- State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Institute of Plant Virology, Ningbo University, Ningbo, China
Cowpea mild mottle virus (CPMMV; genus Carlavirus) can be a destructive pathogen of soybean but there is little information about its distribution on soybean in China. Here, we collected soybean plants with virus-like symptoms from 11 fields widely scattered within China, and used high-throughput sequencing to determine their virome. Most samples (8/11) were co-infected by the well-studied potyvirus soybean mosaic virus (SMV) and CPMMV, and the remaining three samples were singly infected with CPMMV. The near-complete genome sequences of the 11 CPMMV isolates were determined and phylogenetic analysis showed that they constituted a new genetic clade. One recombination event was detected among the CPMMV sequences, and the isolate CPMMV_JL_CC was identified as recombinant. In mechanical inoculation assays, co-infection by CPMMV and SMV resulted in an enhancement of disease symptoms, but decreased the expression level of the genomic RNAs and CP of CPMMV, without significantly affecting SMV accumulation. The interaction between these viruses needs further investigation.
Introduction
Soybean [Glycine max (L.) Merr.], one of the most important oil and cash crops worldwide, provides protein-rich food and feed for humans and animals (Wilson, 2008). Many different pests and pathogens attack soybean crops causing significant losses in yield and quality (Widyasari et al., 2020) and some of the most serious pathogens are viruses (Wrather and Koenning, 2006). Soybean virus diseases typically cause 10–30% yield losses but losses of 50–100% have been reported from severe outbreaks (Hartman et al., 2011; Song et al., 2016). Soybean mosaic virus (SMV) is the most important virus infecting soybean worldwide but a wide variety of other viruses have been reported, including cucumber mosaic virus, cowpea mild mottle virus (CPMMV) and alfalfa mosaic virus (Hill and Whitham, 2014).
SMV, a member of the genus Potyvirus in the family Potyviridae, has a single-strand RNA genome that encodes a single large open reading frame (ORF) (Hajimorad et al., 2018; Gao et al., 2019). The translated polyprotein yields a series of multifunctional proteins through proteolysis, which are commonly named P1, HC-Pro, P3, PIPO (a product of slippage in the P3 coding sequence), 6K1, CI, 6K2, VPg, NIa-Pro, NIb, and CP (Hajimorad et al., 2018; Gao et al., 2019). SMV is naturally transmitted by aphids in a nonpersistent manner but can also be transmitted through infected seeds (Widyasari et al., 2020). Plants infected with SMV usually have mottling on their leaves, stem necrosis and are stunted. Strains of SMV have been recognized based on their genomic similarity and the response of different soybean cultivars. In this way, isolates were classified into seven strains (G1–G7) in the United States (Cho and Goodman, 1979), and more recently into 22 strains (SC1–SC22) in China (Li et al., 2010; Wang et al., 2013).
CPMMV is another economically important virus that causes serious damage to soybean production particularly in Brazil (Zanardo et al., 2014a,b). It was first reported from cowpea in Ghana and subsequently found in other leguminous crops across the world (Brunt and Kenten, 1973; Zanardo and Carvalho, 2017). CPMMV is a positive-strand RNA virus that belongs to the genus Carlavirus in the family Betaflexiviridae. The genome of CPMMV (7.8–8.9 kb) encodes six ORFs (ORF 1-6) that are translated into the corresponding proteins: Replicase including the RNA-dependent RNA polymerase (RdRp; ORF1), triple gene block proteins (TGBs; ORFs2-4), coat protein (CP; ORF5) and a nucleic-acid-binding protein (NABP; ORF6) (Zanardo and Carvalho, 2017). CPMMV is transmitted by the whitefly Bemisia tabaci in a nonpersistent manner, and the predominant symptoms in soybean are mosaic, stem necrosis and dwarfing (Muniyappa, 1983; Zanardo and Carvalho, 2017). The genomes of CPMMV isolates infecting soybean have been characterized from Brazil, Ghana and India (Brunt and Kenten, 1973; Yadav et al., 2014; Zanardo et al., 2014a,b). Recently, we found CPMMV infecting soybean in Anhui province in China (Wei et al., 2020) but the distribution, importance and biological characteristics of CPMMV in China are largely unknown.
In this study, we collected soybean plants with virus-like symptoms from different regions in China and used high-throughput sequencing (HTS) to investigate the presence of viruses. All the samples contained CPMMV, often in co-infections with SMV, and these CPMMV isolates formed a single clade, distinct from previously reported isolates from elsewhere in the world. Plants co-infected with CPMMV and SMV had enhanced disease symptoms, but had reduced expression levels of the genomic RNA and CP of CPMMV compared to those infected by CPMMV alone.
Materials and Methods
Virus Isolates
Leaves of soybean plants with typical virus-like symptoms were collected from 11 fields among six soybean-growing provinces of China (Jilin, Shandong, Henan, Anhui, Jiangsu and Hubei) during September 2019. Pools of leaves from three to five symptomatic plants in each field were wrapped in plastic bags and placed in dry ice.
RNA Sequencing
Total RNA was extracted from each sample by TRIzol Reagent (Invitrogen, United States). Approximately 10 μg of total RNA was used for transcriptome sequencing. The RNA sequencing was performed by Zhejiang Tiangen (Company, Hangzhou, China). The final cDNA library was constructed using the TruSeq Stranded mRNA Library Prep Kit (Illumina, United States) and sequenced on an Illumina HiSeq 4000 (LC Sciences, United States).
Analysis of Sequencing Reads
The reads were generated through the Illumina paired-end RNA-seq approach, the average insert size for the paired-end library was 300 ± 50 bp. Prior to assembly, the low-quality reads were removed. Clean data were assembled using Trinity software (version 2.8.5). The assembled contigs were first compared with Barcode of Life Data (BOLD) Systems1, and searched against NCBI virus RefSeqs2 using a BlastX algorithm with a cutoff E-value of 10–5.
RT-PCR Detection
About 1.5 μg of total RNA used in the RNA sequencing assay was reversely transcribed to cDNA using a reverse transcription kit (Tiangen Company, Beijing, China). The cDNA (0.5 μL) was amplified by PCR using KOD-FX (Toyobo, Osaka, Japan), with the following conditions: 94°C for 3 min, 34 cycles of 98°C for 10 s, 58°C for 30 s, and 72°C for 2 min. Primers used for amplifying sequences of specific viruses (CPMMV and SMV) are listed in Supplementary Table 2. PCR products were separated on a 1.2% agarose gel.
Phylogenetic Analysis
The nearly complete genome sequences of isolates obtained in this study and reported isolates retrieved from NCBI3 were aligned by MUSCLE method in MEGAX. Phylogenetic trees were constructed using Maximum-Likelihood (ML) methods using the best-fitting model: GTR + G + I (General Time Reversible + Gama Distributed With Invariant Sites). To check the reliability of the constructed trees, the bootstrap test with 1,000 bootstrap replications was used. For the CPMMV phylogenetic tree construct, Indian citrus ringspot virus (Mandarivirus, Alphaflexiviridae) was used as the outgroup. Turnip mosaic virus (Potyvirus, Potyviridae) was used as the outgroup for the SMV phylogenetic tree construct.
Recombination Analysis
Putative recombination events amongst CPMMV isolates and SMV isolates were identified using the recombination detection program RDP5 (Martin et al., 2020), and evaluated using different methods: RDP, GENECONV, BOOTSCAN, MaxiChi, Chimera, SiScan, and 3Seq. Alignments of nucleotide sequences produced in MEGAX were run in RDP5 (P-value cut-off of 0.05). Only recombination events that were detected by three or more methods were considered.
Mechanical Transmission Assay
For inoculation of soybean plants, sap from each of the 11 symptomatic plant pools was used to inoculate a susceptible soybean variety (Jiunong 9). Each inoculum was prepared from 1 g of symptomatic field plant leaves homogenized with 10 mL of 0.01 M phosphate-buffered saline, pH 7.0. After mixing with carborundum, inoculation was performed manually before the trifoliate leaves emerged and the inoculum finally rinsed with tap water. Plants inoculated with phosphate-buffered saline were used as controls. Inoculated plants were grown at 25–28°C (16 h light/8 h dark) in an incubator for 7–10 days, and classified into four phenotype classes based on symptoms of 12 plants per inoculation: symptomless, mosaic, crinkling, stem necrosis. Three independent experiments were conducted to provide data for statistical analysis, Values are means ± Standard Deviation (SD).
RNA Extraction and RT-qPCR
Total RNA was extracted from young trifoliate leaves using TRIzol reagent (Invitrogen, United States). About 1.5 μg of total RNA was reversely transcribed to cDNA using a reverse transcription kit (Tiangen Company, Beijing, China). Real-time PCR was conducted using ChamQTM SYBR qPCR Master Mix (Low ROX Premixed) by an ABI7900HT Sequence Detection System (Applied Biosystems, Carlsbad, CA, United States). The RT-qPCR conditions were as follows: 95°C for 4 min; 40 cycles of 95°C for 10 s and 60°C for 30 s. The relative expression levels of genes were determined using the 2–ΔΔC (t) method (Livak and Schmittgen, 2001). The soybean Actin11 gene was used as an internal control. Three biological and two technical replicates were conducted to determine gene expression. The RT-qPCR primer sequences used in this study are listed in Supplementary Table 2.
Western Blot
Approximately 200 mg soybean leaves were homogenized in 0.3 mL sodium dodecylsulfate (SDS) lysis buffer (100 mm Tris-HCl, pH 6.8, 10% SDS and 2.0% β -mercaptoethanol). The crude extracts were centrifuged at 12,000 × g for 10 min at room temperature, and the resulting supernatant (8 μL per sample plus 2 μL 5 × SDS loading buffer) was electrophoresed in 10–12% SDS-PAGE gels. Western blot analysis was done as previously reported (Zhang et al., 2019). Proteins were transferred to polyvinylidene difluoride (PVDF, Millipore, United States) membranes using the Trans-Blot Turbo transfer system (Bio-Rad, United States). Polyclonal rabbit anti-CPMMV-CP and anti-SMV-CP (at 1:3,000 dilution, synthesized by Huabio, China) were used to detect the respective viruses.
Results
Occurrence of SMV and CPMMV in China
Soybean samples with mosaic and crinkling symptoms on their leaves were collected from 11 locations in six provinces (Jilin, Shandong, Henan, Anhui, Jiangsu and Hubei) in the main soybean producing region of China (Figures 1A–C and Table 1). After extraction of total RNA, high-throughput sequencing (HTS) yielded more than 25 M paired-end reads from each sample (ranging from 25.38 to 54.50 M). BLASTX analysis of the assembled contigs searched against the NCBI virus RefSeq database showed that SMV and CPMMV were widely found in these samples. Most samples (8/11) were co-infected by SMV and CPMMV, and the remaining three samples (AH_FY, HN_XC and HB_JZ) were infected by CPMMV alone (Table 1). These results were confirmed by reverse transcription PCR (RT-PCR) using primers specific for SMV or CPMMV (Supplementary Figure 1). These results indicate that CPMMV is distributed in several soybean producing areas in China and is commonly present in co-infections with SMV.
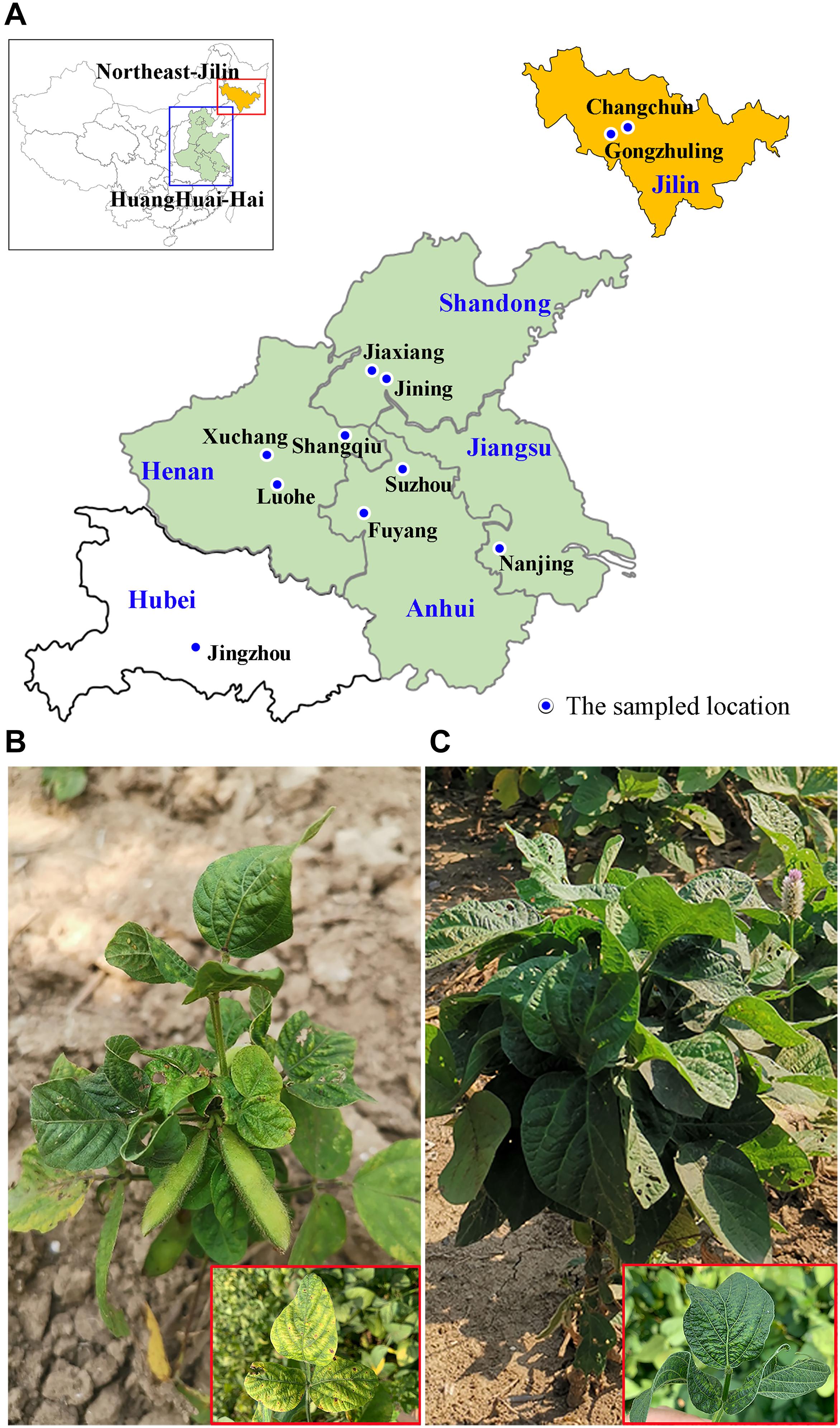
Figure 1. The geographical locations of the symptomatic soybean samples collected in China and the symptoms on leaves of the sampled plants. (A) Map of China (top left corner) showing the study area (red and blue rectangular box). The 11 sampled fields are marked on the enlarged abridged map. (B,C) Leaves of soybean plants displaying typical yellow mosaic (B) and crinkling (C) virus-like symptoms.
Phylogenetic Analysis of SMV and CPMMV Isolates
From the transcriptome sequencing data, the near-complete genome sequences of four SMV isolates and >7kb of four others (Supplementary Figure 2) were assembled and deposited in GenBank (Table 1). To determine the phylogenetic relationships between these eight SMV sequences and other known SMV genomes (G1-G7, United States; SC3, SC7 and SC15, the most important strains in China), we performed phylogenetic analysis based on the nucleotide sequence of the region from P3 to 6K2 using Maximum-Likelihood (ML) methods. The results showed four major clades and that the SMV isolates from China clustered separately from those of USA (Figure 2). Clade IV included all the new SMV isolates from this study (except SMV_JL_GZL from Jilin province) and the SC7 strain, while the isolate SMV_JL_GZL from northeast of China clustered with the SC3 strain. Clades II and III included the USA isolates G1–G7.
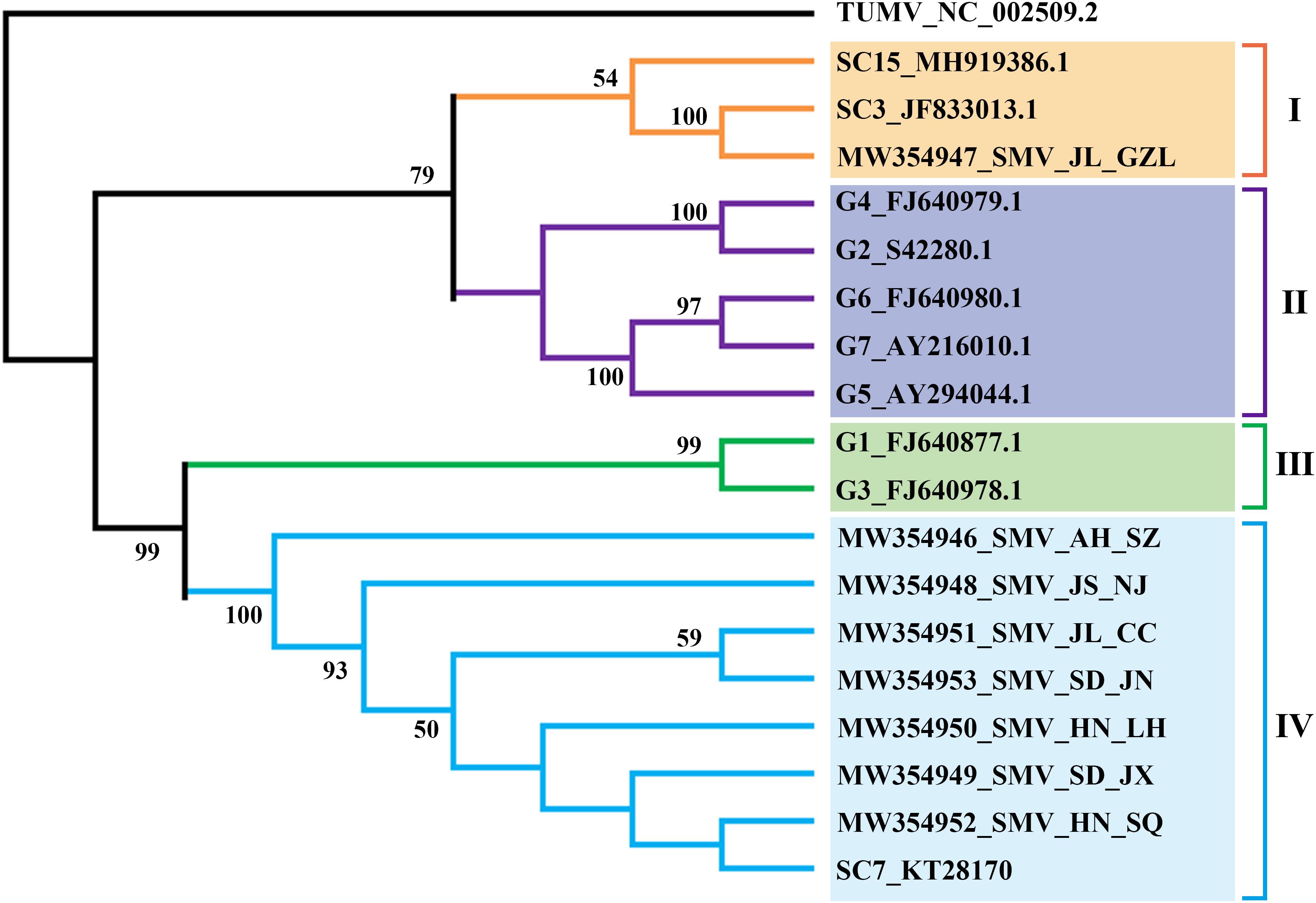
Figure 2. ML-phylogenetic relationships among soybean mosaic virus (SMV) isolates from different countries. The phylogenic tree was constructed using MEGAX. Near-complete genome sequences of SMV isolates were obtained from NCBI (https://www.ncbi.nlm.nih.gov/). The colored bars (blue, orange, green, purple) indicate the different clades observed in the phylogenies.
We assembled the complete CPMMV sequences from each of our 11 samples, deposited them in GenBank (Table 1), and examined the phylogenetic relationships between them and some representative complete CPMMV sequences from Brazil, United States, India, Mexico and Ghana. As shown in Figure 3, there were three lineages. The CPMMV isolates from our samples were closely related to each other within clade III and with a Chinese cowpea isolate (KY420906.1_Hainan). The CPMMV isolates from Brazil (soybean), Mexico (common bean) and Florida (whitefly) were placed in Clade II, while the isolates from Ghana and India were in clade I. Thus, the CPMMV isolates from China form a very distinct cluster. In phylogenetic analyses of each ORF separately, the same pattern of clustering was consistently obtained (Supplementary Table 1 and Supplementary Figure 3).
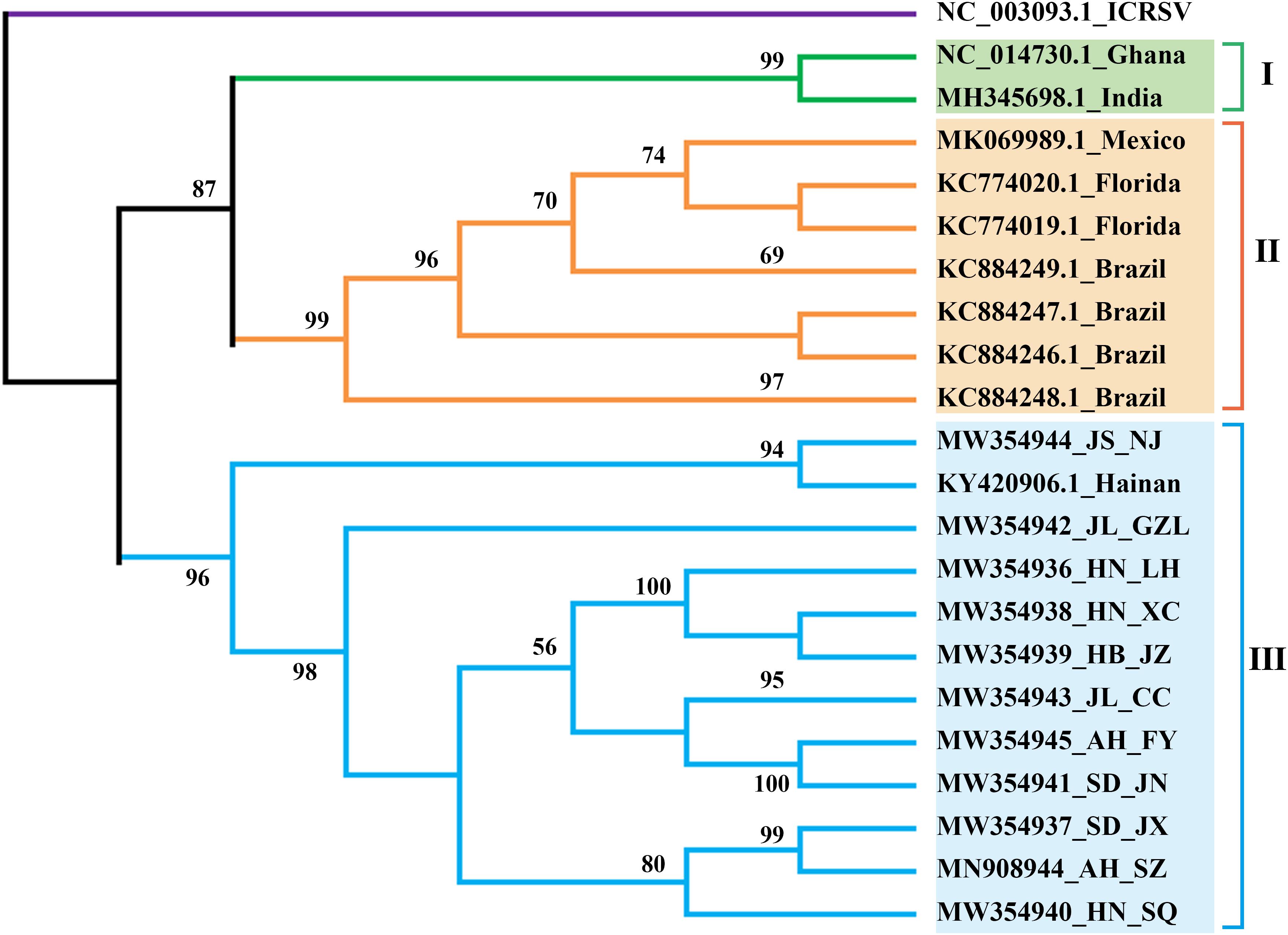
Figure 3. ML-phylogenetic analysis of the complete genomes of cowpea mild mottle virus (CPMMV) isolates from different countries. The phylogenic tree was constructed using MEGAX. Near-complete genome sequences of CPMMV isolates were obtained from NCBI (https://www.ncbi.nlm.nih.gov/). The colored bars (blue, orange, green) indicate the different clades observed in the phylogenies. Blue, China; Orange, Brazil and Florida; Green, India, and Ghana.
Sequence Comparisons of CPMMV Isolates
Because the genetic diversity among CPMMV isolates has been little studied, sequence comparisons were made separately for each ORF of the CPMMV isolates. Among the isolates from China, the nucleotide identity of each ORF was in the range from 95 to 100% but the values were much lower when the Chinese isolates were compared with those from other countries (63.5–82.5%) (Supplementary Figure 4). ORF1 of the Chinese isolates all encoded 1,860 amino acids (Supplementary Table 1) and were very similar to one another (95.3–99.6% nt identity), but had 79.3–81.0% identity with Brazilian isolates and 75 and 64% identity with those from India and Ghana, respectively. In ORF2, the isolates collected in this study had 98.1–99.7% nucleotide identity to one another and the ORF was 11 codons shorter than that of the previously reported CPMMV isolate on cowpea from China. ORF3 and ORF4 were the same size among the isolates from China, Brazil, Florida and India, but not Ghana (Supplementary Table 1). ORF5 encodes the coat protein of 289 aa (Supplementary Table 1), and is the most conserved region among all isolates (Supplementary Figure 4). Among the isolates from China, there was 97.6–100% nt identity and 100% amino acid identity in this ORF, which had 78.3–78.9% nt identity with that of the isolate from Ghana. The ORF6 of Chinese isolates were all the same size and shared 94.9–100% nt identity, but only 59.4–88.5% identity isolates from India and Ghana (Supplementary Figure 4). Overall, our results confirm that the isolates from China are different from those reported in other countries, and represent a unique CPMMV strain.
Recombination Analyses of CPMMV and SMV Isolates
To detect possible recombination events amongst the CPMMV isolates, the complete coding sequences of all known CPMMV isolates in NCBI were analyzed using RDP 5. Four putative recombination events were detected among the isolates but only one among the Chinese isolates (Figure 4A). The isolates CPMMV_HN-SQ and CPMMV_JS-NJ were identified as the respective major and minor parents of CPMMV_JL_CC in a single recombination event identified in the region from nt 2,164 to 3,308 in ORF1 (Figures 4A–C). The beginning and ending breakpoints were identified at nt 2,891 and 3,146. This result was supported by five methods (RDP, p = 2.022 × 10–05; Geneconv, p = 5.858 × 10–04; Bootscan, p = 1.931 × 10–05; MAXchi, p = 2.586 × 10–02; SiScan, p = 2.781 × 10–04) with high confidence and phylogenetic analysis of the putative recombinant and non-recombinant portions also support the conclusion (Supplementary Figure 5).
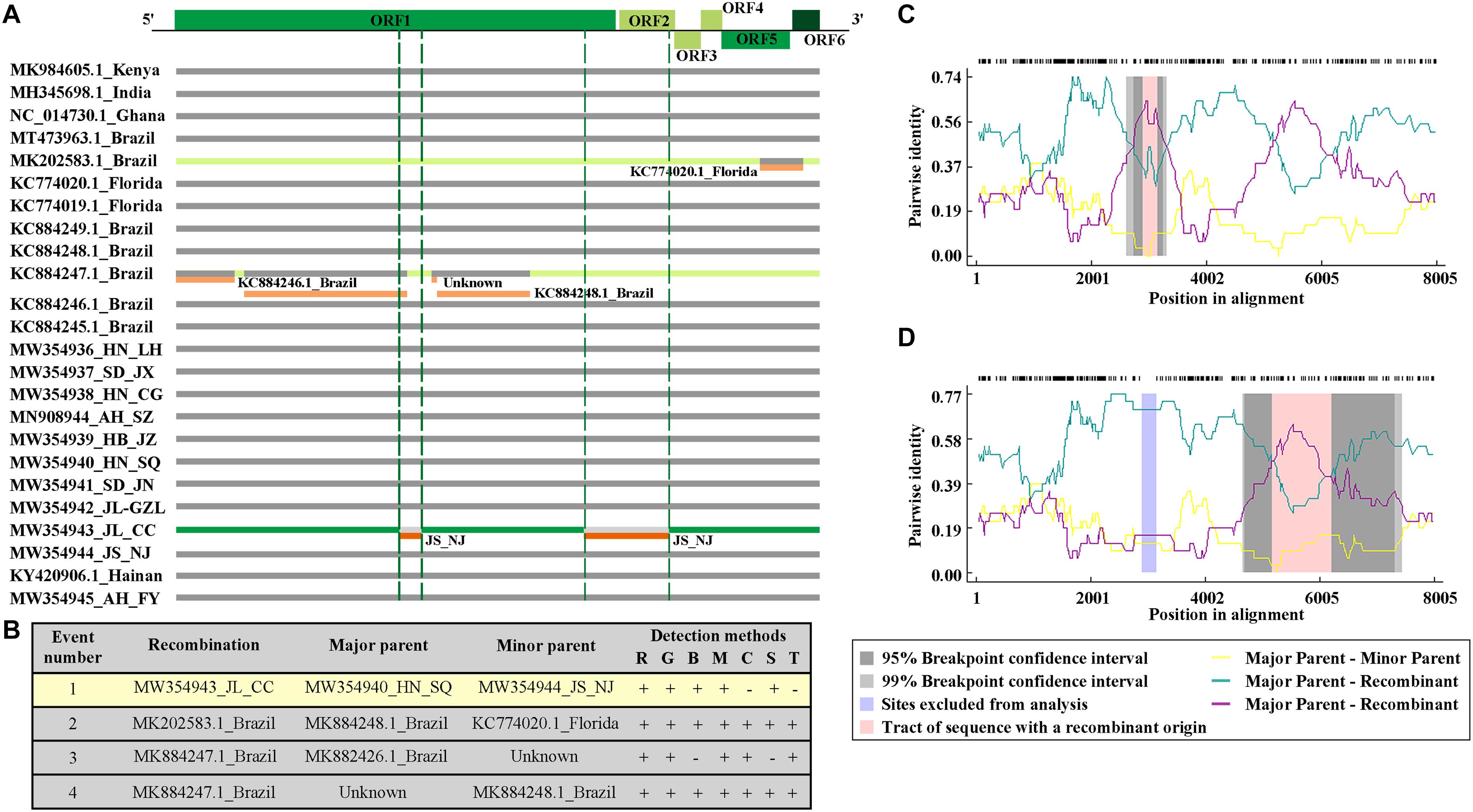
Figure 4. Recombination events among the CPMMV isolates. (A) Schematic representation of the recombination events identified among CPMMV isolates by the RDP 5 program. Each box represents a viral isolate with the recombination events identified by different color (green and orange). The CPMMV genome is shown at the top of the figure. (B) Details of recombination events in the complete coding sequence of CPMMV isolates. Recombination detection methods are represented by letters: R, RDP; G, Genecov; B, Bootscan; M, Maximum v2; C, Chimera; S, Sister scan; 3, 3 Seq. (C,D) Evidence of recombination event in the isolate JL_SMV1.
We also detected six recombination events in the SMV population using the nucleotide sequence covering the region of the genome from P3 to 6K2. Two recombination events occurred in the SMV isolates we identified in this study. Recombination event 1 showed SC7 as the putative major parent and SMV_JL_GZL as the minor parent, which led to recombinant isolate SMV_AH_SZ. SMV_JL_GZL was identified as anther recombinant isolate, with G4 and isolate SMV_AH_SZ as possible parents (Supplementary Figure 6). This result was supported by four to six methods with high confidence.
Symptoms on Mechanically Inoculated Plants
To further characterize the symptoms of CPMMV/SMV caused by the different isolates studied, the samples were used for mechanical transmission assays on susceptible soybean variety “Jiunong 9.” Successful infection was established following inoculation by seven of the mixed sap samples (three to five symptomatic plants) from each field, with systemic symptoms including stem necrosis, mosaic, and crinkled leaves appearing from 7 to 14 days post-inoculation (dpi) (Figure 5A). Most of the plants inoculated with JL_GZL (8.67 ± 1.15/12) developed leaf crinkling while the other plants had leaf mosaic (3.33 ± 1.15/12). About 60% of plants inoculated with sample JL_CC became infected, mainly showing mosaic symptoms (7.00 ± 0.00/12) (Figures 5A,B). RT-qPCR and western-blotting detected RNA and coat protein (CP) of SMV but not CPMMV in these plants (Figures 5C,D) even though the field samples had been co-infected with both viruses. By contrast, plants inoculated with sample SD_JX, developed leaf mosaic (3.33 ± 0.58/12) or stem necrosis (8.67 ± 0.58/12) (Figures 5A,B) and the RT-qPCR and immunoblot analyses showed that CPMMV, but not SMV, was present in the inoculated plants. Inoculation with either JS_NJ or AH_SZ, resulted in co-infection by CPMMV and SMV (Figures 5C,D) with mosaic symptoms on the upper leaves at the early infection stages, followed by systemic necrosis throughout the plants (Figure 5B). HTS results had shown that field sample AH_FY contained only CPMMV (Table 1). Most of the plants mechanically inoculated with AH_FY developed mild stem necrosis symptoms (7.67 ± 1.15/12), while the remainder had leaf mosaic (4.33 ± 1.15/12).
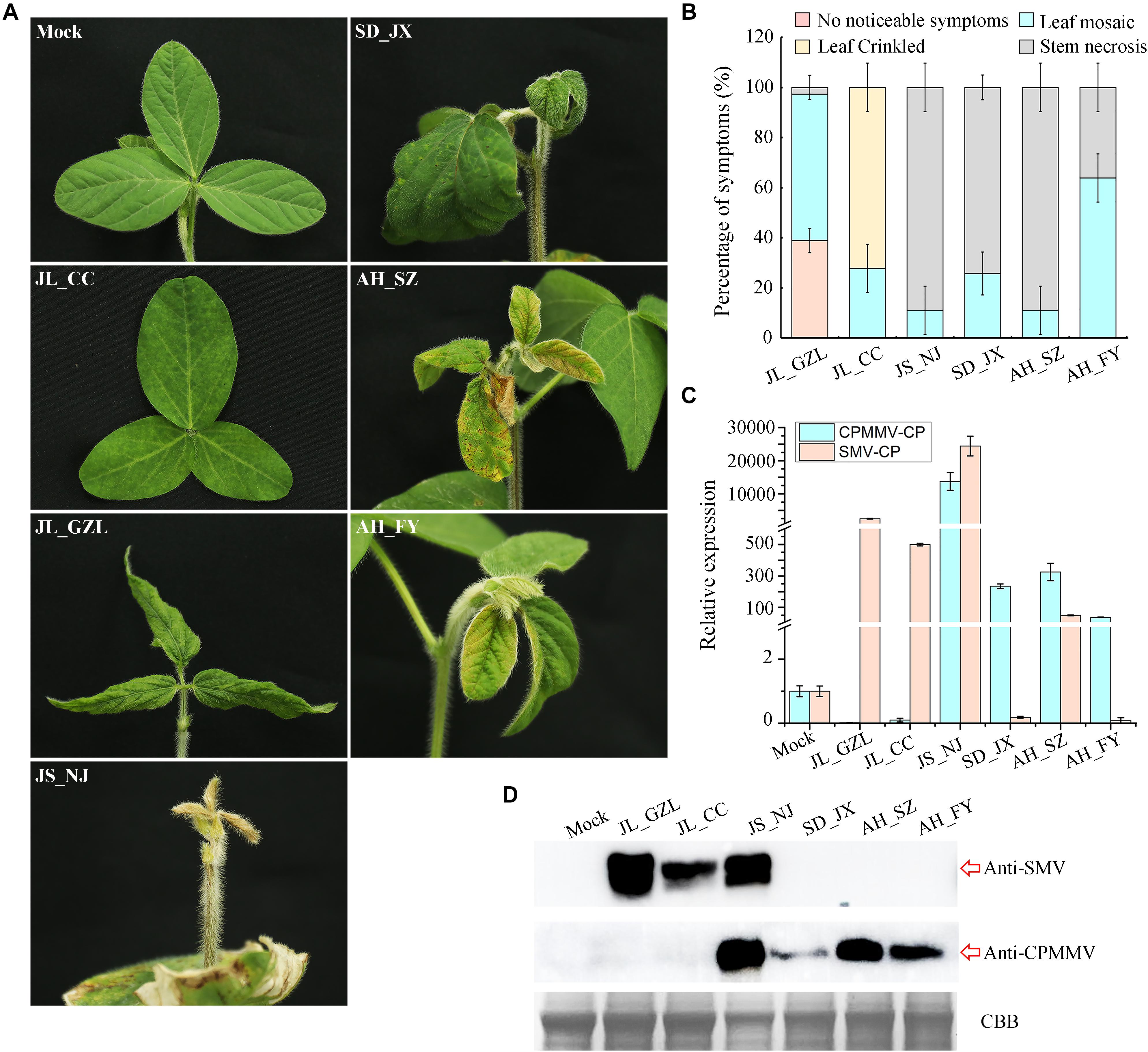
Figure 5. Symptoms of SMV/CPMMV infection on soybean. (A) Photographs of soybean (Jiunong 9) pants 10 days after inoculation with sap from different field locations. (B) The percentages of SMV/CPMMV-infected plants with different grades of disease symptoms, n = 12. (C) SMV and CPMMV RNAs in infected plants as determined by RT-qPCR. Data are shown as means ± SD of three biological replicates. (D) SMV and CPMMV coat protein (CP) protein levels in mock-inoculated and SMV/CPMMV-infected plants as determined by immunoblot analysis. Coomassie brilliant blue (CBB) staining of the same extracts is shown to demonstrate equal loading.
Co-infection of SMV and CPMMV Decreased the Accumulation of CPMMV
To understand the interaction between CPMMV and SMV in co-infection, the sap from mechanically inoculated leaves of isolates AH_FY (CPMMV) and JL_GZL (SMV) were inoculated onto soybean seedlings separately or together. Control (mock) plants were inoculated with phosphate buffer. The plants singly infected with CPMMV or SMV developed mild stem necrosis or mosaic, respectively, at 7 dpi (Figure 6A), while co-infection caused more severe symptoms: systemically infected leaves became crinkled at 5 dpi and developed systemic necrosis at 7 dpi (Figure 6A). RT-qPCR (Figures 6B,C) and western-blotting (Figures 6D,E) at 7 and 14 dpi confirmed the presence of both viruses. Compared to plants inoculated with a single virus, the accumulation of CPMMV genomic RNA and of CP were decreased in co-infected leaves but there were no significant effects on the levels of SMV RNA or CP.
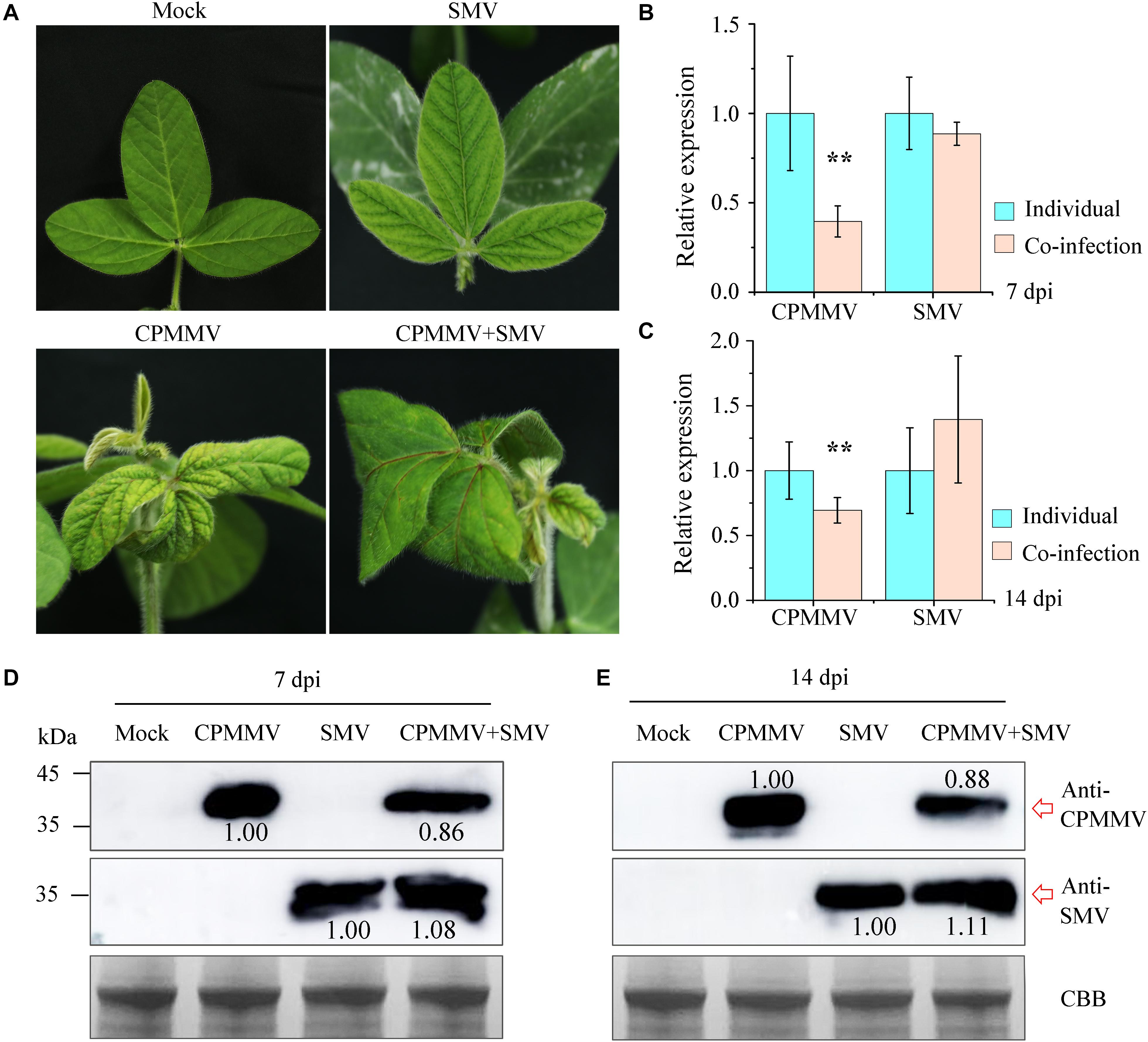
Figure 6. Co-infection of CPMMV and SMV increased the accumulation of SMV. (A) Symptoms on the first newly-emerged leaves 7 days after inoculation with SMV, CPMMV or SMV+CPMMV. (B,C) The relative expression levels of SMV and CPMMV RNAs as determined by RT-qPCR at 7 dpi (B) and 14 dpi (C). Data are shown as means ± SD of three biological replicates, **P < 0.01, Student’s t-tests. (D,E) SMV and CPMMV coat protein (CP) protein levels in mock-inoculated and SMV/CPMMV-infected plants as determined by immunoblot analysis at 7 dpi (D) and 14 dpi (E). Coomassie brilliant blue (CBB) staining of the same extracts is shown to demonstrate equal loading.
Discussion
CPMMV was first discovered on cowpea in Ghana in 1973 (Brunt and Kenten, 1973), and subsequently found infecting soybeans in Thailand and the Ivory Coast in the 1980s (Iwaki, 1982; Thouvenel et al., 1982). CPMMV was considered of minor importance until the occurrence of outbreaks in soybean across Brazil in the 2000s (Zanardo et al., 2014a). CPMMV is transmitted by the widespread whitefly Bemisia tabaci. It induces stem necrosis symptoms, and can cause economic losses in soybean production (Jossey, 2013; Zanardo and Carvalho, 2017). There are few studies on CPMMV: of the 16 complete genomic sequences of CPMMV available in GenBank, most (8/16) are from Brazil. The complete sequences of two CPMMV isolates from China have previously been reported, one from cowpea in Hainan (Yang et al., 2019), and our recent study providing the first report of CPMMV infecting soybean in China showing that it was the cause of leaf mosaic and crinkling symptoms (Wei et al., 2020). We have now demonstrated that CPMMV is widespread in the soybean producing areas in China and was present in all 11 samples collected. Their genomic sequences were obtained through HTS and phylogenetic analysis showed that all the Chinese isolates form a distinct cluster with 95–100% nucleotide identity to one another but rather distantly related to CPMMV isolates from other countries. In the family Betaflexiviridae, viruses are classified in the same species if their CP or replicase proteins have more than 72% nt or 80% aa identity (Adams et al., 2012). The ORF1 (RdRp) and CP of isolates from China have 79.3–81.0% and 83.0–90.0% nt identity to Brazilian isolates and clearly belong to the same species.
In Brazil, CPMMV causes a significant threat to soybean production and has received particular attention (Zanardo and Carvalho, 2017). The molecular variability of Brazilian CPMMV isolates has been investigated and it was concluded that the topology of the phylogenetic tree was not related to the geographical origin of isolates within Brazil (Zanardo et al., 2014a,b). Although the Chinese isolates form a separate branch of the CPMMV tree, there was similarly no evidence of further sub-clades based on their origin within China. This may suggest that CPMMV is a relatively recent introduction in both Brazil and China, or perhaps that the vector is able to rapidly distribute virus variants within the country. In addition, our phylogenetic analysis of SMV isolates also did not show clear relationships with their geographical origin. All SMV isolates (except SMV_JL_GZL from Jilin province) of this study were clustered in one clade with SC7 strain, and distinct from the US strains (G1–G7) (Figure 2). Recombination may explain the position of the SMV_JL_GZL isolate (Supplementary Figure 6). Recombination events have been identified in several viruses (Gagarinova et al., 2008; Zanardo et al., 2014b; Kalyandurg et al., 2017; Chikh-Ali et al., 2019). Our analysis detected one recombination event within the CPMMV isolates and two recombination events within the SMV isolates from China (Figure 4 and Supplementary Figure 6), indicating that recombination is one of the most important factors that contribute to the variability and evolution of SMV and CPMMV.
Previous studies have suggested that virus epidemics in soybean production in China are mainly caused by SMV and co-infection of soybean by SMV and CPMMV has not previously been reported. Our attempts to transmit virus from the co-infected field samples by mechanical inoculation sometimes led to the loss of one of the viruses. The mechanisms of co-infection are diverse in different host-pathogen systems (Bance, 1991; Gonzalez-Jara et al., 2004). Several lines of biochemical and genetic evidence have shown that viral interaction patterns in co-infection may depend on the host cultivar. For example, Wheat streak mosaic virus and Triticum mosaic virus induced cultivar-specific disease synergism in three wheat cultivars (Tatineni et al., 2010). We suppose that the different outcomes of mechanical inoculation in our study may be a consequence of differences in the virus content of field samples and host cultivar-pathogen specificity.
Co-infections by two or more viruses are common in the field. Although co-infections can be either synergistic or antagonistic (Syller, 2012; Bian et al., 2020), they usually cause more severe symptoms and significant damage to crops (Malapi-Nelson et al., 2009; Mahuku et al., 2015; Redinbaugh and Stewart, 2018). In our case, field observation and seedling inoculations suggest that co-infection causes more severe symptoms. However, at the molecular level, there would appear to be some antagonism between the viruses since RT-qPCR and western-blotting results showed that the accumulation level of CPMMV genomic RNAs and CP were decreased in co-infected leaves. This suggests that in co-infection with SMV, CPMMV interacts antagonistically. Similar observations have been reported with SMV and AMV infection in soybean, which showed severe symptoms in doubly infected plants, but the level of SMV accumulation was reduced. Conversely, the level of AMV accumulation was increased indicating that the interaction of AMV with SMV is synergistic (Malapi-Nelson et al., 2009). In the current study, there was no significant effect on the accumulation level of SMV in co-infected leaves. There appears to be a complex mechanism of interaction between SMV and CPMMV that requires further investigation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The sequences of CPMMV and SMV isolates were deposited in The National Center for Biotechnology Information (NCBI) GenBank, https://www.ncbi.nlm.nih.gov/genbank/ and the accession numbers can be found in the article/Supplementary Material. The other raw data supporting the conclusions of this article are available upon request from the corresponding author, without undue reservation.
Author Contributions
ZW and ZS designed the research and performed the experiments. ZW drafted the manuscript. ZS revised the manuscript. CM and CJ supervised the project. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Science Fund for Excellent Young Scholars (32022072), Zhejiang Provincial Natural Science Foundation of China (LQ21C140004), the China National Novel Transgenic Organisms Breeding Project (2019ZX08004-004), and K. C. Wong Magna Fund in Ningbo University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.650773/full#supplementary-material
Supplementary Figure 1 | Detection of CPMMV and SMV in the total RNA of symptomatic field soybean samples as shown by RT-PCR.
Supplementary Figure 2 | The genome organization of SMV isolates.
Supplementary Figure 3 | NJ-phylogenetic relationships among the ORFs of CPMMV isolates from different countries.
Supplementary Figure 4 | Pairwise identity matrix of CPMMV isolates for each open reading frame (ORF).
Supplementary Figure 5 | Phylogenetic trees analysis based on different nucleotide fragments of CPMMV isolates from China.
Supplementary Figure 6 | Recombination events among the SMV isolates.
Supplementary Table 1 | Genomic organization of the 11 CPMMV isolates described in this study.
Supplementary Table 2 | List of primer sequences used in this study.
Footnotes
- ^ https://www.boldsystems.org/
- ^ ftp://ftp.ncbi.nlm.nih.gov/refseq/release/viral
- ^ https://www.ncbi.nlm.nih.gov/
References
Adams, M. J., Candresse, T., Hammond, J., Kreuze, J. F., Yoshikawa, N., et al. (2012). “Family Betaflexiviridae,” in Virus Taxonomy: The Classification and Nomenclature of Viruses. The 9th Report of the ICTV, eds A. M. Q. King, M. J. Adams, E. B. Carstens, and E. J. Lefkowitz (London: Academic Press), 920–941.
Bance, V. B. (1991). Replication of potato virus X RNA is altered in coinfections with potato virus Y. Virology 182, 486–494. doi: 10.1016/0042-6822(91)90589-4
Bian, R. L., Andika, I. B., Pang, T. X., Lian, Z. Q., Wei, S., Niu, E., et al. (2020). Facilitative and synergistic interactions between fungal and plant viruses. Proc. Natl. Acad. Sci. U.S.A. 117, 3779–3788. doi: 10.1073/pnas.1915996117
Brunt, A. A., and Kenten, R. H. (1973). Cowpea mild mottle, a newly recognized virus infecting cowpeas (Vigna unguiculata) in Ghana. Ann. Appl. Biol. 74, 67–74. doi: 10.1111/j.1744-7348.1973.tb07723.x
Chikh-Ali, M., Rodriguez-Rodriguez, M., Green, K. J., Kim, D. J., Chung, S. M., Kuhl, J. C., et al. (2019). Identification and molecular characterization of recombinant Potato Virus Y (PVY) in potato from South Korea, PVY(NTN) strain. Plant Dis. 103, 137–142. doi: 10.1094/PDIS-05-18-0715-RE
Cho, E. K., and Goodman, R. M. (1979). Strains of soybean mosaic virus: classification based on virulence in resistant soybean cultivars. Phytopathology 69, 467–470. doi: 10.1094/Phyto-69-467
Gagarinova, A. G., Babu, M., Stromvik, M. V., and Wang, A. (2008). Recombination analysis of Soybean mosaic virus sequences reveals evidence of RNA recombination between distinct pathotypes. Virol. J. 5:143. doi: 10.1186/1743-422X-5-143
Gao, L., Luo, J. Y., Ding, X. N., Wang, T., Hu, T., Song, P. W., et al. (2019). Soybean RNA interference lines silenced for eIF4E show broad potyvirus resistance. Mol. Plant Pathol. 21, 303–317. doi: 10.1111/mpp.12897
Gonzalez-Jara, P., Tenllado, F., Martinez-Garcia, B., Atencio, F. A., Barajas, D., Vargas, M., et al. (2004). Host-dependent differences during synergistic infection by Potyviruses with potato virus X. Mol. Plant Pathol. 5, 29–35. doi: 10.1111/j.1364-3703.2004.00202.x
Hajimorad, M. R., Domier, L. L., Tolin, S. A., Whitham, S. A., and Saghai Maroof, M. A. (2018). Soybean mosaic virus: a successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 19, 1563–1579. doi: 10.1111/mpp.12644
Hartman, G. L., West, E. D., and Herman, T. K. (2011). Crops that feed the World 2. Soybean—worldwide production, use, and constraints caused by pathogens and pests. Food Secur. 3, 5–17. doi: 10.1007/s12571-010-0108-x
Hill, J. H., and Whitham, S. A. (2014). Control of virus diseases in soybeans. Adv. Virus Res. 90, 355–390. doi: 10.1016/B978-0-12-801246-8.00007-X
Iwaki, M. (1982). Whitefly transmission and some properties of cowpea mild mottle virus on soybean in Thailand. Plant Dis. 66:365. doi: 10.1094/PD-66-365
Jossey, S. (2013). Role of soybean mosaic virus-encoded proteins in seed and aphid transmission in soybean. Phytopathology 103, 941–948. doi: 10.1094/PHYTO-09-12-0248-R
Kalyandurg, P., Gil, J. F., Lukhovitskaya, N. I., Flores, B., Muller, G., Chuquillanqui, C., et al. (2017). Molecular and pathobiological characterization of 61 Potato mop-top virus full-length cDNAs reveals great variability of the virus in the centre of potato domestication, novel genotypes and evidence for recombination. Mol. Plant Pathol. 18, 864–877. doi: 10.1111/mpp.12552
Li, K., Yang, Q. H., Zhi, H. J., and Gai, J. Y. (2010). Identification and distribution of soybean mosaic virus strains in southern China. Plant Dis. 94, 351–357. doi: 10.1094/PDIS-94-3-0351
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mahuku, G., Lockhart, B. E., Wanjala, B., Jones, M. W., Kimunye, J. N., Stewart, L. R., et al. (2015). Maize lethal necrosis (MLN), an emerging threat to maize-based food security in sub-saharan Africa. Phytopathology 105, 956–965. doi: 10.1094/PHYTO-12-14-0367-FI
Malapi-Nelson, M., Wen, R. H., Ownley, B. H., and Hajimorad, M. R. (2009). Co-infection of soybean with soybean mosaic virus and alfalfa mosaic virus results in disease synergism and alteration in accumulation level of both viruses. Plant Dis. 93, 1259–1264. doi: 10.1094/PDIS-93-12-1259
Martin, D. P., Varsani, A., Roumagnac, P., Botha, G., Maslamoney, S., Schwab, T., et al. (2020). RDP5: a computer program for analysing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. veaa087. doi: 10.1093/ve/veaa087
Muniyappa, V. (1983). Transmission of cowpea mild mottle virus by Bemisia tabaci in a nonpersistent manner. Plant Dis. 67, 391–393. doi: 10.1094/PD-67-391
Redinbaugh, M. G., and Stewart, L. R. (2018). Maize lethal necrosis: an emerging, synergistic viral disease. Ann. Rev. Virol. 5, 301–322. doi: 10.1146/annurev-virology-092917-04341
Song, Y. P., Cui, L., Lin, Z., Karthikeyan, A., Na, L., Kai, L., et al. (2016). Disease spread of a popular soybean mosaic virus strain (SC7) in southern China and effects on two susceptible soybean cultivars. Philipp. Agric. Sci. 99, 355–364.
Syller, J. (2012). Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol. Plant Pathol. 13, 204–216. doi: 10.1111/j.1364-3703.2011.00734.x
Tatineni, S., Graybosch, R. A., Hein, G. L., Wegulo, S. N., and French, R. (2010). Wheat cultivar-specific disease synergism and alteration of virus accumulation during co-infection with Wheat streak mosaic virus and Triticum mosaic virus. Phytopathology 100, 230–238. doi: 10.1094/PHYTO-100-3-0230
Thouvenel, J. C., Monsarrat, A., and Fauquet, C. (1982). Isolation of cowpea mild mottle virus from diseased soybeans in the Ivory Coast. Plant Dis. 66, 336–337. doi: 10.1094/PD-66-336
Wang, D. G., Tian, Z., Li, K., Li, H. W., Huang, P. P., Hu, G. Y., et al. (2013). Identification and variation analysis of soybean mosaic virus strains in Shan-Dong, Henan and Anhui Provinces of China. Soybean Sci. 32, 806–809.
Wei, Z. Y., Wu, G. W., Ye, Z. X., Jiang, C., Mao, C. Y., Zhang, H. H., et al. (2020). First report of cowpea mild mottle virus infecting soybean in China. Plant Dis. 104, 2534–2534. doi: 10.1094/PDIS-01-20-0063-PDN
Widyasari, K., Alazem, M., and Kim, K. H. (2020). Soybean resistance to soybean mosaic virus. Plants (Basel) 9:219. doi: 10.3390/plants9020219
Wrather, J. A., and Koenning, S. R. (2006). Estimates of disease effects on soybean yields in the United States 2003 to 2005. J. Nematol. 38, 173–180.
Yadav, M. K., Biswas, K. K., Lal, S. K., Baranwal, V. K., and Jain, R. K. (2014). A distinct strain of cowpea mild mottle virus infecting soybean in India. J. Phytopathol. 161, 739–744. doi: 10.1111/jph.12119
Yang, X., Zhang, D. Y., Zhang, Y., Chen, S., Li, S. S., Gong, Y. N., et al. (2019). Genomic sequences measure and molecular characteristics of cowpea mild mottle virus of hainan isolate. China Vegetables 364, 41–44.
Zanardo, L. G., and Carvalho, C. M. (2017). Cowpea mild mottle virus (Carlavirus, Betaflexiviridae): a review. Trop. Plant Pathol. 42, 417–430. doi: 10.1007/s40858-017-0168-y
Zanardo, L. G., Silva, F. N., Bicalho, A. A. C., Urquiza, G. P. C., Lima, A. T. M., Almeida, A. M. R., et al. (2014a). Molecular and biological characterization of Cowpea mild mottle virus isolates infecting soybean in Brazil and evidence of recombination. Plant Pathol. 63, 456–465. doi: 10.1111/ppa.12092
Zanardo, L. G., Silva, F. N., Lima, A. T., Milanesi, D. F., Castilho-Urquiza, G. P., Almeida, A. M., et al. (2014b). Molecular variability of cowpea mild mottle virus infecting soybean in Brazil. Arch. Virol. 159, 727–737. doi: 10.1007/s00705-013-1879-0
Keywords: high-throughput sequencing, phylogenetic analysis, genetic variation, recombination, co-infection
Citation: Wei Z, Mao C, Jiang C, Zhang H, Chen J and Sun Z (2021) Identification of a New Genetic Clade of Cowpea Mild Mottle Virus and Characterization of Its Interaction With Soybean Mosaic Virus in Co-infected Soybean. Front. Microbiol. 12:650773. doi: 10.3389/fmicb.2021.650773
Received: 08 January 2021; Accepted: 15 March 2021;
Published: 08 April 2021.
Edited by:
Xifeng Wang, Institute of Plant Protection (CAAS), ChinaReviewed by:
Nicolas Bejerman, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaPedro Luis Ramos-González, Biological Institute of São Paulo, Brazil
Copyright © 2021 Wei, Mao, Jiang, Zhang, Chen and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianping Chen, Y2hlbmppYW5waW5nQG5idS5lZHUuY24=; anBjaGVuMjAwMUAxMjYuY29t; Zongtao Sun, c3Vuem9uZ3Rhb0BuYnUuZWR1LmNu
 Zhongyan Wei
Zhongyan Wei Zongtao Sun
Zongtao Sun