
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 17 June 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.658374
This article is part of the Research TopicBacteriophages Isolation From The Environment And Their Antimicrobial Therapeutic Potential, Volume 2View all 12 articles
Due to concerns over the global increase of antibiotic-resistant bacteria, alternative antibacterial strategies, such as phage therapy, are increasingly being considered. However, evolution of bacterial resistance to new therapeutics is almost a certainty; indeed, it is possible that resistance to alternative treatments might result in an evolved trade-up such as enhanced antibiotic resistance. Here, we hypothesize that selection for Escherichia coli bacteria to resist phage T6, phage U115, or albicidin, a DNA gyrase inhibitor, should often result in a pleiotropic trade-up in the form of cross-resistance, because all three antibacterial agents interact with the Tsx porin. Selection imposed by any one of the antibacterials resulted in cross-resistance to all three of them, in each of the 29 spontaneous bacterial mutants examined in this study. Furthermore, cross-resistance did not cause measurable fitness (growth) deficiencies for any of the bacterial mutants, when competed against wild-type E. coli in both low-resource and high-resource environments. A combination of whole-genome and targeted sequencing confirmed that mutants differed from wild-type E. coli via change(s) in the tsx gene. Our results indicate that evolution of cross-resistance occurs frequently in E. coli subjected to independent selection by phage T6, phage U115 or albicidin. This study cautions that deployment of new antibacterial therapies such as phage therapy, should be preceded by a thorough investigation of evolutionary consequences of the treatment, to avoid the potential for evolved trade-ups.
As global concerns grow over the widespread emergence of antibiotic-resistant bacteria, attention has increasingly turned to antibiotic alternatives such as phage therapy: the use of bacteria-specific viruses, bacteriophages (phages), to treat bacterial infections. Although phage therapy is frequently seen as a novel medical technology, the approach originated in the early 20th century soon after phages were discovered (d’Herelle, 1917, 1930). However, roughly a decade after the discovery of phages, penicillin was discovered and focus shifted instead to research and deployment of antibiotics (Fleming, 1929). Recently, Western medicine’s interest in phage therapy has resurged as a tool for treating antibiotic resistant infections (Kortright et al., 2019); however, just as antibiotics select for the evolution of antibiotic resistance, phages select for the evolution of phage resistance (Luria and Delbruck, 1943; Azam and Tanji, 2019). To avoid the historical mistakes that resulted in multi- and even pan-drug resistant strains, the evolutionary consequences of phage therapy need to be considered and investigated prior to its widespread use (Cohan et al., 2020).
While the evolution of phage resistance in bacteria is perhaps inevitable, phage therapy strategies can be devised to leverage the evolution of phage resistance as an asset, rather than a limitation (Kortright et al., 2019). In particular, by using lytic phages that interact with bacterial virulence factors and molecules that confer antibiotic resistance, these phages should kill bacteria while selecting for phage resistance that may compromise—or pleiotropically “trade-off” with—virulence or antibiotic-resistance traits (Kortright et al., 2019). Theory, in vitro experiments and emergency patient treatment provide evidence that certain phages can direct favorable “trade-off” outcomes, where the therapy kills the target pathogen while selecting for reduced virulence or increased antibiotic sensitivity in the remaining bacterial population (Chan et al., 2018; Gurney et al., 2020; Rodriguez-Gonzalez et al., 2020). This approach exemplifies one goal of evolutionary medicine: applying “evolution thinking” to improve the effectiveness of therapy (Stearns, 2012; Turner, 2016).
However, not all bacterial mutations for phage resistance may constitute a genetic change that alters fitness to create a costly trade-off (Burmeister and Turner, 2020). Instead, the opposite may happen whereby evolution of phage resistance results in an additional fitness gain, also known as a “trade-up.” Indeed, it is possible that the fitness effects of evolved phage resistance might pleiotropically trade-up with virulence or antibiotic resistance (Moulton-Brown and Friman, 2018; Burmeister et al., 2020). To examine this possibility, here we studied the interactions of Escherichia coli with two lytic phages and an antibiotic that all require the Tsx porin to enter the cell. Tsx is a substrate-specific outer-membrane porin, which uptakes nucleosides and deoxynucleotides into the periplasmic space (Maier et al., 1988). Phages T6 and U115 are shown to use the Tsx porin as a receptor for binding to E. coli (Kortright et al., 2020). Additionally, the Tsx porin uptakes the antibiotic albicidin, a phytotoxic DNA-gyrase inhibitor with clinical potential produced by Xanthomonas albilineans, an agriculturally important pathogen that causes leaf scald in sugar cane grasses (Birch and Patil, 1987; Birch et al., 1990; Rott et al., 1996; Hashimi et al., 2007; Kretz et al., 2015; von Eckardstein et al., 2017; Rostock et al., 2018; Hashimi, 2019). Earlier studies examined whether phage T6 or albicidin selected for cross resistance of E. coli to the other antibacterials, and found mixed evidence for this idea (Fsihi et al., 1993; Schneider et al., 1993); but these studies looked at small numbers of resistant mutants in a tsx knockout background with exogenous expression of Tsx from a plasmid, making it difficult to discern whether or not evolution of cross-resistance would be a general outcome. Here, we isolated larger collections of spontaneous mutants to test the hypothesis that selection for E. coli resistance to phage T6, phage U115, or albicidin should tend to produce cross-resistance to the other antibacterial agents, due to converging selection at the tsx locus.
Our results confirmed that the predicted pleiotropic trade-ups evolved frequently; selection exerted by any one of the antibacterials led to perfect (100%) cross-resistance of E. coli mutants to all three antibacterials. Moreover, we observed that cross-resistance was generally “cost-free” in the absence of phage and antibiotic selection, evidenced by equivalent growth of bacterial mutants relative to their wild-type ancestor in both high- and low-resource laboratory environments. In addition, we used sequence analysis to show that a wide variety of mutations at the tsx locus of E. coli may govern cross-resistance. Our study suggests that prior characterization of evolutionary consequences of antibacterial treatments, particularly the mechanistic interactions of lytic phages and antibiotics with target bacteria, can be used to inform treatment strategies that potentially avoid the evolution of undesired trade-ups.
Because phages T6 and U115 both rely on the Tsx porin for cell-binding, we predicted that evolution of resistance to one phage should often lead to cross-resistance against the other phage. To test this idea, we used a classic fluctuation analysis (see “Materials and Methods”) to obtain a collection of spontaneous mutants of E. coli that were resistant to each phage individually. We used a paired approach, where each of 10 independently-grown bacterial cultures were used to isolate one T6-resistant and one U115-resistant strain (20 mutants total). Results for efficiency of plaquing (EOP) assays (see “Materials and Methods”) confirmed that growth of phage T6 on each of the 10 T6-resistant mutants (T6R1 through T6R10) was below the limit of detection, compared to normal infectivity of the phage on wild-type bacteria (Supplementary Table 1). Similarly, EOP experiments showed that phage U115 was unable to grow on each of the 10 U115-resistant mutants (U115R1 through U115R10), relative to expected infectivity on the wild-type (Supplementary Table 1). Furthermore, we found positive support for our hypothesis; in all 20 cases, evolved bacterial resistance to phage T6 provided cross-resistance to phage U115 using EOP assays, and vice versa (Supplementary Table 1). We concluded that independent selection for E. coli resistance to one phage provided cross-resistance to the other phage.
We then used antibiotic-resistance assays (see “Materials and Methods”) to test the prediction that evolution of phage resistance should often lead to cross-resistance to the antibiotic albicidin. We first used E. coli strains BW25113 and BW25113ΔicdC as two positive controls, to confirm that these albicidin-sensitive bacteria fail to form confluent lawns (i.e., they show zones of inhibited growth) when exposed to albicidin-producing X. albilineans strain XA23, but grow normally on X. albilineans strain LS126 which does not produce albicidin. Results (Figure 1) showed that the controls behaved as expected, with zones of growth inhibition around XA23 indicating sensitivity of both bacterial strains to albicidin. We estimated that wild-type BW25113 had a mean clearing ratio (a ratio of the zone of clearing divided by the area of the X. albilineans spot) of 4.88 ± 0.47 s.d. in the presence of XA23 albicidin-producing bacteria, and that BW25113ΔicdC bacteria showed a mean clearing ratio of 5.094 ± 0.524 s.d. (Supplementary Figure 1A). As a negative control, the Tsx knockout, BW25113Δtsx, had no zone of inhibited growth on either XA23 or LS126 (Figure 1); in both cases the mean clearing ratio of BW25113Δtsx was 1.0 ± 0.0 s.d. (Supplementary Figure 1A). A test of the hypothesis confirmed our prediction was correct; all 20 phage-resistant mutants showed no zones of growth inhibition around XA23 or LS126 (Figure 1 and Supplementary Figure 1B) and presented mean clearing ratios of 1.0 ± 0.0 s.d. on XA23 and LS126 (Supplementary Figure 1A), indicating that all the phage-resistant mutants were completely resistant to albicidin as well.
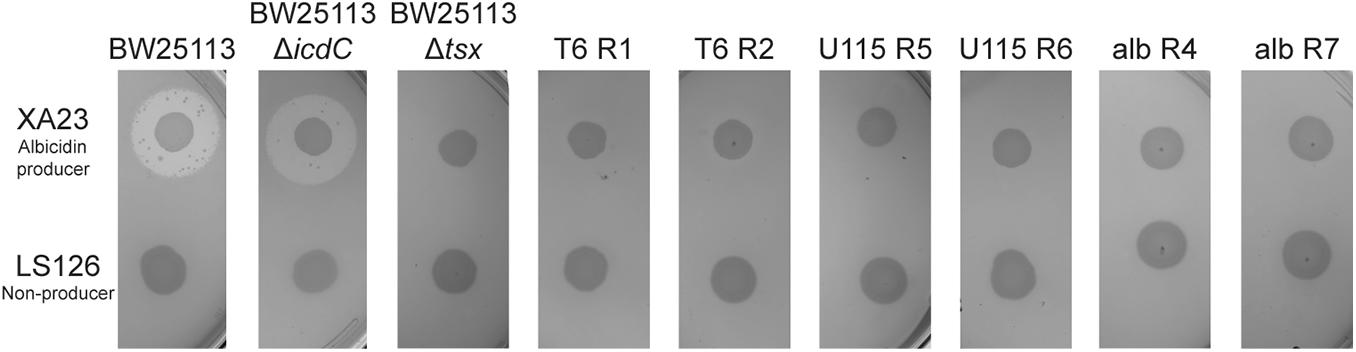
Figure 1. Tests of albicidin resistance for representative spontaneous mutants of E. coli, selected for resistance to phage T6, phage U115, or albicidin. Wild-type BW25113 and the positive-control BW25113ΔicdC showed inhibited growth on agar plates with albicidin-producing X. albilineans (XA23), and normal growth on the non-producer, LS126. In contrast, mutants (T6R1, T6R2, U115R5, U115R6, albR4, and albR7) and negative-control BW25113Δtsx grew equally well in the presence/absence of albicidin-producing bacteria.
We used albicidin selection (see “Materials and Methods”) to isolate nine independent spontaneous mutants of E. coli (albR1 through albR9). Using the above-described growth challenges on X. albilineans strains XA23 and LS126, our results confirmed that each mutant was albicidin resistant (Figure 1 and Supplementary Figures 1A,B). We then tested whether the albicidin-resistant mutants were cross-resistant to infection with phage T6 and phage U115. Results showed that in all 9 strains tested, acquisition of albicidin resistance conferred cross-resistance to both phages T6 and U115 (Supplementary Table 1). We concluded that the evolution of cross-resistance was absolute in this study system, such that evolution of resistance to one of the three antibacterials provided perfect cross-resistance to all of the selective agents.
To further examine the evolution of antibacterial cross-resistance, bacterial growth assays (see “Materials and Methods”) were performed with replication (n = 3) in LB medium. Controls confirmed that wild-type E. coli strain BW25113 showed no discernable growth in the presence of either phage T6 or U115 (Figure 2A), and that growth of the tsx-knockout strain, BW25113Δtsx, was similar in the presence and absence of each phage (Figure 2B). In contrast, all 10 of the T6-resistant mutants grew normally in the presence and absence of phage T6 (representative data shown in Figures 2C,D; see Supplementary Figures 2B–I for all results). Similarly, growth of all 10 U115-resistant mutants was unaffected by presence or absence of phage U115 (representative data in Figures 2E,F; see Supplementary Figures 2J–Q for all results). Lastly, the 9 albicidin-resistant mutants were capable of approximately equivalent growth in the presence and absence of either phage T6 or U115 (representative data in Figures 2G,H; see Supplementary Figures 2R–X for all results).
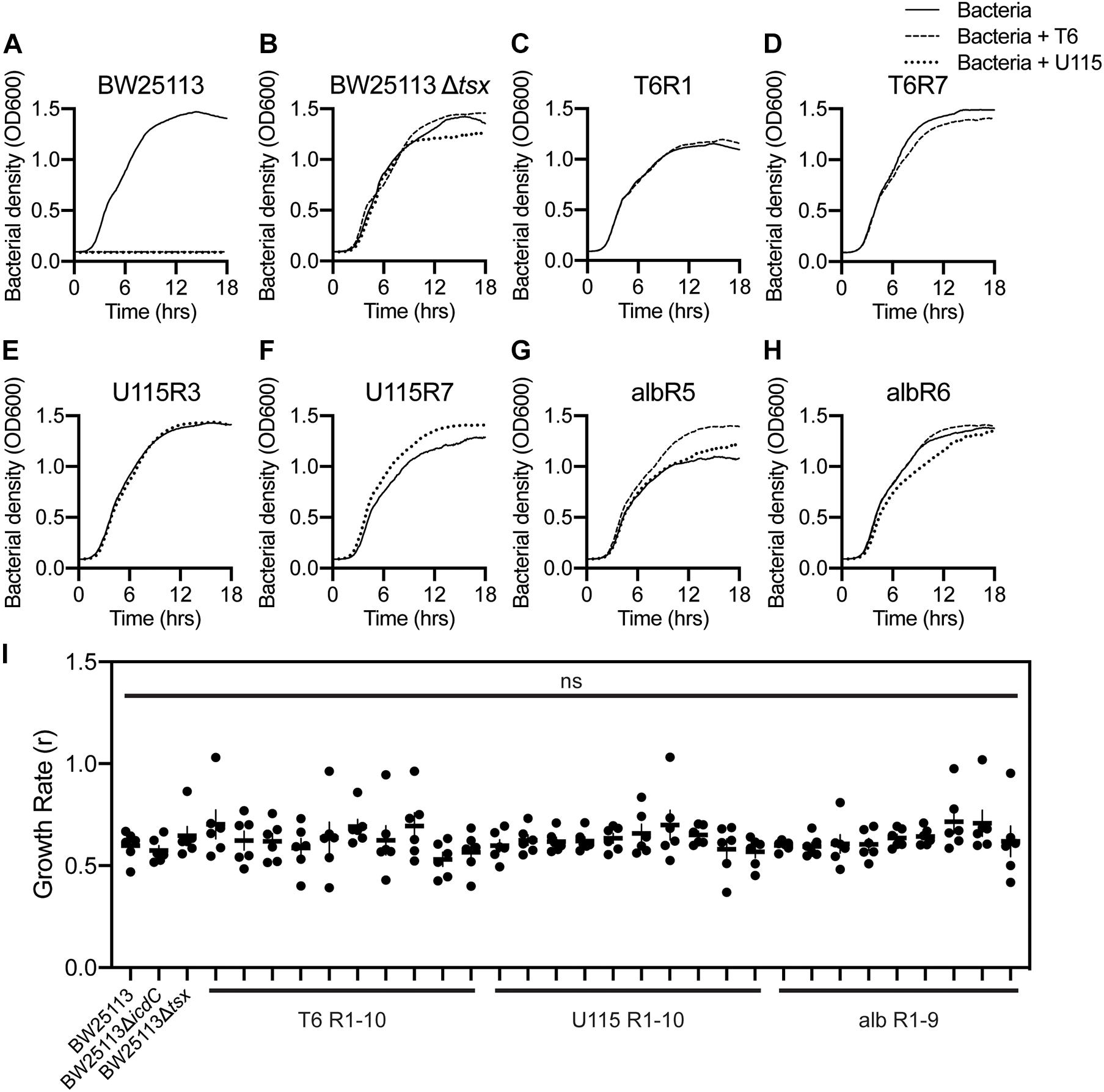
Figure 2. Growth dynamics of representative E. coli mutants selected for resistance to phage T6, phage U115, or albicidin, in environments with and without each phage. (A,B) Wild-type BW25113 bacteria grew only in phage absence (solid line), whereas Tsx knockout strain BW25113Δtsx grew similarly in phage-free as well as T6 (dashed line) and U115 (dotted line) environments. (C–H) Each of the representative resistant mutants grew similarly in the presence/absence of phages T6 and U115, regardless of their prior selection for phage or albicidin resistance. (I) Intrinsic growth rate (r; proxy for fitness) estimates from growth-curve data [(A–H) and Supplementary Figure 2] showed that each strain did not differ statistically from BW25113 (P > 0.05; unpaired t-tests).
A visual comparison of the growth-curve results suggested that all the resistant mutants grew similarly to wild-type strain BW25113 in the absence of each phage (Figure 2 and Supplementary Figure 2). To examine this outcome more closely, we analyzed the growth data with GrowthCurver (Sprouffske and Wagner, 2016) to estimate intrinsic growth rate (r) as a proxy for bacterial fitness (Figure 2I). None of the resistant mutants had an r that differed significantly from that of BW25113. These results suggested that there are no appreciable growth costs associated with bacterial evolution of resistance to phage T6, phage U115 and albicidin. While resistance to any of the antibacterials did not affect bacterial fitness under the tested conditions, we could not eliminate the possibility that our growth assays failed to detect minor fitness differences.
Thus, we conducted additional experiments, in an attempt to measure more subtle fitness differences among bacterial strains. To do so, we performed replicated (n = 3) competition assays (see “Materials and Methods”) for each test strain to gauge its fitness relative to a genetically-marked wild-type strain (BW25113ΔicdC) in the absence of phage and antibiotic selection. Competitive indexes (CIs) were calculated as the ratio of resistant-mutant colony forming units (CFU) to CFU of the common competitor strain, BW25113ΔicdC, to the ratio of BW25113ΔicdC CFU to wild-type, BW25113, CFU. Each ratio was normalized by the starting ratio of each competing strain. Results showed no significant differences in CIs of the resistant mutants as compared to the common competitor when competed in a resource-rich complex LB medium for 72 h (Figure 3). Furthermore, when competed in a resource-poor (minimal glucose) defined M9 medium for 24 h (Figure 3), all of the resistant mutants have CIs higher than the CI of BW25113ΔicdC with albR4 having the only statistically significant CI. Therefore, it appeared that all T6-resistant, U115-resistant and albicidin-resistant mutants suffered no growth deficits, compared to wild-type bacteria, in the absence of phage and antibiotic selection.
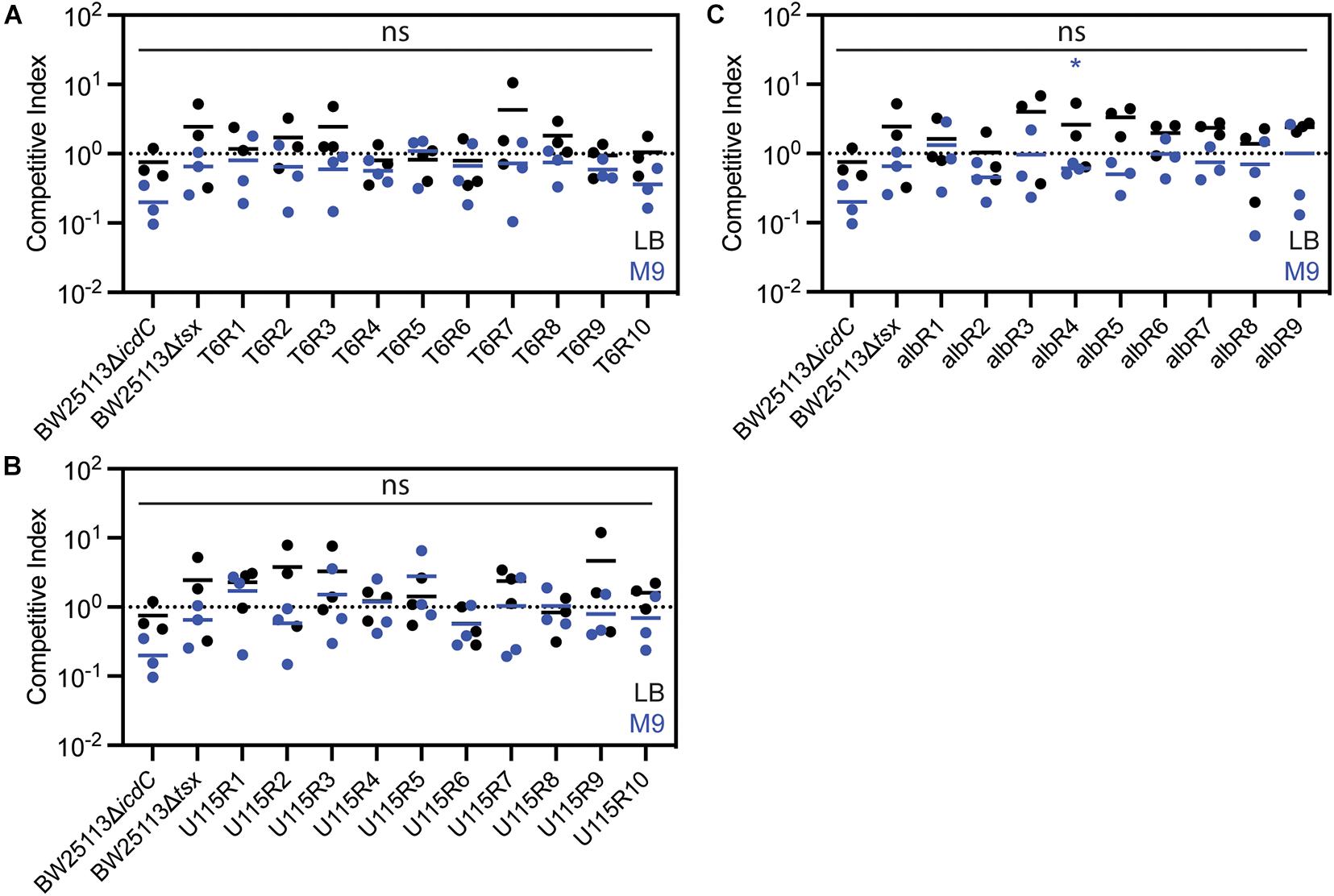
Figure 3. Competition assays in LB media (black) and M9 media (blue) showed that E. coli mutants resistant to phage T6 (A), phage U115 (B), or albicidin (C) did not measurably differ in fitness, relative to a non-resistant wild-type strain. Each point represents results from independent competition assays (n = 3), and statistical significance was determined using a t-test comparing competitive index of the resistant mutant relative to the competitive index of BW25113ΔicdC. None of the competitive indexes of the resistant mutants were statistically significant in LB media. Competitive indexes of resistant mutants that were statistically significant in M9 media are indicated with * (P < 0.05, unpaired t-test).
Owing to the importance of the Tsx porin in the interactions of all three antibacterials with cells, evolution of E. coli cross-resistance suggested that genetic changes in the tsx gene were likely involved. To examine this idea, we conducted whole genome sequencing (WGS) of all 29 strains. While some strains showed some single nucleotide variants (SNVs) and short insertions or deletions (indels) in tsx, many strains had no observable mutations in any genes using multiple variant calling pipelines including GATK (Van der Auwera et al., 2013) and breseq (Deatherage and Barrick, 2014). While WGS and variant calling pipelines are amenable for detecting SNVs and short indels, structural variants (SVs), including movement of transposable elements, are not easily detected (Ewing, 2015). Therefore, we conducted targeted Sanger sequencing of tsx in all 29 resistant mutants. As expected, results for the wild-type showed no mutations in tsx; however, mutations in tsx were identified in all 29 resistant mutants. For the 10 T6-resistant strains we observed the following mutations in the tsx gene: three insertion sequence (IS) elements, three deletions, three nonsense mutations and one missense mutation (Figure 4A and Table 1). For the 10 U115-resistant strains, we documented the following in tsx: four IS elements, two deletions, one insertion, two missense mutations, and two nonsense mutations (Figure 4A and Table 1). Interestingly, of the 10 pairs of phage-resistant mutants isolated from the same parent culture, only three pairs (T6R4/U115R4; T6R8/U115R8; T6R10/U115R10) showed identical mutations in tsx (Figure 4B and Table 1). For the nine albicidin-resistant mutants we observed six IS elements, two deletions, and one missense mutation in tsx (Figure 4A). Of the 29 mutations observed, three were in the promoter (tsxp2) region and four were in the signal sequence peptide of Tsx (Figure 4B and Table 1). In addition, our data showed that eight of the 29 mutations in tsx clustered within the region spanning base pairs 375–435 (Figure 4B).
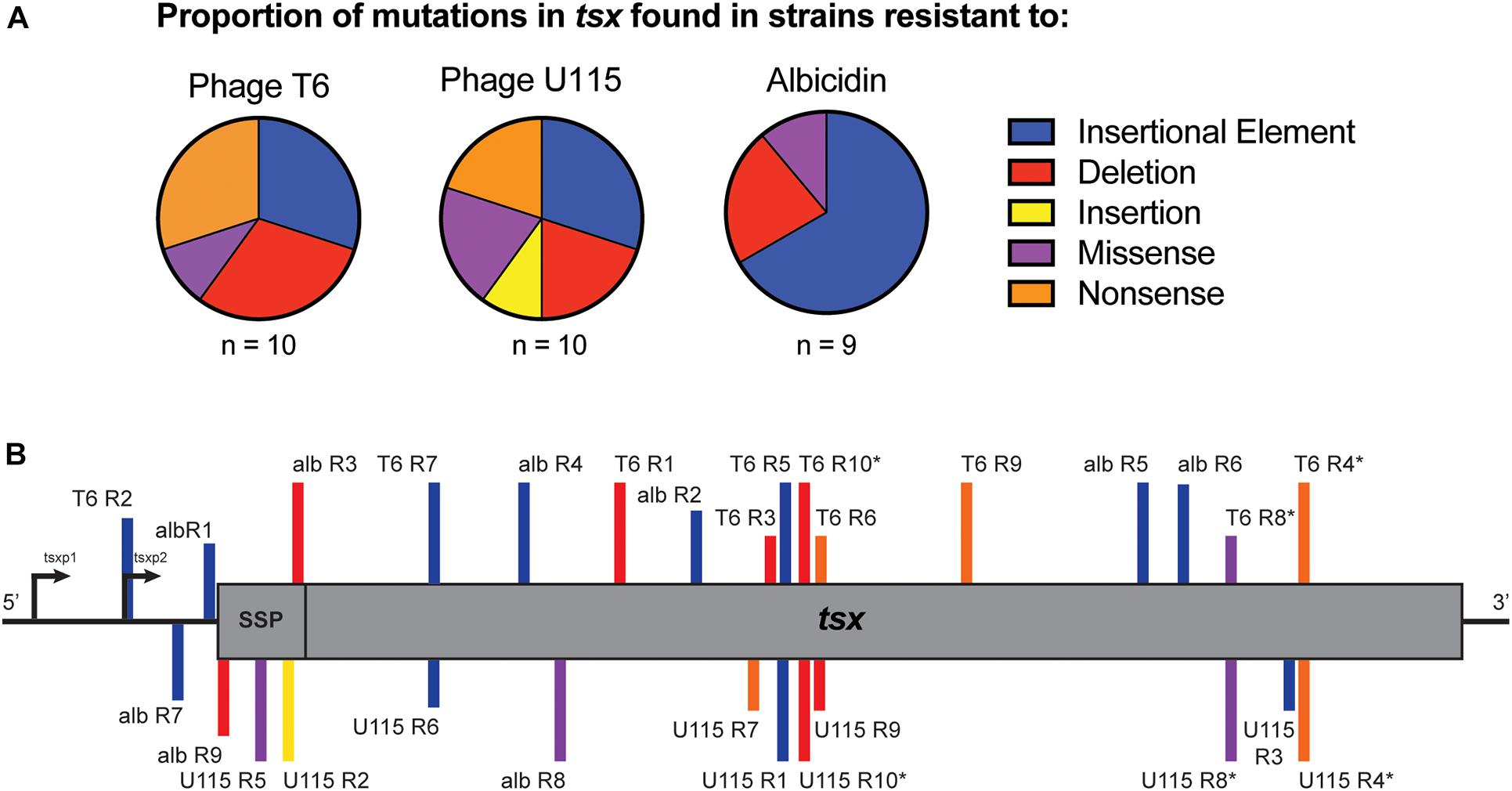
Figure 4. (A) Proportions of tsx mutations observed in sets of E. coli strains, selected for spontaneous resistance to either phage T6, phage U115 or albicidin. Mutations were due to insertion sequence elements (blue), deletions (red), insertions (yellow), missense mutations (purple) and nonsense mutations (orange). (B) Locations of tsx mutations for each resistant mutant (SSP, signal sequence peptide; *, indicates paired mutants).
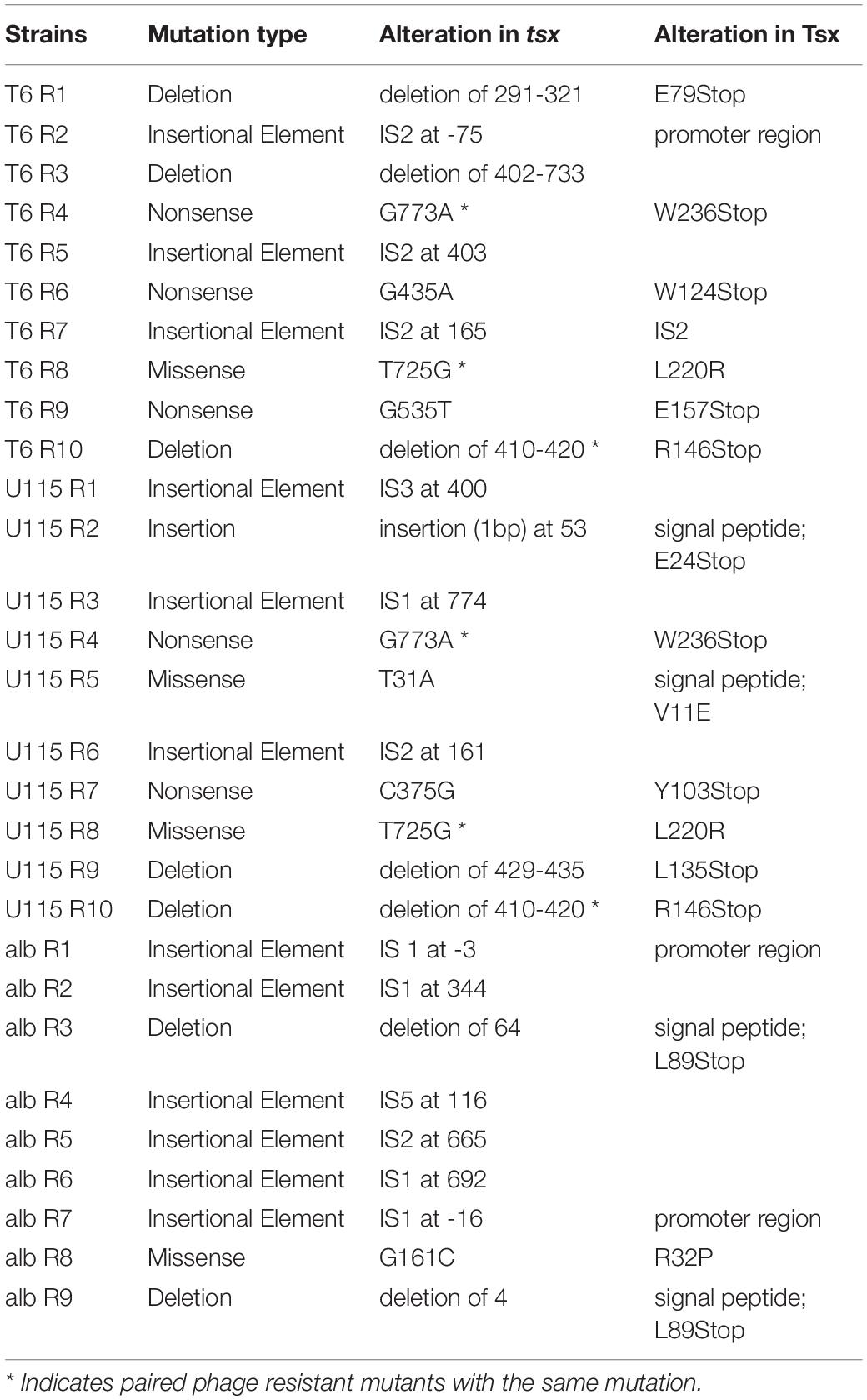
Table 1. Putative resistance mutations in the tsx gene, for each E. coli mutant selected for spontaneous resistance to either phage T6, phage U115, or albicidin.
The different missense mutations observed in this experiment were mapped (Figure 5) onto the crystal structure of Tsx (Ye and van den Berg, 2004) along with previously observed missense mutations (Figure 5 and Supplementary Table 2) that conferred resistance to either phage T6 or albicidin (Fsihi et al., 1993; Schneider et al., 1993). Mutant albR8 had a missense mutation R32P in the same surface exposed loop as three missense mutations (F27L, G28R, and G28E) observed by Fsihi et al. (1993) in mutants that were selected for albicidin resistance (Figure 5). Paired mutants T6R8 and U115R8 had the same missense mutation, L220R, in a beta strand that was in the region of a missense mutation (S217R) observed by Fsihi et al. (1993) in another mutant selected for resistance to albicidin (Figure 5). The third missense mutation observed in this study (V11E) in U115R5 is in the N-terminal signal peptide (Table 1).
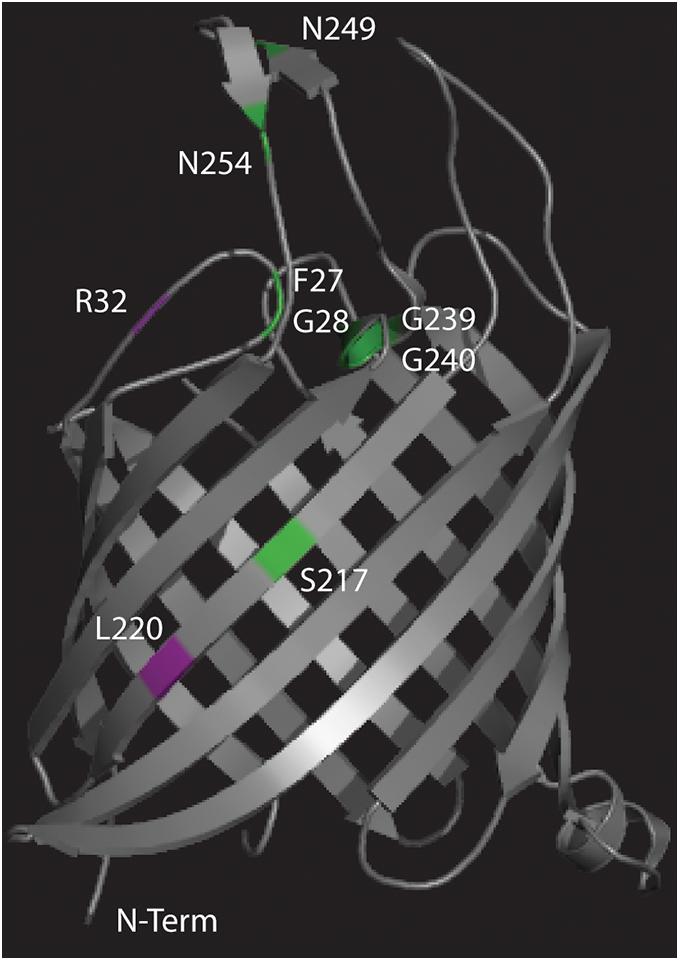
Figure 5. Crystal structure of Tsx (Ye and van den Berg, 2004) with the locations of the missense mutations observed in this study mapped in purple and the locations of missense mutations observed in previous studies (Fsihi et al., 1993; Schneider et al., 1993) mapped in green.
While it is likely that these mutations are conferring resistance to phage T6, phage U115 and albicidin, this was not experimentally confirmed via recombinant genetics because the work fell outside the scope of the current study.
The phage life cycle relies on surface-exposed molecules to initiate infection, and on bacterial metabolism to replicate inside cells. Since phages T6 and U115 both require Tsx as a primary receptor, it is not surprising that spontaneous mutants to one phage confer resistance to the other phage (Kortright et al., 2020). However, it is interesting that all 20 phage resistant mutants are also cross-resistant to albicidin and that all nine albicidin resistant mutants are cross-resistant to both phages. It was previously observed that T6-resistant mutants maintained their sensitivity to colicin-K, another antibiotic that enters via the Tsx porin (Schneider et al., 1993). While all three antibacterials used in the current study require Tsx as a receptor, it is unclear whether replication of phages T6 and U115 and the mechanism of action of albicidin also converge on the same gene to confer resistance. Albicidin is a DNA gyrase inhibitor that blocks topoisomerase II, an enzyme which cleaves both strands of DNA to modulate supercoiling during DNA replication, gene regulation, and transcription (Hashimi et al., 2007). Phages require the DNA replication machinery of the host bacterial cell to make viral progeny within the cell; however, many phages, including T6, encode their own topoisomerases (Huang et al., 1985). Furthermore, the minimum inhibitory concentrations (MICs) of ciprofloxacin (another DNA gyrase inhibitor) and ampicillin for wild-type BW25113 bacteria and for each of the 29 resistant mutants did not differ statistically (Supplementary Table 3). These observations of unchanged phenotypes imply that selection by phage T6, phage U115, and albicidin seems to evolutionarily converge on the tsx locus, rather than involving changes at other loci. Indeed, the results of the targeted sequencing indicated this possibility, because each mutant was observed to undergo a change in the tsx gene (i.e., as opposed to no mutation at this locus, suggesting resistance occurred via a different mechanism). While the identified mutations were not validated as the causative mutations of resistance, it seems likely that the 29 different mutations found in tsx were conferring resistance to phage T6, phage U115, and albicidin. The combination of tsx mutations observed in all 29 resistant mutants and the lack of detectable fitness defects in these strains indicate that Tsx may be superfluous for E. coli in the laboratory environment regardless of media type (high resource, complex versus low resource, defined).
The 29 observed mutations in tsx fall within the promoter region, as well as various other locations along the gene. However, there seems to be a cluster of “hotspots” for mutational changes in the region spanning 332 to 435 base pairs. These residues make up a surface exposed alpha helix and a beta strand that spans the outer membrane of the bacteria (Ye and van den Berg, 2004). All of the mutations in this region were observed in resistant strains resulting from phage selection but not albicidin selection, indicating that perhaps amino acid residues 88 through 123 are likely important for phage binding.
Moreover, of the 10 “paired” phage-resistant mutants, only three of the pairs (T6R4/U115R4; T6R8/U115R8; T6R10/U115R10) showed the same tsx mutation. This means that even in a clonal population of E. coli, there are often multiple spontaneous mutants with different changes in tsx. This finding indicates that the number of resistant mutants observed to arise during fluctuation assays cannot be used to compute a rate of spontaneous mutation; instead, these data should only be used to calculate a rate of spontaneous phenotypic resistance.
Expression and proper localization of Tsx at the outer membrane occurs via two promoters and a signal sequence peptide, respectively (Bremer et al., 1990). Minor promoter, tsxp1, is repressed by transcription factor DeoR while major promoter, tsxp2, is controlled by the cAMP-CRP complex. Of the three mutations that were observed in the promoter region, all three were in the region of tsxp2, the promoter which results in a higher number of tsx transcripts. Likely these mutants (T6R2, albR7, and albR1) are not producing Tsx. Similarly, the four mutants with mutations in the signal sequence peptide region (albR9, U115R5, U115R2, and albR3) likely do not properly express Tsx at the outer membrane.
Indeed, it is probable that many of the resistant mutants do not stably express Tsx in the outer membrane. Mutants with an insertional element disrupting tsx (T6R2, T6R5, T6R7, U115R1, U115R3, U115R6, albR1, albR2, albR4, albR5, albR6, and albR7) or indels and nonsense mutaitons at the beginning of tsx (T6R1, T6R3, T6R6, T6R9, T6R10, U115R7, U115R9, and U115R10) likely do not express Tsx. Furthermore, Tsx is probably not stably expressed at the outer membrane in resistant mutants with nonsense mutations toward the end of the gene (T6R4 and U115R4). However, it is more likely that Tsx is expressed in resistant mutants with missense mutations (T6R8, U115R5, U115R8, and albR8). Interestingly, of the three different missense mutations observed (T6R8 and U115R8 were paired resistant mutants that share the same mutation), two out of three of them were located in regions where previous missense mutations conferring resistance to albicidin—but not to phage T6—had been observed (Fsihi et al., 1993). It is possible that the L220R mutation (T6R8 and U115R8) in one of the membrane-spanning beta strands is sufficient to disrupt the proper folding and insertion of Tsx in the outer membrane. The V11E mutation (U115R5) in the signal sequence likely disrupts the proper localization of Tsx. The third missense mutation, R32P (albR8), is in a surface exposed loop. Since this mutant was resistant to albicidin, phage T6 and phage U115, it is likely that this particular surface exposed loop is essential for binding of these three antimicrobials; future experiments would be needed to confirm this idea.
It is also interesting that the profile of tsx mutations was different for the phage-selected resistant mutants, compared with the antibiotic-selected resistant mutants. There were six albicidin-resistant mutants with an IS element inserted in tsx, while three of each of the phage-resistant mutants showed IS-element changes in tsx. This observation could be due either to a difference in selection pressures of phages versus antibiotics, or to a difference in the method of selection to amass these mutants. Underlying these possibilities are two opposing ideas. It has been previously assumed that insertion and excision of IS elements are stochastic events. However, recent evidence suggests that IS movement may actually be a form of directed mutagenesis in which IS elements target specific chromosomal loci to reduce stress under certain environmental conditions (Humayun et al., 2017). The isolation of phage-resistant mutants in this study relied on spontaneous mutations accumulated in an overnight culture, while the isolation of antibiotic-resistant mutants allowed for both selection of spontaneous resistant mutants as well as evolution in the presence of albicidin. This difference suggests that the larger proportion of IS element insertions in tsx found in the set of albicidin-resistant mutants might be caused by direct targeting of tsx by IS elements under stressful conditions. Without further experiments that use the same method to select for spontaneous phage and albicidin resistant mutants, we cannot rule out that a difference in selection pressures between phages and the antibiotic resulted in the skew of IS-element-insertion changes in tsx found in the albicidin-resistant mutants.
Finally, it is worth noting that all 29 resistant mutants have gained fitness relative to wild-type in the presence of albicidin or phage, with no discernable growth cost when bacteria are grown without phage and antibiotic selection. Bacteria can utilize nucleosides as a carbon source in environments where carbon is limiting (Tozzi et al., 2006; Mulcahy et al., 2010) and nucleoside-uptake mutations are shown to impair bacterial fitness in a low-carbon environment (Gumpenberger et al., 2016). In this study, we used competition assays to show that phage/antibiotic resistant mutants maintained robust growth in both carbon-rich (LB medium) and carbon-limited (M9 medium) environments. However, it is possible that a cost of antibacterial resistance might occur in different lab-media or other environments not examined here. Antibiotic cross-resistance has extreme clinical relevance as selected resistance to a single antibiotic can also result in evolution of multi-drug resistance. Here, we showed that phage resistance can result in antibiotic resistance, and vice versa, at no fitness cost. This highlights a potential pitfall of the future therapeutic use of any antimicrobial whether it be phage or antibiotics. Without a clear understanding of the evolutionary implications of treatment, antimicrobial selection can result in both resistance and cross-resistance at no fitness cost to the bacteria.
Strains in this study are listed in Supplementary Table 4. E. coli strains were cultured for 24 h with shaking incubation at 37°C in lysogeny broth (Luria and Delbruck, 1943). M9 minimal medium (6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl, 1 g NH4Cl, 1 mM MgSO4, 0.1 mM CaCl2, 0.1% (w/v) glucose per liter) was used for competition assays (see below). The Tsx knockout, BW25113Δtsx, was used as a negative control in many experiments. A pseudogene knockout, BW25113ΔicdC, was used as a control for the KanR cassette and as a marked competitor in the competition assays. Where appropriate, LB was supplemented with kanamycin at 30 μg/mL (LB Kan30). X. albilineans strains (provided by D. Gabriel, U Florida) were cultured for 48 h with shaking at 30°C in Modified Wilbrinks (MW) medium (Rott et al., 1994). Lysates of phages T6 and U115 were obtained by mixing each phage with E. coli strain BW25113 in LB medium and culturing for 24 h at 37°C, followed by filtration (0.22 μm) to remove bacteria.
Ten cultures of E. coli strain BW25113 were grown independently as described above, and diluted samples were spread on LB agar (1.5%) plates, pre-saturated with either phage T6 or phage U115. After overnight incubation at 37°C, one colony was chosen randomly from each plate and colony-purified three times. For each of the 20 isolated mutants, phage resistance was confirmed via efficiency of plaquing (EOP) assays, which compared plaquing ability of phage T6 or U115 on the test mutant relative to growth on wild-type strain BW25113.
X. albilineans strain XA23 was cultured as described above, and a 10 μL sample was spotted onto each of ten Sucrose Peptone agar (SPA) plates that were incubated for 5 days at 30°C (Birch and Patil, 1985). A sample from each of ten independently-grown cultures of BW25113 was then overlaid on one of the SPA plates, and incubated overnight at 37°C. Colonies that appeared in the zone of growth inhibition around the XA23 spot represented spontaneous E. coli mutants that were resistant to albicidin. One plate was lost due to contamination; from each of the remaining nine plates, one colony was randomly chosen and colony-purified three times. Albicidin resistance of each mutant was verified by plating a sample of a grown-up culture of the isolate on an XA23 spot, as described above.
Xanthomonas albilineans strains XA23 and LS126 were each cultured for 48 h at 30°C, and their optical densities at wavelength λ = 600 nm (OD600) were estimated via spectrophotometry. Each culture was then diluted in MW medium, to obtain OD600 = 0.25. Then, a 10-μL sample of each diluted culture was spotted on a SPA plate, allowed to dry completely and then incubated for 5 days at 30°C. SPA plates were overlaid with 4 mL of LB top agar (0.75%) with 1 mL of a test E. coli strain at OD600 = 0.25, and incubated overnight at 37°C. The next day, plates were imaged. Images were quantified using ImageJ, by thresholding until either the X. albilineans spots or E. coli clearings were outlined and the area within the outline was quantified. The ratio of the area of the zone of clearing to the area of the X. albilineans spot was used to define albicidin sensitivity: a ratio of greater than 1.0 indicates albicidin sensitivity, while a ratio of 1.0 indicates albicidin resistance.
Minimum inhibitory concentrations (MICs) of ampicillin and ciprofloxacin were determined using the twofold dilution method (MacLowry et al., 1970).
E. coli bacteria were cultured overnight as described above. Dilutions of a test strain in LB medium were placed in wells of a flat-bottomed 96-well plate, in the absence of phage or mixed with phage at a multiplicity-of-infection (MOI; ratio of phage particles to host cells) of ∼10. Plates were incubated with shaking for 18 h in an automated spectrophotometer at 37°C, and OD600 measurements were obtained every 10 min. Growth curve data were fit to a logarithmic curve using GrowthCurver and growth parameters were extracted (Sprouffske and Wagner, 2016). All growth curve data are deposited on Dryad.
Replicated (n = 3) competition assays were conducted in both LB and M9 media by mixing two bacterial strains at a 1:1 initial ratio. Competition assays in LB were serially passaged (1:100 dilution) every 24 h for 72 h total. Competition assays in M9 were measured after 24 h. At the end of each experiment, a diluted sample of each competition was plated on LB agar to measure viable bacterial density (colony-forming units; CFU), and on LB Kan30 agar to estimate the CFU of a KanR competitor; the density of the KanS competitor was estimated by subtraction. Competitive indexes were calculated as the ratio of resistant-mutant CFU to the CFU of BW25113ΔicdC divided by the ratio of BW25113ΔicdC CFU to BW25113 CFU. Each ratio was normalized by the starting ratio of each competing strain.
Sequencing libraries were prepared using Illumina’s Nextera prep kit. Whole genome sequencing was done using paired-end 150 bp reads on the Illumina NextSeq platform or paired-end 300bp reads on the Illumina HiSeq platform. For targeted sequencing, primers (5′-CTGTGAAACGAAACATATTTTTG-3′ and 5′-CGTGCTTTTGTTGGC-3′) were designed ∼100 base pairs upstream and downstream of E. coli gene tsx, and used to amplify tsx of each resistant mutant and the ancestral BW25113 strain. Amplicons were Sanger sequenced at the Yale DNA Analysis Facility on Science Hill. Mutations were identified by comparing sequencing reads to tsx loci of the reference strain GenBank CP009273.1.
Sequence data are available at NCBI SRA accession #PRJNA693868. All other data are available on Dryad (https://doi.org/10.5061/dryad.ghx3ffbmn).
KK and BC designed study. KK and SD-G conducted the experiments. KK, SD-G, BC, and PT wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
PT is a co-founder of Felix Biotechnology Inc., and declares a financial interest in this company that seeks to commercially develop phages for use as therapeutics. PT discloses two provisional patent applications involving phage therapy.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Andrew Goodman, Barbara Kazmierczak, Daniel Weinberger, and James Bull for helpful feedback on this study. We thank Dean Gabriel (U Florida) for kindly providing X. albilineans strains used in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.658374/full#supplementary-material
Azam, A. H., and Tanji, Y. (2019). Bacteriophage-host arm race: an update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl. Microbiol. Biotechnol. 103, 2121–2131. doi: 10.1007/s00253-019-09629-x
Birch, R. G., and Patil, S. S. (1985). Preliminary characterization of an antibiotic produced by Xanthomonas albilineans which inhibits DNA synthesis in Escherichia coli. Microbiology 131, 1069–1075. doi: 10.1099/00221287-131-5-1069
Birch, R. G., and Patil, S. S. (1987). Evidence that an albicidin-like phytotoxin induces chlorosis in sugarcane leaf scald disease by blocking plastid DNA replication. Physiol. Mol. Plant Pathol. 30, 207–214. doi: 10.1016/0885-5765(87)90034-8
Birch, R. G., Pemberton, J. M., and Basnayake, W. V. (1990). Stable albicidin resistance in Escherichia coli involves an altered outer-membrane nucleoside uptake system. J. Gen. Microbiol. 136, 51–58. doi: 10.1099/00221287-136-1-51
Bremer, E., Middendorf, A., Martinussen, J., and Valentinhansen, P. (1990). Analysis of the tsx gene, which encodes a nucleoside-specific channel-forming protein (Tsx) in the outer membrane of Escherichia coli. Gene 96, 59–65. doi: 10.1016/0378-1119(90)90341-n
Burmeister, A. R., and Turner, P. E. (2020). Trading-off and trading-up in the world of bacteria-phage evolution. Curr. Biol. 30, R1120–R1124. doi: 10.1016/j.cub.2020.07.036
Burmeister, A. R., Fortier, A., Roush, C., Lessing, A. J., Bender, R. G., Barahman, R., et al. (2020). Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc. Natl. Acad. Sci. U S A. 117, 11207–11216. doi: 10.1073/pnas.1919888117
Chan, B. K., Turner, P. E., Kim, S., Mojibian, H. R., Elefteriades, J. A., and Narayan, D. (2018). Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 60–66. doi: 10.1093/emph/eoy005
Cohan, F. M., Zandi, M., and Turner, P. E. (2020). Broadscale phage therapy is unlikely to select for widespread evolution of bacterial resistance to virus infection. Virus Evol. 6:veaa060. doi: 10.1093/ve/veaa060
Deatherage, D. E., and Barrick, J. E. (2014). Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188. doi: 10.1007/978-1-4939-0554-6_12
d’Herelle, F. (1917). Sur un microbe invisible antagoniste des bacilles dysentériques. Comptes Rendus Acad. Sci. 165:373.
Ewing, A. D. (2015). Transposable element detection from whole genome sequence data. Mob DNA 6:24. doi: 10.1186/s13100-015-0055-3
Fleming, A. (1929). On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 10, 226–236.
Fsihi, H., Kottwitz, B., and Bremer, E. (1993). Single amino-acid substitutions affecting the substrate specificity of the Escherichia Coli K-12 nucleoside-specific Tsx channel. J. Biol. Chem. 268, 17495–17503. doi: 10.1016/s0021-9258(19)85361-9
Gumpenberger, T., Vorkapic, D., Zingl, F. G., Pressler, K., Lackner, S., Seper, A., et al. (2016). Nucleoside uptake in Vibrio cholerae and its role in the transition fitness from host to environment. Mol. Microbiol. 99, 470–483. doi: 10.1111/mmi.13143
Gurney, J., Brown, S. P., Kaltz, O., and Hochberg, M. E. (2020). Steering phages to combat bacterial pathogens. Trends Microbiol. 28, 85–94. doi: 10.1016/j.tim.2019.10.007
Hashimi, S. M. (2019). Albicidin, a potent DNA gyrase inhibitor with clinical potential. J. Antibiot. 72, 785–792. doi: 10.1038/s41429-019-0228-2
Hashimi, S. M., Wall, M. K., Smith, A. B., Maxwell, A., and Birch, R. G. (2007). The phytotoxin albicidin is a novel inhibitor of DNA gyrase. Antimicrob. Agents Chemother. 51, 181–187. doi: 10.1128/aac.00918-06
Huang, W. M., Wei, L. S., and Casjens, S. (1985). Relationship between bacteriophage T4 and T6 DNA topoisomerases. T6 39-protein subunit is equivalent to the combined T4 39-and 60-protein subunits. J. Biol. Chem. 260, 8973–8977. doi: 10.1016/s0021-9258(17)39444-9
Humayun, M. Z., Zhang, Z., Butcher, A. M., Moshayedi, A., and Saier, M. H. Jr. (2017). Hopping into a hot seat: Role of DNA structural features on IS 5-mediated gene activation and inactivation under stress. PLoS One 12:e0180156. doi: 10.1371/journal.pone.0180156
Kortright, K. E., Chan, B. K., and Turner, P. E. (2020). High-throughput discovery of phage receptors using transposon insertion sequencing of bacteria. Proc. Natl. Acad. Sci. U S A. 117, 18670–18679. doi: 10.1073/pnas.2001888117
Kortright, K. E., Chan, B. K., Koff, J. L., and Turner, P. E. (2019). Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25, 219–232. doi: 10.1016/j.chom.2019.01.014
Kretz, J., Kerwat, D., Schubert, V., Gratz, S., Pesic, A., Semsary, S., et al. (2015). Total synthesis of albicidin: a lead structure from Xanthomonas albilineans for potent antibacterial gyrase inhibitors. Angewandte Chemie Int. Edition 54, 1969–1973. doi: 10.1002/anie.201409584
Luria, S. E., and Delbruck, M. (1943). Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491–511. doi: 10.1093/genetics/28.6.491
MacLowry, J. D., Jaqua, M. J., and Selepak, S. T. (1970). Detailed methodology and implementation of a semiautomated serial dilution microtechnique for antimicrobial susceptibility testing. Appl. Microbiol. 20, 46–53. doi: 10.1128/am.20.1.46-53.1970
Maier, C., Bremer, E., Schmid, A., and Benz, R. (1988). Pore-forming activity of the Tsx protein from the outer membrane of Escherichia coli. Demonstration of a nucleoside-specific binding site. J. Biol. Chem. 263, 2493–2499. doi: 10.1016/s0021-9258(18)69233-6
Moulton-Brown, C. E., and Friman, V. P. (2018). Rapid evolution of generalized resistance mechanisms can constrain the efficacy of phage-antibiotic treatments. Evolut. Applicat. 11, 1630–1641. doi: 10.1111/eva.12653
Mulcahy, H., Charron-Mazenod, L., and Lewenza, S. (2010). Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ. Microbiol. 12, 1621–1629. doi: 10.1111/j.1462-2920.2010.02208.x
Rodriguez-Gonzalez, R. A., Leung, C. Y., Chan, B. K., Turner, P. E., and Weitz, J. S. (2020). Quantitative models of phage-antibiotic combination therapy. mSystems 5, 756–719. doi: 10.1128/mSystems.00756-19
Rostock, L., Driller, R., Gratz, S., Kerwat, D., von Eckardstein, L., Petras, D., et al. (2018). Molecular insights into antibiotic resistance - how a binding protein traps albicidin. Nat. Commun. 9:3095.
Rott, P. C., Costet, L., Davis, M. J., Frutos, R., and Gabriel, D. W. (1996). At least two separate gene clusters are involved in albicidin production by Xanthomonas albilineans. J. Bacteriol. 178, 4590–4596. doi: 10.1128/jb.178.15.4590-4596.1996
Rott, P., Abel, M., Soupa, D., Feldmann, P., and Letourmy, P. (1994). Population dynamics of Xanthomonas albilineans in sugarcane plants as determined with an antibiotic-resistant mutant. Plant Dis. 78, 241–247. doi: 10.1094/pd-78-0241
Schneider, H., Fsihi, H., Kottwitz, B., Mygind, B., and Bremer, E. (1993). Identification of a segment of the Escherichia coli Tsx protein that functions as a bacteriophage receptor area. J. Bacteriol. 175, 2809–2817. doi: 10.1128/Jb.175.10.2809-2817.1993
Sprouffske, K., and Wagner, A. (2016). Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformat. 17:172. doi: 10.1186/s12859-016-1016-7
Stearns, S. C. (2012). Evolutionary medicine: its scope, interest and potential. Proce. R. Soc. B Biol. Sci. 279, 4305–4321. doi: 10.1098/rspb.2012.1326
Tozzi, M. G., Camici, M., Mascia, L., Sgarrella, F., and Ipata, P. L. (2006). Pentose phosphates in nucleoside interconversion and catabolism. FEBS J. 273, 1089–1101. doi: 10.1111/j.1742-4658.2006.05155.x
Turner, P. E. (2016). “Evolutionary medicine,” in How Evolution Shapes Our Lives: Essays on Biology and Society, Chap. 7, eds J. B. Losos and R. E. Lenski (Princeton,NJ: Princeton University Press).
Van der Auwera, G. A., Carneiro, M. O., Hartl, C., Poplin, R., Del Angel, G., Levy-Moonshine, A., et al. (2013). From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 43:bi1110s43. doi: 10.1002/0471250953.bi1110s43
von Eckardstein, L., Petras, D., Dang, T., Cociancich, S., Sabri, S., Gratz, S., et al. (2017). Total synthesis and biological assessment of novel albicidins discovered by mass spectrometric networking. Chem. Eur. J. 23, 15316–15321. doi: 10.1002/chem.201704074
Keywords: phage, antibiotic, resistance, evolution, trade-up
Citation: Kortright KE, Doss-Gollin S, Chan BK and Turner PE (2021) Evolution of Bacterial Cross-Resistance to Lytic Phages and Albicidin Antibiotic. Front. Microbiol. 12:658374. doi: 10.3389/fmicb.2021.658374
Received: 25 January 2021; Accepted: 10 May 2021;
Published: 17 June 2021.
Edited by:
Robert Czajkowski, University of Gdańsk, PolandReviewed by:
Malgorzata Barbara Lobocka, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, PolandCopyright © 2021 Kortright, Doss-Gollin, Chan and Turner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul E. Turner, cGF1bC50dXJuZXJAeWFsZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.