- 1National Risk Assessment Laboratory for Antimicrobial Resistance of Animal Original Bacteria, South China Agricultural University, Guangzhou, China
- 2Laboratory of Veterinary Pharmacology, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 3Guangdong Provincial Key Laboratory of Microbial Safety and Health, Guangdong Institute of Microbiology, Guangdong Academy of Science, Guangzhou, China
- 4Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China
This study aimed to determine the prevalence and transmission characteristics of New Delhi metallo β-lactamase (NDM)-producing Escherichia coli from ducks in Guangdong, China. In this study, a total of 28 NDM-producing E. coli isolates were recovered from 88 unduplicated diseased duck samples (31.8%) from veterinary clinics in Guangzhou, Foshan, Qingyuan, and Huizhou. Two variants, blaNDM−1 and blaNDM−5, were detected and the latter was present in 89.6% of the isolates (25/28). Multilocus sequence typing (MLST) analysis indicated that these E. coli isolates possessed six distinct STs, and ST156 was the most prevalent followed by ST648, ST746, ST354, ST10, and ST162. In addition, phylogenomic analysis found that two of the isolates that were recovered from a single sample possessed different genomes, and the blaNDM-carrying IncX3 plasmids may be horizontal transfer between E. coli isolates in the intestinal tracts of ducks. Whole-genome sequencing (WGS) analysis further revealed that blaNDM co-existed with other 25 types of antimicrobial resistance genes (ARGs), of which 16 ARGs were highly prevalent with detection rates >50%, and a high incidence of coproducing blaNDM and mcr-1 E. coli isolates (22/88, 25.0%) was detected in ducks. This study underscores the importance of surveillance for blaNDM-harboring microbes in ducks.
Introduction
Carbapenems are critically important for the treatment of infections caused by multidrug-resistant Gram-negative bacteria. However, carbapenemase-producing Enterobacteriaceae have become a major global public health threat (Nordmann et al., 2011). In particular, the New Delhi metallo β-lactamase (NDM) was initially found in Escherichia coli and Klebsiella pneumoniae isolates in India in 2018 (Yong et al., 2009). Since then, blaNDM-positive isolates have been found globally (Wu et al., 2019). blaNDM genes have been found in species belonging to 11 bacterial families, and the Enterobacteriaceae are the major hosts of blaNDM (Wu et al., 2019; Zhai et al., 2020).
Carbapenem antibiotics have never been licensed for veterinary use in any country worldwide; however, there have been sporadic reports of blaNDM-positive isolates from a variety of animal hosts. blaNDM-positive E. coli isolates were frequently detected from swine in multiple geographic areas in China (Ho et al., 2019). Similar to the detected rates of blaNDM gene in swine, blaNDM-positive E. coli isolates were highly prevalent in commercial broiler farms (Wang et al., 2017). In addition, several blaNDM-positive E. coli isolates from backyard animals shared closely related core single nucleotide polymorphisms (SNP) with human isolates (Li et al., 2019). A recent study has reported that blaNDM-positive Enterobacteriaceae were detected from migratory birds in China (Liao et al., 2019). Therefore, continued monitoring for blaNDM-positive Enterobacteriaceae in food-producing animals is urgently required.
Duck production has the potential to play a major role in the agricultural economy, and Asian countries alone contribute 84.2% of total duck meat produced in the world (Biswas et al., 2019). According to Food and Agriculture Organization (FAO) data (2019), duck meat production rose from 2.64 to 3.02 million tons from 2015 to 2019 in Asia, while in China, duck meat production rose from 2.19 to 2.50 million tons from 2015 to 2019 in China (FAO, 2019). The data show that China is the largest producer and consumer of cultivated duck in the world (Chen et al., 2020). blaNDM-positive E. coli isolates were highly prevalent along the Chinese poultry production chain, including commercial broiler farms, slaughterhouses, and supermarkets (Wang et al., 2017). Therefore, the prevalence of blaNDM-positive E. coli isolates from duck should be addressed through continued monitoring.
Although we previously reported that the rate of blaNDM-positive E. coli isolates at three duck farms in Guangdong was significantly higher than four other provinces in China, the sample collected was limited in western Guangdong province (Wang et al., 2021). Thus, in this study, we furthermore examined the epidemiology and molecular characterization of blaNDM-positive E. coli isolates recovered from ducks in representative areas for breeding ducks of Guangdong, China.
Materials and Methods
Ethics Statement
The Institutional Review Board of South China Agricultural University (SCAU-IRB) approved the protocols. All animals were sampled under authorization from the Animal Research Committees of South China Agricultural University (SCAU-IACUC).
Sampling Information
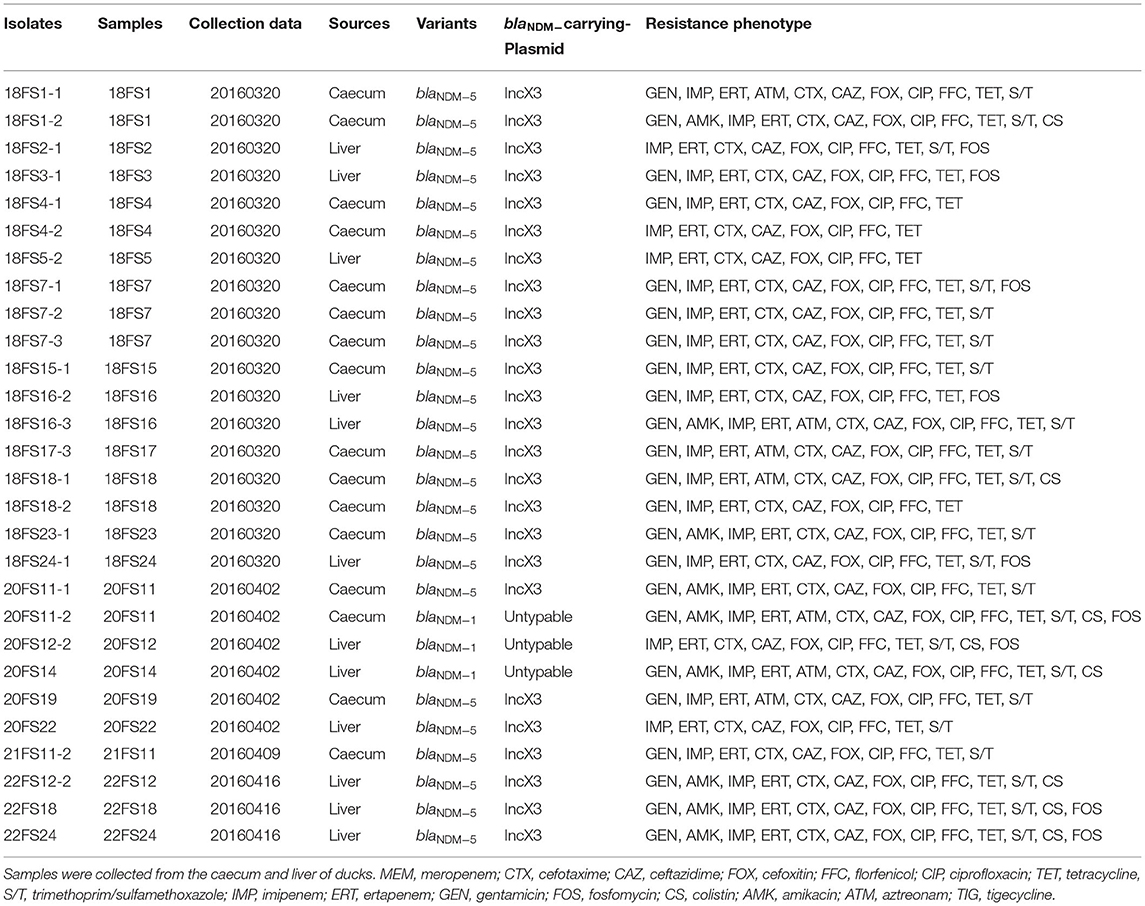
A total of 88 unduplicated samples, including 42 liver samples and 46 caecum samples, were collected from 88 diseased ducks, these diseased ducks were sent to the veterinary clinical diagnostics laboratory in Foshan University from duck farms in Guangzhou, Huizhou, Foshan, and Qingyuan of Guangdong province (Figure 1). Briefly, all sample was added to 1 ml of LB Broth and incubated for 16–18 h in 37°C, followed by inoculating on MacConkey plates containing 2 mg/L meropenem for 12 h. Multiple red clones were selected for identification using MALDI-TOF MS Axima™ (Shimadzu-Biotech Corp., Kyoto, Japan) and 16S rRNA sequencing. For carbapenem-resistant E. coli isolates, five major carbapenem resistance genes, namely, blaKPC, blaNDM, blaIMP, blaOXA−48, and blaVIM, were detected by PCR using previously described primers (Poirel et al., 2011). Samples were collected after obtaining consent from farms and veterinarians.
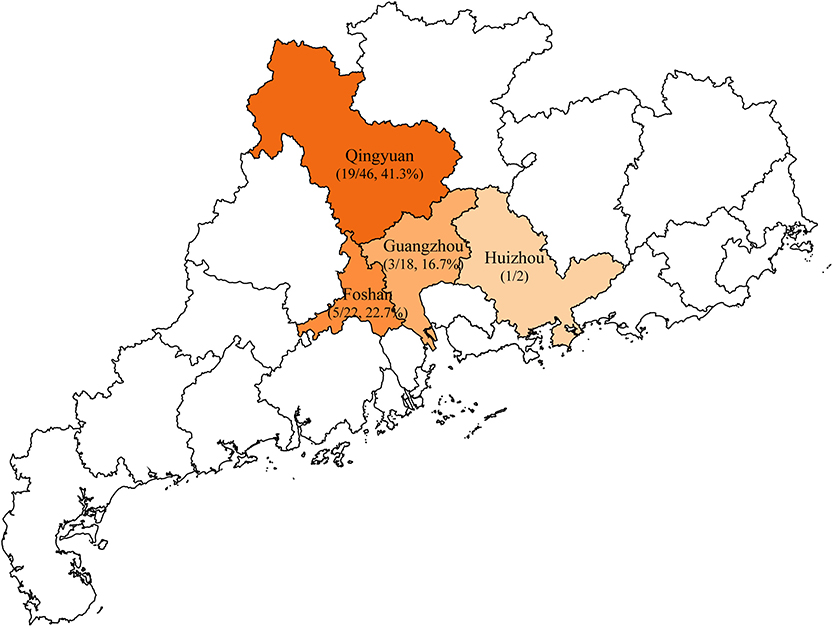
Figure 1. Sample collection areas in Guangdong, China. Municipalities included in the study are shaded in orange and the rate of blaNDM-positive Escherichia coli isolates are indicated by the depth of color.
Antimicrobial Susceptibility Testing
Antibiotic susceptibility testing was performed by the agar dilution method and interpreted according to the Clinical and Laboratory Standards Institute guidelines (CLSI M100-S28) for the following antimicrobials: gentamicin, amikacin, meropenem, imipenem, aztreonam, cefotaxime, ceftazidime, cefoxitin, florfenicol, ciprofloxacin, fosfomycin, trimethoprim-sulfamethoxazole, and tetracycline (CLSI, 2018). Susceptibility to colistin and tigecycline were assessed by broth microdilution as recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST Version 6.0) (EUCAST, 2016). E. coli ATCC 25922 was used as the quality control strain.
Conjugation Assay and Southern Blotting
To investigate the transferability of the resistance genes, a conjugation assay was performed for all blaNDM-positive E. coli isolates with the streptomycin-resistant E. coli C600 as the recipient strain. Donor strains and E. coli C600 were mixed and applied to a 0.22 μm filter in LB agar plates for 16–18 h. The mixture culture was then diluted and spread on selective MacConkey agar plates containing both 1 mg/L of meropenem and 2,000 mg/L of streptomycin to recover transconjugants. Transconjugants were confirmed by PCR and Pulsed Field Gel Electrophoresis (PFGE) patterns. S1-PFGE and Southern blotting were performed to obtain plasmid size, and the Salmonella enterica serotype, Braenderup H9812, was used as the standard size marker.
Whole-Genome Sequencing (WGS)
Total DNA was extracted from blaNDM-producing E. coli isolates using a Genomic DNA Purification Kit (TIANGEN, Beijing, China) as per the instructions of the manufacturer. WGS was performed with the Illumina Hiseq 2500 System (Novogene Guangzhou, China) using the paired-end 2 × 150-bp sequencing protocol. The draft genome was de novo assembled using the SPAdes version 3.9.0 (Bankevich et al., 2012). All genome assemblies of 28 E. coli isolates were deposited in GenBank and are registered with BioProject number PRJNA669620. Then, the sequence types, replicon types, and antibiotic resistance genes of all the isolates were identified by the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/). The sequence comparison of blaNDM-carrying plasmids was performed using Mauve and Brig (Darling et al., 2004; Alikhan et al., 2011). In silico phylotyping of E. coli was carried out using the ClermonTyping method (http://clermontyping.iame-research.center/). Phylogenetic trees for the blaNDM-producing E. coli isolates were structured using CSI Phylogeny (v1.4), and E. coli (18FS1-1) was used as the reference genome (Kaas et al., 2014). The corresponding characteristics of each isolate were visualized using the online tool iTOL (https://itol.embl.de/). The population structure of each phylogenetic tree was defined using rhierbaps (Tonkin-Hill et al., 2018). The genome assemblies were analyzed using a gene-by-gene approach and the allelic distance from the core genome multilocus sequence typing (MLST) (cgMLST) was visualized in a minimum-spanning tree using BacWGSTdb 2.0 (Feng et al., 2021).
Result
Bacterial Isolation and Detection of Carbapenemase Genes
In this study, a total of 28 (31.8%) blaNDM-producing E. coli isolates were recovered from 88 collected samples in the veterinary clinical diagnostic laboratory in Foshan University from duck farms in Guangdong province. The isolation rates of blaNDM-positive E. coli from different districts were 41.3% for Qingyuan (19/46), 22.7% for Foshan (5/22), and 16.7% for Guangzhou (3/18) (Figure 1). Within these groups, we identified only two blaNDM variants: blaNDM−5 (25/28, 89.3%) and blaNDM−1 (3/28, 10.7%) (Table 1).
Antibiotic Susceptibility Testing
All blaNDM-positive E. coli isolates showed reduced susceptibility to meropenem with MICs of 4– >64 mg/L (Supplementary Table 1) and were concurrently resistant to imipenem, ertapenem, cefotaxime, ceftazidime, cefoxitin, florfenicol, ciprofloxacin, and tetracycline (Table 1). Moreover, these isolates exhibited high rates of resistance to trimethoprim/sulfamethoxazole (22/28, 78.6%), gentamicin (23/28, 82.1%), and fosfomycin (12/28, 42.9%) but lower rates for amikacin (9/28, 32.1%), colistin (8/28, 28.6%), and aztreonam (7/28, 25%).
Phylogenetic Analysis of NDM-Positive E. coli Isolates
Whole-genome sequencing data were generated for the 28 blaNDM-positive E. coli isolates. The results of WGS demonstrated that these isolates were divided into six distinct STs: ST156 (8/28, 28.6%), ST648 (7/28, 25.0%), ST746 (5/28, 17.9%), ST354 (3/28, 10.7%), ST10 (3/28, 10.7%), and ST162 (2/28, 7.1%) (Figure 2). Clonotyping revealed seven fumC and fimH (CH) types and exhibited further divergence between clones. The most prevalent clonotypes were C29:H38 (n = 7), C4:H1084 (n = 7), and C7:H54 (n = 5), which belong to ST156, ST648, and ST746, respectively. The remaining clonotypes were C11:H0 (n = 3), C65:H32 (n = 2), C88:H58 (n = 2), C7:H1353 (n = 1), and C88:H27 (n = 1) (Figure 2). The E. coli isolates from the present study were classified using Clermont Typing and the majority belonged to groups B1 (10/28, 35.7%) and F (10/28, 35.7%) from Foshan and Qingyuan, respectively. A phylogenetic tree was established using the 28 blaNDM-positive E. coli isolates. All isolates were classified into four clades lineage, and the major Lineage IV included seven (25%) isolates belonging to ST648 and exhibited high levels of the identity of pairwise single nucleotide polymorphisms (SNP) ≤ 21 (Figure 2). Notably, in five cases, two isolates were possessing a collection of different genomic characteristics that were recovered from the same samples (18FS1, 18FS4, 18FS16, 18FS18, and 20FS11) (Table 1). For instance, both ST354 (18FS1-1) and ST648 (18FS1-2) were recovered from sample 18FS1 and shared 22411 SNPs. This scenario is worrying and indicates the development of diversity in the population of blaNDM-positive E. coli isolates from ducks. To further assess the relationship between the 28 isolates, the genome assemblies were analyzed using a gene-by-gene approach, and the allelic distance from the core genome MLST (cgMLST) was visualized in a minimum-spanning tree using BacWGSTdb 2.0. Based on ST, geographic location, and position in the network, the resulting network shows that the isolates were grouped in accordance with their ST and place of isolation. The MLST type was clustered together, except ST746; 18FS4-1 were more closed to the ST156 cluster (Supplementary Figure 1).

Figure 2. Analysis of blaNDM-positive E. coli isolates from ducks in Guangdong, China. Relationships among 28 blaNDM-positive E. coli isolates are indicated using a maximum likelihood tree. Red-filled squares indicate possession of the indicated antimicrobial resistance genes (ARGs) and brown-filled squares indicated plasmid Inc type.
Antibiotic Resistance Genes
We additionally identified the presence of 25 types of antimicrobial resistance genes (ARGs) that conferred resistance to nine classes of antibiotics including aminoglycosides, β-lactams, MLS (macrolides, lincosamides, and type B streptogramin), fluoroquinolones, sulfonamide, fosfomycin, rifampicin, colistin, and tetracyclines. Among these, 16 ARGs were highly prevalent with detection rates >50%, including aac, aph, blaCTX, blaOXA, blaTEM, cmlA, floR, mdf, mph, oqxA/B, qnrS, dfrA, sul, ARR, mcr-1, and tet (Figure 2).
Plasmid Analysis
Conjugation experiments were performed using the blaNDM-positive E. coli isolates collected from ducks, and all the blaNDM-carrying plasmids were successfully transferred to the recipient strain E. coli C600str. S1-PFGE and hybridization analyses confirmed that blaNDM genes from 25 isolates were located on ~ 50 kb plasmids, and the others genes were located on ~ 140 kb (n = 2) and ~ 200 kb (n = 1) plasmids (Supplementary Figure 2). As shown in Supplementary Figure 3, the ~ 50 kb plasmids carrying blaNDM were the IncX3 incompatibility group and were similar to blaNDM−5-carrying IncX3 plasmid from an ST25 K. pneumoniae isolated from human peritoneal fluid in China (Acc. No. KU761328). In addition, we found that IncX3 plasmids were carried by two different E. coli isolates that were recovered from the same sample in four out of five cases (Table 1). This provides evidence for the horizontal transfer of blaNDM-carrying IncX3 plasmids in the intestines of ducks. WGS analysis demonstrated that 14 different Inc types were present on plasmids in the 28 blaNDM-positive E. coli isolates (Figure 2). Except for the IncX3 plasmid, IncFIB (17/28, 60.7%), IncHI2 (15/28, 53.6%), and IncI1 (10/28, 35.7%) plasmids were highly prevalent in these isolates (Figure 2).
Discussion
In this study, a total of 28 (28/88, 31.8%) blaNDM-producing E. coli isolates were recovered from ducks; this result is similar to our previous report that indicated ahigh prevalence of blaNDM -positive E. coli isolates at duck farms in western Guangdong province. Although there was an overwhelming dominance of blaNDM−1 and blaNDM−5 in the clinical and livestock isolates (Shen et al., 2018; Zhai et al., 2020), blaNDM−5 was more prevalent than blaNDM−1 among the tested duck farms. This finding is similar to the previous report of the high prevalence of blaNDM−5 in the chicken production chains (Wang et al., 2017).
The 28 blaNDM-positive E. coli isolates belonged to six distinct STs (ST156, ST648, ST746, ST354, ST10, and ST162) discussed in the current study, of which, ST156, ST648, and ST746 were the most prevalent. ST156 and ST648 have been associated with the dissemination of blaNDM−5 and mcr-1-producing E. coli isolates (Yang et al., 2016). The blaNDM-positive ST746 and ST354 prevalent in ducks and poultry have become a primary reservoir for blaNDM-positive ST746 E. coli isolates in China (Wang et al., 2021). Phylogenomic analysis found that two of the isolates that were recovered from a single sample possessed different genomes, which indicates the development of diversity in the population of blaNDM-positive E. coli isolates from ducks.
Whole-genome sequencing analysis further revealed that blaNDM coexisted with other 25 types of ARGs, of which 16 ARGs were highly prevalent with detection rates >50%. Of note, mcr-1, conferring resistance to the last-resort antibiotic colistin, was detected in 22 blaNDM-positive E. coli isolates. Some recent studies have reported that blaNDM and mcr-1 coproducing E. coli isolates were recovered from chicken (37/739, 5.0%), swine (16/105, 15.2%), and duck (11/92, 12.0%) farms (Kong et al., 2017; Wang et al., 2017, 2021). There was also a high incidence of the co-harboring of mcr-1 and blaNDM in chickens (21/78, 26.9%) (Liu et al., 2017). In this study, we also discovered a high prevalence of blaNDM and mcr-1 co-carrying E. coli isolates from diseased ducks (22/88, 25.0%) in Guangdong. In addition, the existence of blaNDM−5 is associated with multiple resistances, including aminoglycosides, sulfonamide, and fluoroquinolones. These can further promote the spread and persistence of carbapenem-resistant microbes in the poultry industry (Grönthal et al., 2018; Zhai et al., 2020).
In the current study, all blaNDM−5 genes identified were carried by IncX3 plasmids. IncX3 plasmids may serve as one of the major platforms on which blaNDM genes are evolving with the generation of new NDM variants, such as blaNDM−1/4/5/6/7/13/17/19/20/21 (Wu et al., 2019; Zhai et al., 2020). However, a high prevalence of blaNDM−5-carrying IncX3 plasmid from bacteria of animal farms in China (Zhai et al., 2020). The blaNDM-carrying IncX3 plasmids have a narrow host range and have been mainly found in Enterobacteriaceae worldwide, which may be an association to highly conjugatable and stable and exert no fitness costs on their bacterial hosts (Johnson et al., 2012). In addition, this study found that IncX3 plasmids were carried with two different E. coli isolates recovered from a single sample in four out of five samples. This provides evidence for the horizontal transfer of blaNDM-carrying IncX3 plasmids between E. coli isolates in the intestine of ducks.
Conclusions
In conclusion, we identified 28 blaNDM-positive E. coli isolates from diseased ducks in Guangdong, China. Notably, this is the first study to report the development of diversity in the population of blaNDM-positive E. coli isolates from ducks. WGS analysis further determined that blaNDM coexisted with other ARGs, including mcr-1, and blaNDM-carrying IncX3 plasmids were most likely horizontally transferred between E. coli isolates in the duck intestinal tract. This study underscores the importance of surveillance for blaNDM-harboring microbes in ducks and indicates a high likelihood for the spread of carbapenem resistance from the poultry production chain to humans.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA669620.
Author Contributions
M-GW wrote the first draft of the manuscript. X-PL, YY, and L-XF contributed to conception and design of the study. DW, R-SY, LJ, D-TC, and S-LZ performed the statistical analysis. JS and Y-HL wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was jointly supported by the National Natural Science Foundation of China (31730097 and 31772793), the National Science and Technology Major Project (2018ZX10714002), and the Innovation Team Project of Guangdong University (2019KCXTD001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.677633/full#supplementary-material
Supplementary Figure 1. Minimum spanning tree analysis of the cgMLST profiles of 28 isolates. Numbers on lines correspond to the number of target genes for which allelic differences were detected. Circle colors are used to differentiate the city of origin of the isolates.
Supplementary Figure 2. S1-PFGE and hybridization of the plasmids of blaNDM-positive Escherichia coli isolates. Lane M: XbaI-digested genomic DNA of reference Salmonella enterica serotype Braenderup strain H9812, and the bands on membrane represent the plasmids where blaNDM genes are located on.
Supplementary Figure 3. The sequence comparison and map generation of blaNDM-carrying plasmids with reference IncX3 plasmid (Accession Number: KU761328) from an ST25 Klebsiella pneumoniae isolated from human peritoneal fluid in China.
Supplementary Table 1. Minimum inhibitory concentration of 28 blaNDM-positive E. coli isolates from duck in Guangdong China.
References
Alikhan, N. F., Petty, N. K., Zakour, N. L. B., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genom. 12:402. doi: 10.1186/1471-2164-12-402
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Biswas, S., Banerjee, R., Bhattacharyya, D., Patra, G., Das, A. K., and Das, S. K. (2019). Technological investigation into duck meat and its products - a potential alternative to chicken. Worlds Poultry Sci. J. 75, 609–620. doi: 10.1017/s004393391900062x
Chen, Z., Bai, J., Wang, S., Zhang, X., Zhan, Z., Shen, H., et al. (2020). Prevalence, antimicrobial resistance, virulence genes and genetic diversity of salmonella isolated from retail duck meat in Southern China. Microorganisms 8:444. doi: 10.3390/microorganisms8030444
CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing—Twenty-Eighth Edition: M100.
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
FAO (2019). Food and Agriculture Organization. Available online at: http://www.fao.org/home/en
Feng, Y., Zou, S. M., Chen, H. F., Yu, Y. S., and Ruan, Z. (2021). BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 49, D644–D650. doi: 10.1093/nar/gkaa821
Grönthal, T., Österblad, M., Eklund M, Jalava, J., Nykäsenoja, S., Pekkanen, K., and Rantala, M. (2018). Sharing more than friendship – transmission of NDM-5 ST167 and CTX-M-9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Eurosurveillance 23:1700497. doi: 10.2807/1560-7917.ES.2018.23.27.1700497
Ho, P. L., Wang, Y., Liu, M. C. J., Lai, E. L. Y., Law, P. Y. T., Cao, H. L., et al. (2019). IncX3 epidemic plasmid carrying blaNDM-5 in Escherichia coli from swine in multiple geographic areas in China. Antimicrob. Agents Chemother. 62, e02295–e02217. doi: 10.1128/AAC.02295-17
Johnson, T. J., Bielak, E. M., Fortini, D., Hansen, L. H., Hasman, H., Debroy, C., et al. (2012). Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68, 43–50. doi: 10.1016/j.plasmid.2012.03.001
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., and Lund, O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 9:e104984. doi: 10.1371/journal.pone.0104984
Kong, L. H., Lei, C. W., Ma, S. Z., Jiang, W., Liu, B. H., Wang, Y. X., et al. (2017). Various Sequence Types of Escherichia coli Isolates Coharboring blaNDM-5 and mcr-1 Genes from a Commercial Swine Farm in China. Antimicrob. Agents Chemother. 61, e02167–e02116. doi: 10.1128/AAC.02167-16
Li, J., Bi, Z., Ma, S., Chen, B., Cai, C., He, J., et al. (2019). Inter-host transmission of carbapenemase-producing Escherichia coli among humans and backyard animals. Environ. Health Perspect. 127:107009. doi: 10.1289/EHP5251
Liao, X. P., Yang, R. S., Xia, J., Chen, L., Zhang, R. M., Fang, L. X., et al. (2019). High colonization rate of a novel carbapenem-resistant Klebsiella lineage among migratory birds at Qinghai Lake, China. J. Antimicrob. Chemother. 74, 2895–2903. doi: 10.1093/jac/dkz268
Liu, B. T., Song, F. J., Zou, M., Zhang, Q. D., and Shan, H. (2017). High Incidence of Escherichia coli Strains Coharboring mcr-1 and blaNDM from Chickens. Antimicrob. Agents Chemother. 61, e02347–e02316. doi: 10.1128/AAC.02347-16
Nordmann, P., Naas, T., and Poirel, L. (2011). Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17, 1791–1798. doi: 10.3201/eid1710.110655
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Shen, Z., Hu, Y., Sun, Q., Hu, F., Zhou, H., Shu, L., et al. (2018). Emerging Carriage of NDM-5 and MCR-1 in Escherichia coli from healthy people in multiple regions in China: a cross sectional observational study. EClinicalMedicine 6, 11–20. doi: 10.1016/j.eclinm.2018.11.003
Tonkin-Hill, G., Lees, J. A., Bentley, S. D., Frost, S. D. W., and Corander, J. (2018). RhierBAPS: an R implementation of the population clustering algorithm hierBAPS. Wellcome Open Res. 3:93. doi: 10.12688/wellcomeopenres.14694.1
Wang, M. G., Zhang, R. M., Wang, L. L., Sun, R. Y., Bai, S. C., Han, L., et al. (2021). Molecular epidemiology of carbapenemase-producing Escherichia coli from duck farms in south-east coastal China. J. Antimicrob. Chemother. 76, 322–329. doi: 10.1093/jac/dkaa433
Wang, Y., Zhang, R. M., Li, J. Y., Wu, Z. W., Yin, W. J., Schwarz, S., et al. (2017). Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2:16260. doi: 10.1038/nmicrobiol.2016.260
Wu, W. J., Feng, Y., Tang, G. M., Qiao, F., McNally, A., and Zong, Z. Y. (2019). NDM Metallo-β-Lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32, e00115–00118. doi: 10.1128/CMR.00115-18
Yang, R. S., Feng, Y., Lv, X. Y., Duan, J. H., Chen, J., Fang, L. X., et al. (2016). Emergence of NDM-5- and MCR-1-Producing Escherichia coli Clones ST648 and ST156 from a Single Muscovy Duck (Cairina moschata). Antimicrob. Agents Chemother. 60, 6899–6902. doi: 10.1128/AAC.01365-16
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., and Walsh, T. R. (2009). Characterization of a new metallo-betalactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Keywords: transmission, MCR-1, escherichia coli, duck, blaNDM
Citation: Wang M-G, Yu Y, Wang D, Yang R-S, Jia L, Cai D-T, Zheng S-L, Fang L-X, Sun J, Liu Y-H and Liao X-P (2021) The Emergence and Molecular Characteristics of New Delhi Metallo β-Lactamase-Producing Escherichia coli From Ducks in Guangdong, China. Front. Microbiol. 12:677633. doi: 10.3389/fmicb.2021.677633
Received: 08 March 2021; Accepted: 31 May 2021;
Published: 05 July 2021.
Edited by:
Aloysius Wong, Kean University-Wenzhou, ChinaCopyright © 2021 Wang, Yu, Wang, Yang, Jia, Cai, Zheng, Fang, Sun, Liu and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Ping Liao, eHBsaWFvQHNjYXUuZWR1LmNu
†These authors have contributed equally to this work
 Min-Ge Wang
Min-Ge Wang Yang Yu
Yang Yu Dong Wang1,2
Dong Wang1,2 Liang-Xing Fang
Liang-Xing Fang Jian Sun
Jian Sun Ya-Hong Liu
Ya-Hong Liu Xiao-Ping Liao
Xiao-Ping Liao