
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 13 May 2021
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.669024
This article is part of the Research TopicStructure, Function and Evolution of Complex Cellular Organization in Bacteria and ArchaeaView all 7 articles
Bacterial microcompartments (BMCs) are protein-based organelles that expand the metabolic potential of many bacteria by sequestering segments of enzymatic pathways in a selectively permeable protein shell. Sixty-eight different types/subtypes of BMCs have been bioinformatically identified based on the encapsulated enzymes and shell proteins encoded in genomic loci. BMCs are found across bacterial phyla. The organisms that contain them, rather than strictly correlating with specific lineages, tend to reflect the metabolic landscape of the environmental niches they occupy. From our recent comprehensive bioinformatic survey of BMCs found in genome sequence data, we find many in members of the human microbiome. Here we survey the distribution of BMCs in the different biotopes of the human body. Given their amenability to be horizontally transferred and bioengineered they hold promise as metabolic modules that could be used to probiotically alter microbiomes or treat dysbiosis.
Bacterial Microcompartments (BMCs) are organelles that are functionally similar to those of eukaryotes; they establish and contain a microenvironment that is distinct from the rest of the cell (Kerfeld et al., 2018; Kirst and Kerfeld, 2019). BMCs are bounded by a selectively permeable membrane, however, in contrast to their eukaryotic counterparts, this membrane, —the shell— is composed of proteins. All BMC shells are assembled from homologous building blocks, enabling their bioinformatic identification in genomic sequence data (Axen et al., 2014; Bobik et al., 2015; Zarzycki et al., 2017; Sutter et al., 2021). BMCs are either anabolic, such as the extensively studied carboxysome (Kerfeld and Melnicki, 2016) or catabolic; these are collectively known as metabolosomes (Figure 1A). An aldehyde intermediate is common to the encapsulated chemistry of many metabolosomes and the enzyme generating the aldehyde is referred to as the signature enzyme (Axen et al., 2014; Kerfeld and Erbilgin, 2015). The purpose of the BMC shell in these metabolosomes is to enhance catalysis and sequester toxic aldehyde intermediates (Figure 1B) (Kerfeld et al., 2018). Gut bacteria often have the potential to form ethanolamine utilization (EUT) BMCs because ethanolamine is abundant in the intestine as a breakdown product of phosphatidylethanolamine (Larson et al., 1983). Indeed, the EUT operon is part of the core E. coli genome (Dadswell et al., 2019), allowing the organism to use ethanolamine as a source of both carbon and nitrogen (Kaval and Garsin, 2018). Because it is an environment with a large spectrum of available substrates known to be catabolized within BMCs, they are frequently found in gut microbes (Ravcheev et al., 2019).
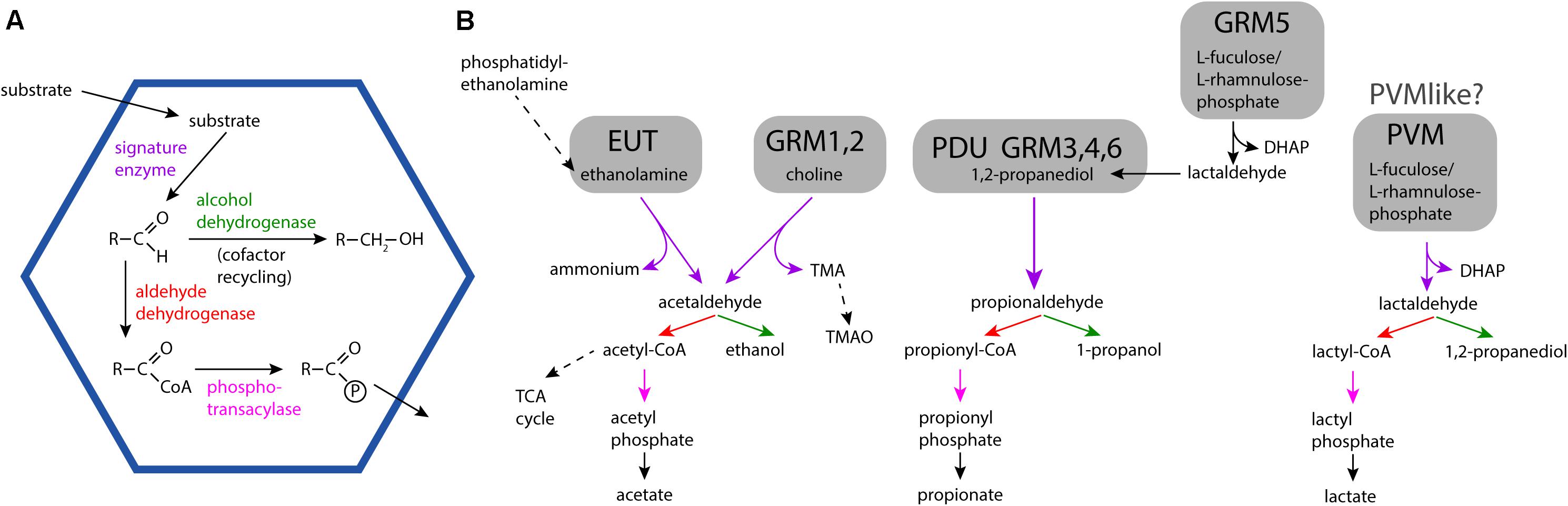
Figure 1. General BMC reaction overview and detailed reaction pathways for different BMC types found in microbiome samples. (A) General encapsulated pathway scheme of metabolosomes. (B) Substrates, intermediates and products of the EUT, PDU, GRM, and PVM/PVM-like BMCs. Solid arrows indicate BMC-associated reactions. TMA: trimethylamine; TMAO: trimethylamine-N-oxide; TCA cycle, tricarboxylic acid cycle; DHAP, dihydroxyacetone phosphate.
Encompassing the gut and beyond, the human microbiome has been defined as an essential organ of the human body given the tremendous effects it has on overall health (Shreiner et al., 2015; Kashyap et al., 2017; Manor et al., 2020; Fan and Pedersen, 2021). Dysbiosis, or disruption of a healthy microbiome, has been implicated in obesity, hypertension, cardiovascular disease, diabetes, cancer and even depression (Ding et al., 2019). Although little is known about the exact mechanism with which the microbiome exerts its influence, it seems plausible that the BMCs could potentially play a role in these outcomes by enabling the dominance of specific taxa. Although yet to be experimentally verified, BMC expression likely exerts some influence on the community composition by conferring the ability to catabolize niche-specific metabolites. Here we survey the available census of human microbiome organisms to identify their BMCs. Our compilation provides the first insights into the prevalence of specific types BMCs in particular niches, and suggests associations with BMC-based metabolism and the nutritional landscape of the sampling site in health and disease.
Tables matching up bacterial strains with body locations were downloaded from the Pathosystems Resource Integration Center (PATRIC)1, the Human Oral Microbiome Database (HOMD)2, and the NIH Human Microbiome Project3. Strain names were the matched with the assigned loci described in (Sutter et al., 2021) and correlated with body sites found in the respective databases (Supplementary Table 1). Duplicates with identical NCBI taxid and body site were removed.
The EUT BMCs allow organisms to utilize ethanolamine as a carbon and nitrogen source by metabolizing it into acetaldehyde and ammonia using the encapsulated ethanolamine ammonia lyase (Tsoy et al., 2009). There are three major types of EUT BMCs, namely EUT1, EUT2, and EUT3. Loci of these three types all encode the signature enzyme ethanolamine ammonia lyase but differ in the genes encoding ancillary proteins, regulatory proteins as well as the type and presence of core metabolosome elements (Sutter et al., 2021). In humans, ethanolamine is obtained through the diet as the product of the breakdown of the common lipid component of plant and animal cell membranes, phosphatidylethanolamine or from the breakdown of phospholipids in normal turnover of epithelial cells. Accordingly, organisms containing EUT BMCs are enriched in the gastrointestinal tract (Figure 2), or other sites with epithelial turnover. Ethanolamine is abundant in the inflamed gut and the presence of tetrathionate as the electron acceptor allows for intestinal pathogens such as Salmonella enterica, Enterococcus faecalis, enterohaemorrhagic Escherichia coli (EHEC), Clostridium difficile to flourish by utilizing EUT BMCs (Bertin et al., 2011; Srikumar and Fuchs, 2011; Thiennimitr et al., 2011; Anderson et al., 2018; Ormsby et al., 2019). Likewise, the ability to derive carbon, nitrogen and energy from ethanolamine is a hallmark of urinary tract infections, in which E. coli inhabit successively the perineum, the urethra and the bladder. The EUT BMC is directly involved in this progression (Sintsova et al., 2018; Dadswell et al., 2019).
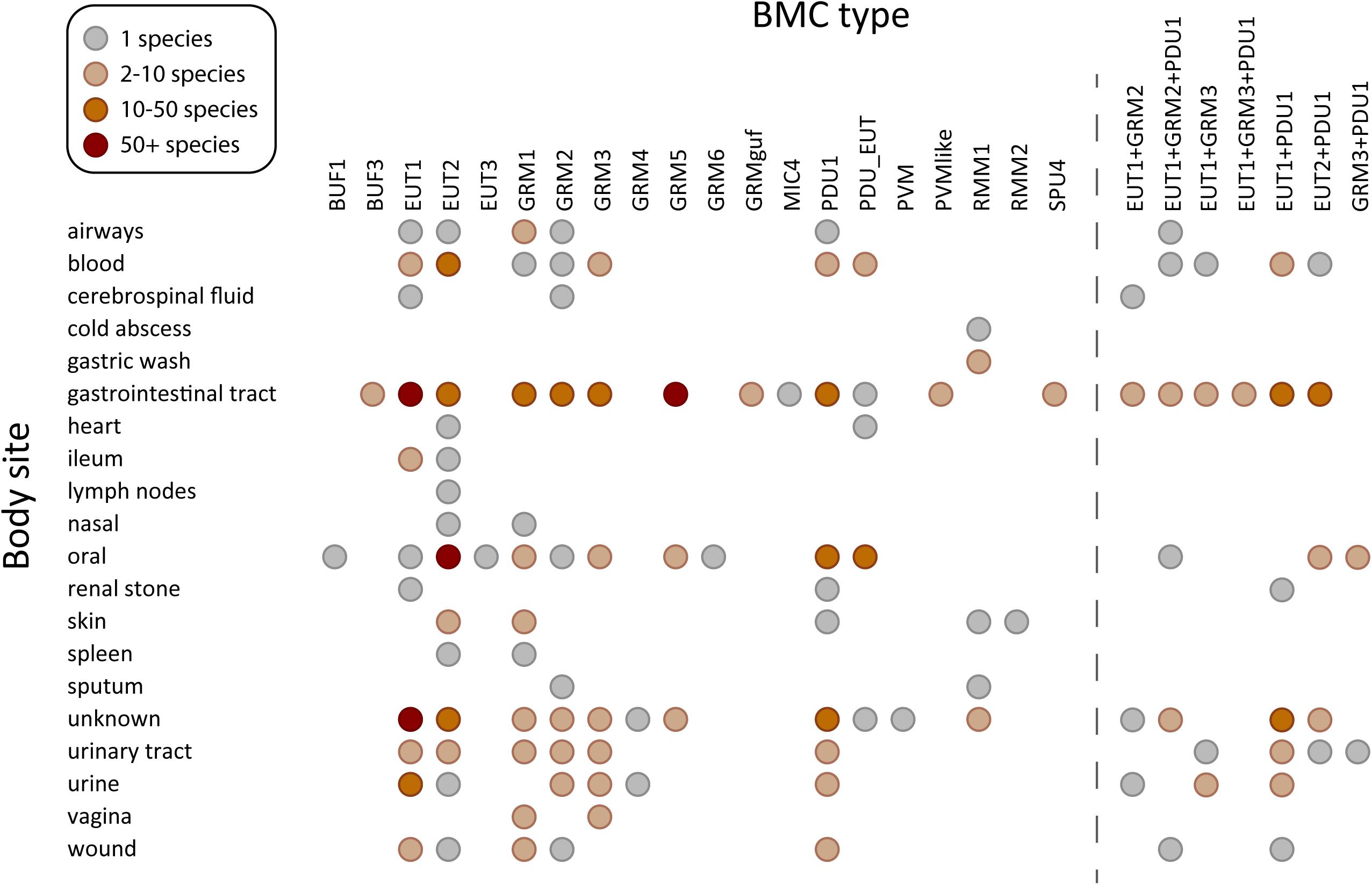
Figure 2. Species containing BMC functional types found in human microbiome sampling sites. BMC types found in sequenced genomes of 625 species in the human body plotted against the site or source from which they were isolated. Acronyms for the BMC functional types (Axen et al., 2014; Sutter et al., 2021): PDU, propane diol utilization; GRM, glycyl radical enzyme containing microcompartment; GRMguf, GRM with unknown function glycyl radical enzyme; EUT, ethanolamine utilization microcompartment; BUF, bacterial microcompartment of unknown function; RMM, Rhodococcus and Mycobacterium microcompartment; PVM/PVM-like, Planctomycete and Verrucomicrobia microcompartment; SPU, sugar phosphate utilizing microcompartment; PDU_EUT, genetic fusion of the PDU/EUT loci; MIC, microcompartments with an incomplete core. BMC types co-occurring in the same species, observed in at least in three species, are shown on the right.
BMCs are also found in organisms in cancer-associated gut dysbiosis. Fusobacterium hwasooki and F. nucleatum (Supplementary Table 1) species contain a EUT2 BMC, and F. nucleatum specifically is suggested to play a role in the progression of colorectal cancer (CRC) as well as oral squamous cell carcinoma (Zhou et al., 2018; Zhang et al., 2019). The microbial community in the oral microenvironment and its imbalance has likewise been implicated in diseases such as periodontitis and in dental caries (Lamont et al., 2018). There are studies showing direct correlation between EUT and periodontitis progression (Kaval and Garsin, 2018). All three major EUT BMC types are found in organisms populating the oral microbiome, underscoring the importance of ethanolamine degradation in this environment (Figure 2). The EUT2 BMC type was the most commonly found EUT subtype present in the genera Leptotrichia, Streptococcus and Fusobacterium (Supplementary Table 1) which are all members of the oral microbiome (Deo and Deshmukh, 2019). Furthermore, it has been shown that in the presence of ethanolamine, there was an increase in the respiratory activity of several of the pathogenic organisms in the oral microbiome (Hernandez-Sanabria et al., 2017) indicating that this is an important metabolite, and the ability to form a EUT BMC would provide a competitive advantage.
PDU (1,2-propanediol utilization) BMCs have a wide distribution comparable to EUT BMCs and they commonly co-occur in many different organisms (Sutter et al., 2021; Figure 2). In some species, such as strains of Streptococcus and Listeria monocytogenes, the two loci are fused (PDU_EUT, Supplementary Table 1). In other organisms, like F. nucleatum, the EUT and PDU BMCs are likely differentially regulated by the locus-encoded regulator, depending on availability of substrates. Via the diet, plant sugars rhamnose and fucose are catabolized by organisms in anaerobic conditions to produce 1,2-propanediol (1,2-PD), which can be utilized by PDU BMCs as a source of carbon and energy (Figure 1B). Lactobacillus panis and Velionella denticariosi, which can form PDU BMCs, were identified in the oral microbiome (Supplementary Table 1) and both are associated with human dental caries (Byun et al., 2004, 2007). Within the gut, Shigella flexneri, Citrobacter sp., and Lactobacillus brevis (Supplementary Table 1) all encode PDU1 BMCs in their genomes. Both Citrobacter and Lactobacillus species are implicated in human gut dysbiosis in patients suffering from irritable bowel syndrome (Ganji et al., 2016). Organisms containing PDU BMCs are also found in the blood and vagina (Figure 2). The presence of 1,2-propanediol in the vagina could be an indicator of a healthy microbiome; a study in 2013 showed that women with bacterial vaginosis had a reduction in the presence of 1,2-propanediol which is formed by the hydrogenation of lactic acid (Yeoman et al., 2013). Lactobacillus, which is a hallmark organism of a healthy vaginal microbiome, was not identified in this survey. F. nucleatum, identified in the vagina harboring the EUT1 and PDU1 BMCs (Supplementary Table 1) has been associated with bacterial vaginosis. This organism has also been implicated in causing preterm birth as well as intrauterine infections (Agarwal et al., 2020).
GRM1 (glycyl radical enzyme containing microcompartment) and GRM2 BMCs produce TMA as a side product of choline metabolism (Figure 1B). TMA can be absorbed and oxidized to trimethylamine-N-oxide (TMAO) by flavin-containing monooxygenases (Fennema et al., 2016). TMAO is known to contribute to cardiovascular disease (Wang et al., 2011; Schiattarella et al., 2017). TMAO is also generated by the gut microbiome after consumption of foods rich in L-carnitine and phosphatidylcholine, such as fish, eggs and red meat (Zeisel et al., 1983; Wang et al., 2011). The abundance of GRM1 and GRM2 BMCs in the gut organisms (Figure 2) reflects that the gastrointestinal tract is rich in choline. Some of the organisms in the gut that contain the GRM2 BMCs include the known pathogen Providencia alcalifaciens, the causative agent of foodborne illnesses (Shah et al., 2019) and Klebsiella sp. which is an opportunistic pathogen (Supplementary Table 1). Additionally, GRM2 was identified in various strains of E. coli (Supplementary Table 1). The presence of the GRM BMCs in the blood and urine could be accounted for by sepsis along with another cardiovascular related health conditions that result in the formation of TMAO. Indeed, GRM1 and GRM2 can be found in the majority of bacterial species associated with urinary tract infections. Recently, a GRM2 BMC, encoded in a pathogenicity island, has been shown to be involved in choline utilization in E. coli UPEC 536 (Herring et al., 2018).
GRM3, GRM4 and GRM6 BMCs are functionally analogous to PDU BMCs (Levin and Balskus, 2018; Ferlez et al., 2019) and consistently show a similar microbiome distribution as species containing PDU BMCs (Figure 2). The GRM5 BMCs have additional enzymes that enable it to process fuculose/rhamnulose phosphate which are typical degradation products of complex polysaccharides. This is consistent with the occurrence of many GRM5-containing species in the gastrointestinal microbiome (Figure 2). One of the GRM5 containing organisms in gut is the anaerobe Ruminococcus gnavus (Supplementary Table 1) that is known to be associated with Crohn’s disease (Henke et al., 2019).
Several newly discovered or less familiar microcompartments such as RMM and the PVM-like have been identified in our human microbiome survey (Figure 2). The RMM organelles are named for representatives found in Rhodococcus and Mycobacterium species (Axen et al., 2014). This BMC has been proposed to metabolize aminoacetone to propionyl-CoA (Mallette and Kimber, 2018). Organisms containing RMM1 were identified in the skin microbiome (Rhodococcus erythropolis SK121) and the gut (various strains of Mycobacteria) (Supplementary Table 1). There have been studies showing the presence of Mycobacterium species with cutaneous infections and conditions such as cold abscess (Franco-Paredes et al., 2018; Bains et al., 2019). It is likely that the ability to utilize aminoacetone could provide a competitive advantage in the disease state. Interestingly, none of the common pathogenic mycobacterial strains such as M. tuberculosis and M. leprae contain the RMM1 BMC. There is a single occurrence of RMM2 in Paracoccus yeei TT13 that was found in a skin sample (Supplementary Table 1) and shown to grow on 1,2-PD as sole carbon and energy source (Lim et al., 2018).
PVM microcompartments encapsulate a class-II aldolase as their signature enzyme with the substrates rhamnose and fucose (Erbilgin et al., 2014; Figure 1B). They are primarily found in environmental samples where they are thought to metabolize algal cell wall degradation products (Sichert et al., 2020; Sizikov et al., 2020). The PVM-like BMC locus contains a claas-II aldolase homolog aldolase and an aldehyde dehydrogenase that is expected to process 1,2-PD (Sutter et al., 2021), so a similar substrate as PVM is likely. Species containing the PVM-like microcompartments can be identified in gastrointestinal tract where they likely also play a role in the breakdown of complex carbohydrates (Figure 2); these organisms include Hungatella hathewayi, Clostridium bolteae, and Clostridium sp. AF18-27 (Supplementary Table 1). One of the PVM-like BMC containing organisms is Faecalibacterium prausnitzii, which has been shown to be beneficial for gut health in a mouse model system (Munukka et al., 2017).
The sugar phosphate utilizing microcompartments (SPU) are emerging as one of the most widespread types of BMCs; they are predicted to be involved in DNA catabolism via the deoxyribose/deoxyribulose 5-phosphate degradation pathway (Axen et al., 2014; Sutter et al., 2021). The degradation of exogenous DNA, a common component of detritus, is a source of carbon and energy (Finkel and Kolter, 2001). Organisms containing SPU4 BMCs such as Anaerotruncus colihominis, Clostridium sp. AF15-17LB and Dorea sp. D27 have been found in the gastrointestinal tract samples (Supplementary Table 1), consistent with availability of nucleic acid from the turnover of resident microorganisms.
BUF microcompartments or Bacterial Microcompartments of Unknown Function loci encode the structural proteins to form the metabolosome shell but not an aldehyde dehydrogenase (Axen et al., 2014). A BUF1 has recently been characterized as a compartment for the potential degradation of xanthine (encapsulating Xanthine dehydrogenase) (Ravcheev et al., 2019). Elevated levels of metabolites including xanthine, hypoxanthine, inosine have been detected in the metabolome of periodontitis and gingivitis associated oral samples (Duran-Pinedo and Frias-Lopez, 2015) and a single BUF1 BMC containing organism (Bacillus sp. 2_A_57_CT2, Supplementary Table 1) has been found in the oral microbiome (Figure 2).
Not much is known about the Microcompartments with Incomplete Core (MIC), except that they contain an aldehyde dehydrogenase (Sutter et al., 2021) and a class II aldolase that may imply a similar function as the PVM BMCs. A single organism (Lachnospiraceae bacterium KGMB03038) containing the MIC4 BMC has been identified in the stool sample of a healthy person (Supplementary Table 1).
With the increasing availability of bacterial genome sequences, including those from culture independent genomic methods and microbiomes, the number and diversity of known BMCs is rapidly increasing (Sutter et al., 2021). In many ecosystems the BMCs employed by community members reflect important characteristics of the nutritional landscape of the environmental niche, such as the importance of the PVM BMC (Planctomycete and Verrucomicrobia microcompartment) for the degradation of complex polysaccharides originating from algae (Erbilgin et al., 2014; Sichert et al., 2020; Sizikov et al., 2020). In the human microbiome, pathogenic bacteria are able to gain a fitness advantage by catabolizing organic compounds that are metabolically unavailable to the native microflora (Passalacqua et al., 2016). For example, numerous studies show the role of the PDU BMC in the proliferation and persistence of pathogens. This compound is naturally present in the gut as a by-product of microbial fermentation of the sugars rhamnose and fucose (Badia et al., 1985; Schardt et al., 2017). During colonization, effector molecules cause inflammation of the intestine subsequently forming tetrathionate (Chowdhury et al., 2014). Tetrathionate is utilized as the electron acceptor by EUT-containing organisms, conferring a distinct competitive advantage, in conjunction with 1,2-propanediol, also found in the gut (Thiennimitr et al., 2011). This allows opportunistic gut pathogens such as Salmonella to survive in anaerobic conditions by not only using tetrathionate as the terminal electron acceptor but also by cobalamin synthesis, which requires anaerobiosis, the expression of which is co-regulated with genes from the PDU locus (Jakobson and Tullman-Ercek, 2016). The PDU gene cluster has been implicated in providing L. monocytogenes with a significant fitness advantage in the gastrointestinal tract as evidenced by faster clearing of infection in murine models infected with pdu deletion mutants (Schardt et al., 2017; Zeng et al., 2019). It is becoming increasingly apparent that many organisms have the potential to form more than one functional type of BMC (Figure 2) (Sutter et al., 2021); such as Salmonella enterica which contains both PDU and EUT BMCs (Stojiljkovic et al., 1995), and organisms associated with urinary tract infections (Sutter et al., 2021).
The types of BMCs in organisms of the human microbiome sampling sites reflect the local nutritional landscape; for example, the EUT, PDU, and GRM BMCs are gut associated. Similarly, the oral microbiome has a predominance of the GRM and EUT BMC types. The GRMs constitute three functionally distinct types based on the substrate of the GRE (Zarzycki et al., 2015; Figure 1B). GRM1 and GRM2 are associated with anaerobic breakdown of choline to trimethylamine (TMA) and acetaldehyde intermediates (Figure 1B). Interestingly, the conversion of choline to TMA is only possible through microbial activity (Craciun and Balskus, 2012; Craciun et al., 2014). The GRM3/4/6 organelles use the GRE 1,2-PD dehydratase to convert 1,2-PD into propanol and propionate (Zarzycki et al., 2017; Schindel et al., 2019) and GRM5 is involved in the anaerobic degradation of rhamnose/fuculose (Petit et al., 2013; Zarzycki et al., 2015). Collectively, we find these BMCs are the most prevalent in available sequence data from human microbiome samples.
In addition, several newly discovered BMCs such as RMM, PVM and PVM-like have been discovered to be associated with human microbiome albeit some of them from undefined sites of the human body. Uncharacterized BMCs such as BUF and MIC were identified in the gut and stool samples. The experimental characterization of these metabolic modules is complementary to metabolomics studies of these microenvironments because the function of the BMCs reflects the metabolic profile of the microenvironments.
The metabolic profile of sampling sites is regulated by several factors; diet, the propensity for host cell breakdown (epithelial layers) and the composition of the microbiome are primary determinants (Valdes et al., 2018; Leeming et al., 2019; Visconti et al., 2019; De Angelis et al., 2020). One study finds increased expression of the EUT genes in Listeria monocytogenes, anaerobically and in the presence of vitamin B12 (Zeng et al., 2020). While this does not provide a direct correlation between change of diet in humans and its effect on BMC gene expression, it does demonstrate that these genes are induced by available substrates.
The catabolic activity of BMCs within the human microbiome likely impacts the metabolic profile of a particular site. Given that the future of personalized medicine likely includes routine site-specific sampling of a patient’s microbiome throughout life, monitoring organism composition, and its metabolic potential may emerge as a means to manage homeostasis and health. Likewise, BMC-based manipulation of the microbiome could offer an approach to treating dysbiosis. A pathogen can colonize a given environment if it has the ability to use a limiting or specific nutrient (Freter et al., 1983). BMCs endow organisms the ability to catabolize substrates metabolically unavailable to commensals for a competitive advantage. In a microbiome-based approach, endowing a probiotic organism with a BMCs used by a pathogen may provide a way to outcompete it.
According to the World Health Organization, as of 2019 communicable diseases such as lower respiratory and diarrheal are among the top 10 causes of death globally. Bacteria responsible for causing lower respiratory illnesses include members of the Streptococcus genera, E. coli, Klebsiella pneumoniae, Mycoplasma pneumoniae, and Mycobacterium tuberculosis (Dasaraju and Liu, 1996). Similarly, urinary tract infections are the primary source for outpatient infections in the United States and are caused by organisms including uropathogenic E. coli (UPEC), K. pneumoniae, and Pseudomonas aeruginosa, Group B Strep (Medina and Castillo-Pino, 2019). Many of the organisms implicated in causing these diseases encode BMCs in their genomes.
The microbiome can be thought of as a pliable ecosystem that can be altered to have immense overall impacts on human health. Altering the microbiome using an individual-based approach to account for the variation may aid in resolving complex issues such as obesity and inflammatory bowel disease (Wang et al., 2020). This can further be improvised by the addition of engineered probiotic strains. BMCs, as metabolic modules encoded by genetic modules, provide a way to introduce by “plug and play” expanded metabolic potential into probiotic organisms. Engineering BMCs for use in microbiome-based therapies can be thought of as an additional approach in the field of precision medicine. BMCs encapsulate necessary enzymes for substrate utilization and can be engineered to include enzymes that will breakdown a disease-causing substrate or even potentially enclosing a toxic intermediate. Indeed, developing engineered BMCs that have a high bacterial host range and are easy to modulate (Kirst and Kerfeld, 2019) may be achievable in the foreseeable future.
CAK conceived the project and wrote the manuscript. KA and MS generated and analyzed the dataset and wrote the manuscript. All authors read and approved the manuscript.
This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID) Grant 1R01AI114975-01.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We appreciate the thoughtful commentary received as review of this manuscript, which was valuable in its revision for publication. This research used resources of the National Energy Research Scientific Computing Center, which is supported by the Office of Science of the U.S Department of Energy under Contract No. DE-AC02-05CH11231.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.669024/full#supplementary-material
Agarwal, K., Robinson, L. S., Aggarwal, S., Foster, L. R., Hernandez-Leyva, A., Lin, H., et al. (2020). Glycan cross-feeding supports mutualism between Fusobacterium and the vaginal microbiota. PLoS Biol. 18:e3000788. doi: 10.1371/journal.pbio.3000788
Anderson, C. J., Satkovich, J., Koseoglu, V. K., Agaisse, H., and Kendall, M. M. (2018). The Ethanolamine Permease EutH promotes vacuole adaptation of Salmonella enterica and Listeria monocytogenes during macrophage infection. Infect. Immun. 86:e00172–18.
Axen, S. D., Erbilgin, O., and Kerfeld, C. A. (2014). A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput. Biol. 10:e1003898. doi: 10.1371/journal.pcbi.1003898
Badia, J., Ros, J., and Aguilar, J. (1985). Fermentation mechanism of fucose and rhamnose in Salmonella typhimurium and Klebsiella pneumoniae. J. Bacteriol. 161, 435–437. doi: 10.1128/jb.161.1.435-437.1985
Bains, L., Lal, P., Chand, T., Gautam, K. K., Beg, M. Y., and Kumar, P. (2019). Isolated primary cold abscess of the sternum: a case report. J. Med. Case Rep. 13:267.
Bertin, Y., Girardeau, J. P., Chaucheyras-Durand, F., Lyan, B., Pujos-Guillot, E., Harel, J., et al. (2011). Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ. Microbiol. 13, 365–377. doi: 10.1111/j.1462-2920.2010.02334.x
Bobik, T. A., Lehman, B. P., and Yeates, T. O. (2015). Bacterial microcompartments: widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol. Microbiol. 98, 193–207. doi: 10.1111/mmi.13117
Byun, R., Carlier, J. P., Jacques, N. A., Marchandin, H., and Hunter, N. (2007). Veillonella denticariosi sp. nov., isolated from human carious dentine. Int. J. Syst. Evol. Microbiol. 57, 2844–2848. doi: 10.1099/ijs.0.65096-0
Byun, R., Nadkarni, M. A., Chhour, K. L., Martin, F. E., Jacques, N. A., and Hunter, N. (2004). Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J. Clin. Microbiol. 42, 3128–3136. doi: 10.1128/jcm.42.7.3128-3136.2004
Chowdhury, C., Sinha, S., Chun, S., Yeates, T. O., and Bobik, T. A. (2014). Diverse bacterial microcompartment organelles. Microbiol. Mol. Biol. Rev. 78, 438–468. doi: 10.1128/mmbr.00009-14
Craciun, S., and Balskus, E. P. (2012). Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. U.S.A. 109, 21307–21312. doi: 10.1073/pnas.1215689109
Craciun, S., Marks, J. A., and Balskus, E. P. (2014). Characterization of choline trimethylamine-lyase expands the chemistry of glycyl radical enzymes. ACS Chem. Biol. 9, 1408–1413. doi: 10.1021/cb500113p
Dadswell, K., Creagh, S., Mccullagh, E., Liang, M., Brown, I. R., Warren, M. J., et al. (2019). Bacterial microcompartment-mediated ethanolamine metabolism in Escherichia coli urinary tract infection. Infect. Immun. 87, e211–e219.
Dasaraju, P. V., and Liu, C. (1996). “Infections of the respiratory system,” in Medical Microbiology, ed. S. Baron (Galveston, TX: University of Texas Medical Branc).
De Angelis, M., Ferrocino, I., Calabrese, F. M., De Filippis, F., Cavallo, N., Siragusa, S., et al. (2020). Diet influences the functions of the human intestinal microbiome. Sci. Rep. 10:4247.
Deo, P. N., and Deshmukh, R. (2019). Oral microbiome: unveiling the fundamentals. J. Oral. Maxillofac. Pathol. 23, 122–128.
Ding, R. X., Goh, W. R., Wu, R. N., Yue, X. Q., Luo, X., Khine, W. W. T., et al. (2019). Revisit gut microbiota and its impact on human health and disease. J. Food Drug Anal. 27, 623–631. doi: 10.1016/j.jfda.2018.12.012
Duran-Pinedo, A. E., and Frias-Lopez, J. (2015). Beyond microbial community composition: functional activities of the oral microbiome in health and disease. Microbes Infect. 17, 505–516. doi: 10.1016/j.micinf.2015.03.014
Erbilgin, O., Mcdonald, K. L., and Kerfeld, C. A. (2014). Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl. Environ. Microbiol. 80, 2193–2205. doi: 10.1128/aem.03887-13
Fan, Y., and Pedersen, O. (2021). Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 19, 55–71. doi: 10.1038/s41579-020-0433-9
Fennema, D., Phillips, I. R., and Shephard, E. A. (2016). Trimethylamine and trimethylamine n-oxide, a flavin-containing monooxygenase 3 (FMO3)-mediated host-microbiome metabolic axis implicated in health and disease. Drug Metab. Dispos. 44, 1839–1850. doi: 10.1124/dmd.116.070615
Ferlez, B., Sutter, M., and Kerfeld, C. A. (2019). Glycyl radical enzyme-associated microcompartments: redox-replete bacterial organelles. mBio 10, e2327–e2318.
Finkel, S. E., and Kolter, R. (2001). DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 183, 6288–6293. doi: 10.1128/jb.183.21.6288-6293.2001
Franco-Paredes, C., Marcos, L. A., Henao-Martinez, A. F., Rodriguez-Morales, A. J., Villamil-Gomez, W. E., Gotuzzo, E., et al. (2018). Cutaneous mycobacterial infections. Clin. Microbiol. Rev. 32:e00069–18.
Freter, R., Brickner, H., Botney, M., Cleven, D., and Aranki, A. (1983). Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect. Immun. 39, 676–685. doi: 10.1128/iai.39.2.676-685.1983
Ganji, L., Alebouyeh, M., Shirazi, M. H., Eshraghi, S. S., Mirshafiey, A., Ebrahimi Daryani, N., et al. (2016). Dysbiosis of fecal microbiota and high frequency of Citrobacter, Klebsiella spp., and Actinomycetes in patients with irritable bowel syndrome and gastroenteritis. Gastroenterol. Hepatol. Bed Bench 9, 325–330.
Henke, M. T., Kenny, D. J., Cassilly, C. D., Vlamakis, H., Xavier, R. J., and Clardy, J. (2019). Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. U.S.A. 116, 12672–12677. doi: 10.1073/pnas.1904099116
Hernandez-Sanabria, E., Slomka, V., Herrero, E. R., Kerckhof, F. M., Zaidel, L., Teughels, W., et al. (2017). In vitro increased respiratory activity of selected oral bacteria may explain competitive and collaborative interactions in the oral microbiome. Front. Cell Infect. Microbiol. 7:235. doi: 10.3389/fcimb.2017.00235
Herring, T. I., Harris, T. N., Chowdhury, C., Mohanty, S. K., and Bobik, T. A. (2018). A Bacterial microcompartment is used for choline fermentation by Escherichia coli 536. J. Bacteriol. 200:e00764–17.
Jakobson, C. M., and Tullman-Ercek, D. (2016). Dumpster diving in the gut: bacterial microcompartments as part of a host-associated lifestyle. PLoS Pathog. 12:e1005558. doi: 10.1371/journal.ppat.1005558
Kashyap, P. C., Chia, N., Nelson, H., Segal, E., and Elinav, E. (2017). Microbiome at the frontier of personalized medicine. Mayo Clin. Proc. 92, 1855–1864. doi: 10.1016/j.mayocp.2017.10.004
Kerfeld, C. A., Aussignargues, C., Zarzycki, J., Cai, F., and Sutter, M. (2018). Bacterial microcompartments. Nat. Rev. Microbiol. 16, 277–290.
Kerfeld, C. A., and Erbilgin, O. (2015). Bacterial microcompartments and the modular construction of microbial metabolism. Trends Microbiol. 23, 22–34. doi: 10.1016/j.tim.2014.10.003
Kerfeld, C. A., and Melnicki, M. R. (2016). Assembly, function and evolution of cyanobacterial carboxysomes. Curr. Opin. Plant Biol. 31, 66–75. doi: 10.1016/j.pbi.2016.03.009
Kirst, H., and Kerfeld, C. A. (2019). Bacterial microcompartments: catalysis-enhancing metabolic modules for next generation metabolic and biomedical engineering. BMC Biol. 17:79. doi: 10.1186/s12915-019-0691-z
Lamont, R. J., Koo, H., and Hajishengallis, G. (2018). The oral microbiota: dynamic communities and host interactions. Nat. Rev. Microbiol. 16, 745–759. doi: 10.1038/s41579-018-0089-x
Larson, T. J., Ehrmann, M., and Boos, W. (1983). Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J. Biol. Chem. 258, 5428–5432. doi: 10.1016/s0021-9258(20)81908-5
Leeming, E. R., Johnson, A. J., Spector, T. D., and Le Roy, C. I. (2019). Effect of diet on the gut microbiota: rethinking intervention duration. Nutrients 11:2862. doi: 10.3390/nu11122862
Levin, B. J., and Balskus, E. P. (2018). Characterization of 1,2-propanediol dehydratases reveals distinct mechanisms for B12-dependent and glycyl radical enzymes. Biochemistry 57, 3222–3226. doi: 10.1021/acs.biochem.8b00164
Lim, J. Y., Hwang, I., Ganzorig, M., Pokhriyal, S., Singh, R., and Lee, K. (2018). Complete genome sequence of paracoccus yeei TT13, isolated from human skin. Microbiol. Resour. Announc. 6:e01514–17. doi: 10.1128/genomea.01514-17
Mallette, E., and Kimber, M. S. (2018). Structure and kinetics of the S-(+)-1-Amino-2-propanol dehydrogenase from the RMM microcompartment of Mycobacterium smegmatis. Biochemistry 57, 3780–3789. doi: 10.1021/acs.biochem.8b00464
Manor, O., Dai, C. L., Kornilov, S. A., Smith, B., Price, N. D., Lovejoy, J. C., et al. (2020). Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 11:5206.
Medina, M., and Castillo-Pino, E. (2019). An introduction to the epidemiology and burden of urinary tract infections. Ther. Adv. Urol. 11:1756287219832172.
Munukka, E., Rintala, A., Toivonen, R., Nylund, M., Yang, B. R., Takanen, A., et al. (2017). Faecalibacterium prausnitzii treatment improves hepatic health and reduces adipose tissue inflammation in high-fat fed mice. ISME J. 11, 1667–1679. doi: 10.1038/ismej.2017.24
Ormsby, M. J., Logan, M., Johnson, S. A., Mcintosh, A., Fallata, G., Papadopoulou, R., et al. (2019). Inflammation associated ethanolamine facilitates infection by Crohn’s disease-linked adherent-invasive Escherichia coli. EBioMedicine 43, 325–332. doi: 10.1016/j.ebiom.2019.03.071
Passalacqua, K. D., Charbonneau, M. E., and O’riordan, M. X. D. (2016). Bacterial metabolism shapes the host-pathogen interface. Microbiol. Spectr. 4:VMBF-0027-2015. doi: 10.1128/microbiolspec.VMBF-0027-2015
Petit, E., Latouf, W. G., Coppi, M. V., Warnick, T. A., Currie, D., Romashko, I., et al. (2013). Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS One 8:e54337. doi: 10.1371/journal.pone.0054337
Ravcheev, D. A., Moussu, L., Smajic, S., and Thiele, I. (2019). Comparative genomic analysis reveals novel microcompartment-associated metabolic pathways in the human gut microbiome. Front. Genet. 10:636. doi: 10.3389/fgene.2019.00636
Schardt, J., Jones, G., Muller-Herbst, S., Schauer, K., D’orazio, S. E. F., and Fuchs, T. M. (2017). Comparison between Listeria sensu stricto and Listeria sensu lato strains identifies novel determinants involved in infection. Sci. Rep. 7:17821.
Schiattarella, G. G., Sannino, A., Toscano, E., Giugliano, G., Gargiulo, G., Franzone, A., et al. (2017). Gut microbe-generated metabolite trimethylamine-N-oxide as cardiovascular risk biomarker: a systematic review and dose-response meta-analysis. Eur. Heart J. 38, 2948–2956. doi: 10.1093/eurheartj/ehx342
Schindel, H. S., Karty, J. A., Mckinlay, J. B., and Bauer, C. E. (2019). Characterization of a glycyl radical enzyme bacterial microcompartment pathway in rhodobacter capsulatus. J. Bacteriol. 201:e00343–18.
Shah, M. M., Odoyo, E., and Ichinose, Y. (2019). Epidemiology and pathogenesis of providencia alcalifaciens infections. Am. J. Trop. Med. Hyg. 101, 290–293. doi: 10.4269/ajtmh.18-0376
Shreiner, A. B., Kao, J. Y., and Young, V. B. (2015). The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 31, 69–75.
Sichert, A., Corzett, C. H., Schechter, M. S., Unfried, F., Markert, S., Becher, D., et al. (2020). Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol. 5, 1026–1039. doi: 10.1038/s41564-020-0720-2
Sintsova, A., Smith, S., Subashchandrabose, S., and Mobley, H. L. (2018). Role of ethanolamine utilization genes in host colonization during urinary tract infection. Infect. Immun. 86:e00542–17.
Sizikov, S., Burgsdorf, I., Handley, K. M., Lahyani, M., Haber, M., and Steindler, L. (2020). Characterization of sponge-associated Verrucomicrobia: microcompartment-based sugar utilization and enhanced toxin-antitoxin modules as features of host-associated Opitutales. Environ. Microbiol. 22, 4669–4688. doi: 10.1111/1462-2920.15210
Srikumar, S., and Fuchs, T. M. (2011). Ethanolamine utilization contributes to proliferation of Salmonella enterica serovar Typhimurium in food and in nematodes. Appl. Environ. Microbiol. 77, 281–290. doi: 10.1128/aem.01403-10
Stojiljkovic, I., Baumler, A. J., and Heffron, F. (1995). Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177, 1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995
Sutter, M., Melnicki, M. R., Schulz, F., Woyke, T., and Kerfeld, C. A. (2021). A catalog of the diversity and ubiquity of metabolic organelles in bacteria. bioRxiv [Preprint] doi: 10.1101/2021.01.25.427685 bioRxiv: 2021.2001.2025.427685
Thiennimitr, P., Winter, S. E., Winter, M. G., Xavier, M. N., Tolstikov, V., Huseby, D. L., et al. (2011). Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U.S.A. 108, 17480–17485. doi: 10.1073/pnas.1107857108
Tsoy, O., Ravcheev, D., and Mushegian, A. (2009). Comparative genomics of ethanolamine utilization. J. Bacteriol. 191, 7157–7164. doi: 10.1128/jb.00838-09
Valdes, A. M., Walter, J., Segal, E., and Spector, T. D. (2018). Role of the gut microbiota in nutrition and health. BMJ 361:k2179. doi: 10.1136/bmj.k2179
Visconti, A., Le Roy, C. I., Rosa, F., Rossi, N., Martin, T. C., Mohney, R. P., et al. (2019). Interplay between the human gut microbiome and host metabolism. Nat. Commun. 10:4505.
Wang, J., Chen, W. D., and Wang, Y. D. (2020). The relationship between gut microbiota and inflammatory diseases: the role of macrophages. Front. Microbiol. 11:1065. doi: 10.3389/fmicb.2020.01065
Wang, Z., Klipfell, E., Bennett, B. J., Koeth, R., Levison, B. S., Dugar, B., et al. (2011). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63. doi: 10.1038/nature09922
Yeoman, C. J., Thomas, S. M., Miller, M. E., Ulanov, A. V., Torralba, M., Lucas, S., et al. (2013). A multi-omic systems-based approach reveals metabolic markers of bacterial vaginosis and insight into the disease. PLoS One 8:e56111. doi: 10.1371/journal.pone.0056111
Zarzycki, J., Erbilgin, O., and Kerfeld, C. A. (2015). Bioinformatic characterization of glycyl radical enzyme-associated bacterial microcompartments. Appl. Environ. Microbiol. 81, 8315–8329. doi: 10.1128/aem.02587-15
Zarzycki, J., Sutter, M., Cortina, N. S., Erb, T. J., and Kerfeld, C. A. (2017). In vitro characterization and concerted function of three core enzymes of a Glycyl radical enzyme – associated bacterial microcompartment. Sci. Rep. 7:42757.
Zeisel, S. H., Wishnok, J. S., and Blusztajn, J. K. (1983). Formation of methylamines from ingested choline and lecithin. J. Pharmacol. Exp. Ther. 225, 320–324.
Zeng, Z., Boeren, S., Bhandula, V., Light, S. H., Smid, E. J., Notebaart, R. A., et al. (2020). Bacterial microcompartments linked to the flavin-based extracellular electron transfer drives anaerobic ethanolamine utilization in Listeria monocytogenes. bioRxiv[Preprint] doi: 10.1101/2020.10.27.358424 bioRxiv: 2020.2010.2027.358424
Zeng, Z., Smid, E. J., Boeren, S., Notebaart, R. A., and Abee, T. (2019). Bacterial microcompartment-dependent 1,2-propanediol utilization stimulates anaerobic growth of Listeria monocytogenes EGDe. Front. Microbiol. 10:2660. doi: 10.3389/fmicb.2019.02660
Zhang, L., Liu, Y., Zheng, H. J., and Zhang, C. P. (2019). The oral microbiota may have influence on oral cancer. Front. Cell Infect. Microbiol. 9:476. doi: 10.3389/fcimb.2019.00476
Keywords: bacterial microcompartments, metabolosomes, pathogenesis, human microbiome, dysbiosis
Citation: Asija K, Sutter M and Kerfeld CA (2021) A Survey of Bacterial Microcompartment Distribution in the Human Microbiome. Front. Microbiol. 12:669024. doi: 10.3389/fmicb.2021.669024
Received: 17 February 2021; Accepted: 13 April 2021;
Published: 13 May 2021.
Edited by:
John A. Fuerst, The University of Queensland, AustraliaReviewed by:
Martin Warren, University of Kent, United KingdomCopyright © 2021 Asija, Sutter and Kerfeld. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheryl A. Kerfeld, Y2tlcmZlbGRAbGJsLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.