- 1School of Life Sciences, Arizona State University, Tempe, AZ, United States
- 2School of Marine and Atmospheric Sciences, Stony Brook University, Stony Brook, NY, United States
- 3Department of Earth Sciences, University of Southern California, Los Angeles, CA, United States
- 4Department of Marine Biology, Leon H. Charney School of Marine Sciences, University of Haifa, Haifa, Israel
- 5College of Veterinary Medicine, Western University of Health Sciences, Pomona, CA, United States
- 6Department of Microbiology, Technical University of Kaiserslautern, Kaiserslautern, Germany
- 7Biosciences & Biotechnology Division, Lawrence Livermore National Laboratory, Livermore, CA, United States
Microbial iron cycling influences the flux of major nutrients in the environment (e.g., through the adsorptive capacity of iron oxides) and includes biotically induced iron oxidation and reduction processes. The ecological extent of microbial iron cycling is not well understood, even with increased sequencing efforts, in part due to limitations in gene annotation pipelines and limitations in experimental studies linking phenotype to genotype. This is particularly true for the marine subseafloor, which remains undersampled, but represents the largest contiguous habitat on Earth. To address this limitation, we used FeGenie, a database and bioinformatics tool that identifies microbial iron cycling genes and enables the development of testable hypotheses on the biogeochemical cycling of iron. Herein, we survey the microbial iron cycle in diverse subseafloor habitats, including sediment-buried crustal aquifers, as well as surficial and deep sediments. We inferred the genetic potential for iron redox cycling in 32 of the 46 metagenomes included in our analysis, demonstrating the prevalence of these activities across underexplored subseafloor ecosystems. We show that while some processes (e.g., iron uptake and storage, siderophore transport potential, and iron gene regulation) are near-universal, others (e.g., iron reduction/oxidation, siderophore synthesis, and magnetosome formation) are dependent on local redox and nutrient status. Additionally, we detected niche-specific differences in strategies used for dissimilatory iron reduction, suggesting that geochemical constraints likely play an important role in dictating the dominant mechanisms for iron cycling. Overall, our survey advances the known distribution, magnitude, and potential ecological impact of microbe-mediated iron cycling and utilization in sub-benthic ecosystems.
Introduction
Iron is the dominant redox-active element in the Earth’s crust and an important nutrient for almost all known life. In many environments, iron cycling is intimately linked to biogeochemical cycling of other elements, including carbon (e.g., CO2, CH4, and organic carbon), nitrogen (Laufer et al., 2016b; McAllister et al., 2020b), and heavy metals (Cooper et al., 2006). Thus, even though biologically available iron is comparatively rare/transient in many ecosystems, its speciation and flux considerably impacts the overall activities and productivity of diverse ecosystems. Research on the genetic basis for microbial iron cycling is in its infancy, and annotation pipelines annotate genes related to iron oxidation or reduction as “hypothetical” or simply “cytochrome c.” Accordingly, the extent of information that can be gained from metagenomes or metagenome-assembled genomes (MAGs), derived from shotgun sequencing, in relation to the potential for microbial iron redox cycling in the environment remains poorly constrained. This is particularly true of the marine subsurface, an extremely remote and difficult to access/sample environment, that is nevertheless significantly influenced by microbe-mineral interactions, particularly those related to iron oxidation and reduction (Edwards et al., 2003a; Roden, 2012).
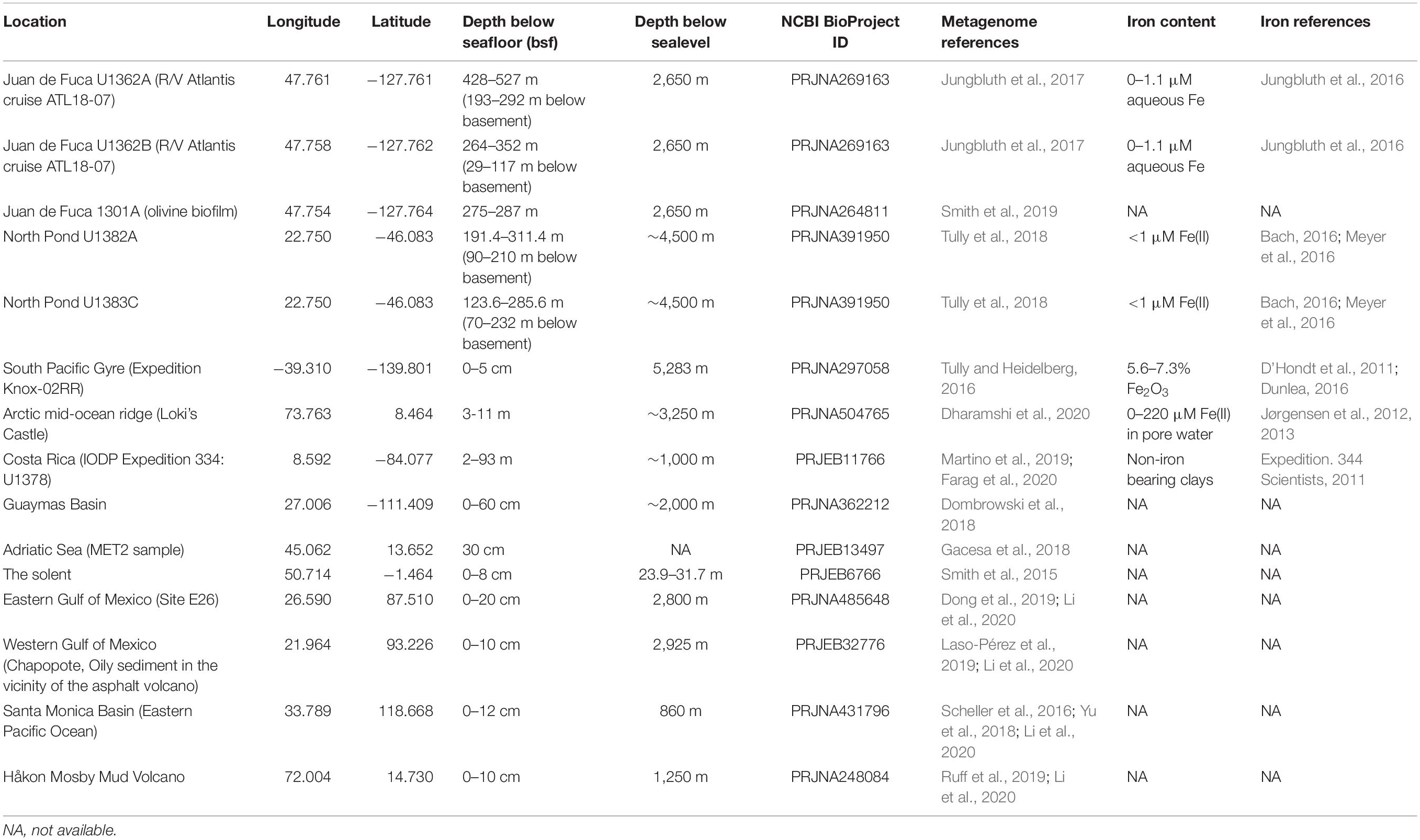
While there have been studies, largely in ecosystems where iron-oxidizing and –reducing bacteria are conspicuously present (Riedinger et al., 2014; Laufer et al., 2016a,b; Beam et al., 2018; Bryce et al., 2018; Aromokeye et al., 2020; McAllister et al., 2020b), the extent of microbial iron cycling in other environments, including marine sediment and sediment-buried crustal aquifers, remains comparatively underexplored. Even though microbes capable of iron redox can form only a small proportion of the community in the latter habitats (e.g., rare biosphere), their influence on iron-cycling has potential to significantly impact the surrounding geochemistry. We previously developed FeGenie (Garber et al., 2020), a database and bioinformatics tool, to aid in annotating the iron redox genes and other genes involved in many aspects of the microbial iron cycle, including iron transport, storage, and regulation; we are also continuously updating the library of iron genes to include more genes and processes, such as siderophore transport and biosynthesis (Neilands, 1995), and fermentative iron reduction (Jones et al., 1984), which will be included in the next release. Herein, we used FeGenie, with an updated set of hidden Markov models (HMMs) for iron redox cycling and iron transport (Supplementary Table 1), to systematically profile the microbial iron cycle in recently published metagenomes representing six marine sediment sites and two sediment-buried crustal aquifer sites (Table 1). These metagenomes, published over the last decade, have recently illuminated the microbial lifestyle under the often harsh conditions imposed by subsurface geochemical regimes. Some of the original analyses of these metagenomes provided evidence for microbial iron oxidation and reduction occurring in the subseafloor, but these conclusions were inferred by using a limited iron redox gene database (Meyer et al., 2016; Tully and Heidelberg, 2016; Tully et al., 2018; Smith et al., 2019). Using FeGenie, we re-analyzed these metagenomes with a focus on iron cycling, using a standardized approach that includes all known genetic markers for dissimilatory iron reduction and oxidation. We note that these metagenomes were generated using a variety of wet-lab and in silico methods, limiting the cross-comparisons that can be carried out. Nonetheless, we highlight the potential for microbial iron cycling in the marine subsurface and demonstrate FeGenie’s capability to provide added valuable insights into iron cycling and acquisition/storage mechanisms in subseafloor habitats.
Materials and Methods
Data Acquisition and Assembly
Metagenome assemblies representing North Pond fluids, which were made available by Tully et al. (2018), were downloaded from figshare (see original publication for figshare link). For the Guaymas Basin metagenome, in lieu of an assembly, we downloaded the 551 MAGs published by Dombrowski et al. (2018), and concatenated the contigs. Thus, due to the great amount of data available from these MAGs, no unbinned fraction from that dataset was evaluated. For all other metagenomic datasets, listed in Table 1, raw metagenome reads were obtained using the SRA Toolkit (release 2.10.0, SRA Toolkit Development Team). Reads were quality trimmed using Trimmomatic v0.36 (minimum length = 36 bp, sliding window = 4 bp, minimum quality score = 15, adaptors used = ILLUMINACLIP:TruSeq3-PE:2:30:10) (Bolger et al., 2014), and assembled using Spades v3.13.0 (default k-mers, Bankevich et al., 2012). Metagenome assemblies were then subjected to FeGenie analysis (Garber et al., 2020). For those metagenomes where metagenome-assembled genomes (MAGs) were publically available (Guaymas Basin, Loki’s Castle, Eastern Gulf of Mexico, Juan de Fuca ridge aquifer fluids, Juan de Fuca ridge olivine biofilms, and North Pond aquifer fluids), those were downloaded and also analyzed with FeGenie.
FeGenie Analysis
We used FeGenie to identify iron genes in metagenome assemblies and MAGs. FeGenie was run with the –meta flag, directing the gene-calling software Prodigal, part of the FeGenie pipeline, to use its metagenomic procedure. Iron redox genes from the FeGenie output files were extracted using a custom python script (Supplementary File 1) and organized into pathways. To identify the closest sequenced relatives of identified iron genes, the protein sequences were extracted from FeGenie and queried (e-value cutoff: 1E-6) against the non-redundant protein database (release 240) from the National Center for Biotechnology Information (NCBI) using DIAMOND v2.0.4.142 (Buchfink et al., 2015). The closest phylogenetic relatives were then inferred from the top 50 DIAMOND matches and summarized in Supplementary File 2.
Siderophore biosynthesis clusters that were identified with FeGenie were confirmed using AntiSMASH v.5 (Blin et al., 2019). Contigs containing putative siderophore cluster genes were extracted using the grep command and subsequently subjected to AntiSMASH analysis (–cb-general, –cb-subclusters, –cb-knownclusters, –clusterhmmer, –asf, all other parameters were default). Results were then visually inspected for confirmation of potential for siderophore synthesis.
The R packages ggplot2 (Wickham et al., 2020), ggdendro (de Vries and Ripley, 2020), and Pvclust (Suzuki and Shimodaira, 2006), were used to generate plots presented in this article.
Site Descriptions
North Pond
The ∼8 Mya North Pond aquifer lies beneath a Mid-Atlantic ridge sediment basin and is composed of basaltic rocks, an important source of iron in this habitat, which are subject to chemical weathering by seawater due to advective fluid flow (Langseth et al., 1992; Meyer et al., 2016). Major solid weathering products include iron and manganese oxides, as well as carbonate minerals. Although classified as an oxic environment, the periodic presence of anaerobes and marker genes for low-O2 metabolisms suggests the occurrence of anaerobic microenvironments and redox oscillations (Tully et al., 2018). The frequency of redox oscillations is likely related to the seawater residence time in fractures, as well as the redox-buffering capacity of the rocks, in which anaerobic zones may occur due to fluid stagnation and consumption of oxygen through reaction with reduced minerals (MacQuarrie and Mayer, 2005; Trinchero et al., 2019). In contrast, intersecting fractures may result in fluid mixing of deep and shallow seawater, leading to microbial hot spots supported by fluctuating O2 concentrations that are ideal for iron-reducing and -oxidizing bacteria, something that has been previously observed in a terrestrial fractured rock aquifer (Bochet et al., 2020).
Juan de Fuca
The ∼3.5 Mya Juan de Fuca (JdF) ridge crustal aquifer is a basalt-hosted habitat located along a mid-ocean ridge flank with hydrothermal fluid circulation (Jungbluth et al., 2016). In comparison to North Pond, the geochemical signature of JdF fluids is more representative of extensive water-rock interactions due to longer fluid residence times and elevated fluid temperatures (Edwards et al., 2012). Seawater recharge occurs slowly due to the impermeability of the surrounding sediment and spreads laterally along fractures. Along the flowpath, fluid is heated and reduced, interacting with the basaltic rock, and is subsequently further altered by diffusive exchange with the overlying sediment pore water. The latter is most intense along the strong redox gradient at the sediment-rock boundary, where fluids are enriched in Fe, Mn, ammonium, Si, and Ca, and depleted in nitrate and oxygen (Wheat et al., 2013). Iron enrichment is particularly substantial and can be approximately 3000-fold higher in concentration relative to seawater and other JdF fluids in the flow path (Wheat et al., 2013).
Marine sediments
The metabolic potential for iron oxidation and reduction in marine sediments, hosting an estimated 0.18–3.6% of Earth’s total living biomass (Kallmeyer et al., 2012), remains understudied. Potential for oxidized iron to serve as an important electron sink and contribute significantly to the breakdown of organic carbon in sediment was recognized as early as 1963 (Kamura et al., 1963; Takai et al., 1963a,b). More recently, it was determined that Zetaproteobacteria are rare within marine sediments (estimated global abundance of 0.11%) but can contribute ∼8 × 1015 g of Fe in sedimentary iron oxides annually (Beam et al., 2018). Here, to survey the genetic capacity for iron cycling across diverse sedimentary regimes, we examine 27 marine sediment metagenomes (from 10 geographical sites, Table 1), including a low productivity oligotrophic site in the South Pacific Gyre, and high productivity sites like the Arctic Mid-Ocean Ridge (Dharamshi et al., 2020), Costa Rica Margin (Martino et al., 2019), and Guaymas Basin (Dombrowski et al., 2018).
Results and Discussion
The Impact of Shifting Redox Conditions on Iron Cycling in the North Pond Aquifer
Recent work, including metagenomic, metatranscriptomic, and colonization/poised-electrode experiments, provide evidence for iron oxidation within the North Pond aquifer. Metagenomic (Tully et al., 2018) and metatranscriptomic (Seyler et al., 2020) studies revealed the presence of genes associated with iron oxidation, specifically, cyc2 [encoding an outer membrane porin-cytochrome fusion; (Appia-Ayme et al., 1998; Castelle et al., 2008; Barco et al., 2015; He et al., 2017, Keffer et al., 2021)] and foxE (encoding a periplasmic cytochrome; Croal et al., 2007; Pereira et al., 2017). Tully et al. (2018) also reconstructed metagenome-assembled genomes (MAGs) affiliated with the iron-oxidizing Zetaproteobacteria, which are known to adapt to fluctuating O2 concentrations and advective flow regimes (Chiu et al., 2017; Blackwell et al., 2020), further solidifying the presence and significant contribution that iron-oxidizing bacteria make to the aquifer community. Recent mineral colonization and current generation on poised electrodes also demonstrated the presence of iron oxidizing bacteria and provided evidence for their ability to utilize insoluble electron acceptors (Jones et al., 2020).
Our survey of 18 metagenome assemblies from the North Pond crustal aquifer (Tully et al., 2018) using the FeGenie library confirmed the presence of previously reported iron oxidases (Figure 1) (Tully et al., 2018; Seyler et al., 2020), which we phylogenetically linked to Zetaproteobacteria (cyc2) and Rhodospirillaceae (foxE) (Supplementary File 2). Linking cyc2 to Zetaproteobacteria is important because this gene is highly diverse and is often encoded by taxa not known to be capable of iron oxidation; thus, only a handful of cyc2 genes have been experimentally shown to be iron oxidases (Castelle et al., 2008; Jeans et al., 2008; Barco et al., 2015; McAllister et al., 2020a; Keffer et al., 2021). Further, we also documented the presence of genes associated with iron reduction [mtrCAB (Pitts et al., 2003; Hartshorne et al., 2007; Edwards et al., 2020) within seven of the timepoints, which span 2 years (Figure 1)]. The mtrCAB genes are most closely related to the dissimilatory iron reducer Shewanella benthica (Supplementary File 2), which was also enriched in mineral colonizations from the North Pond aquifer (Jones et al., 2020). We also identified nine MAGs (Tully et al., 2018), encoding copies of genes linked to respiratory iron oxidation or reduction (Supplementary Table 2). Notably, two MAGs that encode cyc2 belong to the family Mariprofundaceae, known to encompass at least eight isolated strains of iron oxidizers (Emerson and Moyer, 2002; McBeth et al., 2011; Mumford et al., 2016; Barco et al., 2017; Chiu et al., 2017; McAllister et al., 2019). Another MAG, which belongs to the genus Shewanella, encodes two copies of the mtrCAB operon for iron reduction. Other putative iron oxidizers and reducers are shown in Supplementary Table 2. 16S rRNA gene amplicon sequencing presented by Jones et al. (2020), showed that Geobacter-related spp. were also enriched on minerals; however, we did not detect any Geobacter-related gene markers for iron reduction (omcS, omcZ, and type IV aromatic/electroactive pili [t4ap]), suggesting that this lineage may be part of the rare biosphere and undetectable with metagenomic approaches, at least in the aquifer fluids. Additionally, two metagenomes, derived from bottom water sampled near the North Pond aquifer, showed potential for iron oxidation via cyc2, foxE, and sulfocyanin, a blue-copper protein used as a genetic marker for iron oxidation in Archaea (Castelle et al., 2015). However, these sequences were not related to known iron-oxidizing bacteria (e.g., Mariprofundaceae), and no iron reduction genes were detected.
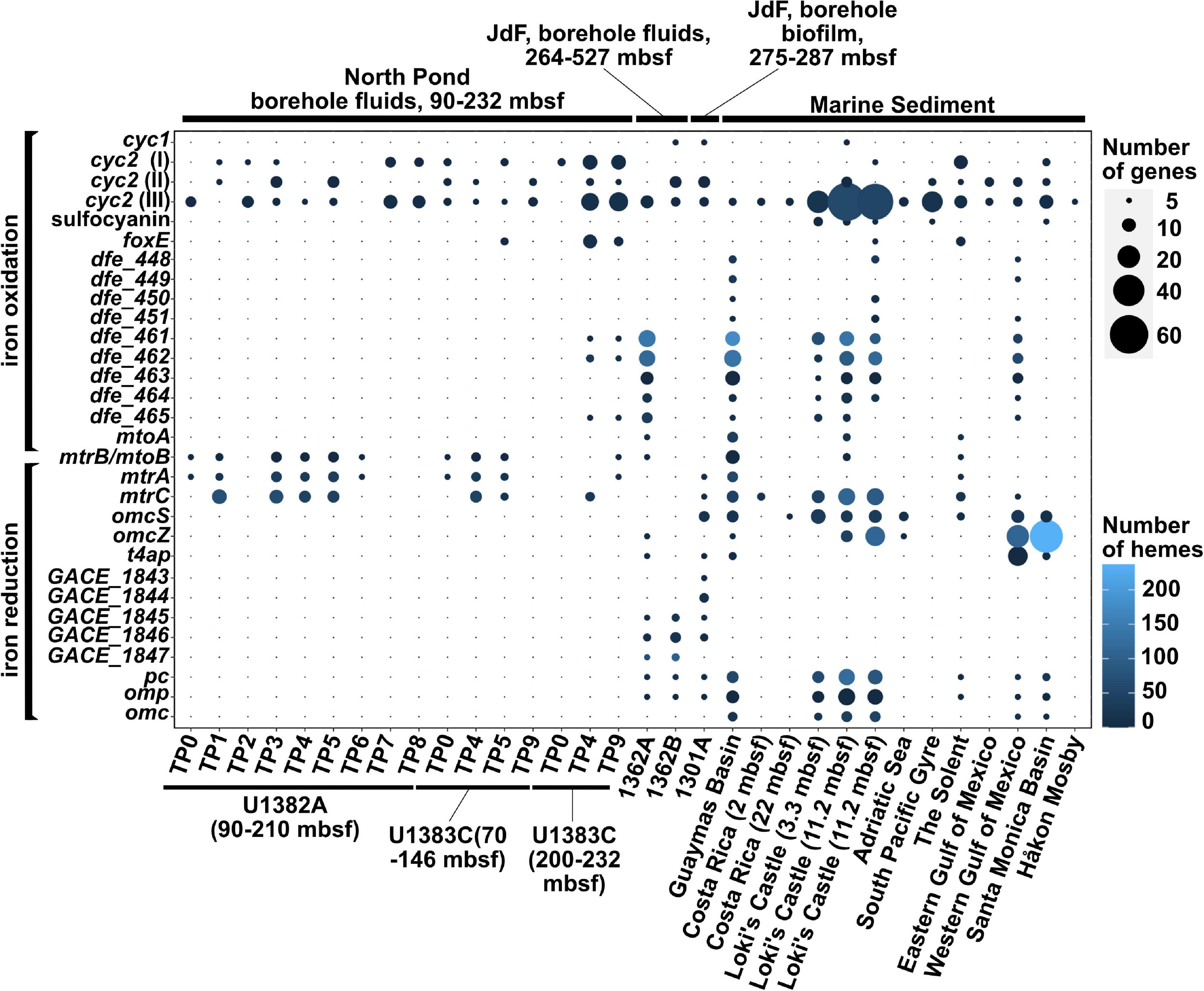
Figure 1. Distribution of iron redox genes among the 32 metagenomes in which potential for iron reduction or oxidation was detected. The gene mtrB/mtoB is deliberately placed in between the iron oxidation and iron reduction categories because it encodes a porin that can be part of both the iron reduction (mtr) or the iron oxidation (mto) pathways.
The co-occurrence of iron oxidizers and reducers in the North Pond aquifer supports potential for coupling of iron redox processes. Mutualistic interactions between iron oxidizers and reducers have been reported in various habitats (Weber et al., 2006; Blöthe and Roden, 2009; Emerson, 2009; Roden et al., 2012; Elliott et al., 2014; Byrne et al., 2015). While the coupling of iron reduction with oxidation may be less apparent in an iron-rich habitat like the Loihi Seamount (Emerson and Moyer, 2002), iron reducers and iron oxidizers may be more dependent on each other’s metabolic by-products in the North Pond aquifer, where dissolved iron and carbon concentrations are much lower. As noted above, the presence of anaerobes within the North Pond aquifer hints at the possibility of sub-oxic microenvironments and varying redox conditions (Tully et al., 2018). This variability may be reflected in the observed fluctuation of iron oxidases and reductases over the 2-year sampling period (Figure 1). Consistent with this, hierarchical clustering of the metagenomes based on the overall iron gene composition (including iron acquisition and storage) demonstrated the dissimilarity between different North Pond timepoints (Figure 2), consistent with the observed temporal separation between iron reducers and oxidizers. Iron-oxidizers and reducers can take advantage of redox oscillations (Coby et al., 2011; Jewell et al., 2016), and in the North Pond aquifer, dominance of either iron reducers and oxidizers may be cyclical.
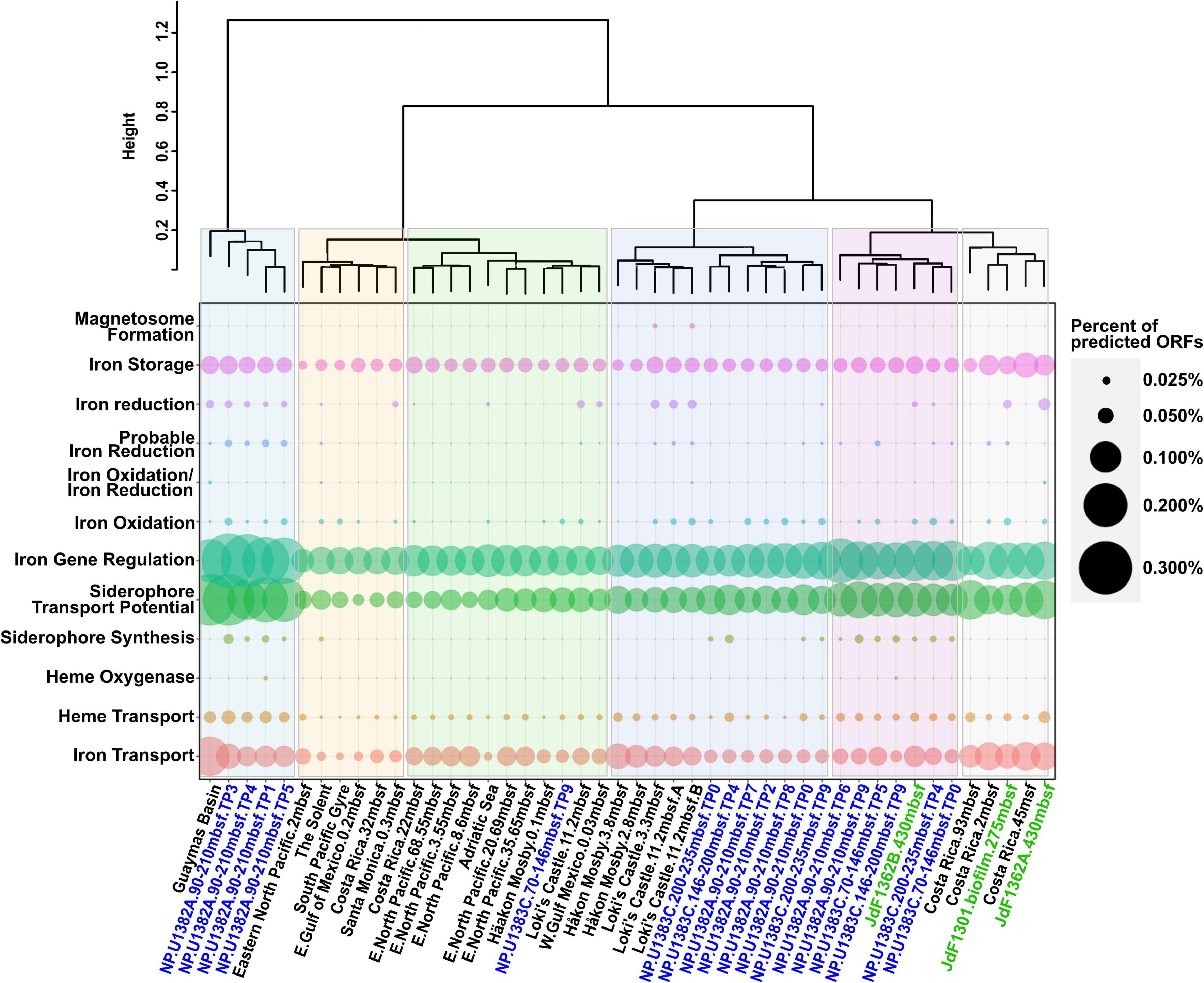
Figure 2. Dotplot summary of iron genes in marine subcrustal aquifers and sediment identified by FeGenie, showing the variation among iron genes from different time points from North Pond. North Pond metagenomes are colored blue, while metagenomes from the Juan de Fuca ridge are colored green. Metagenomes derived from sediment samples are colored black. While certain pathways appear to be universal (iron acquisition and storage), other processes appear to vary in abundance (iron redox cycling and siderophore synthesis), while others still (magnetosome formation and heme oxygenases) appear to be quite rare. “Percent of predicted ORFs” was calculated by dividing the total number of genes identified from each iron gene category by the total number of predicted ORFs from each respective metagenome.
Different Iron Redox Strategies Between Planktonic and Biofilm Communities of the Juan de Fuca Ridge
Evidence for iron oxidation at the JdF first came in the form of a chemolithoautotrophic iron-oxidizing isolate (Edwards et al., 2003b), and then with the identification of biogenic iron oxides resembling those formed by iron-oxidizing bacteria, like Mariprofundus and Leptothrix (Kennedy et al., 2003; Toner et al., 2008). However, this evidence is from seafloor samples collected in the vicinity of the JdF aquifer. For example, the biogenic iron oxides were either (i) found in iron oxide mounds (Kennedy et al., 2003) or (ii) enriched from mineral incubations deployed at the seafloor (Toner et al., 2008). No sequence-based analysis has been performed on these iron oxide-rich samples, and the exact identity and mechanisms for iron-cycling in microbial communities associated with those iron oxides remains unknown. Three metagenomes representing the planktonic (2 samples) and crust-associated (1 sample) microbial communities from the sediment buried crustal aquifer at the JdF have since been published, allowing molecular interrogation of genetic potential for iron-related metabolisms. Two crustal aquifer planktonic metagenomes (Jungbluth et al., 2017) were collected at subseafloor observatories retrofitted with Circulation Obviation Retrofit Kits [CORKs; Edwards et al. (2012)]; one biofilm metagenome is derived from an olivine chip biofilm incubated in situ for 4 years within a Flow-through Osmotic Colonization System [FLOCS; Orcutt et al. (2010)] (Smith et al., 2019).
FeGenie analysis revealed the genetic potential for iron oxidation, via cyc2, in all three JdF metagenomes, while mtoAB, a larger multi-protein porin-cytochrome complex (Jiao and Newman, 2007; Liu J. et al., 2012), was detected in only one of the crustal fluid metagenomes (Figure 1). The JdF crustal biofilm was previously reported to not support iron oxidation through the analysis of MAGs alone (Smith et al., 2019). Analysis of the whole metagenome assembly, including the unbinned fraction (contigs that were not included in the MAGs), identified at least eight cyc2 gene copies. Seven of these cyc2 copies were in the unbinned fraction of the assembly, and one was detected in JdFRolivine-5, a MAG (within the class Clostridia) constructed and published by Smith et al. (2019). Despite its significant homology, the cyc2 copy in JdFRolivine-5 has unclear function, as it is considerably shorter than most other cyc2 genes.
Smith et al. (2019) also reported that the MAG JdFRolivine-10 is related to the known iron reducer Geoglobus (Slobodkina et al., 2009; Mardanov et al., 2015) and encodes some of the cytochromes implicated in iron reduction in Geoglobus acetivorans (Mardanov et al., 2015). Three multiheme cytochromes encoded by JdFRolivine-10 match those in FeGenie’s HMM library (Supplementary Table 2), which were previously linked to extracellular electron transfer: DFE_0449 (14-heme iron oxidase; Deng et al., 2018), GACE_1846 (4-heme iron reductase), and GACE_1847 (22-heme iron reductase) (Mardanov et al., 2015). The outer membrane cytochrome with locus tag GACE_1847 was predicted to encode an outer membrane anchor domain, as well as two hematite-binding sites (Mardanov et al., 2015). These cytochromes are all located on different contigs in JdFRolivine-10’s genome and were identified using the new FeGenie flag –all_results, allowing us to bypass FeGenie’s built-in operon-evaluating algorithm. FeGenie also confirmed the presence of a hematite-binding motif (a new feature in FeGenie’s pipeline, see Supplementary Methods for details). The presence of heme-binding and transmembrane domains encoded on the GACE_1847 homolog provides a clue to one of the possible mechanisms for iron reduction within the JdF aquifer biofilm. Other genes related to iron reduction were also detected in the unbinned fraction of the metagenome from JdF olivine biofilms: omcS (Mehta et al., 2005; Qian, 2011; Wang et al., 2019), mtrC (Hartshorne et al., 2007), mtrA (Pitts et al., 2003), and t4ap (Bray et al., 2020), further supporting the potential for iron reduction within crustal biofilms. However, since olivine contains only reduced iron (Fe2+), it likely has been oxidized first by iron-oxidizing bacteria, supporting a potential cryptic iron cycle on the surface of the olivine mineral (Supplementary Figure 1); and since JdF is an anoxic system, this implicates anaerobic iron oxidation, possibly nitrate-dependent.
In the crustal fluid metagenomes, we detected potential for iron reduction via t4ap (Bray et al., 2020), omcZ (Inoue et al., 2010), and the flavin-based extracellular electron transfer mechanism recently reported in Listeria monocytogenes (Light et al., 2018). Potential for iron oxidation was detected via the dfeEFGHI operon (Deng et al., 2018). Unlike the olivine biofilms, omcS was not detected in the fluid metagenomes. This gene is associated with Geobacter species (Qian, 2011; Wang et al., 2019), which prefer anaerobic environments and less likely to dominate fluids subject to redox gradients (Lin et al., 2004; Lovley et al., 2011; Engel et al., 2020). Indeed, four omcS copies detected in the biofilm were phylogenetically linked to Geoalkalibacter subterraneus and Malonomonas rubra (Supplementary File 2 and Supplementary Table 4), both obligate anaerobes that taxonomically fall within the Desulfuromonadales order, which also contains Geobacter spp. Because the planktonic community in the anoxic crustal fluids is sourced from oxic bottom waters, we hypothesize that these iron reducers, rare in seawater, remain rare in the chemically evolved anoxic fluids. Eventually, these lineages may transition from planktonic to biofilm lifestyles where the activities of previous microbial communities (e.g., iron-oxidizing and sulfur-reducing bacteria) provide them with oxidized iron substrates (Toner et al., 2008; Ramírez et al., 2016). For example, as mentioned above, we did not detect any Geobacter-related gene markers from the North Pond aquifer metagenomes; however, recent mineral incubations enriched an electroactive consortia that includes Geobacter (Jones et al., 2020), supporting the latent presence of this lineage in redox-fluctuating fluids.
A Survey of Iron Redox Potential in Globally Dispered Marine Sediment
Potential for benthic iron reduction is prevalent among the locations under relatively high water-column productivity (e.g., Loki’s Castle, Guaymas Basin, and Western Gulf of Mexico), but is absent in the oligotrophic South Pacific Gyre (SPG) (Figure 1). The lack of iron reduction in SPG is consistent with previous analysis of these sediments (Tully and Heidelberg, 2016). High water column productivity results in benthic conditions ideal for iron reduction (low oxygen and high organic carbon deposition rates). The South Pacific Gyre sediments are far from this ideal, as they are low in carbon and oxic throughout the entire sediment column (D’Hondt et al., 2009). Two of the four cold-seep sediment metagenomes (Håkon Mosby Mud Volcano and Eastern Gulf of Mexico) also lacked any detectable genes for iron reduction. The surveyed metagenome sample from the Håkon Mosby Mud Volcano represents the top cm of young, freshly-erupted mud with ongoing aerobic activities, and, thus, is also not primed for iron reduction (Ruff et al., 2019). It is unclear why we did not detect genes for iron reduction in the Eastern Gulf of Mexico sediment (Dong et al., 2019); this site represent a petroleum seep where benthic communities may be dominated by hydrocarbon-degrading microbes that are not capable of iron reduction. It is important to note that technical (e.g., number of contigs, contig lengths, etc.) and environmental (community richness) differences can significantly influence inferences from broad metagenomic surveys; often, several factors (e.g., quality and concentration of DNA, strain-level diversity, prevalence of repetitive DNA sequences) can play a major role in determining the quality of the metagenome assemblies, as evidenced from the substantial variation in the distribution of contig lengths obtained from different metagenome assemblies (Supplementary File 3). This can affect the capacity of annotation pipelines, like FeGenie or AntiSMASH, to detect certain pathways.
Higher productivity sites demonstrated a diverse array of genes for iron reduction, including omcS, omcZ, and mtrCAB, etc. Geobacter-related iron reductases [e.g., omcS, omcZ, and other porin-cytochrome complexes (Shi et al., 2014)] were more common in the marine sediment sites compared with the surveyed aquifer metagenomes, with only the Guaymas Basin encoding the mtrCAB operon. Numerous copies of the aromatically dense type IV pili (Bray et al., 2020) gene were detected. These are particularly prevalent in marine sediment from the Western Gulf of Mexico (Laso-Pérez et al., 2019).
Potential for iron oxidation was detected in nearly all surveyed marine sediment sites (Figure 1). Specifically, cyc2 [more specifically, cyc2-cluster 3 (McAllister et al., 2020a)] appears to be the most common gene putatively linked to iron oxidation. In the North Pond aquifer, many cyc2 homologs were found to be closely related to those encoded by the iron-oxidizing Mariprofundus. However, in marine sediments, cyc2 homologs appear to have a different phylogenetic distribution, with associated taxa that have not previously been linked to iron oxidation (e.g., Acidobacteria, Desulfobacterales, Ralstonia solanacearum, etc.). In these taxa with no experimental evidence for iron oxidation, the function of cyc2 is less clear. While cyc2 appears to be most widely distributed porin-cytochrome putative iron oxidase, mtoAB is also present in three sediment sites: Guaymas Basin, Loki’s Castle, and The Solent. Other genetic markers for iron oxidation were also detected: sulfocyanin was detected in three of the marine sediment sites (Loki’s Castle, South Pacific Gyre, and Santa Monica Basin); the periplasmic cytochrome-encoding foxE was found in two sites (Loki’s Castle and The Solent). The Solent, which represents the only sediment metagenome from the photic zone, was found to have a variety of different markers for iron oxidation and reduction. Thus, unlike other sites that are well below the photic zone (Table 1), where iron oxidation has potential to fuel chemolithoautotrophy, in The Solent, iron oxidation may fuel photoferrotrophy. While the oxic South Pacific Gyre sediment may support aerobic iron oxidation, the other surficial sediment sites, with higher productivity and lower oxygen content, may support continuous iron oxidation at the surface or during periods of oxygenation [e.g., via bioturbation (Beam et al., 2018)].
Magnetosome Formation at Loki’s Castle, but Absent From Other Sediment and Aquifer Metagenomes
Magnetosomes are organelles of magnetotactic bacteria that contain biomineralized magnetite (Frankel et al., 1979; Uebe and Schüler, 2016). This allows bacteria to sense the Earth’s magnetic field, facilitating orientation along geochemical gradients. We detected nine separate loci encoding genes for magnetosome formation at Loki’s Castle sediment (Figure 2). The genetic loci for magnetosome formation identified at Loki’s Castle presented a mixed phylogenetic distribution, with amino acid identities near 50%, likely indicating a lack of closely related sequenced organisms. However, some of the most closely related sequences in NCBI’s non-redundant protein database appear to be from the Magnetococcales order, which includes Magnetococcus marinus, a marine magnetotactic bacterium isolated from the oxic-anoxic sediment-boundary off the coast of Rhode Island (Bazylinski et al., 2013). Magnetosomes are conceivably advantageous in environments with geochemical gradients, and are considered to be ubiquitous in aquatic habitats (Lefèvre and Bazylinski, 2013). Thus, the apparent lack of magnetosome formation operons in most marine sediment and all aquifer metagenomes is somewhat surprising. It is possible that magenetosome formation is heavily dependent on the availability and speciation of iron, which varies as a function of the spatial distribution of the redox cascade. Alternatively, the dearth of magnetosome formation operons (mam) can be a result of FeGenie’s strict rules for its detection (requiring the presence of at least 5 of the 10 diagnostic genes); while the assembly qualities for most of the metagenomes used in our survey are high (Supplementary File 3), detection of magnetosome formation operons requires contigs greater than 5,000 bases in length, and these make up a relatively small proportion (∼0.1–20%) of contigs in each assembly. In support of the latter hypothesis, we identified a MAG with genetic potential for iron oxidation (via foxE) – this MAG, with a ∼75% complete genome, is closely related to the mam-encoding Magnetovibrio blakemorei, but only has 2/10 of the mam genes, which are found on two different contigs; this is below FeGenie’s detection threshold.
Iron Acquisition and Storage Is Common, but Siderophore Synthesis Is Limited to Subsurface Aquifers and One Sediment Site
In addition to genes for iron redox cycling and magnetosome formation, we also report the distribution of genes for iron acquisition and storage, which are not directly linked to respiration, but are essential functions for the majority of living organisms (Figure 2). As expected, metagenomes examined in this study have the genes necessary for ferrous and ferric iron transport [efeUOB, fbpAB(C), feoAB(C), futABC], iron storage within ferritin (PF00210), as well as iron gene regulation (dtxR, fecR, feoC, and fur) (Supplementary Table 3). Potential for transport of iron-chelating molecules like siderophores (Butler, 2005) and heme via the TonB-ExbB-ExbD system was also detected (Krewulak and Vogel, 2011). We note that TonB-dependent transport is not specific to iron-chelating compounds (Noinaj et al., 2010). Even though heme can also be transported by this pathway (Noinaj et al., 2010; Krewulak and Vogel, 2011), the marine sediment metagenomes were largely devoid of heme oxygenase and storage homologs, except for one copy of pigA (heme oxygenase; Friedman et al., 2003, 2004) and a few copies of hutZ (heme storage; Liu X. et al., 2012) in the North Pond metagenomes.
The North Pond metagenomes harbored several siderophore synthesis gene clusters that share similarity to the known clusters encoding desferrioxamine E, crochelin A, amonabactin P, vicibactin, and vibrioferrin (Supplementary Table 5). The percent of genes in these identified clusters with significant BLAST hits to genes within known siderophore clusters ranged from 33–80%, as determined by AntiSMASH (Blin et al., 2019). Putative siderophore synthesis was also confirmed in several North Pond MAGs (Supplementary Table 2). MAGs taxonomically identified as Pseudomonas tetraodonis, Halomonas alkaliantarctica, and Paracoccus sp. may produce siderophores related to desferrioxamine E (75% gene similarity), crochelin A (46%), and amonabactin P (42%), respectively. Other North Pond MAGs encode siderophore gene clusters similar to aerobactin (33%; Moritella sp000170855), bisucaberin B (100%; Flavobacteriaceae sp.), and unknown siderophores. The presence of siderophore synthesis clusters in the North Pond metagenomes and MAGs with low similarity to known siderophore biosynthesis genes suggests potential for synthesizing structurally related or novel siderophores. Siderophore clusters are genetically diverse and modular systems that are impacted by several evolutionary processes, including gene loss and gene acquisition (e.g., through horizontal gene transfer) (Cruz-Morales et al., 2017; Thode et al., 2018). This leads to variation in siderophore structures (e.g., Seyedsayamdost et al., 2012), which is likely required to prevent uptake by non-siderophore-producing microorganisms, in what is known as siderophore piracy (Hibbing et al., 2010; Butaitë et al., 2017).
We also detected a siderophore synthesis locus in one of the JdF aquifer fluid metagenomes (Supplementary Table 5). Using AntiSMASH, this cluster was confirmed to be related to the acinetoferrin biosynthesis cluster.
Out of the 24 surveyed metagenomes from marine sediment, only one, The Solent, appears to encode a siderophore biosynthesis cluster, confirmed using AntiSMASH, to be most closely related to xanthoferrin. The overall lack of siderophore production in marine sediments may be due to the greater bioavailability of iron in those habitatis, or the potential presence of siderophores derived from the water column. The metagenomes may also harbor siderophore synthesis loci that share no or undetectable similarity to known siderophore synthesis clusters/models. Siderophores are necessary to obtain insoluble Fe(III) from the environment, and ferrisiderophore complexes become an important source of iron when soluble Fe(II) is limited. At Loki’s Castle and Guaymas Basin sediments, pore water Fe(II) concentrations can reach ∼200 μM (Table 1), which may obviate the need to synthesize siderophores. In the South Pacific Gyre sediment, with extremely low productivity and metabolic activities, the synthesis of siderophores may be too energetically costly. In contrast, siderophore synthesis in North Pond and JdF fluids concur with the minimal amounts of bioavailable iron present (Table 1), although JdF fluids have been shown to have highly variable iron concentrations between boreholes (Wheat et al., 2013). Within North Pond, oxic conditions (∼213–216 μM O2) result in the biotic and abiotic removal of soluble iron, leading to extremely low iron concentrations. This is consistent with the relatively high numbers of different siderophore synthesis loci identified there. The apparent lack of siderophore production in iron-limited sediment, like Costa Rica sediments, mainly composed of non-iron bearing clays (Vannucchi et al., 2013), and South Pacific Gyre sediments, which are oxygenated and contain only 5.6–7.3% Fe2O3 (D’Hondt et al., 2011; Dunlea, 2016) suggests dependence on exogenous siderophores, or other iron-chelating molecules (D’Onofrio et al., 2010). For example, in higher-productivity areas, sedimentation of biomass may deliver sufficient amounts of iron and iron-bearing molecules to benthic communities (Boyd and Ellwood, 2010; Gledhill and Buck, 2012; Boiteau et al., 2016). Additionally, release of reduced iron from sediment, which can also be catalyzed by iron-reducing bacteria, can potentially contribute to the iron budget of resident microbes. Alternatively, as mentioned above with regard to the apparent lack of magnetosome formation operons, the lack of siderophore biosynthesis operons may also be due to the fact that, similar to the magnetosome formation operon, siderophore biosynthesis operons are often >10,000 bases in length and involve multiple genes; thus, reliable detection of siderophore biosynthesis requires contigs that are relatively rare in metagenome assemblies.
Concluding Remarks
Our reanalysis of globally distributed marine subsurface metagenomes using FeGenie’s comprehensive iron gene library illuminates the diversity of microbial iron redox mechanisms that can occur across a range of geochemical regimes (Figure 3A). Further, we demonstrate FeGenie’s utility in providing a standardized pipeline for the comparison of iron genes among many large genomic datasets. We note that the data used in our survey were generated from multiple studies; inherent differences in sampling collection and processing, sequencing, and in silico methods, thus, make it difficult to generate overarching conclusions. Despite these potential limitations, our results support a hypothesis that geochemical constraints may influence the distribution of iron redox genes, potentially playing an important role in determining the dominant strategy for iron cycling (Figures 3B–D). At high-productivity sites (e.g., Guaymas Basin, Santa Monica Basin, Western Gulf of Mexico, Costa Rica), iron cycling is likely based on the penetration of oxygen and/or nitrate into surficial sediment, or deeper due to bioturbation, enabling microbial iron oxidation processes until these electron acceptors are diminished and iron reduction dominates. In oxic sediment, such as that underlying areas of low productivity (e.g., South Pacific Gyre), iron cycling is likely dominated by iron oxidation – with no or limited biological iron reduction. In cold and oxic subseafloor aquifers like North Pond, the flux of oxygen and organic carbon are possibly key factors influencing the abundances of iron-oxidizers and iron-reducers (Figure 3C), while in the warm and anoxic waters circulating within the JdF aquifer, iron cycling is dependent on anaerobic metabolisms (e.g., iron reduction and nitrate-dependent iron oxidation) (Figure 3D).
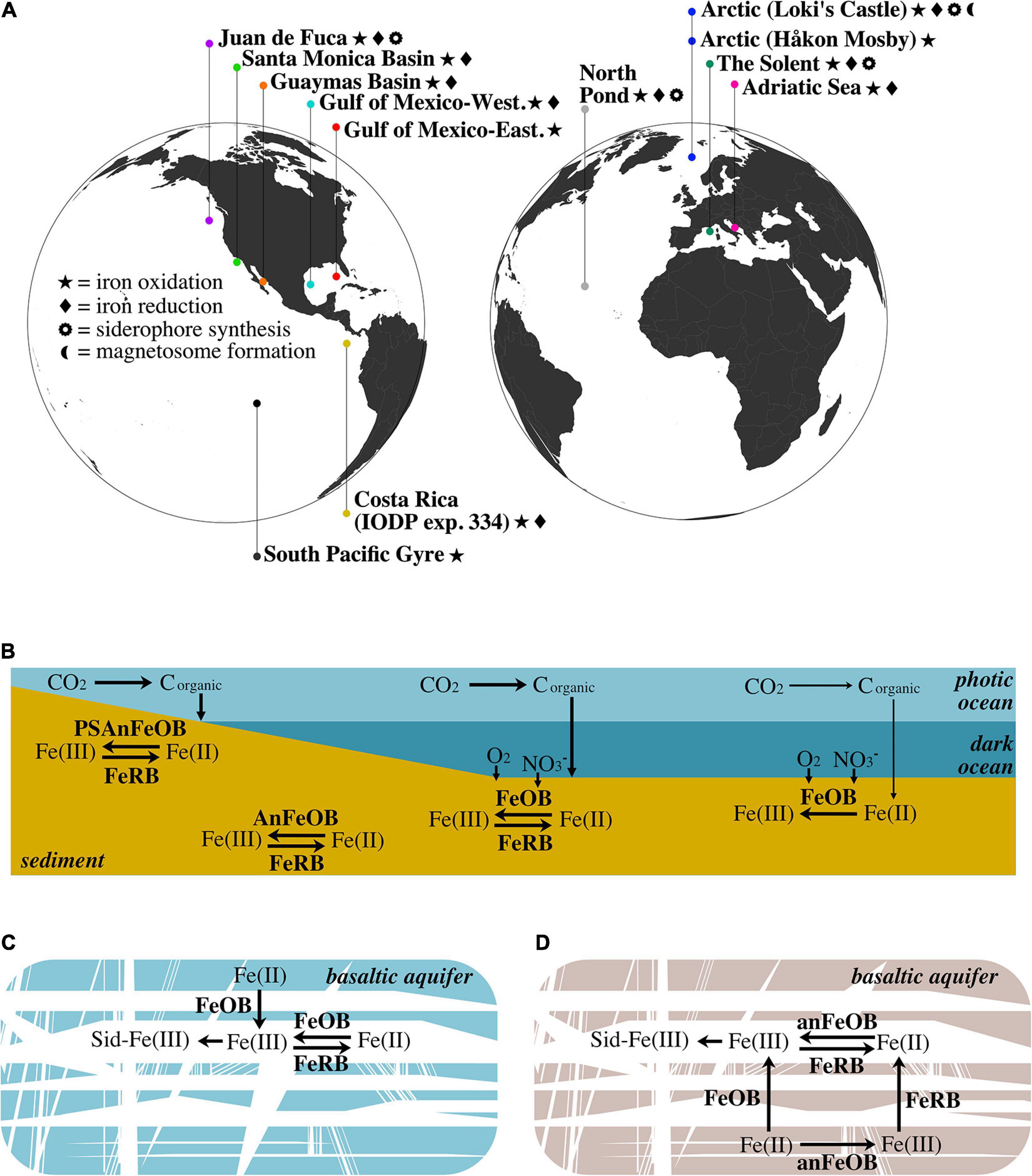
Figure 3. (A) Geographic distribution of marine aquifer and sediment sites from which metagenome samples are derived. (B) Schematic of the iron redox cycle in various types of marine sediment. In shallow sediment within the photic zone (e.g., The Solent), photoferrotrophy can drive iron oxidation. Below the photic zone, aerobic iron oxidation in surficial sediment (e.g., Guaymas Basin, Santa Monica Basin, and Western Gulf of Mexico) is contrasted with anaerobic iron oxidation (e.g., nitrate-dependent) in deep anoxic sediment (e.g., Costa Rica and Loki’s Castle). In these aforementioned sediments, iron reduction is present and represents the other half of the iron cycle. Conversely, in oxic sediments, such as those underlying areas of low productivity (e.g., South Pacific Gyre), aerobic iron oxidation is the dominant process, with no detectable iron reduction. The lower productivity is denoted by thinner arrows that signify carbon fixation and sedimentation. (C) Iron cycling within the cold and oxic North Pond aquifer: iron is released from the young, largely ferrous (Bach and Edwards, 2003), basaltic crust via iron oxidation, where it is free to cycle between the ferric and ferrous forms, and subject to the chelating activity of siderophores. (D) Iron cycling within the warm, anoxic JdF aquifer: since the waters circulating in this aquifer are anoxic, iron oxidation is likely dependent on anaerobic metabolisms. Similar to North Pond, JdF basaltic crust is young and the iron content is largely ferrous (Bach and Edwards, 2003); thus, iron is first released via oxidation. Detection of iron reduction on the surface of an olivine chip suggests that iron oxides may be deposited on the olivine mineral after oxidation by iron-oxidizing microbes. FeOB, aerobic iron-oxidizing bacteria; AnFeOB, anaerobic iron-oxidizing bacteria; PSAnFeOB, photosynthetic anaerobic iron-oxidizing bacteria; FeRB, iron-reducing bacteria; Sid, siderophores.
In nearly every site where iron redox cycling was detected, porin-cytochrome combinations were present and may constitute an important vector for dissimilatory electron transfer to and from soluble and insoluble iron (although porin-cytochrome genes are also the most common genetic markers currently available). For example, the porin-cytochrome fusion encoded by cyc2 appears nearly ubiquitous in our surveyed data; cyc2, whose phylogenetic distribution implies rampant lateral transfer among prokaryotes (McAllister et al., 2020a), may thus represent a widespread mechanism for respiratory iron oxidation. Alternatively, it is possible that Cyc2 acts as an iron detoxification mechanism (Bradley et al., 2020). In addition to porin-cytochromes, other mechanisms for iron redox were detected in diverse habitats. For example, the Geobacter-type omcS, omcZ, and electroactive pili genes demonstrate another strategy that may be largely relegated to sediment and biofilm niches. Although, detection of omcZ and t4ap genes in one of the JdF aquifer plankton metagenomes supports the possibility that this strategy may be utilized in planktonic niches, at least those where anoxic conditions prevail.
Geochemical constraints also seem to impact other aspects of the microbial iron cycle. Unlike the near-ubiquitous occurrence of genes associated with iron transport, iron storage, iron gene regulation, heme transport, and siderophore transport amongst the surveyed metagenomes, siderophore biosynthesis gene clusters were restricted to subseafloor aquifers, with only one sediment site encoding a siderophore biosynthesis cluster. Similarly, magnetosome formation genes were detected in only one of the sediment metagenomes (and none in the subsurface aquifers). It is possible that these biosynthesis operons are in short supply due to geochemical factors influencing the supply of iron or energy needed to synthesize them. However, we also cannot rule out the possibility that siderophore biosynthesis and magnetosome formation are more prevalent but undetected, either due to high level of sequence divergence from pre-existing HMMs used by FeGenie, or due to low abundances in the sequenced samples. For example, FeGenie retains the ability to detect potentially novel types of siderophore biosynthesis clusters, but detection of magnetosome formation depends on gene markers from a single known operon. In summary, our survey provides a comprehensive overview using the currently available genetic markers, generating testable hypotheses and providing insights into the distribution of iron genes in subsurface biomes across the world.
Data Availability Statement
The original contributions generated for this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
AG and NM conceived the study. AG and NM carried out the bioinformatics analysis. AG, NM, AC, GR, RB, TE-B, MG, and KN interpreted the results and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding
MG and TE-B were funded by the German Research Foundation under the SPP1833 programme. Lawrence Livermore National Laboratory is operated by Lawrence Livermore National Security, LLS, for the United States Department of Energy, National Nuclear Security Administration under Contract DE-AC52-07NA27344 (IM #: LLNL-JRNL-819591).
Acknowledgments
We sincerely thank Clara Chan for insights and comments on this manuscript. Thanks to Sean McAllister and Siavash Atashgahi for insightful discussions and input regarding iron cycling. Thanks to Martin Van-Den-Berghe for providing insights regarding siderophore utilization in the deep biosphere. We are grateful to Mike Russell for feedback on this manuscript. Finally, we thank Michał Sitko for making a FeGenie Dockerfile and Natasha Pavlovikj for creating a Conda recipe for FeGenie.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.667944/full#supplementary-material
Supplementary Figure 1 | Cryptic iron cycling on the surface of an olivine mineral incubated within the Juan de Fuca aquifer (borehole 1301A). The green trapezoid represents the olivine mineral with the chemical formula (Mg2+, Fe2+)2SiO4. Oxidation of reduced iron within the olivine mineral by iron-oxidizing bacteria (FeOB) results in the formation of iron(III) oxyhydroxides, and chemical formula Fe(OH)3, a by-product of microbial iron oxidation. The iron oxyhydroxides can then be used as terminal electron acceptors by iron-reducing bacteria (FeRB), releasing reduced iron from the mineral. Cyc2 and GACE_1847 are shown as putative iron oxidases and iron reductases, respectively, in this schematic.
References
Appia-Ayme, C., Bengrine, A., Cavazza, C., Giudici-Orticoni, M.-T., Bruschi, M., Chippaux, M., et al. (1998). Characterization and expression of the co-transcribed cyc1 and cyc2 genes encoding the cytochrome cR (cSSP) and a high-molecular-mass cytochrome c from Thiobacillus ferrooxidans ATCC 33020. FEMS Microbiol. Lett. 167, 171–177.
Aromokeye, D. A., Kulkarni, A. C., Elvert, M., Wegener, G., Henkel, S., Coffinet, S., et al. (2020). Rates and microbial players of iron-driven anaerobic oxidation of methane in methanic marine sediments. Front. Microbiol. 10:3041. doi: 10.3389/fmicb.2019.03041
Bach, W. (2016). Some compositional and kinetic controls on the bioenergetic landscapes in oceanic basement. Front. Microbiol. 7:107. doi: 10.3389/fmicb.2016.00107
Bach, W., and Edwards, K. J. (2003). Iron and sulfide oxidation within the basaltic ocean crust: implications for chemolithoautotrophic microbial biomass production. Geochim. Cosmochim. Acta 67, 3871–3887. doi: 10.1016/s0016-7037(03)00304-1
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barco, R. A., Emerson, D., Sylvan, J. B., Orcutt, B. N., Meyers, M. E. J., Ramírez, G. A., et al. (2015). New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl. Environ. Microbiol. 81, 5927–5937. doi: 10.1128/AEM.01374-15
Barco, R. A., Hoffman, C. L., Ramírez, G. A., Toner, B. M., Edwards, K. J., and Sylvan, J. B. (2017). In-situ incubation of iron-sulfur mineral reveals a diverse chemolithoautotrophic community and a new biogeochemical role for Thiomicrospira. Environ. Microbiol. 19, 1322–1337. doi: 10.1111/1462-2920.13666
Bazylinski, D. A., Williams, T. J., Lefèvre, C. T., Berg, R. J., Zhang, C. L., Bowser, S. S., et al. (2013). Magnetococcus marinus gen. nov., sp. nov., a marine, magnetotactic bacterium that represents a novel lineage (Magnetococcaceae fam. nov., Magnetococcales ord. nov.) at the base of the Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. 63, 801–808. doi: 10.1099/ijs.0.038927-0
Beam, J. P., Scott, J. J., McAllister, S. M., Chan, C. S., McManus, J., Meysman, F. J. R., et al. (2018). Biological rejuvenation of iron oxides in bioturbated marine sediments. ISME J. 12, 1389–1394. doi: 10.1038/s41396-017-0032-6
Blackwell, N., Bryce, C., Straub, D., Kappler, A., and Kleindienst, S. (2020). Genomic Insights into Two Novel Fe(II)-Oxidizing Zetaproteobacteria isolates reveal lifestyle adaption to coastal marine sediments. Appl. Environ. Microbiol. 86:e01160-20. doi: 10.1128/AEM.01160-20
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Blöthe, M., and Roden, E. E. (2009). Microbial iron redox cycling in a circumneutral-pH groundwater seep. Appl. Environ. Microbiol. 75, 468–473. doi: 10.1128/AEM.01817-08
Bochet, O., Bethencourt, L., Dufresne, A., Farasin, J., Pédrot, M., Labasque, T., et al. (2020). Iron-oxidizer hotspots formed by intermittent oxic–anoxic fluid mixing in fractured rocks. Nat. Geosci. 13, 149–155. doi: 10.1038/s41561-019-0509-1
Boiteau, R. M., Mende, D. R., Hawco, N. J., McIlvin, M. R., Fitzsimmons, J. N., Saito, M. A., et al. (2016). Siderophore-based microbial adaptations to iron scarcity across the eastern Pacific Ocean. Proc. Natl. Acad. Sci. U.S.A. 113, 14237–14242. doi: 10.1073/pnas.1608594113
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinform. Oxf. Engl. 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boyd, P. W., and Ellwood, M. J. (2010). The biogeochemical cycle of iron in the ocean. Nat. Geosci. 3, 675–682. doi: 10.1038/ngeo964
Bradley, J. M., Svistunenko, D. A., Wilson, M. T., Hemmings, A. M., Moore, G. R., and Le Brun, N. E. (2020). Bacterial iron detoxification at the molecular level. J. Biol. Chem. 295, 17602–17623. doi: 10.1074/jbc.rev120.007746
Bray, M. S., Wu, J., Padilla, C. C., Stewart, F. J., Fowle, D. A., Henny, C., et al. (2020). Phylogenetic and structural diversity of aromatically dense pili from environmental metagenomes. Environ. Microbiol. Rep. 12, 49–57. doi: 10.1111/1758-2229.12809
Bryce, C., Blackwell, N., Schmidt, C., Otte, J., Huang, Y.-M., Kleindienst, S., et al. (2018). Microbial anaerobic Fe(II) oxidation – Ecology, mechanisms and environmental implications. Environ. Microbiol. 20, 3462–3483. doi: 10.1111/1462-2920.14328
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Butaitë, E., Baumgartner, M., Wyder, S., and Kümmerli, R. (2017). Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities. Nat. Commun. 8:414. doi: 10.1038/s41467-017-00509-4
Butler, A. (2005). Marine siderophores and microbial iron mobilization. Biometals 18, 369–374. doi: 10.1007/s10534-005-3711-0
Byrne, J. M., Klueglein, N., Pearce, C., Rosso, K. M., Appel, E., and Kappler, A. (2015). Redox cycling of Fe(II) and Fe(III) in magnetite by Fe-metabolizing bacteria. Science 347, 1473–1476. doi: 10.1126/science.aaa4834
Castelle, C., Guiral, M., Malarte, G., Ledgham, F., Leroy, G., Brugna, M., et al. (2008). A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J. Biol. Chem. 283, 25803–25811. doi: 10.1074/jbc.M802496200
Castelle, C. J., Roger, M., Bauzan, M., Brugna, M., Lignon, S., Nimtz, M., et al. (2015). The aerobic respiratory chain of the acidophilic archaeon Ferroplasma acidiphilum: a membrane-bound complex oxidizing ferrous iron. Biochim. Biophys. Acta 1847, 717–728. doi: 10.1016/j.bbabio.2015.04.006
Chiu, B. K., Kato, S., McAllister, S. M., Field, E. K., and Chan, C. S. (2017). Novel pelagic iron-oxidizing Zetaproteobacteria from the chesapeake bay oxic–anoxic transition zone. Front. Microbiol. 8:1280. doi: 10.3389/fmicb.2017.01280
Coby, A. J., Picardal, F., Shelobolina, E., Xu, H., and Roden, E. E. (2011). Repeated anaerobic microbial redox cycling of iron. Appl. Environ. Microbiol. 77, 6036–6042. doi: 10.1128/AEM.00276-11
Cooper, D. C., Picardal, F. F., and Coby, A. J. (2006). Interactions between microbial iron reduction and metal geochemistry: effect of redox cycling on transition metal speciation in iron bearing sediments. Env. Sci Technol. 40, 1884–1891. doi: 10.1021/es051778t
Croal, L. R., Jiao, Y., and Newman, D. K. (2007). The fox operon from rhodobacter strain SW2 promotes phototrophic Fe(II) oxidation in Rhodobacter capsulatus SB1003. J. Bacteriol. 189, 1774–1782. doi: 10.1128/JB.01395-06
Cruz-Morales, P., Ramos-Aboites, H. E., Licona-Cassani, C., Selem-Mójica, N., Mejía-Ponce, P. M., Souza-Saldívar, V., et al. (2017). Actinobacteria phylogenomics, selective isolation from an iron oligotrophic environment and siderophore functional characterization, unveil new desferrioxamine traits. FEMS Microbiol. Ecol. 93:fix086. doi: 10.1093/femsec/fix086
de Vries, A., and Ripley, B. D. (2020). ggdendro: Create Dendrograms and Tree Diagrams Using “ggplot2.”. Available online at: https://CRAN.R-project.org/package=ggdendro (accessed February 13, 2021).
Deng, X., Dohmae, N., Nealson, K. H., Hashimoto, K., and Okamoto, A. (2018). Multi-heme cytochromes provide a pathway for survival in energy-limited environments. Sci. Adv. 4:eaao5682. doi: 10.1126/sciadv.aao5682
Dharamshi, J. E., Tamarit, D., Eme, L., Stairs, C. W., Martijn, J., Homa, F., et al. (2020). Marine sediments illuminate chlamydiae diversity and evolution. Curr. Biol. 30, 1032.e7–1048.e7. doi: 10.1016/j.cub.2020.02.016
D’Hondt, S., Inagaki, F., Alvarez Zarikian, C. A., and Expedition 329 Scientists (2011). Expedition 329 Reports. Tokyo: Integrated Ocean Drilling Program Management International, Inc.
D’Hondt, S., Spivack, A. J., Pockalny, R., Ferdelman, T. G., Fischer, J. P., Kallmeyer, J., et al. (2009). Subseafloor sedimentary life in the South Pacific Gyre. Proc. Natl. Acad. Sci. U.S.A. 106, 11651–11656. doi: 10.1073/pnas.0811793106
Dombrowski, N., Teske, A. P., and Baker, B. J. (2018). Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat. Commun. 9:4999. doi: 10.1038/s41467-018-07418-0
Dong, X., Greening, C., Rattray, J. E., Chakraborty, A., Chuvochina, M., Mayumi, D., et al. (2019). Metabolic potential of uncultured bacteria and archaea associated with petroleum seepage in deep-sea sediments. Nat. Commun. 10:1816. doi: 10.1038/s41467-019-09747-0
D’Onofrio, A., Crawford, J. M., Stewart, E. J., Witt, K., Gavrish, E., Epstein, S., et al. (2010). Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem. Biol. 17, 254–264. doi: 10.1016/j.chembiol.2010.02.010
Dunlea, A. G. (2016). Biogeochemistry and Geochemical Paleoceanography of the South Pacific Gyre. Available online at: https://open.bu.edu/handle/2144/19711 (accessed February 12, 2021).
Edwards, K. J., Bach, W., and Rogers, D. R. (2003a). Geomicrobiology of the ocean crust: a role for chemoautotrophic Fe-bacteria. Biol. Bull. 204, 180–185. doi: 10.2307/1543555
Edwards, K. J., Rogers, D. R., Wirsen, C. O., and McCollom, T. M. (2003b). Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing, chemolithoautotrophic α- and γ-Proteobacteria from the Deep Sea. Appl. Environ. Microbiol. 69, 2906–2913. doi: 10.1128/AEM.69.5.2906-2913.2003
Edwards, K. J., Wheat, C. J., Orcutt, B. N., Hulme, S., Becker, K., Bach, W., et al. (2012). Design and Deployment of Borehole Observatories and Experiments During IODP Expedition 336, Mid-Atlantic Ridge flank at North Pond. Tokyo: Integrated Ocean Drilling Program Management International, Inc.
Edwards, M. J., White, G. F., Butt, J. N., Richardson, D. J., and Clarke, T. A. (2020). The crystal structure of a biological insulated transmembrane molecular wire. Cell 181, 665.e10–673.e10. doi: 10.1016/j.cell.2020.03.032
Elliott, A. V. C., Plach, J. M., Droppo, I. G., and Warren, L. A. (2014). Collaborative microbial Fe-redox cycling by pelagic floc bacteria across wide ranging oxygenated aquatic systems. Chem. Geol. 366, 90–102. doi: 10.1016/j.chemgeo.2013.11.017
Emerson, D. (2009). Potential for iron-reduction and iron-cycling in iron oxyhydroxide-rich microbial mats at loihi seamount. Geomicrobiol. J. 26, 639–647. doi: 10.1080/01490450903269985
Emerson, D., and Moyer, C. L. (2002). Neutrophilic Fe-oxidizing bacteria are abundant at the loihi seamount hydrothermal vents and play a major role in Fe oxide deposition. Appl. Environ. Microbiol. 68, 3085–3093. doi: 10.1128/AEM.68.6.3085-3093.2002
Engel, C. E. A., Vorländer, D., Biedendieck, R., Krull, R., and Dohnt, K. (2020). Quantification of microaerobic growth of Geobacter sulfurreducens. PLoS One 15:e0215341. doi: 10.1371/journal.pone.0215341
Expedition. 344 Scientists (2011). Costa Rica Seismogenesis Project (CRISP): sampling and quantifying input to the seismogenic zone and fluid output. Int. Ocean Discov. Program 334:13. doi: 10.2204/iodp.proc.344.2013
Farag, I. F., Biddle, J. F., Zhao, R., Martino, A. J., House, C. H., and León-Zayas, R. I. (2020). Metabolic potentials of archaeal lineages resolved from metagenomes of deep Costa Rica sediments. ISME J. 14, 1345–1358. doi: 10.1038/s41396-020-0615-5
Frankel, R. B., Blakemore, R. P., and Wolfe, R. S. (1979). Magnetite in freshwater magnetotactic bacteria. Science 203, 1355–1356. doi: 10.1126/science.203.4387.1355
Friedman, J., Lad, L., Deshmukh, R., Li, H., Wilks, A., and Poulos, T. L. (2003). Crystal structures of the NO- and CO-bound heme oxygenase from Neisseriae meningitidis. Implications for O2 activation. J. Biol. Chem. 278, 34654–34659. doi: 10.1074/jbc.M302985200
Friedman, J., Lad, L., Li, H., Wilks, A., and Poulos, T. L. (2004). Structural basis for novel delta-regioselective heme oxygenation in the opportunistic pathogen Pseudomonas aeruginosa. Biochemistry 43, 5239–5245. doi: 10.1021/bi049687g
Gacesa, R., Baranasic, D., Starcevic, A., Diminic, J., Korlević, M., Najdek, M., et al. (2018). Bioprospecting for genes encoding hydrocarbon-degrading enzymes from metagenomic samples isolated from northern adriatic sea sediments. Food Technol. Biotechnol. 56, 270–277. doi: 10.17113/ftb.56.02.18.5393
Garber, A. I., Nealson, K. H., Okamoto, A., McAllister, S. M., Chan, C. S., Barco, R. A., et al. (2020). FeGenie: a comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front. Microbiol. 11:37. doi: 10.3389/fmicb.2020.00037
Gledhill, M., and Buck, K. N. (2012). The organic complexation of iron in the marine environment: a review. Front. Microbiol. 3:69. doi: 10.3389/fmicb.2012.00069
Hartshorne, R. S., Jepson, B. N., Clarke, T. A., Field, S. J., Fredrickson, J., Zachara, J., et al. (2007). Characterization of Shewanella oneidensis MtrC: a cell-surface decaheme cytochrome involved in respiratory electron transport to extracellular electron acceptors. JBIC J. Biol. Inorg. Chem. 12, 1083–1094. doi: 10.1007/s00775-007-0278-y
He, S., Barco, R. A., Emerson, D., and Roden, E. E. (2017). Comparative genomic analysis of neutrophilic iron(II) oxidizer genomes for candidate genes in extracellular electron transfer. Front. Microbiol. 8:1584. doi: 10.3389/fmicb.2017.01584
Hibbing, M. E., Fuqua, C., Parsek, M. R., and Peterson, S. B. (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25. doi: 10.1038/nrmicro2259
Inoue, K., Qian, X., Morgado, L., Kim, B.-C., Mester, T., Izallalen, M., et al. (2010). Purification and Characterization of OmcZ, an outer-surface, octaheme c-type cytochrome essential for optimal current production by geobacter sulfurreducens. Appl. Environ. Microbiol. 76, 3999–4007. doi: 10.1128/AEM.00027-10
Jeans, C., Singer, S. W., Chan, C. S., VerBerkmoes, N. C., Shah, M., Hettich, R. L., et al. (2008). Cytochrome 572 is a conspicuous membrane protein with iron oxidation activity purified directly from a natural acidophilic microbial community. ISME J. 2, 542–550. doi: 10.1038/ismej.2008.17
Jewell, T. N. M., Karaoz, U., Brodie, E. L., Williams, K. H., and Beller, H. R. (2016). Metatranscriptomic evidence of pervasive and diverse chemolithoautotrophy relevant to C, S, N and Fe cycling in a shallow alluvial aquifer. ISME J. 10, 2106–2117. doi: 10.1038/ismej.2016.25
Jiao, Y., and Newman, D. K. (2007). The pio operon is essential for phototrophic Fe(II) oxidation in rhodopseudomonas palustris TIE-1. J. Bacteriol. 189, 1765–1773. doi: 10.1128/JB.00776-06
Jones, J. G., Gardener, S., and Simon, B. M. (1984). Reduction of ferric iron by heterotrophic bacteria in lake sediments. Microbiology 130, 45–51. doi: 10.1099/00221287-130-1-45
Jones, R. M., D’Angelo, T., and Orcutt, B. N. (2020). Using cathodic poised potential experiments to investigate extracellular electron transport in the crustal deep biosphere of North Pond, mid-atlantic ridge. Front. Environ. Sci. 8:11. doi: 10.3389/fenvs.2020.00011
Jørgensen, S. L., Hannisdal, B., Lanzen, A., Baumberger, T., Flesland, K., Fonseca, R., et al. (2012). Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc. Natl. Acad. Sci. U.S.A. 109, E2846–E2855. doi: 10.1073/pnas.1207574109
Jørgensen, S. L., Thorseth, I. H., Pedersen, R. B., Baumberger, T., and Schleper, C. (2013). Quantitative and phylogenetic study of the Deep Sea Archaeal Group in sediments of the Arctic mid-ocean spreading ridge. Front. Microbiol. 4:299. doi: 10.3389/fmicb.2013.00299
Jungbluth, S. P., Amend, J. P., and Rappé, M. S. (2017). Metagenome sequencing and 98 microbial genomes from Juan de Fuca Ridge flank subsurface fluids. Sci. Data 4:170037. doi: 10.1038/sdata.2017.37
Jungbluth, S. P., Bowers, R. M., Lin, H.-T., Cowen, J. P., and Rappé, M. S. (2016). Novel microbial assemblages inhabiting crustal fluids within mid-ocean ridge flank subsurface basalt. ISME J. 10, 2033–2047. doi: 10.1038/ismej.2015.248
Kallmeyer, J., Pockalny, R., Adhikari, R. R., Smith, D. C., and D’Hondt, S. (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 109, 16213–16216. doi: 10.1073/pnas.1203849109
Kamura, T., Takai, Y., and Ishikawa, K. (1963). Microbial reduction mechanism of ferric iron in paddy soils (Part I). Soil Sci. Plant Nutr. 9, 5–9. doi: 10.1080/00380768.1963.10431048
Keffer, J. L., McAllister, S. M., Garber, A. I., Hallahan, B. J., Sutherland, M. C., Rozovsky, S., et al. (2021). Iron oxidation by a fused cytochrome-porin common to diverse iron-oxidizing bacteria. mBio. doi: 10.1128/mBio.01074-21 [Epub ahead of print].
Kennedy, C. B., Scott, S. D., and Ferris, F. G. (2003). Characterization of bacteriogenic iron oxide deposits from axial volcano, Juan de Fuca Ridge, Northeast Pacific Ocean. Geomicrobiol. J. 20, 199–214. doi: 10.1080/01490450303873
Krewulak, K. D., and Vogel, H. J. (2011). TonB or not TonB: is that the question? Biochem. Cell Biol. 89, 87–97. doi: 10.1139/O10-141
Langseth, M. G., Becker, K., Herzen, R. P. V., and Schultheiss, P. (1992). Heat and fluid flux through sediment on the western flank of the Mid-Atlantic Ridge: a hydrogeological study of North Pond. Geophys. Res. Lett. 19, 517–520. doi: 10.1029/92GL00079
Laso-Pérez, R., Hahn, C., van Vliet, D. M., Tegetmeyer, H. E., Schubotz, F., Smit, N. T., et al. (2019). Anaerobic degradation of non-methane alkanes by “candidatus methanoliparia” in hydrocarbon seeps of the Gulf of Mexico. mBio 10:e01814-19. doi: 10.1128/mBio.01814-19
Laufer, K., Byrne, J. M., Glombitza, C., Schmidt, C., Jørgensen, B. B., and Kappler, A. (2016a). Anaerobic microbial Fe(II) oxidation and Fe(III) reduction in coastal marine sediments controlled by organic carbon content. Environ. Microbiol. 18, 3159–3174. doi: 10.1111/1462-2920.13387
Laufer, K., Nordhoff, M., Røy, H., Schmidt, C., Behrens, S., Jørgensen, B. B., et al. (2016b). Coexistence of microaerophilic, nitrate-reducing, and phototrophic Fe(II) oxidizers and Fe(III) reducers in coastal marine sediment. Appl. Environ. Microbiol. 82, 1433–1447. doi: 10.1128/AEM.03527-15
Lefèvre, C. T., and Bazylinski, D. A. (2013). Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol. Mol. Biol. Rev. 77, 497–526. doi: 10.1128/MMBR.00021-13
Li, Z., Pan, D., Wei, G., Pi, W., Wang, J.-H., Peng, Y., et al. (2020). Deep sea sediments associated with cold seeps are a subsurface reservoir of viral diversity. bioRxiv [Preprint]. doi: 10.1101/2020.09.08.284018
Light, S. H., Su, L., Rivera-Lugo, R., Cornejo, J. A., Louie, A., Iavarone, A. T., et al. (2018). A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature 562, 140–144. doi: 10.1038/s41586-018-0498-z
Lin, W. C., Coppi, M. V., and Lovley, D. R. (2004). Geobacter sulfurreducens can grow with oxygen as a terminal electron acceptor. Appl. Environ. Microbiol. 70, 2525–2528. doi: 10.1128/aem.70.4.2525-2528.2004
Liu, J., Wang, Z., Belchik, S. M., Edwards, M. J., Liu, C., Kennedy, D. W., et al. (2012). Identification and characterization of MtoA: a decaheme c-type cytochrome of the neutrophilic Fe(II)-oxidizing bacterium Sideroxydans lithotrophicus ES-1. Front. Microbiol. 3:37. doi: 10.3389/fmicb.2012.00037
Liu, X., Gong, J., Wei, T., Wang, Z., Du, Q., Zhu, D., et al. (2012). Crystal structure of HutZ, a heme storage protein from Vibrio cholerae: a structural mismatch observed in the region of high sequence conservation. BMC Struct. Biol. 12:23. doi: 10.1186/1472-6807-12-23
Lovley, D. R., Ueki, T., Zhang, T., Malvankar, N. S., Shrestha, P. M., Flanagan, K. A., et al. (2011). Geobacter: the microbe electric’s physiology, ecology, and practical applications. Adv. Microb. Physiol. 59, 1–100.
MacQuarrie, K. T. B., and Mayer, K. U. (2005). Reactive transport modeling in fractured rock: a state-of-the-science review. Earth Sci. Rev. 72, 189–227. doi: 10.1016/j.earscirev.2005.07.003
Mardanov, A. V., Slododkina, G. B., Slobodkin, A. I., Beletsky, A. V., Gavrilov, S. N., Kublanov, I. V., et al. (2015). The Geoglobus acetivorans genome: Fe(III) reduction, acetate utilization, autotrophic growth, and degradation of aromatic compounds in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 81, 1003–1012. doi: 10.1128/AEM.02705-14
Martino, A., Rhodes, M. E., León-Zayas, R., Valente, I. E., Biddle, J. F., and House, C. H. (2019). Microbial diversity in sub-seafloor sediments from the costa rica margin. Geosciences 9:218. doi: 10.3390/geosciences9050218
McAllister, S. M., Moore, R. M., Gartman, A., Luther, G. W. III, Emerson, D., and Chan, C. S. (2019). The Fe(II)-oxidizing Zetaproteobacteria: historical, ecological and genomic perspectives. FEMS Microbiol. Ecol. 95:fiz015. doi: 10.1093/femsec/fiz015
McAllister, S. M., Polson, S. W., Butterfield, D. A., Glazer, B. T., Sylvan, J. B., and Chan, C. S. (2020a). Validating the Cyc2 neutrophilic iron oxidation pathway using meta-omics of Zetaproteobacteria iron mats at marine hydrothermal vents. mSystems 5:e00553-19. doi: 10.1128/mSystems.00553-19
McAllister, S. M., Vandzura, R., Keffer, J. L., Polson, S. W., and Chan, C. S. (2020b). Aerobic and anaerobic iron oxidizers together drive denitrification and carbon cycling at marine iron-rich hydrothermal vents. ISME J. 15, 1271–1286. doi: 10.1038/s41396-020-00849-y
McBeth, J. M., Little, B. J., Ray, R. I., Farrar, K. M., and Emerson, D. (2011). Neutrophilic iron-oxidizing “Zetaproteobacteria” and mild steel corrosion in nearshore marine environments. Appl. Environ. Microbiol. 77, 1405–1412. doi: 10.1128/AEM.02095-10
Mehta, T., Coppi, M. V., Childers, S. E., and Lovley, D. R. (2005). Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 71, 8634–8641. doi: 10.1128/AEM.71.12.8634-8641.2005
Meyer, J. L., Jaekel, U., Tully, B. J., Glazer, B. T., Wheat, C. G., Lin, H.-T., et al. (2016). A distinct and active bacterial community in cold oxygenated fluids circulating beneath the western flank of the Mid-Atlantic ridge. Sci. Rep. 6:22541. doi: 10.1038/srep22541
Mumford, A. C., Adaktylou, I. J., and Emerson, D. (2016). Peeking under the iron curtain: development of a microcosm for imaging the colonization of steel surfaces by Mariprofundus sp. strain DIS-1, an oxygen-tolerant Fe-oxidizing bacterium. Appl. Environ. Microbiol. 82, 6799–6807. doi: 10.1128/AEM.01990-16
Neilands, J. B. (1995). Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270, 26723–26726. doi: 10.1074/jbc.270.45.26723
Noinaj, N., Guillier, M., Barnard, T. J., and Buchanan, S. K. (2010). TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60. doi: 10.1146/annurev.micro.112408.134247
Orcutt, B., Wheat, C. G., and Edwards, K. J. (2010). Subseafloor ocean crust microbial observatories: development of FLOCS (FLow-through osmo colonization system) and evaluation of borehole construction materials. Geomicrobiol. J. 27, 143–157. doi: 10.1080/01490450903456772
Pereira, L., Saraiva, I. H., Oliveira, A. S. F., Soares, C. M., Louro, R. O., and Frazão, C. (2017). Molecular structure of FoxE, the putative iron oxidase of Rhodobacter ferrooxidans SW2. Biochim. Biophys. Acta BBA Bioenerg. 1858, 847–853. doi: 10.1016/j.bbabio.2017.07.002
Pitts, K. E., Dobbin, P. S., Reyes-Ramirez, F., Thomson, A. J., Richardson, D. J., and Seward, H. E. (2003). Characterization of the Shewanella oneidensis MR-1 decaheme cytochrome MtrA. J. Biol. Chem. 278, 27758–27765. doi: 10.1074/jbc.M302582200
Qian, X. (2011). Biochemical characterization of purified OmcS, a c-type cytochrome required for insoluble Fe(III) reduction in Geobacter sulfurreducens. Biochim. Biophys. Acta 1807, 404–412. doi: 10.1016/j.bbabio.2011.01.003
Ramírez, G. A., Hoffman, C. L., Lee, M. D., Lesniewski, R. A., Barco, R. A., Garber, A., et al. (2016). Assessing marine microbial induced corrosion at santa catalina Island, California. Front. Microbiol. 7:1679. doi: 10.3389/fmicb.2016.01679
Riedinger, N., Formolo, M. J., Lyons, T. W., Henkel, S., Beck, A., and Kasten, S. (2014). An inorganic geochemical argument for coupled anaerobic oxidation of methane and iron reduction in marine sediments. Geobiology 12, 172–181. doi: 10.1111/gbi.12077
Roden, E. E. (2012). Microbial iron-redox cycling in subsurface environments. Biochem. Soc. Trans. 40, 1249–1256. doi: 10.1042/BST20120202
Roden, E. E., McBeth, J. M., Blöthe, M., Percak-Dennett, E. M., Fleming, E. J., Holyoke, R. R., et al. (2012). The microbial ferrous wheel in a neutral pH groundwater seep. Front. Microbiol. 3:172. doi: 10.3389/fmicb.2012.00172
Ruff, S. E., Felden, J., Gruber-Vodicka, H. R., Marcon, Y., Knittel, K., Ramette, A., et al. (2019). In situ development of a methanotrophic microbiome in deep-sea sediments. ISME J. 13, 197–213. doi: 10.1038/s41396-018-0263-1
Scheller, S., Yu, H., Chadwick, G. L., McGlynn, S. E., and Orphan, V. J. (2016). Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351, 703–707. doi: 10.1126/science.aad7154
Seyedsayamdost, M. R., Cleto, S., Carr, G., Vlamakis, H., João Vieira, M., Kolter, R., et al. (2012). Mixing and matching siderophore clusters: structure and biosynthesis of serratiochelins from Serratia sp. V4. J. Am. Chem. Soc. 134, 13550–13553. doi: 10.1021/ja304941d
Seyler, L. M., Trembath-Reichert, E., Tully, B. J., and Huber, J. A. (2020). Time-series transcriptomics from cold, oxic subseafloor crustal fluids reveals a motile, mixotrophic microbial community. ISME J. 15, 1192–1206. doi: 10.1038/s41396-020-00843-4
Shi, L., Fredrickson, J. K., and Zachara, J. M. (2014). Genomic analyses of bacterial porin-cytochrome gene clusters. Front. Microbiol. 5:657. doi: 10.3389/fmicb.2014.00657
Slobodkina, G. B., Kolganova, T. V., Querellou, J., Bonch-Osmolovskaya, E. A., and Slobodkin, A. I. (2009). Geoglobus acetivorans sp. nov., an iron(III)-reducing archaeon from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 59, 2880–2883. doi: 10.1099/ijs.0.011080-0
Smith, A. R., Kieft, B., Mueller, R., Fisk, M. R., Mason, O. U., Popa, R., et al. (2019). Carbon fixation and energy metabolisms of a subseafloor olivine biofilm. ISME J. 13, 1737–1749. doi: 10.1038/s41396-019-0385-0
Smith, O., Momber, G., Bates, R., Garwood, P., Fitch, S., Pallen, M., et al. (2015). Sedimentary DNA from a submerged site reveals wheat in the British Isles 8000 years ago. Science 347, 998–1001. doi: 10.1126/science.1261278
Suzuki, R., and Shimodaira, H. (2006). Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542. doi: 10.1093/bioinformatics/btl117
Takai, Y., Koyama, T., and Kamura, T. (1963a). Microbial metabolism in reduction process of paddy soils (Part 2). Soil Sci. Plant Nutr. 9, 10–14. doi: 10.1080/00380768.1963.10431049
Takai, Y., Koyama, T., and Kamura, T. (1963b). Microbial metabolism in reduction process of paddy soils (Part 3). Soil Sci. Plant Nutr. 9, 1–5. doi: 10.1080/00380768.1963.10431054
Thode, S. K., Rojek, E., Kozlowski, M., Ahmad, R., and Haugen, P. (2018). Distribution of siderophore gene systems on a Vibrionaceae phylogeny: database searches, phylogenetic analyses and evolutionary perspectives. PLoS One 13:e0191860. doi: 10.1371/journal.pone.0191860
Toner, B. M., Santelli, C. M., Marcus, M. A., Wirth, R., Chan, C. S., McCollom, T., et al. (2008). Biogenic iron oxyhydroxide formation at mid-ocean ridge hydrothermal vents: juan de Fuca Ridge. Geochim. Cosmochim. Acta 73, 388–403. doi: 10.1016/j.gca.2008.09.035
Trinchero, P., Sidborn, M., Puigdomenech, I., Svensson, U., Ebrahimi, H., Molinero, J., et al. (2019). Transport of oxygen into granitic rocks: role of physical and mineralogical heterogeneity. J. Contam. Hydrol. 220, 108–118. doi: 10.1016/j.jconhyd.2018.12.001
Tully, B. J., and Heidelberg, J. F. (2016). Potential mechanisms for microbial energy acquisition in oxic deep-sea sediments. Appl. Environ. Microbiol. 82, 4232–4243. doi: 10.1128/AEM.01023-16
Tully, B. J., Wheat, C. G., Glazer, B. T., and Huber, J. A. (2018). A dynamic microbial community with high functional redundancy inhabits the cold, oxic subseafloor aquifer. ISME J. 12, 1–16. doi: 10.1038/ismej.2017.187
Uebe, R., and Schüler, D. (2016). Magnetosome biogenesis in magnetotactic bacteria. Nat. Rev. Microbiol. 14, 621–637. doi: 10.1038/nrmicro.2016.99
Vannucchi, P., Ujiie, K., Stroncik, N., and the IODP Exp. 334 Scientific Party (2013). IODP expedition 334: an investigation of the sedimentary record, fluid flow and state of stress on top of the seismogenic zone of an erosive subduction margin. Sci. Drill. 15, 23–30. doi: 10.5194/sd-15-23-2013
Wang, F., Gu, Y., O’Brien, J. P., Yi, S. M., Yalcin, S. E., Srikanth, V., et al. (2019). Structure of microbial nanowires reveals stacked hemes that transport electrons over micrometers. Cell 177, 361.e10–369.e10. doi: 10.1016/j.cell.2019.03.029
Weber, K. A., Achenbach, L. A., and Coates, J. D. (2006). Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat. Rev. Microbiol. 4, 752–764. doi: 10.1038/nrmicro1490
Wheat, C. G., Hulme, S. M., Fisher, A. T., Orcutt, B. N., and Becker, K. (2013). Seawater recharge into oceanic crust: IODP Exp 327 Site U1363 grizzly bare outcrop. Geochem. Geophys. Geosyst. 14, 1957–1972. doi: 10.1002/ggge.20131
Wickham, H., Chang, W., Henry, L., Pedersen, T. L., Takahashi, K., Wilke, C., et al. (2020). ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. Available online at: https://CRAN.R-project.org/package=ggplot2 (accessed February 13, 2021).
Keywords: metagenomics, marine sediment, marine aquifer, FeGenie, iron cycling, iron acquisition, iron oxidation, iron reduction
Citation: Garber AI, Cohen AB, Nealson KH, Ramírez GA, Barco RA, Enzingmüller-Bleyl TC, Gehringer MM and Merino N (2021) Metagenomic Insights Into the Microbial Iron Cycle of Subseafloor Habitats. Front. Microbiol. 12:667944. doi: 10.3389/fmicb.2021.667944
Received: 15 February 2021; Accepted: 30 July 2021;
Published: 03 September 2021.
Edited by:
Sabine Kasten, Alfred Wegener Institute Helmholtz Centre for Polar and Marine Research (AWI), GermanyReviewed by:
Mark Alexander Lever, ETH Zürich, SwitzerlandKarrie A. Weber, University of Nebraska–Lincoln, United States
Orit Sivan, Ben-Gurion University of the Negev, Israel
Copyright © 2021 Garber, Cohen, Nealson, Ramírez, Barco, Enzingmüller-Bleyl, Gehringer and Merino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arkadiy I. Garber, YWdhcmJlcjRAYXN1LmVkdQ==; Nancy Merino, bWVyaW5vNEBsbG5sLmdvdg==
 Arkadiy I. Garber
Arkadiy I. Garber Ashley B. Cohen
Ashley B. Cohen Kenneth H. Nealson
Kenneth H. Nealson Gustavo A. Ramírez
Gustavo A. Ramírez Roman A. Barco
Roman A. Barco Tristan C. Enzingmüller-Bleyl6
Tristan C. Enzingmüller-Bleyl6 Michelle M. Gehringer
Michelle M. Gehringer Nancy Merino
Nancy Merino