
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 June 2021
Sec. Evolutionary and Genomic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.659116
Staphylococci can cause a wide array of infections that can be life threatening. These infections become more deadly when the isolates are antibiotic resistant and thus harder to treat. Many resistance determinants are plasmid-mediated; however, staphylococcal plasmids have not yet been fully characterized. In particular, plasmids and their contributions to antibiotic resistance have not been investigated within the Arab states, where antibiotic use is not universally regulated. Here, we characterized the putative plasmid content among 56 Staphylococcus aureus and 10 Staphylococcus haemolyticus clinical isolates from Alexandria, Egypt. Putative plasmid sequences were detected in over half of our collection. In total, we identified 72 putative plasmid sequences in 27 S. aureus and 1 S. haemolyticus isolates. While these isolates typically carried one or two plasmids, we identified one isolate—S. aureus AA53—with 11 putative plasmids. The plasmid sequences most frequently encoded a Rep_1, RepL, or PriCT_1 type replication protein. As expected, antibiotic resistance genes were widespread among the identified plasmid sequences. Related plasmids were identified amongst our clinical isolates; homologous plasmids present in multiple isolates clustered into 11 groups based upon sequence similarity. Plasmids from the same cluster often shared antibiotic resistance genes, including blaZ, which is associated with β-lactam resistance. Our analyses suggest that plasmids are a key factor in the pathology and epidemiology of S. aureus in Egypt. A better characterization of plasmids and the role they contribute to the success of Staphylococci as pathogens will guide the design of effective control strategies to limit their spread.
Staphylococci are commensal bacteria in humans and animals with the ability to cause serious infections. Prime among the Staphylococci is Staphylococcus aureus with pathologies that range from superficial skin lesions to bacteremia, pneumonia, and endocarditis, among others (Tong et al., 2015). S. aureus is a major challenge in both the hospital and community settings, with treatment made more difficult given its ability to rapidly acquire antibiotic resistance (Zetola et al., 2005; Grundmann et al., 2006; Foster, 2017). The archetypal and most common antibiotic resistant examples are the Methicillin-resistant S. aureus (MRSA) strains (Foster, 2017), which are usually resistant to most β-lactam antibiotics (Hill-Cawthorne et al., 2014). Strains of Coagulase-Negative Staphylococci (CoNS), including Staphylococcus epidermidis and Staphylococcus haemolyticus, also have acquired antibiotic resistance (Becker et al., 2014). For example, over the past 40 years, Methicillin-resistant S. haemolyticus (MRSH) strains have become more prevalent (Ferreira et al., 2002; Bouchami et al., 2011; Becker et al., 2014; Xu et al., 2018; Al-Tamimi et al., 2020). To treat MRSA and MRSH, clinicians must resort to different classes of antibiotics, but further acquisition of antibiotic resistance by Methicillin-resistant strains is a major problem (Hill-Cawthorne et al., 2014). In Egypt, antibiotic use is not regulated, and most antimicrobial agents are available without the need for a prescription. This unregulated availability increases selective pressure that leads to development of antibiotic resistance. Thus, antibiotic therapy is often inadequate, a problem compounded by the use of the wrong antibiotic and both inappropriate dosage and duration. Consequently, Egypt has seen alarming increases in antibiotic resistance and MRSA/MRSH prevalence (Borg et al., 2007; Falagas et al., 2013; Abouelfetouh, 2017; Mashaly and El-Mahdy, 2017).
Horizontal gene transfer is a major source for acquisition of antibiotic resistance genes in Staphylococci, facilitated by phage transduction and plasmid conjugation (McCarthy et al., 2014). Plasmids can be reservoirs and vectors for antibiotic resistance, virulence, and fitness genes, even in the absence of selective pressure (Kwong et al., 2017). Horizontal transfer of plasmids can occur not only between Staphylococcal strains of the same species but also between species, with CoNS believed to act as reservoirs for antibiotic resistance genes (Otto, 2013). Moreover, recent evidence suggests that plasmids also can be exchanged between Staphylococci and the distantly related Enterococci (Acman et al., 2020). Data on the diversity of plasmid content of Staphylococci is limited (Borg et al., 2007; Abouelfetouh, 2017; Abouelfetouh et al., 2017; Kwong et al., 2017), but it is known that Staphylococcal plasmids can range in size from 1 to 60 kbp and that they can carry resistance genes (or gene clusters) (Kwong et al., 2017). Clinical isolates often contain one or more multi-resistance plasmids (Shearer et al., 2011). Thus, plasmids are key to the epidemiology of Staphylococci, contributing to the rapid spread of antibiotic resistance, and play a major role in determining clinical outcomes (Hisatsune et al., 2013; Shahkarami et al., 2014). While over 1,000 S. aureus plasmid sequences and 15 S. haemolyticus plasmid sequences are publicly available in resources such as PLSDB (Galata et al., 2019), just 6 S. aureus and no S. haemolyticus plasmids are annotated as collected from the Arab states. This knowledge gap motivated our investigation of plasmids from this region, and we hypothesize that plasmids in Egyptian Staphylococcal strains encode for antibiotic resistance.
Large plasmids in bacteria can be classified by their incompatibility groups, as plasmids with similar replication initiation (Rep) proteins cannot be stably maintained in the absence of selective pressure (Jensen et al., 2010; McCarthy and Lindsay, 2012). To date, 7 distinct types of Rep proteins and 15 incompatibility groups have been identified for Staphylococcal plasmids (Kwong et al., 2017). Accordingly, Staphylococcal plasmids can be classified using the sequence of their Rep genes (Jensen et al., 2010). Antibiotic resistance genes may be linked to Rep families; for example, tetracycline resistance has been associated with the rep7 gene and chloramphenicol resistance with the rep7, rep13, and rep27 genes (McCarthy and Lindsay, 2012). Currently, there are limited data on the Rep families present in circulating Staphylococcal isolates, including those from Egypt. Knowledge of the plasmid Rep families present in these clinical isolates could give us insight into Staphylococcal plasmid biology and thus help to better understand Methicillin-resistant strains.
In this study, we utilized genomics to (i) catalog the Rep family classification in the plasmids of Egyptian clinical isolates of S. aureus and S. haemolyticus, (ii) profile the antibiotic resistance genes present in these plasmids, and (iii) assess the prevalence of plasmid-borne antibiotic resistance in Egyptian MRSA isolates. We report on 72 plasmids present in 27 S. aureus and 1 S. haemolyticus isolates from Egypt. We also report on the antibiotic resistance genes in these plasmids, including those from MRSA and MRSH isolates. We furthermore provide bioinformatic and genetic context for these plasmid sequences. We found that the plasmids in Egyptian isolates of S. aureus and S. haemolyticus are diverse and include antibiotic resistance genes for antibiotics used clinically, indicating the potential for plasmids to act as gene reservoirs and thus facilitate the rapid horizontal transfer of antibiotic resistance in Egypt.
Raw sequencing reads for the 56 S. aureus and 10 S. haemolyticus genomes were previously sequenced, annotated, and typed by our group (Montelongo et al., 2021). Here, the raw reads were assembled using plasmidSPAdes (v3.9.0) (Antipov et al., 2016). Plasmid sequences with a length greater than 1,000 bp were selected for further analysis. Each putative plasmid sequence was queried via megablast against the nr/nt database (April 2020). The best hit to a complete plasmid sequence record for each sequence was identified and recorded. PlasmidFinder (v2.1) hosted by the Center for Genomic Epidemiology was queried to identify replicon sequences (Carattoli et al., 2014). BLAST and PlasmidFinder results were considered together to distinguish putative plasmids from other sequences identified by plasmidSPAdes (e.g., bacteriophage). To verify even coverage, reads were mapped to plasmid sequences using the Bowtie2 (v2.3.2) (Langmead and Salzberg, 2012) plug-in through Geneious Prime v2019.1.1 (Biomatters Ltd., Auckland, NZ). To estimate plasmid copy numbers, genome and plasmid coverages were calculated using BBMap v38.471.
Putative plasmid sequences were annotated using PATRIC v3.6.3 (Brettin et al., 2015; Wattam et al., 2017). Putative plasmid sequences also were examined for antibiotic resistance using ResFinder (v3.2) (Zankari et al., 2012), using default parameters (%ID threshold 90%, selected minimum length 60%).
Putative plasmid sequences were clustered using the USEARCH algorithm (v11.0.667) (Edgar, 2010) with the “cluster_fast” option for id = 0.8 to both strands. Each cluster was aligned using MAFFT v7.388 (Katoh and Standley, 2013) and Mauve (Darling et al., 2004) through Geneious Prime and manually inspected. BLAST hits were referenced to verify that plasmids within the same cluster exhibited similarity to related (if not the same) plasmid sequence in the nr/nt database. Plasmid alignments were generated using BLAST Ring Image Generator v0.95 (Alikhan et al., 2011).
Anvi’o (Eren et al., 2015) was used to identify the single copy number genes amongst the S. aureus genome sequences. A phylogenomic tree was created using concatenated amino acid sequences for these single copy number genes. These concatenated sequences were aligned using MAFFT v7.388 (Katoh and Standley, 2013) through Geneious Prime (Biomatters Ltd., Auckland, New Zealand). The tree was derived using FastTree v2 (Price et al., 2010) through Geneious Prime and visualized by iTOL v5.6.1 (Letunic and Bork, 2019).
Upon the examination of 56 S. aureus and 10 S. haemolyticus genomes of Egyptian clinical isolates, we identified 72 putative plasmid sequences in 28 strains: 27 strains of S. aureus and 1 strain of S. haemolyticus. From our previous work (Montelongo et al., 2021), we know that 24 of these isolates are mecA-positive and represent 11 different MLSTs and 2 unknown MLSTs. While 10 of the strains harbored just a single putative plasmid sequence, one strain—AA53—contained 11 (Table 1). The copy number of the plasmid sequences ranged from ∼1 to >300, determined by comparing the coverage of each individual plasmid sequence relative to the average coverage for the assembly (Supplementary Table 1). 50 of the 72 putative plasmid sequences contained replicon proteins identified by PlasmidFinder. Table 2 summarizes these results. The majority of the strains (85%) contained plasmid sequences associated with rep16 (n = 10), rep20 (n = 9), or rep21 (n = 9). The replicons rep22, rep24a, and rep39 were each unique to a single isolate. As expected because of incompatibility, no strain was found to have more than one plasmid for a given replicon protein. Putative plasmid sequences containing a replicon protein frequently showed sequence identity to a Staphylococcus plasmid sequence (Supplementary Table 2).

Table 2. Number and type of replicons identified by PlasmidFinder for plasmid sequences in the Egyptian S. aureus strains and S. haemolyticus strain.
To ascertain if the 22 putative plasmid sequences that did not contain a replicon protein likely represented a plasmid, we referred to the BLAST hits (Supplementary Table 2). These all showed greatest sequence similarity (in some cases, even 100% sequence identity) to a previously reported Staphylococcal plasmid sequence, often one that was significantly longer than our putative plasmid sequence. Of note are the putative plasmid sequences within S. aureus strains AA2, AA3, AA4, and AA45. Each of these strains contained three or four sequences that showed sequence similarity to the same S. aureus plasmid sequence, pGR2A (Accession no. CP010403). For each of these strains, one sequence had a recognizable rep20 replicon. We downloaded the pGR2A record and mapped the raw reads for strains AA2, AA3, AA4, and AA45 to the pGR2A sequence. While the reads mapped to the plasmid sequence, there were two or three regions of low coverage (Figure 1), suggesting that either the plasmid within these strains: (1) has undergone reassortment or expansion, differentiating it from pGR2A; (2) has split into separate extrachromosomal DNAs; (3) contains a region that was not sequenced by the Illumina short-read technology; and/or integrated into the genome. The pGR2A-like plasmid sequences accounted for 14 of the 22 putative plasmid sequences without a recognized replicon protein. Thus, there are just eight putative plasmid sequences in our data set for which a replicon protein was not identified.
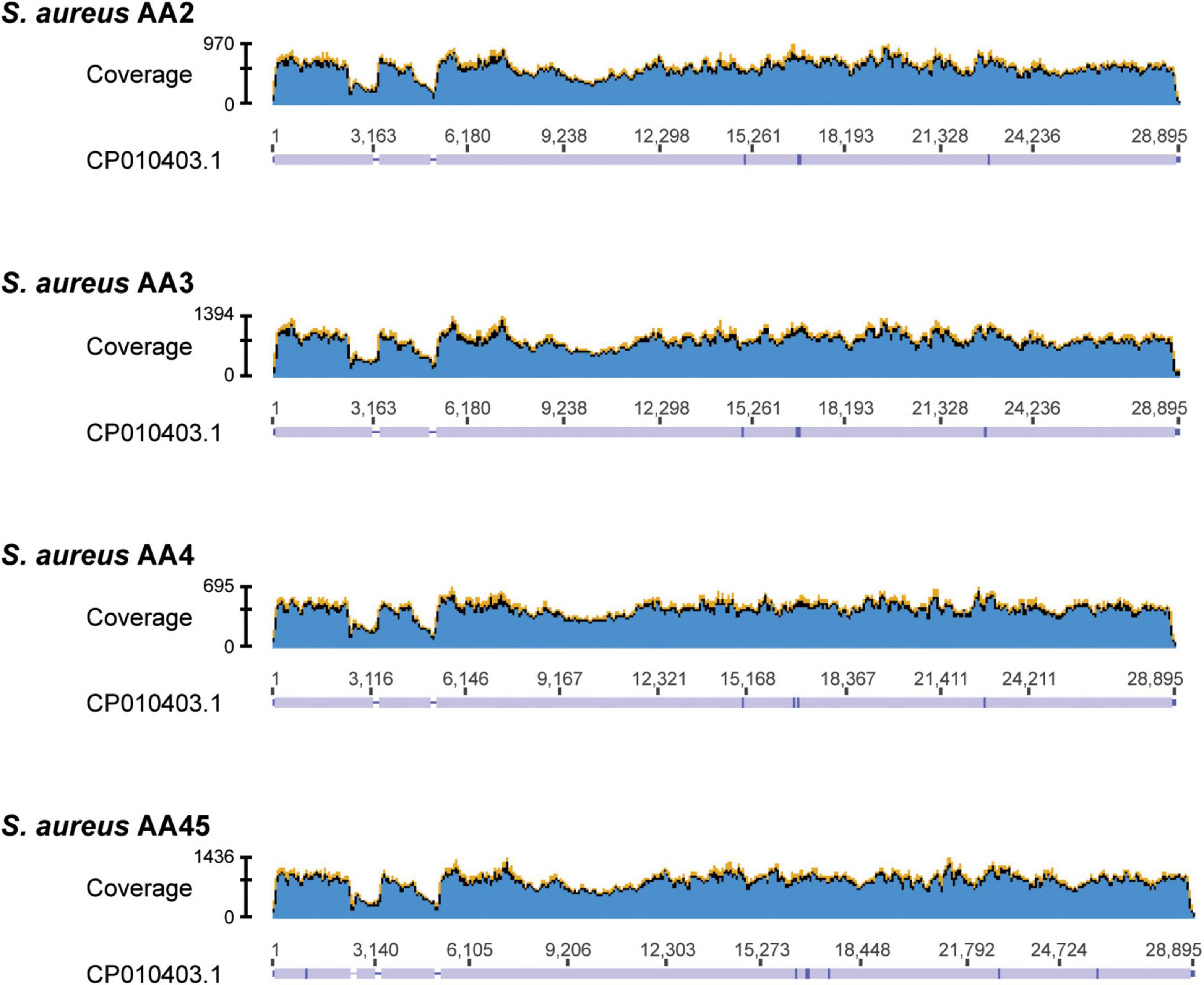
Figure 1. Read coverage maps for the 4 S. aureus strains containing contigs showing homology to plasmid pGR2A (CP010403.1). Raw reads from each of the 4 strains were mapped to the pGR2A sequence and their coverage is shown in the graphs. Below each graph, the consensus sequence is shown. Regions covered appear in lavender. Breaks in this coverage (lavender bar) indicates regions where no reads mapped.
To identify homologous plasmids found in more than one isolate, all 72 putative plasmid sequences were clustered. In total, we identified 11 clusters while 16 plasmids did not show any sequence similarity to another plasmid in our data set. These 16 unclustered plasmids, however, did exhibit sequence similarity to plasmid sequences deposited in GenBank (Supplementary Table 2). This included the two plasmids within the S. haemolyticus A42 strain. Table 3 lists details about these clusters. The largest clusters, plasmid Cluster C and plasmid Cluster J, contained a plasmid sequence found in 10 and 5 different strains, respectively. In some cases, all members of the cluster showed greatest homology to the same GenBank plasmid record, for instance Cluster A (n = 4), Cluster B (n = 4), Cluster D (n = 2), Cluster E (n = 2), and Cluster K (n = 2). In other cases, members of the cluster exhibited greatest sequence similarity to different GenBank plasmid records, reflecting the differences between the homologous plasmids, as well as hinting at diversity in the plasmid group amongst strains of S. aureus. Figure 2 shows that instances of gene acquisition/loss can be detected between homologous plasmids in a cluster. For some clusters, the plasmids are carried by closely related S. aureus strains, while other clusters include plasmids found in strains that span the genomic diversity of the Egyptian isolates (Figure 3).
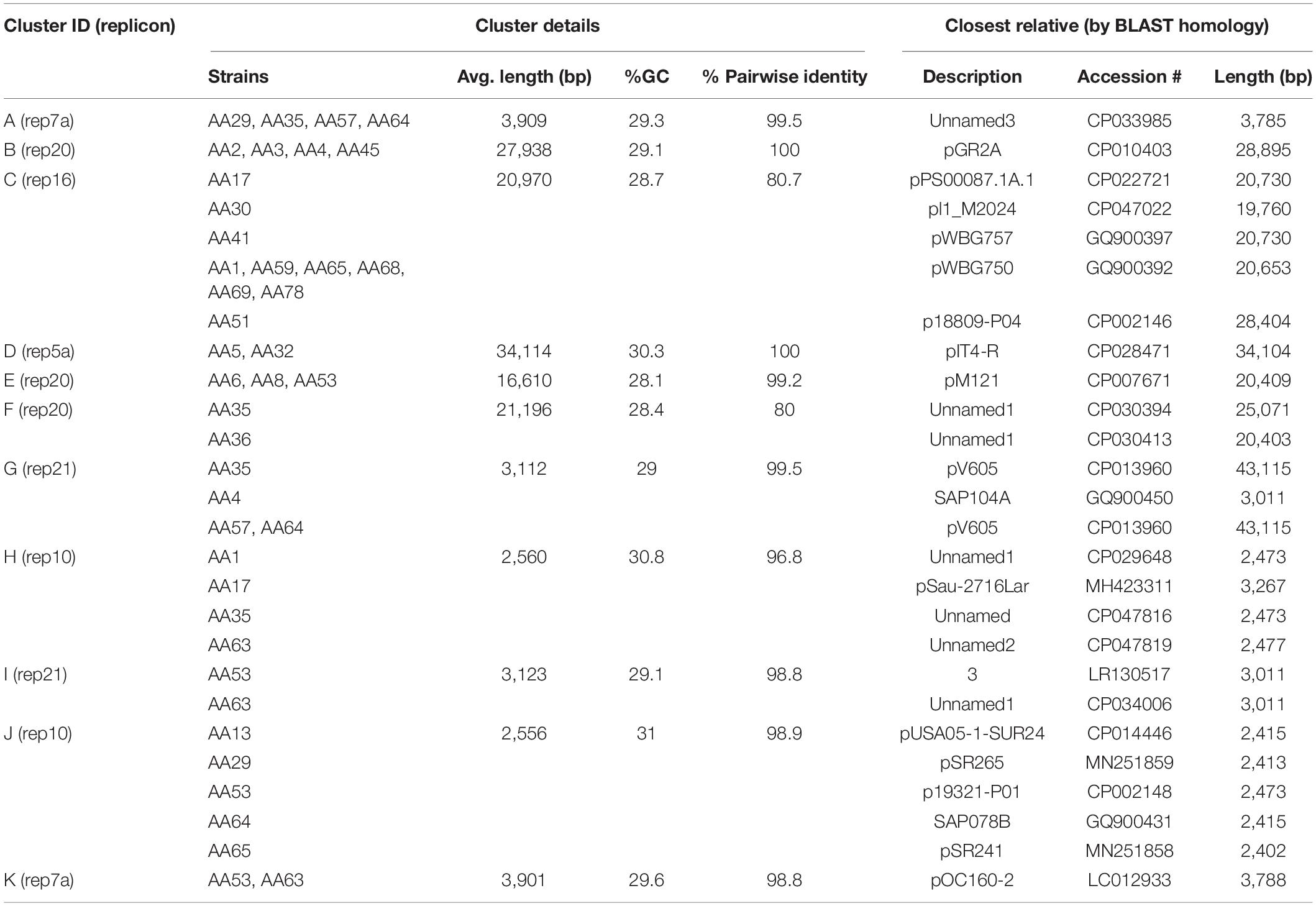
Table 3. Clusters of plasmids based on nucleotide sequence similarity within the Egyptian S. aureus strains.
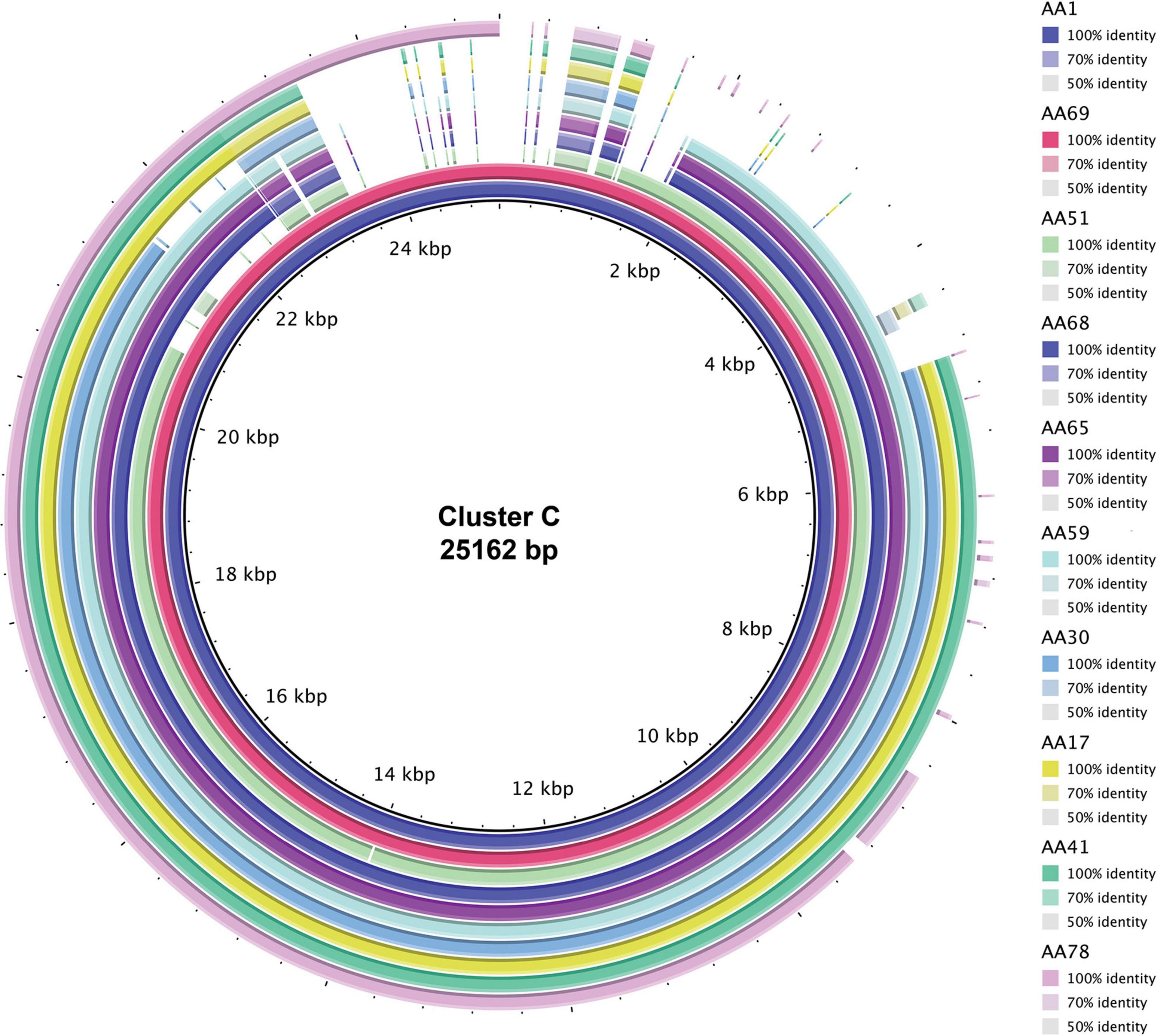
Figure 2. Sequence alignment of the 10 plasmid sequences grouped into Cluster C. The legend indicates the color for the plasmid sequence with the darker tones indicating 100% sequence identity and lighter tones indicative of less sequence identity. Gaps in the alignment signify regions that are not found within the plasmid sequence. The total alignment is 25,162 bp.

Figure 3. Distribution of plasmid clusters across phylogenomic diversity of Egyptian S. aureus strains. The phylogenomic tree for the isolates was generated based upon the concatenated amino acid sequences of the single copy number genes in the core genome. The tree scale indicates the sequence divergence between the core genome sequence. The color blocks show the presence of a plasmid belonging to 1 of the 11 clusters.
We annotated the plasmid sequences using the web service PATRIC (Supplementary Table 3). 38.6% of the coding regions within the 72 plasmid sequences are annotated as hypothetical proteins. PATRIC did identify resistances to metals, e.g., cadmium, copper, cobalt/zinc/cadmium, lead, mercury, and zinc, and antibiotics. Furthermore, the plasmid from S. aureus AA70 also encodes for exotoxin proteins. To further investigate antibiotic resistance genes, each plasmid sequence was examined with the tool ResFinder. Table 4 summarizes the antibiotic resistance determinants predicted by ResFinder, and full results can be found in Supplementary Table 4. 20 of the 27 (74%) S. aureus strains included a putative plasmid encoding blaZ, which is associated with β-lactam resistance. These included all four of the mecA-negative S. aureus strains (AA41, AA59, AA65, and AA68). S. aureus AA69 and AA41 were the only strains carrying a plasmid encoding aminoglycoside resistance. The one plasmid-carrying S. haemolyticus strain (AA42) did not encode blaZ; however, it carried plasmids that encode for lincosamide resistance, macrolide resistance, and streptogramin B resistance. This strain is the only one identified in this study that carries a plasmid that encodes for the lnu(A) gene, which is associated with lincosamide resistance.
Whereas 48.6% of the plasmids carried a single antibiotic resistance gene, others carried multiple antibiotic resistance genes. We therefore examined the plasmid sequences for resistance co-occurrence. Strains harboring plasmids encoding for fusidic acid resistance (fusB) (n = 4) also harbored distinct plasmids that encoded for tetracycline resistance [tet(K)] (n = 5) (r = 0.9). Furthermore, all four strains with plasmids carrying both fusB and tet(K) were MLST-80. Plasmids encoding for tet(L) were only harbored by strains of MLST-1. However, this association between tet(L) and MLST-1 was not observed more broadly; publicly available MLST-1 genomes (Supplementary Table 5) were queried for the tet(L) sequence. Only 8 of the 112 strains queried also coded for tet(L). Interestingly, six of these were Egyptian MRSA isolates (Supplementary Table 5).
Plasmids are key reservoirs for genetic content in Staphylococci and allow the rapid propagation of antibiotic resistance (Jensen et al., 2010; McCarthy and Lindsay, 2012). Here, we screened the genomes of 56 clinical isolates of S. aureus and 10 clinical isolates of S. haemolyticus previously sequenced by our group and found 72 putative plasmid sequences (Table 1). We identified replicon proteins in 50 of the 72 plasmids. Within the S. aureus strains, eight different replicon proteins were detected, representative of six of the seven Staphylococcal replicon protein types, and the S. haemolyticus isolate included a plasmid with an additional replicon protein not found in the S. aureus strains, rep39 (Table 2). This particular plasmid (rep39) was homologous to pLNU4, a plasmid found in an S. chromogenes isolate that was associated with bovine mastitis, and can be acquired by S. aureus via transformation (Lüthje et al., 2007). For strains containing more than 1 plasmid, 11 different combinations of the replicon proteins were found. Strains containing plasmids with rep7a and rep10 were most common (n = 6); whereas one strain, S. aureus AA13, contained just a single rep10 plasmid, no strains contained a single rep7a plasmid. Rep sites were not identified in eight of the putative plasmids, although their sequence reads had homology to plasmid sequences. These eight could represent partial plasmid sequences or may rely on vertical transfer of plasmids, as is the case of smaller plasmids with high copy number (Kwong et al., 2017). However, based on both plasmid copy number (Supplementary Table 1) and BLAST hits (Supplementary Table 2), we believe that these eight sequences are in fact partial plasmid sequences.
S. aureus strains AA2, AA3, AA4, and AA45 had putative plasmid sequences that showed sequence similarity to the S. aureus plasmid pGR2A (Accession no. CP010403). The pGR2A plasmid codes a unique bla system that can suppress oxacillin resistance in MRSA strains (Sabat et al., 2015). The non-functional BlaR1 from pGR2A does not cleave the mecA repressor BlaI; thus, mecA is expressed inefficiently, resulting in a phenotype of penicillin resistance, but oxacillin susceptibility (Sabat et al., 2015). It must be noted that all four strains with plasmid sequences homologous to pGR2A were mecA-positive strains (Tables 1, 3). In the clinical setting, oxacillin-susceptible mecA-positive S. aureus may pose a challenge due to possible misdiagnosis as a typical MRSA (Ikonomidis et al., 2008; Sabat et al., 2015).
In addition to classifying plasmids by their Rep genes, we analyzed plasmid relatedness by clustering putative plasmid-coding regions based on sequence homology (Table 3). We identified 11 plasmid clusters among the S. aureus strains; the two S. haemolyticus plasmid sequences did not resemble any of the S. aureus plasmids. Members of a cluster could have homology to different GenBank plasmid records (C, J), or all could share the same GenBank plasmid record (A, B, D, E, K). The former likely indicates homology of non-identical plasmids and the acquisition/loss of content as indicated by cluster C (Figure 2), while the latter points to multiple S. aureus isolates possessing the same plasmid, potentially due to a shared selective pressure (Lüthje et al., 2007; Kwong et al., 2017). Our results found that Rep genes are not exclusive to a single cluster, concurring with prior findings of frequent gene exchange amongst Staphylococcal plasmids (see review; Kwong et al., 2017). Strains harboring plasmids from the same cluster often shared antibiotic resistance genes, e.g., macrolide resistance in Cluster A plasmids, fusidic acid resistance in Cluster B, and beta-lactam resistance in Cluster C. Thus, while replicon proteins can provide insight into plasmid incompatibilities, they have limited epidemiological information compared to the cluster to which a plasmid belongs.
Of the 28 isolates in which we identified putative plasmid sequences, 24 (86%) were mecA-positive in addition to widespread presence of resistance genes in their plasmid sequences (Table 4). In the clinical setting, the main classes of drugs used against Staphylococcus are those that target the cell wall (e.g., β-lactam antibiotics, glycopeptides), ribosome (e.g., tetracyclines, aminoglycosides, macrolides), and nucleic acid biosynthesis (fluoroquinolones and sulfamethoxazole/trimethoprim) (Foster, 2017). Resistance to these antibiotic classes is a critical concern, especially in the acquisition of multiple resistances (Grundmann et al., 2006; Falagas et al., 2013). There have been reports of over-the-counter antibiotic use in Egypt that could select for resistance to key antibiotics, e.g., penicillins and cephalosporins (Abouelfetouh, 2017; Abouelfetouh et al., 2017). The most common antibiotic resistances in the plasmid sequences were those to macrolides, tetracyclines, fusidic acid, and aminoglycosides, and 20 of our Staphylococcus putative plasmid sequences had genes linked to β-lactam antibiotics resistance (Table 4). While tetracycline resistance can be plasmid- or chromosome-mediated, macrolide resistance is almost exclusively encoded by mobile elements in Staphylococci. Nine different Egyptian S. aureus strains carried plasmids with the 23S rRNA methylase erm(C) (Table 4). Previous studies have found that erm(C) is the most abundant gene conferring resistance to macrolides, as well as to lincosamides and streptogramin B compounds (see review; Feßler et al., 2018). mph(C), found in S. aureus AA41 and S. haemolyticus AA42, mediates resistance to only macrolides, whereas lnu(A), found only in S. haemolyticus AA42, encodes only lincosamide resistance. In addition, our annotation indicated resistance genes for chloramphenicol, clindamycin, and sulfamethoxazole/trimethoprim (Supplementary Table 3). The presence of these antibiotic resistance genes can greatly limit the antibiotic options to treat these clinical isolates (Abouelfetouh, 2017; Foster, 2017; Vestergaard et al., 2019). Of utmost concern is that multiple resistances could be disseminated via plasmids, especially in MRSA (McCarthy and Lindsay, 2012).
We did not identify genes involved in conjugation in our isolates (Supplementary Table 3). It has been noted that S. aureus may rely on vertical transmission of its plasmids, while conjugation genes are relatively rare (McCarthy and Lindsay, 2012). The distribution of plasmids belonging to the same cluster across the Egyptian S. aureus isolates suggests both vertical and horizontal transfer (Figure 3). Horizontal acquisition of the plasmid in our isolates could occur via natural competence or by conjugation systems not yet characterized and therefore not present in our annotation references (Morikawa et al., 2012). It must be highlighted that many of the plasmid sequences presented in these Egyptian isolates were homologous to GenBank entries for plasmids that have not been well-characterized (Table 3); from 37 plasmids homologous to our sequences, only seven had a publication associated with their entry. This underscores the general lack of S. aureus plasmid knowledge. Furthermore, 38.6% of the genes annotated within our plasmids are characterized as hypothetical proteins. Their function and/or the possible genetic advantage that these genes provide the bacterial strains remains an open question. The presence of genes for multiple antibiotic resistance in uncharacterized plasmids indicates not just the relevance of plasmids in the clinical treatment of Egyptian Staphylococcus, but also points to the depth of knowledge still to be discovered in this research area (Abouelfetouh, 2017; Abouelfetouh et al., 2017).
In this study, we utilized genomics to report on the plasmid content of Egyptian Staphylococci, which further adds to our global knowledge of Staphylococcal plasmids. Plasmids and their contributions to antibiotic resistance have not been investigated previously within the Arab states. Putative plasmid sequences were present in over half of isolates analyzed in this study, with widespread presence of antibiotic resistance genes. Since plasmids are reservoirs of and vectors for antibiotic resistance in Staphylococci, plasmids could be a key factor in the pathology and epidemiology of Staphylococci, including MRSA and MRSH (McCarthy and Lindsay, 2012; Shahkarami et al., 2014). Making this knowledge available allows policy makers and infection control officers the chance to design and implement well-informed infection control and antimicrobial stewardship plans. Further characterization of plasmids is necessary to better understand the infectious potential of Staphylococci and devise optimal strategies to curb their spread and virulence.
Data analyzed as part of this study include the following GenBank WGS accession numbers: JAEOUR000000000, JAEOUZ000000000, JAEOVE000000000, JAEOVM000000000, JAEOVQ000000000, JAEOVX000000000, JAEOWM000000000, JAEOUV000000000, JAEOUX000000000, JAEOVD000000000, JAEOVF000000000, JAEOVH000000000, JAEOVJ000000000, JAEOVK000000000, JAEOVN000000000, JAEOWY000000000, JAEOVO000000000, JAEOVR000000000, JAEOVT000000000, JAEOVV000000000, JAEOVW000000000, JAEOWB000000000, JAEOWC000000000, JAEOWD000000000, JAEOWF000000000, JAEOWG000000000, JAEOWH000000000, and JAEOWK000000000.
CM, AJW, and AA designed the study. CRM and CP conducted the analyses. CRM, CM, CP, and AA wrote the initial draft. All authors contributed to the manuscript editing and approved the final version.
We acknowledge funding from NIH (R01 DK104718 awarded to AJW), NSF (1661357 awarded to CP), and USAID (GSP-T85 awarded to AA) and DFG (ZI 665/3-1 awarded to AA). The funders did not play a part in the design or conduct of the study.
AJW discloses membership on the Advisory Boards of Pathnostics and Urobiome Therapeutics. This membership did not play a part in the design of the study, analysis or interpretation of data, writing of the manuscript, or decision to publish the results.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.659116/full#supplementary-material
Supplementary Table 1 | Plasmid vs. genome sequence coverage.
Supplementary Table 2 | BLAST-based similarity of putative plasmid sequences to records in nr/nt database.
Supplementary Table 3 | Annotation of putative plasmid sequences.
Supplementary Table 4 | Antibiotic resistance genes identified for each putative plasmid sequence.
Supplementary Table 5 | Publicly available MLST-1 S. aureus genomes screened for tet(L).
Abouelfetouh, A. (2017). The status of methicillin resistance among Egyptian Staphylococcus aureus isolates: an overview. Infect. Disord. Drug Targets 17, 67–69. doi: 10.2174/1871526516666160802111200
Abouelfetouh, A., Kassem, M., Naguib, M., and El-Nakeeb, M. (2017). Investigation and treatment of fusidic acid resistance among methicillin-resistant staphylococcal isolates from Egypt. Microb. Drug Resist. 23, 8–17. doi: 10.1089/mdr.2015.0336
Acman, M., van Dorp, L., Santini, J. M., and Balloux, F. (2020). Large-scale network analysis captures biological features of bacterial plasmids. Nat. Commun. 11:2452. doi: 10.1038/s41467-020-16282-w
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Al-Tamimi, M., Abu-Raideh, J., Himsawi, N., Khasawneh, A., and Hawamdeh, H. (2020). Methicillin and vancomycin resistance in coagulase-negative Staphylococci isolated from the nostrils of hospitalized patients. J. Infect. Dev. Ctries. 14, 28–35. doi: 10.3855/jidc.11025
Antipov, D., Hartwick, N., Shen, M., Raiko, M., Lapidus, A., and Pevzner, P. A. (2016). plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32, 3380–3387. doi: 10.1093/bioinformatics/btw493
Becker, K., Heilmann, C., and Peters, G. (2014). Coagulase-negative staphylococci. Clin. Microbiol. Rev. 27, 870–926. doi: 10.1128/CMR.00109-13
Borg, M. A., de Kraker, M., Scicluna, E., van de Sande-Bruinsma, N., Tiemersma, E., Monen, J., et al. (2007). Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in invasive isolates from southern and eastern Mediterranean countries. J. Antimicrob. Chemother. 60, 1310–1315. doi: 10.1093/jac/dkm365
Bouchami, O., Achour, W., Mekni, M. A., Rolo, J., and Hassen, A. B. (2011). Antibiotic resistance and molecular characterization of clinical isolates of methicillin-resistant coagulase-negative staphylococci isolated from bacteremic patients in oncohematology. Folia Microbiol. 56, 122–130. doi: 10.1007/s12223-011-0017-1
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Eren, A. M., Esen, ÖC., Quince, C., Vineis, J. H., Morrison, H. G., Sogin, M. L., et al. (2015). Anvi’o: an advanced analysis and visualization platform for ’omics data. PeerJ 3:e1319. doi: 10.7717/peerj.1319
Falagas, M. E., Karageorgopoulos, D. E., Leptidis, J., and Korbila, I. P. (2013). MRSA in Africa: filling the global map of antimicrobial resistance. PLoS One 8:e68024. doi: 10.1371/journal.pone.0068024
Ferreira, R. B. R., Nunes, A. P. F., Kokis, V. M., Krepsky, N., Fonseca, L., de, S., et al. (2002). Simultaneous detection of the mecA and ileS-2 genes in coagulase-negative staphylococci isolated from Brazilian hospitals by multiplex PCR. Diagn. Microbiol. Infect. Dis. 42, 205–212. doi: 10.1016/s0732-8893(01)00345-5
Feßler, A. T., Wang, Y., Wu, C., and Schwarz, S. (2018). Mobile macrolide resistance genes in staphylococci. Plasmid 99, 2–10. doi: 10.1016/j.plasmid.2018.05.001
Foster, T. J. (2017). Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 41, 430–449. doi: 10.1093/femsre/fux007
Galata, V., Fehlmann, T., Backes, C., and Keller, A. (2019). PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res. 47, D195–D202. doi: 10.1093/nar/gky1050
Grundmann, H., Aires-de-Sousa, M., Boyce, J., and Tiemersma, E. (2006). Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368, 874–885. doi: 10.1016/S0140-6736(06)68853-3
Hill-Cawthorne, G. A., Hudson, L. O., El Ghany, M. F. A., Piepenburg, O., Nair, M., Dodgson, A., et al. (2014). Recombinations in staphylococcal cassette chromosome mec elements compromise the molecular detection of methicillin resistance in Staphylococcus aureus. PLoS One 9:e101419. doi: 10.1371/journal.pone.0101419
Hisatsune, J., Hirakawa, H., Yamaguchi, T., Fudaba, Y., Oshima, K., Hattori, M., et al. (2013). Emergence of Staphylococcus aureus carrying multiple drug resistance genes on a plasmid encoding exfoliative toxin B. Antimicrob. Agents Chemother. 57, 6131–6140. doi: 10.1128/AAC.01062-13
Ikonomidis, A., Michail, G., Vasdeki, A., Labrou, M., Karavasilis, V., Stathopoulos, C., et al. (2008). In vitro and in vivo evaluations of oxacillin efficiency against mecA-positive oxacillin-susceptible Staphylococcus aureus. Antimicrob. Agents Chemother. 52, 3905–3908. doi: 10.1128/AAC.00653-08
Jensen, L. B., Garcia-Migura, L., Valenzuela, A. J. S., Løhr, M., Hasman, H., and Aarestrup, F. M. (2010). A classification system for plasmids from enterococci and other Gram-positive bacteria. J. Microbiol. Methods 80, 25–43. doi: 10.1016/j.mimet.2009.10.012
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kwong, S. M., Ramsay, J. P., Jensen, S. O., and Firth, N. (2017). Replication of Staphylococcal Resistance Plasmids. Front. Microbiol. 8:2279. doi: 10.3389/fmicb.2017.02279
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Lüthje, P., von Köckritz-Blickwede, M., and Schwarz, S. (2007). Identification and characterization of nine novel types of small staphylococcal plasmids carrying the lincosamide nucleotidyltransferase gene lnu(A). J. Antimicrob. Chemother. 59, 600–606. doi: 10.1093/jac/dkm008
Mashaly, G. E.-S., and El-Mahdy, R. H. (2017). Vancomycin heteroresistance in coagulase negative Staphylococcus blood stream infections from patients of intensive care units in Mansoura University Hospitals, Egypt. Ann. Clin. Microbiol. Antimicrob. 16:63. doi: 10.1186/s12941-017-0238-5
McCarthy, A. J., and Lindsay, J. A. (2012). The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol. 12:104. doi: 10.1186/1471-2180-12-104
McCarthy, A. J., Loeffler, A., Witney, A. A., Gould, K. A., Lloyd, D. H., and Lindsay, J. A. (2014). Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol. Evol. 6, 2697–2708. doi: 10.1093/gbe/evu214
Montelongo, C., Mores, C. R., Putonti, C., Wolfe, A. J., and Abouelfetouh, A. (2021). Phylogenomic study of Staphylococcus aureus and Staphylococcus haemolyticus clinical isolates from Egypt. bioRxiv [Preprint] doi: 10.1101/2021.05.01.442118
Morikawa, K., Takemura, A. J., Inose, Y., Tsai, M., Nguyen Thi, L. T., Ohta, T., et al. (2012). Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathogens 8:e1003003. doi: 10.1371/journal.ppat.1003003
Otto, M. (2013). Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. BioEssays 35, 4–11. doi: 10.1002/bies.201200112
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Sabat, A. J., Pournaras, S., Akkerboom, V., Tsakris, A., Grundmann, H., and Friedrich, A. W. (2015). Whole-genome analysis of an oxacillin-susceptible CC80 mecA-positive Staphylococcus aureus clinical isolate: insights into the mechanisms of cryptic methicillin resistance. J. Antimicrob. Chemother. 70, 2956–2964. doi: 10.1093/jac/dkv210
Shahkarami, F., Rashki, A., and Ghalehnoo, Z. R. (2014). Microbial susceptibility and plasmid profiles of methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus. Jundishapur J. Microbiol. 7:e16984. doi: 10.5812/jjm.16984
Shearer, J. E. S., Wireman, J., Hostetler, J., Forberger, H., Borman, J., Gill, J., et al. (2011). Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 1, 581–591. doi: 10.1534/g3.111.000760
Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., and Fowler, V. G. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661. doi: 10.1128/CMR.00134-14
Vestergaard, M., Frees, D., and Ingmer, H. (2019). Antibiotic resistance and the MRSA problem. Microbiol. Spectr. 7:GPP3-0057-2018. doi: 10.1128/microbiolspec.gpp3-0057-2018
Wattam, A. R., Davis, J. J., Assaf, R., Boisvert, S., Brettin, T., Bun, C., et al. (2017). Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45, D535–D542. doi: 10.1093/nar/gkw1017
Xu, Z., Shah, H. N., Misra, R., Chen, J., Zhang, W., Liu, Y., et al. (2018). The prevalence, antibiotic resistance and mecA characterization of coagulase negative staphylococci recovered from non-healthcare settings in London, UK. Antimicrob. Resist. Infect. Control 7:73. doi: 10.1186/s13756-018-0367-4
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: antibiotic resistance, Staphylococcus, plasmids, incompatibility group, Rep, horizontal gene transfer
Citation: Mores CR, Montelongo C, Putonti C, Wolfe AJ and Abouelfetouh A (2021) Investigation of Plasmids Among Clinical Staphylococcus aureus and Staphylococcus haemolyticus Isolates From Egypt. Front. Microbiol. 12:659116. doi: 10.3389/fmicb.2021.659116
Received: 27 January 2021; Accepted: 07 May 2021;
Published: 04 June 2021.
Edited by:
Alla Lapidus, Saint Petersburg State University, RussiaReviewed by:
Ben Pascoe, University of Bath, United KingdomCopyright © 2021 Mores, Montelongo, Putonti, Wolfe and Abouelfetouh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alaa Abouelfetouh, QWxhYS5hYm91ZWxmZXRvdWhAcGhhcm1hY3kuYWxleHUuZWR1LmVn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.