- 1Institute of Preventive Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Avian Disease Research Center, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 3Key Laboratory of Animal Disease and Human Health of Sichuan Province, Sichuan Agricultural University, Chengdu, China
Herpesviruses are extremely successful parasites that have evolved over millions of years to develop a variety of mechanisms to coexist with their hosts and to maintain host-to-host transmission and lifelong infection by regulating their life cycles. The life cycle of herpesviruses consists of two phases: lytic infection and latent infection. During lytic infection, active replication and the production of numerous progeny virions occur. Subsequent suppression of the host immune response leads to a lifetime latent infection of the host. During latent infection, the viral genome remains in an inactive state in the host cell to avoid host immune surveillance, but the virus can be reactivated and reenter the lytic cycle. The balance between these two phases of the herpesvirus life cycle is controlled by broad interactions among numerous viral and cellular factors. ICP22/ORF63 proteins are among these factors and are involved in transcription, nuclear budding, latency establishment, and reactivation. In this review, we summarized the various roles and complex mechanisms by which ICP22/ORF63 proteins regulate the life cycle of human herpesviruses and the complex relationships among host and viral factors. Elucidating the role and mechanism of ICP22/ORF63 in virus–host interactions will deepen our understanding of the viral life cycle. In addition, it will also help us to understand the pathogenesis of herpesvirus infections and provide new strategies for combating these infections.
Introduction
Herpesviruses are large DNA viruses that can infect a wide range of hosts, including almost all vertebrates and even invertebrates (McGeoch et al., 1995; Carter et al., 2007). During infection, herpesviruses enter mainly through the skin, mucous membranes, and nerve tissue of hosts, and herpesvirus infections seriously affect the health of humans and other animals (Davison et al., 2009; Azab et al., 2018; Heldwein, 2018). To date, more than 100 herpesviruses have been discovered and classified into α-, β-, and γ-herpesviruses, on the basis of their molecular and biological characteristics (Whitley, 1996; Bigalke and Heldwein, 2017). The members of each subfamily possess the same virion structure, including a linear double-stranded DNA genome, capsid, tegument, and envelope (Griffin, 2007; Loret et al., 2008; Johnson and Baines, 2011; Heming et al., 2017; Liu et al., 2019; Lv et al., 2019; Yang et al., 2020). The life cycle of herpesviruses consists of two phases: lytic infection and latent infection (Stevens, 1989; Knipe and Cliffe, 2008; Dupont and Reeves, 2016; Riaz et al., 2017; Münz, 2019; Cohen, 2020). During lytic infection, shortly after entry into a susceptible cell, virions progress through uncoating, gene transcription, DNA replication, protein translation, assembly, egress, release, etc., to produce progeny virions (Glaunsinger, 2015; Heldwein, 2016; Sathiyamoorthy et al., 2017; Lv et al., 2019; Yan et al., 2019; Cohen, 2020). However, active replication of herpesviruses tends to be limited by the host immune system, leading to lifetime latent infection of the host. During latent infection, the viral genome remains in an inactive state in the host cell to avoid host immune surveillance, but the virus can reactivate and reenter the lytic cycle (Grinde, 2013; Retamal-Diaz et al., 2015; Suazo et al., 2015; Adler et al., 2017; Laing et al., 2018; Tognarelli et al., 2019; Gerada et al., 2020). Immune suppression or exposure to various environmental stimuli has been shown to activate latent herpesvirus to undergo lytic replication (Aneja and Yan, 2017; Forte et al., 2020). Latently infected herpesviruses are periodically activated, reentering the active replication to produce large numbers of infectious virions that can be transmitted to new hosts (Broussard and Damania, 2020). The balance between the lytic and latent phases is an important strategy for herpesvirus survival in vivo and results in lifelong survival of the virus and host-to-host the transmission from host to host (Aneja and Yan, 2017).
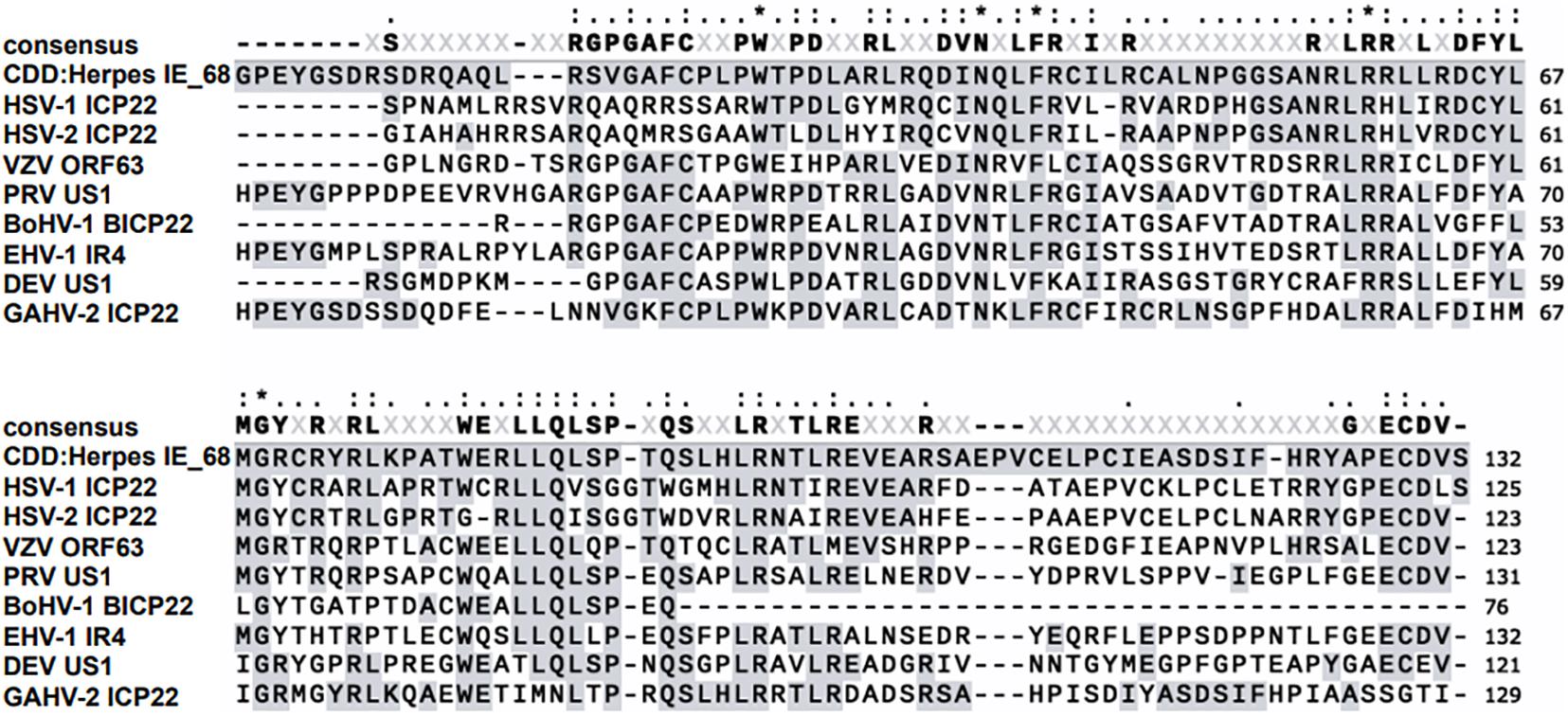
Human herpesviruses are the causative agents of many common diseases, including chickenpox, shingles, mononucleosis, cold sores, and genital herpes (Kimberlin and Rouse, 2004). Eight herpesviruses are known pathogens of humans: herpes simplex virus (HSV) 1 and 2 and varicella zoster virus (VZV) are α-herpesviruses; cytomegalovirus and human herpesvirus (HHV) 6 and 7 are β-herpesviruses, whereas Epstein–Barr virus and HHV8 are γ-herpesviruses infecting humans (Carter et al., 2007; Chayavichitsilp et al., 2009). ICP22 are homologs conserved in all α-herpesviruses; these proteins contain an IE-68 domain conserved in herpesviruses (shown in Figure 1) and are expressed from an immediate–early (IE) gene during the replication cycle of HSV-1/2 and VZV. They have been suggested to play various roles in the viral life cycle that are important for efficient viral replication, latency, and reactivation (summarized in Table 1). HSV-1 ICP22 can generally regulate viral and host gene transcription by changing the phosphorylation status of host RNA polymerase II (RNA pol II) (Bastian and Rice, 2009; Lin et al., 2010; Zaborowska et al., 2014; Fox et al., 2017) and can also facilitate the nuclear egress complex (NEC) accurately locate to the nuclear membrane to promote nuclear budding (Liu et al., 2015), whereas VZV ORF63regulates VZV transcription by destabilizing preinitiation complex (PIC) formation (Di Valentin et al., 2005). In addition, both ICP22 and ORF63 transcripts have been suggested to be important in latency (Kennedy et al., 2015; Depledge et al., 2018a; Matundan et al., 2019; Ouwendijk et al., 2020; Tormanen et al., 2020). By contrast, there are few reports of the participation of ICP22 in the lytic and latent infection of HSV-2. This protein is best known for its function as an E3 ubiquitin ligase involved in the protein ubiquitination pathway. This pathway is related to protein modification and plays a role in the inhibition of innate immunity by preventing interferon (IFN)–mediated signaling. Also, it induces the ubiquitination and degradation of host STAT1, STAT2, and IRF9, which results in the blockade of ISGF3 nuclear translocation (Zhang et al., 2015, 2020). Thus, considering that the main focus of this review is a discussion of the various functions and mechanisms of ICP22/ORF63 during the lytic and latent phases of human herpesvirus infection, we omit HSV-2 ICP22 and focus particularly on the HSV-1 ICP22 and VZV ORF63, which are involved in viral transcription, nuclear egress, and latency and participate in interplay with viral and cellular proteins. We believe that illustrating the functions and underlying mechanisms of ICP22/ORF63 during the human herpesvirus life cycle will help to elucidate the pathogenesis of the related diseases and contribute to the development of new antiviral drugs and vaccines.
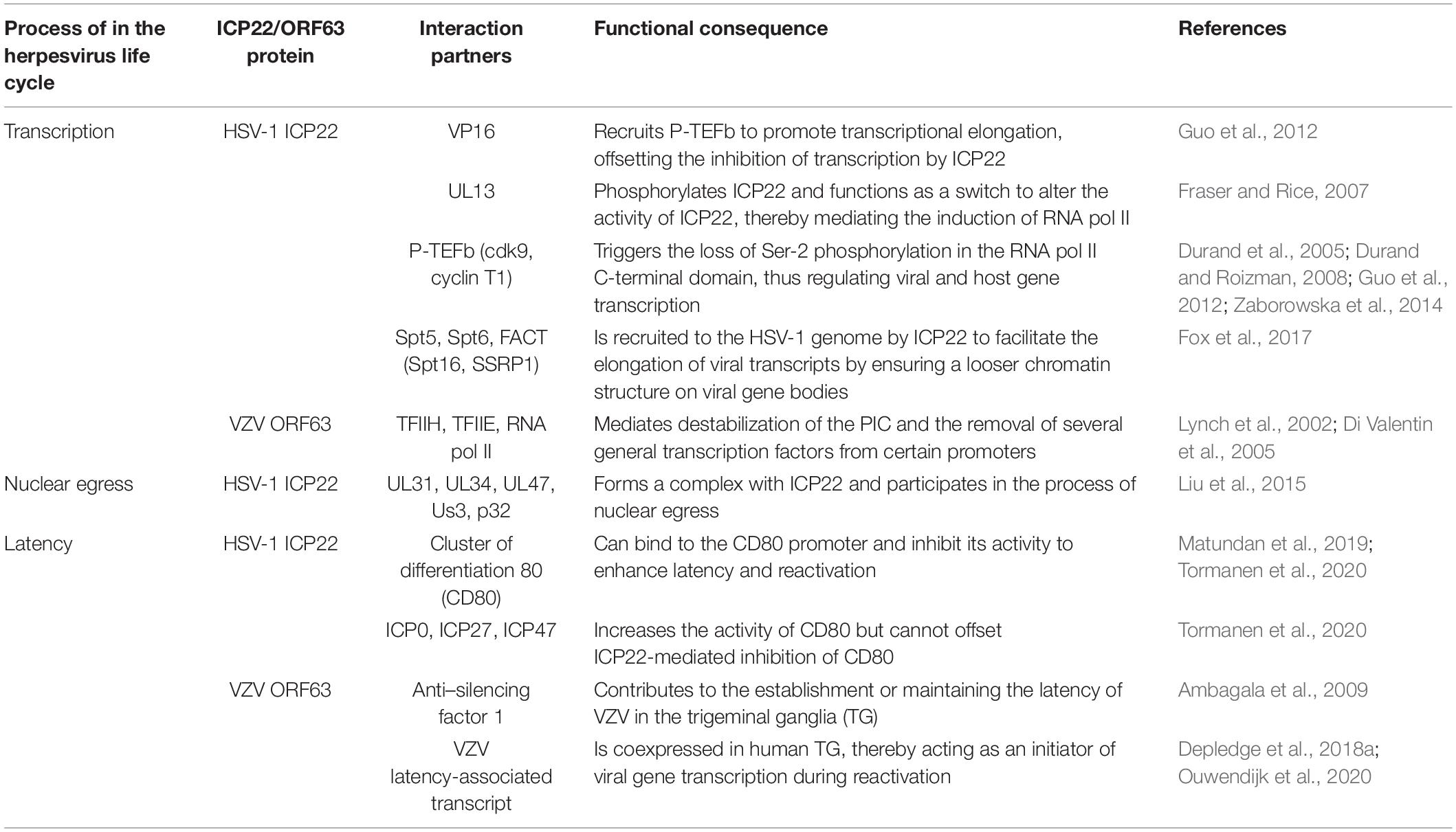
Table 1. Viral and cellular factors that interact with ICP22/ORF63 proteins during the human herpesvirus life cycle.
Roles of ICP22/ORF63 Proteins in Viral and Cellular Transcriptional Regulation
ICP22/ORF63 proteins first attracted attention in virology because of its function as transcriptional regulators of cellular and viral mRNAs. Viruses lack the basic machinery for replication and must hijack the relevant functions of host cells to complete the viral replication cycle in order to produce progeny virions. Like most nuclear-replicating DNA viruses, herpesviruses use the cellular enzyme RNA pol II for the transcription of viral genes (Gruffat et al., 2016). RNA pol II–mediated transcription consists of three steps: initiation, elongation, and termination (Hahn, 2004; Liu et al., 2013; Schier and Taatjes, 2020). During initiation, RNA pol II and general transcription factors (GTFs) bind to gene promoter regions and initiate the synthesis of nascent RNA (Krishnamurthy and Hampsey, 2009; Sainsbury et al., 2015; Haberle and Stark, 2018); during elongation, RNA pol II moves along the template strand to the gene body for extension of nascent RNA (Sims et al., 2004; Chen et al., 2018a); and during termination, upon completion of the processing of the nascent RNA transcript, RNA pol II and the mature RNA are released from the template DNA (Wahle and Keller, 1996; Richard and Manley, 2009; Porrua et al., 2016). Each of these steps requires the involvement of RNA pol II and its regulation by different proteins to ensure the correct transcription and expression of genes (Peck et al., 2019). As discussed below, ICP22/ORF63 are key proteins that participate in each step of RNA pol II–mediated viral gene transcription.
VZV ORF63 Destabilizes the PIC at Certain Promoters
Transcription initiation is the main step in the regulation of gene expression (Haberle and Stark, 2018). RNA pol II requires the proper assembly of at least eight cellular GTFs: TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, RNA pol II, and mediator, to assemble the PICs on core promoter elements (Hahn, 2004; Krishnamurthy and Hampsey, 2009; Sikorski and Buratowski, 2009; Hahn and Young, 2011; Spitz and Furlong, 2012; Nogales et al., 2017; Haberle and Stark, 2018). Once all components precisely assemble and bind, the PIC can “open” the promoter DNA to initiate transcription (Schier and Taatjes, 2020). Transient-transfection assays showed that VZV ORF63 acts as a transcriptional repressor of all genes with a TATA box sequence in the promoter. This protein was found to be immunoprecipitated with TFIIH and, to a lesser extent, TFIIE and RNA pol II, resulting in instability of the PIC and dissociation of GTFs from the promoter (Figure 2). The central and carboxy-terminal domains of IE63 (ORF63 encoded protein) are important for these effects. The transcriptional inhibitory ability of ORF63 has been confirmed in two different cell lines—a Vero cell line with lytic VZV infection and an ND7 cell line with maintained latent VZV infection—demonstrating that its inhibitory ability is independent of the cell type and infection pattern (Di Valentin et al., 2005).
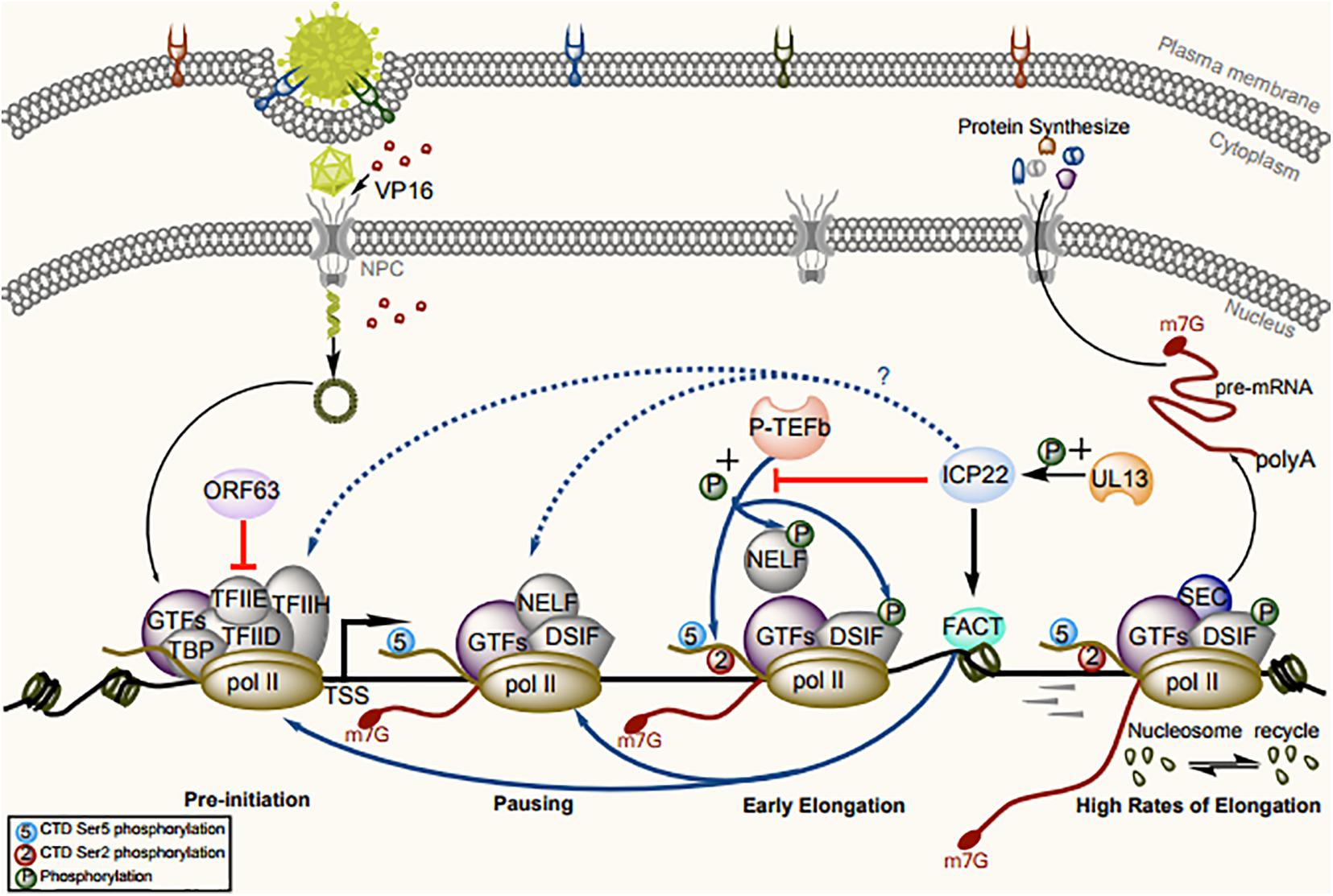
Figure 2. Model for the regulation of RNA pol II–mediated transcription by ICP22/ORF63 and cofactors. Assembly and initiation of the PIC are the first steps in transcription. RNA pol II and GTFs are recruited to the accessible chromatin architecture around the promoter. The CTD of RNA pol II is generally unmodified in the PIC. VZV ORF63 can sequester RNA pol II and TFIIE from promoters to disrupt PIC assembly. After synthesis, a 20- to 60-nt RNA transcript, the pause-inducing factors DSIF and NELF bind the early elongation complex, inhibiting further transcription. The GTF TFIIH complex contains the kinase CDK7, which phosphorylates the RNA pol II CTD at Ser-5, resulting in recruitment of the capping enzyme complex to add a 5′ cap to the nascent RNA. The release of paused RNA pol II is triggered after phosphorylation of RNA pol II CTD Ser-2 and pause-inducing factors by P-TEFb, to dissociate NELF and promote productive elongation. At this stage, ICP22 can trigger loss of phosphorylation of Ser-2 in the RNA pol II CTD by interacting with P-TEFb, and VP16 can recruit P-TEFb into the viral promoter region, thus offsetting ICP22-mediated inhibition of transcription. During elongation, ICP22 can interact with the FACT complex to promote the production of viral progeny by ensuring looser chromatin architecture on viral gene bodies.
HSV-1 ICP22 Generally Regulates Viral and Host Gene Transcription by Changing the Phosphorylation Status of Host RNA Pol II
The greatest difference between RNA pol II, RNA pol I, and RNA pol III is the conserved C-terminal domain (CTD) of Rpb1, which is the largest subunit of RNA pol II (Cramer et al., 2008). The CTD contains a conserved heptapeptide repeat, Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 (Chapman et al., 2008; Egloff and Murphy, 2008). The number of repeats varies across species (Chapman et al., 2008; Eick and Geyer, 2013). The CTD can be used as a platform to recruit transcription factors to RNA pol II, which plays an important role in gene transcriptional regulation (Komarnitsky et al., 2000; Buratowski, 2003; Egloff and Murphy, 2008; Zaborowska et al., 2016; Harlen and Churchman, 2017; Engel et al., 2018). Five amino acid residues in the heptapeptide repeat can be phosphorylated (Hsin et al., 2014), and the dynamic changes in the phosphorylation status during transcription are closely related to the recruitment of specific regulatory proteins (Gomes et al., 2006; Peck et al., 2019). The regulation of transcription elongation is strongly linked to the CTD phosphorylation status (Cadena and Dahmus, 1987; Payne et al., 1989; Harlen and Churchman, 2017). Phosphorylation occurs mainly at Ser-2 and Ser-5 during transcription (McCracken et al., 1997; Schroeder et al., 2000; Licatalosi et al., 2002; Kim et al., 2009; Harlen and Churchman, 2017; Nemec et al., 2019). Pol II pausing is induced when RNA pol II binds to negative elongation factor (NELF) and 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole sensitivity-inducing factor (DSIF), which is a heterodimer formed by SPT4 and SPT5 (Ping and Rana, 2001; Adelman and Lis, 2012; Natarajan et al., 2013; Jonkers and Lis, 2015; Core and Adelman, 2019; Aoi et al., 2020). RNA pol II recruits the positive transcription elongation factor B complex (Peterlin and Price, 2006; Ni et al., 2008; Zhou et al., 2012; Franco et al., 2018), in which cyclin-dependent kinase 9 (CDK9) phosphorylates NELF and DSIF to dissociate NELF from the transcriptional pausing complex and convert DSIF into a positive transcription elongation factor, promoting pausing release and elongation (Peterlin and Price, 2006; Paparidis et al., 2017).
HSV-1 ICP22 encodes an embedded protein, US1.5; which is initiated from methionine 147 of ICP22 and colinear with the remaining portion of that protein, and the translation of each protein is driven by independent promoters (Carter and Roizman, 1996). US1 encodes two sets of functions, one in the amino terminus unique to ICP22 and one shared by ICP22 and US1.5 (Ogle and Roizman, 1999), and these functions are required for efficient HSV-1 replication in most cell types in vitro and in vivo (Orlando et al., 2006). Either protein can functionally enhance the expression of late (L) viral proteins in cell culture and inhibit ICP0-mediated gene expression in transient-transfection assays. However, researchers have not defined a unique function for US1.5 or demonstrated a defect in HSV-1 replication in its absence, which suggests that the viral requirement for US1.5 is conditional and is enhanced only under certain circumstances during the HSV-1 life cycle, further suggesting that US1.5 can function as a biological backup for ICP22 should ribosome read-through be blocked at the translation initiation codon of ICP22 (Bastian and Rice, 2008). Follow-up studies showed that residues 193–256 of ICP22 can interact with CDK9 without affecting its recruitment and inhibit the phosphorylation of Ser-2 in the RNA pol II CTD (Durand et al., 2005; Justyna et al., 2014) but do not directly interact with RNA pol II (Rice et al., 1995; Fraser and Rice, 2005; Guo et al., 2012; Mostafa and Davido, 2013; Rice and Davido, 2013; Figure 2). Expression of the HSV-1 ICP22-related protein US1.5 also triggers loss of Ser-2 phosphorylation in RNA pol II in transfected cells (Fraser and Rice, 2007). Ser-2 phosphorylation induced by CDK9 is necessary for HSV-1 transcription and replication. When CDK9 activity is inhibited, the number of progeny and the expression level of late genes decrease, and these events adversely affect the formation of the viral replication compartment (Durand and Roizman, 2008). In addition, the functions of ICP22 as a general transcriptional regulator can also be regulated by other viral proteins. Previous studies have shown that ICP22-mediated loss of Ser-2 phosphorylation in RNA pol II does not depend on the HSV-1 protein kinase UL13, which can directly or indirectly affect ICP22 and US1.5 phosphorylation (Purves and Roizman, 1992; Asai et al., 2007; Fraser and Rice, 2007). However, in some experiments, the loss of phosphorylation of RNA pol II Ser-2 in the UL13-deficient strain was accelerated compared to that in the wild-type strain, suggesting that the newly expressed UL13 may alter the activity of ICP22, thereby affecting its ability to mediate the phosphorylation of Ser-2 in RNA pol II (Long et al., 1999; Fraser and Rice, 2007). Moreover, when the recruitment of P-TEFb is inhibited by ICP22, the viral transactivation protein VP16 can recruit P-TEFb to promoter region, thus offsetting ICP22-mediated inhibition of transcription (Guo et al., 2012). As P-TEFb is isolated mostly in the 7SK small nuclear ribonucleoprotein (snRNP) complex in an inactive state, it is presumed that VP16 can cause the dissociation of P-TEFb from the 7SK snRNP and functionally activate P-TEFb (Li et al., 2017). However, researchers did not observe loss of RNA pol II Ser-2 phosphorylation in VZV ORF63, the homolog of ICP22 in HSV-1, suggesting that this function is not conserved between HSV-1 and VZV (Fraser and Rice, 2007).
HSV-1 ICP22 Interacts With FACT to Influence Transcription-Coupled Histone Modification
Facilitates chromatin transcription (FACT), which can the ability to destabilize nucleosomes, has long been considered as a transcriptional elongation factor that promotes RNA pol II progression on chromatin templates (LeRoy et al., 1998; Mason and Struhl, 2003; Tettey et al., 2019; Formosa and Winston, 2020). The FACT complex is a heterodimer consisting of two distinct subunits Spt16 and SSRP1 (Orphanides et al., 1999; Formosa, 2012; Zhou et al., 2020) and participates in the assembly and disassembly of nucleosomes encountered by the transcribing polymerase (Belotserkovskaya et al., 2003; Liu et al., 2020). DNA replication requires rapid assembly of large numbers of nucleosomes to form chromatin (Reinberg and Sims, 2006; Formosa, 2012). FACT may be associated with the establishment and maintenance of loose chromatin structure in HSV-1 genomic DNA during replication (Placek and Berger, 2010). Recent studies have suggested that FACT plays a broad role in maintaining chromatin structure and RNA pol II pausing at promoter-proximal pause sites (Tettey et al., 2019). In addition, multiple lines of evidence suggest that FACT also plays a role in transcription initiation. Overexpression subunits of FACT or deficiency of its activity lead to inadequate disinhibition of a transposon-associated promoter in yeast (Clark-Adams et al., 1988; Malone et al., 1991), whereas mutation of FACT results in increased gene transcription of some genes (Pelechano et al., 2009; Feng et al., 2016; Pathak et al., 2018), suggesting a global role of FACT in blocking improper transcription initiation by maintaining chromatin in a particular form (Formosa and Winston, 2020). In accord with previous results (Pathak et al., 2018), depletion of FACT greatly reduces PIC assembly and transcription in vivo (Petrenko et al., 2019). FACT may play an important role in the maintenance or crossing of chromatin barriers outside transcription units prior to the initiation or extension stages of transcription (Belotserkovskaya et al., 2003; Schwabish and Struhl, 2004; Hsieh et al., 2013; Chen et al., 2018b; Formosa and Winston, 2020). After the promoter escapes, RNA pol II must overcome the disruption of transcription induced by nucleosomes, whereas FACT can reshape nucleosomes and reduce this disruption of transcription. In the presence of FACT, the paused state of RNA pol II can be maintained (Bondarenkoa et al., 2015). When FACT is depleted, RNA pol II returns to its elongation activity (Tettey et al., 2019). During HSV-1 transcription, ICP22 interacts with the FACT complex, resulting in altered localization of FACT in the nucleus and the recruitment of two other transcriptional elongation factors encoded in the viral genome (Spt5 and Spt6), thus maintaining a looser chromatin structure in the viral genome and promoting the production of progeny virions (Fox et al., 2017). Whether ICP22 inhibits CDK9 activity or recruits FACT to the transcriptional pause site, we can conclude that ICP22 plays a role during the pausing of RNA pol II, but the effects of the two modes of action are different. The former mode of transcriptional pausing enables FACT to help RNA pol II overcome obstacles and transition transcription from pausing to elongation (Figure 2). Although the mechanism is unclear, the interaction between ICP22 and FACT may be highly important for inhibition or activation at specific stages of the viral life cycle, and this possibility should be the focus of further study.
ICP22 Promotes Primary Envelopment by Interacting With the NEC
During herpesvirus infections, DNA replication and nucleocapsid formation occur in the nucleus of the host cell, whereas further maturation of virions occurs in the cytoplasm. Therefore, the viral capsid must pass through the nuclear envelope to enter the final maturation compartment (Johnson and Baines, 2011; Mettenleiter et al., 2013a; Lv et al., 2019). Transit of the viral nucleocapsid across the nuclear membrane is the first step in early virion maturation. Therefore, an important mechanism by which the viral nucleocapsid traverses the nuclear membrane is the destruction of the nuclear membrane structure (Johnson and Baines, 2011). Nuclear budding of the herpesvirus capsid through the nuclear membrane is mediated by the NEC (Bigalke and Heldwein, 2017). In HSV-1, the NEC is a heterodimer composed of the a type II membrane protein UL34 (Ott et al., 2011) and UL31, and the absence of UL34 or UL31 abolishes nuclear budding and affects the generation of infectious progeny (Chang et al., 1997; Roller et al., 2000; Matundan and Ghiasi, 2018; Takeshima et al., 2019). UL34 is localized mainly on the inner nuclear membrane (INM), outer nuclear membrane (ONM), and endoplasmic reticulum (ER) (Ott et al., 2011; Schuster et al., 2012), whereas UL31 is a nucleophosphoprotein that contains nuclear localization signals and is not membrane anchored (Chang and Roizman, 1993; Arii et al., 2019). When UL31 and UL34 are coexpressed, UL31 can be relocalized to the surface of the INM through the interaction with UL34, and the N-terminal domain of pUL31 was required to prevent their premature interaction in the cytoplasm. Then, the virion and cellular kinases are relocalized, causing some proteins to exert phosphorylation activity and locally degrade the nuclear membrane, after which the capsid in the nucleus can reach the budding site on the INM (Granzow et al., 2001; Mettenleiter, 2004; Roller et al., 2010; Mettenleiter et al., 2013b; Funk et al., 2015; Lorenz et al., 2015a; Yadav et al., 2017). In the absence of assistance from other viral proteins, UL31 and UL34 can guide the formation of vesicles from the INM and promote nuclear budding in vivo (Klupp et al., 2007; Desai et al., 2012; Hagen et al., 2015); further in vitro analysis suggested that the recombinant HSV-1 pUL31 and pUL34 can drive membrane budding and scission of vesicles (Bigalke et al., 2014; Lorenz et al., 2015b). ICP22 has been reported to form a complex with UL31, UL34, UL47, and US3 after HSV-1 infection, resulting in colocalization of ICP22, UL31, and UL34 on the nuclear membrane in infected cells (Mou et al., 2009; Liu et al., 2014; Maruzuru et al., 2014). These proteins have a non-negligible role in the nuclear egress of virions. Therefore, the primary envelopment efficiency of HSV-1 caused by ICP22 deletion or mutation may be altered through its interaction with and regulation of UL31 and UL34. During this process, UL31 guides the recruitment and anchoring of ICP22 to the nuclear membrane, and ICP22 promotes the correct localization of UL31 and UL34 at the nuclear membrane. Indeed, deletion or mutation of ICP22 caused mislocalization of UL31 and UL34 in the ER (Liu et al., 2015). However, although enveloped virions were still detected in the cytoplasm and on the cell surface after ICP22 deletion, the number of enveloped viruses in the perinuclear region was significantly reduced (Figure 3). These results suggest that ICP22 plays a regulatory role in but is not essential for the primary envelopment of HSV-1 virions.
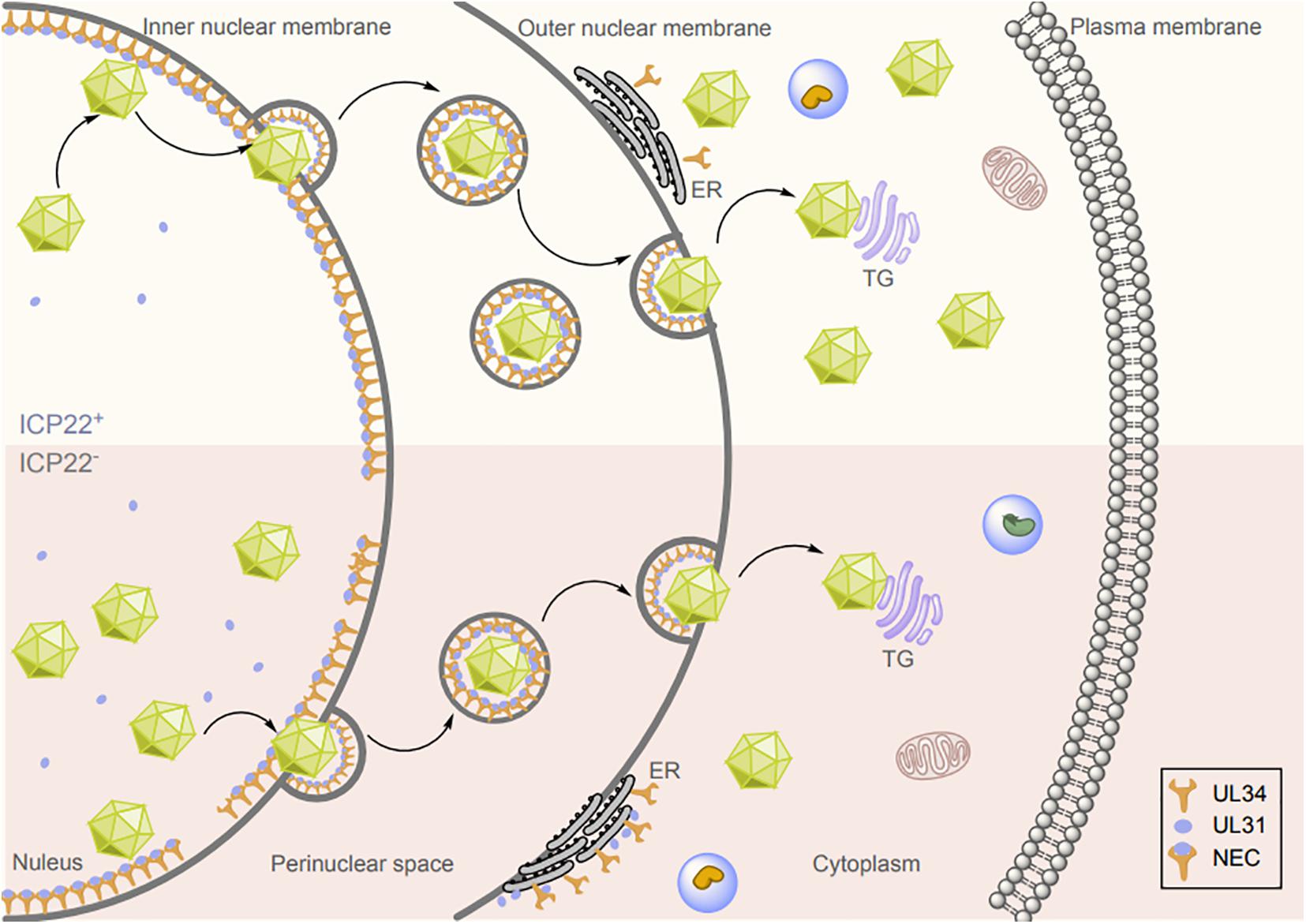
Figure 3. Effects of ICP22 on herpesvirus nuclear budding. Herpesvirus nucleocapsid assembly is completed in the nucleus, and the nucleocapsid is then translocated by budding from the INM to the perinuclear region via the NEC, where it fuses with the ONM to complete the budding and release process. ICP22 promotes HSV-1 nuclear budding by interacting with and regulating UL31 and UL34.
Roles of ICP22/ORF63 in Viral Latency and Reactivation
The balance between the life cycles of lytic and latent herpesvirus infection is a complex and finely tuned process that allows herpesvirus to persist and spread throughout the life of the host (Broussard and Damania, 2020). Lytic infection is critical for host-to-host transmission and pathogenicity of herpesvirus. During latency, the virus enters a state of limited gene expression and does not replicate. In the trigeminal ganglia (TG) of humans, HSV-1 (Yan et al., 2020) and VZV (Depledge et al., 2018a) genomic DNA is not replicated, but exists as continuous (circular) episomes or genome concatemers in latently infected cells, and only limited viral gene transcription occurs to maintain latent infection and evade host immune surveillance (Efstathiou et al., 1986; Clarke et al., 1995; Gary et al., 2006; Muylaert et al., 2011; Nicoll et al., 2012; Depledge et al., 2018a,b). The expression of genes during latency varies considerably among herpesviruses. Latency-associated transcripts (LATs) map antisense to ICP0/RL2 within the long repeat (RL) (Stevens and Cook, 1971; Spivack and Fraser, 1987; Fareed and Spivack, 1994; Wu et al., 1996; Nicoll et al., 2012) and are the only viral transcripts generally detected during HSV-1 latency (Kennedy et al., 2015). However, VZV latency in the TG of humans is characterized by expression of VZV latency-associated transcript (VLT) and the ORF63 transcript (Depledge et al., 2018a). VLT encodes the antisense transcript of ORF61 and is considered a homolog of the LATs encoded by all other well-studied neurotropic α-herpesviruses (Depledge et al., 2018b; Baird et al., 2019). However, VZV appears to be the only α-herpesvirus that has been analyzed in detail and consistently expresses an additional latency transcript, ORF63, in the TG of humans (Depledge et al., 2018b).
ORF63 Plays an Important Role in the Establishment of VZV Latency
Before the discovery of VLT, ORF63 was the first VZV gene shown to be critical for the establishment of latency (Cohrs et al., 1996; Kennedy et al., 1999; Kennedy et al., 2000; Cohen et al., 2004). VZV with ORF63 deletion exhibits impaired replication in melanoma cells and fibroblasts and impaired latency in rodents, but its ability to infect ganglia of the rodents is not impaired (Sadzot-Delvaux et al., 1995; Kennedy et al., 2001; Sommer et al., 2001). During lytic infection of VZV, IE-63 (the protein encoded by ORF63) is detected mainly in the cytoplasm of latently infected sensory neurons (Mahalingam et al., 1996; Lungu et al., 1998), but is expressed mainly in the nucleus during lytic infection in vitro (Debrus et al., 1995; Stevenson et al., 1996; Ambagala et al., 2009). The genome copy numbers and numbers of cotton rats that could establish latent VZV infection were lower for infection with an ORF63 gene deletion strain than for infection with the parental strain (Cohen et al., 2004). Similar studies showed that when animals were inoculated with the wild-type, ORF63 mutant, and revertant strains, the copy numbers in dorsal root ganglia and the frequency of latent infection were the lowest in ORF63 mutant-infected animals. Thus, ORF63 is the key to establishing latent infection but is not necessary for VZV entry into the ganglia (Cohen et al., 2004). Further identification of ORF63 domains revealed that the replacement of five serine or threonine phosphorylation sites in the last 108 amino acids of ORF63 with alanines resulted in impaired viral replication and latency establishment in vitro. This finding suggests that a region of ORF63 important for VZV replication in vitro is required for efficient establishment of latency (Cohen et al., 2005).
In addition, IE63 was found to be able to lock part of anti–silencing factor 1 (ASF1) into the cytoplasm in a model of latent VZV infection of enteric neurons in guinea pigs (Ambagala et al., 2009). ASF1, a member of the H3/H4 family of histone chaperones, is a nucleosome assembly factor that participates in a variety of cellular functions, including DNA replication, gene transcription, and the cellular response to DNA damage by histone removal from and deposition onto DNA (Donham et al., 2011; Zhang et al., 2018, 2019; Cote et al., 2019). In VZV-infected and IE-63–transfected cells, ASF1 colocalized and immunoprecipitated with IE-63. In addition, colocalization of IE-63 and ASF1 was observed in intestinal neurons with lytic and latent VZV infection (Ambagala et al., 2009). Infection with site mutants (such as ROKA63-ACCI and ROKA63-5M) impaired the interaction of IE-63 and ASF1, and infection with a mutant with complete deletion of the IE-63 gene (ROKA63D) led to impaired establishment of VZV latency in rodents (Cohen et al., 2004, 2005). These results suggest that the interaction between IE-63 and ASF1 may contribute to the establishment or maintenance of latent VZV infection.
ICP22 Enhances Latency and Reactivation
HSV-1 evades clearance by the host immune system through a variety of mechanisms to establish lifelong latent infection (Kurt-Jones et al., 2017; Lin and Zheng, 2019; Tognarelli et al., 2019; Zhu and Zheng, 2020). For example, HSV-1 LAT plays a vital role in generating dysfunctional T-cell responses in the TG of ocular infected HSV-1 mice and helps the virus resist host cell apoptosis by inhibiting the type I IFN signaling pathway, thus contributing to the establishment and activation of latent HSV-1 infection (Perng et al., 2000; Branco and Fraser, 2005; Allen et al., 2011; Tormanen et al., 2019; Jaggi et al., 2020). LAT-containing neurons are occasionally surrounded by CD8+ T cells (Theil et al., 2003; Verjans et al., 2007) that are primed in the periphery (Held et al., 2011) and LAT functions in part to protect neurons against granzyme B–induced apoptosis (Allen et al., 2011; Jiang et al., 2011). A recent study reported the novel finding that ICP22 can downregulate host costimulatory molecular cluster of differentiation 80 (CD80) after ocular HSV-1 infection to promote host immune evasion of HSV-1 (Matundan et al., 2019). CD80, as a stimulator of T cells, plays a key role in the activation and proliferation of T cells and can be expressed after ocular infection, thus triggering the immune response and causing corneal scarring (Lanier et al., 1995; Bugeon et al., 2006; Jaggi et al., 2020; Tormanen et al., 2020). Compared with wild-type virus-infected cells, infection of 293 cells with ICP22-deficient viral strain resulted in increased CD80 promoter activity, decreased virus replication and latency, and delayed reactivation from latency (Matundan and Ghiasi, 2019). In vivo experiments showed that the level of the host immune response decreased after ICP22-mediated inhibition of CD80. Mutation of 34 or 116 amino acids in the ICP22 sequence affected the replication and pathogenicity of the virus but did not release the binding of ICP22 to CD80, and the immune response was still suppressed in mice with corneal infection (Matundan et al., 2019). Relevant studies showed that ICP0, ICP27, and ICP47 can increase the activity of the CD80 promoter, but none can offset ICP22-mediated inhibition of CD80 (Lv et al., 2018). A transient reduction in the dendritic cell population through downregulation of CD80 by ICP22 could explain the enhanced latency and reactivation (Tormanen et al., 2020).
VLT-ORF63 Fusion Transcripts Are Potentially Involved in the Transition From Latency to Lytic VZV Infection
Using a highly sensitive enriched RNA sequencing (RNA-Seq) enrichment method, researchers found no evidence for the expression of VZV mRNA other than VLT and ORF63 during latent infection (Depledge et al., 2018a). VLT was defined as a novel 496-nucleotide multiexon transcript transcribed antisense to ORF61 via ultradeep virus-enriched RNA-Seq of latently infected human TG (Depledge et al., 2018a). Further reverse transcriptase–polymerase chain reaction (PCR) and quantitative PCR analyses showed that the latent VZV also transcribes the lytic ORF63 gene at lower levels relative to VLT in a subset of latently infected TG, independent of the latent viral DNA load (Depledge et al., 2018a). Although the expression levels were different, the expression levels of VLT and ORF63 transcripts were significantly correlated, suggesting that they coregulate expression during latent VZV infection (Ouwendijk et al., 2012; Depledge et al., 2018a). This apparent expression of two distinct viral transcripts during latency is unique among well-studied α-herpesviruses, suggesting that these two transcripts and/or their encoded proteins play an important role in the latency and reactivation of VZV (Ouwendijk et al., 2020). In the human induced pluripotent stem cell–derived sensory neuron (HSN) latency model in vitro, the results of coexpression of VLT and two VLT-ORF63 fusion products, VLT63-1 and VLT63-2, showed that the encoded pVLT-ORF63 fusion protein can act as an initiator of VZV reactivation in infected human TG, providing new insights into the mechanism by which VZV establishes latency and reactivation (Ouwendijk et al., 2020). Moreover, ectopic VLT63-1 induces lytic transcription of viral genes in HSN models of latent VZV infection, suggesting that the pVLT-ORF63 fusion protein, not the corresponding transcript, induces transcriptional activation of VZV genes. However, pORF63 did not induce the transcription of any IE, early or late gene, except for the IE promiscuous transactivator pORF61 and the early (E) transcript RNA 16-1 (Moriuchi et al., 1993). These results indicated that gene transcription during VZV reactivation is initiated through VLT-ORF63 transcription/pVLT-ORF63 translation (Ouwendijk et al., 2020). However, whether VLT and ORF63 RNA transcripts are produced by the same or distinct populations of neurons remains unclear, as human TG are composed of dissimilar subtypes of neurons, and how these transcripts may affect ability of VZV to reactivate from latent in neurons is unknown (Depledge et al., 2018b). Thus, more studies such as in situ analyses are required to identify the types of neurons in which coexpression of VLT and ORF63 RNA and the latent VZV genome are present. By contrast, no initiator that drives viral gene expression during reactivation has been reported in HSV-1 (Wilson and Mohr, 2012), suggesting that the mechanisms controlling latent infection and reactivation may differ between HSV and VZV.
Summary
Herpesviruses are extremely successful parasites that have evolved over millions of years to develop a variety of mechanisms to coexist with their hosts and maintain host-to-host transmission and lifelong infection by regulating their life cycles (Adler et al., 2017; Renner and Szpara, 2018). Inhibition of viral replication treats lytic herpesvirus infections but does not cure latent infection (Skoreński and Sieńczyk, 2014; Gershon et al., 2015; van Diemen and Lebbink, 2017; Andrei et al., 2019). An in-depth understanding of the molecular mechanism by which viral proteins regulate the life cycle will offer new insights into the treatment of herpesvirus infections via the destruction of key proteins in the herpesvirus life cycle, thus providing new strategies for the treatment of herpesvirus infections and the resulting diseases (Broussard and Damania, 2020). In this article, we summarized the recent findings on the involvement of the ICP22/ORF63 proteins in the viral life cycle. The broad interplay of HSV-1 ICP22 or VZV ORF63 with cellular and viral proteins contributes to various processes in the herpesvirus life cycle, including transcription, nuclear egress, and establishment of latency and reactivation, which are critical to herpesvirus survival. Any drug that interferes with these steps can disrupt the viral life cycle as a therapeutic strategy for herpesvirus infections. Further research into the processes and underlying mechanisms of the viral life cycle will help to achieve this goal. However, there are very few reports on ICP22/ORF63 homologs in animal herpesviruses, such as duck enteritis virus, Marek disease virus, equine herpesvirus 1, bovine herpesvirus 1, and pseudorabies virus, and researchers must devote more attention to these viruses in the future. The multifaceted roles and complex mechanisms by which of ICP22/ORF63 proteins regulate the life cycle of herpesviruses offer both a foundation and a challenge for our understanding of the mechanisms and for the development of various potential treatment options. As we learn more about these proteins, we can identify the protocols with the greatest potential for herpesvirus treatment and vaccine development.
Author Contributions
MW, RJ, SC, QiaoY, DZ, ML, XZ, SZ, JH, XO, SM, QG, DS, and BT provided ideas contributing to the structure of this manuscript. AC modified the manuscript. All authors listed contributed to the completion of the manuscript, reviewed and approved the final manuscript. YW and QiaoY contributed to the design and writing of the article.
Funding
This research was supported by the National Key Research and Development Program of China (2017YFD0500800) and the China Agricultural Research System (CARS-42-17).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adelman, K., and Lis, J. T. (2012). Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731. doi: 10.1038/nrg3293
Adler, B., Sattler, C., and Adler, H. (2017). Herpesviruses and their host cells: a successful liaison. Trends Microbiol. 25, 229–241. doi: 10.1016/j.tim.2016.11.009
Allen, S. J., Hamrah, P., Gate, D., Mott, K. R., Mantopoulos, D., Zheng, L., et al. (2011). The role of LAT in increased CD8+ T cell exhaustion in trigeminal ganglia of mice latently infected with herpes simplex virus 1. J. Virol. 85, 4184–4197. doi: 10.1128/jvi.02290-10
Ambagala, A. P., Bosma, T., Ali, M. A., Poustovoitov, M., Chen, J. J., Gershon, M. D., et al. (2009). Varicella-zoster virus immediate-early 63 protein interacts with human antisilencing function 1 protein and alters its ability to bind histones h3.1 and h3.3. J. Virol. 83, 200–209. doi: 10.1128/jvi.00645-08
Andrei, G., Trompet, E., and Snoeck, R. (2019). Novel therapeutics for epstein-barr virus. Molecules 24:997. doi: 10.3390/molecules24050997
Aneja, K. K., and Yan, Y. (2017). Reactivation and lytic replication of kaposi’s sarcoma-associated herpesvirus: an update. Front. Microbiol. 8:613. doi: 10.3389/fmicb.2017.00613
Aoi, Y., Smith, E. R., Shah, A. P., Rendleman, E. J., Marshall, S. A., Woodfin, A. R., et al. (2020). NELF regulates a promoter-proximal step distinct from RNA Pol II pause-release. Mol. Cell 78, 261–274.e5.
Arii, J., Takeshima, K., Maruzuru, Y., Koyanagi, N., Kato, A., and Kawaguchi, Y. (2019). Roles of the interhexamer contact site for hexagonal lattice formation of the herpes simplex virus 1 nuclear egress complex in viral primary envelopment and replication. J. Virol. 93:e498–19.
Asai, R., Ohno, T., Kato, A., and Kawaguchi, Y. (2007). Identification of proteins directly phosphorylated by UL13 protein kinase from herpes simplex virus 1. Microbes Infect. 9, 1434–1438. doi: 10.1016/j.micinf.2007.07.008
Azab, W., Dayaram, A., Greenwood, A. D., and Osterrieder, N. (2018). How host specific are herpesviruses? lessons from herpesviruses infecting wild and endangered mammals. Annu. Rev. Virol. 5, 53–68. doi: 10.1146/annurev-virology-092917-043227
Baird, N. L., Zhu, S., Pearce, C. M., and Viejo-Borbolla, A. (2019). Current In Vitro models to study varicella zoster virus latency and reactivation. Viruses 11:103. doi: 10.3390/v11020103
Bastian, T. W., and Rice, S. A. (2008). Identification of sequences in herpes simplex virus type 1 ICP22 that influence RNA polymerase ii modification and viral late gene expression. J. Virol. 83, 128–139. doi: 10.1128/jvi.01954-08
Bastian, T. W., and Rice, S. A. (2009). Identification of sequences in herpes simplex virus type 1 ICP22 that influence RNA polymerase II modification and viral late gene expression. J. Virol. 83, 128–139.
Belotserkovskaya, R., Oh, S., Bondarenko, V. A., Orphanides, G., Studitsky, V. M., and Reinberg, D. (2003). FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093. doi: 10.1126/science.1085703
Bigalke, J. M., and Heldwein, E. E. (2017). Have NEC Coat, will travel: structural basis of membrane budding during nuclear egress in herpesviruses. Adv. Virus Res. 97, 107–141. doi: 10.1016/bs.aivir.2016.07.002
Bigalke, J. M., Heuser, T., Nicastro, D., and Heldwein, E. E. (2014). Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat. Commun. 5:4131.
Bondarenkoa, M. T., Maluchenkob, N. V., and Valievab, M. E. (2015). Structure and function of histone chaperone FACT. Mol. Biol. 49, 796–809.
Branco, F. J., and Fraser, N. W. (2005). Herpes simplex virus type 1 latency-associated transcript expression protects trigeminal ganglion neurons from apoptosis. J. Virol. 79, 9019–9025. doi: 10.1128/jvi.79.14.9019-9025.2005
Broussard, G., and Damania, B. (2020). Regulation of KSHV latency and lytic reactivation. Viruses 12:1034. doi: 10.3390/v12091034
Bugeon, L., Wong, K. K., Rankin, A. M., Hargreaves, R. E., and Dallman, M. J. (2006). A negative regulatory role in mouse cardiac transplantation for a splice variant of CD80. Transplantation 82, 1334–1341. doi: 10.1097/01.tp.0000239343.01775.54
Cadena, D. L., and Dahmus, M. E. (1987). Messenger RNA synthesis in mammalian cells is catalyzed by the phosphorylated form of RNA polymerase II. J. Biol. Chem. 262, 12468–12474. doi: 10.1016/s0021-9258(18)45229-5
Carter, J., Saunders, V., and Saunders, V. A. (2007). Virology:Principles and Applications. Hoboken, NJ: John Wiley & Sons.
Carter, K. L., and Roizman, B. (1996). The promoter and transcriptional unit of a novel herpes simplex virus 1 alpha gene are contained in, and encode a protein in frame with, the open reading frame of the alpha 22 gene. J. Virol. 70, 172–178. doi: 10.1128/jvi.70.1.172-178.1996
Chang, Y. E., and Roizman, B. (1993). The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J. Virol. 67, 6348–6356. doi: 10.1128/jvi.67.11.6348-6356.1993
Chang, Y. E., Van Sant, C., Krug, P. W., Seaherpes, A. E., and Roizman, B. (1997). The null mutant of the U(L)31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71, 8307–8315. doi: 10.1128/jvi.71.11.8307-8315.1997
Chapman, R. D., Heidemann, M., Hintermair, C., and Eick, D. (2008). Molecular evolution of the RNA polymerase II CTD. Trends Genet. 24, 289–296.
Chayavichitsilp, P., Buckwalter, J. V., Krakowski, A. C., and Friedlander, S. F. (2009). Herpes simplex. Pediatr. Rev. 30, 119–129.
Chen, F. X., Smith, E. R., and Shilatifard, A. (2018a). Born to run: control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 19, 464–478. doi: 10.1038/s41580-018-0010-5
Chen, P., Dong, L., Hu, M., Wang, Y. Z., Xiao, X., Zhao, Z., et al. (2018b). Functions of FACT in breaking the nucleosome and maintaining its integrity at the single-nucleosome level. Mol. Cell 71:e284.
Clark-Adams, C. D., Norris, D., Osley, M. A., Fassler, J. S., and Winston, F. (1988). Changes in histone gene dosage alter transcription in yeast. Genes Dev. 2, 150–159. doi: 10.1101/gad.2.2.150
Clarke, P., Beer, T., Cohrs, R., and Gilden, D. H. (1995). Configuration of latent varicella-zoster virus DNA. J. Virol. 69, 8151–8154. doi: 10.1128/jvi.69.12.8151-8154.1995
Cohen, J. I., Cox, E., Pesnicak, L., Srinivas, S., and Krogmann, T. (2004). The varicella-zoster virus open reading frame 63 latency-associated protein is critical for establishment of latency. J. Virol. 78, 11833–11840. doi: 10.1128/jvi.78.21.11833-11840.2004
Cohen, J. I., Krogmann, T., Bontems, S., Sadzot-Delvaux, C., and Pesnicak, L. (2005). Regions of the varicella-zoster virus open reading frame 63 latency-associated protein important for replication in vitro are also critical for efficient establishment of latency. J. Virol. 79, 5069–5077. doi: 10.1128/jvi.79.8.5069-5077.2005
Cohrs, R. J., Barbour, M., and Gilden, D. H. (1996). Varicella-zoster virus (VZV) transcription during latency in human ganglia: detection of transcripts mapping to genes 21, 29, 62, and 63 in a cDNA library enriched for VZV RNA. J. Virol. 70, 2789–2796. doi: 10.1128/jvi.70.5.2789-2796.1996
Core, L., and Adelman, K. (2019). Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 33, 960–982. doi: 10.1101/gad.325142.119
Cote, J. M., Kuo, Y. M., Henry, R. A., Scherman, H., Krzizike, D. D., and Andrews, A. J. (2019). Two factor authentication: Asf1 mediates crosstalk between H3 K14 and K56 acetylation. Nucleic Acids Res. 47, 7380–7391. doi: 10.1093/nar/gkz508
Cramer, P., Armache, K. J., Baumli, S., Benkert, S., Brueckner, F., Buchen, C., et al. (2008). Structure of eukaryotic RNA polymerases. Annu. Rev. Biophys. 37, 337–352.
Davison, A. J., Eberle, R., Ehlers, B., Hayward, G. S., Mcgeoch, D. J., Minson, A. C., et al. (2009). The order herpesvirales. Arch. Virol. 154, 171–177. doi: 10.1007/s00705-008-0278-4
Debrus, S., Sadzot-Delvaux, C., Nikkels, A. F., Piette, J., and Rentier, B. (1995). Varicella-zoster virus gene 63 encodes an immediate-early protein that is abundantly expressed during latency. J. Virol. 69, 3240–3245. doi: 10.1128/jvi.69.5.3240-3245.1995
Depledge, D. P., Ouwendijk, W. J. D., Sadaoka, T., Braspenning, S. E., Mori, Y., Cohrs, R. J., et al. (2018a). A spliced latency-associated VZV transcript maps antisense to the viral transactivator gene 61. Nat. Commun. 9:1167.
Depledge, D. P., Sadaoka, T., and Ouwendijk, W. J. D. (2018b). Molecular aspects of varicella-zoster virus latency. Viruses 10:349. doi: 10.3390/v10070349
Desai, P. J., Pryce, E. N., Henson, B. W., Luitweiler, E. M., and Cothran, J. (2012). Reconstitution of the Kaposi’s sarcoma-associated herpesvirus nuclear egress complex and formation of nuclear membrane vesicles by coexpression of ORF67 and ORF69 gene products. J. Virol. 86, 594–598. doi: 10.1128/jvi.05988-11
Di Valentin, E., Bontems, S., Habran, L., Jolois, O., Markine-Goriaynoff, N., Vanderplasschen, A., et al. (2005). Varicella-zoster virus IE63 protein represses the basal transcription machinery by disorganizing the pre-initiation complex. Biol. Chem. 386, 255–267. doi: 10.1515/bc.2005.031
Donham, D. C. II, Scorgie, J. K., and Churchill, M. E. (2011). The activity of the histone chaperone yeast Asf1 in the assembly and disassembly of histone H3/H4-DNA complexes. Nucleic Acids Res. 39, 5449–5458. doi: 10.1093/nar/gkr097
Dupont, L., and Reeves, M. B. (2016). Cytomegalovirus latency and reactivation: recent insights into an age old problem. Rev. Med. Virol. 26, 75–89. doi: 10.1002/rmv.1862
Durand, L. O., Advani, S. J., Poon, A. P., and Roizman, B. (2005). The carboxyl-terminal domain of RNA polymerase II is phosphorylated by a complex containing cdk9 and infected-cell protein 22 of herpes simplex virus 1. J. Virol. 79, 6757–6762. doi: 10.1128/jvi.79.11.6757-6762.2005
Durand, L. O., and Roizman, B. (2008). Role of cdk9 in the optimization of expression of the genes regulated by ICP22 of herpes simplex virus 1. J. Virol. 82, 10591–10599. doi: 10.1128/jvi.01242-08
Efstathiou, S., Minson, A. C., Field, H. J., Anderson, J. R., and Wildy, P. (1986). Detection of herpes simplex virus-specific DNA sequences in latently infected mice and in humans. J. Virol. 57, 446–455. doi: 10.1128/jvi.57.2.446-455.1986
Egloff, S., and Murphy, S. (2008). Cracking the RNA polymerase II CTD code. Trends Genet. 24, 280–288. doi: 10.1016/j.tig.2008.03.008
Eick, D., and Geyer, M. (2013). The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 113, 8456–8490. doi: 10.1021/cr400071f
Engel, C., Neyer, S., and Cramer, P. (2018). Distinct mechanisms of transcription initiation by RNA polymerases I and II. Annu. Rev. Biophys. 47, 425–446. doi: 10.1146/annurev-biophys-070317-033058
Fareed, M. U., and Spivack, J. G. (1994). Two open reading frames (ORF1 and ORF2) within the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1 are not essential for reactivation from latency. J. Virol. 68, 8071–8081. doi: 10.1128/jvi.68.12.8071-8081.1994
Feng, J., Gan, H., Eaton, M. L., Zhou, H., Li, S., Belsky, J. A., et al. (2016). Noncoding transcription is a driving force for nucleosome instability in spt16 mutant cells. Mol. Cell Biol. 36, 1856–1867. doi: 10.1128/mcb.00152-16
Formosa, T. (2012). The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta 1819, 247–255. doi: 10.1016/j.bbagrm.2011.07.009
Formosa, T., and Winston, F. (2020). The role of FACT in managing chromatin: disruption, assembly, or repair? Nucleic Acids Res. 48, 11929–11941. doi: 10.1093/nar/gkaa912
Forte, E., Zhang, Z., Thorp, E. B., and Hummel, M. (2020). Cytomegalovirus latency and reactivation: an intricate interplay with the host immune response. Front. Cell Infect. Microbiol. 10:130. doi: 10.3389/fcimb.2020.00130
Fox, H. L., Dembowski, J. A., and Deluca, N. A. (2017). A herpesviral immediate early protein promotes transcription elongation of viral transcripts. mBio 8:e745–17.
Franco, L. C., Morales, F., Boffo, S., and Giordano, A. (2018). CDK9: a key player in cancer and other diseases. J. Cell Biochem. 119, 1273–1284. doi: 10.1002/jcb.26293
Fraser, K. A., and Rice, S. A. (2005). Herpes simplex virus type 1 infection leads to loss of serine-2 phosphorylation on the carboxyl-terminal domain of RNA polymerase II. J. Virol. 79, 11323–11334. doi: 10.1128/jvi.79.17.11323-11334.2005
Fraser, K. A., and Rice, S. A. (2007). Herpes simplex virus immediate-early protein ICP22 triggers loss of serine 2-phosphorylated RNA polymerase II. J. Virol. 81, 5091–5101. doi: 10.1128/jvi.00184-07
Funk, C., Ott, M., Raschbichler, V., Nagel, C. H., Binz, A., Sodeik, B., et al. (2015). The herpes simplex virus protein pUL31 escorts nucleocapsids to sites of nuclear egress, a process coordinated by its N-terminal domain. PLoS Pathog. 11:e1004957. doi: 10.1371/journal.ppat.1004957
Gary, L., Gilden, D. H., and Cohrs, R. J. (2006). Epigenetic regulation of varicella-zoster virus open reading frames 62 and 63 in latently infected human trigeminal ganglia. J. Virol. 80, 4921–4926. doi: 10.1128/jvi.80.10.4921-4926.2006
Gerada, C., Campbell, T. M., Kennedy, J. J., Mcsharry, B. P., Steain, M., Slobedman, B., et al. (2020). Manipulation of the innate immune response by varicella zoster virus. Front. Immunol. 11:1. doi: 10.3389/fimmu.2020.00001
Gershon, A. A., Breuer, J., Cohen, J. I., Cohrs, R. J., Gershon, M. D., Gilden, D., et al. (2015). Varicella zoster virus infection. Nat. Rev. Dis. Primers 1:15016.
Glaunsinger, B. A. (2015). Modulation of the translational landscape during herpesvirus infection. Annu. Rev. Virol. 2, 311–333. doi: 10.1146/annurev-virology-100114-054839
Gomes, N. P., Bjerke, G., Llorente, B., Szostek, S. A., Emerson, B. M., and Espinosa, J. M. (2006). Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20, 601–612. doi: 10.1101/gad.1398206
Granzow, H., Klupp, B. G., Fuchs, W., Veits, J., Osterrieder, N., and Mettenleiter, T. C. (2001). Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75, 3675–3684. doi: 10.1128/jvi.75.8.3675-3684.2001
Griffin, B. N. F. D. M. K. P. M. H. D. E. (2007). Fields’ Virology, Fifth Edn. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins.
Grinde, B. (2013). Herpesviruses: latency and reactivation - viral strategies and host response. J. Oral Microbiol. 5:22766. doi: 10.3402/jom.v5i0.22766
Gruffat, H., Marchione, R., and Manet, E. (2016). Herpesvirus late gene expression: a viral-specific pre-initiation complex is key. Front. Microbiol. 7:869. doi: 10.3389/fmicb.2016.00869
Guo, L., Wu, W. J., Liu, L. D., Wang, L. C., Zhang, Y., Wu, L. Q., et al. (2012). Herpes simplex virus 1 ICP22 inhibits the transcription of viral gene promoters by binding to and blocking the recruitment of P-TEFb. PLoS One 7:e45749. doi: 10.1371/journal.pone.0045749
Haberle, V., and Stark, A. (2018). Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 19, 621–637. doi: 10.1038/s41580-018-0028-8
Hagen, C., Dent, K. C., Zeev-Ben-Mordehai, T., Grange, M., Bosse, J. B., Whittle, C., et al. (2015). Structural basis of vesicle formation at the inner nuclear membrane. Cell 163, 1692–1701. doi: 10.1016/j.cell.2015.11.029
Hahn, S. (2004). Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11, 394–403. doi: 10.1038/nsmb763
Hahn, S., and Young, E. T. (2011). Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189, 705–736. doi: 10.1534/genetics.111.127019
Harlen, K. M., and Churchman, L. S. (2017). The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 18, 263–273. doi: 10.1038/nrm.2017.10
Held, K., Junker, A., Dornmair, K., Meinl, E., Sinicina, I., Brandt, T., et al. (2011). Expression of herpes simplex virus 1-encoded microRNAs in human trigeminal ganglia and their relation to local T-cell infiltrates. J. Virol. 85, 9680–9685. doi: 10.1128/jvi.00874-11
Heldwein, E. E. (2016). gH/gL supercomplexes at early stages of herpesvirus entry. Curr. Opin. Virol. 18, 1–8. doi: 10.1016/j.coviro.2016.01.010
Heldwein, E. E. (2018). Up close with herpesviruses. Science (New York, N.Y.) 360, 34–35. doi: 10.1126/science.aat3990
Heming, J. D., Conway, J. F., and Homa, F. L. (2017). Herpesvirus capsid assembly and DNA packaging. Adv. Anat. Embryol. Cell Biol. 223, 119–142. doi: 10.1007/978-3-319-53168-7_6
Hsieh, F. K., Kulaeva, O. I., Patel, S. S., Dyer, P. N., Luger, K., Reinberg, D., et al. (2013). Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 110, 7654–7659. doi: 10.1073/pnas.1222198110
Hsin, J. P., Xiang, K., and Manley, J. L. (2014). Function and control of RNA polymerase II C-terminal domain phosphorylation in vertebrate transcription and RNA processing. Mol. Cell Biol. 34, 2488–2498. doi: 10.1128/mcb.00181-14
Jaggi, U., Matundan, H. H., Tormanen, K., Wang, S., Yu, J., Mott, K. R., et al. (2020). Expression of murine CD80 by herpes simplex virus 1 in place of latency-associated transcript (LAT) can compensate for latency reactivation and anti-apoptotic functions of LAT. J. Virol. 94:e1798–19.
Jiang, X., Chentoufi, A. A., Hsiang, C., Carpenter, D., Osorio, N., Benmohamed, L., et al. (2011). The herpes simplex virus type 1 latency-associated transcript can protect neuron-derived C1300 and Neuro2A cells from granzyme B-induced apoptosis and CD8 T-cell killing. J. Virol. 85, 2325–2332. doi: 10.1128/jvi.01791-10
Johnson, D. C., and Baines, J. D. (2011). Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9, 382–394. doi: 10.1038/nrmicro2559
Jonkers, I., and Lis, J. T. (2015). Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 167–177. doi: 10.1038/nrm3953
Justyna, Z., Sonja, B., Clelia, L., Dawn, O. R., Thomas, P. H., Peter, O. H., et al. (2014). Herpes Simplex Virus 1 (HSV-1) ICP22 protein directly interacts with cyclin-dependent kinase (CDK)9 to inhibit RNA polymerase II transcription elongation. PLoS One 9:e107654.
Kennedy, P. G., Grinfeld, E., and Bell, J. E. (2000). Varicella-zoster virus gene expression in latently infected and explanted human ganglia. J. Virol. 74, 11893–11898. doi: 10.1128/jvi.74.24.11893-11898.2000
Kennedy, P. G., Grinfeld, E., Bontems, S., and Sadzot-Delvaux, C. (2001). Varicella-Zoster virus gene expression in latently infected rat dorsal root ganglia. Virology 289, 218–223. doi: 10.1006/viro.2001.1173
Kennedy, P. G., Grinfeld, E., and Gow, J. W. (1999). Latent Varicella-zoster virus in human dorsal root ganglia. Virology 258, 451–454. doi: 10.1006/viro.1999.9745
Kennedy, P. G., Rovnak, J., Badani, H., and Cohrs, R. J. (2015). A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation. J. Gen. Virol. 96, 1581–1602. doi: 10.1099/vir.0.000128
Kim, M., Suh, H., Cho, E. J., and Buratowski, S. (2009). Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 284, 26421–26426. doi: 10.1074/jbc.m109.028993
Kimberlin, D. W., and Rouse, D. J. (2004). Clinical practice. Genital Herpes. N. Engl. J. Med. 350, 1970–1977.
Klupp, B. G., Granzow, H., Fuchs, W., Keil, G. M., Finke, S., and Mettenleiter, T. C. (2007). Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 7241–7246. doi: 10.1073/pnas.0701757104
Knipe, D. M., and Cliffe, A. (2008). Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 6, 211–221. doi: 10.1038/nrmicro1794
Komarnitsky, P., Cho, E. J., and Buratowski, S. (2000). Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14, 2452–2460. doi: 10.1101/gad.824700
Krishnamurthy, S., and Hampsey, M. (2009). Eukaryotic transcription initiation. Curr. Biol. 19, R153–R156.
Kurt-Jones, E. A., Orzalli, M. H., and Knipe, D. M. (2017). Innate immune mechanisms and herpes simplex virus infection and disease. Adv. Anat. Embryol. Cell Biol. 223, 49–75. doi: 10.1007/978-3-319-53168-7_3
Laing, K. J., Ouwendijk, W. J. D., Koelle, D. M., and Verjans, G. (2018). Immunobiology of varicella-zoster virus infection. J. Infect. Dis. 218, S68–S74.
Lanier, L. L., O’fallon, S., Somoza, C., Phillips, J. H., Linsley, P. S., Okumura, K., et al. (1995). CD80 (B7) and CD86 (B70) provide similar costimulatory signals for T cell proliferation, cytokine production, and generation of CTL. J. Immunol. 154, 97–105.
LeRoy, G., Orphanides, G., Lane, W. S., and Reinberg, D. (1998). Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 282, 1900–1904. doi: 10.1126/science.282.5395.1900
Li, Y., Liu, M., Chen, L. F., and Chen, R. (2017). P-TEFb: finding its ways to release promoter-proximally paused RNA Polymerase II. Transcription 9, 88–94. doi: 10.1080/21541264.2017.1281864
Licatalosi, D. D., Geiger, G., Minet, M., Schroeder, S., Cilli, K., Mcneil, J. B., et al. (2002). Functional interaction of yeast pre-mRNA 3’ end processing factors with RNA polymerase II. Mol. Cell 9, 1101–1111. doi: 10.1016/s1097-2765(02)00518-x
Lin, F. S., Ding, Q., Guo, H., and Zheng, A. C. (2010). The herpes simplex virus type 1 infected cell protein 22. Virol. Sin. 25, 1–7. doi: 10.1007/s12250-010-3080-x
Lin, Y., and Zheng, C. (2019). A tug of war: DNA-sensing antiviral innate immunity and herpes simplex virus type i infection. Front. Microbiol. 10:2627. doi: 10.3389/fmicb.2019.02627
Liu, X., Bushnell, D. A., and Kornberg, R. D. (2013). RNA polymerase II transcription: structure and mechanism. Biochim. Biophys. Acta 1829, 2–8.
Liu, Y., Zhou, K., Zhang, N., Wei, H., Tan, Y. Z., Zhang, Z., et al. (2020). FACT caught in the act of manipulating the nucleosome. Nature 577, 426–431. doi: 10.1038/s41586-019-1820-0
Liu, Y. T., Jih, J., Dai, X., Bi, G. Q., and Zhou, Z. H. (2019). Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome. Nature 570, 257–261. doi: 10.1038/s41586-019-1248-6
Liu, Z., Kato, A., Oyama, M., Kozuka-Hata, H., Arii, J., and Kawaguchi, Y. (2015). Role of Host Cell p32 in herpes simplex virus 1 de-envelopment during viral nuclear egress. J. Virol. 89, 8982–8998. doi: 10.1128/jvi.01220-15
Liu, Z., Kato, A., Shindo, K., Noda, T., Sagara, H., Kawaoka, Y., et al. (2014). Herpes simplex virus 1 UL47 interacts with viral nuclear egress factors UL31, UL34, and Us3 and regulates viral nuclear egress. J. Virol. 88, 4657–4667. doi: 10.1128/jvi.00137-14
Long, M. C., Leong, V., Schaffer, P. A., Spencer, C. A., and Rice, S. A. (1999). ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 73, 5593–5604. doi: 10.1128/jvi.73.7.5593-5604.1999
Lorenz, M., Vollmer, B., Unsay, J. D., Klupp, B. G., García-Sáez, A. J., Mettenleiter, T. C., et al. (2015a). A single herpesvirus protein can mediate vesicle formation in the nuclear envelope. J. Biol. Chem. 290, 6962–6974.
Lorenz, M., Vollmer, B., Unsay, J. D., Klupp, B. G., Garcia-Saez, A. J., Mettenleiter, T. C., et al. (2015b). A single herpesvirus protein can mediate vesicle formation in the nuclear envelope. J. Biol. Chem. 290, 6962–6974. doi: 10.1074/jbc.m114.627521
Loret, S., Guay, G., and Lippe, R. (2008). Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82, 8605–8618. doi: 10.1128/jvi.00904-08
Lungu, O., Panagiotidis, C. A., Annunziato, P. W., Gershon, A. A., and Silverstein, S. J. (1998). Aberrant intracellular localization of Varicella-Zoster virus regulatory proteins during latency. Proc. Natl. Acad. Sci. U.S.A. 95, 7080–7085. doi: 10.1073/pnas.95.12.7080
Lv, Y., Zhou, S., Gao, S., and Deng, H. (2018). Remodeling of host membranes during herpesvirus assembly and egress. Protein Cell 10, 315–326.
Lv, Y., Zhou, S., Gao, S., and Deng, H. (2019). Remodeling of host membranes during herpesvirus assembly and egress. Protein Cell 10, 315–326. doi: 10.1007/s13238-018-0577-9
Lynch, J. M., Kenyon, T. K., Grose, C., Hay, J., and Ruyechan, W. T. (2002). Physical and functional interaction between the varicella zoster virus IE63 and IE62 proteins. Virology 302, 71–82. doi: 10.1006/viro.2002.1555
Mahalingam, R., Wellish, M., Cohrs, R., Debrus, S., Piette, J., Rentier, B., et al. (1996). Expression of protein encoded by varicella-zoster virus open reading frame 63 in latently infected human ganglionic neurons. Proc. Natl. Acad. Sci. U.S.A. 93, 2122–2124. doi: 10.1073/pnas.93.5.2122
Malone, E. A., Clark, C. D., Chiang, A., and Winston, F. (1991). Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell Biol. 11, 5710–5717. doi: 10.1128/mcb.11.11.5710
Maruzuru, Y., Shindo, K., Liu, Z., Oyama, M., Kozuka-Hata, H., Arii, J., et al. (2014). Role of herpes simplex virus 1 immediate early protein ICP22 in viral nuclear egress. J. Virol. 88, 7445–7454. doi: 10.1128/jvi.01057-14
Mason, P. B., and Struhl, K. (2003). The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell Biol. 23, 8323–8333. doi: 10.1128/mcb.23.22.8323-8333.2003
Matundan, H., and Ghiasi, H. (2018). HSV-1 ICP22 suppresses CD80 expression by murine dendritic cells. J. Virol. 93:e1803–18.
Matundan, H., and Ghiasi, H. (2019). Herpes simplex virus 1 ICP22 suppresses cd80 expression by murine dendritic cells. J. Virol. 93, e1803–e1818.
Matundan, H. H., Jaggi, U., Wang, S., and Ghiasi, H. (2019). Loss of ICP22 in HSV-1 elicits immune infiltration and maintains stromal keratitis despite reduced primary and latent virus infectivity. Invest Ophthalmol. Vis. Sci. 60, 3398–3406. doi: 10.1167/iovs.19-27701
McCracken, S., Fong, N., Rosonina, E., Yankulov, K., Brothers, G., Siderovski, D., et al. (1997). 5’-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11, 3306–3318. doi: 10.1101/gad.11.24.3306
McGeoch, D. J., Cook, S., Dolan, A., Jamieson, F. E., and Telford, E. A. (1995). Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247, 443–458. doi: 10.1006/jmbi.1995.0152
Mettenleiter, T. C. (2004). Budding events in herpesvirus morphogenesis. Virus Res. 106, 167–180. doi: 10.1016/j.virusres.2004.08.013
Mettenleiter, T. C., Müller, F., Granzow, H., and Klupp, B. G. (2013b). The way out: what we know and do not know about herpesvirus nuclear egress. Cell. Microbiol. 15, 170–178.
Mettenleiter, T. C., Muller, F., Granzow, H., and Klupp, B. G. (2013a). The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol. 15, 170–178. doi: 10.1111/cmi.12044
Moriuchi, H., Moriuchi, M., Straus, S. E., and Cohen, J. I. (1993). Varicella-zoster virus (VZV) open reading frame 61 protein transactivates VZV gene promoters and enhances the infectivity of VZV DNA. J. Virol. 67, 4290–4295. doi: 10.1128/jvi.67.7.4290-4295.1993
Mostafa, H. H., and Davido, D. J. (2013). Herpes simplex virus 1 ICP22 but not US 1.5 is required for efficient acute replication in mice and VICE domain formation. J. Virol. 87, 13510–13519. doi: 10.1128/jvi.02424-13
Mou, F., Wills, E., and Baines, J. D. (2009). Phosphorylation of the U(L)31 protein of herpes simplex virus 1 by the U(S)3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids. J. Virol. 83, 5181–5191. doi: 10.1128/jvi.00090-09
Münz, C. (2019). Latency and lytic replication in Epstein-Barr virus-associated oncogenesis. Nat. Rev. Microbiol. 17, 691–700. doi: 10.1038/s41579-019-0249-7
Muylaert, I., Tang, K. W., and Elias, P. (2011). Replication and recombination of herpes simplex virus DNA. J. Biol. Chem. 286, 15619–15624. doi: 10.1074/jbc.r111.233981
Natarajan, M., Schiralli Lester, G. M., Lee, C., Missra, A., Wasserman, G. A., Steffen, M., et al. (2013). Negative elongation factor (NELF) coordinates RNA polymerase II pausing, premature termination, and chromatin remodeling to regulate HIV transcription. J. Biol. Chem. 288, 25995–26003. doi: 10.1074/jbc.m113.496489
Nemec, C. M., Singh, A. K., Ali, A., Tseng, S. C., Syal, K., Ringelberg, K. J., et al. (2019). Noncanonical CTD kinases regulate RNA polymerase II in a gene-class-specific manner. Nat. Chem. Biol. 15, 123–131. doi: 10.1038/s41589-018-0194-1
Ni, Z., Saunders, A., Fuda, N. J., Yao, J., Suarez, J. R., Webb, W. W., et al. (2008). P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol. Cell Biol. 28, 1161–1170. doi: 10.1128/mcb.01859-07
Nicoll, M. P., Proenca, J. T., and Efstathiou, S. (2012). The molecular basis of herpes simplex virus latency. FEMS Microbiol. Rev. 36, 684–705. doi: 10.1111/j.1574-6976.2011.00320.x
Nogales, E., Louder, R. K., and He, Y. (2017). Structural Insights into the Eukaryotic Transcription Initiation Machinery. Annu. Rev. Biophys. 46, 59–83. doi: 10.1146/annurev-biophys-070816-033751
Ogle, W. O., and Roizman, B. (1999). Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J. Virol. 73, 4305–4315. doi: 10.1128/jvi.73.5.4305-4315.1999
Orlando, J. S., Astor, T. L., Rundle, S. A., and Schaffer, P. A. (2006). The products of the herpes simplex virus type 1 immediate-early US1/US1.5 genes downregulate levels of S-phase-specific cyclins and facilitate virus replication in S-phase Vero cells. J. Virol. 80, 4005–4016. doi: 10.1128/jvi.80.8.4005-4016.2006
Orphanides, G., Wu, W. H., Lane, W. S., Hampsey, M., and Reinberg, D. (1999). The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400, 284–288. doi: 10.1038/22350
Ott, M., Tascher, G., Hassdenteufel, S., Zimmermann, R., Haas, J., and Bailer, S. M. (2011). Functional characterization of the essential tail anchor of the herpes simplex virus type 1 nuclear egress protein pUL34. J. Gen. Virol. 92, 2734–2745. doi: 10.1099/vir.0.032730-0
Ouwendijk, W. J., Choe, A., Nagel, M. A., Gilden, D., Osterhaus, A. D., Cohrs, R. J., et al. (2012). Restricted varicella-zoster virus transcription in human trigeminal ganglia obtained soon after death. J. Virol. 86, 10203–10206. doi: 10.1128/jvi.01331-12
Ouwendijk, W. J. D., Depledge, D. P., Rajbhandari, L., Lenac Rovis, T., Jonjic, S., Breuer, J., et al. (2020). Varicella-zoster virus VLT-ORF63 fusion transcript induces broad viral gene expression during reactivation from neuronal latency. Nat. Commun. 11:6324.
Paparidis, N. F., Durvale, M. C., and Canduri, F. (2017). The emerging picture of CDK9/P-TEFb: more than 20 years of advances since PITALRE. Mol. Biosyst. 13, 246–276. doi: 10.1039/c6mb00387g
Pathak, R., Singh, P., Ananthakrishnan, S., Adamczyk, S., Schimmel, O., and Govind, C. K. (2018). Acetylation-dependent recruitment of the FACT complex and its role in regulating pol II occupancy genome-wide in saccharomyces cerevisiae. Genetics 209, 743–756. doi: 10.1534/genetics.118.300943
Payne, J. M., Laybourn, P. J., and Dahmus, M. E. (1989). The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J. Biol. Chem. 264, 19621–19629. doi: 10.1016/s0021-9258(19)47159-7
Peck, S. A., Hughes, K. D., Victorino, J. F., and Mosley, A. L. (2019). Writing a wrong: coupled RNA polymerase II transcription and RNA quality control. Wiley Inter. Rev. RNA 10:e1529. doi: 10.1002/wrna.1529
Pelechano, V., Jimeno-Gonzalez, S., Rodriguez-Gil, A., Garcia-Martinez, J., Perez-Ortin, J. E., and Chavez, S. (2009). Regulon-specific control of transcription elongation across the yeast genome. PLoS Genet. 5:e1000614. doi: 10.1371/journal.pgen.1000614
Perng, G. C., Jones, C., Ciacci-Zanella, J., Stone, M., Henderson, G., Yukht, A., et al. (2000). Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 287, 1500–1503. doi: 10.1126/science.287.5457.1500
Peterlin, B. M., and Price, D. H. (2006). Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305. doi: 10.1016/j.molcel.2006.06.014
Petrenko, N., Jin, Y., Dong, L., Wong, K. H., and Struhl, K. (2019). Requirements for RNA polymerase II preinitiation complex formation in vivo. Elife 8:e43654.
Ping, Y. H., and Rana, T. M. (2001). DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 276, 12951–12958. doi: 10.1074/jbc.m006130200
Placek, B. J., and Berger, S. L. (2010). Chromatin dynamics during herpes simplex virus-1 lytic infection. Biochim. Biophys. Acta 1799, 223–227. doi: 10.1016/j.bbagrm.2010.01.012
Porrua, O., Boudvillain, M., and Libri, D. (2016). Transcription termination: variations on common themes. Trends Genet. 32, 508–522. doi: 10.1016/j.tig.2016.05.007
Purves, F. C., and Roizman, B. (1992). The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein alpha 22. Proc. Natl. Acad. Sci. U.S.A. 89, 7310–7314. doi: 10.1073/pnas.89.16.7310
Reinberg, D., and Sims, R. J. III (2006). de FACTo nucleosome dynamics. J. Biol. Chem. 281, 23297–23301. doi: 10.1074/jbc.r600007200
Renner, D. W., and Szpara, M. L. (2018). Impacts of genome-wide analyses on our understanding of human herpesvirus diversity and evolution. J. Virol. 92, e908–e917.
Retamal-Diaz, A. R., Suazo, P. A., Garrido, I., Kalergis, A. M., and Gonzalez, P. A. (2015). [Immune evasion by herpes simplex viruses]. Rev. Chilena Infectol. 32, 58–70.
Riaz, A., Kifayatullah, M. U. H., and Akhtar, N. (2017). Recent understanding of the classification and life cycle of herpesviruses: a review. Sci. Lett. 5, 195–207.
Rice, S. A., and Davido, D. J. (2013). HSV-1 ICP22: hijacking host nuclear functions to enhance viral infection. Future Microbiol. 8, 311–321. doi: 10.2217/fmb.13.4
Rice, S. A., Long, M. C., Lam, V., Schaffer, P. A., and Spencer, C. A. (1995). Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J. Virol. 69, 5550–5559. doi: 10.1128/jvi.69.9.5550-5559.1995
Richard, P., and Manley, J. L. (2009). Transcription termination by nuclear RNA polymerases. Genes Dev. 23, 1247–1269. doi: 10.1101/gad.1792809
Roller, R. J., Bjerke, S. L., Haugo, A. C., and Hanson, S. (2010). Analysis of a charge cluster mutation of herpes simplex virus type 1 UL34 and its extragenic suppressor suggests a novel interaction between pUL34 and pUL31 that is necessary for membrane curvature around capsids. J. Virol. 84, 3921–3934. doi: 10.1128/jvi.01638-09
Roller, R. J., Zhou, Y., Schnetzer, R., Ferguson, J., and Desalvo, D. (2000). Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J. Virol. 74, 117–129. doi: 10.1128/jvi.74.1.117-129.2000
Sadzot-Delvaux, C., Debrus, S., Nikkels, A., Piette, J., and Rentier, B. (1995). Varicella-zoster virus latency in the adult rat is a useful model for human latent infection. Neurology 45, S18–S20.
Sainsbury, S., Bernecky, C., and Cramer, P. (2015). Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 129–143. doi: 10.1038/nrm3952
Sathiyamoorthy, K., Chen, J., Longnecker, R., and Jardetzky, T. S. (2017). The COMPLEXity in herpesvirus entry. Curr. Opin. Virol. 24, 97–104. doi: 10.1016/j.coviro.2017.04.006
Schier, A. C., and Taatjes, D. J. (2020). Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 34, 465–488. doi: 10.1101/gad.335679.119
Schroeder, S. C., Schwer, B., Shuman, S., and Bentley, D. (2000). Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14, 2435–2440. doi: 10.1101/gad.836300
Schuster, F., Klupp, B. G., Granzow, H., and Mettenleiter, T. C. (2012). Structural determinants for nuclear envelope localization and function of pseudorabies virus pUL34. J. Virol. 86, 2079–2088. doi: 10.1128/jvi.05484-11
Schwabish, M. A., and Struhl, K. (2004). Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell Biol. 24, 10111–10117. doi: 10.1128/mcb.24.23.10111-10117.2004
Sikorski, T. W., and Buratowski, S. (2009). The basal initiation machinery: beyond the general transcription factors. Curr. Opin. Cell Biol. 21, 344–351. doi: 10.1016/j.ceb.2009.03.006
Sims, R. J. III, Belotserkovskaya, R., and Reinberg, D. (2004). Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18, 2437–2468. doi: 10.1101/gad.1235904
Skoreński, M., and Sieńczyk, M. (2014). Anti-herpesvirus agents: a patent and literature review (2003 to present). Expert Opin. Ther. Pat. 24, 925–941. doi: 10.1517/13543776.2014.927442
Sommer, M. H., Zagha, E., Serrano, O. K., Ku, C. C., Zerboni, L., Baiker, A., et al. (2001). Mutational analysis of the repeated open reading frames, ORFs 63 and 70 and ORFs 64 and 69, of varicella-zoster virus. J. Virol. 75, 8224–8239. doi: 10.1128/jvi.75.17.8224-8239.2001
Spitz, F., and Furlong, E. E. (2012). Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626. doi: 10.1038/nrg3207
Spivack, J. G., and Fraser, N. W. (1987). Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J. Virol. 61, 3841–3847. doi: 10.1128/jvi.61.12.3841-3847.1987
Stevens, J. G. (1989). Human herpesviruses: a consideration of the latent state. Microbiol. Rev. 53, 318–332. doi: 10.1128/mmbr.53.3.318-332.1989
Stevens, J. G., and Cook, M. L. (1971). Latent herpes simplex virus in spinal ganglia of mice. Science 173, 843–845. doi: 10.1126/science.173.3999.843
Stevenson, D., Xue, M., Hay, J., and Ruyechan, W. T. (1996). Phosphorylation and nuclear localization of the varicella-zoster virus gene 63 protein. J. Virol. 70, 658–662. doi: 10.1128/jvi.70.1.658-662.1996
Suazo, P. A., Ibanez, F. J., Retamal-Diaz, A. R., Paz-Fiblas, M. V., Bueno, S. M., Kalergis, A. M., et al. (2015). Evasion of early antiviral responses by herpes simplex viruses. Mediators Inflamm. 2015:593757.
Takeshima, K., Arii, J., Maruzuru, Y., Koyanagi, N., and Kawaguchi, Y. (2019). Identification of the capsid binding site in the herpes simplex virus 1 nuclear egress complex and its role in viral primary envelopment and replication. J. Virol. 93:e1290–19.
Tettey, T. T., Gao, X., Shao, W., Li, H., Story, B. A., Chitsazan, A. D., et al. (2019). A role for FACT in RNA polymerase ii promoter-proximal pausing. Cell Rep. 27, 3770–3779.e7.
Theil, D., Derfuss, T., Paripovic, I., Herberger, S., Meinl, E., Schueler, O., et al. (2003). Latent herpesvirus infection in human trigeminal ganglia causes chronic immune response. Am. J. Pathol. 163, 2179–2184. doi: 10.1016/s0002-9440(10)63575-4
Tognarelli, E. I., Palomino, T. F., Corrales, N., Bueno, S. M., Kalergis, A. M., and Gonzalez, P. A. (2019). Herpes simplex virus evasion of early host antiviral responses. Front. Cell Infect. Microbiol. 9:127. doi: 10.1155/2015/593757
Tormanen, K., Allen, S., Mott, K. R., and Ghiasi, H. (2019). The latency-associated transcript inhibits apoptosis via downregulation of components of the type i interferon pathway during latent herpes simplex virus 1 ocular infection. J. Virol. 93, e103–e119.
Tormanen, K., Wang, S., and Ghiasi, H. (2020). CD80 plays a critical role in increased inflammatory responses in herpes simplex virus 1-infected mouse corneas. J. Virol. 94, e1511–e1519.
van Diemen, F. R., and Lebbink, R. J. (2017). CRISPR/Cas9, a powerful tool to target human herpesviruses. Cell Microbiol. 19:e12694. doi: 10.1111/cmi.12694
Verjans, G. M., Hintzen, R. Q., Van Dun, J. M., Poot, A., Milikan, J. C., Laman, J. D., et al. (2007). Selective retention of herpes simplex virus-specific T cells in latently infected human trigeminal ganglia. Proc. Natl. Acad. Sci. U.S.A. 104, 3496–3501. doi: 10.1073/pnas.0610847104
Wahle, E., and Keller, W. (1996). The biochemistry of polyadenylation. Trends Biochem. Sci. 21, 247–250. doi: 10.1016/s0968-0004(96)10030-x
Whitley, R. J. (1996). “Herpesviruses,” in Medical Microbiology, ed. S. Baron (Galveston, TX: University of Texas Medical Branch at Galveston).
Wilson, A. C., and Mohr, I. (2012). A cultured affair: HSV latency and reactivation in neurons. Trends Microbiol. 20, 604–611. doi: 10.1016/j.tim.2012.08.005
Wu, T. T., Su, Y. H., Block, T. M., and Taylor, J. M. (1996). Evidence that two latency-associated transcripts of herpes simplex virus type 1 are nonlinear. J. Virol. 70, 5962–5967. doi: 10.1128/jvi.70.9.5962-5967.1996
Yadav, S., Libotte, F., Buono, E., Valia, S., Farina, G. A., Faggioni, A., et al. (2017). EBV early lytic protein BFRF1 alters emerin distribution and post-translational modification. Virus Res. 232, 113–122. doi: 10.1016/j.virusres.2017.02.010
Yan, C., Luo, Z., Li, W., Li, X., Dallmann, R., Kurihara, H., et al. (2020). Disturbed Yin-Yang balance: stress increases the susceptibility to primary and recurrent infections of herpes simplex virus type 1. Acta Pharm. Sin. B 10, 383–398. doi: 10.1016/j.apsb.2019.06.005
Yan, L., Majerciak, V., Zheng, Z. M., and Lan, K. (2019). Towards better understanding of KSHV life cycle: from transcription and posttranscriptional regulations to pathogenesis. Virol. Sin. 34, 135–161. doi: 10.1007/s12250-019-00114-3
Yang, Y., Yang, P., Wang, N., Chen, Z., Su, D., Zhou, Z. H., et al. (2020). Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation. Protein Cell 11, 339–351. doi: 10.1007/s13238-020-00710-0
Zaborowska, J., Baumli, S., Laitem, C., O’reilly, D., Thomas, P. H., O’hare, P., et al. (2014). Herpes simplex virus 1 (HSV-1) ICP22 protein directly interacts with cyclin-dependent kinase (CDK)9 to inhibit RNA polymerase II transcription elongation. PLoS One 9:e107654. doi: 10.1371/journal.pone.0107654
Zaborowska, J., Egloff, S., and Murphy, S. (2016). The pol II CTD: new twists in the tail. Nat. Struct. Mol. Biol. 23, 771–777.
Zhang, L., Serra-Cardona, A., Zhou, H., Wang, M., Yang, N., Zhang, Z., et al. (2018). Multisite substrate recognition in Asf1-dependent acetylation of histone H3 K56 by Rtt109. Cell 174, 818–830.e11.
Zhang, M., Fu, M., Li, M., Hu, H., Gong, S., and Hu, Q. (2020). Herpes Simplex Virus Type 2 Inhibits Type I IFN signaling mediated by the novel E3 ubiquitin protein ligase activity of viral protein ICP22. J. Immunol. 205, 1281–1292. doi: 10.4049/jimmunol.2000418
Zhang, M., Liu, Y., Wang, P., Guan, X., He, S., Luo, S., et al. (2015). HSV-2 immediate-early protein US1 inhibits IFN-beta production by suppressing association of IRF-3 with IFN-beta promoter. J. Immunol. 194, 3102–3115. doi: 10.4049/jimmunol.1401538
Zhang, Y., Tao, H., and Huang, S. Y. (2019). Dynamics and mechanisms in the recruitment and transference of histone chaperone CIA/ASF1. Int. J. Mol. Sci. 20:3325. doi: 10.3390/ijms20133325
Zhou, K., Liu, Y., and Luger, K. (2020). Histone chaperone FACT FAcilitates Chromatin Transcription: mechanistic and structural insights. Curr. Opin. Struct. Biol. 65, 26–32. doi: 10.1016/j.sbi.2020.05.019
Zhou, Q., Li, T., and Price, D. H. (2012). RNA polymerase II elongation control. Annu. Rev. Biochem. 81, 119–143.
Keywords: viral life cycle, latent, lytic, reactivation, ICP22, ORF63
Citation: Wu Y, Yang Q, Wang M, Chen S, Jia R, Yang Q, Zhu D, Liu M, Zhao X, Zhang S, Huang J, Ou X, Mao S, Gao Q, Sun D, Tian B and Cheng A (2021) Multifaceted Roles of ICP22/ORF63 Proteins in the Life Cycle of Human Herpesviruses. Front. Microbiol. 12:668461. doi: 10.3389/fmicb.2021.668461
Received: 16 February 2021; Accepted: 05 May 2021;
Published: 07 June 2021.
Edited by:
Marco De Andrea, University of Turin, ItalyReviewed by:
Paul R. Kinchington, University of Pittsburgh, United StatesKeith William Jarosinski, University of Illinois at Urbana-Champaign, United States
Copyright © 2021 Wu, Yang, Wang, Chen, Jia, Yang, Zhu, Liu, Zhao, Zhang, Huang, Ou, Mao, Gao, Sun, Tian and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Wu, d3V5QHNpY2F1LmVkdS5jbg==; Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=
†These authors have contributed equally to this work
 Ying Wu1,2,3*†
Ying Wu1,2,3*† Qiqi Yang
Qiqi Yang Mingshu Wang
Mingshu Wang Mafeng Liu
Mafeng Liu Xinxin Zhao
Xinxin Zhao Sai Mao
Sai Mao Qun Gao
Qun Gao Anchun Cheng
Anchun Cheng