- 1The M-Lab, Department of Precision Medicine, GROW - School of Oncology, Maastricht University, Maastricht, Netherlands
- 2Exomnis Biotech BV, Oxfordlaan, Maastricht, Netherlands
Despite a history dating back to the 1800s, using Clostridium bacteria to treat cancer has not advanced beyond the observation that they can colonise and partially destroy solid tumours. Progress has been hampered by their inability to eradicate the viable portion of tumours, and an instinctive anxiety around injecting patients with a bacterium whose close relatives cause tetanus and botulism. However, recent advances in techniques to genetically engineer Clostridium species gives cause to revisit this concept. This paper illustrates these developments through the attenuation of C. sporogenes to enhance its clinical safety, and through the expression and secretion of an immunotherapeutic. An 8.6 kb sequence, corresponding to a haemolysin operon, was deleted from the genome and replaced with a short non-coding sequence. The resultant phenotype of this strain, named C. sporogenes-NT, showed a reduction of haemolysis to levels similar to the probiotic strain, C. butyricum M588. Comparison to the parental strain showed no change in growth or sporulation. Following injection of tumour-bearing mice with purified spores of the attenuated strain, high levels of germination were detected in all tumours. Very low levels of spores and vegetative cells were detected in the spleen and lymph nodes. The new strain was transformed with four different murine IL-2-expressing plasmids, differentiated by promoter and signal peptide sequences. Biologically active mIL-2, recovered from the extracellular fraction of bacterial cultures, was shown to stimulate proliferation of T cells. With this investigation we propose a new, safer candidate for intratumoral delivery of cancer immunotherapeutics.
Introduction
Clostridium sporogenes has been an important species in the field of experimental oncology for over 50 years, due to its ability to colonise solid tumours (Gericke and Engelbart, 1964; Heap et al., 2014). The hypoxic and necrotic environment of these tumours is ideal for the germination and growth of proteolytic Clostridium species, such as C. sporogenes. Intravenously injected spores germinate and proliferate exclusively in the hypoxic/necrotic environment of tumour xenografts, while vegetative cells are not detected in non-tumour-bearing control animals (Mowday et al., 2016). Injected spores are very well tolerated in rodent models, indicating low immunogenicity (Theys et al., 2001). While colonisation of tumours can lead to destruction of the necrotic portion of solid tumours, the potency of C. sporogenes alone appears to be inadequate for complete elimination of tumours. The absolute requirement of anoxic conditions, essential for initial colonization, may also explain why the proliferating Clostridium in the centre of a solid tumour are unable to attack the oxygenated fraction of the tumour, which constitutes the viable, proliferating portion of the cancer. A therapeutic agent, secreted by a recombinant C. sporogenes from the anoxic centre of a tumour, could overcome this limitation if it is able to diffuse to the living portion of the tumour and exert its effect.
Progress in understanding the biology of Clostridium species, and the concomitant development of genetic tools and genetic transformation, has enabled researchers to exploit this genus in the industrial and medical settings. Clostridium Directed Enzyme Prodrug Therapy (CDEPT) is an illustration of these developments (Mowday et al., 2021). CDEPT exploits the tumour-specific germination of an engineered C. sporogenes, which expresses a prodrug-converting enzyme from the bacterial chromosome to activate a systemically administered prodrug. Significant therapeutic effect was achieved in a mouse xenograft model (Heap et al., 2014).
C. sporogenes is also an important species for the food industry, where it is utilised as a surrogate for proteolytic Clostridium botulinum to test the efficacy of thermal processing (Brown et al., 2012). This sterilisation technique aims to destroy microbes, including spores, present in the food. Survival of C. botulinum is a concern due to its ability to germinate and produce the botulinum neurotoxin in anaerobic environments. At the nucleotide level, C. sporogenes shares 93.4% sequence identity with proteolytic C. botulinum (group 1), just short of the threshold for classing them as the same species (95%) (Weigand et al., 2015). The near identical phenotype without the risk of exposure to botulinum toxin makes this species an ideal surrogate. Inevitably, the significant similarity between these species has led to the misdiagnosis of pathogenic clostridia as C. sporogenes (Lindstrom et al., 1999). A pubmed search with the query “Clostridium sporogenes[title]” reveals five clinical reports in the last 30 years purporting C. sporogenes as the causative agent of infection. Validation of these cases is not possible due to use of inadequate methods or insufficient detail of diagnostics reported.
C. sporogenes is ubiquitous in the natural environment, and there is a growing weight of evidence to support the notion that this species is a beneficial member of the human gut microbiome. Studies in mice demonstrate its ability to produce a potent antioxidant and to modulate IgA-related immune cells via production of branched short chain fatty acids (SCFAs) (Wikoff et al., 2009; Dodd et al., 2017; Guo et al., 2019). Despite the good press, the presence of a nine gene cluster in C. sporogenes ATCC 15579 with high sequence similarity to the Streptolysin S (SLS) operon of Streptococcus pyogenes is a significant hurdle to its therapeutic use (Gonzalez et al., 2010).
We hypothesised that C. sporogenes NCIMB 10696 could be attenuated by deletion of the SLS operon without loss of the beneficial properties of the parental strain. The resultant strain would be safer for clinical and industrial use. To test this hypothesis, we deleted the SLS operon from this strain using CRISPR-Cas9, and conducted a comparative phenotype analysis with the parental strain. We have named this strain C. sporogenes-NT (non-toxic). In addition, we tested the new strain’s ability to express and secrete the murine cytokine, interleukin-2 (IL-2). This cytokine is the ideal candidate for intratumoral expression, due to its very high ED50 when given intravenously and the frequent occurrence of adverse events in patients (Buchbinder et al., 2019).
Materials and Methods
Bacterial Strains, Growth Conditions and Cell Lines
Details of all bacterial strains used in this study are listed in Supplementary Table 1. C. sporogenes NCIMB 10696 wild type strain was purchased from the NCIMB culture collection, received in a lyophilized state, and processed in accordance with NCIMB specifications. Two Escherichia coli strains (10-beta and S17-1) were used as the cloning and conjugative donor strains, respectively.
Growth of C. sporogenes strains was carried out under anaerobic conditions in an anaerobic cabinet (model MG1000 Mark II, Don Whitley Scientific Ltd.; 80% N2, 10% CO2, 10% H2) at 37°C. C. sporogenes was grown in a bovine-free version of TY media, labelled “Peptone Yeast Thioglycolate” (PYT), supplemented with D-cycloserine (250 μg/ml), and thiamphenicol (15 μg/ml) where appropriate. The CTLL-2 indicator cell line (93042610, ECACC) was cultivated according to the manufacturer’s instructions.
Plasmid Construction and Isolation of C. sporogenes-NT Knockout Strain
Details of all vectors and oligonucleotides used in this study are listed in Supplementary Tables 1, 2. The pMTL82121 vector was digested with restriction enzymes NotI/AscI and the ∼5 kb fragment was used as the backbone for the CRISPR-Cas9 gene editing plasmids (pPME-101-g1 and pPME-101-g2). The construction proceeded as previously reported (Heap et al., 2009; Ingle et al., 2019). The cas9 gene was amplified from S. pyogenes genomic DNA using primers with upstream cloning sites (BsaI/NotI). Expression of the cas9 gene and the gRNA was achieved using the thl and araE gene promoters from Clostridium acetobutylicum, respectively, in a divergent arrangement. The 20-nt targeting sequences (guide 1 and guide 2) were designed using the CRISPR Guide RNA Design tool at the Benchling (2019) website. The gRNA DNA fragment was created using a conserved reverse primer (corresponding to the 3′ of the gRNA handle and terminator) and a forward primer containing the targeting sequence at the 5′ and the beginning of the gRNA handle at the 3′. Complementary sequences between these primers enabled the creation of a product in a template-free “primer dimer” reaction.
The SLS homologue was located in the C. sporogenes NCIMB 10696 genome by blastn alignment, using the equivalent genes in C. sporogenes ATCC 15579 as a query (Gonzalez et al., 2010). Sequences of approximately 700 bp long upstream and downstream of the operon were designated as left homology arm (LHA) and right homology arm (RHA), respectively. These were amplified from genomic DNA using Phusion® high fidelity polymerase, according to the manufacturer’s protocol [M0531, New England Biolabs (NEB)]. A synthetic “bookmark” sequence (BM1) was placed between the homology arms. This was created by dividing the sequence between the LHA reverse and RHA forward primer tails with upstream BsaI restriction sites.
Ligated plasmids were transformed by heat shock into E. coli 10-beta and plated on LB plates supplemented with chloramphenicol. Resultant colonies were PCR-screened for correct assembly and subsequent plasmid samples were confirmed by Sanger sequencing.
Deletion of the haemolysin operon was achieved using the CRISPR-Cas9 genome editing tool (Ingle et al., 2019) and schematically illustrated in Figure 1A. Constructed knockout vectors (pPME-101-g1 and pPME-101-g2) were heat shock transformed into E. coli S17-1 for conjugation to C. sporogenes-WT. Conjugation was carried out as previously described (Heap et al., 2009). Transconjugants were screened by colony PCR using primers that flank the SLS-like operon genome locus. The CRISPR-Cas9 vector was removed from knockout mutants by subculturing in non-selective liquid cultures (24 h) followed by patch plating to selective and non-selective plates to observe loss of chloramphenicol resistance. Plasmid loss was further confirmed by PCR using primers specific for the plasmid-based catP gene (data not shown). Resultant positive clones were sent for sequencing. The haemolytic KO strain has been named C. sporogenes-NT.
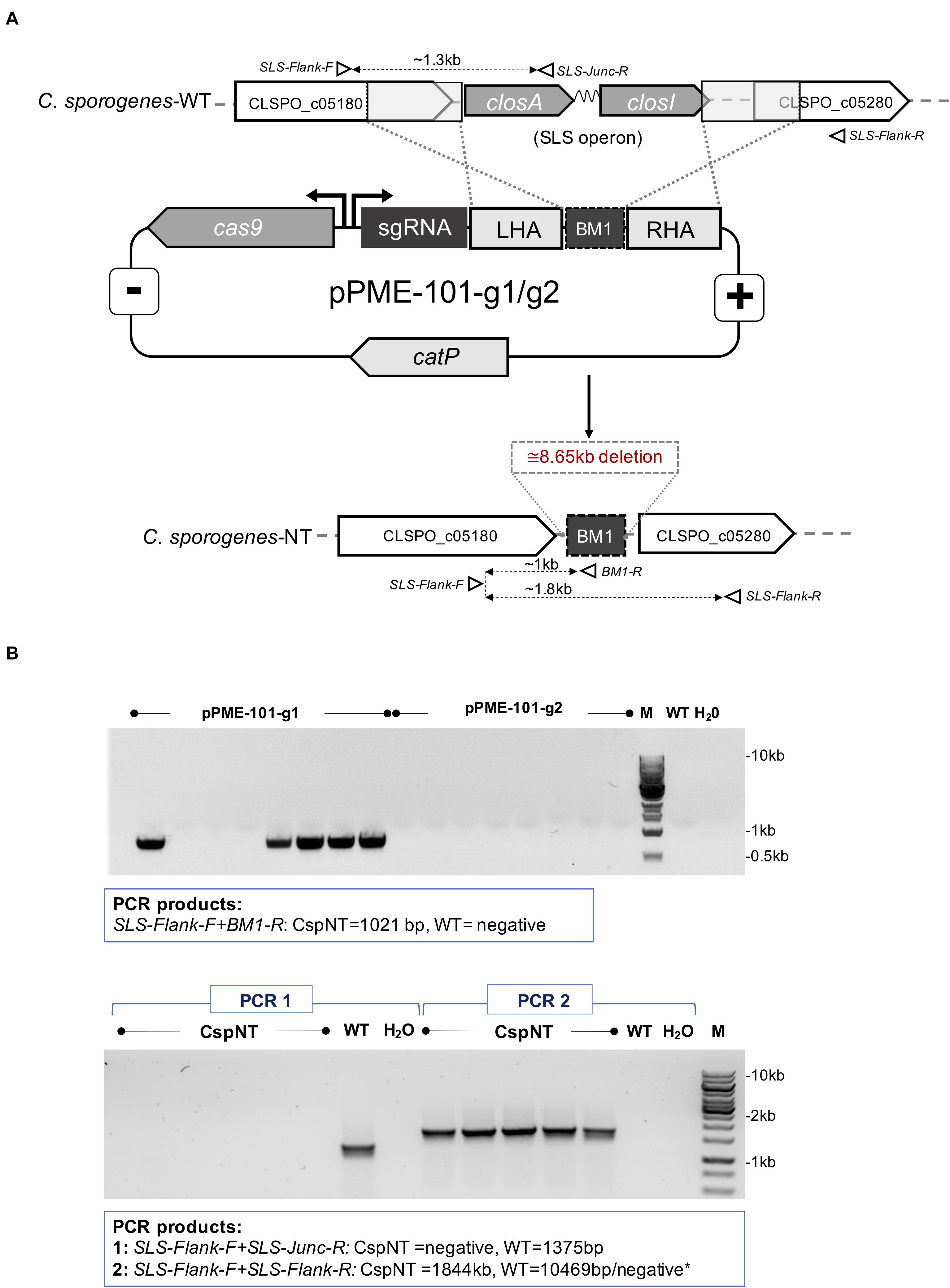
Figure 1. Schematic representation of CRISPR-Cas9 mediated integration and PCR analysis of SLS operon deletion in C. sporogenes-NT strain. (A) The genome editing vectors (pPME-101-g1/g2) containing spCas9, guide RNAs and an editing template for homologous recombination at the target chromosomal locus and integration of a BM1 “bookmark” sequence. Open triangles represent indicative alignment of screening primers as presented in Supplementary Table 2. (B) Gel electrophoresis showing a PCR screen of pPME-101-g1/g2 transconjugant colonies. Top image: Deletion of the operon detected in colonies subjected to a chromosome/bookmark-specific PCR reaction. Bottom image: Subsequent confirmation of SLS operon deletion in five selected clones. “M” denotes DNA marker, “dH2O” water template and “WT” C. sporogenes WT control.
Construction of C. sporogenes-NT Secreting Murine IL2
To construct plasmids harbouring murine IL-2 secretory variants, the DNA sequence of mIL-2 gene was manually codon optimised according to the known codon usage preference of C. sporogenes and based on the reported sequence (UniProtKB: P04351). The FLAG-tag sequence (DYKDDDDK) was added at the C-terminal end of the mIL-2 gene. Five DNA fragments: FLAG-tagged recombinant mIL-2, and four Clostridium-derived promoter-signal sequences were commercially synthesized (IDT) with sites recognized by type IIS restriction enzymes (Supplementary Table 3).
To generate four mIL2 secretion variants, the promoter-signal sequence fragments and murine IL-2 were linearly ligated at BsaI sites, followed by insertion to modified pMTL82121 E. coli-Clostridia shuttle plasmid (named pATB1C) at NotI and XhoI sites. A total of four secretion plasmids containing FLAG-tagged, codon optimised murine IL2 and combinations of promoter (Pfdx or Pptb) and signal sequences (eglA or nprM3) were constructed (Supplementary Table 1). Plasmids were transferred to C. sporogenes-NT by conjugation from an E. coli donor as described previously (Heap et al., 2012). Resultant transconjugants were confirmed by colony PCR using primers specified in Supplementary Table 2 and by Sanger sequencing (Eurofins Genomics).
Preparation of Pure Spore Suspensions
C. sporogenes strains were revived on agar plates and inoculated into 10 ml of growth media. The following day, the overnight cultures were inoculated into 500 ml of fresh media and cultivated under anaerobic conditions for six days. On day seven, cultures were removed from anaerobic cabinet and exposed to oxygen and room temperature for further 24 h. Next, spore-vegetative cell suspensions were centrifuged (10,000 x g, 20°C) for 30 min. Resultant pellets were washed two times with PBS and once with 70% (v/v) ethanol. The final pellet was resuspended in 10 ml PBS, heat treated at 80°C for 20 min to inactivate remaining viable vegetative cells and cooled down. The washed spore-debris suspensions were subjected to a density-gradient purification using HistodenzTM (D2158, Sigma-Aldrich) according to the method of Setlow with minor modifications (Setlow, 2019; Supplementary Material). Spore purification was determined by counting phase bright spores and phase dark cells using a haemocytometer under a light microscope (minimum 200 bodies per purification). Spores still enclosed in the mother cell were counted as impurities.
Phenotype Analysis of C. sporogenes Strains
Growth and Endospore Formation
The ability of C. sporogenes-NT spores to return to vegetative growth was evaluated by measuring the change in OD600 over a period of 24 h. Overnight cultures were inoculated into a fresh PYT media (1:100) and incubated at 37°C under anaerobic conditions.
A sporulation assay was carried out to measure and compare the ability of the C. sporogenes strains to form endospores. Growth media was inoculated with overnight cultures and incubated for 4 h. Following this incubation, fresh sterile broth was inoculated with the 4 h cultures (1:100) and incubated for 120 h. To determine spore titres, samples were taken every 24 h for five days and were heated at 80°C for 20 min to inactivate vegetative cells. Heat-treated samples were serially diluted and plated under anaerobic conditions. After 24 h incubation, colonies were counted, and CFU/ml was calculated to determine sporulation efficiency. A spo0A mutant, in which the master regulator of sporulation has been deleted, was used as a negative control for colony formation after heat treatment.
Measuring Haemolysis
C. sporogenes wild type and knockout strains were assayed for haemolysis by two methods. Plate assays were carried out using columbia sheep blood agar plates (PB0123, Oxoid) according to the ASM haemolysis protocol (Buxton, 2005). In addition, a liquid assay was conducted according to a published method (Totten et al., 1995). Briefly, 200 μl PBS-washed cultures were incubated in a 96-well plate with 20 μl washed whole sheep or horse blood for one hour. The plates were then centrifuged (200 x g, 10 min) and the supernatants transferred to a new sterile 96-well plate. Release of haemoglobin from lysed red blood cells (RBCs) was measured at OD540. Incubation with ultrapure water was used to determine complete lysis and with PBS to determine background lysis. In both assays, C. butyricum MIYAIRI 588 (CBM 588) and S. pyogenes were used as negative and positive haemolysin controls, respectively.
In vivo Colonisation Study
All animal experiments were performed in accordance with local institutional guidelines for animal welfare and were approved by the Animal Ethical Committee of the University of Maastricht (AVD1070020173367, Maastricht, Netherlands). Exponentially growing CT26 mouse colon carcinoma cells (Mus musculus, ATCC CRL-2638TM) syngeneic to the Balb/c mice were cultured in Roswell Park Memorial Institute (RPMI) (Lonza) supplemented with 10% fetal calf serum (FCS) in a humidified 5% CO2 chamber at 37°C. To induce tumours, approximately 8-week-old immunocompetent Balb/c OlaHsd mice (10) were subcutaneously injected with CT26 tumour cells (2 × 106), resuspended in basement membrane matrigel (354234, BD Biosciences). When tumour volumes reached 200 mm3 (determined by measurement of three orthogonal axes with vernier calipers), treated animals (8) were given 1 × 106 purified spores, administered intravenously in the lateral tail vein. Control animals (2) were injected with PBS. Seventy-two hours after spore administration, animals were sacrificed and tumours, spleens and lymph nodes were excised.
Ex vivo Sample Processing
Blood was collected in 10% heparin sodium (5000 I.U./ml, Leo Laboratories Ltd.) and plasma was separated by centrifugation (1600 x g, 4°C, 5 min). Red blood cells were removed from the remaining cell suspension using a RBC lysis buffer (00-4333-57, eBioscience). Single cell suspensions of lymph nodes, spleens and tumours were obtained using a gentleMACS dissociator (130-093-237, Miltenyi Biotec B.V.) and filtered through a 70 μm-pore cell strainer (542070, Greiner Bio-one).
The presence of spores and vegetative cells of C. sporogenes-NT in each tissue was determined by dilution plating on selective media. For total colony forming unit (CFU) counts, single cell suspensions were serially diluted in pre-reduced PBS to 10–7. For each dilution, three 20 μl spots were dispensed on to pre-reduced agar plates. The plates were incubated for 24 h and then observed for single colony growth and counted. Dilution plating was also conducted on heat-treated samples (80°C, 20 min) to quantify spores present. Vegetative cells were quantified by deducting heat-treated counts from non-heat-treated counts. Counts of spores and vegetative cells in tumours were divided by equivalent counts in the same weight of other tissues to give a ratio of the localisation in each tissue. Using bookmark-specific primers, colony PCR was conducted to confirm that the observed CFUs were the C. sporogenes-NT.
Detection of Recombinant mIL-2
The secretion of mIL-2 protein in C. sporogenes-NT-XmIL2F variants was confirmed by Western blotting in DOC-TCA precipitated fractions of culture supernatants using anti-FLAG antibody according to the related literature and manufacturer’s recommendations (Schwarz et al., 2007).
The levels of mIL-2 secreted from C. sporogenes-NT in 7 h culture supernatants were determined by cytokine specific ELISA assay (BMS601, Invitrogen) in accordance with the manufacturer’s instructions. The results were recorded in a microplate reader (BMG Labtech SPECTROstar Omega) and calculated based on recombinant mIL2 standard.
The biological activity of the secreted cytokine was determined in a lymphocyte proliferation assay using the CTLL-2 T cell line. Seven-hour subcultures of mIL2-expressing C. sporogenes-NT were centrifuged (10,000 x g, 10 min), filtered (0.45 μm syringe filter) and the sterile supernatants retained. Supernatant-stimulated CTLL-2 proliferation was measured using the MTT assay according to published methods (Soman et al., 2009) and described in detail in Supplementary Materials. The standard curve was prepared using purified recombinant mIL2 (212-12, Peprotech). Absorbance was recorded in a microplate reader (BMG Labtech SPECTROstar Omega).
Biological Replicates and Statistical Analyses
All data presented in this manuscript represent the results of at least three independent experiments. Statistical evaluations were performed with GraphPad Prism 8 software (San Diego, CA, United States). For the ELISA and lymphocyte proliferation assay, data was analysed using unpaired t-test to compare C. sporogenes-WT with each mIL2 variant. To compare haemolysis in C. sporogenes-WT and C. sporogenes-NT, data was analysed using two-way ANOVA with Dunnett’s multiple comparison test. Values of p < 0.05, p < 0.01, p < 0.001 were considered significant (∗), highly significant (∗∗), or extremely significant (∗∗∗) respectively. Data represent means ± standard deviation (SD).
Results
Deletion of the SLS Homologue Causes a Significant Reduction in Haemolysis
The identification of streptolysin S in the genome of C. sporogenes NCIMB 10696 highlighted a potential risk of using this species as an intratumoral delivery vehicle. We sought to determine whether deletion of this virulence factor affected this strain’s haemolysis capability. Two CRISPR-Cas9 vectors were created with the aim of deleting the SLS operon, distinguished by the presence of two different targeting sequences, guide 1 (g1) and guide 2 (g2) (Figure 1A). Following conjugation, eight colonies for each vector were screened by PCR for chromosomal recombination. For the vector with g1, deletion was detected in five out eight transconjugants (Figure 1B). Deletion of the operon at the SLS locus was confirmed by Sanger sequencing. For transconjugants harbouring the vector with g2, the WT sequence was detected at the SLS locus, indicating that chromosomal recombination had not occurred.
Neither CspWT nor CspNT strains of C. sporogenes displayed strong beta-haemolysis at 24 or 48 h after streaking on blood plates, in contrast to the positive control, S. pyogenes (Figure 2A). A validated liquid assay with horse and sheep blood was employed to quantify haemolysis (Figure 2B). In our experiments, horse blood was more sensitive to haemolysis than that of sheep. Significant differences could not be detected in sheep blood. In horse blood, differences between 4-h cultures of each strain were difficult to distinguish, and differences were not statistically significant. At 8 h, the difference in haemolysis between CspWT and CspNT was most pronounced (CspWT 2.5% ± 0.27 SD; CspNT 0.76% ± 0.23 SD). At this time point CspNT haemolysis was not higher than that of the non-haemolytic control, Cbut-M588 (p = 0.371). At 24 h, CspWT haemolysis remained elevated (0.94% ± 0.09 SD) while for CspNT it was comparable to Cbut-M588. At both 8 and 24 h, the difference between CspWT and CspNT was statistically significant (p < 0.05).
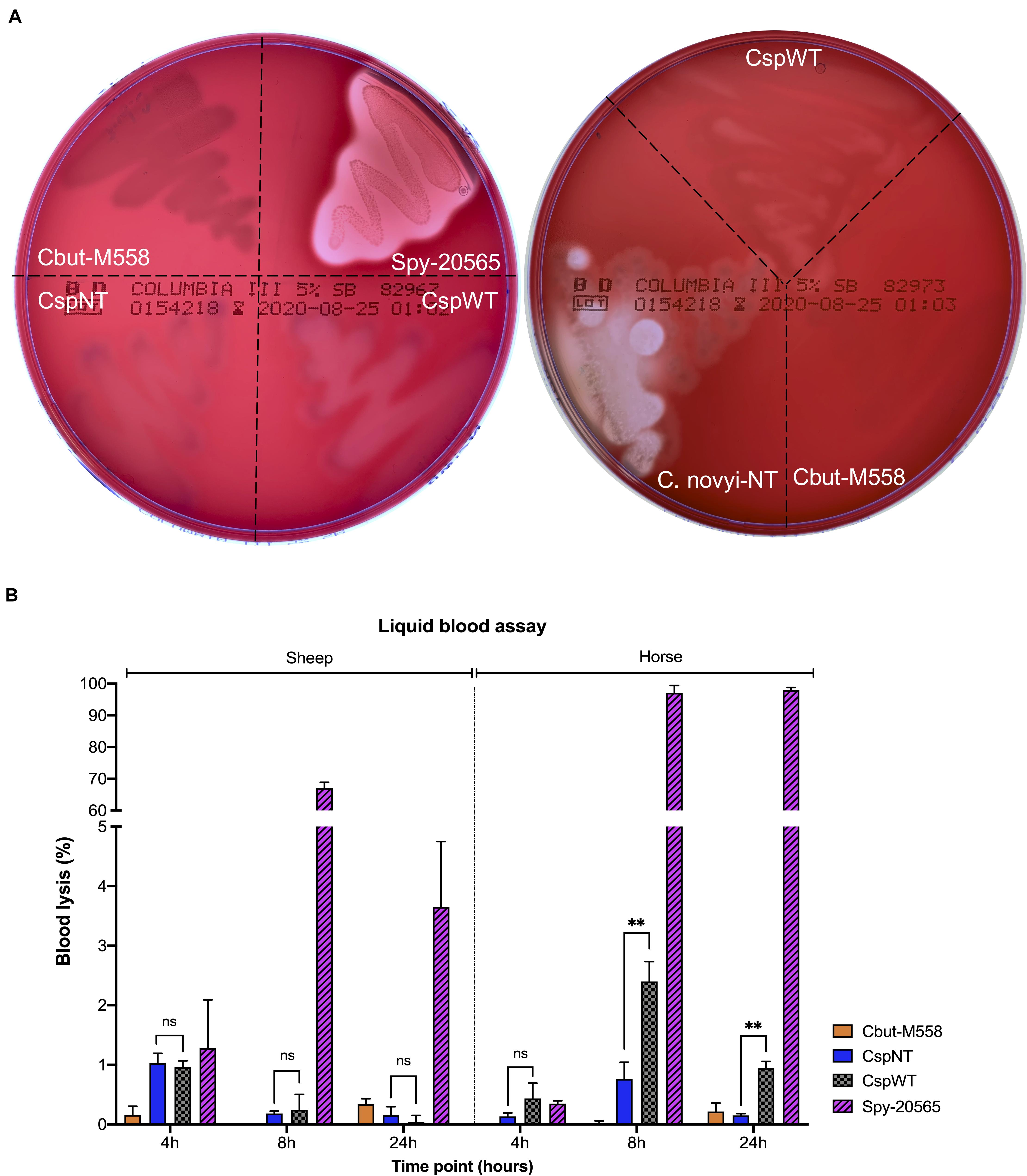
Figure 2. Analysis of haemolytic activity. (A) Five strains were streaked on two Columbia Blood Agar plates (5% sheep blood BD Bioscience) and incubated under anaerobic conditions for 48 h. The strains were: Cbut-M558: C. butyricum MIYAIRI 585 (negative control), Csp-20565: S. pyogenes DSM 20565 (positive control), CspNT: C. sporogenes-NT, CspWT: C. sporogenes-WT and C. novyi-NT. (B) Quantitative analysis of haemolysis in two liquid blood assays (using sheep and horse blood). Samples from liquid cultures were assayed at 4, 8, and 24 h. Haemolysis is presented as a percentage of total lysis (blood incubated with ultrapure water). Assayed strains: Cbut-M558: C. butyricum MIYAIRI 588; CspNT: C. sporogenes-NT; CspWT: C. sporogenes-WT; Spy-20565: S. pyogenes DSM 20565.
Following the announcement of a first in human (FIH) study utilising C. novyi-NT, we sought to benchmark C. sporogenes-NT to compare haemolysis. Growth of C. novyi-NT and C. sporogenes-NT on sheep blood was observed. In contrast to C. sporogenes-NT, C. novyi-NT caused significant beta-haemolysis (Figure 2A). Sequence similarity to the SLS operon could not be detected in the C. novyi-NT genome when using protein (tblastn) or DNA (blastn) as queries.
The ability to form spores is a characteristic feature of C. sporogenes. We sought to determine whether C. sporogenes-NT displayed the same growth and sporulation phenotype as that of the parental strain. The new C. sporogenes-NT strain grew and formed spores at the same rate and to a comparable final titre as the wild type (Figures 3A and 3B). Our data demonstrates that deletion of the SLS operon has no significant impact on growth and germination efficiency over a 24 h period or sporulation over 120 h.
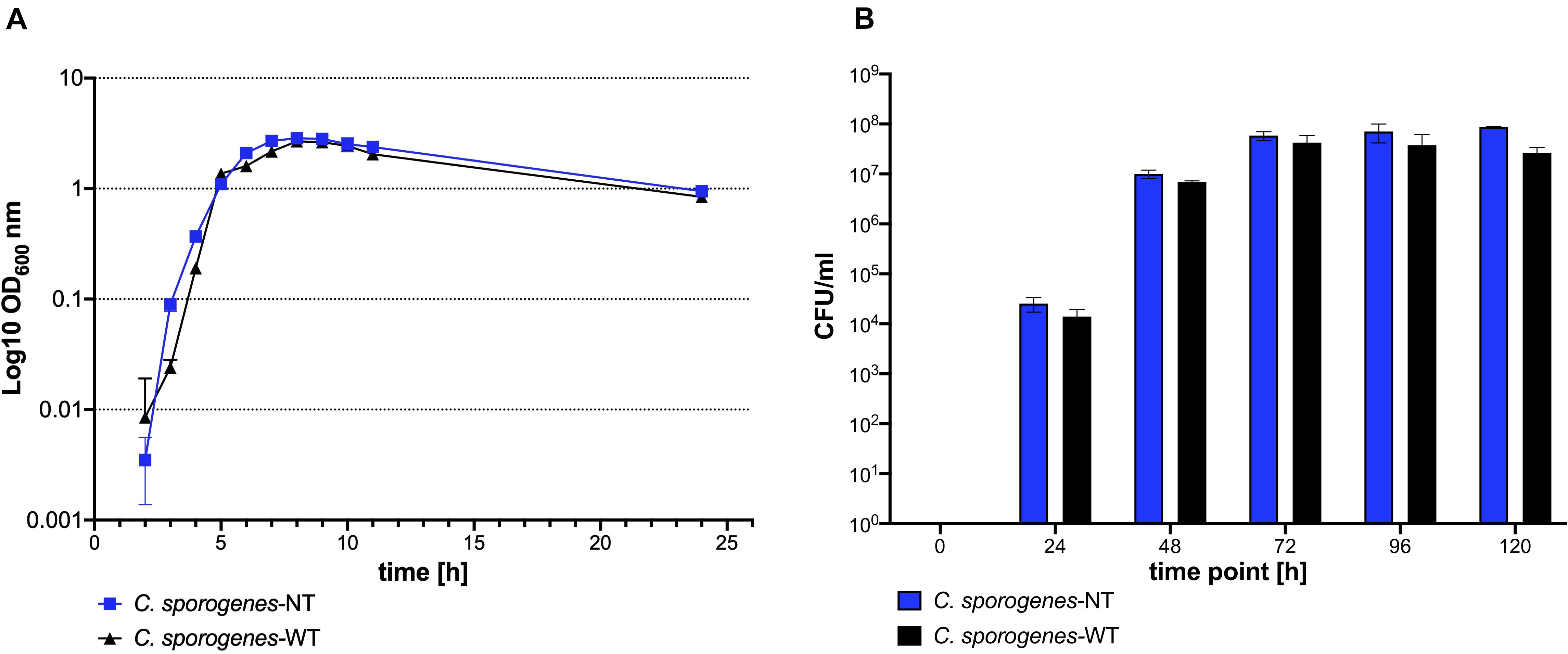
Figure 3. Growth and development of heat resistant colony forming units of C. sporogenes-NT (blue) and C. sporogenes-WT (black) strains. (A) Growth of Clostridium strains was measured as a direct increase in absorbance at 600 nm throughout the course of 24 h bacterial incubation in bovine free medium (PYT). The symbols represent the average of three independent experiments, and error bars indicate the standard errors of the means. (B) Heat-treated bacterial samples (80°C, 20 min) were plated in serial dilution on PYT agar plates and enumerated following 24 h incubation. Bars represent the number of CFU (colony forming unit) per ml of Clostridium culture. The data represent the average of three independent experiments and error bars indicate the standard error of the mean. The sporulation-deficient Clostridium sporogenes-NTΔspo0A mutant was used as a negative control to rule out experimental error. The detection limit for colony counts was 50 CFU/ml.
C. sporogenes-NT Colonises Tumours in Experimental Animals
The ability of C. sporogenes NCIMB 10696 to colonise solid tumours has been demonstrated previously (Heap et al., 2014; Mowday et al., 2021). We sought to determine whether deletion of the SLS operon has an effect on this ability. The CT26 murine colon carcinoma cell line is highly immunogenic. Cells were implanted subcutaneously to allow easy measurement of growing tumours. Tumours were formed in all ten mice injected with CT26 cells. Tumour-bearing animals were sacrificed 72 h after systemic administration of C. sporogenes-NT spores (eight) or PBS (two). An overview of the in vivo experiment is presented in Figure 4A.
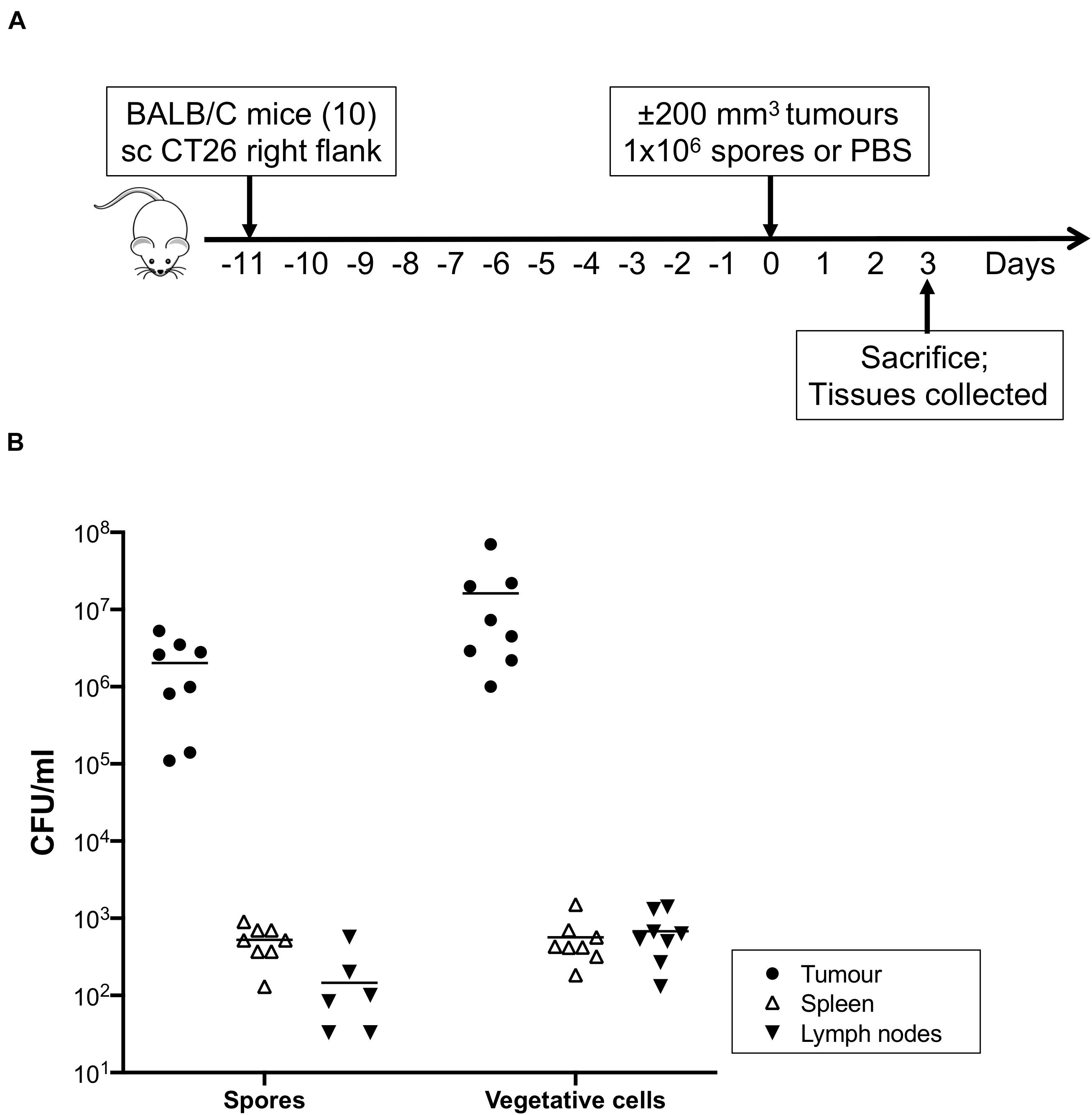
Figure 4. In vivo colonisation study and recovery of bacteria from mouse tissues. (A) Immunocompetent Balb/c mice (ten) were subcutaneously injected with CT26 tumour cells (2 × 106). When tumour volumes reached approximately 200 mm3 eight treated animals were given 1 × 106 purified C. sporogenes-NT spores. Two control animals were injected with PBS. 72 h after administration of spores, animals were sacrificed and tumours, spleens and lymph nodes were excised and subjected to cell counts. (B) The presence of spores and vegetative cells of C. sporogenes-NT in each tissue (tumour, black circles; spleen, open triangles; lymph nodes, inverted black triangles) was determined by dilution plating on selective media and expressed as colony forming unit (CFU).
The presence of spores and vegetative cells in tumours, spleens, lymph nodes and blood was determined (Figure 4B and Supplementary Table 4). Neither spores nor vegetative cells were detected in any tissues from control animals that were injected with PBS instead of spores, or in the blood of spore injected animals. In the tumours of spore-injected animals, spore counts ranged from 1.1 × 105 – 1.7 × 106 per ml. Vegetative cell counts were higher at 1 × 106 – 2.2 × 107 per ml. In the spleens of spore-injected animals, average counts of bacteria were 4,000 and 26,000 times lower than in the equivalent weight of tumour tissue, respectively. For lymph nodes, spores and vegetative cells were 62,000 and 81,000 times lower, respectively. Ratios were determined as outlined in the methods section. For all excised tissues, plate colonies were confirmed to be C. sporogenes-NT by PCR using bookmark-specific primers (Supplementary Table 2, data not shown).
C. sporogenes-NT Secretes Biologically Active Murine IL-2 Cytokine
We tested the ability of C. sporogenes-NT to express and secrete a heterologous protein, exemplified using mIL-2. This cytokine has a short half-life and requires intravenous administration at high doses for clinical efficacy, resulting in adverse events in a high proportion of patients. Four pATB1C-XmIL2 plasmids with different promoter-signal sequence combinations were conjugated into C. sporogenes-NT and verified by colony PCR (Figures 5A–C) and Sanger sequencing. The impact of these variants on expression and secretion of mIL-2 was assessed. Testing multiple expression variants increased the chance of cloning a functional mIL-2 secreting strain without mutation or growth inhibiting host toxicity.
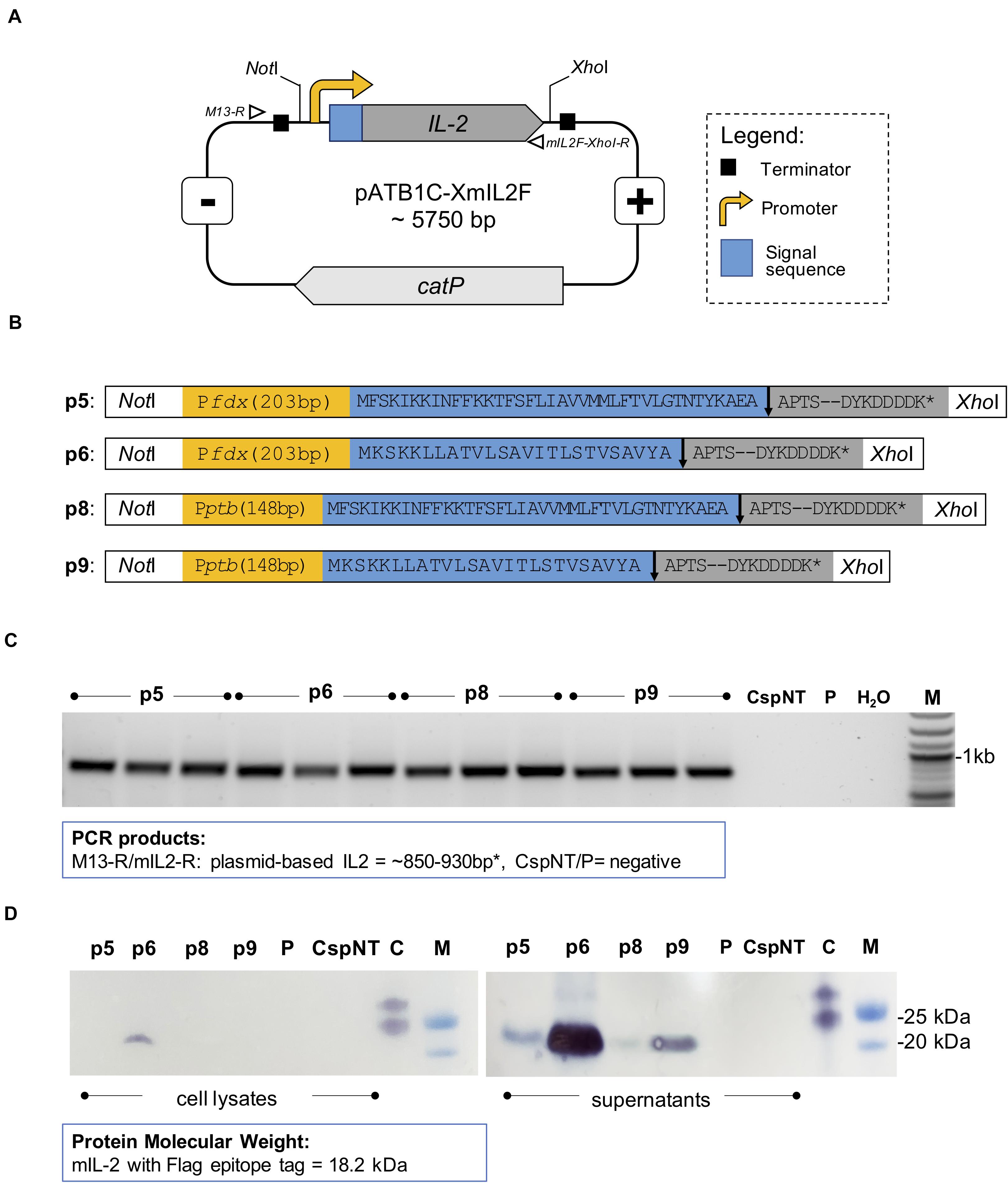
Figure 5. Design and confirmation of C. sporogenes-NT-XmIL2F variants. (A) Schematic illustration of expression vector pATB1C-XmIL2F. The codon optimised and FLAG-tagged murine IL-2 gene was ligated at the BsaI site with relevant promoter and signal sequence variants (Supplementary Table 3) and inserted into pMTL82121 vector at the NotI/XhoI sites. Open triangles indicate alignment of screening primers: M13-R and mIL2F-XhoI-R. (B) Four mIL2 expression modules consist of Pfdx or Pptb promoter (yellow box) followed by eglA or nprM3 signal sequence (blue box). The FLAG-tagged murine IL-2 gene (grey box) was ligated downstream of a relevant signal sequence. The arrow indicates the signal sequence cleavage site. (C) Colony PCR screening of C. sporogenes-NT transconjugants harbouring four variants of plasmid-based mIL-2 gene. For each variant a total of three independent clones have been obtained. Plasmid-based mIL-2 strains were denoted p5, p6, p8 and p9 (Supplementary Table 1), “CspNT” denotes C. sporogenes-NT control, “P” denotes C. sporogenes-NT-pATB1C (empty vector), water (dH20), M: DNA marker. (D) Visualisation of Western immunoblot analysis performed on C. sporogenes-NT-XmIL2F cell lysates and supernatant fractions following the incubation with FLAG-tag antibody. “C” denotes FLAG-tag positive control, “M” denotes protein marker. FLAG-tagged murine IL-2 protein has been visualised on the nitrocellulose membranes with TMB-Blotting 1-Step Solution and corresponds to the size of 18.2 kDa.
Western blot analysis was used to determine the presence of FLAG-tagged protein in the intra- and extra-cellular fractions of recombinant C. sporogenes-NT cultures. mIL-2 was detected in the supernatants of all C. sporogenes-NT variants and was absent in the cellular fraction of three out of four variants, indicating effective and complete secretion (Figure 5D). A small quantity of tagged protein was detected in the cellular fraction of variant p6. These results indicate that nrpM3 is an efficient secretion peptide. For both nprM3 and eglA signal peptides, variants utilising the Pfdx promoter showed the highest expression levels.
Secreted mIL-2 levels in 7-h cultures were quantified in an mIL-2 ELISA. Maximum mIL-2 levels were detected in variant p6 (240 ng/ml, C. sporogenes-NT-p6mIL2F), while the lowest levels were detected in variant p8 (2.9 ng/ml, C. sporogenes-NT-p5mIL2F) (Figure 6A). Next, we sought to confirm that the secreted product was biologically active. The murine cytotoxic T cell line CTLL-2 is dependent on pro-inflammatory cytokines for viability. By stimulating the growth of these cells in the presence of IL-2 standards or bacterial culture supernatant, the biological activity of secreted mIL-2 can be quantified. Supernatants from late-exponential (7 h) cultures of wild-type and mIL-2 expressing C. sporogenes-NT, were applied to washed CTLL-2 cells and incubated for 48 h. The MTT assay was used to quantify T cell proliferation. The final spectrophotometric detection confirmed that mIL-2 levels of 602 ng/ml (3010 U/ml) were measured for “p6”, the most potent recombinant Clostridium variant (C. sporogenes-NT-p6mIL2F), dropping to 65.7 ng/ml (328 U/ml) in variant “p8” (Figure 6B). These results indicate that mIL-2 expressed by recombinant C. sporogenes-NT is efficiently secreted and folds into a native conformation. All results confirmed that haemolysin-free C. sporogenes-NT strain is a suitable vehicle for the secretion of proteins, such as mIL-2 cytokine.
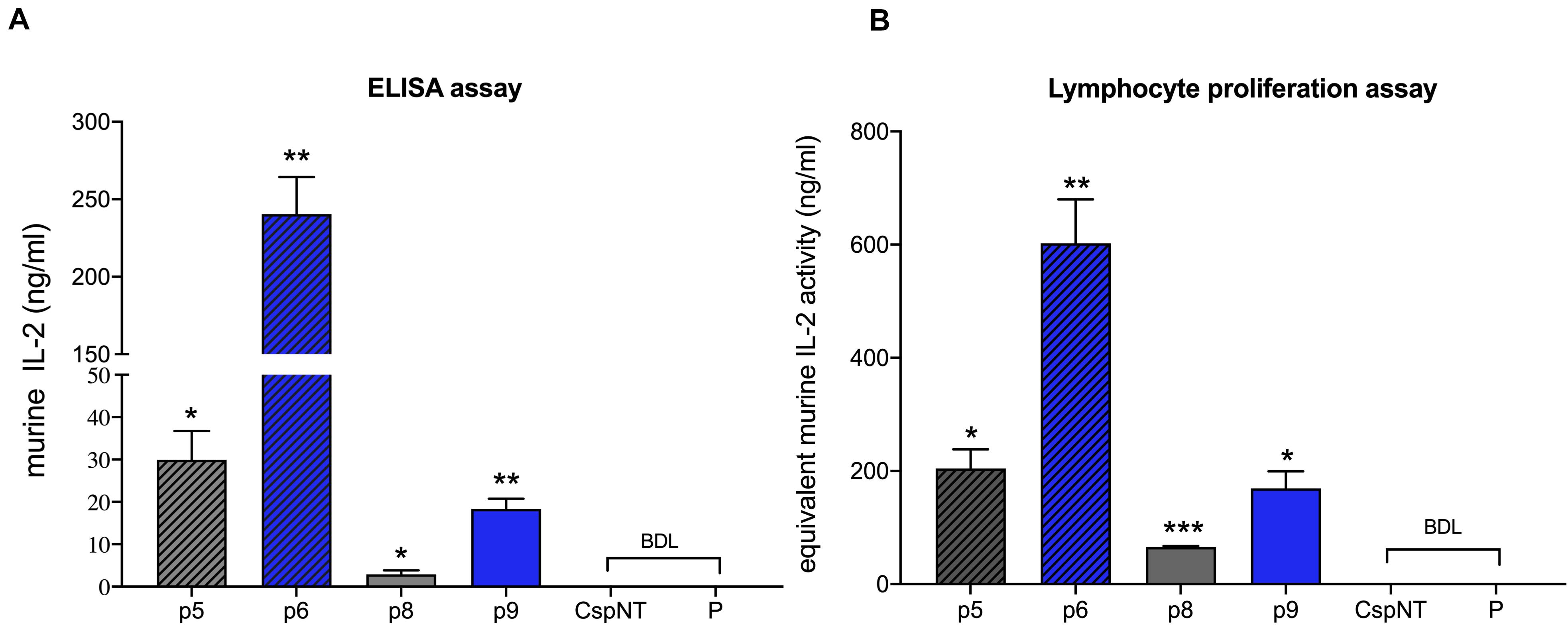
Figure 6. Validation of C. sporogenes-NT variants harbouring plasmid-based mIL-2 in the quantitative and functional assays. (A) Results of commercial ELISA test indicating the quantities of mIL-2 cytokine present in the supernatants of four C. sporogenes-NT-XmIL2F variants following 7-h growth in PYT media. (B) Results of colorimetric MTT functional assay following the incubation of CTLL-2 T-cells in the presence of 7-h culture supernatants of C. sporogenes-NT-XmIL2F variants. Recombinant murine IL2 was used to prepare mIL-2 standard curve. BDL, below detectable levels. Plasmid-based mIL-2 strains were denoted p5, p6, p8, and p9 (Supplementary Table 1), “CspNT” denotes C. sporogenes-NT control, “P” denotes C. sporogenes-NT-pATB1C (empty vector).
Discussion
Haemolysis is a defining feature of many bacterial infections, including necrotising enterocolitis (Clostridium perfringens), haemolytic–uremic syndrome (Escherichia coli) and listeriosis (Listeria monocytogenes). Clinical laboratories routinely determine haemolysis by streaking patient specimens on blood agar plates. A strain is considered beta-haemolytic (displays “true haemolysis”) if it produces a clearing of the red pigment conferred by the blood (Madigan, 2012).
Publication of the genome sequences of C. sporogenes ATCC 15579 and a closely related Group 1 C. botulinum revealed an SLS homologue, which confers a haemolytic phenotype in S. pyogenes (Sebaihia et al., 2007). A subsequent study of this homologue concluded that the genes were functionally equivalent to those of S. pyogenes (Gonzalez et al., 2010). This was demonstrated by the independent deletion of four S. pyogenes M1 SLS genes (sagA-D) followed by complementation with the equivalent genes from C. botulinum. Complementation of sagA and sagD strains restored WT levels of haemolysis, while sagB complementation failed to restore haemolysis and sagC partially restored haemolysis. In contrast to our own results for C. sporogenes NCIMB 10696, this paper reports that C. sporogenes ATCC 15579 exhibits strong beta-haemolysis in blood plate assays. This suggests there is considerable variation in haemolysis between strains of C. sporogenes.
We studied the effect of the SLS homologue in C. sporogenes NCIMB 10696. An 8.6 kb region of the chromosome, corresponding to the SLS operon, was deleted. To accurately quantify haemolysis, we developed a liquid assay that proved considerably more sensitive than the conventional blood plate assay. This method enabled us to detect and quantify the effect of the SLS gene products, and the reduction of haemolysis following operon deletion. This decrease was statistically significant in horse but not in sheep blood. For blood agar cultures in clinical laboratories, sheep blood is commonly used in North America, while horse blood is more common in Europe. We were unable to find relevant studies in the literature that compare haemolysis in the blood of these two animals. Both sources are considered reliable for the detection of haemolysis (Jorgensen et al., 2015). Growth and sporulation were not affected by deletion of the SLS operon.
The ability of Clostridium species, including wild type C. sporogenes NCIMB 10696, to colonise hypoxic/necrotic tumours is well documented (Zhou et al., 2018). The most advanced example of this is a recently reported first-in-human (FIH) study using C. novyi-NT for treatment of treatment-refractory solid tumours (Janku et al., 2020). C. novyi-NT differs from its wild-type parental strain due to the absence of the alpha toxin. In preclinical studies it was determined that intratumoral injection was the best method of administration, due to dose-limiting toxicities observed following intravenous administration. Following intratumoural injection with C. novyi-NT, elevated cytokines as well as local and systemic tumour antigen-specific T cell responses were observed in patients that showed signs of germination. This response was not seen when signs of germination were absent. Three of the 24 patients experienced dose-limiting toxicities, including sepsis and gangrene. The results of the C. novyi-NT FIH study have demonstrated the feasibility of intratumoural Clostridium treatment and has justified further clinical studies (NCT03435952). Intravenous delivery of C. sporogenes secreting clinically approved immunotherapeutics is conceptually different. This method does not rely on the innate tumorolytic ability of the bacteria, rather it is a means to deliver therapeutics. In addition, the less invasive nature of the treatment could be advantageous.
We sought to determine the impact of deleting the SLS-like operon on the ability of C. sporogenes to colonise hypoxic/necrotic tumours in mice. In animals injected with C. sporogenes-NT spores, very high levels of spores and vegetative cells were detected in tumours compared to other healthy tissues (≥ 4000-fold). On average, 90% of the bacteria existed in the metabolically active vegetative form, indicating that the tumour environment promoted germination of the injected spores. Spores and vegetative cells were detected at very low levels in the spleen and lymph nodes. Neither of these tissue types are anoxic and, therefore, are unlikely to support growth of obligately anaerobic bacteria. Detection of spores and vegetative cells in these tissues may be due to accumulation from the initial injection or due to leakage from the tumour. These organs filter the blood and lymphatic system, respectively, so they are likely to concentrate any foreign bacteria present in these systems. Our results are consistent with a previous study of Clostridium localisation following intravenous administration (Lambin et al., 1998; Diaz et al., 2005).
C. sporogenes-NT could be exploited to increase the concentration of an immunotherapeutic at the tumour site without exposing healthy tissues, avoiding the adverse effects caused by systemic delivery. While commercial production of recombinant proteins has become commonplace, adapting these principles for therapeutic delivery raises a number of challenges. The therapeutic must be expressed and secreted from the bacteria before folding correctly to form the functional product. In addition, it must be delivered at levels at or above the effective dose, a function of expression, secretion, protein degradation and proliferation of the bacteria.
Interleukin-2 was one of the earliest immunotherapy treatments, but the significant disadvantages associated with systemic IL-2 treatment have led to more sophisticated versions, capable of targeting the cancer (Rekers et al., 2018; Lieverse et al., 2020). Approved for treatment of renal cell carcinoma and metastatic melanoma, IL-2 is a potent T cell activator (Amin and White, 2014; Payne et al., 2014). Despite preferentially inducing the expansion of immune-suppressing regulatory T cells (Tregs), IL-2 can produce significant clinical responses when administered systemically at high doses (600,000 IU/kg every 8 h) (McDermott et al., 2005). However, objective clinical response is only seen in 15-20% of patients, and immune-related adverse effects (irAEs) are a significant issue that can be treatment limiting (Buchbinder et al., 2019). Attempts to reduce toxicity by using lower doses of IL-2 results in a significant drop in therapeutic effect, due to the dominant effect of Tregs (Ahmadzadeh and Rosenberg, 2006). The high dose requirement and the risks of systemic toxicity make IL-2 an excellent test case for an intratumoral delivery system.
A panel of IL-2 expressing C. sporogenes-NT strains were created using different combinations of native promoters (two) and secretion peptide sequences (two). Expression of recombinant proteins using a single strong promoter carries a risk of failure, due to the potential toxicity to the host bacteria. This can manifest as growth inhibition and the emergence of more competitive mutant variants. Toxicity has been reported for recombinant expression of IL-2 (Mahmoudi Azar et al., 2013). To mitigate this risk, the mIL-2 coding sequence was cloned with two promoters, representing high and low expression. In our own experience, Pfdx is consistantly strong in any genetic context, while Pptb produces significantly lower expression. This is in agreement with other studies (Brunt et al., 2014; Pyne et al., 2014). Secretion from Gram-positive bacteria can be achieved by addition of a secretion peptide to the N terminus of the protein, encoded upstream of the coding sequence. Signal peptides can be functionally interchangeable between species, however the efficiency of protein secretion is strongly determined by these leader sequences. In the context of mIL-2, the native nprM3 precursor greatly improved the efficiency of secretion over that of eglA from Clostridium saccharolyticum. High-level expression combined with inefficient secretion can lead to recombinant product accumulation as inclusion bodies, and corresponding host toxicity (Horga et al., 2018; Li and Rinas, 2020). The performance of signal peptides is coding sequence dependent, and predicting the secretion efficiency based on peptide sequence is not possible (Freudl, 2018). Identifying a range of secretion peptides that perform well in different contexts will facilitate future recombinant protein secretion. In this study, peak mIL-2 production was achieved in strains that utilised the Pfdx-nprM3 promoter/secretion peptide combination. In this construct, a small quantity of tagged protein was detected in the cellular fraction of cultures. This could be a result of contamination of the cell pellet by residual supernatant prior to protein precipitation. Another possibility is that the rate of gene expression with the Pfdx promoter exceeds the rate of secretion, leading to an accumulation of mIL-2 in the cytoplasm. In a T cell proliferation assay, culture supernatants of this strain showed biological activity equivalent to 602 ng/ml (3010 U/ml) while ELISA experiments indicated 240 ng/ml (1200 U/ml). The highly proteolytic nature of C. sporogenes is likely to reduce the level of secreted mIL-2 measured in laboratory batch cultures. In vivo experiments will provide an insight into the different dynamics of protein production and degradation inside a solid tumour. Prior to this research, secretion of recombinant rat IL-2 was demonstrated in C. acetobutylicum, a saccharolytic species from the same Clostridium cluster I. This study reported production of 800 ng/mL active cytokine, although this was measured at a significantly higher culture density (Barbé et al., 2005).
This paper reports a safer version of the well-studied cancer delivery vector, C. sporogenes, by reduction of toxicity through chromosomal gene editing. Our experiments revealed that C. sporogenes NCIMB 10696 was only weakly haemolytic, but the irrefutable presence of the streptolysin S homologue in the genome sequence could undermine confidence of regulatory bodies and patients. Deletion of a significant region of the genome (approx. 8.6kb) did not influence the attenuated strain’s ability to colonise solid tumours.
The new strain was transformed with mIL-2 expression vectors. Using a native gene promoter, significant quantities of mIL-2 were detected in the extracellular fraction of cultures. To improve the performance of this approach, the next step is to optimise recombinant gene expression and secretion, as well as expand the repertoire of therapeutic agents. Further optimisation will minimise the metabolic burden and potential toxic effect on the host bacteria while maximising dose and, by extension, therapeutic effect. In addition, chromosomal integration of optimised expression cassettes will be essential to ensure genetic stability and to eliminate the risk of horizontal gene transfer. Genetically engineered C. sporogenes-NT poses an elegant and highly adaptable solution to the challenge of precise delivery of anti-cancer agents.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by Dier Experimenten Commissie (DEC) Maastricht University Secretariaat DEC/UNS 50/box 48 Postbus 616 6200 MD Maastricht.
Author Contributions
AK and TB co-designed the study, performed the experiments, collected and analysed the data, and wrote the manuscript. PL initiated the project. PL, JT, LD, and TB contribute to the writing of the funding grants. LD and JT contributed to the design and the approval of the in vivo experiments. All authors contributed to data interpretation, discussions and revised the manuscript.
Funding
This project received funding from the EU H2020-INNOSUP Innovation Associate programme under grant number 739756 and project name CMI2T. Authors also acknowledge financial support from ERC advanced grant (ERC-ADG-2015 no. 694812 - Hypoximmuno), ERC-2018-PoC (813200-CL-IO) and KWF (no. 11163/2017-1) and KWF-Alpe d’HuZes (no. 8025/2015).
Conflict of Interest
PL reports, within and outside the submitted work, grants/sponsored research agreements from Varian medical, Oncoradiomics, ptTheragnostic/DNAmito, Health Innovation Ventures. He received an advisor/presenter fee and/or reimbursement of travel costs/external grant writing fee and/or in kind manpower contribution from Oncoradiomics, BHV, Merck, Varian, Elekta, ptTheragnostic and Convert pharmaceuticals. PL has shares in the company Oncoradiomics, Convert pharmaceuticals, MedC2 and LivingMed Biotech, he is co-inventor of two issued patents with royalties on radiomics (PCT/NL2014/050248, PCT/NL2014/050728) licensed to Oncoradiomics and one issue patent on mtDNA (PCT/EP2014/059089) licensed to ptTheragnostic/DNAmito, three non-patented invention (softwares) licensed to ptTheragnostic/DNAmito, Oncoradiomics and Health Innovation Ventures and three non-issues, non-licensed patents on Deep Learning-Radiomics and LSRT (N2024482, N2024889, N2024889). AK is employed by Exomnis Biotech B.V. Exomnis Biotech did not provide funding and was not involved in the design, execution, analysis, interpretation or publication of the study.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Part of the work reported in this publication is the subject of a pending patent.
Acknowledgments
We thank Ph.D. Ludwig Dubois and Ph.D. Alexandra Mowday for writing the work protocol for all in vivo studies at Department of Precision Medicine, and Natasja Lieuwes, Rianne Biemans and Ph.D. Alexandra Mowday for handling experimental animals during in vivo study. We also thank Sebastiaan Huntjens for his sustained support throughout this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.669488/full#supplementary-material
Supplementary Table 1 | Plasmids and strains used in the study.
Supplementary Table 2 | List of oligonucleotides used in the study (restriction enzyme recognition sites underlined).
Supplementary Table 3 | Promoter-signal sequence module for pATB1C-XmIL2F variants.
Supplementary Table 4 | Colony forming unit counts on tissue samples following the in vivo colonisation study.
References
Ahmadzadeh, M., and Rosenberg, S. A. (2006). IL-2 administration increases CD4+CD25hi Foxp3 + regulatory T cells in cancer patients. Blood 2006:2399. doi: 10.1182/blood-2005-06-2399
Amin, A., and White, R. L. (2014). Interleukin-2 in renal cell carcinoma: a has-been or a still-viable option? J. Kidney Cancer VHL 1:74. doi: 10.15586/jkcvhl.2014.18
Barbé, S., Van Mellaert, L., Theys, J., Geukens, N., Lammertyn, E., Lambin, P., et al. (2005). Secretory production of biologically active rat interleukin-2 by Clostridium acetobutylicum DSM792 as a tool for anti-tumor treatment. FEMS Microbiol. Lett. 2005:037. doi: 10.1016/j.femsle.2005.03.037
Brown, J.L., Tran-Dinh, N., and Chapman, B. (2012). Clostridium sporogenes PA 3679 and its uses in the derivation of thermal processing schedules for low-acid shelf-stable foods and as a research model for proteolytic Clostridium botulinum. J. Food Prot. 75, 779–792. doi: 10.4315/0362-028X.JFP-11-391
Brunt, J., Plowman, J., Gaskin, D.J., Itchner, M., Carter, A. T., and Peck, M. W. (2014). Functional characterisation of germinant receptors in Clostridium botulinum and Clostridium sporogenes presents novel insights into spore germination systems. PLoS Pathog. 10:e1004382. doi: 10.1371/journal.ppat.1004382
Buchbinder, E.I., Dutcher, J.P., Daniels, G. A., Curti, B.D., Patel, S.P., Holtan, S.G., et al. (2019). Therapy with high-dose Interleukin-2 (HD IL-2) in metastatic melanoma and renal cell carcinoma following PD1 or PDL1 inhibition. J. ImmunoTher. Cancer. 2019:0522. doi: 10.1186/s40425-019-0522-3
Buxton, R. (2005). Blood Agar Plates and Hemolysis Protocols [Online]. Am. Soc. Microbiol. 2005:2885.
Diaz, L. A.Jr., Cheong, I., Foss, C. A., Zhang, X., Peters, B. A., Agrawal, N., et al. (2005). Pharmacologic and toxicologic evaluation of C. novyi-NT spores. Toxicol Sci. 88, 562–575. doi: 10.1093/toxsci/kfi316
Dodd, D., Spitzer, M. H., Van Treuren, W., Merrill, B. D., Hryckowian, A. J., Higginbottom, S. K., et al. (2017). A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017:24661. doi: 10.1038/nature24661
Freudl, R. (2018). Signal peptides for recombinant protein secretion in bacterial expression systems. Microb. Cell Fact 17:52. doi: 10.1186/s12934-018-0901-3
Gericke, D., and Engelbart, K. (1964). Oncolysis by clostridia. II. experiments on a tumor spectrum with a variety of clostridia in combination with heavy metal. Cancer Res. 24, 217–221.
Gonzalez, D. J., Lee, S. W., Hensler, M. E., Markley, A. L., Dahesh, S., Mitchell, D. A., et al. (2010). Clostridiolysin S, a post-translationally modified biotoxin from Clostridium botulinum. J. Biol. Chem. 285, 28220–28228. doi: 10.1074/jbc.M110.118554
Guo, C. J., Allen, B. M., Hiam, K. J., Dodd, D., Van Treuren, W., Higginbottom, S., et al. (2019). Depletion of microbiome-derived molecules in the host using Clostridium genetics. Science 366:6471. doi: 10.1126/science.aav1282
Heap, J. T., Ehsaan, M., Cooksley, C. M., Ng, Y.-K., Cartman, S.T., Winzer, K., et al. (2012). Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker. Nucleic Acids Res. 40, e59-e59. doi: 10.1093/nar/gkr1321
Heap, J. T., Pennington, O. J., Cartman, S. T., and Minton, N. P. (2009). A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78, 79–85. doi: 10.1016/j.mimet.2009.05.004
Heap, J. T., Theys, J., Ehsaan, M., Kubiak, A. M., Dubois, L., Paesmans, K., et al. (2014). Spores of Clostridium engineered for clinical efficacy and safety cause regression and cure of tumors in vivo. Oncotarget 5, 1761–1769. doi: 10.18632/oncotarget.1761
Horga, L. G., Halliwell, S., Castineiras, T. S., Wyre, C., Matos, C., Yovcheva, D. S., et al. (2018). Tuning recombinant protein expression to match secretion capacity. Microb. Cell Fact 17:199. doi: 10.1186/s12934-018-1047-z
Ingle, P., Groothuis, D., Rowe, P., Huang, H., Cockayne, A., Kuehne, S. A., et al. (2019). Generation of a fully erythromycin-sensitive strain of Clostridioides difficile using a novel CRISPR-Cas9 genome editing system. Sci. Rep. 9:8123. doi: 10.1038/s41598-019-44458-y
Janku, F., Zhang, H. H., Pezeshki, A. M., Goel, S., Murthy, R., Wang-Gillam, A., et al. (2020). Intratumoral injection of Clostridium novyi-NT spores in patients with treatment-refractory advanced solid tumors. Clin. Cancer Res. 2020:2065. doi: 10.1158/1078-0432.CCR-20-2065
Jorgensen, J. H., Pfaller, M. A., and Carroll, K. C. American Society for Microbiology (2015). Manual of Clinical Microbiology. Washington, DC: ASM Press.
Lambin, P., Theys, J., Landuyt, W., Rijken, P., Van Der Kogel, A., Van Der Schueren, E., et al. (1998). Colonisation of Clostridium in the body is restricted to hypoxic and necrotic areas of tumours. Anaerobe 1998:0161. doi: 10.1006/anae.1998.0161
Li, Z., and Rinas, U. (2020). Recombinant protein production associated growth inhibition results mainly from transcription and not from translation. Microb. Cell Fact. 19:83. doi: 10.1186/s12934-020-01343-y
Lieverse, R. I. Y., Van Limbergen, E. J., Oberije, C. J. G., Troost, E. G. C., Hadrup, S. R., Dingemans, A. C., et al. (2020). Stereotactic ablative body radiotherapy (SABR) combined with immunotherapy (L19-IL2) versus standard of care in stage IV NSCLC patients, ImmunoSABR: a multicentre, randomised controlled open-label phase II trial. BMC Cancer 20:557. doi: 10.1186/s12885-020-07055-1
Lindstrom, M. K., Jankola, H. M., Hielm, S., Hyytia, E. K., and Korkeala, H. J. (1999). Identification of Clostridium botulinum with API 20 A, Rapid ID 32 A and RapID ANA II. FEMS Immunol. Med. Microbiol. 24, 267–274. doi: 10.1111/j.1574-695X.1999.tb01293.x
Mahmoudi Azar, L., Mehdizadeh Aghdam, E., Karimi, F., Haghshenas, B., Barzegari, A., Yaghmaei, P., et al. (2013). Influence of foreign DNA introduction and periplasmic expression of recombinant human interleukin-2 on hydrogen peroxide quantity and catalase activity in Escherichia coli. Adv. Pharm. Bull. 3, 395–402. doi: 10.5681/apb.2013.063
McDermott, D. F., Regan, M. M., Clark, J. I., Flaherty, L. E., Weiss, G. R., Logan, T. F., et al. (2005). Randomized phase III trial of high-dose interleukin-2 versus subcutaneous interleukin-2 and interferon in patients with metastatic renal cell carcinoma. J. Clin. Oncol. 2005:206. doi: 10.1200/JCO.2005.03.206
Mowday, A., Guise, C., Ackerley, D., Minton, N., Lambin, P., Dubois, L., et al. (2016). Advancing clostridia to clinical trial: past lessons and recent progress. Cancers 8:63. doi: 10.3390/cancers8070063
Mowday, A. M., Dubois, L. J., Kubiak, A. M., Chan-Hyams, J. V. E., Guise, C. P., Ashoorzadeh, A., et al. (2021). Use of an optimised enzyme/prodrug combination for Clostridia directed enzyme prodrug therapy induces a significant growth delay in necrotic tumours. Cancer Gene Ther. 2021:296. doi: 10.1038/s41417-021-00296-7
Payne, R., Glenn, L., Hoen, H., Richards, B., Smith, J. W., Lufkin, R., et al. (2014). Durable responses and reversible toxicity of high-dose interleukin-2 treatment of melanoma and renal cancer in a Community Hospital Biotherapy Program. J. ImmunoTher. Cancer. 2014:13. doi: 10.1186/2051-1426-2-13
Pyne, M. E., Moo-Young, M., Chung, D. A., and Chou, C. P. (2014). Expansion of the genetic toolkit for metabolic engineering of Clostridium pasteurianum: chromosomal gene disruption of the endogenous CpaAI restriction enzyme. Biotechnol. Biofuels 7:163. doi: 10.1186/s13068-014-0163-1
Rekers, N. H., Olivo Pimentel, V., Yaromina, A., Lieuwes, N. G., Biemans, R., Zegers, C. M. L., et al. (2018). The immunocytokine L19-IL2: An interplay between radiotherapy and long-lasting systemic anti-tumour immune responses. Oncoimmunology 7:e1414119. doi: 10.1080/2162402X.2017.1414119
Schwarz, K., Fiedler, T., Fischer, R. J., and Bahl, H. (2007). A Standard Operating Procedure (SOP) for the preparation of intra- and extracellular proteins of Clostridium acetobutylicum for proteome analysis. J. Microbiol. Methods 68:396–402. doi: 10.1016/j.mimet.2006.09.018
Sebaihia, M., Peck, M. W., Minton, N. P., Thomson, N. R., Holden, M. T., Mitchell, W. J., et al. (2007). Genome sequence of a proteolytic (Group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res. 17, 1082–1092. doi: 10.1101/gr.6282807
Setlow, P. (2019). Observations on research with spores of Bacillales and Clostridiales species. J. Appl. Microbiol. 126, 348–358. doi: 10.1111/jam.14067
Soman, G., Yang, X., Jiang, H., Giardina, S., Vyas, V., Mitra, G., et al. (2009). MTS dye based colorimetric CTLL-2 cell proliferation assay for product release and stability monitoring of Interleukin-15: Assay qualification, standardization and statistical analysis. J. Immunol. Methods 348, 83–94.
Theys, J., Landuyt, W., Nuyts, S., Mellaert, L., Bosmans, E. N., Rijnders, A., et al. (2001). Improvement of Clostridium tumour targeting vectors evaluated in rat rhabdomyosarcomas. FEMS Immunol. Med. Microbiol. 30, 37–41. doi: 10.1111/j.1574-695X.2001.tb01547.x
Totten, P. A., Norn, D. V., and Stamm, W. E. (1995). Characterization of the hemolytic activity of Haemophilus ducreyi. Infect. Immun. 63, 4409–4416.
Weigand, M. R., Pena-Gonzalez, A., Shirey, T. B., Broeker, R. G., Ishaq, M. K., Konstantinidis, K. T., et al. (2015). Implications of genome-based discrimination between Clostridium botulinum group I and Clostridium sporogenes strains for bacterial taxonomy. Appl. Environ. Microbiol. 81, 5420–5429. doi: 10.1128/AEM.01159-15
Wikoff, W. R., Anfora, A. T., Liu, J., Schultz, P. G., Lesley, S. A., Peters, E. C., et al. (2009). Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. U S A 106, 3698–3703. doi: 10.1073/pnas.0812874106
Keywords: streptolysin S, haemolysis, Clostridium, secretion, cytokine, spore, cancer
Citation: Kubiak AM, Bailey TS, Dubois LJ, Theys J and Lambin P (2021) Efficient Secretion of Murine IL-2 From an Attenuated Strain of Clostridium sporogenes, a Novel Delivery Vehicle for Cancer Immunotherapy. Front. Microbiol. 12:669488. doi: 10.3389/fmicb.2021.669488
Received: 03 March 2021; Accepted: 12 May 2021;
Published: 08 June 2021.
Edited by:
Bingyun Li, West Virginia University, United StatesReviewed by:
Prabhat Kumar Talukdar, Washington State University, United StatesAdrianne N. Edwards, Emory University, United States
Copyright © 2021 Kubiak, Bailey, Dubois, Theys and Lambin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aleksandra M. Kubiak, YS5rdWJpYWtAbWFhc3RyaWNodHVuaXZlcnNpdHkubmw=
†These authors have contributed equally to this work and share first authorship
 Aleksandra M. Kubiak
Aleksandra M. Kubiak Tom S. Bailey
Tom S. Bailey Ludwig J. Dubois
Ludwig J. Dubois Jan Theys
Jan Theys Philippe Lambin
Philippe Lambin