- 1College of Veterinary Medicine, Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, Key Laboratory of Zoonosis of Ministry of Agricultural and Rural Affairs, National Risk Assessment Laboratory for Antimicrobial Resistance of Microorganisms in Animals, South China Agricultural University, Guangzhou, China
- 2Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China
- 3Biomedicine Discovery Institute and Department of Microbiology, School of Biomedical Sciences, Monash University, Clayton, VIC, Australia
- 4College of Veterinary Medicine, Center for Emerging and Zoonotic Diseases, South China Agricultural University, Guangzhou, China
Plasmid-mediated colistin resistance gene mcr-1 generally confers low-level resistance. The purpose of this study was to investigate the impact of mcr-1 on the development of high-level colistin resistance (HLCR) in Klebsiella pneumoniae and Escherichia coli. In this study, mcr-1-negative K. pneumoniae and E. coli strains and their corresponding mcr-1-positive transformants were used to generate HLCR mutants via multiple passages in the presence of increasing concentrations of colistin. We found that for K. pneumoniae, HLCR mutants with minimum inhibitory concentrations (MICs) of colistin from 64 to 1,024 mg/L were generated. Colistin MICs increased 256- to 4,096-fold for mcr-1-negative K. pneumoniae strains but only 16- to 256-fold for the mcr-1-harboring transformants. For E. coli, colistin MICs increased 4- to 64-folds, but only 2- to 16-fold for their mcr-1-harboring transformants. Notably, mcr-1 improved the survival rates of both E. coli and K. pneumoniae strains when challenged with relatively high concentrations of colistin. In HLCR K. pneumoniae mutants, amino acid alterations predominately occurred in crrB, followed by phoQ, crrA, pmrB, mgrB, and phoP, while in E. coli mutants, genetic alterations were mostly occurred in pmrB and phoQ. Additionally, growth rate analyses showed that the coexistence of mcr-1 and chromosomal mutations imposed a fitness burden on HLCR mutants of K. pneumoniae. In conclusion, HLCR was more likely to occur in K. pneumoniae strains than E. coli strains when exposed to colistin. The mcr-1 gene could improve the survival rates of strains of both bacterial species but could not facilitate the evolution of high-level colistin resistance.
Introduction
Infections caused by multidrug-resistant pathogens pose significant risks to public health worldwide (Ruhnke et al., 2014; Huang et al., 2018). Owing to a limited number of effective antimicrobial agents, colistin has been reused and is considered to be the last therapeutic agent; however, resistance to colistin has been increasingly reported (Liu et al., 2016; Srinivas and Rivard, 2017). Previously, colistin resistance was mainly caused by mutations in chromosomal genes, including pmrAB, phoPQ, crrAB, and the negative regulator mgrB (Wright et al., 2015; Poirel et al., 2017; Srinivas and Rivard, 2017), which resulted in the modification of lipid A with positively charged residues, such as phosphoethanolamine and/or 4-amino-4-deoxy-L-arabinose. However, plasmid-mediated colistin resistance genes have also emerged recently. Since, we reported the first case of plasmid-mediated colistin resistance gene (mcr-1) in 2015 (Liu et al., 2016), several variants of mcr genes (mcr-2 to mcr-10) have been reported worldwide (Shi et al., 2020; Wang et al., 2020).
Similar to plasmid-mediated quinolone resistance genes, mcr-1 generally confers low-level colistin resistance (2–8 mg/L) (Jeong et al., 2008; Jacoby et al., 2014; Liu et al., 2016). However, it is unknown whether the presence of mcr-1 can facilitate the selection of a higher level of colistin resistance, as reported for qnr family genes (Jeong et al., 2008; Jacoby et al., 2014). Furthermore, the available data show that the prevalence of mcr-1 is significantly higher in Escherichia coli than that in Klebsiella pneumoniae (Quan et al., 2017; Eiamphungporn et al., 2018). Additionally, in clinical K. pneumoniae isolates, colistin resistance is mostly mediated by chromosomal mutations, which usually confer higher levels of resistance (16–1,024 mg/L) than that conferred by mcr (Cannatelli et al., 2014; Liu et al., 2016; Jayol et al., 2017). It seems that K. pneumoniae is more likely to generate chromosomal mutations but is less likely to harbor mcr-1 compared to the phenomenon observed in E. coli. Nevertheless, a previous study showed that after a single overnight exposure to twofold minimum inhibitory concentrations (MICs) of colistin, mcr-1 facilitated the development of high-level colistin resistance (HLCR; MIC ≥ 32 mg/L) mutants in E. coli but did not affect the HLCR mutation rates in K. pneumoniae (Zhang et al., 2019). However, another study reported that MCR-negative strains could be induced to higher level of colistin resistance than MCR-positive strains after step-wise induction (Luo et al., 2019). Thus, the impact of mcr-1 on the development of high level colistin resistance in K. pneumoniae and E. coli, and whether similar probability and frequency of mutations will be observed in E. coli and K. pneumoniae under selection with increasing concentrations of colistin remain unclear.
In this study, we investigated and compared the development of HLCR and chromosomal mutations among strains of E. coli and K. pneumoniae, with or without mcr-1, via multiple passages in the presence of increasing colistin concentrations.
Materials and Methods
Bacterial Strains and Plasmids
Klebsiella pneumoniae P11, HZ7H152, and YX6P94K and E. coli ATCC 25922, C600, and ZYTF186 were used as mcr-1-negative parental strains for this study. The sources and resistance phenotypes of these strains are listed in Supplementary Table 1. To construct an mcr-1-harboring plasmid, mcr-1 was cloned from pHNSHP45 (GenBank accession number KP347127.1) to pHSG575 using the primer pair listed in Supplementary Table 2, which yielded pHSG575-mcr-1 (Takeshita et al., 1987). The recombinant plasmid was transformed into E. coli ATCC 25922 by electroporation, whereas the original plasmid pHNSHP45 was transformed into K. pneumoniae P11, HZ7H152, and YX6P94K and E. coli C600 and ZYTF186. Transformants were selected on Luria–Bertani (LB) medium containing 2 mg/L colistin. The presence of mcr-1 in the transformants was confirmed as per previously described methods (Liu et al., 2016).
Whole-Genome Sequencing and Analysis
The whole genomic DNA of four strains, K. pneumoniae P11, HZ7H152, and YX6P94K and E. coli ZYTF186, was sequenced using Illumina HiSeq 2000 (Illumina, San Diego, CA, United States). Sequence reads were assembled into contigs using SOAPdenovo version 2.04. Resistance genes were explored using ResFinder1. The whole-genome sequences of two laboratory strains, E. coli C600 and ATCC 25922 (GenBank accession numbers NZ_CP031214.1 and NZ_CP009072.1, respectively), were obtained from NCBI and used as reference genomes.
Antimicrobial Susceptibility Testing
Minimum inhibitory concentrations of colistin were determined using Mueller–Hinton (MH) broth microdilution following the recommendations of the Clinical and Laboratory Standards Institute (Matuschek et al., 2018). E. coli ATCC 25922 was used as a reference strain.
Induction of Colistin Resistance by Conducting Serial Passages
Serial passaging was performed as per previously described methods (San Millan et al., 2016), with minor modifications. As shown in Supplementary Figure 1, strains were incubated on MH agar plates at 37°C to obtain single colonies. For each strain, 48 colonies were randomly selected and inoculated in separate wells of 96-well plates containing colistin-free LB broth to obtain overnight cultures. The overnight cultures were inoculated in 0.2 mL of LB broth with colistin (0.5 × MIC) in a new 96-well plate. Following a 24-h incubation at 37°C, the cultures were diluted 1:100 and transferred to a new plate containing a double concentration of colistin relative to that used on the previous day. The serial passages were performed until no surviving populations were observed. Surviving populations were defined considering growth indicated by OD600 (optical density at 600 nm) > 0.10 ± 0.02. The number of surviving populations was recorded for all treatments over time. For each strain, 5–10 surviving populations were selected along the concentration gradient of colistin from low to high, with priority selection of populations that survived at the highest concentration. Subsequently, the populations were separately plated on fresh LB agar to obtain single colonies, and three colonies were randomly selected from each population to determine their susceptibility to colistin and resistance mechanisms.
Genetic Alterations in Colistin-Resistant Mutants
Chromosomal genes (mgrB, pmrAB, phoPQ, and crrAB) associated with colistin resistance were amplified by PCR and sequenced using the primers listed in Supplementary Table 2. Genetic alterations that occurred in colistin-resistant mutants were determined by comparing the resulting sequences to their corresponding parental reference genomes.
In vitro Growth Rate Assay
Forty-four HLCR K. pneumoniae mutants with different mutations, including equal numbers of mcr-1-negative and mcr-1-positive derivatives, were selected incubated in fresh MH broth under shaking (180 rpm) conditions at 37°C. The overnight cultures were inoculated in fresh MH broth, and OD600 was measured using the Multiskan Spectrum microplate spectrophotometer (Thermo Labsystems, Franklin, MA, United States). The growth rates were determined by plotting the logarithm of OD600 versus time. Data analysis was performed using a non-parametric Mann–Whitney U-test, followed by Dunn’s multiple comparison test (Zwietering et al., 1990).
Accession Numbers
The whole genome sequencing data of K. pneumoniae P11, HZ7H152, K. variicola YX6P94K (subtype of K. pneumoniae) and E. coli ZYTF186 have been deposited in GenBank with accession numbers JAFKAD010000000, JAFKAE010000000, JAFKAG010000000, and JAFKAF010000000, respectively.
Results
Development of Resistance to Colistin via Serial Passages of Escherichia coli
Colistin-resistant E. coli mutants were obtained under increasing colistin pressure. The final induction concentrations of colistin, which inhibited the growth of all E. coli strains, ranged from 32 to 64 mg/L. As shown in Figures 1A–C, initially, the surviving populations of the mcr-1-negative parental strains (ATCC 25922, C600, and ZYTF186) plummeted in response to a challenge with 2 mg/L colistin, while those of their corresponding mcr-1-positive derivatives did not show a marked decline until the colistin concentration reached 16 mg/L. Furthermore, when exposed to relatively high concentrations of colistin (16 and 32 mg/L), mcr-1-positive derivatives showed higher survival rates than those of their corresponding mcr-1-negative strains, demonstrating that mcr-1 could enhance the survival rate of E. coli at higher colistin concentrations.
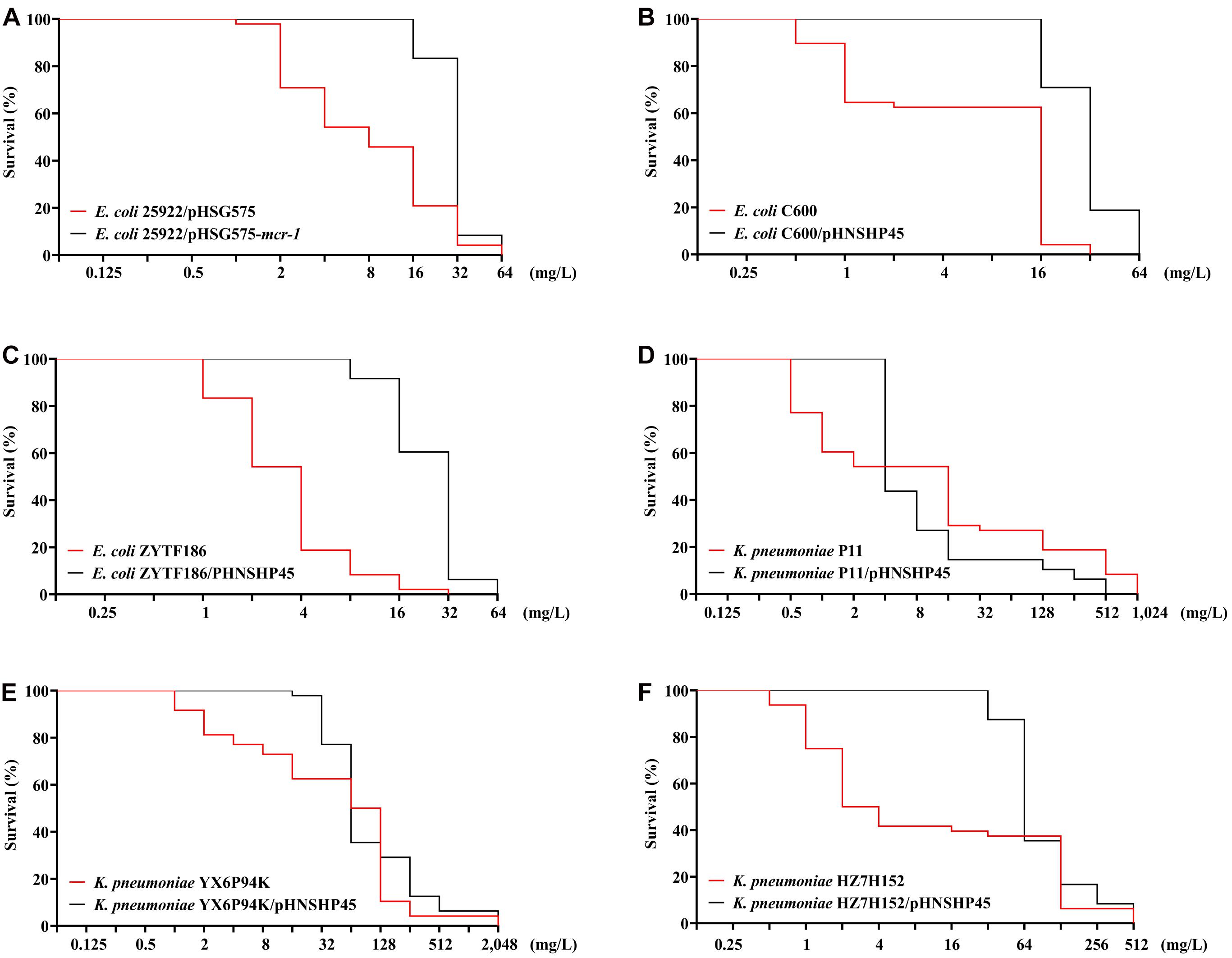
Figure 1. Survival curves of E. coli (A–C) and K. pneumoniae strains (D–F) and their mcr-1-positive transformants in the presence of increasing concentrations of colistin.
After the serial colistin challenge, the highest MICs of colistin for mcr-1-positive E. coli derivatives reached 32 mg/L, while those for mcr-1-negative derivatives only reached 16 mg/L (Figure 2A). Colistin MIC values for all mutants ranged from 2 to 32 mg/L (n = 9), suggesting that it was difficult for E. coli to generate HLCR mutants. Compared with their basal MICs, those for the mcr-1-negative strains (ATCC 25922/pHSG575, C600, and ZYTF186) increased 4- to 64-fold, while the MIC values for their corresponding mcr-1-positive derivatives merely showed 2- to 16-fold increases, suggesting that the presence of mcr-1 did not facilitate the generation of higher colistin resistance in E. coli.
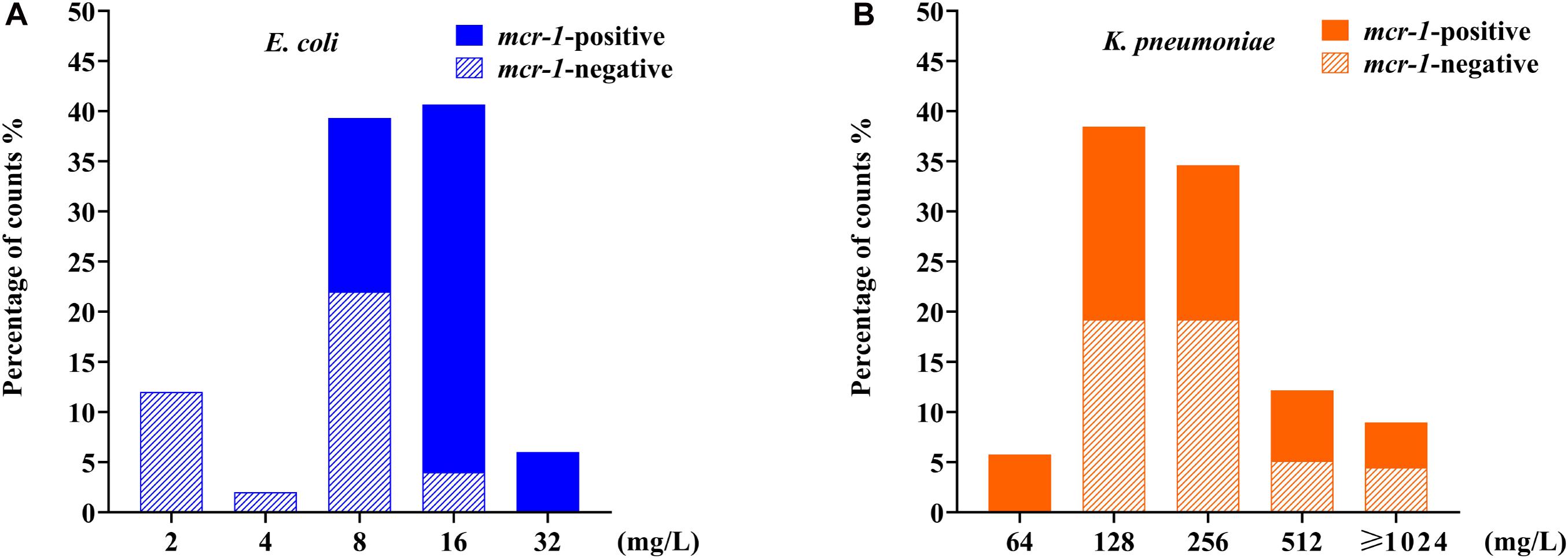
Figure 2. Distributions of colistin minimum inhibitory concentration (MIC) values among E. coli and K. pneumoniae mutants after induction of colistin resistance in vitro. (A) Data for 90 mcr-1-positive and 60 mcr-1-negative E. coli derivatives. (B) Data for 81 mcr-1-positive and 75 mcr-1-negative K. pneumoniae derivatives. E. coli data are indicated in blue, and K. pneumoniae data are indicated in orange.
Development of Resistance to Colistin via Serial Passages of Klebsiella pneumoniae
Compared with E. coli, K. pneumoniae strains, with or without mcr-1, were more likely to generate HLCR mutations. K. pneumoniae strains could survive up to extremely high concentrations of colistin (256–1,024 mg/L) (Figures 1D–F). Notably, the contribution of mcr-1 in improving the survival odds of K. pneumoniae was initially remarkable, however, it was weakened at higher concentrations of colistin. For example, compared with those of the parent strains of K. pneumoniae (YX6P94K and HZ7H152), more populations of their mcr-1-harboringtransformants survived when the concentrations of colistin were lower than 32 mg/L (Figures 1E,F). However, the contribution of mcr-1 gradually diminished in the presence of >64 mg/L colistin. Furthermore, the number of P11/pHNSHP45 subsets was initially higher but decreased to fewer than that of the parent strain P11 upon exposure to more than 4 mg/L colistin (Figure 1D).
After serial passages, the colistin MIC values for K. pneumoniae mutants ranged from 64 to >1,024 mg/L (Figure 2B). Particularly, the colistin MICs increased 256- to 4,096-fold for mcr-1-negative mutants but only 16- to 256-fold for their respective mcr-1-harboring transformants.
Genetic Alterations in pmrAB and phoPQ in Escherichia coli
Although most E. coli strains failed to generate HLCR, 36 derivatives that survived at relatively high concentrations of colistin (16–32 mg/L), including 18 mcr-1-negative and 18 mcr-1-positive derivatives, were selected to determine the presence of mutations in key genes related to colistin resistance (mgrB, pmrAB, and phoPQ). As shown in Supplementary Table 3, alterations mainly occurred in pmrB (21/36, 58.3%), followed by phoQ (15/36, 41.7%), and pmrA (9/36, 25.0%), most of which were associated with different amino acid substitutions, except frameshift mutations found in the pmrB gene (n = 6). No mutations were observed in phoP or mgrB.
Genetic Alterations in mgrB, pmrAB, phoPQ, and crrAB in Klebsiella pneumoniae
A total of 156 HLCR K. pneumoniae mutants, including 75 mcr-1-negative and 81 mcr-1-positive derivatives, were examined for the presence of mutations in the mgrB, pmrAB, phoPQ, and crrAB genes. The HLCR mutants showed a remarkable multiplicity of mutations in these genes (Figure 3, Table 1, and Supplementary Table 4). Among these mutants, genetic changes predominately occurred in crrB (62/156, 39.7%), followed by phoQ (41/156, 26.3%), crrA (35/156, 22.4%), pmrB (28/156, 17.9%), mgrB (27/156, 17.3%), and phoP (23/156, 14.7%). In 55 mutants, mutations were occurred in two or three genes (Table 1). No mutations were observed in pmrA.
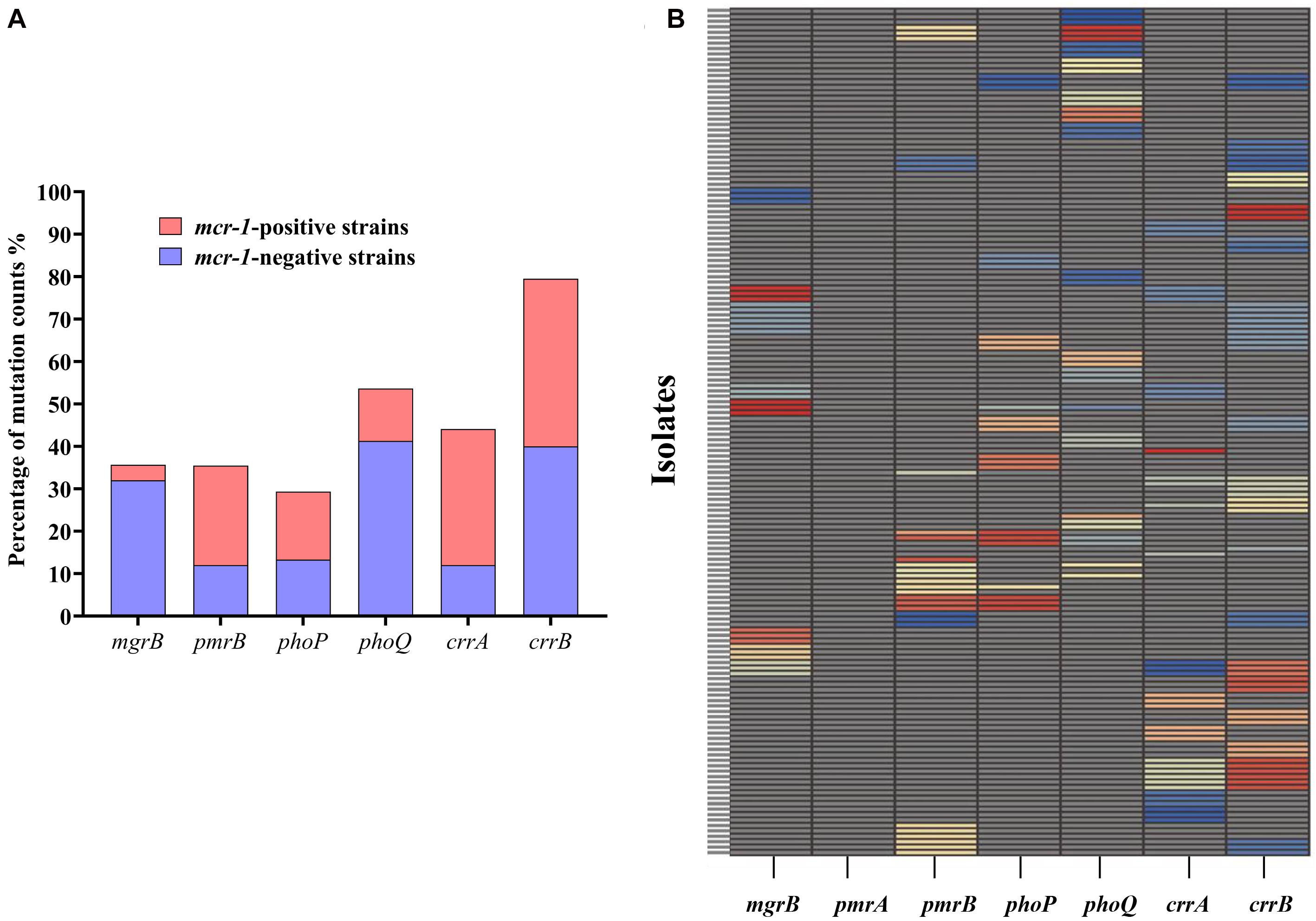
Figure 3. Occurrence of genetic alterations in mgrB, pmrAB, phoPQ, and crrAB in K. pneumoniae. (A) Distribution of mutations in different genes in 81 mcr-1-positive and 75 mcr-1-negative K. pneumoniae derivatives. (B) Profiles of genetic alterations in key genes related to colistin resistance, with 53 unique combinations in the 156 colistin-resistant K. pneumoniae derivatives. Each unique mutation in each colistin resistance gene is represented by a different color. Specific information on the mutations can be found in Supplementary Table 4.
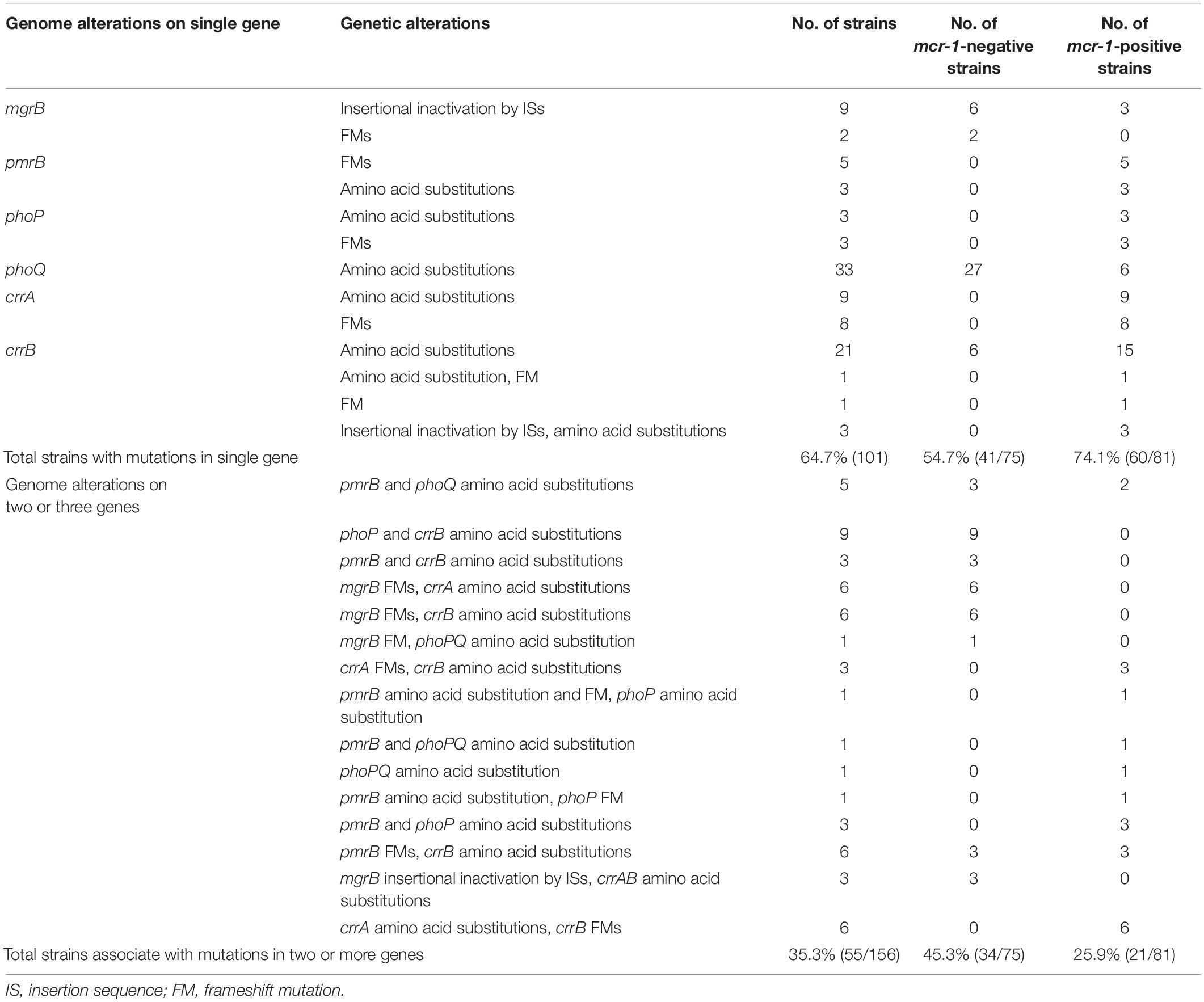
Table 1. Number of K. pneumoniae derivatives with genomic mutation(s) in colistin resistance genes after stepwise induction.
There were evident differences in mutations between the mcr-1-negative and mcr-1-positive isolates (Figure 3A, Table 1, and Supplementary Table 4). phoQ and mgrB alterations were more frequent in the mcr-1-negative derivatives (41.3 and 32.0%, respectively) than in the mcr-1-positive derivatives (12.3 and 3.7%, respectively). By contrast, the mcr-1-positive derivatives predominantly harbored mutations in crrA (26/81, 32.1%) and pmrB (19/81, 23.5%), both of which only occurred in 12.0% (9/75) of the mcr-1-negative derivatives. Moreover, of the 55 mutants with genetic change(s) in two or three genes, 34 (34/75, 45.3%) were mcr-1-negative derivatives and 21 (21/81, 25.9%) were mcr-1-positive derivatives (Table 1).
Regarding mutational patterns, alterations were mainly associated with amino acid substitutions or frameshift mutations (Table 1 and Supplementary Table 4), except for insertion sequences (ISs), which were almost exclusively associated with mgrB (Table 1 and Supplementary Table 4). Among 27 derivatives with alterations in mgrB, 18 (66.7%) were characterized by insertional inactivation of mgrB, which was truncated by one of the ISs, namely ISKpn26 (n = 3), IS903B (n = 6), ISKpn14 (n = 3), ISEcp1 (n = 3), or IS1R (n = 3) (Supplementary Figure 2). Additionally, crrB was first observed to be truncated by IS1R in three derivatives (Supplementary Figure 2).
Impacts of Mutations on the Growth of HLCR Klebsiella pneumoniae Strains
To determine whether the genetic changes in mgrB, pmrAB, phoPQ, and crrAB could impose a fitness cost on K. pneumoniae strains, we determined the growth rates of HLCR mutants harboring different mutations. In the mcr-1-negative groups, most of the HLCR mutants showed subtle advantages in the growth rates compared with those of their wild-type parent strains (Figure 4A), except for one mutant (phoQ V38G), which exhibited a substantial reduction in the growth rate (p < 0.01), and three isolates (13.6%) with different mutations, which showed significant advantages in the growth rates (p < 0.05). Conversely, among the mcr-1-positive HLCR mutants, none exhibited remarkable growth advantages, and most mutants showed slow growth rates compared to their corresponding parent strains (Figure 4B). Additionally, 13 individual mutants (59.1%) exhibited significant biological costs for growth (n = 1, p < 0.05; n = 9, p < 0.01; n = 3, p < 0.001). Overall, most chromosomal mutations alone resulted in no burden on growth of HLCR mutants; however, the fitness cost was observed in most mcr-1-harboring HLCR mutants.
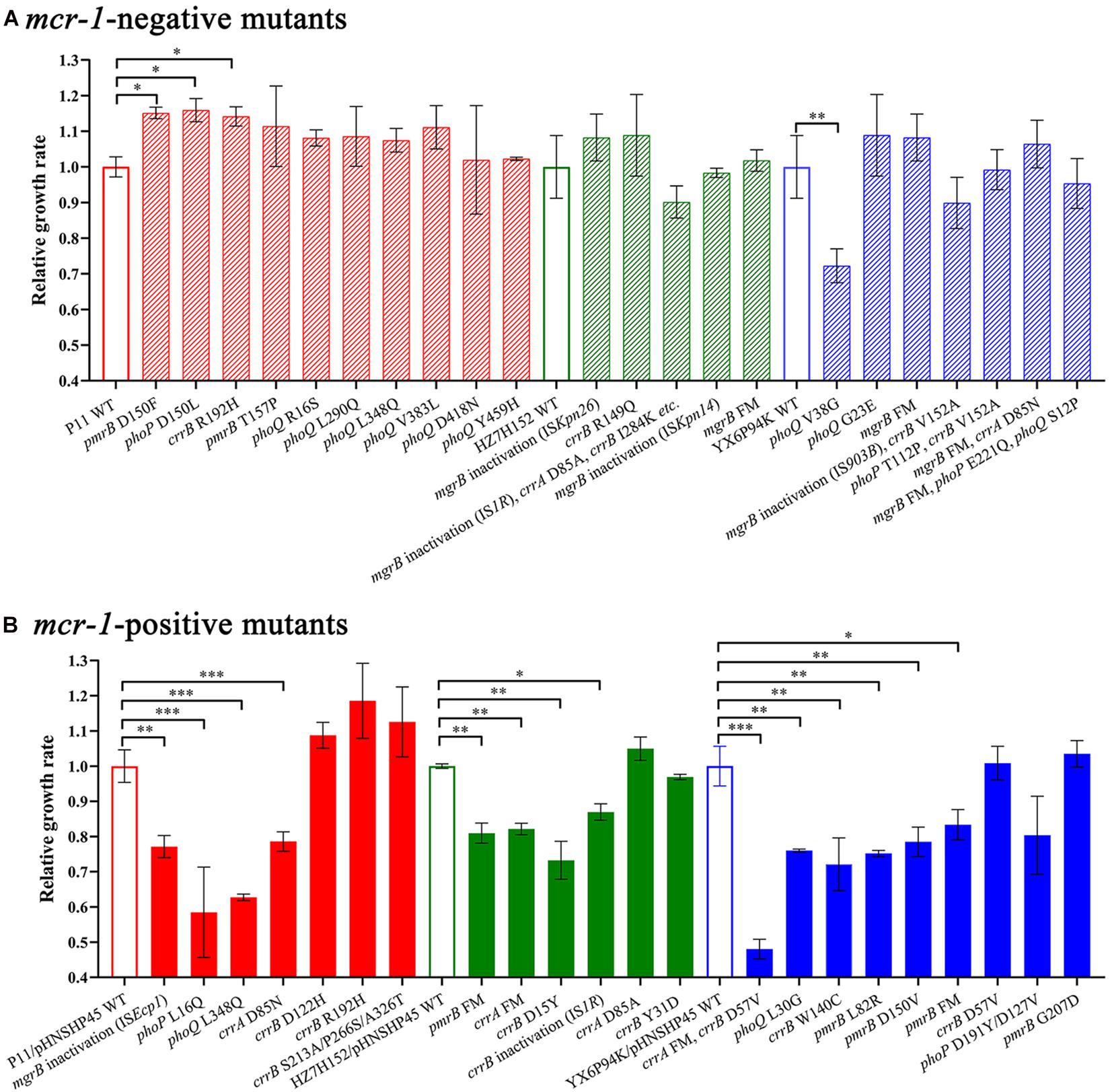
Figure 4. Relative growth rates of mcr-1-negative (A) and mcr-1-positive (B) K. pneumoniae mutants and their parental strains. The maximal growth rates are presented as relative values compared with the wild-type growth rate (set to 1.0). The relevant mutated genes of each strain are indicated below the chart. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Error bars represent standard deviation values. WT, wild-type; FM, frameshift mutation.
Discussion
Although the presence of mcr-1 confers low-level resistance to colistin, there is a concern regarding whether the presence of mcr-1 can accelerate the development of HLCR. In this study, we found that mcr-1 did not potentiate the development of HLCR in E. coli and K. pneumoniae, as the colistin MICs for mcr-1-negative derivatives obtained via serial colistin challenges were similar to those of their corresponding mcr-1-harboring transformants. Moreover, the fold increases in the colistin MICs were higher for mcr-1-negative strains than those for their mcr-1-harboring transformants. Although the presence of mcr-1 could not facilitate the evolution of colistin resistance in these two species of bacteria, mcr-1 could improve their survival rates upon exposure to relatively high concentrations of colistin (16–32 mg/L), which indicated that the presence of mcr-1 might enhance the survival ability of bacteria under clinical colistin pressure, thereby potentially leading to treatment failure.
Previous reports have shown that the mechanisms of resistance to colistin significantly vary between clinical K. pneumoniae and E. coli isolates. In K. pneumoniae, colistin resistance, including HLCR, is usually caused by chromosomal mutations (Cannatelli et al., 2014; Jayol et al., 2017), whereas in E. coli, colistin resistance is mainly associated with the presence of mcr-1 (Huang et al., 2017). Herein, we demonstrated that compared with E. coli, K. pneumoniae strains were more likely to develop HLCR by genetic alterations of the two-component systems (TCSs, e.g., PmrAB and PhoPQ) and their regulators (MgrB and CrrAB) when exposed to increasing concentrations of colistin in vitro. Previous studies have demonstrated that the TCSs play crucial roles in regulating the expression of genes for lipid A modifications in both K. pneumoniae and E. coli strains (Poirel et al., 2017; Srinivas and Rivard, 2017). This result partly explains the clinical phenomenon of HLCR being more common in K. pneumoniae compared to E. coli isolates. However, the mechanism underlying why TCSs and their regulators in K. pneumoniae are more likely to occur genetic alteration under the selective pressure of colistin than those in E. coli is unclear and needs further study.
After colistin exposure, alterations were detected in genes (mgrB, pmrAB, phoPQ, and crrAB) associated with colistin resistance. In E. coli, alterations mainly occurred in pmrB, followed by phoQ, and no mutations were observed in the mgrB or phoP gene. However, in HLCR K. pneumoniae variants, mutations were more complex and diverse and occurred in all the indicated genes, except pmrA. Genetic changes mainly occurred in crrB and phoQ (Figure 3A), in contrast to a previous report which stated that alterations in mgrB played a primary role in colistin resistance in K. pneumoniae (Cannatelli et al., 2014). However, consistent with previous reports (Kim et al., 2019; Yang et al., 2020), in both E. coli and K. pneumoniae, the histidine kinase genes crrB, pmrB, and phoQ seemed to be more common sites for mutations compared to the response regulatory genes crrA, pmrA, and phoP. It is possible that the sensor kinases can directly sense environmental stimuli and are therefore more likely to undergo mutations to confer protection against adverse stimuli, such as colistin pressure (Huang et al., 2020). The detected mgrB-disrupting ISs, ISKpn26, IS903B, ISKpn14, ISEcp1, and IS1R, were similar to those found in other studies (Shamina et al., 2020; Yang et al., 2020). Although recent studies have shown that amino acid substitution mutations in crrB or IS disruptions are responsible for HLCR (Wright et al., 2015; Jayol et al., 2017; McConville et al., 2020), mutations in crrAB in clinical isolates have rarely been reported, possibly due to few tests performed for this newly identified two-component system and the deletion of crrAB in some K. pneumoniae strains (Wright et al., 2015). Moreover, most of the mutations observed in this study have not been previously reported; thus, further studies are warranted to elucidate the potential contributions of these novel mutations to colistin resistance.
Remarkably, there were differences in the occurrence of chromosomal mutations in mcr-1-positive and mcr-1-negative K. pneumoniae isolates. In most (74.1%) mcr-1-positive derivatives, genetic alteration(s) only occurred in single gene involved in colistin resistance, while half of the mcr-1-negative derivatives harbored multiple mutations, thereby implying that the coexistence of chromosomal mutations and mcr-1 might impose a severe fitness burden on K. pneumoniae. To test this hypothesis, we compared the growth rates of mcr-1-positive and mcr-1-negative strains with those of their relevant HLCR mutants and found that HLCR mutants derived from mcr-1-negative strains exhibited almost no fitness cost, while HLCR mutants of mcr-1-positive strains showed an evident fitness cost. Previous studies have shown that the fitness cost attributed to chromosomal mutations or the acquisition of wild-type mcr-1-harboring plasmids seemed to be insignificant (Cannatelli et al., 2015; Wu et al., 2018); however, increased expression of mcr-1 could impose a significant fitness burden on host bacteria (Yang et al., 2017; Liu et al., 2020). Our results demonstrated that the coexistence of chromosomal mutations and mcr-1 was disadvantageous for K. pneumoniae, which might partly explain the lack of clinical strains with both chromosomal mutations and mcr-1 (Nang et al., 2018).
In conclusion, we demonstrated that compared with E. coli, K. pneumoniae was more likely to develop HLCR by acquiring chromosomal mutations. Although mcr-1 could not facilitate the selection of HLCR mutants, it could improve the survival rates of bacteria at relatively high concentrations of colistin, which might result in treatment failure.
Data Availability Statement
The whole genome sequencing data of K. pneumoniae P11, HZ7H152, Klebsiella variicola YX6P94K (subtype of K. pneumoniae) and E. coli ZYTF186 have been deposited at GenBank with accession numbers JAFKAD000000000, JAFKAE000000000, JAFKAG000000000, and JAFKAF000000000, respectively.
Author Contributions
J-HL designed the study. X-QZ, RW, and HX performed the experiments and collected the data. X-QZ, Y-YL, RW, and HX analyzed and interpreted the data. X-QZ and Y-YL wrote the draft of the manuscript. J-HL, Y-YL, X-QZ, JS, JL, and YF edited and revised the manuscript. All authors have read and approved the manuscript. J-HL coordinated the whole project.
Funding
This work was supported in part by the National Natural Science Foundation of China (No. 31830099 and 31625026), the Guangdong Local Innovation Team Program (No. 2019BT02N054), the 111 Project (No. D20008), and the Innovation Team Project of Guangdong University (No. 2019KCXTD001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.666782/full#supplementary-material
Footnotes
References
Cannatelli, A., Giani, T., D’Andrea, M. M., Di Pilato, V., Arena, F., Conte, V., et al. (2014). MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58, 5696–5703. doi: 10.1128/aac.03110-14
Cannatelli, A., Santos-Lopez, A., Giani, T., Gonzalez-Zorn, B., and Rossolini, G. M. (2015). Polymyxin resistance caused by mgrB inactivation is not associated with significant biological cost in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 59, 2898–2900. doi: 10.1128/aac.04998-14
Eiamphungporn, W., Yainoy, S., Jumderm, C., Tan-Arsuwongkul, R., Tiengrim, S., and Thamlikitkul, V. (2018). Prevalence of the colistin resistance gene mcr-1 in colistin-resistant Escherichia coli and Klebsiella pneumoniae isolated from humans in Thailand. J. Glob. Antimicrob. Resist. 15, 32–35. doi: 10.1016/j.jgar.2018.06.007
Huang, J., Li, C., Song, J., Velkov, T., Wang, L., Zhu, Y., et al. (2020). Regulating polymyxin resistance in Gram-negative bacteria: roles of two-component systems PhoPQ and PmrAB. Future Microbiol. 15, 445–459. doi: 10.2217/fmb-2019-0322
Huang, X., Yu, L., Chen, X., Zhi, C., Yao, X., Liu, Y., et al. (2017). High prevalence of colistin resistance and mcr-1gene in Escherichia coli isolated from food animals in China. Front. Microbiol. 8:562. doi: 10.3389/fmicb.2017.00562
Huang, Y. H., Chou, S.-H., Liang, S.-W., Ni, C.-E., Lin, Y.-T., Huang, Y. W., et al. (2018). Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J. Antimicrob. Chemother. 73, 2039–2046. doi: 10.1093/jac/dky164
Jacoby, G. A., Strahilevitz, J., and Hooper, D. C. (2014). Plasmid-mediated quinolone resistance. Microbiol. Spectr. 2:10.
Jayol, A., Nordmann, P., Brink, A., Villegas, M. V., Dubois, V., and Poirel, L. (2017). High-level resistance to colistin mediated by various mutations in the crrB gene among carbapenemase-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 61:e01423-17.
Jeong, J. Y., Kim, E. S., Choi, S. H., Kwon, H. H., Lee, S. R., Lee, S. O., et al. (2008). Effects of a plasmid-encoded qnrA1 determinant in Escherichia coli strains carrying chromosomal mutations in the acrAB efflux pump genes. Diagn. Microbiol. Infect. Dis. 60, 105–107. doi: 10.1016/j.diagmicrobio.2007.07.015
Kim, S., Woo, J. H., Kim, N., Kim, M. H., Kim, S. Y., Son, J. H., et al. (2019). Characterization of chromosome-mediated colistin resistance in Escherichia coli isolates from livestock in Korea. Infect. Drug Resist. 12, 3291–3299. doi: 10.2147/idr.s225383
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Liu, Y. Y., Zhu, Y., Wickremasinghe, H., Bergen, P. J., Lu, J., Zhu, X. Q., et al. (2020). Metabolic perturbations caused by the over-expression of mcr-1 in Escherichia coli. Front. Microbiol. 11:588658. doi: 10.3389/fmicb.2020.588658
Luo, Q., Niu, T., Wang, Y., Yin, J., Wan, F., Yao, M., et al. (2019). In vitro reduction of colistin susceptibility and comparative genomics reveals multiple differences between MCR-positive and MCR-negative colistin-resistant Escherichia coli. Infect. Drug Resist. 12, 1665–1674. doi: 10.2147/idr.s210245
Matuschek, E., Åhman, J., Webster, C., and Kahlmeter, G. (2018). Antimicrobial susceptibility testing of colistin - evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin. Microbiol. Infect. 24, 865–870. doi: 10.1016/j.cmi.2017.11.020
McConville, T. H., Annavajhala, M. K., Giddins, M. J., Macesic, N., Herrera, C. M., Rozenberg, F. D., et al. (2020). CrrB positively regulates high-level polymyxin resistance and virulence in Klebsiella pneumoniae. Cell Rep. 33:108313. doi: 10.1016/j.celrep.2020.108313
Nang, S. C., Morris, F. C., Mcdonald, M. J., Han, M. L., Wang, J., Strugnell, R. A., et al. (2018). Fitness cost of mcr-1-mediated polymyxin resistance in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1604–1610. doi: 10.1093/jac/dky061
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/cmr.00064-16
Quan, J., Li, X., Chen, Y., Jiang, Y., Zhou, Z., Zhang, H., et al. (2017). Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect. Dis. 17, 400–410. doi: 10.1016/s1473-3099(16)30528-x
Ruhnke, M., Arnold, R., and Gastmeier, P. (2014). Infection control issues in patients with haematological malignancies in the era of multidrug-resistant bacteria. Lancet Oncol. 15:e0606-19.
San Millan, A., Escudero, J. A., Gifford, D. R., Mazel, D., and Maclean, R. A. (2016). Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nat. Ecol. Evol. 1:10.
Shamina, O. V., Kryzhanovskaya, O. A., Lazareva, A. V., Alyabieva, N. M., Polikarpova, S. V., Karaseva, O. V., et al. (2020). Emergence of a ST307 clone carrying a novel insertion element MITE Kpn1 in the mgrB gene among carbapenem-resistant Klebsiella pneumoniae from Moscow, Russia. Int. J. Antimicrob. Agents 55:105850. doi: 10.1016/j.ijantimicag.2019.11.007
Shi, X., Li, Y., Yang, Y., Shen, Z., Wu, Y., and Wang, S. (2020). Global impact of mcr-1-positive Enterobacteriaceae bacteria on “one health”. Crit. Rev. Microbiol. 46, 565–577. doi: 10.1080/1040841x.2020.1812510
Srinivas, P., and Rivard, K. (2017). Polymyxin resistance in gram-negative pathogens. Curr. Infect. Dis. Rep. 19:38.
Takeshita, S., Sato, M., Toba, M., Masahashi, W., and Hashimoto-Gotoh, T. (1987). High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61, 63–74. doi: 10.1016/0378-1119(87)90365-9
Wang, C., Feng, Y. A.-O., Liu, L., Wei, L., Kang, M., and Zong, Z. A.-O. X. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microb. Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wright, M. S., Suzuki, Y., Jones, M. B., Marshall, S. H., Rudin, S. D., Van Duin, D., et al. (2015). Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob. Agents Chemother. 59, 536–543. doi: 10.1128/aac.04037-14
Wu, R., Yi, L. X., Yu, L. F., Wang, J., Liu, Y., Chen, X., et al. (2018). Fitness Advantage of mcr-1-bearing IncI2 and IncX4 plasmids in vitro. Front. Microbiol. 9:331. doi: 10.3389/fmicb.2018.00331
Yang, Q., Li, M., Spiller, O. B., Andrey, D. A.-O., Hinchliffe, P., Li, H., et al. (2017). Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat. Commun. 8:2054.
Yang, T. Y., Wang, S. F., Lin, J. E., Griffith, B. T. S., Lian, S. H., Hong, Z. D., et al. (2020). Contributions of insertion sequences conferring colistin resistance in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 55:105894. doi: 10.1016/j.ijantimicag.2020.105894
Zhang, H., Zhao, D., Quan, J., Hua, X., and Yu, Y. (2019). mcr-1 facilitated selection of high-level colistin-resistant mutants in Escherichia coli. Clin. Microbiol. Infect. 25:517.e1-e4.
Keywords: mcr-1, Enterobacterales, colistin, resistance, fitness cost
Citation: Zhu X-Q, Liu Y-Y, Wu R, Xun H, Sun J, Li J, Feng Y and Liu J-H (2021) Impact of mcr-1 on the Development of High Level Colistin Resistance in Klebsiella pneumoniae and Escherichia coli. Front. Microbiol. 12:666782. doi: 10.3389/fmicb.2021.666782
Received: 11 February 2021; Accepted: 26 March 2021;
Published: 26 April 2021.
Edited by:
Shaolin Wang, China Agricultural University, ChinaCopyright © 2021 Zhu, Liu, Wu, Xun, Sun, Li, Feng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Hua Liu, amhsaXVAc2NhdS5lZHUuY24=
†These authors have contributed equally to this work
 Xiao-Qing Zhu
Xiao-Qing Zhu Yi-Yun Liu
Yi-Yun Liu Renjie Wu
Renjie Wu Haoliang Xun
Haoliang Xun Jian Sun
Jian Sun Jian Li
Jian Li Yaoyu Feng
Yaoyu Feng Jian-Hua Liu
Jian-Hua Liu