- 1Department of Infectious Diseases, The Affiliated Xi’an Central Hospital of Xi’an Jiaotong University, Xi’an, China
- 2Storr Liver Centre, The Westmead Institute for Medical Research, The University of Sydney and Westmead Hospital, Sydney, NSW, Australia
- 3School of Medical Science, The University of Sydney, Camperdown, NSW, Australia
- 4Marie Bashir Institute for Infectious Diseases and Biosecurity, University of Sydney, Sydney, NSW, Australia
- 5Centre for Infectious Diseases and Microbiology, Westmead Hospital, Sydney, NSW, Australia
Hepatocellular carcinoma (HCC) is the sixth most common cancer worldwide and the fourth leading cause of cancer-related death. The most common risk factor for developing HCC is chronic infection with hepatitis B virus (HBV). Early stages of HBV-related HCC (HBV-HCC) are generally asymptomatic. Moreover, while serum alpha-fetoprotein (AFP) and abdominal ultrasound are widely used to screen for HCC, they have poor sensitivity. Thus, HBV-HCC is frequently diagnosed at an advanced stage, in which there are limited treatment options and high mortality rates. Serum biomarkers with high sensitivity and specificity are crucial for earlier diagnosis of HCC and improving survival rates. As viral–host interactions are key determinants of pathogenesis, viral biomarkers may add greater diagnostic power for HCC than host biomarkers alone. In this review, we summarize recent research on using virus-derived biomarkers for predicting HCC occurrence and recurrence; including circulating viral DNA, RNA transcripts, and viral proteins. Combining these viral biomarkers with AFP and abdominal ultrasound could improve sensitivity and specificity of early diagnosis, increasing the survival of patients with HBV-HCC. In the future, as the mechanisms that drive HBV-HCC to become clearer, new biomarkers may be identified which can further improve early diagnosis of HBV-HCC.
Introduction
Chronic infection with the Hepatitis B virus (HBV) is the predominant risk factor for primary liver cancer, specifically hepatocellular carcinoma (HCC; Bosch et al., 2004; Kew, 2010; Ozakyol, 2017). Overall, the lifetime incidence of HCC in HBV has been reported to be approximately 10–25% (McGlynn et al., 2015). Moreover, most cases of HBV-associated HCC occur in cirrhotic liver disease, present in 70–90% of cases (Yang et al., 2011). Liver cancer is the fourth most deadly cancer (Bray et al., 2018), with a median survival time as short as 11 months (Greten et al., 2005; Yang and Roberts, 2010). There is also a broad range of indirect health impacts driven by chronic HBV, including anxiety about disease progression, stigma and discrimination, and health care costs associated with treatment (Tu et al., 2020a).
Chronic HBV infection leads to a repeated cycle of liver damage and regeneration, which promotes tumorigenesis (Wang et al., 2006). Treatment of the underlying HBV infection can reduce, but not eliminate HCC risk (Papatheodoridis et al., 2015). Currently, oral nucleo(s/t)ide analogs (NAs) are used as first-line therapy for HBV infection. NA therapy targets the reverse transcriptase of HBV and suppresses HBV DNA replication, reduces progression to end-stage liver disease and improves long-term patient survival (Bitton Alaluf and Shlomai, 2016). NAs suppress viral replication (Ghany and Liang, 2007) but do not target HBV covalently closed circular DNA (cccDNA; Revill et al., 2020; Tu et al., 2020b). cccDNA is the template for HBV replication and expression of viral proteins, so its persistence plays a crucial role in chronic infection, inflammation, and cancer formation.
Current Clinical Detection of HCC
Early screening of HBV-infected patients for HCC reduces mortality (Zhang B.H. et al., 2004). Current AASLD guidelines advise abdominal ultrasound surveillance for HBV-infected patients with advanced fibrosis or cirrhosis at 6-month intervals (Marrero et al., 2018), as marked liver fibrosis is a strong risk factor for HCC. However, ultrasound can miss HCC at early stages [sensitivity 63% (Singal et al., 2009)] and is strongly affected by operator- and patient-dependent factors (Singal et al., 2009; Pocha et al., 2013). Moreover, HCC can occur at any stage of liver fibrosis; hence AASLD guidelines recommend HCC surveillance of people with HBV who are ≥40 (for males) or ≥50 (for females) years old, regardless of fibrosis levels (particularly in those of Asian descent) (Xu et al., 2017). A cohort of studies suggested that liver stiffness measurement using FibroScan can predict HCC development in HBV patients with cirrhosis (Jung et al., 2011; Pesce et al., 2012; Adler et al., 2016) but fails to predict HCC in non-cirrhotic chronic hepatitis B (CHB) patients with liver stiffness measurement <8.0 kPa as well as patients with body mass index >28 kg/m2 and waist circumference ≥102 cm (Foucher et al., 2006; Jung et al., 2011; Cassinotto et al., 2016).
Alpha-fetoprotein (AFP) is the most widely used serum biomarker for the diagnosis of HCC (Marrero et al., 2018). However, elevated serum AFP is only found in 60–70% of HCC patients (Luo et al., 2019). Lectin-reactive AFP (AFP-L3) and des-gamma-carboxy prothrombin (DCP) have also been proven to be useful biomarkers for HCC (Li et al., 2001; Volk et al., 2007) and increase the sensitivity compared to using AFP alone (Marrero et al., 2009; Wang et al., 2020). Unfortunately, considering the low sensitivity (55%) of AFP-L3, HCC detection (particularly in early stages) is still suboptimal (Choi et al., 2019). DCP has a poorer diagnostic power for small HCC compared to AFP, but is better at detecting intermediate and advanced HCC (Nakamura et al., 2006).
Therefore, more sensitive, non-invasive biomarkers for better HCC diagnosis are needed. Here, we review the current knowledge on circulating viral biomarkers to screen for HCC, which may improve detection rates in combination with existing host-derived markers.
HBV Structure, Natural History, and Replication Cycle
Hepatitis B virus is the prototypic member of the Hepadnaviridae family. The HBV virion contains a ∼3.2 kbp double-stranded DNA genome contained in a nucleocapsid composed of hepatitis B core antigen (HBcAg) subunits. The majority (∼90%) of virions contain a relaxed-circular DNA (rcDNA) genome, while a minority contain a double-stranded linear (dslDNA) form of the viral genome (Venkatakrishnan and Zlotnick, 2016). This nucleocapsid is enveloped in a host-derived lipid bilayer studded with hepatitis B surface antigens (HBsAg).
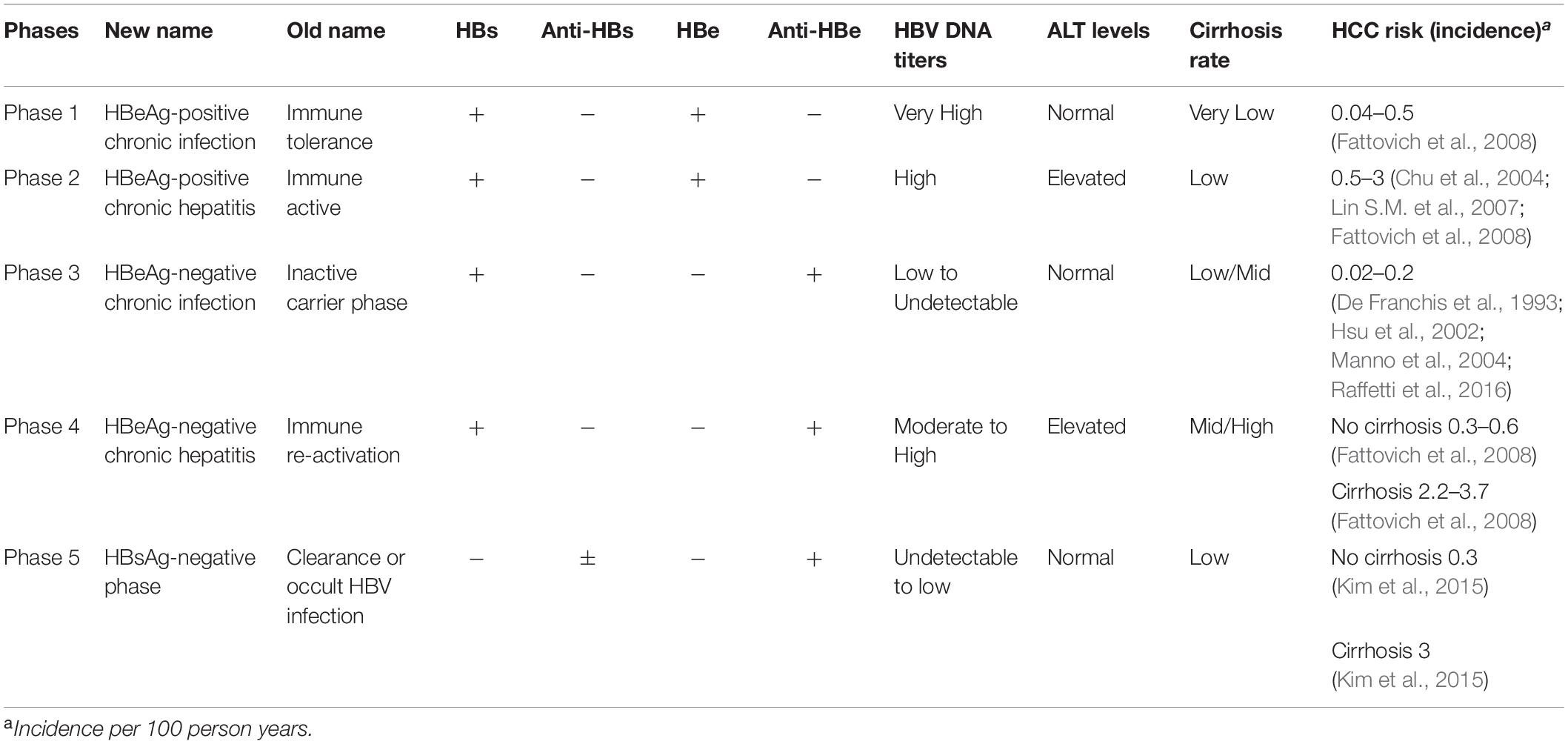
Infection with HBV itself is not cytopathic and the initial infection is usually asymptomatic, despite the production of high levels of virus antigen and viral particles by the liver. After decades of infection, HBV can trigger the immune response, though this is generally insufficient to clear all HBV-infected liver cells and subsequently causes chronic inflammation and liver damage progression. These two phases can be broadly divided serologically by the presence of circulating HBV e antigen (HBeAg, marking a status prior to extensive immune recognition) or antibodies against HBeAg (anti-HBe, present after antiviral clearance of the majority of infected cells). According to EASL 2017 Clinical Practice Guidelines (European Association for the Study of the Liver, 2017), chronic HBV infection can be separated into five clinical phases (Table 1): HBeAg-positive chronic HBV infection, previously termed “immune tolerant”; HBeAg-positive CHB with serum HBeAg positive, high HBV DNA and elevated ALT, termed “immune clearance phase”; HBeAg-negative chronic HBV infection, formerly known as the “inactive carrier” state; HBeAg-negative CHB with positive anti-HBe, persistent or fluctuating levels of HBV DNA and elevated ALT; HBsAg-negative phase, termed “occult HBV infection.”
On a cellular level, the infection of hepatocytes begins with attachment of the virion to the sodium taurocholate co-transporting polypeptide (NTCP), the entry receptor of HBV (Yan et al., 2012, 2014; Ni et al., 2014; Figure 1). After binding and receptor-mediated endocytosis, viral nucleocapsids are transported through the cytoplasm (Yan et al., 2014) to the nuclear membrane, where uncoating and entry of the HBV DNA genome into the nucleus occurs.
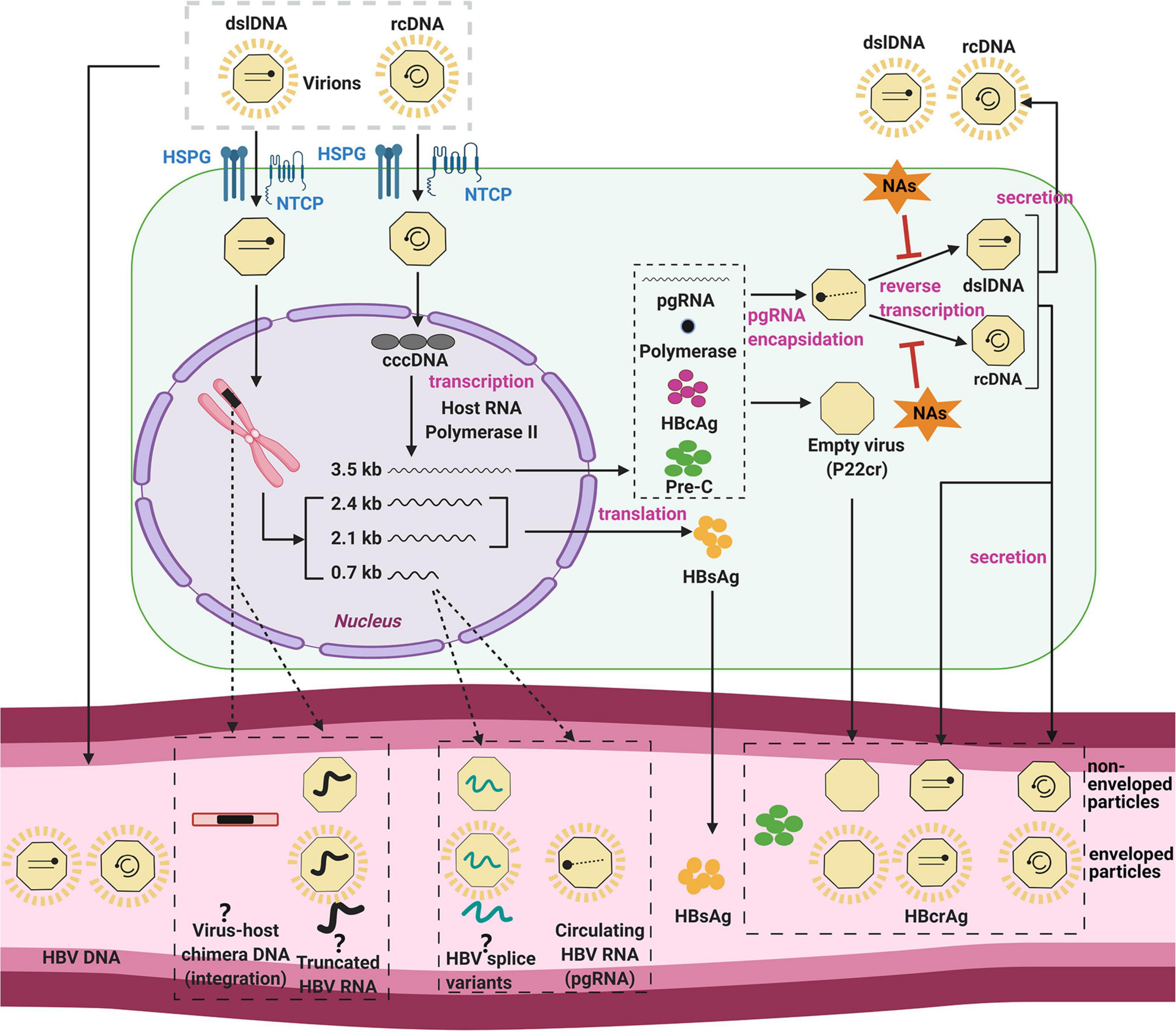
Figure 1. The HBV replication cycle and its secreted products. The HBV virion enters the hepatocyte by NTCP receptor binding, and uncoats prior to entry into the cytoplasm. The viral nucleocapsid is then transported to the nucleus, where it deposits its DNA genome. HBV relaxed-circular DNA (rcDNA) genomes can be repaired and ligated to form cccDNA, the template for all viral RNAs. HBV core antigen (HBcAg) is translated and forms capsids, some of which form around the pregenomic RNA (pgRNA) and viral polymerase. The pgRNA is reverse-transcribed to form either double stranded linear DNA (dslDNA) or rcDNA forms of the virus genome. The mature nucleocapsid is then enveloped by host membranes studded with HBV surface antigen (HBsAg) and secreted at multi-vesicular bodies. Cytoplasmic HBV capsids are recycled at a poor efficiency to the nucleus and do not appear to significantly add to the cccDNA pool (Tu and Urban, 2018; Revill et al., 2020; Tu et al., 2021). In a secondary pathway, HBV dslDNA can integrate into the host genome at host DNA breaks or form defective cccDNA (not shown). Some of these viral components are released in the serum (bottom) by as yet unclear mechanisms (dashed arrows) including within apoptotic bodies of dying hepatocytes, secretion through alternate pathways, or within exosomes. Even the form in which some of these biomarkers exist in the serum is still unknown and controversial (question marks). Figure was generated using Biorender (https://biorender.com/).
Nuclear HBV rcDNA is converted into the HBV cccDNA form using host cell DNA repair enzymes (Königer et al., 2014; Qi et al., 2016). HBV cccDNA is an episomal “mini-chromosome” and acts as a stable template for the 5 viral mRNAs. Each of these transcripts has different 5′ transcription start sites but a common 3′ polyadenylation signal. These mRNAs include the 3.5-kb pregenomic RNA (pgRNA), the 3.5-kb precore RNA, the 2.4-kb/2.1-kb surface mRNAs, and the 0.7-kb X mRNA (Blondot et al., 2016). Moreover, post-transcriptional modification of 3.5-kb species can produce spliced HBV RNA variants (Candotti and Allain, 2017).
The functions of pgRNA are both as the template for reverse transcription and the translation of viral polymerase and core protein. The newly translated viral polymerase binds to the 5′-epsilon region of pgRNA, and is packaged together as sub-viral core particles (Jones and Hu, 2013). Reverse transcription occurs within the HBV nucleocapsid through a series of complicated mechanisms, resulting in the synthesis of rcDNA (major pathway) or dslDNA. These mature nucleocapsids are then enveloped by HBsAg and secreted into the blood at multi-vesicular bodies (Blondot et al., 2016).
Nuclear dslDNA genomes follow separate pathways: these can form replication-defective cccDNA (Yang and Summers, 1998) or integrate into the host cell genome (Yang and Summers, 1999; Tu et al., 2017). While the integrated HBV genome is replication-deficient, but still acts as a template viral antigen expression (e.g. HBsAg and HBx) (Wooddell et al., 2017).
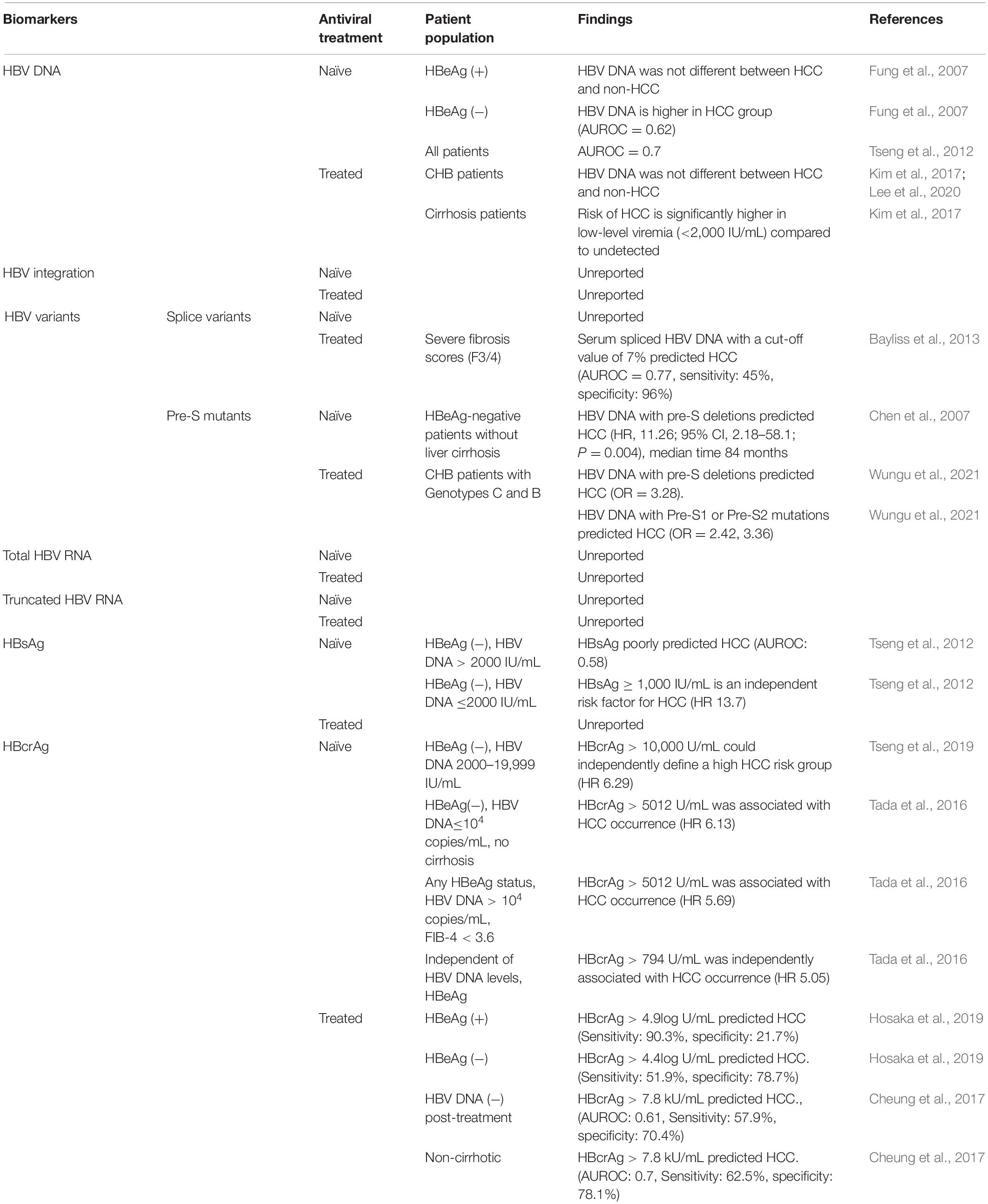
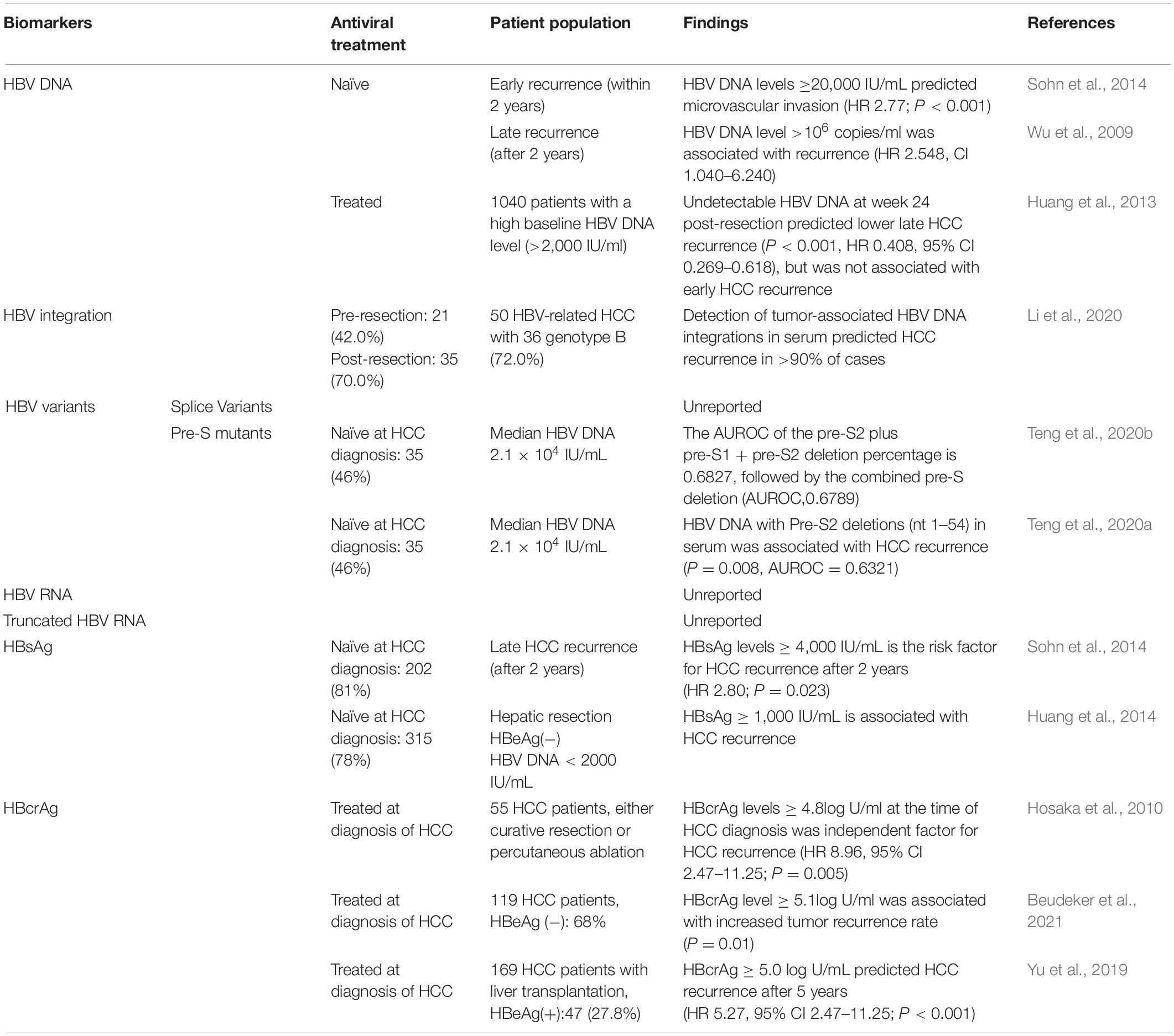
A broad range of components generated by virus-infected cells have been investigated as potential biomarkers for predicting HCC occurrence (summarized in Table 2) and recurrence (summarized in Table 3). For each major serum viral marker that has been investigated, we provide in the next section a description, the mode of quantification, their molecular association with HCC, and their predictive power for HCC occurrence and recurrence.
Serum Viral Biomarkers for HBV Related HCC
HBV DNA
Description
Hepatitis B virus DNA, the genomic nucleic acid of the virus, reflects active viral replication and secretion. There are two forms of HBV genome: rcDNA and dslDNA (as mentioned in the HBV replication cycle) (Lee et al., 2004; Blondot et al., 2016). Quantitative PCR for serum HBV DNA detects both forms of the virus genome and is used as a clinical marker to measure the efficacy of antiviral therapy in people with CHB.
Quantification
Using real-time PCR quantification assays for HBV DNA detection is strongly recommended by EASL (European Association for the Study of the Liver, 2017) and is generally expressed as a WHO-standardized IU/mL (5.26 copies/mL = 1 IU/mL) (Saldanha et al., 2001). At present, with their high sensitivity, specificity, accuracy and broad dynamic range, these techniques are the most widely used assays in clinical practice. The assays include Cobas AmpliPrep/Cobas TaqMan HBV version 2.0 (CAP/CTM HBV 2.0) (Roche Molecular Systems, Pleasanton, CA, United States), with a dynamic range between 105 copies/mL to 9 × 108 copies/mL, and Abbott RealTime HBV assay (Abbott Molecular, Des Plaines, IL, United States), with a dynamic range 50 copies/mL to 5 × 109 copies/mL (Chevaliez et al., 2008, 2010; Yeh et al., 2014). More sensitive pre-clinical tests have also been developed: digital droplet PCR can quantify HBV DNA down to 8 copies/mL (Liu et al., 2017).
The amount of the dslDNA form of HBV (as opposed to rcDNA form) can be measured using quantitative real-time PCR (qPCR) coupled with peptide nucleic acid-mediated clamping (Zhao X.L. et al., 2016). However, this assay is not a standard laboratory test.
Molecular Association With HCC
Serum HBV DNA load in people with CHB has been shown to be closely related to disease activity and progression (Iloeje et al., 2006). Moreover, elevated HBV DNA is considered as a predictive biomarker for HCC, independent of HBeAg and liver cirrhosis (Chen et al., 2006, 2011). HBV DNA is associated with both indirect and direct mechanisms of carcinogenesis. The indirect mechanisms include inducing new HBV infection of hepatocytes, which triggers ongoing liver immune attack, inflammation, and liver injury (Bolukbas et al., 2005; Duygu et al., 2012; Chen and Tian, 2019). Possible mechanisms of direct carcinogenesis include HBV dslDNA integration into the host genome, which reportedly leads to genomic instability, insertional mutagenesis and expression of pro-oncogenic viral proteins (Sze et al., 2013; Zhao L.H. et al., 2016; Gao et al., 2019). Indeed, one study reported that the levels of dslDNA increased to 14% of total serum HBV DNA in people with liver cirrhosis and 20% in those with HCC, compared to 7% in people with CHB alone (Zhao X.L. et al., 2016). However, the utility of dslDNA proportion as a biomarker for HCC has not been examined in clinical trials.
Performance as a Predictor of HCC
HCC occurrence
In NA-naïve patients, two studies in Taiwan have inferred that elevated serum HBV DNA level can be a useful biomarker for monitoring HCC independent of HBeAg and liver cirrhosis (Chen et al., 2006, 2011). A study by Chen et al. (2011) showed that in a cohort of patients with genotype B/C HBV infection aged >30 years, the risk of HCC increased with higher levels of circulating HBV DNA (after excluding patients in immune tolerance phase with HBV DNA >107 copies/mL, as people in this phase have low risk of HCC). In a case-control study of HBeAg-negative CHB patients, levels of HBV DNA were found to be higher in people with HCC than those without (Area Under the Receiver Operating Characteristic curve, AUROC = 0.62) (Fung et al., 2007). Tseng et al. (2012) reported that in a cohort of 2688 treatment-naïve people with CHB, HBV DNA predicts the risk of HCC regardless of HBeAg status [AUROC = 0.7(95% confidence interval (CI): 0.65–0.75)]. Together, this suggests HBV DNA has good predictive strength for HCC risk.
However, HBV DNA titers cannot be used for all patients. NA therapy can reduce levels of serum HBV DNA to an undetectable level, preventing its use as a biomarker in this population that is still susceptible to HCC (Vlachogiannakos and Papatheodoridis, 2013; Varbobitis and Papatheodoridis, 2016). Further, in a Korean cohort of 1,246 patients with CHB who received entecavir, baseline HBV DNA did not predict HCC in non-cirrhotic patients under NA treatment (>5.7 vs. <5.7log IU/mL; P = 0.166) (Kang et al., 2017).
Nevertheless, HBV DNA levels can be used to detect poor response to NAs, which is linked to HCC. In a cohort of 875 patients with CHB treated with entecavir, greater HBV DNA levels were linked to increased HCC risk in patients with cirrhosis (adjusted hazard ratio = 2.20, compared to those with persistently undetectable HBV DNA) (Kim et al., 2017). But, HBV DNA did not predict HCC risk in patients without cirrhosis. HCC incidence was not significantly different between people with persistently detectable HBV DNA and those with undetectable levels (13.3% vs. 8.3%, P = 0.821) (Lee et al., 2020). Thus, in patients treated with NAs, HBV DNA titer is useful in predicting HCC only in cirrhotic patients.
HCC recurrence
In NA-naïve HCC patients, high serum HBV DNA levels were an independent risk factor for HCC recurrence after curative resection or liver transplantation, or percutaneous radiofrequency ablation (Huang et al., 2008; Chuma et al., 2009; Goto et al., 2011; Li et al., 2011). In a study of 248 Korean patients who underwent curative resection for early stage HBV-related HCC, HBV DNA level ≥20,000 IU/mL [hazard ratio (HR) 2.77; P < 0.001] was a risk factor for microvascular invasion and early recurrence (within 2 years) (Sohn et al., 2014). However, Wu et al. (2009) found that HBV DNA level >106 copies/mL (HR 2.548, CI 1.040–6.240) in Taiwan patients with HBV-related HCC was associated with late recurrence (after 2 years). Therefore, the utility of high HBV DNA in predicting HCC recurrence needs further research.
After NA treatment at diagnosis of HCC or follow-up, sustained HBV DNA expression could increase the risk of HCC recurrence (Kim et al., 2008). Moreover, Huang et al. (2013) found that in the 865 HCC patients receiving NAs therapy with a high baseline HBV DNA level (subpopulation of a 1,040 patient cohort), an undetectable HBV DNA before postoperative week 24 (P < 0.001, HR 0.408, 95% CI 0.269–0.618) was a protective factor for late HCC recurrence, but not for early tumor recurrence (P = 0.541, HR 0.946, 95% CI 0.793–1.130). Therefore, detectable HBV DNA level could predict HCC recurrence in patients receiving NA treatment.
HBV Integration
Description
Integration of the dslDNA form of HBV DNA can occur throughout the host genome at double-strand DNA breaks (Bill and Summers, 2004), likely without the help of specific viral proteins (Tu et al., 2019) (instead probably using host DNA repair enzymes). The sites of HBV DNA integration during CHB are randomly distributed across the host genome without strong preference for any specific structural genome features (Budzinska et al., 2018a).
Quantification
Hepatitis B virus integrations can be detected in the serum and tissue of HBV-infected patients as virus-host chimera DNA. Current detection methods for virus-host chimera DNA include whole-exome sequencing, whole-genome sequencing, Alu PCR and inverse-nested PCR (Budzinska et al., 2018b). These have shown less-than-genome length fragments of HBV dslDNA integrate (with terminal truncations of 100s to 1,000s of base pairs being common). Of these detection assays, the only method enabling absolute quantification of HBV integrations is inverse-nested PCR (Mason et al., 2009), though this method is very time-consuming and technically challenging, limiting its clinical utility.
Molecular Association With HCC
While HBV integration sites are randomly distributed across the genome in non-tumor tissue, HBV DNA integrations in HCCs have been reported to be enriched in genes involved in carcinogenesis pathways (i.e., CTNNA2, EGFR, and TERT) and have been found to be preferentially located within CpG islands and close to telomeres (Sung et al., 2012; Zhao L.H. et al., 2016; Li et al., 2019). Even when the HBV infection is cleared (marked by HBsAg seroconversion), HCCs risk remains and 70% of HCCs contain HBV integrations (Wong et al., 2020).
The mechanism behind the association of HCC with HBV integration is currently unknown. Many studies indicate that HBV integration causes genetic damage and chromosomal instability, which has the potential to promote carcinogenic transformation (Scotto et al., 1983; Zhao L.H. et al., 2016; Chen et al., 2019; Jang et al., 2020), or drive downstream host protein expression. HBV DNA can integrate into fragile sites, CpG islands and near telomerase reverse transcriptase, lysine methyltransferase 2B, as well as cyclin A2 (Wong et al., 2020), potentially inducing cancer-initiating genomic instability (Zhao L.H. et al., 2016; Furuta et al., 2018; Wong et al., 2020). However, genomic instability is not evident in many cases of HBV-HCC (Sung et al., 2012). The integrated HBV DNA can also disrupt cellular genes by insertional mutagenesis or drive expression of nearby with viral promotors. Insertion in TERT promoter, CCNE1 (cyclin E1), CCNA2, MLL4 (Myeloid/lymphoid or mixed-lineage leukemia 4), TP53, and CTNNB1 have been repeatedly detected in HCC (Paterlini-Brechot et al., 2003; Sung et al., 2012; Kawai-Kitahata et al., 2016), but these are not present in all tumors.
In addition, mutant HBsAg produced from integrated HBV DNA could contribute to HBV-related HCC by causing endoplasmic reticulum (ER) stress and immune evasion (Hsieh et al., 2004; Wang et al., 2006).
Performance as a Predictor of HCC
HCC occurrence
Specific HBV-host fusion genes created by HBV integrations have been suggested as biomarkers for predicting HCC in people with CHB. In NA-treated patients, a prospective study using liver tissue from people with CHB reported that human ESPL1-HBV S fusion gene was detected in 8 of 12 (66.7%) people with HCC, compared to 0 of 11 (0%) CHB patients without HCC (Hu et al., 2020). Moreover, HBV has been reported to integrate into long interspersed nuclear elements (LINEs), leading to fusion HBx-LINE1 transcripts. HBx-LINE1 can activate β-catenin signaling, reduce E-cadherin and enhance cell migration, which has been suggested to promote HCC progression (Liang et al., 2016). These studies suggest that specific fusion genes could be used as a biomarker for the early detection of HCC in people with CHB, but these have not been able to be repeated independently in other cohorts [for example, in a cohort of Vietnamese patients with HBV-associated HCC (Trung et al., 2019)]. Indeed, the majority of integration sites in tumor samples are randomly distributed across the host genome (Zhao L.H. et al., 2016).
HCC recurrence
Several studies have found that circulating viral-host chimeric DNA (vh-DNA) generated from HBV integration may be a useful biomarker for monitoring HCC recurrence (Wang et al., 2019; Li et al., 2020). A study of 20 people with HBV-HCC found circulating vh-DNA representing 87 different HBV integration sites, which were enriched in genes involved in cancer-related pathways, suggesting they could act as a biomarker for HCC diagnosis (Li et al., 2019). Moreover, Li et al. (2020) detected vh-DNA in 97.7% of people with HBV-related HCC. Two months following HCC resection, the same vh-DNA sequence could be detected in 10 cases (23.3%), nine of whom (90%) experienced HCC recurrence within a year. Thus, vh-DNA of HBV integration could also be a useful circulating biomarker for monitoring HCC recurrence.
HBV Splice Variants
Description
Hepatitis B virus pgRNA has multiple splice donor and acceptor sites and can be spliced by cellular machinery as a post-transcriptional modification. Sixteen spliced pgRNA variants have been identified both in vitro and in tissues of CHB patients (Candotti and Allain, 2017). These splice variants can be encapsidated, reverse-transcribed and secreted into serum as replication-deficient viral particles (Terre et al., 1991).
Spliced viral RNAs can also be translated into HBV spliced proteins. For example, the 2.2-kb singly spliced variant lacking intron 2447/489, can encode hepatitis B spliced protein (HBSP) in the livers of patients with chronic HBV infection (Soussan et al., 2000). The 2447-2901 HBV RNA splice variant can act as the template for a 43 kDa polymerase-surface fusion glycoprotein (P-S FP), which localizes to the ER and is posited to be an HBV structural protein (Huang et al., 2000; Park et al., 2008).
Furthermore, hepatitis B doubly spliced protein and HBSP are respectively encoded by the 2.2-kb doubly spliced pgRNA and the single spliced product 1(SP1) variant (Terre et al., 1991; Huang et al., 2000; Soussan et al., 2000; Lee et al., 2008). However, the specific function of any of these splice variant-derived proteins is currently unclear.
Quantification
Hepatitis B virus splice variants can be quantified by reverse-transcription PCR (RT-PCR). At present, using different combinations of 5′ splice site and 3′ splice site can generate HBV RNA splicing variants, including 16 identified HBV splice variants of pgRNA and 4 splice variants of preS2/S mRNA (Su et al., 1989; Terre et al., 1991; Hass et al., 2005). The 2.2-kb singly spliced variant with a lack of intron 2447/489 which is the most common spliced variant can encode the HBSP, which can be detected by Western blot (Soussan et al., 2003).
Molecular Association With HCC
Some studies have reported increased HBV RNA splicing being associated with HCC (Kremsdorf et al., 2006; Bayliss et al., 2013). The 2.2 kb HBV spliced variant has been reported to be more highly expressed in tumor tissues than in the adjacent-tumor tissues (Lin et al., 2002). Moreover, when full-length (3.2 kb) HBV DNA and 2.2 kb spliced variant are co-transfected into HepG2 cells, the replication signal of the 3.2 kb HBV genome was increased 3–7 times (Lin et al., 2002). This suggests the HBV spliced variant plays a role in increasing HBV, which is a strong risk factor for HCC.
Circulating splice variant DNA is most frequently detected as defective HBV particles (dHBV) derived from reverse transcription of the 2.2-kb singly spliced mRNA, the most common spliced variant (Günther et al., 1997). In NA-naïve patients, the ratio of serum dHBV to wild-type HBV was lower in patients with moderate fibrosis and moderate or no liver necrosis compared to those with severe fibrosis and severe liver necrosis (Soussan et al., 2008). However, the direct clinical relationship between HBV splice variants and HCC remains uncharacterized.
Performance as a Predictor of HCC
HCC occurrence
Many studies have suggested that Pre-S deletion mutants play an important role in HBV-related HCC (Chen et al., 2007; Xie et al., 2010; An et al., 2018; Chen, 2018). In NA-naïve patients, a study enrolled 141 HBeAg-negative patients with CHB, 7 of whom developed HCC with a median time of 84 months. Univariate analysis showed that the presence of pre-S deletions was a significant factor for prediction of HCC (HR 11.26, 95% CI, 2.18-58.1; P = 0.004) (Chen et al., 2007). A recent meta-analysis revealed that pre-S deletions were related to HCC occurrence (OR 3.28, 95% CI 2.32–4.65; P < 0.00001; random-effects model). Both pre-S1 and pre-S2 were risk factors for HCC development, with OR 2.42 (95% CI 1.25–4.68, P = 0.008) and 3.36 (95% CI 2.04–5.55; P < 0.00001), respectively (Wungu et al., 2021).
In a cohort of 165 people with CHB under NA treatment in Australia (58 of whom were diagnosed with HCC), the median level of serum spliced HBV was higher in HCC patients than in non-HCC patients (P < 0.001) (Bayliss et al., 2013). Using a real-time PCR cut-off value of 7% for serum spliced HBV, the AUROC analysis of spliced HBV is 0.77, with a sensitivity of 45% and a specificity of 96%. Multiple regression analysis found that the serum spliced HBV level increased by about 0.1% per year before the diagnosis of HCC, independent of liver fibrosis (Bayliss et al., 2013).
HCC recurrence
Studies have revealed that HBV-related HCC patients with pre-S mutants are at higher risk of HCC recurrence after curative surgery, even when receiving post-surgical NA therapy (Su et al., 2013; Yen et al., 2018; Teng et al., 2020b). Su et al. (2013) analyzed 73 HCC patients without NAs therapy but with pre-S deletion mutants. They found that pre-S deletion mutants were related to a higher rate of HCC recurrence and higher serum HBV DNA levels (P = 0.055) (Su et al., 2013). Moreover, a recent study reported that using next-generation sequencing-based quantitative detection of pre-S mutants in serum can be useful for predicting HCC recurrence (AUROC of either pre-S2/pre-S1 or pre-S2 deletion = 0.683) (Teng et al., 2020b). Teng et al. (2020a) reported that only the presence of pre-S2 deletions (nt 1 to 54) in serum was associated with HCC recurrence (P value = 0.0080) with higher AUROC (0.632, 95% CI 0.556–0.708), compared with the pre-S1 deletion or the pre-S1 + pre-S2 deletion (nt 2,855–2,872, 1–54). In summary, pre-S2 deletion mutants may be a useful biomarker for HCC recurrence.
Circulating HBV RNA
Description
Multiple studies have shown that HBV RNA can be detected both in culture supernatants and in the serum of people with CHB (Hatakeyama et al., 2007; Huang et al., 2015; Wang et al., 2016). Given that HBV RNA exists as pgRNA in virus-like particles [produced by defective or partial reverse transcription (Wang et al., 2016; Prakash et al., 2018)], theoretically, serum HBV RNA is derived only from cccDNA in infected hepatocytes. However, the mechanism of the release of HBV pgRNA viral particles from infected hepatocytes into the circulation is unclear (Lam et al., 2017; Butler et al., 2018; Wang et al., 2018).
Quantification
Serum HBV RNA can be measured by quantitative RT-PCR, and digital droplet PCR (Wang et al., 2016; van Campenhout et al., 2018; Carey et al., 2019). Butler et al. (2018) used quantitative RT-PCR on the m2000 system (Abbott Molecular) to quantify serum HBV RNA detection with a lower limit of quantitation of 45 U/mL (Carey et al., 2020). There is limited standardization between these approaches to HBV RNA quantification, so further work needs to be done to harmonize these assays if they are to be used for routine diagnosis.
Molecular Association With HCC
Serum HBV RNA is closely related to the activity of HBV replication, especially in people with CHB during NA treatment (Giersch et al., 2017; Lu et al., 2017; Wang et al., 2017; Huang et al., 2018). However, there are few data on its predictive power for HCC risk. Halgand et al. reported that HBV pgRNA levels in tumor tissues were correlated with a particular HCC subtype (well-differentiated, non-invasive, and associated with better survival) (Halgand et al., 2018). However, serum levels may not be correlated with this. It is possible that high levels of HBV RNA could be a predictor of HCC in people with CHB under NA treatment (given it is a surrogate of cccDNA activity), but there is no clear clinical evidence for this yet.
HBV Truncated RNA
Description
Hepatitis B virus integration can act as a template for truncated HBV RNA (trRNA) transcripts. Hilger et al. (1991) identified HBV trRNA transcripts that terminated at a non-canonical CATAAA polyadenylation signal within the 3′ end region of the HBx open reading frame in tissue samples from two HBV-HCC patients. This signal can be used when the canonical polyadenylation signal is absent (e.g., when truncated as in the integrated HBV DNA form) (Breitkreutz et al., 2001). Later studies suggested that truncated HBx transcripts with a C-terminal deletion could be transcribed from integrated HBV DNA (Wang et al., 2004).
Quantification
Using specific primers containing a sequence corresponding to the polyadenylated 3′-end of full-length polyadenylated HBV RNA (flRNA) or trRNA, RACE-PCR targets the 3′-ends of the X gene for quantification of all polyadenylated HBV RNA species (Zhang W. et al., 2004; Ou et al., 2020). The assay’s lower limit of detection for HBV RNA was 794 copies/mL with a quantitative range of 800–106 copies/mL.
Molecular Association With HCC
Studies have shown that HBV trRNA, which can be transcribed from integrated HBV DNA, can encode a C-terminal truncated HBx protein (Hilger et al., 1991; Sze et al., 2013; van Bömmel et al., 2015). C-terminal-truncated HBx has been reported to enhance HCC invasion and reduce apoptotic response (Tu et al., 2001; Ma et al., 2008). In vitro studies suggest that C-terminal-truncated HBx promotes HCC through upregulating caveolin-1 to enhance β-catenin-mediated transcription of FRMD5 (FERM domain containing 5) (Ng et al., 2016; Mao et al., 2019). Sze et al. (2013) analyzed clinical data from 50 HBV-HCC patients and found that C-truncated HBx correlated with venous invasion. Also in vitro experiments reported that C-truncated HBx activates matrix metalloproteinase 10 by increasing C-Jun transcriptional activity, resulting in enhanced cell invasion and metastasis (Sze et al., 2013). Moreover, C-terminally truncated middle surface protein MHBst initiates c-Raf-1/Erk-2 signaling, resulting in an increased hepatocyte proliferation rate and dysplastic changes in hepatocytes (Hildt et al., 2002; Wang et al., 2006). Although these suggest possible roles for truncated HBV protein in tumor progression, whether it also plays a role in tumor formation is not clear.
Performance as a Predictor of HCC
Although serum HBV trRNA has been detected and used as a predictor for virological outcomes (van Bömmel et al., 2015), its association with HCC has only been shown in tumor tissues and not serum. A study with 50 people with HCC revealed that C-terminal truncated HBx was detected in 23 of 50 (46%) tumor tissues, and these had more venous invasion compared to tumors expressing only full-length HBx (P = 0.005) (Sze et al., 2013). This is consistent with another study where C-terminal truncated HBx was detected in 88 of 111 (79.3%) HCC tissues, compared with full-length HBx in all 111 non-tumor tissues and 23 of 111 (20.7%) HCC tissues (Ma et al., 2008). However, these studies only detected truncated HBx in confirmed HCC tissues. Whether serum HBV trRNA can predict HCC occurrence or recurrence is still unclear.
Hepatitis B Surface Antigen
Description
Hepatitis B virus sub-genomic mRNA transcripts (2.4- and 2.1-kb surface mRNAs) encode the large, middle, and small variants of the HBV surface antigen, which can assemble at the ER as sub-viral particles (SVP) and be secreted via the Golgi apparatus (Ganem and Schneider, 2001). The majority of circulating HBsAg exists as non-infectious filamentous and spherical SVP, in 1,000- to 100,000-fold excess compared to virions (Wei et al., 2010). HBsAg may be translated from both HBV cccDNA and integrated DNA; the latter especially in HBeAg-negative chronic HBV-infected patients (Hu et al., 2018).
Secreted HBsAg in SVP could play an immunomodulatory role during HBV infection. SVP capture neutralizing anti-HBsAg antibodies and divert host immune recognition away from infectious HBV virions (Carman et al., 1996). The host antiviral immune response is subsequently minimized, allowing HBV persistence (Rydell et al., 2017). HBsAg is also the target of HBV functional cure; if serum HBsAg is eliminated then HBV infection is considered to be cleared (Chen et al., 2016; Al Awaidy and Ezzikouri, 2020).
Quantification
Currently, there are three quantitative assays for HBsAg measurement: Architect HBsAg QT (Abbott Diagnostics), Elecsys HBsAg II Quant (Roche Diagnostics) and DiaSorin Liaison XL. All assays provide measurements that correlate well with each other (Burdino et al., 2014; Liao et al., 2015). The Architect assay is capable of quantifying HBsAg concentrations ranging from 0.4 to 250 IU/mL (Deguchi et al., 2004). The range of the Elecsys II and DiaSorin Liaison XL assays are respectively from 0.05 to 130 IU/mL (sensitivity from 0.017 to 0.022 IU/mL) and 0.03 to 150 IU/mL (sensitivity of 0.03 IU/mL) (Burdino et al., 2014; Cornberg et al., 2017). All three assays have automatic dilution (1:400) to increase the upper limit of detection to over 50,000 IU/mL.
Unfortunately, these assays do not distinguish between the three forms of HBsAg (small, medium, and large). In pre-clinical trials, the ratios and composition of the three HBsAg forms have been reported to predict HBsAg clearance during treatment in patients with HBeAg-positive CHB (Pfefferkorn et al., 2021). Therefore, the quantification of HBsAg variants and monitoring the HBsAg composition throughout treatment could be important to predict the clearance of secreted HBsAg and the associated reduction in HCC risk.
Molecular Association With HCC
Several clinical studies recently reported that high levels of serum HBsAg are associated with an increased risk of HCC (Tseng et al., 2012; Kawanaka et al., 2014). Similarly, HBsAg loss is associated with very low HCC risk (Yip et al., 2017, 2019). While the underlying mechanism is not clear, this may be due to the association of HBsAg with replication levels or the amount of integrated HBV DNA, which are both risk factors for HCC (Xiangji et al., 2011; Yan et al., 2015; Tseng et al., 2019).
Chronic inflammation driven by anti-HBs responses could promote oncogenesis. A chimeric HBV-HCC mouse model was studied by extracting HBsAg-expressing hepatocytes from HBsAg transgenic mice (C57BL/6J) and transferring them into immuno-competent Fah–/– recipient mice (which allow implantation of hepatocytes) (Hao et al., 2021). Persistent HBsAg expression triggered HBsAg-specific CD8+ T cell activation, followed by hepatocyte apoptosis and turnover, progressive chronic inflammation, clonal expansion, and ultimately HCC (Nakamoto et al., 1998; Hao et al., 2021). In in vitro models, HBsAg has been reported to promote HCC invasion through the TLR2/MyD88/NF-kB signaling pathway (Cheng et al., 2017).
Hepatitis B surface antigens with mutations in the Pre-S1 or Pre-S2 regions could be directly oncogenic: these mutated proteins can alter host cell lipid metabolism, lead to ER stress, induce oxidative DNA damage and genomic instability, all of which increase the risk of HCC development (Hsieh et al., 2004; Wang et al., 2006; Yang et al., 2008). HBsAg Pre-S1 and Pre-S2 mutants accumulate intracellularly, forming the characteristic cytopathic effect of ground-glass structures (Roingeard and Sureau, 1998). Ground-glass hepatocytes (GGH) occur as either Type I or Type II GGH containing LHBsAg with mutations in the Pre-S1 or Pre-S2 regions, respectively (Wang et al., 2003). Type I GGH occurs as single hepatocytes during early stages of HBV infection with active HBV replication, while Type II GGH occurs as clusters (suggestive of clonal expansion) during latter stages of reduced HBV replication (Fan et al., 2001; Wang et al., 2003). Type II GGH is associated with cirrhosis and HCC development (Fan et al., 2001). In HBV patients, biopsies of cirrhotic nodules with Type II GGH contained HBV genomes which were integrated and clonally expanded, suggesting that Type II GGH are pre-neoplastic lesions (Fan et al., 2000). Similarly, the presence of Pre-S mutants in the serum of patients with CHB is associated with an increased risk of HCC, with Pre-S1 and Pre-S2 mutants present significantly higher in HCC patients compared to non-HCC carriers (19/64, 29.7% vs 25/202, 12.4%, P = 0.002) (Fan et al., 2001; Lin C.L. et al., 2007). HBsAg mutants may activate ER stress-dependent and -independent pathways to promote genomic instability and cell proliferation needed for HCC tumorigenesis.
Performance as a Predictor of HCC
HCC occurrence
Generally, the predictive value of HBsAg levels for HCC is poorer than HBV DNA or ALT in treatment-naïve patients with CHB (Tseng et al., 2012). In HBeAg-negative patients with HBV DNA levels ≥2,000 IU/mL, HBsAg levels (<1,000 IU/mL or >1,000 IU/mL) were not related to the risk of HCC (AUROC 0.58; P = 0.247) (Tseng et al., 2012). However, multivariate analysis showed that HBsAg ≥1,000 IU/mL was an independent risk factor for HCC development in HBeAg-negative patients with HBV DNA level <2,000 IU/mL (HR 13.7, 95% CI: 4.8–39.3) (Tseng et al., 2012).
The true sensitivity and specificity of HBsAg in predicting HCC in these patients on NA therapy remains unknown and requires larger cohorts. Furthermore, HCC can still occur in patients with CHB who have undergone HBsAg loss (Chen et al., 2016). In one retrospective study, the annual incidence of HCC after HBsAg seroconversion was 2.85 and 0.29% in cirrhotic and non-cirrhotic patients, respectively (Kim et al., 2015). The risk factors associated with HCC development post-HBsAg seroconversion are: age above 50 (HR: 12.14; 95% CI: 1.61–91.68), male gender (HR: 8.96; 95% CI: 1.17–68.80), and infection with HBV genotype C (Kim et al., 2015). Given that HCC can occur in patients with CHB following HBsAg seroconversion, quantitative HBsAg is unlikely to be a suitable standalone biomarker for HCC risk (Kim et al., 2015; Chen et al., 2016).
HCC recurrence
HBsAg can also be used to predict HCC recurrence following curative hepatic resection (HR 1.23, 95% CI: 1.04–1.44, P = 0.01) (Huang et al., 2014; Zhou et al., 2015). Moreover, HBsAg level ≥4,000 IU/mL (HR 2.80; P = 0.023) is a risk factor for late HCC recurrence (after 2 years) (Sohn et al., 2014). Following hepatic resection, in HBeAg-negative patients with HBV DNA level <2,000 IU/mL, HBsAg level determined the risk of HCC recurrence (P = 0.014), while HBV DNA (P = 0.55) and ALT (P = 0.186) were not predictive (Huang et al., 2014). Univariate analyses of patients with HBV-HCC following radiofrequency ablation showed HCC recurrence is associated with HBsAg ≥1,000 IU/mL (Zhang et al., 2017). Similarly, in HBeAg-negative patients following radiofrequency ablation, recurrence-free survival significantly decreased (P = 0.039) as a result of high HBsAg levels; i.e., at 2 years post-treatment, recurrence-free survival decreased from 64% (HBsAg < 1,000 IU/mL) to 50% (HBsAg ≥ 1,000 IU/mL) (Zhang et al., 2017). In summary, high HBsAg may be useful in predicting HCC development and recurrence in HBeAg-seronegative patients with low HBV DNA.
Hepatitis B Core Related Antigen
Description
The 3.5-kb precore RNA derived from the HBV Pre-C/C gene can act as the template for three viral proteins: HBcAg, HBeAg and a truncated 22 kDa precore protein (p22Cr) (Mak et al., 2018). The so-called hepatitis B core related antigen (HBcrAg) consists of these three proteins which share an identical 149 long amino acid sequence. HBcAg forms the viral capsid subunits. HBeAg (164-amino acid protein) is synthesized by removing the C-terminal region of p22 and is secreted from infected cells (Messageot et al., 2003). p22Cr is the pre-core protein with additional post-translational processing at both the N- and C-termini (Kimura et al., 2005).
Quantification
Hepatitis B core related antigen was first measured by a sensitive enzyme immunoassay that denatures antibodies prior to analysis and therefore can detect HBcAg and HBeAg in anti-HBc or anti-HBe antibody-positive patients (Kimura et al., 2002). Currently, a newly chemiluminescence enzyme immunoassay with monoclonal antibodies to HBeAg and HBcAg was developed for the detection of HBcrAg. This assay showed the HBcrAg concentration correlates strongly with the HBV DNA concentration (P < 0.001) over a 5-log range. Moreover, the correlation between HBV load and circulating HBcrAg was not affected in HBeAg-negative patients nor those with pre-core mutations (Rokuhara et al., 2003). Particularly for patients under NA treatment, the HBcrAg assay could be a sensitive and clinically useful surrogate marker of intrahepatic HBV cccDNA levels (Rokuhara et al., 2003).
Molecular Association With HCC
Transcriptional activity of intrahepatic cccDNA is recognized as a risk for HBV-induced HCC under NA therapy (Levrero and Zucman-Rossi, 2016; Chen et al., 2017; Mak et al., 2018; Suzuki et al., 2019; Testoni et al., 2019). Several studies have shown that serum HBcrAg is highly correlated with intrahepatic cccDNA activity (Wong et al., 2017; Mak et al., 2018) as it can only be expressed from cccDNA (unlike HBsAg, which can also be expressed from integrated HBV DNA). Importantly, NA therapy only inhibits reverse transcription of HBV RNA, but does not inhibit protein synthesis from cccDNA (Tong and Revill, 2016). Therefore, HBcrAg is a non-invasive biomarker of active viral replication, which in turn may predict HCC.
Performance as a Predictor of HCC
HCC occurrence
A number of studies have suggested that serum HBcrAg can be a useful viral biomarker for HCC risk (Chen et al., 2018; Hosaka et al., 2019; Suzuki et al., 2019; Tseng et al., 2019; Baudi et al., 2020). A study of 1,031 NA-naïve patients with CHB (78 of whom were diagnosed with HCC during the follow-up period, median duration 10.7 years) revealed that serum HBcrAg was significantly related to the risk of developing HCC. HBcrAg >794 U/mL (HR, 5.05; 95% CI, 2.40–10.63) was associated with the risk of developing HCC, independent of HBV DNA titers. In the subgroup of HBeAg-negative, non-cirrhotic patients with HBV DNA levels ≤10,000 copies/mL, HBcrAg >5,012 U/mL was significantly related to the risk of HCC (HR 6.13, 95% CI 1.71–22.06). However, in the subgroup of CHB patients with HBV DNA levels >10,000 copies/mL, any HBeAg status, and FIB-4 index ≤3.6 (an index of fibrosis), HBcrAg >794 U/mL was associated with the incidence of HCC (HR 5.69, 95% CI 1.37–23.72) (Tada et al., 2016). Another study of 2,666 patients with CHB (of whom 209 developed HCC) reported that baseline HBcrAg levels of >10 kU/mL in HBeAg-negative patients with HBV DNA levels from 2,000 to 19,999 IU/mL are at increased risk of HCC (Tseng et al., 2019). Conversely lower HBcrAg levels (<10 kU/mL) were linked to a low risk of HCC.
Hepatitis B core related antigen has been reported to be superior to HBV DNA or HBsAg in predicting HCC in NA-naïve patients with CHB. Tada et al. (2016) reported that HBcrAg could predict HCC in 2, 5, and 10 years with AUROC curves 0.80, 0.68, 0.70 (compared to HBV DNA at 0.75, 0.63, 0.65, respectively). Moreover, Tseng et al. (2019) found that AUROC of HBcrAg, HBV DNA, HBsAg was 0.73, 0.72, 0.57, respectively with 10 years follow-up, or 0.70, 0.69, 0.56 with 15 years follow-up. These studies show a high correlation between HBV DNA and HBcrAg levels, with HBcrAg being more sensitive than HBsAg in predicting HCC in untreated patients.
In patients with CHB under NA treatment, persistently high HBcrAg levels were associated with HCC development (Kumada et al., 2013; Hosaka et al., 2019). Hosaka et al. (2019) reported that in a study of 1,268 patients treated with NAs for more than 1 year, among the 60 of 667 HBeAg-positive patients, high HBcrAg levels (≥4.9 log U/mL) after 1-year on-treatment was associated with increased HCC incidence within 15 years (HR, 6.15, 95% CI: 1.89–20.0, P = 0.003). Using a HBcrAg cut-off value of 4.9 log U/mL gave positive and negative predictive values of 0.95 and 0.19, with sensitivities and specificities of 0.903 and 0.217, respectively. Moreover, in 601 HBeAg-negative patients, the risk of HCC was higher in those with HBcrAg values >4.4 log U/mL (HR, 2.54, 95% CI: 1.40–4.60; P = 0.002). In this cohort positive and negative predictive values were 0.51 and 0.79, sensitivity and specificity were 0.519 and 0.787 (Hosaka et al., 2019). This result is similar to another study enrolling 76 NA-treated patients with CHB with undetectable HBV DNA diagnosed with HCC and 152 matched controls who did not develop HCC (Cheung et al., 2017). The AUROC of HBcrAg in the HCC group was 0.61 (95% CI: 0.54–0.69) for predicting HCC. Using a cut-off value of ≥7.8 kU/mL, the sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were 57.9, 70.4, 49.4, and 77.0%, respectively, with an odds ratio (OR) of 3.27 (95% CI: 1.84–5.80) for HCC development. HBcrAg was more predictive of HCC in non-cirrhotic patients: AUROC was 0.70 (95% CI: 0.58–0.81) using a HBcrAg cut-off value of ≥7.9 kU/mL, with a sensitivity, specificity, PPV and NPV of 62.5, 78.1, 58.8, 80.6%, respectively, and with an OR of 5.95 (95% CI: 2.35–15.07) for HCC development (Cheung et al., 2017).
HCC recurrence
Hepatocellular carcinoma recurrence after HCC resection is still high, with a rate of ∼50% within two years (Wu et al., 2009). High serum HBcrAg has been reported to predict HCC recurrence: Chen et al. (2018) reported that in 56 of 89 HCC patients with both positive cccDNA and HBcrAg who had been followed up for 5 years, recurrence rates of HCC in patients with high HBcrAg (>5.2 log U/mL) were higher than those with low HBcrAg (≤5.2 log U/mL; P = 0.003).
During NA therapy, higher HBcrAg levels at HCC diagnosis can predict post-treatment recurrence of HCC (Hosaka et al., 2010). In a study of 55 HCC patients with NA treatment at diagnosis of HCC receiving curative surgery, serum HBcrAg levels ≥4.8 log U/mL at the time of HCC diagnosis was an independent risk factor for HCC recurrence with HR of 8.96 (95% CI: 1.94–41.4) (Hosaka et al., 2010). A long-term follow-up study in Netherlands revealed that higher HBcrAg level (≥5.1 log U/mL) was associated with an increased tumor recurrence rate in 53 of 119 HCC patients who were identified with early stage HCC receiving NAs at the time of HCC diagnosis (Beudeker et al., 2021). Moreover, in a cohort of 357 CHB-related HCC patients who underwent liver transplantation followed by NA treatment, HBcrAg ≥5.0 log U/mL was an independent risk factor for HCC recurrence, with a higher 5-year cumulative recurrence rate, compared with an HBcrAg <5.0 log U/mL (37.6 vs 6%, P < 0.001) (HR:5.27, 95% CI 2.47–11.25, P < 0.001) (Yu et al., 2019). In conclusion, HBcrAg may be a useful biomarker for HCC recurrence, however the sensitivity and specificity of HBcrAg in predicting HCC recurrence needs further research.
Future Work and Conclusion
This review has assessed the value of serum viral biomarkers in HBV-related HCC. Of all the potential biomarkers that have been studied, growing evidence supports the use of serum HBcrAg and preS mutations as biomarkers for predicting HCC occurrence in people with CHB, both in NA-naïve patients and in patients receiving NA treatment. In combination with AFP and abdominal ultrasound serum biomarkers might improve HCC screening and increase early diagnosis, although further validation studies are required to confirm their clinical performance in predicting and/or detecting HCC. Moreover, several biomarkers remain to be tested in a clinical setting (e.g., HBV integrations and HBV RNA, both full length and truncated forms), laying the groundwork for future exploratory studies.
Challenges remain in this field of research. Firstly, some of these markers (e.g., HBV RNA and HBcrAg) have no standardized quantification assay. To accurately and robustly compare HCC risk between different studies, equivalent cut-off values need to be used and this can only be done with appropriate reference samples and standardized assays. Moreover, we lack the appropriate laboratory models to investigate new and existing HCC markers in HBV infection. Even if there were a practical experimental animal system that supported HBV infection, no known models recapitulate the decades long-process of HBV-initiated HCC. This makes discovery, characterization, and confirmation of new and existing viral biomarkers difficult.
While this field awaits further developments to enable more in-depth analysis, our review has shown signs of promise in viral biomarkers and their ability to predict HBV-associated HCC occurrence and recurrence. We expect that (in combination with existing markers) viral biomarkers will increasingly become incorporated into HCC risk algorithms, improving health outcomes for the ∼300 million people worldwide living with CHB.
Author Contributions
YL initiated the writing, led the organization, and did the majority of the research for the review. VV and MP produced the figure and contributed to large sections of the review. JG, JF, and MD were involved in editing the manuscript and provided crucial input into several sections of the review. TT was responsible for the conceptualization, coordination, structure, and editing of the review. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Robert W. Storr Bequest to the Sydney Medical Foundation, University of Sydney. TT and MD are supported by grant funding from the Australian Centre for HIV and Hepatitis Virology Research and NHMRC Ideas grant GNT2002565.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adler, M., Larocca, L., Trovato, F. M., Marcinkowski, H., Pasha, Y., and Taylor-Robinson, S. D. (2016). Evaluating the risk of hepatocellular carcinoma in patients with prominently elevated liver stiffness measurements by FibroScan: a multicentre study. HPB 18, 678–683. doi: 10.1016/j.hpb.2016.05.005
Al Awaidy, S. T., and Ezzikouri, S. (2020). Moving towards hepatitis B elimination in Gulf Health Council states: from commitment to action. J. Infect. Public Health 13, 221–227. doi: 10.1016/j.jiph.2019.08.004
An, P., Xu, J., Yu, Y., and Winkler, C. A. (2018). Host and viral genetic variation in HBV-related hepatocellular carcinoma. Front. Genet. 9:261. doi: 10.3389/fgene.2018.00261
Baudi, I., Inoue, T., and Tanaka, Y. (2020). Novel biomarkers of hepatitis B and hepatocellular carcinoma: clinical significance of HBcrAg and M2BPGi. Int. J. Mol. Sci. 21:949. doi: 10.3390/ijms21030949
Bayliss, J., Lim, L., Thompson, A. J., Desmond, P., Angus, P., Locarnini, S., et al. (2013). Hepatitis B virus splicing is enhanced prior to development of hepatocellular carcinoma. J. Hepatol. 59, 1022–1028. doi: 10.1016/j.jhep.2013.06.018
Beudeker, B. J., Groothuismink, Z. M., de Man, R. A., Witjes, C. D., van Der Eijk, A. A., Boonstra, A., et al. (2021). Hepatitis B core-related antigen levels predict recurrence-free survival in patients with HBV-associated early-stage hepatocellular carcinoma: results from a Dutch long-term follow-up study. J. Viral Hepat. 28, 205–208. doi: 10.1111/jvh.13394
Bill, C. A., and Summers, J. (2004). Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc. Natl. Acad. Sci. U.S.A. 101, 11135–11140. doi: 10.1073/pnas.0403925101
Bitton Alaluf, M., and Shlomai, A. (2016). New therapies for chronic hepatitis B. Liver Int. 36, 775–782. doi: 10.1111/liv.13086
Blondot, M.-L., Bruss, V., and Kann, M. (2016). Intracellular transport and egress of hepatitis B virus. J. Hepatol. 64, S49–S59.
Bolukbas, C., Bolukbas, F. F., Horoz, M., Aslan, M., Celik, H., and Erel, O. (2005). Increased oxidative stress associated with the severity of the liver disease in various forms of hepatitis B virus infection. BMC Infect. Dis. 5:95. doi: 10.1186/1471-2334-5-95
Bosch, F. X., Ribes, J., Díaz, M., and Cléries, R. (2004). Primary liver cancer: worldwide incidence and trends. Gastroenterology 127, S5–S16.
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Breitkreutz, R., Zhang, W., Lee, M., Hoffmann, A., Tokus, M., Su, Q., et al. (2001). Hepatitis B virus nucleic acids circulating in the blood: distinct patterns in HBs carriers with hepatocellular carcinoma. Ann. N. Y. Acad. Sci. 945, 195–206. doi: 10.1111/j.1749-6632.2001.tb03886.x
Budzinska, M. A., Shackel, N. A., Urban, S., and Tu, T. (2018a). Cellular genomic sites of hepatitis B virus DNA integration. Genes 9:365. doi: 10.3390/genes9070365
Budzinska, M. A., Shackel, N. A., Urban, S., and Tu, T. (2018b). Sequence analysis of integrated hepatitis B virus DNA during HBeAg-seroconversion. Emerg. Microbes Infect. 7:142.
Burdino, E., Ruggiero, T., Proietti, A., Milia, M. G., Olivero, A., Caviglia, G. P., et al. (2014). Quantification of hepatitis B surface antigen with the novel DiaSorin LIAISON XL Murex HBsAg Quant: correlation with the ARCHITECT quantitative assays. J. Clin. Virol. 60, 341–346. doi: 10.1016/j.jcv.2014.05.013
Butler, E. K., Gersch, J., McNamara, A., Luk, K. C., Holzmayer, V., de Medina, M., et al. (2018). Hepatitis B virus serum DNA and RNA levels in nucleos (t) ide analog-treated or untreated patients during chronic and acute infection. Hepatol. Int. 68, 2106–2117. doi: 10.1002/hep.30082
Candotti, D., and Allain, J.-P. (2017). Biological and clinical significance of hepatitis B virus RNA splicing: an update. Ann. Blood 2, 1–14.
Carey, I., Gersch, J., Wang, B., Moigboi, C., Kuhns, M., Cloherty, G., et al. (2019). Pre-genomic HBV RNA and HBcrAg predict outcomes in HBeAg negative chronic hepatitis B patients suppressed on nucleos (t) ide analogue therapy. Hepatology 72, 42–57. doi: 10.1002/hep.31026
Carey, I., Gersch, J., Wang, B., Moigboi, C., Kuhns, M., Cloherty, G., et al. (2020). Pregenomic HBV RNA and hepatitis B core-related antigen predict outcomes in hepatitis B e antigen-negative chronic hepatitis B patients suppressed on nucleos (t) ide analogue therapy. Hepatology 72, 42–57.
Carman, W. F., Trautwein, C., van Deursen, J., Colman, K., Dornan, E., McIntyre, G., et al. (1996). Hepatitis B virus envelope variation after transplantation with and without hepatitis B immune globulin prophylaxis. Hepatology 24, 489–493. doi: 10.1002/hep.510240304
Cassinotto, C., Boursier, J., de Lédinghen, V., Lebigot, J., Lapuyade, B., Cales, P., et al. (2016). Liver stiffness in nonalcoholic fatty liver disease: a comparison of supersonic shear imaging, FibroScan, and ARFI with liver biopsy. Hepatology 63, 1817–1827. doi: 10.1002/hep.28394
Chen, B.-F. (2018). Hepatitis B virus pre-S/S variants in liver diseases. World J. Gastroenterol. 24, 1507–1520. doi: 10.3748/wjg.v24.i14.1507
Chen, C. F., Lee, W. C., Yang, H. I., Chang, H. C., Jen, C. L., Iloeje, U. H., et al. (2011). Changes in serum levels of HBV DNA and alanine aminotransferase determine risk for hepatocellular carcinoma. Gastroenterology 141, 1240–1248.E2.
Chen, C. H., Hung, C. H., Lee, C. M., Hu, T. H., Wang, J. H., Wang, J. C., et al. (2007). Pre-S deletion and complex mutations of hepatitis B virus related to advanced liver disease in HBeAg-negative patients. Gastroenterology 133, 1466–1474. doi: 10.1053/j.gastro.2007.09.002
Chen, C.-J., Yang, H.-I., Su, J., Jen, C.-L., You, S.-L., Lu, S.-N., et al. (2006). Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 295, 65–73. doi: 10.1001/jama.295.1.65
Chen, E.-Q., Feng, S., Wang, M.-L., Liang, L.-B., Zhou, L.-Y., Du, L.-Y., et al. (2017). Serum hepatitis B core-related antigen is a satisfactory surrogate marker of intrahepatic covalently closed circular DNA in chronic hepatitis B. Sci. Rep. 7:173.
Chen, S., Gao, Y., Li, H., Fang, M., Feng, H., Guan, W., et al. (2018). Clinical evaluation of hepatitis B core-related antigen in chronic hepatitis B and hepatocellular carcinoma patients. Clin. Chim. Acta 486, 237–244. doi: 10.1016/j.cca.2018.07.027
Chen, X.-P., Long, X., Jia, W.-L., Wu, H.-J., Zhao, J., Liang, H.-F., et al. (2019). Viral integration drives multifocal HCC during the occult HBV infection. J. Exp. Clin. Cancer Res. 38:261.
Chen, Y., Jeng, W., Chien, R., Chu, C., and Liaw, Y. (2016). Clinical outcomes after spontaneous and nucleos (t) ide analogue-treated HB sAg seroclearance in chronic HBV infection. Aliment. Pharmacol. Ther. 43, 1311–1318. doi: 10.1111/apt.13630
Chen, Y., and Tian, Z. (2019). HBV-induced immune imbalance in the development of HCC. Front. Immunol. 10:2048. doi: 10.3389/fimmu.2019.02048
Cheng, S., Zhang, B., Du, J.-Y., Jin, Y.-H., Lang, H.-Y., Zeng, L.-H., et al. (2017). Hepatitis B surface antigen promotes the invasion of hepatitis B virus-related hepatocellular carcinoma cells by Upregulation of toll-like receptor 2. Viral Immunol. 30, 232–239. doi: 10.1089/vim.2016.0162
Cheung, K. S., Seto, W. K., Wong, D. K., Lai, C. L., and Yuen, M. F. (2017). Relationship between HB sAg, HB crAg and hepatocellular carcinoma in patients with undetectable HBV DNA under nucleos (t) ide therapy. J. Viral Hepat. 24, 654–661. doi: 10.1111/jvh.12688
Chevaliez, S., Bouvier-Alias, M., Laperche, S., Hézode, C., and Pawlotsky, J.-M. (2010). Performance of version 2.0 of the Cobas AmpliPrep/Cobas TaqMan real-time PCR assay for hepatitis B virus DNA quantification. J. Clin. Microbiol. 48, 3641–3647. doi: 10.1128/jcm.01306-10
Chevaliez, S., Bouvier-Alias, M., Laperche, S., and Pawlotsky, J.-M. J. (2008). Performance of the Cobas AmpliPrep/Cobas TaqMan real-time PCR assay for hepatitis B virus DNA quantification. J. Clin. Microbiol. 46, 1716–1723. doi: 10.1128/jcm.01248-07
Choi, J., Kim, G. A., Han, S., Lee, W., Chun, S., and Lim, Y. S. (2019). Longitudinal assessment of three serum biomarkers to detect very early-stage hepatocellular carcinoma. Hepatology 69, 1983–1994. doi: 10.1002/hep.30233
Chu, C.-M., Hung, S.-J., Lin, J., Tai, D.-I., and Liaw, Y.-F. (2004). Natural history of hepatitis be antigen to antibody seroconversion in patients with normal serum aminotransferase levels. Am. J. Med. 116, 829–834. doi: 10.1016/j.amjmed.2003.12.040
Chuma, M., Hige, S., Kamiyama, T., Meguro, T., Nagasaka, A., Nakanishi, K., et al. (2009). The influence of hepatitis B DNA level and antiviral therapy on recurrence after initial curative treatment in patients with hepatocellular carcinoma. J. Gastroenterol. 44, 991–999. doi: 10.1007/s00535-009-0093-z
Cornberg, M., Wong, V. W.-S., Locarnini, S., Brunetto, M., Janssen, H. L., and Chan, H. L. (2017). The role of quantitative hepatitis B surface antigen revisited. J. Hepatol. 66, 398–411. doi: 10.1016/j.jhep.2016.08.009
De Franchis, R., Meucci, G., Vecchi, M., Tatarella, M., Colombo, M., Del Ninno, E., et al. (1993). The natural history of asymptomatic hepatitis B surface antigen carriers. Ann. Intern. Med. 118, 191–194. doi: 10.7326/0003-4819-118-3-199302010-00006
Deguchi, M., Yamashita, N., Kagita, M., Asari, S., Iwatani, Y., Tsuchida, T., et al. (2004). Quantitation of hepatitis B surface antigen by an automated chemiluminescent microparticle immunoassay. J. Virol. Methods 115, 217–222. doi: 10.1016/j.jviromet.2003.10.002
Duygu, F., Karsen, H., Aksoy, N., and Taskin, A. J. (2012). Relationship of oxidative stress in hepatitis B infection activity with HBV DNA and fibrosis. Ann. Lab. Med. 32, 113–118. doi: 10.3343/alm.2012.32.2.113
European Association for the Study of the Liver (2017). EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 67, 370–398. doi: 10.1016/j.jhep.2017.03.021
Fan, Y. F., Lu, C. C., Chang, Y. C., Chang, T. T., Lin, P. W., Lei, H. Y., et al. (2000). Identification of a pre-S2 mutant in hepatocytes expressing a novel marginal pattern of surface antigen in advanced diseases of chronic hepatitis B virus infection. J. Gastroenterol. Hepatol. 15, 519–528. doi: 10.1046/j.1440-1746.2000.02187.x
Fan, Y.-F., Lu, C.-C., Chen, W.-C., Yao, W.-J., Wang, H.-C., Chang, T.-T., et al. (2001). Prevalence and significance of hepatitis B virus (HBV) pre-S mutants in serum and liver at different replicative stages of chronic HBV infection. Hepatology 33, 277–286. doi: 10.1053/jhep.2001.21163
Fattovich, G., Bortolotti, F., and Donato, F. (2008). Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J. Hepatol. 48, 335–352. doi: 10.1016/j.jhep.2007.11.011
Foucher, J., Castéra, L., Bernard, P.-H., Adhoute, X., Laharie, D., Bertet, J., et al. (2006). Prevalence and factors associated with failure of liver stiffness measurement using FibroScan in a prospective study of 2114 examinations. Eur. J. Gastroenterol. Hepatol. 18, 411–412. doi: 10.1097/00042737-200604000-00015
Fung, J., Yuen, M. F., Yuen, J. C., Wong, D. K., and Lai, C. L. (2007). Low serum HBV DNA levels and development of hepatocellular carcinoma in patients with chronic hepatitis B: a case–control study. Aliment. Pharmacol. 26, 377–382. doi: 10.1111/j.1365-2036.2007.03390.x
Furuta, M., Tanaka, H., Shiraishi, Y., Unida, T., Imamura, M., Fujimoto, A., et al. (2018). Characterization of HBV integration patterns and timing in liver cancer and HBV-infected livers. Oncotarget 9:31789. doi: 10.18632/oncotarget.25960
Ganem, D., and Schneider, R. J. (2001). “Hepadnaviridae: the viruses and their replication,” in Fields Virology, 4th Edn, eds D. M. Knipe and P. M. Howley (Philadelphia, PA: Lippincott Williams & Wilkins), 2923–2969.
Gao, Q., Zhu, H., Dong, L., Shi, W., Chen, R., Song, Z., et al. (2019). Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell 179, 561–577.e22.
Ghany, M., and Liang, T. J. (2007). Drug targets and molecular mechanisms of drug resistance in chronic hepatitis B. Gastroenterology 132, 1574–1585. doi: 10.1053/j.gastro.2007.02.039
Giersch, K., Allweiss, L., Volz, T., Dandri, M., and Lütgehetmann, M. (2017). Serum HBV pgRNA as a clinical marker for cccDNA activity. J. Hepatol. 66, 460–462. doi: 10.1016/j.jhep.2016.09.028
Goto, T., Yoshida, H., Tateishi, R., Enooku, K., Goto, E., Sato, T., et al. (2011). Influence of serum HBV DNA load on recurrence of hepatocellular carcinoma after treatment with percutaneous radiofrequency ablation. Hepatol. Int. 5, 767–773. doi: 10.1007/s12072-011-9255-1
Greten, T., Papendorf, F., Bleck, J., Kirchhoff, T., Wohlberedt, T., Kubicka, S., et al. (2005). Survival rate in patients with hepatocellular carcinoma: a retrospective analysis of 389 patients. Br. J. Cancer 92, 1862–1868. doi: 10.1038/sj.bjc.6602590
Günther, S., Sommer, G., Iwanska, A., and Will, H. (1997). Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology 238, 363–371. doi: 10.1006/viro.1997.8863
Halgand, B., Desterke, C., Rivière, L., Fallot, G., Sebagh, M., Calderaro, J., et al. (2018). Hepatitis B virus pregenomic RNA in hepatocellular carcinoma: a nosological and prognostic determinant. Hepatology 67, 86–96. doi: 10.1002/hep.29463
Hao, X., Chen, Y., Bai, L., Wei, H., Sun, R., and Tian, Z. (2021). HBsAg-specific CD8+ T cells as an indispensable trigger to induce murine hepatocellular carcinoma. Cell. Mol. Immunol. 18, 128–137. doi: 10.1038/s41423-019-0330-1
Hass, M., Hannoun, C., Kalinina, T., Sommer, G., Manegold, C., and Günther, S. J. H. (2005). Functional analysis of hepatitis B virus reactivating in hepatitis B surface antigen-negative individuals. Hepatol. Int. 42, 93–103. doi: 10.1002/hep.20748
Hatakeyama, T., Noguchi, C., Hiraga, N., Mori, N., Tsuge, M., Imamura, M., et al. (2007). Serum HBV RNA is a predictor of early emergence of the YMDD mutant in patients treated with lamivudine. Hepatology 45, 1179–1186. doi: 10.1002/hep.21581
Hildt, E., Munz, B., Saher, G., Reifenberg, K., and Hofschneider, P. H. (2002). The PreS2 activator MHBst of hepatitis B virus activates c-raf-1/Erk2 signaling in transgenic mice. EMBO J. 21, 525–535. doi: 10.1093/emboj/21.4.525
Hilger, C., Velhagen, I., Zentgraf, H., and Schröder, C. J. (1991). Diversity of hepatitis B virus X gene-related transcripts in hepatocellular carcinoma: a novel polyadenylation site on viral DNA. J. Virol. 65, 4284–4291. doi: 10.1128/jvi.65.8.4284-4291.1991
Hosaka, T., Suzuki, F., Kobayashi, M., Fujiyama, S., Kawamura, Y., Sezaki, H., et al. (2019). Impact of hepatitis B core-related antigen on the incidence of hepatocellular carcinoma in patients treated with nucleos (t) ide analogues. Aliment. Pharmacol. 49, 457–471. doi: 10.1111/apt.15108
Hosaka, T., Suzuki, F., Kobayashi, M., Hirakawa, M., Kawamura, Y., Yatsuji, H., et al. (2010). HBcrAg is a predictor of post-treatment recurrence of hepatocellular carcinoma during antiviral therapy. Liver Int. 30, 1461–1470. doi: 10.1111/j.1478-3231.2010.02344.x
Hsieh, Y.-H., Su, I.-J., Wang, H.-C., Chang, W.-W., Lei, H.-Y., Lai, M.-D., et al. (2004). Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis 25, 2023–2032. doi: 10.1093/carcin/bgh207
Hsu, Y.-S., Chien, R.-N., Yeh, C.-T., Sheen, I.-S., Chiou, H.-Y., Chu, C.-M., et al. (2002). Long-term outcome after spontaneous HBeAg seroconversion in patients with chronic hepatitis B. Hepatology 35, 1522–1527. doi: 10.1053/jhep.2002.33638
Hu, B., Huang, W., Wang, R., Zang, W., Su, M., Li, H., et al. (2020). High rate of detection of human ESPL1-HBV S Fusion Gene in Patients With HBV-related Liver Cancer: a Chinese Case–Control Study. Anticancer Res. 40, 245–252. doi: 10.21873/anticanres.13946
Hu, B., Wang, R., Fu, J., Su, M., Du, M., Liu, Y., et al. (2018). Integration of hepatitis B virus S gene impacts on hepatitis B surface antigen levels in patients with antiviral therapy. J. Gastroenterol. 33, 1389–1396. doi: 10.1111/jgh.14075
Huang, G., Lau, W. Y., Zhou, W.-P., Shen, F., Pan, Z.-Y., Yuan, S.-X., et al. (2014). Prediction of hepatocellular carcinoma recurrence in patients with low hepatitis B virus DNA levels and high preoperative hepatitis B surface antigen levels. JAMA Surg. 149, 519–527. doi: 10.1001/jamasurg.2013.4648
Huang, G., Yang, Y., Shen, F., Pan, Z.-Y., Fu, S.-Y., Lau, W. Y., et al. (2013). Early viral suppression predicts good postoperative survivals in patients with hepatocellular carcinoma with a high baseline HBV-DNA load. Ann. Surg. Oncol. 20, 1482–1490. doi: 10.1245/s10434-012-2803-7
Huang, H., Wang, J., Li, W., Chen, R., Chen, X., Zhang, F., et al. (2018). Serum HBV DNA plus RNA shows superiority in reflecting the activity of intrahepatic cccDNA in treatment-naive HBV-infected individuals. J. Clin. Virol. 99, 71–78. doi: 10.1016/j.jcv.2017.12.016
Huang, H.-L., Jeng, K.-S., Hu, C.-P., Tsai, C.-H., Lo, S. J., and Chang, C. (2000). Identification and characterization of a structural protein of hepatitis B virus: a polymerase and surface fusion protein encoded by a spliced RNA. Virology 275, 398–410. doi: 10.1006/viro.2000.0478
Huang, Y., Wang, Z., An, S., Zhou, B., Zhou, Y., Chan, H. L., et al. (2008). Role of hepatitis B virus genotypes and quantitative HBV DNA in metastasis and recurrence of hepatocellular carcinoma. J. Med. Virol. 80, 591–597. doi: 10.1002/jmv.21117
Huang, Y.-W., Takahashi, S., Tsuge, M., Chen, C.-L., Wang, T.-C., Abe, H., et al. (2015). On-treatment low serum HBV RNA level predicts initial virological response in chronic hepatitis B patients receiving nucleoside analogue therapy. Antivir. Ther. 20, 369–375. doi: 10.3851/imp2777
Iloeje, U. H., Yang, H. I., Su, J., Jen, C. L., You, S. L., and Chen, C. J. (2006). Predicting cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology 130, 678–686. doi: 10.1053/j.gastro.2005.11.016
Jang, J. W., Kim, J. S., Kim, H. S., Tak, K. Y., Nam, H., Sung, P. S., et al. (2020). Persistence of intrahepatic HBV DNA integration in patients developing hepatocellular carcinoma after HBsAg seroclearance. Clin. Mol. Hepatol. 27, 207–218. doi: 10.3350/cmh.2020.0115
Jones, S. A., and Hu, J. (2013). Hepatitis B virus reverse transcriptase: diverse functions as classical and emerging targets for antiviral intervention. Emerg. Microbes Infect. 2:e56.
Jung, K. S., Kim, S. U., Ahn, S. H., Park, Y. N., Kim, D. Y., Park, J. Y., et al. (2011). Risk assessment of hepatitis B virus–related hepatocellular carcinoma development using liver stiffness measurement (FibroScan). Hepatology 53, 885–894. doi: 10.1002/hep.24121
Kang, N. K., Chung, J. W., Jang, H. Y., Park, D. W., Jang, E. S., and Kim, J.-W. (2017). PO-021: role of HBsAg Titer and HBV DNA Level in Predicting Hepatocellular Carcinoma in Chronic Hepatitis B Patients Receiving Entecavir Therapy. Korean J. Hepatol. 2017, 94–94.
Kawai-Kitahata, F., Asahina, Y., Tanaka, S., Kakinuma, S., Murakawa, M., Nitta, S., et al. (2016). Comprehensive analyses of mutations and hepatitis B virus integration in hepatocellular carcinoma with clinicopathological features. J. Gastroenterol. 51, 473–486. doi: 10.1007/s00535-015-1126-4
Kawanaka, M., Nishino, K., Nakamura, J., Oka, T., Urata, N., Goto, D., et al. (2014). Quantitative levels of hepatitis B virus DNA and surface antigen and the risk of hepatocellular carcinoma in patients with hepatitis B receiving long-term nucleos (t) ide analogue therapy. Liver Cancer 3, 41–52. doi: 10.1159/000343857
Kew, M. C. (2010). Epidemiology of chronic hepatitis B virus infection, hepatocellular carcinoma, and hepatitis B virus-induced hepatocellular carcinoma. Pathol. Biol. 58, 273–277. doi: 10.1016/j.patbio.2010.01.005
Kim, B. K., Park, J. Y., Kim, D. Y., Kim, J. K., Kim, K. S., Choi, J. S., et al. (2008). Persistent hepatitis B viral replication affects recurrence of hepatocellular carcinoma after curative resection. Liver Int. 28, 393–401. doi: 10.1111/j.1478-3231.2007.01625.x
Kim, G.-A., Lee, H. C., Kim, M.-J., Ha, Y., Park, E. J., An, J., et al. (2015). Incidence of hepatocellular carcinoma after HBsAg seroclearance in chronic hepatitis B patients: a need for surveillance. J. Hepatol. 62, 1092–1099. doi: 10.1016/j.jhep.2014.11.031
Kim, J. H., Sinn, D. H., Kang, W., Gwak, G. Y., Paik, Y. H., Choi, M. S., et al. (2017). Low-level viremia and the increased risk of hepatocellular carcinoma in patients receiving entecavir treatment. Hepatology 66, 335–343. doi: 10.1002/hep.28916
Kimura, T., Rokuhara, A., Sakamoto, Y., Yagi, S., Tanaka, E., Kiyosawa, K., et al. (2002). Sensitive enzyme immunoassay for hepatitis B virus core-related antigens and their correlation to virus load. J. Clin. Microbiol. 40, 439–445. doi: 10.1128/jcm.40.2.439-445.2002
Kimura, T., Ohno, N., Terada, N., Rokuhara, A., Matsumoto, A., Yagi, S., et al. (2005). Hepatitis B virus DNA-negative dane particles lack core protein but contain a 22-kDa precore protein without C-terminal arginine-rich domain. J. Biol. Chem. 280, 21713–21719. doi: 10.1074/jbc.m501564200
Königer, C., Wingert, I., Marsmann, M., Rösler, C., Beck, J., and Nassal, M. (2014). Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc. Natl. Acad. Sci. U.S.A. 111, E4244–E4253.
Kremsdorf, D., Soussan, P., Paterlini-Brechot, P., and Brechot, C. (2006). Hepatitis B virus-related hepatocellular carcinoma: paradigms for viral-related human carcinogenesis. Oncogene 25, 3823–3833. doi: 10.1038/sj.onc.1209559
Kumada, T., Toyoda, H., Tada, T., Kiriyama, S., Tanikawa, M., Hisanaga, Y., et al. (2013). Effect of nucleos (t) ide analogue therapy on hepatocarcinogenesis in chronic hepatitis B patients: a propensity score analysis. J. Hepatol. 58, 427–433. doi: 10.1016/j.jhep.2012.10.025
Lam, A. M., Ren, S., Espiritu, C., Kelly, M., Lau, V., Zheng, L., et al. (2017). Hepatitis B virus capsid assembly modulators, but not nucleoside analogs, inhibit the production of extracellular pregenomic RNA and spliced RNA variants. Antimicrob. Agents Chemother. 61:e00680-17.
Lee, G. H., Wasser, S., and Lim, S. G. (2008). Hepatitis B pregenomic RNA splicing—the products, the regulatory mechanisms and its biological significance. Virus Res. 136, 1–7. doi: 10.1016/j.virusres.2008.05.007
Lee, H. W., Lee, J. I., Kim, S., Kim, S., Chang, H. Y., and Lee, K. S. (2020). Cumulative incidence of hepatocellular carcinoma and hepatitis B surface antigen Seroclearance after Nucleos (t) ide analogue-induced hepatitis B e antigen Seroclearance. BMC Gastroenterol. 20:113. doi: 10.1186/s12876-020-01236-9
Lee, J., Shin, M.-K., Lee, H.-J., Yoon, G., and Ryu, W.-S. J. (2004). Three novel cis-acting elements required for efficient plus-strand DNA synthesis of the hepatitis B virus genome. J. Virol. 78, 7455–7464. doi: 10.1128/jvi.78.14.7455-7464.2004
Levrero, M., and Zucman-Rossi, J. (2016). Mechanisms of HBV-induced hepatocellular carcinoma. J. Hepatol. 64, S84–S101.
Li, C. L., Ho, M. C., Lin, Y. Y., Tzeng, S. T., Chen, Y. J., Pai, H. Y., et al. (2020). Cell-Free Virus-Host Chimera DNA From Hepatitis B virus integration sites as a circulating biomarker of hepatocellular cancer. Hepatology 72, 2063–2076. doi: 10.1002/hep.31230
Li, D., Mallory, T., and Satomura, S. (2001). AFP-L3: a new generation of tumor marker for hepatocellular carcinoma. Clin. Chim. Acta 313, 15–19. doi: 10.1016/s0009-8981(01)00644-1
Li, M.-R., Chen, G.-H., Cai, C.-J., Wang, G.-Y., and Zhao, H. (2011). High hepatitis B virus DNA level in serum before liver transplantation increases the risk of hepatocellular carcinoma recurrence. Digestion 84, 134–141. doi: 10.1159/000324197
Li, W., Cui, X., Huo, Q., Qi, Y., Sun, Y., Tan, M., et al. (2019). Profile of HBV integration in the plasma DNA of hepatocellular carcinoma patients. Curr. Genomics 20, 61–68. doi: 10.2174/1389202919666181002144336
Liang, H.-W., Wang, N., Wang, Y., Wang, F., Fu, Z., Yan, X., et al. (2016). Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J. Hepatol. 64, 278–291. doi: 10.1016/j.jhep.2015.09.013
Liao, C.-C., Hsu, C.-W., Gu, P.-W., Yeh, C.-T., Lin, S.-M., and Chiu, C.-T. (2015). Comparison of the elecsys HBsAg II assay and the architect assay for quantification of hepatitis B surface antigen in chronic hepatitis B patients. Biomed. J. 38, 250–256. doi: 10.4103/2319-4170.143485
Lin, C. L., Liu, C. H., Chen, W., Huang, W. L., Chen, P. J., Lai, M. Y., et al. (2007). Association of pre-S deletion mutant of hepatitis B virus with risk of hepatocellular carcinoma. J. Gastroenterol. Hepatol. 22, 1098–1103. doi: 10.1111/j.1440-1746.2006.04515.x
Lin, S.-M., Yu, M.-L., Lee, C.-M., Chien, R.-N., Sheen, I.-S., Chu, C.-M., et al. (2007). Interferon therapy in HBeAg positive chronic hepatitis reduces progression to cirrhosis and hepatocellular carcinoma. J. Hepatol. 46, 45–52. doi: 10.1016/j.jhep.2006.08.021
Lin, X., Wen, Y., Wan, D., Qian, G., and Gu, J. J. (2002). Structural and functional analysis of 2.2 kb spliced variant of hepatitis B virus genomes isolated from liver tissues from hepatocellular carcinoma patients. Chin. J. Exp. Clin. Virol. 16, 11–15.
Liu, Y., Cathcart, A. L., Delaney, W. E. I. V., and Kitrinos, K. M. (2017). Development of a digital droplet PCR assay to measure HBV DNA in patients receiving long-term TDF treatment. J. Virol. Methods 249, 189–193. doi: 10.1016/j.jviromet.2017.09.015
Lu, F., Wang, J., Chen, X., Xu, D., and Xia, N. (2017). Potential use of serum HBV RNA in antiviral therapy for chronic hepatitis B in the era of nucleos (t) ide analogs. Front. Med. 11:502–508. doi: 10.1007/s11684-017-0590-z
Luo, C.-L., Rong, Y., Chen, H., Zhang, W.-W., Wu, L., Wei, D., et al. (2019). A logistic regression model for noninvasive prediction of AFP-negative hepatocellular carcinoma. Technol. Cancer Res. Treat. 18:1533033819846632.
Ma, N.-F., Lau, S. H., Hu, L., Xie, D., Wu, J., Yang, J., et al. (2008). COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin. Cancer Res. 14, 5061–5068. doi: 10.1158/1078-0432.ccr-07-5082
Mak, L. Y., Wong, D. H., Cheung, K. S., Seto, W. K., Lai, C. L., and Yuen, M. F. (2018). Review article: hepatitis B core-related antigen (HB crAg): an emerging marker for chronic hepatitis B virus infection. Aliment. Pharmacol. 47, 43–54. doi: 10.1111/apt.14376
Manno, M., Cammà, C., Schepis, F., Bassi, F., Gelmini, R., Giannini, F., et al. (2004). Natural history of chronic HBV carriers in northern Italy: morbidity and mortality after 30 years. Gastroenterology 127, 756–763. doi: 10.1053/j.gastro.2004.06.021
Mao, X., Tey, S. K., Ko, F. C. F., Kwong, E. M. L., Gao, Y., Ng, I. O., et al. (2019). C-terminal truncated HBx protein activates caveolin-1/LRP6/β-catenin/FRMD5 axis in promoting hepatocarcinogenesis. Cancer Lett. 444, 60–69. doi: 10.1016/j.canlet.2018.12.003
Marrero, J. A., Feng, Z., Wang, Y., Nguyen, M. H., Befeler, A. S., Roberts, L. R., et al. (2009). α-fetoprotein, des-γ carboxyprothrombin, and lectin-bound α-fetoprotein in early hepatocellular carcinoma. Gastroenterology 137, 110–118. doi: 10.1053/j.gastro.2009.04.005
Marrero, J. A., Kulik, L. M., Sirlin, C. B., Zhu, A. X., Finn, R. S., Abecassis, M. M., et al. (2018). Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American Association for the Study of Liver Diseases. Hepatology 68, 723–750. doi: 10.1002/hep.29913
Mason, W. S., Low, H. C., Xu, C., Aldrich, C. E., Scougall, C. A., Grosse, A., et al. (2009). Detection of clonally expanded hepatocytes in chimpanzees with chronic hepatitis B virus infection. Methods Mol. Biol. 83, 8396–8408. doi: 10.1128/jvi.00700-09
McGlynn, K. A., Petrick, J. L., and London, W. T. (2015). Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability. Clin. Liver Dis. 19, 223–238.
Messageot, F., Salhi, S., Eon, P., and Rossignol, J.-M. (2003). Proteolytic processing of the hepatitis B virus e antigen precursor: cleavage at two furin consensus sequences. J. Biol. Chem. 278, 891–895. doi: 10.1074/jbc.m207634200
Nakamoto, Y., Guidotti, L. G., Kuhlen, C. V., Fowler, P., and Chisari, F. V. (1998). Immune pathogenesis of hepatocellular carcinoma. J. Exp. Med. 188, 341–350.
Nakamura, S., Nouso, K., Sakaguchi, K., Ito, Y. M., Ohashi, Y., Kobayashi, Y., et al. (2006). Sensitivity and specificity of des-gamma-carboxy prothrombin for diagnosis of patients with hepatocellular carcinomas varies according to tumor size. Am. J. Gastroenterol. 101, 2038–2043. doi: 10.1111/j.1572-0241.2006.00681.x
Ng, K.-Y., Chai, S., Tong, M., Guan, X.-Y., Lin, C.-H., Ching, Y.-P., et al. (2016). C-terminal truncated hepatitis B virus X protein promotes hepatocellular carcinogenesis through induction of cancer and stem cell-like properties. Oncotarget 7, 24005–24017. doi: 10.18632/oncotarget.8209
Ni, Y., Lempp, F. A., Mehrle, S., Nkongolo, S., Kaufman, C., Fälth, M., et al. (2014). Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146, 1070–1083. doi: 10.1053/j.gastro.2013.12.024
Ou, Q., Guo, J., Zeng, Y., and Chen, H. (2020). Insights for clinical diagnostic indicators of virus and host in chronic hepatitis B infection. J. Viral Hepat. 27, 224–232. doi: 10.1111/jvh.13260
Ozakyol, A. (2017). Global epidemiology of hepatocellular carcinoma (HCC epidemiology). J. Gastrointest. Cancer 48, 238–240. doi: 10.1007/s12029-017-9959-0
Papatheodoridis, G. V., Chan, H. L.-Y., Hansen, B. E., Janssen, H. L., and Lampertico, P. (2015). Risk of hepatocellular carcinoma in chronic hepatitis B: assessment and modification with current antiviral therapy. J. Hepatol. 62, 956–967. doi: 10.1016/j.jhep.2015.01.002
Park, G.-S., Kim, H.-Y., Shin, H.-S., Park, S., Shin, H.-J., and Kim, K. (2008). Modulation of hepatitis B virus replication by expression of polymerase-surface fusion protein through splicing: implications for viral persistence. Virus Res. 136, 166–174. doi: 10.1016/j.virusres.2008.05.005
Paterlini-Brechot, P., Saigo, K., Murakami, Y., Chami, M., Gozuacik, D., Mugnier, C., et al. (2003). Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene Res. 22, 3911–3916. doi: 10.1038/sj.onc.1206492
Pesce, A., Scilletta, R., Branca, A., Nigro, L., Montineri, A., Larocca, L., et al. (2012). Does transient elastography (FibroScan®) have a role in decision making in hepatocellular carcinoma? HPB 14, 403–408. doi: 10.1111/j.1477-2574.2012.00465.x
Pfefferkorn, M., Schott, T., Böhm, S., Deichsel, D., Felkel, C., Gerlich, W. H., et al. (2021). Composition of HBsAg is predictive of HBsAg loss during treatment in patients with HBeAg-positive chronic hepatitis B. J. Hepatol. 74, 283–292. doi: 10.1016/j.jhep.2020.08.039
Pocha, C., Dieperink, E., McMaken, K., Knott, A., Thuras, P., and Ho, S. (2013). Surveillance for hepatocellular cancer with ultrasonography vs. computed tomography–a randomised study. Aliment. Pharmacol. Ther. 38, 303–312. doi: 10.1111/apt.12370
Prakash, K., Rydell, G. E., Larsson, S. B., Andersson, M., Norkrans, G., Norder, H., et al. (2018). High serum levels of pregenomic RNA reflect frequently failing reverse transcription in hepatitis B virus particles. Virol J. 15:86.
Qi, Y., Gao, Z., Xu, G., Peng, B., Liu, C., Yan, H., et al. (2016). DNA polymerase κ is a key cellular factor for the formation of covalently closed circular DNA of hepatitis B virus. PLoS Pathog. 12:e1005893. doi: 10.1371/journal.ppat.1005893
Raffetti, E., Fattovich, G., and Donato, F. (2016). Incidence of hepatocellular carcinoma in untreated subjects with chronic hepatitis B: a systematic review and meta-analysis. Liver Int. 36, 1239–1251. doi: 10.1111/liv.13142
Revill, P. A., Tu, T., Netter, H. J., Yuen, L. K., Locarnini, S. A., and Littlejohn, M. (2020). The evolution and clinical impact of hepatitis B virus genome diversity. Nat. Rev. Gastroenterol. Hepatol. 17, 618–634. doi: 10.1038/s41575-020-0296-6
Roingeard, P., and Sureau, C. (1998). Ultrastructural analysis of hepatitis B virus in HepG2-transfected cells with special emphasis on subviral filament morphogenesis. Hepatology 28, 1128–1133. doi: 10.1002/hep.510280431
Rokuhara, A., Tanaka, E., Matsumoto, A., Kimura, T., Yamaura, T., Orii, K., et al. (2003). Clinical evaluation of a new enzyme immunoassay for hepatitis B virus core-related antigen; a marker distinct from viral DNA for monitoring lamivudine treatment. J. Viral Hepat. 10, 324–330. doi: 10.1046/j.1365-2893.2003.00437.x
Rydell, G. E., Prakash, K., Norder, H., and Lindh, M. (2017). Hepatitis B surface antigen on subviral particles reduces the neutralizing effect of anti-HBs antibodies on hepatitis B viral particles in vitro. Virology 509, 67–70. doi: 10.1016/j.virol.2017.05.017
Saldanha, J., Gerlich, W., Lelie, N., Dawson, P., Heermann, K., Heath, A., et al. (2001). An international collaborative study to establish a World Health Organization international standard for hepatitis B virus DNA nucleic acid amplification techniques. Vox Sang. 80, 63–71. doi: 10.1046/j.1423-0410.2001.00003.x
Scotto, J., Hadchouel, M., Hery, C., Alvarez, F., Yvart, J., Tiollais, P., et al. (1983). Hepatitis B virus DNA in children’s liver diseases: detection by blot hybridisation in liver and serum. Gut Pathog. 24, 618–624. doi: 10.1136/gut.24.7.618
Singal, A., Volk, M., Waljee, A., Salgia, R., Higgins, P., Rogers, M., et al. (2009). Meta-analysis: surveillance with ultrasound for early-stage hepatocellular carcinoma in patients with cirrhosis. Aliment. Pharmacol. 30, 37–47. doi: 10.1111/j.1365-2036.2009.04014.x
Sohn, W., Paik, Y.-H., Kim, J. M., Kwon, C. H., Joh, J. W., Cho, J. Y., et al. (2014). HBV DNA and HBsAg levels as risk predictors of early and late recurrence after curative resection of HBV-related hepatocellular carcinoma. Ann. Surg. Oncol. 21, 2429–2435. doi: 10.1245/s10434-014-3621-x
Soussan, P., Garreau, F., Zylberberg, H., Ferray, C., Brechot, C., and Kremsdorf, D. (2000). In vivo expression of a new hepatitis B virus protein encoded by a spliced RNA. J. Clin. Invest. 105, 55–60. doi: 10.1172/jci8098
Soussan, P., Pol, J., Garreau, F., Schneider, V., Le Pendeven, C., Nalpas, B., et al. (2008). Expression of defective hepatitis B virus particles derived from singly spliced RNA is related to liver disease. J. Infect. Dis. 198, 218–225. doi: 10.1086/589623
Soussan, P., Tuveri, R., Nalpas, B., Garreau, F., Zavala, F., Masson, A., et al. (2003). The expression of hepatitis B spliced protein (HBSP) encoded by a spliced hepatitis B virus RNA is associated with viral replication and liver fibrosis. J. Hepatol. 38, 343–348. doi: 10.1016/s0168-8278(02)00422-1
Su, C.-W., Chiou, Y.-W., Tsai, Y.-H., Teng, R.-D., Chau, G.-Y., Lei, H.-J., et al. (2013). The influence of hepatitis B viral load and pre-S deletion mutations on post-operative recurrence of hepatocellular carcinoma and the tertiary preventive effects by anti-viral therapy. PLoS One 8:e66457. doi: 10.1371/journal.pone.0066457
Su, T.-S., Lai, C.-J., Huang, J., Lin, L.-H., Yauk, Y., Chang, C., et al. (1989). Hepatitis B virus transcript produced by RNA splicing. J. Virol. 63, 4011–4018. doi: 10.1128/jvi.63.9.4011-4018.1989
Sung, W.-K., Zheng, H., Li, S., Chen, R., Liu, X., Li, Y., et al. (2012). Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 44, 765–769.
Suzuki, Y., Maekawa, S., Komatsu, N., Sato, M., Tatsumi, A., Miura, M., et al. (2019). Hepatitis B virus (HBV)-infected patients with low hepatitis B surface antigen and high hepatitis B core-related antigen titers have a high risk of HBV-related hepatocellular carcinoma. Hepatol. Res. 49, 51–63. doi: 10.1111/hepr.13277
Sze, K. M., Chu, G. K., Lee, J. M., and Ng, I. O. L. (2013). C-terminal truncated hepatitis B virus x protein is associated with metastasis and enhances invasiveness by C-Jun/matrix metalloproteinase protein 10 activation in hepatocellular carcinoma. Hepatol. Int. 57, 131–139. doi: 10.1002/hep.25979
Tada, T., Kumada, T., Toyoda, H., Kiriyama, S., Tanikawa, M., Hisanaga, Y., et al. (2016). HBcrAg predicts hepatocellular carcinoma development: an analysis using time-dependent receiver operating characteristics. J. Hepatol. 65, 48–56. doi: 10.1016/j.jhep.2016.03.013
Teng, C.-F., Li, T.-C., Huang, H.-Y., Chan, W.-L., Wu, H.-C., Shyu, W.-C., et al. (2020a). Hepatitis B virus pre-S2 deletion (nucleotide 1 to 54) in plasma predicts recurrence of hepatocellular carcinoma after curative surgical resection. PLoS One 15:e0242748. doi: 10.1371/journal.pone.0242748
Teng, C.-F., Li, T.-C., Huang, H.-Y., Lin, J.-H., Chen, W.-S., Shyu, W.-C., et al. (2020b). Next-generation sequencing-based quantitative detection of hepatitis B virus pre-S mutants in plasma predicts hepatocellular carcinoma recurrence. Viruses 12:796. doi: 10.3390/v12080796
Terre, S., Petit, M., and Brechot, C. J. (1991). Defective hepatitis B virus particles are generated by packaging and reverse transcription of spliced viral RNAs in vivo. J. Virol. 65, 5539–5543. doi: 10.1128/jvi.65.10.5539-5543.1991
Testoni, B., Lebossé, F., Scholtes, C., Berby, F., Miaglia, C., Subic, M., et al. (2019). Serum hepatitis B core-related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. J. Hepatol. 70, 615–625. doi: 10.1016/j.jhep.2018.11.030
Tong, S., and Revill, P. (2016). Overview of hepatitis B viral replication and genetic variability. J. Hepatol. 64, S4–S16.
Trung, N. T., Giang, D. P., Hoan, P. Q., Binh, M. T., Hoan, N. X., Toan, N. L., et al. (2019). No expression of HBV-human chimeric fusion transcript (HBx-LINE1) among Vietnamese patients with HBV-associated hepatocellular carcinoma. Ann. Hepatol. 18, 404–405. doi: 10.1016/j.aohep.2019.02.002
Tseng, T.-C., Liu, C.-J., Hsu, C.-Y., Hong, C.-M., Su, T.-H., Yang, W.-T., et al. (2019). High Level of Hepatitis B Core–Related Antigen Associated With Increased Risk of Hepatocellular Carcinoma in Patients With Chronic HBV Infection of Intermediate Viral Load. Gastroenterol. Hepatol. 157, 1518–1529.e3.
Tseng, T. C., Liu, C. J., Yang, H. C., Su, T. H., Wang, C. C., Chen, C. L., et al. (2012). High levels of hepatitis B surface antigen increase risk of hepatocellular carcinoma in patients with low HBV load. Gastroenterology 142, 1140–1149.e3.
Tu, H., Bonura, C., Giannini, C., Mouly, H., Soussan, P., Kew, M., et al. (2001). Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 61, 7803–7810.
Tu, T., Block, J. M., Wang, S., Cohen, C., and Douglas, M. W. (2020a). The lived experience of chronic hepatitis B: a broader view of its impacts and why we need a cure. Viruses 12:515. doi: 10.3390/v12050515
Tu, T., Budzinska, M. A., Shackel, N. A., and Urban, S. (2017). HBV DNA integration: molecular mechanisms and clinical implications. Viruses 9:75. doi: 10.3390/v9040075
Tu, T., and Urban, S. (2018). Virus entry and its inhibition to prevent and treat hepatitis B and hepatitis D virus infections. Curr. Opin. Virol. 30, 68–79. doi: 10.1016/j.coviro.2018.04.004
Tu, T., Zehnder, B., Levy, M., Micali, G., Tran, L., Dabere, O., et al. (2019). Hepatitis B virus (HBV) DNA integration is not driven by viral proteins. Z. Gastroenterol. 57:e92.
Tu, T., Zehnder, B., Qu, B., Ni, Y., Main, N., Allweiss, L., et al. (2020b). A novel method to precisely quantify hepatitis B virus covalently closed circular (ccc) DNA formation and maintenance. Antiviral Res. 181:104865. doi: 10.1016/j.antiviral.2020.104865
Tu, T., Zehnder, B., Qu, B., and Urban, S. (2021). De novo synthesis of hepatitis B virus nucleocapsids is dispensable for the maintenance and transcriptional regulation of cccDNA. JHEP Rep. 3:100195. doi: 10.1016/j.jhepr.2020.100195
van Bömmel, F., Bartens, A., Mysickova, A., Hofmann, J., Krüger, D. H., Berg, T., et al. (2015). Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology 61, 66–76. doi: 10.1002/hep.27381
van Campenhout, M. J., van Bömmel, F., Pfefferkorn, M., Fischer, J., Deichsel, D., Boonstra, A., et al. (2018). Host and viral factors associated with serum hepatitis B virus RNA levels among patients in need for treatment. Hepatology 68, 839–847. doi: 10.1002/hep.29872
Varbobitis, I., and Papatheodoridis, G. V. (2016). The assessment of hepatocellular carcinoma risk in patients with chronic hepatitis B under antiviral therapy. Clin. Mol. Hepatol. 22, 319–326. doi: 10.3350/cmh.2016.0045
Venkatakrishnan, B., and Zlotnick, A. (2016). The structural biology of hepatitis B virus: form and function. Annu. Rev. Virol. 3, 429–451. doi: 10.1146/annurev-virology-110615-042238
Vlachogiannakos, J., and Papatheodoridis, G. (2013). Hepatocellular carcinoma in chronic hepatitis B patients under antiviral therapy. World J. Gastroenterol. 19, 8822–8830. doi: 10.3748/wjg.v19.i47.8822
Volk, M. L., Hernandez, J. C., Su, G. L., Lok, A. S., and Marrero, J. A. (2007). Risk factors for hepatocellular carcinoma may impair the performance of biomarkers: a comparison of AFP, DCP, and AFP-L3 1. Cancer Biomark. 3, 79–87. doi: 10.3233/cbm-2007-3202
Wang, H. C., Huang, W., Lai, M. D., and Su, I. J. (2006). Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci. 97, 683–688. doi: 10.1111/j.1349-7006.2006.00235.x
Wang, H.-C., Wu, H.-C., Chen, C.-F., Fausto, N., Lei, H.-Y., and Su, I.-J. (2003). Different types of ground glass hepatocytes in chronic Hepatitis B virus infection contain specific Pre-S Mutants that May induce endoplasmic reticulum stress. Am. J. Pathol. 163, 2441–2449. doi: 10.1016/s0002-9440(10)63599-7
Wang, J., Du, M., Huang, H., Chen, R., Niu, J., Jiang, J., et al. (2017). Reply to:“Serum HBV pgRNA as a clinical marker for cccDNA activity”: consistent loss of serum HBV RNA might predict the “para-functional cure” of chronic hepatitis B. J. Hepatol. 66, 462–463. doi: 10.1016/j.jhep.2016.10.034
Wang, J., Shen, T., Huang, X., Kumar, G. R., Chen, X., Zeng, Z., et al. (2016). Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J. Hepatol. 65, 700–710. doi: 10.1016/j.jhep.2016.05.029
Wang, J., Yu, Y., Li, G., Shen, C., Meng, Z., Zheng, J., et al. (2018). Relationship between serum HBV-RNA levels and intrahepatic viral as well as histologic activity markers in entecavir-treated patients. J. Hepatol. 68, 16–24. doi: 10.1016/j.jhep.2017.08.021
Wang, X., Zhang, Y., Yang, N., He, H., Tao, X., Kou, C., et al. (2020). Evaluation of the Combined Application of AFP, AFP-L3%, and DCP for Hepatocellular Carcinoma Diagnosis: a Meta-analysis. Biomed Res. Int. 2020:5087643.
Wang, Y., Lau, S. H., Sham, J. S., Wu, M.-C., Wang, T., and Guan, X.-Y. (2004). Characterization of HBV integrants in 14 hepatocellular carcinomas: association of truncated X gene and hepatocellular carcinogenesis. Oncogene 23, 142–148. doi: 10.1038/sj.onc.1206889
Wang, Y.-C., Li, C.-L., Ho, M.-C., Lin, Y.-Y., Tzeng, S.-T., Chen, Y.-J., et al. (2019). Cell-Free Junctional DNA Fragment from Hepatitis B Virus Integration in HCC for Monitoring Postresection Recurrence and Clonality. (Alexandria, VA: American Society of Clinical Oncology), 4090–4090.
Wei, Y., Neuveut, C., Tiollais, P., and Buendia, M.-A. (2010). Molecular biology of the hepatitis B virus and role of the X gene. Pathol. Biol. 58, 267–272. doi: 10.1016/j.patbio.2010.03.005
Wong, D., Seto, W. K., Cheung, K. S., Chong, C. K., Huang, F. Y., Fung, J., et al. (2017). Hepatitis B virus core-related antigen as a surrogate marker for covalently closed circular DNA. Liver Int. 37, 995–1001. doi: 10.1111/liv.13346
Wong, D. K., Cheng, S. Y., Mak, L. L., To, E. W., Lo, R. C., Cheung, T.-T., et al. (2020). Among patients with undetectable hepatitis B surface antigen and hepatocellular carcinoma, a high proportion has integration of HBV DNA into hepatocyte DNA and no cirrhosis. Clin. Gastroenterol. Hepatol. 18, 449–456. doi: 10.1016/j.cgh.2019.06.029
Wooddell, C. I., Yuen, M.-F., Chan, H. L.-Y., Gish, R. G., Locarnini, S. A., Chavez, D., et al. (2017). RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci. Transl. Med. 9:eaan0241. doi: 10.1126/scitranslmed.aan0241
Wu, J.-C., Huang, Y.-H., Chau, G.-Y., Su, C.-W., Lai, C.-R., Lee, P.-C., et al. (2009). Risk factors for early and late recurrence in hepatitis B-related hepatocellular carcinoma. J. Hepatol. 51, 890–897. doi: 10.1016/j.jhep.2009.07.009
Wungu, C. D. K., Ariyanto, F. C., Prabowo, G. I., Soetjipto, S., and Handajani, R. (2021). Meta-analysis: association between hepatitis B virus preS mutation and hepatocellular carcinoma risk. J. Viral Hepat. 28, 61–71. doi: 10.1111/jvh.13402
Xiangji, L., Feng, X., Qingbao, C., Weifeng, T., Xiaoqing, J., Baihe, Z., et al. (2011). Knockdown of HBV surface antigen gene expression by a lentiviral microRNA-based system inhibits HBV replication and HCC growth. J. Viral Hepat. 18, 653–660. doi: 10.1111/j.1365-2893.2010.01346.x
Xie, J.-X., Zhao, J., Yin, J.-H., Zhang, Q., Pu, R., Lu, W.-Y., et al. (2010). Association of novel mutations and heplotypes in the preS region of hepatitis B virus with hepatocellular carcinoma. Front. Med. China 4, 419–429. doi: 10.1007/s11684-010-0160-0
Xu, W., Yu, J., and Wong, V. W.-S. (2017). Mechanism and prediction of HCC development in HBV infection. Best Pract. Res. Clin. Gastroenterol. 31, 291–298. doi: 10.1016/j.bpg.2017.04.011
Yan, H., Peng, B., Liu, Y., Xu, G., He, W., Ren, B., et al. (2014). Viral entry of hepatitis B and D viruses and bile salts transportation share common molecular determinants on sodium taurocholate cotransporting polypeptide. J. Virol. 88, 3273–3284. doi: 10.1128/jvi.03478-13
Yan, H., Yang, Y., Zhang, L., Tang, G., Wang, Y., Xue, G., et al. (2015). Characterization of the genotype and integration patterns of hepatitis B virus in early- and late-onset hepatocellular carcinoma. Hepatology 61, 1821–1831. doi: 10.1002/hep.27722
Yan, H., Zhong, G., Xu, G., He, W., Jing, Z., Gao, Z., et al. (2012). Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1:e00049.
Yang, F., Yan, S., He, Y., Wang, F., Song, S., Guo, Y., et al. (2008). Expression of hepatitis B virus proteins in transgenic mice alters lipid metabolism and induces oxidative stress in the liver. J. Hepatol. 48, 12–19. doi: 10.1016/j.jhep.2007.06.021
Yang, J. D., Kim, W. R., Coelho, R., Mettler, T. A., Benson, J. T., Sanderson, S. O., et al. (2011). Cirrhosis is present in most patients with hepatitis B and hepatocellular carcinoma. Clin. Gastroenterol. Hepatol. 9, 64–70. doi: 10.1016/j.cgh.2010.08.019
Yang, J. D., and Roberts, L. R. (2010). Hepatocellular carcinoma: a global view. Nat. Rev. Gastroenterol. Hepatol. 7, 448–458. doi: 10.1038/nrgastro.2010.100
Yang, W., and Summers, J. (1999). Integration of hepadnavirus DNA in infected liver: evidence for a linear precursor. J. Virol. 73, 9710–9717. doi: 10.1128/jvi.73.12.9710-9717.1999
Yang, W., and Summers, J. J. (1998). Infection of ducklings with virus particles containing linear double-stranded duck hepatitis B virus DNA: illegitimate replication and reversion. J. Virol. 72, 8710–8717. doi: 10.1128/jvi.72.11.8710-8717.1998
Yeh, M.-L., Huang, C.-F., Hsieh, M.-Y., Huang, J.-F., Dai, C.-Y., Yu, M.-L., et al. (2014). Comparison of the Abbott RealTime HBV assay with the Roche Cobas AmpliPrep/Cobas TaqMan HBV assay for HBV DNA detection and quantification. J. Clin. Virol. 60, 206–214. doi: 10.1016/j.jcv.2014.04.008
Yen, C. J., Ai, Y. L., Tsai, H. W., Chan, S. H., Yen, C. S., Cheng, K. H., et al. (2018). Hepatitis B virus surface gene pre-S2 mutant as a high-risk serum marker for hepatoma recurrence after curative hepatic resection. Hepatology 68, 815–826. doi: 10.1002/hep.29790
Yip, T. C., Chan, H. L., Wong, V. W., Tse, Y.-K., Lam, K. L., and Wong, G. L. (2017). Impact of age and gender on risk of hepatocellular carcinoma after hepatitis B surface antigen seroclearance. J. Hepatol. 67, 902–908. doi: 10.1016/j.jhep.2017.06.019
Yip, T. C., Wong, G. L., Chan, H. L., Tse, Y.-K., Lam, K. L., Lui, G. C., et al. (2019). HBsAg seroclearance further reduces hepatocellular carcinoma risk after complete viral suppression with nucleos(t)ide analogues. J. Hepatol. 70, 361–370. doi: 10.1016/j.jhep.2018.10.014
Yu, J., Ye, Y., Liu, J., Xu, Y., Lou, B., Zhu, J., et al. (2019). The role of hepatitis B core-related antigen in predicting hepatitis B virus recurrence after liver transplantation. Aliment. Pharmacol. Ther. 50, 1025–1036. doi: 10.1111/apt.15429
Zhang, B.-H., Yang, B.-H., and Tang, Z.-Y. (2004). Randomized controlled trial of screening for hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 130, 417–422.
Zhang, L., Xie, X.-Y., Chen, Y., Ge, N.-L., Chen, R.-X., Gan, Y.-H., et al. (2017). Hepatitis B surface antigen predicts recurrence after radiofrequency ablation in patients with low hepatitis B virus loads. Medicine 96:e9377. doi: 10.1097/md.0000000000009377
Zhang, W., Hacker, H. J., Mildenberger, M., Su, Q., and Schröder, C. H. (2004). Detection of HBV RNA in serum of patients. Methods Mol. Med. 95, 29–40. doi: 10.1385/1-59259-669-x:29
Zhao, L.-H., Liu, X., Yan, H.-X., Li, W.-Y., Zeng, X., Yang, Y., et al. (2016). Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat. Commun. 7:12992.
Zhao, X.-L., Yang, J.-R., Lin, S.-Z., Ma, H., Guo, F., Yang, R.-F., et al. (2016). Serum viral duplex-linear DNA proportion increases with the progression of liver disease in patients infected with HBV. Gut 65, 502–511. doi: 10.1136/gutjnl-2014-308989
Keywords: hepatitis B, hepatocellular carcinoma, biomarkers, HBV surface antigen (HBsAg), HBV DNA integration, HBV RNA, HBcr antigen
Citation: Liu Y, Veeraraghavan V, Pinkerton M, Fu J, Douglas MW, George J and Tu T (2021) Viral Biomarkers for Hepatitis B Virus-Related Hepatocellular Carcinoma Occurrence and Recurrence. Front. Microbiol. 12:665201. doi: 10.3389/fmicb.2021.665201
Received: 07 February 2021; Accepted: 06 May 2021;
Published: 14 June 2021.
Edited by:
Patrick Kennedy, Queen Mary University of London, United KingdomReviewed by:
Anna Salvetti, INSERM U1111, Centre International de Recherche en Infectiologie (CIRI), Université de Lyon (UCBL1), CNRS UMR 5308, FranceLin Deng, Kobe University, Japan
Copyright © 2021 Liu, Veeraraghavan, Pinkerton, Fu, Douglas, George and Tu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Tu, dC50dUBzeWRuZXkuZWR1LmF1
 Yuanyuan Liu
Yuanyuan Liu Vaishnavi Veeraraghavan2,3
Vaishnavi Veeraraghavan2,3 Thomas Tu
Thomas Tu