
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 March 2021
Sec. Evolutionary and Genomic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.652766
 Liping Wang1,2
Liping Wang1,2 Zongze Shao1,2,3*
Zongze Shao1,2,3*Bacteria of Halomonas are widely distributed in various environments and play a substantial role in the nutrient cycle. In this report, 14 strains capable of aerobic denitrification and heterotrophic sulfur oxidation were isolated from different habitats. Based on the phenotypic, genotypic, and chemotaxonomic analyses, these strains were considered to represent six novel species of the genus Halomonas, for which the names Halomonas zhangzhouensis sp. nov. type strain CXT3-11T ( = MCCC 1A11036T = KCTC 72087T), Halomonas aerodenitrificans sp. nov. CYD-9T ( = MCCC 1A11058T = KCTC 72088T), Halomonas sulfidoxydans sp. nov. CYN-1-2T ( = MCCC 1A11059T = KCTC 72089T), Halomonas ethanolica sp. nov. CYT3-1-1T ( = MCCC 1A11081T = KCTC 72090T), Halomonas sulfidivorans sp. nov. NLG_F1ET ( = MCCC 1A13718T = KCTC 72091T), and Halomonas tianxiuensis sp. nov. BC-M4-5T ( = MCCC 1A14433T = KCTC 72092T) are proposed. Intriguingly, they formed a unique group with 11 other species designated as the “H. desiderata group.” To better understand their featured metabolisms, genes involved in denitrification and sulfur oxidation were analyzed, along with 193 other available genomes of the whole genus. Consistently, complete denitrification pathways were confirmed in the “H. desiderata group,” in which napA, narG, nirS, norB, and nosZ genes coexist. Their nitrite reductase NirS formed a unique evolutionary lineage, distinguished from other denitrifiers in Halomonas. In addition, diverse occurrence patterns of denitrification genes were also observed in different phylogenetic clades of Halomonas. With respect to sulfur oxidation, fccAB genes involved in sulfide oxidation commonly exist in the “H. desiderata group,” while sqr genes are diverse and can be found in more species; sqr genes co-occurred with fccAB in eight strains of this study, contributing to more active sulfide oxidation. Besides, the tsdA gene, which encodes an enzyme that oxidizes thiosulfate to tetrathionate, is ubiquitous in the genus Halomonas. The widespread presence of sqr/fccAB, pdo, and tsdA in Halomonas suggests that many Halomonas spp. can act as heterotrophic sulfur oxidizers. These results provide comprehensive insights into the potential of denitrification and sulfur oxidation in the whole genus of Halomonas. With regard to the global distribution of Halomonas, this report implies their unneglectable role in the biogeochemical cycle.
The genus Halomonas, which was established in 1980, belongs to the Halomonadaceae family of the order Oceanospirillales, within the class Gammaproteobacteria. They are a group of Gram-stain-negative, halophilic or halotolerant, aerobic or facultatively anaerobic, and non-sporulated bacteria (Vreeland et al., 1980). At present, more than 100 species in the genus Halomonas have been described1. As a cosmopolitan taxon, members of the genus Halomonas widely inhabit diverse saline environments, such as salt and soda lakes, saline soil, solar salterns, sea ice, seafood, marine invertebrates, effluents, seawater, sea basins, hadal, and hydrothermal vent environments (Kaye and Baross, 2000; Kaye et al., 2004, 2011; Biddle et al., 2005; Inagaki et al., 2006; Arahal et al., 2007; Kim et al., 2013; Nunoura et al., 2015; Salazar et al., 2016; Sanz-Sáez et al., 2020).
The Halomonas bacteria have been described as opportunitrophic microbes, which can readily adapt to various environments and exploit spatially and temporally variable resources, although they are not generally observed to dominate a particular niche (Polz et al., 2006; Kaye et al., 2011; Singer et al., 2011). Halomonas is one of the most abundant genera in the ocean and is commonly isolated, and its success may be attributed to its metabolic and physiological versatility. For instance, some Halomonas species could produce compatible solutes (ectoine, glycine betaine, and hydroxyectoine), biopolymers [exopolysaccharides (EPSs) and polyhydroxyalkanoate (PHA)], and biosurfactants (Cánovas et al., 2000; Amjres et al., 2011; Schwibbert et al., 2011; Kawata et al., 2014); degrade aromatic compounds (García et al., 2004); and oxidize arsenite (Lin et al., 2012; Hamamura et al., 2014), Fe(II), Mn(II) (Homann et al., 2009; Dong et al., 2014), and sulfide (Boltianskaia et al., 2007; Zhang et al., 2020); and are capable of denitrification (González-Domenech et al., 2010; Guo et al., 2013).
Biological denitrification is a facultative respiratory pathway in which nitrate, nitrite, nitric oxide, and nitrous oxide are stepwise reduced to dinitrogen gas, each catalyzed by at least one specific metalloenzyme, with concomitant energy conservation (Zumft, 1997). Aerobic denitrification was first reported in Thiosphaera pantotropha (now known as Paracoccus denitrificans), where oxygen and nitrate were simultaneously utilized as electron acceptors (Robertson and Kuenen, 1984). This important process has been intensively studied during the past three decades. The ability of aerobic denitrification is widely distributed among prokaryotic taxa such as Paracoccus, Pseudomonas, Marinobacter, and Achromobacter, as well as some eukaryotes in various natural and engineered ecosystems (Ji et al., 2015; He et al., 2016; Liu et al., 2016b, 2018; Luque-Almagro et al., 2017). Most of our current knowledge about aerobic denitrification is related to the nitrogen removal characteristics of single strains, the expression patterns of functional genes, and practical applications, as well as electron transfer (Yang et al., 2020). Denitrification has been recognized as an important functional trait of the genus Halomonas, as many species have been described as denitrifiers, and some even as aerobic denitrifiers (González-Domenech et al., 2010). Most studies on denitrifying species within the genus Halomonas are merely restricted to their phylogenetic analysis and phenotypic characterization. Knowledge of their denitrification, especially aerobic denitrification mechanism is quite limited, and is based on only a few functional genes obtained through PCR amplification. There is a lack of genomic information with respect to the denitrification pathway in Halomonas.
Aerobic denitrification has been widely applied in biological nitrogen removal systems due to their high growth rate and their excellent nitrate removal ability. Recently, heterotrophic sulfide-oxidizing nitrate-reducing bacteria, such as Pseudomonas C27 (Chen et al., 2013; Guo et al., 2019; Zhang et al., 2020), have attracted increasing attention. Sulfide oxidation of heterotrophic denitrifiers has also been observed in Halomonas mongoliensis and Halomonas kenyensis, which can oxidize sulfide in the presence of acetate as the carbon source and nitrate or nitrous oxide as the electron acceptor (Boltianskaia et al., 2007). Several studies have shown that Halomonas spp. dominate in bioreactors and play important roles in the removal of sulfide and nitrate (Liu et al., 2016a; Zhang et al., 2020). However, their sulfur oxidation mechanisms are unclear.
In this study, we isolated 14 Halomonas strains from various environments, among which six putative novel species were proposed (Table 1). All these 14 strains could perform aerobic denitrification and heterotrophic sulfide oxidation. In order to establish their robust taxonomic status, polyphasic taxonomic analysis was carried out. To gain more insight into their functional profiles, genome-based analysis of these 14 strains and 193 other publicly available Halomonas genomes was carried out, based on nitrogen metabolism, especially denitrification and the sulfur oxidation pathway. The objectives of this study are to (i) establish their robust taxonomic status, (ii) analyze their aerobic denitrification and sulfide oxidation performance, and (iii) uncover the nitrogen and sulfur metabolic pathways within the genus Halomonas.
Based on 16S rRNA, gyrB, and rpoD gene sequences, the phylogenetic positions of the 14 strains of Halomonas were analyzed, allocating them into six potential novel species, represented by strains MCCC 1A11036T, MCCC 1A11058T, MCCC 1A11059T, MCCC 1A11081T, MCCC 1A14433T, and MCCC 1A13718T. The former five strains shared the highest 16S rRNA gene sequence similarities with Halomonas lactosivorans KCTC 52281T of 98.9, 98.5, 99.4, 98.9, and 99.3%; MCCC 1A13718T shared the highest similarities with Halomonas saliphila LCB169T of 98.8%. For the gyrB and rpoD gene sequences, the six novel species showed 85.3–95.9 and 87.3–97% similarities with the three most closely related species (H. desiderata DSM 9502T, Halomonas daqingensis CGMCC 1.6443T, and H. lactosivorans KCTC 52281T). Therefore, the gyrB gene seems to have a much higher discriminatory power, which has been proposed as a marker gene for species delineation within Halomonas (de la Haba et al., 2012).
These 14 strains formed a closely related group with type strains of H. desiderata DSM 9502T, H. daqingensis CGMCC 1.6443T, H. lactosivorans KCTC 52281T, Halomonas kenyensis DSM 17331T, H. saliphila LCB169T, and Halomonas pellis L5T in the phylogenetic tree based on the concatenation of 16S rRNA, gyrB, and rpoD gene sequences, as well as other species, including Halomonas azerbaijanica TBZ202T, Halomonas campisalis A4T, Halomonas gudaonensis CGMCC 1.6133T, Halomonas endophytica MC28T, Halomonas montanilacus PYC7WT, and Halomonas chromatireducens AGD8-3T (Figure 1). This group could be defined as the “H. desiderata group,” which is composed of at least 18 species. Phylogenomic trees were also constructed using both PhyloPhlAn and UBCG software based on genome sequences, and the phylogenetic relationships of “H. desiderata group” strains were further verified (Figure 2 and Supplementary Figures 1,2).

Figure 1. Neighbor-joining tree based on the concatenated 16S rRNA, gyrB, and rpoD gene sequences showing the phylogenetic positions of six novel strains within the genus Halomonas. The type strain Carnimonas nigrificans CECT 4437T was used as outgroup. Bootstrap values (>50%) are shown at branch points. Bar, 0.01 nucleotide substitution rate (Knuc) units. The phylogeny was inferred using MEGA version 7 and 1,000 bootstrap replicates. Each type strain is marked by a superscript capital T.

Figure 2. Genome-based phylogenomic tree of the 14 strains along with 125 other representative reference genomes in the genus Halomonas and the distribution of genes for nitrogen and sulfur metabolism. The phylogenomic tree was constructed using PhyloPhlAn 3.0. Strains shown in red are organisms isolated in this study. The visualization, annotation, and management of the phylogenomic trees were performed using the web-based tool evolview v3. The type strain Alcanivorax dieselolei B5T was used as outgroup. The branch labeled in red indicates the “H. desiderata group.” The 16 rightmost columns represent the distribution of key functional genes for the denitrification, regulation, and sulfur oxidation pathways.
The general genome properties of the 14 newly sequenced strains are summarized in Supplementary Table 1. Complete genome sequences of strains MCCC 1A11059T, MCCC 1A13718T, and MCCC 1A14433T and draft genomes for 11 other strains were obtained. Their genome sizes ranged from 4.41 to 5.08 Mb; the number of contigs ranged from 1 to 96; the G+C content ranged from 63.3% (MCCC 1A11036T) to 66.2% (MCCC 1A17488), which is within the range of G+C values for Halomonas (Kim et al., 2013).
Pairwise digital DNA–DNA hybridization (dDDH) and average nucleotide identity (ANI) values (ANIb, fastANI, and OrthoANI) were calculated to identify the genomic similarities of the 14 strains to six closely related species within the genus Halomonas (as shown in Figure 3 and Supplementary Table 2). For strains MCCC 1A11036T, MCCC 1A11058T, MCCC 1A11059T, MCCC 1A11081T, MCCC 1A13718T, and MCCC 1A14433T, the dDDH and ANIb values between them and closely related species were below the 70% (Wayne et al., 1987) and 95–96% (Richter and Rosselló-Móra, 2009) threshold values for species definition, ranging at 24.8–53.6 and 84.4–93.2%, respectively, which could represent six novel species of the genus Halomonas.
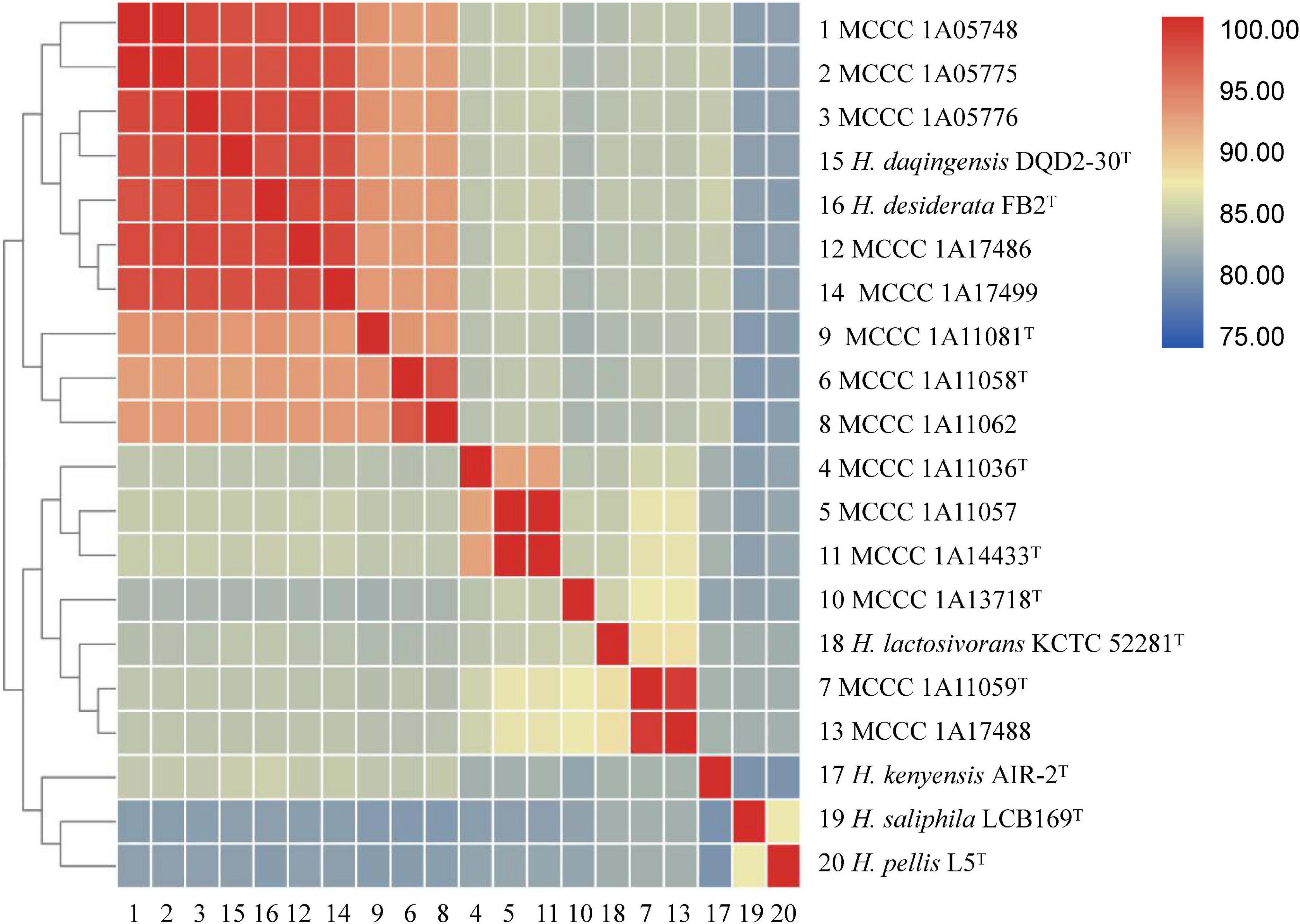
Figure 3. A matrix of the average nucleotide identity between the six novel strains and other closely related species within the genus Halomonas. The ANIb values between each pair of genomes were calculated as described in the Materials and Methods section. The numerical values underlying this matrix are provided in Supplementary Table 2.
All six of the strains (MCCC 1A11036T, MCCC 1A11058T, MCCC 1A11059T, MCCC 1A11081T, MCCC 1A13718T, and MCCC 1A14433T) were Gram-stain-negative and catalase- and oxidase-positive. Their cells were long rods, motile by means of peritrichous flagella (Supplementary Figure 3). All strains grew well on marine agar (MA), with yellow, circular, and smooth colonies. They could grow at 4–50°C, even up to 55°C for strain MCCC 1A13718T. They could tolerate salinity up to 18% (w/v), even up to 20% for strain MCCC 1A13718T. In the API ZYM, API 20NE, and API 20E tests, all the strains were analyzed in parallel with other reference species. They displayed similar physiological characteristics. Differences in phenotypic and physiological characteristics among these strains and other closely related species are shown in Table 2 and in the species descriptions.
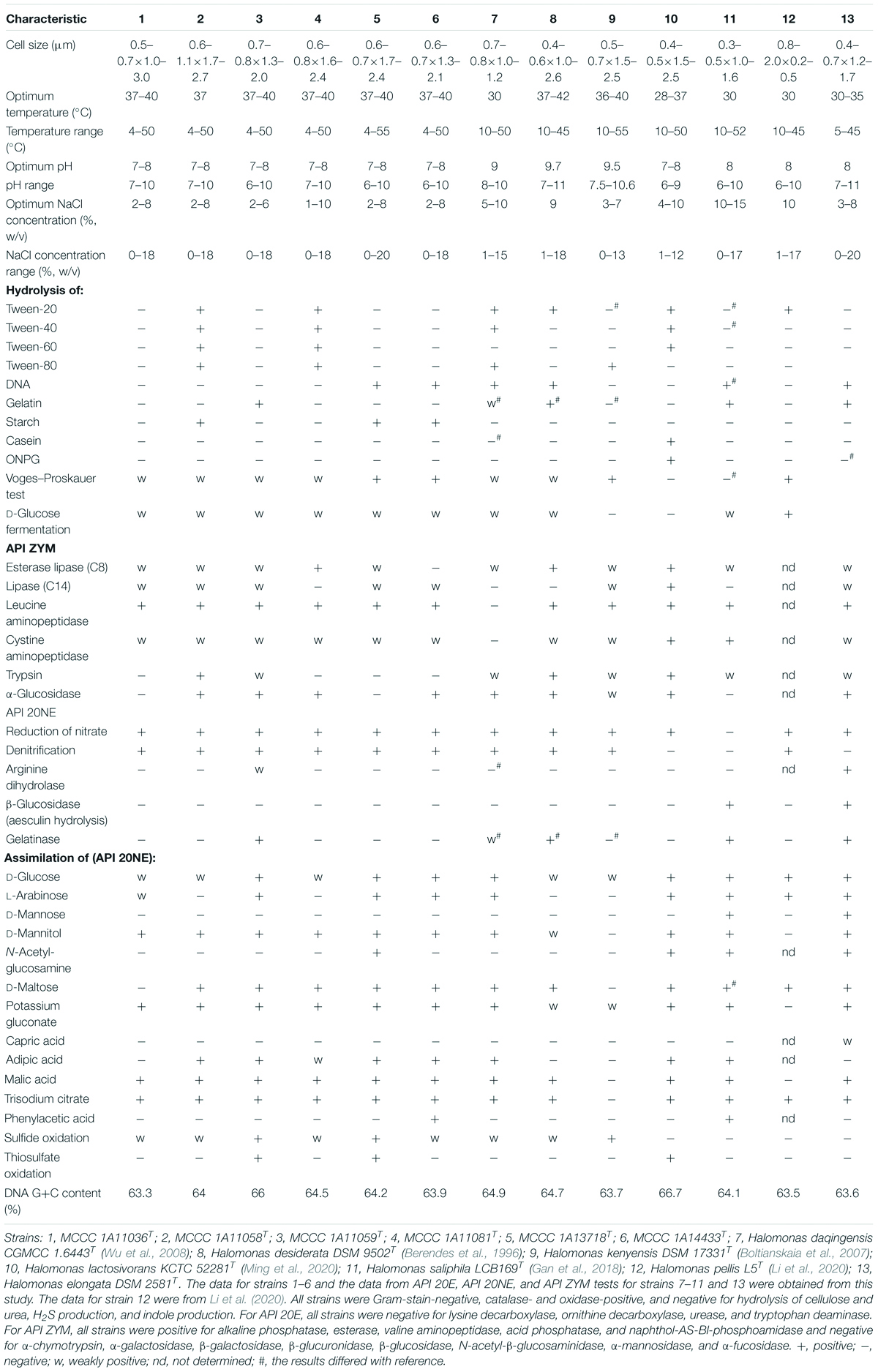
Table 2. Differential characteristics of the six novel species and closely related Halomonas species.
The predominant respiratory quinone of these six species was Q-9, with the minor ubiquinone Q-8 also present, in agreement with previous observations in the genus Halomonas. Cellular fatty acid composition was determined for the six species in parallel with reference type strains, and the results are detailed in Table 3. The principal fatty acids for all of the strains (>10%) were C18:1 ω7c, C16:0, C19:0 cyclo ω8c, summed feature 3 (C16:1 ω7c and/or C16:1 ω6c), and C12:0 3-OH. Although the fatty acid profiles from these strains were similar, their proportions were different from each other. The polar lipids of the six novel strains include diphosphatidylglycerol (DPG), phosphatidylglycerol (PG), phosphatidylethanolamine (PE), and several unidentified phospholipids (PLs) (Supplementary Figure 4). Distinctive features were the presence of aminolipid (AL) in strains MCCC 1A13718T and MCCC 1A11059T and the presence of aminophospholipid (APL) and phosphatidylcholine (PC) in strain MCCC 1A11059T. Phenotypic and physiological characteristics as well as chemotaxonomic features that could distinguish these six novel strains from other closely related strains are presented in Table 2 and in the species descriptions.
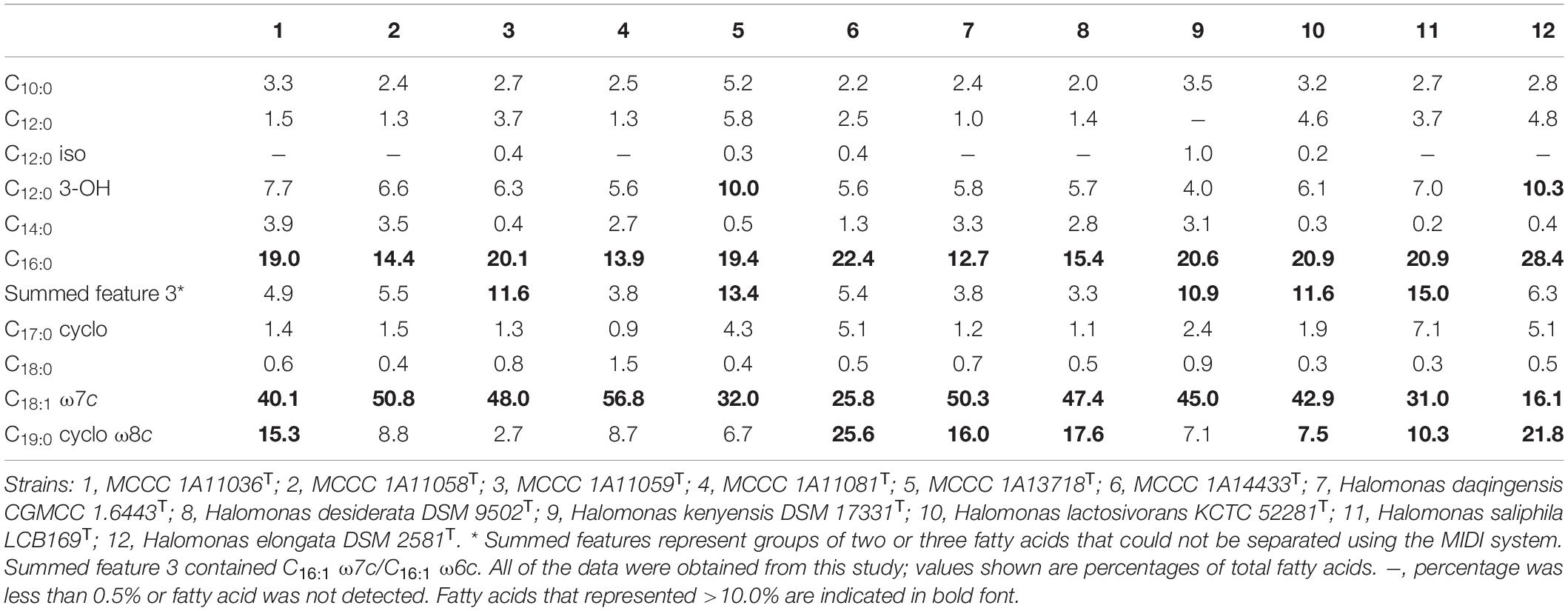
Table 3. Cellular fatty acid compositions of six novel strains compared with other closely related species of the genus Halomonas.
All six of the novel species can perform denitrification using nitrate or nitrite as the sole nitrogen source, under both aerobic and anaerobic conditions. Their aerobic denitrification performance is shown in Figure 4, where nitrate was used as the sole nitrogen source and the O2 percentage in the headspace of the bottles was set at about 25%. As depicted in Figure 4B, their gaseous products were identified to be N2 and N2O. They exhibited efficient removal ability for nitrate; they could remove 15 mM of nitrate completely within 24 h under aerobic conditions, with the removal rate of NO3––N reaching 8.75 mg L–1 h–1, which is much higher than that of other aerobic denitrifiers (Liu et al., 2018). Besides, the percentage of O2 decreased to about 5% within 24 h, suggesting that they could perform co-respiration of nitrate and oxygen during denitrification. Although nitrite accumulated during nitrate reduction, it was removed quickly and was nearly undetectable within 24 h.
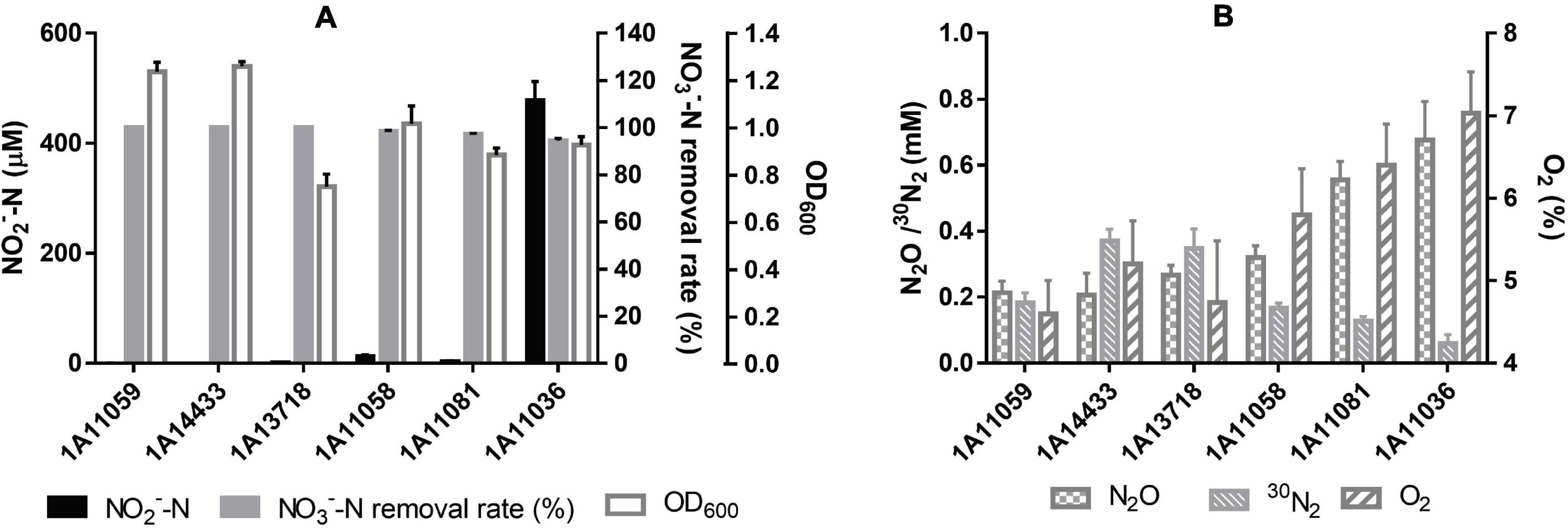
Figure 4. Cell growth and performance of aerobic denitrification by six novel species within 24 h in this study. (A) NO3– –N removal rate, NO2– –N accumulation, and cell OD600 with an initial Na15NO3 concentration of 15 mM. (B) 30N2 and N2O production within 24 h as well as final O2 percentage, with an initial O2 content of 25%. The data are shown as mean value ± SD from three independent measurements.
Acetate, sodium citrate, glucose, sucrose, lactate, glycerol, mannitol, maltose, succinate, and propionate can be used as carbon sources and electron donors to support cell growth coupling aerobic denitrification for all these strains. Moreover, sodium citrate and succinate were the optimal electron donors observed in this study, with the highest nitrate removal efficiency and the lowest nitrite accumulation, as shown in Supplementary Table 3. Besides, strains MCCC 1A11058T, MCCC 1A11081T, MCCC 1A13718T, and MCCC 1A14433T can also utilize ethanol as electron donor to derive energy for aerobic denitrification.
Another important trait of all these isolates was their ability for heterotrophic sulfide oxidation. As shown in Figure 5A and Supplementary Table 4, strains MCCC 1A11059T and MCCC 1A13718T exhibited the strongest sulfide oxidation ability under aerobic conditions; they could completely oxidize 1 mM of Na2S within 1 h, with sulfide oxidation rates reaching 50 and 54 μmol min–1 g–1 of cell dry weight, respectively, which were comparable with those of other reported heterotrophic sulfide oxidizers (Hou et al., 2018). Thiosulfate was identified as one of the main products; however, no sulfite and sulfate could be detected (Figure 5B). These strains varied in sulfide tolerance; for example, strains MCCC 1A11059T and MCCC 1A13718T could grow and aerobically oxidize at least 7 and 3 mM of Na2S, respectively, in the presence of nitrate and sodium citrate; strains MCCC 1A11058T and MCCC 1A11081T could tolerate even 12 mM of Na2S, although their sulfide oxidation ability was weak. In the thiosulfate oxidation test, only strains MCCC 1A11059T and MCCC 1A13718T could oxidize thiosulfate rapidly, producing tetrathionate (Figures 5C,D).
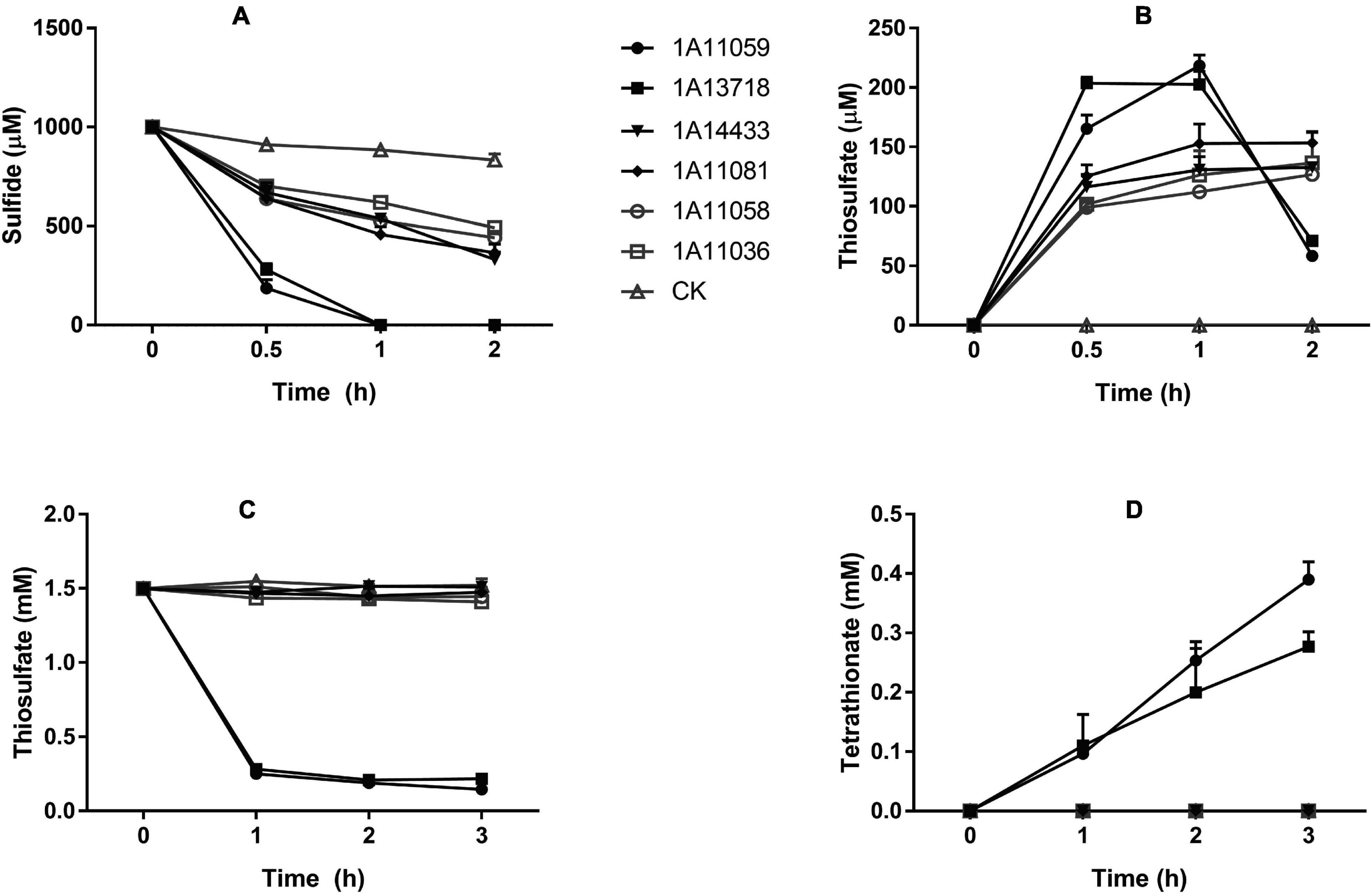
Figure 5. Sulfide and thiosulfate oxidation by six novel strains. (A) Sulfide oxidation by six strains with an initial Na2S concentration of 1 mM. (B) The production of thiosulfate during sulfide oxidation. (C) Thiosulfate oxidation by six strains with an initial Na2S2O3 concentration of 1.5 mM. (D) The production of tetrathionate during thiosulfate oxidation. CK, control containing no cells. The data are shown as mean value ± SD from three independent measurements.
Under anaerobic conditions, all six of the novel strains could oxidize sulfide when nitrate was used as the sole electron acceptor; meanwhile, strains MCCC 1A13718T and MCCC 1A11036T could oxidize sulfide using either nitrate or N2O as the sole electron acceptor. Several heterotrophic denitrifiers were reported previously to oxidize sulfide or thiosulfate, including H. kenyensis AIR-2 and Halomonas mongoliensis Z-7009. The former oxidized sulfide only with nitrate as the sole electron acceptor, while the latter only oxidized sulfide with N2O (Sorokin, 2003; Boltianskaia et al., 2007). A similar trait was also observed in some autotrophic strains such as Thioalkalivibrio and Alkalispirallum (Sorokin et al., 2001, 2006). The oxidation of sulfide combined with the reduction of nitrogen compounds indicated the possible coupling of the nitrogen and sulfur cycles in the natural environment.
Bioinformatic analysis showed that all 14 genomes in this study possessed key genes for the complete denitrification pathways, including both the membrane-bound respiratory nitrate reductase NAR (narG) and the periplasmic nitrate reductase NAP (napA), cytochrome cd1-type nitrite reductase NIR (nirS), cytochrome c nitric oxide reductase NOR (norBC), and nitrous oxide reductase NOS (nosZ). Nearly the same co-occurrence patterns of these denitrification genes were observed in these 14 strains, as well as in other strains belonging to the “H. desiderata group,” except for H. saliphila LCB169T (Figure 2).
One feature of the denitrification pathways of the “H. desiderata group” was the coexistence of two types of dissimilatory nitrate reductases, NAR and NAP, in one organism. Within this group, only two strains, H. endophytica MC28T and Halomonas sp. DX6, lack NAP. In general, the membrane-bound NAR is responsible for anaerobic nitrate respiration, while the periplasmic NAP can reduce nitrate irrespective of O2 (Richardson et al., 2001; Sparacino-Watkins et al., 2014). In the “H. desiderata group,” nap operons were distantly located in genomes, far away from other denitrification gene clusters. On the contrary, narGHJI operons were located within a large chromosomal region that encodes all the other genes involved in the denitrification pathway, including nirS, nor, and nos operons, as well as transporters and transcriptional regulators. Upstream of nar operons was the nor gene cluster norECBQD. The nirS1 gene, which encodes the catalytic subunit of nitrite reductase, was present upstream of the nosRZDFYL operon. At least three nitrate/nitrite transporter-encoding genes were also located in this region. All of these strains can reduce nitrate and nitrite under aerobic and anaerobic conditions, with N2O and N2 as the gaseous products of denitrification, consistent with the existence of nar, nap, nir, nor, and nos operons.
Multiple transcriptional regulators have also been identified nearby the denitrification gene clusters in the “H. desiderata group,” implying their possible roles in the regulation of denitrification. narX-narL, which encodes a two-component system, NarXL, acting as a sensor and a regulator in response to nitrate and/or nitrite (Hartig et al., 1999), was located upstream of narGHJI. This two-component system was found to be closely linked with the nar operon. In addition, at least three genes encoding Crp/Fnr family transcription factors, which have been recognized as major players in the denitrification regulatory network (Spiro, 1994), were also found nearby denitrification gene clusters. One fnr1 gene, encoding an FNR-type regulator, was located downstream of nor operons. Two other genes, dnr1 and dnr2, encoding FNR-like regulators affiliated with the Dnr subgroup of the Crp/Fnr family, were also identified. Among them, dnr1 was found adjacent to nirS1, while dnr2 lied downstream of narGHJI operons. It is interesting to note that dnr1 was almost exclusively present in the “H. desiderata group” strains, showing a significant positive correlation with nirS1, implying a possible unique mechanism of nirS1 regulated by dnr1 in the “H. desiderata group.” Moreover, nearly all the denitrifiers in the genus Halomonas possessed dnr2, except for Halomonas litopenaei and Halomonas titanicae, implying its possible role in the regulation of the denitrification process. Other genes encoding two known dedicated NO sensors, NsrR and NorR (Bodenmiller and Spiro, 2006; Bush et al., 2011), as well as NnrS, which is involved in defense against NO in nitrosative stress (Stern et al., 2013), were present close to the nos gene cluster, while another nnrS gene was located close to the nir operon, indicating a unique sensitive mechanism of NO detoxification. These sensors and regulators might sense different environmental signals and be involved in the complex regulatory network of the denitrification pathway in the “H. desiderata group.”
The “H. desiderata group” formed a distinct clade in the phylogeny of the nitrite reductase NirS, similar to their phylogenomic relationships. Phylogenetic analysis of NirS sequences from the genus Halomonas confirmed the presence of two distinct evolutionary clades, clade I and II, represented by NirS1 and NirS2, respectively (Supplementary Figure 5). Clade I was mainly composed of NirS1-type sequences from nearly all members of the “H. desiderata group,” plus Halomonas alkalicola, Halomonas daqiaonensis, and Halomonas campaniensis, and grouped with sequences from diverse taxonomic members of Gammaproteobacteria and Alphaproteobacteria. Clade II harbored NirS2-type sequences from the remaining denitrifying species of the genus Halomonas and from Marinobacter and Pseudomonas species. Amino acid sequence alignment showed that NirS1-type sequences obtained from the “H. desiderata group” shared only 36–46% identities with NirS2-type sequences of the genus Halomonas. The NirS phylogeny implied that the two clades underwent different evolutionary processes, suggesting that nirS1 might have been horizontally transferred to the “H. desiderata group,” while nirS2 was present due to vertical transfer. The nirS gene is usually found to cluster with ancillary genes, as commonly described for other denitrifiers, such as those from Pseudomonas and Marinobacter (Bali et al., 2014). However, in the “H. desiderata group,” the nirS1 genes were located upstream from the nos operon, far away from the ancillary genes nirCFDLGHJEN, which are essential for electron transfer and maturation of cytochrome cd1 nitrite reductase, whereas the nirS2 gene clustered with ancillary genes (nirCFSDLGHJEN) and was located neighboring with the norBC operon. Obviously, the gene locus of nirS1 in the genome was different from that of nirS2.
The occurrence patterns of nitrite reductase genes differed greatly among the denitrifiers in the “H. desiderata group,” depending on whether two divergent nirS genes coexist or not and the co-occurrence of nirS and nirK. It is worth noting that three strains in the “H. desiderata group,” including MCCC 1A11036T, Halomonas sp. D167-6-1, and H. azerbaijanica TBZ202T, harbored two divergent copies of nirS (nirS1 and nirS2). Previously, the co-occurrence of two divergent nirS genes in one organism was reported in a denitrifying Thauera sp. strain 27, in which one nirS gene was positively regulated by nitrate while the other was constitutively expressed, suggesting an increased competitive ability under different conditions (Etchebehere and Tiedje, 2005). Moreover, the coexistence of the two evolutionary unrelated variants NirK and NirS in one organism was also observed in four genomes, including MCCC 1A11059T, MCCC 1A17488, H. lactosivorans KCTC 52281T, and H. saliphila L5. The nirK gene in the genomes was found as a single gene, not within a larger operon, and far away from other denitrification gene clusters. It has been long considered that both types of Nir are mutually exclusive in one organism (Jones et al., 2008), and communities of denitrifiers with NirS responded more flexibly to environmental gradients than those with NirK, which suggests that these communities occupy different ecological niches (Jones and Hallin, 2010). A recent survey of genomes revealed that a small number of organisms harbor both genes in their genomes (Graf et al., 2014). The co-occurring nirS and nirK in Bradyrhizobium oligotrophicum strain S58 were found to be functionally redundant, while the two types of Nir in Pseudomonas stutzeri strain JM300 were functionally different (Sanchez and Minamisawa, 2018; Wittorf et al., 2018). The presence of two types of Nir or multiple copies of nirS in the same organism is likely to be related with their metabolic needs. Whether these enzymes are truly functional when present in one organism in the genus Halomonas is unclear.
In order to further understand the denitrification potential within the genus Halomonas, a comprehensive comparative analysis was conducted based on 206 Halomonas genomes. The inventory of denitrification genes varied in all these 206 genomes (Supplementary Figure 1 and Supplementary Table 5). Most bacteria (158/206) possess at least one copy of narGHJI, implying the importance of respiratory nitrate reduction for Halomonas. Besides, 53 of 206 genomes carried at least one copy of napA. The coexistence of NAR and NAP in one organism was present in 42 genomes, mostly belonging to the “H. desiderata group” (30/42). They may function in denitrification or dissimilatory nitrate reduction to ammonium (DNRA). Upon inspection of 206 genomes of Halomonas, 66 had nirS, but only 10 possessed nirK, indicating that this genus should be NirS-type denitrifiers. The common occurrence of cytochrome cd1-type nitrite reductase in denitrifiers of Halomonas suggests that this genus might have inherited nirS genes from a common ancestor, and later some species acquired nirK.
Except for the “H. desiderata group,” there are several other groups of denitrifying bacteria within the genus Halomonas that also possess complete or truncated denitrification pathways. Co-occurrence of narG, nirS2, c-nor, and nosZ genes and the lack of a nap gene were found in at least 20 other denitrifying species of Halomonas, including Halomonas denitrificans, Halomonas halodenitrificans, and H. litopenaei. Notably, a majority of these denitrifiers prefer NirS2-type nitrite reductases instead of NirS1, in contrast to the “H. desiderata group,” which prefers NirS1. With respect to nitric oxide reductases, at least five species, including Halomonas xinjiangensis, Halomonas radicis, Halomonas lutea, Halomonas xianhensis, and Halomonas ilicicola, harbored qNor instead of c-Nor. Besides, a truncated denitrification pathway was present in a phylogenetically closely related clade designated as the “H. titanicae group,” including at least four species (H. titanicae, Halomonas olivaria, Halomonas neptunia, and Halomonas alkaliantarctica), which were found dominant in deep sea environments (Kaye et al., 2011; Salazar et al., 2016). Nineteen genomes in this group exhibited incomplete pathways, with the existence of nar, c-Nor, and nos operons. However, nirS and nirK, which are hallmark genes of the denitrification process, were not identified in the genomes of this group, suggesting that this process of NO2– reduction to NO is not carried out, other enzymes play this role, or even chemodenitrification takes place (Onley et al., 2018).
Phylogenies of denitrification functional genes including nirS, napA, narG, norB, and nosZ showed diverse evolutionary clades in the genus Halomonas (Supplementary Figures 5–9). Incongruence of the phylogenies was observed when comparing these functional genes with 16S rRNA gene (Supplementary Figure 10), except for the narG genes, which suggested that the denitrification genes in the genus Halomonas may have undergone different evolutionary processes. According to Petri and Imhoff, respiratory nitrate reductase genes went through a long evolutionary history and already played a key role in energy metabolism during peroxic times (Petri and Imhoff, 2000). Delorme et al. hypothesized that the acquisition of the ability to reduce other nitrogen oxides was probably a more recent event than that of nitrate reduction (Delorme et al., 2003). The diverse co-occurrence patterns as well as different evolutionary lineages of denitrification genes distributed among different phylogenetic clades of the genus Halomonas indicated that they might be involved in the nitrogen cycle in distinct ecological niches.
Genomic analyses further revealed complete nitrate and nitrite assimilation pathways in the genus Halomonas. Assimilatory nitrate reduction gene clusters (nas) encoding NADH-nitrite oxidoreductase (nirBD) and assimilatory nitrate reductase (nasA) were annotated in these genomes, allowing the production of ammonia for L-glutamate biosynthesis and implying the ability of growth under nitrate or nitrite as the sole nitrogen source. Putative regulatory proteins involved in nitrate assimilation were identified in these genomes, such as the transcription antiterminator nasR and the two-component regulatory system NasS–NasT (Luque-Almagro et al., 2011). NasST homologues are common in the “H. desiderata group” strains, while nasR homologues are present in 15 of them, represented by MCCC 1A14433T, MCCC 1A11081T, and MCCC 1A11058T, implicating possible dual regulation. However, the occurrence of NasR and NasST was variable, depending on the organism, and seemed almost mutually exclusive, as distributed in the two major phylogenetic clades within the genus Halomonas, as shown in Figure 2 and Supplementary Figure 1. Although the nasR homologues from these genomes showed low amino acid identities (about 30%) with that from Klebsiella pneumoniae M5al (Goldman et al., 1994), they were composed of an N-terminal NO3–/NO2–-sensing NIT domain and a C-terminal ANTAR signaling domain, indicating their potential role in the regulation of nitrate assimilation. Besides, the ubiquitous pathways for ammonium assimilation and regulation were also widespread in the genus Halomonas. In addition to inorganic nitrogen, urea seemed to be an alternative nitrogen source, due to the presence of the urtABCDE and ureDABCEFGJ gene clusters, which encode the urea ABC transporter and urease, respectively. They might utilize nitroalkane and nitrile based on the presence of nitronate monooxygenase and nitrilase. Their ability to utilize various nitrogen sources would facilitate their adaptation to varied habitats.
In this report, all 14 strains were capable of oxidizing sulfide aerobically. The sulfide oxidization product was thiosulfate instead of sulfite. Correspondingly, the genes responsible for sulfide oxidation were inspected in these strains and compared with other species of the genus Halomonas (Figure 2 and Supplementary Table 5). As a result, fccAB genes encoding flavocytochrome c sulfide dehydrogenase (FCSD) were identified in all the 14 strains, while the sulfide:quinone oxidoreductase (SQR) gene sqr co-occurred with fccAB genes in eight strains, i.e., MCCC 1A05748, MCCC 1A05775, MCCC 1A11059T, MCCC 1A11062, MCCC 1A13718T, MCCC 1A17486, MCCC 1A17488, and MCCC 1A17499. The simultaneous presence of fccAB and sqr genes was also observed in nine other genomes, such as those of H. campisalis MCCC 1A11675T, H. daqingensis DQD2-30T, H. montanilacus PYC7WT, H. endophytica MC28T, H. kenyensis AIR-2T, H. alkalicola JS92-SW72, and Halomonas pantelleriensis AAPT. Upon inspection of 206 genomes in the genus Halomonas, only 38 genomes contained fccAB gene, and most of them (33/38) belonged to the “H. desiderata group.” On the contrary, sqr genes were widely distributed in diverse species through the genus Halomonas (Supplementary Figures 11,12).
In addition, the persulfide dioxygenase gene pdo was commonly present in the genus Halomonas, including the 14 genomes reported here. Heterotrophic bacteria containing sqr/fccAB and pdo genes can oxidize sulfide to sulfite and thiosulfate, which plays a possible role in sulfide detoxification (Xia et al., 2017). The coexistence of sqr/fccAB and pdo in these 14 strains would well explain their ability of sulfide oxidation. SQR is thought to be widespread among prokaryotes and appears to be critical for sulfide oxidation (Reinartz et al., 1998), while FccAB is thought to be more prevalent at low sulfide concentrations (Mussmann et al., 2007). In this study, the eight strains harboring sqr exhibited stronger sulfide oxidation than fccAB-carrying strains, especially strains MCCC 1A11059T, MCCC 1A17488, and MCCC 1A13718T. These putative SQR sequences shared 88.4–100% similarities with each other and grouped together. Although they showed only about 35% sequence identities with that from Pseudomonas putida (UniParc ID: UPI000000E9AA), they grouped with Type II SQRs in the phylogenetic tree and shared conserved amino acid sequences with the biochemically well-characterized SQR from Aquifex aeolicus (UniParc ID: UPI00000567FF) (Sousa et al., 2018; Supplementary Figures 12,13).
In addition to sulfide oxidation, the three strains, MCCC 1A11059T, MCCC 1A17488, and MCCC 1A13718T, can also convert thiosulfate to tetrathionate. Accordingly, the thiosulfate dehydrogenase-encoding gene tsdA was identified in these three strains (Supplementary Figure 14). Their tsdA-like gene was accompanied by the tsdB gene, encoding a diheme cytochrome c. Although these TsdA homologues showed low amino acid sequence identities (only about 35%) with functionally verified TsdA of Allochromatium vinosum (Brito et al., 2015), they contained the characteristic heme groups and conserved amino acid residues, which are essential for the catalytic activity of thiosulfate dehydrogenase (Denkmann et al., 2012; Supplementary Figure 15). TsdA homologues are widespread among prokaryotes, not only in specialized sulfur oxidizers but also in many chemoorganoheterotrophs. The occurrence of tsdA-related genes in Halomonas strains is in agreement with their ability of tetrathionate formation, as they might oxidize thiosulfate as supplemental energy just like in Pseudomonas spp. (Denkmann et al., 2012).
The occurrence pattern of the sulfur oxidation pathways was further compared within the genus Halomonas (Supplementary Figure 1 and Supplementary Table 5). In the 206 genomes searched, 65 genomes of diverse species contained at least one copy of the sqr gene, while four harbored two divergent copies of sqr. SQRs were widespread throughout the genus Halomonas, while FCSD was almost exclusively found in the “H. desiderata group.” The phylogenetic analysis showed that the SQR homologues retrieved from the genus Halomonas were very diverse and divided into at least three different groups of SQRs (Types II, III, and V), according to a previous SQR classification system (Marcia et al., 2010; Sousa et al., 2018; Supplementary Figure 12). A majority of these SQR homologues obtained from Halomonas belonged to Type II SQRs, forming at least three clades in the dendrogram. Clade I showed high amino acid sequence identities with that of P. putida (UniParc ID: UPI000000E9AA) ranging from 57 to 63%, clade II showed 47–50% sequence similarity, and clade III 35–38%. In addition, three sequences obtained from H. denitrificans DSM 18045T, Halomonas nitroreducens 11ST, and Halomonas sp. ES 049 grouped with Type III SQRs, while one from H. alkaliantarctica SAT111 belonged to Type V. The SQR homologues from Halomonas were disperse in the dendrogram, pointing to possible lateral gene transfer events. Some sqr-containing heterotrophic bacteria are reported to harbor sqr and pdo in a gene cluster (Xia et al., 2017); however, the sqr and pdo genes were separate in most Halomonas genomes, with the exception of clade I sqr, which was located adjacent to pdo in the genome.
Besides, almost all 206 of the genomes possessed pdo genes (204/206), 14 of which contained two or three copies of pdo. PDO homologues retrieved in this study showed more than 50% amino acid sequence identities with those from Pseudomonas aeruginosa (NP_251605) and P. putida (ABQ76243) and belonged to Group II PDOs (Supplementary Figure 16). Co-occurrence of SQR/FCSD (or both) and PDO is observed in 81 genomes, indicating that it is quite common in Halomonas spp. to oxidize sulfide to sulfite and thiosulfate. Likewise, TsdA homologues are also ubiquitous in most species of Halomonas (106/206), suggesting the ability of thiosulfate oxidation of this genus. Coexistence of SQR/FCSD, PDO, and TsdA was observed in 37 genomes, distributed in more than 20 Halomonas species, such as Halomonas aestuarii, H. denitrificans, Halomonas fontilapidosi, Halomonas jeotgali, H. halodenitrificans, H. nitroreducens, Halomonas boliviensis, and H. titanicae, in addition to MCCC 1A13718T, MCCC 1A11059T, and MCCC 1A17488, most of which are denitrifying species, indicating that they might be able to remove sulfide, thiosulfate, and nitrate simultaneously. Considering their excellent removal of sulfide and nitrate, Halomonas bacteria might have promising applications in the biotechnology industry.
Sulfide oxidation by heterotrophic denitrifiers has been previously reported in H. mongoliensis and H. kenyensis, as well as several other Halomonas species (Sorokin, 2003; Boltianskaia et al., 2007). In a continuous integrated autotrophic-heterotrophic denitrification bioreactor, H. desiderata was observed to dominate, showing the ability to simultaneously remove sulfide and nitrate (Zhang et al., 2020). Halomonas was also found to dominate; it could enhance sulfide denitrification removal on high-salinity wastewaters (Liu et al., 2016a). Besides, bacterial thiosulfate oxidation was also found in Halomonas isolates from deep sea sediment (Teske et al., 2000; Du et al., 2019). This is the first time the sulfur oxidation pathways in the genus Halomonas are depicted by retrieving their functional genes. Our results support that bacteria of Halomonas having sulfur oxidation potential are more common than previously recognized, implying their ecological contribution to the sulfur cycle.
The heterotrophic lifestyle of the 14 strains was inferred from the existence of organic carbon degradation pathways but lacks key genes for carbon fixation. As predicted from annotation results, all CO2 fixation pathways were absent in these six novel species and other Halomonas genomes used in this study, except for Halomonas heilongjiangensis (with the rTCA cycle and the Calvin cycle) and Halomonas anticariensis (with the Calvin cycle).
Polyhydroxyalkanoates can serve as carbon and energy reserves and can be naturally synthesized by numerous bacterial species. Genes coding for the key enzymes involved in PHA synthesis pathway, including acetyl-CoA acetyltransferase (PhaA), acetoacetyl-CoA reductase (PhaB), poly-β-hydroxybutyrate polymerase (PhaC), and PHA synthesis repressor (PhaR), which belonged to the class I PHA synthases, were identified in all 14 of these genomes as well as other Halomonas genomes (Supplementary Figure 1 and Supplementary Table 5). The PHA biosynthesis genes phaA, phaB, phaC, and phaR were common in the genus Halomonas, and did not cluster in one operon but were distant from each other. This kind of organization was obviously different from that of typical PHA synthase genes, which are usually organized in one operon. Besides, various putative EPS biosynthesis and export gene clusters were identified in these genomes, such as pgaABC (encoding biofilm PGA synthesis protein), lapAB (encoding lipopolysaccharide assembly protein), lptABC (encoding the lipopolysaccharide export system), and genes encoding polysaccharide biosynthesis proteins, and the polysaccharide export protein Wza, implying the ability to produce EPS.
Gene clusters coding for complete phosphonate transport and metabolism pathways, including phnCDEFG (encoding the phosphonate ABC transporter and regulator) and phnHIJKLMNP [encoding a carbon-phosphorus (CP) lyase], were only present in MCCC 1A11036T, MCCC 1A14433T, and MCCC 1A11057 and were absent in 11 other strains (Supplementary Figure 1 and Supplementary Table 5), suggesting that these strains might metabolize ubiquitous phosphonate by CP lyase as a phosphorus or carbon source.
A three-gene cluster (ectA, ectB, and ectC) responsible for the synthesis of ectoine (1,4,5,6-tetrahydro-2-methyl-4-pyrimidinecarboxylic acid), a common osmoprotectant in many halophilic eubacteria (Schwibbert et al., 2011), was identified in these 207 genomes. Other organic osmotic protectants, such as glycine betaine, may also be synthesized via choline dehydrogenase (BetA) and betaine aldehyde dehydrogenase (BetB), similar to Halomonas elongata (Cánovas et al., 2000). The presence of these genes was common in the genus Halomonas (Supplementary Table 5), which might explain the adaptation of Halomonas to high salinity.
These genomes carry genes encoding copper resistance proteins and transporters, implying the tolerance to such bio-toxic metals in the environment. Besides, multiple genes potentially involved in arsenic resistance are identified in these genomes, including two arsenic resistance operons containing genes encoding ArsH and ACR3 and other related genes encoding the arsenical resistance operon repressor ArsR, as well as the arsenite-transporting ATPase ArsA. However, proteins homologous to aerobic arsenite oxidase (AioA and AioB) and anaerobic arsenite oxidase (ArxA) (Lin et al., 2012; Hamamura et al., 2014) could not be identified in these genomes, although they were found in several closely related species (H. campisalis and H. montanilacus), as well as other species, such as H. aestuarii, Halomonas andesensis, and H. boliviensis, as shown in Supplementary Figure 1 and Supplementary Table 5.
Halomonas zhangzhouensis (zhang.zhou.en’sis. N.L. fem. adj. zhangzhouensis, of or pertaining to Zhangzhou, a city in Fujian, China, where the type strain was isolated).
Cells are Gram-stain-negative, facultatively anaerobic, motile by means of peritrichous flagella, rod-shaped, 0.5–0.7 μm in width, and 1.0–3.0 μm in length. Colonies are yellow, circular, smooth, and opaque on MA after 72 h at 32°C. Growth occurs at 4–50°C (optimum, 37–40°C) and in pH 7–10 (optimum, 7–8). The NaCl concentration range for growth is 0–18% (optimum, 2–8%, w/v). Catalase and oxidase are positive. N2 and N2O are the major gaseous products of aerobic and anaerobic denitrification. Bacteria are weak positive for the Voges–Proskauer test and fermentation of D-glucose and negative for the ONPG test, indole production, H2S production, and hydrolysis of Tween-20, -40, -60, -80, gelatin, cellulose, starch, casein, and DNA. In the API ZYM tests, bacteria are positive for alkaline phosphatase, esterase, valine aminopeptidase, acid phosphatase, naphthol-AS-Bl-phosphoamidase, esterase lipase (C8), lipase (C14), leucine aminopeptidase, and cystine aminopeptidase and negative for α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-fucosidase, trypsin, and α-glucosidase. In the API 20NE tests, bacteria are positive for utilization of L-arabinose, D-mannitol, potassium gluconate, malic acid, and trisodium citrate and negative for arginine dihydrolase, β-glucosidase, lysine decarboxylase, ornithine decarboxylase, urease, tryptophan deaminase, and utilization of D-glucose, D-mannose, N-acetyl-glucosamine, D-maltose, capric acid, adipic acid, and phenylacetic acid. The predominant respiratory quinone is Q-9. The principal cellular fatty acids are summed feature 8 (C18:1 ω7c), C16:0, and C19:0 cyclo ω8c. The polar lipids are DPG, PG, PE, PL, and six unknown polar lipids. The genomic G+C content of the type strain is 63.3%.
The type strain CXT3-11T ( = MCCC 1A11036T = KCTC 72087T) was isolated from sediments of a shrimp culture pond, Zhangzhou City, China.
Halomonas aerodenitrificans (a.e.ro.de.ni.tri’fi.cans. Gr. masc. n. aer, aeros, air; N.L. v. denitrificare, to denitrify; N.L. part. adj. aerodenitrificans, denitrifying with or in air).
Cells are Gram-stain-negative, facultatively anaerobic, motile by means of peritrichous flagella, rod-shaped, 0.6–1.1 μm in width, and 1.7–2.7 μm in length. Colonies are yellow, circular, smooth, and opaque on MA after 72 h at 32°C. Growth occurs at 4–50°C (optimum, 37°C) and in pH 7–10 (optimum, 7–8). The NaCl concentration range for growth is 0–18% (optimum, 2–8%, w/v). Catalase and oxidase are positive. N2 and N2O are the major gaseous products of aerobic and anaerobic denitrification. Bacteria are positive for the Voges–Proskauer test, fermentation of D-glucose, and hydrolysis of Tween-20, -40, -60, -80, and starch and negative for the ONPG test, indole production, H2S production, and hydrolysis of gelatin, cellulose, casein, and DNA. In the API ZYM tests, bacteria are positive for alkaline phosphatase, esterase, valine aminopeptidase, acid phosphatase, naphthol-AS-Bl-phosphoamidase, esterase lipase (C8), lipase (C14), leucine aminopeptidase, cystine aminopeptidase, trypsin, and α-glucosidase and negative for α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase. In the API 20NE tests, bacteria are positive for utilization of D-glucose, D-mannitol, D-maltose, potassium gluconate, adipic acid, malic acid, and trisodium citrate and negative for arginine dihydrolase, β-glucosidase, gelatinase, lysine decarboxylase, ornithine decarboxylase, urease, tryptophan deaminase, and utilization of L-arabinose, D-mannose, N-acetyl-glucosamine, capric acid, and phenylacetic acid. The predominant respiratory quinone is Q-9. The principal cellular fatty acids are summed feature 8 (C18:1 ω7c) and C16:0. The polar lipids are DPG, PG, PE, four PLs, and five unknown polar lipids. The genomic G+C content of the type strain is 64%.
The type strain CYD-9T ( = MCCC 1A11058T = KCTC 72088T) was isolated from coastal water from the Taiwan Strait, China.
Halomonas sulfidoxydans (sul.fid.o’xy.dans. N.L. neut. n. sulfidum, sulfide; N.L. part. adj. oxydans, oxidizing; N.L. part. adj. sulfidoxydans, oxidizing sulfides).
Cells are Gram-stain-negative, facultatively anaerobic, motile by means of peritrichous flagella, rod-shaped, 0.7–0.8 μm in width, and 1.3–2.0 μm in length. Colonies are yellow, circular, smooth, and opaque on MA after 72 h at 32°C. Growth occurs at 4–50°C (optimum, 37–40°C) and in pH 6–10 (optimum, 7–8). The NaCl concentration range for growth is 0–18% (optimum, 2–6%, w/v). Catalase and oxidase are positive. N2 and N2O are the major gaseous products of aerobic and anaerobic denitrification. Bacteria are positive for the Voges–Proskauer test, fermentation of D-glucose, and hydrolysis of gelatin and negative for the ONPG test, indole production, H2S production, and hydrolysis of Tween-20, -40, -60, -80, starch, cellulose, casein, and DNA. In the API ZYM tests, bacteria are positive for alkaline phosphatase, esterase, valine aminopeptidase, acid phosphatase, naphthol-AS-Bl-phosphoamidase, esterase lipase (C8), lipase (C14), leucine aminopeptidase, cystine aminopeptidase, trypsin, and α-glucosidase and negative for α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase. In the API 20NE tests, bacteria are positive for arginine dihydrolase, gelatinase, and utilization of D-glucose, L-arabinose, D-mannitol, D-maltose, potassium gluconate, adipic acid, malic acid, and trisodium citrate and negative for β-glucosidase, lysine decarboxylase, ornithine decarboxylase, urease, tryptophan deaminase, utilization of D-mannose, N-acetyl-glucosamine, capric acid and phenylacetic acid. The predominant respiratory quinone is Q-9. The principal cellular fatty acids are summed feature 8 (C18:1 ω7c), summed feature 3 (C16:1 ω7c and/or C16:1 ω6c), and C16:0. The polar lipids are DPG, PG, PE, PC, AL, APL, and two PLs. The genomic G+C content of the type strain is 66%.
The type strain CYN-1-2T ( = MCCC 1A11059T = KCTC 72089T) was isolated from coastal surface sediments of the Taiwan Strait, China.
Halomonas ethanolica (e.tha.no’li.ca N.L. neut. n. ethanol, ethanol; L. fem. suff. -ica, suffix used with various meanings; N.L. fem. adj. ethanolica, belonging to ethanol, in reference to the ability of the species to utilize ethanol as a substrate for growth).
Cells are Gram-stain-negative, facultatively anaerobic, motile by means of peritrichous flagella, rod-shaped, 0.6–0.8 μm in width, and 1.6–2.4 μm in length. Colonies are yellow, circular, smooth, and opaque on MA after 72 h at 32°C. Growth occurs at 4–50°C (optimum, 37–40°C) and in pH 7–10 (optimum, 7–8). The NaCl concentrations range for growth is 0–18% (optimum, 1–10%, w/v). Catalase and oxidase are positive. N2 and N2O are the major gaseous products of aerobic and anaerobic denitrification. Bacteria are positive for the Voges–Proskauer test, fermentation of D-glucose, and hydrolysis of Tween-20, -40, -60, and -80 and negative for the ONPG test, indole production, H2S production, and hydrolysis of starch, cellulose, gelatin, casein, and DNA. In the API ZYM tests, bacteria are positive for alkaline phosphatase, esterase, valine aminopeptidase, acid phosphatase, naphthol-AS-Bl-phosphoamidase, esterase lipase (C8), leucine aminopeptidase, cystine aminopeptidase, and α-glucosidase and negative for α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, lipase (C14), trypsin, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase. In the API 20NE tests, bacteria are positive for utilization of D-glucose, D-mannitol, D-maltose, potassium gluconate, adipic acid, malic acid, and trisodium citrate and negative for arginine dihydrolase, β-glucosidase, gelatinase, lysine decarboxylase, ornithine decarboxylase, urease, tryptophan deaminase, and utilization of L-arabinose, D-mannose, N-acetyl-glucosamine, capric acid, and phenylacetic acid. The predominant respiratory quinone is Q-9. The principal cellular fatty acids are summed feature 8 (C18:1 ω7c) and C16:0. The polar lipids are DPG, PG, PE, and PL. The genomic G+C content of the type strain is 64.5%.
The type strain CYT3-1-1T ( = MCCC 1A11081T = KCTC 72090T) was isolated from sediments of a shrimp culture pond, Zhangzhou City, China.
Halomonas sulfidivorans (sul.fi.di.vo’rans. N.L. neut. n. sulfidum, sulfide; L. pres. part. vorans, devouring; N.L. part. adj. sulfidivorans, sulfide-devouring).
Cells are Gram-stain-negative, facultatively anaerobic, motile by means of peritrichous flagella, rod-shaped, 0.6–0.7 μm in width, and 1.7–2.4 μm in length. Colonies are yellow, circular, smooth, and opaque on MA after 72 h at 32°C. Growth occurs at 4–55°C (optimum, 37–40°C) and in pH 6–10 (optimum, 7–8). The NaCl concentration range for growth is 0–20% (optimum, 2–8%, w/v). Catalase and oxidase are positive. N2 and N2O are the major gaseous products of aerobic and anaerobic denitrification. Bacteria are positive for the Voges–Proskauer test, fermentation of D-glucose, and hydrolysis of starch and DNA and negative for the ONPG test, indole production, H2S production, hydrolysis of Tween-20, -40, -60, -80, gelatin, cellulose, and casein. In the API ZYM tests, bacteria are positive for alkaline phosphatase, esterase, valine aminopeptidase, acid phosphatase, naphthol-AS-Bl-phosphoamidase, esterase lipase (C8), lipase (C14), leucine aminopeptidase, and cystine aminopeptidase and negative for α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-fucosidase, trypsin, and α-glucosidase. In the API 20NE tests, bacteria are positive for utilization of D-glucose, L-arabinose, D-mannitol, N-acetyl-glucosamine, D-maltose, potassium gluconate, adipic acid, malic acid, and trisodium citrate and negative for arginine dihydrolase, β-glucosidase, gelatinase, lysine decarboxylase, ornithine decarboxylase, urease, tryptophan deaminase, and utilization of D-mannose, capric acid, and phenylacetic acid. The predominant respiratory quinone is Q-9. The principal cellular fatty acids are summed feature 8 (C18:1 ω7c), C16:0, summed feature 3 (C16:1 ω7c/C16:1 ω6c), and C12:0 3-OH. The polar lipids are DPG, PG, PE, AL, PL, and five unknown polar lipids. The genomic G+C content of the type strain is 64.2%.
The type strain NLG_F1ET ( = MCCC 1A13718T = KCTC 72091T) was isolated from deep sea sediments, Pacific Ocean.
Halomonas tianxiuensis (tian.xiu.en’sis. N.L. fem. adj. tianxiuensis, pertaining to the Tianxiu Hydrothermal Field, on the Northwest Indian Ridge, from where the type strain was isolated).
Cells are Gram-stain-negative, facultatively anaerobic, motile by means of peritrichous flagella, rod-shaped, 0.6–0.7 μm in width, and 1.3–2.1 μm in length. Colonies are yellow, circular, smooth, and opaque on MA after 72 h at 32°C. Growth occurs at 4–50°C (optimum, 37–40°C) and in pH 6–10 (optimum, 7–8). The NaCl concentration range for growth is 0–18% (optimum, 2–8%, w/v). Catalase and oxidase are positive. N2 and N2O are the major gaseous products of aerobic and anaerobic denitrification. Bacteria are positive for the Voges–Proskauer test, fermentation of D-glucose, and hydrolysis of starch and DNA and negative for the ONPG test, indole production, H2S production, and hydrolysis of Tween-20, -40, -60, -80, gelatin, cellulose, and casein. In the API ZYM tests, bacteria are positive for alkaline phosphatase, esterase, valine aminopeptidase, acid phosphatase, naphthol-AS-Bl-phosphoamidase, lipase (C14), leucine aminopeptidase, cystine aminopeptidase, and α-glucosidase and negative for α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-fucosidase, esterase lipase (C8), and trypsin. In the API 20NE tests, bacteria are positive for utilization of D-glucose, L-arabinose, D-mannitol, D-maltose, potassium gluconate, adipic acid, malic acid, trisodium citrate, and phenylacetic acid and negative for arginine dihydrolase, β-glucosidase, gelatinase, lysine decarboxylase, ornithine decarboxylase, urease, tryptophan deaminase, and utilization of D-mannose, N-acetyl-glucosamine, and capric acid. The predominant respiratory quinone is Q-9. The principal cellular fatty acids are summed feature 8 (C18:1 ω7c), C16:0, and C19:0 cyclo ω8c. The polar lipids are DPG, PG, PE, PL, and six unknown polar lipids. The genomic G+C content of the type strain is 63.9%.
The type strain BC-M4-5T ( = MCCC 1A14433T = KCTC 72092T) was isolated from sulfide samples in Tianxiu hydrothermal vents, on the Northwest Indian Ridge, Indian Ocean.
In the present study, we isolated and established six novel species of Halomonas and classified them into the “H. desiderata group.” All six species as well as other members of this group were featured with aerobic denitrification and heterotrophic sulfide oxidation, attributed to the complete denitrification pathways and the occurrence of the sqr/fccAB genes, which encode sulfide-oxidizing enzymes and were widely distributed in the whole genus. In addition, the widespread occurrence of tsdA genes throughout the genus Halomonas was consistent with their ability of thiosulfate oxidation. This highlights the popularity of aerobic denitrification and heterotrophic sulfur oxidation among Halomonas bacteria, especially the group of “H. desiderata.” These insights help to explain their wide distribution in different environments and highlight their application potential for the removal of nitrogen and sulfide.
Thirteen Halomonas strains from marine environments and one from a municipal wastewater treatment plant were isolated in this study and deposited in the Marine Culture Collection of China (MCCC). As listed in Table 1, five of them are from deep sea sediments of the South China Sea and the Pacific Ocean, four from hydrothermal vent sulfide of the Northwest Indian Ocean, three from coastal water and sediments of the Taiwan Strait, two from sediments of shrimp culture ponds (Zhangzhou, China), and one from sewage of a municipal wastewater treatment plant (Xiamen, China). Unless otherwise specified, strains were grown aerobically on MA 2216 and in marine broth 2216 (BD, Difco) at 32°C. In addition, six type strains were also included. Halomonas desiderata DSM 9502T, Halomonas campisalis DSM 15413T, Halomonas kenyensis DSM 17331T, and Halomonas elongata DSM 2581T were purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). Halomonas daqingensis CGMCC 1.6443T was obtained from the China General Microbiological Culture Collection Center (CGMCC). Halomonas saliphila LCB169T and Halomonas lactosivorans KCTC 52281T were denoted by Yong-qiang Tian and Guo-xing Nie, respectively. These type strains were used for comparison of phenotypic, physiological, and chemotaxonomic characteristics and genomic analysis with the isolated strains in this study.
Genomic DNA was extracted from each strain using the SBS extraction kit (SBS Genetech Co., Ltd., Shanghai, China) according to the manufacturer’s instructions. Purified genomic DNA was quantified by a NanoDrop 2000 spectrophotometer (Thermo Scientific). For strain MCCC 1A14433T, the genome was sequenced using a combination of PacBio RSII and Illumina sequencing platforms by Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China). The assembly was produced using Canu software (Koren et al., 2017). The final assembly generated a circular genome with no gap. For strains MCCC 1A13718T and MCCC 1A11059T, their genome sequences were determined by Tianjin Biochip Corporation using the PacBio sequencing platform. Sequencing reads were assembled using the hierarchical genome assembly process (HGAP 4.0) to obtain circular genomes. Four type strains (H. campisalis DSM 15413T, H. kenyensis DSM 17331T, Halomonas neptunia DSM15720T, and Halomonas utahensis DSM3051T), which lack available genomic information, as well as the other 11 isolated strains (as listed in Table 1), were sequenced on the Illumina HiSeq platform by Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China). The high-quality reads were assembled by the software SPAdes version 3.12.0 with default parameters (Bankevich et al., 2012). All of the assembled genomes of these strains were deposited in the GenBank database, with the accession numbers as listed in Supplementary Table 1. Assembly features, including genome size, the number of contigs, and the G+C content, are summarized in Supplementary Table 1. The tRNA and rRNAs genes were searched using tRNAscan-SE software (v 2.0) (Lowe and Eddy, 1997) and RNAmmer (v 1.2) (Lagesen et al., 2007), respectively.
For the phylogenomic tree construction and comparative genomic analysis, 253 publicly available Halomonas genomes in the GenBank database were collected, 76 of which were type strains. To obtain high-quality genomes, the genomes which were used for further analysis in this study met the criteria of an estimated completeness of >90% (except for type strains of Halomonas sulfidaeris ATCC BAA-803T and Halomonas olivaria TYRC17T), an estimated contamination of <5%, and predicted contigs <200. Quality assessment of these genomes was conducted using the bioinformatic tools of CheckM (v1.0.12) and Quast (v5.0.0) (Gurevich et al., 2013) prior to annotation using Prokka (v1.12) (Seemann, 2014) and the RAST server (Aziz et al., 2008). After the evaluation of quality, a total of 207 genomic sequences were chosen for further comparative analysis. EggNOG-mapper was further used for functional annotation of the 207 genomes (Huerta-Cepas et al., 2017). The complete list of genomes used in this study is provided in Supplementary Table 1.
The 16S rRNA, gyrB, and rpoD gene sequences of the 14 strains were derived from the genomes. Other 16S rRNA, gyrB, and rpoD gene sequences of type strains within the genus Halomonas were obtained from the EzBiocloud database and National Center for Biotechnology Information (NCBI). Phylogenetic trees based on the concatenation of 16S rRNA, gyrB, and rpoD gene sequences were constructed using MEGA version 7 after multiple alignments of the data using the software MUSCLE v3.8.31 (Katoh and Standley, 2013), with distance options according to the Kimura two-parameter model and clustering with the neighbor-joining (NJ) method (Kumar et al., 2016). Bootstrap analysis based on 1,000 replications was used to estimate the confidence level of the tree topologies. Carnimonas nigrificans CECT 4437T was used as outgroup in the phylogenetic analyses.
To determine the phylogenomic positions of the 14 strains, genome-based phylogenetic trees were reconstructed using two methods including PhyloPhlAn 3.0 (Nicola et al., 2013) and an up-to-date bacterial core gene set (UBCG) consisting of 92 genes (Na et al., 2018) using default settings, respectively. The visualization, annotation, and management of the phylogenomic trees were performed using the web-based tool evolview v3 (Subramanian et al., 2019)2.
The similarity between genomes was assessed using the overall genome relatedness index (OGRI), which is useful for species delineation. Thus, dDDH was estimated in silico using the Genome-to-Genome Distance Calculator (GGDC 2.0) with the BLAST method and recommended formula 23 (Meier-Kolthoff et al., 2013). ANIs were calculated by BLAST (ANIb) in JSpeciesWS (Richter et al., 2016), fastANI v1.3 (Jain et al., 2018)4, and the standalone Orthologous Average Nucleotide Identity Tool (OAT) (Lee et al., 2016). The visualization of the numerical matrix for dDDH and ANI values was carried out using TBtools v0.674 (Chen et al., 2020).
To compare the nitrogen metabolism pathways within the genus Halomonas, all 19 of the genomes sequenced in this study and 188 other genomes retrieved from NCBI were analyzed together. In these genomes, genes encoding proteins involved in denitrification (NarGHIJ, NapABCDEFGH, NirSCDEFGHJLN, NirK, NorBCDE, qNor, and NosZYRLFD), nitrate and nitrite assimilation (NasA and NirBD), and nitrate and nitrite transporters (NrtABC and NarK) and regulators (NarXL, Fnr, NasR, and NasST) were identified by a local BLASTP protein similarity search, with the protein sequences listed in Supplementary Table 6 used as queries. For the BLASTP analysis, we used an amino acid similarity cutoff of –40%, alignment coverage >80%, and an e-value cutoff of 1E-5. The genes encoding sulfur oxidation proteins, including SoxXZYABCD (Friedrich et al., 2001), Sqr (SQR) (Marcia et al., 2009), FccAB (FCSD) (Chen et al., 1994), PDO (persulfide dioxygenase) (Liu et al., 2014), and TsdA (thiosulfate dehydrogenase) (Denkmann et al., 2012; Brito et al., 2015) were also analyzed in these genomes. The presence/absence of these genes in the genomes was further checked based on the functional annotation by RAST and EggNOG-mapper.
Putative homologous proteins of NarG, NapA, NirS, NirK, NorB, qNor, NosZ, Sqr, FccA, PDO, and TsdA were retrieved from each query genome (Supplementary Table 7) and then used to construct the respective phylogenetic tree. Reference sequences were obtained from NCBI. Phylogenetic trees based on NarG, NapA, NirS, NorB, NosZ, Sqr, FccA, PDO, and TsdA amino acid sequences were constructed using MEGA version 7 after multiple alignments using the software MUSCLE v3.8.31 (Katoh and Standley, 2013), with distance options according to the Kimura two-parameter model and clustering with the NJ method (Kumar et al., 2016). Bootstrap analysis based on 1,000 replications was used to estimate the confidence level of the tree topologies.
Besides, the obtained putative homologues from the genome were also blasted against the NCBI database to obtain information on the domains present, which was used as an extra confirmation of their functional gene identity. Then, the presence/absence of all these functional genes tested in all of the genomes was used for co-occurrence analysis (Supplementary Table 5).
Cellular and colonial morphology, motility, Gram staining, anaerobic growth of representative strains, and catalase and oxidase activities were examined according to previously described methods (Liu et al., 2017). The ranges and optima of temperature, pH, and NaCl concentration were tested according to previously described methods (Lai et al., 2014). Hydrolysis of protein, starch, casein, DNA, Tweens (20, 40, 60, and 80), and cellulose was conducted using traditional methods described by Mata et al. (2002). Other biochemical tests were carried out using API 20NE, API ZYM, and API 20E strips (bioMérieux) according to the manufacturer’s instructions, with the modification of adjusting the NaCl concentration to 3.0% in all of the tests.
For whole-cell fatty acid analysis, cells were harvested from the third quadrants on MA medium at 32°C. The cells were saponified, methylated, and extracted using the standard MIDI (Sherlock Microbial Identification System, version 6.0B) protocol. The fatty acids were then analyzed by gas chromatography (GC) (Agilent Technologies 6850) and identified using the TSBA6.0 database of the Microbial Identification System. Polar lipids were extracted using a chloroform/methanol system and analyzed by one- and two-dimensional thin-layer chromatography (TLC), as described previously (Kates, 1986). Merck silica gel 60 F254 aluminum-backed thin-layer plates were used in TLC analyses. The plate dotted with sample was subjected to two-dimensional development, with the first solvent being chloroform–methanol–water (65:25:4, by vol.) and the second solvent being chloroform–methanol–acetic acid–water (85:12:15:4, by vol.). Total lipids were detected using molybdatophosphoric acid, and specific functional groups were detected using spray reagents specific for defined functional groups (Kates, 1986). Analysis of the respiratory quinones was carried out according to previously described methods (Collins, 1985).
Denitrification was determined in aerobic and anaerobic conditions, using a denitrification medium containing the following ingredients (per liter distilled water): sodium acetate 1.46 g, sodium succinate 2.40 g, sodium citrate 1.76 g, KH2PO4 0.50 g, Na2HPO4 0.50 g, MgSO4⋅7H2O 0.05 g, FeSO4⋅7H2O 0.002 g, CaCl2 0.02 g, NaCl 30 g, NaNO3 1.275 g (or NaNO2 1.035 g), and trace element solution 1.0 ml, pH 7.5. The composition of trace element solution was described by Joo et al. (2005). The denitrification ability in anaerobic condition was examined in 100-ml sealed serum bottles containing 30 ml of medium, with the headspace filled with 100% He. The aerobic denitrification performance was examined in 500-ml sealed serum bottles containing 50 ml of medium, with the headspace filled with 75% He and 25% O2. The performance of aerobic denitrification of the six novel species was tested at 37°C with 15 mM 15N-labeled NaNO3 or NaNO2 as the sole nitrogen source and sodium citrate as the electron donor, at a C/N ratio of 8.0 and pH 7.5. Nitrate, nitrite, cell OD600, and gaseous nitrogen products (15N2 and N2O) as well as the oxygen percentage were analyzed during the experiment. All of the tests were performed in triplicate. 30N2 was analyzed by GC/isotope ratio mass spectrometry (GC/IRMS), and N2O and O2 were analyzed by GC/mass spectrometry (GC/MS) (Agilent 7890B). Besides, cell growth and the aerobic denitrification ability of the six novel species were tested with different electron donors, including acetate, sodium citrate, glucose, sucrose, lactate, glycerol, mannitol, maltose, ethanol, succinate, propionate, potassium sodium tartrate, sodium formate, pyruvate, methanol, formic acid, and acetic acid.
Sulfide and thiosulfate oxidation tests were carried out according to previously described methods (Hou et al., 2018). Bacteria were cultivated to stationary phase, harvested by centrifugation (6,000 g, 5 min), and resuspended in 50 mM of HEPES buffer (pH 7.5) to a turbidity of 2 at 600 nm. Cell suspension (10 ml) was transferred to a 50-ml centrifuge tube. Freshly prepared sodium sulfide or thiosulfate was added to initiate the reaction. The tube was capped tightly and incubated at 30°C with shaking at 100 rpm. Sulfide and the products were analyzed at various time intervals. All of the tests were performed in triplicate. Sulfide was detected by a colorimetric method (Kamyshny, 2009). Thiosulfate, sulfate, tetrathionate, and sulfite were detected by ion chromatography (ICS-1100 system; Dionex).
Sulfide tolerance tests were also performed using denitrification medium containing NaNO3 as the sole nitrogen source and sodium citrate as the carbon source under aerobic conditions at 28°C, 150 rpm and pH 7.5. The sulfide concentration in the denitrification medium ranged from 1 to 15 mM. Nitrate, nitrite, sulfide, and cell growth were measured during the experiment. The sulfide oxidation ability was also tested under anaerobic conditions using denitrification medium, with sodium citrate as the sole electron donor and 15 mM of NaNO3 or 10% N2O (the headspace of the sealed vial was filled with 90% He and 10% N2O) as the sole electron acceptor.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
LW and ZS designed the experiment and reviewed the manuscript. LW performed the experiment, analyzed the data, and wrote the manuscript. Both authors contributed to the article and approved the submitted version.
This work was financially supported by the National Key R&D Program of China (No. 2018YFC0310701), the COMRA Program (No. DY135-B2-01), the Scientific Research Foundation of the Third Institute of Oceanography, MNR (No. 2019027), and the National Natural Science Foundation of China (No. 41506152).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We greatly appreciate Dr. Yang Liu for the revision of the manuscript. We thank Jianyang Li for providing strain MCCC 1A13718T. We gratefully acknowledge Dr. Qiliang Lai for help with the polar lipid analysis and Dr. Li Gu for help with transmission electron microscopy. We are very grateful to Dr. Ruolin Cheng, Dr. Linfeng Gong, and Dr. Xiaoteng Fu for their help in bioinformatics analysis. We also thank professor Yong-qiang Tian (Sichuan University) and Guo-xing Nie (Henan Normal University) for the donation of type strains Halomonas saliphila LCB169T and Halomonas lactosivorans KCTC 52281T, respectively. We are grateful to Lei Wang for providing the genomes of type strains Halomonas neptunia DSM 15720T and Halomonas utahensis DSM 3051T. We also thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.652766/full#supplementary-material
Supplementary Figure 1 | Genome-based phylogenomic tree of the 14 strains along with 193 other Halomonas genomes and occurrence of genes for nitrogen, sulfur, and other metabolism. Phylogenomic tree was constructed using PhyloPhlAn 3.0. Strains shown in red are organisms isolated in this study. The visualization, annotation, and management of the phylogenomic trees were performed using the web-based tool evolview v3. Type strains of Marinobacter hydrocarbonoclasticus ATCC 49840T, Alcanivorax dieselolei B5T, and Marinospirillum minutulum DSM 6287T were used as outgroup. The 31 right-most columns represent the distribution of key functional genes for nitrogen, sulfur, and other metabolism including carbon metabolism as well as stress responses. Blue represents the denitrification pathway; tan represents regulators for denitrification; green represents assimilation of nitrate and nitrite; fuchsia represents regulators for assimilation of nitrate and nitrite; orange represents the sulfur oxidation pathway; olive represents the PHA synthesis pathway; yellow represents phosphonate transporters and metabolism; dodgerblue represents the ectoine and glycine betaine synthesis pathway; gray represents arsenical oxidation-, reduction-, and resistance-related genes.
Supplementary Figure 2 | Genome-based phylogenomic tree constructed based on the 14 strains along with 193 other Halomonas genomes using an up-to-date bacterial core gene (UBCG) set consisting of 92 genes. Strains shown in red are organisms isolated in this study. The visualization, annotation, and management of the phylogenomic trees were performed using the web-based tool evolview v3. Type strains of Marinobacter hydrocarbonoclasticus ATCC 49840T, Alcanivorax dieselolei B5T, and Marinospirillum minutulum DSM 6287T were used as the outgroup.
Supplementary Figure 3 | Transmission electron micrograph of negatively stained cells of six novel strains grown on MA 2216 (BD) for 12 h. (A) MCCC 1A11036T. (B) MCCC 1A11058T. (C) MCCC 1A11059T. (D) MCCC 1A11081T. (E) MCCC 1A13718T. (F) MCCC 1A14433T.
Supplementary Figure 4 | Polar lipid profile of the six novel strains. (A) MCCC 1A11036T. (B) MCCC 1A11058T. (C) MCCC 1A11059T. (D) MCCC 1A11081T. (E) MCCC 1A13718T. (F) MCCC 1A14433T. DPG, diphosphatidylglycerol; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PL, unidentified phospholipid; PC, phosphatidylcholine; AL, unidentified aminolipid; APL, aminophospholipid; L, unknown polar lipid; F-first dimension of TLC; S, second dimension of TLC.
Supplementary Figure 5 | Neighbor-joining phylogenetic tree based on the cytochrome cd1-type nitrite reductase NirS obtained from 64 Halomonas genomes and reference sequences. Bootstrap values (>50%) are shown at branch points. Bar, 0.1 nucleotide substitution rate (Knuc) units. The phylogeny was inferred using MEGA version 7 and 1000 bootstrap replicates. NirS sequence from Arcobacter trophiarum LMG 25534 was used as outgroup. The label in red font means the sequences from strains isolated in this study; blue font means sequences obtained from genomes of the genus Halomonas.
Supplementary Figure 6 | Neighbor-joining phylogenetic tree based on the periplasmic nitrate reductase NapA obtained from 53 Halomonas genomes and reference sequences. Bootstrap values (>50%) are shown at branch points. Bar, 0.05 nucleotide substitution rate (Knuc) units. The phylogeny was inferred using MEGA version 7 and 1000 bootstrap replicates. The NapA sequence from Magnetospira sp. QH-2 was used as outgroup. The label in red font means the sequences from strains isolated in this study; blue font means sequences obtained from genomes of the genus Halomonas.
Supplementary Figure 7 | Neighbor-joining phylogenetic tree based on the membrane-bound respiratory nitrate reductase NarG obtained from 157 Halomonas genomes and reference sequences. Bootstrap values (>50%) are shown at branch points. Bar, 0.1 nucleotide substitution rate (Knuc) units. The phylogeny was inferred using MEGA version 7 and 1000 bootstrap replicates. The NarG sequence from Haloarcula hispanica ATCC 33960 was used as outgroup. The label in red font means the sequences from strains isolated in this study; blue font means sequences obtained from genomes of the genus Halomonas.
Supplementary Figure 8 | Neighbor-joining phylogenetic tree based on the cytochrome c nitric oxide reductase c-Nor obtained from 87 Halomonas genomes and reference sequences. Bootstrap values (>50%) are shown at branch points. Bar, 0.1 nucleotide substitution rate (Knuc) units. The phylogeny was inferred using MEGA version 7 and 1000 bootstrap replicates. The quinol-dependent nitric oxide reductase sequence from Marinobacter hydrocarbonoclasticus ATCC 49840T was used as outgroup. The label in red font means the sequences from strains isolated in this study; blue font means sequences obtained from genomes of the genus Halomonas.
Supplementary Figure 9 | Neighbor-joining phylogenetic tree based on the nitrous oxide reductase NosZ obtained from 87 Halomonas genomes and reference sequences. Bootstrap values (>50%) are shown at branch points. Bar, 0.05 nucleotide substitution rate (Knuc) units. The phylogeny was inferred using MEGA version 7 and 1000 bootstrap replicates. The NosZ sequence from Azoarcus sp. BH72 was used as outgroup. The label in red font means the sequences from strains isolated in this study; blue font means sequences obtained from genomes of the genus Halomonas.
Supplementary Figure 10 | Phylogenetic tree based on 16S rRNA gene sequences from the 14 strains and type strains within the genus Halomonas as well as 193 other Halomonas genomes. The type strain Marinobacter hydrocarbonoclasticus ATCC 49840T was used as outgroup. Bootstrap values (>50%) are shown at branch points. Bar, 0.01 nucleotide substitution rate (Knuc) units. The phylogeny was inferred using MEGA version 7 and 1000 bootstrap replicates. Each type strain is marked by a superscript capital T.
Supplementary Figure 11 | Neighbor-joining phylogenetic tree based on the flavocytochrome c sulfide dehydrogenase FccA obtained from 38 Halomonas genomes and reference sequences. Bootstrap values (>50%) are shown at branch points. Bar, 20 nucleotide substitution rate (Knuc) units. The phylogeny was inferred using MEGA version 7 and 1000 bootstrap replicates. The FccA sequence from Paracoccus denitrificans was used as outgroup. The label in red font means the sequences from strains isolated in this study; blue font means sequences obtained from genomes of the genus Halomonas.
Supplementary Figure 12 | Neighbor-joining phylogenetic tree based on the sulfide:quinone oxidoreductase SQR obtained from 67 Halomonas genomes and reference sequences. Bootstrap values (>50%) are shown at branch points. Bar, 10 nucleotide substitution rate (Knuc) units. The phylogeny was inferred using MEGA version 7 and 1000 bootstrap replicates. The FccA sequence from Paracoccus denitrificans was used as outgroup. The label in red font means the sequences from strains isolated in this study; blue font means sequences obtained from genomes of the genus Halomonas; bold font represents biochemically characterized SQR sequences.
Supplementary Figure 13 | Partial sequence alignment of SQR homologous proteins from the genus Halomonas and reference species. The Aquifex aeolicus SQR sequence was used as a template for the conserved residues analysis. Strictly conserved residues are marked with asterisks. Reference sequences: sqrIV.Aquifex_aeolicus (UniParc ID: UPI00000567FF), sqrII-Pseudomonas_putida (UniParc ID: UPI000000E9AA), sqrI.Acidithiobacillus_ferrooxidans (UniParc ID: UPI0000562E8C), sqrI.Rhodobacter_capsulatus (UniParc ID: UPI0001D081BD), sqrIII.Caldivirga_maquilingensis (UniParc ID: UPI0000F24E05).
Supplementary Figure 14 | Neighbor-joining phylogenetic tree based on the thiosulfate dehydrogenase TsdA obtained from 105 Halomonas genomes and reference sequences. Bootstrap values (>50%) are shown at branch points. Bar, 10 nucleotide substitution rate (Knuc) units. The phylogeny was inferred using MEGA version 7 and 1000 bootstrap replicates. The sulfur oxidation c-type cytochrome SoxA from Paracoccus denitrificans was used as outgroup. The label in red font means the sequences from strains isolated in this study; blue font means sequences obtained from genomes of the genus Halomonas; bold font represents functionally verified TsdA.
Supplementary Figure 15 | Partial sequence alignment of TsdA homologous proteins from the genus Halomonas and reference species. Heme-binding motifs are indicated by red boxes, putative distal heme ligands are marked with red arrows. Strictly conserved residues are marked with asterisks. The TsdA sequence from Allochromatium vinosum DSM 180T (ADC61061) was used as a template for the conserved domain and residues analysis.
Supplementary Figure 16 | Neighbor-joining phylogenetic tree based on the persulfide dioxygenase PDO obtained from 203 Halomonas genomes and reference sequences. Bootstrap values (>50%) are shown at branch points. Bar, 5 nucleotide substitution rate (Knuc) units. The phylogeny was inferred using MEGA version 7 and 1000 bootstrap replicates. The glyoxalase sequence from Haemophilus influenzae R2846 was used as outgroup. The label in red font means the sequences from strains isolated in this study; blue font means sequences obtained from genomes of the genus Halomonas.
Amjres, H., Bejar, V., Quesada, E., Abrini, J., and Llamas, I. (2011). Halomonas rifensis sp. nov., an exopolysaccharide-producing, halophilic bacterium isolated from a solar saltern. Int. J. Syst. Evol. Microbiol. 61, 2600–2605. doi: 10.1099/ijs.0.027268-0
Arahal, D. R., Vreeland, R. H., Litchfield, C. D., Mormile, M. R., Tindall, B. J., Oren, A., et al. (2007). Recommended minimal standards for describing new taxa of the family Halomonadaceae. Int. J. Syst. Evol. Microbiol. 57, 2436–2446. doi: 10.1099/ijs.0.65430-0
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bali, S., Palmer, D., Schroeder, S., Ferguson, S. J., and Warren, M. J. (2014). Recent advances in the biosynthesis of modified tetrapyrroles: the discovery of an alternative pathway for the formation of heme and heme d 1. Cell. Mol. Life Sci. 71, 2837–2863. doi: 10.1007/s00018-014-1563-x
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Berendes, F., Gottschalk, G., Heine-Dobbernack, E., Moore, E. R. B., and Tindall, B. J. (1996). Halomonas desiderata sp. nov, a new alkaliphilic, halotolerant and denitrifying bacterium isolated from a municipal sewage works. Syst. Appl. Microbiol. 19, 158–167.
Biddle, J. F., House, C. H., and Brenchley, J. E. (2005). Microbial stratification in deeply buried marine sediment reflects changes in sulfate/methane profiles. Geobiology 3, 287–295. doi: 10.1111/j.1472-4669.2006.00062.x
Bodenmiller, D. M., and Spiro, S. (2006). The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188, 874–881. doi: 10.1128/JB.188.3.874-881.2006
Boltianskaia, I., Kevbrin, V. V., Lysenko, A. M., Kolganova, T. V., Turova, T. P., Osipov, G. A., et al. (2007). [Halomonas mongoliensis sp. nov. and Halomonas kenyensis sp. nov., new haloalkaliphilic denitrifiers capable of reducing N2O, isolated from soda lakes]. Mikrobiologiia 76, 834–843.
Brito, J. A., Denkmann, K., Pereira, I. A., Archer, M., and Dahl, C. (2015). Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum: structural and functional insights into thiosulfate oxidation. J. Biol. Chem. 290, 9222–9238. doi: 10.1074/jbc.M114.623397
Bush, M. J., Ghosh, T., Tucker, N. P., Zhang, X., and Dixon, R. (2011). Transcriptional regulation by the dedicated nitric oxide sensor, NorR: a route towards NO detoxification. Biochem. Soc. Trans. 39, 289–293. doi: 10.1042/BST0390289
Cánovas, D., Vargas, C., Kneip, S., Morón, M. A., Ventosa, A., Bremer, E., et al. (2000). Genes for the synthesis of the osmoprotectant glycine betaine from choline in the moderately halophilic bacterium Halomonas elongata DSM 3043. Microbiology 146, 455–463. doi: 10.1099/00221287-146-2-455
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Ho, K. L., Liu, F. C., Ho, M., Wang, A., Ren, N., et al. (2013). Autotrophic and heterotrophic denitrification by a newly isolated strain Pseudomonas sp. C27. Bioresour. Technol. 145, 351–356. doi: 10.1016/j.biortech.2012.12.027
Chen, Z. W., Koh, M., Van Driessche, G., Van Beeumen, J. J., Bartsch, R. G., Meyer, T. E., et al. (1994). The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium. Science 266, 430–432. doi: 10.1126/science.7939681
Collins, M. D. (1985). “Isoprenoid quinone analyses in bacterial classification and identification,” in Chemical Methods in Bacterial Systematics, eds M. Goodfellow and D. E. Minnikin (New York, NY: Academic Press), 267–284.
de la Haba, R. R., Marquez, M. C., Papke, R. T., and Ventosa, A. (2012). Multilocus sequence analysis of the family Halomonadaceae. Int. J. Syst. Evol. Microbiol. 62(Pt 3), 520–538. doi: 10.1099/ijs.0.032938-0
Delorme, S., Philippot, L., Edel-Hermann, V., Deulvot, C., Mougel, C., and Lemanceau, P. (2003). Comparative genetic diversity of the narG, nosZ, and 16S rRNA genes in fluorescent pseudomonads. Appl. Environ. Microbiol. 69, 1004–1012. doi: 10.1128/aem.69.2.1004-1012.2003
Denkmann, K., Grein, F., Zigann, R., Siemen, A., Bergmann, J., van Helmont, S., et al. (2012). Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome. Environ. Microbiol. 14, 2673–2688. doi: 10.1111/j.1462-2920.2012.02820.x
Dong, Y., Kumar, C. G., Chia, N., Kim, P. J., Miller, P. A., Price, N. D., et al. (2014). Halomonas sulfidaeris-dominated microbial community inhabits a 1.8 km-deep subsurface Cambrian Sandstone reservoir. Environ. Microbiol. 16, 1695–1708. doi: 10.1111/1462-2920.12325
Du, R., Yu, M., Cheng, J. G., Zhang, J. J., Tian, X. R., and Zhang, X. H. (2019). Diversity and sulfur oxidation characteristics of cultivable sulfur oxidizing bacteria in hydrothermal fields of Okinawa Trough. Acta Microbiol. Sinica 59, 1036–1049. doi: 10.13343/j.cnki.wsxb.20180353
Etchebehere, C., and Tiedje, J. (2005). Presence of two different active nirS nitrite reductase genes in a denitrifying Thauera sp. from a high-nitrate-removal-rate reactor. Appl. Environ. Microbiol. 71, 5642–5645. doi: 10.1128/AEM.71.9.5642-5645.2005
Friedrich, C. G., Rother, D., Bardischewsky, F., Quentmeier, A., and Fischer, J. (2001). Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67, 2873–2882. doi: 10.1128/AEM.67.7.2873-2882.2001
Gan, L., Long, X., Zhang, H., Hou, Y., Tian, J., Zhang, Y., et al. (2018). Halomonas saliphila sp. nov., a moderately halophilic bacterium isolated from a saline soil. Int. J. Syst. Evol. Microbiol. 68, 1153–1159. doi: 10.1099/ijsem.0.002644
García, M. T., Mellado, E., Ostos, J. C., and Ventosa, A. (2004). Halomonas organivorans sp. nov., a moderate halophile able to degrade aromatic compounds. Int. J. Syst. Evol. Microbiol. 54, 1723–1728.
Goldman, B. S., Lin, J. T., and Stewart, V. (1994). Identification and structure of the nasR gene encoding a nitrate-and nitrite-responsive positive regulator of nasFEDCBA (nitrate assimilation) operon expression in Klebsiella pneumoniae M5al. J. Bacteriol. 176, 5077–5085. doi: 10.1128/jb.176.16.5077-5085
González-Domenech, C. M., Martínez-Checa, F., Béjar, V., and Quesada, E. (2010). Denitrification as an important taxonomic marker within the genus Halomonas. Syst. Appl. Microbiol. 33, 85–93. doi: 10.1016/j.syapm.2009.12.001
Graf, D. R. H., Jones, C. M., and Hallin, S. (2014). Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS One 9:e114118. doi: 10.1371/journal.pone.0114118
Guo, H., Chen, C., and Lee, D. J. (2019). Nitrogen and sulfur metabolisms of Pseudomonas sp. C27 under mixotrophic growth condition. Bioresour. Technol. 293, 122–169. doi: 10.1016/j.biortech.2019.122169
Guo, Y., Zhou, X. M., Li, Y. G., Li, K., Wang, C. X., Liu, J. F., et al. (2013). Heterotrophic nitrification and aerobic denitrification by a novel Halomonas campisalis. Biotechnol. Lett. 35, 2045–2049. doi: 10.1007/s10529-013-1294-3
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hamamura, N., Itai, T., Liu, Y., Reysenbach, A. L., Damdinsuren, N., and Inskeep, W. P. (2014). Identification of anaerobic arsenite-oxidizing and arsenate-reducing bacteria associated with an alkaline saline lake in Khovsgol, Mongolia. Environ. Microbiol. Rep. 6, 476–482. doi: 10.1111/1758-2229.12144
Hartig, E., Schiek, U., Vollack, K., and Zumft, W. G. (1999). Nitrate and nitrite control of respiratory nitrate reduction in denitrifying Pseudomonas stutzeri by a two-component regulatory system homologous to NarXL of Escherichia coli. J. Bacteriol. 181, 3658–3665. doi: 10.1128/JB.181.12.3658-3665.1999
He, T., Li, Z., Sun, Q., Xu, Y., and Ye, Q. (2016). Heterotrophic nitrification and aerobic denitrification by Pseudomonas tolaasii Y-11 without nitrite accumulation during nitrogen conversion. Bioresour. Technol. 200, 493–499. doi: 10.1016/j.biortech.2015.10.064
Homann, V. V., Sandy, M., Tincu, J. A., Templeton, A. S., Tebo, B. M., and Butler, A. (2009). Loihichelins A-F, a suite of amphiphilic siderophores produced by the marine bacterium Halomonas LOB-5. J. Nat. Prod. 72, 884–888. doi: 10.1021/np800640h
Hou, N., Xia, Y., Wang, X., Liu, H., Liu, H., and Xun, L. (2018). H2S biotreatment with sulfide-oxidizing heterotrophic bacteria. Biodegradation 29, 511–524. doi: 10.1007/s10532-018-9849-6
Huerta-Cepas, J., Forslund, K., Coelho, L., Szklarczyk, D., Jensen, L., von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Inagaki, F., Nunoura, T., Nakagawa, S., Teske, A., Lever, M., Lauer, A., et al. (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. U.S.A. 103, 2815–2820. doi: 10.1073/pnas.0511033103
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Ji, B., Yang, K., Zhu, L., Jiang, Y., Wang, H., Zhou, J., et al. (2015). Aerobic denitrification: a review of important advances of the last 30 years. Biotechnol. Bioprocess. Eng. 20, 643–651.
Jones, C. M., and Hallin, S. (2010). Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. ISME J. 4, 633–641. doi: 10.1038/ismej.2009.152
Jones, C. M., Stres, B., Rosenquist, M., and Hallin, S. (2008). Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 25, 1955–1966. doi: 10.1093/molbev/msn146
Joo, H. S., Hirai, M., and Shoda, M. (2005). Characteristics of ammonium removal by heterotrophic nitrification-aerobic denitrification by Alcaligenes faecalis No. 4. J. Biosci. Bioeng. 100, 184–191. doi: 10.1263/jbb.100.184
Kamyshny, A. J. (2009). Improved cyanolysis protocol for detection of zero-valent sulfur in natural aquatic systems. Limnol. Oceanogr. Methods 7, 442–448.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kawata, Y., Ando, H., Matsushita, I., and Tsubota, J. (2014). Efficient secretion of (R)-3-hydroxybutyric acid from Halomonas sp. KM-1 by nitrate fed-batch cultivation with glucose under microaerobic conditions. Bioresour. Technol. 156, 400–403. doi: 10.1016/j.biortech.2013.05.015
Kaye, J. Z., and Baross, J. A. (2000). High incidence of halotolerant bacteria in Pacific hydrothermal-vent and pelagic environments. FEMS Microbiol. Ecol. 32, 249–260. doi: 10.1111/j.1574-6941.2000.tb00718.x
Kaye, J. Z., Márquez, M. C., Ventosa, A., and Baross, J. A. (2004). Halomonas neptunia sp. nov., Halomonas sulfidaeris sp. nov., Halomonas axialensis sp. nov. and Halomonas hydrothermalis sp. nov.: halophilic bacteria isolated from deep-sea hydrothermal-vent environments. Int. J. Syst. Evol. Microbiol. 54, 499–511. doi: 10.1099/ijs.0.02799-0
Kaye, J. Z., Sylvan, J. B., Edwards, K. J., and Baross, J. A. (2011). Halomonas and Marinobacter ecotypes from hydrothermal vent, subseafloor and deep-sea environments. FEMS Microbiol. Ecol. 75, 123–133. doi: 10.1111/j.1574-6941.2010.00984.x
Kim, K. K., Lee, J. S., and Stevens, D. A. (2013). Microbiology and epidemiology of Halomonas species. Future Microbiol. 8, 1559–1573. doi: 10.2217/fmb.13.108
Koren, S., Walenz, B., Berlin, K., Miller, J., Bergman, N., and Phillippy, A. (2017). kCanu: scalable and accurate long-read assembly via adaptive -mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lai, Q., Liu, Y., and Shao, Z. (2014). Bacillus xiamenensis sp. nov., isolated from intestinal tract contents of a flathead mullet (Mugil cephalus). Antonie Van Leeuwenhoek 105, 99–107. doi: 10.1007/s10482-013-0057-4
Lee, I., Kim, Y. O., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Li, X., Gan, L., Hu, M., Wang, S., Tian, Y., and Shi, B. (2020). Halomonas pellis sp. nov., a moderately halophilic bacterium isolated from wetsalted hides. Int. J. Syst. Evol. Microbiol. 70, 5417–5424. doi: 10.1099/ijsem.0.004426
Lin, Y., Fan, H., Hao, X., Johnstone, L., Hu, Y., Wei, G., et al. (2012). Draft genome sequence of Halomonas sp. strain HAL1, a moderately halophilic arsenite-oxidizing bacterium isolated from gold-mine soil. J. Bacteriol. 194, 199–200. doi: 10.1128/JB.06359-11
Liu, C., Zhao, D., Ma, W., Guo, Y., Wang, A., Wang, Q., et al. (2016a). Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp. Appl. Microbiol. Biotechnol. 100, 1421–1426. doi: 10.1007/s00253-015-7039-6
Liu, H., Xin, Y., and Xun, L. (2014). Distribution, diversity, and activities of sulfur dioxygenases in heterotrophic bacteria. Appl. Environ. Microbiol. 80, 1799–1806. doi: 10.1128/AEM.03281-13
Liu, S., Chen, Q., Ma, T., Wang, M., and Ni, J. (2018). Genomic insights into metabolic potentials of two simultaneous aerobic denitrification and phosphorus removal bacteria, Achromobacter sp. GAD3 and Agrobacterium sp. LAD9. FEMS Microbiol. Ecol. 94:fiy020. doi: 10.1093/femsec/fiy020
Liu, Y., Ai, G. M., Miao, L. L., and Liu, Z. P. (2016b). Marinobacter strain NNA5, a newly isolated and highly efficient aerobic denitrifier with zero N2O emission. Bioresour. Technol. 206, 9–15. doi: 10.1016/j.biortech.2016.01.066
Liu, Y., Du, J., Lai, Q., Zeng, R., Ye, D., Xu, J., et al. (2017). Proposal of nine novel species of the Bacillus cereus group. Int. J. Syst. Evol. Microbiol. 67, 2499–2508. doi: 10.1099/ijsem.0.001821
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Luque-Almagro, V. M., Gates, A. J., Moreno-Vivian, C., Ferguson, S. J., Richardson, D. J., and Roldan, M. D. (2011). Bacterial nitrate assimilation: gene distribution and regulation. Biochem. Soc. Trans. 39, 1838–1843. doi: 10.1042/BST20110688
Luque-Almagro, V. M., Manso, I., Sullivan, M. J., Rowley, G., Ferguson, S. J., Moreno-Vivián, C., et al. (2017). Transcriptional and translational adaptation to aerobic nitrate anabolism in the denitrifier Paracoccus denitrificans. Biochem. J. 474, 1769–1787. doi: 10.1042/BCJ20170115
Marcia, M., Ermler, U., Peng, G., and Michel, H. (2009). The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc. Natl. Acad. Sci. U.S.A. 106, 9625–9630. doi: 10.1073/pnas.0904165106
Marcia, M., Ermler, U., Peng, G., and Michel, H. (2010). A new structure-based classification of sulfide:quinone oxidoreductases. Proteins 78, 1073–1083. doi: 10.1002/prot.22665
Mata, J. A., Martínez-Cánovas, J., Quesada, E., and Béjar, V. (2002). A detailed phenotypic characterisation of the type strains of Halomonas species. Syst. Appl. Microbiol. 25, 360–375. doi: 10.1078/0723-2020-00122
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Ming, H., Ji, W. L., Li, M., Zhao, Z. L., Cheng, L. J., Niu, M. M., et al. (2020). Halomonas lactosivorans sp. nov., isolated from salt-lake sediment. Int. J. Syst. Evol. Microbiol. 70, 3504–3512. doi: 10.1099/ijsem.0.004209
Mussmann, M., Hu, F. Z., Richter, M., de Beer, D., Preisler, A., Jørgensen, B. B., et al. (2007). Insights into the genome of large sulfur bacteria revealed by analysis of single filaments. PLoS Biol. 5:e230. doi: 10.1371/journal.pbio.0050230
Na, S. I., Kim, Y. O., Yoon, S. H., Ha, S. M., Baek, I., and Chun, J. (2018). UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285. doi: 10.1007/s12275-018-8014-6
Nicola, S., Daniela, B., Xochitl, C. M., and Curtis, H. (2013). PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 4:2304. doi: 10.1038/ncomms3304
Nunoura, T., Takaki, Y., Hirai, M., Shimamura, S., Makabe, A., Koide, O., et al. (2015). Hadal biosphere: insight into the microbial ecosystem in the deepest ocean on Earth. Proc. Natl. Acad. Sci. U.S.A. 112, E1230–E1236. doi: 10.1073/pnas.1421816112
Onley, J. R., Ahsan, S., Sanford, R. A., and Löffler, F. E. (2018). Denitrification by Anaeromyxobacter dehalogenans, a common soil bacterium lacking the nitrite reductase genes nirS and nirK. Appl. Environ. Microbiol. 84:e1985–17. doi: 10.1128/AEM.01985-17
Petri, R., and Imhoff, J. F. (2000). The relationship of nitrate reducing bacteria on the basis of narH gene sequences and comparison of narH and 16S rDNA based phylogeny. Syst. Appl. Microbiol. 23, 47–57. doi: 10.1016/S0723-2020(00)80045-4
Polz, M. F., Hunt, D. E., Preheim, S. P., and Weinreich, D. M. (2006). Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 2009–2021. doi: 10.1098/rstb.2006.1928
Reinartz, M., Tschäpe, J., Brüser, T., Trüper, H. G., and Dahl, C. (1998). Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch. Microbiol. 170, 59–68. doi: 10.1007/s002030050615
Richardson, D. J., Berks, B. C., Russell, D. A., Spiro, S., and Taylor, C. J. (2001). Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell. Mol. Life Sci. 58, 165–178. doi: 10.1007/PL00000845
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Richter, M., Rosselló-Móra, R., Glöckner, F. O., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Robertson, L. A., and Kuenen, J. G. (1984). Aerobic denitrification-old wine in new bottles? Antonie Van Leeuwenhoek 50, 525–544. doi: 10.1007/bf02386224
Salazar, G., Cornejo-Castillo, F. M., Benítez-Barrios, V., Fraile-Nuez, E., Álvarez-Salgado, X. A., Duarte, C. M., et al. (2016). Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J. 10, 596–608. doi: 10.1038/ismej.2015.137
Sanchez, C., and Minamisawa, K. (2018). Redundant roles of Bradyrhizobium oligotrophicum Cu-type (NirK) and cd1-type (NirS) nitrite reductase genes under denitrifying conditions. FEMS Microbiol. Lett. 365:fny015. doi: 10.1093/femsle/fny015
Sanz-Sáez, I., Salazar, G., Sánchez, P., Lara, E., Royo-Llonch, M., Sà, E. L., et al. (2020). Diversity and distribution of marine heterotrophic bacteria from a large culture collection. BMC Microbiol. 20:207. doi: 10.1186/s12866-020-01884-7
Schwibbert, K., Marin-Sanguino, A., Bagyan, I., Heidrich, G., Lentzen, G., Seitz, H., et al. (2011). A blueprint of ectoine metabolism from the genome of the industrial producer Halomonas elongata DSM 2581 T. Environ. Microbiol. 13, 1973–1994. doi: 10.1111/j.1462-2920.2010.02336.x
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Singer, E., Webb, E., Nelson, W., Heidelberg, J., Ivanova, N., Pati, A., et al. (2011). Genomic potential of Marinobacter aquaeolei, a biogeochemical “opportunitroph”. Appl. Environ. Microbiol. 77, 2763–2771. doi: 10.1128/aem.01866-10
Sorokin, D. (2003). Oxidation of inorganic sulfur compounds by obligatory organotrophic bacteria. Microbiology 72, 725–739.
Sorokin, D. Y., Kuenen, J. G., and Jetten, M. S. M. (2001). Denitrification at extremely high pH values by the alkaliphilic, obligately chemolithoautotrophic, sulfur-oxidizing bacterium Thioalkalivibrio denitrificans strain ALJD. Arch. Microbiol. 175, 94–101. doi: 10.1007/s002030000210
Sorokin, D. Y., Zhilina, T. N., Lysenko, A. M., Tourova, T. P., and Spiridonova, E. M. (2006). Metabolic versatility of haloalkaliphilic bacteria from soda lakes belonging to the Alkalispirillum-Alkalilimnicola group. Extremophiles 10, 213–220. doi: 10.1007/s00792-005-0487-7
Sousa, F. M., Pereira, J. G., Marreiros, B. C., and Pereira, M. M. (2018). Taxonomic distribution, structure/function relationship and metabolic context of the two families of sulfide dehydrogenases: SQR and FCSD. Biochim. Biophys. Acta Bioenerg. 1859, 742–753. doi: 10.1016/j.bbabio.2018.04.004
Sparacino-Watkins, C., Stolz, J. F., and Basu, P. (2014). Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 43, 676–706. doi: 10.1039/c3cs60249d
Spiro, S. (1994). The FNR family of transcriptional regulators. Antonie Van Leeuwenhoek 66, 23–36. doi: 10.1007/BF00871630
Stern, A. M., Liu, B., Bakken, L. R., Shapleigh, J. P., and Zhu, J. (2013). A novel protein protects bacterial iron-dependent metabolism from nitric oxide. J. Bacteriol. 195, 4702–4708. doi: 10.1128/JB.00836-13
Subramanian, B., Gao, S. H., Lercher, M. J., Hu, S. N., and Chen, W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47(W1), W270–W275. doi: 10.1093/nar/gkz357
Teske, A., Brinkhoff, T., Muyzer, G., Moser, D. P., Rethmeier, J., and Jannasch, H. W. (2000). Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl. Environ. Microbiol. 66, 3125–3133. doi: 10.1128/aem.66.8.3125-3133.2000
Vreeland, R. H., Litchfield, C. D., Martin, E. L., and Elliot, E. (1980). Halomonas elongata, a new genus and species of extremely salt-tolerant bacteria. Int. J. Syst. Bacteriol. 30, 485–495. doi: 10.1007/s11274-012-1020-7
Wayne, L. G., Brenner, D., Colwell, R., Grimont, P., Kandler, O., Krichevsky, M. I., et al. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Evol. Microbiol. 37, 463–464.
Wittorf, L., Jones, C. M., Bonilla-Rosso, G., and Hallin, S. (2018). Expression of nirK and nirS genes in two strains of Pseudomonas stutzeri harbouring both types of NO-forming nitrite reductases. Res. Microbiol. 169, 343–347. doi: 10.1016/j.resmic.2018.04.010
Wu, G., Wu, X. Q., Wang, Y. N., Chi, C. Q., Tang, Y. Q., Kida, K., et al. (2008). Halomonas daqingensis sp. nov., a moderately halophilic bacterium isolated from an oilfield soil. Int. J. Syst. Evol. Microbiol. 58, 2859–2865. doi: 10.1099/ijs.0.65746-0
Xia, Y., Lu, C., Hou, N., Xin, Y., Liu, J., Liu, H., et al. (2017). Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 11, 2754–2766. doi: 10.1038/ismej.2017.125
Yang, J. X., Feng, L., Pi, S. S., Cui, D., Ma, F., Zhao, H. P., et al. (2020). A critical review of aerobic denitrification: insights into the intracellular electron transfer. Sci. Total Environ. 20:139080. doi: 10.1016/j.scitotenv.2020.139080
Zhang, R. C., Chen, C., Shao, B., Wang, W., Xu, X. J., Zhou, X., et al. (2020). Heterotrophic sulfide-oxidizing nitrate-reducing bacteria enables the high performance of integrated autotrophic-heterotrophic denitrification (IAHD) process under high sulfide loading. Water Res. 1:115848. doi: 10.1016/j.watres.2020.115848
Keywords: Halomonas, taxonomy, novel species, aerobic denitrification, heterotrophic sulfur oxidation
Citation: Wang L and Shao Z (2021) Aerobic Denitrification and Heterotrophic Sulfur Oxidation in the Genus Halomonas Revealed by Six Novel Species Characterizations and Genome-Based Analysis. Front. Microbiol. 12:652766. doi: 10.3389/fmicb.2021.652766
Received: 13 January 2021; Accepted: 09 February 2021;
Published: 18 March 2021.
Edited by:
Iain Sutcliffe, Northumbria University, United KingdomReviewed by:
Aharon Oren, Hebrew University of Jerusalem, IsraelCopyright © 2021 Wang and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zongze Shao, c2hhb3p6QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.