
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 02 June 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.674560
This article is part of the Research TopicTherapeutics Targeting Intracellular BacteriaView all 7 articles
 Yuanzun Zhong1†
Yuanzun Zhong1† Liyao Tang1†
Liyao Tang1† Qiuhua Deng1
Qiuhua Deng1 Li Jing2
Li Jing2 Jiao Zhang1
Jiao Zhang1 Yao Zhang1
Yao Zhang1 Feng Yu1
Feng Yu1 Yijun Ou1
Yijun Ou1 Shaoju Guo3
Shaoju Guo3 Bin Huang3
Bin Huang3 Hongying Cao1
Hongying Cao1 Ping Huang1*
Ping Huang1* Yifei Xu1,3*
Yifei Xu1,3*The long-term colonization of Helicobacter pylori can cause various gastrointestinal diseases, and its high genetic variability is prone to antibiotic resistance and leads to failure of clinical treatment. Intracellular survival also contributes to the drug tolerance of H. pylori. Patchouli alcohol (PA) shows a highly efficient activity against H. pylori in vitro and in vivo. And this study aims to explore whether PA can reduce the resistance of H. pylori and determine the underlying mechanism. Checkerboard and time–kill bactericidal curve assay reveal that the combination of PA and clarithromycin (CLR) promoted the inhibition and bactericidal effect against H. pylori. Stimulation of CLR leads to the internalization of H. pylori, but PA can effectively inhibit the invasion induced by CLR. Compared with antibiotics, PA remarkably eradicated the intracellular H. pylori, and this intracellular sterilized ability was further improved in combination with antibiotics (CLR and metronidazole). The expression of H. pylori efflux pump genes (hp0605, hp1327, and hp1489) was dose-dependently downregulated by PA. Digital droplet PCR indicated that the H. pylori mutant of A2143G can be inhibited by PA. Cellular uptake and transport assays showed that PA is rapidly absorbed, which promotes its activity against intracellular bacteria. Therefore, PA can act synergistically with CLR as a candidate treatment against drug-resistant H. pylori.
Helicobacter pylori is a Gram-negative spiral-shaped flagellated bacterium colonizing the stomach of more than half of the global population and thus considered as one of the common infecting bacteria worldwide (Salama et al., 2013; Ansari and Yamaoka, 2017; Crowe and Solomon, 2019). H. pylori causes various gastroduodenal diseases, such as chronic atrophic gastritis, gastric ulcer, duodenal ulcer, and gastric cancer (Choi et al., 2020; de Martel et al., 2020; Ford et al., 2020). Effective H. pylori eradication is beneficial for the prevention of gastric cancer (Tsukamoto et al., 2017; Liou et al., 2020). Since the 1990s, standard triple therapy comprising two antibiotics (clarithromycin [CLR], metronidazole [MTZ], or amoxicillin) and a proton pump inhibitor has been used for H. pylori eradication. However, the resistance of H. pylori to CLR and MTZ has increased due to the abuse and misuse of antibiotics. In some highly resistant areas, the resistance rates to MTZ and CLR reached 70–80 and 40–50%, respectively (Thung et al., 2016). Antibiotic resistance has reduced the eradication rate of standard triple therapy to 70% or even lower (Liu et al., 2018; De Francesco et al., 2019).
Antibiotic resistance is a global public health problem in which the sustained increase in antibiotic resistance may eventually surpass the amount of new antibiotics being developed. Bacterial resistance is a complex process between host and bacteria and may be related to the following mechanisms: mutated drug targets, enhanced efflux pump expression, altered membrane permeability, etc. (Gerrits et al., 2006; Gong and Yuan, 2018). Essentially, bacterial mutation occurs during their evolution on some key functional proteins, such as nucleic acid synthesis, redox system, and protein translation, thus contributing to antibiotic resistance (Alba et al., 2017). Alterations on the efflux system (Gerdes and Semsey, 2016) and cell membrane (Garcia-Fernandez et al., 2017) would reduce the intracellular accumulation of antibiotics, thus diminishing the antimicrobial activity. Furthermore, bacteria produce enzymes to inactivate antibiotics or release virulence factors to affect antibiotic activity (Behrens et al., 2020). The above mechanisms do not exist in isolation in the process of bacterial resistance, and recent experiments on Escherichia coli indicated that an increased activity of efflux pump may increase the mutation rate of bacterial resistance genes (El Meouche and Dunlop, 2018). For H. pylori, its cell invasion ability is one of the strategies to avoid the bactericidal effect of antibiotics (Wang et al., 2017).
clarithromycin belongs to the class of macrolide antibiotics (Vazquez-Laslop and Mankin, 2018) and exhibits its antibacterial effect by inhibiting the protein synthesis of bacteria. For some CLR-sensitive H. pylori, the bactericidal effect of CLR can be observed at a minimum inhibitory concentration (MIC) lower than 0.0156 μg/ml; however, for some drug-resistant strains, the anti-H. pylori activity is attained at an MIC of 256 μg/ml (Jung et al., 2018). Point mutations in H. pylori such as A2143G, A2142G, T2182C, C2611A, and T2717C (Seo et al., 2019) contribute to its CLR resistance, change the configuration of the ribosome, and weaken antibiotic binding, thereby leading to drug resistance. Among the point mutations, the A2142G and A2143G accounted for the majority (Matta et al., 2018; Marques et al., 2020).
Pogostemonis Herba, the dried aerial part of Pogostemon cablin (Blanco) Benth. (Labiatae), is a traditional Chinese medicament used to treat gastrointestinal diseases in many Asian countries. The main component of Pogostemonis Herba volatile oil is patchouli alcohol (PA, the chemical structure shown in Figure 1). Our laboratory’s previous experiment revealed that PA can effectively inhibit H. pylori in vivo and in vitro and shows a protective effect on H. pylori-related gastritis (Xu et al., 2017; Lian et al., 2018). A stable MIC against drug-resistant H. pylori and the strong post-antibiotic effect (PAE) of PA were also verified.
The objective of this study is to explore whether the combination of PA and antibiotics (CLR and MTZ) could improve the antibacterial efficiency against H. pylori and to investigate the underlying mechanisms.
Columbia blood agar base, brain heart infusion (BHI), Mueller–Hinton (MH) agar, and H. pylori selective supplement (Dent) SR0147E were obtained from Oxoid, United Kingdom. Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS), Hank’s balanced salt solution (HBSS), MEM nonessential amino acids, TRIzol reagent, and Live/Dead BacLight Bacterial Viability kits (Molecular Probes) were purchased from Thermo Fisher Scientific, United States. Sheep blood was procured from Pingrui Biotechnology, China. CLR was purchased from Dalian Meilun Biotechnology, China. MTZ and saponin were acquired from Sigma, Germany. Fastking gDNA Dispelling RT SuperMix Kit, Talent qPCR PreMix (SYBR Green) Kit, TIANamp Bacteria DNA kit, and 2 × Taq PCR Mix were purchased from TIANGEN, China. Anti-H. pylori antibody ab20459 was obtained from Abcam, United Kingdom. Alexa Fluor 488 and goat anti-rabbit were bought from Southern Biotech, United States.
Patchouli alcohol (purity > 99%) was kindly provided by the Mathematical Engineering Academy of Chinese Medicine, and the quality of PA was confirmed by melting point, infrared spectroscopy, 1H and 13C NMR, and mass spectrometry (Liao et al., 2015). DMSO was used to dissolve PA and served as a control (DMSO < 0.5% in all experiments) in the in vitro study.
The H. pylori reference strain NCTC11637 was purchased from the American Type Culture Collection (ATCC, United States). Clinical drug-resistant strains Hp1869 (MTZ-resistant strain) and Hp1870 (MTZ- and CLR-resistant strain) were obtained from Renji Hospital, Shanghai Jiao Tong University. H. pylori specimens were stored at −80°C in BHI containing 30% glycerol and then cultured on Columbia agar base supplemented with 5% sheep blood at 37°C in a tri-gas incubator (Nuaire Nu-5831E, United States) with 10% CO2, 5% O2, and 85% N2. After 48–72 h proliferation, H. pylori specimens were scraped, suspended in phosphate-buffered saline (PBS), and passaged to fresh Columbia blood agar. Turbidimetry (Zhuhai BaSO Biotechnology Co., Ltd., China) was measured to adjust the McFarland standard of the H. pylori strain when needed.
Human gastric epithelial cells GES-1 and human colon adenocarcinoma cell line Caco-2 were purchased from the BNCC Biotechnology Company (China). The GES-1 cells were cultured in high-glucose DMEM supplemented with 10% FBS, and the Caco-2 cells were cultured in DMEM containing 10% FBS, 1% nonessential amino acids, and 1% penicillin–streptomycin antibiotic solution. Both cell lines were cultured at 37°C in a 5% CO2 humidified incubator (CO150, United States) and passaged when they reached 80–90% confluence at a 1:2 ratio by using 0.25% trypsin.
The checkerboard method was used to determine the potency of the combination of PA with CLR or MTZ on H. pylori. The assay assesses the activities of antimicrobial combinations tested at concentrations in serial two-fold dilutions under MIC. Autoclaved MH agar (5% sheep blood) was prepared for gradient dilutions of PA, CLR, and MTZ. The final concentration of each drug ranges from 1 to 0.25 MIC; the MIC values are shown in Table 1. And according to the checkerboard method, to design different combinations of the two drugs, their combined effects must be observed. H. pylori was then resuspended in PBS, and the concentration was adjusted to McFarland 1 (1 × 108 CFU/ml) through turbidimetry. Bacterial suspension (100 μl) was spread onto the drug-containing MH agar plates and cultured in a tri-gas incubator for 4 days. The fractional inhibitory concentration index (FICI) was calculated as the sum of the MIC of each compound used in combination and divided by the MIC of each compound used alone as follows:
The results were interpreted using the following criteria: synergism (FICI ≤ 0.5), additive (0.5 < FICI ≤ 1), indifference (1 < FICI < 4), and antagonism (FICI ≥ 4) (Krzyzek et al., 2020).
The H. pylori reference strain NCTC11637 and drug-resistant strain Hp1870 were used to measure the killing kinetics of PA and CLR and their combination. H. pylori specimens were resuspended in PBS, and the concentration was adjusted to McFarland 1 through turbidimetry. The bacteria were then mixed with BHI (10% FBS) at a ratio of 1:10 and shaken at 120 rpm in a tri-gas incubator until grown to the logarithmic growth phase (approximately 48–72 h). Different combinations of 1/2 MIC PA and 1/2 MIC CLR (the details were shown as follows) were added to explore the bactericidal effect on H. pylori. NCTC11637 was treated with 12.5 μg/ml PA, 0.007 μg/ml CLR, and 12.5 μg/ml PA + 0.007 μg/ml CLR, and the drug-resistant strain Hp1870 was treated with 12.5 μg/ml PA, 4 μg/ml CLR, and 12.5 μg/ml PA + 4 μg/ml CLR. The same volume of DMSO served as the control. The MIC for the liquid culture strain was tested at OD600 through broth dilution as shown in Table 1. Suitable samples were removed every 24 h for 0–5 days and used for the following assay.
A time–kill bactericidal curve was analyzed to confirm the synergistic activity. Approximately 100 μl of samples was removed, and the gradient dilutions were prepared and flooded on the plate to form colonies that were counted after 4 days of cultivation. The time–kill bactericidal curve was constructed by plotting log10 CFU/ml versus time. In time–kill bactericidal curves, synergy is defined as a ≥2-log10 decrease in CFU/ml between the combination and the most active constituent (Doern, 2014).
For the assay using Live/Dead BacLight Bacterial Viability kits, the samples were removed every 24 h and stained by SYTO 9 and PI (mixed at a ratio of 1:4 before use) to identify live or dead bacteria. Approximately 1 ml of the sample was centrifugated and resuspended in normal saline (NS), and a fluorescence dye was added at a ratio of 3:1,000. After incubation at room temperature in the dark for 15 min, 10 μl of the sample was diluted using 100 μl of NS in a glass-bottom dish and finally photographed by a confocal microscope (LSM 800, Zeiss).
Helicobacter pylori-infected gastric epithelial cells (multiplicity of infection [MOI] = 100:1) were washed three times with PBS, followed by 100 μg/ml gentamicin incubation for 6 h to remove extracellular bacteria. Intracellular bacteria were retrieved from invaded cells by a 15 min incubation with 0.1% saponin in PBS at 37°C. Serial dilutions of bacterial suspensions were prepared using PBS and drop-plated (100 μl drop) onto Columbia blood agar plates. The bacterial colonies were counted after 4 days of cultivation in a tri-gas incubator for CFU determinations.
GES-1 cells (1.5 × 105) were cultured in a glass-bottom dish, allowed to adhere for 6 h, and added with H. pylori NCTC11637 at an MOI of 100:1. After 12 h of co-incubation, unattached bacteria were removed by washing with PBS three times. Then, the cells were treated with 100 μg/ml gentamicin for 6 h to clear extracellular bacteria. The cells were stimulated by PA and antibiotics according to different experiment designs. Next, the cells were fixed with 4% paraformaldehyde for 30 min at room temperature and then washed three times with PBST (PBS + 0.05% Tween 20). PBST washing for three times was performed between each step below. After washing, the cells were permeabilized with PBST containing 0.1% Triton for 30 min. Goat serum (5%) containing PBST was used as a blocking buffer and incubated for 1 h. The intracellular H. pylori was stained by an anti-H. pylori antibody (1:200). The secondary antibody Alexa Fluor 488-conjugated goat anti-rabbit (1:200) was added and incubated at room temperature in the dark for 2 h. Finally, the samples were stained with DAPI, sealed, and placed on an LSM 800 confocal microscope (Carl Zeiss) for image acquisition.
The GES-1 cells (1.5 × 106) were seeded on a six-well plate for 6 h and then added with the H. pylori standard strain NCTC11637 at an MOI of 100:1. H. pylori-infected cells were stimulated by CLR and MTZ at 1/4 MIC and 1/8 MIC doses for 6 h, respectively. A gentamycin protection assay was conducted to measure the CFU of intracellular H. pylori.
H. pylori NCTC11637-infected GES-1 cells (MOI = 100:1) were treated by 6.25, 12.5, and 25 μM PA combined with 1/8 MIC CLR for 6 h to observe the influence of PA on H. pylori invasion promoted by low CLR doses. A gentamicin protection assay was used for CFU determinations, and immunofluorescence was adopted for cell imaging.
The H. pylori reference strain NCTC11637 and clinical drug-resistant strains Hp1869 and Hp1870 were used to compare the ability of various drugs for intracellular H. pylori elimination. The GES-1 cells (1.5 × 106) were seeded on a six-well plate, allowed to adhere for 6 h, and then added with the bacteria at an MOI of 100:1.
The killing ability for intracellular H. pylori was investigated by using PA, MTZ, or CLR alone. After extracellular bacteria were eliminated by gentamicin for 6 h, the H. pylori -infected GES-1 cells were treated with CLR (0.0156, 0.0312, 0.078, 0.156, and 0.312 μg/ml for NCTC11637 and Hp1869; 16, 32, 48, 64, and 72 μg/ml for Hp1870), MTZ (12.5, 25, 50, 75, and 100 μg/ml for all three strains), and PA (25, 30, 35, 40, 45, and 50 μg/ml for all three strains) for 16 h. Intracellular H. pylori survival was examined as described in the gentamycin protection assay.
The combined effects of CLR, MTZ, and PA on intracellular bactericidal effect were further explored. After extracellular bacteria were eliminated by gentamycin for 6 h, the infected GES-1 cells were further incubated with 25 μg/ml PA in combination with antibiotics for 16 h. The doses of CLR were 0.0312, 0.078, and 0.156 μg/ml for NCTC11637 and Hp1869 and 32, 48, and 64 μg/ml for Hp1870. For MTZ, the doses were selected as 25, 50, and 75 μg/ml. Meanwhile, to further study the effect of PA, the combination of CLR and MTZ has also been tested. For NCTC11637 and Hp1869, 0.015 μg/ml CLR combined with 75 μg/ml MTZ or 0.15 μg/ml CLR combined with 25 μg/ml MTZ was used for 16 h. For Hp1870, 32 μg/ml CLR combined with 75 μg/ml MTZ or 64 μg/ml CLR combined with 25 μg/ml MTZ was used. Finally, intracellular H. pylori survival was analyzed as described in the gentamycin protection assay. Relative to the control group, a 99% decrease in the viability of intracellular strains was defined as effective extermination.
Immunofluorescence was used to compare the number of residual intracellular H. pylori. After extracellular bacteria were eliminated by gentamicin, NCTC11637-infected GES-1 cells were treated by CLR (0.312 μg/ml, 20 MIC), MTZ (100 μg/ml, 25 MIC), and PA (50 μg/ml, 2 MIC) for 16 h. The cells were imaged using the immunofluorescence method.
The multidrug-resistant H. pylori strain Hp1870 was used to observe the effect of PA on the expression of the efflux effect gene. Hp1870 with concentration adjusted to 1 × 108 CFU/ml through turbidimetry was cultured, mixed with BHI (10% FBS) at a ratio of 1:10, and classified into a negative control group (DMSO), model group (32 μg/ml CLR), and PA + CLR treatment groups (12.5, 25, and 50 + 32 μg/ml CLR). The bacteria were stimulated for 2 h in each group. An RT-qPCR assay was adopted to measure the gene expression of efflux effect genes as described below.
The time- and dose-dependent effects of PA on the altered expression of hp0605 induced by CLR were explored. Hp1870 specimens were treated with DMSO (negative control), 16 or 32 μg/ml CLR (1/2 MIC or MIC, model group), or CLR + PA (16 or 32 μg/ml CLR + 50 μg/ml PA) for 0.5 or 2 h. An RT-qPCR assay was adopted to measure the hp0605 expression.
After the above stimulation, a TRIzol reagent was used to extract bacterial total RNA. RNA purity and concentration were determined based on A260/280 and A260, respectively, by using a NanoDrop 2000 UV–vis spectrophotometer. The total RNA was reverse transcribed into cDNA using the Fastking gDNA Dispelling RT SuperMix Kit. RT-qPCR was performed using the CFX96 (Bio-Rad, United States) for one cycle at 95°C for 3 min and 40 cycles at 95°C for 5 s and 60°C for 15 s. Data were analyzed using the Pfaffli method with the 16s gene as the internal reference. The primers used in RT-qPCR are shown in Table 2.
The multidrug-resistant H. pylori strain Hp1870 was cultured, and its concentration was adjusted to McFarland 1 using PBS. The bacterial suspension was then added to BHI (10% FBS) at a ratio of 1:10. Five groups were established in this experiment: negative control (DMSO), model group (32 μg/ml CLR), and PA + CLR groups (12.5, 25, and 50 μg/ml + 32 μg/ml CLR). The bacteria were treated for 2 h, and the bacterial total RNA was extracted by a TRIzol reagent and reverse transcribed into cDNA as described above.
PCR experiments were conducted using the 2 × Taq PCR Mix to identify the mutation site of Hp1870 by amplifying and sequencing H. pylori 23S genes. The primers are shown in Table 2. H. pylori -specific droplet digital PCR (ddPCR) assays were performed using a QX200 ddPCR system (Bio-Rad, United States) by Sangon Biotech (China). The primer and probe sequences for the CLR-resistant H. pylori 23S ddPCR assay are listed in Table 3 and quoted from an existing study (Sun et al., 2018). The thermal cycling conditions were as follows: 95°C for 10 min, 40 cycles of 94°C for 30 s and 60°C for 1 min, and one cycle of 98°C for 10 min. Fluorescent amplitudes were analyzed by using the QX200 Droplet Reader (Bio-Rad). Data analysis was performed by using the QuantaSoft software version 1.6.6 (Bio-Rad).
GC–MS was performed on an Agilent 7890A-5975C GC–MS system (Agilent, United States). GC separation was conducted on an HP-5MS capillary column (30 m × 0.25 mm, 0.25 μm). The initial oven temperature was set at 160°C and then programmed to increase to 220°C by a gradient of 6°C/min. The inlet temperature was 230°C. MS conditions were as follows: EI mode, ionization energy 70 eV, ionization source temperature 250°C. The parent ion and product ion mass spectra of the base ion peaks at 222.2 and 150.2 m/z for PA and internal standard cedrol, respectively, were acquired with selected ion monitoring. The split ratio was 1:2, and the inject volume was 1 μl.
For cellular uptake experiments, the Caco-2 cells were seeded onto 12-well plates at 1 × 105 cells per well, grown for 14 days, washed with HBSS (37°C) twice, and incubated with 1 ml of 300, 500, and 700 μM PA in HBSS (pH 4.1, 7.4, or 9.1) for 5–30 min in 37°C or 4°C. After incubation, the cells were washed with HBSS (4°C) three times. Next, the PA within the cells was extracted by ultrasound in 1 ml of cell lysis buffer (HBSS containing 10% methanol). After centrifugation for 10 min at 8,000 g, the supernatant was removed and divided into two parts; one was used to determine the protein concentration by a BCA kit, and the other was used to determine PA concentration by GC–MS after extraction with ethyl acetate.
For cellular transport experiments, the Caco-2 cells were seeded onto polycarbonate filter membranes (pore size 0.4 μm, filter area 1.12 cm2) in 12-well plates at 1 × 105 cells per well and grown for 21 days. Trans-epithelial electrical resistance (TEER) values were measured twice weekly by using a Millicell-ERS volt-ohm meter, and the membranes with monolayer cells that met the criteria with a TEER of above 500 Ω/cm2 were used for the transport studies. Caco-2 cell monolayers were washed twice with HBSS (pH 7.4), and PA (300, 500, and 700 μM) was added to either the apical (AP, 0.5 ml) or basolateral (BL, 1 ml) side. The receiving chamber contained the corresponding volume of HBSS. Verapamil (100 μM) was used to investigate the role of p-glycoprotein (P-gp) in PA absorption. After shaking at 54 rpm for 2 h, the samples were collected from the receiving chamber to assess the drug transport from the AP-to-BL or BL-to-AP direction. The PA content was measured by GC–MS after extraction with ethyl acetate. The apparent permeability coefficient (Papp) was calculated from the following equation.
where ΔQ/Δt is the cumulative transport rate of the compound on the receiving side (μM/s), A is the surface area of the cell monolayer (cm2), and C0 is the initial concentration in the donor compartment (μM/cm).
All results were presented as means ± standard deviation and analyzed using ANOVA and unpaired Student t test in SPSS 24.0. Bonferroni’s test or Dunnett’s test was performed for multiple groups based on the homogeneity of variance to test the significant difference. A t-test was performed for two independent groups. P < 0.05 was considered significant.
The effects of PA combined with CLR and MTZ on the H. pylori reference strain NCTC11637 and the clinical strain Hp1870 (CLR and MTZ resistant) were observed using the checkerboard method (Figure 2A). For the reference strain NCTC11637, the CLR-and-PA combination decreased the MIC of PA from 25 to 6.25 μg/ml (FIC = 0.25) and the MIC of CLR by half (FIC = 0.5). No alteration was induced by the PA-and-MTZ combination (FIC = 1). For the clinical resistant strain Hp1870, the MIC values were reduced by half when PA was combined with CLR or vice versa (FIC = 0.5). For PA combined with CLR, the FICI was 0.75 for the reference strain NCTC11637 and 1 for the clinical resistant strain Hp1870, indicating that both components had an additive effect. For PA combined with MTZ, the FICI was 2 for the reference strain NCTC11637, which is considered as indifferent.
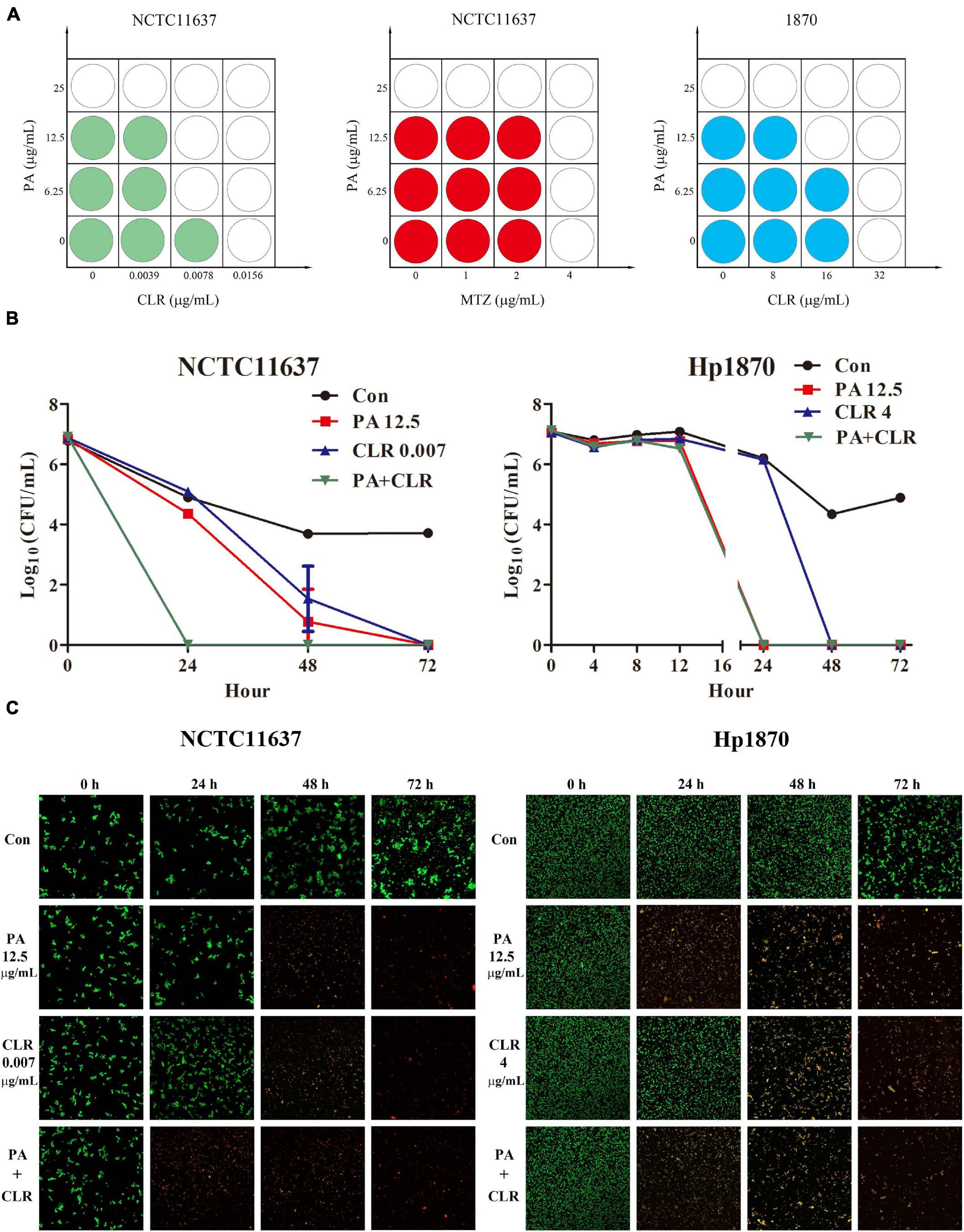
Figure 2. Combined effect of PA and antibiotics against H. pylori. (A) Combined effect tested by the checkerboard method of PA and antibiotics against the H. pylori standard strain NCTC11637 and the drug-resistant strain Hp1870 as determined by FICI (n = 4). (B) Combined effect tested by time–kill curves of PA and CLR against the H. pylori strains NCTC11637 and Hp1870; the synergy is defined as a ≥2-log10 (n = 3). (C) Bacteria imaging by Live/Dead BacLight Bacterial Viability kits to observe the bactericidal capacity of PA and CLR against H. pylori NCTC11637 and Hp1870.
The time–kill bactericidal curve method was adopted to explore the combined efficiency of PA and CLR, and the results are shown in Figure 2B. For the reference strain NCTC11637, PA and CLR alone would eradicate the bacteria in 72 h. Their combination can effectively kill the bacteria in 24 h, indicating successful synergy.
For the drug-resistant strain Hp1870, the number of bacteria in each group did not change significantly after the treatment for 8 h. The number of bacteria in the group treated with PA combined with CLR decreased by approximately 75% compared with that in the control group after 12 h, indicating the faster antibacterial action of this treatment compared with drug use alone. Treatment with PA alone or PA combined with CLR killed H. pylori within 24 h. However, the time had extended to 48 h when the bacteria were treated with CLR only. Figure 2C displays the visible alteration of bacterial death in a time- and dose-dependent manner and corresponds to the results of the time–kill curve.
Figure 3A shows that after 6 h of stimulation by CLR (1/8 MIC), the amount of H. pylori NCTC11637 that invaded into the cells were significantly increased in a concentration-dependent manner compared with that in the control group. For MTZ, no remarkable difference from the control group was observed after 6 h of stimulation.
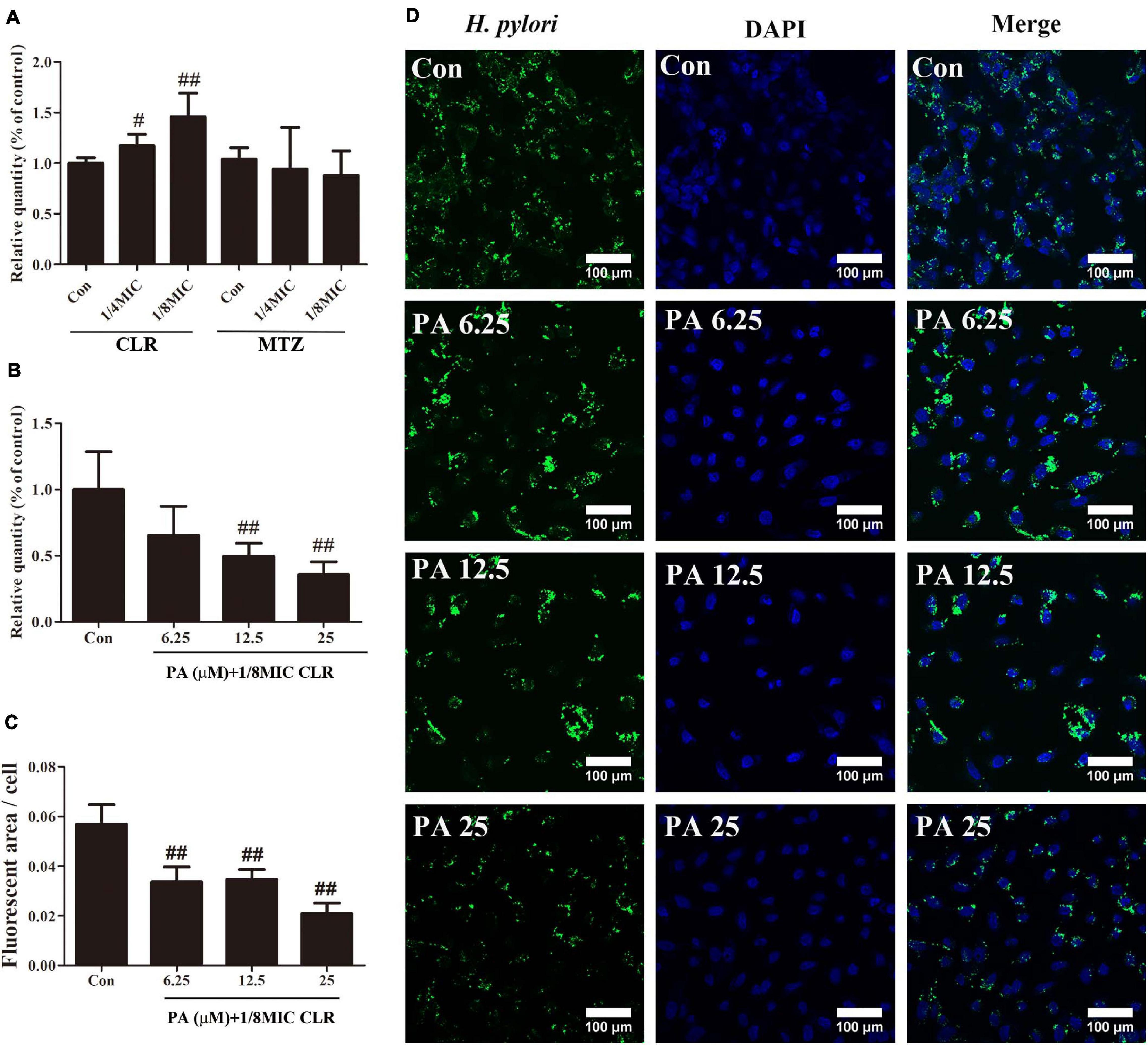
Figure 3. Number of intracellular H. pylori stimulated by antibiotics and PA. (A) Influence of low-dose CLR and MTZ on the number of intracellular H. pylori. (B) Antagonism effect of PA on the increased invasion induced by CLR; a gentamycin protection assay was conducted to measure the CFU of intracellular H. pylori. (C) Quantitative results of the fluorescent area of H. pylori (green area) with treatment of PA and CLR. (D) Representative confocal images of intracellular H. pylori after treatment with PA and CLR. The above results are expressed as the mean ± SD (n = 6). #P < 0.05, and ##P < 0.01 (compared with the results for the control group).
Figure 3B indicates that PA addition effectively and concentration-dependently reduced the increment of the number of invasive H. pylori caused by CLR. Immunofluorescence images also reflected the same trend. Compared with that in the control group, the fluorescent area of H. pylori (green area) was significantly decreased by the PA treatment in different concentrations (Figures 3C,D).
As shown in Figure 4A, PA at a dose of 45 μg/ml (1.8 MIC) effectively eliminated the intracellular H. pylori NCTC11637 and Hp1870, and this dose was reduced to 35 μg/ml for Hp1869 (1.4 MIC). CLR at a dose of 0.156 (10 MIC)–0.312 (20 MIC) showed slight intracellular H. pylori elimination ability for NCTC11637 and Hp1869 strains, which are sensitive to CLR; however, the efficiencies did not reach 99% (two order of magnitude decrease in log10 CFU/ml). For the CLR- and MTZ-resistant strain Hp1870, no alteration was noted even when the CLR dose reached 72 μg/ml. Various MTZ doses also did not induce any changes on these three intracellular strains.
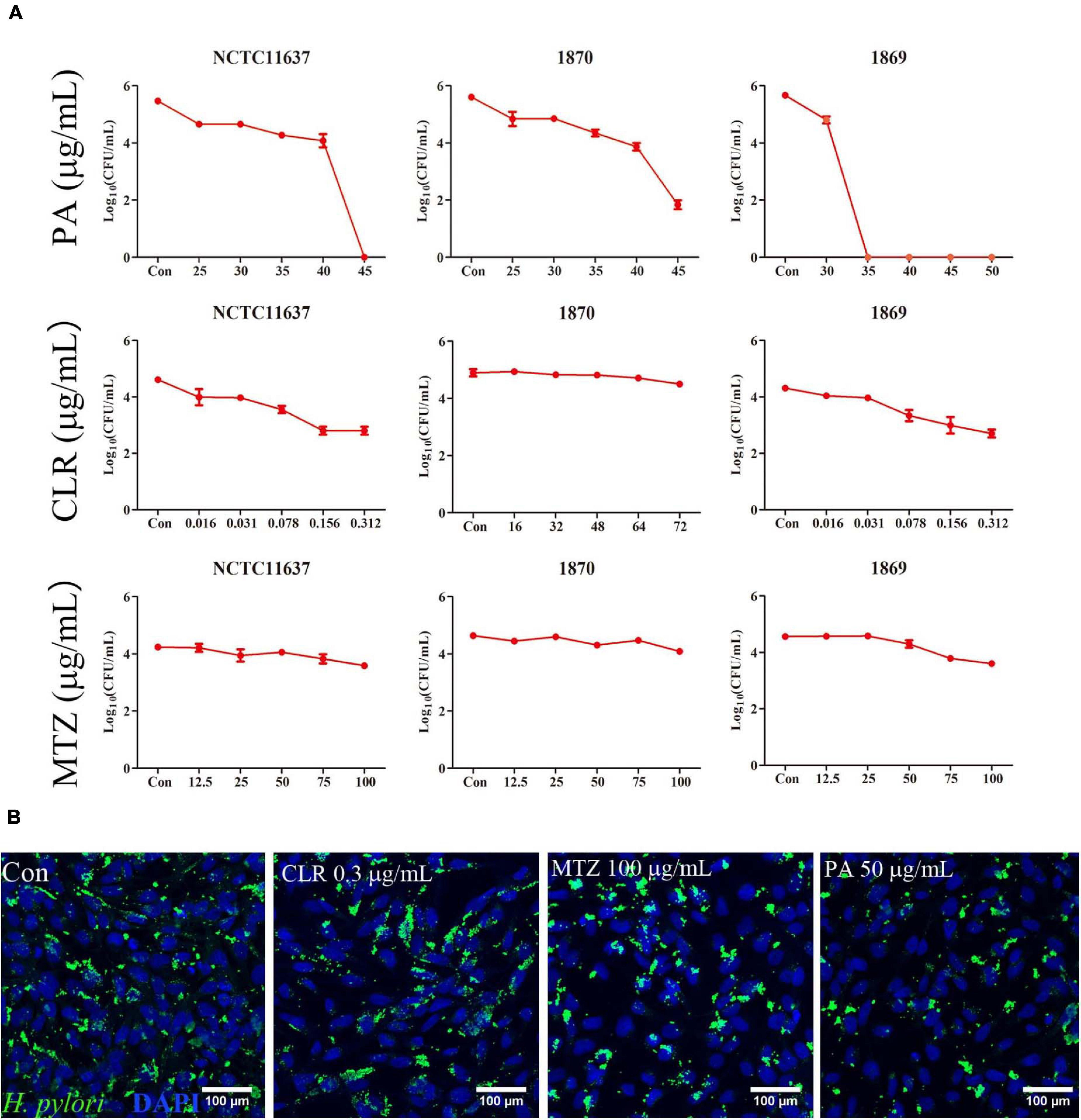
Figure 4. Killing ability for intracellular H. pylori. (A) Intracellular H. pylori standard strain NCTC11637 and drug-resistant strains Hp1869 and Hp1870 were treated with different doses of PA, CLR, and MTZ, and the number of bacteria was presented as CFU/ml (n = 3). Compared with the control group, a 99% decrease in the intracellular strain viability was defined as an effective extermination. (B) Representative confocal images by the immunofluorescence method to observe the ability of PA (2 MIC), CLR (20 MIC), and MTZ (25 MIC) to kill intracellular H. pylori NCTC11637; the bacteria were stained by anti- H. pylori antibody ab20459 (green).
Immunofluorescence was used for visualization to compare the killing ability of CLR, MTZ, and PA for intracellular the H. pylori strain NCTC11637 as shown in Figure 4B. Compared with that in the control group, the reduction of intracellular H. pylori (green area) was observed in 50 μg/ml PA. No significant changes were found in the other groups (0.3 μg/ml CLR and 100 μg/ml MTZ).
As shown in Figure 5A, intracellular H. pylori strains NCTC11637, Hp1869, and Hp1870 can be effectively eliminated by 50 and 75 μg/ml MTZ combined with 25 μg/ml PA. However, 25 μg/ml MTZ combined with 25 μg/ml PA can only effectively eliminate intracellular Hp1870.
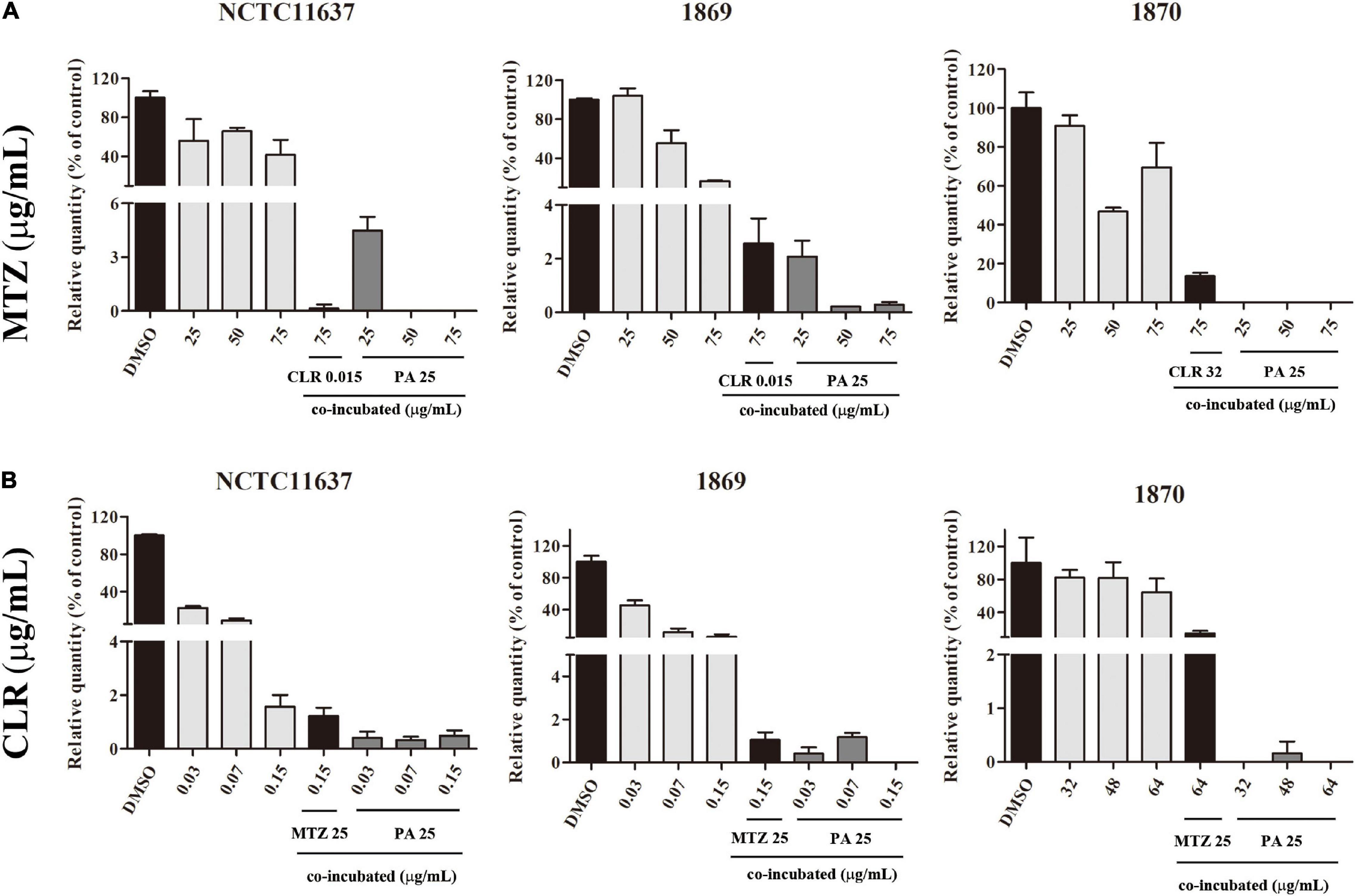
Figure 5. Combined effect of PA and antibiotics against intracellular H. pylori. (A) Killing ability of MTZ alone and in combination with PA (25 μg/ml) against the intracellular H. pylori standard strain NCTC11637 and drug-resistant strains Hp1869 and Hp1870 (n = 3). (B) Killing ability of CLR alone and in combination with PA (25 μg/ml) against intracellular the H. pylori standard strain NCTC11637 and drug-resistant strains Hp1869 and Hp1870 (n = 3). Compared with the control group, a 99% decrease in the intracellular strain viability was defined as an effective extermination.
As shown in Figure 5B, the killing ability of CLR combined with 25 μg/ml PA for intracellular H. pylori was enhanced for the standard strain NCTC11637 and drug-resistant strains Hp1869 and Hp1870 at multiple dosages.
For comparison, the combined use of the two antibiotics showed killing ability against the standard strain NCTC11637. For drug-resistant intracellular H. pylori, except for high-dose CLR combined with MTZ, which is effective for Hp1869, no effective killing was achieved in other cases.
The expression of each efflux pump gene in Hp1870 is shown in Figure 6A. The expression of hp0605 was significantly increased, whereas that of hp1327 was decreased with 32 μg/ml CLR treatment. PA addition at 12.5, 25, and 50 μg/ml significantly and concentration-dependently downregulated the expression of efflux pump genes including hp0605, hp1327, and hp1489.
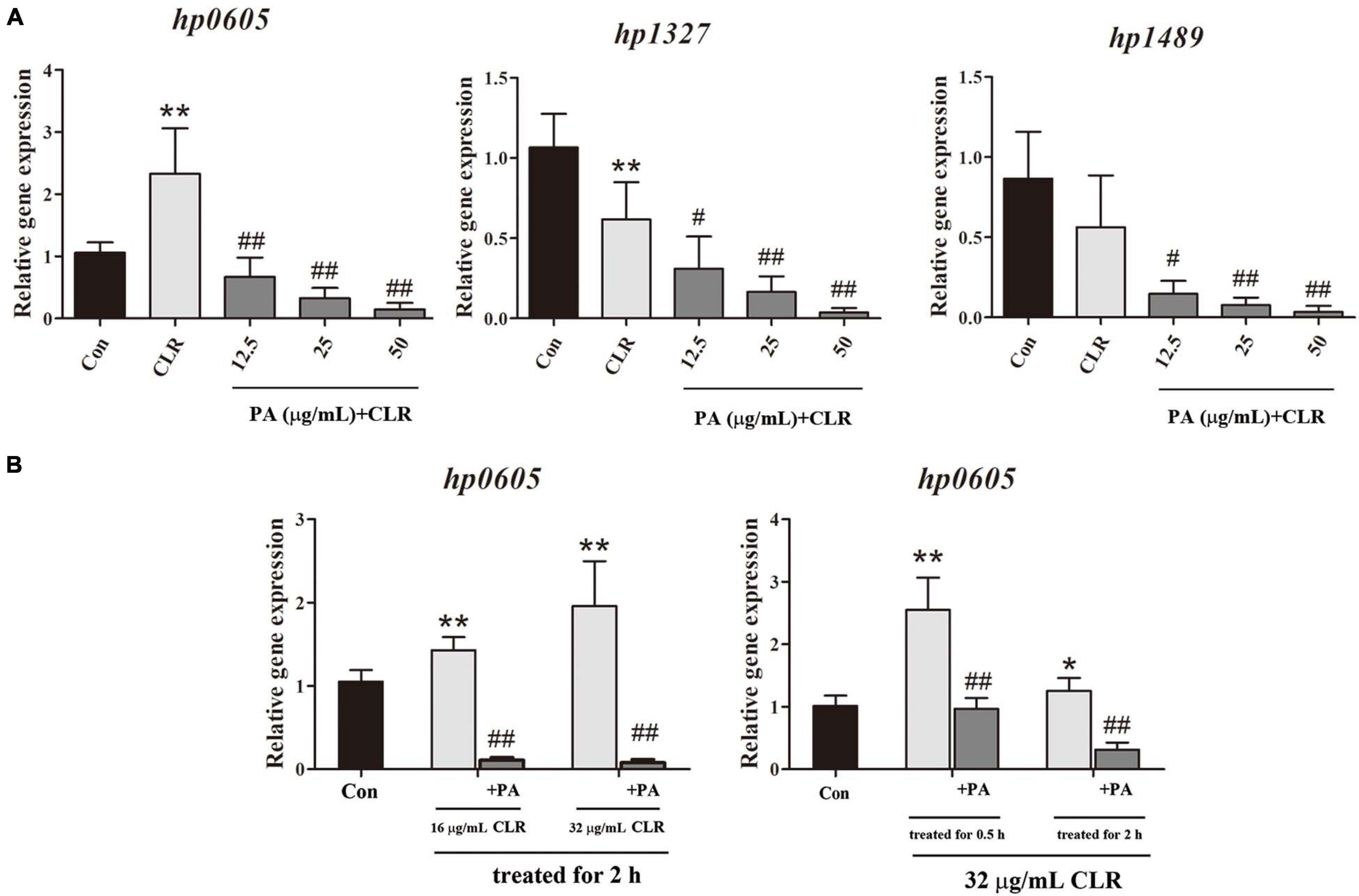
Figure 6. Expression of efflux effect gene. (A) Expression of different efflux effect homolog genes of Hp1870 after being incubated with DMSO, CLR, and PA. (B) Expression of hp0605 after being stimulated with different doses of CLR and PA for different times. The results are expressed as the mean ± SD (n = 6). *P < 0.05 and **P < 0.01, compared with the results for the control group; #P < 0.05 and ##P < 0.01, compared with the results for the model group.
As shown in Figure 6B, the effects of CLR on hp0605 expression at different times and doses were also investigated. After stimulation with the MIC of CLR for 0.5 and 2 h, the hp0605 expression was increased remarkably. In the dose-dependent experiment, 1/2 MIC and MIC of CLR were adopted to stimulate the bacteria for 2 h. The hp0605 expression in CLR groups was higher than that in the control group in a dose-dependent manner and was significantly reduced with PA treatment regardless of times or doses of the CLR stimulation.
The sequencing results of NCTC11637 and Hp1870 indicated that the drug-resistant H. pylori strain Hp1870 is a CLR A2143G mutant strain (Figure 7A). ddPCR was used to detect the rate of mutation of A2143G in each group, and the results are shown in Figure 7B. PA at 25 and 50 μg/ml combined with the MIC of CLR reduced the mutation rate of 23S rRNA in the A2143G position in Hp1870.
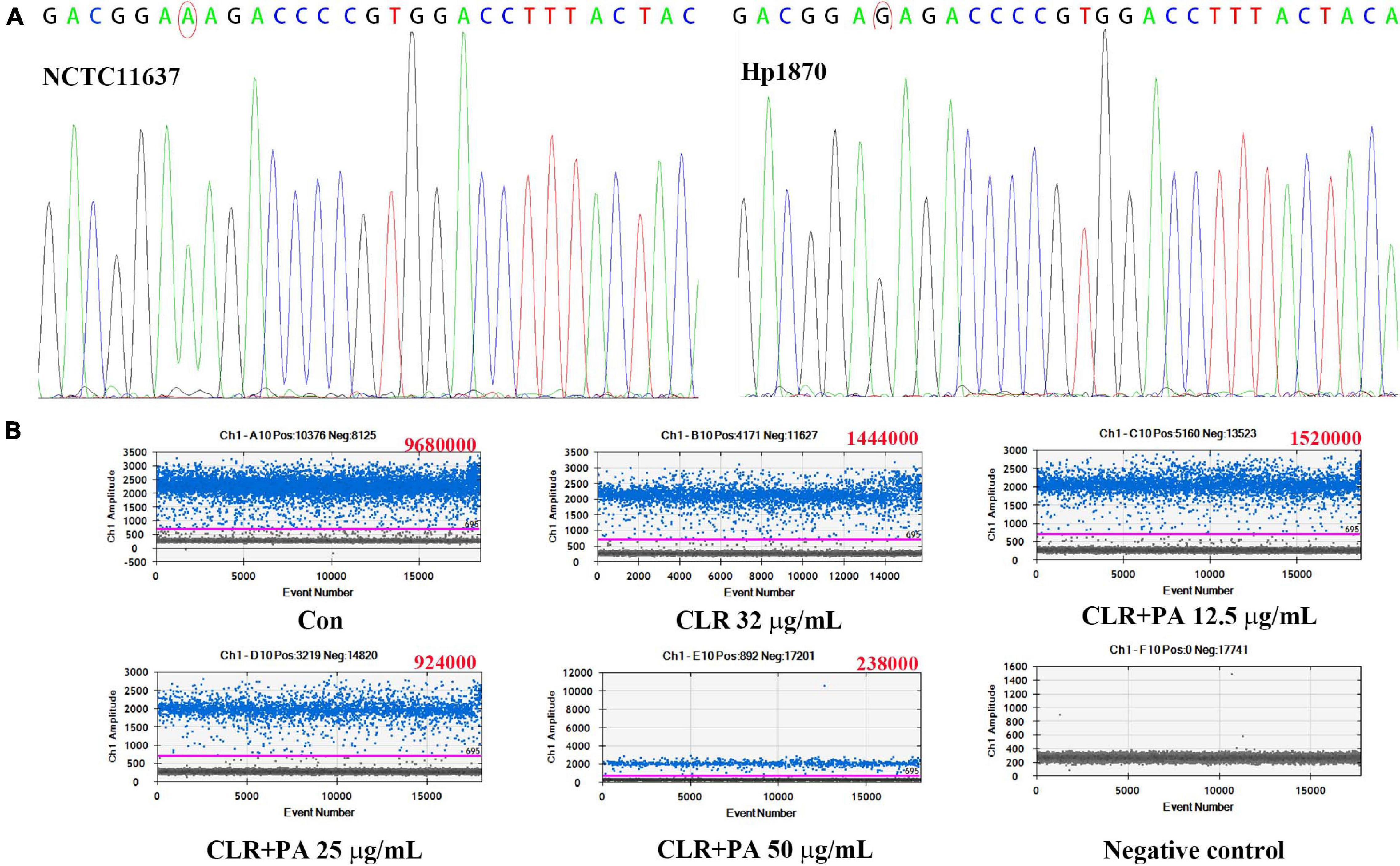
Figure 7. Determination of the mutation site and expression of the mutant gene in the drug-resistant strain Hp1870. (A) First-generation sequencing to determine the mutation of Hp1870. (B) Copy number of A2143G after being treated with DMSO, CLR, and PA. The blue droplets were droplets positive for A2143G, and the copy number value is shown in the upper right corner of each figure.
The methodology of cellular uptake and transport assay was examined, and the results were presented in the Supplementary Materials. Figure 8A indicates that the uptake of PA by Caco-2 cells increased with concentration, time, and temperature and was significantly increased in acidic conditions but not in alkaline conditions as shown in Figure 8B. Table 4 and Figures 8C,D suggested that PA is rapidly absorbed, and its main absorption mechanism is possibly passive transport.
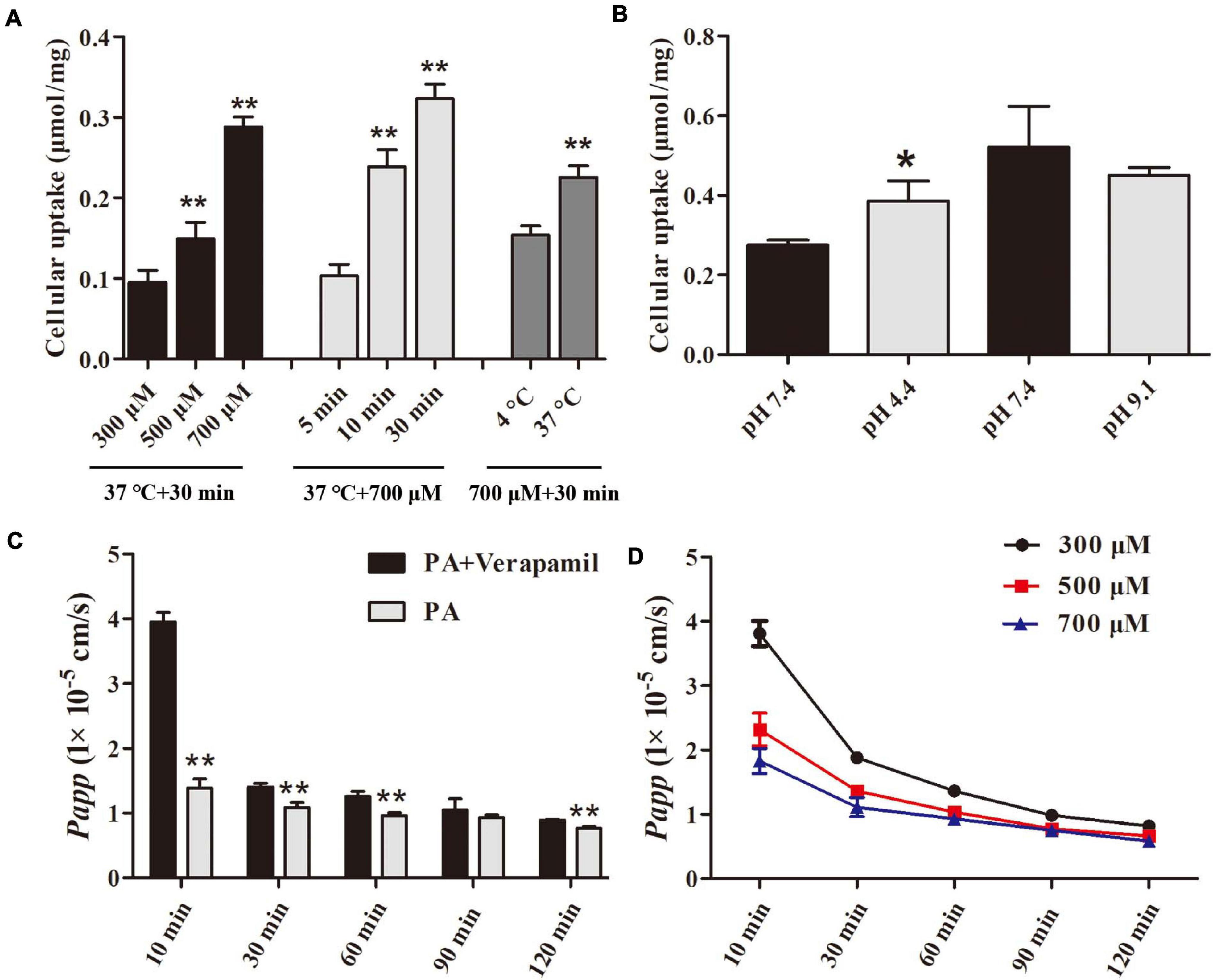
Figure 8. Uptake and transport of PA in Caco-2 monolayers. (A, B) Effect of concentrations, time, temperature, and pH on uptake of PA by Caco-2 cells. (C, D) Effect of time, efflux transporter inhibitor, and concentrations on transport of PA by Caco-2 cells. The data were expressed as the mean ± SD (n = 4). *P < 0.05 and **P < 0.01, compared with control group.
Since antibiotics have been discovered and used to treat infectious diseases in the 20th century, many diseases caused by bacterial infections have been effectively controlled. However, with the indiscriminate use of antibiotics, bacteria developed tolerance to antibiotics and then drug resistance (Levin-Reisman et al., 2017). Bacterial resistance has become a global health issue, which can be solved by developing new antimicrobial agents or reducing bacterial resistance (Alekshun and Levy, 2007). As a commonly resistant specie, H. pylori has a high degree of genetic variability and is prone to produce antibiotic-resistant genes (Wang et al., 2005). This species has a strong resistance to various antibiotics, which often leads to the failure of eradication. Hence, patients have to undergo multiple subsequent treatments, thus increasing the cost and the duration of treatment (Fallone et al., 2019). When the first treatment fails, the strain in the patient is prone to develop resistance after exposure to antibiotics, leading to the increased difficulty of follow-up treatments (Lee et al., 2019).
Common drug-resistant antibiotics of H. pylori include MTZ and CLR. In vitro checkerboard method and time–kill curve experiments showed that a combination with PA can decrease the MIC and enhance the bactericidal efficacy of CLR. These results suggest that PA may increase the sensitivity of H. pylori to CLR. Some studies have suggested that FICI can show synergistic or additive effects when combined with drugs that have the same antibacterial or bactericidal mechanisms (Abstract Culp et al., 2020). Therefore, PA may disturb the protein synthesis of H. pylori or interfere with the resistant mechanism of CLR resistance to H. pylori. The influence of PA on A2143G could be the mechanism improving the sensibility of H. pylori to CLR.
In clinical practice, antibiotic dosage selection should consider efficacy and patient compliance (Suzuki et al., 2020). According to the experiments with low doses of antibiotics to stimulate H. pylori, the use of inappropriate doses of CLR to eliminate H. pylori will increase the number of intracellular H. pylori. Consequently, the usage of CLR should be considered cautiously in clinical treatment. The cellular internalization of H. pylori is one of the strategies for its drug resistance development (Capurro et al., 2019). When PA was combined with CLR for treatment, the amount of H. pylori invading into the cells could be effectively reduced. Additional experiments should be conducted to determine the mechanism. Follow-up experiments should try more antibiotics to summarize the relationship between bacterial internalization and antibiotics.
H. pylori is generally an extracellular, noninvasive bacterium (Capurro et al., 2020); however, evidence suggests that it can alter the tight cellular connections between gastric epithelial cells through complex processes, allowing it to hide in an intracellular niche for protection from antibiotic eradication therapy and thus endowing it with persistence and recolonization. Gentamicin protection experiments showed that several treatments of MIC doses of CLR and MTZ did not have sufficient ability to kill intracellular H. pylori (Sisto et al., 2016). However, the intracellular H. pylori can be effectively killed by PA in low MIC. When the antibiotic is combined with PA, it showed a bactericidal ability against intracellular H. pylori. In the assay of killing intracellular H. pylori, the maximum antibiotic dose was determined based on the antibiotic solubility and cytotoxicity. Although the immunofluorescence method provided visualized images, the anti- H. pylori antibody did not distinguish between dead and live bacteria, and some dead adhered H. pylori cannot be washed off by PBS. Therefore, a small number of dead bacteria were still stained with fluorescence, leading to inconsistent results of gentamycin protection assay.
The efflux pump of bacteria is one of the main mechanisms of bacterial resistance (Bergmiller et al., 2017; Nolivos et al., 2019). Studies on E. coli efflux pumps proved that the RND efflux pumps of Gram-negative bacteria mainly belong to the TolC-AcrAB and OprM-MexAB systems (Li et al., 2015). H. pylori has four TolC-AcrAB homologs (HP0605-HP0607, HP0971-HP0969, HP1327-HP1329, and HP1489-HP1487) (van Amsterdam et al., 2005). With the stimulation of CLR in MIC dose, the efflux pump genes were significant in Hp1870 but not in NCTC11637, which suggests that the efflux pump plays an important role in drug-resistant H. pylori strains. In Hp1870, the expression of hp0605 was significantly increased, whereas those of other homologue genes were decreased. This phenomenon may be related to the main role of the HP0605-HP0607 homolog in Asian H. pylori strains (Monteiro et al., 2000). Nonetheless, the expression of each efflux pump gene was decreased when PA was used. In addition, the expression of the A2143G mutant gene was also decreased by PA treatment. In further research, CLR was used to induce A2143G mutation in the standard strain NCTC11637 to observe the effect of PA on the resistant mutation rate. However, the mutant strain was not successfully produced, and the related mechanism and research need to be further clarified.
The Caco-2 monolayer is applied to preclinical investigations to predict the gastrointestinal permeability of drugs because it expresses transporters, microvilli, and enterocytes of identical characteristics to humans (Ponce de Leon-Rodriguez et al., 2019). The FDA has recommended this model as an integral component of the Biopharmaceutics Classification System (BCS). Therefore, the Caco-2 cell line was chosen as a model to explore the uptake and transport of PA. Verapamil is a classical P-gp transporter inhibitor (Aller et al., 2009). In the verapamil-related transport experiment, the Papp of PA was increased with verapamil treatment, indicating that the P-gp efflux transporter contributes to PA absorption. PA was rapidly absorbed by the cells, which is beneficial for killing intracellular H. pylori. In addition, the rapid absorption rate of PA was observed in acidic conditions, suggesting that the acidic condition in the stomach could promote the permeability of human cells to PA.
On the basis of the above results and the common drug resistance mechanism of H. pylori, the combination of PA and CLR showed a highly efficient bactericidal ability against H. pylori whether in vitro or in vivo. The inhibited expression of the efflux pump gene, A2143G mutation, and high PA absorption by cells contributed to the combined effect. The results reveal that H. pylori invasion is promoted by the inappropriate use of CLR and thus must be considered in clinical treatment. In conclusion, PA can act additively with CLR against H. pylori, indicating its potential as a candidate medication for drug-resistant H. pylori.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
YZ, LT, and QD performed the experiments and wrote the manuscript. LJ revised the manuscript. JZ and YZ summarized and analyzed the data. FY and YO carried out the anti-intracellular H. pylori assay. SG and BH were responsible for the management and coordination of the research activity planning and execution. HC, PH, and YX designed the study and finally revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (no. 81873074); Guangdong Basic and Applied Basic Research Foundation (2019A1515110742); and the Guangdong Provincial Key Research and Development Plan (2020B1111100011).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.674560/full#supplementary-material
Abstract Culp, E. J., Waglechner, N., Wang, W., Fiebig-Comyn, A. A., Hsu, Y. P., Koteva, K., et al. (2020). Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature 578, 582–587. doi: 10.1038/s41586-020-1990-9
Alba, C., Blanco, A., and Alarcon, T. (2017). Antibiotic resistance in Helicobacter pylori. Curr. Opin. Infect. Dis. 30, 489–497.
Alekshun, M. N., and Levy, S. B. (2007). Molecular mechanisms of antibacterial multidrug resistance. Cell 128, 1037–1050. doi: 10.1016/j.cell.2007.03.004
Aller, S. G., Yu, J., Ward, A., Weng, Y., Chittaboina, S., Zhuo, R., et al. (2009). Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323, 1718–1722. doi: 10.1126/science.1168750
Ansari, S., and Yamaoka, Y. (2017). Survival of Helicobacter pylori in gastric acidic territory. Helicobacter 22. doi: 10.1111/hel.12386
Behrens, I.-K., Busch, B., Ishikawa-Ankerhold, H., Palamides, P., Shively, J. E., Stanners, C., et al. (2020). The HopQ-CEACAM interaction controls CagA translocation, phosphorylation, and phagocytosis of in neutrophils. mBio 11:e03256-19. doi: 10.1128/mBio.03256-19
Bergmiller, T., Andersson, A. M. C., Tomasek, K., Balleza, E., Kiviet, D. J., Hauschild, R., et al. (2017). Biased partitioning of the multidrug efflux pump AcrAB-TolC underlies long-lived phenotypic heterogeneity. Science 356, 311–315. doi: 10.1126/science.aaf4762
Capurro, M. I., Greenfield, L. K., Prashar, A., Xia, S., Abdullah, M., Wong, H., et al. (2019). VacA generates a protective intracellular reservoir for Helicobacter pylori that is eliminated by activation of the lysosomal calcium channel TRPML1. Nat. Microbiol. 4, 1411–1423. doi: 10.1038/s41564-019-0441-6
Capurro, M. I., Prashar, A., and Jones, N. L. (2020). MCOLN1/TRPML1 inhibition–a novel strategy used by Helicobacter pylori to escape autophagic killing and antibiotic eradication therapy in vivo. Autophagy 16, 169–170. doi: 10.1080/15548627.2019.1677322
Choi, I. J., Kim, C. G., Lee, J. Y., Kim, Y. I., Kook, M. C., Park, B., et al. (2020). Family history of gastric cancer and Helicobacter pylori treatment. N. Engl. J. Med. 382, 427–436.
Crowe, S. E., and Solomon, C. G. (2019). Helicobacter pylori infection. N. Engl. J. Med. 380, 1158–1165.
De Francesco, V., Zullo, A., Fiorini, G., Saracino, I. M., Pavoni, M., and Vaira, D. (2019). Role of MIC levels of resistance to clarithromycin and metronidazole in Helicobacter pylori eradication. J. Antimicrob. Chemother. 74, 772–774. doi: 10.1093/jac/dky469
de Martel, C., Georges, D., Bray, F., Ferlay, J., and Clifford, G. M. (2020). Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob. Health 8, e180–e190.
Doern, C. D. (2014). When does 2 plus 2 equal 5? A review of antimicrobial synergy testing. J. Clin. Microbiol. 52, 4124–4128. doi: 10.1128/jcm.01121-14
El Meouche, I., and Dunlop, M. J. (2018). Heterogeneity in efflux pump expression predisposes antibiotic-resistant cells to mutation. Science 362, 686–690. doi: 10.1126/science.aar7981
Fallone, C. A., Moss, S. F., and Malfertheiner, P. (2019). Reconciliation of recent Helicobacter pylori treatment guidelines in a time of increasing resistance to antibiotics. Gastroenterology 157, 44–53. doi: 10.1053/j.gastro.2019.04.011
Ford, A. C., Yuan, Y., and Moayyedi, P. (2020). Helicobacter pylori eradication therapy to prevent gastric cancer: systematic review and meta-analysis. Gut 69, 2113–2121. doi: 10.1136/gutjnl-2020-320839
Garcia-Fernandez, E., Koch, G., Wagner, R. M., Fekete, A., Stengel, S. T., Schneider, J., et al. (2017). Membrane microdomain disassembly inhibits MRSA antibiotic resistance. Cell 171, 1354–1367.e20.
Gerdes, K., and Semsey, S. (2016). Microbiology: pumping persisters. Nature 534, 41–42. doi: 10.1038/nature18442
Gerrits, M. M., van Vliet, A. H. M., Kuipers, E. J., and Kusters, J. G. (2006). Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect. Dis. 6, 699–709. doi: 10.1016/s1473-3099(06)70627-2
Gong, Y., and Yuan, Y. (2018). Resistance mechanisms of Helicobacter pylori and its dual target precise therapy. Crit. Rev. Microbiol. 44, 371–392. doi: 10.1080/1040841x.2017.1418285
Jung, D. H., Kim, J. H., Jeong, S. J., Park, S. Y., Kang, I. M., Lee, K. H., et al. (2018). Peptide nucleic acid probe-based analysis as a new detection method for clarithromycin resistance in Helicobacter pylori. Gut Liver 12, 641–647. doi: 10.5009/gnl18111
Krzyzek, P., Paluch, E., and Gosciniak, G. (2020). Synergistic therapies as a promising option for the treatment of antibiotic-resistant Helicobacter pylori. Antibiotics (Basel) 9:658. doi: 10.3390/antibiotics9100658
Lee, J. W., Kim, N., Nam, R. H., Lee, S. M., Kwon, Y. H., Sohn, S. D., et al. (2019). Favorable outcomes of culture-based Helicobacter pylori eradication therapy in a region with high antimicrobial resistance. Helicobacter 24:e12561. doi: 10.1111/hel.12561
Levin-Reisman, I., Ronin, I., Gefen, O., Braniss, I., Shoresh, N., and Balaban, N. Q. (2017). Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830. doi: 10.1126/science.aaj2191
Li, X. Z., Plesiat, P., and Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/cmr.00117-14
Lian, D. W., Xu, Y. F., Ren, W. K., Fu, L. J., Chen, F. J., Tang, L. Y., et al. (2018). Unraveling the novel protective effect of patchouli alcohol against Helicobacter pylori-induced gastritis: insights into the molecular mechanism in vitro and in vivo. Front. Pharmacol. 9:1347.
Liao, J.-B., Liang, Y.-Z., Chen, Y.-L., Xie, J.-H., Liu, W.-H., Chen, J. N., et al. (2015). Novel patchouli alcohol ternary solid dispersion pellets prepared by poloxamers. Iran. J. Pharm. Res. 14, 15–26.
Liou, J. M., Malfertheiner, P., Lee, Y. C., Sheu, B. S., Sugano, K., Cheng, H. C., et al. (2020). Screening and eradication of Helicobacter pylori for gastric cancer prevention: the Taipei global consensus. Gut 69, 2093–2112.
Liu, W. Z., Xie, Y., Lu, H., Cheng, H., Zeng, Z. R., Zhou, L. Y., et al. (2018). Fifth Chinese national consensus report on the management of Helicobacter pylori infection. Helicobacter 23:e12475. doi: 10.1111/hel.12475
Marques, A. T., Vitor, J. M. B., Santos, A., Oleastro, M., and Vale, F. F. (2020). Trends in Helicobacter pylori resistance to clarithromycin: from phenotypic to genomic approaches. Microb. Genom. 6:e000344.
Matta, A. J., Zambrano, D. C., and Pazos, A. J. (2018). Punctual mutations in 23S rRNA gene of clarithromycin-resistant Helicobacter pylori in Colombian populations. World J. Gastroenterol. 24, 1531–1539.
Monteiro, M. A., Zheng, P., Ho, B., Yokota, S., Amano, K., Pan, Z., et al. (2000). Expression of histo-blood group antigens by lipopolysaccharides of Helicobacter pylori strains from asian hosts: the propensity to express type 1 blood-group antigens. Glycobiology 10, 701–713. doi: 10.1093/glycob/10.7.701
Nolivos, S., Cayron, J., Dedieu, A., Page, A., Delolme, F., and Lesterlin, C. (2019). Role of AcrAB-TolC multidrug efflux pump in drug-resistance acquisition by plasmid transfer. Science 364, 778–782. doi: 10.1126/science.aav6390
Ponce de Leon-Rodriguez, M. D. C., Guyot, J. P., and Laurent-Babot, C. (2019). Intestinal in vitro cell culture models and their potential to study the effect of food components on intestinal inflammation. Crit. Rev. Food Sci. Nutr. 59, 3648–3666. doi: 10.1080/10408398.2018.1506734
Salama, N. R., Hartung, M. L., and Muller, A. (2013). Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat. Rev. Microbiol. 11, 385–399. doi: 10.1038/nrmicro3016
Seo, S. I., Do, B. J., Kang, J. G., Kim, H. S., Jang, M. K., Kim, H. Y., et al. (2019). Helicobacter pylori eradication according to sequencing-based 23S ribosomal RNA point mutation associated with clarithromycin resistance. J. Clin. Med. 9:54. doi: 10.3390/jcm9010054
Sisto, F., Scaltrito, M. M., Masia, C., Bonomi, A., Cocce, V., Marano, G., et al. (2016). In vitro activity of artemisone and artemisinin derivatives against extracellular and intracellular Helicobacter pylori. Int. J. Antimicrob. Agents 48, 101–105. doi: 10.1016/j.ijantimicag.2016.03.018
Sun, L., Talarico, S., Yao, L., He, L., Self, S., You, Y., et al. (2018). Droplet digital PCR-based detection of clarithromycin resistance in Helicobacter pylori isolates reveals frequent heteroresistance. J. Clin. Microbiol. 56:e00019-18.
Suzuki, S., Gotoda, T., Kusano, C., Ikehara, H., Ichijima, R., Ohyauchi, M., et al. (2020). Seven-day vonoprazan and low-dose amoxicillin dual therapy as first-line Helicobacter pylori treatment: a multicentre randomised trial in Japan. Gut 69, 1019–1026. doi: 10.1136/gutjnl-2019-319954
Thung, I., Aramin, H., Vavinskaya, V., Gupta, S., Park, J. Y., Crowe, S. E., et al. (2016). Review article: the global emergence of Helicobacter pylori antibiotic resistance. Aliment. Pharmacol. Ther. 43, 514–533.
Tsukamoto, T., Nakagawa, M., Kiriyama, Y., Toyoda, T., and Cao, X. (2017). Prevention of gastric cancer: eradication of Helicobacter Pylori and beyond. Int. J. Mol. Sci. 18:1699. doi: 10.3390/ijms18081699
van Amsterdam, K., Bart, A., and van der Ende, A. (2005). A Helicobacter pylori TolC efflux pump confers resistance to metronidazole. Antimicrob. Agents Chemother. 49, 1477–1482. doi: 10.1128/aac.49.4.1477-1482.2005
Vazquez-Laslop, N., and Mankin, A. S. (2018). How macrolide antibiotics work. Trends Biochem. Sci. 43, 668–684. doi: 10.1016/j.tibs.2018.06.011
Wang, G., Alamuri, P., Humayun, M. Z., Taylor, D. E., and Maier, R. J. (2005). The Helicobacter pylori MutS protein confers protection from oxidative DNA damage. Mol Microbiol. 58, 166–176. doi: 10.1111/j.1365-2958.2005.04833.x
Wang, Y. H., Lv, Z. F., Zhong, Y., Liu, D. S., Chen, S. P., and Xie, Y. (2017). The internalization of Helicobacter pylori plays a role in the failure of H. pylori eradication. Helicobacter 22. doi: 10.1111/hel.12324
Keywords: Helicobacter pylori, patchouli alcohol, resistance, clarithromycin, intracellular
Citation: Zhong Y, Tang L, Deng Q, Jing L, Zhang J, Zhang Y, Yu F, Ou Y, Guo S, Huang B, Cao H, Huang P and Xu Y (2021) Unraveling the Novel Effect of Patchouli Alcohol Against the Antibiotic Resistance of Helicobacter pylori. Front. Microbiol. 12:674560. doi: 10.3389/fmicb.2021.674560
Received: 01 March 2021; Accepted: 20 April 2021;
Published: 02 June 2021.
Edited by:
Eleftherios Mylonakis, Warren Alpert Medical School at Brown University, United StatesReviewed by:
Grażyna Gościniak, Wrocław Medical University, PolandCopyright © 2021 Zhong, Tang, Deng, Jing, Zhang, Zhang, Yu, Ou, Guo, Huang, Cao, Huang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Huang, aHBpbmdAZ3p1Y20uZWR1LmNu; Yifei Xu, MzMwODUxNjM5QHFxLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.