- 1Department of Virology, Nagoya University Graduate School of Medicine, Nagoya, Japan
- 2Medical Genomics Center, Nagoya University Hospital, Nagoya, Japan
- 3PRESTO, Japan Science and Technology Agency, Kawaguchi, Japan
- 4Department of Virology and Parasitology, Fujita Health University School of Medicine, Toyoake, Japan
Epstein–Barr virus (EBV), which encodes >80 genes and nearly 50 non-coding RNAs, is a double-stranded DNA virus. EBV is associated with various types of lymphomas and lymphoproliferative disorders not only of B-cell but also T/NK-cell origin. However, the oncogenic mechanism remains poorly understood, including the EBV receptors expressed on T/NK cells, relationship of EBV with host genes, and epigenetic regulation of EBV and host genes. The roles of host and viral non-coding RNAs during tumorigenesis have been elucidated. EBV encodes at least 49 mature microRNAs (miRNAs), of which 44 are located in BamHI-A rightward transcripts (BARTs) region, and the remaining five are located in BamHI-H rightward fragment 1. BART miRNAs modulate cell differentiation, proliferation, apoptosis, and the cell cycle, and they are considered positive regulators of oncogenesis. We and others have recently reported that EBV-positive lymphomas frequently possess large deletions in BART miRNA clusters, suggesting that some viral miRNAs have suppressive effects on oncogenesis, and that deletion of these miRNAs may aid lymphoma formation.
Introduction
Epstein–Barr virus (EBV), which is the first human oncovirus, was isolated from Burkitt lymphoma by Epstein et al. (1964). EBV is a 170–180 kb double-stranded DNA virus belonging to the herpesvirus family and gammaherpesvirus subfamily, and its genome encodes approximately 80 genes (Longnecker et al., 2013). EBV belongs to the same subfamily as Kaposi’s sarcoma-associated herpesvirus, which is the causative virus of Kaposi’s sarcoma, and both viruses infect B cells and are related to B-cell lymphomas. In addition to Burkitt lymphoma, EBV is associated with a variety of malignancies with B-cell origins, such as Hodgkin lymphoma, diffuse large B-cell lymphoma (DLBCL), and immunodeficiency-related lymphoproliferative disorders detected after organ/hematopoietic stem cell transplantation (Swerdlow et al., 2016).
Epstein–Barr virus is also associated with natural killer (NK)-cell and T-cell neoplastic diseases including extranodal NK/T-cell lymphoma, nasal type (ENKTL) (Chan et al., 2017), chronic active EBV disease (CAEBV) (Quintanilla-Martinez et al., 2017; Cohen et al., 2020), and epithelial tumors such as nasopharyngeal carcinoma and gastric cancer (Cohen, 2000; Longnecker et al., 2013). Although extensive studies have been conducted on the implication of MYC translocation and activation in Burkitt lymphoma (Longnecker et al., 2013), the oncogenic mechanisms (including the T/NK-cell receptors expressed, the relationship of EBV with host genes, and epigenetic regulation of EBV and host genes) associated with other lymphoid tumors are still unclear.
The roles of host and viral non-coding RNAs have been elucidated during tumorigenesis in various malignancies, including EBV-associated diseases (Cai et al., 2006; Qiu et al., 2011; Klinke et al., 2014; Kuzembayeva et al., 2014; Dreyfus, 2017; Iizasa et al., 2020). We and others have recently reported that EBV-positive lymphomas frequently possess large deletions in viral microRNA (miRNA) clusters, suggesting that some miRNAs negatively regulate lymphomagenesis (Peng et al., 2018; Okuno et al., 2019; Mabuchi et al., 2021). This review outlines the role of viral non-coding RNAs in the lymphomagenesis of EBV-associated diseases, focusing on EBV-encoded miRNAs.
Biology of EBV Infection
Epstein–Barr virus can cause either a latent or lytic infection. In a latent infection, EBV exists as an episome in the nucleus. No viral particles are produced in the host cell, and only a limited number of genes are expressed. Latent infections are classified into four patterns, type 0, I, II, and III, depending on the host cell, tissue, and immune status (Longnecker et al., 2013; Munz, 2019). Latency type 0 is seen in the memory B cells of healthy individuals, and few viral proteins are expressed in this type. In type I, EBV nuclear antigen (EBNA) 1 and EBV-encoded small RNAs (EBERs) are expressed. In a type II infection, latent membrane protein (LMP) 1 and LMP2 are additionally expressed. EBNA2, 3s, and LP are expressed in type III infections.
In contrast, immediate-early, early, and late genes are expressed one after another during lytic infection, and virus particles are produced. It was previously considered that virus-infected cells are immediately in a latent state after EBV infects B cells. However, recent evidence suggests that these cells are temporarily in an abortive lytic infection state, and that the expression of EBV lytic genes plays an important role in the immortalization of infected cells (Ma et al., 2011; Murata et al., 2014; Munz, 2019). In fact, these lytic genes are expressed in hydroa vacciniforme-like lymphoproliferative disorder and DLBCL (Yamamoto et al., 2016; Cohen et al., 2018), which suggests that the abortive lytic infection may be involved in lymphomagenesis.
Oncogenesis of EBV and EBV-Related Malignancies
Epstein–Barr virus is used to immortalize human B cells in vitro to produce a lymphoblastoid cell line (LCL). LMP1 is a viral membrane oncoprotein essential for immortalization; it mimics CD40 expressed by T cells and constantly activates downstream NF-κB, PI3K/AKT, JNK, and p38/MAPK pathways, immortalizing infected cells and suppressing apoptosis (Kanda, 2018). Although LMP2A is not essential for immortalization, it is also an oncoprotein that mimics the B-cell receptor expressed by B cells, resulting in constitutive calcium recruitment, protein kinase C activation, cell proliferation, and differentiation suppression (Longnecker et al., 2013). EBNA1, EBNA2, EBNA3A, EBNA3C, and EBNA-LP are nuclear proteins that help transform B cells and maintain latency (Tomkinson et al., 1993; Kanda, 2018). All these latent infection-related genes are expressed in a type III infection where host cell-mediated immunity is suppressed, such as immunodeficiency-related lymphoproliferative disorders. As only a limited number of viral genes are expressed during latent type I and II infections, it is easier to avoid host immunity with these infections compared with a type III infection. However, immortalization and suppression of apoptosis by viral oncoproteins are limited in these latency types (Kimura, 2018).
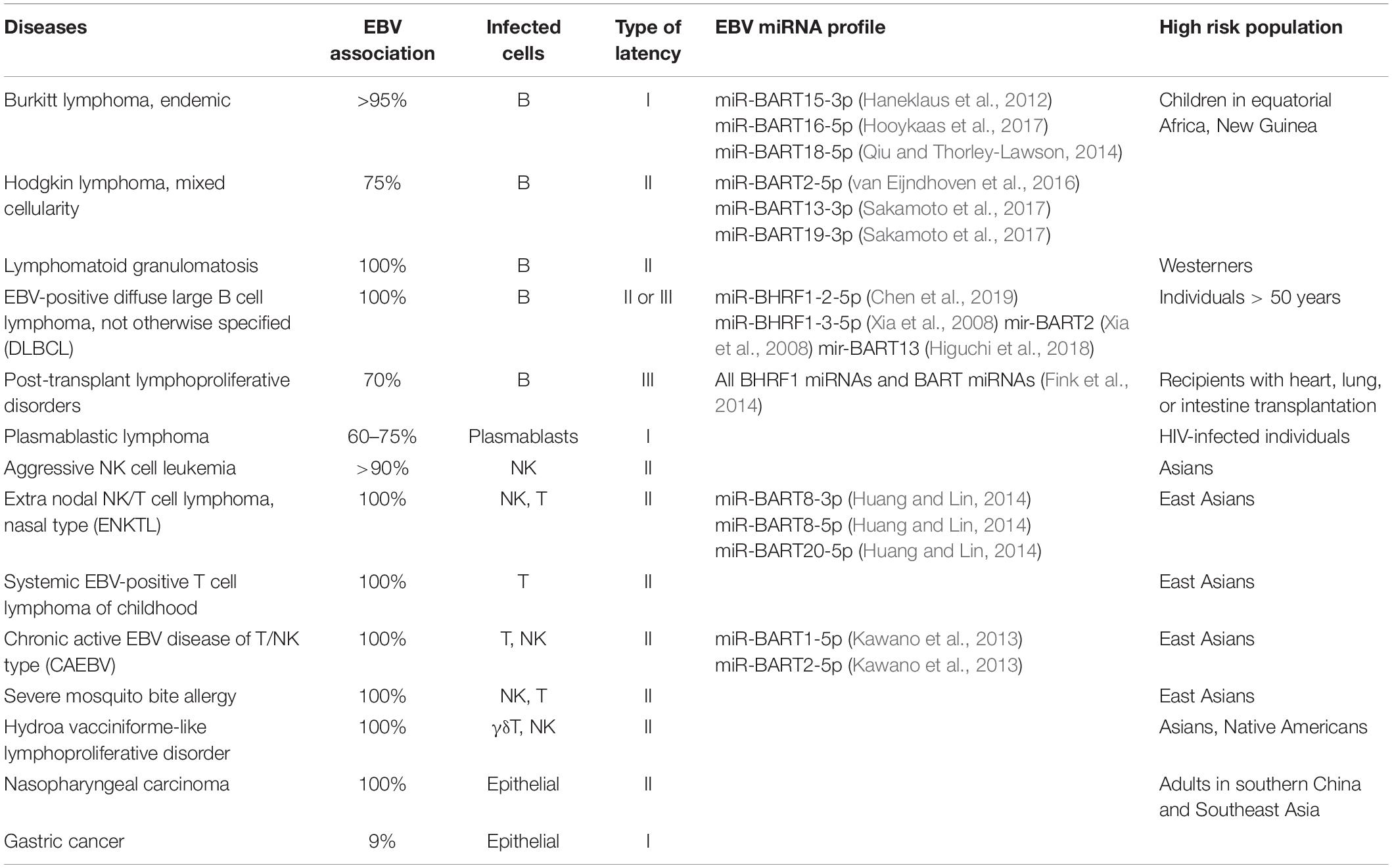
Table 1 summarizes the latent infection patterns of EBV-related malignancies, as well as the degree of EBV involvement, type of cell infected, associated high-risk factors, and EBV miRNA profile. As mentioned above, EBV is associated with a variety of lymphomas and lymphoproliferative diseases, not only of B-cell but also T/NK-cell origin, as well as with epithelial malignancies. All malignancies with T/NK- or epithelial-cell origins are associated with type I and type II latency, in which a limited number of viral genes are expressed and evasion of host immunity is easier (Longnecker et al., 2013). Notably, some EBV-positive tumors have uneven distributions in specific areas as shown in Table 1; whether this is due to genetic variations in the host or differences in the infecting strains remains unclear.
EBV Non-coding RNAs
In addition to viral proteins, EBV encodes many non-coding RNAs that also potentiate oncogenesis. EBER1 and EBER2, which are long non-coding RNAs of 167 and 172 nucleotides, respectively, are expressed most abundantly in EBV latently infected cells at 107 copies per cell (Arrand and Rymo, 1982; Howe and Shu, 1989). EBERs interact with a variety of RNA-binding proteins and optimize B-cell transformation (Yajima et al., 2005; Fok et al., 2006). However, the exact role of EBERs is still unknown (Munz, 2019). EBV also encodes at least 49 mature miRNAs. Of these 44 are located within the intronic regions of BamHI-A rightward transcripts (BART miRNAs), and the remaining five are located in the BamHI-H rightward fragment 1 (BHRF1 miRNAs) (Klinke et al., 2014; Iizasa et al., 2020).
BamHI-H rightward fragment 1 miRNAs are expressed during EBV latency type III and lytic infections. These miRNAs inhibit apoptosis and promote cell cycle progression and proliferation to induce early-phase infection in B cells (Seto et al., 2010). In addition, BHRF1 miRNAs regulate time-restricted expression of the BHRF1 protein (viral BCL-2 homolog) to optimize B-cell transformation (Bernhardt et al., 2016).
BamHI-A rightward transcript miRNAs are expressed in latency types 0, I, II, and III (Amoroso et al., 2011). Their expression is regulated by both the immediate-early protein BZLF1 and feedback loops that involve the host transcription factor NF-κB (Dreyfus, 2017). BART miRNAs are mainly categorized into clusters 1 and 2 (Figure 1; Klinke et al., 2014), which encode eight and 13 pri-miRNAs, respectively. BART miRNAs target host genes and modulate cell differentiation, proliferation, apoptosis, and the cell cycle to establish infection and produce progeny viruses (Klinke et al., 2014). For, example, miR-BART1-3p and miR-BART16 target CASP3 and inhibit apoptosis (Vereide et al., 2014). BIM, a pro-apoptotic protein of the bcl2 family, is targeted by several BART miRNAs (Marquitz et al., 2011). Another BART miRNA (miR-BART5-5p) targets PUMA to promote survival of host cells (Choy et al., 2008). BART miRNAs also help establish EBV infection and transformation by modulating viral and host functions in B cells. Both miR-BART1-3p and miR-BART1-5p suppress adaptive immunity mediated by CD4+ T cells by targeting IL12B and LY75 (Skalsky et al., 2012; Vereide et al., 2014; Tagawa et al., 2016). CD8+ T cell responses are also modulated by miR-BART1-3p and miR-BART17-5p which target IFI30 and TAP2, respectively (Albanese et al., 2016). Thus, BART miRNAs enhance lymphomagenesis. Similarly, BART miRNAs potentiate tumorigenesis in epithelial cells (Cai et al., 2015; Kanda et al., 2015; Qiu et al., 2015; Yang et al., 2017). The notion that BART miRNAs positively regulate oncogenesis is generally accepted.
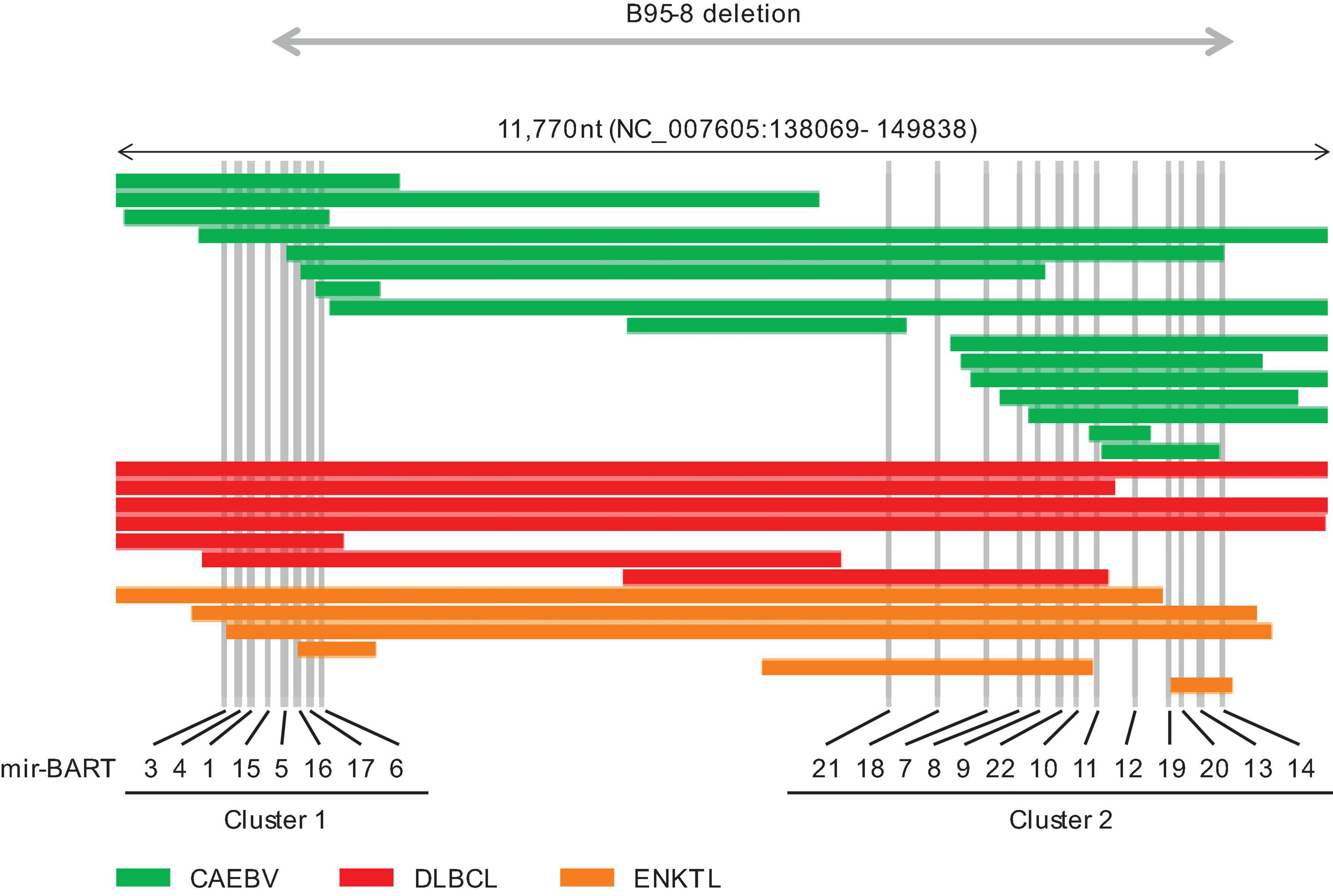
Figure 1. Distribution of intragenic deletions in BamHI-A rightward transcript (BART) microRNA clusters 1 and 2. CAEBV; chronic active EBV disease of T/NK type, DLBCL; diffuse large B cell lymphoma, not otherwise specified, ENKTL; extranodal NK/T cell lymphoma, nasal type.
In DNA viruses such as herpesviruses, exosomes released from virus-infected cells contain virus-derived components that contribute to the growth of the virus itself and the establishment of viral infections by regulating the host’s immune system (Ansari et al., 2013). LMP1, which is an oncoprotein bound to cytoplasmic membranes, is incorporated into exosomes, and plays a role in cancer progression (Nanbo et al., 2013; Sato et al., 2017). EBV miRNAs are also released from infected cells via exosomes to regulate uninfected adjacent cells and promote their growth, which contributes to tumorigenesis (Pegtel et al., 2010; Higuchi et al., 2018; Nanbo et al., 2018; Nkosi et al., 2020).
Deletion of BART miRNA in EBV-Related Lymphomas
It has been reported that the lack of certain genes in human T-cell leukemia virus type 1, human papillomavirus, and Merkel cell polyomavirus results in increased tumorigenicity (Narisawa-Saito and Kiyono, 2007; Moore and Chang, 2010; Kataoka et al., 2015). In contrast, only a few studies have reported specific gene deletions in EBV-associated malignancies (Alfieri and Joncas, 1987; Palser et al., 2015; Mine et al., 2017). However, in this next-generation sequencing era, whole viral genomes are easily sequenced directly from patient samples. We performed whole EBV sequencing by the hybrid capture method using 17,237 probes covering the entire EBV genome in EBV-infected peripheral blood and tumor tissues from patients with various EBV-associated diseases (Okuno et al., 2019; Mabuchi et al., 2021). Interestingly, 22 of 77 cases (35%) of CAEBV, which is a T- or NK-cell lymphoproliferative disease (Kimura et al., 2012; Kimura and Cohen, 2017; Quintanilla-Martinez et al., 2017), had a deletion of 73–49,847 bases in the EBV genome. A similar deletion was found in ENKTL (43%) and EBV-positive DLBCL (71%), which are lymphomas of NK- and B-cell origins, respectively. However, this intragenic deletion was not observed in infectious mononucleosis or post-transplant lymphoma, suggesting that this deletion is a common phenomenon in certain types of EBV-positive lymphoma. The rarity of the intragenic deletion has been reported in healthy individuals and infectious mononucleosis patients (Palser et al., 2015; Yajima et al., 2021).
In addition, EBV deletions were concentrated in the BART miRNA cluster regions (Figure 1). The most frequently deleted miRNAs were mir-BART6-5p and mir-BART6-3p, both of which negatively regulate the EBV immediate-early genes BZLF1 and BRLF1 (Iizasa et al., 2010). mir-BART18-5p and mir-BART20-5p also downregulate these immediate-early genes (Jung et al., 2014; Qiu and Thorley-Lawson, 2014). Mutated EBV lacking this region enhances lymphoma formation by inducing BZLF1 expression in xenograft models (Lin et al., 2015). Elevated BZLF1 due to loss of BART miRNAs may cause abortive lytic infection and promote lymphomagenesis (Munz, 2019).
In our genetic analysis, a group of lytic infection-related genes was also frequently missing outside the BART miRNA clusters, including core replication genes, which are essential for viral replication. We generated a mutant EBV strain lacking BALF5 (viral DNA polymerase catalytic subunit) (Narita et al., 2015) and used it to establish an LCL, which was then transplanted into immunodeficient mice (Okuno et al., 2019). The BALF5-deficient EBV produced lymphoma more frequently compared with the wild-type strain. Additionally, the BALF5-deficient LCL enhanced immediate-early/early gene expression, compared with the wild-type strain. These results suggest that deletion of BALF5, which is a core replication gene, induces the expression of lytic infection-related genes triggered by the immediate-early gene BZLF1 and promotes lymphoma formation. Multiple lytic infection genes, such as BNRF1, BGLF5, and BALF3, are involved in host genome instability (Manners et al., 2018; Xiong et al., 2020), and BHRF1 (viral BCL-2 homolog) and BCRF1 (viral interleukin-10 homolog) also promote cell proliferation (Xu et al., 2001; Zuo et al., 2011). EBV lacking core replication genes cannot produce virus particles or complete a lytic infection, but they can induce an abortive lytic infection and promote oncolytic infection by expressing a viral lytic infection gene (Munz, 2019; Murata et al., 2020). Thus, defective EBV strains may have some advantages during lymphomagenesis.
Interestingly, B95-8, which is the most potent laboratory strain, lacks most of the BART region (Figure 1; Baer et al., 1984; Klinke et al., 2014). Similar large deletions, including BART miRNA clusters, have been reported in patients with ENKTL and Hodgkin lymphoma (Peng et al., 2018; Kawatsuki et al., 2020). However, these defective viruses are relatively rarely associated with epithelial tumors (Cancer Genome Atlas Research Network (CGARN), 2014; Lin et al., 2014). The roles of BART miRNAs may differ between lymphoid and epithelial malignancies. The expression patterns of BART miRNAs depend on the infected cell lineage, and their expression levels vary widely among tumor types, with a 13-fold-increase in nasopharyngeal carcinoma and eightfold increase in gastric cancer relative to LCL and Burkitt lymphoma (Qiu et al., 2011).
Although intragenic deletions involving BART miRNAs have been detected in one-third of CAEBV patients, high expression levels of BART miRNAs are also seen in other patients with CAEBV (Kawano et al., 2013). Furthermore, Higuchi et al. (2018) reported that patients with EBV-positive DLBCL can be stratified according to the miR-BART13 expression level. Moreover, high expression of miR-BART13 was linked to shorter survival time (Higuchi et al., 2018). It is possible that BART miRNAs can contribute to disease progression by suppressing lytic EBV replication and host immune responses. They could also play a role in resistance to DLBCL therapies. On the other hand, loss of BART miRNA expression may also play a pivotal role in lymphoma formation during the early stages (Murata et al., 2020). In other words, cells harboring EBV miRNA deletions may have some advantages during lymphomagenesis as they induce an abortive infection leading to the immortalization of infected cells (Munz, 2019; Murata et al., 2020), at least in certain types of lymphomas. Once lymphoma develops, the loss of BART miRNA expression might not affect its progression and prognoses. Indeed, the overall survival time did not differ between CAEBV patients with and those without intragenic EBV deletions (Okuno et al., 2019).
Conclusion
Epstein–Barr virus is a large DNA virus that encodes >80 genes and nearly 50 non-coding RNAs. Each non-coding RNA performs multiple and different functions, and their expression is regulated differentially according to cell type and micro-environment. EBV miRNAs have various functions and play pivotal roles in oncogenesis. It is clear that some viral miRNAs suppress lymphomagenesis, and that deletion of these miRNAs promotes lymphoma formation. Further research is necessary to elucidate the full roles of EBV miRNAs in tumorigenesis.
Author Contributions
All authors contributed to the concept development process and to the writing and review of the manuscript and also gave final approval of the version to be published.
Funding
This work was supported by grants to HK from Society for the Promotion of Science (19K22560 and 20H03493), the Ministry of Health, Labor, and Welfare of Japan (20FC1004), and the Uehara Memorial Foundation, and the Chemo-Sero-Therapeutic Research Institute.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Albanese, M., Tagawa, T., Bouvet, M., Maliqi, L., Lutter, D., Hoser, J., et al. (2016). Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells. Proc. Natl. Acad. Sci. U.S.A. 113, E6467–E6475. doi: 10.1073/pnas.1605884113
Alfieri, C., and Joncas, J. H. (1987). Biomolecular analysis of a defective nontransforming Epstein-Barr virus (EBV) from a patient with chronic active EBV infection. J. Virol. 61, 3306–3309.
Amoroso, R., Fitzsimmons, L., Thomas, W. A., Kelly, G. L., Rowe, M., and Bell, A. I. (2011). Quantitative studies of Epstein-Barr virus-encoded microRNAs provide novel insights into their regulation. J. Virol. 85, 996–1010. doi: 10.1128/JVI.01528-10
Ansari, M. A., Singh, V. V., Dutta, S., Veettil, M. V., Dutta, D., Chikoti, L., et al. (2013). Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells. J. Virol. 87, 8606–8623. doi: 10.1128/JVI.00805-13
Arrand, J. R., and Rymo, L. (1982). Characterization of the major Epstein-Barr virus-specific RNA in Burkitt lymphoma-derived cells. J. Virol. 41, 376–389. doi: 10.1128/JVI.41.2.376-389.1982
Baer, R., Bankier, A. T., Biggin, M. D., Deininger, P. L., Farrell, P. J., Gibson, T. J., et al. (1984). DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310, 207–211.
Bernhardt, K., Haar, J., Tsai, M. H., Poirey, R., Feederle, R., and Delecluse, H. J. (2016). A viral microRNA cluster regulates the expression of PTEN, p27 and of a bcl-2 homolog. PLoS Pathog. 12:e1005405. doi: 10.1371/journal.ppat.1005405
Cai, L. M., Lyu, X. M., Luo, W. R., Cui, X. F., Ye, Y. F., Yuan, C. C., et al. (2015). EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN. Oncogene 34, 2156–2166. doi: 10.1038/onc.2014.341
Cai, X., Schafer, A., Lu, S., Bilello, J. P., Desrosiers, R. C., Edwards, R., et al. (2006). Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. doi: 10.1371/journal.ppat.0020023
Cancer Genome Atlas Research Network (CGARN) (2014). Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209. doi: 10.1038/nature13480
Chan, J. K. C., Quintanilla-Martinez, L., and Ferry, J. A. (2017). “Extranodal NK/T-cell lymphoma, nasal type,” in WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, revised 4th Edn, eds S. H. Swerdlow, E. Campo, N. L. Harris, E. S. Jaffe, S. A. Pileri, H. Stein, et al. (Lyon: WHO IARC Press), 368–371.
Chen, Y., Fachko, D., Ivanov, N. S., Skinner, C. M., and Skalsky, R. L. (2019). Epstein-Barr virus microRNAs regulate B cell receptor signal transduction and lytic reactivation. PLoS Pathog. 15:e1007535. doi: 10.1371/journal.ppat.1007535
Choy, E. Y., Siu, K. L., Kok, K. H., Lung, R. W., Tsang, C. M., To, K. F., et al. (2008). An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 205, 2551–2560. doi: 10.1084/jem.20072581
Cohen, J. I., Iwatsuki, K., Ko, Y. H., Kimura, H., Manoli, I., Ohshima, K., et al. (2020). Epstein-Barr virus NK and T cell lymphoproliferative disease: report of a 2018 international meeting. Leuk Lymphoma 61, 808–819. doi: 10.1080/10428194.2019.1699080
Cohen, M., Vistarop, A. G., Huaman, F., Narbaitz, M., Metrebian, F., De Matteo, E., et al. (2018). Epstein-Barr virus lytic cycle involvement in diffuse large B cell lymphoma. Hematol. Oncol. 36, 98–103. doi: 10.1002/hon.2465
Dreyfus, D. H. (2017). Genetics and molecular biology of Epstein-Barr Virus-Encoded BART MicroRNA: a paradigm for viral modulation of host immune response genes and genome stability. J. Immunol. Res. 2017:4758539. doi: 10.1155/2017/4758539
Epstein, M. A., Achong, B. G., and Barr, Y. M. (1964). Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet 7335, 702–703. doi: 10.1016/s0140-6736(64)91524-7
Fink, S. E., Gandhi, M. K., Nourse, J. P., Keane, C., Jones, K., Crooks, P., et al. (2014). A comprehensive analysis of the cellular and EBV-specific microRNAome in primary CNS PTLD identifies different patterns among EBV-associated tumors. Am. J. Transplant 14, 2577–2587. doi: 10.1111/ajt.12858
Fok, V., Friend, K., and Steitz, J. A. (2006). Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J. Cell. Biol. 173, 319–325. doi: 10.1083/jcb.200601026
Haneklaus, M., Gerlic, M., Kurowska-Stolarska, M., Rainey, A. A., Pich, D., McInnes, I. B., et al. (2012). Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J. Immunol. 189, 3795–3799. doi: 10.4049/jimmunol.1200312
Higuchi, H., Yamakawa, N., Imadome, K. I., Yahata, T., Kotaki, R., Ogata, J., et al. (2018). Role of exosomes as a proinflammatory mediator in the development of EBV-associated lymphoma. Blood 131, 2552–2567. doi: 10.1182/blood-2017-07-794529
Hooykaas, M. J. G., van Gent, M., Soppe, J. A., Kruse, E., Boer, I. G. J., van Leenen, D., et al. (2017). EBV microrna BART16 suppresses Type I IFN signaling. J. Immunol. 198, 4062–4073. doi: 10.4049/jimmunol.1501605
Howe, J. G., and Shu, M. D. (1989). Epstein-Barr virus small RNA (EBER) genes: unique transcription units that combine RNA polymerase II and III promoter elements. Cell 57, 825–834. doi: 10.1016/0092-8674(89)90797-6
Huang, W. T., and Lin, C. W. (2014). EBV-encoded miR-BART20-5p and miR-BART8 inhibit the IFN-gamma-STAT1 pathway associated with disease progression in nasal NK-cell lymphoma. Am. J. Pathol. 184, 1185–1197. doi: 10.1016/j.ajpath.2013.12.024
Iizasa, H., Kim, H., Kartika, A. V., Kanehiro, Y., and Yoshiyama, H. (2020). Role of viral and host microRNAs in immune regulation of Epstein-Barr virus-associated diseases. Front. Immunol. 11:367. doi: 10.3389/fimmu.2020.00367
Iizasa, H., Wulff, B. E., Alla, N. R., Maragkakis, M., Megraw, M., Hatzigeorgiou, A., et al. (2010). Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J. Biol. Chem. 285, 33358–33370. doi: 10.1074/jbc.M110.138362
Jung, Y. J., Choi, H., Kim, H., and Lee, S. K. (2014). MicroRNA miR-BART20-5p stabilizes Epstein-Barr virus latency by directly targeting BZLF1 and BRLF1. J. Virol. 88, 9027–9037. doi: 10.1128/JVI.00721-14
Kanda, T. (2018). EBV-encoded latent genes. Adv. Exp. Med. Biol. 1045, 377–394. doi: 10.1007/978-981-10-7230-7_17
Kanda, T., Miyata, M., Kano, M., Kondo, S., Yoshizaki, T., and Iizasa, H. (2015). Clustered microRNAs of the Epstein-Barr virus cooperatively downregulate an epithelial cell-specific metastasis suppressor. J. Virol. 89, 2684–2697. doi: 10.1128/JVI.03189-14
Kataoka, K., Nagata, Y., Kitanaka, A., Shiraishi, Y., Shimamura, T., Yasunaga, J., et al. (2015). Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat. Genet. 47, 1304–1315. doi: 10.1038/ng.3415
Kawano, Y., Iwata, S., Kawada, J., Gotoh, K., Suzuki, M., Torii, Y., et al. (2013). Plasma viral MicroRNA profiles reveal potential biomarkers for chronic active Epstein-Barr virus infection. J. Infect. Dis. 208, 771–779. doi: 10.1093/infdis/jit222
Kawatsuki, A., Igawa, T., Urata, T., Tanaka, T., Sato, Y., and Yoshino, T. (2020). Deletion of BART miRNA-encoding cluster in Epstein-Barr virus DNA in classic Hodgkin lymphoma. Pathol. Int. 70, 1032–1033. doi: 10.1111/pin.13022
Kimura, H. (2018). EBV in T-/NK-Cell Tumorigenesis. Adv. Exp. Med. Biol. 1045, 459–475. doi: 10.1007/978-981-10-7230-7_21
Kimura, H., and Cohen, J. I. (2017). Chronic active Epstein-Barr virus disease. Front. Immunol. 8:1867. doi: 10.3389/fimmu.2017.01867
Kimura, H., Ito, Y., Kawabe, S., Gotoh, K., Takahashi, Y., Kojima, S., et al. (2012). EBV-associated T/NK-cell lymphoproliferative diseases in nonimmunocompromised hosts: prospective analysis of 108 cases. Blood 119, 673–686. doi: 10.1182/blood-2011-10-381921
Klinke, O., Feederle, R., and Delecluse, H. J. (2014). Genetics of Epstein-Barr virus microRNAs. Semin. Cancer Biol. 26, 52–59. doi: 10.1016/j.semcancer.2014.02.002
Kuzembayeva, M., Hayes, M., and Sugden, B. (2014). Multiple functions are mediated by the miRNAs of Epstein-Barr virus. Curr. Opin. Virol. 7, 61–65. doi: 10.1016/j.coviro.2014.04.003
Lin, D. C., Meng, X., Hazawa, M., Nagata, Y., Varela, A. M., Xu, L., et al. (2014). The genomic landscape of nasopharyngeal carcinoma. Nat. Genet. 46, 866–871. doi: 10.1038/ng.3006
Lin, X., Tsai, M. H., Shumilov, A., Poirey, R., Bannert, H., Middeldorp, J. M., et al. (2015). The Epstein-Barr virus BART miRNA cluster of the M81 strain modulates multiple functions in primary B cells. PLoS Pathog. 11:e1005344. doi: 10.1371/journal.ppat.1005344
Longnecker, R., Kieff, E., and Cohen, J. I. (2013). “Epstein-Barr virus,” in Fields Virology, 6 Edn, eds D. M. Knipe and P. M. Howley (Philadelphia: Lippincott Williams & Willkins), 1898–1959.
Ma, S. D., Hegde, S., Young, K. H., Sullivan, R., Rajesh, D., Zhou, Y., et al. (2011). A new model of Epstein-Barr virus infection reveals an important role for early lytic viral protein expression in the development of lymphomas. J. Virol. 85, 165–177. doi: 10.1128/JVI.01512-10
Mabuchi, S., Hijioka, F., Watanabe, T., Yanagi, Y., Okuno, Y., Masud, H., et al. (2021). Role of Epstein-Barr virus C promoter deletion in diffuse large B cell lymphoma. Cancers (Basel) 13:561. doi: 10.3390/cancers13030561
Manners, O., Murphy, J. C., Coleman, A., Hughes, D. J., and Whitehouse, A. (2018). Contribution of the KSHV and EBV lytic cycles to tumourigenesis. Curr. Opin. Virol. 32, 60–70. doi: 10.1016/j.coviro.2018.08.014
Marquitz, A. R., Mathur, A., Nam, C. S., and Raab-Traub, N. (2011). The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology 412, 392–400. doi: 10.1016/j.virol.2011.01.028
Mine, S., Hishima, T., Suganuma, A., Fukumoto, H., Sato, Y., Kataoka, M., et al. (2017). Interleukin-6-dependent growth in a newly established plasmablastic lymphoma cell line and its therapeutic targets. Sci. Rep. 7:10188. doi: 10.1038/s41598-017-10684-5
Moore, P. S., and Chang, Y. (2010). Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer 10, 878–889. doi: 10.1038/nrc2961
Munz, C. (2019). Latency and lytic replication in Epstein-Barr virus-associated oncogenesis. Nat. Rev. Microbiol. 17, 691–700. doi: 10.1038/s41579-019-0249-7
Murata, T., Okuno, Y., Sato, Y., Watanabe, T., and Kimura, H. (2020). Oncogenesis of CAEBV revealed: intragenic deletions in the viral genome and leaky expression of lytic genes. Rev. Med. Virol. 30:e2095. doi: 0.1002/rmv.2095
Murata, T., Sato, Y., and Kimura, H. (2014). Modes of infection and oncogenesis by the Epstein-Barr virus. Rev. Med. Virol. 24, 242–253. doi: 10.1002/rmv.1786
Nanbo, A., Katano, H., Kataoka, M., Hoshina, S., Sekizuka, T., Kuroda, M., et al. (2018). Infection of Epstein(-)Barr Virus in Type III latency modulates biogenesis of exosomes and the expression profile of exosomal miRNAs in the Burkitt lymphoma mutu cell lines. Cancers (Basel) 10:237. doi: 10.3390/cancers10070237
Nanbo, A., Kawanishi, E., Yoshida, R., and Yoshiyama, H. (2013). Exosomes derived from Epstein-Barr virus-infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells. J. Virol. 87, 10334–10347. doi: 10.1128/JVI.01310-13
Narisawa-Saito, M., and Kiyono, T. (2007). Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: roles of E6 and E7 proteins. Cancer Sci. 98, 1505–1511. doi: 10.1111/j.1349-7006.2007.00546.x
Narita, Y., Sugimoto, A., Kawashima, D., Watanabe, T., Kanda, T., Kimura, H., et al. (2015). A Herpesvirus specific motif of Epstein-Barr Virus DNA polymerase is required for the efficient lytic genome synthesis. Sci. Rep. 5:11767. doi: 10.1038/srep11767
Nkosi, D., Sun, L., Duke, L. C., Patel, N., Surapaneni, S. K., Singh, M., et al. (2020). Epstein-Barr Virus LMP1 promotes Syntenin-1- and Hrs-Induced extracellular vesicle formation for its own secretion to increase cell proliferation and migration. mBio 11, e589–e520. doi: 10.1128/mBio.00589-20
Okuno, Y., Murata, T., Sato, Y., Muramatsu, H., Ito, Y., Watanabe, T., et al. (2019). Defective Epstein-Barr virus in chronic active infection and haematological malignancy. Nat. Microbiol. 4, 404–413. doi: 10.1038/s41564-018-0334-0
Palser, A. L., Grayson, N. E., White, R. E., Corton, C., Correia, S., Ba Abdullah, M. M., et al. (2015). Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J. Virol. 89, 5222–5237. doi: 10.1128/JVI.03614-14
Pegtel, D. M., Cosmopoulos, K., Thorley-Lawson, D. A., van Eijndhoven, M. A., Hopmans, E. S., Lindenberg, J. L., et al. (2010). Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U.S.A. 107, 6328–6333. doi: 10.1073/pnas.0914843107
Peng, R. J., Han, B. W., Cai, Q. Q., Zuo, X. Y., Xia, T., Chen, J. R., et al. (2018). Genomic and transcriptomic landscapes of Epstein-Barr virus in extranodal natural killer T-cell lymphoma. Leukemia 33, 1451–1462. doi: 10.1038/s41375-018-0324-5
Qiu, J., Cosmopoulos, K., Pegtel, M., Hopmans, E., Murray, P., Middeldorp, J., et al. (2011). A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog. 7:e1002193. doi: 10.1371/journal.ppat.1002193
Qiu, J., Smith, P., Leahy, L., and Thorley-Lawson, D. A. (2015). The Epstein-Barr virus encoded BART miRNAs potentiate tumor growth in vivo. PLoS Pathog. 11:e1004561. doi: 10.1371/journal.ppat.1004561
Qiu, J., and Thorley-Lawson, D. A. (2014). EBV microRNA BART 18-5p targets MAP3K2 to facilitate persistence in vivo by inhibiting viral replication in B cells. Proc. Natl. Acad. Sci. U.S.A. 111, 11157–11162. doi: 10.1073/pnas.1406136111
Quintanilla-Martinez, L., Ko, Y. H., Kimura, H., and Jaffe, E. S. (2017). “EBV–positive T-cell and NK-cell lymphoproliferative diseases of childhood,” in WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, revised 4th Edn, eds S. H. Swerdlow, E. Campo, N. L. Harris, E. S. Jaffe, S. A. Pileri, H. Stein, et al. (Lyon: WHO IARC Press), 355–362.
Sakamoto, K., Sekizuka, T., Uehara, T., Hishima, T., Mine, S., Fukumoto, H., et al. (2017). Next-generation sequencing of miRNAs in clinical samples of Epstein-Barr virus-associated B-cell lymphomas. Cancer Med. 6, 605–618. doi: 10.1002/cam4.1006
Sato, Y., Ochiai, S., Murata, T., Kanda, T., Goshima, F., and Kimura, H. (2017). Elimination of LMP1-expressing cells from a monolayer of gastric cancer AGS cells. Oncotarget 8, 39345–39355. doi: 10.18632/oncotarget.16996
Seto, E., Moosmann, A., Gromminger, S., Walz, N., Grundhoff, A., and Hammerschmidt, W. (2010). Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 6:e1001063. doi: 10.1371/journal.ppat.1001063
Skalsky, R. L., Corcoran, D. L., Gottwein, E., Frank, C. L., Kang, D., Hafner, M., et al. (2012). The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 8:e1002484. doi: 10.1371/journal.ppat.1002484
Swerdlow, S. H., Campo, E., Pileri, S. A., Harris, N. L., Stein, H., Siebert, R., et al. (2016). The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 127, 2375–2390. doi: 10.1182/blood-2016-01-643569
Tagawa, T., Albanese, M., Bouvet, M., Moosmann, A., Mautner, J., Heissmeyer, V., et al. (2016). Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing. J. Exp. Med. 213, 2065–2080. doi: 10.1084/jem.20160248
Tomkinson, B., Robertson, E., and Kieff, E. (1993). Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67, 2014–2025. doi: 10.1128/JVI.67.4.2014-2025.1993
van Eijndhoven, M. A., Zijlstra, J. M., Groenewegen, N. J., Drees, E. E., van Niele, S., Baglio, S. R., et al. (2016). Plasma vesicle miRNAs for therapy response monitoring in Hodgkin lymphoma patients. JCI Insight 1:e89631. doi: 10.1172/jci.insight.89631
Vereide, D. T., Seto, E., Chiu, Y. F., Hayes, M., Tagawa, T., Grundhoff, A., et al. (2014). Epstein-Barr virus maintains lymphomas via its miRNAs. Oncogene 33, 1258–1264. doi: 10.1038/onc.2013.71
Xia, T., O’Hara, A., Araujo, I., Barreto, J., Carvalho, E., Sapucaia, J. B., et al. (2008). EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 68, 1436–1442. doi: 10.1158/0008-5472.CAN-07-5126
Xiong, J., Cui, B. W., Wang, N., Dai, Y. T., Zhang, H., Wang, C. F., et al. (2020). Genomic and transcriptomic characterization of natural killer T cell lymphoma. Cancer Cell. 37, 403.e6–419.e6. doi: 10.1016/j.ccell.2020.02.005
Xu, Z. G., Iwatsuki, K., Oyama, N., Ohtsuka, M., Satoh, M., Kikuchi, S., et al. (2001). The latency pattern of Epstein-Barr virus infection and viral IL-10 expression in cutaneous natural killer/T-cell lymphomas. Br. J. Cancer 84, 920–925.
Yajima, M., Kakuta, R., Saito, Y., Kitaya, S., Toyoda, A., Ikuta, K., et al. (2021). A global phylogenetic analysis of Japanese tonsil-derived Epstein-Barr virus strains using viral whole-genome cloning and long-read sequencing. J. Gen. Virol. 102:001549. doi: 10.1099/jgv.0.001549
Yajima, M., Kanda, T., and Takada, K. (2005). Critical role of Epstein-Barr Virus (EBV)-encoded RNA in efficient EBV-induced B-lymphocyte growth transformation. J. Virol. 79, 4298–4307.
Yamamoto, T., Hirai, Y., Miyake, T., Hamada, T., Yamasaki, O., Morizane, S., et al. (2016). Epstein-Barr virus reactivation is induced, but abortive, in cutaneous lesions of systemic hydroa vacciniforme and hypersensitivity to mosquito bites. J. Dermatol. Sci. 82, 153–159. doi: 10.1016/j.jdermsci.2016.03.001
Yang, Y. C., Liem, A., Lambert, P. F., and Sugden, B. (2017). Dissecting the regulation of EBV’s BART miRNAs in carcinomas. Virology 505, 148–154. doi: 10.1016/j.virol.2017.02.013
Keywords: EBV, microRNA, BART, lymphomagenesis, CAEBV, ENKTL, diffuse large B cell lymphoma
Citation: Kimura H, Okuno Y, Sato Y, Watanabe T and Murata T (2021) Deletion of Viral microRNAs in the Oncogenesis of Epstein–Barr Virus-Associated Lymphoma. Front. Microbiol. 12:667968. doi: 10.3389/fmicb.2021.667968
Received: 15 February 2021; Accepted: 08 June 2021;
Published: 08 July 2021.
Edited by:
Bill Sugden, University of Wisconsin-Madison, United StatesReviewed by:
Yonggang Pei, University of Pennsylvania, United StatesChi-Ju Chen, National Yang-Ming University, Taiwan
Copyright © 2021 Kimura, Okuno, Sato, Watanabe and Murata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hiroshi Kimura, aGtpbXVyYUBtZWQubmFnb3lhLXUuYWMuanA=
 Hiroshi Kimura
Hiroshi Kimura Yusuke Okuno2
Yusuke Okuno2 Yoshitaka Sato
Yoshitaka Sato Takahiro Watanabe
Takahiro Watanabe Takayuki Murata
Takayuki Murata