
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 20 May 2021
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.652546
The EvgS/EvgA two-component signal transduction system in Escherichia coli is activated under mildly acidic pH conditions. Upon activation, this system induces the expression of a number of genes that confer acid resistance. The EvgS histidine kinase sensor has a large periplasmic domain that is required for perceiving acidic signals. In addition, we have previously proposed that the cytoplasmic linker region of EvgS is also involved in the activation of this sensor. The cytoplasmic linker region resembles a Per-ARNT-Sim (PAS) domain, which is known to act as a molecular sensor that is responsive to chemical and physical stimuli and regulates the activity of diverse effector domains. Our EvgS/EvgA reporter assays revealed that under EvgS-activating mildly acidic pH conditions, EvgS was activated only during aerobic growth conditions, and not during anaerobic growth. Studies using EvgS mutants revealed that C671A and C683A mutations in the cytoplasmic PAS domain activated EvgS even under anaerobic conditions. Furthermore, among the electron carriers of the electron transport chain, ubiquinone was required for EvgS activation. The present study proposes a model of EvgS activation by oxidation and suggests that the cytoplasmic PAS domain serves as an intermediate redox switch for this sensor.
Bacteria utilize two-component signal transduction systems (TCSs) to respond and adapt to fluctuating environmental conditions (Stock et al., 2000). A typical TCS consists of a histidine kinase (HK) sensor and its partner, a response regulator (RR). Input of a specific stimulus to the HK sensor causes autophosphorylation of a conserved histidine residue in the catalytic core. This phosphate group is subsequently transferred to the partner RR; the phosphorylated RR functions mainly by regulating gene expression, which is served as the output.
Most HK sensors are membrane proteins that are localized in the cytoplasmic membrane (Cock and Whitworth, 2007). In a prototypical HK sensor, the extracellular sensor region perceives environmental stimuli and transduces the information via the transmembrane segment to the cytoplasmic region, where the catalytic core resides. Additional protein domains such as HAMP (HK, adenylyl cyclases, methyl-accepting proteins, and other prokaryotic signaling proteins) and PAS (Per-ARNT-Sim) are often found between the transmembrane segment and the catalytic core. These domains transmit the N-terminal signal to the catalytic core and/or perceive additional signal inputs (Zschiedrich et al., 2016). Although a typical HK perceives the stimuli at its extracellular sensor domain, some HK sensors lack the extracellular region (sensors consisting of only transmembrane and cytoplasmic regions), while other HK sensors are cytoplasmic proteins. Thus, in addition to the extracellular sensing domain, signals are also perceived at the transmembrane segment and/or cytoplasmic sensing domains. The specific signals that stimulate HK sensors are unknown in many cases. Far less known are the domains through which the signals are perceived. Even when the signal of an HK sensor is well defined, upon close examination, a different signal approaching another domain may be discovered. Such new information may add to our interpretation of how TCS functions.
The present study aimed to understand the activation mechanism of the acid-responding EvgS HK sensor in Escherichia coli. EvgS, together with its cognate EvgA RR, composes the EvgS/EvgA TCS (Utsumi et al., 1994). This system is widely conserved in E. coli and Shigella (Johnson et al., 2014). A homologous system is found in Bordetella pertussis (the etiologic agent of whooping cough) as BvgS/BvgA, which controls a variety of virulence determinants (Arico et al., 1989; Uhl and Miller, 1994), and in Klebsiella pneumoniae as KvgS/KvgA, which is present only in the virulent strains (Lai et al., 2003; Lin et al., 2006). However, EvgS/EvgA is not found in any of the other sequenced bacteria. In E. coli, EvgS/EvgA upregulates a network of acid resistance genes, through a cascade of EvgA-YdeO-GadE regulators (Masuda and Church, 2003; Itou et al., 2009; De Biase and Lund, 2015), and via the SafA-PhoQ/PhoP-IraM-RpoS network, which involves two TCSs, two small TCS-connecting proteins, and a sigma factor (Eguchi et al., 2007, 2011). This upregulation of acid resistance genes confers severe acid resistance to cells in their exponential phase (Ma et al., 2004; Itou et al., 2009; Burton et al., 2010; De Biase and Lund, 2015). EvgS/EvgA also induces the expression of the EmrKY efflux pump via EvgA, which directly binds to the promoter region of the emrKY operon (Kato et al., 2000). The EmrKY pump belongs to the major facilitator superfamily and contributes to bile acid resistance (Nishino and Yamaguchi, 2001). Mutations affecting emrK or emrY gene cause a hypersensitive phenotype to the lethal effect of nalidixic acid, mitomycin C, ultraviolet irradiation, and hydrogen peroxide, suggesting that EmrKY might be pumping out toxic metabolites induced by DNA damage (Han et al., 2010). A recent report highlighted that this pump contributes to the survival of Shigella flexneri (the etiological agent of bacillary dysentery) within the host macrophage (Pasqua et al., 2019). This report also showed that Shigella emrKY is upregulated by EvgS/EvgA within host cells. There are other reports connecting EvgS/EvgA to E. coli pathogenicity. In enteropathogenic E. coli, expression of recombinant EvgA repressed the expression of the type III secretion system, which delivers a set of effector proteins into the host cell cytoplasm (Nadler et al., 2006). Another study showed that in avian pathogenic E. coli, an evgS mutant showed attenuated lung colonization in turkeys (Dziva et al., 2013).
EvgS is one of five unorthodox HK sensors in E. coli and has a large periplasmic domain with two tandem Venus flytrap domains (Sen et al., 2017). The cytoplasmic region consists of three catalytic domains: a HK domain, an intermediate receiver domain, and a Hpt (histidine-containing phosphotransfer) transmitter domain. Upon activation, the conserved histidine residue in the HK domain is autophosphorylated, and the phosphate is subsequently transferred to the aspartate residue of the intermediate receiver domain, then to the histidine residue of the Hpt transmitter (Perraud et al., 1998). This three-step phosphoryl-transfer reactions within the EvgS dimer is carried out in a cis-cis-cis mode, which is different from two other unorthodox HK sensors, ArcB (cis-trans-trans) and BarA (trans-trans-trans) (Kinoshita-Kikuta et al., 2016). From the Hpt transmitter, the phosphate is transferred to the aspartate residue of EvgA. The cytoplasmic linker region, between the transmembrane and HK domains, resembles a PAS domain, which is known to act as a molecular sensor responsive to signals such as oxygen, light, and voltage. Previous studies have shown that mildly acidic pH serves as an activating stimulus for EvgS (Ma et al., 2004; Burton et al., 2010; Eguchi and Utsumi, 2014). This signal is consistent with this system, which induces the expression of genes that confer severe acid resistance (De Biase and Lund, 2015). However, the mildly acidic medium needs to be a synthetic minimal medium, and not a rich medium such as LB (Lysogeny broth). Moreover, at least 150 mM of alkali metals must be included in the acidic medium for EvgS activation (Eguchi and Utsumi, 2014). The periplasmic region of EvgS is necessary for signal perception (Eguchi and Utsumi, 2014; Johnson et al., 2014), as well as the cytoplasmic PAS domain (Eguchi and Utsumi, 2014). Another report suggested that the PAS domain responds to acidic pH (Sen et al., 2017). In a different study, it has been shown that there is a natural variation in the pH-sensing capacity of EvgS, with some strains maintaining the ability to confer acid resistance despite being non-responsive to acidic pH (Roggiani et al., 2017). These reports suggest that there may be more input signals for EvgS.
The ArcB HK sensor of the ArcBA TCS is another member of the five unorthodox HK sensors of E. coli that responds to the changing respiratory conditions of growth. In E. coli, three dominant quinone types are found, which are active in facilitating electron transfer in the respiratory chain. These are ubiquinone (UQ), demethylmenaquinone (DMK), and menaquinone (MK). While DMK and MK are abundant during anaerobic conditions, UQ is the most abundant redox carrier during aerobic growth (Sharma et al., 2012). Under aerobic growth, these quinones silence the kinase activity of ArcB by oxidizing two redox-active cysteine residues in the cytoplasmic PAS domain that form interprotomer disulfide bonds in the ArcB dimer. Upon a shift from aerobic growth conditions to anaerobic, reduced forms of these quinones break the disulfide bonds and activate ArcB kinase activity (Georgellis et al., 2001; Malpica et al., 2004; Bekker et al., 2010; Sharma et al., 2012; Alvarez et al., 2013; van Beilen and Hellingwerf, 2016). Since both BvgS of B. pertussis and EvgS are unorthodox HK sensors and have cytoplasmic PAS domains as in ArcB, the effect of electron carriers on the kinase activities of the cytoplasmic soluble forms of EvgS and BvgS had been examined by Bock and Gross. In their study, oxidized UQ-0 (soluble analog of UQ-8), but not menadione (MK-3, soluble analog of MK-8), strongly inhibited the kinase activities of BvgS and EvgS (Bock and Gross, 2002). However, these in vitro studies were carried out using truncated soluble forms of HK. Whether the respiratory growth conditions affect EvgS activity in E. coli cells has not been examined yet. Therefore, in the present study, we examined in vivo EvgS activation under different respiratory conditions and found that EvgS activation only occurred under oxidative conditions. We propose oxidation as another stimulus perceived by the EvgS sensor.
The E. coli strains and plasmids used in this study are listed in Table 1.
Cultures were grown in LB medium (1% w/v tryptone, 0.5% w/v yeast extract, 1% w/v NaCl, pH 7.5) at 37°C with shaking at 180 rpm, unless stated otherwise. Activation of the EvgS/EvgA system was carried out using M9 medium supplemented with 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) and 100 mM KCl, with the pH adjusted to 5.7 using HCl.
When necessary, selective antibiotics (100 μg/mL ampicillin, 25μg/mL kanamycin, or 25 μg/mL chloramphenicol) were added to the medium. Arabinose was added to the culture at a final concentration of 1% w/v for EvgA overproduction and 0.02% for expression of EvgS and its variants. For ubiA mutants, 1 mM uracil was added to the medium to accelerate growth (personal communication).
The primers used in this study are listed in Table 2. To decrease the background activity of the lacZ-based reporter strains, lacZ of strain MG1655 was disrupted by inserting a chloramphenicol resistance cassette, according to a one-step inactivation method (Datsenko and Wanner, 2000), using primers lacZ-P2-F and lacZ-P1-R. The lacZ deletion was transferred to reporter strains MG1655 ydeP-lacZ and MG1655 emrKY-lacZ by P1 transduction to obtain MG1655 lacZ ydeP-lacZ and MG1655 lacZ emrKY-lacZ, respectively. Deletion of menA was transferred from JW3901 to MG1655 by P1 transduction, followed by removal of the kanamycin resistance cassette with pCP20 plasmid (Datsenko and Wanner, 2000), insertion of ydeP-lacZ, and deletion of lacZ by P1 transduction from MG1655 ydeP-lacZ and MG1655 lacZ, respectively. Deletion of ubiA was transferred from MU1227 to MG1655 ydeP-lacZ by P1 transduction.
Plasmids for expressing evgA and ubiA were constructed by PCR amplified DNA fragments with the primer pairs, evgA-NcoI-F + evgA-XhoI-R and ubiA-NcoI-F + ubiA-XhoI-R, and ligating them to NcoI- and XhoI- (Toyobo, Osaka, Japan) digested pBAD vector. To construct plasmids for expressing EvgS variants, site-directed mutagenesis of C663A, C671A, C683A, C671S, C683S, C671M, and C683M was performed using the primers listed in Table 2 (sites of mutation are underlined), PrimeSTAR® HS DNA polymerase (Takara Bio, Kusatsu, Japan), and pBADevgS plasmid. The PCR product was treated with DpnI (Toyobo) to degrade the template plasmid and transformed into DH5α. Site-directed mutagenesis was also performed to create plasmids for the double mutant C671A C683A. The plasmid for expressing the EvgS cytoplasmic region (553–1197), pBADevgS-cyt, was constructed using EvgS-cyt-F and EvgS-cyt-R primers, PrimeSTAR® HS DNA polymerase, and pBADevgS as the template for PCR. The PCR product was treated with DpnI, phosphorylated at its 5′ end with polynucleotide kinase (Toyobo), and self-ligated. All plasmids were confirmed using DNA sequencing.
A single colony of an E. coli strain was inoculated in 10 mL of LB medium containing appropriate antibiotics and grown overnight with shaking at 37°C. This culture was diluted 100-fold with 10 mL of LB (ampicillin added for transformants) and grown at 37°C with shaking to an optical density at 660 nm (OD660) of 0.6. Three 10 mL cultures of each strain for each sampling time were prepared for the assay. Cells were collected using centrifugation (2,300 × g, 10 min, room temperature) and resuspended in 10 mL of M9 medium (pH 5.7) supplemented with 100 mM MES and 100 mM KCl (EvgS-activation medium). This cell suspension in EvgS-activation medium was allocated in the following three ways. Aerobic condition: 500 μL of the cell suspension was placed into glass test tubes (φ13 mm × 100 mm) with aluminum caps and further shaken (180 rpm) at 37°C. Semi-aerobic condition: 500 μL of the cell suspension was placed into the same glass test tubes with aluminum caps as for aerobic condition, and stood at 37°C. Anaerobic condition: screw-cap tubes were filled up to the rim with the cell suspension (approximately 9.5 mL) and stood at 37°C. At appropriate sampling times, the cultures were subjected to β-galactosidase assays (performed in duplicate), the results for which were expressed in Miller units (Miller, 1972). The data shown are mean and standard deviation of the results from at least three biologically separate cultures. Statistical analyses were performed by Dunnett’s multiple comparisons test with Prism software (version 7.02) (GraphPad, La Jolla, United States), using the time 0 sample as the control.
One milliliter of the cell culture used for the reporter assays was centrifuged (13,000 × g, 3 min, 4°C) and the obtained pellet was resuspended in 1 mL of saline. The OD600 of each suspension was measured and adjusted with saline to obtain equal cell density. Protein denaturation of 500 μL of the adjusted cell suspension was performed by adding 500 μL of 10% trichloroacetic acid (TCA), vortexed, and placed on ice for 20 min. The denatured proteins were precipitated using centrifugation (17,800 × g, 15 min, 4°C). The obtained pellets were washed with 500 μL of acetone followed by centrifugation (17,800 × g, 15 min, 4°C) in order to remove the residual TCA. The pellets were dissolved in 100 μL of 1 × sample buffer for SDS-PAGE, and heated for 5 min at 95°C or for 30 min at 37°C for membrane proteins. Twenty microliters of each sample was subjected to SDS-PAGE and the electrophoresed proteins were transferred to a polyvinylidene difluoride membrane (Immun-Blot® PVDF, Bio-Rad, Hercules, United States). EvgA (His-tagged at the C-terminal end) was probed with an anti-6X His tag antibody (Abcam, Cambridge, United Kingdom), while EvgS and its variants were probed with anti-EvgS antiserum (Eguchi and Utsumi, 2014). Detection was carried out using goat anti-rabbit horseradish peroxidase-linked IgG (Abcam) and ImmobilonTM Western Chemiluminescent HRP Substrate (Merck Millipore, Burlington, United States). Signals were acquired using a MultiImager II Multibox (BioTools, Maebashi, Japan).
The EvgS/EvgA system is activated under mildly acidic conditions, in the presence of alkali metals (Eguchi and Utsumi, 2014). Since the EvgS sensor has a cytoplasmic PAS domain adjacent to the membrane, we examined whether respiratory growth conditions affect EvgS activity in vivo. Three conditions were compared in this study: aerobic (shaking at 180 rpm), semi-aerobic (standing), and anaerobic (standing screw-capped tube filled with cell culture). The EvgS/EvgA reporter strain, MG1655lacZ ydeP-lacZ, with a lacZ insertion immediately downstream of the ydeP gene and deletion of chromosomal lacZ, was grown to the exponential phase in an EvgS-inactivating medium (LB), followed by exchange to an EvgS-activation medium (M9 + MES, pH 5.7), and further incubated at 37°C under aerobic, semi-aerobic, and anaerobic conditions. For each sample, β-galactosidase activity was measured to examine how EvgS/EvgA responded to the different respiratory conditions. As shown in Figure 1A, EvgS/EvgA was activated after 1 and 3 h of aerobic and semi-aerobic growth, with a lower level of activation in the latter condition. No EvgS/EvgA activation was observed under the anaerobic condition. To confirm that the ydeP-lacZ reporter activity represented the EvgS/EvgA activity, and was not influenced by other factors that may respond to the anaerobic condition and shut down ydeP-lacZ expression, we performed the same experiment using the MG1655lacZ emrKY-lacZ reporter strain, which contained a lacZ insertion immediately downstream of the emrKY operon and deletion of chromosomal lacZ. The emrKY operon and ydeP are both directly regulated by the RR EvgA (Kato et al., 2000; Itou et al., 2009), and according to the EcoCyc database (Keseler et al., 2017), these two genes are not regulated by any other common transcriptional factors besides EvgA. As shown in Figure 1A, MG1655lacZ emrKY-lacZ also showed decreased activation under the semi-aerobic condition, and no activation under the anaerobic condition, clearly indicating that the EvgS/EvgA system responded to the availability of oxygen. We also examined EvgS expression in cells used in the reporter assays and found that EvgS levels were fairly constant among the different incubation conditions (Figure 1B, full gel results shown in Supplementary Figure 1). This is in line with our previous results, which suggested that the expression of the evgAS operon is not autoregulated by the EvgS/EvgA system (Eguchi et al., 2003).
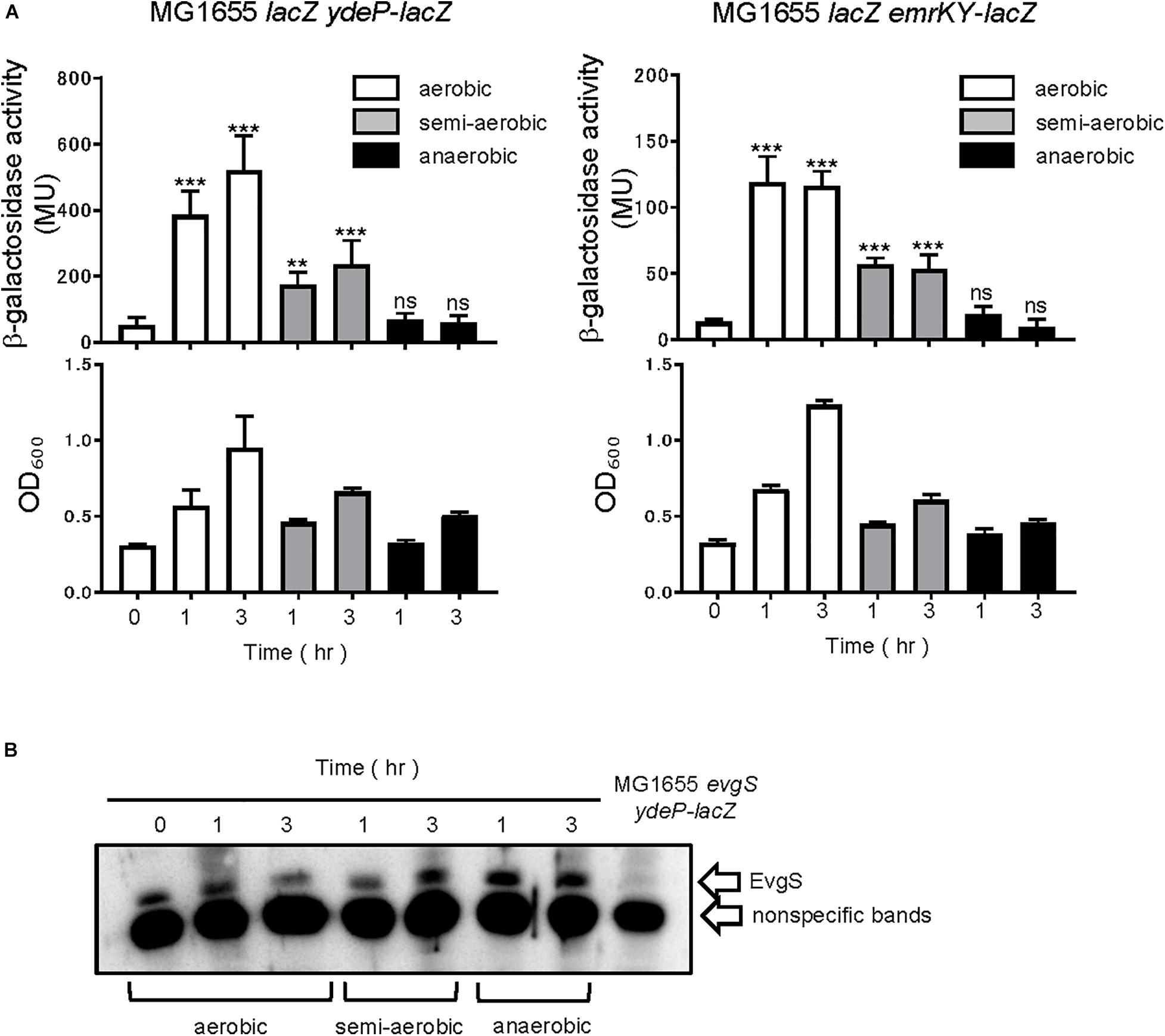
Figure 1. Aeration is required for EvgS/EvgA activation. (A) Activity of the ydeP-lacZ and emrKY-lacZ reporters in different culturing conditions (upper panels). Cells were grown in EvgS-activation medium under aerobic (white bars), semi-aerobic (gray bars), or anaerobic condition (black bars) at 37°C. Cell growth indicated by optical density at 600 nm is shown in the lower panels. Data represent the average of three biologically independent replicates. Error bars indicate the standard deviation, and statistical analyses of each redox condition group were performed using Dunnett’s multiple comparison test with the time 0 sample as the control. ns, not significant; **0.001 ≤ p < 0.01; ***p < 0.001. (B) EvgS expression in MG1655 lacZ ydeP-lacZ. Immunoblotting analysis using anti-EvgS antiserum for EvgS detection is shown. Samples are from the same culture as those subjected to reporter assays. Lane at the right end shows the sample from an evgS deleted strain.
Next, we checked whether the activity estimated by our reporter assays was dependent on EvgS. Deleting evgS resulted in no activation in the EvgS-activation medium, while expressing EvgS using an EvgS-expressing plasmid (pBADevgS) rescued the activation (Figure 2A). Activation was also lost under the anaerobic condition. The small decrease in β-galactosidase activity under anaerobic condition in MG1655 evgS ydeP-lacZ/pBAD vector may be due to the repression of the chromosomal lacZ expression. We confirmed EvgS expression under all conditions (Figure 2B, full gel results shown in Supplementary Figure 2).
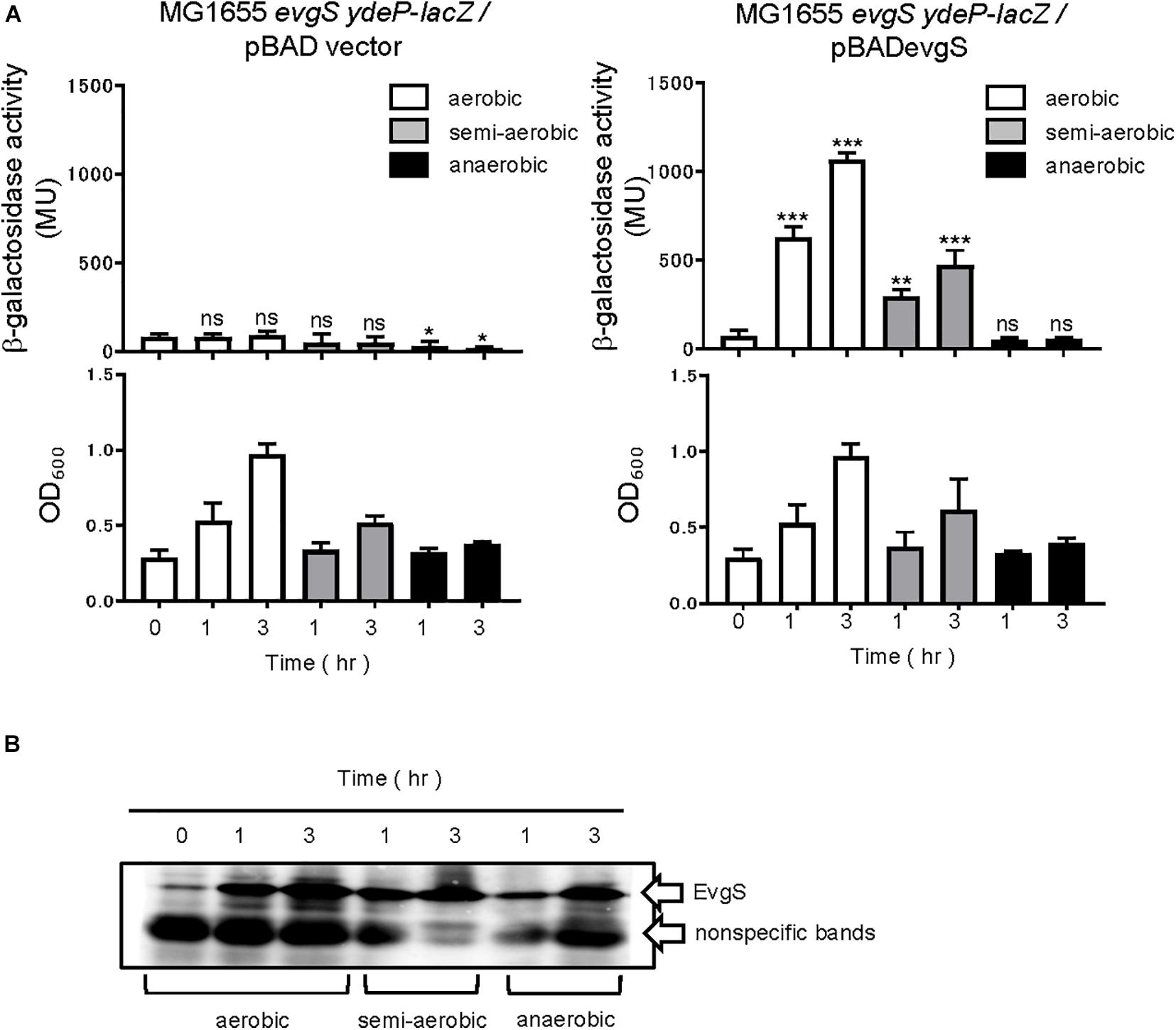
Figure 2. Response to aeration is EvgS-dependent. (A) Activity of the ydeP-lacZ reporter in different culturing conditions (upper panels). Cells were grown in EvgS-activation medium with 0.02% arabinose under aerobic (white bars), semi-aerobic (gray bars), or anaerobic condition (black bars) at 37°C. Cell growth indicated by optical density at 600 nm is shown in the lower panels. Data represent the average of three biologically independent replicates. Error bars indicate the standard deviation, and statistical analyses of each redox condition group were performed as described in Figure 1. ns, not significant; *0.01 ≤ p < 0.05; **0.001 ≤ p < 0.01; ***p < 0.001. (B) EvgS expression in MG1655 evgS ydeP-lacZ/pBADevgS. Immunoblotting analysis using anti-EvgS antiserum for EvgS detection is shown. Samples are from the same culture as those subjected to reporter assays.
Since the anaerobic condition retarded cell growth, it is possible that the decreased cell activity affected lacZ expression, and thus, lowered the β-galactosidase activity. To confirm this, we tested another E. coli reporter strain with a different two-component system, PhoQ/PhoP. This reporter strain, MG1601 (Kato et al., 1999), measures the promoter activity of mgtA, a component of the PhoP regulon. When this strain was first grown until the exponential phase in PhoQ-inactivating medium (LB + 20 mM MgSO4), followed by exchange to a PhoQ-activating medium (LB), the PhoQ/PhoP system showed activation, regardless of the difference in the respiratory conditions (aerobic, semi-aerobic, or anaerobic) (Supplementary Figure 3). Thus, lacZ was still expressed during reduced growth under the anaerobic condition, and EvgS/EvgA inactivation in this condition was not due to growth retardation.
Furthermore, we examined whether the change in respiratory conditions was sensed by the sensor EvgS or by the RR EvgA. Overexpression of RR is often accompanied by the activation of its TCS, which is also true with the overexpression of EvgA (Nishino and Yamaguchi, 2001). RRs may be phosphorylated by small-molecule phosphate donors such as acetyl phosphate. In some cases, increased expression of regulon components due to RR overexpression can take place even in the absence of RR phosphorylation (Bekker et al., 2010). We overexpressed EvgA (His-tagged at the C-terminal) from a pBADevgA plasmid in an evgS-deleted reporter strain, MG1655evgS ydeP-lacZ. Arabinose was added to the EvgS-activation medium at time 0 for EvgA induction, which was confirmed using immunoblotting for the His tag of EvgA. EvgS/EvgA activation was observed under aerobic and semi-aerobic conditions as well as under the anaerobic condition (Figure 3A). Activation of EvgS/EvgA under the anaerobic condition was slow and weak compared to its activation under the aerobic and semi-aerobic conditions. EvgA induction occurred slowly under the anaerobic condition, possibly due to retarded translational activity (Figure 3B, full gel results shown in Supplementary Figure 5) and corresponded to EvgS/EvgA activity. This shows that EvgS/EvgA activation by EvgA overproduction can be performed under anaerobic conditions, which is not what we observed in the reporter assays in Figure 1. Consequently, we claim that only the sensor EvgS, but not EvgA, responds to the change in the redox state, and that oxidation is required for EvgS activation.
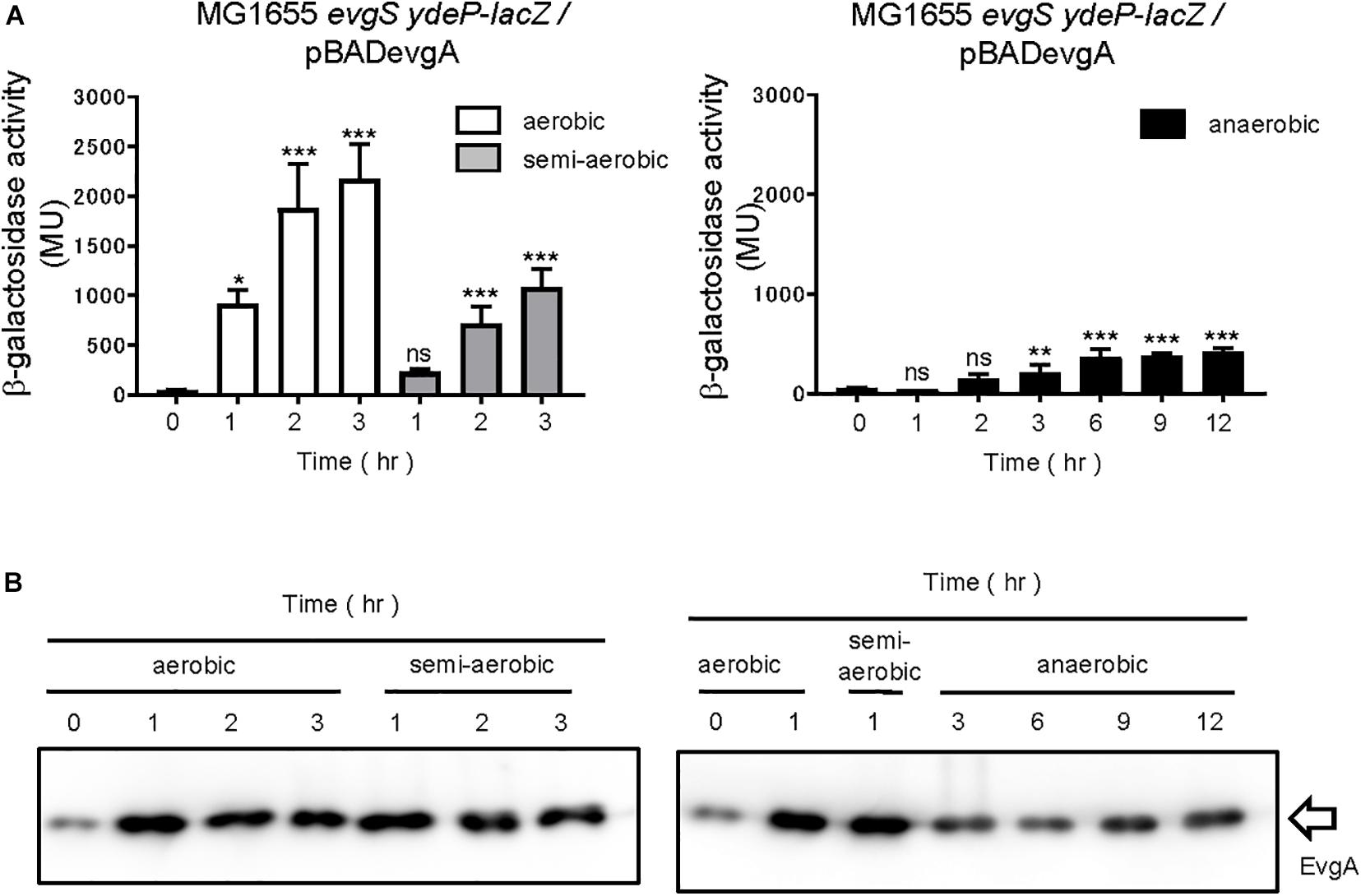
Figure 3. Aeration is not required for activating EvgS/EvgA by EvgA-overproduction. (A) Activity of the ydeP-lacZ reporter by inducing EvgA from pBADevgA. Cells were grown in EvgS-activation medium with 1% arabinose under aerobic (white bars), semi-aerobic (gray bars), or anaerobic condition (black bars) at 37°C. Optical density at 600 nm of the cell cultures subjected to reporter assays is shown in Supplementary Figure 4. Data represent the average of three biologically independent replicates. Error bars indicate the standard deviation, and statistical analyses of each redox condition group were performed as described in Figure 1. ns, not significant; *0.01 ≤ p < 0.05; **0.001 ≤ p < 0.01; ***p < 0.001. (B) EvgA expression in MG1655 evgS ydeP-lacZ/pBADevgA. Immunoblotting analysis using anti-His6 antibody for EvgA-His detection is shown. Samples are from the same culture as those subjected to reporter assays.
EvgS is an unorthodox HK sensor, similar to the anaerobic sensor ArcB. ArcB senses the redox state at two cysteines positioned at 180 and 241 within its cytoplasmic PAS domain (Malpica et al., 2004). We searched for cysteine residues in the PAS domain of EvgS and found three cysteines at the positions 663, 671, and 683 (Figure 4A). We created a homology model of the EvgS PAS domain monomer by the SWISS-MODEL homology-modeling server1 (Waterhouse et al., 2018) using the PAS domain of BvgS of Bordetella pertussis (PDB entry ID: 6ZJ8) as a template (Figure 4B). According to this model, C663 and C671 are in Hβ strand, and C683 in Iβ strand. Alanine mutations were made for each of these cysteines in pBADevgS to express EvgS C663A, EvgS C671A, and EvgS C683A. These EvgS mutants were expressed in an evgS-deleted reporter strain and assayed under conditions similar to those of wild-type EvgS (Figure 2). When compared to the wild-type, EvgS C663A showed similar EvgS/EvgA activity under all the respiratory conditions, with the anaerobic condition shutting down EvgS/EvgA activity (Figure 5A). However, EvgS C671A and EvgS C683A showed enhanced EvgS/EvgA activation, compared to the wild-type, under the aerobic and semi-aerobic conditions. Although mutations within the PAS domain frequently cause a locked-on state of EvgS (Kato et al., 2000; Johnson et al., 2014), C671A and C683A were inactivated during growth in LB (Figures 5B,C, time 0) and were only activated upon transfer to the EvgS-activation medium. Both the mutants showed EvgS/EvgA activation even under the anaerobic condition, indicating that the shutdown of EvgS under anaerobic conditions is alleviated by the C671A and C683A mutations. The double mutant of EvgS (C671A C683A) also showed enhanced activation compared to the wild-type, but not as high as the individual C671A and C683A mutants (Figure 5D). Protein expression of EvgS mutants was confirmed in all reporter strains (Supplementary Figure 7). These results strengthen our hypothesis that EvgS senses the redox state.
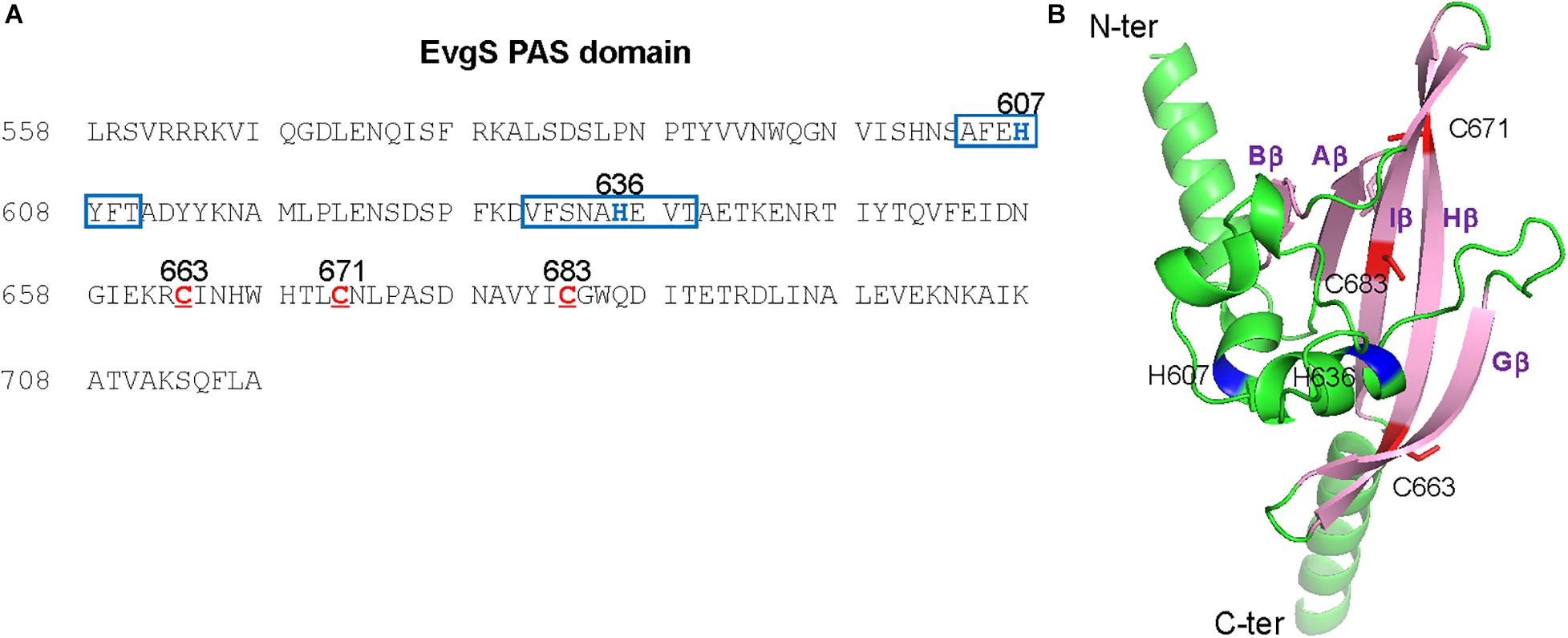
Figure 4. Positions of three cysteine residues and putative quinone-binding motifs in the EvgS PAS domain. (A) The sequence of the EvgS PAS domain is shown with three cysteines (C663, C671, and C683) depicted in red. In addition, the putative quinone-binding motifs are boxed in blue with central histidine residues (H607 and H636) depicted in blue. (B) A homology model of the EvgS PAS domain was created by the SWISS-MODEL homology-modeling server (https://swissmodel.expasy.org, Waterhouse et al., 2018) using the PAS domain of BvgS of Bordetella pertussis (PDB entry ID: 6ZJ8) as a template. The model is presented by a cartoon model with C663, C671 in Hβ strand, and C683 in Iβ strand depicted in red. Side chains of C663, C671, and C683 are shown with sticks. The central histidine residues, H607 and H636, located on two Q-helices, are depicted in blue.
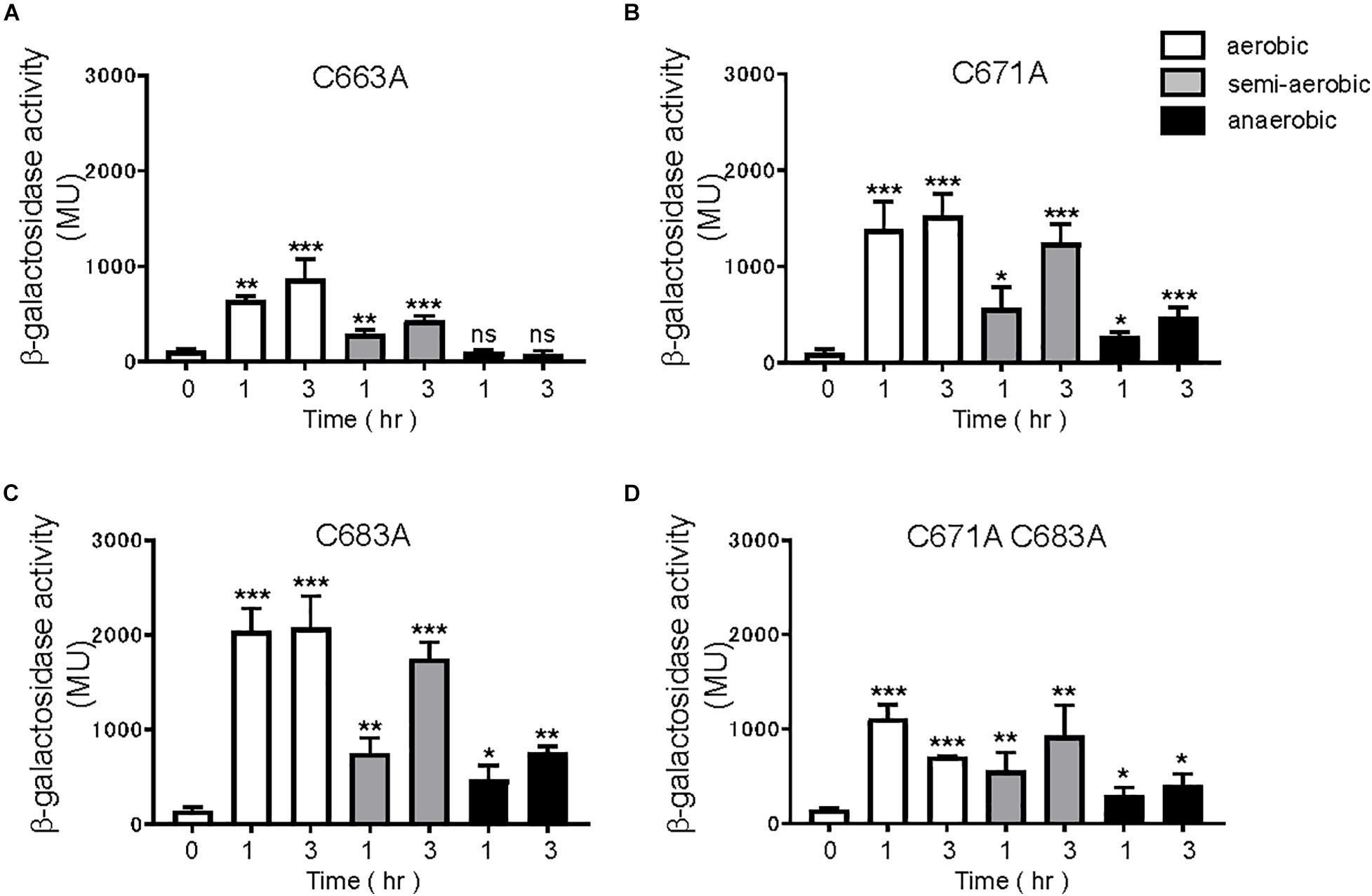
Figure 5. Response to aeration in EvgS-PAS mutants. (A) MG1655 evgS ydeP-lacZ/pBADevgS C663A, (B) MG1655 evgS ydeP-lacZ/pBADevgS C671A, (C) MG1655 evgS ydeP-lacZ/pBADevgS C683A, (D) MG1655 evgS ydeP-lacZ/pBADevgS C671A C683A. Activity of the ydeP-lacZ reporter in different culturing conditions (upper panels). Cells were grown in EvgS-activation medium with 0.02% arabinose under aerobic (white bars), semi-aerobic (gray bars), or anaerobic condition (black bars) at 37°C. Optical density at 600 nm of the cell cultures subjected to reporter assays is shown in Supplementary Figure 6. Data represent the average of three biologically independent replicates. Error bars indicate the standard deviation, and statistical analyses of each redox condition group were performed as described in Figure 1. ns, not significant; *0.01 ≤ p < 0.05; **0.001 ≤ p < 0.01; ***p < 0.001.
A canonical PAS fold comprises of a five-stranded antiparallel β-sheet and several α-helices flanking the sheet (Möglich et al., 2009). According to our homology model of the EvgS PAS domain in Figure 4B, C671 and C683 are positioned in the β-strands Hβ and Iβ, respectively. The side chain of C671 protrudes outside the molecule, while the side chain of C683 faces inside the molecule. Thus, it is assumed that C671 and C683 cannot form intraprotomer disulfide bonds. When EvgS C671S was expressed in MG1655 evgS ydeP-lacZ strain, EvgS activity was lost under all conditions (Figure 6A). This may mimic the breakage of a disulfide bond between C671. However, when EvgS C671M was expressed, EvgS activity was observed under aerobic and semi-aerobic conditions and not under anerobic condition (Figure 6C). The difference in protein expression of EvgS C671S and C671M may also explain the reduced activity in EvgS C671S (Supplementary Figure 9). We also expressed EvgS C683S and C683M in MG1655 evgS ydeP-lacZ strain. Both EvgS variants showed EvgS activity under aerobic and semi-aerobic conditions, and not under anaerobic condition (Figures 6B,D). At present, we cannot propose a mechanism of how C671 and C683 are involved in the redox control of EvgS activity only from our results. Further investigation such as in vivo cross-linking and structural studies of the EvgS PAS domain is necessary for clarification. C671 and C683 are conserved among EvgS PAS domains of different E. coli strains and Shigella species (Supplementary Figure 10). Cysteines were not found at this position in the BvgS-PAS domain.
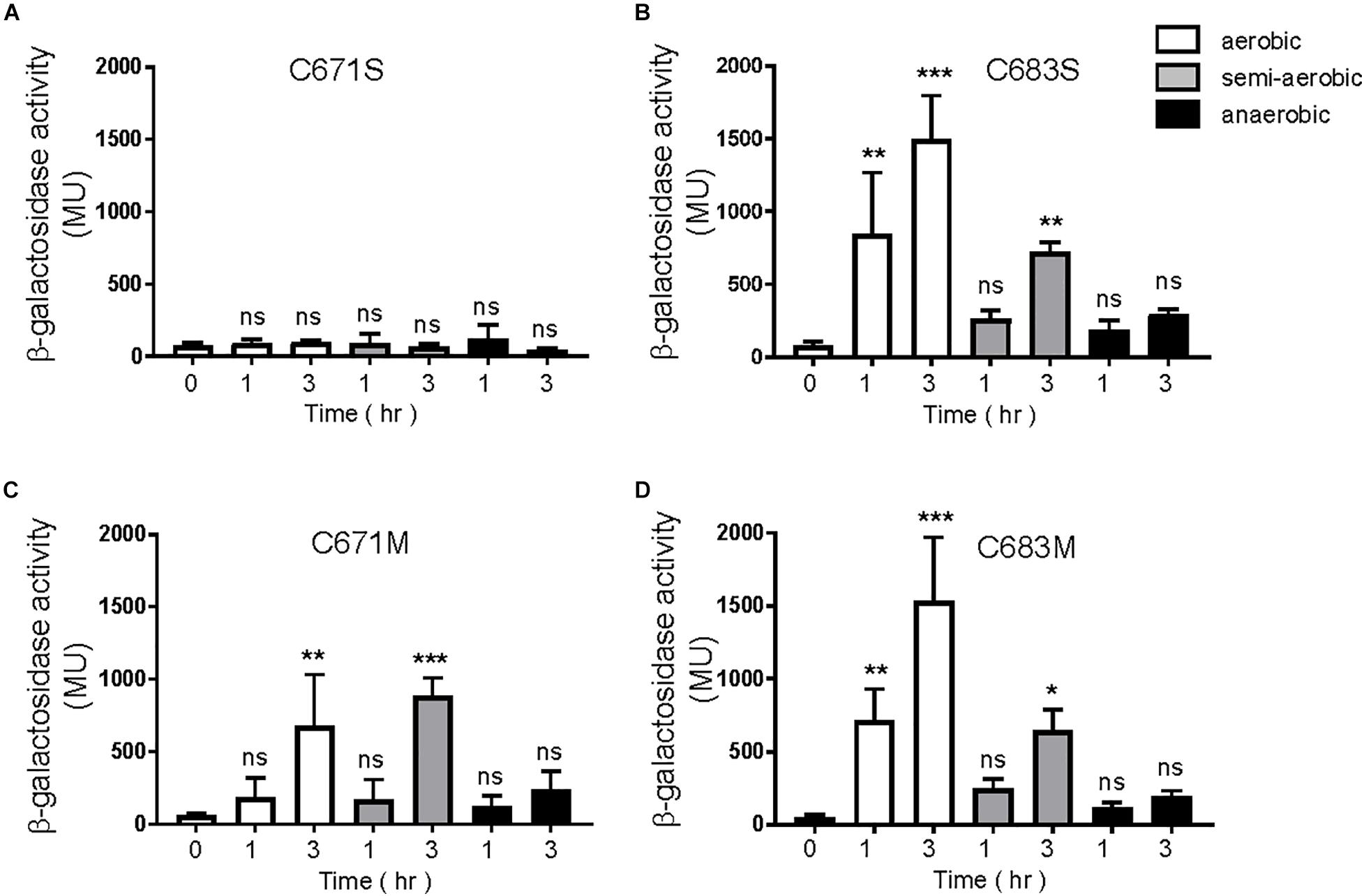
Figure 6. Response to aeration in EvgS-PAS mutants. (A) MG1655 evgS ydeP-lacZ/pBADevgS C671S; (B) MG1655 evgS ydeP-lacZ/pBADevgS C683S; (C) MG1655 evgS ydeP-lacZ/pBADevgS C671M; (D) MG1655 evgS ydeP-lacZ/pBADevgS C683M. Activity of the ydeP-lacZ reporter in different culturing conditions (upper panels). Cells were grown in EvgS-activation medium with 0.02% arabinose under aerobic (white bars), semi-aerobic (gray bars), or anaerobic condition (black bars) at 37°C. Optical density at 600 nm of the cell cultures subjected to reporter assays is shown in Supplementary Figure 8. Data represent the average of three biologically independent replicates. Error bars indicate the standard deviation, and statistical analyses of each redox condition group were performed as described in Figure 1. ns, not significant; *0.01 ≤ p < 0.05; **0.001 ≤ p < 0.01; ***p < 0.001.
Expressing the cytoplasmic region of EvgS also activates the EvgS/EvgA system without EvgS-activating signals (Sen et al., 2017). We examined whether the cytoplasmic region of EvgS retained its response to the redox state. The cytoplasmic region of EvgS was expressed from the pBADevgS-cyt plasmid in an evgS-deleted reporter strain and assayed under conditions similar to those of wild-type EvgS (Figure 2). Expression of the cytoplasmic region of EvgS under aerobic conditions resulted in activation of the EvgS/EvgA system (Figure 7), as previously reported (Sen et al., 2017). This activation was also observed in the semi-aerobic and anaerobic cultures. We confirmed expression of the cytoplasmic region of EvgS under all conditions (Supplementary Figure 12). Although the cytoplasmic region of EvgS comprises the PAS domain, the results shown in Figure 7 indicate that the cytoplasmic region of EvgS did not respond to the redox state. Therefore, it is necessary for EvgS to be anchored to the membrane for responding to anaerobic conditions, suggesting that membrane association promotes the interaction between the cytoplasmic region of the sensor protein and the redox signal. In addition, the loss of the transmembrane domain, which maintains the proper distance between protomers of the linking cytoplasmic region, may prohibit the relevant conformational change required for EvgS inactivation.
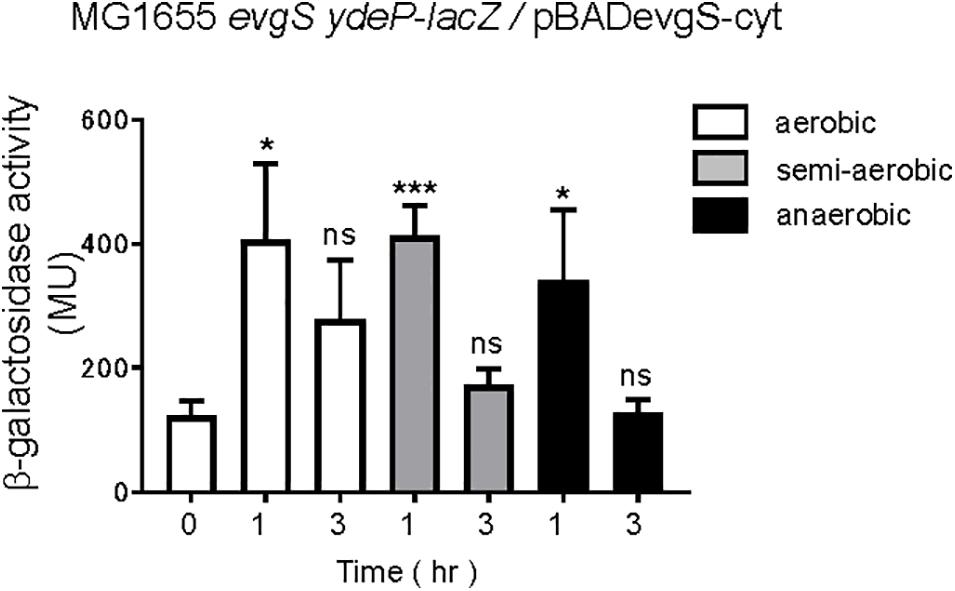
Figure 7. Aeration is not necessary for EvgS/EvgA activation in strains expressing the cytoplasmic region of EvgS (EvgS-Cyt). Activity of the ydeP-lacZ reporter in different culturing conditions (upper panel). Cells were grown in EvgS-activation medium with 0.02% arabinose under aerobic (white bars), semi-aerobic (gray bars), or anaerobic condition (black bars) at 37°C. Optical density at 600 nm of the cell cultures subjected to reporter assays is shown in Supplementary Figure 11. Data represent the average of three biologically independent replicates. Error bars indicate the standard deviation, and statistical analyses of each redox condition group were performed as described in Figure 1. ns, not significant; *0.01 ≤ p < 0.05; ***p < 0.001.
The anaerobic sensor, ArcB, utilizes oxidized and reduced forms of UQ, DMK, and MK to control ArcB activity (Georgellis et al., 2001; Bekker et al., 2010; Alvarez et al., 2013; Sharma et al., 2013; van Beilen and Hellingwerf, 2016). Since membrane localization of EvgS was required for sensing the redox condition, we examined whether these three dominant quinones in E. coli control EvgS activity.
To construct a ydeP-lacZ reporter strain without DMK and MK, menA, which encodes 1,4-dihydroxy-2-naphthoate octaprenyltransferase, was deleted from the reporter strain. Deletion of this gene blocks DMK and MK biosynthesis (Figure 8A). When MG1655 lacZ menA ydeP-lacZ was assayed, EvgS was found to be activated under the aerobic and semi-aerobic conditions, but not under the anaerobic condition, as seen in case of wild-type MG1655 lacZ ydeP-lacZ (Figures 8B,C). This indicates that both DMK and MK are not essential for EvgS to respond to the redox state. However, the small decrease in EvgS activity under aerobic condition in MG1655 lacZ menA ydeP-lacZ suggests involvement of these two quinones for EvgS activation under aerobic condition.
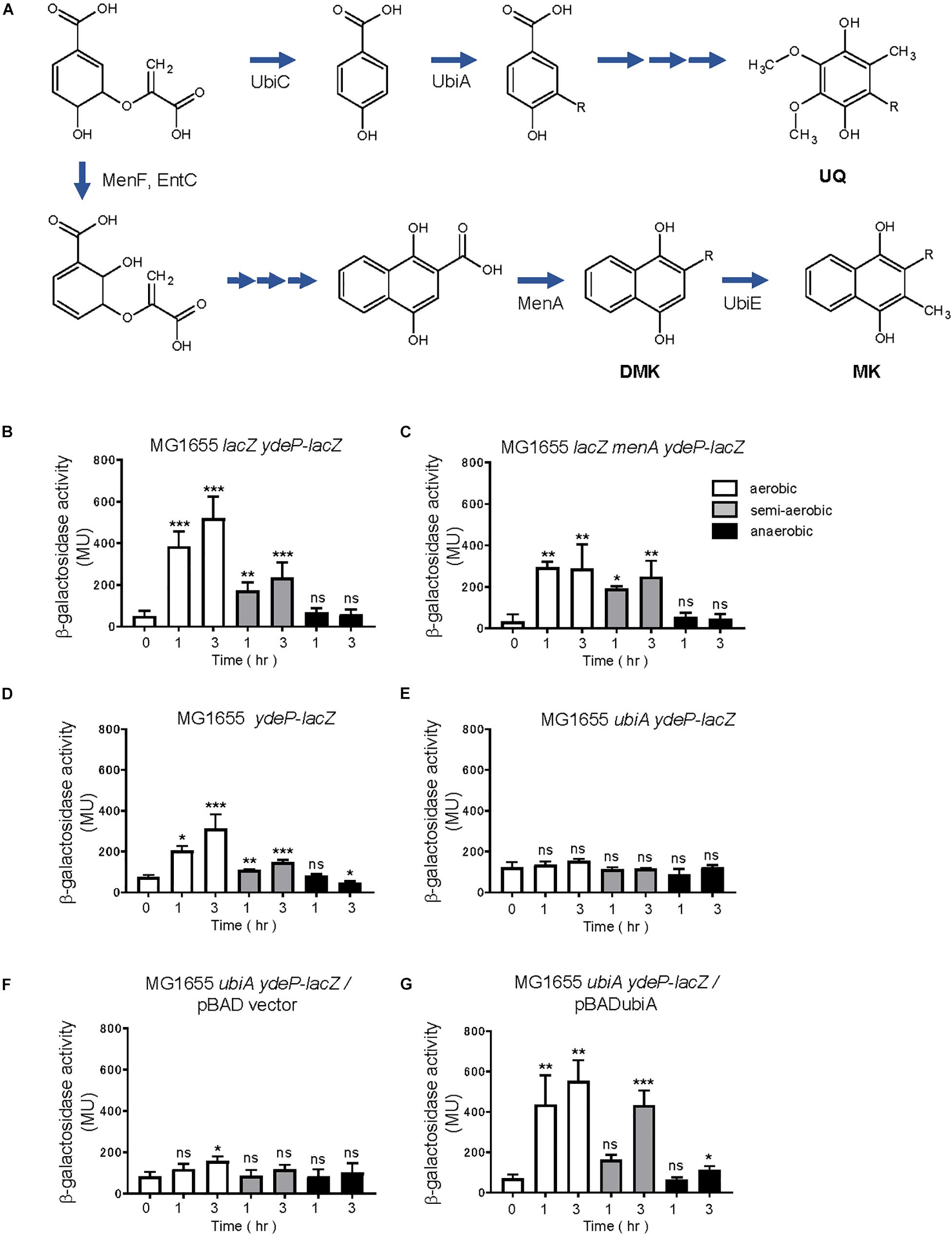
Figure 8. Ubiquinone is required for EvgS/EvgA activation. (A) Biosynthesis of ubiquinone (UQ), demethylmenaquinone (DMK), and menaquinone (MK) in E. coli (arranged from reference, van Beilen and Hellingwerf, 2016). (B–G) Activity of the ydeP-lacZ reporter in different culturing conditions (upper panels). Cells were grown in EvgS-activation medium with 0.02% arabinose under aerobic (white bars), semi-aerobic (gray bars), or anaerobic condition (black bars) at 37°C. Uracil was added to the medium at a concentration of 1 mM for experiments (D–G). Optical density at 600 nm of the cell cultures subjected to reporter assays is shown in Supplementary Figure 13. Data represent the average of three biologically independent replicates. Error bars indicate the standard deviation, and statistical analyses of each redox condition group were performed as described in Figure 1. ns, not significant; *0.01 ≤ p < 0.05; **0.001 ≤ p < 0.01; ***p < 0.001. (B) MG1655 lacZ ydeP-lacZ, (C) MG1655 lacZ menA ydeP-lacZ (no DMK/MK), (D) MG1655 ydeP-lacZ, (E) MG1655 ubiA ydeP-lacZ (no UQ), (F) MG1655 ubiA ydeP-lacZ/pBAD vector (no UQ), (G) MG1655 ubiA ydeP-lacZ/pBADubiA (with UQ).
For construction of a reporter strain without UQ, ubiA, which encodes 4-hydroxybenzoate octaprenyltransferase, was deleted from the reporter strain. Deletion of this gene hinders UQ biosynthesis (Figure 8A), resulting in retarded cell growth. Uracil, at a concentration of 1 mM, was added to the medium to support cell growth for the assay (personal communication). In contrast to the menA-deleted strain, deletion of ubiA resulted in no activation of EvgS (Figures 8D,E) under any of the redox conditions. Expression of ubiA from a UbiA-expressing plasmid, pBADubiA, complemented the ubiA deletion (Figures 8F,G), suggesting that UQ is required for the oxidative activation of EvgS.
Finally, we expressed EvgS, EvgS C671A, EvgS C683A, and EvgS C671A C683A in the ubiA-deleted reporter strain, MG1655 ubiA ydeP-lacZ (the host strain retains its evgS gene). To our surprise, additional expression of the wild type EvgS showed EvgS activity under aerobic and semi-aerobic state, but not under anaerobic state, as seen in the evgS complemented strain in Figure 2A. This indicates that when EvgS is overexpressed, EvgS activation can be controlled without UQ, presumably by DMK and MK. Oxidized UQ may have higher affinity against EvgS than oxidized DMK and MK. When EvgS C671A, EvgS C683A, and EvgS C671A C683A were each expressed in MG1655 ubiA ydeP-lacZ, enhanced level of EvgS activation was also observed under all redox condition. When this result was compared with that in the presence of UQ (Figure 5), higher EvgS activity was found under anaerobic conditions. This suggests that reduced form of UQ may be required for the repression of the activity of EvgS variants. Protein expression of EvgS and EvgS variants was confirmed in all reporter strains. Expression of EvgS and EvgS C671A was weaker than EvgS C683A and EvgS C671A C683A from unknown reasons (Supplementary Figure 15).
In the present in vivo study using an EvgS/EvgA reporter strain, we have shown that activation of EvgS requires a ubiquinone-dependent oxidative condition, in addition to mildly acidic pH. Our interpretation is that the PAS domain serves as an “intermediate redox switch” residing between the periplasmic sensor domain and the cytoplasmic catalytic core. This switch is turned on under aerobic conditions via oxidized UQ and gets switched off under anaerobic conditions via reduced form of UQ. The PAS domain integrates two signals: mildly acidic pH and oxidative conditions. The two naphtoquinones, DMK and MK, are presumed to also activate EvgS, but only when EvgS is overexpressed (Figures 8E, 9A). This suggests that oxidized UQ have higher affinity against EvgS than oxidized DMK and MK. On the other hand, reduced DMK and MK are also presumed to inactivate EvgS (Figure 9A), but not the EvgS variants (Figures 9B–D), suggesting that the reduced form of UQ also have higher affinity against EvgS than DMK and MK.
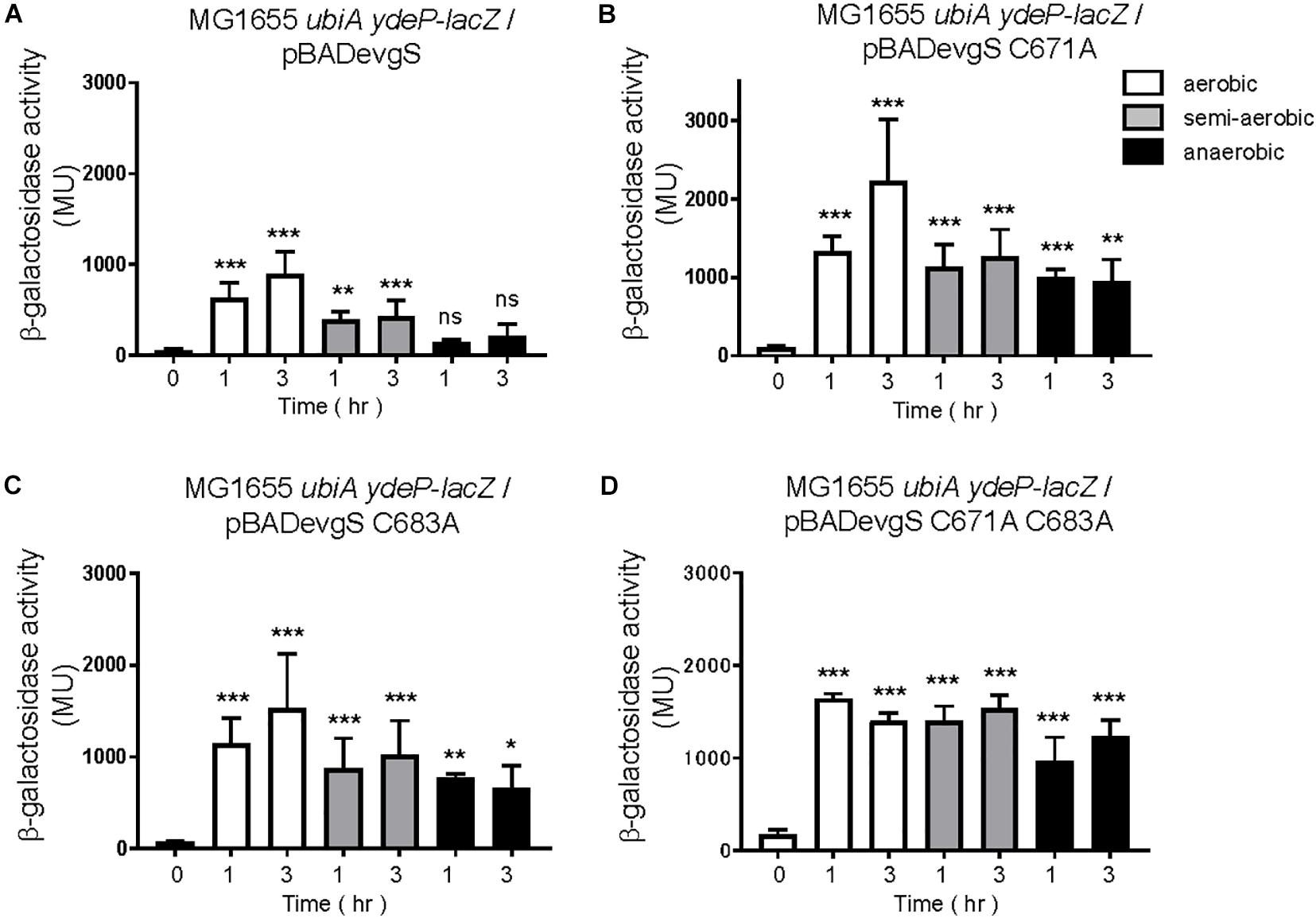
Figure 9. Ubiquinone is not essential for EvgS/EvgA activation in strains overexpressing EvgS and EvgS-PAS mutants. (A–D) Activity of the ydeP-lacZ reporter in different culturing conditions (upper panel). Cells were grown in EvgS activation-medium with 0.02% arabinose and 1 mM uracil under aerobic (white bars), semi-aerobic (gray bars), or anaerobic condition (black bars) at 37°C. Optical density at 600 nm of the cell cultures subjected to reporter assays is shown in Supplementary Figure 14. Data represent the average of three biologically independent replicates. Error bars indicate the standard deviation, and statistical analyses of each redox condition group were performed as described in Figure 1. ns, not significant; *0.01 ≤ p < 0.05; **0.001 ≤ p < 0.01; ***p < 0.001. (A) MG1655 ubiA ydeP-lacZ/pBADevgS, (B) MG1655 ubiA ydeP-lacZ/pBADevgS C671A, (C) MG1655 ubiA ydeP-lacZ/pBADevgS C683A, (D) MG1655 ubiA ydeP-lacZ/pBADevgS C671A C683A.
Previous in vitro studies on EvgS and BvgS (Bock and Gross, 2002) have shown that the purified soluble forms of the cytoplasmic regions of EvgS and BvgS are inhibited by oxidized UQ-0, with half-maximal inhibition occurring at 4 μM (EvgS) and 11 μM (BvgS). Since these results suggested direct binding of UQ to EvgS, we searched for quinone-binding motifs [aliphatic-(X)3-H-(X)2/3-(L/T/S)] (Fisher and Rich, 2000) within the EvgS PAS domain. As shown in Figure 4A, two putative quinone-binding sites were found, with the central histidines located at 607 and 636, although both sites had mismatches in the number of X residues after the aliphatic residue. Most of the ligands of PAS domains bind to the spatially conserved cleft formed by the inner surface of the β-sheet and the helices Eα and Fα (Möglich et al., 2009). Interestingly, His636 is in helix Fα, suggesting that the latter is a stronger candidate for UQ binding. Alignment of EvgS PAS domains of different E. coli strains and Shigella species revealed that they are almost identical, and the two putative quinone-binding sites are conserved (Supplementary Figure 10). As for the PAS domain of BvgS, only one putative quinone-binding site in helix Fα with a central His643 has been found (Bock and Gross, 2002). Whether UQ binds to these putative sites in the EvgS PAS domain will be investigated in our future studies.
The EvgS PAS domain has been considered essential for signal transmission, since many different mutations in the PAS domain led to constitutive activation of the protein (Kato et al., 2000; Johnson et al., 2014). The suggested model was that the non-active state of EvgS is a tight inactive dimer, which upon signal perception, changes to a weak active dimer. The PAS domain mutants are thought to weaken the EvgS dimer and cause EvgS activation (Johnson et al., 2014). The third EvgS state is explained as a weaker inactive dimer. This state has been observed in cytoplasmic EvgS mutants, which lack the periplasmic and transmembrane domains and have mutations in the PAS domain (Sen et al., 2017). We have also confirmed that the cytoplasmic EvgS is weakly active and is no longer under redox control (Figure 7). Sen et al. found that adding mutations, which constitutively activate the full-length EvgS, to the PAS domain of the cytoplasmic EvgS caused inactivation (Sen et al., 2017). We assume that this weaker inactive dimer explains the previous in vitro results of oxidized UQ inhibiting EvgS (Bock and Gross, 2002). The cytoplasmic EvgS, without the periplasmic and transmembrane domains, forms a weak active dimer. Moreover, the cytoplasmic EvgS579–1197 used in their study had a truncated N-terminal helix in the PAS domain. The N-terminal helices form a coiled coil that interacts with the β-sheet of the other protomer; truncation of this N-terminal helix weakens the dimer. When oxidized UQ is added to EvgS579–1197, which is presumed to activate the full-length EvgS in the cell, may further weaken the dimer, resulting in the formation of a weaker inactive dimer.
The present model of EvgS is as follows. Under aerobic conditions, oxidized UQ activates EvgS. MK and DMK can also activate EvgS, but with a lower affinity than UQ. A change to anaerobic conditions inactivates EvgS by reduced UQ. C671 and C683 are presumed to be involved in the redox control. It is of interest that EvgS/EvgA induces the expression of a cytochrome bd-II ubiquinol oxidase (encoded by appCB) via the YdeO transcription factor (Yamanaka et al., 2014). Cytochrome bd is embedded in the prokaryotic cytoplasmic membrane, and produces H2O and oxidized UQ upon O2 oxidization using UQH2 (ubiquinol) as the electron donor (Borisov et al., 2011). The presence of this enzyme and O2 increases the ratio of oxidized UQ to UQH2. Activation of EvgS/EvgA induces the expression of cytochrome bd-II ubiquinol oxidase, and may contribute to the supply of oxidized UQ to maintain the ON state of the EvgS redox switch. Furthermore, Sommer et al. (2013) have reported that EvgS tended to form clusters at the membrane, and that the cytoplasmic PAS domain was required for the clustering.
Recently, another signal has been found for EvgS. Indole, a metabolic product of E. coli and other gut bacteria, acts as an inhibitor of EvgS at the micromolar level (Boon et al., 2020). Directly or indirectly, indole affects EvgS activity, and by doing so inhibits induction of the severe acid resistance systems. It has been suggested that indole could enable E. coli to regulate gene expression, which fits its location in the gut (Boon et al., 2020). The EvgS PAS domain, acting as an intermediate redox switch, can also adjust E. coli to regulate gene expression in accordance with the redox state. Thus, together with the presence of indole, EvgS activity should be repressed in the anaerobic intestine. This is supported by a previous report that showed EvgSA not to have an important role to play under anaerobic conditions (Deng et al., 2013; De Biase and Lund, 2015).
Then, where does EvgS/EvgA function in the environment? There is one report of EvgS/EvgA activation in Shigella flexneri (Pasqua et al., 2019). Inside macrophages, S. flexneri highly induces the EmrKY efflux pump, which is dependent on EvgS/EvgA. This implies that EvgS is activated in macrophages. Similar to E. coli, EvgS in S. flexneri is also activated under mildly acidic conditions, which meets the intracellular pH conditions of macrophages. Since the EvgS PAS domain is almost identical between E. coli and S. flexneri (Supplementary Figure 10), we presume that some kind of oxidative stress that S. flexneri faces inside the macrophage turns on the PAS domain. Moreover, a recent report has shown that EvgS/EvgA activation protected E. coli from killing by gallium nitrate. This was possibly due to the upregulation of genes encoding enzymes involved in ROS detoxification and in the glyoxylate shunt of the TCA cycle (Zeng et al., 2021). This may also occur in S. flexneri and help their survival inside the macrophage. How oxidation of the EvgS PAS domain occurs inside the macrophages is another question that remains to be solved.
Publicly available datasets were analyzed in this study. This data can be found here: [https://www.biocyc.org/].
SI and YE performed the experiments. TO analyzed the structural model of the PAS domain. RU and YE conceived of and supervised the work and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the JSPS KAKENHI Grant Nos. JP16K07681 and JP 20K05796 to YE.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Dr. Peter A. Lund (University of Birmingham, United Kingdom) for critical reading of the manuscript. We thank Prof. Makoto Kawamukai (Shimane University, Japan) for provision of ubiA:cat strain, and to the National BioResource Project (NIG, Japan): E. coli for provision of menA:kan strain. We would also like to thank Editage (www.editage.com) for English language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.652546/full#supplementary-material
Alvarez, A. F., Rodriguez, C., and Georgellis, D. (2013). Ubiquinone and menaquinone electron carriers represent the yin and yang in the redox regulation of the ArcB sensor kinase. J. Bacteriol. 195, 3054–3061. doi: 10.1128/jb.00406-13
Arico, B., Miller, J. F., Roy, C., Stibitz, S., Monack, D., Falkow, S., et al. (1989). Sequences required for expression of Bordetella pertussis virulence factors share homology with prokaryotic signal. Proc. Natl. Acad. Sci. U. S. A. 86, 6671–6675. doi: 10.1073/pnas.86.17.6671
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008.
Bekker, M., Alexeeva, S., Laan, W., Sawers, G., Teixeira de Mattos, M. J., and Hellingwerf, K. J. (2010). The ArcBA two-component system of Escherichia coli is regulatd by the redox state of both the ubiquinone and the menaquinone pool. J. Bacteriol. 192, 746–754. doi: 10.1128/jb.01156-09
Blattner, F. R., Plunkett, G. III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., et al. (1997). The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462. doi: 10.1126/science.277.5331.1453
Bock, A., and Gross, R. (2002). The unorthodox histidine kinases BvgS and EvgS are responsive to the oxidation status of a quinone electron carrier. Eur. J. Biochem. 269, 3479–3484. doi: 10.1046/j.1432-1033.2002.03029.x
Boon, N., Kaur, M., Aziz, A., Bradnick, M., Shibayama, K., Eguchi, Y., et al. (2020). The signaling molecule indole inhibits induction of the AR2 acid resistance system in Escherichia coli. Front. Microbiol. 11:474. doi: 10.3389/fmicb.2020.00474
Borisov, V. B., Gennis, R. B., Hemp, J., and Verkhovsky, M. I. (2011). The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta 1807, 1398–1413. doi: 10.1016/j.bbabio.2011.06.016
Burton, N. A., Johnson, M. D., Antczak, P., Robinson, A., and Lund, P. (2010). Novel aspects of the acid response network of E. coli K-12 are revealed by a study of transcriptional dynamics. J. Mol. Biol. 401, 726–742. doi: 10.1016/j.jmb.2010.06.054
Cock, P. J., and Whitworth, D. E. (2007). Evolution of prokaryotic two-component system signaling pathways: gene fusions and fissions. Mol. Biol. Evol. 24, 2355–2357. doi: 10.1093/molbev/msm170
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97, 6640–6645. doi: 10.1073/pnas.120163297
De Biase, D., and Lund, P. A. (2015). The Escherichia coli acid stress response and its significance for pathogenesis. Adv. Appl. Microbiol. 92, 49–88. doi: 10.1016/bs.aambs.2015.03.002
Deng, Z., Shan, Y., Pan, Q., Gao, S., and Yan, A. (2013). Anaerobic expression of the gadE-mdtEF multidrug efflux operon is primarily regulated by the two-component system ArcBA through antagonizing the H-NS mediated repression. Front. Microbiol. 4:194. doi: 10.3389/fmicb.2013.00194
Dziva, F., Hauser, H., Connor, T. R., van Dieman, P. M., Prescott, G., Langridge, G. C., et al. (2013). Sequencing and functional annotation of avian pathogenic Escherichia coli serogroup O78 strains reveal the evolution of E. coli lineages pathogenic for poultry via distinct mechanisms. Infect. Immun. 81, 838–849. doi: 10.1128/iai.00585-12
Eguchi, Y., Ishii, E., Hata, K., and Utsumi, R. (2011). Regulation of acid resistance by connectors of two-component signal transduction systems in Escherichia coli. J. Bacteriol. 193, 1222–1228. doi: 10.1128/jb.01124-10
Eguchi, Y., Itou, J., Yamane, M., Demizu, R., Yamato, F., Okada, A., et al. (2007). B1500, a small membrane protein, connects the two-component systems EvgS/EvgA and PhoQ/PhoP in Escherichia coli. Proc. Natl. Acad Sci. U. S. A. 104, 18712–18717. doi: 10.1073/pnas.0705768104
Eguchi, Y., Oshima, T., Mori, H., Aono, R., Yamamoto, K., Ishihama, A., et al. (2003). Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology 149, 2819–2828. doi: 10.1099/mic.0.26460-0
Eguchi, Y., and Utsumi, R. (2014). Alkali metals in addition to acidic pH activate the EvgS histidine kinase sensor in Escherichia coli. J. Bacteriol. 196, 3140–3149. doi: 10.1128/jb.01742-14
Fisher, N., and Rich, P. R. (2000). A motif for quinone binding sites in respiratory and photosynthetic systems. J. Mol. Biol. 296, 1153–1162. doi: 10.1006/jmbi.2000.3509
Georgellis, D., Kwon, O., and Lin, E. C. C. (2001). Quinones as the redox signal for the Arc two-component system of bacteria. Science 292, 2314–2316. doi: 10.1126/science.1059361
Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995
Han, X., Dorsey-Oresto, A., Malik, M., Wang, J. Y., Drlica, K., Zhao, X., et al. (2010). Escherichia coli genes that reduce the lethal effects of stress. BMC Microbiol. 10:35. doi: 10.1186/1471-2180-10-35
Itou, J., Eguchi, Y., and Utsumi, R. (2009). Molecular mechanism of transcriptional cascade initiated by the EvgS/EvgA system in Escherichia coli K-12. Biosci. Biotechnol. Biochem. 73, 870–878. doi: 10.1271/bbb.80795
Johnson, M. D., Bell, J., Clarke, K., Chandler, R., Pathak, P., Xia, Y., et al. (2014). Characterization of mutations in the PAS domain of the EvgS sensor kinase selected by laboratory evolution for acid resistance in Escherichia coli. Mol. Microbiol. 93, 911–927. doi: 10.1111/mmi.12704
Kato, A., Ohnishi, H., Yamamoto, K., Furuta, E., Tanabe, H., and Utsumi, R. (2000). Transcription of emrKY is regulated by the EvgA-EvgS two-component system in Escherichia coli K-12. Biosci. Biotechnol. Biochem. 64, 1203–1209. doi: 10.1271/bbb.64.1203
Kato, A., Tanabe, H., and Utsumi, R. (1999). Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J. Bacteriol. 181, 5516–5520. doi: 10.1128/jb.181.17.5516-5520.1999
Keseler, I. M., Mackie, A., Santos-Zavaleta, A., Billington, R., Bonavides-Martinez, C., Caspi, R., et al. (2017). The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 45, D543–D550.
Kinoshita-Kikuta, E., Kinoshita, E., Eguchi, Y., and Koike, T. (2016). Validation of cis and trans modes in multistep phosphotransfer signaling of bacterial tripartite sensor kinases by using Phos-Tag SDS-PAGE. PLoS One 11:e0148294. doi: 10.1371/journal.pone.0148294.g001
Lai, Y. C., Lin, G. T., Yang, S. L., Chang, H. Y., and Peng, H. L. (2003). Identification and characterization of KvgAS, a two-component system in Klebsiella pneumoniae CG43. FEMS Microbiol. Lett. 218, 121–126. doi: 10.1111/j.1574-6968.2003.tb11507.x
Lin, C. T., Huang, T. Y., Liang, W. C., and Peng, H. L. (2006). Homologous response regulators KvgA, KvhA and KvhR regulate the synthesis of capsular polysaccharide in Klebsiella pneumonia CG43 in a coordinated manner. J. Biochem. 140, 429–438. doi: 10.1093/jb/mvj168
Ma, Z., Masuda, N., and Foster, J. W. (2004). Characterization of EvgAS-YdeO-GadE branched regulatory circuit governing glutamate-dependent acid resistance in Escherichia coli. J. Bacteriol. 186, 7378–7389. doi: 10.1128/jb.186.21.7378-7389.2004
Malpica, R., Franco, B., Rodriguez, C., Kwon, O., and Georgellis, D. (2004). Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. U. S. A. 101, 13318–13323. doi: 10.1073/pnas.0403064101
Masuda, N., and Church, G. (2003). Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48, 699–712. doi: 10.1046/j.1365-2958.2003.03477.x
Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Möglich, A., Ayers, R. A., and Moffat, J. (2009). Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17, 1282–1294. doi: 10.1016/j.str.2009.08.011
Nadler, C., Shifrin, Y., Nov, S., Kobi, S., and Rosenshine, I. (2006). Characterization of enteropathogenic Escherichia coli mutants that fail to disrupt host cell spreading and attachment to substratum. Infect. Immun. 74, 839–849. doi: 10.1128/iai.74.2.839-849.2006
Nishino, K., and Yamaguchi, A. (2001). Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 183, 1455–1458. doi: 10.1128/jb.183.4.1455-1458.2001
Pasqua, M., Grossi, M., Scinicariella, S., Aussel, L., Barras, F., Colonna, B., et al. (2019). The MFS efflux pump EmrKY contributes to the survival of Shigella within macrophages. Sci. Rep. 9:2906.
Perraud, A. I., Kimmel, B., Weiss, V., and Gross, R. (1998). Specificity of the BvgAS and EvgAS phosphorelay is mediated by the C-terminal HPt domains of the sensor proteins. Mol. Microbiol. 27, 875–887. doi: 10.1046/j.1365-2958.1998.00716.x
Roggiani, M., Yadavalli, S. S., and Goulian, M. (2017). Natural variation of a sensor kinase controlling a conserved stress response pathway in Escherichia coli. PLoS Genet. 13:e1007101. doi: 10.1371/journal.pgen.1007101
Sen, H., Aggarwal, N., Ishionwu, C., Hussain, N., Parmar, C., Jamshad, M., et al. (2017). Structural and functional analysis of the Escherichia coli Acid-sensing histidine kinase EvgS. J. Bacteriol. 199, e310–e317.
Sharma, P., Stagge, S., Bekker, M., Bettenbrock, K., and Hellingwerf, K. J. (2013). Kinase activity of ArcB from Escherichia coli is subject to regulation by both ubiquinone and demethylmenaquinone. PLoS One 8:e75412. doi: 10.1371/journal.pone.0075412
Sharma, P., Teixeira de Mattos, M. J., Hellingwerf, K. J., and Bekker, M. (2012). On the function of the various quinone species in Escherichia coli. FEBS J. 279, 3354–3373.
Sommer, E., Koler, M., Frank, V., Sourjik, V., and Vaknin, A. (2013). The sensory histidine kinases TorS and EvgS tend to form clusters in Escherichia coli cells. PLoS One 8:e77708. doi: 10.1371/journal.pone.0077708
Stock, A. M., Robinson, V. L., and Goudreau, R. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215.
Suzuki, K., Ueda, M., Yuasa, M., Nakagawa, T., Kawamukai, M., and Matsuda, H. (1994). Evidence that Escherichia coli ubiA product is a functional homolog of yeast COQ2, and the regulation of ubiA gene expression. Biosci. Biotech. Biochem. 58, 1814–1819. doi: 10.1271/bbb.58.1814
Uhl, M. A., and Miller, J. F. (1994). Autophosphorylation and phosphotransfer in the Bordetella pertussis BvgAS signal transduction cascade. Proc. Natl. Acad. Sci. U. S. A. 91, 1163–1167. doi: 10.1073/pnas.91.3.1163
Utsumi, R., Katayama, S., Taniguchi, M., Horie, T., Ikeda, M., Igaki, S., et al. (1994). Newly identified genes involved in the signal transduction of Escherichia coli K-12. Gene 140, 73–77. doi: 10.1016/0378-1119(94)90733-1
van Beilen, J. W. A., and Hellingwerf, K. J. (2016). All three endogenous quinone species of Escherichia coli are involved in controlling the activity of the aerobic/anaerobic response regulator ArcA. Front. Microbiol. 7:1339. doi: 10.3389/fmicb.2016.01339
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303.
Yamanaka, Y., Oshima, T., Ishihama, A., and Yamamoto, K. (2014). Characterization of the YdeO regulon in Escherichia coli. PLoS One 9:e111962. doi: 10.1371/journal.pone.0111962
Zeng, J., Wu, L., Liu, Z., Lv, Y., Feng, J., Wang, W., et al. (2021). Gain-of-function mutations in acid stress response (evgS) protect Escherichia coli from killing by gallium nitrate, an antimicrobial candidate. Antimicrob. Agents Chemother. 65, e1595–e1520.
Keywords: oxidation, redox, acid, ubiquinone, two-component system, Escherichia coli, EvgS, PAS domain
Citation: Inada S, Okajima T, Utsumi R and Eguchi Y (2021) Acid-Sensing Histidine Kinase With a Redox Switch. Front. Microbiol. 12:652546. doi: 10.3389/fmicb.2021.652546
Received: 12 January 2021; Accepted: 19 April 2021;
Published: 20 May 2021.
Edited by:
Daniela De Biase, Sapienza University of Rome, ItalyReviewed by:
Françoise Jacob-Dubuisson, Centre National de la Recherche Scientifique, Center for the National Scientific Research (CNRS), FranceCopyright © 2021 Inada, Okajima, Utsumi and Eguchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoko Eguchi, ZWd1Y2hpQHdha2Eua2luZGFpLmFjLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.