Abstract
HIV/AIDS is still a global threat despite the notable efforts made by the scientific and health communities to understand viral infection, to design new drugs or to improve existing ones, as well as to develop advanced therapies and vaccine designs for functional cure and viral eradication. The identification and analysis of HIV-1 positive individuals that naturally control viral replication in the absence of antiretroviral treatment has provided clues about cellular processes that could interact with viral proteins and RNA and define subsequent viral replication and clinical progression. This is the case of autophagy, a degradative process that not only maintains cell homeostasis by recycling misfolded/old cellular elements to obtain nutrients, but is also relevant in the innate and adaptive immunity against viruses, such as HIV-1. Several studies suggest that early steps of HIV-1 infection, such as virus binding to CD4 or membrane fusion, allow the virus to modulate autophagy pathways preparing cells to be permissive for viral infection. Confirming this interplay, strategies based on autophagy modulation are able to inhibit early steps of HIV-1 infection. Moreover, autophagy dysregulation in late steps of the HIV-1 replication cycle may promote autophagic cell-death of CD4+ T cells or control of HIV-1 latency, likely contributing to disease progression and HIV persistence in infected individuals. In this scenario, understanding the molecular mechanisms underlying HIV/autophagy interplay may contribute to the development of new strategies to control HIV-1 replication. Therefore, the aim of this review is to summarize the knowledge of the interplay between autophagy and the early events of HIV-1 infection, and how autophagy modulation could impair or benefit HIV-1 infection and persistence, impacting viral pathogenesis, immune control of viral replication, and clinical progression of HIV-1 infected patients.
Introduction
A General Overview of HIV: Factors Related to the Control of the Infection and Pathogenesis
According to the Global Human Immunodeficiency Virus (HIV) and Acquired Immune Deficiency Syndrome (AIDS) statistics report published in 2020 by UNAIDS (2020), there were around 38 million people living with HIV and 1.7 million new cases in 2019, and about 26 million people had access to the antiretroviral therapy (ART) at the end of June 2020. Most new infections and deaths are caused by HIV-1 subtype. Advances in science, accumulated experience in the field, along with active community and political commitment, mean that the public health threat of the HIV pandemic could be over by 2030, but there are still many social, economic and healthcare obstacles and inequalities to overcome (Colvin, 2011; UNAIDS, 2015; Poku, 2016). Due to active ART and associated medical achievements, the scientific community considers HIV/AIDS a chronic disease, mainly when access to effective care is assured (Forsyth and Valdiserri, 2012; McGrath et al., 2014).
Regardless of the above-mentioned achievements, ART does not eliminate the virus from latent reservoirs (Chun et al., 1997, 1998; Finzi et al., 1997; Perelson et al., 1997; Wong et al., 1997; Chun and Fauci, 1999; Davey et al., 1999; Hermankova et al., 2003; Siliciano and Greene, 2011; Rosenbloom et al., 2017), such as lymph nodes, gut associated lymphoid tissue (GALT), the central nervous system (CNS) or the most recent discovery of persistence in adipose tissue (Couturier and Lewis, 2018; van Marle et al., 2018). Latent reservoirs favor the emergence of new drug resistances which affect treatment adherence. Moreover, as HIV patients need long-term medication, metabolic concerns, tissue damage and associated aging emerge as a challenge for new drugs and ART regimes (Chawla et al., 2018; Gu et al., 2020). Thus, promising results have been obtained with pre-exposure prophylaxis (PrEP) strategies, which aim to prevent both HIV-1 infection and transmission and are highly dependent on patient adherence (Straubinger et al., 2019), or by using broadly neutralizing antibodies (bnAbs) that could achieve prolonged viral suppression by directly targeting different epitopes from key viral proteins and activating the immune response in order to remove Ab-targeted infected cells (Carrillo et al., 2018; Kumar et al., 2018; Grobben et al., 2019).
HIV-1 entry and early infection is mediated when the virus interacts with a main receptor in target cells, CD4, and a co-receptor, mainly CCR5 or CXCR4 which are involved in HIV-1 pathogenesis (Dalgleish et al., 1984; McDougal et al., 1985, 1986; Cocchi et al., 1995, 1996; Alkhatib et al., 1996; Bleul et al., 1996; Choe et al., 1996; Deng et al., 1996; Doranz et al., 1996, 1999; Dragic et al., 1996; Liu et al., 1996; Samson et al., 1996; Aramori et al., 1997; Davis et al., 1997; Farzan et al., 1997; Weissman et al., 1997; Blanco et al., 2000; Brelot et al., 2000; Clapham and McKnight, 2001; Chanel et al., 2002; Moore et al., 2004; Tuttle et al., 2004; Cavarelli et al., 2008; Cavarelli and Scarlatti, 2011; Gorry and Ancuta, 2011; Wilen et al., 2012; Melikyan, 2014). It should be mentioned that an effective cure for HIV-1 infection has been reported in two patients by CCR5Δ32/Δ32-hematopoietic stem cells (HSCs) transplantation, observing that latently infected cells were eliminated and replaced with HIV-1 resistant donor CCR5–/– cells (Hütter et al., 2009; Gupta et al., 2019). Although it is not possible to universally apply this method (Kordelas et al., 2014), it shows that HIV-1 can be eradicated from the organism (Hütter et al., 2009; Allers et al., 2011; Allers and Schneider, 2015; Haworth et al., 2017; Ambinder et al., 2020). In addition, a plethora of different strategies to keep the latent reservoir under control have been developed, from gene therapy to therapeutic vaccines (Rerks-Ngarm et al., 2009; Elliott et al., 2014; Henrich et al., 2014, 2017; Rasmussen et al., 2014; Luzuriaga et al., 2015; Søgaard et al., 2015; Zhu et al., 2015; Bar et al., 2016; Kaminski et al., 2016a, b; Kuritzkes, 2016; Scheid et al., 2016; Wang et al., 2016; Colby et al., 2018; Davenport et al., 2019).
The relationship between CCR5 or more precisely the lack of CCR5 expression and autophagy in the context of HIV-1 infection is not clear enough. It has been reported that HIV-1 envelope (Env) complex of proteins that targets cells through the cell-surface CCR5 receptor (i.e., R5-tropic Envs) triggers autophagy and cell death in bystander uninfected CD4+ T cells, after long-term virus-cell contacts (Blanco et al., 2004; Espert et al., 2009). However, this autophagy cell death process is triggered by gp41-induced membrane fusion, and is independent of the co-receptor usage (Espert et al., 2009). Additionally, the role of CCR5 seems to differ in CD4+ T cells and monocyte-differentiated macrophages (MDMs). CD4+ T cells are susceptible to cell death induced by CXCR4 and CCR5 using viral strains and MDMs infected by CCR5-using viruses show weak autophagy activation and increased viral replication compared to MDMs infected with viral strains that use CXCR4 (Espert et al., 2009). This is consistent with the fact that CCR5-tropic viral strains are more infectious than CXCX4-tropic viral strains in MDMs (Schweighardt et al., 2004; Willey et al., 2005; Petrov et al., 2013; Wang et al., 2015; Bolduc et al., 2017; Quitadamo et al., 2018; Borrajo et al., 2019), and with the in vivo prevalence of viruses that utilize CCR5 in the majority of infected individuals (De Jong et al., 1992; Roos et al., 1992; Schuitemaker et al., 1992; Connor et al., 1993; Zhu et al., 1993; van’t Wout et al., 1994; Cornelissen et al., 1995; Spijkerman et al., 1995; Huang et al., 1996; Reece et al., 1998; Keele and Derdeyn, 2009; Salazar-Gonzalez et al., 2009). Likewise, functional CCR5 reduced the expression of autophagy genes, such as BECN1, ATG5 and microtubule associated protein 1 light chain 3 (LC3) alpha (MAP1LC3A), and promoted inflammation in an experimental asthmatic mouse model (Liu et al., 2021). Hence, an antagonist peptide of CCR5 enhances the mRNA levels of BECN1, ATG5, and MAP1LC3A and their proteins level, as well as double-membrane autophagic vesicles or vacuoles (AVs), thereby decreasing inflammation (Liu et al., 2021). These AVs are considered hallmarks of autophagy (Arstila and Trump, 1968; Punnonen et al., 1989; Dunn, 1990a, b; Liou et al., 1997; Mizushima et al., 2002; Zhou et al., 2016), the autophagosomes. Therefore, it is plausible that the lack of CCR5 expression could be associated with a fully active autophagy process presenting a protective phenotype against X4-tropic viral strains. Thus, functional CCR5 appears to be involved in autophagy inhibition during HIV infection with R5-tropic strains. However, HIV-1 mediated cell-death autophagy by long-term non-infectious HIV-1 Env/cell contacts is independent of CCR5 (Espert et al., 2009). Therefore, the CCR5/HIV-1/autophagy interplay and its role in viral life cycle is a complex and active subject of research (reviewed in Espert et al., 2008).
One noteworthy discovery in the HIV-1 pandemic history is the existence of infected individuals that naturally control the viral replication for longer than 10 years in the absence of ART, known as long-term non-progressors (LTNP; Lambotte et al., 2005). Briefly, depending on the clinical progression of the disease, there are different groups of LTNP HIV+ individuals: the elite controllers (LTNP-EC), with HIV-1 RNA plasma levels below the level of detection (less than 50 copies of HIV-1 RNA/mL), viremic controllers (LTNP-VC) with HIV-1 RNA levels equal to or below 2,000 copies/mL, non-controllers (LTNP-NC) with levels above 2,000 copies/mL and viremic non-progressors (VNP) with more than 10,000 copies/mL and normal levels of CD4+ T lymphocytes (Lambotte et al., 2005; Diop et al., 2006; Deeks and Walker, 2007; Canducci et al., 2009; Grabar et al., 2009; Lajoie et al., 2009; Limou et al., 2009; Okulicz et al., 2009; Casado et al., 2010, 2018; Rotger et al., 2010; Zhang et al., 2010; Gurdasani et al., 2014; Cabrera-Rodriguez et al., 2019). Nevertheless, despite being controllers, there is evidence of ongoing viral replication in LTNP-EC (Blankson et al., 2007; Lamine et al., 2007; Dinoso et al., 2008; Migueles et al., 2008; Pereyra et al., 2008, 2009; Hatano et al., 2009; Julg et al., 2010; Mens et al., 2010; O’Connell et al., 2010; Salgado et al., 2010; Zaunders et al., 2011; Noel et al., 2016; Ali et al., 2018; Casado et al., 2018; Woldemeskel et al., 2020), and this endorses the importance of periodically monitoring these patients. The causes of this viral replication control are diverse and may be due to both host and viral factors, thereby making it difficult to establish a precise cause for the natural control of HIV. The viral factors include replicative capacity, infectivity, tropism, virulence, mutation rate, viral load or genetic variants in different genes. This is the case of the Australian cohort characterized by invalidating alterations in the nef gene or the Spanish LTNP-C cluster with a heritable conserved non-functional env gene, which is responsible for the low binding of the viral Env complex to CD4, inefficient cell-signaling and subsequent impaired fusion, infection and viral transmission capacities (Zaunders et al., 2011; Casado et al., 2018; Lopez-Galindez et al., 2019; López-Galíndez, 2019). Likewise, LTNP phenotypes are usually associated with genetic factors in the host related to immune control in viral replication and reservoir generation (Pereyra et al., 2010; Poropatich and Sullivan, 2011; Whittall et al., 2011; Buckheit et al., 2012; Bullen et al., 2014; Colomer-Lluch et al., 2018; Goulder and Deeks, 2018; Lopez-Galindez et al., 2019). HLA alleles are well known genetic markers associated with protection or progressive disease, but CCR5-Δ32 allelic deletions or forkhead box O3a (FOXO3) mutations are also linked with the survival of memory CD4+ T cells (de Roda Husman et al., 1997; Eugen-Olsen et al., 1997; Pasi et al., 2000; Dabrowska et al., 2008; van Grevenynghe et al., 2008; Casado et al., 2010; López-Galíndez, 2019). Cell antiviral restriction factors are a first line of defense against viral infection, replication and spreading, such as the tripartite motif-containing protein 5 alpha isoform (TRIM5α), SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 (SAMHD1), tetherin or BST2 (bone marrow stromal cell antigen 2), apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) proteins or histone deacetylase 6 (HDAC6; Valenzuela-Fernandez et al., 2005; Valera et al., 2015; Soliman et al., 2017; Marrero-Hernández et al., 2019). In this regard, proautophagic HDAC6 has recently been reported to be closely linked to HIV-1 control (Valenzuela-Fernandez et al., 2005, 2008; Valera et al., 2015; Casado et al., 2018; Marrero-Hernández et al., 2019), similar to other proteins implicated in the autophagy process (Rotger et al., 2011; Nardacci et al., 2014, 2017), opening up a new pathway to control HIV-1 infection. Thus, recent data are addressed below that indicate that non-functional env-gene of viruses from LTNP-EC individuals is responsible for a defect in Env-mediated productive signaling to overcome the barrier of HDAC6, an autophagy-related factor which blocks HIV-1 fusion, transmission and early infection (Casado et al., 2018). Therefore, the capacity of primary viral Env to signal and overcome this proautophagic restriction factor, at the early stage of the viral life cycle, is related to the progressor phenotype of HIV+ patients (Casado et al., 2018; Cabrera-Rodriguez et al., 2019), whereas non-functional viral Envs are associated with viruses of HIV+ individuals presenting the LTNP-EC phenotype (Casado et al., 2018).
Autophagy and HIV
Autophagy is a complex and orchestrated degradative process that maintains cell homeostasis, eliminating misfolded/old elements and recycling them to obtain nutrients, and is also relevant in the innate and adaptive immunity against pathogen infection, including viruses (Liang et al., 1998, 2014; Meléndez et al., 2003; Pinan-Lucarré et al., 2003; Gutierrez et al., 2004; Nakagawa et al., 2004; Dengjel et al., 2005; Ogawa et al., 2005; Paludan et al., 2005; Birmingham et al., 2006; Hara et al., 2006; Komatsu et al., 2006, 2007; Jounai et al., 2007; Kohda et al., 2007; Lee et al., 2007; Nakai et al., 2007; Nishiyama et al., 2007; Py et al., 2007; Nedjic et al., 2008; Dupont et al., 2009; English et al., 2009; Fujita et al., 2009; Masiero et al., 2009; Yamaguchi et al., 2009; Zhang et al., 2009b; Lee H.K. et al., 2010; Liu et al., 2010; Orvedahl et al., 2010; Ponpuak et al., 2010; Travassos et al., 2010; Levine et al., 2011; Benjamin et al., 2013; Randow and Youle, 2014; Sagnier et al., 2015; Valera et al., 2015; Dasari et al., 2016; Hu et al., 2016; Paul and Münz, 2016; Staring et al., 2017; Viret et al., 2018; Oh and Lee, 2019; Dong et al., 2021).
In mammalian cells autophagosome formation is initiated by signals that activate the mammalian autophagy-related 1 (ATG1) homologs, unc-51 like autophagy activating kinase 1 (ULK1) and ULK2 (Kuroyanagi et al., 1998; Yan et al., 1998, 1999; Tomoda et al., 1999; Young et al., 2006; Chan et al., 2007; Kundu et al., 2008; Egan et al., 2011; Chan, 2012). The mammalian ULK complex is formed by the serin/threonin protein kinase ULK1 and ULK2, ATG13, ATG101, and RB1CC1 (RB1-inducible coiled-coil protein 1) (named FIP200, focal adhesion kinase family interacting protein of 200 kDa) (Hosokawa et al., 2009; Jung et al., 2009; Mercer et al., 2009; Noda and Fujioka, 2015; Qi et al., 2015). This complex initiates the formation of the phagophore, a shaped double membrane structure, which elongates to engulf cytoplasmic components, and finally closes to form an autophagosome (Hara et al., 2008; Chan, 2009; Nakatogawa et al., 2009; Yang and Klionsky, 2010; Weidberg et al., 2011b). The ULK complex is then located in phagophores (Hara et al., 2008) where it regulates the initial events of autophagosome formation together with the class III phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3) complex, consisting of BECN1 (named beclin 1 or ATG6), ATG14 (also named ATG14L), PIK3C3 (named VPS34), and PIK3R4 (phosphoinositide-3-kinase regulatory subunit 4) (named VPS15) (Hara et al., 2008; Itakura et al., 2008; Liang et al., 2008; Ganley et al., 2009; Hosokawa et al., 2009; Jung et al., 2009; Matsunaga et al., 2009; Mercer et al., 2009; Zhong et al., 2009; Yang and Klionsky, 2010). Of note, the ULK complex is negatively regulated by the mechanistic target of rapamycin complex 1 or mammalian target of rapamycin complex 1 (MTORC1, named mTORC1)-mediated phosphorylation of ULK1, ULK2, and ATG13 (Ganley et al., 2009; Hosokawa et al., 2009; Jung et al., 2009), whereas the activity of the BECN1-PIK3C3 complex could be regulated by its interaction with several cofactors, such as UVRAG (UV radiation resistance associated), ATG14 and AMBRA1 (autophagy and beclin 1 regulator 1) (Matsunaga et al., 2009; Zhong et al., 2009; Di Bartolomeo et al., 2010; Mizushima et al., 2011). The elongation of the phagophore needs the recruitment of different factors, such as the WD-repeat protein interacting with phosphoinositides (WIPI) family proteins (yeast ATG18 homologs), ATG2 and the transmembrane protein ATG9 (Lang et al., 2000; Jeffries et al., 2004; Proikas-Cezanne et al., 2004; Young et al., 2006; Obara et al., 2008; Longatti and Tooze, 2009; Polson et al., 2010; Mauthe et al., 2011; Dooley et al., 2014; Grimmel et al., 2015; Gómez-Sánchez et al., 2018; Osawa et al., 2019; Tang et al., 2019; Bozic et al., 2020; Matoba et al., 2020; Sawa-Makarska et al., 2020). The ubiquitin-like (Ubl) conjugation reaction is also required for elongation of the phagophore and involves the conjugation of ATG12 to ATG5 which recruits autophagy related 16 like 1 (ATG16L1) (previous name ATG16L) (Mizushima et al., 1999, 2003; Kuma et al., 2002; Hanada et al., 2007; Walczak and Martens, 2013; Fahmy and Labonté, 2017; Harada et al., 2019; Karow et al., 2020). Furthermore, this complex is able to associate with ATG8 family proteins, thereby allowing ATG8 proteins to conjugate to phosphatidylethanolamine on the phagophore membrane (Nakatogawa et al., 2007). In mammals, there are at least seven ATG8 family members, such as MAP1LC3A and MAP1LC3B, with two isoforms of MAP1LC3B: gamma-aminobutyric acid receptor-associated protein (GABARAP) and GABARAP like 1 and 2 (GABARAPL1 and GABARAPL2) (Xin et al., 2001; He et al., 2003; Weidberg et al., 2010, 2011a; Shpilka et al., 2011; Shvets et al., 2011). Both MAP1LC3A/B and GABARAP subfamily members are conjugated to the phagophore (Kabeya et al., 2000, 2004) and required for the formation of the autophagosome (Weidberg et al., 2010). Additionally, ULK1, ATG13, and RB1CC1 (members of the UKL1 complex) have been reported to interact with members of the MAP1LC3A/B family of proteins (Alemu et al., 2012; Suzuki et al., 2014). Then, phosphorylation of several of these factors by ULK1 initiates the nucleation process and autophagosome biogenesis (Ganley et al., 2009; Hosokawa et al., 2009; Jung et al., 2009). The outer membrane of autophagosomes then fuses with a lysosome allowing lysosomal hydrolytic enzymes to degrade the intra-autophagosomal components together with the inner membrane. Finally, selective autophagy clearance of different cargos is driven by their association with several factors that are recruited to the inner surface of the phagophore via their LC3-interacting region (LIR) motifs, such as sequestosome 1 (SQSTM1)/p62, NBR1 [neighbor of breast cancer 1 (NBR1) autophagy cargo receptor; also named neighbor of BRCA1 gene 1] and OPTN (optineurin) (Pankiv et al., 2007; Ichimura et al., 2008; Kirkin et al., 2009; Wild et al., 2011).
Furthermore, as introduced above, autophagy could act as a first innate immune barrier against invading viruses, clearing them, and also being part of the immune response by means of antigen presentation by major histocompatibility complex (MHC) or human leukocyte antigen (HLA) class I and class II molecules (Brazil et al., 1997; Nimmerjahn et al., 2003; Dengjel et al., 2005; Dörfel et al., 2005; Paludan et al., 2005; Zhou et al., 2005; Zwart et al., 2005; Schmid et al., 2007; Li et al., 2008; Gannage and Münz, 2010; Tey and Khanna, 2012; Bell et al., 2013; Valečka et al., 2018). In this respect, different research studies have tried to develop vaccines in order to promote immune presentation of viral proteins through the autophagy degradation pathway (Andersen et al., 2016). However, autophagy/virus interplay is complex as pathogens have evolved strategies to inhibit or use autophagy to their own benefit. Thus, this process can be either favoring or blocking viral replication (Lee et al., 2007; Denizot et al., 2008; Zhou and Spector, 2008; Espert et al., 2009; Joubert et al., 2009; Kyei et al., 2009; Blanchet et al., 2010; Grégoire et al., 2011, 2012; Levine et al., 2011; Bhatt and Damania, 2012; Chaumorcel et al., 2012; Campbell and Spector, 2013; Liang et al., 2013; Richetta et al., 2013; Jackson, 2015; Valera et al., 2015; Mailler et al., 2019; Mao et al., 2019; Marrero-Hernández et al., 2019; Chen et al., 2020).
Autophagy has become relevant in HIV-1 infection, since it is clearly involved in the late stages of the viral life cycle (i.e., replication and viral egress), preventingviral infection in CD4+ T cells and favoring viral replication in macrophages (Zhou and Spector, 2008; Espert et al., 2009; Kyei et al., 2009; Valera et al., 2015; Mailler et al., 2019; Marrero-Hernández et al., 2019; Chen et al., 2020).
The sections below describe scientific data concerning the role of autophagy in the first steps of HIV-1 infection, presented from two points of view: (i) the ability of the virus to modulate autophagy during viral entry and infection and (ii) the consequences of autophagy modulation, in target cells, for early HIV-1 infection.
The Role of Autophagy in the Early Steps of HIV-1 Infection
The HIV-1 life cycle could be divided into early and late stages, defined by the integration of viral genome into host cells. The early (pre-integration) steps of the infection begin with the attachment of the virion at the cell-surface and end with the integration of the proviral DNA into the host genome (Arhel, 2010; Campbell and Hope, 2015; Deeks et al., 2015; Goodsell, 2015). The late stages (post-integration) initiate with proviral transcription, assembly of viral components at cell-surface, viral egress, and maturation (Stevenson, 2003; Sutton et al., 2013; Deeks et al., 2015; Freed, 2015; Goodsell, 2015; Mailler et al., 2016). The HIV life cycle could end with cell death of infected cells or by HIV-1 Env-mediated cell death of uninfected bystander cells (Mohammadi et al., 2013; Cummins and Badley, 2014; Doitsh and Greene, 2016; Li et al., 2020) which occurs during early infection (Espert et al., 2008, 2009). Hence, one of the most relevant HIV-1 modulators of cell death is the Env glycoprotein (Krüger et al., 1996; Blanco et al., 2001; Vlahakis et al., 2001; Garg and Blumenthal, 2006, 2008; Varbanov et al., 2006; Garg et al., 2009; Sivaraman et al., 2009; Gorry and Ancuta, 2011; Cicala et al., 2016; Joshi et al., 2016; Tsao et al., 2016; Evering, 2018; Daussy et al., 2020).
The HIV-1 env-gene encodes the Env glycoprotein precursor (gp160), which, upon cleavage by the furin cellular protease, results in the generation of the gp120 surface and the gp41 transmembrane glycoproteins that assemble in trimer to form the active Env complex (Decroly et al., 1994, 1997; Moulard et al., 1999; Staropoli et al., 2000; Checkley et al., 2011; Wilen et al., 2012; Chung et al., 2014; Melikyan, 2014; Ward and Wilson, 2017; Schneck et al., 2020). During HIV-1 entry, the viral surface gp120 subunit of the Env binds to CD4 and CCR5 or CXCR4 key co-receptor (Dalgleish et al., 1984; McDougal et al., 1985, 1986; Cocchi et al., 1995, 1996; Alkhatib et al., 1996; Bleul et al., 1996; Choe et al., 1996; Deng et al., 1996; Doranz et al., 1996, 1999; Dragic et al., 1996; Liu et al., 1996; Samson et al., 1996; Aramori et al., 1997; Davis et al., 1997; Farzan et al., 1997; Weissman et al., 1997; Blanco et al., 2000; Brelot et al., 2000; Clapham and McKnight, 2001; Chanel et al., 2002; Moore et al., 2004; Tuttle et al., 2004; Cavarelli et al., 2008; Cavarelli and Scarlatti, 2011; Gorry and Ancuta, 2011; Wilen et al., 2012; Colin et al., 2013, 2018; Melikyan, 2014; Shaik et al., 2019). Hence, HIV-1 Env is responsible for virus attachment to target cells, membrane fusion and subsequent genome-bearing capsid entry into the cytoplasm, thereby initiating the productive infection process (Markosyan et al., 2003; Chang et al., 2005; Dobrowsky et al., 2008; Wilen et al., 2012; Melikyan, 2014; Sood et al., 2017; Stultz et al., 2017; Iliopoulou et al., 2018; Chen, 2019). During these steps, HIV-1 Env-mediated signals prepare cells to be efficiently infected, where membrane dynamics and intracellular organelles and cytoskeleton reorganization could account for permissive control in early infection (Blanco et al., 2003; Jolly and Sattentau, 2004; Jolly et al., 2004; Valenzuela-Fernandez et al., 2005; Jimenez-Baranda et al., 2007; Barrero-Villar et al., 2008, 2009; Yoder et al., 2008; Liu et al., 2009; Puigdomenech et al., 2009; Garcia-Exposito et al., 2011, 2013; Stolp and Fackler, 2011; Vorster et al., 2011; de Armas-Rillo et al., 2016; Casado et al., 2018; Cabrera-Rodriguez et al., 2019). HIV-1 Env-induced autophagy would therefore be a good way to regulate early steps of the viral life cycle.
The interplay between early HIV-1 infection and autophagy is presented below by mainly focusing on the effects driven by the viral Env, and considering different studies performed in permissive cell types and/or tissues, such as T lymphocytes, peripheral blood mononuclear cells (PBMCs), macrophages and dendritic cells (DCs), and in the central nervous system (CNS).
HIV-1 Env-Mediated Regulation of Autophagy and Related Factors During Early Infection in T Lymphocytes
Although HIV-1 induction of apoptosis in bystander CD4+ T cells was reported to be driven by Env glycoproteins, gp120 and gp41 (Blanco et al., 2003), Env-mediated autophagy during HIV-1 early infection was first described in 2006 (Espert et al., 2006). It was reported that Env-transfected or HIV-infected cells expressing the viral Env complex at the cell-surface induced autophagy and accumulation of BECN1 in uninfected CD4+ T lymphocytes via CXCR4 (Espert et al., 2006), where these autophagy events induce apoptosis in bystander CD4+ cells (Espert et al., 2006). The autophagic machinery is activated after long-lasting interaction of uninfected CD4+ T lymphocytes with Env expressed at the cell-surface of X4-tropic HIV-1 infected cells, thus accumulating the BECN1/ATG6 protein which regulates the mechanism interacting with PIK3 class III (Espert et al., 2006, 2007). These reports further highlighted that CD4 signaling and tyrosine-protein kinase Lck (LCK; named LCK proto-oncogene, Src family tyrosine kinase or p56lck) were not required and that autophagy was necessary for apoptosis to be induced. Moreover, CXCR4 or CCR5 signaling does not seem to be involved in HIV-1 Env-induced autophagy, which is dependent on the gp41 fusion capacity (Blanco et al., 2003; Denizot et al., 2008). These results suggest that HIV-mediated autophagic cell-death of bystander CD4+ T lymphocytes is a mechanism that could contribute to immunodeficiency.
It should also be mentioned that autophagy can be also induced by R5-tropic HIV-1 Env upon binding to bystander, uninfected CCR5+/CD4+ T cells, whereas it is inhibited in CD4+ T cells that are infected either by X4-tropic or R5-tropic viral strains (Molina et al., 2007).
Furthermore, after contact between CD4+ T lymphocytes and Env-expressing cells, reactive oxygen species (ROS) are accumulated and its production is known to be involved in autophagy (Molina et al., 2007; Borel et al., 2012). It has recently been reported that peroxisomes, organelles involved in the control of oxidative stress, are targeted by Env-mediated autophagy, inducing oxidative stress and this leads to uninfected CD4+ T cells death (Daussy et al., 2020).
Another aspect of HIV-1 infection and autophagy is that this retrovirus is also able to manipulate the process in order to survive. A new study in CD4+ T lymphocytes has shown that autophagy is rapidly controlled after HIV-1 entry by the viral protein R (Vpr) which reduces the amount of essential autophagy ATG proteins, such as MAP1LC3 and BECN1, and the autophagy-related protein BNIP3 (BCL2 interacting protein 3) and their mRNA, and is further implicated in the degradation of a transcription factor FOXO3 (Alfaisal et al., 2019).
HIV-1 Env Modulates Autophagy and Related Factors Associated With Membrane and Cytoskeleton Dynamics, Key Events for Productive Early Infection
Autophagy is a paradigm of the cellular membrane dynamics driven by cytoskeleton components and other associated factors (Aplin et al., 1992; Fass et al., 2006; Axe et al., 2008; Nakatogawa et al., 2009; Hailey et al., 2010; Lee J.Y. et al., 2010; Mari et al., 2010; Ravikumar et al., 2010; Aguilera et al., 2012; Longatti and Tooze, 2012; Yamamoto et al., 2012; Ge et al., 2013; Knævelsrud et al., 2013; Puri et al., 2013; Zhuo et al., 2013; Coutts and La Thangue, 2015; Kast and Dominguez, 2015; Kast et al., 2015; Mi et al., 2015; de Armas-Rillo et al., 2016; Galluzzi et al., 2017a; Morel, 2020). Similarly, there are data that point to membrane dynamics being essential for successful HIV-1 entry and infection, but the mechanisms behind these events are complex and likely involve several factors. Thus, phosphatidylinositol (4,5)-bisphosphate (PIP2) is a second messenger that has been demonstrated to be important not only in the HIV-1 viral egress but also in the entry, synthetized by phosphatidylinositol-4-phosphate 5-kinase type Iα (PIP5K1A), after Env-gp120 interaction with CD4+ T lymphocyte (Barrero-Villar et al., 2008). Another cell factor that works in membrane dynamics is ADP ribosylation factor 6 (ARF6; Radhakrishna and Donaldson, 1997; Franco et al., 1999; Donaldson, 2003; Naslavsky et al., 2003; Aikawa and Martin, 2005; Donaldson and Honda, 2005), a GTPase which plays a role in the PIP2 generation (Honda et al., 1999; Toda et al., 1999; Brown et al., 2001; Santy and Casanova, 2001; Powner and Wakelam, 2002; Aikawa and Martin, 2005; Bach et al., 2010). ARF6 promotes autophagosome biogenesis, since it works favoring endocytic uptake of the plasma membrane into autophagosome precursors (Moreau et al., 2012), exerting a vital function for effective and productive viral entry (Garcia-Exposito et al., 2011).
In the context of cytoskeleton dynamics, actin adaptors and enzymes that reorganize, stabilize and sever cortical F-actin, such as MSN (moesin), FLNA (filamin A), and GSN (gelsolin) have been involved in early infection, and are responsible for pore fusion and viral entry (Jimenez-Baranda et al., 2007; Barrero-Villar et al., 2009; Garcia-Exposito et al., 2013). Tubulin cytoskeleton and associated factors also play a crucial role in driving membrane dynamics, as reported for cytoplasmic tubulin-deacetylase HDAC6 that controls pore fusion formation and viral entry (Valenzuela-Fernandez et al., 2005, 2008). Viral Env mediates acetylation of α-tubulin in order to allow a reorganization of the tubulin cytoskeleton and efficient pore fusion formation (Valenzuela-Fernandez et al., 2005; Casado et al., 2018). On the contrary, over-expression of HDAC6, which is located in the cytoplasm and regulates deacetylation of α-tubulin in stable microtubules (Hubbert et al., 2002; Valenzuela-Fernandez et al., 2008), inhibits HIV-1 Env-mediated tubulin acetylation, thereby preventing HIV-1 Env-dependent cell fusion and infection (Valenzuela-Fernandez et al., 2005).
Moreover, HDAC6 plays an important role in clearing accumulated protein aggregates, potentially toxic for cells by autophagy (Kawaguchi et al., 2003; Kopito, 2003). The C-terminal ubiquitin-binding ZnF-UBP zinc finger domain (ZnF-UBP or BUZ domain) of HDAC6 binds free ubiquitin as well as mono- and polyubiquitinated proteins with high affinity (Seigneurin-Berny et al., 2001; Hook et al., 2002; Kawaguchi et al., 2003; Boyault et al., 2006), facilitating the transport of ubiquitinated misfolded proteins to form an aggresome in a microtubule network dependent manner (Kawaguchi et al., 2003). In addition, HDAC6 is directly involved in autophagy by promoting the removal of aggresomes (Kawaguchi et al., 2003; Iwata et al., 2005; Pandey et al., 2007; Lee J.Y. et al., 2010). In fact, HDAC6 controls the fusion of autophagosomes and lysosomes by recruiting cortactin, and other key components for actin reorganization, thereby triggering clearance of autophagic substrates (Lee J.Y. et al., 2010). In an HIV-1 context, HDAC6 regulates infectivity of nascent HIV-1 virions by interacting with APOBEC subunit 3G (APOBEC3G; named A3G), stabilizing it and promoting the autophagic degradation of the HIV-1 infectivity factor Vif, thereby impairing the incorporation of Vif in nascent viral particles (Valera et al., 2015; Marrero-Hernández et al., 2019). Moreover, HDAC6 acts as a restriction factor, limiting viral production and infection by driving Pr55Gag and Vif viral proteins to degradation through an aggresome/autophagy route (Marrero-Hernández et al., 2019). Thus, for HIV-1, targeting HDAC6 appears to be critical for assuring viral production and virus infectivity, and this could be a key proinfective function of Nef. Hence, HDAC6 is counteracted by functional Nef which drives its clearance by an acidic, endocytic/lysosomal pathway. In this regard, Nef assures viral production and infection by targeting HDAC6, stabilizing Pr55Gag and Vif, thereby facilitating Pr55Gag location and aggregation at the plasma membrane and subsequent virus production and infection capacity (Marrero-Hernández et al., 2019).
Therefore, the interplay between Nef and HDAC6 may be important to determine the course of HIV-1 infection and pathogenesis in infected individuals, and may contribute to the development of new strategies against HIV.
The Ability of HIV-1 Env of Virus From Infected Individuals to Subvert or Not Autophagy-Associated Factors Is Related to the Patient Phenotype
It has recently been shown that inefficient HIV-1 Env functions and signaling may contribute to the low viral replication capacity and transmissibility of the viruses from natural controllers (LTNP-ECs) (Casado et al., 2013). Furthermore, the contribution of common viral features to the clinical LTNP-EC phenotype and a functional characterization of the initial events of the viral infection demonstrated that Envs from virus of LTNP-EC individuals were ineffective in the binding to CD4 and in the key triggering of actin/tubulin-cytoskeleton modifications, compared to Envs from viruses of HIV-1+ chronic patients (Casado et al., 2018). The viral properties of the cluster viruses result in defective viral fusion, entry, and infection, and these properties were inherited by every virus in the cluster (Casado et al., 2018). Along the same lines, another study that compared Env clones from virus of VNP patients with those from rapid progressors (RPs), revealed that both groups of primary viral Envs induced autophagy in bystander uninfected CD4+ T cells after 3 days of co-culture (Cabrera-Rodriguez et al., 2019). These results were demonstrated after long-lasting cell-to-cell experiments, and were entirely different to first HIV-1-cell contact and signaling to productive early infection (Casado et al., 2018), as this type of late transmission activates different mechanisms such as apoptosis and linked autophagy, leading to an increase in MAP1LC3 and cell-death (Espert et al., 2007; Cabrera-Rodriguez et al., 2019).
Related to the above deadly autophagy activation by HIV-1 Env, it has been reported that viral Env activates peroxisome degradation with a concomitant decrease in the expression of peroxisomal proteins [i.e., CAT (catalase) and PEX14 (peroxisomal biogenesis factor 14; also named peroxin 14)], which appears to be autophagy dependent, since down-regulation of BECN1 and SQSTM1/p62 restores their expression levels (Daussy et al., 2020). It is noteworthy that late autophagy occurrence is reported to be independent of the HIV-1 infected phenotype, VNP or RP, but it strongly correlated with the fusogenic capacity of viral Envs (Cabrera-Rodriguez et al., 2019). This means that functional viral Envs promote deadly late autophagy, under these experimental conditions, regardless of the HIV-1+ patients’ phenotype, and is directly related to the infectious capacity of the HIV-1 Envs. In this regard, inefficient primary HIV-1 Envs from LTNP-EC individuals are not able to signal and surpass the blockade exerted by the proautophagic, tubulin-deacetylase HDAC6 function. However, functional HIV-1 Envs from VNP or RP patients manage to signalize, overcoming the tubulin-deacetylase HDAC6 protective barrier and infect (Casado et al., 2018; Cabrera-Rodriguez et al., 2019). Therefore, this is the first evidence that a defect of the viral Env from HIV-1+ natural controllers to signal and neutralize the proautophagic factor HDAC6 is related to a defect in infection, virus variability and the LTNP-EC phenotype.
Regarding the above-mentioned ideas, the association between autophagy events, viremic control in HIV+ individuals and the progression of the disease suggests a new clinical perspective for HIV/AIDS. In this regard, it has been reported that LTNP and LTNP-EC phenotypes have PBMCs with a higher amount of AVs together with an increased expression of autophagic factors with respect to normal progressors (Nardacci et al., 2014). Additionally, this work shows that treatment of PBMC isolated from LTNP-EC individuals with rapamycin, an inhibitor of the MTOR, promotes autophagy, resulting in the impairment of viral production (Nardacci et al., 2014). Of note, in humanized mouse models, pan-inhibitors of the MTORC1 inhibit HIV-1 infection by interfering with virus entry, reducing CCR5 levels, and with transcription (Heredia et al., 2015). These data suggest that autophagy could contribute to the control of viral pathogenesis in HIV-1 viremic controllers (LTNP and LTNP-EC) and interfere with CCR5-mediated virus transmission, thereby constituting a potential target to drive the development of new therapies (Nardacci et al., 2014).
Autophagy and Early HIV-1 Infection in Macrophages and Dendritic Cells
It is noteworthy that autophagy-associated cell-death is not similarly induced in uninfected macrophages, following exposure to viral particles (van Grol et al., 2010), despite being positive for the presence of autophagosomes (Espert et al., 2009), suggesting that Env-mediated autophagy is a cell-dependent process. However, it is important to bear in mind that during macrophage entry of the virus, other molecules are involved in the reorganization of membrane proteins necessary for HIV-1 entry, such as ceramides, integrins and heparan sulfate proteoglycan, with a demonstrated role of ceramides as inducers of autophagy that could eclipse the pro-autophagic functions of the HIV-1 Env/main receptor interplay (Finnegan et al., 2004; Borel et al., 2012). Furthermore, blockade of macrophage-autophagy appears to be a barrier for HIV-1, since viral replication is favored by inducing autophagy in infected macrophages (Kyei et al., 2009) and autophagy inhibition in infected macrophages abrogates viral replication (Espert et al., 2009; Kyei et al., 2009).
In DCs, HIV-1 induces a rapid shutdown of autophagy as HIV-1 Env activates the MTOR pathways, increasing cell-associated HIV-1 and transfer of the virus to CD4+ T cells and, thus, altering autophagy initiation leading to progressive autophagy exhaustion (Blanchet et al., 2010). In this regard, it has been demonstrated that fusion between endosomes and autophagosomes is inhibited during early HIV-1 infection by activation of MTOR, which is a negative regulator of autophagy (Altfeld et al., 2011). HIV-1 Env can activate the MTORC1 complex resulting in efficient viral replication and viral production. In fact, HIV-1 infection increases MTORC1 activity in both productively infected and bystander cells (Akbay et al., 2020).
On the contrary, it has been reported that autophagy is activated during HIV-1 infection in DCs. HIV-1 enters these cells by using C-type lectins as a receptor, and once inside the cell its RNA is recognized by toll-like receptors (TLRs), inducing the immune response and the beginning of the autophagy process. Although these events are under discussion, there is a clear-cut relationship between this viral recognition and the activation of the immune response together with the antiviral autophagy (Lee et al., 2007; Delgado et al., 2008; Shi and Kehrl, 2008; Blanchet et al., 2010; Altfeld et al., 2011; Nakamoto et al., 2012).
Autophagy and HIV-1 Infection in the Central Nervous System
The CNS is also attacked by HIV-1 and, even under ART, at least half of HIV-1 positive individuals present associated neurocognitive disorders (HAND) with progressive neurotoxicity, neurodegeneration, and a chronic activation of the inflammatory response, but it is difficult to study and is a matter of controversy how HIV-1 manages to reach the CNS and how it infects different cell types (reviewed in Rojas-Celis et al., 2019). Autophagy is involved in both neuroprotection and neurodegeneration, but some research data support the hypothesis that the dysregulation of autophagy during HIV-1 infection is important in the pathogenesis of the neuropathology associated with AIDS.
At a more physiologically relevant level, it has been reported that the levels of several key autophagy factors, such as BECN1, ATG5, ATG7 and the membrane-associated MAP1LC3-phosphatidylethanolamine conjugate (named LC3-II) were higher in postmortem brains presenting HIV-1 encephalitis, compared with HIV-1 patients without encephalitis (Zhou et al., 2011). Additionally, these authors confirmed that overexpression of both X4- or R5-tropic gp120 increased the presence of these autophagy markers. The study further suggests that HIV-1 gp120 induces autophagy in neuron cells, and that the induction of autophagy might be related to the pathogenesis of neuroAIDS (Zhou et al., 2011).
Furthermore, a study carried out on primary human fetal astrocytes revealed that the HIV-1 Env subunit gp120 induced the expression of interleukin 6 (IL6) [named IFNB2 (interferon, beta 2)] through nuclear factor-kappa B (NFKB; named NF-κB) (Shah et al., 2011). This NFKB1-associated signal triggered the expression of some autophagy genes, such as BECN1, ATG5, and MAP1LC3 whose proteins translocate from cytoplasm to the nucleus, and induce the phosphorylation of the inhibitory kappa B (IκB) factor (NFKB inhibitor), which promotes the autophagy degradative pathway (Copetti et al., 2009; Nivon et al., 2009, 2012; Zhang et al., 2009a; Romano et al., 2010; Jiang et al., 2011; Meng and Cai, 2011; Shah et al., 2011; Bhatnagar et al., 2012; Verzella et al., 2020). This event has also been reported in astrocytes, where HIV-1 gp120 increases CXCL8 (named IL8) production involving NFKB (Shah and Kumar, 2010). There have also been some works implicating HIV-1 gp120 in an increase of autophagy proteins, such as the MAP1LC3-phosphatidylethanolamine conjugate, ATG5, ATG7 and a downregulation of MTOR, whose phosphorylated form suppresses autophagy in the astrocyte cell line or in human fetal astrocytes (Cao et al., 2016). A recent study showed that the V3 loop region of HIV-1 gp120 is responsible for inducing autophagy in primary rat hippocampal neurons in a PRKA (protein kinase AMP-activated; named AMPK)/MTOR-dependent pathway (Liu et al., 2019). Moreover, if the autophagy induction is short term, it could inhibit apoptosis. On the contrary, if it lasts for a long time, it could play a detrimental role (Liu et al., 2019).
The relationship between HIV-1 Env and autophagy in the CNS seems to be highly important but little is known about the mechanisms behind the early phases of HIV-1 infection and more studies are needed to elucidate these questions (Rojas-Celis et al., 2019).
Therefore, autophagy is able to play a role as a modulator of HIV-1 infection at the early stages of its viral life cycle (summarized in Table 1 and Figure 1), but all these ideas and conclusions should be appropriately contextualized, since the use of a particular cell model or experimental procedure could condition the results obtained and conclusions drawn (reviewed in Killian, 2012; Leymarie et al., 2017). Taken together, HIV-1-mediated autophagy events could explain, at least in part, the pathophysiology of the viral disease.
TABLE 1
| HIV-1 effect on autophagy or associated factors |
||
| Inhibition | Activation | |
| Productive early infection | MTOR2, HDAC6, APOBEC3G, SQSTM1/p62, ATG5, ATG7, MAP1LC3-phosphatidylethanolamine conjugate, BECN1, BNIP3 | ARF6 |
| Bystander cell death (Late autophagy and cell death) | Class III PIK3s, ceramides1, ROS, peroxisomes proteins (CAT or PEX14), BECN1, ATG5, ATG7, MAP1LC3-phosphatidylethanolamine conjugate | |
| Infected cells/tissue | TLR2, BECN1 3,A,B,C ATG53,A,B,C, ATG73,A,B, MAP1LC3-phosphatidylethanolamine conjugate3,A,B,C, MTOR3,D, NFKB3,D | |
Summary of the HIV-1 effect on autophagy or associated factors.
This table summarizes the effect of HIV-1 on autophagy and associated factors in different HIV-1 scenarios (indicated in left column): (i) during productive infection; (ii) by Env-mediated virus or infected cells contact with bystander target CD4+ cells; and (iii) in persistent infected cells and tissues. The modulatory effect of HIV-1 on autophagy or associated factors that are indicated refer to results obtained in CD4+ T lymphocytes, and when data have also been reported in other cell types and tissues, they are indicated as follows: 1Macrophages; 2DCs; 3studies in CNS. Regarding the latter, the data are indicated in terms of the studied cell/tissue as follows: ANeuroblastoma cells, Bhuman brain tissue, Chuman astrocytes and Drat hippocampal neurons. This information is illustrated in Figure 1.
FIGURE 1
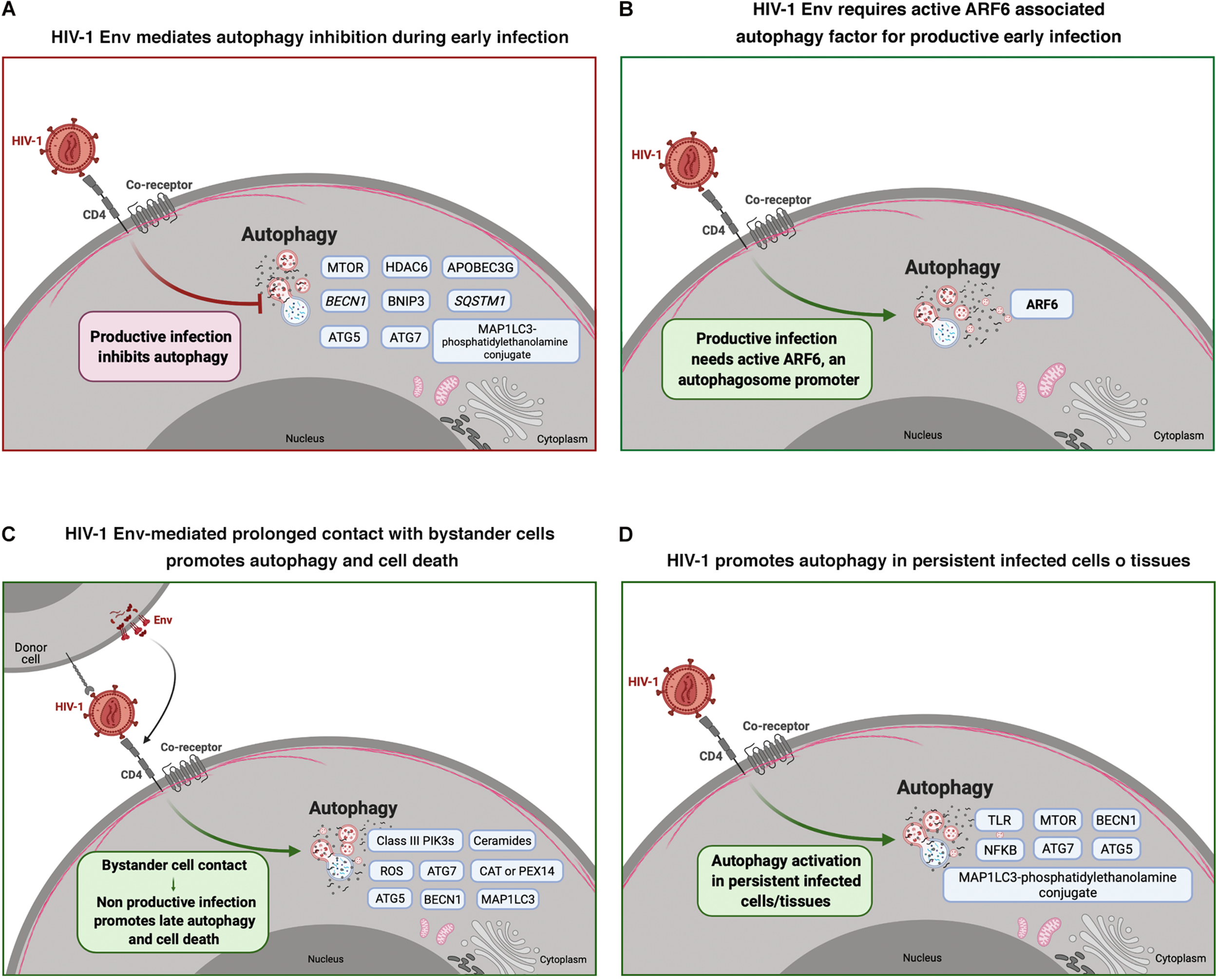
HIV-1 modulation of autophagy during early infection and viral non-productive bystander cell contact, and in persistent infected cells/tissues. Cell scheme summarizing the effect of HIV-1 on autophagy and associated factors in different HIV-1 scenarios. (A) HIV-1 Env mediates autophagy inhibition during early infection in CD4+ T cells. Autophagy associated factors affected by HIV-1 Env signaling are shown in blue boxes. (B) This scheme shows that HIV-1 Env requires active ARF6 associated autophagy factor for productive infection in CD4+ T cells. (C) This panel shows autophagy associated factors (blue boxes) that are activated during non-productive virus-bystander cell contacts. This HIV-1 Env-triggered late autophagy activation leads to CD4+ cell death. (D) This panel shows autophagy associated factors (in blue boxes) that are activated by HIV-1 infection in persistent infected cells and/or tissues. Arrow colors represent the reported positive (green) and negative (red) effects on the different steps of the viral life cycle, with dotted arrows signaling the HIV-1 activation pathway. This figure supports the information summarized in Table 1. Cell schemes were created with BioRender.com.
The Effect of Autophagy Modulation in the Early Steps of HIV-1 Infection
There are other strategies that have shed some light on the involvement of autophagy in HIV-1 early infection, such as the use of autophagy-chemical inhibitors, targeting the expression of autophagy-associated proteins by RNA interference (RNAi) or over-expressing its mutants, or by using the new clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology to abrogate their functions. The CRISPR/Cas system is the most recent genome-edition toolbox of much interest because of its ability to manipulate genomic sequences, enabling the creation of cell lines and animal models for the study of human diseases with promising clinical therapeutic applications, as well as in science and biotechnology (reviewed in Horvath and Barrangou, 2010; Marraffini and Sontheimer, 2010; Zhang et al., 2014; Hille et al., 2018; Leonova and Gainetdinov, 2020).
The inhibition of autophagy during active HIV-1 infection has been considered as a potential supplementary treatment to effective ART for controlling HIV-1 infection (Li et al., 2020).
There are diverse pharmacological ways to inhibit the autophagy process (Vakifahmetoglu-Norberg et al., 2015; Galluzzi et al., 2017b) and some of these have been assayed on HIV-1 infection. The chemical autophagy inhibitor 3-methyladenine (3-MA) that interferes with the PIK3 class III activity to impair autophagosome formation (Seglen and Gordon, 1982; Liang et al., 1999; Klionsky et al., 2008), prevents apoptosis in target CD4+ T cells exposed to Env-expressing effector cells, and monocyte-derived macrophage infection (Espert et al., 2009). Moreover, HDAC6-mediated pro-autophagic and anti-HIV-1 activity is abrogated by 3-MA, thereby stabilizing viral Pr55Gag and Vif, assuring efficient viral production and infectiveness, respectively (Valera et al., 2015; Marrero-Hernández et al., 2019). This fact has been confirmed by using dead-mutants of HDAC6, neutralized in the deacetylase activity of the enzyme and deleted in the proautophagic BUZ C-terminal domain, which are both unable to promote autophagy degradation of these key viral proteins (Valenzuela-Fernandez et al., 2005; Valera et al., 2015; Marrero-Hernández et al., 2019).
The use of constructs that directly encode mutants of proteins, as previously seen with MAP1LC3 (Borel et al., 2015), or indirectly implicated in autophagy membrane dynamics provides some indicators for developing possible strategies against HIV-1 infection. For example, and taking together some of the ideas described above, the expression of kinase-dead mutant D227A (D/A)-PIP5K1A, reduces PIP2 production during first virus-cell contacts impairing HIV-1 infection (Barrero-Villar et al., 2008), and perturbing ARF6/PIP2-plasma membrane bio-regeneration, by using non-functional ARF6 mutants, abrogates HIV-1 Env/CD4-driven viral entry and infection (Garcia-Exposito et al., 2011). An interesting strategy to control autophagy and affect HIV-1 infection has been developed by using Tat-Beclin 1 construct, which is able to interact with the auxiliary viral factor Nef, that inhibits HIV-1 replication in primary human macrophages (Shoji-Kawata et al., 2013).
Bafilomycin A1 (Baf A1) is another candidate to analyze the effect of autophagy inhibition on HIV-1 infection. Baf A1 is an inhibitor of vacuolar H+ ATPases [V-type ATPases (ATP6V)] that blocks acidification of the lysosome and its fusion with autophagosomes, thus inhibiting autophagic degradation (Yamamoto et al., 1998; Klionsky et al., 2012). It has been reported that Baf A1 prevents HDAC6-mediated autophagy degradation of viral Pr55Gag and Vif proteins and stabilizes SQSTM1/p62 protein (Marrero-Hernández et al., 2019), which is a key autophagic factor that fades during active autophagy (Ichimura et al., 2008; Knaevelsrud and Simonsen, 2010; Komatsu and Ichimura, 2010; Rogov et al., 2014; Katsuragi et al., 2015; Valera et al., 2015; Marrero-Hernández et al., 2019).
Infection of macrophages with purified HIV-1 leads to a decrease in SQSTM1/p62, meaning that the autophagy flux is working during the first stages of the infection, and the lysosomal protease inhibitor pepstatin A reverts this situation (Campbell et al., 2015). This study was carried out in MDMs generated from blood of HIV seronegative donors. Another drug, LY 294002 that inhibits PIK3 class I as well as MTORC1, suppresses viral infection of macrophages in post-entry and post-reverse transcription steps prior to HIV-1 gene expression (François and Klotman, 2003). Pan-inhibitors of MTORC1 neutralize HIV-1 infection by interfering with viral entry in humanized mouse in vivo models, since these inhibitors decrease CCR5 levels at the cell-surface of target cells, and also act during viral transcription (Heredia et al., 2015; Akbay et al., 2020). Likewise, other research showed that HIV-1 gp41-induced apoptosis is mediated by caspase-3-dependent mitochondrial production of ROS (Garg and Blumenthal, 2006), which acts as a stimulus to sustain the autophagy process (Levonen et al., 2014; Filomeni et al., 2015). This phenomenon is inhibited by the HIV-1 protease inhibitor nelfinavir (Garg and Blumenthal, 2006).
Trehalose, a natural sugar that inhibits the cellular import of glucose and fructose, with concomitant stimulation of autophagy (Sarkar et al., 2007; Castillo et al., 2013; DeBosch et al., 2016; Mardones et al., 2016) has been found to be involved in controlling early HIV-1 infection (Rawat et al., 2020). Although its action on autophagy is not well elucidated (Yoon et al., 2017), it appears that knockdown of the autophagic ATG5 gene reduces the anti-HIV-1 effect of trehalose in primary human macrophages (Rawat et al., 2020). Likewise, trehalose promotes viral material degradation by MTOR-independent autophagy soon after viral entry. It is noteworthy that trehalose also affects viral entry by down-regulating viral receptors CCR5 and CD4 in CD4+ T cells and macrophages (Rawat et al., 2020).
On the other hand, the ability of HIV-1 Env-early signal to promote autophagy has been confirmed by specific small interfering RNA (siRNA) silencing of BECN1 and ATG7 in CD4+ T lymphocytes, which induce a marked decrease in the Env-mediated cell death (Espert et al., 2006). Similar data have been reported in macrophages and microglia (Kyei et al., 2009; El-Hage et al., 2015). Of note, microglia are the only true CNS parenchymal macrophages and it is thought that they constitute 5-10% of total brain cells (Aguzzi et al., 2013; Li and Barres, 2018). Autophagy inhibition by BECN1 silencing has also been proposed to mitigate neurodegenerative effects mediated by HIV-1 induced inflammation (El-Hage et al., 2015). In this line of action and by using large-scale RNAi screening, several autophagy- and lysosome-related genes have been found to be involved in HIV-1 replication, such as ATG7, ATG12, ATG16L2 (autophagy related 16 like 2), MAP1LC3B, and LAPTM5 (lysosomal protein transmembrane 5) and CLN3 (CLN3 lysosomal/endosomal transmembrane protein, battenin), respectively (Brass et al., 2008; Eekels et al., 2012), as well as PIK3R4 (phosphoinositide-3-kinase regulatory subunit 4), ATG4A (autophagy related 4A cysteine peptidase), ATG5, ATG7 and BECN1, in cell lines or in cells isolated from blood samples (Kyei et al., 2009; Campbell and Spector, 2011, 2012a,b; Eekels et al., 2012). The importance of HIV-1 Env-mediated autophagy has also been identified in HIV-1 infection of CD4+ T cells as this process can selectively degrade the viral transactivator Tat, thus acting as an antiviral process. However, HIV-1 has evolved strategies to inhibit Env-induced autophagy in infected CD4+ T cells, thereby preventing Tat degradation and ensuring viral replication (Sagnier et al., 2015).
A recent study using cell lines suggests that ATG9 is required for HIV-1 infectivity in an Env-dependent way, as the knockout of ATG9 gene does not affect Env incorporation into nascent virions, but it does have an effect on the infectivity, may be because ATG9 is thought to be a lipid transporter and perhaps promotes Env fusion with the host cell (Mailler et al., 2019). Similarly, HIV-1 Env induces a rapid shutdown of autophagy and immunoamphisomes in DCs, and is thus related to an increase in cell-associated HIV-1 and virus transfer to CD4+ T cells (Blanchet et al., 2010). This work reports that downregulation of autophagy by siRNA knock-down of BECN1, ATG5, and MAP1LC3, or by using 3-MA or chloroquine chemical inhibitors does not negatively affect the HIV-1 viral life cycle in cell lines. However, these treatments impede the innate and adaptive immune responses mediated by immunoamphisomes, in a similar way as HIV-1 does during HIV-1 infection in DCs isolated from donors or cell lines (Blanchet et al., 2010).
RNA silencing of the PIP2-membrane recycling driver, ARF6, or the use of mutant constructs of this protein inhibited HIV-1 Env-induced virus-cell membrane fusion, entry in and infection of CD4+ T cells, regardless of viral tropism, highlighting the importance of ARF6 in the regulation of HIV-1 infection (Barrero-Villar et al., 2008; Garcia-Exposito et al., 2011). Furthermore, inhibition of the actin-severing protein gelsolin results in HIV-1 gp120-mediated aberrant pseudopodia formation, a membrane dynamics process regulated by actin cytoskeleton reorganization, with aberrant viral receptors clustering at cell-surface that inhibit early HIV-1 infection (Garcia-Exposito et al., 2013). Other studies demonstrate that specific siRNA inhibition of HDAC6 increases HIV-1 entry due to the increased amount of acetylated α-tubulin that favors virus-cell membrane fusion (i.e., pore fusion formation) and infection (Valenzuela-Fernandez et al., 2005; Valera et al., 2015).
CRISPR/Cas9, a promising technology in the development of strategies against different pathologies, has emerged as a potential anti-HIV-1 tool. One of the first factors targeted by this strategy is TRIM5α, which could be modified to restrict retroviral infection by promoting autophagy against HIV-1, soon after viral entry into the cell (Hatziioannou et al., 2004; Stremlau et al., 2004; Mandell et al., 2014a, b; Richardson et al., 2014; Ribeiro et al., 2016; Saha et al., 2020). Thus, it has been proposed that HSCs harvested from an HIV-positive patient could be transduced with an adeno-associated virus (AAV) vector bearing the Cas9 enzyme together with the single guide RNA (sgRNA) targeting TRIM5α and a repair template to introduce the mutations to target HIV-1 (Weatherley et al., 2017). These transgenic HSCs bearing the mutated TRIM5α would be implanted to achieve long-term cell repopulation in order to generate a durable subset of CD4+ T cells resistant to HIV-1 infection as reported with the CCR5 gene (Xu et al., 2017). Editing of the TRIM5α gene was first reported in cell lines with suboptimal results (Dufour et al., 2018). However, this study suggested that the editing of the TRIM5α gene could be feasible in human cells. In this regard, biallelic CRISPR/Cas9-mediated editing of the TRIM5α gene has recently been associated with the protection of human T lymphocytic cells against infection by HIV-1 (Désaulniers et al., 2020). Furthermore, a link between TRIM5α and several autophagy factors, such as BECN1, ATG7 and ULK1, has been observed by targeting these genes with the CRISPR/Cas9 editing machinery (Saha et al., 2020). This genetic depletion strategy inhibits the anti-HIV-1 proinflammatory function of TRIM5α impairing its capacity to activate adaptor related protein complex 1 (AP1) and NFKB factors that results in the inhibition of the production of the antiviral IFNB1 (named IFN-β) (Saha et al., 2020). All these data are indicative of the importance of the interplay between TRIM5α and autophagy during HIV-1 infection (Saha et al., 2020).
Taken together, all the above studies (summarized in Table 2 and Figure 2), clearly show that autophagy is a potential therapeutic target to control HIV-1 infection and associated pathogenesis.
TABLE 2
| Effect of autophagy modulation by chemical inhibitors* on HIV-1 viral cycle | Effect of autophagy-gene modification (mutants, RNAi or CRISPR/Cas) on HIV-1 viral cycle | |||
| Effect on HIV-1 | Viral entry and Infection | Viral replication | Viral entry and Infection | Viral replication |
| + | 3-MAB, Bafilomycin A | 3-MA, Bafilomycin A | HDAC6 2, ATG52, ATG7A, 2, 3, BECN1A,2,3, MAP1LC3, TRIM5α3, ULK13 | HDAC61, ATG72, ATG122, ATG16L22, MAP1LC3B2, LAPTM52, CLN32, PIK3R42, ATG4A2, ATG52, ATG72, BECN12 |
| - | Pepstatin AA, LY 294002A, Pan-inhibitors of MTORC1, nelfinavir | PIP5K1A1, 2, ARF61,2, TrehaloseA, ATG9B | Tat-Beclin 1 constructA | |
Summary of the effect of autophagy modulation in the early steps of HIV-1 infection.
The table summarizes the autophagy modulatory effect on HIV-1 viral cycle (viral entry and infection or viral replication) exerted either by chemical inhibitors acting on autophagy-associated factors, *which could activate or prevent autophagy, or by acting on autophagy-associated genes by using related mutants, specific RNA interference or CRISPR/Cas editing, as follows: 1 Dead mutant or modified protein; 2 RNAi; 3 CRISPR/Cas9. Left column groups the reported positive (+) and negative (−) effects on the viral cycle. In general, all these data refer to results obtained in CD4+ T lymphocytes, and when data have also been reported in other cell types and tissues, they are indicated as follows: AMacrophages, BDCs and Cstudies in CNS (microglia). This information is illustrated in Figure 2.
FIGURE 2
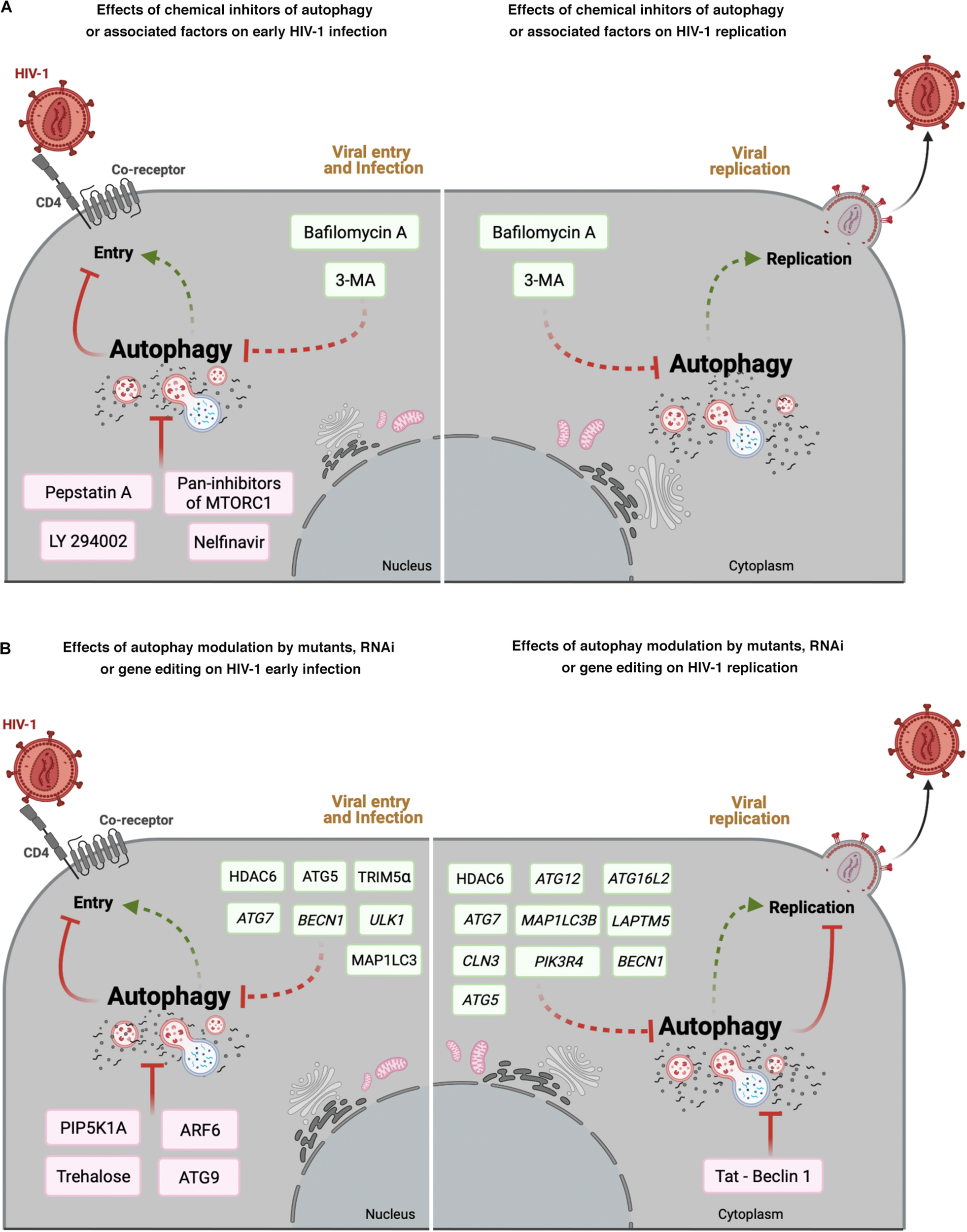
Effect of autophagy modulation on HIV-1 infection. Cell scheme summarizing the autophagy modulatory effect on HIV-1 viral life cycle (early steps: viral entry and infection; late steps: viral replication). (A) This panel shows that chemical inhibitors of autophagy and associated factors could activate (factors indicated in green boxes) or prevent (factors indicated in pink boxes) early HIV- 1 infection (left), or promote viral replication (right). (B) This panel shows the effect of mutants of autophagy associated factors, or specific RNAi or CRISPR/Cas editing acting on autophagy-associated genes in HIV-1 early infection or viral replication. Autophagic factors that activate HIV-1 infection or replication are indicated in green boxes, whereas factors inhibiting HIV-1 are indicated in pink boxes. Arrow colors represent the reported positive (green) and negative (red) effects on the different steps of the viral life cycle, with dotted arrows signaling the HIV-1 activation pathway. This figure supports the information summarized in Table 2. Cell schemes were created with BioRender.com.
Concluding Remarks
One way to block HIV-1 transmission is to inhibit viral spreading into the immune host cells by acting on the viral machinery rather than targeting the intracellular enzymes. However, this is a complex goal, due to viral latency and because viral proteins are highly variable if one only considers the virus as a potential long-term drug-resistant quasi-species (Chun et al., 1998; Schuurman et al., 2002; Zhu et al., 2002; Korn et al., 2003; Belmonte et al., 2007; Buzon et al., 2014; Raymond et al., 2014; Ananworanich et al., 2016; Lorenzo-Redondo et al., 2016; Brenner et al., 2017; Malet et al., 2017; Bandera et al., 2019; Monaco et al., 2020). This is why many of the efforts in basic and clinical research have been aimed at developing strategies to block the virus entry into target cells. Most of the current approaches are based on pharmacological inhibition of the Env proteins, main receptor or co-receptors, as well as the membrane-hemifusion process before viral entry (reviewed in Mostashari Rad et al., 2018). The discovery of LTNP patients (Lambotte et al., 2005) may help to shed some light on identifying cellular mechanisms that are responsible for the control of the disease, not only because of their genetic background but also because of the characteristics of the virus.
Therefore, it is necessary to study the cellular events triggered by HIV-1 infection and to identify the factors implicated in the control of early HIV-1 infection in order to design valid therapies or vaccines. Hence, HIV-1 Env-mediated signals triggered in the cells susceptible to infection become a target of interest, since they induce cellular mechanisms involved in immune response (both innate and adaptive) (Blanco et al., 2003; Altfeld and Gale, 2015) among which is the autophagy process (Espert et al., 2006, 2007; Rotger et al., 2011; Nardacci et al., 2014, 2017). In addition, the recent discovery that functional Env from virus of VNP and RP individuals similarly triggers autophagy shows the importance of this mechanism in HIV-1 early infection and pathogenesis (Nardacci et al., 2014; Cabrera-Rodriguez et al., 2019), and this is supported by the fact that non-functional, signaling-defective primary HIV-1 Envs are associated with the LTNP-EC phenotype (Casado et al., 2018).
The duration of the HIV-1 Env-mediated signals on target cells differentially acts on autophagy functions, having distinct functional roles in HIV-1 infection and pathogenesis. Productive early HIV-1 infection requires the inhibition of autophagy and its related machinery, whereas non-productive signaling in bystander target cells promotes late autophagy and subsequent cell death (Blanco et al., 2003; Valenzuela-Fernandez et al., 2005; Espert et al., 2009; Casado et al., 2018; Cabrera-Rodriguez et al., 2019). In this respect, pro-autophagic and tubulin-deacetylase HDAC6 functions play a crucial role in limiting early HIV-1 entry and infection (Valenzuela-Fernandez et al., 2005, 2008; Valera et al., 2015), which is indicative of the relevance for HIV-1 of abrogating autophagy and factors related to allowing productive infection from the very beginning of HIV-1 fusion and entry. Other studies have analyzed the importance of autophagy after infection or in non-infected bystander cells in long contact with infected Env-expressing cells (Espert et al., 2006, 2007; Denizot et al., 2008; Shah et al., 2011), again confirming an interplay between autophagy and HIV-1 which is key for viral replication achievement and pathogenesis. It should be mentioned that some conclusions may depend on viral and cellular models used to look for this interplay that might bias the reported pro- or anti-autophagy mechanisms.
Moreover, viral factors that could impair or block the autophagy response to infection have also been looked for. In addition, it has been reported that Vif interacts with APOBEC3G promoting its degradation by the proteasome, thus eluding antiviral APOBEC3G activity (Valera et al., 2015). Vif exerts an anti-autophagy role, due to its interaction with MAP1LC3, and that the blockade of the degradative process by Vif is independent of its association with APOBEC3G (Borel et al., 2015). It has recently been demonstrated that Vpr is able to block HIV-1-mediated autophagy soon after its entry into CD4+ T cells (Alfaisal et al., 2019). Vpr down-regulates the expression of essential proteins, such as BECN1 or MAP1LC3, by inducing the proteasomal degradation of the transcription factor FOXO3 which control these autophagy genes. The power of Vpr in controlling autophagy is so strong that even in the presence of autophagy inducers, such as rapamycin or Torin 1, it is still able to block autophagy (Alfaisal et al., 2019).
All the aforementioned autophagy events in favor of and against early HIV-1 infection have led to a longstanding debate about the so-called “dual role” of autophagy in HIV infection. On the one hand, HIV-1 Env produces different signals that promote autophagy events which play a nullifying role in the virus life cycle but, on the other hand, other authors argue that the virus is able to activate or inactivate autophagy for its survival (reviewed in Espert et al., 2008; Killian, 2012; Leymarie et al., 2017). The results need to be carefully interpreted and different conclusions are reached because different strategies have been used, since the use of a construct of a protein, a virus or an Env expressed on a pseudovirus or at the cell-surface may not have the same effect on target cells, as the latter could also differ between independent studies. The case of what happens in the CNS of HIV-1 infected individuals is even more complicated as tissue can only be obtained post-mortem (Zhou et al., 2011; Dever et al., 2015) and the changes observed postmortem might not be present during life.
In summary, it is clear that autophagy has a highly important role in the early stages of HIV-1 infection, and deep characterization of its interplay with HIV-1 Env-triggered functions could play a major role in paving the way for HIV-1 eradication. Thus, more research is needed to address some important questions regarding the relevance of autophagy modulation (i.e., inhibition or activation) by the HIV-1 Env-mediated signaling during the early steps of the viral productive infection or in a context of long virus-target cell contact without productive infection but resulting in bystander cell death (i.e., a critical process that could favor immunodeficiency) (Espert et al., 2006, 2007, 2008, 2009). Elucidating the relationship between HIV-1 Env-mediated signals and autophagy is a priority, after observing that LTNP-EC phenotype is associated with non-functional Env unable to overcome the barrier of autophagy associated factors, such as HDAC6 restriction factor (Valenzuela-Fernandez et al., 2005; Casado et al., 2018; Cabrera-Rodriguez et al., 2019).
Similarly, controlled clinical trials are needed to determine whether addition of autophagy inducers together with ART in HIV-infected individuals may impact HIV-1 reservoirs in vivo. Hence, it is important to analyze the potential universal therapeutic modulation of autophagy in different HIV+ patients, considering factors such as age, coinfections and pharmacological interaction of autophagy drugs with ART. Likewise, there is a growing interest in understanding the complex relationship existing between autophagy functions and HIV-1 pathogenesis in treated patients co-exposed to illicit drugs (Cao et al., 2017). It seems that autophagy is targeted by the use of drugs in HIV+ patients. Thus, the abuse of drugs could favor viral replication not only by increasing inflammation, oxidative and ER stress, but also acting on autophagy (Qi et al., 2011; Dever et al., 2015; El-Hage et al., 2015), thereby affecting the efficiency of ART. On the other hand, autophagy inhibitors could also favor HIV-1 pathogenesis in this scenario, since it has been reported that autophagy inhibitors exacerbated HIV-1 Env/methamphetamine-mediated cell death (Cao et al., 2016). Hence, these data suggest a protective role of autophagy in astrocyte death. Moreover, the use of the autophagy process as a target for HIV-1 therapy should be carefully and deeply studied as this cellular mechanism is implicated in the development of pathogenic reactions, such as cancer or CNS disorders (Aita et al., 1999; Qu et al., 2003; Yue et al., 2003; Ravikumar et al., 2004; Iwata et al., 2005; Hara et al., 2006; Komatsu et al., 2006, 2010; Sarkar et al., 2007; Cherra and Chu, 2008; Koike et al., 2008; Pickford et al., 2008; Martinet et al., 2009; Guo et al., 2011, 2013; Yang et al., 2011, 2014; White, 2012; Levy et al., 2014; Vogl et al., 2014; Wei et al., 2014; Perera et al., 2015; Rohatgi et al., 2015; Tang et al., 2015; Bar-Yosef et al., 2019).
Therefore, it is important to understand how the interplay between autophagy and HIV-1 Env-mediated signal and infection could determine the clinical outcome of HIV-1 patients (i.e., comparing long-term non-progressors that control viremia with progressors) (Espert et al., 2006, 2007, 2008, 2009; Valenzuela-Fernandez et al., 2008; van Grol et al., 2010; Valera et al., 2015; Casado et al., 2018, 2020; Cabrera-Rodriguez et al., 2019) and pathogenesis. In sum, determining the mechanisms underlying HIV/autophagy interplay is probably key to developing novel drug candidates in the challenge of preventing HIV infection and related pathology, thereby offering new insights into HIV-1 functional cure and/or eradication.
Statements
Author contributions
A-VF and R-CR drafted and wrote the manuscript. All authors read, edited, supervised the scientific content and approved the final manuscript.
Funding
AV-F’s Lab was supported by RTI2018-093747-B-100 (“Ministerio de Ciencia, Innovación y Universidades”, Spain) and the European Regional Development Fund (ERDF), ProID2020010093 (RIS-3 Canarias Strategy-“María del Carmen Betancourt y Molina” Program, “Consejería de Economía, Conocimiento y Empleo, Gobierno de Canarias”), UNLL10-3E-783 (ERDF and “Fundación CajaCanarias”), and by the Spanish AIDS network “Red Temática Cooperativa de Investigación en SIDA” RD12/0017/0034 as part of the Plan Nacional R + D + I and co-funded by the Spanish “Instituto de Salud Carlos III (ISCIII)-Subdirección General de Evaluación y el Fondo Europeo de Desarrollo Regional (FEDER)”. RC-R., SP-Y, and DM-A were funded by RD12/0017/0034, M-ULL Ph.D.-training contract (“Contratos predoctorales M-ULL para la Formación de Doctores, Convocatoria 2019, Universidad de La Laguna and Ministerio de Ciencia, Innovación y Universidades”, Spain), “Fundación Doctor Manuel Morales” and “TESIS2017010116” (“Programa Predoctoral de Formación del Personal Investigador, Agencia Canaria de Investigación, Innovación y Sociedad de la Información” and European Social Fund-fellowships). JE-H was funded by the Cabildo of Tenerife “Agustin de Betancourt” 2017 Program. JB and CC are researchers from the Fundació Institut de Recerca en Ciències de la Salut Germans Trias i Pujol supported by the CERCA Programme and the Health Department of the Catalan Government/Generalitat de Catalunya and ISCIII grant numbers PI17/0151 and PI20/00093 (to JB) and PI18/01226 (to CC). LE was supported by institutional funds from the Centre National de la Recherche Scientifique (CNRS) and the Montpellier University and grants from the “Agence Nationale de Recherches sur le SIDA et les Hépatites Virales” (ANRS) and Sidaction. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Acknowledgments
We would like to thank Patrick Dennis for his support. The manuscript was English language edited by Patrick Dennis, an experienced English language editor of scientific manuscripts for international publications.
Conflict of interest
JB is founder and shareholder of AlbaJuna Therapeutics, S.L. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Aguilera M. O. Berón W. Colombo M. I. (2012). The actin cytoskeleton participates in the early events of autophagosome formation upon starvation induced autophagy.Autophagy81590–1603. 10.4161/auto.21459
2
Aguzzi A. Barres B. A. Bennett M. L. (2013). Microglia: scapegoat, saboteur, or something else?Science339156–161. 10.1126/science.1227901
3
Aikawa Y. Martin T. F. (2005). ADP-ribosylation factor 6 regulation of phosphatidylinositol-4,5-bisphosphate synthesis, endocytosis, and exocytosis.Methods Enzymol.404422–431. 10.1016/s0076-6879(05)04037-1
4
Aita V. M. Liang X. H. Murty V. V. Pincus D. L. Yu W. Cayanis E. et al (1999). Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21.Genomics5959–65. 10.1006/geno.1999.5851
5
Akbay B. Shmakova A. Vassetzky Y. Dokudovskaya S. (2020). Modulation of mTORC1 signaling pathway by HIV-1.Cells9:1090. 10.3390/cells9051090
6
Alemu E. A. Lamark T. Torgersen K. M. Birgisdottir A. B. Larsen K. B. Jain A. et al (2012). ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs.J. Biol. Chem.28739275–39290. 10.1074/jbc.m112.378109
7
Alfaisal J. Machado A. Galais M. Robert-Hebmann V. Arnauné-Pelloquin L. Espert L. et al (2019). HIV-1 Vpr inhibits autophagy during the early steps of infection of CD4 T cells.Biol. Cell111308–318. 10.1111/boc.201900071
8
Ali A. Ng H. L. Blankson J. N. Burton D. R. Buckheit R. W. III Moldt B. et al (2018). Highly attenuated infection with a Vpr-deleted molecular clone of human immunodeficiency virus-1.J. Infect. Dis.2181447–1452. 10.1093/infdis/jiy346
9
Alkhatib G. Combadiere C. Broder C. C. Feng Y. Kennedy P. E. Murphy P. M. et al (1996). CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1.Science2721955–1958. 10.1126/science.272.5270.1955
10
Allers K. Hütter G. Hofmann J. Loddenkemper C. Rieger K. Thiel E. et al (2011). Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation.Blood1172791–2799.
11
Allers K. Schneider T. (2015). CCR5Δ32 mutation and HIV infection: basis for curative HIV therapy.Curr. Opin. Virol.1424–29. 10.1016/j.coviro.2015.06.007
12
Altfeld M. Fadda L. Frleta D. Bhardwaj N. (2011). DCs and NK cells: critical effectors in the immune response to HIV-1.Nat. Rev. Immunol.11176–186. 10.1038/nri2935
13
Altfeld M. Gale M. Jr. (2015). Innate immunity against HIV-1 infection.Nat. Immunol.16554–562. 10.1038/ni.3157
14
Ambinder R. F. Capoferri A. A. Durand C. M. (2020). Haemopoietic cell transplantation in patients living with HIV.Lancet HIV7e652–e660.
15
Ananworanich J. Chomont N. Eller L. A. Kroon E. Tovanabutra S. Bose M. et al (2016). HIV DNA set point is rapidly established in acute HIV infection and dramatically reduced by early ART.EBioMedicine1168–72. 10.1016/j.ebiom.2016.07.024
16
Andersen A. N. Landsverk O. J. Simonsen A. Bogen B. Corthay A. Øynebråten I. (2016). Coupling of HIV-1 antigen to the selective autophagy receptor SQSTM1/p62 promotes T-Cell-mediated immunity.Front. Immunol.7:167.
17
Aplin A. Jasionowski T. Tuttle D. L. Lenk S. E. Dunn W. A. Jr. (1992). Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles.J. Cell. Physiol.152458–466. 10.1002/jcp.1041520304
18
Aramori I. Ferguson S. S. Bieniasz P. D. Zhang J. Cullen B. Cullen M. G. (1997). Molecular mechanism of desensitization of the chemokine receptor CCR-5: receptor signaling and internalization are dissociable from its role as an HIV-1 co-receptor.EMBO J.164606–4616. 10.1093/emboj/16.15.4606
19
Arhel N. (2010). Revisiting HIV-1 uncoating.Retrovirology7:96.
20
Arstila A. U. Trump B. F. (1968). Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration.Am. J. Pathol.53687–733.
21
Axe E. L. Walker S. A. Manifava M. Chandra P. Roderick H. L. Habermann A. et al (2008). Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum.J. Cell Biol.182685–701. 10.1083/jcb.200803137
22
Bach A. S. Enjalbert S. Comunale F. Bodin S. Vitale N. Charrasse S. et al (2010). ADP-ribosylation factor 6 regulates mammalian myoblast fusion through phospholipase D1 and phosphatidylinositol 4,5-bisphosphate signaling pathways.Mol. Biol. Cell212412–2424. 10.1091/mbc.e09-12-1063
23
Bandera A. Gori A. Clerici M. Sironi M. (2019). Phylogenies in ART: HIV reservoirs, HIV latency and drug resistance.Curr. Opin. Pharmacol.4824–32. 10.1016/j.coph.2019.03.003
24
Bar K. J. Sneller M. C. Harrison L. J. Justement J. S. Overton E. T. Petrone M. E. et al (2016). Effect of HIV Antibody VRC01 on Viral Rebound after Treatment Interruption.N. Engl. J. Med.3752037–2050.
25
Barrero-Villar M. Barroso-Gonzalez J. Cabrero J. R. Gordon-Alonso M. Alvarez-Losada S. Munoz-Fernandez M. A. et al (2008). PI4P5-kinase Ialpha is required for efficient HIV-1 entry and infection of T cells.J. Immunol.1816882–6888. 10.4049/jimmunol.181.10.6882
26
Barrero-Villar M. Cabrero J. R. Gordon-Alonso M. Barroso-Gonzalez J. Alvarez-Losada S. Munoz-Fernandez M. A. et al (2009). Moesin is required for HIV-1-induced CD4-CXCR4 interaction, F-actin redistribution, membrane fusion and viral infection in lymphocytes.J. Cell Sci.122103–113. 10.1242/jcs.035873
27
Bar-Yosef T. Damri O. Agam G. (2019). Dual role of autophagy in diseases of the central nervous system.Front. Cell Neurosci.13:196.
28
Bell C. English L. Boulais J. Chemali M. Caron-Lizotte O. Desjardins M. et al (2013). Quantitative proteomics reveals the induction of mitophagy in tumor necrosis factor-α-activated (TNFα) macrophages.Mol. Cell. Proteomics122394–2407. 10.1074/mcp.m112.025775
29
Belmonte L. Olmos M. Fanin A. Parodi C. Baré P. Concetti H. et al (2007). The intestinal mucosa as a reservoir of HIV-1 infection after successful HAART.Aids212106–2108. 10.1097/qad.0b013e3282efb74b
30
Benjamin J. L. Sumpter R. Jr. Levine B. Hooper L. V. (2013). Intestinal epithelial autophagy is essential for host defense against invasive bacteria.Cell Host Microbe13723–734. 10.1016/j.chom.2013.05.004
31
Bhatnagar S. Mittal A. Gupta S. K. Kumar A. (2012). TWEAK causes myotube atrophy through coordinated activation of ubiquitin-proteasome system, autophagy, and caspases.J. Cell. Physiol.2271042–1051. 10.1002/jcp.22821
32
Bhatt A. P. Damania B. (2012). AKTivation of PI3K/AKT/mTOR signaling pathway by KSHV.Front. Immunol.3:401.
33
Birmingham C. L. Smith A. C. Bakowski M. A. Yoshimori T. Brumell J. H. (2006). Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole.J. Biol. Chem.28111374–11383. 10.1074/jbc.m509157200
34
Blanchet F. P. Moris A. Nikolic D. S. Lehmann M. Cardinaud S. Stalder R. et al (2010). Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses.Immunity32654–669. 10.1016/j.immuni.2010.04.011
35
Blanco J. Barretina J. Cabrera C. Gutiérrez A. Clotet B. Esté J. A. (2001). CD4(+) and CD8(+) T cell death during human immunodeficiency virus infection in vitro.Virology285356–365. 10.1006/viro.2001.0969
36
Blanco J. Barretina J. Clotet B. Esté J. A. (2004). R5 HIV gp120-mediated cellular contacts induce the death of single CCR5-expressing CD4 T cells by a gp41-dependent mechanism.J. Leukoc. Biol.76804–811. 10.1189/jlb.0204100
37
Blanco J. Barretina J. Ferri K. F. Jacotot E. Gutiérrez A. Armand-Ugón M. et al (2003). Cell-surface-expressed HIV-1 envelope induces the death of CD4 T cells during GP41-mediated hemifusion-like events.Virology305318–329. 10.1006/viro.2002.1764
38
Blanco J. Valenzuela A. Herrera C. Lluis C. Hovanessian A. G. Franco R. (2000). The HIV-1 gp120 inhibits the binding of adenosine deaminase to CD26 by a mechanism modulated by CD4 and CXCR4 expression.FEBS Lett.477123–128. 10.1016/s0014-5793(00)01751-8
39
Blankson J. N. Bailey J. R. Thayil S. Yang H. C. Lassen K. Lai J. et al (2007). Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors.J. Virol.812508–2518. 10.1128/jvi.02165-06
40
Bleul C. C. Farzan M. Choe H. Parolin C. Clark-Lewis I. Sodroski J. et al (1996). The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry.Nature382829–833. 10.1038/382829a0
41
Bolduc J. F. Ouellet M. Hany L. Tremblay M. J. (2017). Toll-like receptor 2 ligation enhances HIV-1 replication in activated CCR6+ CD4+ T cells by increasing virus entry and establishing a more permissive environment to infection.J. Virol.91:e01402-16.
42
Borel S. Espert L. Biard-Piechaczyk M. (2012). Macroautophagy regulation during HIV-1 infection of CD4+ T cells and macrophages.Front. Immunol.3:97.
43
Borel S. Robert-Hebmann V. Alfaisal J. Jain A. Faure M. Espert L. et al (2015). HIV-1 viral infectivity factor interacts with microtubule-associated protein light chain 3 and inhibits autophagy.Aids29275–286. 10.1097/qad.0000000000000554
44
Borrajo A. Ranazzi A. Pollicita M. Bellocchi M. C. Salpini R. Mauro M. V. et al (2019). Different patterns of HIV-1 replication in MACROPHAGES is Led by Co-receptor usage.Medicina55:297. 10.3390/medicina55060297
45
Boyault C. Gilquin B. Zhang Y. Rybin V. Garman E. Meyer-Klaucke W. et al (2006). HDAC6-p97/VCP controlled polyubiquitin chain turnover.EMBO J.253357–3366. 10.1038/sj.emboj.7601210
46
Bozic M. van Den Bekerom L. Milne B. A. Goodman N. Roberston L. Prescott A. R. et al (2020). A conserved ATG2-GABARAP family interaction is critical for phagophore formation.EMBO Rep.21:e48412.
47
Brass A. L. Dykxhoorn D. M. Benita Y. Yan N. Engelman A. Xavier R. J. et al (2008). Identification of host proteins required for HIV infection through a functional genomic screen.Science319921–926. 10.1126/science.1152725
48
Brazil M. I. Weiss S. Stockinger B. (1997). Excessive degradation of intracellular protein in macrophages prevents presentation in the context of major histocompatibility complex class II molecules.Eur. J. Immunol.271506–1514. 10.1002/eji.1830270629
49
Brelot A. Heveker N. Montes M. Alizon M. (2000). Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities.J. Biol. Chem.27523736–23744. 10.1074/jbc.m000776200
50
Brenner B. G. Ibanescu R. I. Oliveira M. Roger M. Hardy I. Routy J. P. et al (2017). HIV-1 strains belonging to large phylogenetic clusters show accelerated escape from integrase inhibitors in cell culture compared with viral isolates from singleton/small clusters.J. Antimicrob. Chemother.722171–2183. 10.1093/jac/dkx118
51
Brown F. D. Rozelle A. L. Yin H. L. Balla T. Donaldson J. G. (2001). Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic.J. Cell Biol.1541007–1017. 10.1083/jcb.200103107
52
Buckheit R. W. III Allen T. G. Alme A. Salgado M. O’Connell K. A. Huculak S. et al (2012). Host factors dictate control of viral replication in two HIV-1 controller/chronic progressor transmission pairs.Nat. Commun.3:716.
53
Bullen C. K. Laird G. M. Durand C. M. Siliciano J. D. Siliciano R. F. (2014). New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo.Nat. Med.20425–429. 10.1038/nm.3489
54
Buzon M. J. Sun H. Li C. Shaw A. Seiss K. Ouyang Z. et al (2014). HIV-1 persistence in CD4+ T cells with stem cell-like properties.Nat. Med.20139–142.
55
Cabrera-Rodriguez R. Hebmann V. Marfil S. Pernas M. Marrero-Hernandez S. Cabrera C. et al (2019). HIV-1 envelope glycoproteins isolated from Viremic Non-Progressor individuals are fully functional and cytopathic.Sci. Rep.9:5544.
56
Campbell E. M. Hope T. J. (2015). HIV-1 capsid: the multifaceted key player in HIV-1 infection.Nat. Rev. Microbiol.13471–483. 10.1038/nrmicro3503
57
Campbell G. R. Rawat P. Bruckman R. S. Spector S. A. (2015). Human immunodeficiency virus type 1 Nef inhibits autophagy through transcription factor EB sequestration.PLoS Pathog.11:e1005018. 10.1371/journal.ppat.1005018
58
Campbell G. R. Spector S. A. (2011). Hormonally active vitamin D3 (1alpha,25-dihydroxycholecalciferol) triggers autophagy in human macrophages that inhibits HIV-1 infection.J. Biol. Chem.28618890–18902. 10.1074/jbc.m110.206110
59
Campbell G. R. Spector S. A. (2012a). Toll-like receptor 8 ligands activate a vitamin D mediated autophagic response that inhibits human immunodeficiency virus type 1.PLoS Pathog.8:e1003017. 10.1371/journal.ppat.1003017
60
Campbell G. R. Spector S. A. (2012b). Vitamin D inhibits human immunodeficiency virus type 1 and Mycobacterium tuberculosis infection in macrophages through the induction of autophagy.PLoS Pathog.8:e1002689. 10.1371/journal.ppat.1002689
61
Campbell G. R. Spector S. A. (2013). Inhibition of human immunodeficiency virus type-1 through autophagy.Curr. Opin. Microbiol.16349–354. 10.1016/j.mib.2013.05.006
62
Canducci F. Marinozzi M. C. Sampaolo M. Berrè S. Bagnarelli P. Degano M. et al (2009). Dynamic features of the selective pressure on the human immunodeficiency virus type 1 (HIV-1) gp120 CD4-binding site in a group of long term non progressor (LTNP) subjects.Retrovirology6:4.
63
Cao L. Fu M. Kumar S. Kumar A. (2016). Methamphetamine potentiates HIV-1 gp120-mediated autophagy via Beclin-1 and Atg5/7 as a pro-survival response in astrocytes.Cell Death Dis.7:e2425. 10.1038/cddis.2016.317
64
Cao L. Glazyrin A. Kumar S. Kumar A. (2017). Role of autophagy in HIV pathogenesis and drug abuse.Mol. Neurobiol.545855–5867. 10.1007/s12035-016-0118-6
65
Carrillo J. Clotet B. Blanco J. (2018). Antibodies and antibody derivatives: new partners in HIV eradication strategies.Front. Immunol.9:2429.
66
Casado C. Colombo S. Rauch A. Martínez R. Günthard H. F. Garcia S. et al (2010). Host and viral genetic correlates of clinical definitions of HIV-1 disease progression.PLoS One5:e11079. 10.1371/journal.pone.0011079
67
Casado C. Galvez C. Pernas M. Tarancon-Diez L. Rodriguez C. Sanchez-Merino V. et al (2020). Permanent control of HIV-1 pathogenesis in exceptional elite controllers: a model of spontaneous cure.Sci. Rep.10:1902.
68
Casado C. Marrero-Hernandez S. Marquez-Arce D. Pernas M. Marfil S. Borras-Granana F. et al (2018). Viral characteristics associated with the clinical nonprogressor phenotype are inherited by viruses from a cluster of HIV-1 elite controllers.mBio9:e02338-17.
69
Casado C. Pernas M. Sandonis V. Alvaro-Cifuentes T. Olivares I. Fuentes R. et al (2013). Identification of a cluster of HIV-1 controllers infected with low replicating viruses.PLoS One8:e77663. 10.1371/journal.pone.0077663
70
Castillo K. Nassif M. Valenzuela V. Rojas F. Matus S. Mercado G. et al (2013). Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons.Autophagy91308–1320. 10.4161/auto.25188
71
Cavarelli M. Karlsson I. Zanchetta M. Antonsson L. Plebani A. Giaquinto C. et al (2008). HIV-1 with multiple CCR5/CXCR4 chimeric receptor use is predictive of immunological failure in infected children.PLoS One3:e3292. 10.1371/journal.pone.0003292
72
Cavarelli M. Scarlatti G. (2011). HIV-1 co-receptor usage: influence on mother-to-child transmission and pediatric infection.J. Transl. Med.9(Suppl. 1), S10.
73
Chan E. Y. (2009). mTORC1 phosphorylates the ULK1-mAtg13-FIP200 autophagy regulatory complex.Sci. Signal.2:e51.
74
Chan E. Y. (2012). Regulation and function of uncoordinated-51 like kinase proteins.Antioxid. Redox. Signal.17775–785. 10.1089/ars.2011.4396
75
Chan E. Y. Kir S. Tooze S. A. (2007). siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy.J. Biol. Chem.28225464–25474. 10.1074/jbc.m703663200
76
Chanel C. Staropoli I. Baleux F. Amara A. Valenzuela-Fernandez A. Virelizier J. L. et al (2002). Low levels of co-receptor CCR5 are sufficient to permit HIV envelope-mediated fusion with resting CD4 T cells.Aids162337–2340. 10.1097/00002030-200211220-00016
77
Chang M. I. Panorchan P. Dobrowsky T. M. Tseng Y. Wirtz D. (2005). Single-molecule analysis of human immunodeficiency virus type 1 gp120-receptor interactions in living cells.J. Virol.7914748–14755. 10.1128/jvi.79.23.14748-14755.2005
78
Chaumorcel M. Lussignol M. Mouna L. Cavignac Y. Fahie K. Cotte-Laffitte J. et al (2012). The human cytomegalovirus protein TRS1 inhibits autophagy via its interaction with Beclin 1.J. Virol.862571–2584. 10.1128/jvi.05746-11
79
Chawla A. Wang C. Patton C. Murray M. Punekar Y. De Ruiter A. et al (2018). A review of long-term toxicity of antiretroviral treatment regimens and implications for an aging population.Infect. Dis. Ther.7183–195. 10.1007/s40121-018-0201-6
80
Checkley M. A. Luttge B. G. Freed E. O. (2011). HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation.J. Mol. Biol.410582–608. 10.1016/j.jmb.2011.04.042
81
Chen B. (2019). Molecular mechanism of HIV-1 entry.Trends Microbiol.27878–891. 10.1016/j.tim.2019.06.002
82
Chen X. Zhang T. Zhang Y. (2020). Endoplasmic reticulum stress and autophagy in HIV-1-associated neurocognitive disorders.J. Neurovirol.26824–833. 10.1007/s13365-020-00906-4
83
Cherra S. J. III Chu C. T. (2008). Autophagy in neuroprotection and neurodegeneration: a question of balance.Future Neurol.3309–323.
84
Choe H. Farzan M. Sun Y. Sullivan N. Rollins B. Ponath P. D. et al (1996). The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates.Cell851135–1148. 10.1016/s0092-8674(00)81313-6
85
Chun T. W. Engel D. Berrey M. M. Shea T. Corey L. Fauci A. S. (1998). Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection.Proc. Natl. Acad. Sci. U.S.A.958869–8873. 10.1073/pnas.95.15.8869
86
Chun T. W. Fauci A. S. (1999). Latent reservoirs of HIV: obstacles to the eradication of virus.Proc. Natl. Acad. Sci. U.S.A.9610958–10961. 10.1073/pnas.96.20.10958
87
Chun T. W. Stuyver L. Mizell S. B. Ehler L. A. Mican J. A. Baseler M. et al (1997). Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy.Proc. Natl. Acad. Sci. U.S.A.9413193–13197. 10.1073/pnas.94.24.13193
88
Chung N. P. Matthews K. Kim H. J. Ketas T. J. Golabek M. De Los Reyes K. et al (2014). Stable 293 T and CHO cell lines expressing cleaved, stable HIV-1 envelope glycoprotein trimers for structural and vaccine studies.Retrovirology11:33. 10.1186/1742-4690-11-33
89
Cicala C. Nawaz F. Jelicic K. Arthos J. Fauci A. S. (2016). HIV-1 gp120: a target for therapeutics and vaccine design.Curr. Drug Targets17122–135. 10.2174/1389450116666150825120735
90
Clapham P. R. McKnight A. (2001). HIV-1 receptors and cell tropism.Br. Med. Bull.5843–59. 10.1093/bmb/58.1.43
91
Cocchi F. Devico A. L. Garzino-Demo A. Arya S. K. Gallo R. C. Lusso P. (1995). Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells.Science2701811–1815. 10.1126/science.270.5243.1811
92
Cocchi F. Devico A. L. Garzino-Demo A. Cara A. Gallo R. C. Lusso P. (1996). The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection.Nat. Med.21244–1247. 10.1038/nm1196-1244
93
Colby D. J. Trautmann L. Pinyakorn S. Leyre L. Pagliuzza A. Kroon E. et al (2018). Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection.Nat. Med.24923–926. 10.1038/s41591-018-0026-6
94
Colin P. Bénureau Y. Staropoli I. Wang Y. Gonzalez N. Alcami J. et al (2013). HIV-1 exploits CCR5 conformational heterogeneity to escape inhibition by chemokines.Proc. Natl. Acad. Sci. U.S.A.1109475–9480. 10.1073/pnas.1222205110
95
Colin P. Zhou Z. Staropoli I. Garcia-Perez J. Gasser R. Armani-Tourret M. et al (2018). CCR5 structural plasticity shapes HIV-1 phenotypic properties.PLoS Pathog.14:e1007432. 10.1371/journal.ppat.1007432
96
Colomer-Lluch M. Ruiz A. Moris A. Prado J. G. (2018). Restriction factors: from intrinsic viral restriction to shaping cellular immunity against HIV-1.Front. Immunol.9:2876.
97
Colvin C. J. (2011). HIV/AIDS, chronic diseases and globalisation.Global Health7:31. 10.1186/1744-8603-7-31
98
Connor R. I. Mohri H. Cao Y. Ho D. D. (1993). Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals.J. Virol.671772–1777. 10.1128/jvi.67.4.1772-1777.1993
99
Copetti T. Bertoli C. Dalla E. Demarchi F. Schneider C. (2009). p65/RelA modulates BECN1 transcription and autophagy.Mol. Cell. Biol.292594–2608. 10.1128/mcb.01396-08
100
Cornelissen M. Mulder-Kampinga G. Veenstra J. Zorgdrager F. Kuiken C. Hartman S. et al (1995). Syncytium-inducing (SI) phenotype suppression at seroconversion after intramuscular inoculation of a non-syncytium-inducing/SI phenotypically mixed human immunodeficiency virus population.J. Virol.691810–1818. 10.1128/jvi.69.3.1810-1818.1995
101
Coutts A. S. La Thangue N. B. (2015). Actin nucleation by WH2 domains at the autophagosome.Nat. Commun.6:7888.
102
Couturier J. Lewis D. E. (2018). HIV persistence in adipose tissue reservoirs.Curr. HIV/AIDS Rep.1560–71. 10.1007/s11904-018-0378-z
103
Cummins N. W. Badley A. D. (2014). Making sense of how HIV kills infected CD4 T cells: implications for HIV cure.Mol. Cell. Ther.2:20. 10.1186/2052-8426-2-20
104
Dabrowska A. Kim N. Aldovini A. (2008). Tat-induced FOXO3a is a key mediator of apoptosis in HIV-1-infected human CD4+ T lymphocytes.J. Immunol.1818460–8477. 10.4049/jimmunol.181.12.8460
105
Dalgleish A. G. Beverley P. C. Clapham P. R. Crawford D. H. Greaves M. F. Weiss R. A. (1984). The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus.Nature312763–767. 10.1038/312763a0
106
Dasari V. Rehan S. Tey S. K. Smyth M. J. Smith C. Khanna R. (2016). Autophagy and proteasome interconnect to coordinate cross-presentation through MHC class I pathway in B cells.Immunol. Cell Biol.94964–974. 10.1038/icb.2016.59
107
Daussy C. F. Galais M. Pradel B. Robert-Hebmann V. Sagnier S. Pattingre S. et al (2020). HIV-1 Env induces pexophagy and an oxidative stress leading to uninfected CD4(+) T cell death.Autophagy[Epub ahead of print].
108
Davenport M. P. Khoury D. S. Cromer D. Lewin S. R. Kelleher A. D. Kent S. J. (2019). Functional cure of HIV: the scale of the challenge.Nat. Rev. Immunol.1945–54. 10.1038/s41577-018-0085-4
109
Davey R. T. Jr. Bhat N. Yoder C. Chun T. W. Metcalf J. A. Dewar R. et al (1999). HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression.Proc. Natl. Acad. Sci. U.S.A.9615109–15114. 10.1073/pnas.96.26.15109
110
Davis C. B. Dikic I. Unutmaz D. Hill C. M. Arthos J. Siani M. A. et al (1997). Signal transduction due to HIV-1 envelope interactions with chemokine receptors CXCR4 or CCR5.J. Exp. Med.1861793–1798. 10.1084/jem.186.10.1793
111
de Armas-Rillo L. Valera M. S. Marrero-Hernández S. Valenzuela-Fernández A. (2016). Membrane dynamics associated with viral infection.Rev. Med. Virol.26146–160. 10.1002/rmv.1872
112
De Jong J. J. De Ronde A. Keulen W. Tersmette M. Goudsmit J. (1992). Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution.J. Virol.666777–6780. 10.1128/jvi.66.11.6777-6780.1992
113
de Roda Husman A. M. Koot M. Cornelissen M. Keet I. P. Brouwer M. Broersen S. M. et al (1997). Association between CCR5 genotype and the clinical course of HIV-1 infection.Ann. Intern. Med.127882–890. 10.7326/0003-4819-127-10-199711150-00004
114
DeBosch B. J. Heitmeier M. R. Mayer A. L. Higgins C. B. Crowley J. R. Kraft T. E. et al (2016). Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis.Sci. Signal.9:ra21. 10.1126/scisignal.aac5472
115
Decroly E. Benjannet S. Savaria D. Seidah N. G. (1997). Comparative functional role of PC7 and furin in the processing of the HIV envelope glycoprotein gp160.FEBS Lett.40568–72. 10.1016/s0014-5793(97)00156-7
116
Decroly E. Vandenbranden M. Ruysschaert J. M. Cogniaux J. Jacob G. S. Howard S. C. et al (1994). The convertases furin and PC1 can both cleave the human immunodeficiency virus (HIV)-1 envelope glycoprotein gp160 into gp120 (HIV-1 SU) and gp41 (HIV-I TM).J. Biol. Chem.26912240–12247. 10.1016/s0021-9258(17)32707-2
117
Deeks S. G. Overbaugh J. Phillips A. Buchbinder S. (2015). HIV infection.Nat. Rev. Dis. Primers1:15035.
118
Deeks S. G. Walker B. D. (2007). Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy.Immunity27406–416. 10.1016/j.immuni.2007.08.010
119
Delgado M. A. Elmaoued R. A. Davis A. S. Kyei G. Deretic V. (2008). Toll-like receptors control autophagy.EMBO J.271110–1121.
120
Deng H. Liu R. Ellmeier W. Choe S. Unutmaz D. Burkhart M. et al (1996). Identification of a major co-receptor for primary isolates of HIV-1.Nature381661–666. 10.1038/381661a0
121
Dengjel J. Schoor O. Fischer R. Reich M. Kraus M. Müller M. et al (2005). Autophagy promotes MHC class II presentation of peptides from intracellular source proteins.Proc. Natl. Acad. Sci. U.S.A.1027922–7927. 10.1073/pnas.0501190102
122
Denizot M. Varbanov M. Espert L. Robert-Hebmann V. Sagnier S. Garcia E. et al (2008). HIV-1 gp41 fusogenic function triggers autophagy in uninfected cells.Autophagy4998–1008. 10.4161/auto.6880
123
Désaulniers K. Ortiz L. Dufour C. Claudel A. Plourde M. B. Merindol N. et al (2020). Editing of the TRIM5 gene decreases the permissiveness of human T lymphocytic cells to HIV-1.Viruses13:24. 10.3390/v13010024
124
Dever S. M. Rodriguez M. Lapierre J. Costin B. N. El-Hage N. (2015). Differing roles of autophagy in HIV-associated neurocognitive impairment and encephalitis with implications for morphine co-exposure.Front. Microbiol.6:653.
125
Di Bartolomeo S. Corazzari M. Nazio F. Oliverio S. Lisi G. Antonioli M. et al (2010). The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy.J. Cell Biol.191155–168. 10.1083/jcb.201002100
126
Dinoso J. B. Kim S. Y. Siliciano R. F. Blankson J. N. (2008). A comparison of viral loads between HIV-1-infected elite suppressors and individuals who receive suppressive highly active antiretroviral therapy.Clin. Infect. Dis.47102–104. 10.1086/588791
127
Diop G. Hirtzig T. Do H. Coulonges C. Vasilescu A. Labib T. et al (2006). Exhaustive genotyping of the interferon alpha receptor 1 (IFNAR1) gene and association of an IFNAR1 protein variant with AIDS progression or susceptibility to HIV-1 infection in a French AIDS cohort.Biomed. Pharmacother.60569–577. 10.1016/j.biopha.2006.08.002
128
Dobrowsky T. M. Zhou Y. Sun S. X. Siliciano R. F. Wirtz D. (2008). Monitoring early fusion dynamics of human immunodeficiency virus type 1 at single-molecule resolution.J. Virol.827022–7033. 10.1128/jvi.00053-08
129
Doitsh G. Greene W. C. (2016). Dissecting how CD4 T cells are lost during HIV infection.Cell Host Microbe19280–291. 10.1016/j.chom.2016.02.012
130
Donaldson J. G. (2003). Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane.J. Biol. Chem.27841573–41576. 10.1074/jbc.r300026200
131
Donaldson J. G. Honda A. (2005). Localization and function of Arf family GTPases.Biochem. Soc. Trans.33639–642. 10.1042/bst0330639
132
Dong X. Yang Y. Zou Z. Zhao Y. Ci B. Zhong L. et al (2021). Sorting nexin 5 mediates virus-induced autophagy and immunity.Nature589456–461. 10.1038/s41586-020-03056-z
133
Dooley H. C. Razi M. Polson H. E. Girardin S. E. Wilson M. I. Tooze S. A. (2014). WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1.Mol. Cell.55238–252. 10.1016/j.molcel.2014.05.021
134
Doranz B. J. Orsini M. J. Turner J. D. Hoffman T. L. Berson J. F. Hoxie J. A. et al (1999). Identification of CXCR4 domains that support coreceptor and chemokine receptor functions.J. Virol.732752–2761. 10.1128/jvi.73.4.2752-2761.1999
135
Doranz B. J. Rucker J. Yi Y. Smyth R. J. Samson M. Peiper S. C. et al (1996). A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors.Cell851149–1158. 10.1016/s0092-8674(00)81314-8
136
Dörfel D. Appel S. Grünebach F. Weck M. M. Müller M. R. Heine A. et al (2005). Processing and presentation of HLA class I and II epitopes by dendritic cells after transfection with in vitro-transcribed MUC1 RNA.Blood1053199–3205. 10.1182/blood-2004-09-3556
137
Dragic T. Litwin V. Allaway G. P. Martin S. R. Huang Y. Nagashima K. A. et al (1996). HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5.Nature381667–673. 10.1038/381667a0
138
Dufour C. Claudel A. Joubarne N. Merindol N. Maisonnet T. Masroori N. et al (2018). Editing of the human TRIM5 gene to introduce mutations with the potential to inhibit HIV-1.PLoS One13:e0191709. 10.1371/journal.pone.0191709
139
Dunn W. A. Jr. (1990a). Studies on the mechanisms of autophagy: formation of the autophagic vacuole.J. Cell Biol.1101923—-1933.
140
Dunn W. A. Jr. (1990b). Studies on the mechanisms of autophagy: maturation of the autophagic vacuole.J. Cell Biol.1101935–1945. 10.1083/jcb.110.6.1935
141
Dupont N. Lacas-Gervais S. Bertout J. Paz I. Freche B. van Nhieu G. T. et al (2009). Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy.Cell Host Microbe6137–149. 10.1016/j.chom.2009.07.005
142
Eekels J. J. Sagnier S. Geerts D. Jeeninga R. E. Biard-Piechaczyk M. Berkhout B. (2012). Inhibition of HIV-1 replication with stable RNAi-mediated knockdown of autophagy factors.Virol. J.9:69. 10.1186/1743-422x-9-69
143
Egan D. F. Shackelford D. B. Mihaylova M. M. Gelino S. Kohnz R. A. Mair W. et al (2011). Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy.Science331456–461. 10.1126/science.1196371
144
El-Hage N. Rodriguez M. Dever S. M. Masvekar R. R. Gewirtz D. A. Shacka J. J. (2015). HIV-1 and morphine regulation of autophagy in microglia: limited interactions in the context of HIV-1 infection and opioid abuse.J. Virol.891024–1035. 10.1128/jvi.02022-14
145
Elliott J. H. Wightman F. Solomon A. Ghneim K. Ahlers J. Cameron M. J. et al (2014). Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy.PLoS Pathog.10:e1004473. 10.1371/journal.ppat.1004473
146
English L. Chemali M. Duron J. Rondeau C. Laplante A. Gingras D. et al (2009). Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection.Nat. Immunol.10480–487. 10.1038/ni.1720
147
Espert L. Codogno P. Biard-Piechaczyk M. (2008). What is the role of autophagy in HIV-1 infection?Autophagy4273–275. 10.4161/auto.5211
148
Espert L. Denizot M. Grimaldi M. Robert-Hebmann V. Gay B. Varbanov M. et al (2006). Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4.J. Clin. Invest.1162161–2172. 10.1172/jci26185
149
Espert L. Denizot M. Grimaldi M. Robert-Hebmann V. Gay B. Varbanov M. et al (2007). Autophagy and CD4+ T lymphocyte destruction by HIV-1.Autophagy332–34. 10.4161/auto.3275
150
Espert L. Varbanov M. Robert-Hebmann V. Sagnier S. Robbins I. Sanchez F. et al (2009). Differential role of autophagy in CD4 T cells and macrophages during X4 and R5 HIV-1 infection.PLoS One4:e5787. 10.1371/journal.pone.0005787
151
Eugen-Olsen J. Iversen A. K. Garred P. Koppelhus U. Pedersen C. Benfield T. L. et al (1997). Heterozygosity for a deletion in the CKR-5 gene leads to prolonged AIDS-free survival and slower CD4 T-cell decline in a cohort of HIV-seropositive individuals.Aids11305–310. 10.1097/00002030-199703110-00007
152
Evering T. H. (2018). Molecular signatures of HIV-1 envelope associated with HIV-associated neurocognitive disorders.Curr. HIV/AIDS Rep.1572–83. 10.1007/s11904-018-0374-3
153
Fahmy A. M. Labonté P. (2017). The autophagy elongation complex (ATG5-12/16L1) positively regulates HCV replication and is required for wild-type membranous web formation.Sci. Rep.7:40351.
154
Farzan M. Choe H. Martin K. A. Sun Y. Sidelko M. Mackay C. R. et al (1997). HIV-1 entry and macrophage inflammatory protein-1beta-mediated signaling are independent functions of the chemokine receptor CCR5.J. Biol. Chem.2726854–6857. 10.1074/jbc.272.11.6854
155
Fass E. Shvets E. Degani I. Hirschberg K. Elazar Z. (2006). Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes.J. Biol. Chem.28136303–36316. 10.1074/jbc.m607031200
156
Filomeni G. De Zio D. Cecconi F. (2015). Oxidative stress and autophagy: the clash between damage and metabolic needs.Cell Death Differ.22377–388. 10.1038/cdd.2014.150
157
Finnegan C. M. Rawat S. S. Puri A. Wang J. M. Ruscetti F. W. Blumenthal R. (2004). Ceramide, a target for antiretroviral therapy.Proc. Natl. Acad. Sci. U.S.A.10115452–15457. 10.1073/pnas.0402874101
158
Finzi D. Hermankova M. Pierson T. Carruth L. M. Buck C. Chaisson R. E. et al (1997). Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy.Science2781295–1300. 10.1126/science.278.5341.1295
159
Forsyth A. D. Valdiserri R. O. (2012). Reaping the prevention benefits of highly active antiretroviral treatment: policy implications of HIV prevention trials network 052.Curr. Opin. HIV AIDS7111–116. 10.1097/coh.0b013e32834fcff6
160
Franco M. Peters P. J. Boretto J. van Donselaar E. Neri A. D’souza-Schorey C. et al (1999). EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization.EMBO J.181480–1491. 10.1093/emboj/18.6.1480
161
François F. Klotman M. E. (2003). Phosphatidylinositol 3-kinase regulates human immunodeficiency virus type 1 replication following viral entry in primary CD4+ T lymphocytes and macrophages.J. Virol.772539–2549. 10.1128/jvi.77.4.2539-2549.2003
162
Freed E. O. (2015). HIV-1 assembly, release and maturation.Nat. Rev. Microbiol.13484–496. 10.1038/nrmicro3490
163
Fujita N. Saitoh T. Kageyama S. Akira S. Noda T. Yoshimori T. (2009). Differential involvement of Atg16L1 in Crohn disease and canonical autophagy: analysis of the organization of the Atg16L1 complex in fibroblasts.J. Biol. Chem.28432602–32609. 10.1074/jbc.m109.037671
164
Galluzzi L. Baehrecke E. H. Ballabio A. Boya P. Bravo-San Pedro J. M. Cecconi F. et al (2017a). Molecular definitions of autophagy and related processes.EMBO J.361811–1836.
165
Galluzzi L. Bravo-San Pedro J. M. Levine B. Green D. R. Kroemer G. (2017b). Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles.Nat. Rev. Drug Discov.16487–511. 10.1038/nrd.2017.22
166
Ganley I. G. Lam Du H. Wang J. Ding X. Chen S. Jiang X. (2009). ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy.J. Biol. Chem.28412297–12305. 10.1074/jbc.m900573200
167
Gannage M. Münz C. (2010). MHC presentation via autophagy and how viruses escape from it.Semin. Immunopathol.32373–381. 10.1007/s00281-010-0227-7
168
Garcia-Exposito L. Barroso-Gonzalez J. Puigdomenech I. Machado J. D. Blanco J. Valenzuela-Fernandez A. (2011). HIV-1 requires Arf6-mediated membrane dynamics to efficiently enter and infect T lymphocytes.Mol. Biol. Cell221148–1166. 10.1091/mbc.e10-08-0722
169
Garcia-Exposito L. Ziglio S. Barroso-Gonzalez J. de Armas-Rillo L. Valera M. S. Zipeto D. et al (2013). Gelsolin activity controls efficient early HIV-1 infection.Retrovirology10:39. 10.1186/1742-4690-10-39
170
Garg H. Blumenthal R. (2006). HIV gp41-induced apoptosis is mediated by caspase-3-dependent mitochondrial depolarization, which is inhibited by HIV protease inhibitor nelfinavir.J. Leukoc. Biol.79351–362. 10.1189/jlb.0805430
171
Garg H. Blumenthal R. (2008). Role of HIV Gp41 mediated fusion/hemifusion in bystander apoptosis.Cell Mol. Life Sci.653134–3144. 10.1007/s00018-008-8147-6
172
Garg H. Joshi A. Blumenthal R. (2009). Altered bystander apoptosis induction and pathogenesis of enfuvirtide-resistant HIV type 1 Env mutants.AIDS Res. Hum. Retroviruses25811–817. 10.1089/aid.2009.0010
173
Ge L. Melville D. Zhang M. Schekman R. (2013). The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis.eLife2:e00947.
174
Gómez-Sánchez R. Rose J. Guimarães R. Mari M. Papinski D. Rieter E. et al (2018). Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores.J. Cell Biol.2172743–2763. 10.1083/jcb.201710116
175
Goodsell D. S. (2015). Illustrations of the HIV life cycle.Curr. Top. Microbiol. Immunol.389243–252. 10.1007/82_2015_437
176
Gorry P. R. Ancuta P. (2011). Coreceptors and HIV-1 pathogenesis.Curr. HIV/AIDS Rep.845–53. 10.1007/s11904-010-0069-x
177
Goulder P. Deeks S. G. (2018). HIV control: is getting there the same as staying there?PLoS Pathog.14:e1007222. 10.1371/journal.ppat.1007222
178
Grabar S. Selinger-Leneman H. Abgrall S. Pialoux G. Weiss L. Costagliola D. (2009). Prevalence and comparative characteristics of long-term nonprogressors and HIV controller patients in the french hospital database on HIV.Aids231163–1169. 10.1097/qad.0b013e32832b44c8
179
Grégoire I. P. Rabourdin-Combe C. Faure M. (2012). Autophagy and RNA virus interactomes reveal IRGM as a common target.Autophagy81136–1137. 10.4161/auto.20339
180
Grégoire I. P. Richetta C. Meyniel-Schicklin L. Borel S. Pradezynski F. Diaz O. et al (2011). IRGM is a common target of RNA viruses that subvert the autophagy network.PLoS Pathog.7:e1002422. 10.1371/journal.ppat.1002422
181
Grimmel M. Backhaus C. Proikas-Cezanne T. (2015). WIPI-mediated autophagy and longevity.Cells4202–217. 10.3390/cells4020202
182
Grobben M. Stuart R. A. van Gils M. J. (2019). The potential of engineered antibodies for HIV-1 therapy and cure.Curr. Opin. Virol.3870–80. 10.1016/j.coviro.2019.07.007
183
Gu S. X. Zhu Y. Y. Wang C. Wang H. F. Liu G. Y. Cao S. et al (2020). Recent discoveries in HIV-1 reverse transcriptase inhibitors.Curr. Opin. Pharmacol.54166–172.
184
Guo J. Y. Chen H. Y. Mathew R. Fan J. Strohecker A. M. Karsli-Uzunbas G. et al (2011). Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis.Genes Dev.25460–470. 10.1101/gad.2016311
185
Guo J. Y. Karsli-Uzunbas G. Mathew R. Aisner S. C. Kamphorst J. J. Strohecker A. M. et al (2013). Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis.Genes Dev.271447–1461. 10.1101/gad.219642.113
186
Gupta R. K. Abdul-Jawad S. Mccoy L. E. Mok H. P. Peppa D. Salgado M. et al (2019). HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation.Nature568244–248.
187
Gurdasani D. Iles L. Dillon D. G. Young E. H. Olson A. D. Naranbhai V. et al (2014). A systematic review of definitions of extreme phenotypes of HIV control and progression.Aids28149–162. 10.1097/qad.0000000000000049
188
Gutierrez M. G. Master S. S. Singh S. B. Taylor G. A. Colombo M. I. Deretic V. (2004). Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages.Cell119753–766. 10.1016/j.cell.2004.11.038
189
Hailey D. W. Rambold A. S. Satpute-Krishnan P. Mitra K. Sougrat R. Kim P. K. et al (2010). Mitochondria supply membranes for autophagosome biogenesis during starvation.Cell141656–667. 10.1016/j.cell.2010.04.009
190
Hanada T. Noda N. N. Satomi Y. Ichimura Y. Fujioka Y. Takao T. et al (2007). The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy.J. Biol. Chem.28237298–37302. 10.1074/jbc.c700195200
191
Hara T. Nakamura K. Matsui M. Yamamoto A. Nakahara Y. Suzuki-Migishima R. et al (2006). Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice.Nature441885–889. 10.1038/nature04724
192
Hara T. Takamura A. Kishi C. Iemura S. Natsume T. Guan J. L. et al (2008). FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells.J. Cell Biol.181497–510. 10.1083/jcb.200712064
193
Harada K. Kotani T. Kirisako H. Sakoh-Nakatogawa M. Oikawa Y. Kimura Y. et al (2019). Two distinct mechanisms target the autophagy-related E3 complex to the pre-autophagosomal structure.eLife8:e43088.
194
Hatano H. Delwart E. L. Norris P. J. Lee T. H. Dunn-Williams J. Hunt P. W. et al (2009). Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy.J. Virol.83329–335. 10.1128/jvi.01763-08
195
Hatziioannou T. Perez-Caballero D. Yang A. Cowan S. Bieniasz P. D. (2004). Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha.Proc. Natl. Acad. Sci. U.S.A.10110774–10779. 10.1073/pnas.0402361101
196
Haworth K. G. Peterson C. W. Kiem H. P. (2017). CCR5-edited gene therapies for HIV cure: closing the door to viral entry.Cytotherapy191325–1338. 10.1016/j.jcyt.2017.05.013
197
He H. Dang Y. Dai F. Guo Z. Wu J. She X. et al (2003). Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B.J. Biol. Chem.27829278–29287. 10.1074/jbc.m303800200
198
Henrich T. J. Hanhauser E. Marty F. M. Sirignano M. N. Keating S. Lee T. H. et al (2014). Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases.Ann. Intern. Med.161319–327. 10.7326/m14-1027
199
Henrich T. J. Hatano H. Bacon O. Hogan L. E. Rutishauser R. Hill A. et al (2017). HIV-1 persistence following extremely early initiation of antiretroviral therapy (ART) during acute HIV-1 infection: an observational study.PLoS Med.14:e1002417.
200
Heredia A. Le N. Gartenhaus R. B. Sausville E. Medina-Moreno S. Zapata J. C. et al (2015). Targeting of mTOR catalytic site inhibits multiple steps of the HIV-1 lifecycle and suppresses HIV-1 viremia in humanized mice.Proc. Natl. Acad. Sci. U.S.A.1129412–9417. 10.1073/pnas.1511144112
201
Hermankova M. Siliciano J. D. Zhou Y. Monie D. Chadwick K. Margolick J. B. et al (2003). Analysis of human immunodeficiency virus type 1 gene expression in latently infected resting CD4+ T lymphocytes in vivo.J. Virol.777383–7392. 10.1128/jvi.77.13.7383-7392.2003
202
Hille F. Richter H. Wong S. P. Bratoviè M. Ressel S. Charpentier E. (2018). The biology of CRISPR-Cas: backward and forward.Cell1721239–1259. 10.1016/j.cell.2017.11.032
203
Honda A. Nogami M. Yokozeki T. Yamazaki M. Nakamura H. Watanabe H. et al (1999). Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation.Cell99521–532. 10.1016/s0092-8674(00)81540-8
204
Hook S. S. Orian A. Cowley S. M. Eisenman R. N. (2002). Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes.Proc. Natl. Acad. Sci. U.S.A.9913425–13430. 10.1073/pnas.172511699
205
Horvath P. Barrangou R. (2010). CRISPR/Cas, the immune system of bacteria and archaea.Science327167–170. 10.1126/science.1179555
206
Hosokawa N. Hara T. Kaizuka T. Kishi C. Takamura A. Miura Y. et al (2009). Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy.Mol. Biol. Cell201981–1991. 10.1091/mbc.e08-12-1248
207
Hu M. M. Yang Q. Xie X. Q. Liao C. Y. Lin H. Liu T. T. et al (2016). Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus.Immunity45555–569. 10.1016/j.immuni.2016.08.014
208
Huang Y. Paxton W. A. Wolinsky S. M. Neumann A. U. Zhang L. He T. et al (1996). The role of a mutant CCR5 allele in HIV-1 transmission and disease progression.Nat. Med.21240–1243. 10.1038/nm1196-1240
209
Hubbert C. Guardiola A. Shao R. Kawaguchi Y. Ito A. Nixon A. et al (2002). HDAC6 is a microtubule-associated deacetylase.Nature417455–458. 10.1038/417455a
210
Hütter G. Nowak D. Mossner M. Ganepola S. Müssig A. Allers K. et al (2009). Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation.N. Engl. J. Med.360692–698. 10.1056/nejmoa0802905
211
Ichimura Y. Kumanomidou T. Sou Y. S. Mizushima T. Ezaki J. Ueno T. et al (2008). Structural basis for sorting mechanism of p62 in selective autophagy.J. Biol. Chem.28322847–22857. 10.1074/jbc.m802182200
212
Iliopoulou M. Nolan R. Alvarez L. Watanabe Y. Coomer C. A. Jakobsdottir G. M. et al (2018). A dynamic three-step mechanism drives the HIV-1 pre-fusion reaction.Nat. Struct. Mol. Biol.25814–822. 10.1038/s41594-018-0113-x
213
Itakura E. Kishi C. Inoue K. Mizushima N. (2008). Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG.Mol. Biol. Cell195360–5372. 10.1091/mbc.e08-01-0080
214
Iwata A. Riley B. E. Johnston J. A. Kopito R. R. (2005). HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin.J. Biol. Chem.28040282–40292. 10.1074/jbc.m508786200
215
Jackson W. T. (2015). Viruses and the autophagy pathway.Virology479-480450–456. 10.1016/j.virol.2015.03.042
216
Jeffries T. R. Dove S. K. Michell R. H. Parker P. J. (2004). PtdIns-specific MPR pathway association of a novel WD40 repeat protein, WIPI49.Mol. Biol. Cell152652–2663. 10.1091/mbc.e03-10-0732
217
Jiang Q. Wang Y. Li T. Shi K. Li Z. Ma Y. et al (2011). Heat shock protein 90-mediated inactivation of nuclear factor-κB switches autophagy to apoptosis through becn1 transcriptional inhibition in selenite-induced NB4 cells.Mol. Biol. Cell221167–1180. 10.1091/mbc.e10-10-0860
218
Jimenez-Baranda S. Gomez-Mouton C. Rojas A. Martinez-Prats L. Mira E. Ana Lacalle R. et al (2007). Filamin-A regulates actin-dependent clustering of HIV receptors.Nat. Cell Biol.9838–846. 10.1038/ncb1610
219
Jolly C. Kashefi K. Hollinshead M. Sattentau Q. J. (2004). HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse.J. Exp. Med.199283–293. 10.1084/jem.20030648
220
Jolly C. Sattentau Q. J. (2004). Retroviral spread by induction of virological synapses.Traffic5643–650. 10.1111/j.1600-0854.2004.00209.x
221
Joshi A. Sedano M. Beauchamp B. Punke E. B. Mulla Z. D. Meza A. et al (2016). HIV-1 Env glycoprotein phenotype along with immune activation determines CD4 T cell loss in HIV patients.J. Immunol.1961768–1779. 10.4049/jimmunol.1501588
222
Joubert P. E. Meiffren G. Grégoire I. P. Pontini G. Richetta C. Flacher M. et al (2009). Autophagy induction by the pathogen receptor CD46.Cell Host Microbe6354–366. 10.1016/j.chom.2009.09.006
223
Jounai N. Takeshita F. Kobiyama K. Sawano A. Miyawaki A. Xin K. Q. et al (2007). The Atg5 Atg12 conjugate associates with innate antiviral immune responses.Proc. Natl. Acad. Sci. U.S.A.10414050–14055. 10.1073/pnas.0704014104
224
Julg B. Pereyra F. Buzón M. J. Piechocka-Trocha A. Clark M. J. Baker B. M. et al (2010). Infrequent recovery of HIV from but robust exogenous infection of activated CD4(+) T cells in HIV elite controllers.Clin. Infect. Dis.51233–238. 10.1086/653677
225
Jung C. H. Jun C. B. Ro S. H. Kim Y. M. Otto N. M. Cao J. et al (2009). ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery.Mol. Biol. Cell201992–2003. 10.1091/mbc.e08-12-1249
226
Kabeya Y. Mizushima N. Ueno T. Yamamoto A. Kirisako T. Noda T. et al (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing.EMBO J.195720–5728. 10.1093/emboj/19.21.5720
227
Kabeya Y. Mizushima N. Yamamoto A. Oshitani-Okamoto S. Ohsumi Y. Yoshimori T. (2004). LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation.J. Cell Sci.1172805–2812. 10.1242/jcs.01131
228
Kaminski R. Bella R. Yin C. Otte J. Ferrante P. Gendelman H. E. et al (2016a). Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study.Gene Ther.23690–695. 10.1038/gt.2016.41
229
Kaminski R. Chen Y. Fischer T. Tedaldi E. Napoli A. Zhang Y. et al (2016b). Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing.Sci. Rep.6:22555.
230
Karow M. Fischer S. Meßling S. Konertz R. Riehl J. Xiong Q. et al (2020). Functional characterisation of the autophagy ATG12~5/16 complex in Dictyostelium discoideum.Cells9:1179. 10.3390/cells9051179
231
Kast D. J. Dominguez R. (2015). WHAMM links actin assembly via the Arp2/3 complex to autophagy.Autophagy111702–1704. 10.1080/15548627.2015.1073434
232
Kast D. J. Zajac A. L. Holzbaur E. L. Ostap E. M. Dominguez R. (2015). WHAMM directs the Arp2/3 complex to the ER for autophagosome biogenesis through an actin comet tail mechanism.Curr. Biol.251791–1797. 10.1016/j.cub.2015.05.042
233
Katsuragi Y. Ichimura Y. Komatsu M. (2015). p62/SQSTM1 functions as a signaling hub and an autophagy adaptor.FEBS J.2824672–4678. 10.1111/febs.13540
234
Kawaguchi Y. Kovacs J. J. Mclaurin A. Vance J. M. Ito A. Yao T. P. (2003). The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress.Cell115727–738. 10.1016/s0092-8674(03)00939-5
235
Keele B. F. Derdeyn C. A. (2009). Genetic and antigenic features of the transmitted virus.Curr. Opin. HIV AIDS4352–357. 10.1097/coh.0b013e32832d9fef
236
Killian M. S. (2012). Dual role of autophagy in HIV-1 replication and pathogenesis.AIDS Res. Ther.9:16. 10.1186/1742-6405-9-16
237
Kirkin V. Lamark T. Sou Y. S. Bjørkøy G. Nunn J. L. Bruun J. A. et al (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates.Mol. Cell.33505–516. 10.1016/j.molcel.2009.01.020
238
Klionsky D. J. Abdalla F. C. Abeliovich H. Abraham R. T. Acevedo-Arozena A. Adeli K. et al (2012). Guidelines for the use and interpretation of assays for monitoring autophagy.Autophagy8445–544.
239
Klionsky D. J. Abeliovich H. Agostinis P. Agrawal D. K. Aliev G. Askew D. S. et al (2008). Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes.Autophagy4151–175.
240
Knaevelsrud H. Simonsen A. (2010). Fighting disease by selective autophagy of aggregate-prone proteins.FEBS Lett.5842635–2645. 10.1016/j.febslet.2010.04.041
241
Knævelsrud H. Søreng K. Raiborg C. Håberg K. Rasmuson F. Brech A. et al (2013). Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation.J. Cell Biol.202331–349. 10.1083/jcb.201205129
242
Kohda T. A. Tanaka K. Konomi M. Sato M. Osumi M. Yamamoto M. (2007). Fission yeast autophagy induced by nitrogen starvation generates a nitrogen source that drives adaptation processes.Genes Cells12155–170. 10.1111/j.1365-2443.2007.01041.x
243
Koike M. Shibata M. Tadakoshi M. Gotoh K. Komatsu M. Waguri S. et al (2008). Inhibition of autophagy prevents hippocampal pyramidal neuron death after hypoxic-ischemic injury.Am. J. Pathol.172454–469. 10.2353/ajpath.2008.070876
244
Komatsu M. Ichimura Y. (2010). Physiological significance of selective degradation of p62 by autophagy.FEBS Lett.5841374–1378. 10.1016/j.febslet.2010.02.017
245
Komatsu M. Kurokawa H. Waguri S. Taguchi K. Kobayashi A. Ichimura Y. et al (2010). The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1.Nat. Cell Biol.12213–223. 10.1038/ncb2021
246
Komatsu M. Waguri S. Chiba T. Murata S. Iwata J. Tanida I. et al (2006). Loss of autophagy in the central nervous system causes neurodegeneration in mice.Nature441880–884. 10.1038/nature04723
247
Komatsu M. Wang Q. J. Holstein G. R. Friedrich V. L. Jr. Iwata J. Kominami E. et al (2007). Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration.Proc. Natl. Acad. Sci. U.S.A.10414489–14494. 10.1073/pnas.0701311104
248
Kopito R. R. (2003). The missing linker: an unexpected role for a histone deacetylase.Mol. Cell.121349–1351.
249
Kordelas L. Verheyen J. Beelen D. W. Horn P. A. Heinold A. Kaiser R. et al (2014). Shift of HIV tropism in stem-cell transplantation with CCR5 Delta32 mutation.N. Engl. J. Med.371880–882. 10.1056/nejmc1405805
250
Korn K. Reil H. Walter H. Schmidt B. (2003). Quality control trial for human immunodeficiency virus type 1 drug resistance testing using clinical samples reveals problems with detecting minority species and interpretation of test results.J. Clin. Microbiol.413559–3565. 10.1128/jcm.41.8.3559-3565.2003
251
Krüger U. Pfeiffer T. Bosch V. (1996). Generation of lymphocyte cell lines coexpressing CD4 and wild-type or mutant HIV type 1 glycoproteins: implications for HIV type 1 Env-induced cell lysis.AIDS Res. Hum. Retroviruses12783–792. 10.1089/aid.1996.12.783
252
Kuma A. Mizushima N. Ishihara N. Ohsumi Y. (2002). Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast.J. Biol. Chem.27718619–18625. 10.1074/jbc.m111889200
253
Kumar R. Qureshi H. Deshpande S. Bhattacharya J. (2018). Broadly neutralizing antibodies in HIV-1 treatment and prevention.Ther. Adv. Vaccines Immunother.661–68. 10.1177/2515135518800689
254
Kundu M. Lindsten T. Yang C. Y. Wu J. Zhao F. Zhang J. et al (2008). Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation.Blood1121493–1502. 10.1182/blood-2008-02-137398
255
Kuritzkes D. R. (2016). Hematopoietic stem cell transplantation for HIV cure.J. Clin. Invest.126432–437. 10.1172/jci80563
256
Kuroyanagi H. Yan J. Seki N. Yamanouchi Y. Suzuki Y. Takano T. et al (1998). Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignment.Genomics5176–85. 10.1006/geno.1998.5340
257
Kyei G. B. Dinkins C. Davis A. S. Roberts E. Singh S. B. Dong C. et al (2009). Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages.J. Cell Biol.186255–268. 10.1083/jcb.200903070
258
Lajoie J. Fontaine J. Tremblay C. Routy J. P. Poudrier J. Roger M. (2009). Persistence of high levels of blood soluble human leukocyte antigen-G is associated with rapid progression of HIV infection.Aids231437–1440. 10.1097/qad.0b013e32832d0825
259
Lambotte O. Boufassa F. Madec Y. Nguyen A. Goujard C. Meyer L. et al (2005). HIV controllers: a homogeneous group of HIV-1-infected patients with spontaneous control of viral replication.Clin. Infect. Dis.411053–1056. 10.1086/433188
260
Lamine A. Caumont-Sarcos A. Chaix M. L. Saez-Cirion A. Rouzioux C. Delfraissy J. F. et al (2007). Replication-competent HIV strains infect HIV controllers despite undetectable viremia (ANRS EP36 study).Aids211043–1045. 10.1097/qad.0b013e3280d5a7ac
261
Lang T. Reiche S. Straub M. Bredschneider M. Thumm M. (2000). Autophagy and the cvt pathway both depend on AUT9.J. Bacteriol.1822125–2133. 10.1128/jb.182.8.2125-2133.2000
262
Lee H. K. Lund J. M. Ramanathan B. Mizushima N. Iwasaki A. (2007). Autophagy-dependent viral recognition by plasmacytoid dendritic cells.Science3151398–1401. 10.1126/science.1136880
263
Lee H. K. Mattei L. M. Steinberg B. E. Alberts P. Lee Y. H. Chervonsky A. et al (2010). In vivo requirement for Atg5 in antigen presentation by dendritic cells.Immunity32227–239. 10.1016/j.immuni.2009.12.006
264
Lee J. Y. Koga H. Kawaguchi Y. Tang W. Wong E. Gao Y. S. et al (2010). HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy.EMBO J.29969–980. 10.1038/emboj.2009.405
265
Leonova E. I. Gainetdinov R. R. (2020). CRISPR/Cas9 technology in translational biomedicine.Cell Physiol. Biochem.54354–370. 10.33594/000000224
266
Levine B. Mizushima N. Virgin H. W. (2011). Autophagy in immunity and inflammation.Nature469323–335.
267
Levonen A. L. Hill B. G. Kansanen E. Zhang J. Darley-Usmar V. M. (2014). Redox regulation of antioxidants, autophagy, and the response to stress: implications for electrophile therapeutics.Free Radic. Biol. Med.71196–207. 10.1016/j.freeradbiomed.2014.03.025
268
Levy J. M. Thompson J. C. Griesinger A. M. Amani V. Donson A. M. Birks D. K. et al (2014). Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors.Cancer Discov.4773–780. 10.1158/2159-8290.cd-14-0049
269
Leymarie O. Lepont L. Berlioz-Torrent C. (2017). Canonical and non-canonical autophagy in HIV-1 replication cycle.Viruses9:270. 10.3390/v9100270
270
Li M. Liu W. Bauch T. Graviss E. A. Arduino R. C. Kimata J. T. et al (2020). Clearance of HIV infection by selective elimination of host cells capable of producing HIV.Nat. Commun.11:4051.
271
Li Q. Barres B. A. (2018). Microglia and macrophages in brain homeostasis and disease.Nat. Rev. Immunol.18225–242. 10.1038/nri.2017.125
272
Li Y. Wang L. X. Yang G. Hao F. Urba W. J. Hu H. M. (2008). Efficient cross-presentation depends on autophagy in tumor cells.Cancer Res.686889–6895. 10.1158/0008-5472.can-08-0161
273
Liang C. Lee J. S. Inn K. S. Gack M. U. Li Q. Roberts E. A. et al (2008). Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking.Nat. Cell Biol.10776–787. 10.1038/ncb1740
274
Liang Q. Chang B. Brulois K. F. Castro K. Min C. K. Rodgers M. A. et al (2013). Kaposi’s sarcoma-associated herpesvirus K7 modulates Rubicon-mediated inhibition of autophagosome maturation.J. Virol.8712499–12503. 10.1128/jvi.01898-13
275
Liang Q. Seo G. J. Choi Y. J. Kwak M. J. Ge J. Rodgers M. A. et al (2014). Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses.Cell Host Microbe15228–238. 10.1016/j.chom.2014.01.009
276
Liang X. H. Jackson S. Seaman M. Brown K. Kempkes B. Hibshoosh H. et al (1999). Induction of autophagy and inhibition of tumorigenesis by beclin 1.Nature402672–676. 10.1038/45257
277
Liang X. H. Kleeman L. K. Jiang H. H. Gordon G. Goldman J. E. Berry G. et al (1998). Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein.J. Virol.728586–8596. 10.1128/jvi.72.11.8586-8596.1998
278
Limou S. Le Clerc S. Coulonges C. Carpentier W. Dina C. Delaneau O. et al (2009). Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS genomewide association study 02).J. Infect. Dis.199419–426. 10.1086/596067
279
Liou W. Geuze H. J. Geelen M. J. Slot J. W. (1997). The autophagic and endocytic pathways converge at the nascent autophagic vacuoles.J. Cell Biol.13661–70. 10.1083/jcb.136.1.61
280
Liu F. Lee J. Y. Wei H. Tanabe O. Engel J. D. Morrison S. J. et al (2010). FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells.Blood1164806–4814. 10.1182/blood-2010-06-288589
281
Liu J. Liang R. Huang H. Zhang Y. Xie A. Zhong Y. (2021). Effect of an antagonistic peptide of CCR5 on the expression of autophagy-related genes and β-arrestin 2 in lung tissues of asthmatic mice.Allergy Asthma Immunol. Res.13106–121. 10.4168/aair.2021.13.1.106
282
Liu R. Paxton W. A. Choe S. Ceradini D. Martin S. R. Horuk R. et al (1996). Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection.Cell86367–377. 10.1016/s0092-8674(00)80110-5
283
Liu S. Xing Y. Wang J. Pan R. Li G. Tang H. et al (2019). The dual role of HIV-1 gp120 V3 loop-induced autophagy in the survival and apoptosis of the primary rat hippocampal neurons.Neurochem. Res.441636–1652. 10.1007/s11064-019-02788-3
284
Liu Y. Belkina N. V. Shaw S. (2009). HIV infection of T cells: actin-in and actin-out.Sci. Signal.2:e23.
285
Longatti A. Tooze S. A. (2009). Vesicular trafficking and autophagosome formation.Cell Death Differ.16956–965. 10.1038/cdd.2009.39
286
Longatti A. Tooze S. A. (2012). Recycling endosomes contribute to autophagosome formation.Autophagy81682–1683. 10.4161/auto.21486
287
López-Galíndez C. (2019). HIV long-term non-progressors elite controllers: an interplay between host, immune and viral factors.Future Med.14287–297. 10.2217/fvl-2018-0207
288
Lopez-Galindez C. Pernas M. Casado C. Olivares I. Lorenzo-Redondo R. (2019). Elite controllers and lessons learned for HIV-1 cure.Curr. Opin. Virol.3831–36. 10.1016/j.coviro.2019.05.010
289
Lorenzo-Redondo R. Fryer H. R. Bedford T. Kim E. Y. Archer J. Pond S. L. K. et al (2016). Persistent HIV-1 replication maintains the tissue reservoir during therapy.Nature53051–56. 10.1038/nature16933
290
Luzuriaga K. Gay H. Ziemniak C. Sanborn K. B. Somasundaran M. Rainwater-Lovett K. et al (2015). Viremic relapse after HIV-1 remission in a perinatally infected child.N. Engl. J. Med.372786–788. 10.1056/nejmc1413931
291
Mailler E. Bernacchi S. Marquet R. Paillart J. C. Vivet-Boudou V. Smyth R. P. (2016). The life-cycle of the HIV-1 Gag-RNA complex.Viruses8:248. 10.3390/v8090248
292
Mailler E. Waheed A. A. Park S. Y. Gershlick D. C. Freed E. O. Bonifacino J. S. (2019). The autophagy protein ATG9A promotes HIV-1 infectivity.Retrovirology16:18.
293
Malet I. Subra F. Charpentier C. Collin G. Descamps D. Calvez V. et al (2017). Mutations located outside the integrase gene can confer resistance to HIV-1 integrase strand transfer inhibitors.mBio8:e00922-17.
294
Mandell M. A. Jain A. Arko-Mensah J. Chauhan S. Kimura T. Dinkins C. et al (2014a). TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition.Dev. Cell30394–409. 10.1016/j.devcel.2014.06.013
295
Mandell M. A. Kimura T. Jain A. Johansen T. Deretic V. (2014b). TRIM proteins regulate autophagy: TRIM5 is a selective autophagy receptor mediating HIV-1 restriction.Autophagy102387–2388. 10.4161/15548627.2014.984278
296
Mao J. Lin E. He L. Yu J. Tan P. Zhou Y. (2019). Autophagy and viral infection.Adv. Exp. Med. Biol.120955–78. 10.1007/978-981-15-0606-2_5
297
Mardones P. Rubinsztein D. C. Hetz C. (2016). Mystery solved: trehalose kickstarts autophagy by blocking glucose transport.Sci. Signal.9:fs2. 10.1126/scisignal.aaf1937
298
Mari M. Griffith J. Rieter E. Krishnappa L. Klionsky D. J. Reggiori F. (2010). An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis.J. Cell Biol.1901005–1022. 10.1083/jcb.200912089
299
Markosyan R. M. Cohen F. S. Melikyan G. B. (2003). HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation.Mol. Biol. Cell14926–938. 10.1091/mbc.e02-09-0573
300
Marraffini L. A. Sontheimer E. J. (2010). CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea.Nat. Rev. Genet.11181–190. 10.1038/nrg2749
301
Marrero-Hernández S. Márquez-Arce D. Cabrera-Rodríguez R. Estévez-Herrera J. Pérez-Yanes S. Barroso-González J. et al (2019). HIV-1 nef targets HDAC6 to assure viral production and virus infection.Front. Microbiol.10:2437.
302
Martinet W. Agostinis P. Vanhoecke B. Dewaele M. De Meyer G. R. (2009). Autophagy in disease: a double-edged sword with therapeutic potential.Clin. Sci.116697–712. 10.1042/cs20080508
303
Masiero E. Agatea L. Mammucari C. Blaauw B. Loro E. Komatsu M. et al (2009). Autophagy is required to maintain muscle mass.Cell Metab.10507–515. 10.1016/j.cmet.2009.10.008
304
Matoba K. Kotani T. Tsutsumi A. Tsuji T. Mori T. Noshiro D. et al (2020). Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion.Nat. Struct. Mol. Biol.271185–1193. 10.1038/s41594-020-00518-w
305
Matsunaga K. Saitoh T. Tabata K. Omori H. Satoh T. Kurotori N. et al (2009). Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages.Nat. Cell Biol.11385–396. 10.1038/ncb1846
306
Mauthe M. Jacob A. Freiberger S. Hentschel K. Stierhof Y. D. Codogno P. et al (2011). Resveratrol-mediated autophagy requires WIPI-1-regulated LC3 lipidation in the absence of induced phagophore formation.Autophagy71448–1461. 10.4161/auto.7.12.17802
307
McDougal J. S. Kennedy M. S. Sligh J. M. Cort S. P. Mawle A. Nicholson J. K. (1986). Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule.Science231382–385. 10.1126/science.3001934
308
McDougal J. S. Mawle A. Cort S. P. Nicholson J. K. Cross G. D. Scheppler-Campbell J. A. et al (1985). Cellular tropism of the human retrovirus HTLV-III/LAV. I. Role of T cell activation and expression of the T4 antigen.J. Immunol.1353151–3162.
309
McGrath J. W. Winchester M. S. Kaawa-Mafigiri D. Walakira E. Namutiibwa F. Birungi J. et al (2014). Challenging the paradigm: anthropological perspectives on HIV as a chronic disease.Med. Anthropol.33303–317. 10.1080/01459740.2014.892483
310
Meléndez A. Tallóczy Z. Seaman M. Eskelinen E. L. Hall D. H. Levine B. (2003). Autophagy genes are essential for dauer development and life-span extension in C. elegans.Science3011387–1391. 10.1126/science.1087782
311
Melikyan G. B. (2014). HIV entry: a game of hide-and-fuse?Curr. Opin. Virol.41–7. 10.1016/j.coviro.2013.09.004
312
Meng Q. Cai D. (2011). Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IkappaB kinase beta (IKKbeta)/NF-kappaB pathway.J. Biol. Chem.28632324–32332. 10.1074/jbc.m111.254417
313
Mens H. Kearney M. Wiegand A. Shao W. Schønning K. Gerstoft J. et al (2010). HIV-1 continues to replicate and evolve in patients with natural control of HIV infection.J. Virol.8412971–12981. 10.1128/jvi.00387-10
314
Mercer C. A. Kaliappan A. Dennis P. B. (2009). A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy.Autophagy5649–662. 10.4161/auto.5.5.8249
315
Mi N. Chen Y. Wang S. Chen M. Zhao M. Yang G. et al (2015). CapZ regulates autophagosomal membrane shaping by promoting actin assembly inside the isolation membrane.Nat. Cell Biol.171112–1123. 10.1038/ncb3215
316
Migueles S. A. Osborne C. M. Royce C. Compton A. A. Joshi R. P. Weeks K. A. et al (2008). Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control.Immunity291009–1021. 10.1016/j.immuni.2008.10.010
317
Mizushima N. Kuma A. Kobayashi Y. Yamamoto A. Matsubae M. Takao T. et al (2003). Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate.J. Cell Sci.1161679–1688. 10.1242/jcs.00381
318
Mizushima N. Noda T. Ohsumi Y. (1999). Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway.EMBO J.183888–3896. 10.1093/emboj/18.14.3888
319
Mizushima N. Ohsumi Y. Yoshimori T. (2002). Autophagosome formation in mammalian cells.Cell Struct. Funct.27421–429. 10.1247/csf.27.421
320
Mizushima N. Yoshimori T. Ohsumi Y. (2011). The role of Atg proteins in autophagosome formation.Annu. Rev. Cell Dev. Biol.27107–132. 10.1146/annurev-cellbio-092910-154005
321
Mohammadi P. Desfarges S. Bartha I. Joos B. Zangger N. Muñ;oz M. et al (2013). 24 hours in the life of HIV-1 in a T cell line.PLoS Pathog.9:e1003161. 10.1371/journal.ppat.1003161
322
Molina L. Grimaldi M. Robert-Hebmann V. Espert L. Varbanov M. Devaux C. et al (2007). Proteomic analysis of the cellular responses induced in uninfected immune cells by cell-expressed X4 HIV-1 envelope.Proteomics73116–3130. 10.1002/pmic.200700306
323
Monaco D. C. Zapata L. Hunter E. Salomon H. Dilernia D. A. (2020). Resistance profile of HIV-1 quasispecies in patients under treatment failure using single molecule, real-time sequencing.Aids342201–2210. 10.1097/qad.0000000000002697
324
Moore J. P. Kitchen S. G. Pugach P. Zack J. A. (2004). The CCR5 and CXCR4 coreceptors–central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection.AIDS Res. Hum. Retroviruses20111–126. 10.1089/088922204322749567
325
Moreau K. Ravikumar B. Puri C. Rubinsztein D. C. (2012). Arf6 promotes autophagosome formation via effects on phosphatidylinositol 4,5-bisphosphate and phospholipase D.J. Cell Biol.196483–496. 10.1083/jcb.201110114
326
Morel E. (2020). Endoplasmic reticulum membrane and contact site dynamics in autophagy regulation and stress response.Front. Cell. Dev. Biol.8:343.
327
Mostashari Rad T. Saghaie L. Fassihi A. (2018). HIV-1 entry inhibitors: a review of experimental and computational studies.Chem. Biodivers.15:e1800159. 10.1002/cbdv.201800159
328
Moulard M. Hallenberger S. Garten W. Klenk H. D. (1999). Processing and routage of HIV glycoproteins by furin to the cell surface.Virus Res.6055–65. 10.1016/s0168-1702(99)00002-7
329
Nakagawa I. Amano A. Mizushima N. Yamamoto A. Yamaguchi H. Kamimoto T. et al (2004). Autophagy defends cells against invading group A Streptococcus.Science3061037–1040. 10.1126/science.1103966
330
Nakai A. Yamaguchi O. Takeda T. Higuchi Y. Hikoso S. Taniike M. et al (2007). The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress.Nat. Med.13619–624. 10.1038/nm1574
331
Nakamoto M. Moy R. H. Xu J. Bambina S. Yasunaga A. Shelly S. S. et al (2012). Virus recognition by Toll-7 activates antiviral autophagy in Drosophila.Immunity36658–667. 10.1016/j.immuni.2012.03.003
332
Nakatogawa H. Ichimura Y. Ohsumi Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion.Cell130165–178. 10.1016/j.cell.2007.05.021
333
Nakatogawa H. Suzuki K. Kamada Y. Ohsumi Y. (2009). Dynamics and diversity in autophagy mechanisms: lessons from yeast.Nat. Rev. Mol. Cell Biol.10458–467. 10.1038/nrm2708
334
Nardacci R. Amendola A. Ciccosanti F. Corazzari M. Esposito V. Vlassi C. et al (2014). Autophagy plays an important role in the containment of HIV-1 in nonprogressor-infected patients.Autophagy101167–1178. 10.4161/auto.28678
335
Nardacci R. Ciccosanti F. Marsella C. Ippolito G. Piacentini M. Fimia G. M. (2017). Role of autophagy in HIV infection and pathogenesis.J. Intern. Med.281422–432. 10.1111/joim.12596
336
Naslavsky N. Weigert R. Donaldson J. G. (2003). Convergence of non-clathrin- and clathrin-derived endosomes involves Arf6 inactivation and changes in phosphoinositides.Mol. Biol. Cell14417–431. 10.1091/mbc.02-04-0053
337
Nedjic J. Aichinger M. Emmerich J. Mizushima N. Klein L. (2008). Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance.Nature455396–400. 10.1038/nature07208
338
Nimmerjahn F. Milosevic S. Behrends U. Jaffee E. M. Pardoll D. M. Bornkamm G. W. et al (2003). Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy.Eur. J. Immunol.331250–1259. 10.1002/eji.200323730
339
Nishiyama J. Miura E. Mizushima N. Watanabe M. Yuzaki M. (2007). Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death.Autophagy3591–596. 10.4161/auto.4964
340
Nivon M. Abou-Samra M. Richet E. Guyot B. Arrigo A. P. Kretz-Remy C. (2012). NF-κB regulates protein quality control after heat stress through modulation of the BAG3-HspB8 complex.J. Cell Sci.1251141–1151. 10.1242/jcs.091041
341
Nivon M. Richet E. Codogno P. Arrigo A. P. Kretz-Remy C. (2009). Autophagy activation by NFkappaB is essential for cell survival after heat shock.Autophagy5766–783. 10.4161/auto.8788
342
Noda N. N. Fujioka Y. (2015). Atg1 family kinases in autophagy initiation.Cell Mol. Life Sci.723083–3096. 10.1007/s00018-015-1917-z
343
Noel N. Peñ;a R. David A. Avettand-Fenoel V. Erkizia I. Jimenez E. et al (2016). Long-term spontaneous control of HIV-1 is related to low frequency of infected cells and inefficient viral reactivation.J. Virol.906148–6158. 10.1128/jvi.00419-16
344
Obara K. Sekito T. Niimi K. Ohsumi Y. (2008). The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function.J. Biol. Chem.28323972–23980. 10.1074/jbc.m803180200
345
O’Connell K. A. Brennan T. P. Bailey J. R. Ray S. C. Siliciano R. F. Blankson J. N. (2010). Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus.J. Virol.847018–7028. 10.1128/jvi.00548-10
346
Ogawa M. Yoshimori T. Suzuki T. Sagara H. Mizushima N. Sasakawa C. (2005). Escape of intracellular Shigella from autophagy.Science307727–731. 10.1126/science.1106036
347
Oh D. S. Lee H. K. (2019). Autophagy protein ATG5 regulates CD36 expression and anti-tumor MHC class II antigen presentation in dendritic cells.Autophagy152091–2106. 10.1080/15548627.2019.1596493
348
Okulicz J. F. Marconi V. C. Landrum M. L. Wegner S. Weintrob A. Ganesan A. et al (2009). Clinical outcomes of elite controllers, viremic controllers, and long-term nonprogressors in the US Department of Defense HIV natural history study.J. Infect. Dis.2001714–1723. 10.1086/646609
349
Orvedahl A. Macpherson S. Sumpter R. Jr. Tallóczy Z. Zou Z. Levine B. (2010). Autophagy protects against Sindbis virus infection of the central nervous system.Cell Host Microbe7115–127. 10.1016/j.chom.2010.01.007
350
Osawa T. Kotani T. Kawaoka T. Hirata E. Suzuki K. Nakatogawa H. et al (2019). Atg2 mediates direct lipid transfer between membranes for autophagosome formation.Nat. Struct. Mol. Biol.26281–288. 10.1038/s41594-019-0203-4
351
Paludan C. Schmid D. Landthaler M. Vockerodt M. Kube D. Tuschl T. et al (2005). Endogenous MHC class II processing of a viral nuclear antigen after autophagy.Science307593–596. 10.1126/science.1104904
352
Pandey U. B. Nie Z. Batlevi Y. Mccray B. A. Ritson G. P. Nedelsky N. B. et al (2007). HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS.Nature447859–863.
353
Pankiv S. Clausen T. H. Lamark T. Brech A. Bruun J. A. Outzen H. et al (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy.J. Biol. Chem.28224131–24145. 10.1074/jbc.m702824200
354
Pasi K. J. Sabin C. A. Jenkins P. V. Devereux H. L. Ononye C. Lee C. A. (2000). The effects of the 32-bp CCR-5 deletion on HIV transmission and HIV disease progression in individuals with haemophilia.Br. J. Haematol.111136–142. 10.1111/j.1365-2141.2000.02325.x
355
Paul P. Münz C. (2016). Autophagy and mammalian viruses: roles in immune response, viral replication, and beyond.Adv. Virus Res.95149–195.
356
Perelson A. S. Essunger P. Ho D. D. (1997). Dynamics of HIV-1 and CD4+ lymphocytes in vivo.Aids11 (Suppl. A), S17–S24.
357
Perera R. M. Stoykova S. Nicolay B. N. Ross K. N. Fitamant J. Boukhali M. et al (2015). Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism.Nature524361–365. 10.1038/nature14587
358
Pereyra F. Addo M. M. Kaufmann D. E. Liu Y. Miura T. Rathod A. et al (2008). Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy.J. Infect. Dis.197563–571. 10.1086/526786
359
Pereyra F. Jia X. Mclaren P. J. Telenti A. De Bakker P. I. Walker B. D. et al (2010). The major genetic determinants of HIV-1 control affect HLA class I peptide presentation.Science3301551–1557. 10.1126/science.1195271
360
Pereyra F. Palmer S. Miura T. Block B. L. Wiegand A. Rothchild A. C. et al (2009). Persistent low-level viremia in HIV-1 elite controllers and relationship to immunologic parameters.J. Infect. Dis.200984–990. 10.1086/605446
361
Petrov V. Funderburg N. Weinberg A. Sieg S. (2013). Human β defensin-3 induces chemokines from monocytes and macrophages: diminished activity in cells from HIV-infected persons.Immunology140413–420. 10.1111/imm.12148
362
Pickford F. Masliah E. Britschgi M. Lucin K. Narasimhan R. Jaeger P. A. et al (2008). The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice.J. Clin. Invest.1182190–2199.
363
Pinan-Lucarré B. Paoletti M. Dementhon K. Coulary-Salin B. Clavé C. (2003). Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina.Mol. Microbiol.47321–333. 10.1046/j.1365-2958.2003.03208.x
364
Poku N. K. (2016). HIV prevention: the key to ending AIDS by 2030.Open AIDS J.1065–77. 10.2174/1874613601610010065
365
Polson H. E. De Lartigue J. Rigden D. J. Reedijk M. Urbé S. Clague M. J. et al (2010). Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation.Autophagy6506–522. 10.4161/auto.6.4.11863
366
Ponpuak M. Davis A. S. Roberts E. A. Delgado M. A. Dinkins C. Zhao Z. et al (2010). Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties.Immunity32329–341. 10.1016/j.immuni.2010.02.009
367
Poropatich K. Sullivan D. J. Jr. (2011). Human immunodeficiency virus type 1 long-term non-progressors: the viral, genetic and immunological basis for disease non-progression.J. Gen. Virol.92247–268. 10.1099/vir.0.027102-0
368
Powner D. J. Wakelam M. J. (2002). The regulation of phospholipase D by inositol phospholipids and small GTPases.FEBS Lett.53162–64. 10.1016/s0014-5793(02)03410-5
369
Proikas-Cezanne T. Waddell S. Gaugel A. Frickey T. Lupas A. Nordheim A. (2004). WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy.Oncogene239314–9325. 10.1038/sj.onc.1208331
370
Puigdomenech I. Massanella M. Cabrera C. Clotet B. Blanco J. (2009). On the steps of cell-to-cell HIV transmission between CD4 T cells.Retrovirology6:89. 10.1186/1742-4690-6-89
371
Punnonen E. L. Pihakaski K. Mattila K. Lounatmaa K. Hirsimäki P. (1989). Intramembrane particles and filipin labelling on the membranes of autophagic vacuoles and lysosomes in mouse liver.Cell Tissue Res.258269–276.
372
Puri C. Renna M. Bento C. F. Moreau K. Rubinsztein D. C. (2013). Diverse autophagosome membrane sources coalesce in recycling endosomes.Cell1541285–1299. 10.1016/j.cell.2013.08.044
373
Py B. F. Lipinski M. M. Yuan J. (2007). Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection.Autophagy3117–125. 10.4161/auto.3618
374
Qi L. Gang L. Hang K. W. Ling C. H. Xiaofeng Z. Zhen L. et al (2011). Programmed neuronal cell death induced by HIV-1 tat and methamphetamine.Microsc. Res. Tech.741139–1144. 10.1002/jemt.21006
375
Qi S. Kim D. J. Stjepanovic G. Hurley J. H. (2015). Structure of the human Atg13-Atg101 HORMA heterodimer: an interaction hub within the ULK1 complex.Structure231848–1857. 10.1016/j.str.2015.07.011
376
Qu X. Yu J. Bhagat G. Furuya N. Hibshoosh H. Troxel A. et al (2003). Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene.J. Clin. Invest.1121809–1820. 10.1172/jci20039
377
Quitadamo B. Peters P. J. Repik A. O’Connell O. Mou Z. Koch M. et al (2018). HIV-1 R5 macrophage-tropic envelope glycoprotein trimers bind CD4 with high affinity, while the CD4 binding site on non-macrophage-tropic, T-tropic R5 envelopes is occluded.J. Virol.92:e00841-17.
378
Radhakrishna H. Donaldson J. G. (1997). ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway.J. Cell Biol.13949–61. 10.1083/jcb.139.1.49
379
Randow F. Youle R. J. (2014). Self and nonself: how autophagy targets mitochondria and bacteria.Cell Host Microbe15403–411. 10.1016/j.chom.2014.03.012
380
Rasmussen T. A. Tolstrup M. Brinkmann C. R. Olesen R. Erikstrup C. Solomon A. et al (2014). Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial.Lancet HIV1e13–e21.
381
Ravikumar B. Moreau K. Jahreiss L. Puri C. Rubinsztein D. C. (2010). Plasma membrane contributes to the formation of pre-autophagosomal structures.Nat. Cell Biol.12747–757. 10.1038/ncb2078
382
Ravikumar B. Vacher C. Berger Z. Davies J. E. Luo S. Oroz L. G. et al (2004). Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease.Nat. Genet.36585–595. 10.1038/ng1362
383
Rawat P. Hon S. Teodorof-Diedrich C. Spector S. A. (2020). Trehalose inhibits human immunodeficiency virus type 1 infection in primary human macrophages and CD4(+) T lymphocytes through two distinct mechanisms.J. Virol.94:e00237-20.
384
Raymond S. Saliou A. Delobel P. Cazabat M. Pasquier C. Jeanne N. et al (2014). Evolution of HIV-1 quasispecies and coreceptor use in cell reservoirs of patients on suppressive antiretroviral therapy.J. Antimicrob. Chemother.692527–2530. 10.1093/jac/dku147
385
Reece J. C. Handley A. J. Anstee E. J. Morrison W. A. Crowe S. M. Cameron P. U. (1998). HIV-1 selection by epidermal dendritic cells during transmission across human skin.J. Exp. Med.1871623–1631. 10.1084/jem.187.10.1623
386
Rerks-Ngarm S. Pitisuttithum P. Nitayaphan S. Kaewkungwal J. Chiu J. Paris R. et al (2009). Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand.N. Engl. J. Med.3612209–2220.
387
Ribeiro C. M. Sarrami-Forooshani R. Setiawan L. C. Zijlstra-Willems E. M. van Hamme J. L. Tigchelaar W. et al (2016). Receptor usage dictates HIV-1 restriction by human TRIM5α in dendritic cell subsets.Nature540448–452. 10.1038/nature20567
388
Richardson M. W. Guo L. Xin F. Yang X. Riley J. L. (2014). Stabilized human TRIM5α protects human T cells from HIV-1 infection.Mol. Ther.221084–1095. 10.1038/mt.2014.52
389
Richetta C. Grégoire I. P. Verlhac P. Azocar O. Baguet J. Flacher M. et al (2013). Sustained autophagy contributes to measles virus infectivity.PLoS Pathog.9:e1003599. 10.1371/journal.ppat.1003599
390
Rogov V. Dötsch V. Johansen T. Kirkin V. (2014). Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy.Mol. Cell.53167–178. 10.1016/j.molcel.2013.12.014
391
Rohatgi R. A. Janusis J. Leonard D. Bellvé K. D. Fogarty K. E. Baehrecke E. H. et al (2015). Beclin 1 regulates growth factor receptor signaling in breast cancer.Oncogene345352–5362. 10.1038/onc.2014.454
392
Rojas-Celis V. Valiente-Echeverría F. Soto-Rifo R. Toro-Ascuy D. (2019). New challenges of HIV-1 infection: how HIV-1 attacks and resides in the central nervous system.Cells8:1245. 10.3390/cells8101245
393
Romano S. D’angelillo A. Pacelli R. Staibano S. De Luna E. Bisogni R. et al (2010). Role of FK506-binding protein 51 in the control of apoptosis of irradiated melanoma cells.Cell Death Differ.17145–157. 10.1038/cdd.2009.115
394
Roos M. T. Lange J. M. De Goede R. E. Coutinho R. A. Schellekens P. T. Miedema F. et al (1992). Viral phenotype and immune response in primary human immunodeficiency virus type 1 infection.J. Infect. Dis.165427–432. 10.1093/infdis/165.3.427
395
Rosenbloom D. I. S. Hill A. L. Laskey S. B. Siliciano R. F. (2017). Re-evaluating evolution in the HIV reservoir.Nature551E6–E9.
396
Rotger M. Dalmau J. Rauch A. Mclaren P. Bosinger S. E. Martinez R. et al (2011). Comparative transcriptomics of extreme phenotypes of human HIV-1 infection and SIV infection in sooty mangabey and rhesus macaque.J. Clin. Invest.1212391–2400. 10.1172/jci45235
397
Rotger M. Dang K. K. Fellay J. Heinzen E. L. Feng S. Descombes P. et al (2010). Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals.PLoS Pathog.6:e1000781. 10.1371/journal.ppat.1000781
398
Sagnier S. Daussy C. F. Borel S. Robert-Hebmann V. Faure M. Blanchet F. P. et al (2015). Autophagy restricts HIV-1 infection by selectively degrading Tat in CD4+ T lymphocytes.J. Virol.89615–625. 10.1128/jvi.02174-14
399
Saha B. Chisholm D. Kell A. M. Mandell M. A. (2020). A non-canonical role for the autophagy machinery in anti-retroviral signaling mediated by TRIM5α.PLoS Pathog.16:e1009017. 10.1371/journal.ppat.1009017
400
Salazar-Gonzalez J. F. Salazar M. G. Keele B. F. Learn G. H. Giorgi E. E. Li H. et al (2009). Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection.J. Exp. Med.2061273–1289. 10.1084/jem.20090378
401
Salgado M. Brennan T. P. O’Connell K. A. Bailey J. R. Ray S. C. Siliciano R. F. et al (2010). Evolution of the HIV-1 nef gene in HLA-B∗57 positive elite suppressors.Retrovirology7:94. 10.1186/1742-4690-7-94
402
Samson M. Libert F. Doranz B. J. Rucker J. Liesnard C. Farber C. M. et al (1996). Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene.Nature382722–725. 10.1038/382722a0
403
Santy L. C. Casanova J. E. (2001). Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D.J. Cell Biol.154599–610. 10.1083/jcb.200104019
404
Sarkar S. Davies J. E. Huang Z. Tunnacliffe A. Rubinsztein D. C. (2007). Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein.J. Biol. Chem.2825641–5652. 10.1074/jbc.m609532200
405
Sawa-Makarska J. Baumann V. Coudevylle N. Von Bülow S. Nogellova V. Abert C. et al (2020). Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation.Science369:eaaz7714. 10.1126/science.aaz7714
406
Scheid J. F. Horwitz J. A. Bar-On Y. Kreider E. F. Lu C. L. Lorenzi J. C. et al (2016). HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption.Nature535556–560. 10.1038/nature18929
407
Schmid D. Pypaert M. Münz C. (2007). Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes.Immunity2679–92. 10.1016/j.immuni.2006.10.018
408
Schneck N. A. Ivleva V. B. Cai C. X. Cooper J. W. Lei Q. P. (2020). Characterization of the furin cleavage motif for HIV-1 trimeric envelope glycoprotein by intact LC-MS analysis.Analyst1451636–1640. 10.1039/c9an02098e
409
Schuitemaker H. Koot M. Kootstra N. A. Dercksen M. W. De Goede R. E. van Steenwijk R. P. et al (1992). Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population.J. Virol.661354–1360. 10.1128/jvi.66.3.1354-1360.1992
410
Schuurman R. Brambilla D. De Groot T. Huang D. Land S. Bremer J. et al (2002). Underestimation of HIV type 1 drug resistance mutations: results from the ENVA-2 genotyping proficiency program.AIDS Res. Hum. Retroviruses18243–248. 10.1089/088922202753472801
411
Schweighardt B. Roy A. M. Meiklejohn D. A. Grace E. J. II Moretto W. J. Heymann J. J. et al (2004). R5 human immunodeficiency virus type 1 (HIV-1) replicates more efficiently in primary CD4+ T-cell cultures than X4 HIV-1.J. Virol.789164–9173. 10.1128/jvi.78.17.9164-9173.2004
412
Seglen P. O. Gordon P. B. (1982). 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes.Proc. Natl. Acad. Sci. U.S.A.791889–1892. 10.1073/pnas.79.6.1889
413
Seigneurin-Berny D. Verdel A. Curtet S. Lemercier C. Garin J. Rousseaux S. et al (2001). Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways.Mol. Cell. Biol.218035–8044. 10.1128/mcb.21.23.8035-8044.2001
414
Shah A. Kumar A. (2010). HIV-1 gp120-mediated increases in IL-8 production in astrocytes are mediated through the NF-κB pathway and can be silenced by gp120-specific siRNA.J. Neuroinflammation7:96. 10.1186/1742-2094-7-96
415
Shah A. Verma A. S. Patel K. H. Noel R. Rivera-Amill V. Silverstein P. S. et al (2011). HIV-1 gp120 induces expression of IL-6 through a nuclear factor-kappa B-dependent mechanism: suppression by gp120 specific small interfering RNA.PLoS One6:e21261. 10.1371/journal.pone.0021261
416
Shaik M. M. Peng H. Lu J. Rits-Volloch S. Xu C. Liao M. et al (2019). Structural basis of coreceptor recognition by HIV-1 envelope spike.Nature565318–323. 10.1038/s41586-018-0804-9
417
Shi C. S. Kehrl J. H. (2008). MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages.J. Biol. Chem.28333175–33182. 10.1074/jbc.m804478200
418
Shoji-Kawata S. Sumpter R. Leveno M. Campbell G. R. Zou Z. Kinch L. et al (2013). Identification of a candidate therapeutic autophagy-inducing peptide.Nature494201–206. 10.1038/nature11866
419
Shpilka T. Weidberg H. Pietrokovski S. Elazar Z. (2011). Atg8: an autophagy-related ubiquitin-like protein family.Genome Biol.12:226. 10.1186/gb-2011-12-7-226
420
Shvets E. Abada A. Weidberg H. Elazar Z. (2011). Dissecting the involvement of LC3B and GATE-16 in p62 recruitment into autophagosomes.Autophagy7683–688. 10.4161/auto.7.7.15279
421
Siliciano R. F. Greene W. C. (2011). HIV latency.Cold Spring Harb. Perspect. Med.1:a007096.
422
Sivaraman V. Zhang L. Meissner E. G. Jeffrey J. L. Su L. (2009). The heptad repeat 2 domain is a major determinant for enhanced human immunodeficiency virus type 1 (HIV-1) fusion and pathogenicity of a highly pathogenic HIV-1 Env.J. Virol.8311715–11725. 10.1128/jvi.00649-09
423
Søgaard O. S. Graversen M. E. Leth S. Olesen R. Brinkmann C. R. Nissen S. K. et al (2015). The depsipeptide romidepsin reverses HIV-1 latency in vivo.PLoS Pathog.11:e1005142. 10.1371/journal.ppat.1005142
424
Soliman M. Srikrishna G. Balagopal A. (2017). Mechanisms of HIV-1 control.Curr. HIV/AIDS Rep.14101–109. 10.1007/s11904-017-0357-9
425
Sood C. Marin M. Chande A. Pizzato M. Melikyan G. B. (2017). SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins.J. Biol. Chem.2926014–6026. 10.1074/jbc.m117.777714
426
Spijkerman I. J. Koot M. Prins M. Keet I. P. van den Hoek A. J. Miedema F. et al (1995). Lower prevalence and incidence of HIV-1 syncytium-inducing phenotype among injecting drug users compared with homosexual men.Aids91085–1092. 10.1097/00002030-199509000-00016
427
Staring J. Von Castelmur E. Blomen V. A. van den Hengel L. G. Brockmann M. Baggen J. et al (2017). PLA2G16 represents a switch between entry and clearance of Picornaviridae.Nature541412–416. 10.1038/nature21032
428
Staropoli I. Chanel C. Girard M. Altmeyer R. (2000). Processing, stability, and receptor binding properties of oligomeric envelope glycoprotein from a primary HIV-1 isolate.J. Biol. Chem.27535137–35145. 10.1074/jbc.m003868200
429
Stevenson M. (2003). HIV-1 pathogenesis.Nat. Med.9853–860.
430
Stolp B. Fackler O. T. (2011). How HIV takes advantage of the cytoskeleton in entry and replication.Viruses3293–311. 10.3390/v3040293
431
Straubinger T. Kay K. Bies R. (2019). Modeling HIV pre-exposure prophylaxis.Front. Pharmacol.10:1514.
432
Stremlau M. Owens C. M. Perron M. J. Kiessling M. Autissier P. Sodroski J. (2004). The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys.Nature427848–853. 10.1038/nature02343
433
Stultz R. D. Cenker J. J. Mcdonald D. (2017). Imaging HIV-1 genomic DNA from entry through productive infection.J. Virol.91:e00034-17.
434
Sutton J. Dotson D. Dong X. (2013). Molecular events in late stages of HIV-1 replication.JSM Microbiol.1:1004.
435
Suzuki H. Tabata K. Morita E. Kawasaki M. Kato R. Dobson R. C. et al (2014). Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.Structure2247–58. 10.1016/j.str.2013.09.023
436
Tang H. Sebti S. Titone R. Zhou Y. Isidoro C. Ross T. S. et al (2015). Decreased BECN1 mRNA expression in human breast cancer is associated with estrogen receptor-negative subtypes and poor prognosis.EBioMedicine2255–263. 10.1016/j.ebiom.2015.01.008
437
Tang Z. Takahashi Y. Wang H. G. (2019). ATG2 regulation of phagophore expansion at mitochondria-associated ER membranes.Autophagy152165–2166. 10.1080/15548627.2019.1666594
438
Tey S. K. Khanna R. (2012). Autophagy mediates transporter associated with antigen processing-independent presentation of viral epitopes through MHC class I pathway.Blood120994–1004. 10.1182/blood-2012-01-402404
439
Toda K. Nogami M. Murakami K. Kanaho Y. Nakayama K. (1999). Colocalization of phospholipase D1 and GTP-binding-defective mutant of ADP-ribosylation factor 6 to endosomes and lysosomes.FEBS Lett.442221–225. 10.1016/s0014-5793(98)01646-9
440
Tomoda T. Bhatt R. S. Kuroyanagi H. Shirasawa T. Hatten M. E. (1999). A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons.Neuron24833–846. 10.1016/s0896-6273(00)81031-4
441
Travassos L. H. Carneiro L. A. Ramjeet M. Hussey S. Kim Y. G. Magalhães J. G. et al (2010). Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry.Nat. Immunol.1155–62. 10.1038/ni.1823
442
Tsao L. C. Guo H. Jeffrey J. Hoxie J. A. Su L. (2016). CCR5 interaction with HIV-1 Env contributes to Env-induced depletion of CD4 T cells in vitro and in vivo.Retrovirology13:22.
443
Tuttle D. L. Coberley C. R. Xie X. Kou Z. C. Sleasman J. W. Goodenow M. M. (2004). Effects of human immunodeficiency virus type 1 infection on CCR5 and CXCR4 coreceptor expression on CD4 T lymphocyte subsets in infants and adolescents.AIDS Res. Hum. Retroviruses20305–313. 10.1089/088922204322996545
444
UNAIDS (2015). Understanding Fast-Track: Accelerating Action To End the AIDS Epidemic by 2030. Available online at: https://www.unaids.org/sites/default/files/media_asset/201506_JC2743_Understanding_FastTrack_en.pdf (accesssed June 2015).
445
UNAIDS (2020). Global HIV & AIDS Statistics. Available online at: https://www.unaids.org/en/resources/fact-sheet (accesssed December 2015).
446
Vakifahmetoglu-Norberg H. Xia H. G. Yuan J. (2015). Pharmacologic agents targeting autophagy.J. Clin. Invest.1255–13. 10.1172/jci73937
447
Valečka J. Almeida C. R. Su B. Pierre P. Gatti E. (2018). Autophagy and MHC-restricted antigen presentation.Mol. Immunol.99163–170. 10.1016/j.molimm.2018.05.009
448
Valenzuela-Fernandez A. Alvarez S. Gordon-Alonso M. Barrero M. Ursa A. Cabrero J. R. et al (2005). Histone deacetylase 6 regulates human immunodeficiency virus type 1 infection.Mol. Biol. Cell165445–5454. 10.1091/mbc.e05-04-0354
449
Valenzuela-Fernandez A. Cabrero J. R. Serrador J. M. Sanchez-Madrid F. (2008). HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions.Trends Cell Biol.18291–297. 10.1016/j.tcb.2008.04.003
450
Valera M. S. de Armas-Rillo L. Barroso-Gonzalez J. Ziglio S. Batisse J. Dubois N. et al (2015). The HDAC6/APOBEC3G complex regulates HIV-1 infectiveness by inducing Vif autophagic degradation.Retrovirology12:53.
451
van Grevenynghe J. Procopio F. A. He Z. Chomont N. Riou C. Zhang Y. et al (2008). Transcription factor FOXO3a controls the persistence of memory CD4(+) T cells during HIV infection.Nat. Med.14266–274. 10.1038/nm1728
452
van Grol J. Subauste C. Andrade R. M. Fujinaga K. Nelson J. Subauste C. S. (2010). HIV-1 inhibits autophagy in bystander macrophage/monocytic cells through Src-Akt and STAT3.PLoS One5:e11733. 10.1371/journal.pone.0011733
453
van Marle G. Church D. L. van der Meer F. Gill M. J. (2018). Combating the HIV reservoirs.Biotechnol. Genet. Eng. Rev.3476–89. 10.1080/02648725.2018.1471641
454
van’t Wout A. B. Kootstra N. A. Mulder-Kampinga G. A. Albrecht-van Lent N. Scherpbier H. J. Veenstra J. et al (1994). Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission.J. Clin. Invest.942060–2067. 10.1172/jci117560
455
Varbanov M. Espert L. Biard-Piechaczyk M. (2006). Mechanisms of CD4 T-cell depletion triggered by HIV-1 viral proteins.AIDS Rev.8221–236.
456
Verzella D. Pescatore A. Capece D. Vecchiotti D. Ursini M. V. Franzoso G. et al (2020). Life, death, and autophagy in cancer: NF-κB turns up everywhere.Cell Death Dis.11:210.
457
Viret C. Rozières A. Faure M. (2018). Autophagy during early virus-host cell interactions.J. Mol. Biol.4301696–1713. 10.1016/j.jmb.2018.04.018
458
Vlahakis S. R. Algeciras-Schimnich A. Bou G. Heppelmann C. J. Villasis-Keever A. Collman R. G. et al (2001). Chemokine-receptor activation by env determines the mechanism of death in HIV-infected and uninfected T lymphocytes.J. Clin. Invest.107207–215. 10.1172/jci11109
459
Vogl D. T. Stadtmauer E. A. Tan K. S. Heitjan D. F. Davis L. E. Pontiggia L. et al (2014). Combined autophagy and proteasome inhibition: a phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma.Autophagy101380–1390. 10.4161/auto.29264
460
Vorster P. J. Guo J. Yoder A. Wang W. Zheng Y. Xu X. et al (2011). LIM kinase 1 modulates cortical actin and CXCR4 cycling and is activated by HIV-1 to initiate viral infection.J. Biol. Chem.28612554–12564. 10.1074/jbc.m110.182238
461
Walczak M. Martens S. (2013). Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation.Autophagy9424–425. 10.4161/auto.22931
462
Wang G. Zhao N. Berkhout B. Das A. T. (2016). CRISPR-Cas9 can inhibit HIV-1 replication but NHEJ repair facilitates virus escape.Mol. Ther.24522–526. 10.1038/mt.2016.24
463
Wang H. W. Zhu B. Hou L. J. Lu G. J. Jiao L. Y. Shen B. S. (2015). An infectious molecular clone in early infection with HIV-1 subtype CRF01_AE strains: construction and biological properties.Mol. Biol. Rep.42329–336. 10.1007/s11033-014-3754-9
464
Ward A. B. Wilson I. A. (2017). The HIV-1 envelope glycoprotein structure: nailing down a moving target.Immunol. Rev.27521–32. 10.1111/imr.12507
465
Weatherley D. A. V. Boswell M. T. Rowland-Jones S. L. (2017). Targeting TRIM5α in HIV cure strategies for the CRISPR-Cas9 Era.Front. Immunol.8:1616.
466
Wei H. Wang C. Croce C. M. Guan J. L. (2014). p62/SQSTM1 synergizes with autophagy for tumor growth in vivo.Genes Dev.281204–1216. 10.1101/gad.237354.113
467
Weidberg H. Shpilka T. Shvets E. Abada A. Shimron F. Elazar Z. (2011a). LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis.Dev. Cell20444–454. 10.1016/j.devcel.2011.02.006
468
Weidberg H. Shvets E. Elazar Z. (2011b). Biogenesis and cargo selectivity of autophagosomes.Annu. Rev. Biochem.80125–156. 10.1146/annurev-biochem-052709-094552
469
Weidberg H. Shvets E. Shpilka T. Shimron F. Shinder V. Elazar Z. (2010). LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis.EMBO J.291792–1802. 10.1038/emboj.2010.74
470
Weissman D. Rabin R. L. Arthos J. Rubbert A. Dybul M. Swofford R. et al (1997). Macrophage-tropic HIV and SIV envelope proteins induce a signal through the CCR5 chemokine receptor.Nature389981–985. 10.1038/40173
471
White E. (2012). Deconvoluting the context-dependent role for autophagy in cancer.Nat. Rev. Cancer12401–410. 10.1038/nrc3262
472
Whittall T. Peters B. Rahman D. Kingsley C. I. Vaughan R. Lehner T. (2011). Immunogenic and tolerogenic signatures in human immunodeficiency virus (HIV)-infected controllers compared with progressors and a conversion strategy of virus control.Clin. Exp. Immunol.166208–217. 10.1111/j.1365-2249.2011.04463.x
473
Wild P. Farhan H. Mcewan D. G. Wagner S. Rogov V. V. Brady N. R. et al (2011). Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth.Science333228–233. 10.1126/science.1205405
474
Wilen C. B. Tilton J. C. Doms R. W. (2012). HIV: cell binding and entry.Cold Spring Harb. Perspect. Med.2:a006866. 10.1101/cshperspect.a006866
475
Willey S. Peters P. J. Sullivan W. M. Dorr P. Perros M. Clapham P. R. (2005). Inhibition of CCR5-mediated infection by diverse R5 and R5X4 HIV and SIV isolates using novel small molecule inhibitors of CCR5: effects of viral diversity, target cell and receptor density.Antiviral Res.6896–108. 10.1016/j.antiviral.2005.07.006
476
Woldemeskel B. A. Kwaa A. K. Blankson J. N. (2020). Viral reservoirs in elite controllers of HIV-1 infection: implications for HIV cure strategies.EBioMedicine62:103118. 10.1016/j.ebiom.2020.103118
477
Wong J. K. Hezareh M. Günthard H. F. Havlir D. V. Ignacio C. C. Spina C. A. et al (1997). Recovery of replication-competent HIV despite prolonged suppression of plasma viremia.Science2781291–1295. 10.1126/science.278.5341.1291
478
Xin Y. Yu L. Chen Z. Zheng L. Fu Q. Jiang J. et al (2001). Cloning, expression patterns, and chromosome localization of three human and two mouse homologues of GABA(A) receptor-associated protein.Genomics74408–413. 10.1006/geno.2001.6555
479
Xu L. Yang H. Gao Y. Chen Z. Xie L. Liu Y. et al (2017). CRISPR/Cas9-mediated CCR5 ablation in human hematopoietic stem/progenitor cells confers HIV-1 resistance in vivo.Mol. Ther.251782–1789. 10.1016/j.ymthe.2017.04.027
480
Yamaguchi H. Nakagawa I. Yamamoto A. Amano A. Noda T. Yoshimori T. (2009). An initial step of GAS-containing autophagosome-like vacuoles formation requires Rab7.PLoS Pathog.5:e1000670. 10.1371/journal.ppat.1000670
481
Yamamoto A. Tagawa Y. Yoshimori T. Moriyama Y. Masaki R. Tashiro Y. (1998). Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells.Cell Struct. Funct.2333–42. 10.1247/csf.23.33
482
Yamamoto H. Kakuta S. Watanabe T. M. Kitamura A. Sekito T. Kondo-Kakuta C. et al (2012). Atg9 vesicles are an important membrane source during early steps of autophagosome formation.J. Cell Biol.198219–233. 10.1083/jcb.201202061
483
Yan J. Kuroyanagi H. Kuroiwa A. Matsuda Y. Tokumitsu H. Tomoda T. et al (1998). Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51.Biochem. Biophys. Res. Commun.246222–227. 10.1006/bbrc.1998.8546
484
Yan J. Kuroyanagi H. Tomemori T. Okazaki N. Asato K. Matsuda Y. et al (1999). Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains.Oncogene185850–5859. 10.1038/sj.onc.1202988
485
Yang A. Rajeshkumar N. V. Wang X. Yabuuchi S. Alexander B. M. Chu G. C. et al (2014). Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations.Cancer Discov.4905–913. 10.1158/2159-8290.cd-14-0362
486
Yang S. Wang X. Contino G. Liesa M. Sahin E. Ying H. et al (2011). Pancreatic cancers require autophagy for tumor growth.Genes Dev.25717–729. 10.1101/gad.2016111
487
Yang Z. Klionsky D. J. (2010). Mammalian autophagy: core molecular machinery and signaling regulation.Curr. Opin. Cell Biol.22124–131. 10.1016/j.ceb.2009.11.014
488
Yoder A. Yu D. Dong L. Iyer S. R. Xu X. Kelly J. et al (2008). HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells.Cell134782–792. 10.1016/j.cell.2008.06.036
489
Yoon Y. S. Cho E. D. Jung Ahn W. Won Lee K. Lee S. J. Lee H. J. (2017). Is trehalose an autophagic inducer? Unraveling the roles of non-reducing disaccharides on autophagic flux and alpha-synuclein aggregation.Cell Death Dis.8:e3091. 10.1038/cddis.2017.501
490
Young A. R. Chan E. Y. Hu X. W. Köchl R. Crawshaw S. G. High S. et al (2006). Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes.J. Cell Sci.1193888–3900. 10.1242/jcs.03172
491
Yue Z. Jin S. Yang C. Levine A. J. Heintz N. (2003). Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor.Proc. Natl. Acad. Sci. U.S.A.10015077–15082. 10.1073/pnas.2436255100
492
Zaunders J. Dyer W. B. Churchill M. (2011). The sydney blood bank cohort: implications for viral fitness as a cause of elite control.Curr. Opin. HIV AIDS6151–156. 10.1097/coh.0b013e3283454d5b
493
Zhang F. Wen Y. Guo X. (2014). CRISPR/Cas9 for genome editing: progress, implications and challenges.Hum. Mol. Genet.23R40–R46.
494
Zhang Y. Wu Y. Wu D. Tashiro S. Onodera S. Ikejima T. (2009a). NF-kappab facilitates oridonin-induced apoptosis and autophagy in HT1080 cells through a p53-mediated pathway.Arch. Biochem. Biophys.48925–33. 10.1016/j.abb.2009.07.017
495
Zhang Y. Yan L. Zhou Z. Yang P. Tian E. Zhang K. et al (2009b). SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans.Cell136308–321. 10.1016/j.cell.2008.12.022
496
Zhang Z. Jiang Y. Zhang M. Liu J. Sun G. Shi W. et al (2010). Alterations of CD4(+) CD25(+) Foxp3(+) regulatory T cells in HIV-infected slow progressors of former blood donors in China.Microbiol. Immunol.54625–633.
497
Zhong Y. Wang Q. J. Li X. Yan Y. Backer J. M. Chait B. T. et al (2009). Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex.Nat. Cell Biol.11468–476. 10.1038/ncb1854
498
Zhou D. Li P. Lin Y. Lott J. M. Hislop A. D. Canaday D. H. et al (2005). Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens.Immunity22571–581. 10.1016/j.immuni.2005.03.009
499
Zhou D. Masliah E. Spector S. A. (2011). Autophagy is increased in postmortem brains of persons with HIV-1-associated encephalitis.J. Infect. Dis.2031647–1657. 10.1093/infdis/jir163
500
Zhou D. Spector S. A. (2008). Human immunodeficiency virus type-1 infection inhibits autophagy.Aids22695–699. 10.1097/qad.0b013e3282f4a836
501
Zhou J. S. Zhao Y. Zhou H. B. Wang Y. Wu Y. F. Li Z. Y. et al (2016). Autophagy plays an essential role in cigarette smoke-induced expression of MUC5AC in airway epithelium.Am. J. Physiol. Lung Cell Mol. Physiol.310L1042–L1052.
502
Zhu T. Mo H. Wang N. Nam D. S. Cao Y. Koup R. A. et al (1993). Genotypic and phenotypic characterization of HIV-1 patients with primary infection.Science2611179–1181. 10.1126/science.8356453
503
Zhu T. Muthui D. Holte S. Nickle D. Feng F. Brodie S. et al (2002). Evidence for human immunodeficiency virus type 1 replication in vivo in CD14(+) monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy.J. Virol.76707–716. 10.1128/jvi.76.2.707-716.2002
504
Zhu W. Lei R. Le Duff Y. Li J. Guo F. Wainberg M. A. et al (2015). The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA.Retrovirology12:22.
505
Zhuo C. Ji Y. Chen Z. Kitazato K. Xiang Y. Zhong M. et al (2013). Proteomics analysis of autophagy-deficient Atg7-/- MEFs reveals a close relationship between F-actin and autophagy.Biochem. Biophys. Res. Commun.437482–488. 10.1016/j.bbrc.2013.06.111
506
Zwart W. Griekspoor A. Kuijl C. Marsman M. van Rheenen J. Janssen H. et al (2005). Spatial separation of HLA-DM/HLA-DR interactions within MIIC and phagosome-induced immune escape.Immunity22221–233. 10.1016/j.immuni.2005.01.006
Summary
Keywords
HIV-1, autophagy, Env signaling, early infection, cell-death
Citation
Cabrera-Rodríguez R, Pérez-Yanes S, Estévez-Herrera J, Márquez-Arce D, Cabrera C, Espert L, Blanco J and Valenzuela-Fernández A (2021) The Interplay of HIV and Autophagy in Early Infection. Front. Microbiol. 12:661446. doi: 10.3389/fmicb.2021.661446
Received
30 January 2021
Accepted
31 March 2021
Published
28 April 2021
Volume
12 - 2021
Edited by
Fatah Kashanchi, George Mason University, United States
Reviewed by
Grant Campbell, University of California, San Diego, United States; Nazira El-Hage, Florida International University, United States
Updates
Copyright
© 2021 Cabrera-Rodríguez, Pérez-Yanes, Estévez-Herrera, Márquez-Arce, Cabrera, Espert, Blanco and Valenzuela-Fernández.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Agustín Valenzuela-Fernández, avalenzu@ull.edu.es
This article was submitted to Virology, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.