- Viral Immunobiology, Institute of Experimental Immunology, University of Zurich, Zurich, Switzerland
Epstein Barr virus (EBV) is one of the most successful pathogens of humans, persistently colonizing more than 95% of the adult human population. At the same time EBV encodes oncogenes that can readily transform human B cells in culture and threaten healthy virus carriers with lymphomagenesis. Cytotoxic lymphocytes have been identified in experimental models and by primary immunodeficiencies as the main protective immune compartments controlling EBV. EBV has reached a stalemate with these cytotoxic T and innate lymphocytes to ensure persistence in most infected humans. Recent evidence suggests that the non-coding RNAs of the virus contribute to viral immune escape to prevent immune eradication. This knowledge might be used in the future to attenuate EBV for vaccine development against this human tumor virus that was discovered more than 55 years ago.
Introduction on EBV and Its Oncogenesis
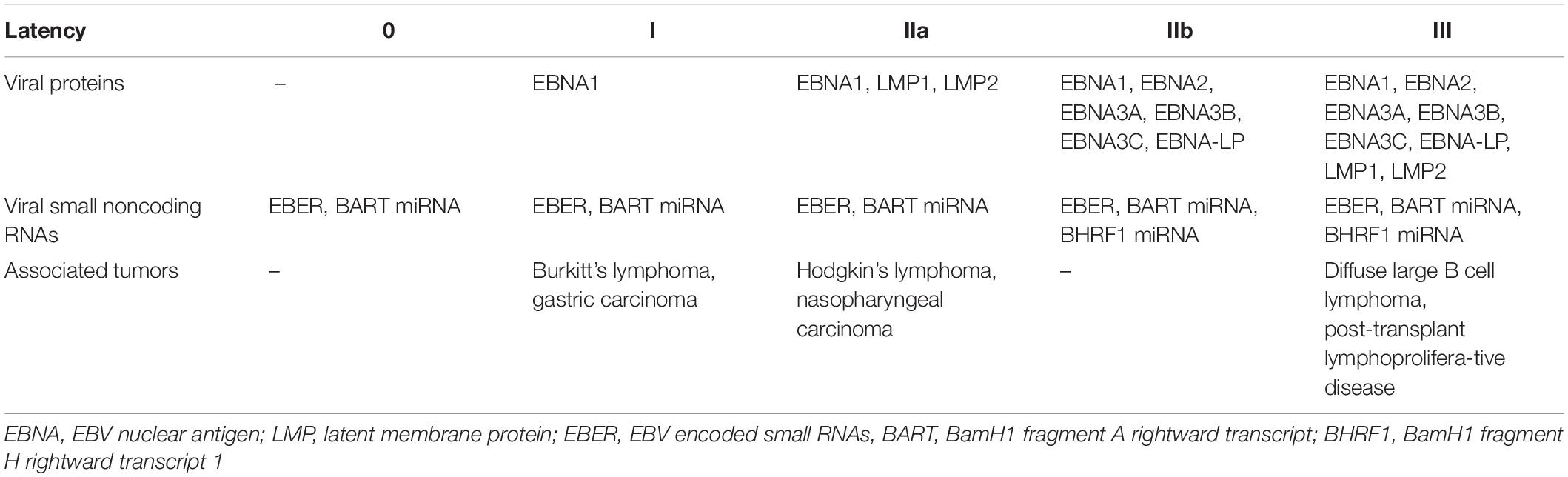
Epstein Barr virus (EBV) that is also called human herpesvirus 4 (HHV4) is a common human γ-herpesvirus establishing persistent infection in more than 95% of the human adult population (Münz, 2019). Characteristic for herpesviruses it exists in latent and lytic infection in its human host. Latent infection allows persistence, while lytic replication generates infectious viral particles for transmission. In the case of EBV, five latent infection programs, namely latency 0, I, IIa, IIb, and III, are thought to establish a latency reservoir in B cells for long-term persistence of this virus (Thorley-Lawson and Gross, 2004; Kempkes and Robertson, 2015; Table 1). For this purpose and after transmission by saliva exchange, EBV is thought to cross mucosal epithelia (Tugizov et al., 2013) for B cell infection in submucosal secondary lymphoid tissues like the tonsils (Farrell, 2019). In naïve B cells successively the latency IIb and III infection programs are established after EBV infection with the expression of all 6 nuclear antigens of EBV (EBNA1, 2, 3A, 3B, 3C, and LP), the small noncoding RNAs EBER1 and 2, 48 miRNAs in latency IIb and for latency III in addition the two latent membrane proteins (LMP1 and 2). The 48 EBV miRNAs originate from 25 miRNA precursors that are encoded in two clusters, namely the BHRF1 cluster (3 miRNA precursors) and the BART cluster (25 miRNA precursors) (Caetano et al., 2021; Pfeffer et al., 2004). The two latency programs IIb and III induce B cell transformation and latency III is also found in lymphoblastoid cell lines (LCLs) that can be generated by EBV infection of B cells in vitro. The associated B cell proliferation and rescue from apoptosis is thought to drive B cells into differentiation in vivo with successive down-regulation of viral transcripts (Babcock et al., 2000). In germinal center B cells only EBNA1, LMP1 and 2, EBERs and BART miRNAs are expressed (latency IIa). This latency can, however, be also reached without prior EBNA2 dependent latency IIb and III in vivo (Li et al., 2020). Latency IIa rescues infected cells from the germinal center reaction for persistence in memory B cells with only EBER and BART miRNA expression (latency 0) or additional EBNA1 expression during homeostatic proliferation (latency I) (Babcock et al., 1998; Hochberg et al., 2004). However, latency 0 persistence can be also achieved from latency IIb directly in the absence of EBNA3C (Murer et al., 2018). From latency 0 and I lytic reactivation of EBV can be induced by B cell receptor cross-linking, suggesting that cognate antigen exposure induces the immediate early transactivation factors BZLF1 and BRLF1 for lytic EBV infection (Kenney and Mertz, 2014). The resulting plasma cell differentiation is also associated with lytic EBV replication in vivo (Laichalk and Thorley-Lawson, 2005; McHugh et al., 2017). More than 80 early and late lytic gene products are then expressed together with EBERs and BHRF1 as well as BART miRNAs to produce infectious EBV particles (McKenzie and El-Guindy, 2015). Such lytic replication resulting in infectious particle production in submucosal secondary lymphoid tissues allows then for mucosal epithelia infection from the basolateral side (Tugizov et al., 2003; Chen et al., 2018; Zhang et al., 2018). Additional lytic replication in epithelia is thought to amplify shedding of EBV that is ideally suited for B cell infection and transmission via saliva exchange (Borza and Hutt-Fletcher, 2002; Hadinoto et al., 2009; Hutt-Fletcher, 2017). In addition to the EBER, BHRF1 and BART small ncRNAs, miRNA independent functions of the BART long ncRNA and EBV derived circular RNAs have been described (Bullard et al., 2018; Ungerleider et al., 2019). Among these the BART long ncRNA contains also a small nucleolar RNA that could be involved in RNA modifications (Hutzinger et al., 2009). However, their functions in EBV infected B cells and the regulation of latent and lytic infection remain largely unclear. Thus, EBV is ideally adapted to the human B cell physiology to achieve both persistence in long-lived memory B cells and lytic replication in plasma cells.
Despite asymptomatic EBV co-existence with its human host in most virus carriers, the above described EBV latency programs are, however, also associated with malignancies that occur more frequently during immune suppression after transplantation or human immunodeficiency virus (HIV) co-infection (Cesarman, 2014; Shannon-Lowe and Rickinson, 2019). In fact EBV causes around 200,000 tumors every year in the human population (Cohen et al., 2011). Latency I is found in Burkitt’s lymphoma and gastric carcinoma, latency IIa in Hodgkin’s lymphoma and nasopharyngeal carcinoma, and latency III in post-transplant lymphoproliferative disease (PTLD) and some diffuse large B cell lymphomas (DLBCL) (Shannon-Lowe and Rickinson, 2019). In this review I will discuss the role of EBV’s non-coding RNAs (ncRNAs), focusing on EBERs and miRNAs, in EBV driven tumorigenesis, transition of latent to lytic replication, and immune control thereof.
Regulation of Viral Transcripts by EBV ncRNAs
EBER1 and 2 are non-polyadenylated RNA polymerase III transcripts of 162 and 172 nucleotide length, respectively (Farrell, 2019). Despite them being with several 1,000 copies per EBV infected cell (Lerner et al., 1981) among the most abundant viral transcripts and due to their expression in all latent and lytic infection program valuable for diagnostics by in situ hybridization (Weiss and Chen, 2013) EBER function still remains enigmatic. Both EBERs are not required for in vitro immortalization of human B cells and do not affect EBV infection of mice with reconstituted human immune system compartments (Gregorovic et al., 2011, 2015). Although deletion of EBER1 or 2 in recombinant EBVs leads to increased LMP2 mRNA in the respective LCLs immortalized with these viruses, LMP2 protein levels are not affected and no changes in the transformation frequency of primary human B cells or the growth of established EBER deficient LCLs were observed. Thus, EBERs seem to have little influence on viral gene expression.
In contrast the RNA polymerase II miRNA precursors that are then processed by Drosha and Dicer, regulate both latent and lytic EBV gene transcripts (Skalsky and Cullen, 2015; Poling et al., 2017). Regarding latent EBV infection regulation it was originally noted that in the absence of BHRF1 miRNAs B cell transformation in vitro and EBV infection in mice with reconstituted immune system components is compromised (Feederle et al., 2011a,b; Wahl et al., 2013). Furthermore, LMP2 is down-regulated by BART miRNA 22 in nasopharyngeal carcinoma cells (Lung et al., 2009). In contrast to this regulation of latency establishment by the EBV miRNAs, BART miRNAs of EBV might be predominantly involved in blocking lytic replication (Figure 1; Cai et al., 2006; Chen et al., 2019; Serrano-Solis et al., 2019). Along these lines it was found that the BART miRNA 2 down-regulates the viral DNA polymerase BALF5 (Barth et al., 2008). This reduced BALF5 protein levels by 50% and infectious virus production by 20%. In addition, BART miRNA 20-5p down-modulates BZLF1 and BRLF1 transcripts (Jung et al., 2014; Lin et al., 2015). This diminishes lytic replication upon BART miRNA 20-5p expression. BART miRNAs are affected by the deletion that is present in the B95-8 laboratory strain of EBV (Yang et al., 2013) and EBV isolates with similar deletions are enriched in DLBCLs (Okuno et al., 2019). Therefore, it was suggested that enhanced early lytic EBV gene expression supports lymphomagenesis, probably via conditioning of the tumor microenvironment (Ma et al., 2011; Münz, 2019). BART miRNAs are expressed during all EBV infection programs, thereby presumably blocking this pro-tumorigenic effect of early lytic EBV gene expression. Furthermore, BART miRNAs are expressed during latency 0 and I, presumably raising the bar for lytic reactivation to maintain persistence in long-lived memory B cells.
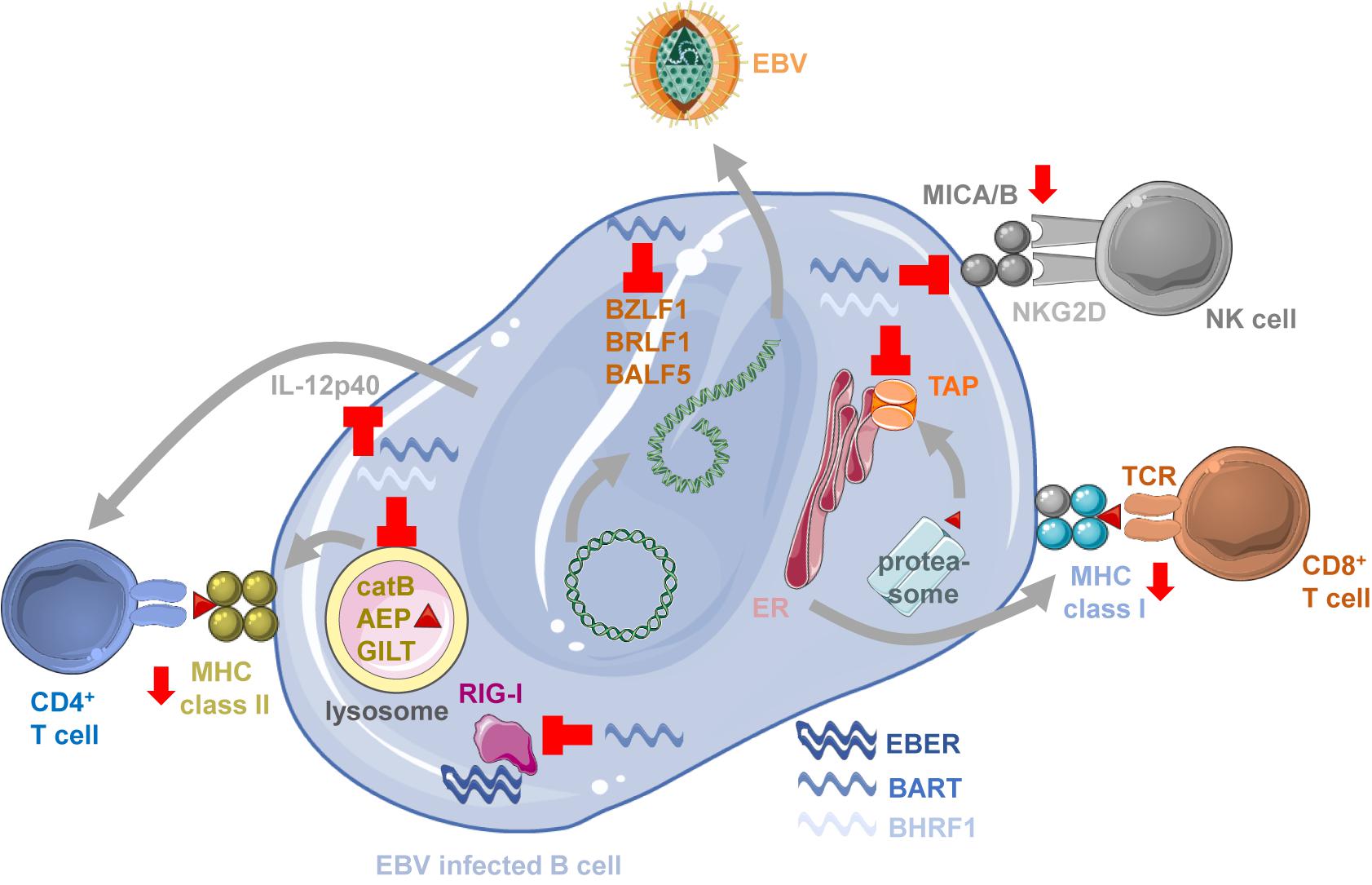
Figure 1. Immune modulation by EBV ncRNAs. BART miRNAs block lytic EBV reactivation by down-regulating BZLF1, BRLF1 and BALF5, limiting viral antigen production for immune responses. BART miRNAs block also MICA/B expression to inhibit NKG2D mediated NK cell recognition of EBV infected B cells. BHRF1 and BART miRNAs compromise antigen presentation by MHC class I molecules by down-regulating the transporter associated with antigen processing (TAP) to load MHC class I complexes with proteasome products in the endoplasmic reticulum (ER) for stimulation of CD8+ T cells. EBERs can be recognized by innate immune sensors of RNA like RIG-I, but the RIG-I pathway is also down-regulated by BART miRNAs. MHC class II ligand generation by lysosomal proteolysis is inhibited by BHRF1 and BART miRNAs down-regulating cathepsin B (catB), asparaginyl endopeptidase (AEP) and IFN-γ–inducible lysosomal thiol reductase (GILT). T cell priming and NK cell activation is also compromised by BHRF1 and BART miRNAs blocking IL-12p40 production. This figure was created in part with modified Servier Medical Art templates, which are licensed under a Creative Commons Attribution 3.0 unported license: https://smart.servier.com.
Regulation of Host Transcripts by EBV ncRNAs
EBV miRNAs do not only compromise lytic EBV replication by targeting the viral DNA polymerase and immediate early transactivators of lytic EBV infection, but also by down-regulating cellular machinery that induces lytic EBV reactivation. BHRF1 miRNA 2-5p and BART miRNA 2-5p have been recently described to target components of B cell receptor signaling (Chen et al., 2019) which is involved in reactivating lytic EBV infection from latency I or 0 (Binne et al., 2002). The targeted transcripts encode GRB2, SOS1, MALT1, RAC1, and INPP5D, and the respective viral miRNAs attenuate lytic EBV reactivation upon BCR cross-linking. Furthermore, caspase 3 dependent apoptosis induction seems also to be required for lytic EBV replication and BART miRNA 20-5p was shown to down-regulate BAD, thereby compromising lytic replication (Lin et al., 2015; Kim et al., 2016). Furthermore, BART miRNAs 1-3p and 16 were reported to target caspase 3 directly (Vereide et al., 2014). Finally, the BART miRNA 18-5p down-regulates MAP kinase kinase kinase 2 (MAP3K2) and thereby also blocks lytic EBV infection (Qiu and Thorley-Lawson, 2014). In contrast, late lytic EBV infection is supported by the BHRF1 miRNA 1 targeting the ubiquitin ligase RNF4 that modifies SUMO-conjugated targets (Li et al., 2017). Expression of miRNA regulation resistant RNF4 attenuates infectious viral particle release. Therefore, multiple miRNAs block induction of lytic EBV infection by downregulating both viral and host factors involved in it, but then might support infectious viral particle production.
In addition to blocking lytic replication BART miRNAs have been also reported to sustain proliferation and block apoptosis. They do so in part by regulating the Wnt/β-catenin pathway. BART miRNA 10-3p has been shown to down-regulate the Dickkopf WNT Signaling Pathway Inhibitor 1 (DKK1) in EBV positive gastric carcinoma cell lines (Min and Lee, 2019). This enhances proliferation and migration of the respective tumor cells. Moreover, BART miRNA 1-3p was reported to down-modulate Disabled Homolog 2 (DAB2) that attenuates the Wnt/β-catenin pathway (Min et al., 2020). The resulting DAB2 down-regulation blocked apoptosis. Furthermore, also the Wnt/β-catenin inhibitor Adenomatous Polyposis Coli (APC) is down-regulated by BART miRNA 19-3p (Zhang et al., 2020). Finally, Mitogen-Activated Protein Kinase 4 (MAP2K4) is down-regulated by BART miRNA 22, stimulating β-catenin dependent transcription (Liu et al., 2019). Thus, BART miRNAs down-regulate inhibitors of the Wnt/β-catenin pathway to enhance proliferation, apoptosis resistance and migration of EBV associated carcinomas.
Additional pathways are targeted by mostly BART miRNAs to increase apoptosis resistance and migration of EBV infected cells. These include p53 mRNA down-regulation by BART miRNA 5-3p (Zheng et al., 2018). This regulation enhanced proliferation of gastric carcinoma cells in vitro and in a xenograft model in mice. In addition, ATM/ATR dependent DNA repair is augmented by BART miRNAs (Zhou et al., 2019). This increases radioresistance of nasopharyngeal carcinoma cells in a xenograft model. Finally, BART miRNA 2-5p targets RND3, a negative regulator of Rho signaling (Jiang et al., 2020). BART miRNA 2-5p expression in EBV negative nasopharyngeal carcinoma cells promoted their migration and metastasis formation in a xenograft model. Many of the studies that argue for a pro-proliferative, anti-apoptotic and migration promoting function of BART miRNAs have been performed with nasopharyngeal or gastric carcinoma cell lines and carcinogenesis in vivo was reported to be increased by BART miRNAs (Qiu et al., 2015), while EBV infection of B cells in mice with reconstituted human immune system components was not significantly altered by BART miRNA deficiency (Murer et al., 2019). However, increased lytic EBV replication in the absence of BART miRNAs might have been missed due to the B95-8 EBV strain that was used and the small as well as transient contribution of lytic EBV replication to viral loads in this model (Antsiferova et al., 2014). Nevertheless, an EBV strain lacking all viral miRNAs replicated to similar viral loads and similar lymphomagenesis in mice with reconstituted immune system components after antibody depletion of T cells (Murer et al., 2019). This suggests that immune escape might be the dominant function of EBV miRNAs, as discussed below.
Similar to their effects on viral gene products, EBER1 and 2 regulation of host gene products remains unclear. While these EBV ncRNAs trap RNA binding proteins like La (SS-B) and rpL22 in the nucleus (Fok et al., 2006; Gregorovic et al., 2011) the role of this relocalization has not been clarified. However, a recent study suggested that EBER1 can substitute for the ncRNA TMER4 in a mouse γ-herpesvirus to allow infected B cells to migrate from secondary lymphoid tissues into the circulation (Hoffman et al., 2019). Such changes in migratory behavior might not have been sufficiently analyzed during EBER deficient EBV infection of mice with reconstituted human immune system components, even so unaltered viral loads were observed in blood (Gregorovic et al., 2015).
Immune Control of EBV
Although most of these pro-tumorigenic functions have been assigned to BART miRNAs of EBV, recombinant EBV that is deficient in BHRF1 miRNAs was found to infect mice with reconstituted human immune system components with a slower kinetic and attenuated systemic dissemination compared to wild-type virus (Wahl et al., 2013; Murer et al., 2019). This seems to be at least in part due to improved cell-mediated immune control (Murer et al., 2019).
Indeed, most adults carry EBV as an asymptomatic persistent infection (Münz, 2019). However, primary or acquired immunodeficiencies predispose for EBV associated malignancies (Damania and Münz, 2019; Latour and Fischer, 2019; Tangye and Latour, 2020). These point toward cytotoxic lymphocytes as the cornerstone of EBV specific immune control (Long et al., 2019). Immune suppression due to human immunodeficiency virus (HIV) co-infection or immune suppressive treatment after transplantation leads to increased EBV associated lymphoma formation (Gottschalk et al., 2005; Totonchy and Cesarman, 2016). This can be modeled by immunosuppressive FK506 treatment and HIV co-infection of EBV infected mice with reconstituted human immune system components (Caduff et al., 2020; McHugh et al., 2020). Both treatments compromise CD4+ T cell help leading to a less functional CD8+ T cell phenotype (Caduff et al., 2020; McHugh et al., 2020) and compromising CD8+ T cell mediated immune control of EBV infection with no further increase of viral loads upon antibody mediated depletion of this cytotoxic T cell subset (McHugh et al., 2020). Moreover, primary immunodeficiencies that predispose for EBV associated pathologies identify molecular requirements for EBV specific immune control. Mutations in perforin and gene products that mediate cytotoxic degranulation (Munc13-4 and 18-2) predispose for uncontrolled EBV infection and identify cytotoxicity as the main effector function during immune control of EBV (Katano et al., 2004; Rohr et al., 2010; Cohen et al., 2015). Furthermore, compromised T cell receptor signaling, predisposing for EBV associated pathologies, identifies these adaptive lymphocytes as main contributors to cell mediated immune control by cytotoxic lymphocytes (Huck et al., 2009; Linka et al., 2012; Angulo et al., 2013; Kuehn et al., 2013; Moshous et al., 2013; Lucas et al., 2014; Salzer et al., 2016; Hoshino et al., 2018; Winter et al., 2018). Accordingly, antibody mediated T cell depletion, and especially cytotoxic CD8+ T cell depletion, compromises immune control of EBV infection in mice with reconstituted human immune system components (Strowig et al., 2009; Yajima et al., 2009; Chijioke et al., 2015; Murer et al., 2019; McHugh et al., 2020). Furthermore, mutations in certain co-receptors on cytotoxic natural killer and T cells are also associated with EBV positive malignancies. This is particularly pronounced for deficiencies in CD27 and its ligand CD70 (Salzer et al., 2012; van Montfrans et al., 2012; Alkhairy et al., 2015; Abolhassani et al., 2017; Izawa et al., 2017; Ghosh et al., 2020). But also, mutations in the SLAM-associated protein (SAP), affecting co-stimulation of receptors like 2B4, predispose for EBV associated pathologies (Coffey et al., 1998; Nichols et al., 1998; Sayos et al., 1998; Sumegi et al., 2000; Pachlopnik Schmid et al., 2011). Accordingly, antibody blocking of 2B4 compromises EBV specific immune control in mice with reconstituted human immune system components but does not do so after CD8+ T cell depletion (Chijioke et al., 2015). In addition to cytotoxic CD8+ T cell mediated immune control also cytotoxic innate lymphocytes seem to contribute to EBV specific immune control. These include natural killer (NK) cells, NKT cells, and γδ T cells, controlling lytic and various stages of latent EBV infection, respectively (Pappworth et al., 2007; Yuling et al., 2009; Chijioke et al., 2013; Chung et al., 2013; Azzi et al., 2014; Xiang et al., 2014; Djaoud et al., 2017; Zumwalde et al., 2017). Therefore, cytotoxic 2B4+CD27+CD8+ T cells and innate lymphocytes seem to be crucial for EBV specific immune control, while type I and type II interferons as well as antibody responses appear to be dispensable (Latour and Fischer, 2019). Maybe not surprisingly EBV ncRNAs have therefore also a crucial function in modulating this immune control, as will be discussed next.
Immune Modulation by EBV ncRNAs
Immune modulatory functions have been described for both EBERs and miRNAs. Indeed, when complete miRNA and BART miRNA knock-out viruses of the B95-8 EBV strain were compared for infection in mice with reconstituted human immune system components, no significant differences between wt and BART miRNA deficient EBV were detected, while complete miRNA deficient virus infection was significantly attenuated (Murer et al., 2019). Both viral loads and lymphomagenesis were lower with the miRNA deficient virus with no tumors detected in the absence of miRNAs. However, upon depletion of CD8+ T cells both viral loads and tumor formation were increased. More than 200-fold increased viral loads for miRNA deficient and more than 40-fold for wt EBV infection were detected after antibody mediated CD8+ T cell depletion, reaching similar levels for both infections. Lymphoma formation was also increased in frequency upon CD8+ T cell depletion with more than 50% of animals developing tumors, up from no tumors in miRNA deficient and 20–30% in wt EBV infected mice. These findings suggest that immune evasion from CD8+ T cell mediated immune control is one of the main functions of EBV miRNAs, especially the BHRF1 cluster.
Indeed, CD8+ T cell recognition of LCLs carrying miRNA deficient EBV is significantly increased (Albanese et al., 2016; Murer et al., 2019). This is associated with down-regulation of the transcripts for TAP2 (Figure 1), one of the two chains of the transporter associated with antigen processing that imports peptides into the endoplasmic reticulum (ER) for MHC class I loading and presentation to CD8+ T cells (Albanese et al., 2016). This diminished peptide import reduces particularly the surface expression of HLA-B molecules by which many immunodominant EBV derived epitopes are restricted (Taylor et al., 2015). TAP2 is targeted by the BHRF1 miRNA 3 and the BART miRNA 17 (Albanese et al., 2016). In addition, CD4+ T cell mediated immune control of LCLs is also compromised by viral miRNAs (Tagawa et al., 2016). CD4+ T cells recognize LCLs with deficiency in miRNA much better than wt EBV transformed B cells. MHC class II ligands for CD4+ T cell stimulation are primarily generated by lysosomal proteolysis (Trombetta and Mellman, 2005). EBV miRNAs downregulate some components of this degradation machinery, namely IFN-γ–inducible lysosomal thiol reductase (GILT) that breaks disulfide bonds prior to protein degradation, and the proteases cathepsin B and asparaginyl endopeptidase (AEP) (Figure 1; Tagawa et al., 2016). GILT is down-regulated by BART miRNAs 1–5p and 1–3p, cathepsin B by BART miRNA 2–5p, and AEP by BHRF1 miRNA 2 and BART miRNA 2–5p (Albanese et al., 2016; Tagawa et al., 2016). Moreover, also the cytokine IL-12 that is instrumental for T cell priming and activation of NK cells is targeted by viral miRNAs (Albanese et al., 2016; Tagawa et al., 2016). BHRF1 miRNA 2, BART miRNA 1–3p, BART miRNA 2–5p, BART miRNA 10-3p and BART miRNA 22 down-regulate IL-12p40 which is part of both IL-12 and IL-23 cytokines (Figure 1). NK cell recognition is further compromised by BART miRNA 2-5p and 7 down-regulation of MICB and MICA, respectively (Figure 1; Nachmani et al., 2009; Wong et al., 2018), two ligands of the activating NK cell receptor NKG2D which has been implicated in the recognition of lytically EBV replicating B cells (Pappworth et al., 2007). As additional immune escape mechanisms EBV miRNAs lead to the up-regulation of PD-L1 and PD-L2, ligands for the inhibitory receptor PD1 on cytotoxic lymphocytes (Cristino et al., 2019; Yoon et al., 2020). Thus, EBV miRNAs compromise immune control by cytotoxic lymphocytes on multiple levels, by targeting antigen presentation, immune stimulation and inhibitory receptor engagement, most likely for efficient virus persistence and transmission. Since, however, CD8+ T cell depletion restores viral load and lymphomagenesis of miRNA deficient EBV infection in mice with reconstituted human immune system components (Murer et al., 2019), regulation of MHC class I presentation might be one of the dominant functions of EBV encoded miRNAs.
EBERs have been suggested to also influence immune responses by being sensed via pathogen associated molecular pattern (PAMP) recognition receptors (PRRs) that detect viral RNAs, including toll-like receptor 3 (TLR3) and 8 (TLR8), RIG-I (Figure 1) and PKR (Vuyisich et al., 2002; McKenna et al., 2006; Samanta et al., 2008; Iwakiri et al., 2009; Li et al., 2019). Such immune stimulation by EBERs might also be transferred via EBER containing exosomes to dendritic cells (Ahmed et al., 2014, 2018; Baglio et al., 2016). BART miRNA 6-3p down-regulates this recognition by attenuating the RIG-I pathway (Figure 1; Lu et al., 2017). Despite this suggested role for innate immune recognition of EBERs, EBER deficient EBV infection of mice with reconstituted human immune system components did not result in a significantly altered immune activation (Gregorovic et al., 2015). The viral DNA is via TLR9 mediated detection at least an equally important immune stimulus and readily sensed by plasmacytoid dendritic cells (Fiola et al., 2010; Severa et al., 2013; Gujer et al., 2019). Thus, the human immune system does not seem to require EBERs for detection of EBV infection.
Conclusion and Outlook
Non-coding RNAs are the ideal tools for viruses to modulate their life cycle, host cell behavior and immune responses, because they do not need the expression of proteins that could serve as antigens for the immune system to detect infection. Along these lines EBV extensively modulates its life cycle, promoting latency and suppressing lytic replication with BART miRNAs. Furthermore, its BHRF1 miRNAs seem to serve a crucial role in immune escape from cytotoxic lymphocytes, compromising anti-viral NK, CD4+ T, and CD8+ T cell responses. This realization offers the opportunity to explore miRNA deficient viruses, probably with additional safety mutations as vaccine candidates. Their superior ability to be recognized by both CD4+ and CD8+ T cell responses should be harnessed to induce both systemic and tissue resident T cell memory to protect from symptomatic acute EBV infection, namely infectious mononucleosis, and the associated risk for EBV positive malignancies and the autoimmune disease multiple sclerosis (MS) (Ruhl et al., 2020).
In contrast, EBERs remain an enigma of EBV biology. Their high expression in all EBV infection programs mocks us, having not identified a non-redundant function for these non-translated RNAs. It might require more sophisticated in vitro and in vivo systems of EBV infection and immune control to reveal the function of EBER1 and 2, possibly with more physiological migration behaviors of EBV infected cells from mucosal sites to secondary lymphoid organs (Hoffman et al., 2019). EBERs are joined by other non-translated RNA species, including small nucleolar RNAs (snoRNAs), long non-coding RNAs (lncRNAs), stable intronic sequence RNAs (sisRNAs) and circular RNAs (circRNAs), for which also functions need to be explored in more detail in the future. Nevertheless, it is already clear that EBV masterfully manipulates the cellular RNA network in addition to its fine-tuned proteins exploiting human B cell differentiation.
Author Contributions
CM wrote the manuscript.
Funding
Research in my laboratory is supported by Cancer Research Switzerland (KFS-4962-02-2020), CRPP-PrecisionMS and HMZ ImmunoTargET of the University of Zurich, the Cancer Research Center Zurich, the Baugarten Foundation, the Sobek Foundation, the Swiss Vaccine Research Institute, Roche, Novartis, Novartis Foundation (20B099), and the Swiss National Science Foundation (310030B_182827, 310030L_197952/1, and CRSII5_180323). The funders were not involved in the writing of this article or the decision to submit it for publication.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abolhassani, H., Edwards, E. S., Ikinciogullari, A., Jing, H., Borte, S., Buggert, M., et al. (2017). Combined immunodeficiency and Epstein-Barr virus-induced B cell malignancy in humans with inherited CD70 deficiency. J. Exp. Med. 214, 91–106. doi: 10.1084/jem.20160849
Ahmed, W., Philip, P. S., Tariq, S., and Khan, G. (2014). Epstein-Barr virus-encoded small RNAs (EBERs) are present in fractions related to exosomes released by EBV-transformed cells. PLoS One 9:e99163. doi: 10.1371/journal.pone.0099163
Ahmed, W., Tariq, S., and Khan, G. (2018). Tracking EBV-encoded RNAs (EBERs) from the nucleus to the excreted exosomes of B-lymphocytes. Sci. Rep. 8:15438.
Albanese, M., Tagawa, T., Bouvet, M., Maliqi, L., Lutter, D., Hoser, J., et al. (2016). Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells. Proc Natl Acad Sci U.S.A. 113, E6467–E6475.
Alkhairy, O. K., Perez-Becker, R., Driessen, G. J., Abolhassani, H., van Montfrans, J., Borte, S., et al. (2015). Novel mutations in TNFRSF7/CD27: clinical, immunologic, and genetic characterization of human CD27 deficiency. J. Allergy Clin. Immunol. 70:e10.
Angulo, I., Vadas, O., Garcon, F., Banham-Hall, E., Plagnol, V., Leahy, T. R., et al. (2013). Phosphoinositide 3-kinase delta gene mutation predisposes to respiratory infection and airway damage. Science 342, 866–871.
Antsiferova, O., Müller, A., Rämer, P., Chijioke, O., Chatterjee, B., Raykova, A., et al. (2014). Adoptive transfer of EBV specific CD8+ T cell clones can transiently control EBV infection in humanized mice. PLoS Pathog. 10:e1004333. doi: 10.1371/journal.ppat.1004333
Azzi, T., Lunemann, A., Murer, A., Ueda, S., Beziat, V., Malmberg, K. J., et al. (2014). Role for early-differentiated natural killer cells in infectious mononucleosis. Blood 124, 2533–2543. doi: 10.1182/blood-2014-01-553024
Babcock, G. J., Decker, L. L., Volk, M., and Thorley-Lawson, D. A. (1998). EBV persistence in memory B cells in vivo. Immunity 9, 395–404. doi: 10.1016/s1074-7613(00)80622-6
Babcock, J. G., Hochberg, D., and Thorley-Lawson, A. D. (2000). The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 13, 497–506. doi: 10.1016/s1074-7613(00)00049-2
Baglio, S. R., van Eijndhoven, M. A., Koppers-Lalic, D., Berenguer, J., Lougheed, S. M., Gibbs, S., et al. (2016). Sensing of latent EBV infection through exosomal transfer of 5’pppRNA. Proc Natl Acad Sci U.S.A. 113, E587–E596.
Barth, S., Pfuhl, T., Mamiani, A., Ehses, C., Roemer, K., Kremmer, E., et al. (2008). Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 36, 666–675. doi: 10.1093/nar/gkm1080
Binne, U. K., Amon, W., and Farrell, P. J. (2002). Promoter sequences required for reactivation of Epstein-Barr virus from latency. J. Virol. 76, 10282–10289. doi: 10.1128/jvi.76.20.10282-10289.2002
Borza, C. M., and Hutt-Fletcher, L. M. (2002). Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8, 594–599. doi: 10.1038/nm0602-594
Bullard, W. L., Flemington, E. K., Renne, R., and Tibbetts, S. A. (2018). Connivance, complicity, or collusion? the role of noncoding rnas in promoting gammaherpesvirus tumorigenesis. Trends Cancer 4, 729–740. doi: 10.1016/j.trecan.2018.09.005
Caduff, N., McHugh, D., Murer, A., Ramer, P., Raykova, A., Landtwing, V., et al. (2020). Immunosuppressive FK506 treatment leads to more frequent EBV-associated lymphoproliferative disease in humanized mice. PLoS Pathog. 16:e1008477. doi: 10.1371/journal.ppat.1008477
Caetano, B. F. R., Jorge, B. A. S., Muller-Coan, B. G., and Elgui de Oliveira, D. (2021). Epstein-Barr virus microRNAs in the pathogenesis of human cancers. Cancer Lett. 499, 14–23. doi: 10.1016/j.canlet.2020.11.019
Cai, X., Schafer, A., Lu, S., Bilello, J. P., Desrosiers, R. C., Edwards, R., et al. (2006). Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. doi: 10.1371/journal.ppat.0020023
Cesarman, E. (2014). Gammaherpesviruses and lymphoproliferative disorders. Annu. Rev. Pathol. 9, 349–372. doi: 10.1146/annurev-pathol-012513-104656
Chen, J., Sathiyamoorthy, K., Zhang, X., Schaller, S., Perez White, B. E., Jardetzky, T. S., et al. (2018). Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nat. Microbiol. 3, 172–180. doi: 10.1038/s41564-017-0081-7
Chen, Y., Fachko, D., Ivanov, N. S., Skinner, C. M., and Skalsky, R. L. (2019). Epstein-Barr virus microRNAs regulate B cell receptor signal transduction and lytic reactivation. PLoS Pathog. 15:e1007535. doi: 10.1371/journal.ppat.1007535
Chijioke, O., Marcenaro, E., Moretta, A., Capaul, R., and Münz, C. (2015). The SAP-dependent 2B4 receptor mediates CD8+ T cell dependent immune control of Epstein Barr virus infection in mice with reconstituted human immune system components. J. Infect. Dis. 212, 803–807. doi: 10.1093/infdis/jiv114
Chijioke, O., Muller, A., Feederle, R., Barros, M. H., Krieg, C., Emmel, V., et al. (2013). Human natural killer cells prevent infectious mononucleosis features by targeting lytic Epstein-Barr virus infection. Cell Rep. 5, 1489–1498. doi: 10.1016/j.celrep.2013.11.041
Chung, B. K., Tsai, K., Allan, L. L., Zheng, D. J., Nie, J. C., Biggs, C. M., et al. (2013). Innate immune control of EBV-infected B cells by invariant natural killer T cells. Blood 122, 2600–2608. doi: 10.1182/blood-2013-01-480665
Coffey, A. J., Brooksbank, R. A., Brandau, O., Oohashi, T., Howell, G. R., Bye, J. M., et al. (1998). Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nat. Genet. 20, 129–135. doi: 10.1038/2424
Cohen, J. I., Fauci, A. S., Varmus, H., and Nabel, G. J. (2011). Epstein-Barr virus: an important vaccine target for cancer prevention. Sci. Transl. Med. 3:107fs7. doi: 10.1126/scitranslmed.3002878
Cohen, J. I., Niemela, J. E., Stoddard, J. L., Pittaluga, S., Heslop, H., Jaffe, E. S., et al. (2015). Late-onset severe chronic active EBV in a patient for five years with mutations in STXBP2 (MUNC18-2) and PRF1 (perforin 1). J. Clin. Immunol. 35, 445–448. doi: 10.1007/s10875-015-0168-y
Cristino, A. S., Nourse, J., West, R. A., Sabdia, M. B., Law, S. C., Gunawardana, J., et al. (2019). EBV microRNA-BHRF1-2-5p targets the 3’UTR of immune checkpoint ligands PD-L1 and PD-L2. Blood 134, 2261–2270. doi: 10.1182/blood.2019000889
Damania, B., and Münz, C. (2019). Immunodeficiencies that predispose to pathologies by human oncogenic gamma-herpesviruses. FEMS Microbiol. Rev. 43, 181–192. doi: 10.1093/femsre/fuy044
Djaoud, Z., Guethlein, L. A., Horowitz, A., Azzi, T., Nemat-Gorgani, N., Olive, D., et al. (2017). Two alternate strategies for innate immunity to Epstein-Barr virus: One using NK cells and the other NK cells and gammadelta T cells. J. Exp. Med. 214, 1827–1841. doi: 10.1084/jem.20161017
Feederle, R., Haar, J., Bernhardt, K., Linnstaedt, S. D., Bannert, H., Lips, H., et al. (2011b). The members of an Epstein-Barr virus microRNA cluster cooperate to transform B lymphocytes. J. Virol. 85, 9801–9810. doi: 10.1128/jvi.05100-11
Feederle, R., Linnstaedt, S. D., Bannert, H., Lips, H., Bencun, M., Cullen, B. R., et al. (2011a). microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 7:e1001294. doi: 10.1371/journal.ppat.1001294
Fiola, S., Gosselin, D., Takada, K., and Gosselin, J. (2010). TLR9 contributes to the recognition of EBV by primary monocytes and plasmacytoid dendritic cells. J. Immunol. 185, 3620–3631. doi: 10.4049/jimmunol.0903736
Fok, V., Friend, K., and Steitz, J. A. (2006). Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J. Cell Biol. 173, 319–325. doi: 10.1083/jcb.200601026
Ghosh, S., Kostel Bal, S., Edwards, E. S. J., Pillay, B., Jimenez-Heredia, R., Rao, G., et al. (2020). Extended clinical and immunological phenotype and transplant outcome in CD27 and CD70 deficiency. Blood 136, 2638–2655.
Gottschalk, S., Rooney, C. M., and Heslop, H. E. (2005). Post-transplant lymphoproliferative disorders. Annu. Rev. Med. 56, 29–44.
Gregorovic, G., Bosshard, R., Karstegl, C. E., White, R. E., Pattle, S., Chiang, A. K., et al. (2011). Cellular gene expression that correlates with EBER expression in Epstein-Barr Virus-infected lymphoblastoid cell lines. J. Virol. 85, 3535–3545. doi: 10.1128/jvi.02086-10
Gregorovic, G., Boulden, E. A., Bosshard, R., Karstegl, C. E., Skalsky, R., Cullen, B. R., et al. (2015). Epstein-Barr viruses deficient in EBER RNAs give higher LMP2 RNA expression in lymphoblastoid cell lines and efficiently establish persistent infection in humanized mice. J. Virol. 89, 11711–11714. doi: 10.1128/jvi.01873-15
Gujer, C., Murer, A., Muller, A., Vanoaica, D., Sutter, K., Jacque, E., et al. (2019). Plasmacytoid dendritic cells respond to Epstein-Barr virus infection with a distinct type I interferon subtype profile. Blood Adv. 3, 1129–1144. doi: 10.1182/bloodadvances.2018025536
Hadinoto, V., Shapiro, M., Sun, C. C., and Thorley-Lawson, D. A. (2009). The dynamics of EBV shedding implicate a central role for epithelial cells in amplifying viral output. PLoS Pathog. 5:e1000496. doi: 10.1371/journal.ppat.1000496
Hochberg, D., Middeldorp, J. M., Catalina, M., Sullivan, J. L., Luzuriaga, K., and Thorley-Lawson, D. A. (2004). Demonstration of the Burkitt’s lymphoma Epstein-Barr virus phenotype in dividing latently infected memory cells in vivo. Proc. Natl. Acad. Sci. U.S.A. 101, 239–244. doi: 10.1073/pnas.2237267100
Hoffman, B. A., Wang, Y., Feldman, E. R., and Tibbetts, S. A. (2019). Epstein-Barr virus EBER1 and murine gammaherpesvirus TMER4 share conserved in vivo function to promote B cell egress and dissemination. Proc. Natl. Acad. Sci. U.S.A. 116, 25392–25394. doi: 10.1073/pnas.1915752116
Hoshino, A., Takashima, T., Yoshida, K., Morimoto, A., Kawahara, Y., Yeh, T. W., et al. (2018). Dysregulation of Epstein-Barr virus infection in hypomorphic ZAP70 mutation. J. Infect. Dis. 218, 825–834. doi: 10.1093/infdis/jiy231
Huck, K., Feyen, O., Niehues, T., Ruschendorf, F., Hubner, N., Laws, H. J., et al. (2009). Girls homozygous for an IL-2-inducible T cell kinase mutation that leads to protein deficiency develop fatal EBV-associated lymphoproliferation. J. Clin. Invest. 119, 1350–1358. doi: 10.1172/jci37901
Hutt-Fletcher, L. M. (2017). The long and complicated relationship between epstein-barr virus and epithelial cells. J. Virol. 91, e01677–16.
Hutzinger, R., Feederle, R., Mrazek, J., Schiefermeier, N., Balwierz, P. J., Zavolan, M., et al. (2009). Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 5:e1000547. doi: 10.1371/journal.ppat.1000547
Iwakiri, D., Zhou, L., Samanta, M., Matsumoto, M., Ebihara, T., Seya, T., et al. (2009). Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J. Exp. Med. 206, 2091–2099. doi: 10.1084/jem.20081761
Izawa, K., Martin, E., Soudais, C., Bruneau, J., Boutboul, D., Rodriguez, R., et al. (2017). Inherited CD70 deficiency in humans reveals a critical role for the CD70-CD27 pathway in immunity to Epstein-Barr virus infection. J. Exp. Med. 214, 73–89. doi: 10.1084/jem.20160784
Jiang, C., Li, L., Xiang, Y. Q., Lung, M. L., Zeng, T., Lu, J., et al. (2020). Epstein-Barr Virus miRNA BART2-5p promotes metastasis of nasopharyngeal carcinoma by suppressing RND3. Cancer Res. 80, 1957–1969. doi: 10.1158/0008-5472.can-19-0334
Jung, Y. J., Choi, H., Kim, H., and Lee, S. K. (2014). MicroRNA miR-BART20-5p stabilizes Epstein-Barr virus latency by directly targeting BZLF1 and BRLF1. J. Virol. 88, 9027–9037. doi: 10.1128/jvi.00721-14
Katano, H., Ali, M. A., Patera, A. C., Catalfamo, M., Jaffe, E. S., Kimura, H., et al. (2004). Chronic active Epstein-Barr virus infection associated with mutations in perforin that impair its maturation. Blood 103, 1244–1252. doi: 10.1182/blood-2003-06-2171
Kempkes, B., and Robertson, E. S. (2015). Epstein-Barr virus latency: current and future perspectives. Curr. Opin. Virol. 14, 138–144. doi: 10.1016/j.coviro.2015.09.007
Kenney, S. C., and Mertz, J. E. (2014). Regulation of the latent-lytic switch in Epstein-Barr virus. Semin. Cancer Biol. 26, 60–68. doi: 10.1016/j.semcancer.2014.01.002
Kim, H., Choi, H., and Lee, S. K. (2016). Epstein-Barr Virus MicroRNA miR-BART20-5p suppresses lytic induction by inhibiting BAD-mediated caspase-3-Dependent apoptosis. J. Virol. 90, 1359–1368. doi: 10.1128/jvi.02794-15
Kuehn, H. S., Niemela, J. E., Rangel-Santos, A., Zhang, M., Pittaluga, S., Stoddard, J. L., et al. (2013). Loss-of-function of the protein kinase C delta (PKCdelta) causes a B-cell lymphoproliferative syndrome in humans. Blood 121, 3117–3125. doi: 10.1182/blood-2012-12-469544
Laichalk, L. L., and Thorley-Lawson, D. A. (2005). Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J. Virol. 79, 1296–1307. doi: 10.1128/jvi.79.2.1296-1307.2005
Latour, S., and Fischer, A. (2019). Signaling pathways involved in the T-cell-mediated immunity against Epstein-Barr virus: Lessons from genetic diseases. Immunol. Rev. 291, 174–189. doi: 10.1111/imr.12791
Lerner, M. R., Andrews, N. C., Miller, G., and Steitz, J. A. (1981). Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. U.S.A. 78, 805–809. doi: 10.1073/pnas.78.2.805
Li, C., Romero-Masters, J. C., Huebner, S., Ohashi, M., Hayes, M., Bristol, J. A., et al. (2020). EBNA2-deleted Epstein-Barr virus (EBV) isolate, P3HR1, causes Hodgkin-like lymphomas and diffuse large B cell lymphomas with type II and Wp-restricted latency types in humanized mice. PLoS Pathog. 16:e1008590. doi: 10.1371/journal.ppat.1008590
Li, J., Callegari, S., and Masucci, M. G. (2017). The Epstein-Barr virus miR-BHRF1-1 targets RNF4 during productive infection to promote the accumulation of SUMO conjugates and the release of infectious virus. PLoS Pathog. 13:e1006338. doi: 10.1371/journal.ppat.1006338
Li, Z., Tsai, M. H., Shumilov, A., Baccianti, F., Tsao, S. W., Poirey, R., et al. (2019). Epstein-Barr virus ncRNA from a nasopharyngeal carcinoma induces an inflammatory response that promotes virus production. Nat. Microbiol. 4, 2475–2486. doi: 10.1038/s41564-019-0546-y
Lin, X., Tsai, M. H., Shumilov, A., Poirey, R., Bannert, H., Middeldorp, J. M., et al. (2015). The Epstein-Barr Virus BART miRNA Cluster of the M81 strain modulates multiple functions in primary B cells. PLoS Pathog. 11:e1005344. doi: 10.1371/journal.ppat.1005344
Linka, R. M., Risse, S. L., Bienemann, K., Werner, M., Linka, Y., Krux, F., et al. (2012). Loss-of-function mutations within the IL-2 inducible kinase ITK in patients with EBV-associated lymphoproliferative diseases. Leukemia 26, 963–971. doi: 10.1038/leu.2011.371
Liu, Y., Jiang, Q., Liu, X., Lin, X., Tang, Z., Liu, C., et al. (2019). Cinobufotalin powerfully reversed EBV-miR-BART22-induced cisplatin resistance via stimulating MAP2K4 to antagonize non-muscle myosin heavy chain IIA/glycogen synthase 3beta/beta-catenin signaling pathway. EBioMedicine 48, 386–404. doi: 10.1016/j.ebiom.2019.08.040
Long, H. M., Meckiff, B. J., and Taylor, G. S. (2019). The T-cell response to Epstein-Barr Virus-New tricks from an old dog. Front. Immunol. 10:2193.
Lu, Y., Qin, Z., Wang, J., Zheng, X., Lu, J., Zhang, X., et al. (2017). Epstein-Barr Virus miR-BART6-3p Inhibits the RIG-I Pathway. J. Innate. Immun. 9, 574–586. doi: 10.1159/000479749
Lucas, C. L., Kuehn, H. S., Zhao, F., Niemela, J. E., Deenick, E. K., Palendira, U., et al. (2014). Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110delta result in T cell senescence and human immunodeficiency. Nat. Immunol. 15, 88–97. doi: 10.1038/ni.2771
Lung, R. W., Tong, J. H., Sung, Y. M., Leung, P. S., Ng, D. C., Chau, S. L., et al. (2009). Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia 11, 1174–1184. doi: 10.1593/neo.09888
Ma, S. D., Hegde, S., Young, K. H., Sullivan, R., Rajesh, D., Zhou, Y., et al. (2011). A new model of Epstein-Barr virus infection reveals an important role for early lytic viral protein expression in the development of lymphomas. J. Virol. 85, 165–177. doi: 10.1128/jvi.01512-10
McHugh, D., Caduff, N., Barros, M. H. M., Rämer, P., Raykova, A., Murer, A., et al. (2017). Persistent KSHV infection increases EBV-associated tumor formation in vivo via enhanced EBV lytic gene expression. Cell Host Microbe. 22, 61–73. doi: 10.1016/j.chom.2017.06.009
McHugh, D., Myburgh, R., Caduff, N., Spohn, M., Kok, Y. L., Keller, C. W., et al. (2020). EBV renders B cells susceptible to HIV-1 in humanized mice. Life Sci. Alliance 3:e202000640.
McKenna, S. A., Kim, I., Liu, C. W., and Puglisi, J. D. (2006). Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs. J. Mol. Biol. 358, 1270–1285. doi: 10.1016/j.jmb.2006.03.003
McKenzie, J., and El-Guindy, A. (2015). Epstein-Barr Virus lytic cycle reactivation. Curr. Top. Microbiol. Immunol. 391, 237–261. doi: 10.1007/978-3-319-22834-1_8
Min, K., Kim, J. Y., and Lee, S. K. (2020). Epstein-Barr virus miR-BART1-3p suppresses apoptosis and promotes migration of gastric carcinoma cells by targeting DAB2. Int. J. Biol. Sci. 16, 694–707. doi: 10.7150/ijbs.36595
Min, K., and Lee, S. K. (2019). EBV miR-BART10-3p promotes cell proliferation and migration by targeting DKK1. Int. J. Biol. Sci. 15, 657–667. doi: 10.7150/ijbs.30099
Moshous, D., Martin, E., Carpentier, W., Lim, A., Callebaut, I., Canioni, D., et al. (2013). Whole-exome sequencing identifies Coronin-1A deficiency in 3 siblings with immunodeficiency and EBV-associated B-cell lymphoproliferation. J. Allergy Clin. Immunol. 131, 1594–1603. doi: 10.1016/j.jaci.2013.01.042
Münz, C. (2019). Latency and lytic replication in the oncogenesis of the Epstein Barr virus. Nat. Rev. Micobiol. 17, 691–700. doi: 10.1038/s41579-019-0249-7
Murer, A., McHugh, D., Caduff, N., Kalchschmidt, J. S., Barros, M. H., Zbinden, A., et al. (2018). EBV persistence without its EBNA3A and 3C oncogenes in vivo. PLoS Pathog. 14:e1007039. doi: 10.1371/journal.ppat.1007039
Murer, A., Ruhl, J., Zbinden, A., Capaul, R., Hammerschmidt, W., Chijioke, O., et al. (2019). MicroRNAs of Epstein-Barr Virus attenuate t-cell-mediated immune control in vivo. MBio 10, e01941–18.
Nachmani, D., Stern-Ginossar, N., Sarid, R., and Mandelboim, O. (2009). Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5, 376–385. doi: 10.1016/j.chom.2009.03.003
Nichols, K. E., Harkin, D. P., Levitz, S., Krainer, M., Kolquist, K. A., Genovese, C., et al. (1998). Inactivating mutations in an SH2 domain-encoding gene in X-linked lymphoproliferative syndrome. Proc. Natl. Acad. Sci. U.S.A. 95, 13765–13770. doi: 10.1073/pnas.95.23.13765
Okuno, Y., Murata, T., Sato, Y., Muramatsu, H., Ito, Y., Watanabe, T., et al. (2019). Defective Epstein-Barr virus in chronic active infection and haematological malignancy. Nat. Microbiol. 4, 404–413.
Pachlopnik Schmid, J., Canioni, D., Moshous, D., Touzot, F., Mahlaoui, N., Hauck, F., et al. (2011). Clinical similarities and differences of patients with X-linked lymphoproliferative syndrome type 1 (XLP-1/SAP deficiency) versus type 2 (XLP-2/XIAP deficiency). Blood 117, 1522–1529. doi: 10.1182/blood-2010-07-298372
Pappworth, I. Y., Wang, E. C., and Rowe, M. (2007). The switch from latent to productive infection in epstein-barr virus-infected B cells is associated with sensitization to NK cell killing. J. Virol. 81, 474–482. doi: 10.1128/jvi.01777-06
Pfeffer, S., Zavolan, M., Grasser, F. A., Chien, M., Russo, J. J., Ju, J., et al. (2004). Identification of virus-encoded microRNAs. Science 304, 734–736. doi: 10.1126/science.1096781
Poling, B. C., Price, A. M., Luftig, M. A., and Cullen, B. R. (2017). The Epstein-Barr virus miR-BHRF1 microRNAs regulate viral gene expression in cis. Virology 512, 113–123. doi: 10.1016/j.virol.2017.09.015
Qiu, J., Smith, P., Leahy, L., and Thorley-Lawson, D. A. (2015). The Epstein-Barr virus encoded BART miRNAs potentiate tumor growth in vivo. PLoS Pathog. 11:e1004561. doi: 10.1371/journal.ppat.1004561
Qiu, J., and Thorley-Lawson, D. A. (2014). EBV microRNA BART 18-5p targets MAP3K2 to facilitate persistence in vivo by inhibiting viral replication in B cells. Proc. Natl. Acad. Sci. U.S.A. 111, 11157–11162. doi: 10.1073/pnas.1406136111
Rohr, J., Beutel, K., Maul-Pavicic, A., Vraetz, T., Thiel, J., Warnatz, K., et al. (2010). Atypical familial hemophagocytic lymphohistiocytosis due to mutations in UNC13D and STXBP2 overlaps with primary immunodeficiency diseases. Haematologica 95, 2080–2087. doi: 10.3324/haematol.2010.029389
Ruhl, J., Leung, C. S., and Münz, C. (2020). Vaccination against the Epstein-Barr virus. Cell Mol. Life Sci. 1045, 477–493.
Salzer, E., Cagdas, D., Hons, M., Mace, E. M., Garncarz, W., Petronczki, O. Y., et al. (2016). RASGRP1 deficiency causes immunodeficiency with impaired cytoskeletal dynamics. Nat. Immunol. 17, 1352–1360. doi: 10.1038/ni.3575
Salzer, E., Daschkey, S., Choo, S., Gombert, M., Santos-Valente, E., Ginzel, S., et al. (2012). Combined immunodeficiency with life-threatening EBV-associated lymphoproliferative disorder in patients lacking functional CD27. Haematologica 98, 473–478. doi: 10.3324/haematol.2012.068791
Samanta, M., Iwakiri, D., and Takada, K. (2008). Epstein-Barr virus-encoded small RNA induces IL-10 through RIG-I-mediated IRF-3 signaling. Oncogene 27, 4150–4160. doi: 10.1038/onc.2008.75
Sayos, J., Wu, C., Morra, M., Wang, N., Zhang, X., Allen, D., et al. (1998). The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM [see comments]. Nature 395, 462–469. doi: 10.1038/26683
Serrano-Solis, V., Carlos, A. C., Maracaja-Coutinho, V., and de Farias, S. T. (2019). Prediction of MicroRNAs in the Epstein-Barr Virus reveals potential targets for the viral self-regulation. Indian J. Microbiol. 59, 73–80. doi: 10.1007/s12088-018-0775-4
Severa, M., Giacomini, E., Gafa, V., Anastasiadou, E., Rizzo, F., Corazzari, M., et al. (2013). EBV stimulates TLR- and autophagy-dependent pathways and impairs maturation in plasmacytoid dendritic cells: implications for viral immune escape. Eur. J. Immunol. 43, 147–158. doi: 10.1002/eji.201242552
Shannon-Lowe, C., and Rickinson, A. (2019). The global landscape of EBV-Associated tumors. Front. Oncol. 9:713.
Skalsky, R. L., and Cullen, B. R. (2015). EBV noncoding RNAs. Curr. Top. Microbiol. Immunol. 391, 181–217.
Strowig, T., Gurer, C., Ploss, A., Liu, Y. F., Arrey, F., Sashihara, J., et al. (2009). Priming of protective T cell responses against virus-induced tumors in mice with human immune system components. J. Exp. Med. 206, 1423–1434. doi: 10.1084/jem.20081720
Sumegi, J., Huang, D., Lanyi, A., Davis, J. D., Seemayer, T. A., Maeda, A., et al. (2000). Correlation of mutations of the SH2D1A gene and epstein-barr virus infection with clinical phenotype and outcome in X-linked lymphoproliferative disease. Blood 96, 3118–3125.
Tagawa, T., Albanese, M., Bouvet, M., Moosmann, A., Mautner, J., Heissmeyer, V., et al. (2016). Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing. J. Exp. Med. 213, 2065–2080. doi: 10.1084/jem.20160248
Tangye, S. G., and Latour, S. (2020). Primary immunodeficiencies reveal the molecular requirements for effective host defense against EBV infection. Blood 135, 644–655. doi: 10.1182/blood.2019000928
Taylor, G. S., Long, H. M., Brooks, J. M., Rickinson, A. B., and Hislop, A. D. (2015). The immunology of Epstein-Barr virus-induced disease. Annu. Rev. Immunol. 33, 787–821.
Thorley-Lawson, D. A., and Gross, A. (2004). Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 350, 1328–1337. doi: 10.1056/nejmra032015
Totonchy, J., and Cesarman, E. (2016). Does persistent HIV replication explain continued lymphoma incidence in the era of effective antiretroviral therapy? Curr. Opin. Virol. 20, 71–77. doi: 10.1016/j.coviro.2016.09.001
Trombetta, E. S., and Mellman, I. (2005). Cell biology of antigen processing in vitro and in vivo. Annu. Rev. Immunol. 23, 975–1028. doi: 10.1146/annurev.immunol.22.012703.104538
Tugizov, S. M., Berline, J. W., and Palefsky, J. M. (2003). Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat. Med. 9, 307–314. doi: 10.1038/nm830
Tugizov, S. M., Herrera, R., and Palefsky, J. M. (2013). Epstein-Barr virus transcytosis through polarized oral epithelial cells. J. Virol. 87, 8179–8194. doi: 10.1128/jvi.00443-13
Ungerleider, N. A., Tibbetts, S. A., Renne, R., and Flemington, E. K. (2019). Gammaherpesvirus RNAs Come Full Circle. mBio 10, e00071–19.
van Montfrans, J. M., Hoepelman, A. I., Otto, S., van Gijn, M., van de Corput, L., and de Weger, R. A. (2012). CD27 deficiency is associated with combined immunodeficiency and persistent symptomatic EBV viremia. J. Allergy Clin. Immunol. 129, 787–793 e6.
Vereide, D. T., Seto, E., Chiu, Y. F., Hayes, M., Tagawa, T., Grundhoff, A., et al. (2014). Epstein-Barr virus maintains lymphomas via its miRNAs. Oncogene 33, 1258–1264. doi: 10.1038/onc.2013.71
Vuyisich, M., Spanggord, R. J., and Beal, P. A. (2002). The binding site of the RNA-dependent protein kinase (PKR) on EBER1 RNA from Epstein-Barr virus. EMBO Rep. 3, 622–627. doi: 10.1093/embo-reports/kvf137
Wahl, A., Linnstaedt, S. D., Esoda, C., Krisko, J. F., Martinez-Torres, F., Delecluse, H. J., et al. (2013). A Cluster of virus-encoded micrornas accelerates acute systemic Epstein-Barr virus infection but does not significantly enhance virus-induced oncogenesis in vivo. J. Virol. 87, 5437–5446. doi: 10.1128/jvi.00281-13
Weiss, L. M., and Chen, Y. Y. (2013). EBER in situ hybridization for Epstein-Barr virus. Methods Mol. Biol. 999, 223–230. doi: 10.1007/978-1-62703-357-2_16
Winter, S., Martin, E., Boutboul, D., Lenoir, C., Boudjemaa, S., Petit, A., et al. (2018). Loss of RASGRP1 in humans impairs T-cell expansion leading to Epstein-Barr virus susceptibility. EMBO Mol. Med. 10, 188–199. doi: 10.15252/emmm.201708292
Wong, T. S., Chen, S., Zhang, M. J., Chan, J. Y., and Gao, W. (2018). Epstein-Barr virus-encoded microRNA BART7 downregulates major histocompatibility complex class I chain-related peptide A and reduces the cytotoxicity of natural killer cells to nasopharyngeal carcinoma. Oncol. Lett. 16, 2887–2892.
Xiang, Z., Liu, Y., Zheng, J., Liu, M., Lv, A., Gao, Y., et al. (2014). Targeted activation of human Vgamma9Vdelta2-T cells controls epstein-barr virus-induced B cell lymphoproliferative disease. Cancer Cell. 26, 565–576. doi: 10.1016/j.ccr.2014.07.026
Yajima, M., Imadome, K., Nakagawa, A., Watanabe, S., Terashima, K., Nakamura, H., et al. (2009). T cell-mediated control of Epstein-Barr virus infection in humanized mice. J. Infect. Dis. 200, 1611–1615.
Yang, H. J., Huang, T. J., Yang, C. F., Peng, L. X., Liu, R. Y., Yang, G. D., et al. (2013). Comprehensive profiling of Epstein-Barr virus-encoded miRNA species associated with specific latency types in tumor cells. Virol. J. 10:314. doi: 10.1186/1743-422x-10-314
Yoon, C. J., Chang, M. S., Kim, D. H., Kim, W., Koo, B. K., Yun, S. C., et al. (2020). Epstein-Barr virus-encoded miR-BART5-5p upregulates PD-L1 through PIAS3/pSTAT3 modulation, worsening clinical outcomes of PD-L1-positive gastric carcinomas. Gastric. Cancer 23, 780–795. doi: 10.1007/s10120-020-01059-3
Yuling, H., Ruijing, X., Li, L., Xiang, J., Rui, Z., Yujuan, W., et al. (2009). EBV-induced human CD8+ NKT cells suppress tumorigenesis by EBV-associated malignancies. Cancer Res. 69, 7935–7944. doi: 10.1158/0008-5472.can-09-0828
Zhang, H., Li, Y., Wang, H. B., Zhang, A., Chen, M. L., Fang, Z. X., et al. (2018). Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat. Microbiol. 3, 1–8.
Zhang, Q., Luo, D., Xie, Z., He, H., and Duan, Z. (2020). The oncogenic role of miR-BART19-3p in Epstein-Barr virus-associated diseases. Biomed. Res. Int. 2020:5217039.
Zheng, X., Wang, J., Wei, L., Peng, Q., Gao, Y., Fu, Y., et al. (2018). Epstein-Barr Virus MicroRNA miR-BART5-3p inhibits p53 expression. J. Virol. 92, e01022–18.
Zhou, X., Zheng, J., Tang, Y., Lin, Y., Wang, L., Li, Y., et al. (2019). EBV encoded miRNA BART8-3p promotes radioresistance in nasopharyngeal carcinoma by regulating ATM/ATR signaling pathway. Biosci. Rep. 39:BSR20190415. doi: 10.1042/BSR20190415
Keywords: T cells, NK cells, antigen processing, MHC presentation, humanized mice
Citation: Münz C (2021) Immune Escape by Non-coding RNAs of the Epstein Barr Virus. Front. Microbiol. 12:657387. doi: 10.3389/fmicb.2021.657387
Received: 22 January 2021; Accepted: 28 May 2021;
Published: 21 June 2021.
Edited by:
Asuka Nanbo, Nagasaki University, JapanReviewed by:
Yonggang Pei, University of Pennsylvania, United StatesTakayuki Murata, Fujita Health University, Japan
Bill Sugden, University of Wisconsin-Madison, United States
Copyright © 2021 Münz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian Münz, Y2hyaXN0aWFuLm11ZW56QHV6aC5jaA==
 Christian Münz
Christian Münz