- 1Biomolecular Engineering Laboratory, School of Food and Nutritional Science, University of Shizuoka, Shizuoka, Japan
- 2Laboratory of Fermentation Microbiology, Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
The filamentous fungus Aspergillus oryzae, also known as yellow koji mold, produces high levels of hydrolases such as amylolytic and proteolytic enzymes. This property of producing large amounts of hydrolases is one of the reasons why A. oryzae has been used in the production of traditional Japanese fermented foods and beverages. A wide variety of hydrolases produced by A. oryzae have been used in the food industry. The expression of hydrolase genes is induced by the presence of certain substrates, and various transcription factors that regulate such expression have been identified. In contrast, in the presence of glucose, the expression of the glycosyl hydrolase gene is generally repressed by carbon catabolite repression (CCR), which is mediated by the transcription factor CreA and ubiquitination/deubiquitination factors. In this review, we present the current knowledge on the regulation of hydrolase gene expression, including CCR, in A. oryzae.
Introduction
The koji mold Aspergillus oryzae is a filamentous fungus that has been used for over a thousand years to manufacture Japanese fermented foods and beverages, such as shoyu (soy sauce), miso (soybean paste), and sake (rice wine) (Machida et al., 2008). The most industrially important characteristic of A. oryzae is the ability to produce large amounts of hydrolytic enzymes such as amylolytic and proteolytic enzymes. This is one of the reasons why A. oryzae has been used in the production of traditional Japanese fermented foods and beverages. Genome sequencing of A. oryzae has revealed that this fungus has more hydrolytic enzyme genes than other related filamentous fungi such as Aspergillus nidulans and Aspergillus fumigatus (Machida et al., 2005; Kobayashi et al., 2007). A wide variety of hydrolases produced by A. oryzae have been used in the food processing and pharmaceutical industries, in addition to the fermentation industry (Gomi, 2014). Moreover, promoters of hydrolytic enzyme genes, especially those of amylolytic genes, are widely used for high expression of homologous and heterologous genes in A. oryzae to produce useful proteins and secondary metabolites (Tanaka and Gomi, 2014; Oikawa, 2020; Jin et al., 2021). Therefore, understanding the regulation of hydrolytic enzyme gene expression in A. oryzae is both scientifically and industrially important. The expression of most hydrolase genes is induced by the presence of certain substrates. For instance, it has long been known that amylolytic gene expression is induced by starch and malto-oligosaccharides (Tonomura et al., 1961; Yabuki et al., 1977). Such substrate-specific gene expression is often regulated by fungal-specific Zn(II)2Cys6-type transcription factors. Several transcription factors that control the expression of hydrolytic enzyme genes in A. oryzae have been identified (Table 1), and the regulatory mechanisms of their activation have recently been elucidated. On the other hand, in the presence of glucose, the expression of hydrolytic enzyme genes is strongly repressed even when inducing substrates are present. This phenomenon is known as carbon catabolite repression (CCR). CCR-regulating factors have been identified, and their function has been analyzed in the model filamentous fungus A. nidulans; however, details on the control mechanism of CCR remain unclear. Recent studies have revealed a part of the molecular mechanism of CCR regulation in A. oryzae.
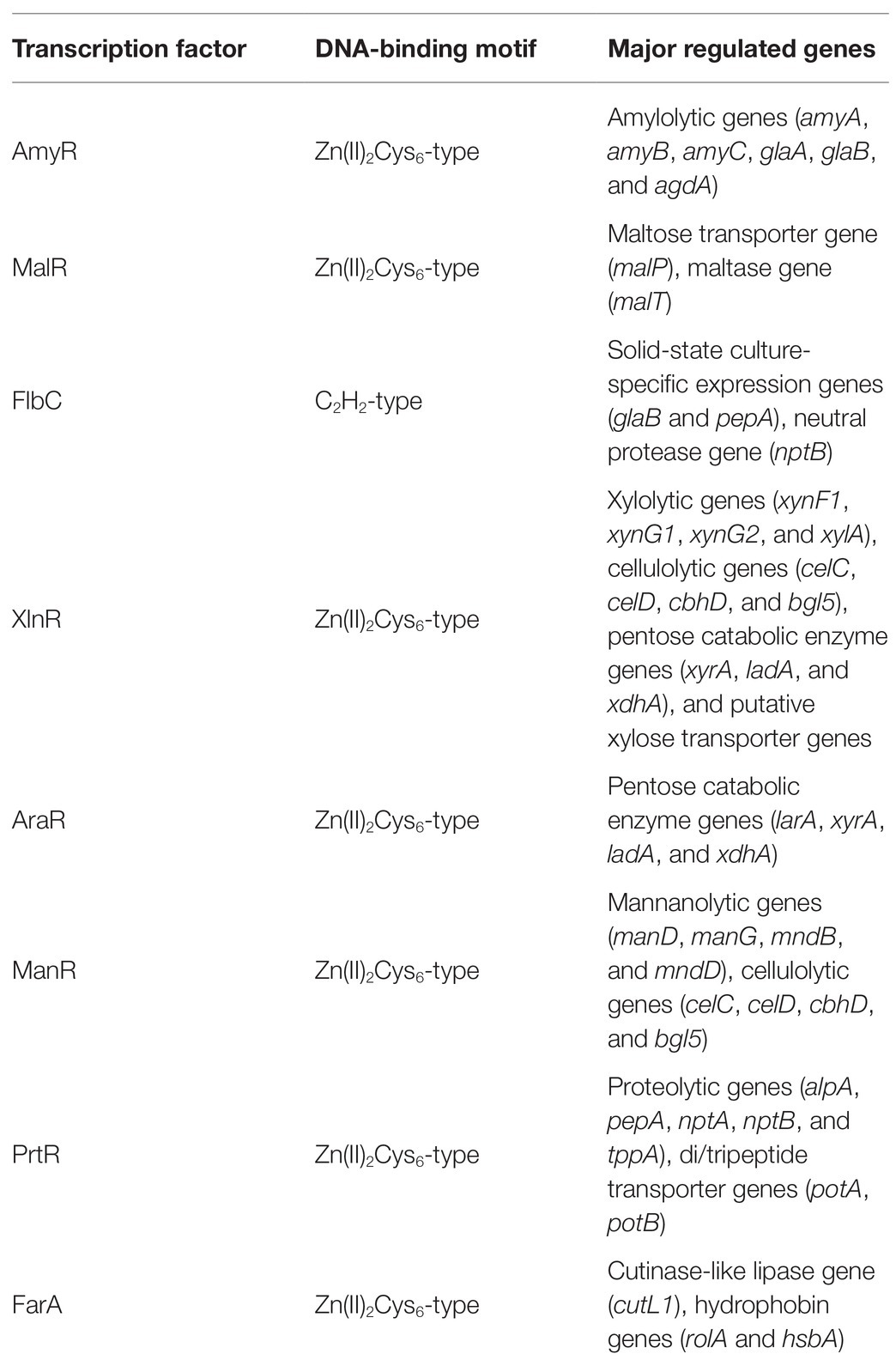
Table 1. Transcription factors involved in induction of hydrolytic gene expression in Aspergillus oryzae.
In this review, we introduce transcription factors that induce the expression of hydrolytic enzyme genes in A. oryzae. In addition, we describe the molecular mechanism of CCR, which was revealed by recent studies on A. oryzae and other filamentous fungi.
Transcription Factors That Induce Gene Expression of Hydrolytic Enzymes in A. Oryzae
AmyR and MalR
The regulation of amylolytic gene expression in A. oryzae has been studied for many years, because amylolytic enzymes are the major hydrolytic enzymes produced by A. oryzae, and are essential for sake production (Gomi, 2019). The expression of amylolytic genes is directly regulated by the transcription factor AmyR (Petersen et al., 1999; Gomi et al., 2000). This Zn(II)2Cys6-type transcription factor binds to the CGGN8(C/A)GG sequence in the promoter of amylolytic genes, such as the α-amylase (amyA/B/C), glucoamylase (glaA and glaB), and α-glucosidase (agdA) genes (Petersen et al., 1999; Ito et al., 2004). Expression of these genes is induced by maltose and isomaltose, and also by glucose when CCR is released (Suzuki et al., 2015). The amyR gene is constitutively expressed regardless of the presence of inducing or non-inducing sugars, and under conditions where amylolytic genes are not expressed; AmyR is localized in the cytoplasm (Suzuki et al., 2015). AmyR is rapidly transferred into the nucleus when isomaltose is added to the medium (Suzuki et al., 2015). Glucose and maltose also induce nuclear transfer of AmyR, but these sugars require higher concentrations and longer time periods than isomaltose to induce AmyR nuclear transfer and amylolytic gene expression (Suzuki et al., 2015). This dynamic of AmyR nuclear transfer is similar to that in the model filamentous fungus A. nidulans (Murakoshi et al., 2012). The nuclear transfer of AmyR depends on nuclear localization signals located within its DNA-binding domain (Suzuki et al., 2015). In contrast, C-terminal truncated AmyR is constitutively localized in the nucleus in both A. nidulans and A. oryzae (Makita et al., 2009; Suzuki et al., 2015). This suggests that the C-terminal region of AmyR is required to keep AmyR in the cytoplasm. C-terminal truncated A. nidulans AmyR retains its transcriptional activation function (Makita et al., 2009), but C-terminal truncation of A. oryzae AmyR leads to the loss of such function (Suzuki et al., 2015). This suggests that the function of the C-terminal region of AmyR differs among Aspergillus species. Therefore, the function of the C-terminal region of AmyR in other Aspergillus species that produce large amounts of amylolytic enzymes, such as Aspergillus niger and Aspergillus luchuensis, should be investigated.
In A. nidulans, maltose utilization is dependent on AmyR (Tani et al., 2001). However, a disruption mutant of A. oryzae amyR was able to assimilate maltose because maltose utilization in A. oryzae is regulated by another Zn(II)2Cys6-type transcription factor, MalR (Hasegawa et al., 2010). MalR is a homolog of the yeast MAL activator, a transcriptional activator of genes encoding maltose transporter and maltase (Needleman, 1991). In contrast to AmyR, MalR is constitutively localized in the nucleus (Suzuki et al., 2015). Similar to the yeast MAL activator gene, malR comprises a gene cluster with the maltase gene (malT) and maltose transporter gene (malP), and the expression of both these genes is regulated by MalR (Hasegawa et al., 2010). The expression of malP and malT is not induced by the addition of isomaltose, but is preceded by amylolytic gene expression upon the addition of maltose (Suzuki et al., 2015). Therefore, MalR is activated prior to AmyR to convert maltose into a physiologically active inducing substrate for AmyR, i.e., isomaltose. This conversion of maltose to isomaltose is probably caused by the glycosyltransferase activity of MalT. In budding yeast, activation of the MAL activator is regulated by the dissociation of its chaperone proteins Hsp70 and Hsp90 (Ran et al., 2008). Although A. oryzae MalR interacts with orthologs of Hsp70 and Hsp90 (Konno et al., 2018), details on the activation mechanism and binding sequence are not known.
Importantly, the α-amylase (amyB) gene promoter is commonly used for high-level expression of heterologous genes in A. oryzae (Jin et al., 2021). Improved promoters PglaA142 (Minetoki et al., 2003) and PenoA142 (Tsuboi et al., 2005), have been constructed by tandem insertion of AmyR-binding sequences (Region III) within the glucoamylase and enolase gene promoters. These improved promoters are now utilized in homologous and heterologous protein production.
FlbC
Aspergillus oryzae has two glucoamylase genes, glaA and glaB, the expression patterns of which are quite different (Hata et al., 1991, 1998). Similar to other amylolytic enzymes, GlaA is produced in both submerged and solid-state cultures (Oda et al., 2006). On the other hand, GlaB is secreted exclusively in solid-state culture and is not produced in submerged culture. Although the expression of both glaA and glaB is regulated by AmyR (Watanabe et al., 2011), a transcription factor that binds to the promoter region of glaB and regulates the expression of specific genes in solid-state culture was expected to be present (Ishida et al., 2000; Hisada et al., 2013). A screening of the A. oryzae disruption mutant library for transcriptional regulators indicated that FlbC is a transcription factor that regulates the expression of glaB (Tanaka et al., 2016). In addition to that of glaB, the expression level of pepA, an aspartic protease gene that is predominantly expressed in solid-state culture, was also significantly reduced by flbC disruption (Tanaka et al., 2016). Therefore, FlbC is presumed to be a transcription factor that regulates the specific expression of hydrolytic enzyme genes in solid-state culture. The expression of the neutral protease gene (nptB) is also regulated by FlbC (Tanaka et al., 2016). FlbC is a C2H2-type transcription factor that was originally identified as one of the regulators of conidiospore development (Wieser et al., 1994; Kwon et al., 2010; Ogawa et al., 2010). Disruption of other transcription factors that control conidiospore development has no effect on the production of GlaB, suggesting that FlbC regulates glaB expression independently of the regulatory mechanism of conidiospore development (Tanaka et al., 2016). Moreover, FlbC probably binds directly to the promoter region of glaB and regulates its expression (Gomi, 2019). Although the FlbC-binding sequence has not been empirically identified, the sequence containing GATC would be a candidate based on recent studies of the FlbC orthologs of Neurospora crassa (Boni et al., 2018) and Magnaporthe oryzae (Matheis et al., 2017). Furthermore, the regulatory mechanism of specific gene expression in solid-state culture should be elucidated. Expression from the glaB promoter is induced by low water activity, high temperature, and physical barriers to hyphal extension (Ishida et al., 1998). Since these inducing factors cause environmental stress in A. oryzae cells, the involvement of stress response pathways, including that of mitogen-activated protein kinase (MAPK) signaling pathways, in FlbC activation should be considered.
XlnR and AraR
XlnR is a Zn(II)2Cys6-type transcription factor that regulates the expression of xylanolytic and cellulolytic genes (Marui et al., 2002). This transcription factor is also involved in the regulation of putative xylose transporter genes and pentose metabolic enzyme genes (Noguchi et al., 2009). In Aspergillus species, the expression of pentose catabolic pathway genes is also regulated by AraR, a paralog of XlnR (Battaglia et al., 2011). Electrophoretic mobility shift assays revealed that XlnR and AraR bind competitively to the CGGNTAAW sequence in the promoter region of pentose catabolic genes such as the xylose dehydrogenase-encoding gene (xdhA) (Ishikawa et al., 2018). Notably, XlnR binds to the CGGNTAAW sequence solely found in the promoter region of pentose catabolic genes as a monomer, whereas it binds to the TTAGSCTAA and TAGSCTA sequences in the promoter region of the xylanase genes (xynF1 and xynG2) as a dimer (Ishikawa et al., 2018).
XlnR is constitutively located in the nucleus, similar to MalR. When xylose is added, XlnR is rapidly phosphorylated. In contrast, XlnR is rapidly dephosphorylated when xylose is removed from the culture medium (Noguchi et al., 2011). This reversible phosphorylation probably regulates the activation of XlnR. Identification of the phosphorylation site of XlnR is important for elucidating the activation mechanism of this transcription factor.
ManR
ManR is a Zn(II)2Cys6-type transcription factor that regulates the expression of mannanolytic enzyme genes. This transcription factor was identified by screening for mutants exhibiting reduced β-mannanase activity from a gene disruptant library of transcriptional regulators (Ogawa et al., 2012). ManR also regulates cellulolytic enzyme genes, such as the cellobiohydrolase, endoglucanase, and β-glucosidase genes (Ogawa et al., 2013). Most of these genes are also regulated by XlnR. Therefore, ManR and XlnR probably regulate the expression of these genes in a coordinated manner (Tani et al., 2014). ManR is an ortholog of N. crassa CLR-2 and A. nidulans ClrB, both of which regulate the expression of cellulase genes. However, the physiological roles of these transcription factors are slightly different (Kunitake and Kobayashi, 2017). ManR binds to the promoter regions of the β-mannanase gene (manG), which contains the CAGAAT sequence that is conserved in the promoter regions of mannanolytic enzyme genes (Ogawa et al., 2012). However, this conserved sequence is quite different from consensus sequences for the binding of CLR-2 and ClrB (Kunitake and Kobayashi, 2017).
PrtR
PrtR is a Zn(II)2Cys6-type transcription factor; the deletion of this transcription factor results in a significant decrease in extracellular protease activity (Mizutani et al., 2008), indicating that it is essential for extracellular proteolytic gene expression. Interestingly, the prtR gene is located adjacent to the amylolytic gene cluster consisting of amyR, agdA, and amyA (Gomi, 2019). Orthologs of PrtR in other Aspergillus species are named PrtT (Punt et al., 2008). Duplication or triplication of the chromosomal region containing the prtR gene by forced translocation of the 1.4 Mb chromosome 2 resulted in a significant increase in alkaline protease and acid carboxypeptidase activity in a solid-state culture of A. oryzae (Takahashi et al., 2018). Over 20 proteolytic genes were upregulated in the duplicated strain. Furthermore, the expression level of an alkaline protease gene (alpA) increased more than 5-fold when prtR was highly expressed from the promoter of the α-amylase gene (Takahashi et al., 2018). In addition, the expression levels of two of the three di/tripeptide transporter genes (potA and potB) also increased upon prtR overexpression and decreased upon prtR disruption (Tanaka et al., 2021). These results suggest that PrtR plays a central role in the regulation of gene expression for the acquisition of nutrients in the presence of protein. In fact, a disruption mutant of prtR showed poor growth in solid-state culture using wheat bran as a substrate. In A. niger and A. fumigatus, PrtT regulates the expression of multiple protease genes and tri/tetrapeptide transporter genes (Sharon et al., 2009; Hartmann et al., 2011; Huang et al., 2020); PrtT orthologs are absent in A. nidulans (Punt et al., 2008). Although the expression of proteolytic genes is induced by proteins or peptides, the direct substrate that induces the activation of PrtR/PrtT is not clear for any Aspergillus species; hence, further studies are required to ascertain this. Disruption of multiple protease genes has resulted in highly effective heterologous protein production in A. oryzae (Jin et al., 2021). Considering that many extracellular protease genes are thought to be regulated by PrtR, the effect of prtR disruption on heterologous protein production is strongly expected.
FarA
Aspergillus oryzae can degrade polyester poly(butylene succinate-co-adipate) (PBSA), a biodegradable plastic. The cutinase-like lipase CutL1 is the major enzyme that degrades PBSA in A. oryzae, and two hydrophobic surface binding proteins (RolA and HsbA) assist in the binding of CutL1 to PBSA (Maeda et al., 2005; Takahashi et al., 2005; Ohtaki et al., 2006). FarA is a Zn(II)2Cys6-type transcription factor that regulates the expression of fatty acid metabolism genes in A. nidulans; its ortholog, CTF1α of Fusarium solani, regulates cutinase gene expression (Li and Kolattukudy, 1997; Hynes et al., 2006). The expression of the cutL1 gene is regulated by FarA, and a disruption mutant of farA abolished PBSA degradation activity in A. oryzae (Garrido et al., 2012). In addition to that of the cutL1 gene, the expression of rolA and hsbA is also repressed by the disruption of farA (Garrido et al., 2012). Although details on the mechanism of FarA regulation of the cutinase gene are unknown for all Aspergillus species, a recent study on A. nidulans showed that FarA-dependent expression of the cutinase genes is affected by CCR (Bermúdez-García et al., 2019).
Carbon Catabolite Repression of Hydrolytic Enzyme Genes
Regulating Factors of CCR
The study of CCR in filamentous fungi began with the identification of regulatory factors in A. nidulans. Four factors involved in the regulation of CCR in filamentous fungi were identified in the 1970s by genetic analysis of A. nidulans (Arst and Cove, 1973; Hynes and Kelly, 1977; Kelly and Hynes, 1977). These four factors were denoted as CreA, CreB, CreC, and CreD. Firstly, the creA gene was identified in an A. nidulans CCR-deficient mutant. This gene encodes a C2H2-type transcription factor that directly regulates CCR (Dowzer and Kelly, 1991). Similar to A. nidulans CreA, A. oryzae CreA binds to the SYGGRG sequence in the promoter region of the α-amylase gene (Kato et al., 1996). The amino acid sequence of the CreA DNA-binding domain is highly homologous to that of Mig1, a CCR-regulating transcription factor in the budding yeast Saccharomyces cerevisiae. However, the homology between CreA and Mig1 in the regions other than the DNA-binding domain is not high. In addition, recent studies have revealed that the functional control mechanisms of Mig1 and CreA are quite different (see Nuclear export-dependent degradation of CreA).
Similar to creA (described above), creB and creC were identified in CCR-deficient mutants as well. CreB is a deubiquitinating enzyme homolog with a particularly high homology to Ubp9 of the fission yeast Schizosaccharomyces pombe (Lockington and Kelly, 2001). Many deubiquitinating enzymes interact with WD40-repeat proteins that regulate the activity and function of such enzymes (Villamil et al., 2013). The creC gene encodes a WD40-repeat protein that shows high homology to fission yeast Bun62 (Lockington and Kelly, 2002) but has no counterpart in budding yeast. In A. nidulans, CreB and CreC form complexes in vivo (Lockington and Kelly, 2002). In fission yeast, Ubp9 interacts with Bun62 and another WD40-repeat protein, Bun107, and is involved in endocytosis, actin dynamics, and cell polarity (Kouranti et al., 2010). However, the function of the CreB and CreC complex in filamentous fungi has not been clarified.
The gene responsible for suppressing the phenotype of creB and creC mutants was identified and named creD (Kelly and Hynes, 1977). CreD contains two arrestin domains and three or four PxY motifs that are involved in protein-protein interactions (Boase and Kelly, 2004). There is a high homology between CreD and yeast Rod1/Art4, an arrestin-related trafficking adaptor (ART) protein that acts as an adaptor for ubiquitin ligase and its target protein. In budding yeast, ART proteins recruit the HECT E3 ubiquitin ligase Rsp5 to cell membrane proteins (Lin et al., 2008; Nikko and Pelham, 2009). Aspergillus oryzae CreD also physically interacts with HulA, an ortholog of yeast Rsp5 (Tanaka et al., 2017). CreD and HulA are involved in the degradation of the maltose transporter MalP in A. oryzae (see Glucose-induced endocytosis of maltose transporter).
Based on the putative function of CreB and CreD, a model was proposed wherein CCR is regulated by the stabilization and degradation of CreA protein mediated by these factors (Lockington and Kelly, 2002; Boase and Kelly, 2004). However, this hypothesis was not supported by several recent studies (see Nuclear export-dependent degradation of CreA).
Improved Production of Hydrolytic Enzymes by Disruption and Mutation of CCR Regulators
The release of CCR is highly effective in improving hydrolytic enzyme production in filamentous fungi. For instance, disruption and mutation of the creA ortholog in Trichoderma reesei significantly increase the production of cellulase (Nakari-Setälä et al., 2009). α-Amylase production in A. oryzae also increases upon the disruption of creA (Ichinose et al., 2014). Disruption of creB also increases the production of α-amylase in A. oryzae (Hunter et al., 2013; Ichinose et al., 2014). Moreover, double disruption of creA and creB further increases amylase production, which is more than 10 times higher than that in the wild-type strain (Ichinose et al., 2014). The transcript level of the α-amylase gene markedly increased in the creA disruption strain, whereas it only slightly increased upon creB disruption (Ichinose et al., 2014). This suggests that the destruction of creA and creB has different effects on α-amylase production. In addition to those of α-amylase, the production levels of xylanase and β-glucosidase significantly increased upon the double disruption of creA and creB (Ichinose et al., 2018). In contrast, creA and creB disruption had no effect on cellulase (endo-β-glucanase) production. In A. nidulans, creA disruption did not result in de-repression of cellulose production; it was de-repressed by the disruption of protein kinase A gene (pkaA) (Kunitake et al., 2019), suggesting that CreA is not relevant to CCR regulation of cellulase production in A. oryzae.
In A. oryzae CreD, two serine residues at positions 402 and 515 were identified as phosphorylation sites (Tanaka et al., 2017). Mutation of these phosphorylation sites to glutamic acid for the phosphorylation mimic repressed the amylolytic enzyme production of the creB disruption mutant in the presence of glucose (Tanaka et al., 2017). In contrast, dephosphorylation mutations of CreD promoted CCR release by creB disruption and increased the production levels of α-amylase (Tanaka et al., 2017). This finding provides a novel approach, the combination of the dephosphorylation mutation of CreD and creB disruption, to improve the production of secretory glycoside hydrolases in filamentous fungi. In addition, these results suggest that CreB targets unknown factor(s) that are recognized by dephosphorylated CreD for ubiquitination. However, CreA is unlikely to be a target factor for CreB and CreD (see Nuclear export-dependent degradation of CreA).
Nuclear Export-Dependent Degradation of CreA
In budding yeast, Mig1 shuttles between the nucleus and cytoplasm in response to the glucose concentration. When green fluorescent protein (GFP) is fused to CreA in A. oryzae, almost all GFP fluorescence is observed in the nucleus in the presence of CCR-inducing sugars such as glucose and mannose. However, when sugars such as maltose and xylose, which induce the expression of glycosyl hydrolase genes, are used as carbon sources, CreA is exported to the cytoplasm. This nuclear export depends on a leucine-rich nuclear export signal (NES) near the C-terminus of CreA. CreA with a 3 × FLAG tag fused to its N-terminus is rapidly degraded when maltose or xylose is used as a carbon source. However, mutations in the NES significantly inhibit the degradation of CreA (Tanaka et al., 2018). These results indicate that CreA is rapidly degraded in the cytoplasm after export from the nucleus under conditions that induce the production of secretory hydrolytic enzymes. The deletion of a 20 amino acid region near the C-terminus significantly stabilizes CreA (Tanaka et al., 2018). This 20 amino acid region is highly conserved in the CreA orthologs of other filamentous fungi.
In A. oryzae, disruption of creB and creC significantly reduces the amount of CreA in the presence of glucose (Tanaka et al., 2018). Similarly, CreA-GFP protein levels also reduced in an A. nidulans creC mutant strain (Ries et al., 2016). In addition, creA transcript levels reduced in the presence of glucose in an A. nidulans creB mutant strain (Strauss et al., 1999). This reduced CreA protein level may have contributed to the release of CCR by the disruption of creB or creC. However, as mentioned above, the increase in transcript levels of the α-amylase gene by disruption of creB was slight, and double disruption of creA and creB resulted in a significant increase in α-amylase production (Ichinose et al., 2014), suggesting that the main reason for CCR release due to creB disruption is independent of the decrease in CreA abundance. In addition, CreA stability was not significantly affected by the disruption of creD (Tanaka et al., 2018). Therefore, there are likely to be other factor(s) involved in the regulation of CreA-independent CCR, the stability of which is controlled by CreB and CreD. Identification of such CreB and CreD target factors would lead to an understanding of CCR regulation in filamentous fungi.
In A. nidulans, the amount of CreA-GFP protein under repressing conditions significantly increased upon the deletion of fbx23, an F-box protein that constitutes the Skp-Cullin-F-box (SCF) ubiquitin ligase complex (de Assis et al., 2018). Therefore, CreA is possibly degraded in an SCF ubiquitin ligase complex-dependent manner in filamentous fungi. However, there is no direct experimental evidence for CreA ubiquitination, although the polyubiquitin precursor Ubi4 was identified as an interacting partner for both CreA and Fbx23 (de Assis et al., 2018). Further investigation of CreA degradation is required for a better understanding of the CCR regulation mechanism in filamentous fungi.
In budding yeast, Mig1 is phosphorylated by the cyclic AMP (cAMP) kinase Snf1 under low-glucose conditions, and exported from the nucleus to the cytoplasm (DeVit and Johnston, 1999). However, the subcellular localization and stability of A. oryzae CreA are not affected by disruption of the SNF1 ortholog (Tanaka et al., 2018). This indicates that the regulatory mechanisms for the subcellular localization of yeast Mig1 and A. oryzae CreA are different. In agreement with this, the purified recombinant protein of a T. reesei Snf1 ortholog phosphorylates yeast Mig1 but not T. reesei Cre1, an ortholog of CreA (Cziferszky et al., 2003). In budding yeast, hexokinase Hxk2 is also phosphorylated by Snf1 and is involved in the regulation of Mig1 nuclear export (Ahuatzi et al., 2007). Aspergillus species have two functional glucose kinases, hexokinase HxkA and glucokinase GlkA (Fleck and Brock, 2010). It would be interesting to investigate the involvement of these glucose kinases in the nuclear export and degradation of CreA in A. oryzae.
Several recent studies have examined the phosphorylation of CreA in A. nidulans. Six serine residues (S262, S277, S288, S289, S312, and S319) in A. nidulans CreA were identified as phosphorylation sites by LC-MS analysis, and three of them (S289, S312, and S319) were phosphorylated only under conditions of growth in a glucose medium (Alam et al., 2017). Another recent study revealed six additional phosphorylation sites in A. nidulans CreA (S176, S268, S281, S284, T308, and S406; de Assis et al., 2021). The replacement of T308 with alanine significantly inhibited the nuclear accumulation of CreA-GFP. In contrast, the replacement of S262 or S268 with alanine increased the nuclear localization of CreA-GFP under de-repressing conditions, although these CreA-GFP mutants were not detectable by western blotting under such conditions (de Assis et al., 2021). Phosphorylation at S319 is lost upon the deletion of pkaA, which encodes a cAMP-dependent protein kinase, whereas CreA is not a direct target of this protein kinase (Ribeiro et al., 2019). Although the replacement of S319 with alanine has no significant effect on CreA subcellular localization, this mutation and the T308A mutation significantly increased CreA-GFP protein levels under repressing conditions (de Assis et al., 2021). However, none of the substitutions of these phosphorylation sites with alanine had a significant effect on CreA degradation under de-repressing conditions (de Assis et al., 2021). A recent study showed that replacement of S388 at the T. reesei Cre1 C-terminus with valine releases CCR (Han et al., 2020). This serine residue is conserved in the Aspergillus CreA proteins. However, deletion of 20 amino acids in the C-terminus, including this conserved serine residue, has no effect on the stability of A. oryzae CreA (Tanaka et al., 2018).
Identification of the protein kinase that phosphorylates CreA is an important issue that remains to be addressed. In T. reesei, S241 within Cre1 (corresponding to S262 in A. nidulans CreA) has been identified as a phosphorylation site, and the phosphorylation of this serine residue positively regulates DNA binding (Cziferszky et al., 2002). In addition, casein kinase II is a strong candidate kinase that phosphorylates Cre1 (Cziferszky et al., 2002). In A. nidulans, casein kinase CkiA has been identified as an interacting partner of Fbx23 (de Assis et al., 2018). Therefore, it is important to elucidate the role of casein kinase in the regulation of CreA function.
Glucose-Induced Endocytosis of Maltose Transporter
The amount of membrane proteins, including transporters involved in nutrient uptake, is tightly controlled. When the extracellular environment changes, unnecessary membrane proteins are internalized into the cell by endocytosis and transported to the vacuole for degradation (Polo and Di Fiore, 2008). This endocytosis is caused by ubiquitination of cell membrane proteins. MalP is the major maltose transporter required for amylolytic enzyme production in A. oryzae (Hasegawa et al., 2010; Hiramoto et al., 2015). When glucose is added, MalP is translocated from the cell membrane to the vacuole via endocytosis (Hiramoto et al., 2015). The addition of mannose, which induces the CCR of amylolytic genes, as well as glucose, also induces MalP endocytosis (Hiramoto et al., 2015). These results suggest that MalP is degraded to inhibit the uptake of maltose, which induces the expression of amylolytic genes. CreD and HulA are essential for this glucose-induced endocytosis of MalP (Tanaka et al., 2017). Therefore, in the presence of glucose, CreD probably induces endocytosis by recruiting HulA to MalP. Although CreD is rapidly dephosphorylated upon the addition of glucose, the phosphorylation state of CreD is not associated with the interaction with HulA and the endocytosis of MalP (Tanaka et al., 2017). The regulatory mechanism by which CreD induces glucose-specific MalP ubiquitination is unknown. In budding yeast, ART proteins, including Rod1/Art4, are also ubiquitinated by Rsp5. This ubiquitination is required for adaptor activation or degradation (Ho et al., 2017; MacDonald et al., 2020). It is important to examine whether CreD is ubiquitinated in A. oryzae.
The inhibition of glucose-induced endocytosis is expected to enhance substrate uptake. In fact, cellobiose consumption and ethanol production were enhanced by stabilizing heterologously expressed cellobiose transporters in budding yeast lacking four ARTs, including Rod1/Art4 (Sen et al., 2016). However, disruption of creD in A. oryzae counteracts the release of CCR by creB disruption and reduces α-amylase production (Tanaka et al., 2017). Therefore, stabilizing MalP without creD disruption, e.g., by introducing mutations in the ubiquitination sites of MalP, is necessary to increase amylase production.
Discussion
As mentioned above, details on the regulatory mechanism of the induction and repression of hydrolytic gene expression in A. oryzae are becoming clearer, particularly for amylolytic genes (Figure 1). The release of CCR is significantly effective in improving the productivity of hydrolytic enzymes. In addition, constitutive activation of transcription factors leads to a reduction in the cost of producing hydrolytic enzymes (Alazi and Ram, 2018). However, the molecular mechanism of transcriptional regulation of hydrolytic enzyme genes in A. oryzae has not been elucidated, even for amylolytic genes. For example, glaB gene expression in solid-state culture requires at least AmyR and FlbC, but another unidentified transcription factor(s) seems to be needed because promoter deletion analysis shows that a GC-rich sequence is important for glaB expression (Ishida et al., 2000; Hisada et al., 2013). The AmyR-binding sequence and putative FlbC-binding sequence are different from this GC-rich sequence, to which a certain transcription factor may bind. Moreover, because recent studies have revealed that the regulatory mechanism of hydrolytic gene expression differs among Aspergillus species; further research on the regulatory mechanism of hydrolytic gene expression in A. oryzae should be accelerated.
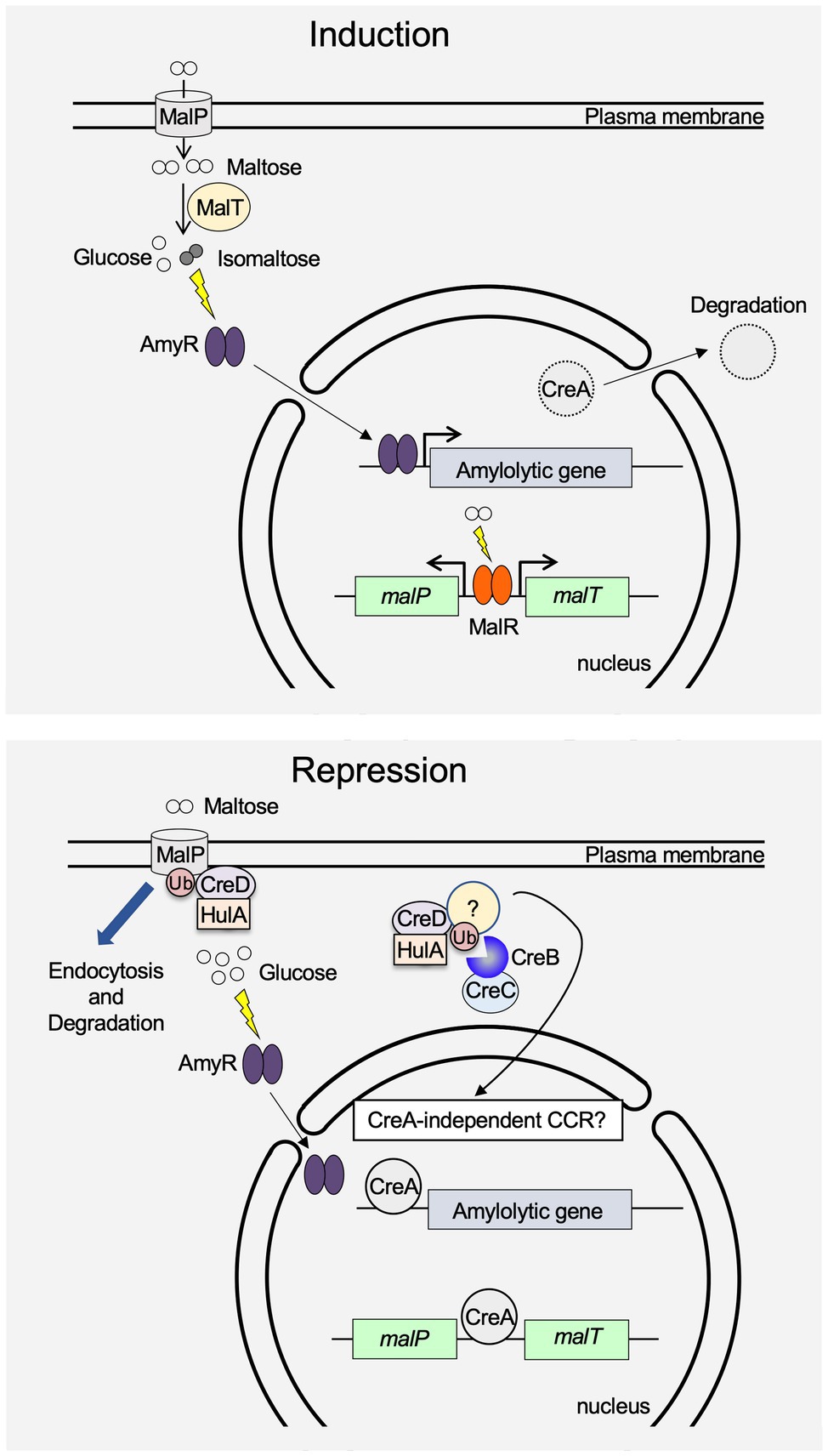
Figure 1. Schematic model for induction and repression of amylolytic gene expression in A. oryzae. When maltose is added to the culture medium, MalR induces malP and malT expression. After the uptake of maltose into the cell by MalP, AmyR nuclear transfer is triggered by isomaltose, which is presumably generated by the glycosyltransferase activity of MalT. CreA is exported from the nucleus to the cytoplasm, where it is degraded, and amylolytic gene expression is induced by activated AmyR. In the presence of glucose, the expression of malP and malT is repressed by CreA. Moreover, MalP protein is brought into the cell by endocytosis mediated by HulA and CreD, and is degraded at the vacuole. Although a high concentration of glucose induces AmyR nuclear transfer, expression of amylolytic genes is strongly repressed by CreA. The unknown protein stabilized by the CreB-CreC complex presumably regulates CreA-independent carbon catabolite repression (CCR).
Autolysis of the A. oryzae mycelium is important for its production of volatile compounds that contribute to soy sauce flavor (Xu et al., 2016). Cell wall degrading enzymes including chitinase and β-1,3-glucanase degrade fungal cell walls, and intracellular proteases and nucleases then degrade proteins and DNA/RNA, respectively, during autolysis. In A. nidulans, two transcription factors, RlmA and XprG, which regulate glucanase and chitinase genes, are involved in autolysis (Katz et al., 2013; Kovács et al., 2013). Given that no intracellular protease or nuclease gene expression regulatory mechanisms have been reported thus far, identification of transcription factors involved in A. oryzae autolyis will aid in development of industrial applications for A. oryzae. In addition, chitin in the A. oryzae cell wall adsorbs secreted α-amylase. This adsorption is inhibited when chitin is masked by α-1,3-glucan (Sato et al., 2011; Zhang et al., 2017). In this regard, regulatory mechanisms for genes encoding cell wall remodeling enzymes are also of interest.
The analysis of gene expression mechanisms in A. oryzae has progressed significantly after the completion of A. oryzae whole genome sequencing. In particular, similar to that for A. fumigatus (Furukawa et al., 2020), a disruption library for transcriptional regulatory genes, including transcription factors and transcription-related machinery, has been constructed by the Japanese A. oryzae research community. This disruption library as well as the overexpression library which covers a part of transcription factor genes has greatly contributed to the identification of novel transcription factors responsible for hydrolytic gene expression, such as FlbC, ManR, and PrtR. Such libraries have also been effectively utilized to mine novel transcription factors, such as AtrR (azole drug resistance and ABC transporter gene regulation; Hagiwara et al., 2017), EcdR (conidiophore development; Jin et al., 2011), and KpeA (kojic acid production and conidiation; Arakawa et al., 2019), that are involved in the regulatory expression of genes other than hydrolytic genes. Since the library of disruption mutant strains contains over 500 genes putatively involved in transcriptional regulation, further identification of novel transcription factors that regulate the expression of hydrolytic genes in A. oryzae is highly expected.
Author Contributions
MT and KG wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was partly supported by Grants-in-Aid for Scientific Research (JSPS KAKENHI, 22248007, 25292044, and 16H04894), the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry, and the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries, and Food Industry.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Editage (www.editage.com) for English language editing.
References
Ahuatzi, D., Riera, A., Peláez, R., Herrero, P., and Moreno, F. (2007). Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J. Biol. Chem. 282, 4485–4493. doi: 10.1074/jbc.M606854200
Alam, M. A., Kamlangdee, N., and Kelly, J. M. (2017). The CreB deubiquitinating enzyme does not directly target the CreA repressor protein in Aspergillus nidulans. Curr. Genet. 63, 647–667. doi: 10.1007/s00294-016-0666-3
Alazi, E., and Ram, A. (2018). Modulating transcriptional regulation of plant biomass degrading enzyme networks for rational design of industrial fungal strains. Front. Bioeng. Biotechnol. 6:133. doi: 10.3389/fbioe.2018.00133
Arakawa, G. Y., Kudo, H., Yanase, A., Eguchi, Y., Kodama, H., Ogawa, M., et al. (2019). A unique Zn(II)2-Cys6-type protein, KpeA, is involved in secondary metabolism and conidiation in Aspergillus oryzae. Fungal Genet. Biol. 127, 35–44. doi: 10.1016/j.fgb.2019.02.004
Arst, H. N., and Cove, D. J. (1973). Nitrogen metabolite repression in Aspergillus nidulans. Mol. Gen. Genet. 126, 111–141. doi: 10.1007/BF00330988
de Assis, L. J., Silva, L. P., Bayram, O., Dowling, P., Kniemeyer, O., Krüger, T., et al. (2021). Carbon catabolite repression in filamentous fungi is regulated by phosphorylation of the transcription factor CreA. MBio 12:e03146–20. doi: 10.1128/mBio.03146-20
de Assis, L. J., Ulas, M., Ries, L., El Ramli, N., Sarikaya-Bayram, O., Braus, G. H., et al. (2018). Regulation of Aspergillus nidulans CreA-mediated catabolite repression by the F-box proteins Fbx23 and Fbx47. MBio 9:e00840–18. doi: 10.1128/mBio.00840-18
Battaglia, E., Visser, L., Nijssen, A., van Veluw, G. J., Wösten, H. A., and de Vries, R. P. (2011). Analysis of regulation of pentose utilisation in Aspergillus niger reveals evolutionary adaptations in Eurotiales. Stud. Mycol. 69, 31–38. doi: 10.3114/sim.2011.69.03
Bermúdez-García, E., Peña-Montes, C., Martins, I., Pais, J., Pereira, C. S., Sánchez, S., et al. (2019). Regulation of the cutinases expressed by Aspergillus nidulans and evaluation of their role in cutin degradation. Appl. Microbiol. Biotechnol. 103, 3863–3874. doi: 10.1007/s00253-019-09712-3
Boase, N. A., and Kelly, J. M. (2004). A role for creD, a carbon catabolite repression gene from Aspergillus nidulans, in ubiquitination. Mol. Microbiol. 53, 929–940. doi: 10.1111/j.1365-2958.2004.04172.x
Boni, A. C., Ambrósio, D. L., Cupertino, F. B., Montenegro-Montero, A., Virgilio, S., Freitas, F. Z., et al. (2018). Neurospora crassa developmental control mediated by the FLB-3 transcription factor. Fungal Biol. 122, 570–582. doi: 10.1016/j.funbio.2018.01.004
Cziferszky, A., Mach, R. L., and Kubicek, C. P. (2002). Phosphorylation positively regulates DNA binding of the carbon catabolite repressor Cre1 of Hypocrea jecorina (Trichoderma reesei). J. Biol. Chem. 277, 14688–14694. doi: 10.1074/jbc.M200744200
Cziferszky, A., Seiboth, B., and Kubicek, C. P. (2003). The Snf1 kinase of the filamentous fungus Hypocrea jecorina phosphorylates regulation-relevant serine residues in the yeast carbon catabolite repressor Mig1 but not in the filamentous fungal counterpart Cre1. Fungal Genet. Biol. 40, 166–175. doi: 10.1016/S1087-1845(03)00082-3
DeVit, M. J., and Johnston, M. (1999). The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 9, 1231–1241. doi: 10.1016/S0960-9822(99)80503-X
Dowzer, C. E., and Kelly, J. M. (1991). Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol. Cell. Biol. 11, 5701–5709. doi: 10.1128/MCB.11.11.5701
Fleck, C. B., and Brock, M. (2010). Aspergillus fumigatus catalytic glucokinase and hexokinase: expression analysis and importance for germination, growth, and conidiation. Eukaryot. Cell 9, 1120–1135. doi: 10.1128/EC.00362-09
Furukawa, T., van Rhijn, N., Fraczek, M., Gsaller, F., Davies, E., Carr, P., et al. (2020). The negative cofactor 2 complex is a key regulator of drug resistance in Aspergillus fumigatus. Nat. Commun. 11:427. doi: 10.1038/s41467-019-14191-1
Garrido, S. M., Kitamoto, N., Watanabe, A., Shintani, T., and Gomi, K. (2012). Functional analysis of FarA transcription factor in the regulation of the genes encoding lipolytic enzymes and hydrophobic surface binding protein for the degradation of biodegradable plastics in Aspergillus oryzae. J. Biosci. Bioeng. 113, 549–555. doi: 10.1016/j.jbiosc.2011.12.014
Gomi, K. (2014). “ASPERGILLUS | Aspergillus oryzae,” in Encyclopedia of food microbiology. 2nd Edn. eds. C. A. Batt and M. L. Tortorello (London, UK: Academic Press), 92–96.
Gomi, K. (2019). Regulatory mechanisms for amylolytic gene expression in the koji mold Aspergillus oryzae. Biosci. Biotechnol. Biochem. 83, 1385–1401. doi: 10.1080/09168451.2019.1625265
Gomi, K., Akeno, T., Minetoki, T., Ozeki, K., Kumagai, C., Okazaki, N., et al. (2000). Molecular cloning and characterization of a transcriptional activator gene, amyR, involved in the amylolytic gene expression in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 64, 816–827. doi: 10.1271/bbb.64.816
Hagiwara, D., Miura, D., Shimizu, K., Paul, S., Ohba, A., Gonoi, T., et al. (2017). A novel Zn2-Cys6 transcription factor AtrR plays a key role in an azole resistance mechanism of Aspergillus fumigatus by coregulating cyp51A and cdr1B expressions. PLoS Pathog. 13:e1006096. doi: 10.1371/journal.ppat.1006096
Han, L., Tan, Y., Ma, W., Niu, K., Hou, S., Guo, W., et al. (2020). Precision engineering of the transcription factor Cre1 in Hypocrea jecorina (Trichoderma reesei) for efficient cellulase production in the presence of glucose. Front. Bioeng. Biotechnol. 8:852. doi: 10.3389/fbioe.2020.00852
Hartmann, T., Cairns, T. C., Olbermann, P., Morschhäuser, J., Bignell, E. M., and Krappmann, S. (2011). Oligopeptide transport and regulation of extracellular proteolysis are required for growth of Aspergillus fumigatus on complex substrates but not for virulence. Mol. Microbiol. 82, 917–935. doi: 10.1111/j.1365-2958.2011.07868.x
Hasegawa, S., Takizawa, M., Suyama, H., Shintani, T., and Gomi, K. (2010). Characterization and expression analysis of a maltose-utilizing (MAL) cluster in Aspergillus oryzae. Fungal Genet. Biol. 47, 1–9. doi: 10.1016/j.fgb.2009.10.005
Hata, Y., Ishida, H., Ichikawa, E., Kawato, A., Suginami, K., and Imayasu, S. (1998). Nucleotide sequence of an alternative glucoamylase-encoding gene (glaB) expressed in solid-state culture of Aspergillus oryzae. Gene 207, 127–134. doi: 10.1016/S0378-1119(97)00612-4
Hata, Y., Tsuchiya, K., Kitamoto, K., Gomi, K., Kumagai, C., Tamura, G., et al. (1991). Nucleotide sequence and expression of the glucoamylase-encoding gene (glaA) from Aspergillus oryzae. Gene 108, 145–150. doi: 10.1016/0378-1119(91)90500-b
Hiramoto, T., Tanaka, M., Ichikawa, T., Matsuura, Y., Hasegawa-Shiro, S., Shintani, T., et al. (2015). Endocytosis of a maltose permease is induced when amylolytic enzyme production is repressed in Aspergillus oryzae. Fungal Genet. Biol. 82, 136–144. doi: 10.1016/j.fgb.2015.05.015
Hisada, H., Sano, M., Ishida, H., Hata, Y., and Machida, M. (2013). Identification of regulatory elements in the glucoamylase-encoding gene (glaB) promoter from Aspergillus oryzae. Appl. Microbiol. Biotechnol. 97, 4951–4956. doi: 10.1007/s00253-012-4622-y
Ho, H. C., MacGurn, J. A., and Emr, S. D. (2017). Deubiquitinating enzymes Ubp2 and Ubp15 regulate endocytosis by limiting ubiquitination and degradation of ARTs. Mol. Biol. Cell 28, 1271–1283. doi: 10.1091/mbc.E17-01-0008
Huang, L., Dong, L., Wang, B., and Pan, L. (2020). The transcription factor PrtT and its target protease profiles in Aspergillus niger are negatively regulated by carbon sources. Biotechnol. Lett. 42, 613–624. doi: 10.1007/s10529-020-02806-3
Hunter, A. J., Morris, T. A., Jin, B., Saint, C. P., and Kelly, J. M. (2013). Deletion of creB in Aspergillus oryzae increases secreted hydrolytic enzyme activity. Appl. Environ. Microbiol. 79, 5480–5487. doi: 10.1128/AEM.01406-13
Hynes, M. J., and Kelly, J. M. (1977). Pleiotropic mutants of Aspergillus nidulans altered in carbon metabolism. Mol. Gen. Genet. 150, 193–204. doi: 10.1007/BF00695399
Hynes, M. J., Murray, S. L., Duncan, A., Khew, G. S., and Davis, M. A. (2006). Regulatory genes controlling fatty acid catabolism and peroxisomal functions in the filamentous fungus Aspergillus nidulans. Eukaryot. Cell 5, 794–805. doi: 10.1128/EC.5.5.794-805.2006
Ichinose, S., Tanaka, M., Shintani, T., and Gomi, K. (2014). Improved α-amylase production by Aspergillus oryzae after a double deletion of genes involved in carbon catabolite repression. Appl. Microbiol. Biotechnol. 98, 335–343. doi: 10.1007/s00253-013-5353-4
Ichinose, S., Tanaka, M., Shintani, T., and Gomi, K. (2018). Increased production of biomass-degrading enzymes by double deletion of creA and creB genes involved in carbon catabolite repression in Aspergillus oryzae. J. Biosci. Bioeng. 125, 141–147. doi: 10.1016/j.jbiosc.2017.08.019
Ishida, H., Hata, Y., Ichikawa, E., Kawato, A., Suginami, K., and Imayasu, S. (1998). Regulation of the glucoamylase-encoding gene (glaB), expressed in solid-state culture (koji) of Aspergillus oryzae. J. Ferment. Bioeng. 86, 301–307. doi: 10.1016/S0922-338X(98)80134-7
Ishida, H., Hata, Y., Kawato, A., Abe, Y., Suginami, K., and Imayasu, S. (2000). Identification of functional elements that regulate the glucoamylase-encoding gene (glaB) expressed in solid-state culture of Aspergillus oryzae. Curr. Genet. 37, 373–379. doi: 10.1007/s002940000118
Ishikawa, K., Kunitake, E., Kawase, T., Atsumi, M., Noguchi, Y., Ishikawa, S., et al. (2018). Comparison of the paralogous transcription factors AraR and XlnR in Aspergillus oryzae. Curr. Genet. 64, 1245–1260. doi: 10.1007/s00294-018-0837-5
Ito, T., Tani, S., Itoh, T., Tsukagoshi, N., Kato, M., and Kobayashi, T. (2004). Mode of AmyR binding to the CGGN8AGG sequence in the Aspergillus oryzae taaG2 promoter. Biosci. Biotechnol. Biochem. 68, 1906–1911. doi: 10.1271/bbb.68.1906
Jin, F., Hu, S., Wang, B., and Jin, L. (2021). Advances in genetic engineering technology and its application in the industrial fungus Aspergillus oryzae. Front. Microbiol. 12:644404. doi: 10.3389/fmicb.2021.644404
Jin, F., Nishida, M., Hara, S., and Koyama, Y. (2011). Identification and characterization of a putative basic helix-loop-helix transcription factor involved in the early stage of conidiophore development in Aspergillus oryzae. Fungal Genet. Biol. 48, 1108–1115. doi: 10.1016/j.fgb.2011.10.001
Kato, M., Sekine, K., and Tsukagoshi, N. (1996). Sequence-specific binding sites in the Taka-amylase A G2 promoter for the CreA repressor mediating carbon catabolite repression. Biosci. Biotechnol. Biochem. 60, 1776–1779. doi: 10.1271/bbb.60.1776
Katz, M. E., Braunberger, K., Yi, G., Cooper, S., Nonhebel, H. M., and Gondro, C. (2013). A p53-like transcription factor similar to Ndt80 controls the response to nutrient stress in the filamentous fungus, Aspergillus nidulans. F1000Res 2:72. doi: 10.12688/f1000research.2-72.v1
Kelly, J. M., and Hynes, M. J. (1977). Increased and decreased sensitivity to carbon catabolite repression of enzymes of acetate metabolism in mutants of Aspergillus nidulans. Mol. Gen. Genet. 156, 87–92. doi: 10.1007/BF00272256
Kobayashi, T., Abe, K., Asai, K., Gomi, K., Juvvadi, P. R., Kato, M., et al. (2007). Genomics of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 71, 646–670. doi: 10.1271/bbb.60550
Konno, Y., Suzuki, K., Tanaka, M., Shintani, T., and Gomi, K. (2018). Chaperone complex formation of the transcription factor MalR involved in maltose utilization and amylolytic enzyme production in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 82, 827–835. doi: 10.1080/09168451.2018.1447359
Kouranti, I., McLean, J. R., Feoktistova, A., Liang, P., Johnson, A. E., Roberts-Galbraith, R. H., et al. (2010). A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS Biol. 8:e1000471. doi: 10.1371/journal.pbio.1000471
Kovács, Z., Szarka, M., Kovács, S., Boczonádi, I., Emri, T., Abe, K., et al. (2013). Effect of cell wall integrity stress and RlmA transcription factor on asexual development and autolysis in Aspergillus nidulans. Fungal Genet. Biol. 54, 1–14. doi: 10.1016/j.fgb.2013.02.004
Kunitake, E., and Kobayashi, T. (2017). Conservation and diversity of the regulators of cellulolytic enzyme genes in Ascomycete fungi. Curr. Genet. 63, 951–958. doi: 10.1007/s00294-017-0695-6
Kunitake, E., Li, Y., Uchida, R., Nohara, T., Asano, K., Hattori, A., et al. (2019). CreA-independent carbon catabolite repression of cellulase genes by trimeric G-protein and protein kinase A in Aspergillus nidulans. Curr. Genet. 65, 941–952. doi: 10.1007/s00294-019-00944-4
Kwon, N. J., Garzia, A., Espeso, E. A., Ugalde, U., and Yu, J. H. (2010). FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 77, 1203–1219. doi: 10.1111/j.1365-2958.2010.07282.x
Li, D., and Kolattukudy, P. E. (1997). Cloning of cutinase transcription factor 1, a transactivating protein containing Cys6Zn2 binuclear cluster DNA-binding motif. J. Biol. Chem. 272, 12462–12467. doi: 10.1074/jbc.272.19.12462
Lin, C. H., MacGurn, J. A., Chu, T., Stefan, C. J., and Emr, S. D. (2008). Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135, 714–725. doi: 10.1016/j.cell.2008.09.025
Lockington, R. A., and Kelly, J. M. (2001). Carbon catabolite repression in Aspergillus nidulans involves deubiquitination. Mol. Microbiol. 40, 1311–1321. doi: 10.1046/j.1365-2958.2001.02474.x
Lockington, R. A., and Kelly, J. M. (2002). The WD40-repeat protein CreC interacts with and stabilizes the deubiquitinating enzyme CreB in vivo in Aspergillus nidulans. Mol. Microbiol. 43, 1173–1182. doi: 10.1046/j.1365-2958.2002.02811.x
Machida, M., Asai, K., Sano, M., Tanaka, T., Kumagai, T., Terai, G., et al. (2005). Genome sequencing and analysis of Aspergillus oryzae. Nature 438, 1157–1161. doi: 10.1038/nature04300
Machida, M., Yamada, O., and Gomi, K. (2008). Genomics of Aspergillus oryzae: learning from the history of Koji mold and exploration of its future. DNA Res. 15, 173–183. doi: 10.1093/dnares/dsn020
MacDonald, C., Shields, S. B., Williams, C. A., Winistorfer, S., and Piper, R. C. (2020). A cycle of Ubiquitination regulates adaptor function of the Nedd4-family ubiquitin ligase Rsp5. Curr. Biol. 30, 465–479. doi: 10.1016/j.cub.2019.11.086
Maeda, H., Yamagata, Y., Abe, K., Hasegawa, F., Machida, M., Ishioka, R., et al. (2005). Purification and characterization of a biodegradable plastic-degrading enzyme from Aspergillus oryzae. Appl. Microbiol. Biotechnol. 67, 778–788. doi: 10.1007/s00253-004-1853-6
Makita, T., Katsuyama, Y., Tani, S., Suzuki, H., Kato, N., Todd, R. B., et al. (2009). Inducer-dependent nuclear localization of a Zn(II)(2)Cys(6) transcriptional activator, AmyR, in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 73, 391–399. doi: 10.1271/bbb.80654
Marui, J., Kitamoto, N., Kato, M., Kobayashi, T., and Tsukagoshi, N. (2002). Transcriptional activator, AoXlnR, mediates cellulose-inductive expression of the xylanolytic and cellulolytic genes in Aspergillus oryzae. FEBS Lett. 528, 279–282. doi: 10.1016/S0014-5793(02)03328-8
Matheis, S., Yemelin, A., Scheps, D., Andresen, K., Jacob, S., Thines, E., et al. (2017). Functions of the Magnaporthe oryzae Flb3p and Flb4p transcription factors in the regulation of conidiation. Microbiol. Res. 196, 106–117. doi: 10.1016/j.micres.2016.12.010
Minetoki, T., Tsuboi, H., Koda, A., and Ozeki, K. (2003). Development of high expression system with the improved promoter using the cis-acting element in Aspergillus species. J. Biol. Macromol. 3, 89–96.
Mizutani, O., Kudo, Y., Saito, A., Matsuura, T., Inoue, H., Abe, K., et al. (2008). A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet. Biol. 45, 878–889. doi: 10.1016/j.fgb.2007.12.010
Murakoshi, Y., Makita, T., Kato, M., and Kobayashi, T. (2012). Comparison and characterization of α-amylase inducers in Aspergillus nidulans based on nuclear localization of AmyR. Appl. Microbiol. Biotechnol. 94, 1629–1635. doi: 10.1007/s00253-012-3874-x
Nakari-Setälä, T., Paloheimo, M., Kallio, J., Vehmaanperä, J., Penttilä, M., and Saloheimo, M. (2009). Genetic modification of carbon catabolite repression in Trichoderma reesei for improved protein production. Appl. Environ. Microbiol. 75, 4853–4860. doi: 10.1128/AEM.00282-09
Needleman, R. (1991). Control of maltase synthesis in yeast. Mol. Microbiol. 5, 2079–2084. doi: 10.1111/j.1365-2958
Nikko, E., and Pelham, H. R. (2009). Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10, 1856–1867. doi: 10.1111/j.1600-0854.2009.00990.x
Noguchi, Y., Sano, M., Kanamaru, K., Ko, T., Takeuchi, M., Kato, M., et al. (2009). Genes regulated by AoXlnR, the xylanolytic and cellulolytic transcriptional regulator, in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 85, 141–154. doi: 10.1007/s00253-009-2236-9
Noguchi, Y., Tanaka, H., Kanamaru, K., Kato, M., and Kobayashi, T. (2011). Xylose triggers reversible phosphorylation of XlnR, the fungal transcriptional activator of xylanolytic and cellulolytic genes in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 75, 953–959. doi: 10.1271/bbb.100923
Oda, K., Kakizono, D., Yamada, O., Iefuji, H., Akita, O., and Iwashita, K. (2006). Proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl. Environ. Microbiol. 72, 3448–3457. doi: 10.1128/AEM.72.5.3448-3457.2006
Ogawa, M., Kobayashi, T., and Koyama, Y. (2012). ManR, a novel Zn(II)2Cys6 transcriptional activator, controls the β-mannan utilization system in Aspergillus oryzae. Fungal Genet. Biol. 49, 987–995. doi: 10.1016/j.fgb.2012.09.006
Ogawa, M., Kobayashi, T., and Koyama, Y. (2013). ManR, a transcriptional regulator of the β-mannan utilization system, controls the cellulose utilization system in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 77, 426–429. doi: 10.1271/bbb.120795
Ogawa, M., Tokuoka, M., Jin, F. J., Takahashi, T., and Koyama, Y. (2010). Genetic analysis of conidiation regulatory pathways in koji-mold Aspergillus oryzae. Fungal Genet. Biol. 47, 10–18. doi: 10.1016/j.fgb.2009.10.004
Ohtaki, S., Maeda, H., Takahashi, T., Yamagata, Y., Hasegawa, F., Gomi, K., et al. (2006). Novel hydrophobic surface binding protein, HsbA, produced by Aspergillus oryzae. Appl. Environ. Microbiol. 72, 2407–2413. doi: 10.1128/AEM.72.4.2407-2413.2006
Oikawa, H. (2020). Reconstitution of biosynthetic machinery of fungal natural products in heterologous hosts. Biosci. Biotechnol. Biochem. 84, 433–444. doi: 10.1080/09168451.2019.1690976
Petersen, K. L., Lehmbeck, J., and Christensen, T. (1999). A new transcriptional activator for amylase genes in Aspergillus. Mol. Gen. Genet. 262, 668–676. doi: 10.1007/s004380051129
Polo, S., and Di Fiore, P. P. (2008). Finding the right partner: science or ART? Cell 135, 590–592. doi: 10.1016/j.cell.2008.10.032
Punt, P. J., Schuren, F. H., Lehmbeck, J., Christensen, T., Hjort, C., and van den Hondel, C. A. (2008). Characterization of the Aspergillus niger prtT, a unique regulator of extracellular protease encoding genes. Fungal Genet. Biol. 45, 1591–1599. doi: 10.1016/j.fgb.2008.09.007
Ran, F., Bali, M., and Michels, C. A. (2008). Hsp90/Hsp70 chaperone machine regulation of the Saccharomyces MAL-activator as determined in vivo using noninducible and constitutive mutant alleles. Genetics 179, 331–343. doi: 10.1534/genetics.107.084921
Ribeiro, L., Chelius, C., Boppidi, K. R., Naik, N. S., Hossain, S., Ramsey, J., et al. (2019). Comprehensive analysis of Aspergillus nidulans PKA phosphorylome identifies a novel mode of CreA regulation. MBio 10:e02825–18. doi: 10.1128/mBio.02825-18
Ries, L. N., Beattie, S. R., Espeso, E. A., Cramer, R. A., and Goldman, G. H. (2016). Diverse regulation of the CreA carbon catabolite repressor in Aspergillus nidulans. Genetics 203, 335–352. doi: 10.1534/genetics.116.187872
Sato, H., Toyoshima, Y., Shintani, T., and Gomi, K. (2011). Identification of potential cell wall component that allows Taka-amylase A adsorption in submerged cultures of Aspergillus oryzae. Appl. Microbiol. Biotechnol. 92, 961–969. doi: 10.1007/s00253-011-3422-0
Sen, A., Acosta-Sampson, L., Alvaro, C. G., Ahn, J. S., Cate, J. H., and Thorner, J. (2016). Internalization of heterologous sugar transporters by endogenous α-arrestins in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 82, 7074–7085. doi: 10.1128/AEM.02148-16
Sharon, H., Hagag, S., and Osherov, N. (2009). Transcription factor PrtT controls expression of multiple secreted proteases in the human pathogenic mold Aspergillus fumigatus. Infect. Immun. 77, 4051–4060. doi: 10.1128/IAI.00426-09
Strauss, J., Horvath, H. K., Abdallah, B. M., Kindermann, J., Mach, R. L., and Kubicek, C. P. (1999). The function of CreA, the carbon catabolite repressor of Aspergillus nidulans, is regulated at the transcriptional and post-transcriptional level. Mol. Microbiol. 32, 169–178. doi: 10.1046/j.1365-2958.1999.01341.x
Suzuki, K., Tanaka, M., Konno, Y., Ichikawa, T., Ichinose, S., Hasegawa-Shiro, S., et al. (2015). Distinct mechanism of activation of two transcription factors, AmyR and MalR, involved in amylolytic enzyme production in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 99, 1805–1815. doi: 10.1007/s00253-014-6264-8
Takahashi, T., Maeda, H., Yoneda, S., Ohtaki, S., Yamagata, Y., Hasegawa, F., et al. (2005). The fungal hydrophobin RolA recruits polyesterase and laterally moves on hydrophobic surfaces. Mol. Microbiol. 57, 1780–1796. doi: 10.1111/j.1365-2958.2005.04803.x
Takahashi, T., Ogawa, M., Sato, A., and Koyama, Y. (2018). Translocated duplication of a targeted chromosomal segment enhances gene expression at the duplicated site and results in phenotypic changes in Aspergillus oryzae. Fungal Biol. Biotechnol. 5:17. doi: 10.1186/s40694-018-0061-6
Tanaka, M., and Gomi, K. (2014). “Strategies for increasing the production level of heterologous proteins in Aspergillus oryzae,” in Microbial Production. eds. H. Anazawa and S. Shimizu (Tokyo: Springer), 149–164.
Tanaka, M., Hiramoto, T., Tada, H., Shintani, T., and Gomi, K. (2017). Improved α-amylase production by dephosphorylation mutation of CreD, an arrestin-like protein required for glucose-induced endocytosis of maltose permease and carbon catabolite derepression in Aspergillus oryzae. Appl. Environ. Microbiol. 83:e00592–17. doi: 10.1128/AEM.00592-17
Tanaka, M., Ichinose, S., Shintani, T., and Gomi, K. (2018). Nuclear export-dependent degradation of the carbon catabolite repressor CreA is regulated by a region located near the C-terminus in Aspergillus oryzae. Mol. Microbiol. 110, 176–190. doi: 10.1111/mmi.14072
Tanaka, M., Ito, K., Matsuura, T., Kawarasaki, Y., and Gomi, K. (2021). Identification and distinct regulation of three di/tripeptide transporters in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 85, 452–463. doi: 10.1093/bbb/zbaa030
Tanaka, M., Yoshimura, M., Ogawa, M., Koyama, Y., Shintani, T., and Gomi, K. (2016). The C2H2-type transcription factor, FlbC, is involved in the transcriptional regulation of Aspergillus oryzae glucoamylase and protease genes specifically expressed in solid-state culture. Appl. Microbiol. Biotechnol. 100, 5859–5868. doi: 10.1007/s00253-016-7419-6
Tani, S., Katsuyama, Y., Hayashi, T., Suzuki, H., Kato, M., Gomi, K., et al. (2001). Characterization of the amyR gene encoding a transcriptional activator for the amylase genes in Aspergillus nidulans. Curr. Genet. 39, 10–15. doi: 10.1007/s002940000175
Tani, S., Kawaguchi, T., and Kobayashi, T. (2014). Complex regulation of hydrolytic enzyme genes for cellulosic biomass degradation in filamentous fungi. Appl. Microbiol. Biotechnol. 98, 4829–4837. doi: 10.1007/s00253-014-5707-6
Tonomura, K., Suzuki, H., Nakamura, N., Kuraya, K., and Tanabe, O. (1961). On the inducers of α-amylase formation in Aspergillus oryzae. Agric. Biol. Chem. 25, 1–6.
Tsuboi, H., Koda, A., Toda, T., Minetoki, T., Hirotsune, M., and Machida, M. (2005). Improvement of the Aspergillus oryzae enolase promoter (P-enoA) by the introduction of cis-element repeats. Biosci. Biotechnol. Biochem. 69, 206–208. doi: 10.1271/bbb.69.206
Villamil, M. A., Liang, Q., and Zhuang, Z. (2013). The WD40-repeat protein-containing deubiquitinase complex: catalysis, regulation, and potential for therapeutic intervention. Cell Biochem. Biophy 67, 111–126. doi: 10.1007/s12013-013-9637-1
Watanabe, J., Tanaka, H., Mogi, Y., Yamazaki, T., Suzuki, K., Watanabe, T., et al. (2011). Loss of Aspergillus oryzae amyR function indirectly affects hemicellulolytic and cellulolytic enzyme production. J. Biosci. Bioeng. 111, 408–413. doi: 10.1016/j.jbiosc.2010.12.006
Wieser, J., Lee, B. N., Fondon, J. W. 3rd., and Adams, T. H. (1994). Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr. Genet. 27, 62–69. doi: 10.1007/BF00326580
Xu, N., Liu, Y., Hu, Y., Zhou, M., Wang, C., and Li, D. (2016). Autolysis of Aspergillus oryzae mycelium and effect on volatile flavor compounds of soy sauce. J. Food Sci. 81, C1883–C1890. doi: 10.1111/1750-3841.13396
Yabuki, M., Ono, N., Hoshino, K., and Fukui, S. (1977). Rapid induction of alpha-amylase by nongrowing mycelia of Aspergillus oryzae. Appl. Environ. Microbiol. 34, 1–6. doi: 10.1128/AEM.34.1.1-6.1977
Keywords: Aspergillus oryzae, hydrolase, carbon catabolite repression, transcription factor, ubiquitination, endocytosis
Citation: Tanaka M and Gomi K (2021) Induction and Repression of Hydrolase Genes in Aspergillus oryzae. Front. Microbiol. 12:677603. doi: 10.3389/fmicb.2021.677603
Edited by:
Kap-Hoon Han, Woosuk University, South KoreaReviewed by:
István Pócsi, University of Debrecen, HungaryShuji Tani, Osaka Prefecture University, Japan
Copyright © 2021 Tanaka and Gomi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mizuki Tanaka, bWl6dWtpLnRhbmFrYUB1LXNoaXp1b2thLWtlbi5hYy5qcA==; Katsuya Gomi, a2F0c3V5YS5nb21pLmE2QHRvaG9rdS5hYy5qcA==
 Mizuki Tanaka
Mizuki Tanaka Katsuya Gomi
Katsuya Gomi