- Department of Nutrition, Dietetics and Food Sciences, Utah State University, Logan, UT, United States
Prior research has suggested that the use of organic acids in the food industry may unintentionally enhance pathogenicity of Listeria monocytogenes strain N1-227 and R2-499. This study explored the connection between habituation to L-lactic acid or acetic acid and virulence in L. monocytogenes strains N1-227 and R2-499 using selected gene expression analysis and the in vivo Galleria mellonella wax worm model for infection. Expression of transcription factors (sigB and prfA) and genes related to acid resistance (gadD2, gadD3, and arcA) and bile resistance (bsh and bilE) or to virulence (inlA, inlB, hly, plcA, plcB, uhpT, and actA) was investigated by quantitative real-time PCR (qRT-PCR), while in vivo virulence was assessed by following the lethal time to 50% population mortality (LT50) of G. mellonella larvae after injection of untreated and habituated L. monocytogenes. Twenty minutes of habituation to the organic acids at pH 6.0 significantly increased expression of key acid and bile stress response genes in both strains, while expression of virulence genes was strain-dependent. The expression of transcription factor sigB was strain-dependent and there was no significant change in the expression of transcription factor prfA in both strains. Habituation to acid increased virulence of both strains as evidenced by decreased LT50 of G. mellonella larvae injected with Listeria habituated to either acid. In summary, habituation of both L. monocytogenes strains to organic acids up-regulated expression of several stress and virulence genes and concurrently increased virulence as measured using the G. mellonella model.
Introduction
The genus Listeria is comprised of Gram-positive, non-spore-forming, rod-shaped, facultative anaerobic bacteria which can be found ubiquitously in the environment (Mélanie et al., 2006; Gahan and Hill, 2014; Lani and Hassan, 2016). Among Listeria species, only L. monocytogenes and L. ivanovii are pathogenic (Robinson and Batt, 1999); L. ivanovii primarily infects animals while L. monocytogenes shows pathogenicity toward both humans and animals (Liu, 2006). During food production, L. monocytogenes can experience several stresses such as low pH and high salt. The ability of Listeria to adapt to these adverse conditions plays a crucial role in food contamination and food-borne infection (Lani and Hassan, 2016).
In response to stress, L. monocytogenes may induce an acid tolerance response and other stress responses mechanisms that allow it to overcome these hurdles (Glass et al., 1995; Silva et al., 2012; Melo et al., 2015). L. monocytogenes is able to utilize a variety of regulators (over 100 different transcriptional regulators have been identified) to survive and grow in different environments (Glaser et al., 2001; Gaballa et al., 2019). Among those regulators, the alternative sigma factor B (σB) and the listeriolysin positive regulatory factor A (PrfA) are two essential transcriptional regulators for stress response and for host infection.
σB, encoded by sigB, is a general stress responsive transcription sigma factor in L. monocytogenes and many other Gram-positive bacteria (Kazmierczak et al., 2005; Chaturongakul et al., 2008). In L. monocytogenes, σB regulates numerous genes that are associated with acid, bile and other physiological stressors (Sue et al., 2004; Zhang et al., 2011; Smith et al., 2012; Melo et al., 2015). The acid stress response systems in L. monocytogenes include the glutamate decarboxylase (GAD) system and an arginine deiminase (ADI) system. The GAD system, which involves genes encoding three glutamate decarboxylase enzymes (gadD1, gadD2 and gadD3) and two gamma aminobutyric acid (GABA) antiporters (gadT1 and gadT2), plays a significant role in pH homeostasis in L. monocytogenes (Cotter et al., 2001; Melo et al., 2015). Expression of the GAD system results in the decarboxylation of glutamate into γ-aminobutyrate with consumption of intracellular protons (Cotter et al., 2001; Karatzas et al., 2012). Additionally, the arginine deiminase (ADI) system also contributes to the stabilization of the bacterial cytoplasmic pH (Melo et al., 2015). The ADI pathway involves the enzymes arginine deiminase, ornithine carbamoyltransferase and carbamate kinase, which are encoded by arcA, arcB, and arcD, respectively (Melo et al., 2015). With respect to bile resistance, one of the most important mechanisms in L. monocytogenes involves the ability to detoxify individual conjugated bile acid through bile salt hydrolase (BSH) (Dussurget et al., 2002; Begley et al., 2005). Another novel bile resistance system in L. monocytogenes is the bile exclusion system (BilE), which acts to exclude bile from bacterial cells (Sleator et al., 2005).
The listeriolysin positive regulatory factor A (PrfA), encoded by prfA, is a bacterial transcription factor that controls and coordinates the expression of key virulence genes in L. monocytogenes associated with cell invasion and the intracellular infection cycle (Kazmierczak et al., 2006; Scortti et al., 2007; de las Heras et al., 2011). Cell invasion is mediated by two surface proteins, internalin A and B (InlA and InlB); after entering the cell, L. monocytogenes are entrapped in a phagocytic vacuole from which they escape by lysing the membrane of the vacuole through the combined actions of the pore-forming toxin listeriolysin O (LLO, encoded by hly) and two phospholipases, PlcA and PlcB (Mélanie et al., 2006). Multiplication and invasion within host cells can then occur with the involvement of the permease UhpT (a hexose phosphate transporter) and the surface protein ActA (propel bacteria through the cytoplasm) (Chico-Calero et al., 2002; Mélanie et al., 2006; Cossart and Toledo-Arana, 2008).
Acid stress resistance has been well studied and observed in various microorganisms such as Escherichia coli (Goodson and Rowbury, 1989) and Salmonella (Foster and Hall, 1990). Prior research by our group has suggested that the use of organic acids in the food industry may unintentionally enhance virulence of some L. monocytogenes strains (Zhang et al., 2014). Those results showed that habituation of two L. monocytogenes strains, N1-227 and R2-499, to organic acid under mildly acidic conditions (pH = 6.0) induced acid and bile resistance, which indicated these treatments could promote virulence by enhancing survival during passage through the gastrointestinal tract (Zhang et al., 2014). It also suggested the increased acid and bile resistance was specifically due to organic acid exposure rather than a decrease in environmental pH (Carpenter and Broadbent, 2009; Zhang et al., 2014). Similar responses were not observed in that study with other pathogenic strains of L. monocytogenes (Zhang et al., 2014), so R2-499 and N1-227 were selected for further study to explore the genetic basis for inducible acid and bile resistance, and to determine if it affected virulence in an in vivo model.
Virulence of Listeria spp. is frequently assessed using a murine model (Lecuit, 2007). However, this model has limitations for studying human pathogenicity of L. monocytogenes because the interaction between InlA and mouse E-cadherin (identified as InlA receptor in human) is poor, which makes L. monocytogenes entry into epithelial cells less efficient (Mengaud et al., 1996; Lecuit et al., 1999). The larvae of Galleria mellonella have also been used as a model for L. monocytogenes virulence (Joyce and Gahan, 2010; Mukherjee et al., 2010, 2013; Banville et al., 2012; Ramarao et al., 2012; Schrama et al., 2013). Compared to the mammalian model and other alternative models, the G. mellonella model offers several significant advantages, including structural and functional similarities with the mammalian immune system (Hoffmann et al., 1999; Strand, 2008). Additionally, the infection process can be performed over a range of temperatures (from 15°C to above 37°C), which enables use of the G. mellonella model to study the virulence of L. monocytogenes human pathogens at 37°C (Jones et al., 2010; Rejasse et al., 2012).
To better understand the molecular basis and potential consequences of induced acid and bile resistance in organic acid habituated strains, we used quantitative real time polymerase chain reaction (qRT-PCR) to measure the expression of key transcription factors and some of their target genes related to acid or bile resistance or virulence in L. monocytogenes strains N1-227 and R2-499 after habituation to lactic acid or acetic acid at pH 6.0. Additionally, the G. mellonella infection model was used to analyze the in vivo virulence of control and acid habituated L. monocytogenes strains.
Materials and Methods
Bacterial Strains and Growth Conditions
Original cultures (Table 1) were stored as frozen stocks at −80°C in tryptic soy broth (TSB, pH 7.4; Becton, Dickinson and Company, Sparks, MD) supplemented with 20% v/v glycerol. Prior to use, cultures were first propagated on tryptic soy agar (TSA; Becton, Dickinson and Company, Sparks, MD) plate and incubated at 37°C for 24 h. A single colony from the TSA plate was transferred into TSB and incubated overnight at 37°C with shaking (220 rpm).
RNA Isolation
Overnight cultures of each strain were harvested by centrifugation (2,500 × g for 10 min; Sorvall RT1, Thermo Scientific, Germany) at 4°C, and then diluted to an optical density at 600 nm (OD600) of 0.03 in TSB. Cells were acid habituated as described by Zhang et al. (2014). A 1% inoculum (v/v) of diluted overnight cultures was transferred into 50 mL of standard TSB (pH 7.4) and incubated at 37°C for 4 h with shaking (220 rpm) to reach mid-log phase as determined by Zhang et al. (2014). The cultures were collected by centrifugation (2,500 × g for 10 min) at 4 °C and then suspended in 50 mL of either standard TSB (pH 7.4, baseline control) or TSB without dextrose (pH 6.0 adjusted with HCl, Becton, Dickinson and Company, Sparks, MD) containing 0 (pH control) or 4.75 mM of either L-lactic acid (Sigma Chemicals, St. Louis, MO) or acetic acid (Johnson Matthey Company, Ward Hill, MA). The cultures were incubated at 37°C for 20 min with shaking (220 rpm). After incubation, 100 mL of RNAprotect Bacteria Reagent (Qiagen, Inc., Valencia, CA) was added to each sample. Cells were incubated at room temperature for 10 min then collected by centrifugation (9,500 × g for 10 min). The supernatant was discarded and cell pellets were suspended in 900 μL of lysozyme solution (Sigma-Aldrich, 20 mg/mL in Tris-EDTA buffer) that contained 20 units of mutanolysin (Sigma-Aldrich). Samples were incubated for 30 min at 37°C on a shaker incubator at 220 rpm, then 20 μL of proteinase K (Omega Bio-Tek Inc., Norcross, GA) (20 mg/mL) was added and the samples were returned to the shaker/incubator for 30 min. Total RNA was isolated using an Aurum total RNA mini kit (Bio-Rad, Hercules, CA) following the vendor’s recommended procedures. Residual DNA was removed using The Ambion® DNA-freeTM DNase Treatment and Removal Reagents. RNA samples were then purified using the GeneJET RNA Cleanup and concentration Micro Kit PCR purification kit (Thermo Fisher Scientific, Lithuania). The amount and quality of the RNA were measured using a NanoDrop Spectrophotometer (Thermo Fisher Scientific, United States) and TapeStation System (Agilent, Santa Clara, CA), respectively.
cDNA Synthesis and Real Time Quantitative PCR (qPCR)
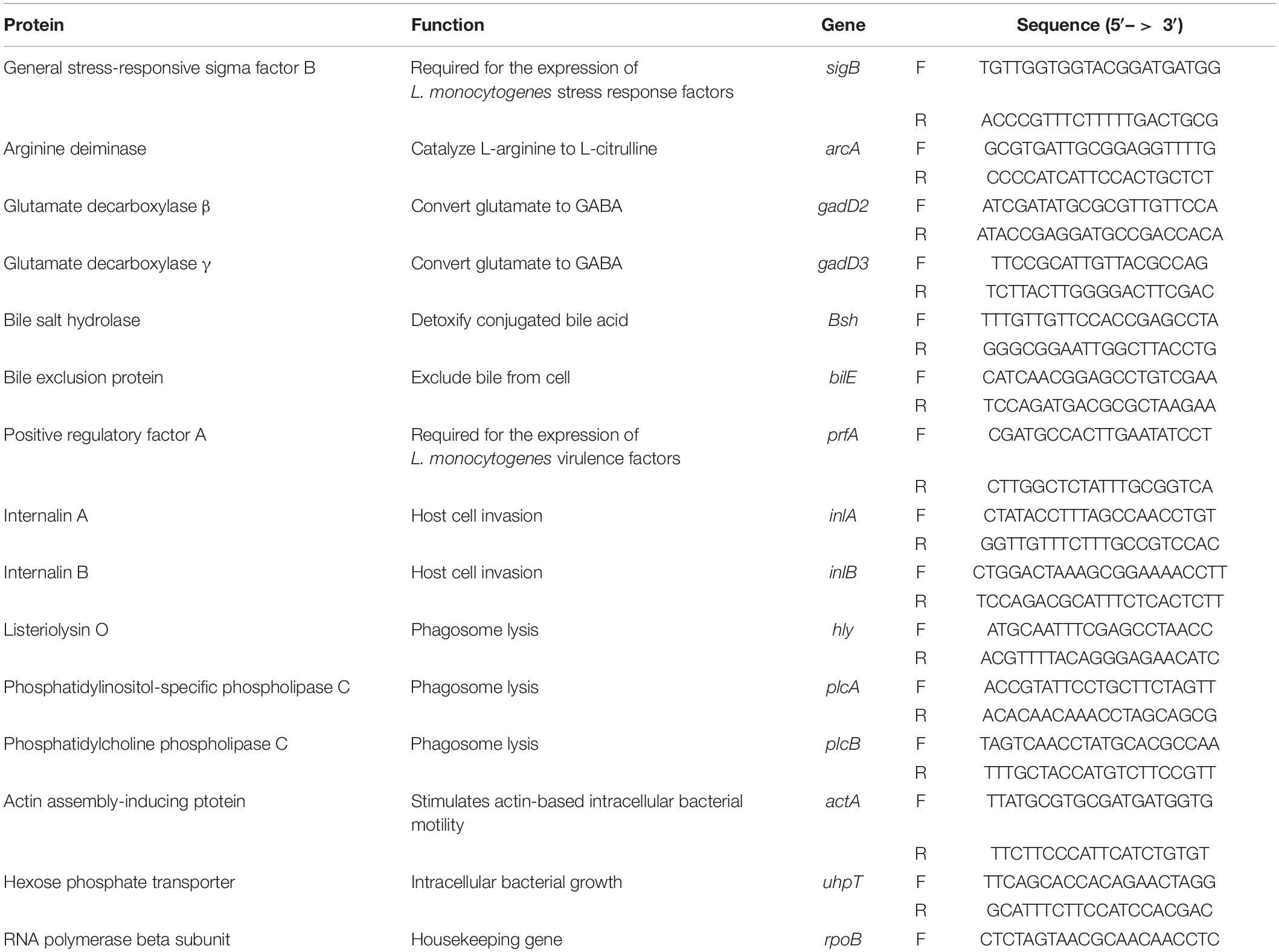
cDNA was synthesized from total RNA using random primers (Invitrogen, Carlsbad, CA) and SuperScript II reverse transcriptase (Invitrogen). The qPCR was carried out using cDNA as template in an Opticon II thermal cycler (MJ Research, Reno, NV) using HotStart-ITTM SYBR Green qPCR Master Mix with UDG kit (Affymetrix, Inc.). Each reaction was performed in triplicate and the relative gene expression of targeted genes was calculated by the Pfaffl Method and normalized by the baseline control (Pfaffl, 2001). The primers used in this study are listed in Table 2 and rpoB was used as a housekeeping gene to normalize the gene expression data (Bookout and Mangelsdorf, 2003; Tasara and Stephan, 2007). The amplification efficiency for each primer was tested by plotting the cycle threshold (Ct) value with different template concentrations and fitting the data to a regression line (Bookout and Mangelsdorf, 2003; Ruijter et al., 2009). The amplification efficiency for all the primers reached 90% or above (Li, 2020).
Galleria mellonella Wax Worm Model
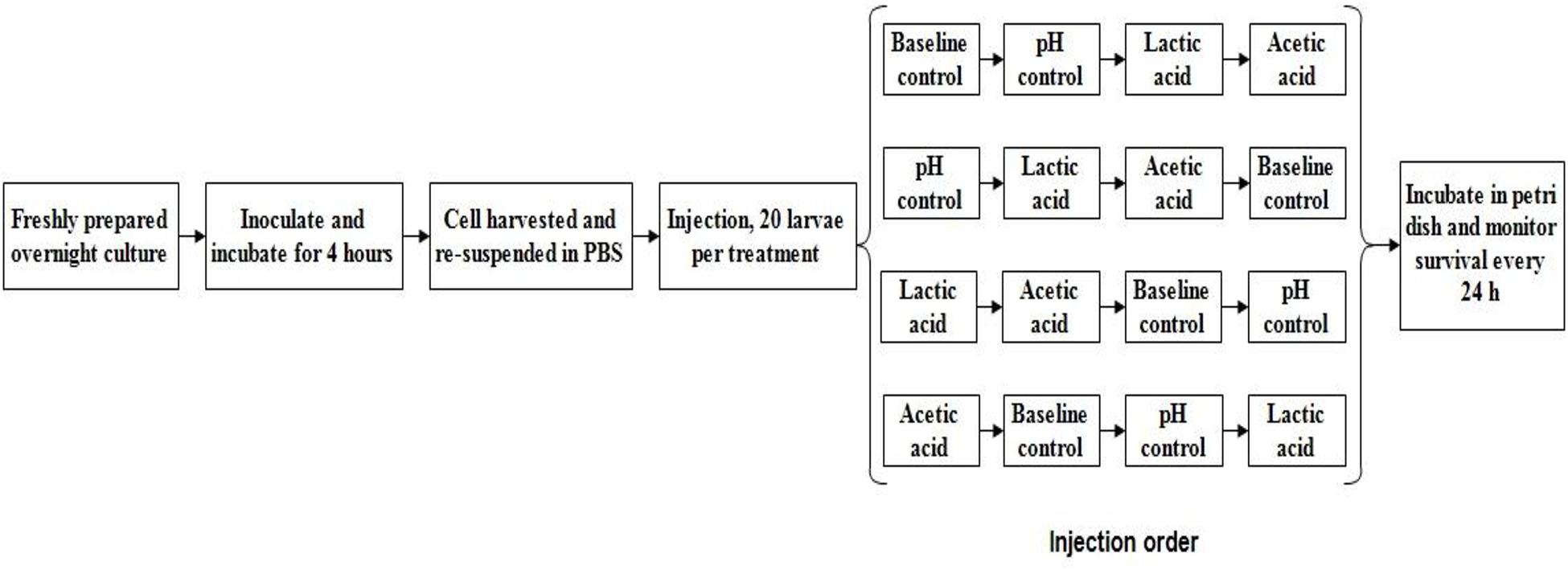
The in vivo virulence of L. monocytogenes strains was determined using the Galleria mellonella wax worm model described by Ramarao et al. (2012). A 1% inoculum (v/v) of freshly prepared L. monocytogenes cells was transferred into 50 mL of either standard TSB (pH 7.4, baseline control) or TSB without dextrose (pH 6.0 with HCl) containing 0 (pH control) or 4.75 mM of either L-lactic acid or acetic acid and incubated at 37°C for 4 h with shaking (220 rpm). The mid-log phase cultures were collected by centrifugation (2,500 × g for 10 min) at 4°C. The bacterial cells were then re-suspended with sterile PBS solution (pH 7.4) and diluted to an optical density at 600 nm (OD600) of 0.25. Ten microliters of 108 cfu/mL L. monocytogenes, either control or acid habituated, was injected into the haemocoel of the wax worms using an automated syringe pump (KDS 100, KD Scientific; 20 larvae per treatment; see Figure 1 for schematic experimental design and injection order. Injection was done in two biological repetition). A PBS-only control injection was also included. The larvae were placed in petri dish (5 per dish) and incubated at 37°C. Larvae survival was evaluated every 24 h for 5 days after injection. The larvae were considered dead when they showed no movement in response to finger touch. Lethal times until 50% population mortality (LT50) for each treatment were then determined by Probit analysis (Bliss, 1934, 1935).
Enumeration of Listeria monocytogenes in Galleria mellonella Wax Worms
L. monocytogenes in G. mellonella larvae was enumerated at 5, 10, 15, and 20 h after injection. At each time point, 5 larvae were collected and homogenized in 9 mL of sterile peptone physiological solution (PPS) in a stomacher. Serial dilutions were made by pipetting 1 mL of diluted sample into 9 mL PPS, then 100 μL of diluted samples was spread on Palcam agar (L. monocytogenes selective media; Oxoid Limited, Hampshire, United Kingdom). Plates were incubated at 37°C for 48 h then L. monocytogenes colonies were enumerated. Microbiological count data were expressed as log10 of colony-forming units per larvae.
Statistical Analysis
The data collected in this study (relative expression ratio of target genes compared with reference genes in three biological repetitions, the survival rate of G. mellonella larvae and the enumeration of L. monocytogenes in Galleria mellonella wax worm in two biological repetitions) were continuous outcome variables for every categorical treatment variable (acidification treatments of L. monocytogenes). Significant differences in each outcome between treatments were assessed using one-way analysis of variance (ANOVA) followed by Tukey’s test to compare means of the gene expression outcome variables between treatments. Differences were considered significant at P < 0.05.
Results
Influence of Acid Habituation on Expression of Acid and Bile Stress Response Genes
Increased expression of gadD3 was observed for strain N1-227 in the pH control relative to the baseline control (P < 0.05, Figure 2). Additionally, acetic acid or lactic acid habituation resulted in significant upregulation of gadD3 as compared to the pH control in both N1-227 and R2-499 (P < 0.05). No significant change of gadD2 expression was observed for both strains in the pH control compared to the baseline control. However, similar to gadD3, acetic acid or lactic acid habituation induced significant and dramatic expression of gadD2 in comparison with pH control in both strains (P < 0.05). The qPCR results for both strains also showed no significant changes in the expression of gene encoding arginine deiminase (arcA) in the pH control relative to the baseline control, and that acetic acid or lactic acid habituation significantly increased arcA expression in both strains (P < 0.05, Figure 2).
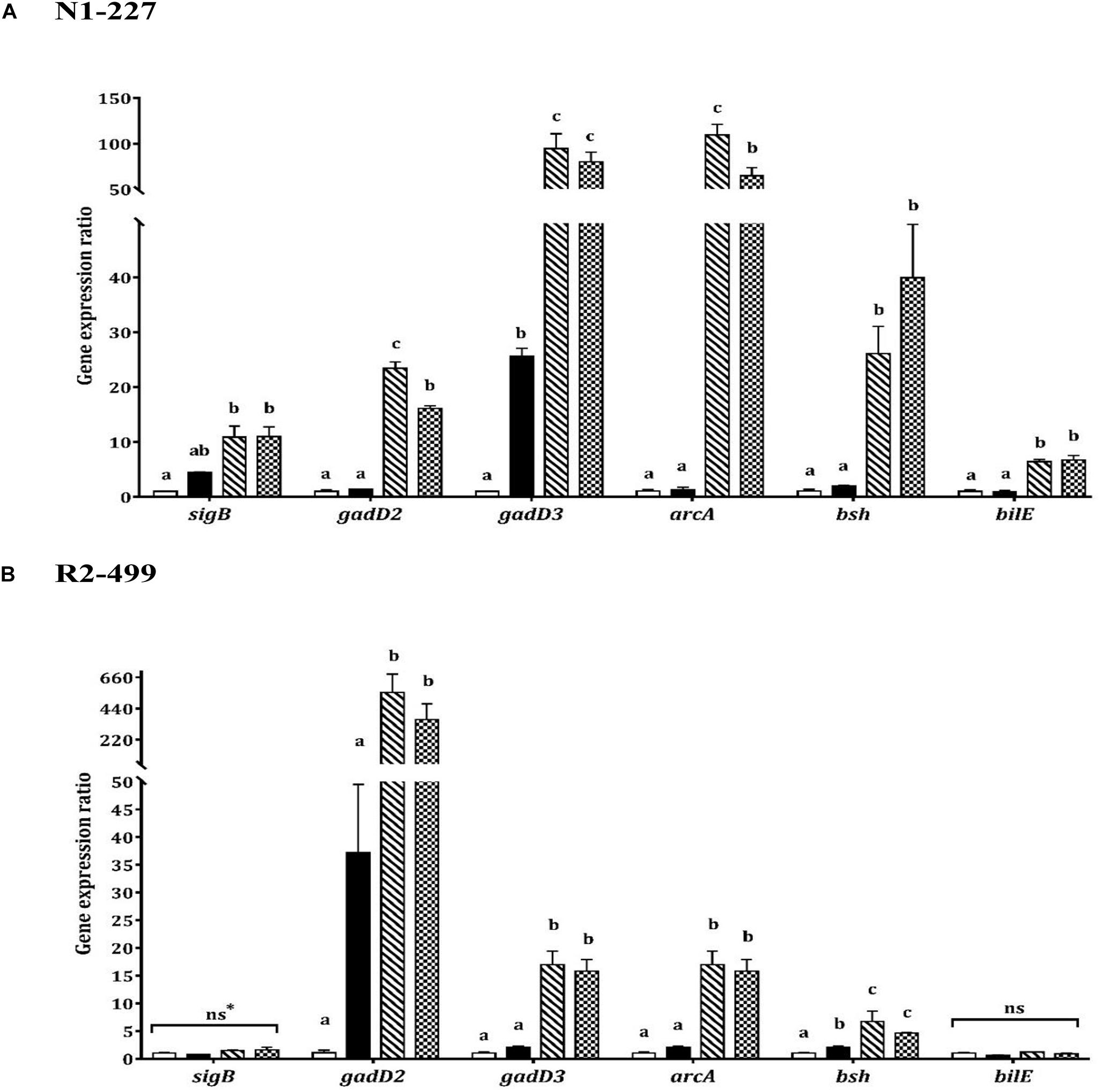
Figure 2. Relative gene expression of acid and bile stress response related genes in 20 min habituated Listeria monocytogenes strains (A) N1-227 and (B) R2-499 cells in comparison with non-habituated cells (baseline control, TSB pH 7.4,  ). Habituated treatments include as follows: TSB pH 6.0 (pH control,
). Habituated treatments include as follows: TSB pH 6.0 (pH control,  ), TSB at pH 6.0 with 4.75 mM of acetic acid (
), TSB at pH 6.0 with 4.75 mM of acetic acid ( ) and TSB at pH 6.0 with 4.75 mM of L-lactic acid (
) and TSB at pH 6.0 with 4.75 mM of L-lactic acid ( ). Error bars represent standard error of mean for two biological trials with three replicates for each trial. Different letters indicate that treatments are significantly different (p < 0.05) as determined by one-way ANOVA with Tukey’s post-hoc tests; *ns, Non-significant.
). Error bars represent standard error of mean for two biological trials with three replicates for each trial. Different letters indicate that treatments are significantly different (p < 0.05) as determined by one-way ANOVA with Tukey’s post-hoc tests; *ns, Non-significant.
In contrast, transcription of genes related to bile tolerance was variable between the strains. Habituation to lactic acid or acetic acid significantly increased bsh gene expression in comparison with the pH control for both strains (P < 0.05). However, the pH control had no significant effect on bsh expression relative to the baseline control in strain N1-227 (Figure 2). Changes in the expression of bilE were also strain-dependent. For strain N1-227, bilE was significantly overexpressed (P < 0.05) when cells were habituated to acetic or L-lactic acid, whereas no significant changes were observed in strain R2-499. Finally, qPCR data showed habituation to L-lactic acid or acetic acid significantly (P < 0.05) induced sigB expression in strain N1-227 cells compared to the baseline control (Figure 2A). However, no significant change on sigB expression was observed between treatments in strain R2-499 (Figure 2B).
Influence of Acid Habituation on Expression of Virulence Genes
As was observed with stress genes, qPCR results showed similarities and differences between strains with respect to virulence gene expression in response to organic acid habituation (Figure 3). The transcription level of prfA or uhpT was not significantly impacted by pH or acid exposure in either strain. However, expression of inlA, inlB and hly increased in both strains when the pH was decreased. Both strains showed significantly (P < 0.05) increased expression of inlA and inlB in organic acid habituated cells compared to the baseline control or pH control (Figure 3). Furthermore, hly expression was significantly (P < 0.05) increased in R2-499 cells habituated to acetic acid or L-lactic acid relative to baseline control and pH control. However, significant (P < 0.05) overexpression of hly in strain N1-227 compared to baseline control was only observed with the pH control and acetic acid habituation treatment.
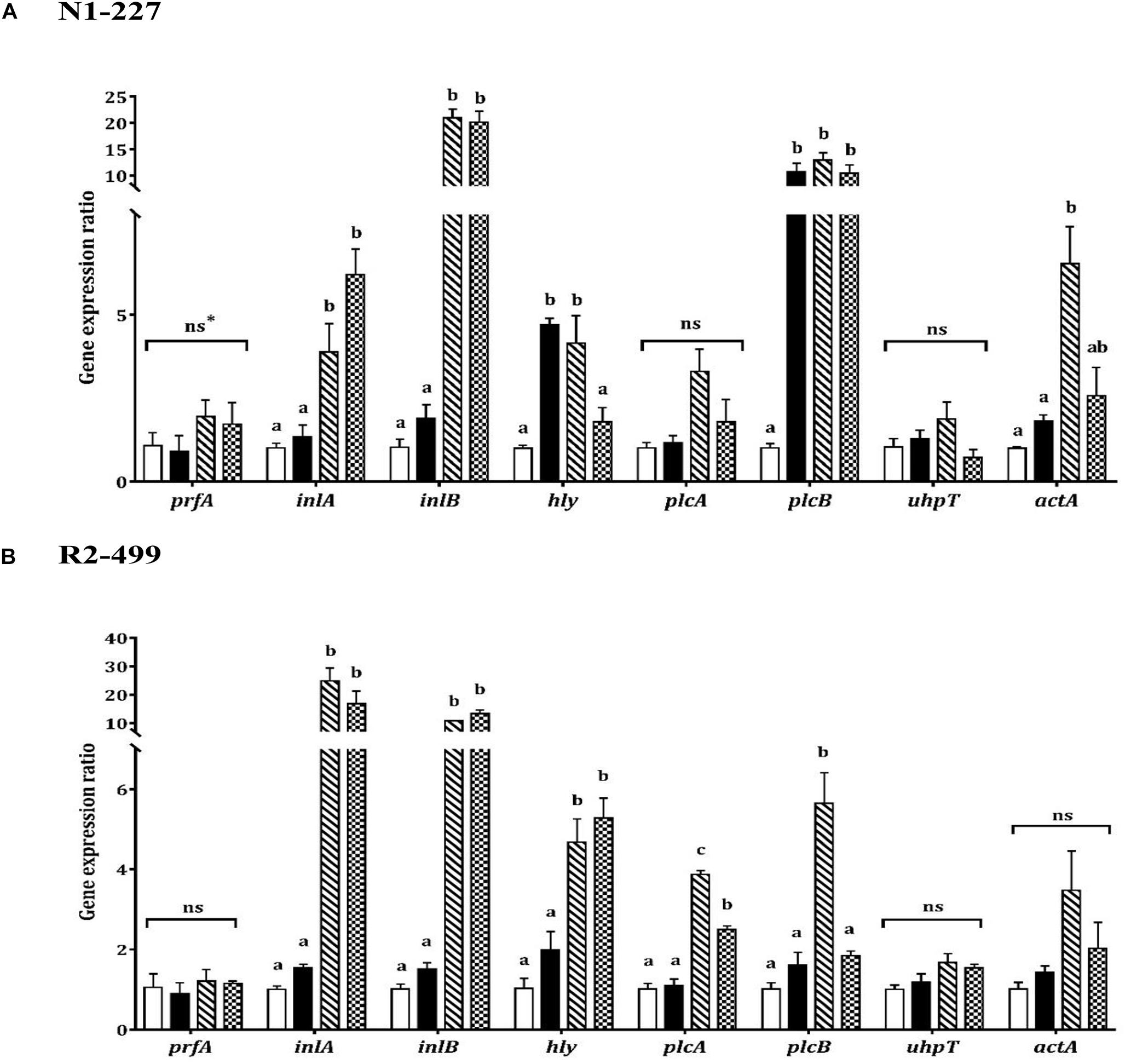
Figure 3. Relative gene expression of virulence related genes in 20 min habituated Listeria monocytogenes (A) N1-227 and (B) R2-499 cells in comparison with non-habituated cells (baseline control, TSB pH 7.4,  ). Habituated treatments include as follows: TSB at pH 6.0 (pH control,
). Habituated treatments include as follows: TSB at pH 6.0 (pH control,  ), TSB at pH 6.0 with 4.75 mM of acetic acid (
), TSB at pH 6.0 with 4.75 mM of acetic acid ( ) and TSB pH 6.0 with 4.75 mM of L-lactic acid (
) and TSB pH 6.0 with 4.75 mM of L-lactic acid ( ). Error bars represent standard error of mean for two biological trails with three replicates for each trail. Different letters indicate that treatments are significantly different (p < 0.05) as determined by one-way ANOVA with Tukey’s post-hoc tests. *ns, Non-significant.
). Error bars represent standard error of mean for two biological trails with three replicates for each trail. Different letters indicate that treatments are significantly different (p < 0.05) as determined by one-way ANOVA with Tukey’s post-hoc tests. *ns, Non-significant.
The qPCR results showed the expression profile for the other virulence genes (plcA, plcB, actA) was also strain-dependent (Figure 3). No significant changes were observed in plcA expression for strain N1-227 (Figure 3A), while organic acid habituation significantly (P < 0.05) increased expression of this gene in strain R2-499 compared to baseline control and pH control (Figure 3B). All three acid treatments (pH control and organic acid habituated cells) significantly (P < 0.05) induced plcB expression compared to the baseline control in strain N1-227, whereas significant induction in strain R2-499 was only observed with the acetic acid treatment. Conversely, no significant differences were recorded in actA expression for strain R2-499, and only acetic acid habituated N1-227 cells showed a significant (P < 0.05) increase in the expression level of this gene (Figure 3).
Effect of Habituation to Organic Acid on Galleria mellonella Survivability
The lethal time to 50% population mortality (LT50) of each treatment for both strains (Table 3) was determined based on the survival of G. mellonella over 5 days post-injection (see Supplementary Figure 1). For both N1-227 and R2-499, the LT50 of larvae injected with L. monocytogenes habituated with HCl (pH control) was lower than that of larvae injected with non-habituated L. monocytogenes (baseline control) and LT50 values decreased considerably more when larvae were injected with organic acid habituated L. monocytogenes (Table 3). The shortest LT50 results were noted with organic acid habituated L. monocytogenes R2-499, which suggests this strain may be more virulent than N1-227.
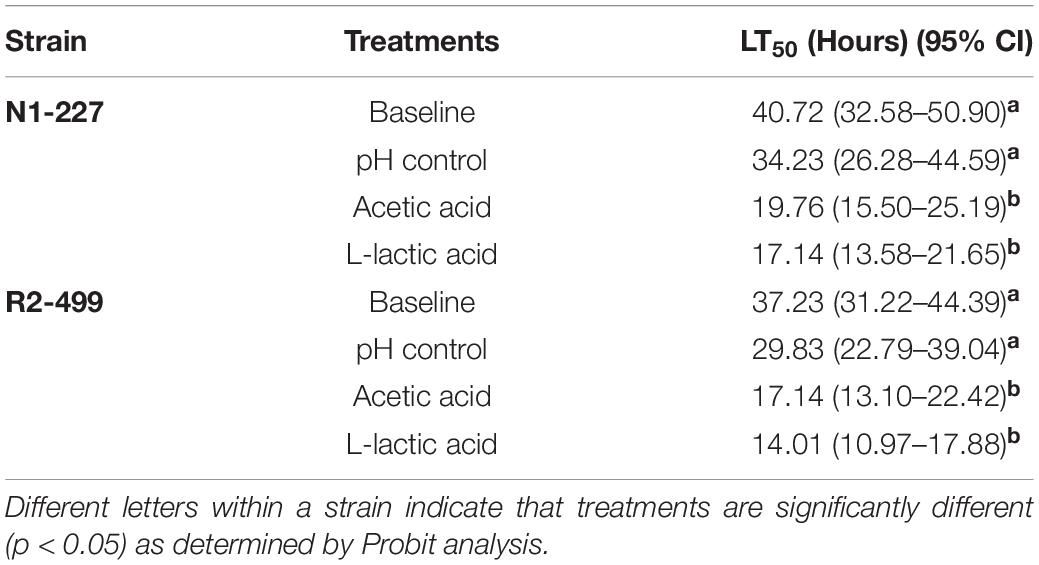
Table 3. Lethal times until 50% population mortality (LT50) for Galleria mellonella larva injected with Listeria monocytogenes strains habituated to various acid treatments.
To test whether the previous organic acid habituation affected the survival or growth of L. monocytogenes in G. mellonella larvae, post-injection bacterial cell numbers were determined over time. The number of L. monocytogenes cells showed a slight decrease for the first 5 h and then remained constant through the 20 h sampling period (see Supplementary Figure 2). Other researchers have also reported that L. monocytogenes cells decreased in number for the first 2 h post-injection (Joyce and Gahan, 2010; Schrama et al., 2013). No statistically significant differences were observed between treatments for either L. monocytogenes strain, indicating that the enhanced virulence observed in organic acid habituated cells is not due to enhanced survival or growth in the larvae.
Discussion
The qPCR experiments showed organic acid habituation impacted the expression of genes encoding important acid and bile stress response mechanisms in both strains of L. monocytogenes. The GAD system serves as a key mechanism of L. monocytogenes survival in acid environments (Cotter et al., 2001; Melo et al., 2015). Karatzas et al. (2012) proposed a model wherein GAD-mediated acid resistance consists of two semi-independent systems: An intracellular system that involves GadD3 acting on intracellular glutamate and an extracellular system that involves GadD2 decarboxylation of glutamate imported by the antiporter GadT2. Interestingly, the differential induction of gadD3 vs. gadD2 in strains N1-227 and R2-499 suggests that gadD3 may play a more prominent role in acid protecting in N1-227, while gadD2 serves as primary defense mechanism in R2-499. Additionally, the fold-change in bilE expression was lower than that of bsh in both strains, which might be a consequence of cell growth phase. Sue et al. (2003) showed that bilE expression is growth phase-dependent, with highest expression level observed in stationary phase cells, and this study used cells collected at mid-log phase.
Infection of host cells by L. monocytogenes can be divided into three stages that require specific virulence factors: Initial cell invasion (InlA and InlB), escape from vacuole (Hly, PlcA, and PlcB) and cell-to-cell spread (ActA and UhpT) (Cossart et al., 1989; Mélanie et al., 2006; Schnupf and Portnoy, 2007; Joyce and Gahan, 2010; Hamon et al., 2012). It has been reported that L. monocytogenes is able to sense different environments and host cell compartments and regulate virulence gene expression accordingly (Freitag and Jacobs, 1999; Gaballa et al., 2019). Other researchers have found that inlA and inlB are induced prior to the cell invasion, while hly, plcB, and plcA are overexpressed within the phagosome and uhpT and actA are expressed in the cytosol (Bubert et al., 1999). In this study, inlA and inlB showed a similar expression pattern in both strains in response to acid exposure (Figure 3). Significant induction of other virulence genes in response to pH or acid was also observed but the patterns were strain-dependent. Additionally, although the transcription level of prfA was not significantly altered by acid exposure, hly transcription is PrfA-dependent (Kazmierczak et al., 2006; Scortti et al., 2007; de las Heras et al., 2011). The observed induction of hly may therefore reflect post-transcriptional control of PrfA activity in these cells.
In summary, RT-qPCR demonstrated that habituation to L-lactic or acetic acids induces statistically significant increases in the expression of several genes associated with acid and bile stress resistance in two L. monocytogenes strains that are known human pathogens. While many of these changes were strain-specific, induction patterns for several stress and virulence genes, including gadD2, arcA, bsh, two internalin genes ilnA and ilnB, in response to acid habituation were similar between N1-227 and R2-499. Future studies might explore the role of nucleotide polymorphism in promoter sequences or in DNA binding motifs in gene expression patterns.
Organic acid habituation also enhanced in situ virulence of both L. monocytogenes strains as evidenced by a reduced the LT50 value in the in vivo G. mellonella infection model. Our finding that HCl or organic acid habituation enhanced virulence of both strains in the G. mellonella model stands in contrast with the report of Schrama et al. (2013), who observed acid or salt adaptation reduced the infectious ability of some L. monocytogenes. However, factors such as different strains and stressors may have contributed to this discrepancy. Our prior research suggests this difference is likely due to strain-specific variation among L. monocytogenes (Zhang et al., 2014), and illustrates the need for further study to determine how widespread this phenomenon is among pathogenic and non-pathogenic strains of L. monocytogenes.
Taken together, these results suggest that exposure to organic acids can increase the pathogenicity of some L. monocytogenes strains by enhancing their ability to survive passage through the gastrointestinal tract while simultaneously priming them for intracellular virulence. While our prior results indicate that this phenomenon may not be universally shared among strains of L. monocytogenes (Zhang et al., 2014), the fact that it does occur in pathogenic strains associated with foodborne outbreaks (Table 1) underscores the potential for organic acids to have unanticipated consequences on food safety and public health. To fully understand the broader impact, future studies are needed to determine how widespread this phenomenon is among additional strains of L. monocytogenes, including both known human pathogens and strains not currently recognized as pathogenic, and to examine the impact of food systems and conditions encountered during processing and storage such as refrigeration temperatures.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
CC and JB: conceptualization, data curation, funding, acquisition, project administration, supervision, validation, and writing—review and editing. ML: formal analysis, investigation, methodology, data collection, software, visualization, and writing—original draft. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Utah Agricultural Experiment Station, Utah State University, and approved as journal paper no. 9366. Additional support was from the Western Dairy Center through the BUILD Dairy program at Utah State University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Martin Wiedmann and the International Life Sciences Institute for supplying the L. monocytogenes strains used in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.675241/full#supplementary-material
References
Banville, N., Browne, N., and Kavanagh, K. (2012). Effect of nutrient deprivation on the susceptibility of Galleria mellonella larvae to infection. Virulence 3, 497–503.
Begley, M., Sleator, R. D., Gahan, C. G. M., and Hill, C. (2005). Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 73, 894–904. doi: 10.1128/iai.73.2.894-904.2005
Bliss, C. I. (1935). The calculation of the dosage-mortality curve. Ann. Appl. Biol. 22, 134–167. doi: 10.1111/j.1744-7348.1935.tb07713.x
Bookout, A. L., and Mangelsdorf, D. J. (2003). Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl. Recept. Signal 1:nrs.01012. doi: 10.1621/nrs.01012
Bubert, A., Sokolovic, Z., Chun, S.-K., Papatheodorou, L., Simm, A., and Goebel, W. (1999). Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol. Gen. Genet. 261, 323–336. doi: 10.1007/pl00008633
Carpenter, C. E., and Broadbent, J. R. (2009). External concentration of organic acid anions and pH: key independent variables for studying how organic acids inhibit growth of bacteria in mildly acidic foods. J. Food Sci. 74, R12–R15.
Chaturongakul, S., Raengpradub, S., Wiedmann, M., and Boor, K. J. (2008). Modulation of stress and virulence in Listeria monocytogenes. Trends Microbiol. 16, 388–396. doi: 10.1016/j.tim.2008.05.006
Chico-Calero, I., Suarez, M., Gonzalez-Zorn, B., Scortti, M., Slaghuis, J., Goebel, W., et al. (2002). Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc. Natl. Acad. Sci. U S A. 99, 431–436. doi: 10.1073/pnas.012363899
Cossart, P., and Toledo-Arana, A. (2008). Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect. 10, 1041–1050. doi: 10.1016/j.micinf.2008.07.043
Cossart, P., Vicente, M. F., Mengaud, J., Baquero, F., Perez-Diaz, J., and Berche, P. (1989). Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect. Immun. 57, 3629–3636. doi: 10.1128/iai.57.11.3629-3636.1989
Cotter, P. D., Gahan, C. G. M., and Hill, C. (2001). A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40, 465–475. doi: 10.1046/j.1365-2958.2001.02398.x
de las Heras, A., Cain, R. J., Bieleckal, M. K., and Vazquez-Boland, J. A. (2011). Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 14, 118–127. doi: 10.1016/j.mib.2011.01.005
Dussurget, O., Cabanes, D., Dehoux, P., Lecuit, M., Buchrieser, C., Glaser, P., et al. (2002). Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45, 1095–1106. doi: 10.1046/j.1365-2958.2002.03080.x
Foster, J. W., and Hall, H. K. (1990). Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 172:771. doi: 10.1128/jb.172.2.771-778.1990
Freitag, N. E., and Jacobs, K. E. (1999). Examination of Listeria monocytogenes intracellular gene expression by using the green fluorescent protein of Aequorea victoria. Infect. Immun. 67, 1844–1852. doi: 10.1128/iai.67.4.1844-1852.1999
Gaballa, A., Guariglia-Oropeza, V., Wiedmann, M., and Boor, K. J. (2019). Cross talk between SigB and PrfA in Listeria monocytogenes facilitates transitions between extra-and intracellular environments. Microbiol. Mol. Biol. Rev. 83, e34–e19.
Gahan, C. G. M., and Hill, C. (2014). Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front. Cell Infect. Microbiol. 4:9. doi: 10.3389/fcimb.2014.00009
Glaser, P., Frangeul, L., Buchrieser, C., Rusniok, C., Amend, A., Baquero, F., et al. (2001). Comparative genomics of Listeria species. Science 294, 849–852.
Glass, K. A., Prasad, B. B., Schlyter, J. H., Uljas, H. E., Farkye, N. Y., and Luchansky, J. B. (1995). Effects of Acid Type and Alta(Tm)2341 on Listeria monocytogenes in a Queso Blanco Type of Cheese. J. Food Prot. 58, 737–741. doi: 10.4315/0362-028x-58.7.737
Goodson, M., and Rowbury, R. J. (1989). Resistance of acid-habituated Escherichia coli to organic acids and its medical and applied significance. Lett. Appl. Microbiol. 8, 211–214. doi: 10.1111/j.1472-765x.1989.tb00250.x
Hamon, M. A., Ribet, D., Stavru, F., and Cossart, P. (2012). Listeriolysin O: the Swiss army knife of Listeria. Trends Microbiol. 20, 360–368. doi: 10.1016/j.tim.2012.04.006
Hoffmann, J. A., Kafatos, F. C., Janeway, C. A., and Ezekowitz, R. A. B. (1999). Phylogenetic perspectives in innate immunity. Science 284, 1313–1318. doi: 10.1126/science.284.5418.1313
Jones, R. T., Sanchez-Contreras, M., Vlisidou, I., Amos, M. R., Yang, G. W., Munoz-Berbel, X., et al. (2010). Photorhabdus adhesion modification protein (Pam) binds extracellular polysaccharide and alters bacterial attachment. BMC Microbiol. 10:141. doi: 10.1186/1471-2180-10-141
Joyce, S. A., and Gahan, C. G. M. (2010). Molecular pathogenesis of Listeria monocytogenes in the alternative model host Galleria mellonella. Microbiology-Sgm 156, 3456–3468. doi: 10.1099/mic.0.040782-0
Karatzas, K. A. G., Suur, L., and O’Byrne, C. P. (2012). Characterization of the intracellular glutamate decarboxylase system: analysis of its function, transcription, and role in the acid resistance of various strains of Listeria monocytogenes. Appl. Environ. Microbiol. 78, 3571–3579. doi: 10.1128/aem.00227-12
Kazmierczak, M. J., Wiedmann, M., and Boor, K. J. (2005). Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69, 527–543. doi: 10.1128/mmbr.69.4.527-543.2005
Kazmierczak, M. J., Wiedmann, M., and Boor, K. J. (2006). Contributions of Listeria monocytogenes sigma B and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology 152, 1827–1838. doi: 10.1099/mic.0.28758-0
Lani, M. N., and Hassan, Z. (2016). Listeria monocytogenes: general overview and its significance to food safety. Terengganu: Penerbit UMT.
Lecuit, M. (2007). Human listeriosis and animal models. Microbes Infect. 9, 1216–1225. doi: 10.1016/j.micinf.2007.05.009
Lecuit, M., Dramsi, S., Gottardi, C., Fedor-Chaiken, M., Gumbiner, B., and Cossart, P. (1999). A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18, 3956–3963. doi: 10.1093/emboj/18.14.3956
Li, M. (2020). Exploring the connection between acid exposure and virulence in Listeria monocytogenes. Ph. D. thesis. Logan: Utah State University.
Liu, D. Y. (2006). Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J. Med. Microbiol. 55, 645–659. doi: 10.1099/jmm.0.46495-0
Mélanie, H., Hélène, B., and Pascale, C. (2006). Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 4, 423–434. doi: 10.1038/nrmicro1413
Melo, J., Andrew, P. W., and Faleiro, M. L. (2015). Listeria monocytogenes in cheese and the dairy environment remains a food safety challenge: The role of stress responses. Food Res. Int. 67, 75–90. doi: 10.1016/j.foodres.2014.10.031
Mengaud, J., Ohayon, H., Gounon, P., Mege, R. M., and Cossart, P. (1996). E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84, 923–932. doi: 10.1016/s0092-8674(00)81070-3
Mukherjee, K., Altincicek, B., Hain, T., Domann, E., Vilcinskas, A., and Chakraborty, T. (2010). Galleria mellonella as a model system for studying Listeria pathogenesis. Appl. Environ. Microbiol. 76, 310–317.
Mukherjee, K., Raju, R., Fischer, R., and Vilcinskas, A. (2013). Galleria Mellonella as a model host to study gut microbe homeostasis and brain infection by the human pathogen Listeria Monocytogenes. Yellow Biotechnol. I Insect Biotechnol. Drug Discov. Preclin. Res. 135, 27–39. doi: 10.1007/10_2013_203
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45–e45.
Ramarao, N., Nielsen-Leroux, C., and Lereclus, D. (2012). The insect Galleria mellonella as a powerful infection model to investigate bacterial pathogenesis. J. Vis. Exp. 70:e4392.
Rejasse, A., Gilois, N., Barbosa, I., Huillet, E., Bevilacqua, C., Tran, S., et al. (2012). Temperature-dependent production of various PlcR-controlled virulence factors in Bacillus weihenstephanensis Strain KBAB4. Appl. Environ. Microbiol. 78, 2553–2561. doi: 10.1128/aem.07446-11
Robinson, R. K., and Batt, C. A. (1999). Encyclopedia of food microbiology. Florida, FL: Academic press.
Ruijter, J., Ramakers, C., Hoogaars, W., Karlen, Y., Bakker, O., Van den Hoff, M., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37, e45–e45.
Schnupf, P., and Portnoy, D. A. (2007). Listeriolysin O: a phagosome-specific lysin. Microbes Infect. 9, 1176–1187. doi: 10.1016/j.micinf.2007.05.005
Schrama, D., Helliwell, N., Neto, L., and Faleiro, M. L. (2013). Adaptation of Listeria monocytogenes in a simulated cheese medium: effects on virulence using the Galleria mellonella infection model. Lett. Appl. Microbiol. 56, 421–427. doi: 10.1111/lam.12064
Scortti, M., Monzo, H. J., Lacharme-Lora, L., Lewis, D. A., and Vazquez-Boland, J. A. (2007). The PrfA virulence regulon. Microbes Infect. 9, 1196–1207. doi: 10.1016/j.micinf.2007.05.007
Silva, E. N. G., Almeida, R. C. C., Figueiredo, A. C. L., and Miranda, F. A. (2012). Effects of salts of organic acids on the survival of Listeria monocytogenes in soft cheeses. Int. J. Food Sci. Technol. 47, 1819–1825. doi: 10.1111/j.1365-2621.2012.03037.x
Sleator, R. D., Wemekamp-Kamphuis, H. H., Gahan, C. G., Abee, T., and Hill, C. (2005). A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol. Microbiol. 55, 1183–1195. doi: 10.1111/j.1365-2958.2004.04454.x
Smith, J. L., Liu, Y., and Paoli, G. C. (2012). How does Listeria monocytogenes combat acid conditions? Can. J. Microbiol. 59, 141–152.
Strand, M. R. (2008). The insect cellular immune response. Insect Sci. 15, 1–14. doi: 10.1111/j.1744-7917.2008.00183.x
Sue, D., Boor, K. J., and Wiedmann, M. (2003). sigma(B)-dependent expression patterns of compatible solute transporter genes opuCA and Imo1421 and the conjugated bile salt hydrolase gene bsh in Listeria monocytogenes. Microbiology-Sgm 149, 3247–3256. doi: 10.1099/mic.0.26526-0
Sue, D., Fink, D., Wiedmann, M., and Boor, K. J. (2004). σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150, 3843–3855. doi: 10.1099/mic.0.27257-0
Tasara, T., and Stephan, R. (2007). Evaluation of housekeeping genes in Listeria monocytogenes as potential internal control references for normalizing mRNA expression levels in stress adaptation models using real-time PCR. FEMS Microbiol. Lett. 269, 265–272. doi: 10.1111/j.1574-6968.2007.00633.x
Zhang, Q., Feng, Y., Deng, L., Feng, F., Wang, L., Zhou, Q., et al. (2011). SigB plays a major role in Listeria monocytogenes tolerance to bile stress. Int. J. Food Microbiol. 145, 238–243. doi: 10.1016/j.ijfoodmicro.2010.12.028
Keywords: organic acid, acid resistance, bile resistance, virulence, gene expression, Listeria monocytogenes, Galleria mellonella
Citation: Li M, Carpenter CE and Broadbent JR (2021) Organic Acid Exposure Enhances Virulence in Some Listeria monocytogenes Strains Using the Galleria mellonella Infection Model. Front. Microbiol. 12:675241. doi: 10.3389/fmicb.2021.675241
Received: 02 March 2021; Accepted: 14 June 2021;
Published: 06 July 2021.
Edited by:
Catherine M. Logue, University of Georgia, United StatesReviewed by:
Taurai Tasara, University of Zurich, SwitzerlandIvan Dubovskiy, Novosibirsk State Agrarian University, Russia
Copyright © 2021 Li, Carpenter and Broadbent. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeff R. Broadbent, amVmZi5icm9hZGJlbnRAdXN1LmVkdQ==
 Minghao Li
Minghao Li Charles E. Carpenter
Charles E. Carpenter Jeff R. Broadbent*
Jeff R. Broadbent*