
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 July 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.658943
This article is part of the Research Topic Antibiotic Resistance and its Continuity in the Environmental Niche View all 19 articles
The tetracycline resistance gene tet(W) encodes a ribosomal protection protein that confers a low level of tetracycline resistance in the probiotic bacterium Bifidobacterium animalis subsp. lactis. With the aim of assessing its phylogenetic origin and potential mobility, we have performed phylogenetic and in silico genome analysis of tet(W) and its flanking genes. tet(W) was found in 41 out of 44 examined B. animalis subsp. lactis strains. In 38 strains, tet(W) was flanked by an IS5-like element and an open reading frame encoding a hypothetical protein, which exhibited a similar GC content (51–53%). These genes were positioned in the same genomic context within the examined genomes. Phylogenetically, the B. animalis subsp. lactis tet(W) cluster in a clade separate from tet(W) of other species and genera. This is not the case for tet(W) encoded by other bifidobacteria and other species where tet(W) is often found in association with transferable elements or in different genomic regions. An IS5-like element identical to the one flanking the B. animalis subsp. lactis tet(W) has been found in a human gut related bacterium, but it was not associated with any tet(W) genes. This suggests that the IS5-like element is not associated with genetic mobility. tet(W) and the IS5 element have previously been shown to be co-transcribed, indicating that co-localization may be associated with tet(W) expression. Here, we present a method where phylogenetic and in silico genome analysis can be used to determine whether antibiotic resistance genes should be considered innate (intrinsic) or acquired. We find that B. animalis subsp. lactis encoded tet(W) is part of the ancient resistome and thereby possess a negligible risk of transfer.
Antibiotic resistance genes are widely spread among bacteria and they pose a serious threat to human health as they can compromise our ability to treat bacterial infections (World Health Organisation (WHO), 2017). Although the extensive use of antibiotics to treat infections in both humans and animals is considered to be the main reason for the development and spread of resistance genes (Levy and Bonnie, 2004; WHO, 2011), they have been present long before the introduction of antibiotics to the clinic (Martínez, 2008; Allen et al., 2010). Antibiotics are naturally produced by environmental microorganisms and the producers often have “self-resistance” encoded by antibiotic resistance genes located in the antibiotic biosynthesis gene clusters (Martínez, 2008). Some antibiotic resistance genes show homology to housekeeping genes such as those involved in protein synthesis suggesting that they may have evolved from such functions and this could explain their prevalence among bacteria (Martínez, 2008; Allen et al., 2010). Antibiotic resistance genes have mainly been studied in clinically relevant bacteria and often in relation to horizontally transferable elements (Shrivastava et al., 2018). In contrast, less attention has been paid to antibiotic resistance in non-pathogenic bacteria (Klare et al., 2007; Agersø et al., 2019; Campedelli et al., 2019), e.g., bacteria ingested via the food chain.
When products contain viable, non-pathogenic bacteria, e.g., fermented food, probiotics or feed additives, it is a requirement from legal authorities [e.g., European Food Safety Authority (EFSA)] that these bacteria do not possess acquired genes encoding resistance toward antimicrobials, which are considered as highly or critically important for treatment of humans and/or animals by the World Health Organization (WHO) (WHO, 2011; EFSA panel on Additives and Products or Substances used in Animal Feed (FEEDAP), 2018). However, some bacteria are intrinsically resistant to some of the antimicrobials (Peterson and Kaur, 2018). Impermeability of the outer membrane provides resistance to vancomycin for Escherichia coli and other Gram-negative bacteria (Arthur and Courvalin, 1993). Bacillus licheniformis and Bacillus paralicheniformis are resistant (or reduced in susceptibility) to erythromycin, chloramphenicol and streptomycin due to putative intrinsic resistance genes (Agersø et al., 2019).
Thus, homology to a known antibiotic resistance gene does not in itself indicate whether a putative resistance gene is acquired or intrinsic. Therefore, analysis of the genetic context and comparison to other genomes within the same species/subspecies are needed, although exact guidance on this is not provided by EFSA (EFSA panel on Additives and Products or Substances used in Animal Feed (FEEDAP), 2018).
Tetracyclines are broad spectrum antibiotics, which have been used for treatment of infections in humans and animals since the early 1950s and resistance toward tetracyclines is widespread. The tet(W) tetracycline resistance gene encodes a protection protein that attaches to the ribosome and causes an alteration of the ribosomal conformation to which tetracycline cannot bind and therefore protein synthesis can proceed (Chopra and Roberts, 2001; Connell et al., 2003). Genes with more than 80% identity to tet(W) have been found in 19 different genera belonging to both Gram-positive and Gram-negative bacteria and thus, it is the most widely spread tetracycline resistance gene class (Chopra and Roberts, 2001). The first tet(W) gene was reported in Butyrivibrio fibrisolvens located on a Tn B1230-like transposable element, which has spread to several different genera due to the broad host range of the element (Scott et al., 1997; Barbosa et al., 1999). Transfer of tet(W) in association with mobile genetic elements has also been reported to occur at low frequencies in Bifidobacterium longum strain F8 (Kazimierczak et al., 2006), Arcanobacterium pyogenes (Billington et al., 2002) and Streptococcus suis (Palmieri et al., 2011).
Several bifidobacterial species carry tet(W) genes, including B. longum, B. thermophilum and B. bifidum (Ammor et al., 2008). tet(W) is widespread and confers a low level of tetracycline resistance in B. animalis subsp. lactis that varies over three two-fold dilutions between different strains (Gueimonde et al., 2010), which has been suggested to be caused by genetic diversity in the miaA gene encoding for a tRNA dimethylallyltransferase (Milani et al., 2013). Furthermore, bile exposure have been shown to induce tet(W) expression (Gueimonde et al., 2010). The widespread nature of tet(W) suggest that it confers a selective advantage, perhaps a physiological function such as improving translation under the stress conditions of the gut. Although unsuccessful transfer studies are often not published, several studies on transferability of tet(W) from B. animalis subsp. lactis to other bacterial species and genera are published and all were unsuccessful (Gueimonde et al., 2010; Naghizadeh Raeisi et al., 2018; Polit et al., 2018). Bifidobacteria are Gram-positive, anaerobic, non-motile and non-spore-forming bacteria, which are commonly found in the gastrointestinal tract of various animals and humans, the human oral cavity and sewage (Milani et al., 2014). Members of the Bifidobacterium genus are among the first microbes to colonize the human gastrointestinal tract of newborns. Multiple health beneficial effects including reduction of diarrhea, colorectal cancer prevention and inhibition of pathogen growth and adherence have been reported for Bifidobacterium spp. (Turroni et al., 2012; O’Callaghan and van Sinderen, 2016). Therefore, many Bifidobacterium spp. are widely used in probiotic products (Garrigues et al., 2010). B. animalis including B. animalis subsp. lactis have had Qualified Presumption of Safety (QPS) status by EFSA since the establishment of the QPS concept in 2007 (Barlow et al., 2007; Koutsoumanis et al., 2020) and specific strains have acquired the Generally Recognized as Safe (GRAS) status from the Food and Drug Administration (FDA) in the United States (O’Callaghan and van Sinderen, 2016).
The aim of this study was to assess the phylogenetic relationship of tet(W) in B. animalis subsp. lactis through phylogenetic analysis, analysis of the genetic context surrounding the gene and core genome analysis. The study will serve as evidence to further establish that tet(W) in B. animalis subsp. lactis is innate; it originates from the ancestral host and has retained the same genomic position ever since. This supports the common perception that tet(W) should be considered an intrinsic and non-transferable gene in B. animalis subsp. lactis.
All publicly available genome sequences of B. animalis subsp. lactis (50 strains including the type strain DSM 10140) and B. animalis subsp. animalis (8 strains including the type strain ATCC 25527) were downloaded from the NCBI microbe genome database on the 21st of November 2019 (Sayers et al., 2019).
Subspecies identification was either obtained from previously published articles (Lugli et al., 2019) or performed by employing the rpoA and 16S ribosomal DNA sequence. A >98% identity to the type strain genes was used as threshold and the genes should furthermore be different from the type strain of a related subspecies, in this case B. animalis subsp. animalis, as shown through a phylogenetic tree (data not shown).
The sequence quality was assessed and sequences with an average coverage of ≥30× and a contig number below 120 were considered acceptable for phylogenetic analysis. The quality of the genomes was also evaluated by checking that the length of the sequenced genome corresponds with the expected length of the genome, based on the type strain (Milani et al., 2014).
Other bifidobacterial species, which have been shown to harbor tet(W) (Ammor et al., 2008; Wang et al., 2017) were also downloaded from the NCBI microbe genome database on the 21st of November 2019 and included B. longum (14 strains, type strain NCTC11818), B. thermophilum (6 strains, type strain DSM 20212), B. bifidum (11 strains, type strain ATCC 29521), B. pseudolongum (4 strains, type strain DSM 20099), B. pseudocatenulatum (3 strains, type strain DSM 20438) and B. breve (41 strains, type strain NCTC 11815). All tet(W) sequences from other genera where the gene have been described (Scott et al., 1997; Chopra and Roberts, 2001; Flórez et al., 2006; Kazimierczak et al., 2006; Ammor et al., 2008; Palmieri et al., 2011; Schröder et al., 2012) and shared identity to the tet(W) gene found in B. animalis subsp. lactis were also downloaded from NCBI on the 21st of November 2019.
ResFinder (Zankari et al., 2012), with a 80% identity threshold, was used to search for the presence of tet(W) in the examined genomes and the Rapid Annotation using Subsystems Technology (RAST) server with default settings was used to annotate the genomes. The annotated genomes were downloaded in GenBank format from the RAST server (Aziz et al., 2008; Overbeek et al., 2014) and imported to CLC Genomics Workbench 20 (Qiagen Bioinformatics, Aarhus, Denmark), where the presence of tet(W), its flanking genes and presence of mobile genetic elements was examined. tet(W) nucleotide and protein sequences was extracted from the annotated genomes for further phylogenetic analysis. GC content of tet(W) and other genes was assessed by employing the DNA/RNA GC Content Calculator at ENDMEMO (Endmemo, 2020).
The blastN tool available at ISFinder (Siguier et al., 2006) with default settings was used to determine the identity of the mobile genetic protein next to tet(W) in B. animalis subsp. lactis and its sequence was used to search for its presence in other genomic regions in the B. animalis subsp. lactis genomes, which was performed in CLC Genomics Workbench 20 (Qiagen Bioinformatics, Aarhus, Denmark).
The phylogenetic analysis of tet(W) included both the nucleotide and protein sequences from B. animalis subsp. lactis (Supplementary Table 1) and tet(W) genes found in other bifidobacterial species and other genera where the presence of tet(W) previously have been published (Table 1) (Scott et al., 1997; Chopra and Roberts, 2001; Flórez et al., 2006; Kazimierczak et al., 2006; Ammor et al., 2008; Palmieri et al., 2011; Schröder et al., 2012). The nucleotide and protein tet(W) sequences was either extracted from the annotated genomes or from NCBI (Sayers et al., 2019).
ClustalX2 (Larkin et al., 2007) was used to perform a pairwise multiple alignment of the tet(W) sequences (Higgins and Sharp, 1988) and BioEdit (Hall, 1999) was used to remove gaps and unpaired ends. The nucleotide phylogeny was built by evolutionary analysis by the Maximum Likelihood method and Tamura-Nei model by MEGA X (Tamura and Nei, 1993; Kumar et al., 2018) and the amino acid phylogeny was built by evolutionary analysis by Maximum Likelihood method and JTT matrix-based model also by MEGA X (Jones et al., 1992; Kumar et al., 2018). Number of single nucleotide polymorphisms (SNPs) and single amino acid polymorphisms (SAPs) was obtained from the multiple alignment output from MEGA X that was used to build the phylogenetic relationships.
The genomes, either fully assembled or contigs were annotated by Prokka, which annotates genomes through the use of different tools including Prodigal (coding sequences), RNAmmer (Ribosomal RNA genes), Aragorn (Transfer RNA genes), SignalP (Signal leader peptides) and Infernal (Non-coding RNA) (Seemann, 2014). Prokka annotation is a requirement for using Roary, since the .gff file (file containing sequences and annotations) provided by Prokka is used by Roary to create a multi-FASTA alignment of all the core genes (Page et al., 2015). Roary was set to perform nucleotide alignment using MAFFT and a Blastp percentage identity at 80% (Katoh, 2002). FastTree was used to produce an approximately maximum-likelihood phylogenetic tree from the core gene alignment file, which was visualized by MEGA X (Price et al., 2009, 2010; Kumar et al., 2018).
A total of 50 publicly available B. animalis subsp. lactis strains including the type strain DSM 10140 were downloaded from NCBI and consisted either of contigs or assembled genomes (Supplementary Table 1). The sequence quality was assessed and sequences with an average coverage of ≥30 fold and a contig number below 120 were considered acceptable. On this basis, six strains (B420, DS1_2, BI-04, IDCC4301, CF3_2, AD011) were excluded from the study. The genomes of CNCM I-2994 (Chervaux et al., 2011) and AD011 (Kim et al., 2009) had both been sequenced by Sanger shotgun sequencing and consist of complete genomes. However, AD011 has previously been shown to exhibit a poor sequence quality and was therefore excluded (Garrigues et al., 2010), CNCM I-2994 was not excluded from the study. A total of 44 genome sequences were therefore acceptable for further phylogenetic analysis.
The B. animalis subsp. lactis genomes exhibited a size of 1.91–2.08 Mb with a GC content of 60.0–60.6% (Supplementary Table 1), which is in agreement with data for the type strain of the subspecies (Milani et al., 2014).
Subspecies identification was either obtained from previously published articles (Lugli et al., 2019) or performed by analysis of the rpoA and 16S ribosomal DNA sequence.
The majority of the B. animalis subsp. lactis strains originated from human feces, but also from food samples, dietary supplements and domestic pigs, chimpanzees, rabbits, vervet monkeys, a barbary macaque, three different dog breeds and one strain, the genomic unique ATCC 27673 (Loquasto et al., 2013) originated from sewage (Supplementary Table 1). Species within the bifidobacterial genera are commonly found in the gastrointestinal tract of various animals, the human oral cavity and sewage (Milani et al., 2014) and the strains in this study therefore represent the most common habitats of bifidobacteria.
Since B. animalis subsp. lactis is included in a wide range of probiotics, it cannot be excluded that the strains isolated from human feces, domestic pigs and dogs originate from ingested products such as probiotics. However, the strain collection also include strains such as Bl12 that has been isolated from a healthy patient, which has not ingested probiotic products (Milani et al., 2013) and rabbits and monkeys have with high likelihood not been exposed to probiotics and these strains are therefore expected to be diverse from the industrially exploited strains. The genome sizes of the different strains also vary, which also indicate that the strains are diverse (Supplementary Table 1). Most of the strains are isolated or submitted to NCBI between year 2006–2018, which reflect the increased focus on probiotics in the last decades (Gogineni, 2013), while the type strain DSM 10140 originates from 1997 (Supplementary Table 1). However, the submission date of the genome sequences to NCBI does not necessarily reflect the time of isolation as some strains are isolated even earlier.
B. animalis subsp. lactis has previously been shown to be a strict monophyletic bifidobacterial taxon that has recently evolved (Milani et al., 2013), however, some diversity is observed between the strains within the subspecies based on the presence of truly unique genes in some of the strains (Lugli et al., 2019). The strains with the highest number of truly unique genes are also included in this study. It is therefore concluded that the strains included in the current study represent the diversity within the subspecies.
A 1920 bp tet(W) gene flanked by genes annotated as mobile element protein (966 bp), with inverted repeats at both ends of 50 bp and a hypothetical protein (HP) of unknown function (183 bp) was found in the majority of the studied B. animalis subsp. lactis strains (38 out of 44). These genes exhibit similar GC content (51.01–53.23%), which is lower than the flanking genes in the genetic region (52.46–62.25%) (Figure 1) and the average of the genome (60.0–60.6%) (Supplementary Table 1). tet(W) genes found in non-bifidobacterial and bifidobacterial species exhibit a GC content of 52.19–53.18%, indicating that tet(W) genes generally exhibit a GC content around 53%.
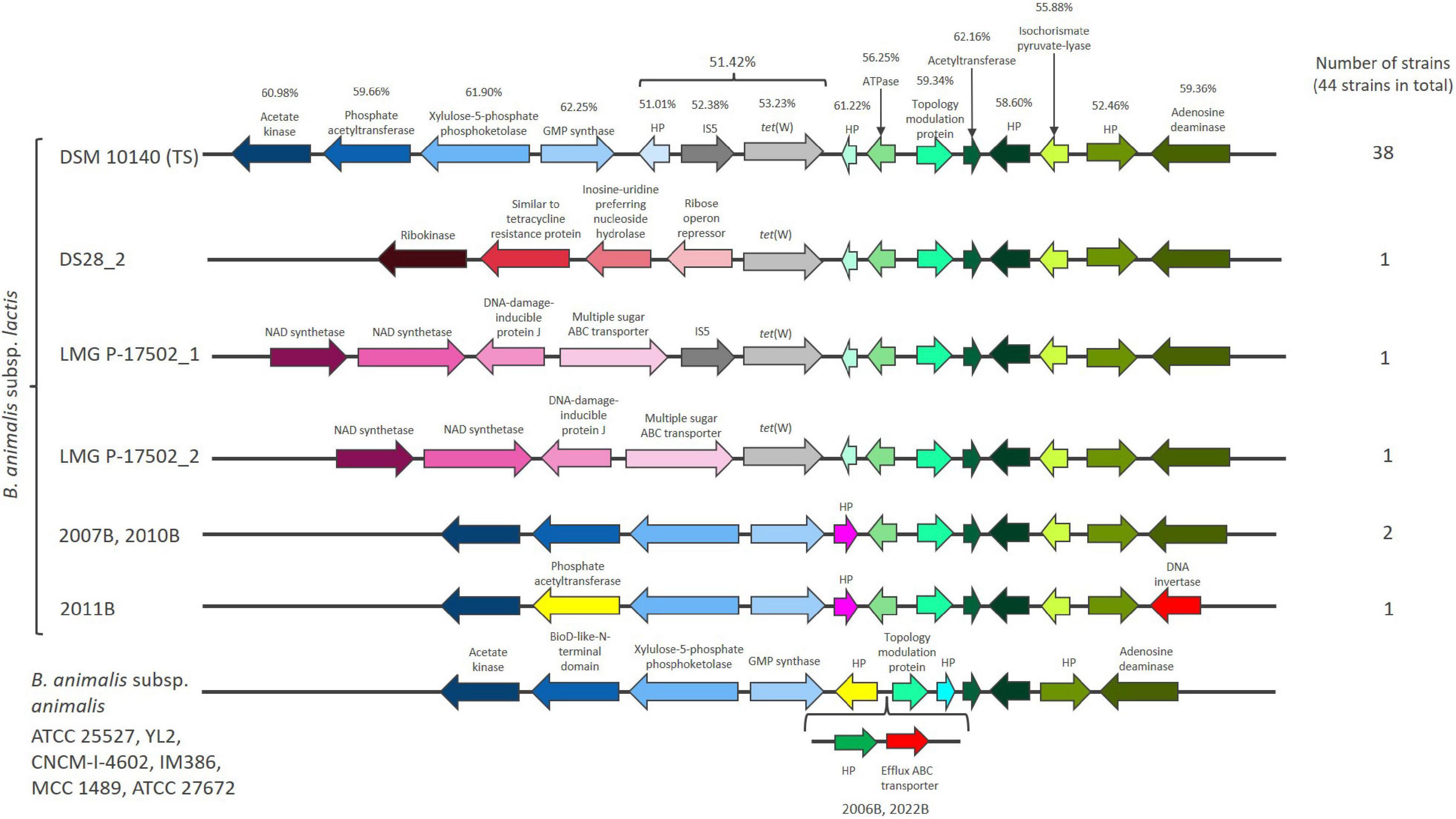
Figure 1. The chromosomal region flanking tet(W) in Bifidobacterium animalis subsp. lactis and the same region in Bifidobacterium animalis subsp. animalis. Hypothetical proteins are designated HP. GC content (%) is provided for the genes found in the B. animalis subsp. lactis type strains (TS) DSM 10140. Genes that are present in the majority of the examined B. animalis subsp. lactis strains (represented by DSM 10140) has the same color in all the shown strains [blue colors downstream of tet(W) and green colors upstream of tet(W)].
The three strains originating from dogs (2007B, 2010B, 2011B) did not encode tet(W), the mobile element protein or the HP (Figure 1 and Supplementary Table 1). Two strains (DS28_2, LMG P-17502_2) only encoded the tet(W) gene, while LMG P-17502 encoded tet(W) and the mobile element protein (Figure 1). UBBLa 70 exhibited a large deletion in the tet(W), with only 117 bp remaining and two strains (ATCC 27673, 1528B) encoded a truncated version of the mobile element protein. This indicate that the three genes have been present originally in B. animalis subsp. lactis but have been subject to deletion in some strains. Despite these differences, the presence of tet(W), the putative mobile element protein and the HP are highly conserved within B. animalis subsp. lactis strains. This conservation was even observed in the strains that are more genomic unique which include ATCC 27673 and 1528B, and the Bl12 strain and the strains isolated from monkeys and rabbits. This suggest that the genetic organization surrounding tet(W) is not only present in the industrially exploited B. animalis subsp. lactis strains.
The tet(W), the mobile element protein and the HP genes were positioned in the same genomic context in the majority of the examined strains, however, in a few strains, alterations downstream (DS28_2, LMG P-17502_1, LMG P-17502_2, 2007B, 2010B, 2011B) and upstream (2011B) (Figure 1) of the three genes were observed. These were the same strains that exhibited complete or partial deletions of the tet(W), the mobile element protein and HP genes.
The genomic position of tet(W) was also reported by Rozman et al. (2020). They suggest that tet(W) and its flanking genes from the HP before the IS element to the HP after isochorismate pyruvate-lyase (Figure 1), based on nucleotide bias and codon usage bias, is part of a putative genomic island that has co-evolved together with B. animalis subsp. lactis and originate from an ancestral host (Guo et al., 2012; Bertelli et al., 2017). The codon usage bias corresponds with the gene GC content being lower in these genes compared to the rest of the genome. Genomic islands are defined as clusters of genes in bacterial genomes of probable horizontal origin and they often provide adaptive traits that has the ability to enhance the fitness of bacteria within a specific niche (Dobrindt et al., 2004). The putative genomic island in B. animalis subsp. lactis encodes for genes involved in cell metabolism and gene regulation and has not been found in other bacteria (Rozman et al., 2020). This could suggest that the putative genomic island including tet(W) encodes for important B. animalis subsp. lactis niche factors, which enable it to survive and compete for nutrients in the gut and has been part of the genome of B. animalis subsp. lactis long before the antibiotic era.
The tet(W), the mobile element protein and the HP gene were absent in all eight B. animalis subsp. animalis strains included in the study (Supplementary Table 1), which otherwise exhibited almost identical gene organization in the genomic region including the genes part of the putative genomic island (Figure 1). This could suggest that the tet(W), the mobile element protein and HP genes have been inserted in an ancestor of the B. animalis subsp. lactis close to subspecies differentiation and most likely lost by the three dog originating strains (2007B, 2010B, 2011B) not carrying tet(W).
The presence of a putative mobile element protein next to tet(W) has previously been reported (Ammor et al., 2008; Gueimonde et al., 2010; Rozman et al., 2020). The sequence encodes a putative DDE transposase gene that is flanked by inverted repeats upstream and downstream of 50 bp, which collectively belong to the insertion sequence (IS) 5-like element ISBian1 family that originate from B. animalis according to ISFinder (Siguier et al., 2006).
DDE transposases are able to catalyze the movement of IS elements and transposons by introducing nicks at each end of the elements (Frost et al., 2005) and are able to move within a genome or horizontally if they are part of mobile genetic element vectors such as plasmids, conjugative transposon and phages (Vandecraen et al., 2017). However, several studies have been unsuccessful in transferring tet(W) from B. animalis subsp. lactis to other species and genera (Gueimonde et al., 2010; Naghizadeh Raeisi et al., 2018; Polit et al., 2018), A BLASTp analysis showed that the IS5-like element ISBian1 family with 99.07% identity was found in the human ileum isolated Angelaksiella massiliensis (Mailhe et al., 2017) and the IS5 element was not associated with tet(W) in this species. The IS5 element was not found in other bifidobacterial species besides B. animalis subsp. lactis. The IS5 element was not found in other positions within the B. animalis subsp. lactis genomes and the inverted repeats flanking the transposase was only flanking the transposase next to tet(W). This indicates that the IS element is stably positioned next to tet(W) and does not mobilize within the B. animalis subsp. lactis genome, which is in accordance with the stable nature of the B. animalis subsp. lactis genome (Morovic et al., 2018).
Besides IS elements involvement in mobilization, IS5 elements are mainly able to modulate the expression of neighboring genes through co-transcription from the transposase promoter located in the terminal inverted repeat if inserted into non-coding regions (Schnetz and Rak, 1992; Luque et al., 2006; Vandecraen et al., 2017). The IS5 element flanking tet(W) in B. animalis subsp. lactis is positioned in a non-coding region meaning it does not cause deletion of other genes (Figure 1) and has previously been shown to be co-transcribed with tet(W) (Gueimonde et al., 2010). This indicates that the IS5 element potentially is involved in modulating the expression of tet(W) rather than mobilization.
All previously published tet(W) genes were included in the analysis. Direct submissions at NCBI also include other tet(W) genes, however, none of these exhibited 100% identity to the subspecies B. animalis subsp. lactis tet(W) and we did not find any variants not represented in the analysis (data not shown). The published tet(W) genes are therefore a good presentation of tet(W).
tet(W) is one of the most widely spread resistance genes and is both found in Gram-positive and -negative bacteria (Chopra and Roberts, 2001). Despite the wide spread nature of tet(W), it was not found to be encoded by all the strains within the examined Gram-positive and -negative species, showing that tet(W) has been acquired by a few strains or lost as compared with B. animalis subsp. lactis where it is a general genetic feature of the subspecies. For both the Gram-positive and -negative bacteria other than B. animalis subsp. lactis, tet(W) was often found to be flanked by mobile genetic elements (Table 1) and in some strains tet(W) was positioned in a genomic region with several mobile genetic elements, e.g., B. longum BG7 and A. pyogenes BBR1. Transfer of tet(W) has been reported for B. longum strain F8 (Kazimierczak et al., 2006), A. pyogenes (Billington et al., 2002), S. suis (Palmieri et al., 2011) and B. fibrosolvens (Scott et al., 1997). Within species, the tet(W) genes in the examined Gram-positive and -negative bacteria were positioned in different genomic regions. Together, this indicates that tet(W) probably has been acquired independently in the examined bacteria in Table 1.
The observation that tet(W) is generally present in B. animalis subsp. lactis strains and is positioned in the same genomic region indicates that tet(W) is conserved and thereby an innate part of the subspecies, while tet(W) only has been acquired by a few strains within the examined Gram-positive and -negative bacterial species.
A phylogenetic analysis was conducted of the tet(W) gene (Supplementary Figure 1) and protein (Figure 2) present in B. animalis subsp. lactis (Supplementary Table 1) and in the examined Gram-positive and -negative bacteria (Table 1).
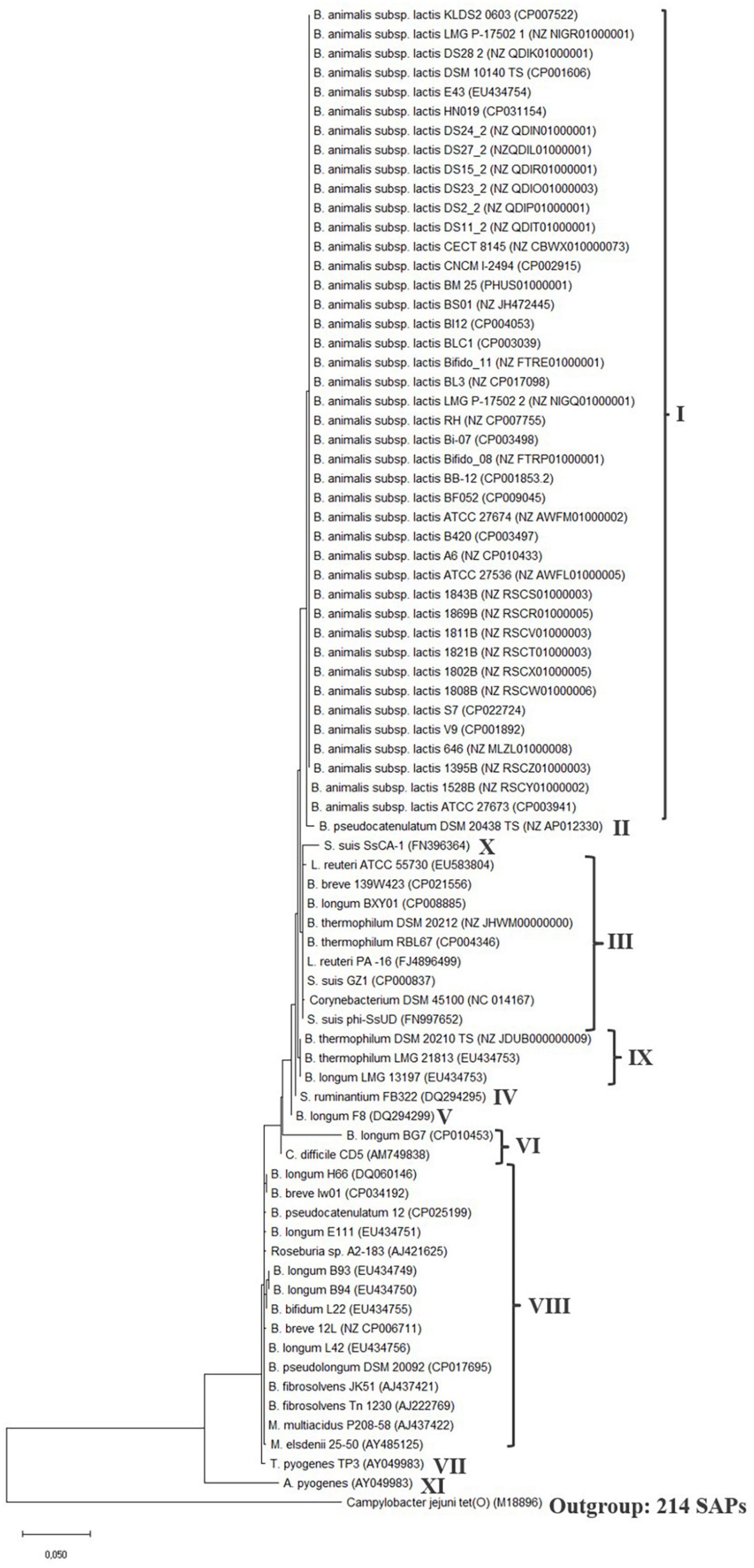
Figure 2. tet(W) protein phylogenetic tree. The tree was built by evolutionary analysis by maximum likelihood method and JTT matrix-based model (Jones et al., 1992; Kumar et al., 2018). The branch lengths are measured in the number of substitutions per site. Strain name and genome or tet(W) gene accession number is provided for the sequences. Type strains (TS) are included for the species, when the type strain encodes tet(W). Clades are defined by the number of SAPs, which can be seen in Table 2. The phylogenetic tree was rooted with the ribosomal protection gene tet(O) from Campylobacter jejuni (M18896) as an outgroup and similar results was obtained with the Streptococcal ribosomal protection gene tet(M) (X04388) (data not shown) (Levy et al., 1999).
The tet(W) genes encoded by the M. elsdenii strains (2–9, 7–11, 4–13) was shorter (1474–1476 bp) and exhibited a GC content (54.61–55.22%) higher compared to the other examined tet(W) genes and was therefore excluded from the phylogenetic analysis. The tet(W) gene of the remaining M. elsdenii strain (25–50) was found to be more similar to the other tet(W) genes and therefore included in the analysis.
Generally, the phylogenetic trees showed a high similarity between the different tet(W) genes and proteins, which is in agreement with previous observations (Aminov and Mackie, 2007), with the number of SNPs ranging from 1 to 46 and single amino acid polymorphisms (SAPs) ranging from 1 to 23 in the coding region compared to the tet(W) genes encoded by B. animalis subsp. lactis. The tet(W) gene encoded by A. pyogenes differed the most from B. animalis subsp. lactis tet(W) (161 SNPs and 69 SAPs). None of the SNPs lead to a premature stop codon. Based on the number of SNPs and SAPs (Table 2), clades were formed in the phylogenetic trees (Figure 2 and Supplementary Figure 1), which follows the phylogeny for B. animalis subsp. lactis but not the other examined Gram-positive and -negative bacteria.
The phylogenetic analysis showed that the tet(W) genes (Supplementary Figure 1) and proteins (Figure 2) from the B. animalis subsp. lactis strains share a high degree of homology and forms a separate clade.
The tet(W) gene and protein in the B. pseudocatenulatum type strain DSM 20438 (Genome GC content 56.40%) was located nearest the B. animalis subsp. lactis tet(W) genes and proteins in the phylogenetic trees and exhibited 12 SNPs and 5 SAPs compared to the tet(W) genes and proteins encoded by B. animalis subsp. lactis. The tet(W) gene encoded by B. pseudocatenulatum DSM 20438 and B. animalis subsp. lactis both exhibit a high identity to tet(W) from S. suis (FN396364). The tet(W) gene encoded by B. pseudocatenulatum strain 12 exhibited 45 SNPs and 22 SAPs and was located in another clade than the DSM 20438 tet(W) gene, indicating that the tet(W) encoded by the two B. pseudocatenulatum strains differ. tet(W) has been shown to be present in 33–41% of B. pseudocatenulatum isolates from human (Aires et al., 2007; Wang et al., 2017), no mobile genetic elements including IS5 elements was found in the flanking regions of tet(W) in the two examined strains (Table 1) and transfer of tet(W) from B. pseudocatenulatum have so far not been shown to occur (Wang et al., 2017). An examination of the flanking sequences of tet(W) in B. pseudocatenulatum type strain DSM 20438 revealed that the downstream genes were organized similarly as the genes downstream of tet(W) in the majority of the studied B. animalis subsp. lactis strains (Figure 1), except that six hypothetical proteins was present between tet(W) and the GMP synthase gene and no IS5-like element was present (Supplementary Figure 2). These genes were also present in B. pseudocatenulatum strain 12 but in another genetic location than tet(W), and in a B. pseudocatenulatum strain (ca_0067, NZ_RCXS00000000) that did not encode tet(W). This indicates that the presence of these genes is independent of the presence of tet(W) and are shared genes between B. animalis subsp. lactis and B. pseudocatenulatum.
The tet(W) genes present in the examined Gram-positive and -negative bacteria including the two B. pseudocatenulatum strains, were scattered over different clades in the phylogenetic tree indicating that the tet(W) genes encoded by these bacteria are diverse, does not follow the phylogeny of the specific species and thereby support the acquired nature of these tet(W) genes.
A core genome phylogenetic analysis was conducted with the examined B. animalis subsp. lactis strains (Supplementary Table 1), the bifidobacterial species from Table 1 and B. animalis subsp. animalis strains from Supplementary Table 1 (Figure 3). For each species, strains were included that both did and did not encode tet(W), except for B. animalis subsp. animalis and B. bifidum.
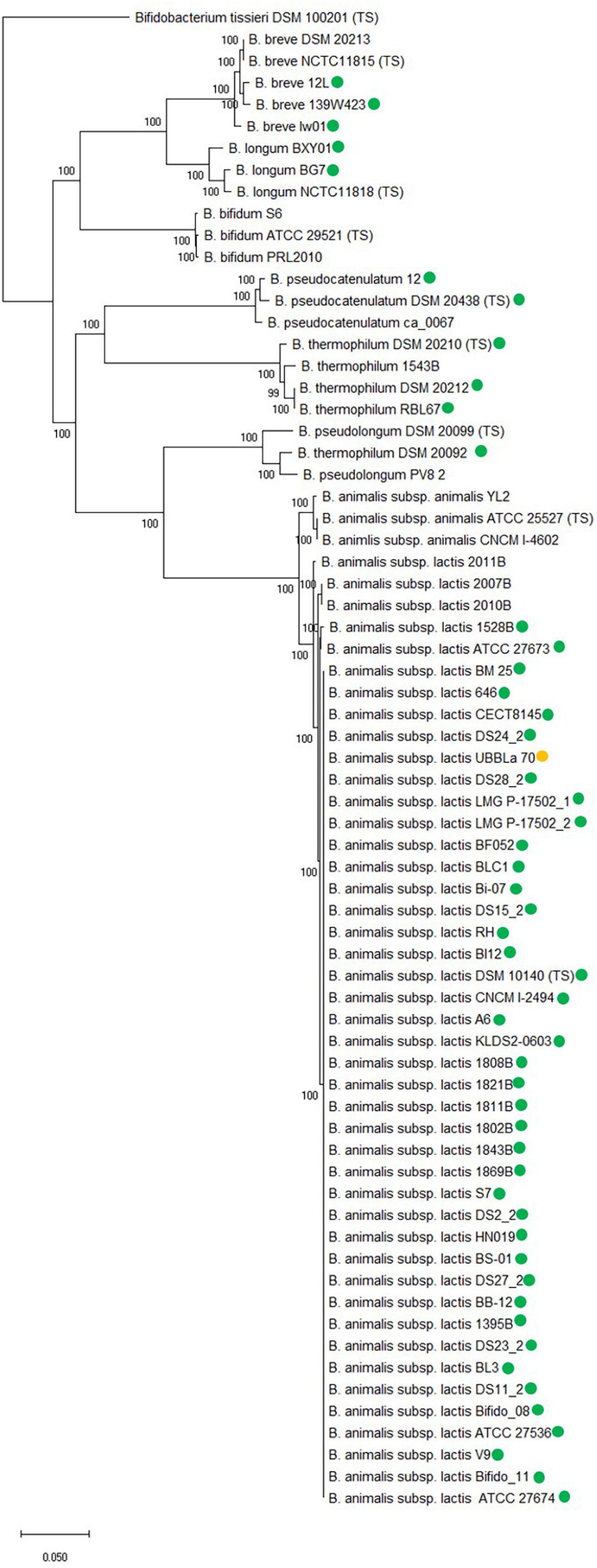
Figure 3. Core genome phylogenetic tree based on 250 core genes which include B. animalis subsp. lactis strains and other related Bifidobacterium species. Type strain has been included for each species, designated TS and strains both with and without tet(W) are included for each species, except for B. animalis subsp. animalis and B. bifidum. tet(W) positive strains are marked with a green circle. B. animalis subsp. lactis UBBLa 70 exhibit a tet(W) gene with large deletions and is marked with a yellow circle. The tree is rooted with the Bifidobacterium tissieri type strain DSM 100201 as an outgroup (Lugli et al., 2018). Bootstrap percentages are shown at node points.
The core genome phylogenetic analysis showed that the bifidobacterial species separated from each other in individual clades and both strains with and without tet(W) clustered together within species, showing that the core genome analysis was able to separate at species and subspecies level.
The fact that the tet(W) gene encoded by the examined B. animalis subsp. lactis strains formed a separate clade in the gene and protein phylogenetic analysis (Supplementary Figure 1 and Figure 2) similar to the one formed in the core genome phylogenetic tree shows that the phylogeny of tet(W) follows the phylogenetic relationship of the subspecies, indicates that tet(W) originates from an ancestral host. This is further supported by the gene being positioned in the same genomic context in the examined strains. For the other examined bifidobacterial species, the tet(W) genes does not follow the phylogeny of the species, indicating that the tet(W) gene has been acquired at different timepoints, which is in line with them being flanked by different mobile genetic elements and positioned in different genomic contexts. This indicates that tet(W) present in B. animalis subsp. lactis is distinct from tet(W) found in other bifidobacterial species and other genera.
The paper presents a method where in silico genome analysis together with phylogenetic analysis can be used to determine whether a gene is innate and thereby not considered a safety concern.
A phylogenetic analysis of tet(W) in B. animalis subsp. lactis, a widely used probiotic bacterium, was performed and shows that tet(W) in this specific subspecies is present in the majority of the strains (41 out of 44), positioned in the same genomic region and is different on the amino acid level from tet(W) genes found in other species. tet(W) is flanked by an IS5-like element, which is known to be present in other human gut related bacteria, however, the IS5-like element was not associated with tet(W) in these bacteria. Previously results show that tet(W) is co-transcribed with the IS5 transposase in B. animalis subsp. lactis, indicating that the expression of tet(W) is regulated by the IS5 transposase. Together with the previous unsuccessful attempts to transfer tet(W), our data suggest that tet(W) is non-transferable and that the flanking IS5 element is not involved in mobilization of tet(W). The phylogenetic analysis showed that tet(W) follows the phylogenetic relationship of the subspecies and is distinct from tet(W) found in other genera and bifidobacterial species.
We conclude that tet(W) in B. animalis subsp. lactis originates from an ancestral host and is therefore an innate part of the subspecies and should be considered as innate (intrinsic) in this subspecies. There is therefore a negligible risk that tet(W) from B. animalis subsp. lactis will add to the pool of mobile resistance genes and thus potentially cause treatment failures in humans and animals.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
KN-M wrote the manuscript, made figures, tables, performed the analysis and was involved in developing the concept and the method. CS was involved in developing the concept, guiding the analysis, discussion, and review and editing. HI was involved in developing the concept, discussion, and review and editing. YA was involved in conceiving the idea, developing and guiding the concept, analysis, design, discussion, and review and editing. All authors have read and approved the submitted manuscript.
This research was funded by Innovation Fund Denmark (Grant no. 9065-00029B) as well as internal funding at Chr. Hansen A/S.
Most authors were employees at Chr. Hansen A/S, a company that produces strains for plant protection, animal and human health as well as for the food and dairy industry. Some of the authors are share-holders in Chr. Hansen A/S. This does not alter our adherence to Frontiers Microbiology policies on sharing data and materials.
We thank Eric Johansen for useful discussions and for contributing with knowledge about Bifidobacterium animalis subsp. lactis. Trademark notice: BB-12® is a trademark of Chr. Hansen A/S.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.658943/full#supplementary-material
Agersø, Y., Bjerre, K., Brockmann, E., Johansen, E., Nielsen, B., Siezen, R., et al. (2019). Putative antibiotic resistance genes present in extant Bacillus licheniformis and Bacillus paralicheniformis strains are probably intrinsic and part of the ancient resistome. PLoS One 14:e0210363. doi: 10.1371/journal.pone.0210363
Aires, J., Doucet-Populaire, F., and Butel, M. J. (2007). Tetracycline resistance mediated by tet(W), tet(M), and tet(O) genes of Bifidobacterium isolates from humans. Appl. Environ. Microbiol. 73, 2751–2754. doi: 10.1128/aem.02459-06
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
Aminov, R. I., and Mackie, R. I. (2007). Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271, 147–161. doi: 10.1111/j.1574-6968.2007.00757.x
Ammor, M. S., Flórez, A. B., Álvarez-Martín, P., Margolles, A., and Mayo, B. (2008). Analysis of tetracycline resistance tet (W) genes and their flanking sequences in intestinal Bifidobacterium species. J. Antimicrob. Chemother. 62, 688–693. doi: 10.1093/jac/dkn280
Arthur, M., and Courvalin, P. (1993). Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 37, 1563–1571. doi: 10.1128/aac.37.8.1563
Aziz, R. K., Bartels, D., Best, A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The rast server: rapid annotations using subsystems technology. BMC Genom. 9:75. doi: 10.1186/1471-2164-9-75
Barbosa, T. M., Scott, K. P., and Flint, H. J. (1999). Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminai bacteria. Environ. Microbiol. 1, 53–64. doi: 10.1046/j.1462-2920.1999.00004.x
Barlow, S., Chesson, A., Collins, J. D., Dybing, E., Flynn, A., Fruijtier-, C., et al. (2007). Introduction of a qualified presumption of safety (QPS) approach for assessment of selected microorganisms referred to efsaopinion of the scientific committee. EFSA J. 5, 1–16.
Bertelli, C., Laird, M. R., Williams, K. P., Lau, B. Y., Hoad, G., Winsor, G. L., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35.
Billington, S. J., Songer, J. G., and Jost, B. H. (2002). Widespread distribution of a tet W determinant among tetracycline-resistant isolates of the animal pathogen Arcanobacterium pyogenes. Antimicrob. Agents Chemother. 46, 1281–1287. doi: 10.1128/aac.46.5.1281-1287.2002
Bottacini, F., Morrissey, R., Roberts, R. J., James, K., Van Breen, J., Egan, M., et al. (2018). Comparative genome and methylome analysis reveals restriction/modification system diversity in the gut commensal Bifidobacterium breve. Nucleic Acids Res. 46, 1860–1877. doi: 10.1093/nar/gkx1289
Campedelli, I., Mathur, H., Salvetti, E., Clarke, S., Rea, M. C., Torriani, S., et al. (2019). Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 85, 1–21.
Chervaux, C., Grimaldi, C., Bolotin, A., Quinquis, B., Legrain-Raspaud, S., van Hylckama Vlieg, J. E. T., et al. (2011). Genome sequence of the probiotic strain Bifidobacterium animalis subsp. lactis CNCM I-2494. J. Bacteriol. 193, 5560–5561. doi: 10.1128/jb.05716-11
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260. doi: 10.1128/mmbr.65.2.232-260.2001
Connell, S. R., Tracz, D. M., Nierhaus, K. H., and Taylor, D. E. (2003). Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob. Agents Chemother. 47, 3675–3681. doi: 10.1128/aac.47.12.3675-3681.2003
Dobrindt, U., Hochhut, B., Hentschel, U., and Hacker, J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424. doi: 10.1038/nrmicro884
EFSA panel on Additives and Products or Substances used in Animal Feed (FEEDAP). (2018). Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 16:e05206
Egervärn, M., Lindmark, H., Olsson, J., and Roos, S. (2010). Transferability of a tetracycline resistance gene from probiotic Lactobacillus reuteri to bacteria in the gastrointestinal tract of humans. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 97, 189–200. doi: 10.1007/s10482-009-9401-0
Egervärn, M., Roos, S., and Lindmark, H. (2009). Identification and characterization of antibiotic resistance genes in Lactobacillus reuteri and Lactobacillus plantarum. J. Appl. Microbiol. 107, 1658–1668.
Endmemo. (2020). DNA/RNA GC Content Calculator. Available online at: http://www.endmemo.com/bio/gc.php
Feßler, A. T., and Schwarz, S. (2017). Antimicrobial Resistance in Corynebacterium spp., Arcanobacterium spp., and Trueperella pyogenes. Microbiol. Spectr. 5.
Flórez, A. B., Ammor, M. S., Álvarez-Martín, P., Margolles, A., and Mayo, B. (2006). Molecular analysis of tet(W) gene-mediated tetracycline resistance in dominant intestinal Bifidobacterium species from healthy humans. Appl. Environ. Microbiol. 72, 7377–7379. doi: 10.1128/aem.00486-06
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Garrigues, C., Johansen, E., and Pedersen, M. B. (2010). Complete genome sequence of Bifidobacterium animalis subsp. lactis BB-12, a widely consumed probiotic strain. J. Bacteriol. 192, 2467–2468. doi: 10.1128/jb.00109-10
Gogineni, V. K. (2013). Probiotics: history and evolution. J. Anc. Dis. Prev. Remedies 1, 1–7. doi: 10.1007/978-94-011-2364-8_1
Gueimonde, M., Flórez, A. B., Van Hoek, A. H. A. M., Stuer-Lauridsen, B., Strøman, P., De Los Reyes-Gavilán, C. G., et al. (2010). Genetic basis of tetracycline resistance in Bifidobacterium animalis subsp. lactis. Appl. Environ. Microbiol. 76, 3364–3369. doi: 10.1128/aem.03096-09
Guo, F. B., Wei, W., Wang, X. L., Lin, H., Ding, H., Huang, J., et al. (2012). Co-evolution of genomic islands and their bacterial hosts revealed through phylogenetic analyses of 17 groups of homologous genomic islands. Genet. Mol. Res. 11, 3735–3743. doi: 10.4238/2012.october.15.5
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Higgins, D. G., and Sharp, P. M. (1988). CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73, 237–244. doi: 10.1016/0378-1119(88)90330-7
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Katoh, K. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kazimierczak, K. A., Flint, H. J., and Scott, K. P. (2006). Comparative analysis of sequences flanking tet(W) resistance genes in multiple species of gut bacteria. Antimicrob. Agents Chemother. 50, 2632–2639. doi: 10.1128/aac.01587-05
Kim, J. F., Jeong, H., Yu, D. S., Choi, S. H., Hur, C. G., Park, M. S., et al. (2009). Genome sequence of the probiotic bacterium bifidobacterium animalis subsp. lactis AD011. J. Bacteriol. 191, 678–679. doi: 10.1128/jb.01515-08
Klare, I., Konstabel, C., Werner, G., Huys, G., Vankerckhoven, V., Kahlmeter, G., et al. (2007). Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J. Antimicrob. Chemother. 59, 900–912. doi: 10.1093/jac/dkm035
Koutsoumanis, K., Allende, A., Alvarez-Ordóñez, A., Bolton, D., Bover-Cid, S., Chemaly, M., et al. (2020). Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 11: suitability of taxonomic units notified to EFSA until September 2019. EFSA J. 18:5965.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kwon, S. K., Kwak, M. J., Seo, J. G., Chung, M. J., and Kim, J. F. (2015). Complete genome sequence of Bifidobacterium longum KCTC 12200BP, a probiotic strain promoting the intestinal health. J. Biotechnol. 214, 169–170. doi: 10.1016/j.jbiotec.2015.09.039
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Levy, S. B., and Bonnie, M. (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10, S122–S129.
Levy, S. B., McMurry, L. M., Barbosa, T. M., Burdett, V., Courvalin, P., Hillen, W., et al. (1999). Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 43, 1523–1524. doi: 10.1128/aac.43.6.1523
Loquasto, J. R., Barrangou, R., Dudley, E. G., Stahl, B., Chen, C., and Roberts, R. F. (2013). Bifidobacterium animalis subsp. lactis ATCC 27673 Is a genomically unique strain within its conserved subspecies. Appl. Environ. Microbiol. 79, 6903–6910. doi: 10.1128/aem.01777-13
Lugli, G. A., Mancino, W., Milani, C., Duranti, S., Mancabelli, L., Napoli, S., et al. (2019). Dissecting the evolutionary development of the species bifidobacterium animalis through comparative genomics analyses. Appl. Environ. Microbiol. 85, 1–16.
Lugli, G. A., Milani, C., Duranti, S., Mancabelli, L., Mangifesta, M., Turroni, F., et al. (2018). Tracking the taxonomy of the genus Bifidobacterium based on a phylogenomic approach. Appl. Environ. Microbiol. 84, 1–14.
Luque, I., Andújar, A., Jia, L., Zabulon, G., De Marsac, N. T., Flores, E., et al. (2006). Regulated expression of glutamyl-tRNA synthetase is directed by a mobile genetic element in the cyanobacterium Tolypothrix sp. PCC 7601. Mol. Microbiol. 60, 1276–1288. doi: 10.1111/j.1365-2958.2006.05170.x
Mailhe, M., Ricaboni, D., Vitton, V., Cadoret, F., Fournier, P. E., and Raoult, D. (2017). ‘Angelakisella massiliensis’ gen. nov., sp. nov., a new bacterial species isolated from human ileum. New Microbes. New Infect. 16, 51–53. doi: 10.1016/j.nmni.2017.01.003
Martínez, J. L. (2008). Antibiotics and antibiotic resistance genes in natural environments. Science 321, 365–367. doi: 10.1126/science.1159483
Milani, C., Duranti, S., Lugli, G. A., Bottacini, F., Strati, F., Arioli, S., et al. (2013). Comparative genomics of Bifidobacterium animalis subsp. lactis reveals a strict monophyletic bifidobacterial taxon. Appl. Environ. Microbiol. 79, 4304–4315. doi: 10.1128/aem.00984-13
Milani, C., Lugli, G. A., Duranti, S., Turroni, F., Bottacini, F., Mangifesta, M., et al. (2014). Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl. Environ. Microbiol. 80, 6290–6302. doi: 10.1128/aem.02308-14
Morita, H., Toh, H., Oshima, K., Nakano, A., Arakawa, K., Takayama, Y., et al. (2015). Complete genome sequence of Bifidobacterium pseudocatenulatum JCM 1200T isolated from infant feces. J. Biotechnol. 210, 68–69. doi: 10.1016/j.jbiotec.2015.06.416
Morovic, W., Roos, P., Zabel, B., Hidalgo-cantabrana, C., Kiefer, A., and Barrangou, R. (2018). Transcriptional and functional analysis of Bifidobacterium animalis subsp. lactis exposure to tetracycline. Appl. Environ. Microbiol. 84:e01999-18.
Naghizadeh Raeisi, S., Ghoddusi, H. B., Juncker Boll, E., Farahmand, N., Stuer-Lauridsen, B., Johansen, E., et al. (2018). Antimicrobial susceptibility of bifidobacteria from probiotic milk products and determination of the genetic basis of tetracycline resistance in Enterococcus species after in vitro conjugation with Bifidobacterium animalis subsp. lactis. Food Control 94, 205–211. doi: 10.1016/j.foodcont.2018.07.016
O’Callaghan, A., and van Sinderen, D. (2016). Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 7:925.
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The seed and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, 206–214.
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Palmieri, C., Princivalli, M. S., Brenciani, A., Varaldo, P. E., and Facinelli, B. (2011). Different genetic elements carrying the tet(W) gene in two human clinical isolates of Streptococcus suis. Antimicrob. Agents Chemother. 55, 631–636. doi: 10.1128/aac.00965-10
Peterson, E., and Kaur, P. (2018). Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 9:2928. doi: 10.3389/fmicb.2018.02928
Polit, A., Yang, H., and Amund, D. (2018). Investigating the transmissibility of tet(W) in bifidobacteria exposed to acid and bile stress. Biosci. Microbiota Food Heal 37, 39–43. doi: 10.12938/bmfh.17-017
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). Fast tree 2approximately maximum-likelihood trees for large alignments. PLoS One 5:9490. doi: 10.1371/journal.pone.0009490
Rbl, S., Jans, C., Lacroix, C., Follador, R., and Stevens, M. J. A. (2013). Complete genome sequence of the probiotic Bifidobacterium thermophilum strain RBL67. Genome Announc. 1:e00191-13.
WHO (2011). World Health Organization. Tackling Antibiotic Resistance from a Food Safety Perspective in Europe. Copenhagen: Copenhagen World Heal Organ, 1–88.
Rozman, V., Mohar Lorbeg, P., Accetto, T., and Bogovič Matijašć, B. (2020). Characterization of antimicrobial resistance in lactobacilli and bifidobacteria used as probiotics or starter cultures based on integration of phenotypic and in silico data. Int. J. Food Microbiol. 314:108388. doi: 10.1016/j.ijfoodmicro.2019.108388
Sayers, E. W., Agarwala, R., Bolton, E. E., Brister, J. R., Canese, K., Clark, K., et al. (2019). Database resources of the national center for biotechnology information. Nucleic Acids Res. 47, D23–D28.
Schnetz, K., and Rak, B. (1992). IS5: a mobile enhancer of transcription in Eschericia coli. Proc. Natl. Acad. Sci. U.S.A. 89, 1244–1248. doi: 10.1073/pnas.89.4.1244
Schröder, J., Maus, I., Meyer, K., Wördemann, S., Blom, J., Jaenicke, S., et al. (2012). Complete genome sequence, lifestyle, and multi-drug resistance of the human pathogen Corynebacterium resistens DSM 45100 isolated from blood samples of a leukemia patient. BMC Genomics 13:141. doi: 10.1186/1471-2164-13-141
Scott, K. P., Barbosa, T. M., Forbes, K. J., and Flint, H. J. (1997). High-frequency transfer of a naturally occurring chromosomal tetracycline resistance element in the ruminal anaerobe Butyrivibrio fibrisolvens. Appl. Environ. Microbiol. 63, 3405–3411. doi: 10.1128/aem.63.9.3405-3411.1997
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shrivastava, S. R., Shrivastava, P. S., and Ramasamy, J. (2018). World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. JMS J. Med. Soc. 32, 76–77. doi: 10.4103/jms.jms_25_17
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36.
Spigaglia, P., Barbanti, F., and Mastrantonio, P. (2008). Tetracycline resistance gene tet(W) in the pathogenic bacterium Clostridium difficile. Antimicrob. Agents Chemother. 52, 770–773. doi: 10.1128/aac.00957-07
Stanton, T. B., and Humphrey, S. B. (2003). Isolation of tetracycline-resistant Megasphaera elsdenii strains with novel mosaic gene combinations of tet(O) and tet(W) from swine. Appl. Environ. Microbiol. 69, 3874–3882. doi: 10.1128/aem.69.7.3874-3882.2003
Sun, Z., Zhang, W., Guo, C., Yang, X., Liu, W., Wu, Y., et al. (2015). Comparative genomic analysis of 45 type strains of the genus bifidobacterium: a snapshot of its genetic diversity and evolution. PLoS One 10:e0117912. doi: 10.1371/journal.pone.0117912
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526.
Turroni, F., Peano, C., Pass, D. A., Foroni, E., Severgnini, M., Claesson, M. J., et al. (2012). Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi: 10.1371/journal.pone.0036957
Vandecraen, J., Chandler, M., Aertsen, A., and Van Houdt, R. (2017). The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 43, 709–730. doi: 10.1080/1040841x.2017.1303661
Wang, L., Wang, Y., Li, Q., Tian, K., Xu, L., Liu, G., et al. (2019). Exopolysaccharide, isolated from a novel strain Bifidobacterium breve lw01 possess an anticancer effect on head and neck cancer - genetic and biochemical evidences. Front. Microbiol. 10:1044. doi: 10.3389/fmicb.2019.01044
Wang, N., Hang, X., Zhang, M., Liu, X., and Yang, H. (2017). Analysis of newly detected tetracycline resistance genes and their flanking sequences in human intestinal bifidobacteria. Sci. Rep. 7, 1–10.
World Health Organisation (WHO). (2017). Global Action Plan on Antimicrobial Resistance. Switzerland: WHO, 1–28.
Keywords: antimicrobial, antibiotic, resistance evolution, non-pathogenic bacteria, ribosomal protection, intrinsic resistance
Citation: Nøhr-Meldgaard K, Struve C, Ingmer H and Agersø Y (2021) The Tetracycline Resistance Gene, tet(W) in Bifidobacterium animalis subsp. lactis Follows Phylogeny and Differs From tet(W) in Other Species. Front. Microbiol. 12:658943. doi: 10.3389/fmicb.2021.658943
Received: 26 January 2021; Accepted: 27 May 2021;
Published: 15 July 2021.
Edited by:
Magdalena Rzewuska, Warsaw University of Life Sciences, PolandReviewed by:
Wanderson Marques da Silva, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaCopyright © 2021 Nøhr-Meldgaard, Struve, Ingmer and Agersø. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yvonne Agersø, REtZVkFHQGNoci1oYW5zZW4uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.