- 1Laboratory of Metabolic Manipulation of Herbivorous Animal Nutrition, College of Animal Science and Technology, Yangzhou University, Yangzhou, China
- 2School of Biomedical Sciences, The University of Western Australia, M Block, Queen Elizabeth II Medical Centre, Nedlands, WA, Australia
- 3College of Animal Science and Technology, Northwest A & F University, Yanglin, China
Long-term supplementation of a high-concentrate diet enhances the accumulation of lactate and decrease in pH in goat rumen, thereby disrupting the composition of microbial community. Studies have shown that incorporation of thiamine in high-concentrate diet increases ruminal pH and decreases rumen lactate concentration. To explore the effects of thiamine supplementation with a high-concentrate diet on alteration of the whole ruminal microbiota and their metabolites, 18 mid-lactating Saanen goats were randomly fed with one of three diets: (1) control diet (CON; n = 6; concentrate:forage 30:70), (2) high-concentrate diet (HG; n = 6; concentrate:forage 70:30), and (3) high-concentrate diet with 200 mg of thiamine/kg of DMI (HGT; n = 6; concentrate:forage 70:30). The goats received experimental diets for 8 weeks. Ruminal samples were collected on the last day of the 8 weeks for 16S rRNA gene sequencing and the liquid chromatograph–mass spectrometer (LC-MS) analysis. The results revealed significant alterations of the ruminal bacterial community structure and diversity in HGT groups compared to HG groups, with an overall dominance of Bacteroidetes at the phylum level and Oribacterium (P < 0.05), Anaerobiospirillum (P < 0.01), and Fibrobacter (P < 0.01) at genus level in the HGT group. The LC-MS analysis revealed that thiamine supplementation resulted in lower levels of propionate (P < 0.05), pyruvate (P < 0.01), lactate (P < 0.05), putrescine (P < 0.05), tyramine (P < 0.05), and histamine (P < 0.01) and higher levels of acetate (P < 0.05), succinates (P < 0.01), oxaloacetic acid (P < 0.01), leucine (P < 0.01), valine (P < 0.05), linoleic acid (P < 0.05), docosahexaenoic acid (P < 0.05), and 4-phenylbutyric acid (P < 0.05) in the HGT group than in the HG group. The decrease in these compounds enhanced homeostasis in the rumen environment and suppressed epithelial inflammation. Correlation analysis revealed the potential relationships between ruminal metabolites and microbial community. These findings demonstrate that thiamine supplementation can alleviate subacute ruminal acidosis (SARA) by stabilizing the microbial community and reducing toxic unnatural compounds.
Introduction
Subacute ruminal acidosis (SARA) is a nutritional metabolic disease that affects the finishing period of feedlot beef cattle, especially during the early and middle lactation periods of dairy cattle. The incidence of SARA is characterized by low fiber and high starch energy intake of diets, while the rumen environment exhibits no adaption to fermentation. It has been shown that short-chain fatty acids (SCFAs) and lactate generated by fermentation can be fully absorbed, thus reducing the ruminal pH, which exceeds the adaptability of animals (Kleen et al., 2003). Long-term supply of a high-concentrate diet has been found to affect the structure of the bacterial community and fermentation characteristics in the rumen (Khafipour et al., 2009; Hook et al., 2011). For instance, gram-negative bacteria die and lyse when the structure of the bacterial community changes and pH drops sharply, leading to the formation of endotoxins and histamine, which are absorbed into the blood, forming endotoxemia acidosis (Slyter, 1976; Andersen and Jarlv, 1990; Duffield et al., 2004). The absorption of histamine and lipopolysaccharide (LPS) in the rumen stimulates its epithelium to present inflammation, aggravates the damage of rumen epithelial cells, and exacerbates the condition of acidosis (Aschenbach et al., 2000; Pawlinski et al., 2003; Gressley, 2014). Recently, numerous studies have focused on the prevention of SARA. Antibiotics can control the yield of lactate by effectively inhibiting the growth of lactate-producing bacteria (Owens et al., 1998). Some studies have shown that inoculating lactate-utilizing bacteria can relieve SARA. For instance, Robinson et al. (1992) inoculated 2 × 1012–3 × 1012 Megasphaera elsdenii 407A in the rumen of bovine, and the concentration of lactate in the rumen decreased significantly. Elsewhere, organic acids (Callaway et al., 1997; Martin et al., 1999) and a buffering agent (Mao et al., 2016) were also applied to prevent SARA. In our previous study, we found that the supply of thiamine may serve as a novel strategy to alleviate SARA (Zhang et al., 2019). Pan et al. (2017a) opined that the supply of thiamine can stabilize the bacterial community as well as increase the pH of the rumen. Besides, the addition of thiamine could relieve the inflammatory response by decreasing endotoxin levels in the rumen and also inhibit the expression of proinflammatory cytokines in the rumen epithelium (Pan et al., 2017b; Zhang et al., 2019).
Presently, 16S rRNA sequencing technology has been widely used in agriculture, medicine, and biology to reveal microbial communities and correlations with environmental factors (Guo, 2008; Zhang H.W. et al., 2010; Zhang Y. et al., 2010; Liu et al., 2011; Patel et al., 2011; Saikia et al., 2015). Metabonomics, based on LC-MS, is an emerging tool for targeted and nontargeted analysis of small-molecule metabolites in organisms (Vuckovic, 2012; Zhao et al., 2014; Caboni et al., 2016). These key technologies have been proven valuable for further studies focusing on all aspects of molecular mechanisms (Boudonck et al., 2009; Saleem et al., 2012; Saro et al., 2018).
Thiamine is crucial to the host’s metabolism, immunity, and health. More importantly, a biologically active form of thiamine, thiamine pyrophosphate (TPP), is essential for energy metabolism (Bettendorff et al., 2014). Most of the thiamine in ruminants are derived from the synthesis of rumen microorganisms and a small part in the decomposition of feed materials (Miller et al., 1986). For ruminants, thiamine is synthesized adequately by ruminal microbes for host demand in most cases from a normal diet. Breves et al. (1981) and Miller et al. (1986) reported that ruminants require no dietary thiamine requirements because rumen microbes can synthesize it. However, Karapinar et al. (2010) and Pan et al. (2016) noted that thiamine deficiency occurred when sheep or cattle had subacute or acute ruminal acidosis, which is associated with increasing thiamine degradation by thiaminase and decreasing microbial thiamine synthesis activity under high-grain-induced SARA (Brent, 1976). Ruminants have been established to acquire subacute or acute ruminal acidosis due to a high-concentrate diet, while the exchange of substances and energy may occur abnormally, causing thiamine deficiency (Karapinar et al., 2010; Pan et al., 2016). More interestingly, thiamine supplementation when feeding high-concentrate diets can increase ruminal pH, followed by a concomitant decrease in ruminal lactate concentration (Pan et al., 2016), thereby increasing the population of ruminal cellulolytic bacteria (Pan et al., 2017a). Our previous study uncovered that thiamine supplementation with a high-concentrate diet can increase ruminal pH along with the reduction of rumen lactate concentration (Pan et al., 2016). In addition, we also have demonstrated that thiamine supplementation with a high-concentrate diet can improve the rumen epithelial activity of TPP-dependent enzymes (unpublished results). In another study, thiamine regulated the structure of the rumen microbial community of SARA cows by reducing the population of Streptococcus bovis, thereby promoting the growth of M. elsdenii (Wang et al., 2015).
Previous studies have used 16S rRNA sequencing technology to explore the correlations between the microbial communities and thiamine in dairy cows during SARA (Pan et al., 2017a). Unfortunately, there is a dearth of information regarding the relationship between the supply of thiamine and rumen metabolites. We thus hypothesize that thiamine supplementation can modulate ruminal fluid microbiota and metabolites of goats fed a high-concentrate diet.
Materials and Methods
Animals, Feedstuffs, and Feeding Regimes
All animal experiments were conducted in line with the Animal Protection Law based on the Guidelines for the Care and Use of Laboratory Animals approved by the Ethics Committee of Yangzhou University (SXXY 2015-0054).
Eighteen mid-lactating (148 ± 3 DIM) Saanen goats (body weight = 36.5 ± 1.99 kg; body condition score = 2.73 ± 0.16, where 0 = emaciated and 5 = obese; Russel et al., 1969) in parity 1 or 2 were used. All goats were randomly fed one of three diets: (1) control diet (CON; n = 6; concentrate:forage 30:70), (2) high-concentrate diet (HG; n = 6; concentrate:forage 70:30), (3) high-concentrate diet with 200 mg of thiamine/kg of DMI (HGT; n = 6; concentrate:forage 70:30). The supplementation dose of thiamine was based on previous studies on lambs (Neville et al., 2010), dairy goats (Zhang H.W. et al., 2010), and cows (Zhang et al., 2014; Pan et al., 2017b). Goats in each group were fed on the respective diets for 8 weeks. Dietary components of dairy goats were formulated according to NRC (2001). The dietary components are presented in Supplementary Table 1, and the data have been published in the Journal of Dairy Science (Zhang et al., 2019). The goats were fed at 7:00 and 18:00 and with one half of the allowed daily dose at each feeding. Thiamine was mixed with the concentrate to be licked by the goats. During the trial period, goats were allowed free access to freshwater.
Collection and Measurement of Rumen Samples
On the last day of week 8, the goats were slaughtered 4 h after feeding, and a representative sample of ruminal digesta (at least 200 ml) was collected from the ruminal ventral sac. Then, 200 ml of rumen fluid was collected from each animal and transferred into a separate sterilized container, immediately frozen with liquid nitrogen, and stored at −80°C for further processing. In total, 18 rumen fluid samples were collected from the goats; however, two rumen fluid samples from the HGT group were damaged, and therefore, metabolomic analysis was only performed for the remaining 16 rumen fluid samples. A pH meter (Sartorius, Göttingen, Germany) was used to measure rumen pH using the collected samples. One hundred milliliters of the fluid samples was filtered through four layers of cheesecloth and centrifuged at 10,000 × g for 15 min at 4°C to obtain the supernatant, which was used to determine the concentration of volatile fatty acids (VFAs) and ammonia nitrogen (NH3-N). The content of thiamine in the rumen fluid was measured by high-performance liquid chromatography (HPLC) according to the Analytical Methods Committee (2000). Another 100 ml of aliquot was stored at −80°C immediately for microbial DNA extraction and metabolomic analysis.
DNA Extraction, 16S rRNA Gene Amplification, and Sequencing
The collected rumen samples were thawed at room temperature, and a 1-ml aliquot was centrifuged at 10,000 × g for 1 min at 4°C, and then the supernatant was discarded. DNA was extracted with a bead-beating method using a mini-bead beater (BioSpec Products, Bartlesville, United States) as per manufacturer’s instructions, followed by phenol–chloroform extraction (Zoetendal et al., 1998). After the solution was precipitated with anhydrous ethanol, the pellets were suspended in the EDTA buffer. The DNA was determined spectrophotometrically using a Nanodrop spectrophotometer (Nyxor Biotech, Paris, France), following staining with QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). DNA was stored at −80°C until further processing.
Next-generation sequencing (NGS) library preparations and Illumina MiSeq sequencing were conducted at Novogene Inc. (Nanjing, China). The V3–V4 regions of the bacterial 16S rRNA gene were amplified by PCR using a forward primer (CCTACGGRRBGCASCAGKVRVGAAT) and reverse primer (GGACTACNVGGGTWTCTAATCC). These primers also contained adapter sequences that allow uniform amplification of highly complex libraries in preparation for downstream NGS on Illumina MiSeq. All PCRs were performed in a 50-μl thermal cycler (Bio-Rad, United States) containing 4 μl of fivefold FastPfu buffer, 10 ng of DNA, 0.8 μM of each primer, 0.5 U of Pfu polymerase, and 2 μl of 2.5 mM dNTPs. PCR conditions utilized for amplification were as follows: first step of 95°C for 3 min, followed by 27 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 45 s, and a final extension at 72°C for 10 min. The amplicons were purified using the AxyPrep DNA gel Extraction Kit (Axygen Biosciences, Union City, CA, United States). Thereafter, amplicon libraries were generated using a TruSeqTM DNA Sample Prep Kit (TransGen Biotech, China) according to the manufacturer’s protocol. Paired-end (2 × 250 bp) sequencing was performed with the Illumina MiSeq platform based on the manufacturer’s guidelines (Caporaso et al., 2010).
Sequence Processing and Analysis
We used Cutadapt V1.9.1 software (Martin, 2011) to trim the low-quality part of sequence reads, while sample data were extracted from the reads obtained using barcodes. Next, we removed barcode and primer sequences to obtain raw reads. Then read sequences (Rognes et al., 2016) were compared against the species annotation database to detect the chimeric sequences, which were subsequently removed (Haas et al., 2011) to obtain the final clean reads. Using UPARSE V7.0 software, clean reads from all samples were clustered (Edgar, 2013), whereby 97% of identity sequences were clustered into operational taxonomic units (OTUs). Then, sequences with the highest occurrence frequency in OTUs were screened out as the representative sequences of OTUs following the principles of the algorithm. Additionally, community richness and diversity were measured using the Chao1 and Shannon indices, respectively. Species annotation analysis was conducted using the Mothur method and SILVA132 (Wang, 2007) SSU rRNA database (Quast and Pruesse, 2013) (the threshold value was set as 0.8–1) so as to obtain taxonomic information and make statistics of the community composition of each sample at various classification levels, including kingdom, phylum, class, order, family, genus, and species. The data of each sample were homogenized. We employed QIIME 1.9.1 and R 2.15.3 software to execute different analyses between alpha diversity index groups.
LC-MS Metabolomics Processing
A total of 16 rumen samples were analyzed using the LC-MS platform as previously reported elsewhere (Chen et al., 2009; Zhu, 2013). Afterward, samples were resuspended with prechilled methanol and 0.1% formic acid by vortexing, followed by incubation on ice for 5 min and centrifugation at 15,000 rpm at 4°C for 5 min. After centrifugation, supernatants were diluted to a final concentration of 60% methanol using LC-MS-grade water. The samples were subsequently transferred to a fresh Eppendorf tube with a 0.22-μm filter and then centrifuged at 15,000 × g at 4°C for 10 min. Finally, the filtrate was injected into the LC-MS system for analysis. All LC-MS analyses were performed using a Vanquish UHPLC system (Thermo Fisher) coupled with an Orbitrap Q Exactive HF-X mass spectrometer (Thermo Fisher). Briefly, samples were injected onto a Hypersil Gold column (100 × 2.1 mm, 1.9 μm) using a 16-min linear gradient at a flow rate of 0.2 ml/min. The eluents for the positive polarity mode were eluent A (0.1% FA in water) and eluent B (methanol), while those of the negative polarity mode were eluent A (5 mM ammonium acetate, pH 9.0) and eluent B (methanol). The solvent gradient were set as follows: 2% B, 1.5 min; 2–100% B, 12.0 min; 100% B, 14.0 min; 100–2% B, 14.1 min; and 2% B, 16 min. Lastly, the Q Exactive HF-X mass spectrometer was operated in positive/negative polarity mode with a spray voltage at 3.2 kV, capillary temperature at 320°C, sheath gas flow rate at 35 arb, and aux gas flow rate at 10 arb.
Metabolomics Data Analysis
To perform peak alignment, peak picking, and quantitation for each metabolite, we processed the raw data files generated by ultra-HPLC–mass spectrometry (UHPLC-MS/MS) using the Compound Discoverer 3.0 (CD 3.0, Thermo Fisher). For data collection, the main parameters were set as follows: retention time tolerance, 0.2 min; actual mass tolerance, 5 ppm; signal intensity tolerance, 30%; signal/noise ratio, 3; and minimum intensity, 100,000. Then, peak intensities were normalized to the total spectral intensity. The normalized data were used to predict the molecular formula based on additive ions, molecular ion peaks, and fragment ions. In order to obtain accurate qualitative and quantitative results, the peaks were matched against the mzCloud and ChemSpider databases.
After Pareto scaling treatment, principal component analysis (PCA) was performed on the peaks extracted from all trial samples. The PLS-DA variable importance in the projection (VIP) values combined with P-values of the t-test were used to highlight significant differential expression metabolites. The screening criteria were as follows: threshold (VIP) > 1.0 and difference multiple (FC) > 2.0 or FC < 0.5 and P-value < 0.05. In addition, significantly differentially expressed metabolites were analyzed for expression pattern clustering using the gplots package in R (Warnes et al., 2016). Correlations between different metabolites and bacterial communities were explored with Spearman’s correlation analysis using the pheatmap package in R (Kolde, 2012). For all analyses, FDR-corrected P-values below 0.05 were considered statistically significant.
Results
Rumen Fermentation Parameters
The fermentation and thiamine characteristics (pH, NH3-N, VFA, LPS, and PDH profile) are detailed in Supplementary Table 2. The data have been published in the Journal of Dairy Science (Zhang et al., 2019). In summary, thiamine altered rumen fermentation characteristics.
Richness, Diversity Estimates, and Rumen Bacteria Composition
A total of 624,283 reads were detected from 12 samples via Illumina sequencing. Read shear filtration and quality control obtained an average of 58,551 effective data, while the quality control effective rate reached 93.88%. Sequences were clustered into OTUs at a 97% similarity level, generating a total of 1,257 OTUs. The Venn diagram shows that the intersection numbers of OTU for the CON, HG, and HGT groups were 764 (Figure 1A). Further, rarefaction curves revealed that the curve flattens out as the sequencing deepens, demonstrating that the OTU of the HGT group was higher than that of the HG group but lower than that of the CON group at the same sequencing depth (Figure 1B). Within the bacterial population, we uncovered three dominant phyla, namely, Bacteroidetes (53.41%), Firmicutes (36.32%), and Proteobacteria (3.36%) (Figure 2).
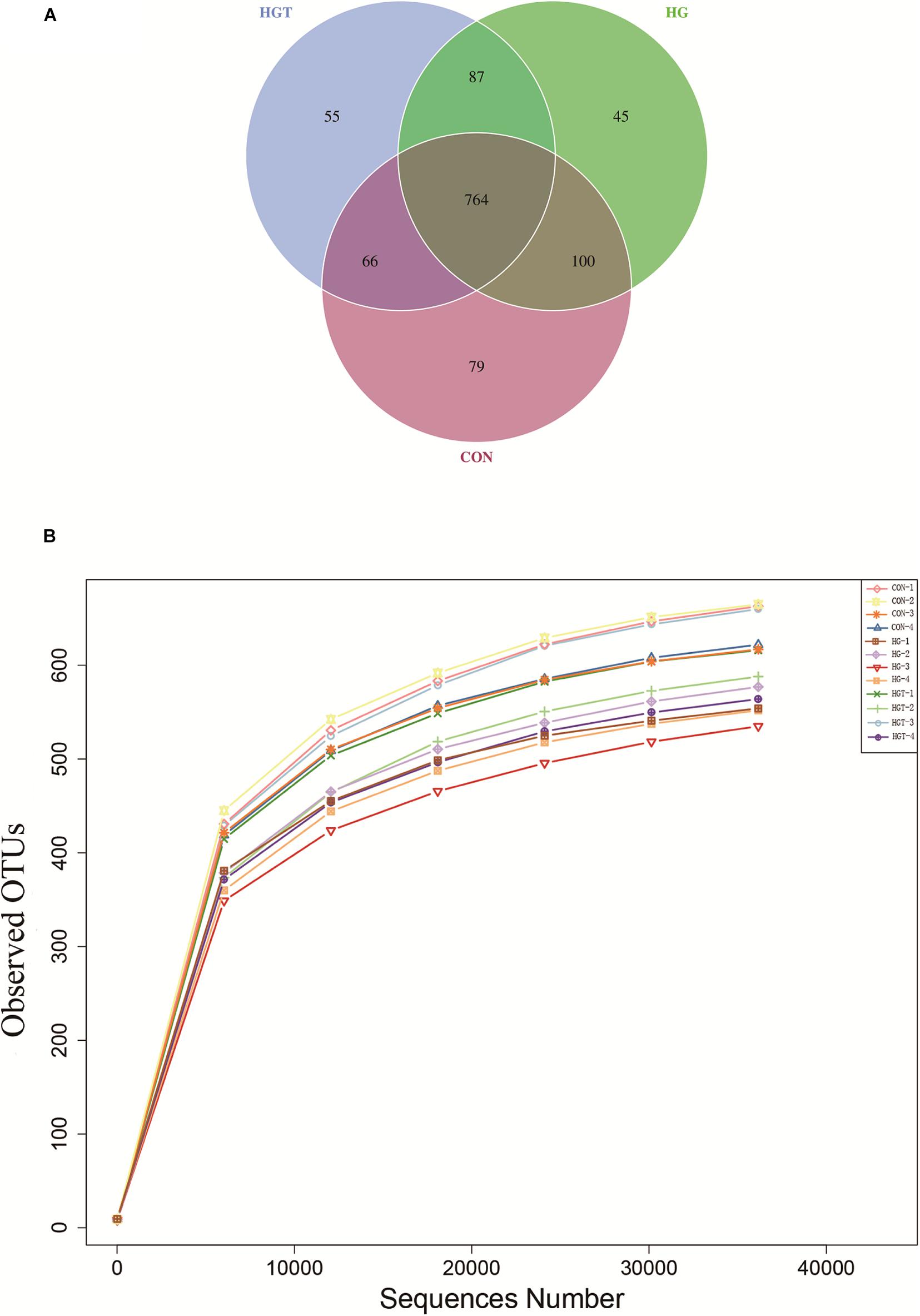
Figure 1. Ruminal microbial operational taxonomic units (OTUs) with different treatments of diets. CON: basal diets without supplement; HG: high-concentrate diet; HGT: high-concentrate diet supplemented with 200 mg of thiamine / kg of DMI. (A) Venn diagram of ruminal bacterial OTUs. The number of unique OTUs were represented by the non-overlapped portion of Venn diagram for each group. (B) Bacterial rarefaction curves based on OTUs were used to assess the depth of coverage for each sample.
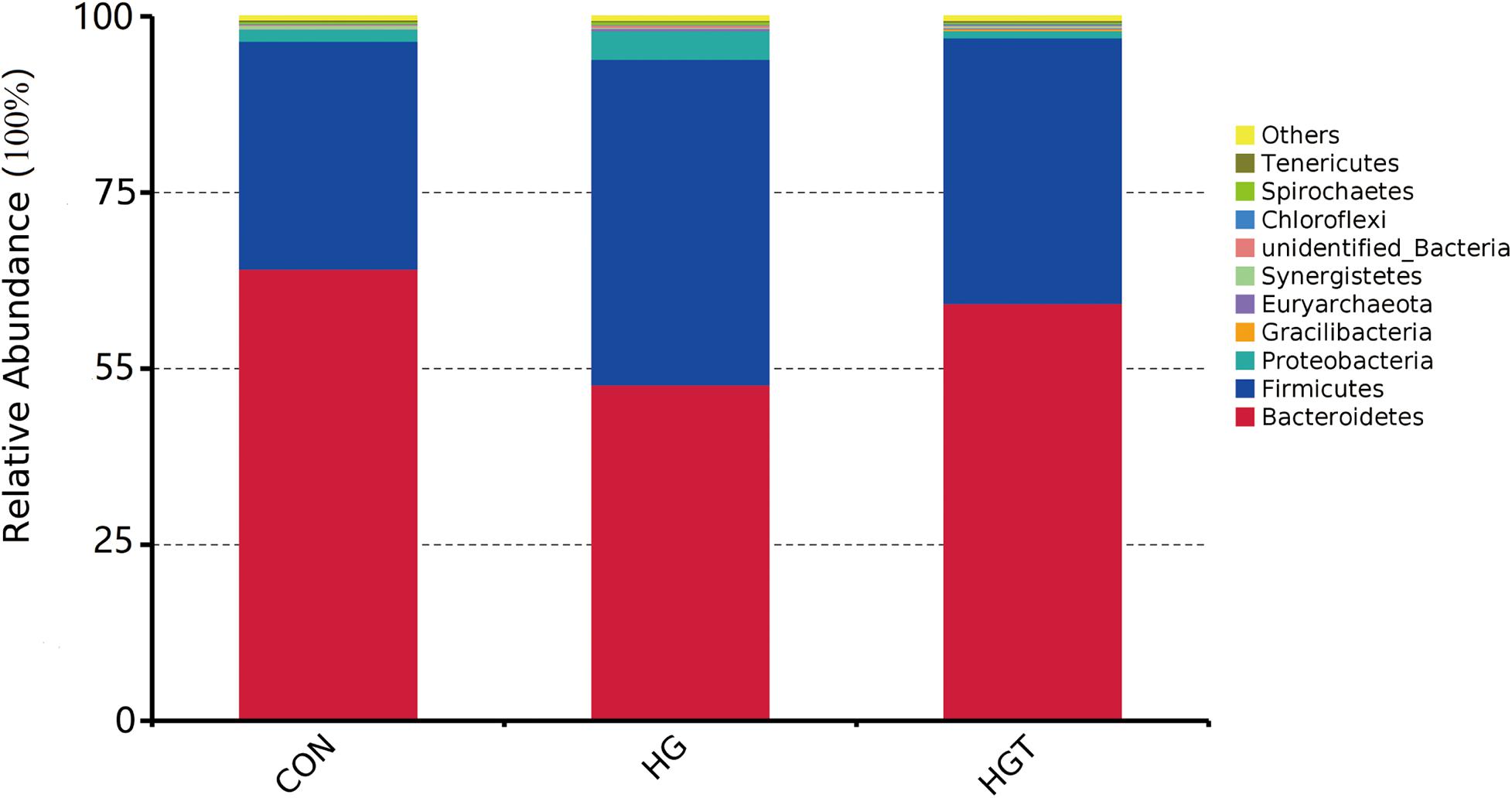
Figure 2. Percentage composition of the top 10 predominant phyla in rumen fluid. CON: basal diets without supplement; HG: high-concentrate diet; HGT: high-concentrate diet supplemented with 200 mg of thiamine / kg of DMI.
In terms of alpha bacterial diversity (Table 1), we observed no significant differences in OTU and Good’s coverage between the CON and HGT groups. Compared with the CON group, the feeding of high-grain diets (HG) markedly decreased Chao1 richness, abundance-based coverage estimator (ACE), and Shannon diversity index. Supplementation with thiamine significantly increased Shannon (P < 0.05), but the increases in ACE, Chao1 value, and Simpson index were insignificant (P > 0.05) in the HGT group compared with the HG group.
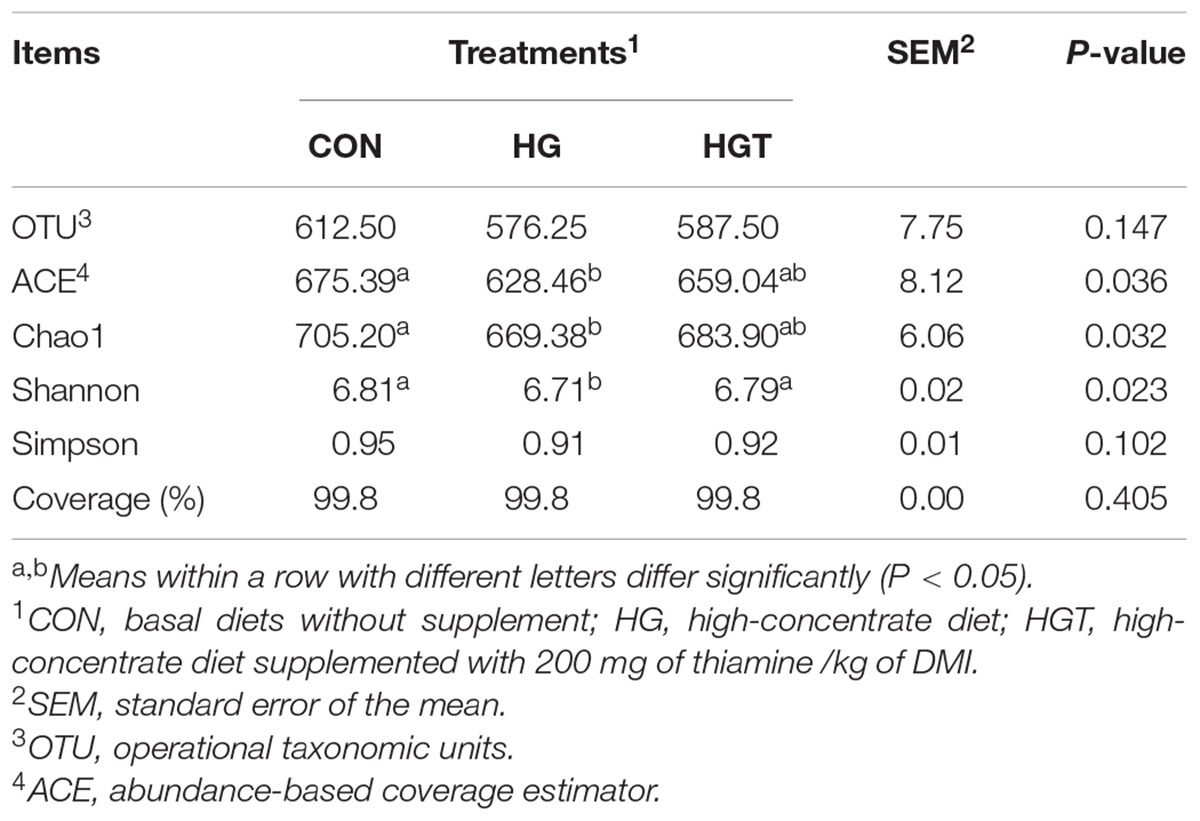
Table 1. Number of observed species, richness, and diversity indices in ruminal samples from each dietary treatment.
PCA revealed that goats fed with HG diet were distinctly separate from goats in the CON and HGT groups (Figure 3). Importantly, principal coordinates 1 and 2 accounted for 13.57 and 11.13% of the total variation, respectively.
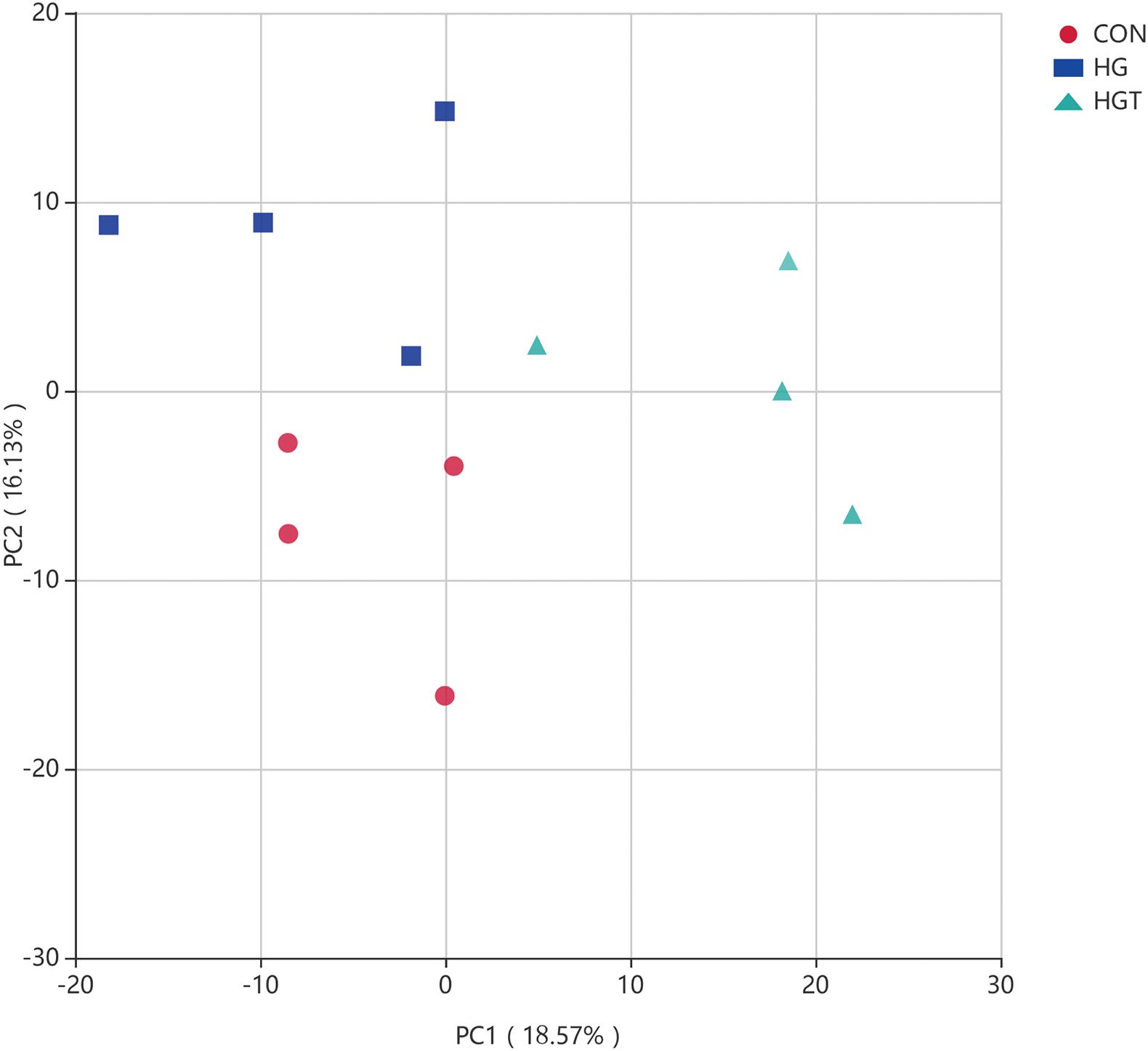
Figure 3. Principal Component Analysis (PCA) of bacterial community structures of the ruminal microbiota in CON (red circles), HG (blue rectangle), and HGT (green triangle) groups. CON: basal diets without supplement; HG: high-concentrate diet; HGT: high-concentrate diet supplemented with 200 mg of thiamine / kg of DMI.
Effect of Thiamine Supplementation on Relative Abundance of Bacterial Communities
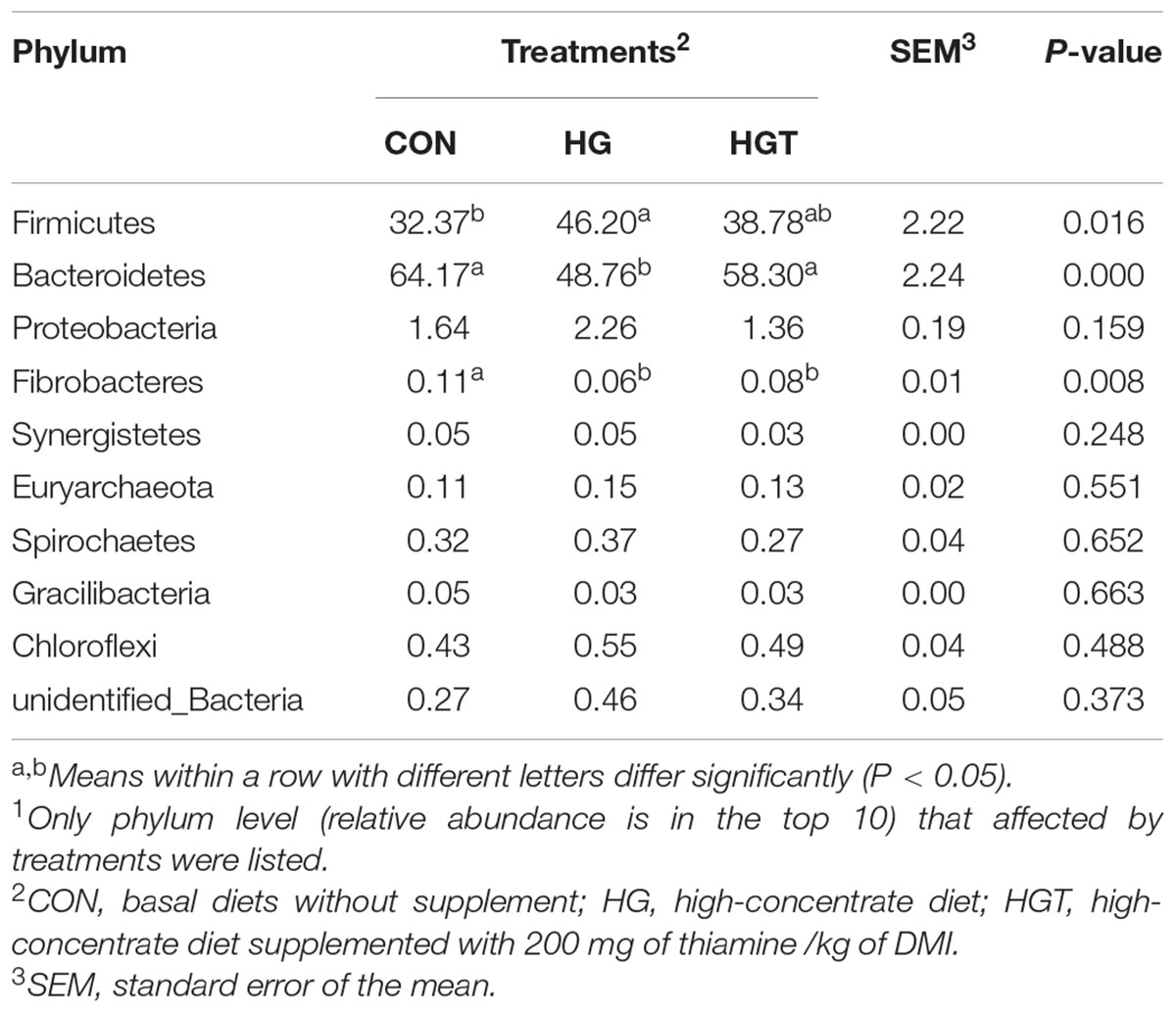
At the phylum level (Table 2), the HG diet significantly decreased the relative abundance of Bacteroidetes (P < 0.01) and Firmicutes (P < 0.05), as well as increased the relative abundance of Firmicutes (P < 0.05) compared with the CON groups. Whereas supplementation with thiamine in the HGT group considerably increased the abundance of Bacteroidetes (P < 0.01), the increase of Fibrobacteres and the decrease of Firmicutes in relative abundance were insignificant (P > 0.05) compared with those in the CON group. We also identified no significant shifts in the relative abundance of Proteobacteria, Synergistetes, Euryarchaeota, Spirochaetes, Gracilibacteria, and Chloroflexi with the supplementation of HGT and thiamine compared with the CON group (P > 0.05).
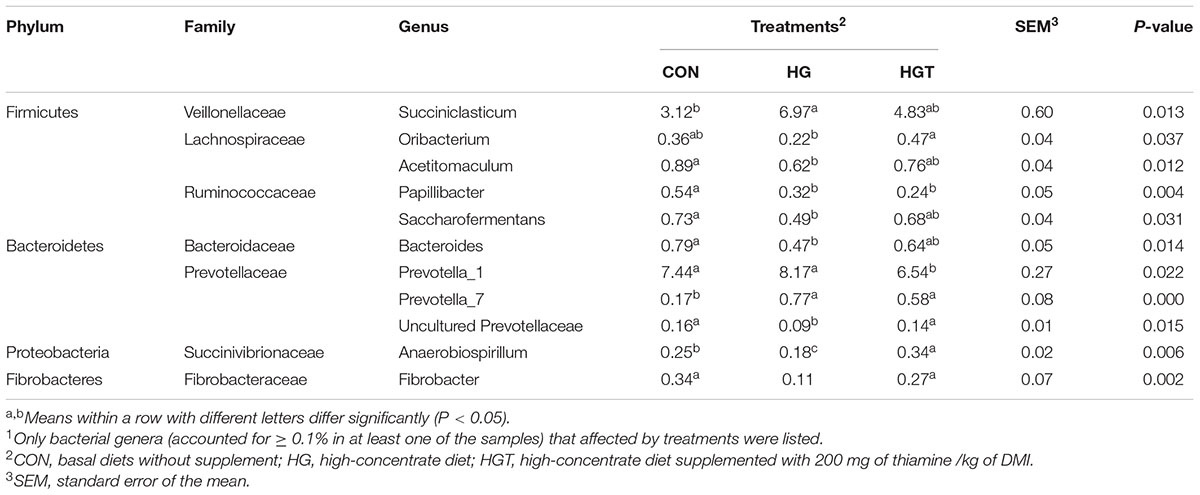
At the genus level (Table 3), compared with the CON group, high-concentrate diet (HG) substantially decreased the relative abundance of Oribacterium, Acetitomaculum, Saccharofermentans, Bacteroides, uncultured Prevotellaceae, Anaerobiospirillum, Papillibacter, and Fibrobacter (P < 0.05) but also increased the abundance of Succiniclasticum, Prevotella_1 (P < 0.05), and Prevotella_7 (P < 0.01). While supplementation with thiamine largely increased the relative abundance of Oribacterium, uncultured Prevotellaceae (P < 0.05), Anaerobiospirillum, and Fibrobacter (P < 0.01), as well as decreased the relative abundance of Prevotella_1 (P < 0.05), but the effect on the relative abundance of Succiniclasticum, Acetitomaculum, Saccharofermentans, Bacteroides, and Prevotella_7 was insignificant compared with that of high-concentrate diet (P < 0.05). Notably, the results of correlation analysis revealed that phenyl propionic acid was positively associated (r > 0.38, P < 0.05) with 11 taxa and negatively associated (r < −0.38, P < 0.05) with four taxa and several ciliate and methanogens.
Rumen Metabolomics Profiling
Sample Quality Control
We further evaluated the stability and reproducibility of data using the QC samples measured during the whole experimental period. The higher the QC sample correlation (R2 close to 1), the better the quality of data. The correlation of QC samples is shown in Supplementary Figure 1. The R2 values of the positive and negative polarity modes were 0.981 and 0.986, respectively.
Differential Metabolite Analysis
The PCA results among the CON group, HG group, and HGT group. Remarkably, PCA provided a satisfactory separation of data among the high-concentrate diet group (HG), high-concentrate diet + thiamine group (HGT), and the CON group (Supplementary Figure 2).
The volcano maps of differential metabolites. We employed volcanic diagrams to show the number of differential metabolites in each comparison. As shown in Figure 4, the gray plot indicates no difference, the red plots show upregulated endogenous metabolites, and the green plots show downregulated endogenous metabolites. There were 14 common differential metabolites between the two comparisons for the HG versus CON groups and HGT versus HG groups that presented different expression trends (Table 4).
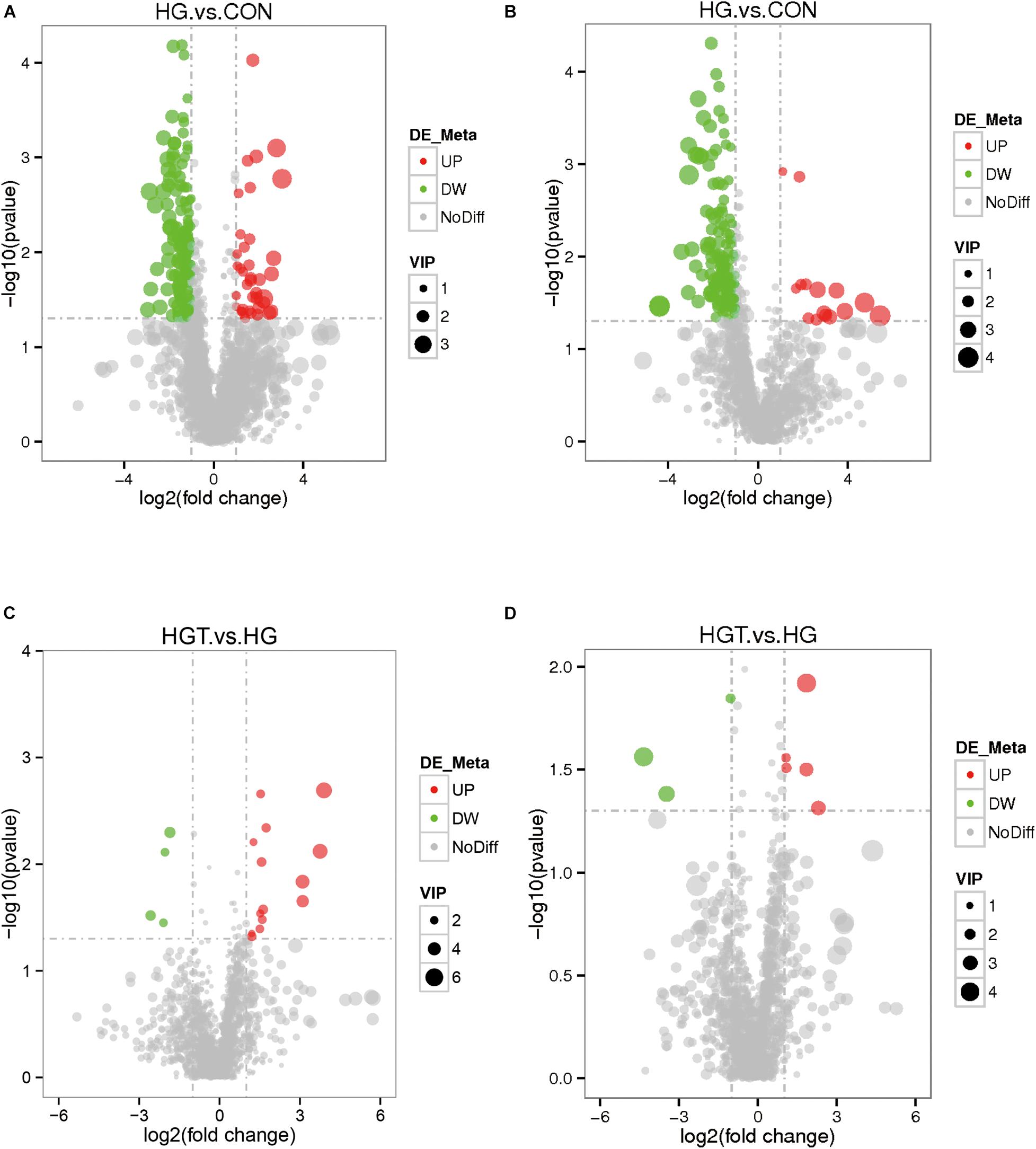
Figure 4. The volcano plot of differential metabolites with different treatments of diets. The gray plot shows there is no difference, the red plots show up-regulated endogenous metabolites, while the green plots show down-regulated endogenous metabolites. VIP value represents the importance projection value of the metabolite obtained in the PLS-DA model compared in this group. CON: basal diets without supplement; HG: high-concentrate diet; HGT: high-concentrate diet supplemented with 200 mg of thiamine / kg of DMI. (A) The volcano plot between the HG and the HGT group in the positive polarity mode. (B) The volcano plot between the HG and the HGT group in the negative polarity mode. (C) The volcano plot between the HGT and the HG group in the positive polarity mode. (D) The volcano plot between the HGT and the HG group in the negative polarity mode.
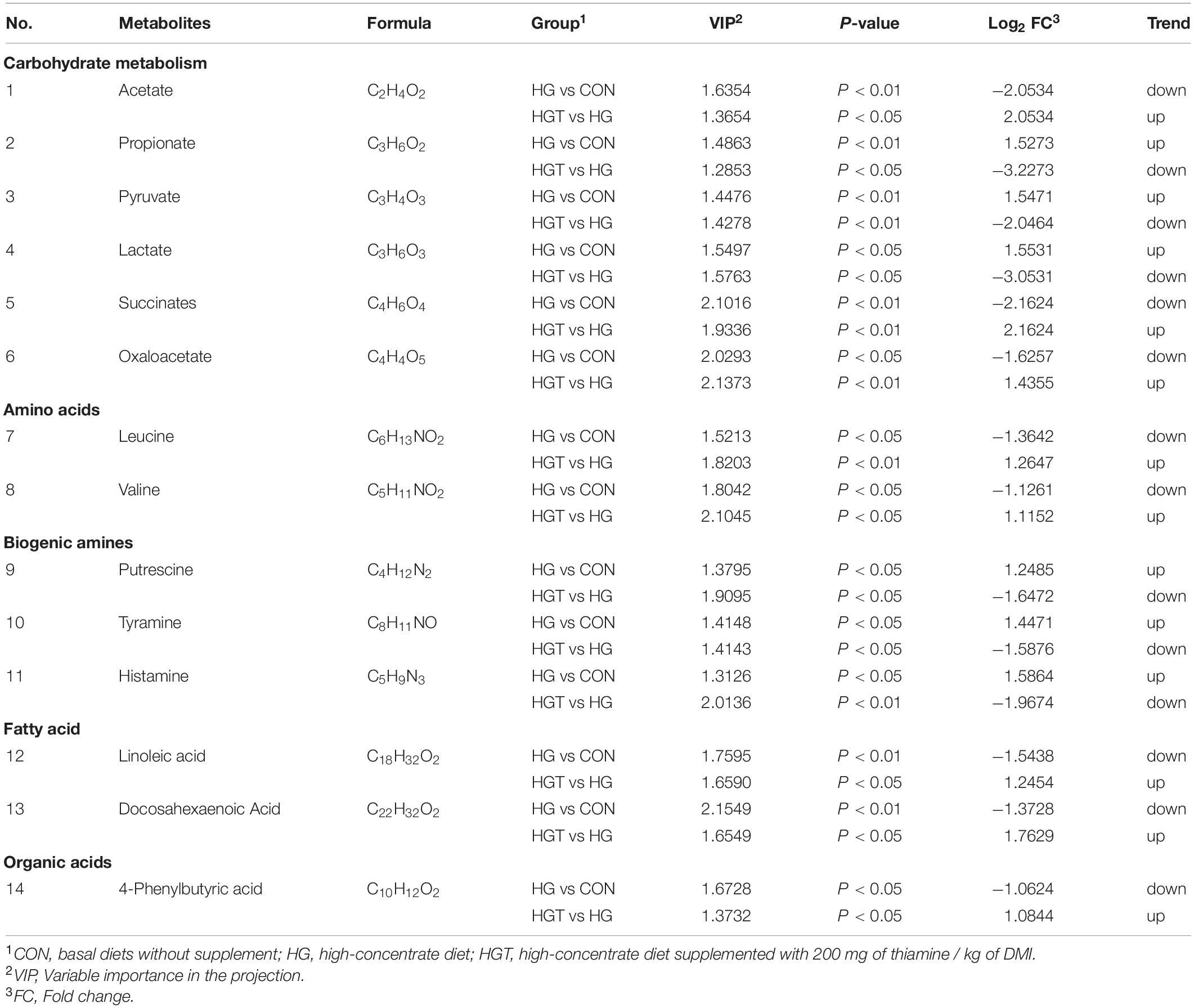
Table 4. Different metabolites content in the rumen of goats fed SARA-induced diet (HG) versus goats fed SARA-induced diet with thiamine supplementation (HGT).
Correlation Analysis Between Genera and Metabolite Concentrations Affected by the Feeding of Thiamine
To explore the functional correlation between the rumen microbiome changes and metabolite perturbations, a Pearson correlation matrix was generated by calculating the Pearson correlation coefficient in high-concentrate diet with thiamine supplement (Figure 5). The results showed a clear correlation between microbial structure and metabolites (r > 0.38 or < 0.38, P < 0.05). More interestingly, correlation analysis revealed that acetate was positively associated with eight genera (r > 0.49, P < 0.05) and negatively associated with one genus (r < −0.62, P < 0.05). Propionate was positively associated with six genera (r > 0.41, P < 0.05) and negatively associated with four genera (r < −0.55, P < 0.05). Pyruvate was positively associated with 12 genera (r > 0.52, P < 0.05), while lactate was positively associated with three genera (r > 0.54, P < 0.01) and negatively associated with six genera (r < −0.45, P < 0.05). Succinates were positively associated with nine genera (r > 0.55, P < 0.05). Likewise, oxaloacetate was positively associated with nine genera (r > 0.47, P < 0.05). Leucine was positively associated with four genera (r > 0.48, P < 0.05) and negatively associated with five genera (r < −0.59, P < 0.05), whereas valine was positively correlated with five genera (r > 0.48, P < 0.05) and negatively associated with two genera (r < −0.62, P < 0.01). Putrescine was positively associated with two genera (r > 0.60, P < 0.05) and negatively associated with seven genera (r < −0.69, P < 0.01). Tyramine was positively associated with two genera (r > 0.76, P < 0.05) and negatively associated with seven genera (r < −0.66, P < 0.05). Histamine was positively associated with one genus (r > 0.78, P < 0.05) and negatively associated with eight genera (r < −0.65, P < 0.05). Linoleic acid was positively associated with three genera (r > 0.62, P < 0.05) and negatively associated with three genera (r < −0.52, P < 0.01). Docosahexaenoic acid was positively associated with four genera (r > 0.62, P < 0.05) and negatively associated with two genera (r < −0.57, P < 0.05). Finally, 4-phenylbutyric acid (4-PBA) was positively associated with one genus (r > 0.86, P < 0.01) and negatively associated with seven genera (r < −0.64, P < 0.05).
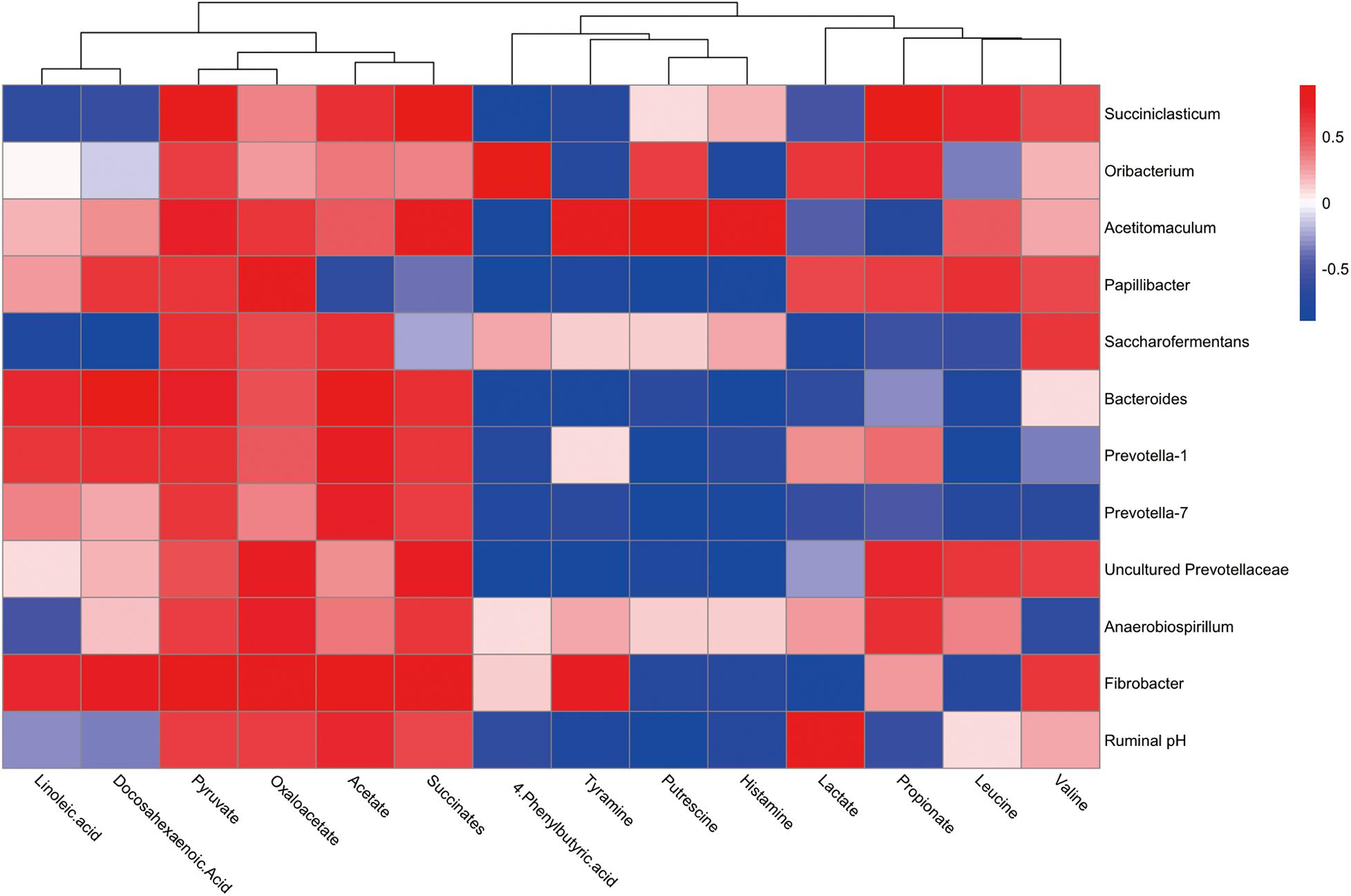
Figure 5. Correlation analysis between genera and metabolite concentrations affected by the feed high-concentrate diet with thiamine supplementation. Each row in the graph represents a genus, each column represents a metabolite, and each lattice represents a Pearson correlation coefficient between a component and a metabolite. Red represents a positive correlation, while blue represents a negative correlation.
Discussion
Effects of Thiamine Supplementation on Ruminal Microbial Community Under High-Concentrate Diet Feeding
In the present manuscript, we have demonstrated the alteration of rumen fermentation characteristics by thiamine (Supplementary Table 2), and the data have been published in the Journal of Dairy Science (Zhang et al., 2019). To the best of our knowledge, the detailed mechanisms by which thiamine supplementation relieves SARA in goats and dairy cows remain largely unknown. Therefore, to determine the potential mechanism of thiamine on rumen fermentation and bacterial community, we analyzed a high-concentrate diet using 16S rRNA gene sequencing technology.
Our PCA results showed differences in the bacterial community among CON, HG, and HGT groups, indicating the impact of thiamine supplementation. Recent studies have shown that a long-term supply of HG diet results in an alteration of rumen microbial populations (Mao et al., 2015; Hua et al., 2017), while thiamine supplementation in our previous study could increase the abundance of rumen bacterial community (Pan et al., 2017a).
At the phyla level, we observed that high-concentrate dietary supplementation with thiamine might significantly improve the abundance of Bacteroidetes. We also noted that the abundance of Firmicutes decreased in the HGT group compared with the HG group, although the change was statistically insignificant. Of note, the phyla Firmicutes and Bacteroidetes are known for polysaccharide fermentation (Wang et al., 2019). Additionally, Bacteroidetes, one of the most abundant gram-negative bacteria in the lumen, are classified into three categories, namely, Bacteroidetes, Flavobacteria, and Sphingobacteria. Zened et al. (2013) reported that an increase in fermentable substrates led to a decrease in pH in the rumen during high-concentrate diet supplementation, which can subsequently lead to death and lysis of gram-negative bacteria (Cole et al., 2005). Firmicutes are predominately composed of gram-positive bacteria (Fernando et al., 2010), which are also insensitive to low pH. Similarly, these bacteria can still degrade easily fermentable carbohydrates under low pH (Wetzels et al., 2015). However, supplementation with thiamine partially reversed this effect.
At the genus level, a high abundance of Succiniclasticum was identified in the high-concentrate diet group than in the control group, whereas thiamine supplementation reversed this change, although this result was insignificant. A similar trend was also reported for Succiniclasticum with a high-concentrate diet elsewhere (Petri et al., 2013). Importantly, Succiniclasticum is an organism that specializes in fermenting succinate by converting it quantitatively to propionate, the most important precursor of glucose in ruminants (Van Gylswyk, 1995). Unfortunately, the function of Succiniclasticum in the rumen remains enigmatic. As shown in Supplementary Table 2, the changing trend of propionate content in the rumen was similar to that of Succiniclasticum. The low energy yield when anaerobes convert succinate to propionate should make this bacterium highly efficient in this conversion (Thauer et al., 1977). Thiamine is a coenzyme of pyruvate dehydrogenase (PDH) (Chen et al., 2016); the increasing thiamine concentration could facilitate the transformation of pyruvate to acetyl CoA, which ultimately might be hydrolyzed to acetate. The change reduced the production of succinate and subsequently decreased the proportion of Succiniclasticum in rumen concentration. Oribacterium has been identified as one of the predominant bacteria in the rumen of cows fed forage-based diets (Kong et al., 2010; Kenters et al., 2011), but its role in the rumen remains unknown. Noticeably, thiamine supplementation increased significantly the abundance of Oribacterium. However, the reasons for the change in abundance of this genus due to thiamine supplementation need further clarification.
The thiamine treatment group exhibited a higher proportion of the Anaerobiospirillum, which can ferment glucose or starch into succinate (Nghiem et al., 1997; Lee et al., 2000). Interestingly, thiamine is associated with the production of acetyl coenzyme A, while acetyl coenzyme A is also related to the production of succinate (Nyström, 2015). The supply of thiamine increased the utilization of the substrate by Anaerobiospirillum, hence promoting its proliferation. Besides, Asma et al. (2013) reported the differences due to dietary starch levels in the relative abundances of Fibrobacteres, whose relative abundance decreased by more than two times when the dietary starch level increased. This changing trend is consistent with our results. At present, two species belonging to the genus Fibrobacter are known, including Fibrobacter succinogenes and Fibrobacter intestinalis (Amann et al., 1992). In particular, F. succinogenes has been found to utilize its incomplete citric acid cycle to produce succinate, the bacterium’s major fermentative end product (Miller, 1978). Additionally, a recent study has demonstrated that F. succinogenes contains both a phosphoenolpyruvate carboxykinase and a pyruvate carboxylase that can reversibly convert pyruvate, respectively, to oxaloacetate, which is sequentially converted to malate, fumarate, and succinate. This degradation process requires thiamine pyrophosphate as a coenzyme factor, which is possibly one of the reasons for the increased content of succinate in the rumen. Moreover, Prevotella_1, a polysaccharide-degrading bacteria belonging to the Prevotella ruminicola family, in the HGT group tended to be lessened by thiamine supplementation that can promote the transformation of some substances in this degradation pathway (Purushe et al., 2010).
Effects of Thiamine Supplementation on Ruminal Metabolites
Thiamine contributes significantly to carbohydrate metabolism and is essential for normal cellular function and growth. For the effects of thiamine supplementation on carbohydrate metabolism, our metabolome data indicated that thiamine supplementation alters fermentation type with a high-concentrate diet in the rumen, implying that the ruminal metabolism pattern might be associated with structural changes in bacterial communities. Previous studies have elucidated that supplementation of nonstructural carbohydrates can affect metabolism (Afzal, 1999). As a result, change in carbohydrate structure affects the bacterial communities in the rumen (Mcallan and Smith, 1974). Carbohydrate metabolism plays a vital role in regulating animal health. For example, Sutton (1985) found that the end products of carbohydrates act as energy substrates, inflammation modulators, and signaling molecules in the rumen. As a metabolite of carbohydrates, VFA can regulate the differentiation of the rumen epithelium, cell apoptosis, and morphology of the rumen papilla (Mentschel et al., 2001; Inagaki and Sakata, 2005; Shen et al., 2005). Our findings unearthed that thiamine supplementation in a high-concentrate diet markedly increased the content of acetate and decreased the content of propionate compared with those in the HG group diet in ruminal fluid. The ratio of acetate to propionate is positively correlated with the content of the concentrate (Lechartier and Peyraud, 2010). Studies have emphasized that the succinate pathway is the principal pathway of propionate production, which is used by Bacteroidetes to generate propionate from a substrate (Louis et al., 2014). More importantly, this may be a plausible explanation for the decrease in the ratio of acetate to propionate with thiamine supplementation relative to the high-concentrate diet group. Besides, as a coenzyme factor of PDH (Bubber et al., 2004), deficiency of thiamine leads to the accumulation of pyruvate, which is then converted to lactate under the catalytic action of lactate dehydrogenase (LDH) (Kumar et al., 2015). The supplement of thiamine can promote the conversion of pyruvate into acetyl-CoA, reducing the accumulation of lactate and pyruvate. Moreover, thiamine is a coenzyme required for the activity of α-ketoglutarate dehydrogenase (α-KGDHC) that is involved in the Krebs cycle (Hanson and Cox, 1967). The increase in oxaloacetate content can be interpreted as the enhancement of the tricarboxylic acid cycle, which promotes the transformation of oxaloacetate.
For amino acid metabolism, amino acids play a pivotal role in the rumen, whereby rumen microbes are associated with amino acid metabolism (Chalupa, 1975). Our data demonstrate that the content of leucine and valine in the rumen was considerably increased by thiamine supplementation in the high-concentrate diet. Amino acids are the degradation products of microbial or dietary proteins and precursors of protein synthesis in the rumen, which can regulate some metabolic pathways (Mariz et al., 2018). Previous studies have shown that L-leucine tended to decrease the level of acetate in the rumen (Menahan and Schultz, 1964). This result concurs with our findings. The relationship between leucine and thiamine has been rarely reported in the rumen. Thiamine supplementation has been noted to promote the transformation of acetyl-CoA, which is key to amino acid metabolism (Schwartau et al., 1984). Furthermore, TPP, as a coenzyme factor of hydroxyl acetate synthase, catalyzes the first step reaction to the biosynthesis of valine (Byrne, 1998), while thiamine supplementation can enhance this metabolic pathway.
For biogenic amine metabolism, high concentrations of bioamines such as histamine, putrescine, and tyramine may induce a series of inflammatory responses in the rumen epithelium (Nocek, 1997; Fusi et al., 2004), which emerges in the rumen because of decarboxylation of precursor amino acids under the degradation of ruminal microbes (Wang et al., 2013). Studies have suggested that the absorption of histamine in the rumen increases the content of histamine in the whole organism accompanied by the damage of the rumen epithelial cells caused by lactate poisoning, which is the main factor that aggravates the condition of ruminal acidosis (Andersen and Jarlv, 1990). In this work, we found that thiamine supplementation with a high-concentrate diet significantly decreased the content of histamine, putrescine, and tyramine. It has been postulated that gram-negative bacteria die and lyse when carbohydrates ferment quickly and pH drops sharply, leading to the release and absorption of LPS and histamine (Duffield et al., 2004; Khafipour et al., 2009). The stimulation of toll-like receptor 4 (TLR4) by LPS induces the release of critical proinflammatory cytokines (Lu et al., 2008), while thiamine might regulate the pathways (Pan et al., 2017b). Besides, competitive inhibition of histamine and thiamine have been reported in the study of bacterial metabolism (Terzl and Keek, 1949), which is likely one of the reasons that thiamine supplementation reduces biogenic amines.
For fatty acid (FA) metabolism, diet composition can modify the content and composition of ruminal bacterial FAs (Bas and Archimède, 2003). Rumen microbes are assumed to play an essential role in the metabolism of trans-FA and conjugated linoleic acid (Kalscheur et al., 1997). Also, a shift in the rumen bacteria community has been reported as dietary NDF content (Dehority and Orpin, 1997). This could have influenced FA composition due to the large differences in FA composition in different community bacterial structures (Minato et al., 1988). In this study, the contents of linoleic acid and docosahexaenoic acid in the rumen were decreased significantly with thiamine supplementation in a high-concentrate diet. A reasonable explanation for FA alteration may be that thiamine supplement perhaps changed the bacterial community structure (Pan et al., 2017a). Moreover, thiamine is a coenzyme factor in the synthesis of acetyl-CoA, which is a precursor for the synthesis of FA; thus, thiamine supplementation can facilitate this pathway.
For organic acid metabolism, the 4-PBA plays an integral role in maintaining proteostasis and apoptosis of cells (Sun et al., 2012; Kolb et al., 2015). Studies have revealed that the 4-PBA reduced cellular damage as well as cell death (Sawada et al., 2008) and attenuated inflammation (Qi et al., 2011). This may be an indication that thiamine supplementation increased the content of 4-PBA, thus alleviating rumen epithelial inflammation. Currently, there are few reports on the association between 4-PBA and bacterial community structure in animals. It is also important to note that the underlying mechanism by which thiamine supplementation affects the metabolism of 4-PBA requires further investigation.
Correlations Between Bacterial Community and Metabolism With Thiamine Supplementation
The relationship between bacterial community and metabolism with thiamine supplementation has been discussed above. In general, there was a utilization or productive association between bacteria and metabolomes in the rumen throughout the study. Mao et al. (2015) concluded that the type of feeding affects the concentration of metabolites by influencing bacterial community structure, hence affecting the rumen function. Taken together, these changes and correlations provide new insights that can reveal the mechanism of action of thiamine on animal health.
Conclusion
We applied Illumina sequencing and metabolomics analysis to reveal thiamine modulation of ruminal microbiota and metabolites in goats fed a high-concentrate diet in this study. As a result, thiamine supplementation of a high-concentrate diet may modify the rumen fermentation, which is partially attributed to the growing abundances of Oribacterium, Anaerobiospirillum, Fibrobacter, and uncultured Prevotellaceae, as well as the decreasing proportions of Prevotella_1. We also found that thiamine supplementation might help alleviate high-grain-induced SARA by modifying bacterial community structure, resulting in changes in the metabolism of carbohydrates, amino acids, FAs, biogenic amines, and organic acids. Overall, our results shed light on understanding the function of thiamine on ruminal metabolism modulation and also provide new guidelines to alleviate SARA.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: PRJNA700131.
Ethics Statement
All management and experimental procedures were conducted according to the Animal Protection Law based on the Guidelines for the Care and Use of Laboratory Animals approved by the Ethics Committee of Yangzhou University (SXXY 2015-0054).
Author Contributions
YM, HZ, and HW designed the research. CW, LY, and LD conducted the research. DG, JY, and HZ analyzed the data. YM wrote the manuscript. YM and CW had the primary responsibility for the final content. All authors read and approved the final manuscript.
Funding
The study was financially supported by the project of National Key Research and Development Program of China (2017YFD0500504, Beijing), the National Natural Science Foundation of China (No. 31872988, Beijing), and the Top Talents Award Plan of Yangzhou University (2016).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank all the members of Hong Rong Wang’s laboratory who contributed to sample determination. We also thank The University of Western Australia and, more specifically the research platform, School of Biomedical Sciences for their cooperation that made this study possible.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.653283/full#supplementary-material
References
Afzal, H. (1999). The Effect of the Form of Supplementary Non-structural Carbohydrate on Metabolism in the Rumen. Cambridge: University of Cambridge.
Amann, R. I., Lin, C., Key, R., Montgomery, L., and Stahl, D. A. (1992). Diversity among fibrobacter isolates: towards a phylogenetic classification. Syst. Appl. Microbiol. 15, 23–31. doi: 10.1016/s0723-2020(11)80133-5
Andersen, P. H., and Jarlv, N. (1990). Investigation of the possible role of endotoxin, TXA2, PGI2 and PGE2 in experimentally induced rumen acidosis in cattle. Acta. Vet. Scand. 31, 27–38. doi: 10.1186/bf03547574
Aschenbach, J. R., Oswald, R., and GABel, G. (2000). Transport, catabolism and release of histamine in the ruminal epithelium of sheep. Pflugers Arch. 440, 171–178. doi: 10.1007/s004240051036
Asma, Z., Sylvie, C., Laurent, C., Mariette, J., Klopp, C., Bouchez, O., et al. (2013). Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol. Ecol. 83, 504–514. doi: 10.1111/1574-6941.12011
Bas, P., and Archimède, H. (2003). Fatty acid composition of mixed-rumen bacteria: effect of concentration and type of forage. J. Dairy. Sci. 86, 2940–2948. doi: 10.3168/jds.s0022-0302(03)73891-0
Bettendorff, L., Lakaye, B., Kohn, G., and Wins, P. (2014). Thiamine triphosphate: a ubiquitous molecule in search of a physiological role. Metab Brain Dis. 29, 1069–1082. doi: 10.1007/s11011-014-9509-4
Boudonck, K. J., Mitchell, M. W., Wulff, J., and Ryals, J. A. (2009). Characterization of the biochemical variability of bovine milk using metabolomics. Metabolomics 5, 375–386. doi: 10.1007/s11306-009-0160-8
Brent, B. E. (1976). Relationship of acidosis to other feedlot ailments. J. Anim. Sci. 43, 930–935. doi: 10.2527/jas1976.434930x
Breves, G. M., Hoeller, H. Q., and Rohr, K. (1981). Flow of thiamine to the duodenum in dairy cows fed different rations. J. Anim. Sci. 96, 587–591. doi: 10.1017/s0021859600034559
Bubber, P., Ke, Z. J., and Gibson, G. E. (2004). Tricarboxylic acid cycle enzymes following thiamine deficiency. Neurochem. Int. 45, 1021–1028. doi: 10.1016/j.neuint.2004.05.007
Byrne, K. (1998). Genetic Analysis of Thiamine Metabolism in Saccharomyces cerevisiae. Leicester: University of Leicester.
Caboni, P., Murgia, A., Porcu, A., Demuru, M., Pulina, G., and Nudda, A. (2016). Gas chromatography-mass spectrometry metabolomics of goat milk with different polymorphism at the αs1-casein genotype locus. J. Dairy. Sci. 99, 6046–6051. doi: 10.3168/jds.2015-10537
Callaway, T. R., Martin, S. A., and Wampler, J. L. (1997). Malate content of forage varieties commonly fed to cattle. J. Dairy. Sci. 80, 1651–1655. doi: 10.3168/jds.s0022-0302(97)76096-x
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., and Bushman, F. D. (2010). QIIME allows analysis of highthroughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chalupa, W. (1975). Rumen bypass and protection of proteins and amino acids. J. Dairy. Sci. 58, 1198–1218. doi: 10.3168/jds.s0022-0302(75)84697-2
Chen, L., Luo, Y., Wang, H., Liu, S., and Wang, M. (2016). Effects of glucose and starchon lactate production by newly isolated Streptococcus bovis S1 from saanen goats. Appl. Environ. Microbiol. 82, 5982–5989 doi: 10.1128/aem.01994-16
Chen, Y., Zhang, R., Song, Y., He, J., Sun, J., Bai, J., et al. (2009). RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: finding potential biomarkers for breast cancer. Analyst 134, 2003–2011. doi: 10.1039/b907243h
Cole, J. R., Chai, B., Farris, R. J., Wang, Q. S., Kulam, A. D., McGarrell, M., et al. (2005). The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic. Acids. Res. 33, D294–D296.
Dehority, B. A., and Orpin, C. G. (1997). “Development of, and natural fluctuations in, rumen mcrobial populations,” in The Rumen Microbial Ecosystem. ed P. N. Hobson, (London: Elsevier Science Publishers), 196–245. doi: 10.1007/978-94-009-1453-7_5
Duffield, T., Plaizier, J. C., and Fairfield, A. (2004). Comparison of techniques for measurement of rumen pH in lactating dairy cows. J. Dairy. Sci. 87, 59–66. doi: 10.3168/jds.s0022-0302(04)73142-2
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Fernando, S. C., Purvis, H. T., Najar, F. Z., Sukharnikov, L. O., Krehbiel, C. R., Nagaraja, T. G., et al. (2010). Rumen microbial population dynamics during adaptation to a high concentrate diet. Appl. Environ. Microbiol. 76, 7482–7490. doi: 10.1128/aem.00388-10
Fusi, E., Rossi, L., Rebucci, R., Cheli, F. D., Giancamillo, A., and Domeneghini, C. (2004). Administration of biogenic amines to Saanen kids: effects on growth performance, meat quality and gut histology. Small. Ruminant. Res. 53, 1–7. doi: 10.1016/j.smallrumres.2003.07.009
Gressley, T. F. (2014). “Inflammatory responses to sub-acute ruminal acidosis,” in Proceedings of the 25th Annual Florida Ruminant Nutrition Symposium (Gainesville, FL) 4, 28–41.
Guo, H. Y. (2008). Application of 16SrRNA gene sequencing in the identification of bacteria in animal digestive tracts. J. Anhui Agricul. Sci. 36, 7152–7154.
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., and Giannoukos, G. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome. Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hanson, R. S., and Cox, D. P. (1967). Effect of different nutritional conditions on the synthesis of tricarboxylic acid cycle enzymes. J. Bacteriol. 93, 1777–1787. doi: 10.1128/jb.93.6.1777-1787.1967
Hook, S. E., Steele, M. A., Northwood, K. S., Dijkstra, J., France, J., and Wright, A. D. (2011). Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol. Ecol. 78, 275–284. doi: 10.1111/j.1574-6941.2011.01154.x
Hua, C., Tian, J., Tian, P., Cong, R., Luo, Y., Geng, Y., et al. (2017). Feeding a high concentration diet induces unhealthy alterations in the composition and metabolism of ruminal microbiota and host response in a goat model. Front. Microbiol. 8:138. doi: 10.3389/fmicb.2017.00138
Inagaki, A., and Sakata, T. (2005). Dose-dependent stimulatory and inhibitory effects of luminal and serosal N-butyric acid on epithelial cell proliferation of pig distal colonic mucosa. J. Nutr. Sci. Vitaminol. 51, 156–160. doi: 10.3177/jnsv.51.156
Kalscheur, K. F., Teter, B. B., Piperova, L. S., and Erdman, R. A. (1997). Effect of fat source on duodenal flow of trans-C18:1, fatty acids and milk fat production in dairy cows. J. Dairy Sci. 80, 2115–2126. doi: 10.3168/jds.s0022-0302(97)76157-5
Karapinar, T., Dabak, M., and Kizil, O. (2010). Thiamine status of feedlot cattle fed a high-concentrate diet. Can. Vet. J. 51, 1251–1253.
Kenters, N., Henderson, G., Jeyanathan, J., Kittelmann, S., and Janssen, P. H. (2011). Isolation of previously uncultured rumen bacteria by dilution to extinction using a new liquid culture medium. J. Microbiol. Methods 84, 52–60. doi: 10.1016/j.mimet.2010.10.011
Khafipour, E., Li, S., Plaizier, J. C., and Krause, D. O. (2009). Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl. Environ. Microb. 75, 7115–7124. doi: 10.1128/aem.00739-09
Kleen, J. L., Hooijer, G. A., Rehage, J., and Noordhuizen, J. P. (2003). Subacute ruminal acidosis (sara): a review. J. Vet. Med. A 50:406. doi: 10.1046/j.1439-0442.2003.00569.x
Kolb, P. S., Ayaub, E. A., Zhou, W., Yum, V., Dickhout, J. G., and Ask, K. (2015). The therapeutic effects of 4-phenylbutyric acid in maintaining proteostasis. Int. J. Biochem. Cell. B 61, 45–52. doi: 10.1016/j.biocel.2015.01.015
Kolde, R. (2012). Pretty Heatmaps. R Package Version 61. Available online at: http://CRAN.R-project.org/package=pheatmap (accessed September 2019).
Kong, Y., Teather, R., and Forster, R. (2010). Composition, spatial distribution, and diversity of the bacterial communities in the rumen of cows fed different forages. FEMS Microbiol. Ecol. 74, 612–622. doi: 10.1111/j.1574-6941.2010.00977.x
Kumar, K. N., Shah, V. R., Parikh, B. K., and Sonde, S. (2015). Reversal of severe lactic acidosis with thiamine in a renal allograft recipient. Ind. J. Crit. Care Med. 19, 425–428. doi: 10.4103/0972-5229.160293
Lechartier, C., and Peyraud, J. L. (2010). The effects of forage proportion and rapidly degradable dry matter from concentrate on ruminal digestion in dairy cows fed corn silage-based diets with fixed neutral detergent fiber and starch contents. J. Dairy. Sci. 93, 666–681. doi: 10.3168/jds.2009-2349
Lee, P. C., Lee, W. G., Lee, S. Y., Chang, H. N., and Chang, Y. K. (2000). Fermentative production of succinic acid from glucose and corn steep liquor by Anaerobiospirillum succiniciproducens. Biotechnol. Bioproc. E 5, 379–381. doi: 10.1007/bf02942216
Liu, C. M., Cosetti, M. K., Aziz, M., Buchhagen, J. L., Contente-Cuomo, T. L., and Price, L. B. (2011). The otologic microbiome: a study of the bacterial microbiota in a pediatric patient with chronic serous otitis media using 16srrna gene-based pyrosequencing. Arch Otolaryngol. 137, 664–668. doi: 10.1001/archoto.2011.116
Louis, P., Hold, G. L., and Flint, H. J. (2014). The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12, 661–672. doi: 10.1038/nrmicro3344
Lu, Y. C., Yeh, W. C., and Ohashi, P. S. (2008). LPS/TLR4 signal transduction pathway. Cytokine 42, 145–151. doi: 10.1016/j.cyto.2008.01.006
Mao, S. Y., Huo, W. J., Liu, J. H., Zhang, R. Y., and Zhu, W. Y. (2016). In vitro effects of sodium bicarbonate buffer on rumen fermentation, levels of lipopolysaccharide and biogenic amine, and composition of rumen microbiota. J. Sci. Food Agric. 97, 1276–1285. doi: 10.1002/jsfa.7861
Mao, S. Y., Huo, W. J., and Zhu, W. Y. (2015). Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 18:525. doi: 10.1111/1462-2920.12724
Mariz, L. D., Amaral, P. M., Filho, S. C., Santos, S. A., Detmann, E., and Marcondes, M. I. (2018). Dietary protein reduction on microbial protein, amino acid digestibility, and body retention in beef cattle: 2. Amino acid intestinal absorption and their efficiency for whole-body deposition. J. Anim. Sci. 96, 670–683. doi: 10.1093/jas/sky018
Martin, S. A., Streeter, M. N., and Nisbet, D. J. (1999). Effects of DL-malate on ruminal metabolism and performance of cattle fed a high-concentrate diet. J. Anim. Sci. 77, 1008–1015. doi: 10.2527/1999.7741008x
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mcallan, A. B., and Smith, R. H. (1974). Carbohydrate metabolism in the ruminant. bacterial carbohydrates formed in the rumen and their contribution to digesta entering the duodenum. Brit. J. Nutr. 31:77. doi: 10.1079/bjn19740010
Menahan, L. A., and Schultz, L. H. (1964). Metabolism of leucine and valine within the rumen1, 2. J. Dairy Sci. 47, 1080–1085. doi: 10.3168/jds.s0022-0302(64)88849-4
Mentschel, J., Leiser, R., Mülling, C., Pfarrer, C., and Claus, R. (2001). Butyric acid stimulates rumen mucosa development in the calf mainly by a reduction of apoptosis. Archiv Fr Tierernaehrung 55, 85–102. doi: 10.1080/17450390109386185
Miller, B. L., Meiske, J. C., and Groodrich, R. D. (1986). Effect of grain source and concentrate level on B-vitamin production and absorption in steers. J. Anim. Sci. 62, 473–483. doi: 10.2527/jas1986.622473x
Miller, T. L. (1978). The pathway of formation of acetate and succinate from pyruvate by Bacteroides succinogenes. Arch. Microbiol. 117, 145–152. doi: 10.1007/BF00402302
Minato, H., Ishibashi, S., and Hamaoka, T. (1988). Cellular fatty acid and sugar composition of representative strains of rumen bacteria. J. Gen. Appl. Microbiol. 34, 303–319. doi: 10.2323/jgam.34.303
Neville, B. W., Schauer, C. S., Karges, K., Gibson, M. L., Thomp-son, M. M., Kirschten, L. A., et al. (2010). Effect of thiamine concentration on animal health, feedlot performance, carcass characteristics, and ruminal hydrogen sulfide concentrations in lambs fed diets based on 60% distillers dried grains plus solubles. J. Anim. Sci. 88, 2444–2455. doi: 10.2527/jas.2009-2607
Nghiem, N. P., Davison, B. H., Suttle. B. E., and Richardson, G. R. (1997). Production of succinic acid by anaerobiospirillum succiniciproducens. Appl. Biochem. Biotech. 63, 565–576. doi: 10.1007/BF02920454
Nocek, J. E. (1997). Bovine acidosis: implications on laminitis. J Dairy Sci. 80, 1005–1028. doi: 10.3168/jds.s0022-0302(97)76026-0
Owens, F. N., Secrist, D. S., and Hill, W. J. (1998). Acidosis in cattle: a review. J. Anim. Sci. 76, 275–286. doi: 10.2527/1998.761275x
Pan, X. H., Xue, F., Nan, X., Tang, Z., Wang, K., Yves, B., et al. (2017a). Illumina sequencing approach to characterize thiamine metabolism related bacteria and the impacts of thiamine supplementation on ruminal microbiota in dairy cows fed high-grain diets. Front. Microbiol. 8:1818. doi: 10.3389/fmicb.2017.01818
Pan, X. H., Yang, L., Beckers, Y., Xue, F. G., Tang, Z. W., Jiang, L. S., et al. (2017b). Thiamine supplementation facilitates thiamine transporter expression in the rumen epithelium and attenuates high-grain-induced inflammation in low-yielding dairy cows. J. Dairy Sci. 100, 5329–5342. doi: 10.3168/jds.2016-11966
Pan, X. H., Yang, L., Xue, F. G., Xin, H. R., Jiang, L. S., and Xiong, B. H. (2016). Relationship between thiamine and subacute rumen acidosis induced by a high concentrate diet in dairy cows. J. Dairy Sci. 99, 8790–8801. doi: 10.3168/jds.2016-10865
Patel, J., Jhala, M. K., Soni, P., Shabir, N., Pandya, P., Singh, K., et al. (2011). Molecular characterization and diversity of rumen bacterial flora in indian goat by 16S rDNA sequencing. VetScan 6:77.
Pawlinski, R., Pedersen, B., and Kehrle, B. (2003). Regulation of tissue factor and inflammatory mediators by Egr-1 in a mouse endotoxemia model. Blood 101, 3940–3947. doi: 10.1182/blood-2002-07-2303
Petri, R. M., Schwaiger, T., Penner, G. B., Beauchemin, K. A., Forster, R. J., and Mckinnon, J. J. (2013). Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Appl. Environ. Microb. 79, 3744–3755. doi: 10.1128/aem.03983-12
Purushe, J., Fouts, D., Morrison, M., White, B., and Mackie, R. (2010). Comparative genome analysis of Prevotella ruminicola and Prevotella bryantii: insights into their environmental niche. Microb Ecol. 60, 721–729. doi: 10.1007/s00248-010-9692-8
Qi, W., Mu, J., Luo, Z. F., Zeng, W., Guo, Y. H., and Pang, Q. (2011). Attenuation of diabetic nephropathy in diabetes rats induced by streptozotocin by regulating the endoplasmic reticulum stress inflammatory response. Metabolism 60, 594–603. doi: 10.1016/j.metabol.2010.07.021
Quast, C., and Pruesse, E. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41, D590–D596.
Robinson, J. A., Amolenski, W. J., and Greening, R. C. (1992). Prevention of acute acidosis and enhancement of feed intake in the bovine by Megasphaera elsdenii407A. J. Anim. Sci. 70:310.
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584.
Russel, A. J. F., Doney, J. M., and Gunn, R. G. (1969). Subjective assessment of body fat in live sheep. J. Agric. Sci. 72, 451–454. doi: 10.1017/s0021859600024874
Saikia, D. P., Kalita, D. J., Borah, P., Sarma, S., and Dutta, R. (2015). Differentiation of sheep and goat species by pcr-rflp of mitochondrial 16s rrna gene. Ind. J. Anim. Res. 5, 213–217. doi: 10.5958/2277-940x.2015.00036.4
Saleem, F., Ametaj, B. N., Bouatra, S., Mandal, R., Zebeli, Q., and Dunn, S. M. (2012). A metabolomics approach to uncover the effectsofgraindietsonrumen health in dairy cows. J. Dairy Sci. 95, 6606–6623. doi: 10.3168/jds.2012-5403
Saro, C., Hohenester, U. M., Bernard, M., Lagrée, M., Martin, C., and Doreau, M. (2018). Effectiveness of interventions to modulate the rumen microbiota composition and function in pre-ruminant and ruminant lambs. Front. Microbiol. 9:1273. doi: 10.3389/fmicb.2018.01273
Sawada, N., Yao, J., Hiramatsu, N., Hayakawa, K., Araki, I., and Takeda, M. (2008). Involvement of hypoxia-triggered endoplasmic reticulum stress in outlet obstruction-induced apoptosis in the urinary bladder. Lab. Investig. 88, 553–563. doi: 10.1038/labinvest.2008.21
Schwartau, M., Doehn, M., and Jürgens, P. (1984). Animal experiment studies of chronic thiamine deficiency. changes in carbohydrate and amino acid metabolism under resting and load conditions. Zeitschrift für Ernährungswissenschaft 23, 206–218.
Shen, Z., Kuhla, S., and Zitnan, R. (2005). Intraruminal infusion of n-butyric acid induces an increase of ruminal papillae size independent of IGF-1 system in castrated bulls. Arch. Anim. Nutr. 59, 213–225. doi: 10.1080/17450390500216894
Sun, Q., Xiang, R. L., and Zhang, W. L. (2012). Effects of 4-phenylbutyric acid on apoptosis of βcells induced by endoplasmic reticulum stress. Acta Anatomica Sinica. 43, 784–788.
Sutton, J. (1985). Digestion and absorption of energy substrates in the lactating cow. J. Dairy Sci. 68, 3376–3393. doi: 10.3168/jds.s0022-0302(85)81251-0
Terzl, J., and Keek, J. (1949). Competitive inhibition by histamine and thiamine of the metabolic influence of antihistamines on bacteria. Nature 164:700. doi: 10.1038/164700a0
Thauer, R. K., Jungermann, K., and Decker, K. (1977). Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41, l00–l180.
Van Gylswyk, N. O. (1995). Succiniclasticum ruminis gen. nov. sp. nov. a Ruminal bacterium converting succinate to propionate as the sole energy-yielding mechanism. Int. J. Systemat. Bacteriol. 45, 297–300. doi: 10.1099/00207713-45-2-297
Vuckovic, D. (2012). Current trends and challenges in sample preparation for global metabolomics using liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 403, 1523–1548. doi: 10.1007/s00216-012-6039-y
Wang, D. S., Zhang, R. Y., Zhu, W. Y., and Mao, S. Y. (2013). Effects of subacute ruminal acidosis challenges on fermentation and biogenic amines in the rumen of dairy cows. Livestock. Ence. 155, 262–272. doi: 10.1016/j.livsci.2013.05.026
Wang, H., Pan, X., Wang, C., Wang, M., and Yu, L. (2015). Effects of different dietary concentrate to forage ratio and thiamine supplementation on the rumen fermentation and ruminal bacterial community in dairy cows. Anim. Prod. Sci. 55:189. doi: 10.1071/an14523
Wang, Q. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 73, 5261–5267. doi: 10.1128/aem.00062-07
Wang, S., Yao, B., Gao, H., Zang, J., Tao, S., Zhang, S., et al. (2019). Combined supplementation of Lactobacillus fermentum and Pediococcus acidilactici promoted growth performance, alleviated inflammation, and modulated intestinal microbiota in weaned pigs. BMC Vet. Res. 15:239. doi: 10.1186/s12917-019-1991-9
Warnes, G. R., Bolker, B., Bonebakker, L., Gentleman, R., Liaw, W. H. A., and Lumley, T. (2016). gplots: Various R Programming Tools for Plotting Data. R Package Version 3.0.1. The Comprehensive R Archive Network. Available online at:https://cran.rproject.org/web/packages/gplots/gplots.pdf (accessed September 2019).
Wetzels, S. U., Mann, E., Metzler-Zebeli, B. U., Wagner, M., Klevenhusen, F., and Zebeli, Q. (2015). Pyrosequencing reveals shifts in the bacterial epimural community relative to dietary concentrate amount in goats. J. Dairy Sci. 98, 5572–5587. doi: 10.3168/jds.2014-9166
Zened, A., Combes, S., Cauquil, L., Mariette, J., Klopp, C., and Bouchez, O. (2013). Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS. Microbiol. Ecol. 83, 504–514. doi: 10.1111/1574-6941.12011
Zhang, H., Peng, A. L., and Zhao, F. F. (2019). Thiamine ameliorates inflammation of the ruminal epithelium of Saanen goats suffering from subacute ruminal acidosis. J. Dairy. Sci. 103, 1931–1943. doi: 10.3168/jds.2019-16944
Zhang, J., Wang, M., Hao, Z., Yu, L., and Wang, H. (2014). Effects of thiamine on concentrations of volatile fatty acids and lactate in culture medium of high concentrate substrate after in vitro rumen fermentation. Chinese J. Anim. Nutr. 26, 489–495.
Zhang, Y., Ma, C. Y., and Ma, L. B. (2010). Molecular phylogenetic relationships of 13 Sciaenidae species in China sea areas based on 16SrRNA fragment sequences. Mar. Coast. Fish. 32, 276–282.
Zhang, H. W., Wang, H. R., Liu, X., Cao, W. W., and Wei, Z. Y. (2010). Effects of thiamin supplementation on rumen fermentation in goat on high-concentration substrate. China Dairy Cattle 3, 3–16.
Zhao, X., Xu, F., Qi, B., Hao, S., Li, Y., and Li, Y. (2014). Serum metabolomics study of polycystic ovary syndrome based on liquid chromatography-mass spectrometry. J. Proteome. Res. 13, 1101–1111. doi: 10.1021/pr401130w
Zhu, Z. J. (2013). Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat Protoc. 8, 451–456. doi: 10.1038/nprot.2013.004
Keywords: thiamine, SARA, 16S rRNA gene sequencing, dairy goats, LC-MS
Citation: Ma Y, Wang C, Zhang H, Yu L, Dong L, Gong D, Yao J and Wang H (2021) Illumina Sequencing and Metabolomics Analysis Reveal Thiamine Modulation of Ruminal Microbiota and Metabolome Characteristics in Goats Fed a High-Concentrate Diet. Front. Microbiol. 12:653283. doi: 10.3389/fmicb.2021.653283
Received: 14 January 2021; Accepted: 05 March 2021;
Published: 07 April 2021.
Edited by:
Marie-Joelle Virolle, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Katie Edwards, Binghamton University, United StatesLucien Bettendorff, University of Liège, Belgium
Copyright © 2021 Ma, Wang, Zhang, Yu, Dong, Gong, Yao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Wang, Y2hhby53YW5nQHJlc2VhcmNoLnV3YS5lZHUuYXU=; Hongrong Wang, aHJ3YW5nQHl6dS5lZHUuY24=
†These authors have contributed equally to this work
 Yi Ma
Yi Ma Chao Wang
Chao Wang Hao Zhang
Hao Zhang Lihuai Yu1
Lihuai Yu1 Daoqing Gong
Daoqing Gong