- 1Department of Molecular Biology and Genetics, Federal Research and Clinical Center of Physical-Chemical Medicine, Moscow, Russia
- 2Department of Molecular Microbiology, State Research Center for Applied Microbiology and Biotechnology, Moscow, Russia
- 3Research Center of Biotechnology of the Russian Academy of Sciences, Winogradsky Institute of Microbiology, Moscow, Russia
Antibiotic resistance is a major public health concern in many countries worldwide. The rapid spread of multidrug-resistant (MDR) bacteria is the main driving force for the development of novel non-antibiotic antimicrobials as a therapeutic alternative. Here, we isolated and characterized three virulent bacteriophages that specifically infect and lyse MDR Klebsiella pneumoniae with K23 capsule type. The phages belonged to the Autographiviridae (vB_KpnP_Dlv622) and Myoviridae (vB_KpnM_Seu621, KpS8) families and contained highly similar receptor-binding proteins (RBPs) with polysaccharide depolymerase enzymatic activity. Based on phylogenetic analysis, a similar pattern was also noted for five other groups of depolymerases, specific against capsule types K1, K30/K69, K57, K63, and KN2. The resulting recombinant depolymerases Dep622 (phage vB_KpnP_Dlv622) and DepS8 (phage KpS8) demonstrated narrow specificity against K. pneumoniae with capsule type K23 and were able to protect Galleria mellonella larvae in a model infection with a K. pneumoniae multidrug-resistant strain. These findings expand our knowledge of the diversity of phage depolymerases and provide further evidence that bacteriophages and phage polysaccharide depolymerases represent a promising tool for antimicrobial therapy.
Introduction
Klebsiella pneumoniae is a widespread Gram-negative, non-motile, facultative anaerobic bacterium naturally occurring in soil, sewage, or plants. These bacteria are traditionally considered as commensals and can be found on human skin and in the gastrointestinal and respiratory tracts (Paczosa and Mecsas, 2016). Despite this, K. pneumoniae is the world’s second most common nosocomial pathogen, capable of causing a wide range of infections such as septicemia, pneumonia, urinary tract infections, and surgical- and catheter-related infections (Podschun and Ullmann, 1998).
The extensive use of antibiotics to treat Klebsiella-associated infections has led to the emergence and spread of drug resistance. According to data from the European Centre for Disease Prevention and Control, nearly 40% of the K. pneumoniae isolates in Europe are resistant to at least one class of antibiotic – fluoroquinolones, third-generation cephalosporins, aminoglycosides, or carbapenems (European Centre for Disease Prevention and Control, 2019). Of these, the isolates carrying extended-spectrum beta-lactamases or carbapenemases encoding genes are considered especially difficult to treat. As previously reported, these isolates cause infections with high morbidity and mortality rates (Cantón et al., 2012; Tzouvelekis et al., 2012; Munoz-Price et al., 2013; Girmenia et al., 2015; Tängdén and Giske, 2015; Lee et al., 2016).
In light of the antibiotic resistance crisis, the development of new approaches to antimicrobial therapy is especially relevant. One promising alternative to the use of antibiotics is therapy with virulent bacteriophages (phages; Górski et al., 2020). Phages are natural killers that can rapidly and selectively infect and lyse pathogenic bacteria, including K. pneumoniae clones associated with antibiotic resistance. Bacteriophage therapy has been clinically used since the beginning of the 20th century, and no known significant side effects have been identified throughout the entire history of use (Payne and Jansen, 2000). Nowadays, the efficiency and safety of bacteriophage therapy have been confirmed in mammal and Galleria mellonella models and in humans (Chhibber et al., 2008; Hung et al., 2011; D’Andrea et al., 2017; Manohar et al., 2019). The natural limitation of the approach is the usually narrow host range of individual bacteriophages, so the phage cocktails have to be employed to combat an unknown pathogen (Clark and March, 2006).
The host range of K. pneumoniae phages shows a good correlation with the type of capsular polysaccharide (CPS; Pires et al., 2016). To date, at least 130 types of CPS have been described, which is the key virulence factor protecting the bacteria from the immune system and the action of some antibiotics (Stewart, 1996; Yu et al., 2015; Wyres et al., 2016). Isolates characterized by the overexpression of capsular polysaccharides often possess an enhanced virulence and have been categorized as a separate group called hypervirulent K. pneumoniae (hvKp). In addition to hyperexpression of the capsule, frequently regulated by the rmpA/rmpA2 gene, hvKp may also carry other virulence factors, such as yersiniabactin, colibactin, aerobactin, and salmochelin. HvKp has been described as a cause of primary purulent liver abscesses in the Asia-Pacific region (Liu et al., 1986; Fang et al., 2005; Turton et al., 2007; Shon et al., 2013).
The main factor that allows phages to infect the K. pneumoniae cell is the presence of a specific receptor-binding protein (RBP) – a processive enzyme with depolymerase activity (Pires et al., 2016). Depolymerases are enzymes that can cleave capsular polysaccharides, clearing the way for phage adsorption to the surface of a bacterial cell, followed by injection of phage DNA. Phage-borne depolymerases are usually presented as a structural component of the phage adsorption apparatus (tail fibers, tail spikes, or base plates; Pires et al., 2016; Knecht et al., 2020). Most of the phage genomes encode only one depolymerase, but some phages have two or more depolymerases that allow them to infect multiple capsular types (Pan et al., 2017; Latka et al., 2019). Characterizing the diversity of phage depolymerases is of particular interest, as this could lead to the development of a new class of antivirulence agents. It has been shown that phage depolymerases could increase the rate of bacterial killing by serum in vitro and significantly increase survival in mice and G. mellonella larvae models (Majkowska-Skrobek et al., 2018; Solovieva et al., 2018; Volozhantsev et al., 2020). Additionally, phage-borne depolymerases can be used for the rapid determination of microbial capsule types or disruption of bacterial biofilms (Scorpio et al., 2008).
Currently, over 30 specific polysaccharide-depolymerases have been characterized (Lin et al., 2014; Majkowska-Skrobek et al., 2016, 2018; Hsieh et al., 2017; Pan et al., 2017, 2019; Latka et al., 2019; Domingo-Calap et al., 2020a,b; Liu et al., 2020; Volozhantsev et al., 2020). However, phages and depolymerases specific for K. pneumoniae K23 capsular type have not been characterized yet. In this study, we described the biological characteristics and performed a genomic analysis of one new podovirus vB_KpnP_Dlv622 and two new myoviruses (vB_KpnM_Seu621 and KpS8) infecting specifically the strains of K. pneumoniae with capsule type K23. In addition, we compared their depolymerases and assayed their protective activity in G. mellonella larvae during infection with a clinical multidrug-resistant (MDR) K. pneumoniae strain.
Materials and Methods
Bacterial Strains and Their Characterization
A collection comprising 32 clinical K. pneumoniae isolates from the Clinical Hospital №123 (Odintsovo, Russia) and 51 strains from the State Collection of Pathogenic Microorganisms and Cell Cultures, SCPM-Obolensk (State Research Center for Applied Microbiology and Biotechnology, Obolensk, Russia) were included in this study (Supplementary Table 3). All bacteria were grown in the Nutrient Medium No. 1 (SRCAMB, Obolensk, Russia), or in the lysogeny broth (LB) medium (Himedia, India) at 37°C. Bacterial identification was performed by MALDI-TOF mass spectrometry as described previously (Kornienko et al., 2016). The antibiotic susceptibility was tested using the disc diffusion method according to Clinical and Laboratory Standards Institute guidelines 28th edition (Clinical and Laboratory Standards Institute, 2018). Multilocus sequence typing (MLST) of K. pneumoniae strains was performed by determining the nucleotide sequences of seven housekeeping genes as described previously (Diancourt et al., 2005). The capsular type was determined by wzi gene sequencing (Brisse et al., 2013).
Bacteriophage Isolation and Purification
Two K. pneumoniae K23 strains (Kp-9068 and KPi4275) were used as hosts for bacteriophage isolation. Phages vB_KpnM_Seu621 and vB_KpnP_Dlv622 were isolated from the Chermyanka river water samples and phage KpS8 was isolated from sewage by a previously described method (Van Twest and Kropinski, 2009), with slight modifications. In brief, 15 ml of sewage or river water samples were centrifuged at 10,000 g for 15 min, and the supernatant was filtered using a 0.22-μm sterile membrane syringe filter (Millipore, United States). Filtered supernatant and 0.2 ml of early log-phase host cultures (OD600nm = 0.3) were added to 15 ml of double-concentrated LB broth and incubated overnight with agitation (200 rpm) at 37°C to amplify the phages. Further, the culture was centrifuged at 10,000 g for 15 min and subsequently filtered through 0.22-μm filters. Obtained lysates were serially diluted in LB and spotted on double-layer agar plates of host-strains for phage detection and isolation. Three rounds of single plaque purification and re-infection of exponentially growing host strains yielded pure bacteriophage suspensions. Bacteriophage titers were determined using a double-agar overlay plaque assay (Mazzocco et al., 2009).
Electron Microscopy of Phage Particles
Purified phage preparations were analyzed by transmission electron microscopy using a JOEL JSM 100 CXII electron microscope (JOEL, Japan) at an acceleration voltage of 100 kV with a Gatan Erlangshem CCD camera (Gatan, Inc.). Carbon-coated grids with collodion supporting film were negatively stained with 1% uranyl acetate in methanol.
Host Range Determination
The host range of phages was determined by the spot test using 83 K. pneumoniae strains. Briefly, 100 μl of log-phase (OD600nm = 0.3) culture of each strain was added to 5 ml of top agar, which was subsequently poured onto a bottom agar plate. Around 5 μl of phage lysate at a titer of 106 PFU/ml were spotted onto freshly seeded lawns of the strains and left to dry before overnight incubation at 37°C. Additionally, the efficiency of plating (EOP) assay was also performed for phage-sensitive strains as previously described (Solovieva et al., 2018). The average EOP value for a phage-bacterium ratio was classified according to Mirzaei and Nilsson (2015): highly productive (EOP ≥ 0.5), medium productive (0.1 ≤ EOP < 0.5), low productive (0.001 < EOP < 0.1), or inefficient (EOP ≤ 0.001; Horváth et al., 2020).
Thermal Tolerance of Phages
The stability of phages at different temperatures was determined by incubating 1 ml of phage lysate (109 PFU/ml) at different temperatures (4, 37, 45, 55, 65, and 75°C) for 1 h, and then the phage titer was determined using the double-layer agar method (Mazzocco et al., 2009). The assay was performed in triplicate.
Adsorption Assay and One-Step Growth Curve
The adsorption rate was estimated as described earlier with a slight modification (Kropinski, 2009). Host strains were grown to exponential phase (OD600nm = 0.3) and mixed with bacteriophage at a multiplicity of infection (MOI) of 0.01. Every 2 min from 2 to 17 min, aliquots were taken and treated with 2% chloroform, shaken briefly and set aside for 10 min at RT. The titers of free phage were quantified by plaque assay. Determining the number of PFU of the unbound phage in the supernatant and subtracting it from the total number of inputs PFU gave phage adsorption percentage.
The dynamic changes in the number of phage particles during a replicative cycle were performed as described earlier (Ciacci et al., 2018). Aliquots of phage lysates were added to an exponential phase host strains (OD600nm = 0.3) in MOI of 0.01 and allowed to adsorb for 10 min at 37°C. The mixture was then centrifuged at 10,000 g for 5 min and the pellet was resuspended in 10 ml of LB broth to remove free phage particles. Aliquots were taken at 15, 20, 40, 50, 60, 70, and 80 min post-infection and treated with 2% chloroform, shaken briefly, set aside for 10 min at RT, and centrifuged. Finally, the titers of the lysates were quantified by plaque assay. The burst size was calculated as the ratio of the final count of released phage particles to the initial count of infected bacterial cells during the latent period. Independent experiments were repeated three times.
DNA Sequencing and Analysis
A standard phenol-chloroform extraction protocol was used for phage DNA isolation as described previously (Green et al., 2012). The whole-genome sequencing of bacteriophages was performed with a high-throughput Illumina HiSeq system. The assembly was performed using SPAdes (v.3.14.0; Bankevich et al., 2012). Terminal repeats were predicted with the PhageTerm tool (Garneau et al., 2017). Open reading frames (ORFs) were predicted using GeneMarkS (version 4.32; Besemer et al., 2001), Phast (Zhou et al., 2011), and VGAS (Zhang et al., 2019). tRNAScan-SE (Chan and Lowe, 2019) and ARAGORN (Laslett and Canback, 2004) were used to predict tRNA. The putative functions of the proteins encoded by each ORF were retrieved manually using BLASTp, HHPred, Phast, and InterPro. ORFs were also compared against Virulence Factors of Pathogenic Bacteria (VFDB; Liu et al., 2019) and Antibiotic Resistance Genes Databases (ARDB; Liu and Pop, 2009) to verify the safety of the phages. The annotated genome sequences of bacteriophages vB_KpnM_Seu621, vB_KpnP_Dlv622, and KpS8 were deposited in the NCBI GenBank database under accession numbers MT939253, MT939252, and MT178275, respectively. Phylogenetic analysis was performed using the amino acid sequences of RNA polymerase for Autographiviridae family phages and major capsid protein for Myoviridae family phages as recommended by the International Committee on Taxonomy of Viruses (ICTV) classification. The phylogenetic tree was constructed with the Genome-BLAST Distance Phylogeny method implemented by the VICTOR webserver (Meier-Kolthoff and Göker, 2017). Depolymerase domain multiple sequences alignment was performed using MAFFT (v.7.475; Katoh and Standley, 2013). ProtTest (v.3.4.2) was used to define the best suitable amino acid substitution model (Darriba et al., 2011). Maximum likelihood trees were inferred with RAxML-NG (v.1.0.2) with Blosum62+G+F substitution model suggested by ProtTest (Kozlov et al., 2019). Trees were visualized with the ggtree package for R (Yu et al., 2017).
Preparation of Recombinant CPS Depolymerases and Determination of Their Activity
To obtain recombinant CPS depolymerases, coding sequences for the genes kps8_053 and dlv622_00059 containing the putative depolymerase domains were amplified by PCR using specific oligonucleotide primer pairs: DepS8F_NcoI 5'-ACGCCATGGACTGGGTCACTCTTGAAAT-3' and DepS8R_XhoI 5'-ATACTCGAGCCCGTTCACCCTTGAAA-3'; Dep622F_NcoI 5'-TACCATGGCTTTGACAAAGTTAGTAC-3' and Dep622R_XhoI 5'-TACTCGAGCACCCCCGTCAACCGC-3'. Amplified fragments were cloned into a pET22b expression vector (Novagen, United States) via the NcoI and XhoI restriction sites and then transformed into Escherichia coli BL21 (DE3). Resulting constructs were quality-checked via Sanger sequencing. Protein expression was performed in LB medium supplemented with ampicillin at 100 mg/L. Transformed cells were grown at 37°C until the optical density reached the value of 0.4 at 600 nm. The medium was cooled to the temperature of 16°C followed by expression induction with 1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG). After further incubation at 16°C overnight, the cells were harvested by centrifugation at 3,700 g for 20 min, 4°C. The cell pellets were resuspended in 1/50 of the original cell volume in buffer A (20 mM Tris pH 8.0, 0.5 M NaCl, 20 mM imidazole) and then lysed by sonication. The cell debris was removed by centrifugation at 16,000 g for 30 min, at 4°C. The supernatants were loaded onto nickel Ni2+-charged 5 ml GE HisTrap columns (GE Healthcare Life Sciences) equilibrated with buffer A, and eluted with a 20–500 mM imidazole linear gradient in buffer A. The fractions containing the target proteins were pooled together and then dialyzed against 20 mM Tris pH 8.0, 200 mM NaCl, 0.5 mM DTT buffer at 4°C. The protein samples were concentrated with Sartorius ultrafiltration devices (molecular weight cutoff of 10,000) and stored at 4°C. The CPS-degrading activity of the recombinant proteins was assayed in а spot test using K. pneumoniae strains of different capsular types.
Phage KpS8 and vB_KpnP_Dlv622 Adsorption Inhibition Assay
Bacterial cell suspension (~2 × 108 CFU/ml) in SM-buffer (8 mM MgCl2, 100 mM NaCl, 50 mM Tris-HCl pH 7.5) with added recombinant depolymerase (400 μg/ml) and a phage sample (~107 PFU/ml) were first brought to 37°C. Then equal volumes of the phage and cell suspension were mixed. After 5 min incubation at 37°C, bacterial cells with adsorbed phages were precipitated by centrifugation at 14,000 g for 1 min and the titer of non-adsorbed phages was determined in the supernatant.
Galleria mellonella Larvae Infection Model
Greater Wax moth larvae (G. mellonella) were obtained from a laboratory culture maintained at State Research Center for Applied Microbiology and Biotechnology, Obolensk. The larvae were reared on an artificial nutrient medium (maize flour – nine parts, wheat flour – four parts, dry brewer’s yeasts, dry milk, beeswax, bee honey, and glycerol – five parts each) at 27°C for 25–27 days and were subsequently selected for the experiments. The larvae were inoculated with a bacterial suspension or bacteria simultaneously with the depolymerase enzyme into the hemocoel using an insulin syringe. Bacterial suspensions of an overnight agar K. pneumoniae Kp-9068 culture in phosphate-buffered saline (PBS) buffer with cell concentration of 3 × 108 CFU per injection, 3 × 107 CFU, 3 × 106 CFU, 3 × 105 CFU, and 3 × 104 CFU were used to infect the larvae. Three larvae groups were used in the experiments: (1) larvae infected with bacteria only; (2) larvae infected with bacteria and DepS8 enzyme; and (3) larvae infected with bacteria and Dep622 enzyme. Each test was performed in triplicate, with 50 larvae per trial (10 larvae for each bacterial dose). The final dose of the enzymes was 2 μg per larvae for all trials. In addition, three control groups were used: larvae injected with PBS and larvae injected with depolymerase DepS8 or Dep622. Infected larvae were incubated at 37°C for 5 days and mortality was recorded daily. The GraphPad Prism software (GraphPad Software, Inc., La Jolla, United States) was used for statistical analysis and graphical presentation of the results. Statistical analysis was performed for pairwise comparisons between larvae infected with bacteria only and larvae infected with bacteria simultaneously with depolymerase DepS8 or Dep622 using log-rank (Mantel-Cox) test. Values of p < 0.05 were considered as statistically significant.
A dose of bacteria that is sufficient to kill 50% of a larvae population (LD50) was calculated from the cumulative mortality observed 5 days after dosing by the Ashmarin-Vorobiev modification of Karber’s method (Ashmarin and Vorobyev, 1962).
Results
Isolation and Phenotypic Characterization of Three Novel Klebsiella pneumoniae Phages
Phages vB_KpnM_Seu621 and vB_KpnP_Dlv622 were isolated in 2018 from the freshwater of the Chermyanka river (Moscow, Russia). Phage KpS8 was isolated in 2016 from sewage samples in the Moscow region, Russia. Klebsiella pneumoniae strain Kp-9068 was used as a host for isolation vB_KpnM_Seu621 and vB_KpnP_Dlv622 phages. The KpS8 phage was isolated using the KPi4275 strain as a host. Both strains had a multidrug-resistant phenotype (resistance to three or more classes of antibiotics), belonged to sequence type 11 (ST11), and had a K23 capsular type (Supplementary Table 3).
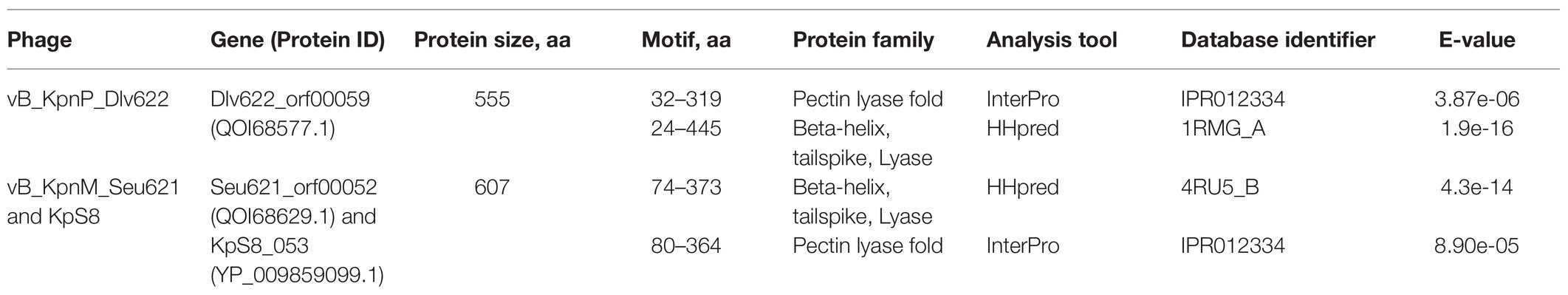
Phage vB_KpnP_Dlv622 had morphological features typical of the Podoviridae family: symmetrical polyhedral head (51 nm in diameter) and a short, noncontractile tail (12 nm long; Figure 1A). Both vB_KpnM_Seu621 and KpS8 phages, had an isometric head of 75–78 nm in diameter and a contractile tail of 104–113 nm in length, with the overgrowths on a terminal side (Figures 1B,C), suggesting that the phages belong to the Myoviridae family.
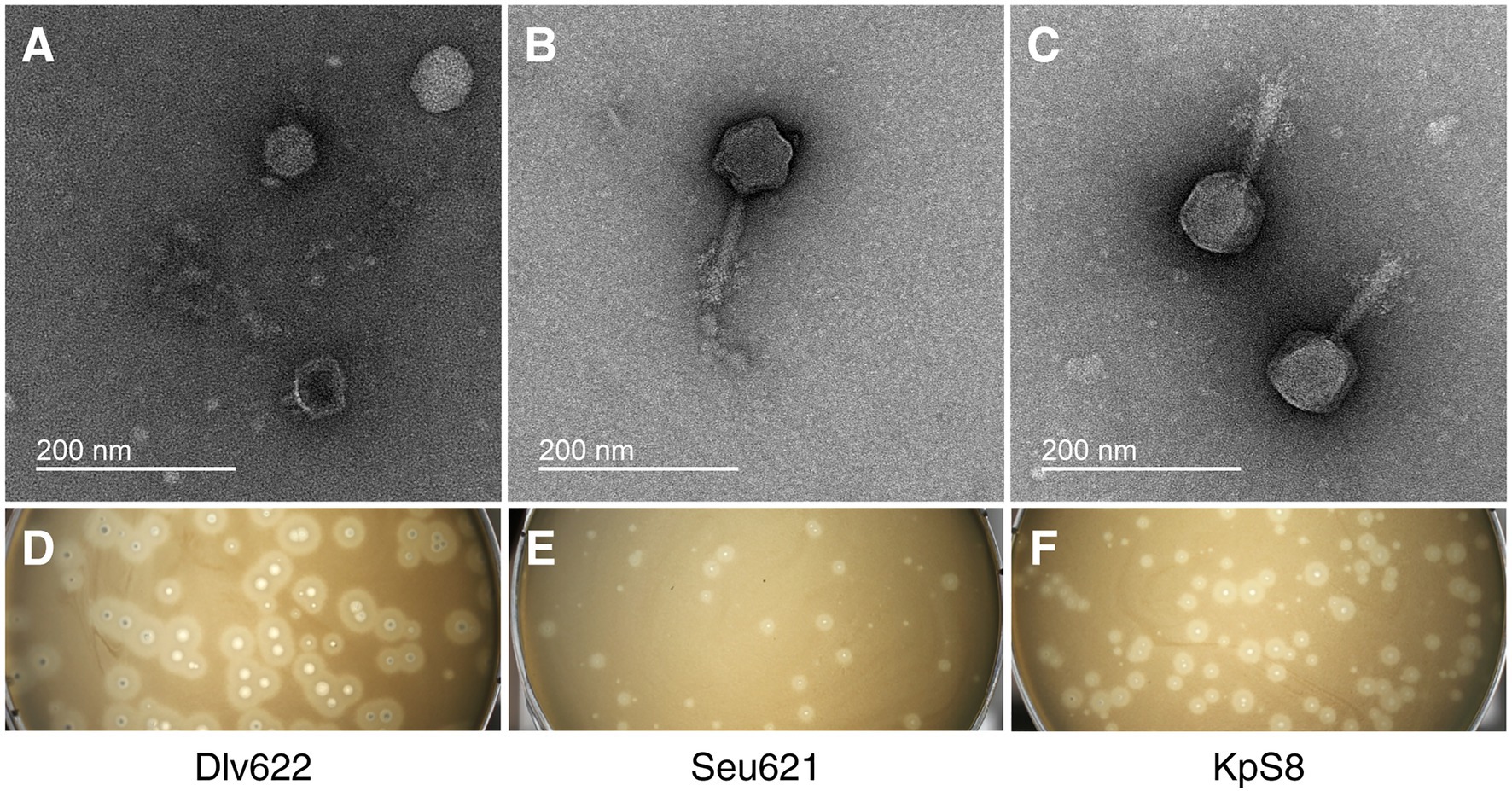
Figure 1. Initial characterization of Klebsiella phages vB_KpnP_Dlv622, vB_KpnM_Seu621, and KpS8. Transmission electron microscopy of phages Dlv622 (A), Seu621 (B), and KpS8 (C). Plaque morphology of phages Dlv622 (D), Seu621 (E), and KpS8 (F).
Different plaque morphology was exhibited by the phages. The vB_KpnP_Dlv622 phage formed clear circular plaques with a diameter of about 2–3 mm with halos (Figure 1D). Phages vB_KpnM_Seu621 and KpS8 were able to form similar circular plaques with a diameter of about 0.5–1 mm, surrounded by hazy halos (Figures 1E,F). Previous studies have suggested that the translucent halos were related to a putative phage-derived polysaccharide depolymerase (Wang et al., 2019).
Studies on phage adsorption have shown that all phages were able to adsorb to the Kp9068 strain by 6–7 min of incubation (Supplementary Figure 1A). The results of a one-step growth assay revealed similar latency and burst periods for all three phages of approximately 30 min (Supplementary Figure 1B). The burst sizes were approximately 66 PFU/cell for the vB_KpnP_Dlv622 phage and 85 and 96 PFU/cell for the vB_KpnM_Seu621 and KpS8, respectively.
The stability test of vB_KpnP_Dlv622, vB_KpnM_Seu621, and KpS8 phages showed that the phages were relatively stable in a range of temperature between 4 and 55°C (Supplementary Figure 2). vB_KpnM_Seu621 and KpS8 phages retained high plaque-forming activity even after incubation at 65°C, indicating good thermal stability. Incubation at a temperature of 75°C demonstrated a significant decrease in titer of all bacteriophages from 109 PFU/ml to 102–105 PFU/ml.
Genome Sequencing and Comparative Genomics
The complete genome of the vB_KpnP_Dlv622 phage was 44,687 bp long, with a G + C content of 54% and two 278 bp long direct repeats located at both ends. Bioinformatic analysis revealed 59 ORFs with a total length of 41,477 bp (coding percentage, 92.8%). BLAST analysis showed that this phage belongs to the Autographiviridae family. To verify this, amino acid sequences of RNA polymerase were used to estimate phylogenetic relationships between vB_KpnP_Dlv622 phage and 46 phages recommended by ICTV and belonging to the Autographiviridae family (last accessed April 23, 2020; Supplementary Figure 3). According to the phylogenetic tree, the vB_KpnP_Dlv622 phage belongs to the Drulisvirus genus of the family Autographiviridae and most closely related to the phage phiKpS2 (accession NC_047857.1, 82% query coverage and 92.6% identity according to BLASTn).
The genome of vB_KpnM_Seu621 was a linear dsDNA molecule of 142,896 bp with a G + C content of 44.64%. A total of 274 ORFs were identified. The complete genome of the KpS8 phage was also a linear dsDNA molecule of 143,800 bp (44.64% G + C content) containing 285 ORFs. Most of the predicted ORFs for both vB_KpnM_Seu621 and KpS8 phages were encoded on the positive strand. According to the Average Nucleotide Identity (ANI; Rodriguez-R and Konstantinidis, 2014) analysis, vB_KpnM_Seu621 and KpS8 phages had high ANI value of 99.48%. According to the BLAST analysis, phages belonged to the Myoviridae family. To verify the taxonomic position of the phages, the phylogenetic analysis was performed using multiple alignments of amino acid sequences of major capsid protein of vB_KpnM_Seu621 and KpS8 phages, 24 phages recommended by ICTV, and four the most related phages from the NCBI database. The results of the phylogenetic analysis showed that our phages are closely related to phage vB_KpnM_KB57 (accession NC_028659.1, 90% query coverage and 98.9% identity according to BLAST) and formed a distinct branch within the clade of Mydovirus genus, Vequintavirinae subfamily (Supplementary Figure 4).
Functional Annotation of K. pneumoniae Phages
According to functional analysis, the proteins of the vB_KpnP_Dlv622 phage could be attributed to five groups: (1) phage structure (11 ORFs); (2) replication, regulation, transcription, and translation (20 ORFs); (3) DNA packaging (two ORFs); (4) host lysis (three ORFs); and (5) hypothetical protein (23 ORFs). vB_KpnP_Dlv622 had typical T7-related genome organization with the presence of lysis cassette composed of spanin-, holin-, and endolysin-encoding genes located next to each other, as well as phage DNA- and RNA-polymerases. No tRNA genes were identified with tRNAScan-SE and ARAGORN toolsets, as well as no significant similarities with known antibiotic resistance determinants, virulence or toxin proteins, and integrase genes were revealed (Supplementary Table 1A).
The genome of vB_KpnP_Dlv622 phage encoded two putative tail fiber and spike proteins dlv622_orf00051 and dlv622_orf00059. The dlv622_orf00051 was 317 aa long and was highly conserved among phages of the Drulisvirus genus. In contrast, dlv622_orf00059 showed more than 80% similarity only to a few phage proteins available in the NCBI database and encoded the potential pectate lyase domain with beta-helix structure characteristic of CPS depolymerases. According to a previous study, this structure of tail fiber and spike proteins means that phage vB_KpnM_Dlv622 belongs to Group A of KP34 viruses containing only one receptor-binding protein dlv622_orf00059 (Table 1; Latka et al., 2019).
For the vB_KpnM_Seu621 and KpS8 phages, a specific putative function (structural proteins, enzymes involved in the replication, regulation, transcription, and translation of DNA, host lysis) could be assigned to products of the same number of ORFs (N = 78). About 24 tRNA were detected in each bacteriophage by tRNAScan-SE and confirmed by ARAGORN. No genes related to phage lysogeny were predicted (Supplementary Tables 1B,C).
Based on the annotations of vB_KpnM_Seu621 and KpS8 phages, five ORFs are likely involved in tail fiber synthesis: (1) putative tail fiber protein (phage621_orf00040 and kps8_orf041), (2) putative tail fiber protein (phage621_orf00045 and kps8_orf046), (3) L-shaped tail fiber (phage621_orf00048 and kps8_orf049), (4) putative tail spike protein (phage621_orf00050 and kps8_orf051), and (5) putative tail spike protein (phage621_orf00052 and kps8_orf053). All the genes involved in tail fiber synthesis were either identical in both phages or had single amino-acid substitutions (phage621_orf00040 and kps8_orf041). It should be noted that among the ORFs described above phage621_orf00052/kps8_orf053 had a high score of similarity with dlv622_orf00059 (98% query coverage and 65.98% identity according to BLASTp; Supplementary Figure 5). In addition, it was revealed that these proteins possess the pectate lyase domain and beta-helix structure characteristic of CPS depolymerases (Table 1).
Host Range of the Three Klebsiella pneumoniae Phages
The panel of 83 K. pneumoniae strains was used to determine the host ranges of the phages. Antimicrobial susceptibility profiles revealed that 53 strains had a multidrug-resistant phenotype (resistance to three or more classes of antibiotics), including at least 19 carbapenem-resistant strains. Results of multilocus sequence typing and wzi gene sequencing showed that K. pneumoniae strains belong to 21 different sequence types and had 26 unique capsular types, respectively (Supplementary Table 3). The most common CPS-types were K2 (n = 14; 16.5%), K57 (n = 11; 12.9%), KL39 (n = 9; 10.6%), and K1 (n = 8; 9.4%).
Isolated phages had a narrow range of lytic activity against K. pneumoniae strains (Supplementary Table 3) and could only infect and lyse strains belonging to the K23 (4/4) capsular types. According to EOP analysis, Myoviridae phages vB_KpnM_Seu621 and KpS8 revealed a high productive infection for three of the four K23 strains, whereas phage vB_KpnP_Dlv622 showed similar effectiveness only on its original host strain (Table 2).
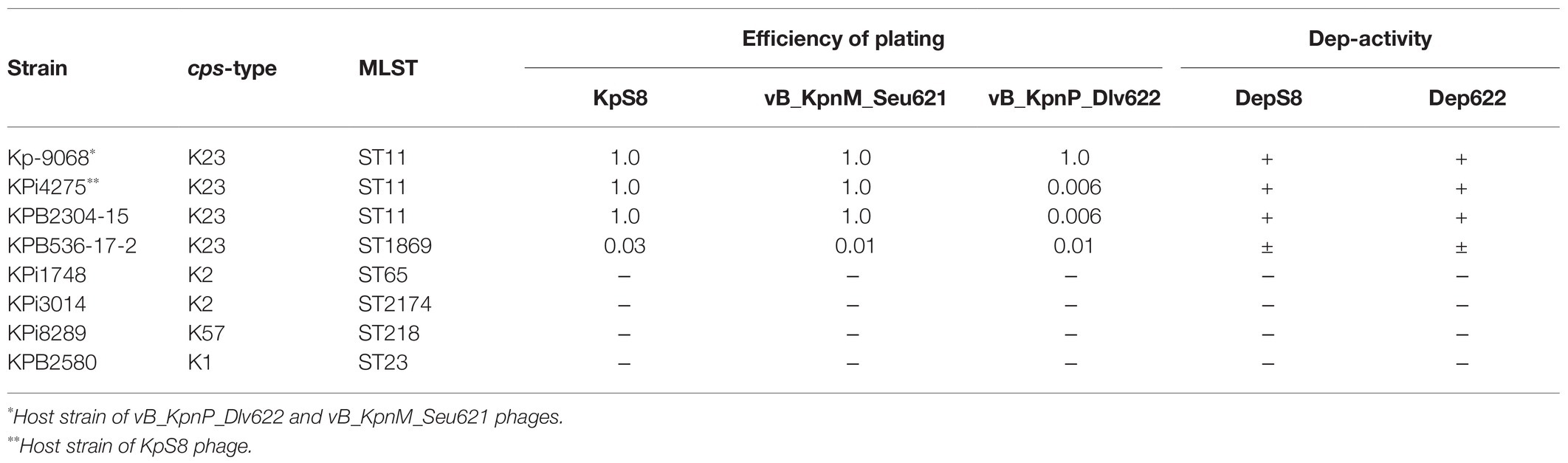
Table 2. The efficiency of plating and depolymerase activity for phages KpS8, vB_KpnM_Seu621, and vB_KpnP_Dlv622 phages and their recombinant depolymerases.
Activity of Phage Depolymerases
To confirm whether the predicted proteins have polysaccharide-degrading activity, we cloned genes kps8_053 (phage KpS8) and dlv622_00059 (phage vB_KpnP_Dlv622) into the pET22b+ expression vector. His-tag fusion proteins DepS8 and Dep622 were expressed and purified.
The activities of DepS8 and Dep622 were tested on four isolates showing sensitivity according to the spot test and four control non-sensitive isolates. Both DepS8 and Dep622 recombinant depolymerases were able to form translucent spots resembling the plaque halo on four K23 isolates and showed a lack of activity for other isolates under investigation (Table 2; Figure 2).
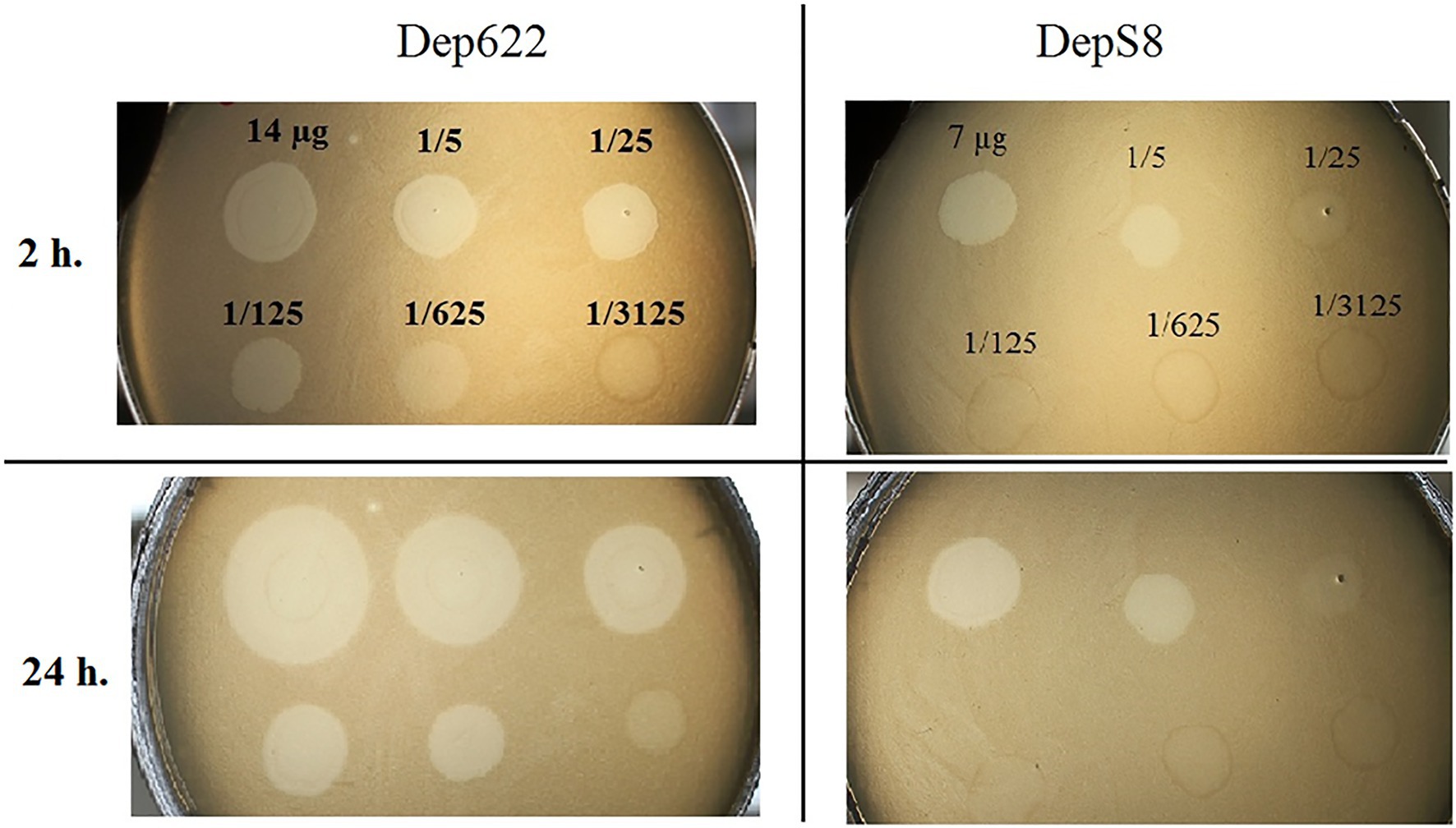
Figure 2. Spot tests of purified depolymerases DepS8 and Dep622 on Klebsiella pneumoniae Kp-9068 lawn.
Depolymerases DepS8 and Dep622 Inhibit the Adsorption of Corresponding Bacteriophages
To study the interaction of bacterial viruses with surface receptors of a bacterial cell and determine whether the CPS depolymerases are the only key components to infect host cells, an adsorption inhibition assay was carried out. We used a competition assay to assess the role of DepS8 and Dep622 in the KpS8 and vB_KpnP_Dlv622 phage-cell interaction at the initial stages of the infection process. The K1 specific depolymerase Dep_kpv71 (Solovieva et al., 2018) was used as a control (Figure 3).
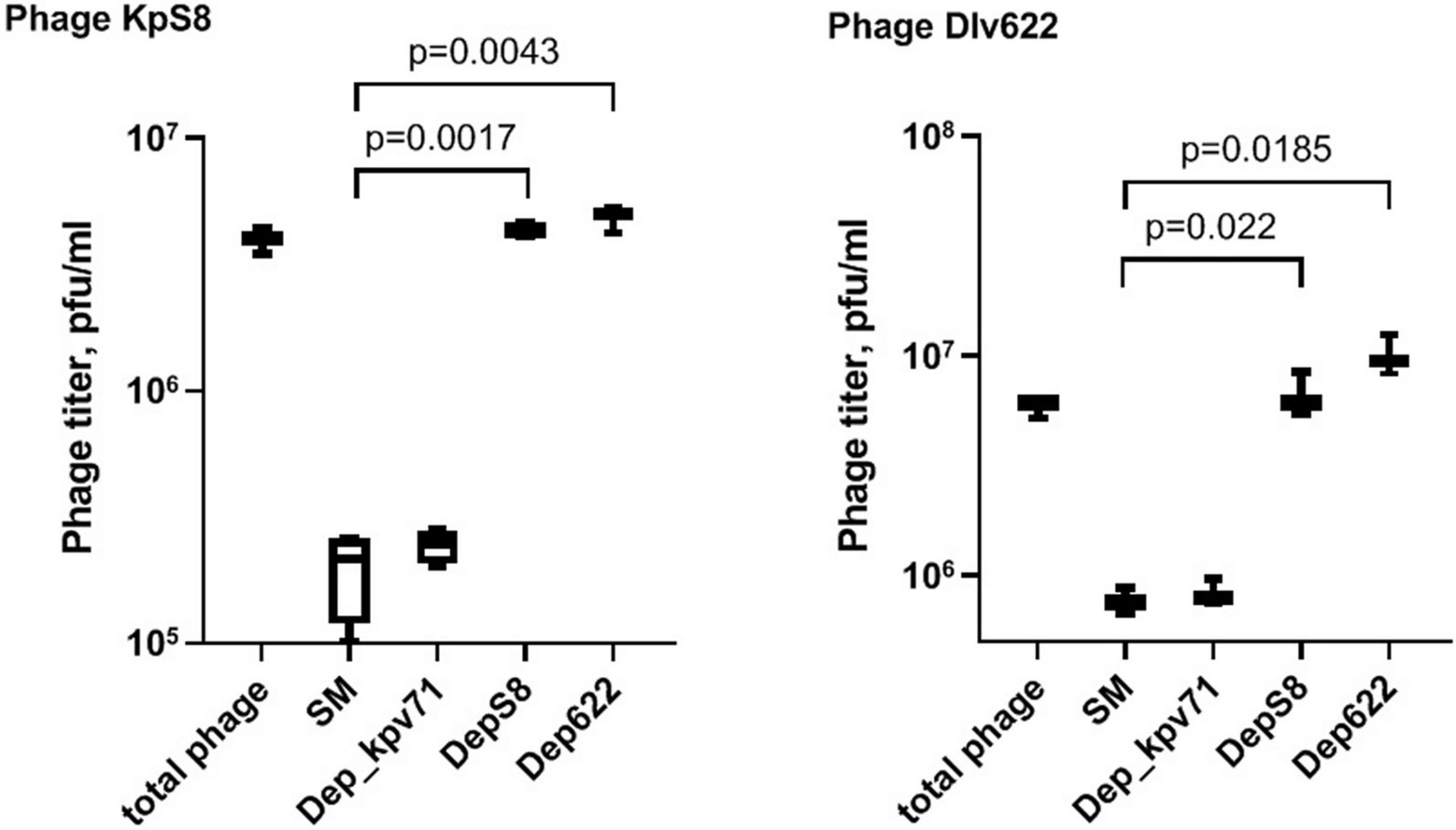
Figure 3. Phage KpS8 and vB_KpnP_Dlv622 adsorption inhibition assay. From left to right, a titer of the total phage sample and titers of the free phage particles after the incubation with K. pneumoniae Kp-9068 cells in SM and in the presence of Dep_kpv71 (K1 specific depolymerase), DepS8, and Dep622 are shown.
The assay showed a 10-fold decrease in phage titer in the control experiments with SM-buffer and with treatment by K1-specific recombinant depolymerase. On the contrary, treating bacterial cells with K23-specific depolymerases DepS8 and Dep622 did not lead to decrease in the titer of free phage. Both DepS8 and Dep622 inhibit the phage adsorption to K. pneumoniae cells, indicating that the phage and depolymerases compete for the same moieties on the host cell surface, which are capsular polysaccharides. These results are consistent with that DepS8 and Dep622 proteins were in silico predicted as putative tail fibers/spikes, responsible for the host cells’ recognition and reversible binding and subsequent degradation of K. pneumoniae cell capsular polysaccharides.
Depolymerase Application Increases the Survival Rate of K. pneumoniae Kp-9068 Infected G. mellonella Larvae
The influence of DepS8 and Dep622 depolymerases on K. pneumoniae Kp-9068 virulence was estimated on the G. mellonella model by two parameters: 50% lethal dose of bacterial cells and survival curves following injection of G. mellonella larvae with fixed doses of K. pneumoniae Kp-9068 (3 × 105 and 3 × 106 CFU per injection).
Experiments have shown a decrease in the virulence of K. pneumoniae Kp-9068 cells that were treated with depolymerases. The median lethal dose (LD50) of untreated strain Kp-9068 was estimated as 1.3 × 105 ± 5.8 × 104 CFU. At the same time, LD50 for bacteria Kp-9068 treated with depolymerase DepS8 or Dep622 was higher by 27.5 and 32.5 times, respectively (Figure 4).
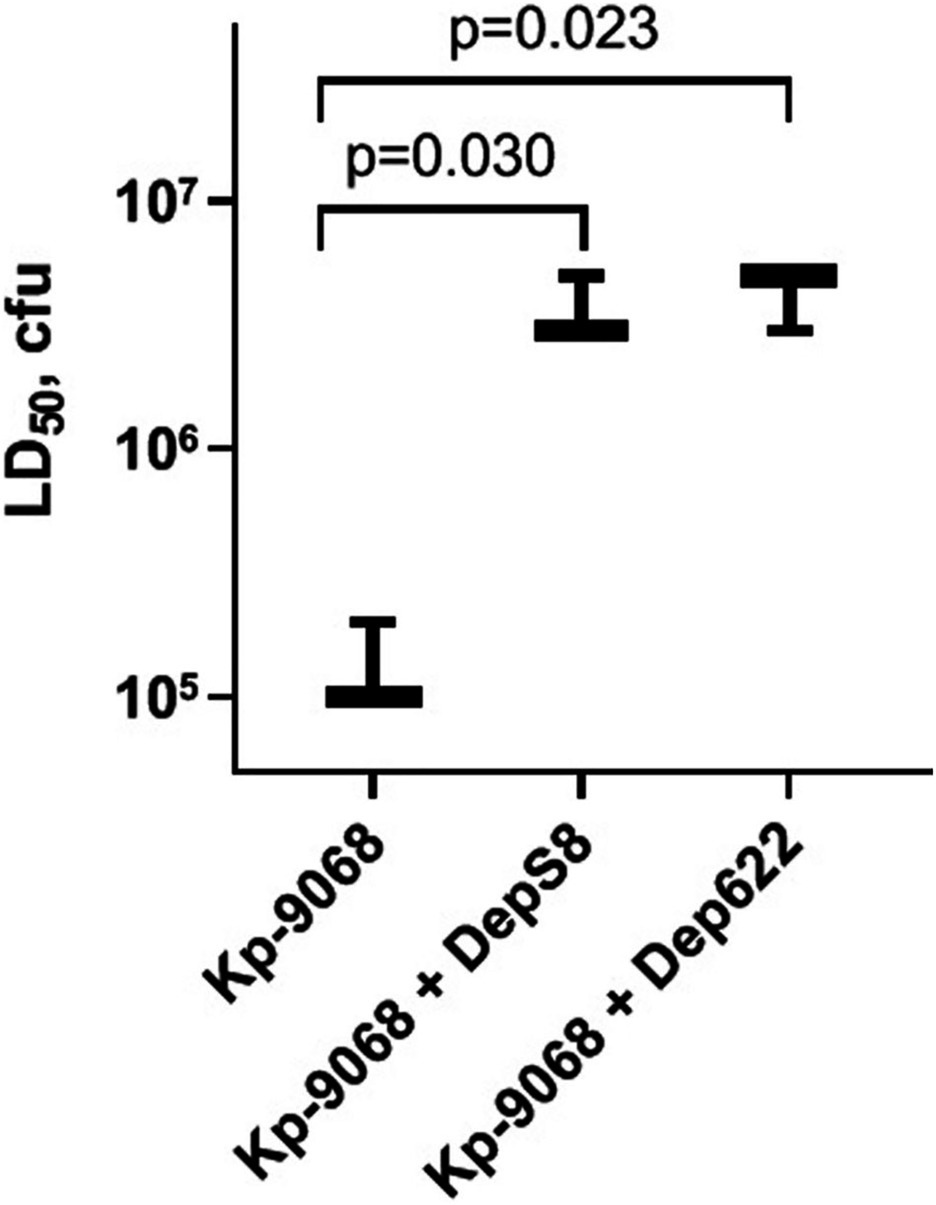
Figure 4. A dose of bacteria K. pneumoniae Kp-9068 is sufficient to kill 50% of the Galleria mellonella larvae population (LD50). From left to right, LD50 of the Kp-9068 untreated with depolymerase enzymes and LD50 of the Kp-9068 cells injected simultaneously with depolymerase DepS8 or Dep622. Average LD50 values from three independent experiments are shown and compared using Student’s t-test analysis. Values of p < 0.05 were considered as statistically significant.
Without the treatment, 100% of the larvae died within 48 h after inoculation of 3 × 106 CFU of K. pneumoniae Kp-9068 and 70% of the larvae died within 5 days after inoculation of 3 × 105 CFU (Figure 5). In contrast, a single dose of each enzyme injected together with bacteria significantly inhibited K. pneumoniae-induced death in a time-dependent manner. Only 10–30% mortality was recorded within 5 days after inoculation of K. pneumoniae Kp-9068 together with depolymerase DepS8 or Dep622 (Figure 5). No mortality of larvae was observed in the controls, upon injection of PBS buffer, DepS8, or Dep622 enzymes alone.
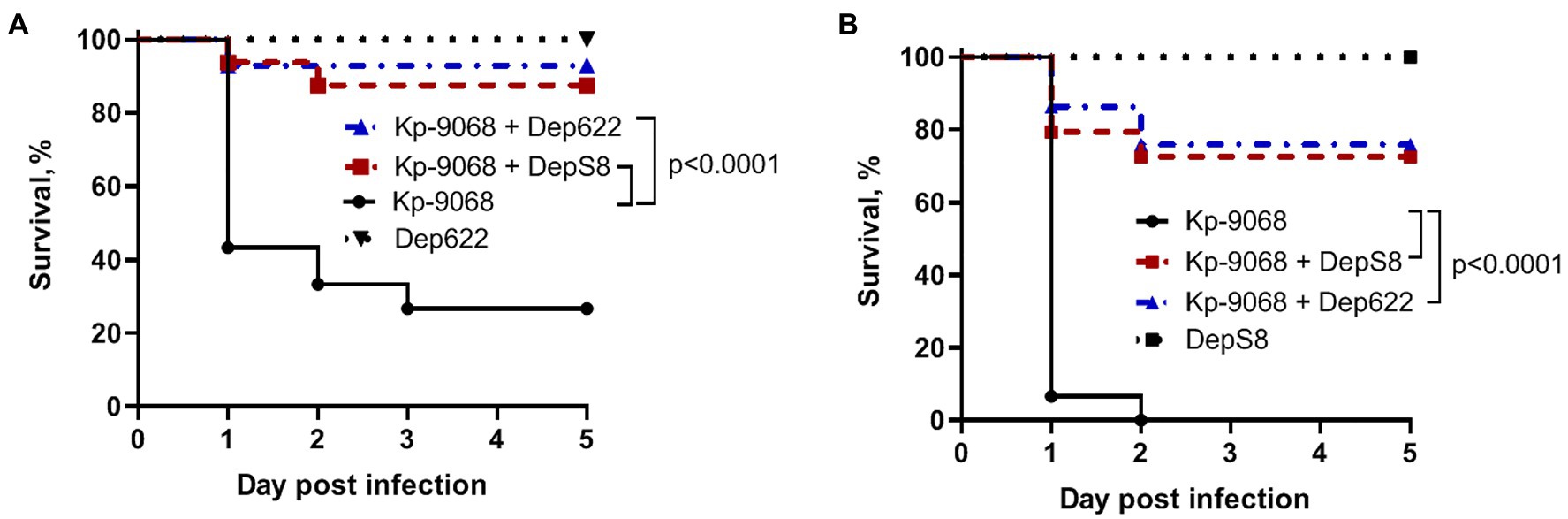
Figure 5. Kaplan–Meier survival curves following injection of G. mellonella larvae with K. pneumoniae Kp-9068 and Kp-9068 simultaneously with depolymerase enzymes. (A) Larvae treatment with 3 × 105 CFU of K. pneumoniae Kp-9068; (B) Larvae treatment with 3 × 106 CFU of K. pneumoniae Kp-9068. Larvae (n = 30) were injected with either bacteria and bacteria simultaneously with enzyme DepS8 or Dep622 (2 μg per larva). The experiments were controlled by observation of phosphate-buffered saline (PBS)-injected larvae and larvae receiving the depolymerase only. Survival for each control group was 100%, so for simplicity, a group of PBS-injected larvae was not included in the figure. Statistically significant differences in survival between larvae infected with bacteria only and larvae infected with bacteria simultaneously with depolymerase DepS8 or Dep622 were estimated by the log-rank (Mantel-Cox) test.
Discussion
Multidrug-resistant K. pneumoniae strains belonging to ST11 were used as host strains for the isolation of bacteriophages. Such isolates are widespread and are often associated with nosocomial outbreaks caused by carbapenem-resistant and hypervirulent strains in China (Gu et al., 2018) and Greece (Voulgari et al., 2014). Furthermore, the K23 capsular type is one of the 10 most common capsular types typically associated with nosocomial K. pneumoniae infection provoked by carbapenem-resistant strains (Follador et al., 2016). Isolates with capsular type K23 are detected on average in 4% of cases; however, among K. pneumoniae associated with the production of carbapenemase, the percentage of such isolates can reach 9–17% (Satlin et al., 2017; Sonda et al., 2018).
Within the context of research for a possible antimicrobial therapeutic agent, three novel lytic K23-specific K. pneumoniae phages were isolated. Phages KpS8 and vB_KpnM_Seu621 belonged to the Myoviridae family and had significant genetic similarities despite different times and sources of isolation (in 2016 from wastewater and in 2018 from river water). In turn, the vB_KpnP_Dlv622 phage belonged to the Autographiviridae family. Despite the considerable phylogenetic distance between the families, the bacteriophages possessed similar depolymerases and their corresponding recombinant proteins Dep622 and DepS8 exhibited the same rate of specificity and activity.
To search for similar cases, we analyzed proteins containing putative depolymerase domains for 76 Klebsiella phages, presented in NCBI (Volozhantsev et al., 2016, 2020; Labudda et al., 2017; Solovieva et al., 2018; Latka et al., 2019; Pan et al., 2019; Teng et al., 2019; Thiry et al., 2019; Wu et al., 2019; Domingo-Calap et al., 2020a,b; Gao et al., 2020; Horváth et al., 2020; Li et al., 2020; Zhang et al., 2020; Supplementary Figure 6). The depolymerase domains of our bacteriophages formed a separate cluster on the tree together with the BIS47_29 protein’s domain (GenBank: YP_009832536.1). Given the fact that according to BLASTp the BIS47_29 protein was similar to kps8_orf053 (100% coverage and 92.09% identity) and dlv622_orf00059 (98% coverage and 65.9% identity), we can assume that BIS47_29 depolymerase also has specificity against capsule type K23.
Confirmed cases in the detection of homologous depolymerases in phages belonging to different families are quite rare (Volozhantsev et al., 2020). Apart from depolymerases specific against the K57 capsule, described in our previous study (Volozhantsev et al., 2020), and depolymerases described in the current article, no similar cases of depolymerases encoded by phages from different families have been reported. In addition to the cases mentioned above, we can assume based on the phylogenetic analysis that similar patterns are observed for at least four other types of depolymerases specific against capsule types K1, K30/K69, K63, and KN2 (Supplementary Figure 6). Although some authors suggest the theoretical possibility of lateral transfer of RBPs or depolymerase domains (Latka et al., 2019), for the time being, there are no reliable ways to distinguish between lateral transfer and convergent evolution.
Due to the dose-dependent effect and the standardizability of application protocols, therapy with phage derivatives is actively discussed on a par with the therapy with virulent bacteriophages (Górski et al., 2020; Volozhantsev et al., 2020). Recombinant tail spike proteins of phages vB_KpnP_Dlv622 and KpS8 significantly reduced the mortality of G. mellonella larvae infected with the host strain supporting infection by these phages (Figure 5). As can be seen from the spot-test data, depolymerases are not less effective than phages directly, and they behave as a potent tool in the treatment of antibiotic-resistant K. pneumoniae infection. Depolymerases cleave the polysaccharides of bacterial cells but, unlike phages, do not possess a lytic activity and do not kill host bacteria. Their therapeutic effect is due to the fact that by cleaving the capsular polysaccharides of K. pneumoniae, depolymerase eliminates the defense of the bacterium against the immune system, thus creating conditions for complement-mediated killing (Lin et al., 2014; Majkowska-Skrobek et al., 2018; Liu et al., 2020; Volozhantsev et al., 2020), and phagocytosis by macrophages (Majkowska-Skrobek et al., 2018).
Conclusion
Three new bacteriophages specific against K. pneumoniae with capsule type K23 were functionally characterized. Their depolymerases had significant similarity, despite the fact that bacteriophages belong to different families. Moreover, apart from the depolymerases specific against the K23 capsule, similar patterns were detected among K1-, K30/K69-, K57-, K63-, and KN2-specific depolymerases. Depolymerases derived from phages KpS8 and vB_KpnP_Dlv622 were equally efficient in cleaving capsular polysaccharides and also significantly reduced the mortality of G. mellonella larvae.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank, MT939253, MT939252, and MT178275; BioProject, PRJNA705078.
Author Contributions
RG, NV, and ES drafted the main manuscript and performed the data analysis. RG, NV, VK, IB, MK, NK, AP, GM, PS, VVV, ED, EK, MM, ID, EI, and ES planned and performed the experiments. RG, NV, AM, VAV, DB, and ES were responsible for bioinformatics analysis of data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the State Assignment on the Development of a personalized approach to the therapy of infections using virulent bacteriophages (Code: Bacteriophage) and the Ministry of Science and Higher Education of the Russian Federation (grant number 075-15-2019-1671).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Federal Research and Clinical Center of Physical-Chemical Medicine of Federal Medical Biological Agency for the opportunity to use computational and sequencing resources. We are grateful to Nadezhda K. Fursova for providing data on antibiotic resistance and sequence types of Klebsiella strains from SCPM-Obolensk.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.669618/full#supplementary-material
References
Ashmarin, I. P., and Vorobyev, A. A. (1962). Statistical Methods in Microbiological Researches. Leningrad: Medgiz.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Besemer, J., Lomsadze, A., and Borodovsky, M. (2001). GenemarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618. doi: 10.1093/nar/29.12.2607
Brisse, S., Passet, V., Haugaard, A. B., Babosan, A., Kassis-Chikhani, N., Struve, C., et al. (2013). Wzi gene sequencing, a rapid method for determination of capsulartype for Klebsiella strains. J. Clin. Microbiol. 51, 4073–4078. doi: 10.1128/JCM.01924-13
Cantón, R., Akóva, M., Carmeli, Y., Giske, C. G., Glupczynski, Y., Gniadkowski, M., et al. (2012). Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 18, 413–431. doi: 10.1111/j.1469-0691.2012.03821.x
Chan, P. P., and Lowe, T. M. (2019). “tRNAscan-SE: searching for tRNA genes in genomic sequences,” in Methods in Molecular Biology ed. M. Kollmar (New York, NY: Humana Press), 1–14. doi: 10.1007/978-1-4939-9173-0_1
Chhibber, S., Kaur, S., and Kumari, S. (2008). Therapeutic potential of bacteriophage in treating Klebsiella pneumoniae B5055-mediated lobar pneumonia in mice. J. Med. Microbiol. 57, 1508–1513. doi: 10.1099/jmm.0.2008/002873-0
Ciacci, N., D’Andrea, M. M., Marmo, P., Demattè, E., Amisano, F., Di Pilato, V., et al. (2018). Characterization of vB_Kpn_F48, a newly discovered lytic bacteriophage for Klebsiella pneumoniae of sequence type 101. Viruses 10:482. doi: 10.3390/v10090482
Clark, J. R., and March, J. B. (2006). Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends Biotechnol. 24, 212–218. doi: 10.1016/j.tibtech.2006.03.003
Clinical and Laboratory Standards Institute (2018). Performance Standards for Antimicrobial Susceptibility Testing M100. Wayne, PA: CLSI.
D’Andrea, M. M., Marmo, P., Angelis, L. H. D., Palmieri, M., Ciacci, N., Di Lallo, G., et al. (2017). φbO1E, a newly discovered lytic bacteriophage targeting carbapenemase-producing Klebsiella pneumoniae of the pandemic clonal group 258 clade II lineage. Sci. Rep. 7:2614. doi: 10.1038/s41598-017-02788-9
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). Prottest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
Diancourt, L., Passet, V., Verhoef, J., Grimont, P. A. D., and Brisse, S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43, 4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005
Domingo-Calap, P., Beamud, B., Mora-Quilis, L., González-Candelas, F., and Sanjuán, R. (2020a). Isolation and characterization of two Klebsiella pneumoniae phages encoding divergent depolymerases. Int. J. Mol. Sci. 21:3160. doi: 10.3390/ijms21093160
Domingo-Calap, P., Beamud, B., Vienne, J., González-Candelas, F., and Sanjuán, R. (2020b). Isolation of four lytic phages infecting Klebsiella pneumoniae K22 clinical isolates from Spain. Int. J. Mol. Sci. 21:425. doi: 10.3390/ijms21020425
European Centre for Disease Prevention and Control (2019). Surveillance of Antimicrobial Resistance in Europe Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2018. Stockholm: ECDC.
Fang, F. C., Sandler, N., and Libby, S. J. (2005). Liver abscess caused by magA+ Klebsiella pneumoniae in North America. J. Clin. Microbiol. 43, 991–992. doi: 10.1128/JCM.43.2.991-992.2005
Follador, R., Heinz, E., Wyres, K. L., Ellington, M. J., Kowarik, M., Holt, K. E., et al. (2016). The diversity of Klebsiella pneumoniae surface polysaccharides. Microb. Genom. 2:e000073. doi: 10.1099/mgen.0.000073
Gao, M., Wang, C., Qiang, X., Liu, H., Li, P., Pei, G., et al. (2020). Isolation and characterization of a novel bacteriophage infecting carbapenem-resistant Klebsiella pneumoniae. Curr. Microbiol. 77, 722–729. doi: 10.1007/s00284-019-01849-8
Garneau, J. R., Depardieu, F., Fortier, L. C., Bikard, D., and Monot, M. (2017). Phageterm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 7:8292. doi: 10.1038/s41598-017-07910-5
Girmenia, C., Viscoli, C., Piciocchi, A., Cudillo, L., Botti, S., Errico, A., et al. (2015). Management of carbapenem resistant Klebsiella pneumoniae infections in stem cell transplant recipients: an Italian multidisciplinary consensus statement. Haematologica 100, e373–e376. doi: 10.3324/haematol.2015.125484
Górski, A., Międzybrodzki, R., Węgrzyn, G., Jończyk-Matysiak, E., Borysowski, J., and Weber-Dąbrowska, B. (2020). Phage therapy: current status and perspectives. Med. Res. Rev. 40, 459–463. doi: 10.1002/med.21593
Green, M., Hughes, H., Sambrook, J., and MacCallum, P. (2012). Molecular Cloning: a Laboratory Manual. New York: Cold Spring Harbor Laboratory Press.
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
Horváth, M., Kovács, T., Koderivalappil, S., Ábrahám, H., Rákhely, G., and Schneider, G. (2020). Identification of a newly isolated lytic bacteriophage against K24 capsular type, carbapenem resistant Klebsiella pneumoniae isolates. Sci. Rep. 10:5891. doi: 10.1038/s41598-020-62691-8
Hsieh, P. F., Lin, H. H., Lin, T. L., Chen, Y. Y., and Wang, J. T. (2017). Two T7-like bacteriophages, K5-2 and K5-4, each encodes two capsule depolymerases: isolation and functional characterization. Sci. Rep. 7:4624. doi: 10.1038/s41598-017-04644-2
Hung, C. H., Kuo, C. F., Wang, C. H., Wu, C. M., and Tsao, N. (2011). Experimental phage therapy in treating Klebsiella pneumoniae-mediated liver abscesses and bacteremia in mice. Antimicrob. Agents Chemother. 55, 1358–1365. doi: 10.1128/AAC.01123-10
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Knecht, L. E., Veljkovic, M., and Fieseler, L. (2020). Diversity and function of phage encoded depolymerases. Front. Microbiol. 10:2949. doi: 10.3389/fmicb.2019.02949
Kornienko, M., Ilina, E., Lubasovskaya, L., Priputnevich, T., Falova, O., Sukhikh, G., et al. (2016). Analysis of nosocomial Staphylococcus haemolyticus by MLST and MALDI-TOF mass spectrometry. Infect. Genet. Evol. 39, 99–105. doi: 10.1016/j.meegid.2015.12.015
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., and Stamatakis, A. (2019). RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. doi: 10.1093/bioinformatics/btz305
Kropinski, A. M. (2009). “Measurement of the rate of attachment of bacteriophage to cells,” in Bacteriophages Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. eds. M. R. J. Clokie and A. M. Kropinski (New York: Humana Press), 151–157.
Labudda, Ł., Strapagiel, D., and Karczewska-golec, J. (2017). Complete annotated genome sequences of four Klebsiella pneumoniae phages isolated from sewage in Poland. Genome Announc. 5, e00919–e01017. doi: 10.1128/genomeA.00919-17
Laslett, D., and Canback, B. (2004). ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16. doi: 10.1093/nar/gkh152
Latka, A., Leiman, P. G., Drulis-Kawa, Z., and Briers, Y. (2019). Modeling the architecture of depolymerase-containing receptor binding proteins in Klebsiella phages. Front. Microbiol. 10:2649. doi: 10.3389/fmicb.2019.02649
Lee, C. R., Lee, J. H., Park, K. S., Kim, Y. B., Jeong, B. C., and Lee, S. H. (2016). Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7:895. doi: 10.3389/fmicb.2016.00895
Li, M., Xiao, Y., Li, P., Wang, Z., Qi, W., Qi, Z., et al. (2020). Characterization and genome analysis of Klebsiella phage P509, with lytic activity against clinical carbapenem-resistant Klebsiella pneumoniae of the KL64 capsular type. Arch. Virol. 165, 2799–2806. doi: 10.1007/s00705-020-04822-0
Lin, T. L., Hsieh, P. F., Huang, Y. T., Lee, W. C., Tsai, Y. T., Su, P. A., et al. (2014). Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment. J. Infect. Dis. 210, 1734–1744. doi: 10.1093/infdis/jiu332
Liu, Y. C., Cheng, D. L., and Lin, C. L. (1986). Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 146, 1913–1916. doi: 10.1001/archinte.1986.00360220057011
Liu, Y., Leung, S. S. Y., Huang, Y., Guo, Y., Jiang, N., Li, P., et al. (2020). Identification of two depolymerases from phage IME205 and their antivirulent functions on K47 capsule of Klebsiella pneumoniae. Front. Microbiol. 11:218. doi: 10.3389/fmicb.2020.00218
Liu, B., and Pop, M. (2009). ARDB—antibiotic resistance genes database. Nucleic Acids Res. 37, 443–447. doi: 10.1093/nar/gkn656
Liu, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, D687–D692. doi: 10.1093/nar/gky1080
Majkowska-Skrobek, G., Łątka, A., Berisio, R., Maciejewska, B., Squeglia, F., Romano, M., et al. (2016). Capsule-targeting depolymerase, derived from Klebsiella KP36 phage, as a tool for the development of anti-virulent strategy. Viruses 8:324. doi: 10.3390/v8120324
Majkowska-Skrobek, G., Latka, A., Berisio, R., Squeglia, F., Maciejewska, B., Briers, Y., et al. (2018). Phage-borne depolymerases decrease Klebsiella pneumoniae resistance to innate defense mechanisms. Front. Microbiol. 9:2517. doi: 10.3389/fmicb.2018.02517
Manohar, P., Tamhankar, A. J., Lundborg, C. S., and Nachimuthu, R. (2019). Therapeutic characterization and efficacy of bacteriophage cocktails infecting Escherichia coli, Klebsiella pneumoniae, and Enterobacter species. Front. Microbiol. 10:574. doi: 10.3389/fmicb.2019.00574
Mazzocco, A., Waddell, T. E., Lingohr, E., and Roger, J. P. (2009). “Enumeration of bacteriophages by the direct plating plaque assay,” in Bacteriophages Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. eds. M. R. J. Clokie and A. M. Kropinski (New York: Humana Press), 77–81.
Meier-Kolthoff, J. P., and Göker, M. (2017). VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 33, 3396–3404. doi: 10.1093/bioinformatics/btx440
Mirzaei, M. K., and Nilsson, A. S. (2015). Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS One 10:e0118557. doi: 10.1371/journal.pone.0118557
Munoz-Price, L. S., Poirel, L., Bonomo, R. A., Schwaber, M. J., Daikos, G. L., Cormican, M., et al. (2013). Clinical epidemiology of the global expansion of kpn carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/S1473-3099(13)70190-7
Paczosa, M. K., and Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/MMBR.00078-15
Pan, Y. J., Lin, T. L., Chen, Y. Y., Lai, P. H., Tsai, Y. T., Hsu, C. R., et al. (2019). Identification of three podoviruses infecting Klebsiella encoding capsule depolymerases that digest specific capsular types. Microb. Biotechnol. 12, 472–486. doi: 10.1111/1751-7915.13370
Pan, Y., Lin, T., Chen, C., and Tsai, Y. (2017). Klebsiella phage ΦK64-1 encodes multiple depolymerases for multiple host capsular types. J. Virol. 91, e02457–e02516. doi: 10.1128/JVI.02457-16
Payne, R. J. H., and Jansen, V. A. A. (2000). Phage therapy: the peculiar kinetics of self-replicating pharmaceuticals. Clin. Pharmacol. Ther. 68, 225–230. doi: 10.1067/mcp.2000.109520
Pires, D. P., Oliveira, H., Melo, L. D. R., Sillankorva, S., and Azeredo, J. (2016). Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 100, 2141–2151. doi: 10.1007/s00253-015-7247-0
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/CMR.11.4.589
Rodriguez-R, L. M., and Konstantinidis, K. T. (2014). Bypassing cultivation to identify bacterial species. Microbe 9, 111–118. doi: 10.1128/microbe.9.111.1
Satlin, M. J., Chen, L., Patel, G., Gomez-Simmonds, A., Weston, G., Kim, A. C., et al. (2017). Multicenter clinical and molecular epidemiological analysis of bacteremia due to carbapenem-resistant Enterobacteriaceae (CRE) in the CRE epicenter of the United States. Antimicrob. Agents Chemother. 61, e02349–e02416. doi: 10.1128/AAC.02349-16
Scorpio, A., Tobery, S. A., Ribot, W. J., and Friedlander, A. M. (2008). Treatment of experimental anthrax with recombinant capsule depolymerase. Antimicrob. Agents Chemother. 52, 1014–1020. doi: 10.1128/AAC.00741-07
Shon, A. S., Bajwa, R. P. S., and Russo, T. A. (2013). Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4, 107–118. doi: 10.4161/viru.22718
Solovieva, E. V., Myakinina, V. P., Kislichkina, A. A., Krasilnikova, V. M., Verevkin, V. V., Mochalov, V. V., et al. (2018). Comparative genome analysis of novel podoviruses lytic for hypermucoviscous Klebsiella pneumoniae of K1, K2, and K57 capsular types. Virus Res. 243, 10–18. doi: 10.1016/j.virusres.2017.09.026
Sonda, T., Kumburu, H., van Zwetselaar, M., Alifrangis, M., Mmbaga, B. T., Lund, O., et al. (2018). Molecular epidemiology of virulence and antimicrobial resistance determinants in Klebsiella pneumoniae from hospitalised patients in Kilimanjaro, Tanzania. Eur. J. Clin. Microbiol. Infect. Dis. 37, 1901–1914. doi: 10.1007/s10096-018-3324-5
Stewart, P. S. (1996). Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob. Agents Chemother. 40, 2517–2522. doi: 10.1128/AAC.40.11.2517
Tängdén, T., and Giske, C. G. (2015). Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J. Intern. Med. 277, 501–512. doi: 10.1111/joim.12342
Teng, T., Li, Q., Liu, Z., Li, X., Liu, Z., Liu, H., et al. (2019). Characterization and genome analysis of novel Klebsiella phage Henu1 with lytic activity against clinical strains of Klebsiella pneumoniae. Arch. Virol. 164, 2389–2393. doi: 10.1007/s00705-019-04321-x
Thiry, D., Passet, V., Danis-wlodarczyk, K., Lood, C., Wagemans, J., De Sordi, L., et al. (2019). New bacteriophages against emerging lineages ST23 and ST258 of Klebsiella pneumoniae and efficacy assessment in Galleria mellonella larvae. Viruses 11:411. doi: 10.3390/v11050411
Turton, J. F., Englender, H., Gabriel, S. N., Turton, S. E., Kaufmann, M. E., and Pitt, T. L. (2007). Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J. Med. Microbiol. 56, 593–597. doi: 10.1099/jmm.0.46964-0
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., and Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
Van Twest, R., and Kropinski, A. M. (2009). “Bacteriophage enrichment from water and soil,” in Bacteriophages Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions eds. M. R. J. Clokie and A. M. Kropinski (New York: Humana Press), 15–23.
Volozhantsev, N. V., Myakinina, V. P., Popova, A. V., Kislichkina, A. A., Komisarova, E. V., Knyazeva, A. I., et al. (2016). Complete genome sequence of novel T7-like virus vB_KpnP_KpV289 with lytic activity against Klebsiella pneumoniae. Arch. Virol. 161, 499–501. doi: 10.1007/s00705-015-2680-z
Volozhantsev, N. V., Shpirt, A. M., Borzilov, A. I., Komisarova, E. V., Krasilnikova, V. M., Shashkov, A. S., et al. (2020). Characterization and therapeutic potential of bacteriophage-encoded polysaccharide depolymerases with β galactosidase activity against Klebsiella pneumoniae K57 capsular type. Antibiotics 9:732. doi: 10.3390/antibiotics9110732
Voulgari, E., Gartzonika, C., Vrioni, G., Politi, L., Priavali, E., Levidiotou-Stefanou, S., et al. (2014). The Balkan region: NDM-1-producing Klebsiella pneumoniae ST11 clonal strain causing outbreaks in Greece. J. Antimicrob. Chemother. 69, 2091–2097. doi: 10.1093/jac/dku105
Wang, C., Li, P., Niu, W., Yuan, X., Liu, H., Huang, Y., et al. (2019). Protective and therapeutic application of the depolymerase derived from a novel KN1 genotype of Klebsiella pneumoniae bacteriophage in mice. Res. Microbiol. 170, 156–164. doi: 10.1016/j.resmic.2019.01.003
Wu, Y., Wang, R., Xu, M., Liu, Y., Zhu, X., Qiu, J., et al. (2019). A novel polysaccharide depolymerase encoded by the phage SH-KP152226 confers specific activity against multidrug-resistant Klebsiella pneumoniae via biofilm degradation. Front. Microbiol. 10:2768. doi: 10.3389/fmicb.2019.02768
Wyres, K. L., Wick, R. R., Gorrie, C., Jenney, A., Follador, R., Thomson, N. R., et al. (2016). Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2:e000102. doi: 10.1099/mgen.0.000102
Yu, Z., Qin, W., Lin, J., Fang, S., and Qiu, J. (2015). Antibacterial mechanisms of polymyxin and bacterial resistance. Biomed Res. Int. 2015:679109. doi: 10.1155/2015/679109
Yu, G., Smith, D. K., Zhu, H., Guan, Y., and Lam, T. T. Y. (2017). Ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. doi: 10.1111/2041-210X.12628
Zhang, K. Y., Gao, Y. Z., Du, M. Z., Liu, S., Dong, C., and Guo, F. B. (2019). Vgas: a viral genome annotation system. Front. Microbiol. 10:184. doi: 10.3389/fmicb.2019.00184
Zhang, R., Zhao, F., Wang, J., Pei, G., Fan, H., Zhangxiang, L., et al. (2020). Biological characteristics and genome analysis of a novel phage vB_KpnP_IME279 infecting Klebsiella pneumoniae. Folia Microbiol. 65, 925–936. doi: 10.1007/s12223-020-00775-8
Keywords: Klebsiella pneumoniae, multidrug-resistance, capsular type, capsule depolymerase, bacteriophage, Galleria mellonella
Citation: Gorodnichev RB, Volozhantsev NV, Krasilnikova VM, Bodoev IN, Kornienko MA, Kuptsov NS, Popova AV, Makarenko GI, Manolov AI, Slukin PV, Bespiatykh DA, Verevkin VV, Denisenko EA, Kulikov EE, Veselovsky VA, Malakhova MV, Dyatlov IA, Ilina EN and Shitikov EA (2021) Novel Klebsiella pneumoniae K23-Specific Bacteriophages From Different Families: Similarity of Depolymerases and Their Therapeutic Potential. Front. Microbiol. 12:669618. doi: 10.3389/fmicb.2021.669618
Edited by:
Robert Czajkowski, University of Gdansk, PolandReviewed by:
Flavia Squeglia, Institute of Biostructure and Bioimaging, National Research Council (CNR), ItalyMarco Maria D’Andrea, University of Rome Tor Vergata, Italy
Hugo Oliveira, University of Minho, Portugal
Copyright © 2021 Gorodnichev, Volozhantsev, Krasilnikova, Bodoev, Kornienko, Kuptsov, Popova, Makarenko, Manolov, Slukin, Bespiatykh, Verevkin, Denisenko, Kulikov, Veselovsky, Malakhova, Dyatlov, Ilina and Shitikov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roman B. Gorodnichev, Z29yb2RuaWNoZXYuci5iQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Roman B. Gorodnichev
Roman B. Gorodnichev Nikolay V. Volozhantsev
Nikolay V. Volozhantsev Valentina M. Krasilnikova2
Valentina M. Krasilnikova2 Alexander I. Manolov
Alexander I. Manolov Dmitry A. Bespiatykh
Dmitry A. Bespiatykh Vladimir A. Veselovsky
Vladimir A. Veselovsky Maja V. Malakhova
Maja V. Malakhova Elena N. Ilina
Elena N. Ilina Egor A. Shitikov
Egor A. Shitikov