- 1Natural Resources Institute Finland, Helsinki, Finland
- 2Department of Biology, University of Oulu, Oulu, Finland
The fungal genus Heterobasidion includes forest pathogenic species hosting a diverse group of partitiviruses. They include the host debilitating Heterobasidion partitivirus 13 strain an1 (HetPV13-an1), which was originally observed in a slowly growing H. annosum strain 94233. In this study, a relatively fast-growing sector strain 94233-RC3 was isolated from a highly debilitated mycelial culture of 94233, and its gene expression and virus transcript quantities as well as the genomic sequence of HetPV13-an1 were examined. The sequence of HetPV13-an1 genome in 94233-RC3 was identical to that in the original 94233, and thus not the reason for the partial phenotypic recovery. According to RNA-seq analysis, the HetPV13-an1 infected 94233-RC3 transcribed eight genes differently from the partitivirus-free 94233-32D. Three of these genes were downregulated and five upregulated. The number of differentially expressed genes was considerably lower and the changes in their expression were small compared to those of the highly debilitated original strain 94233 with the exception of the most highly upregulated ones, and therefore viral effects on the host transcriptome correlated with the degree of the virus-caused debilitation. The amounts of RdRp and CP transcripts of HetPV13-an1 were considerably lower in 94233-RC3 and also in 94233 strain infected by a closely related mildly debilitating virus HetPV13-an2, suggesting that the virus titer would have a role in determining the effect of HetPV13 viruses on their hosts.
Introduction
Fungal viruses (mycoviruses) are found in a broad range of fungal taxa including Ascomycota and Basidiomycota as well as early diverging fungal lineages, such as Chytridiomycota, Blastocladiomycota, Neocallimastigomycota, Zoopagomycota, and Mucoromycota (Ghabrial and Suzuki, 2009; Pearson et al., 2009; Ghabrial et al., 2015; Sutela et al., 2019, 2020; Myers et al., 2020). Viruses are transmitted among fungal strains by cell to cell contacts (hyphal anastomosis) and sexual or asexual spores (Ghabrial and Suzuki, 2009; Vainio et al., 2015b; Kashif et al., 2019). Mycoviruses usually cause asymptomatic infections. Hollings (1962) reported for the first time, debilitated growth of Agaricus bisporus mycelium caused by mycovirus infection on artificial agar media. Recently mycoviruses have also been reported to cause phenotypic alterations and hypovirulence (reduced virulence) in their hosts (Huang and Ghabrial, 1996; Preisig et al., 2000; Márquez et al., 2007; Yu et al., 2010; Hyder et al., 2013; Xiao et al., 2014; Vainio et al., 2018b). The hypovirulence responses have been well studied in Cryphonectria parasitica infected by Cryphonectria hypovirus 1 (Allen and Nuss, 2004; Deng et al., 2007; Dawe and Nuss, 2013; Eusebio-Cope et al., 2015). Genetically different fungal strains may also react differently to the same mycovirus strain (Hyder et al., 2013; Kim et al., 2015; Vainio et al., 2018b).
H. annosum s.lat. Species complex is considered as one of the most destructive groups of fungal pathogens of conifer forests in the Northern Hemisphere where these fungi cause root and butt rot diseases (Garbelotto and Gonthier, 2013). There are two geographical groups in this complex including three European species – H. annosum, H. parviporum, and H. abietinum – causing infections preferably on pine, spruce (Picea abies and others), and fir species, respectively (Niemelä and Korhonen, 1998; Dai and Korhonen, 1999), and two North American species – H. irrugulare and H. occidentale – infecting predominantly pines or overlapping host ranges of fir, spruce, and hemlock, respectively (Otrosina and Garbelotto, 2010; Garbelotto and Gonthier, 2013). For all these species, the primary infection is initiated by fungal spores landing and germinating on newly cut conifer stumps, or stem and root wounds, and continues as secondary infection via root contacts to neighboring trees (Stenlid and Redfern, 1998). They are also capable of both necrotic and saprotrophic growth. H. irregulare was the first Heterobasidion species to be characterized for complete genome sequence (Olson et al., 2012; Garbelotto and Gonthier, 2013), and thereafter also genomes of H. annosum (Sillo et al., 2015; Choi et al., 2017), H. occidentale (Liu et al., 2018), and H. parviporum (Zeng et al., 2018) have been characterized. There are different preventive control methods practiced against these root rot fungi, but truly curative methods are urgently needed. For this purpose, the use of mycoviruses has been suggested as a highly potential biocontrol option (Vainio and Hantula, 2016; Vainio et al., 2018b).
About 15% of Heterobasidion strains host dsRNA mycoviruses (Ihrmark, 2001; Vainio et al., 2015a; Vainio and Hantula, 2018) but the frequency of virus infections is considerably higher at forest sites with numerous Heterobasidion disease centers and thus ubiquitous mycelial contacts (Vainio et al., 2015b; Hyder et al., 2018). Up to 70% of Heterobasidion dsRNA virus infections are caused by Heterobasidion RNA virus 6 (HetRV6) (Vainio et al., 2012) belonging to the virus family Curvulaviridae, but also viruses belonging to genera Alphapartitivirus, Betapartitivirus, and family Mitoviridae have been observed (Vainio and Hantula, 2016; Vainio et al., 2018a,b), and actually partitiviruses form a major share of the viral species diversity in Heterobasidion spp. (Vainio et al., 2011, 2012; Kashif et al., 2015; Vainio et al., 2015a). The dsRNA genomes of partitiviruses are composed of two essential independently encapsidated segments coding for an RNA-dependent RNA polymerase (RdRp) and a coat protein (CP) (Nibert et al., 2014: Vainio et al., 2018a).
Although most Heterobasidion partitiviruses are cryptic, there are also strains causing variable and debilitating phenotypes on at least some of their hosts (Hyder et al., 2013; Jurvansuu et al., 2014). The best described of them is Heterobasidion partitivirus 13 strain an1 (HetPV13-an1) causing up to 90% reduction of its host’s growth rate (Vainio et al., 2018b; Kashif et al., 2019). There is also a closely related virus, HetPV13-an2 (Kashif et al., 2015; Hyder et al., 2018), which does not seem to cause noticeable effects on its natural host H. annosum S45-8 (Hyder et al., 2018).
The mechanisms behind the debilitating effects by HetPV13-an1 are not clear, although transcriptome analysis revealed considerable changes in Heterobasidion gene expression (Vainio et al., 2018b). This study is based on our observation of a fast-growing sector isolate of the original host strain of HetPV13-an1, H. annosum 94233, that seemed to have recovered spontaneously although the virus was still present in its hyphae. We aimed to investigate how the host can recover from the virus-induced growth retardation and the relevance of this phenomenon to the potential use of the virus as a biocontrol agent by (1) sequencing of the virus genome to determine the possibility of viral mutations, (2) analyzing the sector isolate’s gene expression to find changes that had occurred in the host, (3) quantifying viral genomes and transcripts in the recovered and debilitated isolates to determine whether changes had occurred in the virus quantity, and (4) analyzing the ratio of RdRp and CP transcripts of HetPV13-an1 among the isolates to test whether excess amount of viral polymerase transcripts correlates with a debilitated host phenotype as suggested earlier (Jurvansuu et al., 2014). A conspecific but apparently cryptic strain HetPV13-an2 was included in the phenotypical testing and analysis of viral transcript quantities to assess the effects of natural minor sequence variations to symptom severity.
Materials and Methods
Fungi and Viruses
HetPV13-an1 (GenBank accession: KF963177-78) is naturally hosted by H. annosum 94233 (Kashif et al., 2015; Vainio et al., 2015a). 94233-RC3 is a fast-growing sector isolate from 94233 (Figure 1) which was detected and stored in 2013 and then refreshened and restored in 2017 and 2018 at +4°C. The originally stored strain of 94233-RC3 as well as the restored strain in 2017 had lost the virus during the storage by 2021. Fortunately, the recently stored sample 2018 had retained the virus and could be used for further analysis. However, also one of the replicates regrown from this vial was found to have lost HetPV13-an1 and was named as 94233-RC3-0. In addition, two other isolates designated as 94233-RC1 and 94233-RC2 appeared to have recovered but returned quickly to their original slow-growing phenotype and were not analyzed further. The recovered HetPV13-an1 infected sector isolate 94233-RC3 was studied for its growth rate and gene expression in 2013. The strain was then stored at 4°C and moved back to +20°C only recently, after which it was found to have been cured of HetPV13-an1 in two of the three vials available: Vials from 2013 to 2017 had lost the virus by 2021, whereas a vial from 2018 had retained the virus and was selected for further analysis. However, also one of the replicates regrown from this vial was found to have lost HetPV13-an1 and was named as 94233-RC3-0.
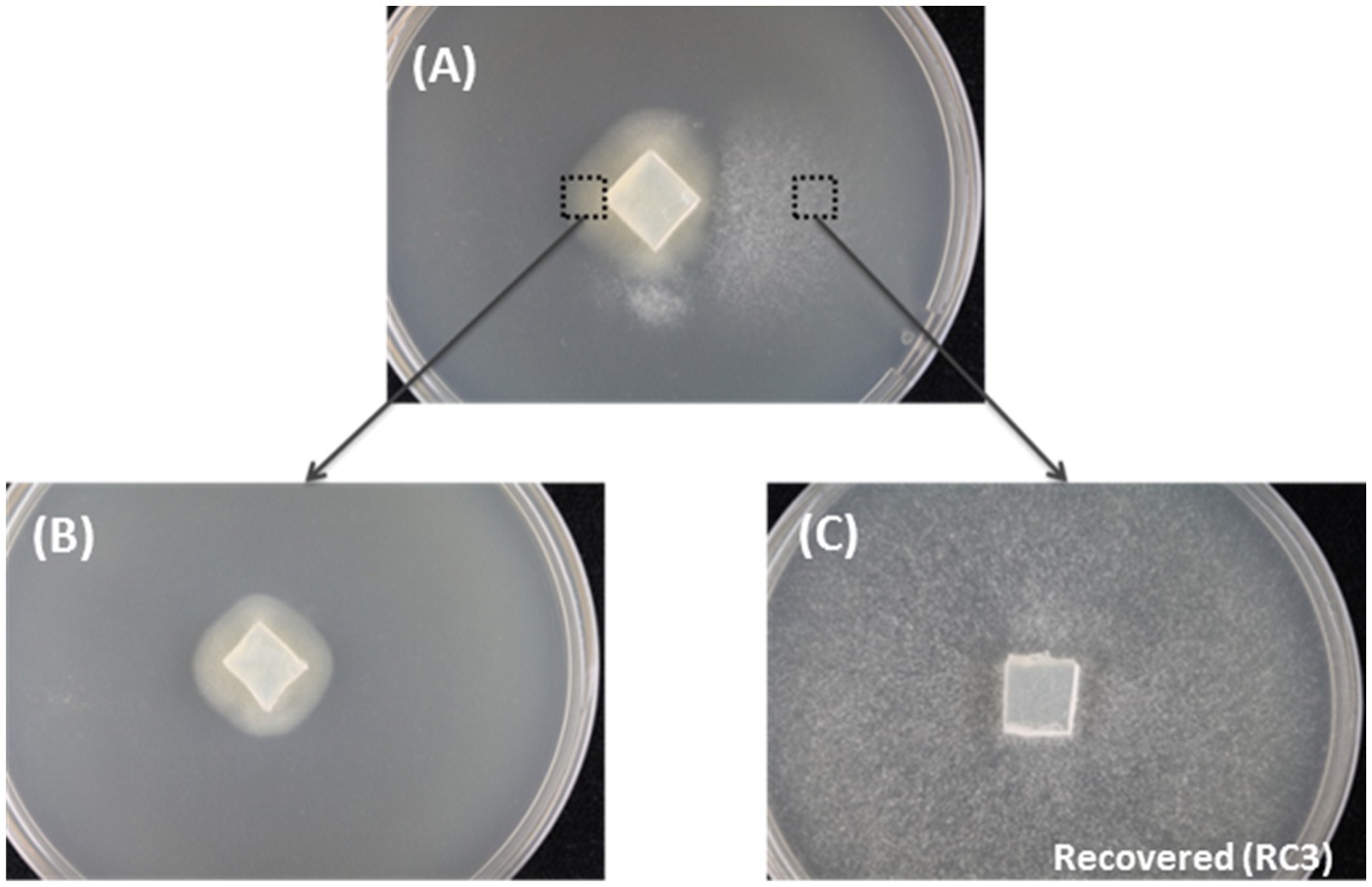
Figure 1. The spontaneous recovery of HetPV13-an1 infected fungal isolate as shown by mycelial morphology. (A) The original mycelium with slow and fast growth on the left and right side of the dish, respectively. (B) Mycelia transferred from the left side of the original mycelium. (C) Recovered 94233-RC3 transferred from the right side of the original mycelium.
H. annosum S45-8 is naturally coinfected by HetPV13-an2 (KF963179-80) (Kashif et al., 2015) and HetPV7-an1-b (KY859975-76) (Hyder et al., 2018). S45-8-PV13-an2 is a derivative of S45-8 cured of HetPV7 by single hyphal tip isolation. 94233-32D and S45-8-0 are previously and newly created partitivirus-free derivatives of 94233 (Vainio et al., 2018b) and S45-8, respectively, from which partitiviruses have been removed by a thermal treatment. Strain 94233-PV13-an2 was created by a transmission of HetPV13-an2 to 94233-32D by hyphal anastomosis on artificial medium.
mRNA Sample Preparation and RNA-Sequencing
We analyzed the effects of virus infection on the gene expression of 94233-RC3 as compared to an isogenic partitivirus-free strain H. annosum 94233-32D. The fungal strains were grown on cellophane covered modified orange serum (MOS) plates (Müller et al., 1994) for 1week. Mycelia were collected and homogenized in TRI-Reagent (Molecular Research Center Inc., United States) using the Fast-Prep FP120 homogenizer (JT Baker, Holland) with quartz sand grains as recommended by the manufactures. The sample was precipitated with isopropanol and total RNA was further purified with E.Z.N.A Fungal RNA Kit (Omega Bio-tek), and RNA was resuspended into DEPC-treated water (G. Biosciences, United States). RNA quantity and quality were checked with NanoVue (GE healthcare, United States) and bioanalyzer 2,100 (Agilent technologies, United States). Each analysis was performed with three biological replicates. For RNA-seq, the TruSeq RNA Library Prep kit v2 (Illumina Inc.) was used to prepare the samples. The process involved poly(A) selection with oligo(dT) beads without the depletion of rRNA. The sequencing was conducted with an Illumina HiSeq2500 instrument using paired-end sequencing chemistry with 50-bp read length (Bioinformatics Core of Turku Centre for Biotechnology, University of Turku and Åbo Akademi University).
dsRNA Extraction, RT-PCR, and Genome Sequence Determination
The dsRNA was extracted from 14days old fungal mycelia cultivated on cellophane membrane-covered MOS agar plates. Isolation of dsRNA was conducted according to Jurvansuu et al. (2014). Briefly, mycelia were homogenized and thereafter, RNA was isolated by phenol–chloroform (1:1) and chloroform–isoamylalcohol (24:1) extractions followed by precipitation by adding ethanol and NaCl at final concentrations of 15% and 100mm, respectively, and then adding cellulose fibers (medium) (Catalog no. C6288, Sigma-Aldrich, St. Louis, MO, United States). The precipitant was then moved to SigmaPrepTM spin columns (Sigma-Aldrich, United States) and washed. After eluting, the dsRNA was precipitated with ethanol. The dsRNA genome segments were excised from the gel and purified with RNAid Kit (MP Biomedicals, United States) and used in RT reaction.
T4RNA adapters were ligated and RT reactions set up using the RevertAid H minus M-MuLV reverse transcriptase (Thermo Scientific). The single primer amplification technique (Lambden et al., 1992) was used with previously described modifications (Tuomivirta and Hantula, 2003; Vainio et al., 2011; Kashif et al., 2015) to determine dsRNA sequences from isolate 94233-RC3. RT-PCR amplification was conducted using DyNAzyme DNA polymerase (Thermo Scientific) and specific primers (Kashif et al., 2015) as described before (Vainio et al., 2011). PCR products were sequenced at Macrogen Inc., South Korea1 using an Applied Biosystems 96-capillary ABI 3730xl DNA analyzer.
Bioinformatics
The gene expression analysis based on RNA-seq (Illumina sequencing) was performed by the Bioinformatics Core of Turku Centre for Biotechnology, University of Turku and Åbo Akademi University as described in Vainio et al. (2018b). The resulting Illumina sequence reads were aligned against the reference genome of H. irregulare, named at the time as H. annosum v.2.0 at the Joint Genome Institute (JGI).2 The read alignment was against the H. annosum v.2.0 reference genome using TopHat version 2.0.10 (Kim et al., 2013). Sequence annotations were based on known genes from H. annosum v.2.0 reference genome using HTSeq tool v.0.5.4p3 (Vainio et al., 2018b).
The data were normalized to make the values comparable across the sample set and the counts were normalized by the TMM normalization method of the edgeR R/Bioconductor package. Summary of the mapping statistics is provided in Supplementary Table S1. The expression level of each gene was analyzed by the number of sequenced reads mapped to the sequence of the reference gene, and differentially expressed genes (DEGs) were determined according to criteria used in Vainio et al. (2018b) based on their fold changes (FC) over a value of 4 and modified t-test value of p (=false-discovery-rate value of p) of 0.001 (p<0.001).
Validation of Gene Expression Data Using RT-qPCR
In order to simultaneously validate the results of RNA-Seq and to make comparison to the results in previous study (Vainio et al., 2018b) on the gene expression differences caused by HetPV13-an1 to strain 94233, the same 28 DEGs encoding various cellular functions as in Vainio et al. (2018b) were subjected for the RT-qPCR analysis of 94233-RC3 (Supplementary Table S2). Three independent biological replicates of each fungal sample (H. annosum 94233/32D and 94233-RC3) were incubated for 1week at 20°C and total RNA was extracted as described above for mRNA sequencing. The complementary DNA (cDNA) was diluted to a half-sample volume of water prior their use in RT-qPCR. The primers have been described in Vainio et al. (2018b), and three internal reference genes were used to normalize the results and primers as described by Raffaello and Asiegbu (2013) for RNA polymerase III transcription factor, Alfa tubulin, and Actin along with the 28 target genes.
RT-qPCR was carried out using EvaGreen® qPCR Mix Plus (Solis BioDyne, Estonia), 2μl of cDNA and 1μl (10μm) of each primer in a total volume of 20μl in the 36-well rotor of Rotor-GeneQ (Qiagen, United States) by following the manufacturer’s instructions. Two replicates for each biological sample were prepared for each RT-qPCR reaction. Cycling conditions were as follows: pre-incubation at 95°C for 15min, denaturation at 95°C for 15s, using specific annealing temperatures for primers (Vainio et al., 2018b) for 20s, and extension at 72°C for 20s. Additionally, melting curve profile was analyzed to test the quality and specificity of the reactions. Relative gene expression (fold change) as expression ratios of samples to controls (normalized with three reference genes) were analyzed with relative expression software tool Rest20093(Pfaffl et al., 2002) based on comparative quantitation analysis data. Relative expression (RT-qPCR) corresponds to log2 fold change of expression ratios measure by take-off values normalized with three reference genes.
Quantification of Viral Transcripts and Genome Segments
Total RNA Isolation and cDNA Synthesis
Total RNA was isolated from fungal mycelia after 1week growth on MOS agar plates as described in Jurvansuu et al. (2014). In short, fungal mycelia were collected and total RNA was extracted by TRI-Reagent. The RNA pellet was eluted into DEPC-treated water and the concentration and purity of the isolated RNA were analyzed by NanoVue (GE healthcare, United States). cDNA was made from 2μg of DNase I treated total RNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, United States) and random hexamer primers (Thermo Scientific, United States).
RT-qPCR Quantification for Virus Transcripts
Diluted cDNAs to half volume were used before using them in quantitative-PCR (qPCR). EvaGreen® dye (Solis BioDyne, Estonia) was used in qPCR on Rotor-GeneQ (Qiagen, United States) as recommended by the manufacturer. PCR primers were used as follows: GAPDH as a reference gene and specific primers based on RdRp and CP genomic segments (proteins) of two virus strains including HetPV13-an1 and HetPV13-an2 (Vainio et al., 2015a) were used for virus transcripts. The purified plasmids of cloned genes of CP and RdRp of these viral strains (Jurvansuu et al., 2014; Kashif et al., 2019) were then used in absolute quantification as shown by Jurvansuu et al. (2014). The normalization of the viral transcript levels was done using the host GAPDH as a reference gene (Raffaello and Asiegbu, 2013) and the absolute quantities of RNA transcripts or virus copy number were calculated using a standard curve.
Phenotype Testing by Growth Rate Experiments
The effects of single viral infections by HetPV13-an1 or HetPV13-an2 were assessed by measuring the growth rate difference between pairs of isogenic HetPV13 infected and uninfected strains on 2% MEA plates at 20°C as described by Kashif et al. (2019). The growth rate effects of HetPV13-an1 were measured using derivatives of host strain 94233 and those of HetPV13-an2 using derivatives of host strains 94233 and S45-8. Each inoculum was a circular plug of 0.5cm diameter, which was picked from a fresh mycelial culture (1–2week old depending on the apparent growth rate), grown on 2% MEA plate and placed at the center of a 2% MEA plate. Twelve independent biological replicates were prepared for each isolate and fungal growth was measured every second day after mycelial growth was initiated 3days post-inoculation and continued until the mycelium covered the plate. The fungal growth was measured with a digital planimeter (Planix 10S, Tamaya) and statistical analysis was done using t-test in Microsoft Excel 2010 (Supplementary Table S3).
Results
Recovered Phenotype of Strain 94233
Three fast-growing sectors of H. annosum strain 94233 hosting HetPV13-an1 were initially observed in year 2013 and isolated as depicted in Figure 1. Two of the isolates returned quickly to their original slow-growing phenotype, but the third one, 94233-RC3, seemed to be stable in room temperature during several months. The presence of HetPV13-an1 in this fast-growing isolate was verified by RT-qPCR with specific primers 13an1RdRpF and 13an1RdRpRev as described earlier (Vainio et al., 2015a). The three mitoviruses previously shown to be hosted by H. annosum 94233 (Vainio et al., 2018b; Vainio, 2019) were also retained in 94233-RC3 as revealed by mapping of the RNA-Seq reads.
Sequencing of the Virus Genome to Determine the Possibility of Virulence Reducing Mutations
The genome sequence of HetPV13-an1 in the H. annosum 94233 sector isolate 94233-RC3 (accession numbers MW115956 and MW115957) was 100% identical with the previously determined sequence for HetPV13-an1 (KF963177-78). Therefore, the observed phenotypical change was not caused by mutations in the virus genome sequence. Figure 2 shows a schematic presentation of the genome characterization scheme used. Each sequence site was covered by direct Sanger sequencing reactions including three PCR reactions each for CP and RdRp in two replicates, respectively.
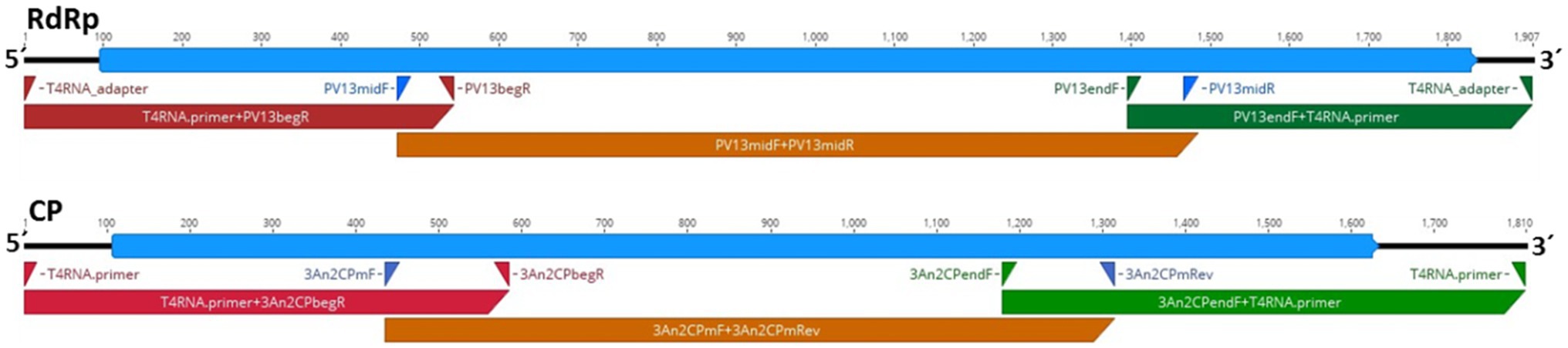
Figure 2. Schematic presentation of the genome organization of HetPV13-an1 (94233-RC3) showing the predicted coding regions for the RNA-dependent RNA polymerase (RdRp) and CP (blue) and the location of primers and corresponding amplification products (red, brown, and green) used for genome sequence determination.
Analysis of Host Gene Expression
The transcriptome analysis of the sector isolate 94233-RC3 was conducted simultaneously with that of the partitivirus-free strain H. annosum 94233/32D and the debilitated original isolate 94233 included in our earlier study (Vainio et al., 2018b). Here, we first compared the expression of the sector isolate to the primary results on the transcriptome of 94233-32D (cured of HetPV13-an1) and thereafter compared the degree of observed differences to the corresponding changes in 94233 (with HetPV13-an1). After those analyses, the sector isolate was stored at 4°C.
The RNA-seq analysis of 94233-RC3 involved three biological replicates each of which produced ~16–22 million reads (Supplementary Table S1). The raw sequencing reads are deposited in the NCBI GenBank SRA data archive under the submission number SAMN16435702 (BioProject PRJNA362289). The annotation of identified transcripts was made based on the reference genome Heterobasidium annosum (v2.0) and resulted in identifying putative functions for eight genes containing five upregulated and three downregulated transcripts based on the same criteria as in Vainio et al., 2018b on the expression ratio (fold change) between the compared sample groups and FDR-adjusted value of p for the comparison between the sample groups (Table 1; Supplementary Table S4).
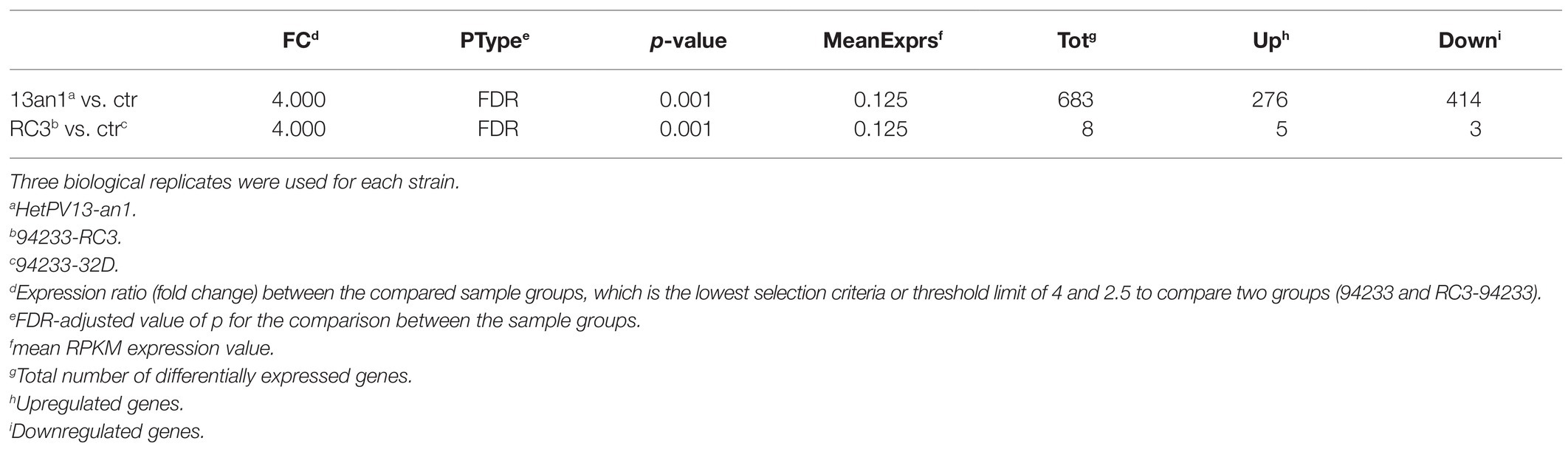
Table 1. Summary of RNA-seq analysis of H. annosum 94233-RC3 and 94233 infected with HetPV13-an1 and 94233/32D, which is a partitivirus-free isogenic strain.
The transcriptome of the sector isolate 94233-RC3 differed much less from that of the 94233-32D than the original virus-hosting strain 94233. Based on the number of reads related to each gene, only a total of eight transcripts were affected by HetPV13-an1 in 94233-RC3 (683 affected in 94233), of which five (276) were up- and three (414) downregulated (Table 1). All of the five genes upregulated in 94233-RC3 were upregulated also in 94233. In the case of the three downregulated genes, however, two of the genes were found to be almost five times less affected in 94233-RC3 than in 94233. Overall, the level of up- and downregulation of the host transcripts due to the HetPV13-an1 infection were considerably lower in 94233-RC3 than in 94233 (Figure 3).
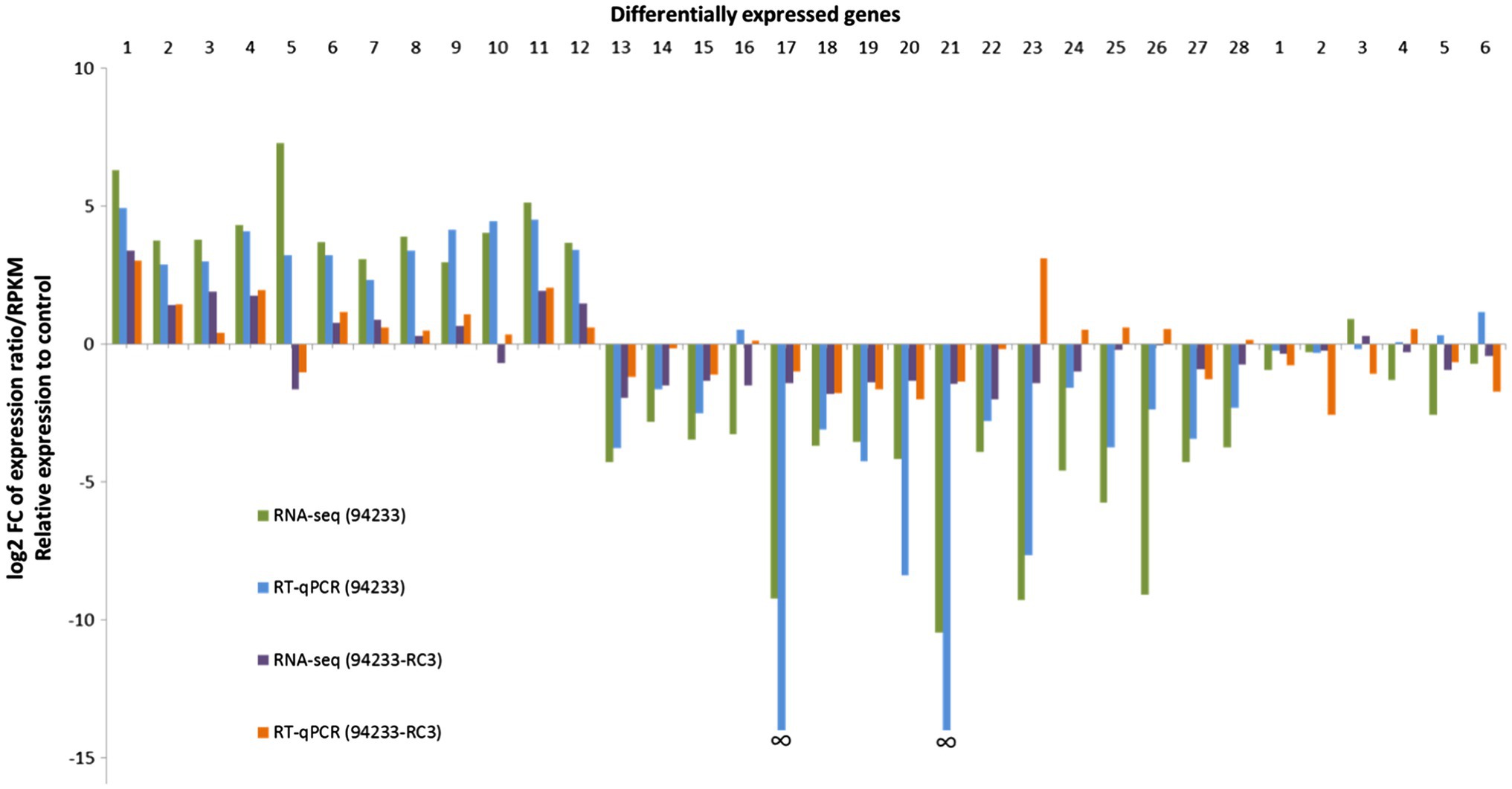
Figure 3. 94233-RC3-transcriptome RNA-seq data and qPCR validation. The columns relate to fold changes in gene expression of HetPV13-an1 hosting H. annosum strains 94233 and 94233-RC3, each was compared to the HetPV13-an1-free control 94233-32D. Names and gene id numbers of the 28 and then 6 RNAi-related target genes are listed (Supplementary Table S2). The relative expression of RNA-Seq corresponds to log2 fold change of reads per kb of transcript per million mapped reads (RPKM). The data for HetRV3-an1 (94233) are taken from Vainio et al. (2018b).
The validation of 94233-RC3 gene expression by RT-qPCR mostly agreed with the RNA-seq data but there was also a significant exception: target gene (TG) 23 with clear down- and upregulation in RNA-seq and RT-qPCR, respectively (Figure 3). In addition, TGs 10, 24, 25, 26, and 28 for 94233-RC3 were found to be slightly downregulated based on RNA-seq but in RT-qPCR validation only negligible upregulation was observed (Figure 3; Supplementary Table S2). Target genes 17 and 21 did not amplify at all from H. annosum 94233 and therefore the fold change was not shown by RT-qPCR in Figure 3 (Vainio et al., 2018b).
Qualitative Comparison of Gene Expressions of 94233-RC3 and 94233-32D in Relation to Biopathways
Probable gene functions affected by HetPV13-an1 in the sector isolate 94233-RC3 were assessed by comparing its transcript levels to the cured strain 94233-32D by RNA-Seq (Supplementary Table S4). The most prominent differences in the gene expression between HetPV13-an1 hosting sector isolate 94233-RC3 and the partitivirus-free strain 94233-32D were the three DEGs estExt_Genewise1Plus.C_080637, Hetan1.EuGene10000523, and e_gw1.12.503.1 that showed significant upregulation with FCs of 588, 387, and 180, respectively. These genes were related to putative Myosin class II heavy chain, Farnesyl cysteine-carboxyl methyltransferase (P450), and Sorbitol dehydrogenase proteins, corresponding to functions related to (1) fungal hyphal growth, septation, and conidial germination, (2) monooxygenase activity, and (3) secondary metabolites biosynthesis, transport, and catabolism, respectively. The functions of the two other upregulated genes are not known.
The difference in downregulated genes in the HetPV13-an1 infected 94233-RC3 was much smaller than in upregulated genes. DEG genesh1_kg.03__767__4245_1_CCOZ_CCPA_CCPB_CCPC_EXTA was downregulated 10-fold, but its function is not known. However, DEGs Hetan1.e_gw1.5.283.1 and Hetan1.estExt_fgenesh2_pg.C_100320 were downregulated less than 5-fold and code for putative L-fuculose-phosphate aldolase (fucA) and phenol 2-monooxygenase, respectively. The first of these functions is related to fructose and mannose metabolism, whereas the second one affects detoxification and aromatic compound metabolism.
RNAi-related genes are of a special interest because their functions are related to defense against mycoviruses. In 94233-RC3, none of these genes were up- or downregulated based on the RNA-Seq according to the criteria used here (and in Vainio et al., 2018b). However, in RT-qPCR analysis, two of the RNAi-related genes had significantly lower FC-values of −5.95 and−2.12, respectively. They code for a dicer (e_gw1.03.2068.1) and an argonaute gene (e_gw1.02.2254.1) (Supplementary Table S2). Thus, the effect of HetPV13-an1 on the viral RNAi-based defense of 94233-RC3 was quite low.
The Most Differentially Expressed Genes in 94233-RC3 Compared to 94233
The gene expression was in general significantly less affected by HetPV13-an1 in 94233-RC3 than in the original 94233 (Vainio et al., 2018b). However, the most differentially expressed genes in the sector isolate, estExt_Genewise1Plus.C_080637, Hetan1.EuGene10000523, and e_gw1.12.503.1, had FCs 588, 387 and 180 in 94233-RC3 and 871, 202 and 225 in 94233, respectively, suggesting that HetPV13-an1 was still able to modify considerably the most seriously affected genes in the sector isolate.
Analysis of the Ratio of HetPV13-an1 RdRp and CP Among the Isolates
The quantification of viral CP and RdRp mRNA transcripts using RT-qPCR revealed that the amounts of HetPV13-an1-encoded transcripts before storing 94233-RC3 were considerably lower in 94233-RC3 than in 94233 (Figures 4A,B) and differed between the two genes. The amounts of RdRp were 3.5 and 13.4 times of that of GAPDH RNA in 94233-RC3 and 94233, and the corresponding values for CP were 0.45 and 1.5, respectively. However, the RdRp and CP amounts reduced to 0.71 and 0.14 times of that of GAPDH RNA after long storage (2013–2021) of 94233-RC3. Moreover, the quantities of transcripts for RdRp and CP in 94233-RC3 were only 26 and 32% in 2013, respectively, of that in 94233, which further reduced in 94233-RC3 to 5 and 9% after long storage, respectively (Figures 4A,B). Similarly, the transcript levels of HetPV13-an2 RdRp and CP were 32 and 36% of that observed for HetPV13-an1 in 94233. However, HetPV13-an2 in its native host S45-8 produced significantly lower titer of the virus-an2 and showed more equal amounts of both transcripts from its genome segments (Figures 4C,D).
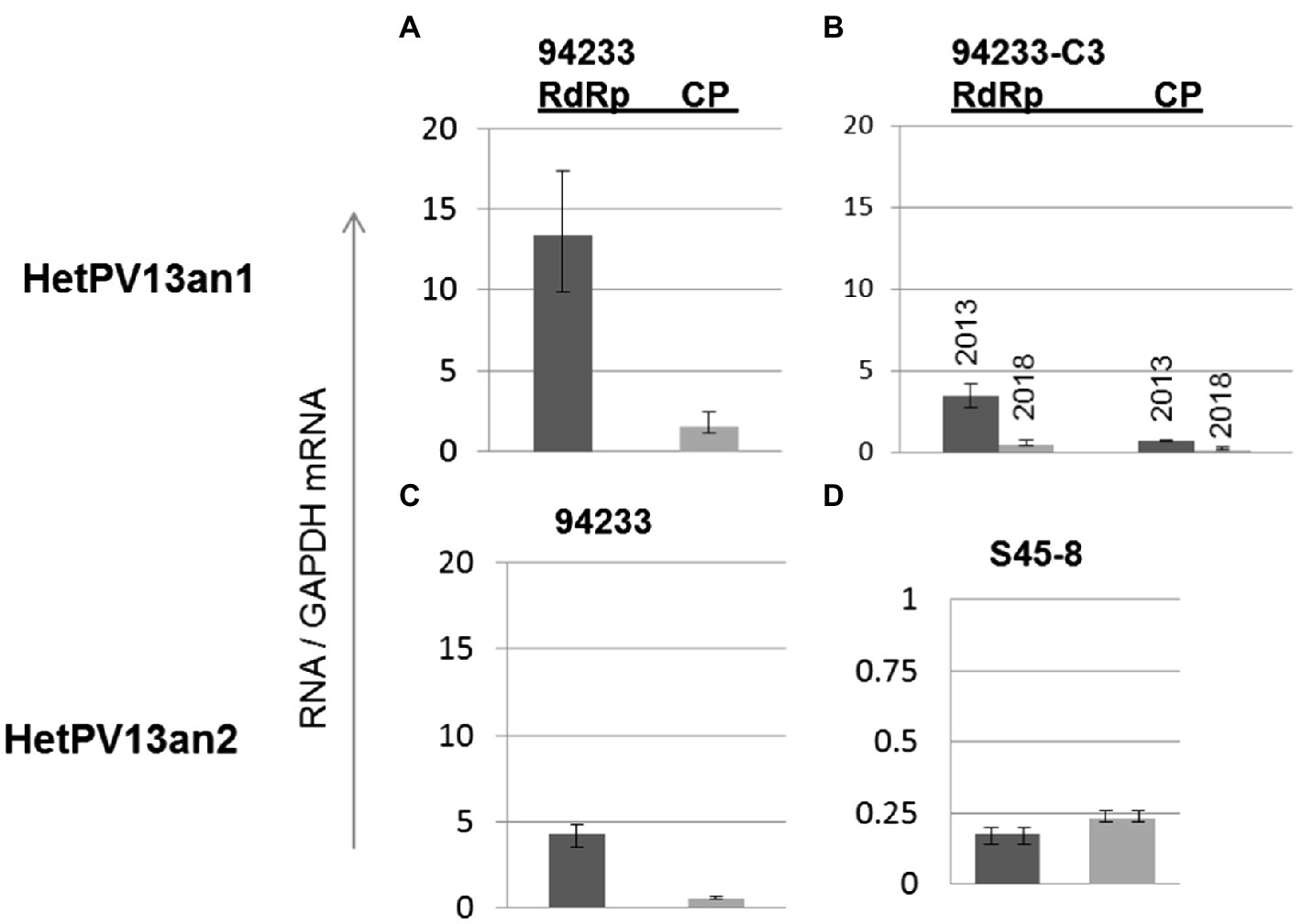
Figure 4. The variation in viral CP and RdRp transcript levels in H. annosum strains infected by partitiviruses. (A) Strain 94233 with its natural viral infection HetPV13-an1, (B) fast-growing sector isolate 94233-RC3 with infection by HetPV13-an1 before (2013) and after storage (2018), and (C,D) the amounts of viral transcripts of HetPV13-an2 in 94233/32D and in its native host (S45-8). Note: The scale for mRNA transcript levels is different in D due to significantly lower levels of expression.
The relative ratios of RdRp to CP were somewhat lower (7.7 and 5.2 before and after the long storage, respectively) in 94233-RC3 and 94233-PV13-an2 (7.7) compared to 94233 (8.7). Moreover, isolates 94233-RC1 and 94233-RC2 that recovered their growth rates temporarily had lowered levels of transcripts amounts (Supplementary Figure 1) and also had a significantly higher RdRp to CP ratios of 27 and 17, respectively (Supplementary Figure 2). In contrast, the ratio of RdRp and CP transcripts of HetPV13-an2 was about 90% lower in its original host S45-8 than in 94233 (Figure 5).
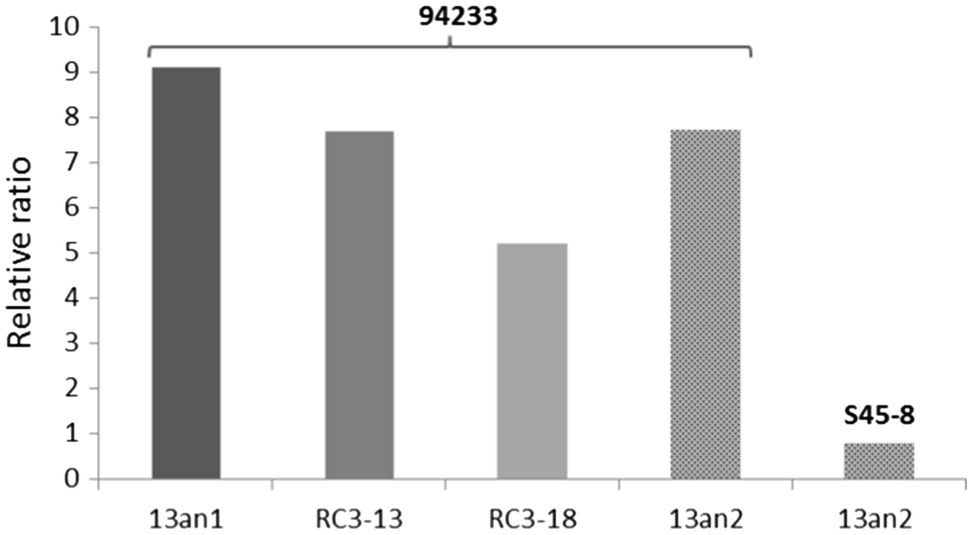
Figure 5. Variation in the ratio of viral RdRp and CP in different host strains. Strains 94233 and 94233-RC3 hosted virus HetPV13-an1, and strains 94233-PV13-an2 and S45-8 hosted virus HetPV13-an2. 13an1=HetPV13-an1 and RC3-13=94233-RC3 before storage; RC3-18=94233-RC3 after storage, 13an2=HetPV13-an2.
Growth Rates
Inoculums made from single 2% MEA plate in order to create 12 independent biological replicates of 94233-RC3 before and after storage 94233-RC3 appeared to be stable (Supplementary Figure 3) and had growth rates that were 45 and 62% of partitivirus-free 94233-32D and 12 and 16 times higher than the original HetPV13-an1 hosting 94233 (Figure 6). The 94233-RC3 strain that had lost HetPV13-an1 during the storage had a very similar growth rate as the virus-hosting strain 94233-RC3 before and after the storage.
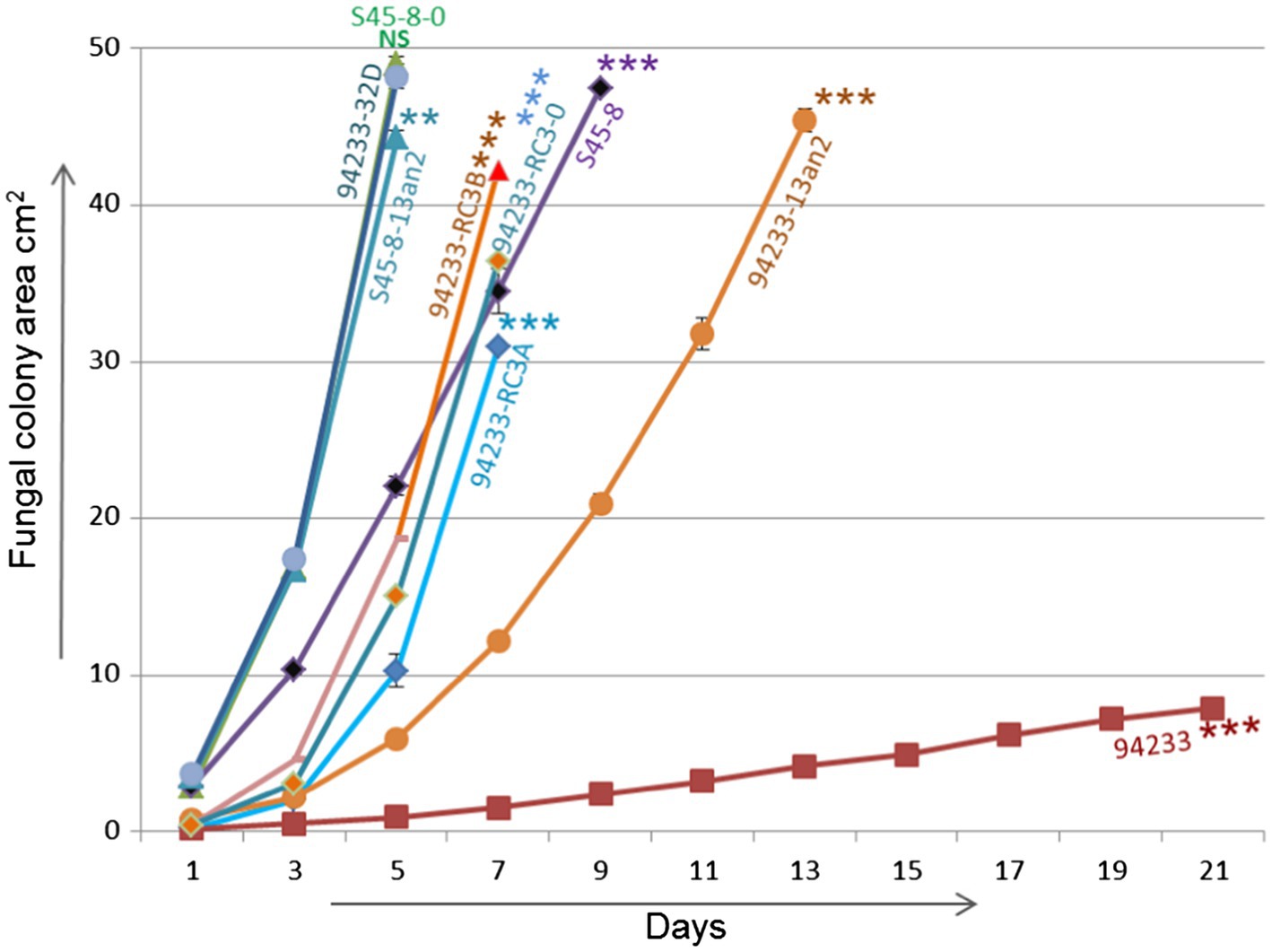
Figure 6. Growth rate analysis. Growth rates of 94233-RC3 and its stored derivative 94233-RC3B infected with HetPV13-an1 including original 94233 with its consistent reduced phenotype and the partitivirus-free control 94233-32D (Kashif et al., 2019). 94233-RC3A=before storage; 94233-RC3B=after storage; 94233 (the original slow-growing strain hosting HetPV13-an1); S45-8-0=virus-free control; S45-8-PV13an2 (single virus infection HetPV13-an2); S45-8 (the original strain with HetPV13-an2 and HetPV7-an1); and 94233-PV13an2 (HetPV13-an2 hosted by 94233 strain). The significance of the differences between the mean growth per day of the virus-free/cured and virus infected fungal isolates is shown by asterisks: *p<0.05; **p<0.001; and ***p<0.0001; #=single replicate; NS=Non-significant.
94233-PV13-an2 showed a significant 64% growth reduction compared to 94233-32D, but S45-8-an2 had only 9.6% growth reduction compared to the cured S45-8-0 and thus grew at the same rate as 94233-32D. Originally HetPV13-an2 was found in coinfection with HetPV7-an1 in its original host S45-8 and showed similar growth rates as 94233-RC3, i.e., up to 45% slower growth compared to the partitivirus-free S45-8-0 (Figure 6).
Discussion
This study was launched based on our observation that a Heterobasidion strain seriously debilitated due to a HetPV13-an1 infection had recovered spontaneously. That accords with some previous studies on other mycoviruses which have shown that coinfections of two fusarivirus strains of F. graminearum are linked with debilitating phenotypic alterations in their fungal hosts and that the effect of Fusarium mycoviruses in the fungal transcriptome is virus-specific (Darissa et al., 2012; Lee et al., 2014). In this study, we originally assumed that the fast-growing hyphal sector had been cured of the virus, but after examining the presence of the virus and its genome sequence, it was evident that there was no change in the virus itself. As mutation in the virus genome did not cause the altered phenotype of 94233-RC3, we conducted analyses of gene expression to investigate possible host-related reasons behind the recovery of our original fast-growing sector isolate. In a previously published study, the debilitating infection of HetPV13-an1 seriously affected many different biochemical pathways and the expression of some genes was even turned down completely (Vainio et al., 2018b). Similar analysis of the sector isolate 94233-RC3 revealed that the fungus had restored most of its gene regulation despite the presence of HetPV13-an1, including the two extremely downregulated (practically knocked out) genes in 94233. However, notably high changes were still observed in single genes related to the formation of fungal hypha, monooxygenase activity, and carbohydrate metabolism, although it remained unclear how and why the expression of these three genes was retained in the sector isolate 94233-RC3.
The recovery of most gene functions in 94233-RC3 was accompanied by a drastic drop in the quantities of viral transcripts, i.e., the viral titer: The amounts of RdRp and CP transcripts of HetPV13-an1 in 94233-RC3 were 74 and 68% lower, respectively, than in seriously debilitated 94233. In F. oxysporum and Lentinula edodes, the viral titer correlates with the degree of debilitation (Kim et al., 2015; Lemus-Minor et al., 2018; Yu and Kim, 2021), but there is also evidence that viral titers depend on host cell physiology (Schoffelen et al., 2013), culture conditions (Hillman et al., 1990), and environmental factors (Bryner and Rigling, 2011). Also RNA silencing could have a role in controlling the virus titers and associated phenotypic changes as, e.g., Mochama et al. (2018) showed that phenotypic alterations caused by virus infection are driven by RNA silencing mechanisms in Sclerotium sclerotiorum and antiviral RNA silencing responses led to phenotypic changes of fungal hosts in Cryphonectria parasitica (Segers et al., 2007) and Colletotrichum higginsianum (Campo et al., 2016). The expression of genes for RNA silencing was only relatively slightly affected by the debilitating HetPV13-an1 infection of 94233 (Vainio et al., 2018b), and the sector isolate 94233-RC3 did not differ much from that. Therefore, our results support the view that RNA silencing is not much affected by an infection of HetPV13-an1.
Jurvansuu et al. (2014) suggested that the ratio of RdRp and CP transcripts of partitiviruses would be more important than their actual quantities in determining the phenotypic effects on the host. In this study, we observed slightly lower ratios of virus-encoded transcripts in 94233-RC3 and also in 94233-PV13-an2 hosting a closely related virus strain HetPV13-an2 causing only mild symptoms. Furthermore, the quantities of the two transcripts were almost equal in the original host S45-8 with a somewhat debilitating coinfection of HetPV13-an2 and HetPV7-an1, and highly similar to HetPV13-an1 in the recovered sector isolate 94233-RC3. These findings may be taken as support for the hypothesis by Jurvansuu et al. (2014), but also the direct transcript quantities correlate with the degree of symptoms. However, it has recently been described that RdRp transcripts in partitiviruses are often more abundant than CP transcripts although these infections seem to lack debilitating effects (Kashif et al., 2019; (Vainio et al., 2015a), which seems to contradict with the hypothesis of Jurvansuu et al. (2014).
94233-RC3 isolates before and after storage (2018) did not show significantly different growth rates. However, one of the replicates from the storage was free of HetPV13-an1, which might have been due to uneven distribution of viruses in the mycelium. The growth rate of this isolate 94233-RC3-0 was similar to 94233-RC3 strains that had retained the virus. This might suggest that the recovered phenotype of 94233-RC3 is due to a genetic or other change in the host mycelium that provides tolerance against the debilitating effects of HetV13-an1 but at the same time reduces the inherent growth rate of the mycelium. Non-uniform occurrence of virus particles within fungal mycelia has been reported by Ghabrial (1980), but in contrast to our observation, virus-like particles were observed more frequently in old than in actively growing hyphal tips by Border et al. (1972) and DeMarini et al. (1977).
A single infection of HetPV13-an2 was cryptic in its native host but caused considerable debilitation in a non-native host. This was in accordance to a previous study (Jurvansuu et al., 2014) where virus strains in exotic hosts seemed to have more significant phenotypic effects than in their native hosts. However, a coinfection with another virus strain HetPV7-an1 (S45-8) slowed down the growth by 45% after inoculation. This accords also with analyses by Kashif et al. (2019), who showed that different strains of HetPV11 affected differently the debilitating effects of HetPV13-an1. Interactions between viral double infections were also studied by Wu et al. (2010), who described suppression of replication of Botrytis cinerea mitovirus 1 (BcMV1) by another related RNA virus (BcMV1-S). Also in Rosellinia necatrix, the phenotypic outcomes were different when the fungus was hosting single or coinfections of a megabirnavirus and a partitivirus (Sasaki et al., 2016). Moreover, phenotypic variations caused by interplay between partitiviruses (Telengech et al., 2020) and in a coinfection of a chrysovirus and a partitivirus in the human pathogen A. fumigatus caused unique phenotypic modifications (Bhatti et al., 2011).
The recovered HetPV13-an1 infected sector isolate 94233-RC3 was originally isolated and studied for its growth rate as well as for fungal and viral gene expressions almost a decade ago (but reported only here). In this study, we found that the virus infection in the sector isolate was low titer and susceptible/vulnerable to storage and culturing conditions. As HetPV13-an1 and other Heterobasidion partitiviruses have previously been stored in similar conditions for several years and even more than half a century (Kashif et al., 2015), this loss of a virus was surprising and suggests that the balance between the host and virus in 94233-RC3 was somewhat less stable than normally. However, loss of fungal viruses during a storage has previously been observed in other fungi, such as C. parasitica and Fusarium circinatum (Springer et al., 2013; Zamora-Ballesteros et al., 2021).
Conclusion
This study showed that Heterobasidion strains infected by a single partitivirus may show different phenotypes with considerably different fungal transcriptomes, viral titers, transcript ratios, and mycelial appearances.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
MK, EV, and JH conceived and designed the experiments. MK, JJ, and RH performed the experiments. MK, JJ, EV, and JH analyzed the data and wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Academy of Finland (grant numbers 322001 and 309896).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Juha Puranen, Tuija Hytönen, and Carita Karenius for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.661554/full#supplementary-material
Footnotes
References
Allen, T. D., and Nuss, D. L. (2004). Specific and common alterations in host gene transcript accumulation following infection of the chestnut blight fungus by mild and severe hypoviruses. J. Virol. 78, 4145–4155. doi: 10.1128/JVI.78.8.4145-4155.2004
Bhatti, M. F., Jamal, A., Petrou, M. A., Cairns, T. C., Bignell, E. M., and Coutts, R. H. A. (2011). The effects of dsRNA mycoviruses on growth and murine virulence of aspergillus fumigatus. Fungal Genet. Biol. 48, 1071–1075. doi: 10.1016/j.fgb.2011.07.008
Border, D. J., Buck, K. W., Chain, E. B., Kempson-Jones, G. F., Lhoas, P., and Ratti, G. (1972). Viruses of Penicillium and aspergillus species. Biochem. J. 127, 4P–6P. doi: 10.1042/bj1270004P
Bryner, S. F., and Rigling, D. (2011). Temperature-dependent genotype-by-genotype interaction between a pathogenic fungus and its hyperparasitic virus. Am. Nat. 177, 65–74. doi: 10.1086/657620
Campo, S., Gilbert, K. B., and Carrington, J. C. (2016). Small RNA-based antiviral defense in the phytopathogenic fungus Colletotrichum higginsianum. PLoS Pathog. 12:e1005640. doi: 10.1371/journal.ppat.1005640
Choi, J., Lee, G. W., Kim, K. T., Jeon, J., Détry, N., Kuo, H. C., et al. (2017). Comparative analysis of genome sequences of the conifer tree pathogen, Heterobasidion annosum. Genom. Data. 14, 106–113. doi: 10.1016/j.gdata.2017.10.003
Dai, Y. C., and Korhonen, K. (1999). Heterobasidion annosum group S identified in northern-eastern China. Eur. J. For. Pathol. 29, 273–279. doi: 10.1046/j.1439-0329.1999.00153.x
Darissa, O., Adam, G., and Schafer, W. (2012). A dsRNA mycovirus causes hypovirulence of Fusarium graminearum to wheat and maize. Eur. J. Plant Pathol. 134, 181–189. doi: 10.1007/s10658-012-9977-5
Dawe, A. L., and Nuss, D. L. (2013). Hypovirus molecular biology: From koch's postulates to host self-recognition genes that restrict virus transmission. Adv. Virus Res. 86, 109–147. doi: 10.1016/B978-0-12-394315-6.00005-2
DeMarini, D. M., DeMarini, D. M., Kurtzman, C. P., and Fennell, D. I. (1977). Transmission of PsVF and PsV S mycoviruses during conidiogenesis of Penicillium stoloniferum. J. Gen. Microbiol. 100, 59–64. doi: 10.1099/00221287-100-1-59
Deng, F., Allen, T. D., Hillman, B. I., and Nuss, D. L. (2007). Comparative analysis of alterations in host phenotype and transcript accumulation following hypovirus and mycoreovirus infections of the chestnut blight fungus Cryphonectria parasitica Eukaryot. Cell 6, 1286–1298. doi: 10.1128/EC.00166-07
Eusebio-Cope, A., Sun, L., Tanaka, T., Chiba, S., Kasahara, S., and Suzuki, N. (2015). The chestnut blight fungus for studies on virus/host and virus/virus interactions: from a natural to a model host. Virology 477, 164–175. doi: 10.1016/j.virol.2014.09.024
Garbelotto, M., and Gonthier, P. (2013). Biology, epidemiology, and control of Heterobasidion species worldwide. Annu. Rev. Phytopathol. 51, 39–59. doi: 10.1146/annurev-phyto-082712-102225
Ghabrial, S. A. (1980). Effects of fungal viruses on their hosts. Annu. Rev. Phytopathol. 18, 441–461. doi: 10.1146/annurev.py.18.090180.002301
Ghabrial, S. A., Castón, J. R., Jiang, D., Nibert, M. L., and Suzuki, N. (2015). 50-plus years of fungal viruses. Virology 479, 356–368. doi: 10.1016/j.virol.2015.02.034
Ghabrial, S. A., and Suzuki, N. (2009). Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47, 353–384. doi: 10.1146/annurev-phyto-080508-081932
Hillman, B. I., Shapira, R., and Nuss, D. L. (1990). Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80, 950–956. doi: 10.1094/Phyto-80-950
Hollings, M. (1962). Viruses associated with a die-Back disease of cultivated mushroom. Nature 196, 962–965. doi: 10.1038/196962a0
Huang, S., and Ghabrial, S. A. (1996). Organization and expression of the double-stranded RNA genome of Helminthosporium victoriae 190S virus, a totivirus infecting a plant pathogenic filamentous fungus. Proc. Natl. Acad. Sci. U. S. A. 93, 12541–12546.
Hyder, R., Pennanen, T., Hamberg, L., Vainio, E. J., Piri, T., and Hantula, J. (2013). Two viruses of Heterobasidion confer beneficial, cryptic or detrimental effects to their hosts in different situations. Fungal Ecol. 6, 387–396. doi: 10.1016/j.funeco.2013.05.005
Hyder, R., Piri, T., Hantula, J., Nuorteva, H., and Vainio, E. J. (2018). Distribution of viruses inhabiting Heterobasidion annosum in a pine-dominated forest plot in southern Finland. Microb. Ecol. 75, 622–630. doi: 10.1007/s00248-017-1027-6
Ihrmark, K., (2001). Double-Stranded RNA Elements in the Root Rot Fungus Heterobasidion Annosum. Uppsala, Sweden: Dissertation. Swedish University of Agricultural Sciences.
Jurvansuu, J., Kashif, M., Vaario, L., Vainio, E., and Hantula, J. (2014). Partitiviruses of a fungal forest pathogen have species-specific quantities of genome segments and transcripts. Virology 462, 25–33. doi: 10.1016/j.virol.2014.05.021
Kashif, M., Hyder, R., De Vega Perez, D., Hantula, J., and Vainio, E. J. (2015). Heterobasidion wood decay fungi host diverse and globally distributed viruses related to Helicobasidium mompa partitivirus V70. Virus Res. 195, 119–123. doi: 10.1016/j.virusres.2014.09.002
Kashif, M., Jurvansuu, J., Vainio, E. J., and Hantula, J. (2019). Alphapartitiviruses of Heterobasidion wood decay fungi affect each other’s transmission and host growth. Front. Cell. Infect. Microbiol. 9:64. doi: 10.3389/fcimb.2019.00064
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Kim, J. M., Song, H. Y., Choi, H. J., Yun, S. H., So, K. K., Ko, H. K., et al. (2015). Changes in the mycovirus (LeV) titer and viral effect on the vegetative growth of the edible mushroom Lentinula edodes. Virus Res. 197, 8–12. doi: 10.1016/j.virusres.2014.11.016
Lambden, P. R., Cooke, S. J., Caul, E. O., and Clarke, I. N. (1992). Cloning of noncultivatable human rotavirus by single primer amplification. J. Virol. 66, 1817–1822. doi: 10.1128/jvi.66.3.1817-1822.1992
Lee, K. M., Cho, W. K., Yu, J., Son, M., Choi, H., Min, K., et al. (2014). A comparison of transcriptional patterns and mycological phenotypes following infection of Fusarium graminearum by four mycoviruses. PLoS One 9:e100989. doi: 10.1371/journal.pone.0100989
Lemus-Minor, C. G., Cañizares, M. C., García-Pedrajas, M. D., and Pérez-Artés, E. (2018). Fusarium oxysporum f. sp. dianthi virus 1 accumulation is correlated with changes in virulence and other phenotypic traits of its fungal host. Phytopathology 108, 957–963. doi: 10.1094/PHYTO-06-17-0200-R
Liu, J. J., Shamoun, S. F., Leal, I., Kowbel, R., Sumampong, G., and Zamany, A. (2018). Characterization of Heterobasidion occidentale transcriptomes reveals candidate genes and DNA polymorphisms for virulence variations. Microb. Biotechnol. 11, 537–550. doi: 10.1111/1751-7915.13259
Márquez, L. M., Redman, R. S., Rodriguez, R. J., and Roossinck, M. J. (2007). A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315, 513–515. doi: 10.1126/science.1136237
Mochama, P., Jadhav, P., Neupane, A., and Marzano, S.-Y. L. (2018). Mycoviruses as triggers and targets of RNA silencing in white mold fungus Sclerotinia sclerotiorum. Viruses 10:214. doi: 10.3390/v10040214
Müller, M. M., Kantola, R., and Kitunen, V. (1994). Combining sterol and fatty acid profiles for the characterization of fungi. Mycol. Res. 98, 593–603. doi: 10.1016/S0953-7562(09)80404-8
Myers, J. M., Bonds, A. E., Clemons, R. A., Thapa, N. A., Simmons, D. R., Carter-House, D., et al. (2020). Survey of early diverging lineages of fungi reveals abundant and diverse mycoviruses. MBio 11, e02027–e02020. doi: 10.1128/mBio.02027-20
Nibert, M. L., Ghabrial, S. A., Maiss, E., Lesker, T., Vainio, E., Jiang, D., et al. (2014). Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res. 188, 128–141. doi: 10.1016/j.virusres.2014.04.007
Niemelä, T., and Korhonen, K. (1998). “Taxonomy of the genus Heterobasidion,” in Heterobasidion Annosum: Biology, Ecology. eds. S. Woodward, J. Stenlid, R. Karjalainen, and A. Hütterman (Impact and Control. C.A.B International), 27–33.
Olson, A., Aerts, A., Asiegbu, F., Belbahri, L., Bouzid, O., Broberg, A., et al. (2012). Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol. 194, 1001–1013. doi: 10.1111/j.1469-8137.2012.04128.x
Otrosina, W. J., and Garbelotto, M. (2010). Heterobasidion occidentale sp. nov. and Heterobasidion irregulare nom. nov.: a disposition of north American Heterobasidion biological species. Fungal Biol. 114, 16–25. doi: 10.1016/j.mycres.2009.09.001
Pearson, M. N., Beever, R. E., Boine, B., and Arthur, K. (2009). Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 10, 115–128. doi: 10.1111/j.1364-3703.2008.00503.x
Pfaffl, M. W., Horgan, G. W., and Dempfle, L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. doi: 10.1093/nar/30.9.e36
Preisig, O., Moleleki, N., Smit, W. A., Wingfield, B. D., and Wingfield, M. J. (2000). A novel RNA mycovirus in a hypovirulent isolate of the plant pathogen Diaporthe ambigua. J. Gen. Virol. 81, 3107–3114. doi: 10.1099/0022-1317-81-12-3107
Raffaello, T., and Asiegbu, F. (2013). Evaluation of potential reference genes for use in gene expression studies in the conifer pathogen (Heterobasidion annosum). Mol. Biol. Rep. 40, 4605–4611. doi: 10.1007/s11033-013-2553-z
Sasaki, A., Nakamura, H., Suzuki, N., and Kanematsu, S. (2016). Characterization of a new megabirnavirus that confers hypovirulence with the aid of a co-infecting partitivirus to the host fungus, Rosellinia necatrix. Virus Res. 219, 73–82. doi: 10.1016/j.virusres.2015.12.009
Schoffelen, A. F., Wensing, A. M., Tempelman, H. A., Geelen, S. P., Hoepelman, A. I., and Barth, R. E. (2013). Sustained virological response on second-line antiretroviraltherapy following virological failure in HIV-infected patients in rural SouthAfrica. PLoS One 8:e58526. doi: 10.1371/journal.pone.0058526
Segers, G. C., Zhang, X., Deng, F., Sun, Q., and Nuss, D. L. (2007). Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. 104, 12902–12906. doi: 10.1073/pnas.0702500104
Sillo, F., Garbelotto, M., Friedman, M., and Gonthier, P. (2015). Comparative genomics of sibling fungal pathogenic taxa identifies adaptive evolution without divergence in pathogenicity genes or genomic structure. Genome Biol. Evol. 7, 3190–3206. doi: 10.1093/gbe/evv209
Springer, J. C., Davelos Baines, A. L., Chansler, M. T., and Jarosz, A. M. (2013). Evaluating the long-term storage of Cryphonectria parasitica. Fungal. Genet. Rep. 60, 11–15. doi: 10.4148/1941-4765.1007
Stenlid, J., and Redfern, D. B. (1998). “Spread within the tree and stand,” in Heterobasidion Annosum: Biology, Ecology, Impact and Control. eds. S. Woodward, J. Stenlid, R. Karjalainen, and A. Hütterman (Wallingford, UK: CAB International), 125–141.
Sutela, S., Forgia, M., Vainio, E. J., Chiapello, M., Daghino, S., Vallino, M., et al. (2020). The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus. Evol. 6:veaa076. doi: 10.1093/ve/veaa076
Sutela, S., Poimala, A., and Vainio, E. J. (2019). Viruses of fungi and oomycetes in the soil environment. FEMS Microbiol. Ecol. 95:fiz119. doi: 10.1093/femsec/fiz119
Telengech, P., Hisano, S., Mugambi, C., Hyodo, K., Arjona-López, J. M., López-Herrera, C. J., et al. (2020). Diverse partitiviruses from the phytopathogenic fungus, Rosellinia necatrix. Front. Microbiol. 11:1064. doi: 10.3389/fmicb.2020.01064
Tuomivirta, T., and Hantula, J. (2003). Two unrelated double-stranded RNA molecule patterns in Gremmeniella abietina type A code for putative viruses of the families Totiviridae and Partitiviridae. Arch. Virol. 148, 2293–2305. doi: 10.1007/s00705-003-0194-6
Vainio, E. J. (2019). Mitoviruses in the conifer root rot pathogens Heterobasidion annosum and H. parviporum. Virus Res. 271:197681. doi: 10.1016/j.virusres.2019.197681
Vainio, E. J., Chiba, S., Ghabrial, S. A., Maiss, E., Roossinck, M., Sabanadzovic, S., et al. (2018a). ICTV virus taxonomy profile: Partitiviridae. J. Gen. Virol. 99, 17–18. doi: 10.1099/jgv.0.000985
Vainio, E. J., Hakanpää, J., Dai, Y., Hansen, E., Korhonen, K., and Hantula, J. (2011). Species of Heterobasidion host a diverse pool of partitiviruses with global distribution and interspecies transmission. Fungal Biol. 115, 1234–1243. doi: 10.1016/j.funbio.2011.08.008
Vainio, E. J., and Hantula, J. (2016). Taxonomy, biogeography and importance of Heterobasidion viruses. Virus Res. 219, 2–10. doi: 10.1016/j.virusres.2015.10.014
Vainio, E. J., and Hantula, J. (2018). “Fungal viruses,” in Viruses of Microorganisms. eds. P. Hyman and S. T. Abedon (UK: Caister Academic Press), 193–209.
Vainio, E. J., Hyder, R., Aday, G., Hansen, E., Piri, T., and Dogmus-Lehtijarvi, T., T., Lehtijarvi, A., Korhonen, K., and Hantula, J. (2012). Population structure of a novel putative mycovirus infecting the conifer root-rot fungus Heterobasidion annomm sensu lato. Virology 422, 366–376. doi: 10.1016/j.virol.2011.10.032
Vainio, E. J., Jurvansuu, J., Hyder, R., Kashif, M., Piri, T., Tuomivirta, T., et al. (2018b). Heterobasidion partitivirus 13 mediates severe growth debilitation and major alterations in the gene expression of a fungal forest pathogen. J. Virol. 12, e01744–e01717. doi: 10.1128/JVI.01744-17
Vainio, E. J., Jurvansuu, J., Streng, J., Rajamäki, M., Hantula, J., and Valkonen, J. P. T. (2015a). Diagnosis and discovery of fungal viruses using deep sequencing of small RNAs. J. Gen. Virol. 96, 714–725. doi: 10.1099/jgv.0.000003
Vainio, E. J., Mueller, M. M., Korhonen, K., Piri, T., and Hantula, J. (2015b). Viruses accumulate in aging infection centers of a fungal forest pathogen. ISME J. 9, 497–507. doi: 10.1038/ismej.2014.145
Wu, M., Zhang, L., Li, G., Jiang, D., and Ghabrial, S. A. (2010). Genome characterization of a debilitation-associated mitovirus infecting the phytopathogenic fungus Botrytis cinerea. Virology 406, 117–126. doi: 10.1016/j.virol.2010.07.010
Xiao, X., Cheng, J., Tang, J., Fu, Y., Jiang, D., Baker, T. S., et al. (2014). A novel partitivirus that confers hypovirulence on plant pathogenic fungi. J. Virol. 88, 10120–10133. doi: 10.1128/JVI.01036-14
Yu, J., and Kim, K. (2021). A phenome-wide association study of the effects of Fusarium graminearum transcription factors on Fusarium graminearum virus 1 infection. Front. Microbiol. 12:622261. doi: 10.3389/fmicb.2021.622261
Yu, X., Li, B., Fu, Y., Jiang, D., Ghabrial, S. A., Li, G., et al. (2010). A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. 107, 8387–8392. doi: 10.1073/pnas.0913535107
Zamora-Ballesteros, C., Wingfield, B. D., Wingfield, M. J., Martín-García, J., and Diez, J. J. (2021). Residual effects caused by a past Mycovirus infection in Fusarium circinatum. Forests 2:11. doi: 10.3390/f12010011
Zeng, Z., Sun, H., Vainio, E. J., Raffaello, T., Kovalchuk, A., Morin, E., et al. (2018). Intraspecific comparative genomics of isolates of the Norway spruce pathogen (Heterobasidion parviporum) and identification of its potential virulence factors. BMC Genomics 19:220. doi: 10.1186/s12864-018-4610-4
Keywords: mycovirus, coinfection, virus transmission, growth rate, conifer pathogen
Citation: Kashif M, Jurvansuu J, Hyder R, Vainio EJ and Hantula J (2021) Phenotypic Recovery of a Heterobasidion Isolate Infected by a Debilitation-Associated Virus Is Related to Altered Host Gene Expression and Reduced Virus Titer. Front. Microbiol. 12:661554. doi: 10.3389/fmicb.2021.661554
Edited by:
Ioly Kotta-Loizou, Imperial College London, United KingdomReviewed by:
Sotaro Chiba, Nagoya University, JapanDae-Hyuk Kim, Jeonbuk National University, South Korea
Copyright © 2021 Kashif, Jurvansuu, Hyder, Vainio and Hantula. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Kashif, bXVoYW1tYWQua2FzaGlmQGx1a2UuZmk=
 Muhammad Kashif
Muhammad Kashif Jaana Jurvansuu2
Jaana Jurvansuu2 Rafiqul Hyder
Rafiqul Hyder Eeva J. Vainio
Eeva J. Vainio Jarkko Hantula
Jarkko Hantula