
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 July 2021
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.658113
This article is part of the Research Topic Plants and Microbial Communities: Diversity, Pathogens and Biological Control View all 28 articles
Phytophthora nicotianae causes black shank, a serious soil-borne disease, in tobacco. In this study, the Bacillus strain GUMT319 was isolated from the rhizosphere of healthy tobacco plants grown in a field in Guizhou with a high incidence of tobacco black shank. Genome sequencing revealed that GUMT319 contained a single circular chromosome 3,940,023 bp in length, with 4,053 predicted genes and an average GC content of 46.6%. Based on phylogenomic analyses, GUMT319 was designated as Bacillus velezensis. The genome of GUMT319 contained more than 60 genes and 13 gene clusters that have previously been found to be active in antifungal mechanisms, biofilm formation, and chemotaxis motility. Additionally, confocal laser scanning microscopy and scanning electron microscopy showed that GUMT319 formed a spatially organized biofilm in vivo. In addition, lauric acid negatively regulated biofilm formation. This is the first study to report that nicotine in tobacco root exudates was a chemoattractant for biocontrol Bacillus strains. In this study, we identified new interactions between beneficial microorganisms and tobacco roots in the rhizosphere. Moreover, dual culture tests in vitro showed that GUMT319 inhibited the growth of P. nicotianae and also displayed inhibitory effects against eight other plant pathogens, namely, Colletotrichum scovillei, Colletotrichum capsici, Fusarium carminascens, Sclerotinia sclerotiorum, Alternaria alternata, Phomopsis sp., Phyllosticta sorghina, and Exserohilum turcicum. Furthermore, GUMT319 exhibited > 70% control efficiency against tobacco black shank in field experiments conducted in 2018–2020. Thus, GUMT319 was more effective in controlling the incidence of tobacco black shank than other treatments including fungicide application. Overall, these results suggested that GUMT319 (B. velezensis) could be used as a potential biocontrol agent against tobacco black shank.
Tobacco black shank, caused by the fungal pathogen Phytophthora nicotianae, is a serious disease of flue-cured tobacco, which is an economically important crop in Guizhou, China. The severity of tobacco black shank continues to increase each year, causing significant economic losses. In the field, tobacco black shank affects the stem base (Han et al., 2016; Guo et al., 2020) and the chemical fungicide metalaxyl is generally used for disease control. However, this practice is not conducive to the development of sustainable tobacco agriculture, as it creates an excessive dependence on chemical agents, leaves fungicide residues, and causes environmental pollution (Han et al., 2016; Guo et al., 2020). As an alternative, biological control agents (BCAs) offer safe alternative methods for controlling tobacco black shank (Zhang et al., 2017a; Srikhong et al., 2018).
Biocontrol has proved to be a promising strategy for the management of many plant diseases (Chowdhury et al., 2015; Han et al., 2016; Zhang et al., 2017c; Fan et al., 2018, Mao et al., 2020). Numerous BCAs have been studied, but only a limited number of strains, such as those of Bacillus species, have been commercially developed (Chowdhury et al., 2015; Fan et al., 2018; Ye et al., 2020). Bacillus spp. are ubiquitous bacteria widely distributed in natural environments, especially in the rhizosphere and plant roots. They are also excellent candidates for BCAs since they produce heat- and desiccation-resistant endospores that are easily stored and transported as stable products (Weng et al., 2013; Fan et al., 2018). However, the use of BCAs remains a challenge since their effect in the field is frequently inconsistent (Weng et al., 2013).
Bacillus spp. employ a number of mechanisms in their effective biocontrol of pathogens, such as antagonism, systemic resistance induction, and plant growth promotion (Chowdhury et al., 2015). In previous studies, Bacillus strains exhibited biological activity against various plant pathogens by producing different types of antimicrobial compounds, such as cell wall-degrading enzymes and non-ribosomally synthesized antibiotics, as well as secondary metabolites that trigger induced systemic resistance (ISR), thus protecting the plants against pathogen attack (Chowdhury et al., 2015; Fan et al., 2018; Yan et al., 2020).
Successful root colonization of BCAs is the key to effective biocontrol and is considered to be the main factor responsible for consistent performance in the field (Weng et al., 2013; Xu et al., 2013; Liu et al., 2014). Root colonization is divided into two steps: chemotaxis toward the root and subsequent biofilm formation on the root surface (Zhang et al., 2017b; Feng et al., 2018, 2019). Bacillus spp. colonize the roots of many plant species and form a biofilm, which contributes to their biocontrol efficacy (Gao et al., 2015a). Chemotaxis-mediated response to root exudates enhances the colonization and beneficial effects of plant growth-promoting Bacillus strains (Feng et al., 2018).
Bacillus velezensis is an important member of the plant growth-promoting rhizobacteria (PGPR), which are known to enhance plant growth and control soil-borne diseases (Chowdhury et al., 2015; Fan et al., 2018). B. velezensis FZB42 was the first strain to be sequenced (Chen et al., 2007). It was initially identified as a Bacillus amyloliquefaciens strain, but was later recognized as a B. velezensis strain and as a model of gram-positive plant growth-promoting and biocontrol rhizobacteria (Fan et al., 2018). Approximately 130 whole genome sequences of B. velezensis have been deposited in GenBank to date. B. velezensis strains have their own specific genomic characteristics, because these strains reside in different host plants and environments (Wang et al., 2019). For instance, B. velezensis SQR9 isolated from the cucumber rhizosphere has been used as a BCA against fungal pathogens, mainly because of its ability to trigger ISR (Borriss et al., 2018; Wu et al., 2018), while B. velezensis HAB-2 isolated from cotton has been used as a biological pesticide against bacterial pathogens (Xu et al., 2020).
In this study, we isolated B. velezensis GUMT319 from the rhizosphere of healthy tobacco plants growing in high-incidence tobacco black shank fields in Guizhou, China. Previous research has shown that GUMT319 produces enzymes with biocontrol activity, such as proteases, cellulases, siderophores, and phosphatases, and inhibits the mycelial growth of P. nicotianae in vitro (Luo et al., 2019). Here, we aimed to investigate the biocontrol effects of GUMT319 in the field and to gain insights into the underlying mechanisms. Genome sequencing of GUMT319 revealed the presence of genes involved in plant growth promotion, biofilm formation, chemotaxis, and antifungal activity. In addition, confocal laser scanning microscopy and scanning electron microscopy analyses of the GUMT319 strain labeled with green fluorescent protein (GFP) revealed its patterns of tobacco root colonization. Overall, this study highlights the excellent potential of GUMT319 as a BCA against the tobacco black shank disease.
Bacillus strain GUMT319 was grown at 37°C in Luria–Bertani medium (LB). The GFP-labeled GUMT319 (GUMT319-gfp) was maintained in LB supplemented with 10 mg/mL tetracycline. The fungal pathogens used in this study included P. nicotianae, Colletotrichum scovillei, Colletotrichum capsici, Fusarium carminascens, Sclerotinia sclerotiorum, Alternaria alternata, Phomopsis spp., Phyllosticta sorghina, and Exserohilum turcicum. These were maintained on potato dextrose agar (PDA) medium at the Department of Plant Pathology, Guizhou University. To store these pathogens, hyphae were sampled from the growing zone of mycelia in agar disks (1 cm diameter), and they were mixed with 15% glycerol and stored in a 4°C freezer.
A single bacterial colony was inoculated in 5 mL of LB broth and grown for 12 h at 37°C with agitation at 200 rpm. Then, 2 mL of the bacterial culture was centrifuged at 10,000 rpm for 1 min, and bacterial genomic DNA was isolated using an Ezup Column Bacteria Genomic DNA Purification Kit [Sangon Biotech (Shanghai) Co., Ltd., China], in accordance with the manufacturer’s protocol. Subsequently, 16S rRNA and gyrA genes were amplified by PCR using sequence-specific primers: 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) for amplifying approximately 1,400 bp of the 16S rRNA gene and gyrA-F (5′-CAGTCAGGAAATGCGTACGTCCTT-3′) and gyrA-R (5′-CAAGGTAATGCTCCAGGCATTGCT-3′) for amplifying approximately 1,000 bp of the gyrA gene (Chen et al., 2016). Each 25-μL PCR mixture contained 1 unit of Pfu DNA Polymerase (Sangon Biotech), 1 × PCR buffer, 1 mM MgCl2, 100 μM dNTPs, 1 μM of each primer, 50 ng of genomic DNA template, and ultrapure water. PCR was performed using the following conditions: initial denaturation at 95°C for 10 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 90 s, with a final extension at 72°C for 10 min. The PCR products were sequenced at Sangon Biotech. Sequences of the 16S rRNA and gyrA gene fragments were searched in the National Center for Biotechnology Information (NCBI) nucleotide database using Blastn to determine the closest taxonomic relatives. Subsequently, phylogenetic analysis of 16S rRNA and gyrA gene sequences was performed in MEGA 6.0 using the maximum likelihood method to estimate the evolutionary position of GUMT319 relative to other Bacillus strains (Supplementary Table 1).
The genomic DNA of GUMT319 was sequenced at Beijing Novogene Bioinformatics Technology Co., Ltd. using an Illumina PE150 system and PacBio RSII high-throughput sequencing technology. Low-quality reads were filtered using SMRT Link v5.0.1, and the filtered reads were assembled into one contig without gaps. The complete genome sequence of GUMT319 was annotated using the Prokaryotic Genomes Annotation Pipeline (PGAP) at NCBI, and gene functions were predicted using five databases, namely, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Clusters of Orthologous Groups (COG), Non-Redundant (NR) protein sequences, and Swiss-Prot, based on whole genome Blast search (E-value < 1e-5; minimal alignment length > 40%) against each database.
The GFP plasmid pGFP78, an Escherichia coli–Bacillus subtilis shuttle vector containing the 78 promoter-controlled GFP gene (Zhang et al., 2017c), was electroporated into electrocompetent cells of strain GUMT319, as described previously (Zhang et al., 2012). Briefly, the cells were grown in liquid LB medium at 37°C and were harvested during the exponential growth phase by centrifugation at 10,000 rpm for 10 min at 4°C. The harvested cells were washed five times with an equal volume of cold electroporation buffer containing 0.5 M sorbitol, 0.5 M mannitol, and 10% glycerol (pH 7.0). Subsequently, 100 μL of competent cells were electroporated on ice with 1 mg of pGFP78 DNA using a 1.8-kV electric shock. After electroporation, the cell suspension was diluted with 900 μL of LB medium and incubated at 37°C on an agitator at 120 rpm for 3 h to allow the expression of antibiotic resistance markers. The cell suspension was then spread on LB agar medium supplemented with tetracycline (10 mg/mL). The GFP-labeled cells were selected for tetracycline for three generations, and this was confirmed by fluorescence microscopy (Gao et al., 2015b).
Seeds of tobacco (Nicotiana tabacum) cultivar Yunyan 87, native to China, were surface-sterilized by soaking in 2% sodium hypochlorite for 15 min and then washed thoroughly with distilled water. The sterilized seeds were sown in axenic tissue culture bottles containing vermiculite, and seedlings grew for 15–20 days in a growth chamber at 25°C day/20°C night temperature and a 16-h light/8-h dark photoperiod. Then, seedlings were aseptically transplanted into 250-mL flasks (one seedling per flask) containing 100 mL sterile liquid 25% sucrose-free Murashige and Skoog (MS) medium, which was renewed every other day during the growth period. The hydroponic system was placed on an agitator and gently shaken at 50 rpm for 2 h each day. Before inoculating the MS medium with B. velezensis GUMT319-gfp, 100-μL aliquots of the MS medium were sampled from each flask and spread onto solid LB medium to verify the absence of contamination (Han et al., 2016).
To perform the root colonization assay, GUMT319-gfp was grown overnight in liquid LB medium and was subsequently resuspended in sterile double-distilled water. Roots of tobacco plants were inoculated with 100 mL of GUMT319-gfp (OD600 = 0.5) via drench application for 2 h; 10 replicates were performed. The plants were then transferred to fresh MS medium and cultivated for an additional 2 days without agitation. Roots were rinsed with sterile double-distilled water. To visualize colonization by GUMT319-gfp and GUMT319, at least 20 root tips were observed under a confocal laser scanning microscope (CLSM) and scanning electron microscope (SEM), as described previously (Weng et al., 2013).
Seeds of tobacco cultivar Yunyan 87 and pepper cultivar Dangwu were germinated, and the seedlings were grown for approximately 15–20 days in MS medium, as described above. Before starting the collection of root exudates, plant roots were washed with sterile double-distilled water to avoid contamination from the nutrient solution. The plants were then transferred to a flask, with the roots submerged in sterile water, and the flasks were placed in a growth chamber for 24 h at 25°C and a 16-h light/8-h dark photoperiod, with gentle agitation (50 rpm). The root exudates collected were filtered through a 0.45-μm membrane and lyophilized. The freeze-dried powder of tobacco root exudates was dissolved in ethanol and concentrated 50-fold. To ensure that the root exudates were free from contamination, 100 μL of the filtered root exudate was plated on LB agar medium, and the plates were incubated at 30°C for 24 h.
GC-TOF-MS (Nanjing Zoonbio Biotechnology Co., Ltd., Nanjing, China) was performed to qualitatively analyze the freeze-dried root exudates using an Agilent 7890 gas chromatograph system coupled with a Pegasus HT time-of-flight mass spectrometer. Chroma TOF 4.3X software (LECO Corporation, St. Joseph, MI, United States) and the LECO-Fiehn Rtx5 database were used to extract raw peaks, data baseline filtering, and calibration of the baseline, peak alignment, deconvolution analysis, peak identification, and integration of the peak area. Both the mass spectrum match and retention index match were considered during metabolite identification.
B. velezensis GUMT319 was grown in LB broth until reaching an OD600 of 0.8. Subsequently, 10 mL of the culture was centrifuged. The cell pellet was washed twice with distilled water and resuspended in 100 μL. Then, 15 mL of LB (0.7% agar) medium was poured into a Petri dish (90 mm diameter), and a 5-μL drop of concentrated bacterial culture was dispensed at the center of the dish. The Petri dish was incubated at 37°C, and the appearance of the bacterial zone was observed after 12 h (de Weert et al., 2002).
B. velezensis GUMT319 was grown and harvested as described above. Then, 15 mL of LB (0.7% agar) medium with the 50-fold concentrated root exudates (or 200 μM different composition of root exudates) was poured into a 90-mm-diameter Petri dish. A 5-μL drop of concentrated bacterial culture was added to the center of the dish and incubated at 37°C for 12 h to check for the appearance of the bacterial zone (de Weert et al., 2002; Weng et al., 2013).
The biofilm formation efficiency of B. velezensis GUMT319 was quantified using the microtiter plate test. Briefly, GUMT319 was grown in 5 mL of LB broth at 37°C until reaching an OD600 value of 0.8. Each well of a sterile 12-well PVC microtiter plate was filled with 2 mL of LB broth and 4 μL of bacterial suspension; 2 mL of LB broth with the 50-fold concentrated root exudate (or 200 μM different composition of root exudates) and 4 μL of bacterial suspension were considered as one treatment. Wells containing only 2 mL of LB broth were used as negative controls. The microtiter plates were incubated at 37°C for 3 days without agitation (Weng et al., 2013).
The effect of GUMT319 on hyphal growth was tested using the dual culture method. Interaction experiments were performed using PDA and LB media. An agar plug (0.5 cm diameter) of actively growing fungi was placed at the center of a PDA plate and incubated for 1 day. GUMT319 was then inoculated at two points, each at a distance of 2 cm from the plug, and plates were photographed after 7 days (Li et al., 2014). This experiment was conducted for five replicates.
To determine the disease control efficacy of GUMT319 under field conditions, experiments were conducted during 2018–2020 in a field naturally infested with tobacco black shank disease. The experimental field was located in Meitan, Zunyi (107°41′N, 27°47′E), Guizhou, China. Roots taken from tobacco seedlings of cultivar Yunyan 87 were soaked in GUMT319 suspension (108 CFU/mL) for 1 h and then transplanted. During growth, each tobacco plant was irrigated with 100 mL of GUMT319 suspension (108 CFU/mL) at 7, 14, and 21 days post-transplantation, and the disease index was measured at 50 and 60 days post-transplantation. Due to frequent rain during the growing season, the GUMT319 suspension was applied more often when appropriate. At 21 days post-transplantation, tobacco plants that had been irrigated with 100 mL of 1:400 of 58% metalaxyl manganese zinc WP (Haixun Agri-Biotech Co., Ltd., Shandong, China) and 1:700 of 1010 CFU/g Bacillus spp. (Green Conway WP, Sino Green Agri-Biotech Co., Ltd., Beijing, China) were used as controls, and plants irrigated with water were designated as blank controls. In 2018 and 2019, the field experiments were performed in an approximately 0.1-ha demonstration area. Tobacco plants were planted in randomized blocks and each block contained 100 plants. Each of the four treatments was replicated four times. In 2020, the field experiments were performed on an approximately 0.67-ha demonstration area and each treatment contained over 2,000 plants. The appearance of tobacco black shank disease symptoms and the cumulative number of infected plants were recorded at 45 days post-transplantation.
The disease incidence was calculated as the percentage of diseased plants relative to the total number of plants growing in each block; evaluation took place when the disease emerged. Disease severity was scored on a scale of 0–9, as follows: 0, no symptoms; 1, less than one-third of the total leaves wilted; 3, one-third to one-half of the total leaves wilted; 5, one-half to two-thirds of the total leaves wilted; 7, more than two-thirds of total leaves wilted; and 9, plant dead. The disease index was calculated using the following equation:
where a, b, c, d, e, and f are the number of plants in each disease category.
Data were statistically analyzed using SPSS v.24.0 (SPSS Inc., Chicago, IL, United States). Mean values of the control and treatment groups were compared using Duncan’s new multiple range test at a significance level of 5% (P < 0.05). All percentage data were subjected to arc-sine transformation before statistical analysis.
The GUMT319 strain was deposited in the China Center for Type Culture Collection (CCTCC Accession No.: M 2018871). Strain GUMT319 was previously classified as B. amyloliquefaciens (Luo et al., 2019). Here, phylogenetic analysis of 16S rRNA and gyrA sequences using the maximum likelihood method placed GUMT319 in a well-supported cluster with B. velezensis FZB42 (Figure 1A). BLASTn analysis of 16S rRNA and gyrA sequences amplified from GUMT319 returned a match to the reference strain B. velezensis FZB42, with >99% identity. Annotation using the NR protein database indicated that sequences amplified from GUMT319 were most similar to sequences of B. velezensis and B. amyloliquefaciens (Figure 1B). Thus, taking into account the 16S rRNA and gyrA gene sequences as well as the whole genome sequence comparisons, GUMT319 was identified as a B. velezensis strain.
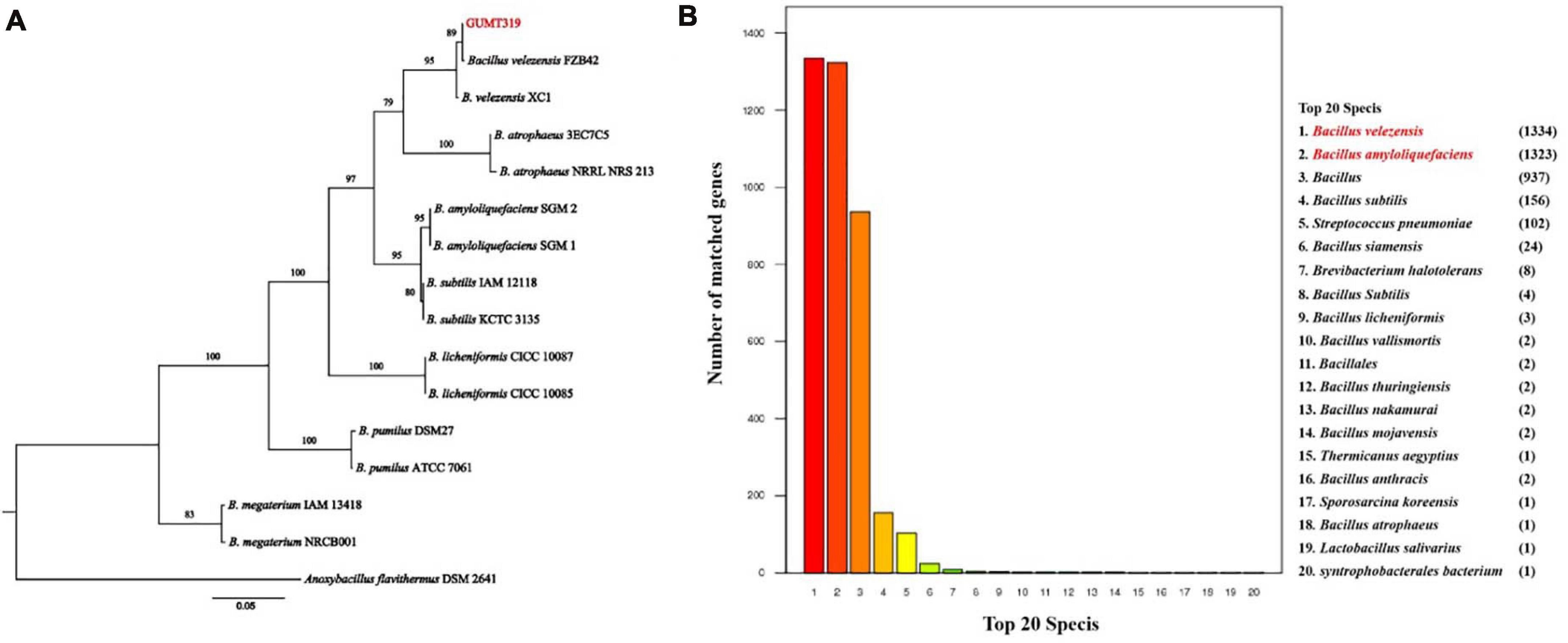
Figure 1. Identification of Bacillus velezensis strain GUMT319. (A) Phylogenetic tree based on the nucleotide sequences of 16S rRNA and gyrA genes. (B) Non-redundant (NR) protein database annotation.
To investigate the biocontrol mechanisms of B. velezensis GUMT319 and its application for sustainable agriculture, its complete genome sequence was determined. The B. velezensis GUMT319 genome consisted of a single circular chromosome 3,940,023 bp in length, with an average GC content of 46.6% (Figure 2), and did not harbor any plasmids. The whole genome of GUMT319 was predicted to contain 4,053 protein-coding genes covering 90.0% of the genome, with an average gene length of 875 bp, as well as 27 rRNAs and 86 tRNAs. Principal features of the genomes of B. velezensis GUMT319 and model strains, B. velezensis FZB42 and SQR9, are summarized in Table 1 (Chen et al., 2007; Zhang et al., 2015). We performed a collinearity analysis to further compare the genomic similarities and differences between the genome of GUMT319 and those of FZB42 and SQR9 (Figure 3). The results showed that the GUMT319 genome displayed different synteny to those of the other strains. GUMT319 showed the highest synteny with B. velezensis FZB42, indicating that their evolutionary stages were the closest, and their genomes were closely related. Amino acid sequence similarity searches against various databases with an E-value threshold of 1e-5 revealed that 3,915 (96.6%), 3,874 (95.6%), 3,293 (81.2%), 2,884 (71.2%), and 2,623 (64.7%) protein-coding genes showed matches in the NR, KEGG, Swiss-Prot, COG, and GO databases, respectively. Abnormal hyphae of plant pathogens were reported in dual cultures with GUMT319, which could be due to the production of secondary metabolites (Luo et al., 2019). Bioinformatics analyses showed that the GUMT319 genome contained 13 putative gene clusters involved in the biosynthesis of secondary metabolites with potential antimicrobial activities (Table 2), most of which were conserved in all B. velezensis strains (bacilysin, surfactin, macrolactin, fengycin, bacillaene, difficidin, and terpene; Supplementary Figure 1; Grady et al., 2019). Among these gene clusters, five encoded non-ribosomal peptide synthetases (NRPSs), four encoded trans-acyl transferase polyketide synthetases (transAT-PKSs), and one encoded type III polyketide synthetase (T3PKS), while two clusters were involved in terpene biosynthesis, and one cluster was involved in lantipeptide biosynthesis (Table 2). In contrast, the clusters predicted to produce lantipeptide had not typically been found in other Bacillus spp. (Supplementary Figure 1).
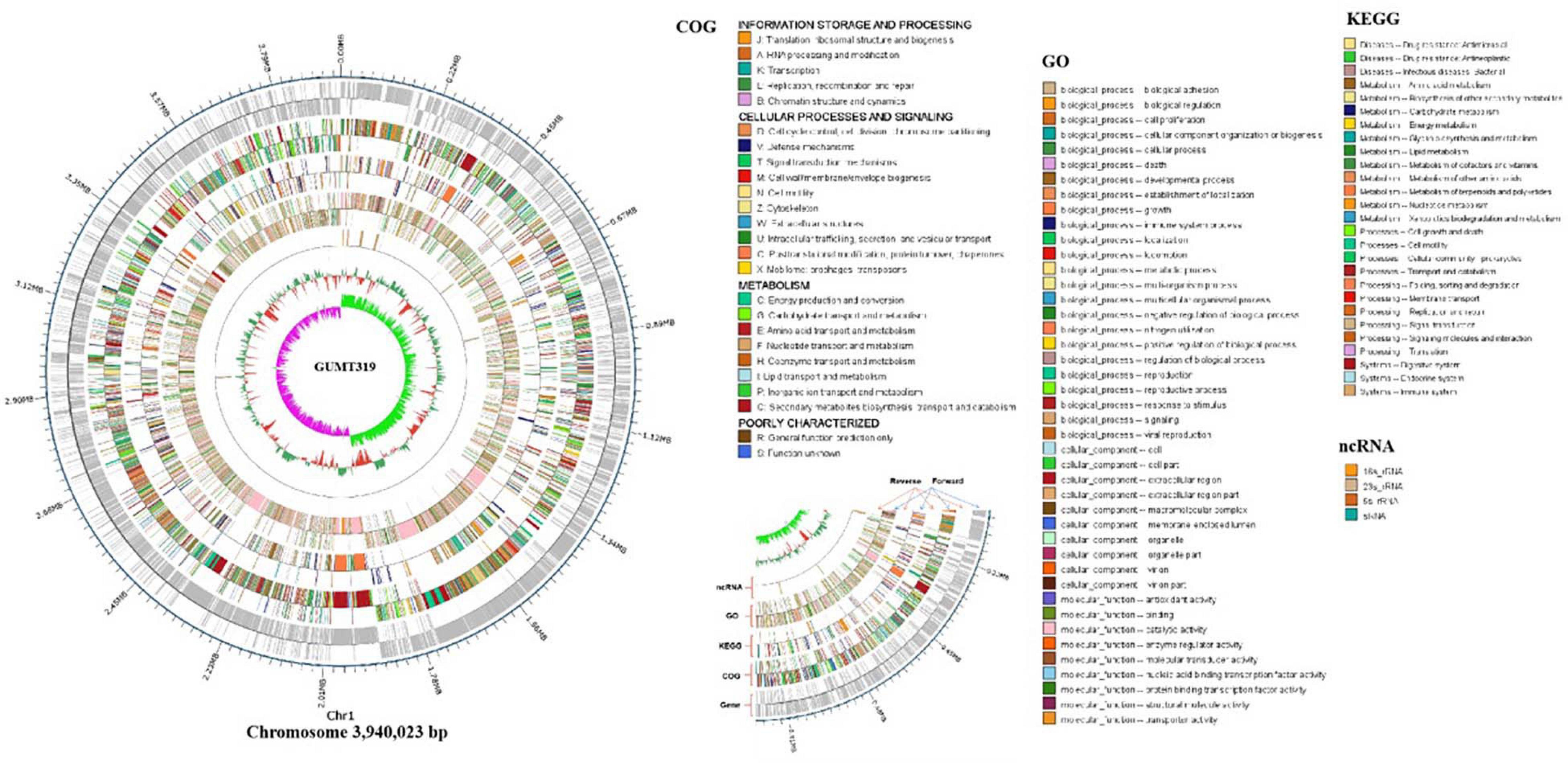
Figure 2. Circular genome map of B. velezensis GUMT319. The scale is shown in the outermost circle. The second and third circles indicate genes in forward and reverse orientations, respectively. The outer to inner rings represent forward and reverse DNA sequences of protein-coding genes annotated according to COG, KEGG, and GO databases and ncRNA. The 10th circle shows the GC content, and the 11th circle shows the GC skew in green and purple.
When B. velezensis GUMT319 was grown in liquid culture without agitation, it formed robust pellicles at the liquid–air interface (Luo et al., 2019). The genome of B. velezensis GUMT319 contained a complete set of genes implicated in biofilm formation, including 15 genes belonging to the exopolysaccharide (EPS) operon epsA-O and tapA-sipW-tasA operon, which are required for the production of EPS and TasA fibers, which hold chains of cells together in bundles (Chen et al., 2007; Gao et al., 2015a; Al-Ali et al., 2018; Table 3). B. velezensis GUMT319 displayed a robust swarming phenotype (Supplementary Figure 2), and the protein encoded by swrA was essential for swarming. The genome of B. velezensis GUMT319 contained a complete set of genes implicated in chemotaxis and motility, including six MCPS genes, 25 flagellin gene clusters, and nine chemotaxis protein genes (Liu et al., 2020; Table 3). A previous study shows that GUMT319 produces siderophores, which facilitate rhizosphere competition and growth promotion (Gu et al., 2020). The genome of B. velezensis GUMT319 contained genes involved in siderophore biosynthesis, such as dhbB, dhbE, dhbF, and entC. GUMT319 also contained master global transcriptional regulators, such as AbrB and Spo0A, as well as the quorum-sensing regulators LuxS/AI-2 and ComA/ComP, which regulate biofilm formation, chemotaxis, and root colonization (Comella and Grossman, 2005; Weng et al., 2013; Yan et al., 2016; Xiong et al., 2020; Table 3).
Root colonization of the rhizosphere by antagonistic bacteria is a prerequisite for effective biological control (Liu et al., 2014). In this study, we investigated the colonization of healthy tobacco and pepper seedling roots by B. velezensis GUMT319. After 2 days of incubation in the hydroponic system, root colonization by B. velezensis GUMT319 and GUMT319-gfp was investigated using SEM and a CLSM. The SEM images showed that GUMT319 cells were rod-shaped (Figures 4A,C). Additionally, a complex biofilm structure consisting of GUMT319 cells (Figure 4C) and GUMT319-gfp cells (Figure 4D) was formed on the tobacco root surface, and colonization occurred preferentially in the elongation region of tobacco roots (Figure 4). We also found that GUMT319 (Figure 4A) and GUMT319-gfp (Figure 4B) could colonize pepper roots, although to a much lesser extent than in tobacco roots (Figure 4).
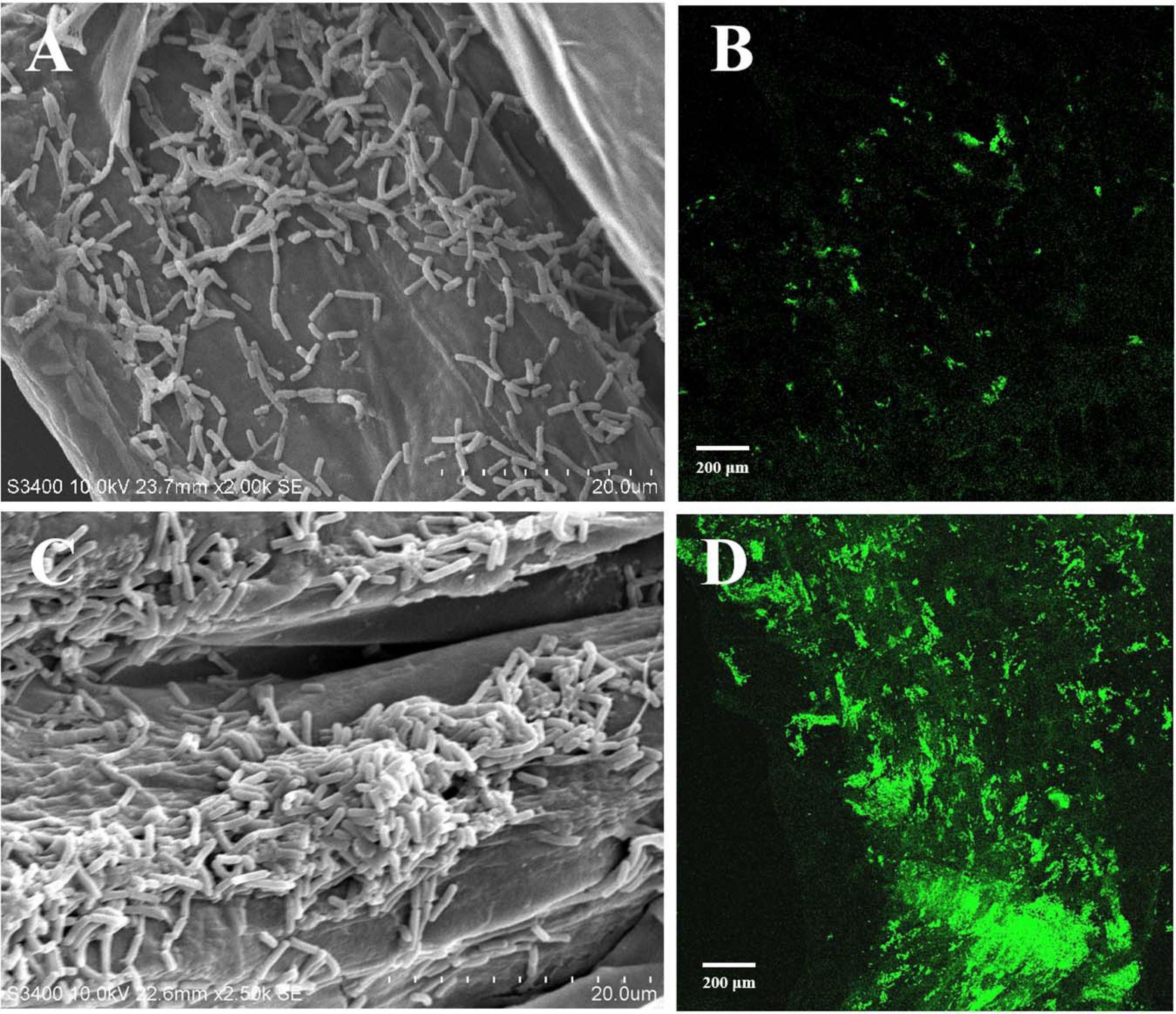
Figure 4. Root colonization assay of B. velezensis GUMT319. (A) Scanning electron microscope (SEM, HITACHI S-3400N) images of pepper seedling roots colonized by strain GUMT319. (B) Confocal laser scanning microscope (CLSM, Olympus FV10i) images of pepper seedling roots colonized by GUMT319-gfp. (C) SEM images of tobacco seedling roots colonized by strain GUMT319. (D) Confocal laser scanning microscope (CLSM) images of tobacco seedling roots colonized by GUMT319-gfp. The green fluorescence signal depicts live cells colonizing the tobacco roots.
Root exudates are known to play an important role in plant–microbe interaction in the rhizosphere. The different compositions of root exudates from different plants are directly related to the chemotaxis reaction, biofilm formation, and colonizing behavior of bacterial strains originating from the respective rhizosphere (Zhang et al., 2014). We wanted to identify the different components of the root exudates that mediate the interaction between biocontrol Bacillus spp. and plant roots, which influenced their root colonization. To this end, root exudates from tobacco and pepper plants were collected and analyzed. Qualitative analyses of the freeze-dried tobacco and pepper root exudates were performed by GC-TOF-MS, and 248 peaks (Supplementary Figure 2) were detected, including amino acids, organic acids, and others (data not shown). Here, some organic acids (cinnamic acid, fumaric acid, phthalic acid, benzoic acid, and lauric acid) and nicotine were selected as the targets for evaluation of their roles on B. velezensis GUMT319 (Zhang et al., 2014; Feng et al., 2018; Ma et al., 2018). Cinnamic acid, fumaric acid, and benzoic acid were detected both in tobacco and pepper root exudates. Phthalic acid and nicotine were found only in tobacco root exudates and lauric acid was found only in pepper root exudates.
To investigate the cause of root colonization by B. velezensis GUMT319, the chemotactic reaction and biofilm formation response of GUMT319 to two root exudates and six components were determined. After the 12-h incubation period, the LB (0.7% agar) plate was almost fully covered by GUMT319 cells (Supplementary Figure 2), indicating that GUMT319 was highly proficient in swarming. We measured the chemotaxis activity of strain GUMT319 using tobacco root exudates, pepper root exudates, organic acids (cinnamic acid, fumaric acid, phthalic acid, benzoic acid, and lauric acid), and nicotine as attractants. The root exudates induced a positive chemotactic response in B. velezensis GUMT319, and cells showed faster migration toward all the attractants than toward the control after a 6-h incubation (Figure 5). The biofilm formation activity of strain GUMT319 was also examined using the qualitative biofilm experiment with tobacco root exudates, pepper root exudates, organic acids (cinnamic acid, fumaric acid, phthalic acid, benzoic acid, and lauric acid), and nicotine as attractants. The results showed that only lauric acid could be involved in the negative regulation of biofilm formation. No obvious difference was observed in biofilm formation among other treatments conducted with or without root exudates. The pellicles formed by GUMT319 were similar in all other groups (Figure 6).
In this study, it was found that GUMT319 displayed strong antagonistic activity against P. nicotianae, C. scovillei, C. capsici, F. carminascens, S. sclerotiorum, A. alternata, Phomopsis sp., P. sorghina, and E. turcicum (Figure 7 and Supplementary Table 2), indicating that B. velezensis GUMT319 was effective against a relatively broad spectrum of oomycetes and ascomycetes.
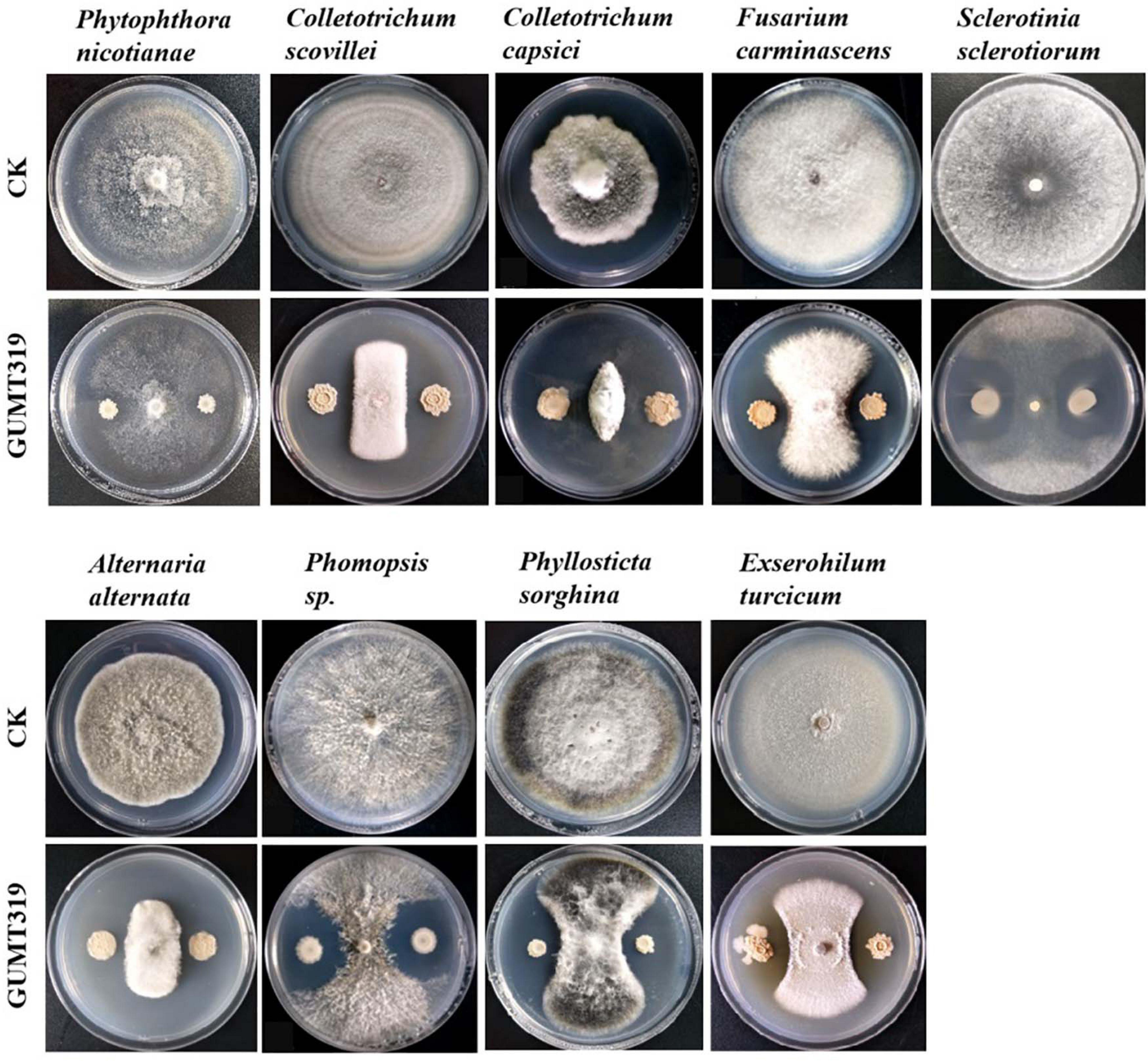
Figure 7. Antagonistic activity of B. velezensis GUMT319 against nine plant pathogens in dual-culture test.
We further investigated the efficiency of control of GUMT319 on tobacco black shank in field experiments. In those conducted in 2018, the disease incidence of tobacco plants treated with sterile water was 20.48%, while the disease incidence of plants treated independently with GUMT319 suspension, 58% metalaxyl manganese zinc WP, and Green Conway WP decreased, having control efficacy on tobacco black shank of 77.15, 76.43, and 65.59%, respectively (Table 3). Tobacco plants treated with strain GUMT319 showed the lowest disease incidence, with a control efficiency significantly higher than those treated with Green Conway WP. In the 2019 field experiments, the disease incidence of tobacco plants treated with water was 15.42%, while the control efficacy on tobacco black shank of those plants treated independently with GUMT319, 58% metalaxyl manganese zinc WP, and Green Conway WP were 71.97, 71.59, and 66.47%, respectively (Table 3). In the 2020 field experiments, the disease incidence of tobacco plants treated with water reached 79.09%, while the control efficacy on tobacco black shank of those plants treated independently with GUMT319, 58% metalaxyl manganese zinc WP, and Green Conway WP were 79.54, 76.98, and 62.31%, respectively (Table 4 and Supplementary Figure 4). Thus, the control effect of GUMT319 suspension was significantly higher than that of 58% metalaxyl manganese zinc WP and Green Conway WP (P < 0.05), suggesting that GUMT319 could be used as a potential BCA for tobacco black shank.
The tobacco black shank control efficiency of several Bacillus strains, such as B. subtilis Tpb55 and B. velezensis Ba168, has been tested previously. Tobacco black shank, caused by P. nicotianae, is destructive to almost all flue-cured tobacco cultivars and is widespread in many tobacco-growing countries. Bacillus strains represent a promising strategy for the management of this soil-borne disease (Han et al., 2016; Zhang et al., 2017a; Srikhong et al., 2018; Guo et al., 2020). In this study, we identified another B. velezensis isolate, GUMT319, from a healthy tobacco rhizosphere in the fields in Guizhou with a high incidence of tobacco black shank. The control efficiency of GUMT319 against tobacco black shank was greater than 70% in field experiments conducted in Meitan in 2018–2020, which was significantly higher than that obtained using other treatments including fungicide application (Table 4 and Supplementary Figure 4). Hence, GUMT319 could potentially be used as a BCA for tobacco black shank in Guizhou.
GUMT319 was previously identified as a B. amyloliquefaciens strain; however, 16S rRNA and gyrA sequences and whole genome sequence comparisons in the current study revealed that GUMT319 is a B. velezensis strain (Figure 1). Similarly, SQR9 and FZB42, which were previously classified as B. amyloliquefaciens strains, are now recognized as B. velezensis strains and considered as the model strains for gram-positive plant growth-promoting and biocontrol rhizobacteria (Chen et al., 2007; Zhang et al., 2015; Fan et al., 2018). We support the theory that many strains currently classified as B. amyloliquefaciens are actually B. velezensis.
In this study, we identified and classified B. velezensis strain GUMT319 based on genomic data generated using second- and third-generation sequencing technologies. Strain GUMT319 contained a single circular chromosome 3,940,023 bp in length, with 4,053 predicted genes and an average GC content of 46.6% (Figure 2). The genome of GUMT319 is smaller than that of SQR9 (4,117,023 bp) but comparable to that of FZB42 (3,918,596 bp). The three strains were identical in terms of the number of protein-coding genes but differed in the number and size of prophage regions as well as gene clusters encoding secondary metabolites. The majority of genes unique to GUMT319 (i.e., absent in SQR9 and FZB42) were phage related (Table 1 and Figure 3).
The ability of B. velezensis GUMT319 to efficiently colonize the surface of tobacco roots was a prerequisite for biocontrol. Many beneficial bacteria form biofilms on plant roots, which play an important role in their antagonistic activities and biocontrol efficacy (Chen et al., 2013). Rhizosphere competence of bacteria is associated with their ability to form sessile, multicellular communities (biofilms) and their chemotaxis motility (Niu et al., 2017; Liu et al., 2020). The genome of GUMT319 contains more than 60 genes and 13 putative gene clusters related to secondary metabolites, which have previously been described as being involved in biofilm formation, chemotaxis motility, growth promotion, and antifungal activity (Tables 2, 3).
B. velezensis has been widely researched because of its potential to produce bacteriostatic secondary metabolites (Fan et al., 2018). B. velezensis strain GUMT319 has been reported to produce cell wall-degrading enzymes such as proteases, cellulases, and phosphatases as well as siderophores, which facilitate rhizosphere competition and plant growth promotion and show strong inhibitory activity against P. nicotianae as well as eight other plant pathogens in vitro (Luo et al., 2019). The GUMT319 genome contained 13 putative gene clusters that were expected to participate in antimicrobial production, including bacilysin, surfactin, macrolactin, fengycin, bacillaene, difficidin, and terpene (Table 3 and Supplementary Figure 1; Grady et al., 2019); the majority of these gene clusters are conserved in all B. velezensis strains. However, clusters predicted to produce lantipeptides were not typical in other Bacillus spp. (Supplementary Figure 3). Lantipeptides are ribosomally synthesized peptides that are extensively post-translationally modified. It is suggested that lantipeptides are the antimicrobial compounds produced by gram-positive bacteria (Grady et al., 2019).
The ability of Bacillus to colonize plant roots and proliferate in the rhizosphere is important for stable, long-lasting disease prevention (Han et al., 2016). To examine the ability of GUMT319 to colonize plant roots, we labeled the strain with GFP. In the present study, GUMT319 and GUMT319-gfp colonized the surface of tobacco roots, forming microcolonies and complex biofilm structures. The root elongation zone was colonized to a greater extent than other areas of the root. We also found that GUMT319 and GUMT319-gfp could colonize on pepper roots, but lesser than that on tobacco roots (Figure 4). As a signal to attract or repel microbes, the root exudates serve as a carbon source for soil microorganisms. In addition, they play significant roles in chemotaxis, biofilm formation, and colonization of Bacillus strains on plant roots (Feng et al., 2018). The composition of root exudates is closely related to plant species, soil environment, climate factors, microorganisms, nutritional status, etc. Among them, plant species is the most important (Zhang et al., 2014). Therefore, we speculated that some substances in tobacco root exudates (not pepper) play a key role. The root exudates from tobacco and pepper plants were collected and analyzed using GC-TOF-MS (Supplementary Figure 3). It was found that organic acids were the most represented class of compounds in both plants. Hence, some organic acids (cinnamic acid, fumaric acid, phthalic acid, benzoic acid, and lauric acid) (Zhang et al., 2014; Feng et al., 2018) and nicotine (Ma et al., 2018) were selected as targets for the evaluation of their roles on B. velezensis GUMT319. Cinnamic acid, fumaric acid, and benzoic acid were detected both in tobacco and pepper root exudates, phthalic acid and nicotine in tobacco root exudates, and lauric acid only in pepper root exudates.
In this study, in tobacco root exudates, pepper root exudates, organic acids (cinnamic acid, fumaric acid, phthalic acid, benzoic acid, and lauric acid), and nicotine as attractants, it was found that GUMT319 moved faster toward all root exudates than toward the control (Figure 5). Lauric acid could be involved in the negative regulation of biofilm formation. There was no obvious difference among other treatments conducted with or without root exudates in biofilm formation (Figure 6). Compared with other root-secreted compounds, it was first reported that nicotine in the root exudates of tobacco was a chemoattractant for B. velezensis GUMT319. Previous research shows that nicotine from tobacco root exudates has the ability to enhance chemotaxis, growth, biocontrol efficiency, and colonization by Pseudomonas aeruginosa NXHG29 (Ma et al., 2018). However, we do not know how B. velezensis GUMT319 recognizes nicotine and regulates chemotaxis and root colonization.
Overall, this study showed that the beneficial function of GUMT319 in controlling tobacco black shank disease is through its ability to inhibit the mycelial growth of plant pathogens and to successfully colonize tobacco roots. Hence, B. velezensis GUMT319 could be used as a potential BCA against tobacco black shank.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP068563.
HD, LP, and ZL designed the research and wrote the manuscript. HD, WM, SY, HC, and LP performed all the experiments. HD and WM analyzed the data. All authors reviewed the final manuscript.
This work was supported by the China Postdoctoral Science Foundation (Grant No. 2020M683658XB), the Development Project of Young Scientific and Technological Talents of Guizhou Province (Grant No. QJH-KY[2018]101), and the Science and Technology Project of Zunyi, Guizhou Tobacco Company (Grant No. 201706).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We gratefully acknowledge Prof. Yong Wang for useful discussions about this article.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.658113/full#supplementary-material
Al-Ali, A., Deravel, J., Krier, F., Béchet, M., Ongena, M., and Jacques, P. (2018). Biofilm formation is determinant in tomato rhizosphere colonization by Bacillus velezensis FZB42. Environ. Sci. Pollut. Res. 25, 29910–29920. doi: 10.1007/s11356-017-0469-1
Borriss, R., Danchin, A., Harwood, C. R., Médigue, C., Rocha, E. P. C., Sekowska, A., et al. (2018). Bacillus subtilis, the model Gram-positive bacterium: 20 years of annotation refinement. Microb. Biotechnol. 11, 3–17. doi: 10.1111/1751-7915.13043
Chen, X., Zhang, Y., Fu, X., Li, Y., and Wang, Q. (2016). Isolation and characterization of Bacillus amyloliquefaciens PG12 for the biological control of apple ring rot. Postharvest Biol. Technol. 115, 113–121. doi: 10.1016/j.postharvbio.2015.12.021
Chen, X. H., Koumoutsi, A., Scholz, R., Eisenreich, A., Schneider, K., Heinemeyer, I., et al. (2007). Comparative analysis of the complete genome sequence of the plant growth–promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 25, 1007–1014. doi: 10.1038/nbt1325
Chen, Y., Yan, F., Chai, Y., Liu, H., Kolter, R., Losick, R., et al. (2013). Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol. 15, 848–864. doi: 10.1111/j.1462-2920.2012.02860.x
Chowdhury, S. P., Hartmann, A., Gao, X., and Borriss, R. (2015). Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 – a review. Front. Microbiol. 6:780. doi: 10.3389/fmicb.2015.00780
Comella, N., and Grossman, A. D. (2005). Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol. Microbiol. 57, 1159–1174. doi: 10.1111/j.1365-2958.2005.04749.x
de Weert, S., Vermeiren, H., Mulders, I. H., Kuiper, I., Hendrickx, N., Bloemberg, G. V., et al. (2002). Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant Microbe Interact. 15, 1173–1180. doi: 10.1094/MPMI.2002.15.11.1173
Fan, B., Wang, C., Song, X., Ding, X., Wu, L., Wu, H., et al. (2018). Bacillus velezensis FZB42 in 2018: the gram-positive model strain for plant growth promotion and biocontrol. Front. Microbiol. 9:2491. doi: 10.3389/fmicb.2018.02491
Feng, H., Zhang, N., Du, W., Zhang, H., Liu, Y., Fu, R., et al. (2018). Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant-growth-promoting rhizobacteria Bacillus amyloliquefaciens SQR9. Mol. Plant-Microbe Inter. 31, 995–1005. doi: 10.1094/MPMI-01-18-0003-R
Feng, H., Zhang, N., Fu, R., Liu, Y., Krell, T., Du, W., et al. (2019). Recognition of dominant attractants by key chemoreceptors mediates recruitment of plant growth-promoting rhizobacteria. Environ. Microbiol. 21, 402–415. doi: 10.1111/1462-2920.14472
Gao, T., Foulston, L., Chai, Y., Wang, Q., and Losick, R. (2015a). Alternative modes of biofilm formation by plant-associated Bacillus cereus. MicrobiologyOpen 4, 452–464. doi: 10.1002/mbo3.251
Gao, T., Greenwich, J., Li, Y., Wang, Q., and Chai, Y. (2015b). The bacterial tyrosine kinase activator TkmA contributes to biofilm formation largely independently of the cognate kinase PtkA in Bacillus subtilis. J. Bacteriol. 197, 3421–3432. doi: 10.1128/JB.00438-15
Grady, E. N., MacDonald, J., Ho, M. T., Weselowski, B., McDowell, T., Solomon, O., et al. (2019). Characterization and complete genome analysis of the surfactin-producing, plant-protecting bacterium Bacillus velezensis 9D-6. BMC Microbiol. 19:5. doi: 10.1186/s12866-018-1380-8
Gu, S., Wei, Z., Shao, Z., Friman, V. P., Cao, K., Yang, T., et al. (2020). Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat. Microbiol. 5, 1002–1010. doi: 10.1038/s41564-020-0719-8
Guo, D., Yuan, C., Luo, Y., Chen, Y., Lu, M., Chen, G., et al. (2020). Biocontrol of tobacco black shank disease (Phytophthora nicotianae) by Bacillus velezensis Ba168. Pestic. Biochem. Physiol. 165:104523. doi: 10.1016/j.pestbp.2020.01.004
Han, T., You, C., Zhang, L., Feng, C., Zhang, C., Wang, J., et al. (2016). Biocontrol potential of antagonist Bacillus subtilis Tpb55 against tobacco black shank. Biocontrol 61, 195–205. doi: 10.1007/s10526-015-9705-0
Li, B., Li, Q., Xu, Z., Zhang, N., Shen, Q., and Zhang, R. (2014). Responses of beneficial Bacillus amyloliquefaciens SQR9 to different soilborne fungal pathogens through the alteration of antifungal compounds production. Front. Microbiol. 5:636. doi: 10.3389/fmicb.2014.00636
Liu, Y., Feng, H., Fu, R., Zhang, N., Du, W., Shen, Q., et al. (2020). Induced root-secreted d-galactose functions as a chemoattractant and enhances the biofilm formation of Bacillus velezensis SQR9 in an McpA-dependent manner. Appl. Microbiol. Biotechnol. 104, 785–797. doi: 10.1007/s00253-019-10265-8
Liu, Y., Zhang, N., Qiu, M., Feng, H., Vivanco, J. M., Shen, Q., et al. (2014). Enhanced rhizosphere colonization of beneficial Bacillus amyloliquefaciens SQR9 by pathogen infection. FEMS Microbiol. Lett. 353, 49–56. doi: 10.1111/1574-6968.12406
Luo, Y., Peng, L., Ding, J., Fu, S., and Li, Y. (2019). Screening and field control effect of biological control Bacillus strains against tobacco black shank. Guizhou Agric. Sci. North. Hort. 47, 59–64.
Ma, L., Zheng, S. C., Zhang, T. K., Liu, Z. Y., Wang, X. J., Zhou, X. K., et al. (2018). Effect of nicotine from tobacco root exudates on chemotaxis, growth, biocontrol efficiency, and colonization by Pseudomonas aeruginosa NXHG29. Antonie Van Leeuwenhoek 111, 1237–1257. doi: 10.1007/s10482-018-1035-7
Mao, T., Chen, X., Ding, H., Chen, X., and Jiang, X. (2020). Pepper growth promotion and Fusarium wilt biocontrol by Trichoderma hamatum MHT1134. Biocontrol Sci. Technol. 30, 1228–1243. doi: 10.1080/09583157.2020.1803212
Niu, B., Paulson, J. N., Zheng, X., and Kolter, R. (2017). Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. U.S.A. 114, E2450–E2459.
Srikhong, P., Lertmongkonthum, K., Sowanpreecha, R., and Rerngsamran, P. (2018). Bacillus sp. strain M10 as a potential biocontrol agent protecting chili pepper and tomato fruits from anthracnose disease caused by Colletotrichum capsici. Biocontrol 63, 833–842. doi: 10.1007/s10526-018-9902-8
Wang, J., Xing, J., Lu, J., Sun, Y., Zhao, J., Miao, S., et al. (2019). Complete genome sequencing of Bacillus velezensis WRN014, and comparison with genome sequences of other Bacillus velezensis strains. J. Microbiol. Biotechnol. 29, 794–808. doi: 10.4014/jmb.1901.01040
Weng, J., Wang, Y., Li, J., Shen, Q., and Zhang, R. (2013). Enhanced root colonization and biocontrol activity of Bacillus amyloliquefaciens SQR9 by abrB gene disruption. Appl. Microbiol. Biotechnol. 97, 8823–8830. doi: 10.1007/s00253-012-4572-4
Wu, G., Liu, Y., Xu, Y., Zhang, G., Shen, Q., and Zhang, R. (2018). Exploring elicitors of the beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 to induce plant systemic resistance and their interactions with plant signaling pathways. Mol. Plant Microbe Interact. 31, 560–567. doi: 10.1094/mpmi-11-17-0273-r
Xiong, Q., Liu, D., Zhang, H., Dong, X., Zhang, G., Liu, Y., et al. (2020). Quorum sensing signal autoinducer-2 promotes root colonization of Bacillus velezensis SQR9 by affecting biofilm formation and motility. Appl. Microbiol. Biotechnol. 104, 7177–7185. doi: 10.1007/s00253-020-10713-w
Xu, P., Xie, S., Liu, W., Jin, P., Wei, D., Yaseen, D. G., et al. (2020). Comparative genomics analysis provides new strategies for bacteriostatic ability of Bacillus velezensis HAB-2. Front. Microbiol. 11:594079. doi: 10.3389/fmicb.2020.594079
Xu, Z., Shao, J., Li, B., Yan, X., Shen, Q., and Zhang, R. (2013). Contribution of bacillomycin D in Bacillus amyloliquefaciens SQR9 to antifungal activity and biofilm formation. Appl. Environ. Microbiol. 79, 808–815. doi: 10.1128/AEM.02645-12
Yan, F., Li, C., Ye, X., Lian, Y., Wu, Y., and Wang, X. (2020). Antifungal activity of lipopeptides from Bacillus amyloliquefaciens MG3 against Colletotrichum gloeosporioides in loquat fruits. Biol. Control 146:104281. doi: 10.1016/j.biocontrol.2020.104281
Yan, F., Yu, Y., Wang, L., Luo, Y., Guo, J., and Chai, Y. (2016). The comer gene plays an important role in biofilm formation and sporulation in both Bacillus subtilis and Bacillus cereus. Front. Microbiol. 7:1025. doi: 10.3389/fmicb.2016.01025
Ye, X., Li, Z., Luo, X., Wang, W., Li, Y., Li, R., et al. (2020). A predatory myxobacterium controls cucumber Fusarium wilt by regulating the soil microbial community. Microbiome 8:49. doi: 10.1186/s40168-020-00824-x
Zhang, C., Gao, J., Han, T., Tian, X., and Wang, F. (2017a). Integrated control of tobacco black shank by combined use of riboflavin and Bacillus subtilis strain Tpb55. Biocontrol 62, 835–845. doi: 10.1007/s10526-017-9849-1
Zhang, G., Wang, W., Deng, A., Sun, Z., Zhang, Y., Liang, Y., et al. (2012). A mimicking-of-DNA-methylation-patterns pipeline for overcoming the restriction barrier of bacteria. PLoS Genet. 8:e1002987. doi: 10.1371/journal.pgen.1002987
Zhang, N., Wang, D., Liu, Y., Li, S., Shen, Q., and Zhang, R. (2014). Effects of different plant root exudates and their organic acid components on chemotaxis, biofilm formation and colonization by beneficial rhizosphere-associated bacterial strains. Plant Soil 374, 689–700. doi: 10.1007/s11104-013-1915-6
Zhang, N., Yang, D., Wang, D., Miao, Y., Shao, J., Zhou, X., et al. (2015). Whole transcriptomic analysis of the plant-beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 during enhanced biofilm formation regulated by maize root exudates. BMC Genomics 16:685. doi: 10.1186/s12864-015-1825-5
Zhang, R., Vivanco, J. M., and Shen, Q. (2017b). The unseen rhizosphere root-soil-microbe interactions for crop production. Curr. Opin. Microbiol. 37, 8–14. doi: 10.1016/j.mib.2017.03.008
Keywords: tobacco black shank, Bacillus velezensis, biocontrol, genome sequencing, antifungal
Citation: Ding H, Mo W, Yu S, Cheng H, Peng L and Liu Z (2021) Whole Genome Sequence of Bacillus velezensis Strain GUMT319: A Potential Biocontrol Agent Against Tobacco Black Shank Disease. Front. Microbiol. 12:658113. doi: 10.3389/fmicb.2021.658113
Received: 25 January 2021; Accepted: 25 May 2021;
Published: 06 July 2021.
Edited by:
Amin Uddin Mridha, University of Chittagong, BangladeshReviewed by:
Khaled Abbas El-Tarabily, United Arab Emirates University, United Arab EmiratesCopyright © 2021 Ding, Mo, Yu, Cheng, Peng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijuan Peng, Mjk2NDMwMDA2QHFxLmNvbQ==; Zuoyi Liu, Z3psaXV6dW95aUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.