- 1Department of Clinical Laboratory Science, Faculty of Health Care, Tenri Health Care University, Tenri, Japan
- 2Faculty of Health Sciences, Kyoto Tachibana University, Kyoto, Japan
- 3Department of Infection Control and Prevention, Osaka City University Hospital, Osaka, Japan
- 4Department of Clinical Laboratory, Naga Municipal Hospital, Wakayama, Japan
- 5Laboratory for Clinical Investigation, Osaka University Hospital, Osaka, Japan
Worldwide spread of Enterobacteriaceae resistant to colistin, a polypeptide antibacterial drug for last-resort treatment of carbapenemase-producing Enterobacteriaceae (CPE) infections, is concerning. This study aimed to elucidate colistin MICs and molecular characteristics of mcr-1 to mcr-9 of ESBL-producing Escherichia coli (ESBL-Ec) and CPE in Japan and clarify the genomic structure of strains harboring mcr genes (especially mcr-9). This study included 168 ESBL-Ec and 126 CPE strains isolated at Japanese medical facilities. Colistin susceptibility testing and multiplex PCR targeting mcr-1 to mcr-9 were performed for all strains with S1-nuclease pulsed-field gel electrophoresis, Southern blot hybridization, and whole-genome sequencing (WGS) with hybrid assembly performed for mcr gene-carrying strains. Two CPE strains showed a MIC ≥ 4 μg/ml in colistin susceptibility testing, with no known resistance mechanism detected. However, PCR conducted on all target strains detected three mcr-9-carrying strains showing colistin susceptibility. The blaCTX–M–62-positive E. coli THUN648 strain simultaneously carried blaCTX–M–62 and mcr-9 on a 275-kbp plasmid. Besides, blaIMP–6 + blaCTX–M–2-positive Klebsiella pneumoniae THUN262 and blaGES–24-positive Enterobacter kobei THUN627 had mcr-9 encoded on the chromosome. Only THUN627 encoded qseB/C, which is suggested to be a regulatory gene for mcr-9, downstream of mcr-9. However, this strain showed no increased expression of these genes in mRNA quantitative analysis under colistin exposure. Colistin MICs of ESBL-Ec and CPE in Japan were all below 2 μg/ml, which is below the epidemiological cutoff (ECOFF) value (https://eucast.org/) or clinical breakpoint (CB) (CLSI M100-S30) reported for colistin, indicating neither “microbiological” nor “clinical” resistance. Several colistin-susceptible Enterobacteriaceae carrying silent mcr-9 encoded on plasmids and chromosomes have already spread worldwide along with other antimicrobial resistance genes. However, the mechanism of colistin resistance by mcr-9 remains unclear.
Introduction
In recent years, the global spread of drug-resistant bacteria has become a concern, and among these bacteria, multidrug-resistant Enterobacteriaceae are remarkable (Nicolas-Chanoine et al., 2014; Mathers et al., 2015). The emergence of carbapenemase-producing Enterobacteriaceae (CPE) that are resistant to carbapenem antibiotics, which are commonly used against severe infections, is a threat to humanity and is currently on the rise both in Japan and abroad (Nakamura et al., 2016; Ohno et al., 2017, 2020). The emergence of Enterobacteriaceae resistant to colistin, a polypeptide antibacterial drug of last resort for the treatment of CPE infections, is being reported, and its global spread is troubling (Liu et al., 2016).
Colistin has been used since the 1950s as a feed additive to treat Gram-negative bacterial infections and to promote healthy growth of food animals (Kawanishi et al., 2017; Poirel et al., 2017). However, the use of colistin in humans was discontinued due to frequent adverse reactions such as renal dysfunction. However, it was reapproved as a last-resort treatment in response to the global increase in multidrug-resistant Gram-negative bacterial infections (Poirel et al., 2017). Therefore, the World Health Organization has listed colistin as an extremely important antibiotic since 2016 (World Health Organization and Who Advisory Group on Integrated Surveillance of Antimicrobial Resistance, 2017). The longtime use of colistin in food animals is thought to be the cause of colistin resistance. Therefore, Japan banned the market sale of colistin as a feed additive in 2018, and many other countries have also banned colistin for this use due to public health considerations (Shen et al., 2018; The Bureau of Investigative Journalism, 2018; Amr Insights, 2019; Market Research.Com, 2019).
There are two main mechanisms of colistin resistance: one is the acquisition of a plasmid-mediated colistin resistance mcr gene, and the other is point mutation or inactivation of chromosomal genes such as PmrAB, PhoPQ, CrrAB, and MgrB (Olaitan et al., 2014; Sato et al., 2018; Mmatli et al., 2020). Mutations in PmrAB, PhoPQ, CrrAB, and MgrB affect the phosphorylation of lipid A in lipopolysaccharides, which leads to colistin resistance. Inactivation of MgrB also affects capsule structure, and various other factors such as efflux hyperexpression and porin downregulation are also involved in colistin resistance (Mmatli et al., 2020). The plasmid-mediated colistin resistance mcr gene is transmitted across bacterial species, and its trends need to be monitored closely.
The mcr gene was first discovered in China in 2015, and many reports have since been published, mainly in Asia (Liu et al., 2016; Cui et al., 2017; Tada et al., 2017; La et al., 2019; Farzana et al., 2020). Cui et al. (2017) reported that 1.4% of Salmonella spp. isolated from human clinical samples in China carried mcr-1. According to the report of La et al. (2019), when culture and PCR were used to screen human fecal material for Enterobacteriaceae carrying mcr-1 in Singapore, 9% of the cultures were positive, indicating the presence of a large number of potential carriers. According to a 2020 report by Farzana et al. (2020), 0.3% of Klebsiella pneumoniae were found to carry mcr-8 in Bangladesh, and transmission of this resistance gene has been a problem since 2017. As for the emergence of strains carrying the mcr gene in Japan, Tada et al. (2017) first reported the presence of Escherichia coli carrying mcr-1 in 2017. Except for a few reports since then, there have been no reports of large-scale surveillance (Tada et al., 2017). Among mcr genes, mcr-9 is a novel colistin resistance gene identified in Salmonella enterica serotype Typhimurium as reported by Carroll et al. (2019). Although this is a silent colistin resistance gene that has been present in the gene database since around 2010, it has not received much attention as it does not confer colistin resistance. This Mcr-9 protein shares 33–65% identity with the plasmid-encoded Mcr-1 to -8, which have been reported to be phenotypically colistin resistance enzymes, and is most similar to Mcr-3 (Carroll et al., 2019; Kieffer et al., 2019). It has been reported that the mRNA levels of mcr-9 increased with exposure to colistin, thus causing the MIC to increase (Carroll et al., 2019). The levels of mcr-9-induced expression are related to the presence of qseB and qseC genes downstream. A significant increase in the mRNA expression level of the qseB/C genes was observed with exposure to colistin, suggesting that overexpression of mcr-9 is associated with the QseC/QseB binary system (Carroll et al., 2019).
The purpose of this study was to elucidate the colistin MICs and molecular characteristics of ESBL-producing E. coli (ESBL-Ec) and CPE harboring mcr-1 to mcr-9 in Japan and to clarify the genomic structure of strains harboring mcr genes.
Materials and Methods
Bacterial Isolates
All 126 strains of CPE isolated from 2001 to 2018 at 18 Japanese medical facilities from Western Japan (17 acute-care hospitals and 1 commercial laboratory) and 168 ESBL-Ec strains extracted randomly were included in the study (Table 1). The 126 strains of CPE were provided in the framework of the Study of Bacterial Resistance in the Kinki Region of Japan (SBRK), of which 37 strains were those used in our previous four studies (Ohno et al., 2017, 2020; Yamasaki et al., 2017; Kuchibiro et al., 2018) and 89 strains were used in the present study. Two of the four studies were epidemiological studies of CPE in a primary care hospital in Tenri, Japan, conducted from 2010 to 2015, and these included fecal isolates (Ohno et al., 2017, 2020). Besides, one study conducted an outbreak survey of CPE (blaGES group) in a primary care hospital in Wakayama City, Japan, in 2009, and the source was blood (Yamasaki et al., 2017). One study was collected for the purpose of epidemiological study of CPE from 2000 to 2016 in 15 facilities in Kinki region, Japan, and these did not include fecal isolates (Kuchibiro et al., 2018). Thirty-seven strains of CPE were randomly selected from these four studies. Besides, 89 strains of CPE were collected for the epidemiological study in SBRK. Moreover, the Association of Japan Community Healthcare Organization (JCHO) hospitals and SBRK provided the 168 strains of ESBL-Ec, all of which were also used in our previous studies (Nakamura et al., 2012, 2016; Shibasaki et al., 2016). One of the three studies were epidemiological studies of ESBL-Ec in a primary care hospital in Tenri, Japan, conducted from 2011 to 2012, and the source is feces (Nakamura et al., 2016). Besides, one study was collected for the purpose of epidemiological study of ESBL-producing Enterobacteriaceae from 2000 to 2009 in 18 facilities in Kinki region, Japan, and these did not include fecal isolates (Nakamura et al., 2012). One study was collected for the purpose of epidemiological study of ESBL-producing Enterobacteriaceae in 2012 in 11 facilities in all Japan, and these did not include fecal isolates (Shibasaki et al., 2016). A total of 168 strains of ESBL-Ec were randomly selected from these three studies. This study was approved by the ethical committee of Tenri Health Care University (approval no. 115). Bacterial species identification except for isolates carrying the mcr gene was performed using MALDI Biotyper (Bruker Daltonik, Bremen, Germany), and WGS with hybrid assembly was also used to confirm species identification of mcr-positive isolates (KmerFinder 3.2)1. Screening of the β-lactamase resistance mechanism and multiplex PCR was performed following previous reports (Nakamura et al., 2016; Ohno et al., 2017; Kuchibiro et al., 2018). Seven carbapenemase gene types (blaIMP–1, blaIMP–2, blaVIM group, blaKPC group, blaGES group, blaNDM group, and blaOXA–48) and six ESBL gene types (blaSHV group, blaTEM group, blaCTX–M–1, blaCTX–M–2, blaCTX–M–8, and blaCTX–M–9) were determined (Nakamura et al., 2016; Kuchibiro et al., 2018; Ohno et al., 2020). In addition, PCR amplicon sequencing (Sanger sequencing) was performed on the entire length of the blaSHV and blaTEM groups, and only those belonging to 2be as per the classification by Bush and Jacoby (2010) are listed in Table 1.

Table 1. Characteristics of carbapenemase-producing Enterobacteriaceae and ESBL-producing Escherichia coli used in this study.
Antimicrobial Susceptibility Testing
The agar dilution method (Sigma-Aldrich, Tokyo, Japan) was implemented using colistin sulfate powder for all strains, based on the Clinical and Laboratory Standards Institute (CLSI) M07 11th edition and M100 30th edition (Clinical and Laboratory Standards Institute, 2018, 2020). We set the measurement range of colistin between 0.03 and 128 μg/ml. The microdilution method and E-test (bioMérieux Japan Ltd., Tokyo, Japan) were additionally performed for strains showing a MIC of 4 μg/ml or higher with the agar dilution method. All methods were quality checked using standard bacterial strains: E. coli ATCC25922, Pseudomonas aeruginosa ATCC27853, and E. coli NCTC13846 (mcr-1 positive).
Molecular Analysis of Plasmid-Mediated mcr-Gene Using Multiplex PCR
All target strains were screened for mcr-1 to mcr-9 by conventional PCR, which was performed based on the multiplex-PCR methods reported by Lescat et al. (2018) for mcr-1 to mcr-5 and Borowiak et al. (2020) for mcr-6 to mcr-9. DNA was extracted using the Cica Genius DNA extraction test (Kanto Chemical Co., Inc., Tokyo, Japan), and PCR target genes were amplified using GoTaq® Green Master Mix (Promega K.K., Tokyo, Japan). Strains harboring plasmid-mediated mcr gene underwent antimicrobial susceptibility testing using the microdilution method and E-test.
S1-Nuclease Pulsed-Field Gel Electrophoresis (S1-PFGE) and Southern Blot Hybridization Using the mcr Gene
Bacterial lysates of strains carrying any of the mcr genes from mcr-1 to mcr-9 that were cultured overnight were enclosed in Seakem® Gold agarose plugs (Lonza Japan Inc., Tokyo, Japan). After agar plugs were prepared, the chromosomal DNA was digested by S1-nuclease (TaKaRa Bio, Shiga, Japan) under an enzyme volume of 5 U and reaction time of 1 h. Migration was performed for 19.5 h using the CHEF-DR-III system (Bio-Rad Laboratories, Inc., Hercules, CA, United States) under a switch time of 5.3–34.9 s and a voltage of 6.0 V/cm. After migration, ethidium bromide staining was performed, followed by imaging with ChemiDoc (Bio-Rad Laboratories, Inc.). Agarose gel obtained by S1-PFGE as mentioned above was transferred overnight using nylon membranes, and Southern blotting was performed with a mcr gene-labeled DIG probe (Roche Diagnostics, Inc., Tokyo, Japan) for 20 h at 40°C. The mcr coding plasmid was detected by ChemiDoc after chemiluminescence using CDP Star (Roche Diagnostics, Inc.), and the plasmid size was confirmed. In addition, the β-lactamase-producing genes (blaIMP–6, blaGES–24, and blaCTX–M–62) of the strains harboring mcr-9 were also subjected to Southern blotting using DIG probe as described above. The primers used for each were those of Borowiak et al. (2020) for mcr-9, Nishio et al. (2004) for blaIMP–6, Weldhagen and Prinsloo (2004) for blaGES–24, and Yagi et al. (1997) for blaCTX–M–62. Southern blotting was performed using the method of Yamasaki et al. (2017).
WGS and Bioinformatic Analysis
Whole-genome sequencing with a hybrid assembly of strains harboring the plasmid-mediated mcr gene and strains with colistin MIC greater than 4 μg/ml was performed using MiSeq (Illumina, Inc., CA, United States) and MinION (Oxford Nanopore Technologies, Oxford, United Kingdom). We used the DNeasy PowerSoil Pro Kit (Qiagen, Hilden, Germany) for DNA extraction. The Nextera DNA sample preparation kit (Illumina, Inc.) was used to tune the library for MiSeq sequencing, and 300-bp paired-end sequencing was performed for the tuned library using Nextera XT Index Kit v2 (Illumina, Inc.) and MiSeq Reagent Kit v3 (Illumina, Inc.). As for MinION sequencing, the library preparation was performed using Ligation Sequencing Kits 1D (Oxford Nanopore Technologies). Long-read sequencing was performed for the tuned library using MinION Flow cell R10.3 (Oxford Nanopore Technologies). The long reads obtained by MinION were assembled using Flye v2.8.1 and Canu v1.7.1, and the Miseq data were used to correct errors in the long-read assemblies using Pilon v1.22.
The complete genome sequencing data obtained by the above methods were checked for the presence of various acquired drug resistance genes, including the mcr gene and their localization with ResFinder 4.0 from the Center for Genomic Epidemiology website2. BLAST search, comparison to similar sequences using BLAST Ring Image Generator (BRIG), and mcr gene peripheral structure analysis by Easyfig were performed for the contig sequences coding the mcr gene.
For strains with a MIC of 4 μg/ml or higher in colistin susceptibility testing, the sequences obtained by WGS with a hybrid assembly were searched for staining of colistin chromosomal resistance mechanisms according to previous reports (Sato et al., 2018; Mmatli et al., 2020): pmrAB, phoPQ, crrB, and mgrB.
RT-qPCR of mcr-9 Gene and qseB/C Gene
In strains carrying mcr-9, mRNA were assayed for the expression of mcr-9 and qseB/C using the method of Kieffer et al. (2019). The bacterial strains were cultured in LB broth containing 0.25 μg/ml and 1 μg/ml of colistin for 4 or 24 h with shaking. RNA was extracted from the culture solution using an RNA Protect Bacteria Reagent (Qiagen) and RNeasy Mini Kit (Qiagen). A StepOnePlus real-time PCR system (ThermoFisher Scientific K.K., Tokyo, Japan) was used to perform RT-PCR, and PCR target genes were amplified using a Power SYBR Green RNA-to-Ct 1-Step Kit (ThermoFisher Scientific K.K). Each assay was performed in duplicate. Quantitative values were calculated by relative quantitative PCR using E. coli ATCC25922 as the reference for the Relative Quantification ratio. We used the GAPDH gene as an internal control (Carey et al., 2008).
Results
Susceptibility Testing of Colistin and WGS of Strains With MIC of 4 μg/ml or Higher
The results of colistin susceptibility testing using the agar dilution method for all target strains are shown in Figure 1. Distribution of the colistin MIC was unimodal, ranging from 0.25 to 4 μg/ml. Two strains, E. coli BPML00000000, positive for blaIMP–6 + blaCTX–M–2 + blaCTX–M–27, and E. coli BPMM00000000, positive for blaIMP–1, showed MICs above the CLSI breakpoint of 4 μg/ml. Table 2 shows the results of antimicrobial susceptibility testing and WGS of these two strains (GenBank accession numbers: BPML00000000: SAMD00334384 and BPMM00000000: SAMD00334385). The colistin MIC of these two strains were 0.5 μg/ml and 0.5 μg/ml by the microdilution method and 0.25 μg/ml and 0.25 μg/ml by E-test, respectively, and they were not carriers of the plasmid-associated mcr gene. They were not observed to have the previously reported chromosomal colistin resistance-associated mutations: amino acid substitutions L105P, I128N, and G144S in pmrA, amino acid substitutions C84Y, D149Y, L10P, G206D, and Δ27–45 in pmrB, and amino acid substitution R6H in phoQ. In addition, these species were E. coli, which also did not harbor crrB and mgrB.
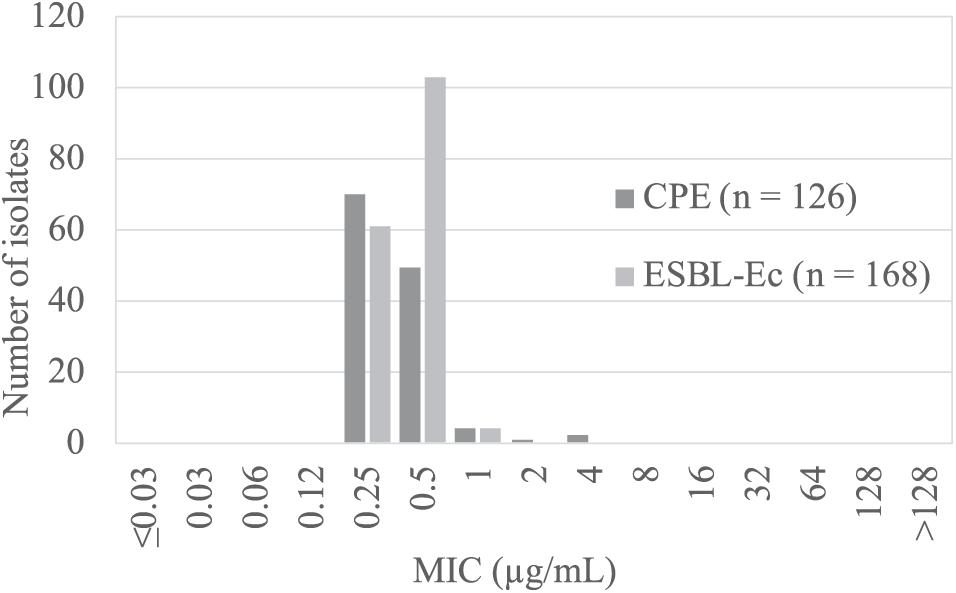
Figure 1. Results of colistin susceptibility testing in carbapenemase- producing Enterobacteriaceae (CPE, dark gray) and ESBL-producing Escherichia coli (ESBL-Ec, light gray).
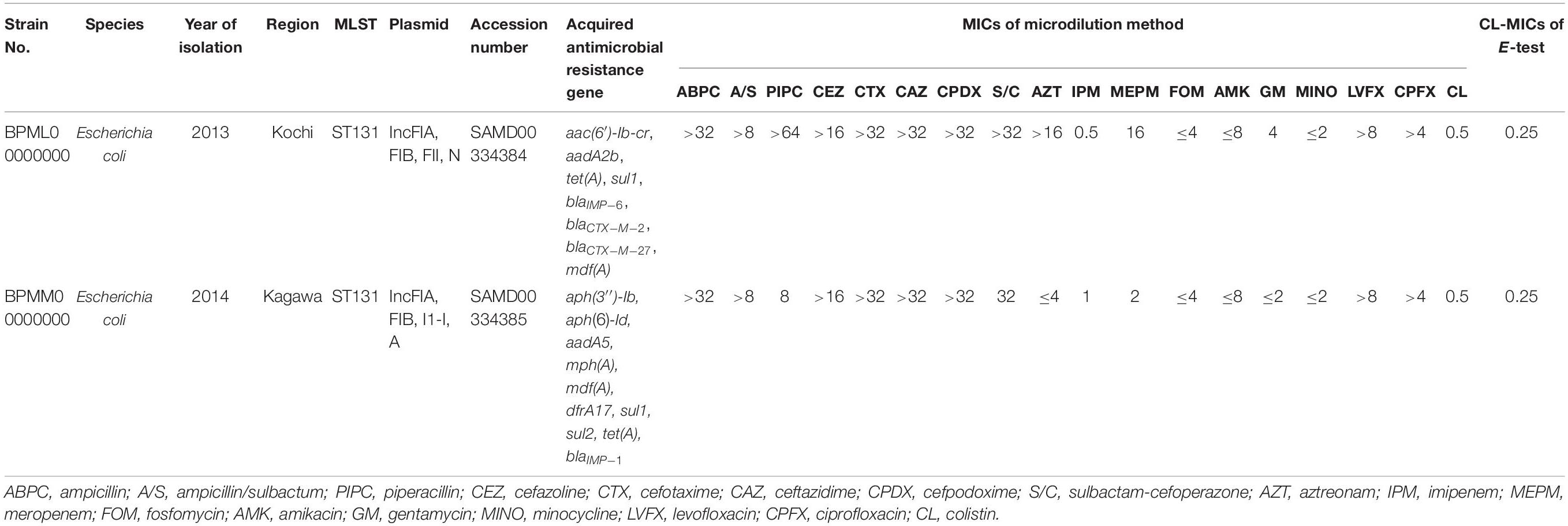
Table 2. Genetic characterization of two strains showing MIC of 4 μg/ml using agar dilution method and MICs using microdilution method and E-test.
Search for mcr-1 to mcr-9 in All Target Strains Using Multiplex PCR
Although all strains were negative for mcr-1 to mcr-8, three strains, K. pneumoniae THUN262 (blaIMP–6 + blaCTX–M–2-positive) and Enterobacter kobei THUN627 (blaGES–24-positive) from the CPE group and E. coli THUN648 (blaCTX–M–62-positive) from the ESBL group, were positive for mcr-9. The colistin MIC of these three strains using the agar dilution method was 0.5 μg/ml (microdilution method = 0.25 μg/ml, E-test = 0.125 μg/ml) for THUN262, 0.25 μg/ml (0.25 μg/ml, 0.125 μg/ml) for THUN627, and 0.5 μg/ml (0.5 μg/ml, 0.125 μg/ml) for THUN648, respectively.
S1-PFGE and Southern Blot Hybridization Targeting mcr-9 and Each bla Gene
Figure 2 shows the results of S1-PFGE and Southern blot hybridization for the three strains carrying mcr-9. The mcr-9 of K. pneumoniae THUN262 and E. kobei THUN627 was not present on the plasmid. However, mcr-9 of E. coli THUN648 was encoded on a 260-kbp plasmid harboring blaCTX–M–62.
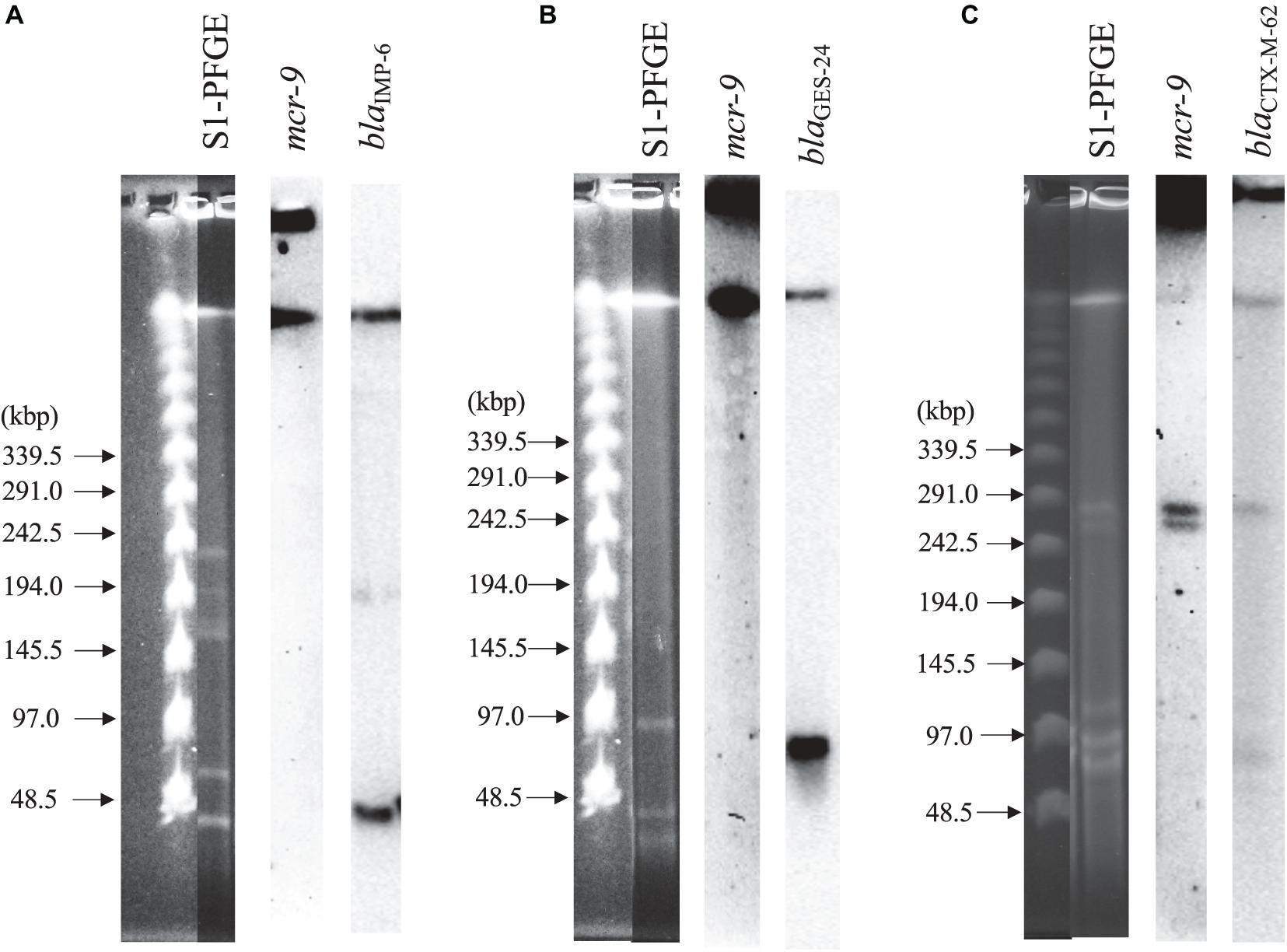
Figure 2. S1-nuclease pulsed-field gel electrophoresis and Southern blot hybridization using three isolates with mcr-9. (A) blaIMP–6 + blaCTX–M–2-positive K. pneumoniae THUN262, (B) blaGES–24-positive E. kobei THUN627, and (C) blaCTX–M–62-positive E. coli THUN648.
WGS and Bioinformatic Analysis of Strains Carrying the mcr-9 Gene
Table 3 shows the results of WGS and antimicrobial susceptibility testing of the three strains harboring mcr-9 and the characteristics of each isolate. The genomic sequences of these three strains showed that the mcr-9 gene was encoded on the chromosome of K. pneumoniae THUN262 (GenBank accession number BNSV01000001) and E. kobei THUN627 (GenBank accession number BNSW01000001) and on the plasmid of E. coli THUN648 (GenBank accession number BNSX01000002). E. coli THUN648 with mcr-9 that was encoded on a 275-kbp plasmid also hosted blaCTX–M–62. Figure 3 shows the results of a BLAST search using the contig sequence that codes mcr-9 for each bacterial strain and the results of a comparison with similar sequences carrying mcr-9 using BRIG. No K. pneumoniae chromosomal gene data encoding mcr-9 on the chromosome, such as that on K. pneumoniae THUN262, were found in the GenBank database. However, there were three sequences that were most similar to the E. kobei THUN627 chromosome sequence: E. hormaechei (accession number CP042571) isolated in Australia (75.1% of query coverage and 88.4% of identity), E. kobei (accession number CP017181) isolated in Japan (85.6% of query coverage and 99.1% of identity), and Enterobacter sp. (accession number CP048736) isolated in China (87.8% of query coverage and 99.2% of identity). In the BLAST search, several sequences similar to the plasmid of E. coli THUN648 encoding mcr-9 and blaCTX–M–62 were found. The most similar sequences were E. hormaechei_pMCR-SCNJ07(accession number MK933279) isolated in China (78.8% of query coverage and 99.9% of identity), K. quasipneumoniae subsp. quasipneumoniae_p17277A_477 (accession number CP043927) isolated in Argentina (53.2% of query coverage and 99.8% of identity), and E. cloacae subsp. cloacae_pNUH14_ECL028_1 (accession number AP019384) isolated in Japan (69.0% of query coverage and 99.7% of identity). No similar plasmid with mcr-9 was found in the species of E. coli in GenBank. The only virulence factor encoded in this plasmid of E. coli THUN648 was terC, a tellurium ion resistance protein.
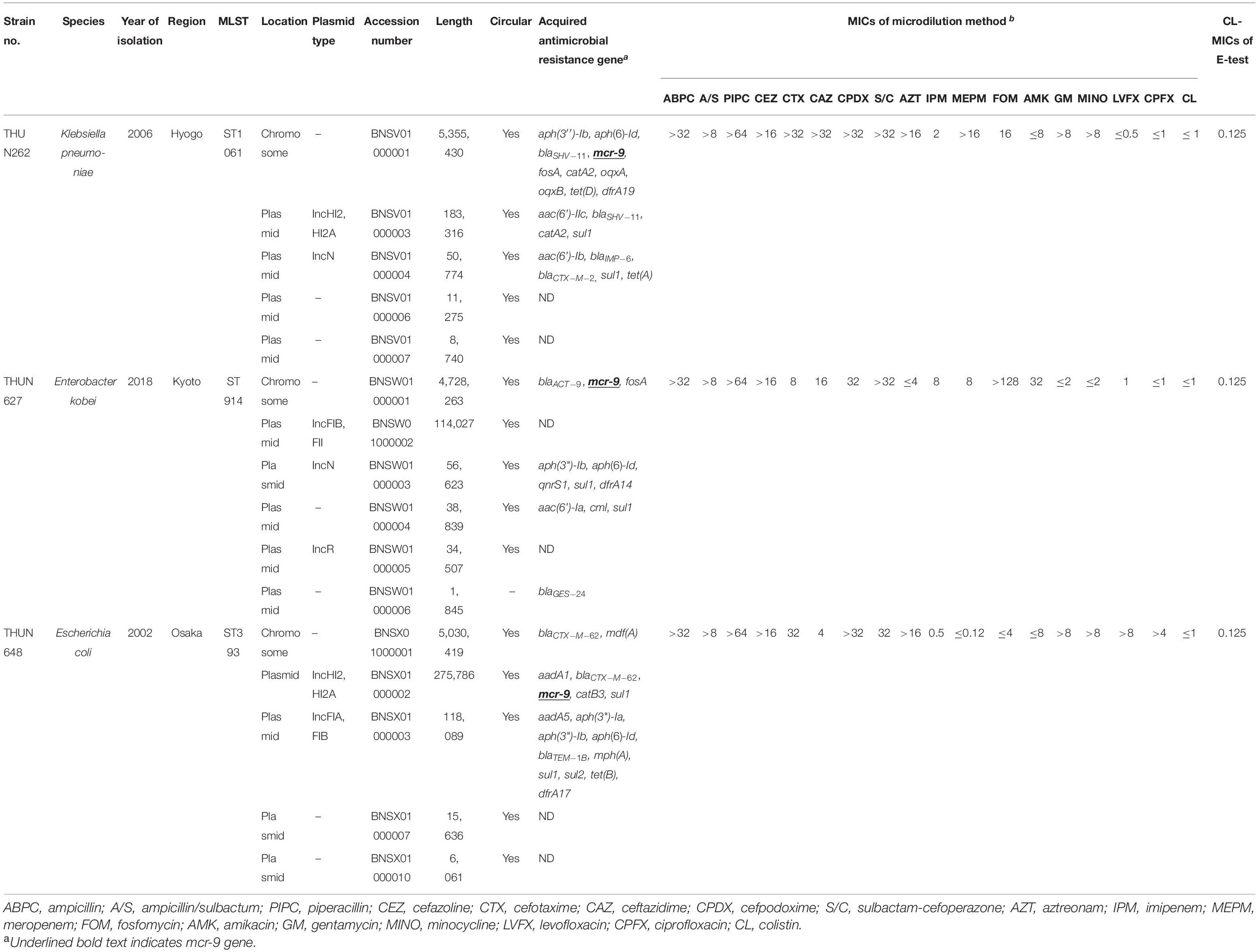
Table 3. Genetic characterization of three strains with mcr-9 and MICs using microdilution method and E-test.
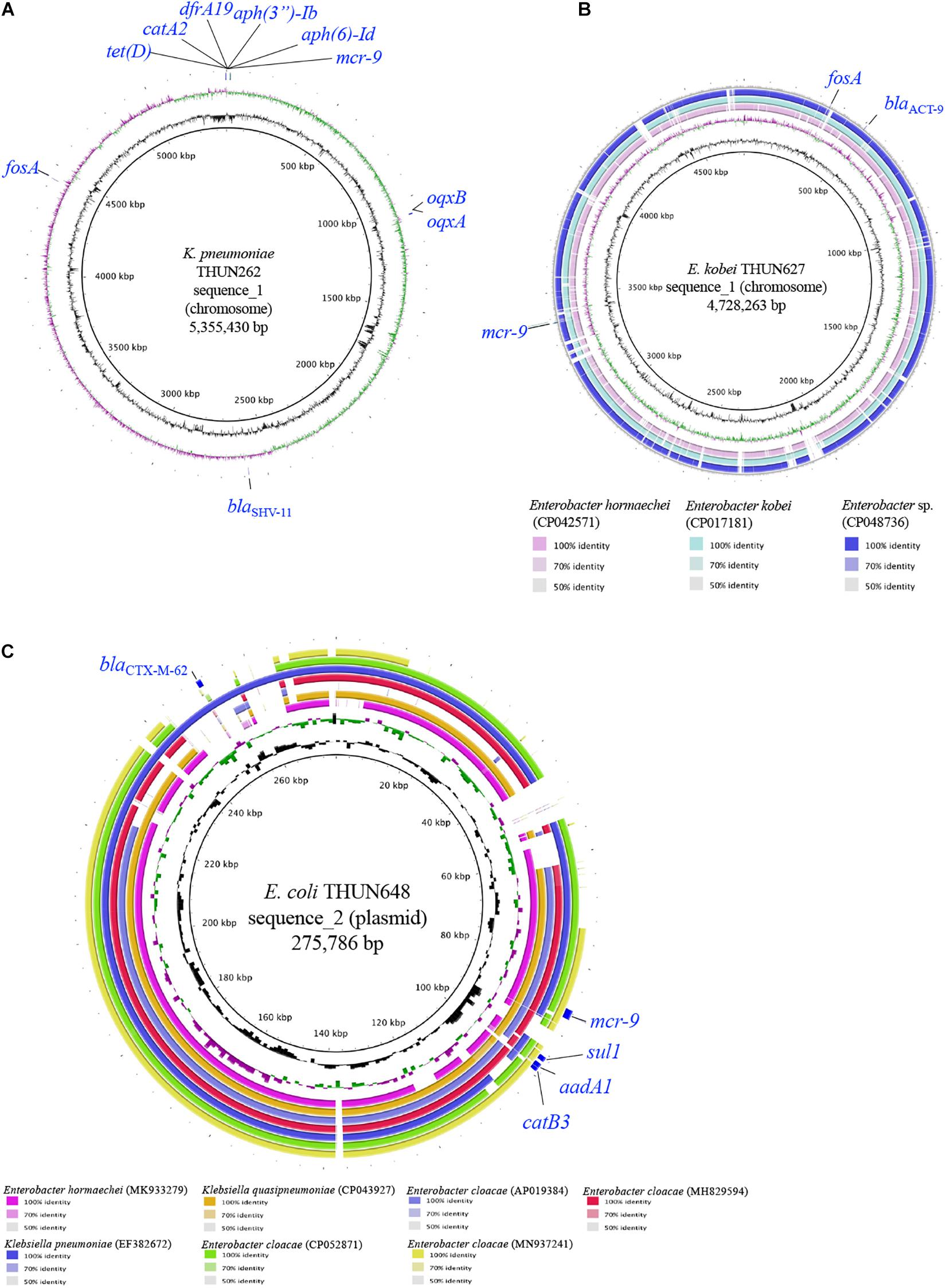
Figure 3. Circular structure of chromosome harboring mcr-9 of K. pneumoniae THUN262 (A), E. kobei THUN627 (B), and plasmid harboring mcr-9 of E. coli THUN648 (C). The green and purple circles indicate GC skew, and the black circles indicate GC content.
Figure 4 shows the results of a comparison of the peripheral structure of mcr-9 for these three strains using Easyfig. The qseB/C gene, which is suggested to be a regulator of mcr-9 expression (Kieffer et al., 2019), is present downstream of mcr-9 only in E. kobei THUN627, and the other two strains are not carriers of the qseB/C gene.
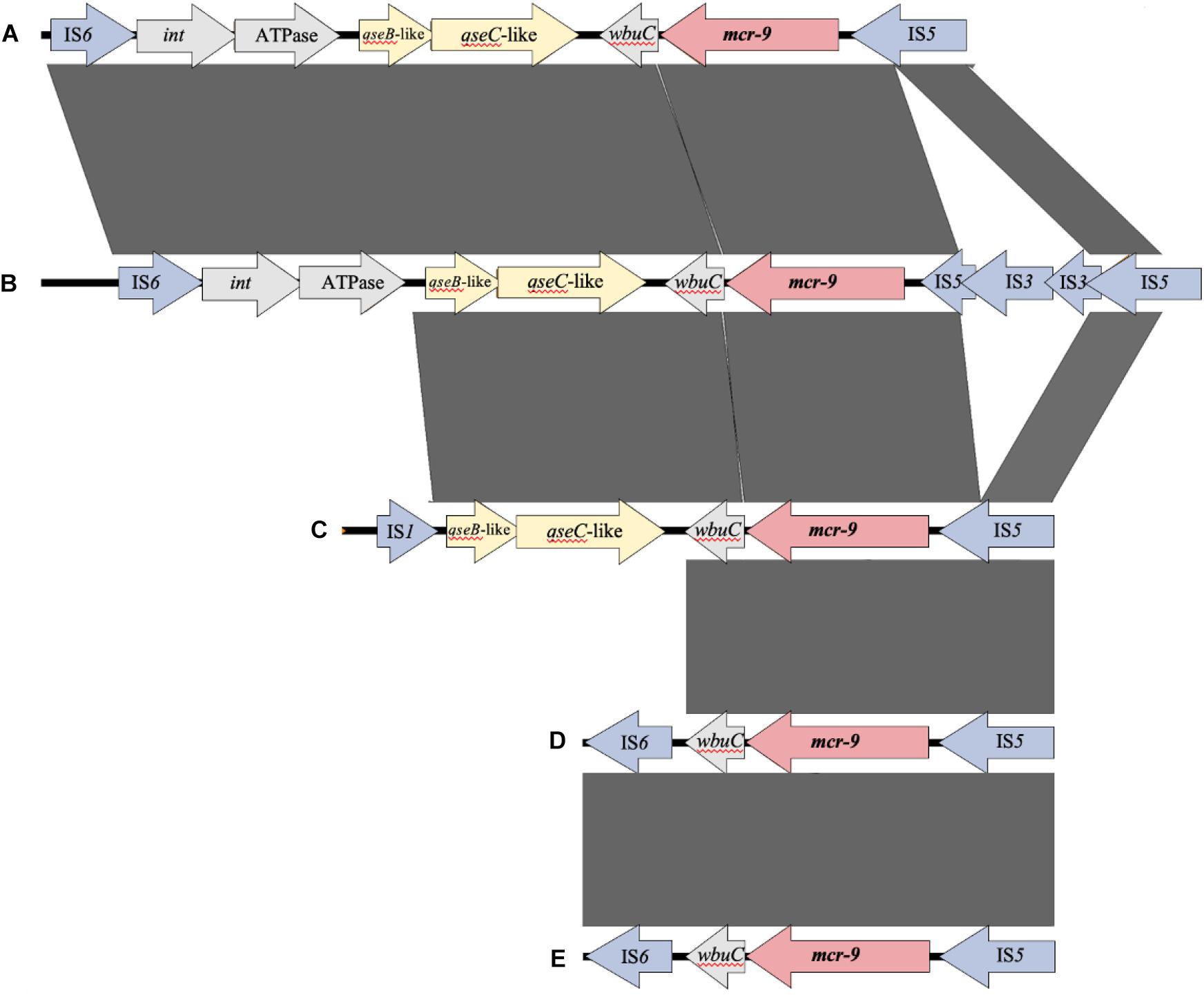
Figure 4. Comparison of the structure around the mcr-9 gene in each isolate. (A) Leclercia sp. (accession number CP031102), (B) Enterobacter kobei THUN627 (this study), (C) Salmonella enterica (accession number CP029037), (D) Enterobacter cloacae (accession number CP020529), and (E) Escherichia coli THUN648 (this study) and Klebsiella pneumoniae THUN262 (this study).
RT-qPCR of the mcr-9 and qseB/C Genes
Figure 5 shows the results of RT-qPCR for the three strains carrying mcr-9. The expression of mcr-9 in all three strains did not increase under colistin exposure or non-exposure and the 4- or 24-h incubation conditions. Also, E. kobei THUN627, which has qseB/C downstream, did not show an increase in the expression level of qseB/C.
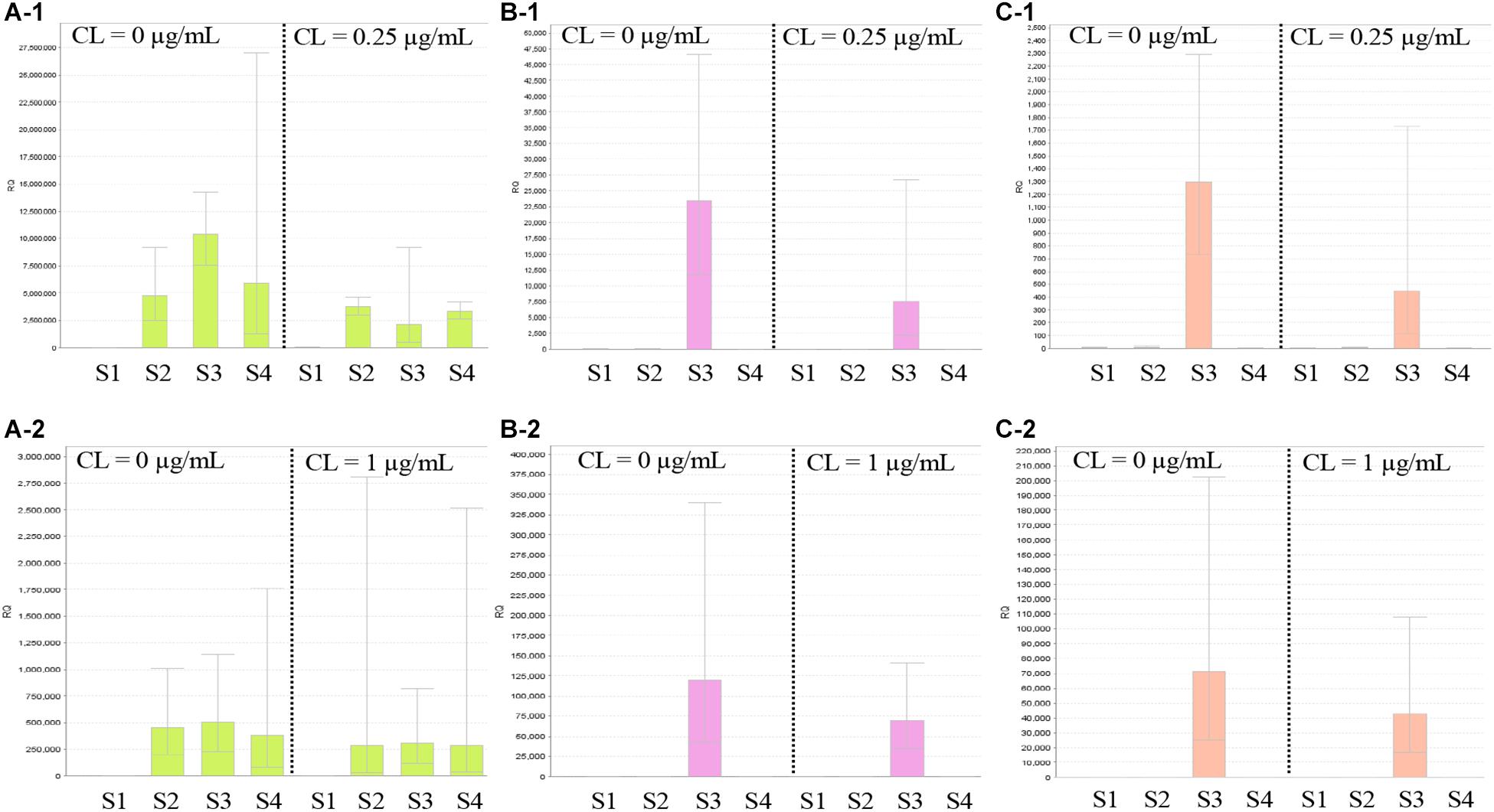
Figure 5. Results of RT-qPCR of mcr-9 and qseB/C genes using three isolates with mcr-9 and E. coli ATCC25922. S1, E. coli ATCC25922 without mcr-9 and qseB/C; S2, K. pneumoniae THUN262 with mcr-9; S3, E. kobei THUN627 with mcr-9 and qseB/C; and S4, E. coli THUN648 with mcr-9. (A-1) RT-qPCR of mcr-9 cultured for 4 h and (A-2) for 24 h. (B-1) RT-qPCR of qseB cultured for 4 h and (B-2) for 24 h. (C-1) RT-qPCR of qseC cultured for 4 h and (C-2) for 24 h. Each assay was performed in duplicate. Quantitative values were calculated by relative quantitative PCR using E. coli ATCC25922 as the reference for the Relative Quantification ratio.
Discussion
The purposes of this study were to elucidate the colistin resistance status of ESBL-Ec and CPE in Japan and to perform a comprehensive search for mcr-1 to mcr-9 to elucidate the genome structure that carries the mcr gene.
Two strains showed MICs of 4 μg/ml using agar plate dilution based on the CLSI method, and these were carbapenemase-producing strains that were isolated in 2013 and 2014. However, these strains showed susceptibility by the microdilution method and E-test. These strains also underwent WGS with hybrid assembly, but no known chromosomal or plasmid resistance mechanisms were detected. Recently, various chromosomal colistin resistance mechanisms have been reported, such as mutation or loss of pmrAB and phoQP, mutation of crrB and presence of mgrB (Mmatli et al., 2020). In the present study, the strains that showed a MIC of 4 μg/ml by agar dilution method did not possess any of these known chromosomal colistin resistance mechanisms and may have unknown mechanisms. However, as the microdilution method and E-test showed susceptibility, the results may be method dependent. In the future, the mechanisms in these strains will need to be investigated further. Besides, in the present study, three strains carrying mcr-9 were detected among the bacterial strains with a susceptibility to colistin (MIC of 1 μg/ml or less).
Umeda et al. (2021) isolated Enterobacter roggenkampii harboring both IncP6 plasmid coding blaIMP–1 and blaGES–5 and IncHI2 plasmid coding blaCTX–M–9 and mcr-9 in Osaka, Japan in 2019. These plasmids could be transferred to other strains by conjugation, but qseBC was not harbored downstream of mcr-9. Besides, Kananizadeh et al. (2020) detected three strains harboring mcr-9 among 32 E. cloacae complex strains showing a MIC ≥ 2 μg/ml collected from 14 hospitals in Japan in 2018. These simultaneously harbored blaIMP–1. In two E. xiangfangensis, mcr-9 was located on a 280/290-kbp plasmid, qseBC was not harbored downstream of mcr-9, and blaIMP–1 was located on another plasmid (62 kbp). The replicon type of mcr-9 harboring plasmid was located on IncHI2, as reported by Umeda et al. (2021). Besides, mcr-9 of one E. asburiae was located on the chromosome simultaneously encoded by blaIMP–1 and blaACT–9, and qseBC was encoded downstream of mcr-9. The mcr-9 detected in this study was coded on the chromosome of the two K. pneumoniae THUN262 and E. kobei THUN627 isolates and on a plasmid of size 276 kbp from E. coli THUN648. As mentioned above, the qseB/C gene currently recognized as a regulator gene is present only in E. kobei THUN627, downstream of mcr-9. Of the two strains with mcr-9 coded on the chromosome, as the chromosome sequence including mcr-9 similar to that of K. pneumoniae THUN262 was not detected in the GenBank database, this chromosome sequence of K. pneumoniae THUN262 would appear to be a novel sequence. However, a few chromosome sequences of E. kobei THUN627 that included mcr-9 were present, and strains isolated from various foreign countries besides Japan were also present. The plasmid sequence of E. coli THUN648, which had mcr-9 coded in the plasmid, was observed in multiple bacterial species such as K. quasipneumoniae and E. cloacae and was also detected in various foreign countries in addition to Japan. This plasmid of E. coli THUN648 also harbored blaCTX–M–62 of the ESBL-producing gene. Therefore, it was confirmed that mcr-9 has already spread worldwide through plasmids along with other antimicrobial resistance genes. In fact, Osei Sekyere et al. (2020) reported that plasmids containing mcr-9 harbored blaCTX–M–15 and blaTEM–1.
RT-qPCR for mcr-9 and qseB/C was performed on three strains carrying mcr-9 after exposure to colistin, followed by relative quantitative comparison. However, no increase in the mRNA expression level was observed in any of the three strains. Although E. kobei THUN627 carried the qseB/C gene downstream of mcr-9, which was suggested to be a regulator gene for mcr-9 in a previous report (Kieffer et al., 2019), this strain did not show an increase in mRNA expression level. Tyson et al. (2020) examined the colistin susceptibility results of more than 100 strains of S. enterica and E. coli carrying mcr-9 that were isolated in the U.S. All strains were reported to have a MIC of 1 μg/ml or less (Tyson et al., 2020), suggesting that qseB/C may not regulate the expression of mcr-9 by itself.
This study has three limitations. First, the ESBL-producing strains investigated for colistin resistance in this study are limited to E. coli. Other Enterobacteriaceae should be examined on a large scale to elucidate the full extent of the mcr gene. Second, the resistance mechanism of the two strains that showed a MIC of 4 μg/ml or higher in this study is still not clear. These strains will be investigated further by bioinformatics analysis using WGS. However, in this study, mcr-9 was found on a plasmid of E. coli isolated in 2002 and was found to be on the same plasmid together with the ESBL-producing gene. This suggested that mcr-9 may have already been spread worldwide together with other antimicrobial resistance genes. Third, screening with agar dilution with a breakpoint of ≥4 μg/ml may have decreased the overall diagnostic sensitivity of the study. In other words, strains with ≤ 2 μg/ml may harbor colistin resistance factors. However, the agar dilution method was reported by Turlej-Rogacka et al. (2018) to result in higher MICs than the broth dilution method. In addition, the colistin agar dilution method is acceptable in CLSI M100-S30. Therefore, we used the agar dilution method to minimize its limitation.
Conclusion
Strains carrying mcr-9 on both plasmid and chromosome existed in Japan prior to 2018, when the use of colistin was banned as a feed additive for food animals, and mcr-9 is already spreading around the world along with other antimicrobial resistance genes. However, the detailed mechanism by which mcr-9 is involved in colistin resistance is still not clear. We need to pay attention to future trends as mcr-9 will be highly expressed by various genetic mutations and insertions and has the potential to become a non-silent colistin resistance gene.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The studies involving human participants were reviewed and approved by Tenri Health Care University. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
AN and MK conceived and designed the experiments. AN, TN, MN, and TK performed the experiments. AN and MK drafted the manuscript. All authors provided critical input and approved the final manuscript.
Funding
This work was supported by JSPS KAKENHI (grant numbers 18K16189 and 21K08499) and internal funding.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to the members of the Association of Japan Community Healthcare Organization (JCHO) hospitals and the Study of Bacterial Resistance in the Kinki Region of Japan (SBRK) Medical Technologists for their help during the collection of the strains.
Footnotes
References
Amr Insights (2019). Argentina: no more Colistin in Veterinary Products. Available online at: https://www.amr-insights.eu/argentina-no-more-colistin-in-veterinary-products/ (accessed April 1, 2021).
Borowiak, M., Baumann, B., Fischer, J., Thomas, K., Deneke, C., Hammerl, J. A., et al. (2020). Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in colistin-resistant Salmonella enterica isolates from environment, feed, animals and food (2011-2018) in Germany. Front. Microbiol. 11:80. doi: 10.3389/fmicb.2020.00080
Bush, K., and Jacoby, G. A. (2010). Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 54, 969–976. doi: 10.1128/aac.01009-09
Carey, C. M., Kostrzynska, M., Ojha, S., and Thompson, S. (2008). The effect of probiotics and organic acids on Shiga-toxin 2 gene expression in enterohemorrhagic Escherichia coli O157:H7. J. Microbiol. Methods 73, 125–132. doi: 10.1016/j.mimet.2008.01.014
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype typhimurium isolate. MBio 10:e853–19.
Clinical and Laboratory Standards Institute. (2018). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard. CLIS Document M07-A11, 11th ed Edn, Vol. 32. Wayne, PA: Clinical and Laboratory Standards Institute.
Clinical and Laboratory Standards Institute. (2020). Performance Standards for Antimicrobial Susceptibility Testing. CLIS document M100, 30th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Cui, M., Zhang, J., Gu, Z., Li, R., Chan, E. W., Yan, M., et al. (2017). Prevalence and molecular characterization of mcr-1-positive Salmonella strains recovered from clinical specimens in China. Antimicrob. Agents Chemother. 61:e2471–16.
Farzana, R., Jones, L. S., Barratt, A., Rahman, M. A., Sands, K., Portal, E., et al. (2020). Emergence of mobile colistin resistance (mcr-8) in a highly successful Klebsiella pneumoniae sequence type 15 clone from clinical infections in Bangladesh. mSphere 5:e23–20.
Kananizadeh, P., Oshiro, S., Watanabe, S., Iwata, S., Kuwahara-Arai, K., Shimojima, M., et al. (2020). Emergence of carbapenem-resistant and colistin-susceptible Enterobacter cloacae complex co-harboring blaIMP-1 and mcr-9 in Japan. BMC Infect Dis. 20:282. doi: 10.1186/s12879-020-05021-7
Kawanishi, M., Abo, H., Ozawa, M., Uchiyama, M., Shirakawa, T., Suzuki, S., et al. (2017). Prevalence of colistin resistance gene mcr-1 and absence of mcr-2 in Escherichia coli isolated from healthy food-producing animals in Japan. Antimicrob. Agents Chemother. 61, e2057–16.
Kieffer, N., Royer, G., Decousser, J. W., Bourrel, A. S., Palmieri, M., Ortiz De La Rosa, J. M., et al. (2019). mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob. Agents Chemother. 63:e965–19.
Kuchibiro, T., Komatsu, M., Yamasaki, K., Nakamura, T., Nishio, H., Nishi, I., et al. (2018). Evaluation of the modified carbapenem inactivation method for the detection of carbapenemase-producing Enterobacteriaceae. J. Infect. Chemother. 24, 262–266.
La, M. V., Lee, B., Hong, B. Z. M., Yah, J. Y., Koo, S. H., Jiang, B., et al. (2019). Prevalence and antibiotic susceptibility of colistin-resistance gene (mcr-1) positive Enterobacteriaceae in stool specimens of patients attending a tertiary care hospital in Singapore. Int. J. Infect. Dis. 85, 124–126. doi: 10.1016/j.ijid.2019.05.029
Lescat, M., Poirel, L., and Nordmann, P. (2018). Rapid multiplex polymerase chain reaction for detection of mcr-1 to mcr-5 genes. Diagn. Microbiol. Infect. Dis. 92, 267–269. doi: 10.1016/j.diagmicrobio.2018.04.010
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168.
Market Research.Com (2019). Global Colistin Sulphate Market Research Report 2019. Available online at: https://www.marketresearch.com/QYResearch-Group-v3531/Global-Colis-tin-Sulphate-Research-12365052/ (accessed 1 April, 2021).
Mathers, A. J., Peirano, G., and Pitout, J. D. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28, 565–591. doi: 10.1128/cmr.00116-14
Mmatli, M., Mbelle, N. M., Maningi, N. E., and Osei Sekyere, J. (2020). Emerging transcriptional and genomic mechanisms mediating carbapenem and polymyxin resistance in Enterobacteriaceae: a systematic review of current reports. mSystems 5:e783–20.
Nakamura, A., Komatsu, M., Noguchi, N., Ohno, Y., Hashimoto, E., Matsutani, H., et al. (2016). Analysis of molecular epidemiologic characteristics of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli colonizing feces in hospital patients and community dwellers in a Japanese city. J. Infect. Chemother. 22, 102–107. doi: 10.1016/j.jiac.2015.11.001
Nakamura, T., Komatsu, M., Yamasaki, K., Fukuda, S., Miyamoto, Y., Higuchi, T., et al. (2012). Epidemiology of Escherichia coli, Klebsiella species, and Proteus mirabilis strains producing extended-spectrum beta-lactamases from clinical samples in the Kinki Region of Japan. Am. J. Clin. Pathol. 137, 620–626.
Nicolas-Chanoine, M. H., Bertrand, X., and Madec, J. Y. (2014). Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/cmr.00125-13
Nishio, H., Komatsu, M., Shibata, N., Shimakawa, K., Sueyoshi, N., Ura, T., et al. (2004). Metallo-beta-lactamase-producing gram-negative bacilli: laboratory-based surveillance in cooperation with 13 clinical laboratories in the Kinki region of Japan. J. Clin. Microbiol. 42, 5256–5263. doi: 10.1128/jcm.42.11.5256-5263.2004
Ohno, Y., Nakamura, A., Hashimoto, E., Matsutani, H., Abe, N., Fukuda, S., et al. (2017). Molecular epidemiology of carbapenemase-producing Enterobacteriaceae in a primary care hospital in Japan, 2010-2013. J. Infect. Chemother. 23, 224–229. doi: 10.1016/j.jiac.2016.12.013
Ohno, Y., Nakamura, A., Hashimoto, E., Noguchi, N., Matsumoto, G., Fukuda, S., et al. (2020). Fecal carriage and molecular epidemiologic characteristics of carbapenemase-producing Enterobacterales in primary care hospital in a Japanese city. J. Infect. Chemother. 26, 928–932. doi: 10.1016/j.jiac.2020.04.012
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Osei Sekyere, J., Maningi, N. E., Modipane, L., and Mbelle, N. M. (2020). Emergence of mcr-9.1 in extended-spectrum-β-lactamase-producing clinical Enterobacteriaceae in Pretoria, South Africa: global evolutionary phylogenomics, resistome, and mobilome. mSystems 5:e148–20.
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/cmr.00064-16
Sato, T., Shiraishi, T., Hiyama, Y., Honda, H., Shinagawa, M., Usui, M., et al. (2018). Contribution of novel amino acid alterations in PmrA or PmrB to colistin resistance in mcr-negative Escherichia coli clinical isolates, including major multidrug-resistant lineages O25b:H4-ST131-H30Rx and Non-x. Antimicrob. Agents Chemother. 62:e864–18.
Shen, Y., Zhou, H., Xu, J., Wang, Y., Zhang, Q., Walsh, T. R., et al. (2018). Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat. Microbiol. 3, 1054–1062. doi: 10.1038/s41564-018-0205-8
Shibasaki, M., Komatsu, M., Sueyoshi, N., Maeda, M., Uchida, T., Yonezawa, H., et al. (2016). Community spread of extended-spectrum beta-lactamase-producing bacteria detected in social insurance hospitals throughout Japan. J. Infect. Chemother. 22, 395–399. doi: 10.1016/j.jiac.2016.03.001
Tada, T., Uechi, K., Nakasone, I., Shimada, K., Nakamatsu, M., Kirikae, T., et al. (2017). Emergence of a colistin-resistant Escherichia coli clinical isolate harboring mcr-1 in Japan. Int. J. Infect. Dis. 63, 21–22. doi: 10.1016/j.ijid.2017.07.023
The Bureau of Investigative Journalism (2018). India Set to Ban use of “Last Hope” Antibiotic to Fatten Livestock After Bureau Story. Available online at: https://www.thebureauinvestigates.com/stories/2018-12-05/indian-government-colistin-ban-bureau-story (accessed April 1, 2021).
Turlej-Rogacka, A., Xavier, B. B., Janssens, L., Lammens, C., Zarkotou, O., Pournaras, S., et al. (2018). Evaluation of colistin stability in agar and comparison of four methods for MIC testing of colistin. Eur. J. Clin. Microbiol. Infect. Dis. 37, 345–353. doi: 10.1007/s10096-017-3140-3
Tyson, G. H., Li, C., Hsu, C. H., Ayers, S., Borenstein, S., Mukherjee, S., et al. (2020). The mcr-9 gene of Salmonella and E. coli is not associated with colistin resistance in the United States. Antimicrob. Agents Chemother. 64:e573–20.
Umeda, K., Nakamura, H., Fukuda, A., Matsumoto, Y., Motooka, D., Nakamura, S., et al. (2021). Genomic characterization of clinical Enterobacter roggenkampii co-harbouring blaIMP-1- and blaGES-5-encoding IncP6 and mcr-9-encoding IncHI2 plasmids isolated in Japan. J. Glob. Antimicrob. Resist. 24, 220–227. doi: 10.1016/j.jgar.2020.11.028
Weldhagen, G. F., and Prinsloo, A. (2004). Molecular detection of GES-2 extended spectrum Beta-lactamase producing Pseudomonas aeruginosa in Pretoria. South Africa. Int. J. Antimicrob. Agents 24, 35–38. doi: 10.1016/j.ijantimicag.2003.12.012
World Health Organization and Who Advisory Group on Integrated Surveillance of Antimicrobial Resistance (2017). Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance Due to Non-Human use, 5th Rev. Geneva: World Health Organization.
Yagi, T., Kurokawa, H., Senda, K., Ichiyama, S., Ito, H., Ohsuka, S., et al. (1997). Nosocomial spread of cephem-resistant Escherichia coli strains carrying multiple Toho-1-like β-lactamases genes. Antimicrob. Agents Chemother. 41, 2606–2611. doi: 10.1128/aac.41.12.2606
Keywords: multidrug-resistant Enterobacteriaceae, colistin, mcr-9, whole-genome sequence, antimicrobial resistance
Citation: Nakamura A, Nakamura T, Niki M, Kuchibiro T, Nishi I and Komatsu M (2021) Genomic Characterization of ESBL- and Carbapenemase-Positive Enterobacteriaceae Co-harboring mcr-9 in Japan. Front. Microbiol. 12:665432. doi: 10.3389/fmicb.2021.665432
Received: 08 February 2021; Accepted: 16 July 2021;
Published: 24 August 2021.
Edited by:
Axel Cloeckaert, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceReviewed by:
John Osei Sekyere, University of Pretoria, South AfricaMichael Brouwer, Wageningen University & Research, Netherlands
Remy A. Bonnin, Université Paris-Saclay, France
Antonio Battisti, Experimental Zooprophylactic Institute of the Lazio and Tuscany Regions (IZSLT), Italy
Séamus Fanning, University College Dublin, Ireland
Copyright © 2021 Nakamura, Nakamura, Niki, Kuchibiro, Nishi and Komatsu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akihiro Nakamura, YS5uYWthbXVyYUB0ZW5yaXlvcm96dS11LmFjLmpw
 Akihiro Nakamura
Akihiro Nakamura Tatsuya Nakamura2
Tatsuya Nakamura2 Isao Nishi
Isao Nishi