- 1Department of Natural Sciences, CREST Center for the Integrated Study of Coastal Ecosystem Processes and Dynamics in the Mid-Atlantic Region, NOAA Living Marine Resources Cooperative Science Center, University of Maryland Eastern Shore, Princess Anne, MD, United States
- 2Department of Agriculture, Food and Resource Sciences, Food and Agricultural Sciences Program, University of Maryland Eastern Shore, Princess Anne, MD, United States
Vibrio vulnificus and V. parahaemolyticus, found naturally in marine and estuarine environments, are the leading cause of seafood associated gastrointestinal illness and death. Consumption of improperly cooked crabs and handling of live crabs are potential routes of exposure to pathogenic bacteria such as V. vulnificus and V. parahaemolyticus. Little information is available on serotype genetic and antimicrobial profiles of V. vulnificus and V. parahaemolyticus recovered from Maryland estuaries. The aim of the present study was to determine the serotype of V. parahaemolyticus, evaluate antimicrobial susceptibility and genetic profiles of V. vulnificus and V. parahaemolyticus isolated from water and blue crab (Callinectes sapidus) samples collected from the Maryland Coastal Bays. One hundred and fifty (150) PCR confirmed V. parahaemolyticus including 52 tdh+ (pathogenic) and 129 V. vulnificus strains were tested for susceptibility to twenty (20) different antibiotics chosen by clinical usage for Vibrio species. The O serogroups were determined using an agglutination test with V. parahaemolyticus antisera. Pulsed-field gel electrophoresis (PFGE) was used for molecular subtyping to investigate the genetic diversity among tested strains. The most prevalent serotypes were O5 (33.3%), O3 (18.7%) and O1 (14.7%). More than 41% of all tested Vibrio isolates were resistant to three or more antibiotics. Cephalothin showed the highest resistance (42% and 61%), followed by cefoxitin (42% and 31%) and ceftazidime (36% and 29%) for V. vulnificus and V. parahaemolyticus, respectively. Most strains (99–100%) were susceptible to ampicillin/sulbactam, levofloxacin, piperacillin, piperacillin/tazobactam, and tetracycline. Fifty percent (50%) of the cephalothin resistant strains were crab isolates. Vibrio vulnificus and V. parahaemolyticus isolates demonstrated a high genetic diversity and 31% of V. vulnificus and 16% of V. parahaemolyticus strains were PFGE untypeable. No correlations were found between the V. parahaemolyticus serotype, pathogenicity, genetic and antimicrobial resistance profiles of both species of Vibrio. The observed high multiple drug resistance of V. vulnificus and V. parahaemolyticus from blue crab and its environment is of public health concern. Therefore, there is a need for frequent antibiotic sensitivity surveillance for Vibrio spp.
Introduction
Vibrio vulnificus and V. parahaemolyticus are Gram negative non-spore forming rod-shaped halophilic bacteria naturally found in warm marine and estuarine environments (Oliver, 2013; Parveen and Tamplin, 2013). These bacteria are the leading cause of seafood-borne illness and mortality in the United States (Scallan et al., 2011). Persons with underlying medical conditions, such as alcoholism, liver disease, cancer, and diabetes may be at increased risk of infection and serious complications (Liu et al., 2006). V. vulnificus can cause septicemia and wound infection in immunocompromised individuals who eat contaminated seafood or have an open wound that is exposed to seawater (Oliver, 2013; Xu et al., 2014; Centers for Disease Control and Prevention (CDC), 2019).
Vibrio species produce many virulence factors including enterotoxins, hemolysins, cytotoxins, proteases, siderophores, adhesive factors, and hemagglutinins. Vibrio parahaemolyticus produces Thermostable Direct Hemolysin (TDH) and Thermostable Related Hemolysin (TRH) which are determinants of virulent strains (Broberg et al., 2011; Zhang and Orth, 2013; Letchumanan et al., 2014). There are two genetic markers for V. vulnificus: a species-specific marker- V. vulnificus hemolysin gene A (vvhA) and virulence correlated gene (vcgC) (Rosche et al., 2005).
Nationwide, approximately 80,000 cases of vibriosis including an average of 287 culture-confirmed cases of V. parahaemolyticus and 111 cases of V. vulnificus, 500 hospitalizations and 100 deaths of which 35 are V. vulnificus related deaths are reported every year (Scallan et al., 2011; Newton et al., 2012). The incidence of Vibrio infections has been increasing in the United States and there has been an increase in infections caused by a specific strain of V. parahaemolyticus due to climate change and rising water temperature. Before 2012, V. parahaemolyticus infections of this strain were rarely associated with shellfish from the United States Atlantic coast. Maryland is among the seven coastal states with increased incidence of vibriosis. In August 2012, a V. parahaemolyticus outbreak involving six persons occurred in Maryland and the outbreak isolates were linked to the O3:K6 pandemic clone of V. parahaemolyticus that have been observed throughout the world (Wang et al., 2017). In 2018, CDC reported a multistate outbreak of V. parahaemolyticus infections linked to eating fresh crab meat imported from Venezuela. Among 26 infected people, 15 (58%) were from Maryland [Centers for Disease Control and Prevention (CDC), 2018; Haendiges et al., 2014]. V. vulnificus is the leading cause of reported human death in the United States caused by the consumption of seafood. The increase of V. vulnificus infection is particularly concerning given the high mortality rate (35%) associated with the pathogen [Newton et al., 2012; Centers for Science in the Public Interest (CSPI), 2020]. For example, in 2016 a man died from V. vulnificus infection four days after cleaning crab pots in Ocean City, Maryland (Washington Post, 2016).
The recommended antibiotics for the treatment of V. vulnificus infections are doxycycline, third-generation cephalosporin (e.g., ceftazidime), fluoroquinolone (such as levofloxacin, ciprofloxacin or gatifloxacin), and for children, trimethoprim-sulfamethoxazole plus an aminoglycoside. The use of quinolones or tetracycline for treatments of V. vulnificus infections is associated with lower mortality than cephalosporin alone. Tetracycline or ciprofloxacin can also be used in severe or prolonged illnesses of V. parahaemolyticus (Wong et al., 2015; Centers for Disease Control and Prevention (CDC), 2019).
During the past few decades, many bacteria have acquired antimicrobial resistance due to the excessive use of antimicrobials in human, agriculture, and aquaculture systems (Park et al., 2018). Antibiotics are one of the “contaminants of emerging concern” that are increasingly occurring in livestock and poultry manure across the United States. According to the United States Environmental Protection Agency (EPA), manure accounts for 19% of nitrogen and 26% of phosphorous entering the Chesapeake Bay (Krikstan, 2013). Manure can contain antibiotics that could facilitate the development of antimicrobial resistance in bacteria. Marine bacteria exposed to antibiotics can develop antimicrobial resistance (Labella et al., 2013) transferable by mobile genetic elements and horizontal gene transfer and can cause changes in the coastal environment (Christaki et al., 2020). Vibrio vulnificus and V. parahaemolyticus are included in the natural microbial flora of the Chesapeake (DaSilva et al., 2012; Elmahdi et al., 2018; Parveen et al., 2020) and Maryland Coastal Bays, and have been isolated from crabs (Callinectes sapidus), oysters (Crassostrea virginica), water and sediment samples (Rodgers et al., 2014). Improperly cooked crabs and handling of live crabs represent a potential route of exposure to pathogenic and antimicrobial resistant strains of V. vulnificus and V. parahaemolyticus. The prevalence of V. vulnificus and V. parahaemolyticus in Maryland Coastal Bays is high, and the highest concentrations were found in crab compared to water and sediment (Rodgers et al., 2014). Despite the fact that the prevalence and ecology of V. vulnificus and V. parahaemolyticus is well documented in Maryland, especially in the Chesapeake Bay, little information is available on the serotype, and genetic and antimicrobial profiles of V. vulnificus and V. parahaemolyticus recovered from crab samples from the Maryland Coastal Bays. The aim of the present study was to determine the phenotypic and genotypic characteristics of V. vulnificus and V. parahaemolyticus strains isolated from crab, seawater and sediment samples collected from the Maryland Coastal Bays.
Materials and Methods
Bacterial Strains and Sample Collection
The bacterial strains were isolated previously (Rodgers et al., 2014) from crab hemolymph, sediment and water samples collected from the Maryland Coastal Bays including Chincoteague Bay (site 8), Newport Bay (site 9), Sinepuxent Bay (site 10), and St. Martin River (site 13) (Figure 1). V. parahaemolyticus and V. vulnificus strains were stored at −80°C in the cryogenic vials for further analysis.
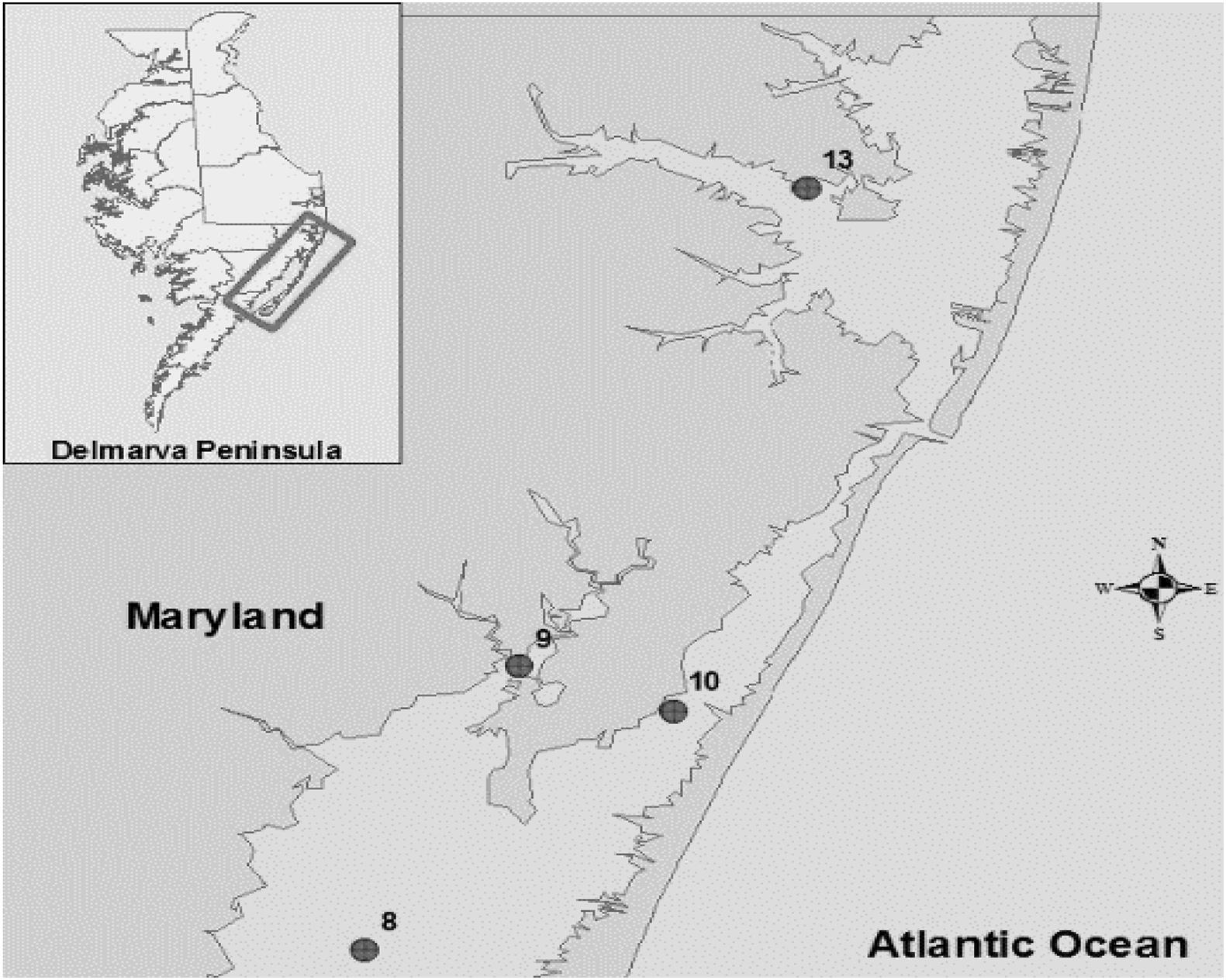
Figure 1. Location of sampling sites in the Maryland Coastal Bays. 8- Chincoteague Bay, 9- Newport Bay, 10- Sinepuxent Bay, 13–St. Martin River. Courtesy of Tracie J. Bishop and Andres G. Morales-Nunez (Rodgers et al., 2014).
Serotyping
The identification of somatic (O) serotype of V. parahaemolyticus strains was performed with 11 commercially available antisera using slide agglutination test according to the manufacturer’s recommendations (Denka Seiken Co., Ltd., Tokyo, Japan).
Antimicrobial Susceptibility Testing
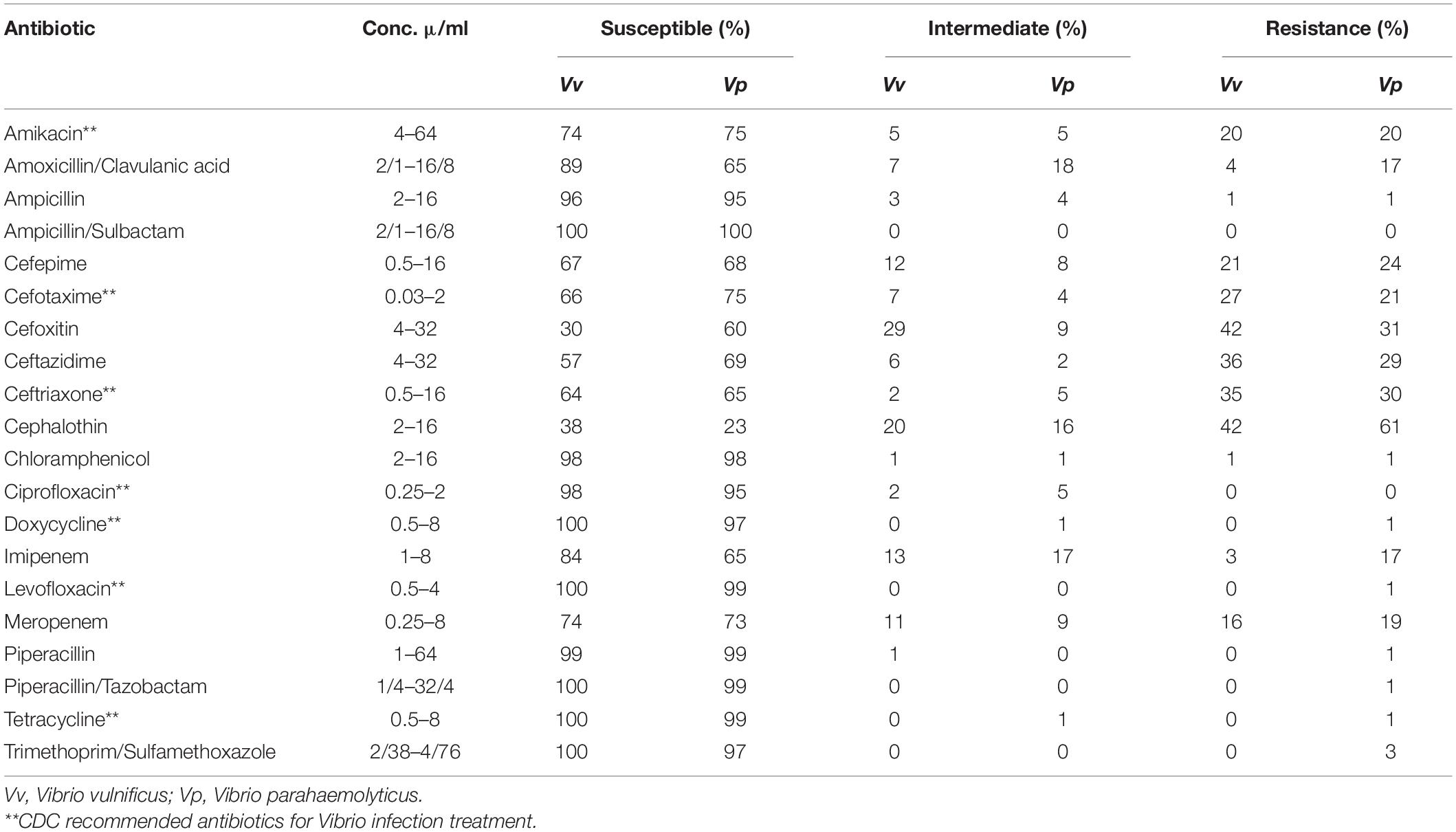
Vibrio isolates were tested for susceptibility to antibacterial drugs using the microbroth dilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). For this testing, twenty different antibiotics were chosen based on clinical usage for V. vulnificus and V. parahaemolyticus. There were amikacin (AMI; 4–64 μ/ml), amoxicillin/clavulanic acid (AMC; 2/1–16/8 μ/ml), ampicillin (AMP; 2–16 μ/ml), ampicillin/sulbactam (SAM; 2/1–16/8 μ/ml), cefepime (FEP; 0.5–16 μ/ml), cefotaxime (FOT; 0.03–2 μ/ml), cefoxitin (FOX; 4–32 μ/ml), ceftazidime (TAZ; 4–32), ceftriaxone (AXO; 0.5–16 μ/ml), cephalothin (CEP; 2–16 μ/ml), ciprofloxacin (CIP; 0.25–2 μ/ml), chloramphenicol (CHL; 2–16 μ/ml), doxycycline (DOX; 0.5–8 μ/ml), imipenem (IMI; 1–8 μ/ml), levofloxacin (LEVO; 0.5–4 μ/ml), meropenem (MERO; 0.25–8 μ/ml), piperacillin (PIP; 1–64 μ/ml), piperacillin/tazobactam (P/T4; 1/4–32/4 μ/ml), tetracycline (TET; 0.5–8 μ/ml), and trimethoprim/sulfamethoxazole (SXT; 2/38–4/76 μ/ml). The Minimal Inhibitory Concentration (MIC) was recorded as the lowest concentration of antimicrobial agent with no visible growth. Multidrug resistance was defined as an absence of susceptibility to two or more classes of antibiotics. Escherichia coli ATCCH25922 and Staphylococcus aureus ATCCH29213 were used as controls [Clinical and Laboratory Standards Institute (CLSI), 2010; Elmahdi et al., 2018].
Pulsed Field Gel Electrophoresis
Pulsed-field gel electrophoresis (PFGE) was used to investigate the genetic variation of the Vibrio isolates with different antibiotic susceptibility profiles. Plug preparation, digestion and separation of DNA fragment and PFGE were performed using the SfiI restriction enzyme as described in the CDC Pulse-Net protocol for V. parahaemolyticus and V. vulnificus (PulseNet, United States 2013; Parsons et al., 2007; Kam et al., 2008; Elmahdi et al., 2018). To improve the typeability of some strains 50 μM of thiourea was added to electrophoresis buffer [0.5X TBE (Tris-Borate EDTA)] (Banerjee and Farber, 2009). The gel was stained with ethidium bromide, and DNA bands were visualized with a UV transilluminator. Cluster analysis of PFGE was performed using bionumerics software (version 7.6, Applied Maths, Austin, TX, United States). Dice coefficient was calculated according to unweighted-pair group method with arithmetic mean. Isolates were assigned to the same macrorestriction profile when they clustered within > 99% pattern similarity (1.2% optimization and 1.5% position tolerance).
Statistical Analysis
One-Way Analysis of Variance (ANOVA) and Pearson’s Chi square test were used to analyze the data and compare the means among sites, sample type, antibiotic resistance patterns of V. vulnificus and V. parahaemolyticus isolates, and to evaluate the differences in the occurrence of antimicrobial susceptibility and correlation between the sample sites, types pathogenicity, Genetic and antimicrobial resistance profiles using SPSS version 26.0 (IBM Corp., NY, United States).
Results
Serotype
Serotype was performed in all 150 V. parahaemolyticus strains. Except for O7 and O9, all serogroups were equally distributed in all four sites in Maryland Coastal Bays (Table 1). The most prevalent serotypes were O5 (33.3%), O3 (18.7%), and O1 (14.7%). Serotypes O2 and O8 were rarely found (Table 1). Twenty-seven (52%) of O5 strains were multidrug resistant and 19 (37%) of these strains were pathogenic (18 tdh+ and 1 tdh+ and trh+). However, there was no correlation between serotype and pathogenicity (r = 0.36, p > 0.05).
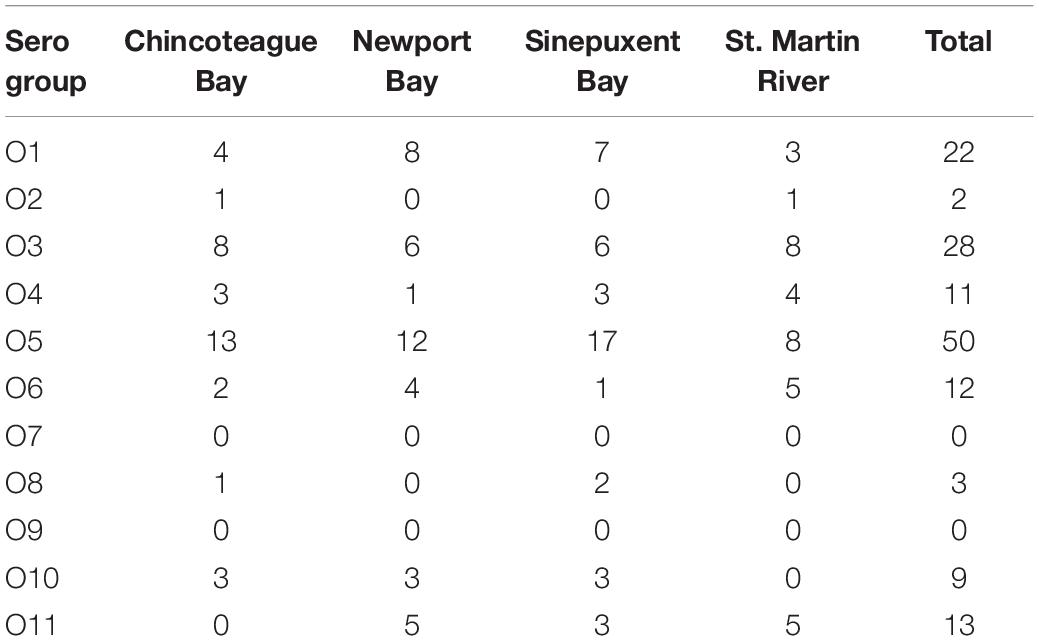
Table 1. Distribution of Vibrio parahaemolyticus O serogroups in four sites of Maryland Coastal Bays.
Antimicrobial Profile
A total of 279 (129 V. vulnificus and 150 V. parahaemolyticus) strains were tested for antibiotic sensitivity. Sixty-five percent (65%) of all tested V. vulnificus and V. parahaemolyticus strains were resistant to one or more classes of antibiotics and 42% were multidrug resistant. The highest resistance was observed for cephalothin, 42% for V. vulnificus and 61% for V. parahaemolyticus, followed by cefoxitin (42 and 31%), ceftazidime (36 and 29%), ceftriaxone (35 and 30%), cefotaxime (27 and 20%), cefepime (21 and 20%), amikacin (20%) and meropenem (16 and 19%) for V. vulnificus and V. parahaemolyticus, respectively. Ninety-five to 100% of both Vibrio species were susceptible to most of the commonly used antibiotics such as ampicillin, ampicillin/sulbactam, doxycycline, levofloxacin, piperacillin, piperacillin/tazobactam, tetracycline and trimethoprim/sulfamethoxazole (Table 2). Isolates recovered from water and sediment had significantly (p ≤ 0.05) higher cephalothin resistance than those from crab meat and hemolymph (Figures 2, 3).
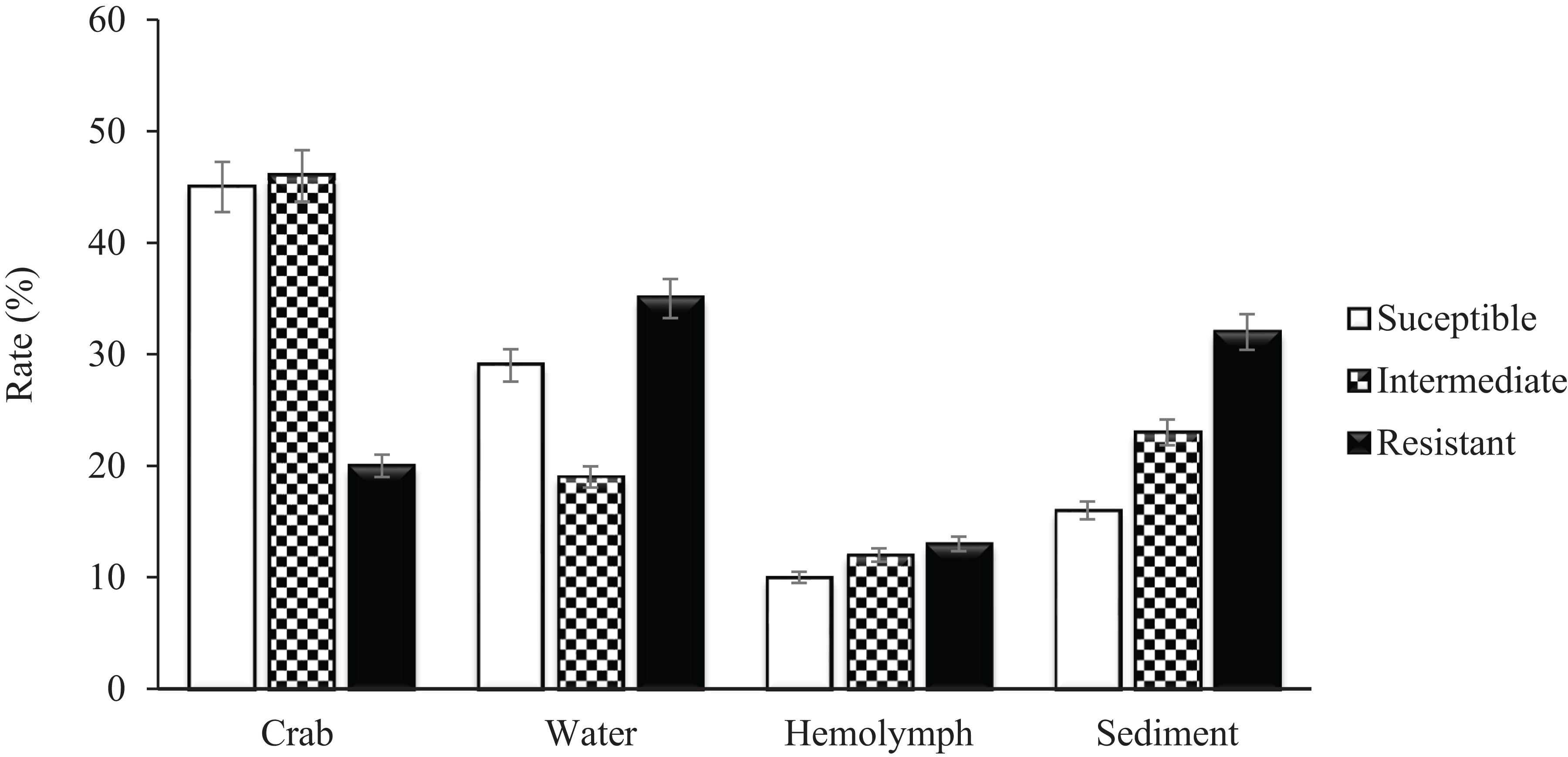
Figure 2. Percentage of Vibrio vulnificus isolates susceptible, intermediate and resistant to Cephalothin.
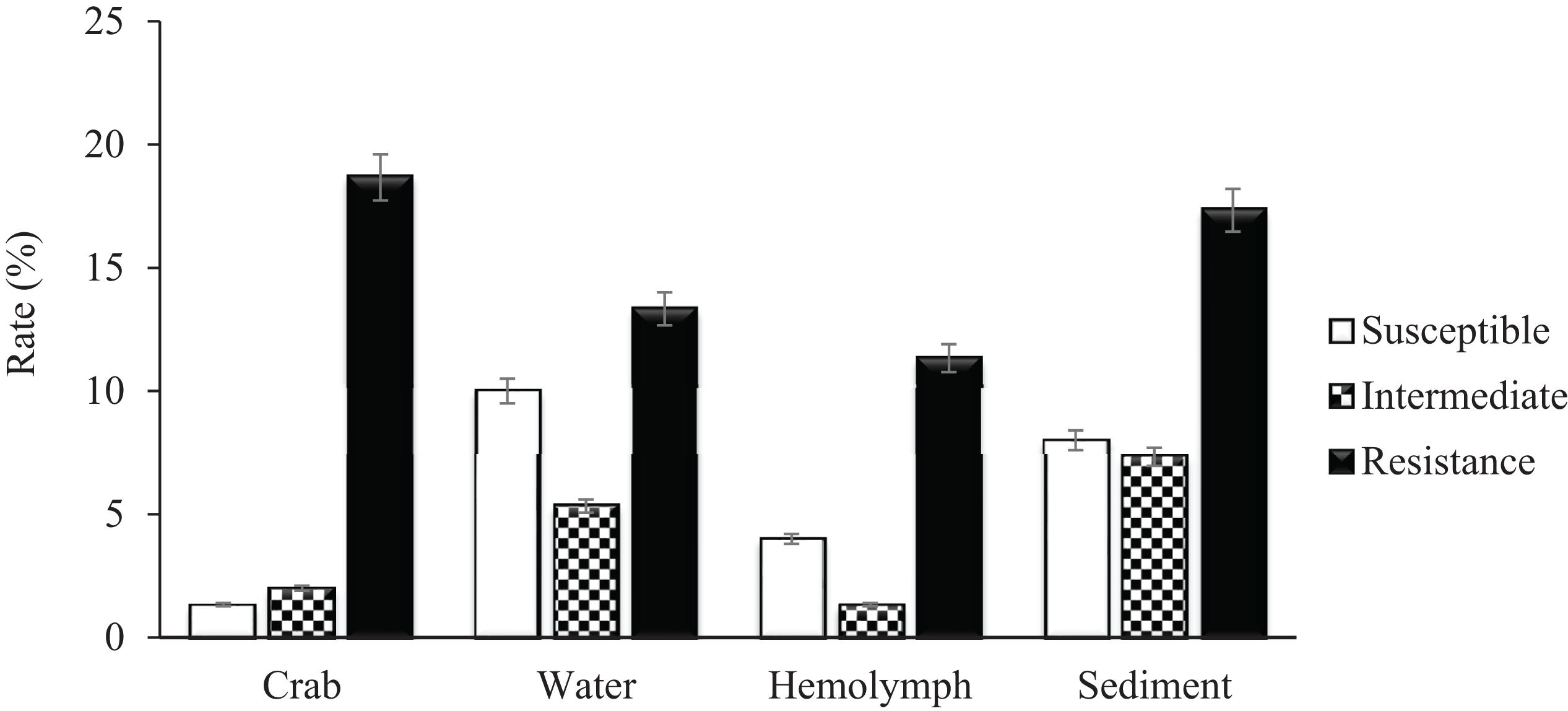
Figure 3. Percentage of Vibrio parahaemolyticus susceptible, intermediate and resistant to cephalothin.
Pulsed Field Gel Electrophoresis
Based on the antimicrobial resistance profiles and typeability, 49 (including 9 vcgC+) V. vulnificus and 92 (including 28 tdh+) V. parahaemolyticus strains from different sources were selected and a dendrogram was constructed based on SfiI PFGE patterns to compare the banding profiles of these bacteria. Thirty-one percent (31%) of V. vulnificus and 16% of V. parahaemolyticus strains were PFGE untypeable and failed to yield discernible patterns. Both vibrio species demonstrated a high genetic diversity and they were grouped into 8 (A to H) V. vulnificus and 19 (A to R) V. parahaemolyticus clusters of related patterns of ≥ 75% similarity. No relationship was observed between the genetic and antimicrobial resistance profiles of both Vibrio species. Five (5) V. vulnificus (number 91, 117, 114, 115, and 22) and 10 V. parahaemolyticus (number 97, 62, 43, 31, 55, 52, 210, 212, 106, and 65) strains showed similar antimicrobial resistance profile, although they were genetically different (Figures 4, 5).

Figure 4. Dendrogram of PFGE profiles of Vibrio vulnificus strains collected form crab and its environment in Maryland Coastal Bays; Site 8- Chincoteague Bay, 9- Newport Bay, 10- Sinepuxent Bay, 13–St. Martin River. Key = stain number. AMI, amikacin; AUG2, amoxicillin/clavulanic acid; AMP, ampicillin; A/S2 ampicillin-sulbactam; FEP, cefepime; FOT, cefotaxime; FOX, cefoxitin; TAZ, ceftazidime; AXO, ceftriaxone; CEP, cephalothin; CIP, ciprofloxacin; CHL, chloramphenicol; DOX, doxycycline; IMI, imipenem; LEVO, levofloxacin; MERO, meropenem; PIP, piperacillin; P/T4, piperacillin/tazobactam; TET, tetracycline; SXT, trimethoprim/sulfamethoxazole. vcgC, virulence-correlating gene type C. Letters A to H on the left represent pulsed-field gel electrophoresis clusters; key represent strain numbers. Minus sign on the resistance and intermediate profiles columns indicate “no resistance” or “no intermediate” profiles and on the Pathogenicity column (-) mean vcgC gene negative.
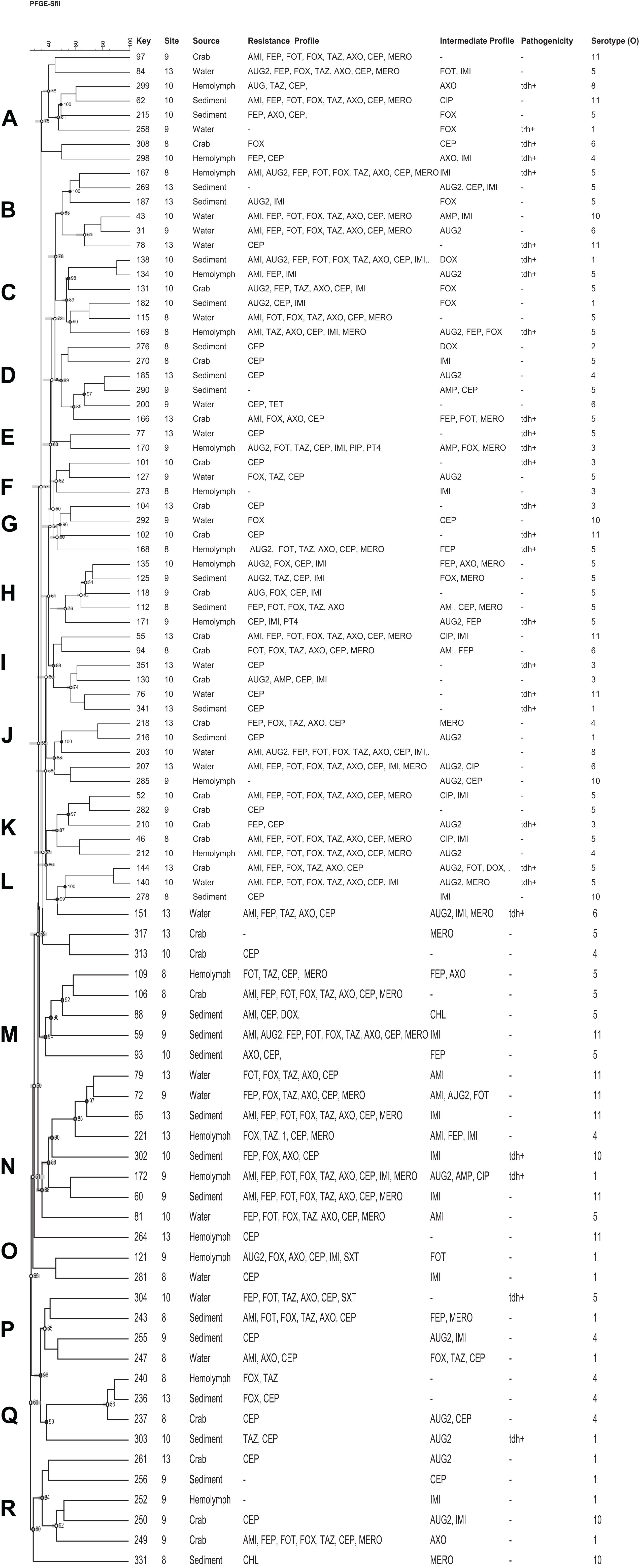
Figure 5. Dendrogram of PFGE profiles of Vibrio parahaemolyticus strains collected form crab and its environment in Maryland Coastal Bays; Site 8–Chincoteague Bay; 9–Newport Bay; 10–Sinepuxent Bay; 13–St. Martin River. Key = stain number. AMI, amikacin; AUG2, amoxicillin/clavulanic acid; AMP, ampicillin; A/S2, ampicillin-sulbactam; FEP, cefepime; FOT, cefotaxime; FOX, cefoxitin; TAZ, ceftazidime; AXO, ceftriaxone; CEP, cephalothin; CIP, ciprofloxacin; CHL, chloramphenicol; DOX, doxycycline; IMI, imipenem; LEVO, levofloxacin; MERO, meropenem; PIP, piperacillin; P/T4, piperacillin/tazobactam; TET, tetracycline; SXT, trimethoprim/sulfamethoxazole; tdh, thermostable direct hemolysin. Letters A to R on the left represent pulsed-field gel electrophoresis clusters; key represent strain numbers; minus sign (-) on the resistance and intermediate profiles columns indicates “no resistance” or “no intermediate” profiles and on the Pathogenicity column (-) indicates tdh and trh negative; Serotype (O) = +O1–O11.
Discussion
For epidemiological perspective, V. parahaemolyticus strains were serotyped. The most prevalent serotypes were O5, O3 group that have been observed around the world, and O1; followed by O6 and O4. Serotypes O2 and O8 were very scarce; whereas O7 and O9 were not found in the Maryland Coastal Bays (Table 1). Similar results were found in the Chesapeake Bay oysters and their environment, where the most isolated groups were O3, O1, O5, and O7, and O9 were not found (de Hernández-Díaz et al., 2015; Chen et al., 2017; Elmahdi et al., 2018). In the present study we did not determine the K type of our strains. In 2009, Chao and his collaborators isolated other serotypes of the pandemic strain (O1:K36, O3:K25, and O3:K68), in China and in 2009 also reported another serotype (O3:K5) in the American continent (Chao et al., 2009; Velazquez-Roman et al., 2014). A recent study conducted by Siddique et al. (2021) on V. parahaemolyticus isolated from fish aquaculture in Bangladesh, the majority of strains contain O8 antigen followed by O5, O11, O3, and O1. Most of the strains could not be typed serologically for K (KUT) antigen using conventional kits. Similar to our results they did not report O7 and O9 serotypes. Besides of O3 group, O4:K12 and O4: K (unknown) are pandemic V. parahaemolyticus strains of the Pacific Northwest associated with outbreaks in New York, Atlantic Coast of Spain in 2012 and in 13 United States Atlantic Coast states in 2013 (Centers for Disease Control and Prevention (CDC), 2013; Newton et al., 2014). It is highly recommended that future studies perform K-typing to identify and enumerate the pandemic serotype in the Maryland Coastal Bays.
Vibrio vulnificus and V. parahaemolyticus strains exhibited similar antimicrobial profiles and contain high percentage of multidrug resistant strains that were distributed along the four sites (Chincoteague Bay, Newport Bay, Sinepuxent Bay and St. Martin River) (p > 0.0%) in the Maryland Coastal Bays. Both Vibrio species showed the highest resistance to cephalothins, one of the recommended antibiotics for vibriosis treatment, followed by cefoxitin, ceftazidime, ceftriaxone, cefotaxime, cefepime, amikacin and meropenem. V. parahaemolyticus strains showed higher resistance to cephalothin compared to V. vulnificus. Resistant to commonly used antibiotic is also elevated in V. parahaemolyticus than V. vulnificus (Table 2). All tested Vibrio strains (100%) were susceptible to ampicillin/sulbactam, and all V. vulnificus strains were susceptible to levofloxacin, piperacillin, piperacillin/tazobactam, tetracycline and trimethoprim/sulfamethoxazole (Table 2). Half of the cephalothin resistant strains were crab isolates. Water and sediment samples had significantly (p ≤ 0.05) higher cephalothin resistant strains compared to crab meat and hemolymph samples (Figures 2, 3); there was no significant difference among the sites. No correlations were found between the serotype, pathogenicity and antimicrobial resistance profiles. Similar antimicrobial profiles for both, V. vulnificus and V. parahaemolyticus were found by Elmahdi et al. (2018) and Shaw et al. (2014). However, Shaw’s team found higher susceptibility of V. vulnificus (95%) and V. parahaemolyticus (82%) to cephalothin versus 38 and 23% for V. vulnificus and V. parahaemolyticus, respectively observed in the present study.
It has been reported that molecular typing is a reliable and useful tool for investigating the genetic diversity and tracking sources of contamination of food and waterborne pathogens in aquatic systems and food processing plants (Mohamed et al., 2014; Elmahdi et al., 2018). In this study, dendrogram of PFGE cluster analysis of 141 V. vulnificus and V. parahaemolyticus strains from crabs and its environments (water, sediment, crab meat and hemolymph) were performed according to the standard PulseNet PFGE protocol for Vibrio spp. with the restriction enzyme SfiI. Though a few V. vulnificus and V. parahaemolyticus strains recovered from seafood in the United States, specifically in Maryland have been genetically characterized by PFGE analysis, this is the first study that reported the PGFE profiles of V. vulnificus and V. parahaemolyticus strains recovered from crabs and its surrounding environments in the Maryland Coastal Bays.
In this study, eight clusters (Figure 4; marked with letter A to H on the left) for V. vulnificus were found with different 80% similarity in banding patterns. Multidrug resistant as well as potential pathogenic strains of both species of Vibrio were distributed in different clusters. Only two strains out of 49 V. vulnificus strains fell into the same cluster, with 100% similarity and exhibited the same multidrug resistant pattern. Moreover, these isolates were from the same site (site 9) and source (crab); however, they differed in intermediate antibiotic profile (strain #24 showed intermediate profile for AUG2 and IMI, whereas strain # 26 showed intermediate profile for CEP). V. vulnificus showed higher percentage (≥80%) of similarity compare to V. parahaemolyticus (≥75%); however, these differences were not statistically significant (p > 0.05). The PFGE profiles of V. parahaemolyticus (Figure 5) were genetically diverse, and no genetic relationship was found among and between the sampling sites, antimicrobial profile, pathogenicity, and serogroups. All recovered V. parahaemolyticus isolates clustered (A to R) at 75% or higher similarity in banding patterns. These results indicate high intraspecific diversity of this species. Our findings are consistent with a previous study that reported high genetic heterogeneity among V. vulnificus and V. parahaemolyticus isolates recovered from oysters and water during a relaying study in the Chesapeake Bay, Maryland (Elmahdi et al., 2018). Moreover Chen et al. (2017) observed a high genetic diversity among V. vulnificus and V. parahaemolyticus isolates recovered from oysters and water collected from the Chesapeake Bay, Maryland. The percentage of untypeable stains of V. vulnificus (31%) was higher than V. parahaemolyticus (16%) despite the fact that 50 μM of thiourea was added to 0.5X TBE (Tris-Borate EDTA) buffer for the repeated PFGE experiments. Only 4 V. parahaemolyticus and 1 of V. vulnificus strains were typeable after the addition of thiourea in the electrophoresis buffer. Fawley and Wilcox (2002) found that of 200 μM of thiourea must be present in both agarose gel and the electrophoresis buffer to ensure minimal DNA degradation. High level of untypeable V. vulnificus strain was also reported in studies of PFGE analysis of V. vulnificus strains isolated from Taiwan and the United States (Wong et al., 2004). Therefore, it is important to analyze these untypeable isolates using a more sensitive and specific method, whole genome sequencing to reveal the reason for the untypeability.
Conclusion
The PFGE profiles of V. vulnificus and V. parahaemolyticus stains isolated from Maryland Coastal Bays were diverse. No genetic relationship was found among the sampling sites, antimicrobial resistance profile, and pathogenicity. The observed high multiple drug resistance of V. vulnificus and V. parahaemolyticus from blue crab and its environment is of public health concern, despite the fact that these bacteria were susceptible to the two CDC recommended antibiotics (Tetracycline 99–100% and Ciprofloxacin 95–98%) for its treatment. Therefore, frequent antibiotic sensitivity surveillance is needed.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without reservation.
Author Contributions
SP, PC and LS contributed to the conception and design of the study. LS and SO were involved in experimental procedures in the laboratory and data collection and analysis. The manuscript was written by LS, and reviewed, edited, and approved by all authors. SP and PC were responsible for the integrity of the work and overall supervision.
Funding
The present study was supported by National Science Foundation (NSF) Center of Excellence in Science and Technology (CREST) award no. 1036586 to UMES, the USDA Capacity Building Grant Program, grant # 2010-02370 and National Oceanic and Atmospheric Administration, Office of Education Educational Partnership Program award (NA11SEC4810002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be taken as a potential conflict of interest.
Acknowledgments
We sincerely thank Candace Rodgers, Sara Elmahdi, Andres Morales-Nunez, John Jacobs, Captain Christopher Daniels, Joan Meredith, and Matt Rhodes for their assistance during the study and sample collection.
References
Banerjee, S. K., and Farber, J. M. (2009). Susceptibility of Vibrio parahaemolyticus to tris-dependent DNA degradation during pulsed-field gel electrophoresis. J. Clin. Microbiol. 47, 870–871. doi: 10.1128/JCM.02306-08
Broberg, C. A., Calder, T. J., and Orth, K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13, 992–1001. doi: 10.1016/j.micinf.2011.06.013
Centers for Science in the Public Interest (CSPI) (2020). Outbreaks and Recalls. Vibrio: Raw Oysters From Hood Canal (WA). Food Safety Outbreak Alert. Washington, DC: Centers for Science in the Public Interest (CSPI).
Centers for Disease Control and Prevention (CDC) (2013). Available online at: https://www.cdc.gov/vibrio/investigations/vibriop-09-13/index.html (accessed January 5, 2021).
Centers for Disease Control and Prevention (CDC) (2018). Available online at: https://www.cdc.gov/vibrio/investigations/vibriop-07-18/index.html (accessed January 5, 2021).
Centers for Disease Control and Prevention (CDC) (2019). Available online at:https://www.cdc.gov/vibrio/diagnosis.html (accessed January 5, 2021).
Chao, G., Jiao, X., Zhou, X., Yang, Z., Huang, J., Pan, Z., et al. (2009). Serodiversity, pandemic O3:K6 clone, molecular typing, and antibiotic susceptibility of foodborne and clinical Vibrio parahaemolyticus isolates in Jiangsu, China. Foodborne Pathog. Dis. 6, 1021–1028. doi: 10.1089/fpd.2009.0295
Chen, A. J., Hasan, N. A., Haley, B. J., Taviani, E., Tarnowski, M., Browan, K., et al. (2017). Characterization of pathogenic Vibrio parahaemolyticus from the chesapeake bay, Maryland. Front. Microbiol. 8:2460. doi: 10.3389/fmicb.2017.02460
Christaki, E., Marcou, M., and Tofarides, A. (2020). Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J. Mol. Evol. 88, 26–40. doi: 10.1007/s00239-019-09914-3
Clinical and Laboratory Standards Institute (CLSI) (2010). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; Approved Guideline. CLSI Document M45-A2, 2nd Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
DaSilva, L., Parveen, S., DePaola, A., Bowers, J., Brohawn, K., and Tamplin, M. L. (2012). Development and validation of a predictive model for the growth of Vibrio vulnificus in postharvest shellstock oysters. Appl. Environ. Microbiol. 78, 1675–1681.
de Hernández-Díaz, J, Leon-Sicairos, L. N., Roman, V. J., Villaseñor, F. H., Guadron-Llanos, A. M., Martinez-Garcia, J. J., et al. (2015). A pandemic Vibrio parahaemolyticus O3:K6 clone causing most associated diarrhea cases in the Pacific Northwest coast of Mexico. Front. Microbiol. 6:221. doi: 10.3389/fmicb.2015.00221
Elmahdi, S., Parveen, S., Ossai, S., DaSilva, L. V., Jahncke, M., Bowers, J., et al. (2018). Vibrio parahaemolyticus and Vibrio vulnificus recovered from oysters during an oyster relay study. Appl. Environ. Microbiol. 84:e01790-17. doi: 10.1128/AEM.01790-17
Fawley, W. N., and Wilcox, M. H. (2002). Pulsed-field gel electrophoresis can yield DNA fingerprints of degradation-susceptible clostridium difficile strains. J. Clin. Microbiol. 40, 3546–3547. doi: 10.1128/jcm.40.9.3546-3547.2002
Haendiges, J., Rock, M., Myers, R. A., Brown, E. W., Evans, P., and Gonzalez-Escalona, N. (2014). Pandemic Vibrio parahaemolyticus, Maryland, USA, 2012. Emerg. Infect. Dis. 20, 718–720. doi: 10.3201/eid2004.130818
Kam, K. M., Luey, C. Y., Parsons, M. B., Cooper, K. L. F., Nair, G. B., Alam, M., et al. (2008). Evaluation and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping Vibrio parahaemolyticus: an international multicenter collaborative study. J. Clin. Microbiol. 46, 2766–2773. doi: 10.1128/JCM.00424-08
Krikstan, C. (2013). Antibiotics, Hormones Increasingly Found in Livestock and Poultry Manure Chesapeake Bay Program. Available online at: https://www.chesapeakebay.net/news/blog/antibiotics_hormones_increasingly_found_in_livestock_and_poultry_manure (accessed December 20, 2021).
Labella, A., Gennari, M., Ghidini, V., Trento, I., Manfrin, A., Borrego, J. J., et al. (2013). High incidence of antibiotic multi-resistant bacteria in coastal areas dedicated to fish farming. Mar. Pollut. Bull. 70, 197–203.
Letchumanan, V., Chan, K., and Lee, L. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Liu, J. W., Lee, I. K., Tang, H. J., Ko, W. C., Lee, H. C., Liu, Y. C., et al. (2006). Prognostic factors and antibiotics in Vibrio vulnificus septicemia. Arch. Intern. Med. 166, 2117–2123. doi: 10.1001/archinte.166.19.2117
Mohamed, T., Zhao, S., White, D., and Parveen, S. (2014). Molecular characterization of antibiotic resistant Salmonella Typhimurium and Salmonella Kentucky isolated from pre- and post-chill whole broilers carcasses. Food Microbiol. 38, 6–15.
Newton, A. E., Garrett, N., Stroika, S. G., Halpin, J. L., Turnsek, M., and Mody, R. K. (2014). Centers for Disease Control and Prevention (CDC). Increase in Vibrio parahaemolyticus infections associated with consumption of Atlantic Coast shellfish–2013. MMWR Morb. Mortal. Wkly. Rep. 63, 335–336.
Newton, A., Kendall, M., Vugia, D. J., Henao, O. L., and Mahon, B. E. (2012). Increasing rates of vibriosis in the United States, 1996-2010: review of surveillance data from 2 systems. Clin. Infect. Dis. 54(Suppl. 5), S391–S395. doi: 10.1093/cid/cis243
Oliver, J. D. (2013). Vibrio vulnificus: death on the half shell. Microb. Ecol. 65, 793–799. doi: 10.1007/s00248-012-0140-9
Park, K., Mok, J. S., Kwon, J. Y., Ryu, A. R., Kim, S. H., and Lee, H. J. (2018). Food-borne outbreaks, distributions, virulence, and antibiotic resistance profiles of Vibrio parahaemolyticus in Korea from 2003 to 2016: a review. Fish. Aquatic Sci. 21:3. doi: 10.1186/s41240-018-0081-4
Parsons, M. B., Cooper, K. L., Kubota, K. A., Puhr, N., Simington, S., Calimlim, P. S., et al. (2007). PulseNet USA standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio parahaemolyticus. Foodborne Pathog. Dis. 4, 285–292. doi: 10.1089/fpd.2007.0089
Parveen, S., Jacobs, J., Ozbay, G., Chintapenta, K., Almuhaideb, E., Meredith, J., et al. (2020). Seasonal and geographical differences in total and pathogenic Vibrio parahaemolyticus and Vibrio vulnificus levels in seawater and oysters from the Delaware and Chesapeake Bays using several methods. Appl. Environ. Microbiol. 86:e01581-20. doi: 10.1128/AEM.01581
Parveen, S., and Tamplin, M. L. (2013). “Vibrio vulnificus, Vibrio parahaemolyticus and Vibrio cholerae,” in Guide to Foodborne Pathogens, eds R. G. Labbe and S. Garcia (Hoboken NJ: Wiley Online Library), 148–176. doi: 10.1002/9781118684856.ch9
Rosche, T. M., Yano, Y., and Oliver, J. D. (2005). A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol. Immunol. 49, 381–389. doi: 10.1111/j.1348-0421.2005.tb03731.x
Rodgers, C., Parveen, S., Chigbu, P., Jacobs, J., Rhodes, M., and Harter-Dennis, J. (2014). Prevalence of Vibrio parahaemolyticus, and Vibrio vulnificus in blue crabs (Callinectes sapidus), seawater and sediments of the Maryland Coastal Bays. J. Appl. Microbiol. 117, 1198–1209. doi: 10.1111/jam.12608
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.09-1101p1
Siddique, A. B., Moniruzzaman, M., Ali, S., Dewan, Md. N, Islam, M. R., and Islam, Md. S, et al. (2021). Characterization of pathogenic Vibrio parahaemolyticus isolated from fish aquaculture of southwest coastal area of Bangladesh. Front. Microbiol. 12:635539. doi: 10.3389/fmicb.2021.635539
Shaw, K. S., Goldstein, R. E. R., He, X., Jacobs, J. M., Crump, B. C., and Sapkota, A. R. (2014). Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Chesapeake Bay and Maryland Coastal Bays. PLoS One 9:e89616. doi: 10.1371/journal.pone.0089616
Velazquez-Roman, J., León-Sicairos, N., de Jesus Hernández-Díaz, L., and Canizalez-Roman, A. (2014). Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front. Cell. Infect. Microbiol. 3:110. doi: 10.3389/fcimb.2013.00110
Wang, H., Tang, X., Su, Y. C., Chen, J., and Yan, J. (2017). Characterization of clinical Vibrio parahaemolyticus strains in Zhoushan. PLoS One 12:e0180335. doi: 10.1371/journal.pone.0180335
Washington Post (2016). V. vulnificus. Available online at: https://www.washingtonpost.com/news/to-your-health/wp/2016/10/23/like-something-out-of-a-horror-movie-flesh-eating-bacteria-infection-kills-man-in-four-days/ (accessed January 5, 2021).
Wong, K. C., Brown, A. M., Luscombe, G. M., Wong, S. J., and Mendis, K. (2015). Antibiotic use for Vibrio infections: important insights from surveillance data. BMC Infect. Dis. 15:226. doi: 10.1186/s12879-015-0959-z
Wong, H. C., Chen, S. Y., Chen, M. Y., Oliver, J. D., Hor, L. I., and Tsai, W. C. (2004). Pulsed-field gel electrophoresis analysis of Vibrio vulnificus strains isolated from Taiwan and the United States. Appl. Environ. Microbiol. 70, 5153–5158. doi: 10.1128/AEM.70.9.5153-5158.2004
Xu, X., Wu, Q., Zhang, J., Cheng, J., Zhang, S., and Wu, K. (2014). Prevalence, pathogenicity, and serotypes of Vibrio parahaemolyticus in shrimp from Chinese retail markets. Food Control 46, 81–85. doi: 10.1016/j.foodcont.2014.04.042
Keywords: Vibrio vulnificus, Vibrio parahaemolyticus, serotype, antimicrobial profile, genetic profile
Citation: da Silva LV, Ossai S, Chigbu P and Parveen S (2021) Antimicrobial and Genetic Profiles of Vibrio vulnificus and Vibrio parahaemolyticus Isolated From the Maryland Coastal Bays, United States. Front. Microbiol. 12:676249. doi: 10.3389/fmicb.2021.676249
Received: 04 March 2021; Accepted: 26 April 2021;
Published: 21 May 2021.
Edited by:
Ilenys M. Perez-Diaz, Food Science Research (USDA-ARS), United StatesReviewed by:
Pendru Raghunath, Dr. VRK Women’s Medical College, IndiaJessica L. Jones, United States Food and Drug Administration, United States
Copyright © 2021 da Silva, Ossai, Chigbu and Parveen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Salina Parveen, c3BhcnZlZW5AdW1lcy5lZHU=
 Ligia V. da Silva
Ligia V. da Silva Sylvia Ossai2
Sylvia Ossai2 Paulinus Chigbu
Paulinus Chigbu Salina Parveen
Salina Parveen