- 1Division of Plant Pathology, ICAR-Indian Agricultural Research Institute, Pusa, New Delhi, India
- 2International Rice Research Institute, Pusa, New Delhi, India
Trichoderma biopriming enhances rice growth in drought-stressed soils by triggering various plant metabolic pathways related to antioxidative defense, secondary metabolites, and hormonal upregulation. In the present study, transcriptomic analysis of rice cultivar IR64 bioprimed with Trichoderma harzianum under drought stress was carried out in comparison with drought-stressed samples using next-generation sequencing techniques. Out of the 2,506 significant (p < 0.05) differentially expressed genes (DEGs), 337 (15%) were exclusively expressed in drought-stressed plants, 382 (15%) were expressed in T. harzianum-treated drought-stressed plants, and 1,787 (70%) were commonly expressed. Furthermore, comparative analysis of upregulated and downregulated genes under stressed conditions showed that 1,053 genes (42%) were upregulated and 733 genes (29%) were downregulated in T. harzianum-treated drought-stressed rice plants. The genes exclusively expressed in T. harzianum-treated drought-stressed plants were mostly photosynthetic and antioxidative such as plastocyanin, small chain of Rubisco, PSI subunit Q, PSII subunit PSBY, osmoproteins, proline-rich protein, aquaporins, stress-enhanced proteins, and chaperonins. The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis states that the most enriched pathways were metabolic (38%) followed by pathways involved in the synthesis of secondary metabolites (25%), carbon metabolism (6%), phenyl propanoid (7%), and glutathione metabolism (3%). Some of the genes were selected for validation using real-time PCR which showed consistent expression as RNA-Seq data. Furthermore, to establish host–T. harzianum interaction, transcriptome analysis of Trichoderma was also carried out. The Gene Ontology (GO) analysis of T. harzianum transcriptome suggested that the annotated genes are functionally related to carbohydrate binding module, glycoside hydrolase, GMC oxidoreductase, and trehalase and were mainly upregulated, playing an important role in establishing the mycelia colonization of rice roots and its growth. Overall, it can be concluded that T. harzianum biopriming delays drought stress in rice cultivars by a multitude of molecular programming.
Introduction
Rice is an important staple crop, feeding more than half of the world’s population. Due to the high demand of water for maintenance of its growth and productivity, the yield of the crop is severely hampered by the limited availability of water.
Drought is one of the major constraints that affect crop productivity up to 9–10% globally due to its adverse effects on plant growth and development (Lesk et al., 2016). Hence, to combat drought stress and bring an environment-friendly solution to the problem, biological management practices have also been explored. Among fungi, Trichoderma harzianum association is also known for its positive effects in combating abiotic and biotic stresses (Contreras-Cornejo et al., 2013). T. harzianum is an antagonistic mycoparasite that colonizes the roots of both monocots and dicots (Schuster and Schmoll, 2010). Additionally, T. harzianum biopriming also leads to enhanced nutrient uptake and increased water-holding capacity (Doni et al., 2014), which ultimately results in better growth and development in drought-stressed soils (Bae et al., 2009). The Trichoderma association helps plants in sustaining drought stress by increasing the expression of antioxidative enzymes, secondary metabolites, and plant hormones (Pandey et al., 2016; Alwhibi et al., 2017). Bashyal et al. (2020) also reported that seed priming with T. harzianum delayed drought stress by 3–5 days.
Trichoderma-mediated improved growth is also due to alterations in plant physiological parameters related to anabolic pathways such as photosynthetic rate, stomatal conductance, and transpiration rate (Doni et al., 2014). The root colonization of Trichoderma is often associated with enhanced photosynthesis as evident through various studies (Doni et al., 2014; Harman et al., 2019). Previous studies suggested that Trichoderma colonization improves the plant biomass and root growth by increasing auxin biosynthesis (Contreras-Cornejo et al., 2009). A few transcriptomic studies have also been carried out to correlate the genomic basis of T. harzianum-induced physiological and biochemical changes in drought tolerance (Huang et al., 2014; Doni et al., 2019). A microarray study showed that genes related to oxidative and osmotic stress are induced upon Trichoderma root colonization to eliminate salinity stress (Brotman et al., 2013). Transcriptomic studies to understand molecular reprogramming in tomato plants treated with T. harzianum revealed that epigenetic responses and alternative splicing play a crucial role in plant growth and defense. Furthermore, a temporal regulation of ethylene/indole acetic acid and reactive oxygen species-mediated upregulation of defense mechanisms were also reported for the maintenance of growth and stress tolerance (Palma et al., 2019).
Previous transcriptome conducted by Ereful et al. (2020) on rice cultivars suggested the upregulation of genes of secondary metabolites interacting with reactive oxygen species and enhancement of antioxidative genes. However, there is a complete lack of study related to transcriptome profiling in the association of T. harzianum with rice cultivars in drought stress modulation. Hence, in the present study, we compared transcriptomic alterations in drought-stressed T. harzianum-inoculated rice plants using next-generation sequencing techniques. The results of this study may contribute to elucidating the mechanisms involved in the rice–microbe–drought interactions and to identify genes that are putatively responsible for the T. harzianum-mediated drought tolerance.
Materials and Methods
Seed Material and Source
Two contrasting rice cultivars were used in this experiment. A drought-resistant (Sahbhagi Dhan) and a drought-susceptible (IR64) genotype of rice and a biocontrol agent T. harzianum 1 (TH1) isolate were provided by the International Rice Research Institute, New Delhi, India. The TH1 strain was selected on the basis of our previous study (Bashyal et al., 2020).
Inoculation and Plant Growth Conditions
The seeds were sown after surface sterilization for 1 min with 1% (v/v) sodium hypochlorite followed by washing three times with sterilized distilled water. The seeds were then bioprimed with powder formulation of T. harzianum at a concentration of 10 g/kg supplied with 0.02% (v/v) Tween 20 as surfactant. The formulation was mixed thoroughly to provide uniform coating and kept in a moist chamber at room temperature (25°C) for 24 h. The control surface-sterilized seeds were mixed with Tween 20 [0.02% (v/v) Tween 20] only and kept in the moist chamber. The seeds were then planted in earthen pots containing sterilized sand:soil (3:1) mixture and grown under day/night temperatures of 30–35°C/18°C and a relative humidity of 80/90%. After 40 days of plant growth, the potted plants were categorized into three groups: one group was used as control with normal irrigation (Sahbhagi Dhan, IR64), the second was drought-stressed (Sahbhagi Dhan drought-stressed and IR64 drought-stressed), and the third was drought- and biocontrol-treated (Sahbhagi Dhan, T. harzianum-treated + drought-stressed and IR64, T. harzianum-treated + drought-stressed). The drought stress was given as described previously by Bashyal et al. (2020). Briefly, moisture was maintained in the potted plants by applying 100 ml of water per pot every alternate day until plants attained the age of 40 days, and at this point, drought treatments were given by altering the water cycle. Watering was stopped for the subsequent days for each drought treatment (4, 7, and 10 days drought stress), while the control seedlings were watered every alternate day. The experiment was conducted in randomized block design with three replications per treatment and 10 seeds per pot. Overall, 90 plants for each cultivar (30 controls, 30 drought-stressed, and 30 T. harzianum-treated + drought-stressed) were observed for one time interval (i.e., 4 days drought stress). The plant samples were harvested after 4, 7, and 10 days of drought treatment and stored immediately using liquid nitrogen at −80°C for transcriptome analysis (Bashyal et al., 2016).
Total RNA Extraction
Total RNA was isolated from the frozen plant samples (IR64, drought-stressed; IR64, T. harzianum-treated + drought-stressed) using TRIzol (TRI reagent, Molecular Research Center, OH, United States) following the manufacturer’s instructions. RNA isolation was done in two replicates from the pooled samples (4, 7, and 10 days). Briefly, a 100-mg seedling of rice was powdered using liquid nitrogen, homogenized in TRIzol, and incubated for 5 min at room temperature. Then, 200 μl of chloroform was added to it and incubated for 10 min at room temperature after shaking vigorously. Samples were centrifuged (Eppendorf AG, Heidelberg, Germany) at 12,000 rpm for 15 min and the upper aqueous phase was separated. Five hundred microliters of isopropanol was added to it and incubated at room temperature for 5 min. RNA was pelleted by centrifuging at 10,000 rpm for 10 min and purified by washing twice with 75–80% (v/v) alcohol. The RNA was dissolved in 40–50 μl of nuclease-free water and kept on a water bath set at 55–60°C. The quality and quantity of the isolated RNAs were checked on denatured RNA agarose gel and NanoDrop (Thermo Fisher Scientific, Wilmington, DE, United States) reading, respectively.
RNA-Seq Library Construction and Sequencing
RNA-Seq paired-end sequencing libraries were prepared from the isolated total RNA using Illumina TruSeq stranded mRNA sample preparation kit (Illumina, San Diego, CA, United States). For this, mRNA was enriched from the total RNA using poly-T attached magnetic beads, followed by enzymatic fragmentation and first strand cDNA was synthesized. The first strand cDNA was then synthesized to second strand using second strand mix and Act-D mix to facilitate RNA-dependent synthesis. The double strand cDNA samples were then purified using Ampure XP beads (New England Biolabs, Ipswich, MA, United States) followed by A-tailing, adapter ligation, and then enriched by a limited number of PCR cycles.
Transcriptome Sequencing, Quality Control, and Mapping
Sequencing was done in a single HiSeq 4000 lane using 150 bp paired-end chemistry. The library preparation and sequencing was done by commercial service providers (NxGenBio Life Sciences, New Delhi, India). The barcoded gDNA libraries were pooled in equal ratios and used for 2 × 150-bp paired-end sequencing on a single lane of the Illumina HiSeq 4000. Illumina clusters were generated and were loaded onto Illumina Flow Cell on Illumina HiSeq 4000 instrument and sequencing was carried out using 2 × 150-bp paired-end chemistry. After sequencing, the samples were demultiplexed and the indexed adapter sequences were trimmed using the CASAVA v1.8.2 software (Illumina Inc.).
Data Analysis
Bioinformatics Analysis
The quality of raw reads was checked by FastQC (version 0.11.8). The high-quality reads were mapped using Minimap (version 2.17) at default parameters against both the reference genome Oryza sativa (NCBI acc. no. PRJNA13141) and T. harzianum 1 (NCBI acc. nos. PRJNA453596, PRJNA207867).
Differential Expression Analysis
The number of reads mapped to genes was calculated using SAMtools (version: 0.1.19). Differential analysis of all possible combinations was done using DESeq (version 1) R package, used to analyze count data from high-throughput sequencing assays such as RNA-Seq and test for differential expression. The functional annotation was done using UniProt and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Expression plots (volcano plot) were made using In House R scripts and heat map using the MeV software.
Gene Expression Analysis by Quantitative Real-Time PCR
Quantitative real-time reverse transcriptase PCR (qRT-PCR) was done to validate the results of the Illumina sequencing experiment. For this, some of the significant candidate genes (Supplementary Table 1) contributing to drought tolerance and exclusively expressed in T. harzianum-treated and drought-stressed rice plants were selected and primers were designed (PrimerQuest tool, Integrated DNA Technologies). All the qRT-PCR experiments were conducted in three biological replicates with three technical replicates. For gene expression analysis, first RNA was extracted as stated above. Then, 3 μg of total RNA was used for cDNA synthesis using Verso cDNA synthesis kit (Thermo Scientific, Wilmington, DE, United States) according to the manufacturer’s protocol. For this, 3–5 μg of total RNA was taken in a microfuge tube. To this, nuclease-free water was added to make up the volume to 9 μl followed by the addition of different reagents in an indicated order as follows: 2 μl random hexamer, 1 μl of an RT enhancer, 4 μl of a 5× cDNA buffer, 2 μl of a 10-mM dNTP mix, and 1 μl of M-MuLV reverse transcriptase; then, it was mixed gently and spun slowly at 1,000 rpm for 15 s. The tubes were incubated at 40°C for 60 min. The reaction was terminated by heating at 70°C for 15 min.
The PCR reaction mix was prepared using primer pairs specific to rice (Supplementary Table 1), and GAPDH was used as internal control (Kumar et al., 2018). The reaction comprises 1 μl of cDNA mixed with 10 μl of SYBR Green PCR master mix (Qiagen GmbH, Hilden, Germany), 5 pmol of a forward primer, and 5 pmol of a reverse primer in a final volume of 20 μl. Template controls were analyzed for all genes. PCR was performed using a MiniOpticon real-time PCR system (Bio-Rad, Hercules, CA, United States) with the following conditions: an initial activation step at 94°C for 4 min, denaturation at 94°C for 15 s, annealing at 58°C for 30 s, and extension at 70°C for 30 s. Melt curve analysis of the PCR product was carried out at 72°C for 1 min and ramped from 75 to 90°C with an increase by 1°C every 5 s. The specificity of the reaction was confirmed by melt curve analysis and gel electrophoresis. Relative gene expressions were calculated in terms of fold changes using the ΔCt method [Fold change = 2–Δ(ΔCt), ΔCttreated = Ct(target) − Ct(normalizer), ΔCtcontrol = Ct(target) − Ct(normalizer), Δ(ΔCt) = ΔCt(treated) – ΔCt(control)]. The results are presented as arithmetic means and standard deviations of the replicates.
Results
Plant Growth
The comparative results of the two cultivars (Sahbhagi Dhan and IR64) showed that T. harzianum treatment has a significant difference in maintaining plant growth under drought conditions. The cultivar IR64 performed better when compared with Sahbhagi Dhan which was evident from plant growth which was assessed by measuring root and shoot length at 4, 7, and 10 days (Supplementary Figure 1). The root growth showed marginal difference on 4 and 7 days of growth; however, a notable difference was observed in shoot growth in T. harzianum-treated drought-stressed IR64 rice plants when compared with drought-stressed IR64. Sahbhagi Dhan recorded a 23% decrease in shoot length, whereas in IR64, a 6% decrease was recorded on 4 days of drought when compared with their respective controls. In fact, on 7 days of drought stress, Sahbhagi Dhan recorded a 60% less growth, whereas IR64 recorded 11% less growth in shoot length in comparison with their controls. Our previous study also indicated that Trichoderma biopriming delays drought stress by 3–5 days in IR64 and Sahbhagi Dhan (Bashyal et al., 2020). Overall, out of the two contrasting cultivars, i.e., Sahbhagi Dhan and IR64, the T. harzianum-inoculated IR64 cultivar performed better under drought stress at 4–7 days of growth (Supplementary Figure 2). Hence, cultivar IR64 was selected for the transcriptome analysis under T. harzianum-treated drought stress condition.
Sequencing Statistics
The libraries of a total of four samples (IR64 drought; IR64 drought + T. harzianum in duplicates) were analyzed from Illumina NextSeq 500 platform. Approximately 20 million for drought-stressed samples and 21–22 million high-quality reads for T. harzianum-treated drought-stressed rice cultivar were obtained (Table 1). The alignment results showed that 86–95% of clean reads were mapped on the reference genome from all the four samples. The assembly of mapped reads resulted in the identification of a total of 33,691 differentially expressed genes (DEGs).
Gene Profile and Differential Expression Study of Rice
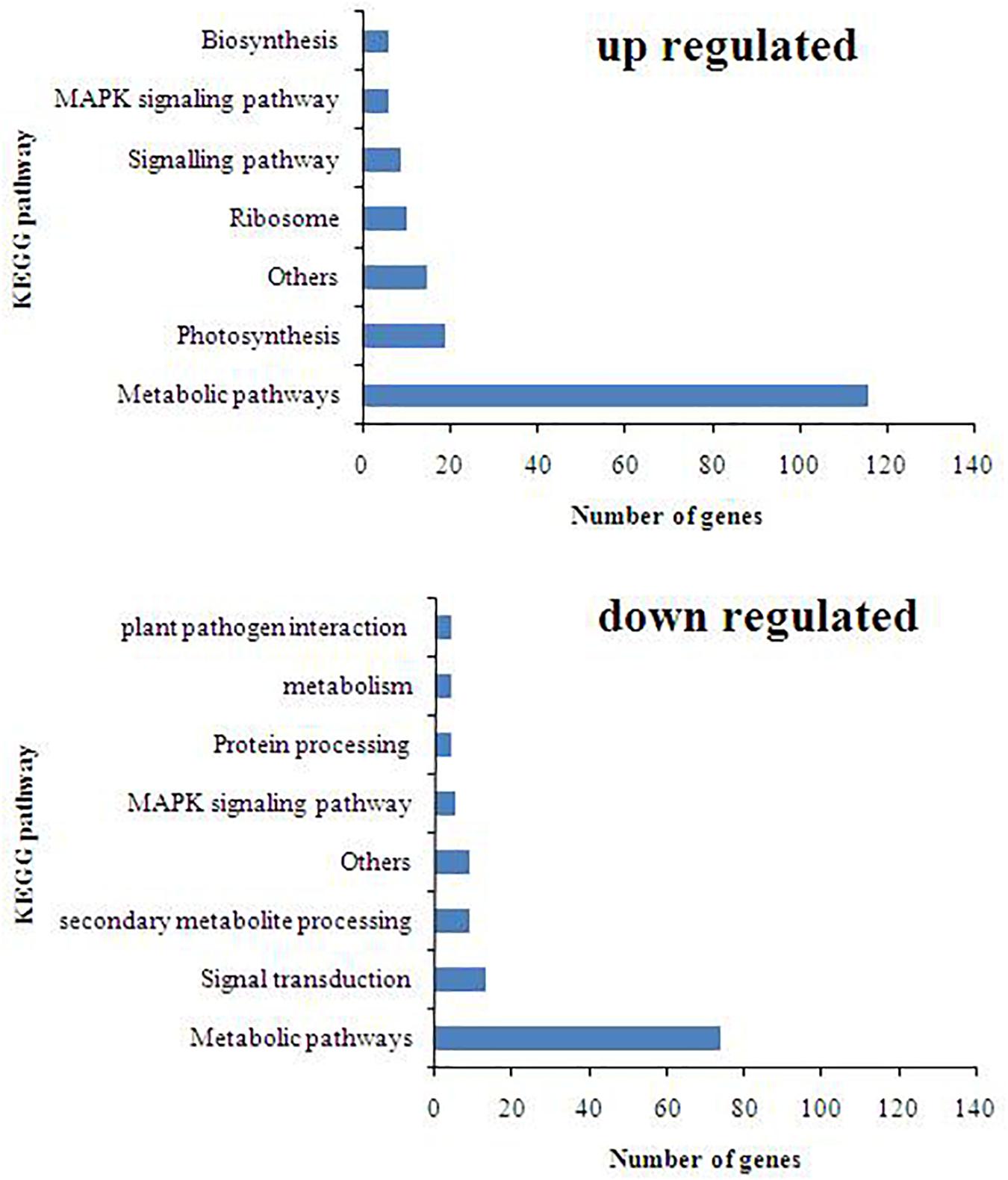
Out of the total of 33,691 DEGs, the highly upregulated and downregulated 10,435 DEGs were considered for further studies excluding the genes lying in the range of log fold change in (−2) to (+2). Volcano plot analysis of DEGs showed remarkable differences in gene distribution patterns, which were aptly delimited in the T. harzianum-treated drought-stressed IR64 cultivar when compared with drought-stressed (Figure 1A). A total of 2,424 genes, i.e., 23%, were exclusively expressed in T. harzianum-treated plants, whereas 2,256 (22%) were expressed in drought-stressed conditions and 5,754 (55%) genes were commonly expressed in both treatments (Figure 1B). The study further stressed on 2,506 significant genes (p < 0.05) out of the highly expressed 10,435 genes; 337 (15%) were exclusively expressed in drought-stressed plants, 382 (15%) were expressed in T. harzianum-treated drought-stressed plants, and 1,787 (70%) were commonly expressed in both. Furthermore, comparative analysis of upregulated and downregulated genes under stressed conditions was also observed in T. harzianum+ drought vs drought-treated rice. The data showed 1,436 genes (57%) were upregulated and 1,070 genes (42%) were downregulated in T. harzianum-treated drought-stressed rice plants when compared with only drought-stressed plants.
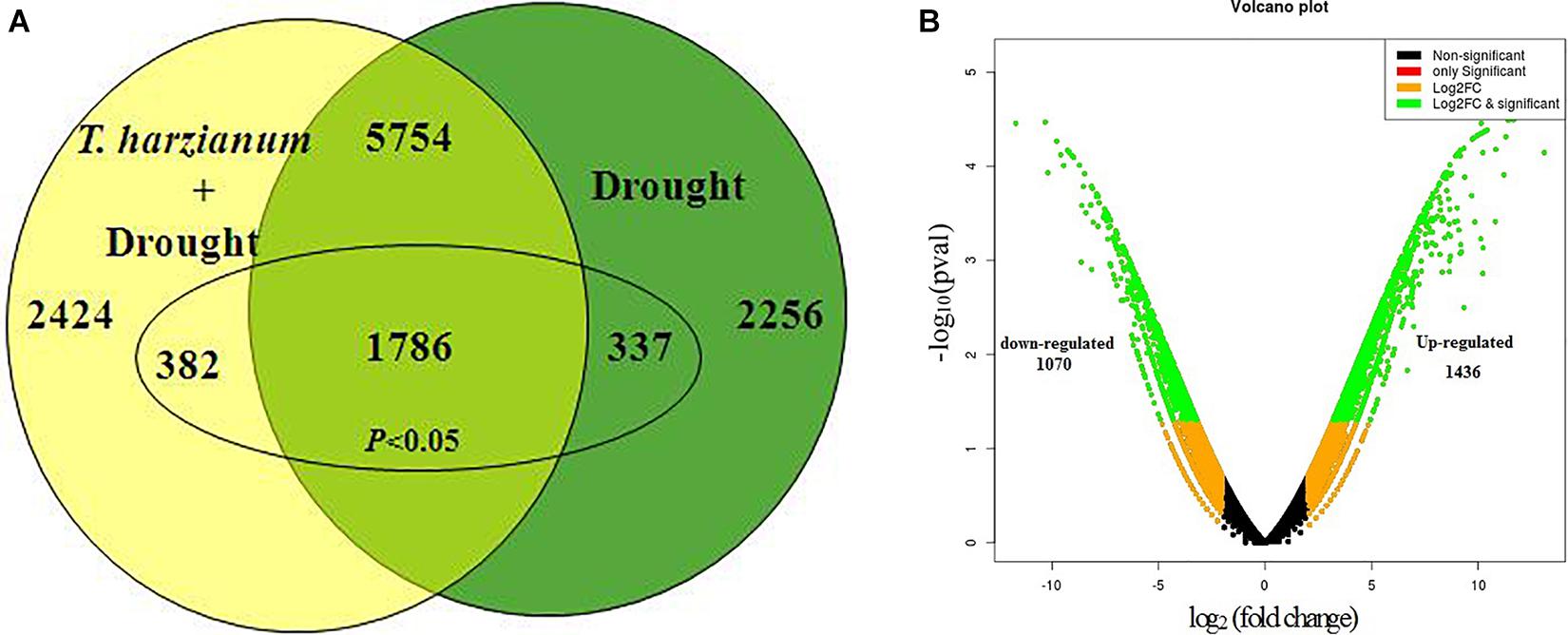
Figure 1. (A) Venn diagram showing the unique (non-overlapping region) and common expressed genes (overlapping region) obtained when drought-stressed rice is compared with T. harzianum-treated + drought-stressed rice. (B) Volcano plot representing significant and non-significant differentially expressed gene (DEG)-based p values. The green dot represents the significant DEGs.
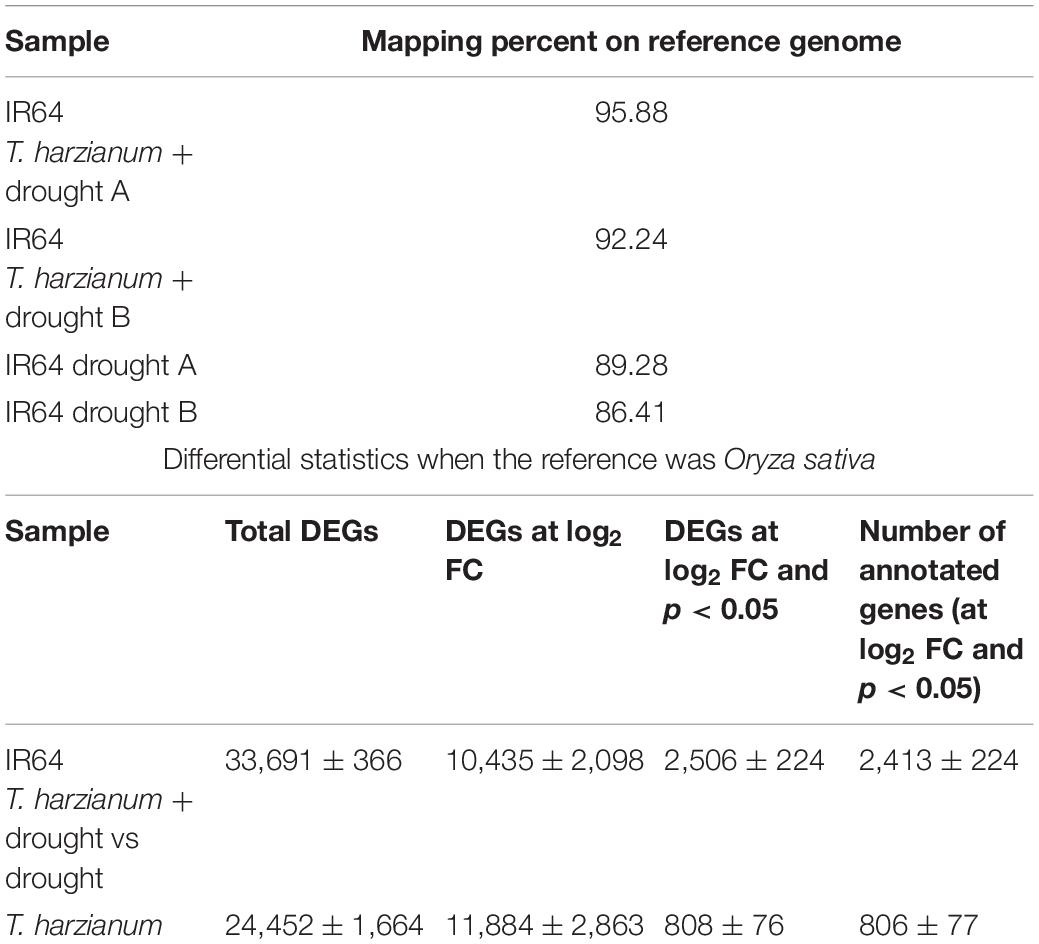
Based on the transcriptome analysis of DEGs under drought-stressed and T. harzianum-bioprimed conditions, the genes were classified into 21 broad categories (Figure 2). Heat map and hierarchical cluster categorization of DEGs were also generated to represent the global view of gene expression patterns and also depict their dynamic differences in T. harzianum-treated, drought-stressed vs drought-stressed (Figure 3). Higher percentages of genes were related to photosynthetic enzymes of both light reaction and dark reaction enzymes and osmotic homeostasis enzymes (Figure 3). The genes exclusively expressed in T. harzianum-treated plants were mostly photosynthetic such as plastocyanin, small chain of Rubisco, PSI subunit Q, and PSII subunit PSBY. Other antioxidative genes included osmoproteins, proline-rich protein, aquaporins, stress-enhanced proteins, chaperonins, peroxidases, and peroxiredoxins.
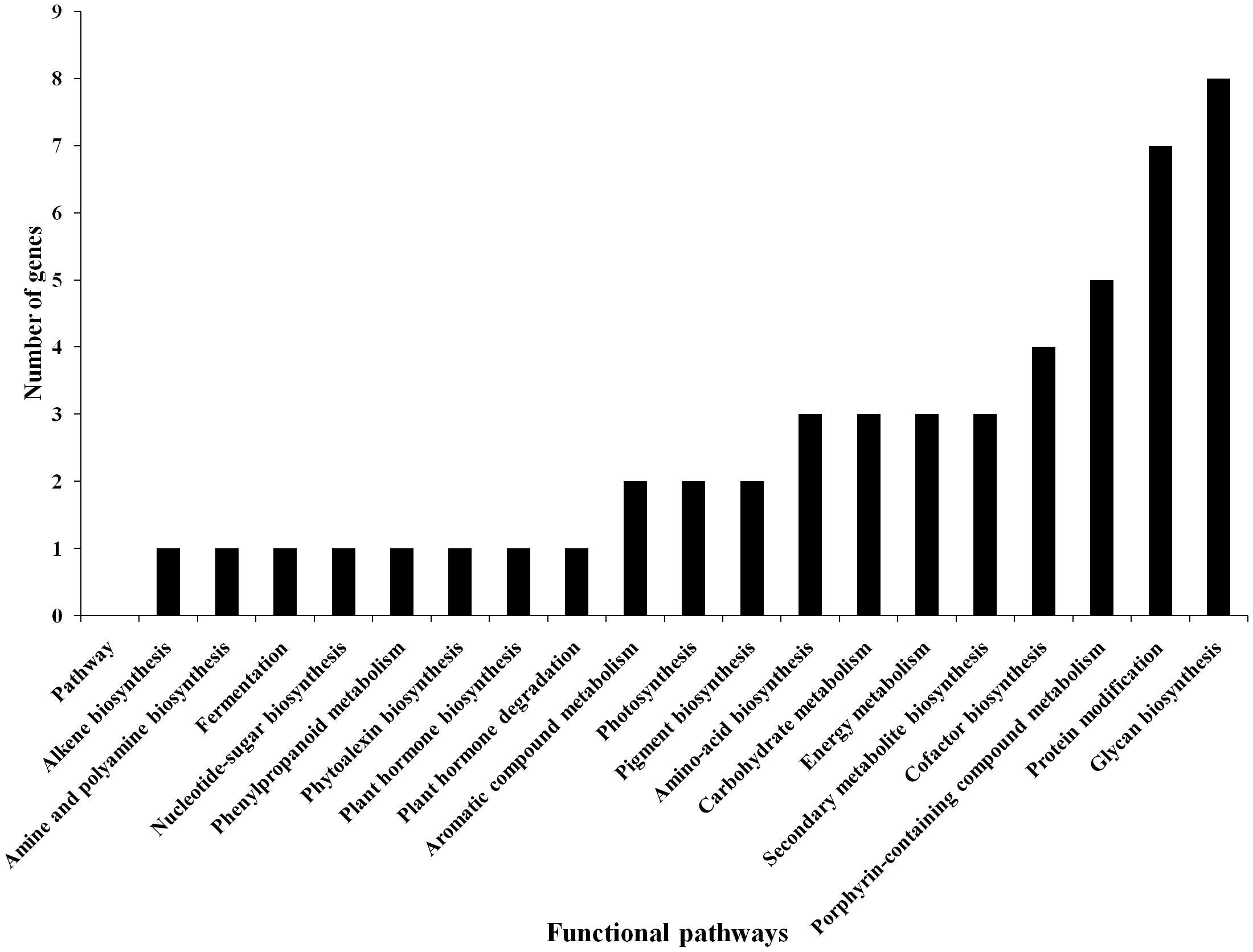
Figure 2. Commonly expressed functional pathway categories in the T. harzianum-treated drought-stressed rice genome vs drought-stressed rice.

Figure 3. Heat map with cluster categorization representing the top 50 significant DEGs at two different comparisons of treatments (T. harzianum-treated + drought-stressed, drought-stressed). Each column represents the DEGs in different samples with two replicates. The red color shows upregulated genes and the green color represents downregulated genes based on highest FPKM values. Each row represents an individual transcript.
Gene Profile and Differential Expression Study of Trichoderma harzianum
To establish rice–T. harzianum interaction, transcriptome analysis of T. harzianum was also carried out. The number of sequences obtained was 532 for a total length of 40,980,648 bp. The N50 length was 2,414,909 and the maximum and minimum contig lengths were 4,089,932 and 1,013 bp, respectively (Table 1). It has been generally accepted that larger values of performance criteria are associated with better assembly performance. An account of the DEGs suggests that there were 808 (3%) significant genes out the total DEGs (Table 1). GO analysis suggested that the annotated genes were functionally related to carbohydrate binding module (GO:0019867; GO:0016021), glycoside hydrolase (GO:0004553; GO:0005975), GMC oxidoreductase (GO:0016614; GO:0050660), and trehalase (GO:0005737). All the genes activated were involved in establishing the mycelia colonization of root and root growth.
Gene Ontology Study
Gene Ontology (GO) analysis revealed GO representation in drought and T. harzianum-primed drought samples. Among the significantly expressed DEGs, 1,657 (66%) GO terms were assigned for T. harzianum-treated drought-stressed vs drought-stressed samples. Cellular processes (39.5%, GO:0009987) and metabolic process (38.5%, GO:0008152) were the most significantly represented groups in the biological process category. Within the cellular component category, cell (50.6%, GO:0005623) and cell part (50.5%; GO:0044464) were the most significantly represented groups, and catalytic activity (49.5%, GO:0003824) was the most significantly represented group within the molecular function category (Figure 4). Supplementary Table 3 provides the list of highly upregulated significant genes expressed in T. harzianum-treated cultivars.
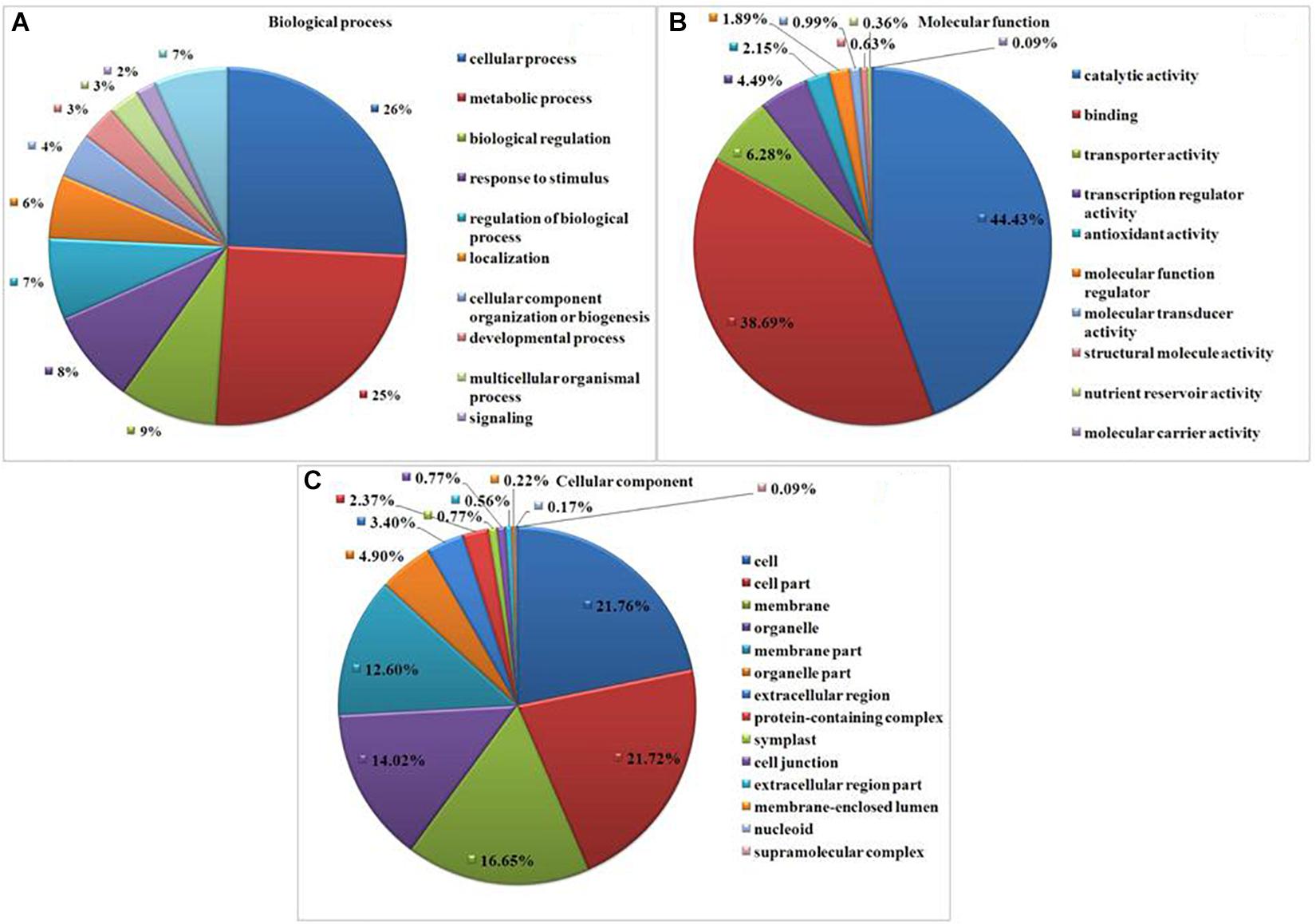
Figure 4. Gene Ontology (GO)-based functional annotation of genes present in the T. harzianum-treated drought-stressed rice genome vs. drought-stressed rice. (A) Biological process domains, (B) molecular function domain, and (C) cellular process domains.
KEGG Pathway and Enrichment Analysis
The KEGG pathway database was used to identify the role of T. harzianum-induced genetic pathway to reduce drought stress in rice plants. It was found that upon T. harzianum priming, a number of genes were upregulated and downregulated in drought-stressed rice plants (Figure 5). The exclusively expressed genes in T. harzianum-inoculated metabolic pathway were mostly related to glutathione metabolism (GO:0004364; GO:0005737; GO:0005576; GO:0006979), steroid biosynthesis (GO:0000254; GO:0005506; GO:0005789), and carbon metabolism (GO:0005739; GO:005960; GO:0019464). The highly upregulated genes included were related to photosynthesis (GO:0009416; GO:0009522; GO:0009523; GO:0009535).
The main pathways enriched in T. harzianum-treated rice relative to drought stress were related to metabolic pathways involved in the synthesis of secondary metabolites followed by pathways related to “photosynthesis.” The proteins of the photosystems and the enzymes of the Calvin cycle were upregulated. Overall, it can be concluded that T. harzianum helped rice plants by increasing the expression of photosynthetic genes, phenyl propanoid pathway genes, glutathione pathway, stress-enhanced osmoproteins, etc. (Figures 6A–C).
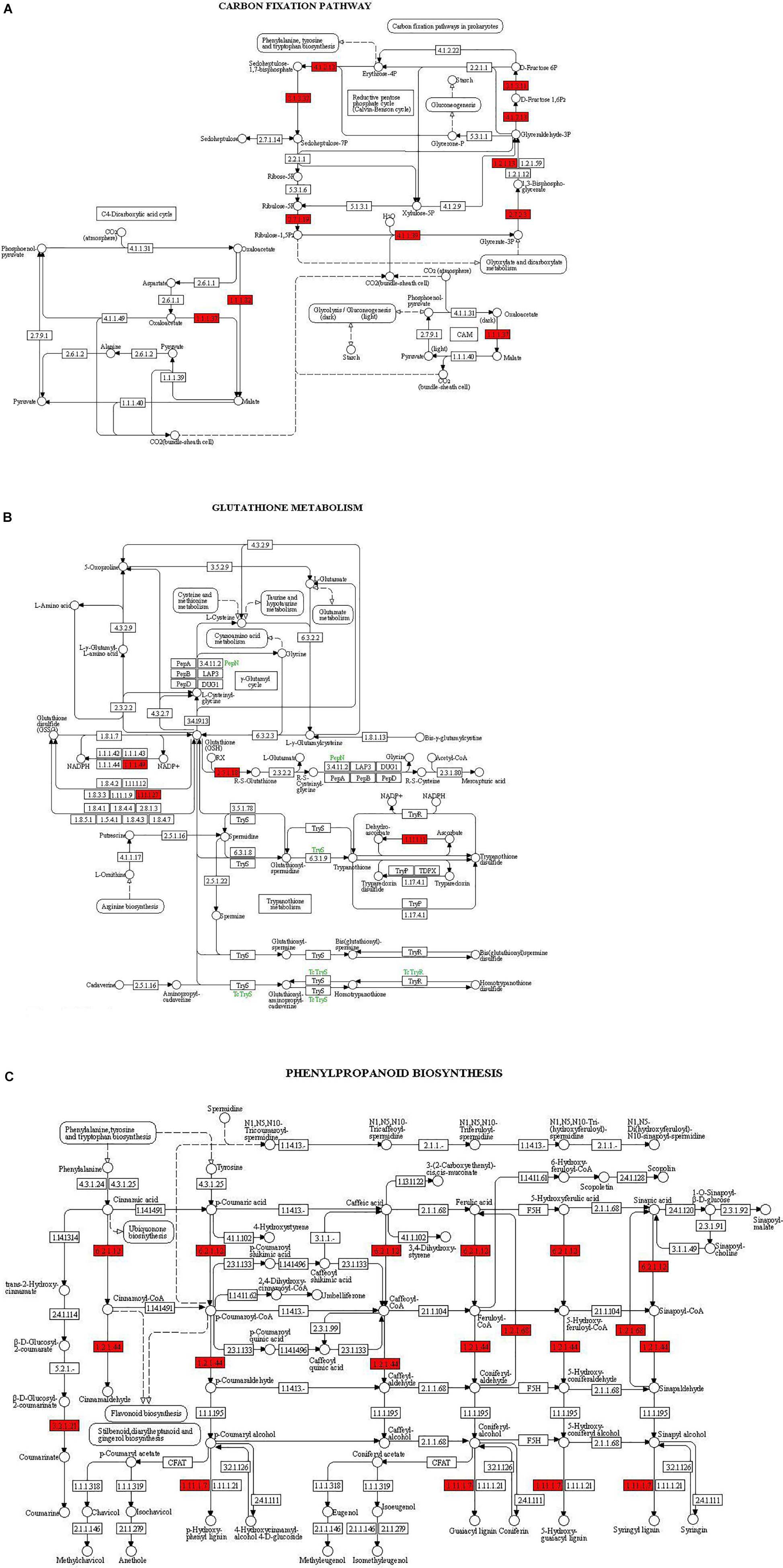
Figure 6. KEGG enrichment for DEGs from the three pathways: (A) carbon fixation, (B) glutathione metabolism, and (C) phenyl propanoid biosynthesis. The red highlights represent the enriched enzymes of the pathways.
To further investigate the DEGs that were involved and enriched in various metabolic pathways, pathway-based analysis was performed using the KEGG pathway database. In this, a total of 468 significant (p < 0.05) genes of the KEGG pathway were found to be enriched. The genes were classified into 13 different categories (Supplementary Table 2). Broadly, the most enriched pathways were metabolic (38%) followed by pathways involved in the synthesis of secondary metabolites (25%), carbon metabolism (6%), phenyl propanoid (7%), and glutathione metabolism (3%).
Validation of Differentially Expressed Genes Through Real-Time PCR
To confirm the accuracy and reproducibility of the Illumina RNA-Seq results, 12 representative genes were chosen to validate the levels of expression in drought and T. harzianum-primed drought-stressed IR64 cultivar by qRT-PCR. The validation results for the selected genes are shown in Figure 7. Out of the 12 genes, five genes—ribulose-bisphosphate carboxylase small chain A, ferredoxin–NADP reductase, chaperonin-like RBCX protein 1, ATP synthase subunit gamma, and photosystem II core complex proteins psbY (Os12g0291100, Os02g0103800, Os08g0425200, Os07g0513000, and Os08g0119800)—were chloroplastic. Three genes such as proline-rich protein (Os10g0150400), osmotin-like protein (Os12g0569500), and aquaporin PIP2 (Os09g0541000) were related to the maintenance of osmotic homeostasis of rice plants. Besides these, PSI subunit (Os04g0414700), peroxidase 43 (Os11g0210100), stress-enhanced protein 1 (Os11g0621400), and auxin-induced protein 15A (Os08g0118500) expression were also assessed. The expressions of all the genes were consistent with RNA-sequencing data.
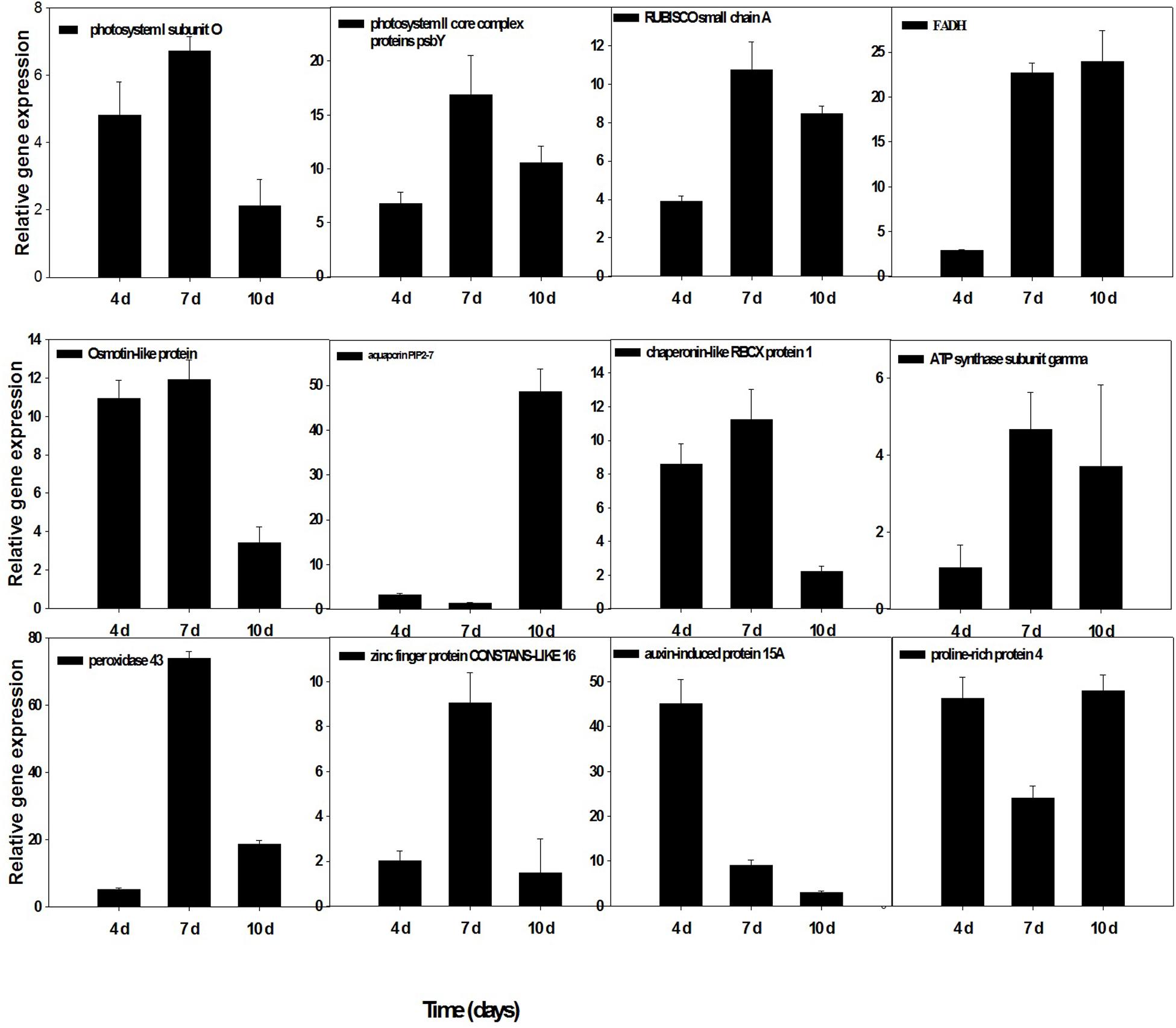
Figure 7. qRT-PCR validation of selected genes showed significant difference in their expression in T. harzianum-treated and drought-stressed when compared with drought-stressed at three different time intervals (4, 7, and 10 days). Error bars show ± SD among the biological replicates.
Discussion
Trichoderma harzianum is an antagonistic biocontrol agent used widely as a plant growth promoter, in alleviation of abiotic stresses such as drought, salinity, and heat stress (Rawat et al., 2012; Ahmad et al., 2015; Angel Contreras-Cornejo et al., 2016). In fact, seed biopriming has shown promising results in decreasing the effect of drought stress as evident from the increase in growth, photosynthetic parameters such as net photosynthetic rate, stomatal conductance, increase in the activity of antioxidative enzymes, secondary metabolites, and related decrease in peroxides (Rawat et al., 2016).
First, our results compared the plant growth of two contrasting cultivars, i.e., IR64 and Sahbhagi Dhan. IR64 was selected for further study due to its better performance. T. harzianum-mediated better growth has been reported in various plants such as rice, tomato, and maize (Morán-Diez et al., 2015; Bashyal et al., 2020). The increase in growth is attributed to the decrease in drought stress by improvement in water use efficiency, osmotic balance by increased proline content, and decrease in MDA content (Pandey et al., 2016; Mishra et al., 2020). The molecular analysis has shown that plant growth promotion might be due to an enhanced expression of carbohydrate binding module family protein (M431DRAFT_521249, M431DRAFT_499139), glycoside hydrolase (M431DRAFT_95191), and polysaccharide lyase (M431DRAFT_92754) as observed in our results. The growth promotion due to T. harzianum colonization may in part be due to cellulose disruption by induction of Tg SWO, carbohydrate binding module, and swollenin protein (Brotman et al., 2008; Meng et al., 2019). The carbohydrate binding module-mediated root elongation is mediated by cellulose breakdown, which results in cellular expansion thereby resulting in cellular signaling for cell growth (Brotman et al., 2008).
Our study showed that T. harzianum biopriming has proved beneficial in withstanding drought stress. The stress tolerance might be correlated to the upregulation of various genes which have been identified to be involved in carbon metabolism and carbon fixation. Among DEGs, some genes which were exclusively expressed or are highly upregulated with respect to drought stress play a significant role in drought stress alleviation. The log fold changes of the exclusively expressed genes such as Os06g0101600 (plastocyanin, chloroplastic-like), Os12g0291100 (ribulose-bisphosphate carboxylase small chain A), Os02g0103800 (ferredoxin–NADP reductase, leaf isozyme 2, chloroplastic-like), Os08g0425200 (chaperonin-like RBCX protein 1, chloroplastic), and Os07g0513000 (ATP synthase subunit gamma, chloroplastic) were infinite as the reads were recorded for T. harzianum-treated rice only and not for drought-treated cultivars (Supplementary Table 3). The log fold changes for highly upregulated genes such as Os04g0414700 (photosystem I subunit O), Os08g0119800 (photosystem II core complex proteins psbY, chloroplastic), Os12g0569500 (osmotin-like protein), Os02g0815200 (28 kDa ribonucleoprotein, chloroplastic), Os04g0490800 (phosphoglycolate phosphatase 1B, chloroplastic), and Os01g0882500 [NAD(P)H-quinone oxidoreductase subunit N, chloroplastic] were 10.2, 10, 9.7, 7, 7.1, and 7.2, respectively (Supplementary Table 3). The increase in expression of photosynthetic genes is consistent with previous results (Azarmi et al., 2011; Alwhibi et al., 2017; Doni et al., 2019). The increase in photosynthetic genes might be due to an increase in phytohormones also observed in our results, for instance Os03g797800 (auxin-responsive protein) and Os01g0883800 (gibberellin oxidase) which are upregulated by 5.41 and 5.29 log fold change. Additionally, the carotenoids protect pigments from photooxidative stress (Strzaka et al., 2003) and our pathway analysis results indicated enriched enzymes of carotenoid biosynthesis.
The exclusive expression of peroxidase and the enrichment of glutathione metabolism pathway in T. harzianum-primed rice cultivar as observed in our study indicate toward the involvement of glutathione peroxidase pathway active in antioxidation ROS formed due to drought stress. The induction of antioxidant defense has also been observed in tomato seedlings and rice in water deficit after T. harzianum interaction (Mastouri et al., 2012; Singh et al., 2020). Similar results were observed when maize seedlings were inoculated with Trichoderma atroviride (Guler et al., 2016).
Besides, the genes involved in photosynthetic pathway, the secondary metabolite pathway genes such as phenylpropanoid, glutathione metabolism, diterpenoid, glyoxylate, nitrogen, cutin, and suberin were also upregulated in T. harzianum-inoculated drought-stressed IR64 rice cultivar. The antioxidative role of secondary metabolites and their effect in decreasing drought stress are well established (Tattini et al., 2014; Kubala et al., 2015). Glutathione is a tripeptide found abundantly in cellular components and gene functions in cell growth and regulation of stress-responsive genes (Sofo et al., 2015).
Additionally, we observed upregulation of osmotin-like proteins (Os12g38170) and aquaporins (Os0232900) in T. harzianum-primed drought-treated rice. Osmotin-like proteins are known to protect plants by maintaining cellular osmolarity by compartmentalization of solutes or by structural and metabolic alterations (Choi et al., 2013). In fact, their overexpression has been correlated with stress tolerance (Chowdhury et al., 2017; Bashir et al., 2020). The aquaporins are channel proteins which play a key role in plant responses to environmental stresses by maintaining the uptake and movement of water in the plant body and maintain cell viability (Kapilan et al., 2018). The upregulation is also linked to drought stress maintenance (Shekoofa and Sinclair, 2018).
Kyoto Encyclopedia of Genes and Genomes enrichment analysis was performed to identify related pathways for 468 genes that were involved and enriched in this study. The most enriched genes in carbon fixation pathway include malate dehydrogenase, sedoheptulose-bisphosphatase, ribulose-5-phosphate kinase, ribulose-bisphosphate carboxylase, fructose bisphosphate aldolase, glyceraldehyde-3-phosphate dehydrogenase, and phosphoglycerate kinase (Os02g0698000, Os03g0129300, Os03g0267300, Os04g0234600, Os04g0459500, Os05g0496200, Os06g0608700, Os07g0630800, Os08g0562100, Os11g0171300, and Os12g0274700). The genes enriched in glutathione pathway were glucose-6-phosphate dehydrogenase (Os07g0406300), glutathione-dependent peroxiredoxin (Os02g0192700), glutathione transferase (Os10g395400), and ascorbate peroxidase (Os02g2553200). The genes found to be enriched in phenyl propanoid pathway are coumarate-CoA ligase (Os02g0697400), cinnamoyl-coA reductase (Os09g0400000), coniferyl aldehyde dehydrogenase (Os04g0540600), beta glucosidase (Os09g0511900), and peroxidase (Os10g0109300).
Hence, overall, T. harzianum biopriming aided rice plants in a multifaceted simultaneous manner by triggering various pathways such as photosynthetic, secondary metabolites, and osmotic balance maintenance contributing to enhanced tolerance to drought stress (Singh et al., 2020).
Conclusion
We provided a comprehensive study of the transcriptome of a drought-challenged IR64 rice cultivar bioprimed with T. harzianum. The complete analyses of DEGs are highlighted with respect to drought treatment. The data revealed that the T. harzianum-mediated drought stress improvement is associated with a synchronous induction of various metabolic pathways involved in photoprotection of photosynthetic apparatus, upregulation of photosynthetic genes, upregulation of antioxidative genes, ascorbate glutathione pathway, and maintenance of osmotic homeostasis by increasing proline, osmotic proteins, and aquaporins. Additionally, the transcriptome analysis of T. harzianum also supported the fact that its association helps in plant growth by the regulation of carbohydrate binding module polysaccharide lyase and glycoside hydrolase. Overall, it can be concluded that T. harzianum biopriming delays drought stress in rice cultivars by a myriad of molecular reprogramming.
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the 373 repositories and accession numbers can be found below: https://www.ncbi.nlm.nih.gov/, SRR12567752, SRR12567751, SRR12567750, and SRR12567749.
Author Contributions
BB, NZ, and RA conceptualized the problem and designed the experiment. PP and BB conducted the experiment and analyzed the data. PP wrote the manuscript. BB, RA, and NZ edited the manuscript. All authors finalized the manuscript.
Funding
We are thankful to ICAR-IARI for providing facilities and the IRRI-EC IFAD project for providing financial support. We are also thankful to the Director, ICAR-Indian Agricultural Research Institute, New Delhi, India, for providing research facilities and financial support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.655165/full#supplementary-material
Supplementary Figure 1 | Effect of T. harzianum treatment in drought stressed rice cultivars’ (IR64 and Shahbhagi Dhan) root length (A–C) and shoot length (D–F) after different days (4, 7, and 10 days) of drought stress.
Supplementary Figure 2 | Effect of T. harzianum treatment on plants of two rice varieties (V1, Shahbhagi Dhan and V2, IR64) after different days (4 and 7 days) of drought stress where a, b, and c represent control, T. harzianum treated + drought stress and drought stressed plants in each image.
Supplementary Table 1 | List of primers used for expression analysis.
Supplementary Table 2 | Enriched KEGG pathways of differentially expressed genes in T. harzianum-rice interaction.
Supplementary Table 3 | List of differentially expressed upregulated genes.
References
Afzal, Z., Howton, T. C., Sun, Y., and Mukhtar, M. S. (2016). The roles of aquaporins in plant stress responses. J. Dev. Biol. 4, 9–30. doi: 10.3390/jdb4010009
Ahmad, P., Hashem, A., Abd-Allah, E. F., Alqarawi, A. A., John, R., Egamberdieva, D., et al. (2015). Role of Trichoderma harzianum in mitigating NaCl stress in Indian mustard (Brassica juncea L.) through antioxidative defense system. Front. Plant Sci. 6:868. doi: 10.3389/fpls.2015.00868
Akoh, C. C., Lee, G. C., Liaw, Y. C., Huang, T. H., and Shaw, J. F. (2004). GDSL family of serine esterases/lipases. Prog. Lipid Res. 43, 534–552. doi: 10.1016/j.plipres.2004.09.002
Alwhibi, M. S., Hashem, A., Fathi, E., Allah, A., Alqarawi, A. A., Soliman, D. W. K., et al. (2017). Increased resistance of drought by Trichoderma harzianum fungal treatment correlates with increased secondary metabolites and proline content. J. Integ. Agric. 16, 1751–1757. doi: 10.1016/S2095-3119(17)61695-2
Azarmi, R., Hajieghrari, B., and Giglou, A. (2011). Effect of Trichoderma isolates on tomato seedling growth response and nutrient uptake. Afric. J. Biotechnol. 10, 5850–5855. doi: 10.5897/AJB10.1600
Bae, H., Sicher, R. C., Kim, M. S., Kim, S. H., Strem, M. D., Melnick, R. L., et al. (2009). The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao. J. Exp. Bot. 60, 3279–3295. doi: 10.1093/jxb/erp165
Bashir, M. A., Silvestri, C., Ahmad, T., Hafiz, I. A., Abbasi, N. A., Manzoor, A., et al. (2020). Osmotin: a cationic protein leads to improve biotic and abiotic stress tolerance in plants. Plants 9, 992–1007. doi: 10.3390/plants9080992
Bashyal, B. M., Aggarwal, R., Sharma, S., Gupta, S., Rawat, K., Singh, D., et al. (2016). Occurrence, identification and pathogenicity of Fusarium spp. associated with bakanae disease of basmati rice in India. Eur. J. Plant Pathol. 144, 457–466. doi: 10.1007/s10658-015-0783-8
Bashyal, B. M., Zaidi, N. W., Singh, U. S., and Aggarwal, R. (2020). Effect of fungal biocontrol agents on enhancement of drought stress tolerance in rice (Oryza sativa L.). Indian J. Biochem. Biophys. 57, 101–108.
Brotman, Y., Briff, E., Viterbo, A., and Chet, I. (2008). Role of swollenin, an expansin-like protein from Trichoderma, in plant root colonization. Plant Physiol. 147, 779–789. doi: 10.1104/pp.108.116293
Brotman, Y., Landau, U., Cuadros-Inostroza, A., Tohge, T., Fernie, A. R., Chet, I., et al. (2013). Trichoderma-plant root colonization: escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance. PLoS Pathog. 9:1003221. doi: 10.1371/annotation/8b818c15-3fe0-4e56-9be2-e44fd1ed3fae
Chen, X., Li, C., Wang, H., and Guo, Z. (2019a). WRKY transcription factors: evolution, binding, and action. Phytopathol. Res. 1, 13–27. doi: 10.1186/s42483-019-0022-x
Chen, X., Wang, H., Li, X., Ma, K., Zhan, Y., and Zeng, F. (2019b). Molecular cloning and functional analysis of 4-Coumarate: CoA ligase 4(4CL-like 1) from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis. BMC Plant Biol. 19:231. doi: 10.1186/s12870-019-1812-0
Chen, Y., Kelly, E. E., Masluk, R. P., Nelson, C. L., Cantu, D. C., and Reilly, P. J. (2011). Structural classification and properties of ketoacyl synthases. Prot. Sci. 20, 1659–1667. doi: 10.1002/pro.712
Choi, D. S., Hong, J. K., and Hwan, B. K. (2013). Pepper osmotin-like protein 1 (CaOSM1) is an essential component for defense response, cell death, and oxidative burst in plants. Planta 238, 1113–1124. doi: 10.1007/s00425-013-1956-3
Chowdhury, S., Basu, A., and Kundu, S. (2017). Overexpression of a new osmotin-like protein gene (sindolp) confers tolerance against biotic and abiotic stresses in sesame. Front. Plant Sci. 8:410. doi: 10.3389/fpls.2017.00410
Ciarkowska, A., Ostrowski, M., Starzyńska, E., and Jakubowska, A. (2019). Plant SCPL acyltransferases: multiplicity of enzymes with various functions in secondary metabolism. Phytochem. Rev. 18, 303–316. doi: 10.1007/s11101-018-9593-x
Contreras-Cornejo, H., Ortiz-Castro, R., and López-Bucio, J. (2013). “Promotion of plant growth and the induction of systemic defence by Trichoderma: physiology, genetics and gene expression,” in Trichoderma: Biology and Applications, eds P. K. Mukherjee, B. A. Horwitz, U. S. Singh, M. Mukherjee, and M. Schmoll (London: CABI International), 175–196.
Contreras-Cornejo, H. A., Macías-Rodríguez, L., Cortés-Penagos, C., and López-Bucio, J. (2009). Trichoderma virens, a plant beneficial fungus enhances biomass production and promotes lateral root growth through an auxin dependent mechanism in Arabidopsis. Plant Physiol. 149, 1579–1592. doi: 10.1104/pp.108.130369
Contreras-Cornejo, H. A., Macías-Rodríguez, L., del-Val, E., and Larsen, J. (2016). Ecological functions of Trichoderma spp. and their secondary metabolites in the rhizosphere: interactions with plants. FEMS Microbiol. Ecol. 92, 36–52. doi: 10.1093/femsec/fiw036
Dao, T. T., Linthorst, H. J., and Verpoorte, R. (2011). Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 10, 397–412. doi: 10.1007/s11101-011-9211-7
Dellero, Y., Jossier, M., Schmitz, J., Maurino, V. G., and Hodges, M. (2016). Photorespiratory glycolate–glyoxylate metabolism. J. Exp. Bot. 67, 3041–3052. doi: 10.1093/jxb/erw090
Dietz, K. J. (2011). Peroxiredoxins in plants and cyanobacteria. Antiox. Redox Signal. 15, 1129–1159. doi: 10.1089/ars.2010.3657
Doni, F., Fathurrahman, F., Mispan, M. S., Suhaimi, N. S. M., Yusoff, W. M. W., and Uphoff, N. (2019). Transcriptomic profiling of rice seedlings inoculated with the symbiotic fungus Trichoderma asperellum SL2. J. Plant Growth Regul. 38, 1507–1515. doi: 10.1007/s00344-019-09952-7
Doni, F., Isahak, A., Zain, C. R. C. M., Ariffin, S. M., Mohamadand, W. N. W., and Yusoff, W. M. W. (2014). Formulation of Trichoderma sp. SL2 inoculants using different carriers for soil treatment in rice seedling growth. SpringerPlus 3, 532–536. doi: 10.1186/2193-1801-3-532
Elena López-Calcagno, P., Omar Abuzaid, A., Lawson, T., and Anne Raines, C. (2017). Arabidopsis CP12 mutants have reduced levels of phosphoribulokinase and impaired function of the Calvin-Benson cycle. J. Exp. Bot. 68, 2285–2298. doi: 10.1093/jxb/erx084
Elhiti, M., and Stasolla, C. (2009). Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal Behav. 4, 86–88. doi: 10.4161/psb.4.2.7692
Ereful, N. C., Liu, L., Greenland, A., Powell, W., Mackay, I., and Leung, H. (2020). RNA-seq reveals differentially expressed genes between two indica inbred rice genotypes associated with drought-yield QTLs. Agronomy 10, 621–639. doi: 10.3390/agronomy10050621
Finnegan, E., and Kovac, K. (2000). Plant DNA methyltransferases. Plant Mol. Biol. 43, 189–201. doi: 10.1023/a:1006427226972
Ghanashyam, C., and Jain, M. (2009). Role of auxin-responsive genes in biotic stress responses. Plant Signal. Behav. 4, 846–848. doi: 10.4161/psb.4.9.9376
Gong, H., Jiao, Y., Hu, W., and Pua, E. C. (2005). Expression of glutathione-S-transferase and its role in plant growth and development in vivo and shoot morphogenesis in vitro. Plant Mol. Biol. 57, 53–66. doi: 10.1007/s11103-004-4516-1
Guler, N. S., Pehlivan, N., Karaoglu, S. A., Guzel, S., and Bozdeveci, A. (2016). Trichoderma atroviride ID20G inoculation ameliorates drought stress-induced damages by improving antioxidant defence in maize seedlings. Acta Physiol. Plant 38, 132–140. doi: 10.1007/s11738-016-2153-3
Gujjar, R. S., Pathak, A. D., Karkute, S. G., and Supaibulwattana, K. (2019). Multifunctional proline rich proteins and their role in regulating cellular proline content in plants under stress. Biol. Plant 63, 448–454. doi: 10.32615/bp.2019.078
Haldrup, A., Naver, H., and Scheller, H. V. (1998). “Characterisation of transgenic Arabidopsis plants lacking the PSI-N subunit of photosystem I,” in Photosynthesis: Mechanisms and Effects, ed. G. Garab (Dordrecht: Springer), 627–630. doi: 10.1007/978-94-011-3953-3_148
Haldrup, A., Naver, H., and Scheller, H. V. (1999). The interaction between plastocyanin and photosystem I is inefficient in transgenic Arabidopsis plants lacking the PSI-N subunit of photosystem I. Plant J. 17, 689–698. doi: 10.1046/j.1365-313x.1999.00419.x
Hardie, D. G. (1999). Plant protein serine/threonine kinases: Classification and functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 97–131.
Harman, G. E., Doni, F., Khadka, R. B., and Uphoff, N. (2019). Endophytic strains of Trichoderma increase plants’ photosynthetic capability. J. Appl. Microbiol. 130, 529–546. doi: 10.1111/jam.14368
Huang, J. Z., Guo-E, Z., Zhang, H. L., and Shu, Q. Y. (2014). Workable male sterility systems for hybrid rice: genetics, biochemistry, molecular biology, and utilization. Rice 7:13. doi: 10.1186/s12284-014-0013-6
Jansson, S. (1994). The light-harvesting chlorophyll a/ b-binding proteins. Biochim. Biophys. Acta 1184, 1–19. doi: 10.1016/0005-2728(94)90148-1
Jansson, S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4, 236–240. doi: 10.1016/s1360-1385(99)01419-3
Jensen, P. E., Bassi, R., Boekema, E. J., Dekker, J. P., Jansson, S., Leister, D., et al. (2007). Structure, function and regulation of plant photosystem I. Biochim. Biophys. Acta 1767, 335–352. doi: 10.1016/j.bbabio.2007.03.004
Jensen, P. E., Haldrup, A., Zhang, S., and Scheller, H. V. (2004). The PSI-O subunit of plant photosystem I is involved in balancing the excitation pressure between the two photosystems. J. Biol. Chem. 279, 24212–24217. doi: 10.1074/jbc.M403147200
Kapilan, R., Vaziri, M., and Zwiazek, J. J. (2018). Regulation of aquaporins in plants under stress. Biol. Res. 51:4. doi: 10.1186/s40659-018-0152-0
Katoh, S. (2003). Early research on the role of plastocyanin in photosynthesis. Photosynth. Res. 76, 255–261. doi: 10.1023/A:1024924711453
Kaur, G., Singh, S., Singh, H., Chawla, M., Dutta, T., Kaur, H., et al. (2015). Characterization of peptidyl-prolyl cis-trans isomerase- and calmodulin-binding activity of a cytosolic Arabidopsis thaliana cyclophilin AtCyp19-3. PLoS One 10:e0136692. doi: 10.1371/journal.pone.0136692
Kossmann, J., and Lloyd, J. (2000). Understanding and influencing starch biochemistry. Crit. Rev. Biochem. Mol. Biol. 35, 141–196. doi: 10.1080/07352680091139204
Kozuleva, M., Goss, T., Twachtmann, M., Rudi, K., Trapka, J., Selinski, J., et al. (2016). Ferredoxin: NADP(H) oxidoreductase abundance and location influences redox poise and stress tolerance. Plant physiol. 172, 1480–1493. doi: 10.1104/pp.16.01084
Kubala, S., Garnczarska, M., Wojtyla, Ł, Clippe, A., Kosmala, A., Zmienko, A., et al. (2015). Deciphering priming-induced improvement of rapeseed (Brassica napus L.) germination through an integrated transcriptomic and proteomic approach. Plant Sci. 231, 94–113. doi: 10.1016/j.plantsci.2014.11.008
Kumar, D., Das, K. P., and Sarmah, B. K. (2018). Reference gene validation for normalization of RT-qPCR assay associated with germination and survival of rice under hypoxic condition. J. Appl. Genet. 59, 419–430. doi: 10.1007/s13353-018-0466-1
Lagercrantz, U., Axelsson, T. (2000). Rapid evolution of the family of CONSTANS LIKE genes in plants. Mol. Biol. Evol. 17, 1499–1507. doi: 10.1093/oxfordjournals.molbev.a026249
Lamkemeyer, P., Laxa, M., Collin, V., Li, W., Finkemeier, I., Schöttler, M. A., et al. (2006). Peroxiredoxin Q of Arabidopsis thaliana is attached to the thylakoids and functions in context of photosynthesis. Plant J. 45, 968–981. doi: 10.1111/j.1365-313X.2006.02665.x
Lesk, C., Rowhani, P., and Ramankutty, N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. doi: 10.1038/nature16467
Lin, C., Choi, H. S., and Cho, H. T. (2011). Root hair-specific EXPANSIN A7 is required for root hair elongation in Arabidopsis. Mol. Cells 31, 393–397. doi: 10.1007/s10059-011-0046-2
Liu, F., Zhang, X., Lu, C., Zeng, X., Li, Y., Fu, D., et al. (2015). Non-specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis. J. Exp. Bot. 66, 5663–5681. doi: 10.1093/jxb/erv313
Liu, H., Wang, X., Zhang, H., Yayun, Y., Xiuchun, G., and Fengming, S. (2008). A rice serine carboxypeptidase-like gene OsBISCPL1 is involved in regulation of defense responses against biotic and oxidative stress. Gene 420, 57–65. doi: 10.1016/j.gene.2008.05.006
Liu, Q., Luo, L., Wang, X., Shen, Z., and Zheng, L. (2017). Comprehensive analysis of rice laccase gene (OsLAC) family and ectopic expression of OsLAC10 enhances tolerance to copper stress in Arabidopsis. Int. J. Mol. Sci. 18, 209–224. doi: 10.3390/ijms18020209
Ma, C., and Subramani, S. (2009). Peroxisome matrix and membrane protein biogenesis. IUBMB Life 61, 713–722. doi: 10.1002/iub.196
Ma, C., Schumann, U., Rayapuram, N., and Subramani, S. (2009). The peroxisomal matrix import of Pex8p requires only PTS receptors and Pex14p. Mol. Biol. Cell 20, 3680–3689. doi: 10.1091/mbc.E09-01-0037
Mastouri, F., Björkman, T., and Harman, G. (2012). Trichoderma harzianum enhances antioxidant defense of tomato seedlings and resistance to water deficit. Mol. Plant Microbe. Interact. 25, 1264–1271. doi: 10.1094/MPMI-09-11-0240
Mellacheruvu, S., Tamirisa, S., Vudem, D. R., and Khareedu, V. R. (2016). Pigeonpea hybrid-proline-rich protein (CcHyPRP) confers biotic and abiotic stress tolerance in transgenic rice. Front. Plant Sci. 6:1167. doi: 10.3389/fpls.2015.01167
Melo, A. M., Bandeiras, T. M., and Teixeira, M. (2004). New insights into type II NAD(P)H: quinone oxidoreductases. Microbiol. Mol. Biol. Rev. 68, 603–616. doi: 10.1128/MMBR.68.4
Meng, X., Miao, Y., Liu, Q., Ma, L., Guo, K., Liu, D., et al. (2019). TgSWO from Trichoderma guizhouense NJAU4742 promotes growth in cucumber plants by modifying the root morphology and the cell wall architecture. Microb. Cell Fact. 18, 148–162. doi: 10.1186/s12934-019-1196-8
Mishra, D., Rajput, R. S., Zaidi, N. W., and Singh, H. B. (2020). Sheath blight and drought stress management in rice (Oryza sativa) through Trichoderma spp. Indian Phytopathol. 73, 71–77. doi: 10.1007/s42360-019-00189-8
Morán-Diez, M. E., Trushina, N., Lamdan, N. L., Rosenfelder, L., Mukherjee, P. K., Kenerley, C. M., et al. (2015). Host-specific transcriptomic pattern of Trichoderma virens during interaction with maize or tomato roots. BMC Genomics 16:8. doi: 10.1186/s12864-014-1208-3
Noordermeer, M. A., Veldink, G. A., and Vliegenthart, J. F. (2001). Fatty acid hydroperoxide lyase: a plant cytochrome p450 enzyme involved in wound healing and pest resistance. Chembiochem 2, 494–504. doi: 10.1002/1439-7633(20010803)2
Oikawa, T., Koshioka, M., Kojima, K., Yoshida, H., and Kawata, M. (2004). A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol. Biol. 55, 687–700. doi: 10.1007/s11103-004-1692-y
Olsson, A. S. B., Engstrom, P., and Soderman, E. (2004). The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 55, 663–677. doi: 10.1007/s11103-004-1581-4
Palma, D. M., Salzano, M., Villano, C., Aversano, R., Lorito, M., Ruocco, M., et al. (2019). Transcriptome reprogramming, epigenetic modifications and alternative splicing orchestrate the tomato root response to the beneficial fungus Trichoderma harzianum. Hortic. Res. 6:5. doi: 10.1038/s41438-018-0079-1
Pandey, V., Ansari, M. W., Tula, S., Yadav, S., Sahoo, R. K., Shukla, N., et al. (2016). Dose-dependent response of Trichoderma harzianum in improving drought tolerance in rice genotypes. Planta 243, 1251–1264. doi: 10.1007/s00425-016-2482-x
Pandey, V. P., Awasthi, M., Singh, S., Tiwari, S., and Dwivedi, U. N. (2017). A comprehensive review on function and application of plant peroxidases. Biochem. Anal. Biochem. 6, 308–323. doi: 10.4172/2161-1009.1000308
Quezada, E. H., García, G. X., Arthikala, M. K., Melappa, G., Lara, M., and Nanjareddy, K. (2019) Cysteine-rich receptor-like kinase gene family identification in the Phaseolus genome and comparative analysis of their expression profiles specific to mycorrhizal and rhizobial symbiosis. Genes 10, 59–79. doi: 10.3390/genes10010059
Rawat, L., Bisht, T., Upadhayay, R., and Kukreti, A. (2016). Selection of salinity tolerant Trichoderma isolates and evaluating their performance in alleviating salinity stress in rice (Oryzae sativa L.). J. Trop. Agric. 34, 1869–1875.
Rawat, L., Singh, Y., Shukla, N., and Kumar, J. (2012). Seed biopriming with salinity tolerant isolates of Trichoderma harzianum alleviates salt stress in rice: Growth, physiological and biochemical characteristics. J. Plant Pathol. 94, 353–365.
Rehman, H. M., Nawaz, M. A., Shah, Z. H., Ludwig-Müller, J., Chung, G., Ahmad, M. Q., et al. (2018). Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 8, 1875–1895. doi: 10.1038/s41598-018-19535-3
Schuster, A., and Schmoll, M. (2010). Biology and biotechnology of Trichoderma. Appl. Microbiol. Biotechnol. 87, 787–799. doi: 10.1007/s00253-010-2632-1
Sharma, N., Sharma, K. P., Gaur, R., and Gupta, V. K. (2011). Role of chitinase in plant defense. Asian J. Biochem. 6, 29–37. doi: 10.3923/ajb.2011.29.37
Shekoofa, A., and Sinclair, T. R. (2018). Aquaporin activity to improve crop drought tolerance. Cells 7, 123–132. doi: 10.3390/cells7090123
Singh, D. P., Singh, V., Gupta, V. K., Shukla, R., Prabha, R., Sarma, B. K., et al. (2020). Microbial inoculation in rice regulates antioxidative reactions and defense related genes to mitigate drought stress. Sci. Rep. 10, 4818–4834. doi: 10.1038/s41598-020-61140-w
Sofo, A., Scopa, A., Nuzzaci, M., and Vitti, A. (2015). Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Int. J. Mol. Sci. 16, 13561–13578. doi: 10.3390/ijms160613561
Strzaka, K., Kostecka-Guga, A., and Latowski, D. (2003). Carotenoids and environmental stress in plants: significance of carotenoid-mediated modulation of membrane physical properties. Russian J. Plant Physiol. 50, 168–172. doi: 10.1023/A:1022960828050
Sun, C. W., Huang, Y. C., and Chang, H. Y. (2009). CIA2 coordinately up-regulates protein import and synthesis in leaf chloroplasts. Plant Physiol. 150, 879–888. doi: 10.1104/pp.109.137240
Sunkar, R., Bartels, D., and Kirch, H. H. (2003). Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J. 35, 452–464. doi: 10.1046/j.1365-313x.2003.01819.x
Suzuki, Y., and Makino, A. (2012). Availability of Rubisco small subunit up-regulates the transcript levels of large subunit for stoichiometric assembly of its holoenzyme in rice. Plant Physiol. 160, 533–540. doi: 10.1104/pp.112.201459
Tambasco-Studart, M., Titiz, O., Raschle, T., Forster, G., Amrhein, N., and Fitzpatrick, T. B. (2005). Vitamin B6 biosynthesis in higher plants. Proc. Natl. Acad. Sci. U.S.A. 102, 13687–13692. doi: 10.1073/pnas.0506228102
Tan, Q. K., and Irish, V. F. (2006). The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol. 140, 1095–1108. doi: 10.1104/pp.105.070565
Tanaka, R., Oster, U., Kruse, E., Rudiger, W., and Grimm, B. (1999). Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol. 120, 695–704. doi: 10.1104/pp.120.3.695
Tattini, M., Velikova, V., Vickers, C., Brunetti, C., Di Ferdinando, M., Trivellini, A., et al. (2014). Isoprene production in transgenic tobacco alters isoprenoid, non-structural carbohydrate and phenylpropanoid metabolism, and protects photosynthesis from drought stress. Plant Cell Environ. 37, 1950–1964. doi: 10.1111/pce.12350
Uematsu, K., Suzuki, N., Iwamae, T., Inui, M., and Yukawa, H. (2012). Increased fructose 1, 6-bisphosphate aldolase in plastids enhances growth and photosynthesis of tobacco plants. J. Exp. Bot. 63, 3001–3009. doi: 10.1093/jxb/ers004
Wang, X., Guan, Y., Zhang, D., Dong, X., Tian, L., and Qu, L. Q. (2017). A β ketoacyl-coa synthase is involved in rice leaf cuticular wax synthesis and requires a CER2-LIKE protein as a cofactor. Plant Physiol. 173, 944–955. doi: 10.1104/pp.16.01527
Zhang, H., and Cramer, W. A. (2004). Purification and crystallization of the cytochrome b6f complex in oxygenic photosynthesis. Methods Mol. Biol. 274, 67–78. doi: 10.1385/1-59259-799-8:067
Zhao, Q., and Liu, C. (2018). Chloroplast chaperonin: an intricate protein folding machine for photosynthesis. Front. Mol. Biosci. 4:98. doi: 10.3389/fmolb.2017.00098
Keywords: Trichoderma harzianum, drought, rice, transcriptome (RNA-seq), DEGs
Citation: Bashyal BM, Parmar P, Zaidi NW and Aggarwal R (2021) Molecular Programming of Drought-Challenged Trichoderma harzianum-Bioprimed Rice (Oryza sativa L.). Front. Microbiol. 12:655165. doi: 10.3389/fmicb.2021.655165
Received: 18 January 2021; Accepted: 24 February 2021;
Published: 13 April 2021.
Edited by:
Samantha Chandranath Karunarathna, Kunming Institute of Botany, Chinese Academy of Sciences, ChinaReviewed by:
Kasun Thambugala, University of Sri Jayewardenepura, Sri LankaBirinchi Kumar Sarma, Banaras Hindu University, India
Copyright © 2021 Bashyal, Parmar, Zaidi and Aggarwal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bishnu Maya Bashyal, YmlzaG51bWF5YWJhc2h5YWxAZ21haWwuY29t
 Bishnu Maya Bashyal
Bishnu Maya Bashyal Pooja Parmar1
Pooja Parmar1