
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 21 July 2021
Sec. Fungi and Their Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.680060
Background: Stephanoascus ciferrii is a heterothallic ascomycetous yeast-like fungus. Recently, the concept of S. ciferrii complex has been proposed and it consists of S. ciferrii, Candida allociferrii, and Candida mucifera. We aimed to identify 32 strains of S. ciferrii complex isolated from patients with chronic suppurative otitis media (CSOM) at the species level and analyze the morphology and antifungal susceptibility profiles of the three species.
Method: The sequencing of the internal transcribed spacer (ITS) region and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) were used to identify S. ciferrii complex species. The SARAMIS software was used for cluster analysis of the mass spectra. All the strains were cultured on Sabouraud dextrose agar (SDA) and CHROM plates for 7 days. In the meantime, colonies of the 32 strains went through Gram staining. The Sensititre YeastOne YO10 colorimetric panel was used for the antifungal susceptibility analysis.
Results: There were 10 strains of C. allociferrii (31.25%), six strains of C. mucifera (18.75%), and 16 strains of S. ciferrii (50%) in the 32 strains of S. ciferrii complex according to the sequencing of the ITS region. MALDI-TOF MS could identify S. ciferrii but showed no results for C. allociferrii and C. mucifera. The cluster analysis of the mass spectra by SARAMIS indicated that the MALDI-TOF MS could distinguish the three species. The morphology characteristics of the three species were similar. As for antifungal susceptibility, S. ciferrii and C. mucifera tended to have high fluconazole MICs compared with C. allociferrii. C. mucifera and C. allociferrii had relatively low flucytosine MICs while S. ciferrii owned high flucytosine MICs. Besides, C. mucifera tended to have a higher MIC value than S. ciferrii for amphotericin B and C. allociferrii for anidulafungin, micafungin, and caspofungin.
Conclusion: The antifungal susceptibility profiles of the three species of S. ciferrii complex had their own characteristics. Besides, more mass spectra of C. allociferrii and C. mucifera are needed to construct the reference database for S. ciferrii complex species, enabling MALDI-TOF MS to identify S. ciferrii complex at species level.
Stephanoascus ciferrii (also called Candida ciferrii or Trichomonascus ciferrii) is a heterothallic ascomycetous yeast-like fungus, which is a teleomorph of Candida ciferrii (de Gentile et al., 1991; Warren et al., 2018). It has been reported to cause various human infections such as endophthalmitis, intraorbital abscess, systemic mycosis, and otitis media (Soki et al., 2010, 2015; Agin et al., 2011; Danielescu et al., 2017). Due to the development of molecular biology techniques for the identification of fungi, more and more species have been discovered. Recently, the concept of microbial complexes is applied. Microbial complex refers to a group of pathogens which are phenotypically indistinguishable but different at the genetic level, such as Acinetobacter calcoaceticus-baumannii complex and Candida parapsilosis complex (Mancilla-Rojano et al., 2020; da Silva et al., 2021). By sequencing of the 18S rRNA gene, Kumiko Ueda-Nishimura and Kozaburo Mikata divided S. ciferrii into three groups and proposed the S. ciferrii complex, which consists of S. ciferrii, Candida allociferrii, and Candida mucifera (Ueda-Nishimura and Mikata, 2002). Usually, the widely used VITEK 2 yeast identification system (bioMérieux, Marcy-l’Étoile, France) just identified the microorganism as S. ciferrii complex and it does not identify the complex at species level (Dave et al., 2018). Also, many clinicians do not distinguish the three species of S. ciferrii complex and they just take it as S. ciferrii. However, as for a rare kind of opportunistic pathogenic fungus, identifying S. ciferrii complex at species level is of great importance in epidemiological studies and the management of empirical antifungal therapy. Recently, matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) has revolutionized microbial diagnostics (Cuenod et al., 2021). Because of its minimal hands-on and turnaround time, low costs, and high accuracy, MALDI-TOF MS has been widely used in the identification of bacterial and fungi species clinically (Angeletti and Ciccozzi, 2019; Cuenod et al., 2021).
Chronic suppurative otitis media (CSOM) is a chronic inflammation, characterized by repeated otorrhea via tympanic membrane perforation and involving polymicrobial infection of the middle ear and mastoid cavity (Bhutta et al., 2020). Usually, the common pathogens causing CSOM are Staphylococcus aureus, Pseudomonas aeruginosa, and coagulase-negative Staphylococcus (Xu et al., 2020). However, the isolation rate of S. ciferrii complex has increased recently in our hospital. In our study, we collected 32 strains of S. ciferrii complex, which were isolated from patients with CSOM and identified by the VITEK 2 yeast identification system. The sequencing of the internal transcribed spacer (ITS) region was used to identify them at species level. Then, the morphology, antifungal susceptibility, and mass spectra from MALDI-TOF MS of the 32 strains of S. ciferrii complex were analyzed and compared, providing experimental evidence and clinical experience for the diagnosis and treatment of the rare fungi at species level.
Thirty-two yeast strains were isolated from the middle ear secretions of patients with CSOM at The First Affiliated Hospital of Sun Yat-sen University from May 2019 to February 2020 and identified as S. ciferrii complex by the VITEK 2 yeast identification system (bioMérieux, France). Our study was approved by the institutional review board of The First Affiliated Hospital.
The strains were cultured at 28°C with shaking in test tubes containing 5 ml YPD broth overnight. One milliliter of overnight yeast culture was centrifuged, and yeast precipitations were collected for the DNA extraction. The Yeast Genomic DNA Rapid Extraction Kit (Sangon Biotech, Shanghai, China) was used for the DNA extraction of collected yeast strains. All experimental operations and experimental conditions were performed according to the manufacturer’s instructions. The sequences of the ITS region rDNA were amplified using the universal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Al Halteet et al., 2020).
The PCR amplification was conducted in a 50-μl reaction mixture containing 10 μl 10 × PCR buffer, 5-μl templates, 1 μl ITS1 primer, 1 μl ITS4 primer, 0.5 μl Taq enzyme, 8 μl dNTP mixture, and 24.5 μl double-distilled water. The PCR amplification condition is as follows: 1 cycle of 95°C 10 min; 40 cycles of 95°C 30 s, 56°C 30 s, 72°C 30 s; 1 cycle of 72°C 10 min. The PCR products were sequenced with the same primers ITS1 and ITS4 (Sangon Biotech, China). The ITS sequences of the 32 yeast strains were identified by comparing the sequencing data with sequences in MycoBank using pairwise alignment of the MycoBank database1.
The 32 yeast strains were inoculated on Sabouraud dextrose agar (SDA) with 0.01% chloramphenicol and chromogenic medium (CHROM agar), respectively. The plates were incubated at 28°C for 7 days, and the growth of the strains on the plates was assessed daily. The macroscopic characteristics and micromorphology of the strains on the plates were observed and determined. Meanwhile, the single colonies on the plates went through Gram staining and the microscopic characteristics were recorded. The Sensititre YeastOne YO10 (Thermo, Waltham, MA, United States) colorimetric panel was used for the antifungal susceptibility analysis of the 32 strains. The operation was performed according to the manufacturer’s instructions, and the interpretation for antifungal susceptibility testing was based on CLSI M59 and CLSI M60 (CLSI, 2020a,b).
Pretreatment of strains: a single colony was picked in a 1.5-ml EP tube containing 1 ml 75% ethanol and 20–30 glass beads, mixed for 2 min. Then the EP tube was centrifuged at 12,000 rpm for 30 s. The supernatant was removed, and 70 μl freshly prepared 70% formic acid was added to the tube and mixed for 30 s. Then, 20 μl acetonitrile was added to the tube and mixed for 30 s. The tube was centrifuged at 12,000 rpm for 2 min. Two microliters of supernatant was added on the spot of the target plate, and 1 μl matrix was added to the same spot after the 2-μl supernatant dried. After the matrix dried, the target plate was taken to the mass spectrometer’s ionization chamber and the mass spectra of the strains were acquired using a VITEK MS Plus (bioMérieux, France) and the Launchpad v2.8 software program (Kratos, Manchester, United Kingdom). Then the cluster analysis of the mass spectra was carried out and dendrograms showing taxonomic relationships were prepared using the SARAMIS software (bioMérieux, France).
For instrument calibration, the Escherichia coli reference strain (ATCC 8739) was applied. The operation procedure was in line with the manufacturer’s instruction.
Kruskal–Wallis H test was used for the S. ciferrii complex species comparisons in the respective antifungal drugs. All results with p value < 0.05 were statistically significant.
The occurrence of S. ciferrii complex in patients with CSOM was 53.13% (17/32) in male and 46.87% (15/32) in female. The age of the patients ranged from 20 to 70 with the median age of 36. The occurrence of S. ciferrii complex was 31.25% (10/32) in patients under 30 years old, 59.38% (19/32) in patients whose age were between 30 and 60 years old, and 9.37% (3/32) in patients above 60 years old. By ITS region alignment in the MycoBank, the 32 yeast strains of S. ciferrii complex were classified at species level. Based on the alignment results, there were 10 strains of C. allociferrii (31.25%), six strains of C. mucifera (18.75%), and 16 strains of S. ciferrii (50%) (Table 1). In our article, the reference strains for S. ciferrii involved CBS5295, CBS6546, and CBS6699. Although different strains of S. ciferrii had different corresponding reference strains (CBS5295, 6546, and 6699), the three reference strains all represented S. ciferrii.
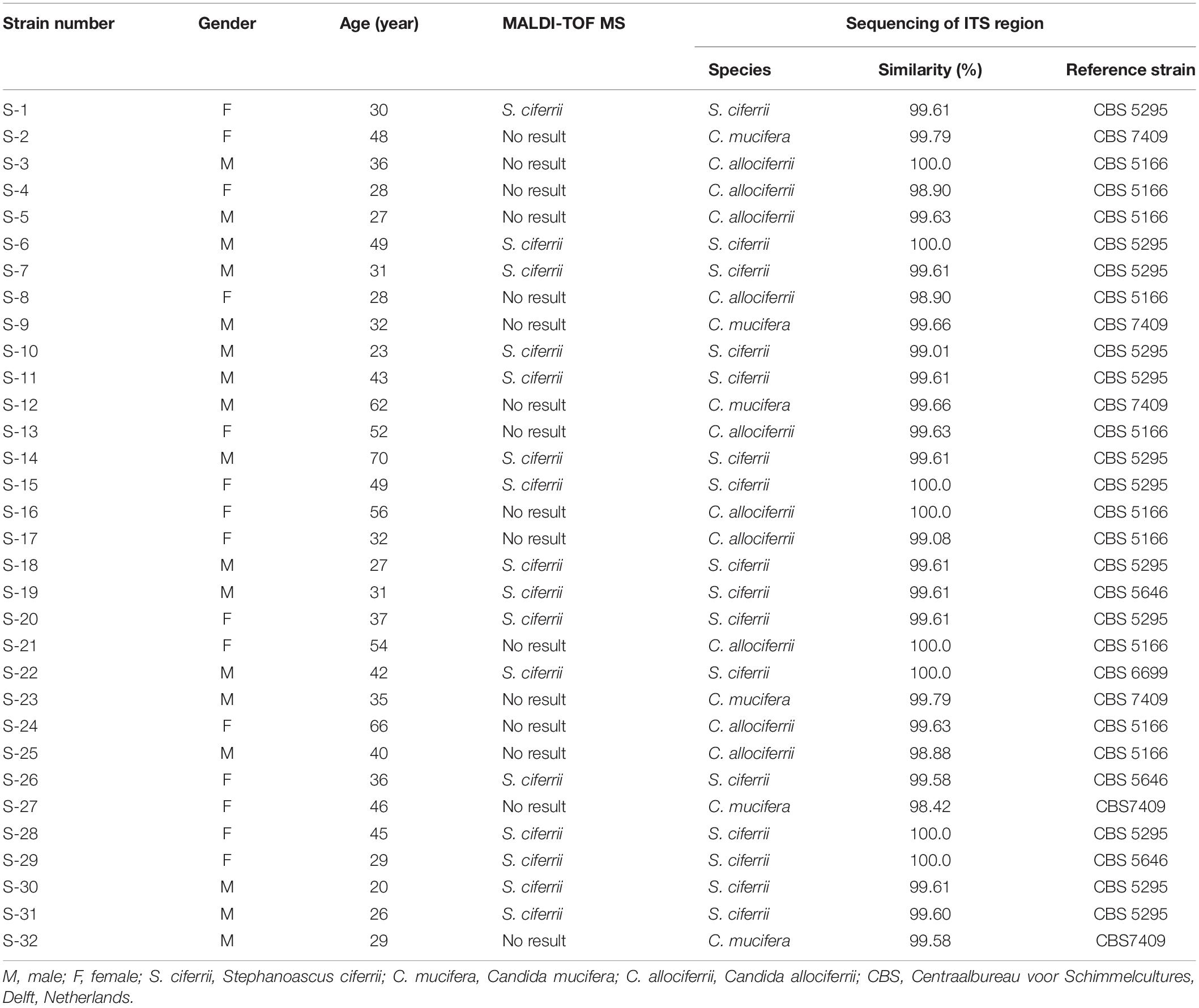
Table 1. The identification of the 32 yeast strains of S. ciferrii complex isolated from CSOM patients using different methods.
The morphology of the three species cultured on the SDA plate and CHROM agar plate at 28°C for 72 h and 7 days is shown in Figures 1A,B, respectively. There were small, round, smooth, and milky white colonies that could be seen after 48 h of culture on the SDA plate. After 72 h, the colonies embedded to the agar and the texture became hard. It was not easy to scrape or grind the colonies from the agar. A week later, creamy or slightly yellowish colonies could be seen on the SDA plate and the center of the colonies was gyrus-like or cauliflower-like. Besides, the colonies of some C. mucifera isolates became sunken. On the CHROM agar plate, the colonies also grew well and they were inlaid into plates 72 h later. The colonies were regular round with a blue center and white edge. The colonies became rough with gyrus-like grooves and irregular edges 7 days later. The Gram staining of the three species is shown in Figure 1C. Under the microscope, there were gram-positive yeast-like fungi and slender hyphae. The spores were single-celled and ovoid, distributing along the periphery of the hyphae. The images about all the 32 isolated species’ appearance on SDA and CHROM agar at 28°C for 3 and 7 days were provided in the Supplementary Material, as well as the Gram staining pictures. Actually, the macroscopic and microscopic characteristics of the three species were similar, making it hard to distinguish them based on morphology.
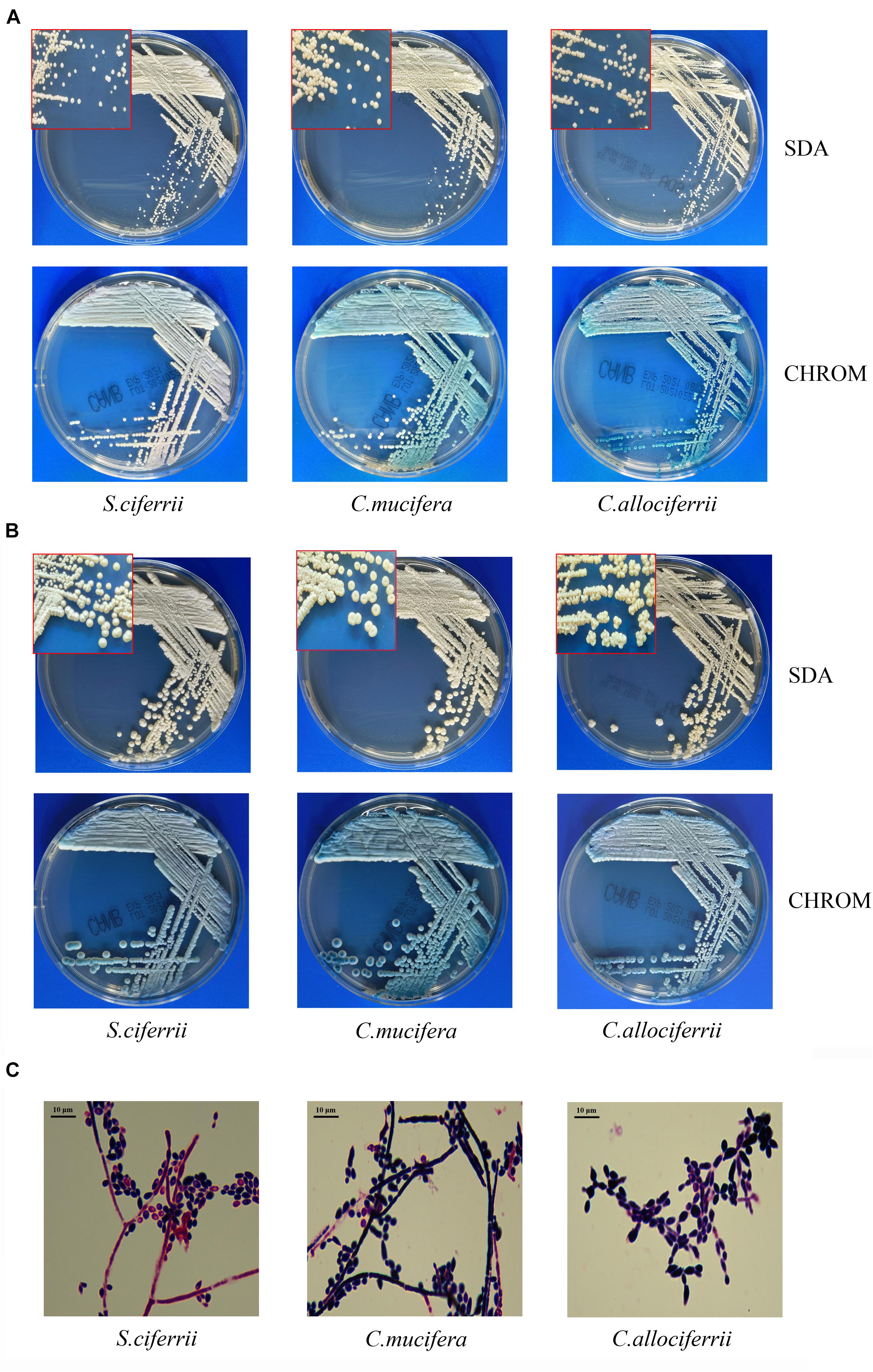
Figure 1. The morphological characteristics of S. ciferrii complex species. (A) Colonies of S. ciferrii complex species cultured on an SDA plate and CHROM agar at 28°C for 72 h. (B) Colonies of S. ciferrii complex species cultured on an SDA plate and CHROM agar at 28°C for 7 days. (C) The Gram staining of colonies of S. ciferrii complex species cultured at 28°C for 72 h, × 1,000.
The MICs of the 32 isolated strains in the antifungal susceptibility testing are shown in Table 2, and the distribution is shown in Figure 2. The MICs for anidulafungin, micafungin, and caspofungin were relatively small. The MICs for anidulafungin, micafungin, and caspofungin ranged between 0.015 and 0.12 μg/ml, 0.008 and 0.25 μg/ml, and 0.006 and 0.06 μg/ml, respectively. There was statistical difference between the MICs of C. allociferrii and those of C. mucifera for anidulafungin, micafungin, and caspofungin (p < 0.05), indicating that C. mucifera tended to have a higher MIC value compared with that of C. allociferrii for the echinocandins. However, the antifungal susceptibility of the 32 strains for azole antifungal drugs differed. The MICs for posaconazole, voriconazole, itraconazole, and fluconazole ranged between 0.06 and 1 μg/ml, 0.12 and 2 μg/ml, 0.06 and 0.5 μg/ml, and 32 and 256 μg/ml, respectively. As for posaconazole, voriconazole, and itraconazole, the 32 strains had relatively low MICs compared with fluconazole and there were no differences among the antifungal susceptibility of the three species for the three antifungal drugs (p < 0.05). For fluconazole, the three species of S. ciferrii complex owned their characteristics. The MICs of C. allociferrii for fluconazole ranged between 32 and 64 μg/ml. Nine of the 10 C. allociferrii strains (90%) had the MICs of 32 μg/ml. The MICs of C. mucifera for fluconazole ranged between 32 and 128 μg/ml, and four of the six C. mucifera strains (66.67%) had MICs of 128 μg/ml. As for S. ciferrii, 50% (8/16) strains had MICs equal or greater than 128 μg/ml and 31.25% (5/16) strains had MICs of 64 μg/ml. The S. ciferrii and C. mucifera tended to have high fluconazole MICs compared with C. allociferrii (p < 0.05). For flucytosine, the MICs of C. allociferrii and C. mucifera ranged from 0.25 to 1 μg/ml and 0.25 to 2 μg/ml, respectively. However, 81.25% (13/16) of the S. ciferrii had MICs of 32 or 64 μg/ml. S. ciferrii tended to have higher MICs for flucytosine compared with C. allociferrii and C. mucifera (p < 0.05). As for amphotericin B, the MICs ranged between 0.12 and 2 μg/ml and there was a difference between the MICs of S. ciferrii and those of C. mucifera (p < 0.05). C. mucifera tended to have a higher MIC value compared with that of S. ciferrii for amphotericin B.
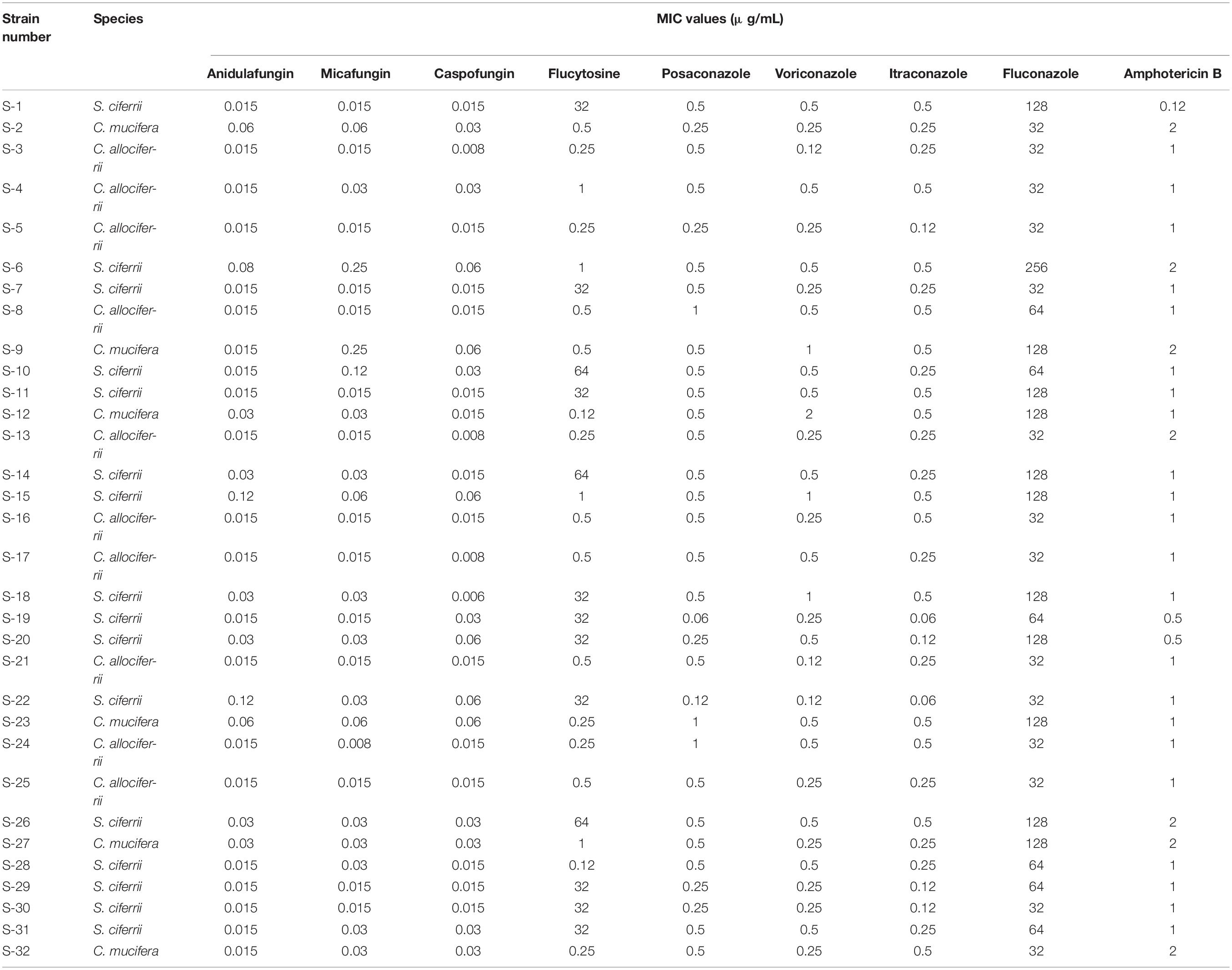
Table 2. The MICs of the 32 isolated strains in the antifungal susceptibility testing with microbroth dilution method.
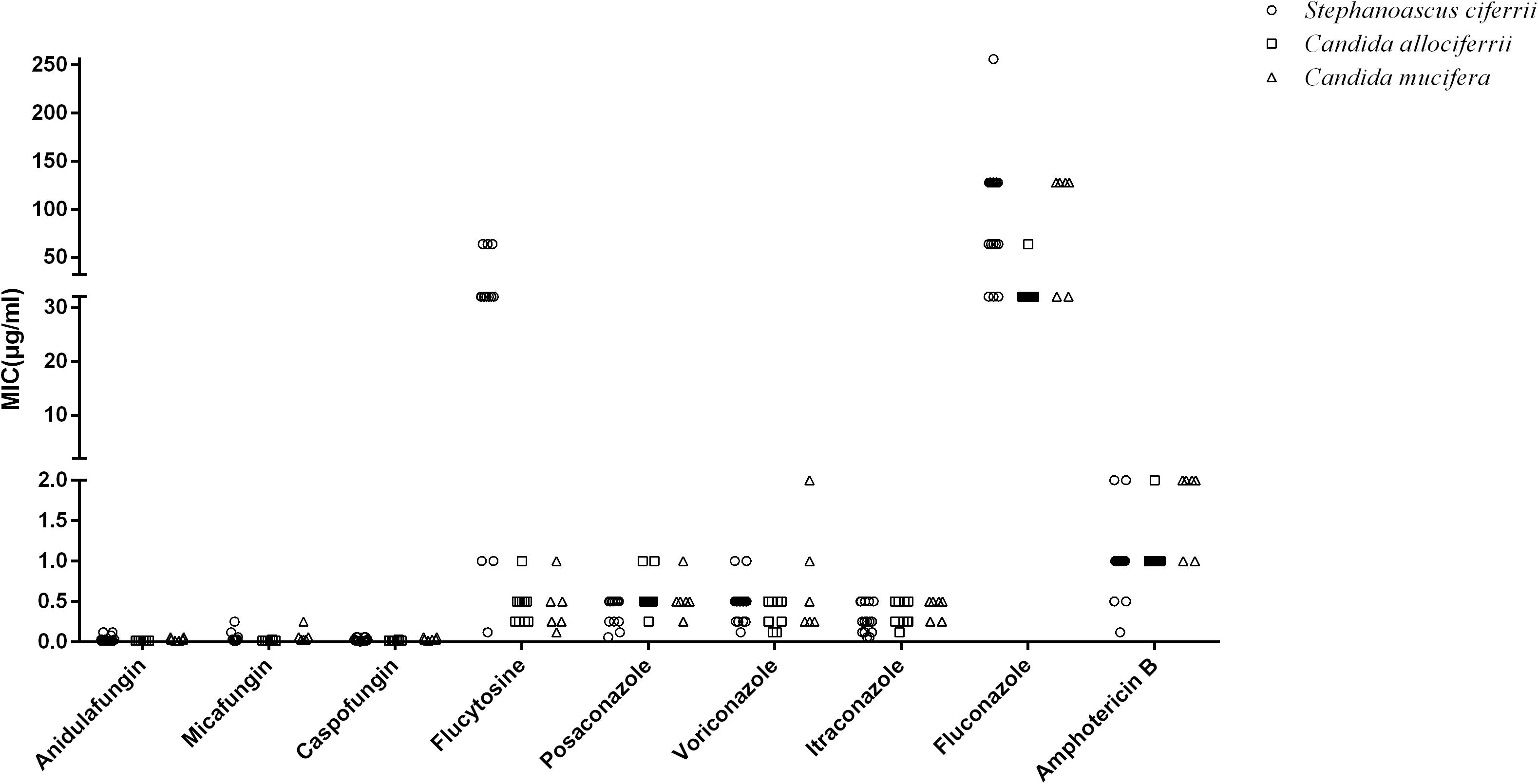
Figure 2. The distribution of the MIC values of S. ciferrii complex species for different antifungal drugs.
Apart from identification by ITS sequencing, the 32 isolated strains all went through identification by MALDI-TOF MS. The 16 strains of S. ciferrii were identified as S. ciferrii with 99.9% similarity by MALDI-TOF MS. However, MALDI-TOF MS was unable to identify the 10 strains of C. allociferrii and six strains of C. mucifera, showing no results (Table 1). For MALDI-TOF MS, the limitation lies in its inability to identify non-clinically validated species or species not included in the MALDI-TOF database (Mesureur et al., 2018). The database of MALDI-TOF MS did not involve the reference database of C. allociferrii and C. mucifera. Thus, the MALDI-TOF MS could not identify the S. ciferrii complex at species level. The cluster analysis of the mass spectra of the 32 isolated strains is shown in Figure 3A. By cluster analysis with SARAMIS, the 32 isolated strains were divided into three clusters. All the S. ciferrii strains were divided into cluster I, all the C. mucifera strains belonged to cluster II, and all the C. allociferrii strains were assigned to cluster III. The mass spectra of two isolates from each species are shown in Figure 3B. The three species of S. ciferrii complex had their own special peaks in the mass spectra, making it possible to identify those using MALDI-TOF MS by constructing reference databases at species level. The mass spectra of all the 32 strains were provided in the Supplementary Material.
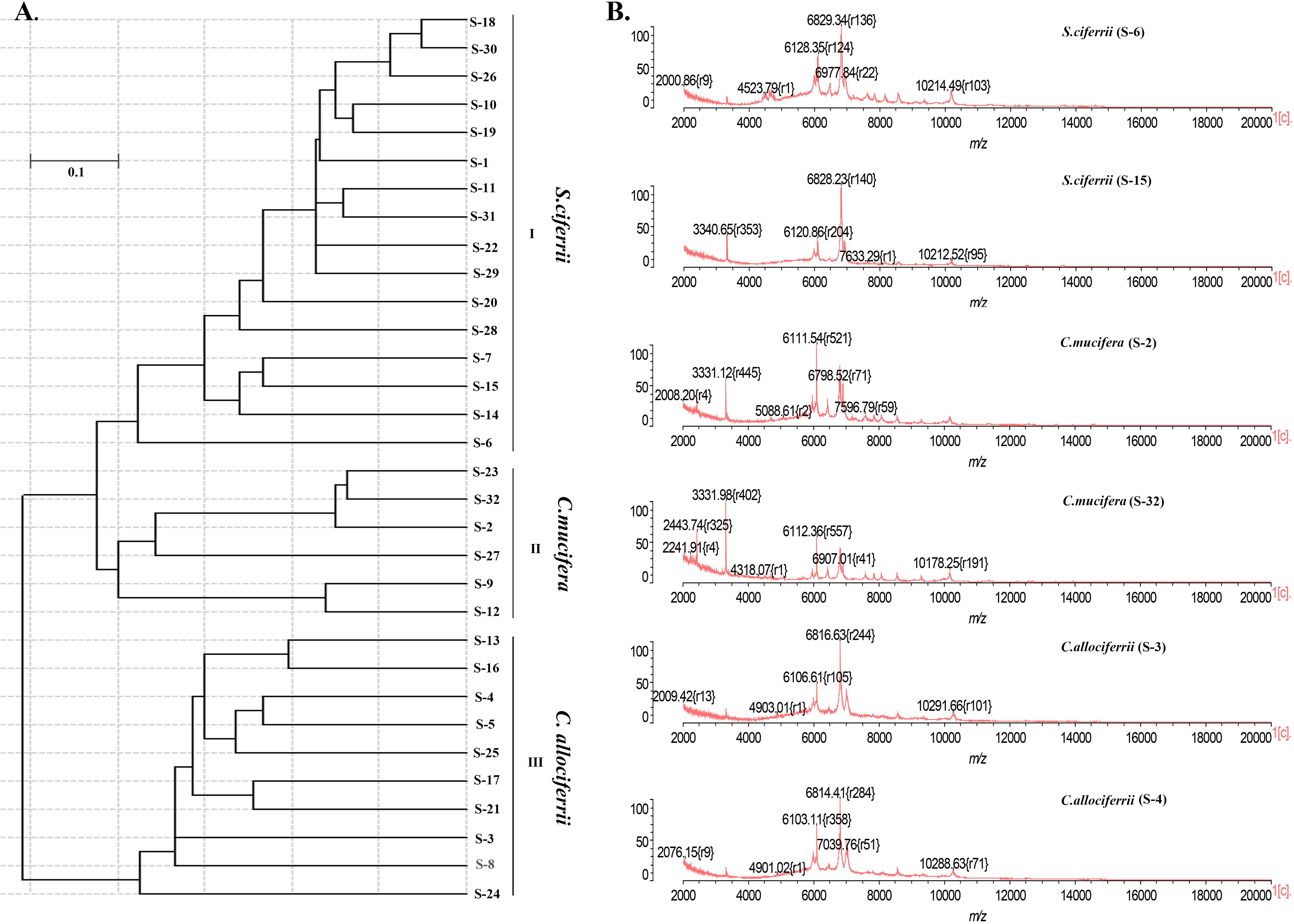
Figure 3. The classification of the 32 isolated S. ciferrii complex strains at species level. (A) The clustering dendrogram of the 32 S. ciferrii complex isolates created by SARAMIS. (B) The mass spectra of six S. ciferrii complex strains.
Stephanoascus ciferrii, which is first discovered in 1965, is an opportunistic rare yeast pathogen with low prevalence (<1% of clinical Candida infections) (Kreger-Van Rij, 1965; Mixao et al., 2019). The literature about S. ciferrii is limited, and it is usually reported in some sporadic cases of infections (Villanueva-Lozano et al., 2016). Reports on S. ciferrii complex are even more limited. Hironobu Soki reported a rare case of intraorbital abscess caused by C. allociferrii, which is the only literature describing S. ciferrii complex since the S. ciferrii complex was proposed (Soki et al., 2015). The clinical isolation rate of S. ciferrii complex is low. Moreover, in the majority of cases, species of the S. ciferrii complex are not identified. CSOM is prevalent in China just like most other developing countries and usually caused by bacteria (Xu et al., 2020). CSOM caused by S. ciferrii complex is rarely reported. In our study, we had collected 32 strains of S. ciferrii complex isolated from patients with CSOM, indicating that more attention should be paid to these species. In our study, the S. ciferrii complex could occur in young, middle-aged, and elderly people. However, the occurrence of S. ciferrii complex mainly focused on the patients whose ages were between 30 and 60 years and showed little relationship with gender. Since the number of clinical isolates at the species level was too small, we did not calculate the occurrence of S. ciferrii complex at species level. Although it is hard to find a firm association from the limited number of clinical isolates in our study, the small sample number is relative and could still be highly informative due to the rare frequency of these species.
By sequencing of the ITS region, the 32 strains were identified as S. ciferrii, C. allociferrii, and C. mucifera. The morphology analysis of the three species shows us the basic phenotypic characteristics of S. ciferrii complex species. The morphology characteristics of the three species are similar, indicating that it is hard to distinguish them based on macroscopic and microscopic characteristics. As for the antifungal susceptibility profiles of S. ciferrii complex, the three species all had low MICs for echinocandins, making this kind of antifungal a good choice to treat S. ciferrii complex infection. As the MICs of S. ciferrii complex species for echinocandins and amphotericin B were relatively low, the MIC differences among the S. ciferrii complex species do not have big effects on the clinical medication. As for the azole antifungal drugs, the MICs for posaconazole, voriconazole, and itraconazole were relatively much smaller than MICs for fluconazole. However, for fluconazole, all the strains had high MICs, which is consistent with the research by Antonio Pérez-Hansen (Perez-Hansen et al., 2019). In the research by Antonio Pérez-Hansen on antifungal susceptibility profiles of rare ascomycetous yeasts, S. ciferrii complex was found to have high fluconazole MICs (>32 μg/ml). Moreover, the antifungal susceptibility to fluconazole at species level revealed that the S. ciferrii and C. mucifera tended to have high fluconazole MICs compared with C. allociferrii. Fluconazole is a triazole antifungal that inhibits fungal cytochrome P450 activity, decreasing ergosterol synthesis and inhibiting cell membrane formation (Hornik et al., 2021). Sasani et al. (2021) reported that the formation of biofilms and the overexpression of ERG11 and UPC2 genes in Candida tropicalis were related with fluconazole resistance. Abbes et al. (2021) found that Mdr gene overexpression in Trichosporon asahii isolates was significantly associated with fluconazole resistance. However, there are no articles reporting why fluconazole resistance is high also in this complex. Perhaps, the fluconazole resistance in S. ciferrii complex is related with the formation of biofilms and the overexpression of some certain genes. In the future study, we plan to explore the mechanism of fluconazole resistance S. ciferrii complex to verify these assumptions. Moreover, the antifungal susceptibility specificity at species level became more obvious for flucytosine. All the strains of C. mucifera and C. allociferrii had relatively low MICs while most of the S. ciferrii owned high flucytosine MICs. However, there were three strains of S. ciferrii with low MICs (≤1 μg/ml). The phenomenon may be worth our attention, and further study to explore the resistance discrepancy is needed.
MALDI-TOF MS, as a rapid, reliable, and high-throughput diagnostic tool, has entered clinical microbiology diagnostics and is wildly used for fungal identification (Delavy et al., 2019). Although ITS region sequencing could identify clinically important yeasts accurately, it is costly and time consuming (Ciardo et al., 2006; Hou et al., 2016). In our study, the strains of C. mucifera and C. allociferrii could not be identified by MALDI-TOF MS while the cluster analysis of the mass spectra divided the 32 strains into three clusters. The three clusters represented the three species of the S. ciferrii complex, suggesting that it is possible to identify S. ciferrii complex species using MALDI-TOF MS. The lack of reference database for C. allociferrii and C. mucifera limits the ability of MALDI-TOF MS in identification, making it essential to construct the reference database of S. ciferrii complex species. Taverna et al. (2019) developed an in-house database of 143 strains belonging to 42 yeast species in the MALDI Biotyper platform, making it possible to differentiate closely related species and distinguish the molecular genotypes of Cryptococcus neoformans and Cryptococcus gattii. Moreover, Veloo et al. (2018) optimized the identification of clinical anaerobic isolates with MALDI-TOF MS using 6,309 anaerobic isolates, thus increasing the number of anaerobic isolates that could be identified with high confidence. Therefore, it is recommended to expand or construct the databases that include less frequently isolated species to identify them quickly and accurately by MALDI-TOF MS, gaining valuable insight into the clinical relevance of the rare microorganism.
In summary, 32 strains of S. ciferrii complex isolated from patients with CSOM were identified at species level by the sequencing of ITS regions. The morphology of the three species was observed in our study in detail. The antifungal susceptibility profiles of three species of S. ciferrii complex exhibited differences with each other. S. ciferrii and C. mucifera tended to have high fluconazole MICs compared with C. allociferrii. C. mucifera and C. allociferrii had relatively low flucytosine MICs while S. ciferrii owned high flucytosine MICs. Besides, C. mucifera tended to have a higher MIC value than S. ciferrii for amphotericin B and C. allociferrii for anidulafungin, micafungin, and caspofungin. Apart from the sequencing of ITS regions, MALDI-TOF MS was used to identify S. ciferrii complex at species level. The MALDI-TOF MS showed no results to C. mucifera and C. allociferrii. Since the cluster analysis of the mass spectra could distinguish the three groups, construction of a reference database of C. mucifera and C. allociferrii is required for subsequent species identification. However, the number of S. ciferrii complex strains in our study is limited and more strains are required to perfect the morphology characteristics of this complex and its antifungal susceptibility profiles. Besides, more mass spectra of C. mucifera and C. allociferrii are needed to construct the reference database, enabling the MALDI-TOF MS to identify S. ciferrii complex at species level.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
PG, KL, and YP participated in research design and data analysis. ZW participated in the performance of the research and data analysis. PL participated in the performance of the research. YC contributed new reagents or analytic tools. YH participated in the writing of the manuscript and data analysis. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.680060/full#supplementary-material
Abbes, S., Sellami, H., Neji, S., Trabelsi, H., Makni, F., and Ayadi, A. (2021). Implication of efflux pumps and ERG11 genes in resistance of clinical Trichosporon asahii isolates to fluconazole. J. Med. Microbiol. 70:1236.
Agin, H., Ayhan, Y., Devrim, I., Gulfidan, G., Tulumoglu, S., and Kayserili, E. (2011). Fluconazole-, amphotericin- B-, caspofungin-, and anidulafungin-resistant Candida ciferrii: an unknown cause of systemic mycosis in a child. Mycopathologia 172, 237–239. doi: 10.1007/s11046-011-9418-6
Al Halteet, S., Abdel-Hadi, A., Hassan, M., and Awad, M. (2020). Prevalence and antifungal susceptibility profile of clinically relevant candida species in postmenopausal women with diabetes. Biomed. Res. Int. 2020:7042 490.
Angeletti, S., and Ciccozzi, M. (2019). Matrix-assisted laser desorption ionization time-of-flight mass spectrometry in clinical microbiology: an updating review. Infect. Genet. Evol. 76:104063. doi: 10.1016/j.meegid.2019.104063
Bhutta, M. F., Head, K., Chong, L. Y., Daw, J., Schilder, A. G. M., Burton, M. J., et al. (2020). Aural toilet (ear cleaning) for chronic suppurative otitis media. Cochrane Database Syst. Rev. 9:CD013057.
Ciardo, D. E., Schar, G., Bottger, E. C., Altwegg, M., and Bosshard, P. P. (2006). Internal transcribed spacer sequencing versus biochemical profiling for identification of medically important yeasts. J. Clin. Microbiol. 44, 77–84. doi: 10.1128/jcm.44.1.77-84.2006
CLSI (2020a). Epidemiological Cutoff Values for Antifungal Susceptibility Testing, 3rd Edn, Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI (2020b). Performance Standards for Antifungal Susceptibility Testing of Yeasts, 2nd Edn, Wayne, PA: Clinical and Laboratory Standards Institute.
Cuenod, A., Foucault, F., Pfluger, V., and Egli, A. (2021). Factors associated with MALDI-TOF mass spectral quality of species identification in clinical routine diagnostics. Front. Cell Infect. Microbiol. 11:646648. doi: 10.3389/fcimb.2021.646648
da Silva, E. M., Sciuniti Benites Mansano, E., de Souza Bonfim-Mendonca, P., Olegário, R., Tobaldini-Valério, F., Fiorini, A., et al. (2021). High colonization by Candida parapsilosis sensu stricto on hands and surfaces in an adult intensive care unit. J. Mycol. Med. 31:101110. doi: 10.1016/j.mycmed.2020.101110
Danielescu, C., Cantemir, A., and Chiselita, D. (2017). Successful treatment of fungal endophthalmitis using intravitreal caspofungin. Arq. Bras. Oftalmol. 80, 196–198.
Dave, V. P., Sharma, S., Dave, P. J., Joseph, J., and Pappuru, R. R. (2018). Clinical presentations, diagnostic dilemma, and management outcomes of chronic postoperative endophthalmitis caused by Stephanoascus ciferrii. Retin. Cases Brief. Rep. 15, 269–274. doi: 10.1097/icb.0000000000000782
de Gentile, L., Bouchara, J. P., Cimon, B., and Chabasse, D. (1991). Candida ciferrii: clinical and microbiological features of an emerging pathogen. Mycoses 34, 125–128. doi: 10.1111/j.1439-0507.1991.tb00632.x
Delavy, M., Dos Santos, A. R., Heiman, C. M., and Coste, A. T. (2019). Investigating antifungal susceptibility in Candida species with MALDI-TOF MS-based assays. Front. Cell Infect. Microbiol. 9:19. doi: 10.3389/fcimb.2019.00019
Hornik, C. D., Bondi, D. S., Greene, N. M., Cober, M. P., and John, B. (2021). Review of fluconazole treatment and prophylaxis for invasive Candidiasis in neonates. J. Pediatr. Pharmacol. Ther. 26, 115–122. doi: 10.5863/1551-6776-26.2.115
Hou, X., Xiao, M., Chen, S. C., Wang, H., Zhang, L., Fan, X., et al. (2016). Sequencer-based capillary gel electrophoresis (SCGE) targeting the rDNA Internal Transcribed Spacer (ITS) regions for accurate identification of clinically important yeast species. PLoS One 11:e0154385. doi: 10.1371/journal.pone.0154385
Kreger-Van Rij, N. J. (1965). Candida ciferrii, a new yeast species. Mycopathol. Mycol. Appl. 26, 49–52. doi: 10.1007/bf02098589
Mancilla-Rojano, J., Ochoa, S. A., Reyes-Grajeda, J. P., Flores, V., Medina-Contreras, O., Espinosa-Mazariego, K., et al. (2020). Molecular epidemiology of Acinetobacter calcoaceticus-Acinetobacter baumannii complex isolated from children at the hospital infantil de mexico federico gomez. Front. Microbiol. 11:576673. doi: 10.3389/fmicb.2020.576673
Mesureur, J., Arend, S., Celliere, B., Courault, P., Cotte-Pattat, P.-J., Totty, H., et al. (2018). A MALDI-TOF MS database with broad genus coverage for species-level identification of Brucella. PLoS Negl. Trop. Dis. 12:e0006874. doi: 10.1371/journal.pntd.0006874
Mixao, V., Saus, E., Hansen, A. P., Lass-Florl, C., and Gabaldon, T. (2019). Genome assemblies of two rare opportunistic yeast pathogens: Diutina rugosa (syn. Candida rugosa) and Trichomonascus ciferrii (syn. Candida ciferrii). G3 9, 3921–3927. doi: 10.1534/g3.119.400762
Perez-Hansen, A., Lass-Florl, C., and Lackner, M. (2019). Rare yeast study g. antifungal susceptibility profiles of rare ascomycetous yeasts. J. Antimicrob. Chemother. 74, 2649–2656. doi: 10.1093/jac/dkz231
Sasani, E., Khodavaisy, S., Rezaie, S., Salehi, M., and Yadegari, M. H. (2021). The relationship between biofilm formation and mortality in patients with Candida tropicalis candidemia. Microb. Pathog. 2020:104889. doi: 10.1016/j.micpath.2021.104889
Soki, H., Abo, K., Yamazaki, K., Kojima, T., Oda, T., Uzawa, Y., et al. (2015). First report of intraorbital abscess caused by Candida allociferrii and specific PCR for differentiating Stephanoascus ciferrii complex species. Med. Mycol. J. 56, E9–E14.
Soki, H., Nagase, Y., Yamazaki, K., Oda, T., and Kikuchi, K. (2010). Isolation of the yeast-like fungus Stephanoascus ciferrii by culturing the aural discharge of a patient with intractable otitis media. Case report. Kansenshogaku Zasshi 84, 210–212. doi: 10.11150/kansenshogakuzasshi.84.210
Taverna, C. G., Mazza, M., Bueno, N. S., Alvarez, C., Amigot, S., Andreani, M., et al. (2019). Development and validation of an extended database for yeast identification by MALDI-TOF MS in Argentina. Med. Mycol. 57, 215–225. doi: 10.1093/mmy/myy021
Ueda-Nishimura, K., and Mikata, K. (2002). Species distinction of the ascomycetous heterothallic yeast-like fungus Stephanoascus ciferrii complex: description of Candida allociferrii sp. nov. and reinstatement of Candida mucifera Kockova-Kratochvilova et Slavikova. Int. J. Syst. Evol. Microbiol. 52(Pt 2), 463–471. doi: 10.1099/00207713-52-2-463
Veloo, A. C. M., Jean-Pierre, H., Justesen, U. S., Morris, T., Urban, E., Wybo, I., et al. (2018). Validation of MALDI-TOF MS Biotyper database optimized for anaerobic bacteria: the ENRIA project. Anaerobe 54, 224–230. doi: 10.1016/j.anaerobe.2018.03.007
Villanueva-Lozano, H., Trevino-Rangel, R. J., Hernandez-Balboa, C. L., Gonzalez, G. M., and Martinez-Resendez, M. F. (2016). An unusual case of Candida ciferrii fungemia in an immunocompromised patient with Crohn’s and Mycobacterium bovis disease. J. Infect. Dev. Ctries 10, 1156–1158. doi: 10.3855/jidc.8228
Warren, W. A., Franco-Palacios, D., King, C. S., Shlobin, O. A., Nathan, S. D., Katugaha, S. B., et al. (2018). A 24-year-old woman with precipitous respiratory failure after lung transplantation. Chest 153:e0053-56.
Keywords: Stephanoascus ciferrii complex, chronic suppurative otitis media, antifungal susceptibility analysis, MALDI-TOF MS, internal transcribed spacer region sequencing
Citation: Guo P, Wu Z, Liu P, Chen Y, Liao K, Peng Y and He Y (2021) Identification and Antifungal Susceptibility Analysis of Stephanoascus ciferrii Complex Species Isolated From Patients With Chronic Suppurative Otitis Media. Front. Microbiol. 12:680060. doi: 10.3389/fmicb.2021.680060
Received: 13 March 2021; Accepted: 07 May 2021;
Published: 21 July 2021.
Edited by:
Marcos Sergio Toledo, Federal University of São Paulo, BrazilReviewed by:
Luciana Lopes Guimaraes, University of Santa Cecília, BrazilCopyright © 2021 Guo, Wu, Liu, Chen, Liao, Peng and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaqin Peng, cHlxZHJlYW1AMTYzLmNvbQ==; Yuting He, aGV5dXRpbmc2MzY2QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.