Koonin, E. V. (2001). Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2:research0033.1. doi: 10.1186/gb-2001-2-9-research0033
Martin, D. P., Murrell, B., Golden, M., Khoosal, A., and Muhire, B. (2015). RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. doi: 10.1093/ve/vev003
Martin, I., Sawatzky, P., Liu, G., Allen, V., Lefebvre, B., Hoang, L., et al. (2016). Decline in decreased cephalosporin susceptibility and increase in azithromycin resistance in Neisseria gonorrhoeae. Can. Emerg. Infect. Dis. 22, 65–67. doi: 10.3201/eid2201.151247
Melnyk, A. H., Wong, A., and Kassen, R. (2015). The fitness costs of antibiotic resistance mutations. Evol. Appl. 8, 273–283. doi: 10.1111/eva.12196
Miyazaki, K., and Tomariguchi, N. (2019). Occurrence of randomly recombined functional 16S rRNA genes in Thermus thermophilus suggests genetic interoperability and promiscuity of bacterial 16S rRNAs. Sci. Rep. 9:11233. doi: 10.1038/s41598-019-47807-z
Moore, S. D., and Sauer, R. T. (2008). Revisiting the mechanism of macrolide-antibiotic resistance mediated by ribosomal protein L22. Proc. Natl. Acad. Sci. U.S.A. 105, 18261–18266. doi: 10.1073/pnas.0810357105
Nielsen, K. M., Bøhn, T., and Townsend, J. P. (2013). Detecting rare gene transfer events in bacterial populations. Front. Microbiol. 4:415. doi: 10.3389/fmicb.2013.00415
O’Connor, M., Gregory, S. T., and Dahlberg, A. E. (2004). Multiple defects in translation associated with altered ribosomal protein L4. Nucleic Acids Res. 32, 5750–5756. doi: 10.1093/nar/gkh913
O’Rourke, M., and Stevens, E. (1993). Genetic structure of Neisseria gonorrhoeae populations: a non-clonal pathogen. J. Gen. Microbiol. 139, 2603–2611. doi: 10.1099/00221287-139-11-2603
Osnes, M. N., Dorp, L., van Brynildsrud, O. B., Alfsnes, K., Schneiders, T., Templeton, K. E., et al. (2021). Antibiotic treatment regimes as a driver of the global population dynamics of a major gonorrhea lineage. Mol. Biol. Evol. 38, 1249–1261. doi: 10.1093/molbev/msaa282
Overballe-Petersen, S., Harms, K., Orlando, L. A. A., Moreno Mayar, J. V., Rasmussen, S., Dahl, T. W., et al. (2013). Bacterial natural transformation by highly fragmented and damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 110, 19860–19865. doi: 10.1073/pnas.1315278110
Pham, C. D., Nash, E., Liu, H., Schmerer, M. W., Sharpe, S., Woods, G., et al. (2020). Atypical Mutation in Neisseria gonorrhoeae 23S rRNA Associated with High-Level Azithromycin resistance. Antimicrob. Agents Chemother. AAC.00885-20 65:e00885-20. doi: 10.1128/AAC.00885-20
Quillin, S. J., and Seifert, H. S. (2018). Neisseria gonorrhoeae host adaptation and pathogenesis. Nat. Rev. Microbiol. 16, 226–240. doi: 10.1038/nrmicro.2017.169
Roberts, M. C., Chung, W. O., Roe, D., Xia, M., Marquez, C., Borthagaray, G., et al. (1999). Erythromycin-resistant Neisseria gonorrhoeae and oral commensal Neisseria spp. carry known rRNA methylase genes. Antimicrob. Agents Chemother. 43, 1367–1372. doi: 10.1128/aac.43.6.1367
Rohl, R., and Nierhaus, K. H. (1982). Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc. Natl. Acad. Sci. U.S.A. 79, 729–733. doi: 10.1073/pnas.79.3.729
Ryan, L., Golparian, D., Fennelly, N., Rose, L., Walsh, P., Lawlor, B., et al. (2018). Antimicrobial resistance and molecular epidemiology using whole-genome sequencing of Neisseria gonorrhoeae in Ireland, 2014–2016: focus on extended-spectrum cephalosporins and azithromycin. Eur. J. Clin. Microbiol. Infect. Dis. 37, 1661–1672. doi: 10.1007/s10096-018-3296-5
Salminen, M. O., Carr, J. K., Burke, D. S., and Mccutchan, F. E. (1995). Identification of breakpoints in intergenotypic recombinants of HIV Type 1 by bootscanning. AIDS Res. Hum. Retroviruses. 11, 1423–1425. doi: 10.1089/aid.1995.11.1423
Sánchez-Busó, L., Golparian, D., Corander, J., Grad, Y. H., Ohnishi, M., Flemming, R., et al. (2019). The impact of antimicrobials on gonococcal evolution. Nat. Microbiol. 4, 1941–1950. doi: 10.1038/s41564-019-0501-y
Schmerer, M. W., Abrams, A. J., Seby, S., Thomas, J. C., Cartee, J., Lucking, S., et al. (2020). Genomic characterization of Neisseria gonorrhoeae strains from 2016 U.S. sentinel surveillance displaying reduced susceptibility to azithromycin. Antimicrob. Agents Chemother. 64:e02420-19. doi: 10.1128/AAC.02420-19
Shafer, W. M. (2018). Mosaic drug efflux gene sequences from commensal neisseria can lead to low-level azithromycin resistance expressed by Neisseria gonorrhoeae clinical isolates. MBio. 9, e01747–e01818. doi: 10.1128/mBio.01747-18
Silva, M., Machado, M. P., Silva, D. N., Rossi, M., Moran-Gilad, J., Santos, S., et al. (2018). chewBBACA: a complete suite for gene-by-gene schema creation and strain identification. Microb. genomics. 4:e000166. doi: 10.1099/mgen.0.000166
Smith, H. O., Gwinn, M. L., and Salzberg, S. L. (1999). DNA uptake signal sequences in naturally transformable bacteria. Res. Microbiol. 150, 603–616. doi: 10.1016/S0923-2508(99)00130-8
St Cyr, S., Barbee, L., Workowski, K. A., Bachmann, L. H., Pham, C., Schlanger, K., et al. (2020). Update to CDC’s treatment guidelines for gonococcal infection, 2020. MMWR. Morb. Mortal. Wkly. Rep. 69, 1911–1916. doi: 10.15585/mmwr.mm6950a6
Tait-Kamradt, A., Davies, T., Cronan, M., Jacobs, M. R., Appelbaum, P. C., and Sutcliffe, J. (2000). Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44, 2118–2125. doi: 10.1128/AAC.44.8.2118-2125.2000
Tapsall, J. W., Ndowa, F., Lewis, D. A., and Unemo, M. (2009). Meeting the public health challenge of multidrug- and extensively drug-resistant Neisseria gonorrhoeae. Exp. Rev. Anti. Infect. Ther. 7, 821–834. doi: 10.1586/ERI.09.63
Thomas, J. C., Seby, S., Abrams, A. J., Cartee, J., Lucking, S., Vidyaprakash, E., et al. (2019). Evidence of recent genomic evolution in gonococcal strains with decreased susceptibility to Cephalosporins or Azithromycin in the United States, 2014-2016. J. Infect. Dis. 220, 294–305. doi: 10.1093/infdis/jiz079
Town, K., Field, N., Harris, S. R., Sánchez-Busó, L., Cole, M. J., Pitt, R., et al. (2020). Phylogenomic analysis of Neisseria gonorrhoeae transmission to assess sexual mixing and HIV transmission risk in England: a cross-sectional, observational, whole-genome sequencing study. Lancet Infect. Dis. 20, 478–486. doi: 10.1016/S1473-3099(19)30610-3
Ühlein, M., Weglöhner, W., Urlaub, H., and Wotmann-Liebold, B. (1998). Functional implications of ribosomal protein L2 in protein biosynthesis as shown by in vivo replacement studies. Biochem. J. 331(Pt. 2), 423–430. doi: 10.1042/bj3310423
Unemo, M., Ross, J., Serwin, A. B., Gomberg, M., Cusini, M., and Jensen, J. S. (2020). 2020 European guideline for the diagnosis and treatment of gonorrhoea in adults. Int. J. STD AIDS 956462420949126. doi: 10.1177/0956462420949126 [Epub ahead of print].
Unemo, M., and Shafer, W. M. (2014). Antimicrobial resistance in Neisseria gonorrhoeae in the 21st Century: past, evolution, and future. Clin. Microbiol. Rev. 27, 587–613. doi: 10.1128/CMR.00010-14
Wadsworth, C. B., Arnold, B. J., Sater, M. R. A., and Grad, Y. H. (2018). Azithromycin resistance through interspecific acquisition of an epistasis-dependent efflux pump component and transcriptional regulator in Neisseria gonorrhoeae. MBio 9, e01419-18. doi: 10.1128/mBio.014
Wan, C., Li, Y., Le, W. J., Liu, Y. R., Li, S., Wang, B. X., et al. (2018). Increasing resistance to azithromycin in Neisseria gonorrhoeae in Eastern Chinese cities: resistance mechanisms and genetic diversity among isolates from Nanjing. Antimicrob. Agents Chemother. 62, e02499-17. doi: 10.1128/AAC.02499-17
Weglöhner, W., Jünemann, R., Von Knoblauchi, K., and Subramanian, A. R. (1997). Different consequences of incorporating chloroplast ribosomal proteins L12 and S18 into the bacterial ribosomes of Escherichia coli. Eur. J. Biochem. 249, 383–392. doi: 10.1111/j.1432-1033.1997.00383.x
Williamson, D. A., Chow, E. P. F., Gorrie, C. L., Seemann, T., Ingle, D. J., Higgins, N., et al. (2019). Bridging of Neisseria gonorrhoeae lineages across sexual networks in the HIV pre-exposure prophylaxis era. Nat. Commun. 10:3988. doi: 10.1038/s41467-019-12053-4
Wind, C. M., De Vries, E., Schim Van Der Loeff, M. F., Van Rooijen, M. S., Van Dam, A. P., Demczuk, W. H. B., et al. (2017). Decreased Azithromycin susceptibility of Neisseria gonorrhoeae isolates in patients recently treated with Azithromycin. Clin. Infect. Dis. 65, 37–45. doi: 10.1093/cid/cix249
Yahara, K., Ma, K. C., Mortimer, T. D., Shimuta, K., Nakayama, S., Hirabayashi, A., et al. (2021). Emergence and evolution of antimicrobial resistance genes and mutations in Neisseria gonorrhoeae. Genome Med. 13:51. doi: 10.1186/s13073-021-00860-8
Yahara, K., Nakayama, S. I., Shimuta, K., Lee, K. I., Morita, M., Kawahata, T., et al. (2018). Genomic surveillance of neisseria gonorrhoeae to investigate the distribution and evolution of antimicrobial-resistance determinants and lineages. Microb. Genom. 4:e000205. doi: 10.1099/mgen.0.000205
Keywords: 50S ribosomal proteins, azithromycin resistance, rplY (L25), rplD (L4), rplB (L2), HGT in Neisseria gonorrhoeae
Citation: Manoharan-Basil SS, Laumen JGE, Van Dijck C, De Block T, De Baetselier I and Kenyon C (2021) Evidence of Horizontal Gene Transfer of 50S Ribosomal Genes rplB, rplD, and rplY in Neisseria gonorrhoeae. Front. Microbiol. 12:683901. doi: 10.3389/fmicb.2021.683901
Received: 22 March 2021; Accepted: 27 April 2021;
Published: 10 June 2021.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Kentaro Miyazaki, National Institute of Advanced Industrial Science and Technology (AIST), JapanJoseph P. Dillard, University of Wisconsin-Madison, United States
Copyright © 2021 Manoharan-Basil, Laumen, Van Dijck, De Block, De Baetselier and Kenyon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheeba Santhini Manoharan-Basil, c2Jhc2lsQGl0Zy5iZQ==
 Sheeba Santhini Manoharan-Basil
Sheeba Santhini Manoharan-Basil Jolein Gyonne Elise Laumen
Jolein Gyonne Elise Laumen Christophe Van Dijck
Christophe Van Dijck Tessa De Block
Tessa De Block Irith De Baetselier1
Irith De Baetselier1