- 1Department of Molecular and Cellular Biochemistry, Indiana University, Bloomington, IN, United States
- 2Department of Biology, Indiana University, Bloomington, IN, United States
Across the three domains of life, B-family DNA polymerases play a variety of roles in both DNA repair and DNA replication processes. We examine the phenotypic consequences of loss of the putative repair polymerases PolB2 and/or PolB3 in the crenarchaeon Sulfolobus islandicus. We detect a modest growth advantage when cells lacking the polymerase are grown in unperturbed conditions. Further, we observe a striking insensitivity of the mutant lines to acute treatment with the oxidizing agent, hydrogen peroxide. In addition, cells lacking PolB3 show enhanced sensitivity to the DNA damaging agent 4-NQO. Our data therefore suggest that these non-essential DNA polymerases may influence DNA repair pathway choice in these hyperthermophilic aerobes.
Introduction
DNA polymerases perform the essential function of genome replication but show striking diversity across phyletic divides. Even within a domain of life there is considerable variation in the representation of different DNA polymerase family members. For example, most archaea possess a signature D-family DNA polymerase, PolD, that has recently been shown to possess a double-psi barrel catalytic fold reminiscent of multi-subunit RNA polymerases. The D-family enzymes are heterodimers and features of the interaction interface between subunits are conserved in eukaryotic multi-subunit DNA polymerases (Tahirov et al., 2009; Raia et al., 2019a, b; Greci and Bell, 2020). Interestingly, the PolD-containing archaea also encode a B-family DNA polymerase, PolB. The available evidence suggests that PolD is responsible for the majority of genome replication with PolB playing a non-essential role, most likely in DNA repair processes (Cubonova et al., 2013; Sarmiento et al., 2013).
The Crenarchaea, which harbor the model organisms of the Sulfolobales lack a D-family polymerase. With a single exception, species within the Sulfolobales typically encode three B-family polymerases as well as a Y-family DNA polymerase that is dedicated to DNA repair (Miyabayashi et al., 2020). Recent work has revealed that of the three PolB enzymes, only PolB1 is essential for viability, while cells lacking PolB2 or PolB3 are able to grow (Martinez-Alvarez et al., 2017). Indeed, a recent characterization of Sulfodiicoccus acidiphilus has revealed that this species naturally lacks PolB3 (Sakai and Kurosawa, 2019). It has long been known that the gene for PolB2 is transcriptionally induced in response to UV-induced stress on cells (Frols et al., 2007; Gotz et al., 2007). However, little is known about the physiological role of PolB3. Notably, PolB3 is the closest sequence relative to the PolB putative DNA repair enzyme in those archaea that possess the PolD replicative DNA polymerase (Makarova et al., 2014). Thus, it seems likely that both PolB2 and PolB3 play roles in the DNA damage response. Recent work has provided evidence to support this proposal, with strains lacking PolB2 or PolB3 demonstrating sensitivity to DNA damaging agents (Feng et al., 2020; Miyabayashi et al., 2020). In the following, we address the roles of PolB2 and/or PolB3 during normal, unperturbed growth and under conditions of acute and chronic treatment with an array of DNA damaging agents.
Materials and Methods
Archaeal Strains
The dpo2 and dpo3 knock-out strains used in these experiments were provided by Prof. Xu Peng, Copenhagen (Martinez-Alvarez et al., 2017). The dpo2/3 knock-out was constructed by a modification of the S. islandicus type I CRISPR-Cas3 system as previously described (Li et al., 2016; see Table 1).
To improve the selection of transformants we modified the pGE1 plasmid (Martinez-Alvarez et al., 2017) to include the Sulfolobus tokodaii gene for HMG-CoA reductase, the over-expression of which confers simvastatin resistance in Sulfolobus species (Zheng et al., 2012), by amplification of the cassette from pSSR (Zheng et al., 2012) using oligonucleotides ForSimR and RevSimR (see Table 3), and insertion into the SmaI site of pGE1 (we note that we have had persistent problems with high background when relying on uracil prototrophy conferred by the original pGE1 vector). Initial transformants were selected using 18 μM simvastatin and subsequently grown without the drug and in the presence of 20 μg/ml 5-Fluoroorotic Acid (5-FOA) to counter select. All strains, plasmids and DNA oligonucleotides used are listed in Tables 1–3. Competent cells were made by washing harvested mid-log phase cells 3 times with 20 mM sucrose at room temperature. Six hundred nanogram of vector were introduced by means of electroporation via a single pulse using a BioRad GenePulserXcell and BioRad Shock Pod with parameters set to 1.2 kV, 25 μF, and 600 Ω.
Growth Conditions
WT and mutant strains were grown in Tryptone-Sucrose-Vitamin-Yeast Extract (TSVY) medium [0.1% tryptone (w/v), 0.2% sucrose (w/v), 1% vitamin solution (v/v), 0.05% Yeast Extract (w/v), pH 3.5], or TSVYu (+20 μg/ml uracil) at 78°C. Transformations were grown on Sucrose-Casamino Acid-Vitamin-Yeast Extract (SCVy) plates with 18 μM simvastatin [0.2% sucrose (w/v), 0.2% Casamino Acids (w/v), 0.004% Yeast Extract (w/v), pH 3.5] at 78°C. All plating was carried out on equivalent medium plates containing 1% Gelrite. For growth curve analysis, 50–100 ml cultures were started at A600 = 0.03 in media pre-warmed to 78°C. The growth was monitored by tracking A600 values with an Eppendorf BioPhotometer at 6 h intervals.
Flow Cytometry
One hundred microliter samples of cell culture were collected at 6 h intervals during growth. The cells were then fixed in 900 μL ice-cold 80% ethanol and stored at 4°C for up to 1 month. 500 μL of fixed cells were subsequently centrifuged for 5 min at 15,000 rpm and washed in 500 μL sterile 10 mM Tris-HCl (pH 7.4), 10 mM MgCl2, followed by a second wash in 250 μL of the same buffer. Cells were centrifuged again and resuspended in 1 mL of the same buffer containing 2.5 μM Sytox Green (Invitrogen) and 5 μg/mL RNase A. Analysis of samples was performed on a BD LSR II flow cytometer using FACSDiva software and a laser excitation of 488-nm. A data set of at least 50,000 events was collected for each sample.
Genomic DNA Preparation and MFA-Seq
Five to twenty milliliter cell culture were harvested and washed 2 times with TEN buffer [50 mM Tris-HCl (pH8.0), 50 mM EDTA, 100 mM NaCl] and resuspended in 1/10 the original volume. 20% SDS solution was added to a final concentration of 0.1% along with RNase A (0.02 mg/ml) and incubated at 37°C for 30 min, followed by the addition of proteinase K (0.02 mg/ml) and a 1 h incubation at 37°C. The lysate was extracted in 1 volume phenol:chloroform:isoamyl alcohol, 25:24:1 (pH 7.8), 2 times, followed by 1 volume of chloroform alone. Genomic DNA was then precipitated in 66% ethanol and analyzed by 0.8% agarose gel electrophoresis. Marker Frequency Analysis (MFA-Seq) was performed by Illumina sequencing of DNA from exponentially growing or stationary phase cells. Between 17 and 22 million reads per sample were mapped to the S. islandicus LAL14/1 reference genome using the program Geneious Prime version 2020.2.2. BAM files generated by Geneious were analyzed using SeqMonk1. Read counts were generated in 2 kb probe windows and filtered to exclude repetitive elements. Read counts per probe were normalized by reference to stationary phase (non-replicating) DNA to account for variation in sequencing efficiency across the genome. SNPs were identified using Geneious.
Drug Preparation
Each drug that was used was prepared from powder or a stable concentrated stock immediately before its addition to the media. Hydrogen Peroxide solution was prepared by diluting 35% H2O2 to a concentration of 127 mM in water. Hydroxyurea was prepared from powder to a working concentration of 1 M in water. 4-Nitroquinoline 1-oxide (4-NQO) was first prepared as a concentrated solution from powder in DMSO, it was then diluted to a working concertation of 130 μM in 1% DMSO. Cisplatin was prepared from powder to a working concentration of 0.5 mg/ml in 0.9% NaCl solution.
Exposure to DNA Damaging Agents and Spot Assays
For acute exposure to Hydrogen Peroxide, Hydroxyurea, and 4-NQO, strains were first grown to A600 = 0.3–0.4, all strains were then normalized to A600 = 0.3 by the addition of pre-warmed media and split into 10 ml cultures before the indicated amount of drug was added. The cells were returned to 78°C and grown with shaking (110 rpm) for 7 h before samples were collected for plating or inoculating new cultures. Spot plating was accomplished by diluting each culture to A600 = 0.1 in pre-warmed TSVY, followed by serial diluting twofold or 10-fold as indicated. Each dilution was kept at 78°C with 5 μl spotted onto TSVYu plates. Plates were grown at 78°C for 7–8 days. For the 4-NQO recovery assay, cells were washed with fresh TSVYu after acute drug exposure as described above, then used to inoculate cultures to A600 = 0.03. The cultures were subsequently grown at 78°C with growth tracked by A600 every 6 h until cell death was observed. UV exposure was accomplished by growing each strain to A600 = 0.1, after which 3 ml were used to cover the bottom of a 100 mm × 15 mm polystyrene petri dish at room temperature. Cells were exposed to 200 and 250 J/m2 UV (additional UV doses were also checked; data not shown) using a SpectroLinker XL-1000 UV Crosslinker (Spectronics Corporation). Plating was carried out as described above in a dark room to prevent photo-reactivation. For chronic exposure to Hydrogen Peroxide, Hydroxyurea, 4-NQO, and Cisplatin, strains were grown to A600 = 0.3–0.4, then diluted to A600 = 0.1 in pre-warmed TSVY. Samples were diluted and spotted as previously described onto TSVYu plates containing the indicated concentration of drug. Plates were grown at 78°C for 7–8 days.
Results
The Impact of Loss of dpo2 and/or dpo3 on Unperturbed Growth
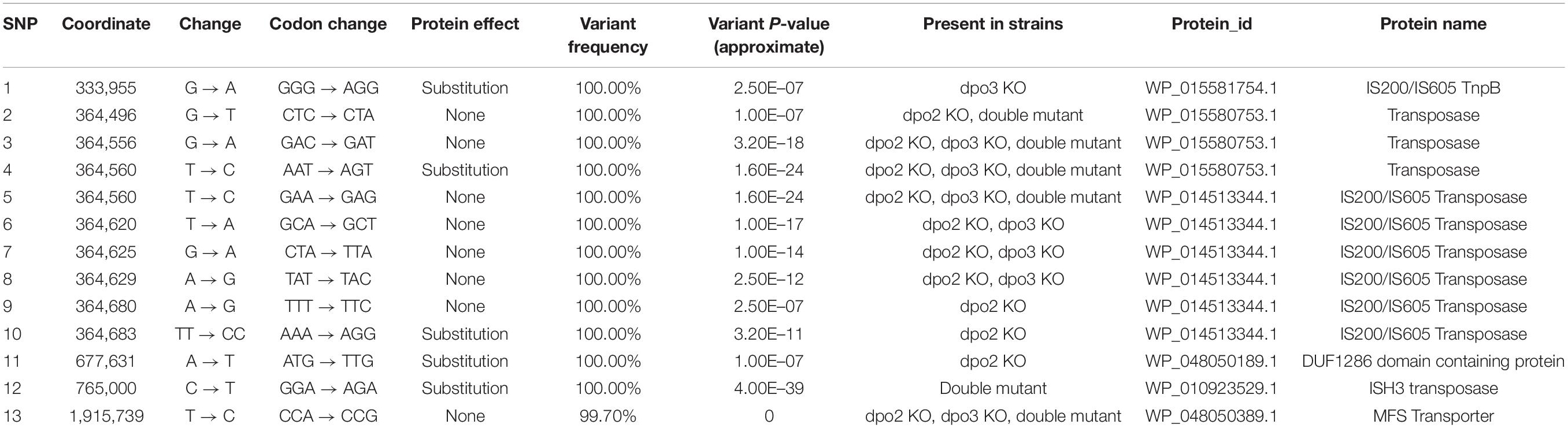
We focused our efforts on the Sulfolobus islandicus strain LAL14/1. Previous work by Peng and colleagues had revealed PolB1 to be essential for viability in this strain (Martinez-Alvarez et al., 2017). However, strains lacking the dpo2 or dpo3 genes, that encode PolB2 or PolB3, respectively, were able to survive. We obtained these mutant strains and, using the endogenous CRISPR system (Li et al., 2016), generated a further strain that lacked both PolB2 and PolB3. We first measured the growth parameters of the wild-type and three mutant strains under standard, non-perturbed conditions in rich medium (Figure 1A). During exponential growth both single mutants showed slightly faster growth rates than wild-type. More specifically, the doubling time of both mutants was 6.3 h, compared to 6.7 h for wild-type (Figures 1A,B). Furthermore, the single mutants entered stationary phase at higher cell densities than did the wild-type cultures and showed considerable variation in cell numbers at analogous times between replicates in stationary phase. The double mutant had the same accelerated growth rate during exponential growth as the single mutants (doubling time of 6.3 h) and entered stationary phase at a higher cell density than observed with the wild-type. We analyzed the cell cycle distribution of the wild-type and mutant lines using flow cytometry but could not observe any reproducible differences in the population of G1, S, and G2 phase cells between wild-type and mutants (Figure 1C). We next analyzed DNA replication profiles by Marker Frequency Analysis as measured by Illumina Sequencing (MFA-Seq). MFA-Seq profiles indicated that all four cell lines initiated DNA replication from the three DNA replication origins conserved in Sulfolobus species (Figures 2A–D). In agreement with our previous analyses on other Sulfolobus species and strains (Duggin et al., 2008; Samson et al., 2013), in wild-type cells the amplitude of the peak corresponding to oriC2 is lower than that for oriC1 and oriC3 (dotted blue line in Figure 2). We have previously demonstrated that this is due to slightly later firing of oriC2 in S-phase (Duggin et al., 2008). Interestingly, examination of the data reveal that the peaks corresponding to initiation at oriC2 are higher than those seen for oriC1 and oriC3 in cells lacking either PolB2 or PolB3. Furthermore, the double mutant has a profile closer to that of wild-type, with all three origins giving peaks of similar amplitude. We speculate that while the gross cell cycle parameters of the mutant cell lines are essentially unaltered, perhaps the accelerated firing of oriC2 is indicative of some subtle mis-regulation linked to the accelerated growth rate of these strains. However, this remains pure speculation at this time. The MFA-Seq data also allowed us to determine if any unanticipated mutations had arisen in any of the strains. Comparison of our laboratory wild-type strain of S. islandicus LAL14/1, obtained from Prof. Xu Peng, Copenhagen, revealed 23 SNPs relative to the reference genome sequence deposited at NCBI (Jaubert et al., 2013; Table 4). A total of 13 further SNPs were detected in the genome of the mutant strains, not including those associated with the targeted mutations (Table 5). Eight of those resulted in synonymous codon changes. Four of the remaining five were in multi-copy transposable element-encoded genes (and could be artifacts due to alignment errors to these repetitive elements) and the final mutation was a conservative mutation in the dpo2 knock-out strain resulting in a M-L substitution in the SiL_0710 open-reading frame. Notably, this same substitution is seen naturally in a second copy of this gene (SiL_0897). Accordingly, we can be confident that the phenotypes we observe arise as a consequence of deletion of the targeted DNA polymerase genes.
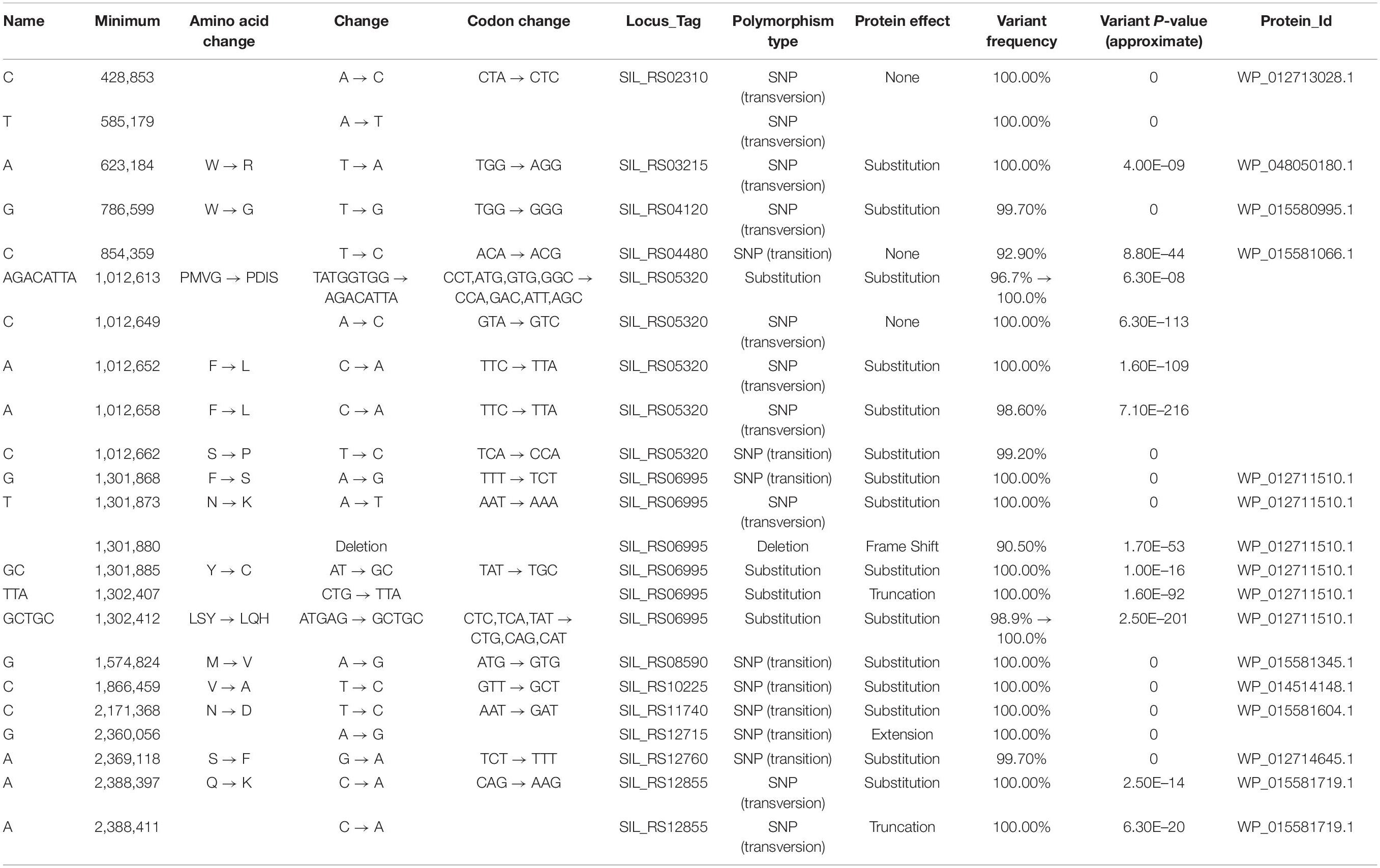
Table 4. SNPs identified in the wild-type strain (compared to the S. islandicus LAL14/1 reference genome: NCBI NC_021058.1).
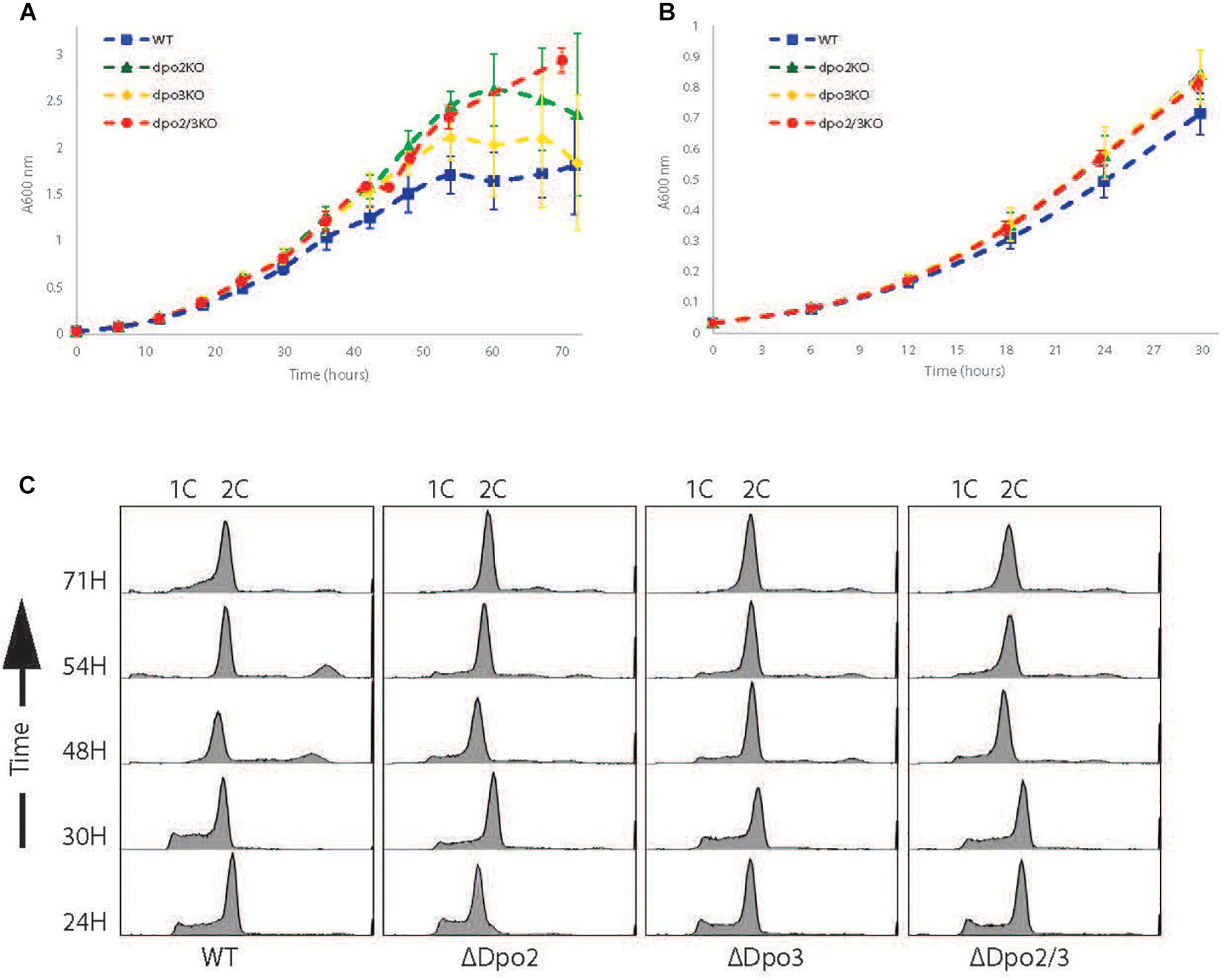
Figure 1. Growth profiles of strains. (A) Growth curve of WT, Dpo2KO, Dpo3KO, and Dpo2/3KO lines over 72 h. Samples were collected for analysis every 6 h. Growth curves for each line were repeated at least 3 times. (B) The first 30 h growth for each line. (C) Flow cytometry profiles of each line. Samples collected simultaneously with growth curve samples.
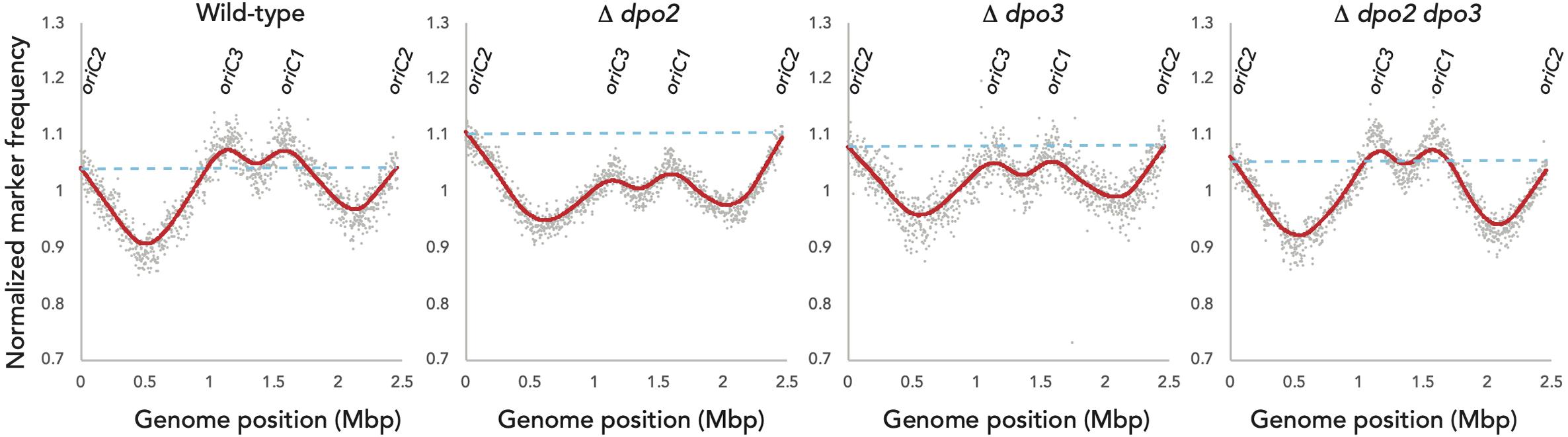
Figure 2. Marker-Frequency Analysis. Marker-frequency analysis of WT, Δdpo2,Δdpo3, andΔdpo2/3 lines. DNA was collected from each culture during exponential phase growth and subjected to Illumina next-generation sequencing. Read counts were generated in 2 kb windows and normalized to stationary phase DNA. Raw values for individual counts are displayed in gray and a LOWESS smoothing is shown in red. The dotted blue line indicates the amplitude of the oriC2 peak.
Sensitivity to Acute DNA Damage
First, we tested the effect of exposure of cells to 0, 200, or 250 J/m2 of UV light of 254 nm (Figure 3A). A modest effect was observed at the highest UV dose, with the dpo2/3 double mutant showing reduced survival in spot tests of serial dilutions of cells following exposure (to prevent photoreactivation these experiments and growth of plates were performed in the dark). We also tested the effect of acute exposure to 4-Nitroquinoline-1 oxide (4-NQO), which, like UV, induces lesions typically repaired by nucleotide excision repair pathways, by administering the drug at 0, 1, 2, or 3 μM) for 7 h (roughly one doubling time) prior to washing the cells and assaying survival by serial dilution and plating. Loss of PolB3 sensitized cells to 4-NQO while loss of PolB2 had no discernable effect. Interestingly, however, the double mutant was more sensitive to 2 and 3 μM 4-NQO than either mutant alone (Figure 3B). We additionally compared growth assays in liquid culture of cells either mock treated or transiently treated with 4-NQO. In agreement with the plating assays, we observed that loss of PolB2 had minimal effects, loss of PolB3 sensitized cells to 4-NQO and the double mutant strain was most sensitive of all (Figure 3C). This suggests that PolB3 plays a primary role in the response to damage induced by acute 4-NQO treatment with PolB2 playing a role in a back-up or ancillary pathway. The lack of sensitivity of the S. islandicus LAL14/1 dpo2 single mutant to either acute or chronic 4-NQO treatment (see below) was surprising in light of a recent report from Feng and colleagues. That study indicated that a dpo2 knock-out mutant in S. islandicus REY15A demonstrated enhanced sensitivity to 4-NQO (Feng et al., 2020). In light of this apparent contradiction, we isolated DNA from the cells plated on the acute 4-NQO treatment plates and confirmed by PCR that the expected genotypes were indeed present (Figures 3D,E). As discussed below, while our data appear at odds with those of Feng and colleagues, they are compatible with a very recent report from Miyabyashi on the phenotypes of DNA polymerase mutants of the congeneric species, Sulfolobus acidocaldarius (Miyabayashi et al., 2020).
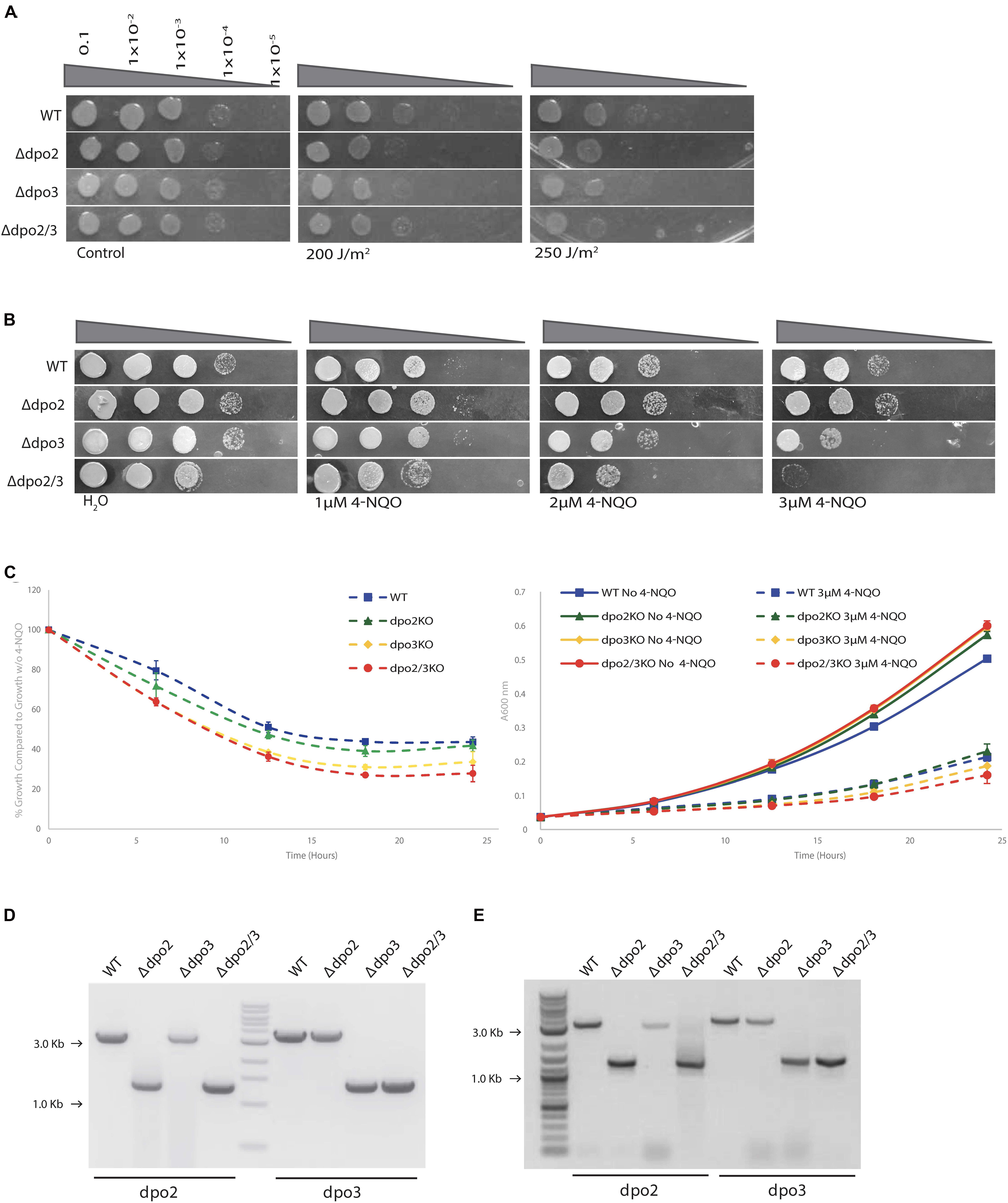
Figure 3. Growth of strains exposed to UV and acute 4-NQO. (A) Serial dilutions of each cell line plated onto TSVYu after exposure to UV (254 nm) at the indicated intensity. Growth conditions and spot plates were repeated at least 3 times. (B) Serial dilutions of each line plated onto TSVYu after growth for 7 h (about 1 doubling time) with 4-NQO at the indicated concentrations. Growth conditions and spot plates were repeated at least 3 times. (C) Growth analysis of cell lines grown in TSVYu media after growth for 7 h with 3 μM 4-NQO. Cultures were grown over a 24 h period starting at A600 = 0.03 with samples collected for analysis ever 6 h. In the left hand panel, A600 values were normalized against growth without 4-NQO. The right hand panel shows the original growth curves. (D) Agarose gel electrophoresis of PCR tests confirming that our freezer stocks possessed anticipated genotype, positions of PCR products indicating amplification of wild-type (WT) or deletion-containing (Δ) loci are indicated (M-DNA markers). (E) As D but with DNA isolated from cells derived from the plates shown (B).
Next, we tested the effect of acute treatment with hydroxyurea (HU). We have previously demonstrated that this compound induced replication stress and DNA damage and that these effects likely arise as a consequence of the HU-mediated inactivation of the iron-sulfur cluster-containing large subunit of the archaeal primase (Liew et al., 2016). While treatment of the double mutant or the strain lacking PolB3 with 5 mM HU resulted in modestly reduced survival, loss of PolB2 resulted in HU sensitivity indistinguishable from the wild-type cells (Figure 4A).
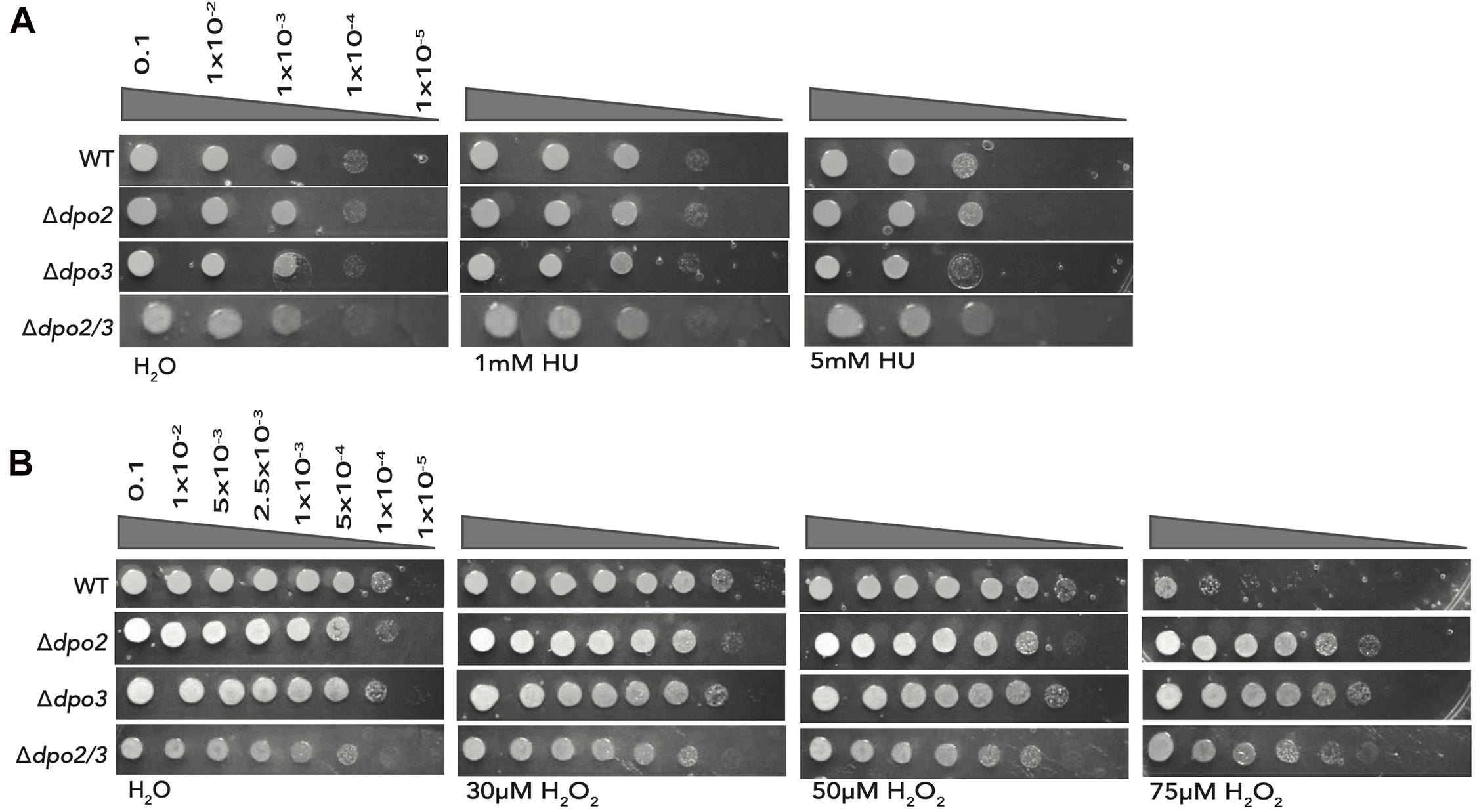
Figure 4. Effect of acute exposure to HU and H2O2 on strain growth. (A) Serial dilutions of each line plated onto TSVYu after growth for 7 h (about 1 doubling time) with HU at the indicated concentrations. Growth conditions and spot plates were repeated at least 3 times. (B) Serial dilutions of each line plated onto TSVYu after growth for 7 h (about 1 doubling time) with H2O2 at the indicated concentrations. Growth conditions and spot plates were repeated at least 3 times.
Finally, we tested the effect of oxidative damage induced by acute treatment with 0, 30, 50, or 75 μM hydrogen peroxide (Figure 4B). Strikingly, loss of either PolB2 or PolB3 caused substantially enhanced survival (~3 orders of magnitude) following treatment with 75 μM H2O2, The double mutant, in this instance, was less resistant to 75 μM H2O2 treatment than either single mutant but still displayed 100-fold enhanced survival compared to the wild-type.
Sensitivity to Chronic DNA Damage
Next, we sought to determine the sensitivity of the wild-type and mutant strains to chronic exposure to the DNA damaging agents tested above. While it was impractical to grow cells under constant exposure to UV light, we tested their ability to grow on plates containing varying concentrations of 4-NQO (Figure 5A). The Δdpo2 mutant was very slightly less sensitive to 120 nM 4-NQO than the wild-type cells, the dpo3 single knock-out was similar to wild-type and the double mutant was more sensitive to 4-NQO than the wild-type.
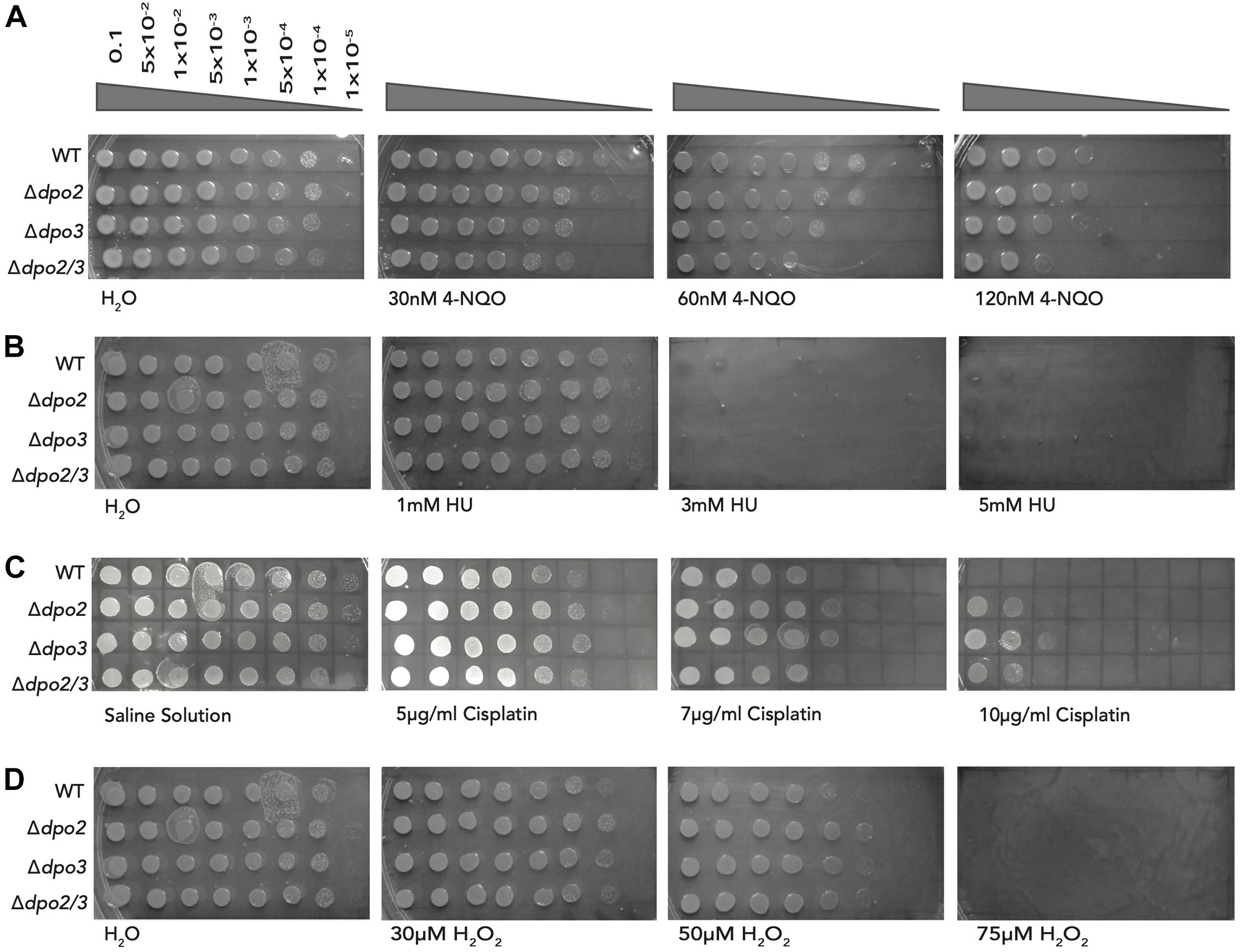
Figure 5. Impact of chronic exposure to 4-NQO, HU, Cisplatin, and H2O2 on the growth of the strains. (A) Serial dilutions of each line plated onto TSVYu containing 4-NQO at the indicated concentrations and grown for 7–8 days. Growth conditions and spot plates were repeated at least 3 times. (B) Serial dilutions of each line plated onto TSVYu containing HU at the indicated concentrations and grown for 7–8 days. The positive control plate presented is the same as that seen in Figure 5D (Chronic H2O2 exposure), both experiments were performed at the same time. Growth conditions and spot plates were repeated at least 3 times. (C) Serial dilutions of each line plated onto TSVYu containing Cisplatin at the indicated concentrations and grown for 7–8 days. Growth conditions and spot plates were repeated at least 3 times. (D) Serial dilutions of each line plated onto TSVYu containing H2O2 at the indicated concentrations and grown for 7–8 days. Note that the positive control plate presented is the same as that seen in Figure 5B (Chronic HU exposure), this plate served as positive control for both of the selected replicates of HU and H2O2 experiments. Growth conditions and spot plates were repeated at least 3 times.
We additionally tested the effect of chronic exposure to hydroxyurea and could not detect any difference in sensitivity to this agent between wild-type and any of the single or double mutant lines. We note that prolonged incubation of hydroxyurea has been reported to yield a number of toxic breakdown products including cyanide, peroxide and nitric oxide (Kuong and Kuzminov, 2009). Thus, interpretation of these chronic exposure data could be complicated by the multifactorial nature of the stresses imposed upon cells exposed to HU during the 7 days of growth of the organisms on plates.
We also assayed the consequence of chronic exposure to the intra-strand crosslinking agent cisplatin we observed that at low doses of the drug (5 μg/ml) cells lacking either of PolB2 or PolB3 showed modestly enhanced survival when compared to wild-type. This enhanced survival is accentuated at higher cisplatin doses. We note that at 10 μg/ml cisplatin, the single and double mutants show reduced sensitivity when compared to the wild-type strain. We note that we did not report the sensitivity to acute exposure to cisplatin as we found high experimental variability between samples. We were confident that the chronic exposure to cisplatin was yielding equivalent exposures to all cells plated on a given plate.
Finally, we assessed the impact of chronic exposure to hydrogen peroxide. As shown above, acute exposure to this agent yielded a striking increase in survival in cells lacking PolB2 and/or PolB3. Under chronic exposure conditions we observed a modest (maximally 10-fold) enhancement of survival in the mutant lines compared to wild-type at 30 and 50 μM hydrogen peroxide. We note that hydrogen peroxide may have undergone breakdown over the time course of growth on the chronic exposure plates, perhaps consequentially attenuating the phenotype observed.
Discussion
In agreement with recent reports, we observe that neither PolB2 or PolB3 are required for viability under unperturbed growth condition (Martinez-Alvarez et al., 2017), indeed we observed a modest enhancement of growth rate in both single and double mutant lines. We therefore add to the body of data confirming that PolB1 is the sole B-family DNA polymerase required for genome replication in Sulfolobus species. Our data regarding the effect of 4-NQO are largely compatible with the recent report from Miyabyashi, describing a comprehensive analysis of the roles of PolB2, PolB3 and the lesion bypass Y-family DNA polymerase, Dbh, from Sulfolobus acidocaldarius (Miyabayashi et al., 2020). However, our data on the role of PolB2 in the response to 4-NQO treatment differ markedly from those of Feng and colleagues, who reported an enhanced sensitivity of the dpo2 mutants of Sulfolobus islandicus REY15A to 4-NQO (Feng et al., 2020). In our experiments in S. islandicus LAL14/1, we do not observe any enhanced sensitivity of the dpo2 mutant strain to either acute or chronic exposure to 4-NQO. In contrast, we actually observe modestly enhanced survival of this mutant line under conditions of chronic exposure to this drug, over a range of concentrations. We note that the predicted amino-acid sequence of PolB2 is identical in the S. islandicus strains REY15A and LAL14/1. Significantly, based upon the sequences provided by Feng and colleagues, the Δdpo2 strain used in their study differs in the scope of the gene deletion to that of the Δdpo2 strain provided to us by Prof. Xu Peng, Copenhagen (Martinez-Alvarez et al., 2017; Feng et al., 2020). The Δdpo2 strain utilized in the study by Feng and colleagues does not completely delete the coding region of dpo2, leaving the first 21 bp (including the start codon) and final 48 bp (including the stop codon) intact, resulting in a 22 amino acid long peptide “MREMEEYLRRVYDNVEEVISRC” the underlined N-terminal seven amino acids correspond to the N-terminal sequences of PolB2 and the 15 residues in bold correspond to PolB2’s C-terminal sequences. It is conceivable that this short peptide could play a trans-dominant negative role (if, for example, the extreme N or C-termini of PolB2 interact with another protein). In contrast, the Δdpo2 strain used in our study only has no start codon and only maintains the final 7 bp of the coding region. These differences between the two strains could account for the discrepancy in the data that we observed. The Δdpo3 strain used by Feng and colleagues is identical to that provided to us by Prof. Xu Peng. Our deep-sequencing analyses confirm that there are no further mutations in our strains that could reasonably account for this discrepancy in the data. We also emphasize that we re-confirmed the genotype of the strains following the 4-NQO experiment, using DNA obtained from cultures derived from the test plates.
Under conditions of acute treatment with 4-NQO, we observe a 10-fold lower survival of the dpo3 mutant and loss of both dpo2 and dpo3 confers clear 4-NQO sensitivity under both acute and chronic exposure conditions. The data therefore suggest that, under the growth conditions we employed, PolB2 plays a minimal role in recovering from 4-NQO-mediated DNA damage and is secondary to PolB3 in this response.
Arguably the most striking phenotype we observe is the dramatic enhancement of the mutants’ survival upon acute exposure to oxidative damage induced by treatment with hydrogen peroxide. Given that the data discussed above suggest a role for these polymerases in the nucleotide excision repair pathway that repairs damage caused by 4-NQO, it is very surprising that loss of these polymerases stimulates the response to oxidative damage. We note, however, that there is precedent for this sort of phenomenon in the archaeal DNA repair literature. More specifically, Allers and colleagues revealed that loss of the rad50 and mre11 genes in Haloferax volcanii resulted in enhanced survival compared to the wild-type strain in the presence of a variety of DNA damaging agents, including UV light, ionizing radiation and the alkylating agent methyl methanesulfonate (Delmas et al., 2009). Significantly, it was noted in that study, while overall survival was higher in the rad50 and mre11 mutant lines, the kinetics of double strand break repair events were significantly slower. Furthermore, it was proposed that the Haloferax RAD50Mre11- containing complex acts to inhibit repair by homologous recombination. We speculate that a conceptually analogous phenomenon of cross-pathway interference may be at play in Sulfolobus.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI SRA BioProject, Accession No: PRJNA716831.
Author Contributions
PB performed the experiments. SB and PB designed the experiments and wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
SB’s lab was funded by the College of Arts and Sciences, Indiana University and by the National Institutes of Health Grants R01GM135178 and R01GM125579.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Bell lab for their comments on this work. We are grateful to the IU Center for Genomics and Bioinformatics for next generation sequencing and to Christiane Hassel of the Indiana University Flow Cytometry Core Facility for assistance with flow cytometry.
Footnotes
References
Cubonova, L., Richardson, T., Burkhart, B. W., Kelman, Z., Connolly, B. A., Reeve, J. N., et al. (2013). Archaeal DNA polymerase D but not DNA polymerase B is required for genome replication in Thermococcus kodakarensis. J. Bacteriol. 195, 2322–2328. doi: 10.1128/jb.02037-12
Delmas, S., Shunburne, L., Ngo, H. P., and Allers, T. (2009). Mre11-Rad50 promotes rapid repair of DNA damage in the polyploid archaeon Haloferax volcanii by restraining homologous recombination. PLoS Genet. 5:e1000552. doi: 10.1371/journal.pgen.1000552
Duggin, I. G., Mccallum, S. A., and Bell, S. D. (2008). Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. U.S.A. 105, 16737–16742. doi: 10.1073/pnas.0806414105
Feng, X., Liu, X., Xu, R., Zhao, R., Feng, W., Liao, J., et al. (2020). A Unique B-Family DNA polymerase facilitating error-prone DNA damage tolerance in crenarchaeota. Front. Microbiol. 11:1585. doi: 10.3389/fmicb.2020.01585
Frols, S., Gordon, P. M., Panlilio, M. A., Duggin, I. G., Bell, S. D., Sensen, C. W., et al. (2007). Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J. Bacteriol. 189, 8708–8718. doi: 10.1128/jb.01016-07
Gotz, D., Paytubi, S., Munro, S., Lundgren, M., Bernander, R., and White, M. F. (2007). Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol. 8:R220.
Jaubert, C., Danioux, C., Oberto, J., Cortez, D., Bize, A., Krupovic, M., et al. (2013). Genomics and genetics of Sulfolobus islandicus LAL14/1, a model hyperthermophilic archaeon. Open Biol. 3:130010. doi: 10.1098/rsob.130010
Kuong, K. J., and Kuzminov, A. (2009). Cyanide, peroxide and nitric oxide formation in solutions of hydroxyurea causes cellular toxicity and may contribute to its therapeutic potency. J. Mol. Biol. 390, 845–862. doi: 10.1016/j.jmb.2009.05.038
Li, Y., Pan, S., Zhang, Y., Ren, M., Feng, M., Peng, N., et al. (2016). Harnessing type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 44:e34. doi: 10.1093/nar/gkv1044
Liew, L. P., Lim, Z. Y., Cohen, M., Kong, Z., Marjavaara, L., Chabes, A., et al. (2016). Hydroxyurea-Mediated Cytotoxicity Without Inhibition of Ribonucleotide Reductase. Cell Rep. 17, 1657–1670. doi: 10.1016/j.celrep.2016.10.024
Makarova, K. S., Krupovic, M., and Koonin, E. V. (2014). Evolution of replicative DNA polymerases in archaea and their contributions to the eukaryotic replication machinery. Front. Microbiol. 5:354. doi: 10.3389/fmicb.2014.00354
Martinez-Alvarez, L., Deng, L., and Peng, X. (2017). Formation of a viral replication focus in sulfolobus cells infected by the rudivirus sulfolobus islandicus rod-shaped virus 2. J. Virol. 91:e00486–17.
Miyabayashi, H., Jain, R., Suzuki, S., Grogan, D. W., and Kurosawa, N. (2020). PolB1 is sufficient for DNA replication and repair under normal growth conditions in the extremely thermophilic crenarchaeon Sulfolobus acidocaldarius. Front. Micro. 11:613375. doi: 10.3389/fmicb.2020.613375
Raia, P., Carroni, M., Henry, E., Pehau-Arnaudet, G., Brule, S., Beguin, P., et al. (2019a). Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases. PLoS Biol. 17:e3000122. doi: 10.1371/journal.pbio.3000122
Raia, P., Delarue, M., and Sauguet, L. (2019b). An updated structural classification of replicative DNA polymerases. Biochem. Soc. Trans.l 47, 239–249. doi: 10.1042/bst20180579
Sakai, H. D., and Kurosawa, N. (2019). Complete genome sequence of the Sulfodiicoccus acidiphilus strain HS-1(T), the first crenarchaeon that lacks polB3, isolated from an acidic hot spring in Ohwaku-dani, Hakone, Japan. BMC Res. Notes 12:444. doi: 10.1186/s13104-019-4488-5
Samson, R. Y., Xu, Y., Gadelha, C., Stone, T. A., Faqiri, J. N., Li, D., et al. (2013). Specificity and function of archaeal DNA replication initiator proteins. Cell Rep. 3, 485–496. doi: 10.1016/j.celrep.2013.01.002
Sarmiento, F., Mrazek, J., and Whitman, W. B. (2013). Genome-scale analysis of gene function in the hydrogenotrophic methanogenic archaeon Methanococcus maripaludis. Proc. Natl. Acad. Sci. U.S.A. 110, 4726–4731. doi: 10.1073/pnas.1220225110
Tahirov, T. H., Makarova, K. S., Rogozin, I. B., Pavlov, Y. I., and Koonin, E. V. (2009). Evolution of DNA polymerases: an inactivated polymerase-exonuclease module in Pol epsilon and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol. Direct 4:11. doi: 10.1186/1745-6150-4-11
Keywords: Sulfolobus, archaea, DNA replication, DNA damage, polymerase
Citation: Bohall PB and Bell SD (2021) Phenotypic Characterization of Sulfolobus islandicus Strains Lacking the B-Family DNA Polymerases PolB2 and PolB3 Individually and in Combination. Front. Microbiol. 12:666974. doi: 10.3389/fmicb.2021.666974
Received: 11 February 2021; Accepted: 29 March 2021;
Published: 22 April 2021.
Edited by:
Hannu Myllykallio, École Polytechnique, FranceCopyright © 2021 Bohall and Bell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen D. Bell, c3RlZGJlbGxAaW5kaWFuYS5lZHU=
 Peter B. Bohall
Peter B. Bohall Stephen D. Bell
Stephen D. Bell