
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 May 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.657353
This article is part of the Research Topic Exposure, Risks, and Drivers of the Mobile Antimicrobial Resistance Genes in the Environment – a Global Perspective View all 17 articles
Development and spread of antimicrobial resistance (AMR) and multidrug resistance (MDR) through propagation of antibiotic resistance genes (ARG) in various environments is a global emerging public health concern. The role of wastewater treatment plants (WWTPs) as hot spots for the dissemination of AMR and MDR has been widely pointed out by the scientific community. In this study, we collected surface water samples from sites upstream and downstream of two WWTP discharge points in an urban watershed in the Bryan-College Station (BCS), Texas area, over a period of nine months. E. coli isolates were tested for resistance to ampicillin, tetracycline, sulfamethoxazole, ciprofloxacin, cephalothin, cefoperazone, gentamycin, and imipenem using the Kirby-Bauer disc diffusion method. Antimicrobial resistant heterotrophic bacteria were cultured on R2A media amended with ampicillin, ciprofloxacin, tetracycline, and sulfamethoxazole for analyzing heterotrophic bacteria capable of growth on antibiotic-containing media. In addition, quantitative real-time polymerase chain reaction (qPCR) method was used to measure eight ARG – tetA, tetW, aacA, ampC, mecA, ermA, blaTEM, and intI1 in the surface water collected at each time point. Significant associations (p < 0.05) were observed between the locations of sampling sites relative to WWTP discharge points and the rate of E. coli isolate resistance to tetracycline, ampicillin, cefoperazone, ciprofloxacin, and sulfamethoxazole together with an increased rate of isolate MDR. The abundance of antibiotic-resistant heterotrophs was significantly greater (p < 0.05) downstream of WWTPs compared to upstream locations for all tested antibiotics. Consistent with the results from the culture-based methods, the concentrations of all ARG were substantially higher in the downstream sites compared to the upstream sites, particularly in the site immediately downstream of the WWTP effluent discharges (except mecA). In addition, the Class I integron (intI1) genes were detected in high amounts at all sites and all sampling points, and were about ∼20 times higher in the downstream sites (2.5 × 107 copies/100 mL surface water) compared to the upstream sites (1.2 × 106 copies/100 mL surface water). Results suggest that the treated WWTP effluent discharges into surface waters can potentially contribute to the occurrence and prevalence of AMR in urban watersheds. In addition to detecting increased ARG in the downstream sites by qPCR, findings from this study also report an increase in viable AMR (HPC) and MDR (E. coli) in these sites. This data will benefit establishment of improved environmental regulations and practices to help manage AMR/MDR and ARG discharges into the environment, and to develop mitigation strategies and effective treatment of wastewater.
Incidences of antimicrobial resistance (AMR) in previously susceptible pathogenic bacteria are on the rise (Jones et al., 2008). Another related issue of serious consequences to public health is the proliferation of multidrug resistance (MDR) within both pathogenic and non-pathogenic bacterial populations (Levy and Marshall, 2004). This has been identified as a critical issue of profound concern by several global organizations such as the World Health Organization, U.S. Center for Disease Control, the National Academy of Science’s Institute of Medicine, the Federal Interagency Task Force on Antimicrobial Resistance, the Infectious Diseases Society of America and numerous other worldwide public health authorities (Spellberg et al., 2008; Allen et al., 2010; Bush et al., 2011; Pruden, 2014). AMR and MDR development in pathogenic bacteria results in several issues concerning public health including limited treatment options, increased morbidity and mortality rates, increased hospital stays, high treatment costs, and the increased necessity for novel antibacterial agents (Goossens et al., 2005; Lim and Webb, 2005; Chopra et al., 2008; Kemper, 2008; Blot et al., 2010; Lye et al., 2012; Nikaido and Pagès, 2012; Naqvi et al., 2013; Worthington and Melander, 2013).
Major contributors to the spread of antibiotic resistance include excessive use in humans and animals, overcrowding and increased rates of transmission between people in communities and hospitals, and the failure of implementing and executing proper hygiene and disinfection practices (Gopal Rao, 2012). AMR and MDR can rapidly spread within bacterial populations of related and unrelated species (Davison, 1999; Pepper and Gentry, 2015) by horizontal gene transfer (Dzidic and Bedeković, 2003) of antibiotic resistance genes (ARG) present in plasmids, transposons, and integrons, or through development of spontaneous mutations (Courvalin, 1994). In recent years, understanding the sources of AMR, MDR and ARG distribution has been deemed critical to eventually control and regulate the spread of ARG (Allen et al., 2010). Yet, enormous gaps still remain in our current knowledge about the occurrence, spread and distribution of AMR, MDR and ARG in the reservoirs found in natural and artificial environments (Allen et al., 2010; Wright, 2010; Rizzo et al., 2013).
While the mechanisms by which antibiotic resistant bacteria (ARB) and ARG are transported and spread through the environment are still not fully understood, previous studies have predicted connections between human activity and the conveyance of resistance traits through agricultural operations, aquatic environments, and sediments (Pei et al., 2006; Baquero et al., 2008; Zhang X. et al., 2009; Huijbers et al., 2015). Pharmaceutical compounds and resistant bacteria may also be introduced to wastewater treatment systems through hospital, industrial, and residential wastewater discharge, and then introduced to the environment (Zuccato et al., 2010; Amador et al., 2015; Verlicchi et al., 2015).
The evolution and development of resistance in clinically important bacteria could be a result of increased opportunities of genetic exchanges with the environmental ARG pool (Bouki et al., 2013; Rizzo et al., 2013; Huijbers et al., 2015, 2019; Chu et al., 2018). Urban WWTPs are increasingly being suspected to be one of the major reservoirs of AMR, MDR and ARG and their mobilization into the environment through effluents (Iwane et al., 2001; Jindal et al., 2006; Szczepanowski et al., 2009; Zhang Y. et al., 2009; Bouki et al., 2013; Korzeniewska et al., 2013; Rizzo et al., 2013; Huijbers et al., 2015; Chu et al., 2018). Contemporary municipal WWTPs are typically incapable of specifically addressing the influx of antibiotics (Adams et al., 2002; Rizzo et al., 2013). Wastewater treatment has also been found to be generally ineffective against certain strains of resistant enterococci, specifically with resistance to ciprofloxacin, erythromycin, and tetracycline (da Silva et al., 2006), with the prevalence of ciprofloxacin resistance actually increasing through the treatment process. The presence of sulfonamide resistance genes in a river environment was found to increase significantly downstream of a swine feedlot WWTP (Hsu et al., 2014). Iwane et al. (2001) found that Escherichia coli isolates obtained along the Tama River in Tokyo, Japan expressed increasing resistance to antibiotic agents as sampling moved downstream, and was attributed to treatment plant discharge. Studies tend to vary with respect to the efficiency in which resistant organisms are removed during the treatment process, the microbial species expressing resistance in the effluent, and the antimicrobial agents to which the organisms express resistance. It also should be noted that different WWTP unit operations will affect the overall removal efficiency and ultimately antibiotic resistant bacteria discharge to the environment (Sayah et al., 2005; Janezic et al., 2013; Hamilton et al., 2020). Czekalski et al. (2012) found that while WWTPs reduced total bacterial loads in the effluent, there was an observed increase in multidrug resistant bacteria and ARG which were then found to accumulate in the sediment of the plant outlet. Aeromonas and Pseudomonas aeruginosa isolates obtained from some water reservoirs were found to express 50 and 100% multi-drug resistance, respectively (Blasco et al., 2008). A recent review also suggests the importance of accounting for stormwater as a key source of ARG propagation considering the cumulative impact stormwater runoff may have as it comes in contact with overflows from untreated wastewater, sewer and sanitary discharges, among others (Hamilton et al., 2020). Understanding of the impacts of urbanization and wastewater effluent on the presence of antibiotic resistance in the environment will aid in future efforts to address antibiotic resistance through treatment plant process design - establishing more informed guidelines and proper regulations surrounding WWTP practices. We investigated the relationship between urban development and the occurrence and persistence of antimicrobial resistance in the surrounding aquatic environment using antimicrobial resistance data produced by culture-based and quantitative PCR methods. In this study, we collected surface water samples from six sites in an urban watershed – upstream and downstream of two wastewater treatment plants in the Bryan - College Station (BCS), Texas, over a period of nine months. Heterotrophic bacteria capable of growing on antibiotic amended media (HPC-Ab) and E. coli were isolated from the six sampling sites and evaluated for resistance to selected antibiotics. Surface water samples were also analyzed and quantified for the presence of eight different ARG targets. Rates of antimicrobial resistance for E. coli isolates and antimicrobial resistant heterotrophic communities were compared by sampling site and their relative position with respect to WWTPs to determine if WWTP discharge may affect the antimicrobial resistance profiles of surface water bacteria in the surrounding environment.
Six sampling sites were established within the boundaries of the Carters Creek watershed in BCS area on the main stems of Carters Creek and Burton Creek (Figure 1). Sites were selected to represent areas up and downstream of two WWTPs. Sites 1, 3, 5, and 6 were located on the main stem of Carters Creek, and sites 2 and 4 located on the main stem of Burton Creek. Most sites (all but site 3) were located at the intersection of the respective creek and an overpassing bridge. Sites 2, 4, 5, and 6 were sampled upstream of the bridge crossing, and site 1 was sampled directly underneath the overpass. Site 3 was sampled on the stream stem of Carters Creek running adjacent to the highway, upstream of its confluence with Burton Creek. Site 2 was located at the outlet of a channelized stretch of Burton Creek, characterized by shallow flow with substantial algal growth on the concrete surface. All of the sampling sites selected in this study are regular water quality monitoring sites for the Texas Commission on Environmental Quality (TCEQ, 2006) since the commencement of an ongoing Carters Creek watershed Total Maximum Daily Load (TMDL) project in August, 2007.
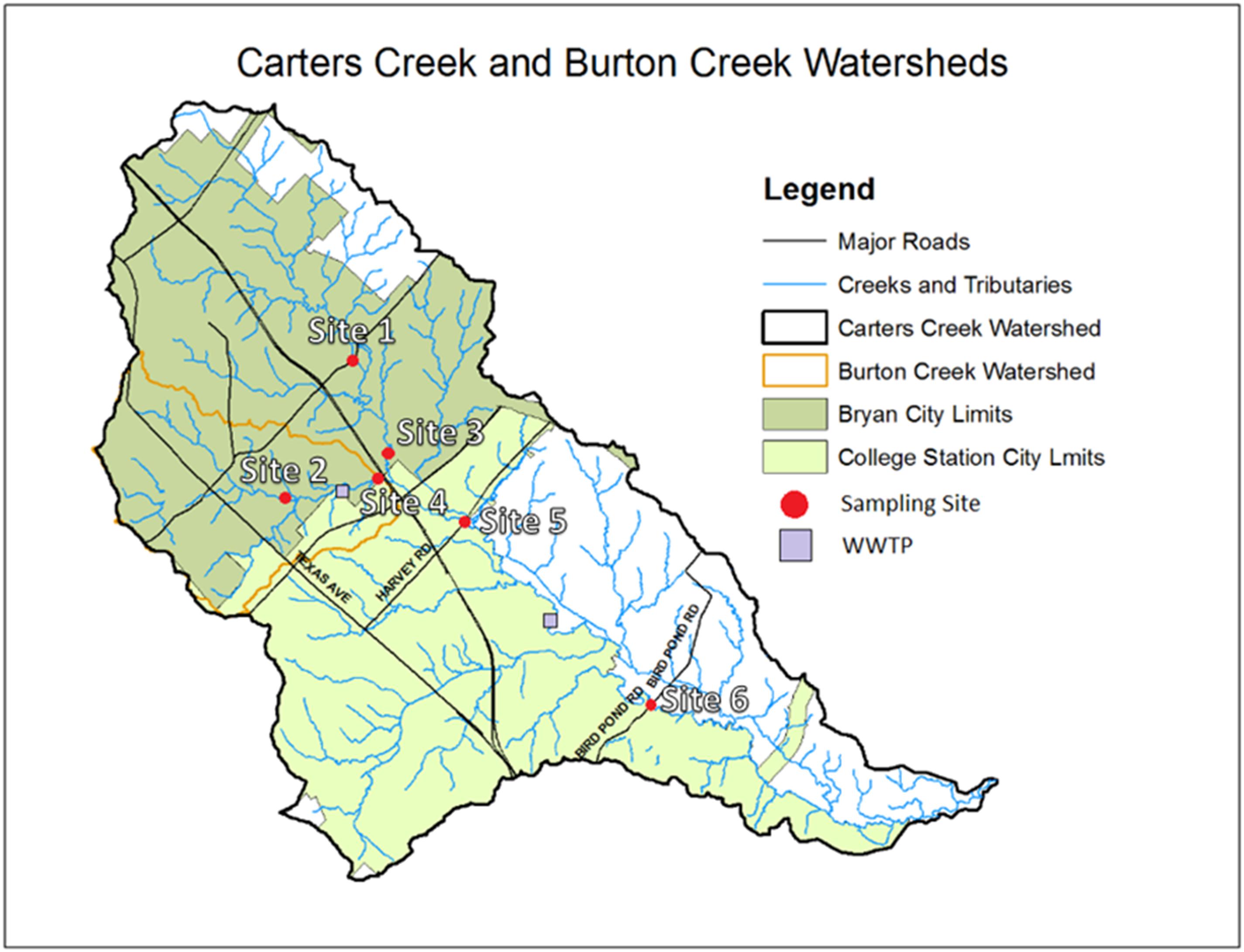
Figure 1. Map of the Carters Creek Watershed and locations of the six sampling sites and the two WWTPs.
The Burton Creek WWTP is located upstream and Carters Creek WWTP downstream. Burton Creek WWTP permitted discharge is 8 MGD. Carters Creek WWTP was built in the late 50’s to treat about 0.6 MGD to meet the population of 6,000 people. Now the treatment capacity is 9.5 MGD, even though it reaches only about 7 MGD maximum. Both the treatment plants use activated sludge process to remove carbonaceous and nitrogenous BOD. They both do not use any tertiary treatments. Burtons Creek WWTP uses chlorination to remove pathogens while Carters Creek WWTP applies UV as a disinfection method. Dewatered sludge is digested anaerobically at Burton Creek and aerobic digestion is used in Carters Creek. To meet the population growth another 2 MGD plant was constructed (Lick Creek WWTP) and in operation since early 90’s. Additional information on this watershed and the water quality standards and compliance is available at https://www.tceq.texas.gov/waterquality/tmdl/85-carterscreek.html. Urbanization-related maps are provided in Supplementary Figure 4. Specific information concerning the sampling sites is included in Supplementary Table 1 (see Supplementary Material).
A total of six separate sampling events were conducted over a nine-month period between July 2015 and April 2016. Surface water samples were collected using ∼500 mL Whirl-Pak® sterile bags (eNasco, Fort Atkinson, WI, United States) attached to a sampling pole. Water samples were collected from the mid-point of the stream flow approximately 3 cm below the surface. Samples were immediately transferred on ice back to the laboratory and processed within 6 h of collection.
Inhibition zone diameters were measured and recorded in millimeters and compared to CLSI (Clinical and Laboratory Standards Institute) standards to determine if each isolate was susceptible or resistant to each antibiotic. Isolates were subsequently confirmed as E. coli through PCR amplification of uidA with E. coli-specific primers (data included in Supplementary Material).
Initially, four concentrations of each water sample were prepared (1.0, 0.1, 0.01, and 0.001) by ten-fold serial dilutions in phosphate-buffered saline solution (PBS). Ten mL of each dilution was then filtered through a 0.45 μm filter membrane (Millipore, Billerica, MA, United States) by vacuum filtration. Filter membranes were placed on 47 mm Difco® Modified mTEC agar plates (Becton, Dickinson and Company, Sparks, MD, United States) and incubated at 35°C for 2 h and then 44.5°C for 24 h in accordance with EPA Method 1603 (USEPA, 2005). Following incubation, ten presumed E. coli (magenta) colonies for each of the six sites were randomly selected, transferred to Difco® Tryptic Soy agar (Becton, Dickinson and Company, Sparks, MD, United States) using a sterile loop, and incubated at 35°C for 24 h. E. coli cell suspensions were prepared by transferring two colonies of each isolate into tubes with 5 mL of BBL® Tryptic Soy Broth (Becton, Dickinson and Company, Sparks, MD, United States) and incubating at 35°C for 3 h while shaking at 150 rpm. Tubes were checked for turbidity against a pre-prepared 0.5 McFarland standard corresponding to a 107–108 CFU/mL bacterial cell count in the broth.
After incubation, sterile swabs were used to inoculate 100 mm Mueller Hinton Agar (MHA) plates (Neogen Corporation, Lansing, MI, United States). Antibiotic resistance of the E. coli isolates was determined by the Kirby-Bauer method for antibiotic susceptibility (Bauer et al., 1966). Eight antibiotic susceptibility discs (Becton, Dickinson and Company, Franklin Lakes, NJ, United States) of tetracycline (30 μg), ampicillin (10 μg), ciprofloxacin (5 μg), imipenem (10 μg), sulfamethoxazole/trimethoprim (23.75/1.25 μg), gentamicin (120 μg), cefoperazone (75 μg), and cephalothin (30 μg) were stamped onto each MHA plate using a BBL® Sensi-Disc® 8-place Dispenser (Becton, Dickinson and Company, Franklin Lakes, NJ, United States). The MHA plates were then incubated at 35°C for 16–24 h and the diameters of the inhibition zones measured to determine resistance or susceptibility of each isolate to the antibiotics according the Clinical and Laboratory Standards Institute (CLSI) standards. Control organisms, E. coli 25922, Staphylococcus aureus 25923, and Pseudomonas aeruginosa 27852, were used to ensure consistency during the antibiotic disc diffusion process.
PCR amplification of the E. coli specific uidA sequence was used to confirm all isolates collected as E. coli (Bower et al., 2005). Cell suspensions of each presumed E. coli isolate were prepared by suspending bacterial growth from the MHA agar in 100 μL of sterile, distilled water. PCR mixtures (50 μL) were prepared consisting of 25 μL of GoTaq® G2 Green Master Mix (Promega, Madison, WI, United States), 1.75 μL (350 nM) each of the forward (uidA1318F) and reverse (uidA1698R) primers (Integrated DNA Technologies, Coralville, IA, United States), 5 μL of cell suspension as the template DNA, and 16.5 μL of sterile nuclease-free water. E. coli 25922 isolates were used for the positive control. Primer sequences, target, and reference are shown in Table 1A.
PCR conditions included one initial heating cycle at 94°C for 4 min; followed by 35 cycles at 94°C for 30 s, 60°C for 30s, and 72°C for 30 s; a final cycle at 72°C for 6 min, and then held at 4°C. DNA electrophoresis was performed in a 2% agarose gel (Amresco, Solon, OH, United States) stained with ethidium bromide (Sigma-Aldrich, St. Louis, MO, United States) and a 1X Tris-Borate-EDTA (TBE) (Fisher BioReagents, Fair Lawn, NJ, United States) buffer solution. A 100 bp ExACTGeneTM DNA ladder (Fisher BioReagents, Fair Lawn, NJ, United States) was used as the marker (Data in Supplementary Material, Section “PCR Isolate Confirmation Results” and Supplementary Figure 1).
Heterotrophic bacteria capable of growth on antibiotic-containing media was analyzed using the following protocol. This method potentially captures both intrinsic and acquired resistant bacteria, but does not differentiate between the populations. Four ten-fold serial dilutions of each of the six water samples were prepared (1.0, 0.1, 0.01, and 0.001) by diluting in PBS. Thirty microliters of each dilution were spread-plated onto five sets of 47 mm plate Bacto® Reasoner’s 2A (R2A) agar (Difco Laboratories, Detroit, MI, United States) amended with the following antibiotics: 32 μg/mL ampicillin (HPC-Am) (Ward’s Science, Rochester, NY, United States), 16 μg/mL tetracycline (HPC-Te) (Alfa Aesar, Ward Hill, MA, United States), 4 μg/mL ciprofloxacin (HPC-Cpr) (TCI America, Portland, OR, United States), 50.4 μg/mL sulfamethoxazole (HPC-Su) (Chem-Impex International Inc., Wood Dale, IL, United States), and un-amended R2A (HPC) with no antibiotic. Antibiotic concentrations in the agar were determined based on prior studies and are generally around half the strength of either the IV or oral dosage concentrations (Pei et al., 2006; Gao et al., 2012; Garcia-Armisen et al., 2013). Plates also contained 200 μg/mL of cycloheximide (Amresco, Solon, OH, United States) as a fungicide to suppress any fungal growth. All plates were incubated at 28°C for 5 days before obtaining bacterial CFU plate counts. The limit of detection was one CFU in 30 μL of undiluted sample, or 1.52 log10 CFU/mL. There were five instances in which no bacteria were culturable within the sample volume and concentration limit; four of the ciprofloxacin-amended plates, and one of the tetracycline-amended plates. These results were reported as below the limit of detection, and were represented as ½ the limit of detection (16.67 CFU/mL) for statistical analysis.
From each sample, 100 mL of water was filtered through sterile 0.22 μm Millipore membrane filters of 47 mm diameter and stored in sealed sterile petri plates with the biomass facing up at −80°C for further analyses. The environmental DNA was extracted from these membrane filters using a MoBio PowerWater® DNA Isolation Kit (currently Qiagen, Germantown, MD, United States), following the manufacturer’s instructions. Biomass from the filters were prepared and lysed using the PowerWater® bead tubes on a MoBio vortex adapter. For DNA extraction, the centrifugation method was used to bind DNA onto the provided spin filter, washed and finally eluted using the kit’s elution buffer. The final elution volume for each DNA sample was 100 μl. The DNA thus obtained was analyzed for concentration using a NanoDrop spectrophotometer (Thermo Fisher Scientific) and used for further quantitative PCR analysis.
The DNA from each sample was analyzed for the presence of eight ARG: intI1, tetA, tetW, ampC, blaTEM, mecA, aacA, and ermA. The primer pairs and conditions used for each qPCR analysis is listed in Table 1. Each primer pair was tested with each qPCR standard for accuracy of product size and annealing temperature by confirming with end point PCR and agarose gel electrophoresis before using for final qPCR analyses. For qPCR analysis, each sample was run in duplicate. Each standard (standard detail listed in Supplementary Table 2, see Supplementary Material) was prepared by (i) extracting DNA from the control cultures using the MoBio microbial DNA isolation kit (currently Qiagen, Germantown, MD), (ii) PCR amplification using respective primers, (iii) confirming purity and amplicon size using agarose gel, and (iv) purifying using the Qiagen PCR purification kit (Germantown, MD, United States). The standard DNA was then quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific) and serially diluted to generate qPCR standard curves for each primer pair. Each qPCR reaction had a final volume of 25 μL and consisted of the following reagents: 12.5 μL of 2X SYBR Green (Applied Biosystems), 1.25 μL of 10 μM of each primer, and 10 μL of environmental DNA from each site. The reactions were run in duplicates on 96-well Eppendorf green-skirted plates (Fisher Scientific), and sealed with a plate sealer using qPCR sealing films (BioRad) before analysis using an Eppendorf Realplex2 Mastercycler system. The results were analyzed using the Eppendorf Realplex2 software and converted into genomic units per 100 mL (GU 100 mL–1) of water sample (Brooks et al., 2014).
Escherichia coli isolate responses to antibiotic susceptibility disc diffusion were categorized as either susceptible or resistant (including intermediate resistance) and assigned a binary value for each response: 1 for resistant and 0 for susceptible. Then, isolates and isolate responses were grouped into a number of various categories and tested for significant associations by chi-square analysis. Groupings were generally done by pairing binary data from two individual sampling sites or two groups of sampling sites, generating two-by-two grids with one degree of freedom. Significant differences were determined by Chi square sums of 3.84 or greater, or p < 0.05 for one degree of freedom. Post hoc multi-comparison tests were carried out for sample site, where appropriate, by conducting pairwise Chi square tests with Bonferonni adjusted p-values. Statistical analysis of the HPC-Ab and box plot generation was done using SAS® University Edition (Cary, NC, United States). Significant differences in the abundance and normalized resistance rates of heterotrophic ARB were evaluated using one-way ANOVA by least-significant-difference (LSD) comparison. Significant differences were checked for homogeneity of variance by Levene’s test. In cases where significant differences in homogeneity were found in the data set, it was (then) determined by Welch’s ANOVA. Relationships were considered to be significant at p < 0.05.
Antibiotic resistance gene levels per 100 mL (GU 100–1 mL) were log10 transformed prior to statistical analysis. Differences in response variables (stream position and creek) was assessed for all measured ARG using the mixed procedure in SAS Enterprise Guide 7.1 (SAS Institute). Creek was used as a random variable in the mixed model. Residuals were normally distributed and means were post-hoc adjusted and compared using least square means. All differences were significant at p < 0.05, unless otherwise noted.
The number of isolates expressing resistance to individual antimicrobial agents by sampling site are displayed in Figure 2 and Supplementary Table 3. Twelve percent of all isolates were susceptible to all 8 antibiotics. A large proportion (84%) of all isolates expressed resistance to cephalothin, with rates of resistance at each individual sampling site falling consistently between 77 and 90% of the isolates collected. The next highest rates of resistance after cephalothin occurred with ampicillin and tetracycline at 15 and 14% of all isolates, respectively. Resistance to ampicillin was expressed in 41 isolates, resistance to tetracycline was expressed in 38 isolates, and resistance to cefoperazone, gentamycin, ciprofloxacin, and sulfamethoxazole/trimethoprim was found in a fewer number of isolates, at rates of 3, 3, 4, and 5%, respectively. All 280 isolates were susceptible to imipenem.
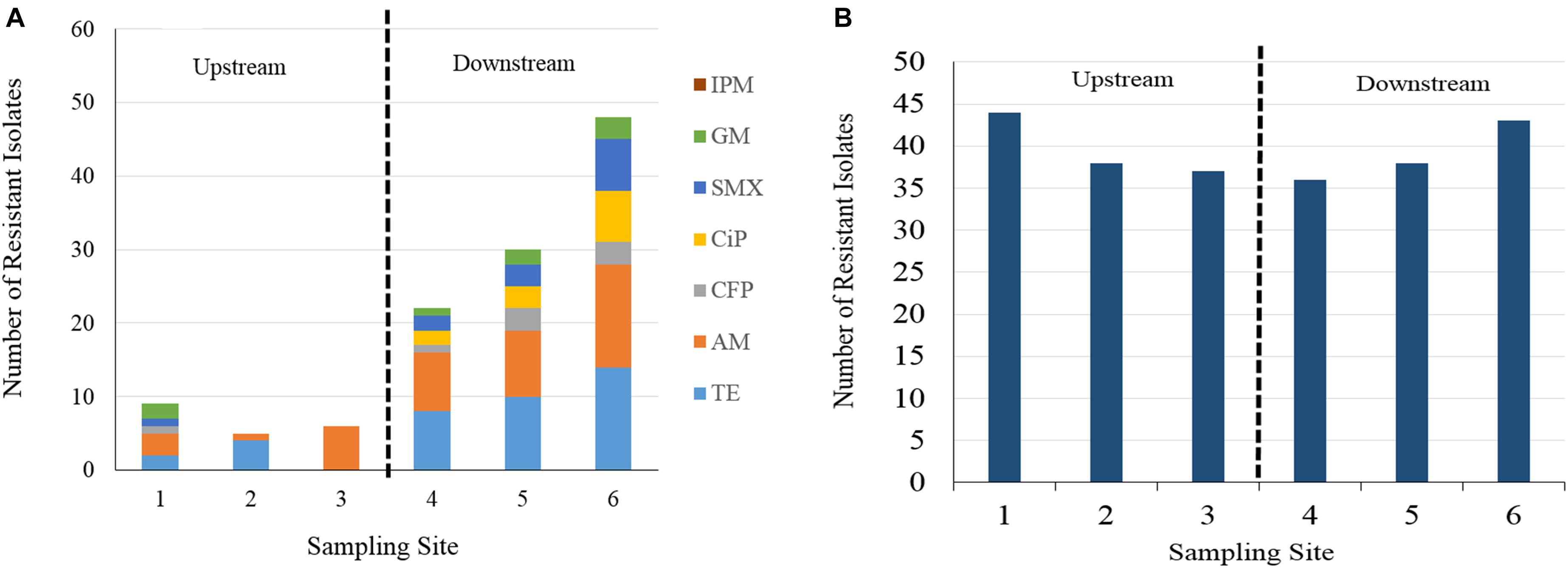
Figure 2. (A) Number of resistant isolate responses to seven of eight antibiotics at different sampling site by Kirby Bauer disc diffusion method. Cephalothin is excluded for visibility of less frequently occurring AB resistances. IPM, imipenem, GM, gentamycin, SMX, sulfamethoxazole, CiP, ciprofloxacin, CFP, cefoperazone, AM, ampicillin, TE, tetracycline. (B) Column chart of cephalothin resistant E. coli isolate responses. Resistance occurred at a greater frequency than other antibiotic resistances, more consistently across upstream and downstream sampling sites.
A column chart of isolate resistance responses by sampling site and antibiotic shows an increase in the total number of resistant responses in the downstream sampling sites (Figure 2). Isolates collected from the downstream sampling sites expressed resistance more frequently and to diverse antimicrobial agents than the upstream sites. Sampling site 1 displays the most diversity in resistance to different agents in the upstream group, due to one isolate sampled during event two expressing resistance to six agents. The number of total resistant responses also appears to increase as the sampling sites are farther away downstream. For ampicillin, resistance rates fell between 2 and 13% for isolates obtained upstream of WWTP discharges and 17–28% for isolates obtained downstream of WWTP discharges. For tetracycline, resistance rates fell between 0 and 9% for isolates obtained upstream of WWTP discharges and 17–28% for isolates obtained downstream of WWTP discharges. Resistance to cefoperazone, gentamycin, ciprofloxacin and sulfamethoxazole were found more frequently in the isolates obtained from downstream sampling sites. Gentamycin resistance was the only instance in which isolate resistance was found to occur more frequently in one of the upstream sites than in one of the downstream sites (site 1 vs. site 4). Cephalothin resistance is presented separately in Figure 2B as to not visually overwhelm the less frequently occurring antibiotic resistances. Cephalothin resistance occurred at a greater frequency and more consistently across all sampling sites than the observed resistance to other antibiotics, irrespective of proximity to WWTP.
Chi-square tests for isolate resistance by individual sampling site (Supplementary Figure 2) showed significant differences (p < 0.003) between at least one pair of sites for ampicillin, sulfamethoxazole, tetracycline, and ciprofloxacin. The majority of these occurred between site pairings in which one site was upstream of a WWTP and the other site was downstream of a WWTP. Only one test reported a significant difference between two sites with the same relative location to a WWTP. This result was reported for the rate of isolate resistance to tetracycline between sites 2 and 3, corresponding to the Burton Creek site upstream of the WWTP and the Carters Creek site upstream of its confluence with Burton Creek, respectively. When sampling sites were categorized into either an upstream (sites 1–3) or downstream (sites 4–6) group, a significant difference (p < 0.05) was found to exist in isolate rates of resistance to ampicillin, tetracycline, cefoperazone, ciprofloxacin, and sulfamethoxazole. While cefoperazone resistance did not increase significantly between any individual sampling sites, there was a significant increase when rates of isolate resistance were categorized and compared between the upstream and downstream.
Binomial resistance values determined by the number of resistant responses of each isolate were tallied, organized by sampling site, and sorted into five groups – isolates resistant to 1, 2, 3, or ≥4 agents (Table 2). Out of the 280 isolates, the majority (88%) showed resistance to at least 1 antibiotic agent. A total of 28 isolates (10% of total) showed resistance to 2 agents, 9 (3% of total) showed resistance to 3 agents, and 17 (6% of total) showed resistance to 4 or more agents. Resistance responses were also sorted by type of antibiotic and number of agents that each isolate was resistant to (Table 3). Cephalothin resistance was again the most frequently occurring (95%) antibiotic resistance in the sample set of multi-drug resistant isolates (resistant to two or more agents). Out of all isolates that were resistant to at least one antibiotic, 74% were only resistant to cephalothin, and cephalothin resistance accounted for 95% of all single-drug resistant isolates. Isolates only resistant to tetracycline, ampicillin, or cefoperazone were found sparingly, each representing less than 2% of the single-drug resistant isolates. No isolates were only resistant to ciprofloxacin, sulfamethoxazole, gentamycin, or imipenem. Isolates resistant to two or more agents were generally resistant to cephalothin and either tetracycline (41%), or ampicillin (48%). Resistance to three agents occurred less frequently than resistance to four or more agents, at only 4% of resistant isolates. All isolates showing resistance to 4 or more antibiotics were resistant to cephalothin, over 80% of these isolates were also resistant to tetracycline, and 90% to ampicillin. Sulfamethoxazole resistance was only found in isolates resistant to three or more agents. Resistance to cefoperazone, ciprofloxacin, sulfamethoxazole, and gentamycin was generally accompanied by several other resistances (Table 3).
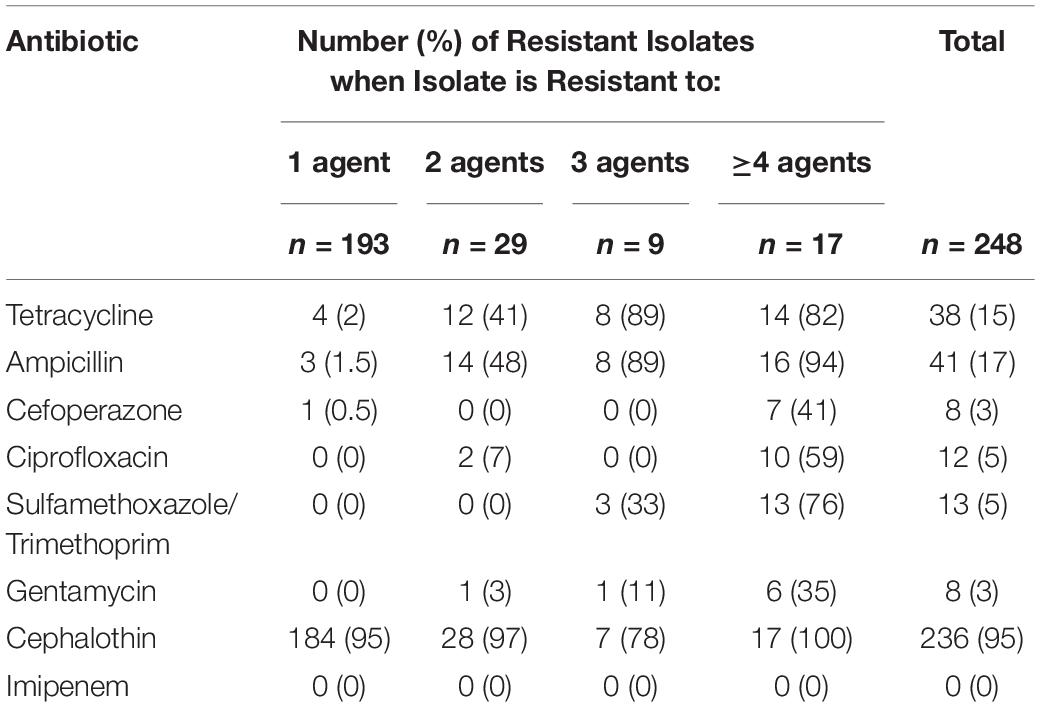
Table 3. Number of E. coli isolates expressing resistance to each antibiotic, grouped by the number of agents the isolate was resistant to.
Of the 54 multi-drug resistant isolates collected (resistant to at least 2 agents), 41 (76%) were obtained from downstream sites (sites 4–6). All isolates resistant to 3 agents and all but one of the isolates that were resistant to four agents were collected from one of the downstream sites.
Chi-square analysis revealed significant associations between several sampling site pairings for when isolates were classified according to multi-drug resistance (Supplementary Figure 3). The most significant factor contributing to differences in isolate multi-drug resistance was associated when isolates were grouped according to relative upstream and downstream position. A significant association (p < 0.001) was found to exist between the number of isolates expressing resistance to 1, 2, 3, and ≥4 antibiotic agents and whether the isolate was collected upstream of any WWTP (sites 1, 2, and 3) vs. downstream of at least 1 WWTP (sites 4, 5, and 6).
Heterotrophic bacterial plate counts were obtained during six sampling events to examine the antibiotic resistance profiles of the culturable, HPC-Ab community in the watershed. The log-transformed bacterial concentrations of each treatment category for all sampling events and sampling sites are displayed in Supplementary Table 4 and Figure 3.
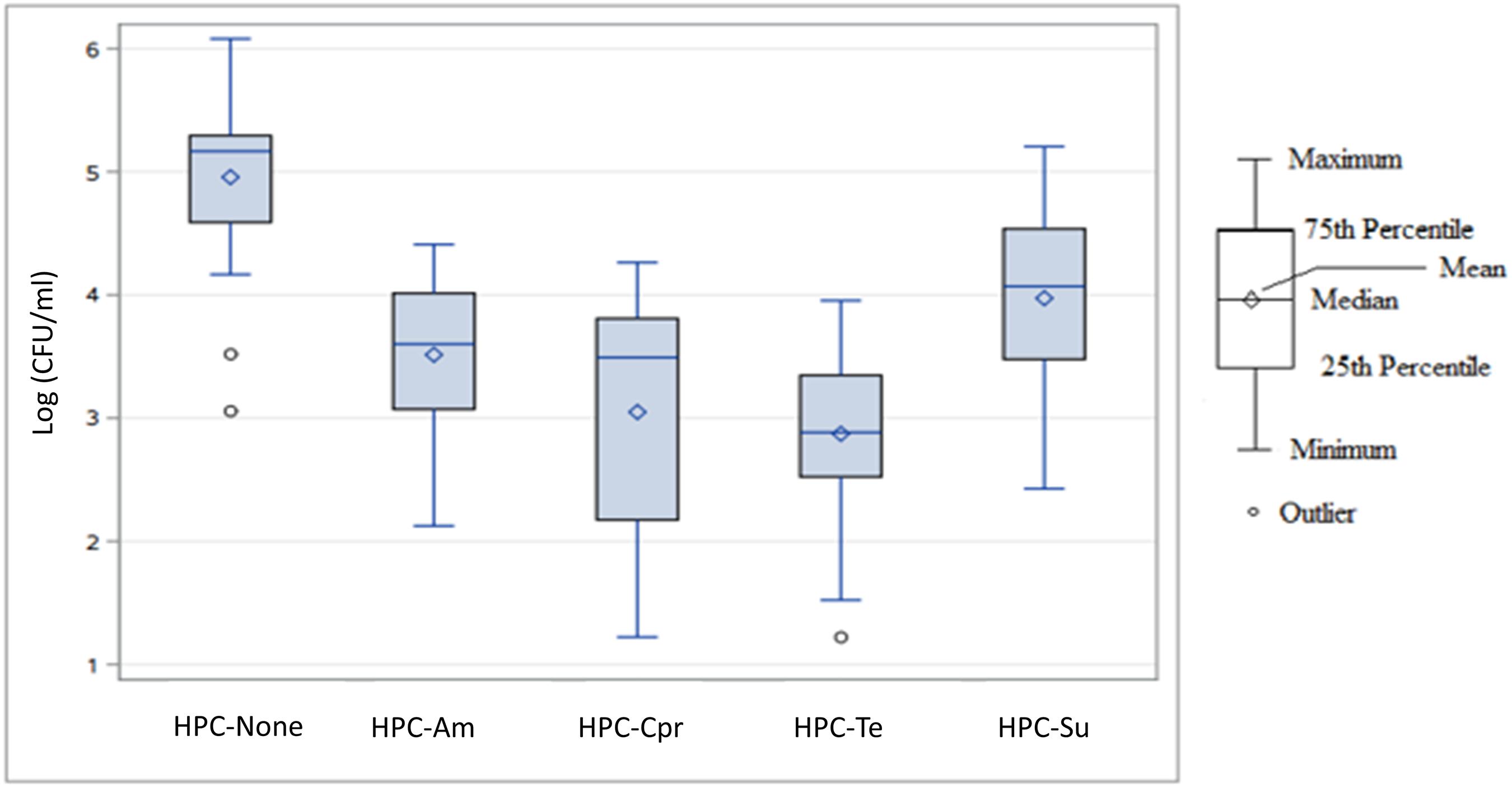
Figure 3. Box plot of log-transformed concentration distributions (log10 CFU/mL) of heterotrophic bacteria capable of growing on antibiotic amended agar by antibiotic agent across all sampling events and sampling sites. HPC-none, no antibiotic added; HPC-Am, HPC on ampicillin amended R2A plate; HPC-Cpr, HPC on ciprofloxacin amended R2A plate; HPC-Te, HPC on tetracycline amended R2A plate; HPC-Su, HPC on sulfamethoxazole amended R2A plate.
For the total concentrations of each subset of heterotrophic bacterial populations, the R2A agar with no antibiotic produced the highest overall concentration with a median value of 1.47 × 105 CFU/mL and a mean value of 1.68 × 105 CFU/mL (Figure 3). Sulfamethoxazole HPC-Su were the next highest with a median concentration of 1.18 × 104 CFU/mL, followed by HPC-Am and HPC-Cpr with median concentrations of 4.00 × 103 CFU/mL and 3.10 × 103 CFU/mL, respectively. HPC-Te had the lowest overall concentration in the study area with a median concentration of 7.67 × 102 CFU/mL. Variance in the total population for each treatment was considerably large, with standard deviations larger than the mean values.
Abundance of HPC-Am varied significantly between sampling sites (p < 0.0001), primarily due to the variance occurring between sites 1, 2, 3, and 6 compared to sites 4 and 5 (Figure 4A). HPC did not vary significantly by sampling event (p > 0.65). HPC-Am were found in significantly greater (p < 0.0001) concentrations in the downstream group relative to WWTP discharge, with mean concentrations of 1.3 × 104 and 1.2 × 104 CFU/mL from sites 4 and 5, respectively.
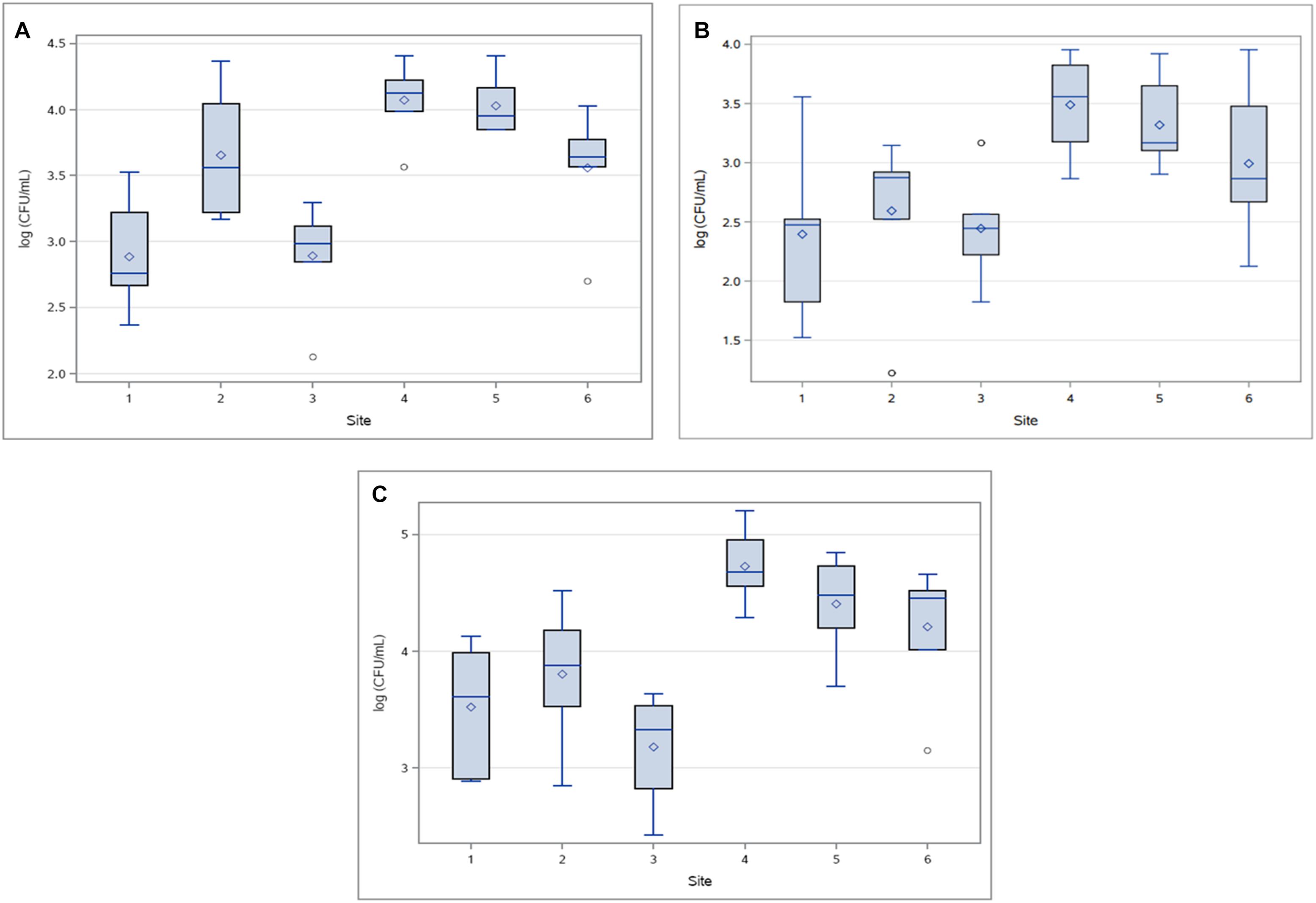
Figure 4. Concentrations (log10 CFU/mL) of (A) HPC-Am, (B) HPC-Tet and (C) HPC-Su across sampling sites for all sampling events.
HPC-Te produced a few outliers due to an atypically compact distribution of concentrations at site 2 (Figure 4B), in contrast to an otherwise expansive distribution and large standard deviations as seen in the other treatments. Standard deviations of HPC-Te concentrations at sites 2 and 3 were lower by one order of magnitude or more than what was typically seen in other resistant bacterial population. The abundance of HPC-Te varied significantly by sampling site (p < 0.007) and sampling event (p < 0.02), mainly due to considerably higher concentrations sampled during event 6.
HPC-Su were the most prominent across all sampling sites in this study with the highest mean concentration of resistant bacteria at any sampling site of 6.67 × 104 CFU/mL (Figure 4C). Sampling site had a significant influence (p < 0.0001) on the concentration of HPC-Ab, mainly due to consistently higher concentrations found at sites downstream from WWTPs.
The mean concentrations HPC-Ab obtained upstream of a WWTP in the tetracycline and sulfamethoxazole amended media were an order of magnitude below the mean concentrations in their respective downstream sites. Significant differences in the abundance HPC-Ab were found to exist between upstream and downstream sites for both the tetracycline (p < 0.0001) and sulfamethoxazole amended media (p < 0.0001).
Overall, analysis of the frequency, distribution, and quantity of the tested ARG reveal a significantly higher measure of ARG in the sites downstream of the WWTP discharge than that found in the upstream sites (Figure 5). Overall, blaTEM, ermA, intI1, tetA, and tetW were significantly greater in downstream compared with upstream sites (p < 0.05). Except for mecA, aacA, emrA, and blaTEM genes at specific sites and sampling events, all other ARG were detected in all sites at all location at all sampling events (Figure 5).
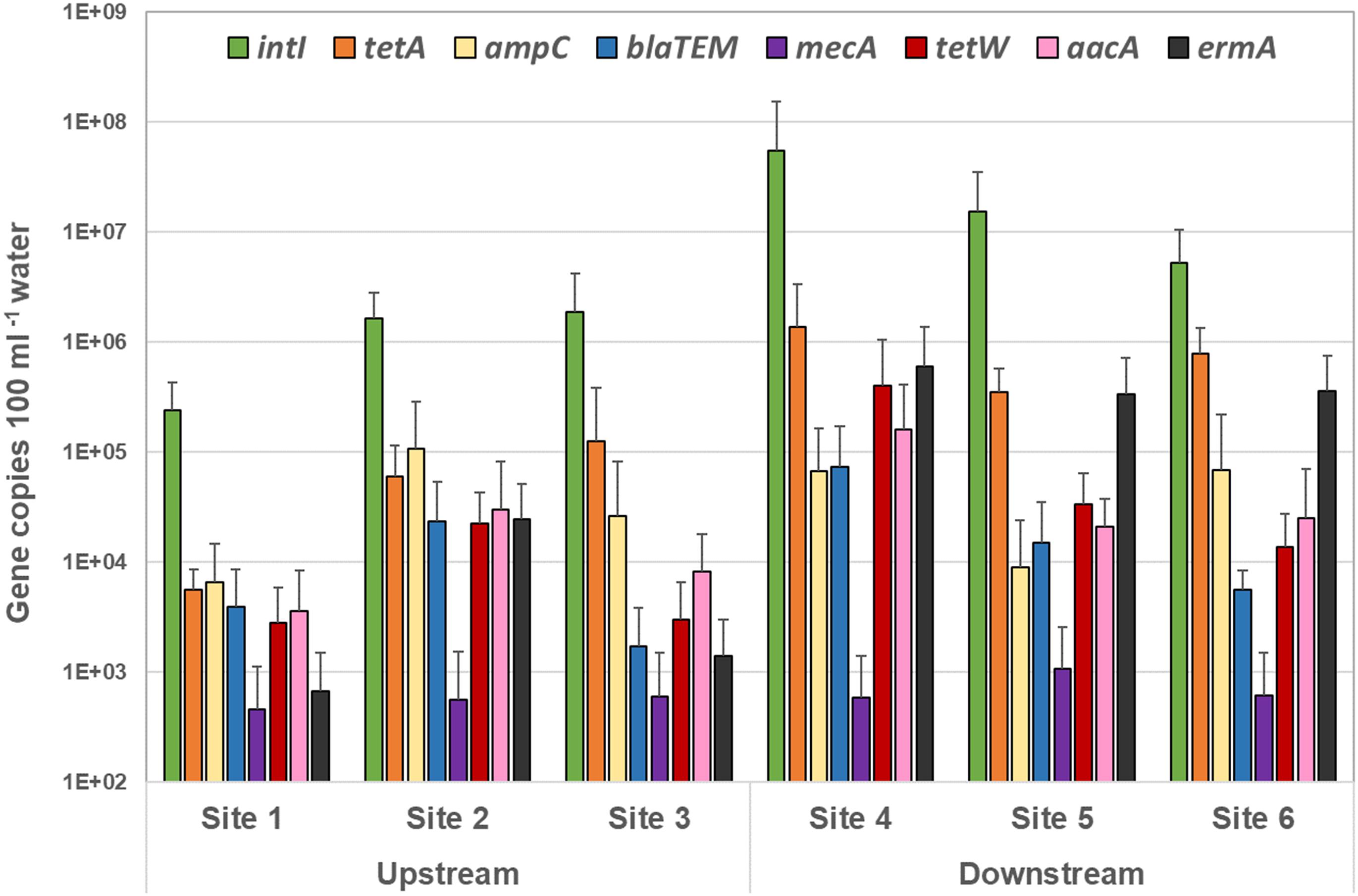
Figure 5. Quantity of each antibiotic resistance gene over all sampling events detected by quantitative PCR in the upstream sites and downstream sites. The target gene values (gene copies 100 ml–1 sample) are displayed in mean of all sampling events, and the standard deviations represent the variation among the six sampling events.
Overall, the highest level of ARG was associated with the combined tetracycline resistant genes (tetA and tetW) – averaged at 7.3 × 104 and 9.9 × 105 copies100 mL–1 surface water in the upstream and downstream sites, respectively (Figure 5 and Supplementary Table 5).
A significantly high concentration of the integrase (intI1) gene was noted amongst all sites – 1.25 × 106 and 2.5 × 107 gene copies 100 mL–1 surface water in the upstream and downstream sites respectively (p < 0.0001) (Supplementary Table 5 and Figure 5). In addition, copies of all ARG tested in this study (except for mecA) were found to be considerably greater in site 4 – the site immediately downstream of the BCWWTP (Burton Creek Wastewater Treatment Plant) (Supplementary Table 5). This is a notable result and may suggest a role of the WWTP in dissemination of the ARG into the immediate surrounding environment. Additionally, the high prevalence in ARG copies within the immediate downstream site taken together with 40 times higher prevalence of intI1 genes in the same site (Supplementary Table 5) also suggest that the WWTP may provide a favorable setting for genetic exchanges that lead to development of AMR and MDR in resident bacterial populations.
Among the other tested ARG, erythromycin resistant genes (ermA) were found to be substantially high in all downstream sites (4.3 × 105 copies 100 mL–1 water) (p < 0.0001), particularly high in site 4 (6 × 105 copies 100 mL–1 water) – 48 times higher when compared to the upstream sites (9 × 103 copies 100 mL–1 water) (Supplementary Table 5). A similar trend, though not significant, was noticed with the broad-spectrum aminoglycoside resistance gene target (aacA) – 1.4 × 104 copies 100 mL–1 water compared to 1.6 × 105 in site 4 (Supplementary Table 5). Similarly, copies of blaTEM genes targeting the TEM type β-lactamases (Bradford, 2001) were significantly greater downstream compared with upstream sites (p = 0.0115), for instance, in site 4 (7.33 × 104 copies 100 mL–1 water) compared to the combined average of all upstream sites (9.6 × 103 copies 100 mL–1 water) (Supplementary Table 5).
Ampicillin, sulfamethoxazole, and ciprofloxacin, or closely related drugs (amoxicillin), are among the top 5 antibiotics prescribed for use for adults in the United States (Shapiro et al., 2013; Van Boeckel et al., 2014; Hicks et al., 2015), and all have been found to occur in WWTPs in varying concentrations and design conditions (Batt et al., 2007). In the current study, a significant association (p < 0.05) was found to exist between the location of sampling sites relative to WWTPs (upstream group vs. downstream group) and isolates expressing resistance to ampicillin, ciprofloxacin, cefoperazone, sulfamethoxazole, and tetracycline. This supports the hypothesis that WWTP effluent may be contributing to the conveyance of antibiotic resistant bacteria downstream from discharge points. The absence of significant associations between rates of isolate resistance among upstream sites indicates that these differences are not solely dependent on variations between all sampling sites, but also their relative location to WWTP discharge points.
The occurrence of antibiotic resistance may not always imply an outside effect and can be an intrinsic property of the natural environment. In this study, cephalothin represented the highest rate (84%) of resistance found in all isolates, irrespective of their collection location. The high rate of resistance to cephalothin and the ubiquity of its presence suggests that this resistance trait may be partially due to natural occurrence in the watershed. This is consistent with results from other studies performed with E. coli isolates obtained from surface waters in Michigan and Illinois, rates of isolate resistance to cephalothin at 80.6 and 80%, respectively (Sayah et al., 2005; Janezic et al., 2013). Cephalothin resistance also represented 95% of the 193 isolates resistant to only 1 antibiotic, dramatically inflating the abundance of isolates classified as resistant to at least 1 antibiotic. If cephalothin was excluded from the AMR data for this study, an additional 184 isolates (66% of total) would be classified as susceptible to all agents.
The rate of isolate resistance to tetracycline (14% of all isolates) was found to be lower than expected when compared to similar research (Jindal et al., 2006; Rajić et al., 2006; Brooks and McLaughlin, 2009; Sullivan and Karthikeyan, 2012). Previous studies have found the occurrence of tetracycline resistance to be prevalent in watersheds associated with agricultural and animal feed lot operations (Jindal et al., 2006; Rajić et al., 2006), with resistance rates of over 90% found in E. coli isolates obtained from swine lagoon effluent (Brooks and McLaughlin, 2009). Sullivan and Karthikeyan (2012) reported the prevalence of tetracycline resistant bacteria and tetracycline resistant genes in sediment and surface water samples collected from Carters Creek watershed; however, the majority were found in greater abundance bound in stream sediment samples than in surface water samples. In another study on the Carters Creek watershed, Sullivan and Karthikeyan (2012) found that while the occurrence of tetracycline resistance genes increased downstream of WWTPs, concentrations of tetracycline resistant bacteria were not significantly affected.
Imipenem is a group 2 carbapenem generally reserved as the last line of defense against particularly resilient Gram-negative pathogens and not widely prescribed (Nicolau et al., 2012). Out of all the isolates collected from all sampling sites, all 280 E. coli isolates were susceptible to imipenem. Imipenem resistance was not detected in the surface water bacteria of the Carters Creek watershed.
A substantial fraction (19%) of all 280 E. coli isolates expressed resistance to two or more antibiotics. Multi-drug resistance was found to increase significantly (p < 0.05) in the sites downstream of a WWTP for isolates resistant to ≥2, ≥3, and ≥4 agents. Other studies have observed high rates in the development of multidrug resistance in E. coli isolates in WWTP processes (Korzeniewska et al., 2013; Amador et al., 2015), found to be primarily driven by the transfer of conjugative plasmids (da Silva et al., 2006). A number of WWTP disinfection practices had negligible effects on reducing rates of multidrug resistant bacteria, and in a number of cases increasing it (Bouki et al., 2013). Even if the WWTP effluents in this study had considerably low levels of multidrug resistant bacteria, in other studies (Rizzo et al., 2013) these low concentrations have been shown to persist and propagate in the environment once discharged. ARG not necessarily bound to culturable organisms are also likely escaping treatment processes and contributing to the development of multidrug resistance (Kümmerer, 2009). Resistance to cefoperazone, sulfamethoxazole, ciprofloxacin, and gentamycin was more frequently found in multidrug resistant isolates, and rarely as the only type of resistance. This suggests that resistance to these agents is either driven by similar modes of defense coded by resistance genes to other agents, or that the acquisition of resistance to these agents usually occurs in tandem with other antibiotic resistances.
The high rates of resistance to cephalothin across all six sampling sites inflated multi-drug resistance rates, present in 95% of the 248 isolates resistant to at least one antibiotic. This increases the importance of the multidrug resistance classifications of isolates resistant to three or more and four or more agents, due to cephalothin resistance effectively acting as a resistance baseline for this data set. While the strictest definition for multidrug resistance is resistant to two or more agents, “resistance to three or more classes” has become increasingly standard for defining multidrug resistance in Gram-positive and Gram-negative bacteria (Magiorakos et al., 2012). Still, a substantial number, 9% of all 280 isolates, expressed resistance to at least 3 antibiotics. This rate is more in line with other reports of the prevalence of multidrug resistant E. coli in surface waters (Blaak et al., 2015), though these rates likely differ considerably as a function of antibiotics tested and sampling site. A large majority (86%) of these isolates were collected downstream of a WWTP, and all significant increases in rates of isolate resistance to three or more agents occurred when comparing an upstream site to a downstream site. While some degree of multidrug resistance appears to exist naturally, the results suggest that WWTPs in the watershed might be contributing significantly to multidrug resistant bacteria in the surface water.
A significant increase in the concentrations of HPC-Ab downstream of WWTP discharge was found for all four agents tested against the total HPC community. Heterotrophic bacteria populations are diverse and possess a considerable amount of intrinsic variability in the way they occur and interact in the environment (Garcia-Armisen et al., 2013). By normalizing the abundance of HPC-Ab in the study area to the total heterotrophic bacteria population, a better understanding can be made concerning the extent of HPC-Ab relative to total numbers. Unfortunately, this diversity also makes it difficult to establish a reliable standard for which to compare resistance rates against. Additionally, the levels of HPC-Am, -Cpr, and -Te bacteria were generally confined to a range of 1–10% of the total heterotrophic community when compared to the control, though in some instances spiking to between 20 and 40% of the total population. However, these large spikes in the ratios of HPC-Ab to the control CFU were generally due to significantly lower counts in the control during a sampling event or at a sampling site and not because the CFU of HPC-Ab increased. In contrast, HPC-Su were frequently found to represent from 20 to 80% of the total heterotrophic population, ratios significantly (p < 0.001) higher than all other HPC-Ab. This same trend in HPC-Su was found to exist throughout numerous processes sampled in a municipal wastewater treatment plant (Gao et al., 2012), also finding that while the total abundance of resistant bacteria were reduced in the effluent, that reduction was consistent with the reduction in total HPC-Ab populations. The similarities in HPC-Su observed in the downstream sites in this study may indicate contribution of resistance traits originating from WWTP effluent.
The occurrence of a significant increase in the concentrations of HPC-Te bacteria downstream of WWTPs in the total heterotrophic populations appears to contradict the findings of Sullivan and Karthikeyan (2012), research also conducted in the Carters Creek watershed. Sullivan and Karthikeyan (2012) found no effect of WWTP location on the prevalence of HPC-Te in surface water, but did see an increase in the abundance of tetracycline ARG. While molar concentrations of tetracycline used in both studies were similar, the discrepancy might be explained by differences in the cultivation media: Sullivan and Karthikeyan (2012) used nutrient-rich agar and this study used nutrient-limited R2A agar. Differences in cultivation media can significantly affect the counts of culturable HPC even from identical samples (Garcia-Armisen et al., 2013). Sullivan and Karthikeyan (2012) also found no seasonal variability in the occurrence of tetracycline resistant genes or bacteria.
While there was a significant increase in the abundance of HPC-Ab in the downstream sites, there was no significant increase when the concentrations were normalized to total heterotrophic bacteria with no antibiotic in the cultivation media. This indicates that while the total amount of resistant bacteria is increasing downstream through the watershed, it is increasing proportionately with the total population. This can be due to several plausible reasons. Viable bacteria from treated effluents may be a reason to increase total abundance without increasing normalized rates of resistance in the watershed. Suspended solids, dissolved organic carbon, and nutrients in WW effluents may facilitate the growth of pre-existing HPC-Ab downstream. Favorable growth conditions and increased total heterotrophic population proportionately increased the abundance of resistant bacteria.
Total heterotrophic population CFUs on control plates during antimicrobial studies can vary dramatically (3 orders of magnitude) (Pei et al., 2006), making it difficult to normalize results of antibiotic bacteria within the population. Additionally, the methodology used to capture HPC-Ab is known to capture both intrinsic and acquired resistance traits (Brooks et al., 2007); thus, these values may represent an over representation of the antibiotic resistant population.
The scope of this project was not limited to only cultivable bacteria, the study also evaluated the prevalence of ARG by qPCR methods. Both the cultivation-based approach and the qPCR approach reveal a major difference in the AMR bacterial numbers in the downstream vs. the upstream sites. The detected dominance of tetracycline resistant genes is not surprising as several other studies that focused on the distribution of ARG in urban waters, wastewaters, and WWTPs have previously found elevated levels of various tetracycline resistance genes (Szczepanowski et al., 2009; Gao et al., 2012; Brooks et al., 2014; Laht et al., 2014; Mao et al., 2015; Makowska et al., 2016; Ng et al., 2016). In addition, these results are in agreement with the culture-based results presented in the previous sections and an earlier study in the Carters Creek watershed where Sullivan et al. (2013) found an abundance of a variety tetracycline resistant genes and tetracycline resistant bacterial groups in several sampling sites within the watershed.
Class I integrons are genetic elements that are tangentially associated with the distribution of AMR and development of MDR among Gram-negative bacteria in a variety of environments (Cambray et al., 2010; Domingues et al., 2012; Gillings, 2014; Strugeon et al., 2016). The integron integrase gene (intI1) is the key fragment of the functional structure of Class I integrons as it is responsible for antibiotic resistance element-containing gene cassettes to be acquired, expressed and disseminated across bacterial species (Stokes and Hall, 1989; Collis and Hall, 1992; Domingues et al., 2012; Strugeon et al., 2016). Several previous studies have linked the presence of intI1 with prevalence of MDR in different environments (Leverstein-van Hall et al., 2002; Stalder et al., 2012; Brooks et al., 2014; Hultman et al., 2018). Furthermore, an elevated quantity of Class I integrase genes has been found to be present in studies that examined the occurrence of intI1 specifically in WWTPs (Makowska et al., 2016; McConell, 2017) and further associated with anthropogenic environmental influence (Gillings et al., 2015). Therefore, the considerably high abundance of the intI1 genes observed in all sites within this study is consistent with the observation of increased instances of AMR and MDR bacterial population in this site. It is important to note that we did not measure 16S rRNA while we measured ARG. Thus, we were not able to normalize our data to 16S rRNA which would provide for proportional context to the data set.
Results from this study find a considerably greater AMR and MDR in the downstream sites of the WWTPs carrying the wastewater effluents compared to the upstream sites. Downstream sites showed an increased resistance to the antibiotics ampicillin, ciprofloxacin, cefoperazone, sulfamethoxazole, and tetracycline, and were more often resistant to a higher number of different antibiotics. These effects were mirrored in the total HPC-Ab community, with a significant increase in the abundance of HPC-Am, -Cpr, -Su, and -Te bacteria in the surface water downstream of WWTP discharge points Quantitative PCR analysis of eight ARG in the samples also revealed similar results. In addition, we also noted in this study that the class 1 integron integrase gene, previously widely reasoned in similar studies to be associated with mobile genetic elements responsible for the movement of AMR and MDR between bacterial species in several environments, was detected at significantly high concentration in all sites at all times. We also detected a markedly higher abundance of most of the tested genes, including the Class I integrons, in the site immediately downstream of the WWTP.
Antibiotic resistance and increased rates of resistance can be attributed by several factors, including WWTP discharges as reported here. A more specific future investigation and more constrained system focusing on the inflows, outflows, and process components at the WWTPs would be beneficial in determining the extent of its contribution to resistance in the environment. Occurrence and persistence of antibiotic resistance and maintenance of the resistome are complex to describe in natural settings such as watersheds with varying hydrology, land use changes, and anthropogenic activities. Several watershed processes including overland runoff, stormwater outflow, and runoff from impermeable surfaces will affect the dissemination of antibiotic resistant bacteria. In watersheds, particularly in urban settings, WWTPs play a critical mitigation point for antibiotic resistance. Effective treatments should decrease further spread of resistance. It is important to note that only a particular fragment of potential ARG were tested in our analysis; additional insights could be obtained into these interpretations with a more extensive antibiotic “resistome” study. Further understanding of the interrelationships among ARB concentrations, ARG concentrations, antibiotic agents, microbial species, and environmental media will help modeling of antibiotic resistance transfer in terrestrial and aquatic environment.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
MM, TG, EL, JB, and RK: conceptualization. MM, TG, EL, and JB: methodology. MM and EL: formal analysis and data curation. MM: writing—original draft preparation. MM, TG, JB, and RK: writing—review and editing. TG: supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding provided by Texas A&M AgriLife Research along with graduate research assistantship support through the Water Management and Hydrological Science Program and Department of Soil and Crop Sciences at Texas A&M University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge Heidi Mjelde for laboratory assistance in this work and Dhanesh Yeganantham (Spatial Sciences Lab, Texas A&M University) for providing land use maps. MM previously presented a subset of these results at ASM South Central Branch Conference in 2019.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.657353/full#supplementary-material
Adams, C., Wang, Y., Loftin, K., and Meyer, M. (2002). Removal of antibiotics from surface and distilled water in conventional water treatment processes. J. Environ. Eng. 128, 253–260. doi: 10.1061/(asce)0733-9372(2002)128:3(253)
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
Amador, P. P., Fernandes, R. M., Prudêncio, M. C., Barreto, M. P., and Duarte, I. M. (2015). Antibiotic resistance in wastewater: occurrence and fate of Enterobacteriaceae producers of Class A and Class C β-lactamases. J. Environ. Sci. Health A 50, 26–39. doi: 10.1080/10934529.2015.964602
Aminov, R. I., Garrigues-Jeanjean, N., and Mackie, R. I. (2001). Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67, 22–32. doi: 10.1128/AEM.67.1.22-32.2001
Baquero, F., Martınez, J. M., and Canto, R. (2008). Antibiotics and antimicrobial resistance in water environments. Curr. Opin. Biotech. 19, 260–265.
Batt, A. L., Kim, S., and Aga, D. S. (2007). Comparison of the occurrence of antibiotics in four full-scale wastewater treatment plants with varying designs and operations. Chemosphere 68, 428–435. doi: 10.1016/j.chemosphere.2007.01.008
Bauer, A., Kirby, W., Sherris, J. C., and Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. American Journal of Clinical Pathology 45, 493–496.
Blaak, H. G., Lynch, R., Italiaander, R. A., Hamidjaja, F. M., and de Roda Husman, A. M. (2015). Multidrug-resistant and extended spectrum beta-lactamase-producing Escherichia coli in Dutch surface water and wastewater. PLoS One 10:e0127752. doi: 10.1371/journal.pone.0127752
Blasco, M. D., Esteve, C., and Alcaide, E. (2008). Multiresistant waterborne pathogens isolated from water reservoirs and cooling systems. J. App. Microbiol. 105, 469–475. doi: 10.1111/j.1365-2672.2008.03765.x
Blot, S., Vandijck, D., Lizy, C., Annemans, L., and Vogelaers, D. (2010). Estimating the length of hospitalization attributable to multidrug antibiotic resistance. Antimicro. Agric. Chemother. 54, 4046–4047. doi: 10.1128/aac.00689-10
Bouki, C., Venieri, D., and Diamadopoulos, E. (2013). Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: a review. Ecotoxicol. Environ. Saf. 91, 1–9. doi: 10.1016/j.ecoenv.2013.01.016
Bower, P. A., Scopel, C. O., Jensen, E. T., Depas, M. M., and McLellan, S. L. (2005). Detection of genetic markers of fecal indicator bacteria in Lake Michigan and determination of their relationship to Escherichia coli densities using standard microbiological methods. App. Environ. Microbiol. 71, 8305–8313. doi: 10.1128/aem.71.12.8305-8313.2005
Bradford, P. A. (2001). Extended-spectrum ß-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951. doi: 10.1128/cmr.14.4.933-951.2001
Brooks, J. P., Adeli, A., and McLaughlin, M. R. (2014). Microbial ecology, bacterial pathogens, and antibiotic resistant genes in swine manure wastewater as influenced by three swine management systems. Wat. Res. 57, 96–103. doi: 10.1016/j.watres.2014.03.017
Brooks, J. P., Maxwell, S. L., Rensing, C., Gerba, C. P., and Pepper, I. L. (2007). Occurrence of antibiotic-resistant bacteria and endotoxin associated with the land application of biosolids. Can. J Microbiol. 53, 616–622. doi: 10.1139/w07-021
Brooks, J. P., and McLaughlin, M. R. (2009). Antibiotic resistant bacterial profiles of anaerobic swine lagoon effluent. J. Environ. Qual. 38, 2431–2437. doi: 10.2134/jeq2008.0471
Bush, K., Courvalin, P., and Dantas, G. (2011). Tackling antibiotic resistance. Nat. Rev. Microbiol. 9, 894–896.
Chen, J., Michel, F. C., Sreevatsan, S., Morrison, M., and Yu, Z. (2010). Occurrence and persistence of erythromycin resistance genes (erm) and tetracycline resistance genes (tet) in waste treatment systems on swine farms. Microb. Ecol. 60, 479–486. doi: 10.1007/s00248-010-9634-5
Chopra, I., Schofield, C., and Everett, M. (2008). Treatment of health-care-associated infections caused by Gram-negative bacteria: a consensus statement. Lancet Infect. Dis. 8, 133–139. doi: 10.1016/s1473-3099(08)70018-5
Chu, B. T. T., Petrovich, M. L., Chaudhary, A., Wright, D., Murphy, B., Wells, G., et al. (2018). Metagenomics reveals the impact of wastewater treatment plants on the dispersal of microorganisms and genes in aquatic sediments. App. Environ. Microbiol. 84:e002168-17.
Collis, C. M., and Hall, R. M. (1992). Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J. Bacteriol. 174, 1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992
Courvalin, P. (1994). Transfer of antibiotic resistance genes between gram-positive and gram-negative bacteria. Antimicro. Agric. Chemother. 38, 1447–1451. doi: 10.1128/aac.38.7.1447
Czekalski, N., Berthold, T., Caucci, S., Egli, A., and Bürgmann, H. (2012). Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland. Front. Microbiol. 3:106. doi: 10.3389/fmicb.2012.00106
da Silva, M. F., Tiago, I., Veríssimo, A., Boaventura, R. A. R., Nunes, O. C., and Manaia, C. M. (2006). Antibiotic resistance of enterococci and related bacteria in an urban wastewater treatment plant. FEMS Microbiol. Ecol. 55, 322–329. doi: 10.1111/j.1574-6941.2005.00032.x
Davison, J. (1999). Genetic exchange between bacteria in the environment. Plasmid 42, 73–91. doi: 10.1006/plas.1999.1421
Domingues, S., da Silva, G. J., and Nielsen, K. M. (2012). Integrons: vehicles and pathways for horizontal dissemination in bacteria. Mob. Genet. Element 2, 211–223. doi: 10.4161/mge.22967
Dzidic, S., and Bedeković, V. (2003). Horizontal gene transfer-emerging multidrug resistance in hospital bacteria. Acta. Pharmacol. Sin. 24, 519–526.
Fan, W., Hamilton, T., Webster-Sesay, S., Nikolich, M. P., and Lindler, L. E. (2007). Multiplex real-time SYBR Green I PCR assay for detection of tetracycline efflux genes of Gram-negative bacteria. Mol. cellular probes 21, 245–256. doi: 10.1016/j.mcp.2006.12.005
Gao, P., Munir, M., and Xagoraraki, I. (2012). Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total. Environ. 421, 173–183. doi: 10.1016/j.scitotenv.2012.01.061
Garcia-Armisen, T., Anzil, A., Cornelis, P., Chevreuil, M., and Servais, P. (2013). Identification of antimicrobial resistant bacteria in rivers: Insights into the cultivation bias. Wat. Res. 47, 4938–4947. doi: 10.1016/j.watres.2013.05.036
Gillings, M. R. (2014). Integrons: past, present, and future. Microbiol Mol. Biol. Rev. 78, 257–277. doi: 10.1128/mmbr.00056-13
Gillings, M. R., Gaze, W. H., Pruden, A., Smalla, K., Tiedje, J. M., and Zhu, Y. G. (2015). Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9, 1269–1279. doi: 10.1038/ismej.2014.226
Goossens, H., Ferech, M., Vander Stichele, R., Elseviers, M., and ESAC Project Group (2005). Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365, 579–587. doi: 10.1016/s0140-6736(05)17907-0
Gopal Rao, G. (2012). Risk factors for the spread of antibiotic-resistant bacteria. Drugs 55, 323–330. doi: 10.2165/00003495-199855030-00001
Hamilton, K. A., Garner, E., Joshi, S., Ahmed, W., Ashbolt, N., Medema, G., et al. (2020). Antimicrobial-resistant microorganisms and their genetic determinants in stormwater: a systematic review. Curr. Opin. Environ. Sci. Health 16, 101–112. doi: 10.1016/j.coesh.2020.02.012
Hardwick, S. A., Stokes, H. W., Findlay, S., Taylor, M., and Gillings, M. R. (2008). Quantification of class 1 integron abundance in natural environments using real-time quantitative PCR. FEMS Microbiol. Lett. 278, 207–212. doi: 10.1111/j.1574-6968.2007.00992.x
Hicks, L. A., Bartoces, M. G., Roberts, R. M., Suda, K. J., Hunkler, R. J., Taylor, T. H., et al. (2015). US outpatient antibiotic prescribing variation according to geography, patient population, and provider specialty in 2011. Clin. Infect Dis. 60, 1308–1316.
Hsu, J. T., Chen, C. Y., Young, C. W., Chao, W. L., Li, M. H., Liu, Y. H., et al. (2014). Prevalence of sulfonamide-resistant bacteria, resistance genes and integron-associated horizontal gene transfer in natural water bodies and soils adjacent to a swine feedlot in northern Taiwan. J. Haz. Mat. 277, 34–40. doi: 10.1016/j.jhazmat.2014.02.016
Huijbers, P. M., Blaak, H., de Jong, M. C., Graat, E. A., Vandenbroucke-Grauls, C. M., and de Roda Husman, A. M. (2015). Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ. Sci. Technol. 49, 11993–12004. doi: 10.1021/acs.est.5b02566
Huijbers, P. M., Flach, C. F., and Larsson, D. J. (2019). A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ. Int. 130:104880. doi: 10.1016/j.envint.2019.05.074
Hultman, J., Tamminen, M., Pärnänen, K., Cairns, J., Karkman, A., and Virta, M. (2018). Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol. Ecol. 94:fiy038.
Iwane, T., Urase, T., and Yamamoto, K. (2001). Possible impact of treated wastewater discharge on incidence of antibiotic resistant bacteria in river water. Wat. Sci. Technol. 43, 91–99. doi: 10.2166/wst.2001.0077
Janezic, K. J., Ferry, B., and Hendricks, E. W. (2013). Phenotypic and genotypic characterization of Escherichia coli isolated from untreated surface waters. Open Microbiol. J. 7, 9–19. doi: 10.2174/1874285801307010009
Jindal, A., Kocherginskaya, S., Mehboob, A., Robert, M., Mackie, R. I., Raskin, L., et al. (2006). Antimicrobial use and resistance in swine waste treatment systems. Appl. Environ. Microbiol. 72, 7813–7820. doi: 10.1128/aem.01087-06
Jones, K. E., Patel, N. G., Levy, M. A., Storeygard, A., Balk, D., Gittleman, J. L., et al. (2008). Global trends in emerging infectious diseases. Nature 451, 990–993.
Kemper, N. (2008). Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indicat. 8, 1–13. doi: 10.1016/j.ecolind.2007.06.002
Korzeniewska, E., Korzeniewska, A., and Harnisz, M. (2013). Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol. Environ. Saf. 91, 96–102. doi: 10.1016/j.ecoenv.2013.01.014
Kümmerer, K. (2009). Antibiotics in the aquatic environment – A review – Part II. Chemosphere 75, 435–441. doi: 10.1016/j.chemosphere.2008.12.006
Lachmayr, K. L., Kerkhof, L. J., DiRienzo, A. G., Cavanaugh, C. M., and Ford, T. E. (2009). Quantifying nonspecific TEM β-lactamase (blaTEM) genes in a wastewater stream. Appl. Environ. Microbiol. 75, 203–211. doi: 10.1128/AEM.01254-08
Laht, M., Karkman, A., Voolaid, V., Ritz, C., Tenson, T., Virta, M., et al. (2014). Abundances of tetracycline, sulphonamide and beta-lactam antibiotic resistance genes in conventional wastewater treatment plants (WWTPs) with different waste load. PLoS One 9:e103705. doi: 10.1371/journal.pone.0103705
Leverstein-van Hall, M. A., Paauw, A., Box, A. T. A., Blok, H. E. M., Verhoef, J., and Fluit, A. C. (2002). Presence of integron-associated resistance in the community is widespread and contributes to multidrug resistance in the hospital. J. Clin. Micobiol. 40, 3038–e3040.
Levy, S. B., and Marshall, B. (2004). Antibacterial resistance worldwide: causes, challenges, and responses. Nat. Med. 10, 122–129.
Lim, S. M., and Webb, S. A. R. (2005). Nosocomial bacterial infections in intensive care units. I: organisms and mechanisms of antibiotic resistance. Anaesthesia 60, 887–902. doi: 10.1111/j.1365-2044.2005.04220.x
Lye, D. C., Earnest, A., Ling, M. L., Lee, T. E., Yong, H. C., Fisher, D. A., et al. (2012). The impact of multidrug resistance in healthcare-associated and nosocomial Gram-negative bacteraemia on mortality and length of stay: cohort study. Clin. Microbiol. Infect 18, 502–508. doi: 10.1111/j.1469-0691.2011.03606.x
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Makowska, N., Koczura, R., and Mokracka, J. (2016). Class 1 integrase, sulfonamide and tetracycline resistance genes in wastewater treatment plant and surface water. Chemosphere 144, 1665–1673. doi: 10.1016/j.chemosphere.2015.10.044
Mao, D., Yu, S., Rysz, M., Luo, Y., Yang, F., Li, F., et al. (2015). Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Wat. Res. 85, 458–466. doi: 10.1016/j.watres.2015.09.010
McConell, M. (2017). Abundance of Antibiotic Resistance Genes in Two Municipal Wastewater Treatment Plants and Receiving Water in Atlantic Canada. Ph. D.thesis. Halifax: Dalhousie University.
Naqvi, S., Kiran, U., Ali, M. I., Jamal, A., Hameed, A., Ahmed, S., et al. (2013). Combined efficacy of biologically synthesized silver nanoparticles and different antibiotics against multidrug-resistant bacteria. Int. J. Nanomed. 8, 3187–3195. doi: 10.2147/ijn.s49284
Ng, C., Tan, B., Jiang, X., Gu, X., Chen, H., Schmitz, B. W., et al. (2016). Metagenomic and resistome analysis of a full-scale municipal wastewater treatment plant in Singapore containing membrane bioreactors. Nat. Microbiol. 161, 1–7.
Nicolau, D. P., Carmeli, Y., Crank, C. W., Goff, D. A., Graber, C. J., Lima, A. L. L., et al. (2012). Carbapenem stewardship: does ertapenem affect Pseudomonas susceptibility to other carbapenems? A review of the evidence. Int. J. Antimicrob. Agric. 39, 11–15. doi: 10.1016/j.ijantimicag.2011.08.018
Nikaido, H., and Pagès, J. M. (2012). Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS. Microbiol. Rev. 36, 340–363. doi: 10.1111/j.1574-6976.2011.00290.x
Pei, R., Kim, S. C., Carlson, K. H., and Pruden, A. (2006). Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Wat. Res. 40, 2427–2435. doi: 10.1016/j.watres.2006.04.017
Pepper, I. L., and Gentry, T. J. (2015). “Chapter 2 - Microorganisms found in the environment,” in Environmental Microbiology, 3rd Edn, eds I. Pepper, C. Gerba, and T. Gentry (San Diego: Academic Press), 9–36. doi: 10.1016/b978-0-12-394626-3.00002-8
Pruden, A. (2014). Balancing water sustainability and public health goals in the face of growing concerns about antibiotic resistance. Environ. Sci. Technol. 48, 5–14. doi: 10.1021/es403883p
Rajić, A., Reid-Smith, R., Deckert, A. E., Dewey, C. E., and McEwen, S. A. (2006). Reported antibiotic use in 90 swine farms in Alberta. Can. Vet. J. 47, 446–452.
Rizzo, L., Mania, C., Merlin, C., Schwartz, T., Dagot, C., Ploy, M. C., et al. (2013). Urban wastewater treatment plant as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci. Total Environ. 447, 345–360. doi: 10.1016/j.scitotenv.2013.01.032
Sabet, N. S., Subramaniam, G., Navaratnam, P., and Sekaran, S. D. (2007). Detection of methicillin-and aminoglycoside-resistant genes and simultaneous identification of S. aureus using triplex real-time PCR Taqman assay. J. Microbiol. Methods 68, 157–162. doi: 10.1016/j.mimet.2006.07.008
Sayah, R. S., Kaneene, J. B., Johnson, Y., and Miller, R. S. (2005). Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl. Envron. Microbiol. 71, 1394–1404. doi: 10.1128/aem.71.3.1394-1404.2005
Schwartz, T., Kohnen, W., Jansen, B., and Obst, U. (2003). Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 43, 325–335. doi: 10.1111/j.1574-6941.2003.tb01073.x
Shapiro, D. J., Hicks, L. A., Pavia, A. T., and Hersh, A. L. (2013). Antibiotic prescribing for adults in ambulatory care in the USA, 2007–09. J. Antimicrob. Chemother. 69, 234–240. doi: 10.1093/jac/dkt301
Silva, J., Castillo, G., Callejas, L., López, H., and Olmos, J. (2006). Frequency of transferable multiple antibiotic resistance amongst coliform bacteria isolated from a treated sewage effluent in Antofagasta, Chile. Electron. J. Biotechnol. 9:8.
Spellberg, B., Guidos, R., Gilbert, D., Bradley, J., Boucher, H. W., Scheld, W. M., et al. (2008). The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 46, 155–164. doi: 10.1086/524891
Stalder, T., Barraud, O., Casellas, M., Dagot, C., and Ploy, M. C. (2012). Integron involvement in environmental spread of antibiotic resistance. Front. Microbiol. 3:119. doi: 10.3389/fmicb.2012.00119
Stokes, H. W., and Hall, R. M. (1989). A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3, 1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x
Strugeon, E., Tilloy, V., Ploy, M. C., and Da Re, S. (2016). The stringent response promotes antibiotic resistance dissemination by regulating integron integrase expression in biofilms. mBio 7:e0868-16.
Sullivan, B. A., Gentry, T. J., and Karthikeyan, R. (2013). Characterization of tetracycline-resistant bacteria in an urbanizing subtropical watershed. J. Appl. Microbiol. 115, 774–785. doi: 10.1111/jam.12283
Sullivan, B. A., and Karthikeyan, R. (2012). Occurrence and prevalence of tetracycline resistant bacteria in a rapidly urbanizing subtropical watershed. J. Nat. Environ. Sci. 2, 25–31.
Szczepanowski, R., Linke, B., Krahn, I., Gartemann, K. H., Gützkow, T., Eichler, W., et al. (2009). Detection of 140 clinically relevant antibioticresistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 155, 2306–2319. doi: 10.1099/mic.0.028233-0
USEPA (2005). Method 1603: Escherichia coli (E. coli) in Water by Membrane Filtration Using Modified Membrane-Thermotolerant Escherichia coli Agar (Modified mTEC). Washington, DC: Environmental Protection Agency.
Van Boeckel, T. P., Gandra, S., Ashok, A., Caudron, Q., Grenfell, B. T., Levin, S. A., et al. (2014). Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lanc. Infect. Dis. 14, 742–750. doi: 10.1016/s1473-3099(14)70780-7
Verlicchi, P., Al Aukidy, M., and Zambello, E. (2015). What have we learned from worldwide experiences on the management and treatment of hospital effluent? — An overview and a discussion on perspectives. Sci. Total Environ. 514, 467–491. doi: 10.1016/j.scitotenv.2015.02.020
Worthington, R. J., and Melander, C. (2013). Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol. 31, 177–184. doi: 10.1016/j.tibtech.2012.12.006
Wright, G. D. (2010). Antibiotic resistance in the environment: a link to the clinic? Curr. Opin. Microbiol. 13, 589–594. doi: 10.1016/j.mib.2010.08.005
Zhang, X., Zhang, T., and Fang, H. H. P. (2009). Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 82, 397–414.
Zhang, Y., Marrs, C. F., Simon, C., and Xi, C. (2009). Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci. Total Environ. 407, 3702–3706. doi: 10.1016/j.scitotenv.2009.02.013
Keywords: antimicrobial resistance, antibiotic resistance genes, wastewater treatment plant, antibiotic resistant bacteria, multidrug resistance
Citation: Mukherjee M, Laird E, Gentry TJ, Brooks JP and Karthikeyan R (2021) Increased Antimicrobial and Multidrug Resistance Downstream of Wastewater Treatment Plants in an Urban Watershed. Front. Microbiol. 12:657353. doi: 10.3389/fmicb.2021.657353
Received: 22 January 2021; Accepted: 27 April 2021;
Published: 24 May 2021.
Edited by:
Christophe Merlin, Université de Lorraine, FranceReviewed by:
Erica Donner, University of South Australia, AustraliaCopyright © 2021 Mukherjee, Laird, Gentry, Brooks and Karthikeyan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maitreyee Mukherjee, TWFpdHJleWVlLk11a2hlcmplZUB1c20uZWR1; bWFpdHJleWVlMjVAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.