
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 May 2021
Sec. Aquatic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.666739
The unicellular N2-fixing cyanobacteria UCYN-A live in symbiosis with haptophytes in the Braarudosphaera bigelowii lineage. Maintaining N2-fixing symbioses between two unicellular partners requires tight coordination of multiple biological processes including cell growth and division and, in the case of the UCYN-A symbiosis, N2 fixation of the symbiont and photosynthesis of the host. In this system, it is thought that the host photosynthesis supports the high energetic cost of N2 fixation, and both processes occur during the light period. However, information on this coordination is very limited and difficult to obtain because the UCYN-A symbiosis has yet to be available in culture. Natural populations containing the UCYN-A2 symbiosis were manipulated to explore the effects of alterations of regular light and dark periods and inhibition of host photosynthesis on N2 fixation (single cell N2 fixation rates), nifH gene transcription, and UCYN-A2 cell division (fluorescent in situ hybridization and nifH gene abundances). The results showed that the light period is critical for maintenance of regular patterns of gene expression, N2 fixation and symbiont replication and cell division. This study suggests a crucial role for the host as a producer of fixed carbon, rather than light itself, in the regulation and implementation of these cellular processes in UCYN-A.
Marine phytoplankton are comprized of diverse eukaryotic algae and cyanobacteria and are responsible for approximately half of the fixation of atmospheric CO2 into organic matter on Earth (Field et al., 1998). This primary productivity is dependent on the biogeochemical cycling of nutrients. In particular, biologically available nitrogen (N) is often in short supply despite the vast reservoir of atmospheric dinitrogen gas (N2). Biological N2 fixation, i.e., the conversion of N2 to biologically available ammonium, is a significant source of new N in the surface ocean that supports primary production (Karl et al., 1997; Shiozaki et al., 2013). It is performed by a limited, albeit diverse, number of Archaea and Bacteria taxa called diazotrophs. Among them, the unicellular cyanobacterium UCYN-A group contributes significantly to oceanic N2 fixation (Church et al., 2005a; Martínez-Pérez et al., 2016). This group encompasses closely related sublineages that are symbiotic with several different species of single celled haptophytes in the Braarudosphaera bigelowii clade (Thompson et al., 2012, 2014; Hagino et al., 2013). Since its discovery, the UCYN-A symbiosis has been found in a wide variety of oceanic habitats (Farnelid et al., 2016) including areas that were previously not considered important for diazotrophy, such as N-rich coastal ecosystems (Mulholland et al., 2012; Tang and Cassar, 2019) and the Arctic Ocean (Harding et al., 2018; Shiozaki et al., 2018).
UCYN-A transfers fixed N to the host in exchange for fixed carbon (C), which was demonstrated with isotope experiments using nanoscale secondary ion mass spectrometry (nanoSIMS) analyses (Thompson et al., 2012; Krupke et al., 2013; Mills et al., 2020). The UCYN-A genome unveiled an extremely streamlined metabolism, missing many important pathways and enzymes that are typical for cyanobacteria and necessary for a free-living lifestyle. UCYN-A lacks the oxygen-evolving photosystem II (PSII), Rubisco and Calvin cycle enzymes, the entire tricarboxylic acid (TCA) cycle and various enzymes for branched chain amino acid and nucleotide synthesis (Zehr et al., 2008; Tripp et al., 2010; Bombar et al., 2014). Thus, it is likely that the host provides several essential metabolites to the symbiont and that the symbiosis is obligatory, at least for the symbiont (Zehr et al., 2016).
In eukaryotic phytoplankton and cyanobacteria, metabolism and growth are coordinated with the light and dark periods of the daily cycle, which is reflected by oscillations in gene expression (Welkie et al., 2019; Becker et al., 2021). In N2-fixing cyanobacteria, such metabolic synchronization is particularly important for light-driven photosynthesis and supply of energy for N2 fixation. N2 fixation is inhibited by oxygen and diazotrophic cyanobacteria typically separate N2 fixation from oxygen-evolving photosynthesis either temporally or spatially in specialized cells called heterocysts (Fay, 1992; Thompson and Zehr, 2013). Most unicellular diazotrophic cyanobacteria, including marine planktonic species, are nighttime N2-fixers (Zehr et al., 2001; Berman-Frank et al., 2003; Tuit et al., 2004; Mohr et al., 2010b; Wilson et al., 2017). In these species, light enables the production of carbohydrates that are later used as energy sources to fuel nighttime N2 fixation (Dron et al., 2012). UCYN-A is unusual among unicellular N2-fixing cyanobacteria because it fixes N2 during the day (Krupke et al., 2013; Zehr et al., 2016). In fact, the whole genome expression pattern in UCYN-A, including genes for N2 fixation, is more similar to that of daytime N2-fixers than of the more closely related unicellular cyanobacteria (Muñoz-Marín et al., 2019). It is not known what controls the unusual N2 fixation timing in UCYN-A. Although it lacks most proteins involved in photosynthesis, UCYN-A retains and expresses photosystem I (PSI) genes in a diel pattern, and depends on its host for photosynthate, which suggests direct and indirect roles for light in N2 fixation and UCYN-A’s metabolism in general. Besides photosynthesis and N2 fixation, the UCYN-A symbiosis faces several physiological challenges with respect to cell growth and division. The host and UCYN-A cells must coordinate growth and cell division to avoid outgrowing one another and losing their partnership. UCYN-A has been shown to express cell division genes at the end of the light period and divide a few hours later, prior to the host cell division (Cabello et al., 2016; Muñoz-Marín et al., 2019), but the mechanisms that control this process are unknown.
In this study, we manipulated coastal microbial communities containing the UCYN-A2 symbiosis to assess the influence of the daily light-dark cycle on the symbiosis growth and metabolism. First, we manipulated light conditions to investigate links between light and daytime N2 fixation by UCYN-A2. Second, we used a photosynthesis inhibitor specific to PSII to examine the response of the symbiont to arrested host C fixation. We sampled at regular time intervals (every 4 h) over a 52-h experimental time course, and combined targeted nifH gene and transcript quantification, microscopic observations, and cell-specific C and N2 fixation rate measurements to determine the responses of the UCYN-A2 symbiosis to altered light-dark cycles and host photosynthesis at high temporal resolution.
Incubations of natural marine microbial communities were set up on the Ellen Browning Scripps Memorial Pier at the Scripps Institution of Oceanography (SIO) in La Jolla, CA, United States in May 2018. Surface water was sampled off the pier using a pneumatic pump, passed through 150-μm nitex mesh (Wildco®, Yulee, FL, United States) and collected into a 40 L polycarbonate (PC) carboy, then gently distributed into 1 and 2 L clear PC bottles. All bottles and tubing were washed with 10% HCl and rinsed three times with milli-Q water, then rinsed three times with seawater immediately before use. Water collection and bottle filling took place between 10 p.m. and 4 a.m. Bottles were placed in clear or opaque incubators (Figure 1 and Supplementary Figure 1) equipped with HOBO data loggers (Onset, Bourne, MA, United States) to monitor light and temperature, and flushed continuously with surface seawater to keep the bottles at in situ temperature. Ambient temperature dropped from an average 19.8 ± 4.2°C during the day to 15.6 ± 0.6°C during the night over the course of the 52-h experiment. In the incubators, average nighttime temperatures (16.1 ± 1.1 and 16.3 ± 1.2°C in clear and opaque incubators, respectively) were only slightly lower than during the day, indicating that temperature in the experimental system was successfully maintained as close as possible to in situ levels (Supplementary Figure 2). Clear containers were fitted with neutral density screening to attenuate incident light to ca. 20% of the surface irradiance, while the opaque containers were closed with an opaque lid to prevent exposure to light. Additionally, bottles in the opaque containers were placed in pairs in dark plastic bags to minimize exposure during sampling. The opaque incubators remained dark, except during brief opening for sampling (Supplementary Figure 3). Light levels in the clear incubators reached an average of 13 ± 10% of incident light during daytime (Supplementary Figure 3).
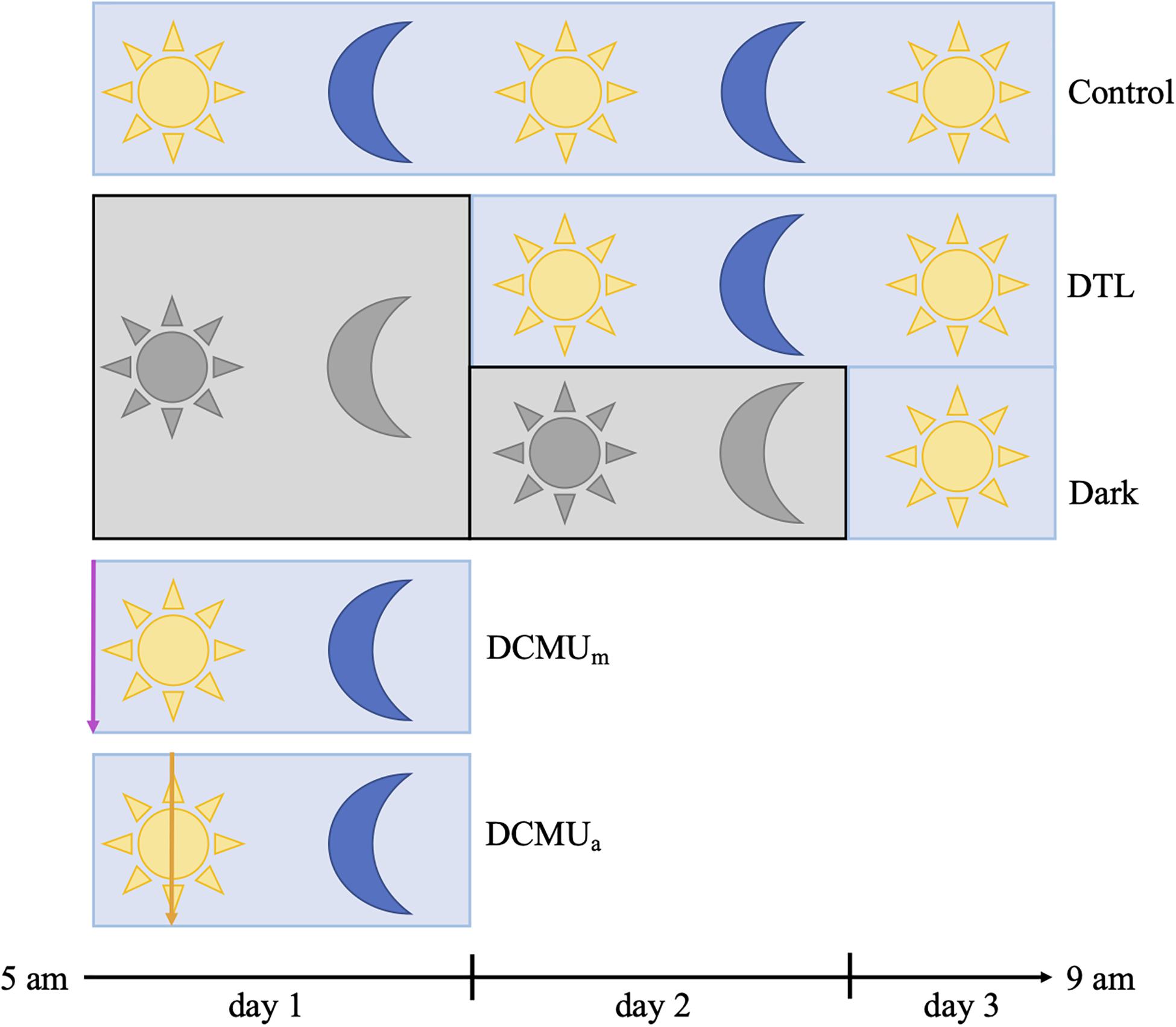
Figure 1. Schematic representation of experimental conditions. Blue rectangles represent clear incubators and gray rectangles represent opaque incubators. Yellow sun and blue moon represent normal light-dark conditions achieved with clear incubators, and gray sun and moon represent complete darkness achieved with opaque incubators. In the two conditions involving a dark phase, bottles were placed in opaque incubators at the beginning of the experiment and then moved to clear incubators at 5 a.m. on day 2 (“Dark then light”, DTL) or on day 3 (Dark). For incubations receiving DCMU, the purple arrow represents DCMU addition before 5 a.m. (DCMUm) and the orange arrow represents DCMU addition at 1 p.m. (DCMUa). See section “Materials and Methods” for additional description of incubation and sampling procedures.
Five experimental conditions, “Control,” “Dark,” “DTL” (dark then light), “DCMUm,” and “DCMUa” were implemented as illustrated in Figure 1 and Supplementary Figure 1. At 5 a.m. on day 1, fourteen 2 L bottles and twelve 1 L bottles were spiked with a solution of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU, Sigma-Aldrich, St. Louis, MO, United States) at a final concentration of 100 μg L–1 and placed in clear incubators (DCMUm). The process was repeated at 1 p.m. on day 1 with twelve 2 L bottles and ten 1 L bottles in the clear incubators (DCMUa).
After the first 24 h of incubation, at 5 a.m. on day 2, sixteen 2 L bottles and fourteen 1 L bottles were moved from opaque incubators to clear incubators (“Dark then Light”). After the first 48 h of incubation, at 5 a.m. on day 3, the two bottles remaining in opaque incubators were moved to a clear incubator (“Dark”). In total, four different conditions were run in parallel during day 1 (“Control,” “Dark,” “DCMUm,” and “DCMUa”) while three different conditions were run during day 2 (“Control”, “DTL,” and “Dark”, Figure 1).
At 5 a.m. on days 1 and 2, duplicate 2 and 1 L bottles from each condition were spiked with 15N2-enriched seawater and NaH13CO3-enriched seawater and incubated for 24 h (Supplementary Figure 1). 15N2-enriched seawater (Mohr et al., 2010a) was prepared based on protocols described in Klawonn et al., 2015 (see also Supplementary Material). Stock NaH13CO3-enriched seawater (353 mM) was prepared by dissolving 6 g NaH13CO3 (Cambridge Isotope Laboratories) into 200 mL of sterile filtered site water. Experimental treatments were spiked with 10% (v/v) 15N2-enriched filtered seawater and 224 μL NaH13CO3-enriched seawater (35 μM final concentration) at the beginning of each 24-h incubation period. This was equivalent to a ca. 2% 13C enrichment. To verify the 15N atom% enrichment, the 15N2-enriched seawater was immediately subsampled (prior to spiking) into duplicate 12 ml Exetainers® (Labco, Lampeter, Ceredigion, United Kingdom), which were stored at room temperature. Nitrogen isotopes (i.e., N masses equivalent to 28, 29, and 30) were measured via membrane inlet mass spectrometer (MIMS) analysis at the University of Hawaii according to Wilson et al. (2012). Calibration of the MIMS was achieved by the analysis of a 1 L reservoir of air-equilibrated filtered (0.2 μm) seawater with a known salinity and a temperature of 23°C (Böttjer et al., 2017). The final 15N atom % enrichment in the seawater incubations averaged 3.9 ± 0.2 (day 1) and 5.7 ± 0.1 (day 2).
Duplicate bottles were retrieved at the onset of the experiment at 5 a.m. on day 1, after which duplicate bottles from all conditions were retrieved every 4 h until 9 a.m. on day 3 (Supplementary Figure 1). For DNA and RNA analysis, 1 L from duplicate 2 L bottles from each experimental condition was gently passed through a Sterivex unit (Millipore, Burlington, MA, United States) using a peristaltic pump. Filtration time never exceeded 15 min. Sterivex units were capped, flash frozen and stored in liquid nitrogen in the field then stored at −80°C until further processing. For catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) analysis, 95 mL subsamples were removed from duplicate 1 L bottles from each experimental condition. At 5 a.m. on days 2 and 3, those samples were taken from the 1 L incubations enriched with 15N/13C 24 h prior to sampling. CARD-FISH samples were fixed with 5 mL 37% formaldehyde (1.85% final concentration) and placed in the dark at 4°C for at least 1 h. Fixed samples were gently filtered onto a PC membrane (25 mm diameter, 0.6 μm pore size, Millipore) using a vacuum pump. Filters were stored at −80°C until further processing.
In parallel with samples for cell-specific rate analysis, 400 mL samples for particulate organic C (POC) and N (PON) were filtered through precombusted (4 h at 450°C) 25 mm Whatman GF/F filters (Millipore) and stored at −20°C. Filters were then dried at 60°C, fumed with concentrated HCl, then dried at 60°C for 24 h, before being packed into tin capsules (Costech Analytical Technologies Inc., Valencia, CA, United States). Samples for the analysis of 15N and 13C enrichment of the particulate organic matter were processed on an Elemental Combustion System (Costech Analytical Technologies) interfaced to a Thermo Finnigan Delta V Advantage isotope ratio mass spectrometer (Thermo Fisher Scientific, Waltham, MA, United States) at the SOEST Biogeochemical Stable Isotope Facility at the University of Hawai’i, Manoa. The limit of detection (LOD), estimates of error, and minimum quantifiable rates (MQR) were calculated as in Gradoville et al. (2017) and are shown in Supplementary Table 1.
Nucleic acids were extracted using a cell lysis protocol followed by the AllPrep DNA/RNA Mini kit (Qiagen, Hilden, Germany). Frozen filters were quickly transferred to bead beating tubes containing a mixture of 0.1- and 0.5-mm glass beads (Biospec Products, Inc., Bartlesville, OK, United States) and 1 mL of pre-warmed RLT+ lysis buffer (Qiagen) containing β-mercaptoethanol (1% final concentration) and RNA standards (ERCC ExFold RNA Spike-In, final concentration 0.04X, InvitrogenTM, Thermo Fisher Scientific). Cell lysis was performed using three freeze-thaw cycles and two 2-min bead beating cycles (Mini-Beadbeater-96; Biospec Products, Inc.). Extraction was then performed following the manufacturer’s protocol, including on-column DNAse treatment (Qiagen) of the RNA fraction. The purity of the isolated DNA and RNA was checked using a Nanodrop (Thermo Fisher Scientific) and concentrations were determined using a Qubit (InvitrogenTM, Thermo Fisher Scientific). The quality and integrity of the RNA were assessed using a Bioanalyzer (Agilent, Santa Clara, CA, United States). A fraction of the RNA was immediately converted into complementary DNA (cDNA) using the High Capacity cDNA Reverse Transcription Kit (Applied BiosystemsTM, Thermo Fisher Scientific). DNA and cDNA samples were stored at −20°C while RNA samples were stored at −80°C until further processing.
Patterns in nifH gene and transcript abundances were investigated using digital droplet PCR (ddPCR), which enables standard-independent absolute quantification. The PCR reaction was prepared by adding DNA or cDNA templates to the ddPCR Supermix (Bio-Rad, Hercules, CA, United States) mixed with forward and reverse primers (0.4 μM final concentration) and a probe (0.2 μM final concentration), all targeting the UCYN-A2 nifH gene (Thompson et al., 2014). All samples were run in triplicates in 96-well microplates along with at least three no-template controls. Additional reactions were also run for the RNA used to generate cDNA which ruled out potential genomic DNA contamination. Plates were sealed, vortexed and centrifuged briefly, and droplets were generated using an Auto Droplet Generator (Bio-Rad) following the manufacturer instructions. The PCR reaction was set up as follows: 10 min at 95°C, then 40 cycles of 30 s at 94°C and 1 min at 64°C, and a final step of 10 min at 98°C. Positive droplet counts were performed using the Automated Droplet Reader (Bio-Rad), and data were processed with the QuantaSoft Analysis Pro software (Bio-Rad). Only runs with no positive droplet counts in the no-template controls were further processed, and sample wells in which the total number of droplets generated was below 10,000 were dropped. Positive droplet counts in each sample were then converted into UCYN-A2 nifH gene copies L–1 or transcripts L–1. After transcript normalization by gene abundance, periodic expression patterns were tested using the RAIN package in R (Thaben and Westermark, 2014).
A subset of fixed, frozen samples were processed for CARD-FISH as described in Cabello et al. (2016) with small modifications (see Supplementary Material). Processed filter sections mounted on slides were kept at −20°C until microscopy observation. Individual symbioses were observed with an epifluorescence microscope (Zeiss Axioplan) using filters to detect the CY-3 signal corresponding to the symbiont, the Alexa-488 signal corresponding to the host, and to visualize DNA through DAPI signal.
Selected samples were transferred to silicon wafer grids for analysis of C and N isotopic composition of individual UCYN-A2 host associations using nanoSIMS according to Dekas and Orphan (2011). The location of at least 10 UCYN-A2-host associations per sample were mapped for analysis on a Cameca NanoSIMS 50 L at the Stanford Nano Shared Facilities. Wafers were loaded in the instrument and mapped symbioses were localized. Each symbiosis was processed individually, and instrument tuning was checked and adjusted between each cell. A 2-min exposure to a cesium ion (Cs+) beam was applied to each cell before analysis to remove contaminants. Sample surfaces were rastered with a 16 keV Cs+ beam and a current between 2 and 4 pA over a 15 × 15 μm area. Data were simultaneously collected for 12C–, 13C–, 12C14N–, and 12C15N– secondary anions along with a secondary electron image, in 30 planes per cell. Raw data processing was performed using look@nanosims (Polerecky et al., 2012) in MATLAB. Symbiosis-specific C and N2 fixation rates were calculated for each association as in Montoya et al. (1996) (see Supplementary Material). The resulting cell-specific rates were compared between conditions with a Mann–Whitney test (α = 0.05).
We investigated the role of light as a direct and indirect (through the photosynthetic activity of the unicellular eukaryotic host) regulator of cellular processes in the diazotrophic cyanobacterial symbiont UCYN-A. Natural microbial populations from coastal surface water containing the UCYN-A2 symbiosis were incubated at in situ temperature and nutrient conditions under several dark/light scenarios using clear and opaque incubators (Figure 1, see also section “Materials and Methods” and Supplementary Figures 1–3). In control conditions, bulk C fixation (BCF) rates were 8 ± 0.4 and 18 mmol C m–3 d–1 during days 1 and 2, respectively (Supplementary Figure 4). When incubations were kept under dark conditions, BCF rates were below the detection limit, indicating broad-scale inhibition of oxygenic photosynthesis at the community level. Switching incubation conditions back to normal day-night cycles after 24 h in the dark (DTL, Figure 1 and Supplementary Figure 1) resulted in recovery of BCF (28 ± 5 mmol C m–3 d–1). In both DCMU conditions, BCF rates were greater than those in the dark and above detection limits, but the rates were negligible compared to control values (Supplementary Figure 4) despite identical light/dark conditions, indicating successful inhibition of oxygenic photosynthesis at the community level. In all conditions, bulk N2 fixation rates were below detection limits, which varied from 0.01 to 0.05 mmol N m–3 d–1 (Supplementary Table 1).
The natural microbial populations present in the waters at the SIO pier in May 2018 contained both UCYN-A1 and UCYN-A2 sublineages (Mills et al., 2020). Preliminary qPCR analyses showed that abundances were 10-fold higher for UCYN-A2 than for UCYN-A1, which constrained our analyses to the UCYN-A2 lineage. UCYN-A2 was present at 2 × 104 nifH gene copies L–1 at the onset of the experiment and remained at this magnitude during the 52-h long experiment in all conditions except when DCMU was added before sunrise (DCMUm, Figure 2). In DCMUm incubations, the nifH-based abundance of UCYN-A2 was maintained for 4 h, then decreased throughout the remaining sampling points. By contrast, when DCMU was added at solar noon (1 p.m.; DCMUa), after the communities had experienced a full morning under control conditions, UCYN-A2 nifH-based abundances stayed close to initial values for the remaining daylight and the ensuing dark period (Figure 2).
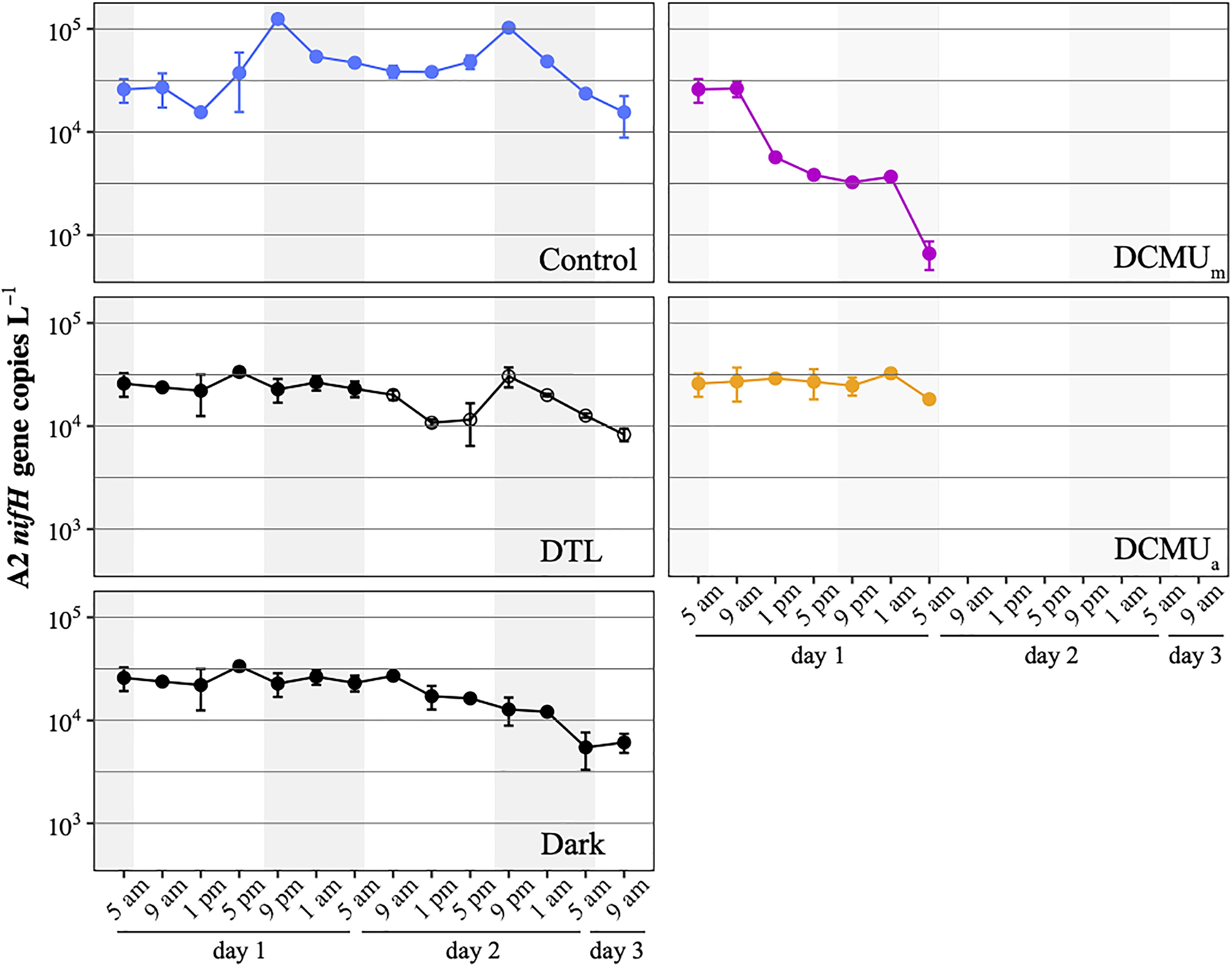
Figure 2. UCYN-A2 nifH gene abundances in control (blue), DTL (black and open circles), Dark (black), DCMUm (purple), and DCMUa (orange) incubations. Mean ± SD from duplicate incubations are shown at each time point. DCMU was spiked at 5 a.m. for the DCMUm condition and at 1 p.m. for the DCMUa condition. Note that the DTL condition data start at 9 a.m. on the second day of the experiment, and values plotted before that are identical to those from the Dark condition.
In control incubations, peak abundance was observed in the early evening at 9 p.m. (Figure 2). CARD-FISH visualization showed a high proportion (up to 74%) of host cells containing replicated UCYN-A2 cells at 9 p.m. compared to 5 p.m. (Figure 3), suggesting that the 9 p.m. increase in nifH gene copies L–1 reflects symbiont DNA replication before cell division, which can be observed with microscopy. The number of host cells containing replicated UCYN-A2 cells declined in the early morning as the hosts presumably divided, returning to a UCYN-A2 to host cell ratio of 1:1 (Figure 3). The pattern was repeated over the two-day experiment, showing that the growth of the UCYN-A2 symbiosis was maintained in control conditions. After replication and division, UCYN-A2 abundances declined in the morning (Figure 2), possibly reflecting grazing losses as previously observed in other cyanobacterial and phytoplankton species such as Prochlorococcus (Ribalet et al., 2015). Contrasting with control conditions, UCYN-A2 replication was absent from all other experimental conditions except DTL (Figures 2, 3). These results indicate that the symbioses did not go through nighttime cell division in the absence of daytime light or when host photosynthesis was inhibited, regardless of the timing of DCMU addition. In the DTL treatment, both the peak in nifH gene copies L–1 and the higher proportion of symbioses with replicated UCYN-A2 were observed at 9 p.m., although at a lower magnitude than in control incubations. These observations show that the return of a normal light period recovered UCYN-A2 DNA replication and cell division.
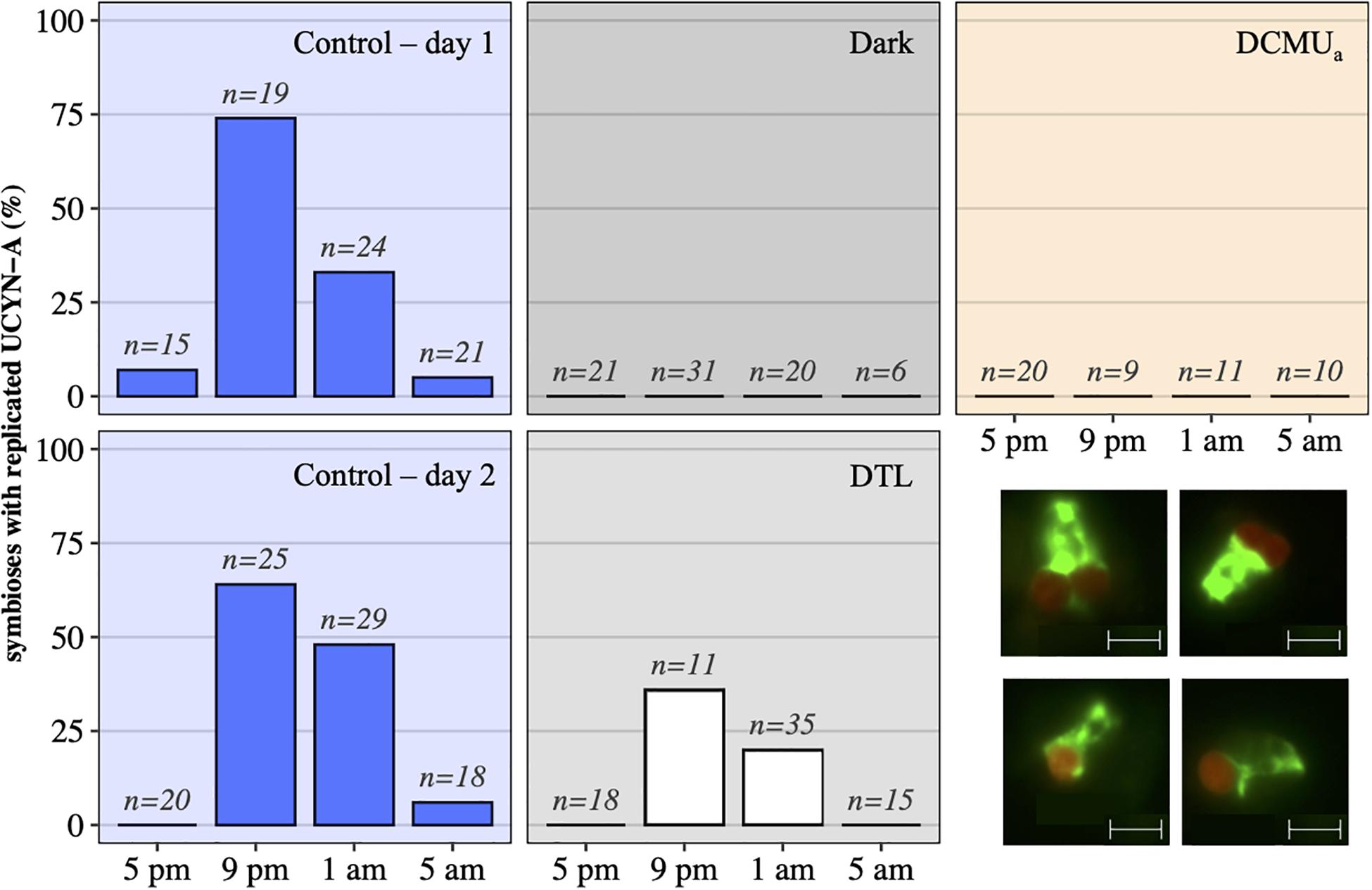
Figure 3. UCYN-A2 replication and division patterns in control (blue – day 1, top and day 2, bottom), dark (black), DCMUa (orange), and DTL (white) experimental conditions, as determined by epifluorescence microscopy. For each time point and condition, n represents the number of individual symbioses analyzed. Bottom right: examples of epifluorescence microscopy images showing individual symbioses where the host (green) is associated with a replicated symbiont (red) as seen in samples collected at 9 p.m. in the control (top row), or symbioses with a unique symbiont as seen in samples collected at 9 p.m. in the dark (bottom row). The scale indicates 5 μm.
In control conditions, UCYN-A2 nifH transcripts per gene copy generally reached maximum values from early morning to afternoon (5 a.m., 9 a.m., and 1 p.m.), and were minimal in the evening (5 p.m., 9 p.m., and 1 a.m.; Figure 4A) on both days. nifH continued to be transcribed under dark conditions but diverged from control patterns. While nifH transcripts per gene copy in control incubations dropped by half between 1 and 5 p.m., in the dark they remained close to expression levels detected in the morning. This resulted in expression levels three times higher in the dark compared to control incubations at 5 p.m. on the first day. nifH transcripts per gene copy in the dark were generally higher during the first 24 h, reflecting different responses to the absence of light after a normal light-dark cycle (day 1) or after 24 h of prolonged darkness (day 2). In the DTL incubations, nifH transcripts per gene copy increased from 5 a.m. to 1 p.m. compared to dark incubations and reached levels similar to those of control incubations (Figure 4A). While nifH transcript abundances varied among the different light-dark experimental conditions, the general diel pattern of nifH expression, i.e., the timing and trajectory of the oscillations, was conserved in all three experimental conditions, which was particularly evident on day 2 (RAIN, p ≤ 0.003, Figure 4C). Minimum expression was observed right after sunset (9 p.m.) on both days in all three experimental conditions.
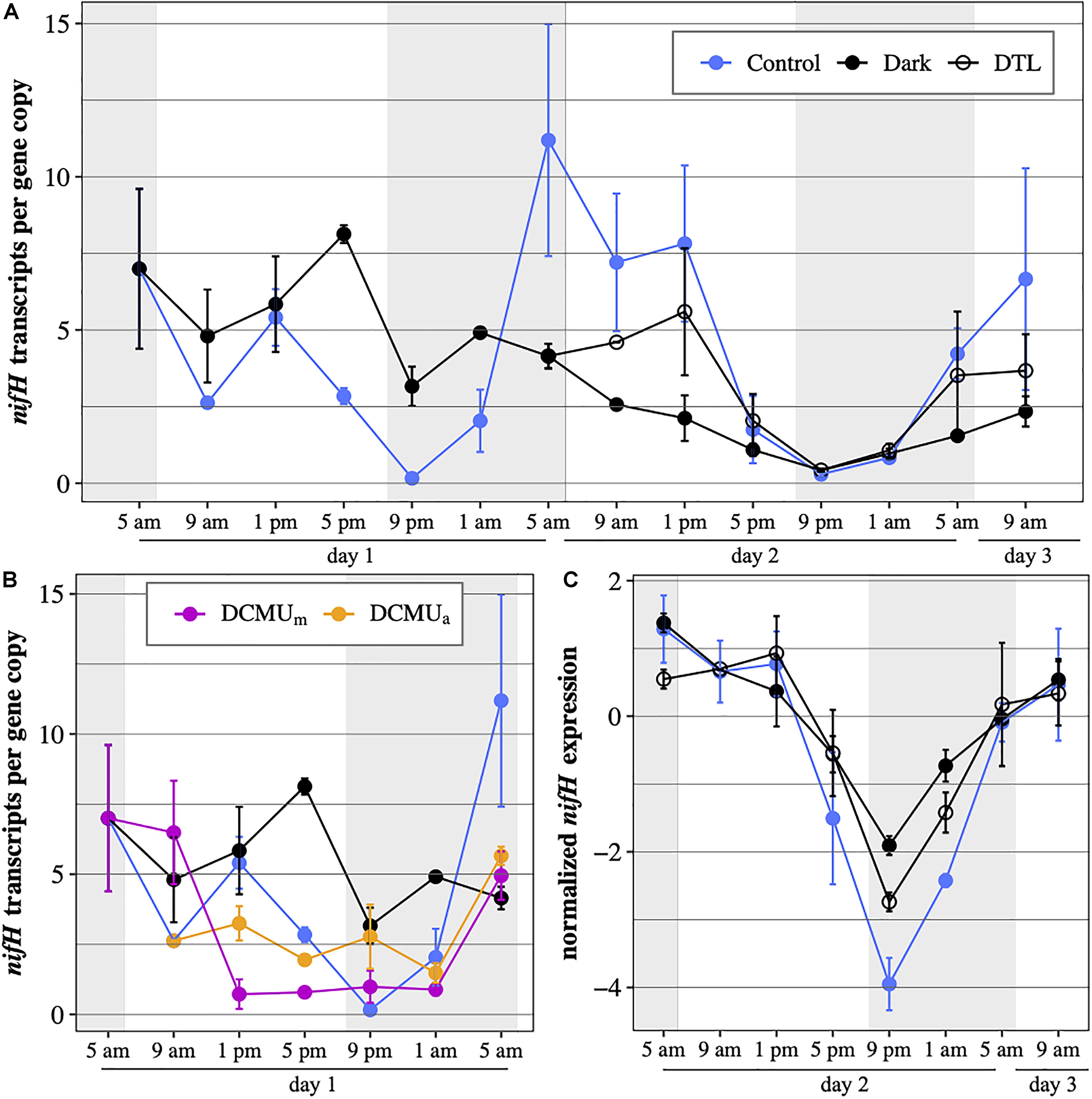
Figure 4. UCYN-A2 nifH gene expression in transcripts per gene copy, (A) during the full 52-h experiment in control incubations (blue), incubations returning to normal light-dark conditions after spending the first 24 h (DTL, open circles) or the first 48 h (Dark, black) of the experiment in the dark and (B) during the first 24 h of the experiment in control incubations (blue), dark incubations (black) and incubations exposed to DCMU added at 5 a.m. (purple) or at 1 p.m. (orange). (C) normalized UCYN-A2 nifH gene expression during the last 28 h of the experiment in control (blue), dark (black), and DTL (open circles) incubations. Expression levels were normalized to the median expression over that period within each condition and log2-transformed. Mean ± SD of biological duplicates are shown. Shaded areas represent nighttime.
In general, DCMU additions resulted in the lowest levels of nifH transcripts per gene copy (Figure 4B). There were minor, but possibly important, differences in the response to DCMU based on the timing of the addition. In DCMUm incubations, nifH transcripts per gene copy at 9 a.m. remained elevated compared to control levels and similar to those measured in dark incubations. After the 9 a.m. time point, DCMUm nifH transcripts per gene copy decreased dramatically, coinciding with the decrease in UCYN-A2 nifH abundance (Figure 2). In contrast, in the DCMUa incubations UCYN-A2 maintained nifH transcription below control values but above those detected in DCMUm. In both DCMU conditions, nifH transcript levels remained relatively constant for 12 h between 1 p.m. and 1 a.m. Interestingly, transcript abundances increased in the late dark period between 1 a.m. and 5 a.m. similarly to the control, although at a smaller magnitude (Figure 4B).
In control conditions, all UCYN-A2 symbioses analyzed were fixing C and N2 during both days 1 and 2 (Figure 5). In the dark, host C fixation was not detected, and UCYN-A2 N2 fixation was negligible compared to control values (Mann–Whitney test, p < 0.00001, Figure 5A). In the DTL treatment, UCYN-A2 symbiosis cell-specific C and N2 fixation rates were comparable to the control rates (Figure 5B), indicating recovery of these processes once the light period resumed. Sufficient numbers of cells could not be found from the DCMUm treatment for CARD-FISH analysis and cell-specific rate measurement.
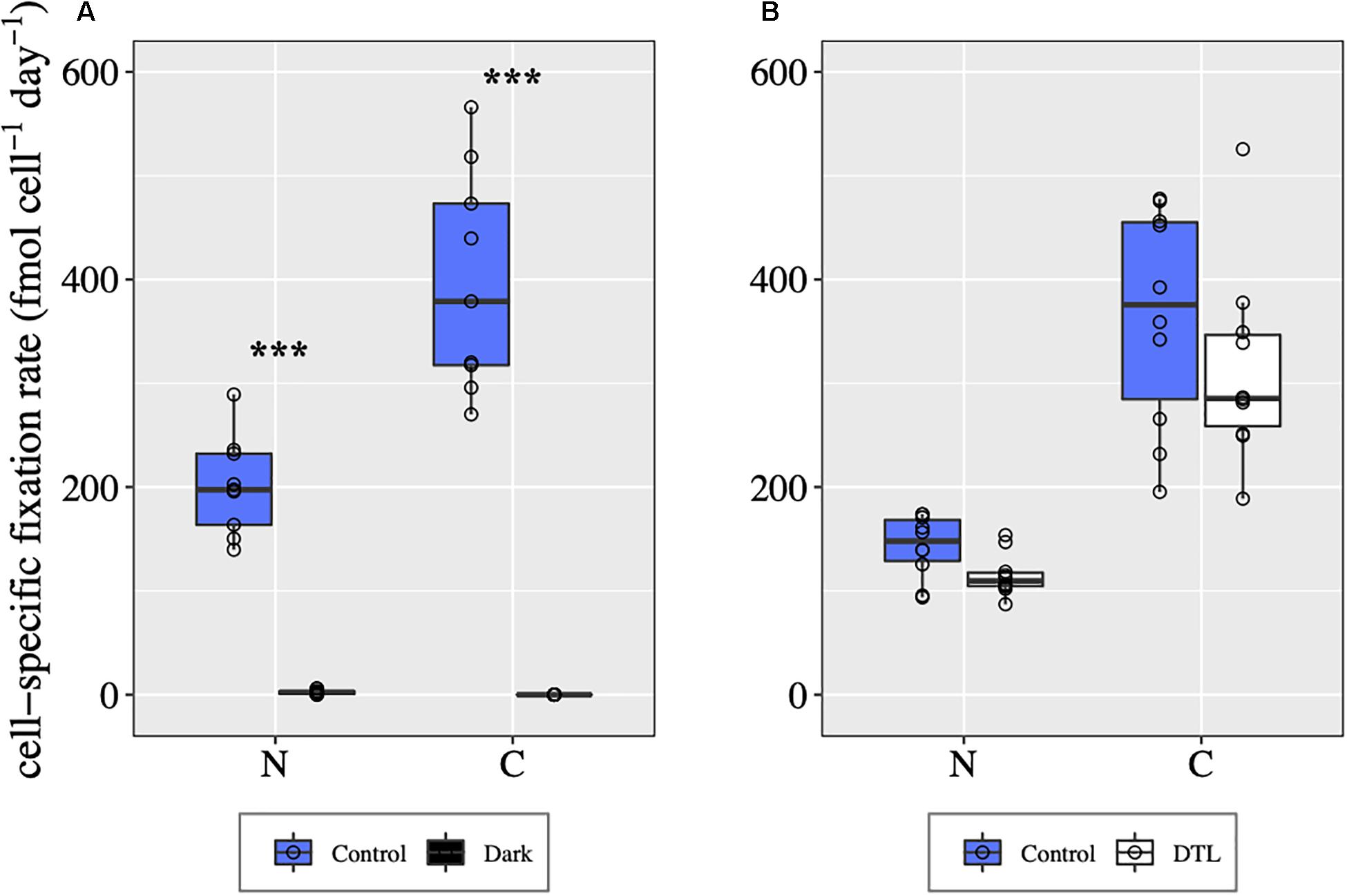
Figure 5. UCYN-A symbiosis cell-specific C and N2 fixation rates in different light-dark conditions during day 1 (A) and day 2 (B), determined for individual UCYN-A2/haptophyte symbiotic assemblages. For each condition, the values shown for C fixation and for N2 fixation were obtained from the same n cells, with n = 10 for each condition except for the control from day 1 for which n = 9. ***indicates significant differences between conditions (p < 0.0001) as determined by a Mann–Whitney test.
This study was designed to investigate mechanisms involved in the daily cycles of N2 fixation, C fixation and cell division in the UCYN-A haptophyte symbiosis, by manipulating the light-dark cycle and interrupting host oxygenic photosynthesis. The experimental setup mimicked natural conditions and results from the control incubations showed steady UCYN-A2 abundances, maximum UCYN-A2 nifH expression and N2 fixation during the day, and symbiont division early during the night, consistent with previous in situ observations on this lineage and others (Church et al., 2005b; Turk et al., 2011; Krupke et al., 2013; Muñoz-Marín et al., 2019). Through alteration of dark-light cycles, we show that light is required to observe typical activity patterns in the symbiont, from molecular processes such as nifH expression patterns, to functional traits such as active diazotrophy and cell replication and division. Using a PSII inhibitor targeting the eukaryotic host we also show that the haptophyte partner, through its photosynthetic activity, could be an important mediator of the symbiont dependence on light for these processes.
Our results demonstrate that light is necessary, as an energy source or as a signal, for N2 fixation in UCYN-A2. The timing of N2 fixation could not be directly assessed since our incubations were run for 24 h. However, (1) under constant dark conditions there was no N2 fixation, and (2) N2 fixation returned to control levels once cells were exposed to light after extended dark incubations. Together these results show a strong temporal connection between N2 fixation in UCYN-A2 and the presence of a light period.
There are several potential explanations for the coupling between light and UCYN-A N2 fixation evidenced in our study. First, light may play a direct role in UCYN-A physiology. Daytime light-dependent N2 fixation by UCYN-A is an unusual feature since closely related, free-living unicellular N2-fixing species only fix during the dark period to avoid photosynthetic oxygen. Although UCYN-A has lost PSII and the Calvin cycle, it has retained the PSI genes which are transcribed in a diel cycle similar to that of nif genes (Muñoz-Marín et al., 2019). The persistence of PSI in this organism appears to be important since similar freshwater symbiotic unicellular diazotrophic cyanobacteria have lost both photosystems (Nakayama et al., 2014; Nakayama and Inagaki, 2017). PSI may play an important role in UCYN-A metabolism to generate energy or reductant to fuel N2 fixation and may provide protection from oxygen using the Mehler reaction (Milligan et al., 2007). Thus, the activity of PSI could be essential to N2 fixation in UCYN-A and explain the atypical timing of this process in this unicellular cyanobacterium.
Our results could also be interpreted as evidence that UCYN-A N2 fixation is dependent on freshly fixed photosynthate or signals from the host. Both sequenced UCYN-A genomes lack the genes necessary for carbon fixation (Tripp et al., 2010; Bombar et al., 2014), implicating a dependence on organic carbon exogenous to the symbiont. Transfer of fixed carbon from the host to the symbiont has been demonstrated in the UCYN-A1 and UCYN-A2 lineages (Thompson et al., 2012; Martínez-Pérez et al., 2016). The timing of transfer of host-derived organic carbon, the chemical form that is transferred, and its contribution to the energy pool supporting N2 fixation by UCYN-A have not yet been resolved. Our study shows parallel responses of C and N2 fixation to continued incubation in the dark, during which both processes are shut down, and to the reintroduction of a light period, where both processes resume to levels comparable to control values. In a study of the freshwater diatom Rhopalodia gibba, light was also found to be indispensable for N2 fixation by the diatom’s cyanobacterial endosymbiont (Prechtl et al., 2004). This organism, which is closely related to UCYN-A (Mareš et al., 2019), does not have the genes required to assemble either photosystem or to perform carbon fixation (Nakayama and Inagaki, 2017). In that case, it is very likely that the symbiont’s requirement for light illustrates a time-sensitive reliance on host photosynthate as a source of energy to fuel daytime N2 fixation. Our study suggests that a similar scenario could be at play for UCYN-A and might be a fundamental characteristic of symbioses involving N2-fixing unicellular cyanobacteria and photosynthetic hosts.
Finally, daytime N2 fixation in UCYN-A could be an adaptation that increases the fitness of the haptophyte host rather than that of the symbiont. It is possible that daytime N2 fixation by UCYN-A provides the host with an ample supply of fixed N necessary to sustain its own daytime metabolic needs, and to maximize C fixation (Krupke et al., 2015). This fits the more general hypothesis that the atypical features of UCYN-A are driven by a tight metabolic embedding with the haptophyte host, which implies that the host at least partially controls some cellular processes in the symbiont. The tight coupling between N2 and C fixation with the presence of light requires mechanisms to regulate the timing and implementation of these processes in UCYN-A and its host.
Results from our study also show that transcriptional activity in UCYN-A is modulated by mechanisms involving light, either directly or through host activity. While both C and N2 fixation were interrupted in the dark treatment, nifH gene transcription continued. However, differences in transcript levels compared to control conditions indicate that the presence of light is also required to observe regular transcriptional patterns in the symbiont. In the dark, nifH transcripts levels measured at dawn persisted throughout daytime, showing that light, either as a direct trigger or through the metabolic activities it enables, is necessary for the decrease observed after the morning peak in control conditions. Dampened transcript levels in the dark on day 2 and the rapid return of control-like levels of nifH gene transcripts in the light after the extended dark period (DTL) also suggest that nifH transcription in UCYN-A is influenced by energy and reductant availability.
UCYN-A2 nifH expression also deviated from control patterns when host photosynthesis was chemically inhibited (DCMUm and DCMUa) while a regular light-dark cycle was maintained for those incubations. Because DCMU is a specific inhibitor of PSII, which UCYN-A lacks, the symbiont response to the presence of DCMU can be attributed to changes in host physiology. In our study, the similarities in the short-term (5 a.m. – 9 a.m.) transcriptional response to darkness and blocked host photosynthesis suggest that in the morning, light-dependent host activity rather than light itself could be an important regulator of the daytime transcription patterns normally observed in the symbiont for nifH, and conceivably other genes. A recent model-based study predicts that a minimum of 28 metabolites synthesized by the host are necessary for growth and activity of the symbiont (Sarkar et al., 2021). The amount provided as well as the timing of delivery of these compounds by the host may act as triggers of transcriptional regulations in the symbiont. Our observation that in the dark UCYN-A2 maintained pre-dawn nifH expression levels until nighttime is consistent with the hypothesis that freshly synthesized and transferred host metabolites are involved in a regulation cascade driving nifH transcript levels down later in the day. Our results provide evidence that UCYN-A activity is controlled in part via host metabolism. However, during the second portion of the daytime, nifH transcriptional patterns differed between dark and in DCMU incubations, suggesting that blocked host photosynthesis doesn’t explain on its own the divergence from control observed in dark incubations. Thus, while the host appears to influence gene expression in the symbiont, other mechanisms may also be at play.
Metabolic activities in cyanobacteria are under the control of a circadian clock that enhances fitness by enabling cells to anticipate daily changes in environmental conditions. In UCYN-A, clear diel oscillations in transcript abundance have been observed for most genes (Muñoz-Marín et al., 2019), a surprising result considering that 2 of the 3 kai genes responsible for the circadian oscillator in cyanobacteria (Ishiura et al., 1998) are missing from the UCYN-A genomes (Tripp et al., 2010; Bombar et al., 2014). In our study, diel oscillations in nifH expression were observed in all experimental conditions, including in dark incubations where nifH expression was lower compared to control conditions on day 2. The typical increase in nifH transcript abundance before sunrise was observed in all incubations, including in DCMU incubations where inhibited host photosynthesis was associated with unusually steady nifH transcript levels during most of the daytime. These observations show a link between the patterns of nifH expression in UCYN-A2 and the time of day regardless of light conditions and host status, suggesting that a circadian clock may still be controlling gene transcription in the symbiont.
It is unclear what mechanisms allow for circadian rhythms in UCYN-A in the absence of the oscillator proteins KaiAB. In the marine cyanobacterium Prochlorococcus, which does not possess the kaiA gene (Garczarek et al., 2001; Rocap et al., 2003), the remaining KaiBC proteins maintain their function through biochemical adaptations that compensate the loss of the KaiA protein (Axmann et al., 2009), leading to a conserved, though less robust, circadian clock (Holtzendorff et al., 2008). Similarly, it is possible that in UCYN-A, the remaining clock components are still operational in the absence of kaiAB through unique adaptations that could be related to the symbiotic lifestyle of UCYN-A. Eukaryotic algae have long been known to also have circadian clocks themselves (Suzuki and Johnson, 2001), and daily cycles in UCYN-A could be under host control. Interestingly, diel variations in energetic status are the primary mechanism through which the cyanobacterial kai oscillator is entrained (Rust et al., 2011; Pattanayak et al., 2014). For example, pulses of glucose are sufficient to entrain the clock in the absence of light cues in the model cyanobacterial strain Synechococcus elongatus (Pattanayak et al., 2015). Thus, if the remaining clock components in UCYN-A are functional, their activity is intrinsically linked to the host as the main provider of fixed carbon to the symbiont. It is probable that multiple parameters, related to the presence of light and variations in host activity and resulting energy levels throughout the day, together contribute to the clock-like orchestration of the daily patterns of transcription and activity in UCYN-A. Future experiments including a constant light treatment are required for definitively evaluating the question of circadian control of gene expression in UCYN-A, and will be facilitated when cultures of the UCYN-A symbiosis become available. Results from our study provide valuable insight and will help form future experiments regarding the existence of such a clock and mechanisms involved.
Our experiment provides the strongest evidence to date that replication and division in UCYN-A are at least partially under host control. As in previous studies (Cabello et al., 2016; Muñoz-Marín et al., 2019), we observed that in control conditions UCYN-A2 cells divided in early evening, going from a single symbiont to two prior to host division later in the night. Incubation in the dark led to no observable replication or division of the symbiont, showing that the cell cycle is disrupted without light. Return to light following extended dark restored division at the expected timing, although the number of cells proceeding to division decreased. The importance of dark-light cycles for cell replication is documented in other microorganisms, such as Prochlorococcus, which replicates in the afternoon and divides at night (Vaulot et al., 1995). In Prochlorococcus, the absence of a light period results in cells arrested in the G1 phase of the cell cycle, before DNA is replicated (Jacquet et al., 2001). In our experiment, the division of UCYN-A2 was also disrupted when host photosynthesis was chemically inhibited in regular dark-light cycles conditions. It is particularly striking that symbiont division was disrupted when DCMU was added midday after a full morning in control conditions. These observations suggest that while light may be involved in cell cycle regulation, it alone is not sufficient for UCYN-A division to proceed; division of the symbiont requires a photosynthetically active host. If the symbiosis between UCYN-A and the haptophyte is obligatory, tight control mechanisms must exist to ensure synchronized growth between the two partners.
Our results provide a detailed picture of how light and the photosynthetic activity of the haptophyte host affect daily processes such as gene transcription, N2 fixation and cell division in the symbiotic cyanobacterium UCYN-A2. Additional experiments in more controlled settings, particularly once cultures of the symbiosis become available, are needed to elucidate the specific mechanisms through which light and the physiology of the haptophyte host control UCYN-A activity. The hypotheses generated by this study about the functioning of the UCYN-A-haptophyte symbiosis are based on results obtained from the UCYN-A2 lineage. At least five ecologically distinct UCYN-A sublineages have been identified to date, with specific global distribution patterns (Turk-Kubo et al., 2017). While functional differences might be identified in the future, existing data indicate strong similarities across the sublineages and a conserved function organized around the exchange of fixed N and C between the cyanobacterial and haptophyte partners (Bombar et al., 2014; Cornejo-Castillo et al., 2016). The existence of core functional traits within the UCYN-A lineage suggests that some, if not most, regulatory mechanisms at play in the UCYN-A2 sublineage are shared by other members of the group. Future work will clarify whether the patterns observed in our study with the UCYN-A2 lineage also hold for other UCYN-A lineages. Our study provides a framework for a better understanding of the unique features that shape this abundant, ecologically important N2-fixing symbiotic association.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
ML and JZ designed the study. ML, KT-K, FC-C, and BH ran the experiments and performed sampling. ML and KT-K performed sample and data analyses. ML, KT-K, and JZ wrote the manuscript with edits from BH and FC-C. All authors contributed to the article and approved the submitted version.
This work was funded by the Simons Foundation (Award ID 545171 to JZ) and partially supported by grants from the National Science Foundation (OCE-1559165) and Simons Collaboration on Ocean Processes and Ecology (SCOPE, Grant 329108).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We gratefully acknowledge F. Azam (UCSD) for providing laboratory facilities for the experiments at SIO, M. R. Gradoville (UCSC), M. Mills (Stanford), and C. Jilly-Rehak (Stanford) for providing nanoSIMS consultations, S. Wilson and D. Karl (UH Manoa) for MIMS measurements, and the SOEST Biogeochemical Stable Isotope Facility at the UH Manoa. Part of this work was performed at the Stanford Nano Shared Facilities (SNSF), supported by the National Science Foundation under award ECCS-1542152. FC-C acknowledges funding from the European Union’s Horizon 2020 research programme under the Marie Sklodowska-Curie grant agreement No. 749380 (UCYN2PLAST).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.666739/full#supplementary-material
Axmann, I. M., Dühring, U., Seeliger, L., Arnold, A., Vanselow, J. T., Kramer, A., et al. (2009). Biochemical evidence for a timing mechanism in Prochlorococcus. J. Bacteriol. 191, 5342–5347. doi: 10.1128/JB.00419-09
Becker, K. W., Harke, M. J., Mende, D. R., Muratore, D., Weitz, J. S., DeLong, E. F., et al. (2021). Combined pigment and metatranscriptomic analysis reveals highly synchronized diel patterns of phenotypic light response across domains in the open oligotrophic ocean. ISME J. 15, 520–533. doi: 10.1038/s41396-020-00793-x
Berman-Frank, I., Lundgren, P., and Falkowski, P. (2003). Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res. Microbiol. 154, 157–164. doi: 10.1016/S0923-2508(03)00029-9
Bombar, D., Heller, P., Sanchez-Baracaldo, P., Carter, B. J., and Zehr, J. P. (2014). Comparative genomics reveals surprising divergence of two closely related strains of uncultivated UCYN-A cyanobacteria. ISME J. 8, 2530–2542. doi: 10.1038/ismej.2014.167
Böttjer, D., Dore, J. E., Karl, D. M., Letelier, R. M., Mahaffey, C., Wilson, S. T., et al. (2017). Temporal variability of nitrogen fixation and particulate nitrogen export at Station ALOHA. Limnol. Oceanogr. 62, 200–216. doi: 10.1002/lno.10386
Cabello, A. M., Cornejo-Castillo, F. M., Raho, N., Blasco, D., Vidal, M., Audic, S., et al. (2016). Global distribution and vertical patterns of a prymnesiophyte–cyanobacteria obligate symbiosis. ISME J. 10, 693–706. doi: 10.1038/ismej.2015.147
Church, M. J., Jenkins, B. D., Karl, D. M., and Zehr, J. P. (2005a). Vertical distributions of nitrogen-fixing phylotypes at Stn ALOHA in the oligotrophic North Pacific Ocean. Aquat. Microb. Ecol. 38, 3–14. doi: 10.3354/ame038003
Church, M. J., Short, C. M., Jenkins, B. D., Karl, D. M., and Zehr, J. P. (2005b). Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl. Environ. Microbiol. 71, 5362–5370. doi: 10.1128/AEM.71.9.5362
Cornejo-Castillo, F. M., Cabello, A. M., Salazar, G., Sánchez-Baracaldo, P., Lima-Mendez, G., Hingamp, P., et al. (2016). Cyanobacterial symbionts diverged in the late Cretaceous towards lineage-specific nitrogen fixation factories in single-celled phytoplankton. Nat. Commun. 7, 11071. doi: 10.1038/ncomms11071
Dekas, A. E., and Orphan, V. J. (2011). “Identification of diazotrophic microorganisms in marine sediment via fluorescence in situ hybridization coupled to nanoscale secondary ion mass spectrometry (fish-nanosims),” in Methods in Enzymology, eds J. Simon, Abelson, M. G. Verdine, and A. Pyle (Cambridge, MA: Academic Press), 281–305. doi: 10.1016/S0076-6879(11)86012-X
Dron, A., Rabouille, S., Claquin, P., Chang, P., Raimbault, V., Talec, A., et al. (2012). Light:dark (12:12 h) quantification of carbohydrate fluxes in Crocosphaera watsonii. Aquat. Microb. Ecol. 68, 43–55. doi: 10.3354/ame01600
Farnelid, H., Turk-kubo, K., Muñoz-marín, M. C., and Zehr, J. P. (2016). New insights into the ecology of the globally significant uncultured nitrogen-fixing symbiont UCYN-A. Aquat. Microb. Ecol. 77, 125–138. doi: 10.3354/ame01794
Fay, P. (1992). Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 56, 340–373. doi: 10.1128/mmbr.56.2.340-373.1992
Field, C. B., Behrenfeld, M. J., Randerson, J. T., and Falkowski, P. (1998). Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240. doi: 10.1126/science.281.5374.237
Garczarek, L., Partensky, F., Irlbacher, H., Holtzendorff, J., Babin, M., Mary, I., et al. (2001). Differential expression of antenna and core genes in Prochlorococcus PCC 9511 (Oxyphotobacteria) grown under a modulated light-dark cycle. Environ. Microbiol. 3, 168–175. doi: 10.1046/j.1462-2920.2001.00173.x
Gradoville, M. R., Bombar, D., Crump, B. C., Letelier, R. M., Zehr, J. P., and White, A. E. (2017). Diversity and activity of nitrogen-fixing communities across ocean basins. Limnol. Oceanogr. 62, 1895–1909. doi: 10.1002/lno.10542
Hagino, K., Onuma, R., Kawachi, M., and Horiguchi, T. (2013). Discovery of an endosymbiotic nitrogen-fixing cyanobacterium UCYN-A in Braarudosphaera bigelowii (Prymnesiophyceae). PLoS One 8:e81749. doi: 10.1371/journal.pone.0081749
Harding, K., Turk-Kubo, K. A., Sipler, R. E., Mills, M. M., Bronk, D. A., and Zehr, J. P. (2018). Symbiotic unicellular cyanobacteria fix nitrogen in the Arctic Ocean. Proc. Natl. Acad. Sci. U.S.A. 115, 13371–13375. doi: 10.1073/pnas.1813658115
Holtzendorff, J., Partensky, F., Mella, D., Lennon, J. F., Hess, W. R., and Garczarek, L. (2008). Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J. Biol. Rhythms 23, 187–199. doi: 10.1177/0748730408316040
Ishiura, M., Kutsuna, S., Aoki, S., Iwasaki, H., Andersson, C. R., Tanabe, A., et al. (1998). Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523. doi: 10.1126/science.281.5382.1519
Jacquet, S., Partensky, F., Marie, D., Casotti, R., and Vaulot, D. (2001). Cell cycle regulation by light in Prochlorococcus strains. Appl. Environ. Microbiol. 67, 782–790. doi: 10.1128/AEM.67.2.782-790.2001
Karl, D., Letelier, R., Tupas, L., Dore, J., Christian, J., and Hebel, D. (1997). The role of nitrogen fixation in biogeochemical cycling in the subtropical North Pacific Ocean. Nature 388, 533–538. doi: 10.1038/41474
Klawonn, I., Lavik, G., Böning, P., Marchant, H. K., Dekaezemacker, J., Mohr, W., et al. (2015). Simple approach for the preparation of 15-15N2-enriched water for nitrogen fixation assessments: evaluation, application and recommendations. Front. Microbiol. 6:1–11. doi: 10.3389/fmicb.2015.00769
Krupke, A., Mohr, W., LaRoche, J., Fuchs, B. M., Amann, R. I., and Kuypers, M. M. (2015). The effect of nutrients on carbon and nitrogen fixation by the UCYN-A–haptophyte symbiosis. ISME J. 9, 1635–1647. doi: 10.1038/ismej.2014.253
Krupke, A., Musat, N., LaRoche, J., Mohr, W., Fuchs, B. M., Amann, R. I., et al. (2013). In situ identification and N2 and C fixation rates of uncultivated cyanobacteria populations. Syst. Appl. Microbiol. 36, 259–271. doi: 10.1016/j.syapm.2013.02.002
Mareš, J., Johansen, J. R., Hauer, T., Zima, J., Ventura, S., Cuzman, O., et al. (2019). Taxonomic resolution of the genus Cyanothece (Chroococcales, Cyanobacteria), with a treatment on Gloeothece and three new genera, Crocosphaera, Rippkaea, and Zehria. J. Phycol. 55, 578–610. doi: 10.1111/jpy.12853
Martínez-Pérez, C., Mohr, W., Löscher, C. R., Dekaezemacker, J., Littmann, S., Yilmaz, P., et al. (2016). The small unicellular diazotrophic symbiont, UCYN-A, is a key player in the marine nitrogen cycle. Nat. Microbiol. 1, 1–7. doi: 10.1038/nmicrobiol.2016.163
Milligan, A. J., Berman-Frank, I., Gerchman, Y., Dismukes, G. C., and Falkowski, P. G. (2007). Light-dependent oxygen consumption in nitrogen-fixing cyanobacteria plays a key role in nitrogenase protection. J. Phycol. 43, 845–852. doi: 10.1111/j.1529-8817.2007.00395.x
Mills, M. M., Turk-Kubo, K. A., van Dijken, G. L., Henke, B. A., Harding, K., Wilson, S. T., et al. (2020). Unusual marine cyanobacteria/haptophyte symbiosis relies on N2 fixation even in N-rich environments. ISME J. 14, 2395–2406. doi: 10.1038/s41396-020-0691-6
Mohr, W., Großkopf, T., Wallace, D. W. R., and LaRoche, J. (2010a). Methodological underestimation of oceanic nitrogen fixation rates. PLoS One 5:e12583. doi: 10.1371/journal.pone.0012583
Mohr, W., Intermaggio, M. P., and LaRoche, J. (2010b). Diel rhythm of nitrogen and carbon metabolism in the unicellular, diazotrophic cyanobacterium Crocosphaera watsonii WH8501. Environ. Microbiol. 12, 412–421. doi: 10.1111/j.1462-2920.2009.02078.x
Montoya, J. P., Voss, M., Kahler, P., and Capone, D. G. (1996). A simple, high-precision, high-sensitivity tracer assay for N2 fixation. Appl. Environ. Microbiol. 62, 986–993. doi: 10.1128/aem.62.3.986-993.1996
Mulholland, M. R., Bernhardt, P. W., Blanco-Garcia, J. L., Mannino, A., Hyde, K., Mondragon, E., et al. (2012). Rates of dinitrogen fixation and the abundance of diazotrophs in North American coastal waters between Cape Hatteras and Georges Bank. Limnol. Oceanogr. 57, 1067–1083. doi: 10.4319/lo.2012.57.4.1067
Muñoz-Marín, M. D. C., Shilova, I. N., Shi, T., Farnelid, H., Cabello, A. M., and Zehr, J. P. (2019). The transcriptional cycle is suited to daytime N2 fixation in the unicellular cyanobacterium Candidatus Atelocyanobacterium thalassa (UCYN-A). mBio 10, 1–17. doi: 10.1128/mBio.02495-18
Nakayama, T., and Inagaki, Y. (2017). Genomic divergence within non-photosynthetic cyanobacterial endosymbionts in rhopalodiacean diatoms. Sci. Rep. 7, 1–8. doi: 10.1038/s41598-017-13578-8
Nakayama, T., Kamikawa, R., Tanifuji, G., Kashiyama, Y., Ohkouchi, N., Archibald, J. M., et al. (2014). Complete genome of a nonphotosynthetic cyanobacterium in a diatom reveals recent adaptations to an intracellular lifestyle. Proc. Natl. Acad. Sci.U.S.A. 111, 11407–11412. doi: 10.1073/pnas.1405222111
Pattanayak, G. K., Lambert, G., Bernat, K., and Rust, M. J. (2015). Controlling the cyanobacterial clock by synthetically rewiring metabolism. Cell Rep. 13, 2362–2367. doi: 10.1016/j.celrep.2015.11.031
Pattanayak, G. K., Phong, C., and Rust, M. J. (2014). Report rhythms in energy storage control the ability of the cyanobacterial circadian clock to reset. Curr. Biol. 24, 1934–1938. doi: 10.1016/j.cub.2014.07.022
Polerecky, L., Adam, B., Milucka, J., Musat, N., Vagner, T., and Kuypers, M. M. M. (2012). Look@NanoSIMS - a tool for the analysis of nanoSIMS data in environmental microbiology. Environ. Microbiol. 14, 1009–1023. doi: 10.1111/j.1462-2920.2011.02681.x
Prechtl, J., Kneip, C., Lockhart, P., Wenderoth, K., and Maier, U. G. (2004). Intracellular spheroid bodies of Rhopalodia gibba have nitrogen-fixing apparatus of cyanobacterial origin. Mol. Biol. Evol. 21, 1477–1481. doi: 10.1093/molbev/msh086
Ribalet, F., Swalwell, J., Clayton, S., Jiménez, V., Sudek, S., Lin, Y., et al. (2015). Light-driven synchrony of Prochlorococcus growth and mortality in the subtropical Pacific gyre. Proc. Natl. Acad. Sci. U.S.A. 112, 8008–8012. doi: 10.1073/pnas.1424279112
Rocap, G., Larimer, F. W., Lamerdin, J., Malfatti, S., Chain, P., Ahlgren, N. A., et al. (2003). Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424, 1042–1047. doi: 10.1038/nature01947
Rust, M. J., Golden, S. S., and O’Shea, E. K. (2011). Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science 331, 220–223. doi: 10.1126/science.1197243
Sarkar, D., Landa, M., Bandyopadhyay, A., Pakrasi, H. B., Zehr, J. P., and Maranas, C. D. (2021). Elucidation of trophic interactions in an unusual single-cell nitrogen-fixing symbiosis using metabolic modeling. PLoS Comput. Biol.
Shiozaki, T., Fujiwara, A., Ijichi, M., Harada, N., Nishino, S., Nishi, S., et al. (2018). Diazotroph community structure and the role of nitrogen fixation in the nitrogen cycle in the Chukchi Sea (western Arctic Ocean). Limnol. Oceanogr. 63, 2191–2205. doi: 10.1002/lno.10933
Shiozaki, T., Kodama, T., Kitajima, S., Sato, M., and Furuya, K. (2013). Advective transport of diazotrophs and importance of their nitrogen fixation on new and primary production in the western Pacific warm pool. Limnol. Oceanogr. 58, 49–60. doi: 10.4319/lo.2013.58.1.0049
Suzuki, L., and Johnson, C. H. (2001). Algae know the time of day: circadian and photoperiodic programs. J. Phycol. 37, 933–942. doi: 10.1046/j.1529-8817.2001.01094.x
Tang, W., and Cassar, N. (2019). Data-driven modeling of the distribution of diazotrophs in the global ocean. Geophys. Res. Lett. 46, 12258–12269. doi: 10.1029/2019GL084376
Thaben, P. F., and Westermark, P. O. (2014). Detecting rhythms in time series with RAIN. J. Biol. Rhythms 29, 391–400. doi: 10.1177/0748730414553029
Thompson, A., Carter, B. J., Turk-Kubo, K., Malfatti, F., Azam, F., and Zehr, J. P. (2014). Genetic diversity of the unicellular nitrogen-fixing cyanobacteria UCYN-A and its prymnesiophyte host. Environ. Microbiol. 16, 3238–3249. doi: 10.1111/1462-2920.12490
Thompson, A. W., Foster, R. A., Krupke, A., Carter, B. J., Musat, N., Vaulot, D., et al. (2012). Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 337, 1546–1551. doi: 10.1126/science.1222700
Thompson, A. W., and Zehr, J. P. (2013). Cellular interactions: lessons from the nitrogen-fixing cyanobacteria. J. Phycol. 49, 1024–1035. doi: 10.1111/jpy.12117
Tripp, H. J., Bench, S. R., Turk, K. A., Foster, R. A., Desany, B. A., Niazi, F., et al. (2010). Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature 464, 90–94. doi: 10.1038/nature08786
Tuit, C., Waterbury, J., and Ravizza, G. (2004). Diel variation of molybdenum and iron in marine diazotrophic cyanobacteria. Limnol. Oceanogr. 49, 978–990. doi: 10.4319/lo.2004.49.4.0978
Turk, K. A., Rees, A. P., Zehr, J. P., Pereira, N., Swift, P., Shelley, R., et al. (2011). Nitrogen fixation and nitrogenase (nifH) expression in tropical waters of the eastern North Atlantic. ISME J. 5, 1201–1212. doi: 10.1038/ismej.2010.205
Turk-Kubo, K. A., Farnelid, H. M., Shilova, I. N., Henke, B., and Zehr, J. P. (2017). Distinct ecological niches of marine symbiotic N2-fixing cyanobacterium Candidatus Atelocyanobacterium thalassa sublineages. J. Phycol. 53, 451–461. doi: 10.1111/jpy.12505
Vaulot, D., Marie, D., Olson, R. J., and Chisholm, S. W. (1995). Growth of Prochlorococcus, a photosynthetic prokaryote, in the equatorial Pacific Ocean. Science 268, 1480–1482. doi: 10.1126/science.268.5216.1480
Welkie, D. G., Rubin, B. E., Diamond, S., Hood, R. D., Savage, D. F., and Golden, S. S. (2019). A hard day’s night: cyanobacteria in diel cycles. Trends Microbiol. 27, 231–242. doi: 10.1016/j.tim.2018.11.002
Wilson, S. T., Aylward, F. O., Ribalet, F., Barone, B., Casey, J. R., Connell, P. E., et al. (2017). Coordinated regulation of growth, activity and transcription in natural populations of the unicellular nitrogen-fixing cyanobacterium Crocosphaera. Nat. Microbiol. 2, 1–9. doi: 10.1038/nmicrobiol.2017.118
Wilson, S. T., Böttjer, D., Church, M. J., and Karl, D. M. (2012). Comparative assessment of nitrogen fixation methodologies, conducted in the oligotrophic north pacific ocean. Appl. Environ. Microbiol. 78, 6516–6523. doi: 10.1128/AEM.01146-12
Zehr, J. P., Bench, S. R., Carter, B. J., Hewson, I., Niazi, F., Shi, T., et al. (2008). Globally distributed uncultivated oceanic N2-fixing cyanobacteria lack oxygenic photosystem II. Science 322, 1110–1112. doi: 10.1126/science.1165340
Zehr, J. P., Shilova, I. N., Farnelid, H. M., Muñoz-Maríncarmen, M. D. C., and Turk-Kubo, K. A. (2016). Unusual marine unicellular symbiosis with the nitrogen-fixing cyanobacterium UCYN-A. Nat. Microbiol. 2, 1–11. doi: 10.1038/nmicrobiol.2016.214
Keywords: UCYN-A, N2-fixing symbiosis, nifH, diel cycle, host photosynthesis, circadian rhythm, light – dark cycles, gene expression
Citation: Landa M, Turk-Kubo KA, Cornejo-Castillo FM, Henke BA and Zehr JP (2021) Critical Role of Light in the Growth and Activity of the Marine N2-Fixing UCYN-A Symbiosis. Front. Microbiol. 12:666739. doi: 10.3389/fmicb.2021.666739
Received: 10 February 2021; Accepted: 08 April 2021;
Published: 05 May 2021.
Edited by:
Mar Benavides, Institut de Recherche Pour le Développement (IRD), FranceReviewed by:
Enrique Flores, Consejo Superior de Investigaciones Científicas (CSIC), SpainCopyright © 2021 Landa, Turk-Kubo, Cornejo-Castillo, Henke and Zehr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonathan P. Zehr, emVocmpAdWNzYy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.