- 1Department of Chemical and Biological Engineering, Korea University, Seoul, South Korea
- 2BK21 Graduate Program, Department of Biomedical Sciences, College of Medicine, Korea University, Seoul, South Korea
Protein production requires a significant amount of intracellular energy. Eliminating the flagella has been proposed to help Escherichia coli improve protein production by reducing energy consumption. In this study, the gene encoding a subunit of FlhC, a master regulator of flagella assembly, was deleted to reduce the expression of flagella-related genes. FlhC knockout in the ptsG-deleted strain triggered significant growth retardation with increased ATP levels and a higher NADPH/NADP+ ratio. Metabolic flux analysis using a 13C-labeled carbon substrate showed increased fluxes toward the pentose phosphate and tricarboxylic acid cycle pathways in the flhC- and ptsG-deleted strains. Introduction of a high copy number plasmid or overexpression of the recombinant protein in this strain restored growth rate without increasing glucose consumption. These results suggest that the metabolic burden caused by flhC deletion was resolved by recombinant protein production. The recombinant enhanced green fluorescent protein yield per glucose consumption increased 1.81-fold in the flhC mutant strain. Thus, our study demonstrates that high-yield production of the recombinant protein was achieved with reduced flagella formation.
Introduction
When a microorganism is used for industrial purposes, its energy metabolism does not have to be similar to that of natural conditions. In an industrial bioreactor, mixing and agitation facilitate nutrient supplements, so that the energy required for movement to search for nutrients would be significantly reduced (Blais et al., 1997). On the other hand, much more energy is needed in the metabolism required for recombinant protein or metabolite production. Although there were attempts to redistribute the energy metabolism optimized for industrial condition through metabolic engineering or genome minimization, it can be said that optimizing complex microorganism metabolism for industrial purposes is still at an early stage (Kolisnychenko et al., 2002; Choe et al., 2019).
The flagellum is a motor that allows bacteria to swim in a liquid and swarm or attach to the surface (Kearns, 2010). Escherichia coli typically consists of multiple flagella that make it possible to take action to reach or move away from specific substances (Macnab, 1999). Oxygen and nutrients are the attractants while aromatic or polyamine compounds are the repellents targeted by flagellar movement (Bibikov et al., 1997; Rebbapragada et al., 1997; Bren and Eisenbach, 2000). Flagella contain a thin and long structured filament composed of many proteins (Macnab, 2003). Therefore, a significant amount of energy is needed to synthesize each flagellum and make it move (Macnab, 1996). Under industrial conditions, where agitation or shaking is active, the need for flagella is considerably lower, so the flagella can be removed to save NADPH or ATP. The saved NADPH or ATP can be used in the generation of target products, such as recombinant proteins (Lim et al., 2002; Lee et al., 2007; Kim et al., 2012).
Previously, we constructed glucose transporter mutants, such as ptsG, in the E. coli W strain and demonstrated that these mutations were beneficial for the improved yield of many target products with reductions in overflow metabolism. When ptsG was knocked out, several metabolic flux responses such as reduced glucose uptake rate, upregulated tricarboxylic acid (TCA) cycle, and suppressed acetate production were observed, resulting in an increased yield of the recombinant protein (Jung et al., 2019). In this study, flhC was deleted in the constructed ptsG knockout mutant. The flhC gene encodes a subunit of the FlhC master regulator for flagellar assembly (Fitzgerald et al., 2014; Sim et al., 2017). The ptsG and flhC double knockout mutants showed growth retardation compared to the ptsG knockout mutant. Interestingly, by transfecting a high copy number plasmid into this mutant, the growth rate was restored. The strains were evaluated using 13C metabolic flux analysis (13C-MFA) in order to explain the restored growth rate and high recombinant protein yield.
Materials and Methods
Strains and Plasmids
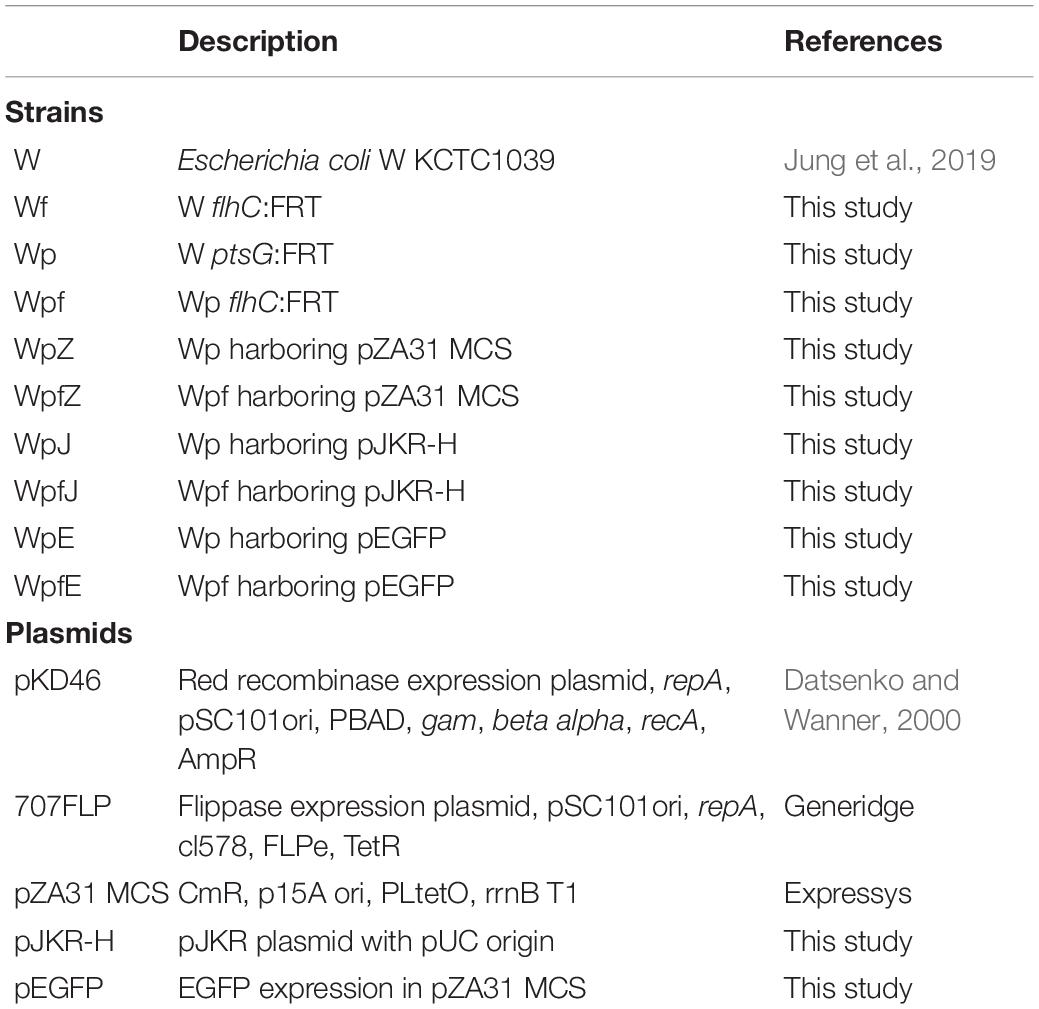
All strains and plasmids used in this study are listed in Table 1. The host strain in this study was E. coli W (KCTC1039), which was provided by the Korean Collection for Type Cultures (KCTC; Jeongeup, Korea). Sugar transporter mutation was performed by deleting ptsG (ADT74705), and flagella mutation was performed by deleting flhC (ADT75524). All deletion methods utilized λ-red recombinase-based homologous recombination (Datsenko and Wanner, 2000). The name of the ptsG knockout mutant was Wp and that of the flhC and ptsG double mutant was Wpf. All deletions were confirmed via polymerase chain reaction using genomic DNA as a template. All oligonucleotides used in this study were synthesized by Bionics (Seoul, Korea) and are listed in Supplementary Table 1.
Medium and Cultivation
During genetic engineering, strains were cultivated and confirmed in Lysogeny broth (10 g/L of tryptone, 10 g/L of NaCl, and 5 g/L of yeast extract). All cultivations for analysis were performed in M9 minimal medium (6 g/L of Na2HPO4, 3 g/L of KH2PO4, 1 g/L of NH4Cl, 0.5 g/L of NaCl, and 0.01% Thiamine–HCl) with 20 g/L of glucose and 1 mL of trace elements [2.86 g/L of H3BO3, 1.81 g/L of MnCl2⋅4H2O, 0.9 g/L of FeCl3⋅6H2O, 0.39 g/L of Na2MoO4⋅2H2O, 0.222 g/L of ZnSO4⋅7H2O, 0.079 g/L of CuSO4⋅5H2O, and 49.4 mg/L of Co(NO3)2⋅6H2O]. The antibiotics used were carbenicillin (100 μg/mL), kanamycin (50 μg/mL), and chloramphenicol (34 μg/mL). All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, United States).
Cultivation was performed in 250 mL Erlenmeyer flasks with 25 mL of working volume at 250 rpm and 37°C under aerobic conditions.
Analytical Methods
Bacterial cell mass was estimated via measurement of optical density at 600 nm (OD600) using a DU730 (Beckman Coulter, Brea, CA, United States). Measurements of glucose and acetate were performed using high-performance liquid chromatography (Waters, Milford, MA, United States) with a Refractive Index Detector 2414 and SH1011 columns (Shodex, Tokyo, Japan). The measurement was carried out at a temperature of 45°C, and 10 mM sulfuric acid was used as the mobile phase and the flow rate was 0.6 mL/min.
Enhanced green fluorescent protein (EGFP) intensity was measured using a microplate reader (Synergy H1; Biotek, Winooski, VT, United States). The cultured bacterial strains were washed with phosphate-buffered saline (PBS) solution and diluted. The measurement result was multiplied by the dilution ratio. The achieved excitation peak was at 479 nm, and the detected emission peak was at 520 nm.
Measurements of the ATP/ADP and NADPH/NADP+ Ratios
For the ATP assay, BacTiter-GloTM (Promega, Fitchburg, United States) was used. The culture broth was harvested in the early exponential phase (OD600nm ≈ 1.0). First, 100 μL of harvested culture broth was mixed with an equal volume of BacTiter-GloTM reagent. The mixed samples were incubated at room temperature for 5 min. Luminescence was measured using a microplate reader (Synergy H1). The luminescence unit was converted to μM using a standard curve, and the result was divided by dry cell weight. Therefore, the ATP concentration was calculated as μmol/gcell.
For the NADPH/NADP+ assay, NADP/NADPH-GloTM (Promega, Fitchburg, United States) was used. Harvested cell of early exponential phase in 50 μL of PBS was lysed in 50 μL of 0.2 M NaOH and 1% (w/v) dodecyl trimethyl ammonium bromide (DTAB). Then, 25 μL of 0.4 M HCl was treated at 60°C for 15 min. Next, 25 μL of 0.5 M Trizma base was added for neutralization, and NADP/NADPH-GloTM reagents were also added. Samples were incubated at room temperature for 30 min. Luminescence was measured using a microplate reader (Synergy H1). PBS, NaOH, DTAB, HCl, and Trizma base were from Sigma-Aldrich (St. Louis, MO, United States).
Metabolite Analysis for 13C-MFA
For 13C-MFA, [1,2-13C]-glucose was added to the M9 medium. The 1 mL of cell broth in the early exponential phase (OD600nm ≈ 1.0) was harvested and centrifuged at 15,000 × g for 10 min at 4°C to remove the supernatant. The obtained pellet was washed twice with distilled water and then fully dried with a freeze dryer (Hail, Gimpo, Korea). Then, 200 μL of 6 N HCl was added and hydrolyzed at 110°C for 24 h. After hydrolysis, sample neutralization was performed by adding 200 μL of 6 N NaOH and filtered using an Amicon Ultra 0.5 mL 10 kDa centrifugation filter (Millipore Corporation, Burlington, MA). Next, each sample was derivatized with 80 μL of N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide and 50 μL of pyridine for 1 h at 70°C.
Samples were measured using gas chromatography-mass spectrometry (Agilent 7890 B GC system) with an HP-5MS column (0.25 μm, 30 m × 0.25 mm; Agilent Technology, Santa Clara, CA, United States). The initial temperature was 80°C, which was raised to 280°C at a rate of 7°C/min and held for 10 min. One microliter of the sample was injected at 270°C in 1:10 split mode, and a flow rate of 1 mL/min of helium was used for the mobile phase. The ion source temperature was 230°C, and the electron impact was ionized at 70 eV. Measurement data were analyzed by single ion monitoring, while MassHunter was used for mass spectra, and mass isotopomer distributions (MIDs) were obtained (Antoniewicz et al., 2007a).
13C-MFA
The network model for the flux analysis construction was conducted based on a previous report (Supplementary Table 2) (Leighty and Antoniewicz, 2013). INCA software, based on the elementary metabolite unit for 13C-MFA, was used (Antoniewicz et al., 2007b; Young, 2014). Flux estimation was performed by minimization of the differences between the simulated ones and the MIDs of the proteinogenic amino acids from experimental measurements using least squares regression. Flux estimations were performed 10 times to find the global solution. Chi-square statistical tests were performed for goodness of fit (Im et al., 2020). The metabolic fluxes were calculated from the MIDs of proteinogenic amino acids with an acceptable sum of squared residuals (SSR) (Supplementary Table 3). The SSRs were 13.1 (expected SSR range: 5.6–26.1) and 34.6 (expected SSR range: 16.8–47.0) in Wp and Wpf, respectively.
Results
Effects of flhC Deletion
Deletion of flhC was expected to reduce unnecessary protein production and ATP consumption (Shachrai et al., 2010). First, we deleted the flhC gene in the E. coli W strain (KCTC1039) and named it Wf. Previously, we demonstrated that the ptsG knockout mutant from the W strain (KCTC1039), Wp, showed improved protein production (Jung et al., 2019). Therefore, the flhC gene was deleted in the Wp strain to construct the strain named Wpf.
Cultivation was performed in a shake flask under aerobic conditions in glucose M9 minimal medium at 37°C. Growth retardations with increased lag phase were observed in Wp and Wpf compared to W and Wf (Figure 1A), although the final ODs were similar after 24 h. The amount of glucose consumption was higher in W and Wf, whereas it was significantly decreased in Wpf. In the Wpf strain, the specific glucose uptake rate was also decreased compared to other strains (Figure 1B). These results indicate that Wpf experienced significant metabolic burden by knocking out both ptsG and flhC. However, the flhC knockout strain in the wild-type strain, Wf, did not show any phenotypic differences from the wild-type strain W. To identify the burden, intracellular cofactors in each strain were measured.
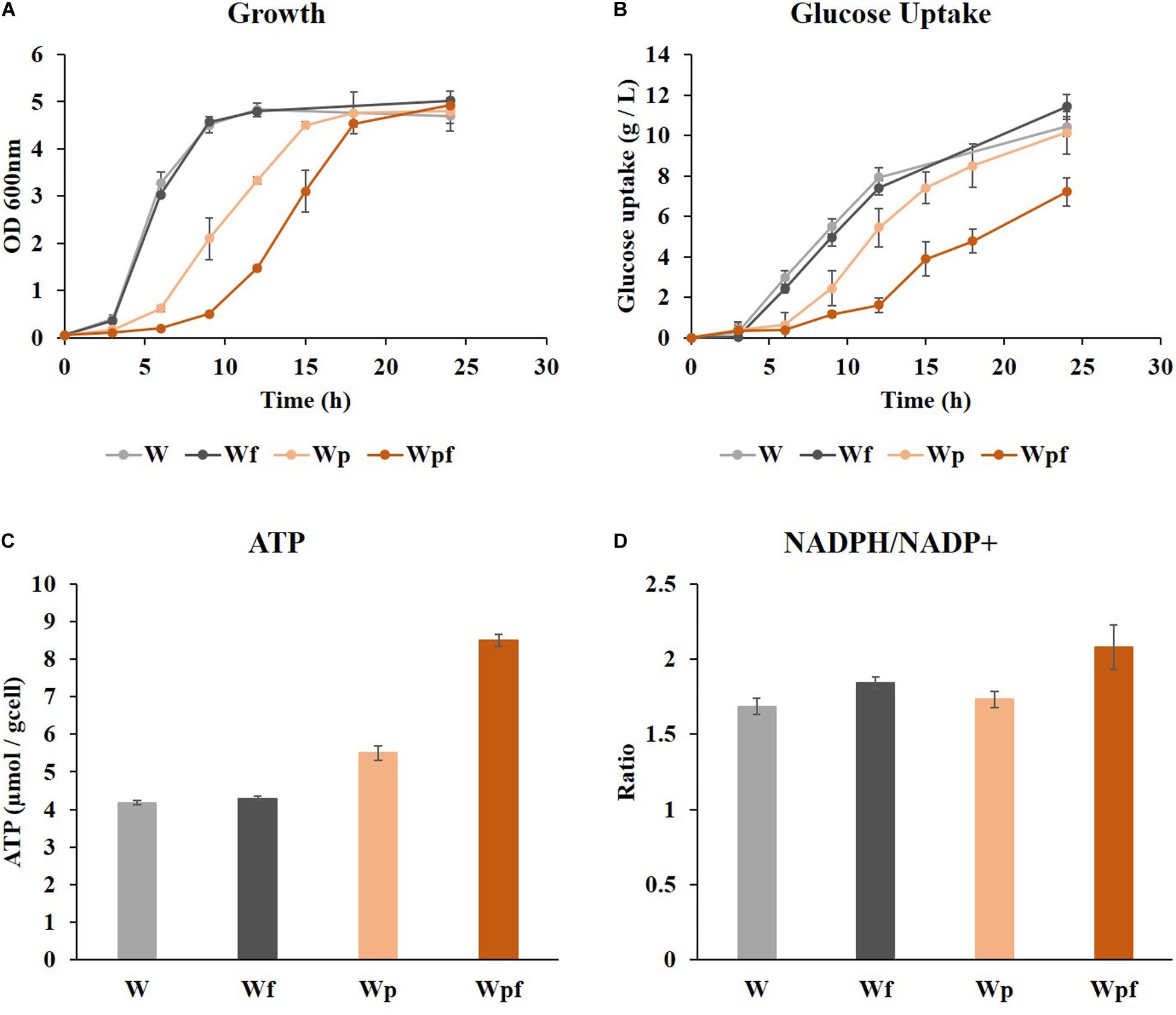
Figure 1. Wild-type (W), flhC (Wf), ptsG (Wp), and ptsG/flhC deletion mutant (Wpf) cultivation in flasks with M9 glucose medium. Growth profiles (A), glucose concentrations (B), specific ATP concentrations (C), and NADPH/NADP+ ratios (D) of the W, Wf, Wp, and Wpf strains are presented. These results are the average values of the experiment repeated three times.
ATP levels and NADPH/NADP+ ratios were measured to see how these factors were affected by the mutations. Slight increases in the ATP concentration and NADPH/NADP+ ratio were observed in the Wf strain compared to those in the W strain (Figures 1C,D). However, the same flhC deletion in the ptsG knockout mutant caused a much larger effect. The ATP level was increased by 1.5-fold, and the NADPH/NADP+ ratio also increased from 1.73 to 2.08 in Wpf compared to Wp. These differences would be significant enough to cause cofactor imbalance, which could result in growth retardation and reduced glucose uptake. The NADH/NAD+ ratios were also measured in these strains, which showed little difference between them. To clarify the cofactor imbalance in more detail, 13C MFA was applied to the Wp and Wpf strains.
13C-MFA of the flhC and ptsG Mutant
13C-MFA was conducted to investigate the metabolic burden of the Wpf strain. The metabolic fluxes of Wpf were compared to those of Wp. The fluxes of the glycolytic pathway were very similar between the two strains. And the fluxes of Entner Doudoroff pathway from 6-phosphogluconate to 2-Keto-3-deoxy-6-phosphogluconate (KDPG) were zero at both two strains. However, the fluxes of the pentose phosphate pathway were changed (Figure 2A). The flux from 6-phosphogluconate to ribose-5-phosphate, the first step in the pentose phosphate pathway, increased from 35.62 to 40.88 in Wpf compared to Wp, and all other fluxes of the subsequent pentose phosphate pathway were increased. Other significant changes in central carbon metabolic fluxes were observed in the TCA pathway. For example, the flux of acetyl CoA to citrate, the first step of the TCA cycle, increased from 55.88 to 71.55 in Wpf compared to Wp, and the fluxes of all other TCA cycle pathways increased in Wpf compared to Wp (Figure 2A).
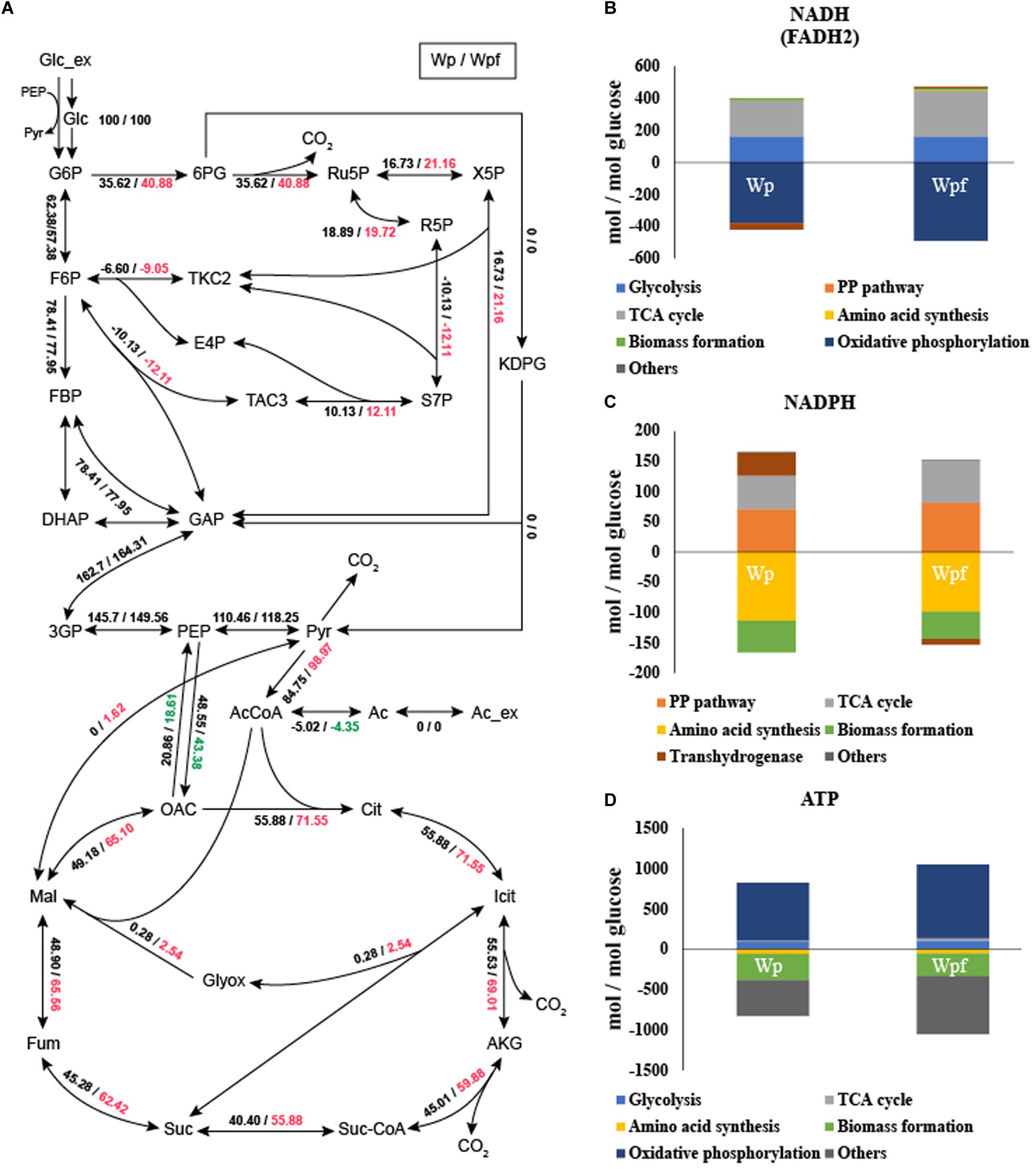
Figure 2. Flux distribution and cofactor balance analysis of the Wp and Wpf strains measured by 13C MFA. The relative carbon fluxes of the two strains (A) were presented as numbers separated by dashes. The generation (upper) and consumption (lower) pathways of NADH (FADH2) (B), NADPH (C), and ATP (D) were calculated by balance analysis. These results are the average values of the three different culture broths. Numbers marked in red and green indicate a value that has increased or decreased by more than 10% compared to Wp strain, respectively.
Based on the carbon flux results, cofactor generation and consumption were also calculated (He et al., 2014; Long et al., 2017, 2018). The results are also showed in Supplementary Table S4. With the increased TCA cycle and pentose phosphate pathway fluxes, the fluxes of NADH (FADH2) and NADPH generation also increased 1.28- and 1.2-fold, respectively (Figures 2B,C). Increased NADH levels were used for oxidative phosphorylation. However, despite the increase in NADPH, the NADPH consumption flux for amino acid synthesis and biomass formation decreased by 0.87-fold. Due to this cellular redox imbalance, the flux of transhydrogenase was also changed (Canonaco et al., 2001). In the case of Wp, the flux for converting NADH to NADPH was 38.0173, whereas for Wpf, the flux for converting NADPH to NADH was observed to be 9.1855 (Figures 2B,C). The significant change in transhydrogenase flux was due to cellular redox imbalance, which might be caused by the excessive accumulation of NADPH in the Wpf strain by not forming flagella.
In addition, due to the increased TCA cycle flux, the ATP generation flux of the TCA cycle changed (Figure 2D). It increased 2.27-fold in Wpf compared to Wp. Subsequently, the increased cofactors were converted to ATP by oxidative phosphorylation. The flux of ATP generated by oxidative phosphorylation increased 1.28-fold in Wpf compared to Wp. Finally, at the section of others that are said to be surplus ATP, the flux increased 1.63-fold in Wpf compared to Wp. Large amounts of ATP resulted from deleting flhC.
The results of 13C-MFA similarly explain the changes in ATP levels and NADPH/NADP+ ratios shown in Figures 1C,D. A previous report showed that artificially increased intracellular ATP retarded cell growth (Na et al., 2015). Thus, we suspected that Wpf with high levels of ATP and NADPH induced growth retardation and reduced glucose consumption.
Growth Restoration by Plasmid Harboring and Overcoming the Metabolic Burden
To reduce the metabolic burden caused by surplus production of ATP and NADPH in the Wpf strain, plasmid replications and protein overexpression were attempted. Plasmids pJKR-H and pZA31 MCS were transformed into the Wpf strain, and the resulting strains were named WpfJ and WpfZ, respectively. In addition, a gene encoding GFP was cloned into pZA31 MCS, and the resulting plasmid was transformed into Wpf, constructing the WpfE strain. The strains were cultivated in a shake flask in M9 glucose medium at 37°C. All three strains, WpfJ, WpfZ, and WpfE, showed growth restoration compared to the Wpf strain (Figure 3A). The growth restoration effect was most significant in the WpfE strain. Interestingly, glucose uptake patterns of those strains were about the same as the Wpf strain, while having much lower glucose uptake than the Wp strain (Figures 1B, 3B).
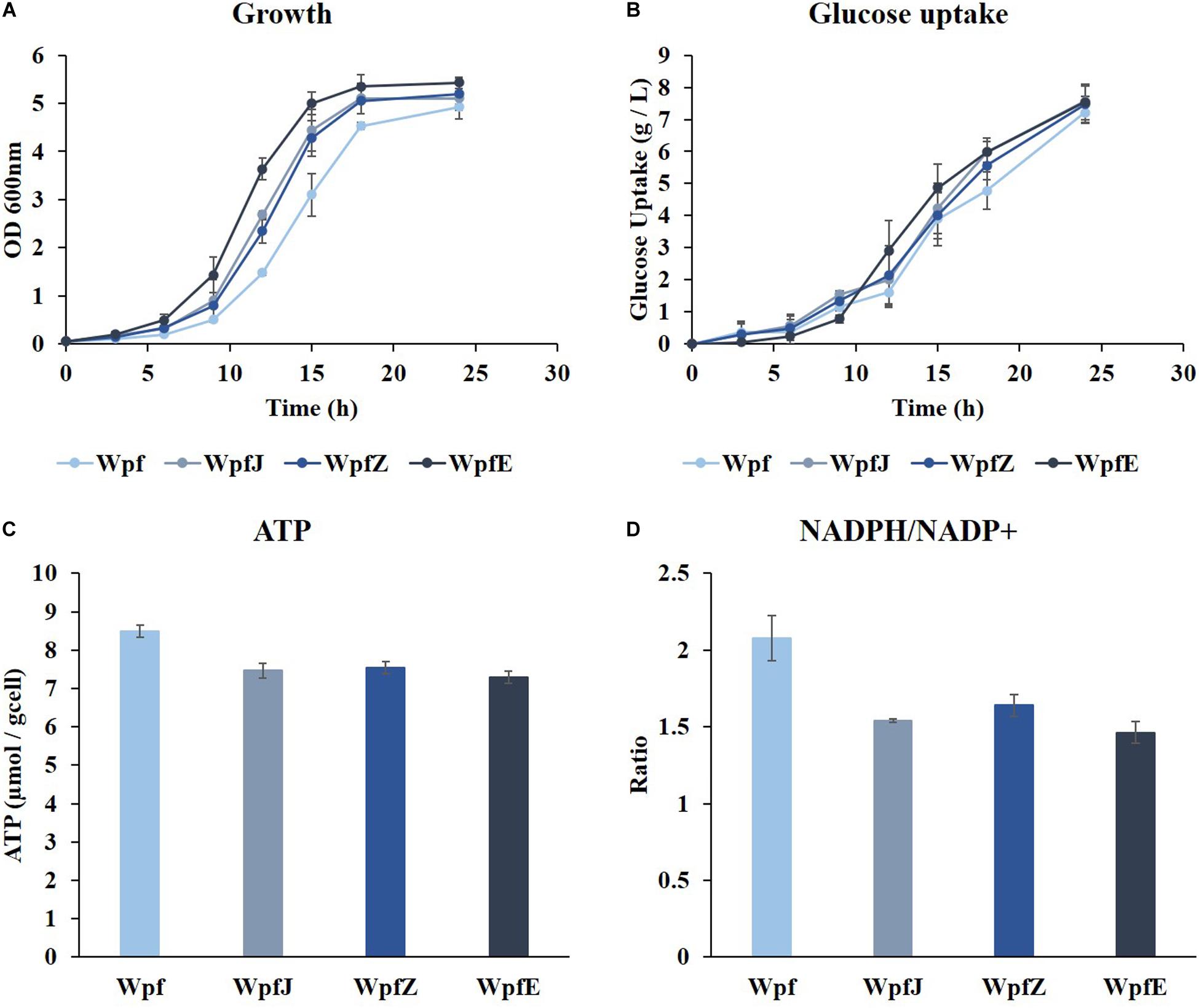
Figure 3. The cultivation results of Wpf, the pJKR-H-harboring strain (WpfJ), the pZA31 MCS-harboring strain (WpfZ), and the pEGFP-harboring strain (WpfE) in flasks with M9 glucose medium. Growth profiles (A), glucose concentrations (B), specific ATP concentrations (C), and NADPH/NADP+ ratios (D) of the Wpf, WpfJ, WpfZ, and WpfE strains are presented. These results are the average values of the experiment repeated three times.
To investigate the metabolic burden of these strains, ATP levels and NADPH/NADP+ ratios were measured. The ATP levels decreased by 0. 88-, 0. 89-, and 0.86-fold in WpfJ, WpjZ, and WpfE, respectively, compared to Wpf (Figure 3C). The NADPH/NADP+ ratios also decreased from 2.08 in the Wpf strain to 1.54, 1.64, and 1.46 in WpfJ, WpjZ, and WpfE, respectively (Figure 3D). It is likely that the growth of the Wpf strain was recovered due to the consumption of excess ATP and NADPH by plasmid replication and heterologous protein expression.
Recombinant Protein Production by the Flagella Knockout Mutant
The excess ATP and NADPH accumulation in cells can be beneficial for recombinant protein production (Lim et al., 2002; Lee et al., 2007; Kim et al., 2012). For example, excess ATP and NADPH were synthesized with a reduced amount of glucose consumption in the Wpf strain. When heterologous protein was expressed in the Wpf strain, EGFP was produced without increased glucose consumption. This means that the yield of protein synthesis per unit of glucose can be increased significantly in the Wpf strain. To verify the effect of flhC knockout on the yield of protein synthesis, the WpE and WpfE strains were constructed by transforming the plasmid-expressing EGFP into the Wp and Wpf strains, which were cultivated in a shake flask in glucose M9 medium at 37°C.
While growth was restored in WpfE compared to Wpf (Figure 3A and Supplementary Figure 1A), it was decreased in WpE compared to Wp (Supplementary Figure 1B). The growth rate of WpfE was comparable to that of WpE with significantly lower glucose consumption (Figures 4A,B). The glucose uptake decreased from 12.49 g/L to 7.54 g/L in WpfE compared to WpE, despite the similar cell densities reached in both strains. These results suggest that the metabolism of WpfE was much more efficient than that of WpE in order to produce biomass and protein with a unit of glucose. The EGFP yield per consumed glucose increased by 1.81-fold in WpfE compared to WpE (Figure 4C). The WpfE strain showed even higher protein production yield than Wp, which outperformed the wild-type strain in terms of protein yield (Jung et al., 2019).
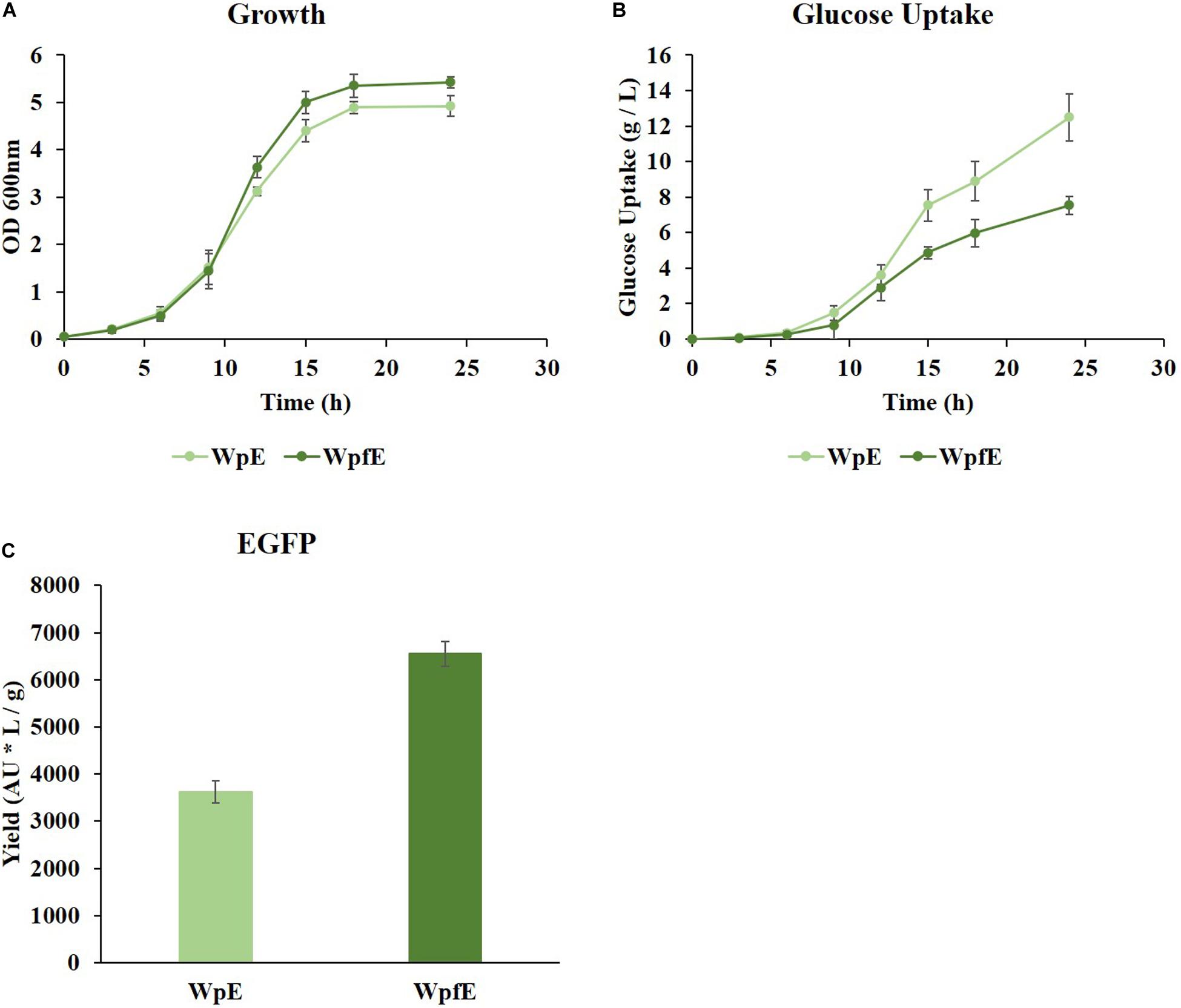
Figure 4. The cultivation results of WpE and WpfE in flasks with M9 glucose medium. The growth profiles (A), glucose concentrations (B), and yields of EGFP production (C) are presented. Each yield was calculated by dividing the fluorescence intensity (AU) of EGFP by the glucose consumption (g/L) at 24 h. These results are the average values of the experiment repeated three times.
Discussion
In this study, flagella were knocked out to construct an efficient E. coli strain for energy utilization. First, the ptsG knockout mutant Wp, confirmed to have increased production of recombinant protein, was constructed (Jung et al., 2019). Additionally, the flhC flagella-related gene was deleted from Wp to create the Wpf strain. Growth retardation was observed in the Wpf strain compared to that in Wp. Using cofactor measurement and 13C MFA, the accumulation of ATP and NADPH saved from flagella assembly was found to be the reason. To address this, several plasmids were transformed to consume ATP and NADPH, and growth was restored (Kaleta et al., 2013). Moreover, when a protein was overexpressed using the plasmids, the protein production yield increased. This result contradicts previous studies showing that a metabolic burden arises when a wild-type strain harbors a plasmid (Diaz Ricci and Hernández, 2000; Englaender et al., 2017). Our strategy can be seen as overcoming the growth retardation and inhibition caused by protein expression or high-copy plasmids that introduced a metabolic burden via flagella deletion (Diaz Ricci and Hernández, 2000; Englaender et al., 2017). In particular, the 13C MFA results and cofactor measurements suggested the metabolic burden and helped to find a strategy to resolve the problem.
Interestingly, no significant differences in phenotypes were found when flhC was knocked out in the wild-type strain. Large amount of ATP level increase and growth retardation and restoration by flhC knockout occurred only in the ptsG knockout strain. We assumed that the reason for this phenomenon is the C-reactive protein (CRP) mechanism. Previous research has revealed that flagellar biosynthesis is activated by CRP-cAMP (Zhao et al., 2007; Fahrner and Berg, 2015). CRP is activated by binding with cAMP when a preferred carbon source, such as glucose, is depleted (Ishizuka et al., 1994). In the ptsG-deleted strain, glucose transport by PTS decreases, and CRP activity rises (Steinsiek and Bettenbrock, 2012). Therefore, in ptsG knockout mutants, flagella synthesis and activities may be increased compared to the wild-type strain. In this situation, flhC deletion can result in energy saving and significant differences in cell growth and cofactor levels. Because of the beneficial effect of ptsG deletion, the mutant is being used more and more widely in industrial strain development (Chou et al., 1994; Chatterjee et al., 2001; Wang et al., 2006). This experiment provides a useful strategy for metabolic engineering based on the ptsG mutant.
The flhC deletion in the ptsG mutant restored the growth rate but not glucose consumption. As a result of 13C MFA, the flhC deletion in the ptsG mutant redistributed the carbon flux from glucose toward the pentose phosphate and TCA cycle pathways. Because more ATP and NAD(P)H could be produced from the same amount of glucose, it was thought that the uptake of glucose was reduced. However, the mechanism of flux redistribution remains unclear. One possibility is that flhC is known to regulate the MglBAC transporter (Prüß et al., 2001), which is one of the major glucose uptake transporters in ptsG mutants (Nikolic et al., 2013). Therefore, glucose uptake may be reduced by suppressing MglBAC expression through flhC deletion.
In conclusion, metabolic perturbation through genetic engineering related to glucose transporter and flagella synthesis was studied, and a strategy for high-yield production of recombinant proteins was proposed. Previously, the production of recombinant proteins in ptsG-deleted strains has been suggested to resolve overflow metabolism and improve production yield. In this experiment, intracellular metabolic flux and cofactor generation were further perturbed by flhC deletion, which resulted in an even higher yield of protein production. This result could make a significant contribution to strain development for protein production.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
J-HH, SJ, and M-KO conceived the idea. J-HH performed the experiment. J-HH and M-KO wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by a the Korea University Grant and the National Research Foundation of Korea, funded by the Korean Government (2012M1A2A2026560).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Jaeseung Hong for help with the experiments and the Matlab program for 13C-MFA.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.655072/full#supplementary-material
References
Antoniewicz, M. R., Kelleher, J. K., and Stephanopoulos, G. (2007a). Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal. Chem. 79, 7554–7559. doi: 10.1021/ac0708893
Antoniewicz, M. R., Kelleher, J. K., and Stephanopoulos, G. (2007b). Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab. Eng. 9, 68–86. doi: 10.1016/j.ymben.2006.09.001
Bibikov, S. I., Biran, R., Rudd, K. E., and Parkinson, J. S. (1997). A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179, 4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997
Blais, B. W., Booth, R. A., Phillippe, L. M., and Yamazaki, H. (1997). Effect of temperature and agitation on enrichment of Escherichia coli O157: H7 in ground beef using modified EC broth with novobiocin. Int. J. Food Microb. 36, 221–225. doi: 10.1016/s0168-1605(97)01274-9
Bren, A., and Eisenbach, M. (2000). How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J. Bacteriol. 182, 6865–6873. doi: 10.1128/jb.182.24.6865-6873.2000
Canonaco, F., Hess, T. A., Heri, S., Wang, T., Szyperski, T., and Sauer, U. (2001). Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microb. Lett. 204, 247–252. doi: 10.1111/j.1574-6968.2001.tb10892.x
Chatterjee, R., Millard, C. S., Champion, K., Clark, D. P., and Donnelly, M. I. (2001). Mutation of the ptsG gene results in increased production of succinate in fermentation of glucose by Escherichia coli. Appl. Env. Microb. 67, 148–154. doi: 10.1128/aem.67.1.148-154.2001
Choe, D., Lee, J. H., Yoo, M., Hwang, S., Sung, B. H., Cho, S., et al. (2019). Adaptive laboratory evolution of a genome-reduced Escherichia coli. Nat. Commun. 10, 1–14.
Chou, C. H., Bennett, G. N., and San, K. Y. (1994). Effect of modified glucose uptake using genetic engineering techniques on high−level recombinant protein production in Escherichia coli dense cultures. Biotechnol. Bioeng. 44, 952–960. doi: 10.1002/bit.260440811
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97, 6640–6645. doi: 10.1073/pnas.120163297
Diaz Ricci, J. C., and Hernández, M. E. (2000). Plasmid effects on Escherichia coli metabolism. Crit. Rev. Biotechnol. 20, 79–108. doi: 10.1016/0147-619x(79)90008-8
Englaender, J. A., Jones, J. A., Cress, B. F., Kuhlman, T. E., Linhardt, R. J., and Koffas, M. A. (2017). Effect of genomic integration location on heterologous protein expression and metabolic engineering in E. coli. ACS Synthetic Biol. 6, 710–720. doi: 10.1021/acssynbio.6b00350
Fahrner, K. A., and Berg, H. C. (2015). Mutations that stimulate flhDC expression in Escherichia coli K-12. J. Bacteriol. 197, 3087–3096.
Fitzgerald, D. M., Bonocora, R. P., and Wade, J. T. (2014). Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet. 10:e1004649. doi: 10.1371/journal.pgen.1004649
He, L., Xiao, Y., Gebreselassie, N., Zhang, F., Antoniewicz, M. R., Tang, Y. J., et al. (2014). Central metabolic responses to the overproduction of fatty acids in Escherichia coli based on 13C−metabolic flux analysis. Biotechnol. Bioeng. 111, 575–585. doi: 10.1002/bit.25124
Im, D. K., Hong, J., Gu, B., Sung, C., and Oh, M. K. (2020). 13C Metabolic flux analysis of Escherichia coli engineered for gamma−aminobutyrate production. Biotechnol. J. 2020:1900346. doi: 10.1002/biot.201900346
Ishizuka, H., Hanamura, A., Inada, T., and Aiba, H. (1994). Mechanism of the down−regulation of cAMP receptor protein by glucose in Escherichia coli: role of autoregulation of the crp gene. EMBO J. 13, 3077–3082. doi: 10.1002/j.1460-2075.1994.tb06606.x
Jung, H.-M., Im, D.-K., Lim, J. H., Jung, G. Y., and Oh, M.-K. (2019). Metabolic perturbations in mutants of glucose transporters and their applications in metabolite production in Escherichia coli. Microb. Cell Fact. 18:170.
Kaleta, C., Schäuble, S., Rinas, U., and Schuster, S. (2013). Metabolic costs of amino acid and protein production in Escherichia coli. Biotechnol. J. 8, 1105–1114. doi: 10.1002/biot.201200267
Kearns, D. B. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microb. 8, 634–644. doi: 10.1038/nrmicro2405
Kim, H.-J., Kwon, Y. D., Lee, S. Y., and Kim, P. (2012). An engineered Escherichia coli having a high intracellular level of ATP and enhanced recombinant protein production. Appl. Microb. Biotechnol. 94, 1079–1086. doi: 10.1007/s00253-011-3779-0
Kolisnychenko, V., Plunkett, G., Herring, C. D., Fehér, T., Pósfai, J., Blattner, F. R., et al. (2002). Engineering a reduced Escherichia coli genome. Genome Res. 12, 640–647. doi: 10.1101/gr.217202
Lee, W.-H., Park, J.-B., Park, K., Kim, M.-D., and Seo, J.-H. (2007). Enhanced production of ε-caprolactone by overexpression of NADPH-regenerating glucose 6-phosphate dehydrogenase in recombinant Escherichia coli harboring cyclohexanone monooxygenase gene. Appl. Microb. Biotech. 76, 329–338. doi: 10.1007/s00253-007-1016-7
Leighty, R. W., and Antoniewicz, M. R. (2013). COMPLETE-MFA: complementary parallel labeling experiments technique for metabolic flux analysis. Metab. Eng. 20, 49–55. doi: 10.1016/j.ymben.2013.08.006
Lim, S.-J., Jung, Y.-M., Shin, H.-D., and Lee, Y.-H. (2002). Amplification of the NADPH-related genes zwf and gnd for the oddball biosynthesis of PHB in an E. coli transformant harboring a cloned phbCAB operon. J. Biosci. Bioeng. 93, 543–549. doi: 10.1016/s1389-1723(02)80235-3
Long, C. P., Gonzalez, J. E., Feist, A. M., Palsson, B. O., and Antoniewicz, M. R. (2017). Fast growth phenotype of E. coli K-12 from adaptive laboratory evolution does not require intracellular flux rewiring. Metab. Eng. 44, 100–107. doi: 10.1016/j.ymben.2017.09.012
Long, C. P., Gonzalez, J. E., Feist, A. M., Palsson, B. O., and Antoniewicz, M. R. (2018). Dissecting the genetic and metabolic mechanisms of adaptation to the knockout of a major metabolic enzyme in Escherichia coli. Proc.Natl. Acad. Sci. 115, 222–227. doi: 10.1073/pnas.1716056115
Macnab, R. (1996). “Flagella and Motility” in Escherichia coli and Salmonella ed Neidhardt, F. C., Washington, DC: American Society for Microbiology, 123–145.
Macnab, R. M. (1999). The bacterial flagellum: reversible rotary propellor and type III export apparatus. J. Bacteriol. 181, 7149–7153. doi: 10.1128/jb.181.23.7149-7153.1999
Macnab, R. M. (2003). How bacteria assemble flagella. A. Rev. Microb. 57, 77–100. doi: 10.1146/annurev.micro.57.030502.090832
Na, Y.-A., Lee, J.-Y., Bang, W.-J., Lee, H. J., Choi, S.-I., Kwon, S.-K., et al. (2015). Growth retardation of Escherichia coli by artificial increase of intracellular ATP. J. Indust. Microb. Biotechnol. 42, 915–924. doi: 10.1007/s10295-015-1609-6
Nikolic, N., Barner, T., and Ackermann, M. (2013). Analysis of fluorescent reporters indicates heterogeneity in glucose uptake and utilization in clonal bacterial populations. BMC Microb. 13:258. doi: 10.1186/1471-2180-13-258
Prüß, B. M., Liu, X., Hendrickson, W., and Matsumura, P. (2001). FlhD/FlhC-regulated promoters analyzed by gene array and lacZ gene fusions. FEMS Microb. Lett. 197, 91–97. doi: 10.1016/s0378-1097(01)00092-1
Rebbapragada, A., Johnson, M. S., Harding, G. P., Zuccarelli, A. J., Fletcher, H. M., Zhulin, I. B., et al. (1997). The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. 94, 10541–10546. doi: 10.1073/pnas.94.20.10541
Shachrai, I., Zaslaver, A., Alon, U., and Dekel, E. (2010). Cost of unneeded proteins in E. coli is reduced after several generations in exponential growth. Mole. Cell 38, 758–767. doi: 10.1016/j.molcel.2010.04.015
Sim, M., Koirala, S., Picton, D., Strahl, H., Hoskisson, P. A., Rao, C. V., et al. (2017). Growth rate control of flagellar assembly in Escherichia coli strain RP437. Sci. Rep. 7, 1–11.
Steinsiek, S., and Bettenbrock, K. (2012). Glucose transport in Escherichia coli mutant strains with defects in sugar transport systems. J. Bacteriol. 194, 5897–5908. doi: 10.1128/jb.01502-12
Wang, Q., Wu, C., Chen, T., Chen, X., and Zhao, X. (2006). Expression of galactose permease and pyruvate carboxylase in Escherichia coli ptsG mutant increases the growth rate and succinate yield under anaerobic conditions. Biotech. Lett. 28, 89–93. doi: 10.1007/s10529-005-4952-2
Young, J. D. (2014). INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics 30, 1333–1335. doi: 10.1093/bioinformatics/btu015
Keywords: flagella, recombinant protein, ATP, NADPH, 13C metabolic flux analysis
Citation: Han J-H, Jung ST and Oh M-K (2021) Improved Yield of Recombinant Protein via Flagella Regulator Deletion in Escherichia coli. Front. Microbiol. 12:655072. doi: 10.3389/fmicb.2021.655072
Received: 18 January 2021; Accepted: 22 February 2021;
Published: 15 March 2021.
Edited by:
Shashi Kant Bhatia, Konkuk University, South KoreaReviewed by:
Sujit Jagtap, University of Illinois at Urbana-Champaign, United StatesVijay Kumar, Institute of Himalayan Bioresource Technology (CSIR), India
Copyright © 2021 Han, Jung and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min-Kyu Oh, bWtvaEBrb3JlYS5hYy5rcg==
 Jae-Ho Han
Jae-Ho Han Sang Taek Jung
Sang Taek Jung Min-Kyu Oh
Min-Kyu Oh