
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 26 April 2021
Sec. Microbiological Chemistry and Geomicrobiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.651103
 Martine A. R. Kox1,2
Martine A. R. Kox1,2 Alfons J. P. Smolders2,3
Alfons J. P. Smolders2,3 Daan R. Speth1†
Daan R. Speth1† Leon P. M. Lamers2
Leon P. M. Lamers2 Huub J. M. Op den Camp1
Huub J. M. Op den Camp1 Mike S. M. Jetten1
Mike S. M. Jetten1 Maartje A. H. J. van Kessel1*
Maartje A. H. J. van Kessel1*Degraded peatlands are often rewetted to prevent oxidation of the peat, which reduces CO2 emission. However, the created anoxic conditions will boost methane (CH4) production and thus emission. Here, we show that submerged Sphagnum peat mosses in rewetted-submerged peatlands can reduce CH4 emission from peatlands with 93%. We were able to mimic the field situation in the laboratory by using a novel mesocosm set-up. By combining these with 16S rRNA gene amplicon sequencing and qPCR analysis of the pmoA and mmoX genes, we showed that submerged Sphagnum mosses act as a niche for CH4 oxidizing bacteria. The tight association between Sphagnum peat mosses and methane oxidizing bacteria (MOB) significantly reduces CH4 emissions by peatlands and can be studied in more detail in the mesocosm setup developed in this study.
Globally, about 15% of peatland area has been drained for agriculture, forestry or bioenergy production, with highest losses in Europe (Joosten and Clarke, 2002; Grootjans et al., 2012). Drainage results in the exposure of the organic peat layer to oxygen, resulting in high CO2 emission (Waddington and Day, 2007; Abdalla et al., 2016; Reumer et al., 2018). As restoration measure, drained peatlands can be rewetted to protect organic matter from fast aerobic degradation (Grootjans et al., 2012; Renou-Wilson et al., 2019). However, the resulting anaerobic conditions create a suitable environment for the production of the potent greenhouse gas methane (CH4), leading to high methane emissions (Harpenslager et al., 2015; Abdalla et al., 2016; Renou-Wilson et al., 2019). The water table is a well-known factor in controlling CH4 cycling in wetlands (Bridgham et al., 2013; Ho et al., 2016); when the water table remains below the field surface, CH4 emissions typically remain low. However, when the water table rises, the oxygen concentration decreases which results in a strong increase in CH4 emission (Smolders et al., 2003; Harpenslager et al., 2015).
The CH4 emission of rewetted peatlands seems to be strongly reduced by the presence of (aquatic) Sphagnum mosses, which harbor CH4-oxidizing microorganisms that consume the produced CH4 (Smolders et al., 2003; Raghoebarsing et al., 2005; Waddington and Day, 2007; Kip et al., 2010). The association between these microorganisms and Sphagnum mosses was shown to be mutually beneficial (Raghoebarsing et al., 2005). By producing CO2, methanotrophs can relieve the CO2 limitation that Sphagnum mosses experience in submerged conditions (Smolders et al., 2001; Raghoebarsing et al., 2005; Kip et al., 2010). In return, MOB can benefit from O2 production and shelter provided by the Sphagnum moss (Kip et al., 2010). Different types of MOB are associated to Sphagnum mosses. Molecular surveys showed that Alphaproteobacterial methanotrophs typically dominate 16S rRNA gene-libraries from Sphagnum mosses (Kip et al., 2011; Kox et al., 2016, 2018). Within the Alphaproteobacteria especially methanotrophs of the family Methylocystaceae (Methylocystis spp.) and the acidophilic methanotrophs of the family Beijerinckiaceae (Methylocella, Methyloferula, and Methylocapsa) are often found and several of these have been isolated from peatlands (Dedysh et al., 2000, 2004; Dedysh, 2011; Vorobev et al., 2011; Danilova et al., 2013). In addition, it has been shown that also gammaproteobacterial methanotrophs are present and active in peatlands (Kip et al., 2010; Esson et al., 2016; Kaupper et al., 2021) with species belonging to Methylomonas, Methylobacter, Methylovulum, and Methylomicrobium being described as active members.
Peatland methane fluxes have been studied in the field, but it is challenging to investigate the association between methanotrophs with Sphagnum mosses in more detail. Therefore, investigating methanotroph-Sphagnum interactions and methane-flux dynamics under laboratory-controlled conditions can provide detailed insights into underlying mechanisms. The goal of our study was to establish the role of Sphagnum mosses and associated methanotrophs in mitigating CH4 fluxes from rewetted peatlands. An excellent case study for a rewetted peatland is the Mariapeel peatland in Netherlands, which has been drained since 1998. The peatland was rewetted for restoration purposes, which resulted in a decrease in CO2 emission, but a strong increase in CH4 emission (Smolders et al., 2003). We developed a new mesocosm set-up (Figure 1) in which methane-oxidation by submerged Sphagnum mosses can be studied in detail in a controlled laboratory setup, without the variability encountered in the field. We hypothesized that the submerged Sphagnum moss layer acts as a biofilter for CH4, thereby reducing CH4-emission to the atmosphere. Furthermore, it was expected that CH4-oxidizing microorganisms are associated with Sphagnum mosses, rather than with peat water. Monitoring of the CH4-flux throughout the mesocosm incubation, as well as CH4 batch-assays and molecular analysis of 16S rRNA and methane monooxygenases (pmoA and mmoX) genes, showed that methanotrophs were highly active and enriched in the newly developed mesocosm setup.
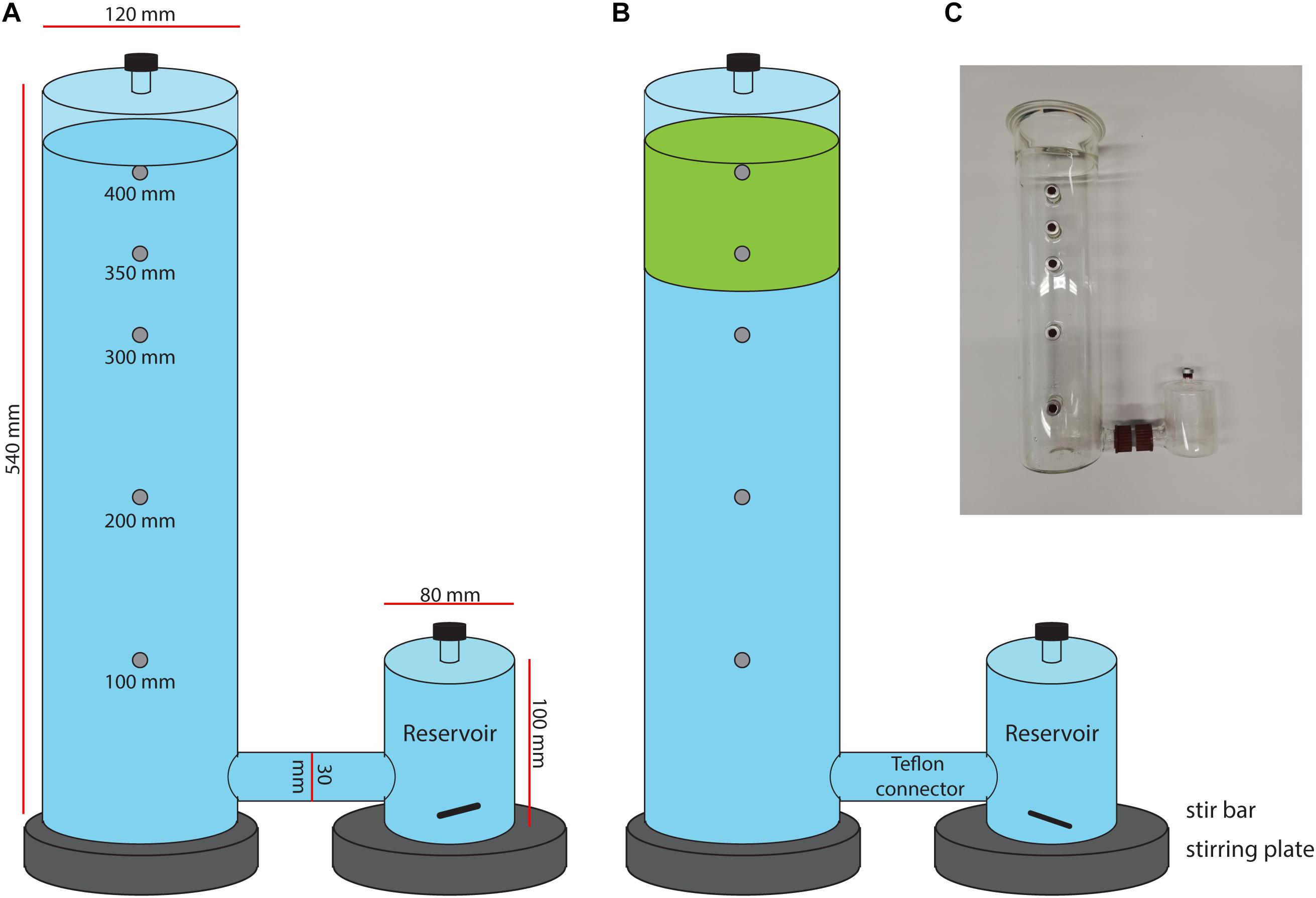
Figure 1. Schematic set up of mesocosm incubation with in (A) control mesocosms containing only filtered peat water (blue) and in (B) the moss mesocosms, containing Sphagnum moss layer (green) in filtered peat water. (C) Picture of the mesocosm setup.
The sampling site was located in the Mariapeel (51°24′28.4″N, 5°55′8″E), a peat bog nature conservation area in the southeast of the Netherlands. This site was visited for measurements and sampling in August 2017. Net diffusive gas fluxes were measured in the field using a fast greenhouse gas analyzer with cavity ringdown spectroscopy (GGA-24EP; Los Gatos Research, United States) connected to a Perspex chamber (15 cm in diameter). The chamber was put on top of the moss for 10 min to measure fluxes of CO2 and CH4. In total three independent measurements were taken within two meter distance from each other. After removal of the moss layer and an equilibration period of 15 min, measurements were repeated. Submerged Sphagnum cuspidatum moss and water was collected after the measurements. Upon arrival in the laboratory, all samples were stored at 4°C until the start of the incubations. The experiment was performed twice; for the first mesocosm experiment, mosses were stored 1 day. For the second mesocosm experiment, this was 35 days.
The mesocosm consists of a glass cylinder with a diameter of 12 cm and a height of 54 cm, with a separate reservoir that was connected with a Teflon connector (see Figure 1 and Supplementary Figure 1). The reservoir volume was 0.5 L, the connector tube volume 0.1 L and the column volume 5.5 L. The liquid level in the mesocosms was maintained at 5.1 L. The column headspace was closed using a greased lid with a sampling port. Several sampling ports (in the reservoir, cylinder headspace and in the cylinder at 10, 20, 30, 35, and 40 cm height) allow for sampling at different heights. Sampling ports were closed using red butyl rubber stoppers and aluminum crimp caps.
The mesocosm incubations were performed in duplicate for 32 days at room temperature. Two mesocosms were incubated simultaneously, one containing 100 Sphagnum cuspidatum plants (6 cm in length each, 120 g fresh weight in total) in filtered peat water and one contained only filtered peat water. Prior to incubation, Sphagnum mosses were carefully rinsed with tap water. Both mesocosms had an acclimatization period of 7 days prior to sampling. CH4 was added via the enclosed reservoir, which was stirred with a 2 cm magnetic stir bar at 250 rpm. Mesocosms were opened for 1 h per day to allow aeration. Directly after aeration, 20 ml of CH4 and 5 ml CO2 was injected in the reservoir. Light (16 h light, 8 h dark) was supplied on top of the mesocosm column using 120 deep red/white LEDs (Philips, Green-Power LED, Poland; 150 μmol m–2 s–1 photosynthetically active radiation at vegetation level). As additional control incubation, 100 plantlets of gnotobiotic Sphagnum fimbriatum (obtained from moss stock center Freiburg, Germany) were incubated.
CH4 concentrations were measured directly after closing (0 h) and just before opening (23 h) the mesocosms by collecting 0.5 ml gas or water samples which were injected into a closed 5.9 ml Exetainer vial (Labco Ltd., Lampeter, United Kingdom). The concentration of dissolved CH4 throughout the column was determined once a week, by sampling water at four different time points during the day (0 h, 3 h, 7 h, and 23 h after closing the headspace). The CH4 concentration in the Exetainers (Labco Ltd., Lampeter, United Kingdom) was measured at least 4 h after sampling by using a gas chromatograph with a flame-ionized detector and a Porapak Q column de Jong et al. (2018). Dissolved CH4 in the water was calculated based on the solubility of CH4 and was accounted for in the flux calculations as well. The daily CH4 flux in the mesocosm was calculated as the change in CH4 concentration in the headspace, divided by the surface area (0.01131 m2).
Prior to and after the mesocosm incubation, moss (3 g fresh weight) and peat water (12 ml, unfiltered) and filtered (0.2 μm) was incubated in 120 ml serum vials that were closed with red-butyl rubber stoppers and metal crimp-caps. Each serum vial received 2 ml CH4 (1.8%) and CH4 concentration was measured as described above. As a control for biological methane oxidation, 6 ml of acetylene was added after 10 h of incubation with CH4.
Water geochemistry was measured for both unfiltered and filtered (2–5 nm pore size, HF80S dialysis filter, Fresenius Medical Care, Homburg, Germany) peat water (see Supplementary Table 9). The pH was measured and elemental composition was determined using ICP-OES as described previously (Kox et al., 2018).
Five grams of moss (fresh weight) was taken from the mesocosm incubations and directly grinded using a pestle and mortar and liquid nitrogen, after which DNA was extracted using the DNeasy PowerSoil DNA extraction kit following manufacturers protocol (Qiagen Benelux B.V., Venlo, Netherlands). DNA quality was checked by gel-electrophoresis and using the Qbit dsDNA HS Assay Kit (Invitrogen, Thermo Fisher Scientific, Carlsbad, CA, United States).
Barcoded Amplicon-sequencing of the amplified V3–V4 region of the bacterial 16s rRNA gene [primers Bact-341f and Bact 785r (Klindworth et al., 2013)] was performed by BaseClear B.V. (Leiden, Netherlands) using Illumina Miseq. The obtained 326045 reads were quality filtered and analyzed using Mothur (v1.36.1), following the Illumina Standard Operating Procedure (SOP, accessed on May 8th 2018, Kozich et al., 2013). Merged reads shorter than 400 bp were discarded, chimeras were removed using the UCHIME algorithm (Edgar et al., 2011) and the remaining sequences were clustered at 97% identity. The resulting OTUs were classified based on the SILVA v132 16s rRNA gene non-redundant database (SSURef_99_v132_SILVA). Non-target sequences (Chloroplasts, Mitochondria, unknown, Archaea, and Eukaryota) were removed from the dataset (see Supplementary Tables 1, 2 for full overview of read processing). The output was analyzed with R [version 3.4.0 by the R Development Core Team (2017)] and Rstudio v1.1.456 (RStudio Team, 2016) using the packages Phyloseq (McMurdie and Holmes, 2013) and Vegan (Oksanen et al., 2016). Singletons were removed, and read libraries of all samples were rarefied by random subsampling (seed: 12345) to 6500 reads per sample (Rarefaction curves are depicted in Supplementary Figure 2). As a follow-up a PcoA plot (Supplementary Figure 4) was created using Phyloseq, based on a Bray-Curtis dissimilarity matrix on rarefied data. All sequencing data can be accessed via GenBank NCBI BioProject PRJNA517391.
Copy numbers of the Bacterial 16S rRNA gene and pmoA and mmoX genes were quantified by qPCR (see Supplementary Table 3 for primers). The qPCR reaction mix consisted of PerfeCTA Quanta master mix (Quanta Biosciences, Beverly, MA, United States) and 0.5 ng sample DNA and 1 μl of each primer (10 μM). qPCR reactions were performed in triplicate with a C1000 Touch thermal cycler equipped with a CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad Laboratories B.V., Veenendaal, Netherlands). Triplicate measurements per sample were averaged prior to statistical analysis. Standard curves were obtained via 10-fold dilution series of a PGEM T-easy plasmid (Promega, Madison, WI, United States) containing the target gene. Data was analyzed using Bio-Rad CFX Manager version 3.0 (Bio-Rad Laboratories B.V., Veenendaal, Netherlands).
Statistics were performed by using R version 3.4.0 by the R Development Core Team (2017). In order to allow for parametrical statistical tests, Shapiro–Wilk’s test was used on the residual (stats-package) to test the normality of the data and Levene’s test (car-package) was used to test for homogeneity of variance. If assumptions of tests were not met, data was log-transformed (ln), which was the case for the field CH4 flux data. A paired T-test was used to test whether the net CH4 flux in the field was affected by the presence of moss (moss field/moss removed). Differences in the potential CH4 oxidation activity in peat water and mosses prior to mesocosm incubation were tested using a non-parametric Kruskal Wallis tests. Within each material (moss/peat water) the effect of treatment (field/washing or filtering) was tested using an independent T-test. Differences in the potential CH4 oxidation activity after mesocosm incubation were tested using a 3-way ANOVA, followed by a Tukey HSD post hoc test. Differences in copy number between each moss sample for each target gene were analyzed using a one-way ANOVA, followed by a Tukey HSD post hoc test. Here, the data for 16S rRNA gene and mmoX gene were log-transformed (ln) prior to analysis.
To estimate diffusive CH4 emissions in the field, flux-chamber measurements were conducted in plots with submerged Sphagnum mosses before and after removal of the moss layer. The CH4 emission in the field situation with the submerged Sphagnum moss layer was 4.1 ± 2.1 mmol CH4 m–2 day–1 (mean ± SEM, n = 3; Figure 2). Removal of the Sphagnum moss layer significantly increased the net CH4 emission [t(2) = −6.1, p < 0.05] to 60 ± 32 mmol CH4 m–2 day–1 (Figure 2).
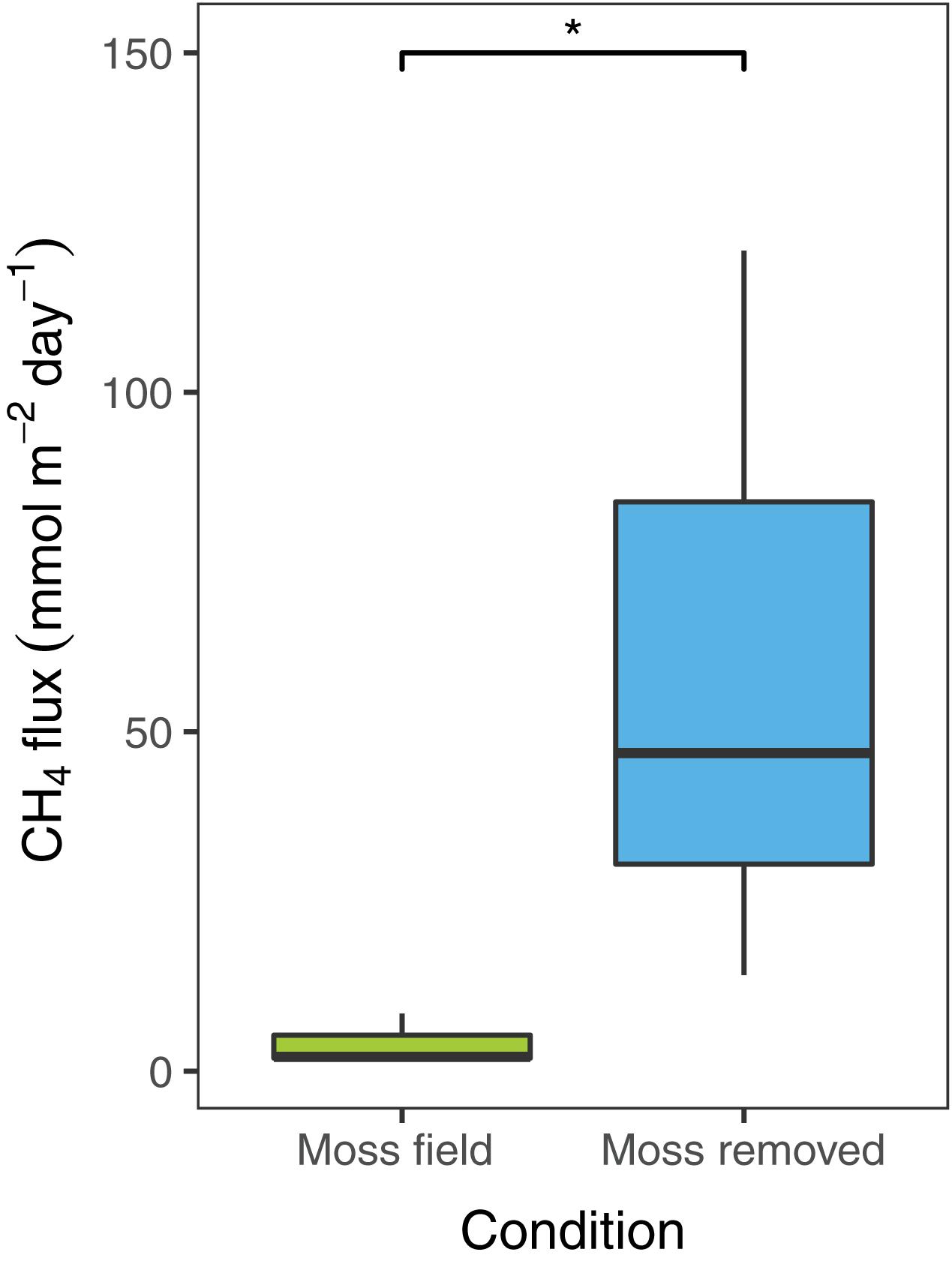
Figure 2. Net CH4 flux (mmol CH4 m– 2 day– 1) measured in the field with Sphagnum moss layer present (green, n = 3) and after moss removal (blue, n = 3). Error bars indicate the standard error of the mean, an asterisks indicates p < 0.05.
The CH4 oxidation rates associated with the Sphagnum moss and peat water were determined prior to incubation in the mesocosm setup by using batch assays (Figure 3). Methane oxidation was clearly associated to Sphagnum mosses, which showed higher CH4 oxidation rates (average rate mosses 143 ± 17 μmol g DW–1 day–1, Figure 3) compared to peat water, which had virtually no activity (0.05 ± 0.06 μmol g DW–1 day–1; χ2 = 7.5, p < 0.01, Supplementary Figure 5 and Supplementary Table 6). Washing of Sphagnum mosses reduced the CH4 oxidation rate by 15% to an average CH4 oxidation rate of 121 μmol g DW–1 day–1; [t(2) = 1.5, p > 0.05, Figure 3], indicating that most MOB were strongly associated with the mosses.
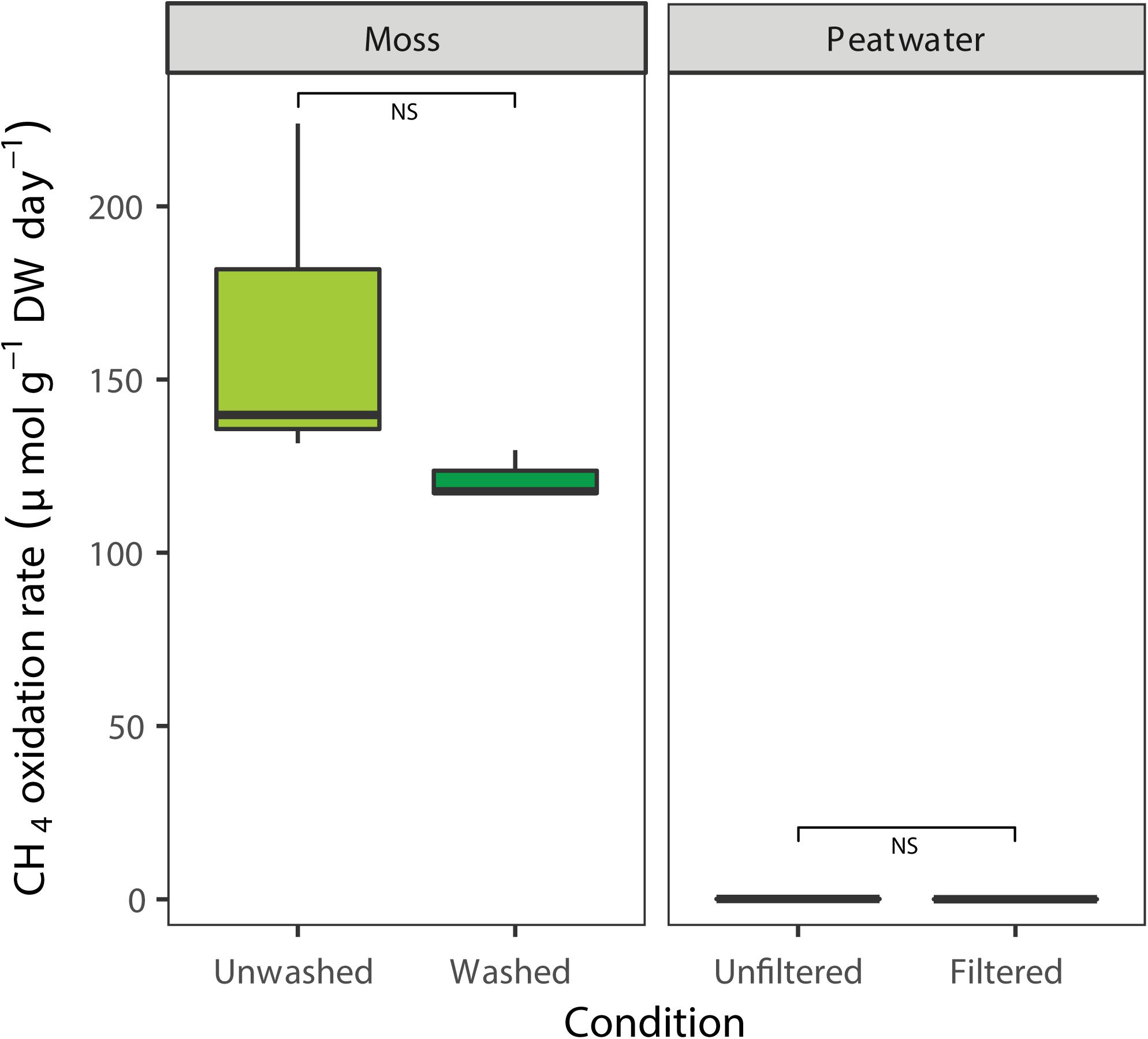
Figure 3. Potential CH4 oxidation rate in batch, associated with field Sphagnum mosses (light green, μmol CH4 g– 1 DW day– 1) or washed Sphagnum mosses (darker colors) and rates in peat water unfiltered or filtered. Error bars indicate the standard error of the mean (n = 3).
The net CH4 flux in the mesocosm showed a similar pattern for moss containing and control mesocosm until day 8 after incubation (Figure 4). Thereafter, the CH4 concentration in the headspace of the Sphagnum moss containing mesocosm was lower compared to the control mesocosm. In addition, the CH4 emission from the Sphagnum moss mesocosm gradually decreased during the 32 days incubation period, which is a strong indication of increasing CH4 oxidation activity. A second replicate of the experiment showed a similar pattern, with lower CH4 emission when Sphagnum mosses were present in the mesocosm (Supplementary Figure 8 and Supplementary Tables 7, 8). In order to test if the methane emission is decreased by activity of microorganisms associated to the moss or by a decreased diffusion of methane from the liquid to the gas phase of the column; a similar experimental setup was performed using Sphagnum mosses without any associated microorganisms (gnotobionts). Net CH4 flux from this mesocosm was about 20% lower than the moss control (Supplementary Figure 9), indicating that both a decreased diffusion rate and microbial activity play a role in the observed decrease in CH4 emission.
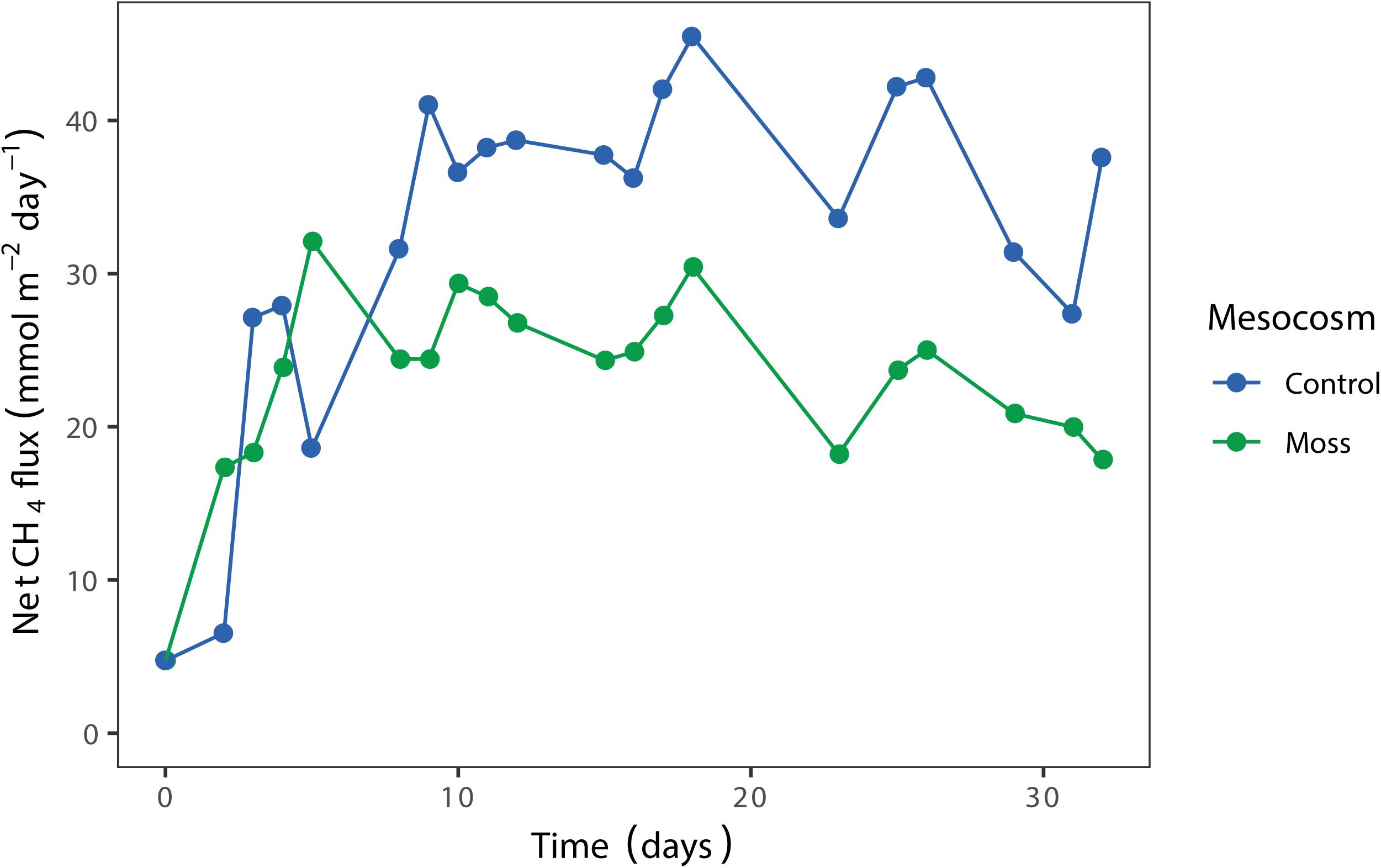
Figure 4. Net CH4 flux (mmol CH4 m– 2 day– 1) from the mesocosms with Sphagnum moss (green) and control mesocosm with only peat water (blue), measured in the headspace over time (days). Each dot represents the mean of two technical replicates.
After 32 days of incubation in the mesocosms, Sphagnum moss and peat water were taken from the mesocosm in order to measure potential CH4 oxidation activity in batch. The activity of the mosses was 189 μmol CH4 g–1 DW day–1 (Table 1). Even after 32 days of incubation, peat water showed no CH4 oxidation activity (R2 < 0.9; see Table 1 and Supplementary Figures 6, 7), again indicating a tight association of the CH4 oxidizing microorganisms with the moss. CH4 oxidation associated with mosses was almost completely inhibited by acetylene [F(1,4) = 981.3, p < 0.001, Table 1], indicating that the CH4 oxidation is indeed entirely performed by methanotrophic microorganisms associated to the moss. During the incubation, the CH4 oxidation activity associated to the moss had increased by 155% (from 121 to 189 μmol g DW–1 day–1; Table 1 and Figure 3).
To quantify the microbial community, qPCR and amplicon sequencing of 16S rRNA genes was performed. Quantification of the bacteria per gram of FW (16S rRNA gene; Figure 5) showed that bacterial copy numbers differed between moss from the field and between moss before and after incubation [F(2,6) = 34.3, p < 0.001]. 98% of presumably loosely attached microorganisms were removed by washing the moss (Tukey HSD p < 0.001). The washing step reduced the abundance of the mmoX-containing methanotrophs from 1010 to 102 copies per g FW (Tukey HSD p < 0.001), whereas pmoA-containing methanotrophs were much less affected (remained around 105 copies per g FW; Tukey HSD p > 0.05). At the end of the incubation time the copy numbers were 97% of the original value (Tukey HSD p < 0.05), indicating regrowth of microorganisms during the incubation in the mesocosm. Quantification of methanotrophic microorganisms by using qPCR targeting the mmoX and pmoA genes showed a similar trend [mmoX F(2,6) = 40.7, p < 0.001; pmoA F(2,6) = 27.1, p < 0.001; Figure 5], although pmoA-containing methanotrophs were overall less abundant than mmoX-containing methanotrophs (resp. 106 vs. 1010 copies per g FW). Upon mesocosm incubation mmoX copies increased from 102 to 108 (Tukey HSD p < 0.001), while pmoA-containing methanotrophs marginally increased in copy number per g FW (Tukey HSD p < 0.01).
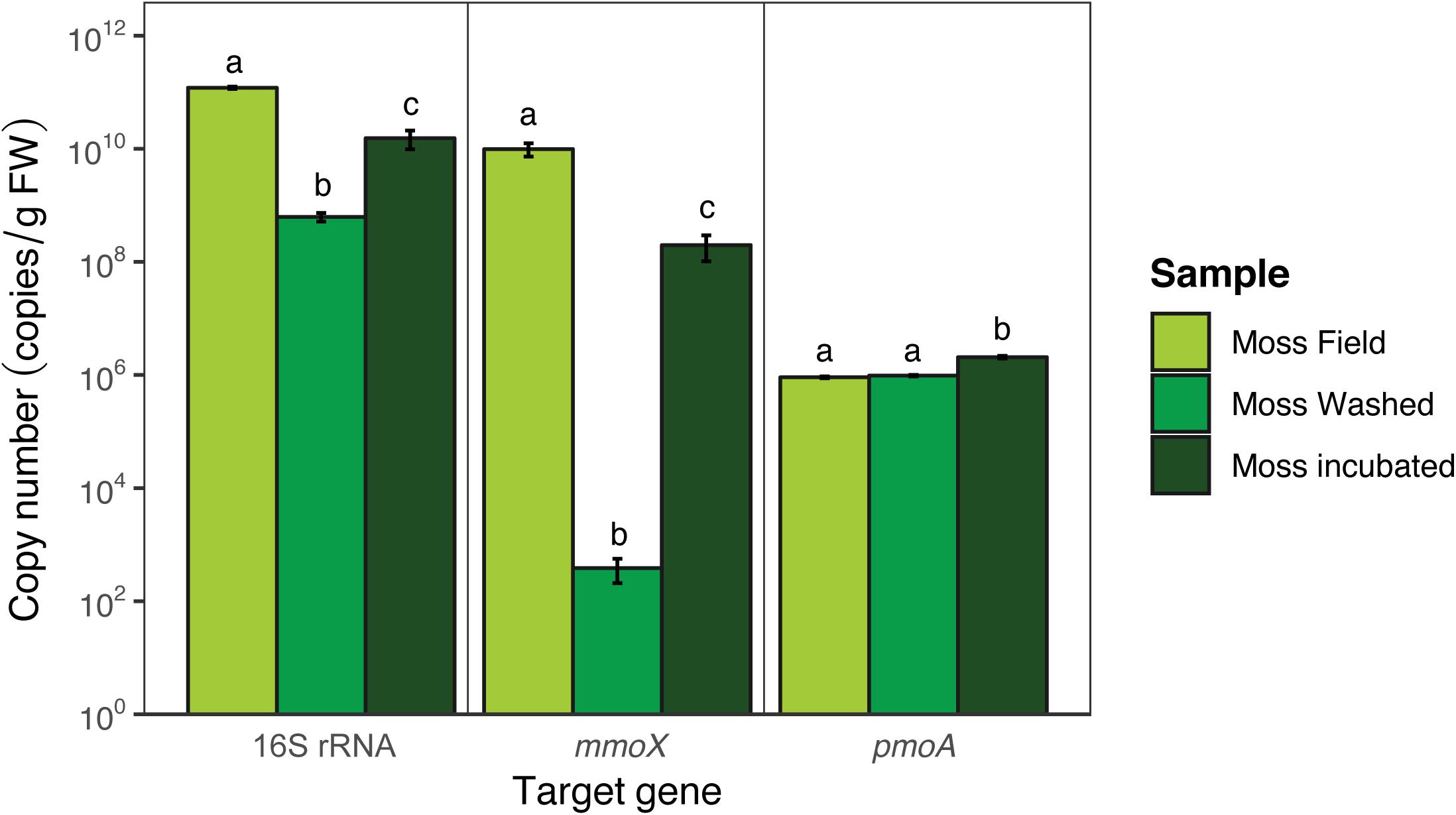
Figure 5. Copy numbers of bacterial 16S rRNA, pmoA, and mmoX genes obtained via qPCR. Error bars indicate the standard error of the mean (n = 3). For each target gene the significant differences between the different samples are indicated by the use of different letters, similar letters indicate no difference.
The microbial community composition associated with the mosses was studied by 16S rRNA gene sequencing of the V3–V4 region. Comparison of the moss microbial community in the field and of the community after washing and incubation in the mesocosm showed a gradual change in microbial community (Figure 6 and Supplementary Figure 4). However, the main classes of microorganisms remained the same throughout the incubation. Furthermore, mesocosm incubation increased the microbial community diversity (Shannon and Chao 1 index, Supplementary Table 4), where Proteobacteria was the most dominant phylum (Figure 6A). The relative abundance of Proteobacteria was not affected by washing, but their relative number increased during incubation in our mesocosm set-up. Furthermore, especially the relative abundance of Pedosphaerales and Opitutales increased upon incubation (Supplementary Table 5). When focusing on the methanotrophic community, the relative abundance of Verrucomicrobial Methylacidiphilales associated to the moss increased after incubation (Figure 6B). Also other methanotrophic bacteria species, such as Methylomonas spp. and Methylocystis spp., increased in relative abundance upon incubation (Figure 6B) indicating that methane oxidation is facilitated by a number of different methanotrophs.
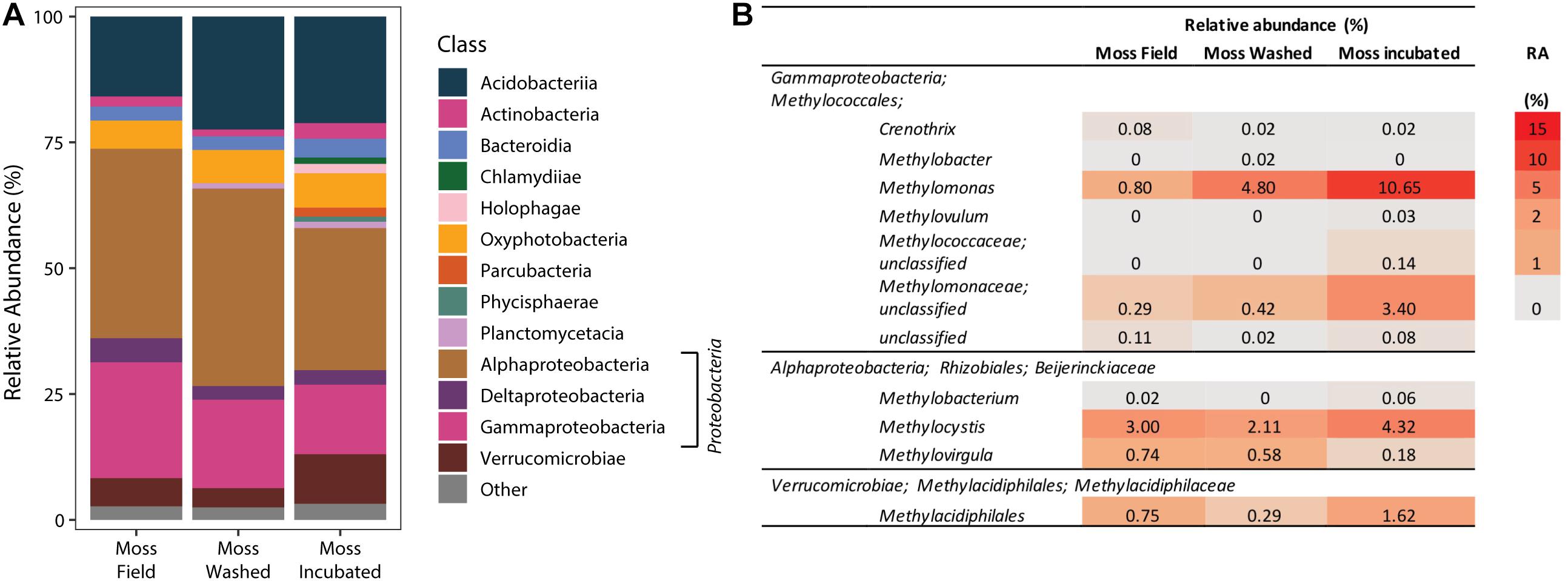
Figure 6. The phylogenetic classification of the bacterial community based on 16S rRNA gene amplification and sequencing (A). Taxonomic groups with a relative abundance >1% are depicted as “other.” In (B), specific relative abundances (RA in %) of methanotrophic bacteria in the bacterial 16S rRNA community profile are shown.
This study aimed to investigate the mitigation of CH4 fluxes in rewetted peatlands by an active, natural biofilter composed of Sphagnum mosses and their associated CH4 oxidizing microorganisms. In our study site, CH4 emission was reduced with 93% by Sphagnum associated methanotrophs (Figure 2). The reduction of methane emission to the atmosphere by Sphagnum-associated methanotrophs could be mimicked in our newly developed mesocosm setup, although the methane emission reduction was less pronounced (31 vs. 93% in the field). Free-floating plants can reduce CH4 emission by up to 70% by a combination of plant-associated CH4 oxidation and decreased flux rates (Kosten et al., 2016). Also in our mesocosm setup, a dense layer of gnotobiotic Sphagnum moss already decreased methane emission, most probably by limiting CH4 diffusion from the surface water to the atmosphere. This results in an increase in CH4 concentration in the porewater, creating ideal conditions for enrichment of CH4 oxidizing microorganisms. All in all, the large decrease of CH4 emission in the presence of both submerged Sphagnum moss and methanotrophs emphasizes their important role in CH4 cycling in peatlands (Basiliko et al., 2004; Kip et al., 2010; Liebner et al., 2011; van Winden et al., 2012, 2020). The tight association between CH4 oxidizers and Sphagnum mosses is further underlined by the fact that washing of the moss and filtering of the peat water had little effect on CH4 oxidation activity.
QPCR revealed that total bacterial copy numbers per g FW decreased after moss washing. The number of sMMO-containing methanotrophs decreased most, indicating that these methanotrophs might be loosely attached epiphytes. However, subsequently they showed the highest increase in copy number (102–108 copies per g FW) upon mesocosm incubation, which indicates that these microorganisms have a very short generation time. The transcription of mmoX gene and activity of sMMO-containing methanotrophs has previously been reported in peatlands (Morris et al., 2002; Chen et al., 2008; Liebner and Svenning, 2012), and together with our findings this suggests that sMMO-containing methanotrophs are relevant for acidic peatland ecosystems.
Surprisingly, the pMMO-containing methanotrophs were initially less abundant compared to sMMO-containing methanotrophs and seemed more tightly associated to Sphagnum moss as washing did not decrease their copy numbers. However, there was hardly any increase in abundance of pMMO-containing methanotrophs upon incubation, which might be explained by the lack of copper in our mesocosm incubations (Murrell et al., 2000). Ultimately, the enrichment of sMMO-containing methanotrophs in the mesocosm incubation shows that this set-up can be used to further study the functioning of sMMO methanotrophs in Sphagnum mosses. Obtaining more insights into their functioning is of great importance since their ecology is less well understood than that of pMMO-containing methanotrophs.
The Sphagnum-associated microbial community in all samples showed high similarity to previous Sphagnum-associated 16S rRNA gene libraries (Bragina et al., 2012, 2014; Kox et al., 2018). The dominant community members found in this study were similar to those in other investigation with dominant phyla being the Proteobacteria (Alpha- and Gammaproteobacteria), Cyanobacteria (Oxyphotobacteria) and Acidobacteria and a relatively high abundance of Verrucomicrobia. Upon mesocosm incubation the microbial diversity increased. The relative abundance of Verrucomicrobia and Planctomycetes increased, whereas the relative abundance of Proteobacteria decreased. Future studies with (micro-) nutrient additions may help to find out what causes these changes in microbial community.
The methanotrophic microbial community profile showed that Methylacidiphilales, Methylocystis, and Methylomonas spp. all were more abundant at the end the mesocosm incubation. The qPCR profiles showed that the abundance of sMMO containing methanotrophs increased most during incubation. Compared to the 16S rRNA gene library, there are few methanotrophs identified known to possess sMMO. The Verrucomicrobial methanotrophic genera Methylacidiphilum and Methylacidimicrobium appear to contain only pMMO (Op den Camp et al., 2018), whereas Methylocystis species typically have solely pMMO, except for the acidophilic Methylocystis isolates Methylocystis bryophila, and Methylocystis heyeri (Dedysh et al., 2007; Belova et al., 2013) that contain both sMMO and pMMO. Methylocella species, facultative methane oxidizers, are the only known organisms containing exclusively sMMO (Dunfield and Dedysh, 2014; Dedysh and Dunfield, 2018). The lack of correlation found in the quantification of pmmo and smmo genes suggests that sMMO-only microorganisms not belonging to Methylocella species are present in our samples. Alternatively, the lack of sMMO-containing methanotrophs in the sequencing analysis could be caused by the coarse taxonomic resolution of the 16S rRNA genes. The presence of sMMO-containing methanotrophs belonging to Beijerinckiaceae can thus not be entirely excluded.
Studying the Sphagnum microbiome in the field is challenging, because the microbial community associated with the moss is influenced by many biotic and abiotic factors which strongly fluctuate in a natural environment. Therefore, we designed a novel mesocosm set-up to mimic a submerged Sphagnum moss ecosystem and operated it under controlled laboratory conditions. The conditions could be even more controlled by supplying methane and air continuously to avoid fluctuations in the concentration of these gasses. This would most probably also reduce the fluctuations observed in methane fluxes (Figure 4). We hypothesized that the submerged Sphagnum cuspidatum moss layer acts as a biofilter for CH4 and expected that the CH4-oxidizing microbial community was mainly associated with Sphagnum moss. Similar to mosses in the field, results of our controlled mesocosm set-up showed a significant reduction (31%) in CH4 emission that was associated with Sphagnum mosses and their microbial community (Figure 4 and Supplementary Figure 8). This CH4 removal was only associated with the mosses; methane oxidation activity was not found in the peat water. However, this water can still contain low numbers of methanotrophs. It has been shown before that peat water can be a potential source for methanotrophs which can colonize Sphagnum moss (Putkinen et al., 2012).
During incubation in the novel mesocosm set-up, methanotrophic activity indeed increased along with an increase in MOB abundance. CH4 oxidation batch-assays revealed a significant increase in methanotrophic activity after incubation (from 121 ± 4 to 189 ± 6 μmol CH4 g–1 DW day–1, resp. Figure 3 and Table 1), indicating MOB involvement in CH4 mitigation. Similarly, qPCR of functional methanotrophic genes (mmoX and pmoA), indicated that significant numbers of CH4-oxidizing bacteria were present in and on the moss and that their numbers increased over the course of the incubation.
The reduction in CH4 emission in the mesocosm set-up was lower than the reduction found in the field, which is most likely due to peat moss density that is much higher in the field (∼50 cm deep in de field compared to 6 cm in the mesocosm). Mesocosm incubations were terminated after 32 days; we believe that the CH4 mitigation by the moss-associated methanotrophs in the mesocosm will increase even further by prolonging the incubation time. In addition, an increased Sphagnum moss density is expected to increase the CH4 oxidation even further. Furthermore, the mesocosm set-up could be improved by a continuous supply system for CH4 and air, which results in a system that is more comparable to the natural situation.
The large organic matter stocks in peatlands are a potential source for CO2. Restoration measures aimed at preventing CO2 emission often involve hydrological measures (rewetting; Lamers et al., 2002; Smolders et al., 2003), which result in high CH4 production rates (Abdalla et al., 2016). Since peatland degradation affects the presence and abundance of Sphagnum (Gorham, 1991; Frolking et al., 2011), care should be taken to bring back and facilitate Sphagnum mosses in restored peatlands. Stimulation of the current population or even reintroduction of Sphagnum in peatland restoration projects can thereby strongly mitigate the resultant CH4 emissions.
Sphagnum mosses have many key roles in peat ecosystems (Rydin and Jeglum, 2006), and our study shows that their microbiome and specifically their associated methanotrophs are crucial to reduce CH4 emissions from peatlands. Peatland restoration practices involving rewetting typically result in high CH4 emissions and should therefore simultaneously aim to stimulate the presence of Sphagnum mosses. With the development of our mesocosm setup, CH4 mitigation by Sphagnum mosses and their associated methanotrophs can be studied in great detail, providing essential knowledge that can be used for restoration practices and climate research in the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA517391.
MARK, AJPS, and MAHJvK collected the samples. MARK and MAHJvK processed the samples and wrote the manuscript with input from all authors. MARK, DRS, MSMJ, and MAHJvK designed the research. MARK performed the experiments. HJMOdC, MSMJ, LPML, and AJPS were involved in project discussion and data interpretation. All authors contributed to the article and approved the submitted version.
MARK was supported by the European Research Council Advanced Grant Ecomom 339880 to MSMJ, who was further supported by the Netherlands Organisation for Scientific Research (SIAM Gravitation grant 024 002 002 and Spinoza Award). MAHJvK was supported by NWO Veni grant (016.veni.192.062). HJMOdC was supported by European Research Council Advanced Grant VOLCANO 669371.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Nardy Kip for her support with the initial design of the mesocosms, Tijs van den Bosch for helping with 16S rRNA sequencing and the Department of General Instrumentation at the Faculty of Science at Radboud University, especially Paul van der Ven and Sebastian Krosse, for measuring elemental composition of the water samples and determining stable isotope contents. Dr. Silvia Coolen is thanked for critical reviewing of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.651103/full#supplementary-material
Abdalla, M., Hastings, A., Truu, J., Espenberg, M., Mander, Ü, and Smith, P. (2016). Emissions of methane from northern peatlands: a review of management impacts and implications for future management options. Ecol. Evol. 6, 7080–7102. doi: 10.1002/ece3.2469
Basiliko, N., Knowles, R., and Moore, T. R. (2004). Roles of moss species and habitat in methane consumption potential in a northern peatland. Wetlands 24, 178–185.
Belova, S. E., Kulichevskaya, I. S., Bodelier, P. L. E., and Dedysh, S. N. (2013). Methylocystis bryophila sp. nov., a facultatively methanotrophic bacterium from acidic Sphagnum peat, and emended description of the genus Methylocystis (ex Whittenbury et al. 1970) Bowman et al. 1993. Int. J. Syst. Evol. Microbiol. 63, 1096–1104. doi: 10.1099/ijs.0.043505-0
Bragina, A., Berg, C., Cardinale, M., Shcherbakov, A., Chebotar, V., and Berg, G. (2012). Sphagnum mosses harbour highly specific bacterial diversity during their whole lifecycle. ISME J. 6, 802–813. doi: 10.1038/ismej.2011.151
Bragina, A., Oberauner-Wappis, L., Zachow, C., Halwachs, B., Thallinger, G. G., Müller, H., et al. (2014). The Sphagnum microbiome supports bog ecosystem functioning under extreme conditions. Mol. Ecol. 23, 4498–4510. doi: 10.1111/mec.12885
Bridgham, S. D., Cadillo-Quiroz, H., Keller, J. K., and Zhuang, Q. (2013). Methane emissions from wetlands: biogeochemical, microbial, and modeling perspectives from local to global scales. Glob. Change Biol. 19, 1325–1346. doi: 10.1111/gcb.12131
Chen, Y., Dumont, M. G., McNamara, N. P., Chamberlain, P. M., Bodrossy, L., Stralis-Pavese, N., et al. (2008). Diversity of the active methanotrophic community in acidic peatlands as assessed by mRNA and SIP-PLFA analyses. Environ. Microbiol. 10, 446–459. doi: 10.1111/j.1462-2920.2007.01466.x
Danilova, O. V., Kulichevskaya, I. S., Rozova, O. N., Detkova, E. N., Bodelier, P. L. E., Trotsenko, Y. A., et al. (2013). Methylomonas paludis sp. nov., the first acid-tolerant member of the genus Methylomonas, from an acidic wetland. Int. J. Syst. Evol. Microbiol. 63, 2282–2289. doi: 10.1099/ijs.0.045658-0
de Jong, A. E. E., in’t Zandt, M. H., Meisel, O. H., Jetten, M. S. M., Dean, J. F., Rasigraf, O., et al. (2018). Increases in temperature and nutrient availability positively affect methane-cycling microorganisms in Arctic thermokarst lake sediments. Environ. Microbiol. 20, 4314–4327. doi: 10.1111/1462-2920.14345
Dedysh, S. N. (2011). Cultivating uncultured bacteria from northern wetlands: knowledge gained and remaining gaps. Front. Microbiol. 2:184. doi: 10.3389/fmicb.2011.00184
Dedysh, S. N., Belova, S. E., Bodelier, P. L. E., Smirnova, K. V., Khmelenina, V. N., Chidthaisong, A., et al. (2007). Methylocystis heyeri sp. nov., a novel type II methanotrophic bacterium possessing “signature” fatty acids of type I methanotrophs. Int. J. Syst. Evol. Microbiol. 57, 472–479. doi: 10.1099/ijs.0.64623-0
Dedysh, S. N., Berestovskaya, Y., Vasylieva, L., Belova, S., Khmelenina, V., Suzina, N. E., et al. (2004). Methylocella tundrae sp. nov., a novel methanotrophic bacterium from acidic tundra peatlands. Int. J. Syst. Evol. Microbiol. 54, 151–156. doi: 10.1099/ijs.0.02805-0
Dedysh, S. N., and Dunfield, P. F. (2018). Facultative Methane Oxidizers, in: Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes. Cham: Springer International Publishing, 1–20.
Dedysh, S. N., Liesack, W., Khmelenina, V. N., Suzina, N. E., Trotsenko, Y. A., Semrau, J. D., et al. (2000). Methylocella palustris gen . nov ., sp . nov ., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50, 955–969.
Dunfield, P. F., and Dedysh, S. N. (2014). Methylocella: a gourmand among methanotrophs. Trends Microbiol. 22, 368–369. doi: 10.1016/j.tim.2014.05.004
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Esson, K. C., Lin, X., Kumaresan, D., Chanton, J. P., Murrell, J. C., and Kostka, J. E. (2016). Alpha- and gammaproteobacterial methanotrophs codominate the active methane-oxidizing communities in an acedic boreal peat bog. Appl. Environ. Microbiol. 82, 2363–2371. doi: 10.1128/AEM.03640-15
Frolking, S., Talbot, J., Jones, M. C., Treat, C. C., Kauffman, J. B., Tuittila, E. S., et al. (2011). Peatlands in the Earth’s 21st century climate system. Environ. Rev. 19, 371–396.
Gorham, E. (1991). Northern peatlands: role in the carbon cycle and probable responses to climatic warming. Ecol. Appl. 1, 182–195.
Grootjans, A. P., Van Diggelen, R., Joosten, H., and Smolders, J. P. (2012). “Restoration of mires,” in Restoration Ecology: The New Frontier, eds J. van Andel, and J. Aronson, (Hoboken, NJ: Wiley), 203–213. doi: 10.1002/9781118223130.ch16
Harpenslager, S. F., van den Elzen, E., Kox, M. A. R., Smolders, A. J. P., Ettwig, K. F., and Lamers, L. P. M. (2015). Rewetting former agricultural peatlands: topsoil removal as a prerequisite to avoid strong nutrient and greenhouse gas emissions. Ecol. Eng. 84, 159–168. doi: 10.1016/j.ecoleng.2015.08.002
Ho, A., van den Brink, E., Reim, A., Krause, S. M. B., and Bodelier, P. L. E. (2016). Recurrence and frequency of disturbance have cumulative effect on methanotrophic activity, abundance, and community structure. Front. Microbiol. 6:1493. doi: 10.3389/fmicb.2015.01493
Joosten, H., and Clarke, D. (2002). Wise Use of Mires and Peatlands. Helsinki: International Mire Conservation Group and International Peat Society.
Kaupper, T., Mendes, L. W., Harnisz, M., Krause, S. M. B., Horn, M. A., and Ho, A. (2021). Recovery of methanotrophic activity is not reflected in the methane-driven interaction network after peat mining. Appl. Environ. Microbiol. 87:e02355-20. doi: 10.1128/AEM.02355-20
Kip, N., Dutilh, B. E., Pan, Y., Bodrossy, L., Neveling, K., Kwint, M. P., et al. (2011). Ultra-deep pyrosequencing of pmoA amplicons confirms the prevalence of methylomonas and methylocystis in sphagnum mosses from a dutch peat bog. Environ. Microbiol. Rep. 3, 667–673. doi: 10.1111/j.1758-2229.2011.00260.x
Kip, N., van Winden, J. F., Pan, Y., Bodrossy, L., Reichart, G.-J., Smolders, A. J. P., et al. (2010). Global prevalence of methane oxidation by symbiotic bacteria in peat-moss ecosystems. Nat. Geosci. 3, 617–621.
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, 1–11. doi: 10.1093/nar/gks808
Kosten, S., Piñeiro, M., de Goede, E., de Klein, J., Lamers, L. P. M., and Ettwig, K. (2016). Fate of methane in aquatic systems dominated by free-floating plants. Water Res. 104, 200–207. doi: 10.1016/j.watres.2016.07.054
Kox, M. A. R., Aalto, S. L., Penttilä, T., Ettwig, K. F., Jetten, M. S. M., and van Kessel, M. A. H. J. (2018). The influence of oxygen and methane on nitrogen fixation in subarctic Sphagnum mosses. AMB Express 8:76. doi: 10.1186/s13568-018-0607-2
Kox, M. A. R., Lüke, C., Fritz, C., van den Elzen, E., van Alen, T., Op den Camp, H. J. M., et al. (2016). Effects of nitrogen fertilization on diazotrophic activity of microorganisms associated with Sphagnum magellanicum. Plant Soil 406, 83–100. doi: 10.1007/s11104-016-2851-z
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Lamers, L. P. M., Smolders, A. J. P., and Roelofs, J. G. M. (2002). The restoration of fens in the Netherlands. Hydrobiologia 478, 107–130. doi: 10.1023/A:1021022529475
Liebner, S., and Svenning, M. M. (2012). Environmental transcription of mmoX by methane-oxidizing Proteobacteria in a subarctic palsa Peatland. Appl. Environ. Microbiol. 79, 701–706. doi: 10.1128/AEM.02292-12
Liebner, S., Zeyer, J., Wagner, D., Schubert, C., Pfeiffer, E. M., and Knoblauch, C. (2011). Methane oxidation associated with submerged brown mosses reduces methane emissions from Siberian polygonal tundra. J. Ecol. 99, 914–922. doi: 10.1111/j.1365-2745.2011.01823.x
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:0061217. doi: 10.1371/journal.pone.0061217
Morris, S. A., Radajewski, S., Willison, T. W., and Murrell, J. C. (2002). Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68, 1446–1453. doi: 10.1128/AEM.68.3.1446-1453.2002
Murrell, J. C., McDonald, I. R., and Gilbert, B. (2000). Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol. 8, 221–225. doi: 10.1016/S0966-842X(00)01739-X
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2016). Vegan: Community Ecology Package. R Package 2.3–3.
Op den Camp, H. J. M., Mohammadi, S. S., Pol, A., and Dunfield, P. F. (2018). “Verrucomicrobial methanotrophs,” in Methane Biocatalysis: Paving the Way to Sustainability, eds M. G. Kalyuzhnaya, and X.-H. Xing, (Cham: Springer International Publishing), 43–55. doi: 10.1007/978-3-319-74866-5_3
Putkinen, A., Larmola, T., Tuomivirta, T., Siljanen, H. M. P., Bodrossy, L., Tuittila, E.-V., et al. (2012). Water dispersal of methanotrophic bacteria maintains functional methane oxidation in Sphagnum mosses. Front. Microbiol. 3:15. doi: 10.3389/fmicb.2012.00015
R Development Core Team, (2017). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Raghoebarsing, A. A., Smolders, A. J. P., Schmid, M. C., Rijpstra, W. I. C., Wolters-Arts, M., Derksen, J., et al. (2005). Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature 436, 1153–1156. doi: 10.1038/nature03802
Renou-Wilson, F., Moser, G., Fallon, D., Farrell, C. A., Müller, C., and Wilson, D. (2019). Rewetting degraded peatlands for climate and biodiversity benefits: results from two raised bogs. Ecol. Eng. 127, 547–560. doi: 10.1016/j.ecoleng.2018.02.014
Reumer, M., Harnisz, M., Lee, H. J., Reim, A., Grunert, O., Putkinen, A., et al. (2018). Impact of peat mining and restoration on methane turnover potential and methane-cycling microorganisms in a northern bog. Appl. Environ. Microbiol. 84, 1–17. doi: 10.1128/AEM.02218-17
Smolders, A. J. P., Tomassen, H. B. M., Pijnappel, H. W., Lamers, L. P. M., and Roelofs, J. G. M. (2001). Substrate-derived CO2 is important in the development of Sphagnum spp. New Phytol. 152, 325–332.
Smolders, A. J. P., Tomassen, H. B. M., Van Mullekom, M., Lamers, L. P. M., and Roelofs, J. G. M. (2003). Mechanisms involved in the re-establishment of Sphagnum-dominated vegetation in rewetted bog remnants. Wetlands Ecol. Manag. 11, 403–418. doi: 10.1023/B:WETL.0000007195.25180.94
van Winden, J. F., Reichart, G.-J., McNamara, N. P., Benthien, A., and Sinninghe Damsté, J. S. (2012). Temperature-induced increase in methane release from peat bogs: a mesocosm experiment. PLoS One 7:e39614. doi: 10.1371/journal.pone.0039614
van Winden, J. F., Talbot, H. M., Reichart, G.-J., McNamara, N. P., Benthien, A., and Sinninghe Damsté, J. S. (2020). Influence of temperature on the δ13C values and distribution of methanotroph-related hopanoids in Sphagnum-dominated peat bogs. Geobiology 18, 497–507.
Vorobev, A. V., Baani, M., Doronina, N. V., Brady, A. L., Liesack, W., Dunfield, P. F., et al. (2011). Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int. J. Syst. Evol. Microbiol. 61, 2456–2463. doi: 10.1099/ijs.0.028118-0
Keywords: methanotrophy, peatland restoration, Sphagnum moss, methane cycle, mesocosm
Citation: Kox MAR, Smolders AJP, Speth DR, Lamers LPM, Op den Camp HJM, Jetten MSM and van Kessel MAHJ (2021) A Novel Laboratory-Scale Mesocosm Setup to Study Methane Emission Mitigation by Sphagnum Mosses and Associated Methanotrophs. Front. Microbiol. 12:652486. doi: 10.3389/fmicb.2021.651103
Received: 11 January 2021; Accepted: 26 March 2021;
Published: 26 April 2021.
Edited by:
Sukhwan Yoon, Korea Advanced Institute of Science and Technology, South KoreaReviewed by:
Adrian Ho, Leibniz University Hannover, GermanyCopyright © 2021 Kox, Smolders, Speth, Lamers, Op den Camp, Jetten and van Kessel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maartje A. H. J. Van Kessel, bWFhcnRqZS52YW5rZXNzZWxAc2NpZW5jZS5ydS5ubA==
†Present address: Daan R. Speth, Division of Geological and Planetary Sciences, California Institute of Technology, Pasadena, CA, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.