
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 July 2021
Sec. Extreme Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.679245
This article is part of the Research Topic Extremophiles: Microbial Genomics and Taxogenomics View all 26 articles
 Galina Slobodkina1*
Galina Slobodkina1* Maxime Allioux2
Maxime Allioux2 Alexander Merkel1
Alexander Merkel1 Marie-Anne Cambon-Bonavita2
Marie-Anne Cambon-Bonavita2 Karine Alain2
Karine Alain2 Mohamed Jebbar2
Mohamed Jebbar2 Alexander Slobodkin1
Alexander Slobodkin1Hyperthermophilic archaea of the genus Archaeoglobus are the subject of many fundamental and biotechnological researches. Despite their significance, the class Archaeoglobi is currently represented by only eight species obtained as axenic cultures and taxonomically characterized. Here, we report the isolation and characterization of a new species of Archaeoglobus from a deep-sea hydrothermal vent (Mid-Atlantic Ridge, TAG) for which the name Archaeoglobus neptunius sp. nov. is proposed. The type strain is SE56T (=DSM 110954T = VKM B-3474T). The cells of the novel isolate are motile irregular cocci growing at 50–85°C, pH 5.5–7.5, and NaCl concentrations of 1.5–4.5% (w/v). Strain SE56T grows lithoautotrophically with H2 as an electron donor, sulfite or thiosulfate as an electron acceptor, and CO2/HCO3− as a carbon source. It is also capable of chemoorganotrophic growth by reduction of sulfate, sulfite, or thiosulfate. The genome of the new isolate consists of a 2,115,826 bp chromosome with an overall G + C content of 46.0 mol%. The whole-genome annotation confirms the key metabolic features of the novel isolate demonstrated experimentally. Genome contains a complete set of genes involved in CO2 fixation via reductive acetyl-CoA pathway, gluconeogenesis, hydrogen and fatty acids oxidation, sulfate reduction, and flagellar motility. The phylogenomic reconstruction based on 122 conserved single-copy archaeal proteins supported by average nucleotide identity (ANI), average amino acid identity (AAI), and alignment fraction (AF) values, indicates a polyphyletic origin of the species currently included into the genus Archaeoglobus, warranting its reclassification.
Over several decades since their discovery, thermophilic archaea have been the subject of an increasing number of studies related to microbial ecology, biogeochemistry, origin of life, and evolution of the biosphere (Offre et al., 2013). It was long thought that hyperthermophilic archaea represented the deepest and shortest phylogenetic branches of the tree of life and could be considered the first living organisms on Earth since most of them have a chemolithoautotrophic mode of nutrition (Stetter, 2006; Martin and Sousa, 2016; Weiss et al., 2016), but this hypothesis is now being questioned. Indeed, numerous archaeal lineages, with new complete genomes or high-quality metagenomes-assembled genomes (MAGs) and single-amplified genomes (SAGs), have been discovered thanks to remarkable advances in sequencing techniques and in data processing capabilities, allowing the construction of robust phylogenetic trees based on several genetic markers, which demonstrate that the root is far from being resolved (Baker et al., 2020). Dissimilatory sulfate reduction is a microbial process with significant ecological and biogeochemical implication (Rabus et al., 2015). It is mainly related to the marine environments because of the high sulfate concentrations. In the marine ecosystems with temperatures above 80°C, the only conclusively proven actors of this process are the representatives of the genus Archaeoglobus that was established by Stetter et al. (1987) and Stetter (1988). The type strain, A. fulgidus VC-16T, is one of the best studied Archaea, as it was the first described hyperthermophilic sulfate-reducing archaea, and one of the first archaea whose genome has been sequenced (Stetter, 1988; Klenk et al., 1997). In addition to physiological properties given in the original description, new metabolic features were found based on genome sequencing data analysis, e.g., growth coupled with carbon monoxide oxidation and sulfate reduction or acetogenesis (Henstra et al., 2007); the ability for anaerobic oxidation of fatty acids, n-alkenes, and n-alkanes (Khelifi et al., 2010, 2014); and growth with (per)chlorate reduction combining biotic and abiotic reactions (Liebensteiner et al., 2013). Along with the type strain, A. fulgidus 7324 has been isolated from North Sea oilfield waters and its genome has been sequenced (Beeder et al., 1994; Birkeland et al., 2017).
Archaeoglobus fulgidus became a source and a model organism for the study of structure and function of many thermostable enzymes. A good example is A. fulgidus ferritin (AfFt) whose unique structure and properties make it useful in clinical therapy (Palombarini et al., 2020). Archaeoglobus fulgidus is also a promising candidate to clean up oil-contaminated environments at low cost and with high efficiency due to its high resistance to anaerobic conditions, high temperatures, and high salts concentrations, and its ability to degrade alkanes (Park and Park, 2018).
At the time of writing, the genus Archaeoglobus includes five species with validly published names: A. fulgidus (Stetter, 1988), A. profundus (Burggraf et al., 1990), A. veneficus (Huber et al., 1997), A. infectus (Mori et al., 2008). and A. sulfaticallidus (Steinsbu et al., 2010). Together with related genera, Geoglobus and Ferroglobus, they compose the family Archaeoglobaceae belonging to the order Archaeoglobales, class Archaeoglobi (Parte et al., 2020; http://www.bacterio.net/index.html). It is notable that class Archaeoglobi is represented by only eight species obtained as axenic cultures and taxonomically characterized in detail. All known representatives of Archaeoglobi are strict anaerobes and hyperthermophiles isolated from marine hydrothermal systems and off-shore oil reservoirs. With the exception of A. infectus and A. profundus, all species of the order Archaeoglobales are capable of chemolithoautotrophic growth. Members of this order differ significantly in terms of the electron acceptors they use. All known species of the genus Archaeoglobus are able to grow by reduction of sulfite and thiosulfate. Archaeoglobus fulgidus, A. profundus, and A. sulfaticallidus can also reduce sulfate with organic carbon source, but only A. sulfaticallidus is capable of lithoautotrophic growth with sulfate as a terminal electron acceptor. Species of the genus Geoglobus are obligate iron-reducers (Kashefi et al., 2002; Slobodkina et al., 2009). The only representative of the genus Ferroglobus, F. placidus, can use nitrate, thiosulfate, and Fe3+ as electron acceptors (Hafenbradl et al., 1996; Tor et al., 2001).
Over the last decade, genomic data have increased exponentially and revealed many phylogenetic inconsistencies that require revision and reclassification of existing prokaryotic taxonomy and nomenclature. These taxonomic issues were recently addressed by proposing a standardized taxonomy for Bacteria and Archaea referred to as the Genome Taxonomy DataBase (GTDB; Parks et al., 2018; https://gtdb.ecogenomic.org/), based on average nucleotide criteria to delineate species and on sets of protein markers to infer a taxonomic position and define genomic clusters. Archaea were recognized as a separate domain in 1977 on the basis of small subunit (SSU) ribosomal RNA (rRNA) gene sequences analysis (Woese and Fox, 1977) and this phylogenetic marker was used for their classification afterward. Therefore, compared to Bacteria, which classification has long been relied on phenotypic properties, the Archaea are less affected by historical misclassifications (Zuo et al., 2015). The Archaeoglobus species were classified into the genus based mostly on phylogenetic analyses of 16S rRNA gene sequences. According to the GTDB, the genus Archaeoglobus encompasses four distinct clusters of genomic sequences. In this paper, we describe a novel thermophilic and facultative lithoautotrophic strain SE56T, belonging to the genus Archaeoglobus. To further understand the physiological features and genome characteristics of this novel species, the whole-genome of strain SE56T was sequenced and analyzed. An advanced phylogenomic reconstruction based on 122 conserved single-copy archaeal protein markers and supported by Average Nucleotide Identity (ANI), Alignment Fraction (AF), and Average Amino Acid Identity (AAI) values clearly indicated a polyphyletic origin of the species currently affiliated to the genus Archaeoglobus.
Strain SE56T was isolated from a hydrothermal diffuser fragment (from the sample BIC2-PL1917-11-PBT1-02) collected in 2018 during the BICOSE 2 oceanographic cruise,1 using the manned operated submersible Nautile, at the TAG vent field (26°13'69"N 44°82'61"W, 3,625 m water depth) on the Mid-Atlantic Ridge. Onboard, the samples were immediately subsampled under sterile conditions. Small rock fragments were placed into 50 ml glass flasks with in situ seawater under N2 flow, closed with a rubber stopper and aluminum cap, and were stored at 4°C.
The medium for enrichment and isolation contained (per liter distilled water): 0.33 g NH4Cl, 0.33 g KCl, 0.33 g CaCl2∙6H2O, 0.33 g KH2PO4, 18.0 g NaCl, 4.33 g MgCl2∙6H2O, 2.0 g NaHCO3, 0.001 g resazurin, 0.5 g Na2S·9H2O, 1 ml trace element solution (Slobodkin et al., 2012) and 1 ml vitamin solution (Wolin et al., 1963). The isolation medium had a pH of 6.5–6.8 (measured at 25°C). The medium was dispensed in 10 ml portions into 17 ml Hungate tubes and heat-sterilized at 121°C for 30 min; the gas phase consisted of H2/CO2 (20: 80 v/v). Sodium sulfite from a sterile anoxic stock solution was added as an electron acceptor to a final concentration of 5 mM. All transfers and sampling of cultures were performed with syringes. All incubations were at 80°C unless otherwise noted.
Cell growth was determined after acridine orange staining, by direct cell counting using a phase-contrast microscope (Olympus CX-41, Olympus Corp., Japan). The morphological properties of strain SE56T were observed using light microscopy and transmission electron microscopy (JEM-100, JEOL, Japan). For Gram-staining cells from exponential and stationary phases of growth were used (Gerhardt et al., 1994). Determination of temperature, pH, and salinity ranges for growth was carried out as described previously (Slobodkina et al., 2016) in the same medium used for isolation, with pyruvate (10 mM) instead of molecular hydrogen. The influence of temperature on growth of strain SE56T was determined in the range 40–95°C with an interval of 5°C. The effect of pH on growth was investigated at 80°C in the range 4.0–9.5 with a 0.5 pH unit interval. The salt requirement was tested at 80°C and pH 6.5 in the range 0–7% NaCl (w/v) with an interval of 0.5% (w/v). All experiments were performed in duplicate, whereas testing the effect of pH on growth was performed in triplicate. Soluble electron donors and acceptors were added from sterile anoxic stock solutions before inoculation. Elemental sulfur was added directly into each test tube with liquid medium prior to tindallisation. Fe(III) was provided in the form of amorphous iron(III) oxide (ferrihydrite) at about 90 mmol Fe(III) L−1. The ferrihydrite was synthesized by titrating a solution of FeCl3, with 10% (w/v) NaOH to pH 9.0. The effect of O2 on growth of strain SE56T was tested in 60 ml flasks containing 10 ml of the medium The flasks were sealed with a rubber stopper and aluminum screw cap. To check microaerobic growth, anoxic medium with pyruvate (10 mM) and without an electron acceptor, reducing agent, and resazurin was used. The headspace of the flask was filled with CO2 (100%). Various amounts of sterile air were injected by syringes in the headspace to obtain oxygen concentrations of 0.5, 2.0, or 5.0% (v/v). The ability of the strain to aerobic growth was determined in the same medium prepared in air. The strain was regarded as utilizing the added electron acceptors and donors if growth was sustained after at least three subsequent transfers into fresh medium.
For genomic DNA extraction, the strain was cultivated at 80°C with pyruvate (10 mM) and sodium sulfite (5 mM) as electron donor and acceptor, respectively. Cells were harvested in the late exponential phase of growth by centrifugation at 4°C (15,000 × g, 20 min). The DNA was extracted using a FastDNA™ Spin Kit (MP Biomedicals, United States) according to the manufacturer’s instructions. The whole genome was sequenced using the Illumina nanoMiSeq technology (Fasteris, Switzerland; 2 × 150 paired-reads, Nano V2 chemistry). Clean reads from short-reads sequencing were assembled and circularized using SPAdes v1.13.1 assembly pipeline (Bankevich et al., 2012; https://github.com/ablab/spades). Average coverage was calculated using BBtools (BBMap – Bushnell B., v38.70 – sourceforge.net/projects/bbmap/). Contamination and completeness were assessed by СheckM v1.1.2 (Parks et al., 2015; https://github.com/Ecogenomics/CheckM). Gene search and annotation were performed by means of the Rapid Annotation using Subsystem Technology (RAST)/SEED v2.0 pipeline (Overbeek et al., 2014; https://rast.nmpdr.org/), the Integrated Microbial genomes (IMGs)/M v.5.0 analysis system (Chen et al., 2019; https://img.jgi.doe.gov/), and National Center of Biotechnology Information (NCBI) integrated prokaryotic genome annotation pipeline (PAGP; Tatusova et al., 2016). Identification and classification of the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas system was performed by the CRISPRCas Finder web server (Couvin et al., 2018; https://crispr.i2bc.paris-saclay.fr/Server/). The prediction of laterally transferred gene clusters [genomic islands (GI)] was performed with the IslandViewer4 web server (Bertelli et al., 2017; http://www.pathogenomics.sfu.ca/islandviewer/). Genome and gene clusters visualization was made with the CGView program (Grant and Stothard, 2008; https://paulstothard.github.io/cgview/) and Gene Graphics web application (Harrison et al., 2018; https://katlabs.cc/genegraphics/app). Analysis of the clusters of orthologous genes (COG) functional categories was performed with the eggNOG-Mapper v2 (Huerta-Cepas et al., 2017; http://eggnog-mapper.embl.de/). Hydrogenase classification has been checked using the HydDB webtool (Søndergaard et al., 2016; https://services.birc.au.dk/hyddb/).
For phylogenetic analysis, the 16S rRNA gene sequence of the isolate (retrieved from the whole-genome sequencing) was compared with other sequences in GenBank (Benson et al., 1999) using the BLAST program (Altschul et al., 1990; http://www.ncbi.nlm.nih.gov/BLAST/) and by means of the EzBio-Cloud server (Yoon et al., 2017; https://www.ezbiocloud.net/) to identify its closest relatives. Alignment with a representative set of related 16S rRNA gene sequences from the GenBank database was carried out with the CLUSTALW program implemented in the MEGA software package v7.0 (Kumar et al., 2016). Evolutionary analysis and phylogenetic tree reconstruction used the Maximum-Likelihood algorithm based on the Tamura–Nei model and the Neighbor-Joining methods (Saitou and Nei, 1987; Tamura and Nei, 1993) provided by MEGA v7.0. ANI were calculated using three methods: (i) OrthoANIu, the orthologous ANI algorithm using the USEARCH program (Edgar, 2010) provided by the EzBioCloud ANI calculator2; (ii) ANIb, the algorithm using BLASTN (Goris et al., 2007) provided by JSpeciesWS Online Service (Richter et al., 2016; http://jspecies.ribohost.com/jspeciesws/#analyse); and (iii) gANI obtained by the Microbial Species Identifier (MiSI) method (Varghese et al., 2015) using ANIcalculator implemented in the IMG/M system.3 The alignment fraction (AF) values were also obtained via the Pairwise ANI tool implemented in the IMG/M online service. Digital DNA–DNA hybridizations (dDDH) were performed using the genome-to-genome distance (GGDC) method with the GGDC 2.0 blast+ model provided by the Genome-to-Genome Distance Calculator (Meier-Kolthoff et al., 2013; https://www.dsmz.de/). The AAI between the selected genomes was calculated using the aai.rb script from the enveomics collection.4 The list of 122 archaeal marker genes used for phylogenetic inference was taken from the GTDB (Parks et al., 2018). These marker genes were extracted from genomes using GTDB-Tk v1.3.0 (Chaumeil et al., 2019; https://github.com/Ecogenomics/GTDBTk), aligned using mafft v7.475 (Katoh and Standley, 2013), trimmed using trimAl 1.2 (Capella-Gutiérrez et al., 2009), and concatenated. The phylogenomic tree was built using PhyML v3.3.2 (Guindon et al., 2010) and the Bayesian like transformation of approximate likelihood-ratio test for branches (Anisimova et al., 2011). LG was selected as the best substitution model by the SMS algorithm (Le and Gascuel, 2008; Lefort et al., 2017).
An enrichment culture was initiated by inoculation of the mixture of small rock fragments and sea water [at 10% (w/v)] into anoxically prepared, bicarbonate-buffered, sterile liquid medium with molecular hydrogen as an electron donor, sulfite as an electron acceptor, and CO2/HCO3− as a carbon source. After incubation in the dark for 10 days at 80°C, growth of irregular-shaped cells was observed. After three subsequent transfers and following serial 10-fold dilutions in the same medium, only one morphological type was observed in the highest growth-positive dilution (10−6). A pure culture of strain SE56T was obtained by performing multiple dilution-to extinction series in the same medium. Purity of the isolate was confirmed by routine microscopic examination on various media, and sequencing of the 16S rRNA gene and genome.
Phylogenetic analysis based on comparison of 16S rRNA gene sequences showed that the strain SE56T belonged to the genus Archaeoglobus (Figure 1; Supplementary Figure S1) and had the highest sequence similarity to A fulgidus VC-16T (98.6%). Sequence similarities between the 16S rRNA gene sequence of the isolate and those of other representatives in the order Archaeoglobales were 96.3–94.4%.
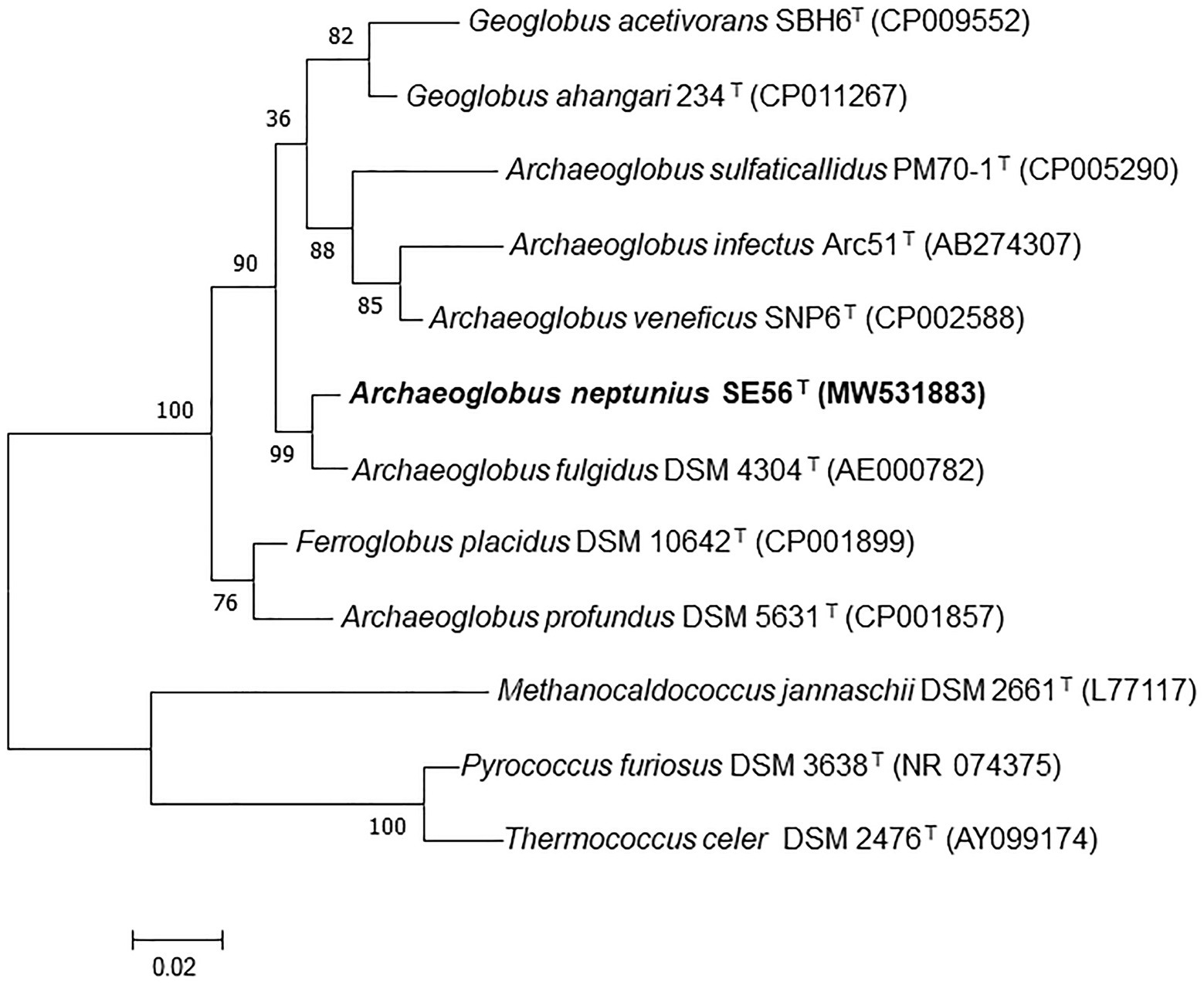
Figure 1. Maximum-likelihood phylogenetic tree based on 16S rRNA gene sequences showing the position of the strain SE56T among the other members of the class Archaeoglobi. Bootstrap values based on 1,000 replicates are shown at branch nodes. Methanocaldococcus jannaschii DSM 2661T, Pyrococcus furiosus DSM 3638T, and Thermococcus celer DSM 2476T were used as outgroup. Bar, 0.02 substitutions per nucleotide position.
Cells of strain SE56T were motile irregular cocci (approximately 0.6–0.8 μm in diameter), usually occurring singly (Figure 2). Cells stained Gram-negative and showed blue-green fluorescence when exposed to UV under a fluorescence microscope.
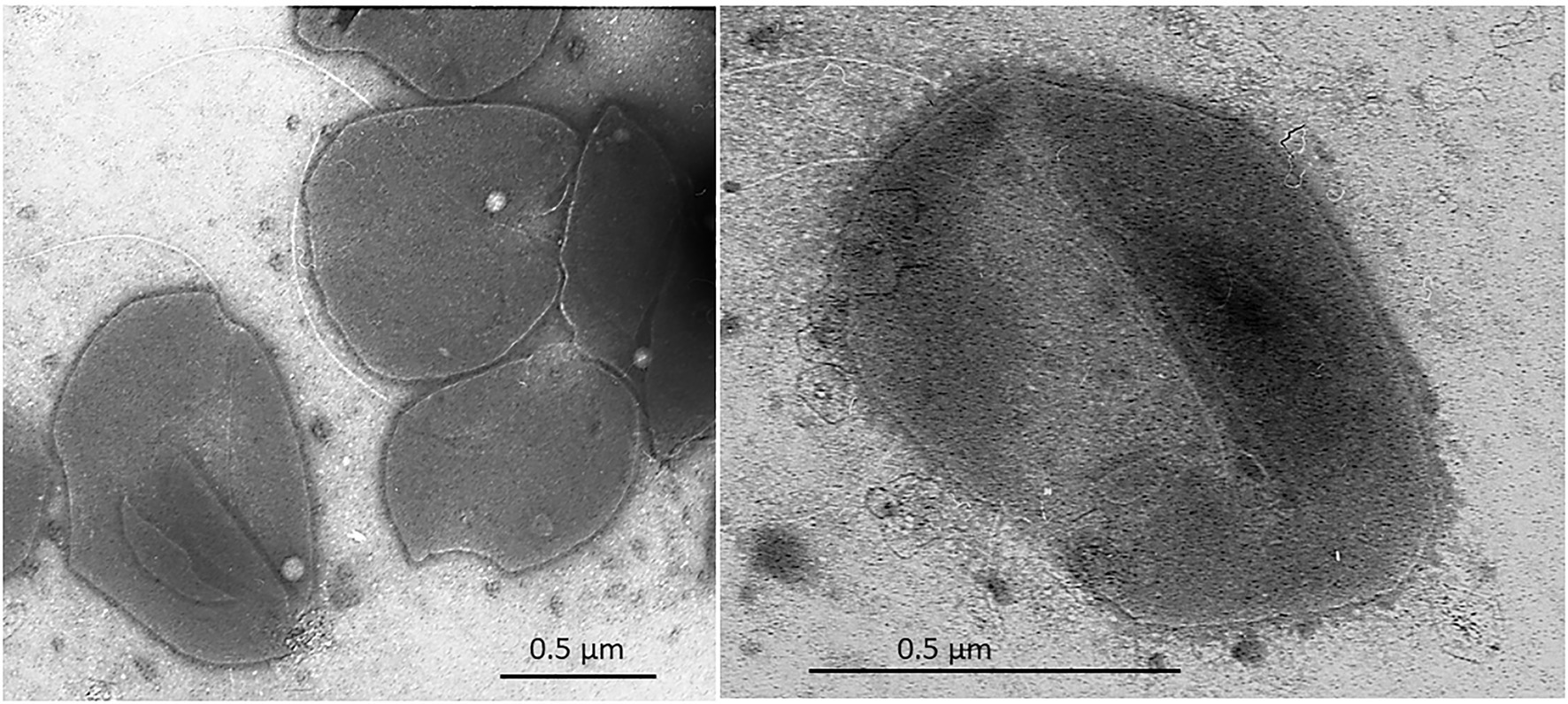
Figure 2. Transmission electron micrograph (TEM) of negatively stained cells of Archaeoglobus neptunius SE56T sp. nov. shows an overall cell morphology. Bar, 0.5 μm.
The temperature range for the growth of strain SE56T was 50–85°C, with an optimum at 80°C. No growth was detected at 90°C or above, neither at 45°C or below, after 3 weeks of incubation. The pH range for growth was 5.5–7.5, with an optimum at pH 6.5. No growth was observed at pH 5.0 or below or at pH 8.0 or above. Growth of the isolate was observed at NaCl concentrations ranging from 1.5 to 4.5% (w/v), with an optimum at 2.0–2.5% (w/v). No growth was evident neither at 5.0% NaCl (w/v) or above, nor at 1.0% NaCl (w/v) and below (Supplementary Figure S2). The doubling time under optimal conditions was about 2 h. Yeast extract was not necessary for growth and did not stimulate it.
Growth with potential organic substrates was tested with sulfite as a terminal electron acceptor. The best growth (final cell density 2–4∙108 cells ml−1) was observed with pyruvate (10 mM), but growth was not observed after 7 days of incubation in the same conditions with the addition of elemental sulfur (5 g L−1). Acetate propionate, butyrate, fumarate, succinate (10 mM each), stearate (1 mM), peptone, tryptone, and yeast extract (2 g L−1 each) also sustained growth of the isolate (final cell density 5–9∙107 cells ml−1). No growth was observed with formate, methanol, ethanol, isopropanol, glycerol, citrate, lactate, malate (10 mM each), fructose, glucose, sucrose, maltose, xylose, starch, and alginate (2 g L−1 each) during 4 weeks of incubation under optimal growth conditions.
Potential electron acceptors were tested with molecular hydrogen [H2/CO2 (20:80, v/v)], pyruvate (10 mM), or in simultaneous presence of hydrogen and pyruvate as energy and/or carbon sources. Strain SE56T could grow lithoautotrophically with molecular hydrogen as an electron donor, sulfite (5 mM) or thiosulfate (15 mM) as a terminal electron acceptor, and CO2/HCO3− as carbon source. Under lithoautrotrophic conditions with sulfate (15 mM) as a terminal electron acceptor, no growth was observed. Growth with sulfate (15 mM), sulfite (5 mM), or thiosulfate (15 mM) was observed with pyruvate (10 mM) or with hydrogen combined with pyruvate. Elemental sulfur (5 g L−1), fumarate, nitrate (10 mM each), nitrite (2.5 mM), chlorate, perchlorate (5 mM each), and ferrihydrite [90 mmol Fe(III) L−1] did not support growth under any tested conditions. Strain SE56T did not grow aerobically or when oxygen (0.5, 2.0, or 5.0%, v/v) was added to the gas phase. No growth was observed with pyruvate (10 mM), yeast extract, or peptone (2 g L−1 each) in the absence of an electron acceptor.
The novel isolate shares general features with the other members of the genus, such as morphology, growth conditions, capacity to reduce oxidized sulfur compounds, and inability to grow through iron or nitrate reduction. The differential characteristics of strain SE56T and other species of the genus Archaeoglobus are summarized in Table 1.
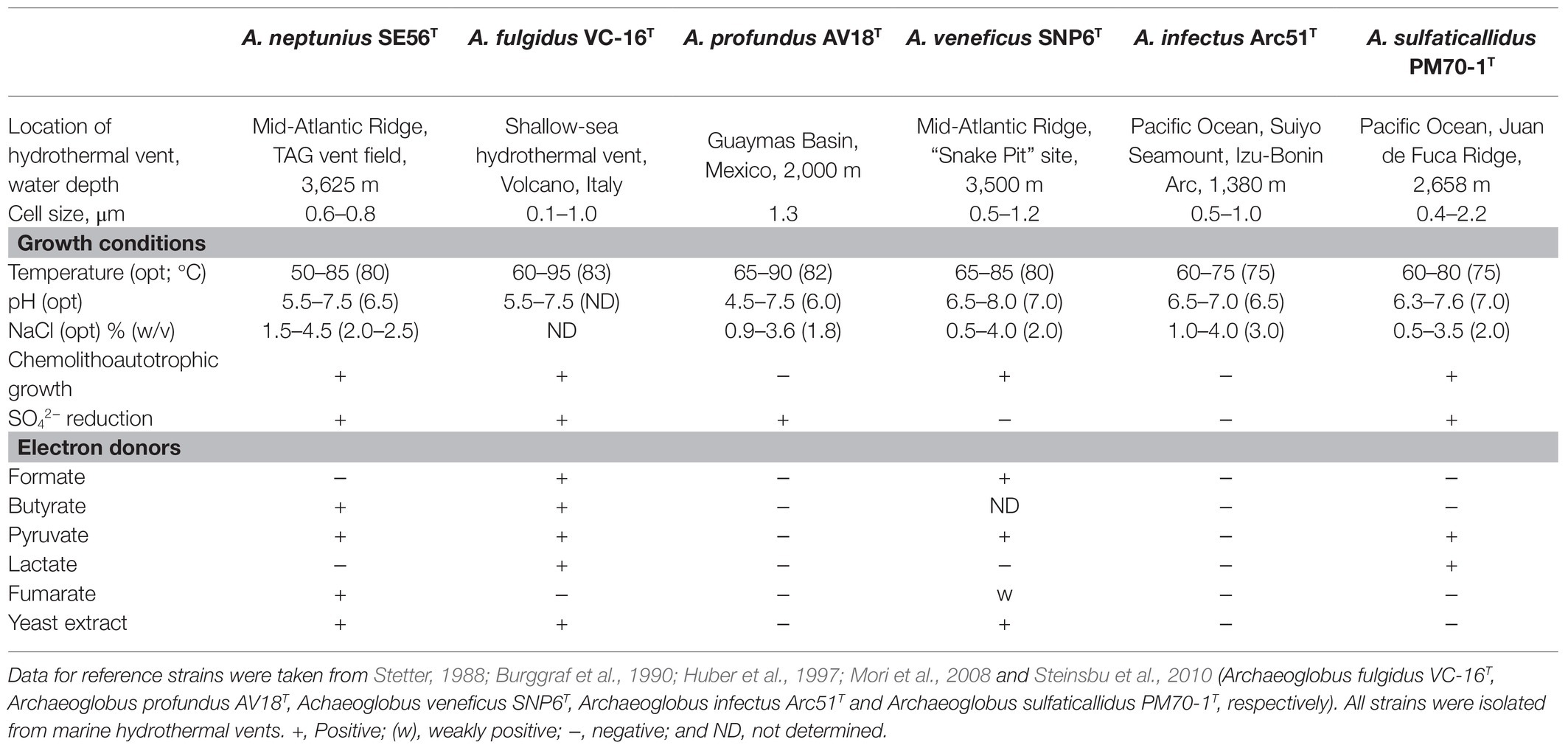
Table 1. Differential characteristics of strain SE56T and the type stains of the species of the genus Archaeoglobus.
The draft genome of strain SE56T was assembled into 32 contigs with genome size of 2,115,826 bp and a N50 value of 138,848 bp. Final assembly coverage was 153.537×. Based on CheckM analysis of single copy-core genes, the estimate of genome completeness was 100% and the estimate of contamination was 0%. The genomic DNA G + C content was 46.0 mol%. Annotation with prokaryotic genome annotation pipeline (PGAP) resulted in prediction of 2,471 genes, 2,386 of which are protein-coding sequences (CDSs) that cover about 96.5% of the entire genome. Genome also contained one copy of 5, 16, and 23S rRNA genes and 47 tRNA genes for all 20 standard amino acids. Two CRISPR loci were found (Figure 3).
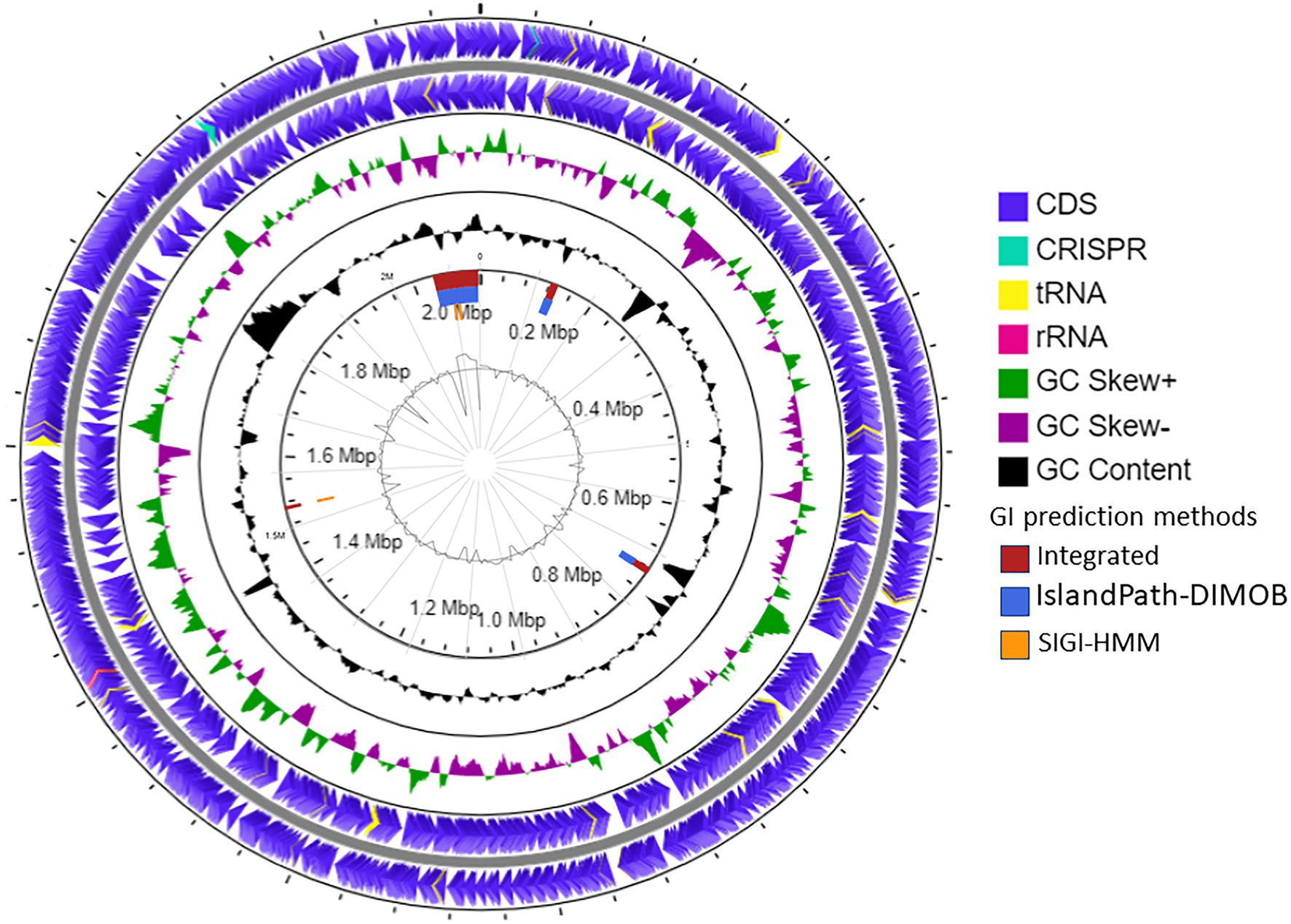
Figure 3. Schematic representation of the genome of A. neptunius SE56T. Labeling from the outside to the center is as follows: circle 1, genes on the forward strand; circle 2, genes on reverse strand [tRNAs yellow, ribosomal RNAs (rRNAs) lilac pink, and clustered regularly interspaced short palindromic repeats (CRISPR) cyan blue]; circle 3, G + C skew; circle 4, G + C content; circle 5, genomic islands shown as red, blue, and orange rectangles attributed, respectively, to integrated, IslandPath-DIMOB and SIGI-HMM prediction genomic islands methods; and circle 6, IslandViewer4 automatic calculation of the G + C content.
Majority of the CDSs (72.3%) could be assigned to at least one COG group. The main predicted COG categories (encompassing more than 100 CDSs) were energy production and conversion (11.73%); amino acid transport and metabolism (8.60%); translation, ribosomal structure, and biogenesis (8.60%); transcription (6.73%); lipid transport and metabolism (5.87%); and coenzyme transport and metabolism (5.72%) and replication, recombination, and repair (5.36%).
The prediction of laterally transferred genes showed that the genome of strain SE56T possesses four GI with a total length of 112.5 kb (Figure 3). Few genes were found encoding for enzymes related to archaeal chromosome replication, segregation, and cell division, such as ribonucleotide reductase, DNA helicase, transcription factor B, chromosome partitioning protein ParA, and cell division protein FtsH. Genes for 5'-nucleotidase SurE, AMP phosphorylase, and ribose-5-phosphate isomerase A were present as well, while the majority of CDSs located on the genomic islands encoded for hypothetical proteins (Supplementary Table S1).
Strain SE56T is capable of autotrophic growth using CO2 as a carbon source. Like all Archaeoglobi species except A. profundus, strain SE56T has the complete set of genes for the CO2 fixation via reductive acetyl-CoA (Wood-Ljungdahl) pathway (Table 2; Klenk et al., 1997; von Jan et al., 2010; Anderson et al., 2011; Stokke et al., 2013; Manzella et al., 2015; Mardanov et al., 2015). The genome also contains a gene coding for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO, WP_202319886). This enzyme belongs to form III RubisCO, which does not participate in autotrophy in Archaea (Berg, 2011). Many other enzymes of the Calvin-Benson cycle are missing. The key enzymes of other known microbial carbon fixation pathways, namely, ATP-citrate lyase and citryl-CoA lyase (two variants of the reductive tricarboxylic acid cycle), malonyl-CoA reductase (3-hydroxypropionate bi-cycle), and most of enzymes participating in 3-hydroxypropionate/4-hydroxybutyrate and dicarboxylate/4-hydroxybutyrate cycles are also absent.
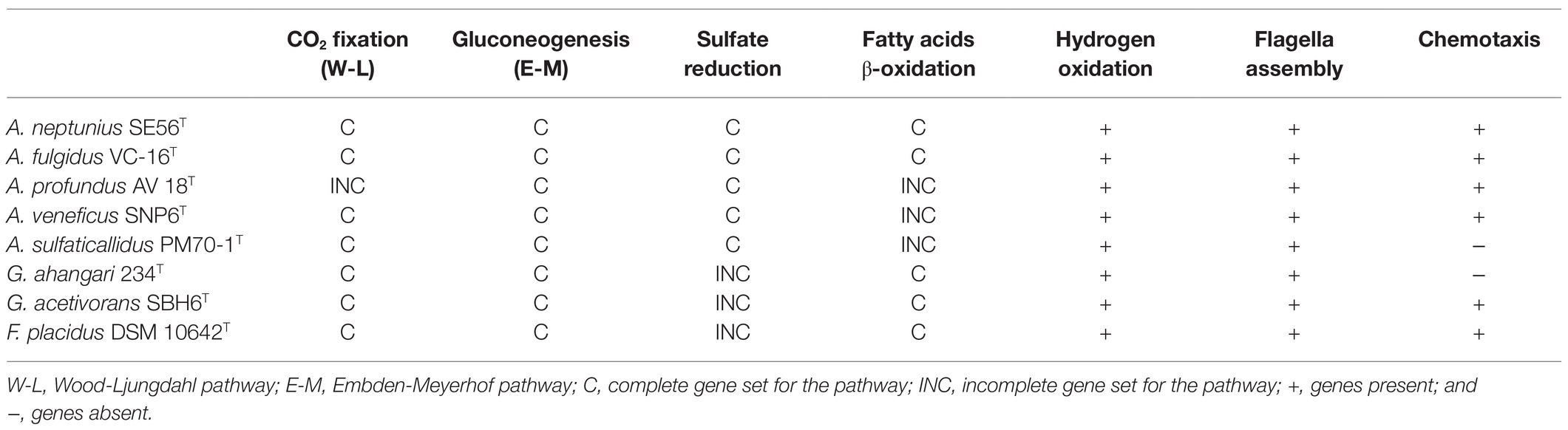
Table 2. Gene sets encoding various physiological properties in genomes of type strains of Archaeoglobi.
The genome of SE56T contains most of the genes encoding the Embden-Meyerhof pathway enzymes (Table 2), with the exception of the enzymes that catalyze irreversible reactions of glycolysis, pyruvate kinase, and 6-phosphofructokinase. Instead, there are genes encoding fructose-bisphosphatase (WP_202319097) and phosphoenolpyruvate synthase (WP_202318379) that enable glucose production from pyruvate. Thus, the Embden-Meyerhof pathway in this organism operates in reverse direction, toward gluconeogenesis.
Although strain SE56T lacks the key enzymes of the “standard” tricarboxylic acid cycle (TCA) cycle, aconitase and 2-oxoglutarate dehydrogenase complex, most of the genes involved in this cycle including citrate synthase (WP_202320798) are present. In this case, 2-oxoglutarate synthase may substitute the missing 2-oxoglutarate dehydrogenase. At least, two genes coding for 2-oxoglutarate synthase (oorCBAD) were found in the genome of strain SE56T. As well as the majority of Euryarchaeota whose genomes are sequenced, the genome of strain SE56T does not encode orthologs of aconitase A or members of other aconitase families. A putative family of aconitases, aconitase X, has been proposed on the basis of comparative-genomic analysis (Makarova and Koonin, 2003). The predicted aconitase X consists of two proteins that function together as a TCA cycle enzyme catalyzing the citrate to isocitrate isomerization. Experimental validation of this prediction has not yet been received. Nevertheless, a complete oxidative TCA is declared to function in the cells of class Archaeoglobi members (A. fulgidus, A. profundus, F. placidus, Geoglobus ahangari, and Geoglobus acetivorans). In the genome of strain SE56T, two genes (WP_202319516 and WP_202319760) were found encoding proteins that have 70% similarity with the proteins of the putative aconitase X from A. fulgidus (AF2333 and AF0055). Alternatively, incomplete TCA cycle can be used in the isolate SE56T to provide intermediates for anabolic pathways. Acetyl-CoA formed in Wood-Ljungdahl pathway or from acetate can be converted to pyruvate by a pyruvate synthase POR (porBADG, WP_202318910–WP_202318913). Pyruvate is converted to oxaloacetate in an irreversible reaction catalyzed by pyruvate carboxylase (pycAB, WP_202320749, and WP_202320510). Further formation of the necessary biosynthetic intermediates, in particular, succinyl-CoA and 2-oxoglutarate, is enabled by the presence of corresponding enzymes. This pathway has been experimentally shown in methanogenic archaea (Fuchs and Stupperich, 1978; Shieh and Whitman, 1987). Each of Archaeoglobi’s available genome contains homologs of all the genes involved in this incomplete TCA cycle.
The capacity of strain SE56T to utilize propionate is confirmed by a number of genes putatively encoding enzymes involved in propionate oxidation via the methylmalonyl-CoA pathway. This includes glutaconate CoA-transferase (WP_202318740 and WP_202318741) and a short mmc cluster (WP_202319622–WP_202319626) consisting of propionyl-CoA carboxylase, methylmalonyl-CoA epimerase and methylmalonyl-CoA mutase. Several enzymes such as succinyl-CoA synthase (SCS; WP_202320107 and WP_202320108), succinate dehydrogenase (SDH; WP_202319710, WP_202319660-, and WP_202319663), and fumarate hydratase (FHT; WP_202319682 and WP_202319683) can participate in both propionate oxidation and TCA cycle. The NAD-dependent malic enzyme (maeA, WP_202320482), pyruvate ferredoxin oxidoreductase (porBADG, WP_202318910–WP_202318913), and/or formate acetyltransferase (WP_202319769) can be involved in final steps of propionate oxidation to acetyl-CoA.
The genome contains a large number of genes coding for fatty acid utilization enzymes via β-oxidation pathway (Table 2), including long-chain acyl-CoA synthetases, acyl-CoA dehydrogenases, enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase, and acetyl-CoA acyltransferases. The genome also possesses 13 genes encoding acetyl-CoA C-acetyltransferase that are involved in fatty acid metabolism too, mainly in butyrate oxidation. This is consistent with the ability of strain SE56T to utilize butyrate and stearate. Archaeoglobus fulgidus, as well as the species of the related genus Geoglobus, are known to use fatty acids as electron donors (Kashefi et al., 2002; Slobodkina et al., 2009; Khelifi et al., 2010). In the genomes of these organisms and in the genome of Ferroglobus placidus, a large number of genes encoding the core enzymes of fatty acid β-oxidation have been found (Klenk et al., 1997; Anderson et al., 2011; Manzella et al., 2015; Mardanov et al., 2015). In contrast, in the genome of A. veneficus, we found only one gene of long-chain acyl-CoA synthetase and two genes of acetyl-CoA acetyltransferase. The genome of A. sulfaticallidus which was reported as unable to utilize butyrate (Steinsbu et al., 2010), lacks genes of two of the key enzymes for β-oxidation: enoyl-CoA hydratase and 3-hydroxyacyl-CoA dehydrogenase. The same genes are missing in the genome of A. profundus, as previously reported (von Jan et al., 2010).
Our attempts to grow strain SE56T with lactate as the electron donor and sulfite or sulfate as the electron acceptor were unsuccessful, although two known strains of A. fulgidus are capable of oxidizing lactate with sulfate. The genes for D-lactate and L-lactate dehydrogenases (dld, AF0394 and lldD, AF0807) were annotated in the genome of the type strain A. fulgidus (Klenk et al., 1997). The transcriptomic study proposed that lldD (AF0807) is part of a cluster and operates along with L-lactate permease (AF0806), monomeric dld (AF0808), and oligomeric LldEFG (AF0809−AF0811) lactate dehydrogenases (Hocking et al., 2014). This study also suggested to consider two more oxidoreductases (AF0507 and AF0867) as putative lactate dehydrogenases. In the genome of strain SE56T, we have found close homologs (62–78%) of proteins encoded by AF0394, AF0507, and AF0867 and the homologs of the subunits of LldEFG lactate dehydrogenase. Yet, the homologs of L-lactate permease and lactate dehydrogenases lldD and dld, encoded by the genes AF0806−AF0808 were absent. Thus, the reason for the absence of growth of strain SE56T with lactate is unclear. This may be because the missing enzymes are essential for lactate metabolism, or because the conditions for growth of strain SE56T with lactate are not yet identified.
A cluster of genes encoding a membrane-bound hydrogen uptake [NiFe] hydrogenase (Vht), a cytoplasmic [NiFe] hydrogenase:heterodisulfide (Mvh:HdrABC) complex, and a set of hydrogenase maturation proteins (WP_202319836−WP_202319849) provides strain SE56T the ability to utilize molecular hydrogen (Table 2). This cluster is syntenic to the one encoding the hydrogenases in A. fulgidus (AF1365−AF1381). Large catalytic subunits of Vht hydrogenase (vhtA) and Mvh hydrogenase (mvhA) of strain SE56T have 83 and 63% of amino acids similarities with corresponding proteins in A. fulgidus. According to a model of hydrogen metabolism proposed for A. fulgidus (Hocking et al., 2014), Vht hydrogenase is required for energy conservation which is achieved by electron transfer from hydrogen to sulfite via DsrMK/HdrDE complexes, where a particular putative heterodisulfide reductase HdrDE (AF0755) may play an important role. We detected close homolog of this HdrDE (WP_202318846) with 75% amino acid similarity in strain SE56T. Thus, the same model of hydrogenotrophy might be operative in A. neptunius SE56T cells.
Genome of strain SE56T contains the complete set of genes involved in dissimilatory sulfate reduction (Table 2), including sulfate adenylyltransferase sat and adenylyl-sulfate reductase aprAB (WP_202320080 and WP_202320082–WP_202320083), manganese-dependent inorganic pyrophosphatase ppaC (WP_202318845), dissimilatory sulfite reductase dsrABD (WP_202320149–WP_202320151), and dsrC which is more distantly located (WP_202320007). Genes coding for the electron transfer complexes dsrMKJOP (WP_202318457–WP_202318460) and QmoABC (WP_202319400–WP_202319402) are also present. Growth through sulfate reduction is a distinctive characteristic among species of the genus Archaeoglobus. Archaeoglobus veneficus and A. infectus were reported to be incapable to grow with sulfate as an electron acceptor (Huber et al., 1997; Mori et al., 2008). Meanwhile, the genome of A. veneficus contains a complete set of genes for dissimilatory sulfate reduction (Table 2), and A. infectus has aprA and dsrAB genes. A number of microorganisms did not demonstrate the growth due to dissimilatory sulfate reduction, despite the presence of complete set of genes for this pathway in their genomes (Finster et al., 2013; Slobodkin and Slobodkina, 2019). The reason of this is unclear; probably, special cultivation conditions are required.
In genome of the type species of the genus Archaeoglobus, several molybdopterin-binding oxidoreductases have been found, including a gene cluster (AF0174–AF0176) to which the function of a nitrate reductase has been assigned (Klenk et al., 1997; Richardson et al., 2001). Later, growth related to perchlorate reduction was demonstrated in this strain, and a two-stage model was proposed for this pathway (Liebensteiner et al., 2013). Based on genomic and proteomic evidence, it has been shown that perchlorate is reduced to chlorite by the molybdopterin-binding oxidoreductase encoded by the gene cluster AF0174–AF0176. Tetrathionate reductase (AF0157−AF0159) and an enzyme coded by the genes AF2384–AF2386 have been proposed as candidates for the second step of perchlorate reduction. Later transcriptome analysis showed upregulation of genes AF2384−AF2386 during growth with thiosulfate vs sulfate indicating that corresponding enzyme is a thiosulfate reductase (Hocking et al., 2014). In the genome of strain SE56T, we found a gene cluster (WP_202320693–WP_202320695) coding for proteins very similar to subunits of the perchlorate reductase of A. fulgidus (AF0174–AF0176). The amino acid similarity of the catalytic subunit (AF0176) to its homolog in strain SE56T (WP_202320693) was 79%. However, the genome of SE56T does not have close homologs to the genes of clusters AF0157–AF0159 and AF2384–AF2386, which could explain the inability of the strain to grow by (per)chlorate reduction. We did not find the close homologs to genes AF0157–AF0159 and AF2384–AF2386 in the genomes of the other Archaeoglobi representatives (Figure 4). Aside from the strain SE56T, we found close homologs to the catalytic subunit AF0176 (79% amino acids similarity) in the genomes of Ferroglobus placidus DSM 10642T (Ferp_0124) and Geoglobus ahangari 234T (GAH_01285). In both genomes, they are part of clusters coding for subunits of molybdopterin oxidoreductases. In G. ahangari, this cluster (GAH_01285–01288) has been previously designated as a nitrate reductase NarGHIJ (Manzella et al., 2015). The genome of F. placidus additionally contains another cluster, referenced Ferp_0311–0314, encoding three subunits (NarGHI) of the Nar-type respiratory nitrate reductase and the chaperone NarJ, which could explain why nitrate respiration could be demonstrated experimentally (Hafenbradl et al., 1996; Anderson et al., 2011). Our analysis showed that the alpha subunit of the nitrate reductase has the highest similarity with the nitrate reductase subunit alpha of Nitrospirae bacterium (71% amino acids similarity) and Deltaproteobacteria (67–68%). The other Archaeoglobi do not have close homologs to this enzyme and are unable to carry out dissimilatory nitrate reduction. The ability of F. placidus to reduce nitrate seems to have been acquired by horizontal transfer and to correspond to a quite recent evolutionary event.
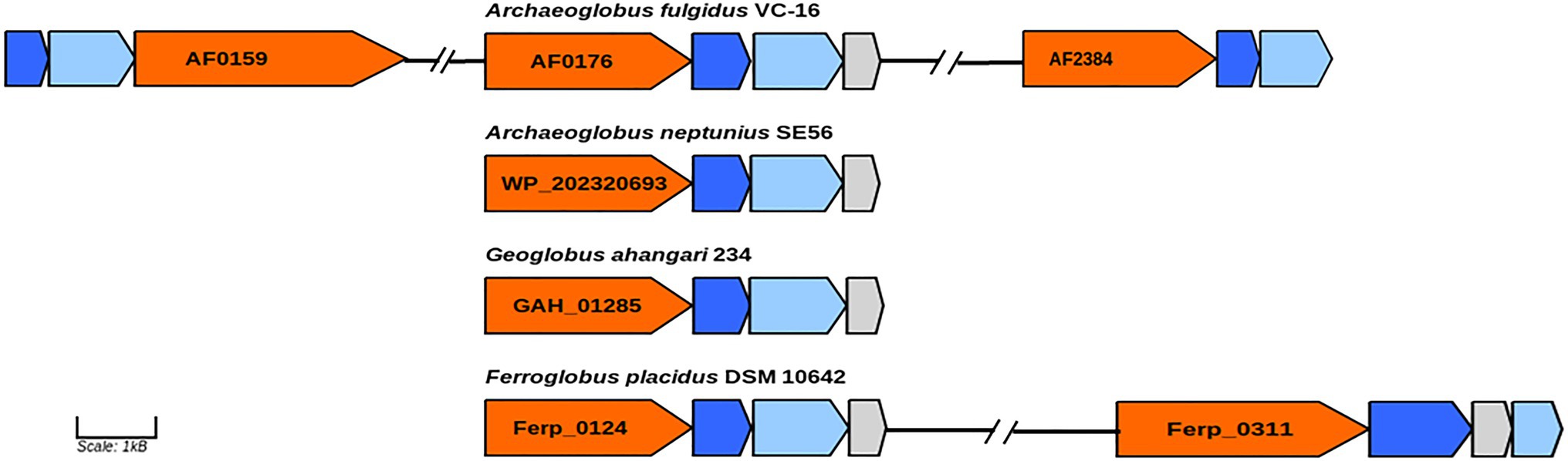
Figure 4. Genes of molybdopterin oxidoreductases probably involved in perchlorate and nitrate reduction. Orange, molybdopterin oxidoreductase molybdopterin binding subunit; blue, molybdopterin-containing oxidoreductase family iron–sulfur binding subunit; light blue, molybdopterin-containing oxidoreductase family membrane subunit; and gray: molybdenum cofactor insertion protein. GenBank accession numbers of catalytic subunits of each enzyme are shown.
Strain SE56T is motile and has a complete set of genes involved in proper assembly and function of flagella. The flagella apparatus operon flaB1-2B1-1D/EGFHIJ (WP_202320707–WP_202320713) found in the genome of strain SE56T is characteristic of Euryarcheota (Albers and Jarrell, 2015). The gene encoding preflagellin peptidase (FlaK; WP_202320792), essential for the flagella assembly, is not in the same cluster in the assembly used for this study. The isolate also has the chemotaxis system to direct the movement towards more favorable location, including an operon cheWFYBACDR (WP_202319867–WP_202319871) and genes encoding homologs of several methyl-accepting chemotaxis (MCP) proteins. Unlike A. fulgidus, A. profundus and A. veneficus, flagella- and chemotaxis-coding genes do not cluster together in the strain SE56T. Among members of the genus Archaeoglobus, A. profundus and A. sulfaticallidus have been reported to be non-motile (Burggraf et al., 1990; Steinsbu et al., 2010). Our analysis of genome data of A. sulfaticallidus revealed the presence of a complete fla genes cluster as well as putative flaK gene (AGK60851–AGK60859; AGK61636). However, it lacks the chemotaxis system che genes that might explain the unobserved motility (Table 2). In contrast, A. profundus possesses both fla and che genes clusters which are virtually identical in content, order and orientation to corresponding genes of the motile Archaeoglobi. This discrepancy between genotype and phenotype has been noted earlier, and it was assumed that A. profundus can be motile in some specific conditions, or additional unknown factors are needed for motility (von Jan et al., 2010).
The 16S rRNA gene-based phylogenetic analysis showed that strain SE56T belongs to the genus Archaeoglobus. Further clarification of the taxonomic position of strain SE56T was carried out using genome-based methods, ANI, AF, dDDH and AAI. At the time of writing, all genome sequences of Archaeoglobales type strains were available in public databases, with the exception of the type strain of A. infectus. The properties and statistics of genome sequences are presented in Table 3. The pairwise OrthoANIu value between genomes of strain SE56T and its closest relative organism, A. fulgidus VC-16T (NC_000917.1), was 73.49%. The OrthoANIu values with other available species were below 68.5%. Similar results were obtained by means of ANIb and MiSI algorithms (Table 3). The AF value for strain SE56T and A. fulgidus VC-16T was 0.62, whereas these values for strain SE56T and other representatives of the order Archaeoglobales were 0.21–0.30 (Table 3). The predicted in silico DNA–DNA hybridization (dDDH) values between strain SE56T and members of the order Archaeoglobales varied from 31.7% (A. profundus DSM 5631T) to 18.6% (A. fulgidus VC-16T). All these values are lower than the proposed threshold criteria for prokaryotic species delineation (95–96% ANI, 0.6–0.7 AF and 70% dDDH; Goris et al., 2007; Richter and Rosselló-Móra, 2009; Varghese et al., 2015).
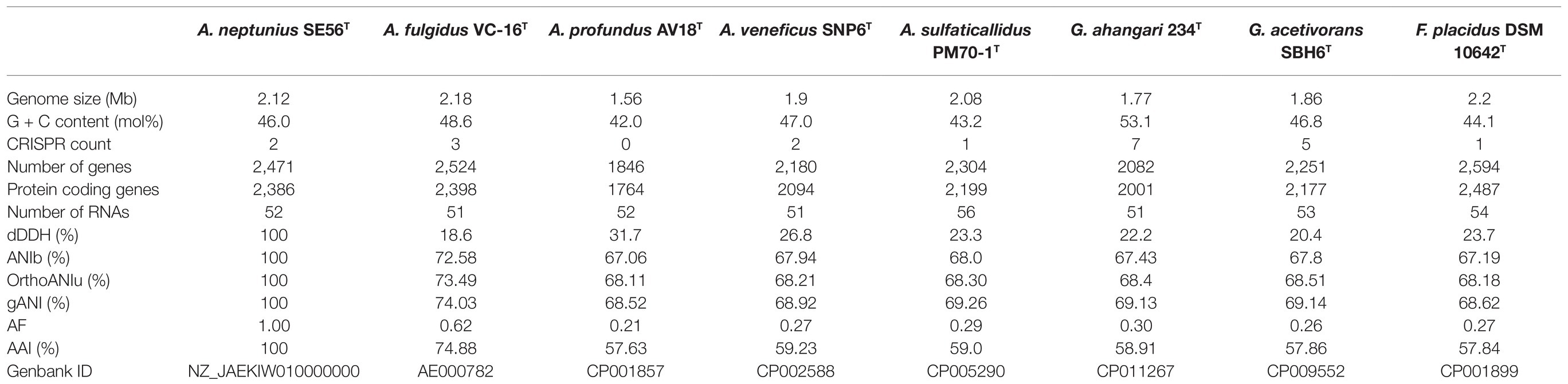
Table 3. General genomic features and digital DNA–DNA hybridization (dDDH), average nucleotide identity (ANI), AAI, and alignment fraction (AF) values of strain Archaeoglobus neptunius SE56T and the type strains of the related species.
Additionally, we studied the taxonomic position of strain SE56T based on the alignment of a concatenated set of 122 single-copy partial amino acid sequences of conserved archaeal proteins. The phylogenomic reconstruction confirmed the position of strain SE56T as a novel species within the genus Archaeoglobus. At the same time, it clearly indicated a polyphyletic origin of the species currently included into the genus Archaeoglobus (Figure 5). The AAI value between strain SE56T and its closest relative, A. fulgidus VC-16T, was 74.88%. The AAI values between the other members of the genus Archaeoglobus were in the range of 57–60% (Figure 6). Corresponding ANI and AF values were in the range of 68–70% and 0.21–0.29, respectively (Supplementary Tables S2 and S3). At present, the AAI thresholds proposed for taxonomic delineation are 45–65% for the same family, 65–95% for the same genus and 95–100% for the same species (Konstantinidis et al., 2017). Other proposed genus demarcation criteria are 73.34–74.62% ANI and 0.308–0.354 AF (Barco et al., 2020). On this basis, strain SE56T and A. fulgidus represent different species of one genus, while each of the other three known species of Archaeoglobus represents an individual genus, distinct from the first and distinct from each other. Among the other members of the class Archaeoglobi, only G. ahangari and G. acetivorans have AAI, ANI and AF values above the proposed genus threshold, that confirms the placement of these species into the same genus. Except strain SE56T and A. fulgidus, as well as Geoglobus species, the AAI, ANI and AF values between all species of the Archaeoglobi class were in the range of 56–64%, 68–71%, and 0.21–0.39, respectively (Figure 6; Supplementary Tables S2 and S3), proving that they belong to different genera of the same family.
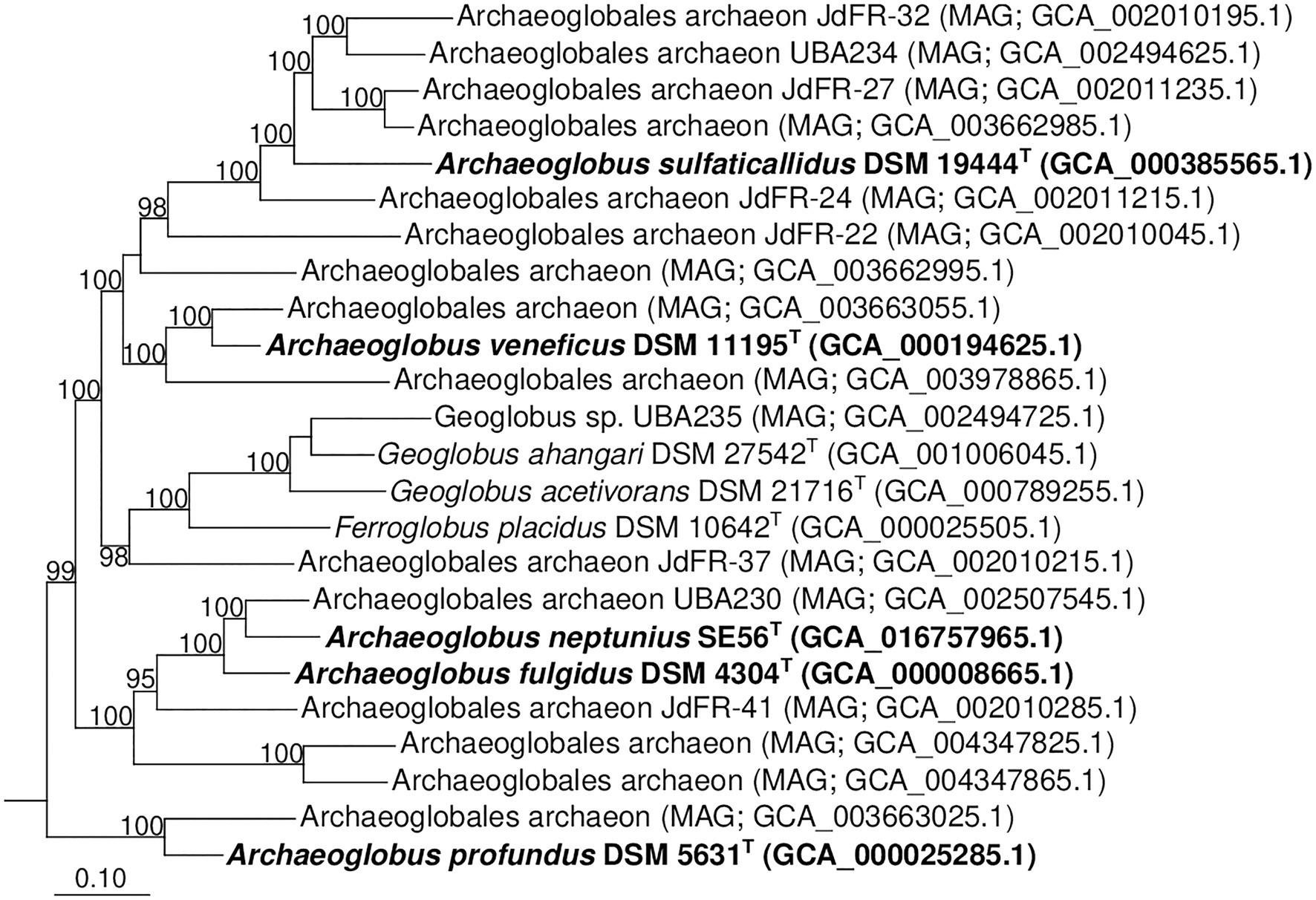
Figure 5. Phylogenomic placement of A. neptunius and other species of family Archaeoglobaceae based on concatenated partial amino acid sequences of 122 archaeal conservative proteins [Parks et al., 2018; taxonomic designations correspond with Genome Taxonomy DataBase (GTDB)]. The tree was built using the PhyML 3.0 program (Guindon et al., 2010). For rooting, the tree sequences from 62 genomes of type strains of type species of genera of “Halobacteriota” phylum (correspond with GTDB) were taken. All assemblages of Archaeoglobaceae family according to GTDB Release 05-RS95 are shown on the tree. Bootstrap values above 90% are shown at the nodes. Bar, 0.10 changes per position.
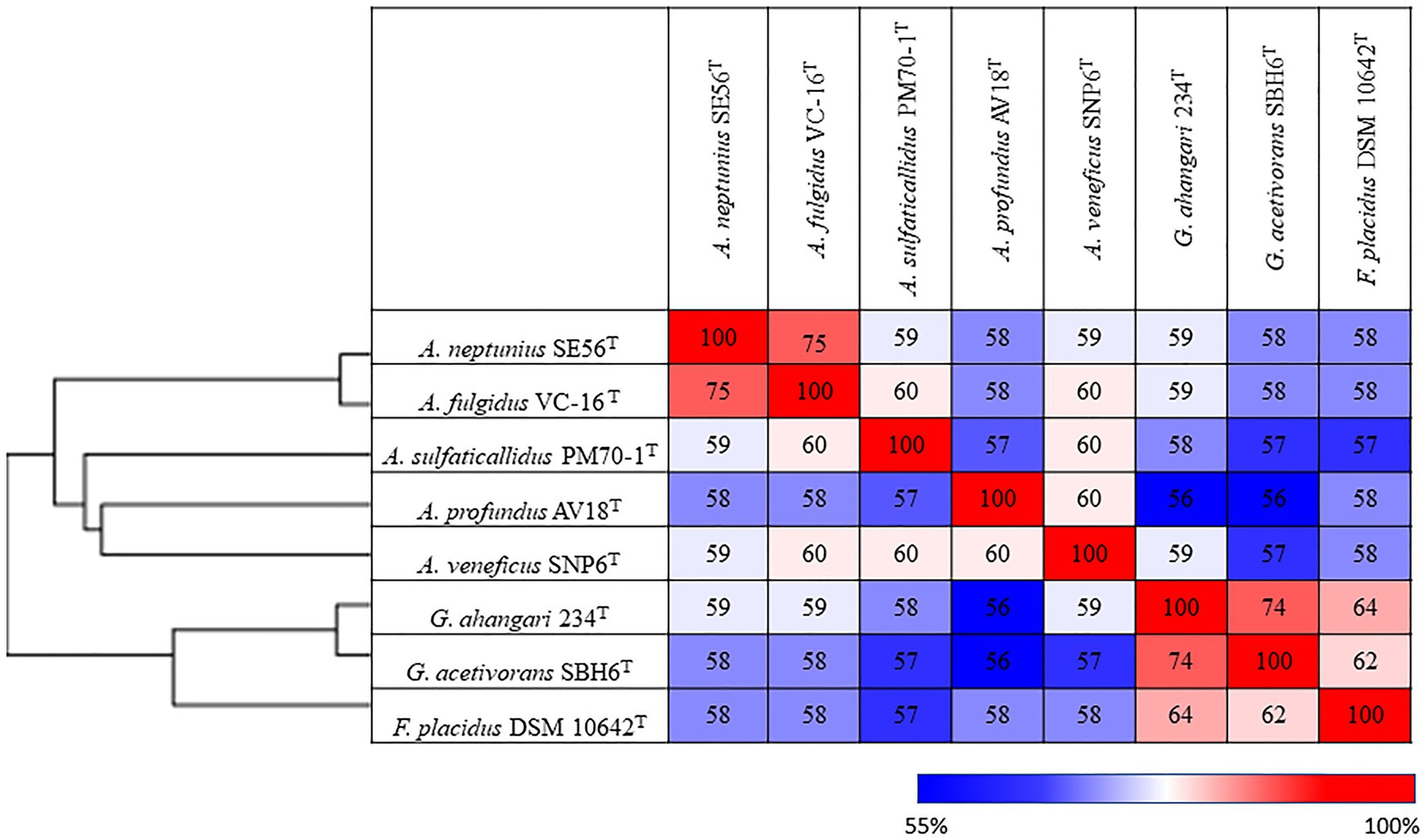
Figure 6. Pairwise average amino acid identity (AAI) determined for members of the class Archaeglobi.
We therefore conclude, that, according to phylogenomic differentiation supported by ANI, AF and AAI values, the genus Archaeoglobus should be divided into four distinct genera. The genus name Archaeoglobus should be retained for the monophyletic group containing the type species, A. fulgidus. We propose to consider the strain SE56T as a new species of the genus Archaeoglobus, Archaeoglobus neptunius sp. nov. All four proposed genera belong to the family Archaeoglobaceae and share the main morphological, physiological and metabolic features. Representatives of these genera are hyperthermophilic strict anaerobes with respiratory type of metabolism that use sulfur compounds as the terminal electron acceptors. All known species were isolated from marine ecosystems. In addition to the phylogenetic distance, each of the proposed genera has the distinctive phenotypic properties (Tables 1 and 2). The genus Archaeoglobus sensu stricto differs from the other three genera in its ability to utilize fatty acids provided by the presence of all the genes necessary for β-oxidation of fatty acids. A key distinguishing feature of the genus represented by A. profundus is the inability to grow lithoautotrophically caused by the incompleteness of Wood-Ljungdahl pathway. The genus represented by A. sulfaticallidus can use sulfate as an electron acceptor for lithoautotrophic growth with molecular hydrogen, whereas members of other genera can perform dissimilatory sulfate reduction only with organic carbon source. In addition, cells of A. sulfaticallidus are non-motile, lack genes encoding chemotaxis and have lower optimum growth temperature. The genus encompassing A. veneficus is distinguished by its inability to reduce sulfate although it has all the necessary genes. Further work, including the sequencing of the genome of A. infectus and core- and pan-genome analysis is required for the formal proposal of the reclassification of the genus Archaeoglobus.
Archaeoglobus neptunius (nep.tu’ni.us L. masc. adj. neptunius, pertaining to Neptunius (Roman god of the sea), referring to the habitat of the microorganism).
Cells are Gram-negative irregular motile cocci of 0.6–0.8 μm in diameter usually occurring singly. Blue-green fluorescence under UV light. Strictly anaerobic. Growth occurs at 50–85°C (optimum at 80°C), pH 5.5–7.5 (optimum pH 6.5) and NaCl concentrations of 1.5–4.5% [w/v; optimum 2.0–2.5% (w/v) NaCl]. With sulfite or thiosulfate as an electron acceptor, it grows chemolithoautotrophically with molecular hydrogen as an electron donor and CO2/HCO3− as a carbon source. Chemoorganotrophic growth on acetate, propionate, butyrate, fumarate, pyruvate, succinate, peptone, tryptone and yeast extract with sulfate, sulfite or thiosulfate as electron acceptor. Growth is inhibited by elemental sulfur. Formate, methanol, ethanol, iso-propanol, glycerol, lactate, malate, citrate, fructose, glucose, sucrose, maltose, xylose, starch and alginate are not utilized. Elemental sulfur, fumarate, nitrate, nitrite, chlorate, perchlorate and Fe(III) are not used as electron acceptors.
The type strain is SE56T (=DSM 110954T = VKM B-3474T), isolated from a deep-sea hydrothermal vent of the Mid-Atlantic Ridge (TAG vent field: 26°13'69"N 44°82'61"W, 3,625 m water depth). The genome consists of a 2,115,826 bp chromosome with an overall G + C content of 46.0 mol%.
GenBank/EMBL/DDBJ accession (16S rRNA gene): MW531883.
GenBank/EMBL/DDBJ accession (genome): NZ_JAEKIW010000000.
The Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAEKIW000000000. The version described in this paper is version JAEKIW010000000. The BioSample data is available in the NCBI BioSample database (http://www.ncbi.nlm.nih.gov/biosample/) under the accession number SAMN17126929. The BioProject data is available in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) with BioProject ID: PRJNA686870. The 16S rRNA gene sequence of A. neptunius SE56T (retrieved from the whole-genome sequencing) has been deposited in GenBank/EMBL under accession number MW531883. The strain is available in the German Collection of Microorganisms and Cell Cultures (DSMZ) and All-Russian Collection of Microorganisms (VKM) under the accession numbers DSM 110954T and VKM B-3474T.
AS, GS, and MJ: conceptualization. GS, MA, AM, and KA: investigation. M-AC-B: resources. GS: writing – original draft preparation. AS, KA, AM, M-AC-B, and MA: writing – review and editing. GS and AM: visualization. AS and MJ: supervision and funding acquisition. All authors contributed to the article and approved the submitted version.
This research was funded by the Russian Foundation for Basic Research, grant number 18-54-15008, by the Ministry of Science and Higher Education of the Russian Federation, by the French–Russian collaborative project (CNRS/RFBR) Neptune (PRC Russie 2017 n°281295), and by the IRP 1211 MicrobSea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer TN declared a shared affiliation with one of the authors GS, AM, and AS to the handling editor at the time of the review.
The authors thank the captains and crews of the R/V Pourquoi pas? and the Nautile submersible team for their efficiency, as well as the chief scientists and scientific parties of the BICOSE 2 cruise (http://dx.doi.org/10.17600/18000004).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.679245/full#supplementary-material
AAI, Average amino acid identity; AF, Alignment fraction; ANI, Average nucleotide identity; bp, Base pair; CDS, Coding DNA sequences; COGs, Clusters of orthologous groups; CRISPR, Clustered regularly interspaced short palindromic repeats; dDDH, digital DNA–DNA hybridization; DNA, deoxyribonucleic acid; EMBL, European Molecular Biology Laboratory; IMG/M, Integrated microbial genomes and microbiomes; GI, Genomic islands; GTDB, Genome taxonomy database; NCBI, National Center for Biotechnology Information; PGAP, Prokaryotic genome annotation pipeline; rRNA, Ribosomal ribonucleic acid; TCA, Tricarboxylic acid cycle.
Albers, S. V., and Jarrell, K. F. (2015). The archaellum: how Archaea swim. Front. Microbiol. 6:23. doi: 10.3389/fmicb.2015.00023
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anderson, I., Risso, C., Holmes, D., Lucas, S., Copeland, A., Lapidus, A., et al. (2011). Complete genome sequence of Ferroglobus placidus AEDII12DO. Stand. Genomic Sci. 5, 50–60. doi: 10.4056/sigs.2225018
Anisimova, M., Gil, M., Dufayard, J. F., Dessimoz, C., and Gascuel, O. (2011). Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 60, 685–699. doi: 10.1093/sysbio/syr041
Baker, B. J., De Anda, V., Seitz, K. W., Dombrowski, N., Santoro, A. E., and Lloyd, K. G. (2020). Diversity, ecology and evolution of Archaea. Nat. Microbiol. 5, 887–900. doi: 10.1038/s41564-020-0715-z
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barco, R. A., Garrity, G. M., Scott, J. J., Amend, J. P., Nealson, K. H., and Emerson, D. (2020). A genus definition for Bacteria and Archaea based on a standard genome relatedness index. MBio 11, e02475–e02419. doi: 10.1128/mBio.02475-19
Beeder, J., Nilsen, R. K., Rosnes, J. T., Torsvik, T., and Lien, T. (1994). Archaeoglobus fulgidus isolated from hot North Sea oil field waters. Appl. Environ. Microbiol. 60, 1227–1231. doi: 10.1128/aem.60.4.1227-1231.1994
Benson, D. A., Boguski, M. S., Lipman, D. J., Oullette, B. F. F., Rapp, B. A., and Wheeler, D. L. (1999). GenBank. Nucleic Acids Res. 27, 12–17. doi: 10.1093/nar/27.1.12
Berg, I. A. (2011). Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl. Environ. Microbiol. 77, 1925–1936. doi: 10.1128/AEM.02473-10
Bertelli, C., Laird, M. R., Williams, K. P., Simon Fraser University Research Computing Group, Lau, B. Y., Hoad, G., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343
Birkeland, N. K., Schönheit, P., Poghosyan, L., Fiebig, A., and Klenk, H. P. (2017). Complete genome sequence analysis of Archaeoglobus fulgidus strain 7324 (DSM 8774), a hyperthermophilic archaeal sulfate reducer from a North Sea oil field. Stand. Genomic Sci. 12:79. doi: 10.1186/s40793-017-0296-5
Burggraf, S., Jannasch, H. W., Nicolaus, B., and Stetter, K. O. (1990). Archaeoglobus profundus sp. nov., represents a new species within the sulfur-reducing Archaebacteria. Syst. Appl. Microbiol. 13, 24–28. doi: 10.1016/S0723-2020(11)80176-1
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2019). GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Chen, I.-M. A., Chu, K., Palaniappan, K., Pillay, M., Ratner, A., Huang, J., et al. (2019). IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, 666–677. doi: 10.1093/nar/gky901
Couvin, D., Bernheim, A., Toffano-Nioche, C., Touchon, M., Michalik, J., Néron, B., et al. (2018). CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46, W246–W251. doi: 10.1093/nar/gky425
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Finster, K. W., Kjeldsen, K. U., Kube, M., Reinhardt, R., Mussmann, M., Amann, R., et al. (2013). Complete genome sequence of Desulfocapsa sulfexigens, a marine deltaproteobacterium specialized in disproportionating inorganic sulfur compounds. Stand. Genomic Sci. 8, 58–68. doi: 10.4056/sigs.3777412
Fuchs, G., and Stupperich, E. (1978). Evidence for an incomplete reductive carboxylic acid cycle in Methanobacterium thermoautotrophicum. Arch. Microbiol. 118, 121–125. doi: 10.1007/BF00406084
Gerhardt, P., Murray, R. G. E., Wood, W. A., and Krieg, N. R. (1994). Methods for General and Molecular Bacteriology. Washington, DC: American Society for Microbiology.
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Grant, J. R., and Stothard, P. (2008). The cgview server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36, W181–W184. doi: 10.1093/nar/gkn179
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hafenbradl, D., Keller, M., Dirmeier, R., Rachel, R., Roßnagel, P., Burggraf, S., et al. (1996). Ferroglobus placidus gen. Nov., sp. nov., a novel hyperthermophilic archaeum that oxidizes Fe2+ at neutral pH under anoxic conditions. Arch. Microbiol. 166, 308–314. doi: 10.1007/s002030050388
Harrison, K. J., de Crécy-Lagard, V., and Zallot, R. (2018). Gene graphics: a genomic neighborhood data visualization web application. Bioinformatics 34, 1406–1408. doi: 10.1093/bioinformatics/btx793
Henstra, A. M., Dijkema, C., and Stams, A. J. M. (2007). Archaeoglobus fulgidus couples CO oxidation to sulfate reduction and acetogenesis with transient formate accumulation. Environ. Microbiol. 9, 1836–1841. doi: 10.1111/j.1462-2920.2007.01306.x
Hocking, W. P., Stokke, R., Roalkvam, I., and Steen, I. H. (2014). Identification of key components in the energy metabolism of the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus by transcriptome analyses. Front. Microbiol. 5:95. doi: 10.3389/fmicb.2014.00095
Huber, H., Jannasch, H., Rachel, R., Fuchs, T., and Stetter, K. O. (1997). Archaeoglobus veneficus sp. nov., a novel facultative chemolithoautotrophic hyperthermophilic sulfite reducer, isolated from abyssal black smokers. Syst. Appl. Microbiol. 20, 374–380. doi: 10.1016/S0723-2020(97)80005-7
Huerta-Cepas, J., Forslund, S. K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Kashefi, K., Tor, J. M., Holmes, D. E., Gaw Van Praagh, C. V., Reysenbach, A.-L., and Lovley, D. R. (2002). Geoglobus ahangari gen. nov., sp. nov., a novel hyperthermophilic archaeon capable of oxidizing organic acids and growing autotrophically on hydrogen with Fe(III) serving as the sole electron acceptor. Int. J. Syst. Evol. Microbiol. 52, 719–728. doi: 10.1099/00207713-52-3-719
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Khelifi, N., Amin Ali, O., Roche, P., Grossi, V., Brochier-Armanet, C., Valette, O., et al. (2014). Anaerobic oxidation of long-chain n-alkanes by the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus. ISME J. 8, 3057–3060. doi: 10.1038/ismej.2014.58
Khelifi, N., Grossi, V., Hamdi, M., Dolla, A., Tholozan, J. L., Ollivier, B., et al. (2010). Anaerobic oxidation of fatty acids and alkenes by the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus. Appl. Environ. Microbiol. 76, 3057–3060. doi: 10.1128/AEM.02810-09
Klenk, H. P., Clayton, R. A., Tomb, J. F., White, O., Nelson, K. E., Ketchum, K. A., et al. (1997). The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390, 364–370. doi: 10.1038/37052
Konstantinidis, K. T., Rosselló-Móra, R., and Amann, R. (2017). Uncultivated microbes in need of their own taxonomy. ISME J. 11, 2399–2406. doi: 10.1038/ismej.2017.113
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Le, S. Q., and Gascuel, O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. doi: 10.1093/molbev/msn067
Lefort, V., Longueville, J. E., and Gascuel, O. (2017). SMS: smart model selection in PhyML. Mol. Biol. Evol. 34, 2422–2424. doi: 10.1093/molbev/msx149
Liebensteiner, M. G., Pinkse, M. W. H., Schaap, P. J., Stams, A. J. M., and Lomans, B. P. (2013). Archaeal (per)chlorate reduction at high temperature: an interplay of biotic and abiotic reactions. Science 340, 85–87. doi: 10.1126/science.1233957
Makarova, K. R., and Koonin, E. V. (2003). Filling a gap in the central metabolism of archaea: prediction of a novel aconitase by comparative-genomic analysis. FEMS Microbiol. Lett. 227, 17–23. doi: 10.1016/S0378-1097(03)00596-2
Manzella, M. P., Holmes, D. E., Rocheleau, J. M., Chung, A., Reguera, G., and Kashefi, K. (2015). The complete genome sequence and emendation of the hyperthermophilic, obligate iron-reducing archaeon “Geoglobus ahangari” strain 234T. Stand. Genomic Sci. 10:77. doi: 10.1186/s40793-015-0035-8
Mardanov, A. V., Slobodkina, G. B., Slobodkin, A. I., Beletsky, A. V., Gavrilov, S. N., Kublanov, I. V., et al. (2015). The Geoglobus acetivorans genome: Fe(III) reduction, acetate utilization, autotrophic growth, and degradation of aromatic compounds in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 81, 1003–1012. doi: 10.1128/AEM.02705-14
Martin, W. F., and Sousa, F. L. (2016). Early microbial evolution: the age of anaerobes. Cold Spring Harb. Perspect. Biol. 8:a018127. doi: 10.1101/cshperspect.a018127
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 14:60. doi: 10.1186/1471-2105-14-60
Mori, K., Maruyama, A., Urabe, T., Suzuki, K., and Hanada, S. (2008). Archaeoglobus infectus sp. nov., a novel thermophilic, chemolithoheterotrophic archaeon isolated from a deep-sea rock collected at Suiyo Seamount, Izu-Bonin Arc, western Pacific Ocean. Int. J. Syst. Evol. Microbiol. 58, 810–816. doi: 10.1099/ijs.0.65422-0
Offre, P., Spang, A., and Schleper, C. (2013). Archaea in biogeochemical cycles. Annu. Rev. Microbiol. 67, 437–457. doi: 10.1146/annurev-micro-092412-155614
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Palombarini, F., Di Fabio, E., Boffi, A., Macone, A., and Bonamore, A. (2020). Ferritin nanocages for protein delivery to tumor cells. Molecules 25:825. doi: 10.3390/molecules25040825
Park, C., and Park, W. (2018). Survival and energy producing strategies of alkane degraders under extreme conditions and their biotechnological potential. Front. Microbiol. 9:1081. doi: 10.3389/fmicb.2018.01081
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, R., Skarshewski, A., Chaumeil, P.-A., et al. (2018). Standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Parte, A. C., Sardà Carbasse, J., Meier-Kolthoff, J. P., Reimer, L. C., and Göker, M. (2020). List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Rabus, R., Venceslau, S. S., Wöhlbrand, L., Voordouw, G., Wall, J. D., and Pereira, I. A. (2015). A post-genomic view of the ecophysiology, catabolism and biotechnological relevance of sulphate-reducing prokaryotes. Adv. Microb. Physiol. 66, 55–321. doi: 10.1016/bs.ampbs.2015.05.002
Richardson, D. J., Berks, B. C., Russell, D. A., Spiro, S., and Taylor, C. J. (2001). Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell. Mol. Life Sci. 58, 165–178. doi: 10.1007/PL00000845
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Richter, M., Rosselló-Móra, R., Glöckner, F. O., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Shieh, J. S., and Whitman, W. B. (1987). Pathway of acetate assimilation in autotrophic and heterotrophic methanococci. J. Bacteriol. 169, 5327–5329. doi: 10.1128/jb.169.11.5327-5329.1987
Slobodkin, A. I., Reysenbach, A. L., Slobodkina, G. B., Baslerov, R. V., Kostrikina, N. A., Wagner, I. D., et al. (2012). Thermosulfurimonas dismutans gen. Nov., sp. nov., an extremely thermophilic sulfur-disproportionating bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 62, 2565–2571. doi: 10.1099/ijs.0.034397-0
Slobodkin, A. I., and Slobodkina, G. B. (2019). Diversity of sulfur-disproportionating microorganisms. Microbiology 88, 509–522. doi: 10.1134/S0026261719050138
Slobodkina, G. B., Baslerov, R. V., Novikov, A. A., Viryasov, M. B., Bonch-Osmolovskaya, E. A., and Slobodkin, A. I. (2016). Inmirania thermothiophila gen. Nov., sp. nov., a thermophilic, facultatively autotrophic, sulfur-oxidizing gammaproteobacterium isolated from shallow-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 66, 701–706. doi: 10.1099/ijsem.0.000773
Slobodkina, G. B., Kolganova, T. V., Querellou, J., Bonch-Osmolovskaya, E. A., and Slobodkin, A. I. (2009). Geoglobus acetivorans sp. nov., an iron(III)-reducing archaeon from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 59, 2880–2883. doi: 10.1099/ijs.0.011080-0
Søndergaard, D., Pedersen, C. N. S., and Greening, C. (2016). HydDB: A web tool for hydrogenase classification and analysis. Sci. Rep. 6:34212. doi: 10.1038/srep34212
Steinsbu, B. O., Thorseth, I. H., Nakagawa, S., Inagaki, F., Lever, M. A., Engelen, B., et al. (2010). Archaeoglobus sulfaticallidus sp. nov., a thermophilic and facultatively lithoautotrophic sulfate-reducer isolated from black rust exposed to hot ridge flank crustal fluids. Int. J. Syst. Evol. Microbiol. 60, 2745–2752. doi: 10.1099/ijs.0.016105-0
Stetter, K. O. (1988). Archaeoglobus fulgidus gen. Nov., sp. nov.: a new taxon of extremely thermophilic Archaebacteria. Syst. Appl. Microbiol. 10, 172–173. doi: 10.1016/S0723-2020(88)80032-8
Stetter, K. O. (2006). Hyperthermophiles in the history of life. Philos. Trans. R. Soc. B 361, 1837–1843. doi: 10.1098/rstb.2006.1907
Stetter, K. O., Lauerer, G., Thomm, M., and Neuner, A. (1987). Isolation of extremely thermophilic sulfate reducers: evidence for a novel branch of archaebacteria. Science 236, 822–824. doi: 10.1126/science.236.4803.822
Stokke, R., Hocking, W. P., Steinsbu, B. O., and Steen, I. H. (2013). Complete genome sequence of the thermophilic and facultatively chemolithoautotrophic sulfate reducer Archaeoglobus sulfaticallidus strain PM70-1T. Genome Announc. 1, 4–5. doi: 10.1128/genomeA.00406-13
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526. doi: 10.1093/oxfordjournals.molbev.a040023
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Tor, J. M., Kashefi, K., and Lovley, D. R. (2001). Acetate oxidation coupled to Fe(III) reduction in hyperthermophilic microorganisms. Appl. Environ. Microbiol. 67, 1363–1365. doi: 10.1128/AEM.67.3.1363-1365.2001
Varghese, N. J., Mukherjee, S., Ivanova, N., Konstantinidis, K. T., Mavrommatis, K., Kyrpides, N. C., et al. (2015). Microbial species delineation using whole genome sequences. Nucleic Acids Res. 43, 6761–6771. doi: 10.1093/nar/gkv657
von Jan, M., Lapidus, A., Del Rio, T. G., Copeland, A., Tice, H., Cheng, J. F., et al. (2010). Complete genome sequence of Archaeoglobus profundus type strain (AV18). Stand. Genomic Sci. 2, 327–346. doi: 10.4056/sigs.942153
Weiss, M. C., Sousa, F. L., Mrnjavac, N., Neukirchen, S., Roettger, M., Nelson-Sathi, S., et al. (2016). The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1:16116. doi: 10.1038/nmicrobiol.2016.116
Woese, C. R., and Fox, G. E. (1977). Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U. S. A. 74, 5088–5090. doi: 10.1073/pnas.74.11.5088
Wolin, E. A., Wolin, M. J., and Wolfe, R. S. (1963). Formation of methane by bacterial extracts. J. Biol. Chem. 238, 2882–2886. doi: 10.1016/S0021-9258(18)67912-8
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblie. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Keywords: sulfate reduction, chemolithoautotroph, anaerobe, hyperthermophile, Archaea
Citation: Slobodkina G, Allioux M, Merkel A, Cambon-Bonavita M-A, Alain K, Jebbar M and Slobodkin A (2021) Physiological and Genomic Characterization of a Hyperthermophilic Archaeon Archaeoglobus neptunius sp. nov. Isolated From a Deep-Sea Hydrothermal Vent Warrants the Reclassification of the Genus Archaeoglobus. Front. Microbiol. 12:679245. doi: 10.3389/fmicb.2021.679245
Received: 11 March 2021; Accepted: 30 June 2021;
Published: 16 July 2021.
Edited by:
Rafael R. de la Haba, University of Sevilla, SpainReviewed by:
Chaomin Sun, Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaCopyright © 2021 Slobodkina, Allioux, Merkel, Cambon-Bonavita, Alain, Jebbar and Slobodkin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Galina Slobodkina, Z3Nsb2JvZGtpbmFAbWFpbC5ydQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.