
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 14 May 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.662575
This article is part of the Research TopicBacterial Pathogen Genomics: Recent Achievements, Current Applications and Future ChallengesView all 16 articles
 Elias Eger1
Elias Eger1 Stefan E. Heiden1
Stefan E. Heiden1 Katja Korolew2
Katja Korolew2 Claude Bayingana3
Claude Bayingana3 Jules M. Ndoli3,4
Jules M. Ndoli3,4 Augustin Sendegeya3,4
Augustin Sendegeya3,4 Jean Bosco Gahutu3,4†
Jean Bosco Gahutu3,4† Mathis S. E. Kurz5
Mathis S. E. Kurz5 Frank P. Mockenhaupt5
Frank P. Mockenhaupt5 Julia Müller1
Julia Müller1 Stefan Simm2
Stefan Simm2 Katharina Schaufler1*
Katharina Schaufler1*Multi-drug resistant (MDR), gram-negative Enterobacteriaceae, such as Escherichia coli (E. coli) limit therapeutic options and increase morbidity, mortality, and treatment costs worldwide. They pose a serious burden on healthcare systems, especially in developing countries like Rwanda. Several studies have shown the effects caused by the global spread of extended-spectrum beta-lactamase (ESBL)-producing E. coli. However, limited data is available on transmission dynamics of these pathogens and the mobile elements they carry in the context of clinical and community locations in Sub-Saharan Africa. Here, we examined 120 ESBL-producing E. coli strains from patients hospitalized in the University Teaching Hospital of Butare (Rwanda), their attending caregivers as well as associated community members and livestock. Based on whole-genome analysis, the genetic diversification and phylogenetics were assessed. Moreover, the content of carried plasmids was characterized and investigated for putative transmission among strains, and for their potential role as drivers for the spread of antibiotic resistance. We show that among the 30 different sequence types (ST) detected were the pandemic clonal lineages ST131, ST648 and ST410, which combine high-level antimicrobial resistance with virulence. In addition to the frequently found resistance genes blaCTX–M–15, tet(34), and aph(6)-Id, we identified csg genes, which are required for curli fiber synthesis and thus biofilm formation. Numerous strains harbored multiple virulence-associated genes (VAGs) including pap (P fimbriae adhesion cluster), fim (type I fimbriae) and chu (Chu heme uptake system). Furthermore, we found phylogenetic relationships among strains from patients and their caregivers or related community members and animals, which indicates transmission of pathogens. Also, we demonstrated the presence and potential transfer of identical/similar ESBL-plasmids in different strains from the Rwandan setting and when compared to an external plasmid. This study highlights the circulation of clinically relevant, pathogenic ESBL-producing E. coli among patients, caregivers and the community in Rwanda. Combining antimicrobial resistance with virulence in addition to the putative exchange of mobile genetic elements among bacterial pathogens poses a significant risk around the world.
The versatility of Escherichia coli (E. coli) is based on the diversity of genetic substructures within this species (Whittam et al., 1983). In addition to commensal strains, which are an essential part of the non-anaerobic intestinal microflora of humans, other mammals and birds, pathogenic variants occur. The dissimilarity of these pathotypes depends also on their virulence attributes, resulting in a wide range of pathologies in both humans and animals. The intestinal pathogenic E. coli (InPEC) express characteristic virulence factors that allow to adhere and invade intestinal cells, causing specific enteric and diarrheal diseases. While InPEC are obligate pathogens, extraintestinal pathogenic E. coli (ExPEC) are part of the intestinal microbiome but exhibit a heterogeneous composition of virulence factors to colonize niches such as the urinary tract (Kaper et al., 2004). They can thus cause infections in almost any organ or non-intestinal site, regardless of the state of the host’s immune system (Russo and Johnson, 2000). However, a strict differentiation of pathogenic and commensal E. coli is difficult, provided by their rapid geno- and phenotypic adaptation to changing environmental conditions, for example through horizontal gene transfer (Pallen and Wren, 2007). Despite the plasticity of the genome, phylogenetic studies have shown some clonality within the population structure of E. coli, from which seven distinct phylogenetic groups were derived (Jaureguy et al., 2008; Touchon et al., 2009; Clermont et al., 2013). Usually, commensal strains and obligatory pathogens belong to the phylogroups A and B1, whereas strains with extended virulent attributes (mainly ExPEC) are part of the phylogroups B2, D, and F, with the latter as a sister group of B2 (Escobar-Páramo et al., 2004; Clermont et al., 2013). Multi-locus sequence typing (MLST) allows additional classification and several phylogenetic studies suggest the spread of pandemic high-risk clonal lineages including primarily sequence type (ST) 131 (Nicolas-Chanoine et al., 2008; Ewers et al., 2010; Hussain et al., 2012), ST648 (Ewers et al., 2014; Schaufler et al., 2019), ST410 (Schaufler et al., 2016b; Zurita et al., 2020), putatively ST405 (Manges et al., 2019), and others.
The management of zoonotic infections caused by antibiotic-resistant bacteria has become a multidisciplinary challenge for all modern healthcare systems and is nowadays often approached in a holistic One Health context. Bacterial pathogens spread through direct contact among humans and animals, indirectly by (environmental) pollution and also through non-living and living vectors (Rahman et al., 2020). One example for the latter are houseflies, which have been demonstrated to carry antibiotic-resistant pathogens including extended-spectrum beta-lactamases-(ESBL)-producing E. coli (Rahuma et al., 2005; Heiden et al., 2020b; Tufa et al., 2020) non-susceptible to third-generation cephalosporins (e.g., cefotaxime) and monobactams (e.g., aztreonam) (Bevan et al., 2017). Notably, ESBL enzyme production is often accompanied by cross- and co-resistances (Cantón and Coque, 2006; Hidron et al., 2008; Pitout, 2012) resulting in multi-drug resistant (MDR) representatives (Beceiro et al., 2013).
Both pandemic clonal lineages including the aforementioned ST131, ST648 and others, as well as mobile genetic elements (i.e., ESBL-plasmids) drive the spread of antibiotic resistance and virulence-associated genes (VAGs) (Cantón and Coque, 2006). Interestingly, previous studies suggest that ESBL-plasmid carriage does not ineluctably reduce bacterial fitness, which seems particularly true for specific clonal lineages (McNally et al., 2016; Schaufler et al., 2016a; Ranjan et al., 2018; Monárrez et al., 2019; Schaufler et al., 2019). The combination of MDR with high-level bacterial virulence and fitness leads to the emergence of these pandemic, high-risk clonal lineages and subsequently contributes to treatment failures, increasing morbidity, and mortality (Melzer and Petersen, 2007; Schwaber and Carmeli, 2007; Tumbarello et al., 2007; Beceiro et al., 2013; Schaufler et al., 2019; Heiden et al., 2020a).
The One Health concept—addressing human, animal and environmental health—encounters some challenges, especially in low-income countries like Sub-Saharan Africa/Rwanda. On the one hand, the lack of surveillance systems may result in inadequate establishment and implementation of hygienic strategies and therapy guidelines (Muvunyi et al., 2011; Ntirenganya et al., 2015; Carroll et al., 2016). On the other hand, uncontrolled over-the-counter sale of partially counterfeit and substandard antibiotic drugs (Kayumba et al., 2004; Carroll et al., 2016) as well as close human-livestock contact and household crowding might contribute to the broad occurrence and interspecies transmission of MDR bacteria in Sub-Saharan Africa.
This study aimed to investigate whether (i) ESBL-producing E. coli circulate among patients, caregivers, the community, and animals in Rwanda, (ii) some of these belong to pandemic high-risk clonal lineages and how they are phylogenetically related, (iii) they demonstrate virulence-associated features, (iv) their mobile genetic elements contribute to the spread of antibiotic resistance.
The E. coli strains investigated in this study were sampled over a time period of 8 weeks at the University Teaching Hospital of Butare (Rwanda) in 2014 (previously described by Kurz et al., 2017). Rectal swabs (Sarstedt AG & Co. KG, Nümbrecht, Germany) were collected from patients and caregivers at admission and discharge as well as from several community members and animals. Each patient had their own caregiver, who were usually relatives accompanying the patient upon admission. They stayed in the patient’s room and were involved in personal care of the patient and food preparation. This is a common practice in African hospitals (Hoffman et al., 2012; Ugochukwu, 2013). Sample groups consisting of a patient and related caregiver, and associated family members, neighbors and/or pets were included in the same study-ID. The samples were plated onto chromogenic agar (CHROMagar-ESBL, Mast Diagnostica GmbH, Reinfeld, Germany) supplemented with 2 μg/mL cefotaxime (Cayman Chemical Company, Ann Arbor, United States) and incubated at 37°C. For putative ESBL-positive colonies, the production of ESBL and/or ampicillinase (AmpC) was verified (ESBL-AmpC-Detection Test, Mast Diagnostica GmbH, Reinfeld, Germany) and all strains positive for AmpC only were excluded. The strains were stored at –80°C in LB broth (Carl Roth GmbH & Co. KG, Karlsruhe, Germany) supplemented with 20% (V/V) glycerol (Merck KGaA, Darmstadt, Germany). Originally, we have obtained overall 289 ESBL-producing E. coli strains (from patients, caregivers, community members, and animals), with 120 selected strains (based on related study-IDs) that were whole-genome sequenced. Additionally, flies caught with fly traps at different wards of the hospital examined in a previous study (Heiden et al., 2020b) were partly included in this study (Supplementary Table 1).
One single E. coli colony was cultured in LB broth supplemented with 2 μg/mL cefotaxime overnight and the total DNA was extracted using the MasterPureTM DNA Purification Kit for Blood, Version II (Lucigen, Middleton, United States) according to the manufacturer’s instructions. DNA was purity-controlled and quantified using NanoDropTM 2000 (Thermo Fisher Scientific Inc., Waltham, United States). WGS was performed in collaboration with LGC (LGC Genomics GmbH, Berlin, Germany) with 150 bp paired-end-reads using Illumina NextSeq 500/550 V2.
Raw reads were quality-trimmed, adapter-trimmed and contaminant-filtered using BBDuk from BBTools v. 38.861. After de novo assembly (at a maximum coverage of 100×) using shovill v. 1.1.02 in combination with SPAdes v. 3.14.1 (Bankevich et al., 2012), draft genomes were polished by mapping all trimmed reads back to the contigs with bwa v. 0.7.17 (Li and Durbin, 2009), processing SAM/BAM files marking optical duplicates with Samtools v. 1.10 (Li et al., 2009) and calling variants with Pilon v. 1.23 (Walker et al., 2014) (Supplementary Table 2).
Plasmid sequences of all strains were manually extracted using similarity searches (BLASTN Megablast) against the NCBI nucleotide collection for visualization in BRIG v. 0.95-dev.0004 (Alikhan et al., 2011). The in silico MLST, antibiotic resistance/virulence gene and single-nucleotide polymorphism (SNP) detection was carried out using mlst v. 2.19.03, ABRicate v. 1.0.04, and snippy v. 4.6.05. We inferred core SNP phylogenies for pandemic sequence types. Alignments were filtered for recombinations using Gubbins v. 2.4.1 (Croucher et al., 2015) and core SNPs extracted using snp-sites v. 2.5.1 (Page et al., 2016) [core SNP sites filtered out: 1,219 (ST131); 0 (ST354); 5,370 (ST405); 2,940 (ST410); 3,564 (ST648)]. The final core SNP alignments contained 208 (ST131; reference: PBIO440), 20 (ST354; reference: PBIO388), 177 (ST405; reference: PBIO397), 154 (ST410; reference: PBIO289), and 135 (ST648; reference: PBIO368) sites (Supplementary Table 3). Maximum likelihood trees were inferred with RAxML-NG v. 1.0.0 (Kozlov et al., 2019) using GTR + G (discrete GAMMA model of rate heterogeneity with 4 categories) and searching from 500 random and 500 parsimony-based starting trees. The best-scoring maximum likelihood tree supplemented with support values from 1,000 non-parametric bootstrap replicates was midpoint-rooted and visualized with iTOL v. 5.7 (Letunic and Bork, 2019). To assess the population structure of all available genomes, we inferred an additional phylogeny constructed with JolyTree v 2.0.19092ac (Criscuolo, 2019) of all 120 strains from this study and 13 housefly isolates previously published (Heiden et al., 2020b). A synteny plot comparing ST38 chromosome- and plasmid-derived contigs was created with genoPlotR v 0.8.9 (Luan and Li, 2004).
When the genotype was positive for colistin resistance (presence of mcr genes), we evaluated the resistance phenotype by determining the minimum inhibitory concentration (MIC) using MICRONAUT MIC-Strip Colistin (Merlin Diagnostika GmbH, Bornheim, Germany) according to the manufacturer’s instructions and interpreted the results according to the published breakpoints of EUCAST (The European Committee on Antimicrobial Susceptibility Testing, 2021). Experiments were performed thrice.
The largest fraction of the 120 ESBL-producing E. coli originated from rectal swabs of patients (50.8%; 61/120), followed by caregivers (38.3%; 46/120), neighbors (5.0%; 6/120), family members (4.2%; 5/120), and animals (1.7%; 2/120) (Figure 1 and Supplementary Table 1). The majority of strains belonged to phylogroup A (30.8%; 37/120), which usually comprises commensal strains. The remaining strains were distributed among the phylogroups D (21.7%; 26/120), F (20.0%; 24/120), and B2 (14.2%; 17/120) as well as phylogroups B1 (6.7%; 8/120) and C (6.7%; 8/120), based on Clermont’s revised E. coli phylotyping method (Clermont et al., 2013).
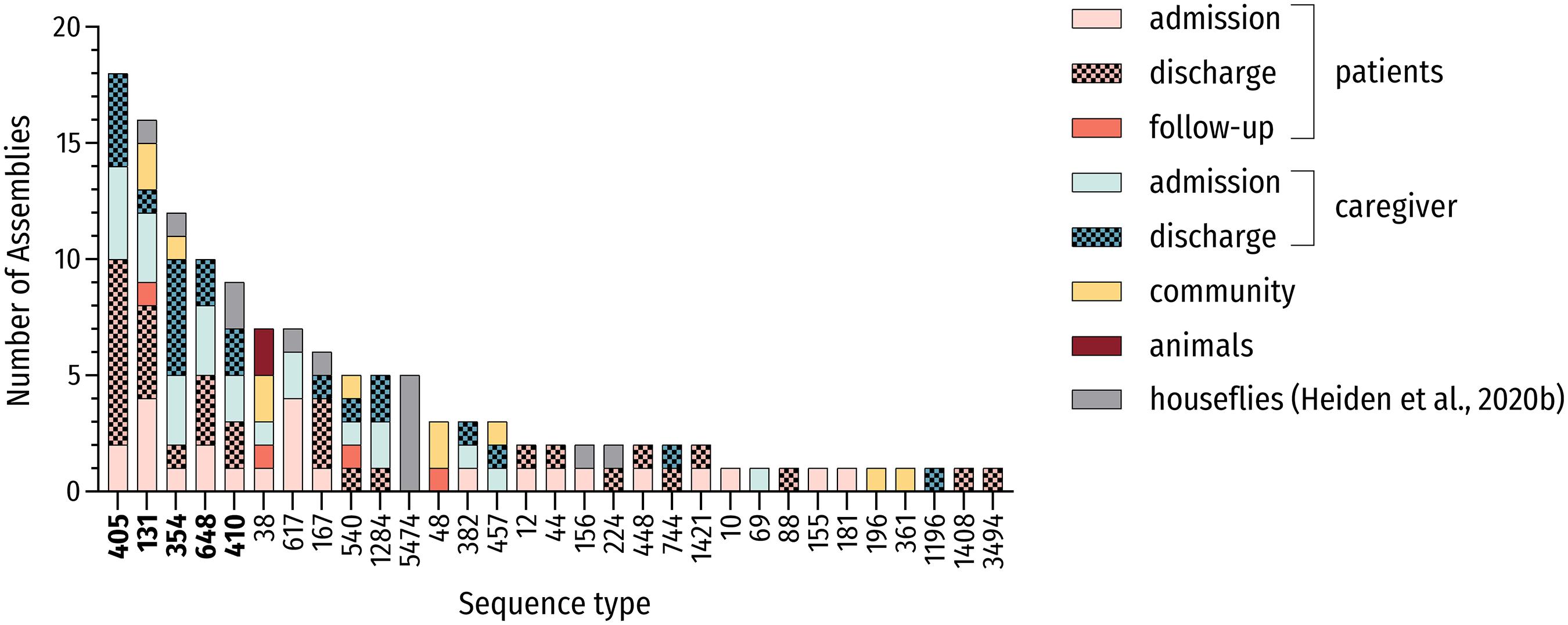
Figure 1. MLST-based distribution of investigated strains. Distribution of all investigated strains including genomes of ESBL-producing E. coli carried by houseflies (Heiden et al., 2020b; n = 133) and associated sequence types (STs). The five most prevalent STs are in bold.
Multi-locus sequence typing (MLST) from genomic data revealed 30 different STs. In total, sixty percent (18/30) of identified STs were present more than once and 10 or more strains belonged to 4 main STs (13.3%; 4/30) including the high-risk ST131 and ST648 E. coli lineages. These most frequently encountered STs were ST405 (15.0%; 18/120), ST131 (12.5%, 15/120), ST354 (9.2%; 11/120) and ST648 (8.3%; 10/120), which accounted for 45.0% (54/120) of all strains. Including the pandemic ST410 lineage, more than half of all strains (50.8%; 61/120) belonged to one of the five most prevalent STs in this study (Figure 1). These sequence types belonged to phylogroups B2 (ST131), D (ST405), and F (ST354, ST648) as well as phylogroup C (ST410), thus underlining the heterogeneity of phylogenetic backgrounds among ESBL-producing E. coli.
To elucidate phylogenetic relationships, we constructed a tree in an alignment-free manner for all investigated genomes (Supplementary Figure 1). Additionally, we inferred phylogenies, which are based on SNPs in the core genome of strains belonging to the five predominant STs of this study to assess potential transmission scenarios (Figure 2). For comparative reasons, we included 13 genomes of ESBL-producing E. coli isolated from houseflies (Heiden et al., 2020b) originating from the same Rwandan hospital.
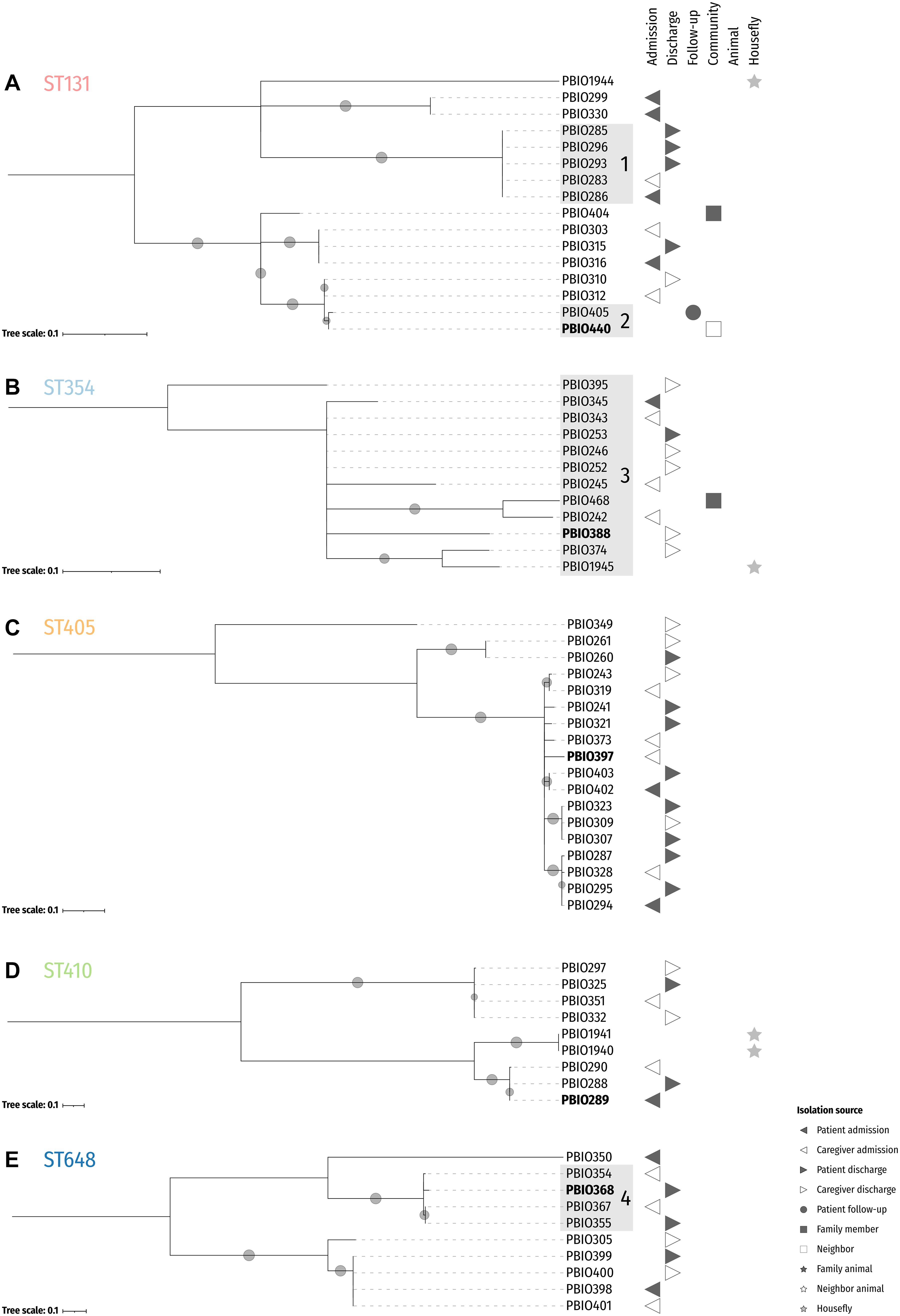
Figure 2. Midpoint-rooted maximum likelihood core SNP phylogenies of the five dominating sequence types. Reference isolates for ST131 (PBIO440; A), ST354 (PBIO388; B), ST405 (PBIO397; C), ST410 (PBIO289; D), and ST648 (PBIO368; E) are highlighted in bold. The trees are based on alignments with 208 (A), 20 (B), 177 (C), 154 (D), and 135 (E) sites. Sub-clades are highlighted in gray. Circles at branches display bootstrap proportions ≥50% (1,000 replicates). Symbols besides the tree depict isolation sources as given in the legend.
The phylogenetic analysis (Supplementary Figure 1) shows that the E. coli strains were distributed among six distinct phylogroups and grouped into several clades according to their sequence type. Within these ST-associated phylogenies, several sub-clades were defined with genomes interspersed in patients, caregivers, related community members as well as animals and flies, which suggests common phylogenetic backgrounds (Clermont et al., 2011) potentially based on interspecies transmission. For example, PBIO458 (study-ID 60) and PBIO459 [(study-ID 133)—both ST38 isolated from two animals—clustered with four different strains from patients, family members and neighbors, PBIO272 (patient admission, study-ID 60), PBIO451 (neighbor, study-ID 60), PBIO455 (family member, study-ID 60), and PBIO467 (patient follow-up, study-ID 60)]. These findings are corroborated by results of two of our previous studies, where we demonstrated the likely transmission of ESBL-producing E. coli ST38 among humans and animals (Guenther et al., 2017; Schaufler et al., 2018). Note, however, that in this current study, only one representative of ST38 [PBIO302 (caregiver admission, study-ID 131)] carried a chromosomally encoded blaCTX–M–15 gene and the before mentioned strains carried plasmid-encoded ESBLs (Supplementary Figure 1), which is contrary to our previous findings. We then compared the blaCTX–M–15 gene-carrying chromosomal contig of PBIO302 to two of the plasmid-encoded blaCTX–M–15 sequences of ST38 (PBIO272 and PBIO459; Supplementary Figure 2). The chromosomal sections of PBIO302, PBIO272, and PBIO459 were highly similar, except the chromosomal insertion of blaCTX–M–15 in PBIO302. This resistance gene was flanked by transposable elements, as described below.
In addition, strains with the numbers PBIO1939, PBIO1942, PBIO1945, PBIO1946, and PBIO1947 from houseflies were in the same sub-clade as strains isolated from different human sources indicating the potential role of living vectors in the spread of pathogenic bacteria.
For the phylogenetic trees of the five predominant STs of this study (Figure 2) it is interesting to notice that some genomes stemming from different sources were more closely related than genomes from the same source. For example, PBIO283 [(study-ID 92) Figure 2A, ST131, sub-clade 1], which originates from a caregiver at admission differed in 0.2 ± 0.0003 SNPs/Mbp with strains isolated from the related patient at admission [PBIO286 (study-ID 92)] as well as discharge [PBIO285 (study-ID 92)] and unrelated patients at discharge [PBIO293 (study-ID 114) and PBIO296 (study-ID 117)]. Moreover, PBIO440 [(study-ID 133) Figure 2A, ST131, sub-clade 2], isolated from a community member, only varied in 0.2 SNPs/Mbp compared to a follow-up strain of an already discharged patient [PBIO405 (study-ID 434)]. Notably, all strains belonging to ST354 (Figure 2B, sub-clade 3) only differed in 1.0 ± 0.4 SNPs/Mbp including one strain carried by a housefly (PBIO1945). Also interesting was the difference between PBIO368 [(study-ID 335) Figure 2E, ST648, sub-clade 4], originating from a patient at discharge, and strains of distinct sources [PBIO354 (caregiver admission, study-ID 265), PBIO355 (patient discharge, study-ID 265), and PBIO367 (caregiver admission, study-ID 288)], differing in 0.9 ± 0.1 SNPs/Mbp. These numbers of SNPs were up to 10-fold lower than described for clonal EHEC strains during an outbreak in Germany (1.8 SNPs/Mbp) (Grad et al., 2012; Been et al., 2014), suggesting the circulation of only a handful of sequence types in this African setting, which interestingly happen to mostly be international high-risk clonal lineages. In addition, some strains from identical STs were carried by both flies and humans, for example PBIO1945 and PBIO374 (caregiver discharge, study-ID 401; Figure 2B), again indicating the role of flies in the spread of antibiotic-resistant pathogens.
The predominant ESBL-gene was blaCTX–M–15 (87.5%; 105/120). Furthermore, 76 strains (63.3%; 76/120) carried blaOXA–1 and eight (6.7%; 8/120) blaCTX–M–27. Previous studies reported the co-occurrence of blaCTX–M–15 and blaOXA–1, whereas other members of the CTX-M family (e.g., blaCTX–M–27) and blaOXA–1 appear to be mutually exclusive (Schaufler et al., 2016b; Livermore et al., 2019; Bodendoerfer et al., 2020). Here, almost all strains carrying the blaOXA–1 gene (98.7%; 75/76) carried the blaCTX–M–15 gene in addition and none showed the combination of blaOXA–1 and another CTX-M gene. The co-occurence of distinct CTX-M genes was not detected. For the majority of blaCTX–M-positive strains (71.1%; 81/114), the corresponding gene was encoded on a plasmid (Supplementary Figure 1). Interestingly, all strains of phylogroup F (20.0%; 24/120) were carriers of blaCTX–M–15 genes, of which 87.5% (21/24) were located on the chromosome (all strains belonging to ST648 and ST354). These chromosomally encoded genes were flanked by an insertion sequence ISEcp1 upstream and a Tn2 downstream. Also note that the strains PBIO242 and PBIO245 (ST354) showed two of these motifs consecutively. The blaCTX–M–15 gene of the ST410 strains PBIO288, PBIO289, and PBIO290 were found in proximity to an upstream-located ISEcp1 only. Previous studies have already demonstrated the diversity and global distribution of these complex transposable units in enterobacteria (Poirel et al., 2005; Lartigue et al., 2006; Decano and Downing, 2019; Ludden et al., 2020; Yoon et al., 2020).
Genes encoding for carbapenem-hydrolyzing enzymes, like blaOXA–48 or blaNDM–1, were not present.
A growing body of ESBL-producers is MDR (Cantón and Coque, 2006; Hidron et al., 2008; Pitout, 2012; Beceiro et al., 2013). In our study, all investigated strains (100.0%; 120/120) were carriers of genes conferring resistance to three or more different classes of antimicrobial agents, thus exhibiting a MDR genotype (Magiorakos et al., 2012). In total, 120 strains (100.0%; 120/120) carried resistance genes to tetracyclines (tet), followed by genes encoding for aminoglycoside (aac, aad, and aph [98.3%; 118/120]), sulfonamide [sul (90.0%; 108/120)], trimethoprim [dfr (89.2%; 107/120)], chloramphenicol [cat (72.5%; 87/120)], and fluoroquinolone [aac(6′)-Ib-cr and qnr (71.7%; 86/120)] resistances. Two strains (1.7%; 2/120) belonging to ST181 and ST540 were carriers of mcr-9 but showed phenotypic susceptibility to colistin, which is a last-resort antibiotic (MIC of both strains: 0.5 μg/mL). Interestingly, previous studies suggest that this latest member of the mcr gene family does not always confer phenotypic resistance to colistin in clinical isolates, although overexpression in E. coli TOP10 cells leads to increased MICs (Carroll et al., 2019; Kieffer et al., 2019; Tyson et al., 2020).
In addition to the resistances described, genes encoding for efflux pumps were found frequently, with mdfA in all (120/120) and acrB in 85.8% (103/120) of all genomes.
As previously demonstrated by us and others, the combination of MDR and high-level bacterial virulence seems to be a hallmark of pandemic high-risk clonal lineages (Hussain et al., 2012; Ewers et al., 2014; Shaik et al., 2017; Schaufler et al., 2019). To investigate the strains’ genetic virulome, we analyzed the genomes of the predominant ST131 (n = 15), ST648 (n = 10), ST354 (n = 11), ST405 (n = 18), and ST410 (n = 7) for these features (Figure 3).
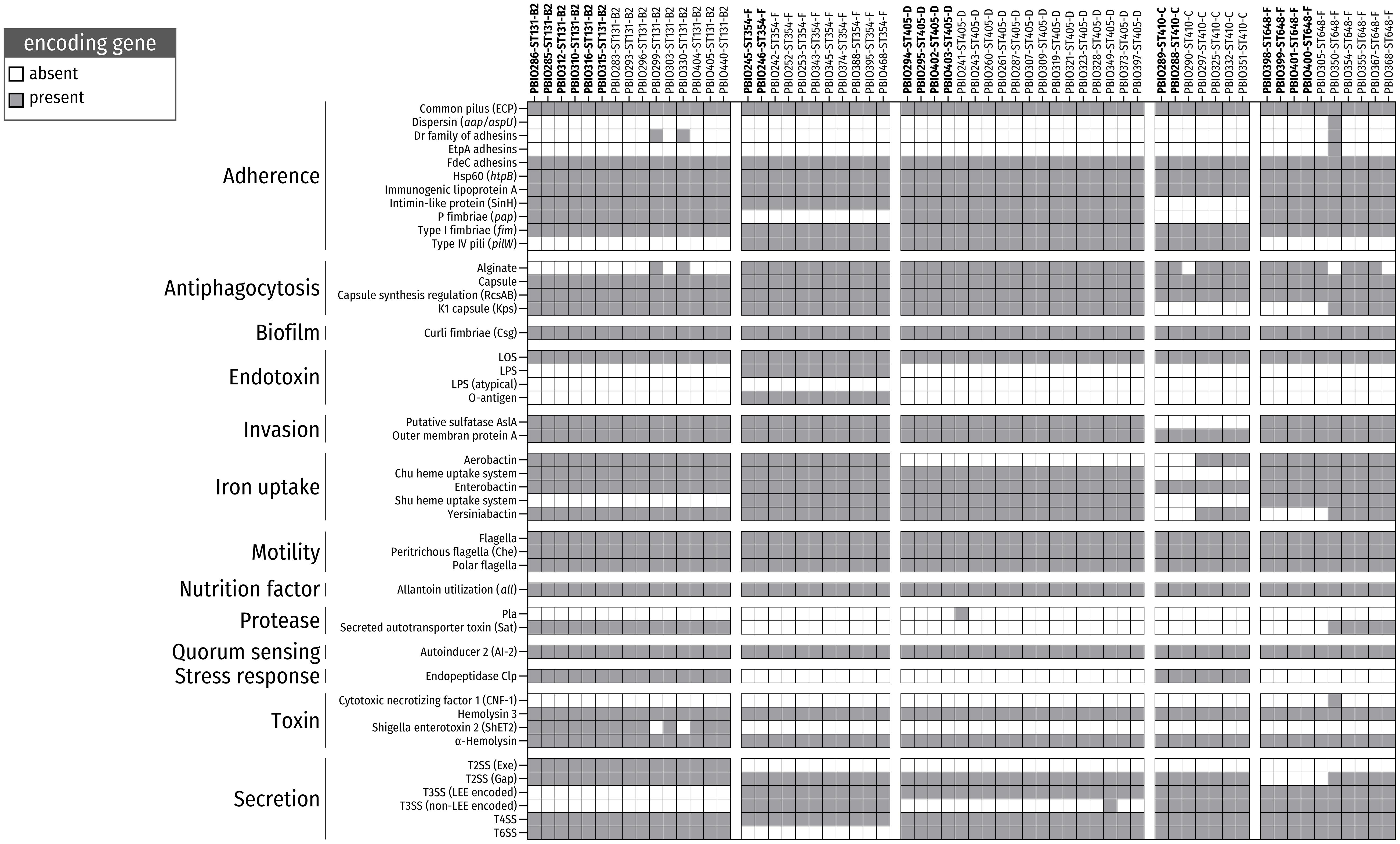
Figure 3. Presence/absence of virulence factors (VAGs) among ESBL-producing E. coli belonging to the five most prevalent sequence types (STs). Color-filled boxes show the presence of genes (coverage and identity ≥ 65%) encoding for different VAGs.
The strains belonging to the five predominant STs carried several VAGs, mainly associated with adherence, antiphagocytosis, biofilm formation, invasion, iron uptake and bacterial secretion. The ability to attach to surfaces/cells and form biofilms is a common strategy used by bacterial populations to resist antibiotic treatment and host defense mechanisms as well as cause infection (Moser et al., 2017; Amanatidou et al., 2019). In particular, genes for the P fimbriae adhesion cluster [pap operon (70.5%; 43/61)], Dr family of adhesins (4.9%; 3/61) and type I fimbriae [fim (100%; 61/61)], which are necessary for uroepithelia cell adhesion and invasion, and, thus, for causing urinary tract infection (Mulvey, 2002), were frequently found. Notably, strains belonging to ST354 and ST410 showed a lack of pap genes, which is consistent with previous findings (Vangchhia et al., 2016; Zogg et al., 2018; Schaufler et al., 2019). Furthermore, we detected several members of the csg gene family in all genomes (100%; 120/120). These genes encode curli fibers, which are essential components of bacterial biofilms (Hammar et al., 1995; Evans and Chapman, 2014).
The ability to acquire intracellular heme and hemolysin, which is based on the expression of iron uptake-associated genes [e.g., Chu heme uptake system (88.5%; 54/61), yersiniabactin (86.9%; 53/61), and aerobactin (65.6%; 40/61)], is an effective strategy for iron utilization during infection (Fischbach et al., 2006) and is another important virulence-associated feature in the repertoire of these bacteria.
We next investigated the occurrence and circulation/transfer of ESBL-plasmids among strains and thus their contribution to the spread of antibiotic resistance in the Rwandan setting.
In total, 107 strains (89.5%; 107/120) carried plasmids with incompatibly (Inc) group FIB, followed by IncFIA (75.8%; 91/120) and IncFII (70.8%; 85/120). In particular IncF plasmids are frequently associated with genes encoding ESBL-genes, other resistances as well as virulence features important for iron acquisition (Han et al., 2012), serum resistance (Ranjan et al., 2018), and biofilm formation (Schaufler et al., 2016a).
To better assess similar plasmid backgrounds, we compared the plasmid sequences of all strains that carried a plasmid-borne blaCTX–M–15 gene against the plasmid sequence of PBIO241 (ST405; patient discharge; study-ID 159; Figure 4) as a representative for the most prevalent ST. Keep in mind, however, that the selection of the reference biases the visual representation when a large number of plasmid sequences is absent in the query sequences. On the other hand, plasmid sequences, not present in the reference but in queries, are missing in this approach. The plasmid backbones of other ST405 strains were highly similar to the reference. In contrast, plasmids from strains of other pandemic STs (ST131 and ST410) only showed few similarities when compared to the reference plasmid underlining their genetic diversity. Interestingly, in addition, the plasmid sequences of four ST167 strains [PBIO276 (patient discharge, study-ID 78), PBIO277 (patient admission, study-ID 78), PBIO301 (caregiver discharge, study-ID 131), and PBIO306 (patient discharge, study-ID 131)] were highly similar to the reference. Five other strains [PBIO256 (ST12, patient admission, study-ID 40), PBIO257 (ST12, patient discharge, study-ID 40), PBIO258 (ST1421, patient admission, study-ID 40), PBIO1948 (ST5474, housefly), and PBIO1949 (ST5474, housefly)] showed identical plasmid sequences in parts of the BRIG visualization that were absent in most of the other sequences.
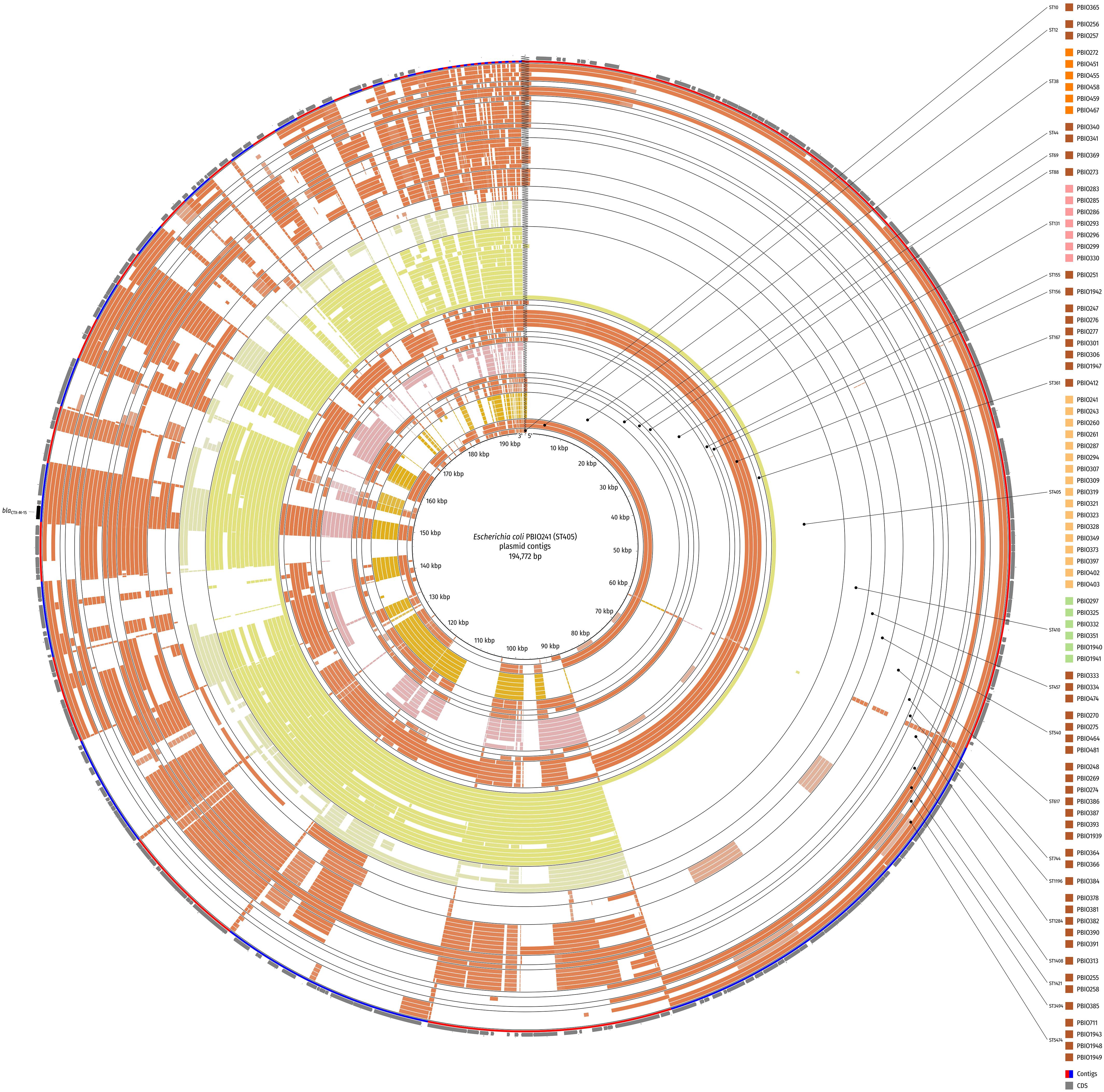
Figure 4. Circular comparison of the plasmid background of strains with plasmid-borne blaCTX–M–15 gene. The BLAST-classified plasmid contigs were searched against plasmid contigs of PBIO241 (ST405) as a reference. Strains are ordered by ST and name in ascending order and depicted in concentric rings from inner to outer. Red and blue arcs show contig boundaries and the outermost ring contains coding sequences (CDS; gray arrows) with the blaCTX–M–15 gene highlighted in black. The comparison was created with BLAST Ring Image Generator (BRIG).
We also compared some of our plasmid sequences of representative strains (criteria: top BLAST-hits) against a publicly available reference plasmid pIV_IncHI2_CTX_M_15 (Marchetti et al., 2020; Supplementary Figure 3). This blaCTX–M–15-containing plasmid has been originally obtained from an E. coli ST58 strain causing a deadly puppy infection in Italy in 2019. Interestingly, the plasmid backbone of PBIO251 (ST155, patient admission, study-ID 153) was nearly identical when compared to this external Italian plasmid. The plasmids of PBIO275 (ST540, caregiver admission, study-ID 78), PBIO385 (ST3494, patient discharge, study-ID 434), and PBIO1943 (ST5474, housefly) also showed high similarities to the reference and thus among each other.
The presence of similar resistance plasmids in strains of distinct sequence types and sample groups indicates the potential transmission of mobile genetic elements and the growing prevalence of successful plasmid families (Carattoli, 2011).
In contrast to Europe, the United States of America and Australia, only little information is available concerning the exact characteristics and distribution of ESBL-producing E. coli in Africa. This study reveals the broad occurrence and potential circulation of several international high-risk clonal lineages in a hospital and associated locations in Rwanda, following up on a publication by Kurz et al. (2017). Despite the potential bias due to the analysis of both admission and discharge strains of the same patient and/or caregiver, it is remarkable that our sample set was dominated by ST131, ST648, and ST410, which have been frequently reported from humans, animals and the environment (Nicolas-Chanoine et al., 2008; Ewers et al., 2010, 2014; Hussain et al., 2012; Schaufler et al., 2016b, 2019) and which are “classic” pandemic, high-risk clonal lineages. In addition to these, we found ST354 to be the third most detected sequence type in this study. Its emergence has been previously described globally except for the African continent (Manges et al., 2019). This locally restricted accumulation of one single ST in combination with a small number of other STs indicates re-entering and circulation of dominant bacterial lineages in the Rwandan hospital and among family members/neighbors and animals.
Interestingly, within the ST-associated clades, some genomes without genetic differences were interspersed in humans (hospitalized patients and caregivers as well as community members) and animals. This putative lack of host adaptation and the close phylogenetic relationships indicate the colonization and rapid transmission of several clones within the community and the potential transmission into the clinical setting and vice versa, underlined by the high acquisition rates of ESBL-producing E. coli during hospitalization as described previously (Kurz et al., 2017).
The resistance genes found in this study confer resistances to antibiotics frequently used in veterinary medicine and/or in sub-therapeutic doses as food supplements and growth promotors in Africa (Eagar et al., 2012; Adesokan et al., 2015; Mainda et al., 2015; Manishimwe et al., 2017). When also considering the zoonotic character of ESBL-producing E. coli, it is not surprising that we found clonal strains with similar patterns of resistance features in the different sample groups. Transmission likely occurred among patients and caregivers/family members and was also influenced by livestock animals due to close human-animal contact (Klous et al., 2016).
Two strains of this study carried the mcr-9 gene but were phenotypically susceptible to colistin. This phenomenon was first reported in 2019 (Carroll et al., 2019; Kieffer et al., 2019). Due to the structural heterogeneity compared to other mcr genes (65% amino acid identity with the most closely related mcr-3 gene) and the weak inactivation of colistin, the clinical importance of mcr-9 is unknown (Tyson et al., 2020).
Notably, some of the strains showed extensive, chromosomally encoded virulence-associated features. The CNF-1-encoding gene (cnf1) detected in strain PBIO350 (ST648), for example, is associated with causing neonatal meningitis (Khan et al., 2002). In addition to the major virulence factors of meningitis-associated and uropathogenic E. coli (like P fimbriae adhesion cluster, K1 capsule, heme utilization systems, and the secreted autotransporter toxin), the strains showed VAGs typical for InPEC (especially the various bacterial secretion systems) underlining the clinical relevance of these pathogens.
Finally, we demonstrate that similar plasmid sequences were present in strains from different sample groups, thus likely indicating mobile genetic element transmission, and underlining the importance of plasmid-driven spread of antimicrobial resistance independent of the host’s phylogenetic background (Schaufler et al., 2016a; Ranjan et al., 2018). Interesting in addition to similarities among strains from the Rwandan setting is in particular the close relationship to an external plasmid, which has been obtained only recently (Marchetti et al., 2020). This highlights the sometimes global spread of such mobile genetic elements and their bacterial hosts.
In this study, we investigated and identified the presence of clinically relevant ESBL-producing E. coli that circulate among patients, caregivers, the community and animals in a Rwandan setting. The findings contribute to the understanding of the global dissemination of bacterial high-risk clonal lineages, their virulence features as well as plasmid transmissions. They also underline the potential role of houseflies in this harmful dynamic.
The data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB42795 (https://www.ebi.ac.uk/ena/browser/view/PRJEB42795).
KS and EE designed the study. EE and JM performed the laboratory and phenotypic experiments. SH, SS, and KK performed the bioinformatics analyses. KS, EE, SH, KK, CB, JN, AS, JG, MK, FM, and SS analyzed the data. KS, EE, and SH wrote the manuscript and prepared the tables and figures. All authors read and approved the final version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.662575/full#supplementary-material
Adesokan, H. K., Akanbi, I. O., Akanbi, I. M., and Obaweda, R. A. (2015). Pattern of antimicrobial usage in livestock animals in south-western Nigeria: the need for alternative plans. Onderstepoort J. Vet. Res. 82:816. doi: 10.4102/OJVR.V82I1.816
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genom. 12:402. doi: 10.1186/1471-2164-12-402
Amanatidou, E., Matthews, A. C., Kuhlicke, U., Neu, T. R., McEvoy, J. P., and Raymond, B. (2019). Biofilms facilitate cheating and social exploitation of β-lactam resistance in Escherichia coli. NPJ Biofilms Microb. 5:36. doi: 10.1038/s41522-019-0109-2
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Beceiro, A., Tomás, M., and Bou, G. (2013). Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 26, 185–230. doi: 10.1128/CMR.00059-12
Been, M., de Lanza, V. F., Toro, M., de, Scharringa, J., Dohmen, W., et al. (2014). Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet. 10:e1004776. doi: 10.1371/journal.pgen.1004776
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Bodendoerfer, E., Marchesi, M., Imkamp, F., Courvalin, P., Böttger, E. C., and Mancini, S. (2020). Co-occurrence of aminoglycoside and β-lactam resistance mechanisms in aminoglycoside- non-susceptible Escherichia coli isolated in the zurich area, switzerland. Int. J. Antimicrob. Agents 56:106019. doi: 10.1016/j.ijantimicag.2020.106019
Cantón, R., and Coque, T. M. (2006). The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9, 466–475. doi: 10.1016/j.mib.2006.08.011
Carattoli, A. (2011). Plasmids in gram negatives: molecular typing of resistance plasmids. Int. J. Med. Microbiol. 301, 654–658. doi: 10.1016/j.ijmm.2011.09.003
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype typhimurium isolate. mBio 10, e819–e853. doi: 10.1128/mBio.00853-19
Carroll, M., Rangaiahagari, A., Musabeyezu, E., Singer, D., and Ogbuagu, O. (2016). Five-year antimicrobial susceptibility trends among bacterial isolates from a tertiary health-care facility in kigali, Rwanda. Am. J. Trop Med. Hyg. 95, 1277–1283. doi: 10.4269/ajtmh.16-0392
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65.
Clermont, O., Olier, M., Hoede, C., Diancourt, L., Brisse, S., Keroudean, M., et al. (2011). Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 11, 654–662.
Criscuolo, A. (2019). A fast alignment-free bioinformatics procedure to infer accurate distance-based phylogenetic trees from genome assemblies. Res. Ideas Outcomes 5:e36178. doi: 10.3897/rio.5.e36178
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using gubbins. Nucleic Acids Res. 43:e15. doi: 10.1093/nar/gku1196
Decano, A. G., and Downing, T. (2019). An Escherichia coli ST131 pangenome atlas reveals population structure and evolution across 4,071 isolates. Sci. Rep. 9:17394. doi: 10.1038/s41598-019-54004-5
Eagar, H., Swan, G., and van Vuuren, M. (2012). A survey of antimicrobial usage in animals in south africa with specific reference to food animals. J. S. Afr. Vet. Assoc. 83:16. doi: 10.4102/jsava.v83i1.16
Escobar-Páramo, P., Clermont, O., Blanc-Potard, A.-B., Bui, H., Le Bouguénec, C., and Denamur, E. (2004). A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21, 1085–1094. doi: 10.1093/molbev/msh118
Evans, M. L., and Chapman, M. R. (2014). Curli biogenesis: order out of disorder. Biochim. Biophys. Acta 1843, 1551–1558. doi: 10.1016/j.bbamcr.2013.09.010
Ewers, C., Bethe, A., Stamm, I., Grobbel, M., Kopp, P. A., Guerra, B., et al. (2014). CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J. Antimicrob. Chemother. 69, 1224–1230. doi: 10.1093/jac/dkt516
Ewers, C., Grobbel, M., Stamm, I., Kopp, P. A., Diehl, I., Semmler, T., et al. (2010). Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum-beta-lactamase-producing Escherichia coli among companion animals. J. Antimicrob. Chemother. 65, 651–660. doi: 10.1093/jac/dkq004
Fischbach, M. A., Lin, H., Liu, D. R., and Walsh, C. T. (2006). How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol. 2, 132–138. doi: 10.1038/nchembio771
Grad, Y. H., Lipsitch, M., Feldgarden, M., Arachchi, H. M., Cerqueira, G. C., Fitzgerald, M., et al. (2012). Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in europe, 2011. Proc. Natl. Acad. Sci. U S A 109, 3065–3070. doi: 10.1073/pnas.1121491109
Guenther, S., Semmler, T., Stubbe, A., Stubbe, M., Wieler, L. H., and Schaufler, K. (2017). Chromosomally encoded ESBL genes in Escherichia coli of ST38 from mongolian wild birds. J. Antimicrob. Chemother. 72, 1310–1313. doi: 10.1093/jac/dkx006
Hammar, M. R., Arnqvist, A., Bian, Z., Olsén, A., and Normark, S. (1995). Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18, 661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x
Han, J., Lynne, A. M., David, D. E., Tang, H., Xu, J., Nayak, R., et al. (2012). DNA sequence analysis of plasmids from multidrug resistant Salmonella enterica serotype heidelberg isolates. PLoS One 7:e51160. doi: 10.1371/journal.pone.0051160
Heiden, S. E., Hübner, N.-O., Bohnert, J. A., Heidecke, C.-D., Kramer, A., Balau, V., et al. (2020a). A Klebsiella pneumoniae ST307 outbreak clone from germany demonstrates features of extensive drug resistance, hypermucoviscosity, and enhanced iron acquisition. Genome Med. 12:113. doi: 10.1186/s13073-020-00814-6
Heiden, S. E., Kurz, M. S. E., Bohnert, J., Bayingana, C., Ndoli, J. M., Sendegeya, A., et al. (2020b). Flies from a tertiary hospital in Rwanda carry multidrug-resistant gram-negative pathogens including extended-spectrum beta-lactamase-producing E. coli sequence type 131. Antimicrob. Resist. Infect. Control 9:34. doi: 10.1186/s13756-020-0696-y
Hidron, A. I., Edwards, J. R., Patel, J., Horan, T. C., Sievert, D. M., Pollock, D. A., et al. (2008). NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2006-2007. Infect. Control Hosp. Epidemiol. 29, 996–1011. doi: 10.1086/591861
Hoffman, M., Mofolo, I., Salima, C., Hoffman, I., Zadrozny, S., Martinson, F., et al. (2012). Utilization of family members to provide hospital care in malawi: the role of hospital guardians. Malawi Med. J. 24, 74–78.
Hussain, A., Ewers, C., Nandanwar, N., Guenther, S., Jadhav, S., Wieler, L. H., et al. (2012). Multiresistant uropathogenic Escherichia coli from a region in India where urinary tract infections are endemic: genotypic and phenotypic characteristics of sequence type 131 isolates of the CTX-M-15 extended-spectrum-β-lactamase-producing lineage. Antimicrob. Agents Chemother. 56, 6358–6365. doi: 10.1128/AAC.01099-12
Jaureguy, F., Landraud, L., Passet, V., Diancourt, L., Frapy, E., Guigon, G., et al. (2008). Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genom. 9:560. doi: 10.1186/1471-2164-9-560
Kaper, J. B., Nataro, J. P., and Mobley, H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Kayumba, P. C., Risha, P. G., Shewiyo, D., Msami, A., Masuki, G., Ameye, D., et al. (2004). The quality of essential antimicrobial and antimalarial drugs marketed in rwanda and tanzania: influence of tropical storage conditions on in vitro dissolution. J. Clin. Pharm. Ther. 29, 331–338. doi: 10.1111/j.1365-2710.2004.00568.x
Khan, N. A., Wang, Y., Kim, K. J., Chung, J. W., Wass, C. A., and Kim, K. S. (2002). Cytotoxic necrotizing factor-1 contributes to Escherichia coli K1 invasion of the central nervous system. J. Biol. Chem. 277, 15607–15612.
Kieffer, N., Royer, G., Decousser, J.-W., Bourrel, A.-S., Palmieri, M., La Ortiz De Rosa, J.-M., et al. (2019). mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob. Agents Chemother. 63, e1819–e1866. doi: 10.1128/AAC.00965-19
Klous, G., Huss, A., Heederik, D. J., and Coutinho, R. A. (2016). Human–livestock contacts and their relationship to transmission of zoonotic pathogens, a systematic review of literature. One Health 2, 65–76. doi: 10.1016/j.onehlt.2016.03.001
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., and Stamatakis, A. (2019). RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. doi: 10.1093/bioinformatics/btz305
Kurz, M. S. E., Bayingana, C., Ndoli, J. M., Sendegeya, A., Durst, A., Pfüller, R., et al. (2017). Intense pre-admission carriage and further acquisition of ESBL-producing Enterobacteriaceae among patients and their caregivers in a tertiary hospital in Rwanda. Trop Med Int Health 22, 210–220. doi: 10.1111/tmi.12824
Lartigue, M.-F., Poirel, L., Aubert, D., and Nordmann, P. (2006). In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring beta-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob Agents Chemother. 50, 1282–1286. doi: 10.1128/AAC.50.4.1282-1286.2006
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Livermore, D. M., Day, M., Cleary, P., Hopkins, K. L., Toleman, M. A., Wareham, D. W., et al. (2019). OXA-1 β-lactamase and non-susceptibility to penicillin/β-lactamase inhibitor combinations among ESBL-producing Escherichia coli. J. Antimicrob. Chemother. 74, 326–333. doi: 10.1093/jac/dky453
Luan, Y., and Li, H. (2004). Model-based methods for identifying periodically expressed genes based on time course microarray gene expression data. Bioinformatics 20, 332–339. doi: 10.1093/bioinformatics/btg413
Ludden, C., Decano, A. G., Jamrozy, D., Pickard, D., Morris, D., Parkhill, J., et al. (2020). Genomic surveillance of Escherichia coli ST131 identifies local expansion and serial replacement of subclones. Microb. Genom. 6, e000352. doi: 10.1099/mgen.0.000352
Magiorakos, A.-P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mainda, G., Bessell, P. R., Muma, J. B., McAteer, S. P., Chase-Topping, M. E., Gibbons, J., et al. (2015). Prevalence and patterns of antimicrobial resistance among Escherichia coli isolated from zambian dairy cattle across different production systems. Sci. Rep. 5:12439. doi: 10.1038/srep12439
Manges, A. R., Geum, H. M., Guo, A., Edens, T. J., Fibke, C. D., and Pitout, J. D. D. (2019). Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin. Microbiol. Rev. 32, e118–e135. doi: 10.1128/CMR.00135-18
Manishimwe, R., Nishimwe, K., and Ojok, L. (2017). Assessment of antibiotic use in farm animals in Rwanda. Trop Anim. Health Prod. 49, 1101–1106. doi: 10.1007/s11250-017-1290-z
Marchetti, M. V., Bitar, I., Mercato, A., Nucleo, E., Marchesini, F., et al. (2020). Deadly puppy infection caused by an MDR Escherichia coli O39 blaCTX–M–15, blaCMY-2, blaDHA-1, and aac(6)-Ib-cr - positive in a breeding kennel in central italy. Front. Microbiol. 11:584. doi: 10.3389/fmicb.2020.00584
McNally, A., Oren, Y., Kelly, D., Pascoe, B., Dunn, S., Sreecharan, T., et al. (2016). Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. PLoS Genet 12:e1006280. doi: 10.1371/journal.pgen.1006280
Melzer, M., and Petersen, I. (2007). Mortality following bacteraemic infection caused by extended spectrum beta-lactamase (ESBL) producing E. coli compared to non-ESBL producing E. coli. J. Infect. 55, 254–259. doi: 10.1016/j.jinf.2007.04.007
Monárrez, R., Braun, M., Coburn-Flynn, O., Botelho, J., Odetoyin, B. W., Otero-Vera, J. I., et al. (2019). A large self-transmissible resistance plasmid from nigeria contains genes that ameliorate a carrying cost. Sci. Rep. 9:19624. doi: 10.1038/s41598-019-56064-z
Moser, C., Pedersen, H. T., Lerche, C. J., Kolpen, M., Line, L., Thomsen, K., et al. (2017). Biofilms and host response - helpful or harmful. APMIS 125, 320–338. doi: 10.1111/apm.12674
Mulvey, M. A. (2002). Adhesion and entry of uropathogenic Escherichia coli. Cell Microbiol. 4, 257–271. doi: 10.1046/j.1462-5822.2002.00193.x
Muvunyi, C. M., Masaisa, F., Bayingana, C., Mutesa, L., Musemakweri, A., Muhirwa, G., et al. (2011). Decreased susceptibility to commonly used antimicrobial agents in bacterial pathogens isolated from urinary tract infections in Rwanda: need for new antimicrobial guidelines. Am. J. Trop Med. Hyg. 84, 923–928. doi: 10.4269/ajtmh.2011.11-0057
Nicolas-Chanoine, M.-H., Blanco, J., Leflon-Guibout, V., Demarty, R., Alonso, M. P., Caniça, M. M., et al. (2008). Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61, 273–281. doi: 10.1093/jac/dkm464
Ntirenganya, C., Manzi, O., Muvunyi, C. M., and Ogbuagu, O. (2015). High prevalence of antimicrobial resistance among common bacterial isolates in a tertiary healthcare facility in Rwanda. Am. J. Trop. Med. Hyg. 92, 865–870. doi: 10.4269/ajtmh.14-0607
Page, A. J., Taylor, B., Delaney, A. J., Soares, J., Seemann, T., Keane, J. A., et al. (2016). SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2:e000056. doi: 10.1099/mgen.0.000056
Pallen, M. J., and Wren, B. W. (2007). Bacterial pathogenomics. Nature 449, 835–842. doi: 10.1038/nature06248
Pitout, J. D. D. (2012). Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front. Microbiol. 3:9. doi: 10.3389/fmicb.2012.00009
Poirel, L., Lartigue, M.-F., Decousser, J.-W., and Nordmann, P. (2005). ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49, 447–450. doi: 10.1128/AAC.49.1.447-450.2005
Rahman, M. T., Sobur, M. A., Islam, M. S., Ievy, S., Hossain, M. J., El Zowalaty, M. E., et al. (2020). Zoonotic diseases: etiology, impact, and control. Microorganisms 8:1405. doi: 10.3390/microorganisms8091405
Rahuma, N., Ghenghesh, K. S., Ben Aissa, R., and Elamaari, A. (2005). Carriage by the housefly (Musca domestica) of multiple-antibiotic-resistant bacteria that are potentially pathogenic to humans, in hospital and other urban environments in misurata, libya. Ann. Trop Med. Parasitol. 99, 795–802.
Ranjan, A., Scholz, J., Semmler, T., Wieler, L. H., Ewers, C., Müller, S., et al. (2018). ESBL-plasmid carriage in E. coli enhances in vitro bacterial competition fitness and serum resistance in some strains of pandemic sequence types without overall fitness cost. Gut. Pathog. 10:24. doi: 10.1186/s13099-018-0243-z
Russo, T. A., and Johnson, J. R. (2000). Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181, 1753–1754. doi: 10.1086/315418
Schaufler, K., Nowak, K., Düx, A., Semmler, T., Villa, L., Kourouma, L., et al. (2018). Clinically relevant ESBL-producing K. pneumoniae ST307 and E. coli ST38 in an urban west african rat population. Front. Microbiol. 9:150. doi: 10.3389/fmicb.2018.00150
Schaufler, K., Semmler, T., Pickard, D. J., Toro, M., de La Cruz, F., de, et al. (2016a). Carriage of extended-spectrum beta-lactamase-plasmids does not reduce fitness but enhances virulence in some strains of pandemic E. coli lineages. Front. Microbiol. 7:336. doi: 10.3389/fmicb.2016.00336
Schaufler, K., Semmler, T., Wieler, L. H., Wöhrmann, M., Baddam, R., Ahmed, N., et al. (2016b). Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410–another successful pandemic clone? FEMS Microbiol. Ecol. 92:fiv155. doi: 10.1093/femsec/fiv155
Schaufler, K., Semmler, T., Wieler, L. H., Trott, D. J., Pitout, J., Peirano, G., et al. (2019). Genomic and functional analysis of emerging virulent and multidrug-resistant Escherichia coli lineage sequence type 648. Antimicrob. Agents Chemother. 63, e219–e243. doi: 10.1128/AAC.00243-19
Schwaber, M. J., and Carmeli, Y. (2007). Mortality and delay in effective therapy associated with extended-spectrum beta-lactamase production in Enterobacteriaceae bacteraemia: a systematic review and meta-analysis. J. Antimicrob. Chemother. 60, 913–920. doi: 10.1093/jac/dkm318
Shaik, S., Ranjan, A., Tiwari, S. K., Hussain, A., Nandanwar, N., Kumar, N., et al. (2017). Comparative genomic analysis of globally dominant ST131 clone with other epidemiologically successful extraintestinal pathogenic Escherichia coli (ExPEC) lineages. mBio 8, e1517–e1596. doi: 10.1128/mBio.01596-17
The European Committee on Antimicrobial Susceptibility Testing. (2021). Breakpoint tables for interpretation of MICs and zone diameters: Version 11.0. Available online at: http://www.eucast.org
Touchon, M., Hoede, C., Tenaillon, O., Barbe, V., Baeriswyl, S., Bidet, P., et al. (2009). Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. doi: 10.1371/journal.pgen.1000344
Tufa, T. B., Fuchs, A., Wienemann, T., Eggers, Y., Abdissa, S., Schneider, M., et al. (2020). Carriage of ESBL-producing Gram-negative bacteria by flies captured in a hospital and its suburban surroundings in ethiopia. Antimicrob. Resist. Infect. Control 9:175. doi: 10.1186/s13756-020-00836-0
Tumbarello, M., Sanguinetti, M., Montuori, E., Trecarichi, E. M., Posteraro, B., Fiori, B., et al. (2007). Predictors of mortality in patients with bloodstream infections caused by extended-spectrum-beta-lactamase-producing Enterobacteriaceae: importance of inadequate initial antimicrobial treatment. Antimicrob Agents Chemother. 51, 1987–1994. doi: 10.1128/AAC.01509-06
Tyson, G. H., Li, C., Hsu, C.-H., Ayers, S., Borenstein, S., Mukherjee, S., et al. (2020). The mcr-9 gene of Salmonella and Escherichia coli is not associated with colistin resistance in the united states. Antimicrob. Agents Chemother. 64, e520–e573. doi: 10.1128/AAC.00573-20
Ugochukwu, G. (2013). Roles of nurses in sub-saharan african region. Int. J. Nurs. Midwifery 5, 117–131. doi: 10.5897/IJNM2013.0104
Vangchhia, B., Abraham, S., Bell, J. M., Collignon, P., Gibson, J. S., Ingram, P. R., et al. (2016). Phylogenetic diversity, antimicrobial susceptibility and virulence characteristics of phylogroup F Escherichia coli in australia. Microbiology 162, 1904–1912.
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Whittam, T. S., Ochman, H., and Selander, R. K. (1983). Geographic components of linkage disequilibrium in natural populations of Escherichia coli. Mol. Biol. Evol. 1, 67–83. doi: 10.1093/oxfordjournals.molbev.a040302
Yoon, E.-J., Gwon, B., Liu, C., Kim, D., Won, D., Park, S. G., et al. (2020). Beneficial chromosomal integration of the genes for CTX-M extended-spectrum β-lactamase in Klebsiella pneumoniae for stable propagation. mSystems 5, e420–e459. doi: 10.1128/mSystems.00459-20
Zogg, A. L., Simmen, S., Zurfluh, K., Stephan, R., Schmitt, S. N., and Nüesch-Inderbinen, M. (2018). High prevalence of extended-spectrum β-lactamase producing Enterobacteriaceae among clinical isolates from cats and dogs admitted to a veterinary hospital in switzerland. Front. Vet. Sci. 5:62. doi: 10.3389/fvets.2018.00062
Keywords: ESBL—E. coli, whole-genome sequencing, Rwanda, virulence factors, phylogenetic analysis
Citation: Eger E, Heiden SE, Korolew K, Bayingana C, Ndoli JM, Sendegeya A, Gahutu JB, Kurz MSE, Mockenhaupt FP, Müller J, Simm S and Schaufler K (2021) Circulation of Extended-Spectrum Beta-Lactamase-Producing Escherichia coli of Pandemic Sequence Types 131, 648, and 410 Among Hospitalized Patients, Caregivers, and the Community in Rwanda. Front. Microbiol. 12:662575. doi: 10.3389/fmicb.2021.662575
Received: 01 February 2021; Accepted: 20 April 2021;
Published: 14 May 2021.
Edited by:
Guido Werner, Robert Koch Institute (RKI), GermanyReviewed by:
Tim Downing, Dublin City University, IrelandCopyright © 2021 Eger, Heiden, Korolew, Bayingana, Ndoli, Sendegeya, Gahutu, Kurz, Mockenhaupt, Müller, Simm and Schaufler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katharina Schaufler, a2F0aGFyaW5hLnNjaGF1ZmxlckB1bmktZ3JlaWZzd2FsZC5kZQ==
†Deceased
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.