
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 13 April 2021
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.659464
This article is part of the Research Topic Flagellar Motors and Force Sensing in Bacteria View all 12 articles
The bacterial flagellar motor (BFM) is a rotary molecular motor embedded in the cell membrane of numerous bacteria. It turns a flagellum which acts as a propeller, enabling bacterial motility and chemotaxis. The BFM is rotated by stator units, inner membrane protein complexes that stochastically associate to and dissociate from individual motors at a rate which depends on the mechanical and electrochemical environment. Stator units consume the ion motive force (IMF), the electrochemical gradient across the inner membrane that results from cellular respiration, converting the electrochemical energy of translocated ions into mechanical energy, imparted to the rotor. Here, we review some of the main results that form the base of our current understanding of the relationship between the IMF and the functioning of the flagellar motor. We examine a series of studies that establish a linear proportionality between IMF and motor speed, and we discuss more recent evidence that the stator units sense the IMF, altering their rates of dynamic assembly. This, in turn, raises the question of to what degree the classical dependence of motor speed on IMF is due to stator dynamics vs. the rate of ion flow through the stators. Finally, while long assumed to be static and homogeneous, there is mounting evidence that the IMF is dynamic, and that its fluctuations control important phenomena such as cell-to-cell signaling and mechanotransduction. Within the growing toolbox of single cell bacterial electrophysiology, one of the best tools to probe IMF fluctuations may, ironically, be the motor that consumes it. Perfecting our incomplete understanding of how the BFM employs the energy of ion flow will help decipher the dynamical behavior of the bacterial IMF.
Several bacteria propel themselves by rotating their flagella (Berg and Anderson, 1973). The bacterial flagellar motor (BFM) is the powerful molecular nanomachine at the base of each flagellum responsible for such rotation. One of the few known examples of biological rotatory machines, the BFM is unique in its remarkable power and efficiency in converting free energy into mechanical work. Flagellar rotation is driven by the ion-motive force (IMF), the electro-chemical potential difference built across the membrane during cellular respiration. The demonstration that the motor is driven not by the energy of ATP hydrolysis but by the flux of ions across the membrane (Larsen et al., 1974; Manson et al., 1977) came a few years after the realization that flagella rotate (Berg and Anderson, 1973). Given an energy source that is a charged quantized particle, moving along an electric field, both the experimental and theoretical treatments of the BFM energization are challenging. For example, under physiological conditions, the energy provided by the translocation of a single ion is about three times smaller than that released by ATP hydrolysis. As a consequence, important quantities like the elementary step size and mechanisms like the force generation are difficult to resolve experimentally and model theoretically. Ions move according to both electrical and concentration gradients. The IMF is defined as the sum of these two contributions by
where ΔΨ = Ψint−Ψext is the difference in membrane potential Ψ, built up by the ensemble of charges separated by the membrane, is the difference in the specific ion (I) potential, [I] is the ion concentration, R is the gas constant, F is the Faraday constant, and T is the temperature (2.3RT/F ≃ 60 mV at T = 300K). The IMF is maintained out of equilibrium in respiring cells by transmembrane (TM) complexes that actively pump ions outward across the inner membrane. External ions can diffuse and finally fall along the energy potential to be consumed by the BFM and by other TM complexes such as the ATP synthase.
Despite differences in ion selectivity and a rich diversity of evolved structural details (Rossmann and Beeby, 2018), the core structure of the BFM (see Figure 1) is well conserved. In Escherichia coli, the rotor is about 45 nm in diameter and composed of two main rings, the inner membrane embedded MS-ring and the cytoplasmic C-ring. The rotor couples rotation to the flagellum via the rod and then the extracellular hook. The stator consists of multiple units anchored to the peptidoglycan (PG) at the rotor's periphery. Each stator unit acts as an ion channel through which ions translocate and transfer their energy to generate the force, which rotates the rotor. Ion consumption of the stator unit is specific: MotAB stator units (e.g., in E. coli or Salmonella enterica) consume the cellular proton motive force (PMF), whereas PomAB stators (e.g., in Vibrio alginolyticus) consume the sodium ion motive force (SMF) Sowa and Berry (2008). Importantly, at least in enteric bacteria such as E. coli and S. enterica, stator units are not always bound to the motor complex, but they are observed to dynamically exchange between an inactive unbound state, diffusing in the inner membrane, and a motor-bound active state (Leake et al., 2006). Moreover, such exchange is mechanosensitive, as the motor can adapt the number of bound stator units depending on the external viscous load, incorporating up to a dozen (Lele et al., 2013; Tipping M. J. et al., 2013; Chawla et al., 2017; Nord et al., 2017; Terahara et al., 2017). Several studies suggest that stators may also be capable of sensing IMF, and that stator assembly is dependent upon the driving ion (Fukuoka et al., 2009; Tipping J. M. et al., 2013).
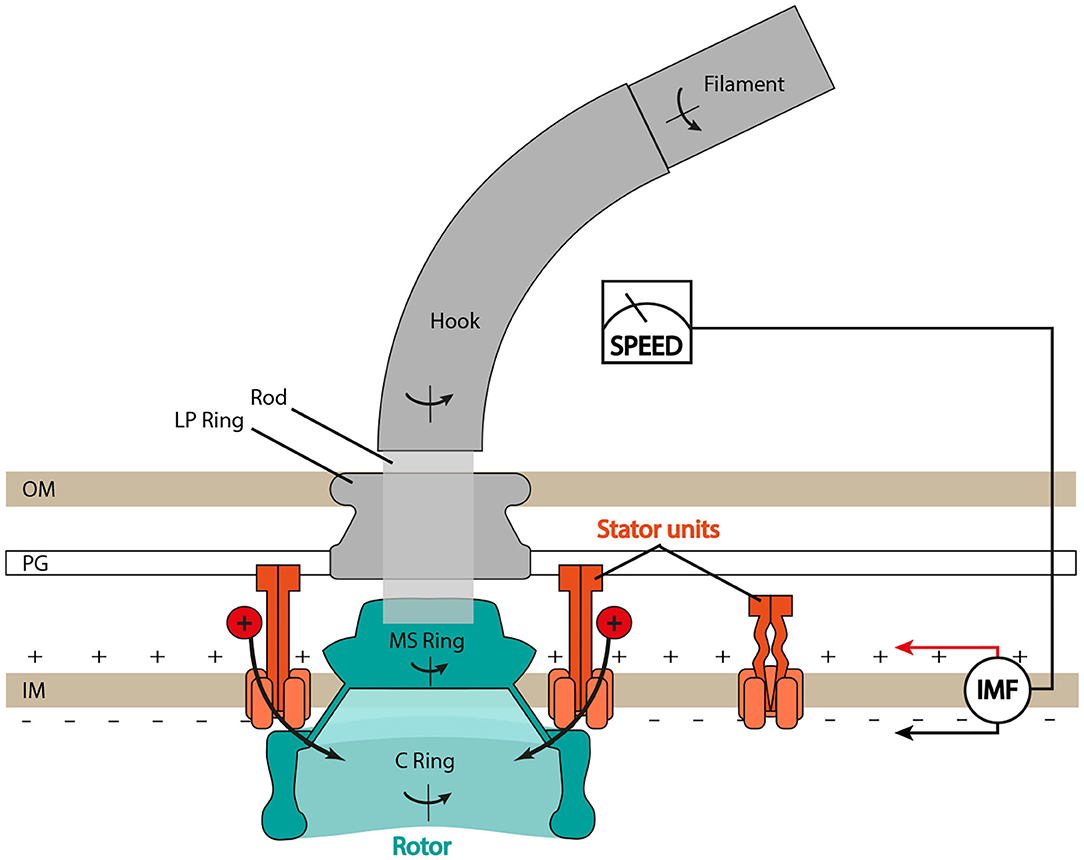
Figure 1. Schematic of the bacterial flagellar motor (BFM) in Gram-negative bacteria. The flagellar filament is connected to the rotor (in blue) through the hook, the universal joint, and the rod, the central driveshaft that connects the hook to the MS ring. The LP ring is assembled around the rod and acts as a bushing. Stator units bind to the peptidoglycan (PG) layer, translocate ions from the periplasm to the cytoplasm, and generate torque by interacting with the C ring. IM, inner membrane; OM, outer membrane. Type III export apparatus not shown.
In this review, we present and discuss some of the major results that shed light on the dynamic bacterial IMF, in particular with respect to the structure and activity of the flagellar motor and its stator units.
Understanding the relations that couple IMF, ΔΨ, ΔpI, and motor activity (measured via the swimming speed of cells or the angular speed and torque of individual motors) is of great interest, particularly as it provides accurate constraints for physical mechanistic models of the motor. Unfortunately, drawing a conclusive general scheme on how the motor activity depends on the IMF components is complicated by the dependence of membrane and ion potentials on intrinsic cellular homeostasis, large cell-to-cell variability, and differences among strains. Measuring the activity of single motors under controlled conditions helps to obtain the full distribution of behaviors.
A natural parameter to vary when studying the motor behavior as a function of IMF is the concentration of the coupling ion in the external medium, affecting pIext, as illustrated in Figure 2 for three published works. For proton-consuming motors (e.g., E. coli, B. subtilis, Streptococcus), changing pHext affects not only motor activity, but also several other cellular mechanisms. This results in a coupling between the components of the PMF. In B. subtilis (Figure 2A) (Shioi et al., 1980) and E. coli (Figure 2B) (Minamino et al., 2003), an increase in pHext is accompanied by an increase in pHint (linear in E. coli, with a saturation in B. subtilis), with pHint > pHext up to pHext ~ 7.5. As ΔpH decreases, ΔΨ increases in a compensatory manner, resulting in a PMF that does not change dramatically (within ~ 50 mV) in the range of pHext = 5−8. The coupling between pIext and ΔΨ has been cleverly avoided by expressing chimeric Na+-driven stators in E. coli (Lo et al., 2007) (Figure 2C). In this case, varying pNaext has very little effect on ΔΨ. Therefore, the SMF follows ΔpNa, decreasing with increasing pNaext, given that pNaint increases less rapidly than pNaext. Moreover, pHext can also be used to control the SMF, as it linearly affects ΔΨ but is almost entirely decoupled from ΔpNa.
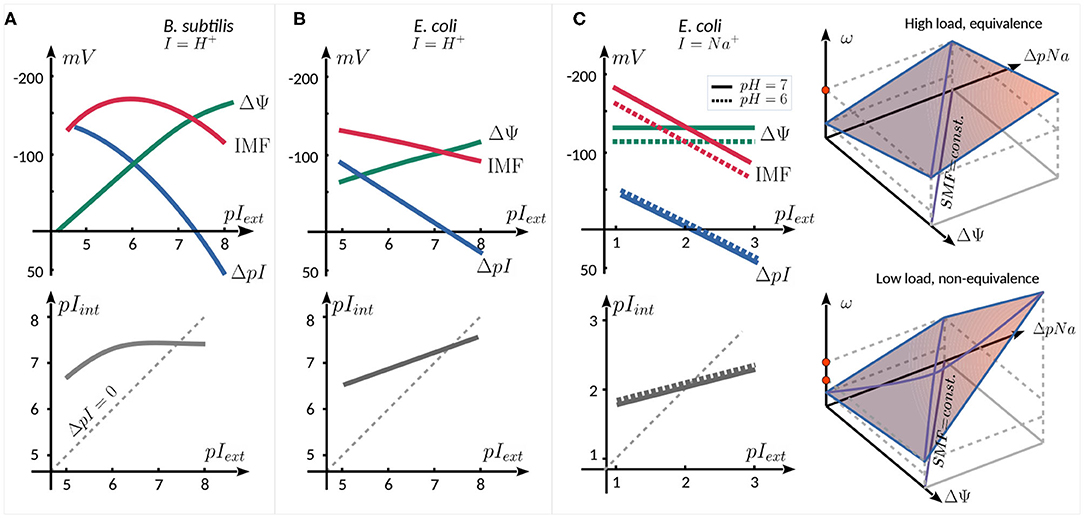
Figure 2. Schematic relation between ΔΨ, ΔpI, IMF (top panels), and pIint (bottom panels) as a function of external ion concentration. The graphs are reproduced from the data published in (A) (Shioi et al., 1980), for the proton consuming motor of B. subtilis, (B) Minamino et al. (2003), for the proton consuming motor of E. coli, and (C) Lo et al. (2007) for the sodium consuming chimeric motor of E. coli employing chimeric Na+ stators. The two rightmost panels in (C) show schematically the equivalence of the SMF components at high load (top), and their nonequivalence (bottom panel, see text).
In the Na+-driven chimeric motor of E. coli, at high load the speed of the motor is linearly proportional to SMF, with an equivalent dependence upon ΔΨ (controlled by pHext) and ΔpNa (controlled by pNaext). At lower load (0.35 μm beads), the equivalence between the two components breaks down (Figure 2C, right panels). For instance, if ΔΨ is decreased and ΔpNa is increased so as to keep the SMF constant, one observes an increase in motor speed. The non-equivalence is graphically shown in the space (Δψ, ΔpNa, ω) in Figure 2C: : while at high load, the same speed (red dot) can be reached by increasing either Δψ or ΔpNa, at low load, this is no longer true (red dots), as the slope of ω, seen as a function of one component, depends on the other component. The leading hypothesis to explain this non-equivalence is that the rate-limiting step at low load and low [Na+]ext conditions is the diffusion-limited binding of Na+ ions to the stator units (Lo et al., 2007).
Despite some descriptions of nonlinearities (threshold, saturation, and the non-equivalence of the IMF components; Khan and Macnab, 1980; Shioi et al., 1980; Lo et al., 2007), reflecting the complexity of the measurements and of their interpretation, numerous results contribute to a general consensus that there exists a linear relationship between the speed of the motor and IMF. Table 1 summarizes some of the published works that have described this proportionality and the range of IMF for which it holds, covering the physiological range. In Lo et al. (2007), the authors investigated motor speed per stator unit in Na+-powered motors, finding that the linear relationship between speed and IMF holds true at the level of an individual stator unit. However, the proportionality breaks down with acidification of the cytoplasm, at least in H+-powered motors. In the presence of a weak acid (which dissociates upon crossing the cytoplasmic membrane, lowering pHint and partially collapsing ΔpH), a decrease of pHext from 7 to 5 leads to a total decrease in PMF of only ~ 10 mV, yet causes a strong decline in motor speed. From these results, it was hypothesized that high internal proton concentration hinders proton unbinding from the stators in the cytosol, hampering their function (Minamino et al., 2003). Therefore, the motor seem to respond to the entire IMF, unless pHint is lowered substantially (Gabel and Berg, 2003).
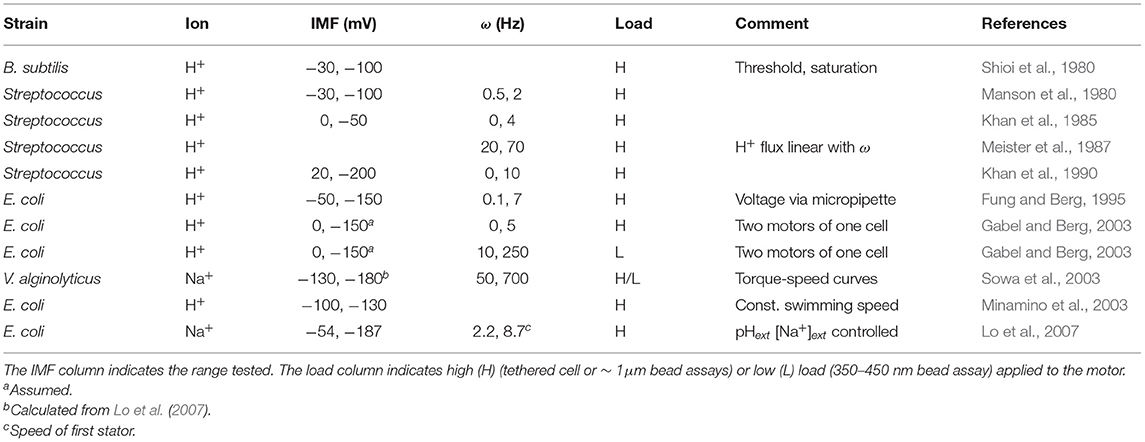
Table 1. Published linearity ranges between motor speed ω and ion motive force (IMF) in different strains.
When considering the linearity between IMF and speed, one should also keep in mind the associated time scale. Because of technical limitations, mainly in the dynamical measurement of IMF, the linear relationship has been experimentally demonstrated only at low frequency, i.e., in ensemble averages of ω and IMF acquired over relatively long periods of time. While it is now possible to measure ω with ~ 10 ms resolution in bead assays, the IMF is often measured on a scale of several seconds to minutes, and often averaged over the entire cell population. However, at least two assays have managed to directly and dynamically manipulate the IMF. By applying a varying external voltage directly to a trapped and permeabilized cell within a micropipette, the linearity between ω and IMF was demonstrated with a resolution of a few seconds (Fung and Berg, 1995). By expressing and exciting the proton pump proteorhodopsin to trigger proton efflux (Walter et al., 2007; Tipping J. M. et al., 2013), the IMF could be dynamically perturbed (though not measured) while measuring ω with sub-second resolution. Below, we discuss recent techniques for dynamic IMF quantification. When observing motor speed and IMF at higher frequency (i.e., averaging on sub-second time intervals), the analysis is complicated by fluctuations, and resolving the relationship between fast IMF fluctuations and fast speed variations remains challenging. Finally, as discussed further below, dynamic stator assembly and exchange must also be considered (or controlled) when studying the relation between IMF and speed.
The observation that the ion flow through the motor is directly proportional to speed, together with a linear relation between IMF and speed, contributed to a widely used model wherein ion translocation is tightly coupled to rotation, i.e., a fixed number of ions yields a fixed angular displacement (Manson et al., 1980; Meister et al., 1987). The IMF-speed linearity is a necessary condition for the tight-coupling model, and it has been validated by several measurements, as described above. However, it is not a sufficient condition. Given its importance for BFM mechanistic modeling, as well as other microbiological mechanisms (e.g., mechanosensing and biofilm initiation; Belas, 2013), the tight-coupling hypothesis warrants continued scrutiny, ideally at the single motor level.
Contrary to the long-held assumption that, once assembled, the structure of the BFM was static, it is now well established that multiple motor components dynamically exchange. The most well-studied example of this are the stator units, which exchange between an active motor-bound form and an inactive membrane diffusing form on a timescale of seconds to minutes (Leake et al., 2006). The rate of exchange and the steady-state number of stators is dependent upon the viscous load on the motor (Lele et al., 2013; Tipping M. J. et al., 2013; Chawla et al., 2017; Nord et al., 2017), a property that allows the bacterium to adapt to changes in local viscosity. However, there are multiple clues that suggest that stator exchange and assembly is sensitive not only to the local mechanical environment, but also to the local electrochemical environment. Such a capacity would allow strains that couple multiple ions via separate stators to tune their motor composition to the prevailing conditions (Thormann and Paulick, 2010).
One of the first experiments to suggest that stator assembly was sensitive to IMF was Fung and Berg's micropipette experiment (Fung and Berg, 1995). Once a filamentous cell of E. coli was installed in the micropipette, they dissipated the PMF via an ionophore, which halted motor rotation, as expected. But, when the membrane voltage was reinstated, they observed a delay before motors began rotating, after which speed increased in step-wise increments. This suggested that stator units inactivated, in some way, upon PMF collapse, then individually reactivated or reassembled upon reinstatement of the PMF. In an attempt to hold the number of stator units constant while assessing the linearity between speed and IMF, the authors avoided total PMF collapse by maintaining a nonzero DC voltage. However, IMF-dependent stator association was less evident in later experiments of the activity of two motors on the same E. coli cell during slow collapse of the IMF. The two motors reached zero speed at the same time, potentially suggesting that the stator units did not dissociate or inactivate at vanishing IMF (Gabel and Berg, 2003), though the low temporal resolution (~ 5 s) leaves room for uncertainty.
Tipping J. M. et al. (2013) used fluorescent protein (FP)-tagged stators to show that, upon collapse of the PMF in E. coli, stators dissociate from the motor on a timescale of minutes. However, these results are in disagreement with two separate studies that, also using FP-tagged stators, observe that the PMF is not necessary for stator assembly in either S. enterica or E. coli (Morimoto et al., 2010; Suzuki et al., 2019). While it seems well established that the ΔpH component of the PMF is not necessary for assembly in E. coli or S. enterica (Fung and Berg, 1995; Morimoto et al., 2010; Suzuki et al., 2019), recent work has observed that assembly increases at lower external pH (Suzuki et al., 2019). Thus, the concentration of accessible protons seems to be more important than the gradient across the membrane. Yet, surprisingly, mutation of a conserved protonatable residue in MotB, which renders the stator nonfunctional, has shown that proton translocation is not necessary for stator assembly in E. coli or S. enterica (Zhou et al., 1998; Kojima and Blair, 2001; Morimoto et al., 2010; Suzuki et al., 2019).
For the Na+ stator, SMF is required for assembly in V. alginolyticus (Fukuoka et al., 2009), and contrary to the H+ stator, Na+ conduction is also required for assembly in V. alginolyticus and B. subtilis (Fukuoka et al., 2009; Terahara et al., 2017). And while the concentration of external Na+ is crucial for stator assembly in V. alginolyticus, B. subtilis, and the Na+-driven hybrid stator in E. coli (Fukuoka et al., 2009; Sowa et al., 2014; Terahara et al., 2017), it is not necessary for Na+-stator assembly in S. oneidensis (Paulick et al., 2015).
We summarize the studies to date which probe the ion-related conditions necessary for stator assembly in Table 2. There is not yet a coherent universal picture of stator ion sensing; as with other behaviors, it may prove ion and species dependent, though the external concentration of coupling ion seems generally important. It remains to be shown whether this is due to an ion sensing capability of the stator. Alternatively, if stator binding to the PG is stabilized by force (Chawla et al., 2017; Nord et al., 2017), it may be simply the case that increasing ion concentration and rate of ion conduction increases, on average, the stability of the stator-PG bond. The fact that ion conduction in not required for H+ stator assembly argues against this explanation in the case of the H+-stator.
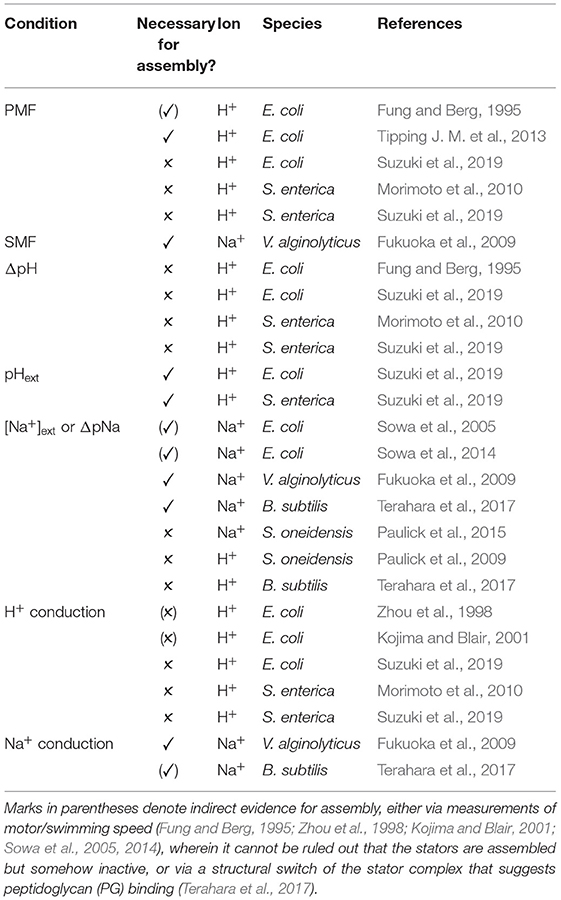
Table 2. Ion motive force (IMF) and ion related conditions shown to be necessary, ✓, or not, ✗, for stator assembly.
Studies that have found both motor speed and single stator speed to be linearly dependent upon IMF seem to make sense only under the assumption that the number of assembled stators is constant. Thus, the fact that stator assembly depends, in some yet-to-be-determined manner, on IMF complicates the interpretation of the linear dependence of speed on IMF. There exists an unresolved contradiction, as a linear relationship between IMF and speed should not be observed if both stator number and speed per stator increase simultaneously. It may prove that the relationship between stator number and IMF is sub-linear, and that stator disassembly mostly occurs only at very low IMF values. Future studies of stator dynamics as a function of IMF will be key to understanding how these pieces fit together.
In most bacteria, stators are powered by a single species of cation, mostly commonly protons or sodium ions. Typically, MotAB-type stators are H+-coupled and PomAB and MotPS-type stators are Na+-coupled. Yet some bacteria are capable of coupling multiple types of cations. One way in which this is accomplished is via multiple sets of genes that encode multiple types of stator complexes. For example, B. subtilis, V. alginolyticus, and S. oneidensis all encode both an H+-driven MotAB stator and an Na+-driven MotPS or PomAB stator (Atsumi et al., 1992; Asai et al., 2000; Ito et al., 2005; Paulick et al., 2009). In V. alginolyticus, PomAB stators are constitutively expressed and power the polar flagellar motor, whereas MotAB stators are expressed only under certain environmental conditions and exclusively power lateral flagella motors (Belas et al., 1986; McCarter et al., 1988; Atsumi et al., 1992; Kawagishi et al., 1995). In B. subtilis and S. oneidensis, both types of stators are constitutive. They incorporate into the same motor, working in unison, though their affinity to the motor depends upon the chemical and physical conditions of the environment (Ito et al., 2005; Paulick et al., 2009). A systematic survey of bacterial genomic data has shown that at least 65 species of bacteria possess two or more putative stators (Thormann and Paulick, 2010). Other species couple multiple cations via a single stator complex. For example, the MotAB stator of Bacillus clausi can couple either H+ or Na+ (Terahara et al., 2008), and MotPS in Bacillus alcalophilus couples Na+, K+, or Rb+ (Terahara et al., 2012). While it was reported that MotAB in Paenibacilus sp. was unique in its ability to couple divalent cations Mg2+ and Ca2+ (Imazawa et al., 2016), recent results suggest that this stator is actually powered by monovalent ions, likely protons (Onoe et al., 2020). Regardless of the coupling ion, a universally conserved aspartate residue at the N-terminal side of the TM region of MotB (D32 in E. coli) is believed to serve as the site of ion binding (Zhou et al., 1998).
Many hybrid stators have been created in order to probe the question of what in the stator determines ion specificity. Early work demonstrated that the combination of MotA (R. Sphaeroidis) and PomB (V. alginolyticus) produced a Na+-driven stator in V. alginolyticus. This hybrid required MotX and MotY proteins in order to function, components which are necessary for PomAB incorporation in V. alginolyticus (Terashima et al., 2013). This work ruled out the A subunit as the ion-decisive component (Asai et al., 1999). In B. subtilis, hybrid stators MotAS and MotPB were shown to be Na+ and H+-coupled, respectively, suggesting that the B/S subunit is the dominant determinant of ion-selectivity (Ito et al., 2005). However, it was observed that MotA and MotP subunits confer an H+ or Na+ responsiveness, respectively (Ito et al., 2005). This suggests that either the A/P subunit plays a secondary role in ion-specificity, or, given the evidence presented above for stator sensing of IMF, it may be the case that the A/P subunit is a determining factor of H+ and Na+-dependent stator assembly. It was also recently proposed that the P subunit is critical for K+ ion selectivity in B. alcalophilus and Bacillus trypoxylicola (Naganawa and Ito, 2020).
To explore which portion of the B subunit determines ion specificity, a chimeric subunit, termed MomB, was constructed from the N-terminus portion of MotB (R. Sphaeroidis) and the C-terminus portion of PomB (V. alginolyticus). MomB and PomA produced an Na+ stator in V. alginolyticus, and interestingly, the position of the MotB/PomB junction within the conserved TM region affected the Na+/Li+ specificity. It was thus hypothesized that the periplasmic region of PomB proximal to the inner membrane plays a role in ion specificity, potentially by changing the size of the TM pore (Asai et al., 2000). Certain MomB constructs also produced a Na+-driven stator with MotA in V. alginolyticus, albeit its function was dependent upon the presence of MotX and MotY (Asai et al., 2000, 2003). The reverse construction, the N terminus of PomB combined with the C terminus of MotB, named PotB, functioned with PomA as a Na+-powered stator in V. alginolyticus or E. coli, regardless of presence of MotXY (Asai et al., 2003). Thus, while the periplasmic portion of PomB is not necessary for Na+ coupling in the PomAB stator, it is sufficient to convert the MotAB stator from H+- to Na+-coupling.
Mutations near the surface of the conserved TM segment of the B subunit can cause the dual-ion coupling stator of B. clausi to prefer either H+ or Na+, and mutations in the same region can confer a dual-ion coupling capacity to B. subtilis stators (Terahara et al., 2008). A single mutation in the TM region of MotS of B. alcalophilus changed it from multi-ion coupling to single ion coupling (Terahara et al., 2012). Sequence alignment revealed that H+-coupled MotB stators from many bacteria contain a conserved valine in the middle of the TM segment (residue 43 in E. coli), whereas Na+-coupled PomB and MotS stators contain a conserved leucine at this position, and the Na+/K+/RB+-coupled MotS of B. alcalophilus contains a methionine (Terahara et al., 2012). Steered molecular dynamic simulations of an atomic model of MotAB constructed based on disulfide cross-linking and tryptophan scanning mutations showed that the size of the ion channel at its narrowest point was sensitive to mutations at this TM residue, suggesting size exclusion as the mechanism for ion-selectivity (Nishihara and Kitao, 2015).
Recently, the structure of the stator has been determined via cryoelectron microscopy (Deme et al., 2020; Santiveri et al., 2020). In Figure 3A, we show portions of a multiple sequence alignment, initially performed with the Clustal Omega alignment program (Madeira et al., 2019), then refined and edited using ViTO (Catherinot and Labesse, 2004), taking into account the published structures (PDB: 6YKP, 3S0Y). These alignments highlight the differentially conserved residues within the transmembrane helix (TMH) of MotB, but they also identify multiple differentially conserved residues in TMH3 and TMH4 of Mot/PomA that map to the A/B interface, which is very compact and largely hydrophobic in its most central part (Deme et al., 2020; Santiveri et al., 2020). The differentially conserved residues generally localize to three rings, as shown in Figure 3B, with no tight connections. Near the periplasmic interface, residue EcMotBV43 faces residues EcMotAI202 and EcMotAA187 within TMH4 and TMH3, respectively (Figure 3C). While this ring is fully buried and corresponds to hydrophobic substitutions, differences between the H+ and Na+ stators here may yield an overall change of size or flexibility of the region, as previously hypothesized (Nishihara and Kitao, 2015). Further into the channel, residues MotBY30 and MotBF33 face residues 178–181 of MotA TMH3, and with more accessibility, this presents another region that may control ion specificity (Figure 3D). Substitutions at that ring will impact a network of interactions involving hydrogen bonds to the essential aspartate MotBD32. Indeed, an imperfect correlation between MotB residue 30 and ion specificity was previously noted (Ishida et al., 2019; Islam et al., 2020), and selection may prove to depend partly upon the facing TMH3 region. Finally, residues 217–219 of MotA TMH4 face the conserved MotBW26 residue (Figure 3), which forms an H-bond with MotAY217 in H+ motors. In sodium MotP/PomA stators, a smaller and more flexible asparagine is observed, instead. As a MotBW26A mutation abolishes motility (Deme et al., 2020), this contact is important, though we see no obvious mechanism of ion selection in this open region.
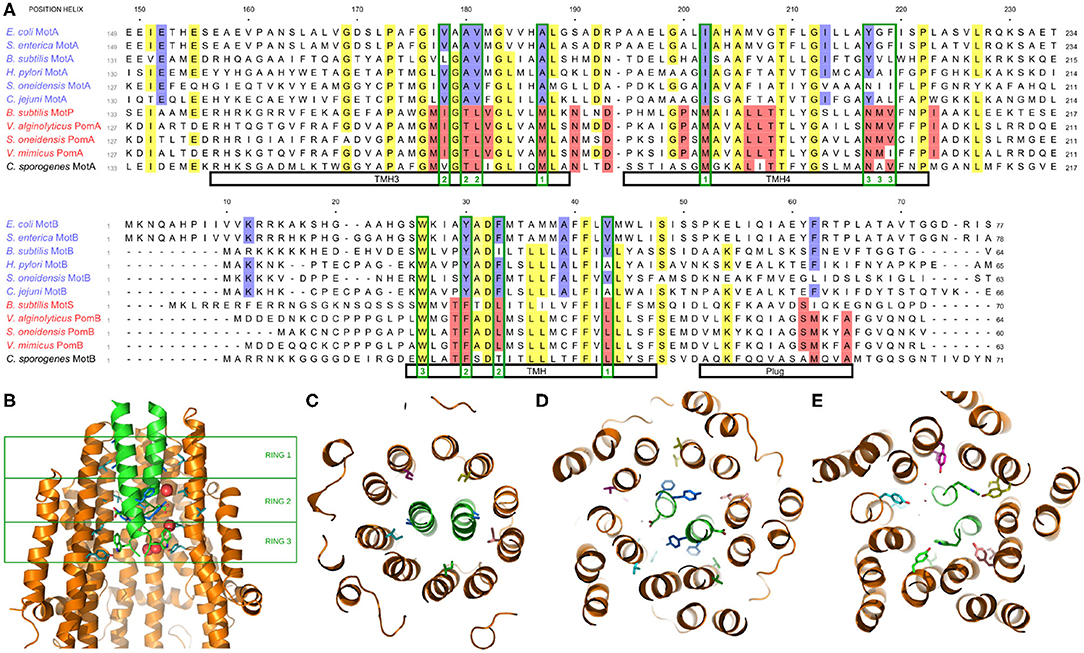
Figure 3. (A) Multiple species sequence alignment of the A and B stator subunits. Highlighting denotes >70% conservation across either all species (yellow) or differentially across only the H+ or Na+ stators (blue and red, respectively; the coupling ion of C. sporogenes remains unclear). Residues mentioned in the text which map to one of the three rings of differentially conserved residues are outlined in green with the corresponding ring noted at the bottom. (B) Side view of the stator, showing the two MotB and three of the five MotA (side chains shown for MotB and two MotA units). The three rings of differentially conserved residues are marked by the green boxes. (C) Bottom view (looking from the cytoplasm) of Ring 1. (D) Bottom view of Ring 2. (E) Bottom view of Ring 3.
The electrochemical gradient across the bacterial cell membrane is central not only for motility, but for other physiological processes from ATP synthesis (Mitchell, 1961; Maloney et al., 1974) to cell division (Strahl and Hamoen, 2010; Chimerel et al., 2012). For this reason, it was traditionally considered to be homeostatic. However, recent work has shown that the IMF can be dynamic, is integrally tied to the cell's physiological state, and plays a role in information signaling. IMF-mediated intra- and intercellular electrical signalings have recently been shown to regulate physiological processes such as mechanosensation and metabolic coordination, sometimes over long distances, either among or between communities (Liu et al., 2015; Bruni et al., 2017). We discuss here a few examples where spatiotemporal variation of the IMF has been observed, both at the single cell and population level.
As a pioneering work in the burgeoning field of bacterial electrophysiology, Kralj et al. (2011) developed a voltage-sensitive fluorescent membrane protein (PROPS) to probe voltage dynamics on the single cell level. They observed that, while cells show great heterogeneity in behavior, many cells displayed quasi-periodic fluctuations in their membrane voltage. Predictably, these transient depolarizations correlated to transient decreases in motor speed. The physiological role of electrical spiking was unclear at the time; as the intensity and frequency were affected by the power of the imaging laser, the authors hypothesized that such fluctuations may be a signature of a bacterial stress response. When PROPS was combined with a genetically encoded calcium sensor, it was observed that transient membrane depolarizations induced transient influxes of calcium, reminiscent of neuronal action potentials (Bruni et al., 2017). Evidence that such a phenomenon is a signature of signaling came from the observation that transient voltage-induced Ca+ influxes increased upon mechanical stimulation, and that such stimulation led to changes in protein concentrations at the level of individual cells (Bruni et al., 2017).
IMF dynamics have recently been shown to be important for surface-attached communities of bacteria, called biofilms. At the population level, a multi-electrode array system has shown that the intensity of electrical activity correlates with biofilm formation (Masi et al., 2015). With single cell level resolution, it has been shown that biofilms produce spatially propagating waves of oscillating membrane potential, and that these waves correlate with propagating waves of extracellular potassium. There is substantial evidence to support a model in which membrane depolarization is triggered by metabolic stress at the interior of a biofilm, which triggers the release of intracellular K+, which subsequently depolarizes neighboring cells. Thus, membrane potential is used to conduct long-range electrical signaling to coordinate the metabolic states within a growing biofilm, thereby increasing the fitness of the biofilm as a whole (Prindle et al., 2015). Signals transmitted by these propagating waves of potassium extend beyond the biofilm community. Spatially separated biofilms can capitalize upon this electrochemical signaling to coordinate a time-sharing behavior of resource consumption (Liu et al., 2017), and the swimming behavior of distant planktonic cells can be modified in a manner to recruit cells to join a biofilm. While the majority of this work has been done on B. subtilis, the mechanisms may well be generic among bacteria; electrical signaling from a B. subtilis biofilm attracts swimming cells from the evolutionary distant P. aeruginosa (Humphries et al., 2017).
There appears to be great cell-to-cell heterogeneity in both the steady-state and dynamic behavior of cellular IMF (Kralj et al., 2011). Recent evidence has shown that both the steady-state membrane voltage and the dynamic behavior in response to electrical stimulation is dependent upon a cell's proliferative capacity (Stratford et al., 2019). Additionally, electrical polarization of the cell plays a role in successful spore formation (Sirec et al., 2019). While it may seem that such cell-to-cell heterogeneity may interfere with long-distance electrical signaling, a model based upon percolation theory proposes that the number of cells capable of transmitting the electrical signal is consistent with a predicted phase transition. Thus, a dense biofilm composed of a heterogeneous mixture of signaling and non-signaling cells manages to propagate signals over long distances while minimizing the cost at the single-cell level (Larkin et al., 2018).
While the developing field of bacterial electrophysiology takes its inspiration from the advanced field of eukaryotic electrophysiology, the reduced bacterial cell size and the complexity of the membrane present novel challenges. Techniques employed in the study of eukaryotic cells, such as microelectrode and patch clamp approaches, so far remain impractical, lack single-cell resolution (Masi et al., 2015), or come at the expense of great physical perturbation (Martinac et al., 2008). While bacterial bioenergetics has historically been investigated via bulk population measurements (Kashket, 1985), emerging techniques are enabling measurements of both the electric and chemical components of the IMF at the single bacterial cell level.
Measurements of ΔΨ are most commonly made with Nernstian sensors (Lo et al., 2006, 2007; Kralj et al., 2011; Prindle et al., 2015; Sirec et al., 2019; Stratford et al., 2019; Mancini et al., 2020), small fluorescent molecules that diffuse across the membrane in accordance with the Nernst equation. Fluorescent measurements of the ratio of the extracellular to intracellular concentration at equilibrium reports upon ΔΨ. However, their permeability to Gram-negative bacteria is low, yielding a slow dynamic response (~ minutes), and so they are not suitable to report upon rapid fluctuations in ΔΨ. Additionally, while high concentrations are desired for improved signal-to-noise ratio, increasing concentrations of Nernstian dyes actually perturb membrane voltage (Sirec et al., 2019; Mancini et al., 2020). Thus, their application requires careful calibration (Mancini et al., 2020). As mentioned above, an alternative to probe ΔΨ is a genetically encoded voltage sensor called PROPS, engineered from green-absorbing proteorhodopsin (Kralj et al., 2011; Bruni et al., 2017). PROPS provides a significantly faster temporal response (~ 10 ms; Kralj et al., 2011), without the need for a dye-loading step, though at the cost of lower signal-to-noise ratio.
Measurements of the chemical component of the IMF require a reporter for the intracellular concentration of the coupling ion. Intracellular pH was first quantified on the single cell level using membrane-permeable pH-dependent fluorescent dyes (Siegumfeldt et al., 2000; Rasmussen et al., 2008; Kurre et al., 2013; Chao et al., 2017). A more recent approach, which obviates the need for a dye loading step and its potential invasive effects (Han and Burgess, 2010), uses genetically encoded fluorescent proteins, such as pH-sensitive derivatives of GFP (Miesenböck et al., 1998; Martinez et al., 2012; Kurre et al., 2013; Morimoto et al., 2016; Rupprecht et al., 2017; Arce-Rodríguez et al., 2019). Aside from excellent temporal (<1 s) and pH (<0.1) resolution (Kralj et al., 2011), a fusion between pHluorin and FliG of the BFM has shown local pH measurements with sub-cellular spatial resolution (Morimoto et al., 2016). PHluorin can also be combined with PROPS for simultaneous single cell measurements of ΔpH and membrane potential (Kralj et al., 2011; Bruni et al., 2017). Quantitative single cell measurements of intracellular sodium have been demonstrated using membrane-permeable fluorescent Na+ dyes and applied to studies of the hybrid Na+-driven motor of E. coli (Lo et al., 2006, 2007; Minamino et al., 2016). Relative measurements of cytoplasmic Ca2+ have been demonstrated in E. coli via a genetically encoded fluorescent Ca2+ sensor (Bruni et al., 2017).
The fact that the speed of the BFM is proportional to IMF provides an intriguing opportunity to exploit it as a measuring device for the IMF of the coupling ion. Recently, it has been shown that the BFM of E. coli can measure dynamic changes in the PMF during transient exposures to an ionophore as well as dynamic light-induced photodamage to the cell membrane (Krasnopeeva et al., 2019). As the temporal resolution of such measurements are theoretically limited only by the acquisition rate and the relaxation time of the particle attached to the flagellar filament (~ ms), the BFM presents as an exciting future tool to quantitatively explore rapid electrophysiological dynamics on the single cell level. However, as highlighted above, such measurements will be affected by stochastic stator dynamics, as well as other factors, such as second messenger-mediated binding of cytoplasmic protein YcgR to the rotor, which slows motor speed in E. coli (Boehm et al., 2010; Fang and Gomelsky, 2010; Paul et al., 2010).
In species investigated thus far, the speed of the BFM is linear with IMF. It remains to be fully elucidated when and to what extent stator assembly depends on IMF, as well as the underlying mechanism of this electrical or chemical sensing. Future work will also need to decode how the linearity between speed and IMF arises if both single stator speed and stator assembly are proportional to IMF. Such details will contribute to models of the still elusive mechanism of torque generation, which will be greatly aided by recently solved structures of the stator complex (Deme et al., 2020; Santiveri et al., 2020). These structures will also open the way for future simulations to shed light on ion specificity and ion-dependent assembly.
With recent advances in single-cell electrophysiology techniques, the field is poised to learn a great deal about IMF dynamics and its intrinsic relation to diverse physiological processes, on scales from a single motor complex up to an entire bacterial colony. One of the best tools to investigate IMF dynamics may, ironically, be the motor that consumes it; yet, our understanding of the IMF and its rate of consumption by the BFM remains incomplete.
AB-B, FP, GL, and AN researched and wrote this review. All authors contributed to the article and approved the submitted version.
This work was supported by the ANR FlagMotor project grant ANR-18-CE30-0008 of the French Agence Nationale de la Recherche and by the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INBS-0005. The CBS is a member of the France-BioImaging (FBI) and the French Infrastructure for Integrated Structural Biology (FRISBI), 2 national infrastructures supported by the French National Research Agency (ANR-10-INBS-04-01 and ANR-10-INBS-05, respectively).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Arce-Rodríguez, A., Volke, D. C., Bense, S., Häussler, S., and Nikel, P. I. (2019). Non-invasive, ratiometric determination of intracellular pH in Pseudomonas species using a novel genetically encoded indicator. Microb. Biotechnol. 12, 799–813. doi: 10.1111/1751-7915.13439
Asai, Y., Kawagishi, I., Sockett, R. E., and Homma, M. (1999). Hybrid motor with H+ and Na+ driven components can rotate Vibrio polar flagella by using sodium ions. J. Bacteriol. 181, 6332–6338. doi: 10.1128/JB.181.20.6332-6338.1999
Asai, Y., Kawagishi, I., Sockett, R. E., and Homma, M. (2000). Coupling ion specificity of chimeras between H+- and Na+-driven motor proteins, MotB and PomB, in Vibrio polar flagella. EMBO J. 19, 3639–3648. doi: 10.1093/emboj/19.14.3639
Asai, Y., Yakushi, T., Kawagishi, I., and Homma, M. (2003). Ion-coupling determinants of Na+-driven and H+-driven flagellar motors. J. Mol. Biol. 327, 453–463. doi: 10.1016/S0022-2836(03)00096-2
Atsumi, T., McCarter, L., and Imae, Y. (1992). Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355, 182–184. doi: 10.1038/355182a0
Belas, R. (2013). When the swimming gets tough, the tough form a biofilm. Mol. Microbiol. 90, 1–5. doi: 10.1111/mmi.12354
Belas, R., Simon, M., and Silverman, M. (1986). Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J. Bacteriol. 167, 210–218. doi: 10.1128/JB.167.1.210-218.1986
Berg, H. C., and Anderson, R. A. (1973). Bacteria swim by rotating their flagellar filaments. Nature 245, 380–382. doi: 10.1038/245380a0
Boehm, A., Kaiser, M., Li, H., Spangler, C., Kasper, C. A., Ackermann, M., et al. (2010). Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141, 107–116. doi: 10.1016/j.cell.2010.01.018
Bruni, G., Weekley, A., Dodd, B., and Kralj, J. (2017). Voltage-gated calcium flux mediates Escherichia coli mechanosensation. Proc. Natl. Acad. Sci. U.S.A. 114, 9445–9450. doi: 10.1073/pnas.1703084114
Catherinot, V., and Labesse, G. (2004). ViTO: tool for refinement of protein sequence-structure alignments. Bioinformatics 20, 3694–3696. doi: 10.1093/bioinformatics/bth429
Chao, J., Song, K., Wang, H., Li, Z., Zhang, Y., Yin, C., et al. (2017). A colorimetric and fluorescent pH probe for imaging in E. coli cells. RSC Adv. 7, 964–970. doi: 10.1039/C6RA24885C
Chawla, R., Ford, K. M., and Lele, P. P. (2017). Torque, but not FliL, regulates mechanosensitive flagellar motor-function. Sci. Rep. 7:5565. doi: 10.1038/s41598-017-05521-8
Chimerel, C., Field, C. M., Pi nero-Fernandez, S., Keyser, U. F., and Summers, D. K. (2012). Indole prevents Escherichia coli cell division by modulating membrane potential. Biochim. Biophys. Acta 1818, 1590–1594. doi: 10.1016/j.bbamem.2012.02.022
Deme, J. C., Johnson, S., Vickery, O., Muellbauer, A., Monkhouse, A., Griffiths, T., et al. (2020). Structure of the stator complex that drives rotation of the bacterial flagellum. Nat. Microbiol. 5, 553–1564. doi: 10.1038/s41564-020-0788-8
Fang, X., and Gomelsky, M. (2010). A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 76, 1295–1305. doi: 10.1111/j.1365-2958.2010.07179.x
Fukuoka, H., Wada, T., Kojima, S., Ishijima, A., and Homma, M. (2009). Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol. Microbiol. 71, 825–835. doi: 10.1111/j.1365-2958.2008.06569.x
Fung, D. C., and Berg, H. C. (1995). Powering the flagellar motor of Escherichia coli with an external voltage source. Nature 375, 809–812. doi: 10.1038/375809a0
Gabel, C. V., and Berg, H. C. (2003). Speed of the flagellar rotary motor of Escherichia coli varies linearly with protonmotive force. Proc. Natl. Acad. Sci. U.S.A. 100, 8748–8751. doi: 10.1073/pnas.1533395100
Han, J., and Burgess, K. (2010). Fluorescent indicators for intracellular pH. Chem. Rev. 110, 2709–2728. doi: 10.1021/cr900249z
Humphries, J., Xiong, L., Liu, J., Prindle, A., Yuan, F., Arjes, H. A., et al. (2017). Species-independent attraction to biofilms through electrical signaling. Cell 168, 200–209.e12. doi: 10.1016/j.cell.2016.12.014
Imazawa, R., Takahashi, Y., Aoki, W., Sano, M., and Ito, M. (2016). A novel type bacterial flagellar motor that can use divalent cations as a coupling ion. Sci. Rep. 6:19773. doi: 10.1038/srep19773
Ishida, T., Clark, J., Matzke, N. J., Sowa, Y., and Baker, M. A. B. (2019). Sodium-powered stators of the bacterial flagellar motor can generate torque in the presence of phenamil with mutations near the peptidoglycan-binding region. Mol. Microbiol. 111, 1689–1699. doi: 10.1111/mmi.14246
Islam, M. I., Lin, A., Lai, Y.-W., Matzke, N.-J., and Baker, M. A. B. (2020). Ancestral sequence reconstructions of MotB are proton-motile and require MotA for motility. Front. Mircrobiol. 11:3271. doi: 10.3389/fmicb.2020.625837
Ito, M., Terahara, N., Fujinami, S., and Krulwich, T. A. (2005). Properties of motility in Bacillus subtilis powered by the H+-coupled MotAB flagellar stator, Na+-coupled MotPS or hybrid stators MotAS or MotPB. J. Mol. Biol. 352, 396–408. doi: 10.1016/j.jmb.2005.07.030
Kashket, E. (1985). The proton motive force in bacteria: a critical assessment of methods. Annu. Rev. Micro. 39, 219–42. doi: 10.1146/annurev.mi.39.100185.001251
Kawagishi, I., Maekawa, Y., Atsumi, T., Homma, M., and Imae, Y. (1995). Isolation of the polar and lateral flagellum-defective mutants in Vibrio alginolyticus and identification of their flagellar driving energy sources. J. Bacteriol. 177, 5158–5160. doi: 10.1128/JB.177.17.5158-5160.1995
Khan, S., Dapice, M., and Humayun, I. (1990). Energy transduction in the bacterial flagellar motor. Effects of load and pH. Biophys. J. 57, 779–796. doi: 10.1016/S0006-3495(90)82598-4
Khan, S., and Macnab, R. M. (1980). Proton chemical potential, proton electrical potential and bacterial motility. J. Mol. Biol. 138, 599–614. doi: 10.1016/S0022-2836(80)80019-2
Khan, S., Meister, M., and Berg, H. C. (1985). Constraints on flagellar rotation. J. Mol. Biol. 184, 645–656. doi: 10.1016/0022-2836(85)90310-9
Kojima, S., and Blair, D. F. (2001). Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40, 13041–13050. doi: 10.1021/bi011263o
Kralj, J. M., Hochbaum, D. R., Douglass, A. D., and Cohen, A. E. (2011). Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein. Science 333, 345–348. doi: 10.1126/science.1204763
Krasnopeeva, E., Lo, C.-J., and Pilizota, T. (2019). Single-cell bacterial electrophysiology reveals mechanisms of stress-induced damage. Biophys. J. 116, 2390–2399. doi: 10.1016/j.bpj.2019.04.039
Kurre, R., Kouzel, N., Ramakrishnan, K., and Oldewurtel, E. R. (2013). Speed switching of gonococcal surface motility correlates with proton motive force. PLoS ONE 8:e67718. doi: 10.1371/journal.pone.0067718
Larkin, J. W., Zhai, X., Kikuchi, K., Redford, S. E., Prindle, A., Liu, J., et al. (2018). Signal percolation within a bacterial community. Cell Syst. 7, 137–145. doi: 10.1016/j.cels.2018.06.005
Larsen, S. H., Adler, J., Gargus, J. J., and Hogg, R. W. (1974). Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc. Natl. Acad. Sci. U.S.A. 71, 1239–1243. doi: 10.1073/pnas.71.4.1239
Leake, M. C., Chandler, J. H., Wadhams, G. H., Bai, F., Berry, R. M., and Armitage, J. P. (2006). Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443, 355–358. doi: 10.1038/nature05135
Lele, P. P., Hosu, B. G., and Berg, H. C. (2013). Dynamics of mechanosensing in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U.S.A. 110, 11839–11844. doi: 10.1073/pnas.1305885110
Liu, J., Martinez-Corral, R., Prindle, A., Lee, D.-Y. D., Larkin, J., Gabalda-Sagarra, M., et al. (2017). Coupling between distant biofilms and emergence of nutrient time-sharing. Science 356, 638–642. doi: 10.1126/science.aah4204
Liu, J., Prindle, A., Humphries, J., Gabalda-Sagarra, M., Asally, M., Lee, D.-Y. D., et al. (2015). Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature 523, 550–554. doi: 10.1038/nature14660
Lo, C. J., Leake, M. C., and Berry, R. M. (2006). Fluorescence measurement of intracellular sodium concentration in single Escherichia coli cells. Biophys. J. 90, 357–365. doi: 10.1529/biophysj.105.071332
Lo, C. J., Leake, M. C., Pilizota, T., and Berry, R. M. (2007). Nonequivalence of membrane voltage and ion-gradient as driving forces for the bacterial flagellar motor at low load. Biophys. J. 93, 294–302. doi: 10.1529/biophysj.106.095265
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Maloney, P., Kashket, E., and Hastings Wilson, T. (1974). A proton motive force drives ATP synthesis in bacteria. Proc. Natl. Acad. Sci. U.S.A. 71, 3896–3900. doi: 10.1073/pnas.71.10.3896
Mancini, L., Terradot, G., Tian, T., Pu, Y., Li, Y., Lo, C.-J., et al. (2020). A general workflow for characterization of nernstian dyes and their effects on bacterial physiology. Biophys. J. 118, 4–14. doi: 10.1016/j.bpj.2019.10.030
Manson, M. D., Tedesco, P., Berg, H. C., Harold, F. M., and Van der Drift, C. (1977). A protonmotive force drives bacterial flagella. Proc. Natl. Acad. Sci. U.S.A. 74, 3060–3064. doi: 10.1073/pnas.74.7.3060
Manson, M. D., Tedesco, P. M., and Berg, H. C. (1980). Energetics of flagellar rotation in bacteria. J. Mol. Biol. 138, 541–561. doi: 10.1016/S0022-2836(80)80017-9
Martinac, B., Saimi, Y., and Kung, C. (2008). Ion channels in microbes. Physiol. Rev. 88, 1449–1490. doi: 10.1152/physrev.00005.2008
Martinez, K. A., Kitko, R. D., Mershon, J. P., Adcox, H. E., Malek, K. A., Berkmen, M. B., et al. (2012). Cytoplasmic pH response to acid stress in individual cells of Escherichia coli and Bacillus subtilis observed by fluorescence ratio imaging microscopy. Appl. Environ. Microbiol. 78, 3706–3714. doi: 10.1128/AEM.00354-12
Masi, E., Ciszak, M., Santopolo, L., Frascella, A., Giovennetti, L., Marchi, E., et al. (2015). Electrical spiking in bacterial biofilms. J. R. Soc. Interface 12:20141036. doi: 10.1098/rsif.2014.1036
McCarter, L., Hilmen, M., and Silverman, M. (1988). Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54, 345–351. doi: 10.1016/0092-8674(88)90197-3
Meister, M., Lowe, G., and Berg, H. C. (1987). The proton flux through the bacterial flagellar motor. Cell 49, 643–650. doi: 10.1016/0092-8674(87)90540-X
Miesenböck, G., De Angelis, D. A., and Rothman, J. E. (1998). Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192–195. doi: 10.1038/28190
Minamino, T., Imae, Y., Oosawa, F., Kobayashi, Y., and Oosawa, K. (2003). Effect of intracellular pH on rotational speed of bacterial flagellar motors. J. Bacteriol. 185, 1190–1194. doi: 10.1128/JB.185.4.1190-1194.2003
Minamino, T., Morimoto, Y. V., Hara, N., Aldridge, P. D., and Namba, K. (2016). The bacterial flagellar type III export gate complex is a dual fuel engine that can use both H+ and Na+ for flagellar protein export. PLoS Pathog. 12:e1005495. doi: 10.1371/journal.ppat.1005495
Mitchell, P. (1961). Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191, 144–148. doi: 10.1038/191144a0
Morimoto, V. Y., Nakamura, S., Kami-ike, N., Namba, K., and Minamino, T. (2010). Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol. Microbiol. 78, 1117–1129. doi: 10.1111/j.1365-2958.2010.07391.x
Morimoto, Y. V., Kami-ike, N., Miyata, T., Kawamoto, A., Kato, T., Namba, K., et al. (2016). High-resolution pH imaging of living bacterial cells to detect local pH differences. mBio 7, e01911–16. doi: 10.1128/mBio.01911-16
Naganawa, S., and Ito, M. (2020). Motp subunit is critical for ion selectivity and evolution of a k+-coupled flagellar motor. Biomolecules 10:691. doi: 10.3390/biom10050691
Nishihara, Y., and Kitao, A. (2015). Gate-controlled proton diffusion and protonation-induced ratchet motion in the stator of the bacterial flagellar motor. Proc. Natl. Acad. Sci. U.S.A. 112, 7737–7742. doi: 10.1073/pnas.1502991112
Nord, A. L., Gachon, E., Perez-Carrasco, R., Nirody, J. A., Barducci, A., Berry, R. M., et al. (2017). Catch bond drives stator mechanosensitivity in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U.S.A. 114, 12952–12957. doi: 10.1073/pnas.1716002114
Onoe, S., Yoshida, M., Terahara, N., and Sowa, Y. (2020). Coupling Ion specificity of the flagellar stator proteins MotA1/MotB1 of Paenibacillus sp. TCA20. Biomolecules 10:1078. doi: 10.3390/biom10071078
Paul, K., Nieto, V., Carlquist, W. C., Blair, D. F., and Harshey, R. M. (2010). The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38, 128–139. doi: 10.1016/j.molcel.2010.03.001
Paulick, A., Delalez, N. J., Brenzinger, S., Steel, B. C., Berry, R. M., Armitage, J. P., et al. (2015). Dual stator dynamics in the Shewanella oneidensis MR-1 flagellar motor. Mol. Microbiol. 96, 993–1001. doi: 10.1111/mmi.12984
Paulick, A., Koerdt, A., Lassak, J., Huntley, S., Wilms, I., Narberhaus, F., et al. (2009). Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol. Microbiol. 71, 836–850. doi: 10.1111/j.1365-2958.2008.06570.x
Prindle, A., Liu, J., Asally, M., Ly, S., Garcia-Ojalvo, J., and Süel, G. M. (2015). Ion channels enable electrical communication in bacterial communities. Nature 527, 59–63. doi: 10.1038/nature15709
Rasmussen, M. B., Oddershede, L. B., and Siegumfeldt, H. (2008). Optical tweezers cause physiological damage to Escherichia coli and listeria bacteria. Appl. Environ. Microbiol. 74, 2441–2446. doi: 10.1128/AEM.02265-07
Rossmann, F. M., and Beeby, M. (2018). Insights into the evolution of bacterial flagellar motors from high-throughput in situ electron cryotomography and subtomogram averaging. Acta Crystallogr. Sect. D 74, 585–594. doi: 10.1107/S2059798318007945
Rupprecht, C., Wingen, M., Potzkei, J., Gensch, T., Jaeger, K.-E., and Drepper, T. (2017). A novel FbFP-based biosensor toolbox for sensitive in vivo determination of intracellular pH. J. Biotechnol. 258, 25–32. doi: 10.1016/j.jbiotec.2017.05.006
Santiveri, M., Roa-Eguiara, A., Kuhne, C., Wadhwa, N., Hu, H., Berg, H. C., et al. (2020). Structure and function of stator units of the bacterial flagellar motor. Cell 183, 1–14. doi: 10.1016/j.cell.2020.08.016
Shioi, J. I., Matsuura, S., and Imae, Y. (1980). Quantitative measurements of proton motive force and motility in Bacillus subtilis. J. Bacteriol. 144, 891–897. doi: 10.1128/JB.144.3.891-897.1980
Siegumfeldt, H., Rechinger, B., and Jakobsen, M. (2000). Dynamic changes of intracellular pH in individual lactic acid bacterium cells in response to a rapid drop in extracellular pH. Appl. Environ. Microbiol. 66, 2330–2335. doi: 10.1128/AEM.66.6.2330-2335.2000
Sirec, T., Benarroch, J. M., Buffard, P., Garcia-Ojalvo, J., and Asally, M. (2019). Electrical polarization enables integrative quality control during bacterial differentiation into spores. iScience 16, 378–389. doi: 10.1016/j.isci.2019.05.044
Sowa, Y., and Berry, R. M. (2008). Bacterial flagellar motor. Q. Rev. Biophys. 41, 103–132. doi: 10.1017/S0033583508004691
Sowa, Y., Homma, M., Ishijima, A., and Berry, R. M. (2014). Hybrid-fuel bacterial flagellar motors in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 111, 3436–3441. doi: 10.1073/pnas.1317741111
Sowa, Y., Hotta, H., Homma, M., and Ishijima, A. (2003). Torque-speed relationship of the Na+-driven flagellar motor of Vibrio alginolyticus. J. Mol. Biol. 327, 1043–1051. doi: 10.1016/S0022-2836(03)00176-1
Sowa, Y., Rowe, A. D., Leake, M. C., Yakushi, T., Homma, M., Ishijima, A., et al. (2005). Direct observation of steps in rotation of the bacterial flagellar motor. Nature 437, 916–919. doi: 10.1038/nature04003
Strahl, H., and Hamoen, L. W. (2010). Membrane potential is important for bacterial cell division. Proc. Natl. Acad. Sci. U.S.A. 107, 12281–12286. doi: 10.1073/pnas.1005485107
Stratford, J. P., Edwards, C. L. A., Ghanshyam, M. J., Malyshev, D., Delise, M. A., Hayashi, Y., et al. (2019). Electrically induced bacterial membrane-potential dynamics correspond to cellular proliferation capacity. Proc. Natl. Acad. Sci. U.S.A. 116, 9552–9557. doi: 10.1073/pnas.1901788116
Suzuki, Y., Morimoto, Y. V., Oono, K., Hayashi, F., Oosawa, K., Kudo, S., et al. (2019). Effect of the MotA(M206I) mutation on torque generation and stator assembly in the Salmonella H+-driven flagellar motor. J. Bacteriol. 201:e00727-18. doi: 10.1128/JB.00727-18
Terahara, N., Kodera, N., Uchihashi, T., Ando, T., Namba, K., and Minamino, T. (2017). Na-induced structural transition of MotPS for stator assembly of the flagellar motor. Sci. Adv. 3:4119. doi: 10.1126/sciadv.aao4119
Terahara, N., Krulwich, T. A., and Ito, M. (2008). Mutations alter the sodium versus proton use of a Bacillus clausii flagellar motor and confer dual ion use on Bacillus subtilismotors. Proc. Natl. Acad. Sci. U.S.A. 105, 14359–14364. doi: 10.1073/pnas.0802106105
Terahara, N., Sano, M., and Ito, M. (2012). Bacillus flagellar motor that can use both Na+ and K+ as a coupling ion is converted by a single mutation to use only Na+. PLoS ONE 7:e46248. doi: 10.1371/journal.pone.0046248
Terashima, H., Li, N., Sakuma, M., Koike, M., Kojima, S., Homma, M., et al. (2013). Insight into the assembly mechanism in the supramolecular rings of the sodium-driven Vibrio flagellar motor from the structure of FlgT. Proc. Natl. Acad. Sci. U.S.A. 110, 6133–6138. doi: 10.1073/pnas.1222655110
Thormann, K. M., and Paulick, A. (2010). Tuning the flagellar motor. Microbiology 156(Pt 5), 1275–1283. doi: 10.1099/mic.0.029595-0
Tipping, J. M., Steel, C. B., Delalez, J. N., Berry, M. R., and Armitage, P. J. (2013). Quantification of flagellar motor stator dynamics through in vivo proton-motive force control. Mol. Microbiol. 87, 338–347. doi: 10.1111/mmi.12098
Tipping, M. J., Delalez, N. J., Lim, R., Berry, R. M., and Armitage, J. P. (2013). Load-dependent assembly of the bacterial flagellar motor. mBio 4, e00551–13. doi: 10.1128/mBio.00551-13
Walter, J. M., Greenfield, D., Bustamante, C., and Liphardt, J. (2007). Light-powering Escherichia coli with proteorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 104, 2408–2412. doi: 10.1073/pnas.0611035104
Keywords: ion motive force, bacterial flagellar motor, stator, bacterial electrophysiology, subunit exchange, fluctuations, ion specificity, cell-to-cell signaling
Citation: Biquet-Bisquert A, Labesse G, Pedaci F and Nord AL (2021) The Dynamic Ion Motive Force Powering the Bacterial Flagellar Motor. Front. Microbiol. 12:659464. doi: 10.3389/fmicb.2021.659464
Received: 27 January 2021; Accepted: 02 March 2021;
Published: 13 April 2021.
Edited by:
Masahiro Ito, Toyo University, JapanCopyright © 2021 Biquet-Bisquert, Labesse, Pedaci and Nord. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashley L. Nord, YXNobGV5Lm5vcmRAY2JzLmNucnMuZnI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.