
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 31 March 2021
Sec. Virology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.650849
This article is part of the Research Topic Phage Biology and Phage Therapy View all 13 articles
 Mwila Kabwe1
Mwila Kabwe1 Terri Meehan-Andrews1
Terri Meehan-Andrews1 Heng Ku1
Heng Ku1 Steve Petrovski2
Steve Petrovski2 Steven Batinovic2
Steven Batinovic2 Hiu Tat Chan2,3
Hiu Tat Chan2,3 Joseph Tucci1*
Joseph Tucci1*Enterococcus faecalis is an opportunistic pathogen in the gut microbiota that’s associated with a range of difficult to treat nosocomial infections. It is also known to be associated with some colorectal cancers. Its resistance to a range of antibiotics and capacity to form biofilms increase its virulence. Unlike antibiotics, bacteriophages are capable of disrupting biofilms which are key in the pathogenesis of diseases such as UTIs and some cancers. In this study, bacteriophage EFA1, lytic against E. faecalis, was isolated and its genome fully sequenced and analyzed in silico. Electron microscopy images revealed EFA1 to be a Siphovirus. The bacteriophage was functionally assessed and shown to disrupt E. faecalis biofilms as well as modulate the growth stimulatory effects of E. faecalis in a HCT116 colon cancer cell co-culture system, possibly via the effects of ROS. The potential exists for further testing of bacteriophage EFA1 in these systems as well as in vivo models.
Understanding the interactions between host and microbe is cardinal to the development of treatment modalities to curb pathogen associated disease. In the human body, the number of bacteria cells is relatively similar to that of human cells (Sender et al., 2016), with the gut being the largest reservoir (Kho and Lal, 2018). Enterococcus species which are Gram-positive facultative anaerobes form approximately 1% of all microbiota in the gut and are readily isolated from human and animal feces as well as from the environment (Dubin and Pamer, 2014). Enterococci, including E. faecium and E. faecalis are the most predominant species and have traditionally been considered normal flora and sometimes used as probiotics to treat gut infections (Franz et al., 2003). Previous recommendations for use as probiotics stemmed from studies that explored the capacity for Enterococcus spp. to attach to epithelial cells thereby preventing colonization of more pathogenic bacteria (Kropec et al., 2005; Nueno-Palop and Narbad, 2011; Tinrat et al., 2018; Wang J. et al., 2020). However, emerging trends show that Enterococcus spp. are a major cause of a range of difficult to treat nosocomial infections, with implicated isolates displaying both acquired and intrinsic multi-antibiotic resistance (Franz et al., 2003; Hollenbeck and Rice, 2012). Enterococcus spp. survive on inanimate objects (e.g., intravenous catheters, handrails and bed frames) as fomites (Zervos et al., 1987; Hota, 2004) creating an opportunity for the transmission of antibiotic resistant Enterococci among patients in hospitals via their attending health workers (Dubin and Pamer, 2014; Drees et al., 2008). Although both E. faecium and E. faecalis are predominant Enterococci in the normal gut microbiota (De Lastours et al., 2017), the latter has been implicated in a higher frequency of Hospital Acquired Infections (HAI) (Weiner-Lastinger et al., 2020). In the American national surveillance program report from 2015 to 2017 involving more than 2,400 hospitals, E. faecalis was the fifth most common cause of HAI after Escherichia coli, Staphylococcus aureus, Klebsiella spp. and Pseudomonas aeruginosa while it was the most frequently reported pathogen in oncology units (Weiner-Lastinger et al., 2020).
Although E. faecalis has traditionally been considered a normal constituent of the gut microbiome, recent data suggests that it is intrinsically linked to colorectal cancer (CRC) (Lennard et al., 2016). A study quantifying bacterial prevalence in feces of CRC patients found an increased level of E. faecalis compared to normal controls (Balamurugan et al., 2008) while transcriptional modeling of host genes expressed in E. faecalis colonized CRC showed activation of pathways related to tumor invasion and cancer metastasis (Lennard et al., 2016). Further, E. faecalis has been shown to colonize the murine gastrointestinal tract by formation of bacterial biofilms (Barnes et al., 2017), promote aneuploidy and tetraploidy, induce colonic epithelial DNA double-strand breaks and arrest cell cycle (Wang et al., 2008) via increased reactive oxygen species (ROS) production (Huycke et al., 2002; Wang and Huycke, 2007; Wang et al., 2008). The bacterial microbiome is now considered an important aspect of the tumor microenvironment. The recent highlighting of the role of oncobacteria in tumorigenesis has added weight to the argument that tumor microbiota manipulation could be key to unlocking the potential of cancer chemotherapy and immunotherapy (Bhatt et al., 2017; Rajagopala et al., 2017; Helmink et al., 2019; Parhi et al., 2020).
The abundance of bacteria in humans and the environment pales in comparison to their viral predators, the bacteriophages. For instance, bacterial numbers are estimated at 1011 cells per gram weight of fecal material compared to approximately 1012 bacteriophages in the same amount (Cani, 2018). Bacteriophages have been shown to drive the composition of the gut microbiome (Moreno-Gallego et al., 2019) by specifically targeting gut bacteria and creating an evolutionary arms race between bacterial host and virus (Azam and Tanji, 2019; Wang G. et al., 2020). Further, studies have shown the capacity of bacteriophages to disrupt biofilms (Hansen et al., 2019; Lusiak-Szelachowska et al., 2020), kill intracellular bacteria (Jonczyk-Matysiak et al., 2015), and modulate the immune system (Van Belleghem et al., 2018), all key to counteracting virulence mechanisms that oncobacteria use to promote tumor growth (Holt and Cochrane, 2017). Bacteriophage against Fusobacterium nucleatum, a classic oncobacterium implicated in various cancers (Mitsuhashi et al., 2015; Yamamura et al., 2016; Al-Hebshi et al., 2017; Zhao et al., 2017; Yang S. F. et al., 2018; Gaiser et al., 2019; Kharrat et al., 2019; Yachida et al., 2019), have shown the potential to augment chemotherapy in the treatment of F. nucleatum associated CRC (Zheng et al., 2019). In this study, we demonstrate the augmentation of CRC cell growth in vitro by E. faecalis, and how these effects can be modulated by treatment with the E. faecalis specific bacteriophage EFA1, possibly via an increase of ROS production. Bacteriophage EFA1 was isolated from wastewater, phenotypically characterized and its whole genome sequenced. The whole genome sequence of EFA1 bacteriophage has been deposited in GenBank ® (Benson et al., 2000) under accession number MT857001.
Study protocols were approved by the La Trobe University Ethics Committee under reference number S17-112 and all methods performed in accordance with the La Trobe University Ethics, Biosafety and Integrity guidelines, and regulations.
Human colonic HCT116 cells (ATCC CCL-247) were maintained in RPMI-1640 medium with L-glutamine and sodium bicarbonate (Sigma-Aldrich ®, Australia) that was supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich ®, Australia) and passaged by detachment using 0.5% trypsin (Sigma-Aldrich ®, Australia). HCT116 cells were maintained in humidified 5% CO2 at 37°C.
The E. faecalis strains used in these experiments were isolated from wastewater. They were cultured at 37°C in Brain-Heart infusion media (BHI; OxoidTM, Australia) under anaerobic conditions. Anaerobic conditions were generated using the AnaeroGen pack (OxoidTM, Australia). For confirmation of E. faecalis strain identity, 16s rRNA PCR gene amplification was performed and the amplicons purified using the QIAquick ® PCR purification kit (Qiagen, Australia) before Sanger sequencing at the Australian Genome Research Facility. The primers used for the 16s rRNA PCR and sequencing were U27F: 5′AGAGTTTGATCMTGGCTCAG3′ and U492R: 5′AAGGAG GTGWTCCARCC 3′ under thermocycling conditions: 95°C for 3 min, 32 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 90 s, with a final extension at 72°C for 10 min (Kabwe et al., 2019).
Bacteriophage isolation was carried out as previously described (Kabwe et al., 2020). Wastewater from Victoria, Australia was collected and filtered using 0.2 μm cellulose acetate filters (Advantec, Australia). Filtered wastewater was then added to 108 colony forming units/mL (CFU/mL) of E. faecalis in BHI broth at 1% (v/v). The E. faecalis-wastewater enrichment culture was then incubated at 37°C anaerobically for 4 d before 0.2 μm cellulose acetate filtration. Serially diluted enrichment filtrates (10 μL) were placed on fresh lawns of E. faecalis on BHI containing 1% agar and plates incubated for 24 h. Any potential bacteriophage clearing was excised (along with a portion of agar) and resuspended in 500 μL of media, before centrifugation (12,000 × g for 5 min) and a 10-fold serial dilution was completed. Then 10 μL of each dilution was placed on a bacterial lawn such that plaques could be observed after an overnight anaerobic incubation at 37°C. This serial dilution purification was repeated five times to ensure single virion infection. The concentration of the bacteriophage suspensions was determined and calculated as number of plaque forming units (PFU)/mL.
Host range was assessed by making 10-fold serial dilutions of the bacteriophage stock (1 × 108 PFU/mL) and spotting 10 μL of the serial dilution aliquots onto freshly plated lawns of oral and enteric bacteria including, F. nucleatum, Solobacterium moorei, Streptococcus mutans, Lactobacillus Casei, Aeromonas hydrophila, and Escherichia coli. All plates were incubated at 37°C under anaerobic conditions except for E. coli and A. hydrophila that were incubated under aerobic conditions. A lack of individual plaques indicated that the bacteriophage EFA1 did not target these strains.
To determine the one-step growth curve (OSGC), 1 mL of E. faecalis in exponential growth phase at the concentration of 1 × 108 CFU/mL was centrifuged at 12, 000 × g for 5 min. The cell pellet was then resuspended in 900 μL of cold BHI broth before adding 100 μL of EFA1 bacteriophage at multiplicity of infection (MOI) of 0.1 as previously described (Kabwe et al., 2020). The bacteria-bacteriophage mixture was incubated at 4°C for 5 min to allow for bacteriophage adsorption onto bacteria. The mixture was then centrifuged at 12,000 × g for 10 min and unadsorbed bacteriophages assayed. Adsorbed bacteriophage were together collected with bacteria in a pellet and resuspended in BHI broth at 50 × dilution, incubated anaerobically at 37°C and bacteriophage concentration determined every 5 min. A graph of bacteriophage concentration (y-axis) against time (x-axis) was plotted and burst size (PFU/bacterial cell) calculated as a fraction of burst of newly released bacteriophage out of the total number of infecting bacteriophages.
Visualization of the bacteriophage phenotype was achieved using a JEOL JEM-2100 transmission electron microscope (TEM) operated at 200 kV as previously described (Kabwe et al., 2020). Using a 400-mesh formvar and carbon copper grids (ProScieTech, Australia), EFA1 bacteriophage particles were adsorbed for 1 min before rinsing the grids with milli-Q water and negatively staining for 20 s with 2% (w/v) uranyl acetate (Sigma-Aldrich®, Australia). Excess uranyl acetate was removed, and the copper grids allowed to air dry at room temperature for 30 min. The Gatan Orius SC200D 1 wide-angle camera coupled to the Gatan Microscopy Suite and Digital Micrograph Imaging software (Version 2.3.2.888.0) was used to take the TEM images before being exported to Image J (Version 1.8.0_112) for further analysis.
Using an established method (Kabwe et al., 2019), 10 mL of 1 × 108 PFU/mL bacteriophage solution in phosphate buffered saline (PBS, pH 7.4) was prepared for genomic DNA extraction. In brief, the bacteriophage solution was treated for 30 min at room temperature with 5 mmol/L of MgCl2 (Sigma-Aldrich®, Australia), and 10 μg/mL RNAse A and DNAse I (Promega, Australia). Bacteriophage particles were then precipitated at 4°C with 10% (w/v) Polyethylene glycol (PEG-8000) and 1 g/L sodium chloride, and resuspended in 50 μL nuclease-free water (Promega, Australia) before viral proteins were digested with 50 μg/mL of proteinase K, 20 mmol/L EDTA (Sigma-Aldrich®, Australia) and 0.5% (v/v) of sodium dodecyl sulfate (Sigma-Aldrich®, Australia) for 1 h at 55°C. An equal volume of phenol-chloroform-isoamyl alcohol (29:28:1) (Sigma-Aldrich®, Australia) was added to the DNA/bacteriophage protein mixture and their columns separated by centrifugation at 12,000 × g for 10 min to collect an aqueous phase with bacteriophage DNA. The bacteriophage DNA was then precipitated out using 70% ethanol and collected by 12,000 × g centrifugation before resuspension in 30 μL nuclease-free water (Promega, Australia).
Bacteriophage whole genome sequencing was performed on the Illumina MiSeq® technology using the NEBNext® UltraTM II DNA Library Prep Kit (NEB) and a MiSeq® V3 600 cycle reagent kit (Illumina, Australia) according to manufacturer’s instructions. Generated reads were trimmed using Trim Galore v0.6.4 with the default settings (Q scores of ≥ 20, with automatic adapter detection) and assembled using the Unicycler de novo assembly pipeline (Wick et al., 2017). PhageTerm was used to reorient the phage genome to begin at its pac site (Garneau et al., 2017) before exporting to Geneious (Version 11.0.5)1. Translated open reading frames (ORFs) were mapped onto the National Centre for Biotechnology Institute (NCBI) database using BLASTP (Mount, 2007) and annotated sequence submitted to GenBank®. To allocate taxa for bacteriophage EFA1, Viral Proteomic Tree (ViPTree) webserver was used to construct a viral proteomic tree with other related bacterial viral genomes in the reference database (Nishimura et al., 2017). The generated proteomic tree was annotated using the Interactive Tree of Life (iTOL) (Letunic and Bork, 2019). Amino acids of putative beta-lactamase protein in bacteriophage EFA1 and bacteria (E. faecalis) were aligned by a pair-wise progressive alignment in CLC genomics workbench version 9.5.4.
The E. faecalis mono-biofilms were grown in 96 well polystyrene plates as described previously (Kabwe et al., 2020). Briefly, 100 μL of 108 CFU/mL E. faecalis in BHI broth supplemented with 5% glucose (Sigma-Aldrich®, Australia) was added to sterile BHI (100 μL) and incubated anaerobically for 4 d at 37°C and 120 rpm shaking (Ratek Medium Orbital shaking incubator). Bacteriophage was added at concentration 108 PFU/mL and biofilm bacteriophage mixture allowed to incubate anaerobically at 37°C. The biofilm mass was determined at 2, 4, 6, 8, and 24 h post bacteriophage treatment (Merritt et al., 2005). Biofilms were rinsed in milli-Q water for 5 min and stained with 200 μL of 1% aqueous crystal violet for 10 min. Excess crystal violet was rinsed off in milli-Q water. Adherent crystal violet was solubilized in 70% ethanol and absorbance read on the FlexStation 3 plate reader (Molecular Devices, United States) at 600 nm wavelength.
In order to visualize the E. faecalis biofilm and the disruptive effects of bacteriophage EFA1, the biofilms were cultured on glass slides and stained with SYBR® gold (Eugene, United States) and Propidium Iodide (PI), each at final concentration of 2 μg/mL. The biofilms were visualized on the Olympus Fluoview Fv10i-confocal laser-scanning microscope (Olympus Life Science, Australia). The excitation wavelength was 485 nm, and emission wavelengths of 535 nm (green) and 635 nm (red) to indicate membrane intact and compromised cells, respectively.
For the HCT116 and E. faecalis co-culture experiments, E. faecalis in exponential growth phase was resuspended in RPMI 1640 media with 10% FBS (Sigma-Aldrich, Australia) before adding to a suspension of HCT116 cells at MOI of 10. For treatment with bacteriophages, a single plaque was purified and concentrated to make a working stock that was further purified by removing lipoteichoic acid and other cell debris as performed by Branston and colleagues (Branston et al., 2015) with minor modification. Briefly, after treating with 10 μg/mL DNase I and RNase A to digest any naked nucleic acids, bacteriophage was precipitated in PEG and NaCl at 4°C for 10 min. Precipitated bacteriophage particles were treated with 2% v/v Triton®, Australia) and washed 3 times in PBS (pH 7.4) by centrifugation at 12,000 × g for 15 min. The precipitation, Triton® X-100 treatment and PBS (pH 7.4) wash steps were repeated 3 times before precipitated bacteriophage was resuspended in RPMI 1640. E. faecalis present in HCT116 co-cultures were treated with the bacteriophage at an MOI of 0.1 and incubated for 48 h in humidified 5% CO2 at 37°C.
Colon cancer cells and co-cultures were incubated at 37°C for 48 h in humidified 5% CO2 and proliferation determined by Sulforhodamine B (SRB) assay (Skehan et al., 1990). Briefly, cells were fixed in 10% (w/v) trichloroacetic acid (Thermo Fisher Scientific, Australia) for 30 min at 4°C. Fixed cells were then washed in milli-Q water 5 times before staining with 1% (w/v) SRB (Sigma-Aldrich®) in glacial acetic acid (Thermo Fisher Scientific, Australia). Excess SRB was washed off in 1% (v/v) glacial acetic acid before using 10 mM of unbuffered TRIS base to bring the cell associated SRB into solution. Absorbance at 540 nm was measured using the FlexStation 3 plate reader (Molecular Devices, United States).
Reactive oxygen species activity was evaluated using the ROS-GloTM Hydrogen Peroxide (H2O2) assay kit (Promega, Australia), according to the manufacturer’s instructions. Briefly, 48 h co-cultures in 96-well plates were treated with H2O2 substrate solution at a final concentration of 25 μM. The treated co-cultures were then further incubated at 37°C, 5% CO2 for 6 h before adding an equal volume of ROS-GloTM detection solution. This was then incubated for 20 min before reading relative luminescence units (RLU) on the FlexStation 3 plate reader (Molecular Devices, United States).
To assess bacteriophage capacity to disrupt biofilms and to modulate E. faecalis effects on the colon cancer cell line, Shapiro-Wilk was used to test for the normality of the data. For data that were determined to be normally distributed, the means between two groups were compared using paired T-test whilst one-way analysis of variance (ANOVA) was used to compare means of more than two groups. Data on biofilms and cell proliferation were normally distributed while luminescence data were not. In this case, median of two different groups were compared using the Mann-Whitney U test. All data were visualized as box plots illustrating the five-number statistic comprising median, 25th, 75th percentile and upper (Q3 + 1.5 × IQR) and lower limit (Q1-1.5 × IQR). All statistical analysis was performed using the Statistical Package for Social Sciences (SPSS, Inc., United States) with P-values less than 0.05 considered statistically significant.
After combining the filtrate of the wastewater and E. faecalis 4 d enrichment, pinprick-sized plaques were visualized on agar plates. Image analysis revealed plaques to be approximately 0.3 mm in diameter (Figure 1A). A concentrated bacteriophage stock assessed by TEM indicated Siphoviridae viral particles with a tail length approximately 223 ± 7 nm and capsid diameter approximately 58 ± 3 nm (Figure 1B). The host range of EFA1 extended to both of the E. faecalis strains isolated in this study but not other oral/enteric bacteria such as F. nucleatum, Solobacterium moorei, Streptococcus mutans, Lactobacillus Casei, Aeromonas hydrophila, or Escherichia coli. The OSGC analysis revealed EFA1 on the host E. faecalis to have a latent period of 20 min. Initial release up to the point where no new bacteriophage were released (burst) took 10 min with each bacterium releasing 120 plaque forming units (PFU)/cell (Figure 1C).
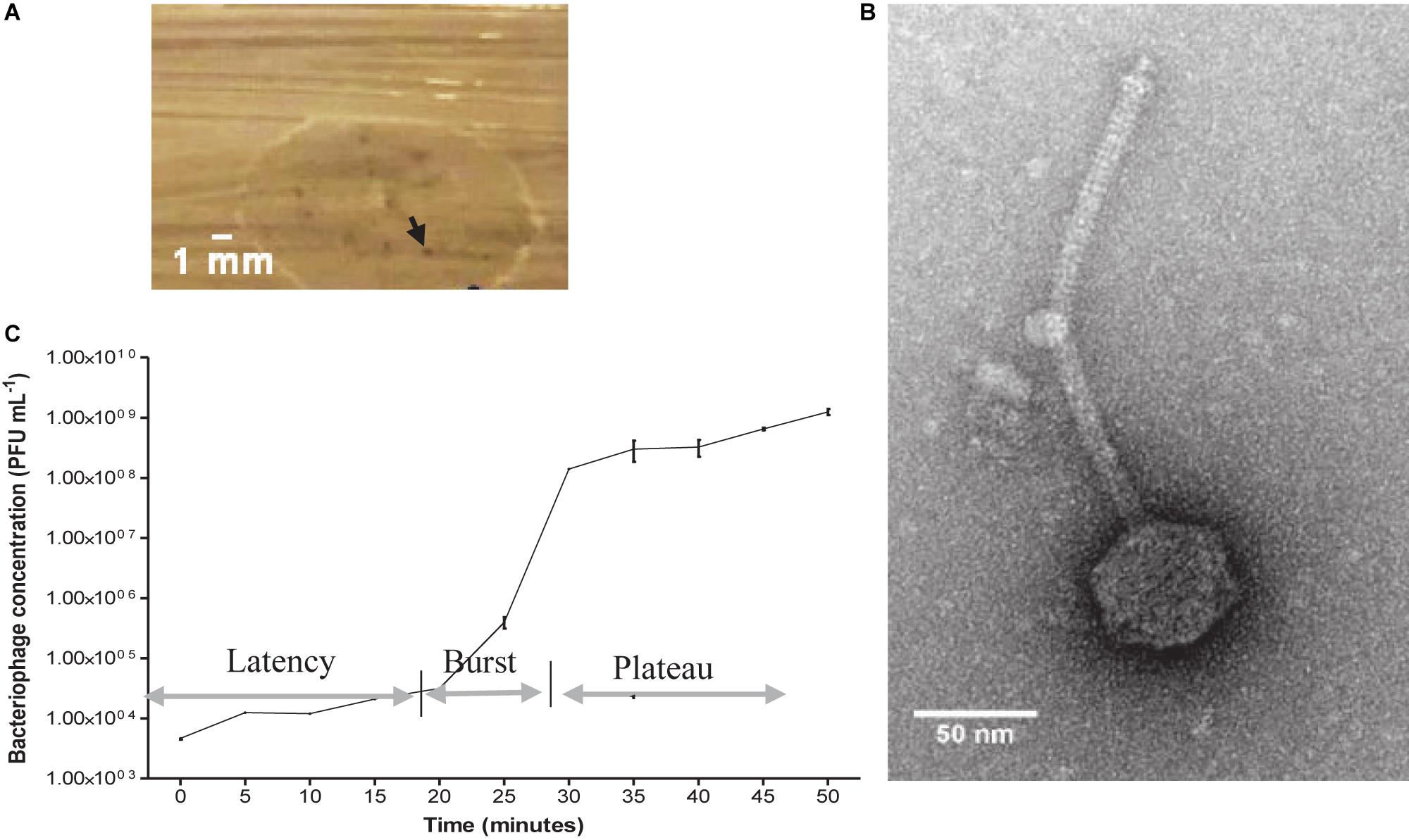
Figure 1. EFA1 plaque morphology, TEM image and growth kinetics. Pin prick plaques with diameter approximately 0.3 mm on E. faecalis host (A). TEM revealed Siphoviridae morphology with capsid diameter of 58 ± 3 nm and tail length of 223 ± 7 nm (B). EFA1 OSGC: latent phase of 20 min; burst lasting 10 min; plateau phase occurs after approximately 30 min (C).
Whole genome sequencing revealed a linear dsDNA genome of 40,454 bp with GC content of 34.8% and 68 predicated ORFs. The ORFs were bi-directional with the predicted genes encoding packaging, structural and lysis putative proteins orientated from left to right and putative genes encoding DNA manipulation functions orientated in the opposite direction (Figure 2). After arrangement of the genome so that the putative terminase gene marked the start (Garneau et al., 2017), putative terminase and portal proteins were followed by putative capsid genes then genes predicated to connect the capsid to tail and putative tail genes. The putative autolysin was located after the putative tail genes and was the last gene in the left-to-right orientation, before putative glutaredoxin in the opposite direction (Figure 2). The putative Glutaredoxin was then followed by putative DNA modification, DNA polymerase and interestingly, a putative beta-lactamase superfamily domain in ORF 33 (Figure 2). However, alignment of the putative protein sequence of ORF 33 and the E. faecalis metallo-beta-lactamase protein, revealed low homology (Figure 3). Analysis of the EFA1 genome using InterProScan (Mitchell et al., 2019) and Prosite (Sigrist et al., 2013) did not reveal any active sites or beta-lactamase domains required for catalysis (Moali et al., 2003). These findings were confirmed by the Comprehensive Antibiotic Resistance Database (CARD) (Jia et al., 2017) program, which predicted that the EFA1 genome did not carry any functional antibiotic resistance genes. The EFA1 genome also contained genes coding for putative DNA manipulation enzymes including putative endonucleases, NUMOD4 motif protein, helicase and primase. Putative hypothetical genes made up 57.4% (39/68) of EFA1 ORFs. There were no tRNAs, tmRNAs (Laslett and Canback, 2004; Lowe and Chan, 2016), CRISPR sequences (Grissa et al., 2007), or genes predicted to code for integrase found in the bacteriophage EFA1 genome.
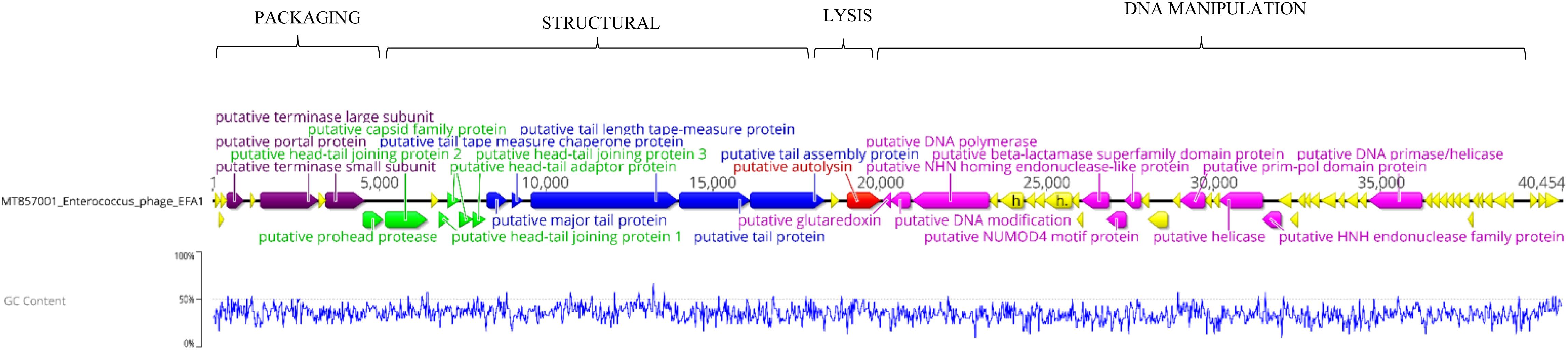
Figure 2. Bacteriophage EFA1 genome orientated according to bacteriophage termini (PAC site). Putative genes coding for DNA packaging (purple), capsid genes (green), tail genes (blue) and putative endolysin (red) are orientated in the left-to-right direction, while putative DNA manipulation genes (pink) are in the opposite direction. Other putative genes whose functionality could not be predicted were considered hypothetical genes (yellow).

Figure 3. Amino acid alignment of the E. faecalis beta-lactamase protein (EPH86145) with EFA1 bacteriophage putative beta-lactamase protein (ORF number 33). Red colored regions highlight areas of identical amino acids [47/272 (17.3%)] between the bacterial and bacteriophage putative beta-lactamase proteins, whereas the non-colored areas define regions where amino acids were non-identical.
Using the ViPTree server, bacteriophages that clustered with EFA1 were selected to generate a proteomic tree based on genome wide sequence similarities. This was comprised of bacteriophages targeting Enterococcus, Lactobacillus, Streptococcus, Oenococcus, and Staphylococcus bacteria. Proteomic analysis revealed that EFA1 is part of a monophyletic group with the E. faecalis bacteriophage SANTOR1 (Subfamily Efquatrovirus), with which it is most closely related (nucleotide similarity of 93.42% over 79% of the whole genome and 87% amino acid similarity). These two are part of a clade of 11 Enterococcus bacteriophages which target E. faecalis or E. faecium, and which also form a larger clade with bacteriophages infecting Lactobacillus and Streptococcus hosts, but cluster more distantly from other Siphoviridae, Herelleviridae and Podoviridae bacteriophages which target E. faecalis hosts (Figure 4).
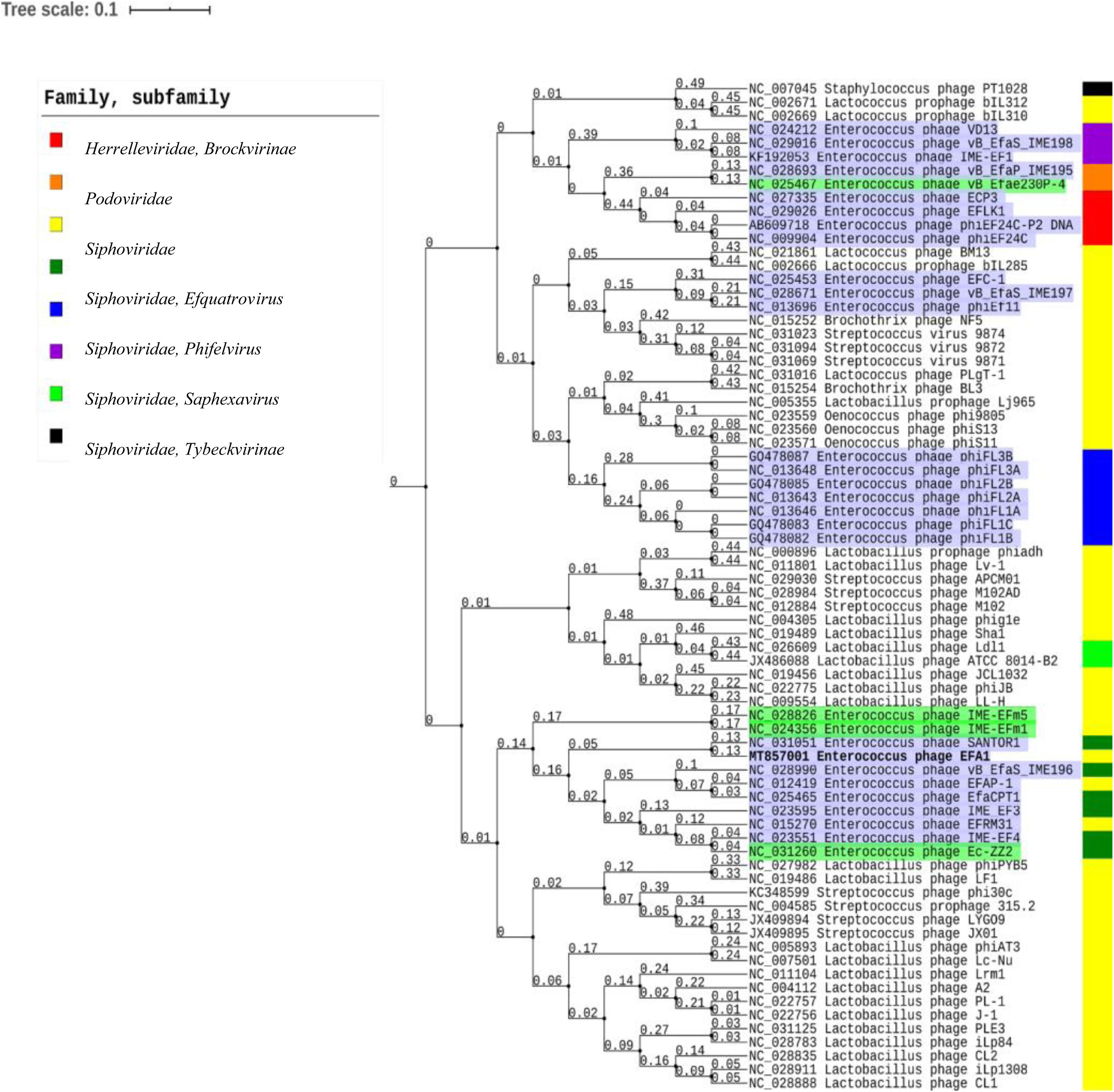
Figure 4. Proteomic tree showing the genome-wide proteomic diversity between bacteriophage EFA1 (highlighted in bold font) and related bacteriophage genomes. Bacteriophages targeting E. faecalis are highlighted in purple while those targeting E. faecium are highlighted in green. Color coded bar represents the family and subfamily of the bacteriophage (key is at top left).
To demonstrate the capacity of bacteriophage EFA1 to disrupt E. faecalis biofilms, confocal imaging was used to visualize biofilm mass on glass slides. Untreated E. faecalis biofilm mass (Figure 5A) and that with 2 h bacteriophage EFA1 treatment (Figure 5B) were stained with SYBR Gold® and PI to indicate membrane intact bacterial cells (green color) as proxy for live cells and dead/membrane compromised cells indicated by the red color. Images indicated a densely populated biofilm mass in the untreated control (Figure 5A) with both live and dead cells remaining attached to the glass slide while a sparse population of membrane intact bacteria remained on the EFA1 treated biofilm (Figure 5B).
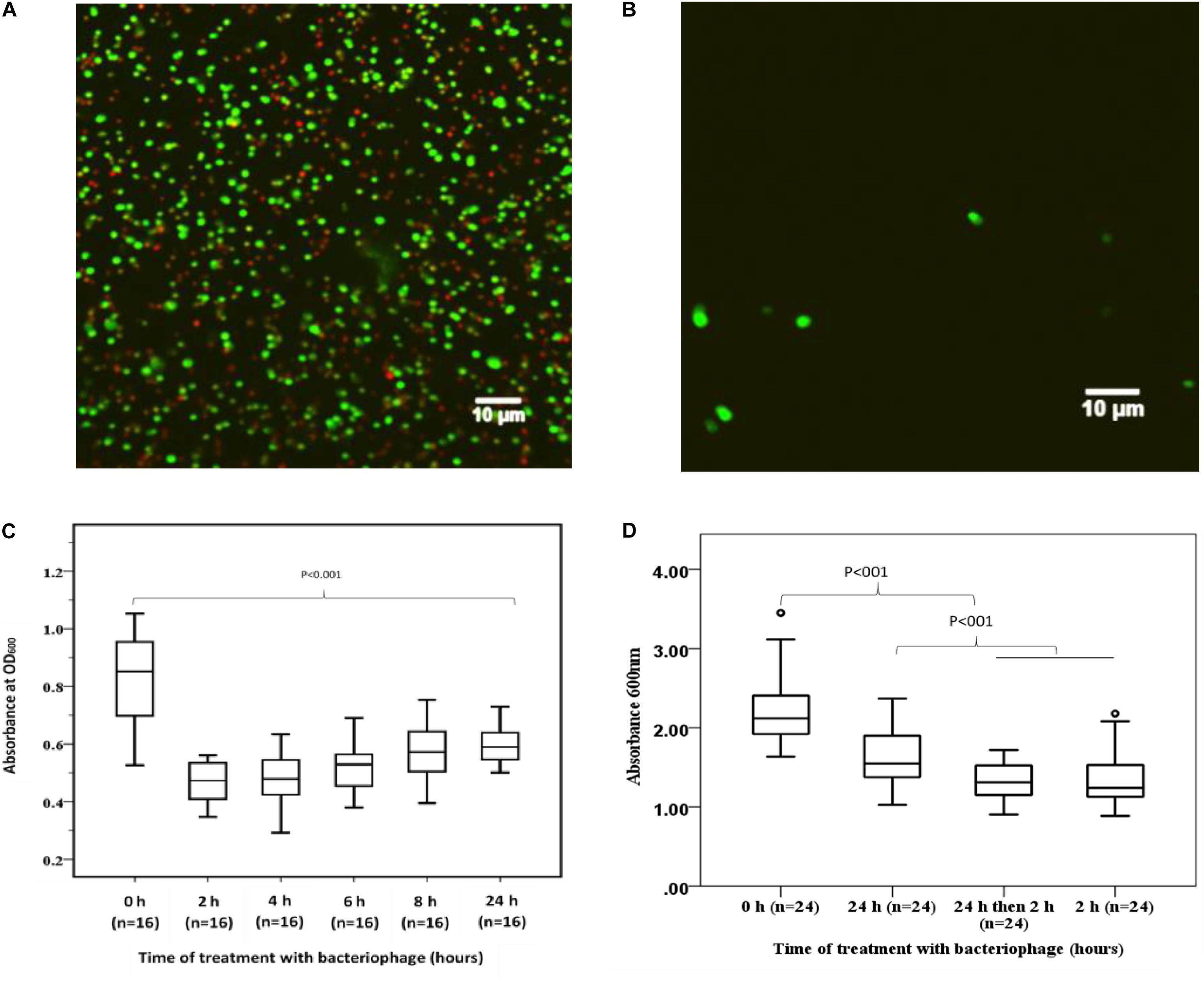
Figure 5. Effects of EFA1 bacteriophage on E. faecalis biofilm viability and mass. (A) Untreated E. faecalis biofilm showing bacteria attached to slide were predominantly membrane intact cells (green), with some bacteria with leaky membranes (red/brown). (B) Bacteriophage EFA1 treated biofilm with scant cells remaining attached to slide. (C) Quantification of biofilm at time 0 h (without EFA1 treatment) is significanlty greater than any other biofilm treated with bacteriophage EFA1. The lowest amount of biofilm mass was seen after 2 h treatment. Biofilm mass steadly increased with exposure to EFA1. (D) 5 days biofilm treated with EFA1 for 24, 2, and 24 h + 2 h, showing similar disruption at 24 + 2 h exposure as that for 2 h.
In order to quantify the effect of bacteriophage EFA1 on E. faecalis mono-biofilms, these were grown in polystyrene 96 well plates (Greiner Bio-one, Australia) for 4 d to yield biofilm mass of mean absorbance (SE) at OD600nm of 0.86 (0.05). The bacteriophage EFA1 significantly lowered the mean biofilm mass at all treatments from 2 to 24 h (p < 0.001) (Figure 5C). The mean (SE) biomass following treatment with EFA1 bacteriophage ranged from 0.47 (0.02) for a 2 h treatment to 0.6 (0.07) for a 24 h treatment. While there was a significant reduction in biofilm following treatments at all time points, exposure to EFA1 for 2 h resulted in the greatest decrease in biofilm mass, and the biofilm mass was shown to increase with the length of EFA1 treatment (p < 0.001). Paired T-tests revealed statistically increased mean (SE) absorbance of biofilm mass between EFA1 treatments for 2 h [0.47 (0.02)] and 6 h [0.52 (0.02)], 4 h [0.48 (0.02)] and 8 h [0.58 (0.02)]; 6 h [0.52 (0.02)] and 24 h [0.6 (0.07)], 8 h [(0.58 (0.02)], and 24 h [0.6 (0.07)]. In order to assess whether the bacteria was developing resistance to EFA1 with the longer treatments, a 5 days biofilm was exposed to EFA1 for (a) 24 h (b) 2 h, and (c) 24 h + 2 h (where after 24 h, media was replaced with fresh media and EFA1, then left for a further 2 h). In these experiments, the biofilm reduction with the 24 h + 2 h treatment was similar to that for 2 h, suggesting that E. faecalis had not developed resistance to EFA1 (Figure 5D).
As SRB stoichiometrically binds to protein under mild acidic conditions, the absorbance of bound SRB is proportional to cell mass and hence used as proxy for cell proliferation (Orellana and Kasinski, 2016). The greatest proliferation of HCT116 colon cancer cells was seen when co-cultured with E. faecalis (Figure 6A). The crudely prepared bacteriophage EFA1 caused an increased proliferation of HCT116 cells with mean (SE) SRB absorbance of 0.46 (0.02), p < 0.001 (Figure 6A), while the mean (SE) SRB absorbance of purified EFA1 treated HCT116 colon cancer cells of 0.31 (0.02) was comparable to that of untreated cells: 0.34 (0.01) (p = 0.709). Both the untreated HCT116 cells and those treated with purified EFA1 had significantly less proliferation than HCT116 treatment with unpurified EFA1 and the E. faecalis/HCT116 colon cancer cell co-culture [0.57 (0.02), p < 0.001] (Figure 6A). The least cell proliferation was seen in HCT116 cells that were co-cultured with E. faecalis and purified bacteriophage EFA1 with mean (SE) SRB absorbance of 0.13 (0.02) equivalent to a 77.2% reduction in cell proliferation (p < 0.001) (Figure 6A). The RPMI growth media and E. faecalis growing in RPMI showed minimal SRB absorbance.
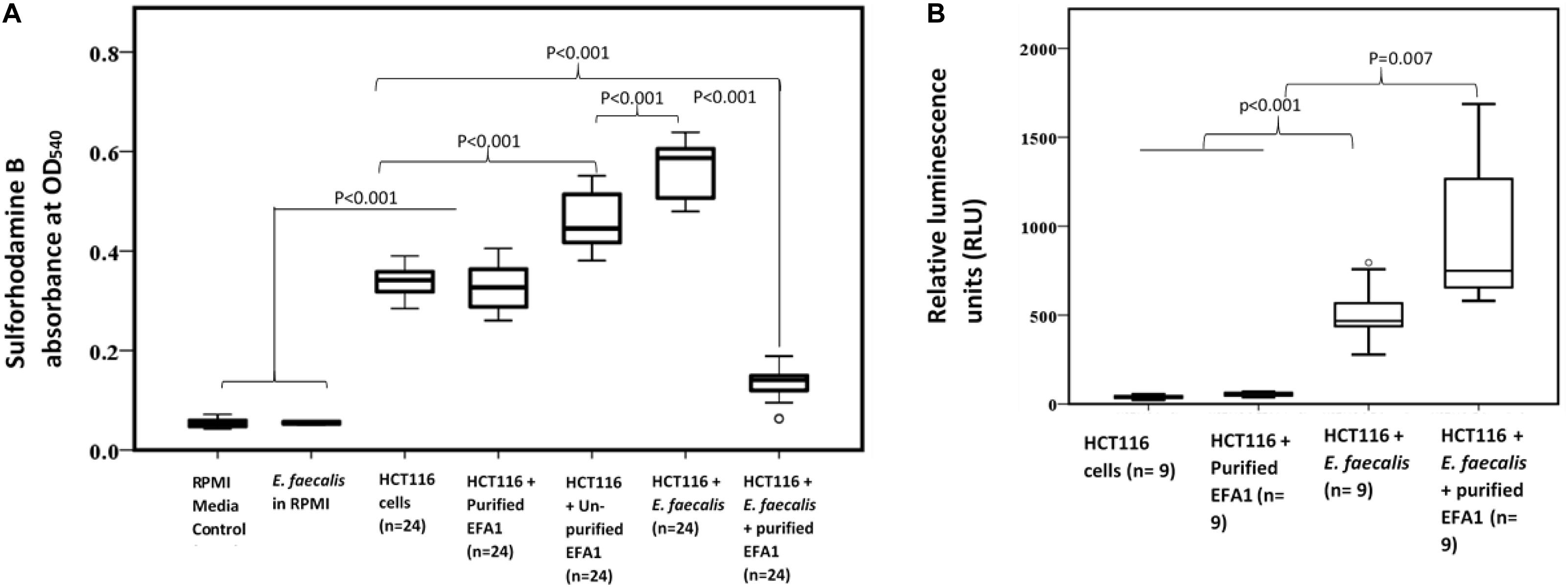
Figure 6. Bacteriophage EFA modulates the effects of E. faecalis on HCT116 cancer cell line by increasing ROS production and attenuating cancer cell proliferation. (A) Cancer cell proliferation was significantly increased in E. faecalis co-cultured HCT116 cells compared to all other cultures. The RPMI growth media and E. faecalis growing in RPMI showed minimal SRB absorbance. HCT116 cells grown with purified bacteriophage EFA1, and the HCT116 cells grown on their own, had similar levels of proliferation. Treatment of the HCT116 cells/E. faecalis co-culture with purified bacteriophage EFA1 resulted in significant reduction of cancer cell growth, to levels below those of the control HCT116 culture. (B) ROS production was significantly increased in the HCT116/E. faecalis co-culture with purified EFA1, and to a lesser extent in the HCT116/E. faecalis co-culture, compared to HCT116 cells on their own. There was no statistical difference in ROS production between untreated HCT116 and those grown with purified bacteriophage EFA1.
To investigate the ROS production in co-culture, RLU as proxy for ROS production were compared between HCT116 colon cancer cells alone and those in co-culture with E. faecalis (Figure 6B). HCT116 colon cancer cells alone had similar RLU to HCT116 colon cancer cells with added purified bacteriophage EFA1 (P = 0.200). The median (Q1–Q3) RLU for HCT116 cells and the HCT116 with EFA1 were 45.2 (31.2–106.1) and 59.5 (45.5–65.7), respectively. The HCT116 colon cancer cells that were co-cultured with E. faecalis had higher RLU than both non-treated HCT116 cancer cells and those with purified EFA1 (P < 0.001) (Figure 6B). The median (Q1–Q3) RLU for HCT116 co-cultured with E. faecalis was 467.0 (365.7–662.3). The RLU of HCT116 colon cancer cells co-cultured with E. faecalis and purified bacteriophage EFA1 was significantly increased over that of the HCT116 colon cancer cell pure culture, HCT116 colon cancer cells with EFA1 (P < 0.001), and HCT116 colon cancer cells co-cultured with E. faecalis (p = 0.007) (Figure 6B). The mean (Q1-Q3) RLU of the HCT116 colon cancer cells co-cultured with E. faecalis and bacteriophage EFA1 was 748.8 (643.8–1475.1).
In this study we describe the isolation and characterization of the lytic bacteriophage EFA1 which targets E. faecalis. We also describe its modulation of the growth stimulating effects of E. faecalis in a colon cancer cell co-culture. There have not been any other studies investigating the actions of Enterococcus bacteriophages in colon cancer cells and E. faecalis bacterial co-cultures. EFA1 belongs to the family Siphoviridae. The bacteriophage host range did not extend to the other bacterial species tested. As such, EFA1 has the potential to minimize impact on other microbes if used in a diverse microbial environment. In terms of its replication kinetics, the bacteriophage had a latency of 20 min, and a burst size of 120 PFU/bacterial cell. This compares to a latency of 30 min and burst size of 116 for the E. faecium bacteriophage IME-Efm1 (Wang et al., 2014), which is the closest related bacteriophage whose replication kinetics are known. To date there are at least 80 Enterococcus bacteriophage genomic sequences deposited in NCBI GenBank (Accessed 31 July 2020). The diversity among these viruses is very broad. EFA1 is most closely related to the E. faecalis bacteriophage Santor1. These two are part of a monophyletic group of 11 bacteriophages which target E. faecalis or E. faecium, and which also form a larger clade with bacteriophages infecting Lactobacillus and Streptococcus hosts, but which cluster more distantly from other bacteriophages which target E. faecalis hosts. The genomic arrangement and predicted putative genes found in the bacteriophage EFA1 genome infers a lytic bacteriophage lifestyle and therefore supports its potential suitability to be used in therapy. This was further supported by the lack of toxins or factors that may enhance bacterial virulence such as antibiotic resistance (Mathew, 2016). Some bacteriophages have been shown to carry antibiotic resistant genes that may be transferred between biomes (Muniesa et al., 2013). While genomic analysis of EFA1 did not show any putative genes coding for such bacterial virulence enhancing proteins, ORF33 in EFA1 coded for a putative beta-lactamase protein. However, amino-acid alignment with the bacterial beta-lactamase sequence showed very little identity between the two proteins. Further, screening with the antibiotic resistance CARD database did not show any positive hits for putative antibiotic resistance genes (Jia et al., 2017) in the EFA1 genome. The InterProScan (Mitchell et al., 2019) and Prosite (Sigrist et al., 2013) did not reveal any active sites or beta-lactamase domains required for catalysis (Moali et al., 2003) in the EFA1 genome either. While the ORF 33 genetic element is currently apparently non-functional as a beta-lactamase, it may have allowed the transfer of E. faecalis antibiotic resistance by bacteriophages in the past. ORF33 is located among genes coding for putative DNA manipulation, and it is possible its function is in this area, or otherwise unknown, rather than that of a bacterial beta-lactamase. Most of the Enterococcus bacteriophages that form a clade with EFA1 also contain a genetic element with some similarity to ORF 33, and so this may represent an evolutionary link between closely related viruses.
Functionally, bacteriophage EFA1 was capable of disrupting E. faecalis biofilms within 2 h of treatment. This was evidenced using confocal microscope imaging that revealed sparse cells on bacteriophage treated biofilm compared to untreated biofilm. Quantification of the biofilm mass supported these findings. With increased exposure to the bacteriophage, the biofilm mass began to steadily increase. However, it is likely that this consistent increase was not due to bacterial resistance to EFA1, as adding fresh media and EFA1 after 24 h resulted in biofilm reduction similar to that seen after exposure for 2 h. In this connection, studies with the E. coli bacteriophage T4 in E. coli mono-biofilms revealed increased bacterial growth after prolonged treatment from 4 to 6 h (Corbin et al., 2001). This phenomenon was not seen in planktonic cultures but considered to be unique to biofilms and suggested to be important in maintenance of the bacteria and bacteriophage ecosystem (Sutherland et al., 2004). These findings raise important issues with respect to potential applications of bacteriophages such as EFA1 in therapy. For instance, they highlight the need for further experiments to more precisely define these host-parasite interactions and kinetics, so that timing of bacteriophage treatment regimens results in optimal biofilm degradation.
Since E. faecalis has been shown to be closely linked to CRC, we investigated the effect of this bacteria on the proliferation of HCT116 colon cancer cells. Similar to the effects of oncobacteria such as F. nucleatum (Rubinstein et al., 2013, 2019), E. faecalis increased the proliferation of HCT116 colon cancer cells. We found that when we exposed HCT116 cells to unpurified EFA1 (which did not have treatment to rid remnant lipoteichoic acid and/or other cell debris), there was a mitogenic effect and stimulation of cancer cell growth. This effect was lost when EFA1 was purified to remove bacterial cell debris. When treated with purified bacteriophage EFA1, E. faecalis induced HCT116 colon cancer cell proliferation was significantly attenuated to levels below that of untreated HCT116 colon cancer cells. To investigate this further we assayed for ROS production. In HCT116 cells co-cultured with E. faecalis there was a significant increase, while exposure of HCT116 to purified EFA1 alone resulted in no change, in ROS levels. Our findings also showed significantly increased ROS production, to levels higher than in the HCT116/E. faecalis co-culture, when purified bacteriophage EFA1 was added to the HCT116/E. faecalis co-culture. We could not find any reports investigating the effects of bacteriophage and associated bacteria on ROS production in cancer cells or mammalian cells other than in leukocytes (Przerwa et al., 2006; Miedzybrodzki et al., 2008; Miernikiewicz et al., 2013). In these studies, there was a decrease in ROS when bacteriophage T4 treated granulocytes were co-cultured with E. coli, in contrast with our findings. Therefore, it may be that this difference in ROS induction is dependent on cell type.
The role of ROS in cancer (Liou and Storz, 2010) and in cancer therapy (Trachootham et al., 2009) has been reviewed. It is known that cells express antioxidants that neutralize ROS while maintaining sufficient levels needed for cellular signaling (Gonzalez et al., 2002). In some tumorigenic events, increased intracellular ROS promotes tumor progression (Jackson, 1994). However, high and excessive ROS levels are induced by chemotherapy agents leading to cell cycle arrest and cell death (Yang H. et al., 2018). Although the exact mechanism and effect of ROS production in our system is unknown, and requires further investigation, the excessive ROS production could contribute to the significantly lower cancer cell proliferation seen when the HCT116/E. faecalis co-culture was treated with EFA1.
The bacteriophage EFA1 isolated from wastewater was fully characterized and its whole genome sequenced. EFA1 was capable of disrupting E. faecalis mono-biofilms. Further, while E. faecalis enhanced the proliferation of HCT116 colon cancer cells, in the presence of EFA1 the proliferation was significantly inhibited with an associated increase in ROS production. EFA1 alone did not significantly alter the proliferation or ROS production of HCT116 cells. Such findings may lead to further testing of bacteriophages such as EFA1 in the control of oncobacteria induced cancer cell growth.
The authors declare that all relevant data supporting the findings of the study are available in this article and the Supplementary Figure file, or from the corresponding author upon reasonable request. The complete genome sequence of bacteriophage EFA1 has been submitted to NCBI GenBank under accession number MT857001.
JT and MK: conceptualization, data analysis, and writing—original draft. MK and TM-A: mammalian cell and bacterial co-culture. MK and SB: genomic. MK and HK: electron microscope imaging and biofilm. JT, HC, and SP: supervision. All authors contributed to writing, reviewing, and editing of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to acknowledge La Trobe University, the Olivia Newton John Cancer Research Institute, and the La Trobe Institute for Molecular Science Bioimaging facility.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.650849/full#supplementary-material
Al-Hebshi, N. N., Nasher, A. T., Maryoud, M. Y., Homeida, H. E., Chen, T., Idris, A. M., et al. (2017). Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci. Rep. 7:1834.
Azam, A. H., and Tanji, Y. (2019). Bacteriophage-host arm race: an update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl. Microbiol. Biotechnol. 103, 2121–2131. doi: 10.1007/s00253-019-09629-x
Balamurugan, R., Rajendiran, E., George, S., Samuel, G. V., and Ramakrishna, B. S. (2008). Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J. Gastroenterol. Hepatol. 23, 1298–1303.
Barnes, A. M. T., Dale, J. L., Chen, Y., Manias, D. A., Greenwood Quaintance, K. E., Karau, M. K., et al. (2017). Enterococcus faecalis readily colonizes the entire gastrointestinal tract and forms biofilms in a germ-free mouse model. Virulence 8, 282–296. doi: 10.1080/21505594.2016.1208890
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., Rapp, B. A., and Wheeler, D. L. (2000). GenBank. Nucleic Acids Res. 28, 15–18.
Bhatt, A. P., Redinbo, M. R., and Bultman, S. J. (2017). The role of the microbiome in cancer development and therapy. Ca Cancer J. Clin. 67, 326–344. doi: 10.3322/caac.21398
Branston, S. D., Wright, J., and Keshavarz-Moore, E. (2015). A non-chromatographic method for the removal of endotoxins from bacteriophages. Biotechnol. Bioeng. 112, 1714–1719. doi: 10.1002/bit.25571
Cani, P. D. (2018). Human gut microbiome: hopes, threats and promises. Gut 67, 1716–1725. doi: 10.1136/gutjnl-2018-316723
Corbin, B. D., Mclean, R. J., and Aron, G. M. (2001). Bacteriophage T4 multiplication in a glucose-limited Escherichia coli biofilm. Can. J. Microbiol. 47, 680–684. doi: 10.1139/w01-059
De Lastours, V., Maugy, E., Mathy, V., Chau, F., Rossi, B., Guerin, F., et al. (2017). Ecological impact of ciprofloxacin on commensal enterococci in healthy volunteers. J. Antimicrob. Chemother. 72, 1574–1580. doi: 10.1093/jac/dkx043
Drees, M., Snydman, D. R., Schmid, C. H., Barefoot, L., Hansjosten, K., Vue, P. M., et al. (2008). Prior environmental contamination increases the risk of acquisition of vancomycin-resistant enterococci. Clin. Infect. Dis. 46, 678–685. doi: 10.1086/527394
Dubin, K., and Pamer, E. G. (2014). Enterococci and their interactions with the intestinal microbiome. Microbiol. Spectr. 5:10.1128/microbiolsec.BAD–0014–2016. doi: 10.1128/microbiolspec.BAD-0014-2016
Franz, C. M., Stiles, M. E., Schleifer, K. H., and Holzapfel, W. H. (2003). Enterococci in foods–a conundrum for food safety. Int. J. Food. Microbiol. 88, 105–122. doi: 10.1016/s0168-1605(03)00174-0
Gaiser, R. A., Halimi, A., Alkharaan, H., Lu, L., Davanian, H., Healy, K., et al. (2019). Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut 68, 1–9. doi: 10.1007/978-1-62703-287-2_1
Garneau, J. R., Depardieu, F., Fortier, L. C., Bikard, D., and Monot, M. (2017). PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 7:8292.
Gonzalez, C., Sanz-Alfayate, G., Agapito, M. T., Gomez-Nino, A., Rocher, A., and Obeso, A. (2002). Significance of Ros in oxygen sensing in cell systems with sensitivity to physiological hypoxia. Respir Physiol. Neurobiol. 132, 17–41. doi: 10.1016/s1569-9048(02)00047-2
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). Crisprfinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57.
Hansen, M. F., Svenningsen, S. L., Røder, H. L., Middelboe, M., and Burmølle, M. (2019). Big impact of the tiny: bacteriophage–bacteria interactions in biofilms. Trends Microbiol. 27, 739–752. doi: 10.1016/j.tim.2019.04.006
Helmink, B. A., Khan, M. A. W., Hermann, A., Gopalakrishnan, V., and Wargo, J. A. (2019). The microbiome, cancer, and cancer therapy. Nat. Med. 25, 377–388.
Hollenbeck, B. L., and Rice, L. B. (2012). Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 3, 421–433. doi: 10.4161/viru.21282
Holt, R. A., and Cochrane, K. (2017). Tumor potentiating mechanisms of fusobacterium nucleatum, a multifaceted microbe. Gastroenterology 152, 694–696. doi: 10.1053/j.gastro.2017.01.024
Hota, B. (2004). Contamination, disinfection, and cross-colonization: are hospital surfaces reservoirs for nosocomial infection? Clin. Infect. Dis. 39, 1182–1189. doi: 10.1086/424667
Huycke, M. M., Abrams, V., and Moore, D. R. (2002). Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell Dna. Carcinogenesis 23, 529–536. doi: 10.1093/carcin/23.3.529
Jackson, J. H. (1994). Potential molecular mechanisms of oxidant-induced carcinogenesis. Environ. Health Perspect. 102(Suppl. 10), 155–157. doi: 10.2307/3432232
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). Card 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573.
Jonczyk-Matysiak, E., Lusiak-Szelachowska, M., Klak, M., Bubak, B., Miedzybrodzki, R., Weber-Dabrowska, B., et al. (2015). The effect of bacteriophage preparations on intracellular killing of bacteria by phagocytes. J. Immunol. Res. 2015:482863.
Kabwe, M., Brown, T., Speirs, L., Ku, H., Leach, M., Chan, H. T., et al. (2020). Novel bacteriophages capable of disrupting biofilms from clinical strains of Aeromonas hydrophila. Front. Microbiol. 11:194. doi: 10.3389/fmicb.2020.00194
Kabwe, M., Brown, T. L., Dashper, S., Speirs, L., Ku, H., Petrovski, S., et al. (2019). Genomic, morphological and functional characterisation of novel bacteriophage Fnu1 capable of disrupting Fusobacterium nucleatum biofilms. Sci. Rep. 9:9107.
Kharrat, N., Assidi, M., Abu-Elmagd, M., Pushparaj, P. N., Alkhaldy, A., Arfaoui, L., et al. (2019). Data mining analysis of human gut microbiota links Fusobacterium spp. with colorectal cancer onset. Bioinformation 15, 372–379. doi: 10.6026/97320630015372
Kho, Z. Y., and Lal, S. K. (2018). The human gut microbiome - a potential controller of wellness and disease. Front. Microbiol. 9:1835. doi: 10.3389/fmicb.2018.01835
Kropec, A., Hufnagel, M., Zimmermann, K., and Huebner, J. (2005). In vitro assessment of the host response against Enterococcus faecalis used in probiotic preparations. Infection 33, 377–379. doi: 10.1007/s15010-005-5063-7
Laslett, D., and Canback, B. (2004). Aragorn, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16. doi: 10.1093/nar/gkh152
Lennard, K. S., Goosen, R. W., and Blackburn, J. M. (2016). Bacterially-associated transcriptional remodelling in a distinct genomic subtype of colorectal cancer provides a plausible molecular basis for disease development. PLoS One 11:e0166282. doi: 10.1371/journal.pone.0166282
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259.
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-Se On-line: integrating search and context for analysis of transfer Rna genes. Nucleic Acids Res. 44, W54–W57.
Lusiak-Szelachowska, M., Weber-Dabrowska, B., and Gorski, A. (2020). Bacteriophages and lysins in biofilm control. Virol Sin. 35, 125–133. doi: 10.1007/s12250-019-00192-3
Mathew, S. (2016). A need for careful consideration of bacteriophage therapy. Indian J. Med. Res. 144, 482–483. doi: 10.4103/0971-5916.198691
Merritt, J. H., Kadouri, D. E., and O’toole, G. A. (2005). Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 1, Unit 1B.1.
Miedzybrodzki, R., Switala-Jelen, K., Fortuna, W., Weber-Dabrowska, B., Przerwa, A., Lusiak-Szelachowska, M., et al. (2008). Bacteriophage preparation inhibition of reactive oxygen species generation by endotoxin-stimulated polymorphonuclear leukocytes. Virus Res. 131, 233–242. doi: 10.1016/j.virusres.2007.09.013
Miernikiewicz, P., Dabrowska, K., Piotrowicz, A., Owczarek, B., Wojas-Turek, J., Kicielinska, J., et al. (2013). T4 phage and its head surface proteins do not stimulate inflammatory mediator production. PloS One 8:e71036. doi: 10.1371/journal.pone.0071036
Mitchell, A. L., Attwood, T. K., Babbitt, P. C., Blum, M., Bork, P., Bridge, A., et al. (2019). InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360.
Mitsuhashi, K., Nosho, K., Sukawa, Y., Matsunaga, Y., Ito, M., Kurihara, H., et al. (2015). Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget 6, 7209–7220. doi: 10.18632/oncotarget.3109
Moali, C., Anne, C., Lamotte-Brasseur, J., Groslambert, S., Devreese, B., Van Beeumen, J., et al. (2003). Analysis of the importance of the metallo-beta-lactamase active site loop in substrate binding and catalysis. Chem. Biol. 10, 319–329. doi: 10.1016/s1074-5521(03)00070-x
Moreno-Gallego, J. L., Chou, S. P., Di Rienzi, S. C., Goodrich, J. K., Spector, T. D., Bell, J. T., et al. (2019). Virome diversity correlates with intestinal microbiome diversity in adult monozygotic twins. Cell Host. Microbe. 25, 261e5–272e5.
Muniesa, M., Colomer-Lluch, M., and Jofre, J. (2013). Could bacteriophages transfer antibiotic resistance genes from environmental bacteria to human-body associated bacterial populations? Mob. Genet. Elements 3:e25847. doi: 10.4161/mge.25847
Nishimura, Y., Yoshida, T., Kuronishi, M., Uehara, H., Ogata, H., and Goto, S. (2017). Viptree: the viral proteomic tree server. Bioinformatics 33, 2379–2380. doi: 10.1093/bioinformatics/btx157
Nueno-Palop, C., and Narbad, A. (2011). Probiotic assessment of Enterococcus faecalis Cp58 isolated from human gut. Int. J. Food Microbiol. 145, 390–394. doi: 10.1016/j.ijfoodmicro.2010.12.029
Orellana, E. A., and Kasinski, A. L. (2016). Sulforhodamine B (Srb) assay in cell culture to investigate cell proliferation. Bio. Protoc. 6:e1984.
Parhi, L., Alon-Maimon, T., Sol, A., Nejman, D., Shhadeh, A., Fainsod-Levi, T., et al. (2020). Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 11:3259.
Przerwa, A., Zimecki, M., Switala-Jelen, K., Dabrowska, K., Krawczyk, E., Luczak, M., et al. (2006). Effects of bacteriophages on free radical production and phagocytic functions. Med. Microbiol. Immunol. 195, 143–150. doi: 10.1007/s00430-006-0011-4
Rajagopala, S. V., Vashee, S., Oldfield, L. M., Suzuki, Y., Venter, J. C., Telenti, A., et al. (2017). The human microbiome and cancer. Cancer Prev. Res. (Phila) 10, 226–234.
Rubinstein, M. R., Baik, J. E., Lagana, S. M., Han, R. P., Raab, W. J., Sahoo, D., et al. (2019). Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/beta-catenin modulator Annexin A1. Embo Rep. 20:e47638.
Rubinstein, M. R., Wang, X., Liu, W., Hao, Y., Cai, G., and Han, Y. W. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host. Microbe. 14, 195–206. doi: 10.1016/j.chom.2013.07.012
Sender, R., Fuchs, S., and Milo, R. (2016). Are we really vastly outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell 164, 337–340. doi: 10.1016/j.cell.2016.01.013
Sigrist, C. J., De Castro, E., Cerutti, L., Cuche, B. A., Hulo, N., Bridge, A., et al. (2013). New and continuing developments at Prosite. Nucleic Acids Res. 41, D344–D347.
Skehan, P., Storeng, R., Scudiero, D., Monks, A., Mcmahon, J., Vistica, D., et al. (1990). New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 82, 1107–1112. doi: 10.1093/jnci/82.13.1107
Sutherland, I. W., Hughes, K. A., Skillman, L. C., and Tait, K. (2004). The interaction of phage and biofilms. Fems Microbiol. Lett. 232, 1–6. doi: 10.1016/s0378-1097(04)00041-2
Tinrat, S., Khuntayaporn, P., Thirapanmethee, K., and Chomnawang, M. T. (2018). In vitro assessment of Enterococcus faecalis Mtc 1032 as the potential probiotic in food supplements. J. Food Sci. Technol. 55, 2384–2394. doi: 10.1007/s13197-018-3155-5
Trachootham, D., Alexandre, J., and Huang, P. (2009). Targeting cancer cells by Ros-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 8, 579–591. doi: 10.1038/nrd2803
Van Belleghem, J. D., Dabrowska, K., Vaneechoutte, M., Barr, J. J., and Bollyky, P. L. (2018). Interactions between bacteriophage, bacteria, and the mammalian immune system. Viruses 11:10. doi: 10.3390/v11010010
Wang, G., Liu, Q., Pei, Z., Wang, L., Tian, P., Liu, Z., et al. (2020a). The diversity of the Crispr-cas system and prophages present in the genome reveals the co-evolution of Bifidobacterium pseudocatenulatum and phages. Front. Microbiol. 11:1088. doi: 10.3389/fmicb.2020.01088
Wang, J., Da, R., Tuo, X., Cheng, Y., Wei, J., Jiang, K., et al. (2020b). Probiotic and safety properties screening of Enterococcus faecalis from healthy chinese infants. Probiotics Antimicrob. Proteins 12, 1115–1125. doi: 10.1007/s12602-019-09625-7
Wang, X., Allen, T. D., May, R. J., Lightfoot, S., Houchen, C. W., and Huycke, M. M. (2008). Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 68, 9909–9917. doi: 10.1158/0008-5472.can-08-1551
Wang, X., and Huycke, M. M. (2007). Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology 132, 551–561. doi: 10.1053/j.gastro.2006.11.040
Wang, Y., Wang, W., Lv, Y., Zheng, W., Mi, Z., Pei, G., et al. (2014). Characterization and complete genome sequence analysis of novel bacteriophage Ime-Efm1 infecting Enterococcus faecium. J. Gen. Virol. 95, 2565–2575. doi: 10.1099/vir.0.067553-0
Weiner-Lastinger, L. M., Abner, S., Edwards, J. R., Kallen, A. J., Karlsson, M., Magill, S. S., et al. (2020). Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: summary of data reported to the national healthcare safety network, 2015-2017. Infect. Control Hosp. Epidemiol. 41, 1–18. doi: 10.1086/668770
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Yachida, S., Mizutani, S., Shiroma, H., Shiba, S., Nakajima, T., Sakamoto, T., et al. (2019). Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 25, 968–976. doi: 10.1038/s41591-019-0458-7
Yamamura, K., Baba, Y., Nakagawa, S., Mima, K., Miyake, K., Nakamura, K., et al. (2016). Human microbiome Fusobacterium Nucleatum in esophageal cancer tissue is associated with prognosis. Clin. Cancer Res. 22, 5574–5581. doi: 10.1158/1078-0432.ccr-16-1786
Yang, H., Villani, R. M., Wang, H., Simpson, M. J., Roberts, M. S., Tang, M., et al. (2018a). The role of cellular reactive oxygen species in cancer chemotherapy. J. Exp. Clin. Cancer Res. 37:266.
Yang, S. F., Huang, H. D., Fan, W. L., Jong, Y. J., Chen, M. K., Huang, C. N., et al. (2018b). Compositional and functional variations of oral microbiota associated with the mutational changes in oral cancer. Oral Oncol. 77, 1–8. doi: 10.1016/j.oraloncology.2017.12.005
Zervos, M. J., Terpenning, M. S., Schaberg, D. R., Therasse, P. M., Medendorp, S. V., and Kauffman, C. A. (1987). High-level aminoglycoside-resistant enterococci. Colonization of nursing home and acute care hospital patients. Arch. Intern. Med. 147, 1591–1594. doi: 10.1001/archinte.147.9.1591
Zhao, H., Chu, M., Huang, Z., Yang, X., Ran, S., Hu, B., et al. (2017). Variations in oral microbiota associated with oral cancer. Sci. Rep. 7:11773.
Keywords: Enterococcus faecalis, bacteriophage, genomics, biofilm, colon cancer proliferation, reactive oxygen species
Citation: Kabwe M, Meehan-Andrews T, Ku H, Petrovski S, Batinovic S, Chan HT and Tucci J (2021) Lytic Bacteriophage EFA1 Modulates HCT116 Colon Cancer Cell Growth and Upregulates ROS Production in an Enterococcus faecalis Co-culture System. Front. Microbiol. 12:650849. doi: 10.3389/fmicb.2021.650849
Received: 08 January 2021; Accepted: 08 March 2021;
Published: 31 March 2021.
Edited by:
Shuai Le, Army Medical University, ChinaReviewed by:
Alan J. Wolfe, Loyola University Chicago, United StatesCopyright © 2021 Kabwe, Meehan-Andrews, Ku, Petrovski, Batinovic, Chan and Tucci. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joseph Tucci, ai50dWNjaUBsYXRyb2JlLmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.