- 1State Key Laboratory of Biobased Material and Green Papermaking, Shandong Academy of Sciences, Qilu University of Technology, Jinan, China
- 2School of Bioengineering, Shandong Academy of Sciences, Qilu University of Technology, Jinan, China
- 3Shandong Provincial Key Laboratory of Microbial Engineering, Shandong Academy of Sciences, Qilu University of Technology, Jinan, China
- 4Haixia Institute of Science and Technology, Fujian Agriculture and Forestry University, Fuzhou, China
- 5State Key Laboratory of Microbial Technology, School of Life Sciences, National Glycoengineering Research Center, Shandong University, Qingdao, China
Homeodomain-containing transcription factors (Htfs) play important roles in animals, fungi, and plants during some developmental processes. Here, a homeodomain-containing transcription factor PoHtf1 was functionally characterized in the cellulase-producing fungi Penicillium oxalicum 114-2. PoHtf1 was shown to participate in colony growth and conidiation through regulating the expression of its downstream transcription factor BrlA, the key regulator of conidiation in P. oxalicum 114-2. Additionally, PoHtf1 inhibited the expression of the major cellulase genes by coordinated regulation of cellulolytic regulators CreA, AmyR, ClrB, and XlnR. Furthermore, transcriptome analysis showed that PoHtf1 participated in the secondary metabolism including the pathway synthesizing conidial yellow pigment. These data show that PoHtf1 mediates the complex transcriptional-regulatory network cascade between developmental processes and cellulolytic gene expression in P. oxalicum 114-2. Our results should assist the development of strategies for the metabolic engineering of mutants for applications in the enzymatic hydrolysis for biochemical production.
Introduction
Penicillium oxalicum 114-2 produces diverse lignocellulolytic enzymes, which have been widely used in the effective degradation of agricultural biomass. In the research for hyper-production of cellulases, P. oxalicum 114-2 has been used as a model fungus to understand the cellulase regulation systems. A single-gene disruption library of 470 transcription factors has been constructed, and 20 main transcription factors that play putative roles in the activation or repression of cellulase synthesis were identified (Li et al., 2015). PDE_07199 is a cellulase transcription factor involved in both the regulation of cellulase expression and developmental process of P. oxalicum 114-2. However, its functional mechanism has not been studied systematically in P. oxalicum 114-2.
PDE_07199 amino acid sequence analysis using the Simple Modular Architecture Research Tool (SMART) (Letunic et al., 2021) showed the presence of a homeodomain. Homeodomain-containing proteins have been proved to function as transcription factors and play important roles in animals, fungi, and plants during some developmental processes, such as development and differentiation (Burglin, 2011; Miksiunas et al., 2020). In fungi, homeodomain transcriptional factors (Htfs) play a crucial role in regulating developmental processes. pah1 from Podospora anserina was the first homeobox gene identified in filamentous ascomycetes and was considered to be a repressor of genes involved in the conidiation process. Moreover, pah1 was also involved in hyphal branching and possibly in the development of female organs (Arnaise et al., 2001). In Neurospora crassa, the homolog of pah1, kal-1, was uncovered as an important regulator of asexual growth and development. kal-1 mutation lead to substantial changes in colony morphology and conidial development (Colot et al., 2006). In the ascomycete fungus Magnaporthe oryzae, eight Htfs were characterized, each of which functions as a stage-specific regulator for conidial shape, hyphal growth, conidiation, appressorium development, and invasive growth during M. oryzae development (Kim et al., 2009; Liu et al., 2010). Zheng et al. (2012) reported that a conserved Htf1 is required for phialide development and conidiogenesis in Fusarium species. Moreover, Htfs are also involved in fruiting body development in several members of mushroom forming fungi, including Schizophyllum commune and Volvariella volvacea straw mushroom (Pelkmans et al., 2017; Wang et al., 2019). Aspergillus nidulans is a model filamentous fungus that is commonly used for understanding fungal development. Several transcription factors are involved in conidiation in A. nidulans including BrlA, FluG, and FlbA-E (Lee and Adams, 1996; Emri et al., 2005; Kwon et al., 2010a, b; Arratia-Quijada et al., 2012; Chang et al., 2012; Oiartzabal-Arano et al., 2015). Recently, two homeodomain proteins, HbxA and HbxB, were functionally characterized in A. nidulans. And the two proteins play crucial roles in the conidiophore production and secondary metabolism of A. nidulans (Son et al., 2020).
Previously reported Htfs play regulatory roles during different developmental stages in fungi. In P. oxalicum 114-2, a homeodomain-containing protein called PDE_07199 (PoHtf1) was identified. PoHtf1 is both crucial for development and in the regulation of cellulase expression. Our results showed that PoHtf1 mediates the complex transcriptional-regulatory network cascade between developmental processes and cellulolytic gene expression.
Materials and Methods
Strains and Culture Conditions
The P. oxalicum 114-2 (CGMCC 5302) wild-type (WT) strain was stored in our laboratory. Spores of P. oxalicum strains were cultured on wheat bran agar medium at 30°C for 4 days and harvested using sterile water. The final concentration was over 1010 spores/mL. For cellulase production, 108/mL spores were inoculated in Vogel’s salts liquid medium containing 2% glucose and incubated at 30°C with shaking at 200 rpm for 24 h. Then, the mycelia were collected by vacuum pump filtration. Exactly 0.5 g of mycelia was transferred into 50 mL of cellulase production medium containing Vogel’s salts liquid medium, 1% wheat bran, and 1% microcrystalline cellulose. Cellulase production was performed in a 300 mL flask at 30°C with shaking at 200 rpm.
Construction of Pohtf1 Deletion and Complement Strains
Pohtf1 gene deletion was performed using the homologous recombination method. The up- and down-stream homologous flanks of Pohtf1 were amplified from P. oxalicum 114-2 genomic DNA with primers 7199-F1/7199-ptraR and 7199-ptraF/7199-R1. The selected marker gene, ptrA, was amplified from the pME2892 plasmid with ptrA-F1/ptrA-R1 primers. The three fragments were fused using double-joint PCR (Yu et al., 2004), and the full-length deletion cassette was amplified by primer 7199-F2/7199-R2. The deletion cassette was transformed into P. oxalicum 114-2 using the PEG-mediated method (Li et al., 2010) to obtain the Pohtf1 gene knock-out strain ΔPohtf1.
For Pohtf1 gene complementation, the integrated expression cassette was amplified from P. oxalicum 114-2 genomic DNA with primers 7199-hphF/7199-hphR. Hygromycin resistance gene (hph) was amplified from the pSilent-1 plasmid using primers hph-F/hph-R. The two fragments were fused, and the complement cassette was amplified using primers 7199-F4/7199-R4. The cassette was transformed into strain ΔPohtf1 as described above, and the complement strain CPohtf1 was constructed.
Transformants were analyzed using full-length amplifying primers. All primers used in the construction of strains ΔPohtf1 and CPohtf1 are listed in Supplementary Table 1.
Transcriptome Analysis
For transcriptome analysis, 108/mL spores were inoculated in Vogel’s salts liquid medium containing 2% glucose and incubated at 30°C with shaking at 200 rpm for 24 h. Then, the mycelia were collected by vacuum pump filtration and transferred into Vogel’s salts liquid medium lacking a carbon source. After 2 h incubation, the mycelia were collected and 0.5 g of mycelia were transferred into 50 mL of Vogel’s salts liquid medium including 2% microcrystalline cellulose and incubated at 30°C with shaking at 200 rpm for 4 h. The RNA of P. oxalicum 114-2 and ΔPohtf1 was extracted using TRIzol reagent (Invitrogen, United States) according to the manufacturer’s protocol. Digital gene expression profiling experiments, based on RNA-Seq, were performed using the Illumina HiSeq 2000 System (Beijing Genomics Institute, China). Gene expression levels were normalized to reads per kb per million reads (RPKM). Significantly differentially expressed genes were filtered with combined thresholds using a false discovery rate (FDR) ≤ 0.001 and fold change ≥ 2. The raw RNA-Seq data were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) with the series reference number GSE160881.
Phenotype Analysis
Phenotype analysis was performed on Vogel’s medium plates containing 2% glucose or 1% cellulose and on 10% wheat bran medium plates (Li et al., 2015). Exactly 1 μL of fresh spores with concentration of 108 spores/mL was inoculated into the center of the plates and incubated for 3 days at 30°C.
Spore production capacities of P. oxalicum 114-2 and its mutants were measured on Vogel’s medium plates containing 2% glucose. Exactly 250 μL of fresh spores with concentration of 108 spores/mL was spread across the plates. The plates were incubated at 30°C, and the spore numbers in a fixed area were counted at 24, 30, 36, 48, 60, and 72 h.
Enzyme Activities Assay
For enzyme activities assay, strains P. oxalicum 114-2, ΔPohtf1 and CPohtf1 were fermented in the cellulase production medium under the conditions described above for 6 days. Fermentation broths were sampled and measured every 24 h from day 3 to day 6. WhatmanTM 1 filter papers, sodium carboxymethyl cellulose (CMC-Na, Sigma), p-nitrophenyl-β-D-cellobioside (pNPC, Sigma), and p-nitrophenyl-β-D-glucopyranoside (pNPG, Sigma) were used as substrates for filter paper activity (FPA), endoglucanase, cellobiohydrolase, and beta-glucosidase activities assays, respectively (Chen et al., 2013). One unit of enzyme activity was defined as the amount of enzyme required to produce 1 μmol glucose or p-nitrophenyl (pNP, Sigma) per minute under the assayed conditions.
Real-Time Quantitative PCR Analysis
For Real-time quantitative PCR (qRT-PCR), 0.5 g preincubated mycelia of the three strains was induced in 50 mL of cellulase production medium for 4 h. Then, the mycelia were collected for RNA extraction as previously described (Li et al., 2015). The synthesis of cDNA and qRT-PCR reaction were performed using the PrimeScriptTM RT reagent kit with gDNA Eraser (Perfect Real Time) and TB Green® Premix Ex TaqTM II (Tli RNaseH Plus) (TAKARA) following the manufactures protocols. qRT-PCR reaction procedure was performed on the LightCycler®480 System (Roche) with cycling conditions of: 95°C for 2 min, and 40 cycles of 95°C for 10 s, and 61°C for 30 s. The melting curves were measured with a temperature gradient of 0.1°C per second from 65°C to 95°C. The expression levels of all genes were calculated using the method of relative quantification using actin as the reference gene. Primers used for qRT-PCR are listed in Supplementary Table 2. Three biological replicates were performed for all reactions.
Results
Sequence and Phylogenetic Analysis of PoHtf1
The PoHtf1 amino acid sequence was analyzed by SMART. A homeodomain of 63 amino acids (amino acid 67–129) was identified in the PoHtf1 sequence. Homeodomains proteins performs its regulatory function by binding DNA through the helix-turn-helix structure (Dorn et al., 1994). The PoHtf1 homologs of PoHtf1 were identified using the Basic Local Alignment Search Tool (Blast) in the National Center of Biotechnology Information (NCBI) database. The PoHtf1 amino acid sequence was closely related to those of other Penicillium species (Figure 1A). PoHtf1 shared the highest identity of 67.37% with the P. brasilianum homolog (OOQ90489.1), and shared identities of 41.51 and 45.35% with the A. nidulans homolog, HbxA (AN1217.2, XP_658821.1) and Trichoderma reesei QM6a homolog (XP_006963962.1), respectively. However, PoHtf1 was distantly related to several reported Htfs that function in developmental processes in filamentous fungi. PoHtf1 only shared identities of 17 and 18% with the first identified Htf in P. anserine, Pah1 (CAC16792.1) and its homolog in N. crassa OR74a, Kal-1 (NCU03593, EAA32084.1), respectively. Htfs from M. oryzae (MGG_00184, XP_003718936) and F. graminearum (FGSG_07097, XP_011326799.1), which are involved in hyphal growth and conidiogenesis (Liu et al., 2010; Zheng et al., 2012), shared 40.94 and 22.00% identities, respectively, with PoHtf1.
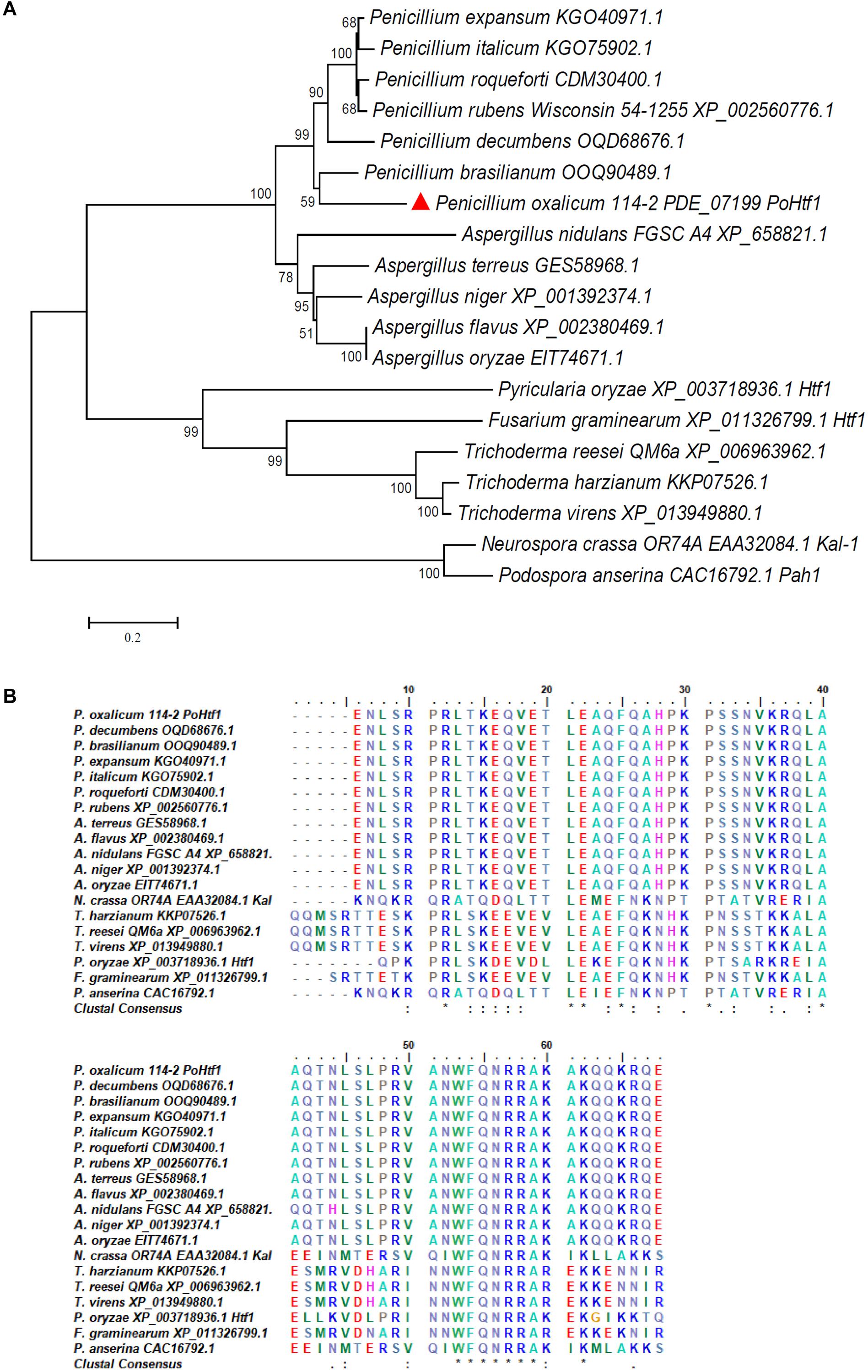
Figure 1. Analysis of the complete and homeodomain sequences of Htfs homologs. (A) Phylogenetic analysis of PoHtf1 homologs. The complete sequences of Htfs from Penicillium sp., Aspergillus sp., Trichoderma sp., Neurospora crassa, Fusarium graminearum, Pyricularia oryzae, and Podospora anserina were downloaded from NCBI. The phylogenetic tree was generated by MEGA 5.0 software using the neighbor-joining (NJ) method. (B) Sequence alignment of the Htfs homolog homeodomains. The alignment was performed by ClustalW Multiple Alignment function in the Bioedit tool. The consensus residues are labeled by asterisk (*).
The homeodomain sequences of the Htf homologs were aligned by ClustalW Multiple Alignment. The Htf homeodomains from Penicillium and Aspergillus species were highly conserved. However, the PoHtf1 homeodomain shared a low level of identity with homeodomains from other strains (Figure 1B). These data indicate that PoHtf1 probably has conserved functions in developmental processes, like those in Aspergillus species. However, PoHtf1 might execute its regulatory functions differently than related proteins do in F. graminearum and M. oryzae.
Comparative Transcriptome Analysis
To investigate the functions of PoHtf1 at the whole genome level, PoHtf1 was deleted in the P. oxalicum 114-2 strain using homologous recombination. PoHtf1 deletion was verified by PCR, Southern blot, and qRT-PCR (Figure 2). The effects of PoHtf1 deletion were analyzed by comparing the transcriptomes of P. oxalicum 114-2 and ΔPohtf1 using RNA-Seq (Li et al., 2015). After PoHtf1 deletion, there were 158 up-regulated and 237 down-regulated genes. Moreover, the differentially expressed genes were mainly involved in starch and sucrose metabolism (11 genes), glycolysis (8 genes), and tyrosine metabolism (6 genes) pathways (Supplementary Figure 1).
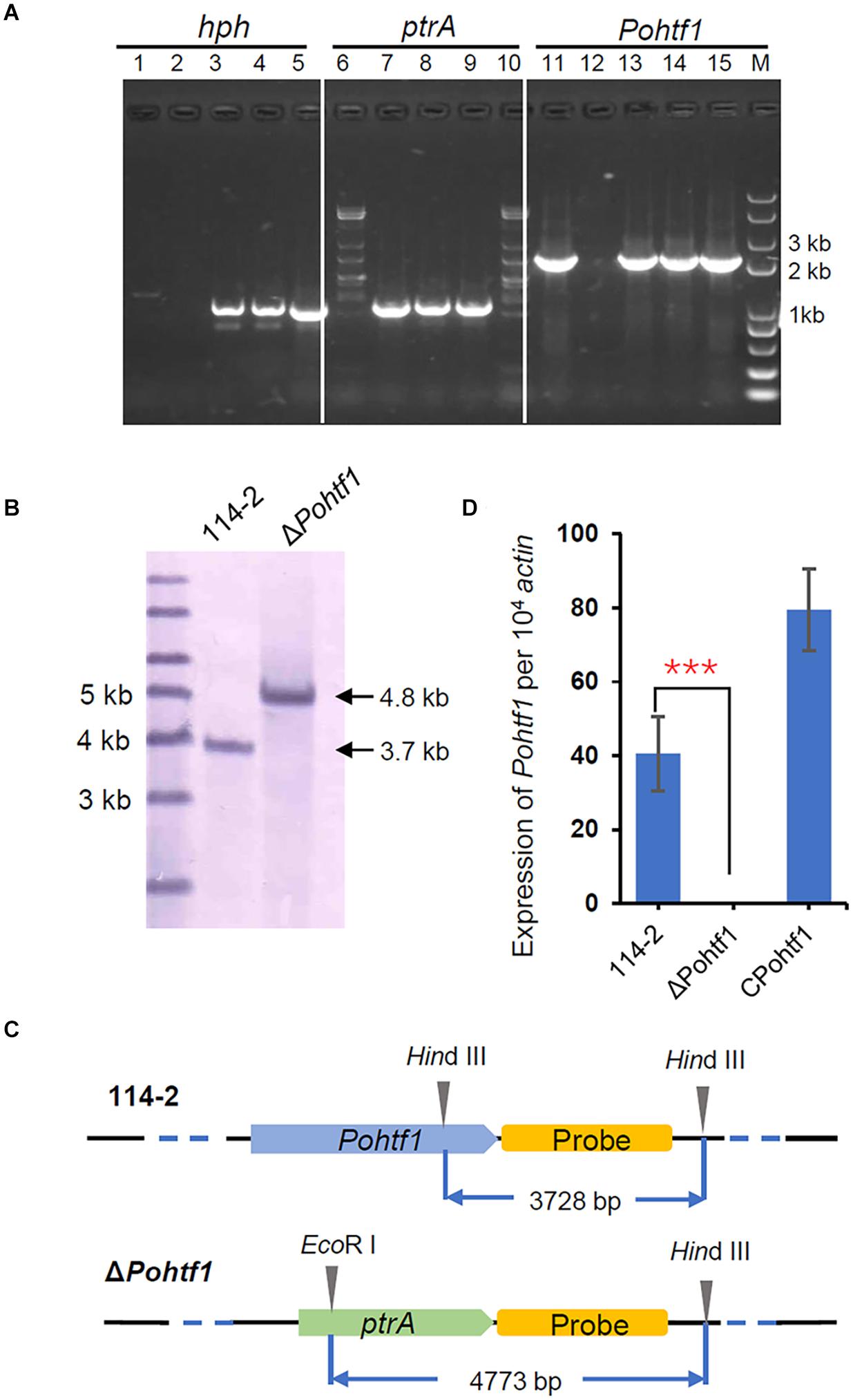
Figure 2. Deletion and verification of PoHtf1. (A) PCR analysis of Pohtf1 deletion and complement. The confirmation of Pohtf1 deletion and complement was performed using three pairs of primers: primers inside the selected marker genes hph (hph-YZ-F/R) (lane 1–5) and ptrA (ptrA-YZ-F/R) (lane 6–10) and primers amplifying the target gene Pohtf1 (7199-hphF/7199-hphR) (lane 11–15). Primer sequences are listed in Supplementary Table 1. Lane 1, 6, 11 are the results of the WT strain P. oxalicum 114-2. Lane 2, 7, 12 are the results of Pohtf1-deletion strain. Lane 3–4, 8–9, and 13–14 are the results of two Pohtf1-complement strains. (B) Southern blot analysis of ΔPohtf1. After deletion of Pohtf1, a 4.8 kb-length fragment was hybridized in the ΔPohtf1 genome by Southern blot, while the fragment length in P. oxalicum 114-2 was 3.7 kb. (C) Southern blot strategy. A 2 kb-length fragment downstream of the target gene was amplified as the hybridization probe. P. oxalicum 114-2 genomic DNA was digested by a single restriction endonuclease, HindIII, and a 3.7 kb-length fragment containing the complete probe sequence was generated. The ΔPohtf1 genomic DNA was double digested with HindIII and EcoRI to generate a 4.8 kb-length fragment. (D) qRT-PCR analysis of Pohtf1 expression levels in the mutant strains. Pohtf1 expression levels in P. oxalicum 114-2, ΔPohtf1, and CPohtf1 were tested using primers RT-7199-F/R inside Pohtf1 (Supplementary Table 2).
Given the potential of PoHtf1 to regulate cellulases, the expression levels of 80 annotated cellulolytic genes were analyzed. The results showed that the expression levels of most of cellulase and hemicellulase genes were substantially up-regulated in ΔPohtf1 (Figure 3). Among these genes, the major cellobiohydrolase gene cbh1, endo-β-1,4-glucanase gene eg1 and intracellular β-glucosidase gene bgl2 were up-regulated in ΔPohtf1, with expression levels nearly 3. 7-, 3. 1-, and 2.1-fold higher, respectively, than those in P. oxalicum 114-2. However, bgl1 which encodes the major extracellular constitutive beta-glucosidase was not differentially expressed between strains 114-2 and ΔPohtf1.
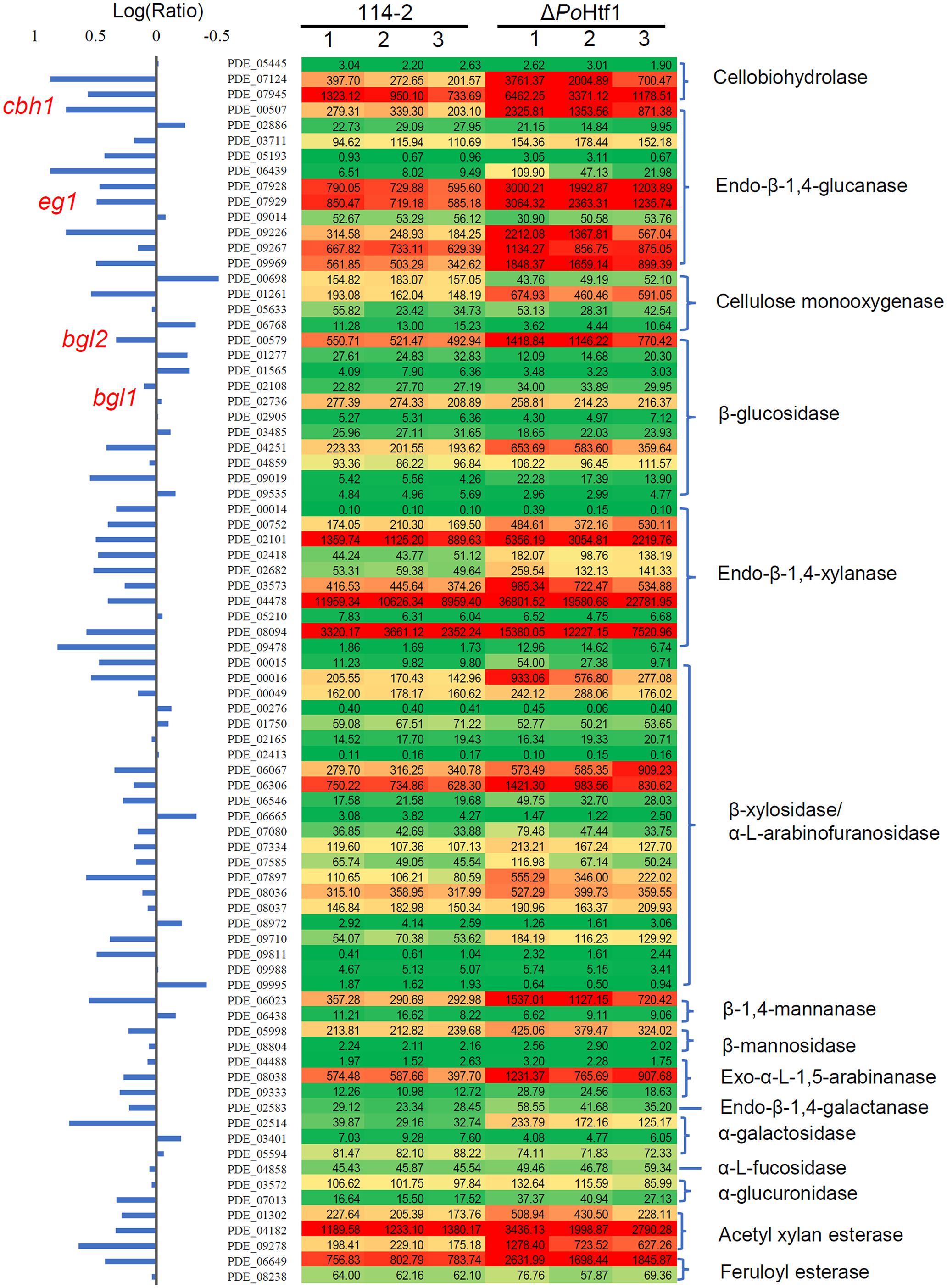
Figure 3. Expression profile of 80 annotated cellulose hydrolyase genes in P. oxalicum 114-2 and ΔPohtf1. The expression levels (RPKM) of the 80 genes are labeled by green-yellow-red color scales in excel. The minimum value is 0, the median is 100, and the maximum value is 36,801.
Moreover, secondary metabolism was also influenced by the absence of PoHtf1. Several secondary metabolic gene clusters were substantially up- or down-regulated in ΔPohtf1 (Supplementary Table 3). Comparative transcriptome results showed that two predicted gene clusters that synthesized oxaline (Newmister et al., 2016) and aspyridones (Klejnstrup et al., 2012) were down-regulated. Similarly, expression of the gene cluster generating conidial yellow pigment (Viggiano et al., 2018) also decreased, possibly altering the ΔPohtf1 conidium and colony phenotype. Furthermore, five unknown secondary metabolic gene clusters were down-regulated, and two unknown gene clusters were up-regulated in ΔPohtf1 (Supplementary Table 3).
The comparative transcriptome results indicate that PoHtf1 is not only crucial in development, but also for the expression of cellulose hydrolyases and secondary metabolism in P. oxalicum 114-2.
PoHtf1 Functions in Development and Conidiation in Penicillium oxalicum 114-2
The influence of PoHtf1 on P. oxalicum 114-2 development was assessed by measuring colony phenotypes and conidiation. The phenotypes on glucose plates showed that PoHtf1 deletion restrained colony growth and conidiation (Figure 4), and ΔPohtf1 colonies were much smaller than those of the P. oxalicum 114-2 parental strain. The hydrolysis halo of ΔPohtf1 on cellulose plates was much more obvious than that of P. oxalicum 114-2, indicating enhanced cellulase production in ΔPohtf1.
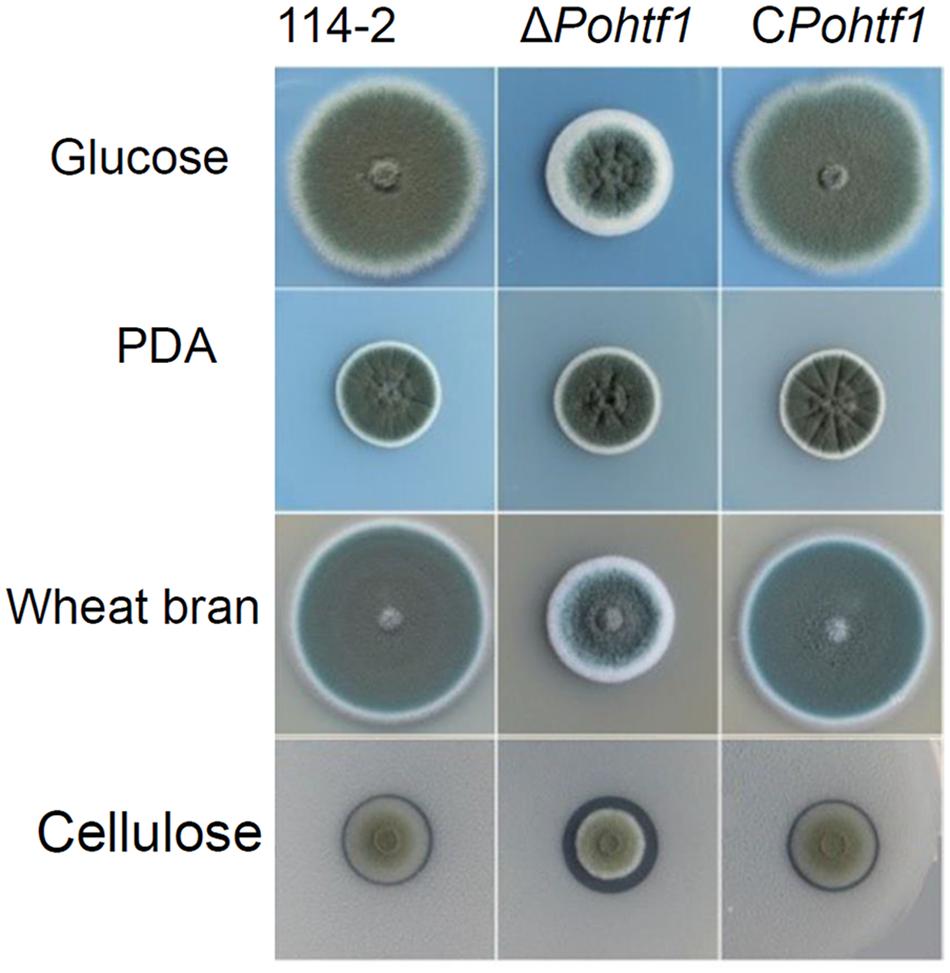
Figure 4. Colony phenotypes of strains P. oxalicum 114-2, ΔPohtf1 and CPohtf1. Phenotype analysis was performed on Vogel’s medium plates containing 2% glucose or 1% cellulose, PDA plates and 10% wheat bran medium plates. The plates were incubated for 3 days at 30°C.
The conidiation of ΔPohtf1 was delayed and visibly restricted on glucose plates. The conidium of ΔPohtf1 was detected after incubation for 36 h on glucose medium plates, which is 6 h later than that of the parental strain P. oxalicum 114-2 (Figure 5A). Furthermore, ΔPohtf1 produced fewer conidium, by nearly three orders of magnitude, than P. oxalicum 114-2 did after incubation for 72 h on glucose medium plates.
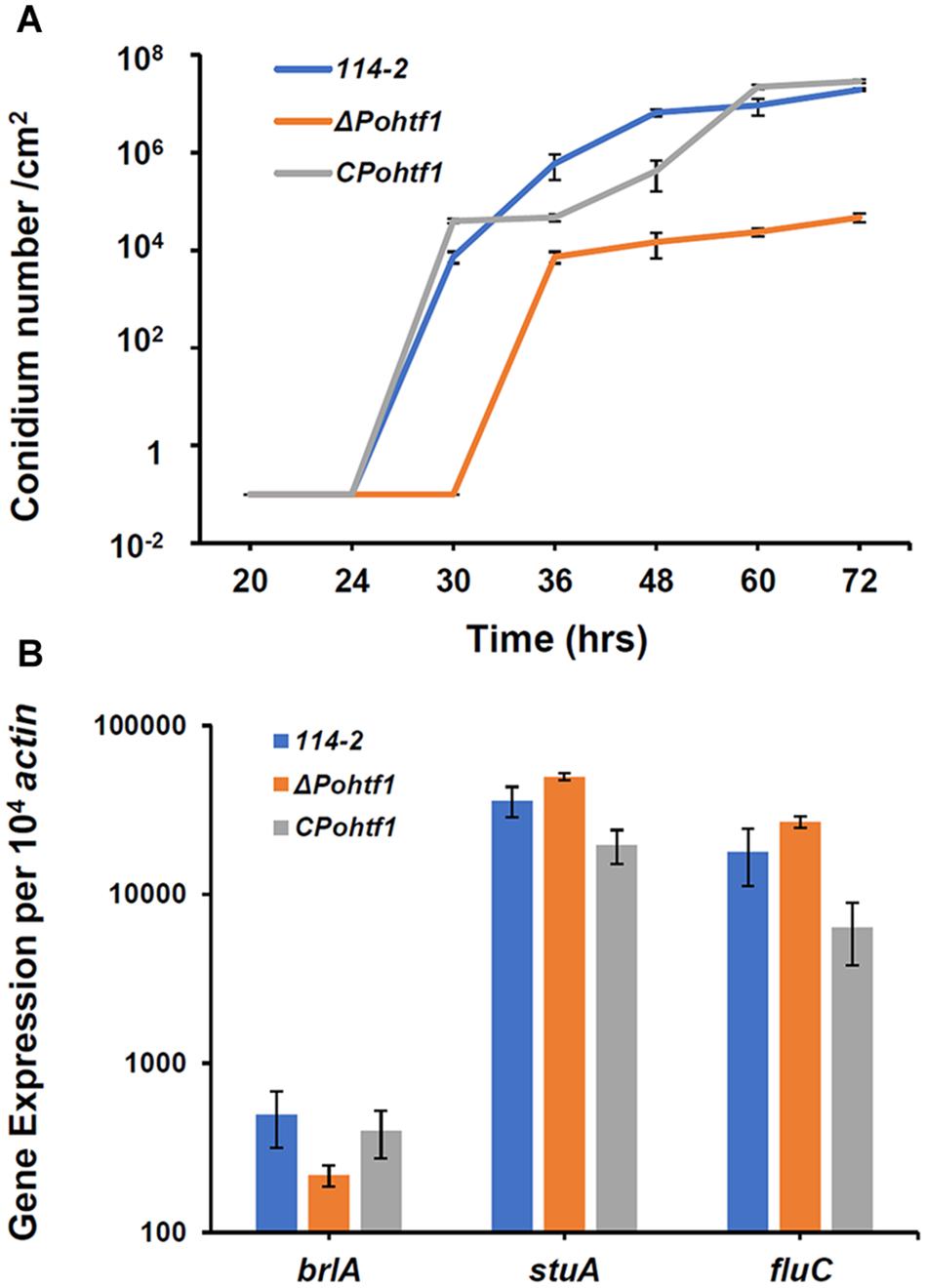
Figure 5. The effects of PoHtf1 on conidiation. (A) The conidiation curve of P. oxalicum 114-2, ΔPohtf1 and CPohtf1 on glucose plates. The plates were incubated at 30°C, and the spore numbers in a fixed area were counted at 24, 30, 36, 48, 60, and 72 h. (B) Expression of the major asexual development transcriptional regulators. All strains were incubated in the cellulase production medium for 4 h. The expression levels of brlA, stuA and fluC were measured by qRT-PCR.
To investigate the mechanism by which PoHtf1 regulates conidiation, we analyzed how PoHtf1 affects the genes that function in the development and conidiation of P. oxalicum 114-2. In P. oxalicum 114-2, three key transcriptional regulators of asexual development have been identified, BrlA, FlbC, and StuA. These proteins regulate the expression levels of pigmentation-related and spore wall protein-related genes (Qin et al., 2013; Yao et al., 2016; Li et al., 2019). Transcription levels of brlA, flbC, and stuA were analyzed in all strains by RT-PCR. After deletion of Pohtf1, the expression levels of brlA in ΔPohtf1 were 44% of that in P. oxalicum 114-2 (Figure 5B). The expression levels of flbC and stuA were almost unchanged. BrlA is crucial in conidiation, and deletion of brlA completely blocks the conidiation of P. oxalicum 114-2 (Qin et al., 2013). Therefore, we assumed that PoHtf1 functions in conidiation by regulating the transcription levels of the key regulator, BrlA.
PoHtf1 Deletion Promotes the Expression of Cellulases
Besides regulating colonial growth and conidiation, PoHtf1 also inhibits cellulase expression in P. oxalicum 114-2. The effect PoHtf1 on the expression of cellulases was determined by assessing the activities toward filter paper (indicating overall cellulase activity), CMC-Na (endoglucanase), pNPC (cellobiohydrolase activity), and pNPG (β-glucosidase). ΔPohtf1 showed the highest FPA after fermentation for 5 days, increasing from 0.6 IU/mL in P. oxalicum 114-2 to 2.0 IU/mL in ΔPohtf1 (Figure 6A). Similarly, the endoglucanase activity increased to nearly fivefold of that of P. oxalicum 114-2 on the 5th day (Figure 6B). The cellobiohydrolase activity continued to increase on the 6th day and was almost fivefold of that of P. oxalicum 114-2 (Figure 6C). The maximum β-glucosidase activity was observed on the 4th day, and it increased to 9.8 IU/mL in ΔPohtf1 from 1.1 IU/mL in P. oxalicum 114-2 (Figure 6D). Together, the activity analysis showed that the absence of Pohtf1 promoted cellulase production.
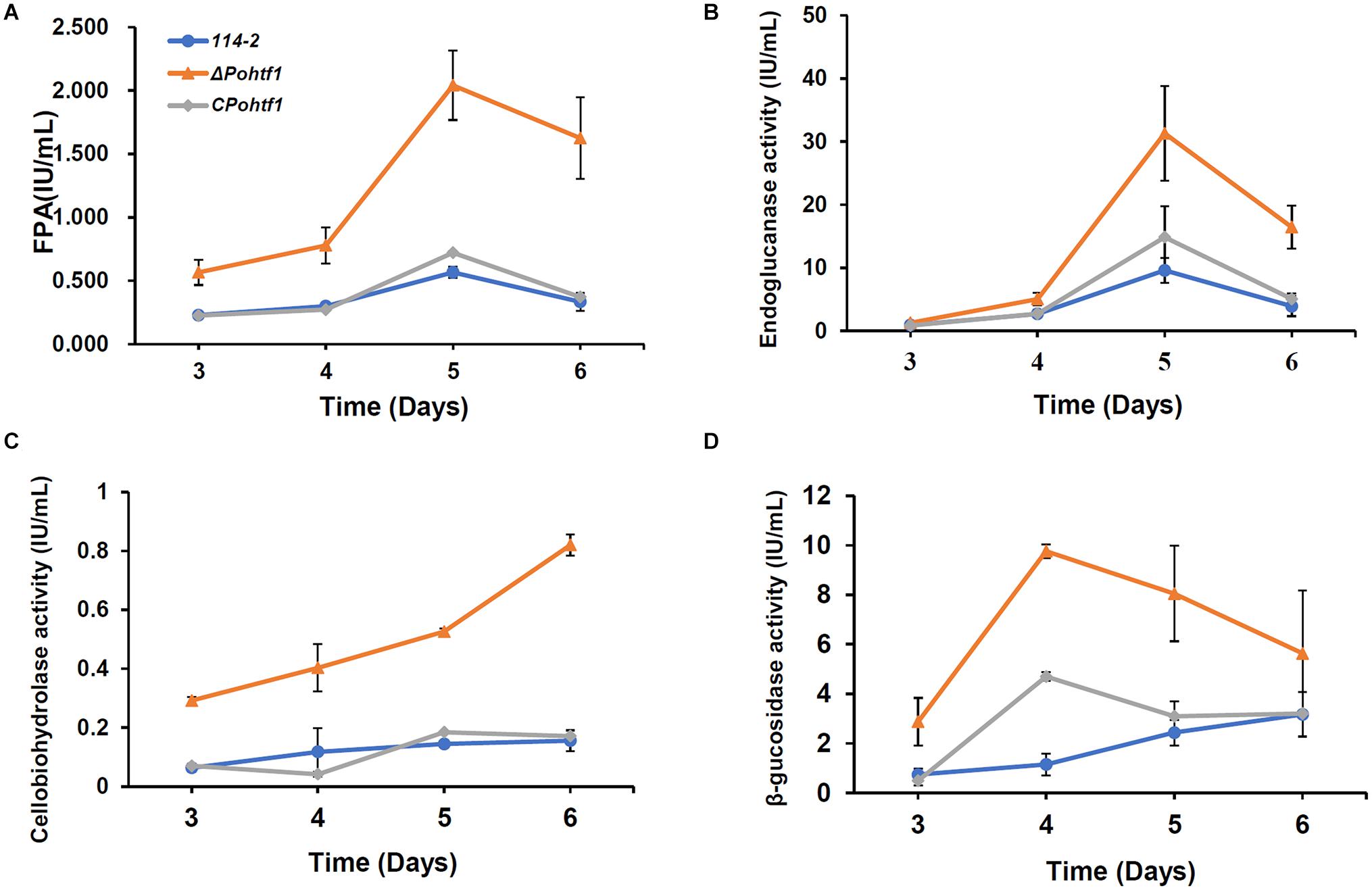
Figure 6. Analysis of the cellulase activities of P. oxalicum 114-2, ΔPohtf1 and CPohtf1. (A) Filter Paper activity (FPA), (B) Endoglucanase, (C) Cellobiohydrolase and (D) β-glucosidase activities of strains P. oxalicum 114-2, ΔPohtf1 and CPohtf1 were assessed. All the strains were sampled and measured every 24 h from day 3 to day 6.
To investigate the mechanism of PoHtf1 in regulating cellulase expression, we analyzed the transcriptional levels of the main cellulase genes and regulators. Mycelium of all the strains were induced in cellulase production medium for 4 h. The expression levels of four major cellulase genes, cbh1, eg1, bgl1, and bgl2, were measured by qRT-PCR. In ΔPohtf1, the expression levels of the major cellobiohydrolase and endoglucanase genes cbh1 and eg1 were 7.5- and 19.2-fold higher, respectively, than those observed in P. oxalicum 114-2 (Figure 7A). This is consistent with the observed increased FPA, cellobiohydrolase, and endoglucanase activities in ΔPohtf1. Additionally, the expression of major intracellular β-glucosidase gene bgl2 was twofold higher than that observed in P. oxalicum 114-2, while the expression level of the main extracellular β-glucosidase gene bgl1 of ΔPohtf1 did not increase. These results are consistent with the RNA-Seq comparison of strains ΔPohtf1 and 114-2.
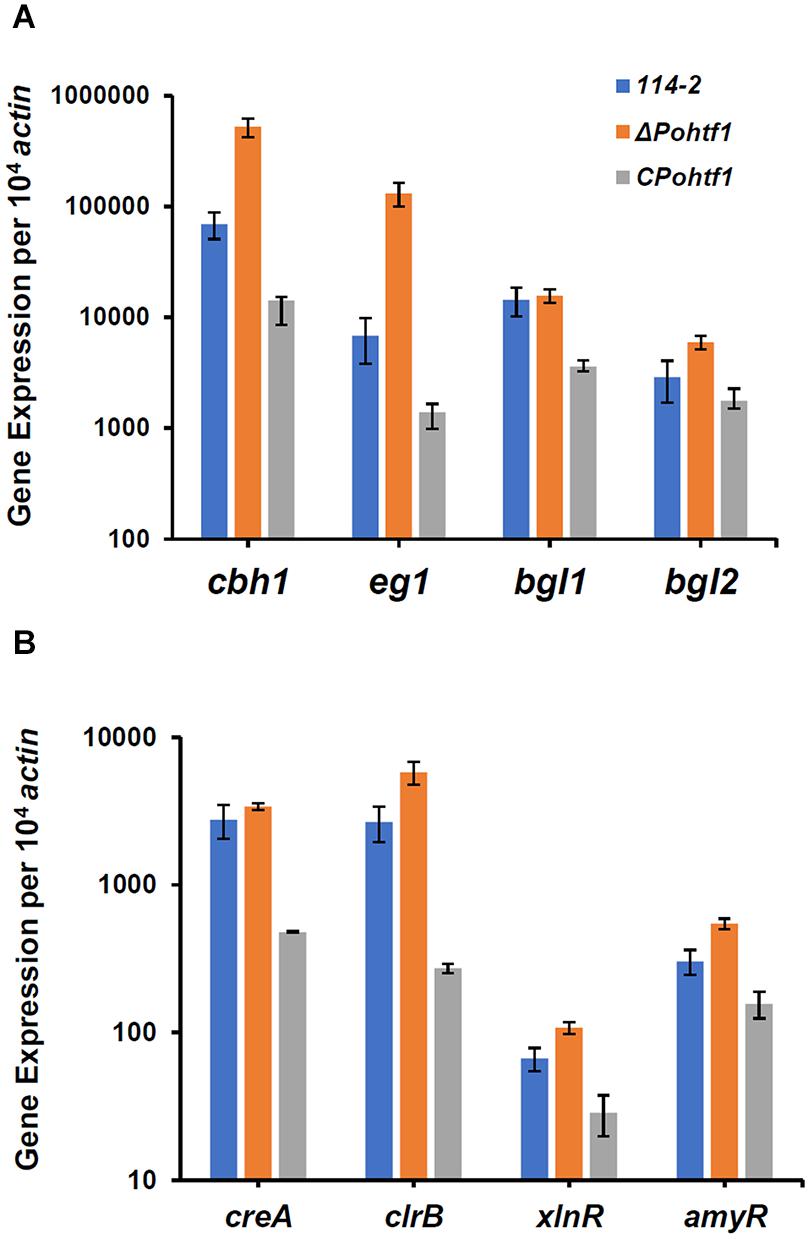
Figure 7. qRT-PCR analysis of the major cellulases and transcriptional regulators in P. oxalicum 114-2, ΔPohtf1 and CPohtf1. (A) Expression levels of the four cellulase genes, cbh1, eg1, bgl1, and bgl2; (B) Expression levels of the four transcriptional regulator genes, creA, clrB, xlnR, and amyR. All strains were incubated in the cellulase production medium for 4 h. The gene expression levels were measured by qRT-PCR.
To investigate whether the up-regulation of cellulase genes following PoHtf1 deletion was occurred through regulating cellulolytic transcriptional factors, we measured the expression levels of CreA, AmyR, ClrB, and XlnR in ΔPohtf1 (Figure 7B). CreA is the most important cellulase transcription repressor and can seriously limit cellulase expression under inducing conditions in P. oxalicum 114-2. AmyR is the major activator of amylases and can repress the expression of cellulases. ClrB and XlnR are the major activators of cellulases and xylanases, respectively, and both can markedly enhance the expression of cellulases (Li et al., 2015). Our results showed that the expression of the two repressors CreA and AmyR and two activators ClrB and XlnR were enhanced 23, 80, 117, and 62%, respectively, by the deletion of Pohtf1 when compared with the expression levels in P. oxalicum 114-2 (Figure 7B). These results indicate that regulation of PoHtf1 toward the four transcription factors was not specific, and that the inhibiting effect of PoHtf1 on cellulases was the result of coordinated regulation of the transcription factors.
Discussion
In this study, we identified a conserved homeodomain-containing transcriptional regulator PoHtf1, an important regulator of genes involved in colonial growth and conidiation in P. oxalicum 114-2. After deletion of PoHtf1, colony growth and conidiation were substantially limited (Figures 4, 5). In P. oxalicum 114-2, there are five other PoHtfs (PDE_00454, PDE_03945, PDE_03970, PDE_04931, and PDE_05741) annotated. However, only PoHtf1 displayed developmental regulatory function (Li et al., 2015). This is mainly because PoHtf1 is the major PoHtf in P. oxalicum 114-2, as its transcription level is much higher than that of other PoHtfs (Supplementary Figure 2). After deletion of PoHtf1, the other five PoHtfs could not compensate for function because their transcription levels were almost unchanged in ΔPohtf1 (Supplementary Figure 2). Furthermore, there were only five highly conserved residues in the homeodomain sequences of the six PoHtfs (Supplementary Figure 3). This suggested that the six PoHtfs might participate in different developmental processes in P. oxalicum 114-2.
Conidiation is the most common reproductive mode of many filamentous fungi. Htf homologs are necessary for conidiation in other fungi including A. nidulans, M. oryzae, and F. graminearum (Liu et al., 2010; Zheng et al., 2012; Son et al., 2020). In M. oryzae, HTF1 is the essential transcription factor for conidiogenesis, and the htf1 mutant strain failed to differentiate conidia (Liu et al., 2010). Some evidence revealed that the G protein-cAMP signaling pathway is implicated in conidiation through regulating HTF1 expression. Mutation of trimeric G-protein β subunit (mgb1) and cAMP-dependent protein kinase gene (cpkA) significantly downregulated HTF1 expression (Zhou et al., 2009). However, HTF1 is not a direct regulator of conidiation. Transcription factors ACR1 and CON7 participate in conidiation and conidium morphology in M. oryzae (Lau and Hamer, 1998; Odenbach et al., 2007). And Htf1 may regulate conidiation by interacting with ACR1, as ACR1 is significantly regulated by HTF1 (Liu et al., 2010). In A. nidulans, HbxA and HbxB were identified to participate in conidia production in A. nidulans (Son et al., 2020). BrlA is a key regulator of conidiation (Adams et al., 1988, 1998), and HbxA effected conidiation by regulating brlA expression (Son et al., 2020).
The PoHtf1 homeodomain sequence is highly conserved with that of HbxA in A. nidulans (Figure 1B), suggesting that PoHtf1 and HbxA have identical conidiation regulation mechanisms. There are three key transcriptional regulators of asexual development, BrlA, FlbC, and StuA, which are involved in conidiation and conidium morphology in P. oxalicum 114-2 (Qin et al., 2013; Yao et al., 2016; Li et al., 2019). BrlA is the key regulator of conidiation, and deletion of brlA completely blocks conidiation. PoHtf1 deletion significantly represses brlA expression (Figure 5B), indicating that PoHtf1 might be the upstream regulator of BrlA. FlbC is also an upstream regulator of BrlA, and flbC deletion leads to significant downregulation of brlA and impaired conidiation (Yao et al., 2016). However, PoHtf1 do not appear to interact with FlbC or StuA, as deletion of PoHtf1 barely caused any effect on the expression levels of FlbC and StuA (Figure 5B). Therefore, we assumed that PoHtf1 regulated conidiation via BrlA, similar to the regulation pattern observed in A. nidulans (Son et al., 2020).
Penicillium oxalicum produces diverse cellulolytic enzymes, which are coordinately regulated by the combinations of many transcription factors. Regulators of asexual development, such as BrlA and FlbC, are involved in the regulation of cellulase expression (Qin et al., 2013; Yao et al., 2016). Deletion of brlA upregulates the expression of most of the major cellulases (Qin et al., 2013). While the absence of flbC reduces cellulase and hemicellulase production (Yao et al., 2016). In this study, deletion of PoHtf1 also increased cellulase production in P. oxalicum 114-2 (Figure 6). These results suggest that there is some cross-regulation between cellulase expression and asexual development. To investigate the mechanism of PoHtf1 in regulating the expression of cellulases, we determined the expression levels of four major identified transcription factors involved in regulating cellulase expression, CreA, AmyR, ClrB, and XlnR, in ΔPohtf1. CreA is the most important transcription repressor of cellulases in P. oxalicum 114-2. AmyR is the major activator of amylases and can repress the expression of cellulases. ClrB and XlnR are the major activators of cellulases and xylanases, respectively, and both can enhance the expression of cellulases. Deletion of Pohtf1 led to a 117% increase in ClrB expression (Figure 7B), indicating that PoHtf1 might mediate the complex transcriptional-regulatory network cascade between developmental processes and cellulolytic gene expression mainly through regulating expression of ClrB in P. oxalicum 114-2.
Given the regulatory function of PoHtf1 in development and cellulase expression, PoHtf1 might act as a general regulator. To analyze the regulation patterns of PoHtf1 at the whole genome level, we performed differential transcriptome analysis of P. oxalicum 114-2 and ΔPohtf1. The target genes regulated by PoHtf1 were widely distributed in the differentially expressed pathways, including starch and sucrose metabolism, glycolysis, and tyrosine metabolism (Supplementary Figure 1). In addition to the verified developmental regulator genes and cellulose hydrolyase genes, several secondary metabolism gene clusters were also influenced by PoHtf1 (Supplementary Table 3). These results revealed the general regulation role of PoHtf1 in the whole genome of P. oxalicum 114-2.
In this research, the functions of the homeodomain-containing protein PoHtf1 were characterized in P. oxalicum 114-2. PoHtf1 was not only crucial in colonial growth and conidiation but also an important transcriptional inhibitor of cellulases. Comparative transcriptome analysis also showed that PoHtf1 participated in several other metabolic pathways, including secondary metabolism.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ZoL conceived and designed the experiments. RW, HG, GX, ZiL, MY, and ZJ performed the experiments. ZoL, MC, XB, and YQ analyzed the data. MC and ZoL drafted the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (31670079), the Natural Science Foundation of Shandong Province (ZR2020MC013), the Project of Shandong Province Higher Educational Science and Technology Program (J18KA159), the Foundation of State Key Laboratory of Biobased material and Green papermaking (ZZ20190307), and the Qilu University of Technology of Cultivating Subject for Biology and Biochemistry (202006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.671089/full#supplementary-material
References
Adams, T. H., Boylan, M. T., and Timberlake, W. E. (1988). brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54, 353–362. doi: 10.1016/0092-8674(88)90198-5
Adams, T. H., Wieser, J. K., and Yu, J. H. (1998). Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62, 35–54. doi: 10.1128/mmbr.62.1.35-54.1998
Arnaise, S., Zickler, D., Poisier, C., and Debuchy, R. (2001). pah1: a homeobox gene involved in hyphal morphology and microconidiogenesis in the filamentous ascomycete Podospora anserina. Mol. Microbiol. 39, 54–64. doi: 10.1046/j.1365-2958.2001.02163.x
Arratia-Quijada, J., Sanchez, O., Scazzocchio, C., and Aguirre, J. (2012). FlbD, a Myb transcription factor of Aspergillus nidulans, is uniquely involved in both asexual and sexual differentiation. Eukaryot Cell 11, 1132–1142. doi: 10.1128/EC.00101-12
Burglin, T. R. (2011). Homeodomain subtypes and functional diversity. Subcell. Biochem. 52, 95–122. doi: 10.1007/978-90-481-9069-0_5
Chang, P. K., Scharfenstein, L. L., Mack, B., and Ehrlich, K. C. (2012). Deletion of the Aspergillus flavus orthologue of A. nidulans fluG reduces conidiation and promotes production of sclerotia but does not abolish aflatoxin biosynthesis. Appl. Environ. Microbiol. 78, 7557–7563. doi: 10.1128/AEM.01241-12
Chen, M., Qin, Y., Cao, Q., Liu, G., Li, J., Li, Z., et al. (2013). Promotion of extracellular lignocellulolytic enzymes production by restraining the intracellular beta-glucosidase in Penicillium decumbens. Bioresour. Technol. 137, 33–40. doi: 10.1016/j.biortech.2013.03.099
Colot, H. V., Park, G., Turner, G. E., Ringelberg, C., Crew, C. M., Litvinkova, L., et al. (2006). A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U.S.A. 103, 10352–10357. doi: 10.1073/pnas.0601456103
Dorn, A., Affolter, M., Gehring, W. J., and Leupin, W. (1994). Homeodomain proteins in development and therapy. Pharmacol. Ther. 61, 155–184. doi: 10.1016/0163-7258(94)90061-2
Emri, T., Molnar, Z., Pusztahelyi, T., Varecza, Z., and Pocsi, I. (2005). The fluG-BrlA pathway contributes to the initialisation of autolysis in submerged Aspergillus nidulans cultures. Mycol. Res. 109, (Pt 7) 757–763. doi: 10.1017/s0953756205003023
Kim, S., Park, S. Y., Kim, K. S., Rho, H. S., Chi, M. H., Choi, J., et al. (2009). Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet. 5:e1000757. doi: 10.1371/journal.pgen.1000757
Klejnstrup, M. L., Frandsen, R. J., Holm, D. K., Nielsen, M. T., Mortensen, U. H., Larsen, T. O., et al. (2012). Genetics of polyketide metabolism in Aspergillus nidulans. Metabolites 2, 100–133. doi: 10.3390/metabo2010100
Kwon, N. J., Garzia, A., Espeso, E. A., Ugalde, U., and Yu, J. H. (2010a). FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 77, 1203–1219. doi: 10.1111/j.1365-2958.2010.07282.x
Kwon, N. J., Shin, K. S., and Yu, J. H. (2010b). Characterization of the developmental regulator FlbE in Aspergillus fumigatus and Aspergillus nidulans. Fungal Genet. Biol. 47, 981–993. doi: 10.1016/j.fgb.2010.08.009
Lau, G. W., and Hamer, J. E. (1998). Acropetal: a genetic locus required for conidiophore architecture and pathogenicity in the rice blast fungus. Fungal Genet. Biol. 24, 228–239. doi: 10.1006/fgbi.1998.1053
Lee, B. N., and Adams, T. H. (1996). FluG and flbA function interdependently to initiate conidiophore development in Aspergillus nidulans through brlA beta activation. EMBO J. 15, 299–309. doi: 10.1002/j.1460-2075.1996.tb00360.x
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, Y., Hu, Y., Zhao, K., Pan, Y., Qu, Y., Zhao, J., et al. (2019). The indispensable role of histone methyltransferase PoDot1 in extracellular glycoside hydrolase biosynthesis of Penicillium oxalicum. Front. Microbiol. 10:2566. doi: 10.3389/fmicb.2019.02566
Li, Z., Yao, G., Wu, R., Gao, L., Kan, Q., Liu, M., et al. (2015). Synergistic and dose-controlled regulation of cellulase gene expression in Penicillium oxalicum. PLoS Genet. 11:e1005509. doi: 10.1371/journal.pgen.1005509
Li, Z. H., Du, C. M., Zhong, Y. H., and Wang, T. H. (2010). Development of a highly efficient gene targeting system allowing rapid genetic manipulations in Penicillium decumbens. Appl. Microbiol. Biotechnol. 87, 1065–1076. doi: 10.1007/s00253-010-2566-7
Liu, W., Xie, S., Zhao, X., Chen, X., Zheng, W., Lu, G., et al. (2010). A homeobox gene is essential for conidiogenesis of the rice blast fungus Magnaporthe oryzae. Mol. Plant. Microb. Interact. 23, 366–375. doi: 10.1094/MPMI-23-4-0366
Miksiunas, R., Mobasheri, A., and Bironaite, D. (2020). Homeobox genes and homeodomain proteins: new insights into cardiac development, degeneration and regeneration. Adv. Exp. Med. Biol. 1212, 155–178. doi: 10.1007/5584_2019_349
Newmister, S. A., Gober, C. M., Romminger, S., Yu, F., Tripathi, A., Parra, L. L., et al. (2016). OxaD: a versatile indolic nitrone synthase from the marine-derived fungus Penicillium oxalicum F30. J. Am. Chem. Soc. 138, 11176–11184. doi: 10.1021/jacs.6b04915
Odenbach, D., Breth, B., Thines, E., Weber, R. W., Anke, H., and Foster, A. J. (2007). The transcription factor Con7p is a central regulator of infection-related morphogenesis in the rice blast fungus Magnaporthe grisea. Mol. Microbiol. 64, 293–307. doi: 10.1111/j.1365-2958.2007.05643.x
Oiartzabal-Arano, E., Garzia, A., Gorostidi, A., Ugalde, U., Espeso, E. A., and Etxebeste, O. (2015). Beyond asexual development: modifications in the gene expression profile caused by the absence of the Aspergillus nidulans transcription factor FlbB. Genetics 199, 1127–1142. doi: 10.1534/genetics.115.174342
Pelkmans, J. F., Patil, M. B., Gehrmann, T., Reinders, M. J., Wosten, H. A., and Lugones, L. G. (2017). Transcription factors of Schizophyllum commune involved in mushroom formation and modulation of vegetative growth. Sci. Rep. 7:310. doi: 10.1038/s41598-017-00483-3
Qin, Y., Bao, L., Gao, M., Chen, M., Lei, Y., Liu, G., et al. (2013). Penicillium decumbens BrlA extensively regulates secondary metabolism and functionally associates with the expression of cellulase genes. Appl. Microbiol. Biotechnol. 97, 10453–10467. doi: 10.1007/s00253-013-5273-3
Son, S. H., Son, Y. E., Cho, H. J., Chen, W., Lee, M. K., and Kim, L. H. (2020). Homeobox proteins are essential for fungal differentiation and secondary metabolism in Aspergillus nidulans. Sci. Rep. 10:6094. doi: 10.1038/s41598-020-63300-4
Viggiano, A., Salo, O., Ali, H., Szymanski, W., Lankhorst, P. P., Nygard, Y., et al. (2018). Pathway for the biosynthesis of the pigment chrysogine by Penicillium chrysogenum. Appl. Environ. Microbiol. 84:e02246-17. doi: 10.1128/AEM.02246-17
Wang, W., Wang, L., Chen, B., Mukhtar, I., Xie, B., Li, Z., et al. (2019). Characterization and expression pattern of homeobox transcription factors in fruiting body development of straw mushroom Volvariella volvacea. Fungal Biol. 123, 95–102. doi: 10.1016/j.funbio.2018.10.008
Yao, G., Li, Z., Wu, R., Qin, Y., Liu, G., and Qu, Y. (2016). Penicillium oxalicum PoFlbC regulates fungal asexual development and is important for cellulase gene expression. Fungal Genet. Biol. 86, 91–102. doi: 10.1016/j.fgb.2015.12.012
Yu, J. H., Hamari, Z., Han, K. H., Seo, J. A., Reyes-Dominguez, Y., and Scazzocchio, C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. doi: 10.1016/j.fgb.2004.08.001
Zheng, W., Zhao, X., Xie, Q., Huang, Q., Zhang, C., Zhai, H., et al. (2012). A conserved homeobox transcription factor Htf1 is required for phialide development and conidiogenesis in Fusarium species. PLoS One 7:e45432. doi: 10.1371/journal.pone.0045432
Keywords: homeodomain-containing transcription factors, development, conidiation, cellulase, Penicillium oxalicum
Citation: Guo H, Xu G, Wu R, Li Z, Yan M, Jia Z, Li Z, Chen M, Bao X and Qu Y (2021) A Homeodomain-Containing Transcriptional Factor PoHtf1 Regulated the Development and Cellulase Expression in Penicillium oxalicum. Front. Microbiol. 12:671089. doi: 10.3389/fmicb.2021.671089
Received: 23 February 2021; Accepted: 14 May 2021;
Published: 10 June 2021.
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaReviewed by:
Xinqing Zhao, Shanghai Jiao Tong University, ChinaDavid Canovas, University of Sevilla, Spain
Copyright © 2021 Guo, Xu, Wu, Li, Yan, Jia, Li, Chen, Bao and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonghai Li, bHpobHpoQHZpcC4xMjYuY29t; Mei Chen, Y2hlbm1laV8xNkAxNjMuY29t
†These authors have contributed equally to this work
 Hao Guo1,2,3†
Hao Guo1,2,3† Mei Chen
Mei Chen Yinbo Qu
Yinbo Qu