
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 March 2021
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.653354
This article is part of the Research Topic Genome-wide Analyses of Pectobacterium and Dickeya Species View all 7 articles
 Chao Yu1†
Chao Yu1† Doan-Phuong Nguyen1†
Doan-Phuong Nguyen1† Fenghuan Yang1
Fenghuan Yang1 Jia Shi1
Jia Shi1 Yiming Wei1
Yiming Wei1 Fang Tian1
Fang Tian1 Xiuxiang Zhao2*
Xiuxiang Zhao2* Huamin Chen1*
Huamin Chen1*σ54 factor (RpoN) plays a crucial role in bacterial motility, virulence, growth, and other biological functions. In our previous study, two homologous σ54 factors, RpoN1 and RpoN2, were identified in Xanthomonas oryzae pv. oryzae (Xoo), the causative agent of bacterial leaf blight in rice. However, their functional roles, i.e., whether they exert combined or independent effects, remain unknown. In the current study, rpoN1 or rpoN2 deletion in Xoo significantly disrupted bacterial swimming motility, flagellar assembly, and virulence. Transcriptome analysis led to the identification of 127 overlapping differentially expressed genes (DEGs) regulated by both RpoN1 and RpoN2. Furthermore, GO and KEGG classification demonstrated that these DEGs were highly enriched in flagellar assembly, chemotaxis, and c-di-GMP pathways. Interestingly, ropN1 deletion decreased ropN2 transcription, while rpoN2 deletion did not affect ropN1 transcription. No interaction between the rpoN2 promoter and RpoN1 was detected, suggesting that RpoN1 indirectly regulates rpoN2 transcription. In addition, RpoN1-regulated DEGs were specially enriched in ribosome, carbon, and nitrogen metabolism pathways. Besides, bacterial growth was remarkably repressed in ΔrpoN1 but not in ΔrpoN2. Taken together, this study demonstrates the overlapping and unique regulatory roles of RpoN1 and RpoN2 in motility, virulence, growth and provides new insights into the regulatory mechanism of σ54 factors in Xoo.
Transcription regulates a myriad of biological processes. In bacteria, σ factors are the most widely occurring transcriptional regulators that reversibly bind RNA polymerase (RNAP) and regulate transcription of numerous functional genes. σ factors initiate the RNA synthesis process by guiding RNAP holoenzyme binding to specific promoters, melting double-stranded promoter DNA strands, and stabilizing it as a single-stranded open complex. Till now, only two families of σ factors, i.e., σ70 and σ54, have been reported (Kazmierczak et al., 2005). Unlike the major σ70 class, σ54 factors interact with other transcriptional activators, as also named enhancer-binding proteins (EBPs), to induce gene expression (Studholme and Dixon, 2003; Davis et al., 2017). Furthermore, σ54 factor binding sites are typically located at the −24/−12 region of the highly conserved sequence, i.e., GGN10GC, whereas that of σ70 is located at the −35/−10 region (Barrios et al., 1999; Yang et al., 2015). Due to the indispensable role of σ54 factor in the transcription process, it is essential to identify its target genes and discern its regulatory pathways.
σ54 factor, alternatively named as RpoN, has been implicated as a multifunctional regulator of multiple vital biological processes, such as nitrogen assimilation, swimming motility, biofilm formation, extracellular polysaccharides (EPS) production, virulence, and type III secretion system (Kustu et al., 1989; Yang et al., 2009; Hao et al., 2013; Lee et al., 2016). Recent studies have focused on RpoN as a major regulator of bacterial growth, carbohydrate metabolism, twitching motility, quorum sensing, and type VI secretion system (Cai et al., 2015; Hayrapetyan et al., 2015; Ray et al., 2015; Shao et al., 2018). In addition, RpoN has been associated with environmental adaptation and tobramycin resistance (Viducic et al., 2017; Xu et al., 2019). Thus, it is necessary to thoroughly understand the complex regulatory network of bacterial RpoN.
The rpoN gene is widely distributed in bacteria. The majority of bacterial species contain only one copy of the rpoN gene; however, certain bacterial species contain two or more copies of the rpoN gene. Four rpoN genes were identified in Rhodobacter sphaeroides with distinct biological functions (Poggio et al., 2002). Two functionally distinct σ54 factors, i.e., RpoN1 and RpoN2, were studied in Ralstonia solanacearum. The outcomes showed that only RpoN1 but not RpoN2 was required for virulence, twitching motility, natural competence, and growth (Lundgren et al., 2015; Ray et al., 2015). In Xanthomonas campestris, σ54 factor RpoN2 regulates flagellar biosynthesis, swimming motility, biofilm formation, EPS, and virulence. In contrast, its homologous protein RpoN1 is involved in branched-chain fatty acid production and quorum sensing (Li et al., 2020). These studies demonstrated specific regulatory roles and biological functions of σ54 factors in pathogenic bacteria.
Xanthomonas oryzae pv. oryzae (Xoo), a gram-negative bacterium and causative agent of bacterial blight, leads to severe economic loss in the field. Besides, it is used as an ideal model to study the molecular mechanisms of bacterial pathogenicity in monocot plants (Nino-Liu et al., 2006). Xoo contains several genes related to motility, chemotaxis, EPS, biofilm, and cyclic dimeric guanosine monophosphate (c-di-GMP), controlled by diverse transcriptional factors (Das et al., 2009; White and Yang, 2009; Yang et al., 2019). Two σ54 factors, RpoN1 and RpoN2, were identified in Xoo in our previous work (Tian et al., 2015). The rpoN2 gene is located in the flagellar regulon and transcribed in an operon with fleQ, encoding one of σ54 EBPs. RpoN2 controls flagellar movements and motility by regulating gene expression and virulence through an unknown mechanism (Tian et al., 2015). In addition, RpoN2 and FleQ regulate the post-translational modification of flagellin (Yu et al., 2018a). However, the unique biological functions of RpoN1 and overlapping functions of RpoN1 and RpoN2 in Xoo remain unclear.
In this study, RNA sequencing (RNA-seq) was employed to discern the regulatory networks and functional roles of RpoN1 and RpoN2 in Xoo swimming motility, virulence, and growth. Unlike σ54 factors in Ralstonia solanacearum and Xanthomonas campestris, in Xoo, RpoN1 and RpoN2 play important regulatory roles in flagellar assembly, swimming motility, chemotaxis, and c-di-GMP signal system. The results of this study will extend our understanding of the biological functions and regulatory mechanisms of bacterial σ54 factors.
Details of the bacterial strains and plasmids employed in this study are listed in Supplementary Table 1. Xoo strains were cultured in M210 liquid medium (0.8% casein enzymatic hydrolyzate, 0.4% yeast extract, 0.5% sucrose, 17.2 mM K2HPO4, 1.2 mM MgSO4⋅7H2O), PSA medium (1% tryptone, 0.5% sucrose, 0.1% glutamate, and 1.5% agar, pH 7.0), and XOM2 medium [0.18% D-(+) xylose, 14.7 mM KH2PO4, 10 mM sodium L-(+) glutamate, 5 mM MgCl2, 670 μM L-methionine, 240 μM Fe(III)-EDTA, and 40 μM MnSO4, pH 6.5] and incubated at 28°C for 24 h. Escherichia coli strains were cultured in Luria-Bertani medium (1% peptone, 1% NaCl, and 0.5% yeast extract) and incubated at 37°C for 24 h. The antibiotics ampicillin (Ap) and kanamycin (Km) were used at the concentration of 100 and 50 μg/mL, respectively.
Mutants derived from Xoo MAFF 311018 strain were generated by homologous recombination, as described previously (Yu et al., 2018a). Concisely, to amplify the left and right arms of rpoN1, fliC, fliD, fliS, fleQ, fliA, and flgRR genes, Xoo genomic DNA was subjected to PCR amplification using relevant F/R primers (Supplementary Table 2). The amplified PCR products were ligated into suicide vector pKMS1, and resulting plasmid vectors were referred to as pKM-rpoN1, pKM-fliC, pKM-fliD, pKM-fliS, pKM-fleQ, pKM-fliA, and pKM-flgRR. Later, these cloned plasmid vectors were electrophoretically transformed into Xoo. The transformants were first selected using NAN agar (1% tryptone, 0.3% peptone, 0.1% yeast extract, and 1.5% agar) containing Km, followed by continuous transfer culture in NBN broth (NAN without agar) for three times. The candidates were screened on NAS medium (NBN plus 10% sucrose). The Km sensitive mutants that grew on NAS medium were identified by PCR analysis. To generate the double mutant of rpoN1 and rpoN2, the pKM-rpoN1 plasmids were transformed into ΔrpoN2 through electroporation method, and double mutants were screened as mentioned above. To obtain the complemented strains, the full-length of rpoN1 and rpoN2 were amplified using appropriate F/R primers (Supplementary Table 2) and ligated into the pBBR1MCS-4 vector. The resulting recombinant plasmids, pBBR-rpoN1, and pBBR-rpoN2, were electroporated into the relevant mutants. Lastly, the complemented strains were validated using PCR.
Xoo cells were cultured in M210 medium at 28°C till the culture reached the optical density of 1.0 at 600 nm (OD600). The bacterial culture was centrifuged at 8,000 × g for 5 min. The bacterial cell pellet was resuspended in an equal volume of XOM2 medium and cultured at 28°C for 1 h. The bacterial medium was again centrifuged at 12,000 × g for 5 min to harvest bacterial cells. Total RNA was isolated from resulting bacterial cells using an RNAprep Pure Bacteria Kit (Tiangen, Beijing, China), and total RNA was stored at −80°C for RNA sequencing.
RNA sequencing was performed by Novogene (Novogene, Beijing, China). The sequencing libraries were generated using NEBNext® UltraTM Directional RNA Library Prep Kit for Illumina® (New England Biolabs, Ipswich, MA, United States), as described previously (Fan et al., 2019) and as per manufacturer’s instructions. To assign sequences to each sample, the index codes were added. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS. The resulting library was sequenced on an Illumina Hiseq platform to generate paired-end reads.
Raw data in the FASTQ format were processed using in-house Perl scripts. To obtain clean reads, reads containing adapter or ploy-N, and low-quality reads were removed. Bowtie 2-2.2.3 was used to align clean reads to the Xoo reference genome (Langmead and Salzberg, 2012), and HTSeq version 0.6.1 was used to count the number of reads mapped to each gene. The expected number of FPKM (Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced) of each gene was calculated based on the length of the gene and read count mapped to the gene (Trapnell et al., 2009). The RNA-Seq data were deposited in NCBI Sequence Read Archive (SRA) with the SRA Series accession number: PRJNA684983. DESeq R package version 1.18.0 was employed to identify differentially expressed genes in two samples. The threshold with P ≤ 0.05 and the absolute value of log2Ratio ≥ 1 was used to determine the significance of differential gene expression.
GOseq R package1 was employed for the Gene Ontology (GO) enrichment analysis of DEGs. The GO terms with corrected p-value < 0.05 were considered significantly enriched. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed as described previously (Mao et al., 2005), and KOBAS software was used to determine the significant enrichment of DEGs in KEGG pathways.
Bacterial strains were cultured, and cells were collected as mentioned above. Total RNA was extracted from bacterial cells using the RNAprep pure Cell/Bacteria Kit (Tiangen Biotech), and cDNA was synthesized using HiScript II RT SuperMix kit (Vazyme, Nanjing, China). The qRT-PCR primers were designed using Primer Premier version 5.0 software (PREMIER Biosoft, Palo Alto, CA, United States) (Supplementary Table 2) with the gyrB as a reference gene. Finally, qRT-PCR was performed as described previously (Yu et al., 2020).
Xoo strains were cultured, as mentioned above. The bacterial medium was centrifuged at 12,000 × g, resuspended in distilled water, and OD600 was adjusted to 0.8. 2 μl of bacterial suspension was inoculated onto semisolid medium plates (0.03% peptone, 0.03% yeast extract, and 0.25% agar) and incubated at 28°C. The swimming zones of these bacteria were evaluated after 4 days. The assays were repeated three times.
The flagella of Xoo strains were visualized as described previously (Yu et al., 2018a). Briefly, Xoo strains were cultured on PSA plates at 28°C for 48 h, suspended in distilled water, and the bacterial suspension was transferred to Formvar coated grids (Standard Technology, Ormond Beach, FL, United States). The bacterial cells were stained with 2% uranyl acetate for 30 s, dried at room temperature, and visualized using the transmission electron microscope (H-7500, Hitachi, Tokyo, Japan).
Xoo inoculation and bacterial growth assay were performed as described previously (Yu et al., 2018b). Briefly, Xoo strains were cultured overnight in M210 medium, and the bacterial medium was centrifuged at 7,000 × g, resuspended in distilled water, and OD600 was adjusted to 1.0. The Xoo strain suspension was applied on susceptible rice (Oryza sativa L. ssp. Indica IR24) by leaf clipping method, and the lesion lengths of 10 leaves were measured at 2 weeks after inoculation for every strain. For bacterial population assay, three Xoo strain inoculated rice leaves were weighed and ground with distilled water followed by its dilution to attain specific optical concentration, and later spread onto PSA plates. The Xoo colonies were counted after 72 h incubation. These experiments were performed three times independently.
The DNA fragment of rpoN1 was amplified with specific primer pairs N1F/R, and the amplified PCR product was cloned into pColdSUMO, resulting in plasmid pC-rpoN1. This recombinant plasmid was transformed into E. coli BL21 strain. RpoN1 protein expression and purification were performed as described previously (Yu et al., 2020). For EMSA assay, rpoN2, and glnA promoters (−300 to +50 upstream or downstream of the translation start [+1]) were PCR amplified using 5′ end FAM-labeled primers. 2 μM of labeled rpoN2 or glnA promoter, 5 μM RpoN1 protein, and 1X EMSA/GelShift Binding Buffer (Beyotime, Shanghai, China) were mixed, and resolved using 4% native (wt/vol) polyacrylamide gel and electrophoresed for approximately 1 h at 100 V. The outcome of electrophoresis was detected on Typhoon FLA-5100 (Fuji Film, Tokyo, Japan) at 488 nm. All primers used in this experiment are listed in Supplementary Table 2.
Xoo strains were inoculated into M210 medium, and bacterial cell concentration was adjusted to obtain OD600 of 0.05 and incubated at 28°C under 200 rpm. The bacterial population was measured after every 6 h. For bacterial population assay, the bacterial cells were spread onto PSA plats after optional diluted, and cultured at 28°C for 3 days, the bacterial colonies then were counted. These experiments were repeated three times independently.
Swimming zones, disease lesion length, bacterial growth measurement, and relative gene expression were presented as means ± standard deviation. Two-tail Student’s t-test was performed with statistical significance set to 0.05 confidence level.
To elucidate the function of σ54 factor in Xoo, the protein sequence of Xoo σ54 factors was aligned against Pseudomonas syringae pv. tomato DC3000 (Pst) and X. campestris pv. campestris (Xcc) σ54 factor (RpoN) protein sequences. The outcomes indicated that RpoN1Xoo protein shared 41, 94, and 40% identical residues with RpoNPst, RpoN1Xcc, and RpoN2Xcc, respectively (Supplementary Figure 1). RpoN2Xoo protein sequence was aligned against protein sequence of RpoNPst, RpoN1Xcc, RpoN2Xcc, and it showed 39, 40, and 93% sequence similarity, respectively (Supplementary Figure 1). Interestingly, RpoN1Xoo protein shared only 39% identical residues with RpoN2Xoo, but both contained conserved σ54 factor domains, including activator interacting domain (AID) and DNA binding domain (DBD) (Supplementary Figure 1).
σ54 factors affect the flagellar assembly and virulence by regulating the transcription of multiple genes (Yang et al., 2009; Ray et al., 2015). In this study, the gene knockout strains ΔrpoN1 and ΔrpoN2 and the double-mutant strain ΔrpoN1N2 were constructed through homologous recombination to investigate the regulatory roles of RpoN1 and RpoN2 on Xoo’ motility and virulence. Firstly, the swimming zones of mutated Xoo strains on semisolid medium plates were measured. As the result showed, swimming motility of ΔrpoN1 was significantly reduced, and that of ΔrpoN2, ΔrpoN1N2 was nearly lost compared to the wild-type strain (Figures 1A,B). Later, flagella of these strains were visualized using the scanning electron microscope. As speculated, ΔrpoN1 strain showed abnormal flagella, but flagella were absent in ΔrpoN2 and ΔrpoN1N2 (Figure 1C). In addition, the swimming motility and flagellum synthesis of these mutants were restored in the relevant complemented strains (Figure 1). The results suggested that RpoN1 and RpoN2 positively regulated Xoo swimming motility by regulating the flagellar assembly. Finally, the virulence of these Xoo strains for the susceptible rice was estimated through leaf-clipping inoculation. The outcomes of our analysis showed that the bacterial blight symptoms and lesion lengths were significantly reduced in ΔrpoN1, ΔrpoN2, and ΔrpoN1N2 than the wild-type strain. These diseased phenotypes were restored in the complemented strains (Figures 2A,B). Similarly, the ΔrpoN1, ΔrpoN2, and ΔrpoN1N2 growth in rice leaf tissues were significantly reduced than the wild-type strain (Figure 2C). Thus, these results indicated that RpoN1 and RpoN2 positively regulated the virulence of Xoo in rice.
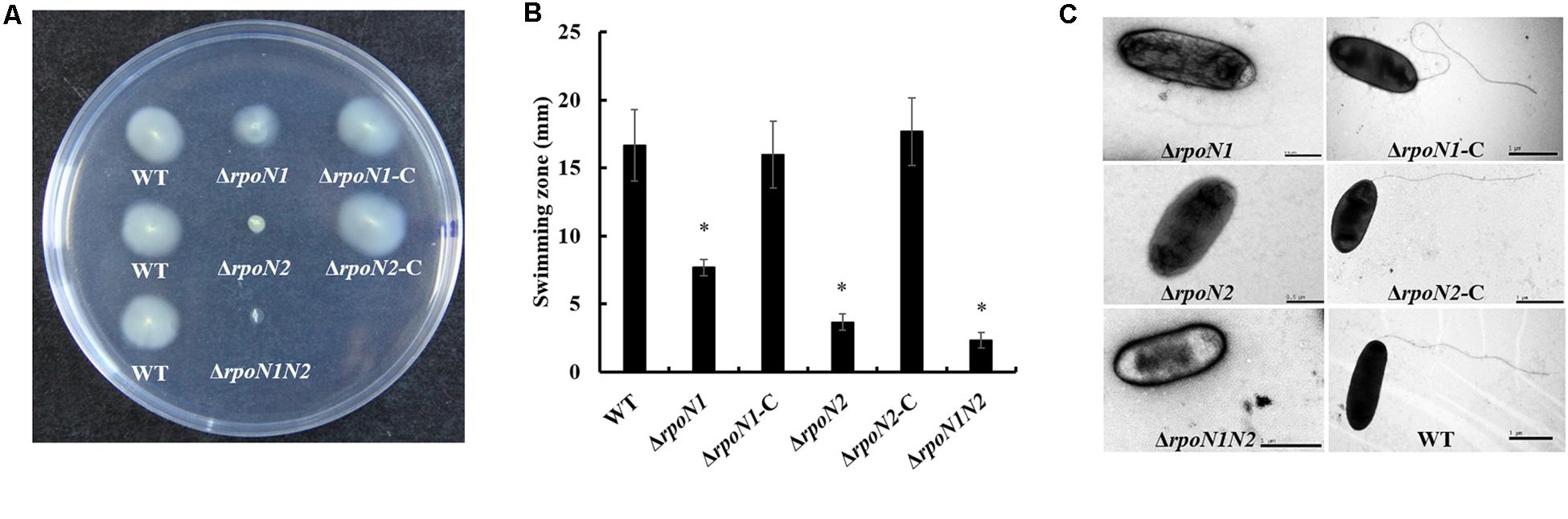
Figure 1. RpoN1 and RpoN2 for swimming motility and flagellar assembly in Xanthomonas oryzae pv. oryzae (Xoo). (A) Schematic representation and (B) quantitative analysis of swimming motility assay. The values represent the average of three independent experiments. Asterisks indicate significant differences between wild-type and mutants (P < 0.05) (Student’s t test). (C) Visualization of flagellar filament using scanning electron microscopy. WT, wild-type; ΔrpoN1, rpoN1 gene deletion mutant; ΔrpoN1-C, the complemented strain of ΔrpoN1; ΔrpoN2, rpoN2 gene deletion mutant; ΔrpoN2-C, the complemented strain of ΔrpoN2; ΔrpoN1N2, double gene (rpoN1 and rpoN2) deletion mutant.

Figure 2. RpoN1 and RpoN2 positively regulate the virulence in Xoo. (A) Disease symptoms, (B) disease lesion lengths, and (C) bacterial population. Disease symptoms and lesion lengths were photographed and measured in the second week after inoculation. For bacterial population assay, three leaves were collected and mixed as one sample for each strain. The experiments were performed three times independently. Asterisks indicate significant differences between wild-type and mutants (P-value < 0.05) (Student’s t test).
To apprehend RpoN1 and RpoN2 regulated motility and virulence of Xoo, RNA-Seq analysis of the Xoo wild-type and rpoN mutant strains was performed. DEGs at each mutant were identified as per the standards below: the absolute value of log2 (Fold Change) > 1 and adjusted P-value < 0.05. A total of 338 up-regulated and 325 down-regulated DEGs were identified in ΔrpoN1 (Figure 3A), and in the ΔrpoN2, 49 up-regulated, and 170 down-regulated DEGs were identified as compared to wild-type strain (Figure 3B). Besides, 575 up-regulated and 731 down-regulated DEGs were identified in the double mutant ΔrpoN1N2 compared to the wild-type strain (Figure 3C). Additionally, the Venn diagram was employed to analyze transcriptomic profiles, which showed 127 overlapping DEGs in different mutant strains (Figure 3D). To validate the reliability of RNA-seq results, the expression levels of 40 randomly selected DEGs were analyzed using the qRT-PCR assay. The results indicated that the gene expression patterns were in line with the RNA-seq analysis, indicating the reliability of RNA-seq results (Supplementary Figure 2).
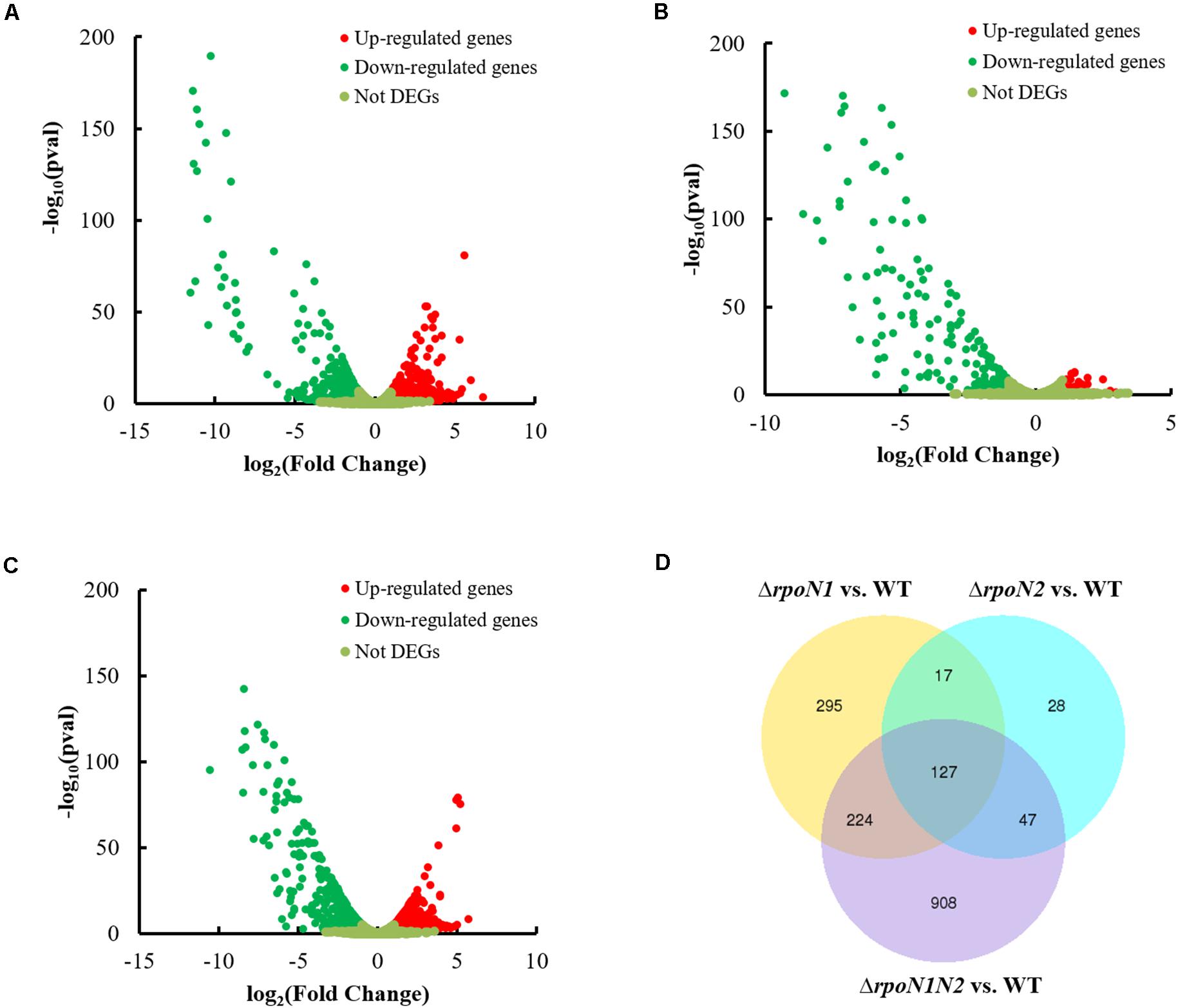
Figure 3. Differentially-expressed genes (DEGs) in ΔrpoN1, ΔrpoN2, and ΔrpoN1N2. DEGs in (A) ΔrpoN1, (B) ΔrpoN2, and (C) ΔrpoN1N2. Red and green dots represent up- and down-regulated genes, respectively. (D) RpoN1 and RpoN2 regulated unique and overlapping DEGs. Data represent the number of DEGs. DEGs at each mutant were selected according to the standards below: the absolute value of log2(Fold Change) > 1 and adjusted P-value < 0.05.
The DEGs were categorized into three functional groups: biological process, cellular component, and molecular function, as per the GO enrichment analysis. The majority of the DEGs in ΔrpoN1, ΔrpoN2, and ΔrpoN1N2 were enriched in the biological process category (Figure 4). In the biological process category, up-regulated genes were mostly enriched in single-organism localization and transport, biological regulation, and regulation of the cellular process, whereas down-regulated genes were mostly enriched in biological regulation, locomotion, and cell motility in ΔrpoN1, ΔrpoN2, and ΔrpoN1N2 (Figure 4). As speculated, most down-regulated genes associated with bacterial motility were significantly enriched in the GO database’s biological process category (Figures 4B,D,F). In the cellular component category, down-regulated genes in ΔrpoN1N2 were highly enriched in ribosomal associated categories (Figure 4F). It indicated that RpoN1 and RpoN2 regulate DEG expression by influencing ribosomal functions.
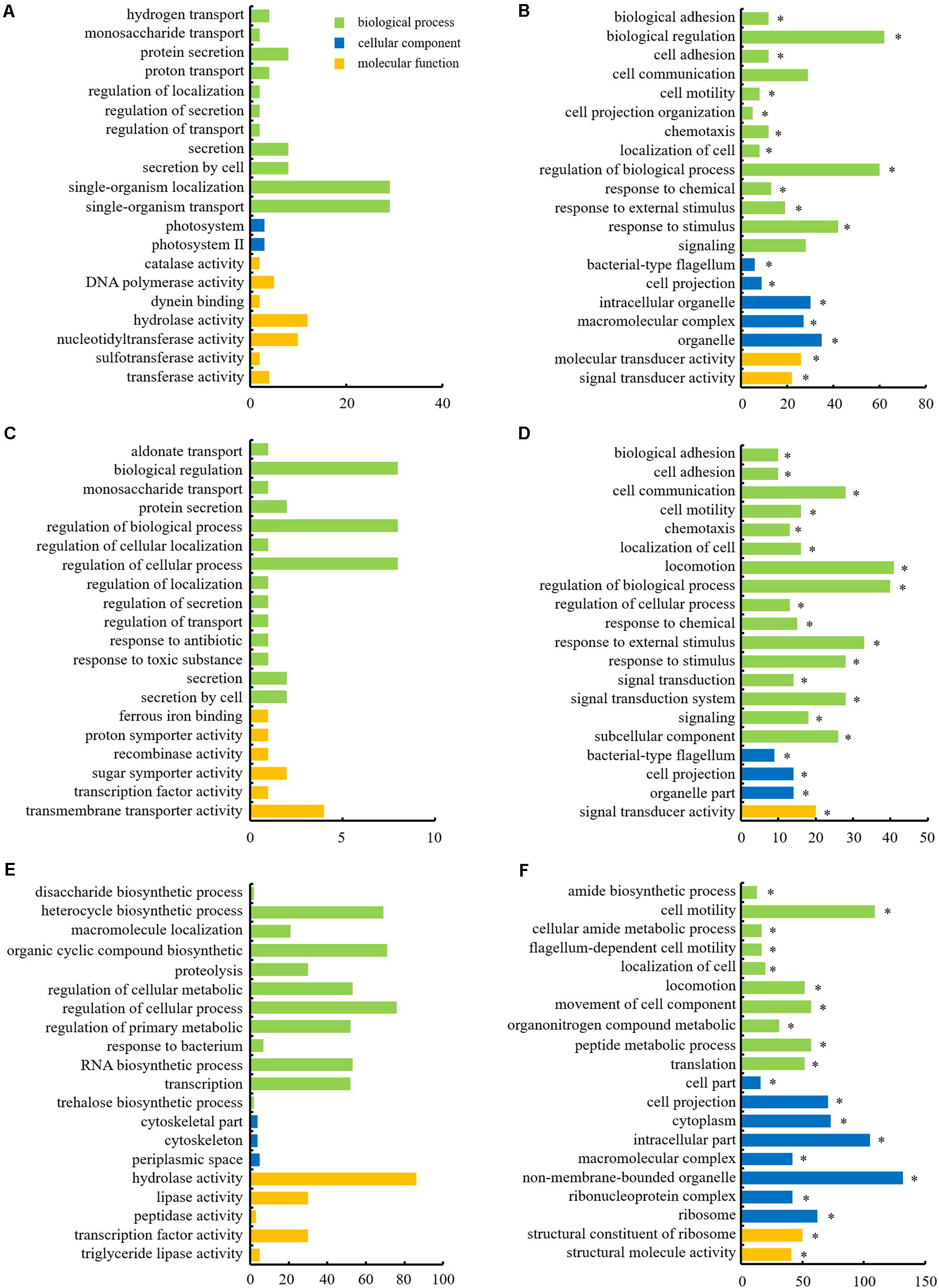
Figure 4. Histograms depicting GO analysis of up- and down-regulated genes in mutants. (A) Up-regulated and (B) down-regulated genes in ΔrpoN1. (C) Up-regulated and (D) down-regulated genes in ΔrpoN2. (E) Up-regulated and (F) down-regulated genes in ΔrpoN1N2. The abscissa axis represents the number of DEGs, and the ordinate axis represents the GO category. Asterisks indicate significant enrichment (P < 0.05).
To discern if RpoN1 and RpoN2 regulate Xoo motility and virulence through overlapping pathways, a total of 127 RpoN1 and RpoN2 regulated DEGs were analyzed using KEGG pathway analysis. These DEGs were significantly enriched in flagellar assembly, bacterial chemotaxis, and two-component system groups, suggesting the potential co-regulatory functions of RpoN1 and RpoN2 in Xoo (Figure 5).
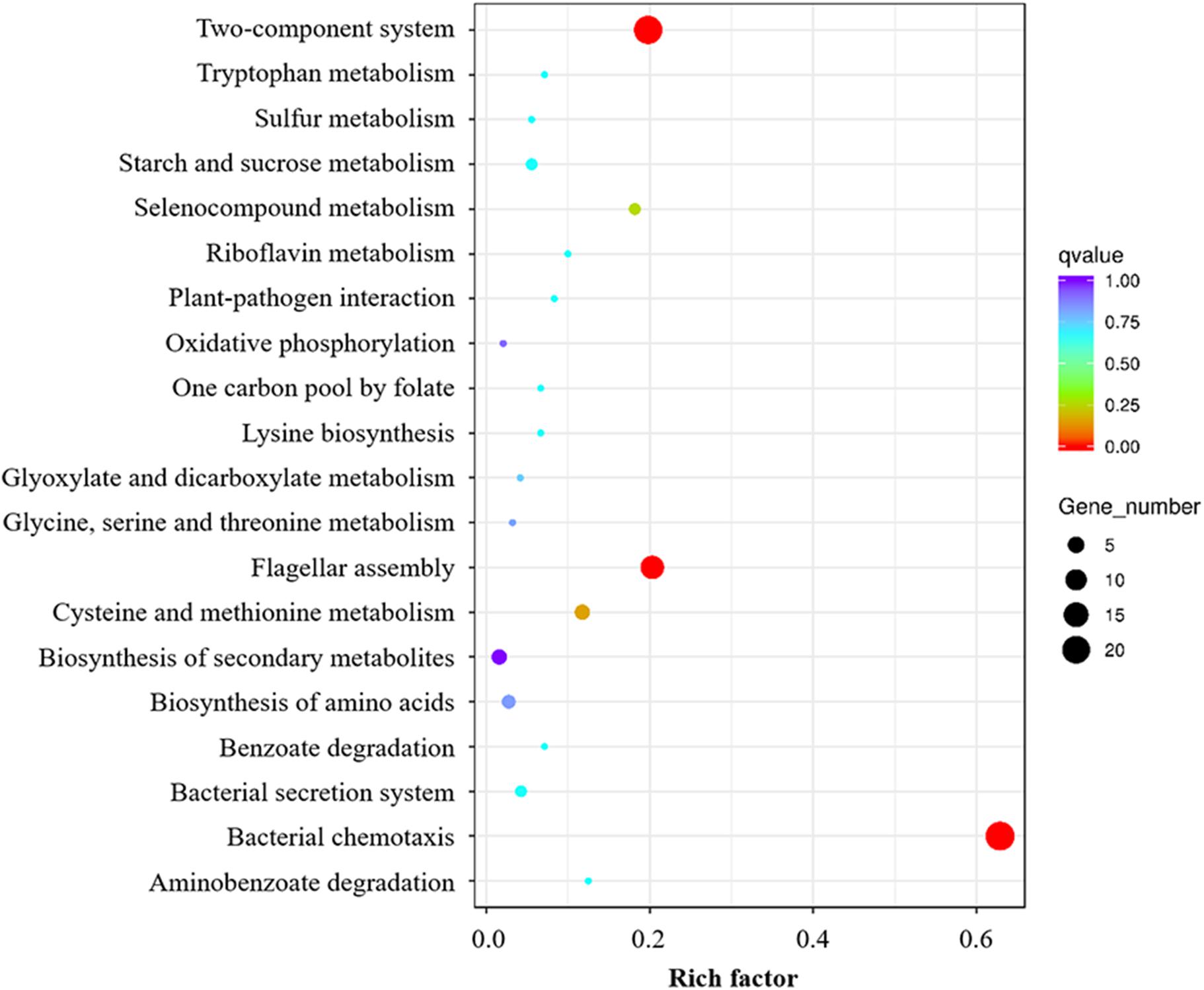
Figure 5. KEGG enrichment analysis of overlapping pathways regulated by both RpoN1 and RpoN2. Different colors represent the enriched pathways, and the size of the circle represents the number of DEGs.
In our previous work, the gene cluster associated with flagellar assembly in Xoo was identified (Tian et al., 2015). In this study, 35 genes encoding flagellar structural components and regulatory factors in this gene cluster were down-regulated by RpoN1 and RpoN2 (Figure 6A). Most of these gene’s expression levels were significantly decreased in ΔrpoN2 than ΔrpoN1 (Figure 6A). It indicated that RpoN2 plays a more crucial regulatory role in flagellar assembly than RpoN1. Expression levels of three flagellar structural component genes, fliC, fliD, and fliS, and three regulatory factor genes, fleQ, fliA, and flgRR were validated using qRT-PCR. As per the outcomes of PCR based analysis, expression levels of these genes were down-regulated in ΔrpoN1 and ΔrpoN2 and restored in the relevant complemented strains (Figure 6B). Lastly, swimming motility and virulence of these gene deletion mutants were also determined. Interestingly, swimming zones were significantly decreased in ΔfliC, ΔfliD, ΔfliS, ΔfleQ, and ΔfliA, but not in ΔflgRR as compared to wild-type strain (Figure 6C). However, except for ΔflgRR, virulence between the wild-type strain and these mutants did not differ significantly, displaying reduced lesion length (Figure 6D).
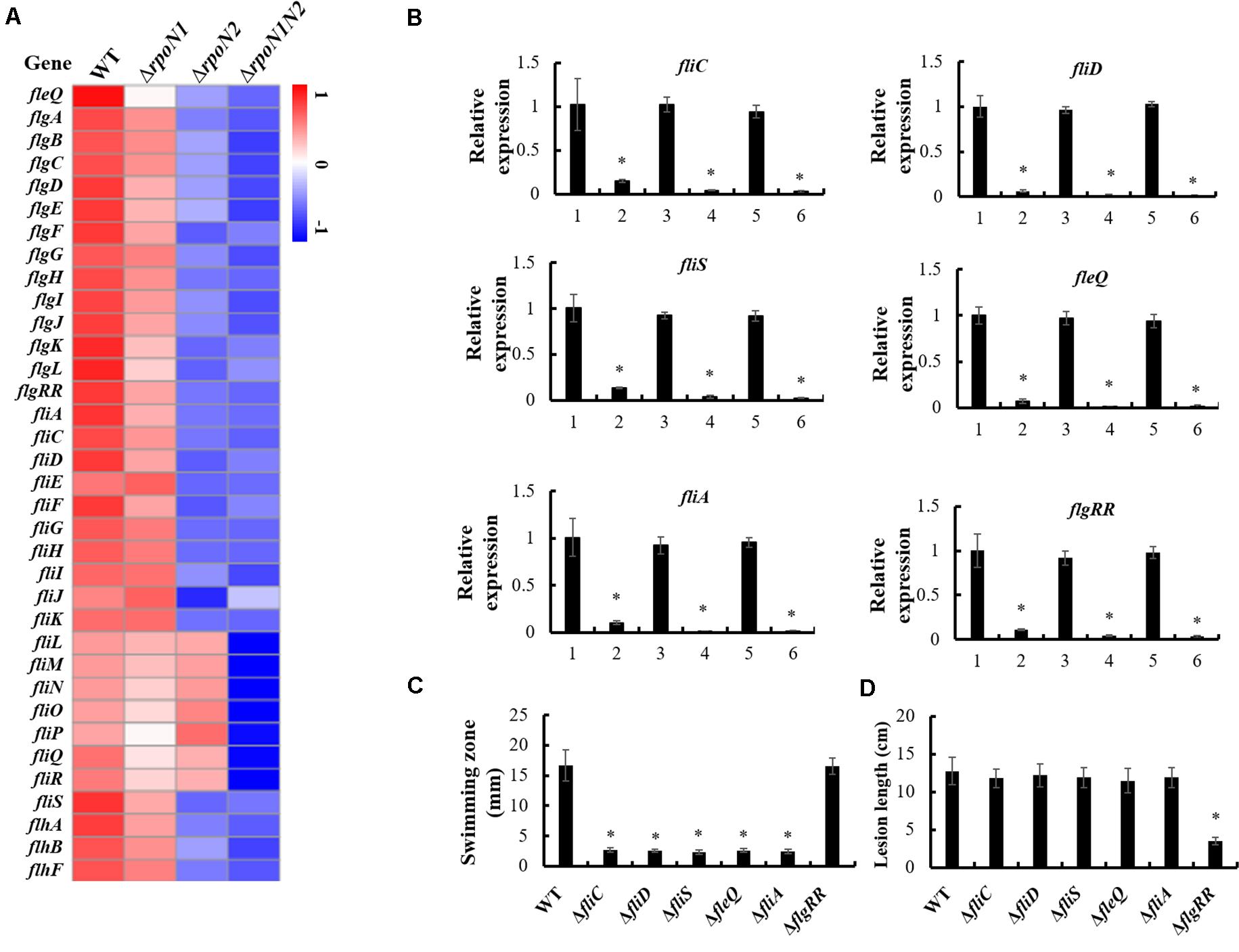
Figure 6. RpoN1 and RpoN2 regulated flagellar assembly. (A) Heat map of flagellar assembly associated genes expression. (B) The expression patterns of fliC, fliD, fliS, fleQ, fliA, and flgRR in ΔrpoN1, ΔrpoN2, and ΔrpoN1N2 were validated using qRT-PCR. 1, WT; 2, ΔrpoN1; 3, ΔrpoN1-C; 4, ΔrpoN2; 5, ΔrpoN2-C; 6, ΔrpoN1N2. (C) Swimming motility was tested in ΔfliC, ΔfliD, ΔfliS, ΔfleQ, ΔfliA, and ΔflgRR. (D) Virulence was detected in ΔfliC, ΔfliD, ΔfliS, ΔfleQ, ΔfliA, and ΔflgRR. Asterisks indicate significant differences between WT and mutants (P-value < 0.05) (Student’s t test).
Multiple DEGs were highly enriched in bacterial chemotaxis. Also, expression patterns of 26 genes encoding the core chemotaxis components, auxiliary proteins, and putative chemoreceptors in the RNA-seq data were specifically inspected. Similar to the flagellar assembly genes, RpoN1 and RpoN2 down-regulated the chemotaxis genes, and their expression levels were remarkably reduced in ΔrpoN2 than ΔrpoN1 (Figure 7A). Also, the expression patterns of four core chemotaxis components genes, cheA2, cheR2, cheW2, and cheY1, were validated using qRT-PCR. As per the outcomes, transcription levels of these genes were significantly down-regulated in ΔrpoN1 and ΔrpoN2 and restored in the relevant complemented strains (Figure 7B).
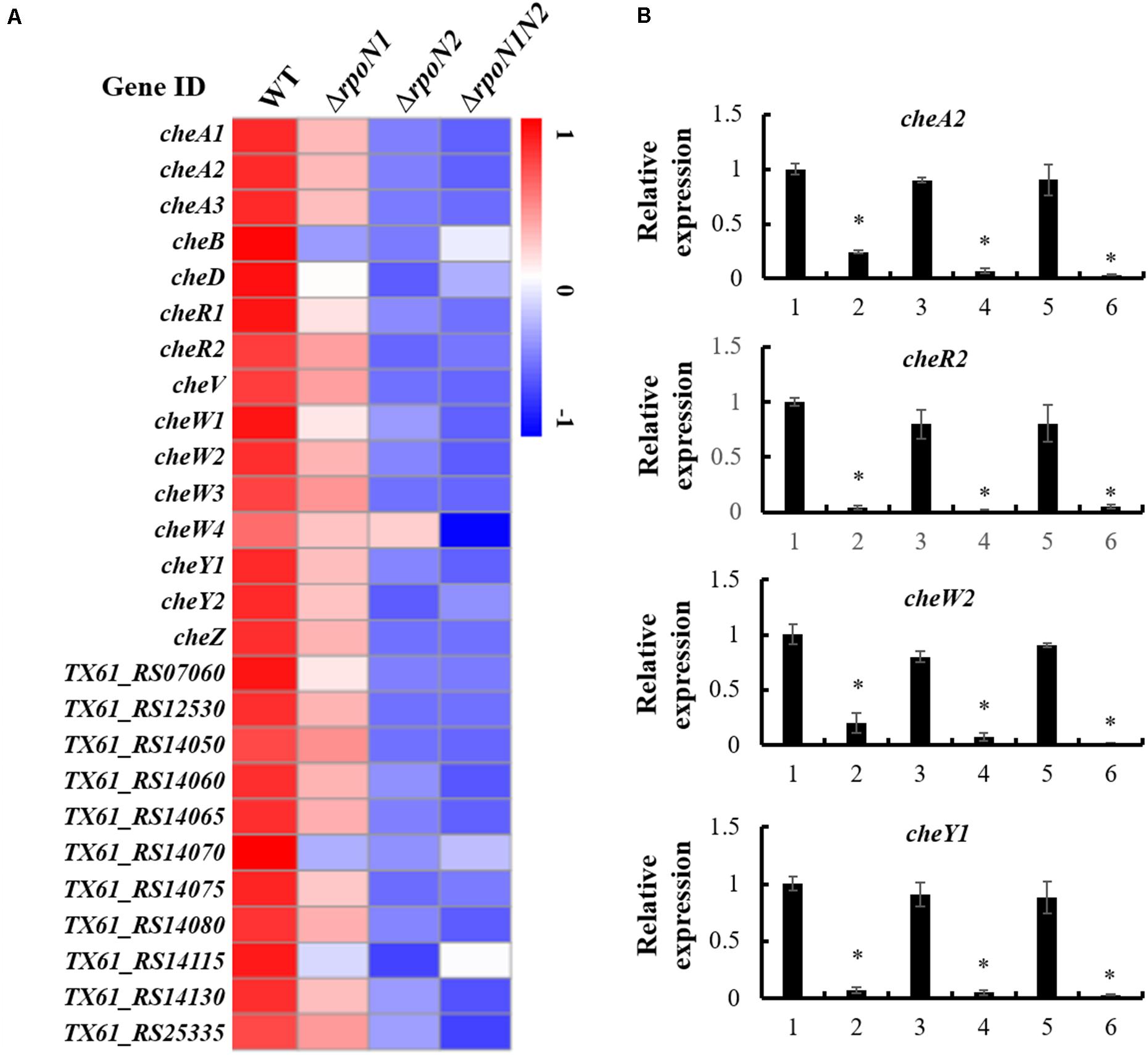
Figure 7. RpoN1 and RpoN2 regulated chemotaxis associated genes. (A) Heat map of gene expression of chemotaxis genes. (B) The expression patterns of cheA2, cheR2, cheW2, and cheY1 in ΔrpoN1, ΔrpoN2, and ΔrpoN1N2 were validated using qRT-PCR. 1, WT; 2, ΔrpoN1; 3, ΔrpoN1-C; 4, ΔrpoN2; 5, ΔrpoN2-C; 6, ΔrpoN1N2. Asterisks indicate significant differences between WT and mutants (P-value < 0.05) (Student’s t test).
c-di-GMP, the widespread second messenger in pathogenic bacteria, plays a crucial role in regulating Xoo motility and virulence (Yang et al., 2019). Diguanylate cyclase (DGC) enzyme contains a conserved GGDEF domain, which regulates c-di-GMP synthesis, whereas phosphodiesterase (PDE) enzyme-containing conserved EAL or HD-GYP domains controls c-di-GMP degradation. To elucidate if RpoN1 and RpoN2 regulate the c-di-GMP synthesis or degradation in Xoo, the expression patterns of 24 genes encoding proteins with GGDEF, EAL, or HD-GYP domains in the RNA-seq data were analyzed. The outcomes indicated that six of nine genes encoding GGDEF domain-containing proteins, six of ten genes encoding GGDEF and EAL domains-containing proteins, one of two genes encoding EAL domain-containing proteins, and one of three genes encoding HD-DYP domain-containing proteins were down-regulated in ΔrpoN1 and ΔrpoN2 (Figure 8A). In addition, TX61_RS09105, the gene encoding GGDEF domain-containing protein, was up-regulated in ΔrpoN1 and ΔrpoN2 (Figure 8A). Three of these genes were selected and their expression patterns were validated using qRT-PCR based analysis (Figure 8B). It indicated that RpoN1 and RpoN2 might regulate the c-di-GMP synthesis and degradation by regulating DGC and PDE enzyme transcription in Xoo.
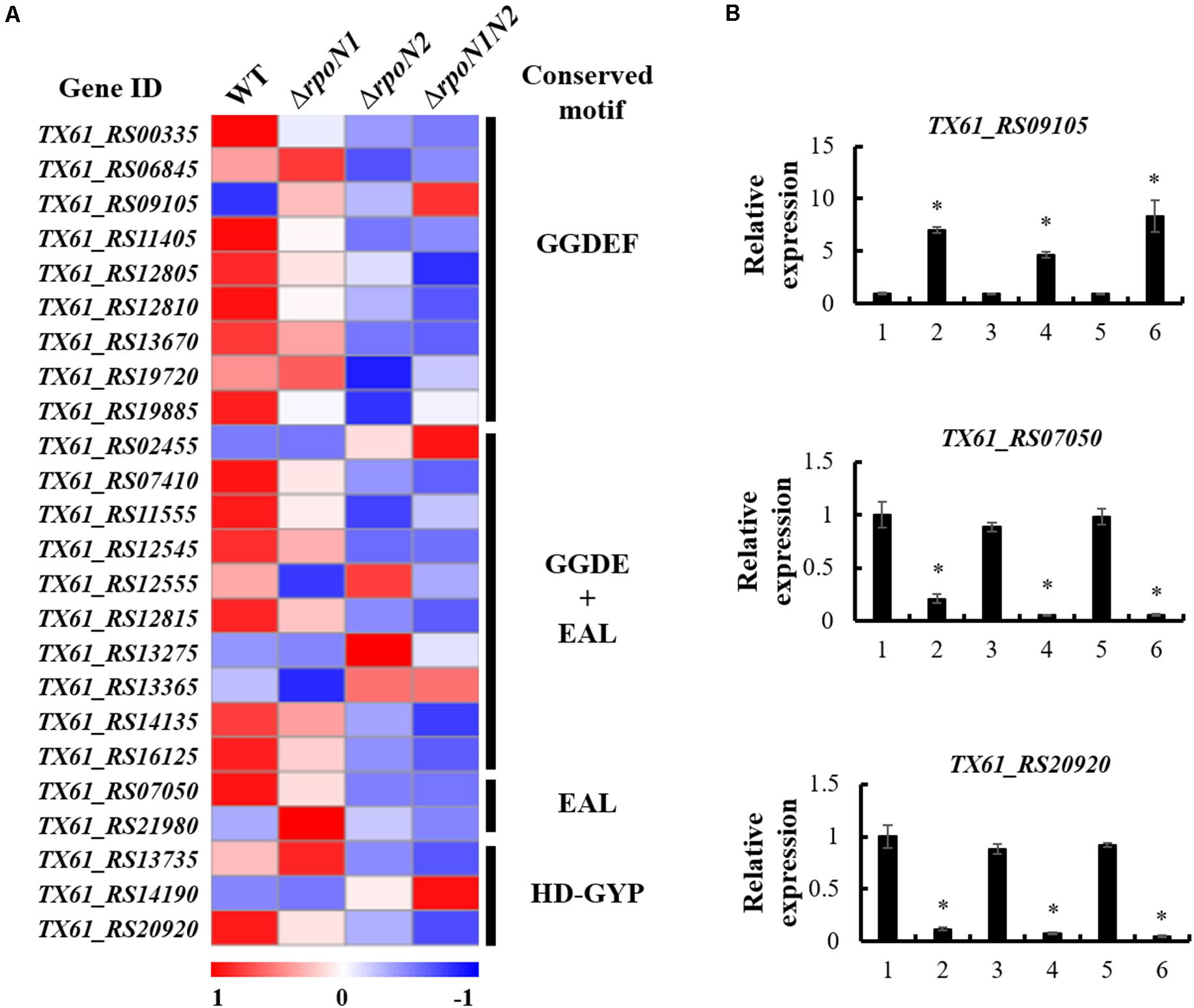
Figure 8. RpoN1 and RpoN2 regulated the expression of c-di-GMP associated genes. (A) Heat map of c-di-GMP associated gene expression. (B) The expression patterns of TX61_RS09105, TX61_RS07050, TX61_RS20920 in ΔrpoN1, ΔrpoN2, and ΔrpoN1N2 were validated by qRT-PCR. 1, WT; 2, ΔrpoN1; 3, ΔrpoN1-C; 4, ΔrpoN2; 5, ΔrpoN2-C; 6, ΔrpoN1N2. Asterisks indicate significant differences between WT and mutants (P-value < 0.05) (Student’s t test).
In the RNA-seq data, the rpoN2 expression level was decreased in ΔrpoN1, but the rpoN1 expression level did not differ significantly in ΔrpoN2 (data not shown). These expression patterns were validated using qRT-PCR (Figure 9A). However, as per EMSA, direct binding of RpoN1 to the promoter of rpoN2 was not observed (Figure 9B). It indicated that RpoN1 indirectly regulated the rpoN2 expression in Xoo.
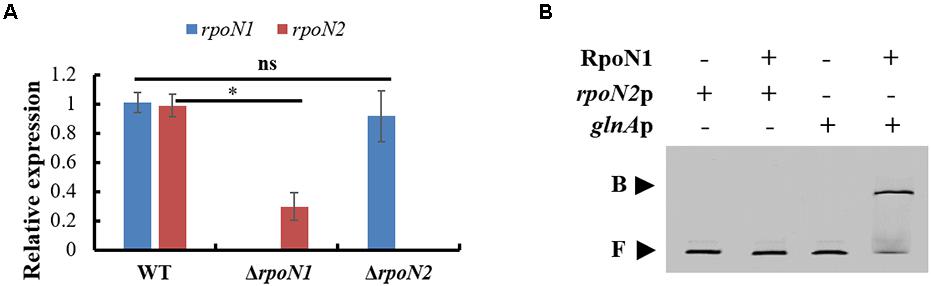
Figure 9. RpoN1 indirectly regulates the expression of rpoN2. (A) The expression patterns of rpoN1 and rpoN2 in ΔrpoN1 and ΔrpoN2 were investigated using qRT-PCR. (B) EMSA analysis to assess the interaction between RpoN1 and rpoN2 promoter. rpoN2p, the promoter of rpoN2; glnAp, the promoter of glnA; B, binding probe; F, free probe. In RNA-seq data, the glnA encoded glutamine synthetase expression was significantly decreased in ΔrpoN1 (data not shown). Thus, glnAp was selected as a positive control. Asterisk and ns indicate significant and insignificant differences, respectively (P-value < 0.05) (Student’s t test).
To apprehend the distinct roles of RpoN1 and RpoN2 in regulating various biological pathways in Xoo, RpoN1, or RpoN2 regulated DEGs were analyzed using KEGG. The top ten KEGG pathways enriching these DEGs were recorded. It showed that majority of RpoN1 regulated DEGs were enriched in metabolic pathways, followed by the ribosome (Figure 10A), whereas the majority of RpoN2 regulated DEGs were enriched in flagellar assembly followed by the two-component system (Figure 10B). Interestingly, the majority of both RpoN1 and RpoN2 regulated DEGs were enriched in metabolic pathways, followed by the ribosome and microbial metabolism (Figure 10C). The bacterial metabolic process is closely associated with bacterial growth. In this study, we assessed the growth curves of wild-type strain, ΔrpoN1, ΔrpoN1-C, ΔrpoN2, ΔrpoN2-C, and ΔrpoN1N2 strains. As the result showed, ΔrpoN1significantly reduced bacterial growth, and ΔrpoN1N2 showed more serious inhibition of bacterial growth than ΔrpoN1, whereas ΔrpoN2, ΔrpoN1-C, ΔrpoN2-C, and wild-type bacterial strain showed similar growth curves (Figure 10D). These results suggested that the function of RpoN2 in Xoo growth might be complemented by RpoN1.
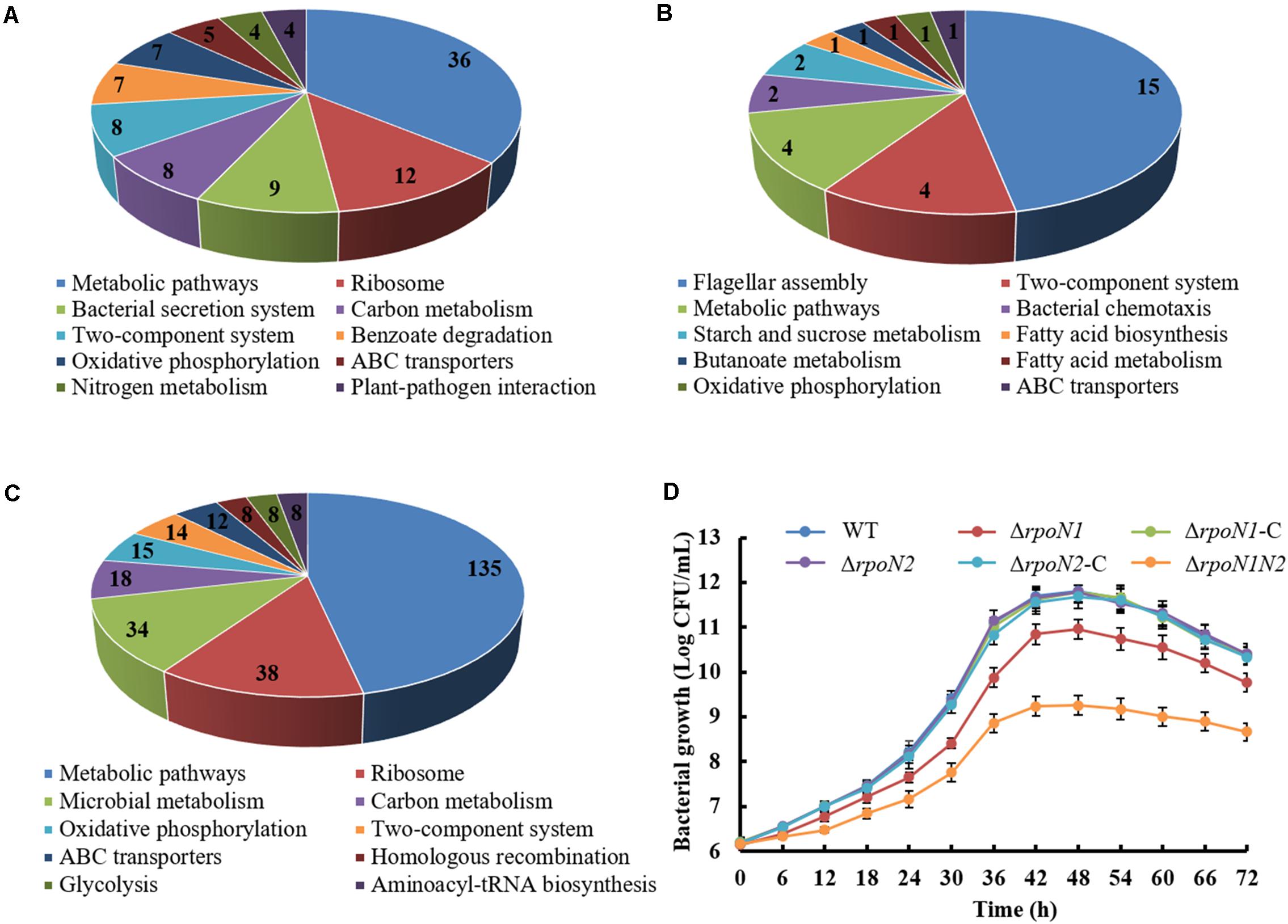
Figure 10. Analysis of DEGs specifically regulated by RpoN1 or RpoN2. DEGs specifically regulated by (A) rpoN1, (B) rpoN2, (C) ΔrpoN1N2. The number represents the number of DEGs enriched in KEGG pathways. (D) Growth curves of Xoo strain cultured using rich media M210. The growth curves were constructed three times, independently. Error bars indicate standard deviation.
σ54 factors control a myriad of crucial biological functions in bacteria, such as motility, nitrogen metabolism, stress resistance, and virulence factor production (Dasgupta et al., 2003; Kazmierczak et al., 2005; Viducic et al., 2017). In Xoo, two σ54 factors, RpoN1 and RpoN2, were identified, whereas their overlapping or unique regulatory roles remain unknown (Tian et al., 2015). In this study, RNA-seq was employed to analyze the transcriptome-wide regulome of RpoN1 and RpoN2 and elucidate their co-regulatory and specific-regulatory pathways in Xoo. In previous reports, the regulatory roles of two homologous σ54 factors were found to be no overlapping in Xanthomonas campestris (Li et al., 2020). However, in Xoo, both RpoN1 and RpoN2 were involved in regulating flagellar assembly, chemotaxis and c-di-GMP synthesis, and degradation. In addition, the function of RpoN2 in Xoo growth might be complemented by RpoN1, and the rpoN2 transcription level was indirectly regulated by RpoN1, while the rpoN1 expression was not regulated by RpoN2, indicating the different regulatory roles of RpoN1 and RpoN2 in Xoo. Thus, these findings could extend our understanding of the overlapping and unique regulatory roles of σ54 factor homologous in bacteria.
Flagellar assembly is a highly organized process requiring the temporal expression of multiple genes, regulated in a hierarchical manner. In P. aeruginosa, a four-tiered transcriptional cascade including σ54-dependent transcriptional activator FleQ (class I), two-component system FleSR (class II), σ28 factor FliA (class III), and flagellin gene fliC (class IV) was identified (Dasgupta et al., 2003). Our previous report showed that RpoN2/FleQ regulated the fliC gene by regulating the transcription of fliA in Xoo (Tian et al., 2015). In this work, more than thirty flagellar-related genes, including flagellar structural component genes fliC, fliD, and fliS, and regulatory factor genes fleQ, fliA, and flgRR that were positively regulated by RpoN1 and RpoN2 were identified (Figures 6A,B). The innate immune response triggering fliC gene-encoded flagellin protein has been well studied as a pathogen-associated molecular pattern in Arabidopsis (Chinchilla et al., 2006). The fliC gene deletion blocked swimming motility, but it did not affect the Xoo pathogenicity in rice significantly (Figures 6C,D; Wang et al., 2015). Impaired swimming motility and similar pathogenicity in rice were also found in ΔfliD, ΔfliS, ΔfleQ, and ΔfliA mutants as compared to wild-type strain (Figures 6C,D). It indicated that RpoN1 and RpoN2 controlled bacterial virulence in a manner different from their regulatory role in flagellar motility of Xoo. In X. citri, the response regulator VemR, the homologous protein of FlgRR, functions as a RpoN2 cognate activator to positively regulate motility and virulence (Wu et al., 2019). In this study, RpoN1 and RpoN2 regulated flgRR expression, and flgRR deletion did not reduce Xoo’s swimming motility but remarkably decreased its virulence (Figure 6). The outcomes of this study suggested that RpoN1 and RpoN2 might regulate the bacterial swimming motility and virulence in Xoo by regulating the expression of flagellar synthesis genes and flgRR, respectively.
In bacteria, chemotaxis regulates directional motility, and thus it plays a crucial role in the adaptation of bacteria to changing environmental conditions. The basic chemotaxis mechanism of the signal transduction system was discerned in E. coli (Wadhams and Armitage, 2004; Parkinson et al., 2015). Briefly, activated chemoreceptor and phosphorylated cytoplasmic sensor kinase CheA recognizes the chemosensory signal. Subsequently, adaptor protein CheW mediates the transfer of phosphate radical of CheA to the response regulator CheY. Thus, CheY regulates flagellar motor activity. Meanwhile, methy-transferase CheR binds to CheA, CheW, and CheY and constitutes the core components of the chemotactic signal transduction pathway. A previous study also showed that in Xoo, core elements of the chemotaxis signal transduction system, including CheA2, CheR2, CheW2, and CheY1, entailed several paralogs of chemotaxis components (Kumar Verma et al., 2018). In addition, deficiency of these core components significantly mitigated virulence-associated functions, such as attachment and iron homeostasis (Kumar Verma et al., 2018). In this study, more than twenty RpoN1 and RpoN2 regulated chemotaxis components and putative chemoreceptors genes were identified (Figure 7A). The expression patterns of cheA2, cheR2, cheW2, and cheY1 were validated using qRT-PCR in ΔrpoN1 and ΔrpoN2 (Figure 7B). The results of the current study indicated that RpoN1 and RpoN2 might regulate the virulence by regulating the chemotaxis pathway in Xoo.
c-di-GMP plays a vital role in regulating multiple biological functions, such as motility, biofilm formation, EPS production, environment adaptation, and virulence (Hengge, 2009). The antagonistic enzymatic actions of DGC and PDE enzymes regulate the synthesis and degradation of intracellular c-di-GMP, respectively. DGCs contain a conserved GGDEF domain, while PDEs contain conserved EAL or HD-GYP domains (Yang et al., 2019). A previous study reported that two GGDEF domain proteins, GdpX1 from Xoo strain PXO99A and DgcA from Xoo strain KACC10331, negatively regulates bacterial virulence, EPS production, and motility (Su et al., 2016; Yang et al., 2016). EdpX1, an EAL domain protein, exhibited PDE activity, positive regulation of c-di-GMP level and virulence in Xoo strain PXO99A (Xue et al., 2018). In this study, a putative DGC protein containing GGDEF domain encoded by TX61_RS09105 was overexpressed in ΔrpoN1 and ΔrpoN2. Besides, two putative PDEs containing EAL and HD-GYP domain encoded by TX61_RS07050 and TX61_RS20920, respectively, were down-regulated in ΔrpoN1 and ΔrpoN2 (Figure 8A). The results suggested that RpoN1 and RpoN2 control the synthesis and degradation of c-di-GMP by regulating DGCs and PDEs in Xoo.
Although two or more rpoN copies were identified in certain bacteria, the difference in these RpoN proteins’ regulatory roles was rarely reported. In R. solanacearum, rpoN1, but not rpoN2, is necessary for virulence, twitching motility, natural transformation, and growth on nitrate (Ray et al., 2015). However, in X. campestris, RpoN1 regulates branched-chain fatty acid production and diffusible signal synthesis, whereas RpoN2 regulates swimming motility, biofilms, EPS production, and virulence (Li et al., 2020). Unlike these studies, in the current study, both RpoN1 and RpoN2 were involved in regulating Xoo swimming motility and virulence (Figures 1, 2). Interestingly, the transcription levels of flagellar assembly-related genes were substantially decreased in ΔrpoN2 than ΔrpoN1 (Figure 6A). Also, RpoN1 indirectly regulated the rpoN2 expression (Figure 9). It indicated that in Xoo, RpoN1 might regulate motility by regulating rpoN2 expression. Moreover, RpoN1 regulated multiple DEGs related to ribosome, carbon, and nitrogen metabolism whereas only a few DEGs related to metabolic pathways were specifically regulated by RpoN2 in Xoo (Figures 10A,B). Importantly, RpoN1 is more important than RpoN2 for Xoo growth in rich medium (Figure 10D). These results suggested that as compared to RpoN2, RpoN1 plays a more crucial role in regulating basal metabolism and growth in Xoo.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
CY and D-PN designed and performed the experiments and wrote the manuscript. FY, JS, and YW analyzed the data and commented on the manuscript. FT commented on the manuscript. XZ and HC designed the experiments, supervised the research, and finalized the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (No. 2017YFD0200900), the National GMO New Variety Breeding Program of China (No. 2016ZX08001-002), and the National Natural Science Foundation of China (Nos. 31600105 and 31870120).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank all the medical staff for their brave fight against the COVID-19 and protecting us. Without them we can’t finish this work smoothly.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.653354/full#supplementary-material
Supplementary Figure 1 | Conserved σ54 factors in P. syringae pv. tomato DC3000 (Pst), X. campestris pv. campestris (Xcc), and Xoo strains. The σ54 factor sequences were downloaded from the National Center for Biotechnology Information database and aligned using DNAMAN based on identical residues. Activator interacting domain (AID) and DNA binding domain (DBD) were highly conserved among σ54 proteins from different species.
Supplementary Figure 2 | Correlation analysis between qRT-PCR and RNA-seq data of forty selected DEGs. Analysis of the expression patterns of DEGs in (A) ΔrpoN1, (B) ΔrpoN2, and (C) ΔrpoN1N2.
Supplementary Table 1 | Bacterial strains and plasmids used in this study.
Supplementary Table 2 | Primers used in this study.
Supplementary Table 3 | Expression patterns of selected DEGs in ΔrpoN1, ΔrpoN2, and ΔrpoN1N2 from RNA-seq data.
Barrios, H., Valderrama, B., and Morett, E. (1999). Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic. Acids. Res. 27, 4305–4313. doi: 10.1093/nar/27.22.4305
Cai, Z., Liu, Y., Chen, Y., Yam, J. K., Chew, S. C., Chua, S. L., et al. (2015). RpoN regulates virulence factors of Pseudomonas aeruginosa via modulating the PqsR quorum sensing regulator. Int. J. Mol. Sci. 16, 28311–28319. doi: 10.3390/ijms161226103
Chinchilla, D., Bauer, Z., Regenass, M., Boller, T., and Felix, G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18, 465–476. doi: 10.1105/tpc.105.036574
Das, A., Rangaraj, N., and Sonti, R. V. (2009). Multiple adhesin-like functions of Xanthomonas oryzae pv. oryzae are involved in promoting leaf attachment, entry, and virulence on rice. Mol. Plant Microbe. Interact. 22, 73–85. doi: 10.1094/MPMI-22-1-0073
Dasgupta, N., Wolfgang, M. C., Goodman, A. L., Arora, S. K., Jyot, J., Lory, S., et al. (2003). A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50, 809–824. doi: 10.1046/j.1365-2958.2003.03740.x
Davis, M. C., Kesthely, C. A., Franklin, E. A., and MacLellan, S. R. (2017). The essential activities of the bacterial sigma factor. Can. J. Microbiol. 63, 89–99. doi: 10.1139/cjm-2016-0576
Fan, S., Tian, F., Fang, L., Yang, C. H., and He, C. (2019). Transcriptional responses of Xanthomonas oryzae pv. oryzae to type III secretion system inhibitor ortho-coumaric acid. BMC Microbiol. 19:163. doi: 10.1186/s12866-019-1532-5
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/s0022-2836(83)80284-8
Hao, B., Mo, Z. L., Xiao, P., Pan, H. J., Lan, X., and Li, G. Y. (2013). Role of alternative sigma factor 54 (RpoN) from Vibrio anguillarum M3 in protease secretion, exopolysaccharide production, biofilm formation, and virulence. Appl. Microbiol. Biotechnol. 97, 2575–2585. doi: 10.1007/s00253-012-4372-x
Hayrapetyan, H., Tempelaars, M., Nierop Groot, M., and Abee, T. (2015). Bacillus cereus ATCC 14579 RpoN (Sigma 54) is a pleiotropic regulator of growth, carbohydrate metabolism, motility, biofilm formation and toxin production. PLoS One 10:e0134872. doi: 10.1371/journal.pone.0134872
Hengge, R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273. doi: 10.1038/nrmicro2109
Kazmierczak, M. J., Wiedmann, M., and Boor, K. J. (2005). Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69, 527–543. doi: 10.1128/MMBR.69.4.527-543.2005
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Kumar Verma, R., Samal, B., and Chatterjee, S. (2018). Xanthomonas oryzae pv. oryzae chemotaxis components and chemoreceptor Mcp2 are involved in the sensing of constituents of xylem sap and contribute to the regulation of virulence-associated functions and entry into rice. Mol. Plant Pathol. 19, 2397–2415. doi: 10.1111/mpp.12718
Kustu, S., Santero, E., Keener, J., Popham, D., and Weiss, D. (1989). Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol. Rev. 53, 367–376.
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lee, J. H., Sundin, G. W., and Zhao, Y. (2016). Identification of the HrpS binding site in the hrpL promoter and effect of the RpoN binding site of HrpS on the regulation of the type III secretion system in Erwinia amylovora. Mol. Plant Pathol. 17, 691–702. doi: 10.1111/mpp.12324
Li, K., Wu, G., Liao, Y., Zeng, Q., Wang, H., and Liu, F. (2020). RpoN1 and RpoN2 play different regulatory roles in virulence traits, flagellar biosynthesis, and basal metabolism in Xanthomonas campestris. Mol. Plant Pathol. 21, 907–922. doi: 10.1111/mpp.12938
Li, Y. R., Zou, H. S., Che, Y. Z., Cui, Y. P., Guo, W., Zou, L. F., et al. (2011). A novel regulatory role of HrpD6 in regulating hrp-hrc-hpa genes in Xanthomonas oryzae pv. oryzicola. Mol. Plant Microbe. Interact. 24, 1086–1101. doi: 10.1094/MPMI-09-10-0205
Lundgren, B. R., Connolly, M. P., Choudhary, P., Brookins-Little, T. S., Chatterjee, S., Raina, R., et al. (2015). Defining the metabolic functions and roles in virulence of the rpoN1 and rpoN2 genes in Ralstonia solanacearum GMI1000. PLoS One 10:e0144852. doi: 10.1371/journal.pone.0144852
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Nino-Liu, D. O., Ronald, P. C., and Bogdanove, A. J. (2006). Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324. doi: 10.1111/j.1364-3703.2006.00344.x
Parkinson, J. S., Hazelbauer, G. L., and Falke, J. J. (2015). Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 23, 257–266. doi: 10.1016/j.tim.2015.03.003
Poggio, S., Osorio, A., Dreyfus, G., and Camarena, L. (2002). The four different sigma(54) factors of Rhodobacter sphaeroides are not functionally interchangeable. Mol. Microbiol. 46, 75–85. doi: 10.1046/j.1365-2958.2002.03158.x
Ray, S. K., Kumar, R., Peeters, N., Boucher, C., and Genin, S. (2015). rpoN1, but not rpoN2, is required for twitching motility, natural competence, growth on nitrate, and virulence of Ralstonia solanacearum. Front. Microbiol. 6:229. doi: 10.3389/fmicb.2015.00229
Shao, X., Zhang, X., Zhang, Y., Zhu, M., Yang, P., Yuan, J., et al. (2018). RpoN-dependent direct regulation of quorum sensing and the type VI secretion system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 200, e205–e218. doi: 10.1128/JB.00205-18
Studholme, D. J., and Dixon, R. (2003). Domain architectures of sigma54-dependent transcriptional activators. J. Bacteriol. 185, 1757–1767. doi: 10.1128/jb.185.6.1757-1767.2003
Su, J., Zou, X., Huang, L., Bai, T., Liu, S., Yuan, M., et al. (2016). DgcA, a diguanylate cyclase from Xanthomonas oryzae pv. oryzae regulates bacterial pathogenicity on rice. Sci. Rep. 6:25978. doi: 10.1038/srep25978
Tian, F., Yu, C., Li, H., Wu, X., Li, B., Chen, H., et al. (2015). Alternative sigma factor RpoN2 is required for flagellar motility and full virulence of Xanthomonas oryzae pv. oryzae. Microbiol. Res. 170, 177–183. doi: 10.1016/j.micres.2014.07.002
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Viducic, D., Murakami, K., Amoh, T., Ono, T., and Miyake, Y. (2017). RpoN promotes Pseudomonas aeruginosa survival in the presence of tobramycin. Front. Microbiol. 8:839. doi: 10.3389/fmicb.2017.00839
Wadhams, G. H., and Armitage, J. P. (2004). Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5, 1024–1037. doi: 10.1038/nrm1524
Wang, S., Sun, Z., Wang, H., Liu, L., Lu, F., Yang, J., et al. (2015). Rice OsFLS2-mediated perception of bacterial flagellins is evaded by Xanthomonas oryzae pv. oryzae and oryzicola. Mol. Plant 8, 1024–1037. doi: 10.1016/j.molp.2015.01.012
White, F. F., and Yang, B. (2009). Host and pathogen factors controlling the rice-Xanthomonas oryzae interaction. Plant Physiol. 150, 1677–1686. doi: 10.1104/pp.109.139360
Wu, W., Zhao, Z., Luo, X., Fan, X., Zhuo, T., Hu, X., et al. (2019). Response regulator VemR regulates the transcription of flagellar rod gene flgG by interacting with sigma(54) factor RpoN2 in Xanthomonas citri ssp. citri. Mol. Plant Pathol. 20, 372–381. doi: 10.1111/mpp.12762
Xu, T., Yu, M., Liu, J., Lin, H., Liang, J., and Zhang, X. H. (2019). Role of RpoN from Labrenzia aggregata LZB033 (Rhodobacteraceae) in formation of flagella and biofilms, motility, and environmental adaptation. Appl. Environ. Microbiol. 85, e02844–18. doi: 10.1128/AEM.02844-18
Xue, D., Tian, F., Yang, F., Chen, H., Yuan, X., Yang, C. H., et al. (2018). Phosphodiesterase EdpX1 promotes Xanthomonas oryzae pv. oryzae virulence, exopolysaccharide production, and biofilm formation. Appl. Environ. Microbiol. 84, e1717–e1718. doi: 10.1128/AEM.01717-18
Yang, F., Qian, S., Tian, F., Chen, H., Hutchins, W., Yang, C. H., et al. (2016). The GGDEF-domain protein GdpX1 attenuates motility, exopolysaccharide production and virulence in Xanthomonas oryzae pv. oryzae. J. Appl. Microbiol. 120, 1646–1657. doi: 10.1111/jam.13115
Yang, F., Xue, D., Tian, F., Hutchins, W., Yang, C. H., and He, C. (2019). Identification of c-di-GMP signaling components in Xanthomonas oryzae and their orthologs in Xanthomonads involved in regulation of bacterial virulence expression. Front. Microbiol. 10:1402. doi: 10.3389/fmicb.2019.01402
Yang, T. C., Leu, Y. W., Chang-Chien, H. C., and Hu, R. M. (2009). Flagellar biogenesis of Xanthomonas campestris requires the alternative sigma factors RpoN2 and FliA and is temporally regulated by FlhA. FlhB, and FlgM. J. Bacteriol. 191, 2266–2275. doi: 10.1128/JB.01152-08
Yang, Y., Darbari, V. C., Zhang, N., Lu, D., Glyde, R., Wang, Y. P., et al. (2015). Transcription. structures of the RNA polymerase-sigma54 reveal new and conserved regulatory strategies. Science 349, 882–885. doi: 10.1126/science.aab1478
Yu, C., Chen, H., Tian, F., Yang, F., Yuan, X., Yang, C. H., et al. (2018a). A ten gene-containing genomic island determines flagellin glycosylation: implication for its regulatory role in motility and virulence of Xanthomonas oryzae pv. oryzae. Mol. Plant Pathol. 19, 579–592. doi: 10.1111/mpp.12543
Yu, C., Chen, Y., Cao, Y., Chen, H., Wang, J., Bi, Y. M., et al. (2018b). Overexpression of miR169o, an overlapping microRNA in response to both nitrogen limitation and bacterial infection, promotes nitrogen use efficiency and susceptibility to bacterial blight in rice. Plant Cell Physiol. 59, 1234–1247. doi: 10.1093/pcp/pcy060
Keywords: σ54 factor, Xanthomonas oryzae pv. oryzae, transcriptome analysis, motility, virulence, growth
Citation: Yu C, Nguyen D-P, Yang F, Shi J, Wei Y, Tian F, Zhao X and Chen H (2021) Transcriptome Analysis Revealed Overlapping and Special Regulatory Roles of RpoN1 and RpoN2 in Motility, Virulence, and Growth of Xanthomonas oryzae pv. oryzae. Front. Microbiol. 12:653354. doi: 10.3389/fmicb.2021.653354
Received: 14 January 2021; Accepted: 15 February 2021;
Published: 04 March 2021.
Edited by:
Robert Czajkowski, University of Gdańsk, PolandReviewed by:
Yong-Qiang He, Guangxi University, ChinaCopyright © 2021 Yu, Nguyen, Yang, Shi, Wei, Tian, Zhao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiuxiang Zhao, emhhb3h4MDc3MkBzeWF1LmVkdS5jbg==; Huamin Chen, Y2hlbmh1YW1pbkBjYWFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.